- 1College of Life Science and Technology, Huazhong Agricultural University, Wuhan, China
- 2CAS Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 3Laboratory of Marine Ecology and Environmental Science, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 4Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 5College of Marine Science, University of Chinese Academy of Sciences, Beijing, China
- 6Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, BC, Canada
Chaetoceros is a species-rich diatom genus with broad distribution and plays an important role in global carbon cycle and aquatic ecosystems. However, genomic information of Chaetoceros species is limited, hindering advanced researches on Chaetoceros biodiversity and their differential impact on ecology. In this study, we constructed full-length chloroplast genomes (cpDNAs) for seven Chaetoceros species, including C. costatus, C. curvisetus, C. laevisporus, C. muelleri, C. pseudo-curvisetus, C. socialis, and C. tenuissimus. All of these cpDNAs displayed a typical quadripartite structure with conserved genome arrangement and specific divergence. The sizes of these cpDNAs were similar, ranging from 116,421 to 119,034 bp in size, and these cpDNAs also displayed similar GC content, ranging from 30.26 to 32.10%. Despite extensive synteny conservation, discrete regions showed high variations. Divergence time estimation revealed that the common ancestor of Chaetoceros species, which formed a monophyletic clade at approximately 58 million years ago (Mya), split from Acanthoceras zachariasii at about 70 Mya. The availability of cpDNAs of multiple Chaetoceros species provided valuable reference sequences for studying evolutionary relationship among Chaetoceros species, as well as between Chaetoceros species and other diatom species.
Introduction
Diatoms (Bacillariophyta) are one of the most diverse lineages of phytoplankton on earth, with approximately 200,000 species (Mann and Droop, 1996; Malviya et al., 2016). As primary producers, they play an important role in aquatic food webs and in biogeochemical cycles (Smetacek, 1998; Armbrust, 2009).
Chaetoceros Ehrenberg is a species-rich genus of the class Mediophyceae (Rine, 1988; Kooistra et al., 2010; Malviya et al., 2016; Gaonkar et al., 2018; De Luca et al., 2019a) with 232 taxonomically accepted species (accessed on June 2021) (Guiry and Guiry, 2021). As one of the largest genera of planktonic diatom, Chaetoceros plays an important role in global carbon cycle and aquatic ecosystems (Nelson et al., 1995). Chaetoceros species play an important role in ecological systems as an important component of natural food webs. As such, some Chaetoceros species are often cultivated to serve as feed for aquaculture of shellfish, shrimp, and fish because of its high nutrition content (Göksan et al., 2003; Liang et al., 2020). Additionally, some Chaetoceros species have been used as biological indicators for studying marine environmental change (Wang et al., 2010). Furthermore, many Chaetoceros species have been applied to remove certain antibiotics from wastewater (Mojiri et al., 2021), and C. muelleri has been exploited as a renewable precursor to liquid fuels or as a lipid source because of its high growth rate, tolerance to a broad range of temperatures, and specific conductance and large quantity of intracellular lipid (McGinnis et al., 1997; López-Elías et al., 2005; Yin and Hu, 2021).
Nevertheless, many Chaetoceros species can also pose negative impact on environment by inducing harmful algal blooms (HABs) under certain circumstances. HABs caused by various Chaetoceros species have been reported in many countries including Japan (Oyama et al., 2008; Tomaru et al., 2011; Tomaru et al., 2017), Spain (Trigueros et al., 2002), the United States (Montresor et al., 2013), India (Begum et al., 2015), and China (Lv et al., 1993; Han et al., 2004; Liu et al., 2006; Wang et al., 2010). Some Chaetoceros species can also negatively impact aquaculture and fisheries (Albright et al., 1993; Treasurer et al., 2003; Begum et al., 2015). For example, Chaetoceros densus has been found to impact the Porphyra yezoensis cultures in Japan (Oyama et al., 2008), and C. convolutus and C. concavicornis can cause fish mortality by anchoring the setae to the sensitive gill tissue (Albright et al., 1993; Treasurer et al., 2003; Wang et al., 2008).
Chaetoceros species are generally easily recognized among diatom species by the chain-forming cells that are separated by apertures, and the long setae protruding from each of the four corners of the cells. A small minority are solitary in their growth form (Round et al., 1990; Li et al., 2017). Nevertheless, Chaetoceros species could not be accurately characterized due primarily to their high morphological similarities. New species (Marino et al., 1991; Rines et al., 2010; Yang et al., 2015) and cryptic species (Chamnansinp et al., 2013, 2015; Balzano et al., 2017; Li et al., 2017) are being uncovered, suggesting that a considerable part of the diversity in the Chaetoceros is still to be revealed.
Molecular markers have been applied to distinguish and describe taxa, including species in Chaetocerotaceae. Gaonkar et al. (2018) used the full-length or partial 18S rDNA and partial 28S rDNA as molecular markers to enable phylogenetic inference of species in Chaetocerotaceae, but often could not be used to accurately distinguish different species of a same genus. Although concatenated alignment of multiple molecular markers such as 18S rDNA, partial 28S rDNA, rbcL, psbA, and partial COI could enhance resolution capability (De Luca et al., 2019b), molecular markers with even higher resolution are urgently needed to enrich public databases for research on biodiversity and evolution.
Chloroplast genomes (cpDNAs) have been used as “super-barcode” for comparative genomics analysis (Fu et al., 2019; Ji et al., 2019). cpDNA has a typical quadripartite structure consisting of one large single copy region (LSC), one small single copy region (SSC), and a pair of inverted repeats (IRs) (Bendich, 2004; Daniell et al., 2016). The complete cpDNAs have been shown to be valuable in inferring evolutionary relationships as an accessible genetic resource (Yu et al., 2018). With the recent development of DNA sequencing technologies, a growing number of cpDNAs of species in Bacillariophyta have been fully constructed (Yu et al., 2018; Yao et al., 2021; Zhang et al., 2021). Comparative analysis of cpDNAs can help us understand the complex evolutionary relationships of algal species. In addition, comparative analysis of cpDNAs can also be applied as an effective method to develop high-resolution molecular markers (Ji et al., 2019; Song et al., 2020).
Fossil evidence suggests that diatoms originated in the late Jurassic period (Finkel et al., 2005; Lewitus et al., 2018). Chaetocerotaceae spores sink out of the water column and can remain dormant in the sediment for prolonged periods, so species in this family exhibit extensive fossil records (Suto, 2006). However, the biodiversity of existent Chaetoceros species has not been adequately explored. Comparative analysis of fossil records and cpDNAs of Chaetoceros species may provide valuable insight into the understanding of origin and evolution of Chaetoceros species.
By now, cpDNAs of only two Chaetoceros species (i.e., C. muelleri and C. simplex) have been constructed (Sabir et al., 2014; Li and Deng, 2021). In this study, we constructed full-length cpDNAs for seven Chaetoceros species, including C. costatus, C. curvisetus, C. laevisporus, C. muelleri, C. pseudo-curvisetus, C. socialis, and C. tenuissimus, all of which were isolated from coastal waters in China. Comparative analysis of these cpDNAs revealed extensive gene and synteny conservation, as well as the identification of several variation hotspots. We also explored phylogenetic analysis and divergence time for Chaetoceros species and other species in the diatom.
Materials and Methods
Strain Isolation and Whole Genome Sequencing
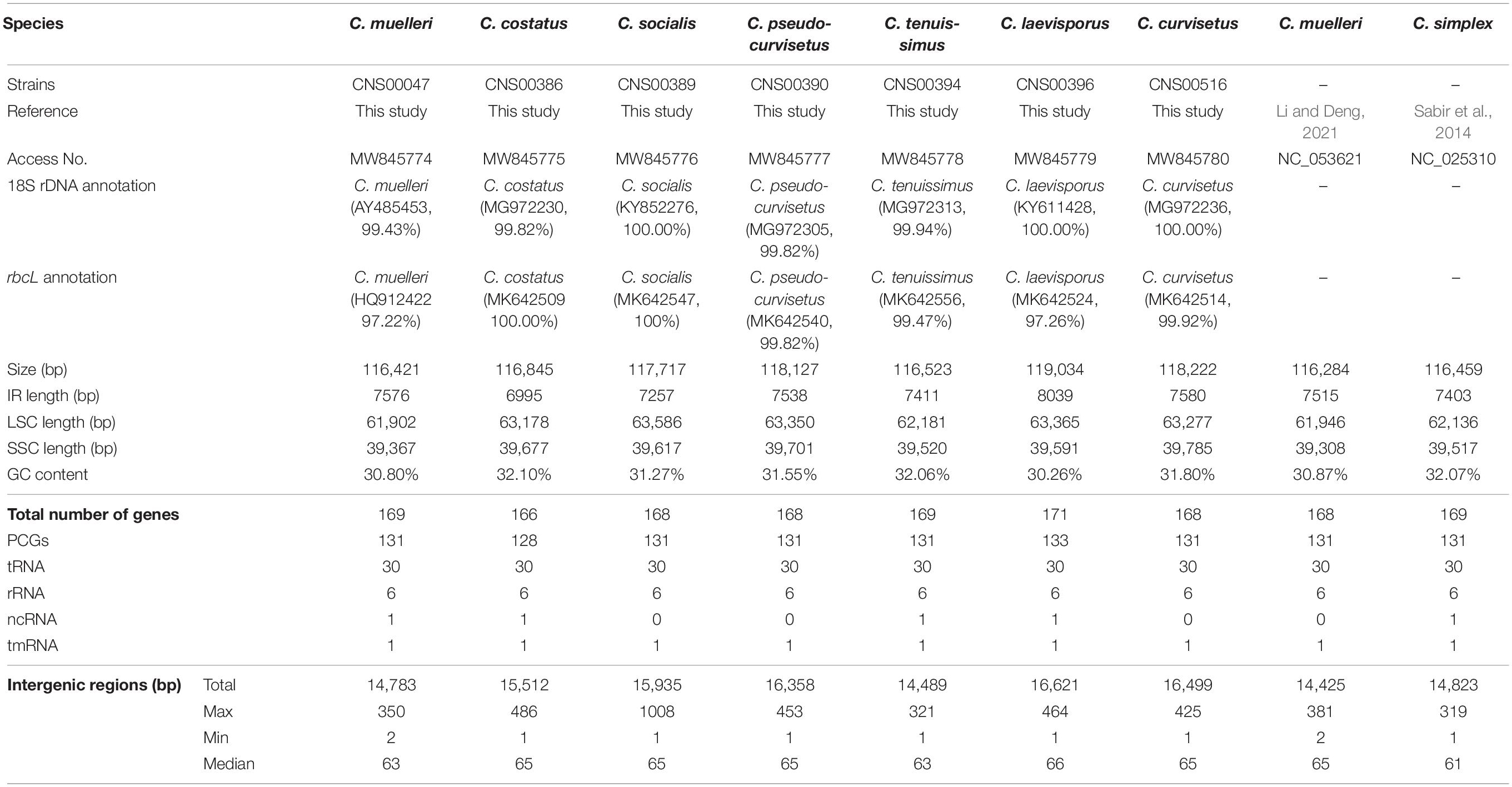
Seven candidate Chaetoceros species studied in this project were isolated from water samples collected during multiple expeditions in Chinese coastal waters, among which CNS00389, CNS00390, and CNS00516 were isolated from the Jiaozhou Bay (July and August, 2019) on the research vehicle “Chuangxin,” CNS00386 and CNS00396 were isolated from the Changjiang Estuary (July, 2019) on the research vehicle “Zheyu 2,” and CNS00394 was isolated from the East China Sea (May, 2019) on the research vehicle “Xiang Yang Hong 18” (Figure 1A and Supplementary Table 1). The Chaetoceros cells were individually isolated using a micropipette, followed by multiple washes before transferring each single cell to 24-well culture dishes for growth and characterization. These Chaetoceros strains were cultured in L1 medium with 1 ‰ volume fraction Na2SiO3 with H2O added (Guillard and Hargreaves, 1994). The culture temperature was set at 19 ± 1°C, and the illumination intensity was from 2000Lx to 3000Lx at the photoperiod of 12 h light-12 h dark.
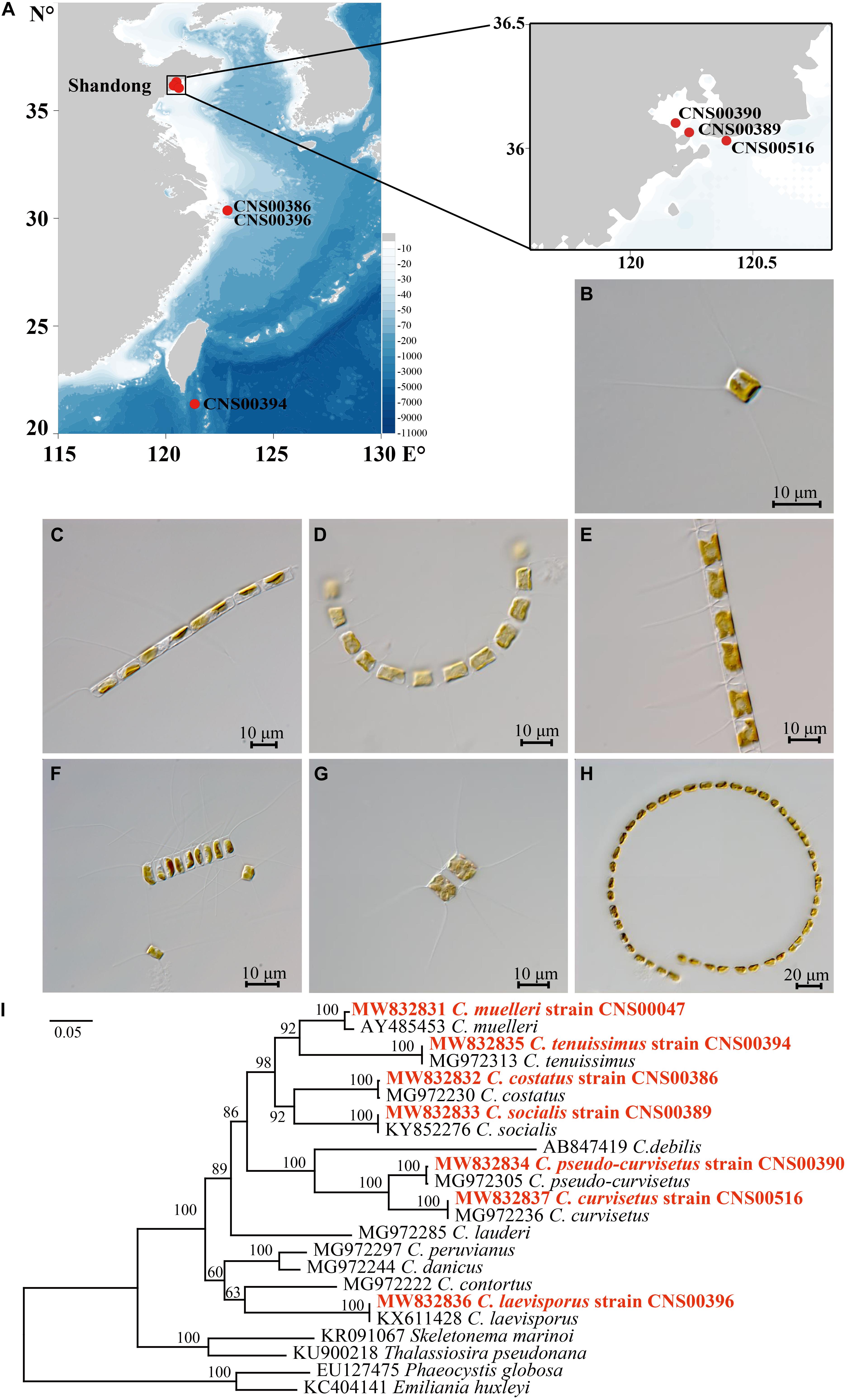
Figure 1. (A) Sampling locations of Chaetoceros species. (B–H) The micrographs of Chaetoceros species. (B) C. muelleri (strain CNS00047). (C) C. costatus (strain CNS00386). (D) C. socialis (strain CNS00389). (E) C. pseudo-curvisetus (strain CNS00390). (F) C. tenuissimus (strain CNS00394). (G) C. laevisporus (strain CNS00396). (H) C. curvisetus (strain CNS00516). (I) The phylogenetic analysis of Chaetoceros species and outgroups (Phaeocystis globosa and Emiliania huxleyi) using full length of 18S rDNA gene.
Identification of the cultured Chaetoceros strains was done according to both microscopic morphological characters and phylogenetic analysis using universal markers, including full-length 18S rDNA, rbcL, and 28S rDNA D1-D3. The morphological features of the Chaetoceros species were observed using ZEISS Axio Imager 2 (ZEISS, Germany). Molecular marker sequences were assembled using Illumina reads with SPAdes (Bankevich et al., 2012) and GetOrganelle (Jin et al., 2020), with publicly available molecular marker sequences of Chaetoceros species as reference sequences. The assembled sequences were validated by the following steps. (1) Reads were aligned to the assembled sequences using BWA (0.7.17) (Li and Durbin, 2009). (2) Alignment results were extracted using SAMtools (1.10) (Li et al., 2009). (3) Resulting alignments were inspected for validation and error correction using IGV (Thorvaldsdottir et al., 2013). Phylogenetic trees based on molecular markers were constructed using MEGAX (Kumar et al., 2018). Phylogenetic relationships were inferred using the Maximum Likelihood (ML) (Tamura et al., 2004). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was shown next to the branches (Felsenstein, 1985).
For DNA library preparation for whole genome sequencing, Chaetoceros cells were collected by centrifugation, and algae mud samples were stored in liquid nitrogen for subsequent DNA extraction. Total DNA was extracted for each sample by using DNAsecure Plant Kit (Tiangen Biotech, Beijing, China). The integrity and purity of DNA were examined by 1% agarose gel electrophoresis and DNA concentration was accurately quantified by Qubit 2.0 Flurometer (Life Technologies, CA, United States). DNA libraries were prepared using NEB NextTM Ultra® DNA Library Prep Kit for Illumina (NEB, United States). PCR products were purified using AMPure XP system (Beckman Coulter, Beverly, MA, United States), libraries were analyzed for size distribution using NGS3K/Caliper and quantified using real-time PCR (Qubit®3.0 Flurometer, Invitrogen, United States). Qualified libraries were sequenced using NovaSeq PE150 (Illumina, San Diego, CA, United States) at Novogene (Beijing, China).
Chloroplast Genome Assembly and Annotation
We obtained an average of 5.85 Gb of Illumina paired-end clean sequencing data from genomic DNA of seven Chaetoceros strains. An average of 19,511,552 paired-end reads were retrieved from each sample, with a sequence length of 150 bp. Raw reads in FASTQ format were first processed through a series of quality control (QC) procedures to obtain clean reads, according to method described previously (Song et al., 2020). Complete cpDNAs were assembled using the GetOrganelle (Jin et al., 2020) with clean reads. Chloroplast genome sequences were verified using the same method used for verifying molecular markers described above in 2.1. Annotation of cpDNA was made using MFannot1 using genetic code of Bacterial, Archaeal, and Plant chloroplast. For genes whose lengths were different from expected, whose start and stop codons were non-canonical, or open reading frames (orfs) that did not show similarity to known genes, Open Reading Frame Finder (ORF finder)2 was applied to examine and edit gene models. Annotated results were further validated and formatted using NCBI’s Sequin15.103. cpDNAs in the Genbank format of the cpDNAs were converted into genome maps by using Organellar Genome DRAW (OGDRAW) online software (Greiner et al., 2019).
Phylogenetic Analysis
Phylogenetic tree based on protein-coding genes (PCGs) was constructed using extracting 95 PCGs (Supplementary Table 2) shared by published Bacillariophyta cpDNAs (Supplementary Table 3) including seven Chaetoceros cpDNAs constructed in this study. The amino acid sequences of each of the 95 PCGs from different diatom cpDNAs were individually aligned using MAFFT v7.310 (–auto) (Katoh and Standley, 2013). Regions that were ambiguously aligned in each alignment were deleted and all amino acid sequences were concatenated using PhyloSuite v1.2.2 (Zhang et al., 2020). Phylogenetic tree was constructed using IQ-TREE v1.6.1 with SH-aLRT support (%)/aBayes support/ultrafast bootstrap support (%) (parameters: -st AA -m TEST -bb 1000 -alrt 1000 -abayes) (Trifinopoulos et al., 2016). Triparma laevis (AP014625) in Ochrophyta was included as out-group taxa.
Genome Comparison
Alignment of Chaetoceros cpDNAs were performed by using Mauve v2.4.0 (Darling et al., 2010) with default parameters. The cpDNAs borders were analyzed to show the IR expansions and contractions using irscope_pack.3.1 (modified from IRscope) (Amiryousefi et al., 2018).
Identification of Variation Hotspot Regions
Chaetoceros cpDNAs were aligned using MAFFT v7.310 (Katoh and Standley, 2013). Nucleotide diversity (Pi), which could be used to estimate the degree of nucleotide sequence variations, which could be used as potential molecular markers, was calculated using the software DnaSP v6.12.03 (Rozas et al., 2017) and cpDNA alignment as input. The window size was set to 600 bp and the step size was 50 bp.
Divergence Time Estimations
Divergence time estimation was performed by 95 PCGs (Supplementary Table 2) shared by the published 55 Bacillariophyta cpDNAs (Supplementary Table 3) and seven Chaetoceros cpDNAs constructed in this study using MCMCTree in PAML v4.8a (Yang, 2007). Branch lengths, gradient (g) and Hessian (H) were estimated using maximum likelihood estimates (MLE) and GTR + G substitution model (model = 7) with independent rates clock model (clock = 1). Three calibration points4 were included in this analysis (Supplementary Table 4), including the calibration point between Ectocarpus siliculosus and diatoms [176.0–202.0 Million years ago (Mya)], the calibration point between Rhizosolenia setigera and Skeletonema pseudocostatum (90.5–91.5 Mya), and the calibration point between Pseudo-nitzschia multiseries and Fragilariopsis cylindrus (10.0–35.3 Mya). The phylogenetic tree was displayed using FigTree v1.4.3 and visualized with 95% highest posterior density interval (HPD) for each node.
Results
Morphological and Molecular Identification of Seven Chaetoceros Species
All seven Chaetoceros species studied in this project formed chains in which cells were separated by apertures, with long setae protruding from each of the four corners of the cells. These Chaetoceros strains all displayed substantial morphological variations (Li et al., 2017; Xu et al., 2020). The strain CNS00047 was annotated as C. muelleri because these cells were rectangular with long setae, with valve diameter varying from 4.5 to 20.0 μm (Figure 1B), similar to previous description of C. muelleri (Reinke, 1984). Phylogenetics analysis of full-length 18S rDNA sequences of these candidate Chaetoceros species and reference sequences of known Chaetoceros species confirmed that the strain CNS00047 was C. muelleri because its 18S rDNA sequence (MW832831) clustered well with that (AY485453) of C. muelleri (Damste et al., 2004) with high percentage identity (PID) 99.43% (Figure 1I and Table 1). This annotation was also supported by phylogenetic analysis using another molecular marker rbcL (Supplementary Figure 1A and Table 1). The strain CNS00386 was annotated as Chaetoceros costatus, which contained a single, lobed plastid, formed straight chains (Figure 1C; Kooistra et al., 2010). The strains CNS00389 was annotated as C. socialis, which was fan-shaped, with one of the four setae longer than the others and the long setae of adjacent cells joining together (Figure 1D; Degerlund et al., 2012; Pelusi et al., 2019). The strain CNS00390 was annotated as C. pseudo-curvisetus, whose chains were curved, with a large aperture between adjacent cells, and the aperture was large in the middle and small on the sides (Figure 1E; Oku and Kamatani, 1990). The strain CNS00394 was annotated as C. tenuissimus, whose cells were very small, being square to rectangular, with setae being narrow, arising from the two poles of the valve at an angle of 45° to its apical axis (Figure 1F; Sar et al., 2002). The strain CNS00396 was annotated as C. laevisporus, whose cells contained multiple plastids, were rectangular in broad girdle view and formed straight chains (Figure 1G; Chen et al., 2019). The strain CNS00516 was annotated as C. curvisetus, whose chains were helical, with a large elliptical aperture between adjacent cells. Each cell contained only a single plastid, and all setae curve toward the convex side of the chain (Figure 1H). The strains CNS00386, CNS00389, CNS00390, CNS00394, CNS00396, and CNS00516 were further confirmed as C. costatus (Gaonkar et al., 2018), C. socialis (Gaonkar et al., 2017), C. pseudo-curvisetus (Gaonkar et al., 2018), C. tenuissimus (Gaonkar et al., 2018), C. laevisporus (Li et al., 2017), and C. curvisetus (Gaonkar et al., 2018), respectively, according to their molecular features (Figure 1, Supplementary Figure 1, and Table 1).
Construction and Comparative Analysis of Chloroplast Genomes
We constructed full-length cpDNAs for seven Chaetoceros species, among which cpDNAs of six Chaetoceros species (C. costatus, C. curvisetus, C. laevisporus, C. pseudo-curvisetus, C. socialis, and C. tenuissimus) were constructed for the first time. Together with two cpDNAs of C. muelleri and C. simplex that have been previously published (Sabir et al., 2014; Li and Deng, 2021), altogether nine cpDNAs representing eight Chaetoceros species (Table 1) were analyzed in this project. The sizes of these nine Chaetoceros cpDNAs were rather similar, ranging from 116,284 bp (C. muelleri; NC_053621) to 119,034 bp (C. laevisporus; MW845779) (Figure 2A, Supplementary Figure 2, and Table 1). The GC contents of these cpDNAs were also similar (30.26–32.10%). These Chaetoceros cpDNAs all formed typical quadripartite structure with two inverted repeats regions (IRa, IRb), a large single copy (LSC) region, and a small single copy (SSC) region (Figure 2 and Supplementary Figure 2). The lengths of LSC regions of these cpDNAs were similar (ranging from 61,902 to 63,586 bp), so were their SSC regions (ranging from 39,367 to 39,785 bp). In contrast, the lengths of IR regions showed larger variations among these cpDNAs, with the shortest being 6995 bp (C. costatus), while the longest being 8039 bp (C. laevisporus) (Table 1).
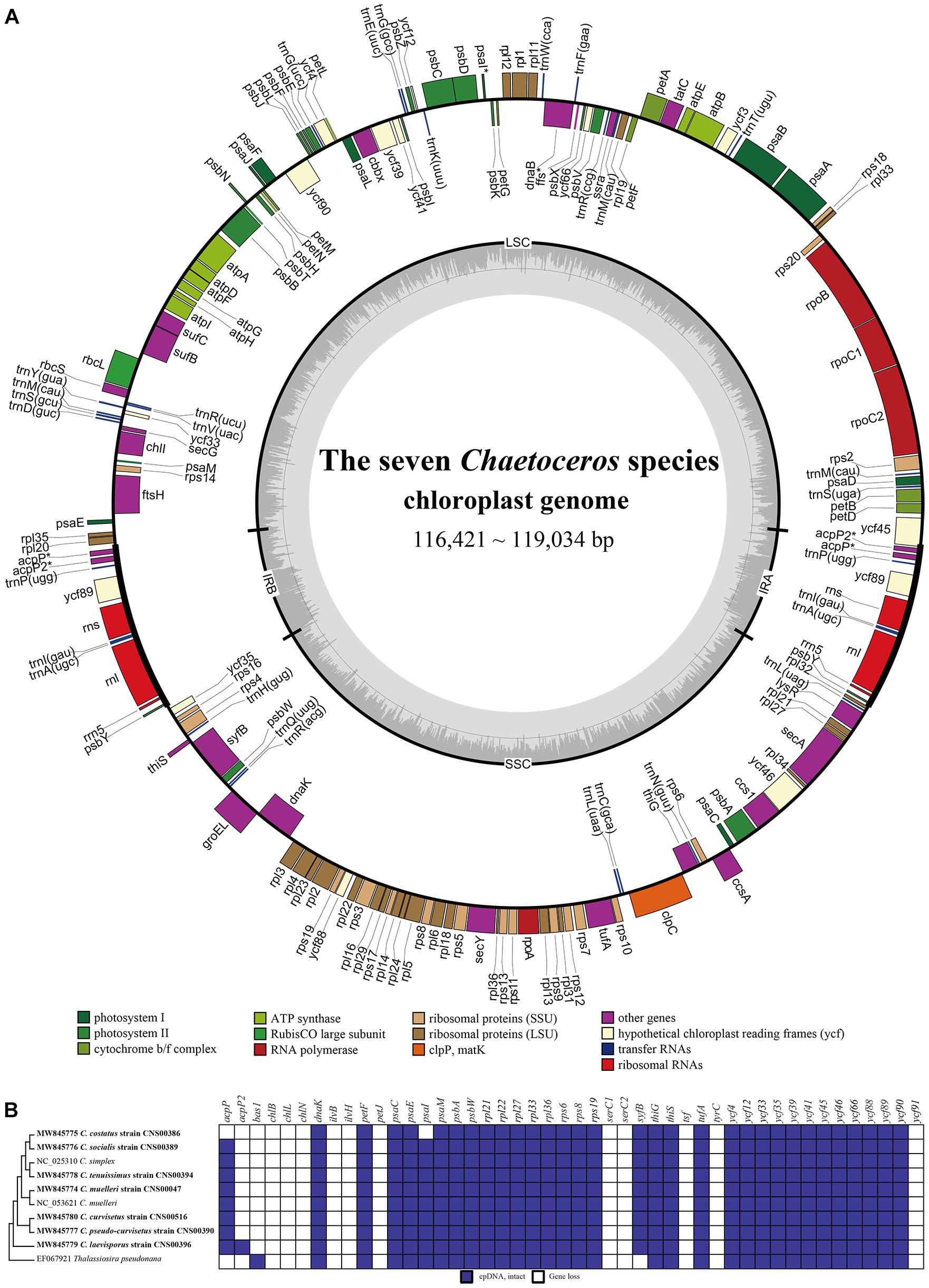
Figure 2. Gene structural and evolutionary patterns of genes of the Chaetoceros cpDNAs. (A) Map of cpDNAs of the seven Chaetoceros. Dashed area in the inner circle indicates the GC content of the cpDNA. Genes belonging to different functional groups are color-coded as indicated by icons on the lower. The asterisk represents differences between the seven Chaetoceros. (B) Evolutionary patterns of loss, and gain of genes in Chaetoceros cpDNAs. The matrix shows 46 genes variably present among the sequenced genomes. Taxa in boldface identify genomes sequenced for this study. Phylogenetic relationships based on concatenated amino acid sequences of protein-coding genes using Maximum likelihood (ML) methods. T. pseudonana was used as out-group taxa.
Annotation of these cpDNAs revealed that the cpDNAs of five species, including C. curvisetus, C. muelleri, C. pseudo-curvisetus, C. socialis, and C. tenuissimus, each contained 131 PCGs. In contrast, the cpDNAs of C. costatus and C. laevisporus contained different numbers of PCGs, and C. costatus and C. laevisporus contained 128 and 133 genes, respectively (Figure 2A, Supplementary Figure 2, and Table 1). All Chaetoceros cpDNAs contained 30 tRNA and six non-coding rRNA genes (rns, rnl, and rrn5 in IRs) (Figure 2A, Supplementary Figure 2, and Table 1). No introns were found in any of the cpDNAs of these seven Chaetoceros species, which was consistent to previous findings that no introns were identified in cpDNAs of C. simplex (NC_025310) and C. muelleri (NC_053621).
A comparison of 46 genes variably present among the diatom cpDNAs between these nine Chaetoceros strains and Thalassiosira pseudonana revealed many instances of gene gains and losses (Figure 2B). Except for acpP, acpP2, and psaI, the presence or absence of genes in Chaetoceros was generally consistent. These events included peroxiredoxin gene (bas1), three genes encoding subunits of protochlorophyllide reductase (chlB/L/N), the large and small subunits of acetolactate synthase (ilvB/H), cytochrome C6 gene (petJ), two putative serine recombinase genes (serC1 and serC2), putative tyrosine recombinase gene (tyrC), florigen genes (tsf), and hypothetical protein ycf91 (Figure 2B).
These cpDNAs were rather compact, with small intergenic regions, and the median lengths of intergenic regions ranged from 63 to 66 bp. The total lengths of intergenic regions of these nine cpDNAs were similar, ranging from 14,425 bp (12.4% of the total cpDNA of C. muelleri; NC_053621) to 16,621 bp (14.0% of the total cpDNA of C. laevisporus).
Phylogenetic Analysis of Chaetoceros Chloroplast Genomes
To explore the evolutionary relationship between Chaetoceros species and other diatom species, we constructed a phylogenetic tree using 95 PCGs (Supplementary Table 2) that were shared by 62 cpDNAs constructed for Bacillariophyta species (including six sp.). The cpDNA of T. laevis (AP014625), which belonged to the class Bolidophyceae in phylum Ochrophyta, was included as an out-group taxon (Figure 3). All Bacillariophyta species were clustered into three major clades in the phylogenetic tree, corresponding to three classes including Mediophyceae, Bacillariophyceae, and Coscinodiscophyceae, respectively (Figure 3). As expected, all nine cpDNAs of the Chaetoceros species clustered into a single clade. In particular, the cpDNA of C. muelleri (MW845774) and that of C. muelleri (NC_053621) clustered together, and the cpDNA of C. tenuissimus (MW845778) clustered closely with that of C. simplex (NC_025310) (Gaonkar et al., 2018; Figure 3). Additionally, cpDNAs of C. curvisetus and C. pseudo-curvisetus clustered closely, which was consistent with previous report (Gaonkar et al., 2018). In contrast, cpDNAs of C. laevisporus formed an independent clade (Li et al., 2017). The Chaetoceros clade clustered closely with Acanthoceras zachariasii (Chaetocerotaceae), which was consistent with previous study (Yu et al., 2018; Li and Deng, 2021).
Figure 3. The phylogenetic tree based on concatenated amino acid sequences of 95 shared protein-coding genes using Maximum likelihood (ML) methods. Triparma laevis was used as out-group taxa. Numbers on the branches represent SH-aLRT support (%), aBayes support, and ultrafast bootstrap support (%), respectively. Support values are shown only for branches that did not come with high confidence level (100/1/100). Thick branches indicate high confidence level (100/1/100).
Synteny Analysis of Chaetoceros Chloroplast Genomes
Comparative analysis of Chaetoceros cpDNAs revealed near perfect synteny among nine cpDNAs of eight Chaetoceros species (Figure 4A). All genes in the nine cpDNAs exhibited nearly identical gene order (Figure 4B), with only four minor differences identified. First, while a single gene acpP (234–246 bp) was found between rpl20 and trnP in the IRb in the cpDNAs of C. curvisetus, C. muelleri (both MW845774 and NC_053621), C. pseudo-curvisetus, C. simplex, C. socialis, and C. tenuissimus, two genes acpP (240 bp) and acpP2 (267 bp) were found in the corresponding region in the cpDNA of C. laevisporus, and no genes were found in the same region in the cpDNA of C. costatus. Second, the same difference was also found in the IRa. Third, while a single protein-associated ncRNA gene ffs (109 bp) was found between the two genes psbX and trnF in the cpDNAs of C. costatus, C. laevisporus, C. muelleri (MW845774), C. simplex, and C. tenuissimus, it was not found in the corresponding region in the cpDNAs of C. curvisetus, C. muelleri (NC_053621), C. pseudo-curvisetus, and C. socialis. Notably, cpDNAs of two C. muelleri strains were different at this site as well, while ffs was found in C. muelleri (MW845774), it was not found in the cpDNA of C. muelleri (NC_053621), suggesting that this site was highly polymorphic. Lastly, the gene psaI was found in all cpDNAs studied in this project, except the C. costatus cpDNA.
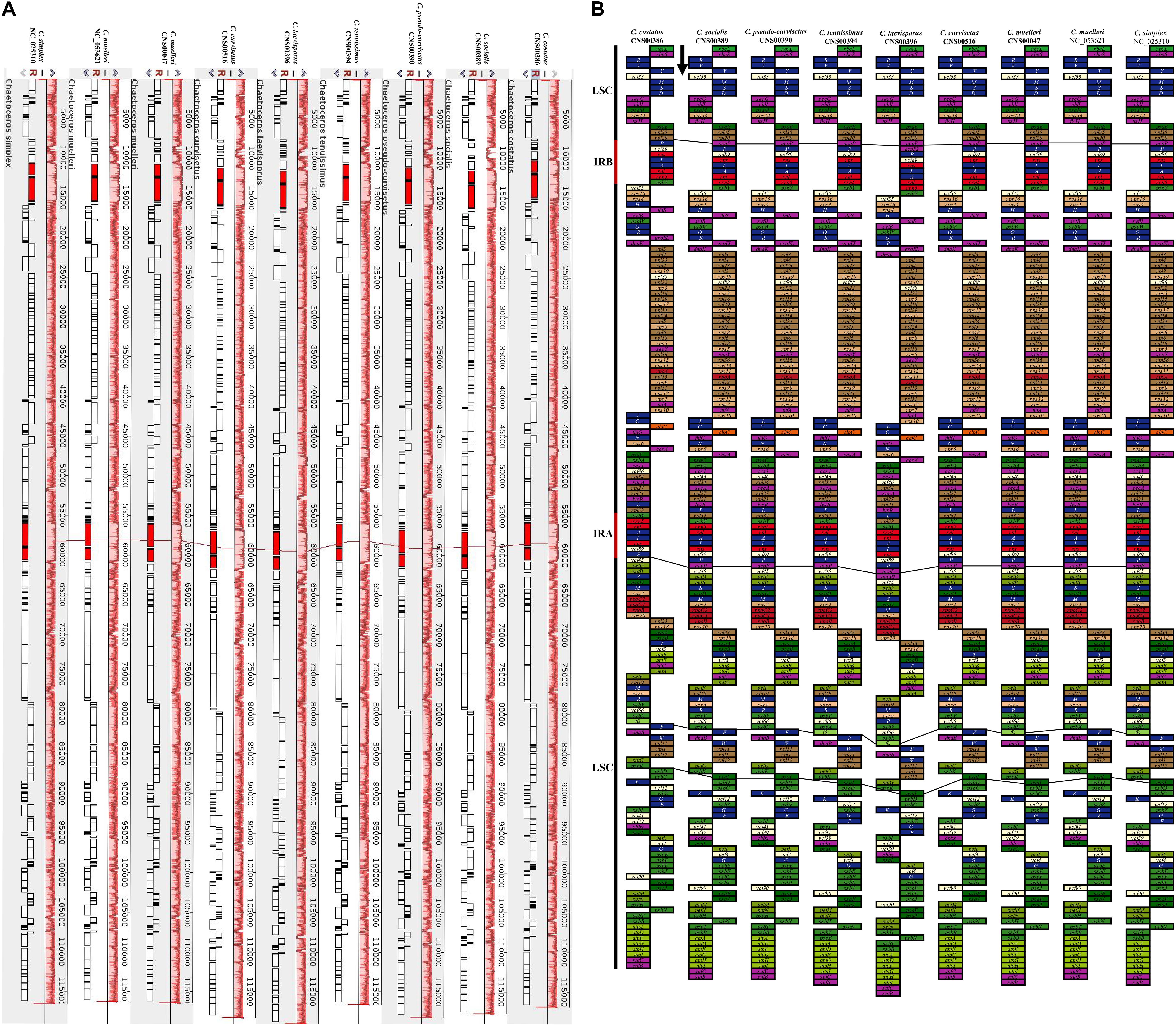
Figure 4. Synteny relationships and gene arrangements of Chaetoceros. (A) Synteny relationships among Chaetoceros cpDNAs. (B) Gene arrangements of Chaetoceros species. Blocks with the same color represent the same type of genes. The black arrow indicates transcription direction.
Expansion and Contraction of Inverted Regions
Comparative analysis of the nine Chaetoceros cpDNAs revealed that the IR regions were generally similar, but with important differences (Figure 5). In particular, the IRa/LSC and IRb/SSC boundaries were different among these cpDNAs. Except for the C. costatus cpDNA, the distances between rpl20 and the LSC/IRb boundaries ranged from 38 to 956 bp, while the distances between acpP and the LSC/IRb boundaries ranged from 52 to 263 bp. The distances between psbY and the SSC/IRb boundaries ranged from 54 to 176 bp, while the distances between ycf35 and the SSC/IRb boundaries ranged from 0 to 117 bp. Except for the C. simplex cpDNA, the C. socialis cpDNA, and the C. tenuissimus cpDNA, rpl32 was located at the SSC/IRa boundaries. Except for the C. costatus cpDNA and the C. socialis cpDNA, ycf45 was located at the SSC/IRa boundaries (Figure 5). Because of the loss of the acpP gene in the C. costatus cpDNA, the boundary between the LSC/IRb shifted, causing a contraction of both IRa and IRb regions. The IRa and IRb of the C. costatus cpDNA each contained eight genes (trnP-UGG, ycf89, rns, trnI-GAU, trnA-UGC, rnl, rrn5, and psbY), compared to nine genes in the cpDNAs of C. curvisetus, C. muelleri, C. pseudo-curvisetus, C. simplex, C. socialis, and C. tenuissimus. In contrast, the IRa and IRb of the C. laevisporus cpDNA each contained 10 genes.

Figure 5. Comparison of the junctions between the LSC, SSC, and IR regions among Chaetoceros cpDNAs.
Variation Hotspots in the Chloroplast Genomes of Chaetoceros Species
Although these nine Chaetoceros cpDNAs showed generally high collinearity, local regions of these cpDNA sequences showed substantial variations at the DNA level. To quantify sequence divergence in these Chaetoceros cpDNAs, we calculated and compared the nucleotide diversity (Pi) values of the Chaetoceros cpDNAs with a window size of 600 bp and a step size of 50 bp. Pi values ranged from 0.0031 to 0.3442. In this analysis, 62 windows were found to have high nucleotide diversity, with Pi greater than 0.25. The main regions contained syfB, ropC2, rpoB genes, and acpP-ycf45 (Figure 6A). Based on the total number of single nucleotide variations (SNVs) and gaps in each window and the ability to distinguish different Chaetoceros species, we identified a hotspots region with 354 SNVs and no gaps (position: 19,025–19,624 bp in C. muelleri cpDNA), which contained a gene syfB (Figure 6B).
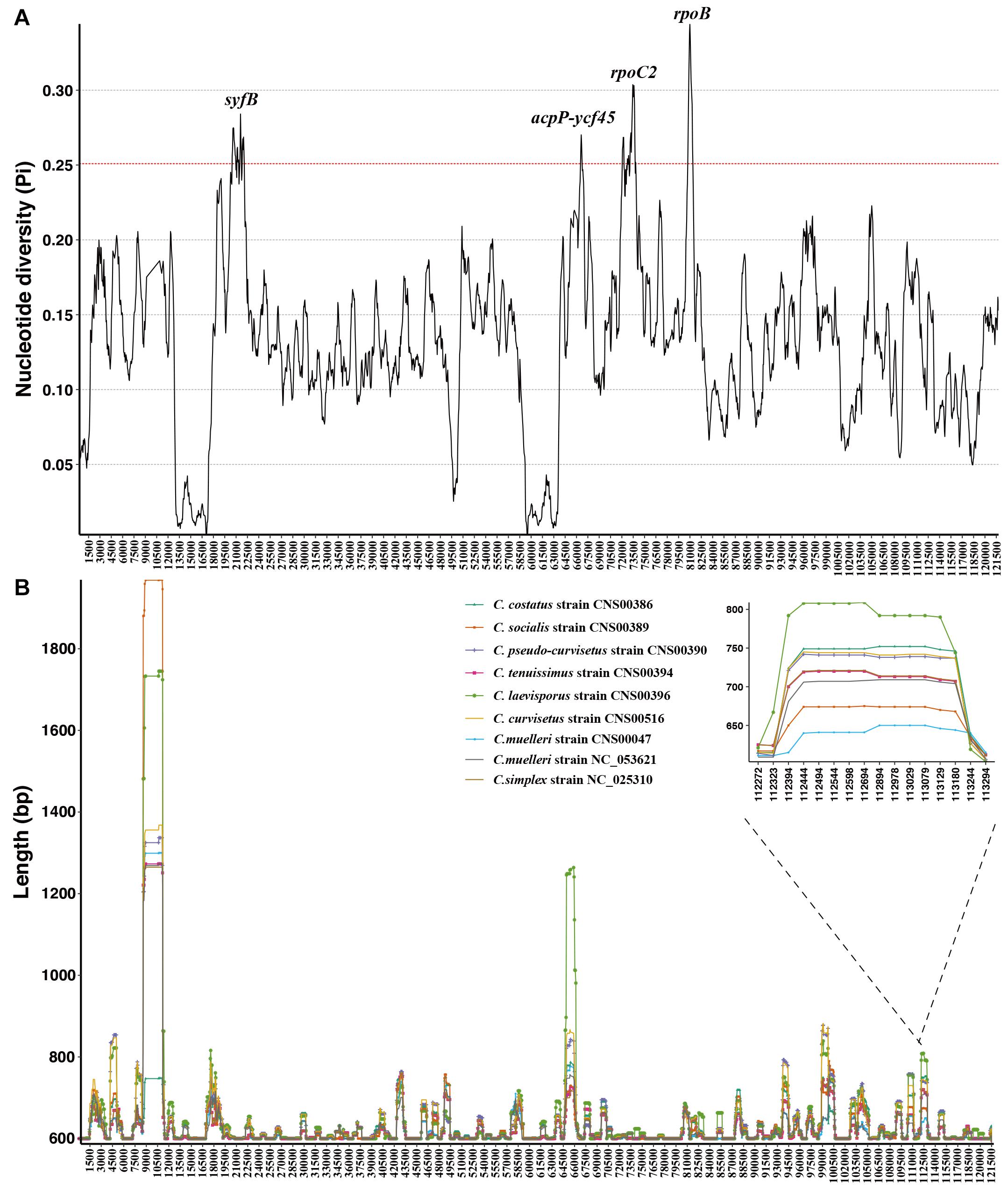
Figure 6. Sliding window analysis of aligned whole cpDNAs of the Chaetoceros. Window length, 600 bp; step size, 50 bp. (A) Based on nucleotide diversity values and (B) the distribution of indel.
To evaluate the resolution power of the hotspots region as a molecular marker, we carried out phylogenetic analysis using it and the result showed that the hotspots region could be used as molecular markers to distinguish different Chaetoceros species (Supplementary Figures 3A, 4A).
Furthermore, based on the sliding window analysis, we also showed distribution of variations of the Chaetoceros cpDNAs. We calculated the actual sequence length of each Chaetoceros cpDNA in each window. We identified a region with high presence and absence variations (ranging from 806 to 961 bp), which corresponded to the region spanning 106,895–107,700 bp in the C. muelleri cpDNA. Phylogenetic analysis of this region showed that it represented a mutation hotspot, which could be used as a potential molecular marker to distinguish different Chaetoceros species (Supplementary Figures 3B, 4B).
Divergence Time Estimation Based on Protein-Coding Genes of Chloroplast Genomes
Divergence time estimation of the Chaetoceros species was achieved by analyzing DNA sequences of 95 PCGs shared by 62 cpDNAs (Figure 7). The branching of the class Coscinodiscophyceae was estimated to have occurred 131 Million years ago (Mya). The two classes Mediophyceae and Bacillariophyceae were estimated to have separated from their common ancestor 101 Mya. Furthermore, divergence time estimation revealed that the common ancestor of Chaetoceros species, which formed a monophyletic clade at approximately 58 Mya, split from A. zachariasii (Chaetocerotaceae) at about 70 Mya. Among the Chaetoceros species, the age estimate for C. laevisporus was 58 Mya. The divergence time between C. costatus and C. socialis was inferred to have occurred at 33 Mya. The branching of C. muelleri was estimated to have occurred 37 Mya and diverged into different strains at 14 Mya. And the divergence time between C. curvisetus and C. pseudo-curvisetus was inferred to have occurred at about 18 Mya. While the divergence time between C. simplex and C. tenuissimus was estimated at 5 Mya. Taken together, the majority of these Chaetoceros species arose within 50 Mya.
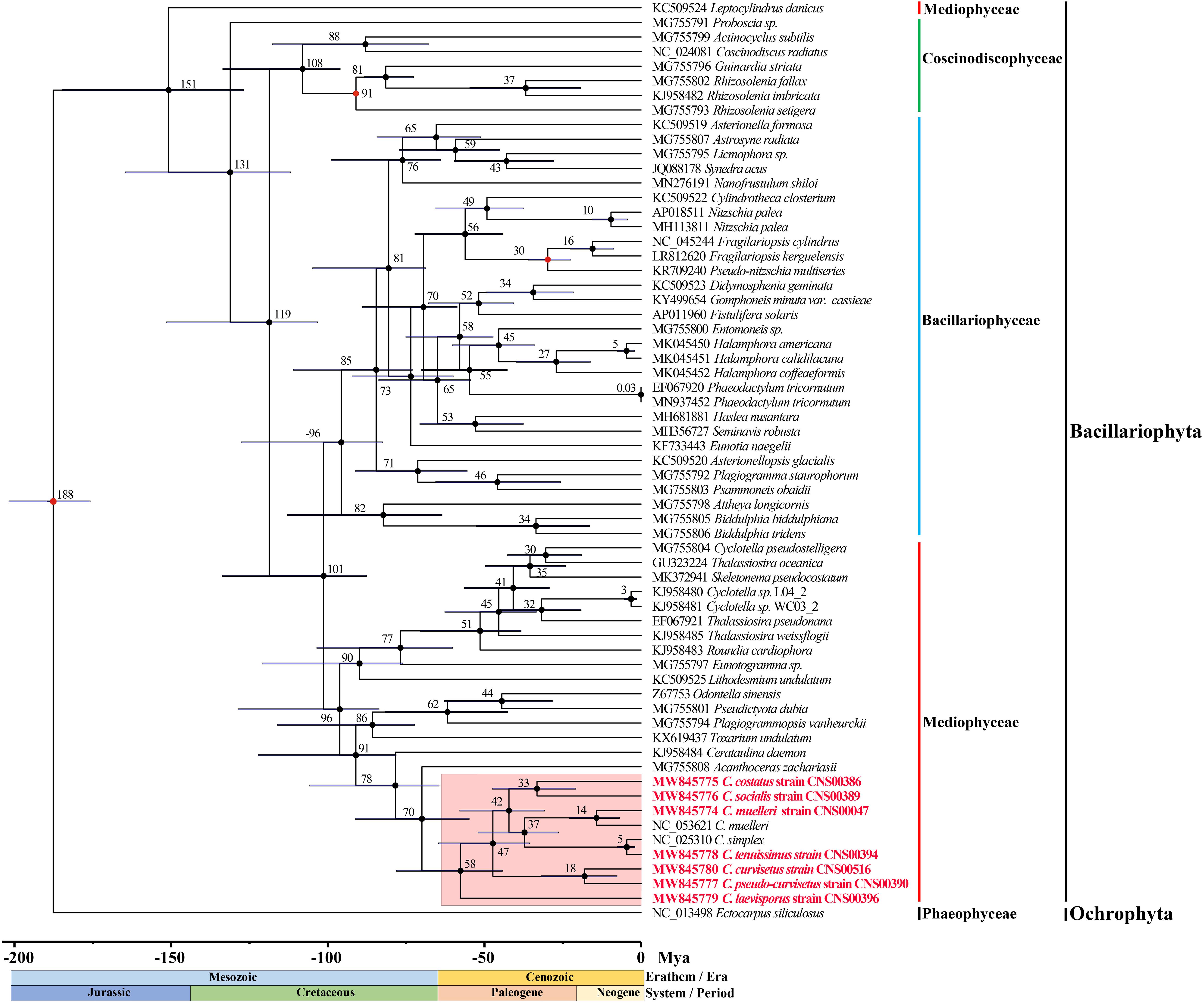
Figure 7. Time-calibrated phylogeny of 62 species based on 95 shared PCGs in the diatoms and outgroup (Ectocarpus siliculosus). The red dots represent calibration point and the 95% highest posterior density interval for node ages are shown with translucent black bars.
Discussion
In this study, cpDNAs of seven Chaetoceros species were constructed for the first time, increased the number of Chaetoceros cpDNAs from two to nine. This research represented a major step toward in-depth understanding of biodiversity, ecology, and speciation of Chaetoceros, which is a species-rich, widespread and abundant diatom genus and plays an important role in global carbon cycle and aquatic ecosystems (Nelson et al., 1995; De Luca et al., 2019a).
General features of cpDNAs of Chaetoceros species constructed in this study were comparable to that of cpDNAs of other diatom species, whose cpDNAs vary widely in size, ranging from 111,539 bp in Pseudo-nitzschia multiseries (Cao et al., 2016) to 201,816 bp in Plagiogramma staurophorum (Yu et al., 2018). The sizes of cpDNAs of Chaetoceros species were generally similar, ranging from 116,421 bp to 119,034 bp. IR contraction and expansion, gene loss and gain, presence and absence of introns, and the variation of intergenic regions are the major factors contributing to variations in the sizes of cpDNAs (Zhu et al., 2016). Comparative analysis revealed that the variation of Chaetoceros cpDNA lengths was mainly driven by the variations of IR lengths and intergenic regions. The IR regions of the Chaetoceros cpDNA varied from 6995 bp in C. costatus to 8039 bp in C. laevisporus (Table 1). Moreover, the total length of intergenic regions of the Chaetoceros cpDNA ranged from 14,489 bp in C. tenuissimus to 16,621 bp in C. laevisporus. No introns were found in any of these Chaetoceros cpDNAs, which was consistent to previous reports that introns are rare in diatom cpDNAs (Ruck et al., 2014).
In addition to variations in the IR and intergenic regions, multiple instances of genes were found to be variable in the Chaetoceros cpDNAs. Compared to these five cpDNAs with 131 PCGs, the cpDNA of C. costatus lost psaI from LSC and acpP from each of its two IRs. The loss of the photosynthetic gene psaI was an rare event but has been reported for other photosynthetic organisms including R. imbricate (Sabir et al., 2014) and R. fallax (Yu et al., 2018), both of which are species in the class Coscinodiscophyceae. The loss of acpP is a common event which has been reported in Thalassiosira species, Cyclotella species, and Synedra acus (Galachyants et al., 2011; Sabir et al., 2014; Yu et al., 2018). In contrast, the C. laevisporus cpDNA gained an extra acpP2 gene in each of the two IRs, which encoded proteins with low percentage identify (34.72%), suggesting an ancient duplication event, similar to acpP and acpP2 reported in the cpDNAs in the cpDNAs of Lithodesmium undulatum, Asterionella formosa, and Eunotia naegelii (Ruck et al., 2014).
The presence or absence of genes in Chaetoceros were generally consistent, suggesting that these events may have occurred in the common ancestors of Chaetoceros species. The synteny of complete Chaetoceros cpDNAs was highly conserved, which was not unexpected because a previous study found high synteny conservation between the cpDNAs of Thalassiosirales species and non-Thalassiosirales species (Sabir et al., 2014). Our analysis found that Chaetoceros cpDNAs contained similar numbers of PCGs and non-coding genes with only minor exceptions. The C. costatus cpDNA lacked acpP (in IR) and psaI, while the C. laevisporus cpDNA had an extra acpP2 gene (in IR). It is well known that cpDNA genes tend to undergo a sequential process of transfer from the chloroplast to the nucleus (Yu et al., 2018). BLASTP searches of acpP (77 aa) and psaI (36 aa) in the assembled nuclear genome of C. costatus (CNS00386) and identified two putative hits with PID of 54.7% and 50.7% to acpP in the nuclear genome assembly, respectively, one putative hit with PID of 80.6% to psaI in the nuclear genome. The absence of acpP and psaI from the C. costatus cpDNA and the presence of their potential homologs in the nuclear genome suggested that these genes could have been transferred to the host genome.
Despite high synteny of the Chaetoceros cpDNAs, some high variation regions were found in DNA sequences (Figure 6). Such a region (corresponding to 19,025–19,624 bp in C. muelleri cpDNA) with great sequence difference might be the relatively ideal marker to distinguish Chaetoceros species (Supplementary Figure 3A). Another region (corresponding to 106,895–107,700 bp in C. muelleri cpDNA) could also be applied as a molecular marker for distinguishing Chaetoceros species (Supplementary Figure 3B). These potential molecular markers could be valuable because even though Chaetoceros species are usually easily recognized to genus level for their morphological features, precise species identification can be challenging because of morphological variations (Li et al., 2017; Xu et al., 2020). Common molecular markers including full-length 18S rDNA usually do not have adequate resolution for distinguishing Chaetoceros species, molecular markers with higher resolution and specificity are urgently needed. Thus, these variable regions identified in this study could be applied used as potential molecular markers that have both high specificity to Chaetoceros species and high resolution for distinguishing closely related Chaetoceros species.
Based on the phylogenetic tree of species in diatoms of 95 core PCGs in cpDNAs, we found that the first event of diversification within the diatoms occurred 188 Mya (95% HPD: 175.8–201.8 Mya) (Figure 7). Previous research suggests that diatoms arose in the lower Triassic period, perhaps as early as 250 Mya according to the molecular clock estimate (Sims et al., 2006; Lewitus et al., 2018). Other studies have suggested that the first diatom lineage is likely to have evolved any time between 183 –250 Mya ago based on 18S rDNA gene (Sorhannus, 2007), which was between the Early Triassic and Early Jurassic. Furthermore, the results suggest that most diatoms occurred Paleogene period (28–66 Mya) with many Chaetoceros species arose within 50 Mya. The Chaetoceros species were closely related to the A. zachariasii (Chaetocerotaceae), which was consistent with previous studies (Matari and Blair, 2014; Yu et al., 2018; Li and Deng, 2021). The branching of Chaetoceros species was estimated to have occurred 58 Mya. However, previous studies have reported that Chaetoceros species was estimated to have occurred at around 90 Mya with the research based on 18S rDNA gene (Sorhannus, 2007). Among the Chaetoceros genus, the strains CNS00047 and NC_053621 (Li and Deng, 2021) identified as C. muelleri was sister clade as expect, but we also found genetic distance between the two strains (Figure 3). The branching of C. muelleri was estimated to have occurred 37 Mya and diverged into different strains at 14 Mya (95% HPD: 6.9–22.0 Mya) (Figure 7), suggesting that these two C. muelleri strains could represent two distinct Chaetoceros species. Chaetoceros simplex diverged from C. tenuissimus approximately 5 Mya (95% HPD: 1.9–7.6 Mya). This study provided the divergence time among the Chaetoceros species based on the cpDNAs for the first time.
Conclusion
In this study, we successfully constructed the full-length cpDNAs for seven Chaetoceros species. The Chaetoceros cpDNAs ranged from 116,421 to 119,034 bp in size and displayed similar GC content of 30.26–32.10%. Comparative analysis of these cpDNAs revealed extensive gene and synteny conservation, as well as the presence of hotspot regions with high variations. Moreover, our study explored phylogenetic and divergence times for Chaetoceros species and other species in the diatom.
Data Availability Statement
The original contributions presented in the study are publicly available. These data can be found here: https://www.ncbi.nlm.nih.gov/sra/PRJNA745567 and https://www.ncbi.nlm.nih.gov/nuccore/MW845774; https://www.ncbi.nlm.nih.gov/nuccore/MW845775; https://www.ncbi.nlm.nih.gov/nuccore/MW845776; https://www.ncbi.nlm.nih.gov/nuccore/MW845777; https://www.ncbi.nlm.nih.gov/nuccore/MW845778; https://www.ncbi.nlm.nih.gov/nuccore/MW845779; https://www.ncbi.nlm.nih.gov/nuccore/MW845780; https://www.ncbi.nlm.nih.gov/nuccore/MW832831; https://www.ncbi.nlm.nih.gov/nuccore/MW832832; https://www.ncbi.nlm.nih.gov/nuccore/MW832833; https://www.ncbi.nlm.nih.gov/nuccore/MW832834; https://www.ncbi.nlm.nih.gov/nuccore/MW832835; https://www.ncbi.nlm.nih.gov/nuccore/MW832836; https://www.ncbi.nlm.nih.gov/nuccore/MW832837; https://www.ncbi.nlm.nih.gov/nuccore/MZ267682; https://www.ncbi.nlm.nih.gov/nuccore/MZ267683; https://www.ncbi.nlm.nih.gov/nuccore/MZ267684; https://www.ncbi.nlm.nih.gov/nuccore/MZ267685; https://www.ncbi.nlm.nih.gov/nuccore/MZ267686; https://www.ncbi.nlm.nih.gov/nuccore/MZ267687.
Author Contributions
NC conceived of the project and revised the manuscript. ZC carried out strain selection, cultivation, DNA preparation, and organized genome sequencing. QX carried out genome assembly, annotation, quality control, and comparative analysis of cpDNAs. QX and ZC wrote the manuscript. All authors read and approved the final version of the manuscript.
Funding
This research was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDB42000000), Chinese Academy of Sciences Pioneer Hundred Talents Program (to NC), Taishan Scholar Project Special Fund (to NC), Qingdao Innovation and Creation Plan (Talent Development Program – 5th Annual Pioneer and Innovator Leadership Award to NC, 19-3-2-16-zhc), and National Key Research and Development Program of China (2017YFC1404300).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are thankful to all members of the Marine Ecological and Environment Genomics Research Group at Institute of Oceanology, Chinese Academy of Sciences.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.742554/full#supplementary-material
Supplementary Figure 1 | The phylogenetic analysis of Chaetoceros species using full length of (A) rbcL gene and (B) partial 28S rDNA.
Supplementary Figure 2 | Gene structural map of the seven Chaetoceros cpDNAs. (A) C. muelleri (strain CNS00047), (B) C. costatus (strain CNS00386), (C) C. socialis (strain CNS00389), (D) C. pseudo-curvisetus (strain CNS00390), (E) C. tenuissimus (strain CNS00394), (F) C. laevisporus (strain CNS00396), and (G) C. curvisetus (strain CNS00516).
Supplementary Figure 3 | Phylogenetic trees constructed using different hotspots regions. (A) Position: 19,025–19,624 bp in C. muelleri cpDNA. (B) 106,895–107,700 bp in C. muelleri cpDNA.
Supplementary Figure 4 | (A,B) The DNA alignment information of hotspots region (position: 19,025–19,624 bp and 106,895–107,700 bp in C. muelleri cpDNA) for Chaetoceros species.
Footnotes
- ^ https://github.com/BFL-lab/Mfannot
- ^ https://www.ncbi.nlm.nih.gov/orffinder
- ^ https://www.ncbi.nlm.nih.gov/projects/Sequin/
- ^ http://www.timetree.org/
References
Albright, L. J., Yang, C. Z., and Johnson, S. (1993). Sub-lethal concentrations of the harmful diatoms, Chaetoceros concavicornis and C. convolutus, increase mortality rates of penned Pacific salmon. Aquacultrue 117, 215–225. doi: 10.1016/0044-8486(93)90321-O
Amiryousefi, A., Hyvonen, J., and Poczai, P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Armbrust, E. V. (2009). The life of diatoms in the world’s oceans. Nature 459, 185–192. doi: 10.1038/nature08057
Balzano, S., Percopo, I., Siano, R., Gourvil, P., Chanoine, M., Marie, D., et al. (2017). Morphological and genetic diversity of Beaufort Sea diatoms with high contributions from the Chaetoceros neogracilis species complex. J. Phycol. 53, 161–187. doi: 10.1111/jpy.12489
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Begum, M., Sahu, B. K., Das, A. K., Vinithkumar, N. V., and Kirubagaran, R. (2015). Extensive Chaetoceros curvisetus bloom in relation to water quality in Port Blair Bay, Andaman Islands. Environ. Monit. Assess. 187:226. doi: 10.1007/s10661-015-4461-2
Bendich, A. J. (2004). Circular chloroplast chromosomes: the grand illusion. Plant Cell 16, 1661–1666. doi: 10.1105/tpc.160771
Cao, M., Yuan, X. L., and Bi, G. (2016). Complete sequence and analysis of plastid genomes of Pseudo-nitzschia multiseries (Bacillariophyta). Mitochondrial DNA A DNA Mapp. Seq. Anal. 27, 2897–2898. doi: 10.3109/19401736.2015.1060428
Chamnansinp, A., Li, Y., Lundholm, N., and Moestrup, O. (2013). Global diversity of two widespread, colony-forming diatoms of the marine plankton, Chaetoceros socialis (syn. C. radians) and Chaetoceros gelidus sp. nov. J. Phycol. 49, 1128–1141. doi: 10.1111/jpy.12121
Chamnansinp, A., Moestrup, J., and Lundholm, N. (2015). Diversity of the marine diatom Chaetoceros (Bacillariophyceae) in Thai waters – revisiting Chaetoceros compressus and Chaetoceros contortus. Phycologia 54, 161–175. doi: 10.2216/14-074.1
Chen, Z., Xu, X., Zhu, S., Zhai, M., and Li, Y. (2019). Species diversity and geographical distribution of the Chaetoceros lorenzianus complex along the coast of China. Biodivers. Sci. 27, 149–158. doi: 10.17520/biods.2018261
Damste, J. S., Muyzer, G., Abbas, B., Rampen, S. W., Masse, G., Allard, W. G., et al. (2004). The rise of the rhizosolenid diatoms. Science 304, 584–587. doi: 10.1126/science.1096806
Daniell, H., Lin, C. S., Yu, M., and Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17:134. doi: 10.1186/s13059-016-1004-2
Darling, A. E., Mau, B., and Perna, N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147
De Luca, D., Kooistra, W., Sarno, D., Gaonkar, C. C., and Piredda, R. (2019a). Global distribution and diversity of Chaetoceros (Bacillariophyta, Mediophyceae): integration of classical and novel strategies. PeerJ 7:e7410. doi: 10.7717/peerj.7410
De Luca, D., Sarno, D., Piredda, R., and Kooistra, W. (2019b). A multigene phylogeny to infer the evolutionary history of Chaetocerotaceae (Bacillariophyta). Mol. Phylogenet. Evol. 140:106575. doi: 10.1016/j.ympev.2019.106575
Degerlund, M., Huseby, S., Zingone, A., Sarno, D., and Landfald, B. (2012). Functional diversity in cryptic species of Chaetoceros socialis Lauder (Bacillariophyceae). J. Plankton Res. 34, 416–431. doi: 10.1093/plankt/fbs004
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Finkel, Z. V., Katz, M. E., Wright, J. D., Schofield, O. M., and Falkowski, P. G. (2005). Climatically driven macroevolutionary patterns in the size of marine diatoms over the Cenozoic. Proc. Natl. Acad. Sci. U.S.A. 102, 8927–8932. doi: 10.1073/pnas.0409907102
Fu, C. N., Wu, C. S., Ye, L. J., Mo, Z. Q., Liu, J., Chang, Y. W., et al. (2019). Prevalence of isomeric plastomes and effectiveness of plastome super-barcodes in yews (Taxus) worldwide. Sci. Rep. 9:2773. doi: 10.1038/s41598-019-39161-x
Galachyants, Y. P., Morozov, A. A., Mardanov, A. V., Beletsky, A. V., Ravin, N. V., Petrova, D. P., et al. (2011). Complete chloroplast genome sequence of freshwater araphid pennate diatom alga Synedra acus from Lake Baikal. Int. J. Biol. 4, 27–35. doi: 10.5539/ijb.v4n1p27
Gaonkar, C. C., Kooistra, W., Lange, C. B., Montresor, M., and Sarno, D. (2017). Two new species in the Chaetoceros socialis complex (Bacillariophyta): C. sporotruncatus and C. dichatoensis, and characterization of its relatives, C. radicans and C. cinctus. J. Phycol. 53, 889–907. doi: 10.1111/jpy.12554
Gaonkar, C. C., Piredda, R., Minucci, C., Mann, D. G., Montresor, M., Sarno, D., et al. (2018). Annotated 18S and 28S rDNA reference sequences of taxa in the planktonic diatom family Chaetocerotaceae. PLoS One 13:e0208929. doi: 10.1371/journal.pone.0208929
Göksan, T., Durmaz, Y., and Gökpınar, S. (2003). Effects of light path lengths and initial culture density on the cultivation of Chaetoceros muelleri (Lemmermann, 1898). Aquac. Res. 217, 431–436. doi: 10.1016/S0044-8486(01)00854-7
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Guillard, R. R. L., and Hargreaves, P. E. (1994). Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia 33:66.
Guiry, M. D., and Guiry, G. M. (2021). AlgaeBase. Available online at: http://www.algaebase.org (accessed March 30, 2021).
Han, X., Zou, J., and Zhang, Y. (2004). Harmful algae bloom species in Jiaozhou Bay and the features of distribution. Mar. Sci. 28, 49–54.
Ji, Y., Liu, C., Yang, Z., Yang, L., He, Z., Wang, H., et al. (2019). Testing and using complete plastomes and ribosomal DNA sequences as the next generation DNA barcodes in Panax (Araliaceae). Mol. Ecol. Resour. 19, 1333–1345. doi: 10.1111/1755-0998.13050
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., dePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:241. doi: 10.1186/s13059-020-02154-5
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kooistra, W. H., Sarno, D., Hernández-Becerril, D., Assmy, P., Prisco, C. D., and Montresor, M. (2010). Comparative molecular and morphological phylogenetic analyses of taxa in the Chaetocerotaceae (Bacillariophyta). Phycologia 49, 471–500. doi: 10.2216/09-59.1
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lewitus, E., Bittner, L., Malviya, S., Bowler, C., and Morlon, H. (2018). Clade-specific diversification dynamics of marine diatoms since the Jurassic. Nat. Ecol. Evol. 2, 1715–1723. doi: 10.1038/s41559-018-0691-3
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, Y., Boonprakob, A., Gaonkar, C. C., Kooistra, W. H., Lange, C. B., Hernandez-Becerril, D., et al. (2017). Diversity in the globally distributed diatom genus Chaetoceros (Bacillariophyceae): three New Species from Warm-Temperate Waters. PLoS One 12:e0168887. doi: 10.1371/journal.pone.0168887
Li, Y., and Deng, X. (2021). The complete chloroplast genome of the marine microalgae Chaetoceros muellerii (Chaetoceroceae). Mitochondrial DNA B Resour. 6, 373–375. doi: 10.1080/23802359.2020.1869608
Liang, C., Zhang, Y., Wang, L., Shi, L., Xu, D., Zhang, X., et al. (2020). Features of metabolic regulation revealed by transcriptomic adaptions driven by long-term elevated pCO2 in Chaetoceros muelleri. Phycol. Res. 68, 236–248. doi: 10.1111/pre.12423
Liu, S., Huang, L. M., Huang, H., Lian, J. S., Long, A. M., and Li, T. (2006). Ecological response of phytoplankton to the operation of Daya Bay nuclear power station. Mar. Environ. Sci. 25, 9–12.
López-Elías, J. A., Voltolina, D., Enríquez-Ocaña, F., and Gallegos-Simental, G. (2005). Indoor and outdoor mass production of the diatom Chaetoceros muelleri in a Mexican commercial hatchery. Aquac. Eng. 33, 181–191. doi: 10.1016/j.aquaeng.2005.01.001
Lv, S., Qi, Y., Qian, H., and Liang, S. (1993). Studies on phytoplankton and red tide organisms in embayments on central Guangdong coast-II. Guanghai Bay. Mar. Sci. Bull. 12, 63–66.
Malviya, S., Scalco, E., Audic, S., Vincent, F., Veluchamy, A., Poulain, J., et al. (2016). Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. U.S.A. 113, E1516–E1525. doi: 10.1073/pnas.1509523113
Mann, D. G., and Droop, S. J. M. (1996). Biodiversity, biogeography and conservation of diatoms. Hydrobiologia 336, 19–32. doi: 10.1007/BF00010816
Marino, D., Giuffré, G., Montresor, M., and Zingone, A. (1991). An electron microscope investigation on Chaetoceros minimus (Levander) comb. nov. and new observations on Chaetoceros throndsenii (Marino. Montresor and Zingone) comb. nov. Diatom Res. 6, 317–326. doi: 10.1080/0269249X.1991.9705177
Matari, N. H., and Blair, J. E. (2014). A multilocus timescale for oomycete evolution estimated under three distinct molecular clock models. BMC Evol. Biol. 14:101. doi: 10.1186/1471-2148-14-101
McGinnis, K. M., Dempster, T. A., and Sommerfeld, M. R. (1997). Characterization of the growth and lipid content of the diatom Chaetoceros muelleri. J. Appl. Phycol. 9, 19–24. doi: 10.1023/A:1007972214462
Mojiri, A., Baharlooeian, M., and Zahed, M. A. (2021). The potential of Chaetoceros muelleri in bioremediation of antibiotics: performance and optimization. Int. J. Environ. Res. Public Health 18:977. doi: 10.3390/ijerph18030977
Montresor, M., Di Prisco, C., Sarno, D., Margiotta, F., and Zingone, A. (2013). Diversity and germination patterns of diatom resting stages at a coastal Mediterranean site. Mar. Ecol. Prog. Ser. 484, 79–95. doi: 10.3354/meps10236
Nelson, D. M., Tréguer, P., Brzezinski, M. A., Leynaert, A., and Quéguiner, B. (1995). Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob. Biogeochem. Cycles 9, 359–372. doi: 10.1029/95GB01070
Oku, O., and Kamatani, A. (1990). Resting spore formation and biochemical composition of the marine planktonic diatom Chaetoceros pseudocurvisetus in culture: ecological significance of decreased nucleotide content and activation of the xanthophyll cycle by resting spore formation. Mar. Biol. 135, 425–436. doi: 10.1007/s002270050643
Oyama, K., Yoshimatsu, S., Honda, K., Abe, Y., and Fujisawa, T. (2008). Bloom of a large diatom Chaetoceros densus in the coastal area of Kagawa Prefecture from Harima-Nada to Bisan-Seto, the Seto Inland Sea, in February 2005: environmental features during the bloom and influence on Nori Porphyra yezoensis cultures. Nippon Suisan Gakkaishi 74, 660–670. doi: 10.2331/suisan.74.660
Pelusi, A., Santelia, M. E., Benvenuto, G., Godhe, A., and Montresor, M. (2019). The diatom Chaetoceros socialis: spore formation and preservation. Eur. J. Phycol. 55, 1–10. doi: 10.1080/09670262.2019.1632935
Reinke, D. C. (1984). Ultrastructure of Chaetoceros muelleri (Bacillariophyceae): auxospore, resting spore and vegetative cell morphology. J. Phycol. 20, 153–155. doi: 10.1111/j.0022-3646.1984.00153.x
Rine, J. E. (1988). The Chaetoceros Ehrenberg (Bacillariophyceae) flora of Narragansett Bay. Port Jervis, NY: Lubrecht & Cramer.
Rines, J., Boonruang, P., and Theriot, E. C. (2010). Chaetoceros phuketensis sp. nov. (Bacillariophyceae): a new species from the Andaman Sea. Phycol. Res. 48, 161–168. doi: 10.1111/j.1440-1835.2000.tb00212.x
Round, F. E., Crawford, R. M., and Mann, D. G. (1990). The Diatoms. Biology & Morphology of the Genera. Cambridge: Cambridge University Press.
Rozas, J., Ferrer-Mata, A., Sanchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Ruck, E. C., Nakov, T., Jansen, R. K., Theriot, E. C., and Alverson, A. J. (2014). Serial gene losses and foreign DNA underlie size and sequence variation in the plastid genomes of diatoms. Genome Biol. Evol. 6, 644–654. doi: 10.1093/gbe/evu039
Sabir, J. S., Yu, M., Ashworth, M. P., Baeshen, N. A., Baeshen, M. N., Bahieldin, A., et al. (2014). Conserved gene order and expanded inverted repeats characterize plastid genomes of Thalassiosirales. PLoS One 9:e107854. doi: 10.1371/journal.pone.0107854
Sar, E. A., Hernández-Becerril, D. U., and Sunesen, I. (2002). A morphological study of Chaetoceros tenuissimus Meunier, a little-known planktonic diatom, with a discussion of the section simplicia, subgenus Hyalochaete. Diatom Res. 17, 327–335. doi: 10.1080/0269249X.2002.9705552
Sims, P. A., Mann, D. G., and Medlin, L. K. (2006). Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45, 361–402. doi: 10.2216/05-22.1
Song, H., Liu, F., Li, Z., Xu, Q., Chen, Y., Yu, Z., et al. (2020). Development of a high-resolution molecular marker for tracking Phaeocystis globosa genetic diversity through comparative analysis of chloroplast genomes. Harmful Algae 99:101911. doi: 10.1016/j.hal.2020.101911
Sorhannus, U. (2007). A nuclear-encoded small-subunit ribosomal RNA timescale for diatom evolution. Mar. Micropaleontol. 65, 1–12. doi: 10.1016/j.marmicro.2007.05.002
Suto, I. (2006). The explosive diversification of the diatom genus Chaetoceros across the Eocene/Oligocene and Oligocene/Miocene boundaries in the Norwegian Sea. Mar. Micropaleontol. 58, 259–269. doi: 10.1016/j.marmicro.2005.11.004
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 101, 11030–11035. doi: 10.1073/pnas.0404206101
Thorvaldsdottir, H., Robinson, J. T., and Mesirov, J. P. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. doi: 10.1093/bib/bbs017
Tomaru, Y., Fujii, N., Oda, S., Toyoda, K., and Nagasaki, K. (2011). Dynamics of diatom viruses on the western coast of Japan. Aquat. Microb. Ecol. 63, 223–230. doi: 10.3354/ame01496
Tomaru, Y., Toyoda, K., and Kimura, K. (2017). Occurrence of the planktonic bloom-forming marine diatom Chaetoceros tenuissimus Meunier and its infectious viruses in western Japan. Hydrobiologia 805, 221–230. doi: 10.1007/s10750-017-3306-0
Treasurer, J. W., Hannah, F., and Aquaculture, D. C. J. (2003). Impact of a phytoplankton bloom on mortalities and feeding response of farmed Atlantic salmon, Salmo salar, in west Scotland. Aquaculture 218, 103–113. doi: 10.1016/S0044-8486(02)00516-1
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Trigueros, J. M., Orive, E., and Arriluzea, J. (2002). Observations on Chaetoceros salsugineus (Chaetocerotales, Bacillariophyceae): first record of this bloom-forming diatom in a European estuary. Eur. J. Phycol. 37, 571–578. doi: 10.1017/S0967026202003906
Wang, C., Huang, C., and Du, H. (2008). Seasonal variation of Chaetoceros community in Zhelin Bay of eastern Guangdong. Acta Ecol. Sin. 28, 237–245.
Wang, Y., Nie, R., Li, Y., and Lv, S. (2010). Species diversity and geographical distribution of Chaetoceros in Guangdong coastal waters. Adv. Mar. Sci. 28, 342–352.
Xu, X., Lundholm, N., and Li, Y. (2020). A Study of Chaetoceros debilis Sensu Lato Species (Bacillariophyceae), with Ememdation of C. debilis and Description of C. galeatus Sp. Nov. J. Phycol. 56, 784–797. doi: 10.1111/jpy.12982
Yang, L., Zhu, S., Lundholm, N., and Lü, S. (2015). Morphology and molecular phylogeny of Chaetoceros dayaensis sp. nov. (Bacillariophyceae), characterized by two 90° rotations of the resting spore during maturation. J. Phycol. 51, 469–479. doi: 10.1111/jpy.12290
Yang, Z. (2007). PAML 4: phylogenetic analysis by paximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yao, Y., Liu, F., and Chen, N. (2021). Complete mitochondrial genome of Rhizosolenia setigera (Coscinodiscophyceae, Bacillariophyta). Mitochondrial DNA B Resour. 6, 2319–2321. doi: 10.1080/23802359.2021.1950059
Yin, W., and Hu, H. (2021). High-efficiency transformation of a centric diatom Chaetoceros muelleri by electroporation with a variety of selectable markers. Algal Res. 55:102274. doi: 10.1016/j.algal.2021.102274
Yu, M., Ashworth, M. P., Hajrah, N. H., Khiyami, M. A., Sabir, M. J., Alhebshi, A. M., et al. (2018). Evolution of the plastid genomes in diatoms. Adv. Botan. Res. 129–155. doi: 10.1016/bs.abr.2017.11.009
Zhang, D., Gao, F., Jakovliæ, I., Zou, H., Zhang, J., Li, W., et al. (2020). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang, M., Cui, Z., Liu, F., and Chen, N. (2021). Complete chloroplast genome of Eucampia zodiacus (Mediophyceae, Bacillariophyta). Mitochondrial DNA B Resour. 6, 2194–2197. doi: 10.1080/23802359.2021.1944828
Keywords: Chaetoceros species, chloroplast genome, comparative genomics, variation hotspots, divergence time
Citation: Xu Q, Cui Z and Chen N (2021) Comparative Analysis of Chloroplast Genomes of Seven Chaetoceros Species Revealed Variation Hotspots and Speciation Time. Front. Microbiol. 12:742554. doi: 10.3389/fmicb.2021.742554
Received: 16 July 2021; Accepted: 11 October 2021;
Published: 03 November 2021.
Edited by:
Hongbin Liu, Hong Kong University of Science and Technology, Hong Kong SAR, ChinaReviewed by:
Sunil Kumar Sahu, Beijing Genomics Institute (BGI), ChinaChengxu Zhou, Ningbo University, China
Hongtao Xiao, University of Electronic Science and Technology of China, China
Shiou Yih Lee, INTI International University, Malaysia
Pan Li, Zhejiang University, China
Copyright © 2021 Xu, Cui and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nansheng Chen, Y2hlbm5AcWRpby5hYy5jbg==
 Qing Xu
Qing Xu Zongmei Cui
Zongmei Cui Nansheng Chen
Nansheng Chen