- 1National Engineering Laboratory for Efficient Utilization of Soil and Fertilizer Resources, College of Resources and Environment of Shandong Agricultural University, Taian, China
- 2Marine Agriculture Research Center, Tobacco Research Institute of Chinese Academy of Agricultural Sciences, Qingdao, China
- 3Oak Ridge National Laboratory, Biosciences Division, Oak Ridge, TN, United States
- 4Key Laboratory of Coastal Environmental Processes and Ecological Remediation, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
Bacterial signal transduction pathways are important for a variety of adaptive responses to environment, such as two-component systems (TCSs). In this paper, we reported the characterization of a transcriptional regulator in Azorhizobium caulinodans ORS571, ActR, with an N-terminal receiver domain and one C-terminal OmpR/PhoB-type DNA binding domain. Sequence analysis showed that ActR shared a high similarity with FtcR regulator of Brucella melitensis 16M known to be involved in flagellar regulation. The structural gene of this regulator was largely distributed in Alphaproteobacteria, in particular in Rhizobiales and Rhodobacterales, and was located within clusters of genes related to motility functions. Furthermore, we studied the biological function of ActR in A. caulinodans grown at the free-living state or in association with Sesbania rostrata by constructing actR gene deletion mutant. In the free-living state, the bacterial flagellum and motility ability were entirely deleted, the expression of flagellar genes was downregulated; and the exopolysaccharide production, biofilm formation, and cell flocculation decreased significantly compared with those of the wild-type strain. In the symbiotic state, ΔactR mutant strain showed weakly competitive colonization and nodulation on the host plant. These results illustrated that FtcR-like regulator in A. caulinodans is involved in flagellar biosynthesis and provide bacteria with an effective competitive nodulation for symbiosis. These findings improved our knowledge of FtcR-like transcriptional regulator in A. caulinodans.
Introduction
Legumes obtain nitrogen through rhizobia residing in root nodules. Sesbania rostrata, one of the most valuable green manure legumes, could grow rapidly in waterlogged conditions and represent a high rate of nitrogen fixation. Azorhizobium caulinodans establishes a specific symbiosis with S. rostrata, which specially induces effective nodules on the stems of S. rostrata besides root nodulation. It also could infect cereal crop wheat (Triticum aestivum L.) and form para-nodules to provide 16–23% nitrogen for host wheat (Kennedy et al., 1997). What makes it more unique is that A. caulinodans could fix N2 in the free-living state (outside of the nodule) (Dreyfus et al., 1983). The wide adaptability and applicability in agriculture of A. caulinodans are reasons for its great attractions.
Two-component systems (TCSs) are the main paradigm for bacterial signal transduction, which are responsible for sensing and responding to a variety of physical and chemical signals (Mitrophanov and Groisman, 2008). A well-known example is one of chemotactic TCSs, CheA/Y. In A. caulinodans ORS571, the chemotaxis ability of cheY deletion mutant was defective, and its colonization on host plant was impaired (Liu et al., 2020a). TCS NtrX/Y was identified in A. caulinodans, which was responsible for nitrogen metabolism in free-living state and nodulation in symbiosis (Pawlowski et al., 1991). TCSs also play a crucial role in the process of flagellar synthesis and motility. For example, the TCS FleS/R was required for transcription of 20 or more flagellar biosynthetic genes in Pseudomonas aeruginosa (Dasgupta et al., 2003). In the monoflagellate Shewanella oneidensis, the TCS FlrB/C was related to regulating the expression of flaA and flaB genes related to flagellar motility (Shi et al., 2014). Bacterial flagella are complex rotary engines like a propeller embedded in the cell envelope, which participate in a variety of processes, including motility, biofilm formation (Pratt and Kolter, 2010), colonization (Shen et al., 2018), and cell invasion (Horstmann et al., 2020).
Rhizobia manifest a range of behaviors leading to symbiotic association with host plants, which mainly consist of chemotaxis and motility, adhesion, colonization, infection, and nodulation. For the mechanisms of symbiosis, we have always focused on the effect of chemotaxis and flagellar motility on symbiosis, because they are the first and key steps in establishing plant–microbe interaction (Brencic and Winans, 2005; Jiang et al., 2016; Yang et al., 2017; Liu W. et al., 2018; Shen et al., 2018; Liu et al., 2019, 2020a). The flagellar motor proteins FliM and FliN in A. caulinodans were involved in flagellum synthesis, bacterial motility, and biofilm formation (Shen et al., 2018). The genome of A. caulinodans contains the che (chemotaxis) gene cluster (cheA, cheW, cheY1, cheB, and cheR); and their relevant mutants were detected for defection in competitive colonization and nodulation with S. rostrata (Liu W. et al., 2018). Latterly, it was found that cheZ-like gene of A. caulinodans was a key gene in influencing root colonization by regulating exopolysaccharide (EPS) production (Liu X. et al., 2018; Liu et al., 2019). Besides cheY1 (in the che gene cluster), A. caulinodans possesses another gene encoding CheY protein, cheY2 (outside the che gene cluster). Both of them were involved in chemotaxis, while they had different regulatory mechanisms, and CheY2 had a more remarkable role (Liu et al., 2020a). The whole-genome sequence of A. caulinodans has been obtained (Lee et al., 2008), AZC_0619 is closely located upstream of cheY2 and cheZ, and it is a homolog of ftcR gene of Brucella melitensis 16M. FtcR, a two-component response regulator, regulated flagellar gene expression (Léonard et al., 2007). The biological functions of the ftcR in B. melitensis raise the questions of whether ftcR-like gene in A. caulinodans exhibits similar regulation in flagellar genes expression or has specific symbiotic properties.
In this study, the main aim was to elucidate the biological functions of ActR in A. caulinodans ORS571 under free-living and symbiotic conditions. We found that ActR protein regulated motility, EPS production, biofilm formation, cell flocculation, root colonization, and competitive nodulation on the stems of S. rostrata. This study provides important insights on the two-component regulatory FtcR-like protein in bacterial motility and symbiosis of ORS571.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
All bacterial strains, plasmids, and primers used in this study are shown in Tables 1, 2. A. caulinodans ORS571 was grown in TY or L3 media with ampicillin and nalidixic acid at 37°C. Escherichia coli DH5α strain was grown in LB medium at 37°C.
Antibiotics were used as follows: nalidixic acid (Nal), 25 μg/ml; kanamycin (Kan), 30 μg/ml; gentamicin (Gen), 30 μg/ml; tetracycline (Tc), 10 μg/ml; and ampicillin (Amp), 100 μg/ml. TY medium contains tryptone (5 g/L), yeast extract (3 g/L), and CaCl2 (0.6 g/L). The L3 medium contains KH2PO4 (1.36 mg/L), NH4Cl (0.53 mg/L), carbon source (adjusting the carbon according to the need, 10 mM), MgSO4 (100 mg/L), NaCl (50 mg/L), CaCl2 (40 mg/L), FeCl3 (5.4 mg/L), Na2MoO4 (5 mg/L), biotin (2 mg/L), nicotinic acid (4 mg/L), and pantothenic (4 mg/L). LB medium contains tryptone (10 g/L), yeast extract (5 g/L), and NaCl (10 g/L).
Sequence Analysis
The domains of ActR (AZC_0619) were predicted by InterPro annotation (Mitchell et al., 2019). The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was searched for the protein sequences of FtcR-like proteins (accession number: K21603) (Kanehisa et al., 2021). Alignments of the protein sequences of FtcR-like protein were done using COBALT with the default parameters (Papadopoulos and Agarwala, 2007). The maximum likelihood (ML) tree of FtcR-like protein sequences was reconstructed using the MEGAX with the LG amino acid substitution model and gamma distributed with invariant sites (G+I) (Kumar et al., 2018). Furthermore, complete deletion of gaps and missing data was carried out to exclude highly variable regions. The AZC_0619 and selected proteins were shown in a logo that was generated by Weblogo with the default parameters (Crooks et al., 2004) and further analyzed with ESPript 3.0 (Robert and Gouet, 2014).
Construction of the Deletion Mutant and Complemented Strain
Genomic DNA was extracted from A. caulinodans ORS571, and the actR deletion mutant was constructed using a methodology of allelic exchange mutagenesis. First, fragments of upstream and downstream genes of actR were amplified from the genomic DNA of A. caulinodans ORS571 with two pairs of primers (actR-up-F and actR-up-R, and actR-down-F and actR-down-R), carrying KpnI, NdeI, AgeI, and SacI restriction enzyme sites, respectively (Table 1). These fragments were cloned into vector pEASY-Blunt Simple to generate plasmid pEASY::actR up and pEASY::actR down and then, respectively, linked with pCM351 to generate pCM351::actR up–down by restriction enzyme digestion. Then, pCM351::actR up–down was transformed into E. coli DH5α competent cells. Finally, pCM351::actR up–down was introduced into A. caulinodans ORS571 via allelic exchange, and actR gene was replaced. Mutations were selected in TY medium with Nal, Amp, and Gen and were verified by PCR using a pair of primers: actR-up-F and actR-down-R. The mutant with a deleted actR gene was named ΔactR.
To construct the complemented strain of ΔactR (ΔactR-C), the fragment of the entire open reading frame and predicted promoter of actR was amplified, then was digested with Xho I and BamH I, and cloned into plasmid pBBR1MCS-2 (Kovach et al., 1995). The recombinant plasmid pBBR-actR was introduced into ΔactR mutant by triparental conjugation. The complemented strain was named as ΔactR-C. The ActR overexpressed strain, ΔactR with empty plasmid pBBR1MCS-2, and wild type (WT) with empty plasmid pBBR1MCS-2 were constructed in the same way and was designated as WT-O, ΔactR-P, and WT-P, respectively.
The Growth Curve
WT and ΔactR mutant strains were cultured overnight with TY medium containing antibiotics. The overnight cultures were collected and adjusted to an optical density (OD) at OD600 of 0.05. Normalized cultures measuring 500 μl of the WT and mutant were inoculated into 50 ml of TY medium and then were cultured in a rotary shaker at 37°C. The value of OD600 was measured every 2 h. The data were recorded as means and SDs from three repetitions.
Swimming Assay
The swimming assay was analyzed on L3 soft agar (0.3% agar) plates containing succinate as the sole carbon source and 10 mM of NH4Cl. Overnight cultures were adjusted to a density at OD600 of 1.0. Aliquots measuring 5 μl of cell suspensions of the WT, ΔactR, ΔactR-C, ΔactR-P, WT-P, and WT-O were inoculated on L3 soft agar plates for 48 h at 37°C.
Transmission Electron Microscopy
The bacterial strains were cultured in TY medium for 24 h at 37°C. The overnight cultures were collected and washed twice with sterile phosphate-buffered saline (PBS), spotted on copper grids, and negatively stained with 2% phosphotungstic acid. The transmission electron microscopy (TEM) images of the WT and ΔactR mutant were taken at random with grid.
Quantitative Real-Time PCR
The total RNA was isolated from A. caulinodans ORS571 using TransZol Up Plus RNA kit. cDNA was generated through TransScript One-Step gDNA removal kit according to the manufacturer’s instructions. The synthesized cDNA was diluted 500-fold and used as template to analyze relative gene expression. The quantitative qPCR was performed with GoTaq® qPCR Master Mix kit using gene specific primer pairs (Table 2). The quantitative PCR program consisted of an initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 1 min. To evaluate the gene expression, the copy number of each gene was normalized to that of the 16S rRNA. The analysis of its results was performed using the comparative cycle threshold method (Schmittgen and Livak, 2008).
Biofilm Formation Assay
WT and ΔactR mutant were incubated overnight in the L3 medium at 37°C, and then the culture was adjusted to OD600 of 2. Bacterial suspensions measuring 15 μl were added into glass tubes including 1.5 ml of L3 medium and were incubated for 3 days at 37°C. The L3 medium contained succinic acid (10 mM) as a sole carbon source, with nitrogen. After 3 days’ incubation, the glass tubes were gently washed three times with PBS (pH = 7.2) to remove free-floating bacteria and then stained with 2 ml of crystal violet (CV; 0.1%, w/v) for 20 min. CV was gently removed by washing three times with deionized water. Finally, biofilm formation was quantified by ethanol-solubilized CV from glass tube biofilms. The OD value of each tube was determined after CV staining with a wavelength of 590 nm.
Exopolysaccharide Production
EPS production was estimated using the method described by Jiang et al. (2016) with the following modifications. The WT and ΔactR mutant were incubated in the L3 medium at 37°C. Then, bacterial cultures were adjusted to an OD600 of 0.8. Ten-microliter aliquots of bacterial suspensions were spotted on L3 solid agar (0.8% agar) plates with three different carbon sources (sodium lactate, glycerol, and malic acid) and incubated at 37°C. The L3 plates included 10 mM of carbon source, 10 mM of NH4Cl, and 40 μg/ml of Congo red. Photographs were taken after 4 days of incubation. The quantitative analysis of EPS content was measured by a colorimetric method using anthrone and sulfuric acid and was evaluated by normalizing to OD600 of the bacterial suspension.
Flocculation
Flocculation was measured using the method described by Jiang et al. (2016). Overnight cultures were normalized to an OD600 of 1.0, and then 200 μl of the normalized cultures was inoculated into 10 ml of L3 medium containing 10 mM of sodium lactate as the carbon source and 0.5 mM of NH4Cl as the nitrogen source. Photographs of the non-flocculated cells and flocculated cells were taken after 48 h of incubation. For the quantitative analysis, the bacterial suspensions were left to stand for 30 min. The non-flocculated cells were removed from the tube, and the OD600 of the suspension (ODs) was measured. The flocculated cells that settled to the bottom of the tube were dispersed using a tissue homogenizer and thoroughly mixed with free cells; the density of the mixed culture was also measured at 600 nm and named as ODt. The percent flocculation was calculated as follows: percent flocculation = (1 – ODs/ODt) × 100.
Rhizosphere Colonization Assay
Seeds of S. rostrata were treated with concentrated sulfuric acid for 30 min, followed by rinsing five times with sterilized water. Sterile seedlings were germinated in the dark at 37°C for 72 h. WT and mutant strains were grown overnight in sterile TY medium; then overnight cultures were collected and adjusted to a density of 0.8. For competitive colonization, suspensions of the WT and mutant were mixed at 1:1, 1:5, and 1:10. Germinated S. rostrata seeds were inoculated with the mixed cultures for 1 h. The surfaces of seedlings were washed seven times using sterilized water. And then the root tips were vortexed, and bacteria were reisolated on TY agar plates with ampicillin. A total of 100 of the colony-forming units (CFUs) were selected, and the number of the WT and ΔactR was counted by plate streaking in TY agar plates with ampicillin and gentamicin. In addition, the WT and mutant colonies were also identified by PCR.
Competitive Nodulation Assay
Surface-sterilized seedlings of S. rostrata were grown in soil supplemented with sterilized vermiculite and low nitrogen plant nutrient solution for 4 weeks. The bacterial suspension (OD600 = 0.6) of the WT and ΔactR mutant strains was mixed at ratios of 1:1 and 1:5; then the mixed cultures were used to paint the stem surface of S. rostrata. After 30 days’ inoculation at 27°C in the greenhouse, nodules were harvested, and bacteria were reisolated and plated on TY agar medium from stem nodules. The WT and mutant bacteria were identified and distinguished by PCR using the primer pair actR in-F and actR in-R, and the ratios between the WT and mutant were further counted.
Statistical Analysis
Mean and standard errors were measured based on each experiment repeated three times. The Statistical Package for the Social Sciences (SPSS version 20) was used to analyze the least significant difference test (p < 0.05).
Results
The Genome of Azorhizobium caulinodans Encodes a Homolog of the Motility Regulator FtcR
From the genome sequence of A. caulinodans ORS571, we searched for the presence of FtcR homolog, AZC_0619. Only one open reading frame encoding for the FtcR-like protein was identified, which was named as ActR. Sequence analysis showed that ActR contains 222 amino acids, encoding one N-terminal receiver domain (IPR001789) and one C-terminal OmpR/PhoB-type DNA binding domain (IPR001867) (Supplementary Figure 1). A BLASTp analysis exhibited that AZC_0619 shared about 53.2% of identity with a flagellar master regulator FtcR of B. melitensis 16M. actR gene was located downstream of two chemotactic genes (cheY and cheZ), mltE gene encoded for transmembrane protein, and three flagellar genes (motB, motC, and fliK) in A. caulinodans (Figure 1A). And flagellar genes were found in the proximity of ftcR-like gene in most species (Figure 1A), indicating that FtcR-like protein may play a role in the regulating flagellar motility among Rhizobiales.
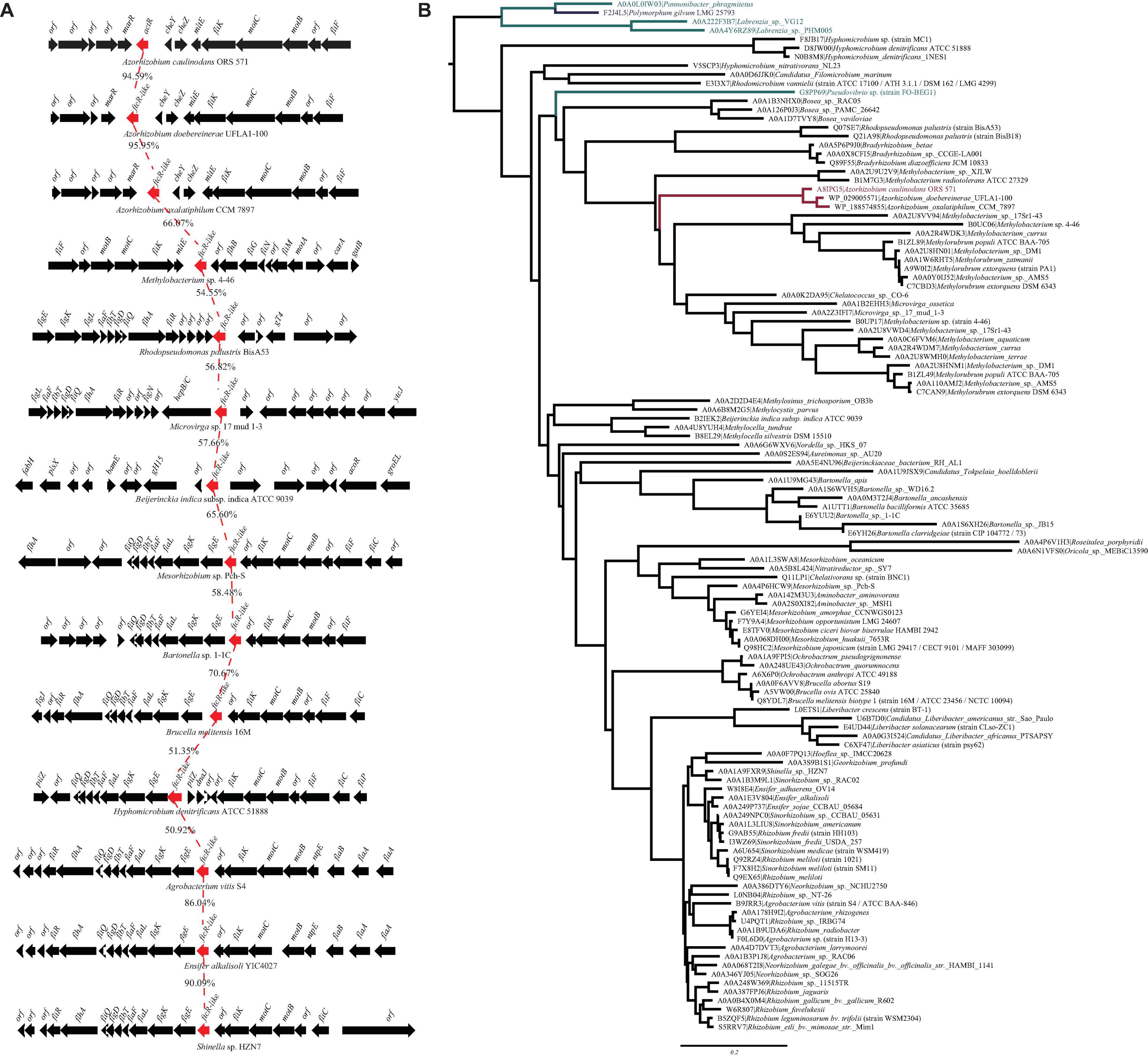
Figure 1. (A) Genetic organization of gene clusters in the genomic region containing the ftcR. The frcR coding sequences (CDS) is indicated in red. Percentage amino acid identities of the FtcR are shown. (B) Maximum likelihood phylogeny of the protein sequences of FtcR homologs. The clades of the order Rhizobiales, Rhodobacterales, and Polymorphum are indicated in black, turquoise, and blue, respectively. The Azorhizobium clade and A8IPG5 (ActR; Azorhizobium caulinodans ORS571) are indicated in red.
To clarify the evolutionary relationships of the FtcR-like proteins, a phylogenetic tree was generated. As shown in Figure 1B, the FtcR-like tree exhibited extensive genetic diversity. FtcR-like proteins among Rhizobiales members were monophyletic and exhibited topology that was generally congruent with the species taxonomy [KEGG Organisms in the National Center for Biotechnology Information (NCBI) Taxonomy], suggesting that FtcR-like proteins originated from a last common Rhizobiales ancestor and underwent divergent evolution during species differentiation. Within this phylogenetic tree, we found that FtcR-like proteins in Azorhizobium formed a monophyletic subclade with long branch lengths (Figure 1B). Notably, FtcR-like proteins from A. caulinodans ORS571 and B. melitensis 16M are clustered into separated clades.
We then systematically examined the prevalence and evolution of FtcR. Based on the KEGG orthology database (KO: K21603) (Kanehisa et al., 2021), FtcR-like proteins were present in orders Rhizobiales, Rhodobacterales, and Polymorphum of Alphaproteobacteria. FtcR-like proteins were widespread in most genus/species among Rhizobiales and had a sporadic distribution among Rhodobacterales (genera Pseudovibrio, Pannonibacter, and Labrenzia). A total of 117 protein sequences of FtcR-like were recovered from the KEGG database (KO: K21603) and exhibited a significant divergence (more than 35.4% identity) at the amino acid level (Figure 2). The FtcR-type sequences are conserved in genus Azorhizobium (A. caulinodans, Azorhizobium doebereinerae, and Azorhizobium oxalatiphilum) with the pairwise sequence identities in the range of 93.24–95.59% (Figures 1A, 2); this result was consistent with that of Azorhizobium FtcR proteins that formed a monophyletic subclade in Figure 1B. Our analysis indicated that FtcR-like protein is conserved and that it may play an important role among Azorhizobium. Although Léonard et al. (2007) provided insights on the role of flagellar master regulator FtcR in Brucella, the function of its homology in rhizobia has remained unclear. Therefore, we will investigate the biological function of ActR in A. caulinodans ORS571.
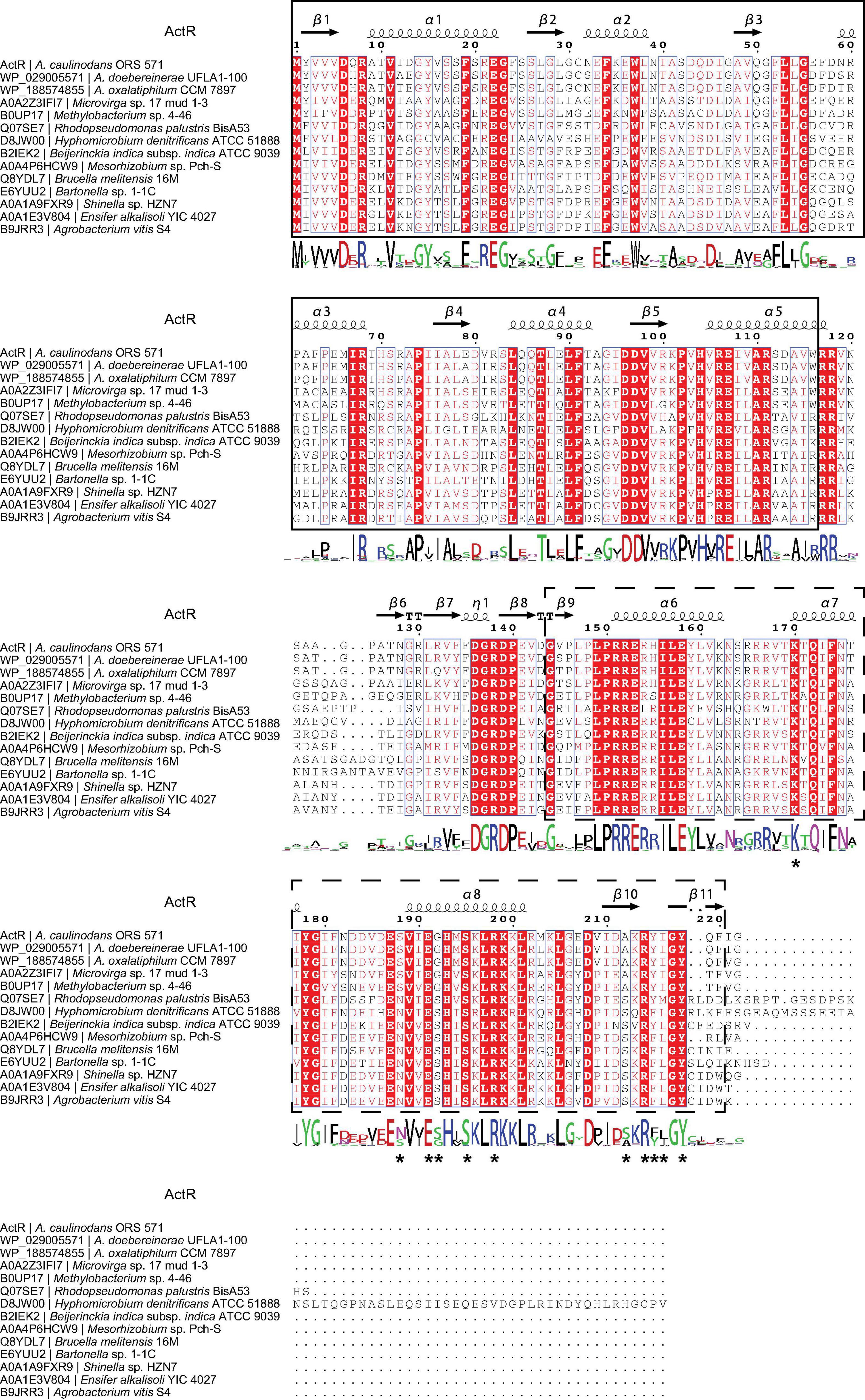
Figure 2. Analysis of the FtcR-like sequence. Protein sequences were aligned using the COBALT with the default parameters. The picture of the alignment was performed by ESPript 3.0. The predicted secondary structure of ActR is shown above the alignment. Conserved residues are shown with red background, and similar residues are shown in red and boxed. The sequence logo is shown below the alignment. The bigger the logo, the more conserved the residue. The ligand binding site residues of ActR predicted by the COFACTOR and COACH programs in I-TASSER are marked with asterisks. The solid line frame and the dashed line frame indicate the response regulator domain and the transcriptional regulatory domain (OmpR/PhoB-type) as recognized in InterPro annotation.
ActR Regulates Cell Motility and the Expression of Flagellar Genes
ActR was predicted to be a flagellar two-component response regulator. To characterize the role of ActR in regulating motility, ΔactR mutant strain was constructed through allelic exchange mutagenesis (see section “Materials and Methods,” Supplementary Figure 2), and the swimming motility behavior of the ΔactR mutant and complementary strain were compared with those of the WT (Figures 3A,B). We found that the mutant (ΔactR-P) with an empty plasmid pBBR1MCS-2 was devoid of motility ability, which could be rescued by introducing the pBBR1MCS-2 carrying WT actR gene and its native promoter into the mutant (ΔactR-C) (Figure 3A). We also found that an overexpressing strain (WT-O) exhibited significant increase of swimming motility when compared with that of the WT-P. The WT-O and WT-P were constructed by the introduction of an empty plasmid pBBR1MCS-2 and a pBBR1MCS-2 carrying WT actR gene and its native promoter, respectively. The quantitative data clearly confirmed the result, too (Figure 3B).
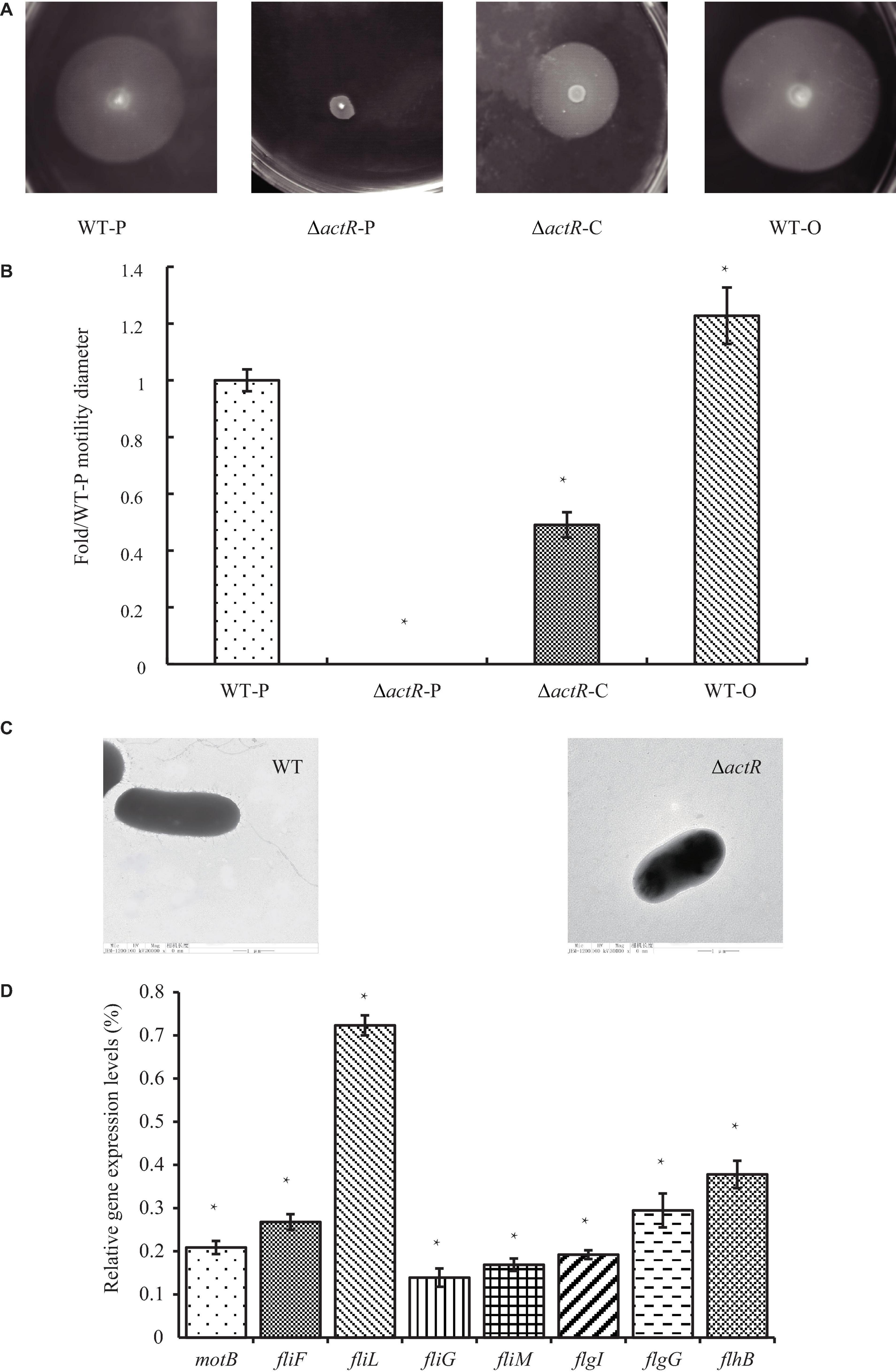
Figure 3. ActR was required for swimming motility and flagellar biosynthesis in Azorhizobium caulinodans. (A) Representative images of the bacterial motility rings in soft agar plates, including the wild-type (WT) strain (WT-P), the ΔactR strain (ΔactR-P), the complemented strain (ΔactR-C), and the actR-overexpressing strain (WT-O). (B) Quantitative analysis of the swimming motility abilities. Bar plot represents the mean and SD of fold change about the bacterial motility ring diameter compared with that of WT-P. (C) Representative images obtained by transmission electron microscopy using cells from the WT and ΔactR strains. Scale bar represents 1 μm. (D) Relative expression of flagellar genes (motB, fliF, fliL, fliG, fliM, flgI, flgG, and flhB) in the ΔactR mutant strain compared with that of WT. Bar plot represents the mean and SD of the gene expression level. Means and SDs of fold change and gene expression were obtained from at least three independent experiments. Student’s t-test; *represents significant difference at p < 0.05.
To explore the reason for lost motility ability, it was first checked that growth properties of the mutant strain was not impaired (Supplementary Figure 3). What is really interesting was that TEM of the WT and the mutant strains revealed the absence of flagella in the mutant strain (Figure 3C).
Next, we tested whether ActR regulator regulated the expression of flagellar synthesis-related genes by quantitative real-time PCR. It is true that the deletion of any flagellar component is sufficient to abolish flagellar synthesis, such as FliM and FliN (Shen et al., 2018). The results showed that the expression levels of eight flagellar genes (motB, fliF, fliL, fliG, fliM, flgI, flgG, and flhB) in ΔactR mutant were significantly downregulated than those in the WT (Figure 3D). For example, the expression values of fliL and fliG declined 27 and 88%, respectively. These results suggested that ActR was closely associated with the synthesis of flagella and positively regulated the motility of A. caulinodans ORS571.
ActR Positively Regulates Biofilm Formation and Exopolysaccharide Production
Biofilm formation and motility are modulated directly by flagellar power (Subramanian and Kearns, 2019), and biofilm forming on plant surfaces is important for bacterial symbiotic interaction with host plant (Xu et al., 2019). To investigate whether the deletion of actR has an influence on bacterial biofilm formation, we tested the biofilm of the WT and the mutant. As shown in Figure 4A, the mutant produced less biofilm than the WT in the L3 medium with nitrogen (L3+N medium), and quantitative data confirmed the results. In the L3+N medium, the biomass of biofilm in ΔactR mutant was 32% less than that of the WT strain (Figure 4B).
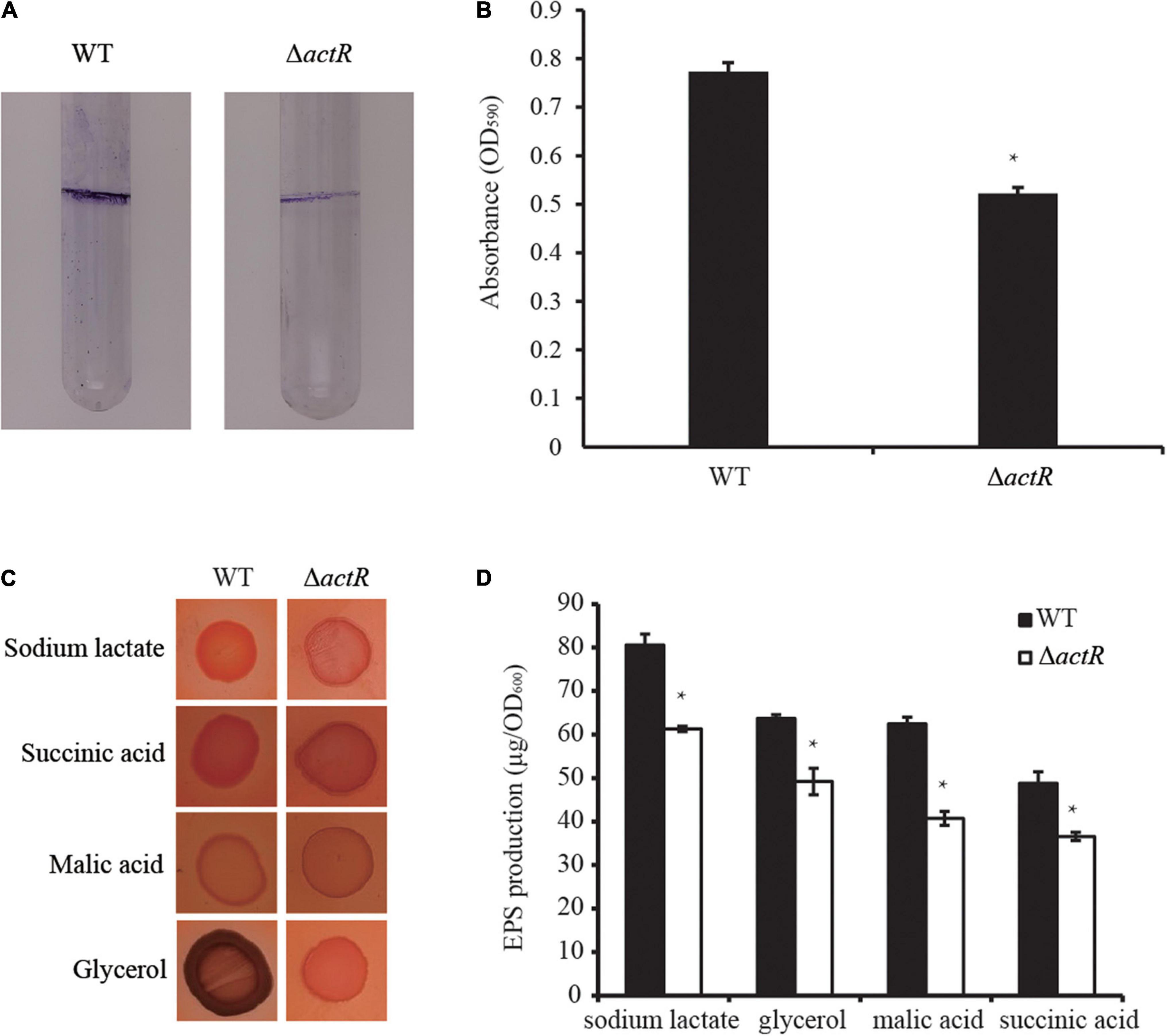
Figure 4. Effects of ActR on the biofilm formation and exopolysaccharide (EPS) production of Azorhizobium caulinodans ORS571. (A) Representative images of biofilms formed by the wild type (WT) and ΔactR. (B) Quantitative analysis of biofilm formation. Bar plot represents the mean and SD of the optical density at 590 nm. (C) Representative images of colony morphologies of the WT strain and ΔactR mutant spotted on the L3 plates with Congo red staining and four different carbon sources (sodium lactate, glycerol, malic acid, and succinic acid). Photographs were taken after 4 days of incubation. (D) Quantitative analysis of the EPS production. Bar plot represents the mean and SD of the optical density at 600 nm of EPS production. Means and SDs of biofilm formation and EPS production were obtained from at least three independent experiments. Student’s t-test; *represents significant difference at p < 0.05.
We next tested the production of EPS by Congo red staining. EPS is essential for the maintenance of biofilm formation in bacteria (Cugini et al., 2019). And it is involved in bacterial symbiotic nodulation with S. rostrata (Sun et al., 2020). To study the EPS production ability of ΔactR, the colony morphologies and quantitative analysis of the EPS were examined using minimal medium plates containing different carbon sources (sodium lactate, glycerol, malic acid, and succinic acid). A significant difference between the WT and the mutant was observed. The colonies of the WT cells produced more EPS than ΔactR cells regardless of the kind of carbon sources were assayed (Figures 4C,D). These data indicated that ActR positively affected EPS production and biofilm formation in A. caulinodans ORS571.
ActR Contributes to Cell Flocculation
Flocculating substances are secreted by many microorganisms in the culture broth, which comprise polysaccharides, proteins, and lipids (Salehizadeh and Yan, 2014). To study whether ActR influences flocculation in A. caulinodans ORS571, we tested the flocculation morphologies between the WT and mutant strains in the L3 medium with 5 mM of NH4Cl as a nitrogen source. Figure 5A shows that flocs formed by the WT were larger and more abundant than those of the mutant strain. Quantitative analysis suggested that the WT flocculated more than the mutant strain and that the flocculation formation of both strains increased over time (Figure 5B). These results indicated that ActR was involved in regulating flocculation formation.
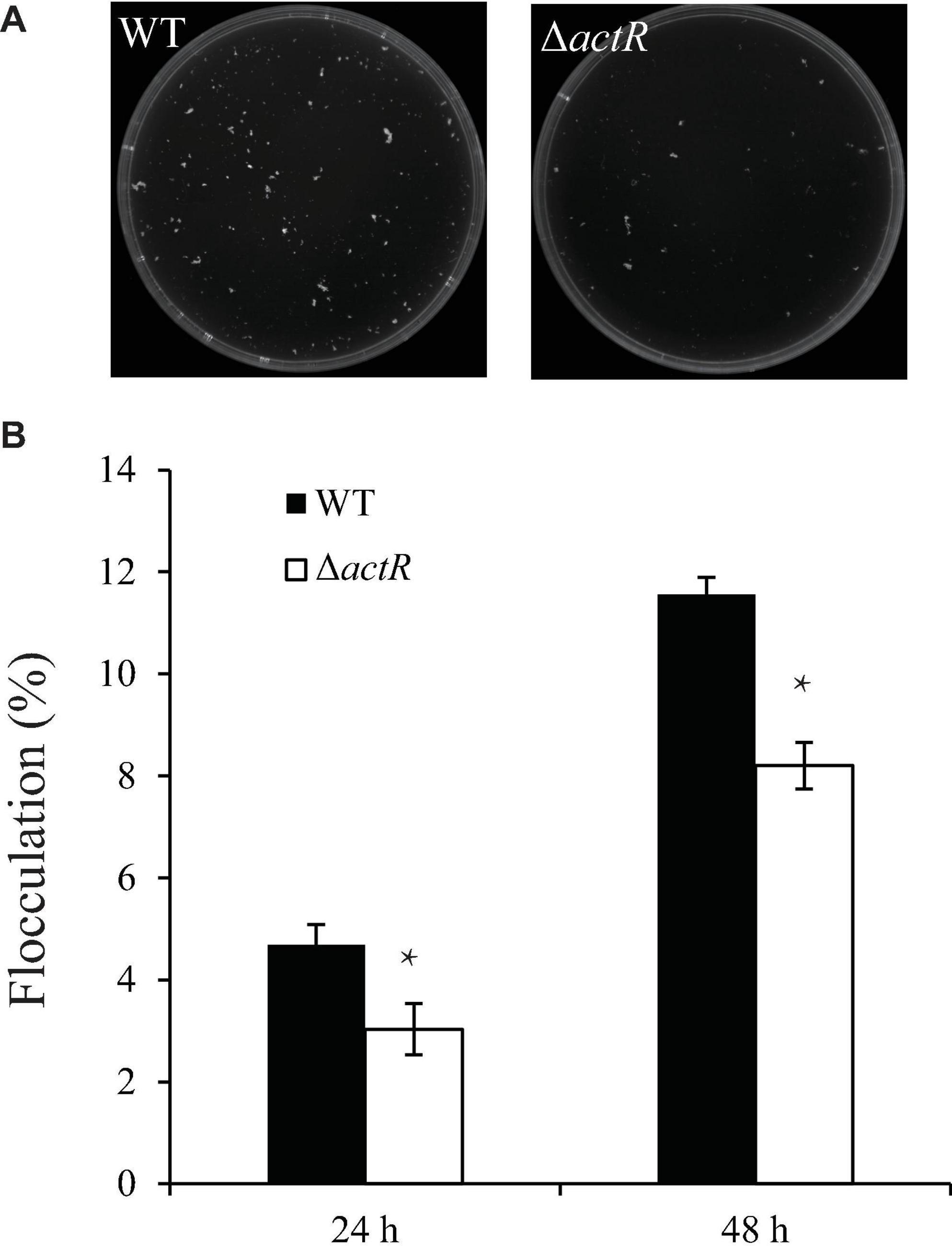
Figure 5. Effect of ActR on the flocculation generated by wild-type (WT) and mutant strains. (A) Representative images of flocculation morphologies produced by the WT and ΔactR mutant. Strains were inoculated with L3+1/2N liquid medium, and photographs were taken after 48 h of incubation. (B) Quantitative analysis of the flocculation cell mass. Bar plot represents the mean and SD of the percent density of flocculation generated by each strain. Means and SDs were obtained from at least three independent experiments. Student’s t-test; *represents significant difference at p < 0.05.
ActR Is Involved in Root Colonization and Nodule Formation by Azorhizobium caulinodans ORS571
The colonization on the surface of the host plant is a key step for successfully establishing symbiosis. To investigate the symbiotic role of ActR in A. caulinodans, competitive colonization experiments were performed. We counted the number of cells reisolated from the seedlings to verify the efficiency of competitive colonization. As shown in Figure 6A, the ΔactR mutant was less competitive than the WT. When the WT and the mutant cells were mixed in equal proportion, 100% of the bacteria reisolated from the root surface belonged to the WT, showing that the mutant could not compete. When the proportions between the WT and mutant were 1:5 and 1:10, a few of the cells of ΔactR mutant strains (14.3 and 23.1%, respectively) were reisolated from the root system. This result suggested that the deletion of actR did not disable bacterial colonization ability but affected A. caulinodans with an effective competitive ability for root colonization.
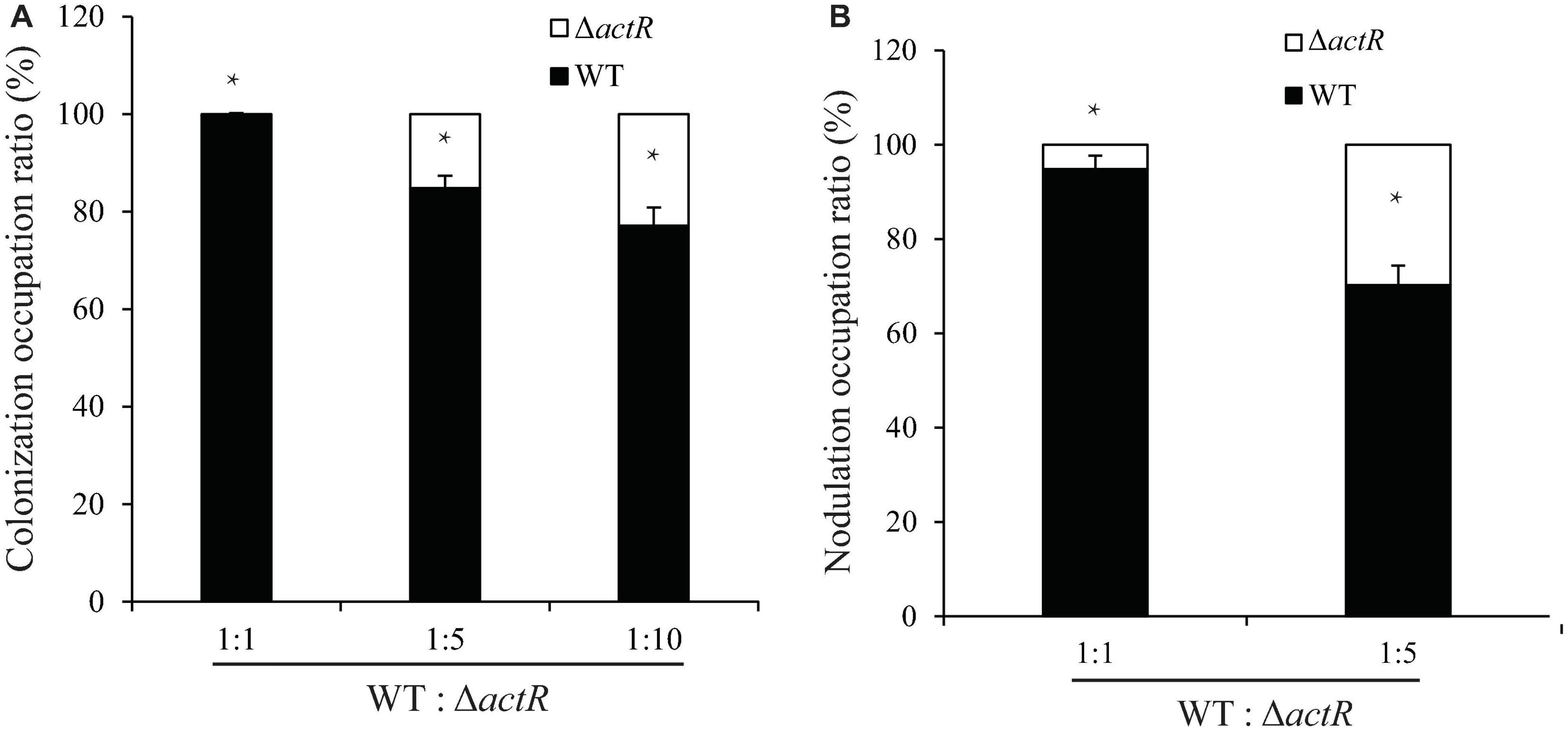
Figure 6. The mutant reduced colonization and nodulation competitiveness on Sesbania rostrata. (A) Competitive colonization of wild type (WT) and ΔactR mutant strains on S. rostrata root. Ratios of 1:1, 1:5, and 1:10 represent the ratio of WT to the mutant. (B) Competitive nodulation of WT and ΔactR on the stems of S. rostrata. Ratios of 1:1 and 1:5 represent the WT to the mutant. Bar plot represents the mean and SD of the relative colonization ratio and the relative nodulation occupation ratio of the WT (black bar) and ΔactR mutant (white bar). Means and SDs were obtained from at least three independent experiments. Student’s t-test; *represents significant difference at p < 0.05.
Induction of nodule morphogenesis is associated with the EPS production and colonization of the root system (Liu and Xie, 2019). To study whether ActR plays an important role in nodule formation, the competitive nodulation assay was performed by counting the number of cells (WT and mutant strains) reisolated from the stem nodules. As shown in Figure 6B, the ratios of ΔactR mutant were 8 and 31% when the WT and mutant strains were mixed at ratios of 1:1 and 1:5, respectively, indicating that the ΔactR mutant could still nodulate S. rostrata but not compete with the WT strain. Taken together, these results indicated that ActR could provide A. caulinodans an effectively competitive ability for symbiosis.
Discussion
The FtcR-like regulator is widely distributed across genus/species within Rhizobiales, such as A. caulinodans, Ensifer meliloti, and Agrobacterium rhizogenes. The phylogeny general congruence with species taxonomy and sequence divergence between homologs indicated that FtcR-like proteins in Rhizobiales were predominantly vertically inherited from a common ancestor and underwent divergent evolution during species differentiation. In this study, AZC_0619 in A. caulinodans (named actR) was characterized, which was a homolog of ftcR gene of B. melitensis 16M. This gene is generally in close proximity to chemotaxis and flagellum-related genes, indicating that ActR protein may play an important role in motility and symbiosis with the host plant. By the construction of ΔactR mutant, we found that mutant strain had no flagella and was devoid of motility. The improperly formed flagellum is a major cause of defective motility. The structure of a flagellum is constructed by a highly ordered process and consisted of about 30 flagellar proteins upon various stoichiometries (Altegoer and Bange, 2015). The flagellar gene fliM deletion mutant of A. caulinodans was confirmed to have no flagellum (Shen et al., 2018). RT-PCR revealed that the genes (motB, fliF, fliL, fliG, fliM, flgI, flgG, and flhB) involved in flagellar synthesis were downregulated in the ΔactR mutant. Therefore, ActR regulator in A. caulinodans ORS571 was involved in the flagellar assembly and further affected bacterial motility.
ActR protein in A. caulinodans was involved in bacterial symbiosis with the host plant. The ΔactR mutant still formed colonization and nodulation on S. rostrata, but its competitiveness was obviously eroding, which is to say that ActR hardly affected the formation of nodules, but what is the reason for the competitive disadvantage of this mutant? There are two possible explanations: first, the colonization and nodulation are related to flagella and motility. Bacterial flagella could function as adhesion molecules (Friedlander et al., 2015), and non-mobile mutant could not move away from the initial inoculation region (Liu et al., 2019). So it was unsurprising that non-flagellated ΔactR mutant had narrower colonization region and lower nodulation than the WT strain. It was confirmed that the impaired motility of ΔaclR1 and ΔacfR mutants was consistent with the weak nodulation abilities in A. caulinodans (Liu et al., 2020b,2021). Second, EPS production and biofilm formation influence root colonization. Previous studies indicated that EPS and biofilm were essential for attachment to the root surface (Ma et al., 2006; Al-Ali et al., 2018). When the EPS biosynthesis genes of Rhizobium sp. IRBG74 were disrupted, the mutant exhibited defective colonization and nodulation (Mitra et al., 2016). Take the case of A. caulinodans: the oac2 gene deletion mutant had decreased the EPS production ability, and its symbiosis with host plant was disabled (Gao et al., 2001). It was also proposed that EPS production regulated bacterial early colonization and the A. caulinodans-S. rostrata nodulation process (Liu et al., 2019). In this study, it was showed that the biofilm formation and EPS production of ΔactR mutant was decreased (Figure 4). Therefore, we conjectured that ActR probably helped cells in forming a symbiotic association with host plant by modulating EPS production and biofilm formation in Rhizobiales.
Based on the observation of non-flagellated morphology and decreased EPS formation of ΔactR mutant (Figures 3C, 5), two hypotheses about the decrease of biofilm formation were made. First, ΔactR mutant reduced biofilm formation due to the lack of flagella. Flagella are a component of the biofilm matrix and are vital in stabilizing biofilm by acting as bacterial biofilm scaffolds (Hathroubi et al., 2018). The non-flagellated mutants (ΔfliM and ΔfliN) in A. caulinodans showed the decrease of biofilm biomass compared with the wild-type strain (Shen et al., 2018). Liu et al. (2020) further verified that ΔfliN mutant reduced the biofilm formation after 12 h. Second, the EPS production of ΔactR mutant may affect biofilm formation. EPS in biofilm has vital role in maintaining the biofilm structure and providing biofilm cells with nutrients (Altaf and Ahmad, 2016). The actR gene deletion mutant had less EPS production regardless of what carbon source was used, which may be another reason for the decrease of biofilm formation. The positive correlation between biofilm formation and EPS was shown in ΔcheZ mutant of A. caulinodans (Liu X. et al., 2018). Meanwhile, we found that ΔactR mutant formed less flocculation than the WT. Flocs was encysting bacteria surrounded by EPS (Sadasivan and Neyra, 1985). In A. caulinodans, Sun et al. (2020) indicated that the increase of flocculation was correlated with the increase of EPS production. So we proposed that the decrease of EPS production contributed to less formation of biofilm and flocculation. As for the decrease of EPS content in ΔactR mutant, the possible reason is that the genes related to EPS production in ΔactR mutant strain did not express like that in the WT strain. Lauriano et al. (2004) reported that sodium-driven motors (mot), such as motA, motB, motX, and motY, were involved in EPS production, and any of the mot gene deletion mutants of flaA MO10 strain reduced EPS expression. It was observed that motB gene was downregulated in the ΔactR mutant, so flagellar motors may play a similar role in regulating EPS expression. In addition, Belas (2014) also described that EPS production was involved in the stimulation of mechanosensing signals caused by flagellar rotation. A non-flagellated ΔactR mutant might decrease EPS expression by reducing signal stimulation of flagellar rotation. On the other hand, ActR of A. caulinodans may have a specific role, not like known regulatory mechanisms of EPS expression. There is little evidence that FtcR-like protein regulates EPS production and biofilm formation in other bacteria; thus, the regulatory mechanisms of ActR to EPS production at the molecular level need to be verified further.
This study further deepens our understanding of the role of OmpR domain-containing transcriptional regulators in A. caulinodans. ActR not only can regulate bacterial motility but also can influence bacterial symbiosis with S. rostrata. However, the specific regulatory mechanism of ActR needs to be further studied.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
LS: data curation, formal analysis, investigation, methodology, visualization, writing—original draft, and writing—review and editing. DW: data curation, visualization, writing—original draft, and writing—review and editing. ZY: formal analysis and writing—review and editing. CZ and AB: writing—review and editing. ZX: funding acquisition, project administration, and writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was financed by the NSFC-Shandong Joint Fund Key Projects (U1806206), the National Natural Science Foundation of China (31870020), and National Key Research and Development Program (2019YFD1002702).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Claudine Elmerich for her valuable suggestion and Rimin Shen for her helpful work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.744268/full#supplementary-material
References
Al-Ali, A., Deravel, J., Krier, F., Béchet, M., Ongena, M., and Jacques, P. (2018). Biofilm formation is determinant in tomato rhizosphere colonization by Bacillus velezensis FZB42. Environ. Sci. Pollut. Res. Int. 25, 29910–29920. doi: 10.1007/s11356-017-0469-1
Altaf, M. M., and Ahmad, I. (2016). “Biofilm formation on plant surfaces by rhizobacteria: impact on plant growth and ecological significance,” in The Handbook of Microbial Bioresources, eds V. K. Gupta et al. (Wallingford: CAB International).
Altegoer, F., and Bange, G. (2015). Undiscovered regions on the molecular landscape of flagellar assembly. Curr. Opin. Microbiol. 28, 98–105. doi: 10.1016/j.mib.2015.08.011
Belas, R. (2014). Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 22, 517–527. doi: 10.1016/j.tim.2014.05.002
Brencic, A., and Winans, S. C. (2005). Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol. Mol. Biol. Rev. 69, 155–194. doi: 10.1128/MMBR.69.1.155-194.2005
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Cugini, C., Shanmugam, M., Landge, N., and Ramasubbu, N. (2019). The role of exopolysaccharides in oral biofilms. J. Dent. Res. 98, 739–745. doi: 10.1177/0022034519845001
Dasgupta, N., Wolfgang, M. C., Goodman, A. L., Arora, S. K., Jyot, J., Lory, S., et al. (2003). A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50, 809–824. doi: 10.1046/j.1365-2958.2003.03740.x
Dreyfus, B., Garcia, J. L., and Gillis, M. (1988). Characterization of Azorhizobium caulinodans gen. nov., sp. nov., a stem-nodulating nitrogen-fixing bacterium isolated from Sesbania rostrata. Int. J. Syst. Bacteriol 38, 89–98. doi: 10.1099/00207713-38-1-89
Dreyfus, B. L., Elmerich, C., and Dommergues, Y. R. (1983). Free-living Rhizobium strain able to grow on N2 as the sole nitrogen source. Appl. Environ. Microbiol. 45, 711–713. doi: 10.1128/aem.45.2.711-713.1983
Figurski, D. H., and Helinski, D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652. doi: 10.1073/pnas.76.4.1648
Friedlander, R. S., Vogel, N., and Aizenberg, J. (2015). Role of flagella in adhesion of Escherichia coli to abiotic surfaces. Langmuir 31, 6137–6144. doi: 10.1021/acs.langmuir.5b00815
Gao, M., D’Haeze, W., De, R. R., Wolucka, B., and Holsters, M. (2001). Knockout of an azorhizobial dTDP-L-rhamnose synthase affects lipopolysaccharide and extracellular polysaccharide production and disables symbiosis with Sesbania rostrata. Mol. Plant Microbe Interact. 14, 857–866. doi: 10.1094/MPMI.2001.14.7.857
Hathroubi, S., Zerebinski, J., and Ottemann, K. M. (2018). Helicobacter pylori biofilm involves a multigene stress-biased response, including a structural role for flagella. mBio 9:e01973-18. doi: 10.1128/mBio.01973-18
Horstmann, J. A., Lunelli, M., Cazzola, H., Heidemann, J., and Erhardt, M. (2020). Methylation of Salmonella typhimurium flagella promotes bacterial adhesion and host cell invasion. Nat. Commun. 11:2013. doi: 10.1038/s41467-020-15738-3
Jiang, N., Liu, W., Li, Y., Wu, H., Zhang, Z., Alexandre, G., et al. (2016). A chemotaxis receptor modulates nodulation during the Azorhizobium caulinodans-Sesbania rostrata symbiosis. Appl. Environ. Microbiol. 82, 3174–3184. doi: 10.1128/aem.00230-16
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M., and Tanabe, M. (2021). KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545–D551. doi: 10.1093/nar/gkaa970
Kennedy, I. R., Pereg-Gerk, L. L., Wood, C., Deaker, R., Gilchrist, K., and Katupitiya, S. (1997). Biological nitrogen fixation in non-leguminous field crops: facilitating the evolution of an effective association between Azospirillum and wheat. Plant Soil 194, 65–79. doi: 10.1023/A:1004260222528
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., and Roop, R. M. II, et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lauriano, C. M., Ghosh, C., Correa, N. E., and Klose, K. E. (2004). The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 186, 4864–4874. doi: 10.1128/jb.186.15.4864-4874.2004
Lee, K. B., Backer, D. P., Aono, T., Liu, C. T., Suzuki, S., Suzuki, T., et al. (2008). The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics 9:271. doi: 10.1186/1471-2164-9-271
Léonard, S., Ferooz, J., Haine, V., Danese, I., Fretin, D., Tibor, A., et al. (2007). FtcR is a new master regulator of the flagellar system of Brucella melitensis 16M with homologs in Rhizobiaceae. J. Bacteriol. 189, 131–141. doi: 10.1128/jb.00712-06
Liu, W., Bai, X., Li, Y., Min, J., Kong, Y., and Hu, X. (2020a). CheY1 and CheY2 of Azorhizobium caulinodans ORS571 regulate chemotaxis and competitive colonization with the host plant. Appl. Environ. Microbiol. 86:e00599-20. doi: 10.1128/aem.00599-20
Liu, W., Bai, X., Li, Y., Zhang, H., and Hu, X. (2021). FixJ family regulator AcfR of Azorhizobium caulinodans is involved in symbiosis with the host plant. BMC Microbiol. 21:80. doi: 10.1186/s12866-021-02138-w
Liu, W., Li, Y., Bai, X., Wu, H., Bian, L., and Hu, X. (2020b). LuxR-type regulator AclR1 of Azorhizobium caulinodans regulates cyclic di-GMP and numerous phenotypes in free-living and symbiotic states. Mol. Plant Microbe Interact. 33, 528–538. doi: 10.1094/MPMI-10-19-0306-R
Liu, W., Sun, Y., Shen, R. M., Dang, X. X., Liu, X. L., Sui, F., et al. (2018). A chemotaxis-like pathway of Azorhizobium caulinodans controls flagella-driven motility, which regulates biofilm formation, exopolysaccharide biosynthesis, and competitive nodulation. Mol. Plant Microbe Interact. 31, 737–749. doi: 10.1094/MPMI-12-17-0290-R
Liu, X., Liu, W., Sun, Y., Xia, C., Elmerich, C., and Xie, Z. (2018). A cheZ-like gene in Azorhizobium caulinodans is a key gene in the control of chemotaxis and colonization of the host plant. Appl. Environ. Microbiol. 84:e01827-17. doi: 10.1128/aem.01827-17
Liu, X., and Xie, Z. (2019). Inactivation of the phosphatase CheZ alters cell-surface properties of Azorhizobium caulinodans ORS571 and symbiotic association with Sesbania rostrata. Mol. Plant Microbe Interact. 32, 1547–1556. doi: 10.1094/mpmi-05-19-0143-r
Liu, X., Xie, Z., Wang, Y., Sun, Y., Dang, X., and Sun, H. (2019). A dual role of amino acids from Sesbania rostrata seed exudates in the chemotaxis response of Azorhizobium caulinodans ORS571. Mol. Plant Microbe Interact. 32, 1134–1147. doi: 10.1094/mpmi-03-19-0059-r
Liu, X., Zhang, K., Liu, Y., Zou, D., Wang, D., and Xie, Z. (2020). Effects of calcium and signal sensing systems on Azorhizobium caulinodans biofilm formation and host colonization. Front. Microbiol. 11:563367. doi: 10.3389/fmicb.2020.563367
Ma, L., Jackson, K. D., Landry, R. M., Parsek, M. R., and Wozniak, D. J. (2006). Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188, 8213–8221. doi: 10.1128/jb.01202-06
Marx, C. J., and Lidstrom, M. E. (2002). Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33, 1062–1067. doi: 10.2144/02335rr01
Mitchell, A. L., Attwood, T. K., Babbitt, P. C., Blum, M., Bork, P., Bridge, A., et al. (2019). InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360. doi: 10.1093/nar/gky1100
Mitra, S., Mukherjee, A. Wiley-Kalil, A., Das, S., Owen, H., Reddy, P. M., et al. (2016). A rhamnose-deficient lipopolysaccharide mutant of Rhizobium sp. IRBG74 is defective in root colonization and beneficial interactions with its flooding-tolerant hosts Sesbania cannabina and wetland rice. J. Exp. Bot. 67, 5869–5884. doi: 10.1093/jxb/erw354
Mitrophanov, A. Y., and Groisman, E. A. (2008). Signal integration in bacterial two-component regulatory systems. Genes Dev. 22, 2601–2611. doi: 10.1101/gad.1700308
Papadopoulos, J. S., and Agarwala, R. (2007). COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079. doi: 10.1093/bioinformatics/btm076
Pawlowski, K., Klosse, U., and Bruijn, F. J. (1991). Characterization of a novel Azorhizobium caulinodans ORS571 two-component regulatory system, NtrY/NtrX, involved in nitrogen fixation and metabolism. Mol. Gen. Genet. 231, 124–138. doi: 10.1007/BF00293830
Pratt, L. A., and Kolter, R. (2010). Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30, 285–293. doi: 10.1046/j.1365-2958.1998.01061.x
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Sadasivan, L., and Neyra, C. A. (1985). Flocculation in Azospirillum brasilense and Azospirillum lipoferum: exopolysaccharides and cyst formation. J. Bacteriol. 163, 716–723. doi: 10.1128/jb.163.2.716-723.1985
Salehizadeh, H., and Yan, N. (2014). Recent advances in extracellular biopolymer flocculants. Biotechnol. Adv. 32, 1506–1522. doi: 10.1016/j.biotechadv.2014.10.004
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shen, R. M., Liu, W., Sun, Y., Li, R., and Xie, Z. H. (2018). Function analysis of flagellar genes fliN and fliM in Azorhizobium caulinodans ORS571. Acta Miicrobiol. Sin. 58, 882–892. doi: 10.13343/j.cnki.wsxb.20170477
Shi, M., Gao, T., Ju, L., Yao, Y., and Gao, H. (2014). Effects of FlrBC on flagellar biosynthesis of Shewanella oneidensis. Mol. Microbiol. 93, 1269–1283. doi: 10.1111/mmi.12731
Subramanian, S., and Kearns, D. B. (2019). Functional regulators of bacterial flagella. Annu. Rev. Microbiol. 73, 225–246. doi: 10.1146/annurev-micro-020518-115725
Sun, Y., Liu, Y., Liu, X., Dang, X., Dong, X., and Xie, Z. (2020). Azorhizobium caulinodans c-di-GMP phosphodiesterase Chp1 involved in motility, EPS production, and nodulation of the host plant. Appl. Microbiol. Biotechnol. 104, 2715–2729. doi: 10.1007/s00253-020-10404-6
Xu, Z., Zhang, H., Sun, X., Liu, Y., Yan, W., Xun, W., et al. (2019). Bacillus velezensis wall teichoic acids are required for biofilm formation and root colonization. Appl. Environ. Microbiol. 85:e2116-18. doi: 10.1128/AEM.02116-18
Keywords: Azorhizobium caulinodans ORS571, transcriptional regulator, flagella, exopolysaccharide, biofilm, symbiosis
Citation: Sun L, Wang D, Yin Z, Zhang C, Bible A and Xie Z (2021) The FtcR-Like Protein ActR in Azorhizobium caulinodans ORS571 Is Involved in Bacterial Motility and Symbiosis With the Host Plant. Front. Microbiol. 12:744268. doi: 10.3389/fmicb.2021.744268
Received: 20 July 2021; Accepted: 20 October 2021;
Published: 19 November 2021.
Edited by:
Robert Czajkowski, University of Gdańsk, PolandReviewed by:
Saswat S. Mohapatra, Berhampur University, IndiaDavid Salvador Zamorano Sanchez, National Autonomous University of Mexico, Mexico
Copyright © 2021 Sun, Wang, Yin, Zhang, Bible and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihong Xie, emhpaG9uZ3hpZTIxMUAxNjMuY29t; Chengsheng Zhang, emhhbmdjaGVuZ3NoZW5nQGNhYXMuY24=
†These authors have contributed equally to this work and share first authorship
 Li Sun1†
Li Sun1† Dandan Wang
Dandan Wang Zhiqiu Yin
Zhiqiu Yin Chengsheng Zhang
Chengsheng Zhang Amber Bible
Amber Bible Zhihong Xie
Zhihong Xie