- 1Department of Cardiovascular and Metabolic Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, United States
- 2Cleveland Clinic Lerner College of Medicine, Case Western Reserve University, Cleveland, OH, United States
- 3Center for Microbiome and Human Health, Cleveland Clinic, Cleveland, OH, United States
The profound impact of the gut microbiome on host health has led to a revolution in biomedical research, motivating researchers from disparate fields to define the specific molecular mechanisms that mediate host-beneficial effects. The advent of genomic technologies allied to the use of model microbiomes in gnotobiotic mouse models has transformed our understanding of intestinal microbial ecology and the impact of the microbiome on the host. However, despite incredible advances, our understanding of the host-microbiome dialogue that shapes host physiology is still in its infancy. Progress has been limited by challenges associated with developing model systems that are both tractable enough to provide key mechanistic insights while also reflecting the enormous complexity of the gut ecosystem. Simplified model microbiomes have facilitated detailed interrogation of transcriptional and metabolic functions of the microbiome but do not recapitulate the interactions seen in complex communities. Conversely, intact complex communities from mice or humans provide a more physiologically relevant community type, but can limit our ability to uncover high-resolution insights into microbiome function. Moreover, complex microbiomes from lab-derived mice or humans often do not readily imprint human-like phenotypes. Therefore, improved model microbiomes that are highly defined and tractable, but that more accurately recapitulate human microbiome-induced phenotypic variation are required to improve understanding of fundamental processes governing host-microbiome mutualism. This improved understanding will enhance the translational relevance of studies that address how the microbiome promotes host health and influences disease states. Microbial exposures in wild mice, both symbiotic and infectious in nature, have recently been established to more readily recapitulate human-like phenotypes. The development of synthetic model communities from such “wild mice” therefore represents an attractive strategy to overcome the limitations of current approaches. Advances in microbial culturing approaches that allow for the generation of large and diverse libraries of isolates, coupled to ever more affordable large-scale genomic sequencing, mean that we are now ideally positioned to develop such systems. Furthermore, the development of sophisticated in vitro systems is allowing for detailed insights into host-microbiome interactions to be obtained. Here we discuss the need to leverage such approaches and highlight key challenges that remain to be addressed.
Introduction
As a species, humans are surrounded by and inhabited by trillions of microorganisms, encompassing bacteria, fungi, archaea, other eukaryotic organisms such as parasites and protists, as well as viruses (Gill et al., 2006; Parfrey et al., 2011; Hallen-Adams and Suhr, 2017; Koskinen et al., 2017; Nkamga et al., 2017; Gregory et al., 2020) that are collectively referred to as the microbiome. Decades of work have established the profound role of the microbiome in shaping host physiology and its capacity to regulate a wide variety of health and disease states. The rapid growth of microbiome research, spurred by technological innovations, has resulted in remarkable discoveries that have altered our conceptualization of the role played by this complex ecosystem in host health. Furthermore, these efforts have uncovered several features that highlight the therapeutic potential of the microbiome. First, dysfunction of the microbiome or host responses to the microbiome have been implicated in the pathogenesis of myriad human diseases, including, undernutrition and its associated maladies (Smith M. I. et al., 2013; Kau et al., 2015; Blanton et al., 2016; Charbonneau et al., 2016; Wagner et al., 2016; Cowardin et al., 2019), metabolic diseases such as obesity (Ley et al., 2005; Turnbaugh et al., 2006, 2008, 2009b; Hildebrandt et al., 2009; Ridaura et al., 2013), cardiovascular disease (Kelly et al., 2016), cancer and its susceptibility to treatment (Arthur et al., 2012; Buc et al., 2013; Sivan et al., 2015; Vétizou et al., 2015), food allergy (Feehley et al., 2019), multiple sclerosis (Berer et al., 2017; Cekanaviciute et al., 2017), and inflammatory bowel disease (IBD) (Manichanh et al., 2006; Frank et al., 2007; Gevers et al., 2014; Britton et al., 2019). Second, there is significant inter-personal variation in microbiome composition and/or function across individuals (Turnbaugh et al., 2009a; Qin et al., 2010; Schloissnig et al., 2013) that can impact host phenotypes, and thus, microbiome composition represents a personalized risk factor for the development of disease (Smith M. I. et al., 2013; Subramanian et al., 2014; Alavi et al., 2020). Third, microbiota repair, where specific microbial taxa or microbial consortia are introduced to communities lacking these microbes, has proven effective in restoration of beneficial microbiome-mediated effects (van Nood et al., 2013; Buffie et al., 2015; Blanton et al., 2016; Caballero et al., 2017; Di Luccia et al., 2020), underscoring the potential of microbiome manipulation for therapy.
This has prompted a flurry of exploration from researchers across a wide-array of disciplines to provide a systematic understanding of the microbiome and its interaction with the host, especially in defining the features that shape microbiome composition and function, as well as uncovering how the microbiome imparts its beneficial or deleterious effects on host physiology. Investigation of these processes has typically followed a trajectory beginning with identifying disruptions to microbiome composition, commonly referred to as dysbiosis, followed by in vivo animal studies whereby transplantation of microbiomes from donors exhibiting a phenotype of interest is used to assess how much, if any, of the donor phenotype can be transmitted by the microbiome. These studies are essential to establish a causal role for the microbiome and microbe(s) in question. This process is exemplified by studies of malnutrition (obesity and undernutrition) and IBD. Obesity is associated with an altered gut microbiome composition (Ley et al., 2005; Turnbaugh et al., 2008, 2009b; Hildebrandt et al., 2009), and the gut microbiome from these individuals or obese mice promotes increased adiposity and metabolic dysfunction upon transplantation to germ-free recipient mice relative to healthy donor controls (Turnbaugh et al., 2006; Ridaura et al., 2013). Likewise, individuals suffering from undernutrition have disrupted gut microbiomes, and transplantation of fecal microbiomes from such donors recapitulates features of weight loss/cachexia in recipient mice relative to control donors (Smith M. I. et al., 2013; Kau et al., 2015; Blanton et al., 2016; Wagner et al., 2016). Although there remains some debate about whether or not IBD patients have distinct microbiome compositions, gut microbiomes from IBD patients elicit more severe intestinal inflammation in gnotobiotic IBD models than those from healthy controls (Gevers et al., 2014; Britton et al., 2019; Lloyd-Price et al., 2019). In addition to establishing a causal role for the microbiome in such diseases, model systems have also been leveraged to elucidate how specific microbiome members impact the progression or prevention of diseases. These efforts have yielded detailed insights that would have been likely impossible without a tractable model system. For example, particular strains of E. coli containing the pathogenicity island pks have been identified as being enriched in patients with colorectal cancer, and gnotobiotic mouse models have been utilized to demonstrate causality for these specific strains in the disease (Arthur et al., 2012; Buc et al., 2013). Enterotoxigenic strains of B. fragilis have also been linked to colorectal cancer development (Toprak et al., 2006; Wu et al., 2009), as well as aspects of undernutrition (Wagner et al., 2016). Species of Clostridium and the Bacteroides have also been implicated in limiting the severity of food allergy (Atarashi et al., 2011; Stefka et al., 2014; Abdel-Gadir et al., 2019). Animal model systems have thus proven essential in determining causal roles for the microbiome in shaping disease susceptibility and facilitating the precise delineation of host-microbiome interactions that mediate these effects on host physiology. As such, they remain an irreplaceable component of the microbiome researcher’s toolkit.
While the advances to date have been captivating, there is still a dearth of information regarding the specific gut microbes that mediate the effects of the microbiome on the host. Moreover, despite the enormous progress that they have facilitated, current models have deficiencies that limit their translational relevance. As targeted and rational microbiome manipulation becomes an increasingly attractive approach for therapy, tractable and physiologically relevant model systems to interrogate host-microbiome interactions are needed. Here we will discuss current challenges and describe a path to addressing this need in microbiome research through the creation of new and improved model systems to interrogate host-microbiome interactions. We will focus our attention primarily on the effects of gut bacteria on host physiology. However, it is increasingly clear that intestinal fungi, viruses, archaea, and other eukaryotic species can profoundly impact host phenotypes, such as promoting intestinal immune system maturation and regulating disease susceptibility, often able to imprint phenotypic responses equivalent to gut bacteria (Kernbauer et al., 2014; Chudnovskiy et al., 2016; Escalante et al., 2016; Lin et al., 2020; Yeung et al., 2020; Dallari et al., 2021). Moreover, these agents do not act in isolation, and their direct or indirect interactions may regulate host health as has been demonstrated in murine models of inflammatory bowel disease (IBD) and parasitic infection (Cadwell et al., 2010; Hayes et al., 2010). It would therefore be remiss to ignore the contributions of these oft-overlooked microbiome members in our conceptualization of the gut ecosystem and its effects on the host, as highlighted by others (Norman et al., 2014; Reynolds et al., 2015; Runge and Rosshart, 2021).
Mouse Models for the Study of Host-Microbiome Interactions
Model systems have been widely employed by researchers going all the way back to the days of Gregor Mendel’s use of pea plants to study inherited traits (Ellis et al., 2011). In order to define paradigms of host-microbiome mutualism, researchers have utilized a variety of organisms (Reyniers and Sacksteder, 1958; Smith et al., 2007; Ericsson, 2019), ranging from Drosophila (Lee and Brey, 2013), Hydra (Augustin et al., 2012), zebrafish (Kanther and Rawls, 2010; Stagaman et al., 2020) and squid (Nyholm and McFall-Ngai, 2004; McFall-Ngai, 2014) to mice and rats (Reyniers and Sacksteder, 1958; Smith et al., 2007), and pigs (Vlasova et al., 2018; Gehrig et al., 2019). This has enabled fundamental insights into the relationship between the host and the resident microbiome and the identification of features that typify these interactions, akin to Koch’s postulates that describe the paradigm that defines microbial pathogenesis (Neville et al., 2018). Systems like Drosophila, Hydra, squid, and zebrafish offer numerous advantages including the relative ease of husbandry, the ability to study large numbers of offspring, less complex and more readily cultivated microbiomes for study than higher organisms, the availability of whole-organism imaging, etc. Pioneering studies in these systems have uncovered principles that govern host-microbiome interactions, including (but not limited to): (i) a role for gut symbionts in the coordination of tissue developmental programs and the microbial components responsible for these effects (particularly microbial cell wall products such as peptidoglycan and LPS, as well as microbial metabolites like acetate) (Koropatnick et al., 2004; Buchon et al., 2009; Shin et al., 2011; Troll et al., 2018), (ii) host adaptations to the microbiome that limit the inflammatory potential of microbiota-derived factors (Bates et al., 2007; Lhocine et al., 2008; Rader et al., 2012), (iii) host regulation of microbiome composition (Rawls et al., 2006; Ryu et al., 2008; Fraune et al., 2009), (iv) gut symbiont factors driving host adaptation (Rawls et al., 2007; Koehler et al., 2018), and (v) microbiome contributions to growth and nutrient acquisition (Storelli et al., 2011; Semova et al., 2012; Schwarzer et al., 2016). Gnotobiotic pigs have now become more widely utilized, which has allowed the study of these processes in an animal system with physiology more similar to that of humans than provided by commonly used murine models. While pig models present many challenges due to their size, they offer several advantages over more commonly used model systems, including more human-like physiology, susceptibility to many human-relevant infectious agents, greater microbiome complexity, and therefore they offer important insights of more translational relevance (Vlasova et al., 2018; Ericsson, 2019; Gehrig et al., 2019). Although less commonly employed due to the more challenging and expensive nature of their husbandry, gnotobiotic pig models are proving to be a highly valuable component of microbiome research.
Despite the utility of these other model systems, the mouse has reigned supreme in biomedical research, especially for microbiome studies. The emergence of the mouse as a model organism can be traced to the early 1900s with the house mouse, Mus musculus being used to study Mendelian genetics (Castle and Little, 1910), followed shortly thereafter by the development of the first inbred Mus musculus strain in 1929 by C.C. Little at what is now known as Jackson labs (Phifer-Rixey and Nachman, 2015). Although models in other small animals have been widely used, including rats (Modlinska and Pisula, 2020), hamsters (Miao et al., 2019), and gerbils (Bleich et al., 2010), none are quite as adapted for the breadth of study possible with mice. Mice offer several advantages that include their relatively quick gestation period, their size, which allows for easier housing and manipulation, the plethora of tools for phenotypic assessment of the mouse, the availability of sophisticated tools for genetic modification to interrogate the role played by distinct genes and cell types, and most importantly the availability of approaches to raise mice in germ-free settings. Furthermore, the availability of inbred strains of mice and standardized, albeit imperfect, housing and husbandry that helps to minimize unwanted variation, allows for easier comparison of data from different researchers.
However, mirroring the interpersonal variation in human microbiomes, model organisms display significant variation in the composition of their microbiomes, which in turn contributes to phenotypic variation reported in mouse models, especially in studies associated with immune activation. Several notable examples highlight how microbiome variation can impact phenotypes in murine models: (i) microbiome mediated spontaneous colitis and metabolic dysfunction has been reported in TLR5-/- mice by some, but not by others (Vijay-Kumar et al., 2007, 2010; Letran et al., 2011); (ii) the aggravation of colitis and development of communicable disease in NLRP6-/- mice is critically dependent on the microbiome context in which it is studied (Elinav et al., 2011; Lemire et al., 2017; Mamantopoulos et al., 2017); (iii) IL-10-/- mice develop a spontaneous colitis in some animal facilities, yet they remain largely free of disease in others depending on the presence or absence of select microbes (Kuhn et al., 1993; Nagalingam et al., 2013; Seregin et al., 2017); (iv) DSS-induced colitis models display significantly varied kinetics and severity depending on microbiome composition (Forster et al., 2021); (v) diabetes in the Non-Obese Diabetic (NOD) type 1 diabetes model is regulated by various parameters, including the presence of specific gut microbes or infectious agents (Wilberz et al., 1991; Takei et al., 1992; Pozzilli et al., 1993; Kriegel et al., 2011; Markle et al., 2013; Yurkovetskiy et al., 2013). Although not an exhaustive list, these examples demonstrate that variation in the gut microbiome can lead to disparate phenotypic outcomes in mice. Experimental variation, whether it be technical or biological in origin, has long been seen as a thorn in the side of researchers, contributing in part to issues of reproducibility in science (von Kortzfleisch et al., 2020). To limit the impact of overt pathogens, and the large-scale microbiome variation across different animal facilities, efforts were made to create a more standardized murine system that would limit issues of reproducibility. Thus, the concept of Specific Pathogen Free (SPF) mice was born. SPF mice are free of certain (but not all) pathogenic organisms or microorganisms capable of interfering with experimental outcomes (Lane-Petter, 1962). These mice provide the advantage of controlling the health status of the animal and allowing for better standardization between experiments, labs, and institutions. While the SPF mouse was adopted with the intention of allowing for more reproducible results, it has been shown that microbiome and phenotypic variability also exists between SPF mouse colonies from commercial vendors, as SPF only determines what is excluded, but not what should be present (Smith et al., 2007; Ivanov et al., 2009; Denning et al., 2011; Rosshart et al., 2017).
Box 1. Common terminology used to describe the colonization status/microbiome communities commonly found in murine model systems.
Germ-free (GF)-Mice that are raised devoid of all known microbes.
Gnotobiotic-Term used to denote GF mice that are now colonized with a defined community of microbes where all members are known such as the Altered Schaedler Flora and the use of synthetic bacterial communities.
Specific Pathogen Free (SPF)-Conventional mice that are devoid of particular known pathogens such as bacterial, viral, fungal, and parasitic inhabitants that could affect the health of the mouse colony and the validity of experimental outcomes.
Conventional mice-Laboratory mice that are raised in the presence of a gut microbiome, and are not necessarily considered free of pathogens (they may have inhabitants such as murine norovirus and Helicobacter hepaticus) but are generally considered healthy.
Wild mice-These are mice that have been captured in a non-lab environment (in the wild) and then transferred to a lab for study. Such mice can be mated with other wild-caught mice for study over several generations. Inbred lines of mice harboring wild-mouse microbiomes, often referred to as “WildR mice” (Rosshart et al., 2017), can be generated through microbial transfer from wild mice to inbred strains of lab mice to limit the effects of genetic variation.
Synthetic communities-Communities constructed from cultured isolates from naturally occurring complex microbiomes. These communities may represent isolates from a single donor, or isolates obtained from many different donors. Moreover, they may be constructed from subsets of all cultured isolates.
Model Gut Microbiomes
To counteract the effects of microbiome variation researchers have turned to the use of defined reference communities. Beginning in the 1960s, Dubos et al. (1965), Schaedler et al. (1965), and Dewhirst et al. (1999) developed small model communities that were used to standardize the microbiome of animal models, mostly to be used in conventionally raised mice (Box 1). In an effort to move toward systems with greater control over community composition an ever-increasing number of researchers have begun to adopt germ-free/gnotobiotic models first established more than 60 years ago (Trexler and Reynolds, 1957; Gordon and Pesti, 1971). Such models allow the study of communities of interest without unwanted invasion by microbes present in the environment. Pioneering studies using small communities, ranging from mono-associations (colonization with a single microbe) that establish the roles of individual genes and metabolic pathways in bacteria (Rey et al., 2010; Koppel et al., 2018; Glowacki et al., 2020) to more complex communities with up to 20 members (Mahowald et al., 2009; Faith et al., 2011, 2014; Geuking et al., 2011; Cahenzli et al., 2013; McNulty et al., 2013; Sefik et al., 2015; Brugiroux et al., 2016; Geva-Zatorsky et al., 2017; Becattini et al., 2021), reduced the complexity of the system (Figure 1). These simplified models have provided high-resolution insights into the ecological, transcriptional, and metabolic responses of microbes to environmental variations (e.g., diet, inflammation) (McNulty et al., 2013; Ridaura et al., 2013; Becattini et al., 2021), uncovered microbe-microbe interactions that shape community function (Mahowald et al., 2009; Caballero et al., 2017), and highlighted a role for microbiome members in host growth (Blanton et al., 2016; Charbonneau et al., 2016; Schwarzer et al., 2016), weight gain and metabolic health (Fei and Zhao, 2013; Ridaura et al., 2013), pathogen resistance (Fukuda et al., 2011; Hsiao et al., 2014; Alavi et al., 2020), as well as specific enzymatic functions that directly impact host health (Skye et al., 2018; Song et al., 2020). Thus, model communities studied in germ-free/gnotobiotic mice have provided key insights into the several facets of microbiome function and host-microbiome interactions.
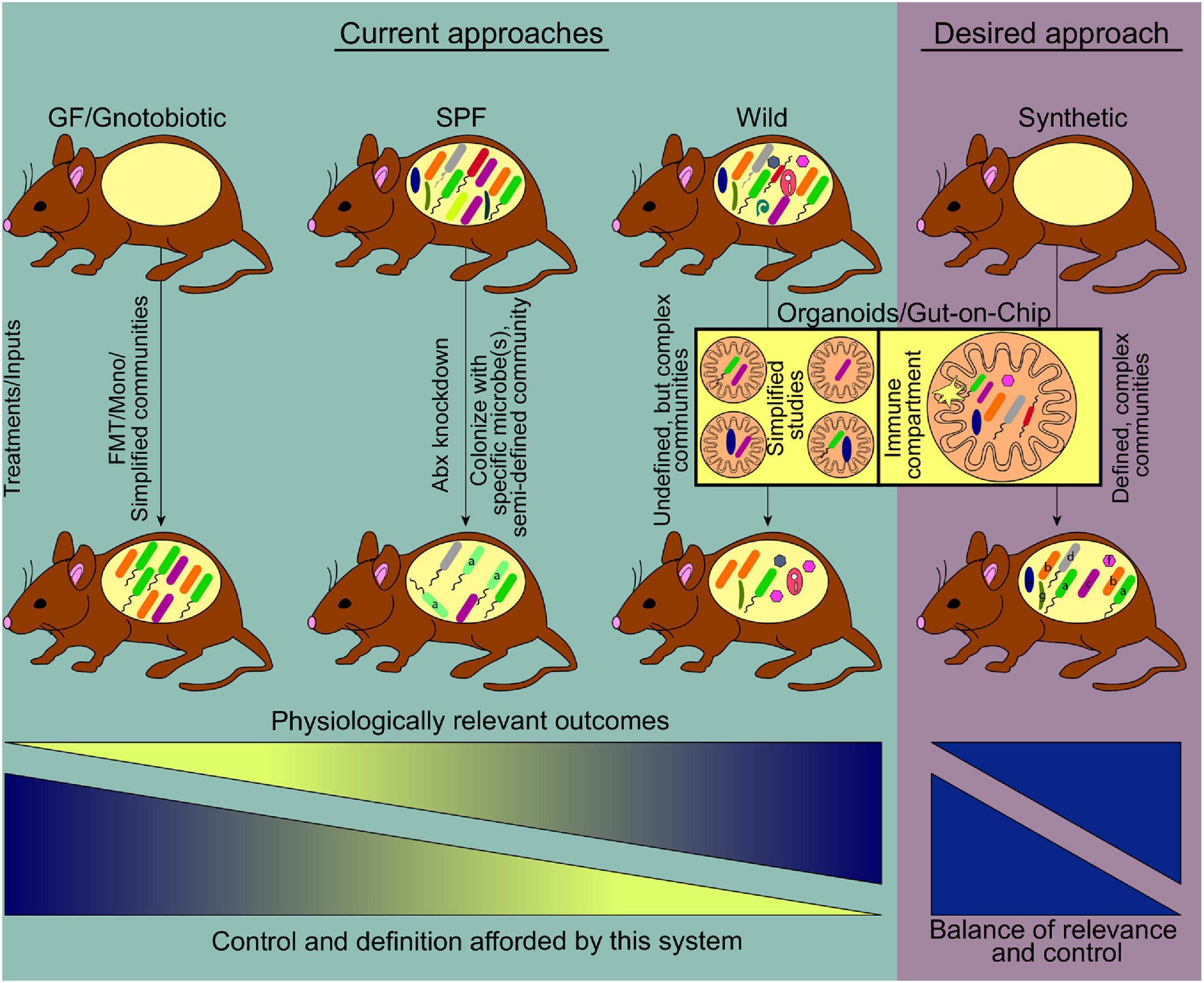
Figure 1. Current and emerging systems to study host-gut microbiome interactions. Prevalent model systems in use to study physiological effects of gut microbiome-host interactions and representative community complexity within each model are shown (teal shaded area). The general usefulness of these systems based on physiologically relevant outcomes, increases left to right. At the same time, the ability to manipulate or define the community structure of these models decreases along the gradient, driving the need for a medium where physiologic relevance and tractability reach an optimum. Current technologies striving toward this desired model include the use of germ free mice where synthetic communities of known microorganisms are used for colonization, as well as the use of emerging lab-on-chip approaches (pink shaded area) (FMT, fecal microbiome transplant; Mono, monocolonization; Abx, antibiotics).
The power of the germ-free mouse as a model system may be best exemplified by the advances they have facilitated in understanding immune-microbiome mutualism (reviewed extensively in Honda and Littman, 2016; Ost and Round, 2018; McCoy et al., 2019; Ahern and Maloy, 2020; Ansaldo et al., 2021). These studies have revealed the profound impact of the gut microbiome on immune function and provided detailed insights into the mechanisms that underlie these interactions. The absence of a microbiome leads to the development of a drastically altered intestinal immune system with striking defects in adaptive immune function including reductions in lymphocyte numbers and activation within the intestine and mesenteric lymph nodes, and an enhanced susceptibility to infection by certain pathogens (Abrams and Bishop, 1966; Thompson and Trexler, 1971; Imaoka et al., 1996; Macpherson and Harris, 2004). Notably, the microbiome has also been implicated in the shaping of the innate immune compartment, including aspects of trained immunity (reviewed in McCoy et al., 2019; Negi et al., 2019). Intensive efforts have subsequently identified specific microbial taxa/microbial consortia, and their derived molecules that influence the intestinal immune system of gnotobiotic mice, especially the intestinal T cell compartment. Colonizing germ-free mice with microbial consortia like the Altered Schaedler flora (ASF) and select Clostridium species, or mono-colonization with specific members of the Bacteroides genus (Bacteroides thetaiotaomicron, Bacteroides caccae, and Bacteroides fragilis) or Bifidobacteria (B. bifidum), coordinates the development of colonic regulatory T cells, an anti-inflammatory population of CD4+ T cells that maintain intestinal homeostasis (Round and Mazmanian, 2010; Atarashi et al., 2011, 2013; Geuking et al., 2011; Faith et al., 2014; Sefik et al., 2015; Verma et al., 2018; Wegorzewska et al., 2019). Furthermore, cellular products derived from these microbes, like polysaccharide A (PSA) (Round and Mazmanian, 2010) or β-glucan/galactan polysaccharides (Verma et al., 2018), short-chain fatty acids (SCFA; gut microbial fermentation products) (Arpaia et al., 2013; Furusawa et al., 2013; Smith P. M. et al., 2013) or microbially-transformed bile acids (Song et al., 2020) also regulate the size and function of colonic regulatory T cell pools. Likewise, Th17 cell differentiation can be coordinated by distinct microbes, most notably segmented filamentous bacteria (SFB) (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009) in addition to Bifidobacterium adolescentis (Tan et al., 2016) or particular strains of E. coli (Britton et al., 2020), which in turn can improve protective immunity against pathogens (Ivanov et al., 2009). Intraepithelial T cell populations are also impacted by gut microbes, with SFB able to shape the activation status of this immune compartment (Umesaki et al., 1999), while Lactobacillus reuteri promotes the development of CD4+ CD8αα + intraepithelial lymphocytes (IELs) by agonizing the aryl hydrocarbon pathway (Cervantes-Barragan et al., 2017).
Despite the enormous power of these systems, they have several limitations. First, the physiologic relevance of model communities can be hard to decipher. Second, due to their low diversity, they may fail to identify redundancy in effector functions that exist in larger communities, inappropriately attributing essential roles to particular microbes. Third, most groups study a limited number of strains of each species, which ignores the enormous strain-level variation present in the gut microbiome. To overcome these limitations, models where mice are colonized with human gut microbiomes from healthy or diseased individuals (“humanized” mice) that could transmit features of their donor’s health status (Raibaud et al., 1980; Turnbaugh et al., 2009b; Ridaura et al., 2013; Smith M. I. et al., 2013; Kau et al., 2015; Berer et al., 2017; Cekanaviciute et al., 2017; Britton et al., 2019; Feehley et al., 2019) have been widely adopted. The microbiomes of such humanized mice are typically more diverse than model communities and comprising distinct strains depending on the individual donor. Moreover, they represent a system with clear translational relevance, albeit with less defined community membership. For example, microbiome transplantation from individuals suffering from IBD (Britton et al., 2019), food allergy (Feehley et al., 2019), undernutrition (Smith M. I. et al., 2013; Kau et al., 2015; Wagner et al., 2016), obesity (Ridaura et al., 2013), and multiple sclerosis (Berer et al., 2017; Cekanaviciute et al., 2017), among other conditions, could enhance the susceptibility to such diseases in recipient gnotobiotic mice. However, despite all their advantages, significant caveats to the use of human-derived microbiomes exist that suggest a need for new and improved approaches. First, these communities are typically neither defined nor cultured (with exceptions; Wagner et al., 2016; Britton et al., 2019, 2020), limiting the establishment of causality for specific microbes and/or microbial products. Second, while it is clear that microbes derived from humans can modulate particular facets of the murine immune system, host-specificity in such interactions (Atarashi et al., 2015) means that human-derived microbiota may not shape immune responses equivalent to murine-derived microbes (Gaboriau-Routhiau et al., 2009; Chung et al., 2012; Lundberg et al., 2020). Third, human-derived microbiomes are not as well adapted to the murine intestine as mouse-derived communities. Elegant studies using gnotobiotic mice colonized with either human or murine-derived microbiomes demonstrated that the murine microbiome exhibited superior fitness in the mouse intestine, and could displace many members of a human microbiome from a stably colonized mouse upon co-housing (Seedorf et al., 2014). Finally, the overall community composition and structure in the recipient mouse may vary significantly from the donor, both in terms of membership and microbial abundance, potentially over or understating the contributions of particular microbes (Turnbaugh et al., 2009b; Chung et al., 2012; Krych et al., 2013; Xiao et al., 2015; Lundberg et al., 2020). While there is obvious value to these approaches, and a wealth of information has been obtained from their use, it is important to consider that key elements of host-microbiome interactions may be missed by studying microbes outside of their natural environment (i.e., human microbes in the mouse).
While no single system is perfectly suited to address all goals, an optimized model to study host-microbiome interactions should encompass as many of the following features as possible: (i) Completely defined microbiome with high-quality reference genomes for each organism. Such systems allow high-resolution strain quantification and gene expression profiling with strain-level gene expression assessment; (ii) culturable and genetically manipulable strains. If all strains have been captured in culture in a clonally arrayed format, it allows for the construction of consortia of defined membership to determine how specific members impact the phenotype being studied (Goodman et al., 2011; Ahern et al., 2014; Faith et al., 2014), commonly referred to as synthetic communities. Moreover, the availability of tools for genetic modification allows for unambiguous assessment of the role for genes of interest, which has contributed to the significant insights afforded by members of the Bacteroides, for which sophisticated tools are available (Anderson and Salyers, 1989; Koropatkin et al., 2008; Goodman et al., 2009; Mimee et al., 2015; Wu et al., 2015; Lim et al., 2017); (iii) a genetically tractable host that allows for the interrogation of host pathways that mediate the effects of specific microbes/microbial products; (iv) a germ-free host that allows for high-level control over the composition of the community. The utility of germ-free mice for advancing microbiome studies is hard to overstate, and coupled with the ability to generate host mutants to dissect pathways of host-microbiome interactions is invaluable; and (v) microbiome whose members and imprinted host responses mirror the human population being modeled. Such a system would allow a high-resolution examination of complex communities that imprint human-like phenotypic variation, overcoming the shortcomings of the systems currently in vogue. An idealized system will capture all the advantages of the systems described above and is represented in Figure 1. While a model that captures all these parameters remains aspirational, advances in microbial culturing and isolation, allied to genomic approaches that continue to decrease in cost while increasing in output mean that large libraries of cultured isolates can now be generated and characterized in wild-type and genetically modified gnotobiotic mice.
One of the primary challenges has been the identification of murine microbiomes that promote human-like phenotypes. Recent ground-breaking studies from a small number of labs have revealed that the microbial inhabitants of mice in the wild (or in pet stores) promote the acquisition of a human-like immune system in lab mice (Figure 1), recapitulating the activated and antigen-experienced phenotype found in humans (Beura et al., 2016; Rosshart et al., 2017, 2019; Lin et al., 2020; Yeung et al., 2020) by contrast with the typical immune system of lab mice that has a phenotype more akin to that of neonates (Beura et al., 2016). Wild mouse microbiomes lead to a profound reshaping of the host immune system, promoting a more human-like phenotype, most potently with respect to boosting T cells with effector/memory phenotypes. These alterations in immune phenotype are characterized by an expansion in systemic and tissue-resident memory CD8+ T cell populations, increases in effector CD4+ T cells (Th1, Th2, Th17, and Tregs), and innate immune populations such as innate-lymphoid cells and neutrophils (Beura et al., 2016; Lin et al., 2020; Yeung et al., 2020). Moreover, the levels of serum immunoglobulins and select cytokines are also increased (Beura et al., 2016). Consistent with this, these animals were found to be more resistant to viral infections (Influenza A), bacterial infections (Listeria monocytogenes), and colon cancer (DSS plus azoxymethane model) (Beura et al., 2016; Rosshart et al., 2017). Conversely, they are more susceptible to surgery-associated sepsis, likely due to increased inflammatory tone and/or increased reactivity to microbial products (Huggins et al., 2019). Thus, wild microbiome-driven enhancement of host immunity is associated with improved immune-mediated resistance to a variety of infectious diseases and cancer, and enhanced susceptibility to other inflammatory diseases, linking microbiome-mediated immunomodulation to organismal health.
Notably, the transcriptional responses that distinguish neonates and adults are reminiscent of those that differentiate SPF mice and those harboring a wild/pet store microbiome. These features are communicable to lab mice following co-housing, suggesting they are microbially-driven (Beura et al., 2016; Rosshart et al., 2017), although whether this is attributable to pathobionts, or the distinct strain composition of these communities is unknown. Moreover, the genomic diversity of wild mice, and its impact on host immune responses and disease susceptibility, may additionally contribute to distinct phenotypic features of wild mice, representing an area ripe for further exploration. Some of these features are likely attributable to infectious agents like pathogenic viruses (Reese et al., 2016), but notably, may also be due to specific endogenous bacterial or fungal members of the gut microbiome (Lin et al., 2020; Yeung et al., 2020). Indeed, wild mice that retained SPF status (i.e., free of all pathogens excluded under SPF guidelines) also induced distinct phenotypic variation relative to lab-raised SPF counterparts, and this could be transmitted to lab mice through gut microbiome transplantation (Rosshart et al., 2017). These data suggest that differences in the specific strain composition of the gut microbiome of wild mice, rather than pathogen exposure, are responsible. More recently, a study revealed that wild mice more faithfully recapitulate the outcome of clinical trials targeting the immune system (Rosshart et al., 2019), by contrast with their conventional lab animals, reinforcing the notion that such models are of greater translational relevance. Thus, wild mice microbiomes provide an opportunity to improve the utility of mouse model systems.
However, while we and others (Hamilton et al., 2020; Graham, 2021; Kuypers et al., 2021) posit that wild microbiome-elicited phenotypes create a murine system with a more human-like phenotype, different approaches to wilding the microbiome including the specific donor material used, or the creation of environments that more accurately mimic the natural environment of the mouse (Arnesen et al., 2020; Lin et al., 2020; Yeung et al., 2020) vs. co-housing under controlled laboratory conditions (Beura et al., 2016; Rosshart et al., 2017), may lead to disparate phenotypic outcomes in recipients. The use of wild microbiomes thus may not fully humanize the murine response or be fully representative of human phenomena. For example, in the case of allergy development and the hygiene hypothesis, one study (Ottman et al., 2019) suggests that lack of microbe diversity drives such allergic states, while (Ma et al., 2021) contradicts these claims. Undoubtedly, the approach of using wild mice and/or their derived microbiomes does not fully address the nuances of in vivo mouse models vs. humans as discussed in depth elsewhere (Hamilton et al., 2020; Graham, 2021; Kuypers et al., 2021). To address these limitations, more studies are needed with comparative phenotyping of adult human and wild microbiome-exposed murine immune systems to determine whether these responses truly reflect the development of a more human-like response, as opposed to a response that is simply distinct from lab mice. With the advent of approaches that allow for detailed assessment of the non-lymphoid immune compartment in humans (Szabo et al., 2019), such studies are now possible. Moreover, as we discuss below, detailed knowledge of the microorganisms that coordinate the phenotypic features shared between mice harboring wild microbiomes and humans will advance efforts to generate improved murine model systems.
Synthetic Wild Communities, an Optimized System to Study Immune-Microbiome Interactions
While wild microbiomes offer the potential to shed new light on host-microbiome interactions, currently there is limited information regarding the effector microbes within these communities. There is therefore a need to determine which microbes are responsible for mediating the human-like phenotypic variation that they induce. These efforts need not focus solely on non-pathogenic members of the microbiome but should include controlled pathogen as well as non-pathogen exposures. While defining these effector microbes is a daunting challenge, we and others have described effective strategies to do so in a systematic and efficient manner (Goodman et al., 2011; Ahern et al., 2014; Faith et al., 2014; Palm et al., 2014; Kau et al., 2015; Surana and Kasper, 2017). The generation of culture libraries in arrayed format (i.e., where individual wells of multi-well plates contain distinct microbiome members) from human donor microbiomes that retain the effector functions of the donor community has allowed mechanistic insight into host-microbiome interactions of biological relevance (Ridaura et al., 2013; Faith et al., 2014; Wagner et al., 2016). Such strategies allow the precise delineation of the effects of individual community members, whether they operate in concert with other members or in isolation (Ahern et al., 2014; Faith et al., 2014). Similarly, the isolation in pure culture of the constituents of the wild mouse microbiome represents a key first step toward the generation of more complete synthetic communities that recapitulate wild microbiome imprinted functions and allow for greater manipulability. Likewise, in defining these wild microbiomes, it is important to appreciate that for the past several decades, non-bacterial residents of the gut (fungi, archaea, viruses, parasites, and other non-fungal eukaryotic members) have received scant attention, mostly attributable to technical challenges. Such impediments include the variability of internal transcribed spacer regions (ITS) within fungal ribosomal genes and a general lack of reference genomes to compare species prevalence in metagenomic samples (Paterson et al., 2017). Sequencing challenges have also dampened the ability to get a fully representative picture of the gut virome. Most techniques for nucleic acid isolation and sequencing are biased toward DNA viruses, largely missing RNA viruses, while isolation and propagation of gut viruses is also a challenge that needs to be overcome (Wang, 2020; Khan Mirzaei et al., 2021). This lack of cultivation methods extends to archaeal members as well, with archaea often requiring specific culture conditions (Borrel et al., 2020) which can hamper in vivo studies. The genesis of libraries of isolates of all microbiome-member types in arrayed format from wild-microbiomes that are known to impact host phenotypes, such that consortia of individual library members can be compiled to study their effects on the host will form an essential component moving forward in defining complex host-microbiome interactions (Goodman et al., 2011; Ahern et al., 2014; Faith et al., 2014; Palm et al., 2014). Moreover, this will facilitate the dissemination to other researchers for implementation in their studies. The use of a common library of microorganisms freely available to all researchers that can be leveraged to understand host-microbiome interactions at multiple scales will advance efforts to uncover mechanistic insights into the operations of large diverse communities that reflect the breadth of microbial taxa and viruses that characterize humans and their associated phenotypic effects.
Despite our call for more standardized gut microbiomes, it would be foolish to demand complete homogenization across institutions, or even within an institution. Microbiome variation can itself represent a form of “natural experiment” that can present a challenge to researchers, but that has also proved a rich source of information regarding how the microbiome can mediate interpersonal variation among a population. For example, the varied presence of Th17 cells in the small intestinal lamina propria in C57BL/6 mice from different commercial vendors led to the identification of a single microbe, SFB, which was differentially represented in the microbiomes of these animals and was both required and sufficient for the development of intestinal Th17 cells (Ivanov et al., 2008, 2009; Gaboriau-Routhiau et al., 2009). Similarly, the varying presence of Lactobacillus reuteri in different animal facilities within the same institution led to its identification as a potent modulator of CD4+ CD8αα+ IEL development (Cervantes-Barragan et al., 2017). More recently, the fungus Debaryomyces hansenii was highlighted as a mediator of impaired intestinal healing, which again was differentially represented among different colonies of lab mice at the same institution and directly regulated the intestinal healing potential of mice (Jain et al., 2021). Others have also linked different microbiome composition to the phenotype of various animal models of infectious and autoimmune disease (Wu et al., 2010; Lee et al., 2014; Hilbert et al., 2017; Moskowitz et al., 2019; Velazquez et al., 2019). What these studies highlight is that phenotypic variance can be leveraged, even embraced (Ivanov et al., 2008, 2009; Cervantes-Barragan et al., 2017; Jain et al., 2021), to uncover novel host-microbiome interactions that shape host responses. Consequently, although variation poses challenges for microbiome research, total standardization is itself not without issue. Instead, the utility of such variance is linked to an ability to measure and define the causes of the variation, and the reporting of the microbiome composition that is associated with a phenotype will be of enormous value in linking specific microbes to phenotypes of interest. Such an approach will reveal contextualized roles for host phenotypes that may manifest only in the presence of particular community types. Thus, there remains a prominent place for non-standardized models in illuminating fundamentally important host-microbiome interactions.
Alternatives to in vivo Mouse Models
In spite of all the advantages of the in vivo mouse models we describe, it is ultimately a system with limitations that demands alternative approaches that augment our understanding. Fundamental differences between mice and humans mean that key aspects of host-microbiome interactions may not be modeled in a murine system. Indeed, the specificity in molecular aspects of host-microbe interactions (Lecuit et al., 1999; Atarashi et al., 2015) demands systems to study human-derived microbes in the context of human cells. The advent of sophisticated in vitro/ex vivo approaches that use human-derived cells that can themselves be genetically manipulated represent attractive alternatives that can be used in parallel to murine models. In addition to overcoming shortfalls in murine systems, these approaches help with the continued efforts to replace, reduce, and refine animals in research.
Organoids
Human intestinal organoids (HIOs) or enteroids (HIEs) remove the need for a live model organism, and instead rely on primary cells derived from human biopsies or stem cells. This technique was originally pioneered from the use of ex vivo tissue explants of human intestines (Browning and Trier, 1969). Growth factors are used to drive differentiation of Lgr5+ intestinal stem cells into intestinal cell types that mimic the 3D spatial and functional environment of the intestine, allowing for simultaneous differentiation into discrete cell types (Ootani et al., 2009; Sato et al., 2009). HIOs have some advantages over other in vitro systems as they maintain the crypt-villus architecture and allow for multiple columnar cell types to be generated (Hill and Spence, 2017). HIOs permit the study of phenomena that have proved challenging in other in vivo systems, including tight junctions of non-enterocyte cells of the small intestine (Pearce et al., 2018); IBD models of infection and inflammatory processes (Angus et al., 2019; Sarvestani et al., 2021); and models of infection such as human norovirus (Ettayebi et al., 2016) and rotavirus (Finkbeiner et al., 2012), for which no in vivo model organism exists, and in vitro culture efforts had not been successful at the time. Additionally, microinjection of bacterial and parasitic pathogens including C. difficile (Leslie et al., 2015), S. enterica Typhimurium (Forbester et al., 2015; Wilson et al., 2015), E. coli (In et al., 2016; Karve et al., 2017; Rajan et al., 2018), and Cryptosporidium (Heo et al., 2018) have all been performed. This platform has allowed for controlled studies into the interactions these pathogens have with the intestine, however, there are several key challenges that remain. Although bacteria can be injected into these structures, the process has drawbacks such that the specialized technique of microinjection is required to avoid compromising the organoid structure and the lumen within the organoid contains a growth-limiting concentration of nutrients that generally can only support the growth of bacteria for less than 24 h. This nutrient limitation also significantly hinders the diversity of microorganisms that can be cultured together in poly-microbial communities. Other constraints involve the physical structure of organoids, and the lack of immune cells, calling into question how well the system recapitulates in vivo biology with the absence of such features (Blutt et al., 2018). Despite the advantages that organoids provide, the noted limitations suggest that the organoid model is not yet advanced to the point of being able to replicate all aspects found in vivo. Instead, these systems are likely more useful as tools to study parameters such as the permeability of the mucosa, drug kinetics, and bacterial interactions in disease states using tissue derived from patients with IBD or related conditions.
Gut-on-Chip Technologies
A relatively new approach to studying microbe-microbe and intestinal cell-microbe interactions are “lab-on-chip” technologies. Although systems such as Transwell plates (two-dimensional technology) and Ussing chambers (three-dimensional) have been used for decades (Ussing and Zerahn, 1951; Hidalgo et al., 1989) and have been used to study bacterial-host epithelium interactions and diseases of the intestine (Kurkchubasche et al., 1998; Thomson et al., 2019), both have known limitations. With respect to the microbiome, these technologies are not well-suited for maintenance of both aerobic and anaerobic compartments except in limited circumstances (Ulluwishewa et al., 2015; Jafari et al., 2016). This makes studying gut microbiome-host interactions with obligate anaerobes a challenge. Additional limitations involve the duration in which bacteria can be co-cultured before either they or the epithelial cells die, with most ranging from hours to a few days due to the non-peristaltic nature of these devices (Sadaghian Sadabad et al., 2015). Although other devices, such as the mucosal simulator of the human intestinal microbial ecosystem (M-SHIME), or its derivative, the Host-Microbiota Interaction (HMI)-module, have shown promise in incorporating peristalsis-like flow; they suffer from similar problems of short co-culture incubation times and rely on artificial mucus layers (Van den Abbeele et al., 2012; Marzorati et al., 2014). The SHIME reactors are also large, expensive to produce, and not easily scalable.
Using technology pioneered by lithography of computer chip manufacturing (Bhatia and Ingber, 2014), microfluidic devices may be a happy medium that yields more information on bacterial-gut epithelial-immune system interactions. These devices allow for a 3-D spatial reconstruction of the in vivo environment. Specifically for bacteria, these platforms have been used to study bacterial quorum sensing (Osmekhina et al., 2018), the response to antibiotics, and other chemicals in a complex community (Hsu et al., 2019), and taxis and motility (Gurung et al., 2020). Adoption of these devices has led to the generation of new platforms that are being used to study the human gut ecosystem and the human gut microbiota (von Martels et al., 2017; Tan and Toh, 2020). These devices overcome a substantial amount of the need for using in vivo models and tissue explants to maintain an environment necessary to study long-duration, complex, multi-species, and multiple cell type interactions. Although there are at least 12 different microfluidic devices in use, the vast majority rely on the colorectal carcinoma cell line, Caco-2 cells, to establish an epithelium, and have only managed to culture one species of bacteria at a time; several of these devices have previously been reviewed for benefits and drawbacks (Bein et al., 2018; Tan and Toh, 2020). Recent advancements include the nBioChip which supports the co-culture of both bacteria (Staphylococcus aureus and Pseudomonas aeruginosa) together with the fungus, Candida albicans (Srinivasan et al., 2017). Perhaps the largest advancement is the Intestine Chip, with the ability to maintain over 200 operational taxonomic units (OTUs) of bacteria directly from human feces with both obligate anaerobes and aerobic bacteria established along a hypoxia gradient (Jalili-Firoozinezhad et al., 2019). The Intestine Chip can also support stable colonization periods of up to or beyond 1 week due to its peristalsis-like flow of media. However, like the platform of the two previous iterations of this specific device (Kim et al., 2012, 2016), Caco-2 cells are used to develop the epithelial compartment rather than primary cells, which limits some downstream applications due to these cells not being representative of the primary cells of the intestinal tract.
Recent advancements have merged organoid and gut-on-chip technologies. First described as an early version of the Intestine Chip, the use of matured organoids containing villus structures and multiple cell types were enzymatically broken down and used to seed extracellular matrix (ECM)-coated membranes of a microfluidic chip (Kasendra et al., 2018). The other half of the chip was then seeded with human intestinal microvascular endothelial cells to examine cell-cell interactions, thus creating a multi-system organ on a chip. Through RNA-sequencing, confocal microscopy, and tissue staining it was shown that the Intestine Chip recapitulates key features of the signaling pathways, cellular differentiation, mucus production, and epithelial-endothelial interactions seen in the human duodenum. A similar platform termed the gut microbiome physiome (GuMI) has been developed, specifically for the culture of extremely oxygen-sensitive microbes such as Faecalibacterium prausnitzii (Zhang et al., 2021). Similar to the Intestine Chip, the gut microbiome (GuMI) ECM is impregnated with cells derived from organoids and the device has inlets for sampling and injection of bacteria. While many other chip-based devices are fabricated using polydimethylsiloxane (PMDS), the GuMI uses polysulfone, which can be autoclaved for sterility and is less permeable to oxygen, allowing for more strict control of oxygen gradients (Shin et al., 2019). Lastly, this chip allows for the independent culture of six different bacteria within the luminal portion. Using organoid-derived cells, this three-layer-chip contains an ECM seeded with both intestinal cells and monocyte-derived macrophages and recapitulates features observed in IBD patients (Beaurivage et al., 2020). While the presence of immune cells is an important advancement, the generation of systems that can maintain interaction with a complex immune system is essential to boost the translational relevance of these systems. Nevertheless, the combination of gut-on-chip and organoids is a promising step forward toward the goal of having a tunable system to interrogate complex interactions that are difficult to perform in vivo (Figure 1).
Discussion/Prospectus
A wealth of knowledge about gut microbiome-host interactions has been gained through the use of the model systems discussed in this review. Conventionally raised mice (both SPF and non-SPF), and gnotobiotic mice, have been and continue to be essential tools to study interactions between the gut microbiome in host health and disease. While there is a continued use for these models, their limitations hinder efforts to gain the mechanistic insights required to target the microbiome for therapeutic purposes. Development of synthetic, wild mouse gut microbiome communities comprising cultured and genome-sequenced microbiome members derived from wild mice provides an opportunity to gain mechanistic understanding of specific microbe-host phenotypes that recapitulate the interactions of humans with their microbiomes and the associated microbiome imprinted phenotypes. Used in conjunction with ever-improving in vitro/ex vivo model systems that facilitate high-resolution studies of complex host-microbiome interactions, these technologies will advance our understanding of the range of microbiome members that shape host physiology and help define the nature of the interactions that underlie these phenomena.
Conclusion
In conclusion, model systems to study gut microbiome-host interactions continue to evolve. The incorporation of synthetic, wild microbiomes into the suite of model systems provides an opportunity to increase mechanistic insight and translatability. The use of these advanced mouse models and ever-improving alternative model systems to study gut microbiome-host interactions will increase our understanding of the functionality of specific microbes in human physiology and disease, advancing efforts to target the microbiome for therapeutic purposes.
Author Contributions
RWPG, MJE, and PPA performed relevant literature searches, critical appraisals of the literature, and wrote the review. All authors contributed to the article and approved the submitted version.
Funding
The authors are supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (R01DK126772).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdel-Gadir, A., Stephen-Victor, E., Gerber, G. K., Noval Rivas, M., Wang, S., Harb, H., et al. (2019). Microbiota therapy acts via a regulatory T cell MyD88/RORgammat pathway to suppress food allergy. Nat. Med. 25, 1164–1174. doi: 10.1038/s41591-019-0461-z
Abrams, G. D., and Bishop, J. E. (1966). Effect of the normal microbial flora on the resistance of the small intestine to infection. J. Bacteriol. 92, 1604–1608. doi: 10.1128/jb.92.6.1604-1608.1966
Ahern, P. P., Faith, J. J., and Gordon, J. I. (2014). Mining the human gut microbiota for effector strains that shape the immune system. Immunity 40, 815–823. doi: 10.1016/j.immuni.2014.05.012
Ahern, P. P., and Maloy, K. J. (2020). Understanding immune-microbiota interactions in the intestine. Immunology 159, 4–14. doi: 10.1111/imm.13150
Alavi, S., Mitchell, J. D., Cho, J. Y., Liu, R., Macbeth, J. C., and Hsiao, A. (2020). Interpersonal Gut Microbiome Variation Drives Susceptibility and Resistance to Cholera Infection. Cell 181, 1533–1546. doi: 10.1016/j.cell.2020.05.036
Anderson, K. L., and Salyers, A. A. (1989). Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 171, 3199–3204. doi: 10.1128/jb.171.6.3199-3204.1989
Angus, H. C. K., Butt, A. G., Schultz, M., and Kemp, R. A. (2019). Intestinal Organoids as a Tool for Inflammatory Bowel Disease Research. Front. Med. 6:334. doi: 10.3389/fmed.2019.00334
Ansaldo, E., Farley, T. K., and Belkaid, Y. (2021). Control of Immunity by the Microbiota. Annu. Rev. Immunol. 39, 449–479. doi: 10.1146/annurev-immunol-093019-112348
Arnesen, H., Knutsen, L. E., Hognestad, B. W., Johansen, G. M., Bemark, M., Pabst, O., et al. (2020). A Model System for Feralizing Laboratory Mice in Large Farmyard-Like Pens. Front. Microbiol. 11:615661. doi: 10.3389/fmicb.2020.615661
Arpaia, N., Campbell, C., Fan, X., Dikiy, S., van der Veeken, J., deRoos, P., et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. doi: 10.1038/nature12726
Arthur, J. C., Perez-Chanona, E., Mühlbauer, M., Tomkovich, S., Uronis, J. M., Fan, T. J., et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123. doi: 10.1126/science.1224820
Atarashi, K., Tanoue, T., Ando, M., Kamada, N., Nagano, Y., Narushima, S., et al. (2015). Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380. doi: 10.1016/j.cell.2015.08.058
Atarashi, K., Tanoue, T., Oshima, K., Suda, W., Nagano, Y., Nishikawa, H., et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236. doi: 10.1038/nature12331
Atarashi, K., Tanoue, T., Shima, T., Imaoka, A., Kuwahara, T., Momose, Y., et al. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341. doi: 10.1126/science.1198469
Augustin, R., Fraune, S., Franzenburg, S., and Bosch, T. C. (2012). Where simplicity meets complexity: hydra, a model for host-microbe interactions. Adv. Exp. Med. Biol. 710, 71–81. doi: 10.1007/978-1-4419-5638-5_8
Bates, J. M., Akerlund, J., Mittge, E., and Guillemin, K. (2007). Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe 2, 371–382. doi: 10.1016/j.chom.2007.10.010
Beaurivage, C., Kanapeckaite, A., Loomans, C., Erdmann, K. S., Stallen, J., and Janssen, R. A. J. (2020). Development of a human primary gut-on-a-chip to model inflammatory processes. Sci. Rep. 10:21475. doi: 10.1038/s41598-020-78359-2
Becattini, S., Sorbara, M. T., Kim, S. G., Littmann, E. L., Dong, Q., Walsh, G., et al. (2021). Rapid transcriptional and metabolic adaptation of intestinal microbes to host immune activation. Cell Host Microbe 29, 378–393e375. doi: 10.1016/j.chom.2021.01.003
Bein, A., Shin, W., Jalili-Firoozinezhad, S., Park, M. H., Sontheimer-Phelps, A., Tovaglieri, A., et al. (2018). Microfluidic Organ-on-a-Chip Models of Human Intestine. Cell Mol. Gastroenterol. Hepatol. 5, 659–668. doi: 10.1016/j.jcmgh.2017.12.010
Berer, K., Gerdes, L. A., Cekanaviciute, E., Jia, X., Xiao, L., Xia, Z., et al. (2017). Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. U S A 114, 10719–10724. doi: 10.1073/pnas.1711233114
Beura, L. K., Hamilton, S. E., Bi, K., Schenkel, J. M., Odumade, O. A., Casey, K. A., et al. (2016). Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516. doi: 10.1038/nature17655
Bhatia, S. N., and Ingber, D. E. (2014). Microfluidic organs-on-chips. Nat. Biotechnol. 32, 760–772. doi: 10.1038/nbt.2989
Blanton, L. V., Charbonneau, M. R., Salih, T., Barratt, M. J., Venkatesh, S., Ilkaveya, O., et al. (2016). Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351:6275. doi: 10.1126/science.aad3311
Bleich, E. M., Martin, M., Bleich, A., and Klos, A. (2010). The Mongolian gerbil as a model for inflammatory bowel disease. Int. J. Exp. Pathol. 91, 281–287. doi: 10.1111/j.1365-2613.2009.00701.x
Blutt, S. E., Crawford, S. E., Ramani, S., Zou, W. Y., and Estes, M. K. (2018). Engineered Human Gastrointestinal Cultures to Study the Microbiome and Infectious Diseases. Cell Mol. Gastroenterol. Hepatol. 5, 241–251. doi: 10.1016/j.jcmgh.2017.12.001
Borrel, G., Brugère, J. F., Gribaldo, S., Schmitz, R. A., and Moissl-Eichinger, C. (2020). The host-associated archaeome. Nat. Rev. Microbiol. 18, 622–636. doi: 10.1038/s41579-020-0407-y
Britton, G. J., Contijoch, E. J., Mogno, I., Vennaro, O. H., Llewellyn, S. R., Ng, R., et al. (2019). Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORgammat(+) Regulatory T Cells and Exacerbate Colitis in Mice. Immunity 50, 212–224e214. doi: 10.1016/j.immuni.2018.12.015
Britton, G. J., Contijoch, E. J., Spindler, M. P., Aggarwala, V., Dogan, B., Bongers, G., et al. (2020). Defined microbiota transplant restores Th17/RORgammat(+) regulatory T cell balance in mice colonized with inflammatory bowel disease microbiotas. Proc. Natl. Acad. Sci. U S A 117, 21536–21545. doi: 10.1073/pnas.1922189117
Browning, T. H., and Trier, J. S. (1969). Organ culture of mucosal biopsies of human small intestine. J. Clin. Invest. 48, 1423–1432. doi: 10.1172/JCI106108
Brugiroux, S., Beutler, M., Pfann, C., Garzetti, D., Ruscheweyh, H. J., Ring, D., et al. (2016). Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol. 2:16215. doi: 10.1038/nmicrobiol.2016.215
Buc, E., Dubois, D., Sauvanet, P., Raisch, J., Delmas, J., Darfeuille-Michaud, A., et al. (2013). High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 8:e56964. doi: 10.1371/journal.pone.0056964
Buchon, N., Broderick, N. A., Chakrabarti, S., and Lemaitre, B. (2009). Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 23, 2333–2344. doi: 10.1101/gad.1827009
Buffie, C. G., Bucci, V., Stein, R. R., McKenney, P. T., Ling, L., Gobourne, A., et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. doi: 10.1038/nature13828
Caballero, S., Kim, S., Carter, R. A., Leiner, I. M., Susac, B., Miller, L., et al. (2017). Cooperating Commensals Restore Colonization Resistance to Vancomycin-Resistant Enterococcus faecium. Cell Host Microbe 21, 592–602e594. doi: 10.1016/j.chom.2017.04.002
Cadwell, K., Patel, K. K., Maloney, N. S., Liu, T. C., Ng, A. C., Storer, C. E., et al. (2010). Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145. doi: 10.1016/j.cell.2010.05.009
Cahenzli, J., Koller, Y., Wyss, M., Geuking, M. B., and McCoy, K. D. (2013). Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe 14, 559–570. doi: 10.1016/j.chom.2013.10.004
Castle, W. E., and Little, C. C. (1910). On a Modified Mendelian Ratio among Yellow Mice. Science 32, 868–870. doi: 10.1126/science.32.833.868
Cekanaviciute, E., Yoo, B. B., Runia, T. F., Debelius, J. W., Singh, S., Nelson, C. A., et al. (2017). Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. U S A 114, 10713–10718. doi: 10.1073/pnas.1711235114
Cervantes-Barragan, L., Chai, J. N., Tianero, M. D., Di Luccia, B., Ahern, P. P., Merriman, J., et al. (2017). Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science 357, 806–810. doi: 10.1126/science.aah5825
Charbonneau, M. R., O’Donnell, D., Blanton, L. V., Totten, S. M., Davis, J. C., Barratt, M. J., et al. (2016). Sialylated Milk Oligosaccharides Promote Microbiota-Dependent Growth in Models of Infant Undernutrition. Cell 164, 859–871. doi: 10.1016/j.cell.2016.01.024
Chudnovskiy, A., Mortha, A., Kana, V., Kennard, A., Ramirez, J. D., Rahman, A., et al. (2016). Host-Protozoan Interactions Protect from Mucosal Infections through Activation of the Inflammasome. Cell 167, 444–456e414. doi: 10.1016/j.cell.2016.08.076
Chung, H., Pamp, S. J., Hill, J. A., Surana, N. K., Edelman, S. M., Troy, E. B., et al. (2012). Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149, 1578–1593. doi: 10.1016/j.cell.2012.04.037
Cowardin, C. A., Ahern, P. P., Kung, V. L., Hibberd, M. C., Cheng, J., Guruge, J. L., et al. (2019). Mechanisms by which sialylated milk oligosaccharides impact bone biology in a gnotobiotic mouse model of infant undernutrition. Proc. Natl. Acad. Sci. U S A 116, 11988–11996. doi: 10.1073/pnas.1821770116
Dallari, S., Heaney, T., Rosas-Villegas, A., Neil, J. A., Wong, S. Y., Brown, J. J., et al. (2021). Enteric viruses evoke broad host immune responses resembling those elicited by the bacterial microbiome. Cell Host Microbe 29, 1014–1029e1018. doi: 10.1016/j.chom.2021.03.015
Denning, T. L., Norris, B. A., Medina-Contreras, O., Manicassamy, S., Geem, D., Madan, R., et al. (2011). Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J. Immunol. 187, 733–747. doi: 10.4049/jimmunol.1002701
Dewhirst, F. E., Chien, C. C., Paster, B. J., Ericson, R. L., Orcutt, R. P., Schauer, D. B., et al. (1999). Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl. Environ. Microbiol. 65, 3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999
Di Luccia, B., Ahern, P. P., Griffin, N. W., Cheng, J., Guruge, J. L., Byrne, A. E., et al. (2020). Combined Prebiotic and Microbial Intervention Improves Oral Cholera Vaccination Responses in a Mouse Model of Childhood Undernutrition. Cell Host Microbe 27, 899–908e895. doi: 10.1016/j.chom.2020.04.008
Dubos, R., Schaedler, R. W., Costello, R., and Hoet, P. (1965). Indigenous, normal, and autochthonous flora of the gastrointestinal tract. J. Exp. Med. 122, 67–76. doi: 10.1084/jem.122.1.67
Elinav, E., Strowig, T., Kau, A. L., Henao-Mejia, J., Thaiss, C. A., Booth, C. J., et al. (2011). NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757. doi: 10.1016/j.cell.2011.04.022
Ellis, T. H., Hofer, J. M., Timmerman-Vaughan, G. M., Coyne, C. J., and Hellens, R. P. (2011). Mendel, 150 years on. Trends Plant Sci. 16, 590–596. doi: 10.1016/j.tplants.2011.06.006
Ericsson, A. C. (2019). The use of non-rodent model species in microbiota studies. Lab. Anim. 53, 259–270. doi: 10.1177/0023677219834593
Escalante, N. K., Lemire, P., Cruz Tleugabulova, M., Prescott, D., Mortha, A., Streutker, C. J., et al. (2016). The common mouse protozoa Tritrichomonas muris alters mucosal T cell homeostasis and colitis susceptibility. J. Exp. Med. 213, 2841–2850. doi: 10.1084/jem.20161776
Ettayebi, K., Crawford, S. E., Murakami, K., Broughman, J. R., Karandikar, U., Tenge, V. R., et al. (2016). Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393. doi: 10.1126/science.aaf5211
Faith, J. J., Ahern, P. P., Ridaura, V. K., Cheng, J., and Gordon, J. I. (2014). Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci. Transl. Med. 6:220ra211. doi: 10.1126/scitranslmed.3008051
Faith, J. J., McNulty, N. P., Rey, F. E., and Gordon, J. I. (2011). Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science 333, 101–104. doi: 10.1126/science.1206025
Feehley, T., Plunkett, C. H., Bao, R., Choi Hong, S. M., Culleen, E., Belda-Ferre, P., et al. (2019). Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 25, 448–453. doi: 10.1038/s41591-018-0324-z
Fei, N., and Zhao, L. (2013). An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 7, 880–884. doi: 10.1038/ismej.2012.153
Finkbeiner, S. R., Zeng, X. L., Utama, B., Atmar, R. L., Shroyer, N. F., and Estes, M. K. (2012). Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio 3, e159–e112. doi: 10.1128/mBio.00159-12
Forbester, J. L., Goulding, D., Vallier, L., Hannan, N., Hale, C., Pickard, D., et al. (2015). Interaction of Salmonella enterica Serovar Typhimurium with Intestinal Organoids Derived from Human Induced Pluripotent Stem Cells. Infect. Immun. 83, 2926–2934. doi: 10.1128/IAI.00161-15
Forster, S. C. S., Beresford-Jones, B. S., Harcourt, K., Notley, G., Stares, M., Kumar, N., et al. (2021). Novel gut pathobionts confound results in a widely used mouse model of human inflammatory disease. bioRxiv 2021:430393. doi: 10.1101/2021.02.09.430393
Frank, D. N., St Amand, A. L., Feldman, R. A., Boedeker, E. C., Harpaz, N., and Pace, N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U S A 104, 13780–13785. doi: 10.1073/pnas.0706625104
Fraune, S., Abe, Y., and Bosch, T. C. (2009). Disturbing epithelial homeostasis in the metazoan Hydra leads to drastic changes in associated microbiota. Environ. Microbiol. 11, 2361–2369. doi: 10.1111/j.1462-2920.2009.01963.x
Fukuda, S., Toh, H., Hase, K., Oshima, K., Nakanishi, Y., Yoshimura, K., et al. (2011). Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547. doi: 10.1038/nature09646
Furusawa, Y., Obata, Y., Fukuda, S., Endo, T. A., Nakato, G., Takahashi, D., et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. doi: 10.1038/nature12721
Gaboriau-Routhiau, V., Rakotobe, S., Lecuyer, E., Mulder, I., Lan, A., Bridonneau, C., et al. (2009). The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689. doi: 10.1016/j.immuni.2009.08.020
Gehrig, J. L., Venkatesh, S., Chang, H. W., Hibberd, M. C., Kung, V. L., Cheng, J., et al. (2019). Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science 365. doi: 10.1126/science.aau4732
Geuking, M. B., Cahenzli, J., Lawson, M. A., Ng, D. C., Slack, E., Hapfelmeier, S., et al. (2011). Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34, 794–806. doi: 10.1016/j.immuni.2011.03.021
Geva-Zatorsky, N., Sefik, E., Kua, L., Pasman, L., Tan, T. G., Ortiz-Lopez, A., et al. (2017). Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 168, 928–943e911. doi: 10.1016/j.cell.2017.01.022
Gevers, D., Kugathasan, S., Denson, L. A., Vazquez-Baeza, Y., Van Treuren, W., Ren, B., et al. (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392. doi: 10.1016/j.chom.2014.02.005
Gill, S. R., Pop, M., Deboy, R. T., Eckburg, P. B., Turnbaugh, P. J., Samuel, B. S., et al. (2006). Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359. doi: 10.1126/science.1124234
Glowacki, R. W. P., Pudlo, N. A., Tuncil, Y., Luis, A. S., Sajjakulnukit, P., Terekhov, A. I., et al. (2020). A Ribose-Scavenging System Confers Colonization Fitness on the Human Gut Symbiont Bacteroides thetaiotaomicron in a Diet-Specific Manner. Cell Host Microbe 27, 79–92e79. doi: 10.1016/j.chom.2019.11.009
Goodman, A. L., Kallstrom, G., Faith, J. J., Reyes, A., Moore, A., Dantas, G., et al. (2011). Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. U S A 108, 6252–6257. doi: 10.1073/pnas.1102938108
Goodman, A. L., McNulty, N. P., Zhao, Y., Leip, D., Mitra, R. D., Lozupone, C. A., et al. (2009). Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6, 279–289. doi: 10.1016/j.chom.2009.08.003
Gordon, H. A., and Pesti, L. (1971). The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol. Rev. 35, 390–429. doi: 10.1128/br.35.4.390-429.1971
Graham, A. L. (2021). Naturalizing mouse models for immunology. Nat. Immunol. 22, 111–117. doi: 10.1038/s41590-020-00857-2
Gregory, A. C., Zablocki, O., Zayed, A. A., Howell, A., Bolduc, B., and Sullivan, M. B. (2020). The Gut Virome Database Reveals Age-Dependent Patterns of Virome Diversity in the Human Gut. Cell Host Microbe 28, 724–740e728. doi: 10.1016/j.chom.2020.08.003
Gurung, J. P., Gel, M., and Baker, M. A. B. (2020). Microfluidic techniques for separation of bacterial cells via taxis. Microb Cell 7, 66–79. doi: 10.15698/mic2020.03.710
Hallen-Adams, H. E., and Suhr, M. J. (2017). Fungi in the healthy human gastrointestinal tract. Virulence 8, 352–358. doi: 10.1080/21505594.2016.1247140
Hamilton, S. E., Badovinac, V. P., Beura, L. K., Pierson, M., Jameson, S. C., Masopust, D., et al. (2020). New Insights into the Immune System Using Dirty Mice. J. Immunol. 205, 3–11. doi: 10.4049/jimmunol.2000171
Hayes, K. S., Bancroft, A. J., Goldrick, M., Portsmouth, C., Roberts, I. S., and Grencis, R. K. (2010). Exploitation of the intestinal microflora by the parasitic nematode Trichuris muris. Science 328, 1391–1394. doi: 10.1126/science.1187703
Heo, I., Dutta, D., Schaefer, D. A., Iakobachvili, N., Artegiani, B., Sachs, N., et al. (2018). Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 3, 814–823. doi: 10.1038/s41564-018-0177-8
Hidalgo, I. J., Raub, T. J., and Borchardt, R. T. (1989). Characterization of the Human Colon Carcinoma Cell Line (Caco-2) as a Model System for Intestinal Epithelial Permeability. Gastroentrerology 96, 736–748.
Hilbert, T., Steinhagen, F., Senzig, S., Cramer, N., Bekeredjian-Ding, I., Parcina, M., et al. (2017). Vendor effects on murine gut microbiota influence experimental abdominal sepsis. J. Surg. Res. 211, 126–136. doi: 10.1016/j.jss.2016.12.008
Hildebrandt, M. A., Hoffmann, C., Sherrill-Mix, S. A., Keilbaugh, S. A., Hamady, M., Chen, Y. Y., et al. (2009). High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 137, .e1711–.e1712. doi: 10.1053/j.gastro.2009.08.042
Hill, D. R., and Spence, J. R. (2017). Gastrointestinal Organoids: Understanding the Molecular Basis of the Host-Microbe Interface. Cell Mol. Gastroenterol. Hepatol. 3, 138–149. doi: 10.1016/j.jcmgh.2016.11.007
Honda, K., and Littman, D. R. (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84. doi: 10.1038/nature18848
Hsiao, A., Ahmed, A. M., Subramanian, S., Griffin, N. W., Drewry, L. L., and Petri, W. A. Jr. (2014). Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515, 423–426. doi: 10.1038/nature13738
Hsu, R. H., Clark, R. L., Tan, J. W., Ahn, J. C., Gupta, S., Romero, P. A., et al. (2019). Microbial Interaction Network Inference in Microfluidic Droplets. Cell Syst. 9, 229–242e224. doi: 10.1016/j.cels.2019.06.008
Huggins, M. A., Sjaastad, F. V., Pierson, M., Kucaba, T. A., Swanson, W., Staley, C., et al. (2019). Microbial Exposure Enhances Immunity to Pathogens Recognized by TLR2 but Increases Susceptibility to Cytokine Storm through TLR4 Sensitization. Cell Rep. 28, 1729–1743e1725. doi: 10.1016/j.celrep.2019.07.028
Imaoka, A., Matsumoto, S., Setoyama, H., Okada, Y., and Umesaki, Y. (1996). Proliferative recruitment of intestinal intraepithelial lymphocytes after microbial colonization of germ-free mice. Eur. J. Immunol. 26, 945–948. doi: 10.1002/eji.1830260434
In, J., Foulke-Abel, J., Zachos, N. C., Hansen, A. M., Kaper, J. B., Bernstein, H. D., et al. (2016). Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell Mol. Gastroenterol. Hepatol. 2, 48–62e43. doi: 10.1016/j.jcmgh.2015.10.001
Ivanov, I. I., Atarashi, K., Manel, N., Brodie, E. L., Shima, T., Karaoz, U., et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. doi: 10.1016/j.cell.2009.09.033
Ivanov, I. I., Frutos Rde, L., Manel, N., Yoshinaga, K., Rifkin, D. B., Sartor, R. B., et al. (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349. doi: 10.1016/j.chom.2008.09.009
Jafari, N. V., Kuehne, S. A., Minton, N. P., Allan, E., and Bajaj-Elliott, M. (2016). Clostridium difficile-mediated effects on human intestinal epithelia: Modelling host-pathogen interactions in a vertical diffusion chamber. Anaerobe 37, 96–102. doi: 10.1016/j.anaerobe.2015.12.007
Jain, U., Ver Heul, A. M., Xiong, S., Gregory, M. H., Demers, E. G., Kern, J. T., et al. (2021). Debaryomyces is enriched in Crohn’s disease intestinal tissue and impairs healing in mice. Science 371, 1154–1159. doi: 10.1126/science.abd0919
Jalili-Firoozinezhad, S., Gazzaniga, F. S., Calamari, E. L., Camacho, D. M., Fadel, C. W., Bein, A., et al. (2019). A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 3, 520–531. doi: 10.1038/s41551-019-0397-0
Kanther, M., and Rawls, J. F. (2010). Host-microbe interactions in the developing zebrafish. Curr. Opin. Immunol. 22, 10–19. doi: 10.1016/j.coi.2010.01.006
Karve, S. S., Pradhan, S., Ward, D. V., and Weiss, A. A. (2017). Intestinal organoids model human responses to infection by commensal and Shiga toxin producing Escherichia coli. PLoS One 12:e0178966. doi: 10.1371/journal.pone.0178966
Kasendra, M., Tovaglieri, A., Sontheimer-Phelps, A., Jalili-Firoozinezhad, S., Bein, A., Chalkiadaki, A., et al. (2018). Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 8:2871. doi: 10.1038/s41598-018-21201-7
Kau, A. L., Planer, J. D., Liu, J., Rao, S., Yatsunenko, T., Trehan, I., et al. (2015). Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci. Transl. Med. 7:276ra224. doi: 10.1126/scitranslmed.aaa4877
Kelly, T. N., Bazzano, L. A., Ajami, N. J., He, H., Zhao, J., Petrosino, J. F., et al. (2016). Gut Microbiome Associates With Lifetime Cardiovascular Disease Risk Profile Among Bogalusa Heart Study Participants. Circ. Res. 119, 956–964. doi: 10.1161/CIRCRESAHA.116.309219
Kernbauer, E., Ding, Y., and Cadwell, K. (2014). An enteric virus can replace the beneficial function of commensal bacteria. Nature 516, 94–98. doi: 10.1038/nature13960
Khan Mirzaei, M., Xue, J., Costa, R., Ru, J., Schulz, S., Taranu, Z. E., et al. (2021). Challenges of Studying the Human Virome - Relevant Emerging Technologies. Trends Microbiol. 29, 171–181. doi: 10.1016/j.tim.2020.05.021
Kim, H. J., Huh, D., Hamilton, G., and Ingber, D. E. (2012). Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab. Chip 12, 2165–2174. doi: 10.1039/c2lc40074j
Kim, H. J., Li, H., Collins, J. J., and Ingber, D. E. (2016). Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. U S A 113, E7–E15. doi: 10.1073/pnas.1522193112
Koehler, S., Gaedeke, R., Thompson, C., Bongrand, C., Visick, K. L., Ruby, E., et al. (2018). The model squid-vibrio symbiosis provides a window into the impact of strain- and species-level differences during the initial stages of symbiont engagement. Environ. Microbiol. 2018:14392. doi: 10.1111/1462-2920.14392
Koppel, N., Bisanz, J. E., Pandelia, M. E., Turnbaugh, P. J., and Balskus, E. P. (2018). Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins. Elife 7:33953. doi: 10.7554/eLife.33953
Koropatkin, N. M., Martens, E. C., Gordon, J. I., and Smith, T. J. (2008). Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16, 1105–1115. doi: 10.1016/j.str.2008.03.017
Koropatnick, T. A., Engle, J. T., Apicella, M. A., Stabb, E. V., Goldman, W. E., and McFall-Ngai, M. J. (2004). Microbial factor-mediated development in a host-bacterial mutualism. Science 306, 1186–1188. doi: 10.1126/science.1102218
Koskinen, K., Pausan, M. R., Perras, A. K., Beck, M., Bang, C., Mora, M., et al. (2017). First Insights into the Diverse Human Archaeome: Specific Detection of Archaea in the Gastrointestinal Tract, Lung, and Nose and on Skin. mBio 8:6. doi: 10.1128/mBio.00824-17
Kriegel, M. A., Sefik, E., Hill, J. A., Wu, H. J., Benoist, C., and Mathis, D. (2011). Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. U S A 108, 11548–11553. doi: 10.1073/pnas.1108924108
Krych, L., Hansen, C. H., Hansen, A. K., van den Berg, F. W., and Nielsen, D. S. (2013). Quantitatively different, yet qualitatively alike: a meta-analysis of the mouse core gut microbiome with a view towards the human gut microbiome. PLoS One 8:e62578. doi: 10.1371/journal.pone.0062578
Kuhn, R., Lohler, J., Rennick, D., Rajewsky, K., and Muller, W. (1993). Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274. doi: 10.1016/0092-8674(93)80068-p
Kurkchubasche, A. G., Cardona, M., Watkins, S. C., Smith, S. D., Albanese, C. T., Simmons, R. L., et al. (1998). Transmucosal passage of bacteria across rat intestinal epithelium in the Ussing chamber: effect of nutritional factors and bacterial virulence. Shock 9, 121–127. doi: 10.1097/00024382-199802000-00008
Kuypers, M., Despot, T., and Mallevaey, T. (2021). Dirty mice join the immunologist’s toolkit. Microbes Infect. 23:104817. doi: 10.1016/j.micinf.2021.104817
Lane-Petter, W. (1962). The Provision and Use of Pathogen-free Laboratory Animals. Proc. R. Soc. Med. 55, 253–263.
Lecuit, M., Dramsi, S., Gottardi, C., Fedor-Chaiken, M., Gumbiner, B., and Cossart, P. (1999). A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18, 3956–3963. doi: 10.1093/emboj/18.14.3956
Lee, H., Jin, B. E., Jang, E., Lee, A. R., Han, D. S., Kim, H. Y., et al. (2014). Gut-residing Microbes Alter the Host Susceptibility to Autoantibody-mediated Arthritis. Immune Netw. 14, 38–44. doi: 10.4110/in.2014.14.1.38
Lee, W. J., and Brey, P. T. (2013). How microbiomes influence metazoan development: insights from history and Drosophila modeling of gut-microbe interactions. Annu Rev. Cell Dev. Biol. 29, 571–592. doi: 10.1146/annurev-cellbio-101512-122333
Lemire, P., Robertson, S. J., Maughan, H., Tattoli, I., Streutker, C. J., Platnich, J. M., et al. (2017). The NLR Protein NLRP6 Does Not Impact Gut Microbiota Composition. Cell Rep. 21, 3653–3661. doi: 10.1016/j.celrep.2017.12.026
Leslie, J. L., Huang, S., Opp, J. S., Nagy, M. S., Kobayashi, M., Young, V. B., et al. (2015). Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun. 83, 138–145. doi: 10.1128/IAI.02561-14
Letran, S. E., Lee, S. J., Atif, S. M., Flores-Langarica, A., Uematsu, S., Akira, S., et al. (2011). TLR5-deficient mice lack basal inflammatory and metabolic defects but exhibit impaired CD4 T cell responses to a flagellated pathogen. J. Immunol. 186, 5406–5412. doi: 10.4049/jimmunol.1003576
Ley, R. E., Bäckhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D., and Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U S A 102, 11070–11075. doi: 10.1073/pnas.0504978102
Lhocine, N., Ribeiro, P. S., Buchon, N., Wepf, A., Wilson, R., Tenev, T., et al. (2008). PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe 4, 147–158. doi: 10.1016/j.chom.2008.07.004
Lim, B., Zimmermann, M., Barry, N. A., and Goodman, A. L. (2017). Engineered Regulatory Systems Modulate Gene Expression of Human Commensals in the Gut. Cell 169, 547–558e515. doi: 10.1016/j.cell.2017.03.045
Lin, J. D., Devlin, J. C., Yeung, F., McCauley, C., Leung, J. M., Chen, Y. H., et al. (2020). Rewilding Nod2 and Atg16l1 Mutant Mice Uncovers Genetic and Environmental Contributions to Microbial Responses and Immune Cell Composition. Cell Host Microbe 27, 830–840e834. doi: 10.1016/j.chom.2020.03.001
Lloyd-Price, J., Arze, C., Ananthakrishnan, A. N., Schirmer, M., Avila-Pacheco, J., Poon, T. W., et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662. doi: 10.1038/s41586-019-1237-9
Lundberg, R., Toft, M. F., Metzdorff, S. B., Hansen, C. H. F., Licht, T. R., Bahl, M. I., et al. (2020). Human microbiota-transplanted C57BL/6 mice and offspring display reduced establishment of key bacteria and reduced immune stimulation compared to mouse microbiota-transplantation. Sci. Rep. 10:7805. doi: 10.1038/s41598-020-64703-z
Ma, J., Classon, C. H., Stark, J. M., Li, M., Huang, H. J., Vrtala, S., et al. (2021). Laboratory mice with a wild microbiota generate strong allergic immune responses. bioRxiv doi: 10.1101/2021.03.28.437143
Macpherson, A. J., and Harris, N. L. (2004). Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4, 478–485. doi: 10.1038/nri1373
Mahowald, M. A., Rey, F. E., Seedorf, H., Turnbaugh, P. J., Fulton, R. S., Wollam, A., et al. (2009). Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. U S A 106, 5859–5864. doi: 10.1073/pnas.0901529106
Mamantopoulos, M., Ronchi, F., Van Hauwermeiren, F., Vieira-Silva, S., Yilmaz, B., Martens, L., et al. (2017). Nlrp6- and ASC-Dependent Inflammasomes Do Not Shape the Commensal Gut Microbiota Composition. Immunity 47, 339–348e334. doi: 10.1016/j.immuni.2017.07.011
Manichanh, C., Rigottier-Gois, L., Bonnaud, E., Gloux, K., Pelletier, E., Frangeul, L., et al. (2006). Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55, 205–211. doi: 10.1136/gut.2005.073817
Markle, J. G., Frank, D. N., Mortin-Toth, S., Robertson, C. E., Feazel, L. M., Rolle-Kampczyk, U., et al. (2013). Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084–1088. doi: 10.1126/science.1233521
Marzorati, M., Vanhoecke, B., De Ryck, T., Sadaghian Sadabad, M., Pinheiro, I., Possemiers, S., et al. (2014). The HMI module: a new tool to study the Host-Microbiota Interaction in the human gastrointestinal tract in vitro. BMC Microbiol. 14:133. doi: 10.1186/1471-2180-14-133
McCoy, K. D., Burkhard, R., and Geuking, M. B. (2019). The microbiome and immune memory formation. Immunol. Cell Biol. 97, 625–635. doi: 10.1111/imcb.12273
McFall-Ngai, M. J. (2014). The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu. Rev. Microbiol. 68, 177–194. doi: 10.1146/annurev-micro-091313-103654
McNulty, N. P., Wu, M., Erickson, A. R., Pan, C., Erickson, B. K., Martens, E. C., et al. (2013). Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 11:e1001637. doi: 10.1371/journal.pbio.1001637
Miao, J., Chard, L. S., Wang, Z., and Wang, Y. (2019). Syrian Hamster as an Animal Model for the Study on Infectious Diseases. Front. Immunol. 10:2329. doi: 10.3389/fimmu.2019.02329
Mimee, M., Tucker, A. C., Voigt, C. A., and Lu, T. K. (2015). Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota. Cell Syst. 1, 62–71. doi: 10.1016/j.cels.2015.06.001
Modlinska, K., and Pisula, W. (2020). The Norway rat, from an obnoxious pest to a laboratory pet. Elife 9:50651. doi: 10.7554/eLife.50651
Moskowitz, J. E., Andreatta, F., and Amos-Landgraf, J. (2019). The gut microbiota modulates differential adenoma suppression by B6/J and B6/N genetic backgrounds in Apc(Min) mice. Mamm. Genome 30, 237–244. doi: 10.1007/s00335-019-09814-3
Nagalingam, N. A., Robinson, C. J., Bergin, I. L., Eaton, K. A., Huffnagle, G. B., and Young, V. B. (2013). The effects of intestinal microbial community structure on disease manifestation in IL-10-/- mice infected with Helicobacter hepaticus. Microbiome 1:15. doi: 10.1186/2049-2618-1-15
Negi, S., Das, D. K., Pahari, S., Nadeem, S., and Agrewala, J. N. (2019). Potential Role of Gut Microbiota in Induction and Regulation of Innate Immune Memory. Front. Immunol. 10:2441. doi: 10.3389/fimmu.2019.02441
Neville, B. A., Forster, S. C., and Lawley, T. D. (2018). Commensal Koch’s postulates: establishing causation in human microbiota research. Curr. Opin. Microbiol. 42, 47–52. doi: 10.1016/j.mib.2017.10.001
Nkamga, V. D., Henrissat, B., and Drancourt, M. (2017). Archaea: Essential inhabitants of the human digestive microbiota. Hum. Microb. J. 3, 1–8.
Norman, J. M., Handley, S. A., and Virgin, H. W. (2014). Kingdom-agnostic metagenomics and the importance of complete characterization of enteric microbial communities. Gastroenterology 146, 1459–1469. doi: 10.1053/j.gastro.2014.02.001
Nyholm, S. V., and McFall-Ngai, M. J. (2004). The winnowing: establishing the squid-vibrio symbiosis. Nat. Rev. Microbiol. 2, 632–642. doi: 10.1038/nrmicro957
Ootani, A., Li, X., Sangiorgi, E., Ho, Q. T., Ueno, H., Toda, S., et al. (2009). Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 15, 701–706. doi: 10.1038/nm.1951
Osmekhina, E., Jonkergouw, C., Schmidt, G., Jahangiri, F., Jokinen, V., Franssila, S., et al. (2018). Controlled communication between physically separated bacterial populations in a microfluidic device. Commun. Biol. 1:97. doi: 10.1038/s42003-018-0102-y
Ost, K. S., and Round, J. L. (2018). Communication Between the Microbiota and Mammalian Immunity. Annu. Rev. Microbiol. 72, 399–422. doi: 10.1146/annurev-micro-090817-062307
Ottman, N., Ruokolainen, L., Suomalainen, A., Sinkko, H., Karisola, P., Lehtimaki, J., et al. (2019). Soil exposure modifies the gut microbiota and supports immune tolerance in a mouse model. J. Allergy Clin. Immunol. 143, 1198–1206e1112. doi: 10.1016/j.jaci.2018.06.024
Palm, N. W., de Zoete, M. R., Cullen, T. W., Barry, N. A., Stefanowski, J., Hao, L., et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010. doi: 10.1016/j.cell.2014.08.006
Parfrey, L. W., Walters, W. A., and Knight, R. (2011). Microbial eukaryotes in the human microbiome: ecology, evolution, and future directions. Front. Microbiol. 2:153. doi: 10.3389/fmicb.2011.00153
Paterson, M. J., Oh, S., and Underhill, D. M. (2017). Host-microbe interactions: commensal fungi in the gut. Curr. Opin. Microbiol. 40, 131–137. doi: 10.1016/j.mib.2017.11.012
Pearce, S. C., Al-Jawadi, A., Kishida, K., Yu, S., Hu, M., Fritzky, L. F., et al. (2018). Marked differences in tight junction composition and macromolecular permeability among different intestinal cell types. BMC Biol. 16:19. doi: 10.1186/s12915-018-0481-z
Phifer-Rixey, M., and Nachman, M. W. (2015). Insights into mammalian biology from the wild house mouse Mus musculus. Elife 4:5959. doi: 10.7554/eLife.05959
Pozzilli, P., Signore, A., Williams, A. J., and Beales, P. E. (1993). NOD mouse colonies around the world–recent facts and figures. Immunol. Today 14, 193–196. doi: 10.1016/0167-5699(93)90160-M
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Rader, B. A., Kremer, N., Apicella, M. A., Goldman, W. E., and McFall-Ngai, M. J. (2012). Modulation of symbiont lipid A signaling by host alkaline phosphatases in the squid-vibrio symbiosis. mBio 3:3. doi: 10.1128/mBio.00093-12
Raibaud, P., Ducluzeau, R., Dubos, F., Hudault, S., Bewa, H., and Muller, M. C. (1980). Implantation of bacteria from the digestive tract of man and various animals into gnotobiotic mice. Am. J. Clin. Nutr. 33, 2440–2447. doi: 10.1093/ajcn/33.11.2440
Rajan, A., Vela, L., Zeng, X. L., Yu, X., Shroyer, N., Blutt, S. E., et al. (2018). Novel Segment- and Host-Specific Patterns of Enteroaggregative Escherichia coli Adherence to Human Intestinal Enteroids. mBio 9:1. doi: 10.1128/mBio.02419-17
Rawls, J. F., Mahowald, M. A., Goodman, A. L., Trent, C. M., and Gordon, J. I. (2007). In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc. Natl. Acad. Sci. U S A 104, 7622–7627. doi: 10.1073/pnas.0702386104
Rawls, J. F., Mahowald, M. A., Ley, R. E., and Gordon, J. I. (2006). Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127, 423–433. doi: 10.1016/j.cell.2006.08.043
Reese, T. A., Bi, K., Kambal, A., Filali-Mouhim, A., Beura, L. K., Burger, M. C., et al. (2016). Sequential Infection with Common Pathogens Promotes Human-like Immune Gene Expression and Altered Vaccine Response. Cell Host Microbe 19, 713–719. doi: 10.1016/j.chom.2016.04.003
Rey, F. E., Faith, J. J., Bain, J., Muehlbauer, M. J., Stevens, R. D., Newgard, C. B., et al. (2010). Dissecting the in vivo metabolic potential of two human gut acetogens. J. Biol. Chem. 285, 22082–22090. doi: 10.1074/jbc.M110.117713
Reyniers, J. A., and Sacksteder, M. R. (1958). The use of germfree animals and techniques in the search for unknown etiological agents. Ann. N. Y. Acad. Sci. 73, 344–356.
Reynolds, L. A., Finlay, B. B., and Maizels, R. M. (2015). Cohabitation in the Intestine: Interactions among Helminth Parasites, Bacterial Microbiota, and Host Immunity. J. Immunol. 195, 4059–4066. doi: 10.4049/jimmunol.1501432
Ridaura, V. K., Faith, J. J., Rey, F. E., Cheng, J., Duncan, A. E., Kau, A. L., et al. (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1241214. doi: 10.1126/science.1241214
Rosshart, S. P., Herz, J., Vassallo, B. G., Hunter, A., Wall, M. K., Badger, J. H., et al. (2019). Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365:6452. doi: 10.1126/science.aaw4361
Rosshart, S. P., Vassallo, B. G., Angeletti, D., Hutchinson, D. S., Morgan, A. P., Takeda, K., et al. (2017). Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 171, 1015–1028e1013. doi: 10.1016/j.cell.2017.09.016
Round, J. L., and Mazmanian, S. K. (2010). Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. U S A 107, 12204–12209. doi: 10.1073/pnas.0909122107
Runge, S., and Rosshart, S. P. (2021). The Mammalian Metaorganism: A Holistic View on How Microbes of All Kingdoms and Niches Shape Local and Systemic Immunity. Front. Immunol. 12:702378. doi: 10.3389/fimmu.2021.702378
Ryu, J. H., Kim, S. H., Lee, H. Y., Bai, J. Y., Nam, Y. D., Bae, J. W., et al. (2008). Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782. doi: 10.1126/science.1149357
Sadaghian Sadabad, M., von Martels, J. Z., Khan, M. T., Blokzijl, T., Paglia, G., Dijkstra, G., et al. (2015). A simple coculture system shows mutualism between anaerobic faecalibacteria and epithelial Caco-2 cells. Sci. Rep. 5:17906. doi: 10.1038/srep17906
Sarvestani, S. K., Signs, S., Hu, B., Yeu, Y., Feng, H., Ni, Y., et al. (2021). Induced organoids derived from patients with ulcerative colitis recapitulate colitic reactivity. Nat. Commun. 12:262. doi: 10.1038/s41467-020-20351-5
Sato, T., Vries, R. G., Snippert, H. J., van de Wetering, M., Barker, N., Stange, D. E., et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. doi: 10.1038/nature07935
Schaedler, R. W., Dubs, R., and Costello, R. (1965). Association of germfree mice with bacteria isolated from normal mice. J. Exp. Med. 122, 77–82. doi: 10.1084/jem.122.1.77
Schloissnig, S., Arumugam, M., Sunagawa, S., Mitreva, M., Tap, J., Zhu, A., et al. (2013). Genomic variation landscape of the human gut microbiome. Nature 493, 45–50. doi: 10.1038/nature11711
Schwarzer, M., Makki, K., Storelli, G., Machuca-Gayet, I., Srutkova, D., Hermanova, P., et al. (2016). Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science 351, 854–857. doi: 10.1126/science.aad8588
Seedorf, H., Griffin, N. W., Ridaura, V. K., Reyes, A., Cheng, J., Rey, F. E., et al. (2014). Bacteria from diverse habitats colonize and compete in the mouse gut. Cell 159, 253–266. doi: 10.1016/j.cell.2014.09.008
Sefik, E., Geva-Zatorsky, N., Oh, S., Konnikova, L., Zemmour, D., McGuire, A. M., et al. (2015). Mucosal immunology. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 349, 993–997. doi: 10.1126/science.aaa9420
Semova, I., Carten, J. D., Stombaugh, J., Mackey, L. C., Knight, R., Farber, S. A., et al. (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12, 277–288. doi: 10.1016/j.chom.2012.08.003
Seregin, S. S., Golovchenko, N., Schaf, B., Chen, J., Pudlo, N. A., Mitchell, J., et al. (2017). NLRP6 Protects Il10(-/-) Mice from Colitis by Limiting Colonization of Akkermansia muciniphila. Cell Rep. 19, 733–745. doi: 10.1016/j.celrep.2017.03.080
Shin, S. C., Kim, S. H., You, H., Kim, B., Kim, A. C., Lee, K. A., et al. (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674. doi: 10.1126/science.1212782
Shin, W., Wu, A., Massidda, M. W., Foster, C., Thomas, N., Lee, D. W., et al. (2019). A Robust Longitudinal Co-culture of Obligate Anaerobic Gut Microbiome With Human Intestinal Epithelium in an Anoxic-Oxic Interface-on-a-Chip. Front. Bioeng. Biotechnol. 7:13. doi: 10.3389/fbioe.2019.00013
Sivan, A., Corrales, L., Hubert, N., Williams, J. B., Aquino-Michaels, K., Earley, Z. M., et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089. doi: 10.1126/science.aac4255
Skye, S. M., Zhu, W., Romano, K. A., Guo, C. J., Wang, Z., Jia, X., et al. (2018). Microbial Transplantation With Human Gut Commensals Containing CutC Is Sufficient to Transmit Enhanced Platelet Reactivity and Thrombosis Potential. Circ. Res. 123, 1164–1176. doi: 10.1161/CIRCRESAHA.118.313142
Smith, K., McCoy, K. D., and Macpherson, A. J. (2007). Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19, 59–69. doi: 10.1016/j.smim.2006.10.002
Smith, M. I., Yatsunenko, T., Manary, M. J., Trehan, I., Mkakosya, R., Cheng, J., et al. (2013). Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554. doi: 10.1126/science.1229000
Smith, P. M., Howitt, M. R., Panikov, N., Michaud, M., Gallini, C. A., Bohlooly, Y. M., et al. (2013). The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573. doi: 10.1126/science.1241165
Song, X., Sun, X., Oh, S. F., Wu, M., Zhang, Y., Zheng, W., et al. (2020). Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature 577, 410–415. doi: 10.1038/s41586-019-1865-0
Srinivasan, A., Torres, N. S., Leung, K. P., Lopez-Ribot, J. L., and Ramasubramanian, A. K. (2017). nBioChip, a Lab-on-a-Chip Platform of Mono- and Polymicrobial Biofilms for High-Throughput Downstream Applications. mSphere 2:3. doi: 10.1128/mSphere.00247-17
Stagaman, K., Sharpton, T. J., and Guillemin, K. (2020). Zebrafish microbiome studies make waves. Lab. Anim. 49, 201–207. doi: 10.1038/s41684-020-0573-6
Stefka, A. T., Feehley, T., Tripathi, P., Qiu, J., McCoy, K., Mazmanian, S. K., et al. (2014). Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. U S A 111, 13145–13150. doi: 10.1073/pnas.1412008111
Storelli, G., Defaye, A., Erkosar, B., Hols, P., Royet, J., and Leulier, F. (2011). Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 14, 403–414. doi: 10.1016/j.cmet.2011.07.012
Subramanian, S., Huq, S., Yatsunenko, T., Haque, R., Mahfuz, M., Alam, M. A., et al. (2014). Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421. doi: 10.1038/nature13421
Surana, N. K., and Kasper, D. L. (2017). Moving beyond microbiome-wide associations to causal microbe identification. Nature 552, 244–247. doi: 10.1038/nature25019
Szabo, P. A., Miron, M., and Farber, D. L. (2019). Location, location, location: Tissue resident memory T cells in mice and humans. Sci. Immunol. 4:34. doi: 10.1126/sciimmunol.aas9673
Takei, I., Asaba, Y., Kasatani, T., Maruyama, T., Watanabe, K., Yanagawa, T., et al. (1992). Suppression of development of diabetes in NOD mice by lactate dehydrogenase virus infection. J. Autoimmun. 5, 665–673. doi: 10.1016/0896-8411(92)90184-r
Tan, H. Y., and Toh, Y. C. (2020). What can microfluidics do for human microbiome research? Biomicrofluidics 14:051303. doi: 10.1063/5.0012185
Tan, T. G., Sefik, E., Geva-Zatorsky, N., Kua, L., Naskar, D., Teng, F., et al. (2016). Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. U S A 113, E8141–E8150. doi: 10.1073/pnas.1617460113
Thompson, G. R., and Trexler, P. C. (1971). Gastrointestinal structure and function in germ-free or gnotobiotic animals. Gut 12, 230–235. doi: 10.1136/gut.12.3.230
Thomson, A., Smart, K., Somerville, M. S., Lauder, S. N., Appanna, G., Horwood, J., et al. (2019). The Ussing chamber system for measuring intestinal permeability in health and disease. BMC Gastroenterol. 19:98. doi: 10.1186/s12876-019-1002-4
Toprak, N. U., Yagci, A., Gulluoglu, B. M., Akin, M. L., Demirkalem, P., Celenk, T., et al. (2006). A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin. Microbiol. Infect. 12, 782–786. doi: 10.1111/j.1469-0691.2006.01494.x
Trexler, P. C., and Reynolds, L. I. (1957). Flexible film apparatus for the rearing and use of germfree animals. Appl. Microbiol. 5, 406–412. doi: 10.1128/am.5.6.406-412.1957
Troll, J. V., Hamilton, M. K., Abel, M. L., Ganz, J., Bates, J. M., Stephens, W. Z., et al. (2018). Microbiota promote secretory cell determination in the intestinal epithelium by modulating host Notch signaling. Development 145:4. doi: 10.1242/dev.155317
Turnbaugh, P. J., Bäckhed, F., Fulton, L., and Gordon, J. I. (2008). Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223. doi: 10.1016/j.chom.2008.02.015
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., et al. (2009a). A core gut microbiome in obese and lean twins. Nature 457, 480–484. doi: 10.1038/nature07540
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
Turnbaugh, P. J., Ridaura, V. K., Faith, J. J., Rey, F. E., Knight, R., and Gordon, J. I. (2009b). The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1:6ra14. doi: 10.1126/scitranslmed.3000322
Ulluwishewa, D., Anderson, R. C., Young, W., McNabb, W. C., van Baarlen, P., Moughan, P. J., et al. (2015). Live Faecalibacterium prausnitzii in an apical anaerobic model of the intestinal epithelial barrier. Cell Microbiol. 17, 226–240. doi: 10.1111/cmi.12360
Umesaki, Y., Setoyama, H., Matsumoto, S., Imaoka, A., and Itoh, K. (1999). Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect. Immun. 67, 3504–3511. doi: 10.1128/IAI.67.7.3504-3511.1999
Ussing, H. H., and Zerahn, K. (1951). Active transport of sodium as the source of electric current in the short-circuited isolated frog skin. Acta Physiol. Scand. 23, 110–127. doi: 10.1111/j.1748-1716.1951.tb00800.x
Van den Abbeele, P., Roos, S., Eeckhaut, V., MacKenzie, D. A., Derde, M., Verstraete, W., et al. (2012). Incorporating a mucosal environment in a dynamic gut model results in a more representative colonization by lactobacilli. Microb. Biotechnol. 5, 106–115. doi: 10.1111/j.1751-7915.2011.00308.x
van Nood, E., Vrieze, A., Nieuwdorp, M., Fuentes, S., Zoetendal, E. G., de Vos, W. M., et al. (2013). Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415. doi: 10.1056/NEJMoa1205037
Velazquez, E. M., Nguyen, H., Heasley, K. T., Saechao, C. H., Gil, L. M., Rogers, A. W. L., et al. (2019). Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection. Nat. Microbiol. 4, 1057–1064. doi: 10.1038/s41564-019-0407-8
Verma, R., Lee, C., Jeun, E. J., Yi, J., Kim, K. S., Ghosh, A., et al. (2018). Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3(+) regulatory T cells. Sci. Immunol. 3:28. doi: 10.1126/sciimmunol.aat6975
Vétizou, M., Pitt, J. M., Daillère, R., Lepage, P., Waldschmitt, N., Flament, C., et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084. doi: 10.1126/science.aad1329
Vijay-Kumar, M., Aitken, J. D., Carvalho, F. A., Cullender, T. C., Mwangi, S., Srinivasan, S., et al. (2010). Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231. doi: 10.1126/science.1179721
Vlasova, A. N., Rajashekara, G., and Saif, L. J. (2018). Interactions between human microbiome, diet, enteric viruses and immune system: novel insights from gnotobiotic pig research. Drug Discov. Today Dis. Models. 28, 95–103. doi: 10.1016/j.ddmod.2019.08.006
Vijay-Kumar, M., Sanders, C. J., Taylor, R. T., Kumar, A., Aitken, J. D., Sitaraman, S. V., et al. (2007). Deletion of TLR5 results in spontaneous colitis in mice. J. Clin. Invest. 117, 3909–3921. doi: 10.1172/JCI33084
von Kortzfleisch, V. T., Karp, N. A., Palme, R., Kaiser, S., Sachser, N., and Richter, S. H. (2020). Improving reproducibility in animal research by splitting the study population into several ‘mini-experiments’. Sci. Rep. 10:16579. doi: 10.1038/s41598-020-73503-4
von Martels, J. Z. H., Sadaghian Sadabad, M., Bourgonje, A. R., Blokzijl, T., Dijkstra, G., Faber, K. N., et al. (2017). The role of gut microbiota in health and disease: In vitro modeling of host-microbe interactions at the aerobe-anaerobe interphase of the human gut. Anaerobe 44, 3–12. doi: 10.1016/j.anaerobe.2017.01.001
Wagner, V. E., Dey, N., Guruge, J., Hsiao, A., Ahern, P. P., Semenkovich, N. P., et al. (2016). Effects of a gut pathobiont in a gnotobiotic mouse model of childhood undernutrition. Sci. Transl. Med. 8:366ra164. doi: 10.1126/scitranslmed.aah4669
Wang, D. (2020). 5 challenges in understanding the role of the virome in health and disease. PLoS Pathog. 16:e1008318. doi: 10.1371/journal.ppat.1008318
Wegorzewska, M. M., Glowacki, R. W. P., Hsieh, S. A., Donermeyer, D. L., Hickey, C. A., Horvath, S. C., et al. (2019). Diet modulates colonic T cell responses by regulating the expression of a Bacteroides thetaiotaomicron antigen. Sci. Immunol. 4:32. doi: 10.1126/sciimmunol.aau9079
Wilberz, S., Partke, H. J., Dagnaes-Hansen, F., and Herberg, L. (1991). Persistent MHV (mouse hepatitis virus) infection reduces the incidence of diabetes mellitus in non-obese diabetic mice. Diabetologia 34, 2–5. doi: 10.1007/BF00404016
Wilson, S. S., Tocchi, A., Holly, M. K., Parks, W. C., and Smith, J. G. (2015). A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal. Immunol. 8, 352–361. doi: 10.1038/mi.2014.72
Wu, H. J., Ivanov, I. I., Darce, J., Hattori, K., Shima, T., Umesaki, Y., et al. (2010). Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827. doi: 10.1016/j.immuni.2010.06.001
Wu, M., McNulty, N. P., Rodionov, D. A., Khoroshkin, M. S., Griffin, N. W., Cheng, J., et al. (2015). Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science 350:aac5992. doi: 10.1126/science.aac5992
Wu, S., Rhee, K. J., Albesiano, E., Rabizadeh, S., Wu, X., Yen, H. R., et al. (2009). A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15, 1016–1022. doi: 10.1038/nm.2015
Xiao, L., Feng, Q., Liang, S., Sonne, S. B., Xia, Z., Qiu, X., et al. (2015). A catalog of the mouse gut metagenome. Nat. Biotechnol. 33, 1103–1108. doi: 10.1038/nbt.3353
Yeung, F., Chen, Y. H., Lin, J. D., Leung, J. M., McCauley, C., Devlin, J. C., et al. (2020). Altered Immunity of Laboratory Mice in the Natural Environment Is Associated with Fungal Colonization. Cell Host Microbe 27, 809–822e806. doi: 10.1016/j.chom.2020.02.015
Yurkovetskiy, L., Burrows, M., Khan, A. A., Graham, L., Volchkov, P., Becker, L., et al. (2013). Gender bias in autoimmunity is influenced by microbiota. Immunity 39, 400–412. doi: 10.1016/j.immuni.2013.08.013
Keywords: microbiome, model system, synthetic communities, gnotobiotic, wild mice, translation, model microbial communities
Citation: Glowacki RWP, Engelhart MJ and Ahern PP (2021) Controlled Complexity: Optimized Systems to Study the Role of the Gut Microbiome in Host Physiology. Front. Microbiol. 12:735562. doi: 10.3389/fmicb.2021.735562
Received: 02 July 2021; Accepted: 24 August 2021;
Published: 27 September 2021.
Edited by:
Jean-François Brugère, Université Clermont Auvergne, FranceReviewed by:
Aaron Conrad Ericsson, University of Missouri, United StatesStephan Rosshart, University of Freiburg Medical Center, Germany
Preben Boysen, Norwegian University of Life Sciences, Norway
Copyright © 2021 Glowacki, Engelhart and Ahern. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philip P. Ahern, YWhlcm5wQGNjZi5vcmc=
†These authors have contributed equally to this work
 Robert W. P. Glowacki
Robert W. P. Glowacki Morgan J. Engelhart
Morgan J. Engelhart Philip P. Ahern
Philip P. Ahern