
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 October 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.740348
This article is part of the Research Topic Zoonotic Microorganisms and Spread of Acquired Polymyxin Resistance Determinants View all 15 articles
 Emmanuel O. Ngbede1†
Emmanuel O. Ngbede1† Folasade Adekanmbi2†
Folasade Adekanmbi2† Anil Poudel2
Anil Poudel2 Anwar Kalalah2
Anwar Kalalah2 Patrick Kelly3
Patrick Kelly3 Yi Yang4
Yi Yang4 Andrew M. Adamu5
Andrew M. Adamu5 Salem T. Daniel6
Salem T. Daniel6 Alex A. Adikwu7
Alex A. Adikwu7 Chinedu A. Akwuobu1
Chinedu A. Akwuobu1 Paul O. Abba8
Paul O. Abba8 Levi M. Mamfe1
Levi M. Mamfe1 Nanven A. Maurice9
Nanven A. Maurice9 Mohammed I. Adah10
Mohammed I. Adah10 Olivia Lockyear2
Olivia Lockyear2 Patrick Butaye3,11
Patrick Butaye3,11 Chengming Wang2*
Chengming Wang2*Resistance to last resort drugs such as carbapenem and colistin is a serious global health threat. This study investigated carbapenem and colistin resistance in 583 non-duplicate Enterobacteriaceae isolates utilizing phenotypic methods and whole genome sequencing (WGS). Of the 583 isolates recovered from humans, animals and the environment in Nigeria, 18.9% (110/583) were resistant to at least one carbapenem (meropenem, ertapenem, and imipenem) and 9.1% (53/583) exhibited concurrent carbapenem-colistin resistance. The minimum inhibitory concentrations of carbapenem and colistin were 2–32 μg/mL and 8 to >64 μg/mL, respectively. No carbapenem resistant isolates produced carbapenemase nor harbored any known carbapenemase producing genes. WGS supported that concurrent carbapenem-colistin resistance was mediated by novel and previously described alterations in chromosomal efflux regulatory genes, particularly mgrB (M1V) ompC (M1_V24del) ompK37 (I70M, I128M) ramR (M1V), and marR (M1V). In addition, alterations/mutations were detected in the etpA, arnT, ccrB, pmrB in colistin resistant bacteria and ompK36 in carbapenem resistant bacteria. The bacterial isolates were distributed into 37 sequence types and characterized by the presence of internationally recognized high-risk clones. The results indicate that humans and animals in Nigeria may serve as reservoirs and vehicles for the global spread of the isolates. Further studies on antimicrobial resistance in African countries are warranted.
Antimicrobial resistance, particularly in Gram-negative bacteria, challenges the ability to treat common infections and is one of the greatest threats to global public health systems (Breijyeh et al., 2020). Resistance is more worrisome in resource-limited countries such as in sub-Saharan Africa where infections are common and last-resort antimicrobial agents are scarce and/or unaffordable (Tompkins et al., 2021).
Carbapenems and colistin play a significant role as “last resort” antibiotics in the treatment of infections caused by an extended spectrum of β-lactamase producing Enterobacteriaceae and multidrug-resistant Enterobacteriaceae (including carbapenem-resistant isolates), respectively. Categorized as the highest priority critically important drugs, the emergence of resistance to them, both individually and concurrently, is a serious source of healthcare concern (World Health Organization [WHO], 2018). Globally, resistance to colistin and carbapenem has been increasingly reported in isolates from animals and humans. The concurrent resistance of Enterobacteriaceae to carbapenems and colistin has been reported with increasing frequency in some parts of the world (Du et al., 2016; Yao et al., 2016; Lomonaco et al., 2018). Concurrent resistance determinants are usually located on conjugative plasmids and can thereby be co-transferred, setting the stage for pandrug resistance (Long et al., 2019).
Carbapenem resistant Enterobacteriaceae (CRE) comprise both carbapenemase producing (CP-CRE) and non-carbapenemase producing CRE (non-CP-CRE) strains. While CPE produce carbapenemases to hydrolyze carbapenem, non-CP-CRE have β-lactamase (ESBLs and AmpC enzymes) activity combined with structural mutations of the outer membrane protein and drug efflux pumps (Logan and Weinstein, 2017).
Colistin resistance in Enterobacteriaceae can be due to structural modifications of the bacterial lipopolysaccharide, such as the addition of 4-amino-4-deoxy-l-arabinose (L-Ara4N) or phosphoethanolamine (pEtN) (intrinsic resistance). Acquired resistance may result from chromosomal mutations in genes encoding the PhoPQ and PmrAB a two-component regulatory system, the mgrB, a negative regulator of PhoPQ, or the plasmid-borne mobile colistin resistance genes (mcr-1 to mcr-10) encoding a group of pEtN transferases (Aires et al., 2016; Zafer et al., 2019; Wang C. et al., 2020).
We recently reported the occurrence of mcr-mediated colistin resistance among 99 colistin resistant isolates; Escherichia coli (67/99), Klebsiella pneumoniae (30/99), Citrobacter werkmanii (1/99), and Alcaligenes faecalis (1/99) from humans and animals in Nigeria (Ngbede et al., 2020). Studies have also reported a high prevalence, up to 52%, of carbapenem resistance mediated by carbapenemase producing genes in samples from humans in Nigeria (Ogbolu and Webber, 2014; Jesumirhewe et al., 2017; Olowo-okere et al., 2019; Otokunefor et al., 2019; Ogbolu et al., 2020; Olalekan et al., 2020; Olowo-Okere et al., 2020; Shettima et al., 2020). These findings suggest that resistance to the last resort drugs carbapenem and colistin is a significant problem in Nigeria. Despite these increasing reports on colistin and carbapenem resistant bacteria emanating from sub Saharan Africa, the occurrence of co-resistance to both drugs and detailed insight into the molecular mechanism associated with this resistance phenotype has not been a major focus of such studies. Similarly, majority of these studies focus mainly on PCR detection of the most commonly and previously reported mechanisms including those mediated by blaKPC, blaNDM, blaIMP genes for carbapenem and the mcr- gene for colistin and rarely followed by WGS analyses of the isolates which is usually targeted at strain typing. This is in spite of the increasing non-detection of these genes in some of the isolates resistant to these antibiotics. Understanding the mechanisms of resistance is crucial to countering the mounting burden of infections caused by multidrug resistant bacteria, and whole-genome sequencing (WGS) has been shown to play a significant role in the rapid and accurate detection and characterization of known and emerging resistance determinants. Rarely, studies from sub Saharan Africa utilized whole genome sequencing to understand the detailed underlying genetic mechanism for carbapenem-colistin co-resistance in Enterobacteriaceae to the best of our knowledge. Such information is critical in formulating strategies for the clinical management and control of infections caused by multidrug-resistant Enterobacteriaceae. This study aimed to investigate the prevalence and genetic mechanisms underlying colistin-carbapenem co-resistance among E. coli and Klebsiella species (two important nosocomial pathogens) recovered from animals, humans, and the environment in Nigeria.
This study utilized a total of 583 non-duplicate Enterobacteriaceae acquired between 2016 and 2019. Isolates comprised 487 E. coli, 87 Klebsiella species, and nine Citrobacter species and were recovered from human clinical sample: stool (n = 60) and urine (n = 35), human hospital environment (n = 15), rectal swabs of camels (n = 40), cattle (n = 36), dogs (n = 42), pigs (n = 65), and cloacal swabs of poultry (n = 250) and clinical samples (liver) of poultry (n = 40) were used in this study (Ngbede et al., 2020). Procedures for sample collection, isolation, and identification have been previously reported (Ngbede et al., 2020). Ethical approval for the collection of samples from humans was provided by the Health Research and Ethics Committee of the facilities (FMH/FMC/MED.108/VOL.I/X and BSUTH/MKD/HREC/2013B/2018/0027).
Minimum inhibitory concentrations (MICs) of carbapenem (meropenem, ertapenem, and imipenem) were determined on the 583 isolates using the broth dilution method as recommended by the Clinical and Laboratory Standards Institute (CLSI) (Clinical and Laboratory Standards Institute, 2020). Briefly, colonies of each isolate from an overnight growth on tryptone soya agar were suspended in 5 mL normal saline to make an inoculum the equivalent of a 0.5 MacFarland standard. The turbidity of the inoculum was measured using a densitometer (BioScan). A 20 μL volume of the inoculum was dispensed into 2 mL of Mueller–Hinton broth containing different concentrations of the respective carbapenems and incubated at 35°C for 24 h. E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 served as controls.
The Carbapenem inactivation method (CIM) was used to screen the isolates for carbapenemase production as described (van der Zwaluw et al., 2015). Briefly, a loopful (≈10 μL) of the isolate was suspended in 400 μL of double distilled water and followed by a 10 μg meropenem disk (Oxoid, United Kingdom) which was immersed in the suspension and incubated for 2 h at 35°C. The disk was removed from the suspension with an inoculation loop and placed on a Mueller–Hinton agar plate inoculated with a 0.5 McFarland standard E. coli strain ATCC 29522 (a susceptible indicator strain) using a sterile cotton swab; it was subsequently incubated at 35°C for 24 h. A positive results was provided when isolates with carbapenemase production inactivates meropenem in the disk, allowing uninhibited growth of the susceptible indicator strain. A negative result occurred when meropenem disks were incubated in suspensions of isolates without carbapenemases, yielding a clear inhibition zone of the susceptible indicator strain (van der Zwaluw et al., 2015).
Antimicrobial susceptibility profiles of the isolates were determined by the disk diffusion method (Clinical and Laboratory Standards Institute, 2020) using 10 antimicrobial agents sourced from Oxoid, United Kingdom: Amoxycillin (10 μg), amoxicillin/clavulanic acid (30 μg), cefoxitin (30 μg), ceftriaxone (30 μg), ciprofloxacin (5 μg), chloramphenicol (10 μg), gentamicin (10 μg), doxycycline (30 μg), enrofloxacin (5 μg), and sulfamethoxazole-trimethoprim (25 μg). The results were interpreted based on the guidelines of the CLSI (Clinical and Laboratory Standards Institute, 2020).
Fifty out of the 53 isolates co-resistant to colistin and carbapenem, were randomly selected and subjected to WGS. Only 50 isolates were randomly selected for the WGS due to its associated cost and the selection of isolates represented the different species and susceptibility patterns encountered among the 53 co-resistant (carbapenem-colistin) isolates recovered from humans, animals and the environment. Genomic DNA (gDNA) was extracted from overnight cultures of the isolates using the Wizard Genomic DNA Purification Kit (Promega, United States) following the manufacturer’s recommendations. Isolates were sequenced using next-generation sequencing on an Illumina MiSeq platform (OE Biotech, Shanghai, China) using the V2 paired-end chemistry (2 × 250 bp).
Quality of the sequencing was assessed using QUAST v5.11 (Gurevich et al., 2013) before de novo assembly using the SeqMan Pro v.11.2.1 (DNASTAR, United States) followed by annotation using PROKKA v1.14.5. Species identity of the isolates was further confirmed with the WGS data using the KmerFinder v3.22 (Hasman et al., 2014; Larsen et al., 2014; Clausen et al., 2018). Genetic relatedness of isolates was analyzed by phylogroup (E. coli only), multilocus sequence types (MLST) and core genome MLST (cgMLST). MLST for each isolate was predicted using the pubMLST3 while the assignment of core genome MLST and phylogroups for the E. coli isolates were carried out using the cgMLSTFinder v1.14 (Zhou et al., 2020) and the online Clermont tool v20.035 (Beghain et al., 2018; Clermont et al., 2019).
In silico serotyping for the E. coli and Klebsiella isolates was performed using the SeroFinder 2.06 (Joensen et al., 2015) and Kaptive7 (Wick et al., 2018).
The presence of acquired antimicrobial resistance genes were investigated using the ResFinder v4.08 (Bortolaia et al., 2020) and CARD RGI 5.2.09 (Alcock et al., 2020). Plasmids were identified using PlasmidFinder 2.110 (Carattoli et al., 2014) and MGEFinder v1.0.311 (Johansson et al., 2021). Virulence genes were identified using the VirulenceFinder 2.0 (Lui et al., 2019).
Mutations in the genes previously identified as responsible for resistance to colistin (mgrB, prmAB, phoPQ, arnT, ccrB) and carbapenem (ompC, ompF, ompK/35/36/37, marR, acrR, ramR) were investigated by alignment with wild type reference genomes of E. coli strain K-12 substrain MG1655 (NC_000913.3) and K. pneumonia subspecies pneumoniae MGH 78578 (NC_009648.1). PROVEAN v1.1.312 was employed to predict the possible role/effect of observed amino acid substitutions (mutation/alteration) on protein functions i.e., colistin or carbapenem resistance (Choi and Chan, 2015). The standard PROVEAN cutoff score of ≤-2.5 and >-2.5 was used to categorize the mutation with deleterious and/or neutral effects on protein function, respectively (Choi et al., 2012; Choi and Chan, 2015).
The conjugative transferability of colistin and carbapenem resistance determinants was assessed using the solid mating conjugation assay with sodium azide-resistant E. coli J53 as the recipient (Ojo et al., 2016). Overnight cultures of the donor and recipient were mixed in a 1:4 ratio in tryptone soya broth and centrifuged at 14,000 × g for 1 min. The pelleted cells were resuspended in 15 μL of 0.85% NaCl, spotted onto MHA, incubated at 37°C for 20–24 h. Bacteria growing on the MHA were resuspended in 1 mL of 0.85% NaCl and a 100 μL serially diluted aliquot. Each dilution was placed on Brain Heart Infusion (BHI) agar supplemented with sodium azide (150 μL/mL) + colistin (2 μg/mL), meropenem (2 μg/mL) + sodium azide (150 μL/mL), colistin (2 μg/mL) + meropenem (2 μg/mL) + sodium azide (150 μL/mL) for the selection of transconjugant. The conjugation, or transfer, the frequency was measured based on the ratio of the observed transconjugant CFU (T) divided by the recipient CFU (T/R).
Of the 583 isolates tested, 110 (18.9%) comprising 53 E. coli and 57 Klebsiella species were resistant to at least one of three carbapenems (meropenem, 18.7%; 109/583; ertapenem, 16.8%; 98/583; imipenem, 18.2%; 106/583) and originated from pigs (n = 14), poultry (n = 39), humans (n = 33), environment (n = 3), cattle (n = 10), camel (n = 1), and dogs (n = 10). Based on colistin resistance findings determined in a former study (Ngbede et al., 2020) and this study, 53 isolates (9.1%; 53/583) were considered resistant to both colistin and at least one of the carbapenems. Of the 53 isolates expressing concurrent carbapenem-colistin resistance, 50 were randomly selected for further investigation: 23 human isolates (16 K. pneumoniae, 2 K. quasipneumoniae, and 5 E. coli), one hospital environmental isolate (K. quasipneumoniae), and 26 animal isolates (1 K. pneumoniae, 24 E. coli, and 1 C. werkmanii). The MIC of carbapenems ranged between 2 and 32 μg/mL (Supplementary Table 1).
The resistance profile is shown in Table 1 for the Citrobacter and E. coli isolates and Table 2 for the Klebsiella isolates. The 50 carbapenem/colistin resistant strains were negative for carbapenemase production, with 98% of the isolates resistant to the β-lactam antibiotics. This particularly included amoxicillin and amoxicillin/clavulanic acid as all the isolates except one E. coli, were resistant to amoxycillin. Similarly, 48, 66, and 78% of the isolates were resistant to aminoglycosides, sulphamethaxazole/trimethoprim, and fluoroquinolones, respectively (Tables 1, 2).
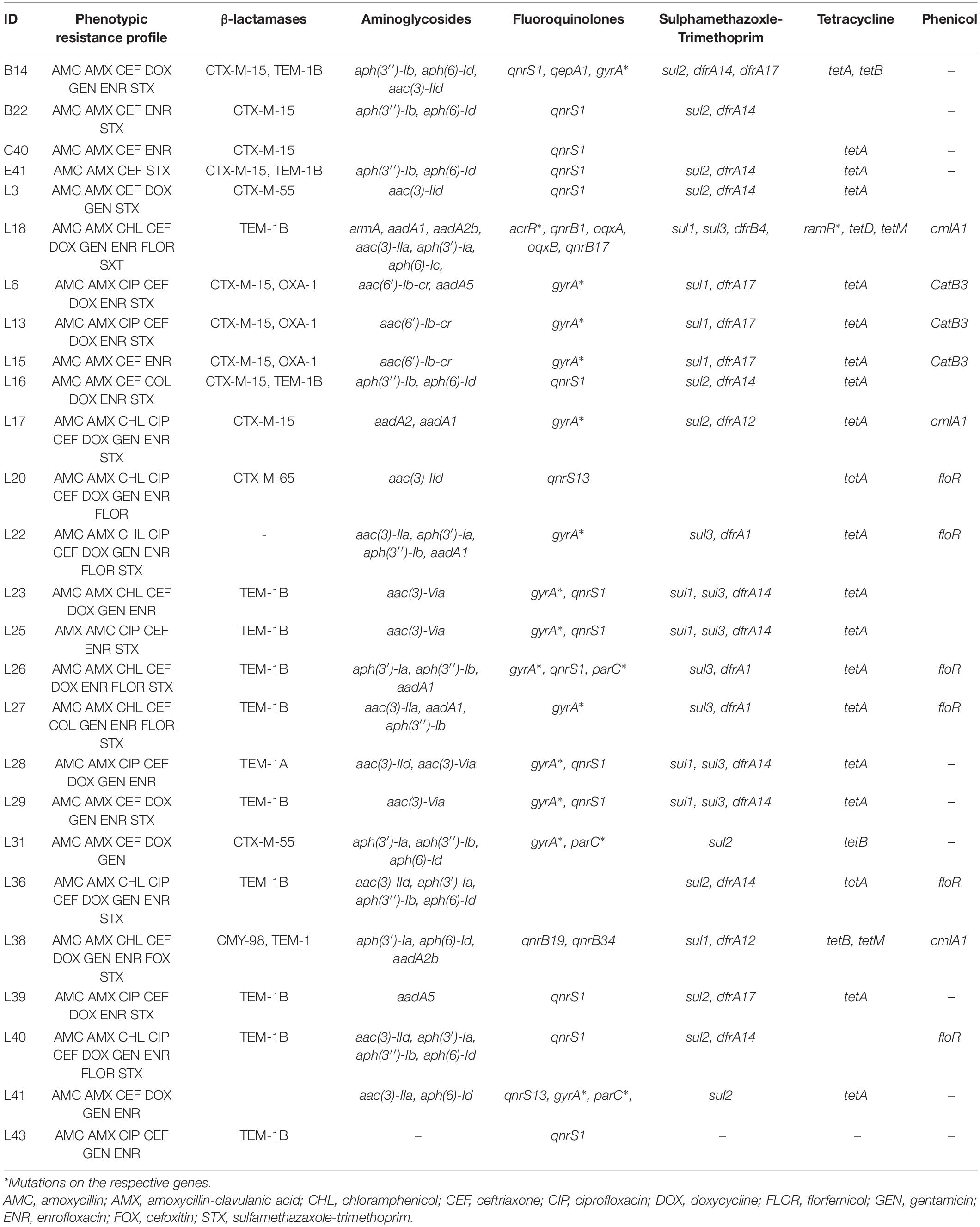
Table 1. Phenotypic and genotypic resistance profile of Enterobacteriaceae of animal origin expressing concurrent carbapenem-colistin resistance from Nigeria.
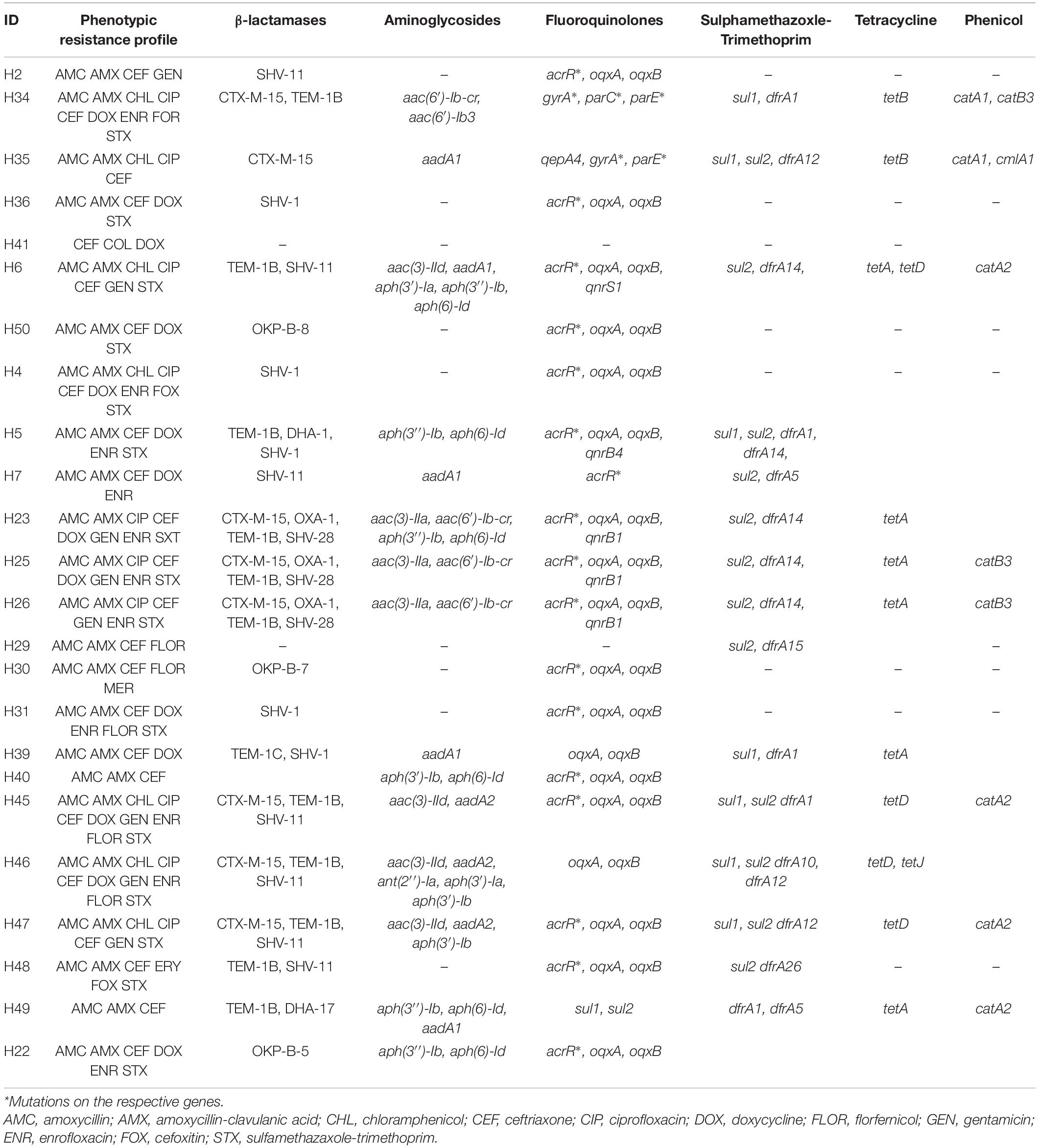
Table 2. Phenotypic and genotypic resistance profile of Enterobacteriaceae of human origin expressing concurrent carbapenem-colistin resistance from Nigeria.
Quality assessment of the genome reads revealed the Citrobacter isolate had a total genome length of 5.0 MB distributed over 60 contigs with an N50 and average GC content of 48,746 bp and 52, respectively. The total genome length of the E. coli isolates ranged from 3.7 to 4.2 MB distributed over 50–1465 contigs with an N50 length of 2,642–296,122 bp and an average GC content (mol%) of 50 while the Klebsiella isolates had an average GC content (mol%) of 50, total genome and N50 length of 5.0–5.6 MB and 126,035–462,504 bp, respectively distributed over 24–195 contigs. The WGS data further confirmed the identity of the 50 isolates as C. werkmanii (n = 1), E. coli (n = 29), K. pneumoniae (n = 17), K. quasipneumoniae (n = 3).
The WGS data confirmed that none of the isolates harbored any known carbapenemase genes. However, we found novel deletions, including the deletion of 24 amino acids from Met1 to Val24 (M1_V24del) in the ompC gene of all the E. coli isolates (29/29) (Table 3). Previously known substitutions in the ompC gene mediating carbapenem resistance were also detected among E. coli isolates (D192G/K in 17/29 isolates; N47D in 5/29 isolates) but, no mutations associated with carbapenem resistance were detected in the ompF gene. Among the Klebsiella isolates, we detected a HYTH insertion between amino acid Met233 and Thr234 (M233_T234insHYTH) (10/20) insertion in the ompK37, substitution A217S (15/20) and N218H (6/20) in ompK36, and substitution I70M (20/20), I128M (20/20), N230G (10/20) and T261A (1/20) in ompK37 (Table 4).
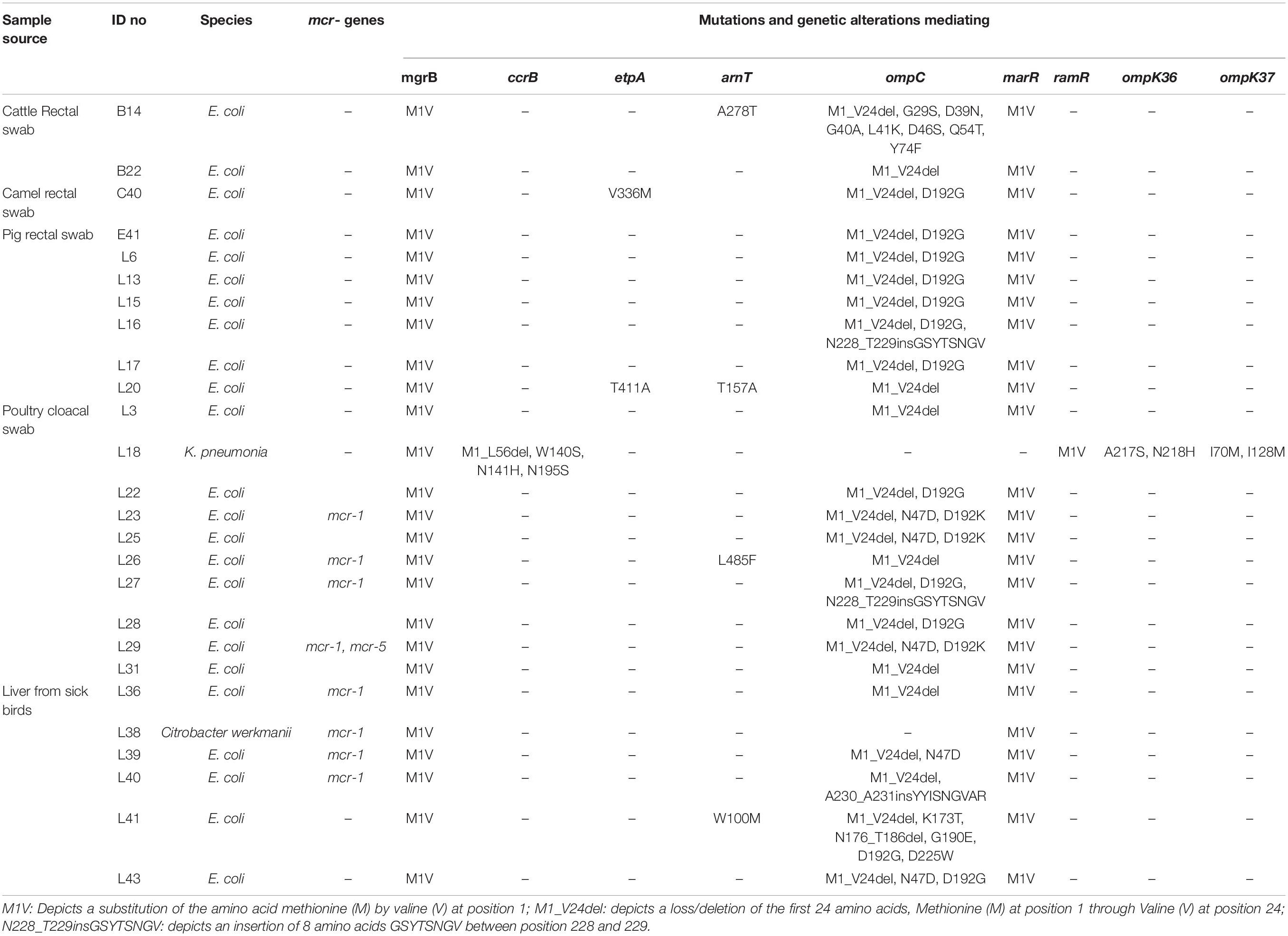
Table 3. Characteristics and resistance mechanisms associated with concurrent carbapenem-colistin among Enterobacteriaceae isolated from animals in Nigeria.
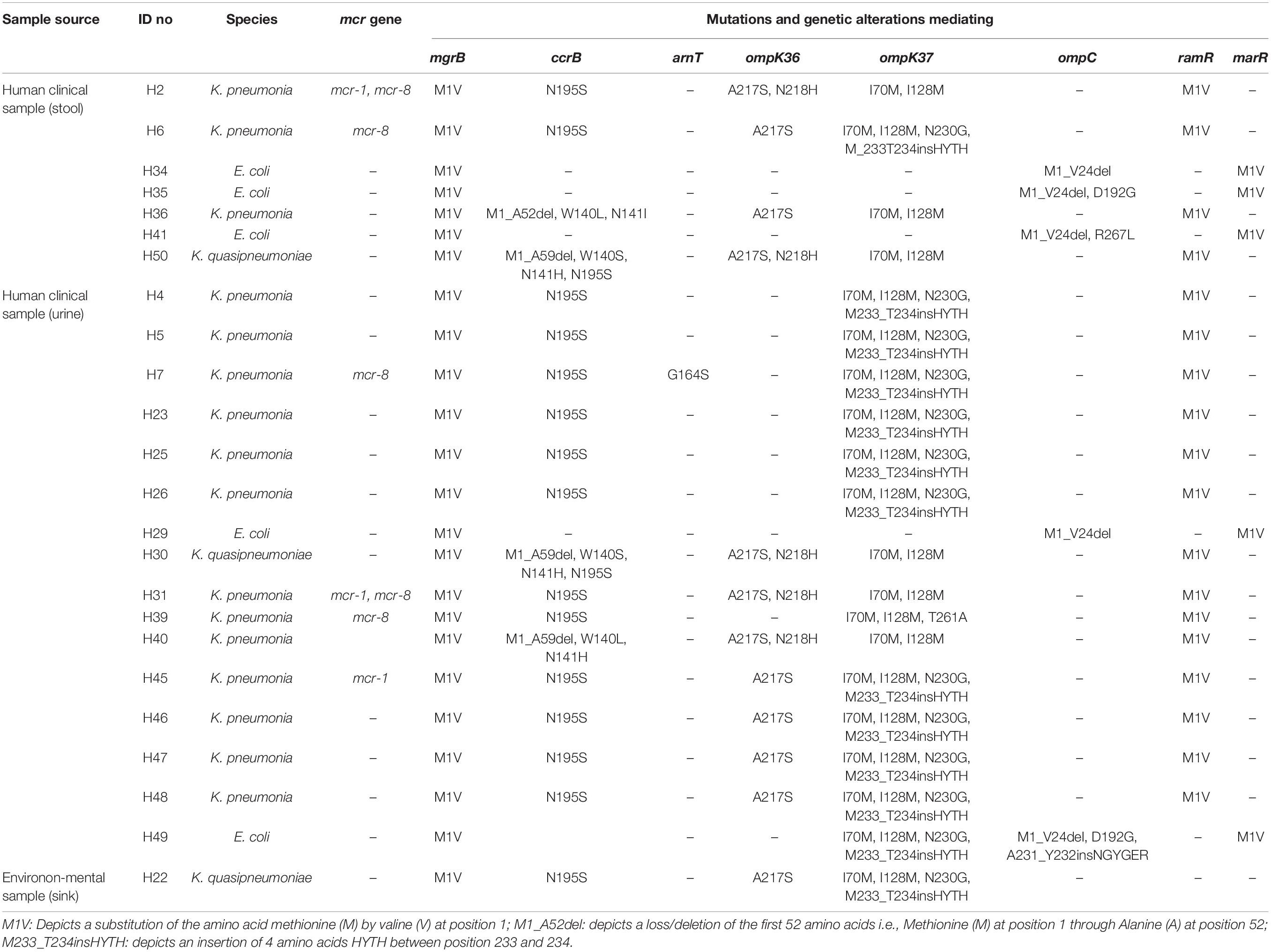
Table 4. Characteristics and resistance mechanisms associated with concurrent carbapenem-colistin among Enterobacteriaceae isolated from human sources in Nigeria.
Multiple substitutions were detected in the global regulator proteins marR and ramR, but not in the regulators AcrR, MarA, RamA, and SoxR. Only the M1V substitution in E. coli marR and K. pneumoniae ramR was associated with carbapenem resistance (Tables 3, 4).
Sixteen (one C. werkmanii, eight E. coli, and seven K. pneumoniae isolates) of the 50 carbapenem-colistin resistant isolates harbored the plasmid mediated colistin resistant genes: mcr-1 (n = 9), mcr-1 and +mcr-5 (n = 1), mcr-8 (n = 4), mcr-1, and +mcr-8 (n = 2) (Tables 3, 4). No point mutations associated with colistin resistance were detected in the pmrABC and phoPQ genes. Alterations in the mgrB were detected in all the isolates viz E. coli (29/29), Klebsiella (20/20), and Citrobacter (1/1) (Tables 3, 4). Novel substitutions were found in some the E. coli isolates, including V336M and T411I in the etpA and A278T, T157A, L485F, and W100M in the arnT (Table 3). The majority of the Klebsiella isolates harbored the N195S amino acid substitution (18/20), and some harbored the N141I/H (5/20) and W140L/S (5/20) substitutions in the ccrB, which were previously reported to mediate colistin resistance (Cheng et al., 2016). Finally, novel mutations as predicted to mediate colistin resistance, G164S in arnT, and the M1_L56del deletion were detected in one Klebsiella isolate (Tables 3, 4).
The 50 sequenced isolates harbored a plethora of resistant genes including β-lactamases (blaCTX–M, blaSHV, blaOXA–1, blaTEM–1, blaCMY, blaDHA), aminoglycosides [aac(3)-IIa, aac(6′)-Ib3, aac(3)-VIa, aph(3′)-Ia, aph(3″)-Ib, aph(6)-Ic, aph(6)-Id, armA, aadA1, aadA2b, aadA5, aac(6′)-Ib-cr], fluoroquinolones [qepA1, qepA4, oqxAB, qnrS1, qnrS13, qnrB4, qnrB17, qnrB19, qnrB34, aac(6′)-Ib-cr], sulfonamides (sul1, sul2, sul3), trimethoprim (dfrA1, dfrA10, dfrA12, dfrA14, dfrA15, dfrA17 dfrB4), phenicols (catA2, catB3, cmlA1, floR), tetracyclines [tet(A), tet(B), tet(D), tet(J), tet(M)] as well as mutations in acrR, gyrA, and parC for fluoroquinolones and ramR for tetracyclines particularly tigecycline (Tables 1, 2).
Genes blaTEM (15/30) and blaCTX–M (14/30) were found to be the predominant beta-lactamases among the E. coli isolates (Table 1) while blaSHV was frequently observed among the Klebsiella isolates (15/20) (Table 2). Some isolates (19/50) harbored more than one bla in different combinations. The single Citrobacter isolate in the study harbored the only AmpC type bla detected i.e., CMY-98 genotype (Table 1). Fluoroquinolone resistance was mostly mediated by plasmid mediated quinolone resistance qnr (16/29) and chromosomal mutations (S83L, D87N) of the gyrA (15/29) in the E. coli. Amongst the Klebsiella isolates, quinolone resistance was mostly due to a mutation of the acrR gene (18/20). Seven isolates harbored the aac(6)-Ib-cr gene which confers resistance to both aminoglycosides and fluoroquinolones.
Gene tet(A) (20/29) was the most common tetracycline resistance gene carried by the E. coli isolates while tet(A) (5/20) and tet(D) (n = 5/20) were the most common among the Klebsiella species. The tet(M) commonly restricted to Gram-positive bacteria was detected in one E. coli and a K. pneumoniae isolated from a cloacal swab from poultry. Additionally, one of the Klebsiella isolates (L18) harbored the ramR mutation (A19V) responsible for tigecycline resistance.
Multilocus sequence and phylogroup typing showed that the isolates were polyclonal and genetically diverse (Supplementary Tables 2, 3). The E. coli isolates clustered within six of the eight known phylogroups: A, B1, B2, C, D, E with phylogroup B1 been the predominantly occurring (12/29) based on the Clermont typing profile. MLST assigned the isolates into 23 sequence types (STs) (and one1 unknown ST) with four of the STs occurring more than once: ST191 (n = 3), ST224 (n = 2), ST410 (n = 3), and ST2485 (n = 3) (Supplementary Table 2). cgMLST assigned them into 24 different cgSTs due to genetic differences between the two ST224 strains which were identified as cgST72904 (isolate L17) and cgST38401 (isolate L22) (Supplementary Table 3).
In silico serotyping using the WGS data assigned the isolates into 23 serotypes, O128ab/ac:H20 (n = 2). Although the “O”-group antigen of eight isolates were unknown, the isolates belonged to the H9 (n = 3), H21 (n = 1), H23 (n = 1), H30 (n = 1), H37 (n = 1), and H45 (n = 1).
Similarly, the Klebsiella isolates were distributed into 14 different multilocus sequence types (STs) with three STs occurring more than once: ST45 (n = 2), ST307 (n = 3), and ST340 (n = 3). We were however unable to infer the ST for one of the K. quasipneumoniae isolate (Supplementary Table 3). In silico serotyping of the Klebsiella isolates assigned them into 15 different serotypes with the most commonly occurring serotype being KL102:O2v2 (n = 4).
Thirty different plasmid replicon types were detected via analyses of the WGS data using PlasmidFinder (Supplementary Tables 2, 3). The majority of the isolates harbored at least two plasmid replicon types with the most abundant being the IncFIB (AP001918) in E. coli (n = 17) and IncFIB(K) in Klebsiella (n = 16). Other dominant replicon types were IncFII and IncFIA in E. coli (n = 9 each), IncFII(K) and IncR in Klebsiella (n = 11 each). No plasmids were detected in three E. coli and two Klebsiella isolates.
Although, the mcr-1 was located in a unique contig that had the same sequence as the IncX4 plasmid backbone, no other resistance genes were found on the IncX4 plasmid in this study. Other resistance genes harbored on plasmids include sul2 and aph(3″)-Ib on IncQ1, blaTEM-1B on IncX1, blaTEM-1C on IncFII(K).
A total of 42 different virulence genes were detected across the 50 isolates (Supplementary Tables 2, 3). Carriage of extra-intestinal pathogenic E. coli (ExPEC) virulence associated genes (VAGs): cva, cvi, hylF, iroN, iss, iutA, ompT, sitA, traT was widespread among the E. coli isolates (Supplementary Table 2). Pathotype specific VAGs including astA (EAEC), afaD, hlyA, ihA, fyuA (UPEC), vat (APEC), and papC (APEC/UPEC) were also detected (Supplementary Table 2).
The conjugation assay confirmed transfer of the colistin resistant determinant from two isolate (H2 and L40) to the recipient E. coli J53 with conjugation frequencies of 3.4 × 10–1 cfu/recipient cfu and 4.6 × 10–1 cfu/recipient cfu, respectively. None of the isolates transferred the carbapenem resistant determinant. The colistin MIC for the two transconjugants was 4 μg/mL compared with 0.5 μg/mL for the E. coli J53. Furthermore, the transconjugants expressed two resistant phenotypes (L40: AMC-AMX-CIP-CEF-DOX-GEN; H2: AMC-AMX-CEF-GEN).
The last two decades have witnessed a significant rise in infections caused by multidrug resistant Enterobacteriaceae and resulted in an increase in the use of carbapenems and colistin as last resort drugs (Peyclit et al., 2019). Concurrent resistance to last resort drugs represents a serious health concern globally (Chaudhary, 2016; Serwecinska, 2020) and our study has provided evidence of high levels of concurrent resistance to colistin and carbapenem in Enterobacteriaceae in Nigeria with a prevalence of 9.1%. This is of major concern as Nigeria is a low-income country with minimal antimicrobial surveillance. This also indicates the emergence and establishment of potentially pandrug resistant strains, and creates limitations in the ability to control common infections.
While transferable mobile genetic elements such as mcr- and carbapenemase genes have generated much interest and are the target of most studies, our results show that concurrent carbapenem–colistin resistance in the E. coli and K. pneumoniae isolates is also linked to previously reported and novel mechanisms. These mechanisms include chromosomal mutations/disruptions affecting regulatory and non-regulatory genes controlling efflux-reflux pumps and membrane permeability. Such mutations/disruptions may play a greater role in resistance to carbapenem and colistin resistance than previously suspected.
Surprisingly, we found no evidence for carbapenemase producing genes in both our phenotypic and genomic investigations (WGS). This is in contrast to some studies on resistance in the region that reported carbapenemase genes such as blaNDM and blaOXA–181 are widespread (Jesumirhewe et al., 2017; Olalekan et al., 2020; Olowo-Okere et al., 2020; Shettima et al., 2020).
Consistent with previous reports on their role in colistin/carbapenem resistance, we detected the plasmid mediated colistin resistance genes; mcr-1 mcr-5, and mcr-8, and alterations/mutations in the mgrB, ccrB, that mediates colistin resistance as well as alterations/mutations in the OmpC, OmpK36, OmpK37 which mediates carbapenem resistance (Olaitan et al., 2014; Poirel et al., 2015; Cheng et al., 2016). We also identified multiple potential novel mutations/alteration associated with colistin resistance: arnT (W100M, T157A, G164S, A278T, L485F), etpA (V336M, T411H), pmrB (G164S, R256G), ccrB (M1_L56del, M1_KA52del, M1_A78del, M1_M59del) and carbapenem resistance: ompC (M1_V24del, K173T, N228_T229insGSYTSNGV, A231_Y232insNGYGER, A230_A231insYYISNGVAR), ompK37 (K27Q, D28Q, G29V, N30G, K31S, D33T, M1_Y25del, M233_T234insHYTH). Colistin resistance is mediated by LPS modification, which is encoded by the pmrHFIJKLM operon and the pmrC locus, and regulated by PhoPQ and PmrAB. This modification decreases the negative charge of the outer membrane, reducing its interaction with colistin (Trimble et al., 2016). Alterations in the genes of the two component regulatory systems (2CRS) particularly mgrB, phoP/phoQ, pmrA, pmrB, pmrC, and crrABC mediate colistin resistance (Wright et al., 2015; Cheng et al., 2016). Genetic alteration in the mgrB gene, which was common among the isolates, results in a disruption of the negative feedback loop of the PhoP/PhoQ, overexpression of the PhoP-regulated genes leading to the up-regulation of the pmrHFIJKLM operon and an abnormally high levels of lipid A modification, and ultimately a low susceptibility to colistin. Similarly, crrB mutations consistent with those detected in this study were shown to mediate reduced colistin susceptibility (Cheng et al., 2016) via induction of CrrC expression, which induces an elevated expression of the pmrHFIJKLM operon and pmrC via the PmrAB two-component system, loss of regulation of the crrAB gene that encodes a glycosyltransferase-like protein, which in turn leads to modification of lipid A and increased autophosphorylation of ccrB which leads to colistin resistance (Wright et al., 2015; Cheng et al., 2016; Aghapour et al., 2019). Porins are outer membrane proteins associated with the modulation of cellular permeability and antibiotic resistance. OmpF and OmpA in E. coli and OmpK35/36/37 in Klebsiella play a major role in antibiotic transport into the bacteria cell. Alterations in the gene encoding these proteins have been reported to infer with protein configuration and thus entry of antibiotics, particularly carbapenems (Fernández and Hancock, 2012). We hypothesized that the observed disruptions in these gene as observed among our isolates and identified as deleterious by PROVEAN may be responsible for the high carbapenem MIC and resistance in this study.
While the significance of these mutations and their impact on the MIC and resistance to carbapenem and colistin will require expression level analyses including transcriptomics and complementation assays, we hypothesize that the observed concurrent carbapenem–colistin resistance resulted from a combination of the chromosomal mutations/alterations in the mgrB (M1V) ompC (M1_V24del)ompK37 (I70M, I128M) and the regulatory efflux pump genes (marR (M1V), and ramR (M1V). Other studies have reported that mutations in the efflux system, particularly the global regulator marR and ramR as found in our study, β-lactamase production, and porin deficiency could play major roles in carbapenem resistance (Girlich et al., 2009; Findlay et al., 2012; Shin et al., 2012; Adler et al., 2013; Tsai et al., 2013; Chetri et al., 2019, 2020). An important finding in this study that we also wish to highlight, is the high probability for an isolate that is colistin resistant to also be carbapenem resistant, and vice versa.
Reports emanating particularly from Asia demonstrate a significant reduction in the rate of colistin resistant isolates due to the effect of the ban on colistin use in the wake of the detection of the plasmid mediated mcr-1 gene (Walsh and Wu, 2016; EMA/AMEG, 2021; Usui et al., 2021; Wang Y. et al., 2020). Similarly, countries such as the United States, Canada, and United Kingdom which have never approved colistin usage in animal production have continually reported lower rates of colistin resistant strains (EMA/AMEG, 2021). However, this is not the situation in Nigeria and other African countries where there is a rise in the numbers of colistin resistant strains, as demonstrated in this study. No such ban or regulation on colistin use exist in Nigeria where its use is currently widespread and is a common active ingredient in most antibiotic combinations/preparation used in livestock production for the purpose of prophylaxis and therapy. Although carbapenem is rarely used in food animals in Nigeria, the inappropriate use of colistin has been shown to provide selective pressure for the emergence of colistin and multidrug (including carbapenem) resistant strains (Napier et al., 2013). We, therefore, hypothesize that the high rates of co-resistance observed in our study may be connected to the widespread use of colistin, particularly in livestock production in Nigeria.
Majority of the isolates expressed multidrug resistance profiles, including high resistance to amoxicillin-clavulanic acid (98%), fluoroquinolones (78%), and gentamicin (48%) which are widely used in the treatment of infections in Nigerian hospitals. The high resistance rates we recorded are in accordance with data in other reports on human and animal isolates from Nigeria and Sub-Saharan Africa (Ojo et al., 2016; Chah et al., 2018; Aworh et al., 2019; Olalekan et al., 2020; Shettima et al., 2020). Most of our isolates also carried the blaCTX–M–15 which is consistent with the increasing reports of this genotype from animal and human sources in Nigeria (Chah et al., 2018; Okpara et al., 2018; Olowo-Okere et al., 2020). There is thus growing evidence that this genotype is expanding rapidly and might become the dominant mechanism mediating resistance to the β-lactams.
The significant clonal diversity i.e., polyclonality observed amongst our isolates is consistent with previous reports, particularly those of carbapenem and colistin resistant isolates in Nigeria where the population structure is diverse (Ngbede et al., 2020; Olalekan et al., 2020). Some of the animal and human isolates in our study had similar STs with previously reported ″high-risk″ clones including K. pneumoniae ST11, ST17, ST45, ST340 (human isolates) and E. coli ST58, ST744, ST410 (animal isolates). These high risk STs are known for their global dissemination, ease of transmission between different hosts, ability to cause disease and acquire genetic determinants such as virulence factors, epidemic plasmids and antibiotic resistance that provide them with a competitive advantage over other bacterial clones (Lee et al., 2016; Roer et al., 2018; Feng et al., 2019; Nadimpalli et al., 2019; Patiño-Navarrete et al., 2020). The E. coli ST58, ST744, ST410 virulence profile categorized them to the UPEC, EAEC and DAEC pathotypes. Similarly, the ST11 and ST340 detected in our human isolates are closely related to ST258, all belonging to clonal complex CC258 which has been associated with outbreaks, pandemics and mass dissemination of KPC K. pneumoniae (Netikul and Kiratisin, 2015; Sui et al., 2018; Cienfuegos-Gallet et al., 2019; Fu et al., 2019; Zhao et al., 2019).
In this study, we report evidence for the occurrence of multidrug resistant Enterobacteriaceae with concurrent carbapenem-colistin resistance in 9.1% of the isolates. The genetic mechanism underlying this concurrent resistance phenotype was majorly novel and previously known chromosomal alterations (deletion, insertions, and substitutions). The plasmid-mediated colistin resistance gene mcr- in combination with these chromosomal alterations accounted for colistin resistance in few of the isolates. Some of the E. coli and Klebsiella isolates expressing concurrent carbapenem-colistin resistance in this study belonged to the internationally recognized “high-risk” clones. The combination of diverse drug resistance genes and sequence types highlight the considerable genome plasticity and polyclonality that characterize the population structure of both clinical and non-clinical colistin and CRE in Nigeria. The polyclonality might create considerable problems during outbreak tracing and source attribution.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by Research and Ethics Committee of the facilities (FMH/FMC/MED.108/VOL.I/X and BSUTH/MKD/HREC/2013B/2018/0027). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
EN and CW: conceptualization and funding acquisition. EN, FA, AP, and CW: methodology. EN, AMA, SD, AAA, CA, PA, LM, NM, and MA: sampling/investigation. EN, FA, AP, AK, YY, and PK: data curation. EN, MA, and CW: supervision. EN and FA: writing—original draft preparation. EN, YY, PK, OL, PB, and CW writing—review and editing. All authors read and agreed to the published version of the manuscript.
This work was supported by the International Society for Infectious Diseases (ISID) 2018 Research Grant, USDA-ARS program (58-6040-9-017), and by Alabama Agricultural Experimental Station and the USDA National Institute of Food and Agriculture, Hatch project (ALA052-1-17026).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.740348/full#supplementary-material
Adler, M., Anjum, M., Andersson, D. I., and Sandegren, L. (2013). Influence of acquired β-lactamases on the evolution of spontaneous carbapenem resistance in Escherichia coli. J. Antimicrob. Chemother. 68, 51–59. doi: 10.1093/jac/dks368
Aghapour, Z., Gholizadeh, P., Ganbarov, K., Bialvaei, A. Z., Mahmood, S. S., Tanomand, A., et al. (2019). Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 12, 965–975. doi: 10.2147/IDR.S199844
Aires, C. A., Pereira, P. S., Asensi, M. D., and Carvalho-Assef, A. P. (2016). mgrB mutations mediating polymyxin B resistance in Klebsiella pneumoniae isolates from rectal surveillance swabs in Brazil. Antimicrob. Agents Chemother. 60, 6969–6972. doi: 10.1128/AAC.01456-16
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Aworh, M. K., Kwaga, J., Okolocha, E., Mba, N., and Thakur, S. (2019). Prevalence and risk factors for multi-drug resistant Escherichia coli among poultry workers in the Federal Capital Territory, Abuja, Nigeria. PLoS One 14:e0225379. doi: 10.1371/journal.pone.0225379
Beghain, J., Bridier-Nahmias, A., Le Nagard, H., Denamur, E., and Clermont, O. (2018). ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 4:e000192. doi: 10.1099/mgen.0.000192
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Breijyeh, Z., Jubeh, B., and Karaman, R. (2020). Resistance of Gram-negative bacteria to current antibacterial agents and approaches to resolve It. Molecules 25:1340. doi: 10.3390/molecules25061340
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chah, K. F., Ugwu, I. C., Okpala, A., Adamu, K. Y., Alonso, C. A., Ceballos, S., et al. (2018). Detection and molecular characterisation of extended-spectrum β-lactamase-producing enteric bacteria from pigs and chickens in Nsukka, Nigeria. J. Glob. Antimicrob. Resist. 15, 36–40. doi: 10.1016/j.jgar.2018.06.002
Chaudhary, A. S. (2016). A review of global initiatives to fight antibiotic resistance and recent antibiotics discovery. Acta Pharm Sinica B 6, 552–556. doi: 10.1016/j.apsb.2016.06.004
Cheng, Y. H., Lin, T. L., Lin, Y. T., and Wang, J. T. (2016). Amino acid substitutions of CrrB responsible for resistance to colistin through CrrC in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 60, 3709–3716. doi: 10.1128/AAC.00009-16
Chetri, S., Bhowmik, D., Paul, D., Pandey, P., Chanda, D. D., Chakravarty, A., et al. (2019). AcrAB-TolC efflux pump system plays a role in carbapenem non-susceptibility in Escherichia coli. BMC Microbiol. 19:210. doi: 10.1186/s12866-019-1589-1
Chetri, S., Das, B. J., Bhowmik, D., Chanda, D. D., Chakravarty, A., and Bhattacharjee, A. (2020). Transcriptional response of mar, sox and rob regulon against concentration gradient carbapenem stress within Escherichia coli isolated from hospital acquired infection. BMC Res. Notes 13:168. doi: 10.1186/s13104-020-04999-2
Choi, Y., and Chan, A. P. (2015). PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747. doi: 10.1093/bioinformatics/btv195
Choi, Y., Sims, G. E., Murphy, S., Miller, J. R., and Chan, A. P. (2012). Predicting the functional effect of amino acid substitutions and indels. PLoS One 7:e46688. doi: 10.1371/journal.pone.0046688
Cienfuegos-Gallet, A. V., Ocampo de Los Ríos, A. M., Sierra Viana, P., Ramirez Brinez, F., Restrepo Castro, C., Roncancio Villamil, G., et al. (2019). Risk factors and survival of patients infected with carbapenem-resistant Klebsiella pneumoniae in a KPC endemic setting: a case-control and cohort study. BMC Infect Dis. 19:830. doi: 10.1186/s12879-019-4461-x
Clausen, P. T. L. C., Aarestrup, F. M., and Lund, O. (2018). Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics 19:307. doi: 10.1186/s12859-018-2336-6
Clermont, O., Dixit, O. V. A., Vangchhia, B., Condamine, B., Dion, S., Bridier-Nahmias, A., et al. (2019). Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol. 21, 3107–3117. doi: 10.1111/1462-2920.14713
Clinical and Laboratory Standards Institute (2020). Performance Standards for Antimicrobial Susceptibility Testing; M100-ED30, 30th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Du, H., Chen, L., Tang, Y. W., and Kreiswirth, B. N. (2016). Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect. Dis. 16, 287–288. doi: 10.1016/S1473-3099(16)00056-6
EMA/AMEG (2021). Updated Advice on the Use of Colistin Products in Animals within the European Union: Development of Resistance and Possible Impact on Human and Animal Health. Available: https://www.ema.europa.eu/en/documents/scientific-guideline/updated-advice-usecolistin-products-animals-within-european-union-development-resistance-possible_en-0.pdf (accessed August 10, 2021).
Feng, Y., Liu, L., Lin, J., Ma, K., Long, H., Wei, L., et al. (2019). Key evolutionary events in the emergence of a globally disseminated, carbapenem resistant clone in the Escherichia coli ST410 lineage. Commun. Biol. 2:322. doi: 10.1038/s42003-019-0569-1
Fernández, L., and Hancock, R. E. (2012). Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 25, 661–681. doi: 10.1128/CMR.00043-12
Findlay, J., Hamouda, A., Dancer, S. J., and Amyes, S. G. (2012). Rapid acquisition of decreased carbapenem susceptibility in a strain of Klebsiella pneumoniae arising during meropenem therapy. Clin. Microbiol. Infect. 18, 140–146. doi: 10.1111/j.1469-0691.2011.03515.x
Fu, P., Tang, Y., Li, G., Yu, L., Wang, Y., and Jiang, X. (2019). Pandemic spread of blaKPC-2 among Klebsiella pneumoniae ST11 in China is associated with horizontal transfer mediated by IncFII-like plasmids. Int. J. Antimicrob. Agents 54, 117–124. doi: 10.1016/j.ijantimicag.2019.03.014
Girlich, D., Poirel, L., and Nordmann, P. (2009). CTX-M expression and selection of ertapenem resistance in Klebsiella pneumoniae and Escherichia coli. Antimicrob. Agents Chemother. 53, 832–834. doi: 10.1128/AAC.01007-08
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hasman, H., Saputra, D., Sicheritz-Ponten, T., Lund, O., Svendsen, C. A., Frimodt-Møller, N., et al. (2014). Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol. 52, 139–146. doi: 10.1128/JCM.02452-13
Jesumirhewe, C., Springer, B., Lepuschitz, S., Allerberger, F., and Ruppitsch, W. (2017). Carbapenemase-producing Enterobacteriaceae isolates from Edo State, Nigeria. Antimicrob. Agents Chemother. 61:e00255-17. doi: 10.1128/AAC.00255-17
Joensen, K. G., Tetzschner, A. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy In Silico serotyping of Escherichia coli isolates by use of Whole-Genome Sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Johansson, M. H. K., Bortolaia, V., Tansirichaiya, S., Aarestrup, F. M., Roberts, A. P., and Petersen, T. N. (2021). Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 76, 101–109. doi: 10.1093/jac/dkaa390
Larsen, M. V., Cosentino, S., Lukjancenko, O., Saputra, D., Rasmussen, S., Hasman, H., et al. (2014). Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 52, 1529–1539. doi: 10.1128/JCM.02981-13
Lee, C. R., Lee, J. H., Park, K. S., Kim, Y. B., Jeong, B. C., and Lee, S. H. (2016). Global Dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7:895. doi: 10.3389/fmicb.2016.00895
Logan, L. K., and Weinstein, R. A. (2017). The Epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215, (Suppl._1) S28–S36. doi: 10.1093/infdis/jiw282
Lomonaco, S., Crawford, M. A., Lascols, C., Timme, R. E., Anderson, K., Hodge, D. R., et al. (2018). Resistome of carbapenem- and colistin-resistant Klebsiella pneumoniae clinical isolates. PLoS One 13:e0198526. doi: 10.1371/journal.pone.0198526
Long, H., Feng, Y., Ma, K., Liu, L., McNally, A., and Zong, Z. (2019). The co-transfer of plasmid-borne colistin-resistant genes mcr-1 and mcr-3.5, the carbapenemase gene blaNDM-5 and the 16S methylase gene rmtB from Escherichia coli. Sci. Rep. 9:696. doi: 10.1038/s41598-018-37125-1
Lui, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comprehensive pathogenomic platform with an interactive web interface. Nucleic Acid Res. 47, D687–D692. doi: 10.1093/nar/gky1080
Nadimpalli, M. L., de Lauzanne, A., Phe, T., Borand, L., Jacobs, J., Fabre, L., et al. (2019). Escherichia coli ST410 among humans and the environment in Southeast Asia. Int. J. Antimicrob. Agents 54, 228–232. doi: 10.1016/j.ijantimicag.2019.05.024
Napier, B. A., Burd, E. M., Satola, S. W., Cagle, S. M., Ray, S. M., McGann, P., et al. (2013). Clinical use of colistin induces cross-resistance to host antimicrobials in Acinetobacter baumannii. mBio 4:e000210-13. doi: 10.1128/mBio.00021-13
Netikul, T., and Kiratisin, P. (2015). Genetic characterization of carbapenem-resistant Enterobacteriaceae and the spread of carbapenem-resistant Klebsiella pneumonia ST340 at a university hospital in Thailand. PLoS One 10:e0139116. doi: 10.1371/journal.pone.0139116
Ngbede, E. O., Poudel, A., Kalalah, A., Yang, Y., Adekanmbi, F., Adikwu, A. A., et al. (2020). Identification of mobile colistin resistance genes (mcr-1.1, mcr-5 and mcr-8.1) in Enterobacteriaceae and Alcaligenes faecalis of human and animal origin, Nigeria. Int. J. Antimicrob. Agents 56:106108. doi: 10.1016/j.ijantimicag.2020.106108
Ogbolu, D. O., Piddock, L. J. V., and Webber, M. A. (2020). Opening Pandora’s box: high-level resistance to antibiotics of last resort in Gram-negative bacteria from Nigeria. J. Glob. Antimicrob. Resist. 21, 211–217. doi: 10.1016/j.jgar.2019.10.016
Ogbolu, D. O., and Webber, M. A. (2014). High-level and novel mechanisms of carbapenem resistance in Gram-negative bacteria from tertiary hospitals in Nigeria. Int. J. Antimicrob. Agents 43, 412–417. doi: 10.1016/j.ijantimicag.2014.01.014
Ojo, O. E., Schwarz, S., and Michael, G. B. (2016). Detection and characterization of extended-spectrum β-lactamase-producing Escherichia coli from chicken production chains in Nigeria. Vet. Microbiol. 194, 62–68. doi: 10.1016/j.vetmic.2016.04.022
Okpara, E. O., Ojo, O. E., Awoyomi, O. J., Dipeolu, M. A., Oyekunle, M. A., and Schwarz, S. (2018). Antimicrobial usage and presence of extended-spectrum β-lactamase-producing Enterobacteriaceae in animal-rearing households of selected rural and peri-urban communities. Vet. Microbiol. 218, 31–39. doi: 10.1016/j.vetmic.2018.03.013
Olaitan, A. O., Diene, S. M., Kempf, M., Berrazeg, M., Bakour, S., Gupta, S. K., et al. (2014). Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int. J. Antimicrob. Agents 44, 500–507. doi: 10.1016/j.ijantimicag.2014.07.020
Olalekan, A., Onwugamba, F., Iwalokun, B., Mellmann, A., Becker, K., and Schaumburg, F. (2020). High proportion of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae among extended-spectrum β-lactamase-producers in Nigerian hospitals. J. Glob. Antimicrob. Resist. 21, 8–12. doi: 10.1016/j.jgar.2019.09.007
Olowo-okere, A., Abdullahi, M. A., Ladidi, B. K., Suleiman, S., Tanko, N., Ungokore, H. Y., et al. (2019). Emergence of metallo-b-lactamase producing gram-negative bacteria in a hospital with no history of carbapenem usage in northwest Nigeria. Ife J. Sci. 21, 323–331. doi: 10.4314/ijs.v21i2.6
Olowo-Okere, A., Ibrahim, Y. K. E., Olayinka, B. O., Ehinmidu, J. O., Mohammed, Y., Nabti, L. Z., et al. (2020). Phenotypic and genotypic characterization of clinical carbapenem-resistant Enterobacteriaceae isolates from Sokoto, northwest Nigeria. New Microbes New Infect. 37:100727. doi: 10.1016/j.nmni.2020.100727
Otokunefor, K., Tamunokuro, E., and Amadi, A. (2019). Molecular detection of mobilized colistin resistance (mcr-1) gene in Escherichia coli isolates from Port Harcourt, Nigeria. J. Appl. Sci. Environ. Manag. 23, 401–405.
Patiño-Navarrete, R., Rosinski-Chupin, I., Cabanel, N., Gauthier, L., Takissian, J., Madec, J. Y., et al. (2020). Stepwise evolution and convergent recombination underlie the global dissemination of carbapenemase-producing Escherichia coli. Genome Med. 12:10. doi: 10.1186/s13073-019-0699-6
Peyclit, L., Baron, S. A., and Rolain, J. M. (2019). Drug repurposing to fight colistin and carbapenem-resistant bacteria. Front. Cell Infect. Microbiol. 9:193. doi: 10.3389/fcimb.2019.00193
Poirel, L., Jayol, A., Bontron, S., Villegas, M. V., Ozdamar, M., Türkoglu, S., et al. (2015). The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 70, 75–80. doi: 10.1093/jac/dku323
Roer, L., Overballe-Petersen, S., Hansen, F., Schønning, K., Wang, M., Røder, B. L., et al. (2018). Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere 3:e00337-18. doi: 10.1128/mSphere.00337-18
Serwecinska, L. (2020). Antimicrobials and antibiotic-resistant bacteria: a risk to the environment and to public health. Water 12:3313. doi: 10.3390/w12123313
Shettima, S. A., Tickler, I. A., Dela Cruz, C. M., and Tenover, F. C. (2020). Characterisation of carbapenem-resistant Gram-negative organisms from clinical specimens in Yola, Nigeria. J. Glob. Antimicrob. Resist. 21, 42–45. doi: 10.1016/j.jgar.2019.08.017
Shin, S. Y., Bae, I. K., Kim, J., Jeong, S. H., Yong, D., Kim, J. M., et al. (2012). Resistance to carbapenems in sequence type 11 Klebsiella pneumoniae is related to DHA-1 and loss of OmpK35 and/or OmpK36. J. Med. Microbiol. 61,(Pt 2) 239–245. doi: 10.1099/jmm.0.037036-0
Sui, W., Zhou, H., Du, P., Wang, L., Qin, T., Wang, M., et al. (2018). Whole genome sequence revealed the fine transmission map of carbapenem-resistant Klebsiella pneumonia isolates within a nosocomial outbreak. Antimicrob. Resist. Infect. Control. 7:70. doi: 10.1186/s13756-018-0363-8
Tompkins, K., Juliano, J. J., and van Duin, D. (2021). Antimicrobial resistance in Enterobacterales and its contribution to sepsis in Sub-saharan Africa. Front. Med. 8:615649. doi: 10.3389/fmed.2021.615649
Trimble, M. J., Mlynárčik, P., Kolář, M., and Hancock, R. E. (2016). Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 6:a025288. doi: 10.1101/cshperspect.a025288
Tsai, Y. K., Liou, C. H., Fung, C. P., Lin, J. C., and Siu, L. K. (2013). Single or in combination antimicrobial resistance mechanisms of Klebsiella pneumoniae contribute to varied susceptibility to different carbapenems. PLoS One 8:e79640. doi: 10.1371/journal.pone.0079640
Usui, M., Nozawa, Y., Fukuda, A., Sato, T., Yamada, M., Makita, K., et al. (2021). Decreased colistin resistance and mcr-1 prevalence in pig-derived Escherichia coli in Japan after banning colistin as a feed additive. J. Glob. Antimicrob. Resist. 24, 383–386. doi: 10.1016/j.jgar.2021.01.016
van der Zwaluw, K., de Haan, A., Pluister, G. N., Bootsma, H. J., de Neeling, A. J., and Schouls, L. M. (2015). The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS One 10:e0123690. doi: 10.1371/journal.pone.0123690
Walsh, T. R., and Wu, Y. (2016). China bans colsitin as a feed additive for animals. Lancet Infect. Dis. 16, 1102–1103. doi: 10.1016/s1473-3099(16)30329-2
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., and Zong, Z. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020). Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/s1473-3099(20)30149-3
Wick, R. R., Heinz, E., Holt, K. E., and Wyres, K. L. (2018). Kaptive Web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol. 56:e00197-18. doi: 10.1128/JCM.00197-18
World Health Organization [WHO] (2018). Critically Important Antimicrobials for Human Medicine 6th Revision. Geneva: WHO.
Wright, M. S., Suzuki, Y., Jones, M. B., Marshall, S. H., Rudin, S. D., van Duin, D., et al. (2015). Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob. Agents Chemother. 59, 536–543. doi: 10.1128/AAC.04037-14
Yao, X., Doi, Y., Zeng, L., Lv, L., and Liu, J. H. (2016). Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis. 16, 288–289. doi: 10.1016/S1473-3099(16)00057-8
Zafer, M. M., El-Mahallawy, H. A., Abdulhak, A., Amin, M. A., Al-Agamy, M. H., and Radwan, H. H. (2019). Emergence of colistin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli strains isolated from cancer patients. Ann. Clin. Microbiol. Antimicrob. 18:40. doi: 10.1186/s12941-019-0339-4
Zhao, Y., Zhang, X., Torres, V. V. L., Liu, H., Rocker, A., Zhang, Y., et al. (2019). An outbreak of carbapenem-resistant and hypervirulent Klebsiella pneumoniae in an intensive care unit of a major teaching Hospital in Wenzhou, China. Front. Public Health 7:229. doi: 10.3389/fpubh.2019.00229
Keywords: concurrent carbapenem-colistin resistance, Enterobacteriaceae, high-risk clones, Nigeria, Africa, whole genome sequencing
Citation: Ngbede EO, Adekanmbi F, Poudel A, Kalalah A, Kelly P, Yang Y, Adamu AM, Daniel ST, Adikwu AA, Akwuobu CA, Abba PO, Mamfe LM, Maurice NA, Adah MI, Lockyear O, Butaye P and Wang C (2021) Concurrent Resistance to Carbapenem and Colistin Among Enterobacteriaceae Recovered From Human and Animal Sources in Nigeria Is Associated With Multiple Genetic Mechanisms. Front. Microbiol. 12:740348. doi: 10.3389/fmicb.2021.740348
Received: 12 July 2021; Accepted: 09 September 2021;
Published: 06 October 2021.
Edited by:
Azucena Mora Gutiérrez, University of Santiago de Compostela, SpainReviewed by:
Mehmet Demirci, Kırklareli University, TurkeyCopyright © 2021 Ngbede, Adekanmbi, Poudel, Kalalah, Kelly, Yang, Adamu, Daniel, Adikwu, Akwuobu, Abba, Mamfe, Maurice, Adah, Lockyear, Butaye and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengming Wang, d2FuZ2NoZUBhdWJ1cm4uZWR1
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.