
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 October 2021
Sec. Evolutionary and Genomic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.736565
This article is part of the Research Topic The CRISPR/Cas System in Pathogen Resistance, Virulence, Diagnosis and Typing View all 8 articles
Staphylococcus aureus (S. aureus), which is one of the most important species of Staphylococci, poses a great threat to public health. Clustered regularly interspaced short palindromic repeats (CRISPR) and their CRISPR-associated proteins (Cas) are an adaptive immune platform to combat foreign mobile genetic elements (MGEs) such as plasmids and phages. The aim of this study is to describe the distribution and structure of CRISPR-Cas system in S. aureus, and to explore the relationship between CRISPR and horizontal gene transfer (HGT). Here, we analyzed 67 confirmed CRISPR loci and 15 companion Cas proteins in 52 strains of Staphylococci with bioinformatics methods. Comparing with the orphan CRISPR loci in Staphylococci, the strains harboring complete CRISPR-Cas systems contained multiple CRISPR loci, direct repeat sequences (DR) forming stable RNA secondary structures with lower minimum free energy (MFE), and variable spacers with detectable protospacers. In S. aureus, unlike the orphan CRISPRs away from Staphylococcal cassette chromosome mec (SCCmec), the complete CRISPR-Cas systems were in J1 region of SCCmec. In addition, we found a conserved motif 5′-TTCTCGT-3′ that may protect their downstream sequences from DNA interference. In general, orphan CRISPR locus in S. aureus differed greatly from the structural characteristics of the CRISPR-Cas system. Collectively, our results provided new insight into the diversity and characterization of the CRISPR-Cas system in S. aureus.
Staphylococcus is a genus of Gram-positive bacteria that can be found in the air, water, dust, and the skin and mucous membranes of humans and other organisms. Most of them are harmless and a few can cause diseases, for example, Staphylococcus pseudintermedius, Staphylococcus schleiferi, Staphylococcus argenteus, Staphylococcus epidermidis, and Staphylococcus lugdunensis. Staphylococcus aureus (S. aureus), as the most noticeable species in Staphylococci, is an important pathogen that can cause a series of diseases, including food poisoning, pneumonia, endocarditis, and sepsis, thus causing property losses and public health problems (Lowy, 1998; Hoffman et al., 2015; Vega and Dowzicky, 2017). The emergence and evolution of healthcare-associated methicillin-resistant S. aureus (HA-MRSA), community-associated MRSA (CA-MRSA), and livestock-associated MRSA (LA-MRSA) have greatly contributed to the pressure of human response to S. aureus infection (DeLeo et al., 2010; Gray et al., 2010; Aires-de-Sousa, 2017). Notably, in recent years, numerous studies have reported the detection of MRSA or multidrug-resistant S. aureus (MDRSA) in retail food worldwide (de Lencastre et al., 2017; Ge et al., 2017; Tang et al., 2017; Wang et al., 2017), indicating a shift to non-special locations. This is directly related to the popularization of antibiotics and the frequent horizontal transfer of mobile genetic elements (MGEs), such as phages, plasmids, transposons, and Staphylococcal cassette chromosomes (SCC) (Holden et al., 2013; Lindsay, 2014; Strommenger et al., 2014). It is obvious that these exogenous genes provide additional advantages for the survival of bacteria, but they also cause unavoidable problems, such as bacterial lysis caused by bacteriophage infection.
As prokaryotes evolved to meet challenges in vivo and in vitro, they developed an adaptive immune system called clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (CRISPR-Cas) system to combat foreign MGEs, such as plasmids and phages, thus seeking the continuation of relative balance of species in continuous biological evolution. These systems consist of CRISPR loci, including highly conservative direct repeat sequences (DR) and spacer sequences derived from invading genes, and companion cas genes. According to the composition and structural characteristics of Cas protein complex, CRISPR-Cas systems are divided into 2 classes, 6 types, and 30 subtypes (Couvin et al., 2018; Makarova et al., 2020). Class 1 is defined by the presence of a multisubunit crRNA-effector complex, including type I, III, and IV, such as the effector module composed of Cas10, small subunit (SS) protein, Cas5, and severe paralogous Cas7 proteins in type III (Watson et al., 2021). Class 2 is defined by the presence of a single subunit crRNA-effector module, including type II, V, and VI, such as Cas9 in type II (Makarova et al., 2015). The unique signature proteins of these 6 types are Cas3 for type I, Cas9 for type II, Cas10 for type III, Csf1 for type IV, Cpf1 for type V, and Cas13 for VI (Makarova et al., 2011; Shmakov et al., 2015; Zetsche et al., 2015; Jung et al., 2016). At present, the subtypes reported in S. aureus are III-A and II-A (Golding et al., 2010; Ran et al., 2015; Aires-de-Sousa, 2017; Guan et al., 2017; Larsen et al., 2017). These type III-A CRISPR-Cas systems are either located in Staphylococcal cassette chromosome mec (SCCmec) or adjacent to SCCmec, supporting this hypothesis that the CRISPR-Cas systems can be transferred by MGE (Yang et al., 2015). Watson et al. (2018) proved bacteria could acquire an entire chromosomal CRISPR-Cas system through transduction.
Clustered regularly interspaced short palindromic repeats immunity is in 3 main stages: adaptation (spacer acquisition), expression (crRNAs formation), and interference (invading nucleic acids degradation). There are some slight differences among the various CRISPR-Cas types. The exogenous DNA sequence corresponding to spacer captured by the CRISPR-Cas system is called a protospacer (Moreb and Lynch, 2021). The main difference between a protospacer in invading nucleic acids and a spacer in CRISPR is the peripheral sequence. To distinguish self from non-self, type I and type II systems rely on 2–4 nt protospacer-adjacent motif (PAM), which is a short sequence adjacent to a protospacer and highly conserved that is not present in the host’s own CRISPR repeats (Wang et al., 2019). While, in type III and VI, typical PAM sequences were not found (Leenay and Beisel, 2017), they rely on the crRNA “tag,” an 8-nt sequence derived from the CRISPR repeats located at the 5’ flank of mature crRNA (Wang et al., 2019). PAM sequences play an important role in the acquisition of adaptive spacers and in the recognition and targeted cleavage of foreign DNA. Thus, phages can escape CRISPR attack by mutating PAM sequences (Collias and Beisel, 2021). The primed CRISPR adaptation (priming), as a pathway to spacer acquisition, was observed in type I systems, which resulted in spacer acquisition from location near the site of confirmed protospacers, indicating spacer acquisition is not random (Jackson et al., 2017). Furthermore, Nicholson et al. (2018) found evidence of a priming-like pathway in type II systems using bioinformatics methods. As opposed to type I and II systems, priming is not found in type III.
With the development of life sciences, the research of the CRISPR-Cas system has deepened. Nowadays, the CRISPR-Cas system is widely used in expression regulation, gene editing, nucleic acid detection, cell imaging, DNA assembly, and other fields (Ma et al., 2020; Ahmed et al., 2021; Ghaffari et al., 2021; Khajanchi and Saha, 2021; Luthra et al., 2021). Wang et al. (2020) developed a novel one-pot toolbox with precision and ultra-sensitivity platform for foodborne pathogen detection based on Cas12a/crRNA. Curti et al. (2020) demonstrated that Cas12a was able to detect DNA target sequences corresponding to carbapenemases resistance genes such as KPC, NDM, and OXA.
Bioinformatics analysis can help us understand CRISPR more comprehensively. S. aureus has SCCmec, which is obviously different from other bacteria, so the characteristics of the CRISPR-Cas system may be different from other systems. However, systematic bioinformatics analysis of S. aureus genome sequence structure is rarely reported. The main purpose of this study is to describe the distribution and structure of the CRISPR-Cas system in S. aureus, and to explore the relationship between CRISPR and horizontal gene transfer (HGT). We analyzed 67 confirmed CRISPR loci and 15 subtype III-A Cas proteins in 52 strains of Staphylococci. The structural characteristics and possible functions of CRISPR in Staphylococci were investigated by bioinformatics methods. We also analyzed the relationships between CRISPR loci and multilocus sequence typing (MLST), S. aureus protein A gene (spa), and SCCmec. The bioinformatics tools were used to explore the RNA secondary structures of DRs, the protospacers and potential PAM of spacers, and the phylogenetic tree of Cas proteins. We further studied the composition of spacers from each CRISPR locus and III-A Cas proteins.
We downloaded 325 complete genomes of Staphylococci from the National Center for Biotechnology Information (NCBI) nucleotide database1 with default parameters (updated before August 30, 2018). The confirmed CRISPR loci were searched by the CRISPR Finder2 and cas genes by the CRISPRCas Finder3.
The confirmed CRIPSR loci were found in 42 strains of S. aureus and the complete CRISPR-Cas system was identified in 15 strains of Staphylococci by the CRISPR Finder. To further verify the accuracy of the CRISPR array, CRISPRone was used for CRISPR array analysis of 15 strains of Staphylococci with type III-A system identified by the CRISPR Finder4 (Zhang and Ye, 2017). The phylogenetic tree of cas genes was performed by MAGE-X. The information about MLST, spa, and SCCmec of S. aureus was obtained from the Center for Genomic Epidemiology5. Consensus direct repeat sequences (CDRs) of CRISPR loci were recruited by removing redundance and not forming RNA secondary structures. The RNA secondary structures and minimum free energy (MFE) of CDRs were predicted by the RNA fold Web server, the output option is set to default6. Spacers in the CRISPR loci flanking the complete Cas cluster were analyzed. The difference between two spacer sequences below 5% is a homologous sequence by BLAST7. CRISPRTarget was used to identify protospacers in plasmids and phage database and to learn about the products encoded by the genes in which they reside8. The bases at both ends of the protospacer were collected according to the number of bases in the hairpin rings of the RNA secondary structure of the repeated sequences.
There were 52 strains of Staphylococci detected to carry 67 confirmed CRISPR loci with 3 or 4 Cas3 proteins in 325 strains of Staphylococci, including 42 strains of S. aureus (Supplementary Tables 1, 2). There were 15 strains found to contain complete III-A Cas proteins by the CRISPR Finder and CRISPRone (Supplementary Table 3). They were S. aureus 08BA02176, S. aureus JS395, S. aureus AR_0472, S. aureus AR_0470, S. aureus AR_0473, S. pseudintermedius 063228, S. schleiferi TSCC54, S. equorum KS1039, S. argenteus MSHR1132, S. argenteus XNO62, S. argenteus XNO106, S. epidermidis FDAARGOS_153, S. lugdunensis N920143, S. lugdunensis HKU09-01, and S. epidermidis RP62A (with two questionable CRISPR loci). As shown in Supplementary Table 2, 41 strains of Staphylococci contained only 1 CRISPR locus, 6 strains contained 2 CRISPR loci, and the remaining contained 3 loci. Also, 19 ST, 20 spa types, and 8 SCCmec types were found among the 42 strains of S. aureus, of which the most representative was ST398-t034-Vc. Strain AR_0472, AR_0470, AR_0473, AR466, and AR_0471 cannot be assigned to the SCCmec type because of the lack of SCCmec components. In addition, as to the position of CRISPRs in S. aureus, unlike the orphan CRISPRs away from SCCmec, the complete CRISPR-Cas systems were in the J1 region (Figure 1).

Figure 1. The position of CRISPR in MRSA. Strain 19_LA_388 is an example of strains carrying orphan CRISPR. The complete CRISPR-Cas systems in strain 08BA02176, JS395, AR_0472, AR_0470, and AR_0473 are in the J1 region.
Repeat sequences always maintain a high degree of similarity and even are identical in one CRISPR locus. Therefore, the CDR of each CRISPR locus was chosen as the representative to predict the RNA secondary structure and MFE. A total of 25 CDRs were recruited by removing redundance and not forming RNA secondary structures from 67 confirmed CRISPR loci in 52 strains of Staphylococci (Supplementary Table 4). The length of CDRs was concentrated in 23–37 bp. Compared with CDRs from an orphan CRISPR array, CDRs from a complete CRISPR-Cas system were more likely to form stable RNA secondary structures with lower MFE (Figure 2). Notably, group 13–21 had different CDRs length, their MFE were all −7.8 kcal/mol, and the stem of their secondary structure was 4 “G-C” base pairs. The secondary structure of group 22 and group 25 was complicated with 3 rings and 2 stems. Comparing with other CDRs forming conservative dumbbell-shaped RNA secondary structures, the CDRs of these two groups were overlength and the bases forming the stem were distributed at one end of the sequences.
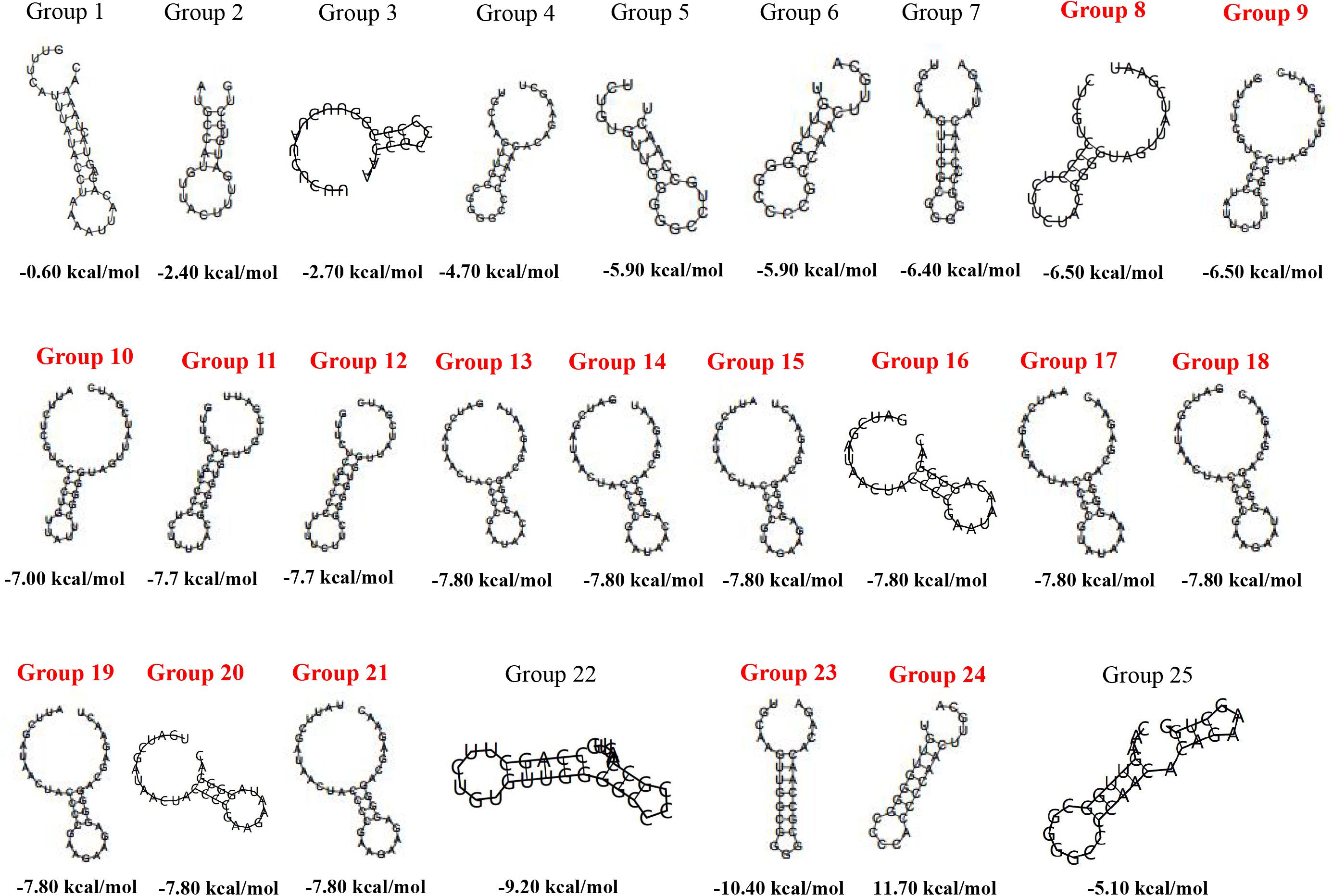
Figure 2. The RNA secondary structures and the MFE of the 25 group of CDRs. The CDRs from the CRISPR-Cas system are marked in red.
Among the 67 confirmed CRISPR loci, a total of 313 spacers were found. The difference between two spacer sequences below 5% was the same spacer sequence. Based on this principle, a total of 130 unique spacers were generated. In the process, we identified that the spacers in some CRISPR loci appeared to be highly homologous, all of which belonged to the strains harboring orphan CRISPR array. The length of spacers was concentrated in 19–53 bp. Since some DRs from a CRISPR locus could not form an RNA secondary structure, 127 spacers remained and then were employed as queries to search protospacers in phage and plasmid database by CRISPRTarget. Finally, 59 spacers hit 782 protospacers. These spacers belonged to the genomes carrying the complete CRISPR-Cas system and their length was concentrated in 29–43 bp. Furthermore, the provenance of the detected protospacers was examined. About 70% of protospacers mapped to hypothetical protein or unannotated genes. A clear provenance was shown in 239 protospacers of 31 spacers (Supplementary Table 5). These genes containing protospacers were DNA binding protein, encapsidation protein, terminase, transposase, and so on. A small fraction of spacers targeted open reading frames (ORF). Notably, two spacers targeted phi PVL-like protein in phage StauST398-2 and phage phi 13.
By drawing the graphic representation of spacers in CRISPR loci flanking Cas proteins, we realized the spacers that make up the S. aureus 08BA02176 CRISPR array were also frequently found in other S. aureus CRISPR arrays, even in S. pseudintermedius 063228 and S. schleiferi TSCC54, especially the CRISPR loci composed of spacer 16–18 (Figure 3).
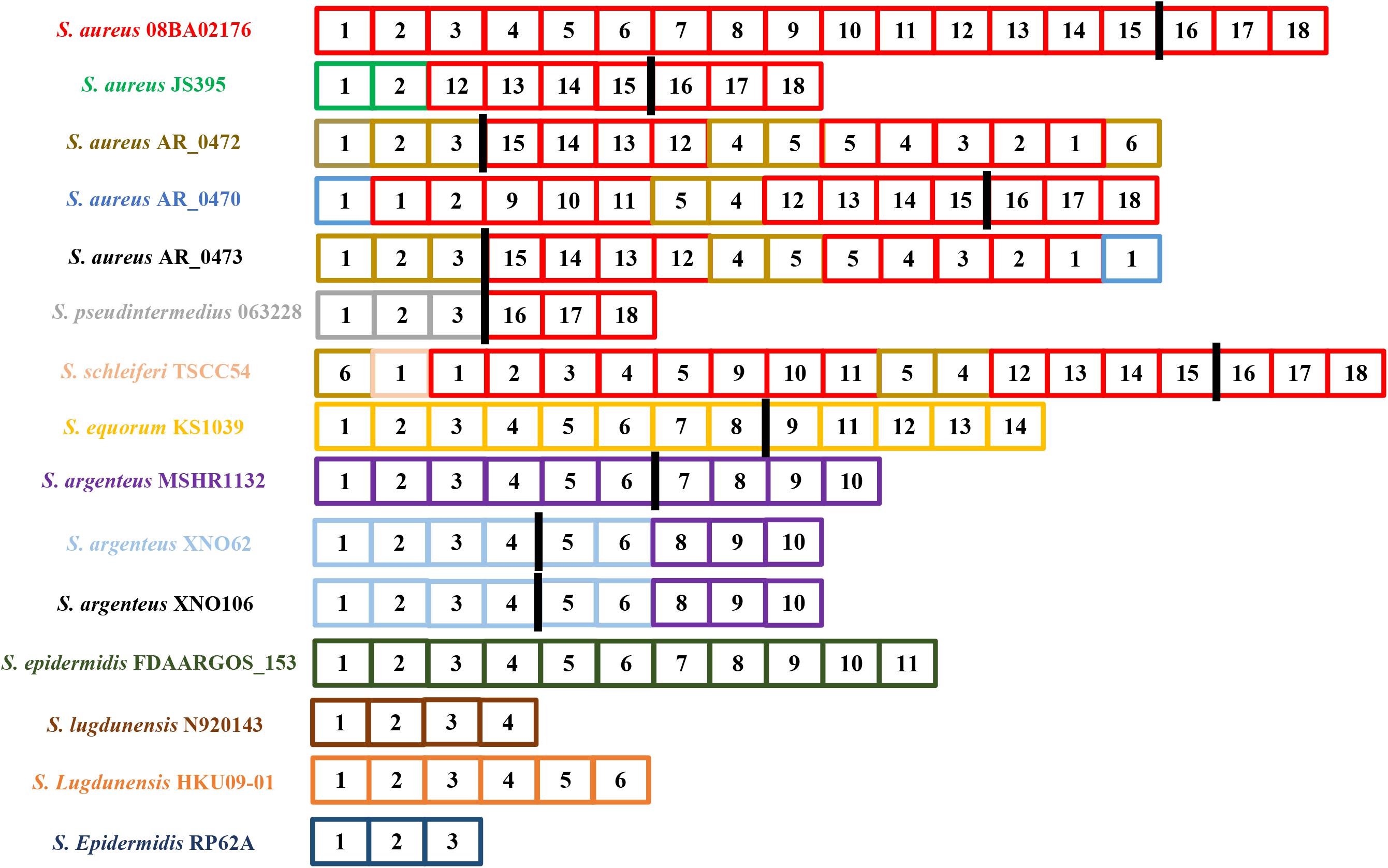
Figure 3. Graphic representation of spacers in the CRISPR loci flanking the complete Cas proteins. The strains shown in the figure all contain a complete type III-A CRISPR system. Repeats are not included. Two spacers are the same one when their diversity in DNA sequence is less than 5%. Each spacer consists of a small square, a number (continuously numbered in a strain), and a specific background color. The black rectangle represents the Cas cluster. The bacteria on the left are labeled as the color of the spacer as it originally appeared in its CRISPR locus. The strain labeled black indicates that there is no new spacer at its CRISPR locus.
In addition to exploring protospacer, CRISPRTarget is a rapid tool to identify potential PAM sequences. The portion of the crRNA consisting of DRs was presumed based on the secondary structure of the DRs. The length of crRNA 5′ handle was concentrated in 2–12 nt and crRNA was 40–62 nt. The nucleotides at both ends of the protospacers were extracted according to the number of nucleotides at the corresponding end of the crRNA. By removing redundance, a total of 176 PAMs were incorporated into the base pairing analysis. No complete base pairing between crRNA and PAM was found. We found that 51% (90/176) of crRNA 5′ handle contain the conserved motif 5′-ACGAGAA-3′ (shown in the Supplementary Materials). Therefore, we thought that the conserved motif 5′-ACGAGAA-3′ was widely distributed in crRNA 5′ handle. In S. lugdunensis N920143, the five base pairs at −8, −4, −3, −2, and −1 were the motifs with the most base pairs between crRNA and PAM (Figure 4).
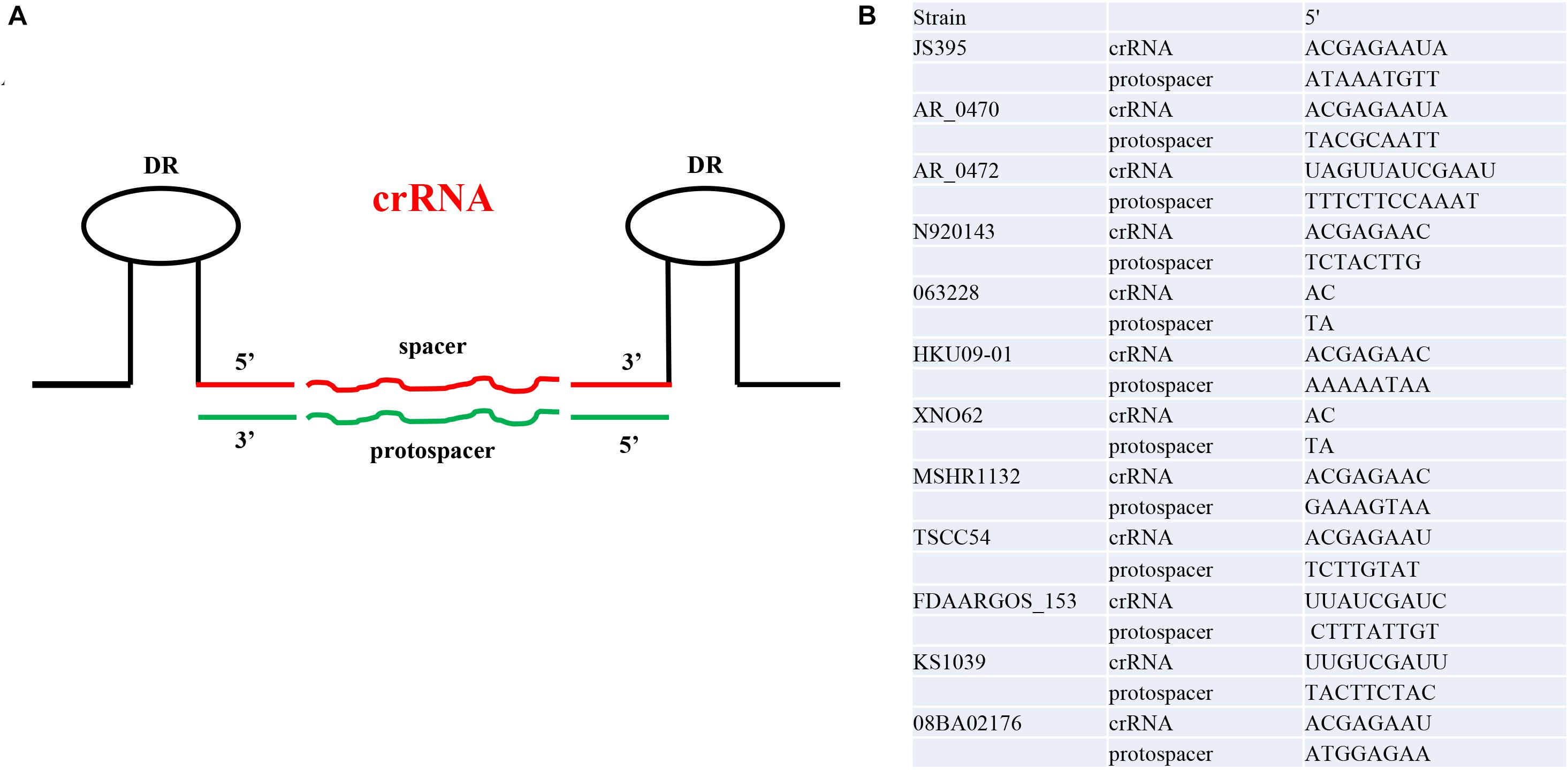
Figure 4. The schematic diagram and examples of the crRNA 5’ handle and protospacer 3’ end. (A) The schematic diagram of crRNA and protospacer. (B) Complementarity of crRNA 5’ handle and protospacer 3’ end. One Staphylococci has a representative.
In this study, 15 strains of Staphylococci were found carrying a complete CRISPR-Cas system and these cas gene clusters appeared to be highly homologous. As shown in Figure 5, the architecture of complete III-A Cas protein genes is cas1, cas2, cas10, csm2, csm3, csm4, csm5, csm6, and cas6. However, in S. epidermidis FDAARGOS_153, S. lugdunensis N920143, S. lugdunensis HKU09-01, and S. epidermidis RP62A, csm5 has been replaced by csm3, whose length was the same as that of csm5 in this position. Moreover, in S. lugdunensis HKU09-01, csm6 disappeared. A complete II CRISPR-Cas system was found in S. schleiferi TSCC54, presumably belonging to subtype IIC. There was no difference in the overall topology of phylogenetic trees among cas1, cas2, and complete cas genes, due to the highly homologous (Supplementary Figure 1).
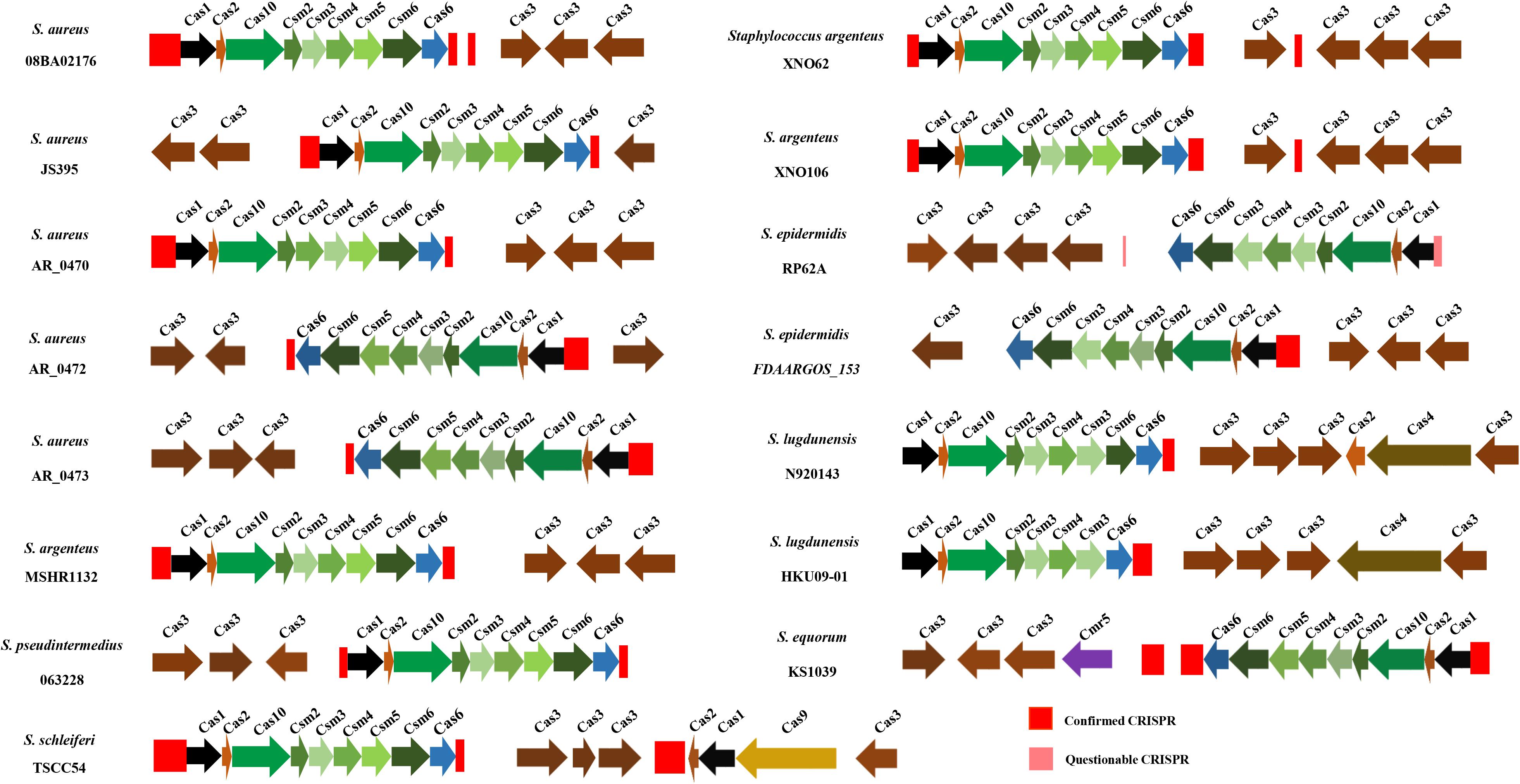
Figure 5. The distribution of CRISPR and cas genes in 15 Staphylococci. The red and pink rectangles represent confirmed CRISPRs and questionable CRISPRs, respectively.
As an adaptive prokaryotic immune system, the CRISPR-Cas system exists widely in nature. However, there are relatively few confirmed CRISPR loci in Staphylococcus. In this study, only 42 strains (12.92%) of S. aureus among 325 strains of Staphylococci carried confirmed CRISPR loci, far lower than other bacteria did (Watson et al., 2021). However, during the process of study, we found that there were several questionable CRISPR loci in each strain of S. aureus. We speculated that there were several reasons for this phenomenon. One is the lack of a unified discriminating standard, which leads to confusion in the confirmed and questionable CRISPR loci. For example, S. epidermidis RP62A is a model for Type III-A CRISPR-Cas systems (Makarova et al., 2015), but only two questionable CRISPR loci were found by CRISPR Finder. Another is no experimental evidence to prove whether these questionable CRISPR loci can play a role in the fighting against exogenous genes. Finally, it is possible to discover new CRISPR-Cas types (Nethery et al., 2021).
In this study, we analyzed the structures of 67 confirmed CRISPR loci in 52 strains of Staphylococci. Unlike the spacers in the orphan CRISPR loci, spacers in a complete CRISPR-Cas system showed high diversity in its CRISPR locus, consistent with the necessary for bacterial self-defense systems, which to some extent indicated that the acquisition of the new spacer was not affected by the pre-existing spacers. This was quite different from other subtypes in class 1, such as I-B, I-C, I-E, and I-F where the acquisition of the new spacer was often influenced by older spacers present in the same host (Nicholson et al., 2018). Some experimental studies have proved that this was not random, and pre-existing spacers were necessary in the acquisition process of new spacers (Li et al., 2014; Staals et al., 2016). By further analysis of these spacers, we found that the horizontal transfer and recombination of CRISPR loci within the same species were very frequent, like in S. aureus and S. argenteus. We also observed the same phenomena between different but closely related species, like among S. aureus, S. pseudintermedius, and S. schleiferi. Our previous study speculated this may be related to the SCCmec elements (Yang et al., 2015). We further observed that these CRISPRs were in the J1 region (Golding et al., 2010). Regions other than the ccr gene complex and mec gene complex are regarded as joining (J) regions. The J1 region often includes several ORFs and regulator genes (Lakhundi and Zhang, 2018). This suggests that these CRISPR loci are on SCCmec, and SCCmec can move from strain to strain, which may move along with SCCmec in horizontal transfer. Cao et al. (2016) analyzed six clinical isolates of S. aureus with subtype III-A CRISPR-Cas systems and found that four ST630 strains had similar CRISPR-Cas systems with that of ST398 S. aureus 08BA02176. Combined with our results, the link between MLST typing and CRISPR requires further studies.
Short palindromic sequences exist in direct repeats. Kunin et al. (2007) have shown that CRISPR repeats were structurally uneven, so the RNA secondary structures that may be formed during these direct repeats are studied. This structure can be combined with crRNA transcribed from the entire CRISPR sequence to form a bimodal structure to guide Cas protein to target the site (Yosef et al., 2012). Kunin et al. (2007) indicated that the stem-loop structure of some repeats may contribute to recognition-mediated contact between a gap-targeted exogenous RNA or DNA and a Cas-encoded protein, suggesting that the stability of RNA secondary structure may affect CRISPR function (He et al., 2012). Repeats from the CRISPR-Cas system were more likely to form stable RNA secondary structures with small MFE, while repeats from isolated CRISPR loci were more likely to form complex RNA secondary structures with unstable MFE (Wang et al., 2021). Our results showed that repeats from the CRISPR-Cas system were more likely to form stable RNA secondary structures with small MFE, while repeats from orphan CRISPR loci were more likely to form complex RNA secondary structures with unstable MFE. This further suggested that RNA secondary structure stability may enhance the function of CRISPR loci.
Spacers as genetic memories can show the history of past challenges from mobile genetic elements, such as bacteriophages and plasmids (Zhao et al., 2018). In this study, 59 spacers hit 782 protospacers. The vast majority of the protospacers originated from phage, indicating that those Staphylococci had been subjected to high frequency phage invasion and phage played an important role in the evolution of Staphylococcus. Levin (2010) have suggested that CRISPR immune bacteria and their phages are engaged in a co-evolutionary arms race, with host accumulation of spacers and phage accumulation point mutations. In recent years, this CRISPR-phage co-evolution model has been modified based on more refined models (Weissman et al., 2018; Common et al., 2019). Gophna et al. (2015) showed the inhibitory effect of CRISPR-Cas on HGT was undetectable on evolutionary timescales. Later, Watson et al. (2018) proved CRISPR-Cas-mediated phage resistance enhanced horizontal gene transfer by transduction. Notably, more protospacers have been detected with the development and popularity of next-generation sequencing technologies in recent years (Watson et al., 2018). Nevertheless, in a recent study, researchers found that protospacers were detectable for 1% to about 19% of the spacers and subtype III-A was 2% (Shmakov et al., 2017). More than half of the spacers targeted hypothetical protein, and for the rest, the enzymes and proteins necessary for bacterial growth and survival were the main targets.
Protospacer-adjacent motif plays a dual role: first, it initiates crRNA binding to the invading target sequence enabling interference, and second, prevents self-targeting (Gleditzsch et al., 2019; Collias and Beisel, 2021). The PAM allows the nuclease to discriminate between subsequent infection by the invader (non-self) from the invader-derived spacer sequence encoded in the CRISPR array (self) in type I, II, and V. Without the PAM requirement, CRISPR Cas systems would target their CRISPR arrays, leading to a potentially catastrophic autoimmune response (Collias and Beisel, 2021). Virtually, all CRISPR nucleases require a PAM in one form or another. However, the recognized PAM sequences are not shared by all Cas nucleases and instead vary widely, with different sequences, lengths, complexities, orientations, and distances from the target (Yosef et al., 2012). Therefore, the CRISPR Cas system will evolve and maintain PAM recognition as an absolute requirement for immune function under strict selection pressures (Collias and Beisel, 2021). Different CRISPR-Cas systems use diverse mechanisms to check the presence of the PAM sequence in the pre-spacer (Gleditzsch et al., 2019). In the E. coli subtype I-E system, this role is fulfilled by the Cas1 (Gleditzsch et al., 2019). Type III and VI evaluate the extent of complementarity between the flanking portions of the gRNA and target. Previous experimental researches suggested subtype III-A systems differentiated self and non-self sequences based on the complementarity between the crRNA 5′ handle and target 3′ terminal (Marraffini and Sontheimer, 2010; Samai et al., 2015). In the case of Type III and Type VI systems, limited evidence suggests that the PAM is located within the target RNA (Elmore et al., 2016). Because of this unique location, the PAM for these systems was renamed the RNA PAM (rPAM) or the protospacer-flanking sequence (PFS), respectively (Elmore et al., 2016). Both Deng et al. (2013) and Peng et al. (2015) observed DNA sequences downstream of pentanucleotide 5′-GAGAC-3′ and 5′-GAAAG-3′ could avoid DNA interference in subtype III-B Cmr systems. So, in this study, we focused on base pairing between the crRNA 5′ handle and target 3′ terminal and conserved motifs that were widely distributed in crRNA 5′ handle. We speculated that 5′-ACGAGAA-3′ seemed to be the conserved motif, then 5′-TTCTCGT-3′ was the protective motif, but further experimental studies are needed to verify our current findings. The potential impact of crRNA 5′ handle site mutations has also been studied in some studies. Cao et al. (2016) found that the first five nucleotides in the 5′ end did impact the CRISPR immunity in subtype III-A systems. Marraffini and Sontheimer (2010) found that protection of the CRISPR locus would disappear with at least two consecutive mismatches from position −4 to −2. Recently, Pyenson et al. (2017) indicated that PAM or seed sequences are not required for targeting in S. epidermidis type III-A CRISPR-Cas system. In subtype VI systems, researchers showed that repeat nucleotides at positions 1, 2, and 3 are the most important for the protective effects of a repeat sequence placed downstream of the protospacer (Meeske and Marraffini, 2018).
In type III systems, the adaptation module (Cas1 and Cas2) and repeat-associated mysterious protein (Cas6) are dispensable (Makarova et al., 2015). All strains of Staphylococci possessed these proteins in our study. As the best conserved Cas protein, Cas1 is mainly characterized by horizontal evolution in type III (Makarova et al., 2011; Vestergaard et al., 2014). Furthermore, the phylogeny of Cas1 and reverse transcriptase (RT) phylogeny suggested that the two proteins (or domains in fusion proteins) generally co-evolve (Silas et al., 2017). As for Cas2, other than the function in the adaptation module in Legionella pneumophila, it has been reported that Cas2 nuclease activity is critical for promoting infection in amoebae (Gunderson et al., 2015). Later, Dixit et al. (2016) found Cas2 possesses divalent metal-independent RNase activity in Leptospira interrogans. In type III systems, Cas6 is not a subunit of the effector complex. In a recent review, authors proposed a bivalent trapping and an unwinding mechanism for CRISPR-Cas6 to interact with the relaxed and the tight repeat RNA, respectively (Sefcikova et al., 2017). All type III systems encode the signature protein Cas10, SS protein, one Cas5 protein, and typically several paralogous Cas7 proteins (Makarova et al., 2015). During the interference stage, Cas-crRNA complexes function as sequence-specific nucleases to identify and degrade invading genetic material. Recent studies have shown that this is carried out by the Cas10 subunit in type III systems (Estrella et al., 2016; Kazlauskiene et al., 2016). By studying the interaction between a type III-A ribonucleoprotein complex and various RNA substrates, researchers found that the central role of the internal dynamics of CRISPR-Cas complexes in self- vs. non-self-discrimination and target specificity (Wang et al., 2019). Considering the case in our study, Csm5 was replaced by Csm3 in four strains of Staphylococci and one strain did not harbor Csm6. However, these CRISPR loci were active and no anti-CRISPR proteins, therefore a simpler csm complex could serve as the original function, or would CRISPR have a narrow target? For Csm6, as an RNase, it is essential for immunity when the target is located in late-expressed genes or contains mismatches to the spacer (Jiang et al., 2016).
In conclusion, this study primarily analyzed the structure of DRs and spacers, as well as the companion Cas proteins. The results showed that orphan CRISPR loci in S. aureus differed greatly from the structural characteristics of the CRISPR-Cas system. The strains harboring complete CRISPR-Cas systems contained multiple CRISPR loci, and whose DRs were likely to form the stable RNA secondary structures with lower MFE and spacers were variable with detectable protospacers. In addition, these CRISPRs were in the J1 region. The transfer of the CRISPR locus was frequent in Staphylococcus. Again, a conserved motif 5′-TTCTCGT-3′ that may protect their downstream sequences from DNA interference was found.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
TM designed this study. YW and TM finished the experiments, collected the data, and wrote the manuscripts together. YW, YL, TM, WX, XL, GD, and HY revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
This work was funded by the National Natural Science Foundation of China (No. 81973105). The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.736565/full#supplementary-material
Ahmed, M. Z., Badani, P., Reddy, R., and Mishra, G. (2021). CRISPR/Cas advancement in molecular diagnostics and signal readout approaches. J. Mol. Diagn. S1525-S1578:00261. doi: 10.1016/j.jmoldx.2021.07.025
Aires-de-Sousa, M. (2017). Methicillin-resistant Staphylococcus aureus among animals: current overview. Clin. Microbiol. Infect. 23, 373–380. doi: 10.1016/j.cmi.2016.11.002
Cao, L., Gao, C. H., Zhu, J., Zhao, L., Wu, Q., Li, M., et al. (2016). Identification and functional study of type III-A CRISPR-Cas systems in clinical isolates of Staphylococcus aureus. Int. J. Med. Microbiol. 306, 686–696. doi: 10.1016/j.ijmm.2016.08.005
Collias, D., and Beisel, C. L. (2021). CRISPR technologies and the search for the PAM-free nuclease. Nat. Commun. 12:555. doi: 10.1038/s41467-020-20633-y
Common, J., Morley, D., Westra, E. R., and van Houte, S. (2019). CRISPR-Cas immunity leads to a coevolutionary arms race between Streptococcus thermophilus and lytic phage. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374:20180098. doi: 10.1098/rstb.2018.0098
Couvin, D., Bernheim, A., Toffano-Nioche, C., Touchon, M., Michalik, J., Néron, B., et al. (2018). CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 46, W246–W251. doi: 10.1093/nar/gky425
Curti, L. A., Pereyra-Bonnet, F., Repizo, G. D., Fay, J. V., Salvatierra, K., Blariza, M. J., et al. (2020). CRISPR-based platform for carbapenemases and emerging viruses detection using Cas12a (Cpf1) effector nuclease. Emerg. Microbes Infect. 9, 1140–1148. doi: 10.1080/22221751.2020.1763857
de Lencastre, H., Sato, T., Usui, M., Konishi, N., Kai, A., Matsui, H., et al. (2017). Closely related methicillin-resistant Staphylococcus aureus isolates from retail meat, cows with mastitis, and humans in Japan. PLoS One 12:e0187319. doi: 10.1371/journal.pone.0187319
DeLeo, F. R., Otto, M., Kreiswirth, B. N., and Chambers, H. F. (2010). Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375, 1557–1568. doi: 10.1016/s0140-6736(09)61999-1
Deng, L., Garrett, R. A., Shah, S. A., Peng, X., and She, Q. (2013). A novel interference mechanism by a type IIIB CRISPR-Cmr module in sulfolobus. Mol. Microbiol. 87, 1088–1099. doi: 10.1111/mmi.12152
Dixit, B., Ghosh, K. K., Fernandes, G., Kumar, P., Gogoi, P., and Kumar, M. (2016). Dual nuclease activity of a Cas2 protein in CRISPR-Cas subtype I-B of leptospira interrogans. FEBS Lett. 590, 1002–1016. doi: 10.1002/1873-3468.12124
Elmore, J. R., Sheppard, N. F., Ramia, N., Deighan, T., Li, H., Terns, R. M., et al. (2016). Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR-Cas system. Genes Dev. 30, 447–459. doi: 10.1101/gad.272153.115
Estrella, M. A., Kuo, F.-T., and Bailey, S. (2016). RNA-activated DNA cleavage by the Type III-B CRISPR–Cas effector complex. Genes Dev. 30, 460–470. doi: 10.1101/gad.273722.115
Ge, B., Mukherjee, S., Hsu, C. H., Davis, J. A., Tran, T. T. T., Yang, Q., et al. (2017). MRSA and multidrug-resistant Staphylococcus aureus in U.S. retail meats, 2010-2011. Food Microbiol. 62, 289–297. doi: 10.1016/j.fm.2016.10.029
Ghaffari, S., Khalili, N., and Rezaei, N. (2021). CRISPR/Cas9 revitalizes adoptive T-cell therapy for cancer immunotherapy. J. Exp. Clin. Cancer Res. 40:269. doi: 10.1186/s13046-021-02076-5
Gleditzsch, D., Pausch, P., Müller-Esparza, H., Özcan, A., Guo, X., Bange, G., et al. (2019). PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biol. 16, 504–517. doi: 10.1080/15476286.2018.1504546
Golding, G. R., Bryden, L., Levett, P. N., McDonald, R. R., Wong, A., Wylie, J., et al. (2010). Livestock-associated methicillin-resistant Staphylococcus aureus sequence type 398 in humans, Canada. Emerg. Infect. Dis. 16, 587–594. doi: 10.3201/eid1604.091435
Gophna, U., Kristensen, D. M., Wolf, Y. I., Popa, O., Drevet, C., and Koonin, E. V. (2015). No evidence of inhibition of horizontal gene transfer by CRISPR-Cas on evolutionary timescales. ISME J. 9, 2021–2027. doi: 10.1038/ismej.2015.20
Gray, R. R., Tatem, A. J., Johnson, J. A., Alekseyenko, A. V., Pybus, O. G., Suchard, M. A., et al. (2010). Testing spatiotemporal hypothesis of bacterial evolution using methicillin-resistant Staphylococcus aureus ST239 genome-wide data within a bayesian framework. Mol. Biol. Evol. 28, 1593–1603. doi: 10.1093/molbev/msq319
Guan, J., Wang, W., and Sun, B. (2017). Chromosomal targeting by the Type III-A CRISPR-cas system can reshape genomes in Staphylococcus aureus. mSphere 2:e00403. doi: 10.1128/mSphere.00403-17
Gunderson, F. F., Mallama, C. A., Fairbairn, S. G., Cianciotto, N. P., and Fang, F. C. (2015). Nuclease activity of Legionella pneumophila Cas2 promotes intracellular infection of amoebal host cells. Infect. Immun. 83, 1008–1018. doi: 10.1128/iai.03102-14
He, L., Fan, X., and Xie, J. (2012). Comparative genomic structures of Mycobacterium CRISPR-Cas. J. Cell Biochem. 113, 2464–2473. doi: 10.1002/jcb.24121
Hoffman, S. J., Outterson, K., Rottingen, J. A., Cars, O., Clift, C., Rizvi, Z., et al. (2015). An international legal framework to address antimicrobial resistance. Bull. World Health Organ. 93:66. doi: 10.2471/BLT.15.152710
Holden, M. T., Hsu, L. Y., Kurt, K., Weinert, L. A., Mather, A. E., Harris, S. R., et al. (2013). A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 23, 653–664. doi: 10.1101/gr.147710.112
Jackson, S. A., McKenzie, R. E., Fagerlund, R. D., Kieper, S. N., Fineran, P. C., and Brouns, S. J. (2017). CRISPR-Cas: adapting to change. Science 356:eaal5056. doi: 10.1126/science.aal5056
Jiang, W., Samai, P., and Marraffini, L. A. (2016). Degradation of phage transcripts by CRISPR-associated RNases enables Type III CRISPR-Cas immunity. Cell 164, 710–721. doi: 10.1016/j.cell.2015.12.053
Jung, T.-Y., Park, K.-H., An, Y., Schulga, A., Deyev, S., Jung, J.-H., et al. (2016). Structural features of Cas2 from Thermococcus onnurineusin CRISPR-cas system type IV. Protein Sci. 25, 1890–1897. doi: 10.1002/pro.2981
Kazlauskiene, M., Tamulaitis, G., Kostiuk, G., Venclovas, Č, and Siksnys, V. (2016). Spatiotemporal Control of Type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol. Cell 62, 295–306. doi: 10.1016/j.molcel.2016.03.024
Khajanchi, N., and Saha, K. (2021). Controlling CRISPR with small molecule regulation for somatic cell genome editing. Mol. Ther. 2021:578. doi: 10.1016/j.ymthe.2021.06.014
Kunin, V., Sorek, R., and Hugenholtz, P. (2007). Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8:R61. doi: 10.1186/gb-2007-8-4-r61
Lakhundi, S., and Zhang, K. (2018). Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 31:e00020. doi: 10.1128/cmr.00020-18
Larsen, J., Andersen, P. S., Winstel, V., and Peschel, A. (2017). Staphylococcus aureus CC395 harbours a novel composite staphylococcal cassette chromosome mec element. J. Antimicrob. Chemother. 72, 1002–1005. doi: 10.1093/jac/dkw544
Leenay, R. T., and Beisel, C. L. (2017). Deciphering, communicating, and engineering the CRISPR PAM. J. Mol. Biol. 429, 177–191. doi: 10.1016/j.jmb.2016.11.024
Levin, B. R. (2010). Nasty viruses, costly plasmids, population dynamics, and the conditions for establishing and maintaining CRISPR-mediated adaptive immunity in bacteria. PLoS Genet. 6:e1001171. doi: 10.1371/journal.pgen.1001171
Li, M., Wang, R., Zhao, D., and Xiang, H. (2014). Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 42, 2483–2492. doi: 10.1093/nar/gkt1154
Lindsay, J. A. (2014). Staphylococcus aureus genomics and the impact of horizontal gene transfer. Int. J. Med. Microbiol. 304, 103–109. doi: 10.1016/j.ijmm.2013.11.010
Lowy, F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. doi: 10.1056/NEJM199808203390806
Luthra, R., Kaur, S., and Bhandari, K. (2021). Applications of CRISPR as a potential therapeutic. Life Sci. 284:119908. doi: 10.1016/j.lfs.2021.119908
Ma, P., Meng, Q., Sun, B., Zhao, B., Dang, L., Zhong, M., et al. (2020). MeCas12a, a highly sensitive and specific system for COVID-19 detection. Adv. Sci. 7:2001300. doi: 10.1002/advs.202001300
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J., Charpentier, E., Horvath, P., et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477. doi: 10.1038/nrmicro2577
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi: 10.1038/nrmicro3569
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J. J., et al. (2020). Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83. doi: 10.1038/s41579-019-0299-x
Marraffini, L. A., and Sontheimer, E. J. (2010). Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571. doi: 10.1038/nature08703
Meeske, A. J., and Marraffini, L. A. (2018). RNA guide complementarity prevents self-targeting in type VI CRISPR systems. Mol. Cell 71, 791–801. doi: 10.1016/j.molcel.2018.07.013
Moreb, E. A., and Lynch, M. D. (2021). Genome dependent Cas9/gRNA search time underlies sequence dependent gRNA activity. Nat. Commun. 12:5034. doi: 10.1038/s41467-021-25339-3
Nethery, M. A., Korvink, M., Makarova, K. S., Wolf, Y. I., Koonin, E. V., and Barrangou, R. (2021). CRISPRclassify: repeat-based classification of CRISPR Loci. CRISPR J. 4, 558–574. doi: 10.1089/crispr.2021.0021
Nicholson, T. J., Jackson, S. A., Croft, B. I., Staals, R. H. J., Fineran, P. C., and Brown, C. M. (2018). Bioinformatic evidence of widespread priming in type I and II CRISPR-Cas systems. RNA Biol. 16, 566–576. doi: 10.1080/15476286.2018.1509662
Peng, W., Feng, M., Feng, X., Liang, Y. X., and She, Q. (2015). An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 43, 406–417. doi: 10.1093/nar/gku1302
Pyenson, N. C., Gayvert, K., Varble, A., Elemento, O., and Marraffini, L. A. (2017). Broad targeting specificity during bacterial Type III CRISPR-Cas immunity constrains viral escape. Cell Host Microbe 22, 343–353. doi: 10.1016/j.chom.2017.07.016
Ran, F. A., Cong, L., Yan, W. X., Scott, D. A., Gootenberg, J. S., Kriz, A. J., et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191. doi: 10.1038/nature14299
Samai, P., Pyenson, N., Jiang, W., Goldberg, G. W., Hatoum-Aslan, A., and Marraffini, L. A. (2015). Co-transcriptional DNA and RNA cleavage during Type III CRISPR-Cas immunity. Cell 161, 1164–1174. doi: 10.1016/j.cell.2015.04.027
Sefcikova, J., Roth, M., Yu, G., and Li, H. (2017). Cas6 processes tight and relaxed repeat RNA via multiple mechanisms: a hypothesis. Bioessays 39:1700019. doi: 10.1002/bies.201700019
Shmakov, S., Abudayyeh, O. O., Makarova, K. S., Wolf, Y. I., Gootenberg, J. S., Semenova, E., et al. (2015). Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 60, 385–397. doi: 10.1016/j.molcel.2015.10.008
Shmakov, S. A., Sitnik, V., Makarova, K. S., Wolf, Y. I., Severinov, K. V., and Koonin, E. V. (2017). The CRISPR spacer space is dominated by sequences from species-specific mobilomes. mBio 8, e1397–e1317. doi: 10.1128/mBio.01397-17
Silas, S., Makarova, K. S., Shmakov, S., Páez-Espino, D., Mohr, G., Liu, Y., et al. (2017). On the origin of reverse transcriptase-using CRISPR-Cas systems and their hyperdiverse, enigmatic spacer repertoires. mBio 8:e00897. doi: 10.1128/mBio.00897-17
Staals, R. H. J., Jackson, S. A., Biswas, A., Brouns, S. J. J., Brown, C. M., and Fineran, P. C. (2016). Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR–Cas system. Nat. Commun. 7:12853. doi: 10.1038/ncomms12853
Strommenger, B., Bartels, M. D., Kurt, K., Layer, F., Rohde, S. M., Boye, K., et al. (2014). Evolution of methicillin-resistant Staphylococcus aureus towards increasing resistance. J. Antimicrob. Chemother. 69, 616–622. doi: 10.1093/jac/dkt413
Tang, Y., Larsen, J., Kjeldgaard, J., Andersen, P. S., Skov, R., and Ingmer, H. (2017). Methicillin-resistant and -susceptible Staphylococcus aureus from retail meat in Denmark. Int. J. Food Microbiol. 249, 72–76. doi: 10.1016/j.ijfoodmicro.2017.03.001
Vega, S., and Dowzicky, M. J. (2017). Antimicrobial susceptibility among gram-positive and gram-negative organisms collected from the Latin American region between 2004 and 2015 as part of the tigecycline evaluation and surveillance trial. Anna. Clin. Microbiol. Antimicrob. 16:50. doi: 10.1186/s12941-017-0222-0
Vestergaard, G., Garrett, R. A., and Shah, S. A. (2014). CRISPR adaptive immune systems of Archaea. RNA Biol. 11, 156–167. doi: 10.4161/rna.27990
Wang, L., Mo, C. Y., Wasserman, M. R., Rostøl, J. T., Marraffini, L. A., and Liu, S. (2019). Dynamics of Cas10 govern discrimination between self and non-self in type III CRISPR-Cas immunity. Mol. Cell 73, 278–290. doi: 10.1016/j.molcel.2018.11.008
Wang, L., Wang, L., Liu, Y., Wang, Z., Chen, Q., Liu, Z., et al. (2021). Characterization of type I-F CRISPR-Cas system in Laribacter hongkongensis isolates from animals, the environment and diarrhea patients. Int. J. Food Microbiol. 346:109153. doi: 10.1016/j.ijfoodmicro.2021.109153
Wang, W., Baloch, Z., Jiang, T., Zhang, C., Peng, Z., Li, F., et al. (2017). Enterotoxigenicity and antimicrobial resistance of Staphylococcus aureus isolated from retail food in China. Front. Microbiol. 8:2256. doi: 10.3389/fmicb.2017.02256
Wang, Y., Ke, Y., Liu, W., Sun, Y., and Ding, X. (2020). A one-pot toolbox based on Cas12a/crRNA enables rapid foodborne pathogen detection at attomolar level. ACS Sens. 5, 1427–1435. doi: 10.1021/acssensors.0c00320
Watson, B. N. J., Staals, R. H. J., Fineran, P. C., Bondy-Denomy, J., and Gilmore, M. S. (2018). CRISPR-Cas-mediated phage resistance enhances horizontal gene transfer by transduction. mBio 9:e02406. doi: 10.1128/mBio.02406-17
Watson, B. N. J., Steens, J. A., Staals, R. H. J., Westra, E. R., and van Houte, S. (2021). Coevolution between bacterial CRISPR-Cas systems and their bacteriophages. Cell Host Microbe 29, 715–725. doi: 10.1016/j.chom.2021.03.018
Weissman, J. L., Holmes, R., Barrangou, R., Moineau, S., Fagan, W. F., Levin, B., et al. (2018). Immune loss as a driver of coexistence during host-phage coevolution. ISME J. 12, 585–597. doi: 10.1038/ismej.2017.194
Yang, S., Liu, J., Shao, F., Wang, P., Duan, G., and Yang, H. (2015). Analysis of the features of 45 identified CRISPR loci in 32 Staphylococcus aureus. Biochem. Biophys. Res. Commun. 464, 894–900. doi: 10.1016/j.bbrc.2015.07.062
Yosef, I., Goren, M. G., and Qimron, U. (2012). Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576. doi: 10.1093/nar/gks216
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas System. Cell 163, 759–771. doi: 10.1016/j.cell.2015.09.038
Zhang, Q., and Ye, Y. (2017). Not all predicted CRISPR-Cas systems are equal: isolated cas genes and classes of CRISPR like elements. BMC Bioinformatics 18:92. doi: 10.1186/s12859-017-1512-4
Keywords: Staphylococcus aureus, CRISPR-Cas, protospacer, crRNA, gene transfer
Citation: Wang Y, Mao T, Li Y, Xiao W, Liang X, Duan G and Yang H (2021) Characterization of 67 Confirmed Clustered Regularly Interspaced Short Palindromic Repeats Loci in 52 Strains of Staphylococci. Front. Microbiol. 12:736565. doi: 10.3389/fmicb.2021.736565
Received: 05 July 2021; Accepted: 27 September 2021;
Published: 22 October 2021.
Edited by:
Ruiting Lan, University of New South Wales, AustraliaReviewed by:
Haridha Shivram, Genentech, Inc., United StatesCopyright © 2021 Wang, Mao, Li, Xiao, Liang, Duan and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiyan Yang, eWh5QHp6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.