
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 November 2021
Sec. Microbial Symbioses
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.727202
This article is part of the Research Topic Exploring the Insect Microbiome: The Potential Future Role in Biotechnology Industry View all 8 articles
In the present study, we identified a novel, positive-sense single-stranded RNA virus in the Chinese black cutworm, Agrotis ipsilon. It has a genome length of 11,312 nucleotides, excluding the poly(A) tails, and contains five open reading frames. The ORF2 encodes the conserved domains of RNA helicase and RNA-dependent RNA polymerase, while ORF4 and 5 encode three viral proteins. Herein, the A. ipsilon virus was clustered with a Helicoverpa armigera Nora virus and was thus provisionally named “Agrotis ipsilon Nora virus” (AINV). AINV was successfully transmitted into a novel host, Spodoptera frugiperda, through injection, causing a stable infection. This found the possibility of horizontal AINV transmission among moths belonging to the same taxonomic family. Nonetheless, AINV infection was deleterious to S. frugiperda and mainly mediated by antiviral and amino acid metabolism-related pathways. Furthermore, the infection significantly increased the S. frugiperda larval period but significantly reduced its moth eclosion rate. It suggests that AINV is probably to be a parasitic virus of S. frugiperda.
The second-generation sequencing technology has expanded our understanding of the diversity of insect viruses (Xu et al., 2014; Liu et al., 2017; Shi et al., 2018; Yang et al., 2019). Recently, picorna-like symbiotic viruses are commonly identified in insects (Shi et al., 2016; Yang et al., 2016; Cholleti et al., 2018). Taxonomically, picorna-like viruses belong to the order Picornavirales, which share some properties with members of the family Picornaviridae (Le Gall et al., 2008). Generally, picorna-like viruses possess a (+) ssRNA genome (positive-sense, single-stranded RNA), and their translated polyproteins are cleaved into structural and non-structural proteins. Their pathogenicity is diverse, ranging from lethal to symbiotic (de Miranda et al., 2010; Jakubowska et al., 2014).
The Nora virus was first reported in Drosophila melanogaster and then proposed to represent a new picorna-like virus family (Habayeb et al., 2006). Subsequent studies have revealed that Nora viruses interfere with the D. melanogaster RNAi system (van Mierlo et al., 2012, 2014). Recently, additional members of the Nora virus clade have been identified in arthropods through high-throughput sequencing technologies (Shi et al., 2016). Agrotis ipsilon (Hufnagel) is a worldwide pest that causes significant damage to vegetables and grains. Despite its interactions with its endosymbionts being thoroughly studied (Shi et al., 2013), its viral landscape remains ambiguous. Herein, we identified a novel Nora virus in Chinese A. ipsilon through RNA sequencing. The virus was provisionally named as “Agrotis ipsilon Nora virus” (AINV). The complete AINV genome was subsequently determined, and its phylogenetic position and gene expression patterns were uncovered among the A. ipsilon developmental stages. AINV was also introduced into wild Spodoptera frugiperda, an invasive pest in China (Sun et al., 2021), to evaluate its effects on fitness and gene expressions.
Colonies of A. ipsilon and S. frugiperda were established with caterpillars collected in Shanghai, China, in 2018 and Zhengzhou, China, in 2020, respectively. The caterpillars were fed with artificial diets at 25°C (±1°C) under a 14/10h light/dark cycle. Adult moths were fed with 10% honey water.
Total RNA was extracted from A. ipsilon samples using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s protocol. The samples were collected at four developmental stages, i.e., egg and larvae (n=30), pupae (n=30), and adults (n=30) in both male and female insects. RNA quality and quantity were assessed on an Agilent 2,100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States) and RNase-free agarose gel electrophoresis. The mRNA in the RNA samples was enriched by removing rRNA using the Ribo-ZeroTM Magnetic Kit (Epicentre, Madison, WI, United States). The mRNA libraries were subsequently prepared by BGI (Shenzhen, China) and sequenced on an Illumina Hiseq 2000 platform. De novo transcriptome assemblies were performed using the SOAPdenovo software with default parameters (Xie et al., 2014). Protein sequences of sRNA viruses (txid 439,488), dsRNA viruses (txid 35,325), and Delta viruses (txid 39,759) were retrieved from the NCBI RefSeq database (Reference Sequences) and employed as subjects in local blastx searches to uncover possible viral fragments in assembled unigenes. The non-virus originating hits were filtered by performing online blastx searches against the NCBI nr database (non-redundant protein sequences). An e-value threshold of 1×10−5 was used in these searches.
Full-length cDNA sequences of the AINV were obtained using the 3′ and 5′ rapid amplification of cDNA ends (RACE) system (Life Technologies, Carlsbad, CA, United States), following the manufacturer’s instructions. The sequences were subsequently verified through amplification and sequencing using specific primers. Total RNA for specific RT-PCR was extracted using the TaKaRa MiniBEST Universal RNA Extraction Kit (Takara, Dalian, China), followed by RT-PCR amplification of the AINV fragments using PrimeScript™ One-Step RT-PCR Kit Ver.2 (Dye Plus; Takara). The gene-specific primers used herein were listed in Table 1. The standard genetic code was employed to predict the open reading frames of the AINV using the NCBI online ORF finder program.1 Conserved domains within the ORFs were subsequently predicted using the NCBI conserved domain database v3.17,2 with threshold of 1×10−2.
The RNA-dependent RNA polymerase (RdRp) protein sequences from taxa within Picornavirales and Caliciviridae were retrieved to uncover the phylogenetic position of AINV. The sequences were aligned using the MUSCLE program in MEGA 7.0 (Kumar et al., 2016) and then trimmed using trimAl to remove the poorly aligned regions (Capella-Gutierrez et al., 2009). Phylogenetic analysis was performed in IQ-TREE 1.6.6 (Nguyen et al., 2015), followed by resampling 1,000 ultrafast bootstraps to assess the support for each node. The substitution model based on the Bayesian information criterion in ModelFinder (Kalyaanamoorthy et al., 2017) was selected to improve the accuracy of phylogenetic estimates.
An AINV infected liquid was formulated following the methods described by Xu et al. (2020). The AINV infected fourth A. ipsilon larvae were ground in liquid nitrogen and then homogenized with 1ml PBS buffer (0.01M, pH 7.4). The homogenate was then centrifuged at 6500×g for 15min at 4°C, followed by filtration of the liquid supernatant through 0.2μm syringe filters (Pall Corporation, New York, United States). The filtrate (10μl) was subsequently injected into the third S. frugiperda larvae using a Hamilton Microliter syringe (705N) to introduce the AINV into the host. Control hosts were injected with an equal volume of sterile water. The S. frugiperda larvae were reared in room, followed by detection of AINV infections in the moths using AINV specific primers, AINV_72F and AINV_44R (Table 1). The S. frugiperda larvae were reared in room, followed by detection of AINV infections in the moths using AINV specific primers, AINV_72F and AINV_44R (Table 1). AINV titers in S. frugiperda were quantified based on the ORF2 (polyprotein) copies and then normalized with S. frugiperda β-actin gene copies. The qPCR reactions were performed in a CFX384 Touch™ Real-Time PCR Detection System (Bio-Rad, United States) using the SYBR Green method. The cDNA used in qPCR reactions was synthesized using the PrimeScript™ RT reagent Kit with gDNA Eraser (Takara). The reactions were carried out in a 10μl volume containing 5 ul of 2×TB Green® Premix Ex Taq™ (Takara), 0.4μl of each primer (10μm), 1μl of cDNA, and 3.2μl sterile water. The qPCR primers used are listed in Table 1. AINVQF and AINVQR2 amplified a 102bp fragment of the AINV, while SFACTINF and SFACTINR amplified a 156bp fragment of the S. frugiperda β-actin gene. The qPCR reactions for quantifying AINV titers in infected male and female S. frugiperda larvae were done in triplicate. Four replicates of AINV infected larvae were used in these qPCRs.
Six fitness indices, including larval period, pupal period, pupal weight, pupation rate, eclosion rate, and sex ratio, were measured in AINV positive and negative S. frugiperda populations to uncover the effects of AINV infection on the novel host. The GraphPad Prism 8.3.0 software was then employed to analyze and visualize the data for the six indices.
RNA-seq was also employed to reveal the effects of AINV infection on the gene expression of female S. frugiperda adults. The first day hatched moths were subjected to RNA-seq, and three replicates of AINV positive and negative females were used following the aforementioned sequencing procedure. The HISAT2 (2.0.4; Kim et al., 2015) alignment program was used to map the obtained reads to the S. frugiperda published genome (ZJU_Sfru_1.0, GCF_011064685.1). Then, the mapped reads were assembled using the StringTie (Version: 1.3.4) sequence assembler (Pertea et al., 2015), followed by merging of the transcripts using gffcompare (Version: 0.9.8).3 The ballgown R package4 was subsequently used to estimate the expression levels of all transcripts and to detect the differentially expressed mRNAs by calculating FPKM (Fragments Per Kilobase Million; FPKM = [total exon fragments/mapped reads(millions)×exon length(kB)]). The edgeR R package (Robinson et al., 2010) was employed to filter the differentially expressed genes (DEGs) at a threshold absolute log2 (FC, fold change)≥1 and false discovery rate (FDR)<0.05. The DEGs were shown with volcano plot, which was visualized using the ggplot2, ggrepel, gridExtra, and ggthemes R packages. Principal component analysis (PCA) of the samples was further determined with the gene expression matrix. The analysis was then visualized using the stats R package. GO and KEGG enrichment results of the DEGs were visualized using the clusterProfiler R package (Yu et al., 2012), while the heat maps were visualized using TBtools (Chen et al., 2020).
There were 62,933 unigenes within the A. ipsilon transcriptome. Two virus-originated fragments were detected as: unigene-23,909 (5,239 nucleotides) and unigene-23,910 (6,052 nucleotides). Blastp searches revealed that both unigenes were similar to the protein sequences of a Nora virus isolated in Helicoverpa armigera (HaNV, accession no: MK033133). These findings suggested that the unigenes probably originated from a Nora virus member and was tentatively named AINV. Supplementary Figure S1 showed the alignment of the two unigenes. The overlapping region between unigene-23,909 and unigene-23,910 was subsequently amplified, and the sequences were manually verified using specific primers (Table 1). Furthermore, following the methods described by Yang et al., (2019), we enriched the virus particles in AINV positive population and then observed them with a transmission electron microscope. The result was provided in Supplementary Figure S3.
The RACE alignments (data not shown) indicated that the complete genome of AINV was 11,312 nucleotides in length, excluding the poly(A) tails (Figure 1A). The locations of unigene-23,909 and unigene-23,910 on AINV genome were position 25 to position 5,263, and position 5,261 to position 11,312, respectively. The genome shared 85.69% nucleotide identity with HaNV with a 96% coverage. The ORF finder results suggested that AINV had a (+) ssRNA genome containing five ORFs. The conserved replication polyprotein domains were in ORF2. Comparison of the replication polyproteins of AINV and HaNV revealed a 14.60% genetic divergence between them. In the same line, sequences homologous to the three structural proteins of Drosophila Nora viruses, named viral protein (VP) 4A, VP4B, and VP4C (Ekstrom et al., 2011), were also detected in ORF4 and ORF5 of the AINV.
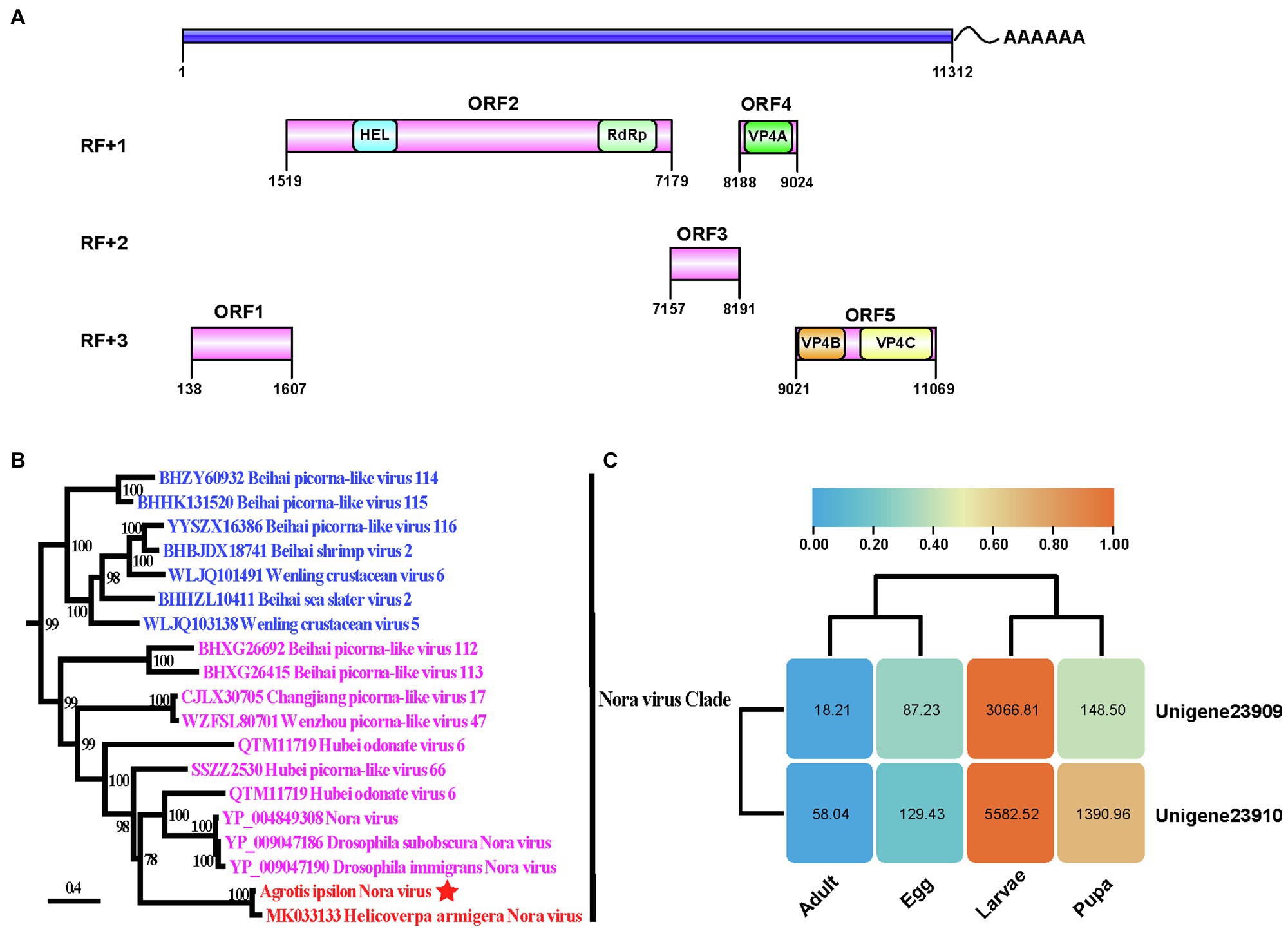
Figure 1. The genome characteristic and phylogenetic analysis of Agrotis ipsilon Nora virus (AINV). (A) The genome organization of A. ipsilon Nora virus. HEL, RNA helicase; RdRp, RNA-dependent RNA polymerase, and VP, structural protein. The genome structure is visualized by IBS program (Liu et al., 2015). (B) The maximum likelihood phylogeny of the Nora virus clade. A. ipsilon Nora virus is underlined. Numbers near nodes indicate ultrafast bootstrap values. The bar indicates the estimated number of substitutions per site. (C) The expression patterns of unigene-23,909 and unigenes-23,910 among A. ipsilon developmental stages. The numbers in the round rectangle indicate the FPKM (Fragments Per Kilobase Million) values. Log2 FPKM values are indicated in the clustered heat map.
The LG substitution model with unequal amino acid frequencies (+F) and rate variation among sites (+R10) was selected in the phylogenetic analysis. The results strongly supported the monophyly of the Nora virus clade (UFBoot=99; Supplementary Figure S2). Furthermore, AINV shared a sister relationship with HaNV (UFBoot=100) within the Nora virus clade (Figure 1B) and further clustered with Nora viruses detected in Drosophila and Odonata (UFBoot=78). These findings confirmed that AINV was a new Nora virus member.
Both unigenes exhibited the same expression pattern in A. ipsilon, with higher expression levels in larvae and pupae than in eggs and adults (Figure 1C). These findings suggested that AINV titers varied among the developmental stages of A. ipsilon, with the larvae and adults having the highest and lowest AINV titers, respectively.
The AINV liquid was injected into 45 third S. frugiperda larvae, while another 45 larvae were injected with sterile water to act as the control. The adults of these larvae were then harvested and subjected to AINV detection using specific primers. Notably, all the adults in the AINV liquid injected group were positive for AINV, but none was positive in the controls. These findings suggested that AINV had a highly accurate horizontal transmission rate even between different hosts with the liquid injection method. Further quantification of the AINV based on the ORF2 polyprotein copies and subsequent normalization with S. frugiperda β-actin gene copies revealed the highest AINV titers in the larvae (1.77±0.32, n=4). In addition, male (1.56±0.05) and female (1.57±0.07) S. frugiperda had almost similar AINV titers. Similarly, there was no significant difference in AINV titer between the larvae and adults (F=1.086, p=0.388; Figure 2).
AINV infection significantly increased the larval (p<0.05) and pupal growth periods and decreased the pupal weight (Figure 3A). There were no significant differences in the pupae rate and sex ratio between the AINV positive and negative populations (Figures 3B,C). However, the AINV negative population had a higher pupae rate than the positive population. Similarly, the moth eclosion rate was significantly decreased in the AINV positive population (p<0.05; Figure 3D). Moreover, we also observed that the AINV positive females laid about average 30 eggs that are much less than the AINV negative females, which can averagely lay more than 500 eggs under room conditions. It indicated that AINV infection would significantly affect the reproductive capacity of S. frugiperda females.
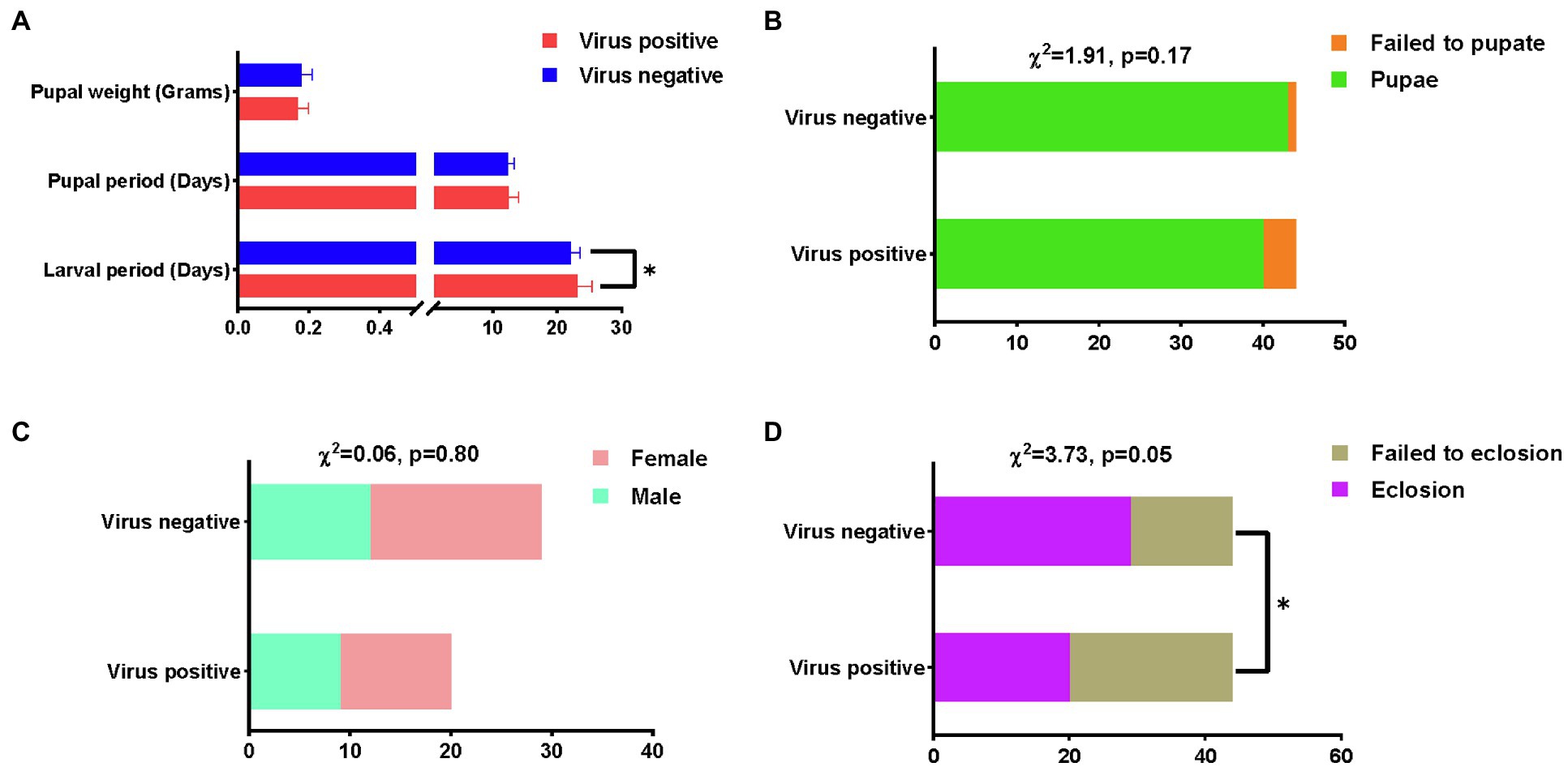
Figure 3. The comparative results of fitness measurements between AINV positive and AINV negative Spodoptera frugiperda populations. (A) The measurements of the larval period, pupal period, and pupal weight. (B) The measurements of pupal rate. (C) The measurements of sex ratio. (D) The measurements of eclosion rate. * Indicates value of p lower than 0.05.
PCA results of AINV positive and negative adult subjected to RNA-seq revealed that the AINV positive and negative replicates were well distinguished in females (Figure 4A), but not in the males (Figure 5A). This finding suggested that AINV infection strongly changed the gene expression pattern in the S. frugiperda females, nor the males. Hence, in this study, we mainly uncovered the effects of AINV on the gene expression patterns in S. frugiperda females. However, the analyses of males were provided in Figure 5.
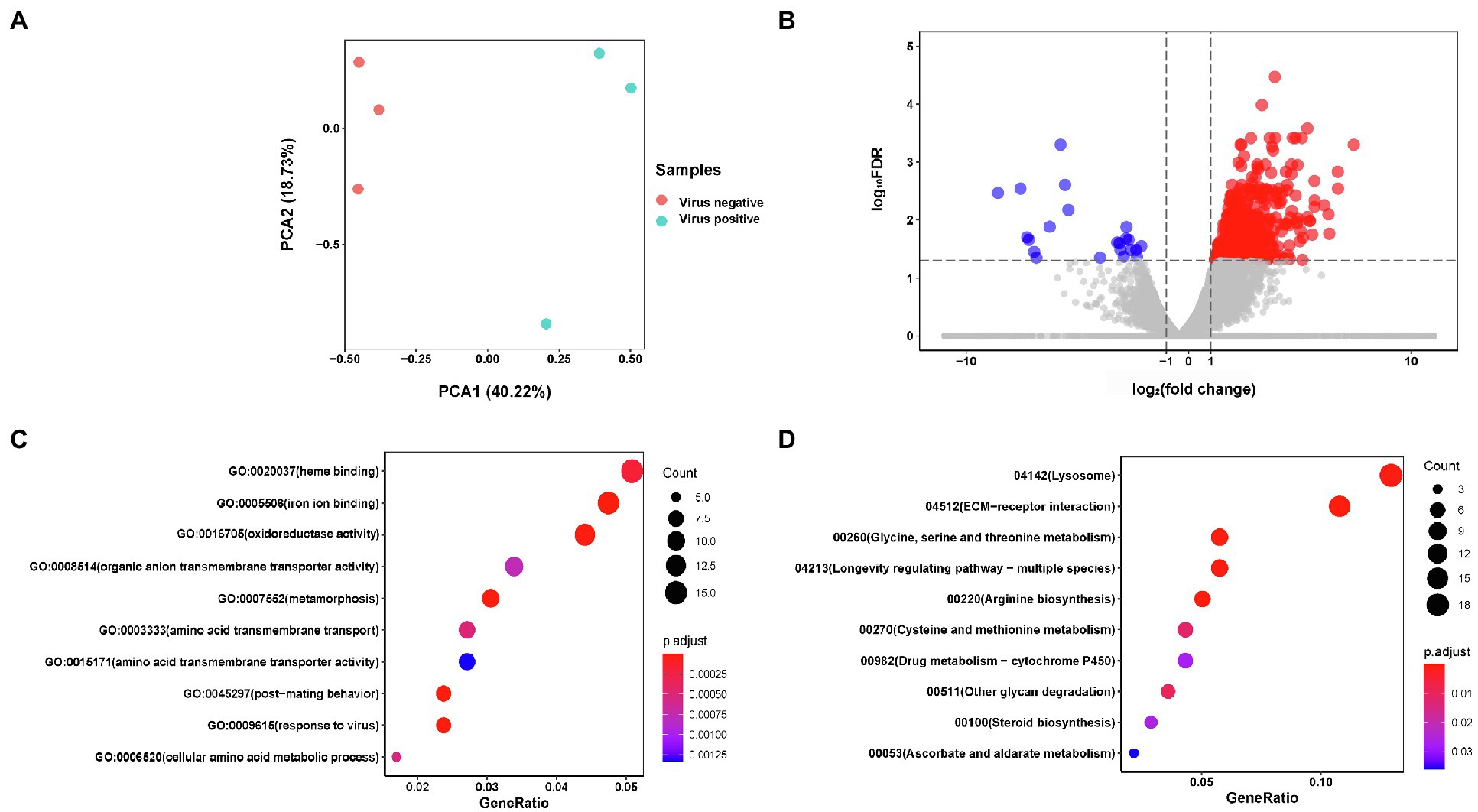
Figure 4. The summary of RNA-seq results in AINV positive and AINV negative Spodoptera frugiperda females. (A) Principal component analysis (PCA) of the samples using the gene expression matrix. (B) Volcano plot of differentially expressed genes (DEGs). (C) The GO enrichment of DGEs. (D) The KEGG enrichment of DEGs.
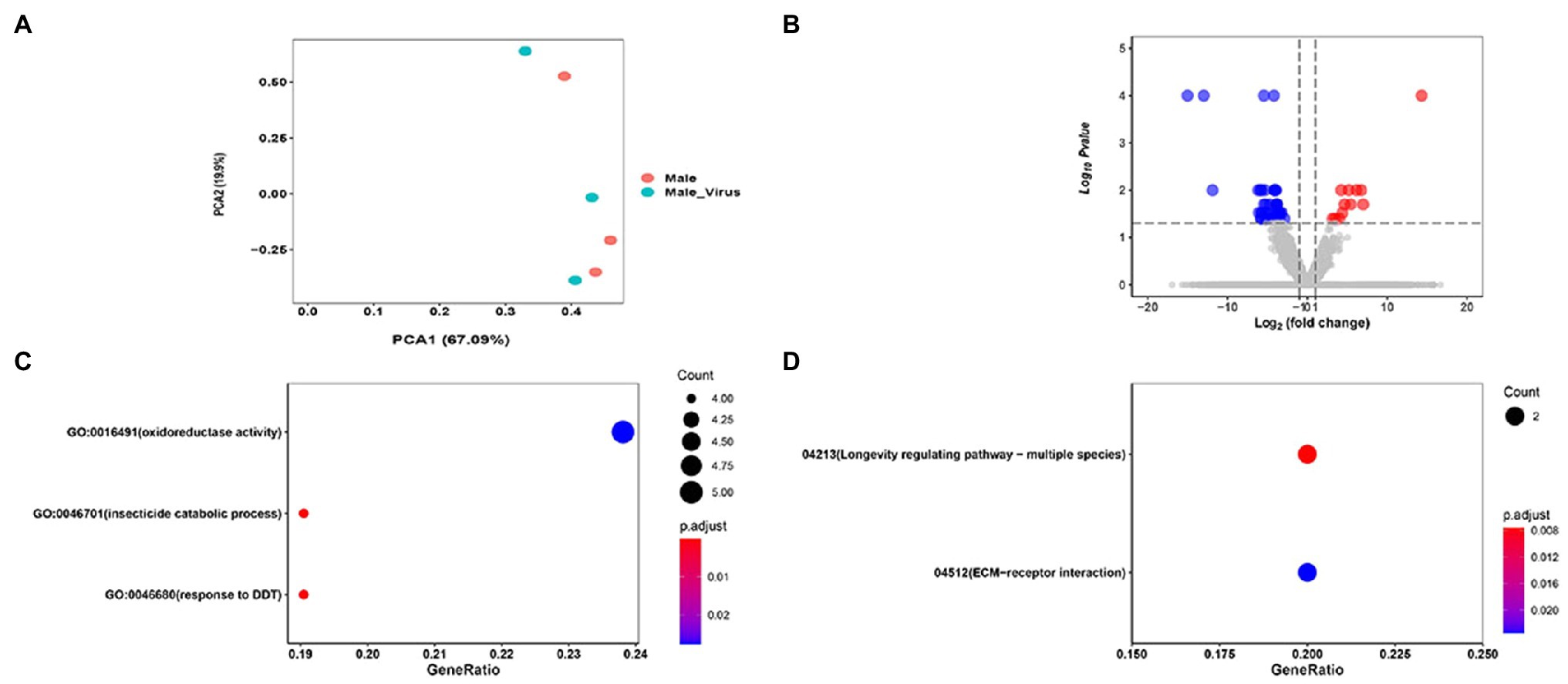
Figure 5. The summary of RNA-seq results in AINV positive and AINV negative Spodoptera frugiperda males. (A) PCA of the samples using the gene expression matrix. (B) Volcano plot of DEGs. (C) The GO enrichment of DGEs. (D) The KEGG enrichment of DEGs.
Based on the log2FC>1 or log2FC<−1 and FDR<0.05 threshold, 627 unigenes were filtered as DEGs. Among them, 26 were significantly downregulated while 601 were upregulated in AINV positive females compared to the AINV negative females (Figure 4B). The enriched GO terms of the DEGs included a response to virus infection, iron-binding, amino acid metabolism and transport, and insect metamorphosis and mating process (FDR<0.05; Figure 4C). Similarly, the enriched KEGG pathways of the DEGs included the lysosome, ECM-receptor interaction, amino acids biosynthesis and metabolism, and longevity regulating pathways (FDR<0.05; Figure 4D). Moreover, all the DEGs grouped into these KEGG pathways were upregulated in AINV positive females except DEG118270764 (Figure 6).
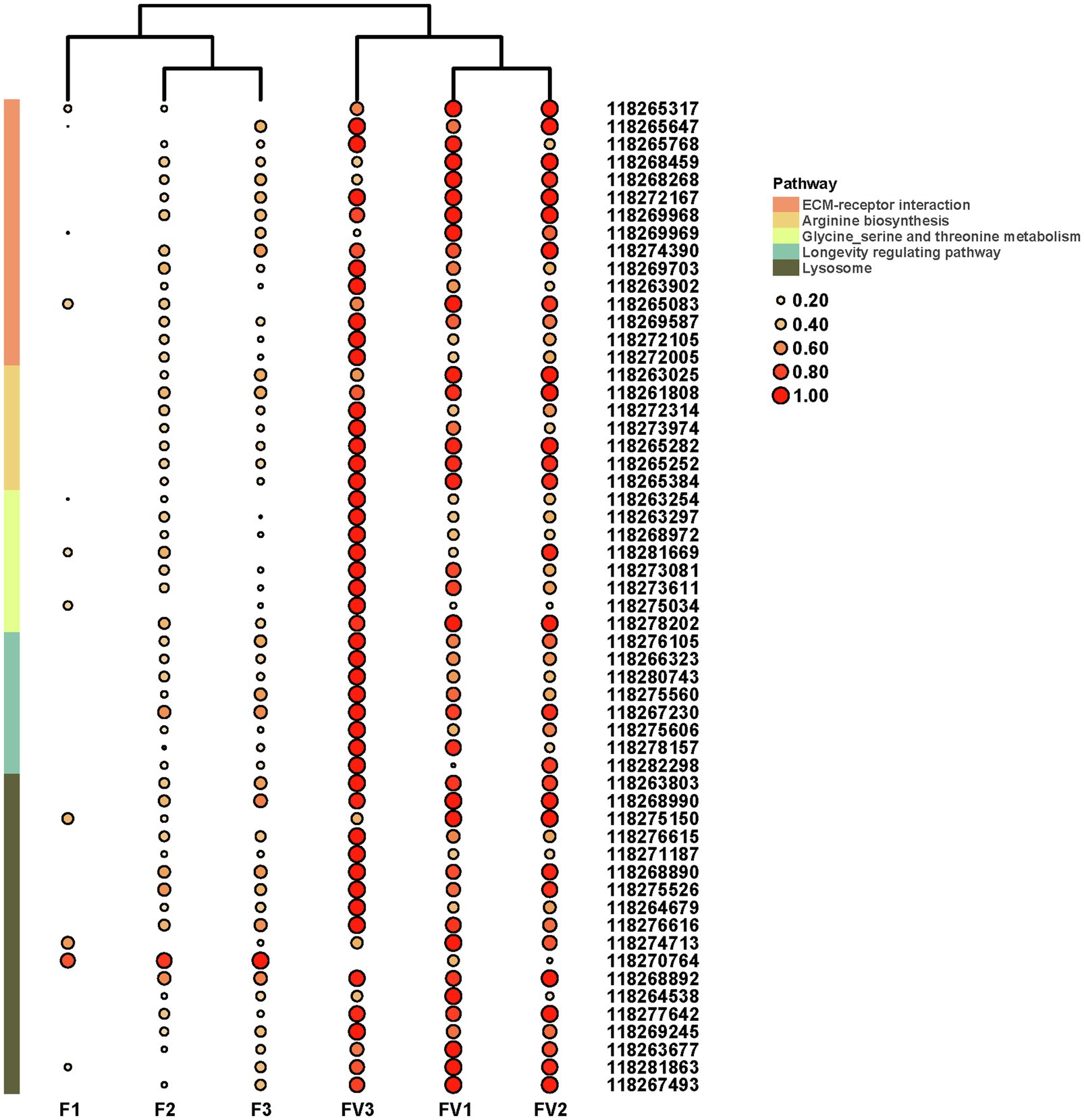
Figure 6. The expression patterns of DEGs related to the significantly enriched pathways. F1-3, AINV negative S. frugiperda females; FV1-3, AINV positive S. frugiperda females. Log2 FPKM values are shown by the color and area of the circles. Log2 FPKM values are further scaled by row with zero to one method, and then shown by the color and area of the circles.
The Nora virus is a single-stranded, positive-sense RNA virus that was first reported in two Drosophila species: D.melanogaster and Drosophila simulans (Habayeb et al., 2006). Thereafter, metagenomic analyses have revealed novel Nora-like viruses in Drosophila (van Mierlo et al., 2014; Medd et al., 2018), Nasonia (Oliveira et al., 2010), honey bee (Remnant et al., 2017), and diverse environmental samples (Shi et al., 2016). Recently, Nora-like virus infections were also detected in two lepidopteran pests: S. exigua and H. armigera (Yang et al., 2019). Herein, AINV clustered with the H. armigera Nora virus with robust supports and grouped into the Nora virus clade suggesting that A. ipsilon is the third lepidopteran natural host of Nora virus. Nora virus shares a conserved picornavirus-like helicase-protease-replicase (H-P-Rep) cassette and has a unique genome organization pattern (Habayeb et al., 2006; Ekstrom et al., 2011). The taxonomic position of the Nora virus is further confirmed by the recent structure analyses of its capsid proteins (Laurinmaki et al., 2020). The virus has a relatively large RNA genome, comprising more than 11,000 nucleotides in length (Habayeb et al., 2006; Laurinmaki et al., 2020). AINV has 11,312 nucleotides and which include five ORFs. The ORF2 of AINV encodes the picornavirus-like replicative cassette, while ORF4 and ORF5 encode the three structural proteins. The AINV ORFs have short overlaps, indicating that ribosomal frameshifting maybe involved in the expression of its proteins (Dreher and Miller, 2006).
Nora virus can cause persistent infections in Drosophila, and these infections are not affected by the Drosophila RNAi pathways (Habayeb et al., 2009b). The Drosophila Nora virus is horizontally transmitted through the fecal-oral route, causing mild effects on the fitness of infected flies (Habayeb et al., 2009a). A recent study postulated that geotaxis dysfunction is a phenotypic hallmark of Drosophila Nora virus infection, which causes the infected flies to significantly lose their climbing ability (Rogers et al., 2020). Notably, the Nora virus is horizontally and vertically transmitted within H. armigera populations, with unclear pathogenic effects on the infected moths (Yang et al., 2019). Similarly, AINV stably infects the A. ipsilon population, with no clear negative effects on the fitness of A. ipsilon (data not shown). As such, AINV is probably a mutualistic virus in A. ipsilon. Novel partiti-like viruses can horizontally transfer among congener lepidopteran hosts and subsequently cause deleterious effects on the new hosts (Xu et al., 2020). These findings strongly suggest that the novel viruses are potential biological resources for pest management. S. frugiperda is a destructive crop pest discovered in China in 2018 (Sun et al., 2021). Herein, AINV was injected into S. frugiperda and established stable infections, indicating that AINV can horizontally transfer among the Noctuidae hosts. Our previous study indicates that relative to the oral transmitted pathway, microinjection has its own advantage in the horizontal transmission of novel insect viruses (Xu et al., 2020). Hence, in this study, we establish the AINV infection S. frugiperda population by microinjection. However, we also notice that Nora virus can horizontally transmitted in its original host with perfect transmitted rate, by sharing the virus contamination food (Yang et al., 2019). It implies that AINV is probably can also be transmitted among A. ipsilon individuals through food-borne transmission route. AINV titers in A. ipsilon and S. frugiperda varied among its host developmental stages, with the larvae having the highest titer. AINV infection also had some side effects on the fitness of its new host, S. frugiperda. It significantly extended the S. frugiperda larval period but significantly reduced its moth eclosion rate. Moreover, after AINV infection, S. frugiperda pupal weight loss, it indicates that AINV and S. frugiperda lack a long co-evolutionary history which causes AINV to be a parasitic virus of S. frugiperda.
Previous studies postulate that the Drosophila Nora virus structural protein has RNAi suppressive activity in its natural host (van Mierlo et al., 2012, 2014). Herein, RNA-seq analysis revealed a higher expression of Argonaute-2 gene (AGO2) in AINV positive S. frugiperda females than the negative ones (Fold change=1.47, FDR>0.05), indicating that AINV stimulates the RNAi antiviral defense in the new host. Nora virus infection upregulates immune genes in Drosophila (Cordes et al., 2013; Lopez et al., 2018). GO and KEGG enrichments herein revealed no immune-related pathway in response to AINV infection. However, some potential antiviral pathways, such as ko04142 (lysosome), were uncovered (Du and Jin, 2017). Notably, ko04512 (glycine, serine, and threonine metabolism; Ding et al., 2019) and ko00260 (ECM-receptor interaction; Guo et al., 2015) have been reported to respond to animal virus infections. Herein, the DEGs are mainly enriched in the amino acid-related pathways, indicating that AINV infection affects the amino acid metabolism in S. frugiperda.
This study reports a new Nora virus infecting A. ipsilon and provisionally names it AINV. Its genome has 11,312 nucleotides, which include five ORFs. AINV was successfully transmitted into a novel host, S. frugiperda, through injection, causing stable infection, suggesting horizontal AINV transmission among moths of the same taxonomic family. Furthermore, the fitness measurements and RNA-seq analysis showed that the AINV infection was deleterious to S. frugiperda and mainly mediated by antiviral and amino acid metabolism-related pathways.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at https://www.ncbi.nlm.nih.gov/, PRJNA742481 and PRJNA76097.
LT: formal analysis, investigation, data curation, and writing—original draft. GR: formal analysis and writing—original draft. WY: resources. CS: investigation. YG: supervision. MX: project administration. LH: conceptualization, formal analysis, data curation, and writing—review and editing. All authors read and approved the final manuscript.
This work was supported by the Fund for Distinguished Young Scholars from the Henan Academy of Agricultural Sciences (grant no. 2020JQ05) and the National Natural Science Foundation of China (31772520; 31702057; and 31601897).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fmicb.2021.727202/full#supplementary-material
Supplementary Figure S1 | The sequence alignment of unigene-23909 and unigene-23910.
Supplementary Figure S2 | Maximum likelihood phylogeny of Picornavirales and Caliciviridae with RdRp protein sequences. The Nora virus clade was highlighted. Numbers near nodes indicate the ultrafast bootstrap values. The bar indicates the estimated number of substitutions per site.
Supplementary Figure S3 | Electron microscopy of purified AINV particles.
1. ^https://www.ncbi.nlm.nih.gov/orffinder/
2. ^https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
3. ^http://ccb.jhu.edu/software/stringtie/gffcompare.shtml
4. ^http://www.bioconductor.org/packages/release/bioc/html/ballgown.html
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). Trimal: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cholleti, H., Hayer, J., Fafetine, J., Berg, M., and Blomstrom, A. L. (2018). Genetic characterization of a novel picorna-like virus in Culex spp. mosquitoes from Mozambique. Virol. J. 15, 1–10. doi: 10.1186/s12985-018-0981-z
Cordes, E. J., Licking-Murray, K. D., and Carlson, K. A. (2013). Differential gene expression related to Nora virus infection of Drosophila melanogaster. Virus Res. 175, 95–100. doi: 10.1016/j.virusres.2013.03.021
de Miranda, J. R., Dainat, B., Locke, B., Cordoni, G., Berthoud, H., Gauthier, L., et al. (2010). Genetic characterization of slow bee paralysis virus of the honeybee (Apis mellifera L.). J. Gen. Virol. 91, 2524–2530. doi: 10.1099/vir.0.022434-0
Ding, F., Cheng, J., Fu, Y., Chen, T., Li, B., Jiang, D., et al. (2019). Early transcriptional response to DNA virus infection in Sclerotinia sclerotiorum. Viruses 11:278. doi: 10.3390/v11030278
Dreher, T. W., and Miller, W. A. (2006). Translational control in positive strand RNA plant viruses. Virology 344, 185–197. doi: 10.1016/j.virol.2005.09.031
Du, Z. Q., and Jin, Y. H. (2017). Comparative transcriptome and potential antiviral signaling pathways analysis of the gills in the red swamp crayfish, Procambarus clarkii infected with white spot syndrome virus (WSSV). Genet. Mol. Biol. 40, 168–180. doi: 10.1590/1678-4685-gmb-2016-0133
Ekstrom, J. O., Habayeb, M. S., Srivastava, V., Kieselbach, T., Wingsle, G., and Hultmark, D. (2011). Drosophila Nora virus capsid proteins differ from those of other picorna-like viruses. Virus Res. 160, 51–58. doi: 10.1016/j.virusres.2011.05.006
Guo, R., Wang, S., Xue, R., Cao, G., Hu, X., Huang, M., et al. (2015). The gene expression profile of resistant and susceptible Bombyx mori strains reveals cypovirus-associated variations in host gene transcript levels. Appl. Microbiol. Biotechnol. 99, 5175–5187. doi: 10.1007/s00253-015-6634-x
Habayeb, M. S., Cantera, R., Casanova, G., Ekström, J.-O., Albright, S., and Hultmark, D. (2009a). The drosophila Nora virus is an enteric virus, transmitted via feces. J. Invertebr. Pathol. 101, 29–33. doi: 10.1016/j.jip.2009.02.003
Habayeb, M. S., Ekengren, S. K., and Hultmark, D. (2006). Nora virus, a persistent virus in drosophila, defines a new picorna-like virus family. J. Gen. Virol. 87, 3045–3051. doi: 10.1099/vir.0.81997-0
Habayeb, M. S., Ekström, J. O., and Hultmark, D. (2009b). Nora virus persistent infections are not affected by the RNAi machinery. PLoS One 4:e5731. doi: 10.1371/journal.pone.0005731
Jakubowska, A. K., D'Angiolo, M., Gonzalez-Martinez, R. M., Millan-Leiva, A., Carballo, A., Murillo, R., et al. (2014). Simultaneous occurrence of covert infections with small RNA viruses in the lepidopteran Spodoptera exigua. J. Invertebr. Pathol. 121, 56–63. doi: 10.1016/j.jip.2014.06.009
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Laurinmaki, P., Shakeel, S., Ekstrom, J. O., Mohammadi, P., Hultmark, D., and Butcher, S. J. (2020). Structure of Nora virus at 2.7 A resolution and implications for receptor binding, capsid stability and taxonomy. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-76613-1
Le Gall, O., Christian, P., Fauquet, C. M., King, A. M., Knowles, N. J., Nakashima, N., et al. (2008). Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch. Virol. 153, 715–727. doi: 10.1007/s00705-008-0041-x
Liu, S. J., Chen, Y. T., and Bonning, B. C. (2017). RNA virus discovery in insects. Curr. Opin. Insect. Sci. 8, 54–61. doi: 10.1016/j.cois.2014.12.005
Liu, W. Z., Xie, Y. B., Ma, J. Y., Luo, X. T., Nie, P., Zuo, Z. X., et al. (2015). IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361. doi: 10.1093/bioinformatics/btv362
Lopez, W., Page, A. M., Carlson, D. J., Ericson, B. L., Cserhati, M. F., Guda, C., et al. (2018). Analysis of immune-related genes during Nora virus infection of Drosophila melanogaster using next generation sequencing. AIMS. Microbiol. 4, 123–139. doi: 10.3934/microbiol.2018.1.123
Medd, N. C., Fellous, S., Waldron, F. M., Xuereb, A., Nakai, M., Cross, J. V., et al. (2018). The virome of Drosophila suzukii, an invasive pest of soft fruit. Virus. Evol. 4:vey009. doi: 10.1093/ve/vey009
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Oliveira, D. C., Hunter, W. B., Ng, J., Desjardins, C. A., Dang, P. M., and Werren, J. H. (2010). Data mining cDNAs reveals three new single stranded RNA viruses in Nasonia (hymenoptera: Pteromalidae). Insect Mol. Biol. 19(Suppl 1), 99–107. doi: 10.1111/j.1365-2583.2009.00934.x
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Remnant, E. J., Shi, M., Buchmann, G., Blacquiere, T., Holmes, E. C., Beekman, M., et al. (2017). A diverse range of novel RNA viruses in geographically distinct honey bee populations. J. Virol. 91, e00158–e00117. doi: 10.1128/JVI.00158-17
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rogers, A., Towery, L., McCown, A., and Carlson, K. A. (2020). Impaired geotaxis as a novel phenotype of Nora virus infection of Drosophila melanogaster. Scientifica 2020:6. doi: 10.1155/2020/1804510
Shi, M., Lin, X. D., Chen, X., Tian, J. H., Chen, L. J., Li, K., et al. (2018). The evolutionary history of vertebrate RNA viruses. Nature 556, 197–202. doi: 10.1038/s41586-018-0012-7
Shi, M., Lin, X. D., Tian, J. H., Chen, L. J., Chen, X., Li, C. X., et al. (2016). Redefining the invertebrate RNA virosphere. Nature 540, 539–543. doi: 10.1038/nature20167
Shi, W., Xie, S., Chen, X., Sun, S., Zhou, X., Liu, L., et al. (2013). Comparative genomic analysis of the microbiome [corrected] of herbivorous insects reveals eco-environmental adaptations: biotechnology applications. PLoS Genet. 9:e1003131. doi: 10.1371/annotation/91a25db3-8127-42c7-baa0-ce398a2857a6
Sun, X. X., Hu, C. X., Jia, H. R., Wu, Q. L., Shen, X. J., Zhao, S. Y., et al. (2021). Case study on the first immigration of fall armyworm, Spodoptera frugiperda invading into China. J. Integr. Agr. 20, 664–672. doi: 10.1016/S2095-3119(19)62839-X
van Mierlo, J. T., Bronkhorst, A. W., Overheul, G. J., Sadanandan, S. A., Ekstrom, J. O., Heestermans, M., et al. (2012). Convergent evolution of argonaute-2 slicer antagonism in two distinct insect RNA viruses. PLoS Path. 8:e1002872. doi: 10.1371/journal.ppat.1002872
van Mierlo, J. T., Overheul, G. J., Obadia, B., van Cleef, K. W., Webster, C. L., Saleh, M. C., et al. (2014). Novel drosophila viruses encode host-specific suppressors of RNAi. PLoS Path. 10:e1004256. doi: 10.1371/journal.ppat.1004256
Xie, Y., Wu, G., Tang, J., Luo, R., Patterson, J., Liu, S., et al. (2014). SOAPdenovo-trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30, 1660–1666. doi: 10.1093/bioinformatics/btu077
Xu, P., Liu, Y., Graham, R. I., Wilson, K., and Wu, K. (2014). Densovirus is a mutualistic symbiont of a global crop pest (Helicoverpa armigera) and protects against a baculovirus and Bt biopesticide. PLoS Path. 10:e1004490. doi: 10.1371/journal.ppat.1004490
Xu, P., Yang, L., Yang, X., Li, T., Graham, R. I., Wu, K., et al. (2020). Novel partiti-like viruses are conditional mutualistic symbionts in their normal lepidopteran host, African armyworm, but parasitic in a novel host Fall armyworm. PLoS Path. 16:e1008467. doi: 10.1371/journal.ppat.1008467
Yang, Y. T., Nai, Y. S., Lee, S. J., Lee, M. R., Kim, S., and Kim, J. S. (2016). A novel picorna-like virus, Riptortus pedestris virus-1 (RiPV-1), found in the bean bug, R. pedestris, after fungal infection. J. Invertebr. Pathol. 141, 57–65. doi: 10.1016/j.jip.2016.11.007
Yang, X., Xu, P., Yuan, H., Graham, R. I., Wilson, K., and Wu, K. (2019). Discovery and characterization of a novel picorna-like RNA virus in the cotton bollworm Helicoverpa armigera. J. Invertebr. Pathol. 160, 1–7. doi: 10.1016/j.jip.2018.11.003
Keywords: Agrotis ipsilon, Spodoptera frugiperda, Nora virus, virus genome, fitness
Citation: Li T, Guan R, Wu Y, Chen S, Yuan G, Miao X and Li H (2021) The Novel Agrotis ipsilon Nora Virus Confers Deleterious Effects to the Fitness of Spodoptera frugiperda (Lepidoptera: Noctuidae). Front. Microbiol. 12:727202. doi: 10.3389/fmicb.2021.727202
Received: 18 June 2021; Accepted: 11 October 2021;
Published: 15 November 2021.
Edited by:
Hongjie Li, Ningbo University, ChinaReviewed by:
Jianglong Guo, Hebei Academy of Agricultural and Forestry Sciences, ChinaCopyright © 2021 Li, Guan, Wu, Chen, Yuan, Miao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuqing Wu, eXVxaW5nd3UzNkBob3RtYWlsLmNvbQ==; Haichao Li, bGloYWljaGFvQGNlbXBzLmFjLmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.