
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 04 October 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.736464
This article is part of the Research TopicGlobal Dissemination and Evolution of Epidemic Multidrug-Resistant Gram-Negative Bacterial Pathogens: Surveillance, Diagnosis and TreatmentView all 11 articles
 Razib Mazumder1†
Razib Mazumder1† Arif Hussain1†
Arif Hussain1† Ahmed Abdullah1
Ahmed Abdullah1 Md. Nazrul Islam2
Md. Nazrul Islam2 Md. Tuhin Sadique2
Md. Tuhin Sadique2 S. M. Muniruzzaman2
S. M. Muniruzzaman2 Anika Tabassum2
Anika Tabassum2 Farhana Halim2
Farhana Halim2 Nasrin Akter2
Nasrin Akter2 Dilruba Ahmed2*
Dilruba Ahmed2* Dinesh Mondal1*
Dinesh Mondal1*Background: Escherichia coli is a major extended-spectrum β-lactamase (ESBL)–producing organism responsible for the rapid spread of antimicrobial resistance (AMR) that has compromised our ability to treat infections. Baseline data on population structure, virulence, and resistance mechanisms in E. coli lineages from developing countries such as Bangladesh are lacking.
Methods: Whole-genome sequencing was performed for 46 ESBL–E. coli isolates cultured from patient samples at the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b)-Dhaka. Sequence data were analyzed to glean details of AMR, virulence, and phylogenetic and molecular markers of E. coli lineages.
Results: Genome comparison revealed presence of all major high-risk clones including sequence type 131 (ST131) (46%), ST405 (13%), ST648 (7%), ST410 (4.3%), ST38 (2%), ST73 (2%), and ST1193 (2%). The predominant ESBL gene and plasmid combination were blaCTX–M–15 and FII-FIA-FIB detected in diverse E. coli phylogroups and STs. The blaNDM–5 (9%) gene was present in prominent E. coli STs. One (2%) mcr-1–positive ST1011 E. coli, coharboring blaCTXM–55 gene, was detected. The extraintestinal pathogenic E. coli genotype was associated with specific E. coli lineages. The single nucleotide polymorphism (SNP)-based genome phylogeny largely showed correlation with phylogroups, serogroups, and fimH types. Majority of these isolates were susceptible to amikacin (93%), imipenem (93%), and nitrofurantoin (83%).
Conclusion: Our study reveals a high diversity of E. coli lineages among ESBL-producing E. coli from Dhaka. This study suggests ongoing circulation of ST131 and all major non-ST131 high-risk clones that are strongly associated with cephalosporin resistance and virulence genes. These findings warrant prospective monitoring of high-risk clones, which would otherwise worsen the AMR crises.
Infections with Escherichia coli that produce extended-spectrum β-lactamases (ESBLs) present an increasing clinical and public health threat (Mathers et al., 2015). These bacteria are resistant to several new-generation cephalosporin agents and render them ineffective for treating infections (Pitout and Laupland, 2008). Infections of ESBL E. coli causes increased morbidity, high mortality, longer hospital stays, and increased health care costs in comparison to infections caused by non-ESBL E. coli (Paterson and Bonomo, 2005). Treatment of such infections has been further complicated by the emergence of carbapenem resistance in ESBL–E. coli; these carbapenem-resistant isolates are often found resistant to all the available antibiotics (Doi, 2019).
Several strains of ESBL-producing E. coli are pathogenic; they commonly cause urinary tract infections (UTIs) in otherwise healthy people (Calbo et al., 2006; Franz et al., 2015). Moreover, when certain pathogenic bacterial clones horizontally acquire ESBL genes, they can emerge and reemerge rapidly within the population through clonal dissemination and thereby gain local or even global predominance as international high-risk clones (Price et al., 2013). Examples of such epidemiological successful extraintestinal pathogenic E. coli (ExPEC) lineages include sequence type 131 (ST131), ST410, ST38, ST73, ST405, and ST648, which are associated with both nosocomial and community-acquired infections and are being increasingly detected from multiple origins, worldwide (Baquero et al., 2013; Shaik et al., 2017; Manges et al., 2019). Since the last decade, CTX-M–type enzymes have become the most predominant ESBLs in E. coli and other Gram-negative bacteria of clinical significance (Cantón and Coque, 2006). Similarly, the transposable elements containing the New Delhi metallo-β-lactamase (NDM) gene have been identified to be responsible for the rapid dissemination of carbapenem resistance in E. coli (Kumarasamy et al., 2010).
Currently, a high number of ESBL-producing E. coli infections are linked to the pandemic E. coli lineage ST131 (Nicolas-Chanoine et al., 2008). Moreover, it is shown that E. coli ST131 strains are strongly associated with CTX-M-15–type ESBL and are predominantly responsible for causing bladder infections, kidney infections, and urosepsis worldwide including Southeast Asia (Nicolas-Chanoine et al., 2014; Chen et al., 2019). For instance, in India, it was reported that 93% of ST131 E. coli isolates carried CTX-M-15. Studies have also reported the presence of the H30 subclone that is reportedly responsible for the clonal dissemination of ST131 E. coli (Hussain et al., 2012, 2014; Ranjan et al., 2015; Shaik et al., 2017). A study from Bangladesh reported that 71% of ESBL-producing E. coli was linked to the ST131 clone (Begum and Shamsuzzaman, 2016). Another study from Bangladesh identified a clonal cluster of clinical E. coli isolates belonging to serotype O25:H4, which indicates the widespread circulation of the ST131 E. coli lineage (Lina et al., 2014).
High-resolution studies on ESBL-producing E. coli strains from Bangladesh are lacking compared with the rest of the world, which limit our understanding of the population structure, emerging antimicrobial resistance (AMR) and their potential to spread. Herein, we report the findings of a genomic epidemiological investigation of the recent ESBL–E. coli isolates, especially the CTX-M–type ESBL producers associated with extraintestinal infections in Dhaka, Bangladesh. We analyzed their resistance genes, virulence genes, plasmid types, phylogenetic relatedness, and molecular features. Our study suggests ongoing circulation of ST131 and all major non-ST131 E. coli high-risk clones in Dhaka, Bangladesh. Studies such as these will provide important insights into the evolution of pathogens and will inform novel interventional strategies.
Bacterial isolates were collected randomly as part of routine (1%) surveillance (128 E. coli isolates) between March 2018 and July 2019 from the Clinical Microbiology and Immunology Laboratory of the International Center for Diarrheal Disease Research, Bangladesh (icddr,b) (Mazumder et al., 2020a). From this collection of 128 E. coli isolates, 46 randomly selected ESBL E. coli spanning over the entire 16-month period (representing 1–4 isolates per month) were whole genome sequenced. Of these 46 isolates, 34 were obtained from urine and 12 from pus specimen (Table 1). Identification of bacterial isolates was performed using standard biochemical methods. Antimicrobial susceptibility testing by disk diffusion was performed against the following antibiotics: gentamicin (10 μg), amikacin (30 μg), cotrimoxazole (25 μg), ciprofloxacin (5 μg), nitrofurantoin (200 μg), cefuroxime (30 μg), ceftriaxone (30 μg), cefixime (5 μg), cefepime (30 μg), and imipenem (10 μg). E. coli ATCC 25922 was used for quality control (Clinical and Laboratory Standards Institute, 2015).
Genomic DNA from an overnight bacterial culture was extracted and purified using QIAamp DNA Mini kit (Qiagen, Germany). The purity of the genomic DNA was assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific, United States), and quantification was performed using a Qubit 2.0 fluorometer (Life Technologies). DNA libraries were prepared using the Nextera XT DNA library Prep kit (Illumina) (Mazumder et al., 2020b). The pooled library was sequenced at the genomic sequencing facility of icddr,b on an Illumina Nextseq500 system using the 150-base-paired-end Mid-output v2.5 sequencing kit. The sequence reads were trimmed and filtered using Trimmomatic 0.39 (Bolger et al., 2014), and de novo assemblies of the resulting reads were generated using SPAdes 3.11.1 (Bankevich et al., 2012), and the quality check was done using QUAST v5.02 (Mikheenko et al., 2018). The genomes of our study were annotated using Prokka de novo (Seemann, 2014). The program was run on fast mode, and the genus-specific BLAST database was used. The genome features and associated metadata are listed in Table 1.
Phylotyping was done using the ClermonTyping online tool (Beghain et al., 2018). Other molecular features were identified using the Center for Genomic Epidemiology’s Bioinformatics tools using default parameters unless otherwise stated. Specifically, MLST 2.0 (Larsen et al., 2012) was used to identify STs, SerotypeFinder 1.1 (Joensen et al., 2015) for detecting serotypes, FimTyper 1.0 (Roer et al., 2017) to determine fimH types, and PlasmidFinder 2.1 (Carattoli et al., 2014) for identification of plasmid incompatibility groups. Comprehensive Antibiotic Resistance Database (McArthur et al., 2013) was used for screening the presence of acquired AMR genes; virulence genes were detected using a custom-made database of 50 genes from the Virulence Factors Database (Chen et al., 2005) belonging to different categories as described previously (Stoesser et al., 2015), a threshold, ≥ 90% identity; minimum coverage, ≥ 70% was used for both. AMR-related chromosomal point mutations were identified in the draft genomes using PointFinder (Zankari et al., 2017). The genetic context of blaCTX–M–15, blaNDM–5, and mcr-1 with respect to plasmid was determined by BLAST searching of particular contigs against the GenBank database. Genomes having three or more ExPEC-associated genes were classified as ExPEC according to Johnson’s criteria (Johnson and Stell, 2000). From virulence and resistance gene presence and absence CSV files, heat plots were obtained, using the Python module seaborn and matplotlib.
The reference (E. coli 0154:H7 Sakai strain) guided multi-fasta consensus alignment of 46 E. coli genomes was obtained using the Snippy v4.4.0 pipeline (Seemann, 2015). Putative recombination regions were detected and masked using Gubbins v2.3.4 (Croucher et al., 2015). Finally, the SNP- based phylogeny was inferred using RaxML v8.2.12 (Stamatakis, 2014) using GTR (Generalized Time Reversible) substitution model and GAMMA distribution as the model of rate heterogeneity.
Using SPSS statistics software (version 25.0), continuous variables were compared with the non-parametric Mann–Whitney U-test, and proportions were analyzed using the χ2-test; p-values with a threshold of < 0.05 were considered statistically significant. Accordingly, the p-values are indicated within text where appropriate.
The sequence diversity of the study isolates as analyzed by in silico MLST demonstrates that the 46 ESBL–E. coli isolates belonged to 19 different STs (Figure 1). The pandemic ST131 lineage was the most predominant ST identified, comprising 46% (21/46) of the isolates. Apart from ST131, the other significant STs included the presence of isolates belonging to the international high-risk clones, such as ST405 (13% 6/46), ST648 (7% 3/46), ST410 (4.3% 2/46) ST38 (2%), ST73 (2%), and ST1193 (2%). Phylogrouping identified a total of six phylogroups, namely, A, B1, B2, C, D, and F, in the ESBL–E. coli isolates (Figure 1). The phylogroup B2 (24/46) was the major phylogroup in our collection, which predominantly consisted of ST131 isolates (88%, 21/24). Phylogroup D was the second largest phylogroup with 20% isolates (n = 9, 6 ST405, 1 ST38, 1 ST1011, 1 ST1884), which was followed by group F (n = 4, 3 ST648, 1 ST354), B1 (n = 3, 1 ST3748, 1 ST162, 1 ST2178), C (n = 3, 2 ST410, 1 ST2851), and A (n = 3, 1 ST167, 1 ST617, 1 ST48). Serotyping revealed that out of the 21 ST131 E. coli, 19 had serotype O25:H4, and two had O16:H5 serogroup (Figure 1). All 19 ST131 E. coli with O25; H4 serogroup had fimH 30 alleles, whereas the two ST131 E. coli isolates with O16:H5 serogroup had fimH41 allele, whereas the non-ST131 isolates exhibited diverse serotypes that included 19 different serotypes. Among them, the six emerging urosepsis pathogenic isolates belonging to ST405 had O102:H6 serogroup with fimH 29 allele except for one strain, which had fimH 27. The two isolates of high-risk clone ST410 had O8:H21 serogroup with fimH 24 allele. However, the three isolates belonging to the emerging pandemic lineage ST648 were not associated with any distinct sero or fimH group. Similar to the results of serogroups, the non-ST131 isolates exhibited diverse fimH (10 different) types.
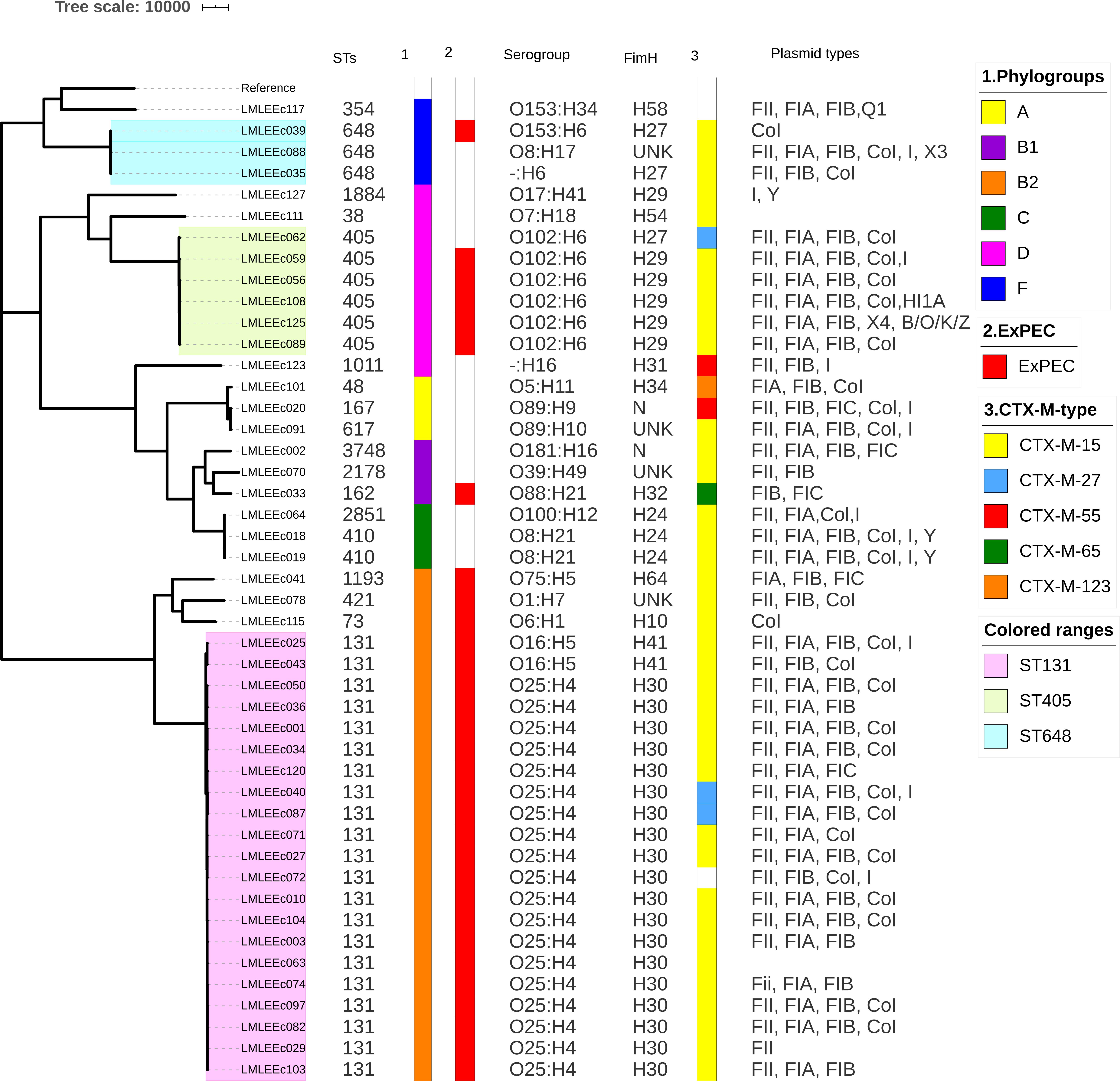
Figure 1. Core-genome SNP-based phylogenetic tree representing the population structure and comparison of genetic traits observed in 46 ESBL E. coli isolates. Sequence types, phylogroups, ExPEC status, serogroups, fimH types, CT-X-M types, and plasmid replicon types are shown next to the tree.
Majority of isolates were susceptible to amikacin (93%), imipenem (93%), and nitrofurantoin (83%) (Figure 2). On the other hand, a majority of isolates were resistant to the common, empirically used antibiotics such as cefixime (96%), cefuroxime (96%), ceftriaxone (98%), and ciprofloxacin (78%). Besides, a moderate number of isolates were also resistant to cotrimoxazole (70%), gentamicin (43%), and cefepime (67%). In contrast to the non-ST131 E. coli, isolates belonging to the ST131 lineage had higher resistance prevalence as the aggregate resistance score [median (range)] for ST131 isolates [6 (4–8)] was higher than that of non-ST131 isolates [5.5 (0–10)]. However, the difference was not statistically significant (p > 0.5 Mann–Whitney U-test).
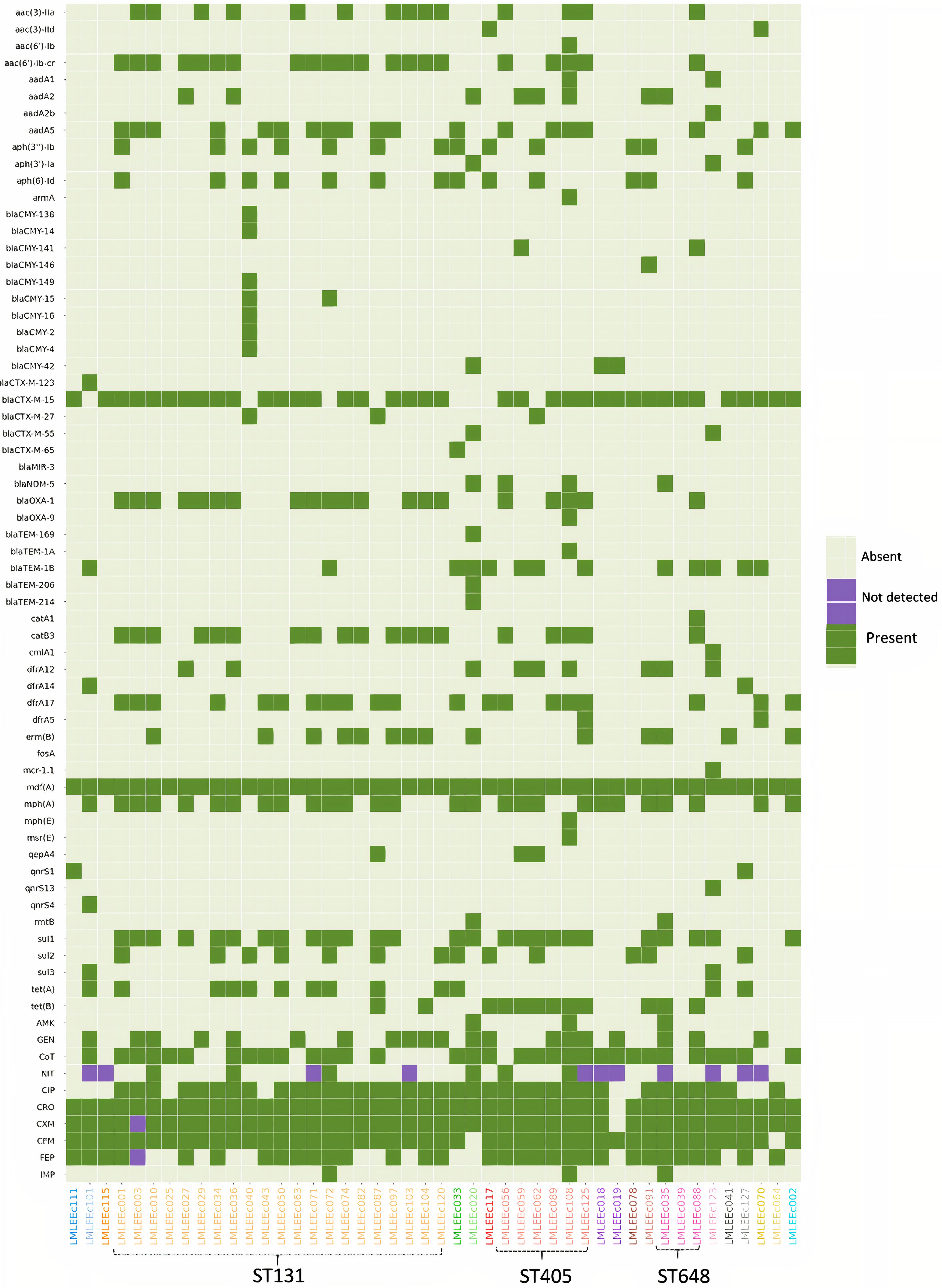
Figure 2. Heatmap showing the distribution of acquired AMR genes and AMR phenotypic profiles within 46 E. coli isolates. Gene names are listed on the left, and strain names are listed below the image. The presence of gene is indicated by colored blocks, and the gene absence is indicated by gray blocks. CRO, ceftriaxone; CXM, cefuroxime; CFM, cefixime; FEP, cefepime; NIT, nitrofurantoin; AMK, amikacin; CIP, ciprofloxacin; CoT, cotrimoxazole; IMP, imipenem; GEN, gentamicin.
Whole-genome sequencing (WGS) analysis identified 60 acquired AMR genes that are known to encode proteins conferring resistance to different classes of antibiotics including β-lactams, aminoglycosides, chloramphenicol, tetracycline, sulfonamides, trimethoprim, fluoroquinolone, and colistin (Figure 2).
We identified 24 genes associated with β-lactam resistance including 13 ESBL, 10 AmpC β-lactamase, and 1 carbapenemase gene. The 13 ESBL genes included blaOXA–1 (43%), blaTEM–1B (28%), blaCTX–M–27 (7%), and blaCTX–M–55 (4%). Thirty-eight of 46 isolates (81%) carried the blaCTX–M–15 gene. Of 38 isolates carrying the blaCTX–M–15 gene, 36 were resistant to ceftriaxone and cefixime; therefore, this gene was strongly associated with third-generation cephalosporin resistance (p < 0.001). Moreover, the gene blaCTX–M–15 was detected in isolates of diverse genetic backgrounds affiliated to 6 different phylogroups and 13 different STs, including the high-risk clones—ST131 (18/21), ST405 (5/6), ST648 (3/3), ST410 (2/2), ST38 (1/1), ST73 (1/1), and ST1193 (1/1). The blaCTX–M–15 gene was located on a plasmid for 23 of 38 isolates. Isolates also harbored other ESBL genes that included blaCTX–M–123, blaCTX–M–65, blaMIR–3, blaOXA–9, blaTEM–169, blaTEM–1A, blaTEM–1B, blaTEM–206, and blaTEM–214 in lesser proportion. Similarly, a low prevalence of AmpC β-lactamase genes was detected (Figure 2).
We identified 12 genes known to confer resistance to aminoglycosides (Figure 2). These included aac(6′)-Ib-cr (45%), aadA5 (40%), aac(3)-IIa (30%), aph(3′′)-Ib (30%), aph(6)-Id (30%), and aadA2 (17%). Of these six prominent aminoglycoside genes, only two, aac(3)-IIa (14/13) and aac(6′)-Ib-cr (21/14), were strongly associated with gentamicin resistance (p < 0.001). None of the aminoglycoside genes was associated with amikacin resistance. Approximately 4% of the isolates harbored aac(3)-IId, aadA1, and aph(3′)-Ia; other genes of this class included aac(6′)-Ib (one isolate), aadA2b (one isolate), and armA (one isolate).
Three genes known to encode sulfonamide resistance were identified: sul1 in 26/46 (57%) isolates, sul2 in 14/46 (30%) isolates, and sul3 in 2/46 (4%) isolates. The presence of sul1 was positively correlated with phenotypic resistance to cotrimoxazole (23/26) (p < 0.001). Similarly, of the four genes responsible for trimethoprim resistance [dfrA17 (43%), dfrA12 (19%), dfrA14 (4%), dfrA5 (4%)], the gene dfrA17 was strongly associated with cotrimoxazole resistance (p < 0.001).
Using PointFinder, we identified two mutations in gyrA including S83L (serine to leucine) and D87N (aspartic acid to asparagine). The mutation S83L was detected in 13% (6/46) of isolates. In contrast, 80% (37/46) of isolates had both the mutations (S83L and D87N); the dual gyrA mutants were all resistant to ciprofloxacin. However, isolates (4/46) having a single mutation (S83L) were all susceptible to ciprofloxacin. Similarly, in the parC amino acid product, we identified two substitutions at codon position 80 (serine to isoleucine) and codon 84 (glutamic acid to valine). The mutation S80I was present in 33% (15/46) of isolates, and the mutation E84V was detected in one isolate, whereas 22 of 46 isolates harbored both of these mutations, and they were all resistant to ciprofloxacin. Three substitutions were also detected in parE gene, with I529L being predominant (20/46) followed by S458A (11/46) and L416F (1/46). All isolates (20/46) carrying I529L mutation were ciprofloxacin-resistant, and this mutation was found in the majority of ST131 strains (18/21 ST131 strains). Additionally, the plasmid-mediated quinolone resistance gene qnrs1 was detected in two isolates followed by qnrS4 and qnrS13 in one isolate each. Except for qnrs13, isolates carrying qnrS genes were susceptible to ciprofloxacin.
NDM-5 carbapenemase was detected in four E. coli isolates (8.5%). All four isolates belonged to the prominent clonal groups: two ST405 (phylogroup D), one ST648 (phylogroup F), and one ST167 (phylogroup A). All four blaNDM–5 genes were found to be located on plasmids (4/4 isolates). Except for one (ST167), all three isolates coharbored a blaCTX–M–15 gene. Phenotypic resistance to imipenem was detected in only two of four blaNDM–5–positive isolates. Nonetheless, all four blaNDM–5–positive E. coli were pan–drug-resistant as resistance was detected to at least 7 of 10 antibiotics tested. We detected one mcr-1–positive E. coli belonging to ST1011; the mcr-1 gene was detected on a plasmid. This strain was coharboring a blaCTXM–55 gene and showed phenotypic resistance to cotrimoxazole, ciprofloxacin, ceftriaxone, cefuroxime, and cefixime and was notably susceptible to imipenem, gentamicin, and amikacin.
Screening of plasmid replicons among 46 E. coli isolates using PlasmidFinder database detected a total of 12 plasmid replicons, which included FII, FIA, FIB, FIC, Col, I, X3, X4, Q1, B/O/K/Z, HI, and Y (Figure 1). FII was the predominant replicon identified in 83% (38/46) of isolates followed by FIB, Col, and FIA replicon types, with 80% (37/46), 72% (33/46), and 70% (32/46), respectively. The mcr-1–positive E. coli harbored FII and FIB plasmid replicon types.
Figure 3 shows the distribution of virulence genes among the 46 sequenced E. coli isolates. A total of 32 of the 46 E. coli isolates were classified as ExPEC, which comprised 21/21 ST131 isolates (100%) and 11/26 non-ST131 isolates (42%). The ExPEC isolates identified were enriched with virulence genes such as pap, fim, sat, tia, hlyE, iutA, sitA, and fyuA. Isolates belonging to the ST131 clone showed higher aggregate virulence scores [median (range)], 22 (18–27) compared with non-ST131 isolates 14.5 (6–33). The only isolate that carried the highest virulence genes (33) belonged to ST73.
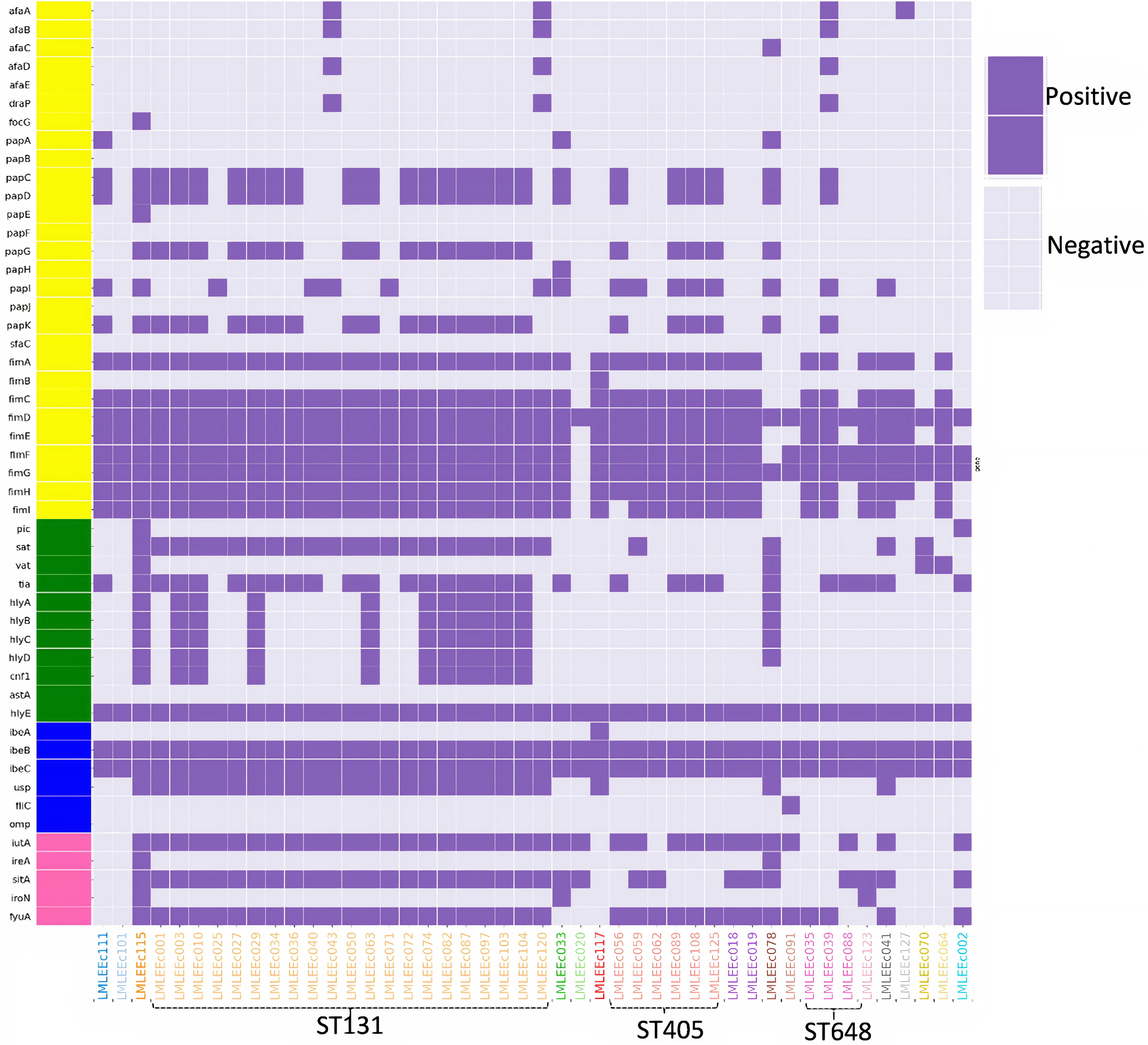
Figure 3. Distribution of virulence gene profiles within 46 E. coli isolates are represented in the heatmap, with colored blocks indicating the gene presence and gray-colored blocks indicating the gene absence. Further, the virulence genes are vertically colored coded by functional categories (yellow, adhesins; blue, toxins; pink, miscellaneous; green, siderophores).
To investigate the relatedness of 46 ESBL E. coli isolates, single nucleotide polymorphism (SNP)-based core genome phylogenetic tree was constructed (Figure 1). The 46 E. coli genomes clustered with a good concordance to their phylogroups. Particularly, strains belonging to phylogroups B2, D, and F formed distinct clades, whereas strains belonging to phylogroups A, B1, and C were clustered together under a large clade; 24 of these 46 E. coli isolates formed a large cluster of B2 phylogroup all qualified as ExPEC and had the most common plasmid combination of FII, FIA, and FIB; this clade mainly comprised (21/24) ST131 isolates. Further, it was identified that 16 of 21 ST131 E. coli belonged to the most virulent, widespread H30Rx subclone, three belonged to H30R, and two belonged to fluoroquinolone-susceptible fimH 41, CTX-M-15–positive ST131 E. coli. The 21 ST131 E. coli isolates showed little or no difference in the SNP analysis; these isolates were collected over 15 months, which indicates widespread dissemination of this particular lineage. The sister clade of ST131 clustered E. coli lineages that included ST1193, ST73, and ST421. When the branches of non-ST131 isolates were assessed closely, high clonality was observed between individual isolates in a few cases. For instance, six strains of ST405 formed an identical cluster; these were collected from November 2018 to July 2018, and five of six isolates carried blaCTX–M–15 and had identical serogroup and fimH types with almost common plasmid profile. However, two of these six isolates carried blaNDM–5 gene in addition to the shared profiles. Similarly, ST410 isolates formed identical clusters with shared molecular profiles. However, ST648 strains formed an identical cluster but demonstrated diverse molecular profiles.
ESBL-producing E. coli is responsible for a significant number of nosocomial and community-acquired infections (Pana and Zaoutis, 2018). In this study, we used a WGS approach to analyze 46 ESBL–E. coli isolates collected over a year (16 months) from a referral diagnostic center in Dhaka, Bangladesh, a Southeast Asian country from which data such as these are acutely lacking.
Our study revealed that the ST131 lineage is by far the most prevalent lineage (45%) represented particularly by H30Rx isolates (34%). This dominance of ST131 E. coli in our study isolates is possibly because of our focus on ESBL–E. coli. However, the non-ST131 (55%) E. coli isolates showed high diversity with respect to phylogroups, phylogenetic clustering, STs, serotypes, and fimH types, which is consistent with previous reports (Hussain et al., 2014; Musicha et al., 2017). Furthermore, we demonstrated that ESBLs, in particular, the CTX-M–type ESBLs, in Bangladesh have emerged across major high-risk international clones that comprise ST38, ST73, ST405, ST410, ST648, and ST1193 lineages, in addition to isolates of the pandemic E. coli ST131.
Molecular typing revealed that a majority of CTX-M–type ESBL producers belonged to the ST131 lineage, which constituted the most clonal group in the population structure of the studied isolates. The predominance of this lineage and its association with CTX-M-15 is in line with the reports from worldwide studies (Nicolas-Chanoine et al., 2014), including those from Asia (Hussain et al., 2012; Ranjan et al., 2015). Further, we documented a greater proportion (76%) of H30Rx subclones within the ST131 E. coli isolates, indicating a scenario of endemic circulation of H30Rx strains in the current setting, as these strains have been recovered between the span of April 2018 to June 2019. Clonal expansion of H30RX subclone might be the reason for the high prevalence and transmission of CTX-M-15–associated ST131 E. coli isolates in this setting. The H30Rx subclone is also reported in the neighboring country, India, in 2015 (Ranjan et al., 2015). The H30Rx isolates detected in this study were all qualified as ExPEC, which harbored a broad range of virulence genes with a relatively high virulence score, median (range): 27 (18–27). These isolates exhibited a high prevalence of a combination of IncFII, IncFIA, and IncFIB plasmid replicons, which is consistent with a recent WGS study from North Carolina (Kanamori et al., 2017). The gene aac(6’)-lb-cr encoding the aminoglycoside and fluoroquinolone-modifying enzyme was strongly associated with H30Rx strains. In addition to the blaCTX–M–15 gene, they exhibited a high prevalence of blaOXA–1 and high resistance to third-generation cephalosporin antibiotics (ceftriaxone and cefixime). The association of H30Rx subclone with extended-spectrum cephalosporin and fluoroquinolone resistance might have contributed to their predominance and epidemiological success over other E. coli lineages.
In this study, we have further shown that the blaCTX–M–15 gene exists in genetically diverse strains including the prominent non-ST131 uropathogenic E. coli lineages such as ST38, ST73, ST405, ST410, ST648, and ST1193. Isolates of ST38 were reported to be evolving and is described to be increasingly detected in UTIs, which was previously considered to be a gut pathogen because they harbor genes for both EAEC and ExPEC (Chattaway et al., 2014). It is noticed that the ST38 isolate identified in this study was moderately lower in virulence and AMR gene content; particularly, it lacked the fluoroquinolone resistance and plasmids. Nonetheless, they were identified as ExPEC. ST73 is another frequently isolated ExPEC from UTI and bloodstream infections (Riley, 2014). The ST73 isolate in our study was found to carry an extensive array of virulence genes, whereas it was susceptible to most of the antibiotics and carried only IncCoI plasmid type, in contrast to other studies, which detected FII, FIA, and FIB plasmids; the reason for this could be that the ST73 strains are not expanding clonally as also suggested by others (Bogema et al., 2020).
ST405 E. coli lineage has been implicated as drivers of blaCTXM–15 and is often associated with blaNDM genes and extensive virulence repertoire similar to the ST131 (Devanga Ragupathi et al., 2020). All six ST405 isolates from our study demonstrated high AMR rates and specifically carried a set of common plasmid replicons, including IncFII, IncFIA, IncFIB, and IncCoI. Five of these six ST405 strains qualified as ExPEC with extensive virulence gene profiles, consistent with other reports (Zhang et al., 2018; Devanga Ragupathi et al., 2020). Two of these six ST405 isolates also harbored blaNDM–5 gene. ST410 is considered another emerging “high-risk” clone that is resistant to fluoroquinolones, cephalosporins, and sometimes carbapenems (Roer et al., 2018). The two ST410 isolates in our collection showed moderate virulence and resistance profiles; they harbored blaCTX–M–15 gene with a set of five plasmid replicon types and fimH 24 allele (Figure 1). The lineage ST648 has been predicted to become another internationally circulating clone that will worsen infection treatment possibilities because of its AMR (Schaufler et al., 2019). The three ST648 isolates detected in this study carried blaCTX–M–15 and demonstrated high AMR rates; all the three strains consistently carried IncCoI plasmids. One ST648 strain was also positive for the blaNDM–5 gene. ST1193 is the latest pandemic multidrug-resistant uropathogenic clonal group (Johnson et al., 2019), which is usually detected among non-lactose fermenters (Johnson et al., 2019). The ST1193 isolate in our collection was qualified as ExPEC; it showed high resistance rates, with particular resistance to fluoroquinolones, cephalosporins, and cotrimoxazole. It harbored IncFIA, IncFIB, and IncCoI plasmid types.
Several WGS studies have described the features of E. coli ST131. In contrast, not many have addressed the molecular epidemiology of non-ST131 E. coli worldwide. In this study, we have described that the ST131 E. coli is currently the most prevalent CTX-M–type ESBL producer in our community. However, we have also drawn attention to the emergence of significant clonal groups among non-ST131 E. coli. Further work is warranted in Bangladesh to accurately estimate the prevalence of these clonal groups and systematically analyze them in a global context, which is pertinent from public health and clinical standpoint.
Taken together, our study suggests that the CTX-M–type ESBLs and particularly the CTX-M-15 are prevalent among diverse E. coli STs circulating in Dhaka, Bangladesh. Further, our findings confirm the striking predominance of ST131 lineage and also revealed the presence of several major high-risk non-ST131 ExPEC clonal lineages, such as ST38, ST73, ST405, ST410, ST648, and ST1193, all associated with cephalosporin resistance and virulence. Further genomic epidemiological studies are needed to keep track of significant virulent/multiresistant E. coli clones in Bangladesh. Such studies are needed to inform us about the evolutionary dynamics of pathogenicity and resistance in emerging E. coli lineages of public health concern which will aid in evidence-based infection control and antibiotic-prescribing policies.
The datasets generated for this study can be found in the GenBank (Bioproject Accession: PRJNA654992). The GenBank accession numbers of 46 genomes sequenced in this study can be found in Table 1.
AH and RM designed the study, performed genome sequencing, and drafted the manuscript. AA, AH, and RM carried out the bioinformatics analyses and interpretation of results. MI, MS, SM, AT, FH, and NA were involved in the culture and AST of bacteria isolates. DA and DM supervised the study. All authors have read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This research study was funded by core donors who provide unrestricted support to icddr,b for its operations and research. Current donors providing unrestricted support include the governments of Bangladesh, Canada, Sweden, and the United Kingdom. We gratefully acknowledge these donors for their support and commitment to icddr,b’s research efforts.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Baquero, F., Tedim, A. P., and Coque, T. M. (2013). Antibiotic resistance shaping multi-level population biology of bacteria. Front. Microbiol. 4:15. doi: 10.3389/fmicb.2013.00015
Beghain, J., Bridier-Nahmias, A., Le Nagard, H., Denamur, E., and Clermont, O. (2018). ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genomics 4:192. doi: 10.1099/mgen.0.000192
Begum, N., and Shamsuzzaman, S. M. (2016). Emergence of CTX-M-15 producing E. coli O25b-ST131 clone in a tertiary care hospital of Bangladesh. Malays. J. Pathol. 38, 241–249.
Bogema, D. R., McKinnon, J., Liu, M., Hitchick, N., Miller, N., Venturini, C., et al. (2020). Whole-genome analysis of extraintestinal Escherichia coli sequence type 73 from a single hospital over a 2 year period identified different circulating clonal groups. Microb. Genom. 6:e000255. doi: 10.1099/mgen.0.000255
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Calbo, E., Romaní, V., Xercavins, M., Gómez, L., Vidal, C. G., Quintana, S., et al. (2006). Risk factors for community-onset urinary tract infections due to Escherichia coli harbouring extended-spectrum β-lactamases. J. Antimicrob. Chemother. 57, 780–783. doi: 10.1093/jac/dkl035
Cantón, R., and Coque, T. M. (2006). The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9, 466–475.
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. doi: 10.1093/nar/gki008
Chen, S. L., Ding, Y., Apisarnthanarak, A., Kalimuddin, S., Archuleta, S., Omar, S. F. S., et al. (2019). The higher prevalence of extended spectrum beta-lactamases among Escherichia coli ST131 in Southeast Asia is driven by expansion of a single, locally prevalent subclone. Sci. Rep. 9:13245. doi: 10.1038/s41598-019-49467-5
Clinical and Laboratory Standards Institute (2015). Performance Standards for Antimicrobial Susceptibility Testing; 20th Informational Supplement M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute.
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43:e15. doi: 10.1093/nar/gku1196
Doi, Y. (2019). Treatment options for carbapenem-resistant gram-negative bacterial infections. Clin. Infect. Dis. 69, S565–S575. doi: 10.1093/cid/ciz830
Franz, E., Veenman, C., van Hoek, A. H. A. M., Husman, A., de, R., Blaak, H., et al. (2015). Pathogenic Escherichia coli producing extended-spectrum β-lactamases isolated from surface water and wastewater. Sci. Rep. 5:14372. doi: 10.1038/srep14372
Hussain, A., Ewers, C., Nandanwar, N., Guenther, S., Jadhav, S., Wieler, L. H., et al. (2012). Multiresistant uropathogenic Escherichia coli from a region in India where urinary tract infections are endemic: genotypic and phenotypic characteristics of sequence type 131 isolates of the CTX-M-15 extended-spectrum-β-lactamase-producing lineage. Antimicrob. Agents Chemother. 56, 6358–6365. doi: 10.1128/AAC.01099-12
Hussain, A., Ranjan, A., Nandanwar, N., Babbar, A., Jadhav, S., and Ahmed, N. (2014). Genotypic and phenotypic profiles of Escherichia coli isolates belonging to clinical sequence type 131 (ST131), clinical non-ST131, and fecal non-ST131 lineages from India. Antimicrob. Agents Chemother. 58, 7240–7249. doi: 10.1128/AAC.03320-14
Joensen, K. G., Tetzschner, A. M. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Johnson, J. R., and Stell, A. L. (2000). Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181, 261–272. doi: 10.1086/315217
Johnson, T. J., Elnekave, E., Miller, E. A., Munoz-Aguayo, J., Figueroa, C. F., Johnston, B., et al. (2019). Phylogenomic analysis of extraintestinal pathogenic Escherichia coli sequence type 1193, an emerging multidrug-resistant clonal group. Antimicrob. Agents Chemother. 63:e01913-18. doi: 10.1128/AAC.01913-18
Kanamori, H., Parobek, C. M., Juliano, J. J., Johnson, J. R., Johnston, B. D., Johnson, T. J., et al. (2017). Genomic analysis of multidrug-resistant Escherichia coli from north carolina community hospitals: ongoing circulation of CTX-M-producing ST131-H30Rx and ST131-H30R1 strains. Antimicrob. Agents Chemother. 61:e00912-17. doi: 10.1128/AAC.00912-17
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. doi: 10.1016/S1473-3099(10)70143-2
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Lina, T. T., Khajanchi, B. K., Azmi, I. J., Islam, M. A., Mahmood, B., Akter, M., et al. (2014). Phenotypic and molecular characterization of extended-spectrum beta-lactamase-producing Escherichia coli in Bangladesh. PLoS One 9:e108735. doi: 10.1371/journal.pone.0108735
Chattaway, M. A., Jenkins, C., Ciesielczuk, H., Day, M., DoNascimento, V., Day, M., et al. (2014). Evidence of evolving extraintestinal enteroaggregative Escherichia Coli ST38 clone. Emerg. Infect. Dis. 20, 1935–1937. doi: 10.3201/EID2011.131845
Manges, A. R., Geum, H. M., Guo, A., Edens, T. J., Fibke, C. D., and Pitout, J. D. D. (2019). Global extraintestinal pathogenic escherichia coli (Expec) lineages. Clin. Microbiol. Rev. 32:e00135-18. doi: 10.1128/CMR.00135-18
Mathers, A. J., Peirano, G., and Pitout, J. D. D. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28, 565–591. doi: 10.1128/CMR.00116-14
Mazumder, R., Abdullah, A., Ahmed, D., and Hussain, A. (2020a). High prevalence of blactx-m-15 gene among extended-spectrum β-lactamase-producing escherichia coli isolates causing extraintestinal infections in bangladesh. Antibiotics 9:796. doi: 10.3390/antibiotics9110796
Mazumder, R., Abdullah, A., Hussain, A., Ahmed, D., and Mondal, D. (2020b). Draft genome sequence of chromobacterium violaceum RDN09, isolated from a patient with a wound infection in bangladesh. Microbiol. Resour. Announc. 9:e00957-20. doi: 10.1128/mra.00957-20
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother 57, 3348–3357. doi: 10.1128/AAC.00419-13
Mikheenko, A., Prjibelski, A., Saveliev, V., Antipov, D., and Gurevich, A. (2018). Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34, i142–i150. doi: 10.1093/bioinformatics/bty266
Musicha, P., Feasey, N. A., Cain, A. K., Kallonen, T., Chaguza, C., Peno, C., et al. (2017). Genomic landscape of extended-spectrum β-lactamase resistance in Escherichia coli from an urban African setting. J. Antimicrob. Chemother. 72, 1602–1609. doi: 10.1093/jac/dkx058
Nicolas-Chanoine, M.-H., Bertrand, X., and Madec, J.-Y. (2014). Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/CMR.00125-13
Nicolas-Chanoine, M. H., Blanco, J., Leflon-Guibout, V., Demarty, R., Alonso, M. P., Caniça, M. M., et al. (2008). Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61, 273–281. doi: 10.1093/jac/dkm464
Ragupathi, N. K. D., Veeraraghavan, B., Sethuvel, D. P. M., Anandan, S., Vasudevan, K., Neerav, A. R., et al. (2020). First Indian report on genome-wide comparison of multidrug-resistant Escherichia Coli from blood stream infections. PLoS One 15:e0220428. doi: 10.1371/JOURNAL.PONE.0220428
Pana, Z. D., and Zaoutis, T. (2018). Treatment of extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBLs) infections: what have we learned until now? F1000 Res. 7:F1000 Faculty Rev-1347. doi: 10.12688/F1000RESEARCH.14822.1
Paterson, D. L., and Bonomo, R. A. (2005). Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686. doi: 10.1128/CMR.18.4.657-686.2005
Pitout, J. D., and Laupland, K. B. (2008). Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8, 159–166. doi: 10.1016/S1473-3099(08)70041-0
Price, L. B., Johnson, J. R., and Aziz, M. (2013). The epidemic of extended-spectrum- β -lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone. H 30-Rx. MBio 4:e00377-13. doi: 10.1128/mBio.00377-13.Editor
Ranjan, A., Shaik, S., Hussain, A., Nandanwar, N., Semmler, T., Jadhav, S., et al. (2015). Genomic and functional portrait of a highly virulent, CTX-M-15-producing H 30-Rx subclone of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 59, 6087–6095. doi: 10.1128/AAC.01447-15
Riley, L. W. (2014). Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 20, 380–390. doi: 10.1111/1469-0691.12646
Roer, L., Overballe-Petersen, S., Hansen, F., Schønning, K., Wang, M., Røder, B. L., et al. (2018). Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere 3:e00337-18. doi: 10.1128/MSPHERE.00337-18
Roer, L., Tchesnokova, V., Allesoe, R., Muradova, M., Chattopadhyay, S., Ahrenfeldt, J., et al. (2017). Development of a web tool for Escherichia coli subtyping based on fimh alleles. J. Clin. Microbiol. 55, 2538–2543. doi: 10.1128/JCM.00737-17
Schaufler, K., Semmler, T., Wieler, L. H., Trott, D. J., Pitout, J., Peirano, G., et al. (2019). Genomic and functional analysis of emerging virulent and multidrug-resistant Escherichia coli lineage sequence type 648. Antimicrob. Agents Chemother. 63:e00243-19. doi: 10.1128/AAC.00243-19
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seemann, T. (2015). Snippy: Rapid Haploid Variant Calling and Core SNP Phylogeny. San Francisco, CA: GitHub.
Shaik, S., Ranjan, A., Tiwari, S. K., Hussain, A., Nandanwar, N., Kumar, N., et al. (2017). Comparative genomic analysis of globally dominant ST131 clone with other epidemiologically successful extraintestinal pathogenic Escherichia coli (ExPEC) lineages. MBio 8:e01596-17. doi: 10.1128/mBio.01596-17
Stamatakis, A. (2014). RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies.
Stoesser, N., Sheppard, A. E., Moore, C. E., Golubchik, T., Parry, C. M., Nget, P., et al. (2015). Extensive within-host diversity in fecally carried extended-spectrum-beta-lactamase-producing Escherichia coli Isolates: Implications for transmission analyses. J. Clin. Microbiol. 53, 2122–2131. doi: 10.1128/JCM.00378-15
Zankari, E., Allesøe, R., Joensen, K. G., Cavaco, L. M., Lund, O., and Aarestrup, F. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768. doi: 10.1093/jac/dkx217
Keywords: CTX-M, NDM, MCR, extraintestinal pathogenic E. coli (ExPEC), carbapenem resistance, whole-genome sequencing, epidemiological successful clones, ST131 and non-ST131 lineages
Citation: Mazumder R, Hussain A, Abdullah A, Islam MN, Sadique MT, Muniruzzaman SM, Tabassum A, Halim F, Akter N, Ahmed D and Mondal D (2021) International High-Risk Clones Among Extended-Spectrum β-Lactamase–Producing Escherichia coli in Dhaka, Bangladesh. Front. Microbiol. 12:736464. doi: 10.3389/fmicb.2021.736464
Received: 05 July 2021; Accepted: 03 September 2021;
Published: 04 October 2021.
Edited by:
Ziad Daoud, Central Michigan University, United StatesCopyright © 2021 Mazumder, Hussain, Abdullah, Islam, Sadique, Muniruzzaman, Tabassum, Halim, Akter, Ahmed and Mondal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dilruba Ahmed, ZGFobWVkQGljZGRyYi5vcmc=; Dinesh Mondal, ZGluNjNkQGljZGRyYi5vcmc=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.