- 1State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Department of Laboratory Medicine, College of Medicine, The First Affiliated Hospital, Zhejiang University, Hangzhou, China
- 3Department of Infectious Diseases, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 4Department of Neurological Surgery, College of Medicine, The First Affiliated Hospital, Zhejiang University, Hangzhou, China
Intracranial infections caused by multidrug-resistant Gram-negative bacterium have led to considerable mortality due to extremely limited treatment options. Herein, we firstly reported a clinical carbapenem-resistant Escherichia coli isolate coharboring blaNDM–5 and blaCTX–M–65 from a patient with post-craniotomy meningitis. The carbapenem-resistant Escherichia coli strain CNEC001 belonging to Sequence Type 410 was only susceptible to amikacin and tigecycline, both of which have poor penetration through the blood-brain barrier (BBB). The blaCTX–M–65 gene was expressed on a 135,794 bp IncY plasmid. The blaNDM–5 gene was located on a genomic island region of an IncX3-type plasmid pNDM5-CNEC001. Based on the characteristics of the strain, we presented the successful treatment protocol of intravenous (IV) tigecycline and amikacin combined with intrathecal (ITH) amikacin in this study. Intracranial infection caused by Escherichia coli coharboring blaNDM–5 and blaCTX–M–65 is rare and fatal. Continuous surveillance and infection control measures for such strain need critical attention in clinical settings.
Introduction
Intracranial infection caused by carbapenem-resistant Enterobacteriaceae (CRE) is one of the most devastating complications following neurosurgery and also a serious nosocomial infection with high mortality (Fang et al., 2017). Limited antibiotics options due to the high resistance profile of CRE and poor blood-brain barrier (BBB) penetration complicate the treatment of CRE-related meningitis/encephalitis. The most common mechanism in CRE is carbapenemases production (Berger, 2011). New Delhi metallo-beta-lactamase (NDM) is a recently discovered metallo-beta-lactamase with enhanced carbapenem hydrolysis activity, especially NDM-5, and it can hydrolyze almost all beta-lactams except monobactams (Hornsey et al., 2011). NDM-5-producing Escherichia coli can lead to severe infections in diverse anatomical locations. Previous studies in China have shown several sporadic cases of clinical infections caused by blaNDM–5-positive E. coli including intestinal disease, urinary tract infections, invasive bloodstream and pulmonary infections (Zhu et al., 2016). Currently, intracranial infections due to E. coli producing NDM-5 and the related treatment experience are scarce. In this study, we report a case of secondary meningitis caused by an E. coli ST410 strain coproducing NDM-5 and CTX-M-65 for the first time, which was successfully treated by intravenous (IV) tigecycline and amikacin in combination with intrathecal (ITH) amikacin. Meanwhile, genomic and phenotypic characteristics of the strain are described in detail.
Case Presentation
A 67-year-old male, weighing 62 kg, was treated with intracranial hematoma clearance and bone flap decompression due to severe craniocerebral trauma caused by a traffic accident. The patient was transferred to the intensive care unit (ICU) after emergency surgery. The patient developed a fever of 38.1°C the day after surgery. Ten days later, consciousness disturbance was deteriorated; the culture of the cerebrospinal fluid (CSF) yielded carbapenem-resistant E. coli. Systematic tigecycline and intrathecal polymyxin B were used but did not improve CSF findings. The patient was transferred to our hospital for further treatment. The first CSF examination after admission revealed severe leukocytic pleocytosis (5,700 cells/μL, neutrophil accounted for 93%), elevated protein (2.82 g/L), and low glucose (0.7 mmol/L). Lumbar cistern drainage was performed immediately. The CSF exhibited pale yellow with turbidity. Antibiotics were changed to polymyxin B 75 mg × every 12 h (q12 h) IV + tigecycline 50 mg × every 12 h (q12 h) IV + polymyxin B 5 mg × once daily (qd) ITH. On day 6 after admission, we got the CSF bacterial culture and susceptibility results that E. coli was the pathogen, being resistant to many classes of antibiotics (including carbapenem) while only susceptible to tigecycline and amikacin. Due to severe neurotoxicity (newly emergent seizure) induced by polymyxin B, it was discontinued and was replaced with amikacin 800 mg × qd IV. Given that systemic anti-infection therapy alone may not achieve the effective concentration for antimicrobial activity in central nervous system (CNS), the patient was commenced on concurrent intrathecal administration of amikacin 50 mg × qd. As the poor activity of tigecycline to cross the BBB, we applied a higher dose of 100 mg × q12 h IV. From day 7, repeated CSF cultures were negative. The highest temperature during treatment was 39°C. From day 9, the patient had no fever. CSF cell count and protein content all followed a declining trend during the treatment process. A wide range of bacteria can produce biofilm on prosthetic implants and survive antimicrobial therapy. Hence, on day 15, the lumbar cistern drain was removed and intrathecal amikacin was adjusted to 50 mg × every other day (qod) ITH. The timeline of treatment options, disease course, and laboratory findings is shown in Figure 1. After further consolidation therapy, the patient recovered well and no recurrence was observed during the 1-year follow-up period.
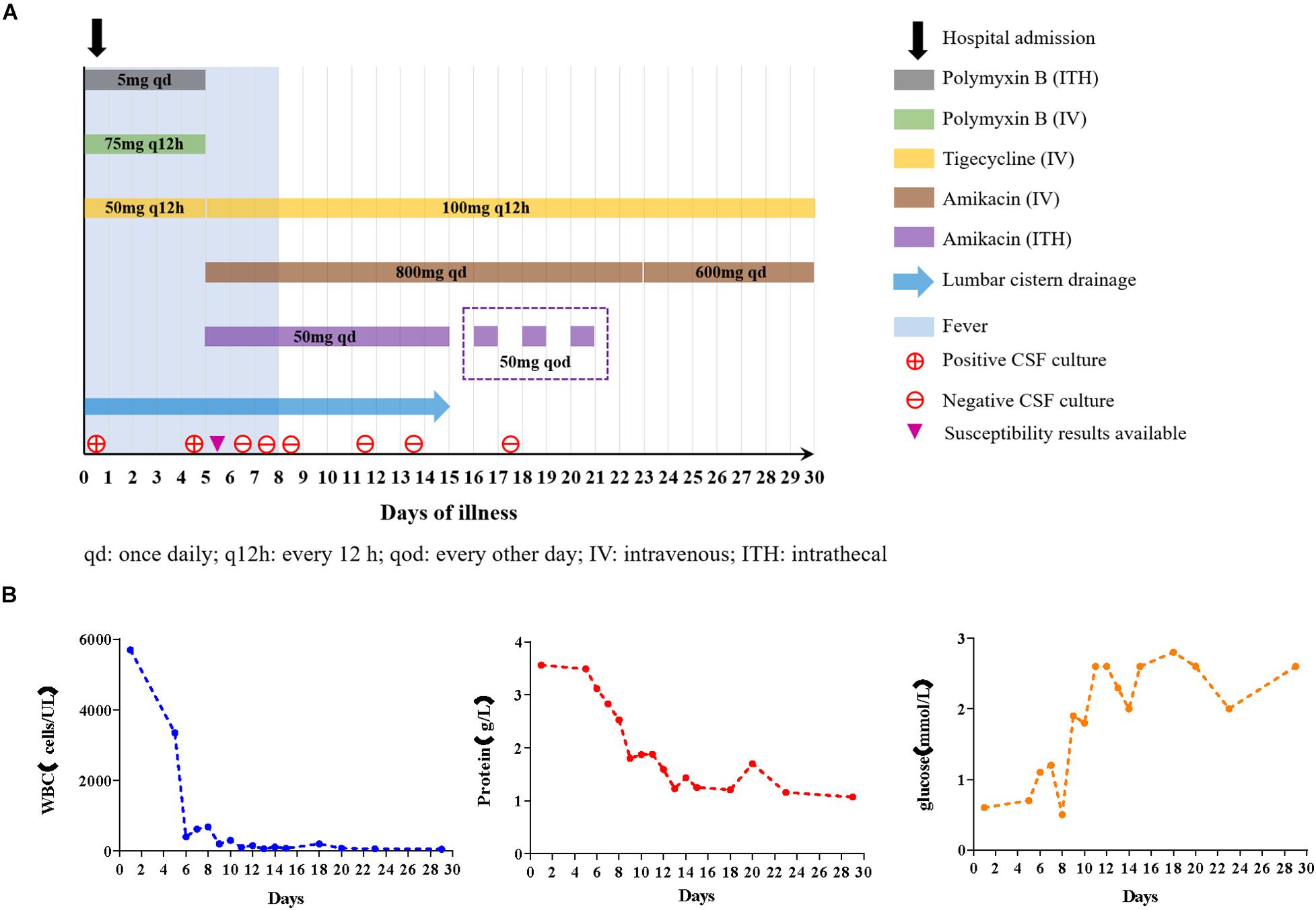
Figure 1. (A) Diagram of antibiotics administration, fever, and CSF cultures in the case during hospitalization. The contents in the box refer to the usage and dosage of each antibiotic at different periods. (B) Trends of CSF analytes after treatment.
Materials and Methods
Collection and Identification of Isolate
Strain CNEC001 was isolated from the patients’ cerebrospinal fluid sample and identified by an automated Vitek 2 compact system (bioMérieux, France).
Antibiotic Susceptibility Testing
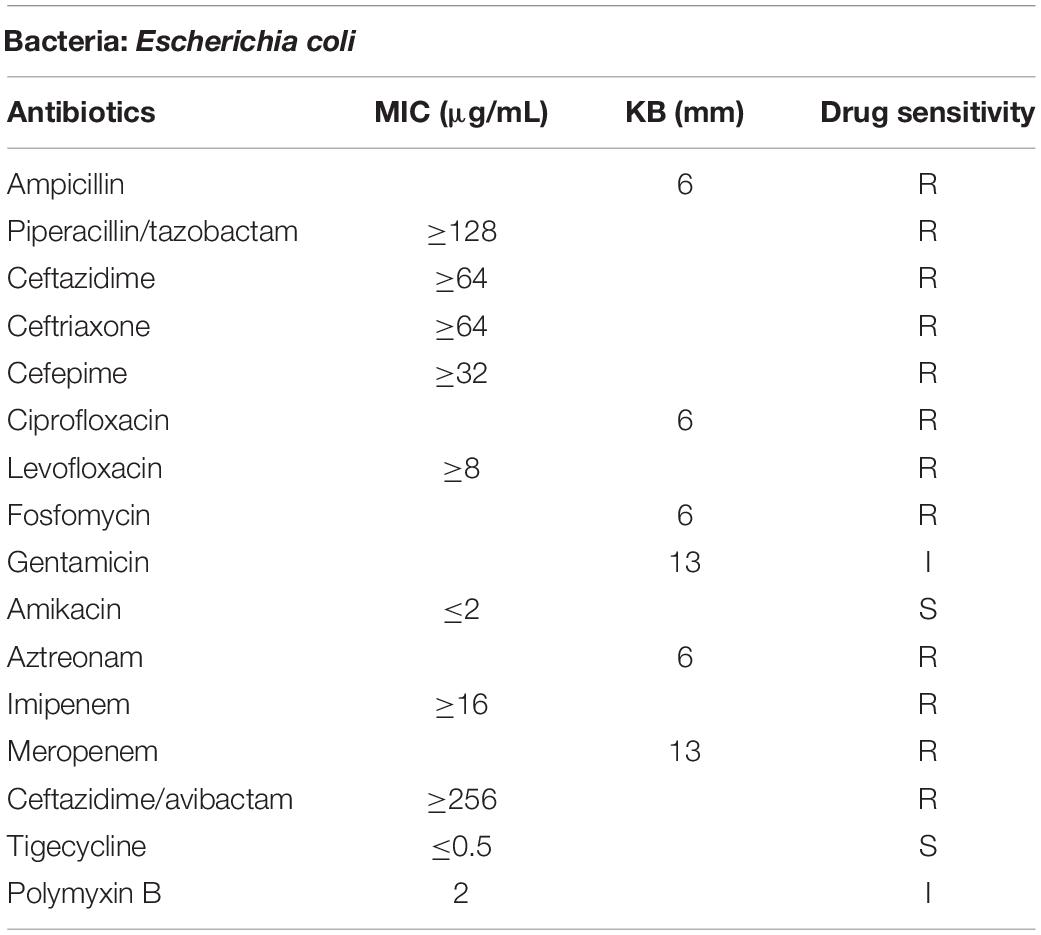
Susceptibility testings to tigecycline, ceftazidime/avibactam and polymyxin B were conducted by using the reference Clinical and Laboratory Standards Institute (CLSI) broth microdilution method (Clinical and Laboratory Standards Institute, 2020), while other antimicrobial agents were performed by Vitek 2 compact system and the Kirby--Bauer disk diffusion assay. Antibiotics tested in the study were ampicillin, piperacillin/tazobactam, ceftazidime, ceftriaxone, cefepime, ciprofloxacin, levofloxacin, fosfomycin, gentamicin, amikacin, aztreonam, imipenem, meropenem, ceftazidime/avibactam, tigecycline, and polymyxin B. The breakpoint of tigecycline was interpreted according to the criterion and recommendation from European Committee on Antimicrobial Susceptibility Testing1 while the remaining antimicrobial results were determined in accordance with Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute, 2020). E. coli ATCC 25922 was used as a control strain.
Whole Genome Sequencing (WGS) and Bioinformatic Analysis
Whole DNA of the strain was extracted using a QIAamp DNA MiniKit (Qiagen, Valencia, CA, United States) according to the manufacturer’s protocol. Purified DNA samples were submitted to next-generation high-throughput sequencing (NGS) on the HiSeq2000TM platform (Illumina, San Diego, United States) with 2∗100-bp paired-end reads. The overall genome coverage was 108 times and the N50 value of the contigs was 94.9 kbp, after assembling by the Illumina sequencing solely. The strain was further subjected to long-read high-throughput sequencing (LRS) on the MinION platform (Nanopore, Oxford, United Kingdom). Sequencing, libraries were prepared using the SQK-LSK109 Ligation Sequencing kit in conjunction with the PCR-Free ONT EXP-NBD104 and 114 Native Barcode Expansion kit, without optional shearing steps to select for long reads. Individual libraries were quantitated through Qubit. At last, the library was sequenced on a MinION platform. The overall genome coverage of the MinION sequencing was 176 times. We did not obtain the N50 value only for long read data because we used hybrid assembly method and the long reads were only utilized for closing the gap between the contigs that short reads assembled. We used Unicycler v0.4.8, an assembly pipeline, for our bacterial genome assembly with both short and long reads. Unicycler will first use SPAdes to make a short-read assembly graph, and then it will use various methods to scaffold that graph with the long reads. Several mapping strategies were used in Unicycler assembly pipeline for after-polishing to improve the assembly quality, including using the Racon to polish the assembly (Wick et al., 2017). Contigs were annotated by Prokka (version 1.14.5) combined with BLAST searches.2 Antibiotic resistance genes, acquired virulence genes, multi-locus sequence typing (MLST) and plasmid incompatibility groups were all detected using bioinformatics tools available on the CGE Server.3 Plasmid DNA sequences were collinearly analyzed using Easyfig 2.2.5 (Sullivan et al., 2011). Gene organization diagrams were drawn in the CGView Server (Larsen, 2013).
Ethical Approval
This study has been reviewed and approved by the ethical research committee of the First Affiliated Hospital, College of Medicine, Zhejiang University. The ethics committee approved the waiver of the patient’s informed consent, with the justification that this was a retrospective study whose information was obtained from medical records and that the data were de-identified and anonymously analyzed.
Results
Antibiotic Susceptibility Results
Strain CNEC001 was identified as E. coli by the Vitek 2 automated system. Antimicrobial susceptibility test revealed that CNEC001 was resistant to the majority of antimicrobial agents, including ampicillin, piperacillin/tazobactam, ceftazidime, ceftriaxone, cefepime, ciprofloxacin, levofloxacin, fosfomycin, aztreonam, imipenem, meropenem, and ceftazidime/avibactam, intermediate to polymyxin B and gentamicin, while only susceptible to tigecycline and amikacin (Table 1).
Genomic Characteristics of CNEC001
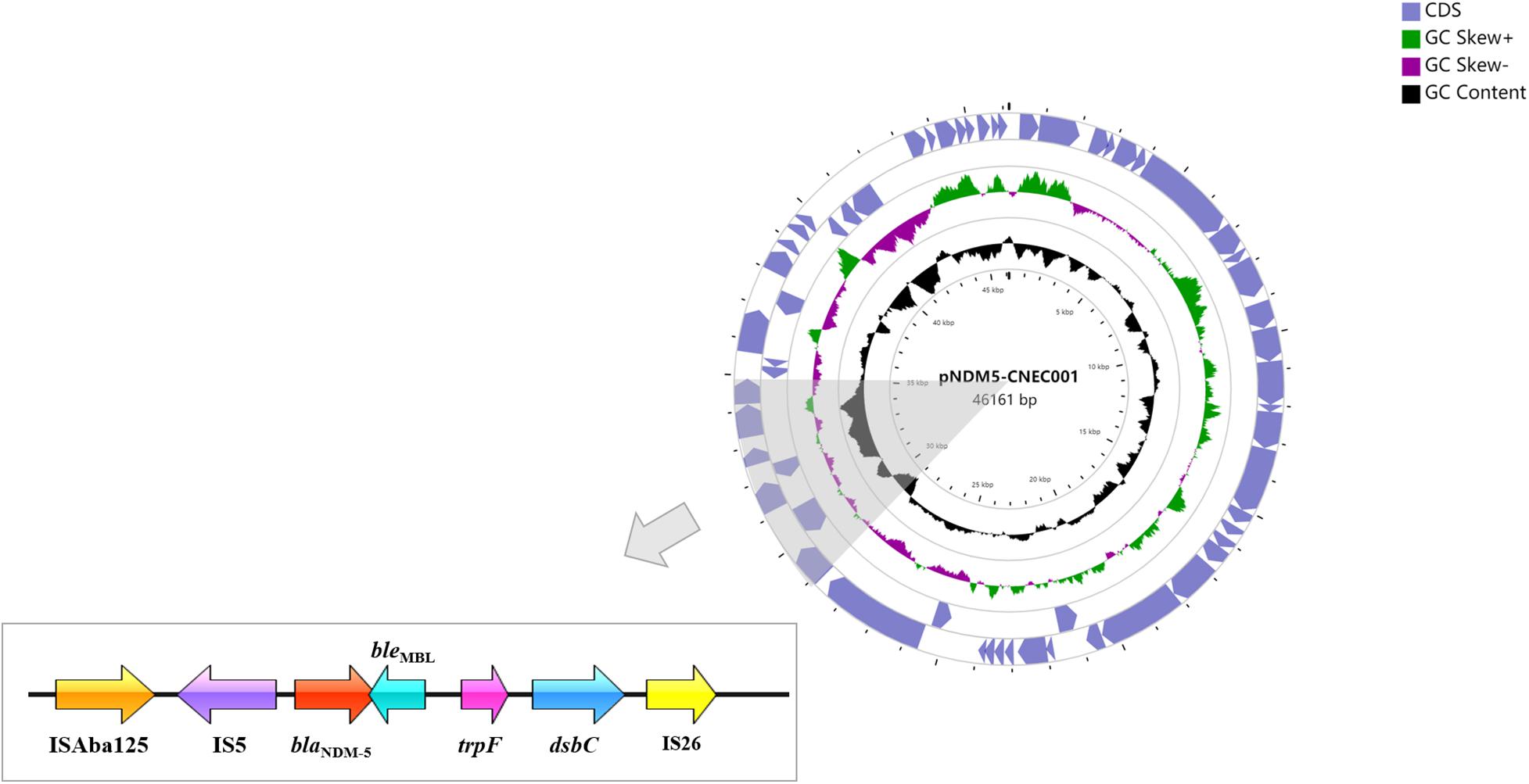
Strain CNEC001 belonged to sequence type 410 (ST410). The genome size of CNEC001 was 5,034,429 bp, with 50.6% GC content. This strain carried four different plasmids and harbored multiple resistance genes, including three β-lactam resistance genes (blaNDM–5, blaCTX–M–65, blaCMY–2), two aminoglycoside resistance genes [aph (4)- Ia, aadA5], three sulphonamide resistance genes (sul1, sul2, dfrA17), one macrolide resistance gene [mph(A)] and one tetracycline resistance gene [tet (B)]. Most of these genes [blaCTX–M–65, sul1, sul2, dfrA17, aph (4)- Ia, aadA5, and mph(A)] were found on a 135,794 bp IncY plasmid. blaNDM–5 was located on a 46,161 bp IncX3 plasmid, which here was assigned pNDM5-CNEC001. blaCMY–2 was the only resistance gene mediated by chromosome. Virulence gene terC (Tellurium ion resistance protein) may be correlated with significant pathogenic potential of E. coli.
Characteristics of the BlaNDM–5-Harboring Plasmid pNDM5-CNEC001
Plasmid pNDM5-CNEC001 was 46161 bp in size with average G+C content of 46.7%, encoding 63 predicted open reading frames (ORFs). It belonged to the IncX3 incompatibility group based on PlasmidFinder analysis and carried the sole resistance gene blaNDM–5. Sequence alignments on BLAST revealed that pNDM5-CNEC001 was highly similar to pNDM-QD28 (GenBank accession number KU167608.1), a plasmid carried by an NDM-5-producing E. coli ST167 in China, with more than 99.99% identities and 100% query coverage (Figure 2). Gene blaNDM–5 was within the typical structure ISAba125-IS5-blaNDM–5-bleMBL-trpF-dsbC-IS26 (Figure 3). pNDM5-CNEC001-like plasmids could be found in various Enterobacteriaceae isolates among which E. coli appeared to be particularly common, followed by Klebsiella pneumoniae.
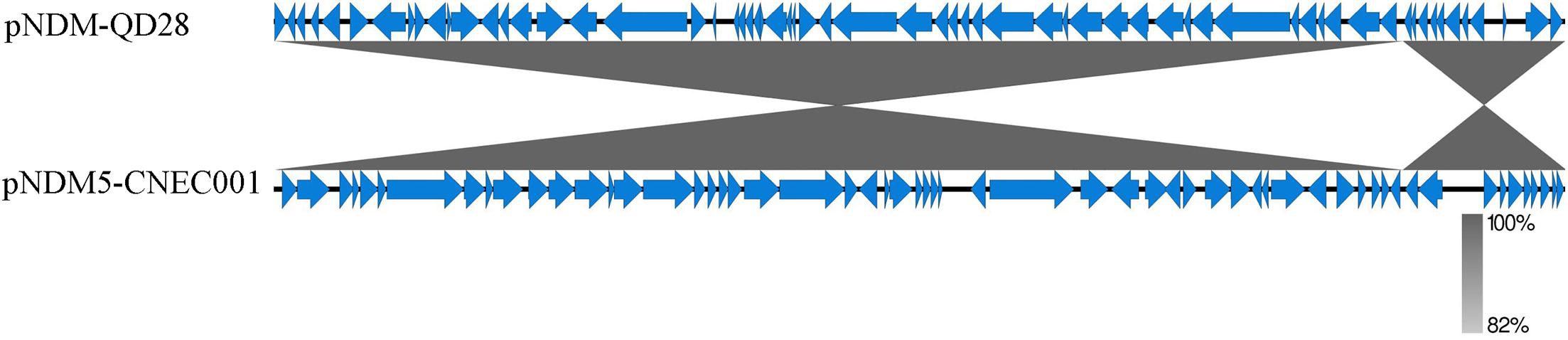
Figure 2. Comparison of the genetic organization between pNDM5-CNEC001 and pNDM-QD28 (GenBank accession number KU167608.1). Genes are denoted by arrows. Shading indicates regions of homology.
Nucleotide Sequence Accession Number
The complete genome sequence of strain CNEC001 has been deposited at DDBJ/ENA/GenBank under the accession JAIELB000000000. The version described in this paper is version JAIELB010000000. The complete nucleotide sequence of plasmid pNDM5-CNEC001 was submitted to GenBank with the accession number MZ270636.
Discussion
To date, E. coli ST167 is classified as the predominant clonal lineage spreading the blaNDM–5 gene in China (Bi et al., 2018; Xu et al., 2019; Li et al., 2020). E. coli ST410 has been increasingly reported as NDM-5 or OXA-181-type carbapenemase producer, this lineage has been categorized as an internationally emerging “high-risk” clone evidenced by its expression of various antimicrobial resistance determinants (ESBLs, pAmpCs, carbapenemases gene, colistin resistance genes), effective interspecies transmission, persistence in hosts and enhanced pathogenicity (Roer et al., 2018). Humans, food animals, and the environments are all important sources of ST410 E. coli (Feng et al., 2019). Comparison between NDM-5-producing ST167 and ST410 E. coli isolates previously reported in China reveals that NDM-5-producing ST167 E. coli strains were recovered from more clinical samples (including ascites, sputum, urine, blood, pus, CSF, bile, rectal swab). NDM-5-producing ST410 E. coli have rarely reported in CSF (Hammerum et al., 2015; Krishnaraju et al., 2015; Wailan et al., 2015; Zhu et al., 2016; Li et al., 2018). Moreover, ST410 E. coli were more common to carry blaCMY.
In this study, according to the guideline established by Clinical and Laboratory Standards Institute (2020) which eliminated the “susceptible” interpretive category for the polymyxins, the ST410 E. coli strain CNEC001 was intermediate to polymyxin B and gentamicin, only susceptible to tigecycline and amikacin, and possessed virulence geneterC that enabled it to cause disease in the host. The tellurite resistance gene is widely spread among pathogenic species. Ter proteins play an important role in phagocytosis inhibition, allowing infective pathogens to evade the neutrophil responses (Bai et al., 2012). Apart from carbapenem resistance, CNEC001 showed aztreonam and cephalosporins resistance, which could be attributed to blaNDM–5 and blaCTX–M–65. Carbapenemase-encoding gene blaNDM–5 of strain CNEC001was detected on an IncX3-type plasmid pNDM5-CNEC001. IncX3-type plasmid is self-transmissible and has been proved as a common vehicle facilitating the horizontal transmission of either blaNDM–5 or blaOXA–181 between diverse species and hosts (Yang et al., 2014). BLAST analysis revealed that the IncX3 plasmid carrying blaNDM–5 in our study was highly similar to each other isolated from different countries and host sources, suggesting its ability to be an efficient vehicle for blaNDM–5 dissemination among humans, animals, foods and the environments, potentially indicating its role in the rapid spread of blaNDM–5-harboring isolates. The blaCTX–M–65 gene is also encoded on an easily transferable plasmid carrying multiple resistance genes of other classes of antibiotics, which is consistent with the results of previous studies (Bevan et al., 2017; Karami et al., 2021), leading to multidrug resistance if the plasmid is horizontally spread. CTX-M beta-lactamases are the most common types of Extended spectrum beta-lactamases (ESBLs), which can inactivate penicillins, cephalosporins, and aztreonam. The blaNDM genes generally coexist with blaCTX–M genes (Suay-Garcia and Perez-Gracia, 2019; Tooke et al., 2019). A study of the epidemiology of carbapenem-resistant E. coli in Chinese patients showed that almost all (>96%) NDM-producing strains encoded CTX-M-type β-lactamases (Tian et al., 2020). Once this occurs, it will lead to increased difficulty of antibiotic treatment, aztreonam being ineffective. There are few relevant reports on E. coli harboring the blaCTX–M–65 gene isolated from human in China. blaCTX–M–65 gene is widely found in E. coli isolated from food-producing animals and no sequence type related to the widespread popularity of CTX-M-65-type ESBL has been found. Previous study showed that E. coli ST410 encoding blaCTX–M–65 had been obtained from healthy broiler chickens (Liu et al., 2020). It is very likely that poultry will be sources of blaCTX–M–65 and transfer this resistant gene to human via food supply chain. We previously reported a community-acquired renal abscess caused by a ST410 E. coli strain coharboring blaNDM–5 and blaCTX–M–65 in an outpatient (Hu et al., 2020). However, to our knowledge, this is the first report of a clinical E. coli ST410 co-harboring blaNDM–5, blaCTX–M–65 and blaCMY–2 isolated from CSF. In this study, our patient had a history of head trauma that was treated with craniotomy and developed meningitis soon after emergent surgery, thus, we speculate that strain CNEC001 may originate from the surgical site colonized by the pathogenic bacteria which translocated into CNS during surgery and thus caused the infection. However, community-acquired resistant isolates might become epidemic in healthcare settings and cause hospital outbreaks, presenting a serious threat to inpatients’ health. Increased surveillance is still urgently required to prevent the emergence and further dissemination of the “superbug” which has co-production of ESBLs and NDM.
The optimal antimicrobial treatment regimen at present for MDR bacterial meningitis still stands unclear but should be based on in vitro drug sensitivity testing. Currently, drugs with inability to adequately permeate through the BBB (including amikacin, polymyxin B) are clinically used for intrathecal therapy of CNS infections caused by MDR Gram-negative pathogens (Tunkel et al., 2017; Molinaro et al., 2018; Nau et al., 2020). Intrathecal/intraventricular polymyxins combined with systemic administration were recommended to treat intracranial infection caused by CRE in recent several studies (Chen et al., 2020; Zhong et al., 2020). However, nephrotoxicity and neurotoxicity remain major problems for the clinical use of polymyxins. Intrathecal/intraventricular polymyxins therapy has been associated with serious neurological adverse effects including seizures, chemical meningitis and cauda equina syndrome. In order to improve clinical outcomes, polymyxins should be chosen in combination with other active antimicrobials whenever possible in treatment of CNS infections (Paul et al., 2018; van Duin et al., 2018; Pogue et al., 2020). When alternative drugs are available, it is strongly recommended to prioritize non–polymyxin drugs with antibacterial activity against CRE in vitro, including ceftazidime/avibactam (ineffective against bacteria producing metallo-beta-lactamase), dual carbapenems, aminoglycosides and tigecycline. Aminoglycosides gentamicin and amikacin are effective against approximately 50% of CRE isolates (Satlin et al., 2011) and still considered first-line therapy for CRE (Suay-Garcia and Perez-Gracia, 2019), however, they have disadvantages of nephrotoxicity and low penetration of CSF.
In our study, the pathogen was intermediate to gentamicin and polymyxin B and only susceptible to amikacin and tigecycline. The patient developed new adverse reaction (epilepsy) caused by polymyxin B during therapy, so treatment regimen we performed was i.v. tigecycline and i.v. amikacin plus intrathecal amikacin. Tigecycline is characterized as a time-dependent agent with concentration-dependent killing and drug-induced prolonged effects. The recommended intravenous regimen of tigecycline is an initial dose of 100 mg, followed by 50 mg every 12 h. Despite its high effectiveness against MDR pathogens, tigecycline is currently not recommended in cases of intracranial infection, as CSF concentrations ranged from only 0.035 to 0.048 mg/L with the usual intravenous dose of 100 mg/day in the absence of meningeal inflammation (Nau et al., 2020). We used a higher dose of 100 mg IV q12 h in order to obtain tigecycline concentration in excess of the MIC (0.5 μg/ml) for E. coli in CSF. Unfortunately, we did not monitor therapeutic drug concentrations while on therapy. Several cases previously reported failed to achieve desired target site concentrations (those exceeding the MIC) in CNS by utilizing double the recommended dosage (i.e.,100 mg IV Q12 h) (Chen et al., 2007; Ray et al., 2010). Given the strain was also susceptible to amikacin, hence, intravenous combined with intrathecal administration of amikacin were added to improve therapeutic effect. Amikacin is concentration-dependent antibiotic with prolonged post-antibiotic effect. CSF concentrations with uninflamed meninges are close to the MICs of moderately susceptible bacteria under usual intravenous dose (600 mg qd) (Nau et al., 2018). In our case, a higher intravenous dose of amikacin (800 mg × qd) and intrathecal administration (50 mg × qd) were performed in the initial stages of treatment to improve the effective concentration for antimicrobial activity in CNS. A systematic review covering the years 1946–2015 reported intrathecal doses ranging from 5 to 50-mg amikacin daily to be effective and well tolerated, though the optimum dosage still remains unclear (Nau et al., 2018).
Monotherapy with intrathecal drugs rarely induces durable responses. Intravenous combined with intrathecal administration can provide a high local drug concentration at the site of infection and has been demonstrated to be effective by recent research with successful outcomes (De Bonis et al., 2016; Nau et al., 2018). Combined antibiotic therapy can increase antimicrobial activity and yield better results than monotherapy. Early indwelling drainage of CSF is beneficial in alleviating intracranial infection, however, it is crucial for successful treatment that removing the prostheses as soon as possible after the CSF cultures have been negative as well as CSF and clinical parameters have improved.
Conclusion
In conclusion, this is the first report of a clinical E. coli ST410 co-harboring blaNDM–5, blaCTX–M–65, and blaCMY–2 responsible for intracranial infection in China, which renders therapeutic intervention quite difficult and leads to poor outcome. This study also provides treatment experience that may apply to patients under similar circumstances, and unravels possible mechanisms of dissemination of antibiotic resistance genes. Constant and careful surveillance for NDM and ESBL-producing strains is urgently warranted in clinical settings. Prompt and appropriate infection control measures should be taken after the first isolation of such species.
Data Availability Statement
The data presented in the study are deposited in the GenBank repository, accession numbers JAIELB000000000 and MZ270636. Our data is publicly available.
Author Contributions
QY and P-pZ conceived the idea and performed the experiments. QY, P-pZ, and X-jZ analyzed the data. YJ helped with materials and reagents. QY wrote the manuscript. T-tQ and MZ reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the research grants from the National Natural Science Foundations of China (Nos. NSFC81871689 and NSFC 82073610).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ http://www.eucast.org/clinical_breakpoints
- ^ www.ncbi.nlm.nih.gov/blast/
- ^ https://cge.cbs.dtu.dk/services/
References
Bai, X.-n., Zhao, A.-l., Meng, Q., Xu, J.-g., and Xiong, Y.-w. (2012). Analysis of tellurite resistance level and resistance gene cluster in non-O157 Shiga toxin-producing Escherichia coli isolates. Zhonghua Weishengwuxue He Mianyixue Zazhi 32, 585–588. doi: 10.3760/cma.j.issn.0254-5101.2012.07.003
Berger, R. E. (2011). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study editorial comment. J. Urol. 185:154. doi: 10.1016/j.juro.2010.09.058
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M beta-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Bi, R., Kong, Z., Qian, H., Jiang, F., Kang, H., Gu, B., et al. (2018). High prevalence of blaNDM variants among carbapenem-resistant Escherichia coli in Northern Jiangsu Province, China. Front. in Microbiol. 9:2704. doi: 10.3389/fmicb.2018.02704
Chen, H. W., Guo, X. C., Xie, D. C., Dong, X. W., Niu, J. X., and Chen, G. Q. A. (2020). Clinical study on the use of intraventricular polymyxin B supplemented by continuous external ventricular drainage in the treatment of drug-resistant gram-negative bacilli intracranial infection. Infect. Drug Resist. 13, 2963–2970. doi: 10.2147/idr.S261510
Chen, J. L., Orsini, J., and Killu, C. (2007). Poor central nervous system penetration of tigecycline in a patient with sepsis and ventriculitis caused by multidrug-resistant Klebsiella pneumoniae. J. Pharm. Technol. 23, 344–348.
Clinical and Laboratory Standards Institute (CLSI) (2020). Performance Standards for Antimicrobial Susceptibility Testing, CLSI supplement M100, 30th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
De Bonis, P., Lofrese, G., Scoppettuolo, G., Spanu, T., Cultrera, R., Labonia, M., et al. (2016). Intraventricular versus intravenous colistin for the treatment of extensively drug resistant Acinetobacter baumannii meningitis. Eur. J. Neurol. 23, 68–75. doi: 10.1111/ene.12789
Fang, C., Zhu, T., Zhang, P., Xia, L., and Sun, C. (2017). Risk factors of neurosurgical site infection after craniotomy: a systematic review and meta-analysis. Am. J. Infect. Control. 45, E123–E134. doi: 10.1016/j.ajic.2017.06.009
Feng, Y., Liu, L., Lin, J., Ma, K., Long, H., Wei, L., et al. (2019). Key evolutionary events in the emergence of a globally disseminated, carbapenem resistant clone in the Escherichia coli ST410 lineage. Commun. Biol. 2:322. doi: 10.1038/s42003-019-0569-1
Hammerum, A. M., Littauer, P., and Hansen, F. (2015). Detection of Klebsiella pneumoniae co-producing NDM-7 and OXA-181, Escherichia coli producing NDM-5 and Acinetobacter baumannii producing OXA-23 in a single patient. Int. J. Antimicrob. Agents 46, 597–598. doi: 10.1016/j.ijantimicag.2015.07.008
Hornsey, M., Phee, L., and Wareham, D. W. (2011). A novel variant, NDM-5, of the New Delhi metallo-beta-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55, 5952–5954. doi: 10.1128/aac.05108-11
Hu, H., Mao, J., Chen, Y., Wang, J., Zhang, P., Jiang, Y., et al. (2020). Clinical and microbiological characteristics of community-onset Carbapenem-resistant Enterobacteriaceae isolates. Infect. Drug Resist. 13, 3131–3143. doi: 10.2147/idr.S260004
Karami, N., Kk, S., Yazdanshenas, S., Lin, Y.-L., Jaen-Luchoro, D., Ekedahl, E., et al. (2021). Identity of blaCTX-M carrying plasmids in sequential ESBL-E. coli isolates from patients with recurrent urinary tract infections. Microorganisms 9:1138. doi: 10.3390/microorganisms9061138
Krishnaraju, M., Kamatchi, C., Jha, A. K., Devasena, N., Vennila, R., Sumathi, G., et al. (2015). Complete sequencing of an IncX3 plasmid carrying bla(NDM-5) allele reveals an early stage in the dissemination of the bla(NDM) gene. Indian J. Med. Microbiol. 33, 30–38. doi: 10.4103/0255-0857.148373
Larsen, M. V. (2013). Internet-based solutions for analysis of next-generation sequence data. J. Clin. Microbiol. 51:3162. doi: 10.1128/jcm.01348-13
Li, J., Yu, T., Tao, X.-Y., Hu, Y.-M., Wang, H.-C., Liu, J.-L., et al. (2020). Emergence of an NDM-5-producing Escherichia coli sequence type 410 clone in infants in a children’s Hospital in China. Infect. Drug Resist. 13, 703–710. doi: 10.2147/idr.S244874
Li, X., Fu, Y., Shen, M. Y., Huang, D. Y., Du, X. X., Hu, Q. F., et al. (2018). Dissemination of bla(NDM-5) gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob. Resist. Infect. Control. 7:9. doi: 10.1186/s13756-018-0349-6
Liu, X., Wei, X., Liu, L., Feng, X., Shao, Z., Han, Z., et al. (2020). Prevalence and characteristics of extended-spectrum beta-lactamases-producing Escherichia coli from broiler chickens at different day-age. Poult. Sci. 99, 3688–3696. doi: 10.1016/j.psj.2020.04.015
Molinaro, M., Morelli, P., De Gregori, M., De Gregori, S., Giardini, I., Tordato, F., et al. (2018). Efficacy of intraventricular amikacin treatment in pan-resistant Pseudomonas aeruginosa postsurgical meningitis. Infect. Drug Resist. 11, 1369–1372. doi: 10.2147/idr.S169271
Nau, R., Blei, C., and Eiffert, H. (2020). Intrathecal antibacterial and antifungal therapies. Clin. Microbiol. Rev. 33:e00190-19. doi: 10.1128/CMR.00190-19
Nau, R., Seele, J., Djukic, M., and Eiffert, H. (2018). Pharmacokinetics and pharmacodynamics of antibiotics in central nervous system infections. Curr. Opin. Infect. Dis. 31, 57–68. doi: 10.1097/qco.0000000000000418
Paul, M., Daikos, G. L., Durante-Mangoni, E., Yahav, D., Carmeli, Y., Benattar, Y. D., et al. (2018). Colistin alone versus colistin plus meropenem for treatment of severe infections caused by carbapenem-resistant Gram-negative bacteria: an open-label, randomised controlled trial. Lancet Infect. Dis. 18, 391–400. doi: 10.1016/s1473-3099(18)30099-9
Pogue, J. M., Kaye, K. S., Veve, M. P., Patel, T. S., Gerlach, A. T., Davis, S. L., et al. (2020). Ceftolozane/tazobactam vs polymyxin or aminoglycoside-based regimens for the treatment of drug-resistant Pseudomonas aeruginosa. Clin. Infect. Dis. 71, 304–310. doi: 10.1093/cid/ciz816
Ray, L., Levasseur, K., Nicolau, D. P., and Scheetz, M. H. (2010). Cerebral spinal fluid penetration of tigecycline in a patient with Acinetobacter baumannii cerebritis. Ann. Pharmacother. 44, 582–586. doi: 10.1345/aph.1M480
Roer, L., Overballe-Petersen, S., Hansen, F., Schonning, K., Wang, M., Roder, B. L., et al. (2018). Escherichia coli sequence type 410 is causing new international high-risk clones. Msphere 3:e00337-18. doi: 10.1128/mSphere.00337-18
Satlin, M. J., Kubin, C. J., Blumenthal, J. S., Cohen, A. B., Furuya, E. Y., Wilson, S. J., et al. (2011). comparative effectiveness of aminoglycosides, polymyxin B, and tigecycline for clearance of carbapenem-resistant Klebsiella pneumoniae from urine. Antimicrob. Agents Chemother. 55, 5893–5899. doi: 10.1128/aac.00387-11
Suay-Garcia, B., and Perez-Gracia, M. T. (2019). Present and future of carbapenem-resistant Enterobacteriaceae (CRE) infections. Antibiotics Basel 8:16. doi: 10.3390/antibiotics8030122
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tian, X. B., Zheng, X. K., Sun, Y., Fang, R. C., Zhang, S. Q., Zhang, X. C., et al. (2020). Molecular mechanisms and epidemiology of carbapenem-resistant Escherichia coli isolated from Chinese patients during 2002-2017. Infect. Drug Resist. 13, 501–512. doi: 10.2147/idr.S232010
Tooke, C. L., Hinchliffe, P., Bragginton, E. C., Colenso, C. K., Hirvonen, V. H. A., Takebayashi, Y., et al. (2019). beta-Lactamases and beta-lactamase inhibitors in the 21st century. J. Mol. Biol. 431, 3472–3500. doi: 10.1016/j.jmb.2019.04.002
Tunkel, A. R., Hasbun, R., Bhimraj, A., Byers, K., Kaplan, S. L., Scheld, W. M., et al. (2017). 2017 infectious diseases society of America’s clinical practice guidelines for healthcare-associated ventriculitis and meningitis. Clin. Infect. Dis. 64, E34–E65. doi: 10.1093/cid/ciw861
van Duin, D., Lok, J. J., Earley, M., Cober, E., Richter, S. S., Perez, F., et al. (2018). Colistin versus ceftazidime-avibactam in the treatment of infections due to carbapenem-resistant Enterobacteriaceae. Clin. Infect. Dis. 66, 163–171. doi: 10.1093/cid/cix783
Wailan, A. M., Paterson, D. L., Caffery, M., Sowden, D., and Sidjabat, H. E. (2015). Draft genome sequence of NDM-5-producing Escherichia coli sequence type 648 and genetic context of bla(NDM-5) in Australia. Microbiol. Resour. Announc. 3:e00194-15. doi: 10.1128/genomeA.00194-15
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Xu, L., Wang, P., Cheng, J., Qin, S., and Xie, W. (2019). Characterization of a novel bla NDM-5-harboring IncFII plasmid and an mcr-1-bearing IncI2 plasmid in a single Escherichia coli ST167 clinical isolate. Infect. Drug Resist. 12, 511–519. doi: 10.2147/idr.S192998
Yang, P., Xie, Y., Feng, P., and Zong, Z. (2014). bla(NDM-5) carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob. Agents Chemother. 58, 7548–7552. doi: 10.1128/aac.03911-14
Zhong, L., Shi, X.-Z., Su, L., and Liu, Z.-F. (2020). Sequential intraventricular injection of tigecycline and polymyxin B in the treatment of intracranial Acinetobacter baumannii infection after trauma: a case report and review of the literature. Mil. Med. Res. 7:23. doi: 10.1186/s40779-020-00253-9
Keywords: intracranial infection, blaCTX–M–65, treatment protocol, blaNDM–5, carbapenem-resistant Escherichia coli
Citation: Yang Q, Zhang P-p, Jiang Y, Zheng X-j, Zheng M and Qu T-t (2021) Successful Treatment of Severe Post-craniotomy Meningitis Caused by an Escherichia coli Sequence Type 410 Strain Coharboring blaNDM–5 and blaCTX–M–65. Front. Microbiol. 12:729915. doi: 10.3389/fmicb.2021.729915
Received: 24 June 2021; Accepted: 18 August 2021;
Published: 08 September 2021.
Edited by:
Axel Cloeckaert, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceReviewed by:
Mariana Carmen Chifiriuc, University of Bucharest, RomaniaGhassan M. Matar, American University of Beirut, Lebanon
Henrik Hasman, Statens Serum Institut (SSI), Denmark
Copyright © 2021 Yang, Zhang, Jiang, Zheng, Zheng and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting-ting Qu, cXV0aW5ndGluZ0B6anUuZWR1LmNu; Min Zheng, bWluemhlbmdAemp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Qing Yang1,2†
Qing Yang1,2† Piao-piao Zhang
Piao-piao Zhang Yan Jiang
Yan Jiang Min Zheng
Min Zheng Ting-ting Qu
Ting-ting Qu