
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 07 October 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.742040
This article is part of the Research Topic New Insights into the Transmission Dynamics and Control of Antimicrobial Resistance to Last-resort Antibiotics View all 22 articles
 Yixin Chen1†
Yixin Chen1† Ya Shi2†
Ya Shi2† Weifen Zhu1†
Weifen Zhu1† Jiaxing You3
Jiaxing You3 Jie Yang3
Jie Yang3 Yaping Xie4
Yaping Xie4 Hanxin Zhao1
Hanxin Zhao1 Hongye Li3
Hongye Li3 Shunwu Fan3
Shunwu Fan3 Lin Li1*
Lin Li1* Chao Liu3*
Chao Liu3*Introduction: Diabetic foot infections (DFIs) pose a huge challenge for clinicians. Staphylococcus aureus, including methicillin-resistant S. aureus (MRSA), is one of the most significant pathogens of DFI. Early pathogen identification will greatly benefit the diagnosis and treatment of the disease. However, existing diagnostic methods are not effective in early detection.
Methods: We developed an assay that coupled loop-mediated isothermal amplification (LAMP) and clustered regularly interspaced short palindromic repeats (CRISPR) techniques to enable quick and specific detection of Staphylococcus aureus and differentiate MRSA in samples from patients with DFI. Furthermore, the results were compared using a reference culture, quantitative real-time polymerase chain reaction (qRT-PCR), and metagenomics next generation sequencing (mNGS).
Results: The CRISPR-LAMP assay targeting nuc and mecA successfully detected S. aureus strains and differentiated MRSA. The limit of detection (LoD) of the real-time LAMP for nuc and mecA was 20 copies per microliter reaction in comparison to two copies per μL reaction for the qRT-PCR assay. The specificity of the LAMP-CRISPR assay for nuc was 100%, without cross-reactions with non-S. aureus strains. Evaluating assay performance with 18 samples from DFI patients showed that the assay had 94.4% agreement (17/18 samples) with clinical culture results. The results of mNGS for 8/18 samples were consistent with those of the reference culture and LAMP-CRISPR assay.
Conclusion: The findings suggest that the LAMP-CRISPR assay could be promising for the point-of-care detection of S. aureus and the differentiation of MRSA in clinical samples. Furthermore, combining the LAMP-CRISPR assay and mNGS provides an advanced platform for molecular pathogen diagnosis of DFI.
Diabetic foot ulcers, one of the most critical complications of diabetes mellitus, has become a significant public health problem. When patients with diabetic foot ulcers first consult physicians, over half of them have complications with diabetic foot infections (DFIs) (Prompers et al., 2006; Jia et al., 2017). Treatment of DFI requires precise evaluation of infection conditions, pathogen confirmation of infectious ulcers, appropriate selection of antibiotics, and surgical intervention, if necessary (Lipsky et al., 2012). DFI patients are often challenged by the complexity of intricate ulceration and moderate to severe infections, even sepsis. Broad-spectrum empirical antimicrobials would be the primary option for clinicians to hinder the further deterioration of infectious conditions and lower medical risks, which increases antibiotic exposure and selective pressure that contribute to the development of multi-drug resistant microorganisms. Nevertheless, a rapid, accessible, and accurate diagnostic test for pathogenic identification is valuable for clinical procedures.
Staphylococcus aureus is the dominant pathogen of DFI. The resistance rate of S. aureus to methicillin fluctuated between 15–30% (Eleftheriadou et al., 2010). It has been shown that infections caused by methicillin-resistant S. aureus (MRSA) are linked to higher mortality rates and greater consumption of medical resources than infections caused by methicillin-sensitive S. aureus (MSSA) (Cosgrove et al., 2003; Filice et al., 2010). Bacterial culture, quantitative real-time polymerase chain reaction (qRT-PCR) (Buchan et al., 2015; Ellem et al., 2015; Silbert et al., 2017), and metagenomics next generation sequencing (mNGS) (Charalampous et al., 2019; Kalan et al., 2019; Zou et al., 2020) are typically used to identify S. aureus and discriminate between MSSA and MRSA. Although traditional culture is the gold standard for pathogen diagnosis, it is time-consuming, and delayed results cannot provide timely clinical guidance. qRT-PCR has a shortened detection time; however, it has not been widely applied because of its high requirements for equipment and laboratory staff. mNGS, a novel detection method, is still not commonly accessible because of its high cost in terms of time, funding, instrumentation, and technical expertise.
In recent years, the application of isothermal amplification combined with clustered regularly interspaced short palindromic repeats (CRISPR) has been developed for pathogen diagnosis (Li et al., 2019). An innovative discovery reported that RNA-guided DNA binding activated Cas12a for both site-specific double-stranded DNA (dsDNA) cis-cleavage and indiscriminate single-stranded DNA (ssDNA) trans-cleavage. With the guidance of a single CRISPR RNA (crRNA), Cas12a recognizes a TTTN protospacer-adjacent motif (PAM) and binds to crRNA-complementary dsDNA (Li et al., 2018a,b). This indicates that the high specificity of the CRISPR/Cas12a assay is determined by PAM and crRNA. The coupling of isothermal amplification and trans-ssDNA repetitive cutting results in double-magnifying biosensing signals, indicating the high sensitivity of the assay. It has been proven to enable a rapid and accurate detection of the human papillomavirus and a new coronavirus (resulting in the disease referred to as COVID-19) in patient samples (Chen et al., 2018; Broughton et al., 2020).
In this study, we developed an assay coupling loop-mediated isothermal amplification (LAMP) and trans-cleavage of Cas12a for the quick and specific detection of S. aureus and the differentiation of MRSA in samples of patients with DFI. Furthermore, the results were compared with standard methods, including bacterial culture, qRT-PCR, and mNGS.
DNA extraction from the tissues of DFI patients was performed using QuickExtractTM DNA Extraction Solution (Lucigen, Beijing, China). Quick DNA extractions were finished after incubation at 65°C for 10 min and 98°C for 2 min. The sequences of nuc (S. aureus-specific gene) and mecA (encoding penicillin-binding protein-2a) were obtained from a public database. LAMP primers were designed against regions of nuc and mecA using PrimerExplorer v.5. with compatible gRNAs (Supplementary Table 1). The gRNA was designed with a TTTN structure (PAM) at the right end. The length of gRNA was 20-25bp, and the GC content was 40–60%. The design was assured to avoid the combination of gRNA and primer dimer (Table 1). The primers and gRNAs were obtained from Shanghai General Biotech Co., Ltd. (Shanghai, China).
A 20-μL system was used to conduct the real-time PCR assay for the detection of nuc and mecA with the following specifications: Forward primer for nuc gene: 5′-ACTGTAACTTTGGCACTGG-3′; Reverse primer for nuc gene: 5′-GCAGATACCTCATTACCTGC-3′; Probe: 5′-[6FAM]- ATCGCAACGACTGGCGCTA-[BHQ1]-3′ [12]; Forward primer for mecA gene: AAAACTAGGTGTTGGTGAAGATATACC; Reverse primer for mecA gene: GAAAGGATCTGTACTGGGTTAATCAG; Probe: [6FAM]-TTCACCTTGTCCGTAACCTGAATCAGCT-[BHQ1] (Sabet et al., 2007). Real-time PCR assays were performed using the Premix Ex TaqTM (Probe qPCR) Kit (TaKara Biomedical Technology Co., Ltd., Beijing, China). Ten microliters Premix Ex Taq (Probe qPCR) (2×), 0.2 μM forward primer, 0.2 μM reverse primer, 0.1 μM probe, and 2 μL DNA templates of different concentrations were included in the reaction system. The temperature cycling conditions were 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, and 60°C for 30 s. The reaction was performed using the StepOneTMPlus Real-Time PCR System (Applied Biosystems, Inc., Carlsbad, CA, United States).
The real-time LAMP assay was conducted in a 25-μL mixture containing 1 × Bst DNA polymerase buffer (New England Biolabs, Beijing, China), 0.4 μM FIP primers, 0.4 μM BIP primers, 0.2 μM LF primers, 0.2 μM LB primers, 0.1 μM F3 primers F3, 0.1 μM B3 primers, 1.4 μM dNTPs (New England Biolabs), 8U Bst DNA polymerase (New England Biolabs), 8 mM MgSO4 (New England Biolabs), 1 × SYBR Green I (Thermo Fisher Scientific, Inc., Waltham, MA, United States), and 2 μL of DNA templates of different concentrations. Real-time LAMP assays of nuc and mecA were conducted in the same reaction system, except for the primers. The LAMP reaction solution was covered with 40 μL mineral oil. The reaction was performed at 65°C for 60 min in a StepOneTM Plus Real-Time PCR System (Applied Biosystems, Inc., Carlsbad, CA, United States) and terminated at 80°C for 5 min. The fluorescence information was collected every 30 s during the reaction.
The LAMP system contained the same mixture as the real-time LAMP assay. The fluorescent dyes were added into the system and the LAMP reagents covered with 40 μL of mineral oil. The LAMP reaction was followed by the admixture of CRISPR reaction solution (preloaded inside the lid) to the LAMP amplification solution by centrifugation. The 20 μL CRISPR reaction solution contained 1 × NEBuffer 2.1, reaction buffer, 0.2 μM EnGen® Lba Cas12a (Cpf1) (New England Biolabs), 0.6 μM gRNA, 2.5 μM CRISPR probes, and 1 U/μL RNA inhibitor (Thermo Fisher Scientific, Inc., Waltham, MA, United States). Sequence information of gRNA is shown in Table 1. The CRISPR reaction was conducted in a ProFlex PCR System (Applied Biosystems, Inc., Carlsbad, CA, United States) at 37°C for 5 min. After the reaction, the visualized results were observed using a simple fluorescence reader.
Amplification of S. aureus genomic DNA, including nuc and mecA, was performed using outer primers (F3 and B3, Supplementary Table 1). The lengths of the PCR products of nuc and mecA were 248 bp and 208 bp, respectively. The PCR product was purified using the TAKARA gel DNA fragment recovery kit, and the purified product was ligated to the pMD18-T vector (2692 bp). The recombinant plasmids were transformed into Escherichia coli DH5α competent cells. LB growth medium containing 10 μg/mL ampicillin was used to screen positive colonies. Plasmids of E. coli that comprised the pMD18-T vector and nuc (248 bp)/mecA (208 bp) were extracted using the TaKaRa MiniBEST Plasmid Purification Kit (TaKaRa, 9760). We then measured the concentration of plasmids and calculated the DNA copy number [6.02 × 1023 × concentration (ng/μL) × 109/(DNA sequence length × 660)]. The concentration of plasmids containing nuc was 3.26 ng/μL. The DNA copy number of nuc was 1.0 × 109 copies per μl, calculated according to the above formula. The concentration of plasmids containing mecA was 12 ng/μL, and the copy number was 3.78 × 109 copies per μl. The plasmids were serially diluted to 1:10. Isothermal amplifications of the dilutions were repeatedly performed three times, and the limit of detection (LoD) was determined as the minimum DNA copy number of the diluted plasmids that could amplify the target genomic DNA.
Genomic DNA from 14 strains of staphylococci species was analyzed using the LAMP-CRISPR assay to examine the specificity of the LAMP assay. The strains were S. epidermidis, S. hominis, S. cohnii, S. warneri, S. haemolyticus, S. aureus, S. capitis, S. saprophyticus, S. pasteuri, S. saprophyticus ATCC® 15305, S. haemolyticus ATCC ®29970, S. epidermidis ATCC ®14990, S. caprae, and S. aureus subsp. sureus ATCC ®12600.
After DNA extractions using a DNeasy Blood and Tissue Kit (Qiagen, 69504, Shenzhen, China), DNA was fragmented using a Bioruptor Pico instrument to generate 200–300-bp fragments (Bioruptor Pico protocols). Then, libraries were constructed by MGIEasy (MGIEasy universal DNA library prep kit) as follows: first, the DNA fragments were subjected to end-repair sequencing, and A-tailing was added to one tube. Subsequently, the resulting DNA was ligated with bubble-adaptors that included barcode sequences and then amplified by the PCR method. Quality control was carried out using bioanalyzer (Agilent 2100, Agilent Technologies, Santa Clara, CA, United States) to assess DNA concentrations and fragment size. Qualified libraries were pooled together to make a single strand DNA circle (ssDNA circle), and then, DNA nanoballs (DNB) were generated by rolling circle replication (RCA). The final DNBs were loaded into the sequence chip and sequenced on the BGISEQ platform using 50 bp/single-end sequencing. Finally, optical signals were collected using a high-resolution imaging system, and then, these signals were transformed into digital information, which can be decoded into DNA sequence information.
The sample culture broths were plated on blood agar and incubated for 18–48 h. Identification of S. aureus and examination of antimicrobial susceptibility were conducted using a fully automatic vitek-2 compact system (Biomerieux, Shanghai, China). The results were interpreted on the basis of the guidelines (M100, 28th edition) established by the Clinical and Laboratory Standards Institute (CLSI, 2018).
Tissues of infected ulcers were acquired from DFI patients at their first visit to the outpatient clinic or the Department of Emergency at Sir Run Run Shaw Hospital. After the wound was cleaned with saline solution, DFU tissue with the size of a rice grain was taken out with a blade and immediately sent to the laboratory for DNA extraction. The study was approved by the Human Research Ethics Committee of Sir Run Run Shaw Hospital.
All reactions were repeated three times, the CT value of the real-time PCR is shown as “mean ± standard deviation” (x ± s).
We conducted a real-time LAMP assay for amplifying nuc and mecA with seven dilutions. The amplification curves of the dilutions with serial concentrations are shown in Figure 1. The LAMP assay was able to amplify genomic DNA targets. There was no stable fluorescent signal obtained when the genomic DNA template was at two copies per μL reaction.
Next, we used the CRISPR/Cas12a system to realize the visual readout of the targeted amplicons, based on the collateral DNA cleavage activity of the Cas12a effector. The cleaved ssDNA reporters produced fluorescent signals, which could be seen through a portable fluorescence reader (Figure 2). The entire procedure and explanation of the results are presented in Figure 3. Pre-loading of the CRISPR reagents inside the lid avoided the need to open the cap during the process. We performed this assay for detecting nuc and mecA with serial dilutions. Green fluorescence was observed when the DNA sample was 2 × 105, 2 × 104, 2 × 103, 2 × 102, and 2 × 101 copies, which was referred to as a positive result. No green fluorescence signal was seen for a sample of 2 × 100 copies, one copy, and the no template control (NTC) (Figure 4). The interpretation of the genetic results for bacterial identification are illustrated in Figure 4.
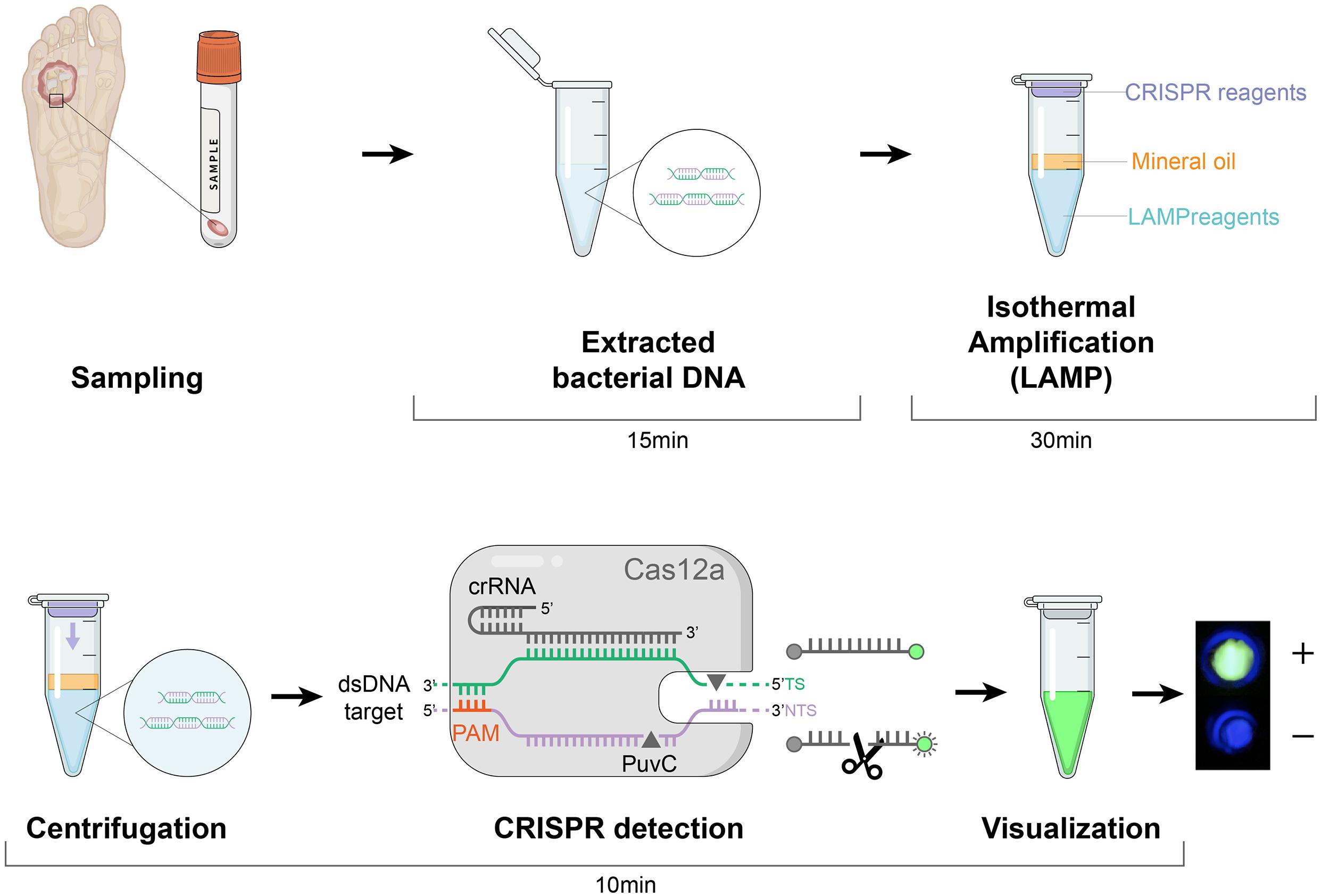
Figure 3. Schematic of the LAMP-CRISPR assay workflow. Quick DNA extraction was used as an input to the LAMP-CRISPR system (LAMP preamplification and Cas12-based detection for nuc and mecA). The result is visualized by a fluorescence reader.
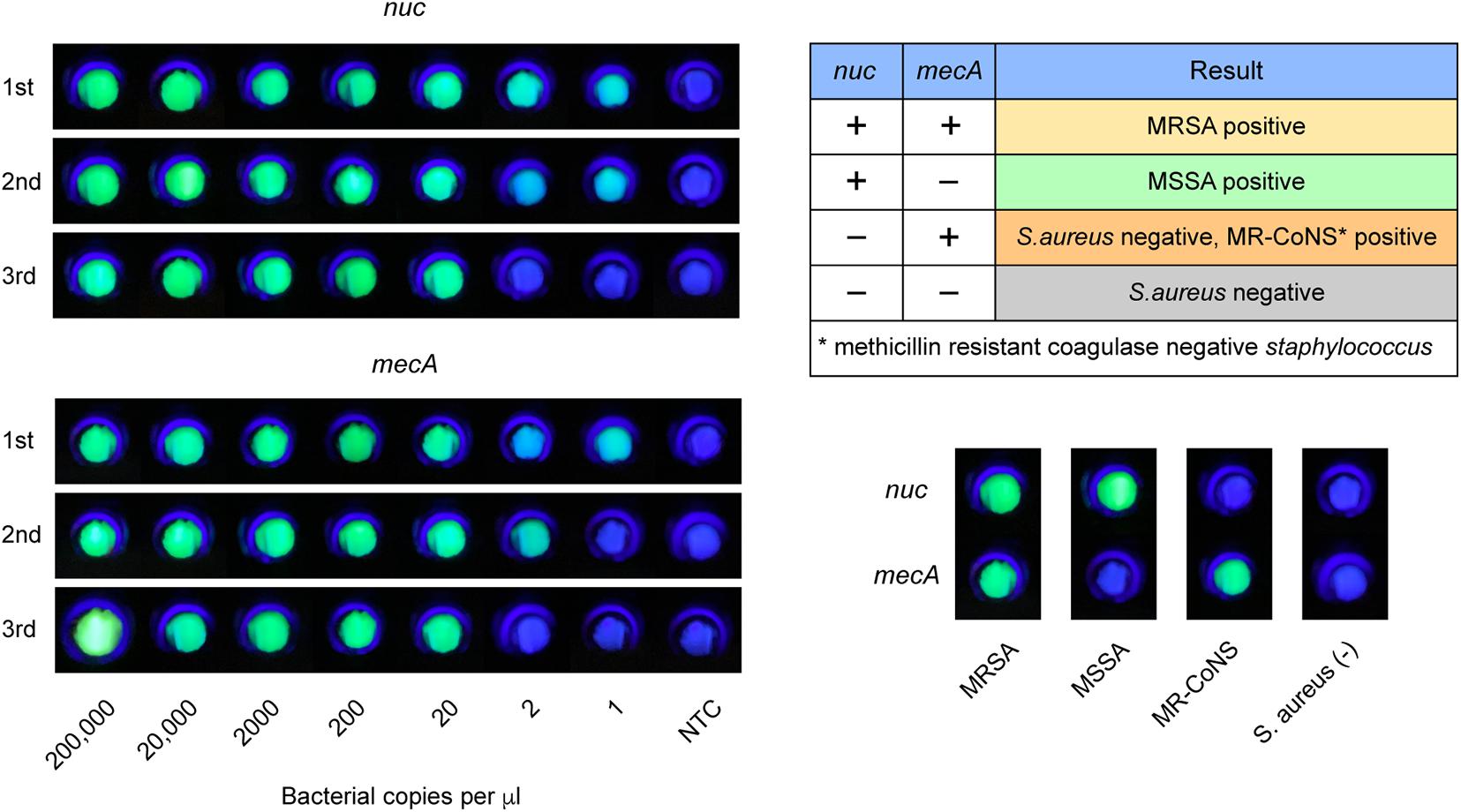
Figure 4. LAMP-CRISPR assay readout. Fluorescent results of MRSA, MSSA, MR-CoNS, and S. aureus are presented at the bottom right. Nuc detection indicates S. aureus positive. MRSA identification requires positive for both nuc and mecA.
We compared the LoD of the LAMP assay relative to the qRT- PCR assay for the detection of S. aureus and MRSA. A standard scatter plot was plotted using seven serial dilutions with three replicates at each dilution (Figure 5). The LoD of the LAMP assay for nuc was 20 copies per μL reaction in comparison to two copies per μL reaction for the qRT-PCR assay. For mecA, the LoD of the LAMP assay was 20 copies per μL reaction, versus two copies per μL reaction for the qRT-PCR assay.
We tested the extracted DNA templates from strains of several staphylococcus species using the LAMP-CRISPR assay with primers for nuc. These strains included ten isolates, including S. epidermidis, S. hominis, S. cohnii, S. warneri, S. haemolyticus, S. aureus, S. capitis, S. saprophyticus, S. pasteuri, and S. caprae, and four purchased strains, including S. saprophyticus ATCC® 15305, S. haemolyticus ATCC ®29970, S. epidermidis ATCC ®14990, and S. aureus subsp. aureus ATCC ®12600. Positive results were generated only for DNA samples from S. aureus (Figure 6). It was confirmed that the LAMP-CRISPR assay for nuc could specifically detect S. aureus, with no cross-reactions to other staphylococcal families.
Clinical validation of this assay was performed using 18 samples of isolated strains from DFI ulcers. Confirmatory qRT-PCR was used to establish target gene detected by the LAMP-CRISPR assay. The results of the two molecular methods were compared to those of the gold-standard culture method (Table 2). For S. aureus detection, six samples were nuc-positive as per the CRISPR-LAMP assay and S. aureus-positive by culture. For MRSA detection, two samples were positive for both nuc and mecA as per the CRISPR-LAMP assay and MRSA-positive by culture. One discordant sample (No. 8) was positive for both nuc and mecA as per the LAMP-CRISPR assay and MRSA-negative by culture. The results showed that MSSA was mixed with methicillin-resistant Staphylococcus pippi.
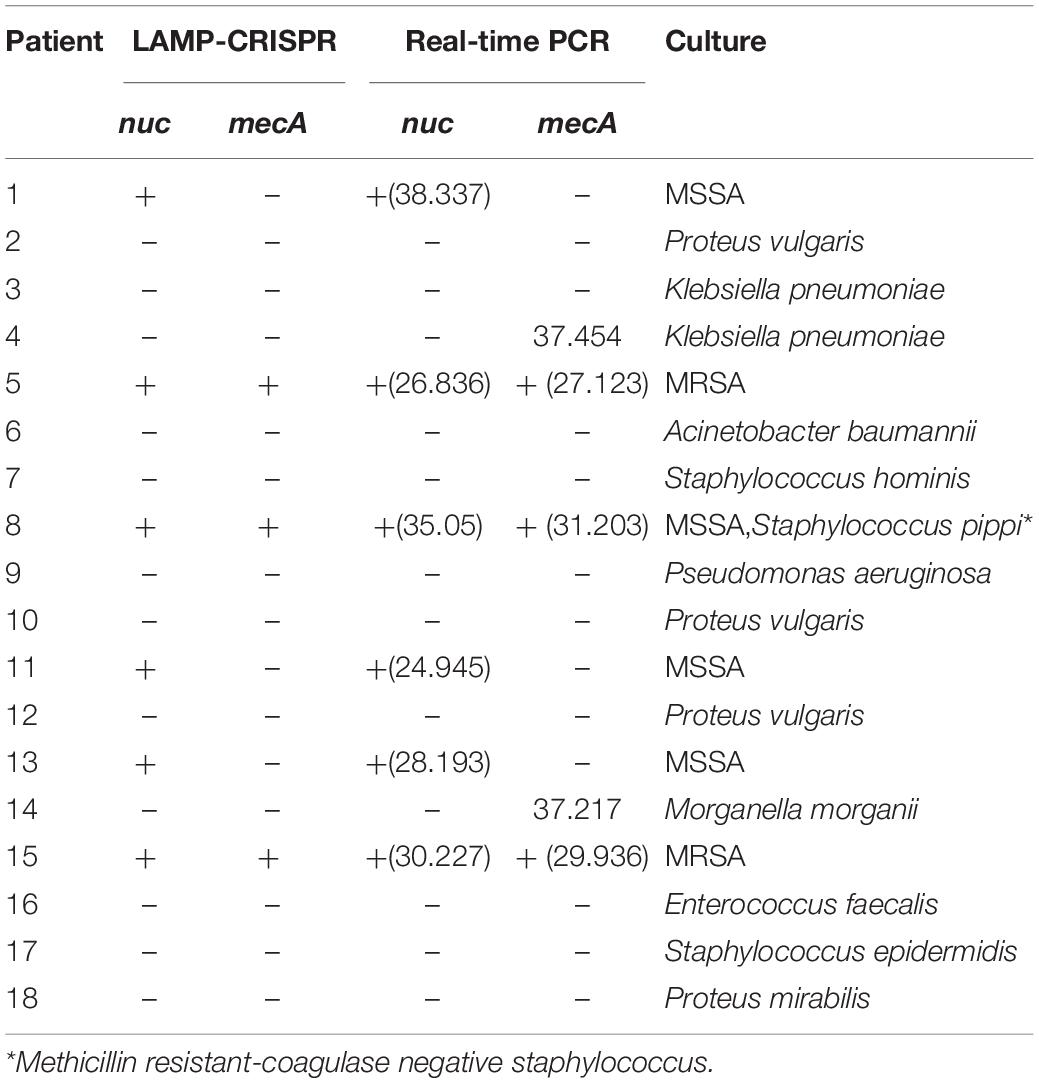
Table 2. Results of 18 samples from DFI patients using the LAMP-CRISPR assay, real-time PCR, and the reference culture.
After LAMP-CRISPR and qRT-PCR analysis, 8/18 samples were selected for mNGS. The culture growth of these eight samples indicated pathogens commonly seen in clinical practice, such as S. aureus, Enterococcus faecalis, Pseudomonas aeruginosa, and Klebsiella pneumoniae. The results of the mNGS analysis are listed in Supplementary Material (Supplementary Table 2). The mNGS results were consistent with the reference culture (Figure 7).
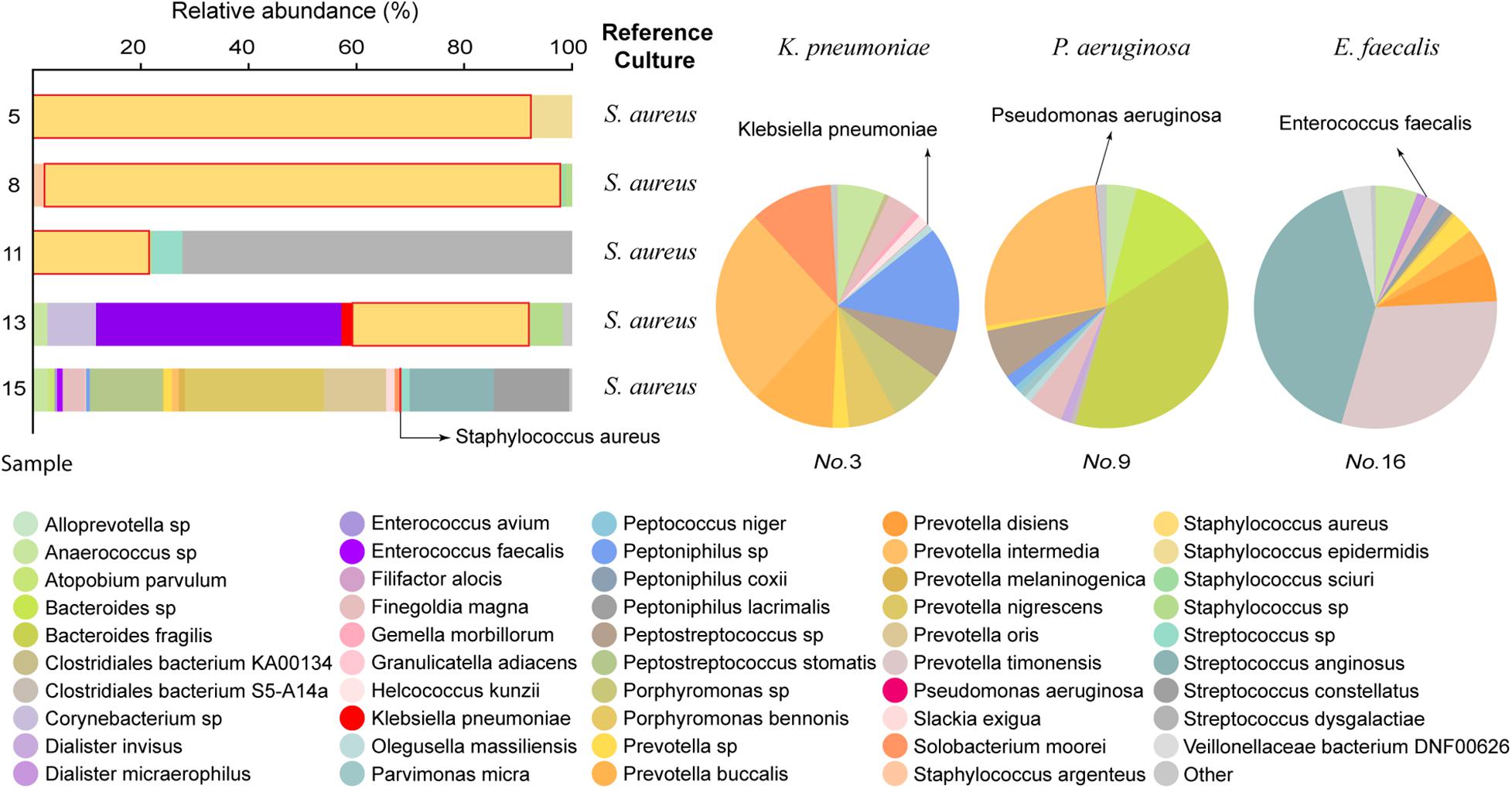
Figure 7. Comparison of mNGS data to the reference culture. In the left 100% stacked columns, the column representing S. aureus is framed by a red line.
Here, we combined the CRISPR-Cas12a-based technique with LAMP to develop a rapid diagnostic assay for the detection of S. aureus in samples directly from patients with DFI. The CRISPR-LAMP assay targeting nuc and mecA successfully identified S. aureus strains and differentiated MRSA from MSSA. The entire procedure was brief and contamination-free. Visualization of the results through a simple device also made them easy to interpret. This molecular assay could be promising for the point-of-care detection of S. aureus from clinical samples at some underprovided hospitals and areas.
MRSA can be transmitted by contact between the medical staff and patients. Infection-control precautions, such as the primary isolation of patients with MRSA infection and disinfection of the clinic environment after every diabetic foot patient with MRSA infection, can prevent cross-transmission of MRSA (Lecornet et al., 2007). Early detection of MRSA can alert hospital staff to carry out the necessary precautions to reduce the spread of MRSA among patients. With the worldwide prevalence of MRSA isolates and frequent exposure to vancomycin, the appearance of vancomycin-intermediate S. aureus (Centers for Disease Control and Prevention (CDC), 2002; Chang et al., 2003) as well as vancomycin-resistant S. aureus (Centers for Disease Control and Prevention (CDC), 1997) strains have been identified globally. Early discrimination between MSSA and MRSA could reduce the inappropriate use of broad-spectrum antimicrobials and decrease the selective pressure that initiates the emergence and spread of resistance in bacteria (Gardete and Tomasz, 2014).
A potential weakness of this assay was the partial disagreement between the phenotype and genotype of microorganisms, which was also observed in other molecular strategies such as GeneOhm MRSA or Xpert MRSA assays (Buchan et al., 2015). Microbes that were mecA-negative might contain mecC, which was recently discovered and identified as a novel resistance gene in MRSA (Lakhundi and Zhang, 2018). DFI is often caused by multiple bacteria (Lipsky et al., 2012), which indicates that the culture growth of a clinical sample could be a mixture of S. aureus and coagulase-negative Staphylococcus, both of which can carry the mecA gene. Therefore, apart from being caused MRSA alone, the double positivity of nuc and mecA could be explained by mixed Staphylococcal isolates. This discrepancy was observed in one patient (No. 8). Using long-range PCR may encompass the amplicons, including both nuc and mecA, from a single strain that can uniquely identify MRSA (McClure et al., 2020). This will compensate for the current deficiency of the LAMP or regular PCR assays and improve specificity.
We then compared this novel LAMP-CRISPR assay with the reference culture, using qRT-PCR and mNGS methods (Figure 8). Compared to other diagnostic technologies, the CRISPR-LAMP assay enabled the detection of targeted genes at very low DNA concentrations within 1 h, which is faster than qRT-PCR and no complex infrastructure was required (Table 3). DFI is a rapidly progressing and relatively critical disease that may lead to disability and mortality. CRISPR-LAMP detection could aid clinicians in selecting targeted antibiotics and prevent the spread of multi-drug resistant microbes. Application of the CRISPR-LAMP assay optimizes the early treatment of DFI and complements the current paradigm for clinical diagnostics.
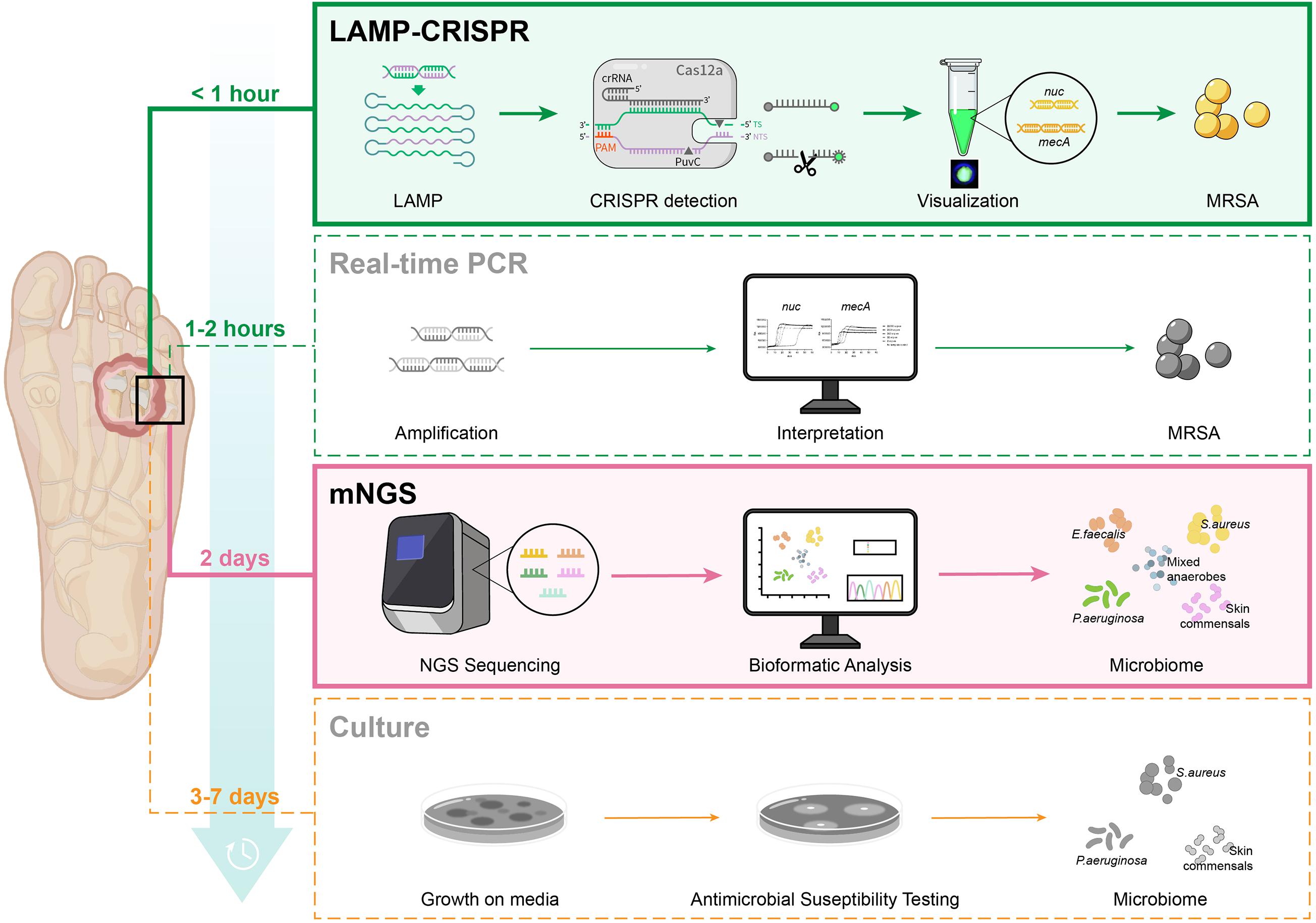
Figure 8. Procedure of four diagnostic methods: LAMP-CRISPR assay, real-time PCR, mNGS, and reference culture.
We also chose several representative samples for mNGS. mNGS can unbiasedly identify multiple microbes and address the limitations in pathogen width. It provides comprehensive information on the microbial profile of DFIs, including some rare pathogens that cannot be detected by conventional methods. mNGS can also provide quantitative data on the microbial concentration in the sample by counting sequenced reads, which is useful for polymicrobial samples (Wei et al., 2019). Study of the microbiome can potentially improve the diagnosis and treatment of DFIs. This revolutionary genetic approach further advances the management procedure of DFIs, which is becoming a rising trend in the clinical detection of infectious diseases. With the rapid determination of pathogens via CRISPR-LAMP and comprehensive supplementation of the bacterial spectrum by mNGS, a combination of these two methods can realize a timely and accurate pathogenic diagnosis of DFIs.
Targeting both nuc and mecA, our LAMP-CRISPR assay has proven to be a rapid (within 1 h) and effective method for detecting S. aureus and distinguishing MRSA from MSSA in DFI samples. Due to its convenient visualized readout and portable nature, this method is promising for improving point-of-care detection. mNGS can provide comprehensive information for the microbiome study of DFI samples. Combining the LAMP-CRISPR technique and mNGS expanded and improved the pathogen diagnostic routine of DFI.
The data presented in this study are deposited in the EBI Metagenomics repository, accession numbers ERR6769676–ERR6769683.
The studies involving human participants were reviewed and approved by Human Research Ethics Committee of Sir Run Run Shaw Hospital. The patients/participants provided their written informed consent to participate in this study.
CL, LL, and YC conceived and designed the study. YC was the major contributor in manuscript writing. YS, YC, and WZ completed the laboratory procedures. JaY, JeY, HL, and SF collected the samples of DFI patients and managed the diagnosis and treatment of patients. HZ analyzed the mNGS Data. YX proofread the first draft of the manuscript. CL and LL reviewed the manuscript. All authors agree to be accountable for the content of the work.
Zhejiang Province Medical and Health Science Project (grant numbers 2021454695, 2021KY881, and 2019323752), Hangzhou Municipality Medical and Health Science Project (grant number A20200297).
YS was employed by Hangzhou Digital-Micro Biotech Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.742040/full#supplementary-material
Supplementary Table 1 | The sequences of the LAMP primers for nuc and mecA, and the CRISPR probe.
Supplementary Table 2 | The results of the mNGS analysis. The highlights in the table represent the consistent microbials detected both by the mNGS and reference culture.
DFI, diabetic foot infections; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-sensitive Staphylococcus aureus; LAMP, loop-mediated isothermal amplification; CRISPR, clustered regularly interspaced short palindromic repeats; mNGS, metagenomics next generation sequencing; qRT-PCR, quantitative real-time polymerase chain reaction; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; crRNA, CRISPR RNA; PAM, protospacer-adjacent motif; LoD, limit of detection; DNB, DNA nanoballs; RCA, rolling circle replication; NTC, no template control.
Prompers, L., Huijberts, M., Apelqvist, J., Jude, E., Piaggesi, A., Bakker, K., et al. (2006). High prevalence of ischaemia, infection and serious comorbidity in patients with diabetic foot disease in Europe. Baseline results from the Eurodiale study. Diabetologia 50, 18–25.
Jia, L., Parker, C. N., Parker, T. J., Kinnear, E. M., Derhy, P. H., Alvarado, A. M., et al. (2017). Incidence and risk factors for developing infection in patients presenting with uninfected diabetic foot ulcers. PLoS One 12:e0177916. doi: 10.1371/journal.pone.0177916
Lipsky, B. A., Berendt, A. R., Cornia, P. B., Pile, J. C., Peters, E. J. G., Armstrong, D. G., et al. (2012). 2012 Infectious Diseases Society of America clinical practice guideline for the diagnosis and treatment of diabetic foot infections. Clin. Infect. Dis. 54, e132–e173.
Eleftheriadou, I., Tentolouris, N., Argiana, V., Jude, E., and Boulton, A. J. (2010). Methicillin-resistant Staphylococcus aureus in diabetic foot infections. Drugs 70, 1785–1797. doi: 10.2165/11538070-000000000-00000
Cosgrove, S. E., Sakoulas, G., Perencevich, E. N., Schwaber, M. J., Karchmer, A. W., and Carmeli, Y. (2003). Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 36, 53–59.
Filice, G. A., Nyman, J. A., Lexau, C., Lees, C. H., Bockstedt, L. A., Como-Sabetti, K., et al. (2010). Excess costs and utilization associated with methicillin resistance for patients with Staphylococcus aureus infection. Infect. Control Hosp. Epidemiol. 31, 365–373. doi: 10.1086/651094
Buchan, B. W., Allen, S., Burnham, C.-A. D., McElvania TeKippe, E., Davis, T., Levi, M., et al. (2015). Comparison of the next-generation Xpert MRSA/SA BC assay and the GeneOhm StaphSR assay to routine culture for identification of Staphylococcus aureus and methicillin-resistant S. aureus in positive-blood-culture broths. J. Clin. Microbiol. 53, 804–809. doi: 10.1128/jcm.03108-14
Ellem, J. A., Olma, T., and O’Sullivan, M. V. N. (2015). Rapid detection of methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus directly from positive blood cultures by use of the BD Max StaphSR assay. J. Clin. Microbiol. 53, 3900–3904. doi: 10.1128/jcm.02155-15
Silbert, S., Gostnell, A., Kubasek, C., and Widen, R. (2017). Evaluation of the BD Max StaphSR assay for detecting methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-susceptible S. aureus (MSSA) in ESwab-collected wound samples. J. Clin. Microbiol. 55, 2865–2867. doi: 10.1128/jcm.00641-17
Kalan, L. R., Meisel, J. S., Loesche, M. A., Horwinski, J., Soaita, I., Chen, X., et al. (2019). Strain- and species-level variation in the microbiome of diabetic wounds is associated with clinical outcomes and therapeutic efficacy. Cell Host Microbe 25, 641–645. doi: 10.1016/j.chom.2019.03.006
Zou, M., Cai, Y., Hu, P., Cao, Y., Luo, X., Fan, X., et al. (2020). Analysis of the composition and functions of the microbiome in diabetic foot osteomyelitis based on 16S rRNA and metagenome sequencing technology. Diabetes 69, 2423–2439. doi: 10.2337/db20-0503
Charalampous, T., Kay, G. L., Richardson, H., Aydin, A., Baldan, R., Jeanes, C., et al. (2019). Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat. Biotechnol. 37, 783–792. doi: 10.1038/s41587-019-0156-5
Li, Y., Li, S., Wang, J., and Liu, G. (2019). CRISPR/Cas Systems towards next-generation biosensing. Trends Biotechnol. 37, 730–743. doi: 10.1016/j.tibtech.2018.12.005
Li, S.-Y., Cheng, Q.-X., Liu, J.-K., Nie, X.-Q., Zhao, G.-P., and Wang, J. (2018a). CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 28, 491–493.
Li, S.-Y., Cheng, Q.-X., Wang, J.-M., Li, X.-Y., Zhang, Z.-L., Gao, S., et al. (2018b). CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 4:20.
Chen, J. S., Ma, E., Harrington, L. B., Da Costa, M., Tian, X., Palefsky, J. M., et al. (2018). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439. doi: 10.1126/science.aar6245
Broughton, J. P., Deng, X., Yu, G., Fasching, C. L., Servellita, V., Singh, J., et al. (2020). CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 395, 470–478.
Sabet, N. S., Subramaniam, G., Navaratnam, P., and Sekaran, S. D. (2007). Detection of mecA and ermA genes and simultaneous identification of Staphylococcus aureus using triplex real-time PCR from Malaysian S. aureus strain collections. Int. J. Antimicrob. Agents 29, 582–585. doi: 10.1016/j.ijantimicag.2006.12.017
CLSI (2018). Performance standards for antimicrobial susceptibility testing; twenty-sixth informational supplement. CLSI document M100-S28. Wayne: Clinical and Laboratory Standards Institute.
Lecornet, E., Robert, J., Jacqueminet, S., Van Georges, H., Jeanne, S., Bouilloud, F., et al. (2007). Preemptive isolation to prevent methicillin-resistant Staphylococcus aureus cross-transmission in diabetic foot. Diabetes Care 30, 2341–2342. doi: 10.2337/dc07-0743
Centers for Disease Control and Prevention (CDC) (2002). Staphylococcus aureus resistant to vancomycin–United States, 2002. MMWR Morb. Mortal. Wkly. Rep. 51, 565–567.
Chang, S., Sievert, D. M., Hageman, J. C., Boulton, M. L., Tenover, F. C., Downes, F. P., et al. (2003). Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348, 1342–1347.
Centers for Disease Control and Prevention (CDC) (1997). Staphylococcus aureus with reduced susceptibility to vancomycin–United States, 1997. MMWR Morb. Mortal. Wkly. Rep. 46, 765–766.
Gardete, S., and Tomasz, A. (2014). Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Invest. 124, 2836–2840. doi: 10.1172/jci68834
Lakhundi, S., and Zhang, K. (2018). Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 31, 5–103.
McClure, J. A., Conly, J. M., Obasuyi, O., Ward, L., Ugarte-Torres, A., Louie, T., et al. (2020). A novel assay for detection of methicillin-resistant Staphylococcus aureus directly from clinical samples. Front. Microbiol. 11:1295. doi: 10.3389/fmicb.2020.01295
Keywords: diabetic foot infections, Staphylococcus aureus, loop-mediated isothermal amplification, clustered regularly interspaced short palindromic repeats, metagenomics next generation sequencing
Citation: Chen Y, Shi Y, Zhu W, You J, Yang J, Xie Y, Zhao H, Li H, Fan S, Li L and Liu C (2021) Combining CRISPR-Cas12a-Based Technology and Metagenomics Next Generation Sequencing: A New Paradigm for Rapid and Full-Scale Detection of Microbes in Infectious Diabetic Foot Samples. Front. Microbiol. 12:742040. doi: 10.3389/fmicb.2021.742040
Received: 15 July 2021; Accepted: 14 September 2021;
Published: 07 October 2021.
Edited by:
Shaolin Wang, China Agricultural University, ChinaReviewed by:
Polly H. M. Leung, Hong Kong Polytechnic University, Hong Kong, SAR ChinaCopyright © 2021 Chen, Shi, Zhu, You, Yang, Xie, Zhao, Li, Fan, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Li, MzMxMjAxMkB6anUuZWR1LmNu; Chao Liu, bGl1Y2hhb3pqdUB6anUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.