
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 01 October 2021
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.746632
This article is part of the Research TopicTechnological Advances in Microbiological Risk AssessmentView all 11 articles
 Yang Qu1,2
Yang Qu1,2 Chao Wei3
Chao Wei3 Xiaohang Dai3
Xiaohang Dai3 Yalong Bai1
Yalong Bai1 Xin Zhao4
Xin Zhao4 Qingkuo Lan4
Qingkuo Lan4 Wenbo Wang5
Wenbo Wang5 Yuanjuan Wu5
Yuanjuan Wu5 Min Gao6
Min Gao6 Weihao Tang6
Weihao Tang6 Changyan Zhou1,2*
Changyan Zhou1,2* Yujuan Suo1,2*
Yujuan Suo1,2*Bacillus cereus is a well-characterized human pathogen that produces toxins associated with diarrheal and emetic foodborne diseases. To investigate the possible transmission of B. cereus on lettuce farms in China and determine its enterotoxicity, (I) a total of 524 samples (lettuce: 332, soil: 69, water: 57, manure: 57, pesticide: 9) were collected from 46 lettuce farms in five Chinese provinces, (II) multilocus sequence typing (MLST) was used to classify B. cereus isolates and for trace analysis, and (III) the presence of toxin genes and enterotoxins (Hbl and Nhe) was detected in 68 strains. The results showed that one hundred and sixty-one lettuce samples (48.5%) tested positive for B. cereus at levels ranging from 10 to 5.3 × 104 CFU/g. Among the environmental sample categories surveyed, the highest positive rate was that of the pesticide samples at 55.6%, followed by soil samples at 52.2% and manure samples at 12.3%. Moreover, one hundred isolates of B. cereus yielded 68 different sequence types (STs) and were classified into five phylogenetic clades. Furthermore, Nhe toxin genes (nheA, nheB, nheC) were broadly distributed and identified in all 68 strains (100%), while Hbl toxin genes (hblA, hblC, hblD) were present in 61 strains (89.7%), entFM was detected in 62 strains (91.2%), and cytK was found in 29 strains (42.6%). All strains were negative for ces. As for the enterotoxin, Nhe was observed in all 68 isolates carrying nheB, while Hbl was present in 76.5% (52/68) of the strains harboring hblC. This study is the first report of possible B. cereus transmission and of its potential enterotoxicity on lettuce farms in China. The results showed that soil and pesticides are the main sources of B. cereus on lettuce farms in China, and the possible transmission routes are as follows: soil-lettuce, manure-lettuce, pesticide-lettuce, manure-soil-lettuce, and water-manure-soil-lettuce. Furthermore, the B. cereus isolates, whether from lettuce or the environment, pose a potential risk to health.
The World Health Organization recommends a daily intake of 400 g of fresh vegetables for improved health (Park et al., 2018). Lettuce is the most widely consumed fresh vegetable and is usually eaten raw with no or minimal processing, increasing the occurrence of lettuce-related foodborne outbreaks, which have gained attention among government agencies, industries, and the public (Bozkurt et al., 2021). Raw lettuce has been reported to harbor foodborne pathogens, such as Bacillus cereus, Escherichia coli O157:H7, Salmonella, and Listeria monocytogenes (Abadias et al., 2008; Park et al., 2018; Yu et al., 2019). A total of 597 outbreaks caused by B. cereus toxins involving 6,221 cases were reported in European Member States (MSs) from 2014 to 2016 due to the diarrhea and emesis (vomiting) caused by B. cereus (Osimani et al., 2018). Thus, the safety of B. cereus strains in lettuce should be considered.
Bacillus cereus is a spore-forming gram-positive species (Ceuppens et al., 2013) that is distributed among seven phylogenetic clades with nine species (Guinebretière et al., 2008), including (I) B. cereus sensu stricto and B. cytotoxicus, which cause foodborne illness (Dierick et al., 2005; Guinebretière et al., 2008); (II) B. weihenstephanensis and B. mycoides, which cause food spoilage (Meer et al., 1991); (III) B. anthracis, which causes anthrax in both humans and animals (Ivanova et al., 2003); (IV) B. thuringiensis, which is used as an insecticide in agriculture (Höfte and Whiteley, 1989); (V) B. pseudomycoides, which is considered a non-pathogenic environmental microorganism; (VI) B. toyonensis, which has recently been recognized as a putative probiotic species (Jiménez et al., 2013); and (VII) B. wiedmannii, which is a psychrotolerant cytotoxic species (Miller et al., 2016). Therefore, it is difficult to identify B. cereus isolates, and multilocus sequence typing (MLST) is recommended in combination with traditional methods to differentiate diverse species of B. cereus (Jung et al., 2011; Maiden et al., 2013; Otlewska et al., 2013; Castiaux et al., 2014; Zhuang et al., 2019).
Farm-to-fork supply chains are considered to be the source of B. cereus in lettuce, including field production, harvest, processing, packaging, transportation, retail, and home storage (Pang et al., 2017), and farm environments are the core source (Drewnowska et al., 2020). Moreover, MLST is a powerful method for tracing analysis based on genetic evolution, which can be determined by comparative analysis of alleles (Forsythe et al., 2014; Hammerum et al., 2015). Processing environments and packing areas have been found to be the sources most likely to be associated with B. cereus contamination in powdered infant formula production (Zhuang et al., 2019). The transmission of B. cereus in lettuce farms must be detected and monitored to promote food safety and human health.
In this study, the prevalence of B. cereus in lettuce and farm environments distributed in China was described. Genetic methods were applied to examine the phylogenetically diverse B. cereus isolates from lettuce and farm environments, and their associations were examined to identify the possible transmission of B. cereus in lettuce farms. In addition, the safety of B. cereus isolates was investigated by identifying virulence factors and enterotoxins from lettuce and in farm environments.
A total of 524 samples were collected from 46 lettuce farms in five Chinese provinces from April 2019 to November 2020 (Figure 1). These samples were classified as lettuce (Lactuca sativa L.) (n = 332), soil (n = 69), water (n = 57), manure (n = 57), and pesticide (n = 9) samples; more details about the samples can be found in Table 1. The samples were transported to the laboratory in a cold box (4°C) within 2 h.
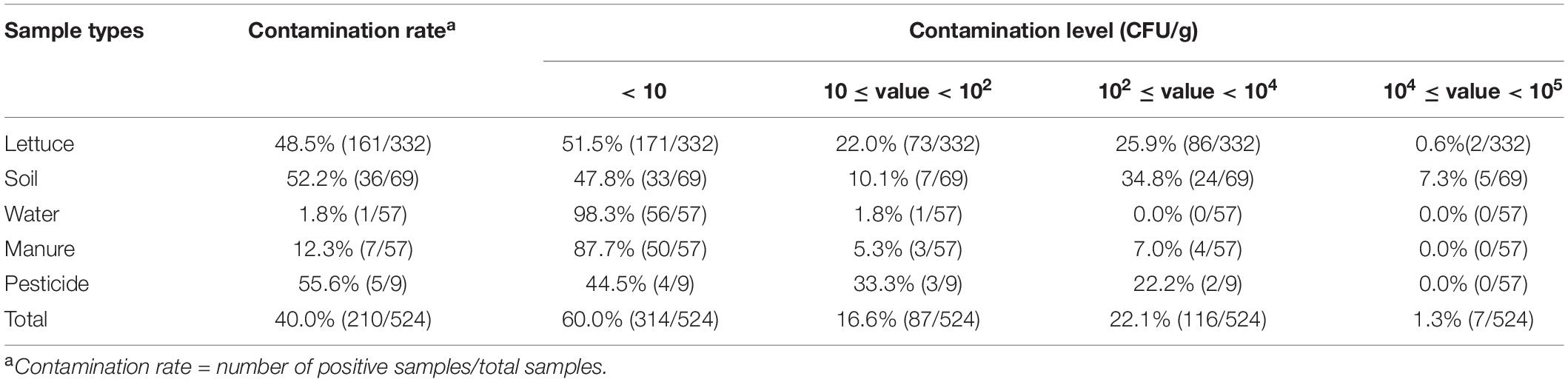
Table 1. Prevalence and contamination level of B. cereus in different samples among 46 lettuce farms.
The B. cereus strains were isolated in accordance with GB4789.14-2014, the National Food Safety Standard for Food Microbiological Examination used in China. Genomic DNA was extracted from overnight cultures of B. cereus isolates in Luria-Bertani (LB) broth using a TIANamp Bacteria DNA Extraction Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s protocol for gram-positive bacteria.
Isolates were characterized by using seven housekeeping genes (glp, gmk, ilvD, pta, pur, pycA, and tpi) with different primers and conditions (Table 2), which are provided in the B. cereus PubMLST database1. The PCR products were sequenced by Sangon Biotech (Shanghai, China) and submitted to the B. cereus PubMLST database to obtain the allele number. By combination of allele numbers for all seven housekeeping genes, a sequence type (ST) clonal complex could be obtained. New allele sequences and STs were submitted to the B. cereus PubMLST database and assigned by the MLST website administrator.
The phylogenetic study included all the isolates and nine references obtained from the NCBI database2 (B. anthracis Ames Ancestor, B. cereus ATCC 1457, B. cytotoxicus NVH 391-98, B. mycoides DSM 2048, B. pseudomycoides DSM 12442, B. thuringiensis ATCC 10792, B. toyonensis BCT-7112, B. weihenstephanensis WSBC 10204, B. wiedmannii FSL W8-1069) (Miller et al., 2018). The phylogenetic tree was constructed using the neighbor-joining (NJ) method in Molecular Evolutionary Genetic Analysis (MEGA-X) based on the concatenated sequences (2,829 bp) of the seven housekeeping genes (Kumar et al., 2018). In addition, 1,000 bootstrap replicates were used for branch quality.
To analyze the relationship between different STs and sample sources, a minimum spanning tree was constructed with PHYLOViZ 2.0 software (Instituto de Microbiologia, Portugal) (Francisco et al., 2012) with the goeBURST algorithm and 1,000 bootstrap resamplings (Feil et al., 2004).
Nine virulence genes, namely, hblA, hblC, hblD, nheA, nheB, nheC, entFM, cytK, and ces, were identified according to a previous study (Stenfors and Granum, 2001; Guinebretière et al., 2002; Ehling-Schulz et al., 2005; Kim et al., 2011), and the primers and conditions are listed in Table 2. The 20 μL PCR mixture consisted of 10 μL of TaqTM PCR Premix (Takara, China), 1 μL of diluted DNA, and 0.2 mM each primer.
A Duopath® Cereus Enterotoxins kit (Merck, Kenilworth, NJ, United States) was used to detect Hbl (the lytic component L2, a subunit of toxin Hbl) and NheB (the binding component of the Nhe toxin), the detection limits for which were 20.0 and 6.0 ng/mL, respectively. Isolates were maintained on brain heart infusion (BHI; Co. CM1135, Oxoid, Hampshire, United Kingdom) agar plates at 4°C. Single colonies of bacteria were aseptically picked and cultured in 1 mL of CGY broth (with 1% glucose) and incubated for 4 h at 37°C. For testing, the cultures and Duopath® kit were both cooled to room temperature (20°C), and 150 μL of culture was pipetted into the circular sample port on the Duopath® kit. The results could be observed 30 min after applying the culture to the kit at room temperature (Krause et al., 2010).
In this study, B. cereus was detected in 210 of the 524 (40.0%) samples from 46 lettuce farms (Table 1) distributed in five Chinese provinces (Figure 1). The positivity rates of B. cereus were 48.5% (161/332) for lettuce, ranging from 10 to 5.3 × 104 lg CFU/g, and the farm environment also contained B. cereus. Of these environmentally positive samples, the highest frequency of B. cereus was found in pesticides (55.6%), followed by soil (52.2%), manure (12.3%), and water (1.8%). Enumeration of B. cereus showed that 60.0% of the lettuce farm samples had counts below 10 CFU/g, 16.6% had counts in the range from 10–102 CFU/g, 22.1% had counts in the range from 102–104 CFU/g, and 1.3% had counts above 104 CFU/g. High counts (>104 CFU/g) were most frequently found in soil and lettuce.
Considering the effect of source, location and sampling time, strains collected from the same lettuce or soil on the same farm during different sampling seasons were removed, and only 100 isolates that from lettuce, soil, pesticides, manure and water were selected for genetic structure analysis. A total of 65 different STs and seven clonal complexes (CCs) were identified among the isolates (Figure 2). Thirty-seven isolates were assigned to 31 new STs. Forty-seven of the 65 (72.3%) STs included only one isolate; however, the remaining 18 STs included more than one isolate. The most frequent ST was ST1000, comprising five isolates from lettuce and soil samples, which were found in Shanghai, Beijing, and Tianjin provinces.
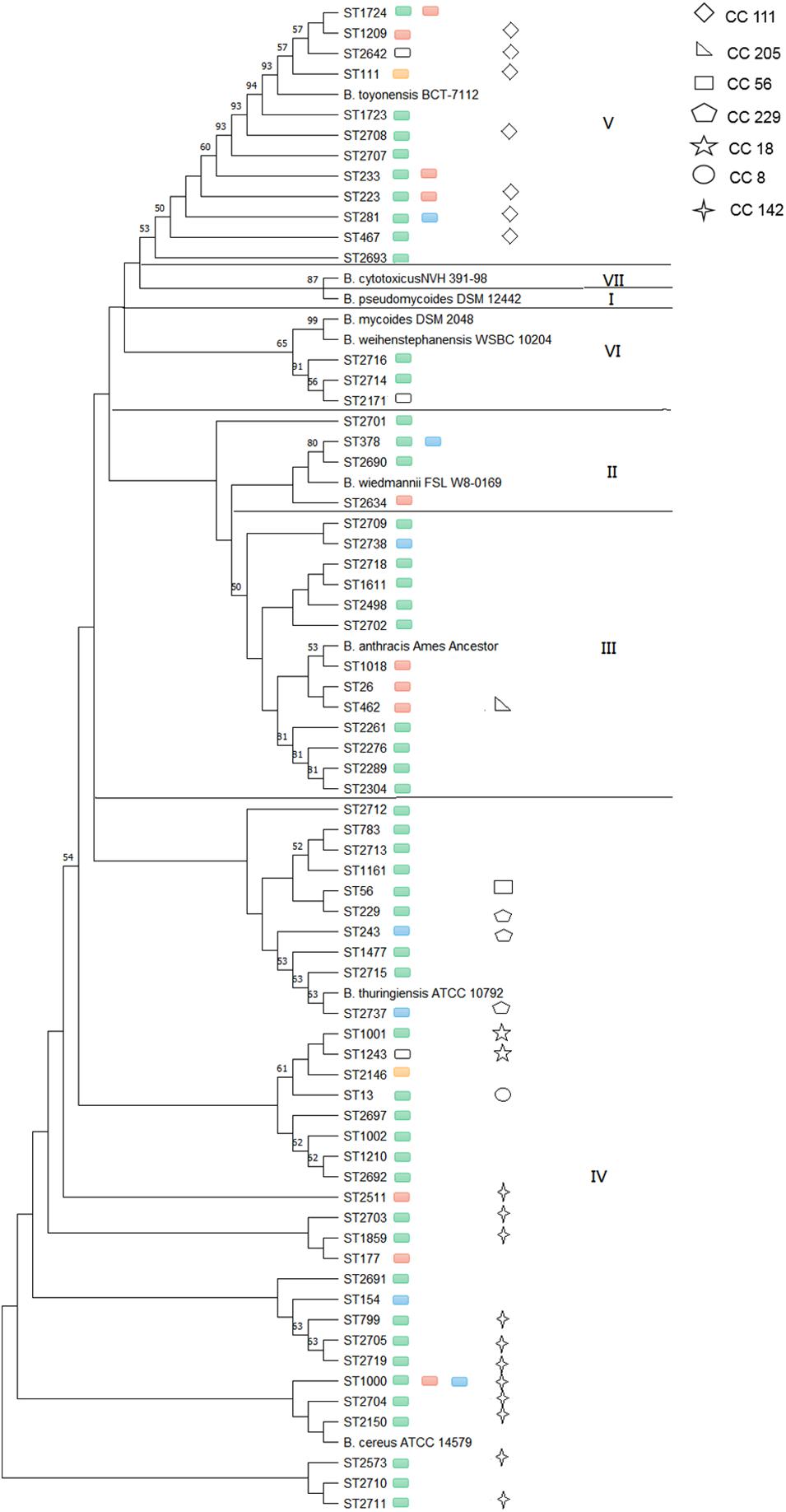
Figure 2. Phylogenetic relatedness of B. cereus strains from different provinces within particular phylogenetic groups. Green boxes represent isolates from Shanghai, red boxes represent isolates from Beijing, blue boxes represent isolates from Tianjin, yellow boxes represent isolates from Shandong, and white boxes represent isolates from Sichuan.
The phylogenetic study was based on MLST, which clustered the STs into five phylogenetic clades (II to VI) (Guinebretière et al., 2008), and nine additional B. cereus clade species were included in phylogenetic analyses to serve as a reference (Miller et al., 2018). The most common clade was clade IV (50.8%), followed by clade III (20.0%), clade V (18.5%), clade II (6.2%), and clade VI (4.6%). Clade IV included both B. cereus sensu stricto ATCC 14579T (ST4) and B. thuringiensis ATCC 10792T as well as 33 STs obtained from five provinces that consisted of five CCs: CC8, CC18, CC56, CC229, and CC142. Frequent types of ST1000 and CC142 (11 STs) were observed in this clade. Clade III with 13 STs, in addition to the B. anthracis Ames ancestor, was represented by Shanghai (69.2%), Beijing (23.1%), and Tianjin (7.7%). Only CC205 (1 ST) was included in this clade. Clade V, with the B. toyonensis BCT-7112T reference strain (ST111), contained 12 STs in five provinces and strains mainly from Shanghai (56.2%) and Beijing (25.0%), while other geographic locations were represented by less than 7.0% of isolates. ST1724, ST233, ST223, and ST281 were found in two different provinces, and isolates in clade V were clustered together in CC111 (7 STs). Clade II included the B. wiedmannii FSL W8-0169T (ST1081) reference strain and four STs from Shanghai, Beijing, and Tianjin, while ST378 was found in both Shanghai and Tianjin. Clade VI contained both the B. mycoides DSM 2048T and B. weihenstephanensis WSBC 10204T reference strains as well as three STs in the Shanghai and Sichuan provinces. The isolates in clades II and VI were singletons. None of the B. cereus isolate strains were clustered with B. pseudomycoides DSM 12442T within clade I or with B. cytotoxicus NVH 391-98T within clade VII.
Among the 100 B. cereus isolates, 53 were collected from lettuce, 19 from soil, 14 from pesticide, 13 from manure, and 1 from water. A minimum spanning tree-like structure was drawn to show the link between the sample sources and different STs of B. cereus (Figure 3). The ST type in lettuce was associated with the environmental source, and there was no crossover between environmental samples. Isolates from lettuce and soil had seven identical STs, while manure had five STs, identical to lettuce. Pesticide and water also had one ST, consistent with lettuce. Five STs were associated with lettuce and soil, namely, ST223, ST229, ST378, ST799, and ST1000, while three STs (ST233, ST1724, and ST2692) were commonly recovered from lettuce and manure. Only ST2150 originated from lettuce and pesticide. Moreover, there were other STs from additional source categories: one major ST2498 was a prevalent ST type in lettuce, soil and manure. In addition, ST1210 was found in lettuce, soil, manure, and water.
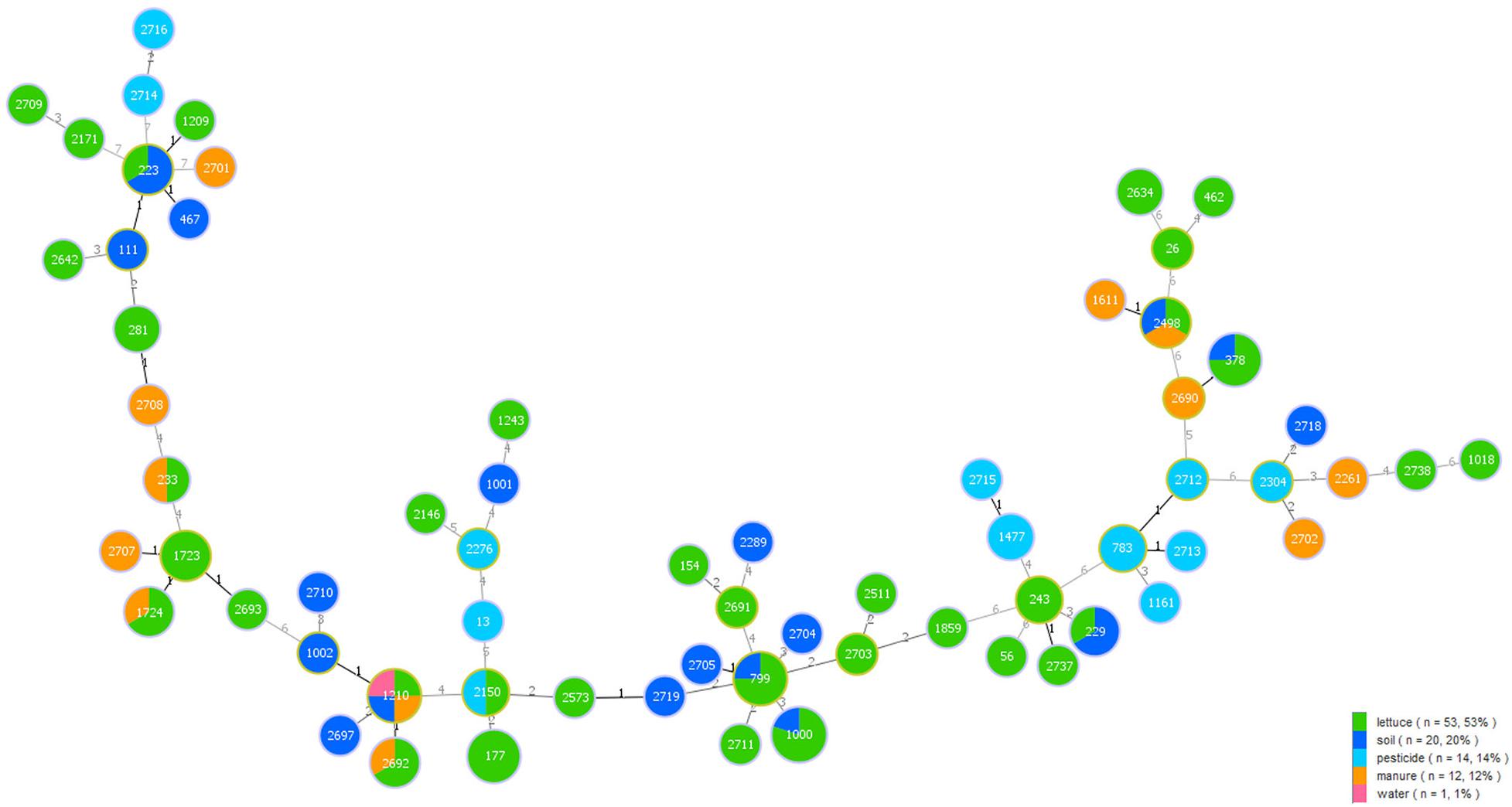
Figure 3. Minimum spanning tree analysis of 100 B. cereus isolates collected from different sample categories on lettuce farms. The size of each circle indicates the number of STs. Different sample categories are represented by different colors, and the size of each color block represents the number of STs in that source.
Sixty-eight strains were selected from lettuce farms, corresponding to sources and phylogenetic groups, for further enterotoxic potential (Figure 4). All isolates were tested via PCR for the presence of key toxin genes. (1) The most frequently distributed genes were those encoding the enterotoxin Nhe, namely, the nheB, nheA and nheC genes, which were detected in 100.0, 91.2, and 80.9% of the tested strains, respectively. While these nheABC genes (the strains were positive for nheA, nheB, and nheC at the same time) genes were all detected in 54 isolates in clades III and IV, nheA and nheC were detected less frequently in clades II, VI, and VI (23.1%∼80.0%). Isolates from lettuce, soil, water, manure, and pesticides all harbored the Nhe toxin genes (Figures 3, 4). (2) Genes encoding Hbl (hblA, hblC, and hblD) were detected in 89.7% of the tested strains, and sixty-one Hbl PCR-positive isolates were distributed in clade II (100.0%), clade V (100.0%), clade IV (100.0%), clade IV (97.4%), and clade III (33.3%). hblD was the most commonly detected Hbl toxin genes (89.7%), while hblA and hblC were detected individually in 76.5 and 73.5% of the isolates, respectively. Fifty strains harbored hblACD (the strains were positive for hblA, hblC, and hblD at the same time). Among the sources, 100.0% of the water and pesticides isolates (1/1 water isolate), 92.0% of the lettuce isolates, 88.9% of the soil isolates, and 75.0% of the manure isolates harbored the Hbl toxin genes. (3) The entFM gene, encoding enterotoxin FM, was also broadly distributed (91.2%). All six PCR-negative isolates were present in clade IV and clade V. In addition, the entFM gene was detected in 100% of the isolates from all sources except lettuce and pesticides. Thus, the positive detection rate was still high in lettuce and pesticides, with values of 80.0 and 91.7%, respectively. (4) The cytK gene, encoding cytotoxin K, was detected in 42.6% of the strains. All PCR isolates positive for cytK represented clades III (88.9%), IV (51.3%), and II (20.0%). The strains harboring the cytK gene were isolated from all five sources. The highest percentage of cytK detected was 100.0%, which was observed in water (1/1 isolate), followed by 58.3% in pesticides, 50.0% in soil, 36.0% in lettuce, and 25.0% in manure. (5) The ces gene, encoding emetic toxin, known as cerulide synthetase, was not detected in the isolates.
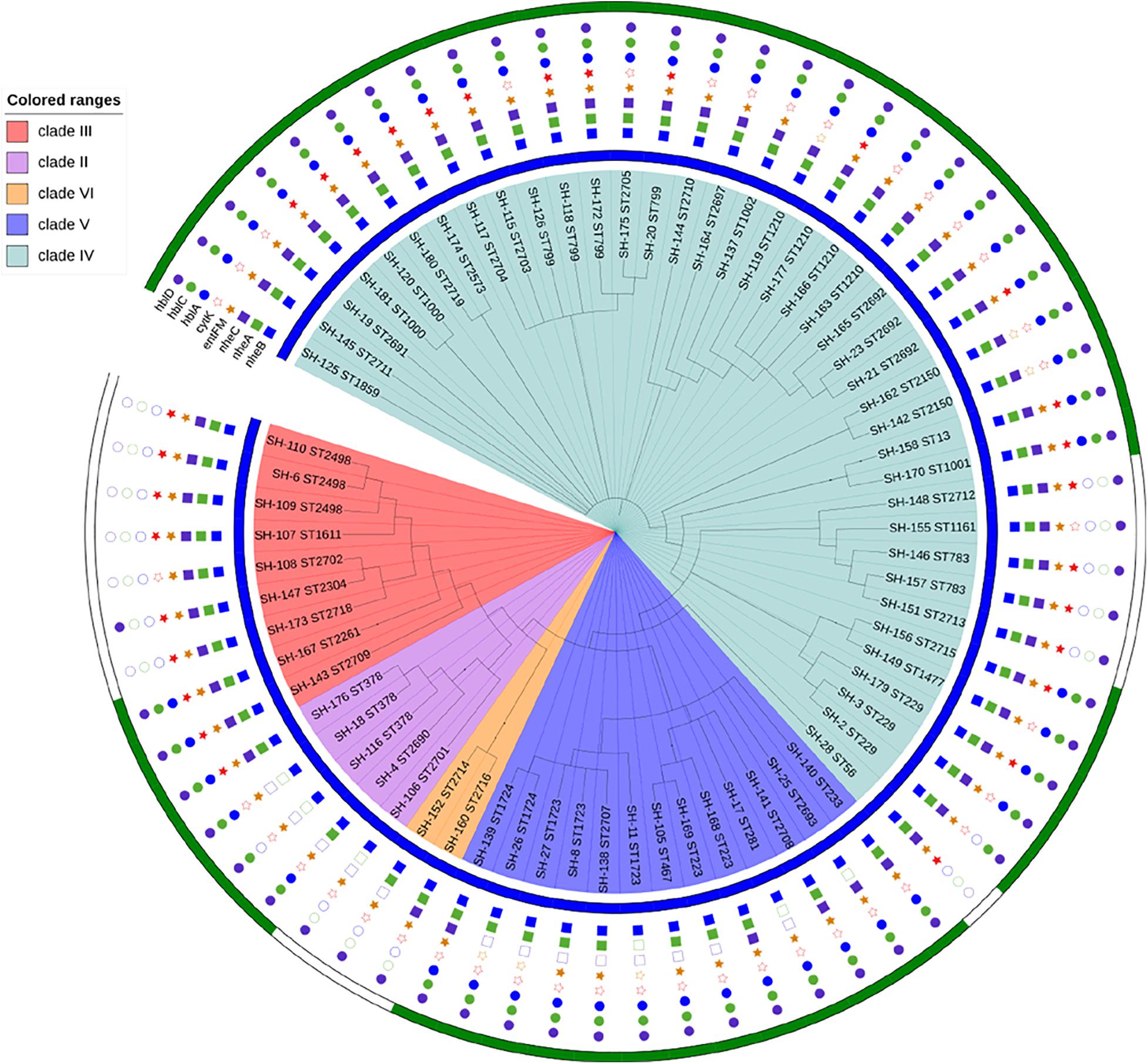
Figure 4. Enterotoxic potential of 68 B. cereus strains. The blue color strip represents Nhe (enterotoxin), while green represents Hbl (enterotoxin). The blue square represents NheB (gene), the green square represents nheA (gene), the purple square represents nheC (gene), the orange star represents entFM (gene), the red star represents cytK (gene), the blue circle represents hblA (gene), the green circle represents hblC (gene), and the purple circle represents hblD (gene).
All B. cereus isolates used in this study carried at least 3 of the 8 enterotoxin genes tested. The distribution of virulence genes was divided into 17 different profiles. Eighteen isolates possessed all eight virulence genes, which was one of the main gene profiles. The other main gene profile (18/68 of all isolates) was hblA-hblC-hblD-nheA-nheB-nheC-entFM. Only one isolate, SH-106 (ST2701), harbored the smallest virulence gene profile (nheB-hblD-entFM).
Nhe and Hbl, the most notable enterotoxins, were analyzed to obtain a broader view of the potential B. cereus enterotoxicity (Figure 4). Nhe was observed in the 68 isolates isolated from five sources and all phylogenetic groups, while Hbl was present in 76.5% (52/68) of the strains that were clustered into groups, with the exception of clade VI. The percentages of strains harboring Hbl were 100.0% in clade V, 84.6% in clade IV, 80.0% in clade II, and 22.2% in clade III. Among the sources, 100.0% of the water isolates (1/1), 92.0% of the lettuce isolates, 88.9% of the soil isolates, 66.7% of the manure isolates, and 33.3% of the pesticide isolates harbored Hbl. Figure 4 also reveals the relationship between virulence genes and enterotoxins: (1) Nhe was expressed when the nheB gene was present, and (2) Hbl was detected when the strain harbored the hblC gene.
Bacillus cereus is widely distributed around the world, and some studies have evaluated B. cereus in vegetables (Gdoura-Ben Amor et al., 2018; Park et al., 2018; Yu et al., 2019), while few studies have examined it in planting environments (Drewnowska et al., 2020). In this study, we determined the occurrence of B. cereus on lettuce farms in five Chinese provinces and determined the possible contamination pathways. Moreover, the potential of the isolates to cause foodborne disease was evaluated based on their production of diarrheal and emetic toxins.
The overall prevalence (48.5%) of B. cereus observed in the current study was somewhat less than previous research (57.7%) about markets in China (Yu et al., 2019), but much less than that reported in some studies conducted in other countries, e.g., 84.0% in Mexico City (Flores-Urbán et al., 2014), 81.3% in Korea (Park et al., 2018), and 80.0% in Glasgow (Altayar and Sutherland, 2006). These results indicate that B. cereus is common in lettuce. Emetic syndrome and diarrheal diseases are often associated with B. cereus counts of at least 105 CFU/g (Osimani et al., 2018). In our study, concentrations below 105 CUF/g were found in all lettuce samples. Thus, the loads at all levels were considered to be safe for consumption.
Multilocus sequence typing was used to evaluate the evolution and population diversity of B. cereus. The 100 B. cereus isolates from 46 lettuce farms represented 65 STs and seven CCs and were subtyped into five phylogenetic groups. ST233, ST378, ST1724, CC111, and CC142, which were crossed with different provinces, and each province had unique STs. Two strains isolated from lettuce and soil were assigned to ST26 and ST111, which were the same molecular types as those of the clinical isolates E6345 and F4794 (Hoffmaster et al., 2008). Eleven strains, except for those from pesticides, contained ST233, ST243, ST1001, ST1002, ST1210, and ST2171, which are listed as blood isolates in the B. cereus PubMLST database3. These results reveal a potential risk when consuming these lettuces directly or with minimal processing. ST1000 was the most frequent type in the 100 isolates and in lettuce, and ST177 was also a frequent type in lettuce, thus, these findings were different from the STs in vegetables on the Chinese market, in which ST26, ST770, and ST1605 were the most frequent types (Yu et al., 2019). Group IV was one of the three largest groups in this study, similar to its predominance in soil (Drewnowska et al., 2020). These results indicate the high genetic diversity of B. cereus isolates.
Soil and pesticides were the most frequently detected sources of B. cereus in our study, but the concentration in soil was higher than that in pesticides. In total, 7.3% of the soil samples had concentrations ranging from 2 × 104 to 5 × 105 CFU/g, which was consistent with previous studies (Drewnowska et al., 2020). Tracing analysis showed that soil, manure, pesticide and water had the same STs as lettuce, but soil and manure had more of these STs, indicating that B. cereus in lettuce was mainly from soil and manure. Additionally, the possible mechanism by which B. cereus spreads during lettuce planting were inferred as follows: soil-lettuce, manure-lettuce, pesticide-lettuce, manure-soil-lettuce, and water-manure-soil-lettuce. Quantitative microbial risk assessment of E. coli O157:H7 in lettuce revealed that bacterial concentration in soil, soil transfer by irrigation, and bacterial concentration in water were the most important input factors during lettuce preharvest (Pang et al., 2017; Bozkurt et al., 2021), which were partly consistent with our study and these indicated that the planting process needs to be controlled.
It has been demonstrated that B. cereus is the causative agent of two types of gastrointestinal diseases, namely, emetic syndrome and diarrheal syndrome (Osimani et al., 2018). The emetic form occurs due to a heat-stable toxin (cerulide) that is preformed in the food, while diarrhea is caused by the ingestion of viable cells, which produce enterotoxins in the small intestine (Osimani et al., 2018). In this study, in all the strains isolated from lettuce farms, only enterotoxins were detected, which was consistent with previous research (Park et al., 2018; Drewnowska et al., 2020).
Among the key toxin genes in B. cereus isolates, (1) the genes encoding the Nhe toxin were detected in 100.0% of the isolates in this study, especially nheB, which was distributed broadly. In addition, it has been shown that nearly 100.0% of B. cereus food poisoning outbreak strains harbor the Nhe toxin genes in Austria (Jessberger et al., 2019). (2) The genes encoding the Hbl toxin were present in 89.7% of the 68 isolates, which is somewhat higher than the value in Korea (Park et al., 2018), in which the hblACD genes were detected in 35.7% of lettuce isolates. In previous studies (Chon et al., 2015; Yu et al., 2019), the Hbl toxin genes were detected in 60.0% of B. cereus vegetable and soil isolates. (3) EntFM was detected in 91.2% of the strains, consistent with the results showing that 90.0 to 100.0% of B. cereus isolates harbored the entFM gene in B. cereus outbreak isolation (Jessberger et al., 2019). In total, 42.6% of the B. cereus isolates harbored the cytK gene. In Korea, 71.4% of the strains isolated from lettuce harbored cytK (Park et al., 2018), and cytK was present in 8.0 to 91.0% of the soil isolates (Drewnowska et al., 2020).
The enterotoxins Nhe and Hbl were detected by the Duopath® Cereus Enterotoxins test. Nhe was detected in 100.0% of the 68 isolates, and Hbl was observed in 76.5% of the isolates, thus the rates were higher than those of dairy-associated isolates among which tested positive for Nhe and 30.8% for Hbl (Miller et al., 2018). Moreover, 92.0% of the lettuce isolates harbored Hbl. Additionally, in this study, we found that (1) Nhe was expressed when the nheB gene was present. (2) Hbl was detected when the strain harbored the hblC gene. The Duopath® Cereus Enterotoxins test tracks Nhe by detecting the NheB component (encoded by nheB) in a sandwich complex with immobilized specific antibodies (Krause et al., 2010). NheA and NheC expression did not correlate with toxicity, while the expression of NheB showed a rough correlation with strain toxicity (Jessberger et al., 2019). In addition, the Duopath® Cereus Enterotoxins test detected Hbl based on gold-labeled monoclonal antibodies of Hbl lytic component L2 (Hbl-L2; encoded by hblC).
Taken together, the results of the present study showed that despite the high presence of B. cereus in lettuce and farm environment samples, the loads were at levels considered to be safe for consumption. However, pathogenic STs (ST26 and ST111) were detected in lettuce and soil, and all B. cereus strains tested in this study carried at least 3 of the 8 enterotoxin genes. Nhe and Hbl enterotoxins were the major toxins among the B. cereus strains tested. Additionally, B. cereus present in planting environments (soil, manure, pesticide, and water) could be easily transferred to lettuce. Our study provides useful information for improving the microbiological safety of fresh lettuce.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
YQ, YS, and CZ conceived and designed the experiments. YQ, CW, XD, XZ, QL, WW, YW, MG, and WT performed the experiments. YQ analyzed the data and wrote the draft. YB contributed in the reagents/materials/analysis tools. YS and CZ supervised the research. YS revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the Special Project of Agriculture Produce Quality Safety Risk Assessment (Grant No. GJFP2019005) from Ministry of Agriculture and Rural Affairs of the People’s Republic of China and Youth Development Project (Grant No. 2020. No.1-7) from Institute for Agri-Food Standards and Testing Technology, Shanghai Academy of Agricultural Sciences.
YQ, CZ, and YS are employed by Institute for Agro-Food Standards and Testing Technology, Shanghai Academy of Agricultural Sciences, Shanghai, China and Shanghai Co-Elite Agro-Food Testing Service Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Shenghao Yu from the Information Application Research Center of Shanghai Municipal Administration of Market Supervision and Yiwei Dong from Changsha Tunkia Co., Ltd., for their assistance with the experiments.
Abadias, M., Usall, J., Anguera, M., Solsona, C., and Viñas, I. (2008). Microbiological quality of fresh, minimally-processed fruit and vegetables, and sprouts from retail establishments. Int. J. Food Microbiol. 123, 121–129. doi: 10.1016/j.ijfoodmicro.2007.12.013
Altayar, M., and Sutherland, A. D. (2006). Bacillus cereus is common in the environment but emetic toxin producing isolates are rare. J. Appl. Microbiol. 100, 7–14. doi: 10.1111/j.1365-2672.2005.02764.x
Bozkurt, H., Bell, T., van Ogtrop, F., Phan-Thien, K. Y., and McConchie, R. (2021). Assessment of microbial risk during Australian industrial practices for Escherichia coli O157:H7 in fresh cut-cos lettuce: A stochastic quantitative approach. Food Microbiol. 95:103691. doi: 10.1016/j.fm.2020.103691
Castiaux, V., N’Guessan, E., Swiecicka, I., Delbrassinne, L., Dierick, K., and Mahillon, J. (2014). Diversity of pulsed-field gel electrophoresis patterns of cereulide-producing isolates of Bacillus cereus and Bacillus weihenstephanensis. FEMS Microbiol. Lett. 353, 124–131. doi: 10.1111/1574-6968.12423
Ceuppens, S., Boon, N., and Uyttendaele, M. (2013). Diversity of Bacillus cereus group strains is reflected in their broad range of pathogenicity and diverse ecological lifestyles. FEMS Microbiol. Ecol. 84, 433–450. doi: 10.1111/1574-6941.12110
Chon, J. W., Yim, J. H., Kim, H. S., Kim, D. H., Kim, H., Oh, D. H., et al. (2015). Quantitative prevalence and toxin gene profile of Bacillus cereus from ready-to-eat vegetables in South Korea. Foodborne Pathog. Dis. 12, 795–799. doi: 10.1089/fpd.2015.1977
Dierick, K., Van Coillie, E., Swiecicka, I., Meyfroidt, G., Devlieger, H., Meulemans, A., et al. (2005). Fatal family outbreak of Bacillus cereus-associated food poisoning. J. Clin. Microbiol. 43, 4277–4279. doi: 10.1128/jcm.43.8.4277-4279.2005
Drewnowska, J. M., Stefanska, N., Czerniecka, M., Zambrowski, G., and Swiecicka, I. (2020). Potential enterotoxicity of phylogenetically diverse Bacillus cereus sensu lato soil isolates from different geographical locations. Appl. Environ. Microbiol. 86:19. doi: 10.1128/aem.03032-19
Ehling-Schulz, M., Vukov, N., Schulz, A., Shaheen, R., Andersson, M., Märtlbauer, E., et al. (2005). Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl. Environ. Microbiol. 71, 105–113. doi: 10.1128/aem.71.1.105-113.2005
Feil, E. J., Li, B. C., Aanensen, D. M., Hanage, W. P., and Spratt, B. G. (2004). eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186, 1518–1530. doi: 10.1128/jb.186.5.1518-1530.2004
Flores-Urbán, K. A., Natividad-Bonifacio, I., Vázquez-Quiñones, C. R., Vázquez-Salinas, C., and Quiñones-Ramírez, E. I. (2014). Detection of toxigenic Bacillus cereus strains isolated from vegetables in Mexico City. J. Food Prot. 77, 2144–2147. doi: 10.4315/0362-028x.Jfp-13-479
Forsythe, S. J., Dickins, B., and Jolley, K. A. (2014). Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genomics 15:1121. doi: 10.1186/1471-2164-15-1121
Francisco, A. P., Vaz, C., Monteiro, P. T., Melo-Cristino, J., Ramirez, M., and Carriço, J. A. (2012). PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinform. 13:87. doi: 10.1186/1471-2105-13-87
Gdoura-Ben Amor, M., Siala, M., Zayani, M., Grosset, N., Smaoui, S., Messadi-Akrout, F., et al. (2018). Isolation, identification, prevalence, and genetic diversity of Bacillus cereus group bacteria from different foodstuffs in Tunisia. Front. Microbiol. 9:447. doi: 10.3389/fmicb.2018.00447
Guinebretière, M.-H., Broussolle, V., and Nguyen-The, C. (2002). Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J. Clin. Microbiol. 40, 3053–3056. doi: 10.1128/jcm.40.8.3053-3056.2002
Guinebretière, M. H., Thompson, F. L., Sorokin, A., Normand, P., Dawyndt, P., Ehling-Schulz, M., et al. (2008). Ecological diversification in the Bacillus cereus group. Environ. Microbiol. 10, 851–865. doi: 10.1111/j.1462-2920.2007.01495.x
Hammerum, A. M., Hansen, F., Skov, M. N., Stegger, M., Andersen, P. S., Holm, A., et al. (2015). Investigation of a possible outbreak of carbapenem-resistant Acinetobacter baumannii in Odense, Denmark using PFGE, MLST and whole-genome-based SNPs. J. Antimicrob. Chemother. 70, 1965–1968. doi: 10.1093/jac/dkv072
Hoffmaster, A. R., Novak, R. T., Marston, C. K., Gee, J. E., Helsel, L., Pruckler, J. M., et al. (2008). Genetic diversity of clinical isolates of Bacillus cereus using multilocus sequence typing. BMC Microbiol. 8:191. doi: 10.1186/1471-2180-8-191
Höfte, H., and Whiteley, H. R. (1989). Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol. Rev. 53, 242–255.
Ivanova, N., Sorokin, A., Anderson, I., Galleron, N., Candelon, B., Kapatral, V., et al. (2003). Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423, 87–91. doi: 10.1038/nature01582
Jessberger, N., Kranzler, M., Da Riol, C., Schwenk, V., Buchacher, T., Dietrich, R., et al. (2019). Assessing the toxic potential of enteropathogenic Bacillus cereus. Food Microbiol. 84:103276. doi: 10.1016/j.fm.2019.103276
Jiménez, G., Urdiain, M., Cifuentes, A., López-López, A., Blanch, A. R., Tamames, J., et al. (2013). Description of Bacillus toyonensis sp. nov., a novel species of the Bacillus cereus group, and pairwise genome comparisons of the species of the group by means of ANI calculations. Syst. Appl. Microbiol. 36, 383–391. doi: 10.1016/j.syapm.2013.04.008
Jung, M. Y., Kim, J. S., Paek, W. K., Lim, J., Lee, H., Kim, P. I., et al. (2011). Bacillus manliponensis sp. nov., a new member of the Bacillus cereus group isolated from foreshore tidal flat sediment. J. Microbiol. 49, 1027–1032. doi: 10.1007/s12275-011-1049-6
Kim, J. B., Kim, J. M., Cho, S. H., Oh, H. S., Choi, N. J., and Oh, D. H. (2011). Toxin genes profiles and toxin production ability of Bacillus cereus isolated from clinical and food samples. J. Food Sci. 76, T25–T29. doi: 10.1111/j.1750-3841.2010.01958.x
Krause, N., Moravek, M., Dietrich, R., Wehrle, E., Slaghuis, J., and Märtlbauer, E. (2010). Performance characteristics of the Duopath® cereus enterotoxins assay for rapid detection of enterotoxinogenic Bacillus cereus strains. Int. J. Food Microbiol. 144, 322–326. doi: 10.1016/j.ijfoodmicro.2010.10.008
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Maiden, M. C., Jansen van Rensburg, M. J., Bray, J. E., Earle, S. G., Ford, S. A., Jolley, K. A., et al. (2013). MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 11, 728–736. doi: 10.1038/nrmicro3093
Meer, R. R., Baker, J., Bodyfelt, F. W., and Griffiths, M. W. (1991). Psychrotrophic Bacillus spp. in fluid milk products: a review. J. Food Prot. 54, 969–979. doi: 10.4315/0362-028x-54.12.969
Miller, R. A., Beno, S. M., Kent, D. J., Carroll, L. M., Martin, N. H., Boor, K. J., et al. (2016). Bacillus wiedmannii sp. nov., a psychrotolerant and cytotoxic Bacillus cereus group species isolated from dairy foods and dairy environments. Int. J. Syst. Evol. Microbiol. 66, 4744–4753. doi: 10.1099/ijsem.0.001421
Miller, R. A., Jian, J., Beno, S. M., Wiedmann, M., and Kovac, J. (2018). Intraclade Variability in toxin production and cytotoxicity of Bacillus cereus group type strains and dairy-associated isolates. Appl. Environ. Microbiol. 84:17. doi: 10.1128/aem.02479-17
Osimani, A., Aquilanti, L., and Clementi, F. (2018). Bacillus cereus foodborne outbreaks in mass catering. Int. J. Hosp. Manag. 72, 145–153. doi: 10.1016/j.ijhm.2018.01.013
Otlewska, A., Oltuszak-Walczak, E., and Walczak, P. (2013). Differentiation of strains from the Bacillus cereus group by RFLP-PFGE genomic fingerprinting. Electrophoresis 34, 3023–3028. doi: 10.1002/elps.201300246
Pang, H., Lambertini, E., Buchanan, R. L., Schaffner, D. W., and Pradhan, A. K. (2017). Quantitative microbial risk assessment for Escherichia coli O157:H7 in fresh-cut lettuce. J. Food Prot. 80, 302–311. doi: 10.4315/0362-028x.Jfp-16-246
Park, K. M., Jeong, M., Park, K. J., and Koo, M. (2018). Prevalence, enterotoxin genes, and antibiotic resistance of Bacillus cereus isolated from raw vegetables in korea. J. Food Prot. 81, 1590–1597. doi: 10.4315/0362-028x.Jfp-18-205
Stenfors, L. P., and Granum, P. E. (2001). Psychrotolerant species from the Bacillus cereus group are not necessarily Bacillus weihenstephanensis. FEMS Microbiol. Lett. 197, 223–228. doi: 10.1111/j.1574-6968.2001.tb10607.x
Yu, P., Yu, S., Wang, J., Guo, H., Zhang, Y., Liao, X., et al. (2019). Bacillus cereus isolated from vegetables in china: incidence, genetic diversity, virulence genes, and antimicrobial resistance. Front. Microbiol. 10:948. doi: 10.3389/fmicb.2019.00948
Keywords: Bacillus cereus, lettuce farm, multilocus sequence typing (MLST), tracing analysis, enterotoxicity
Citation: Qu Y, Wei C, Dai X, Bai Y, Zhao X, Lan Q, Wang W, Wu Y, Gao M, Tang W, Zhou C and Suo Y (2021) The Possible Transmission and Potential Enterotoxicity of Bacillus cereus on Lettuce Farms in Five Chinese Provinces. Front. Microbiol. 12:746632. doi: 10.3389/fmicb.2021.746632
Received: 24 July 2021; Accepted: 13 September 2021;
Published: 01 October 2021.
Edited by:
Jun Wang, Qingdao Agricultural University, ChinaReviewed by:
Moutong Chen, Guangdong Institute of Microbiology, Guangdong Academy of Sciences, ChinaCopyright © 2021 Qu, Wei, Dai, Bai, Zhao, Lan, Wang, Wu, Gao, Tang, Zhou and Suo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changyan Zhou, emhvdWNoYW5neWFuQHNhYXMuc2guY24=; Yujuan Suo, c3VveXVqdWFuQHNhYXMuc2guY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.