
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 27 September 2021
Sec. Microbiotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.730807
This article is part of the Research TopicBioactive Compounds with Potential Medicinal Properties Derived from Fungi: Recent and Future Developments in Microbial BiotechnologyView all 15 articles
 Xiaoyan Pang1,2,3
Xiaoyan Pang1,2,3 Weihao Chen1
Weihao Chen1 Xin Wang4
Xin Wang4 Xuefeng Zhou1,2
Xuefeng Zhou1,2 Bin Yang1,2
Bin Yang1,2 Xinpeng Tian1,2
Xinpeng Tian1,2 Junfeng Wang1,2*
Junfeng Wang1,2* Shihai Xu3*
Shihai Xu3* Yonghong Liu1,2*
Yonghong Liu1,2*Three new tetramic acid derivatives (1–3) and a new polyketide (4) along with eight known compounds (5–12) were isolated from cultures of the deep-sea-derived fungus Penicillium sp. SCSIO06868. Four new structures were elucidated by analysis of one-dimensional/two-dimensional nuclear magnetic resonance (NMR) data and high-resolution electrospray ionization mass spectrometry. Their absolute configurations were established by X-ray crystallography analysis and comparison of the experimental and reported electronic circular dichroism (ECD) values or specific optical rotation. Compound 3 exhibited potent, selective inhibitory activities against Staphylococcus aureus and methicillin-resistant S. aureus with minimum inhibitory concentration values of both 2.5 μg/ml. Also, compound 3 showed weak antiviral activity against severe acute respiratory syndrome coronavirus 2 main protease, which was responsible for the coronavirus disease 2019 pandemic.
Natural products bearing a tetramic acid structural fragment (pyrrolidine-2,4-dione) are isolated from various terrestrial and marine organisms, such as bacteria, cyanobacteria, fungi, and sponges (Mo et al., 2014; Jiang et al., 2020). Tetramic acids showed a remarkable diversity of bioactivities, including antitumor (Lin et al., 2008; Fan et al., 2020), antiviral (Sun et al., 2015), antibacterial (Nord et al., 2020; Wingen et al., 2020), larvicidal (Mao et al., 2019), and herbicidal (Schrey et al., 2019) activities (Schobert and Schlenk, 2008; Mo et al., 2014; Jiang et al., 2020). Among the different marine sources, marine fungi mainly containing Aspergillus, Penicillium, and Cladosporium species are the dominant sources of the rapidly increasing numbers of tetramic acids (Jiang et al., 2020). With the development of sampling techniques and the possibility to culture organisms from deep-sea even in conventional standard microbiological laboratories, deep-sea-derived fungi have recently received a wide concern as a new area for bioprospecting (Jin et al., 2016; Pang et al., 2020). As part of our ongoing research for bioactive secondary metabolites from deep-sea-derived fungi (Chen et al., 2016; Wang et al., 2016; Pang et al., 2021), the fungus Penicillium sp. SCSIO06868 was studied. Three new tetramic acid derivatives (1–3) and a new polyketide (4) along with eight known compounds (5–12) (Figure 1) were isolated from the deep-sea-derived fungus Penicillium sp. SCSIO06868, which was cultured on a liquid medium. The coronavirus disease 2019 (COVID-19) pandemic has left a mark in more than 180 countries, with more than 2.0 billion cases worldwide and over 4.4 million deaths in total (until August 2021). The COVID-19 is an infectious disease caused by a novel strain of coronavirus [severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)] (Zehra et al., 2020; Shamsi et al., 2021). SARS-CoV-2 main proteinase (MPro), a key protease of CoV-2, mediates viral replication and transcription. SARS-CoV-2 MPro has emerged as an attractive target for SARS-CoV-2 drug design and development (Sabbah et al., 2021). All isolated compounds (1–12) were tested for their antiviral activities against SARS-CoV-2 Mpro in vitro. Molecular docking research was performed to mimic the interactions between the bioactive compound and SARS-CoV-2 Mpro. Herein, we described the isolation, structure elucidation, and bioactivity evaluation of the 12 compounds.
One-dimensional and two-dimensional (2D) nuclear magnetic resonance (NMR) spectra were measured on a Bruker Avance 700 MHz NMR spectrometer (Fällanden, Switzerland) with Tetramethylsilane as an internal standard. High-resolution electrospray ionization mass spectrometry (HRESIMS) data were recorded on a maXis Q-TOF mass spectrometer in positive ion mode (Bruker, Fällanden, Switzerland). Electronic circular dichroism (ECD) and ultraviolet (UV) spectra were measured with a Chirascan circular dichroism spectrometer (Applied Photophysics). Optical rotations were measured using an MCP-500 polarimeter (Anton, Austria). High-performance liquid chromatography (HPLC) was performed on Hitachi Primaide with YMC ODS SERIES column (YMC-Pack ODS-A, YMC Co. Ltd., Kyoto, 250 × 10 mm I.D., S-5 μm, 12 nm). Column chromatography was carried out on silica gel (200–300 mesh, Jiangyou Silica Gel Development Co., Yantai, China), YMC Gel ODS-A (12 nm, S-50 μm YMC, MA, United States), and Sephadex LH-20 (40–70 μm, Amersham Pharmacia Biotech AB, Uppsala, Sweden). Spots were detected under UV light by heating after spraying with the mixed solvent of saturated vanillin and 5% sulfuric acid in water. The thin layer chromatography plates with silica gel GF254 (0.4–0.5 mm, Qingdao Marine Chemical Factory, Qingdao, China) were used for analysis and preparation.
The strain SCSIO06868 was isolated from the deep-sea sediment collected from the Indian Ocean (94°37.377′E; 2°59.853′S; depth 4,762 m). The internal transcribed spacer sequences of SCSIO06868 (494 base pairs, GenBank accession no. MZ277624) have 99% sequence identity to that of Penicillium citrinum DUCC5728 (GenBank accession no. 582768). Then, it was designated as a member of Penicillium sp. and named as Penicillium sp. SCSIO06868. The strain SCSIO06868 was stored on methylene blue agar (malt extract 15 g, agar 16 g, sea salt 10 g, water 1 L, pH 7.4–7.8) slants at 4°C and deposited at Key Laboratory of Tropical Marine Bio-resources and Ecology, Chinese Academy of Sciences.
The mass fermentation of this fungus was carried out in 1-L Erlenmeyer flasks. The fungus was inoculated in a liquid medium (2% maltose, 2% mannitol, 1% monosodium glutamate, 1% glucose, 0.3% yeast extract, 0.05% monopotassium phosphate, 0.03% MgSO4⋅7H2O, and 300-ml tap water/flask, 93 flasks, 28 L total) at 25°C under static condition for 35 days. After 35 days, the fermentation was soaked in ethyl acetate (500 ml/flask), and the mycelia were cut into small pieces and sonicated for 20 min. The ethyl acetate solution was concentrated under reduced pressure to gain a brown crude extract (59.0 g).
The crude extract was subjected to silica gel column chromatography, which was eluted with dichloromethane and methanol (MeOH) mixed solvent in a step gradient (100:0–5:1, v/v) and separated into seven fractions (Fr-1–Fr-7). Fr-1 (3.2 g) was applied to Sephadex LH-20 column eluted with MeOH, reversed-phase C18 medium pressure liquid chromatography (MPLC) eluted with MeOH/water (H2O) (10:90–100:0, v/v), and semipreparative HPLC [72% CH3OH/H2O with 0.3‰ trifluoroacetic acid (TFA), 2 ml/min] to afford compounds 7 (2.2 mg, tR = 20.2 min) and 8 (18.4 mg, tR = 22.2 min). Fr-2 (6.3 g) was subjected to Sephadex LH-20 column eluted with MeOH, reversed-phase C18 MPLC eluted with MeOH/H2O (10:90–100:0, v/v), and semipreparative HPLC (2 ml/min) to gain compounds 3 (4.0 mg, 54% CH3CN/H2O, tR = 31.0 min), 5 (4.5 mg, 35% CH3CN/H2O with 0.3‰ TFA, tR = 33.6 min), 6 (7.6 mg, 35% CH3CN/H2O with 0.3‰ TFA, tR = 35.0 min), and 10 (6.4 mg, 33% CH3CN/H2O, tR = 22.0 min). Fr-3 (4.6 g) was purified with Sephadex LH-20 column eluted with MeOH, reversed-phase C18 MPLC eluted with MeOH/H2O (10:90–100:0, v/v), and semipreparative HPLC (2 ml/min) to obtain compounds 4 (2.9 mg, 72% CH3OH/H2O with 0.3‰ TFA, tR = 8.2 min) and 9 (42.2 mg, 70% CH3OH/H2O with 0.3‰ TFA, tR = 14.6 min). Fr-4 (3.7 g) was purified with Sephadex LH-20 column eluted with MeOH, reversed-phase C18 MPLC eluted with MeOH/H2O (10:90–100:0, v/v), and semipreparative HPLC (75% CH3CN/H2O with 0.3‰ TFA, 2 ml/min) to yield compound 1 (30.5 mg, tR = 12.2 min). Fr-5 (1.3 g) was applied to Sephadex LH-20 column eluted with MeOH, reversed-phase C-18 MPLC eluted with MeOH/H2O (10:90–100:0, v/v), and semipreparative HPLC (2 ml/min) to get compounds 11 (76.5 mg, 40% CH3CN/H2O with 0.3‰ TFA, tR = 11.4 min) and 12 (60% CH3CN/H2O, tR = 15.2 min, 6.0 mg). Fr-6 (1.6 g) was subjected to Sephadex LH-20 column eluted with MeOH, reversed-phase C18 MPLC eluted with MeOH/H2O (10:90–100:0, v/v), and semipreparative HPLC (75% CH3CN/H2O with 0.3‰ TFA, 2 ml/min) to obtain compound 2 (2.5 mg, tR = 12.8 min).
Penicillenol G1 (1): Pale white solid; –156.6 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 219 (3.21), and 278 (2.79) nm; ECD (1.06 mM, MeOH) λmax (Δε) 209 (+ 5.26), 227 (–5.27), and 281 (−4.38) nm; 1H and 13C NMR data (Table 1); HRESIMS m/z 284.1866 [M + H]+ (calcd for C15H26NO4, 284.1856).
Penicillenol G2 (2): Yellowish oil; + 46.0 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 242 (2.87) and 279 (3.12) nm; ECD (1.06 mM, MeOH) λmax (Δε) 213 (–3.08), 238 (+ 2.50), and 271 (2.89) nm; 1H and 13C NMR data (Table 1); HRESIMS m/z 284.1862 [M + H]+ (calcd for C15H26NO4, 284.1856).
Penicillenol H (3): Yellow oil; –21.0 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 227 (2.81) and 285 (3.04) nm; ECD (0.71 mM, MeOH) λmax (Δε) 213 (2.87), 231 (–2.76), and 289 (–1.27) nm; 1H and 13C NMR data (Table 1); HRESIMS m/z 284.1858 [M + H]+ (calcd for C15H26NO4, 284.1856).
Coniochaetone N (4): Yellow solid powder; + 46.4 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 203 (4.11), 226 (4.15), 243 (4.22), and 342 (3.56) nm; 1H and 13C NMR data (Table 2); HRESIMS m/z 285.0373 [M + Na]+ (calcd for C13H10NaO6, 285.0370).
X-Ray crystallographic analysis of penicillenol G1 (1): Moiety formula: C15H25NO4 (M = 283.36 g/mol), colorless needle, crystal size = 0.6 × 0.03 × 0.03 mm3, trigonal, space group C2; unit cell dimensions: a = 20.4710(5) Å, b = 4.85100(10) Å, c = 33.3706(10) Å, V = 3154.94(15) Å3, Z = 8, ρcalcd = 1.193 g cm–3, T = 101(2) K, μ(Cu Kα) = 0.698 mm–1. A total of 31,816 reflections were measured with 6,225 independent reflections (Rint = 0.0496, Rsigma = 0.0341). Final R indices [I > 2σ (I)]: R1 = 0.0348, wR2 = 0.0872. Final R indexes [all date]: R1 = 0.0407, wR2 = 0.0896, Flack parameter = 0.07(8). Largest diff. peak and hole = 0.18 and –0.20 eÅ–3.
The molecular docking was conducted by AutoDockTools (Version 1.5.6) (Morris et al., 2008). The crystal structure of SARS-CoV-2 main protease (PDB ID: 6LU7) was retrieved from the Protein DataBank1 (Jin et al., 2020). The structures were generated in ChemBio3D Ultra 14.0 (ChemBioOffice version 14.0), followed by an MM2 calculation to minimize the conformation energy. The original ligand and crystal water were removed before the docking calculation. The hydrogens were added to the structure of 6LU7, and Kollman united partial charges were assigned. A Lamarckian genetic algorithm was applied as a default search algorithm and set the grid box within the size of 46 × 44 × 46 Å, with the spacing of 0.375 Å. During the docking, the default parameters were used if it was not mentioned. The docking pose that had the lowest binding energy was represented as the most favorable binding conformation.
All compounds (1–12) were tested for antibacterial activities against five pathogenic bacteria using the method of agar filter paper diffusion. Compounds that had inhibition zone were evaluated in 96-well plates using a modification of the broth microdilution method (Pang et al., 2018). Ampicillin and gentamicin were used as a positive control for Gram-positive and Gram-negative bacteria, respectively.
The antiviral activities of all compounds (1–12) against SARS-CoV-2 Mpro were evaluated through the method mentioned in the previous report (Li et al., 2020). Hydroxychloroquine showed potent inhibitory activity against SARS-CoV-2 Mpro with Ki = 0.36 μm and was used as a positive control.
Penicillenol G1 (1) possessed the elemental composition of C15H25NO4 with 4 degrees of unsaturation as established by its 13C NMR data and a protonated molecule at m/z 284.1866 in the HRESIMS spectrum. Its 1D NMR data (Table 1) displayed three methyls [δC/H 20.4/1.29 (d, J = 7.0 Hz, CH3-7), 17.3/1.16 (d, J = 6.3 Hz, CH3-16), and 14.4/0.89 (t, J = 7.7 Hz, CH3-15)], three sp3 methines [(δC/H 69.0/3.73 (brs, CH-5), 68.0/4.10 (dq, J = 6.3, 2.8 Hz, CH-6), and 37.5/3.58–3.67 (m, CH-9)], four sp3 methenes (δC/H 28.3–34.8/1.23–1.74), and four sp2 non-protonated carbons (δC 178.0 C-2, 102.3 C-3, 196.2 C-4, and 193.8 C-8). The 1H-1H COSY correlations (Figure 2) of H3-16/H-9/H2-10/H2-11 and H3-15/H2-14, along with four overlapping sp3 methenes in the 1H NMR, indicated the presence of a 2-isooctyl group. The 1H-1H COSY correlations of H3-7/H-6/H-5 verified that there was a 1-hydroxyethyl group directly connected to C-5. Comparison of the NMR data of 1 with those of penicillenol A1 (5) (Lin et al., 2008; Yoda et al., 2010) showed that they only differed by an absence of the singlet methyl in 1. The planar structure of 1 was further confirmed by its heteronuclear multiple bond correlation (HMBC) correlations (Figure 2) of H-6 to C-4, H3-7 to C-5, H2-10 to C-8 and C-12, and H3-16 to C-8 and C-10. The configuration of a double bond at C-3 was determined as Z based on that the chemical shift of acylamino (δC 178.0, C-2) was in a lower field than those of normal ones, which caused by the hydrogen bond between the oxygen atom at C-2 and hydroxy at C-8 (Aoki et al., 2000). Thus, the ECD of 1 (Figure 3) displayed a positive Cotton effect at 209 nm (Δε = + 18.39), a negative Cotton effect at 227 nm (Δε = –15.33), and a negative Cotton effect at 281 nm (Δε = –4.38), and the trend of which was consistent with that of 5. Thus, the absolute configuration of C-5 in 1 was determined as S. Mosher’s method was tried to confirm the absolute configuration of C-6 but failed. Fortunately, the single crystal of 1 was obtained, and the absolute configuration of 1 was established as 5S, 6R, 9S by analyzing the X-ray crystallographic data (Figure 4). Compound 1 was named as penicillenol G1.
The molecular formula of penicillenol G2 (2), which was the same as 1, was established as C15H25NO4 by its NMR data (Table 1) and a protonated molecule at m/z 284.1862 in the HRESIMS. Its NMR data were nearly the same as those of 1, and only the chemical shifts of C-5 (δC/H 68.1/3.94, brs), C-6 [δC/H 68.7/4.08 (qd, J = 6.3, 3.5 Hz)], and C-7 [δC/H 17.0/1.10 (d, J = 6.3 Hz)] in 2 have some small differences. Compound 2 had the same planar structure with 1, which was confirmed by the 1H-1H COSY and HMBC spectra (Figure 2). Thereby, the differences mentioned earlier might be caused by their different configurations. The ECD spectrum of 2 showed a negative Cotton effect at 213 nm (Δε = –3.08), a positive Cotton effect at 238 nm (Δε = + 2.5), and a positive Cotton effect at 271 nm (Δε = + 2.89), which exhibited a reverse trend with 1 and 5, but was consistent with penicillenol A2 (6) (Lin et al., 2008; Sengoku et al., 2012). Therefore, the absolute configuration of C-5 in 2 was determined as R. Some synthetic chemists demonstrated that the natural penicillenol A1 (5) (Yoda et al., 2010), penicillenol A2 (6) (Sengoku et al., 2012), and penicillenol C1 (7) (Kempf et al., 2013) were all have the same absolute configurations of 6R and 9S, and the absolute configuration of compound 1 was also determined as 6R and 9S by the X-ray diffraction study. Thus, considering the same biosynthetic pathway (Yin et al., 2019) and comparison of the spectroscopic data with 1, the absolute configurations of C-6 and C-9 in 2 were deduced as 6R, 9S.
Penicillenol H (3) was obtained as a yellow oil. The 13C NMR data and a protonated molecule at m/z 284.1858 in the HRESIMS of 3 suggested that its molecular formula was C15H25NO4 with 4 degrees of unsaturation. The NMR data (Table 1) of 3 were similar to those of 1, except that the 1-hydroxyethyl group in 1 was displaced by oxygenated methylene (δC/H 59.3/3.94, qd, J = 12.6, 2.8 Hz, CH2-6) and an N-methyl group (δC/H 26.8/3.03, s, CH3-17) was added in 3. The extinction was determined by its 1H-1H COSY cross-peak of H2-6/H-5 and HMBC correlations of H2-6 to C-4 and H3-17 to C-2 and C-5. The planar structure of 3 was further established by its 2D NMR (Figure 2). The configuration of a double bond at C-3 was designated as Z by the chemical shift of C-2 (δC 178.0) (Aoki et al., 2000). The absolute configuration of C-5 and C-9 in 3 were determined as S and R same as compound 1 by their similar ECD spectrum (Figure 3), of which 3 showed a positive Cotton effect at 213 nm (Δε = + 2.87), two negative Cotton effects at 231 nm (Δε = – 2.76), and 289 nm (Δε = – 1.27), respectively. Compound 3 was named penicillenol H.
The molecular formula of compound 4 was C13H10O8, established by its 13C NMR data (Table 2) and a sodium adduct ion peak at m/z 285.0373 in the HRESIMS spectrum. Its 1H NMR data were simple and showed two aromatic protons (δH 7.54, brs, CH-7; 7.32, brs, CH-9) at meta-position, an oxygenated methine (δH 5.30, d, J = 7.0 Hz, CH-1), and two sp3 methylenes (δH 2.45–2.53, m, 2.00, brt, J = 11.2 Hz, CH2-2; 3.19, dt, J = 17.5, 7.7 Hz, 2.89, ddd, J = 18.2, 9.1, 2.8 Hz, CH2-3). Besides the corresponding carbons (δC 109.7, CH-7; 113.1, CH-9; 71.5, CH-1; 34.4, CH2-2; 30.7, CH2-3), there were eight sp2 non-protonated carbons in its 13C NMR, which indicated the presences of an α,β-unsaturated ketone (δC 182.2, C-12; 122.9, C-13; 176.1, C-4) and a carboxyl (δC, 167.9, C-14). Its NMR data were very similar to those of coniochaetone L (Guo et al., 2019), except that the methoxy group at C-1 in coniochaetone L was replaced by hydrogen in 4. The speculation was further confirmed by its HMBC and 1H-1H COSY spectra (Figure 2). The positive specific rotation of 4 ( + 46.4, MeOH) that was consistent with that of coniochaetone L ( + 25.3, MeOH) suggested that the configuration of C-1 was R. Thus, compound 4 was established as R-1,8-dihydroxy-9-oxo-1,2,3,9- tetrahydrocyclopenta[b]chromene-6-carboxylic acid and named as coniochaetone N.
In addition, the eight known compounds (5–12) (Figure 1) were identified as penicillenol A1 (5) (Lin et al., 2008; Yoda et al., 2010), penicillenol A2 (6) (Lin et al., 2008; Sengoku et al., 2012), penicillenol C1 (7) (Lin et al., 2008; Kempf et al., 2013), penicillenol C2 (8) Lin et al., 2008), scalusamide C (9) (Tsuda et al., 2005), (E)-7-(3-methyl-4-oxo-6,7,8,8a-tetrahydro-4H-pyrrolo[2,1-b][1,3]oxazin-2-yl)hept-2-enoic acid (10) (Lai et al., 2013), terretrione D (11) (Shaala and Youssef, 2015), and (2R)-2,3-dihydro-7-hydroxy-6,8-dimethyl-2-[(E)-prop-1-enyl] chromen-4-one (12) (Li et al., 2007; Zhang et al., 2019) by comparison of their physical and spectroscopic data with those in the literature.
All isolated compounds (1–12) were tested for their antiviral activities against SARS-CoV-2 Mpro in vitro through the method mentioned in the reported literature (Li et al., 2020). Compound 3 showed weak inhibitory activity against Mpro enzyme, which was responsible for the COVID-19 pandemic. When treated with 50 μm of 3, the relative enzyme activity of SARS-CoV-2 Mpro was 46.64%, and that of the positive control hydroxychloroquine was 5.28% with the same concentration. To better understand the interactions between compounds and SARS-CoV-2 Mpro, molecular docking research was performed to mimic the interactions between compound 3 and Mpro enzyme of SARS-CoV-2 (PDB ID: 6LU7) by utilizing the AutoDockTools. Molecular docking results demonstrated that compound 3 could interact with the SARS-CoV-2 Mpro enzyme at the entrance of the catalytic pocket, with the calculated binding affinities of –4.98 kcal/mol. The 2D binding model for 3 (Figure 5) showed two hydrogen bonds and two intermolecular hydrophobic interactions. Two hydrogen bonds were formed between the carbonyl group at C-4 and Thr-26, as well as between the hydroxyl group at C-6 and Thr-24. The lengths of the two hydrogen bonds were 2.0 and 1.8 Å, respectively. These results suggested that compound 3 could insert into the active site of the enzyme and bind tightly to the catalytic amino acid residues by different types of interactions to inhibit SARS-CoV-2 Mpro.
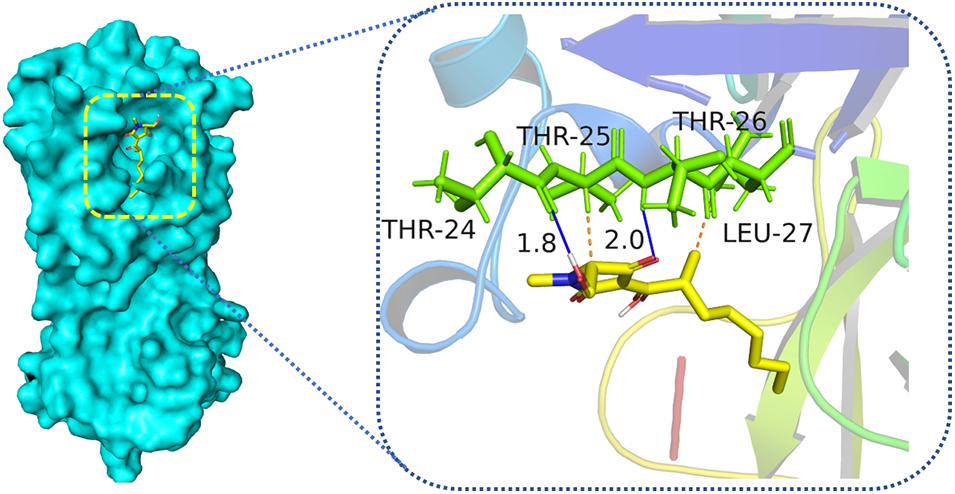
Figure 5. Low-energy binding conformations the complex between compound 3 and SARS-CoV-2 Mpro by virtual docking. Blue solid line means hydrogen bonds, and orange dotted lines mean hydrophobic interactions.
Compounds 1–12 were evaluated their antibacterial activities against five pathogenic bacteria Escherichia coli (ATCC 25922), Enterococcus faecalis (ATCC 29212), Klebsiella pneumonia (ATCC 13883), Staphyloccocus aureus (ATCC 29213), and methicillin-resistant S. aureus (MRSA). Compounds 1, 3, 5, and 6 with 50 μg/disc showed inhibition zones against S. aureus. Compounds 1, 3, and 5 with 50 μg/disc showed an inhibition zone against MRSA (Supplementary Figure 1). Furthermore, their minimum inhibitory concentrations (MICs) were tested, and the results are shown in Table 3. Compound 3 displayed potent inhibitory activities against S. aureus and MRSA with MIC values of both 2.5 μg/ml. Ampicillin was used as a positive control against S. aureus and MRSA with MIC values of 1.56 and 0.39 μg/ml, respectively.
In summary, we reported the isolation and identification of three new tetramic acid derivatives (1–3) and a new polyketide (4) along with eight known compounds (5–12) from cultures of the deep-sea-derived fungus Penicillium sp. SCSIO06868. The absolute configurations of new compounds were established by X-ray crystallography analysis and comparison of the experimental and reported ECD value or specific optical rotation. Compound 3 displayed potent inhibitory activities against S. aureus and MRSA with MIC values of both 2.5 μg/ml. Compound 3 showed weak inhibitory activity against Mpro enzyme of SARS-CoV-2, which was responsible for the COVID-19 pandemic. Molecular docking was performed to mimic the interactions between compound 3 and SARS-CoV-2 Mpro. The molecular docking results indicated that compound 3 could be inserted into the active site of the enzyme and bind tightly to the catalytic amino acid residues by different types of interactions to inhibit SARS-CoV-2 Mpro.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found in the article/Supplementary Material.
XP, JW, and YL contributed to conception and design of the study. XP performed the experiments, analyzed the data, and wrote the manuscript. XW performed the SARS-CoV-2 Mpro inhibition test. XT did the isolation of the fungus. All authors contributed to manuscript revision, review, and approved the submitted version.
This research was financially supported by the Finance Science and Technology Project of Hainan Province (ZDKJ202018), the National Natural Science Foundation of China (Nos. 21772210, 41776169, 41876145, and 42006084), the National Key Research and Development Program of China (No. 2019YFC0312503), the China Postdoctoral Science Foundation (No. 2019M663368), Project from the Institution of South China Sea Ecology and Environmental Engineering, CAS (No. ISEE2018PY04), and the Special Funds for Promoting Economic Development (Marine Economic Development) of Guangdong Province [Nos. GDOE (2019)A28 and (2020)037].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We gratefully acknowledge the assistance of Dr. Xiao, Dr. Zheng, Ms. Sun, Ms. Zhang, and Ms. Ma in the analytical facility center of the SCSIO for recording spectroscopic data.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.730807/full#supplementary-material
Aoki, S., Higuchi, K., Ye, Y., Satari, R., and Kobayashi, M. (2000). Melophlins A and B, novel tetramic acids reversing the phenotype of ras-transformed cells, from the marine sponge melophlus sarassinorum. Tetrahedron 56, 1833–1836. doi: 10.1016/S0040-4020(00)00092-2
Chen, S. T., Wang, J. F., Lin, X. P., Zhao, B. X., Wei, X. Y., Li, G. Q., et al. (2016). Chrysamides A-C, three dimeric nitrophenyl trans-epoxyamides produced by the deep-sea-derived fungus Penicillium chrysogenum SCSIO41001. Org. Lett. 18, 3650–3653. doi: 10.1021/acs.orglett.6b01699
Fan, B., Dewapriya, P., Li, F., Blumel, M., and Tasdemir, D. (2020). Pyrenosetins A-C, new decalinoylspirotetramic acid derivatives isolated by bioactivity-based molecular networking from the seaweed-derived fungus Pyrenochaetopsis sp. FVE-001. Mar. Drugs 18:47. doi: 10.3390/md18010047
Guo, C., Lin, X. P., Liao, S. R., Yang, B., Zhou, X. F., Yang, X. W., et al. (2019). Two new aromatic polyketides from a deep-sea fungus Penicillium sp. SCSIO 06720. Nat. Prod. Res. 34, 1197–1205. doi: 10.1080/14786419.2018.1553880
Jiang, M. H., Chen, S. H., Li, J., and Liu, L. (2020). The biological and chemical diversity of tetramic acid compounds from marine-derived microorganisms. Mar. Drugs 18:114. doi: 10.3390/md18020114
Jin, L. M., Quan, C. S., Hou, X. Y., and Fan, S. D. (2016). Potential pharmacological resources: natural bioactive compounds from marine-derived fungi. Mar. Drugs 14:76. doi: 10.3390/md14040076
Jin, Z. M., Du, X. Y., Xu, Y. C., Deng, Y. Q., Liu, M. Q., Zhao, Y., et al. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293. doi: 10.1038/s41586-020-2223-y
Kempf, K., Raja, A., Sasse, F., and Schobert, R. (2013). Synthesis of penicillenol C1 and of a bis-azide analogue for photoaffinity labeling. J. Org. Chem. 78, 2455–2461. doi: 10.1021/jo3026737
Lai, D., Brotz-Oesterhelt, H., Muller, W. E. G., Wray, V., and Proksch, P. (2013). Bioactive polyketides and alkaloids from Penicillium citrinum, a fungal endophyte isolated from Ocimum tenuiflorum. Fitoterapia 91, 100–106. doi: 10.1016/j.fitote.2013.08.017
Li, D. H., Cai, S. X., Tian, L., Lin, Z. J., Zhu, T. J., Fang, Y. C., et al. (2007). Two new metabolites with cytotoxicities from deep-sea fungus, Aspergillus sydowi YHll-2. Arch. Pharm. Res. 33, 1051–1054. doi: 10.1007/BF02980236
Li, Z., Li, X., Huang, Y. Y., Wu, Y., Liu, R., Zhou, L., et al. (2020). Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. Proc. Natl. Acad. Sci. U. S. A. 117, 27381–27387. doi: 10.1073/pnas.2010470117
Lin, Z. J., Lu, Z. Y., Zhu, T. J., Fang, Y. C., Gu, Q. Q., and Zhu, W. M. (2008). Penicillenols from Penicillium sp. GQ-7, an endophytic fungus associated with Aegiceras corniculatum. Chem. Pharm. Bull. 56, 217–221.
Mao, Z. L., Wang, W. X., Su, R. X., Gu, G., Liu, Z. L., Lai, D., et al. (2019). Hyalodendrins A and B, new decalin-type tetramic acid larvicides from the endophytic fungus Hyalodendriella sp. Ponipodef12. Molecules 25:114. doi: 10.3390/molecules25010114
Mo, X. H., Li, Q. L., and Ju, J. H. (2014). Naturally occurring tetramic acid products: isolation, structure elucidation and biological activity. RSC Adv. 4, 50566–50593. doi: 10.1039/c4ra09047k
Morris, G. M., Huey, R., and Olson, A. J. (2008). Using AutoDock for ligand-receptor docking. Curr. Protoc. Bioinformatics 24, 8–14. doi: 10.1002/0471250953.bi0814s24
Nord, C., Levenfors, J. J., Bjerketorp, J., Guss, B., Oberg, B., and Broberg, A. (2020). Tetramic acid based alkaloids from Aspergillus amoenus Roberg strain UP197-antibiotic properties and new pyranterreones. Nat. Prod. Res. doi: 10.1080/14786419.2020.1855643 [Epub Online ahead of print].
Pang, X. Y., Lin, X. P., Yang, J., Zhou, X. F., Yang, B., Wang, J. F., et al. (2018). Spiro-phthalides and isocoumarins isolated from the marine-sponge-derived fungus Setosphaeria sp. SCSIO41009. J. Nat. Prod. 81, 1860–1868. doi: 10.1021/acs.jnatprod.8b00345
Pang, X. Y., Lin, X. P., Zhou, X. F., Yang, B., Tian, X. P., Wang, J. F., et al. (2020). New quinoline alkaloid and bisabolane-type sesquiterpenoid derivatives from the deep-sea-derived fungus Aspergillus sp. SCSIO06786. Fitoterapia 140:104406. doi: 10.1016/j.fitote.2019.104406
Pang, X. Y., Zhou, X. F., Lin, X. P., Yang, B., Tian, X. P., Wang, J. F., et al. (2021). Structurally various sorbicillinoids from the deep-sea sediment derived fungus Penicillium sp. SCSIO06871. Bioorg. Chem. 107:104600. doi: 10.1016/j.bioorg.2020.104600
Sabbah, D. A., Hajjo, R., Bardaweel, S. K., and Zhong, H. A. (2021). An updated review on SARS-CoV-2 main proteinase (MPro): protein structure and small-molecule inhibitors. Curr. Top. Med. Chem. 21, 442–460. doi: 10.2174/1568026620666201207095117
Schobert, R., and Schlenk, A. (2008). Tetramic and tetronic acids: an update on new derivatives and biological aspects. Bioorg. Med. Chem. 16, 4203–4221. doi: 10.1016/j.bmc.2008.02.069
Schrey, H., Backenkohler, J., Kogler, H., Plaumann, M., and Spiteller, P. (2019). Aminotenuazonic acid: isolation, structure elucidation, total synthesis and herbicidal activity of a new tetramic acid from fruiting bodies of Laccaria species. Chemistry 25, 10333–10341. doi: 10.1002/chem.201901405
Sengoku, T., Nagae, Y., Ujihara, Y., Takahashi, M., and Yoda, H. (2012). A synthetic approach to diverse 3-acyltetramic acids via O- to C-acyl rearrangement and application to the total synthesis of penicillenol series. J. Org. Chem. 77, 4391–4401. doi: 10.1021/jo300527f
Shaala, L. A., and Youssef, D. T. (2015). Identification and bioactivity of compounds from the fungus Penicillium sp. CYE-87 isolated from a marine tunicate. Mar. Drugs 13, 1698–1709. doi: 10.3390/md13041698
Shamsi, A., Mohammad, T., Anwar, S., Amani, S., Khan, M. S., Husain, F. M., et al. (2021). Potential drug targets of SARS-CoV-2: from genomics to therapeutics. Int. J. Biol. Macromol. 177, 1–9. doi: 10.1016/j.ijbiomac.2021.02.071
Sun, Y. L., Wang, J., Wang, Y. F., Zhang, X. Y., Nong, X. H., Chen, M. Y., et al. (2015). Cytotoxic and antiviral tetramic acid derivatives from the deep-sea-derived fungus Trichobotrys effuse DFFSCS021. Tetrahedron 71, 9328–9332. doi: 10.1016/j.tet.2015.10.010
Tsuda, M., Sasaki, M., Mugishima, T., Komatsu, K., Sone, T., Tanaka, M., et al. (2005). Scalusamides A-C, new pyrrolidine alkaloids from the marine-derived fungus Penicillium citrinum. J. Nat. Prod. 68, 273–276. doi: 10.1021/np049661q
Wang, J. F., He, W. J., Huang, X. L., Tian, X. P., Liao, S. R., Yang, B., et al. (2016). Antifungal new oxepine-containing alkaloids and xanthones from the deep-sea-derived fungus Aspergillus versicolor SCSIO 05879. J. Agric. Food Chem. 64, 2910–2916. doi: 10.1021/acs.jafc.6b00527
Wingen, L. M., Rausch, M., Schneider, T., and Menche, D. (2020). Synthesis of tetramic acid fragments derived from Vancoresmycin showing inhibitory effects towards S. aureus. Chem. Med. Chem. 15, 1390–1393. doi: 10.1002/cmdc.202000241
Yin, X., Liu, Y., Pan, J., Ye, H. L., Sun, Y., Zhao, D. Y., et al. (2019). Melongenaterpenes A-L, vetispirane-type sesquiterpenoids from the roots of Solanum melongena. J. Nat. Prod. 82, 3242–3248. doi: 10.1021/acs.jnatprod.9b00206
Yoda, H., Sengoku, T., Wierzejska, J., and Takahashi, M. (2010). First stereoselective synthesis of penicillenol A1 via novel O- to C-acyl rearrangement of O-acyltetramic acid. Synlett 2010, 2944–2946. doi: 10.1055/s-0030-1259045
Zehra, Z., Luthra, M., Siddiqui, S. M., Shamsi, A., Gaur, N. A., and Islam, A. (2020). Corona virus versus existence of human on the earth: a computational and biophysical approach. Int. J. Biol. Macromol. 161, 271–281. doi: 10.1016/j.ijbiomac.2020.06.007
Keywords: deep-sea-derived fungus, Penicillium sp., secondary metabolites, antibacterial, antiviral
Citation: Pang X, Chen W, Wang X, Zhou X, Yang B, Tian X, Wang J, Xu S and Liu Y (2021) New Tetramic Acid Derivatives From the Deep-Sea-Derived Fungus Penicillium sp. SCSIO06868 With SARS-CoV-2 Mpro Inhibitory Activity Evaluation. Front. Microbiol. 12:730807. doi: 10.3389/fmicb.2021.730807
Received: 25 June 2021; Accepted: 27 August 2021;
Published: 27 September 2021.
Edited by:
Paola Angelini, University of Perugia, ItalyCopyright © 2021 Pang, Chen, Wang, Zhou, Yang, Tian, Wang, Xu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junfeng Wang, d2FuZ2p1bmZlbmdAc2NzaW8uYWMuY24=; Shihai Xu, dHh1c2hAam51LmVkdS5jbg==; Yonghong Liu, eW9uZ2hvbmdsaXVAc2NzaW8uYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.