- 1BRGM, DEPA/GME, Orléans, France
- 2ISTO, UMR 7327, CNRS-Université d’Orléans-Brgm, Orléans, France
In the context of climate change and biodiversity loss, rehabilitation of degraded urban soils is a means of limiting artificialization of terrestrial ecosystems and preventing further degradation of soils. Ecological rehabilitation approaches are available to reinitiate soil functions and enhance plant development. However, little is known about the long-term stability of rehabilitated soils in terms of soil functions when further natural or anthropogenic perturbations occur. Based on rehabilitated urban soils, the present study sought to evaluate the resistance and resilience of soil functions linked to carbon cycling and phosphate dynamics in addition to nitrogen cycling and related microbial communities after a heat and drought stress. A laboratory experiment was conducted in microcosms under controlled temperature conditions, with four contrasted soils collected from a rehabilitated urban brownfield; an initial, non-rehabilitated soil (IS), a technosol with a high organic matter level (HO), and two technosols with less organic matter (LO1 and LO2), together with their respective controls (no stress). Changes in potential denitrification (PDR), nitrification (PNR) rates, and their interactive relationships with soil microbial activities and soil physicochemical properties were determined following a combined heat (40°C) and drought stress period of 21 days. Measurements were carried out immediately after the stress (resistance), and then also 5, 30, and 92 days after soil rewetting at 60% water holding capacity (resilience). Microbial activities involved in soil functions such as carbon cycling and phosphate dynamics proved to be of low resistance in all soils except for IS; however, they were resilient and recovered rapidly after rewetting. On the other hand, the microbial activities and gene abundances that were measured in relation to nitrogen cycling processes showed that for denitrification, activities were more rapidly resilient than gene abundances whereas for nitrification the activities and gene abundances were resilient in the same way. Results suggest that, unless the soils contain high amounts of organic matter, microbial communities in imported soils can be more vulnerable to environmental pressures such as drought and heat than communities already present. This should be considered when rehabilitating degraded soils.
Introduction
Anthropized soils have increased significantly in France since the first industrial revolution in the eighteenth century and the increasing urban sprawl. This has led to soil artificialization, which is defined as the process by which soils change use from an agricultural or natural state to a constructed building or a parking lot, for example (Pascual Aguilar et al., 2011), leading to the degradation of its ecological functions. Soil artificialization has occurred especially at the cost of agricultural lands [87% of artificialized soils in France between 2006 and 2015 used to be agricultural lands (CGDD, 2015)]. Additionally, brownfields with degraded soils can cover vast surfaces in urban and peri-urban areas. These are usually characterized by a low to moderate chemical contamination and limited fertility preventing their reuse without a prior rehabilitation process (Vincent et al., 2018). In light of the rising challenge to limit urban expansion at the expense of agricultural and natural terrestrial ecosystems, urban planning needs to take into consideration the requalification of brownfields and promote their productive potential. Moreover, the reuse of abandoned urban spaces and soil rehabilitation are key factors for protecting vital ecosystem services (Vincent et al., 2018).
Many studies have taken an interest in the rehabilitation of agricultural soils (De Noni and Viennot, 1993; Song et al., 2017; Nesper et al., 2019; Norton and Ouyang, 2019). However, to our knowledge, few studies have investigated soil ecological functions in rehabilitated urban soils. Lorenz and Lal (2009) reported that there are both similarities and differences concerning carbon (C) and nitrogen (N) biogeochemical cycling between urban and natural soils. These authors stated that the main differences reside in modifications of pH, compaction, moisture, and organic matter content in urban soils due to human activities, which influence critical soil biogeochemical processes like nitrification and denitrification. In addition, microbial communities in urban soils are substantially affected by human activities through the disturbance of biotic and abiotic soil properties (Li et al., 2018). Urbanization also affects nutrient cycling and their distribution, leading to increased N and P contents in soils (Sardans et al., 2006; Hobbie et al., 2017) as well as the accumulation of nutrients (Noe and Hupp, 2005). Moreover, urban environments may also suffer heat island effects, thus increasing soil temperature (Oke, 1989) which can significantly affect microbial activity (Carreiro et al., 2009) and affect major biogeochemical (C, N, and P) cycles in soils (Bapiri et al., 2010; Gleeson et al., 2010; Kaurin et al., 2018). Microorganisms are the main drivers for N-cycling due to their various adaptive strategies and their capacity to use various energy sources (Philippot et al., 2013). Nitrogen is an essential element for biota growth in soil (Jackson et al., 2008). N-cycling in terrestrial ecosystems is a complex process because N exists under gaseous, dissolved, and particulate forms. Nevertheless, in soils, N pathways are regulated by five interlinked microbial processes: fixation (N2 → NH4+), mineralization (Norg → NH4+), nitrate ammonification (NO3– → NH4+), nitrification (NH4+ → NO3–), and denitrification (NO3– → N2) (Hallin et al., 2009). Previous studies reported that soil’s physicochemical properties and abundances of related microbial genes are key factors in determining soil nitrification and denitrification (Jia and Conrad, 2009; Li et al., 2009; Hynes and Germida, 2012; Zhang et al., 2017; Zhang and Ji, 2018).
The increase in drought frequency and duration, which are typically combined with heat waves and interspersed rewetting, is raising questions concerning the capacity of microorganisms to adapt to such events in Mediterranean regions for example (Gibelin and Déqué, 2003; Guillot et al., 2019). Moreover, especially for agricultural soils, soil microbial communities can be resistant and even resilient to drought, as they are adapted to seasonal climatic extremes (Griffiths and Philippot, 2013; Kaurin et al., 2018). Therefore, in the context of soil rehabilitation it is essential to understand the response of N-cycling functions to drought periods, and the eventual impact on related microbial communities’ structure.
This article addresses knowledge gaps concerning rehabilitated urban soils in terms of the stability (resistance and resilience) of key microbial functions to heat and drought stress. The study investigates the effects of such a stress on global microbial activity and N-cycling functions (denitrification and nitrification) in rehabilitated urban soils. In addition, the effect of stress on specific microbial activities (basal respiration, phosphatase and β-glucosidase activities, and molecular microbial biomass) was also investigated. We hypothesized that soil’s N-cycling functions and microbial activities would be more resistant and resilient to heat (40°C) and drought stress in a soil with a high organic matter level. Finally, through this evaluation of the stability of several key soil microbial processes, this study also aims to compare three rehabilitation itineraries, two with low organic matter technosols with different plantations (LO1 and LO2) and an imported technosol with a high organic matter contents (HO), with the initial soil (IS) to give a first account of the best choice of rehabilitation.
Materials and Methods
Site Description and Soil Sampling
The experimental site of Pierre Bénite is located in Lyon metropolis in France. It has a semi-continental climate with an average annual temperature of 12°C and average annual rainfall of 1,015 mm. Summers are typically hot, and temperatures can reach 40°C. This site located along the Rhône riverbank was initially filled with alluvial sand and gravel then used for several industrial activities. A part of the brownfield was rehabilitated in 2016 during the REBU project (REBU, 2016), with the objective of developping vegetation cover and soil biota by restoring major biogeochemical cycles in soils (C, N, and P) and evaluating the best cost-effective approach. Several experimental modalities were set up using different imported or on-site materials and then received a mixture of PGPR (plant growth-promoting rhizobacteria) involved in nitrogen fixation and mycorrhiza combined with a seed composition adapted to the pedo-climatic conditions and containing a mixture rich in Fabaceae and Poaceae, as well as other species found on site. Selection of seed compositions and microbial inoculations were provided and carried out by Valorhiz (Montferrier-sur-Lez, France).1 In April 2019, 3 years after their rehabilitation, four of these modalities were chosen for the present experiment: the initial, non-rehabilitated soil consisting of a sandy material (IS modality); two modalities that had received the same imported low organic matter content technosol but with two different seed mixtures (LO1 and LO2 modalities, young plants and an accompanying seed composition and a “convalescent” seed composition, respectively); and an imported technosol rich in organic matter (HO) that received the same seed composition as LO1.
Surface soils (0–20 cm) were sampled nine times from each plot following an “M” pattern with an auger and put in sterile sampling bags (Whirl-Pak®). Composite samples were sieved at 4 mm in the laboratory and stored at 20°C for a few days until the experiment.
Soil textures were determined using a LaMotte 1067 soil texture kit (LaMotte Co., Chestertown, MD, United States), following the manufacturer’s protocol. IS is a loamy sand soil (80.7% sand, 16.2% silt, and 3.1% clay). LO1 (40% sand, 45.1% silt, and 14.7% clay) and LO2 (41.3% sand, 43.6% silt, and 15.1% clay) are loamy soils, and HO is a sandy loam soil (53.7% sand, 31.7% silt, and 14.8% clay).
Experimental Setup
Microcosms consisted of 200 ml polystyrene pots filled with 100 g of equivalent dry soil and humidified to 60% water holding capacity (WHC), prepared in triplicates with soils from the four plots (IS, LO1, LO2, and HO, respectively). Half of the microcosms were controls, and the other half was used for stressing conditions. Before starting the experiment, all microcosms were pre-incubated for 2 weeks prior to the stress period to stabilize the microbial communities.
Then, several steps were carried out as described in Figure 1.
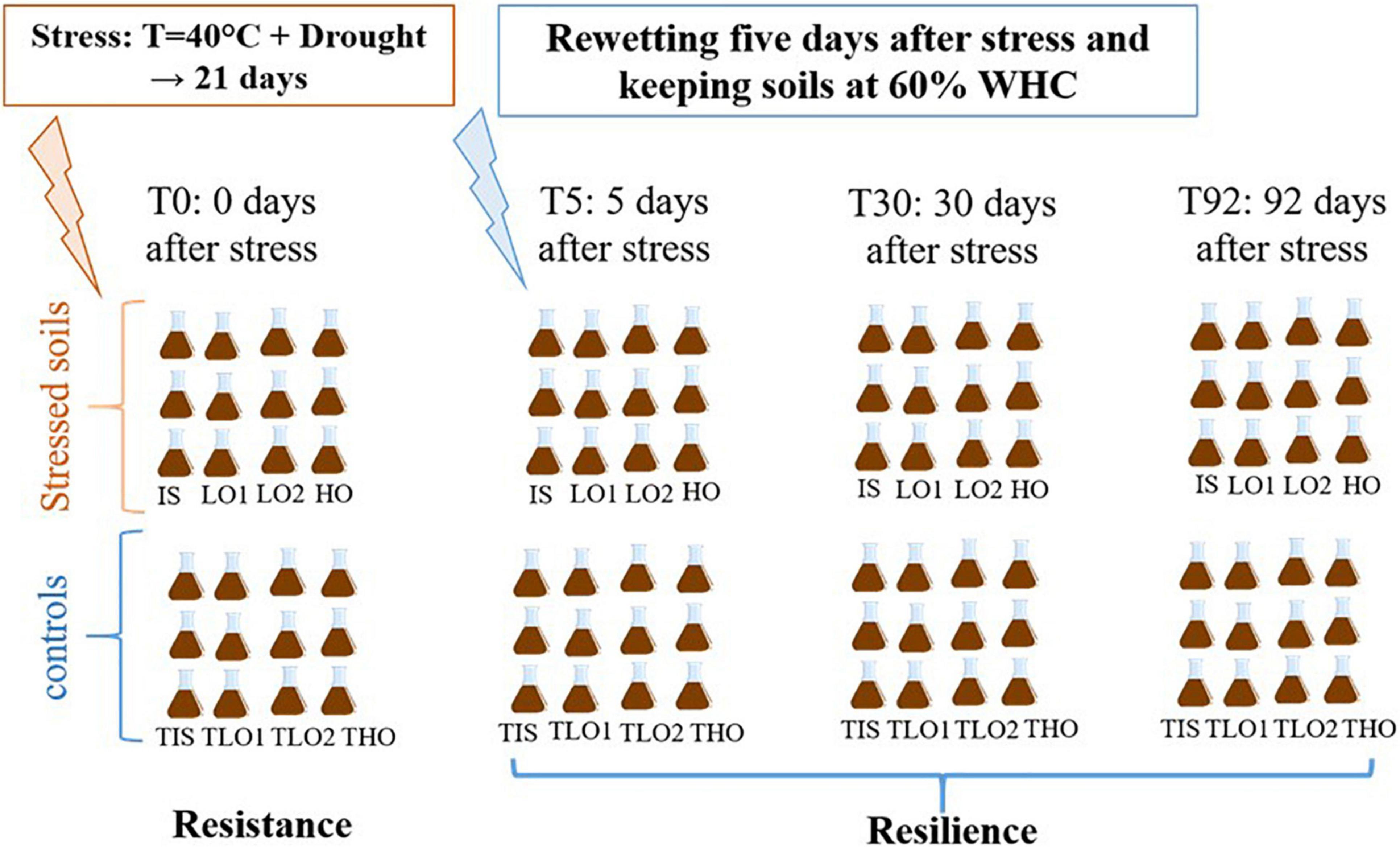
Figure 1. Experimental design conceptualized for the stress experiment for initial soil (IS), low organic matter soil 1 (LO1), low organic matter soil 2 (LO2), and high organic matter soil (HO).
Step 1: a heat (40°C) and drought stress period was applied to soils during 21 days; T0 corresponds to the end of this period for stressed soils and controls.
Step 2: the stressed soils were re-humidified to 60% WHC, and another sampling took place 5 days later (T5).
Step 3: Two more sampling operations were performed 30 days (T30), and 92 days after the stress ended (T92).
Controls and stressed soils from step 2 onward were incubated in a phytotron (plant growth chamber, Memmert HPP750 IPP PLUS, Büchenbach, Germany) at 20°C in the dark with 60% air–water saturation. Soil moisture was maintained at 60% WHC by weighing the soils once a week and, if necessary, adjusting the water contents with Mont Roucous mineral water (Ca2+ 2.9 mg⋅l–1; Mg2+ 0.5 mg⋅l–1; Na+ 3 mg⋅l–1; NO3– 2 mg⋅l–1; SO42– 3 mg⋅l–1) of which the composition is comparable to rainwater. Basal respiration and phosphatase and β-glucosidase activities were measured at every sampling point, as described below. N-cycling functions were assessed by measuring potential denitrification rates (PDR) and potential nitrification rates (PNR), while N-cycling microbial communities were characterized by measuring gene abundances.
Soil samples for enzyme activities and microbial DNA extractions were frozen at –80 and –20°C, respectively. For N transformation rates, soil samples were dried at 60°C and kept in sealed containers for total N and total C measurements.
Physicochemical Parameters
Soil pHwater was measured following the norm NF ISO 10390 (ISO, N. 10390, 2005). Total C, total H, and total N concentrations were measured in soil samples (5 mg of dry soil, ground and sieved at 160 μm to which ∼2 mg of vanadium pentoxide was added to accelerate the reaction) using an elemental Flash pyrolysis (Flash 2000, Thermo Fisher Scientific, Waltham, MA, United States). Total organic C was measured using the Rock-Eval pyrolysis technique (Rock-Eval 6 Turbo, Vinci Technologies, Nanterre, France).
Soil Microbial Community Characterization
Microbial Basal Respiration
Microbial basal respiration was measured for all samples at every sampling time using the soil respiration system (MicroResp™ James Hutton Ltd., Aberdeen, United Kingdom, according to Campbell et al., 2003), following the manufacturer’s protocol.2 MicroResp™ is a colorimetric method based on CO2 release detection in a 96-well plate by a gel containing a colored indicator (cresol red) that changes color from purple to yellow when in contact with CO2. The CO2 emission by soil microorganisms was estimated before and after a 6 h incubation period at 25°C. The colorimetric assessment was performed at λ = 570 nm by an Omega SPECTROstar (BMG Labtech, Ortenberg, Germany) microplate spectrophotometer.
Enzymatic Activities
The activities of two microbial enzymes involved in C and P cycles were measured at each sampling time: ß-glucosidase (ß-Glu) which acts on the osidic bonds of glucose and degrades cellulose, and phosphatase (Phos) which degrades phosphatase esters and is responsible for catalyzing the phosphoester bond resulting in the liberation of phosphate. Measurements were carried out according to the norm (ISO, N. 20130, 2018), which uses a colorimetric method to detect the intensity of enzyme activities during an incubation period in microplates. Different substrates were used for the measurements of enzyme activities: 4-nitrophenyl β-D-glucopyranoside (CAS no. 2492-87-7) for ß-Glu quantification and 4-nitrophenylphosphate disodium salt hexahydrate (CAS no. 333338-18-4) for Phos quantification. Enzyme activity is measured by detecting the formation of para-nitrophenol (PNP) which was read at λ = 405 nm using an Omega SPECTROstar (BMG Labtech) microplate spectrophotometer. Results were expressed in mU.g–1dry soil (namely, nmol⋅min–1⋅g–1dry soil).
Soil DNA Extraction and Quantitative Real-Time PCR
Soil DNA extractions were carried out at each sampling time on subsamples of between 0.4 and 0.8 g dry soil with the FastDNA® Spin Kit for soil and the FastPrep-24™ instrument following the manufacturer’s (MP Biomedicals, Santa Ana, CA, United States) protocol and the following specific conditions: samples were homogenized for 30 s at a speed setting of 5.0 m⋅s–1, then centrifuged for 20 min at 14,000 rpm. DNA was quantified using the Quantus™ fluorometer (Promega, Charbonnières-les-Bains, France) with the Promega QuantiFluor® dsDNA System, following the manufacturer’s recommendation.
Quantitative real-time PCR (qPCR) was used to measure bacterial abundance in soils by quantifying 16S rRNA gene copies. All samples were run in duplicates in a CFX96 Optical Real-Time detection System (Bio-Rad Laboratories, Inc., Hercules, CA, United States) in a 20 μl final volume containing 10 μl of SsoAdvanced Supermix (Bio-Rad), 0.16 μl of forward primer 341F (5′-CCTACGGGAGGCAGCAG-3′) (50 μM), 0.16 μl reserve primer 515R (5′-ATTACCGCGGCTGCTGGCA-3′) (50 μM), 0.2 μl T4 GP 32 (500 ng⋅μl–1; MP Biomedicals), 2 μl DNA at 1 ng⋅μl–1, and ultrapure water. Thermocycling conditions were as follows: 3 min at 95°C; 35 cycles of 30 s at 95°C, 30 s at 60°C, 30 s at 72°C, and 30 s at 80°C. N-cycling’s functional gene abundances were also measured using qPCR in a CFX96 system, in a 20-μl final volume with 10 μl of SsoAdvanced Supermix (Bio-Rad), 0.2 μl of each primer (50 μM) (Table 1), 0.2 μl T4 GP32 (500 ng⋅μl–1) (only for the narG gene), and 2 μl of DNA at 1 ng⋅μl–1, completed with ultrapure water. Thermal cycling conditions were as follows: for the nosZ gene: 2 min at 95°C; 40 cycles of 15 s at 95°C, 15 s at 62°C, 30 s at 72°C, and 30 s at 80°C; for the narG gene: 3 min at 95°C; 6 cycles consisting of 30 s at 95°C and 30 s at 63°C with a touchdown at –1°C by cycle and 30 s at 72°C; and 34 cycles consisting of 30 s at 95°C, 30 s at 58°C, 30 s at 72°C, and 30 s at 80°C; and for the amoA gene: 5 min at 95°C; 40 cycles consisting of 15 s at 95°C, 30 s at 55°C, and 30 s at 72°C, and 30 s at 80°C. At the end of each qPCR, a melting curve analysis was generated by applying a final step of 0.5°C temperature increment every 10 s, from 65 to 95°C.
For each gene, calibration curves were obtained using serial dilutions of a known quantity of linearized plasmids containing known copy numbers of the gene. Blank controls without DNA with ultrapure water were carried out in every qPCR analysis. The amplification efficiencies of target genes ranged between 87.3 and 96.3%, with R2 values between 0.99 and 1.
Determination of Soil N Transformation Rates
PDR were determined by the acetylene block technique of Petersen et al. (2012) and later used by Zhang et al. (2018). The method consists of measuring N2O emissions by gas chromatography (TRACE™ 13,000) after adding acetylene to block its transformation to N2 in anaerobic conditions at the end of a 24 h incubation period.
PNR were determined by placing soils (5 g equivalent dry soil) into 70 ml containers. Soils were further treated with 100 mgN⋅kg–1 (dry weight) (NH4)2SO4. Samples were adjusted to 60% WHC using deionized water and were incubated at 20°C for 7 days. At the end of the incubation period, samples were extracted with a 2 M KCl solution (5:1 solution of soil ratio). Extracts were shaken vertically at 200 rpm for 1 h, then later filtered with 0.2 μm acetate cellulose ClearLine® filters. Extracts were analyzed for N-NH4, N-NO2, and N-NO3 concentrations using colorimetric techniques, Thermo Fisher Gallery™ for ammonium (references 984362 and 984363) and nitrite (reference 984371), and a Merck kit adapted for high salinity because of the extractant used (KCl) (Reference 1.14942.0001). Results were expressed in mgN⋅day–1⋅kg–1dry soil between day 0 and day 7.
Statistical Analysis
Kruskal–Wallis multiple comparison tests were conducted to compare mean values at the 5% level using R software R-3.6.1 (R Core Team, 2017), agricolae package. Differences were considered significant for p-values <0.05. Data of all soils at all measurement periods were analyzed by principal component analysis also using R software, FactoMineR and factoextra packages.
Results
Initial Soil Physicochemical Properties
All soil pH ranged between 7.6 and 9.1, and IS had the highest pH value, followed by HO, LO2, and LO1 (Table 2). HO and IS had the highest total C contents followed by LO1 and then LO2. HO’s TOC content was significantly higher (∼7-fold) than all other soils, followed by LO1, then LO2 and finally IS. Total N contents ranged between 0.5 and 1.6 g⋅kg–1dry soil for all soils, and LO1 and HO had the highest values, followed by LO2, then IS. HO’s C/N was significantly higher (∼3-fold) than all other soils, followed by LO2 and LO1, then IS. HO’s H content was significantly higher (∼2-fold) than all other soils, followed by LO1, then LO2 and finally IS. HO and LO2 N-NH4 contents were significantly higher (∼5-fold the minimal value) than LO1 and IS. N-NO3 contents ranged between 6.9 mgN⋅kg–1 and 17.7 (mgN⋅kg–1); LO1 had the highest value, followed by HO, LO2, and IS.
Characterization of Soil Microbial Communities and Activities
Changes in Total Microbial Biomass and Enzymatic Activities
Basal respiration (BR) was measured at all sampling times to assess the level of microbial activity in the microcosms (Figure 2A). Just after the stress at T0, BRs of LO1 and LO2 were significantly lower than those of controls TLO1 and TLO2 (∼2- and 2.5-fold, respectively). In contrast, BRs in soils HO and IS (6.1 and 3.6 μgC-CO2⋅h–1⋅g–1dry soil, respectively) were not affected by the stress as no significant differences were found compared to BRs of controls THO (5.4 and 4.8 μgC-CO2⋅h–1⋅g–1dry soil, respectively). After rewetting at T5, only the BR of soil HO was found to be significantly lower than that of control THO (∼4-fold), while no significant differences were found for all the other soils. At T30, HO’s BR was the only measurement that was significantly lower than its control THO (∼3-fold). No significant differences were found between treatments and controls at T92. However, we observed that overall BR declined between T0 and T92 for all unstressed soils.
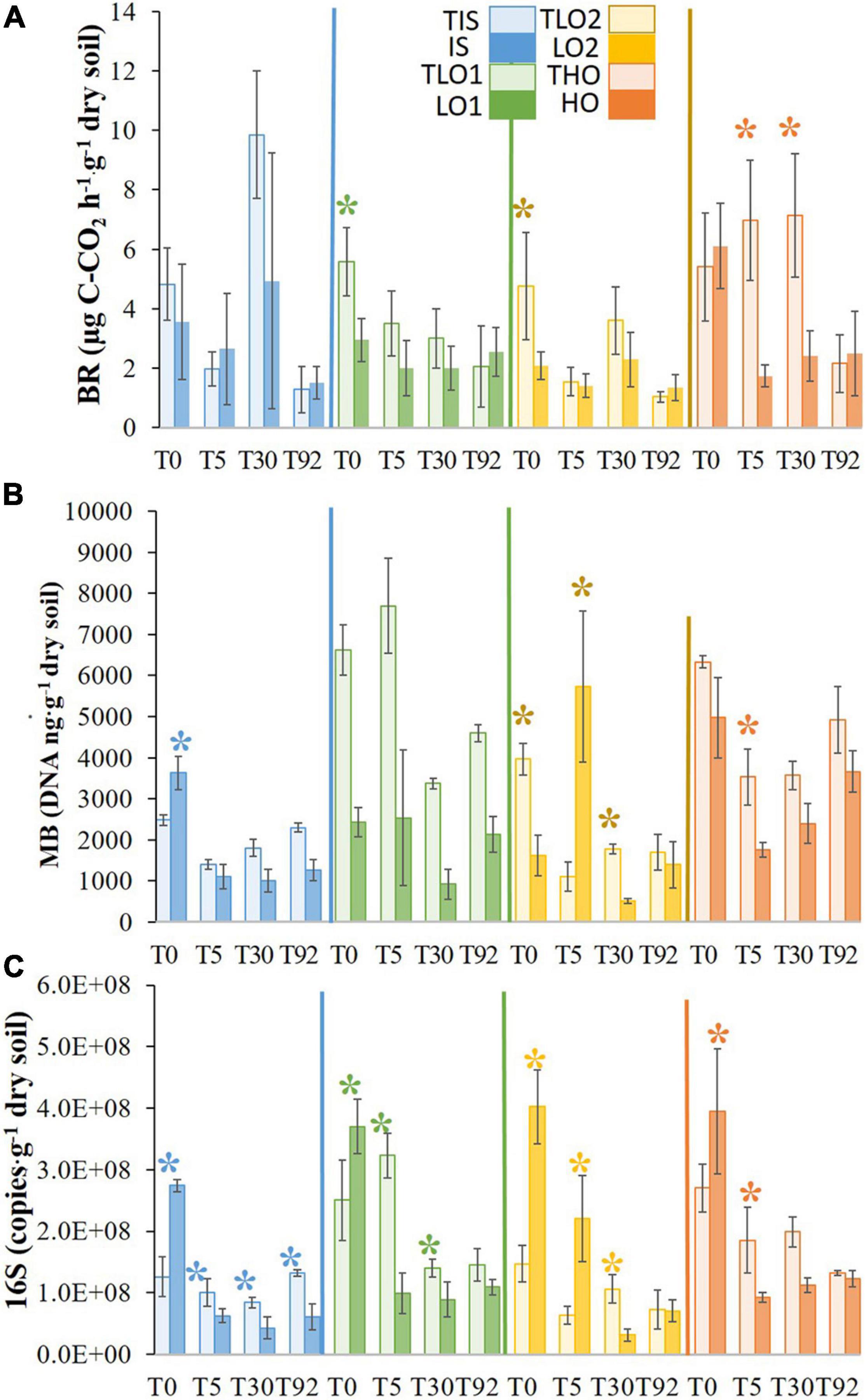
Figure 2. Means of basal respiration (A), molecular biomass (B), and bacterial abundance (C) for initial soil (IS), low organic matter soil 1 (LO1), low organic matter soil 2 (LO2), and high organic matter soil (HO), and their controls TIS, TLO1, THO, and TLO2 at different sampling times (T0, T5, T30, and T92). Statistical tests were performed using the Kruskal–Wallis test. The asterisks indicate a significant difference (p< 0.05) between a stressed soil and its control at the same sampling time. Error bars represent the error of means.
The heat and drought stress significantly decreased the molecular biomass (MB) of LO1 and LO2 soils compared to controls at T0 (∼3- and 2.5-fold, respectively) (Figure 2B). In contrast, MB in HO was not significantly affected, and MB in IS increased significantly (∼1.5-fold) compared to controls at T0. Five days after rewetting, MB in LO2 had increased significantly (∼5-fold) compared to control. In contrast, IS, LO1, and HO’s MB were significantly lower (∼1.3-, 3-, and 3-fold) than controls at T5. MBs in LO1 and LO2 were significantly lower than those of controls (∼4- and 3.5-fold) at T30; in contrast, no significant differences were found between HO and its control. Finally, at T92, no significant differences were found between stressed soils and controls. The stresses also significantly increased bacterial abundance (16S rRNA gene copies) in soils compared to controls at T0 (Figure 2C). However, 5 days (T5) after rewetting the soils, bacterial abundance decreased significantly for the IS, LO1, and HO soils (∼4-fold) compared to controls and LO2 was less affected (∼2-fold decrease). At T30, 16S rRNA gene abundance was still significantly lower in stressed soils than in the controls, except for HO. At the final sampling time T92, 16S rRNA gene abundance was approximately the same as controls in all soils except for IS where it was still significantly lower. Finally, results showed that 16S rRNA gene abundance had a tendency to decrease with the incubation time, as abundances were significantly lower at T92 compared to T0 for all soils.
After the stress at T0, phosphatase activities in LO1, HO, and LO2 decreased significantly (∼5-, ∼2-, and ∼5-fold, respectively) compared to the controls TLO1, THO, and TLO2 (Figure 3A). On the contrary, activity increased significantly (∼2.5-fold) compared to its control TIS. At T5, 5 days after rewetting the stressed soils, phosphatase activities in LO1 and LO2 remained significantly lower (∼4- and ∼5-fold, respectively) than controls TLO1 and TLO2. On the contrary, no significant differences were found between IS and HO and their controls TIS and THO at T5. Furthermore, only one significant difference was found at T30 which was LO1 being lower than TLO1 (∼4-fold). In all stressed soils at T92, phosphatase activities were significantly lower than in controls at T92 except for IS. IS showed the lowest activity compared to other soils.
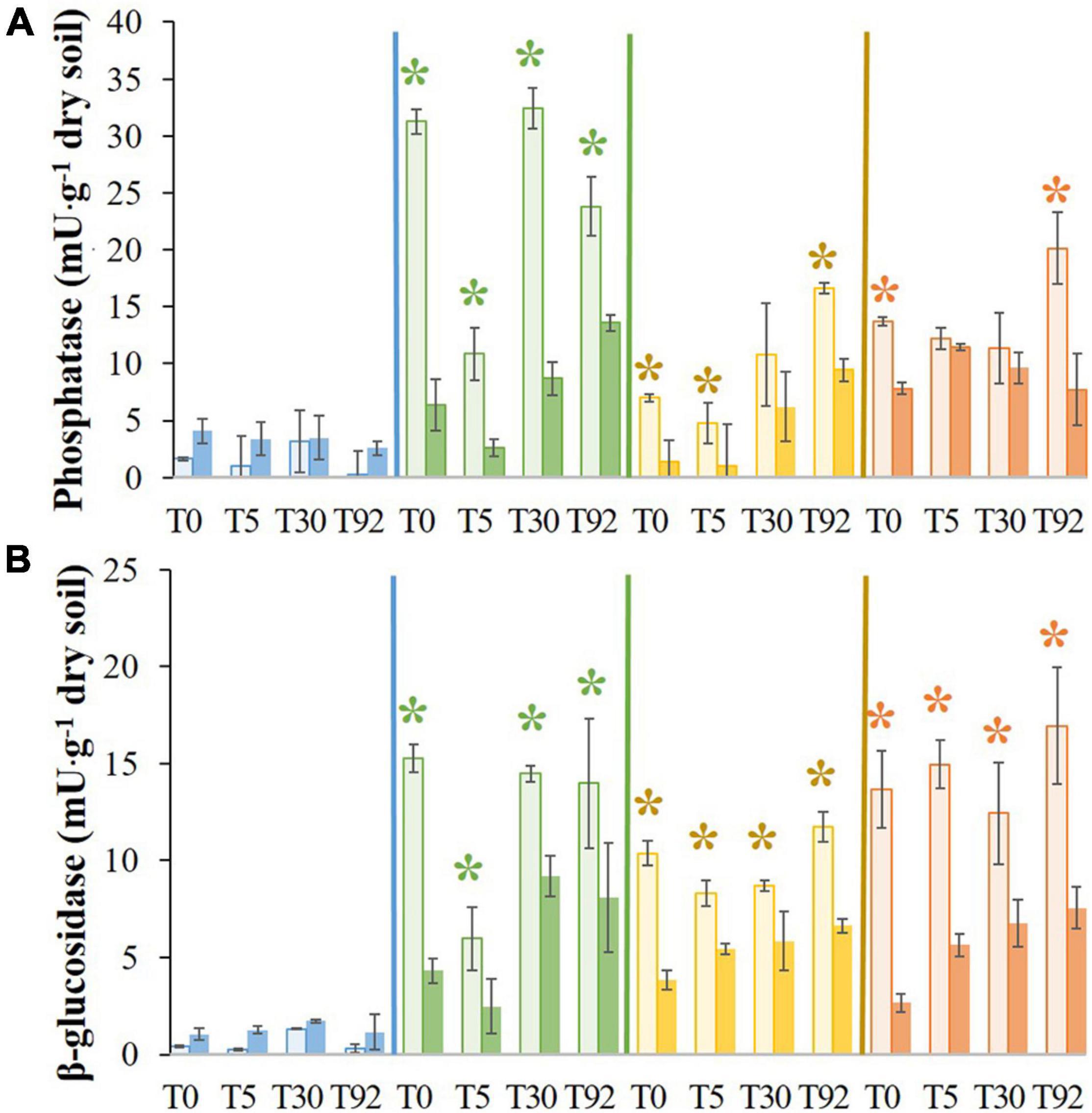
Figure 3. Means of phosphatase activity (A) and β-glucosidase (B), for initial soil (IS), low organic matter soil 1 (LO1), low organic matter soil 2 (LO2), and high organic matter soil (HO), and their controls TIS, TLO1, THO, and TLO2 at different sampling times (T0, T5, T30, and T92). Statistical tests were performed using the Kruskal–Wallis test. The asterisks indicate a significant difference (p< 0.05) between a stressed soil and its control at the same sampling time. Error bars represent the error of means.
Results presented in Figure 3B show that at T0, β-glucosidase activities in LO1, LO2, and HO were significantly lower (∼3.5-, ∼3-, and ∼5-fold, respectively) than the controls TLO1, TLO2, and THO. Then, the β-glucosidase activity in these soils remained significantly lower than in controls all along the experiment at T5 (∼2.4-, 1.5-, and 2.6-fold, respectively), T30 (∼6-, 1.5-, and 2-fold, respectively), and T92 (1.1-, 1.7-, and 2.2-fold, respectively). In contrast, no significant differences were found for IS between controls and treatments after stress, although its activity was the lowest compared to other soils.
Changes in Denitrification and Nitrification Functions
After the stress, PDR decreased significantly in all soils compared to the respective controls (Figure 4A). Five days after the stress and rewetting (T5), all previously affected soils recovered PDR at the same order of magnitude as their respective controls. Furthermore, no significant differences in PDR were found between soils and controls at T30 and T92 either.
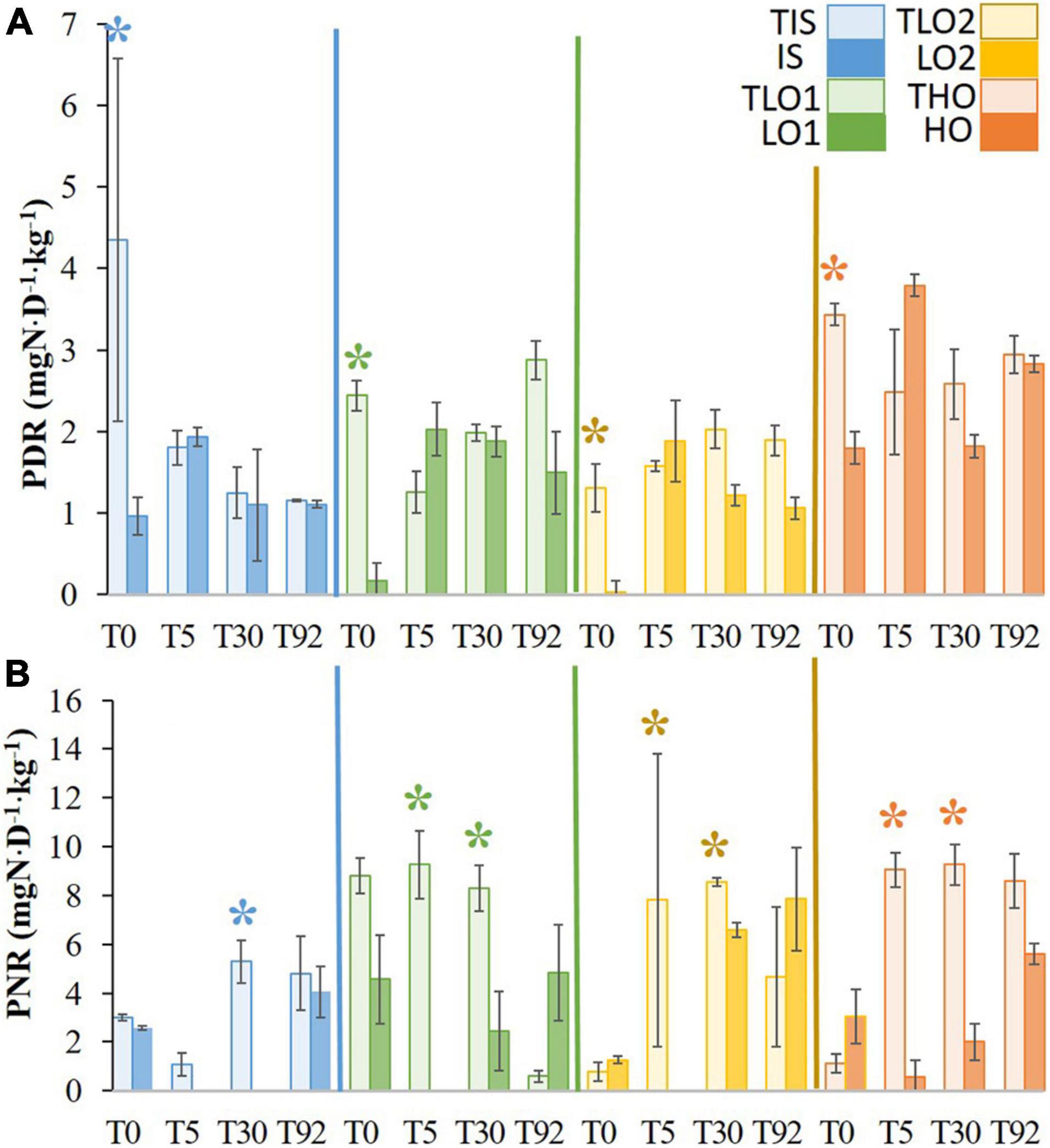
Figure 4. Means of potential denitrification rates (PDR) (A) and potential nitrification rates (PNR) (B) for initial soil (IS), two soils with less organic matter (LO1 and LO2), and high organic matter soil (HO) and their respective controls (TIS, TLO1, TLO2, and THO). Statistical tests were performed using the Kruskal–Wallis test. The asterisks indicate a significant difference (p< 0.05) between a stressed soil and its control at the same sampling time. Error bars represent the error of means.
At T0 just after the stress, no significant differences in PNR were found between treatments and controls (Figure 4B). After rewetting (T5), PNR decreased significantly for all stressed soils compared to controls except for IS. PNR were significantly lower in stressed soils, compared to controls at T30 for all soils. At T92, no significant differences in PNR were found between stressed soils and controls.
Changes in Denitrifying and Nitrifying Microbial Communities
At T0, the stress significantly decreased the narG gene copy abundances in LO1, LO2, and HO soils (∼4-, 7-, and 1.6-fold, respectively) compared to controls TLO1, TLO2, and THO (Figure 5A). In contrast, the stress significantly increased the abundance of narG gene copies in IS at T0 (∼1.3-fold) compared to control TIS. Five days after rewetting at T5, narG gene abundances for all soils were significantly lower than controls except for IS. Thirty days after the stress period (T30), narG gene abundances were significantly lower than controls in all soils. At the final sampling time T92, narG gene copy numbers in IS and LO1 were significantly lower (∼2- and 3-fold, respectively) than controls TIS and TLO1. In contrast, no significant differences were found between HO, LO2, and their controls THO and TLO2 at T92.
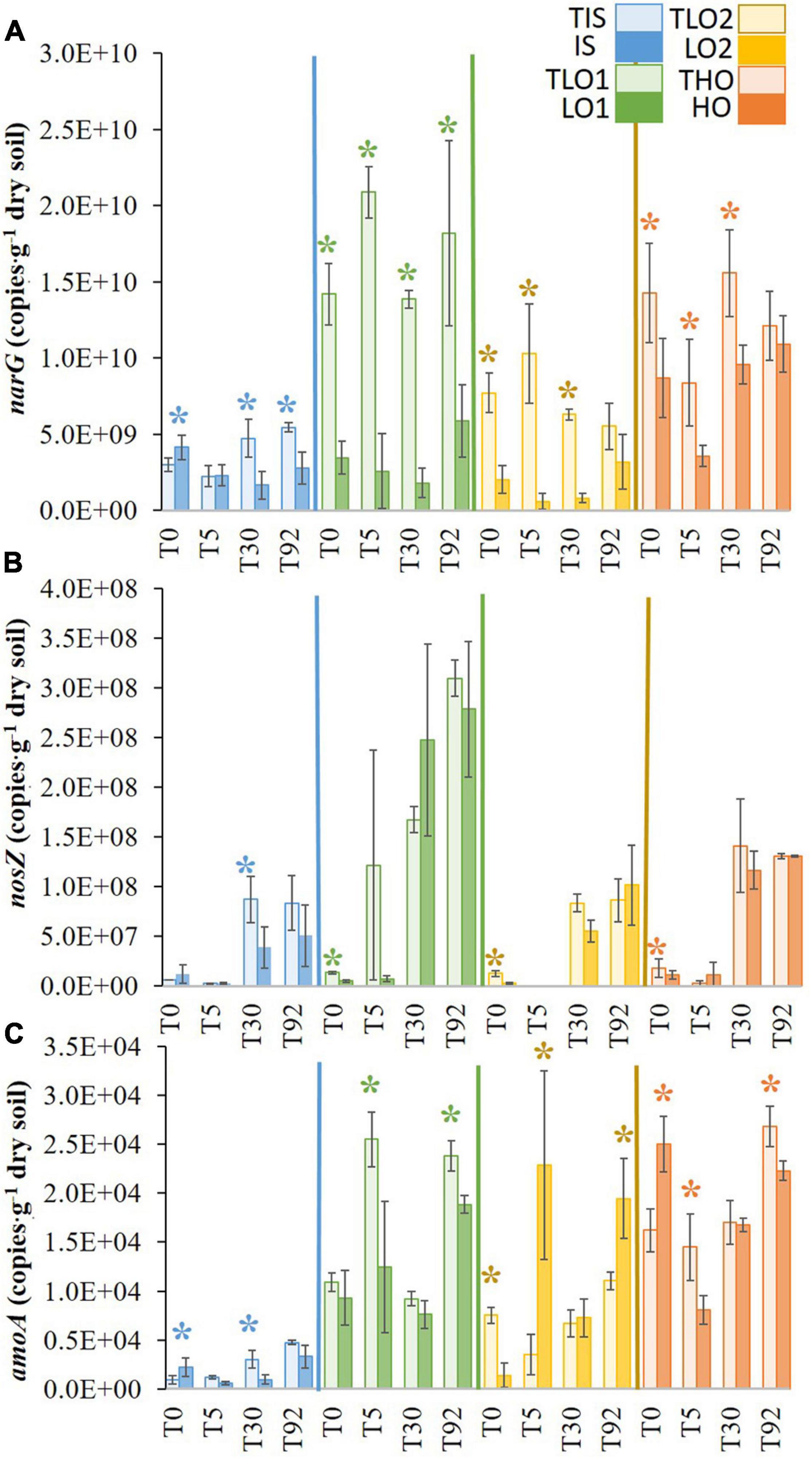
Figure 5. Means of narG (A), nosZ (B) and amoA (C) gene abundances in the soils at different sampling times for initial soil (IS), high organic matter soil (HO), and two soils with less organic matter (LO1 and LO2) and their respective controls (TIS, THO, TLO1, and TLO2). Statistical tests were performed using Kruskal–Wallis. The stars indicate a significant difference (p< 0.05) between soil and control at the same sampling time. Error bars represent the error of means.
Although nosZ gene copy abundances were low in all soils at T0, significant differences were found between the stressed soils and their controls for LO1, HO, and LO2 (∼2.6-, 1.6-, and 4.6-fold, respectively) compared to controls TLO1, THO, and TLO2 (Figure 5B). Five days after rewetting (T5), no significant differences were found between soils and controls. Thirty days after the stress (T30), nosZ gene abundance in IS was significantly lower (∼2-fold) than the control TIS, whereas no significant differences were found between all other soils and controls at T30. At the final sampling time T92, no significant differences were found between treatments and controls for all soils. The results showed that nosZ gene abundances were significantly higher at T30 and T92 compared to T0 and T5 and that the number of nosZ gene copies increased significantly after T5 for all soils and controls.
After the stress period at T0, both IS and HO had significantly higher (∼2- and 1.5-fold, respectively) amoA gene abundances than controls TIS and THO. No significant difference was found between TLO1 and LO1, while LO2 was significantly lower (∼2-fold) than TLO1 (Figure 5C). Five days after rewetting (T5), amoA gene abundances were significantly higher compared to controls for LO2 (∼6-fold) whereas they were lower for LO1 and HO (∼2- and 1.8-fold, respectively) and not significantly different for IS. Thirty days after the stress (T30), amoA gene abundances were significantly lower (∼3-fold) than controls in IS, whereas no significant differences were observed for the other soils. At the final sampling date (T92), amoA gene abundances were significantly higher than controls for LO2 (∼2-fold) whereas they were comparable to controls for for LO1, HO and IS.
Principal Component Analysis of Biological Parameters
PCA was carried out on the following parameters: PDR, PNR, phosphatase activity (Phos), β-glucosidase activity (β-Glu), BR, BM, and nosZ, narG, amoA, and 16S rRNA gene abundances for all soils at all sampling times. Results indicated that PC1 and PC2 accounted for 43.8% of the total variance (Figure 6). THO and TLO2 soils were positioned on the right side of PC1, mainly explained by narG gene abundances, Phos and PNR. The PCA also showed that the positions of soils and controls at T0 tended to be in the lower part of the plot along the second axis, whereas soils and controls from other sampling times (T5, T30, and T92) were positioned higher up along the second axis, which is mainly driven by 16S rRNA gene abundances, MB and β-Glu (Figure 6). No clear correlations between PNR, PDR, and corresponding amoA gene abundances and nosZ and narG gene abundances could be distinguished from the PCA. Furthermore, linear regressions (data not shown) did not demonstrate any significant correlations either between PDRs and functional gene abundances (narG and nosZ), or between PNR and amoA gene abundances.
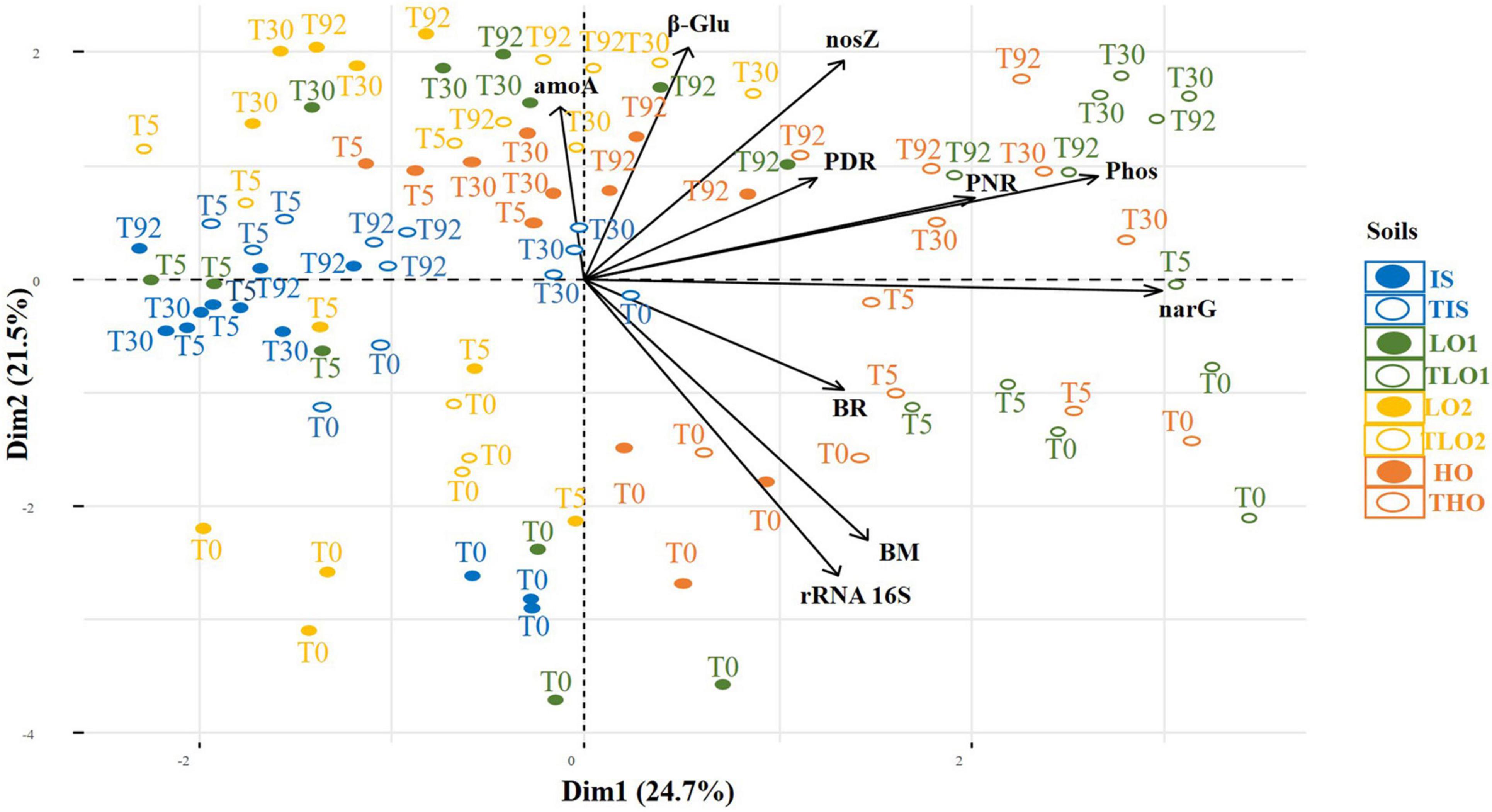
Figure 6. Principal component analysis (PCA) of soil initial soil (IS), two soils with less organic matter (LO1 and LO2), and high organic matter soil (HO), and their respective controls (TIS, THO, TLO1, and TLO2), at different sampling times (T0, T5, T3, and T92). The data analyzed are PNR (potential nitrification rates), PDR (potential denitrification rates), BR (basal respiration), phosphatase and β-glucosidase activities, MB (molecular biomass), phosphatase and nosZ, narG, and amoA gene abundances.
Discussion
Heat and Drought Stress Affected Carbon Mineralization and Nutrient Dynamics on the Short Term
Carbon Mineralization and Phosphate Dynamics Were Not Resistant to Stress
The microbial process of carbon mineralization, which contributes to the ecological function of organic matter stock and dynamics, was evaluated by measuring microbial MB, bacterial abundance, soil respiration, and β-glucosidase activity and generally decreased following heat and drought stress. Microbial biomass was measured using MB, the quantity of DNA extracted from soil which is an indicator of the biological state of an environment (Dequiedt et al., 2011; Bouchez et al., 2016) and was durably impacted by stress in the present study. BR was the most affected in LO1 and LO2 after the stress period (T0). Previous work made similar observations where heat and drought stress drastically affected BR in French agricultural soils (Bérard et al., 2012), in tropical soils, and in Australian agricultural soils (Liang et al., 2014; Yu et al., 2014, respectively). The reasons behind this decrease may be a physiological effect on microorganisms, as drought can cause cell desiccation and lysis (Göransson et al., 2013), or the thermal denaturation of microbial enzymes (Bérard et al., 2011). However, bacterial abundance appeared to increase in all soils directly after stress (T0) which was in contradiction with the other results. β-Glucosidase is an enzyme responsible for the breakdown of several forms of carbohydrates, especially cellulose, which make its role fundamental for nutrient availability and cycling (Singhania et al., 2013). Compared to their respective controls, β-glucosidase activity (β-glu) decreased significantly after the stress (T0) in all soils except IS, indicating a low resistance of this enzymatic activity to heat and drought stress. Our results are comparable to other authors who have also reported a decrease in soil β-glu due to drought stress in Mediterranean grasslands and subtropical acrisol (Sardans and Peñuelas, 2005; Liang et al., 2014, respectively), grass plantations (Sanaullah et al., 2011), and Mediterranean semiarid soils (Hueso et al., 2011). Heat and drought stress can induce a critical situation for soil microbial communities causing cell death, and thus an alteration of enzymatic activities involved in C-cycling (Lamersdorf et al., 1998).
Phosphatase activity is involved in the transformation of phosphorous compounds (Amador et al., 1997), and its activity is essential in P-cycling. Its activity contributes to the process of phosphate cycling and the ecological function of nutrient availability to plants and soil organisms. Phosphatase activities were significantly lower in all soils at T0 in the stressed soils compared to the controls except in IS, showing a low resistance of the activity to drought and heat stress. These findings are in accordance with studies showing that experimental drought stress reduced alkaline phosphatase activity in Quercus ilex Mediterranean forest soils (Sardans et al., 2008) and in Mediterranean terrestrial ecosystems (Sardans and Peñuelas, 2005).
Thus, heat and drought stress impacted microbial functions involved in carbon mineralization and phosphate dynamics in rehabilitated urban soils, demonstrating the vulnerability of these soils.
Carbon Mineralization and Phosphate Dynamics’ Resilience to Stress
The following sampling times, T5, T30, and T92 after soil rewetting, enabled to monitor the bacterial communities’ resilience to the heat and drought stress for the different ecological functions. Overall, except for MB in LO1, carbon mineralization was resilient for MB, BR, and bacterial abundance after 92 days, as was found by Bérard et al. (2012), in which BR recovered after 96 days. Microorganisms have evolved defense mechanisms allowing them to recover after perturbation. Some heat-tolerant bacteria like Actinobacteria can survive desiccation stress by increasing synthesized ribosomes to grow quickly once conditions become favorable (Barnard et al., 2013). This could have been the case for our soils as microorganisms could have adopted this preparation strategy, accelerated their cell division, and increased their biomass after re-humidification. The Firmicutes phylum also includes bacteria with heat and drought tolerance mechanisms (De Vos et al., 2009). Additionally, if bacteria from phyla such as Firmicutes and Actinobacteria were favored during the stress period, this may explain the increase in the copy numbers of the gene coding for 16s rRNA as both these phyla have been shown to possess multiple copies of this gene in their genome (Sun et al., 2013; Větrovský and Baldrian, 2013).
However, the heat and drought stress irreversibly altered β-glu and phosphatase activities. Previous studies have reported that drought can strongly limit enzymatic activities (Henry, 2013; Siebielec et al., 2020). In a previous study involving a heat stress (50°C), β-glu activity decreased by twofold in French agricultural soils (Riah-Anglet et al., 2015), which is somewhat comparable with HO where the activity decreased by 1.7-fold. Drought also reduced phosphatase activity in forest soils in Spain (Sardans and Peñuelas, 2010). This could be explained by the fact that heat stress can cause a decrease in enzyme synthesis and secretion (Allison, 2005). Our results showed that with the exception of IS, none of the soil β-glu and phosphatase activities had recovered after 92 days. Siebielec et al. (2020) found more contrasting results as phosphatase activity had a full recovery in loamy agricultural soils after a 1-month drought period.
Although microbial functions involved in carbon cycling and phosphate dynamics were poorly resistant to the drought and temperature stress, they appeared to recover rapidly as discussed. However, future work could question how many stress events such soils could support before irreversibly affecting these functions, in turn altering nutrient availability and disrupting the cycling of essential elements.
Organic Matter Contents in the Rehabilitated Soils and Acclimation in Existing Soils Could Explain the Resilience of Carbon Mineralization
IS was the initial, un-rehabilitated soil and did not undergo any modifications during the initial REBU project. Therefore, although overall biomass and activities were lower in this soil compared to the others, its intrinsic microbial communities could be more adapted to drought and heat conditions by regularly undergoing seasonal changes. Similar findings were reported in a study where heat and drought stress (21-day period) had less impact on the BR of soils that were pre-exposed to stress, compared to those which were not pre-exposed in French agricultural soils (Bérard et al., 2012). Furthermore, it has already been evidenced that for soil microbial communities, a preselection by drying and rewetting reduces the impact of additional drought stress (Fierer et al., 2003; Williams, 2007; Bouskil et al., 2013; Zeglin et al., 2013; Wang et al., 2014; Bérard et al., 2015). This is probably because drought causes changes in the ecophysiology of microbial communities, and a pre-exposure to stress favored a fraction of microbial communities best adapted to heat and drought conditions (Anderson and Domsch, 1985, 1993).
Soil HO showed the highest total organic carbon content (Table 2). On the contrary to LO1 and LO2, HO’s BR was not affected by heat and drought stress. This might be explained by its higher content of available C making microbial communities more resistant to stress (Sanaullah et al., 2011; Canarini et al., 2017). It is known that organic C is an important driver of BR in soils (Romero-Freire et al., 2016), and drought affects soil processes indirectly by altering the substrate’s availability to microorganisms (Schimel, 2018). This can be explained by the decline of diffusion rates when drought increases, as microorganisms become resource deprived which limits their capacity of using acclimation methods that require carbon and energy (Schimel et al., 2007). This suggestion is further supported by the fact that HO’s MB was resistant to drought, in contrast to LO1 and LO2, which declined. This can be explained by microorganisms resorting to dormancy to avoid stress, therefore reducing growth efficiency and allocating more resources to survival instead of reproduction (Schimel et al., 2007). Higher organic matter content in soil can improve water retention, therefore mitigating water availability for microorganisms. In a study conducted in US maize fields, results showed that higher organic matter content in soils was associated with higher crop yields. Analyses indicated that this positive association of soil organic matter and yields is partially explained by positive effects of soil OM on available water capacity (Kane et al., 2021). This is crucial for microbial survival as water loss can cause a loss of cell turgor (Harris, 2015), which can be catastrophic for microorganisms and result in interferences with physiological functions, reduce metabolic processes, and eventually lead to cell death (Schimel, 2018).
In turn, low organic matter contents could affect other soil microbial processes such as phosphatase activity and nutrient availability (Bonmati et al., 1991; Olander and Vitousek, 2000), which could explain the overall low values of phosphatase activity in IS, correlating with the low N and TOC values, whereas HO’s phosphatase activity was resilient. IS also had the lowest β-glu activity compared to other soils, which might be due to less available organic substrates for β-glu such as cellulose (Hueso et al., 2011). This finding is in accordance with other studies where the increase in organic carbon enhanced soil β-glu activity (Eivazi and Tabatabai, 1990; Piotrowska and Koper, 2010).
Nitrogen Cycling Processes Denitrification and Nitrification Were Affected Differently by Drought and Heat Stress
Denitrification Activities Were More Rapidly Resilient Than Gene Abundance
The effects of drought and heat stress on the nitrogen cycle were evaluated by determining both the nitrification and denitrification potentials in soils and the abundances of functional genes involved in soil N-cycling. All stressed soils showed low resistance for PDRs to drought and heat stress. Many studies have shown that drought decreases the denitrification process in different terrestrial ecosystems like tropical forests (Almaraz et al., 2019), grasslands (Hartmann et al., 2013; Keil et al., 2015), and agricultural fields (Homyak et al., 2017). However, even though they were impacted by the stress, PDR in all soils recovered quickly after rewetting and were resilient in the short, mid, and long terms (T5, T30, and T92). Denitrifying enzymes originate from an enzyme pool known to be tolerant of extended drought periods, which would explain the rapid recovery of the denitrification process after rewetting (Peterjohn, 1991). A recent experiment on a managed grassland, conducted by Harris et al. (2021), also monitored an increase in N2O fluxes that the authors attributed to denitrification when rewetting the soils after a drought period. Davidson (1992) reported that denitrification could be resilient in response to drought stress as it can be activated within minutes to hours after soil rewetting and the flush of available C and N caused by the rewetting of stressed soils can lead to the resuscitation of dormant denitrifiers (Six et al., 2002). Another sign that the denitrifying bacterial communities were affected by stress was the decrease at T0 of the abundances of narG and nosZ genes in LO1, HO, and LO2. Unlike PDR, the abundances of genes involved in denitrification were not resilient at T5 and T30 following rewetting. Consequently, during extreme dry conditions, we might expect a decrease in denitrification and denitrifying bacterial populations as shown by Bremner and Blackmer (1978) and later by Butterbach-Bahl and Dannenmann (2011). However, in the present study we chose to monitor narG and nosZ gene copy numbers as indicators of the impact of the heat and drought stress on denitrification, although other genes are also key functional markers of denitrification; in particular, nirK and nirS genes play an important role in an intermediary step of denitrification, the production of NO which is rapidly further reduced to N2O. These two genes are in the genome of nitrite-reducing denitrifiers, which consists of a highly phylogenetically diverse functional guild (Wei et al., 2015; Bonilla-Rosso et al., 2016). Thus, the resilience of PDR in our experiment could be attributed to nitrite-reducing activity, implying that it was the final denitrification step and reduction of N2O to N2 that was highly impacted by the stress. Further work would enable to specifically distinguish impacts of heat and drought stress on the different steps of the denitrification process.
Nitrification Activities Were Resistant, as Were Gene Abundances
PNR showed no significant differences between stressed soils and controls for all soils at T0, suggesting that the nitrifying communities in all soils were resistant to stress. Hartmann et al. (2013) reported similar findings in grassland soils where PNR were not affected by drought in experimental plots where precipitations were blocked for 41 days. Furthermore, nitrification potentials were reported to be higher in dry and hot summer seasons compared to wet seasons in grasslands in California United States (Parker and Schimel, 2011). Drought actually increased the abundance of amoA genes in IS soil, which is in accordance with Bremner and Blackmer (1978) and Kaurin et al. (2018), who reported that during extreme conditions an increase in nitrifiers and nitrification is expected. Davidson (1992) also reported drought-resistant nitrifying microbial communities that are adapted to survive long dry summer seasons in grasslands. However, PNR in LO1 and HO decreased significantly 5 days after rewetting (T5), which is in accordance with results reported by Tan et al. (2018) showing that nitrification rates decreased significantly with soil moisture increase, contrarily to denitrification that increases with moisture. However, overall amoA gene abundances in soils were lower than abundances reported by other studies in German arable soil and in rice paddy fields (Radl et al., 2015; Zhang et al., 2018, respectively). This may be due to the domination of nitrifying archaeal over bacterial groups especially under drought conditions in arable soils (Radl et al., 2015) and in urban soils (Wang et al., 2017). In contrast, our results were somewhat comparable to mountain abandoned meadow soils (Fuchslueger, 2014). Overall the N-cycling function seemed less affected by stress than the C-cycling and the phosphate dynamics-function as PDR was resilient and PNR was resistant.
No Significant Correlations Were Found Between Soil N-Cycling Processes and Functional Genes Abundances
In the present study, we found no significant correlations between narG and nosZ functional genes abundances and corresponding N-cycling functions (data not shown). Although some studies have found that denitrifier and nitrifier community compositions are important factors in the regulation of denitrification and nitrification processes (Petersen et al., 2012; Zhang et al., 2017, 2018; Zhang and Ji, 2018), others did not suggest any relationship between the two (Hallin et al., 2009; Attard et al., 2011; Graham et al., 2014). One likely reason for these differences is that many microorganisms carry denitrification genes at any given moment but only a subset of the total community is active and carries out denitrification. Another reason for the lack of a relationship in the present work could be the interference of other parameters such as abiotic characteristics, environmental conditions, or the lack of data to show a clear trend. In a study conducted by Graham et al. (2016), the authors suggested that N-cycling processes are more driven by the edaphic characteristics of soils than by the abundances of functional genes in tropical rainforest soil samples using only edaphic factors. In addition, models containing only edaphic factors data explained N-cycling variation more than models using both gene abundance data. Rocca et al. (2015) published a review where they conducted a meta-analysis of the relationships between gene abundances and corresponding process rates linked to the C- or N-cycling. Authors reported that within the 59 chosen studies, there was a significant but weak positive relationship between gene abundance and the corresponding process (r = 0.3, p< 0.0001, n = 189) using the Pearson product–moment test. They analyzed studies exclusively using gene abundances measured by qPCR and including the bacterial amoA gene in relation to nitrification, and narG and nosZ genes to denitrification. Thus, the non-significant correlations found in our study might also be due to dataset dimensions. Again, further analysis of other functional genes encoding different steps of nitrogen cycling, such as nirK and nirS as mentioned previously, could bring more insight on the links between activity measurements and potentials.
Conclusion—The Pros and Cons of Soil Rehabilitation on Carbon Mineralization and Nitrogen Cycling
Heat and drought stress affected both carbon mineralization and phosphate dynamics by altering soil respiration and microbial enzymatic activities, in some cases causing irreversible shifts in the activity’s state. This is probably due to stress causing cell desiccation, lysis, and thermal denaturation of enzymes. Soil respiration showed resilience after 96 days, possibly due to desiccation-resistant bacteria such as Actinobacteria or Firmicutes. Furthermore, the non-rehabilitated soil IS showed resistance of microbial activity involved in nutrient cycling functions, suggesting a previous adaptation of its microbial communities to drought and heat stress as the soil was not rehabilitated. Thus, it can be suggested that an initial soil could be more resistant and resilient in terms of soil functioning as its microbial communities may have acquired a greater flexibility to environmental conditions. However, this soil is characterized with a low level of activity compared to other soils, which highlights the importance of organic matter contents in stimulating microbial activities. Furthermore, microbial activities involved in carbon cycling and nutrient availability in HO were more resilient to stress than LO1 and LO2, highlighting this rehabilitation trajectory as the most optimal on the short term. Our findings highlight the importance of organic matter content and pre-exposition to stress in determining the performance of soil carbon cycling and phosphate dynamics in rehabilitated urban soils.
Microbial activities and genetic potentials regarding processes involved in nitrogen cycling were more resilient. Denitrification processes were sensitive to heat and drought as they decreased significantly in all soils following the stress. However, they recovered quickly due to the resilience of denitrification enzymes and their tolerance to drought. Furthermore, denitrifying bacterial communities were also affected by stress as narG and nosZ gene abundances decreased significantly but did not recover after rewetting, suggesting that rewetting soils either boosts denitrifying communities’ activity, not abundance, or boosts nitrite-reducing denitrifiers increasing N2O production but not N2. Further measurements of nirK and nirS gene abundance, which encode nitrite reduction, are required to elucidate this. In contrast, PNR were not affected by stress, and nitrifying bacterial communities showed resistance to stress as amoA gene abundances were not affected. Moreover, no correlations were found between functional gene abundances and corresponding denitrification and nitrification rates, which is probably due to the interference of other environmental parameters or simply because of our dataset dimensions.
In conclusion, although the poor initial state of the existing soil IS was confirmed by the low activities we measured, it was resistant and resilient to the heat and drought stress, probably due to its pre-exposure to these stress in situ. Among the rehabilitation solutions tested in the present study, i.e., either an organic-matter-rich technosol (HO) or a less rich organic-matter technosol (LO1 and LO2), although the LO soils were the cheaper option, they suffered more from the heat and drought stress, suggesting that even though the initial cost was more important for HO, it may be the better solution on the long run. Also, considering the LO1 and LO2 soils, it appeared that globally in the control soils during the experiment, LO1 activities and gene abundances were overall higher than in LO2 control soils. Future work is required to understand the impact of the types of plants seeded on site on these differences. Finally, an in-between solution may be to amend the IS with organic matter or adapted seeds and microbial inoculation as the bacterial communities in this soil are already resistant and resilient to temperature and drought stress.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
MF, CJ, MM-H, M-PN, and JH: conceptualization, methodology, validation, writing—review and editing, and supervision. MF and M-PN: experiment, sampling, microbiological analysis, and investigation. MF: writing original draft preparation and data curation. CJ, MM-H, and JH: project administration and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
The research was supported by a Ph.D. grant accorded to MF and co-financed by the ADEME and the BRGM.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.727468/full#supplementary-material
Footnotes
References
Allison, S. D. (2005). Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol. Lett. 8, 626–635. doi: 10.1111/j.1461-0248.2005.00756.x
Almaraz, M., Groffman, P. M., and Porder, S. (2019). Effects of changes in nitrogen availability on nitrogen gas emissions in a tropical forest during a drought. J. Geophys. Res. Biogeosci. 124, 2917–2926. doi: 10.1029/2018JG004851
Amador, J., Glucksman, A., Lyons, J., and Görres, J. (1997). Spatial distribution of soil phosphatase activity within a riparian forest1. Soil Sci. 162, 808–825. doi: 10.1097/00010694-199711000-00005
Anderson, T., and Domsch, K. H. (1985). Determination of ecophysiological maintenance carbon requirements of soil microorganisms in a dormant state. Biol. Fertil. Soils 1, 81–89. doi: 10.1007/BF00255134
Anderson, T. H., and Domsch, K. H. (1993). The metabolic quotient for CO2 (qCO2) as a specific activity parameter to assess the effects of environmental conditions, such as pH, on the microbial biomass of forest soils. Soil Biol. Biochem. 25, 393–395. doi: 10.1016/0038-0717(93)90140-7
Attard, E., Recous, S., Chabbi, A., Berranger, C., Guillaumaud, N., Labreuche, J., et al. (2011). Soil environmental conditions rather than denitrifier abundance and diversity drive potential denitrification after changes in land uses. Glob. Chang. Biol. Bioenergy 17, 1975–1989. doi: 10.1111/j.1365-2486.2010.02340.x
Bapiri, A., Bååth, E., and Rousk, J. (2010). Drying-rewetting cycles affect fungal and bacterial growth differently in an arable soil. Microb. Ecol. 60, 419–428. doi: 10.1007/s00248-010-9723-5
Barnard, R. L., Osborne, C. A., and Firestone, M. K. (2013). Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 7, 2229–2241. doi: 10.1038/ismej.2013.104
Bérard, A., Ben Sassi, M., Kaisermann, A., and Renault, P. (2015). Soil microbial community responses to heat wave components: drought and high temperature. Clim. Res. 66, 243–264. doi: 10.3354/cr01343
Bérard, A., Boucher, T., Sévenier, G., Pablo, A. L., and Gros, R. (2011). Resilience of soil microbial communities impacted by severe drought and high temperature in the context of Mediterranean heat waves. Eur. J. Soil Sci. 47, 333–342. doi: 10.1016/j.ejsobi.2011.08.004
Bérard, A., Bouchet, T., Sévenier, G., Pablo, A. L., and Gros, R. (2012). Severe drought-induced community tolerance to heat wave. An experimental study on soil microbial processes. J. Soils Sediments 12, 513–518. doi: 10.1007/s11368-012-0469-1
Bonilla-Rosso, G., Wittorf, L., Jones, C. M., and Hallin, S. (2016). Design and evaluation of primers targeting genes encoding NO-forming nitrite reductases: implications for ecological inference of denitrifying communities. Sci. Rep. 6:39208. doi: 10.1038/srep39208
Bonmati, M., Ceccanti, B., and Nanniperi, P. (1991). Spatial variability of phosphatase, urease, protease, organic carbon and total nitrogen in soil. Soil Biol. Biochem. 23, 391–396. doi: 10.1016/0038-0717(91)90196-Q
Bouchez, T., Blieux, A. L., Dequiedt, S., Domaizon, I., Dufresne, A., Ferreira, S., et al. (2016). Molecular microbiology methods for environmental diagnosis. Environ. Chem. Lett. 14, 423–441.
Bouskil, N., Lim, H. C., Borglin, S., Salve, R., Wood, T. E., Silver, W. L., et al. (2013). Pre-exposure to drought increases the resistance of tropical forest soil bacterial communities to extended drought. ISME J. 7, 384–394. doi: 10.1038/ismej.2012.113
Bremner, J. M., and Blackmer, A. M. (1978). Nitrous oxide: emission from soils during nitrification of fertilizer nitrogen. Science 199, 295–296. doi: 10.1126/science.199.4326.295
Bru, D., Sarr, A., and Philippot, L. (2007). Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl. Environ. Microbiol. 73, 5971–5974. doi: 10.1128/aem.00643-07
Butterbach-Bahl, K. B., and Dannenmann, M. (2011). Denitrification and associated soil N2O emissions due to agricultural activities in a changing climate. Curr Opin Environ Sustain 3, 389–395. doi: 10.1016/j.cosust.2011.08.004
Campbell, C., Chapman, S., and Davidson, M. (2003). A rapid microtiter plate method to measure dioxide evolved from carbon substrate amendments so as to determine the physiological profiles of soil microbial communities by using whole soil. Appl. Environ. Microbiol. 69, 3593–3599. doi: 10.1128/AEM.69.6.3593-3599.2003
Canarini, A., Kiær, L. P., and Dijkstra, F. A. (2017). ‘Soil carbon loss regulated by drought intensity and available substrate: a meta-analysis’. Soil Biol. Biochem. 112, 90–99. doi: 10.1016/j.soilbio.2017.04.020
Carreiro, M. M., Pouyat, R., Tripler, C., and Zhu, W. X. (2009). Carbon and Nitrogen Cycling in Soils of Remnant Forests Along Urban–Rural Gradients: Case Studies in the New York Metropolitan Area and Louisville, Kentucky, in Ecology of Cities and Towns: A Comparative Approach. Cambridge: Cambridge University Press, 308–328. doi: 10.1017/CBO9780511609763.020
CGDD (2015). Le point sur L’occupation des Sols en France, L’occupation des sols en France?: Progression plus Modérée de L’artificialisation Entre 2006 et 2012., (219). Available online at: https://www.actu-environnement.com/media/pdf/news-26026-soes-artificialisation-sols.pdf (accessed April, 2019).
Davidson, E. A. (1992). Sources of Nitric Oxide and Nitrous Oxide following Wetting of Dry Soil. Soil Sci. Soc. Am. J. 56, 95–102. doi: 10.2136/sssaj1992.03615995005600010015x
De Noni, G., and Viennot, M. (1993). Mutations recentes de l’agriculture equatorienne et consequences sur la durabilite des agrosystemes andins. Pédol 28, 277–288.
Dequiedt, S., Saby, N. P. A., Lelievre, M., Jolivet, C., Thioulouse, J., Toutain, B., et al. (2011). Biogeographical patterns of soil molecular microbial biomass as influenced by soil characteristics and management. Glob. Ecol. Biogeogr. 20, 641–652. doi: 10.1111/j.1466-8238.2010.00628.x
De Vos, P., Garrity, G. M., Jones, D., Krieg, N. R., Ludwig, W., Rainey, F. A., et al. (2009). “The Firmicutes,” in Bergey’s Manual of Systematic Bacteriology, eds P. De Vos, G. M. Garrity, D. Jones, N. R. Krieg, W. Ludwig, F. A. Rainey, et al. (New York, NY: Springer).
Fierer, N., Schimel, J. P., and Holden, P. A. (2003). Influence of drying – rewetting frequency on soil bacterial community structure. Microb. Ecol. 45, 63–71. doi: 10.1007/s00248-002-1007-2
Fuchslueger, L. (2014). Effects of drought on nitrogen turnover and abundances of ammonia-oxidizers in mountain grassland. Biogeosci. Discuss. 11, 9183–9214. doi: 10.5194/bgd-11-9183-2014
Gibelin, A. L., and Déqué, M. (2003). Anthropogenic climate change over the Mediterranean region simulated by a global variable resolution model. Clim. Dyn. 20, 327–339. doi: 10.1007/s00382-002-0277-1
Gleeson, D. B., Müller, C., Banerjee, S., Ma, W., Siciliano, S. D., and Murphy, D. V. (2010). Response of ammonia oxidizing archaea and bacteria to changing water filled pore space. Soil Biol. Biochem. 42, 1888–1891. doi: 10.1016/j.soilbio.2010.06.020
Göransson, H., Godbold, D. L., Jones, D. L., and Rousk, J. (2013). Bacterial growth and respiration responses upon rewetting dry forest soils: impact of drought-legacy. Soil Biol. Biochem. 57, 477–486. doi: 10.1016/j.soilbio.2012.08.031
Graham, E. B., Knelman, J. E., Schindlbacher, A., Siciliano, S., Breulmann, M., Yannarell, A., et al. (2016). Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes? Front. Microbiol. 7:214. doi: 10.3389/fmicb.2016.00214
Graham, E. B., Wieder, W. R., Leff, J. W., Weintraub, S. R., Townsend, A. R., Cleveland, C. C., et al. (2014). Do we need to understand microbial communities to predict ecosystem function? A comparison of statistical models of nitrogen cycling processes. Soil Biol. Biochem. 68, 279–282. doi: 10.1016/j.soilbio.2013.08.023
Griffiths, B. S., and Philippot, L. (2013). Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol. Rev. 37, 112–129. doi: 10.1111/j.1574-6976.2012.00343.x
Guillot, E., Hinsinger, P., Dufour, L., Roy, J., and Bertrand, I. (2019). With or without trees: resistance and resilience of soil microbial communities to drought and heat stress in a Mediterranean agroforestry system. Soil Biol. Biochem. 129, 122–135. doi: 10.1016/j.soilbio.2018.11.011
Hallin, S., Jones, C. M., Schloter, M., and Phillipot, L. (2009). Relationship between n-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 3, 597–605. doi: 10.1038/ismej.2008.128
Harris, E., Diaz-Pines, E., Stoll, E., Schloter, M., Schulz, S., Duffner, C., et al. (2021). Denitrifying pathways dominate nitrous oxide emissions from managed grassland during drought and rewetting. Sci. Adv. 7:eabb7118. doi: 10.1126/sciadv.abb7118
Harris, R. F. (2015). Effect of water potential on microbial growth and activity. Water Potential Relat. Soil Microbiol. 9, 23–95. doi: 10.2136/SSSASPECPUB9.C2
Hartmann, A. A., Barnard, R. L., Marhan, S., and Niklaus, P. A. (2013). Effects of drought and N-fertilization on N cycling in two grassland soils. Oecologia 171, 705–717. doi: 10.1007/s00442-012-2578-3
Henry, H. A. L. (2013). Reprint of “Soil extracellular enzyme dynamics in a changing climate”’. Soil Biol. Biochem. 56, 53–59. doi: 10.1016/j.soilbio.2012.10.022
Henry, S., Bru, D., Stres, S., Hallet, S., and Philippot, L. (2006). Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72, 5181–5189. doi: 10.1128/AEM.00231-06
Hobbie, S. E., Finlay, J. C., Janke, B. D., Nidzgorski, D. A., Millet, D. B., and Baker, L. A. (2017). Correction: contrasting nitrogen and phosphorus budgets in urban watersheds and implications for managing urban water pollution. Proc. Natl. Acad. Sci. U.S.A. 114, E4116. doi: 10.1073/pnas.1706049114
Homyak, P. M., Allison, S. D., Huxman, T. E., Goulden, M. L., and Treseder, K. K. (2017). Effects of drought manipulation on soil nitrogen cycling: a meta-analysis. J. Geophys. Res. Biogeosci. 122, 3260–3272. doi: 10.1002/2017JG004146
Hueso, S., Hernández, T., and García, C. (2011). Resistance and resilience of the soil microbial biomass to severe drought in semiarid soils: the importance of organic amendments. Appl. Soil Ecol. 50, 27–36. doi: 10.1016/j.apsoil.2011.07.014
Hynes, H. M., and Germida, J. J. (2012). Relationship between ammonia oxidizing bacteria and bioavailable nitrogen in harvested forest soils of central Alberta. Soil Biol. Biochem. 46, 18–25. doi: 10.1016/j.soilbio.2011.10.018
Jackson, E. L., and Burger, C., and Cavagnaro, T. R. (2008). Roots, nitrogen transformations, and ecosystem services. Annu. Rev. Plant Biol. 59, 341–363. doi: 10.1146/annurev.arplant.59.032607.092932
Jia, Z., and Conrad, R. (2009). Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11, 1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x
Kane, D. A., Bradford, M. A., Fuller, E., Oldfield, E. E., and Wood, S. (2021). Soil organic matter protects US maize yields and lowers crop insurance payouts under drought. Environ. Res. Lett. 16:044018. doi: 10.1088/1748-9326/abe492
Kaurin, A., Mihelič, R., Kastelec, D., Grčman, H., Bru, D., and Philipot, L. (2018). Resilience of bacteria, archaea, fungi and N-cycling microbial guilds under plough and conservation tillage, to agricultural drought. Soil Biol. Biochem. 120, 233–245. doi: 10.1016/j.soilbio.2018.02.007
Keil, D., Niklaus, P. A., Von Riedmatten, L., Boeddinghaus, R. S., Dormann, C., and Scherer-Lorenzen, M. (2015). Effects of warming and drought on potential N2O emissions and denitrifying bacteria abundance in grasslands with different land-use. FEMS Microbiol. Ecol. 91, 1–9. doi: 10.1093/femsec/fiv066
Lamersdorf, N. P., Beier, K. B., Blanck, K., Bredemeier, M., Cummins, T., Farrell, E. P., et al. (1998). Effect of drought experiments using roof installations on acidification/nitrification of soils. For. Ecol. Manag. 101, 95–109. doi: 10.1016/S0378-1127(97)00128-X
Li, G., Sun, X. G., Ren, Y., Luo, X. S., and Zhu, Y. G. (2018). Urban soil and human health: a review. Eur. J. Soil Sci. 69, 196–215. doi: 10.1111/ejss.12518
Li, X., Zhu, Y. G., Cavagnaro, T. R., Chen, M., Sun, J., Chen, X., et al. (2009). Do ammonia-oxidizing archaea respond to soil Cu contamination similarly asammonia-oxidizing bacteria? Plant Soil. 324, 209–217. doi: 10.1007/s11104-009-9947-7
Liang, C., Zhu, X., Fu, S., Méndez, A., Gasco, G., and Ferreiro, J. P. (2014). Biochar alters the resistance and resilience to drought in a tropical soil. Environ. Res. Lett. 9:064013. doi: 10.1088/1748-9326/9/6/064013
Lorenz, K., and Lal, R. (2009). Biogeochemical C and N cycles in urban soils. Environ. Int. 35, 1–8. doi: 10.1016/j.envint.2008.05.006
Nesper, M., Kueffer, C., Krishnan, S., Kushalappa, C. G., and Ghazoul, J. (2019). Simplification of shade tree diversity reduces nutrient cycling resilience in coffee agroforestry. J. Appl. Ecol. 56, 119–131. doi: 10.1111/1365-2664.13176
Noe, G. B., and Hupp, C. R. (2005). Carbon, nitrogen, and phosphorus accumulation in floodplains of Atlantic Coastal Plain rivers, USA. Ecol. Appl. 15, 1178–1190. doi: 10.1890/04-1677
Norton, J., and Ouyang, Y. (2019). Controls and adaptive management of nitrification in agricultural soils. Front. Microbiol. 10:1931. doi: 10.3389/fmicb.2019.01931
Oke, T. R. (1989). Micromet urban forest. R. Soc. Open Sci. 324, 335–349. doi: 10.1098/rstb.1989.0051
Olander, L. P., and Vitousek, P. M. (2000). Regulation of soil phosphatase and chitinase activity by N and P availability. Biogeochemistry 49, 175–190. doi: 10.1023/A:1006316117817
Parker, S. S., and Schimel, J. P. (2011). Soil nitrogen availability and transformations differ between the summer and the growing season in a California grassland. Appl. Soil Ecol. 48, 185–192. doi: 10.1016/j.apsoil.2011.03.007
Pascual Aguilar, J. A., Sanz García, J., de Bustamante Gutierrez, I., and Kallache, M. (2011). “Using environmental metrics to describe the spatial and temporal evolution of landscape structure and soil hydrology and fertility,” in Proceedings of the Spatial2: Spatial Data Methods for Environmental and Ecological processes. (Foggia), 1–5.
Peterjohn, W. T. (1991). Denitrification: enzyme content and activity in desert soils. Soil Bio. Biochem. 23, 845–855. doi: 10.1016/0038-0717(91)90096-3
Petersen, D. G., Blazewicz, S., Firestone, M., Herman, D. J., Turetsky, M., and Waldrop, M. (2012). Abundance of microbial genes associated with nitrogen cycling as indices of biogeochemical process rates across a vegetation gradient in Alaska. Environ. Microbiol. 14, 993–1008. doi: 10.1111/j.1462-2920.2011.02679.x
Philippot, L., Spor, A., Hénault, C., Bru, D., Bizouard, F., Jones, C. M., et al. (2013). Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 7, 1609–1619. doi: 10.1038/ismej.2013.34
Piotrowska, A., and Koper, J. (2010). Soil β-glucosidase activity under winter wheat cultivated in crop rotation systems depleting and enriching the soil in organic matter. J. Elem. 15, 593–600. doi: 10.5601/jelem.2010.15.3.593-600
R Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
Radl, V., Kindler, R., Welzl, G., Albert, A., Wilke, B. M., Amelung, W., et al. (2015). Drying and rewetting events change the response pattern of nitrifiers but not of denitrifiers to the application of manure containing antibiotic in soil. Appl. Soil Ecol. 95, 99–106. doi: 10.1016/j.apsoil.2015.06.016
REBU (2016). Available online at: moz-extension://d71dad57-548d-4ff9-b868-35ab0f8e7a51/enhanced-reader.html?openApp&pdf=https%3A%2F%2Fwww.ademe.fr%2Fsites%2Fdefault%2Ffiles%2Fassets%2Fdocuments%2Ffiche-laureat-rebu-valorhiz.pdf (Accessed January 18, 2021).
Riah-Anglet, W., Trinsoutrot-Gattina, I., Martin-Laurent, F., Laroche-Ajzenberga, E., Norinia, M.-P., Latourc, X., et al. (2015). Soil microbial community structure and function relationships: a heat stress experiment. Appl. Soil Ecol. 86, 121–130. doi: 10.1016/j.apsoil.2014.10.001
Rocca, J. D., Hall, E. K., Lennon, J. T., Evans, S. E., Waldrop, M. P., Cotner, J. B., et al. (2015). Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed. J. ISME 9, 1693–1699.
Romero-Freire, A., Sierra Aragon, M., Martinez Garzón, F. J., and Martín Peinado, F. J. M. (2016). Is soil basal respiration a good indicator of soil pollution? Geoderma 263, 132–139. doi: 10.1016/j.geoderma.2015.09.006
Rotthauwe, J. H., Witzel, K. P., and Liesack, W. (1997). The ammonia monooxygenase structural gene amoa as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997
Sanaullah, M., Blagodatskaya, E., Chabbi, A., Rumpel, C., and Kuzyakov, Y. (2011). Drought effects on microbial biomass and enzyme activities in the rhizosphere of grasses depend on plant community composition. Appl. Soil Ecol. 48, 38–44. doi: 10.1016/j.apsoil.2011.02.004
Sardans, J., and Peñuelas, J. (2005). Drought decreases soil enzyme activity in a Mediterranean Quercus ilex L. forest. Soil Biol. Biochem. 37, 455–461. doi: 10.1016/j.soilbio.2004.08.004
Sardans, J.Peñuelas, J. (2010). Soil enzyme activity in a mediterranean forest after six years of drought. Soil Sci. Soc. Am. J. 74, 838–851. doi: 10.2136/sssaj2009.0225
Sardans, J., Peñuelas, J., and Estiarte, M. (2006). Warming and drought alter soil phosphatase activity and soil P availability in a Mediterranean shrubland. Plant Soil 289, 227–238. doi: 10.1007/s11104-006-9131-2
Sardans, J., Peñuelas, J., and Ogaya, R. (2008). Experimental drought reduced acid and alkaline phosphatase activity and increased organic extractable P in soil in a Quercus ilex Mediterranean forest. Eur. J. Soil Biol. 44, 509–520. doi: 10.1016/j.ejsobi.2008.09.011
Schimel, J., Balser, T. C., and Wallenstein, M. (2007). Microbial stress response physiology and its implications for ecosystem function. Ecology 88, 1386–1394. doi: 10.1890/06-0219
Schimel, J. P. (2018). Life in dry soils: effects of drought on soil microbial communities and processes. Annu. Rev. Ecol. Evol. Syst. 49, 409–432. doi: 10.1146/annurev-ecolsys-110617-062614
Siebielec, S., Siebielec, G., Klimkowicz-Pawlas, A., Gałązka, A., Grządziel, J., and Stuczyński, T. (2020). Impact of water stress on microbial community and activity in sandy and loamy soils. Agronomy 10:1429. doi: 10.3390/agronomy10091429
Singhania, R. R., Patel, A. K., Sukumaran, R. K., Larroche, C., and Pandey, A. (2013). Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresour. Technol. 127, 500–507. doi: 10.1016/j.biortech.2012.09.012
Six, J., Conant, R. T., Paul, E. A., and Paustian, K. (2002). Stabilization mechanisms of soil organic matter: implications for C-saturation of soils. Plant Soil 241, 155–176. doi: 10.1023/A:1016125726789
Song, A., Fan, F., Yin, C., Wen, S., Zhang, Y., Fan, X., et al. (2017). The effects of silicon fertilizer on denitrification potential and associated genes abundance in paddy soil. Biol. Fertil. Soils 53, 627–638. doi: 10.1007/s00374-017-1206-0
Sun, D.-L., Jiang, X., Wu, Q. L., and Zhou, N.-Y. (2013). Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl. Environ. Microbiol. 79, 5962–5969. doi: 10.1128/AEM.01282-13
Eivazi, F., and Tabatabai, M. A. (1990). Factors affecting glucosidase and galactosidase activities in soils. Soil Biol. Biochem. 22, 891–897. doi: 10.1016/0038-0717(90)90126-K
Tan, X., Shao, D., and Gu, W. (2018). Effects of temperature and soil moisture on gross nitrification and denitrification rates of a Chinese lowland paddy field soil. Paddy Water Environ. 16, 687–698. doi: 10.1007/s10333-018-0660-0
Větrovský, T., and Baldrian, P. (2013). The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8:e57923. doi: 10.1371/journal.pone.0057923
Vincent, Q., Auclerc, A., Beguiristain, T., and Leyval, C. (2018). Assessment of derelict soil quality: abiotic, biotic and functional approaches. Sci. Total Environ. 613, 990–1002. doi: 10.1016/j.scitotenv.2017.09.118
Wang, H., Marshall, C. W., Cheng, M., Xu, H. Li, H., Yang, X., et al. (2017). Changes in land use driven by urbanization impact nitrogen cycling and the microbial community composition in soils. Sci. Rep. 7:44049. doi: 10.1038/srep44049
Wang, H., Yang, J. P., Yang, S. H., Yang, Z. C., and Lv, Y. M. (2014). Effect of a 10°C-elevated temperature under different water contents on the microbial community in a tea orchard soil. Eur. J. Soil Sci. 62, 113–120. doi: 10.1016/j.ejsobi.2014.03.005
Wei, W., Isobe, K., Nishizawa, T., Zhu, L., Shiratori, Y., Ohte, N., et al. (2015). Higher diversity and abundance of denitrifying microorganisms in environments than considered previously. ISME J. 9, 1954–1965. doi: 10.1038/ismej.2015.9
Williams, M. A. (2007). Response of microbial communities to water stress in irrigated and drought-prone tallgrass prairie soils. Soil Biol. Biochem. 39, 2750–2757. doi: 10.1016/j.soilbio.2007.05.025
Yu, Z., Wang, G., and Marschner, P. (2014). Drying and rewetting - Effect of frequency of cycles and length ofmoist period on soil respiration and microbial biomass. Eur. J. Soil Biol. 62, 132–137. doi: 10.1016/j.ejsobi.2014.03.007
Zeglin, L. H., Bottomley, P. J., Jumpponen, A., Rice, C. W., Arango, M., Lindsley, A., et al. (2013). Altered precipitation regime affects the function and composition of soil microbial communities on multiple time scales. Ecology 94, 2334–2345. doi: 10.1890/12-2018.1
Zhang</snm>, Y., and Ji, G. (2018). Quantitative responses of potential nitrification and denitrification rates to the size of microbial communities in rice paddy soils. Chemosphere 211, 970–977. doi: 10.1016/j.chemosphere.2018.08.047
Zhang, M., Bai, S. H., Tang, L., Zhang, Y., Teng, Y., and Xu, Z. (2017). Linking potential nitrification rates, nitrogen cycling genes and soil properties after remediating the agricultural soil contaminated with heavy metal and fungicide. Chemosphere 184, 892–899. doi: 10.1016/j.chemosphere.2017.06.081
Keywords: soil functions, resistance, resilience, N-cycling, bacteria, denitrification, nitrification, rehabilitation
Citation: Fikri M, Joulian C, Motelica-Heino M, Norini M-P and Hellal J (2021) Resistance and Resilience of Soil Nitrogen Cycling to Drought and Heat Stress in Rehabilitated Urban Soils. Front. Microbiol. 12:727468. doi: 10.3389/fmicb.2021.727468
Received: 18 June 2021; Accepted: 12 November 2021;
Published: 22 December 2021.
Edited by:
Dimitrios Georgios Karpouzas, University of Thessaly, GreeceReviewed by:
Katharina Kujala, University of Oulu, FinlandMyrto Tsiknia, Agricultural University of Athens, Greece
Copyright © 2021 Fikri, Joulian, Motelica-Heino, Norini and Hellal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer Hellal, ai5oZWxsYWxAYnJnbS5mcg==
 Mehdi Fikri
Mehdi Fikri Catherine Joulian
Catherine Joulian Mikael Motelica-Heino
Mikael Motelica-Heino Marie-Paule Norini1
Marie-Paule Norini1 Jennifer Hellal
Jennifer Hellal