
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 November 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.738896
This article is part of the Research Topic The Development and Utilization of Novel Antibiotic Alternatives View all 15 articles
 Geyin Zhang1†
Geyin Zhang1† Yunqiao Yang1†
Yunqiao Yang1† Fareed Uddin Memon1†
Fareed Uddin Memon1† Kaiyuan Hao1
Kaiyuan Hao1 Baichang Xu1
Baichang Xu1 Shuaiyang Wang1
Shuaiyang Wang1 Ying Wang1
Ying Wang1 Enyun Wu1
Enyun Wu1 Xiaogang Chen1
Xiaogang Chen1 Wenguang Xiong2
Wenguang Xiong2 Hongbin Si1*
Hongbin Si1*The objective of this study was to evaluate the antibacterial mechanisms of phenolic acids as natural approaches against multi-drug resistant Escherichia coli (E. coli). For that purpose, five phenolic acids were combined with each other and 31 combinations were obtained in total. To select the most potent and effective combination, all of the obtained combinations were examined for minimum inhibitory concentration (MIC) and it was found that the compound phenolic acid (CPA) 19 (protocatechuic acid, hydrocinnamic acid, and chlorogenic acid at concentrations of 0.833, 0.208, and 1.677 mg/mL, respectively) showed better efficacy against E. coli compared to other combinations. Furthermore, based on tandem mass tag (TMT) proteomics, the treatment of CPA 19 significantly downregulated the proteins associated with resistance (Tsr, Tar, CheA, and CheW), OmpF, and FliC of multidrug-resistant E. coli. At the same time, we proved that CPA 19 improves the sensitivity of E. coli to antibiotics (ceftriaxone sodium, amoxicillin, fosfomycin, sulfamonomethoxine, gatifloxacin, lincomycin, florfenicol, cefotaxime sodium, and rifampicin), causes the flagellum to fall off, breaks the structure of the cell wall and cell membrane, and leads to macromolecules leaks from the cell. This evidence elaborated the potential therapeutic efficacy of CPA 19 and provided a significant contribution to the discovery of antibacterial agents.
About 78 years ago, penicillin and other antibiotics were discovered as antimicrobial agents and introduced in various clinical treatments because of their positive influences in preventing bacterial infections (Paitan, 2018). However, Abraham and Chain (1988) reported the emergence of antimicrobial resistance.
Among these resistant bacterial strains, species of E. coli have emerged against the variety of antibiotic agents used in clinical practice, which not only damages the production of the animal industry, but also seriously affects the health of human beings. Therefore, there is an urgent need for the search of safe and effective approaches to overcome single as well as multidrug-resistant bacterial infections (Roth et al., 2019).
In recent years, more and more researchers have been devoted to the study of the inhibition of plant polyphenols because of their natural and broad antibacterial properties (Yun-Seok et al., 2010; Sidhu et al., 2014; Yang et al., 2020). Phenolic acids are major plant metabolites, which occur in all parts of the plant: shells, leaves, seeds, fruits, and wood parts (Efenberger et al., 2021). Previous studies have demonstrated that phenolic compounds possess a variety of biological functions including anti-carcinogenic, anti-inflammatory, and anti-oxidant properties. Moreover, some phenolic compounds have been proven to be effective in inhibiting various pathogenic bacteria such as E. coli (Cushnie and Lamb, 2005; Almajano et al., 2007; Martina, 2010), however, the antibacterial effect was not satisfactory, and their studies only assessed the level of the simple antibacterial mechanism. What is more, the components in plants are complex and diverse, it is impossible to determine effective specific antibacterial ingredients, and it is difficult to explain the mechanism of phenolic acids.
Therefore, in this study, five phenolic acids were combined with each other in order to obtain one of the most effective combinations against multidrug-resistant E. coli, which may provide a significant contribution to the field of antibiotic agent discovery.
Seven strains of E. coli (E1, E2, E3, E4, E5, E6, and E7) from poultry were stored in Guangxi University (Nanning, China), which were confirmed by the analysis of 16S rDNA sequencing (Kim et al., 2010). The resistance genes of the seven E. coli strains are shown in Table 1 (identification by the 25 μL PCR reaction system).
The five phenolic acids were dissolved with maximum solubility (salicylic acid, CAS: 69-72-7, 0.22 g/100 g H2O; protocatechuic acid, CAS: 99-50-3, 2 g/100 g H2O; gallic acid, GAS: 149-91-7, 1.14 g/100 g H2O; hydrocinnamic acid, GAS: 501-52-0, 0.5 g/100 g H2O; chlorogenic acid, GAS: 327-97-9, 4 g/100 g H2O), and 5% dimethyl sulfoxide (DMSO) was added to promote the solubility of phenolic acids. Finally, the phenolic acids were mixed with each other in equal volumes to obtain CPAs with different compositions.
The microdilution method (Smekalova et al., 2016) was used to detect the MIC of each CPA on E. coli, and the results were statistically analyzed. Mueller-Hinton Broth (MHB) was used as the incubation medium. The CPAs (100 μL) were serially diluted in a 96-well plate by 100 μL of E. coli inoculum (approximately 1.5 × 105 CFU/mL) for 12 times, and the final concentrations of CPAs were 50, 25, 12.5, 6.15, 3.125, 1.5625, 0.78125, 0.39065, 0.1953125, 0.09765625, 0.048828125, and 0.0244140625% of the initial concentration, respectively. After incubation at 37°C for 12–16 h, MICs were measured by visual inspection of the turbidity of broth in tubes. That is to say, if the test tube was still clear and transparent (un-cloudy) after incubation, the cells could not grow at that concentration and then the MIC value was obtained. Florfenicol (in China, florfenicol is an approved animal-specific antibiotic, but it is banned in laying hens) was used as a positive anti-E. coli control; MHB was used as a blank control.
Escherichia coli were cultured at 37°C until the logarithmic growth phase at 220 r/min. The optical density value (OD600) of the bacterial fluid was diluted to 0.6, and then evenly divided into two groups, each group was tested in triplicate. Group A was treated with CPA at the concentration of 1/2 MIC, while same amount of H2O was added in group B and kept as the control group. The mixture was incubated at 37°C for 60 min at 220 r/min, centrifuged for 15 min (4°C, 5000 r/min), and the supernatant was discarded. After repeated washing with RNA-free PBS three times, the samples were immediately frozen in liquid nitrogen, and then stored at −80°C for further use.
The samples were lysed by ultrasound using four times the volume of lysis buffer (8 M urea, 1% protease inhibitor). Following centrifugation at 12000 g for 10 min (to remove the cell debris), the supernatant was collected to determine the protein concentration using a BCA kit (A045-4-2, NanJing JianCheng Bioengineering Institute Co., Ltd., Nanjing, China).
For digestion, the protein solution was reduced with 5 mM of dithiothreitol for 30 min at 56 °C and alkylated with 11 mM of iodoacetamide for 15 min at room temperature in darkness. The protein sample was then diluted by adding 100 mM of TEAB to a urea concentration less than 2 M. Finally, trypsin was added at a 1:50 trypsin-to-protein mass ratio for the first digestion overnight and a 1:100 trypsin-to-protein mass ratio for a second 4-h digestion.
After trypsin digestion, the peptide was desalted by a Strata X C18 SPE column (Phenomenex) and vacuum-dried. The peptide was reconstituted in 0.5 M of TEAB and processed according to the manufacturer’s protocol for the TMT kit. Briefly, one unit of TMT reagent was thawed and reconstituted in acetonitrile. The peptide mixtures were then incubated for 2 h at room temperature and pooled, desalted, and dried by vacuum centrifugation.
The tryptic peptides were fractionated into fractions by high pH reverse-phase HPLC using a Thermo Betasil C18 column (5 μm particles, 10 mm ID, 250 mm length). Briefly, peptides were first separated with a gradient of 8 to 32% acetonitrile (pH 9.0) over 60 min into 60 fractions. Then, the peptides were combined into six fractions and dried by vacuum centrifuging. LC-MS/MS Analysis.
The tryptic peptides were dissolved in 0.1% formic acid (solvent A), directly loaded onto a homemade reversed-phase analytical column (15-cm length, 75 μm i.d.). The gradient was comprised of an increase from 6 to 23% solvent B (0.1% formic acid in 98% acetonitrile) over 26 min, 23 to 35% in 8 min and climbing to 80% in 3 min then holding at 80% for the last 3 min, all at a constant flow rate of 400 nL/min on an EASY-nLC 1000 UPLC system. The peptides were subjected to NSI source followed by tandem mass spectrometry (MS/MS) in Q Exactive TM Plus (Thermo Fisher Scientific) coupled online to the UPLC. The electrospray voltage applied was 2.0 kV. The m/z scan range was 350 to 1800 for full scan, and intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were then selected for MS/MS using the NCE setting at 28 and the fragments were detected in the Orbitrap at a resolution of 17,500. A data-dependent procedure that alternated between one MS scan followed by 20 MS/MS scans with 15.0 s dynamic exclusion was used. Automatic gain control (AGC) was set at 5E4. The fixed first mass was set as 100 m/z.
The resulting MS/MS data were processed using the Maxquant search engine (v.1.5.2.8). Tandem mass spectra were searched against the human uniprot database concatenated with a reverse decoy database. Trypsin/P was specified as the cleavage enzyme allowing up to four missing cleavages. The mass tolerance for precursor ions was set as 20 ppm in the first search and 5 ppm in the main search, and the mass tolerance for fragment ions was set as 0.02 Da. Carbamidomethyl on Cys was specified as the fixed modification, and acetylation modification and oxidation on Met were specified as variable modifications. FDR was adjusted to < 1% and the minimum score for modified peptides was set > 40.
Parallel reaction monitoring (PRM) mass spectrometric analysis was performed using tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo Fisher Scientific). The liquid chromatography parameters, electrospray voltage, scan range, and Orbitrap resolution were the same as the TMT methods. Automatic gain control (AGC) was set at 3 × 106 for full MS and 1 × 105 for MS/MS. The maximum IT was set at 20 ms for full MS and auto for MS/MS. The isolation window for MS/MS was set at 2.0 m/z. After the quantitative information was normalized by the heavy isotope-labeled peptide, a relative quantitative analysis (three biological replications) was performed on the target peptides.
Modified Lorian (2005) method was used to determine the interaction between antibiotics and CPAs. The CPA was diluted vertically, and the antibiotic was diluted horizontally in a 96-well plate. The following formula was used to calculate FICI:
The combined antimicrobial action of CPAs and antibiotics was determined according to FICI, the combined antibacterial effects were considered to be synergy, additivity, indifference, or oppositive when the FCIC was ≤ 0.5, > 0.5 to ≤ 1, > 1 to ≤ 2, and > 2, respectively (Kurek et al., 2012).
The CPA was added in a bacterial suspension (106 CFU/mL) with a concentration of 1/2 MIC. The same volume of H2O was used as the control group. The mixture was cultured at 37°C with shaking at 220 r/min. Samples were collected separately from 0 to 18 h every hour. The determination of extracellular soluble protein was measured by a protein quantitative test kit (A045-2, NanJing JianCheng Bioengineering Institute Co., Ltd., Nanjing, China). Each group test was repeated three times.
The CPA was added to E6 at the logarithmic growth stage, and the final concentration was adjusted to 1/2 MIC, incubated at 37°C and 220 r/min for 4 h, and the same amount of H2O was used as a blank control. After centrifugation of the mixture (3 000 r/min, 15 min), the precipitation was collected and washed twice with PBS. A total of 1 mL of 3% glutaraldehyde was added to the bacterial plate and kept at 4°C overnight. The liquid was centrifuged (3 000 r/min, 30 min) and the supernatant was collected. After separation, the supernatant was washed with PBS three times, fixed with 1% osmium tetroxide, and kept at 4°C for 2 h. After this, the supernatant was uninterruptedly dehydrated using 30, 50, 75, 95, and 100% ethanol for dehydration. Epoxy resin was embedded at 60°C for 48 h and sliced (with 50-nm thickness) (Ultra 45°, Daitome). The samples were placed on a 400-mesh copper network and stained with 2.0% uranium dioxane acetate and lead citrate for 30 min. After washing with distilled water, the samples were dried at 37°C. Finally, the ultrastructural changes were observed under TEM (TECNAI G2 20 TWIN, FEI).
A total of 31 combinations of CPAs were obtained; all components and their concentrations in each CPA are presented in Table 2. In our subsequent study, the initial concentration of each CPA was recorded as 1, which was not only convenient for recording, but also more intuitive to reflect the antibacterial effect. The specific calculation method of actual MIC values of each CPA in subsequent experiments is described in the second sentence of the next section.
Seven strains of E. coli were used to assess the antibacterial efficacy of all CPAs by the microdilution method. The results are shown in Table 3, taking E1 as an example, the MIC of CPA 1 on E1 was 25% of its initial concentration, at this time, the concentration of each component in CPA1 = 25%*1 (salicylic acid 1.1 mg/mL,protocatechuic acid 10 mg/mL), which meant 0.275 mg/mL of salicylic acid and 2.5 mg/mL of protocatechuic acid.
As shown in Table 3, all of the CPAs showed certain inhibitory effects on E. coli. The antibacterial effect of each combination was different due to different components and E. coli strains. However, compared to all other CPAs, CPA 19 showed better efficacy against all seven strains of E. coli and the MIC was reached at 12.5%. In other words, the concentrations of each component in the MIC (12.5% CPA 19) were 0.834 mg/mL (protocatechuic acid), 0.208 mg/mL (hydrocinnamic acid), and 1.67 mg/mL (chlorogenic acid). Therefore, CPA 19 was selected as a potential antimicrobial agent and its mechanisms were further explored against E6 E. coli.
After treatment with a subinhibitory concentration of CPA 19, 2688 proteins were detected and identified, and 2560 were quantified. The detailed data (the protein score, coverage percentage, number of peptides matching individual proteins, and accession number assigned to each identified protein) of related proteins are shown in Supplementary Table 1. The MS proteomic results reported in this paper have been deposited in the OMIX, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences, under accession number OMIX382 which is accessible at https://ngdc.cncb.ac.cn/omix.
There were 268 differentially expressed proteins after treatment with a subinhibitory concentration of CPA 19, including 84 upregulated proteins and 184 downregulated proteins, when the fold change was defined as greater than 1.3 or lower than 1/1.3, and P < 0.05 (Figure 1). Among them, there were 25/51 significantly up/downregulated proteins with more than two fold changes.
The 268 identified proteins were annotated by GO secondary classification, and the effect of the subinhibitory concentration of CPA 19 on multi-drug resistant E. coli was preliminarily analyzed (Supplementary Table 2). The distribution statistics of differentially expressed proteins in the three major categories of GO secondary annotations are shown in Figure 2. Biological process (Figure 2A) was mainly enriched with the proteins involved in cellular processes (29%), metabolic processes (27%), and response to stimulus (16%); in the classification of cell composition (Figure 2B), most of the differential proteins were distributed in cells (60%), membranes (29%), and the protein-containing complex (9%); in the molecular function classification (Figure 2C), there were mainly differentially expressed proteins in catalytic activity (44%), binding (39%), and transport activity (12%).
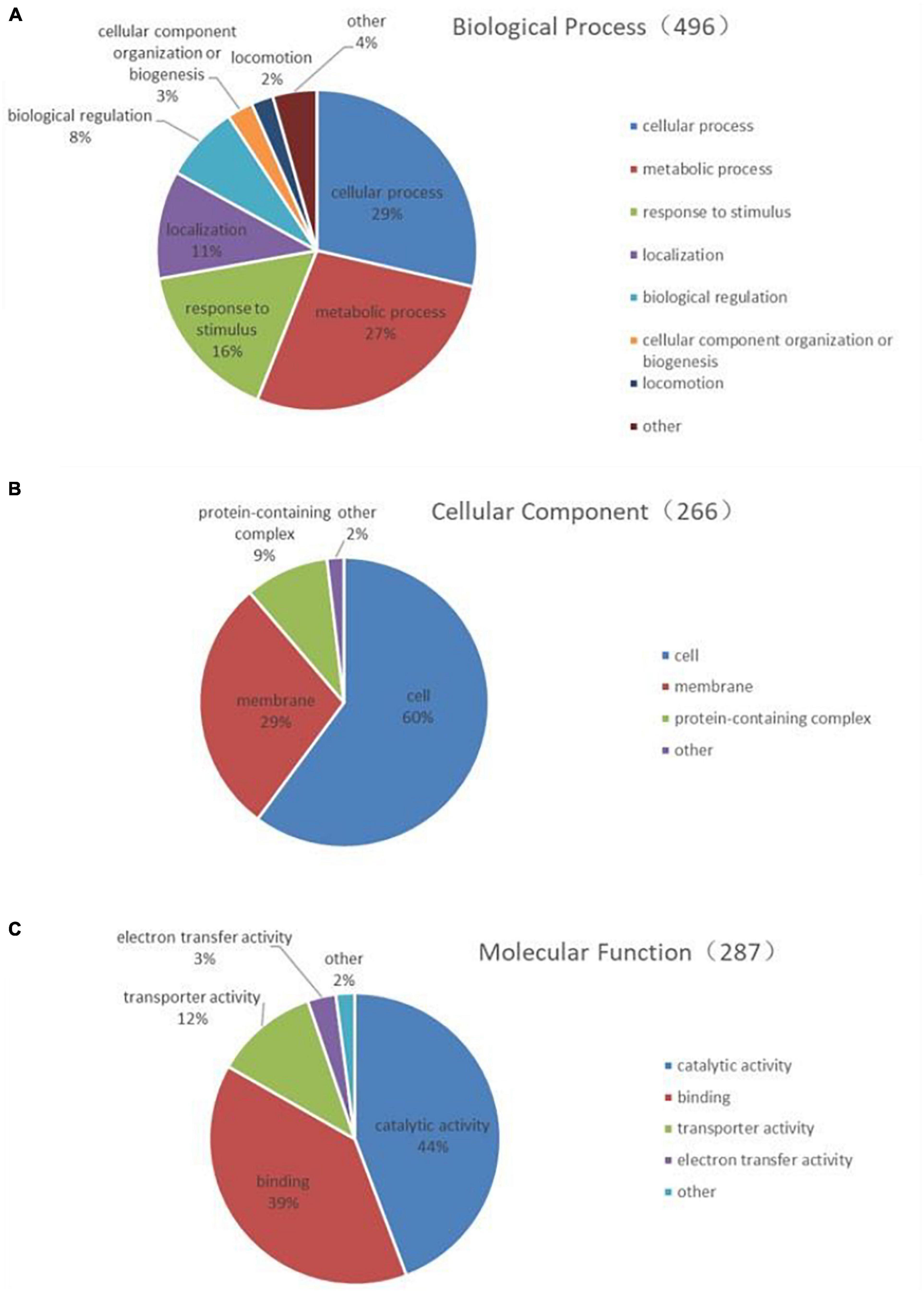
Figure 2. Functional classification of differentially expressed proteins (A) biological process, (B) cell composition, and (C) molecular function.
In order to further understand the effect of CPA 19 on multi-drug resistant E. coli, KEGG pathway enrichment analysis was performed on these 268 differentially expressed proteins (Supplementary Table 3). There were 11 significantly enriched (P < 0.05) KEGG upregulation pathways: Degradation of aromatic compounds, phenylalanine metabolism, sulfur metabolism, microbial metabolism in diverse environments, dioxin degradation, xylene degradation, monobactam biosynthesis, two-component system, selenocompound metabolism, benzoate degradation, and nitrotoluene degradation. And there were 18 significantly enriched (P < 0.05) KEGG downregulation pathways: Bacterial chemotaxis, arginine and proline metabolism, lysine metabolism, ABC transporter, propionate lipid metabolism, microbial metabolism in different environments, phenylalanine metabolism, beta-alanine metabolism, valine, leucine, and isoleucine degradation, fatty acid degradation, geraniol degradation, benzoic acid degradation, quorum sensing, limonene and pinene degradation, caprolactam degradation, tryptophan metabolism, two-component system, and methyl butyrate metabolism. The top 20 enriched pathways are shown in Figure 3.
As can be seen from Figure 4, the expression of some important enzymes (long-chain acyl-CoA synthetase [EC:6.2.1.3], enoyl-CoA hydratase [EC:4.2.1.17], 3-hydroxyacyl-CoA dehydrogenase [EC:1.1.1.35], acetyl-CoA acyltransferase [EC:2.3.1.16]) regulating β oxidation were significantly downregulated in the fatty acid degradation pathway, indicating that the fatty acid degradation of multidrug-resistant E. coli was inhibited after treatment with the subinhibitory concentration of CPA 19, and leading to blocked generation of acetyl-CoA. Similarly, in the pathway of phenylalanine metabolism (Figure 5) and lysine metabolism (Figure 6), the metabolic process of phenylalanine and lysine were also stressed by CPA 19, which led to the obstruction of the conversion of acetyl-CoA.
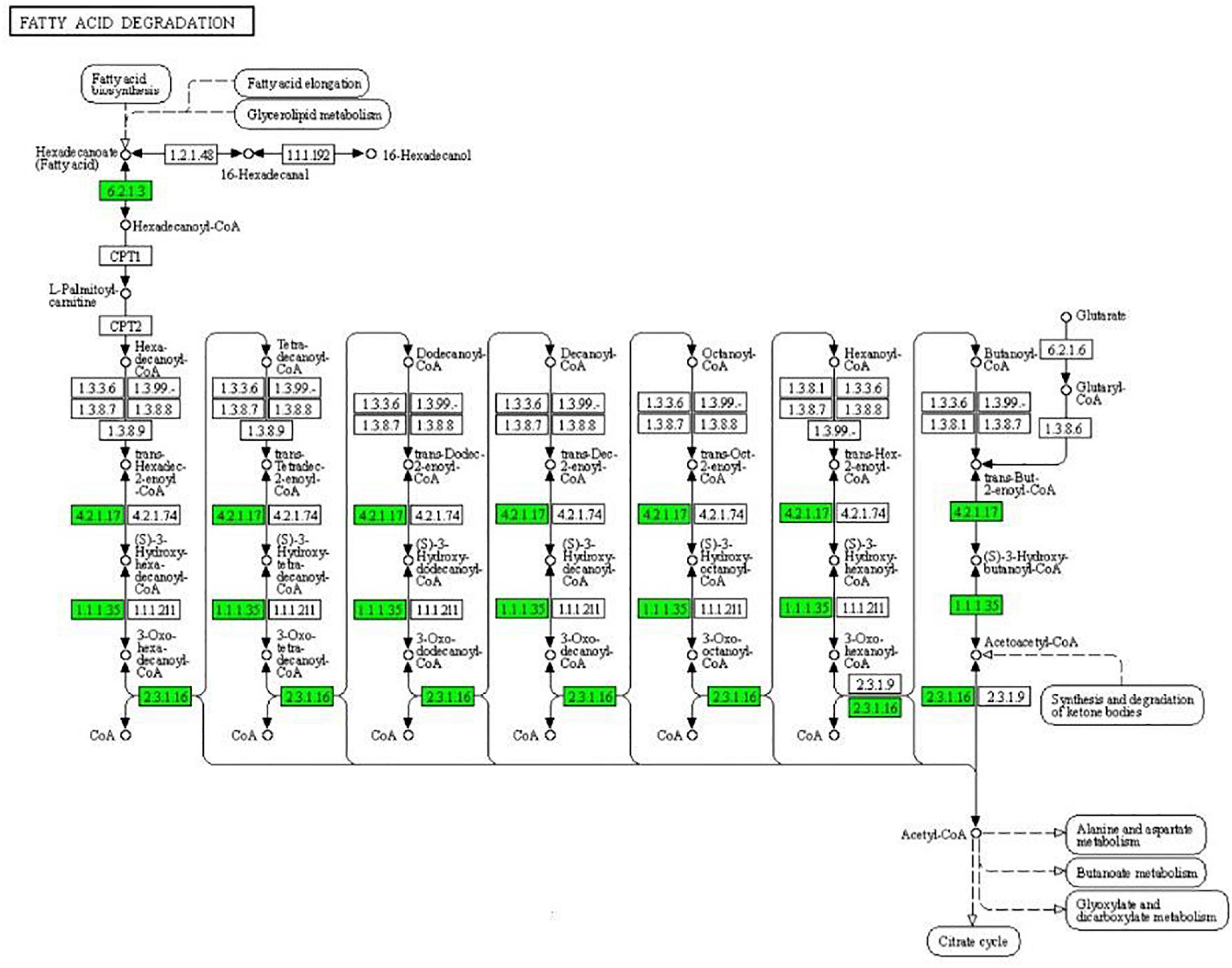
Figure 4. Significantly enriched KEGG pathway: Fatty acid degradation (from the KEGG database, green frames represent downregulation, and red frames represent upregulation).
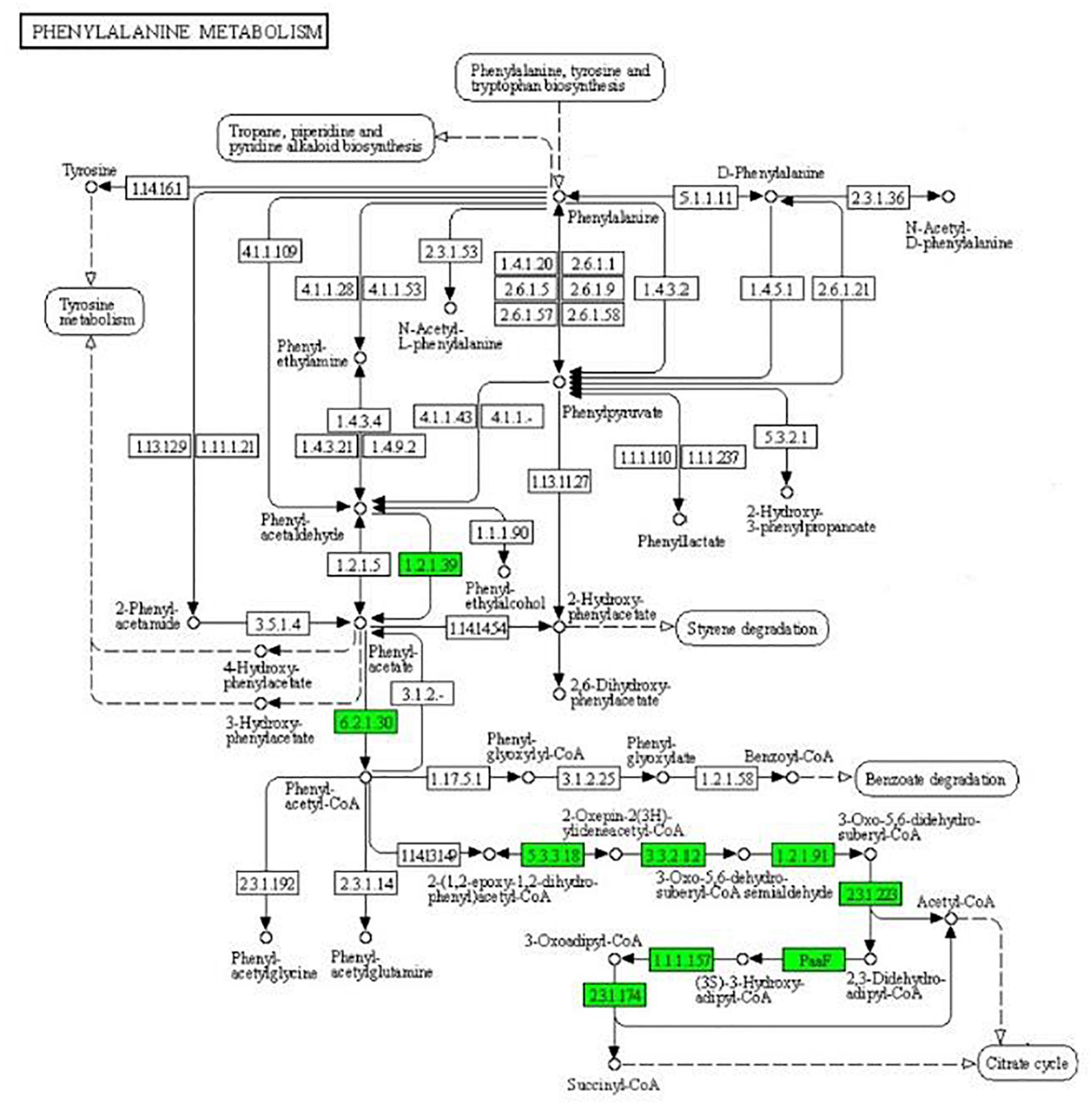
Figure 5. Significantly enriched KEGG pathway: Phenylalanine metabolism (from the KEGG database, green frames represent downregulation, and red frames represent upregulation).
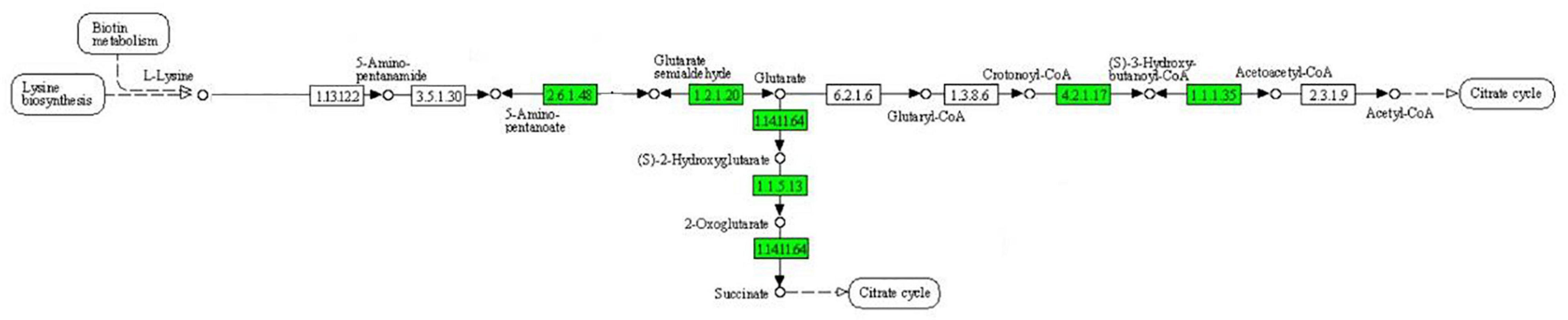
Figure 6. Significantly enriched KEGG pathway: Lysine metabolism (from the KEGG database, green frames represent downregulation, and red frames represent upregulation).
In the two-component system (Figure 7), the expression of outer membrane pore protein F (OmpF) and flagellin (FliC) in the OmpR family, as well as methyl-accepting chemotaxis protein (MCP), purine-binding chemotaxis protein CheW (CheW), sensor kinase (CheA), chemotaxis protein CheY (CheY), and glutaminase (CheB) in the chemotactic family were significantly downregulated. MCP, CheW, CheA, CheY, and CheB were also significantly enriched in the bacterial chemotaxis pathway (Figure 8). At the same time, the expression of serine sensor receptor (Tsr) and aspartate sensor receptor (Tar) in the bacterial chemotaxis pathway were significantly downregulated.
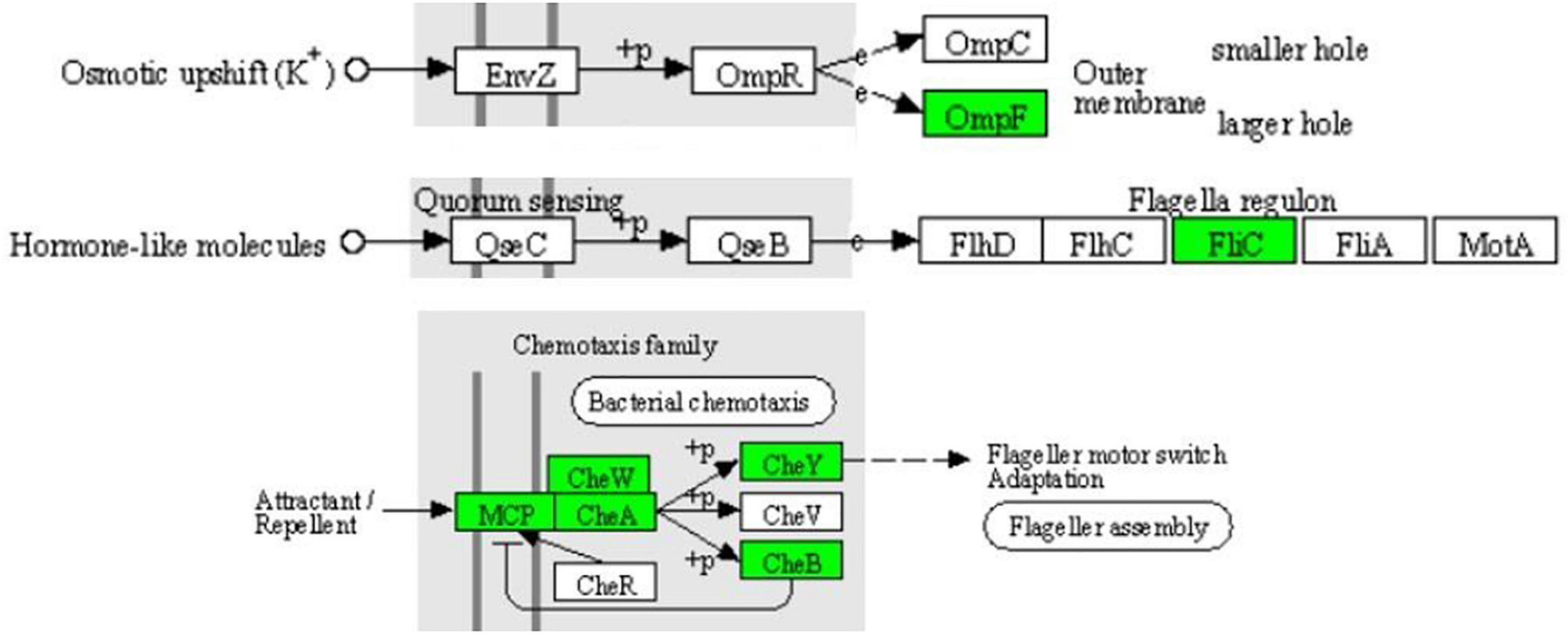
Figure 7. Significantly enriched KEGG pathway: Two-component system (from the KEGG database, green frames represent downregulation, and red frames represent upregulation).
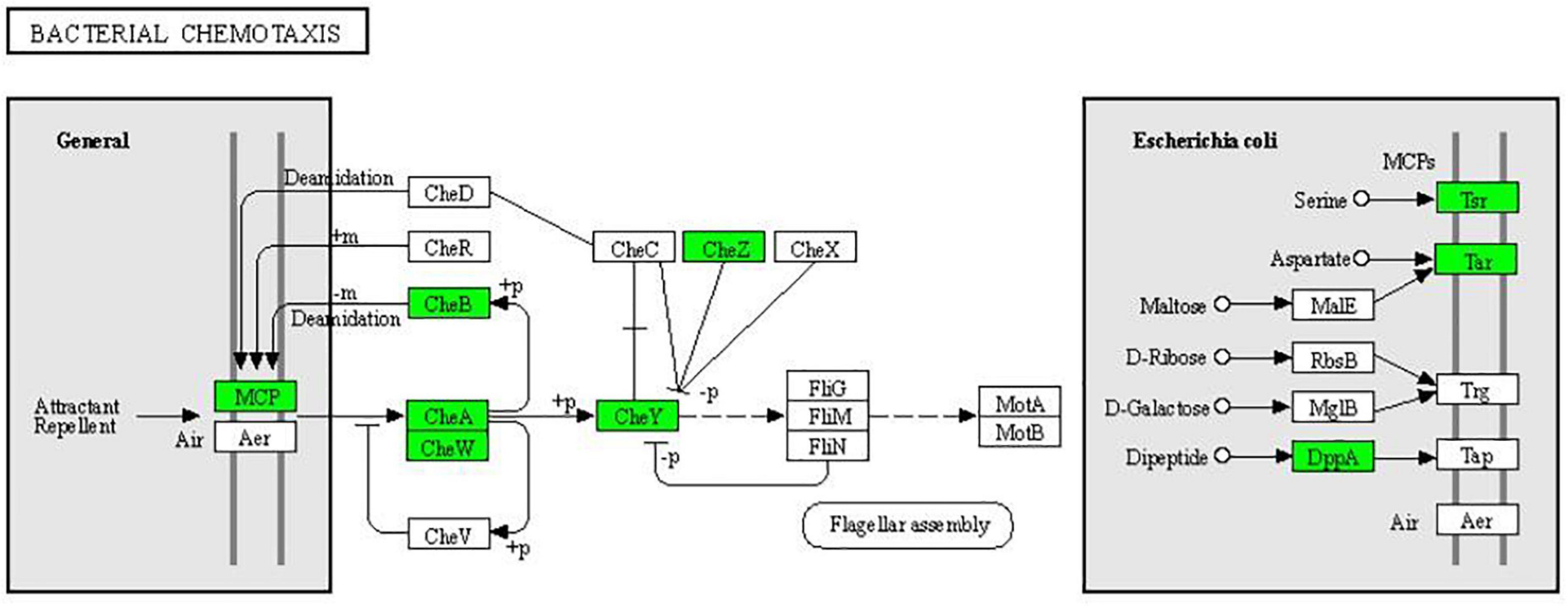
Figure 8. Significantly enriched KEGG pathway: Bacterial chemotaxis (from the KEGG database, green frames represent downregulation, and red frames represent upregulation).
We performed PRM verification on the significantly different expressed proteins in multi-drug resistant E. coli treated with the subinhibitory concentration of CPA 19. After evaluation of 50 differential proteins, 27 candidate proteins met the conditions, and 20 (10 upregulated, 10 downregulated) candidate proteins were selected for further verification. The results of PRM are shown in Table 4 and prove that all 20 target proteins were quantitative and consistent with the change trend of proteomics results, which supports the credibility and reliability of proteomics.
It can be seen from Table 5 that the FICI of CPA 19 and ceftiofur sodium was 1.5, which meant the interaction was indifferent, while the interaction of CPA 19 and azithromycin was oppositive. In addition, when combined with ceftriaxone sodium, amoxicillin, fosfomycin, sulfamonomethoxine, gatifloxacin, lincomycin, florfenicol, cefotaxime sodium, and rifampicin, additive effects were expressed. The MICs of several antibiotics (ceftriaxone sodium, amoxicillin, fosfomycin, sulfamonomethoxine, gatifloxacin, lincomycin, florfenicol, cefotaxime sodium, and rifampicin) after the combination with CPA 19 were all reduced by four times or more, which meant the antibacterial activities in vitro were significantly enhanced.
As shown in Figure 9, the concentration of soluble protein in the culture medium of the 1/2 MIC-treated group was significantly higher than in the culture medium of control group for the previous 12 h. During 13–18 h of incubation, the extracellular soluble protein tended to decline, which might be due to the antibacterial components gradually losing their effects with the lengthening of time.
The morphological changes of E. coli cells are shown in Figure 10. The cell wall of the control group was compact, complete, smooth with uniform cytoplasm, and flagellated (Figure 10A). After the treatment of CPA 19 with 1/2 MIC, significant changes were observed in the bacterial cell wall, the surface area of the cell wall became rough and the boundary between the cell wall and cell membrane became fuzzy, in addition, there was no flagellum on the surface of the bacterial cell (Figure 10B).
Phenolic acids, a major component in plants, have been shown to have antioxidant and antimicrobial activity against Gram-positive and Gram-negative bacteria (Shen et al., 2020). In addition, the order of antibacterial activity of the active ingredients in plants is: phenol > aldehyde > ketone > alcohol > ether > hydrocarbon (Nguyen et al., 2018). In this study, 31 CPAs of five phenolic acids were used to explore new and natural antimicrobial agents against multi-resistant E. coli. Our results of the MICs revealed the antibacterial effects of each CPA. However, the inhibitory effect of each CPA was different against all examined strains of E. coli. A previous study reported that the better therapeutic action of phenolic acids can be achieved from the synergistic effect of complex plant phenolic acid mixtures rather than from individual phenolic acid compounds (Coman and Vodnar, 2020). Moreover, the composition of plants is too complex and diverse to explain its antibacterial mechanism, which greatly limits its further development in clinical applications. Therefore, five phenolic acids were selected to explore the new antimicrobial agents, which could not only make the composition clear, but also retain the better antibacterial effect of plant phenolic acid mixtures when compared with single phenolic acids. Compared to all other CPAs, CPA 19 showed the best antibacterial efficacy. These findings implied that CPAs are better than individual compounds, and the best antibacterial effect of combination 19 not only elaborated the synergism of phenolic acids, but also represented its potential role against all examined multidrug-resistant E. coli.
The tricarboxylic acid cycle (TAC) is the final metabolic pathway through which the three nutrients (carbohydrates, lipids, and amino acids) provide and interconvert energy. These three nutrients can be metabolized to produce acetyl-CoA and then participate in TAC (Fan et al., 2021). Lipids are degraded by fatty acid degradation to generate acetyl-CoA followed by β-oxidation in the cytoplasm by activation of prokaryotic fatty acids. β-oxidation is a cyclic reaction of dehydrogenation, hydration, dehydrogenation, and thiolysis until the acyl-CoA is completely cleaved to acetyl-CoA (Yunqiao Yang et al., 2020). After CPA 19 treatment, the expression of some key enzymes in the β-oxidation process was downregulated, and the downregulation of these key proteins resulted in the decrease of acetyl-CoA. In the meanwhile, the conversion process of acetyl-CoA in the phenylalanine metabolism pathway and lysine metabolism pathway also could be impeded. These findings suggested that the CPA treatment may inhibit E. coli by perturbing the production of acetyl-CoA, and thus interrupt the material conversion and energy metabolism of E. coli.
Proteomic technology was used to analyze the differentially expressed proteins between multidrug-resistant E. coli and ATCC25922, which found that resistance of E. coli is associated with bacterial chemotaxis. Compared with ACCC25922, the expressions of Tsr, Tar, CheA, and CheW in resistant E. coli were significantly upregulated, indicating that these proteins are involved in the resistance of multidrug-resistant E. coli (Xia et al., 2020). What is more, Tsr, CheA, CheW, and CheR have also been shown to be related to the resistance of Bacillus and Aeromonas hydrophila (Wada et al., 2016). In our experiment, after treatment with CPA 19, the expression of Tsr, Tar, CheA, and CheW was significantly downregulated, suggesting that treatment with CPA 19 may restore the sensitivity of multidrug-resistant E. coli to antibiotics.
Combination with antibiotics as a synergist is also an important direction for the development of new antimicrobial agents. Yan (2020) reported that the combination of honokiol and polymyxin showed synergistic effects on Enterobacteriaceae. Further investigation has shown that honokiol could directly bind to the active region of MCR-1 and had competition for the specific binding of MCR-1 to the substrate, thus restoring the sensitivity of MCR-1-positive strains to polymyxin. In our results, CPA 19 also showed additive effects when combined with ceftriaxone sodium, amoxicillin, and cefotaxime sodium, which suggested that treatment with CPA 19 can indeed enhance the sensitivity of multidrug-resistant E. coli to antibiotics.
There have been studies that reported that bacterial chemotaxis had a regulatory effect on bacterial flagella, especially CheA and CheY (Yuhai et al., 2008; Yuan et al., 2012). In this study, not only were CheA and CheY downregulated, but also FliC. The downregulation of these proteins was also corroborated by flagella shedding on the surface of E. coli treated with phenolic acid 19 which was observed via TEM.
Destruction of the cell membrane is the major target of various antimicrobial agents, including phenolic acids (Fang et al., 2018). The antibacterial effect of phenolic acids may be due to the mechanism of the phenol-membrane interaction, isolation of metal ions, and inhibition of enzyme activity (Mirjana et al., 2005; Kemperman et al., 2010; Wang et al., 2010). As an important component of the cell membrane, OmpF crosses the phospholipid bilayer which is closely related to bacterial resistance of E. coli and the entry and exit of intracellular molecules (Jo-Anne and Elizabeth, 1997). In the two-component system, the expression of OmpF was significantly downregulated by the treatment of CPA 19, suggesting that the integrity of the membrane of the E. coli may be damaged. Macromolecular proteins originally present in the cell membrane and cytoplasm, which can be used to evaluate the integrity of the cell membrane (Vivek et al., 2013; Yunbin et al., 2016). The determination of extracellular soluble protein after CPA 19 treatment confirmed the downregulation of OmpF and cell membrane damage. These findings were in great agreement with Chenjie (Chenjie et al., 2015), they reported that the treatment of lactic acid perforated cell membranes, leaked intracellular material, and changed the morphology of E. coli. Phenolic acids can alter the permeability of cell membranes, damage the integrity of cells, not only causing the leakage of macromolecular substances, but also making it easier for harmful substances outside the cell to enter, which will eventually lead to cell death (Moreno et al., 2006). It could be seen from the results of this study that the damage of CPA 19 to the cell membrane might be the reason for the increased sensitivity of multidrug-resistant E. coli to antibiotics, and the antibiotics found it easier to enter the cell interior to play a role. In short, the cell membrane was also one of the targets of CPA 19.
Compound phenolic acid 19, composed of protocatechuic acid, hydrocinnamic acid, and chlorogenic acid, had a good antibacterial effect against multi-drug resistant E. coli. When combined with antibiotics (ceftriaxone sodium, amoxicillin, fosfomycin, sulfamonomethoxine, gatifloxacin, lincomycin, florfenicol, cefotaxime sodium, and rifampicin), CPA 19 increased the susceptibility of E. coli to antibiotics. CPA 19 could shed the flagellum of multidrug-resistant E. coli, break the structure of the cell membrane, and cause the macromolecules to leak out from the cell. These data suggested that the natural compounds represented by CPA 19 in this study can be further developed as novel antimicrobial agents, and the use of CPA 19 in combination with antibiotics can be further explored for expression patterns of proteins involved in E. coli resistance.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
HS designed the work. All authors reviewed and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31760746) and the Key Research and Development Plan of Guangxi, China (AB19245037).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.738896/full#supplementary-material
Abraham, E. P., and Chain, E. (1988). An enzyme from bacteria able to destroy penicillin. 1940. Rev. Infect. Dis. 10, 677–678.
Almajano, M. P., Rosa, C., Jiménez, J. A. L., and Michael, H. G. (2007). Antioxidant and antimicrobial activities of tea infusions. Food Chem. 108, 55–63.
Chenjie, W., Tong, C., Hong, Y., and Min, C. (2015). Antibacterial mechanism of lactic acid on physiological and morphological properties of Salmonella Enteritidis, Escherichia coli and Listeria monocytogenes. Food Control 47, 231–236. doi: 10.1016/j.foodcont.2014.06.034
Coman, V., and Vodnar, D. C. (2020). Hydroxycinnamic acids and human health: recent advances. J. Sci. Food Agric. 100, 483–499. doi: 10.1002/jsfa.10010
Cushnie, T. P., and Lamb, A. J. (2005). Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 26, 343–356. doi: 10.1016/j.ijantimicag.2005.09.002
Efenberger, S. M., Nowak, A., and Czyzowska, A. (2021). Plant extracts rich in polyphenols: antibacterial agents and natural preservatives for meat and meat products. Crit. Rev. Food Sci. Nutr. 61, 149–178.
Fan, J., Zhou, J., Su, S., Yu, Y., Chen, L., Le, Y., et al. (2021). Quantitative detection and metabolic profile analysis of metabolites of tricarboxylic acid cycle and amino acids in psoriasis serum before and after receiving monoclonal antibody treatment by one-pot GC-MS derivatization. Int. J. Mass Spectrom. 460:116478. doi: 10.1016/j.ijms.2020.116478
Fang, L., Fengting, W., Lihui, D., Tong, Z., Michael, P. D., Daoying, W., et al. (2018). Antibacterial and antibiofilm activity of phenyllactic acid against Enterobacter cloacae. Food Control 84, 442–448. doi: 10.1016/j.foodcont.2017.09.004
Jo-Anne, H., and Elizabeth, W. (1997). Molecular characterization of the Serratia marcescens OmpF porin, and analysis of S. marcescens OmpF and OmpC osmoregulation. Microbiology 143, 2797–2806. doi: 10.1099/00221287-143-8-2797
Kemperman, R. A., Bolca, S., Roger, L. C., and Vaughan, E. E. (2010). Novel approaches for analysing gut microbes and dietary polyphenols: challenges and opportunities. Microbiology (Reading) 156(Pt 11), 3224–3231. doi: 10.1099/mic.0.042127-0
Kim, T., Kim, Y., Kim, S., Lee, J., Park, C., and Kim, H. (2010). Identification and distribution of Bacillus species in doenjang by whole-cell protein patterns and 16S rRNA gene sequence analysis. J. Microbiol. Biotechnol. 20, 1210–1214.
Kurek, A., Nadkowska, P., Pliszka, S., and Wolska, K. I. (2012). Modulation of antibiotic resistance in bacterial pathogens by oleanolic acid and ursolic acid. Phytomedicine 19, 515–519. doi: 10.1016/j.phymed.2011.12.009
Lorian, V. (2005). Antibiotics in Laboratory Medicine. Philadelphia, PA: Lippincott Williams & Wilkins.
Martina, B. (2010). Comparison of the antioxidant capacity and the antimicrobial activity of black and green tea. Food Res. Int. 43, 1379–1382.
Mirjana, A., John, V. C., Bruno, D. M., Griet, D., Carmen, S., Marc, V., et al. (2005). Iron-chelation properties of phenolic acids bearing catechol and galloyl groups. Food Chem. 98, 23–31. doi: 10.1016/j.foodchem.2005.05.044
Moreno, S., Scheyer, T., Romano, C. S., and Vojnov, A. A. (2006). Antioxidant and antimicrobial activities of rosemary extracts linked to their polyphenol composition. Free Radic. Res. 40, 223–231. doi: 10.1080/10715760500473834
Nguyen, H. V., Meile, J. C., Lebrun, M., Caruso, D., Chu-Ky, S., and Sarter, S. (2018). Litsea cubeba leaf essential oil from Vietnam: chemical diversity and its impacts on antibacterial activity. Lett. Appl. Microbiol. 66, 207–214. doi: 10.1111/lam.12837
Paitan, Y. (2018). “Current trends in antimicrobial resistance of Escherichia coli,” in Escherichia coli, A Versatile Pathogen, eds G. Frankel and E. Z. Ron (Cham: Springer International Publishing), 181–211.
Roth, N., Kasbohrer, A., Mayrhofer, S., Zitz, U., Hofacre, C., and Domig, K. J. (2019). The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: a global overview. Poult. Sci. 98, 1791–1804. doi: 10.3382/ps/pey539
Shen, H., Tong, X., Yang, J., Yu, L., Zhou, H., Wang, Y., et al. (2020). Biotransformation of natural hydroxycinnamic acids by gut microbiota from normal and cerebral ischemia-reperfusion injured rats: a comparative study. Food Funct. 11, 5389–5395. doi: 10.1039/d0fo00775g
Sidhu, N. S., Schreiber, K., Propper, K., Becker, S., Uson, I., Sheldrick, G. M., et al. (2014). Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA. Acta Crystallogr. D Biol. Crystallogr. 70(Pt 5), 1321–1335. doi: 10.1107/S1399004714002739
Smekalova, M., Aragon, V., Panacek, A., Prucek, R., Zboril, R., and Kvitek, L. (2016). Enhanced antibacterial effect of antibiotics in combination with silver nanoparticles against animal pathogens. Vet. J. 209, 174–179. doi: 10.1016/j.tvjl.2015.10.032
Vivek, K. B., Ajay, S., and Kwang-Hyun, B. (2013). Antibacterial mode of action of Cudrania tricuspidata fruit essential oil, affecting membrane permeability and surface characteristics of food-borne pathogens. Food Control 32, 582–590. doi: 10.1016/j.foodcont.2013.01.032
Wada, K., Kobayashi, J., Furukawa, M., Doi, K., Ohshiro, T., and Suzuki, H. (2016). A thiostrepton resistance gene and its mutants serve as selectable markers in Geobacillus kaustophilus HTA426. Biosci. Biotechnol. Biochem. 80, 368–375. doi: 10.1080/09168451.2015.1079478
Wang, W., Du, Y., and Song, H. (2010). α-Glucosidase and α-amylase inhibitory activities of guava leaves. Food Chem. 123, 6–13. doi: 10.1016/j.foodchem.2010.03.088
Xia, Z., Rui, L., Haixia, G., Wenjing, W., Jun, X., Ruiliang, Z., et al. (2020). Proteomics difference between Escherichia coli multidrug resistant and susceptible strains. J. Northwest. A F Univ. (Nat. Sci. Ed.) 48, 40–48. doi: 10.13207/j.cnki.jnwafu.2020.01.006
Yan, G. (2020). Extraction of Honokiol and Its Enhancement of the Antibacterial Activity of Polymyxin Against mcr-1-Positive Bacteria. Master’s thesis. Jilin: Jilin University.
Yang, Y., Chen, Y., Zhang, G., Sun, J., Guo, L., Jiang, M., et al. (2020). Transcriptomic analysis of Staphylococcus aureus under the stress condition caused by Litsea cubeba L. Essential oil via RNA sequencing. Front. Microbiol. 11:1693. doi: 10.3389/fmicb.2020.01693
Yuan, J., Branch, R. W., Hosu, B. G., and Berg, H. C. (2012). Adaptation at the output of the chemotaxis signalling pathway. Nature 484, 233–236. doi: 10.1038/nature10964
Yuhai, T., Thomas, S. S., and Howard, C. B. (2008). Modeling the chemotactic response of Escherichia coli to time-varying stimuli. Proc. Natl. Acad. Sci. U.S.A. 105, 14855–14860. doi: 10.1073/pnas.0807569105
Yunbin, Z., Xiaoyu, L., Yifei, W., Pingping, J., and SiewYoung, Q. (2016). Antibacterial activity and mechanism of cinnamon essential oil against Escherichia coli and Staphylococcus aureus. Food Control 59, 282–289. doi: 10.1016/j.foodcont.2015.05.032
Keywords: compound phenolic acids, Escherichia coli, TMT, antibacterial mechanism, new antimicrobial agent
Citation: Zhang G, Yang Y, Memon FU, Hao K, Xu B, Wang S, Wang Y, Wu E, Chen X, Xiong W and Si H (2021) A Natural Antimicrobial Agent: Analysis of Antibacterial Effect and Mechanism of Compound Phenolic Acid on Escherichia coli Based on Tandem Mass Tag Proteomics. Front. Microbiol. 12:738896. doi: 10.3389/fmicb.2021.738896
Received: 09 July 2021; Accepted: 01 November 2021;
Published: 29 November 2021.
Edited by:
Wang Jiajun, Northeast Agricultural University, ChinaReviewed by:
Ranjith Kumar Manoharan, Yeungnam University, South KoreaCopyright © 2021 Zhang, Yang, Memon, Hao, Xu, Wang, Wang, Wu, Chen, Xiong and Si. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongbin Si, c2hiMjAwOUBneHUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.