- 1Key Laboratory of Cultivation and Protection for Non-Wood Forest Trees, Ministry of Education, Central South University of Forestry and Technology, Changsha, China
- 2Department of Agricultural Soil Science, Department of Soil Science of Temperate Ecosystems, Georg-August-Universität Göttingen, Göttingen, Germany
- 3Agro-Technological Institute, RUDN University, Moscow, Russia
- 4Institute of Environmental Sciences, Kazan Federal University, Kazan, Russia
- 5College of Food and Agriculture Sciences, King Saud University, Riyadh, Saudi Arabia
- 6Texas A&M AgriLife Research and Extension Center at Dallas, Texas A&M University, Dallas, TX, United States
Oil tea (Camellia spp.) is endemic to the hilly regions in the subtropics. Camellia yuhsienensis is resistant to diseases such as anthracnose and root rot, while Camellia oleifera is a high-yield species but susceptible to these diseases. We hypothesize that differences in the rhizosphere microbial communities and functions will elucidate the resistance mechanisms of these species. We used high-throughput sequencing over four seasons to characterize the rhizosphere microbiome of C. oleifera (Rhizo-Sus) and C. yuhsienensis (Rhizo-Res) and of the bulk soil control (BulkS). In Rhizo-Res, bacterial richness and diversity (Shannon index) in autumn and winter were both higher than that in Rhizo-Sus. In Rhizo-Res, fungal richness in autumn and winter and diversity in summer, autumn, and winter were higher than that in Rhizo-Sus. The seasonal variations in bacterial community structure were different, while that of fungal community structure were similar between Rhizo-Res and Rhizo-Sus. Gram-positive, facultatively anaerobic, and stress-tolerant bacteria were the dominant groups in Rhizo-Sus, while Gram-negative bacteria were the dominant group in Rhizo-Res. The significant differences in bacterial and fungal functions between Rhizo-Sus and Rhizo-Res were as follows: (1) in Rhizo-Sus, there were three bacterial and four fungal groups with plant growth promoting potentials, such as Brevibacterium epidermidis and Oidiodendron maius, and one bacterium and three fungi with pathogenic potentials, such as Gryllotalpicola sp. and Cyphellophora sessilis; (2) in Rhizo-Res, there were also three bacteria and four fungal groups with plant-growth-promoting potentials (e.g., Acinetobacter lwoffii and Cenococcum geophilum) but only one phytopathogen (Schizophyllum commune). In summary, the rhizosphere microbiome of disease-resistant C. yuhsienensis is characterized by a higher richness and diversity of microbial communities, more symbiotic fungal communities, and fewer pathogens compared to the rhizosphere of high-yield but disease-susceptible C. oleifera.
Introduction
The rhizosphere, where the soil volume is directly affected by roots, is a narrow zone with high abundance of microorganisms and is one of the most complex ecosystems on Earth (Kuzyakov and Razavi, 2019; Vetterlein et al., 2020). The function of many microbial groups is to facilitate plant growth and rhizosphere health through their functions. Four important beneficial microbial groups are commonly found in the rhizosphere: (i) nitrogen-fixing bacteria, (ii) mycorrhizal fungi, (iii) plant-growth-promoting rhizobacteria (PGPR), and (iv) biocontrol microorganisms, with the ability to protect plant roots against pathogens through excretion of metabolites or via competition for niche (Mendes et al., 2013). Deleterious microorganisms can invade root tissues, cause diseases or retard root growth, and reduce yield through the excretion of toxic metabolites (Kremer, 2006). Both beneficial and deleterious microorganisms coexist in the rhizosphere. For example, Bacillus megaterium is present in the rhizosphere of Camellia sinensis and has the ability to promote plant growth by solubilizing phosphate, producing indole acetic acid (IAA) and siderophores, and protecting plants against brown root rot disease, caused by rhizosphere fungus Fomes lamaoensis (Chakraborty et al., 2006; Huang et al., 2019).
Rhizosphere microorganisms are affected directly by the host plant and indirectly by seasonal rainfall, temperature, and light intensity, as these factors modify the rhizodeposition (Kuzyakov and Cheng, 2001; Taketani et al., 2016; Zhang X. et al., 2020). Plant diseases caused by microorganisms are often associated with extreme drought or rainfall (Meisner and Boer, 2018). Pathogens can tolerate the negative effects of drought better than plants and infect the stressed plants through synthesis of toxins and cell-wall-degrading enzymes (Dikilitas et al., 2016). Most bacteria that are better adapted to droughts and other resource-limiting conditions are Gram-positive and are K-strategists or oligotrophs. In contrast, Gram-negative bacteria (mainly r-strategists) have an advantage under nutrient-rich conditions in wet soil (Chen et al., 2016).
Oil tea is an indispensable woody oilseed plant that is endemic to the hilly areas in subtropical climate (Liu et al., 2017), growing on soils with low pH, low organic matter, and limited phosphorous content (Li et al., 2019). Low yields of Camellia, caused by barren ground, nutrient shortage, and plant diseases and pests, have been the predominant limiting factor for the development of the oil tea industry for many years. After years of breeding, some nutrient-efficient and disease-resistant species or cultivars have been selected. Camellia oleifera “Huashuo” is a widely planted oil tea species with high yield (Tan et al., 2011), but it is disease sensitive and susceptible to anthracnose, root knot nematode, and root rot diseases (Wang et al., 2017; Xu et al., 2020; Zhu et al., 2020). Camellia yuhsienensis, another important oil tea species, is resistant to these diseases (Yang et al., 2004; Wei et al., 2013). According to previous studies, the yield of C. oleifera is much higher than C. yuhsienensis (Zou, 2010; Tan et al., 2011), while the disease resistance of C. yuhsienensis is much better than C. oleifera (Supplementary Table 1; Yang et al., 2004; Jin et al., 2009; Chen et al., 2021). Nutrient absorption from the soil and plant disease resistance is also affected by the soil microbiome, especially the rhizosphere microorganisms (Bob et al., 1987; Jansson and Hofmockel, 2020). Although the rhizosphere microbial communities have been investigated in C. oleifera (Zhang P. et al., 2020) and C. yuhsienensis (Li et al., 2020), it remains difficult to compare differences in rhizosphere microbial community structures and functions due to inconsistent climate, soil conditions, and management practices. More importantly, none of the previous studies have reported on the microbial functions and their role in oil tea disease susceptibility and growth.
Because of the self-pollination incompatibility in oil tea species, multiple cultivars are planted in the same orchard (Zhou et al., 2020). Plants are able to shape or modify their rhizosphere microbiome through the excretion of specific secondary metabolites, even when grown in one planation (Berendsen et al., 2012). Therefore, we hypothesized that, in one planation (same climate, soil type, and management), C. oleifera and C. yuhsienensis each developed unique rhizosphere microbial communities, and the differences in rhizosphere microbial diversity, structure, and functions explain the different resistances to disease in these two Camellia species. High-throughput sequencing of taxonomic composition and functions of the rhizosphere microbiome were used to verify our hypotheses.
Materials and Methods
Site Description
The sampling site was in Wangcheng district, Changsha City, Hunan, China (N 28°30′, E 113°20′). To study the effect of mixed planation on growth, health, and production of oil tea, two 1-year-old Camellia oil species, namely, C. yuhsienensis and C. oleifera “Huashuo,” were planted in 2011 at the sampling site at a 1:1 ratio. Plant spacing was 2 m within rows, and row spacing was 3 m. The climate is a subtropical monsoon with mean annual rainfall and temperature of 1,370 mm and 17°C, respectively. The soil at the experimental site is a Quaternary red clay (classified as Lixisols in the World Reference Base for Soil Resources) with a pH of 4.3. Total organic carbon content (TOC) was 11.5 ± 0.6 g⋅kg–1, and total nitrogen (TN) content was 870 ± 25 mg⋅kg–1. Available phosphorus (AP) content was 4.9 ± 1.7 mg⋅kg–1 and available potassium (AK) content was 134 ± 11 mg⋅kg–1 (Supplementary Table 2). The understory contains some wild weeds, which were mowed twice a year.
Experimental Design and Sampling
Growth characteristics of C. oleifera and C. yuhsienensis were collected 7 years after planting (Supplementary Table 1). Soil samples were collected at the fruit enlargement stage (October 23, 2018, autumn, Au), dormancy stage (January 19, 2019, winter, Wi), spring shoot growth stage (April 5, 2019, spring, Sp), and flower bud rapid growth stage (July 22, 2019, summer, Su). Three separate plots of 20 m × 20 m were selected in the mixed plantation. Five healthy trees of each species were selected using an “S” type design from the center of each quadrat. Four points in each quadrat were selected at 0.5–1 m distance from the trunk, and samples of the soil rhizosphere of C. oleifera (Rhizo-Sus) and C. yuhsienensis (Rhizo-Res) were collected as described by Koranda et al. (2011). Samples of a corresponding bulk soil (BulkS) at 0–20 cm depth were also collected as a reference control in the middle of the rows, approximately 1.5 m distance from the trunk. Subsamples of the rhizosphere samples or the bulk soil were mixed well and combined into one sample. Soil samples were stored in dry ice and transported back to the laboratory for further processing. After removal of debris and roots, samples were mixed well, ground, and ran through a sieve (<2 mm). One portion of each soil sample was used to determine the physicochemical properties after air-drying in the shade, while the remainder portion was stored at −80°C for high-throughput sequencing at Genedenovo Biotechnology Co., Ltd. (Guangzhou, China).
The soil physicochemical properties were measured according to Li et al. (2020). Soil temperature at 0–20 cm depth was measured using a Wdsen Electronic temperature recorder (iButton DS1925) during the entire experimental period (Supplementary Figure 1).
Bacterial and Fungal Community Assessment
DNA Extraction and PCR Amplification
A soil sample of 2–3 g dry weight was used for microbial DNA extraction using HiPure Soil DNA Kits (Magen, Guangzhou, China) according to manufacturer’s protocol. The 16S rDNA V3–V4 region of the ribosomal RNA gene was amplified by PCR using primer 341F, 5′-CCTACGGGNGGCWGCAG-3′, and 806R, 5′-GGACTACHVGGGTATCTAAT-3′. The PCR amplification of 16S rDNA was conducted as described by Li et al. (2018). The internal transcribed spacer (ITS) rDNA region of the ribosomal RNA gene was amplified by PCR using primers ITS3-KYO2 (F), 5′-GATGAAGAACGYAGYAA-3′, and ITS4 (R), 5′-TCCTCCGCTTATTGATATGC-3′ (Toju et al., 2012). The ITS region of the eukaryotic ribosomal RNA gene was amplified by PCR (95°C for 2 min, followed by 27 cycles at 98°C for 10 s, 62°C for 30 s, and 68°C for 30 s, and a final extension at 68°C for 10 min). The PCR were performed in triplicate using 50-μl mixtures containing 5 μl of 10 × KOD buffer, 5 μl of 2 mM dNTPs, 3 μl of 25 mM MgSO4, 1.5 μl of each primer (10 μM), 1 μl of KOD polymerase, and 100 ng of template DNA.
Illumina Novaseq6000 Sequencing
Amplicons were extracted from 2% agarose gels and purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) according to the manufacturer’s instructions. The qualified amplicon mixture was then sequenced on the Illumina Novaseq6000 platform to generate 2 × 250 bp paired-end reads. The raw reads were deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (accession number PRJNA742848). The link of this BioProject is https://dataview.ncbi.nlm.nih.gov/object/PRJNA742848?reviewer=haouuog1lqfrksad66m99e11p5.
Statistical and Bioinformatics Analysis
Raw reads were further filtered using FASTP by removing reads containing more than 10% unknown nucleotides (N) and reads with < 50% of bases with quality scores (Q-value) > 20 (Chen et al., 2018). Paired-end clean reads were merged as raw tags using FLASH (version 1.2.11) with a minimum overlap of 10 bp and mismatch error rates of 2% (Magoè and Salzberg, 2011). The effective tags were clustered into operational taxonomic units (OTUs) of ≥ 97% similarity using the UPARSE (version 9.2.64) pipeline (Edgar, 2013). The tag sequence with the highest abundance was selected as the representative sequence within each cluster. The tags and OTUs of the bacteria and fungi are presented in Supplementary Tables 3, 4. To ensure the reproducibility and validity of the microbial data, the data extraction flat and dilution curves were processed before analysis (Liang et al., 2017). The dilution curve of the Sobs index indicated the presence of more bacteria and fungi if sequencing was continued, but the dilution curve plateau of the Shannon’s index was reached early, indicating that the number of reads was sufficient for this research (Supplementary Figure 2).
Venn analysis, used to identify unique and common OTUs and species among different compartments, was performed using the R “VennDiagram” package (version 1.6.16) (Chen and Boutros, 2011). The OTU rarefaction curves and rank abundance curves were plotted using the R “ggplot2” package (version 2.2.1) (Wickham, 2006). Alpha diversities of bacteria and fungi (Sobs, Shannon, and Chao 1 index) were calculated in QIIME (version 1.9.1) (Caporaso et al., 2010), and differences in the alpha diversities of bacteria and fungi among treatments were calculated using one-way analysis of variance (ANOVA). Principal coordinates analysis (PCoA), permutational multivariate analysis of variance (MANOVA) (Permanova), and cluster dendrogram based on unweighted uniFrac distances were used to evaluate the influence of plant roots and seasonal dynamics on the bacterial and fungal community structures. The unweight uniFrac distance matrix and Permanova was calculated in R project GuniFrac package (version 1.0) and Vegan package (version 2.5.3), respectively. Ape and Vegan packages were used for PCoA analysis, and ggplot2 package was used for visualization in R. A least discriminant analysis (LDA) effect size (LEfSe) taxonomic cladogram was used to identify specific (LDA > 3.0) bacteria and fungi (LDA > 3.0) in the treatments using LEfSe software (Segata et al., 2011). Ternary plots detected by Kruskal–Wallis H analysis were used to determine the specific species among Rhizo-Sus, Rhizo-Res, and BulkS during the entire year using the R “labdsv” package (version 2.0-1) and R “ggplot” (Roberts, 2016). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the OTUs for prediction of bacterial functions was inferred using Tax4Fun (version 1.0) (Aßhauer et al., 2015). Microbiome phenotypes of bacteria were classified using BugBase (Ward et al., 2017). The functional group (Guild) of the fungi (relative abundance > 0.01% in the rhizosphere) was inferred using FUNGuild (version 1.0) (Nguyen et al., 2016). The bacterial functions and the fungal trophic modes among Rhizo-Sus, Rhizo-Res, and BulkS were calculated by Wilcoxon rank sum test. The fungal functional groups (guild) among Rhizo-Sus, Rhizo-Res, and BulkS were calculated using the Kruskal–Wallis H test.
Among treatments were calculated using one-way analysis of variance was used to determine the differences in soil physicochemical properties among Rhizo-Sus, Rhizo-Res, and BulkS during the entire year. Pearson’s correlation coefficients between soil physicochemical properties and alpha diversities were calculated by Omicshare tools, a free online platform for data analysis (Denovo, 2021).
Results
Soil Microbial Community Structure
Microbial Composition and Diversity
Regardless of whether the soil sampled was bulk or rhizosphere, the dominant bacteria phyla were Chloroflexi (14–43% of total bacteria sequencing), Acidobacteria (8–35%), Proteobacteria (12–28%), Actinobacteria (9–29%), and Planctomycetes (5–13%) (Supplementary Figure 3A). The dominant fungi were Ascomycota (61–96% of total fungi sequencing) and Basidiomycota (2–34%) (Supplementary Figure 3B).
Bacterial richness and diversity were higher in Rhizo-Res than in Rhizo-Sus in autumn and winter (p < 0.05, Supplementary Table 5). Fungal diversity was higher in Rhizo-Res than in Rhizo-Sus in summer and autumn (p < 0.05). Over a 1-year period, the fungal diversity in Rhizo-Res and BulkS was higher than in Rhizo-Sus (p < 0.05) (Figure 1F).
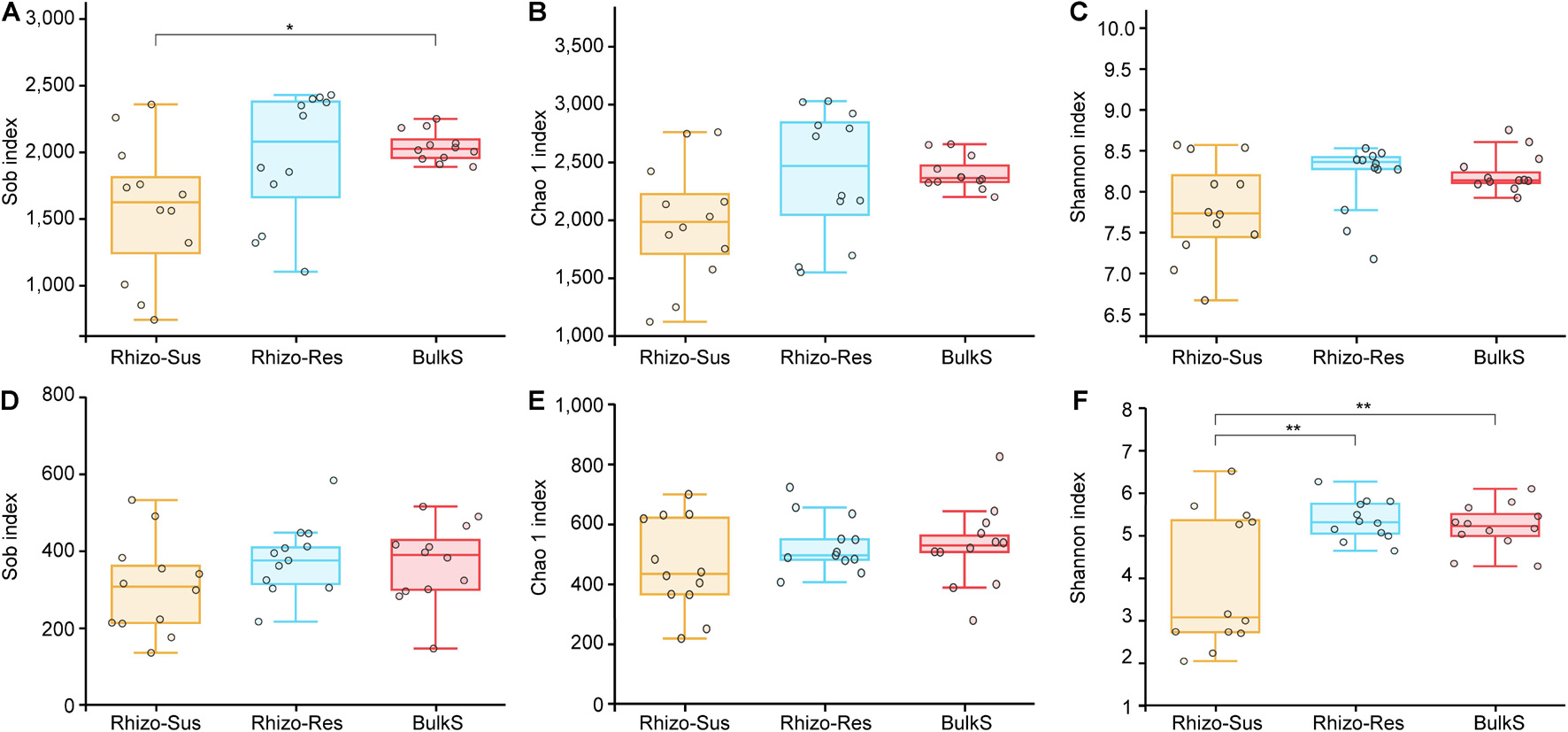
Figure 1. Box plots of bacterial (A–C) and fungal (D–F) Sobs and Chao 1 and Shannon index in disease susceptible C. oleifera rhizosphere (Rhizo-Sus, yellow), disease resistant C. yuhsienensis rhizosphere (Rhizo-Res, blue), and bulk soil (BulkS, red). The line in the box indicates the median of each compartment. The line over and below the box indicates the maximum and minimum value, respectively. The top and bottom line of the box indicates the upper and lower quartile, respectively. The circles indicate each sample. The * and ** over the boxes indicate the significant differences based on Tukey’s HSD test among compartments at p < 0.05 and p < 0.01, respectively.
Both season and Camellia species had a marked influence on bacterial and fungal community structure (p < 0.05) (Figure 2 and Supplementary Figure 4). According to the cluster dendrogram (Figure 2), the bacterial community structure in Rhizo-Sus was different from Rhizo-Res and BulkS in winter. In summer, the bacterial community in Rhizo-Res was different from Rhizo-Sus and BulkS. The fungal community structures were similar among Rhizo-Sus, Rhizo-Res, and BulkS during the year, except in winter (Supplementary Figure 4). Over the whole year, the trend of bacterial community development in Rhizo-Res was similar to BulkS (along with axis PCO2) but different from that in Rhizo-Sus (along with axis PCO1). The trend of fungal community structure in Rhizo-Res was similar to Rhizo-Sus (along with axis PCO1, Supplementary Figure 4).
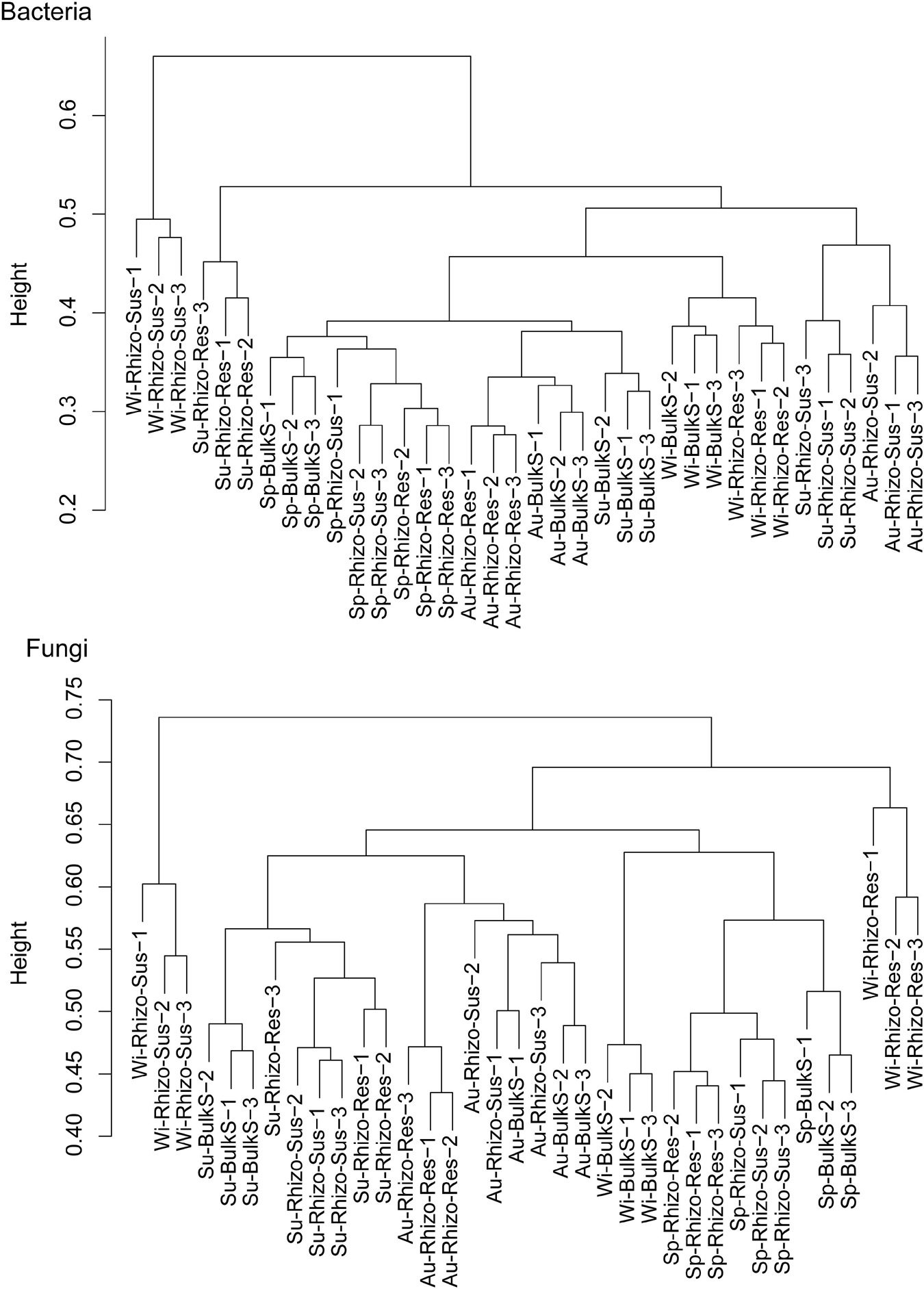
Figure 2. Cluster dendrogram of bacterial and fungal communities based on unweighted uniFrac distance. Sp, Su, Au, and Wi indicates spring, summer, autumn, and winter, respectively. Rhizo-Sus, Rhizo-Res, and BulkS indicate disease-susceptible C. oleifera rhizosphere, disease-resistant C. yuhsienensis rhizosphere, and bulk soil, respectively, N = 3. Height in cluster dendrogram indicates the distance of microbial communities among each sample.
Specific Microbial Species in the Rhizosphere of Oil Tea Plants
Species with higher relative abundance in one rhizosphere or bulk soil (p < 0.05) (Figure 3) or species unique to Rhizo-Sus, Rhizo-Res, and BulkS (Figure 4) are defined as specific microorganisms. According to Ternary plots (Figure 3), the specific species across the year in Rhizo-Sus were Bdellovibrionales bacterium RBG 16 40 8 and Humibacter ginsengisoli bacteria, and Oidiodendron maius, Oidiodendron truncatum, Pyrenochaetopsis leptospora, and Verticillium leptobactrum fungi. The specific bacteria in Rhizo-Res were Burkholderia sp., Rhizobiales bacterium GAS113, and Rudaea cellulosilytica DSM 22992, and Bifiguratus adelaidae fungus.
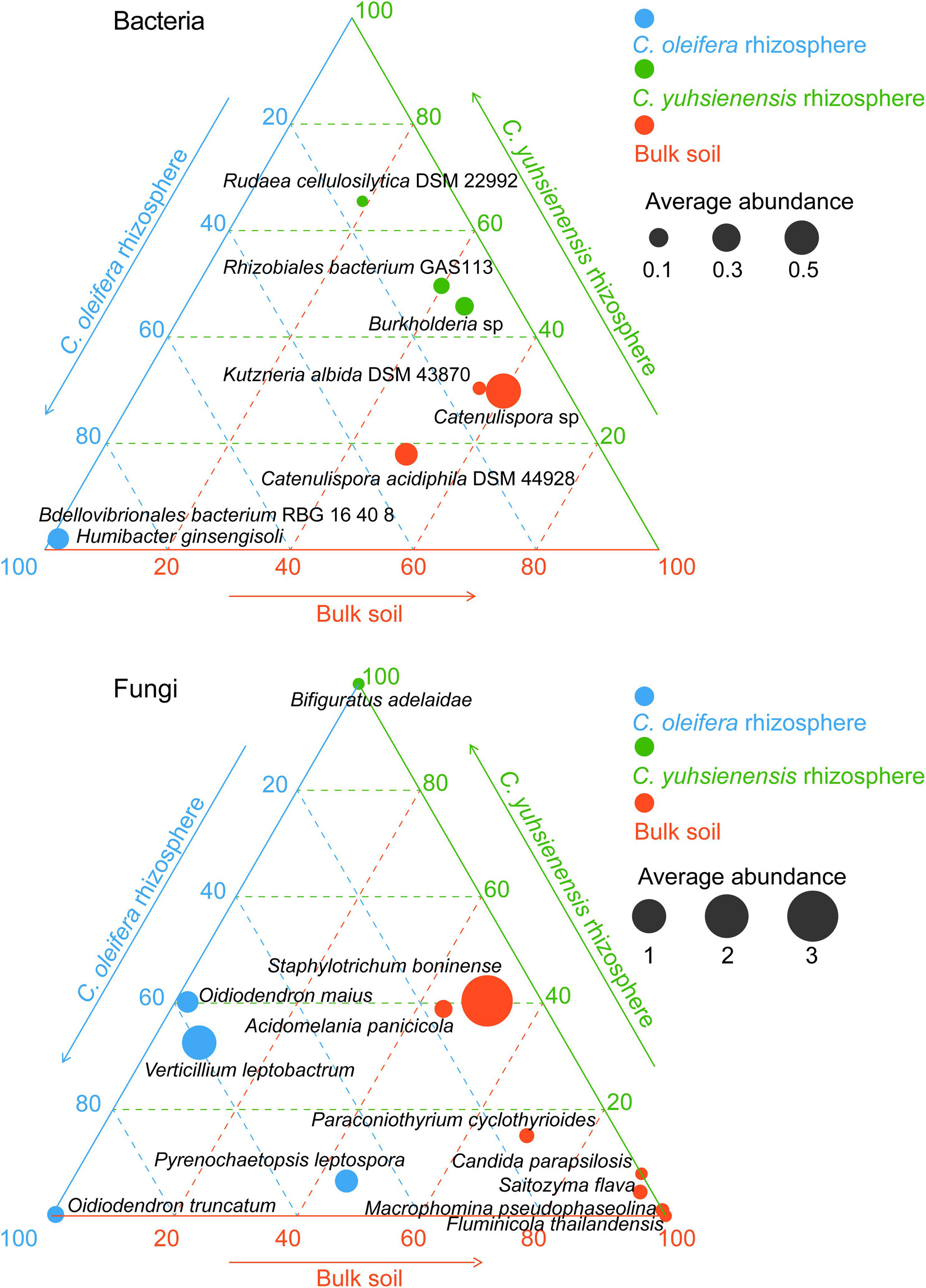
Figure 3. Distribution of specific bacterial and fungal species across disease-susceptible C. oleifera rhizosphere (blue), disease-resistant C. yuhsienensis rhizosphere (green), and bulk soil (red) in ternary plots. The circle size and position indicates the average relative abundance (%) and the ratio of relative abundance of each microbial group with C. oleifera rhizosphere, C. yuhsienensis rhizosphere, and bulk soil (conducted using Kruskal–Wallis analysis, p < 0.05), respectively, N = 12.
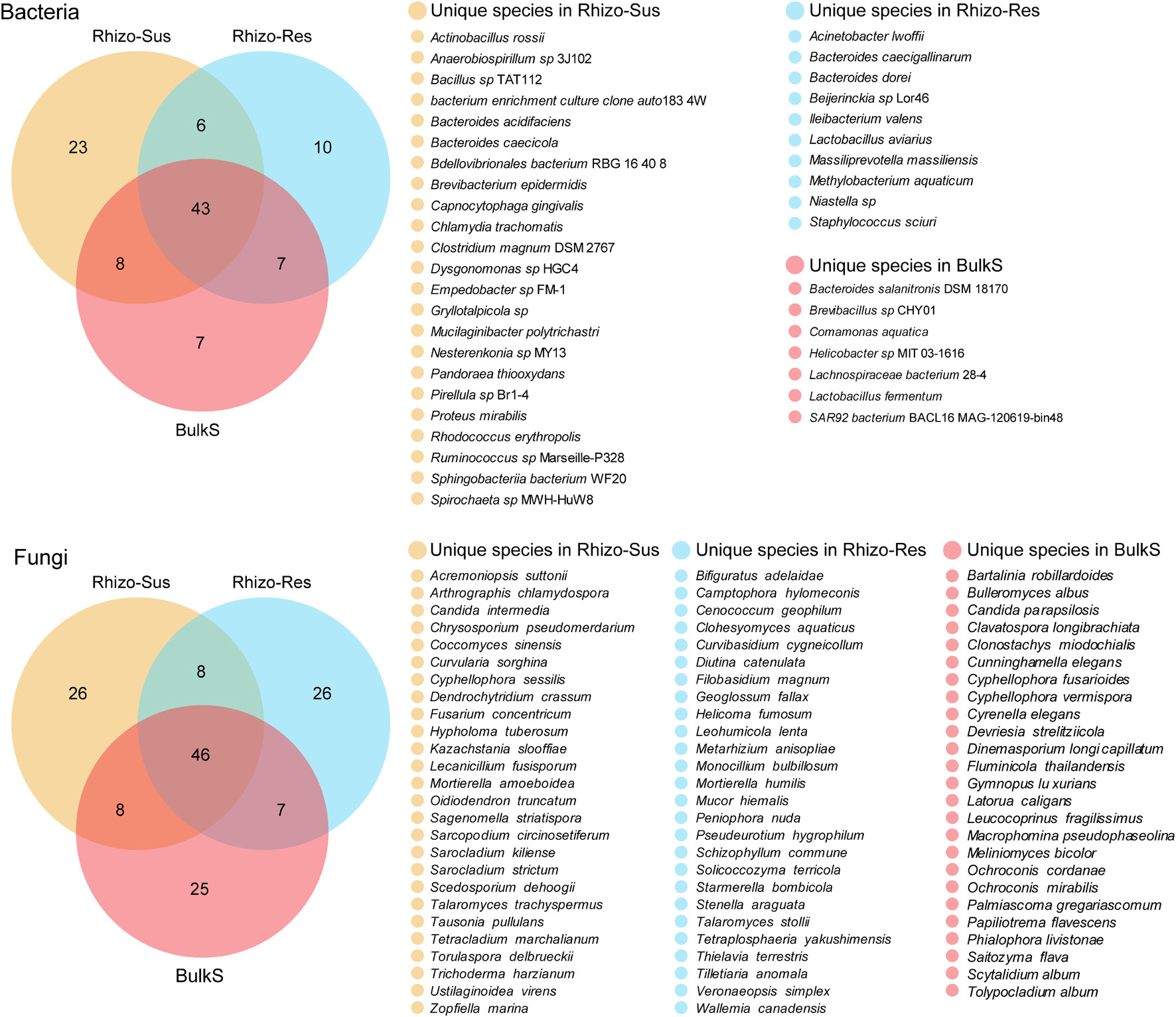
Figure 4. Venn diagram of bacteria and fungi at species level among C. oleifera rhizosphere (Rhizo-Sus), C. yuhsienensis rhizosphere (Rhizo-Res), and bulk soil (BulkS), N = 12. Yellow, blue, and red color indicate the unique species that are only presented in Rhizo-Sus, Rhizo-Res, or BulkS, respectively. The cross-color block indicates the shared species between Rhizo-Sus and Rhizo-Res, or Rhizo-Sus and BulkS, or Rhizo-Res and BulkS, or among Rhizo-Sus, Rhizo-Res, and BulkS. The quantity of unique and shared species was written in each color block of Venn diagram, respectively.
Analysis of Microbial Functions
Microbial functions in the rhizosphere and bulk soil were defined based on KEGG and BugBase. We use the BugBase database term “phenotype,” which predicts organism-level microbiome phenotypes partly corresponding to microbial functional groups. The KEGG and phenotype heatmaps indicated that the functions and phenotypes of the bacteria in Rhizo-Res were like those in BulkS but different from those in Rhizo-Sus (Figure 5). The relative abundance of genes responsible for carbohydrate metabolism (starch and sucrose, pyruvate, and fructose and mannose) and for energy metabolism (methane metabolism) were higher in Rhizo-Sus than in Rhizo-Res (p < 0.05). The relative abundance of genes responsible for oxidative phosphorylation was higher in Rhizo-Res than in Rhizo-Sus. The microbiome phenotype heatmap based on BugBase showed that Gram-negative group in Rhizo-Res was higher than in Rhizo-Sus (p < 0.05). Gram-positive and stress-tolerant and facultatively anaerobic groups were higher in Rhizo-Sus than in Rhizo-Res (p < 0.05).
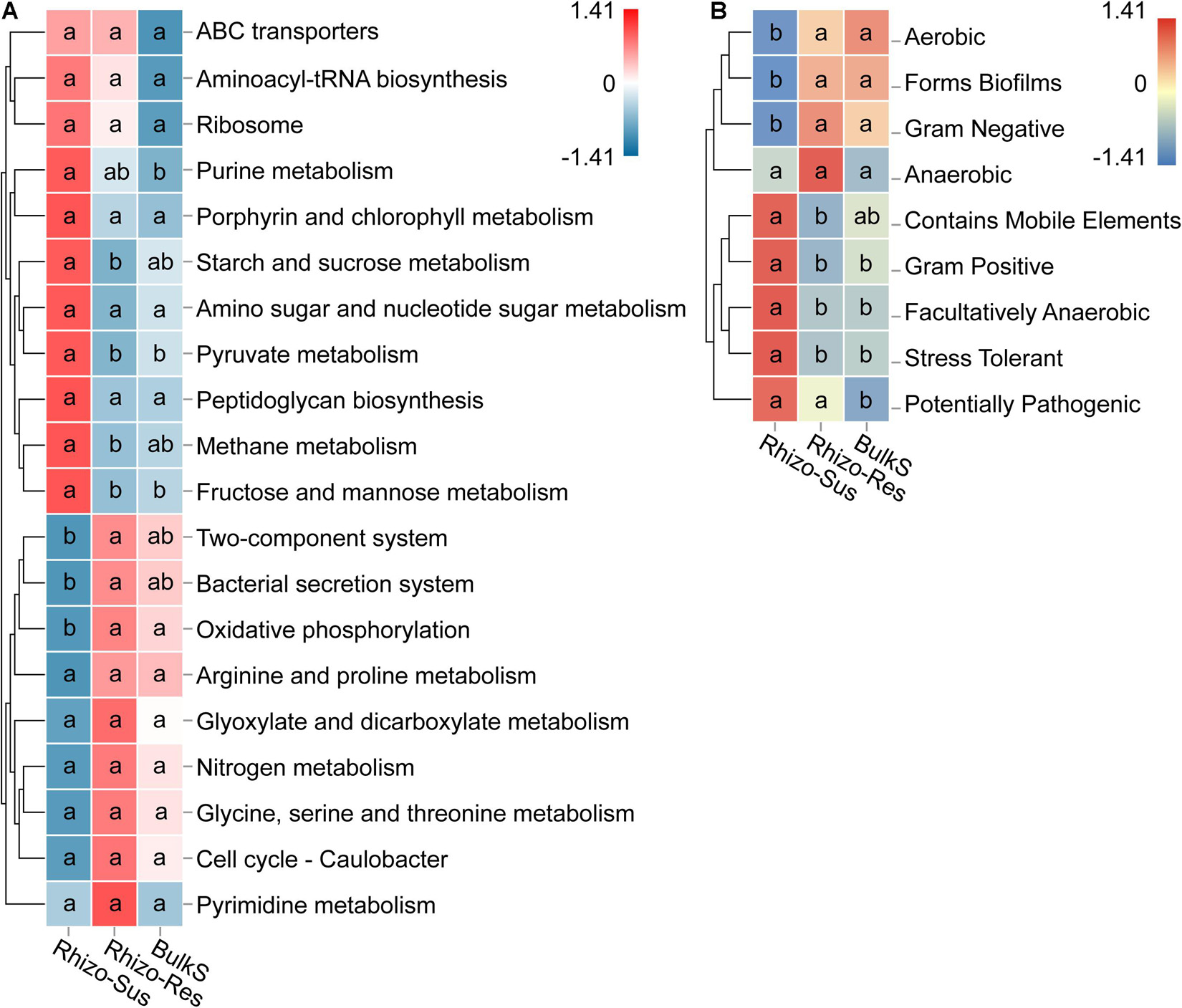
Figure 5. Heatmap of main functions based on relative abundance of bacterial KEGG prediction (A) using Tax4Fun and phenotype (B) using BugBase in rhizosphere of C. oleifera (Rhizo-Sus) and C. yuhsienensis (Rhizo-Res) and bulk soil (BulkS) samples. The abundance of functions was normalized by Z-score method. The connecting lines on the left side describe the clustering of each function. The closer the color to red, the more dominant the function is. The lowercase in each row of heatmap indicated the significant differences among Rhizo-Sus, Rhizo-Res, and BulkS (conducted using Wilcoxon rank sum test, p < 0.05), N = 12.
The analysis of fungal functional groups (guilds) and trophic mode showed that endophytes, plant pathogens, and mycorrhizal guilds were present in both Rhizo-Sus and Rhizo-Res (Figure 6). The symbiotrophs and saprotrophs in Rhizo-Res were higher in Rhizo-Sus (p < 0.05), but pathotrophs were similar between Rhizo-Sus and Rhizo-Res (Figure 6).
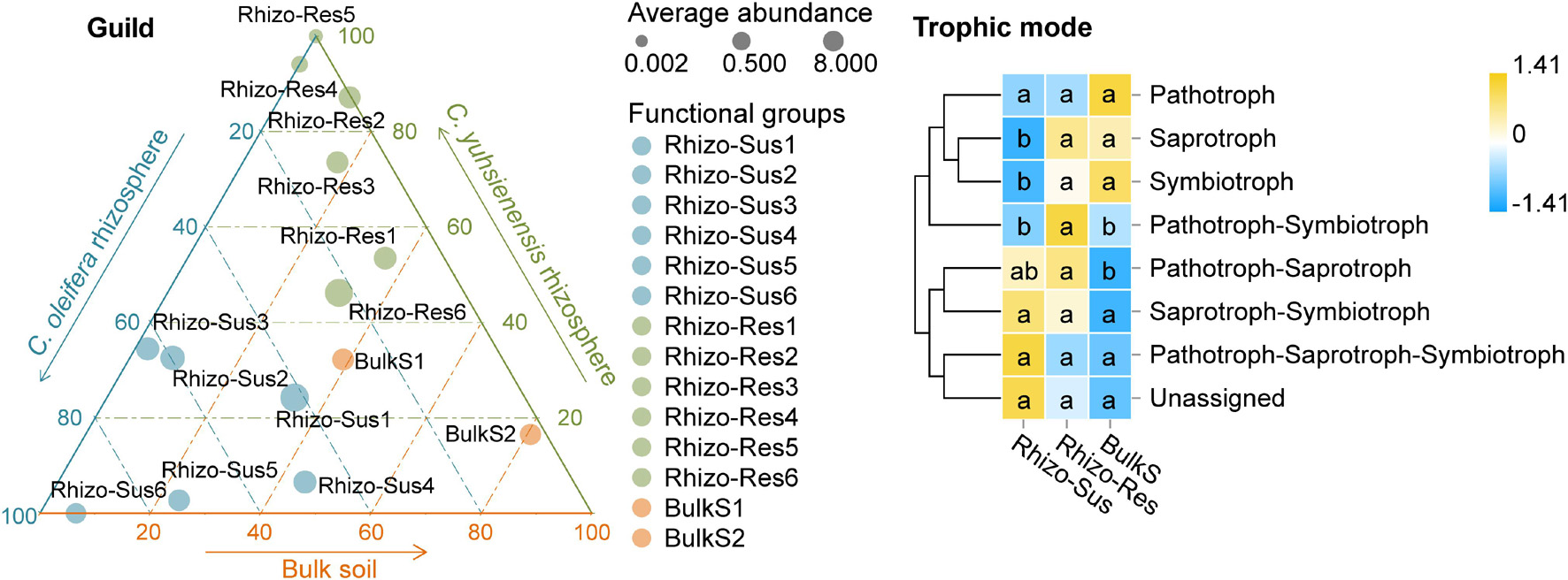
Figure 6. Distribution of fungal functional groups (guild) and heatmap of fungal trophic modes in rhizosphere and bulk soil. Ternary plot of guild plotted based C. oleifera rhizosphere (Rhizo-Sus), C. yuhsienensis rhizosphere (Rhizo-Res), and bulk soil (BulkS) specificity. The circle size, color, and position indicate the average abundance, guild, and affiliation with Rhizo-Sus, Rhizo-Res, and BulkS (detected by Kruskal–Wallis analysis, p < 0.05), respectively. Rhizo-Sus 1–6 are Undefined Saprotroph, Endophyte–Fungal Parasite–Plant Pathogen, Ericoid Mycorrhizal, Endophyte–Lichen Parasite–Undefined Saprotroph, Endomycorrhizal–Plant Pathogen–Undefined Saprotroph, and Animal Pathogen–Endophyte–Plant Saprotroph–Soil Saprotroph, respectively. Rhizo-Res 1–6 are Arbuscular Mycorrhizal, Undefined Saprotroph-Undefined Biotroph, Endophyte–Plant Pathogen, Lichenized, Animal Pathogen–Endophyte–Lichen Parasite–Plant Pathogen–Wood Saprotroph, and Unassigned, respectively. BulkS 1 and 2 are Animal Pathogen–Fungal Parasite–Undefined Saprotroph and Dung Saprotroph–Undefined Saprotroph, respectively. The abundance of functions was normalized by Z-score method. The connecting lines on the left side describe the clustering of each function. The closer the color to yellow, the more dominant the function is. Pathotroph = receiving nutrients by harming host cells; symbiotroph = receiving nutrients by exchanging resources with host cells; saprotroph = receiving nutrients by breaking down dead host cells. The lowercase in each row of heatmap indicated the significant differences among Rhizo-Sus, Rhizo-Res, and BulkS (conducted using Wilcoxon rank sum test, p < 0.05), N = 12.
Effects of Physicochemical Properties on Microbial Communities
The relationships between microbial communities and physiochemical properties were not consistent among Rhizo-Sus, Rhizo-Res, and BulkS (Supplementary Tables 6, 7). Alkaline hydrolyzable nitrogen (AMN) and AK decreased (p < 0.01) with increasing bacterial alpha diversity in Rhizo-Sus and Rhizo-Res, respectively (Supplementary Table 6). Soil moisture had no effects on bacterial alpha diversity in Rhizo-Sus (p > 0.05). The effects of moisture on bacterial alpha diversity in Rhizo-Res was positive (p < 0.05), whereas it was negative in bulk soils (p < 0.05).
Available phosphorous increased with (p < 0.05) fungal richness in all soil conditions (Supplementary Table 7). Total nitrogen increased with fungal richness in Rhizo-Sus and BulkS and fungal diversity in Rhizo-Res (p < 0.05). C/P ratio increased (p < 0.05) with fungal diversity in Rhizo-Sus and Rhizo-Res but not in BulkS.
Discussion
Specific Microorganisms in Each Camellia spp. Rhizosphere
Specific Bacteria in Each Camellia spp. Rhizosphere and Their Functions
Despite partial overlap of bacterial communities in the rhizosphere of susceptible C. oleifera, resistant C. yuhsienensis, and bulk soil, some bacterial communities were specific and were distributed solely under one of the Camellia spp. (Figure 4 and Table 1). Most B. epidermidis, Proteus mirabilis, and Rhodococcus erythropolis were specific bacteria found in the rhizosphere of susceptible C. oleifera and are PGPR and biocontrol bacteria (Figure 3 and Table 1). B. epidermidis is a very important N2-fixing and P-solubilizing bacterium living in the rhizosphere (Karagoz et al., 2012). B. epidermidis can increase length and dry weight of canola roots by producing indole acetic acid (IAA) and 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase and by fixing N2 (Siddikee et al., 2010). P. mirabilis is a PGPR by solubilizing P and K, fixing N2, and producing IAA and phytase in cabbage and Foeniculum vulgare rhizosphere (Motamedi et al., 2016; Dhiman et al., 2019). R. erythropolis protects plant well from Gram-negative soft-rot bacteria by degrading their N-acyl-homoserine lactone signaling molecules (Latour et al., 2013). However, some deleterious bacteria were observed in Rhizo-Sus such as Gryllotalpicola sp., a specific species in the rhizosphere of resistant C. yuhsienensis, commonly found in the gut of wood-feeding insects and is associated with pinewood nematode and pine wilt disease (Fang et al., 2015; Guo et al., 2020). Consequently, pathogenic insects are common in the rhizosphere of C. oleifera.
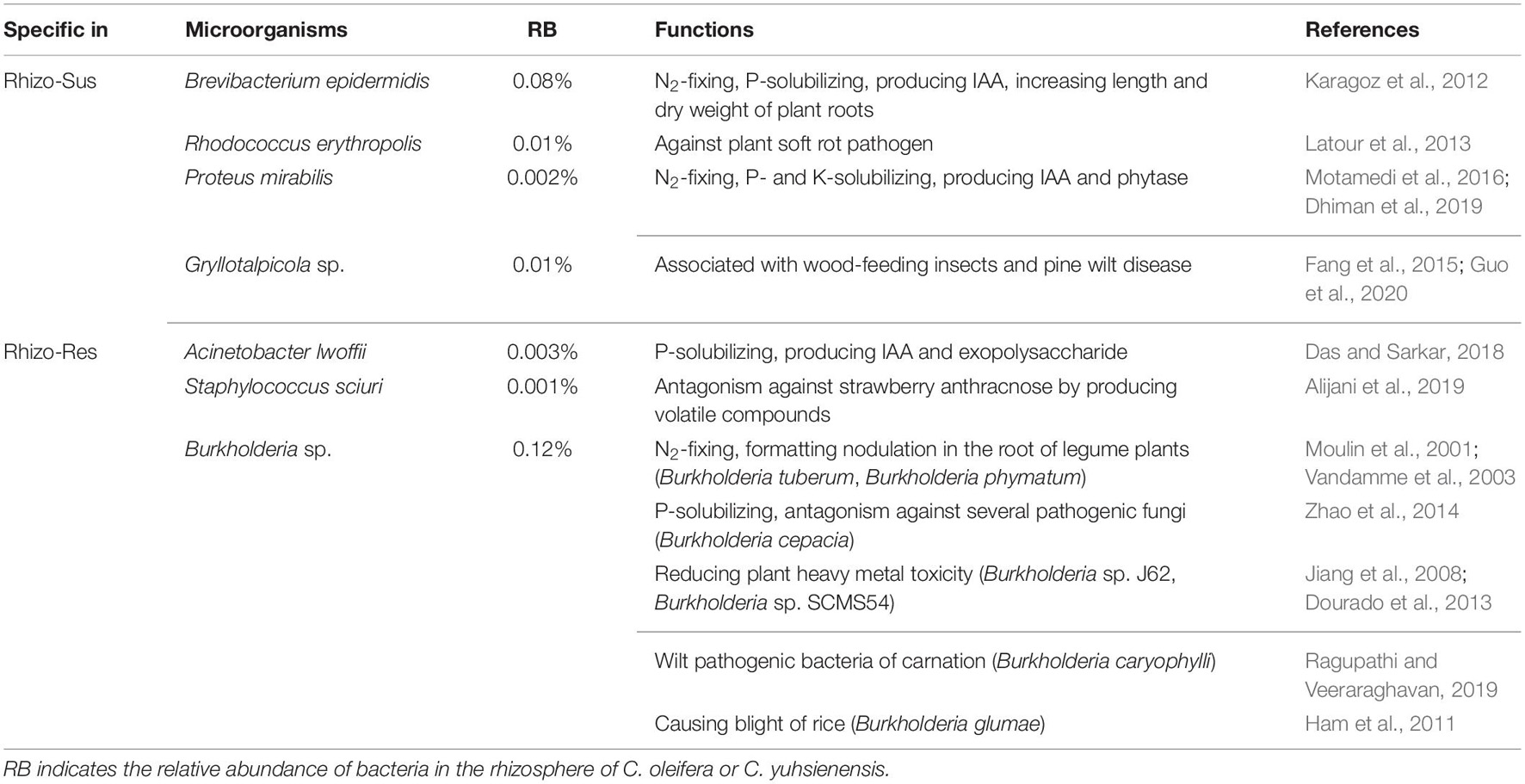
Table 1. Main known functions of bacteria specific for rhizosphere of C. oleifera (Rhizo-Sus) and C. yuhsienensis (Rhizo-Res).
Acinetobacter lwoffii and Staphylococcus sciuri, the specific bacteria in Rhizo-Res, are PGPR and biocontrol bacteria. A. lwoffii can produce IAA and exopolysaccharide (EPS), which are capable of solubilizing P to promote mung bean growth (Das and Sarkar, 2018). S. sciuri is a biocontrol bacterium of strawberry anthracnose, as it produces volatile compounds (VOCs) that can suppress mycelial growth and conidial germination of Colletotrichum nymphaeae (Alijani et al., 2019). Burkholderia sp. has wide ecological niches. Some species in this genus are plant pathogens. For instance, Burkholderia caryophylli is the wilt pathogenic bacteria of carnation (Ragupathi and Veeraraghavan, 2019). Burkholderia glumae is one of the major pathogens of rice that causes blight (Ham et al., 2011). Some are reported as PGPR (Moulin et al., 2001; Vandamme et al., 2003) and have the ability to control plant pathogens (Coenye and Vandamme, 2003; Zhao et al., 2014) and reduce heavy metal toxicity, such as cadmium and palladium (Jiang et al., 2008; Dourado et al., 2013). These results indicate that the specific bacteria in rhizosphere of susceptible C. oleifera and resistant C. yuhsienensis have potential positive and negative influence on plants.
Specific Fungi in Each Camellia spp. Rhizosphere and Their Functions
The major known functions of specific fungi in the rhizosphere of susceptible C. oleifera and resistant C. yuhsienensis are presented in Table 2. Most specific fungi in Rhizo-Sus can increase plant growth and control pathogens. O. maius is an important ericoid mycorrhizal fungus in rhizosphere of Rhododendron spp. (Vohník et al., 2005). O. maius can increase Rhododendron fortune fresh and dry weight by upregulating nitrate transporters, ammonium transporter, glutamine synthetase, and glutamate synthase in plants to increase N uptake (Wei et al., 2016). V. leptobactrum protects plants from root knot nematodes by suppressing growth of eggs and second-stage juveniles (Regaieg et al., 2011; Hajji et al., 2017). Candida intermedia is able to control strawberry fruit rot by producing volatile organic compounds to suppress conidial germination and mycelial growth of B. cinerea (Huang et al., 2011). C. intermedia also has the ability to reduce anthracnose incidence in avocado fruits by restraining mycelia growth of Colletotrichum gloeosporioides (Campos-Martínez et al., 2016). Chrysosporium pseudomerdarium is an endophyte that can increase plant shoot length and chlorophyll content by producing gibberellins to promote plant growth (Hamayun et al., 2009; Waqas et al., 2014). Lecanicillium fusisporum is an important biocontrol endophytic fungus against wheat disease Septoria tritici blotch caused by Zymoseptoria tritici (Latz et al., 2020).
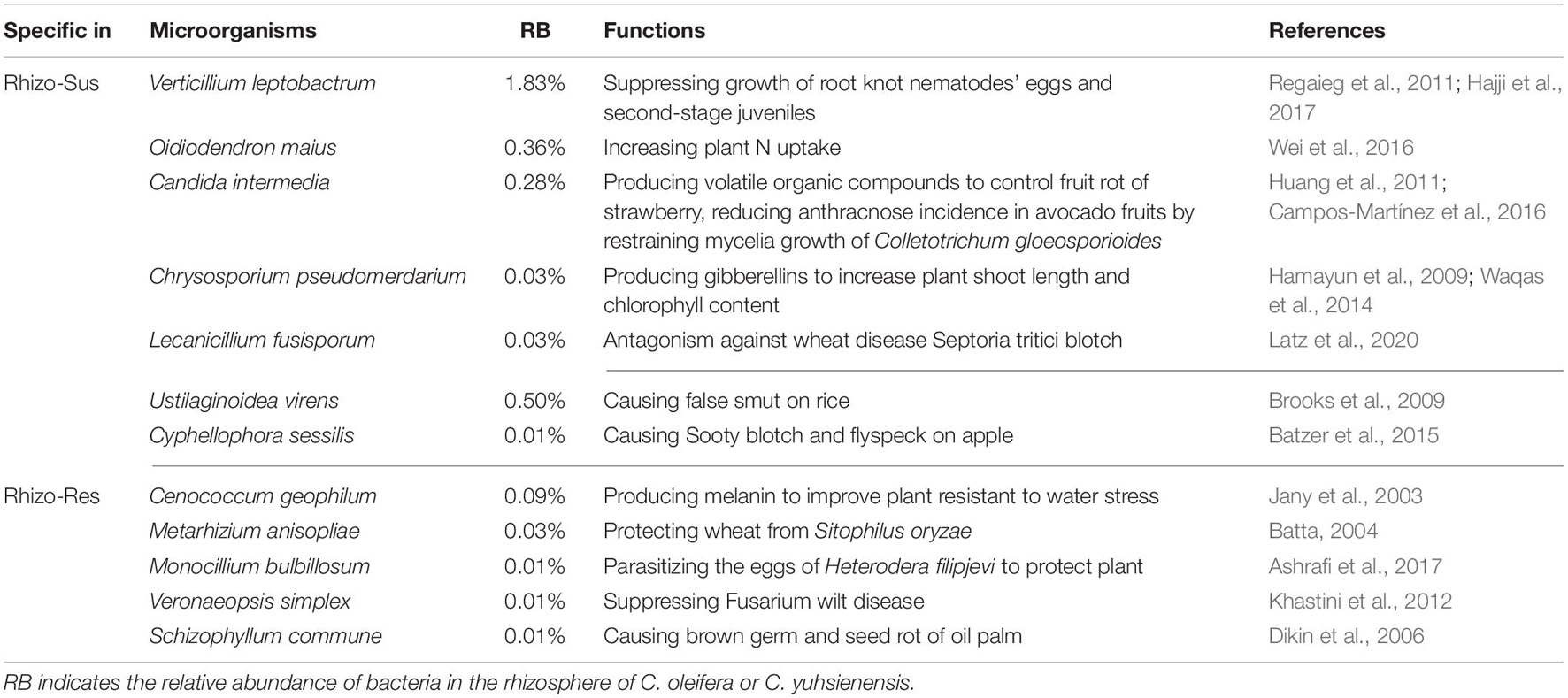
Table 2. Main known functions of fungi specific for rhizosphere of C. oleifera (Rhizo-Sus) and C. yuhsienensis (Rhizo-Res).
Phytopathogens were also observed in Rhizo-Sus. Cyphellophora sessilis (Batzer et al., 2015) and Ustilaginoidea virens (Brooks et al., 2009) are common phytopathogens on apple and rice, respectively. Fusarium concentricum causes rot disease in several plants, such as pepper fruit rot and Paris polyphylla stem rot (Wang et al., 2013; Xiao et al., 2019). On the phylogenetic tree, F. concentricum is very close to Fusarium proliferatum, which is a root rot phytopathogen of oil tea (Li et al., 2008).
Most specific fungi in Rhizo-Res are plant-growth-promoting species. Cenococcum geophilum is an important ectomycorrhizal fungus that maintain the physiological integrity of beech roots facing drought stress (Jany et al., 2003). Melanin, produced by C. geophilum, improved plant resistant to water stress (Fernandez and Koide, 2013). Metarhizium anisopliae and Monocillium bulbillosum are important biocontrol fungi for insect pest. M. anisopliae conidia with oven ash, chalk powder, charcoal, and wheat flour resulted in 73–87% mortality of adult Sitophilus oryzae (Batta, 2004). M. bulbillosum protected plants from nematodes by parasitizing the eggs of Heterodera filipjevi (Ashrafi et al., 2017). Veronaeopsis simplex is a dark septate endophytic fungus with the ability to suppress Fusarium wilt disease in Chinese cabbage (Khastini et al., 2012). There was also a phytopathogen in Rhizo-Res, named Schizophyllum commune, that causes brown germ and seed rot of oil palm (Dikin et al., 2006).
In summary, the abundance of specific pathogenic microorganisms in the rhizosphere of resistant C. yuhsienensis was too low to have similar negative effects on plant growth and health compared to susceptible C. oleifera (Tables 1, 2).
Microbial Community Association With Camellia spp. Growth and Health
Roots and rhizodeposition of Camellia species are important factors of bacterial and fungal abundance, diversity, structure, and function (Figures 1, 2, Tables 1, 2, Supplementary Figures 3, 4, and Supplementary Table 5; Zhang P. et al., 2020). Although the relationship between roots and rhizosphere microorganisms is mostly mutualistic (Qiang et al., 2012; Mendes et al., 2013), competitive relationships do exist in many cases, such as the competition for nitrogen (Hodge et al., 2000; Kuzyakov and Xu, 2013) especially under stress conditions (Xu et al., 2011). Bacteria compete with plants by assimilating and immobilizing P and N using organic carbon (Zhang et al., 2014), and this competition gets more severe due to the root exudates, including available carbon substrates for bacteria (Kuzyakov and Xu, 2013; Kuzyakov et al., 2019).
The BugBase prediction showed that the abundance of Gram-positive, facultatively anaerobic, and stress-tolerant bacteria were higher in Rhizo-Sus than in Rhizo-Res and BulkS, while Gram-negative bacteria had the opposite trend (p < 0.05) (Figure 5B). Gram-positive bacteria are generally considered as environmental stress-tolerant bacteria compared with Gram-negative bacteria (Schimel et al., 2007). Our study (Supplementary Table 2) agrees with previous studies (Liu et al., 2018), which indicated that growth of oil tea is limited by insufficient nutrients in the red soil area. The productivity of C. oleifera is higher than C. yuhsienensis, which means that C. oleifera has a greater ability to mine and absorb nutrients (Tan et al., 2011). The larger crown width, trunk diameter, and yield of C. oleifera also indicated that its nutrient utilization efficiency is higher than that of C. yuhsienensis (Supplementary Table 1). Finally, C. oleifera formed a microbial community of lower richness and diversity but of greater activity of carbohydrate metabolism. In contrast, more bacteria with oxidative phosphorylation and more Gram-negative bacteria indicated that the microbial communities acquired more high-quality substrates in the rhizosphere of C. yuhsienensis. More facultatively anaerobic bacteria implied a poorer water permeability condition in the soil under C. oleifera (Figure 5). Based on these results, we speculate that C. oleifera formed competitive relationships with the rhizosphere microbial communities, compared to the symbiotic interactions of C. yuhsienensis in the rhizosphere. These differences in relationships may be one of the most important factors for specific effects of microorganisms on oil tea growth and health.
The rhizosphere microbial diversity is an important factor in plant health and ecosystem function (Xue et al., 2020), excluding the negative effects of pathotrophs fungi plant health (Mendes et al., 2013; Nguyen et al., 2016). The bacterial and fungal diversity and the relative abundance of Acidobacteria were higher in C. yuhsienensis rhizosphere than in C. oleifera (Supplementary Table 5, Figure 1, and Supplementary Figure 3). Bacterial richness and diversity increase in soil if pathogens are suppressed. This was clearly shown in soils with suppressed Fusarium disease than in soils with serious Fusarium wilt disease (Shen et al., 2015). By studying single and mixed strains of four bacterial species antagonism on plant pathogens, Boer et al. (2007) concluded that (1) more bacteria in soil can lead to stronger competition with pathogens for resources and (2) interactions among bacteria increase antifungal activity. When biocontrol bacteria or fungi meet other microorganisms with similar functions, these microorganisms increased the production of antibiotics, such as 2,4-diacetylphloroglucinol (DAPG) (Lutz et al., 2004; Maurhofer et al., 2004). In other words, a microbial community of higher diversity has a higher potential to antagonize pathogens. Therefore, according to bacterial and fungal diversity, we state that the microbial community in C. yuhsienensis rhizosphere better protects the host from pathogens than the microbial community in C. oleifera rhizosphere (Supplementary Table 5).
Conclusion
The dominant bacteria in the rhizosphere of oil tea (Camellia spp.) were Chloroflexi, Acidobacteria, Proteobacteria, Actinobacteria, and Planctomycetes, and the dominant fungi were Ascomycota and Basidiomycota. Camellia species plays a crucial role in microbial richness, diversity, and community structure of its rhizosphere. The rhizosphere of both Camellia spp. contains beneficial and deleterious microorganisms. Numbers and abundance of deleterious microorganisms were more in C. oleifera than that in C. yuhsienensis. The bacterial groups and functions in the rhizosphere of disease-resistant C. yuhsienensis were similar to those in bulk soil but much different from those in the rhizosphere of disease susceptible C. oleifera. More Gram-negative bacteria were in the rhizosphere of C. yuhsienensis, while more Gram-positive, facultatively anaerobic, and stress-tolerant bacteria were in the rhizosphere of C. oleifera. There was higher bacterial and fungal alpha diversity in the rhizosphere of C. yuhsienensis than C. oleifera. Our results indicate that the more abundant and diverse microbial community in C. yuhsienensis rhizosphere better protects the host from pathogens compared to those in C. oleifera rhizosphere.
Data Availability Statement
The data presented in the study are deposited in the NCBI Sequence Read Archive (SRA) database, accession number: PRJNA742848.
Author Contributions
JY and JL: conceptualization and validation. JL: methodology, formal analysis, data curation, and writing – original draft preparation. JL, CZ, XQ, and ZL: investigation. JL, SL, YK, HA, JY, and GN: writing, review, and editing. JL and YK: visualization. JY and GN: supervision. JY, JL, and YK: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Provincial Science and Technology Major Project of Hunan, China under Grant (grant number 2018NK1030), the Forestry Science and Technology Innovation Project of Hunan, China under Grant (grant number XLK201987), Scientific Innovation Fund for Post-graduates of Central South University of Forestry and Technology (grant number CX20191002), and Hunan Provincial Innovation Foundation for Postgraduate (grant number CX20190598). The authors thank the support of the “RUDN University Strategic Academic Leadership Program” and the Program of Competitive Growth of Kazan Federal University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.732905/full#supplementary-material
References
Alijani, Z., Amini, J., Ashengroph, M., and Bahramnejad, B. (2019). Antifungal activity of volatile compounds produced by Staphylococcus sciuri strain MarR44 and its potential for the biocontrol of Colletotrichum nymphaeae, causal agent strawberry anthracnose. Int. J. Food Microbiol. 307:108276. doi: 10.1016/j.ijfoodmicro.2019.108276
Ashrafi, S., Stadler, M., Dababat, A. A., Richert-Pöggeler, K. R., Finckh, M. R., and Maier, W. (2017). Monocillium gamsii sp. nov. and Monocillium bulbillosum: two nematode-associated fungi parasitising the eggs of Heterodera filipjevi. MycoKeys 27, 21–38. doi: 10.3897/mycokeys.27.21254
Aßhauer, K. P., Wemheuer, B., Daniel, R., and Meinicke, P. (2015). Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31, 2882–2884. doi: 10.1093/bioinformatics/btv287
Batta, Y. A. (2004). Control of rice weevil (Sitophilus oryzae L., Coleoptera: Curculionidae) with various formulations of Metarhizium anisopliae. Crop Prot. 23, 103–108. doi: 10.1016/j.cropro.2003.07.001
Batzer, J. C., Weber, R. W. S., Mayfield, D. A., and Gleason, M. L. (2015). Diversity of the sooty blotch and flyspeck complex on apple in Germany. Mycol. Prog. 15:2. doi: 10.1007/s11557-015-1145-9
Berendsen, R., Pieterse, C., and Bakker, P. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Bob, S., Bakker, A. W., and Bakker, P. A. (1987). Interactions of deleterious and beneficial rhizosphere microorganisms and the effect of cropping practices. Annu. Rev. Phytopathol. 25, 339–358. doi: 10.1146/annurev.py.25.090187.002011
Boer, W. D., Wagenaar, A.-M., Gunnewiek, P., and Veen, J. (2007). In vitro suppression of fungi caused by combinations of apparently non-antagonistic soil bacteria. FEMS Microbiol. Ecol. 59, 177–185. doi: 10.1111/j.1574-6941.2006.00197.x
Brooks, S. A., Anders, M. M., and Yeater, K. M. (2009). Effect of cultural management practices on the severity of false smut and kernel smut of rice. Plant Dis. 93, 1202–1208. doi: 10.1094/pdis-93-11-1202
Campos-Martínez, A., Velázquez-del Valle, M. G., Flores-Moctezuma, H. E., Suárez-Rodríguez, R., Ramírez-Trujillo, J. A., and Hernández-Lauzardo, A. N. (2016). Antagonistic yeasts with potential to control Colletotrichum gloeosporioides (Penz.) Penz. & Sacc. and Colletotrichum acutatum J.H. Simmonds on avocado fruits. Crop Prot. 89, 101–104. doi: 10.1016/j.cropro.2016.07.001
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chakraborty, U., Chakraborty, B., and Basnet, M. (2006). Plant growth promotion and induction of resistance in Camellia sinensis by Bacillus megaterium. J. Basic Microbiol. 46, 186–195. doi: 10.1002/jobm.200510050
Chen, H., and Boutros, P. (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12:35. doi: 10.1186/1471-2105-12-35
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 884–890. doi: 10.1093/bioinformatics/bty560
Chen, X., Liu, C., Liu, J. A., and Zhou, G. Y. (2021). First report of Colletotrichum fructicola causing anthracnose on Camellia yuhsienensis Hu in China. Plant Dis. doi: 10.1094/pdis-04-21-0772-pdn [Epub ahead of print].
Chen, Y., Chen, G., Robinson, D., Yang, Z., Guo, J., Xie, J., et al. (2016). Large amounts of easily decomposable carbon stored in subtropical forest subsoil is associated with r-strategy-dominated soil microbes. Soil Biol. Biochem. 95, 233–242. doi: 10.1016/j.soilbio.2016.01.004
Coenye, T., and Vandamme, P. (2003). Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5, 719–729. doi: 10.1046/j.1462-2920.2003.00471.x
Das, J., and Sarkar, P. (2018). Remediation of arsenic in mung bean (Vigna radiata) with growth enhancement by unique arsenic-resistant bacterium Acinetobacter lwoffii. Sci. Total Environ. 624, 1106–1118. doi: 10.1016/j.scitotenv.2017.12.157
Denovo, G. (2021). Genedenovo. Available online at: https://www.genedenovo.com (accessed October 5, 2021).
Dhiman, S., Dubey, R. C., Baliyan, N., Kumar, S., and Maheshwari, D. (2019). Application of potassium-solubilising Proteus mirabilis MG738216 inhabiting cattle dung in improving nutrient use efficiency of Foeniculum vulgare mill. Environ. Sustain. 2, 401–409. doi: 10.1007/s42398-019-00088-8
Dikilitas, M., Karakas, S., Hashem, A., Allah, E. F. A., and Ahmad, P. (2016). “Oxidative stress and plant responses to pathogens under drought conditions,” in Water Stress and Crop Plants: A Sustainable Approach, 1st Edn, ed. P. Ahmad (Hoboken, NJ: John Wiley & Sons, Ltd), 102–123. doi: 10.1002/9781119054450.ch8
Dikin, A., Sijam, K., Kadir, J., and Abu Seman, I. (2006). Antagonistic bacteria against Schizophyllum Commune FR. in Peninsular Malaysia. Biotropia 13, 111–121. doi: 10.11598/btb.2006.13.2.221
Dourado, M. N., Martins, P. F., Quecine, M. C., Piotto, F. A., Souza, L. A., Franco, M. R., et al. (2013). Burkholderia sp. SCMS54 reduces cadmium toxicity and promotes growth in tomato. Ann. Appl. Biol. 163, 494–507. doi: 10.1111/aab.12066
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10:996. doi: 10.1038/nmeth.2604
Fang, H., Lv, W., Huang, Z., Liu, S.-J., and Yang, H. (2015). Gryllotalpicola reticulitermitis sp. nov., isolated from a termite gut. Int. J. Syst. Evol. Microbiol. 65(Pt 1), 85–89. doi: 10.1099/ijs.0.062984-0
Fernandez, C. W., and Koide, R. T. (2013). The function of melanin in the ectomycorrhizal fungus Cenococcum geophilum under water stress. Fungal Ecol. 6, 479–486. doi: 10.1016/j.funeco.2013.08.004
Guo, Y., Lin, Q., Chen, L., Carballar-Lejarazú, R., Zhang, A., Shao, E., et al. (2020). Characterization of bacterial communities associated with the pinewood nematode insect vector Monochamus alternatus Hope and the host tree Pinus massoniana. BMC Genomics 21:337. doi: 10.1186/s12864-020-6718-6
Hajji, L., Hlaoua, W., Regaieg, H., and Horrigue-Raouani, N. (2017). Biocontrol potential of Verticillium leptobactrum and Purpureocillium lilacinum against Meloidogyne javanica and Globodera pallida on potato (Solanum tuberosum). Am. J. Potato Res. 94, 178–183. doi: 10.1007/s12230-016-9554-0
Ham, J. H., Melanson, R. A., and Rush, M. C. (2011). Burkholderia glumae: next major pathogen of rice? Mol. Plant Pathol. 12, 329–339. doi: 10.1111/j.1364-3703.2010.00676.x
Hamayun, M., Khan, S. A., Iqbal, I., Na, C. I., Khan, A. L., Hwang, Y. H., et al. (2009). Chrysosporium pseudomerdarium produces gibberellins and promotes plant growth. J. Microbiol. 47, 425–430. doi: 10.1007/s12275-009-0268-6
Hodge, A., Robinson, D., and Fitter, A. (2000). Are microorganisms more effective than plant at competing for nitrogen? Trends Plant Sci. 5, 304–308. doi: 10.1016/S1360-1385(00)01656-3
Huang, F., Zhang, Y., Zhang, L., Wang, S., Feng, Y., and Rong, N. (2019). Complete genome sequence of Bacillus megaterium JX285 isolated from Camellia oleifera rhizosphere. Comput. Biol. Chem. 79, 1–5. doi: 10.1016/j.compbiolchem.2018.12.024
Huang, R., Li, G., Zhang, J., Yang, L., Che, H., Jiang, D., et al. (2011). Control of postharvest botrytis fruit rot of strawberry by volatile organic compounds of Candida intermedia. Phytopathology 101, 859–869. doi: 10.1094/PHYTO-09-10-0255
Jansson, J. K., and Hofmockel, K. S. (2020). Soil microbiomes and climate change. Nat. Rev. Microbiol. 18, 35–46. doi: 10.1038/s41579-019-0265-7
Jany, J. L., Martin, F., and Garbaye, J. (2003). Respiration activity of ectomycorrhizas from Cenococcum geophilum and Lactarius sp. in relation to soil water potential in five beech forests. Plant Soil 255, 487–494. doi: 10.1023/A:1026092714340
Jiang, C., Sheng, X., Qian, M., and Wang, Q. (2008). Isolation and characterization of a heavy metal-resistant Burkholderia sp. from heavy metal-contaminated paddy field soil and its potential in promoting plant growth and heavy metal accumulation in metal-polluted soil. Chemosphere 72, 157–164. doi: 10.1016/j.chemosphere.2008.02.006
Jin, A., Zhou, G. Y., and Li, H. (2009). Progress, problem and prospect of oil camelliae anthracnose (Colletotrichum gloeosporioides) research. Forest Pest Dis. 28, 27–31. doi: 10.3969/j.issn.1671-0886.2009.02.010
Karagoz, K., Ateş, F., Karagöz, H., and Çakmakçı, R. (2012). Characterization of plant growth-promoting traits of bacteria isolated from the rhizosphere of grapevine grown in alkaline and acidic soils. Eur. J. Soil Biol. 50, 144–150. doi: 10.1016/j.ejsobi.2012.01.007
Khastini, R. O., Ohta, H., and Narisawa, K. (2012). The role of a dark septate endophytic fungus, Veronaeopsis simplex Y34, in Fusarium disease suppression in Chinese cabbage. J. Microbiol. 50, 618–624. doi: 10.1007/s12275-012-2105-6
Koranda, M., Schnecker, J., Kaiser, C., Fuchslueger, L., Kitzler, B., Stange, C. F., et al. (2011). Microbial processes and community composition in the rhizosphere of European beech–the influence of plant C exudates. Soil Biol. Biochem. 43, 551–558. doi: 10.1016/j.soilbio.2010.11.022
Kremer, R. J. (2006). “Deleterious rhizobacteria,” in Plant-Associated Bacteria, ed. S. S. Gnanamanickam (Dordrecht: Springer), 335–361. doi: 10.1007/1-4020-4538-7_10
Kuzyakov, Y., and Cheng, W. (2001). Photosynthesis controls of rhizosphere respiration and organic matter decomposition. Soil Biol. Biochem. 33, 1915–1925. doi: 10.1016/S0038-0717(01)00117-1
Kuzyakov, Y., Horwath, W. R., Dorodnikov, M., and Blagodatskaya, E. (2019). Review and synthesis of the effects of elevated atmospheric CO2 on soil processes: no changes in pools, but increased fluxes and accelerated cycles. Soil Biol. Biochem. 128, 66–78. doi: 10.1016/j.soilbio.2018.10.005
Kuzyakov, Y., and Razavi, B. S. (2019). Rhizosphere size and shape: temporal dynamics and spatial stationarity. Soil Biol. Biochem. 135, 343–360. doi: 10.1016/j.soilbio.2019.05.011
Kuzyakov, Y., and Xu, X. (2013). Competition between roots and microorganisms for nitrogen: mechanisms and ecological relevance. New Phytol. 198, 656–669. doi: 10.1111/nph.12235
Latour, X., Barbey, C., Chane, A., Groboillot, A., and Burini, J.-F. (2013). Rhodococcus erythropolis and its γ-lactone catabolic pathway: an unusual biocontrol system that disrupts pathogen quorum sensing communication. Agronomy 3, 816–838. doi: 10.3390/agronomy3040816
Latz, M. A. C., Jensen, B., Collinge, D. B., and Lyngs Jørgensen, H. J. (2020). Identification of two endophytic fungi that control Septoria tritici blotch in the field, using a structured screening approach. Biol. Control 141:104128. doi: 10.1016/j.biocontrol.2019.104128
Li, C., Zhao, Q., Gao, T., Wang, H., Zhang, Z., Liang, B., et al. (2018). The mitigation effects of exogenous melatonin on replant disease in apple. J. Pineal Res. 65:e12523. doi: 10.1111/jpi.12523
Li, H., Zhou, G., and He, M. (2008). Study on biological characteristics and molecular identification of pathogens causing root rot of Camellia oleifera. J. Southwest Forest. Coll. 4:56. doi: 10.3969/j.issn.2095-1914.2008.05.013
Li, J., Luo, Z., Zhang, C., Qu, X., Chen, M., Song, T., et al. (2020). Seasonal variation in the rhizosphere and non-rhizosphere microbial community structures and functions of Camellia yuhsienensis Hu. Microorganisms 8:1385. doi: 10.3390/microorganisms8091385
Li, J., Wu, Z., and Yuan, J. (2019). Impact of agro-farming activities on microbial diversity of acidic red soils in a Camellia Oleifera forest. Rev. Bras. Cienc. Solo 43:e0190044. doi: 10.1590/18069657rbcs20190044
Liang, X., Ren, H., Li, S., Leng, X., and Yao, X. (2017). Soil bacterial community structure and co-occurrence pattern during vegetation restoration in karst rocky desertification area. Front. Microbiol. 8:2377. doi: 10.3389/fmicb.2017.02377
Liu, J., Wu, L., Chen, D., Yu, Z., and Wei, C. (2018). Development of a soil quality index for Camellia oleifera forestland yield under three different parent materials in Southern China. Soil Tillage Res. 176, 45–50. doi: 10.1016/j.still.2017.09.013
Liu, J., Wu, L., Chena, D., Lib, M., and Weib, C. (2017). Soil quality assessment of different Camellia oleifera stands in mid-subtropical China. Appl. Soil Ecol. 113, 29–35. doi: 10.1016/j.apsoil.2017.01.010
Lutz, M. P., Simone, W., Monika, M., Geneviève, D., and Brion, D. (2004). Signaling between bacterial and fungal biocontrol agents in a strain mixture. FEMS Microbiol. Ecol. 48, 447–455. doi: 10.1016/S0168-6496(04)00077-7
Magoè, T., and Salzberg, S. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Maurhofer, M., Baehler, E., Notz, R., Martinez, V., and Keel, C. (2004). Cross talk between 2,4-diacetylphloroglucinol-producing biocontrol pseudomonads on wheat roots. Appl. Environ. Microbiol. 70, 1990–1998. doi: 10.1128/AEM.70.4.1990-1998.2004
Meisner, A., and Boer, W. D. (2018). Strategies to maintain natural biocontrol of soil-borne crop diseases during severe drought and rainfall events. Front. Microbiol. 9:2279. doi: 10.3389/fmicb.2018.02279
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Motamedi, H., Aalivand, S., Varzi, H. N., and Mohammadi, M. (2016). Screening cabbage rhizosphere as a habitat for isolation of phosphate-solubilizing bacteria. Environ. Exp. Biol. 14, 173–181. doi: 10.22364/eeb.14.24
Moulin, L., Munive, A., Dreyfus, B., and Boivin-Masson, C. (2001). Nodulation of legumes by members of the β-subclass of Proteobacteria. Nature 411, 948–950. doi: 10.1038/35082070
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Qiang, X., Weiss, M., Kogel, K. H., and Schafer, P. (2012). Piriformospora indica-a mutualistic basidiomycete with an exceptionally large plant host range. Mol. Plant Pathol. 13, 508–518. doi: 10.1111/j.1364-3703.2011.00764.x
Ragupathi, N. K. D., and Veeraraghavan, B. (2019). Accurate identification and epidemiological characterization of Burkholderia cepacia complex: an update. Ann. Clin. Microbiol. Antimicrob. 18:7. doi: 10.1186/s12941-019-0306-0
Regaieg, H., Ciancio, D. A., Horrigue-Raouani, N., and Rosso, L. (2011). Detection and biocontrol potential of Verticillium leptobactrum parasitizing Meloidogyne spp. World J. Microbiol. Biotechnol. 27, 1615–1623. doi: 10.1007/s11274-010-0615-0
Roberts, D. W., (2016). labdsv: Ordination and Multivariate Analysis for Ecology [WWW document]. R package. Availble online at: http://cran.r-project.org/package=labdsv (accessed October 5, 2021).
Schimel, J., Balser, T. C., and Wallenstein, M. (2007). Microbial stress-response physiology and its implications for ecosystem function. Ecology 88, 1386–1394. doi: 10.1890/06-0219
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, 1–18. doi: 10.1186/gb-2011-12-6-r60
Shen, Z., Ruan, Y., Xue, C., Zhong, S., Li, R., and Shen, Q. (2015). Soils naturally suppressive to banana Fusarium wilt disease harbor unique bacterial communities. Plant Soil 393, 21–33. doi: 10.1007/s11104-015-2474-9
Siddikee, M. A., Chauhan, P., Anandham, R., Han, G.-H., and Sa, T. (2010). Isolation, characterization, and use for plant growth promotion under salt stress, of ACC deaminase-producing halotolerant bacteria derived from coastal soil. J. Microbiol. Biotechnol. 20, 1577–1584. doi: 10.4014/jmb.2017.2709.1724
Taketani, R. G., Lançoni, M. D., Kavamura, V. N., Durrer, A., Andreote, F. D., and Melo, I. S. (2016). Dry season constrains bacterial phylogenetic diversity in a semi-arid rhizosphere system. Microb. Ecol. 73, 153–161. doi: 10.1007/s00248-016-0835-4
Tan, X., Yuan, D., Yuan, J., Zou, F., Xie, P., Su, Y., et al. (2011). An elite variety: Camellia oleifera ‘Huashuo’. Sci. Silvae Sin. 47, 184–184. doi: 10.11707/j.1001-7488.20111228
Toju, H., Tanabe, A. S., Yamamoto, S., and Sato, H. (2012). High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One 7:e40863. doi: 10.1371/journal.pone.0040863
Vandamme, P., Holmes, B., Coenye, T., Goris, J., Mahenthiralingam, E., LiPuma, J. J., et al. (2003). Burkholderia cenocepacia sp. nov.–a new twist to an old story. Res. Microbiol. 154, 91–96. doi: 10.1016/s0923-2508(03)00026-3
Vetterlein, D., Carminati, A., Kögel-Knabner, I., Bienert, G. P., Smalla, K., Oburger, E., et al. (2020). Rhizosphere spatiotemporal organization–a key to rhizosphere functions. Front. Agron. 2:8. doi: 10.3389/fagro.2020.00008
Vohník, M., Albrechtová, J., and Vosatka, M. (2005). The inoculation with Oidiodendron maius and Phialocephala fortinii alters phosphorus and nitrogen uptake, foliar C:N ratio and root biomass distribution in Rhododendron cv. Azurro. Symbiosis 40, 87–96.
Wang, J. H., Feng, Z. H., Han, Z., Song, S. Q., Lin, S. H., and Wu, A. B. (2013). First report of pepper fruit rot caused by Fusarium concentricum in China. Plant Dis. 97, 1657–1657. doi: 10.1094/pdis-03-13-0325-pdn
Wang, Y., Chen, J., Li, D.-W., Zheng, L., and Huang, J. (2017). CglCUT1 gene required for cutinase activity and pathogenicity of Colletotrichum gloeosporioides causing anthracnose of Camellia oleifera. Eur. J. Plant Pathol. 147, 103–114. doi: 10.1007/s10658-016-0983-x
Waqas, M., Khan, A. L., and Lee, I.-J. (2014). Bioactive chemical constituents produced by endophytes and effects on rice plant growth. J. Plant Interact. 9, 478–487. doi: 10.1080/17429145.2013.860562
Ward, T., Larson, J., Meulemans, J., Hillmann, B., Lynch, J., Sidiropoulos, D., et al. (2017). BugBase predicts organism-level microbiome phenotypes. bioRxiv[Preprint]. doi: 10.1101/133462
Wei, W., Zhang, Z., and Ye, H. (2013). Resistance of five oil-tea species to Meloidogyne incognita. Chin. J. Trop. Agric. 33, 57–61. doi: 10.3969/j.issn.1009-2196.2013.02.013
Wei, X., Chen, J., Zhang, C., and Pan, D. (2016). A new Oidiodendron maius strain isolated from Rhododendron fortunei and its effects on nitrogen uptake and plant growth. Front. Microbiol. 7:1327. doi: 10.3389/fmicb.2016.01327
Wickham, H. (2006). An Introduction to ggplot: An Implementation of the Grammar of Graphics in R. Statistics, 1–8. Available online at: http://cran.r-project.org/web/packages/ggplot2/index.html
Xiao, R. F., Wang, J. P., Zheng, M. X., Su, H. L., Zhu, Y. J., and Liu, B. (2019). First report of Fusarium concentricum causing stem rot disease on the medicinal plant Paris polyphylla var. chinensis in China. Plant Dis. 103, 1418–1418. doi: 10.1094/pdis-10-18-1810-pdn
Xu, J. X., Li, Z. Y., Lv, X., Yan, H., Zhou, G. Y., Cao, L. X., et al. (2020). Isolation and characterization of Bacillus subtilis strain 1-L-29, an endophytic bacteria from Camellia oleifera with antimicrobial activity and efficient plant-root colonization. PLoS One 15:e0232096. doi: 10.1371/journal.pone.0232096
Xu, X., Ouyang, H., Richter, A., Wanek, W., Cao, G., and Kuzyakov, Y. (2011). Spatio-temporal variations determine plant–microbe competition for inorganic nitrogen in an alpine meadow. J. Ecol. 99, 563–571. doi: 10.1111/j.1365-2745.2010.01789.x
Xue, Y., Tian, J., Quine, T. A., Powlson, D., Xing, K., Yang, L., et al. (2020). The persistence of bacterial diversity and ecosystem multifunctionality along a disturbance intensity gradient in karst soil. Sci. Total Environ. 748:142381. doi: 10.1016/j.scitotenv.2020.142381
Yang, G., Shu, Q., Duan, L., Chen, C., and Zheng, H. (2004). Resistance of main cultivars of oil tea to Colletotrichum gloeosporioides. J. Anhui Agric. Univ. 31, 480–483. doi: 10.1109/JLT.2003.821766
Zhang, L., Ding, X., Chen, S., He, X., Zhang, F., and Feng, G. (2014). Reducing carbon: phosphorus ratio can enhance microbial phytin mineralization and lessen competition with maize for phosphorus. J. Plant Interact. 9, 850–856. doi: 10.1080/17429145.2014.977831
Zhang, P., Cui, Z., Guo, M., and Xi, R. (2020). Characteristics of the soil microbial community in the forestland of Camellia oleifera. PeerJ 8:e9117. doi: 10.7717/peerj.9117
Zhang, X., Kuzyakov, Y., Zang, H., Dippold, M. A., Shi, L., Spielvogel, S., et al. (2020). Rhizosphere hotspots: root hairs and warming control microbial efficiency, carbon utilization and energy production. Soil Biol. Biochem. 148:107872. doi: 10.1016/j.soilbio.2020.107872
Zhao, K., Penttinen, P., Zhang, X., Ao, X., Liu, M., Yu, X., et al. (2014). Maize rhizosphere in Sichuan, China, hosts plant growth promoting Burkholderia cepacia with phosphate solubilizing and antifungal abilities. Microbiol. Res. 169, 76–82. doi: 10.1016/j.micres.2013.07.003
Zhou, J., Lu, M., Yu, S., Liu, Y., Yang, J., and Tan, X. (2020). In-depth understanding of Camellia oleifera self-incompatibility by comparative transcriptome, proteome and metabolome. Int. J. Mol. Sci. 21:1600. doi: 10.3390/ijms21051600
Zhu, J. C., Liu, J. A., and Zhou, G. Y. (2020). First report of Meloidogyne enterolobii on Camellia oleifera in China. Plant Dis. 104:1563. doi: 10.1094/pdis-06-19-1162-pdn
Keywords: Camellia yuhsienensis, Camellia oleifera, rhizosphere microbiome and functions, plant growth promoting microorganisms, soilborne phytopathogens
Citation: Li J, Zhang C, Qu X, Luo Z, Lu S, Kuzyakov Y, Alharbi HA, Yuan J and Niu G (2021) Microbial Communities and Functions in the Rhizosphere of Disease-Resistant and Susceptible Camellia spp. Front. Microbiol. 12:732905. doi: 10.3389/fmicb.2021.732905
Received: 29 June 2021; Accepted: 23 September 2021;
Published: 18 October 2021.
Edited by:
Wanting Ling, Nanjing Agricultural University, ChinaCopyright © 2021 Li, Zhang, Qu, Luo, Lu, Kuzyakov, Alharbi, Yuan and Niu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Yuan, eXVhbmp1bkBjc3VmdC5lZHUuY24=; Genhua Niu, R2VuaHVhLk5pdUBhZy50YW11LmVkdQ==
 Jun Li
Jun Li Chenhui Zhang1
Chenhui Zhang1 Yakov Kuzyakov
Yakov Kuzyakov Genhua Niu
Genhua Niu