- 1Collaborative Innovation Center of Yangtze River Delta Region Green Pharmaceuticals, College of Pharmaceutical Sciences, Institue of Engineering Biology and Health, Zhejiang University of Technology, Hangzhou, China
- 2Shanghai Key Laboratory of Medical Epigenetics, Institutes of Biomedical Sciences, Fudan University, Shanghai, China
Mitochondrial antiviral signaling protein (MAVS) functions as a “switch” in the immune signal transduction against most RNA viruses. Upon viral infection, MAVS forms prion-like aggregates by receiving the cytosolic RNA sensor retinoic acid-inducible gene I-activated signaling and further activates/switches on the type I interferon signaling. While under resting state, MAVS is prevented from spontaneously aggregating to switch off the signal transduction and maintain immune homeostasis. Due to the dual role in antiviral signal transduction and immune homeostasis, MAVS has emerged as the central regulation target by both viruses and hosts. Recently, researchers show increasing interest in viral evasion strategies and immune homeostasis regulations targeting MAVS, especially focusing on the post-translational modifications of MAVS, such as ubiquitination and phosphorylation. This review summarizes the regulations of MAVS in antiviral innate immune signaling transduction and immune homeostasis maintenance.
Introduction
The battle between humans and viruses is never ending. In recent years, new emerging viruses, such as SARS-CoV-2 (also referred to as 2019-nCoV), pose a tremendous threat to public health. Our bodies developed different immune systems to defend ourselves against viral infection. Innate immunity is the body’s first line of defense against foreign pathogens. It consists of tissue barriers (skin and mucous membranes), phagocytes (macrophages and neutrophils), dendritic cells (DC), and killer cells (Thompson and Locarnini, 2007). Innate immunity produces interferons (IFNs) and pro-inflammatory factors to inhibit and eliminate the invading pathogens and maintain the hots immune homeostasis. Meanwhile, IFNs have immunomodulatory effects and can activate acquired immunity through antigen presentation. In addition, innate immunity also plays a key role in inhibiting tumor growth and metastasis.
The Structure and Function of Rlrs and Mavs
The activation of the innate immune system requires pattern recognition receptors (PRRs) to recognize pathogen-associated molecular patterns (Kawai and Akira, 2010; Broz and Monack, 2013; Cao, 2016). retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) are the main PRRs in the cytoplasm that sense and respond to viral RNA (Hiscott et al., 2010), including RIG-I, melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2; Yoneyama et al., 2004; Katze et al., 2008), which all belong to the family of RNA helicases containing DExD/H characteristic domains. RIG-I and MDA5 contain three identical basic domains. The N-terminal caspase-recruitment domains (CARDs) are responsible for transducing signals to downstream protein factors and then activate nuclear factor κB (NF-κB) and INF regulatory factor 3/7 (IRF3/7)-related signaling pathways. A central DExD/H-box helicase domain and a C-terminal domain (CTD) can bind to the 5′ppp-RNA tail of viral RNA (Yoneyama et al., 2004; Hornung et al., 2006). RIG-I recognizes short-stranded viral RNA, while MDA5 recognizes long-stranded viral DNA with the help of LGP2. RLRs play critical roles in the antiviral innate immunity system via the induction of type I INF and its downstream INF-stimulated genes.
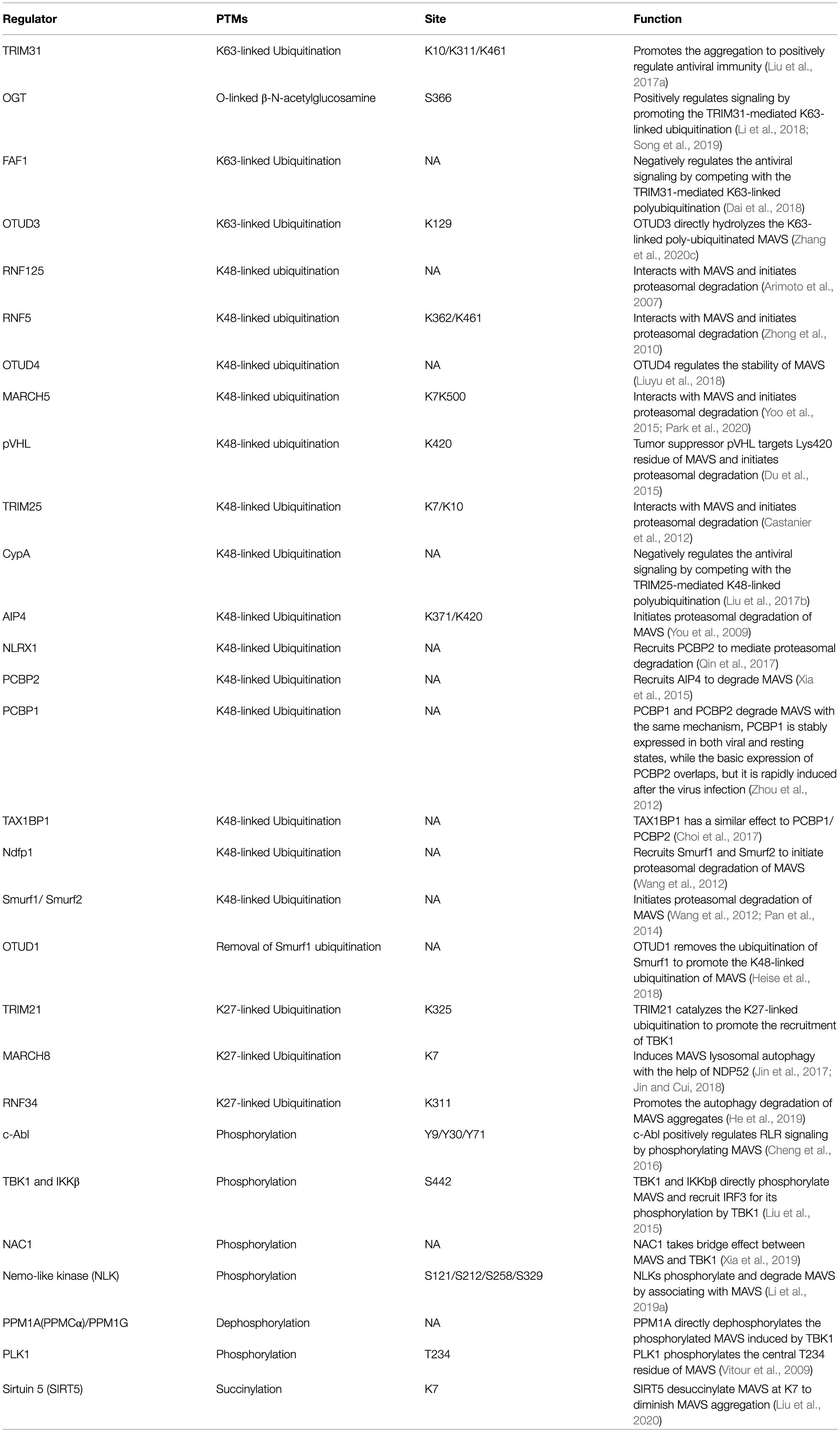
Mitochondrial antiviral signaling protein (MAVS; also known as IPS-1/VISA/Cardif) functions as a platform for antiviral innate immune signal transduction (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). MAVS consists of 540 amino acids. The N-terminal CARD can interact with the CARD of RIG-I/MDA5. The C-terminal is the transmembrane domain (TM) which locates MAVS on the outer mitochondrial membrane. Moreover, a proline-rich region contains three active motif binds to the downstream E3 ubiquitin ligase TRAFs (Seth et al., 2005). Upon viral infection, MAVS forms prion-like aggregates by receiving the signal from the cytosolic RNA sensor RIG-I and subsequently activates downstream NF-κB and IRF3/7-related signal pathways, switching on the type I IFN signaling to produce type I IFNs (e.g., IFN-α and IFN-β) and other cytokines (e.g., TNF-α and interleukins) through a series of cascade reactions (Castanier et al., 2012; Liu et al., 2017a). Figure 1 shows the framework of this review.
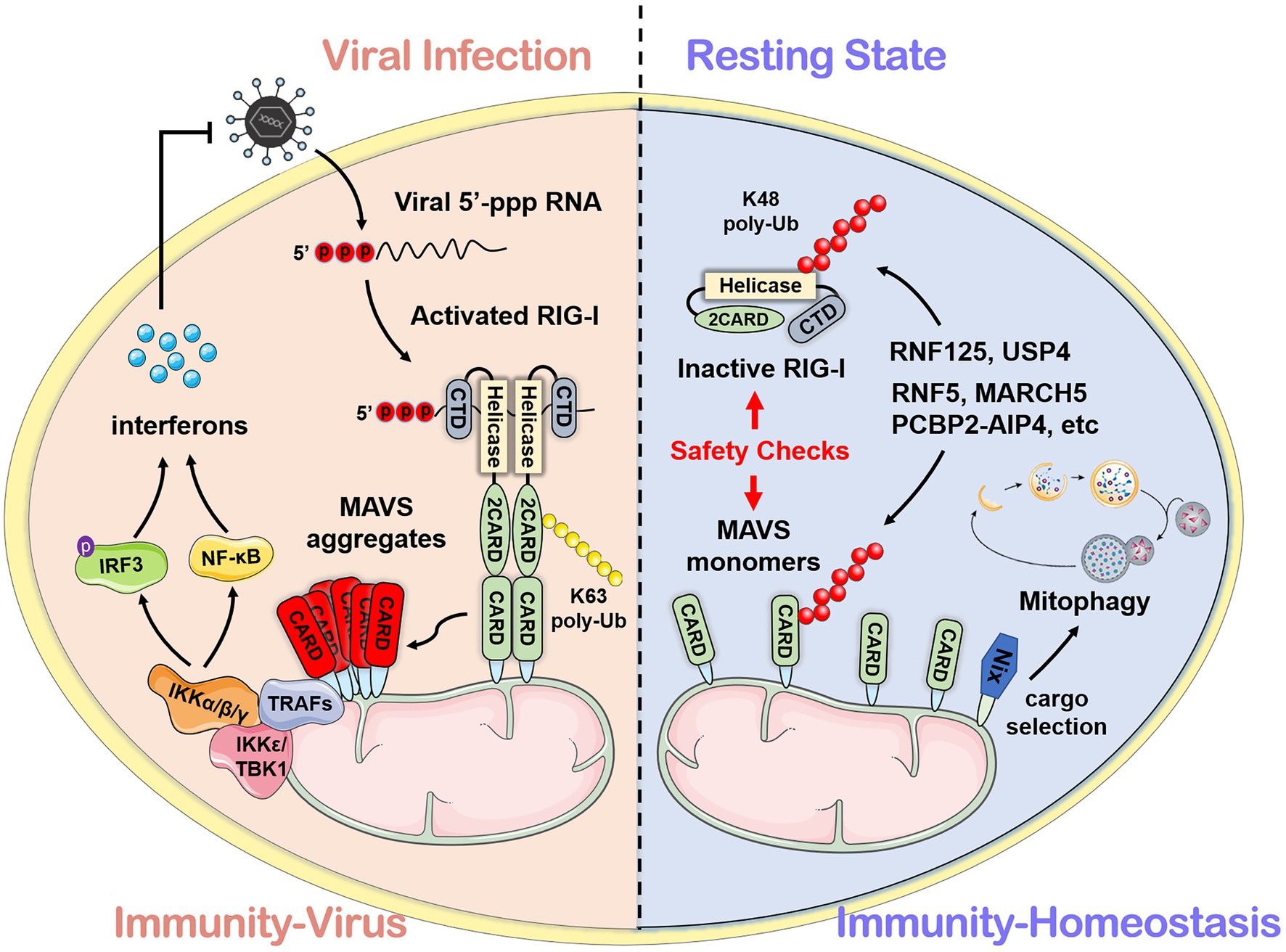
Figure 1. The mechanism mediated by mitochondrial antiviral signaling protein (MAVS) during viral infection and homeostasis. After recognizing viral RNA, the caspase-recruitment domain (CARD) of retinoic acid-inducible gene I interacts with the N-terminal CARD domain of MAVS and subsequently activates downstream antiviral signals. In the resting state, MAVS protein maintains immune homeostasis through self-inhibition and the ubiquitin-proteasome system.
The current research and exploration on the regulation mechanism of MAVS mainly focus on four aspects. The first is the regulation of the molecular activity of MAVS by protein interactions. For example, LGP2 interacts with the TM domain of MAVS to prevent MAVS from recruiting E3 ligase TRAF3. The second is the regulation of mitochondrial polymorphism on the molecular activity of MAVS. Studies have shown that factors affecting the physical state of mitochondria, such as mitochondrial fusion, changes in membrane potential, and the level of reactive oxygen species (ROS), also alter the formation of MAVS aggregates (de Brito and Scorrano, 2008; Onoguchi et al., 2010; Zhao et al., 2012). Thirdly, post-translational modification (PTM) on MAVS molecular activity has become a pivotal host antiviral innate immune signaling regulation. Lastly, cells have evolved many automated mechanisms to maintain immune homeostasis to balance the activation and suppression of the innate immune response.
Ptms Control of Mavs
Brief Conception of PTMs
Post-translational modifications of proteins can dynamically regulate the partitioning, transport, and physical interaction of pivotal molecules in the immune process (Diskin et al., 2021). It is emerging as a key mechanism by which intracellular metabolites can modulate immunity (Deribe et al., 2010; Liu et al., 2016). The precursor protein is generally inactive and often requires a series of post-translational processing to become a functional mature protein. The stability and activity of the protein are regulated by covalently binding with new functional groups, such as phosphate, methyl, and acetate. It has been demonstrated that traditional aluminum electrolytic phosphorylation and ubiquitination and non-traditional modifications, such as carbonylation and hydroxylation (Yang et al., 2017; Boeynaems and Gitler, 2018; Strowitzki et al., 2019), can target inflammatory responses related to PRRs through natural immune signaling pathways. These reversible modifications are catalyzed by specific enzymes. PTMs of MAVS are listed in Table 1, with the bested-studied phosphorylation and ubiquitination illustrated in Figure 2.
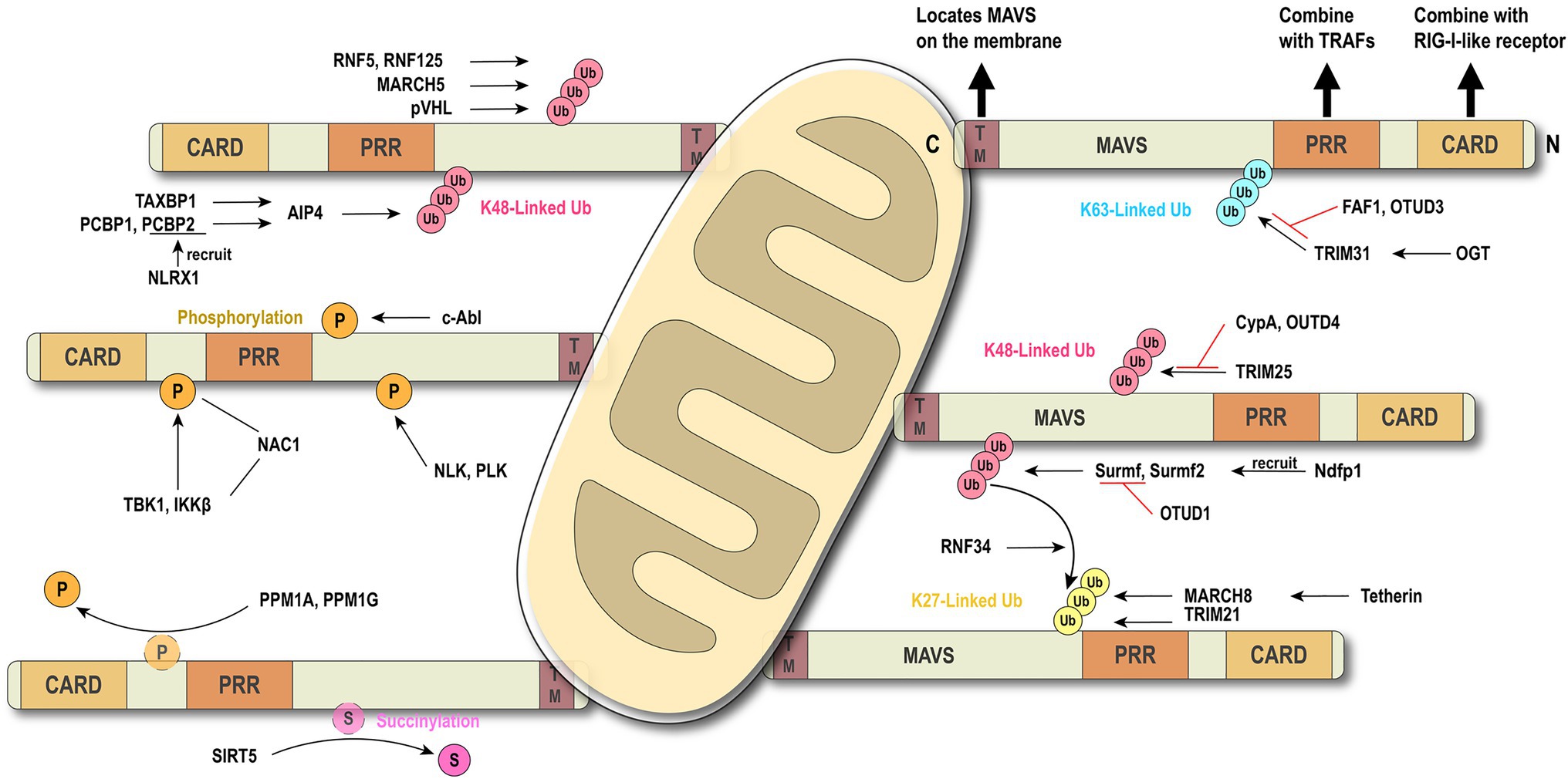
Figure 2. Brief conception of ubiquitination and phosphorylation in MAVS regulation. MAVS contains three key motifs and locates on the outer membrane of mitochondria. Ubiquitination and phosphorylation are the most common types of post-translational modifications (PTMs) which regulate MAVS in the innate immune signaling pathway.
MAVS Regulation by Ubiquitination
Ubiquitination is a common PTM in the RLR signaling pathway. E3 ubiquitin ligase TRIM31-mediated K63-linked ubiquitination plays a positive role in the antiviral immune pathway. Upon viral infection, TRIM31 is recruited to MAVS and catalyze the K63-linked polyubiquitination at K10, K311, and K461, leading to the higher efficient formation of MAVS prion-like aggregates (Liu et al., 2017a). The O-GlcNAcylation of MAVS on S366 mediated by OGT promotes TRIM31-mediated ubiquitination, thereby facilitating the activation of IRF3 and the production of IFN-β. This discovery clarifies the important role of the glucose metabolism pathway in antiviral immunity (Li et al., 2018; Song et al., 2019). Conversely, scaffold protein FAF1 negatively regulates the antiviral signal by competing with the TRIM31-mediated K63-linked polyubiquitination and diminishing the prion-like aggregation of MAVS. After viral infection, IKKε mediates FAF1 phosphorylation and, following acetylation, degradation and consequent enhancement of the MAVS antiviral signaling (Dai et al., 2018). As an acetylation-dependent deubiquitinase, OTUD3 can restrict IFN signaling by directly hydrolyzing the K63-linked poly-ubiquitinated MAVS at K129 residue (Zhang et al., 2020c).
Several lines of evidence suggest that K48-linked ubiquitination catalyzed by different E3 ligases is also critical in antiviral innate immunity (Zhong et al., 2010; Du et al., 2015; Yoo et al., 2015; Liuyu et al., 2018; Park et al., 2020). Compared with the K63-linked ubiquitination that positively regulates MAVS aggregation, TRIM25-mediated K48-linked ubiquitination negatively regulates the RLR signaling pathway through MAVS degradation by the proteasome (Castanier et al., 2012). However, this phenomenon can be competitively suppressed by CypA (Liu et al., 2017b). It has been reported that nucleotide-binding oligomerization domain-like receptor X1 (NLRX1) inhibits the production of IFN (Qin et al., 2017), and poly(rC) binding protein 2 (PCBP2) recruits the E3 ligase AIP4 (also known as ITCH) containing the HECT domain to assist NLRX1 initiating MAVS ubiquitination and degradation (You et al., 2009; Xia et al., 2015). Further research shows that PCBP1 and PCBP2 degrade MAVS with the same mechanism. The difference is that PCBP1 is stably expressed both in viral and resting states, while the basic expression of PCBP2 overlaps, but it is rapidly induced after the virus infection (Zhou et al., 2012). At the same time, TAX1BP1 has also been reported to have a similar effect to PCBP1 and PCBP2 (Choi et al., 2017). Smurf1 and Smurf2 are recruited by Ndfp1, a member of the NEDD4 family, and also mediate the K48-linked ubiquitination and degradation of MAVS (Wang et al., 2012; Pan et al., 2014), whereas OTUD1 inhibits RLR signaling by removing the Smurf1-mediated ubiquitination (Heise et al., 2018). Membrane-associated RING-cysteine-histidine (MARCH) proteins also can directly or indirectly regulate the ubiquitination of MAVS (Zheng, 2021). MARCH5 mediates K48-linked ubiquitination and induces the proteasomal degradation of MAVS by transferring the ubiquitin to K7 and K500 of MAVS (Yoo et al., 2015).
Besides K63-linked and K48-linked ubiquitination, TRIM21 can catalyze the K27-linked ubiquitination to promote the recruitment of TBK1 through the interaction with MAVS, positively regulating the antiviral signaling (Xue et al., 2018). After viral infection, E3 ubiquitin ligase MARCH8 is recruited by antiviral factor Tetherin (BST2/CD317) and mediates K27-linked ubiquitination of MAVS at K7. NDP52 induces proteasomal degradation after recognizing the K27-linked ubiquitination signal (Jin et al., 2017), and NDP52 can also trigger CALCOCO2-directed autophagic degradation (Jin and Cui, 2018). NDP52 is the bridge that connects the ubiquitinated protein and the autophagy-mediated component. Another study has reported that NDP52 recognizes the K63-to-K27 linked ubiquitination transition signal mediated by RNF34 on the K311 of MAVS, efficiently promoting the autophagy degradation of MAVS aggregates (He et al., 2019).
MAVS Regulation by Phosphorylation
Phosphorylation and dephosphorylation have been reported to play a critical role in antiviral innate immunity. c-Abl positively regulates RLR signaling by phosphorylating MAVS at Y9, Y30, and Y71 (Cheng et al., 2016). After phosphorylation by the kinases TBK1 and IKKs at residue S442, pMAVS binds to IRF3, thereby recruiting IRF3, which is phosphorylated and activated by TBK1 to activate antiviral response (Liu et al., 2015). Xia et al. have identified NAC1, a BTB/POZ family member, as a bridge between MAVS and TBK1 (Xia et al., 2019). Conversely, dephosphorylation functions as a switch to turn off antiviral signaling. PPM1A (also known as PP2Cα), a phosphatase, directly dephosphorylates the phosphorylated MAVS induced by TBK1 and subsequently silences the cytoplasmic RNA sensor signal. Similar to PPM1A, PPM1G also dephosphorylates MAVS (Xiang et al., 2016). A Nemo-like kinase is reported to phosphorylate and induce MAVS degradation by associating with MAVS at 451 to 500 of the C-terminal region (Li et al., 2019a). Polo-like kinase PLK1 phosphorylates the central T234 residue of MAVS, while polo-box domain of the PLK1 phospho-independently associates the C terminus of MAVS to destroy the combination of MAVS and downstream partners like TRAF3 (Vitour et al., 2009).
Protein succinylation caused by succinyl-CoA is a newly discovered novel PTM (Yang and Gibson, 2019). Research shows that MAVS is succinylated under viral stimulation. Sirtuin 5 desuccinylates MAVS at K7 to diminish and antagonize the MAVS aggregation and antiviral response (Liu et al., 2020). Although the phosphorylation and ubiquitination of MAVS have been extensively studied, further investigations are still needed to discover new regulators and mechanisms.
Virus Immune Evasion Against Rig-I-Mavs
The global outbreak of COVID-19 raised the scientific interest in how the virus escapes the host rigorous immune system to survive and duplicate. A study found that the Borna disease virus (BDV) genome trimming turns triphosphate end into monophosphate, effectively preventing recognition by RIG-I (Schneider et al., 2007). Compelling evidence shows that the virus has evolved various mechanisms to antagonize the innate immune response (Figure 3). The virus immune evasion methods in the RIG-I-MAVS signaling pathway can be roughly divided into two categories.
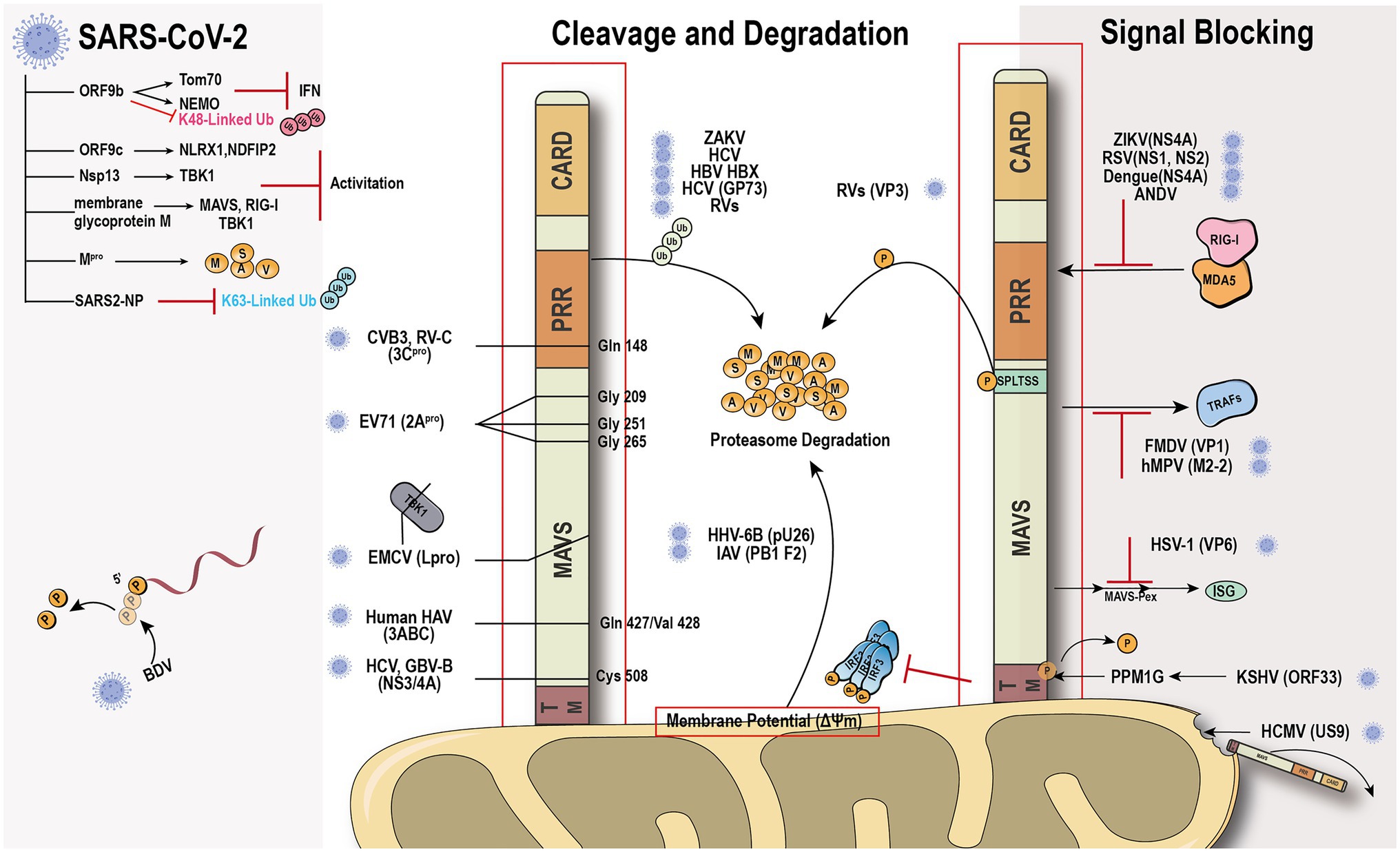
Figure 3. Viral immune evasion mechanism against MAVS. Viruses have evolved multiple immune evasion mechanisms targeting and sequestering MAVS from the signaling pathway directly.
Viral protein directly cleaves or degrades MAVS to avoid the activation of immune signal transduction. Hepatitis C virus (HCV) and GB virus B serine protease NS3/4A can dislodge the N-terminal fragment of MAVS from the mitochondria and endogenous cleavage of MAVS at Cys508 (Chen et al., 2007; Xu et al., 2020). Encephalomyocarditis virus (EMCV) lead protease (Lpro) cleaves the RLR signaling proteins MAVS and TBK1 (Jackson et al., 2020). Coxsackievirus B3 and Rhinovirus C cleaves MAVS at Gln-148 occurred within its proline-rich region (Pang et al., 2017). Enterovirus 71 proteinase 2Apro cleaves MAVS at Gly209, Gly251, and Gly265, between the proline-rich and transmembrane domains (Coyne et al., 2013). 2Apro has the same function as 3Cpro (Feng et al., 2014). The difference is that 2Apro cleaves MDA5 and MAVS, while 3Cpro cleaves RIG-I and MAVS. The cleavage site of viral protease is different depending on the species. HAV 3ABC protease derived from bats cleaves MAVS at Glu463/Gly464, while the protease from human cleaves MAVS at Gln427/Val428 (Feng et al., 2019). Besides direct cleavage, some viruses, such as ZIKV and HCV, can mediate MAVS degradation through ubiquitin-proteasome systems (Wei et al., 2010; Li et al., 2019b). Viral factor pU26 of human herpesvirus 6B can affect the mitochondrial membrane potential and target MAVS for degradation. The rotaviruses (RVs) RNA methyl- and guanylyl transferase (VP3) proteasomal degrade MAVS by the phosphorylation of SPLTSS motif (Ding et al., 2018b). In addition to VP3 of RVs, VP2 of EMCV and the Golgi protein 73 of HCV can interact with MAVS to induce proteasomal degradation (Zhang et al., 2017; Han et al., 2021). PB1-F2 protein of avian influenza A (H7N9) viruses inhibits MAVS aggregation, resulting in the accumulation and degradation of unaggregated MAVS on the mitochondrial membrane (Cheung et al., 2020).
Another “smart” strategy for viral evasion is to sequester MAVS and thus block the antiviral signal transduction through the RIG-I-MAVS pathway. Many non-structural proteins of the virus, such as NS1 and NS2 of respiratory syncytial virus (Ling et al., 2009), NS4A of dengue virus (He et al., 2016), a non-structural protein of unknown function from Andes orthohantavirus (Vera-Otarola et al., 2020), and NS1, NS2B/3, NS4A, NS4B, and NS5, have been shown to block the MAVS signaling transduction platform to inhibit INF production (Wu et al., 2017; Ma et al., 2018; Ding et al., 2018a; Hu et al., 2019; Lundberg et al., 2019; Schilling et al., 2020). The ORF33 of Kaposi’s sarcoma-associated herpesvirus binds to the TM domain of MAVS through the dephosphatase PPM1G, not only inhibiting the phosphorylation of MAVS but also dephosphorylating MAVS to prevent IRF3 recruitment (Yu et al., 2020). Human cytomegalovirus glycoprotein US9 induces the leakage of MAVS into the cytoplasm/ER by disrupting the potential and integrity of the mitochondrial membrane (Choi et al., 2018). Herpes simplex virus 1 tegument protein VP16 dampens the MAVS-Pex signaling and the expression of the immediate-early ISGs (Zheng and Su, 2017). Moreover, tegument protein VP1 of foot-and-mouth disease virus and M2-2 protein of human metapneumovirus block the specific recruitment of TARFs to MAVS (Mossman et al., 2013; Ekanayaka et al., 2020).
SARS-CoV-2 has caused worldwide financial loss and social disruption. Like other positive-sense RNA viruses, SARS-CoV-2 and its counterpart SARS-CoV employ various viral proteins to escape host immune surveillance (Gatti et al., 2020). The accessory protein ORF9b from SARS-CoV has been identified as an IFN antagonist that targets MAVS and promotes its proteasomal degradation, whereas SARS-CoV-2 ORF9b interacts with another mitochondrial protein TOM70 to interfere with the IFN signaling (Shi et al., 2014; Jiang et al., 2020; Kreimendahl and Rassow, 2020; Gordon et al., 2020a,b). Our recent evidence shows that SARS-CoV-2 ORF9b can also target the NF-κB essential modulator NEMO upon viral infection and inhibits its K63-linked ubiquitination, thus disrupting the canonical IKKα/β/γ-NF-κB signaling and subsequent IFN production (Wu et al., 2021). Further study found that MAVS activation can be regulated through the interaction of ORF9c and MAVS signal negative regulators (NLRX1, NDFIP2) or Nsp13 and MAVS effector TBK1 (Gordon et al., 2020b). The membrane glycoprotein M inhibits multiprotein complex formation by interacting with RIG-I, MAVS, and TBK1, and its TM1/2 domains are essential for this inhibitory effect (Zheng et al., 2020; Fu et al., 2021). Another study showed that the Mpro shows similar proteasome functionality as the main protease of SARS-CoV-2 (Wu et al., 2020). Recently, a study identified that SARS-CoV-2 nucleocapsid protein induces innate immune evasion by inhibiting K63-linked poly-ubiquitination and aggregation of MAVS (Wang et al., 2021). These works indicate MAVS plays an important role in the host against SARS-Cov-2 infection.
Although many proteins synthesized by viruses to suppress host antiviral activity have been recognized, ways for viruses to evade the surveillance of the innate immune system remain largely unknown. Recently, a study demonstrated that the Hepatitis B virus activates glycolysis and promotes lactate binding to MAVS to prevent aggregation and mitochondrial localization (Zhou et al., 2021). Another article reported that lactate is a natural suppressor of RLR signaling targeting MAVS (Zhang et al., 2019). These data indicated that a new field of virus immune evade mechanism may have been discovered.
Mavs Immune Homeostasis Maintenance
RIG-I is wrapped by the CTD and maintains a “self-inhibiting” spatial conformation in the resting state. When RIG-I detects a foreign invading RNA virus, its CTD specifically binds to RNA molecular signals to change the spatial conception to release self-inhibition conformation and activates the antiviral immune signaling pathway. Accumulating evidence indicates that the phosphorylation of IRF3 and the production of IFN-β under viral stimulation need MAVS form prion-like aggregates under viral stimulation (Hou et al., 2011). However, MAVS will spontaneously accumulate in its natural state to form active aggregates without stimulating foreign viruses or other factors and activate downstream signaling pathways. The imbalance of antiviral immune response may lead to many autoimmune diseases, such as systemic lupus erythematosus and psoriasis (Pothlichet et al., 2011; Buskiewicz et al., 2016; Zhang et al., 2016). Therefore, in addition to the negative post-translational regulation of MAVS after the viral infection, in the resting state, the cell must have a strict regulation mechanism to prevent the spontaneous aggregation of MAVS and maintain its monomeric form, thereby ensuring the “closed” state of the RIG-I-MAVS signaling pathway (Figure 4).
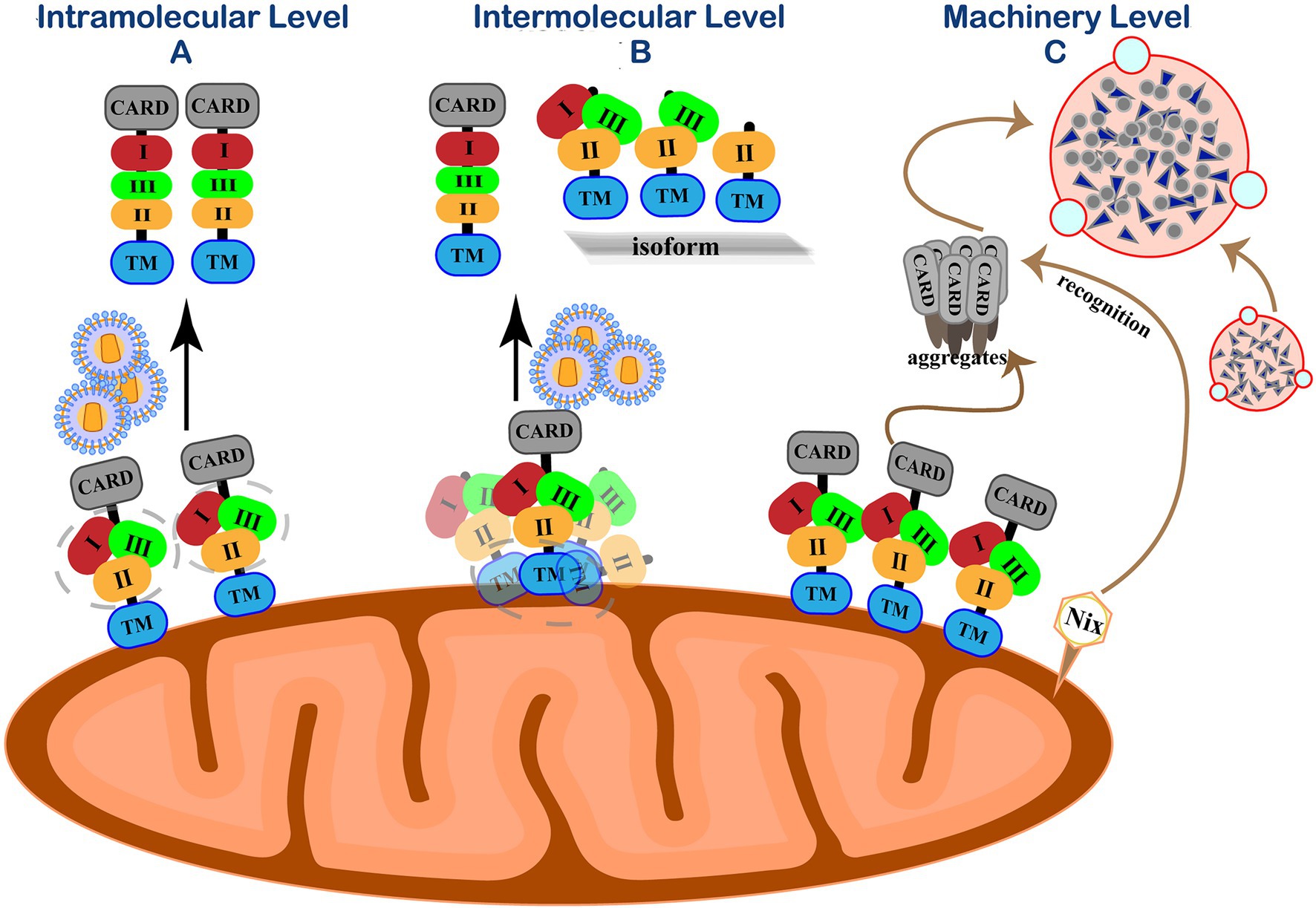
Figure 4. Self-inhibition mechanism of MAVS in resting state. (A) Intramolecular level. Three functional areas of MAVS are wrapped by its adjacent regions to maintain immune homeostasis. (B) Intermolecular level. N-terminally truncated isoforms interact with MAVS by their TM domain homotypic interactions to spatially isolating MAVS monomer molecules from each other on the outer mitochondrial membrane. (C) Machinery level. Eliminate accumulated MAVS through Nix-mediated selective autophagy.
PCBP1 has a certain regulatory effect on MAVS, and it persists in resting cells to prevent MAVS from accumulating (Zhou et al., 2012). RNF115 is related to the MAVS protein regulation in resting cells. RNF115 can continuously catalyze the K48-linked ubiquitination of MAVS and regulate the steady-state MAVS level in uninfected cells (Zhang et al., 2020b). Shi et al. revealed a self-inhibition mechanism that regulates the activity of MAVS in unstimulated cells (Shi et al., 2015). They identified three MAVS active regions with different functions, a region consisting of 401–450 amino acids (region III) for TBK1/IRF3 activation and two MAVS regions responsible for IKK/NF-κB activation (region I aa-138-152 and area II aa-451-465). Similar to the self-inhibition of RIG-I, the three functional areas of MAVS are wrapped by its adjacent regions to avoid the auto-activation of MAVS.
The polycistronic transcript of human MAVS generates a full-length MAVS from ORF1. Unlike full-length MAVS, its ORF2 can generate an N-terminal 141-amino acid truncated isoform that exerts dominant-negative effects on MAVS aggregation and its activity (Brubaker et al., 2014; Minassian et al., 2015). N-terminally truncated isoforms cannot form aggregates by themselves as the N-terminal CARD domain by which MAVS aggregation is mediated. Instead, these isoforms interact with full-length MAVS by their TM domain homotypic interactions, thus spatially isolating MAVS monomer molecules from each other on the outer mitochondrial membrane and preventing their spontaneous aggregation (Qi et al., 2017). In addition to the main ORF1 and ORF2, three short upstream open reading frames (uORFs) exist in the 5′ untranslated mRNA regions (5′UTR) of the MAVS gene transcript mRNA. The uORF can reduce the translation initiation efficiency of downstream genes through various regulatory mechanisms and cause ribosome delay or trigger mRNA degradation, thereby inhibiting protein expression (Xu et al., 2017). Our group demonstrated that uORFs are cis-acting elements of MAVS transcripts that repress the downstream ORF translation by the leaky ribosomal scanning mechanism, limiting MAVS expression and preventing its spontaneous aggregation (Shi et al., 2020).
On the other hand, host cells maintain innate homeostasis by degrading protein aggregates, damaged organelles, or intracellular pathogens through autophagy. Mitophagy is a typical selective autophagy process and is mediated when the mitochondrial kinase PINK1 and E3 ubiquitin ligase Parkin promote mitochondrial outer membrane proteins’ ubiquitination (Lazarou et al., 2015). The formation of MAVS aggregates can increase intracellular ROS (Zhao et al., 2012). After recognizing this signal in resting cells, the mitochondrial protein Nix can directly bind to the ATG8 family member on the phagocytic protein to mediate PINK-PARKIN mitophagy and clear MAVS aggregates (Randow and Youle, 2014). Our group further discovered that in the absence of viral infection, endogenous MAVS produced by internal ORF-deleted transcripts could spontaneously aggregate and activate the IFN signaling pathway (Qi et al., 2017). Nix-mediated selective autophagy is initiated to prevent abnormal protein aggregation and maintain natural immune homeostasis (Shi et al., 2020).
Conclusion and Future Perspectives
As one of the important signaling adaptors, MAVS has attracted extensive attention from researchers for its critical role in the antiviral signaling pathway. MAVS acts as an adaptor in RNA-sensing signaling pathways to induce IFN production by forming prion-like aggregates after receiving upstream signals from PRRs. This linking role and status determine that MAVS is an indispensable ingredient. After viral infection, MAVS regulates the immune response and inflammation levels in the cells through PTMs, such as phosphorylation and ubiquitination. Compelling evidence has shown that viruses have developed various strategies to evade the host’s innate immune surveillance by targeting MAVS. This phenomenon may be a crisis signal, or it may be a chance for a new field. The mitochondria where MAVS is localized are pivotal for energy metabolism and have emerged as vital platforms for MAVS-mediated antiviral signaling. O-linked β-N-acetylglucosamine mentioned in the previous article has proved that glucose metabolism has the MAVS regulation function (Li et al., 2018). Mitochondrial fission factor (Mff) can provide energy for MAVS disorganization. Acute antiviral immunity is mediated by Mff, which senses mitochondrial energy status and regulates the disorganization of MAVS clusters (Hanada et al., 2020). Still, the current research is mostly focused on the regulation of MAVS by mitochondrial dynamics. Whether the mitochondrial metabolic process might contribute to MAVS regulation in host-virus crosstalk is largely unknown and imperative to be elucidated. In summary, studying the mechanisms by which viruses counteract the MAVS-mediated immune response may provide new insights for the therapeutic strategy of viral infections.
As we know, the cGAS-STING pathway plays an important role in anti-tumor immunity. Targeting cGAS-STING has shown great progress in cancer therapy. Researches on the relevance between the RIG-I-MAVS signaling pathway in tumorigenesis and development are also in full swing. Recent reports have shown a significant reduction of MAVS expression in several human cancers and demonstrate its potential anti-tumor prospect (Hou et al., 2014). MAVS interacts with the tumor suppressor P53, maintaining the stability of P53 by inhibiting its ubiquitination and the formation of its P53-MDM2 complex, thereby inhibiting the development of tumors (Zhang et al., 2020a). Otherwise, epigenetic therapy is a novel cancer treatment method. This method can induce virus infection phenomenon to activate the MAVS signaling pathway and target cancer-initiating cells, resulting in a “viral mimicry” state with anti-tumor effects (Roulois et al., 2015). This work indicated that more roles of MAVS in tumorigenesis would be needed to reveal in future work.
Accidental and continuous activation of the immune system may lead to chronic inflammation and even autoimmune diseases. In recent years, the self-inhibition mechanism of how the immune system prevents the spontaneous aggregation of MAVS has gradually been clarified. Our knowledge of the regulatory mechanisms of immune homeostasis will remarkably improve our understanding of the immune system and provide new clues to the pathogenesis of chronic inflammation, autoimmune diseases, and cancer. Although the current investigations on the regulation of MAVS immune homeostasis are still limited, the advance of the field will undoubtedly benefit innate immunity studies. Based on these breakthroughs, this field is still worthy of further investigation.
Author Contributions
NQ and YC conceived and wrote the first draft of the manuscript. YS provided substantial comments on the manuscript that were incorporated. JW depicted the illustrations. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2018YFA0900404), National Natural Science Foundation of China (31870864), Fundamental Research Funds for the Provincial Universities of Zhejiang (RF-B2020003), and Zhejiang Provincial Natural Science Foundation of China (LY20C080003). All sources of funding received for the research being submitted.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arimoto, K., Takahashi, H., Hishiki, T., Konishi, H., Fujita, T., and Shimotohno, K. (2007). Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. U. S. A. 104, 7500–7505. doi: 10.1073/pnas.0611551104
Boeynaems, S., and Gitler, A. D. (2018). Phosphorylation leads the way for protein aggregate disassembly. Dev. Cell 45, 279–281. doi: 10.1016/j.devcel.2018.04.017
Broz, P., and Monack, D. M. (2013). Newly described pattern recognition receptors team up against intracellular pathogens. Nat. Rev. Immunol. 13, 551–565. doi: 10.1038/nri3479
Brubaker, S. W., Gauthier, A. E., Mills, E. W., Ingolia, N. T., and Kagan, J. C. (2014). A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell 156, 800–811. doi: 10.1016/j.cell.2014.01.021
Buskiewicz, I. A., Montgomery, T., Yasewicz, E. C., Huber, S. A., Murphy, M. P., Hartley, R. C., et al. (2016). Reactive oxygen species induce virus-independent MAVS oligomerization in systemic lupus erythematosus. Sci. Signal. 9:ra115. doi: 10.1126/scisignal.aaf1933
Cao, X. (2016). Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 16, 35–50. doi: 10.1038/nri.2015.8
Castanier, C., Zemirli, N., Portier, A., Garcin, D., Bidere, N., Vazquez, A., et al. (2012). MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol. 10:44. doi: 10.1186/1741-7007-10-44
Chen, Z., Benureau, Y., Rijnbrand, R., Yi, J., Wang, T., Warter, L., et al. (2007). GB virus B disrupts RIG-I signaling by NS3/4A-mediated cleavage of the adaptor protein MAVS. J. Virol. 81, 964–976. doi: 10.1128/JVI.02076-06
Cheng, J., Liao, Y., Xiao, L., Wu, R., Zhao, S., Chen, H., et al. (2016). Autophagy regulates MAVS signaling activation in a phosphorylation-dependent manner in microglia. Cell Death Differ. 24, 276–287. doi: 10.1038/cdd.2016.121
Cheung, P. H., Lee, T. T., Kew, C., Chen, H., Yuen, K. Y., Chan, C. P., et al. (2020). Virus subtype-specific suppression of MAVS aggregation and activation by PB1-F2 protein of influenza A (H7N9) virus. PLoS Pathog. 16:e1008611. doi: 10.1371/journal.ppat.1008611
Choi, H. J., Park, A., Kang, S., Lee, E., Lee, T. A., Ra, E. A., et al. (2018). Human cytomegalovirus-encoded US9 targets MAVS and STING signaling to evade type I interferon immune responses. Nat. Commun. 9:125. doi: 10.1038/s41467-017-02624-8
Choi, Y. B., Shembade, N., Parvatiyar, K., Balachandran, S., and Harhaj, E. W. (2017). TAX1BP1 Restrains Virus-Induced Apoptosis by Facilitating Itch-Mediated Degradation of the Mitochondrial Adaptor MAVS. Mol. Cell. Biol. 37:e00422-16. doi: 10.1128/mcb.00422-16
Coyne, C. B., Wang, B., Xi, X., Lei, X., Zhang, X., Cui, S., et al. (2013). Enterovirus 71 Protease 2Apro Targets MAVS to Inhibit Antiviral Type I Interferon Responses. PLoS Pathog. 9:e1003231. doi: 10.1371/journal.ppat.1003231
Dai, T., Wu, L., Wang, S., Wang, J., Xie, F., Zhang, Z., et al. (2018). FAF1 Regulates Antiviral Immunity by Inhibiting MAVS but Is Antagonized by Phosphorylation upon Viral Infection. Cell Host Microbe 24, 776.e5–790.e5. doi: 10.1016/j.chom.2018.10.006
de Brito, O. M., and Scorrano, L. (2008). Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610. doi: 10.1038/nature07534
Deribe, Y. L., Pawson, T., and Dikic, I. (2010). Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17, 666–672. doi: 10.1038/nsmb.1842
Ding, Q., Gaska, J. M., Douam, F., Wei, L., Kim, D., Balev, M., et al. (2018a). Species-specific disruption of STING-dependent antiviral cellular defenses by the Zika virus NS2B3 protease. Proc. Natl. Acad. Sci. U. S. A. 115, E6310–E6318. doi: 10.1073/pnas.1803406115
Ding, S., Zhu, S., Ren, L., Feng, N., Song, Y., Ge, X., et al. (2018b). Rotavirus VP3 targets MAVS for degradation to inhibit type III interferon expression in intestinal epithelial cells. Elife 7:e39494. doi: 10.7554/eLife.39494
Diskin, C., Ryan, T. A. J., and O'Neill, L. A. J. (2021). Modification of Proteins by Metabolites in immunity. Immunity 54, 19–31. doi: 10.1016/j.immuni.2020.09.014
Du, J., Zhang, D., Zhang, W., Ouyang, G., Wang, J., Liu, X., et al. (2015). pVHL Negatively Regulates Antiviral Signaling by Targeting MAVS for Proteasomal Degradation. J. Immunol. 195, 1782–1790. doi: 10.4049/jimmunol.1500588
Ekanayaka, P., Lee, S. Y., Herath, T. U. B., Kim, J. H., Kim, T. H., Lee, H., et al. (2020). Foot-and-mouth disease virus VP1 target the MAVS to inhibit type-I interferon signaling and VP1 E83K mutation results in virus attenuation. PLoS Pathog. 16:e1009057. doi: 10.1371/journal.ppat.1009057
Feng, Q., Langereis, M. A., Lork, M., Nguyen, M., Hato, S. V., Lanke, K., et al. (2014). Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J. Virol. 88, 3369–3378. doi: 10.1128/JVI.02712-13
Feng, H., Sander, A.-L., Moreira-Soto, A., Yamane, D., Drexler, J. F., and Lemon, S. M. (2019). Hepatovirus 3ABC proteases and evolution of mitochondrial antiviral signaling protein (MAVS). J. Hepatol. 71, 25–34. doi: 10.1016/j.jhep.2019.02.020
Fu, Y. Z., Wang, S. Y., Zheng, Z. Q., Yi, H., Li, W. W., Xu, Z. S., et al. (2021). SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell. Mol. Immunol. 18, 613–620. doi: 10.1038/s41423-020-00571-x
Gatti, P., Ilamathi, H. S., Todkar, K., and Germain, M. (2020). Mitochondria Targeted Viral Replication and Survival Strategies—Prospective on SARS-CoV-2. Front. Pharmacol. 11:578599. doi: 10.3389/fphar.2020.578599
Gordon, D. E., Hiatt, J., Bouhaddou, M., Rezelj, V. V., Ulferts, S., Braberg, H., et al. (2020a). Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 370:eabe9403. doi: 10.1126/science.abe9403
Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., et al. (2020b). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. doi: 10.1038/s41586-020-2286-9
Han, Y., Xie, J., Xu, S., Bi, Y., Li, X., Zhang, H., et al. (2021). Encephalomyocarditis virus abrogates the interferon beta signaling pathway via its structural protein vp2. J. Virol. 95:e01590-20. doi: 10.1128/JVI.01590-20
Hanada, Y., Ishihara, N., Wang, L., Otera, H., Ishihara, T., Koshiba, T., et al. (2020). MAVS is energized by Mff which senses mitochondrial metabolism via AMPK for acute antiviral immunity. Nat. Commun. 11:5711. doi: 10.1038/s41467-020-19287-7
He, Z., Zhu, X., Wen, W., Yuan, J., Hu, Y., Chen, J., et al. (2016). Dengue Virus Subverts Host Innate Immunity by Targeting Adaptor Protein MAVS. J. Virol. 90, 7219–7230. doi: 10.1128/JVI.00221-16
He, X., Zhu, Y., Zhang, Y., Geng, Y., Gong, J., Geng, J., et al. (2019). RNF34 functions in immunity and selective mitophagy by targeting MAVS for autophagic degradation. EMBO J. 38:e100978. doi: 10.15252/embj.2018100978
Heise, M. T., Zhang, L., Liu, J., Qian, L., Feng, Q., Wang, X., et al. (2018). Induction of OTUD1 by RNA viruses potently inhibits innate immune responses by promoting degradation of the MAVS/TRAF3/TRAF6 signalosome. PLoS Pathog. 14:e1007067. doi: 10.1371/journal.ppat.1007067
Hiscott, J., Nakhaei, P., Paz, S., Vilasco, M., Goubau, D., Xu, Z.-Y., et al. (2010). CS2-1 RIG-I like receptors: sensing and responding to RNA virus infection. Cytokine 52:7. doi: 10.1016/j.cyto.2010.07.032
Hornung, V., Ellegast, J., Kim, S., Brzozka, K., Jung, A., Kato, H., et al. (2006). 5′-triphosphate RNA is the ligand for RIG-I. Science 314, 994–997. doi: 10.1126/science.1132505
Hou, F., Sun, L., Zheng, H., Skaug, B., Jiang, Q. X., and Chen, Z. J. (2011). MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146, 448–461. doi: 10.1016/j.cell.2011.06.041
Hou, J., Zhou, Y., Zheng, Y., Fan, J., Zhou, W., Ng, I. O., et al. (2014). Hepatic RIG-I predicts survival and interferon-alpha therapeutic response in hepatocellular carcinoma. Cancer Cell 25, 49–63. doi: 10.1016/j.ccr.2013.11.011
Hu, Y., Dong, X., He, Z., Wu, Y., Zhang, S., Lin, J., et al. (2019). Zika virus antagonizes interferon response in patients and disrupts RIG-I–MAVS interaction through its CARD-TM domains. Cell Biosci. 9:46. doi: 10.1186/s13578-019-0308-9
Jackson, W. T., Visser, L. J., Aloise, C., Swatek, K. N., Medina, G. N., Olek, K. M., et al. (2020). Dissecting distinct proteolytic activities of FMDV Lpro implicates cleavage and degradation of RLR signaling proteins, not its deISGylase/DUB activity, in type I interferon suppression. PLoS Pathog. 16:e1008702. doi: 10.1371/journal.ppat.1008702
Jiang, H. W., Zhang, H. N., Meng, Q. F., Xie, J., Li, Y., Chen, H., et al. (2020). SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 17, 998–1000. doi: 10.1038/s41423-020-0514-8
Jin, S., and Cui, J. (2018). BST2 inhibits type I IFN (interferon) signaling by accelerating MAVS degradation through CALCOCO2-directed autophagy. Autophagy 14, 171–172. doi: 10.1080/15548627.2017.1393590
Jin, S., Tian, S., Luo, M., Xie, W., Liu, T., Duan, T., et al. (2017). Tetherin Suppresses Type I Interferon Signaling by Targeting MAVS for NDP52-Mediated Selective Autophagic Degradation in Human Cells. Mol. Cell 68, 308.e4–322.e4. doi: 10.1016/j.molcel.2017.09.005
Katze, M. G., Fornek, J. L., Palermo, R. E., Walters, K. A., and Korth, M. J. (2008). Innate immune modulation by RNA viruses: emerging insights from functional genomics. Nat. Rev. Immunol. 8, 644–654. doi: 10.1038/nri2377
Kawai, T., and Akira, S. (2010). The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat. Immunol. 11, 373–384. doi: 10.1038/ni.1863
Kawai, T., Takahashi, K., Sato, S., Coban, C., Kumar, H., Kato, H., et al. (2005). IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6, 981–988. doi: 10.1038/ni1243
Kreimendahl, S., and Rassow, J. (2020). The Mitochondrial Outer Membrane Protein Tom70-Mediator in Protein Traffic, Membrane Contact Sites and Innate Immunity. Int. J. Mol. Sci. 21:7262. doi: 10.3390/ijms21197262
Lazarou, M., Sliter, D. A., Kane, L. A., Sarraf, S. A., Wang, C., Burman, J. L., et al. (2015). The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314. doi: 10.1038/nature14893
Li, T., Li, X., Attri, K. S., Liu, C., Li, L., Herring, L. E., et al. (2018). O-GlcNAc Transferase links glucose metabolism to MAVS-Mediated Antiviral Innate Immunity. Cell Host Microbe 24, 791.e6–803.e6. doi: 10.1016/j.chom.2018.11.001
Li, W., Li, N., Dai, S., Hou, G., Guo, K., Chen, X., et al. (2019b). Zika virus circumvents host innate immunity by targeting the adaptor proteins MAVS and MITA. FASEB J. 33, 9929–9944. doi: 10.1096/fj.201900260R
Li, S.-Z., Shu, Q.-P., Song, Y., Zhang, H.-H., Liu, Y., Jin, B.-X., et al. (2019a). Phosphorylation of MAVS/VISA by Nemo-like kinase (NLK) for degradation regulates the antiviral innate immune response. Nat. Commun. 10:3233. doi: 10.1038/s41467-019-11258-x
Ling, Z., Tran, K. C., and Teng, M. N. (2009). Human respiratory syncytial virus nonstructural protein ns2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J. Virol. 83, 3734–3742. doi: 10.1128/JVI.02434-08
Liu, S., Cai, X., Wu, J., Cong, Q., Chen, X., Li, T., et al. (2015). Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347:aaa2630. doi: 10.1126/science.aaa2630
Liu, W., Li, J., Zheng, W., Shang, Y., Zhao, Z., Wang, S., et al. (2017b). Cyclophilin A-regulated ubiquitination is critical for RIG-I-mediated antiviral immune responses. Elife 6:e24425. doi: 10.7554/eLife.24425
Liu, J., Qian, C., and Cao, X. (2016). Post-translational modification control of innate immunity. Immunity 45, 15–30. doi: 10.1016/j.immuni.2016.06.020
Liu, B., Zhang, M., Chu, H., Zhang, H., Wu, H., Song, G., et al. (2017a). The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat. Immunol. 18, 214–224. doi: 10.1038/ni.3641
Liu, X., Zhu, C., Zha, H., Tang, J., Rong, F., Chen, X., et al. (2020). SIRT5 impairs aggregation and activation of the signaling adaptor MAVS through catalyzing lysine desuccinylation. EMBO J. 39:e103285. doi: 10.15252/embj.2019103285
Liuyu, T., Yu, K., Ye, L., Zhang, Z., Zhang, M., Ren, Y., et al. (2018). Induction of OTUD4 by viral infection promotes antiviral responses through deubiquitinating and stabilizing MAVS. Cell Res. 29, 67–79. doi: 10.1038/s41422-018-0107-6
Lundberg, R., Melén, K., Westenius, V., Jiang, M., Österlund, P., Khan, H., et al. (2019). Zika virus non-structural protein NS5 inhibits the RIG-I pathway and interferon lambda 1 promoter activation by targeting IKK epsilon. Viruses 11:1024. doi: 10.3390/v11111024
Ma, J., Ketkar, H., Geng, T., Lo, E., Wang, L., Xi, J., et al. (2018). Zika Virus Non-structural Protein 4A Blocks the RLR-MAVS Signaling. Front. Microbiol. 9:1350. doi: 10.3389/fmicb.2018.01350
Meylan, E., Curran, J., Hofmann, K., Moradpour, D., Binder, M., Bartenschlager, R., et al. (2005). Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172. doi: 10.1038/nature04193
Minassian, A., Zhang, J., He, S., Zhao, J., Zandi, E., Saito, T., et al. (2015). An internally Translated MAVS Variant Exposes Its Amino-terminal TRAF-Binding Motifs to Deregulate Interferon Induction. PLoS Pathog. 11:e1005060. doi: 10.1371/journal.ppat.1005060
Mossman, K. L., Bao, X., Kolli, D., Ren, J., Liu, T., Garofalo, R. P., et al. (2013). Human Metapneumovirus Glycoprotein G Disrupts Mitochondrial Signaling in Airway Epithelial Cells. PLoS One 8:e62568. doi: 10.1371/journal.pone.0062568
Onoguchi, K., Onomoto, K., Takamatsu, S., Jogi, M., Takemura, A., Morimoto, S., et al. (2010). Virus-infection or 5'ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog. 6:e1001012. doi: 10.1371/journal.ppat.1001012
Pan, Y., Li, R., Meng, J. L., Mao, H. T., Zhang, Y., and Zhang, J. (2014). Smurf2 negatively modulates RIG-I-dependent antiviral response by targeting VISA/MAVS for ubiquitination and degradation. J. Immunol. 192, 4758–4764. doi: 10.4049/jimmunol.1302632
Pang, L. L., Yuan, X. H., Shao, C. S., Li, M. Z., Wang, Y., Wang, H. M., et al. (2017). The suppression of innate immune response by human rhinovirus C. Biochem. Biophys. Res. Commun. 490, 22–28. doi: 10.1016/j.bbrc.2017.05.169
Park, Y. J., Oanh, N. T. K., Heo, J., Kim, S. G., Lee, H. S., Lee, H., et al. (2020). Dual targeting of RIG-I and MAVS by MARCH5 mitochondria ubiquitin ligase in innate immunity. Cell. Signal. 67:109520. doi: 10.1016/j.cellsig.2019.109520
Pothlichet, J., Niewold, T. B., Vitour, D., Solhonne, B., Crow, M. K., and Si-Tahar, M. (2011). A loss-of-function variant of the antiviral molecule MAVS is associated with a subset of systemic lupus patients. EMBO Mol. Med. 3, 142–152. doi: 10.1002/emmm.201000120
Qi, N., Shi, Y., Zhang, R., Zhu, W., Yuan, B., Li, X., et al. (2017). Multiple Truncated isoforms of MAVS prevent its spontaneous aggregation in antiviral innate immune signalling. Nat. Commun. 8:15676. doi: 10.1038/ncomms15676
Qin, Y., Xue, B., Liu, C., Wang, X., Tian, R., Xie, Q., et al. (2017). NLRX1 mediates MAVS degradation to attenuate the hepatitis C Virus-induced innate immune response through PCBP2. J. Virol. 91:e01264-17. doi: 10.1128/JVI.01264-17
Randow, F., and Youle, R. J. (2014). Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe 15, 403–411. doi: 10.1016/j.chom.2014.03.012
Roulois, D., Loo Yau, H., Singhania, R., Wang, Y., Danesh, A., Shen, S. Y., et al. (2015). DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973. doi: 10.1016/j.cell.2015.07.056
Schilling, M., Bridgeman, A., Gray, N., Hertzog, J., Hublitz, P., Kohl, A., et al. (2020). RIG-I plays a dominant role in the induction of transcriptional changes in zika virus-infected cells, which Protect from virus-induced cell death. Cell 9:1476. doi: 10.3390/cells9061476
Schneider, U., Martin, A., Schwemmle, M., and Staeheli, P. (2007). Genome trimming by Borna disease viruses: viral replication control or escape from cellular surveillance? Cell. Mol. Life Sci. 64, 1038–1042. doi: 10.1007/s00018-007-6545-9
Seth, R. B., Sun, L., Ea, C. K., and Chen, Z. J. (2005). Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122, 669–682. doi: 10.1016/j.cell.2005.08.012
Shi, C. S., Qi, H. Y., Boularan, C., Huang, N. N., Abu-Asab, M., Shelhamer, J. H., et al. (2014). SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 193, 3080–3089. doi: 10.4049/jimmunol.1303196
Shi, Y., Wu, J., Zhong, T., Zhu, W., She, G., Tang, H., et al. (2020). Upstream ORFs Prevent MAVS Spontaneous Aggregation and Regulate Innate Immune Homeostasis. iScience 23:101059. doi: 10.1016/j.isci.2020.101059
Shi, Y., Yuan, B., Qi, N., Zhu, W., Su, J., Li, X., et al. (2015). An autoinhibitory mechanism modulates MAVS activity in antiviral innate immune response. Nat. Commun. 6:7811. doi: 10.1038/ncomms8811
Song, N., Qi, Q., Cao, R., Qin, B., Wang, B., Wang, Y., et al. (2019). MAVS O-GlcNAcylation Is Essential for Host Antiviral Immunity against Lethal RNA Viruses. Cell Rep. 28, 2386.e5–2396.e5. doi: 10.1016/j.celrep.2019.07.085
Strowitzki, M. J., Cummins, E. P., and Taylor, C. T. (2019). Protein Hydroxylation by Hypoxia-Inducible Factor (HIF) Hydroxylases: Unique or Ubiquitous? Cell 8:384. doi: 10.3390/cells8050384
Thompson, A. J., and Locarnini, S. A. (2007). Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol. Cell Biol. 85, 435–445. doi: 10.1038/sj.icb.7100100
Vera-Otarola, J., Solis, L., Lowy, F., Olguín, V., Angulo, J., Pino, K., et al. (2020). The Andes Orthohantavirus NSs protein antagonizes the type I interferon response by inhibiting MAVS signaling. J. Virol. 94:e00454-20. doi: 10.1128/JVI.00454-20
Vitour, D., Dabo, S., Ahmadi Pour, M., Vilasco, M., Vidalain, P.-O., Jacob, Y., et al. (2009). Polo-like Kinase 1 (PLK1) Regulates Interferon (IFN) Induction by MAVS. J. Biol. Chem. 284, 21797–21809. doi: 10.1074/jbc.M109.018275
Wang, S., Dai, T., Qin, Z., Pan, T., Chu, F., Lou, L., et al. (2021). Targeting liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat. Cell Biol. 23, 718–732. doi: 10.1038/s41556-021-00710-0
Wang, Y., Tong, X., and Ye, X. (2012). Ndfip1 Negatively Regulates RIG-I–Dependent Immune Signaling by Enhancing E3 Ligase Smurf1-Mediated MAVS Degradation. J. Immunol. 189, 5304–5313. doi: 10.4049/jimmunol.1201445
Wei, C., Ni, C., Song, T., Liu, Y., Yang, X., Zheng, Z., et al. (2010). The hepatitis B virus X Protein Disrupts Innate Immunity by Downregulating Mitochondrial Antiviral Signaling Protein. J. Immunol. 185, 1158–1168. doi: 10.4049/jimmunol.0903874
Wu, Y., Liu, Q., Zhou, J., Xie, W., Chen, C., Wang, Z., et al. (2017). Erratum: Zika virus evades interferon-mediated antiviral response through the co-operation of multiple non-structural proteins in vitro. Cell Discov. 3:17014. doi: 10.1038/celldisc.2017.14
Wu, Y., Ma, L., Zhuang, Z., Cai, S., Zhao, Z., Zhou, L., et al. (2020). Main protease of SARS-CoV-2 serves as a bifunctional molecule in restricting type I interferon antiviral signaling. Signal Transduct. Target. Ther. 5:221. doi: 10.1038/s41392-020-00332-2
Wu, J., Shi, Y., Pan, X., Wu, S., Hou, R., Zhang, Y., et al. (2021). SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 34:108761. doi: 10.1016/j.celrep.2021.108761
Xia, P., Wang, S., Xiong, Z., Ye, B., Huang, L. Y., Han, Z. G., et al. (2015). IRTKS negatively regulates antiviral immunity through PCBP2 sumoylation-mediated MAVS degradation. Nat. Commun. 6:8132. doi: 10.1038/ncomms9132
Xia, Z., Xu, G., Nie, L., Liu, L., Peng, N., He, Q., et al. (2019). NAC1 Potentiates Cellular Antiviral Signaling by Bridging MAVS and TBK1. J. Immunol. 203, 1001–1011. doi: 10.4049/jimmunol.1801110
Xiang, W., Zhang, Q., Lin, X., Wu, S., Zhou, Y., Meng, F., et al. (2016). PPM1A silences cytosolic RNA sensing and antiviral defense through direct dephosphorylation of MAVS and TBK1. Sci. Adv. 2:e1501889. doi: 10.1126/sciadv.1501889
Xu, L. G., Wang, Y. Y., Han, K. J., Li, L. Y., Zhai, Z., and Shu, H. B. (2005). VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19, 727–740. doi: 10.1016/j.molcel.2005.08.014
Xu, L., Yu, D., Yao, Y. L., Gu, T., Zheng, X., Wu, Y., et al. (2020). Tupaia MAVS Is a Dual Target during Hepatitis C Virus Infection for Innate Immune Evasion and Viral Replication via NF-kappaB. J. Immunol. 205, 2091–2099. doi: 10.4049/jimmunol.2000376
Xu, G., Yuan, M., Ai, C., Liu, L., Zhuang, E., Karapetyan, S., et al. (2017). uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545, 491–494. doi: 10.1038/nature22372
Xue, B., Li, H., Guo, M., Wang, J., Xu, Y., Zou, X., et al. (2018). TRIM21 promotes innate immune response to RNA viral infection through Lys27-linked polyubiquitination of MAVS. J. Virol. 92:e00321-18. doi: 10.1128/JVI.00321-18
Yang, Y., and Gibson, G. E. (2019). Succinylation links metabolism to protein functions. Neurochem. Res. 44, 2346–2359. doi: 10.1007/s11064-019-02780-x
Yang, Y., He, Y., Wang, X., Liang, Z., He, G., Zhang, P., et al. (2017). Protein SUMOylation modification and its associations with disease. Open Biol. 7:170167. doi: 10.1098/rsob.170167
Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., et al. (2004). The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737. doi: 10.1038/ni1087
Yoo, Y. S., Park, Y. Y., Kim, J. H., Cho, H., Kim, S. H., Lee, H. S., et al. (2015). The mitochondrial ubiquitin ligase MARCH5 resolves MAVS aggregates during antiviral signalling. Nat. Commun. 6:7910. doi: 10.1038/ncomms8910
You, F., Sun, H., Zhou, X., Sun, W., Liang, S., Zhai, Z., et al. (2009). PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat. Immunol. 10, 1300–1308. doi: 10.1038/ni.1815
Yu, K., Tian, H., and Deng, H. (2020). PPM1G restricts innate immune signaling mediated by STING and MAVS and is hijacked by KSHV for immune evasion. Sci. Adv. 6:eabd0276. doi: 10.1126/sciadv.abd0276
Zhang, Z., Fang, X., Wu, X., Ling, L., Chu, F., Li, J., et al. (2020c). Acetylation-Dependent Deubiquitinase OTUD3 Controls MAVS Activation in innate Antiviral Immunity. Mol. Cell 79, 304.e7–319.e7. doi: 10.1016/j.molcel.2020.06.020
Zhang, W., Gong, J., Yang, H., Wan, L., Peng, Y., Wang, X., et al. (2020a). The Mitochondrial Protein MAVS Stabilizes p53 to suppress tumorigenesis. Cell Rep. 30, 725.e4–738.e4. doi: 10.1016/j.celrep.2019.12.051
Zhang, L. J., Sen, G. L., Ward, N. L., Johnston, A., Chun, K., Chen, Y., et al. (2016). Antimicrobial Peptide LL37 and MAVS Signaling Drive Interferon-β Production by Epidermal Keratinocytes during Skin Injury. Immunity 45, 119–130. doi: 10.1016/j.immuni.2016.06.021
Zhang, W., Wang, G., Xu, Z.-G., Tu, H., Hu, F., Dai, J., et al. (2019). Lactate is a natural suppressor of RLR signaling by targeting MAVS. Cell 178, 176.e15–189.e15. doi: 10.1016/j.cell.2019.05.003
Zhang, Z.-D., Xiong, T.-C., Yao, S.-Q., Wei, M.-C., Chen, M., Lin, D., et al. (2020b). RNF115 plays dual roles in innate antiviral responses by catalyzing distinct ubiquitination of MAVS and MITA. Nat. Commun. 11:5536. doi: 10.1038/s41467-020-19318-3
Zhang, X., Zhu, C., Wang, T., Jiang, H., Ren, Y., Zhang, Q., et al. (2017). GP73 represses host innate immune response to promote virus replication by facilitating MAVS and TRAF6 degradation. PLoS Pathog. 13:e1006321. doi: 10.1371/journal.ppat.1006321
Zhao, Y., Sun, X., Nie, X., Sun, L., Tang, T. S., Chen, D., et al. (2012). COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. PLoS Pathog. 8:e1003086. doi: 10.1371/journal.ppat.1003086
Zheng, C. (2021). The emerging roles of the MARCH ligases in antiviral innate immunity. Int. J. Biol. Macromol. 171, 423–427. doi: 10.1016/j.ijbiomac.2020.12.207
Zheng, C., and Su, C. (2017). Herpes simplex virus 1 infection dampens the immediate early antiviral innate immunity signaling from peroxisomes by tegument protein VP16. Virol. J. 14:35. doi: 10.1186/s12985-017-0709-5
Zheng, Y., Zhuang, M. W., Han, L., Zhang, J., Nan, M. L., Zhan, P., et al. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct. Target. Ther. 5:299. doi: 10.1038/s41392-020-00438-7
Zhong, B., Zhang, Y., Tan, B., Liu, T. T., Wang, Y. Y., and Shu, H. B. (2010). The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J. Immunol. 184, 6249–6255. doi: 10.4049/jimmunol.0903748
Zhou, L., He, R., Fang, P., Li, M., Yu, H., Wang, Q., et al. (2021). Hepatitis B virus rigs the cellular metabolome to avoid innate immune recognition. Nat. Commun. 12:98. doi: 10.1038/s41467-020-20316-8
Keywords: antiviral signal transduction, immune homeostasis, mitochondrial antiviral signaling protein, post-translational modification, viral evasion
Citation: Chen Y, Shi Y, Wu J and Qi N (2021) MAVS: A Two-Sided CARD Mediating Antiviral Innate Immune Signaling and Regulating Immune Homeostasis. Front. Microbiol. 12:744348. doi: 10.3389/fmicb.2021.744348
Edited by:
Chunfu Zheng, University of Calgary, CanadaReviewed by:
Wei Xiong, Houston Methodist Research Institute, United StatesHongjie Xia, University of Texas Medical Branch at Galveston, United States
Copyright © 2021 Chen, Shi, Wu and Qi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Qi, cWluYW5Aemp1dC5lZHUuY24=orcid.org/0000-0002-1423-7709
†These authors have contributed equally to this work
 Yunqiang Chen
Yunqiang Chen Yuheng Shi2†
Yuheng Shi2† Nan Qi
Nan Qi