
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 September 2021
Sec. Microbe and Virus Interactions with Plants
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.746799
Apple replant disease (ARD) is a common problem in major apple planting areas, and biological factors play a leading role in its etiology. Here, we isolated the bacterial strain QSB-6 from the rhizosphere soil of healthy apple trees in a replanted orchard using the serial dilution method. Strain QSB-6 was provisionally identified as Bacillus amyloliquefaciens based on its morphology, physiological and biochemical characteristics, carbon source utilization, and chemical sensitivity. Maximum likelihood analysis based on four gene sequences [16S ribosomal RNA gene (16S rDNA), DNA gyrase subunit A (gyrA), DNA gyrase subunit B (gyrB), and RNA polymerase subunit B (rpoB)] from QSB-6 and other strains indicated that it had 100% homology with B. amyloliquefaciens, thereby confirming its identification. Flat standoff tests showed that strain QSB-6 had a strong inhibitory effect on Fusarium proliferatum, Fusarium solani, Fusarium verticillioides, Fusarium oxysporum, Alternaria alternata, Aspergillus flavus, Phoma sp., Valsa mali, Rhizoctonia solani, Penicillium brasilianum, and Albifimbria verrucaria, and it had broad-spectrum antibacterial characteristics. Extracellular metabolites from strain QSB-6 showed a strong inhibitory effect on Fusarium hyphal growth and spore germination, causing irregular swelling, atrophy, rupture, and cytoplasmic leakage of fungal hyphae. Analysis of its metabolites showed that 1,2-benzenedicarboxylic acid and benzeneacetic acid, 3- hydroxy-, methyl ester had good inhibitory effects on Fusarium, and increased the length of primary roots and the number of lateral roots of Arabidopsis thaliana plantlet. Pot experiments demonstrated that a QSB-6 bacterial fertilizer treatment (T2) significantly improved the growth of Malus hupehensis Rehd. seedlings. It increased root length, surface area, tips, and forks, respiration rate, protective enzyme activities, and the number of soil bacteria while reducing the number of soil fungi. Fermentation broth from strain QSB-6 effectively prevented root damage from Fusarium. terminal restriction fragment length polymorphism (T-RFLP) and quantitative PCR (qPCR) assays showed that the T2 treatment significantly reduced the abundance of Fusarium in the soil and altered the soil fungal community structure. In summary, B. amyloliquefaciens QSB-6 has a good inhibitory effect on Fusarium in the soil and can significantly promote plant root growth. It has great potential as a biological control agent against ARD.
As a widespread agricultural problem, apple replant disease (ARD) is a serious threat to the major fruit-growing regions of the world (Jaffee et al., 1982; Winkelmann et al., 2018). In Washington state, ARD typically results in a $40,000-per-acre reduction in gross returns over a 10-year period (Smith, 1995; Hewavitharana et al., 2019). Chinese apple cultivation area, total output, and export volume rank first in the world, but most orchards are now in an aging period. Due to the limited land resources, it is inevitable to replant the apple trees when the old orchard is renewed, which leads to the occurrence of ARD (Greene, 1915; Willett et al., 1994; Granatstein et al., 2016; Wang et al., 2016). ARD refers to the common management condition in which poor growth of fruit trees occurs after replanting on a site that has previously supported the same or a closely related species; this phenomenon has been broadly termed “replant disease” or “replant disorder” (Mazzola, 1998; Mazzola and Manici, 2012; Nicola et al., 2018). Specific ARD symptoms in apple trees include weak growth, reduced plant height, impaired root system activity, and poor fruit quality (Tewoldemedhin et al., 2011a, b; Weiß et al., 2017; Nicola et al., 2018; Grunewaldt-Stöcker et al., 2019). It is therefore important to explore new measures for the control of ARD.
Apple replant disease has been attributed to a variety of biotic and abiotic causal factors, but current studies suggest that biotic factors such as fungi (Rhizoctonia, Fusarium, and Cylindrocarpon), Oomycetes (Pythium, Humicola, and Phytophthora), and nematodes (Pratylenchus) play a leading role in disease development (Utkhede et al., 1992; Van Schoor et al., 2009; Tewoldemedhin et al., 2011a, b; Kelderer et al., 2012; Tilston et al., 2018, 2020; Yim et al., 2020). This has been widely demonstrated in other studies via soil pasteurization and the application of biocides (Weller et al., 2002; Garbeva et al., 2004). The pathogens in replanted orchard soils from different regions vary in population characteristics. Previous studies have suggested that Fusarium is one of the main pathogens that cause ARD in China (Yin et al., 2017; Wang G. S. et al., 2018; Sheng et al., 2020). Van Schoor et al. (2009) found that Fusarium, Cylindrosporium, and Pythium are the main harmful fungi that contribute to the occurrence of ARD in South Africa. Kelderer et al. (2012) found that F. solani and F. oxysporum are the main pathogenic fungi associated with the occurrence of ARD in Italy. Currently, the major control strategy for ARD is the use of soil fumigants such as methyl bromide, metam sodium, and chloropicrin (Utkhede and Smith, 2000; Zasada et al., 2010). Although chemical disinfection can effectively control replant diseases, the use of these chemical disinfectants has a number of disadvantages, including difficulty in application, high cost, and potential impacts on the environment and human health (Brown et al., 2000; Leroux, 2003; Chen et al., 2014). Biological control is considered to be an alternative and more sustainable strategy for plant disease control because of its lower cost and environmental friendliness (Podile and Prakash, 1996; Hoitink and Boehm, 1999; Zalila-Kolsi et al., 2016; Chowdhury et al., 2020). Therefore, the development of biological agents is critical for the biological control of ARD.
Bacillus is a new class of biological control agent (Nagórska et al., 2007; Fan et al., 2017b) whose metabolism can produce a variety of antibacterial substances (fengycin, surfactin, etc.), enzymes (amylase, protease, etc.), and nutritional factors, thereby effectively inhibiting the reproduction of pathogens and promoting plant growth (Stein, 2005; Cazorla et al., 2007; Frikha-Gargouri et al., 2017; Zhang C. et al., 2018). Bacillus also produces resting spores during its growth and development. These have the advantage of resistance to high temperatures, acids, salt, drugs, and radiation. When the spores are exposed to favorable conditions they resume growth as vegetative cells (Hallmann et al., 1999; Piggot and Hilbert, 2004; McKenney et al., 2013; Zhang X. et al., 2014). Bacillus has a high reproductive rate, can easily be mass produced and processed, is convenient to transport, and has a high survival rate during storage (Collins and Jacobsen, 2003). Fan et al. (2017a) isolated B. subtilis 9407 from healthy apples in an infested orchard; it can produce antifungal compounds such as fengycin and can effectively prevent apple ring rot disease. Mutaz Al-Ajlani and Hasnain (2010) determined that 54 of 118 Bacillus strains isolated from soil samples had antagonistic activities toward at least two strains from a panel of pathogenic and non-pathogenic microorganisms. Zhang M. et al. (2016) isolated Bacillus amyloliquefaciens IBFCBF-1 that can effectively control Phytophthora blight and promote the growth of pepper. Singh et al. (2008) found that B. subtilis BN1 isolated from the chir pine (Pinus roxburghii) rhizosphere exhibited strong antagonistic activity toward F. oxysporum and R. solani. Cavaglieri et al. (2005) found that B. subtilis CE1 reduced F. verticillioides colonization of the rhizoplane and endorhizosphere at all inoculum levels found with maize roots. Therefore, the use of Bacillus is a potential sustainable alternative for the biological control of ARD.
In this study, we isolated bacteria with biological control effects from healthy apple trees in a replanted orchard in which apple trees showed ARD symptoms. We identified the bacterial isolates using biochemical and molecular approaches. We then tested their ability to produce metabolites with antagonistic activities toward fungal strains and characterized their antifungal compounds by column chromatography and GC–MS. Finally, we evaluated the ability of a bacterial fertilizer to control ARD by measuring plant and root growth and characterizing the soil microbial community, with the aim of providing a new method for ARD control.
In May 2016, samples were collected from four orchards that were replanted 4–5 years previously at a pre-existing apple orchards (more than 25 years old). Apple trees with ARD symptoms (twig growth retardation or death) were common in the sampled orchards. Six healthy apple trees were randomly selected from each orchard, and roots, stems, leaves, fruits, and rhizosphere soil were collected from the trees using a sterile spatula for a total of 120 samples. The samples were packed in clean, dry, sterile polyethylene bags, which were placed on dry ice in the field and taken to the National Key Laboratory of Crop Biology as soon as possible. The geographic coordinates of the sampled sites are presented in Supplementary Table 1. The sampled orchards primarily used Malus × robusta (CarriŠre) Rehder as the rootstock, and Malus pumila Mill was the main cultivated scion variety.
Bacteria were isolated by the serial dilution method (Filippi et al., 2011), with some modifications. The soil was filtered through a sieve (diameter 3–4 mm) to remove detritus. Five grams of rhizosphere soil were placed in 45 mL of sterilized water and shaken at 180 rpm for 30 min, then allowed to stand for 5 min to make a stock suspension of soil bacteria. The suspension was diluted to 10–4, and a 100-μL aliquot of dilution was plated on Luria–Bertani (LB) agar (10 g tryptone, 5 g yeast extract, 10 g NaCl, 15 g agar, pH 7.0) overnight at 37°C. Each treatment was repeated three times. Distinct single colonies were picked and subcultured to purity on LB agar medium. Each purified strain was inoculated into liquid LB medium with constant shaking at 180 rpm for 12 h at 30°C and stored at −80°C in 15% glycerol-containing liquid LB medium (Frikha-Gargouri et al., 2017). Roots, stems, leaves, and fruits were washed with tap water to remove impurities. After natural air drying and rinsing with distilled water three times, the tissues were surface-sterilized in 75% ethanol (v/v) for 30 s, submerged in 1% NaOCl (w/v) for 10 s, and rinsed three times in sterile distilled water (Dongzhen et al., 2020). One hundred microliters of sterile water from the last rinse were spread on LB medium and cultivated for 24 h to verify that the surface disinfection of the plants was complete. The surface-disinfected plants were cut into small pieces with a sterile scalpel, placed in a sterile mortar, and ground in an appropriate amount of sterile PBS buffer. The grinding solution was diluted to 10–4, and the bacteria were isolated as described above for the rhizosphere soil.
The tested pathogens were all maintained in our laboratory (National Key Laboratory of Crop Biology). The pathogens were grown on potato dextrose agar (PDA) for 7 days at 28°C prior to use. A dual-culture test was conducted to examine whether the isolated strains could antagonize the growth of plant fungal pathogens according to the methods described by Yu et al. (2011), with some modifications. A mycelial disk, Φ1.0 cm in diameter, of a pure culture of each fungal pathogen was placed in the center of a PDA plate, and then the isolated strains was inoculated in four symmetrical spots around the mycelium disk. Each treatment was repeated three times. The plates were incubated for 7–10 days at 28°C. The plates were scanned once a day to monitor the formation of an inhibition zone and the growth of fungal pathogens. The width of the inhibition zones were measured and then averaged. The strain showing the strongest antifungal effect was selected for further studies.
Selected bacterial strains were streaked onto LB agar plates, and single colonies were inoculated into LB broth (100 mL in a 250-mL Erlenmeyer flask) and cultivated with constant shaking at 150 rpm for 48 h at 28°C. Each bacterial culture was centrifuged at 10,000 rpm for 20 min to pellet bacterial cells, then the cells were suspended in PBS buffer, and the cell density was adjusted to the required bacterial density [1 × 108 colony forming units (CFU/ml)] (Chen et al., 2014). Freshly prepared bacterial suspension was used for each experiment. Antifungal activity against pathogens was evaluated using the dual culture assay in PDA medium, Fresh mycelial plugs (10 mm diameter) were cut from the margins of each fungal pathogen colony and were transferred to the center of PDA plate. Four pieces of sterile round filter paper containing 2 μL bacterial cell suspension were placed 2.5 cm from the plugs, and sterilized distilled water (SDW) was used as a control according to the methods described by Erdogan and Benlioglu (2010), with some modifications. After culturing Penicillium brasilianum and Albifimbria verrucaria on the PDA plate for 7 days, the PDA plate was rinsed with sterile distilled water. The density of the conidial suspension was measured using a hemocytometer and the inoculated spore culture was diluted using sterile distilled water to obtain a final concentration of 1 × 106 spores/mL. Add the conidia suspension to the PDA medium, mix and invert the plate, and then perform the antibacterial test. Plates were incubated at 28°C until control plates covered the entire surface. The percentage inhibition was calculated using the formula. I = (C − T)/(C − D) × 100. Where I:% inhibition, C: the colony diameter of the control (mm), T: colony diameter of the treatment (mm), and D: plug diameter (mm). Three replicates of each treatment were performed, and the assays were repeated three times (Yu et al., 2011; Li et al., 2020).
Isolates were cultured on LB agar at 37°C for 24 h, and their morphological traits were observed. Gram’s staining and endospore staining by Schaeffer–Fulton method was performed as described by Benson (2002) and Bartholomew and Mittwer (1950). The shape and size of the bacteria were observed with a Nikon BX-51 fluorescence microscope and an SU-8010 scanning electron microscope.
Physiological and biochemical characteristics were assessed according to the methods described in Bergey’s Manual of Systematic Bacteriology (2nd edition) and the Common Bacterial Identification Manual (Garrity, 2007; Vos et al., 2011).
A Biolog GEN III microplate system (Biolog Inc., Hayward, CA, United States) was used to analyze carbon source utilization and chemical sensitivity of strain QSB-6 as described in Hu et al. (2010).
Strain QSB-6 was inoculated into liquid LB medium and cultivated at 30°C for 12 h with shaking at 200 rpm/min. Genomic DNA was extracted using the EasyPure Bacteria Genomic DNA Kit (Del Sal et al., 1988). To confirm the species identity of strain QSB-6, we obtained the DNA sequences of the 16S ribosomal RNA gene (16S rDNA), DNA gyrase subunit A (gyrA), DNA gyrase subunit B (gyrB), and RNA polymerase subunit B (rpoB) (Gardes and Bruns, 1993; Yamamoto and Harayama, 1995; Chun and Bae, 2000; Zalila-Kolsi et al., 2016; Somerville et al., 2020). PCR amplification was performed in a final volume of 50 μL that contained 2.0 μL of genomic DNA (20 ng/μL), 1.0 μL of each primer (10 μM), 0.5 μL of PrimeSTAR HS DNA polymerase (2.5 U/μL), 10.0 μL of 5× PrimeSTAR Buffer (Mg2+ Plus), 4.0 μL of dNTP Mixture (2.5 mM each), and 31.5 μL of ddH2O (Weisburg et al., 1991). PCR amplification was performed in an Applied Biosystems 2720 Thermal Cycler (Applied Biosystems Inc., United States) with an initial denaturation step at 94°C for 5 min, followed by denaturation at 94°C for 40 s, annealing for 40 s, and extension at 72°C for 1 min. Thirty-five cycles of amplification and a final extension at 72°C for 10 min were performed (Zhang Q. et al., 2014). The primers and annealing temperatures are shown in Supplementary Table 2. After the reaction was complete, 3 μL of PCR product were used for 1% agarose gel electrophoresis to confirm the PCR-amplified fragments. Amplified PCR products were purified using a DNA Gel Recovery Kit (AxyPrep, Hangzhou, China) and sequenced on an ABI 3730XL system (Applied Biosystems Inc., United States) at Personalbio Gene Biotech Co., Ltd. (Qingdao, China).
For phylogenetic analysis, 16S rDNA, gyrA, gyrB, and rpoB sequences closely related to our sequences were retrieved from GenBank based on BLAST results from the National Center for Biotechnology Information.1 Maximum likelihood (ML) phylogenies were constructed from the sequence datasets using RAxML-HPC2 run on XSEDE (8.2.12) (Stamatakis, 2014) through the CIPRES Science Gateway2 to obtain multiple measures of branch support. The parameters Maximum Hours to Run and Number of Patterns were modified according to the dataset, and other parameters were set to default values (Lombard et al., 2019; Medeiros Araújo et al., 2021). The trees were visualized using FigureTree v1.4.3 and Adobe Illustrator CS6.
Strain QSB-6 was cultured on LB agar at 37°C for 24 h, and a single colony was inoculated into LB broth (100 mL in a 250-mL Erlenmeyer flask) and grown in a shaker incubator at 180 rpm for 12 h. It was then inoculated into optimized liquid fermentation medium (20 g sucrose, 15 g yeast extract, 1 g MnSO4, 2.0 g NaH2PO4⋅2H2O, and 4.0 g Na2HPO4⋅2H2O in 1 L) at a ratio of 5% and grown in a shaker incubator at 200 rpm for 48 h. The cell-free culture filtrate was obtained as described in Azabou et al. (2020) with some modifications. Culture was then centrifuged at 10,000 rpm for 15 min at 4°C. The supernatant was collected and filtrated through 0.22-μm Nylon 66 microporous membrane, and stored at 4°C for later use.
A thin layer of PDA medium was poured onto a sterile plate, and a sterile glass slide was placed in the center of the plate after the medium had solidified. Then approximately 200 μL of PDA medium was spread onto the glass slide, and a sterile 1-mL pipette tip was used to place a Fusarium cake onto the slide. An Oxford cup was placed at each end of the slide; 200 μL of cell-free culture filtrate was added to one cup as the treatment, and sterile distilled water was added to the other cup as the control. After incubating for 3 days, the slide was observed under a Nikon BX-51 fluorescence microscope. The hyphae at the edge of the bacteriostatic area were picked with an inoculation needle and placed in a sterile centrifuge tube that contained 2.5% glutaraldehyde fixative for 24 h. The control area was sampled similarly, and the samples were sent to Keshang Biotech Co., Ltd., for scanning electron microscope observation.
Fusarium was cultured on VBC medium (1 g KH2PO4, 1 g KNO3, 0.5 g sucrose, vitamin B1 tablet, vitamin C tablet, and 20 g agar in 1 L) at 28°C for 7 days, rinsed thoroughly with sterile water, and shaken evenly. The density of the conidial suspension was measured using a hemocytometer, and the suspension was diluted with sterile distilled water to a final concentration of 1 × 106 spores/mL (Gabrekiristos et al., 2018). It was mixed with fermentation broth and cell-free culture filtrate on a concave glass slide at a ratio of 1:1 and mixed with sterile water as a control (Rahman et al., 2007). The slide was kept moist and incubated at 28°C for 24 h in order to measure spore germination rate. Spores were scored as germinated if the germ tube length equaled or exceeded half the diameter of the spore. The percentage inhibition was calculated using the formula. I = G/T × 100%. Where I:% spore germination rate, G: the number of germinated spores and T: the total number of spores. The total number of spores under investigation should be greater than 200, three replicates were conducted for each treatment, and the experiment was repeated twice (Li et al., 2021).
As described in Podile and Prakash (1996) and Omar and Abd-Alla (1998), the fermentation broth was mixed with PDB to make a liquid medium with a concentration of 250 μL/mL (50 mL in a 250-mL Erlenmeyer flask). A 1-mL pipette tip was used to obtain a Fusarium cake and place it in a triangular flask to serve as the treatment group. The control group was cultured in sterile water and PDB liquid base mix. After culturing on a constant temperature shaker at 28°C and 180 rpm/min for 6, 12, or 24 h, the mycelium was placed on a pre-weighed Whatman No. 1 filter paper to remove the liquid medium from its surface, then dried in a 65°C oven for 24 h and weighed with an electronic balance. These steps were repeated using mixtures of fermentation broth and PDB with concentrations of 20, 100, and 200 μL/mL, and weights were obtained after 24 h of incubation.
Five milliliters of cell-free culture filtrate in a 10-mL centrifuge tube were placed in a constant temperature water bath at 50, 60, 70, 80, 90, or 100°C for 0.5 h to test the thermal stability of the cell-free culture filtrate. The pH of cell-free culture filtrate was adjusted to 1.0, 3.0, 5.0, 7.0, 9.0, 11.0, or 13.0 with 3 M NaOH or 1 M HCl, and the broth was allowed to stand for 2 h to test its acid-base stability. Broth samples were placed under a 20 W UV lamp and irradiated for 0.5, 1.0, 1.5, 2.0, 2.5, or 3.0 h to assess their UV sensitivity. Samples were also placed in a light incubator (4500 ± 500 lx) for 1, 2, 4, 8, 12, 16, or 32 h to test their light stability. Untreated cell-free culture filtrate samples were used as the control group, and each treatment was replicated three times. Antibacterial activity was measured by the filter paper method, and the antibacterial rate was calculated (Hu et al., 2010).
Malus hupehensis Rehd. seedlings were selected as the test material. For treatment 1, the roots were soaked in sterile distilled water for 24 h. For treatment 2, the roots were first soaked for 12 h in fermentation broth from strain QSB-6, then treated with a suspension of Fusarium spores for 12 h. For treatment 3, the root system was first soaked in sterile distilled water for 12 h, then treated with a suspension of Fusarium spores for 12 h. Each treatment was replicated six times. The roots were placed in a sterile centrifuge tube that contained 2.5% glutaraldehyde fixative and sent to Keshang Biotech Co., Ltd., for paraffin sectioning and Periodic Acid Schiff (PAS) staining (Shao et al., 2020).
Twenty-four liters of active fermentation broth were obtained from strain QSB-6 as described above. After centrifugation, the supernatant was obtained, and the fermentation products were extracted using equal volumes of n-butanol, ethyl acetate, chloroform, and petroleum ether. After the extract phase was dried over anhydrous sodium sulfate, it was concentrated under reduced pressure using a Rotary Evaporator N-1300D-WB (Tokyo, Japan) to obtain a crude extract. The in vitro antibacterial activity of the crude extract was tested by the filter paper method (Frikha-Gargouri et al., 2017), and the organic phase after extraction with ethyl acetate was found to have better antibacterial activity (Supplementary Table 3). After filtering the crude ethyl acetate extract to remove impurities that were insoluble in methanol, the next purification work was performed.
An appropriate amount of ethyl acetate extract was dissolved in a small amount of methanol. The crude active fraction was separated on a GF254 silica gel thin layer chromatography plate (Yantai Ocean Chemical Technology Co., Ltd., China) using methanol:dichloromethane 1:1, 2:1, 3:1, 4:1, or 5:1 (v/v) as the solvent system. After separation, the plate was visualized under UV light at 254and 364 nm, and the Rf values were recorded. The solvent ratio that best resolved the active compounds was determined to be methanol:dichloromethane = 5:1, and the Rf value was ∼0.7. This solvent ratio was used to configure the flow ratio for silica gel column chromatography (Chen et al., 2016; Chowdhury et al., 2020).
The silica gel powder used in this experiment was 200–300 mesh. After extraction, the sample was mixed with an appropriate amount of silica gel powder and use a Rotary Evaporator N-1300D-WB (EYELA Tokyo Rikakikai, Japan) to spin-evaporate to powder, then loaded into the column. The column was first eluted with methanol:dichloromethane 5:1) as the mobile phase, and the same amount of effluent was collected after the mobile phase flow had completed. The effluent was eluted with a mobile phase of methanol:dichloromethane 1:1) to flush out active components that remained in the silica gel column, and finally the column was cleaned with pure methanol. The collected effluent was concentrated and analyzed by thin-layer chromatography (TLC). After comparing the bands, the effluent could be divided into six components (Supplementary Table 4). In vitro antibacterial activity was measured by the filter paper method, and the antibacterial activity of components IV and V was strongest. The activity of components II and VI was relatively weak, and components I and III showed no antibacterial activity. A spore germination assay showed that components IV and V had the best inhibitory effects. A small amount of methanol was added to dissolve component IV and V, the dissolved extracts were passed through a Nylon 66 0.22-μm filter membrane, and the filtered extracts were stored in a refrigerator at 4°C for later use.
The compounds in the active extract were identified by gas chromatography–mass spectrometry (GC–MS) followed by an NIST17 database search. The GC–MS analysis was performed on a GC–MS-QP2010 Plus instrument (Shimadzu, Japan). The peak area normalization method was used to calculate the relative content of each component. The chromatographic conditions were: capillary column HP-5 (60 m length, 0.25 mm ID, 0.25 μm film, 325°C maximum temperature), injection volume 1 μL, carrier gas He (99.999%), column flow rate 1.0 mL/min, splitless injection, program temperature rise, injection port temperature 280°C, column starting temperature 50°C for 2 min, temperature increase to 180°C at 10°C/min, 1 min at 180°C, temperature increase to 270°C at 6°C/min, and 15 min at 270°C. The mass spectrometry conditions were full scan acquisition mode, relative value EMV mode, full scan acquisition mass range 50–550 amu, ion source EI 70 eV, interface temperature 230°C, and ion source temperature 200°C.
Metabolites were analyzed using the test-tube detection method and the TLC chromogenic method described in the experimental methods for quality and chemical composition of traditional Chinese medicines (Zhong et al., 2012).
A conidial suspension of Fusarium was obtained as described above and mixed with PDA medium. The plate was inverted, and two Oxford cups were placed on the left and right, one containing 200 μL of sterile distilled water and the other containing 200 μL of pure product (10 μg/L). The plate was incubated at 28°C for 5 days, and the zone of inhibition was measured as described in Zhang et al. (2013), with some modifications. Effect of 1,2-benzenedicarboxylic acid on mycelial growth of plant fungal pathogens was evaluated as the description of Li et al. (2021) with some modifications. Briefly, 1,2-benzenedicarboxylic acid with methanol were diluted to different concentrations (10, 50, 100, and 1000 μg/L) and added to the sterilized PDA medium, respectively. Equal amount of methanol was used as a control. A 5 mm-diameter fungal disc was placed on the plate center. Plates were incubated at 28°C until control plates covered the entire surface. The percentage inhibition was calculated using the above formula. Basic information on the pure product is provided in Supplementary Table 5.
The plant growth promotion activities of the two compounds were measured by the modified method described by Wu et al. (2019). Briefly, three 2−day−old germinated Arabidopsis thaliana Col−0 seedlings were transferred to the one side of the I−plate containing 0.5 × MS, 0.8% sucrose, and 1% Bacto agar. Then, the two synthetic compounds were diluted separately in ethanol, and 20 μL of the resulting suspension was applied to a sterile filter paper disk on the other side of the I−plate. A total of 10, 100, 500, and 1000 μg doses of each pure product were tested. Each treatment was repeated for three times. The fresh weight of the A. thaliana Col−0 seedlings was measured after 10 days.
Soil from a 31-year-old apple orchard was collected in Xiaowangzhuang Village, Manzhuang Town, Taian, China (Lon: 117.081039, Lat: 36.06682). Soil samples were collected 80 cm away from the trunk and 20–40 cm from the soil surface. Samples were collected randomly at multiple points and mixed. The basic soil conditions are shown in Supplementary Table 6. Microbial fertilizer was produced by Chuangdi Microbial Resources Co., Ltd., Dezhou, China. The bacterial manure carrier was cow manure: straw (3:1), and the bacterial density was 2.1 × 109 CFU per gram. The available nitrogen content was 0.36 mg⋅g–1, available phosphorus was 1.49 mg⋅kg–1, and available potassium was 1.03 mg⋅kg–1.
The pot experiment was performed at the National Apple Engineering Experiment Center of the Horticultural Science and Engineering College of Shandong Agricultural University and the State Key Laboratory of Crop Biology (Lon: 117.156540, Lat: 36.164443). In March 2017, Malus hupehensis Rehd. seedlings were transplanted to the trays. When the seedlings had grown a third true leaf, seedlings of similar growth potential were transplanted into clay pots (38 cm × 28 cm × 26 cm). Each pot contained 75.43 kg of soil, and there were two seedlings per pot and 20 pots in each treatment group. The pots were divided into the following treatments: untreated soil from a 31-year-old orchard (CK1), the same soil fumigated with methyl bromide (CK2), the same soil treated with the manure carrier only (T1), and the same soil treated with the B. amyloliquefaciens strain QSB-6 manure treatment (T2). The application amount of bacterial fertilizer and manure carrier accounted for about 1% of the soil weight (Ma et al., 2018). All experiments were performed with normal water and manure management. Soil samples were collected from three randomly selected pots per treatment on July 15, August 15, and September 15, 2017. Soil was removed around the surface layer and basin, and three basins for each treatment were randomly selected as three replicates. Impurities were removed using a 2-mm sieve, and the soil samples were divided and stored in two sealed pockets. One sealed pocket was stored in a refrigerator at 4°C for measurement of soil microorganisms; the other was stored at −80°C for extraction of soil DNA, quantitative PCR (qPCR), and terminal restriction fragment length polymorphism (T-RFLP) analysis. Three seedlings per treatment were harvested and washed for biomass measurement in the laboratory.
Organic matter, nitrogen, phosphorus, and potassium contents were measured as described in the third edition of Agrochemical Analysis of Soils by Bao Shidan (Bao, 2000). Soil pH was measured using a soil to water ratio of 1:2.5 (w/v) with a PHS-3E digital pH meter (LEICI, Shanghai). Soil particle size distribution (the percentage of clay, silt, and sand) was determined by the hydrometer method (Avery, 1973).
Referring to the method of Zhang Q. et al. (2014), the populations of soil microbes (bacteria, fungi, and actinomycetes) were assessed using the dilution method of plate counting. The bacteria, fungi, and actinomycetes were incubated with beef broth peptone substrate, PDA (Difco), and Gause No. 1 substrate, respectively. Five plates per dilution were measured for each parameter for each soil sample (Guo et al., 2015; Wang et al., 2015). The CFUs per gram of dry soil was used as the unit of the populations of bacteria, fungi, and actinomycetes.
The height and ground diameter of M. hupehensis Rehd. seedlings were measured with a rice meter and a vernier caliper (Sangon Biotech, Shanghai); fresh and dry weights were measured with an electronic balance (OLABO, China).
A Microtek ScanMaker i800 Plus scanner (Shanghai Zhongjing Technology Co., Ltd., China) was used to scan the root system. A Wanshen LA-S series plant root analysis system was used to process the sample images; total root length, surface area, forks, and tips were measured. Respiratory rate was measured by the methods of Gao et al. (2010). Roots of uniform diameter were selected, cut into 2-mm lengths (∼0.1 g), and placed in buffer in the measurement cuvette of the OXY-LAB oxygen electrode system (Hansatech, United Kingdom) at 25°C for respiration measurement.
Superoxide dismutase (SOD) activity was measured as described in Zhang et al. (2009), peroxidase (POD) activity as described in Omran (1980), catalase (CAT) activity as described in Singh et al. (2010), and malondialdehyde (MDA) content as described in Hu et al. (2012).
For each replicate pot described above, a 5 g soil sample was obtained and DNA was extracted using the PowerMax soil DNA isolation kit (MO BIO Laboratories Inc., Carlsbad, CA, United States). The quality and quantity of DNA was determined using an EPPENDORF BioPhotometer nuclei acid and protein analyzer (Eppendorf, Germany). The abundance of Fusarium was quantified by qPCR following a modified method (Zhao et al., 2014; Huang et al., 2015; Shen et al., 2018). The primers and annealing temperatures are presented in Supplementary Table 2. Quantitative PCR amplifications for standard and environmental DNA samples were performed with a total volume of 20 μL in each reaction using the SYBR® Premix Ex TaqTM (TaKaRa, Japan) and a CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, United States). Each PCR reaction contained 2 μL of the target DNA, 10 μL of SYBR Green premix Ex Taq, 0.4 μL of each primer, and 7.2 μL sterile distilled water. Thermal cycling conditions consisted of 30 s at 95°C followed by 40 amplification cycles of 5 s at 94°C, annealing for 30 s, and extension at 72°C for 1 min. Each sample was performed with three parallels, and the results were expressed as log copy numbers per gram of dry soil.
Terminal restriction fragment length polymorphism analysis was used as a tool to rapidly and qualitatively compare fungal community structure across the different treatments. DNA was amplified using the universal primers ITS1F-FAM/ITS4R (Supplementary Table 2) that target the fungal ITS region between the 18S and 28S rRNA regions, respectively. The forward primers were labeled at the 5’end with 6-carboxyfluorescein (FAM) which was synthesized by Sangon Biotech (Shanghai, China). The specific steps refer to the method of Quéric and Soltwedel (2012) and Xu et al. (2019). The 50 μL PCR mixture contained 0.6 μL of 5 U/uL of Ex Taq (TaKaRa, Japan), 5 μL of 10× Ex Taq Buffer, 1 μL of 2.5 mM dNTP mixture, 2 μL of 0.5 mM of forward and reverse primer, 12.6 μL of ddH2O and 2.0 μL of 100 ng of the extracted DNA as template. All PCR amplification was carried on an Applied Biosystems 2720 Thermal Cycler (Applied Biosystems Inc., United States). The PCR conditions consisted of 5 min at 95°C, followed by 30 cycles of denaturing at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min, and a final 10 min extension at 72°C. PCR products were cleaned by EZNA PCR purification kit (Omega Bio-Tek, United States), following the manufacturer’s instruction and quantified using a DNA master nuclei acid and protein analyzer (Dynamica, United Kingdom). The enzyme reaction system for enzyme digestion was 17.5 μL of purified PCR product (500 ng), 2 μL of Buffer, and 0.5 μL of HinfI enzymes (TaKaRa, Japan) at 37°C in the dark for 6 h (Yu et al., 2014; Zhang Q. X. et al., 2018), and sequenced by Sangon Biotech Co., Ltd., China.
Statistical analyses were performed using IBM SPSS 20.0 (IBM SPSS Statistics, IBM Corporation, United States) via a one-way analysis of variance (ANOVA). The figures were plotted with Microsoft Excel 2013 (Microsoft Corporation) and Graphpad prism 7.0 (GraphPad software, Inc., United States). The standard deviation (SD) was illustrated using an error bar. The significant differences among treatment-groups and controls were illustrated using the different letters above the columns at the p < 0.05 level via the least significant difference (LSD) test.
Terminal restriction fragment length polymorphism profiles were analyzed using the Peak ScannerTM Software v1.0 (Thermo Fisher Scientific, Wilmington, United States) with an exclusion of, T-RFs less than 50 bp in length or contributing less than 0.5% of peak area in each sample for subsequent analyses. The apparent T-RFs sizes in capillary electrophoresis are compared against MiCA database to analysis the phylotype. The R statistical platform (v.4.1.1) was used for principal coordinates (PCoA) and cluster analysis to study the difference of sample community composition. The differences between samples were calculated by Bray–Curtis, and analysis of similarity (ANOSIM) was conducted to identify the significant differences between the fungal communities (Liu et al., 2019; Xu et al., 2020). Richness index (SR) and evenness index (E) were calculated by Bio-dap software were calculated by Bio-dap software (Fierer et al., 2003; Jun and Dongyan, 2004; Shyu et al., 2007; Yuan et al., 2007; Percent et al., 2008).
Based on colony morphology, size, color, smoothness, and other properties, 509 bacterial strains of differing morphologies were isolated from healthy apple trees in replanted orchards. Antibacterial testing was performed by the flat stand-off and filter paper methods, and 256 strains of biocontrol bacteria were re-screened (Supplementary Table 1). The strain QSB-6 showed a particularly strong broad-spectrum inhibitory effect, and therefore, this strain was selected for further study.
The strain QSB-6 was cultured on LB agar at 37°C for 24 h; the surface of the colony was opaque and yellowish and showed the formation of folds (Supplementary Figure 1A). It can form biofilms when grown in liquid medium. The bacteria exhibited a blue–purple color after Gram staining and were identified as Gram-positive. When observed under a fluorescence microscope (100×/1.30 oil lens), individual bacteria were short and rod-shaped; they formed ovoid spores (0.6–0.8 × 1.0–1.4 μm), mesophytic, or subterminal, and the cyst was not enlarged (Supplementary Figures 1B,C). Rod-shaped cells of approximately 0.7–0.9 × 1.8–3.0 μm were observed under an SU-8010 scanning electron microscope (Supplementary Figures 1D–I).
The strain QSB-6 was able to utilize D-fructose, D-cellobiose, sucrose, α-D-glucose, gelatin, L-lactic acid, and citric acid as carbon sources. It was sensitive to pH 5, pH 6, 1% NaCl, 4% NaCl, 8% NaCl, 1% sodium lactate, and sodium butyrate (Supplementary Table 7). Physiological and biochemical tests showed that strain QSB-6 could produce hydrogen peroxide and hydrogen sulfide, hydrolyze starch, and reduce nitrate. Contact enzyme, arginine dihydrolase, methyl red reaction, Voges–Proskauer reaction, and gelatin hydrolysis enzyme tests were positive, whereas indole enzyme and urea enzyme reaction tests were negative. According to Bergey’s Manual of Systematic Bacteriology (2nd edition) and the Common Bacterial Identification Manual, the physiological and biochemical properties of strain QSB-6 matched those of B. amyloliquefaciens (Supplementary Table 8).
Approximately 554–929 bases were sequenced for rpoB and gyrA, 1452 bases for 16S rDNA, and 1066 bases for gyrB. Congruency analysis revealed no conflict between the 16S rDNA, gyrA, gyrB, and rpoB sequences, and the sequences were therefore combined. The combined sequence dataset included 38 ingroup taxa, with Paenibacillus polymyxa (BLB267) as the outgroup taxon. ML analysis of identities based on the four gene sequence alignments revealed that strain QSB-6 had the highest homology with B. amyloliquefaciens (Figure 1). In summary, strain QSB-6 was identified as B. amyloliquefaciens.
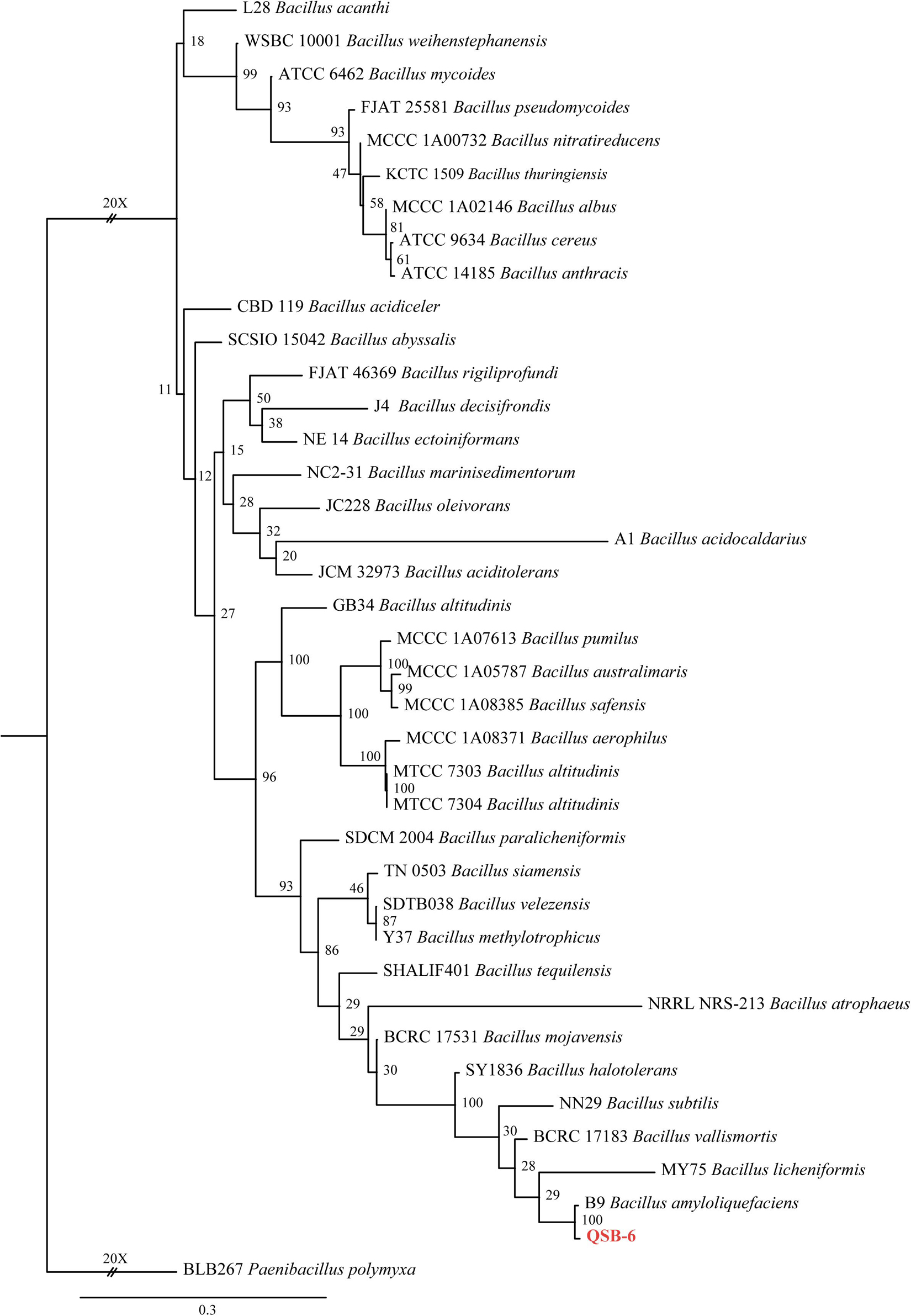
Figure 1. The ML consensus tree inferred from the combined 16S rDNA, gyrA, gyrB, and rpoB sequence alignment. Support for each branch in the inferred tree was evaluated using 1000 bootstrap replications. Support values (ML bootstrap and posterior probability values) are indicated at the branches. The scale bar indicates 0.3 expected changes per site. Clade numbers and Latin name are provided on the right of the tree and these are used for reference in the treatment of the species. The tree is rooted to Paenibacillus polymyxa (BLB267). Strains QSB-6 are indicated in bold and red.
Strain QSB-6 had a strong inhibitory effect on the mycelial growth and spore germination of Fusarium (Figures 2, 3). The inhibitory rate of strain QSB-6 against F. proliferatum was highest, reaching 87% in the PDA plate. The spore germination rate after treatment with fermentation broth and cell-free culture filtrate was 76% lower than that of the control (Table 1 and Figure 3). QSB-6 also had an inhibitory effect on F. oxysporum, F. verticillioides, and F. solani, with inhibition rates of 74, 73, and 84%, and the spore germination rate after treatment dropped by more than 60% compared with the control (Table 1 and Figure 3). The biocontrol bacteria QSB-6 also had an inhibitory effect on 7 common pathogenic fungi, and the inhibitory effect was greater than 48% (Table 1). In particular, the inhibition rate of A. alternata reached 87%, followed by the inhibition rates of V. mali, R. solani, and Phoma sp., which also reached greater than 68%. All pathogens exhibited an inhibition zone at the colony junction. These preliminary results showed that strain QSB-6 had a broad-spectrum inhibitory effect on plant pathogens and underscored its potential for development as a broad-spectrum agricultural biocontrol agent.
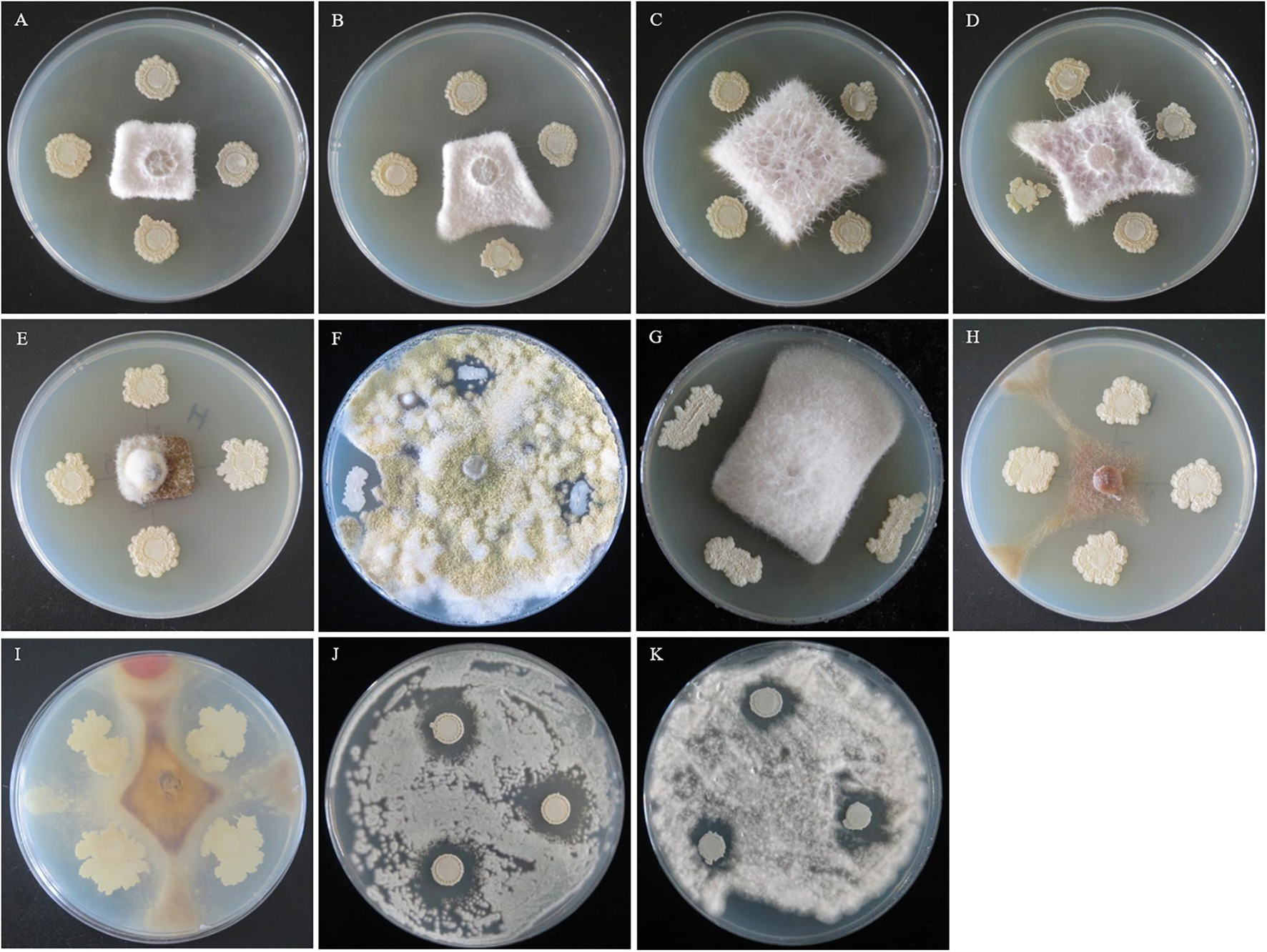
Figure 2. The flat standoff test between strain QSB-6 and pathogenic fungi (PDA medium). (A) Fusarium proliferatum, (B) Fusarium solani, (C) Fusarium verticillioides, (D) Fusarium oxysporum, (E) Alternaria alternata, (F) Aspergillus flavus, (G) Phoma sp., (H) Valsa mali, (I) Rhizoctonia solani, (J) Penicillium brasilianum, and (K) Albifimbria verrucaria.
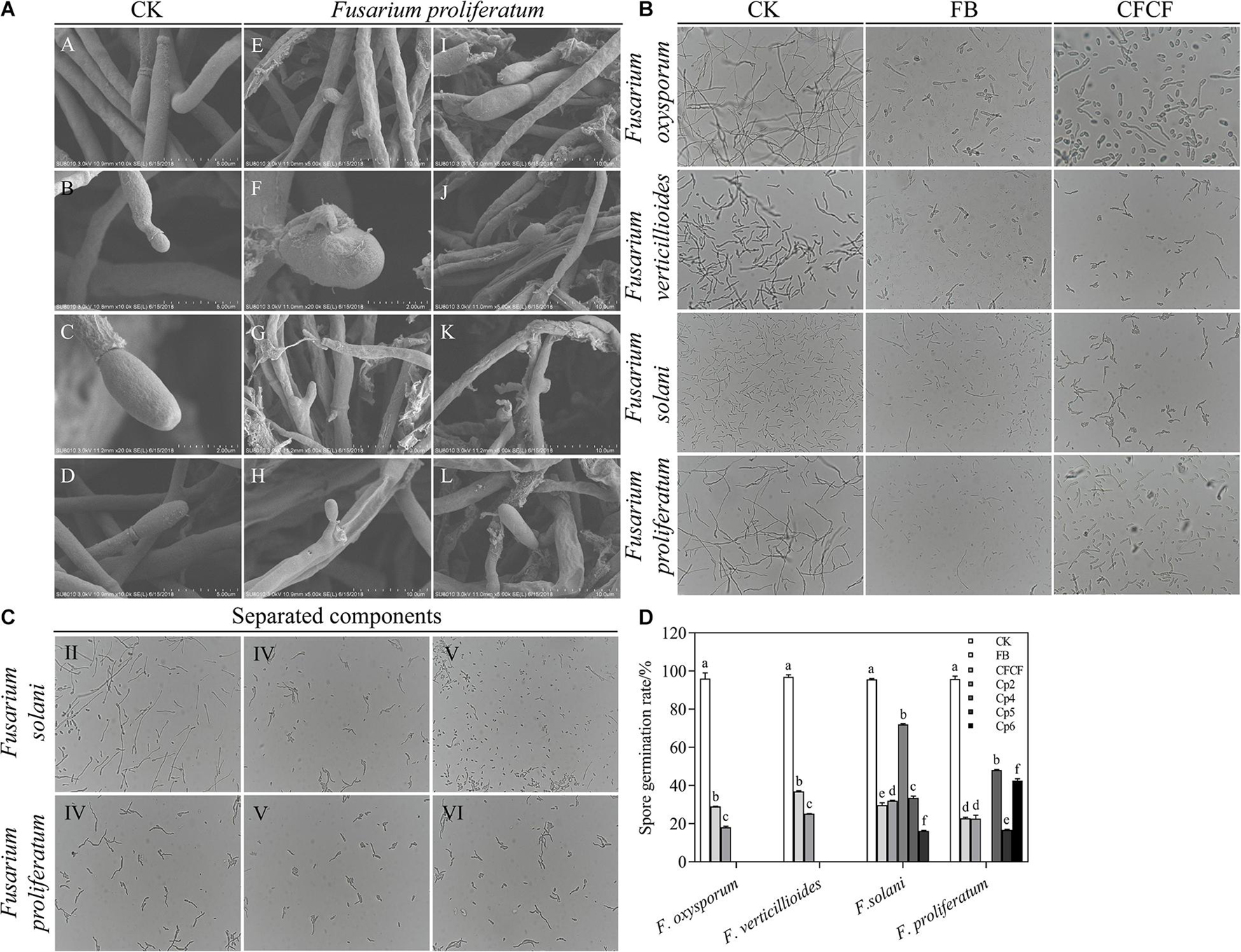
Figure 3. (A) The mycelia and spore morphology of Fusarium proliferatum under the scanning electron microscope. (A–D) Was the normal mycelium and spore, (E–L) was the mycelium treated with cell-free culture filtrate. The mycelium showed irregularly reticulated and uneven in thickness (E,G,J), shrinkage (E–L), twisted (K,L), thinned (E,I–L), broken (E–L), the spore cell wall was broken and deformed (H–L), and the mycelium were dissolved, ruptured and overflowed cell contents (J–L). (B) The effect of strain QSB-6 on the spore germination of Fusarium. CK Fusarium spore suspension was mixed with sterile water at 1:1, FB Fusarium spore suspension was mixed with fermentation broth at 1:1, CFCF Fusarium spore suspension was mixed with cell-free culture filtrate at 1:1. (C) The effect of separated components of fermentation broth on spore germination of Fusarium solani and Fusarium proliferatum. (D) Spore germination rate of different treatments, Cp2, Cp4, Cp5, Cp6 represent the separated substances. Lowercase letters above the columns indicate a significant difference at p < 0.05. Values are mean ± SD.
The control Fusarium mycelium was uniform in thickness and slender with fewer branches; it was full of spores, exhibited a complete structure, and appeared to be in a good growth state (Supplementary Figures 2A,B and Figures 3A, A–D). The mycelium treated with the cell-free culture filtrate was irregularly reticulated, swollen, and uneven in thickness; its hyphae appeared to be curved at the tip, twisted, thinned, broken, shrunken, and shriveled. The mycelia were dissolved and ruptured, and their cell contents had overflowed (Supplementary Figures 2C–H and Figures 3A, E–L). The dry weight of Fusarium hyphae decreased significantly in response to treatment with fermentation broth and, was reduced by 55.87, 58.45, and 81.14% after 6, 12, and 24 h of treatment relative to the control group (Supplementary Figure 3). As the concentration of fermentation broth increased, the mycelial dry weight decreased significantly and then stabilized. These results indicated that metabolites produced by QSB-6 during fermentation affected the normal growth of Fusarium mycelium.
As the temperature of the cell-free culture filtrate rose, its antibacterial rate clearly decreased (Supplementary Figure 4A), perhaps owing to the denaturation and loss of biological activity of antibacterial substances (proteins, enzymes, etc.) in the broth under high temperatures. As pH increased, the antibacterial rate first increased and then decreased; it was close to the antibacterial rate of the control group at pH 7 (Supplementary Figure 4C). The physiological activity of the antibacterial substance(s) in the cell-free culture filtrate may have been inhibited under acidic or alkaline conditions. The antibacterial rate of the cell-free culture filtrate after UV irradiation or exposure to a light intensity of 4500 ± 500 lx did not differ from that of unirradiated broth and always remained above 70% (Supplementary Figures 4B,D). The antagonistic active substance in the cell-free culture filtrate was therefore not sensitive to light.
The results obtained by the test tube method and by thin layer chromatography were essentially the same, confirming the accuracy of the analysis results. Metabolites produced by strain QSB-6 consisted of steroid, lactones, coumarins and their glycosides, flavonoids, cardiac glycosides, amino acids, peptides and proteins, sugars, polysaccharides, phenols, alkaloids, organic acid, and saponin (Supplementary Tables 9, 10). After chromatographic separation, components IV and V were found to have a significant inhibitory effect on the growth of pathogenic fungi and the germination of Fusarium spores (Figures 3C,D and Supplementary Figure 5). After GC–MS chromatographic detection and analysis, the main antibacterial substances with Area% >1.0 and retention index RI >1000 was selected from component V (Table 2 and Supplementary Figure 10), and the main antibacterial substances with Area% >0.5 and retention index RI >1000 was selected from component IV (Table 3 and Supplementary Figure 10). Most substances identified from the two components were organic acids and esters. NIST spectral database analysis of the compounds’ mass spectra identified them as terephthalic acid, 1,2-benzenedicarboxylic acid, isopropyl methyl phthalate, phthalic acid, benzeneacetic acid, carbamic acid, methyl 4-hydroxyphenylacetate, sec-butyl 3,5-dinitrobenzoate, 4-(2-methoxyethyl)phenol (Figure 4). The mass spectra of the compounds are shown in Supplementary Figure 6. An in vitro antibacterial test demonstrated that terephthalic acid, diisopropyl ester, 1,2-benzenedicarboxylic acid, carbamic acid (4-aminophenyl)-, methyl ester, and benzeneacetic acid, 3- hydroxy-, methyl ester had varying degrees of inhibitory effect on Fusarium, among which 1,2-benzenedicarboxylic acid has the best inhibitory effect on Fusarium, and as the concentration increases, the inhibitory effect on 11 strains of pathogenic fungi was stronger (Figure 5 and Supplementary Figure 7). At a concentration of 1000 μg/L, the inhibition rates of F. proliferatum, F. verticillioides, F. oxysporum, and F. solani were 77.74, 65.52, 68.85, and 47.61%, respectively (Supplementary Table 11). Of the four pure compounds, visual inspection revealed that plant shoot and root growth, when compared with the control treatment (water and ethanol), was only enhanced by 1,2-benzenedicarboxylic acid (100 μg) and benzeneacetic acid, 3- hydroxy-, methyl ester (500 μg). The length of primary root of Arabidopsis seedlings was increased approximately 1.50−fold by the presence of 1,2-benzenedicarboxylic acid and 4.03−fold by benzeneacetic acid, 3- hydroxy-, methyl ester, compared to ethanol treatment, respectively (Supplementary Figure 9). These results indicate that the principle antifungal compounds produced by strain QSB-6 are organic acid esters with benzene ring. It plays important roles in disease control and in growth promoting processes.
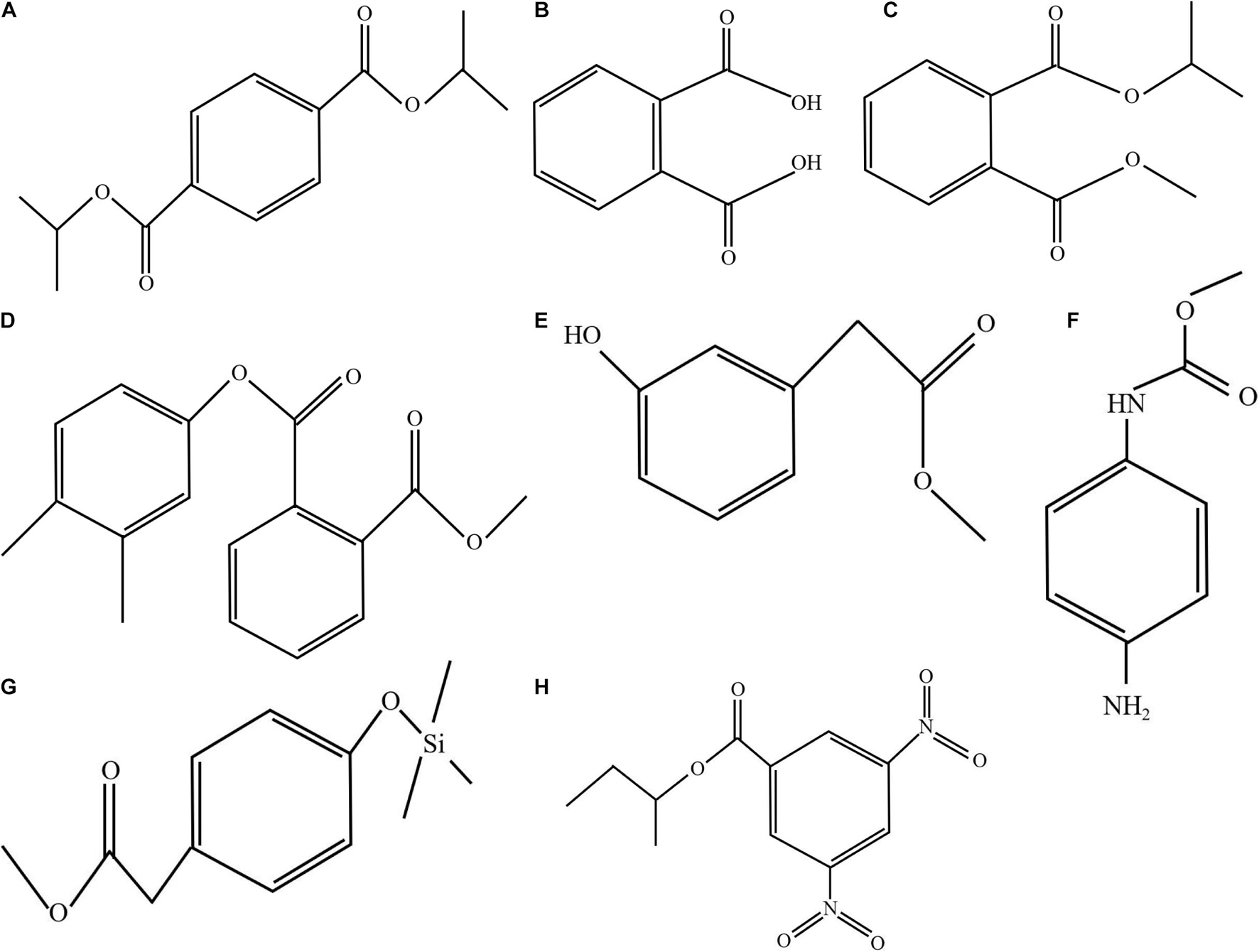
Figure 4. Molecular structure of the representative compounds. Mass spectrum of the compound obtained by GC–MS analysis was compared with the NIST17 spectral database. (A) Terephthalic acid, diisopropyl ester (GC RT 28.92, 30.022, 29.634, and 29.835 min); (B) 1,2-benzenedicarboxylic acid (RT 22.28 and 23.058 min); (C) isopropyl methyl phthalate (RT 26.132, 26.542, and 27.051 min); (D) phthalic acid, 3,4-dimethylphenyl methyl ester (RT 27.647 min); (E) benzeneacetic acid, 3- hydroxy-, methyl ester (RT 27.122, 26.117, 31.463, and 31.223 min); (F) carbamic acid (4-aminophenyl)-, methyl ester (RT 26.698 and 25.615 min); (G) methyl 4-hydroxyphenylacetate, TMS derivative (RT 25.895); and (H) sec-butyl 3,5-dinitrobenzoate (RT 27.55 min).
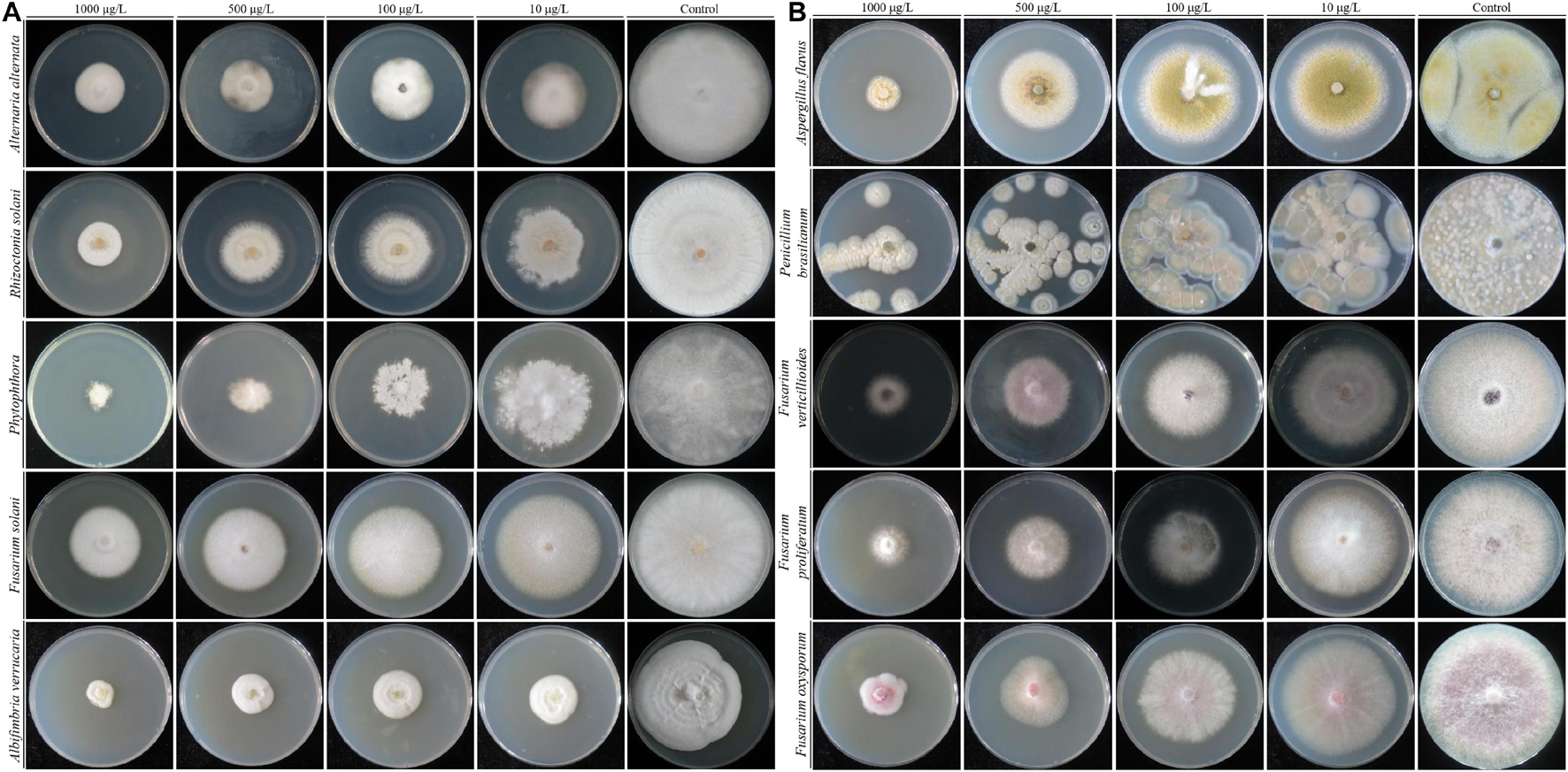
Figure 5. Growth inhibition of plant fungal pathogens on the PDA medium after treated with different gradient 1,2-benzenedicarboxylic acid. (A) The inhibitory effect on the growth inhibition of Alternaria alternata, Rhizoctonia solani, Phytophthora, Fusarium solani and Albifimbria verrucaria. (B) The inhibitory effect on the growth inhibition of Aspergillus flavus, Penicillium brasilianum, Fusarium verticillioides, Fusarium proliferatum and Fusarium oxysporum.
Observation of plant root sections showed that the root system was mainly composed of three parts: (from outside to inside) the epidermis, the cortex (the outer cortex, the cortical parenchyma, the Kjeldahl belt), and the vascular column (the central sheath, phloem, and xylem). Root epidermal cells infected by Fusarium were deformed, broken, detached, and/or irregularly arranged. Fungal hyphae invaded the cortical cells through the intercellular layer, then entered the vascular column. New hyphae were visible mainly close to the cell wall, and mature hyphae were scattered in the cells. There was also a large amount of cell contents (viscous substances and starch granules) in the cortical cells and vascular column (Figure 6 and Supplementary Figure 8). The epidermal and cortical cells treated with both strain QSB-6 and Fusarium appeared slightly shrunken and ruptured, and the conidia and hyphae of Fusarium were attached only to the root epidermal cells (Figures 6A–D). Root systems from the control treatment were complete and neatly arranged (Figure 6, Mock).
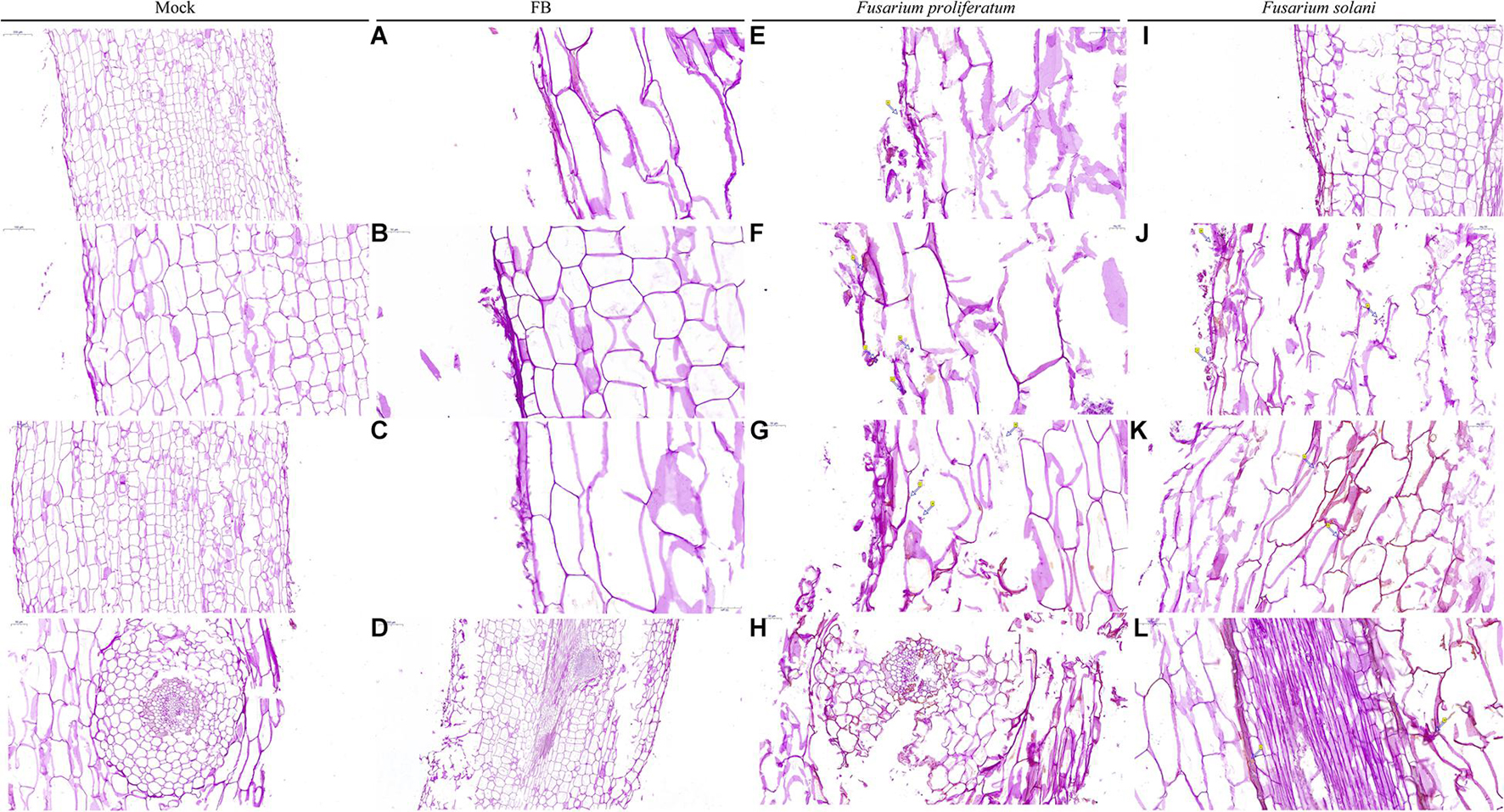
Figure 6. Microscopic observation of the plant root stained by PAS. Mock: sterile distilled water treatment, FB fermentation broth treatment. Fusarium proliferatum and Fusarium solani: the roots of the plants were soaked with the conidia suspension for 12 h. PAS staining turns the polysaccharides on the fungal wall purple-red. Mock: the root tissue was intact, the cell boundaries are clear and neatly arranged. FB: The conidia and hyphae of Fusarium were attached to the epidermis, the epidermis and cortex cells were slightly broken, deformed, and the internal tissue structure is intact (A–D). (E,G,I) Dense mycelium appeared in the epidermis of the root system, and the epidermal and cortical cells appeared ruptured, and deformed (arrows), and the cells were arranged irregularly. (F) Cauliflower-like structure appeared in the infected root areas (arrows). (H,J–L) The conidia and hyphae of Fusarium appeared in the cortex and vascular column.
The CK2, T2, and T1 treatments significantly promoted the growth of plants in July, August, and September. The relative treatment effects were ranked from high to low: CK2 > T2 > T1 > CK1 (Table 4). In September, plant height, ground diameter, aboveground fresh weight, belowground fresh weight, aboveground dry weight, and belowground dry weight were 1.66, 1.32, 1.67, 1.39, 2.53, and 2.49 times higher in the T2 treatment that in the T1 treatment.
Root structure clearly differed among the different treatments (Figure 7A). In July, there were no significant differences in length, surface area, tips, and forks among the different treatments. In September, the plants had grown considerably, and root length, surface area, tips, and forks were significantly lower in the CK1 treatment than in the CK2 and T2 treatments. The length, surface area, tips, and forks were 1.74, 3.32, 6.08, and 2.17 times higher in the T2 treatment that in the CK1 treatment (Figures 7B–E). The root respiration rate and the SOD, POD, and CAT activities increased from July to August and September in all treatment groups. These increases were significantly higher in the T2 treatment than in CK1, whose values were close to those of CK2 (Table 5). In September, the root respiration rate of T2 was 1067.27 μmol O2⋅g–1 FW⋅min–1, which was 1.53 and 1.36 times higher than those of CK1 and T1. The activities of SOD, POD, and CAT in the T2 treatment were 1.50, 2.87, and 1.74 times higher than those in CK1. MDA content showed the opposite trend: compared with CK1, MDA content of T2 decreased by 37.65, 59.82, and 78.10% in July, August, and September, respectively.
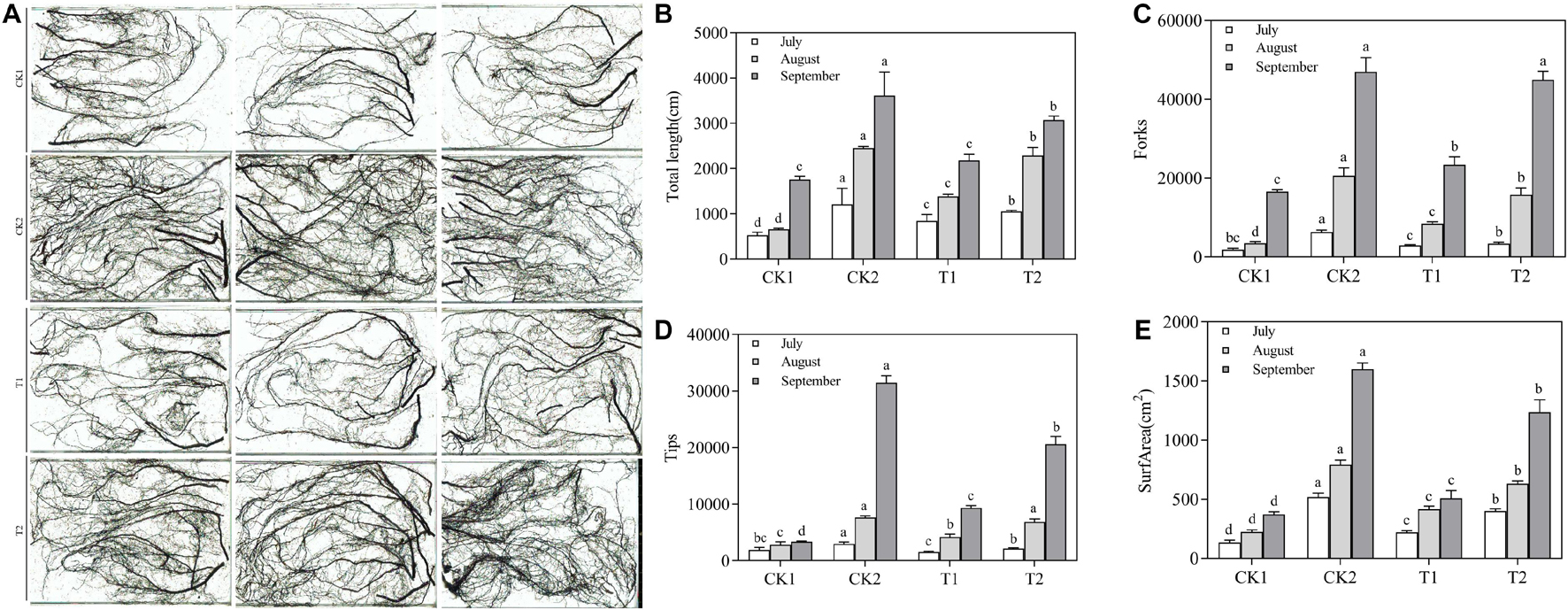
Figure 7. Effect of different treatments on the root architecture of Malus hupehensis Rehd. seedlings. (A) Root system scan obtained by Microtek ScanMaker i800 Plus, (B) total root length, (C) the number of root bifurcation, and (D) root tips, (E) root surface area.
The number of soil bacteria increased significantly after T2 treatment in July, August, and September; their numbers were 12.73, 9.72, and 9.64 times higher in T2 than in CK1 (Table 6). In September, the number of soil fungi differed significantly among treatments. Soil fungal numbers were reduced by 85.58, 17.31, and 81.74% in CK2, T1, and T2 compared with CK1. The number of soil fungi after T1 treatment increased by 13.19% in September compared with July. The number of actinomycetes in the soil and the ratio of soil bacteria/fungi were significantly higher in the T2 treatment than in CK1. The soil bacteria/fungi ratios in July, August, and September could be ranked T2 > CK2 > T1 > CK1. The real-time quantitative PCR results indicated there was a significant difference (p > 0.05) in CFU levels of F. proliferatum, F. verticillioides, F. oxysporum, and F. solani in the soil of different treatments (Table 6). Compared with CK1, the abundance of the four Fusarium species were significantly lower in the CK2 and T2 treatments in July, August, and September. In September, The abundance of F. proliferatum, F. verticillioides, F. oxysporum, and F. solani decreased by 81.30, 71.08, 59.26, and 50.88% in the T2 treatment compared with CK1, respectively. The results of PCoA (Figure 8A) and cluster analysis (Figure 8B) showed that the fungal community structures of the T2 and CK2 treatments differed significantly from that of CK1, and the fungal community structures of T1 and CK1 were similar. These results showed that strain QSB-6 also had a good inhibitory effect on the growth of Fusarium in the soil environment.
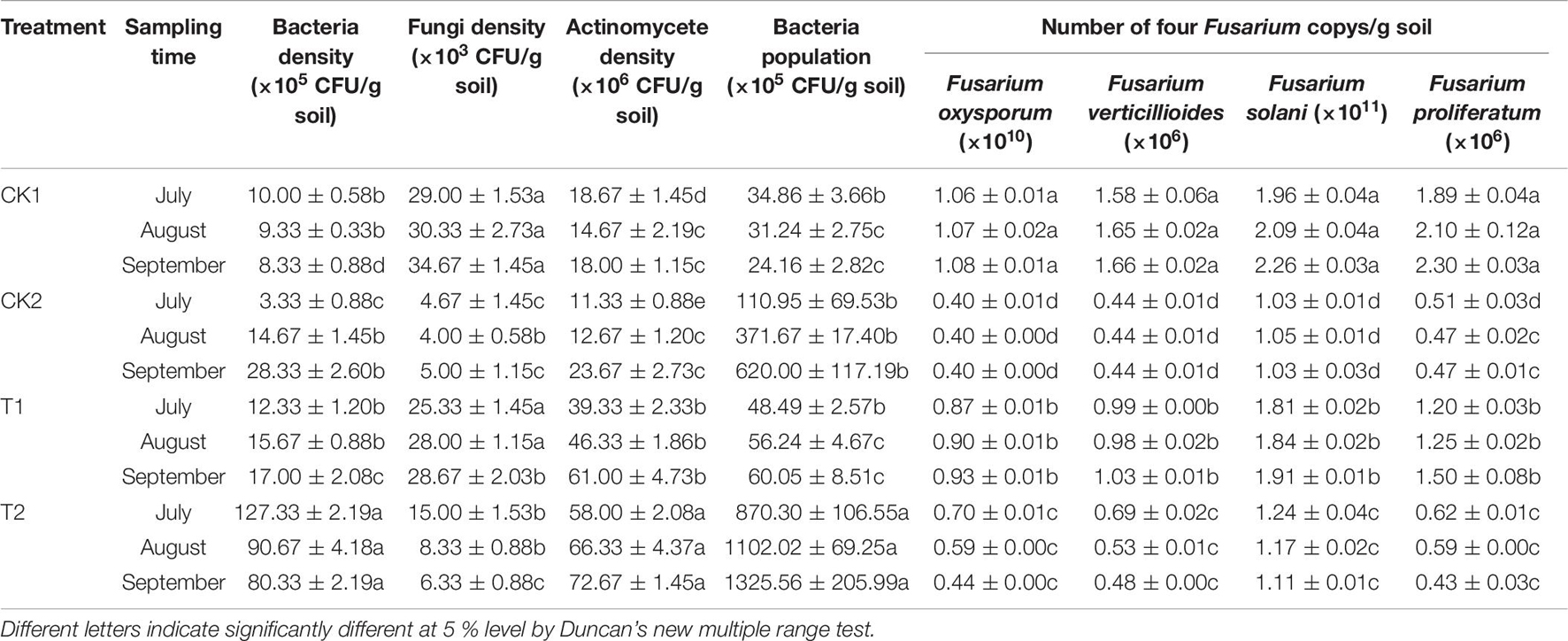
Table 6. The effect of strain QSB-6 on the density of microorganisms in the rhizosphere of Malus hupehensis Rehd. seedlings.
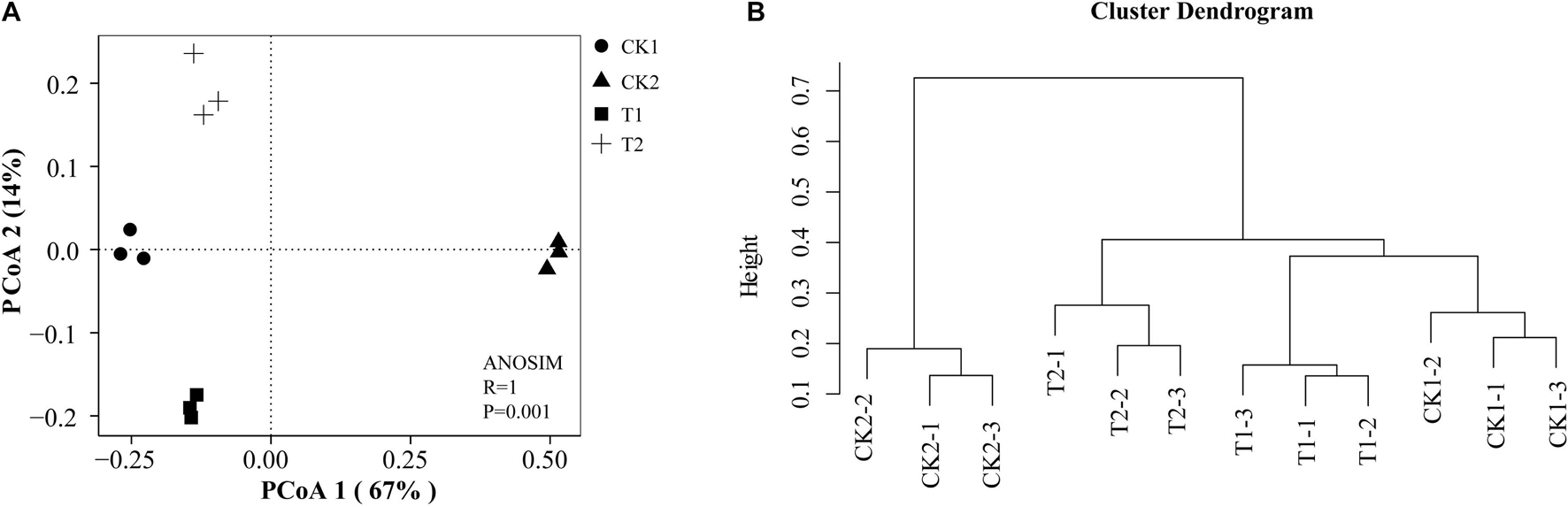
Figure 8. Fungi community structures in the different treatments. Principal coordinate analysis (PCoA) (A) and cluster analysis (B) plot based on the OTUs of Bray–Curtis distance. R = 1 > 0, p < 0.05 means that the difference between groups is greater than the difference within groups, and there are significant differences between different treatments. Each treatment contains three repetitions.
Fusarium spp. include a large number of complex fungi and ascomycete teleomorphs; many Fusarium species produce toxic secondary metabolites and/or cause serious plant diseases (Nelson et al., 1983). F. verticillioides and F. proliferatum are harmful to corn due to their production of fumonisin mycotoxins (Marín et al., 2010). Edel-Hermann and Lecomte (2019) found that Fusarium spp. – particularly formae speciales of F. oxysporum – were important vascular wilt pathogens of many agricultural crops such as ornamental plants and garden crops. Fusarium has been identified as a dominant pathogen in rhizosphere soil from continuously cropped soybean and potato fields (Meng et al., 2012; Bai et al., 2015), and also has been reported to be one of the potential soil pathogens that cause ARD (Van Schoor et al., 2009; Kelderer et al., 2012; Wang G. S. et al., 2018). At present, biological control methods have been widely used to control Fusarium spp., and their main mechanisms include antagonism, competition, and induction (Weller et al., 2002; Mazurier et al., 2009; Kejela et al., 2016; Gómez Expósito et al., 2017). The measurement of antagonistic activity (i.e., the dual−culture plate assay) is the most commonly used method for screening biocontrol bacteria (Hermosa et al., 2000; Gómez Expósito et al., 2017). Agarwal et al. (2017) used the above method to isolate a B. pumilus MSUA3 from higher altitude of Himalayan ranges capable, which can strongly inhibited the growth of R. solani and F. oxysporum by producing chitinolytic enzymes and an antibiotic surfactin, causing lysis, and deformities in the hyphae. Chang et al. (2007) isolated a strain of B. cereus QQ308 that can grow on shellfish chitin wastes, which can secrete a complex of hydrolytic enzymes, including chitinase, chitosanase, and protease, inhibited spore germination and germ tube elongation of F. oxysporum, F. solani, and P. ultimum.
In this study, B. amyloliquefaciens strain QSB-6, which showed good inhibitory effects toward a variety of pathogens, was screened by the flat standoff test. Fusarium spore germination rate was reduced by more than 60% relative to the control following treatment with fermentation broth and cell-free culture filtrate. Strain QSB-6 also can strongly inhibited the growth of Fusarium by producing antibacterial substances, causing lysis and deformities in the hyphae. The above results indicated that strain QSB-6 is a good biological control agent against Fusarium. However, Many biocontrol bacterial strains are sensitive to external conditions (i.e., light, high temperature, soil pH extremes), resulting in poor control efficacy in the field (Alabouvette et al., 2009; Armin et al., 2021). We therefore investigated the physical and chemical properties of extracellular metabolites from strain QSB-6. It showed good thermal and acid-base stability and was not sensitive to ultraviolet and visible light, indicating that strain QSB-6 has good development potential and application prospects.
The metabolites produced by microorganisms are very complex, and their specific molecular structures are also very diverse. The antibacterial substances produced by specific strains also differ. Antibacterial substances that inhibit pathogens tend not to act alone, but instead two or more antibacterial substances act synergistically to produce antibacterial effects (Hyakumachi et al., 2014; Carrión et al., 2019; Joo et al., 2020; Newman and Cragg, 2020; Armin et al., 2021). 2,4-Diacetylphloroglucinol (DAPG) and phenazines (PHZ) produced by Pseudomonas species have been used as antibiotics to control disease suppressive soils and Fusarium wilt (Weller et al., 2002; Haas and Defago, 2005; Mazurier et al., 2009; Raaijmakers and Mazzola, 2012). Kejela et al. (2016) isolated B. amyloliquefaciens strain BT42 from the root rhizosphere and demonstrated that it can antagonize Colletotrichum gloeosporioides and F. oxysporum and promote plant growth. Its main secondary metabolite that inhibits pathogenic fungi is harmine (β-carboline alkaloids). Jeong et al. (2017) found that B. licheniformis MH48 and its metabolite, benzoic acid, clearly show antifungal activity against R. solani and C. gloeosporides. Zhao et al. (2010) isolated an antifungal compound from the n-butanol extract of culture filtrate of B. vallismortis ZZ185 and identified as Bacillomycin D (n-C14 and iso-C15), which has strong in vitro inhibition activity against plant pathogens such as F. graminearum, A. alternata, and R. solani.
In the present experiment, we used test-tube detection and thin-layer chromatography chromogenic methods to characterize the extract phase of fermentation broth from strain QSB-6 and found that its constituent metabolites included steroid, lactones, coumarins and their glycosides, flavonoids, cardiac glycosides, amino acids, peptides and proteins, sugars, polysaccharides and glycosides, phenols, alkaloids, organic acid, coumarins, and saponin. Most are reported to be antibacterial active ingredients in plant extracts, with no occur residual and toxic side effects, Some are used as anti-bacterial antiseptic additives (Demirtas et al., 2013; Djeussi et al., 2013; Desta et al., 2015; Móricz et al., 2015). The toxic components to Fusaria were identified by GC–MS as organic acid esters produced by strain QSB-6. Among them, 1,2-benzenedicarboxylic acid and benzeneacetic acid, 3- hydroxy-, methyl ester had the strongest inhibitory effects on Fusarium growth. This result provides a theoretical foundation for the development of new microbial fungicides.
Bacillus can produce antagonistic substances to inhibit the growth of plant pathogens by colonizing the plant rhizosphere or body surface. It may also compete with pathogens for space and/or nutrition or induce plant resistance, thereby achieving the purpose of biological control (Junaid et al., 2013; Jambhulkar et al., 2015; Gómez Expósito et al., 2017). Many studies have shown that plant resistance involves the accumulation of reactive oxygen species, antioxidant enzymes, and lignin (Pshibytko et al., 2006; Dodds and Rathjen, 2010; Li et al., 2016). The plant antioxidant enzyme system includes SOD, POD, CAT, and other enzymes (Sharma and Dubey, 2007; Radwan et al., 2010; Zhang Y. et al., 2016). The enhancement of plant antioxidant enzyme activity can reduce the accumulation of MDA in response to oxidative stress, maintaining the stability of the cell membrane and enhancing plant tolerance (Balal et al., 2012). At the same time, Bacillus can also promote plant growth by degrading organic matter to release plant nutrients, improve root vitality, and optimize root architecture (Glick, 1995; Nagórska et al., 2007). Root morphological parameters play an important role in plant development because nutrient uptake and water absorption capacity is more dependent on root length and root surface area than on total root biomass (Boot, 1989; Bresson et al., 2013; Syed-Ab-Rahman et al., 2019). The results of this study are consistent with these proposed mechanisms. QSB-6 fertilizer treatment (T2) promoted significant increases in plant root respiration rate and protective enzyme activities (SOD, POD, and CAT); MDA content was reduced, and root length, surface area, tips, and forks were significantly increased, thereby promoting greater plant growth. The promotion of root growth may also be related to the metabolites produced by strain QSB-6. The length of primary roots and the number of lateral roots of A. thaliana plantlet treated with metabolites (1,2-benzenedicarboxylic acid and benzeneacetic acid, 3- hydroxy-, methyl ester) increased significantly. Plant roots treated with strain QSB-6 also showed less damage from Fusarium, suggesting that QSB-6 had colonized the host plant surface or the infection site, forming a biofilm on the root epidermis and preventing pathogen intrusion (Raupach and Kloepper, 1998; Romero et al., 2004; Ji et al., 2008), consistent with the results of Chen et al. (2012). Large amounts of viscous substances and starch granules were also produced in the infected root cortical cells and vascular cylinders. Viscous substances present a strong mechanical barrier that inhibits microbial invasion and can also prevent the longitudinal expansion of microbial infection by blocking the xylem vessel (Daayf et al., 1995; Lagopodi et al., 2002). Fusarium can be classified morphologically as Nectria-like. Intracellular condensed nodular fungal structures with the shape of a cauliflower head are reported to occur in infected root areas (Grunewaldt-Stöcker et al., 2020), and similar structures were observed in this experiment.
In recent years, Bacillus species have been considered as promising agents for the control of diverse plant pathogens. Bacillus are applied primarily in the form of a solid or liquid fertilizers (Shahena et al., 2021), and they have shown a good control effect in cucumber (Fusarium wilt, downy mildew, etc.), pepper (bacterial wilt, soft rot, etc.), rice (rice blast, sheath blight, etc.), wheat (root rot, leaf blight, etc.), and other crop diseases (Baker et al., 1983; Wilson and Pusey, 1985; Lin et al., 2001; Chan et al., 2003; Souto et al., 2004; Ouhaibi-Ben Abdeljalil et al., 2016; Zalila-Kolsi et al., 2016; Zhang M. et al., 2016; Fan et al., 2017b; Gómez Expósito et al., 2017; Qin et al., 2017; Barratt et al., 2018; Wang X. Q. et al., 2018). Ramírez et al. (2021) isolated a B. cereus MH778713 from root nodules of Prosopis laevigata, and the volatile hentriacontane and 2,4-di-tert-butylphenol produced by it can effectively inhibited the radial growth of F. oxysporum and protected tomato plants against Fusarium wilt. Huang et al. (2012) developed a novel bio-organic fertilizer (BIO) by fermenting mature compost with B. pumilus N43 can effectively control of Rhizoctonia solani damping-off disease in cucumber. The QSB-6 bacterial fertilizer used in this experiment adopts solid state fermentation (cow manure: straw = 3:1), which has the advantages of inexpensive substrates, continuality of the process, low waste volumes, and beneficial to the survival of microorganisms (Wang et al., 2009; Dubey and Dubey, 2010; Duan et al., 2020). In September, the effects of QSB-6 bacterial fertilizer and methyl bromide fumigation treatment (Mbr) were almost as good, and both significantly promoted the growth of plant seedlings. When applying in the field, band placement (planting tree row) and localized placement (tree hole) is used and the application rate is about 240-300 g/m2 (400–460 USD/acre). Compared with the common fumigants dimethyl disulfide (DMDS) (60 g/m2), Methyl bromide (568.50 USD/acre), and 1,3-dichloropropene (600 USD/acre) in production, the cost is lower and it is more conducive to farmers’ income (Raski et al., 1976; Pecchia et al., 2017). The above results indicated that strain QSB-6 has the potential to become an excellent biological control agent to replace fumigants to control ARD.
Previous studies have shown that the occurrence of ARD is closely related to changes in the composition of the microbial community in orchard soils. Long-term replant cropping will lead to an imbalance in the rhizosphere microbial population, reducing beneficial microbes and increasing the abundance of soil-borne fungal pathogens, ultimately leading to a decrease in crop yield (Mazzola, 1998; Yim et al., 2013; Franke-Whittle et al., 2015; Spath et al., 2015). At present, T-RFLP, qPCR, and DNA stable isotope probing (DNA-SIP) techniques are often used to study soil microbial community structure (Cretoiu et al., 2013; Gómez Expósito et al., 2017; Moein et al., 2019). Shennan et al. (2018) found that when rice bran was utilized as the ASD carbon input, significant changes in bacterial and fungal community composition were observed in the soil as characterized by T-RFLP analysis. Here, we used T-RFLP and qPCR to show that QSB-6 fertilizer treatment can significantly change the soil fungal community structure, reduce the number of soil fungi, and reduce the abundance of Fusarium. A similar finding was reported in a biocontrol study of Watermelon Fusarium wilt (Zhao et al., 2014). The above results showed that adding QSB-6 bacterial fertilizer to the soil had a better inhibitory effect on the fungi in the soil, especially Fusarium, and the effect was almost as good as the Mbr treatment with the extension of time. The number of bacteria in the soil has also increased significantly, and the soil essentially became a high-fertilizer “bacterial” soil. Strain QSB-6 may consume large amounts of soil nutrients, increase its own growth rate, and thereby reduce the level of nutrients needed for the survival of plant pathogens, inhibiting their growth (Gómez Expósito et al., 2017; Shi et al., 2020). It shows that the application of QSB-6 bacterial fertilizer can provide a healthy soil microbial environment for the growth of plant seedlings. Strain QSB-6 is classified as first level (i.e., exempt from toxicological testing) according to the general technical guidelines for microbial fertilizer safety. This safe and non-pathogenic microorganism can be used as a green, environmentally friendly biocontrol agent for ARD.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
ZM and CY contributed to conception and design of the study. YD organized the database. YD, RC, RZ, and WJ performed the statistical analysis. YD wrote the first draft of the manuscript and contributed in the GC–MS analysis. RC and RZ wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was financially supported by China Agriculture Research System of MOF and MARA (CARS-27); the National Natural Science Foundation of China (No. 32072510); Shandong Agricultural Major Applied Technology Innovation Project (No. SD2019ZZ008); Taishan Scholar Funded Project (No. ts20190923); Qingchuang Science and Technology Support Project of Shandong Colleges and Universities (No. 2019KJF020); and Natural Science Foundation of Shandong Province (No. ZR2020MC131). We declare that the funding bodies had no role in the design of the study, in the collection, analysis, and interpretation of data, or in writing the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the teachers, students, and non-academic Staff of National Key Laboratory of Crop Biology, College of Horticulture Science and Engineering, Shandong Agricultural University, for their help with the wet lab experiments and data analysis. We would also like to thank Jiwen Xia for their guidance in developing phylogenetic trees and Zhentao Wang for their guidance in separation and identification of secondary metabolites.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.746799/full#supplementary-material
Agarwal, M., Dheeman, S., Dubey, R. C., Kumar, P., Maheshwari, D. K., and Bajpai, V. K. (2017). Differential antagonistic responses of Bacillus pumilus MSUA3 against Rhizoctonia solani and Fusarium oxysporum causing fungal diseases in Fagopyrum esculentum Moench. Microbiol. Res. 205, 40–47. doi: 10.1016/j.micres.2017.08.012
Alabouvette, C., Olivain, C., Migheli, Q., and Steinberg, C. (2009). Microbiological control of soil-borne phytopathogenic fungi with special emphasis on wilt-inducing Fusarium oxysporum. New Phytol. 184, 529–544. doi: 10.1111/j.1469-8137.2009.03014.x
Armin, R., Zühlke, S., Mahnkopp-Dirks, F., Winkelmann, T., and Kusari, S. (2021). Evaluation of apple root-associated endophytic Streptomyces pulveraceus strain ES16 by an OSMAC-assisted metabolomics approach. Front. Sustain. Food Syst. 5:643225. doi: 10.3389/fsufs.2021.643225
Avery, B. W. (1973). Soil classification in the soil survey of england and wales. J. Soil Sci. 243, 324–338. doi: 10.1111/j.1365-2389.1973.tb00769.x
Azabou, M. C., Gharbi, Y., Medhioub, I., Ennouri, K., Barham, H., Tounsi, S., et al. (2020). The endophytic strain Bacillus velezensis OEE1: an efficient biocontrol agent against Verticillium wilt of olive and a potential plant growth promoting bacteria. Biol. Control 142:104168. doi: 10.1016/j.biocontrol.2019.104168
Bai, L., Cui, J., Jie, W., and Cai, B. (2015). Analysis of the community compositions of rhizosphere fungi in soybeans continuous cropping fields. Microbiol. Res. 180, 49–56. doi: 10.1016/j.micres.2015.07.007
Baker, C. J., Stavely, J. R., Thomas, C. A., Sasser, M., and MacFall, J. S. (1983). Inhibitory effect of Bacillus subtilis on Uromyces phaseoli and on development of rust pustules on bean leaves. Phytopathology 73, 1148–1152. doi: 10.1094/phyto-73-1148
Balal, R. M., Khan, M. M., Shahid, M. A., Mattson, N. S., Abbas, T., Ashfaq, M., et al. (2012). Comparative studies on the physiobiochemical, enzymatic, and ionic modifications in salt-tolerant and salt-sensitive citrus rootstocks under NaCl stress. J. Am. Soc. Hortic. Sci. 137, 86–95. doi: 10.21273/jashs.137.2.86
Bao, S. D. (2000). Soil Agrochemical Analysis (in Chinese). Beijing: China Agricultural Press, 265–267.
Barratt, B. I. P., Moran, V. C., Bigler, F., and Van Lenteren, J. C. (2018). The status of biological control and recommendations for improving uptake for the future. BioControl. 63, 155–167. doi: 10.1007/s10526-017-9831-y
Bartholomew, J. W., and Mittwer, T. O. D. (1950). A simplified bacterial spore stain. Stain Technol. 25, 153–156. doi: 10.3109/10520295009110979
Benson, H. J. (2002). Microbiological applications: laboratory manual in general microbiology. McGrawHill 85, 173–174.
Boot, R. G. A. (1989). “The significance of size and morphology of root systems for nutrient acquisition and competition,” in Causes and Consequences of Variation in Growth Rate and Productivity of Higher Plants, ed. H. Lambers (The Hague: SPB Academic Publishing), 299–311.
Bresson, J., Varoquaux, F., Bontpart, T., Touraine, B., and Vile, D. (2013). The PGPR strain Phyllobacterium brassicacearum STM196 induces a reproductive delay and physiological changes that result in improved drought tolerance in Arabidopsis. N. Phytol. 200, 558–569. doi: 10.1111/nph.12383
Brown, G. S., Barnes Smith, S., Gibson, L., Kitchener, A., Potter, K., Jennings, D., et al. (2000). A Study of Apple Replant Syndrome. HRDC final report project AP97005. Sydney: Horticulture Australia Limited.
Carrión, V. J., Perez-Jaramillo, J., Cordovez, V., Tracanna, V., De Hollander, M., Ruiz-Buck, D., et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612. doi: 10.1126/science.aaw9285
Cavaglieri, L., Orlando, J. R. M. I., Rodríguez, M. I., Chulze, S., and Etcheverry, M. (2005). Biocontrol of Bacillus subtilis against Fusarium verticillioides in vitro and at the maize root level. Res. Microbiol. 156, 748–754. doi: 10.1016/j.resmic.2005.03.001
Cazorla, F. M., Romero, D., Pérez-García, A., Lugtenberg, B. J. J., Vicente, A. D., and Bloemberg, G. (2007). Isolation and characterization of antagonistic Bacillus subtilis strains from the avocado rhizoplane displaying biocontrol activity. J. Appl. Microbiol. 103, 1950–1959. doi: 10.1111/j.1365-2672.2007.03433.x
Chan, Y. K., McCormick, W. A., and Seifert, K. A. (2003). Characterization of an antifungal soil bacterium and its antagonistic activities against Fusarium species. Can. J. Microbiol. 49, 253–262. doi: 10.1139/w03-033
Chang, W. T., Chen, Y. C., and Jao, C. L. (2007). Antifungal activity and enhancement of plant growth by Bacillus cereus grown on shellfish chitin wastes. Bioresour. Technol. 98, 1224–1230. doi: 10.1016/j.biortech.2006.05.005
Chen, X., Zhang, Y., Fu, X., Li, Y., and Wang, Q. (2016). Isolation and characterization of Bacillus amyloliquefaciens PG12 for the biological control of apple ring rot. Postharvest Biol. Technol. 115, 113–121. doi: 10.1016/j.postharvbio.2015.12.021
Chen, Y., Gao, X., Chen, Y., Qin, H., Huang, L., and Han, Q. (2014). Inhibitory efficacy of endophytic Bacillus subtilis EDR4 against Sclerotinia sclerotiorum on rapeseed. Biol. Control 78, 67–76. doi: 10.1016/j.biocontrol.2014.07.012
Chen, Y., Yan, F., Liu, H., Chai, Y., and Guo, J. (2012). Biofilm formation of Bacillus subtilis on tomato roots enhances biocontrol efficacy against tomato bacterial wilt disease caused by Ralstonia solanacearum. Phytopathology 102, 21–22.
Chowdhury, S. K., Majumdar, S., and Mandal, V. (2020). Application of Bacillus sp. LBF-01 in Capsicum annuum plant reduces the fungicide use against Fusarium oxysporum. Biocatal. Agric. Biotechnol. 27:101714. doi: 10.1016/j.bcab.2020.101714
Chun, J., and Bae, K. S. (2000). Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyrA gene sequences. Antonie Van Leeuwenhoek. 78, 123–127. doi: 10.1023/A:1026555830014
Collins, D. P., and Jacobsen, B. J. (2003). Optimizing a Bacillus subtilis isolate for biological control of sugar beet Cercospora leaf spot. Biol. Control 26, 153–161. doi: 10.1016/s1049-9644(02)00132-9
Cretoiu, M. S., Korthals, G. W., Visser, J. H., and van Elsas, J. D. (2013). Chitin amendment increases soil suppressiveness toward plant pathogens and modulates the actinobacterial and oxalobacteraceal communities in an experimental agricultural field. Appl. Environ. Microbiol. 79, 5291–5301. doi: 10.1128/aem.01361-13
Daayf, F., Nicole, M., and Geiger, J. P. (1995). Differentiation of Verticillium dahliae populations on the basis of vegetative compatibility and pathogenicity on cotton. Eur. J. Plant Pathol. 101, 69–79. doi: 10.1007/bf01876095
Del Sal, G., Manfioletti, G., and Schneider, C. (1988). A one-tube plasmid DNA mini-prepara-tion suitable for sequencing. Nucleic Acids Res. 16:9878. doi: 10.1093/nar/16.20.9878
Demirtas, I., Erenler, R., Elmastas, M., and Goktasoglu, A. (2013). Studies on the antioxidant potential of flavones of Allium vineale isolated from its water-soluble fraction. Food Chem. 136, 34–40. doi: 10.1016/j.foodchem.2012.07.086
Desta, K. T., Kim, G. S., Hong, G. E., Kim, Y. H., Lee, W. S., Lee, S. J., et al. (2015). Dietary-flavonoid-rich flowers of Rumex nervosus Vahl: liquid chromatography with electrospray ionization tandem mass spectrometry profiling and in vitro anti-inflammatory effects. J. Separat. Sci. 38, 3345–3353. doi: 10.1002/jssc.201500737
Djeussi, D. E., Noumedem, J. A., Seukep, J. A., Fankam, A. G., Voukeng, I. K., Tankeo, S. B., et al. (2013). Antibacterial activities of selected edible plants extracts against multidrug-resistant Gram-negative bacteria. BMC Complement. Alternat. Med. 13:164. doi: 10.1186/1472-6882-13-164
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Dongzhen, F., Xilin, L., Xiaorong, C., Wenwu, Y., Yunlu, H., Yi, C., et al. (2020). Fusarium species and Fusarium oxysporum species complex genotypes associated with yam wilt in South-Central China. Front. Microbiol. 11:1964. doi: 10.3389/fmicb.2020.01964
Duan, M., Zhang, Y., Zhou, B., Qin, Z., Wu, J., Wang, Q., et al. (2020). Effects of Bacillus subtilis on carbon components and microbial functional metabolism during cow manure–straw composting. Bioresour. Technol. 303:122868. doi: 10.1016/j.biortech.2020.122868
Dubey, A., and Dubey, D. (2010). Evaluation of Cost Effective Organic Fertilizers. India: Kilpest India Ltd.
Edel-Hermann, V., and Lecomte, C. (2019). Current status of Fusarium oxysporum formae speciales and races. Phytopathology. 109, 512–530. doi: 10.1094/phyto-08-18-0320-rvw
Erdogan, O., and Benlioglu, K. (2010). Biological control of Verticillium wilts on cotton by the use of Fluorescent Pseudomonas spp. under field conditions. Biol. Control 5, 39–45. doi: 10.1016/j.biocontrol.2009.11.011
Fan, H., Ru, J., Zhang, Y., Wang, Q., and Li, Y. (2017a). Fengycin produced by Bacillus subtilis 9407 plays a major role in the biocontrol of apple ring rot disease. Microbiol. Res. 199, 89–97. doi: 10.1016/j.micres.2017.03.004
Fan, H., Zhang, Z., Li, Y., Zhang, X., Duan, Y., and Wang, Q. (2017b). Biocontrol of bacterial fruit blotch by Bacillus subtilis 9407 via surfactin-mediated antibacterial activity and colonization. Front. Microbiol. 8:1973. doi: 10.3389/fmicb.2017.01973
Fierer, N., Schimel, J. P., and Holden, P. A. (2003). Influence of drying–rewetting frequency on soil bacterial community structure. Microb. Ecol. 45, 63–71. doi: 10.1007/s00248-002-1007-2
Filippi, M. C. C., Da Silva, G. B., Silva-Lobo, V. L., Côrtes, M. V. C., Moraes, A. J. G., and Prabhu, A. S. (2011). Leaf blast (Magnaporthe oryzae) suppression and growth promotion by rhizobacteria on aerobic rice in Brazil. Biol. Control 58, 160–166. doi: 10.1016/j.biocontrol.2011.04.016
Franke-Whittle, I. H., Manici, L. M., Insam, H., and Stres, B. (2015). Rhizosphere bacteria and fungi associated with plant growth in soils of three replanted apple orchards. Plant Soil 395, 317–333. doi: 10.1007/s11104-015-2562-x
Frikha-Gargouri, O., Ben Abdallah, D., Bhar, I., and Tounsi, S. (2017). Antibiosis and bmyB gene presence as prevalent traits for the selection of efficient Bacillus biocontrol agents against crown gall disease. Front. Plant Sci. 8:1363. doi: 10.3389/fpls.2017.01363
Gabrekiristos, E., Yesuf, M., and Ayana, G. (2018). An optimized inoculation method of Fusarium wilt (Fusarium oxysporum f. sp. Cubense) causal agent of banana wilt disease in ethiopia. Greener J. Breed. Crop Sci. 6, 7–14.
Gao, X. B., Zhao, F. X., Xiang, S. H. E. N., Hu, Y. L., Hao, Y. H., Yang, S. Q., et al. (2010). Effects of cinnamon acid on respiratory rate and its related enzymes activity in roots of seedlings of Malus hupehensis Rehd. Agric. Sci. China 9, 833–839. doi: 10.1016/s1671-2927(09)60161-9
Garbeva, P. V., Van Veen, J. A., and Van Elsas, J. D. (2004). Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 42, 243–270. doi: 10.1146/annurev.phyto.42.012604.135455
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x
Garrity, G. (2007). Bergey’s Manual§of Systematic Bacteriology: Volume 2: The Proteobacteria, Part B: The Gammaproteobacteria. Berlin: Springer Science & Business Media.
Glick, B. R. (1995). The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 41, 109–117. doi: 10.1139/m95-015
Gómez Expósito, R., De Bruijn, I., Postma, J., and Raaijmakers, J. M. (2017). Current insights into the role of rhizosphere bacteria in disease suppressive soils. Front. Microbiol. 8:2529. doi: 10.3389/fmicb.2017.02529
Granatstein, D., Kirby, E., Ostenson, H., and Willer, H. (2016). Global situation for organic tree fruits. Sci. Hortic. 208, 3–12. doi: 10.1016/j.scienta.2015.12.008
Grunewaldt-Stöcker, G., Mahnkopp, F., Popp, C., Maiss, E., and Winkelmann, T. (2019). Diagnosis of apple replant disease ARD: microscopic evidence of early symptoms in fine roots of different apple rootstock genotypes. Sci. Hortic. 243, 583–594. doi: 10.1016/j.scienta.2018.09.014
Grunewaldt-Stöcker, G., Popp, C., Baumann, A., Fricke, S., Menssen, M., Winkelmann, T., et al. (2020). Observations on early fungal infections with relevance for replant disease in fine roots of the rose rootstock Rosa corymbifera ‘Laxa’. Sci. Rep. 10, 1–17.
Guo, P., Zhu, L., Wang, J., Wang, J., and Liu, T. (2015). Effects of alkyl-imidazolium ionic liquid [Omim] Cl on the functional diversity of soil microbial communities. Environ. Sci. Pollut. Res. 22, 9059–9066. doi: 10.1007/s11356-014-4052-8
Haas, D., and Defago, G. (2005). Biological control of soil-borne pathogens by fluorescent Pseudomonads. Nat. Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Hallmann, J., Rodrıguez-Kábana, R., and Kloepper, J. W. (1999). Chitin-mediated changes in bacterial communities of the soil, rhizosphere and within roots of cotton in relation to nematode control. Soil Biol. Biochem. 31, 551–560. doi: 10.1016/s0038-0717(98)00146-1
Hermosa, M. R., Grondona, I., and Iturriaga, E. A. (2000). Molecular characterization and identification of biocontrol isolates of Trichoderma spp. Appl. Environ. Microbiol. 66, 1890–1898. doi: 10.1128/aem.66.5.1890-1898.2000
Hoitink, H. A. J., and Boehm, M. J. (1999). Biocontrol within the context of soil microbial communities: a substrate-dependent phenomenon. Annu. Rev. Phytopathol. 37, 427–446.
Hu, M., Shi, Z., Zhang, Z., Zhang, Y., and Li, H. (2012). Effects of exogenous glucose on seed germination and antioxidant capacity in wheat seedlings under salt stress. Plant Growth Regul. 68, 177–188. doi: 10.1007/s10725-012-9705-3
Hu, X. J., Li, Z. J., Cao, Y. C., Zhang, J., Gong, Y. X., and Yang, Y. F. (2010). Isolation and identification of a phosphate-solubilizing bacterium Pantoea stewartii subsp. stewartii g6, and effects of temperature, salinity, and pH on its growth under indoor culture conditions. Aquac. Int. 18, 1079–1091. doi: 10.1007/s10499-010-9325-8
Huang, X., Zhang, N., Yong, X., Yang, X., and Shen, Q. (2012). Biocontrol of Rhizoctonia solani damping-off disease in cucumber with Bacillus pumilus SQR-N43. Microbiol. Res. 167, 135–143. doi: 10.1016/j.micres.2011.06.002
Huang, X. Q., Wen, T., Zhang, J. B., Meng, L., Zhu, T. B., and Liu, L. L. (2015). Control of soil-borne pathogen Fusarium oxysporum by biological soil disinfestation with incorporation of various organic matters. Eur. J. Plant Pathol. 143, 223–235. doi: 10.1007/s10658-015-0676-x
Hyakumachi, M., Takahashi, H., Matsubara, Y., Someya, N., Shimizu, M., Kobayashi, K., et al. (2014). Recent studies on biological control of plant diseases in Japan. J. Gen. Plant Pathol. 80, 287–302. doi: 10.1007/s10327-014-0524-4
Jaffee, B. A., Abawi, G. S., and Mai, W. F. (1982). Fungi associated with roots of apple seedlings grown in soil from an apple replant site. Plant Dis. 66, 942–944. doi: 10.1094/PD-66-942
Jambhulkar, P. P., Sharma, M., Lakshman, D., and Sharma, P. (2015). “Natural mechanisms of soil suppressiveness against diseases caused by Fusarium, Rhizoctonia, Pythium, and Phytophthora,” in Organic Amendments and Soil Suppressiveness in Plant Disease Management, eds M. K. Meghvansi and A. Varma (Cham: Springer), 95–123. doi: 10.1007/978-3-319-23075-7_5
Jeong, M. H., Lee, Y. S., Cho, J. Y., Ahn, Y. S., Moon, J. H., Hyun, H. N., et al. (2017). Isolation and characterization of metabolites from Bacillus licheniformis MH48 with antifungal activity against plant pathogens. Microb. Pathog. 110, 645–653. doi: 10.1016/j.micpath.2017.07.027
Ji, X., Lu, G., Gai, Y., Zheng, C., and Mu, Z. (2008). Biological control against bacterial wilt and colonization of mulberry by an endophytic Bacillus subtilis strain. FEMS Microbiol. Ecol. 65, 565–573. doi: 10.1111/j.1574-6941.2008.00543.x
Joo, H. S., Deyrup, S. T., and Shim, S. H. (2020). Endophyte-produced antimicrobials: a review of potential lead compounds with a focus on quorum-sensing disruptors. Phytochem. Rev. 20, 543–568. doi: 10.1007/s11101-020-09711-7
Jun, S., and Dongyan, L. (2004). The application of diversity indices in marine phytoplankton studies. Acta Oceanol. Sin. 26, 62–75.
Junaid, J. M., Dar, N. A., Bhat, T. A., Bhat, A. H., and Bhat, M. A. (2013). Commercial biocontrol agents and their mechanism of action in the management of plant pathogens. Int. J. Modern Plant Anim. Sci. 1, 39–57.
Kejela, T., Thakkar, V. R., and Thakor, P. (2016). Bacillus species (BT42) isolated from Coffea arabica L. rhizosphere antagonizes Colletotrichum gloeosporioides and Fusarium oxysporum and also exhibits multiple plant growth promoting activity. BMC Microbiol. 16:277. doi: 10.1186/s12866-016-0897-y
Kelderer, M., Manici, L. M., Caputo, F., and Thalheimer, M. (2012). Planting in the ‘inter-row’ to overcome replant disease in apple orchards: a study on the effectiveness of the practice based on microbial indicators. Plant Soil 357, 381–393. doi: 10.1007/s11104-012-1172-0
Lagopodi, A. L., Ram, A. F., Lamers, G. E., Punt, P. J., Van den Hondel, C. A., Lugtenberg, B. J., et al. (2002). Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis-lycopersici revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol. Plant Microbe Interact. 15, 172–179. doi: 10.1094/MPMI.2002.15.2.172
Leroux, P. (2003). Modes of action of agrochemicals against plant pathogenic organisms. Comptes Rendus Biol. 326, 9–21.
Li, X., Li, K., Zhou, D., Zhang, M., Qi, D., Jing, T., et al. (2021). Biological control of banana wilt disease caused by Fusarium oxyspoum f. sp. Cubense using Streptomyces sp. H4. Biol. Control 155, 104–524.
Li, Y., Feng, X., Wang, X., Zheng, L., and Liu, H. (2020). Inhibitory effects of Bacillus licheniformis BL06 on Phytophthora capsici in pepper by multiple modes of action. Biol. Control 144:104210. doi: 10.1016/j.biocontrol.2020.104210
Li, Y., Lu, H., Cheng, Q., Li, R., He, S., and Li, B. (2016). Changes of reactive oxygen species and scavenging enzymes of persimmon fruit treated with CO2 deastringency and the effect of hydroxyl radicals on breakdown of cell wall polysaccharides in vitro. Sci. Hortic. 199, 81–87. doi: 10.1016/j.scienta.2015.12.040
Lin, D., Qu, L. J., Gu, H., and Chen, Z. (2001). A 3′ 1-kb genomic fragment of Bacillus subtilis encodes the protein inhibiting growth of Xanthomonas oryzae pv. oryzae. J. Appl. Microbiol. 91, 1044–1050.
Liu, H., Pan, F., Han, X., Song, F., Zhang, Z., Yan, J., et al. (2019). Response of soil fungal community structure to long-term continuous soybean cropping. Front. Microbiol. 9:3316. doi: 10.3389/fmicb.2018.03316
Lombard, L., Sandoval-Denis, M., Lamprecht, S. C., and Crous, P. W. (2019). Epitypification of Fusarium oxysporum-clearing the taxonomic chaos. Persoonia 43:1. doi: 10.3767/persoonia.2019.43.01
Ma, L., Zhang, H. Y., Zhou, X. K., Yang, C. G., Zheng, S. C., Duo, J. L., et al. (2018). Biological control tobacco bacterial wilt and black shank and root colonization by bio-organic fertilizer containing bacterium Pseudomonas aeruginosa NXHG29. Appl. Soil Ecol. 129, 136–144. doi: 10.1016/j.apsoil.2018.05.011
Marín, P., Magan, N., Vázquez, C., and González-Jaén, M. T. (2010). Differential effect of environmental conditions on the growth and regulation of the fumonisin biosynthetic gene FUM1 in the maize pathogens and fumonisin producers Fusarium verticillioides and Fusarium proliferatum. FEMS Microbiol. Ecol. 73, 303–311. doi: 10.1111/j.1574-6941.2010.00894.x
Mazurier, S., Corberand, T., Lemanceau, P., and Raaijmakers, J. M. (2009). Phenazine antibiotics produced by fluorescent Pseudomonads contribute to natural soil suppressiveness to Fusarium wilt. ISME J. 3, 977–991.
Mazzola, M. (1998). Elucidation of the microbial complex having a causal role in the development of Apple replant diseases in Washington. Phytopathology. 88, 930–938. doi: 10.1094/phyto.1998.88.9.930
Mazzola, M., and Manici, L. M. (2012). Apple replant disease: role of microbial ecology in cause and control. Annu. Rev. Phytopathol. 50, 45–65. doi: 10.1146/annurev-phyto-081211-173005
McKenney, P. T., Driks, A., and Eichenberger, P. (2013). The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 11, 33–44. doi: 10.1038/nrmicro2921
Medeiros Araújo, M. B., Moreira, G. M., Nascimento, L. V., Nogueira, G. D. A., Nascimento, S. R. D. C., Pfenning, L. H., et al. (2021). Fusarium rot of melon is caused by several Fusarium species. Plant Pathol. 703, 712–721. doi: 10.1111/ppa.13328
Meng, P. P., Liu, X., Qiu, H. Z., Zhang, W. M., Zhang, C. H., Wang, D., et al. (2012). Fungal population structure and its biological effect in rhizosphere soil of continuously cropped potato. J. Appl. Ecol. 23, 3079–3086.
Moein, S., Mazzola, M., Spies, C. F. J., and McLeod, A. (2019). Evaluating different approaches for the quantification of oomycete apple replant pathogens, and their relationship with seedling growth reductions. Eur. J. Plant Pathol. 154, 243–257. doi: 10.1007/s10658-018-01652-4
Móricz, ÁM., Fornal, E., Jesionek, W., Majer-Dziedzic, B., and Choma, I. M. (2015). Effect-directed isolation and identification of antibacterial Chelidonium majus L. alkaloids. Chromatographia 78, 707–716. doi: 10.1007/s10337-015-2870-6
Mutaz Al-Ajlani, M., and Hasnain, S. (2010). Bacteria exhibiting antimicrobial activities; screening for antibiotics and the associated genetic studies. Open Conf. Proc. J. 1, 230–238. doi: 10.2174/2210289201001010230
Nagórska, K., Bikowski, M., and Obuchowski, M. (2007). Multicellular behaviour and production of a wide variety of toxic substances support usage of Bacillus subtilis as a powerful biocontrol agent. Acta Biochim. Polonica 54, 495–508. doi: 10.18388/abp.2007_3224
Nelson, P. E., Toussoun, T. A., and Marasas, W. F. O. (1983). Fusarium Species: An Illustrated Manual for Identification. Pennsylvania: Pennsylvania State University Press.
Newman, D. J., and Cragg, G. M. (2020). Plant endophytes and epiphytes: burgeoning sources of known and “unknown” cytotoxic and antibiotic agents? Planta Med. 86, 891–905. doi: 10.1055/a-1095-1111
Nicola, L., Insam, H., Pertot, I., and Stres, B. (2018). Reanalysis of microbiomes in soils affected by apple replant disease (ARD): old foes and novel suspects lead to the proposal of extended model of disease development. Appl. Soil Ecol. 129, 24–33. doi: 10.1016/j.apsoil.2018.04.010
Omar, S. A., and Abd-Alla, M. H. (1998). Biocontrol of fungal root rot diseases of crop plants by the use of rhizobia and bradyrhizobia. Folia Microbiol. 43, 431–437. doi: 10.1007/bf02818587
Omran, R. G. (1980). Peroxide levels and the activities of catalase, peroxidase, and indoleacetic acid oxidase during and after chilling cucumber seedlings. Plant Physiol. 65, 407–408. doi: 10.1104/pp.65.2.407
Ouhaibi-Ben Abdeljalil, N., Vallance, J., Gerbore, J., Rey, P., and Daami-Remadi, M. (2016). Bio-suppression of Sclerotinia stem rot of tomato and biostimulation of plant growth using tomato-associated rhizobacteria. J. Plant Pathol. Microbiol. 7:2. doi: 10.4172/2157-7471.1000331
Pecchia, S., Franceschini, A., Santori, A., Vannacci, G., and Myrta, A. (2017). Efficacy of dimethyl disulfide (DMDS) for the control of chrysanthemum Verticillium wilt in Italy. Crop Protect. 93, 28–32. doi: 10.1016/j.cropro.2016.11.019
Percent, S. F., Frischer, M. E., Vescio, P. A., Duffy, E. B., Milano, V., McLellan, M., et al. (2008). Bacterial community structure of acid-impacted lakes: what controls diversity? Appl. Environ. Microbiol. 74:1856. doi: 10.1128/AEM.01719-07
Piggot, P. J., and Hilbert, D. W. (2004). Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7, 579–586. doi: 10.1016/j.mib.2004.10.001
Podile, A. R., and Prakash, A. P. (1996). Lysis and biological control of Aspergillus niger by Bacillus subtilis AF 1. Can. J. Microbiol. 42, 533–538. doi: 10.1139/m96-072
Pshibytko, N. L., Zenevich, L. A., and Kabashnikova, L. F. (2006). Changes in the photosynthetic apparatus during Fusarium wilt of tomato. Russ. J. Plant Physiol. 53, 25–31. doi: 10.1134/S1021443706010031
Qin, Y., Shang, Q., Zhang, Y., Li, P., and Chai, Y. (2017). Bacillus amyloliquefaciens L-S60 reforms the rhizosphere bacterial community and improves growth conditions in cucumber plug seedling. Front. Microbiol. 8:2620. doi: 10.3389/fmicb.2017.02620
Quéric, N. V., and Soltwedel, T. (2012). Benthic prokaryotic community dynamics along the Ardencaple Canyon, Western Greenland Sea. Sediment. Geol. 263, 36–44. doi: 10.1016/j.sedgeo.2011.10.004
Raaijmakers, J. M., and Mazzola, M. (2012). Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 50, 403–424. doi: 10.1146/annurev-phyto-081211-172908
Radwan, D. E. M., Fayez, K. A., Mahmoud, S. Y., and Lu, G. (2010). Modifications of antioxidant activity and protein composition of bean leaf due to Bean yellow mosaic virus infection and salicylic acid treatments. Acta Physiol. Plant. 32, 891–904. doi: 10.1007/s11738-010-0477-y
Rahman, M. A., Kadir, J., Mahmud, T. M. M., Rahman, A. R., and Begum, M. M. (2007). Screening of antagonistic bacteria for biocontrol activities on Colletotrichum gloeosporioides in papaya. Asian J. Plant Sci. 6, 12–20. doi: 10.3923/ajps.2007.12.20
Ramírez, V., Martínez, J., del Rocio Bustillos-Cristales, M., Catañeda-Antonio, D., Munive, J. A., and Baez, A. (2021). Bacillus cereus MH778713 elicits tomato plant protection against Fusarium oxysporum. J. Appl. Microbiol. [Epub ahead of print]. doi: 10.1111/jam.15179
Raski, D., Jones, N., Kissler, J., and Luvisi, D. (1976). Soil fumigation: one way to cleanse nematode-infested vineyard lands. Calif. Agric. 30, 4–7.
Raupach, G. S., and Kloepper, J. W. (1998). Mixtures of plant growth-promoting rhizobacteria enhance biological control of multiple cucumber pathogens. Phytopathology. 88, 1158–1164. doi: 10.1094/PHYTO.1998.88.11.1158
Romero, D., Pérez-García, A., Rivera, M. E., Cazorla, F. M., and De Vicente, A. (2004). Isolation and evaluation of antagonistic bacteria towards the cucurbit powdery mildew fungus Podosphaera fusca. Appl. Microbiol. Biotechnol. 64, 263–269. doi: 10.1007/s00253-003-1439-8
Shahena, S., Rajan, M., Chandran, V., and Mathew, L. (2021). “Conventional methods of fertilizer release,” in Controlled Release Fertilizers for Sustainable Agriculture, eds F.B Lewu, T. Volova, S. Thomas, and R.K. Rakhimol (Cambridge, MA: Academic Press), 1–24. doi: 10.1016/B978-0-12-819555-0.00001-7
Shao, J., Song, Y., Zhou, Y., Wan, Z., Li, R., and Yu, J. (2020). Diagnostic value of fluorescein-labeled chitinase staining in formalin-fixed and paraffin-embedded tissues of fungal disease. Med. Mycol. 58, 66–70. doi: 10.1093/mmy/myz035
Sharma, P., and Dubey, R. S. (2007). Involvement of oxidative stress and role of antioxidative defense system in growing rice seedlings exposed to toxic concentrations of aluminum. Plant Cell Rep. 26, 2027–2038. doi: 10.1007/s00299-007-0416-6
Shen, Z., Penton, C. R., Lv, N., Xue, C., Yuan, X., Ruan, Y., et al. (2018). Banana Fusarium wilt disease incidence is influenced by shifts of soil microbial communities under different monoculture spans. Microb. Ecol. 75, 739–750. doi: 10.1007/s00248-017-1052-5
Sheng, Y., Wang, H., Wang, M., Li, H., Xiang, L., Pan, F., et al. (2020). Effects of soil texture on the growth of young apple trees and soil microbial community structure under replanted conditions. Hortic. Plant J. 63, 123–131. doi: 10.1016/j.hpj.2020.04.003
Shennan, C., Muramoto, J., Koike, S., Baird, G., Fennimore, S., Samtani, J., et al. (2018). Anaerobic soil disinfestation is an alternative to soil fumigation for control of some soilborne pathogens in strawberry production. Plant Pathol. 67, 51–66. doi: 10.1111/ppa.12721
Shi, G., Sun, H., Calderón-Urrea, A., Jia, X., Yang, H., and Su, G. (2020). Soil fungal diversity loss and appearance of specific fungal pathogenic communities associated with the consecutive replant problem (CRP) in lily. Front. Microbiol. 11:1649. doi: 10.3389/fmicb.2020.01649
Shyu, C., Soule, T., Bent, S. J., Foster, J. A., and Forney, L. J. (2007). MiCA: a web-based tool for the analysis of microbial communities based on terminal-restriction fragment length polymorphisms of 16S and 18S rRNA genes. Microb. Ecol. 53, 562–570. doi: 10.1007/s00248-006-9106-0
Singh, B. K., Sharma, S. R., and Singh, B. (2010). Antioxidant enzymes in cabbage: variability and inheritance of superoxide dismutase, peroxidase and catalase. Sci. Hortic. 124, 9–13. doi: 10.1016/j.scienta.2009.12.011
Singh, N., Pandey, P., Dubey, R. C., and Maheshwari, D. K. (2008). Biological control of root rot fungus Macrophomina phaseolina and growth enhancement of Pinus roxburghii (Sarg.) by rhizosphere competent Bacillus subtilis BN1. World J. Microbiol. Biotechnol. 24, 1669–1679. doi: 10.1007/s11274-008-9680-z
Smith, T. G. (1995). Orchard update: Washington State University Cooperative Extension Bulletin. Pullman, WA.
Somerville, T. F., Corless, C. E., Sueke, H., Neal, T., and Kaye, S. B. (2020). 16S ribosomal RNA PCR versus conventional diagnostic culture in the investigation of suspected bacterial keratitis. Transl. Vision Sci. Technol. 9:2. doi: 10.1167/tvst.9.13.2
Souto, G. I., Correa, O. S., Montecchia, M. S., Kerber, N. L., Pucheu, N. L., Bachur, M., et al. (2004). Genetic and functional characterization of a Bacillus sp. strain excreting surfactin and antifungal metabolites partially identified as iturin-like compounds. J. Appl. Microbiol. 97, 1247–1256. doi: 10.1111/j.1365-2672.2004.02408.x
Spath, M., Insam, H., Peintner, U., Kelderer, M., Kuhnert, R., and Franke-Whittle, I. H. (2015). Linking soil biotic and abiotic factors to apple replant disease: a greenhouse approach. J. Phytopathol. 163, 287–299. doi: 10.1111/jph.12318
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 10:1093. doi: 10.1093/bioinformatics/btu033
Stein, T. (2005). Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 56, 845–857. doi: 10.1111/j.1365-2958.2005.04587.x
Syed-Ab-Rahman, S. F., Xiao, Y., Carvalhais, L. C., Ferguson, B. J., and Schenk, P. M. (2019). Suppression of Phytophthora capsici infection and promotion of tomato growth by soil bacteria. Rhizosphere 9, 72–75.7. doi: 10.1016/j.rhisph.2018.11.007
Tewoldemedhin, Y. T., Mazzola, M., Labuschagne, I., and McLeod, A. (2011a). A multi-phasic approach reveals that apple replant disease is caused by multiple biological agents, with some agents acting synergistically. Soil Biol. Biochem. 43, 1917–1927. doi: 10.1016/j.soilbio.2011.05.014
Tewoldemedhin, Y. T., Mazzola, M., Mostert, L., and McLeod, A. (2011b). Cylindrocarpon species associated with apple tree roots in South Africa and their quantification using real-time PCR. Eur. J. Plant Pathol. 129, 637–651. doi: 10.1007/s10658-010-9728-4
Tilston, E. L., Deakin, G., Bennett, J., Passey, T., Harrison, N., Fernández, F., et al. (2020). Effect of fungal, oomycete and nematode interactions on apple root development in replant soil. CABI Agric. Biosci. 1, 1–18. doi: 10.1186/s43170-020-00014-7
Tilston, E. L., Deakin, G., Bennett, J., Passey, T., Harrison, N., O’Brien, F., et al. (2018). Candidate causal organisms for apple replant disease in the United Kingdom. Phytobiomes J. 24, 261–274. doi: 10.1094/PBIOMES-11-18-0050-R
Utkhede, R. S., and Smith, E. M. (2000). Impact of chemical, biological and cultural treatments on the growth and yield of apple in replant-disease soil. Austral. Plant Pathol. 29, 129–136. doi: 10.1071/AP00021
Utkhede, R. S., Vrain, T. C., and Yorston, J. M. (1992). Effects of nematodes, fungi and bacteria on the growth of young apple trees grown in apple replant disease soil. Plant Soil 139, 1–6. doi: 10.1007/BF00012835
Van Schoor, L., Denman, S., and Cook, N. C. (2009). Characterisation of apple replant disease under South African conditions and potential biological management strategies. Sci. Hortic. 119, 153–162. doi: 10.1016/j.scienta.2008.07.032
Vos, P., Garrity, G., Jones, D., Krieg, N. R., Ludwig, W., Rainey, F. A., et al. (2011). Bergey’s Manual of Systematic Bacteriology: Volume 3: The Firmicutes. Cham: Springer Science & Business Media.
Wang, F., Zhu, L., Wang, X., Wang, J., and Wang, J. (2015). Impact of repeated applications of metalaxyl on its dissipation and microbial community in soil. Water Air Soil Pollut. 226, 1–14. doi: 10.1007/s11270-015-2686-x
Wang, G., Gavala, H. N., Skiadas, I. V., and Ahring, B. K. (2009). Wet explosion of wheat straw and codigestion with swine manure: effect on the methane productivity. Waste Manag. 29, 2830–2835. doi: 10.1016/j.wasman.2009.07.004
Wang, G. S., Yin, C. M., Pan, F. B., Wang, X. B., Xiang, L., Wang, Y. F., et al. (2018). Analysis of the fungal community in apple replanted soil around Bohai Gulf. Hortic. Plant J. 4, 175–181. doi: 10.1016/j.hpj.2018.05.003
Wang, N., Wolf, J., and Zhang, F. S. (2016). Towards sustainable intensification of apple production in China-Yield gaps and nutrient use efficiency in apple farming systems. J. Integr. Agric. 15, 716–725. doi: 10.1016/S2095-3119(15)61099-1
Wang, X. Q., Zhao, D. L., Shen, L. L., Jing, C. L., and Zhang, C. S. (2018). “Application and mechanisms of Bacillus subtilis in biological control of plant disease,” in Role of Rhizospheric Microbes in Soil, ed. V. S. Meena (Singapore: Springer), 225–250. doi: 10.1007/978-981-10-8402-7_9
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Weiß, S., Bartsch, M., and Winkelmann, T. (2017). Transcriptomic analysis of molecular responses in Malus domestica ‘M26’ roots affected by apple replant disease. Plant Mol. Biol. 943, 303–318. doi: 10.1007/s11103-017-0608-6
Weller, D. M., Raaijmakers, J. M., Gardener, B. B. M., and Thomashow, L. S. (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40, 309–348. doi: 10.1146/annurev.phyto.40.030402.110010
Willett, M., Smith, T. J., Peterson, A. B., Hinman, H., Stevens, R. G., Ley, T., et al. (1994). Growing profitable apple orchards in replant sites: an interdisciplinary team approach in Washington State. HortTechnology 4, 175–181. doi: 10.21273/HORTTECH.4.2.175
Wilson, C. L., and Pusey, P. L. (1985). Potential for biological control of postharvest plant diseases. Plant Dis. 69, 375–378. doi: 10.1094/PD-69-375
Winkelmann, T., Smalla, K., Amelung, W., Baab, G., Grunewaldt-Stöcker, G., Kanfra, X., et al. (2018). Apple replant disease: causes and mitigation strategies. Curr. Issues Mol. Biol. 30, 89–106. doi: 10.21775/9781912530007.06
Wu, Y., Zhou, J., Li, C., and Ma, Y. (2019). Antifungal and plant growth promotion activity of volatile organic compounds produced by Bacillus amyloliquefaciens. MicrobiologyOpen 8:e00813. doi: 10.1002/mbo3.813
Xu, F., Chen, Y., Cai, Y., Gu, F., and An, K. (2020). Distinct roles for bacterial and fungal communities during the curing of vanilla. Front. Microbiol. 11:552388. doi: 10.3389/fmicb.2020.552388
Xu, H. J., Bai, J., Li, W. Y., Zhao, L. X., and Li, Y. T. (2019). Removal of persistent DDT residues from soils by earthworms: a mechanistic study. J. Hazard. Mater. 365, 622–631. doi: 10.1016/j.jhazmat.2018.11.043
Yamamoto, S., and Harayama, S. (1995). PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microbiol. 61, 1104–1109. doi: 10.1128/aem.61.3.1104-1109.1995
Yim, B., Baumann, A., Grunewaldt-Stöcker, G., Liu, B., Beerhues, L., Zühlke, S., et al. (2020). Rhizosphere microbial communities associated to rose replant disease: links to plant growth and root metabolites. Hortic. Res. 71, 1–16. doi: 10.1038/s41438-020-00365-2
Yim, B., Smalla, K., and Winkelmann, T. (2013). Evaluation of apple replant problems based on different soil disinfection treatments-links to soil microbial community structure? Plant Soil 366, 617–631. doi: 10.1007/s11104-012-1454-6
Yin, C., Xiang, L., Wang, G., Wang, Y., Shen, X., Chen, X., et al. (2017). Phloridzin promotes the growth of Fusarium moniliforme (Fusarium verticillioides). Sci. Hortic. 214, 187–194. doi: 10.1016/j.scienta.2016.11.035
Yu, H. Y., Wang, Y. K., Chen, P. C., Li, F. B., Chen, M. J., and Hu, M. (2014). The effect of ammonium chloride and urea application on soil bacterial communities closely related to the reductive transformation of pentachlorophenol. J. Hazard. Mater. 272, 10–19. doi: 10.1016/j.jhazmat.2014.02.037
Yu, X., Ai, C., Xin, L., and Zhou, G. (2011). The siderophore-producing bacterium, Bacillus subtilis CAS15, has a biocontrol effect on Fusarium wilt and promotes the growth of pepper. Eur. J. Soil Biol. 47, 138–145. doi: 10.1016/j.ejsobi.2010.11.001
Yuan, S. Q., Xue, Y. F., Gao, P., Wang, W. D., Ma, Y. H., Li, X. M., et al. (2007). Microbial diversity in shengli petroleum reservoirs analyzed by T-RFLP. Acta Microbiol. Sin. 47, 290–294.
Zalila-Kolsi, I., Mahmoud, A. B., Ali, H., Sellami, S., Nasfi, Z., Tounsi, S., et al. (2016). Antagonist effects of Bacillus spp. strains against Fusarium graminearum for protection of durum wheat (Triticum turgidum L. subsp. durum). Microbiol. Res. 192, 148–158. doi: 10.1016/j.micres.2016.06.012
Zasada, I. A., Halbrendt, J. M., Kokalis-Burelle, N., LaMondia, J., McKenry, M. V., and Noling, J. W. (2010). Managing nematodes without methyl bromide. Annu. Rev. Phytopathol. 48, 311–328. doi: 10.1146/annurev-phyto-073009-114425
Zhang, C., Wang, J., Zhu, L., Du, Z., Wang, J., Sun, X., et al. (2018). Effects of 1-octyl-3-methylimidazolium nitrate on the microbes in brown soil. J. Environ. Sci. 67, 249–259. doi: 10.1016/j.jes.2017.09.002
Zhang, M., Li, J., Shen, A., Tan, S., Yan, Z., Yu, Y., et al. (2016). Isolation and Identification of Bacillus amyloliquefaciens IBFCBF-1 with potential for biological control of Phytophthora blight and growth promotion of pepper. J. Phytopathol. 164, 1012–1021. doi: 10.1111/jph.12522
Zhang, Q., Zhu, L., Wang, J., Xie, H., Wang, J., Wang, F., et al. (2014). Effects of fomesafen on soil enzyme activity, microbial population, and bacterial community composition. Environ. Monit. Assess. 186, 2801–2812. doi: 10.1007/s10661-013-3581-9
Zhang, Q. X., Zhang, Y., He, L. L., Ji, Z. L., and Tong, Y. H. (2018). Identification of a small antimycotic peptide produced by Bacillus amyloliquefaciens 6256. Pesticide Biochem. Physiol. 150, 78–82. doi: 10.1016/j.pestbp.2018.07.006
Zhang, X., Gao, J., Zhao, F., Zhao, Y., and Li, Z. (2014). Characterization of a salt-tolerant bacterium Bacillus sp. from a membrane bioreactor for saline wastewater treatment. J. Environ. Sci. 26, 1369–1374. doi: 10.1016/S1001-0742(13)60613-0
Zhang, X., Li, B., Wang, Y., Guo, Q., Lu, X., Li, S., et al. (2013). Lipopeptides, a novel protein, and volatile compounds contribute to the antifungal activity of the biocontrol agent Bacillus atrophaeus CAB-1. Appl. Microbiol. Biotechnol. 97, 9525–9534. doi: 10.1007/s00253-013-5198-x
Zhang, Y., Han, X., Chen, X., Jin, H., and Cui, X. (2009). Exogenous nitric oxide on antioxidative system and ATPase activities from tomato seedlings under copper stress. Sci. Hortic. 123, 217–223. doi: 10.1016/j.scienta.2009.08.015
Zhang, Y., Shi, X., Li, B., Zhang, Q., Liang, W., and Wang, C. (2016). Salicylic acid confers enhanced resistance to Glomerella leaf spot in apple. Plant Physiol. Biochem. 106, 64–72. doi: 10.1016/j.plaphy.2016.04.047
Zhao, S., Liu, D., Ling, N., Chen, F., Fang, W., and Shen, Q. (2014). Bio-organic fertilizer application significantly reduces the Fusarium oxysporum population and alters the composition of fungi communities of watermelon Fusarium wilt rhizosphere soil. Biol. Fertil. Soils 50, 765–774. doi: 10.1007/s00374-014-0898-7
Zhao, Z., Wang, Q., Wang, K., Brian, K., Liu, C., and Gu, Y. (2010). Study of the antifungal activity of Bacillus vallismortis ZZ185 in vitro and identification of its antifungal components. Bioresour. Technol. 101, 292–297.
Keywords: apple replant disease, Fusarium spp., Bacillus amyloliquefaciens, metabolites, root, identification
Citation: Duan Y, Chen R, Zhang R, Jiang W, Chen X, Yin C and Mao Z (2021) Isolation, Identification, and Antibacterial Mechanisms of Bacillus amyloliquefaciens QSB-6 and Its Effect on Plant Roots. Front. Microbiol. 12:746799. doi: 10.3389/fmicb.2021.746799
Received: 27 July 2021; Accepted: 26 August 2021;
Published: 16 September 2021.
Edited by:
Tofazzal Islam, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshReviewed by:
Lazarovits George, A&L Biologicals Inc., CanadaCopyright © 2021 Duan, Chen, Zhang, Jiang, Chen, Yin and Mao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengmiao Yin, eWluY2hlbmdtaWFvQDE2My5jb20=; Zhiquan Mao, bXpoaXF1YW5Ac2RhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.