- 1Shandong Provincial Key Laboratory of Synthetic Biology, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao, China
- 2Shandong Energy Institute, Qingdao, China
- 3Qingdao New Energy Shandong Laboratory, Qingdao, China
- 4College of Life Science, University of Chinese Academy of Sciences, Beijing, China
- 5Marine Biology and Biotechnology Laboratory, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 6Key Laboratory of Biofuels, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao, China
Micafungin is an important echinocandin antifungal agent for the treatment of invasive fungal infections. In industry, micafungin is derived from the natural product FR901379, which is a non-ribosomal cyclic hexapeptide produced by the filamentous fungus Coleophoma empetri. The difficulty of genetic manipulation in C. empetri restricts the clarification of FR901379 biosynthetic mechanism. In this work, we developed an efficient genetic manipulation system in the industrial FR901379-producing strain C. empetri MEFC009. Firstly, a convenient protoplast-mediated transformation (PMT) method was developed. Secondly, with this transformation method, the essential genetic elements were verified. Selectable markers hph, neo, and nat can be used for the transformation, and promotors Ppgk, PgpdA, and PgpdAt are functional in C. empetri MEFC009. Thirdly, the frequency of homologous recombination was improved from 4 to 100% by deleting the ku80 gene, resulting in an excellent chassis cell for gene-targeting. Additionally, the advantage of this genetic manipulation system was demonstrated in the identification of the polyketide synthase (PKS) responsible for the biosynthesis of dihydroxynapthalene (DHN)-melanin. This genetic manipulation system will be a useful platform for the research of FR901379 and further genome mining of secondary metabolites in C. empetri.
Introduction
Echinocandins are a family of cyclic lipohexapeptides with the excellent activity of anti-Candida and anti-Aspergillus, which are considered to be the most potential antifungal agents clinically (Denning, 2003; Patil and Majumdar, 2017). To date, three semisynthetic echinocandins, caspofungin, micafungin, and anidulafungin have been approved for the treatment of invasive fungal infections (IFIs). Among them, micafungin is the only sulfated echinocandin agent (Hashimoto, 2009). The biosynthetic pathway of the lead compounds of caspofungin and anidulafungin have been identified in 2012 and 2013, respectively (Cacho et al., 2012; Chen et al., 2013; Hüttel, 2017). Compound FR901379 produced by filamentous fungi Coleophoma empetri is the lead compound of micafungin. The biosynthesis pathway of FR901379 is still cryptic, because of the difficulty of genetic manipulation of C. empetri. In 2009, Masato Yamada developed Agrobacterium tumefaciens-mediated transformation method (ATMT) in the C. empetri F11899, only the fragment of hygromycin B resistant gene was successfully integrated into the genome by random insertion (Yamada et al., 2009). More efficient and systematic genetic engineering tools would be conducive to functional genomics studies and are also fundamental to the metabolic engineering of industrial strains. Therefore, it is still necessary to explore more effective transformation methods and available genetic elements.
Several fundamental points must be considered to construct an efficient genetic manipulation system in filamentous fungi. The primary one is a convenient and efficient genetic transformation method. The protoplast-mediated transformation (PMT) and ATMT are the two most commonly used genetic transformation methods in filamentous fungi (Meyer, 2008). Among them, PMT method is usually more preferred, because it is not necessary to construct a binary vector and the corresponding A. tumefaciens strain (Michielse et al., 2008). The second is an efficient gene-targeting system. Disrupting the nonhomologous end-joining (NHEJ) pathways is a general way to improve the frequency of gene-targeting in filamentous fungi (Ninomiya et al., 2004). Even in the CRISPR/Cas9 genome editing system, the chassis cell of NHEJ deficient is also inevitable (Wei et al., 2020). In addition, the elements of promoter and selectable marker are also indispensable, especially for the sequential metabolic engineering of industrial strains (Blumhoff et al., 2013).
In this work, a systematic and useful genetic manipulation system was established in an excellent FR901379 producing strain C. empetri MEFC009. It includes four key points: efficient protoplast-mediated transformation method, available promoters (PgpdA, PgpdAt, and Ppgk), confirmed selectable markers (hph, neo, and nat), and ku80 deficient chassis cell with high homologous recombination frequency. Furthermore, the polyketide synthase responsible for the biosynthesis of melanin was efficiently identified using this genetic manipulation system. This genetic manipulation system will be significantly conducive to the study of FR901379 biosynthetic pathway and further sequential metabolic engineering.
Materials and Methods
Chemicals and Reagents
Antibiotics were purchased from Solarbio (Beijing China). calcofluor white (CFW) was obtained from Sigma-Aldrich (United States), 4',6-diamidino-2-phenylindole (DAPI) was obtained from Beyotime (Shanghai, China). Lysis enzymes are obtained from Sigma (United States), snailase is obtained from Solarbio (Beijing China), and lywallzyme is obtained from Guangdong Microbial Culture Collection Center (Guangdong, China).
Strains, Medium, and Cultivation Conditions
All strains used in this study are listed in Table 1. The C. empetri MEFC009 and all mutant strains were grown on PDA. Cultivation in shake flasks was carried out in seed medium (MKS: soluble starch 15g/L, sucrose 10g/L, cotton seed meal 5g/L, peptone 10g/L, KH2PO4 1g/L, CaCO3 2g/L, and pH 6.5) and FR901379 production medium (MKF: glucose 10g/L, corn starch 30g/L, peptone 10g/L, (NH4)2SO4 6g/L, KH2PO4 1g/L, FeSO4 0.3g/L, ZnSO4 0.01g/L, CaCO3 2g/L, and pH 6.5).
Staining Nuclei and Cell Wall of the Hyphae
The cell wall and nuclei of the hyphae of C. empetri MEFC009 were stained by CFW and DAPI, respectively (Chazotte, 2011). The hyphae of C. empetri MEFC009 was immersed in 200μl phosphate buffered saline (PBS pH 7.4, 0.1mol/L), then stained by 0.8μg/ml of DAPI or 0.05μg/ml of CFW for 5min. The hyphae were washed twice by PBS to remove the stain and observed by confocal microscopy (FluoView FV1000). In addition, 0.8μg/ml of DAPI and 0.05μg/ml of CFW were added together to stain the nuclei and cell wall of the hyphae simultaneously. The excitation and emission wavelength of CFW and DAPI is 345/405nm with blue fluorescence.
Binary Vector Assembly and Transformation of A. tumefaciens
All primers used in this study are listed in Supplementary Table S1. The selectable marker gene nat was amplified from pPK2natGFPD by PCR using primers PtrpC/nat-R (Gu et al., 2018). The coding DNA sequence (CDS) of neo was synthesized fused with promoter Ppgk and terminator Tpgk by fusion PCR. The markers nat and neo were cloned into the binary vector pCambia1300 using a One-Step Cloning Kit (Vazyme, Nanjing, China) to construct the recombinant plasmids pPM-3, and pPM-4 (Hršelová et al., 2015). The recombinant A. tumefaciens LBA4404 harboring plasmids pCambia1300, pPM-3, and pPM-4 were constructed. The Agrobacterium-mediated transformation of C. empetri MEFC009 was performed as previously described (Yamada et al., 2009).
DNA Manipulation for Cassettes Construction
To evaluate the function of promoters PgpdA, Ppgk, and PgpdAt, the cassettes containing the above promoters were constructed, respectively. The DNA cassettes of PgpdA-sgfp-TtrpC-hph and PgpdAt-sgfp-TtrpC-hph were amplified from plasmid pAN52-4 (Punt et al., 1991) and pXH2-1 (Huang et al., 2014) by PCR using primer pairs PgpdA-F/TtrpC-R and PgpdAt-F/TtrpC-R, respectively. The fragment of sgfp of the cassette of Ppgk-sgfp-Tpgk-hph was amplified from plasmid pXH2-1, and then fused with Ppgk, Tpgk, and hph by fusion PCR, resulting in cassette Ppgk-sgfp-Tpgk-hph.
To knock out the ku80 gene, the flanking 5' and 3' DNA of the ku80 gene were amplified by PCR from the genome of C. empetri MEFC009 using primer pairs Uku80-F/Uku80-(hph)-R and Dku80-(hph)-F/Dku80-R, and fused with hph marker by fusion PCR. The gene-targeting cassette was amplified using primers Uku80-CS-F/Dku80-CS-R. The ku80 deletion cassette using neo marker was constructed through the same approach. The flanking 5' and 3' DNA of pks11.2 gene were amplified by PCR from the genome of C. empetri MEFC009 using primer pairs Upks11.2-F/Upks11.2-R and Dpks11.2-F/Dpks11.2-R and fused with hph marker by fusion PCR. The gene-targeting cassette was amplified by primers Upks11.2-CS-F/Dpks11.2-CS-R.
Protoplast-Mediated Transformation
The transformation mediated by protoplast was developed in C. empetri MEFC009 according to the protocol previously described with modification (Marcone et al., 2010). The fresh hyphae from the PDA plate were broken by Superfine homogenizers (Fluko) and cultivated in 50ml of MKS medium for 2days, 220rpm, 25°C. Then hyphae were broken again and inoculated in a new shack flask with 10% proportion, cultivated for 1day at 25°C. The hyphae were harvested by filtration through Miracloth (Calbiochem), washed with 0.6M MgSO4, and immersed in the enzymatic solution (0.6% of snailase, 0.6% lysis enzymes, and 0.7% of lywallzyme) for 4h at 30°C to prepare protoplasts. The protoplasts were collected by filtered through Miracloth and centrifuged at 1,500rpm at 4°C for 20min. After washed with 1M D-sorbitol buffer and STC (1M D-sorbitol, 10mM Tris-HCl pH 8.0, 50mM CaCl2) successively, the protoplasts were resuspended in STC used for transformation.
To optimize the regeneration medium for protoplasts, the protoplasts suspension was diluted to 104cells/ml with STC and H2O, respectively. A 100μl portion of the protoplast dilution was mixed with the regeneration medium and pour into PDAS plates with different concentrations of D-sorbitol: 0, 0.4, 0.6, 0.8, and 1.2M.
As for transformation, approximately 1μg of DNA fragments were mixed with 100μl protoplast solution (1.5×107 protoplasts/ml) and 50μl of ice-cold PSTC (40% PEG 4000, 1M D-sorbitol, 10mM Tris-HCl pH 8.0, and 50mM CaCl2), incubating on ice for 25min. Then 1ml of PSTC was added and incubated for 20min at room temperature. Thereafter, the protoplasts mixture was mixed with 20ml of liquid top agar (PDB with 0.5% agarose and 0.8M D-sorbitol), spread on PDAS amended with the antibiotic, and cultured for 5–7days at 30°C.
Fermentation and HPLC Measurement
C. empetri MEFC009 and mutant C. empetri MEFC009-Δku80 were inoculated in 50ml of MKF medium at 25°C for 10days. Then 1ml of fermentation culture was taken every 2days and extracted with equal volumes of methanol by ultrasonic crushing for 1h. Four independents were set for each experiment. The amount of FR901379 was determined by high performance liquid chromatography (HPLC) monitored at 210nm. The separation was carried out on a C18 reversed phase column (Agilent ZORBAX SB-C18 column, 4.6×150mm, 5μm) at a flow rate of 1ml/min at 30°C, using acetonitrile (ACN)/H2O as elution solvents. A linear gradient from 5 to 100% ACN in H2O containing 0.1% (v/v) TFA in 20min was used.
Results and Discussion
Morphology of Vegetative Hyphae
Hyphae are the most common resource for preparing protoplast. The morphology of vegetative hyphae is the key factor related to the difficulty of protoplast transformation, such as septate to aseptate and uninucleate to multinucleate. In the genetic engineering of the fungi species with aseptate multinucleate hyphae, heterokaryons are spontaneously formed during transformation. And, it is difficult to screen out the homozygote with engineered nucleus (Kaminskyj and Hamer, 1998). The cell wall of the vegetative hyphae of C. empetri MEFC009 was stained by CFW. As shown in Figure 1, the hyphae of C.empetri are branched and septate. The length of each cell is about 7–8 μm. The nuclei were stained in bright blue by DAPI. We can observe that the nuclei were equidistantly distributed in hyphae. When stained with DAPI and CFW simultaneously, the nuclei and diaphragm of the hyphae could be observed. It indicated that only one nucleus was present in each septate hyphae cell. This type of vegetative hyphae is the most suitable for genetic transformation.
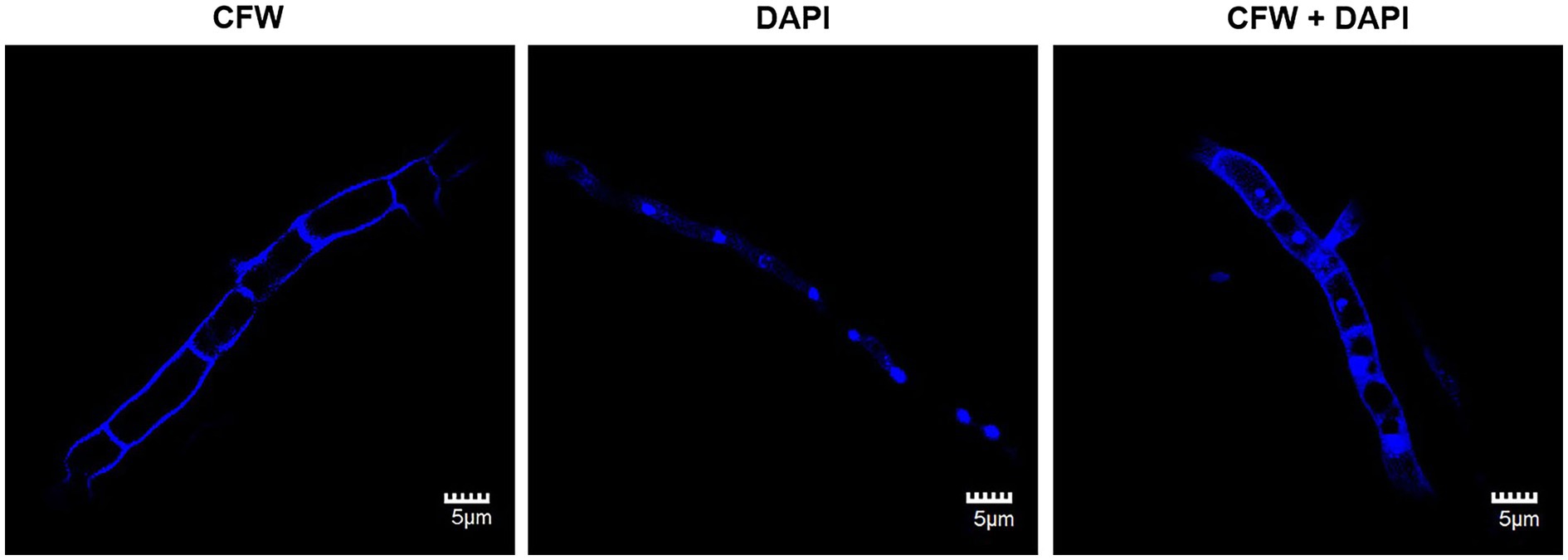
Figure 1. Fluorescence microscopy images of the stained vegetative hyphae of C. empetri MEFC009. CFW: CFW-stained image of hyphae. DAPI: DAPI-stained image of hyphae. CFW+DAPI: CFW and DAPI-stained image of hyphae. All scale bars represent 5μm.
Verification of Selectable Markers
A selectable marker is the essential element for genetic transformation, which is usually also a limiting factor for genetic engineering in filamentous fungi. There are a few selectable markers that have been developed in the genetic transformation of filamentous fungi (Ramsden et al., 2011). However, these selectable markers are not always universal in different species because of the different sensitivity to antibiotics. The resistance gene of hygromycin B, hph, has been confirmed functional for the transformation of C. empetri F-11899 (Yamada et al., 2009). To verify the selectable markers applicable to C. empetri MEFC009, the sensitivity to four selectable antibiotics were tested. The growth of C. empetri MEFC009 were inhibited on potato dextrose agar (PDA) individually supplemented with 100μg/ml of hygromycin B, geneticin, and nourseothricin, but grew normally on plates supplemented with 500μg/L of pyrithiamine. It demonstrated that C. empetri MEFC009 is sensitive to hygromycin B, geneticin, and nourseothricin.
The plasmids containing the resistant genes of hygromycin B (hph), geneticin (neo), or nourseothricin (nat) were respectively transformed into C. empetri MEFC009 using ATMT method as previously described (Yamada et al., 2009). The transformants were successfully screened out on PDA plates containing 100μg/ml of corresponding antibiotics (Figure 2). The results of PCR showed that the resistant genes hph, neo, and nat were integrated into the chromosome of the transformants, respectively, (Supplementary Figure S1). Therefore, the resistant genes hph, neo, and nat are available selectable markers for the transformation of C. empetri.
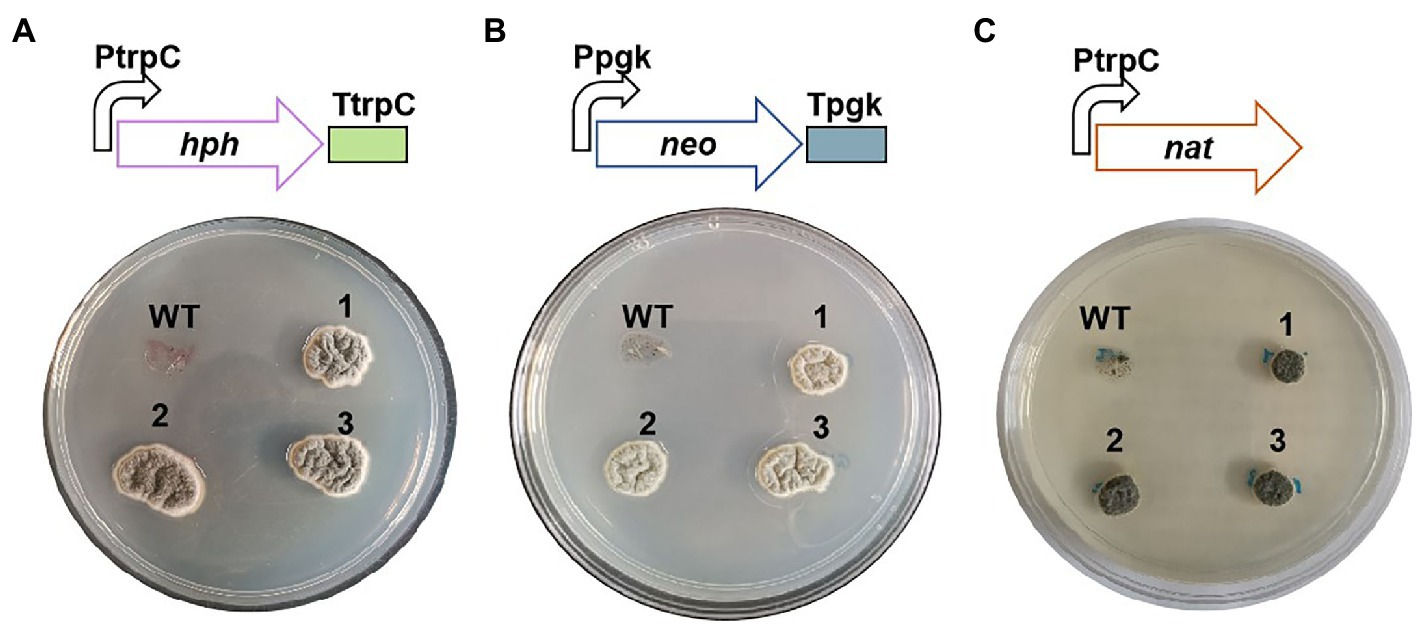
Figure 2. Antibiotics sensitive tests of wild-type C. empetri MEFC009 and mutant strains. Mutant strains with resistance genes of hph (A), neo (B), and nat (C). WT: C. empetri MEFC009, 1–3: mutant strains.
Development of Protoplast-Mediated Transformation Method
Although the genetic transformation of C. empetri MEFC009 was achieved through ATMT method, it is still inefficient, no more than 10 transformants obtained in each experiment. In addition, the transformation process is laborious, time-consuming, and poorly repeatable. Protoplasts-mediated transformation is the most convenient and popular method for the transformation of filamentous fungi (Prabha and Punekar, 2004). The generation of high-quality protoplasts is the major challenge to achieve efficient transformation (Roth and Chilvers, 2019). However, enzymatic digestion conditions of the cell walls are different for each fungal strain. The details should be optimized for each case such as hydrolase composition, isotonic solution, regeneration medium, etc. (Ruiz-Díez, 2002).
We optimized the protoplast transformation according to the previously developed method (Marcone et al., 2010). The hyphae of C. empetri MEFC009 were cultivated in PDB medium using a two-step inoculation method. The optimized enzyme solution of 0.6% of snailase, 0.6% lysis enzymes, and 0.7% of lywallzyme was used for cell wall digestion, which produced 1.5×106 protoplasts/ml at 30°C for 2–4h (Figure 3A). The protoplasts were purified and resuspended at 1.5×107 protoplasts/ml in STC solution. In the following PEG-CaCl2 mediated transformation, 1μg of DNA fragment of hph-sgfp was mixed and incubated with 100μl protoplasts solution. The regeneration medium was optimized, and the protoplasts could regenerate on the PDA plates with 0.8M and 1.0M D-sorbitol (Figure 3B). Beginning with 1.5×106 protoplasts, approximately 100–120 positive transformants were obtained in a reaction using hph marker (Figure 3C). The efficiency is significantly higher than that of the ATMT method and is sufficient for functional genomic research and metabolic engineering.
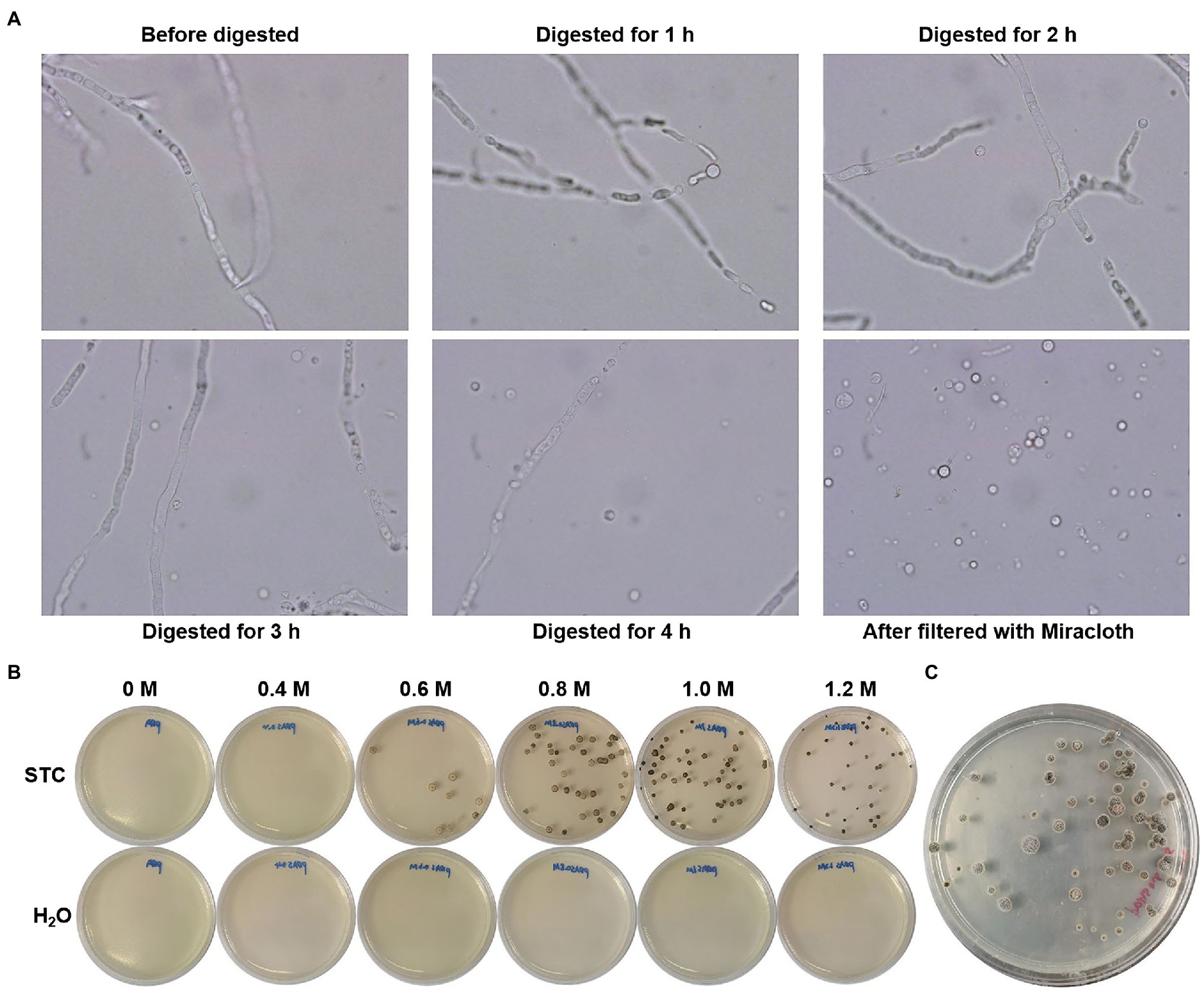
Figure 3. Protoplast-mediated transformation. (A) Images of hyphae and protoplasts incubated in the enzyme solution (100×objective lens). (B) Optimization of regeneration medium with different concentrations of D-sorbitol; STC: protoplasts were diluted with STC; H2O: protoplasts were diluted with H2O. (C) Original transformation plates of hph-sgfp.
Evaluation of Promoters
For a specific fungal host, the efficient promoters are the key elements to achieve high-level expression of target genes (Blumhoff et al., 2013). The promoters of glyceraldehyde-3-phosphate dehydrogenase (gpd) and phosphoglycerate kinase (pgk) of A. nidulans have widely been used in numerous filamentous fungi (Streatfield et al., 1992; Cao et al., 2012). The function of promoters Ppgk, PgpdA (from A. nidulans), and PgpdAt (from A. terreus; Huang et al., 2014) were evaluated in C. empetri MEFC009 using sGFP (synthetic green fluorescent protein) as the reporter. As shown in Figure 4, the DNA cassettes containing the promoters (PgpdA, PgpdAt, or Ppgk), sgfp, TtrpC terminator, and hph were transformed into C. empetri MEFC009, respectively. The integration of DNA cassettes in the genome was confirmed by genomic PCR. The hyphae of transformants were randomly picked out and observed under the fluorescence microscope. The transformants harboring PgpdA, PgpdAt, and Ppgk evaluation cassettes displayed significant green fluorescent, while the parental strain did not show any visible fluorescence (Figure 4). These results demonstrated that promoters PgpdA, PgpdAt, and Ppgk successfully drove sgfp expression in C. empetri MEFC009. According to the intensity of fluorescence, PgpdA and PgpdAt showed similar promoter activity, which is significantly stronger than that of Ppgk.
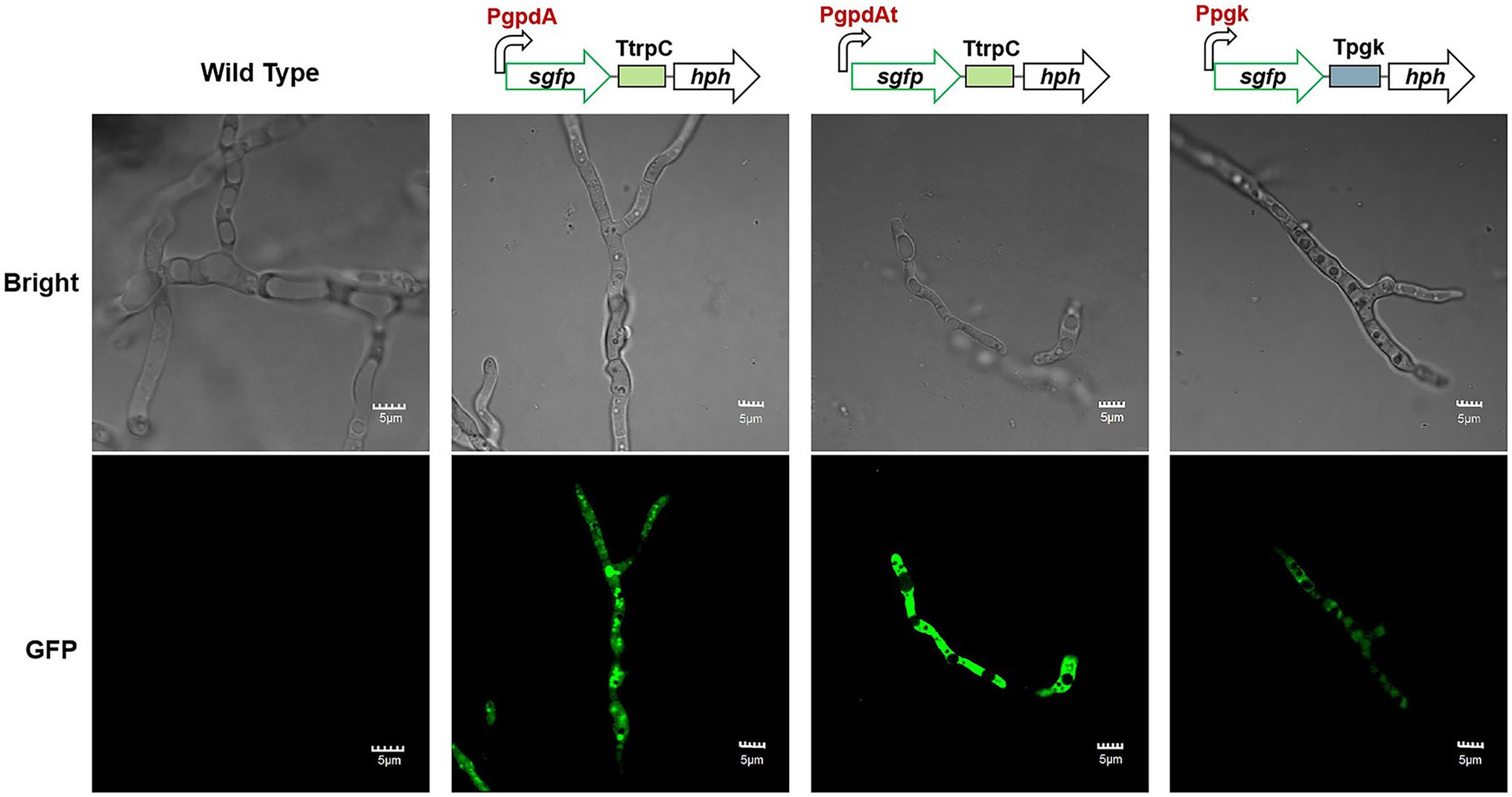
Figure 4. Microscopic features of wild type and representative mutant strains with promoters of PgpdA, PgpdAt, and Ppgk. Bright: Bright-field images. GFP: Fluorescence images. All scale bars represent 5μm.
Construction of the Chassis Cell With High Gene-Targeting Frequency
Gene-targeting is the basis for various genetic engineering techniques, including gene knockout, promoter replacement, and site-specific expression of heterogeneous genes, etc. (Ninomiya et al., 2004). However, homologous recombination (HR) efficiency in filamentous fungi is extremely low because the NHEJ pathway dominates the repair of DNA double-strand breaks (Huang et al., 2016). Learning from previous research, we tried to improve the frequency of gene targeting by disrupting the NHEJ pathway. The ku80 gene was identified according to the annotation of genome sequencing of C. empetri MEFC009. As shown in Figure 5A, 1.5kb region in ku80 gene was replaced by the hph selectable marker. The transformants were screened using PDAS-H plates (PDA with 0.8M D-sorbitol and 100 μg/ml of hygromycin B). Forty-six transformants with hygromycin B resistance were randomly selected for further genotyping. Only in two transformants, the gene-targeting cassette was directly integrated at the ku80 loci (Figure 5B). Therefore, the frequency of HR was about 4% in C. empetri.
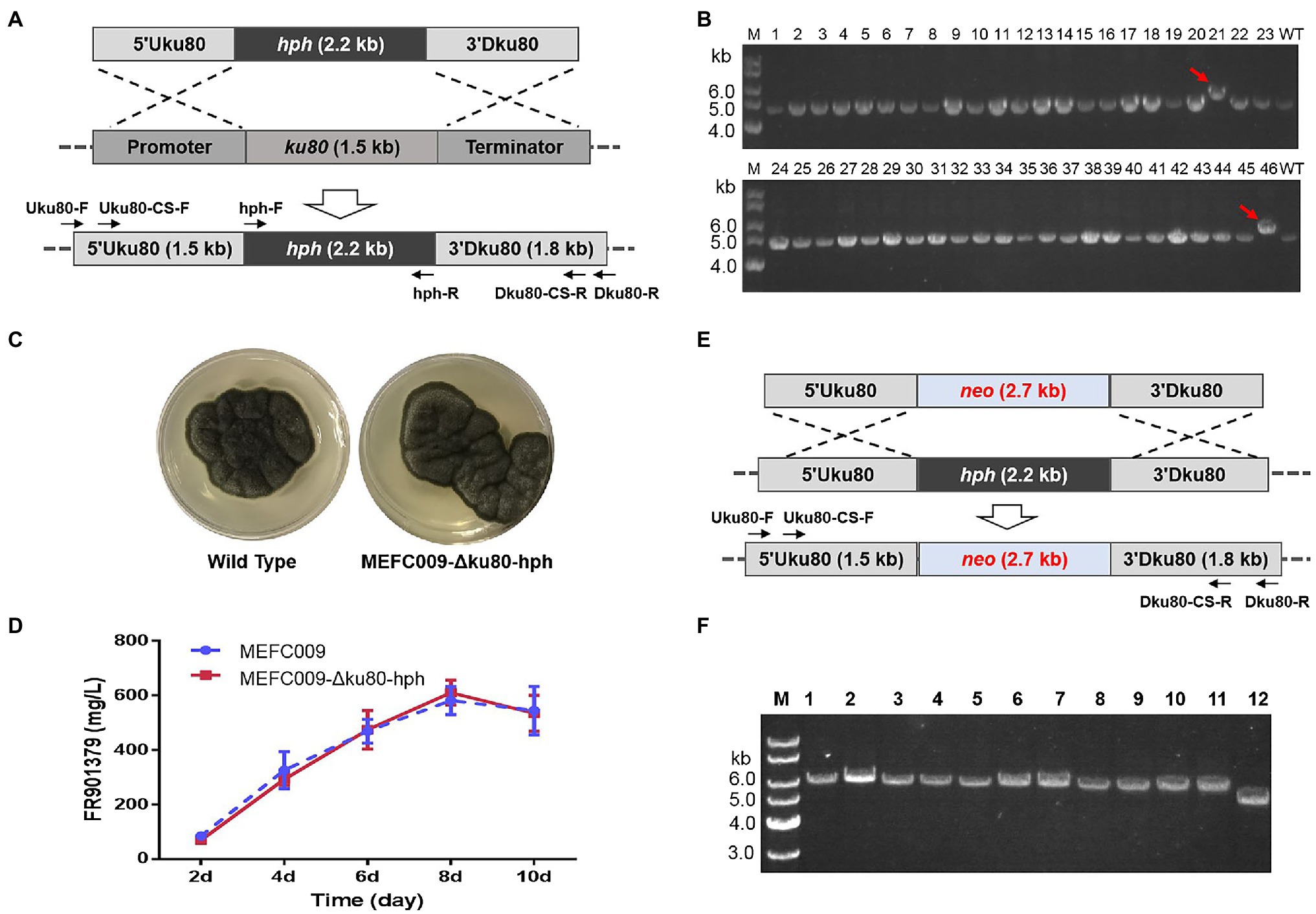
Figure 5. Construction of the chassis cell with high gene-targeting frequency. (A) Strategy for deleting ku80 gene in C. empetri MEFC009. (B) Genotype verification of transformants using primers Uku80-F/Dku80-R. Lane M: 1kb DNA marker; lane 1–46: transformants; lane WT: C. empetri MEFC009. The gene-targeting cassette was integrated at ku80 loci only in transformants NO.21 and NO.46. (C) Comparison of cell growth on PDA plates. (D) Comparison of the FR901379 production. (E) Strategy for replacing the hph marker in C. empetri MEFC009-Δku80-hph with neo. (F) Genotype verification of C. empetri MEFC009-Δku80-neo. Lane M: 1kb DNA marker; lane 1–11: transformants; lane 12: parent strain C. empetri MEFC009-Δku80-hph.
The phenotypic characteristics of C. empetri MEFC009-Δku80-hph were assessed to evaluate the influences of ku80 deletion. The ku80 deletants showed the same growth rates, colonial morphology, and pigmentation on PDA plates (Figure 5C). Furthermore, no deviation in FR901379 productivity was observed in the shake flask fermentation (Figure 5D). To evaluate the HR frequency in the ku80 deletant, the hph integrated in ku80 loci was replaced with neo through another round of gene-targeting (Figure 5E). The transformants were screened by PDAS containing 100μg/ml of geneticin and 11 transformants were randomly selected for genotyping. The results of genomic PCR showed that the hph sequence was successfully replaced with neo sequence in all 11 transformants (Figure 5F). These results demonstrated that the HR frequency has been improved from 4 to 100% by deleting ku80 in C. empetri.
Identification of the DHN-Melanin PKS
The 1,8-dihydroxynaphthalene (DHN) melanin is the most common type of conidial pigments in ascomycetous fungi, which is polymerized from polyketide naphthopyrone (Eisenman and Casadevall, 2012). The colony of C. empetri MEFC009 displays black color on PDA plates. Identifying the synthesis gene of melanin would be a perfect practice for testing the efficiency of genetic modification. According to the results of antiSMASH analysis, there are 18 PKSs in the genome of C. empetri MEFC009 (Blin et al., 2019). Among them, PKS11.2 was predicted to be responsible for the synthesis of melanin, which has four potential catalytic domains: beta-ketoacyl synthase (KS), an acyltransferase (AT), two acyl carrier sites (P1 and P2), and a thioesterase (TE)/Claisen cyclase (CYC; Figure 6A). The phylogenetic tree indicates that PKS11.2 appears highly homologous to other identified DHN-melanin PKSs, BcPKS13 of Botrytis cinerea (Schumacher, 2016), WdPKS1 of Wangiella (Exophiala) dermatitidis (Feng et al., 2001), and BoPKS1 of Bipolaris oryzae (Moriwaki et al., 2004; Figure 6B). To identify the function of pks11.2, the gene-targeting cassette harboring hph selectable marker was transformed in the NHEJ-deficient mutant C. empetri MEFC009-Δku80-neo. All the transformants growing on selection plates exhibited white colonies (Supplementary Figure S2; Figure 6C). It indicated that the pks11.2 was indeed involved in the biosynthesis of melanin and was disrupted by gene-targeting in all transformants. The genotype was further confirmed by genomic PCR, that the selectable marker hph was indeed directly integrated at pks11.2 loci in all eight randomly selected transformants (Figure 6D). Therefore, C. empetri MEFC009-Δku80-neo was indeed an excellent chassis cell for gene-targeting.
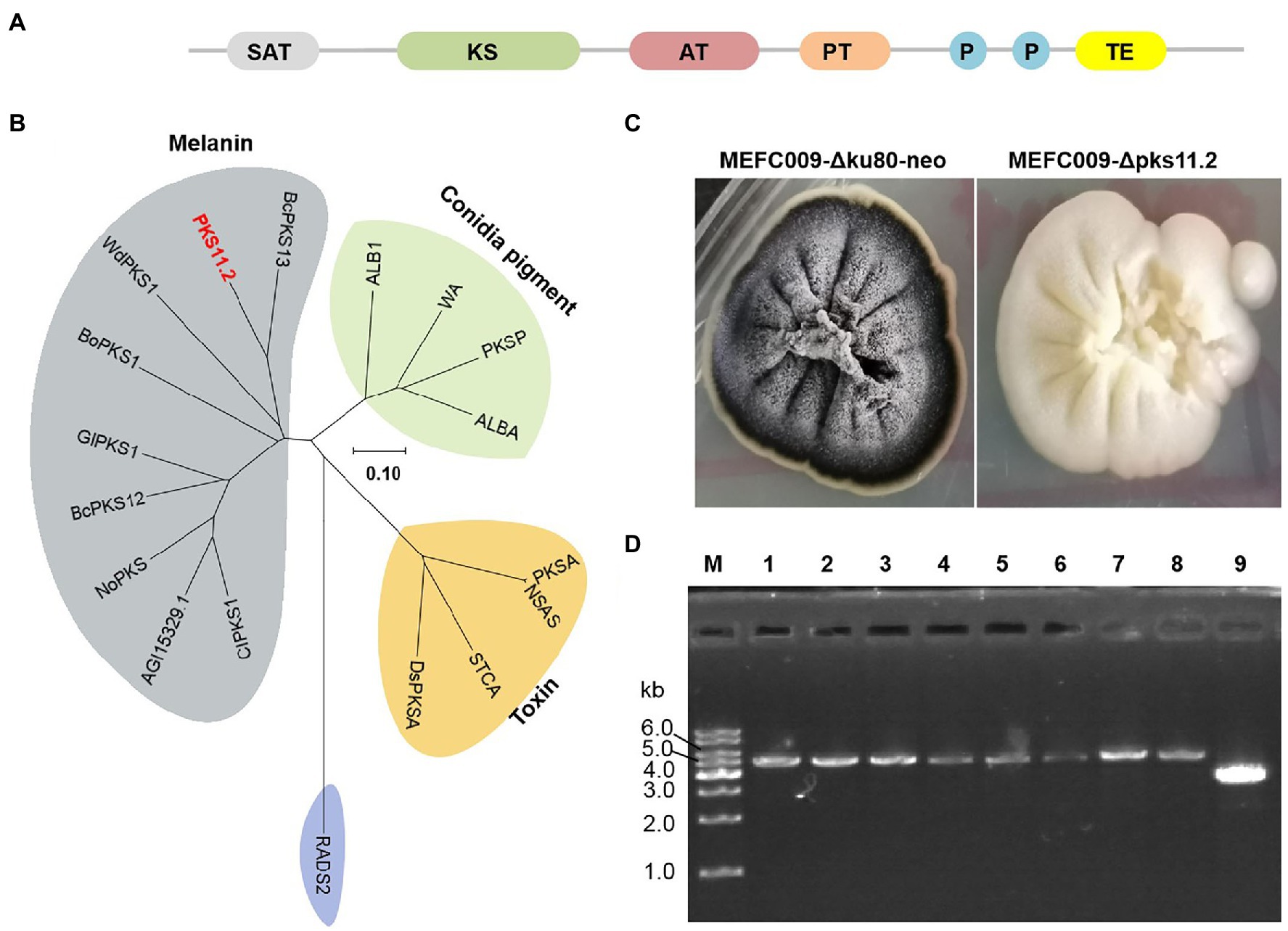
Figure 6. Identification of DHN-melanin PKS in C. empetri MEFC009. (A) Domain structure of PKS11.2. (B) Phylogenetic analysis of PKS11.2. PKS11.2 grouped with the melanin biosynthesis PKSs. The accession numbers of all used PKSs were listed in Supplementary Table S3. (C) Colonial morphology of C. empetri MEFC009-Δku80-neo and C. empetri MEFC009-Δpks11.2. (D) Genotype verification of transformants using primers Upks11.2-F/Dpks11.2-R. Lane M: 1kb DNA marker; lane 1–8: C. empetri MEFC009-Δpks11.2; lane 9: parent strain C. empetri MEFC009-Δku80-neo.
Conclusion
C. empetri was applied in the industrial production of FR901379, the precursor compound of antifungal agent micafungin. The studies of FR901379 biosynthesis were hampered by the poor genetic transformation system. Given that, we developed an efficient genetic manipulation system in the industrial FR901379-producing strain C. empetri MEFC009, including protoplast-mediated transformation method, essential genetic elements of selectable markers and promoters, and chassis cell with high gene-targeting frequency. Based on this genetic manipulation system, we identified the biosynthetic pks gene of DHN-melanin in C. empetri. This efficient genetic manipulation system will facilitate the research of FR901379 biosynthetic mechanism and further sequential metabolic engineering. And, it is also an indispensable platform for the genome mining of secondary metabolites in C. empetri.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XL and XH conceived the project and supervised the research. XH and PM designed the experiments. PM, MW, and JL performed the fungal genetic and fermentation experiments. XL, XH, and PM analyzed all data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (nos. U2032139 and 32070056). XL and XH are supported by the Shandong Taishan Scholarship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.734780/full#supplementary-material
References
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Blumhoff, M., Steiger, M. G., Marx, H., Mattanovich, D., and Sauer, M. (2013). Six novel constitutive promoters for metabolic engineering of Aspergillus niger. Appl. Microbiol. Biotechnol. 97, 259–267. doi: 10.1007/s00253-012-4207-9
Cacho, R. A., Jiang, W., Chooi, Y. H., Walsh, C. T., and Tang, Y. (2012). Identification and characterization of the echinocandin B biosynthetic gene cluster from Emericella rugulosa NRRL 11440. J. Am. Chem. Soc. 134, 16781–16790. doi: 10.1021/ja307220z
Cao, Y., Jiao, R., and Xia, Y. (2012). A strong promoter, PMagpd, provides a tool for high gene expression in entomopathogenic fungus, Metarhizium acridum. Biotechnol. Lett. 34, 557–562. doi: 10.1007/s10529-011-0805-3
Chazotte, B. (2011). Labeling nuclear DNA using DAPI. Cold Spring Harb Protoc 2011:pdb.prot5556. doi: 10.1101/pdb.prot5556
Chen, L., Yue, Q., Zhang, X., Xiang, M., Wang, C., Li, S., et al. (2013). Genomics-driven discovery of the pneumocandin biosynthetic gene cluster in the fungus Glarea lozoyensis. BMC Genomics 14:339. doi: 10.1186/1471-2164-14-339
Denning, D. W. (2003). Echinocandin antifungal drugs. Lancet 362, 1142–1151. doi: 10.1016/s0140-6736(03)14472-8
Eisenman, H. C., and Casadevall, A. (2012). Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 93, 931–940. doi: 10.1007/s00253-011-3777-2
Feng, B., Wang, X., Hauser, M., Kaufmann, S., Jentsch, S., Haase, G., et al. (2001). Molecular cloning and characterization of WdPKS1, a gene involved in dihydroxynaphthalene melanin biosynthesis and virulence in Wangiella (Exophiala) dermatitidis. Infect. Immun. 69, 1781–1794. doi: 10.1128/IAI.69.3.1781-1794.2001
Gu, S., Li, J., Chen, B., Sun, T., Liu, Q., Xiao, D., et al. (2018). Metabolic engineering of the thermophilic filamentous fungus Myceliophthora thermophila to produce fumaric acid. Biotechnol. Biofuels 11:323. doi: 10.1186/s13068-018-1319-1
Hashimoto, S. (2009). Micafungin: a sulfated echinocandin. J. Antibiot. 62, 27–35. doi: 10.1038/ja.2008.3
Hršelová, H., Hujslová, M., and Gryndler, M. (2015). Genetic transformation of extremophilic fungi Acidea extrema and Acidothrix acidophila. Folia Microbiol. 60, 365–371. doi: 10.1007/s12223-015-0398-7
Huang, X., Chen, M., Li, J., and Lu, X. (2016). Establishing an efficient gene-targeting system in an itaconic-acid producing Aspergillus terreus strain. Biotechnol. Lett. 38, 1603–1610. doi: 10.1007/s10529-016-2143-y
Huang, X., Lu, X., and Li, J. J. (2014). Cloning, characterization and application of a glyceraldehyde-3-phosphate dehydrogenase promoter from Aspergillus terreus. J. Ind. Microbiol. Biotechnol. 41, 585–592. doi: 10.1007/s10295-013-1385-0
Hüttel, W. (2017). Structural diversity in echinocandin biosynthesis: the impact of oxidation steps and approaches toward an evolutionary explanation. Z. Naturforsch. C J. Biosci. 72, 1–20. doi: 10.1515/znc-2016-0156
Kaminskyj, S. G., and Hamer, J. E. (1998). hyp loci control cell pattern formation in the vegetative mycelium of Aspergillus nidulans. Genetics 148, 669–680. doi: 10.1093/genetics/148.2.669
Marcone, G. L., Carrano, L., Marinelli, F., and Beltrametti, F. (2010). Protoplast preparation and reversion to the normal filamentous growth in antibiotic-producing uncommon actinomycetes. J. Antibiot. 63, 83–88. doi: 10.1038/ja.2009.127
Meyer, V. (2008). Genetic engineering of filamentous fungi--progress, obstacles and future trends. Biotechnol. Adv. 26, 177–185. doi: 10.1016/j.biotechadv.2007.12.001
Michielse, C. B., Hooykaas, P. J., van den Hondel, C. A., and Ram, A. F. (2008). Agrobacterium-mediated transformation of the filamentous fungus Aspergillus awamori. Nat. Protoc. 3, 1671–1678. doi: 10.1038/nprot.2008.154
Moriwaki, A., Kihara, J., Kobayashi, T., Tokunaga, T., Arase, S., and Honda, Y. (2004). Insertional mutagenesis and characterization of a polyketide synthase gene (PKS1) required for melanin biosynthesis in Bipolaris oryzae. FEMS Microbiol. Lett. 238, 1–8. doi: 10.1016/j.femsle.2004.07.007
Ninomiya, Y., Suzuki, K., Ishii, C., and Inoue, H. (2004). Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. U. S. A. 101, 12248–12253. doi: 10.1073/pnas.0402780101
Patil, A., and Majumdar, S. (2017). Echinocandins in antifungal pharmacotherapy. J. Pharm. Pharmacol. 69, 1635–1660. doi: 10.1111/jphp.12780
Prabha, V. L., and Punekar, N. S. (2004). Genetic transformation in Aspergilli: tools of the trade. Indian J. Biochem. Biophys. 41, 205–215.
Punt, P. J., Zegers, N. D., Busscher, M., Pouwels, P. H., and van den Hondel, C. A. (1991). Intracellular and extracellular production of proteins in Aspergillus under the control of expression signals of the highly expressed Aspergillus nidulans gpdA gene. J. Biotechnol. 17, 19–33. doi: 10.1016/0168-1656(91)90024-P
Ramsden, R., Arms, L., Davis, T. N., and Muller, E. G. (2011). An intein with genetically selectable markers provides a new approach to internally label proteins with GFP. BMC Biotechnol. 11:71. doi: 10.1186/1472-6750-11-71
Roth, M. G., and Chilvers, M. I. (2019). A protoplast generation and transformation method for soybean sudden death syndrome causal agents Fusarium virguliforme and F. brasiliense. Fungal Biol. Biotechnol. 6:7. doi: 10.1186/s40694-019-0070-0
Ruiz-Díez, B. (2002). Strategies for the transformation of filamentous fungi. J. Appl. Microbiol. 92, 189–195. doi: 10.1046/j.1365-2672.2002.01516.x
Schumacher, J. (2016). DHN melanin biosynthesis in the plant pathogenic fungus Botrytis cinerea is based on two developmentally regulated key enzyme (PKS)-encoding genes. Mol. Microbiol. 99, 729–748. doi: 10.1111/mmi.13262
Streatfield, S. J., Toews, S., and Roberts, C. F. (1992). Functional analysis of the expression of the 3'-phosphoglycerate kinase pgk gene in Aspergillus nidulans. Mol. Gen. Genet. 233, 231–240. doi: 10.1007/BF00587584
Wei, T. Y., Wu, Y. J., Xie, Q. P., Tang, J. W., Yu, Z. T., Yang, S. B., et al. (2020). CRISPR/Cas9-based genome editing in the filamentous fungus Glarea lozoyensis and its application in manipulating gloF. ACS Synth. Biol. 9, 1968–1977. doi: 10.1021/acssynbio.9b00491
Keywords: protoplast transformation, promoter, marker, nonhomologous end-joining, Coleophoma empetri, melanin biosynthesis, micafungin
Citation: Men P, Wang M, Li J, Geng C, Huang X and Lu X (2021) Establishing an Efficient Genetic Manipulation System for Sulfated Echinocandin Producing Fungus Coleophoma empetri. Front. Microbiol. 12:734780. doi: 10.3389/fmicb.2021.734780
Edited by:
Lei Chen, Tianjin University, ChinaReviewed by:
Hua Yuan, Shanghai Normal University, ChinaWolfgang Hüttel, University of Freiburg, Germany
Copyright © 2021 Men, Wang, Li, Geng, Huang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuefeng Lu, bHZ4ZkBxaWJlYnQuYWMuY24=; Xuenian Huang, aHVhbmd4bkBxaWJlYnQuYWMuY24=
 Ping Men
Ping Men Min Wang
Min Wang Jinda Li
Jinda Li Ce Geng
Ce Geng Xuenian Huang
Xuenian Huang Xuefeng Lu
Xuefeng Lu