
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 22 September 2021
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.726483
This article is part of the Research TopicWine Microbiology: Current Trends and ApproachesView all 8 articles
Wine production in Cyprus has strong cultural ties with the island’s tradition, influencing local and foreign consumers’ preferences and contributing significantly to Cyprus’ economy. A key contributor to wine quality and sensorial characteristics development is the microbiota that colonizes grapes and performs alcoholic fermentation. Still, the microbial patterns of wines produced in different geographic regions (terroir) in Cyprus remain unknown. The present study investigated the microbial diversity of five terroirs in Cyprus, two from the PGI Lemesos region [Kyperounta (PDO Pitsilia) and Koilani (PDO Krasochoria)], and three from the PGI Pafos region [Kathikas (PDO Laona Akamas), Panayia, and Statos (PDO Panayia)], of two grape varieties, Xynisteri and Maratheftiko, using high-throughput amplicon sequencing. Through a longitudinal analysis, we examined the evolution of the bacterial and fungal diversity during spontaneous alcoholic fermentation. Both varieties were characterized by a progressive reduction in their fungal alpha diversity (Shannon index) throughout the process of fermentation. Additionally, the study revealed a distinct separation among different terroirs in total fungal community composition (beta-diversity) for the variety Xynisteri. Also, Kyperounta terroir had a distinct total fungal beta-diversity from the other terroirs for Maratheftiko. Similarly, a significant distinction was demonstrated in total bacterial diversity between the PGI Lemesos region and the PGI Pafos terroirs for grape juice of the variety Xynisteri. Pre-fermentation, the fungal diversity for Xynisteri and Maratheftiko was dominated by the genera Hanseniaspora, Aureobasidium, Erysiphe, Aspergillus, Stemphylium, Penicillium, Alternaria, Cladosporium, and Mycosphaerella. During and post-fermentation, the species Hanseniaspora nectarophila, Saccharomyces cerevisiae, Hanseniaspora guilliermondii, and Aureobasidium pullulans, became the predominant in most must samples. Regarding the bacterial diversity, Lactobacillus and Streptococcus were the predominant genera for both grape varieties in all stages of fermentation. During fermentation, an increase was observed in the relative abundance of some bacteria, such as Acetobacter, Gluconobacter, and Oenococcus oeni. Finally, the study revealed microbial biomarkers with statistically significant higher relative representation, associated with each geographic region and each grape variety, during the different stages of fermentation. The present study’s findings provide an additional linkage between the grape microbial community and the wine terroir.
Cyprus has been a wine-producing country for centuries, and nowadays, the local wine industry is ranked among the 50 greatest wine producers worldwide (Wine Production (ton), 2015) with an annual production of 10 ml, more than 100 small and four big wineries exist in the island, located in hillside villages. Additionally, there are about 15 indigenous grape varieties, from which Xynisteri, Mavro, Maratheftiko, and Ofthalmo are the most extensively cultivated. The majority of the Cyprus wines are produced by native varieties, contributing to their unique and distinct aroma and flavors. The “terroir approach” is now more evident, with “Single Vineyard” wines released by the producers.
The exclusive sensorial characteristics of wines are influenced by the contribution of regional environmental factors, including topography, climatic conditions, soil, different varieties, and microbial patterns, generally known as “terroir” (Bokulich et al., 2013; Anagnostopoulos et al., 2019). Differences in topoclimate, soil, and vines distinguish a plethora of terroirs in Cyprus, providing a specific character in different terroir’ produced wines (Kokkinofta et al., 2017). The characterization of Cyprus wines’ typicity is essential for defining protected designation of origin (PDO) and protected geographical indication (PGI) wines’ designation. The European Union (EU) has established the PDO and PGI labels to safeguard local products authenticity. In Cyprus, there are five PDO recognized regions: (i) Commandaria, (ii) Krasochoria Lemesou, and (iii) Pitsilia, located in the Lemesos PGI region, (iv) Laona Akamas, and (v) Vouni Panayia-Ampelitis, located in the Paphos PGI region (Figure 1). These labels are critical contributors to consumers’ preferences and the economic appreciation of local wines. The winemakers and the market need to identify and maintain the microbial community that influence terroir and affect wines’ sensorial characteristics and quality. This is especially important for spontaneously fermented wines, in which the native microbiota contributes to the wines’ unique flavors and aromas (Ballantyne et al., 2019).
A key contributor to wine terroir is the grape microbiota. Specific members of the regional microbial communities guide alcoholic fermentation (AF) producing metabolites that affect the wines’ sensorial characteristics and quality. The process of fermentation is dynamic, leading to modifications in microbial diversity (Wang et al., 2021). Traditionally, the spontaneous fermentation process has been applied in winemaking, according to which no extra microbes or chemical compounds are added pre-fermentation. Under these conditions, the wine microbiota is considered unpredictable. Nowadays, apart from the native microbiota, selected species are added as starter cultures to control the processes of AF, such as Saccharomyces cerevisiae, and of malolactic fermentation (MLF), such as Oenococcus oeni and Lactobacillus plantarum (De Benedictis et al., 2011; Berbegal et al., 2016, 2017; Tristezza et al., 2016; Petruzzi et al., 2017). More recently, mixtures of yeasts (S. cerevisiae and non-Saccharomyces) are commercially available, introducing a controlled additional aromatic diversity to the final products (Jolly et al., 2014; Lleixà et al., 2016). The application of the whole genome sequencing methodology assisted in the selection of possible starters, allowing the identification of the total genome sequences of several S. cerevisiae strains (Goffeau et al., 1996; Birkeland et al., 2010; Otero et al., 2010; Akao et al., 2011; Borneman et al., 2016), other Saccharomyces species (Naseeb et al., 2018), non-Saccharomyces yeasts, such as Hanseniaspora guilliermondii (Seixas et al., 2019); Hanseniaspora vineae (Giorello et al., 2014); Torulaspora delbrueckii (Tondini et al., 2018), and malolactic bacteria, such as O. oeni (Mills et al., 2005). As a result, genes associated with metabolic traits that may influence grapes and wine quality have been revealed.
Over the last decade, several High Throughput sequencing (HTS) studies have been conducted to identify the microbial communities developed in wine and to reveal the contribution of the microbial communities in wines taste and aroma (Bokulich et al., 2012, 2016; Knight et al., 2015; Piao et al., 2015; Pinto et al., 2015; Belda et al., 2016). The studies indicated that pre-fermentation factors associated with the regional terroir shape the microbial diversity of wine grapes. During AF, regional fungi, including S. cerevisiae, metabolize sugars producing alcohol and secondary metabolites that contribute to wine’s sensorial characteristics (Swiegers et al., 2005; Lambrechts and Pretorius, 2019). In addition to AF, native bacteria, such as Oenococcus, Lactobacillus, Pediococcus, and Leuconostoc, convert malic acid to lactic acid, influencing wines’ aromas and tastes (Renouf et al., 2005). Post-fermentations, the microbial diversity is formed by species detected in low relative abundances pre-fermentation but can survive this stressful microenvironment. The regional distinction based on the evolution of the microbiological patterns, as revealed through these HTS, increased our understanding regarding the spatiotemporal contribution of the grapes’ microbiome to wines’ typicity.
The determination of the native grape microbial diversity throughout AF unfolds a new horizon for identifying Cyprus wines’ authenticity. Kokkinofta et al. (2017) proved significant discrimination of different grape varieties and produced in different PDOs Cyprus wines based on regional isotopes and elements, using inductively coupled plasma atomic emission spectroscopy (ICP-AES). Our study is the first to discriminate grape wines produced: (i) in diverse PDO regions; and (ii) from the varieties Xynisteri and Maratheftiko, throughout the fermentation process, based on the distinct microbiological patterns.
Wine grapes from the varieties Xynisteri (white) and Maratheftiko (black) were harvested from August until October 2019 from two terroirs in Kyperounta village (PDO Pitsilia) and Koilani village (PDO Krasochoria), both located in Lemesos PGI region, and three terroirs in Kathikas (PDO Laona Akamas), Panayia and Statos (PDO Panayia), located in Paphos PGI region (Figure 1). Among the five terroirs, Kyperounta is at the highest altitude, in 1,134 m elevation above the sea level, followed by Statos in 913 m, Panayia in 900 m, Koilani in 820 m, and finally Kathikas in 655 m. Noteworthy, Panayia and Statos are neighboring villages. From each terroir, four samples were collected, two from one vineyard and another two from a neighbor vineyard. The weight of the samples was about 5 kg grapes/vineyard, and about 8–12 bunches were used. Grapes were fully ripe at 22–23 Brix. Information about samples’ sugar concentration, yeast assimilable nitrogen, pH and total acidity are shown in Supplementary Table 1. Both vineyards were in a radius of less than 1 km apart, showing similar soil and environmental conditions. All vineyards are bush vines, and goblet trained more than 40 years old, with no specific row orientation. Also, the topoclimate between the area samples is the same. The two neighboring vineyards had the same training, microclimate, soil, and crop management (conventional). Some potential differences in crop management, such as spraying, may be attributed to different owners’ practices. Generally, treatments follow very similar patterns as growers follow common local expert advice. Samples were placed in sterile plastic bags and transported to Kyperounta Winery for processing.
Whole bunches of grapes were aseptically crushed without any destemming and left for 24 h at 18°C. A 50 ml “pre-fermentation” sample was taken and stored at −80°C until processing. Must was separated from the skins and stems on the second day of fermentation and kept at 18°C for the whole course of the fermentation, for both the varieties. The process of spontaneous fermentation was conducted without sulfites addition. The fermentation course was monitored by daily density measurement with a DMA 35 portable density meter. “During fermentation” sampling was performed when the alcohol production was 4% ABV. The sampling at this stage indicates the exponential phase of fermentation when the yeast population reached its maximum growth (Olivares-Marin et al., 2018). “Post fermentation” sample was taken after the alcohol concentration was higher than 10%, and when for three consecutive days the density readings were stable.
All samples were collected in a sterile 50 ml container and immediately were frozen with dry ice and stored at −80°C until processing.
Juice and must samples were centrifuged at 14,000 g for 30 min at 4°C and washed twice in 2 ml and 300 μl TE buffer, respectively. The DNA pellet was suspended in 450 μl DNeasy® PowerFood® Solution MBL supplemented with 40 mg/ml lysozyme and incubated at 37°C for 30 min, followed by incubation at 65°C for 10 min and 95°C for 10 min. Then the DNA isolation was performed according to DNeasy® PowerFood® Microbial Kit (MoBio Laboratories Inc., Carlsbad, CA, United States) manufacturer’s instructions. The extracted DNA was stored at −20°C until processing. If the concentration of the extracted DNA was lower than 5 ng/μl, or the 260/230 ratio was lower than 1.9, indicating the presence of contaminants, then the process of DNA extraction was repeated with the addition of a small concentration of polivinilpirrolidone and 20 μl of β−mercaptoethanol followed by 1 h incubation at 60°C, before the addition of the lysis buffer. The addition of polivinilpirrolidone removes tannins and polyphenols, whereas β−mercaptoethanol of proteins from the extract.
The total DNA isolated from the juice and must samples was quantified fluorometrically with Qubit 4.0 fluorometer (Invitrogen, Carlsbad, CA) using Qubit dsDNA HS Assay Kit (Invitrogen). The purity of the DNA was evaluated by measuring the ratio of absorbance A260/280 nm and A260/230 nm using a spectrophotometer (NanoDrop Thermo Scientific, United States).
The 16S rRNA bacterial gene amplification, the Illumina paired-completion library preparation and sequencing was performed as described previously (Kamilari et al., 2020b). The 16S rRNA bacterial gene was amplified using the primers V3: 50-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-30 and V4: 50-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-30, whereas the fungal internal transcribed spacer 1 (ITS1) loci using the primers BITS (5′-NNNNNNNNCTACCTGC GGARGGATCA-3′) and B58S3 (5′-GAGATCCRTTGYTRAAA GTT-3′) with the addition of the overhang adapter sequence, as described by Bokulich et al. (2016). For fungal ITS1 loci amplification and sequencing the “Fungal Metagenomic Sequencing Demonstrated Protocol” provided by Illumina was applied.1 The PCR reaction mixture for each reaction was: 2.5 μL template DNA (5 ng/μL); 5 μL of each forward and reverse primers (1 μM) and 12.5 μL 2 × KAPA HiFi HotStart Ready Mix (KAPA Biosystems, United States). PCR amplification was performed in PCR Thermocycler (Bio-Rad, United States) using the following procedure: (a) Denaturation: 95°C for 3 min; (b) Denaturation: 95°C for 30 s; (c) Annealing: 55°C for 30 s; (d) Elongation: 72°C for 30 s; (e) repeat of steps b–d for 25 cycles; (f) Extension: 72°C for 5 min; (g) Hold at 4°C. PCR amplicons purification, evaluation of DNA quantity and quality and amplicons normalization was conducted as describe previously (Papademas et al., 2020). For the sequencing run, both bacterial 16S rRNA gene and fungal ITS1 loci were loaded on MiSeq 600 cycle Reagent Kit v3 (Illumina, United States) (5% PhiX) and run on a MiSeq Illumina sequencing platform.
Raw fastq sequences were quality filtered, and the diversity indexes Shannon, Simpson, and Chao1 regarding alpha diversity and Bray Curtis dissimilarity regarding beta diversity, were performed using Qiime 2 version 2020.2 (Bolyen et al., 2019), as previously described (Kamilari et al., 2020a). Before diversity analyses, samples were rarefied (Supplementary Table 9). Comparisons of alpha and beta diversity indexes were evaluated separately for the two wine grapes varieties, Xynisteri and Maratheftiko, and for the three stages of the fermentation, pre-, during-, and post-fermentation. To visualize the separation of the bacterial and fungal communities into clusters based on Bray–Curtis dissimilarity distances, the Principal Coordinate Analysis (PCoA) was performed using q2−diversity after samples were rarefied as previously described (Kamilari et al., 2021). To estimate whether sample categories (i) Xynisteri and Maratheftiko; (ii) pre-, during-, and post-fermentation) had statistical differences in their beta microbial diversity, non-parametric permutational analysis of variance (PERMANOVA) (Anderson, 2008) with 999 permutations was applied. To evaluate if the sample categories had differences in alpha and beta diversity, rejecting the null hypothesis, Kruskal-Wallis tests were performed. Additionally, to detect differences in alpha diversity among stages of fermentation, analysis of variance (one-way ANOVA) was applied using the SPSS 20 software (StatSoft Inc., Tulsa, OK, United States). The test used was Least Significant Difference (LSD) at the significance level of 0.05. To assign the taxonomy to the 16S rDNA sequences into OTU the q2−feature−classifier (Bokulich et al., 2018) against the Greengenes 13_8 99% OTUs reference sequences (McDonald et al., 2012) was used, whereas, for fungal ITS loci, the UNITE fungal internal transcribed spacer (ITS) database (8.2 release) (Abarenkov et al., 2020) was applied. The sequences were filtered to remove incomplete taxonomies that failed to be identified to the genus level. For biomarkers discovery, the LEfSe algorithm was used (Segata et al., 2011), with Linear discriminant analysis (LDA) scores greater than 2.0. Taxa with relative abundance < 1% per sample were considered not important to be mentioned.
All raw sequence data in read-pairs format were deposited to the National Centre for Biotechnology Information (NCBI) in Sequence Read Archive (SRA) under BioProject PRJNA731461.
The present study investigated the contribution of the microbial diversity composition in the distinction of Cyprus wines regional terroir, as well as of the wine grape varieties Xynisteri and Maratheftiko, by applying the HTS approach.
Wine grape samples collected from Kyperounta village (PDO Pitsilia) and Koilani village (PDO Krasochoria) (PGI Lemessos region), as well as Kathikas, Panayia, and Statos villages (PDO Panayia, PGI Pafos region) were analyzed separately regarding: (i) the varieties: (a) X ynisteri and (b) Maratheftiko; (ii) The stage of must fermentations: (a) pre-; (b) during, and (c) post-fermentation; and (iii) the microbial communities: (a) bacterial and (b) fungal. In total, from 240 samples, nineteen were excluded from the analysis. Specifically, two pre-fermentation samples from Panayia terroir for the variety Xynisteri had a low amount of 16S rDNA during the Library Quantification and Normalization process. The remaining samples were excluded because of a low number of reads passing filter after 16S rDNA library sequencing (eleven samples) and after ITS library sequencing (eight samples).
To evaluate the bacterial diversity of Xynisteri, pre-, during, and post-fermentation, fifty-five examined samples were used as input to the Illumina MiSeq to produce: 2,787,651 high-quality sequencing reads, with an average of 50,684.6 sequencing reads per sample (range = 12,837–405,275, STD = 53,186.7; Supplementary Table 2). High quality sequences were grouped into average number 110.43 OTUs (range = 60–234, SD = 32.8). About the fungal diversity of Xynisteri from fifty-six examined samples 1,445,265 high quality sequencing reads, were produced with an average of 25,808.3 sequencing reads per sample (range = 6,131–430,689, STD = 56,022.2; Supplementary Table 3). High quality sequences were grouped into average number 60.55 OTUs (range = 31–175, SD = 27.7). Results based on the alpha diversity indexes (Shannon, Simpson, and Chao1 estimators) are shown in Supplementary Table 2 for bacterial and Supplementary Table 3 for fungal diversity.
For estimating the bacterial diversity of Maratheftiko, pre-, during, and post-fermentation fifty-one examined samples were sequenced to generate 2,450,489 high-quality sequencing reads, with an average of 48,048.80 sequencing reads per sample (range = 13,475–429,110, STD = 57,464.9; Supplementary Table 4). High-quality sequences were grouped into average number 102.21 OTUs (range = 59–206, SD = 23.8). Regarding the fungal diversity of Maratheftiko from fifty-eight examined samples 1,908,997 high quality sequencing reads, were generated with an average of 32,913.7 sequencing reads per sample (range = 8,586–104,602, STD = 18,964.7; Supplementary Table 5). High quality sequences were grouped into average number 60.93 OTUs (range = 26–146, SD = 26). Results concerning the alpha diversity indexes (Shannon, Simpson, and Chao1 estimators) are shown in Supplementary Table 4 for bacterial and Supplementary Table 5 for fungal diversity.
Initially, we compared the fungal alpha diversity (Shannon index) of wine grapes growing in the five terroirs of Cyprus, during the three different stages of the fermentation, for the two wine grape varieties, Xynisteri and Maratheftiko. According to Figures 2A,B, pre-fermentation Koilani demonstrated the lowest alpha diversity for both wine grapes varieties. The difference was significant for the variety Xynisteri, based on the Kruskal-Wallis test (p = 0.02; Supplementary Table 6). During and post-fermentation, a significant reduction in the alpha diversity was observed in almost all areas for both Xynisteri and Maratheftiko varieties (p < 0.05; Figures 3A,B and Supplementary Table 6). Post-fermentation, for the variety Xinisteri, the alpha diversity of Panayia was significantly higher than Kathikas and Koilani, whereas, for the variety Maratheftiko, of Kathikas, Koilani and Panayia compared to Kyperounta, based on the Kruskal-Wallis test (p = 0.02; Supplementary Table 6).
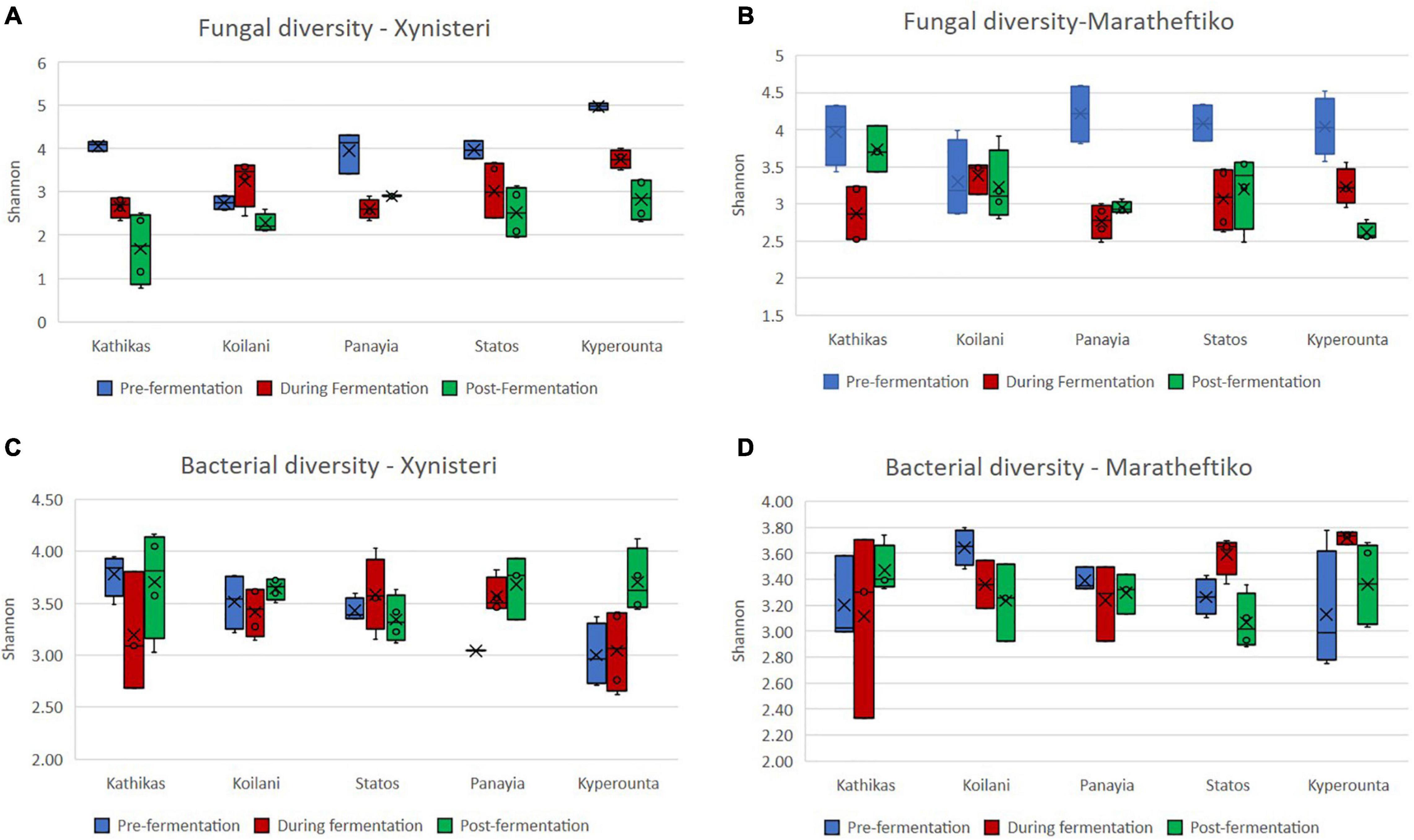
Figure 2. Investigation of the microbial alpha diversity based on the Shannon index in grape juice and must samples for the terroirs Kathikas, Koilani, Kyperounta, Panayia, and Statos during the three stages of fermentation (pre-fermentation, during fermentation, and post-fermentation). (A) Fungal diversity of the variety Xynisteri; (B) fungal diversity of the variety Maratheftiko; (C) bacterial diversity for the variety Xynisteri; (D) bacterial diversity for the variety Maratheftiko. The blue box-plots represent the fungal or bacterial diversity pre-fermentation. The red box-plots represent the fungal or bacterial diversity during fermentation. The green box-plots represent the fungal or bacterial diversity post-fermentation. The o symbols represent the outliers from the median and x symbols the brand means. Statistical analysis was performed using Kruskal-Wallis test.

Figure 3. Comparison of the microbial alpha diversity based on the Shannon index during the three stages of fermentation (pre-fermentation, during fermentation, and post-fermentation). (A) Fungal diversity of the variety Xynisteri; (B) fungal diversity of the variety Maratheftiko; (C) bacterial diversity for the variety Xynisteri; (D) bacterial diversity for the variety Maratheftiko. The o symbol represents the outliers from the median.
Additionally, we compared the bacterial alpha diversity (Shannon index) of the five terroirs, pre-, during, and post-fermentation, for Xynisteri and Maratheftiko (Figures 2C,D). Pre-fermentation, for the variety Xynisteri, significantly higher bacterial alpha diversity was indicated for the areas with the lowest altitude, including Kathikas and Koilani, compared to Kyperounta, the village with the highest altitude, based on the Shannon index, by Kruskal-Wallis test (p = 0.02). Similarly, Koilani showed the most elevated and Kyperounta the lowest bacterial diversity for the wine grape variety Maratheftiko. During and post-fermentation, no significant differences were detected among the five terroirs for both grape varieties and compared to pre-fermentation, based on the Kruskal-Wallis test (Figures 3C,D).
To evaluate the existence of different microbial patterns among wine grapes growing in geographically distant areas of Cyprus with different altitude and weather conditions, we tested the microbial beta diversity of the two wine grapes varieties, Xynisteri and Maratheftiko pre-fermentation. In addition, we investigated the presence of distinct bacterial and fungal patterns during and post-fermentation to identify whether the different areas could be discriminated based on their microbiological composition after the completion of the process. To detect differences, we compared the microbiota structure applying the Bray-Curtis dissimilarity.
Principal Coordinate analysis plot based on Bray-Curtis distance was developed to depict the separation of the individual terroir based on the similarities in the microbial communities. Comparison of the fungal beta diversity for the variety of Xynisteri revealed that pre-fermentation, all the PDO regions were distinct (Figure 4A). PERMANOVA test indicated that the differences were significant (p < 0.05; Supplementary Table 7). Noteworthy, Statos and Panayia that belong to the PDO Panayia didn’t show significant differences. The distinction among the different terroirs remained significant during the rest of the stages of fermentation for almost all terroirs (Figures 4B,C and Supplementary Table 7). Regarding the variety Maratheftiko, the fungal beta-diversity separated Kyperounta, the area found in the higher altitude, from the other terroir, pre-, during, and post-fermentation (Figures 4D–F and Supplementary Table 6). PERMANOVA test confirmed a significant difference between Kyperounta and the other terroir pre-, during, and post-fermentation (p < 0.05). However, the bacterial diversity sufficed to differentiate significantly only the terroir Kyperounta, from the rest terroirs, for the stage pre-fermentation, variety Xynisteri (Figures 5A–F and Supplementary Table S7). No other significant differences were detected among the individual terroirs pre-, during, and post-fermentation for the varieties Xynisteri and Maratheftiko. Furthermore, comparing the Xynisteri and Maratheftiko microbial beta diversity revealed that both bacterial and fungal composition differed significantly during pre- and post-fermentation stages (Supplementary Table 8).
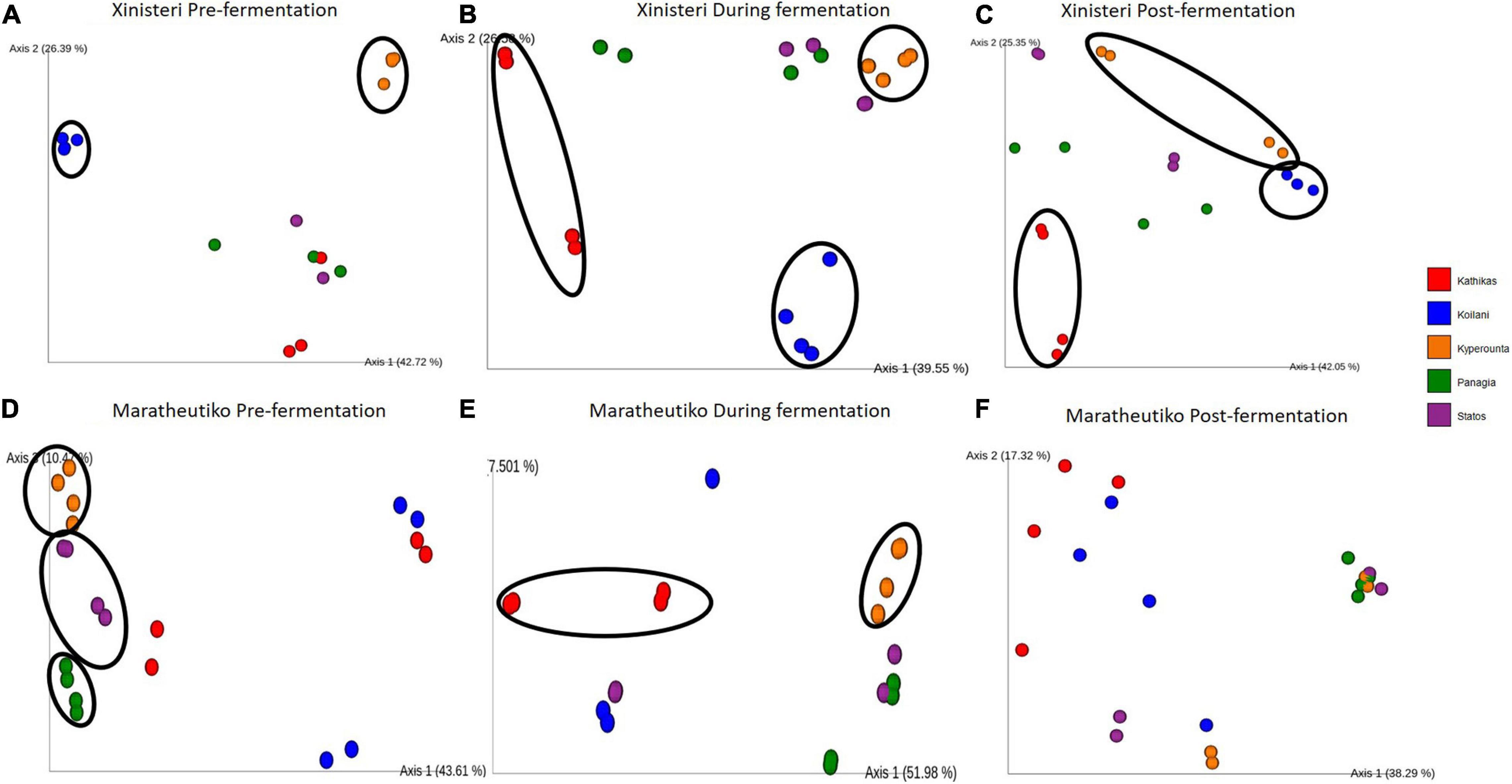
Figure 4. Principal coordinate analysis (PCoA) plot, showing the similarities in the fungal beta diversity according to the Bray-Curtis distance among the individual terroir: (A) for the variety Xynisteri pre-fermentation; (B) for the variety Xynisteri during fermentation; (C) for the variety Xynisteri post-fermentation; (D) for the variety Maratheftiko pre-fermentation; (E) for the variety Maratheftiko during fermentation; (F) for the variety Maratheftiko post-fermentation. Red circles represent Kathikas, blue circles represent Koilani, yellow circles represent Kyperounta, green circles represent Panayia and purple circles represent Statos. Ellipses show the separated terroir.
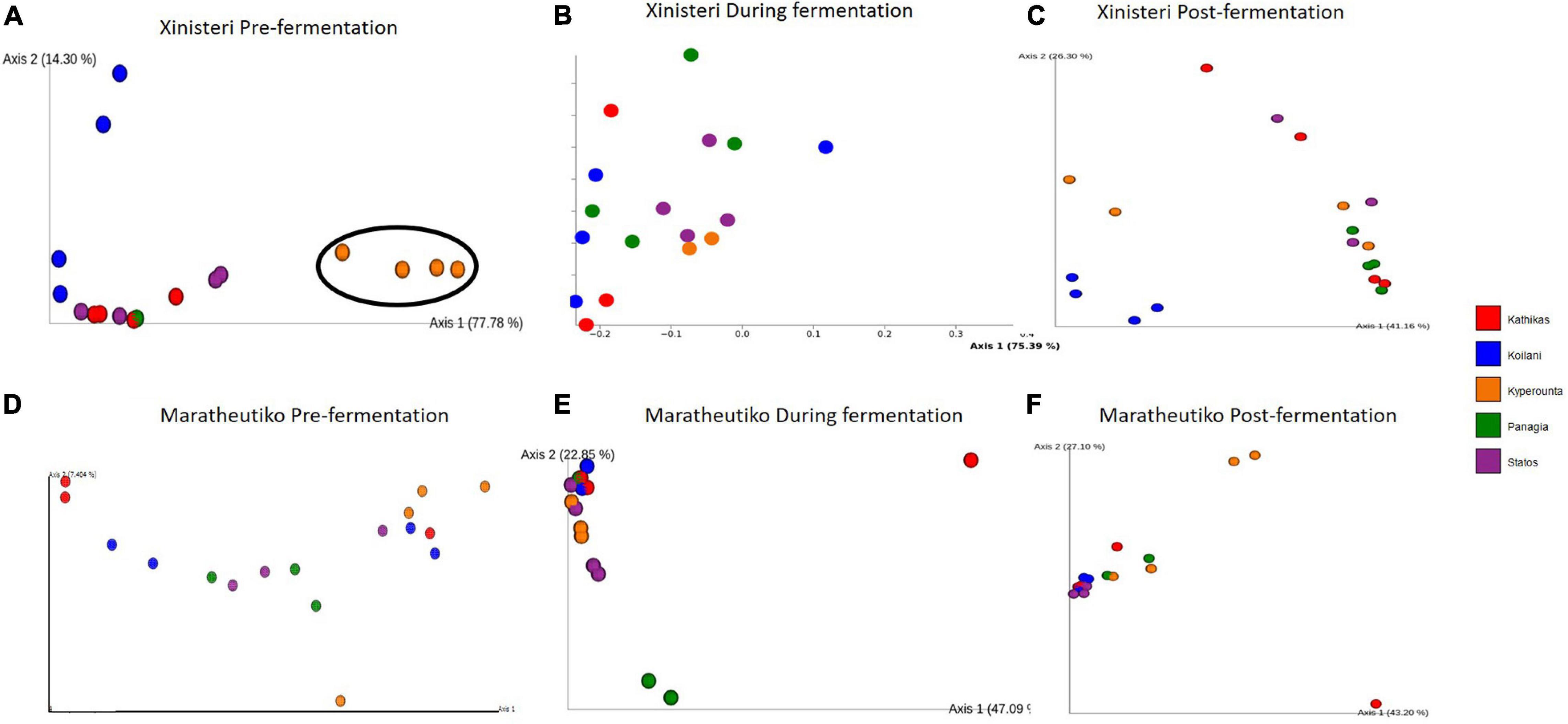
Figure 5. Principal coordinate analysis plot, showing the similarities in the bacterial beta diversity according to the Bray-Curtis distance among the individual terroir: (A) for the variety Xynisteri pre-fermentation; (B) for the variety Xynisteri during fermentation; (C) for the variety Xynisteri post-fermentation; (D) for the variety Maratheftiko pre-fermentation; (E) for the variety Maratheftiko during fermentation; (F) for the variety Maratheftiko post-fermentation. Red circles represent Kathikas, blue circles represent Koilani, yellow circles represent Kyperounta, green circles represent Panayia and purple circles represent Statos. Ellipses show the separated terroir.
The fungal communities of the Cyprus terroir for both varieties, Xynisteri and Maratheftiko, from grape juice until the completion of fermentation, according to ITS1 loci sequencing, were characterized mainly by members of the phylum Ascomycota and in lower relative abundances of the phylum Basidiomycota. The most commonly detected species in grape juice belonged to the genera Hanseniaspora, including Hanseniaspora nectarophila and H. guilliermondii, Aureobasidium including Aureobasidium pullulans, Erysiphe including E. necator, Aspergillus, Stemphylium, Penicillium including Penicillium spinulosum, P. bilaiae, and P. canescens, Alternaria including Alternaria alternata and A. metachromatica, Cladosporium including C. tenuissimum and Mycosphaerella including M. tassiana (Figures 6A,D). During fermentation, the genera Hanseniaspora, including the species H. nectarophila and H. guilliermondii and Aureobasidium, including the species A. pullulans, remained the most dominant taxa (Figures 6B,E). Most samples from both varieties demonstrated an increase in the relative abundance of the genus Saccharomyces, for the species S. cerevisiae and Saccharomyces paradoxus. Post-fermentation, H. nectarophila remained the predominant species in most samples, followed by the genus Saccharomyces, with representative species the S. cerevisiae, H. guilliermondii, and A. pullulans (Figures 6C,F). Interestingly, some samples from Kyperounta were characterized by an increase in the relative abundance of the species Schwanniomyces occidentalis, were some from Kathikas, Panayia, and Statos of the species Lachancea thermotolerans. Furthermore, specifically for the terroir from Kathikas, for the variety Maratheftiko, an increase in the relative abundance of Verticillium leptobactrum, Malassezia restricta, Cladosporium sphaerospermum, Meyerozyma guilliermondii, Blumeria graminis, Candida diddensiae, and Debaryomyces prosopidis was observed.
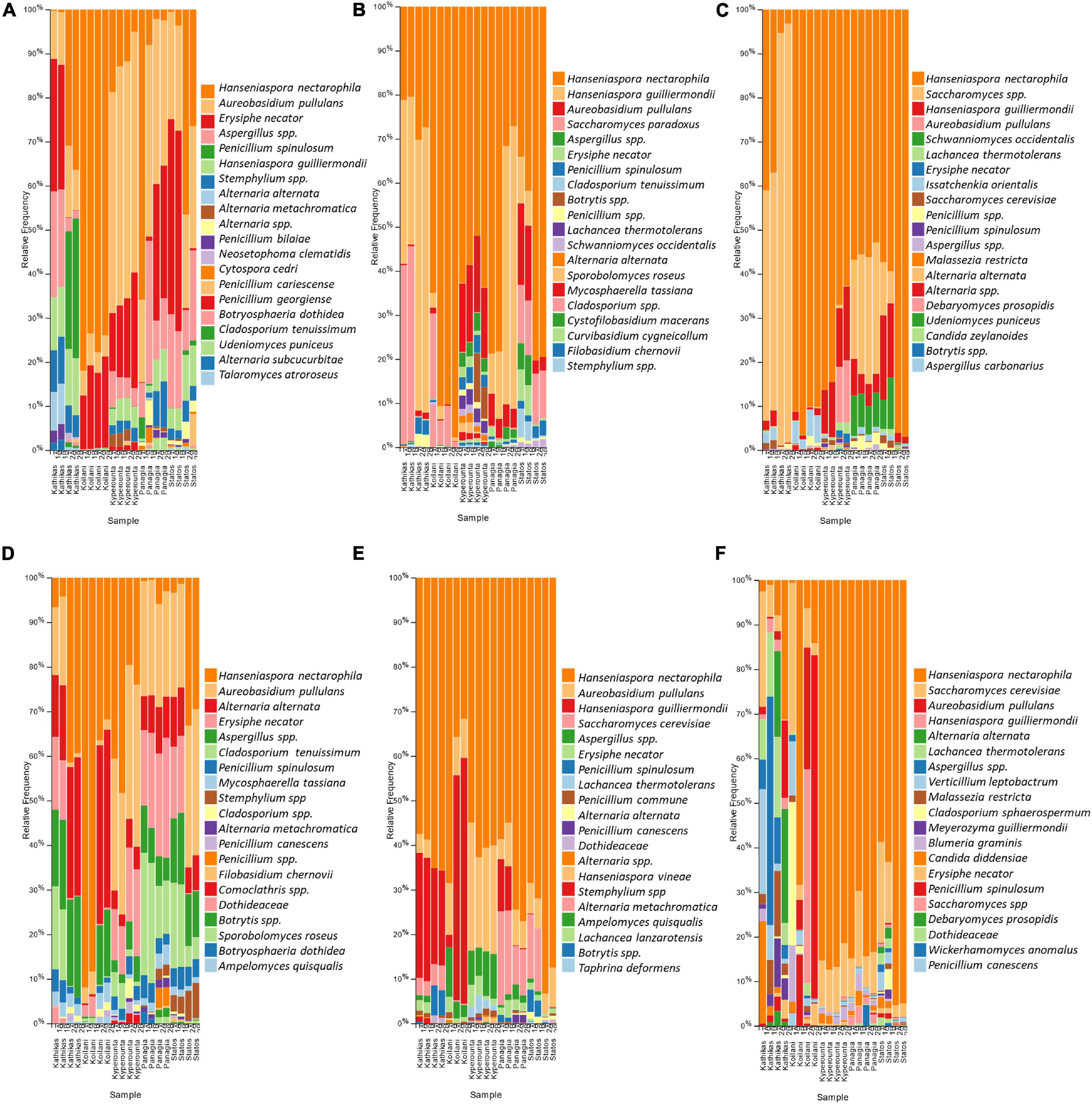
Figure 6. The relative abundance of the twenty most abundant fungi identified at the species level based on ITS loci sequencing for the terroir Kathikas, Koilani, Kyperounta, Panayia, and Statos: (A) for the variety Xynisteri pre-fermentation; (B) for the variety Xynisteri during fermentation; (C) for the variety Xynisteri post-fermentation; (D) for the variety Maratheftiko pre-fermentation; (E) for the variety Maratheftiko during fermentation; (F) for the variety Maratheftiko post-fermentation.
The bacterial communities of the Cyprus terroirs for both varieties, Xynisteri and Maratheftiko, in grape juice, as revealed by 16S rRNA gene sequencing, were mainly represented by members of the phyla Firmicutes followed by Proteobacteria and in lower abundances of Bacteroides, Actinobacteria, and Verrucomicrobia. Lactobacillus species were the most predominant, represented by the species Lactobacillus delbrueckii, followed by Companilactobacillus paralimentarius and Levilactobacillus brevis. Other dominant lactic acid bacteria (LAB) in both grape varieties included the genera Streptococcus and Lactococcus (Figures 7A,D). Additional detected genera, but in lower relative abundances, included Tatumella, represented by the species T. saanichensis, Salinivibrio, represented by the species S. costicola, Staphylococcus, Cloacibacterium, and Rothia, represented by the species R. nasimurium, Mannheimia, and Sphingomonas Pseudomonas, represented by the species P. stutzeri and P. mexicana, Weissella, Methyloversatilis, and Leuconostoc. During fermentation, Lactobacillus, Streptococcus, and Lactococcus were the predominant among the bacterial communities for both varieties (Figures 7B,E). Some samples were characterized by an increase in the relative abundance of the genus Gluconobacter. Also, for Xynisteri, some samples were characterized by an increase in the relative abundance of the species O. oeni, whereas for the variety Maratheftiko of the genus Acetobacter. Post-fermentation, apart from the species L. delbrueckii and the genera Streptococcus and Lactococcus, an increase in the relative abundance on the genus Acetobacter was observed, both for Xynisteri and Maratheftiko (Figures 7C,F). Also, some samples showed a further increase in the relative representation of O. oeni.
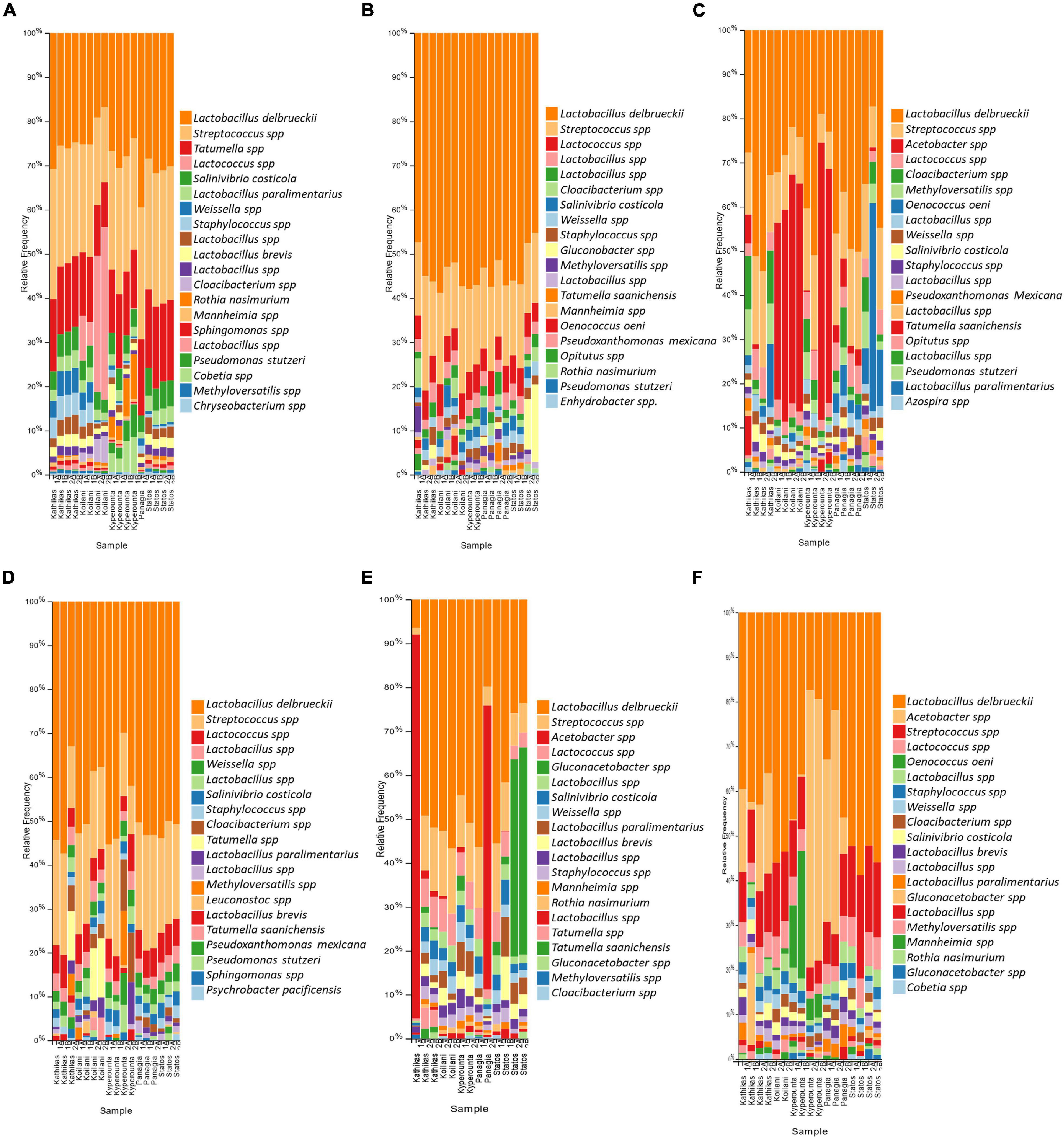
Figure 7. The relative abundance of the twenty most abundant bacteria identified at the species level based on 16S rDNA sequencing for the terroir Kathikas, Koilani, Kyperounta, Panayia, and Statos: (A) for the variety Xynisteri pre-fermentation; (B) for the variety Xynisteri during fermentation; (C) for the variety Xynisteri post-fermentation; (D) for the variety Maratheftiko pre-fermentation; (E) for the variety Maratheftiko during fermentation; (F) for the variety Maratheftiko post-fermentation.
Comparison of the microbial diversity between pre-fermentation samples obtained from the same vineyard indicated differences in the relative representation of microbes. For instance, for the variety Xynisteri, the relative representation of A. pullulans was different between samples from the same vineyard, for both Kathikas (5 and 10%) and Panayia terroir (15 and 29%). Similarly, differences were observed in the relative representation of the genus Aspergillus for Panayia (13 and 22%) and Statos terroir (19 and 39%). Additionally, some differences in the relative representation of microbial species were detected for samples obtained for neighbor vineyards of the same terroir. For example, for both varieties, the relative representation of Penicillium_spinulosum for Kathikas terroir was between 13 and 18% in samples from the one vineyard and about 0–1% for samples from the neighbor vineyard. Similarly, Erysiphe_necator indicated a relative representation of about 23% for samples from the one vineyard of Kathikas terroir and around 0.05% for samples from the neighbor vineyard. Similar results were observed for Erysiphe_necator for Panayia and Statos terroir. Also, for the variety Maratheftiko, for Koilani terroir, the relative representation of Cladosporium_tenuissimum and Alternaria_alternata was about 0.4% for samples obtained from one vineyard and about 11 and 31%, respectively, for samples from the neighbor vineyard.
Comparison of the microbial diversity of post-fermentation samples obtained from the same terroir also indicated differences in the relative microbial representation. Specifically, for the variety Xynisteri in Kathikas terroir, the relative representation of Saccharomyces was about 48% in samples obtained from one vineyard and about 4% in samples obtained from the neighbor vineyard. Similarly, for H. guilliermondii, the relative representation was about 26% in the one vineyard and about 86% in the neighbor vineyard. Moreover, for the variety Maratheftiko in Kyperounta terroir, the relative representation of Lactobacillus delbrueckii was about 15% in samples from the one vineyard, and about 26% from samples from the neighbor vineyard. Additionally, differences were observed between samples obtained from the same vineyard. For instance, the relative representation of A._pullulans and Blumeria_graminis in Koilani terroir for the variety Maratheftiko was 3 and 1%, respectively, in one sample and 30% for both species in the other sample.
The LEfSe algorithm was applied to analyze the identified relative abundances of microbial taxa and determine whether their values were differentially distributed among the five analyzed geographic regions. The analysis was performed from class until species level. The statistical comparisons were executed separately: (i) for the fungal and bacterial diversity; (ii) for the three stages of fermentation: pre- during and post-fermentation; and (iii) for the varieties Xynisteri and Maratheftiko.
Pre-fermentation, for the variety Xynisteri, the Kyperounta terroir indicated significantly a higher relative representation for the genus Penicillium and the species A. pullulans (Figure 8A and Table 1). The genus Stemphylium and the species A. alternata were associated with Kathikas terroir. Also, the species P. spinulosum indicated a higher relative representation in the Paphos PGI region terroirs. For the variety Maratheftiko, in agreement with Xynisteri, the Kyperounta terroir had a significantly higher representation of the species A. pullulans (Figure 8D and Table 1). In addition, Cladosporium tennuisium was associated with Kathikas terroir and Mycosphaerella tassiana and Penicillium canescent with Statos terroir. Finally, Koilani terroir had a significantly higher representation of the species H. nectarophila.
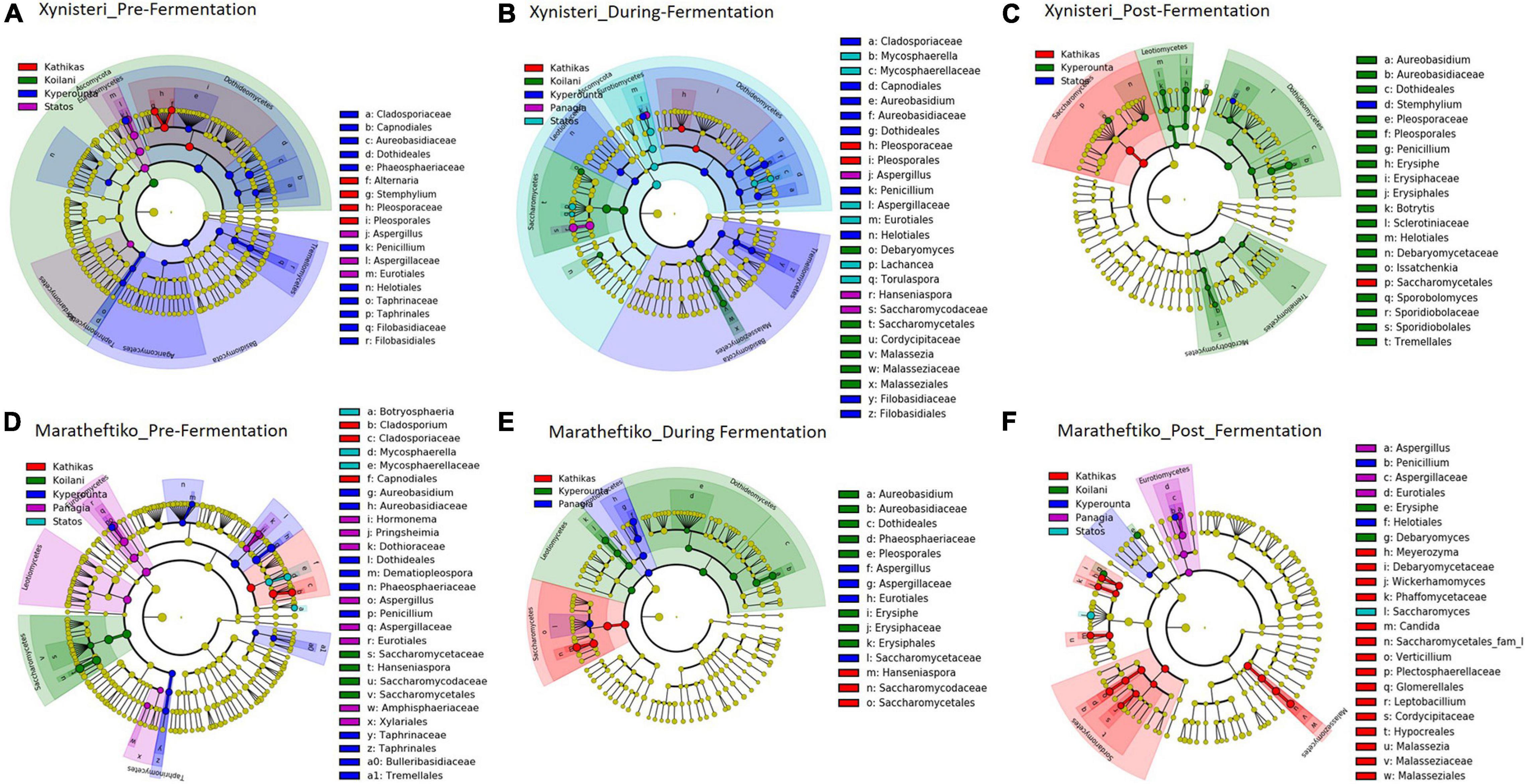
Figure 8. LEfSe analyses of taxon abundances of over-represented fungal taxa among grape wine varieties and among stages of fermentation. (A) Variety Xynisteri pre-fermentation; (B) variety Xynisteri during fermentation; (C) variety Xynisteri post-fermentation; (D) variety Maratheftiko pre-fermentation; (E) variety Maratheftiko during fermentation; (F) variety Maratheftiko post-fermentation.
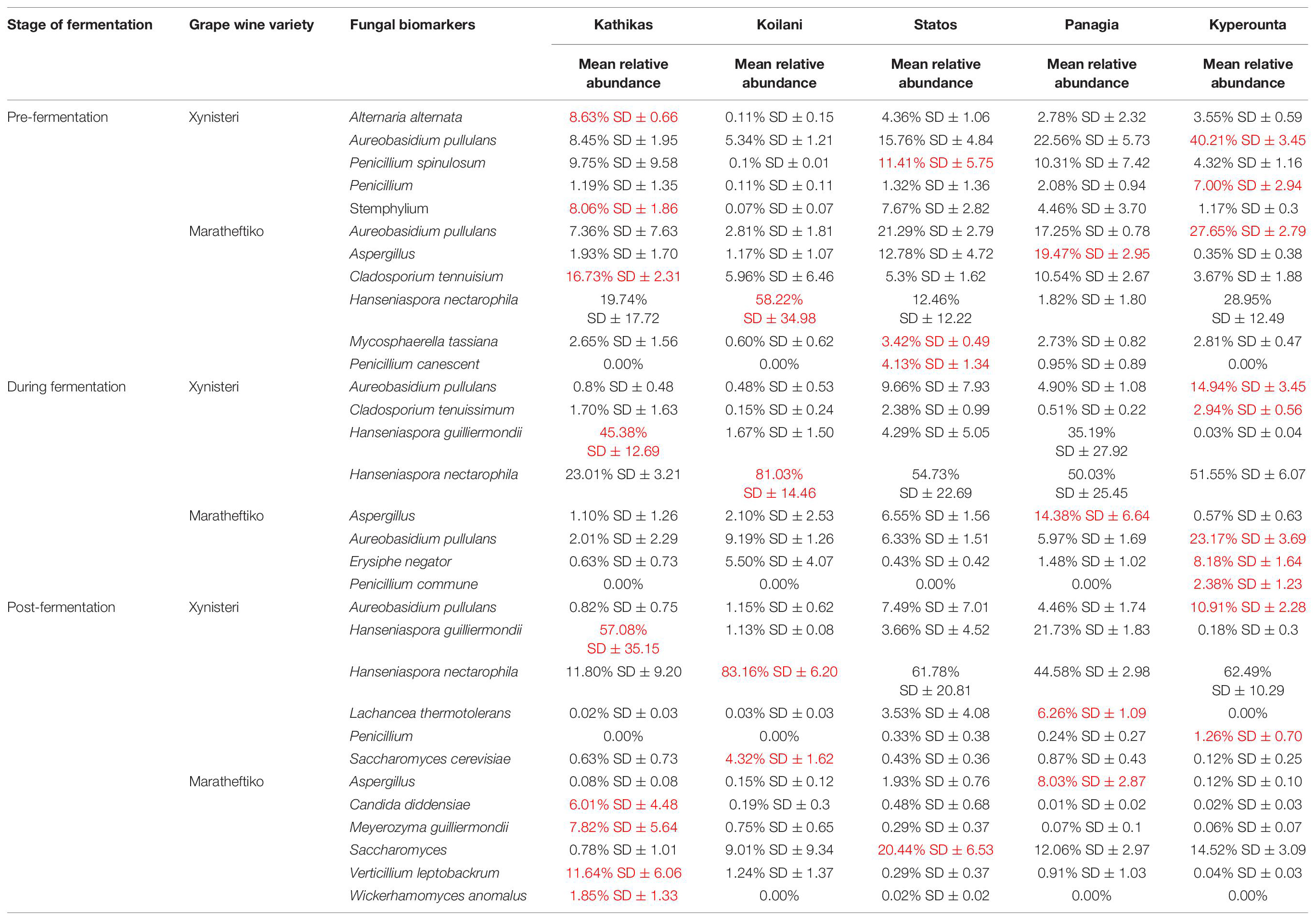
Table 1. Fungal taxa with significantly higher relative representation in a specific terroir (highlighted with red) for the two grape wine varieties during the process of fermentation.
During fermentation, for the variety Xynisteri, in agreement with pre-fermentation, Kyperounta terroir indicated a significantly higher representation of A. pullulans (Figure 8B and Table 1). In addition, the species Cladosporium tenuissimum was associated with Kyperounta. Kathikas had a higher relative representation of S. paradoxus and H. guilliermondii. Finally, Koilani was correlated with the species H. nectarophila. For the variety Maratheftiko, Kyperounta was also associated with A. pullulans (Figure 8E and Table 1). In addition, it was associated with the species Penicillium commune and E. necator. Finally, in agreement with pre-fermentation, Panayia was correlated with Aspergillus.
Post-fermentation analysis revealed that the species A. pullulans were a biomarker of Kyperounta terroir throughout the evolution of the fermentation, especially for the variety Xynisteri. Additionally, some taxa that were associated with specific terroir during fermentation indicated also significantly higher representation post-fermentation. These included, for the variety Xynisteri H. guilliermondii for the Kathikas and H. nectarophila for the Koilani terroir (Figure 8C), and for the variety Maratheftiko, Aspergillus for the Panayia terroir (Figure 8F). Furthermore, some new associations were detected. These included, for the variety Xynisteri Hanseniaspora guilliermondii for the Kathikas and Hanseniaspora nectarophila for the Koilani terroir (Figure 8C) and for the variety Maratheftiko, Aspergillus for the Panayia terroir (Figure 8F). For instance, for the variety Xynisteri, the species L. thermotolerans was correlated with Panayia and the species S. cerevisiae with Koilani terroir. In addition, the species Meyerozyma guilliermondii, Wickerhamomyces anomalus, Verticillium leptobackrum, and Candida diddensiae were associated with Kathikas terroir and Saccharomyces with the Statos terroir.
Pre-fermentation, for the variety Xynisteri, in Kyperounta terroir, the genera Cloacibacterium, Sphingomonas, Azospira and the species Pseudoxanthomonas mexicana were significantly more abundant compare to the other terroirs (Figure 9A and Table 2). Also, Statos terroir had a significantly higher representation of the genera Weissella and Lactococcus and the species L. delbrueckii (Figure 9A and Table 2). Post-fermentation, for the variety Xynisteri, the genus Acetobacter was associated with Koilani terroir (Figure 9C and Table 2). Additionally, for the variety Maratheftiko, Panayia had higher relative representation of L. brevis and C. paralimentarius and Kyperounta of O. oeni (Figure 9F and Table 2). No significant associations were detected Maratheftiko pre-fermentation (Figure 9D) and for Xynisteri (Figure 9B) and Maratheftiko (Figure 9E) during fermentation.
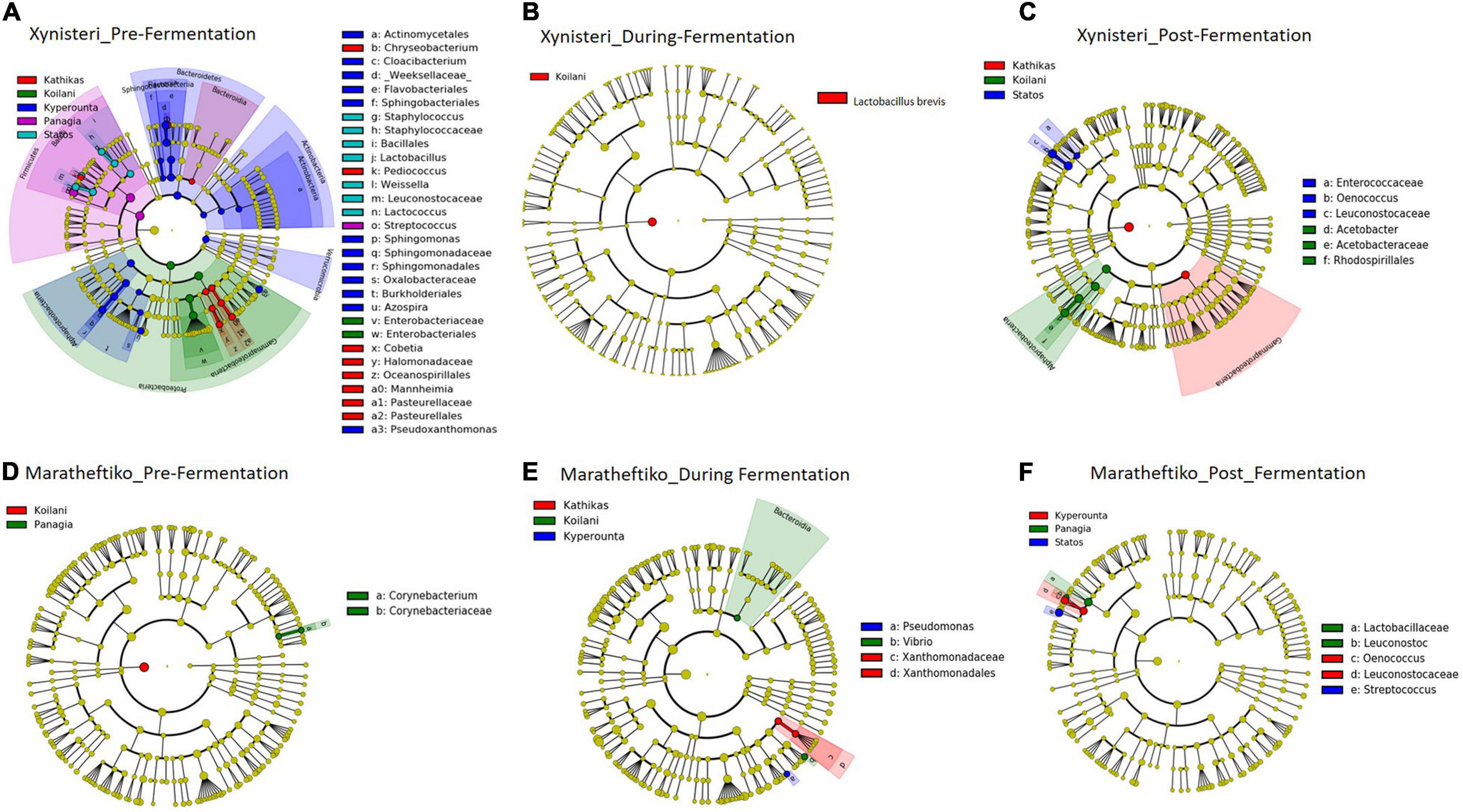
Figure 9. LEfSe analyses of taxon abundances of over-represented bacterial taxa among grape wine varieties and among stages of fermentation. (A) Variety Xynisteri pre-fermentation; (B) variety Xynisteri during fermentation; (C) variety Xynisteri post-fermentation; (D) variety Maratheftiko pre-fermentation; (E) variety Maratheftiko during fermentation; (F) variety Maratheftiko post-fermentation.
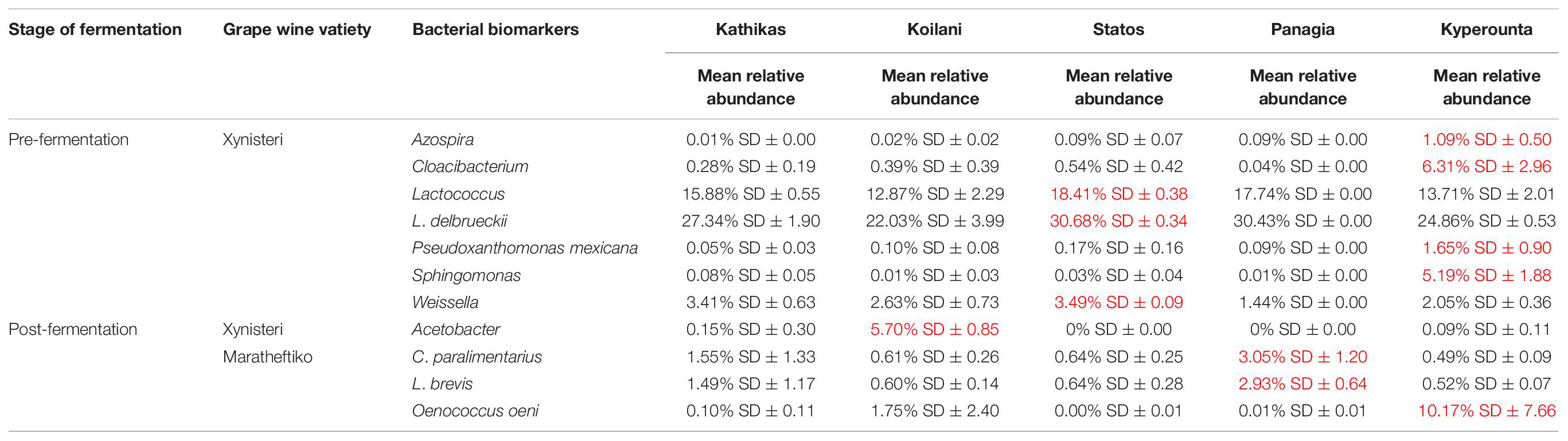
Table 2. Bacterial taxa with significantly higher relative representation in a specific terroir (highlighted with red) for the two grape wine varieties during the process of fermentation.
Finally, we investigated the presence of microbial biomarkers associated with the varieties Xynisteri and Maratheftiko, in the relative abundance of more than 1%. The analysis revealed that none of the analyzed fungal taxa were correlated with either Xynisteri or Maratheftiko during the process of fermentation (pre-, during, and post-fermentation). Regarding bacteria, the detected differences were during fermentation. Specifically, Xynisteri had a higher relative representation of the LAB Lactococcus, Weissella, and the species L. delbrueckii, whereas Maratheftiko with Gluconacetobacter, L. brevis, and C. paralimentarius (Table 3).
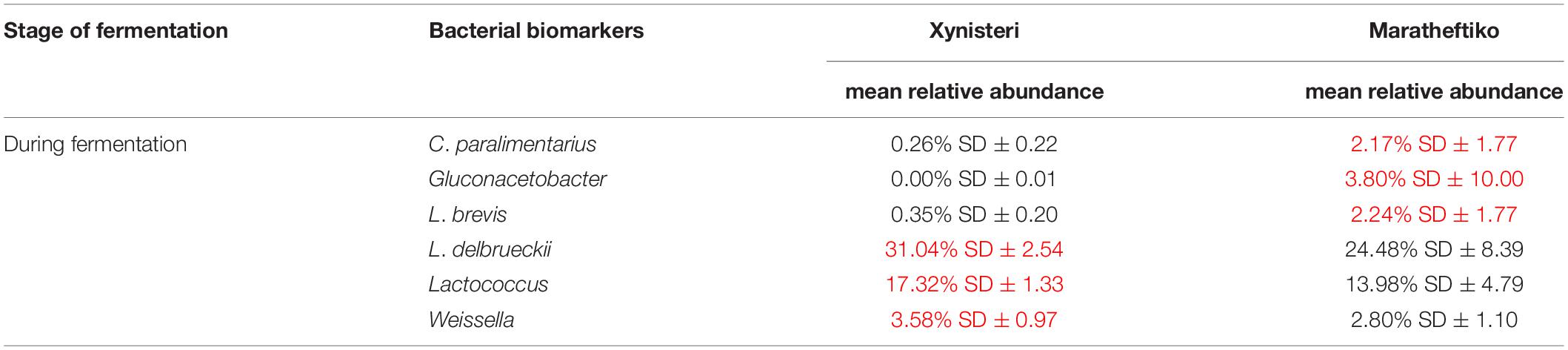
Table 3. Bacterial taxa with significantly higher relative representation in Xynisteri or Maratheftiko grape wine variety (highlighted with red) during the process of fermentation.
Substantial evidence suggests that grape−associated distinguishable microbial patterns (microbial terroir) contribute to the regional identity of the produced wine (Pinto et al., 2015; Bokulich et al., 2016; Anagnostopoulos et al., 2019; Gobbi et al., 2020). The present study was conducted to establish an association between microbial diversity and the different Cyprus wine regions (terroirs) during spontaneous wine fermentation of the grape wine varieties Xynisteri and Maratheftiko. To demonstrate this, we applied HTS for the primary in-depth characterization of Cyprus wines microbial diversity, mapping the dissimilarity in the microbial community of five grape wine terroirs. The study revealed a distinction in alpha and beta diversity among the five terroirs, among the different stages of fermentation and between the varieties Xynisteri and Maratheftiko. Additionally, we identified microbial biomarkers associated with each terroir among the different stages of fermentation and for the different wine grape varieties.
The process of AF contributed to a progressive reduction in the fungal alpha diversity, both for Xynisteri and Maratheftiko grape wine varieties. This observation is in agreement with a recent study comparing the fungal diversity of typical Vitis vinifera L. var. Sauvignon blanc from Chile between pre-fermented and fermented musts (Mandakovic et al., 2020) and a metataxonomic study of Vino Santo mycobiota in the Italian Alps (Stefanini et al., 2016). During AF, yeast species such as S. cerevisiae ferment sugars producing ethanol (Stefanini et al., 2016). The stressful environment for some yeast species created by the high alcohol leads to their elimination (Stefanini and Cavalieri, 2018). Therefore, post-fermentation, the microbial consortia are comprised of specific microbial species (Campisano et al., 2014; Bokulich et al., 2016). Apart from AF, MLF (conversion of malic acid into lactic acid) is conducted, generally by LAB (Renouf et al., 2005). Based on our results, the bacterial alpha diversity that comprised the two grape wine varieties remained stable during the process. Pre-fermentation, the areas with the lowest altitude, including Kathikas and Koilani, indicated significantly higher bacterial alpha diversity than Kyperounta, the area with the highest altitude. High altitude environments select specific microbes able to adapt and maintain their metabolic activities in low temperature, high UV irradiation, reduced atmospheric pressure and limited soil nutrients (Kumar et al., 2019). Therefore, the selective pressure of the stressful highest altitude ecosystem of Kyperounta may be responsible for the lower bacterial diversity compared to the other terroirs. However, for the variety Xynisteri, the fungal diversity of Kyperounta was significantly higher compared to Koilani, indicating that additional factors might affect these terroirs fungal communities’ composition, for instance, cooler climatic conditions and rainfall (Bokulich et al., 2013; Grangeteau et al., 2017).
The Bray-Curtis dissimilarity distance metric defined that the microbial community contributes to distinguishing musts originated from different terroirs and grape varieties. This is in line with additional studies based on HTS, revealing microbiological patterns associated with viticulture areas (Bokulich et al., 2012; Pinto et al., 2015; Burns et al., 2016; Portillo Mdel et al., 2016; Mezzasalma et al., 2017). Taylor et al. (2014) identified a distinct difference in the fungal community composition of Central Otago compare to the other four main regions in New Zealand. The distinction we observed was grape wine variety depended. Specifically, the fungal beta diversity of the variety Xynisteri was significantly different among almost all the different terroirs from pre- until post-fermentation. For the variety Maratheftiko, the fungal beta diversity of Kyperounta was significantly different from the other terroir pre-, during and post-fermentation. Kyperounta was also distinguished from the other terroir based on the bacterial diversity, in the pre-fermentation stage, for the variety Xynisteri. In addition, PERMANOVA analysis revealed significant differences in the microbial beta diversity between Xynisteri and Maratheftiko for the stages pre- and post-fermentation. Bokulich et al. (2016) also demonstrated that the distinction of the different viticultural areas and individual vineyards across California depends on the wine grape variety, with Chardonnay showing more powerful microbial composition separation ability than Cabernet Sauvignon.
Pre-fermentation, the fungal diversity was dominated by non-Saccharomyces yeasts. These included both for Xynisteri and Maratheftiko the genera Hanseniaspora, represented by the species H. nectarophila and H. guilliermondii, Aureobasidium, including A. pullulans, Erysiphe represented by E. necator, Aspergillus, Stemphylium, and Penicillium including P. spinulosum, P. bilaiae, and P. canescens, Alternaria including A. alternata and A. metachromatica, Cladosporium including C. tenuissimum and Mycosphaerella represented by M. tassiana. Similarly, pre-fermentation samples from different wine appellations in Portugal were characterized by a higher relative representation of A. pullulans and Hanseniaspora, but H. uvarum instead of H. nectarophila and H. guilliermondii that we detected (Pinto et al., 2015). The analyzed Cyprus terroir fungal diversity of Xynisteri and Maratheftiko differed from the Italian grape varieties Greco and Aglianico analyzed by De Filippis et al. (2017). Specifically, the abundant species they discovered included Hanseniaspora osmophila, Hanseniaspora uvarum, Candida stellata, Pichia kluyveri, Metschnikowia pulcherrima, Zygosaccharomyces rouxii, Issatchenkia terricola, Pichia occidentalis, Botrytis cinerea, Cladosporium bruhnei, and Aspergillus niger. In agreement with our findings, the genus Hanseniaspora was the predominant pre-fermentation for the variety Grenache from Catalonia, Spain (Portillo Mdel and Mas, 2016). Several common genera were detected in an HTS study of unfermented must of the variety V. vinifera L. var. Sauvignon blanc in Chile, in which a commercial starter culture of S. cerevisiae was added (Mandakovic et al., 2020). Apart from the genus Saccharomyces the genera Hanseniaspora, Torularospora, Botrytis, Penicillium, Cladosporium, Candida, and Aureobasidium were the most dominant in agreement with most of our findings. The genera Aureobasidium, Penicillium, Erysisphe Aspergillus, and Alternaria, were among the predominant genera in an amplicon sequencing study performed in grape juice of the variety Cabernet sauvignon in China (Wei et al., 2018).
The bacterial diversity of pre-fermented samples for Xynisteri and Maratheftiko was dominated by LAB, mostly species of the genera Lactobacillus including L. delbrueckii, followed by C. paralimentarius and L. brevis, Streptococcus and Lactococcus. The genera Tatumella, Salinivibrio, Staphylococcus, Cloacibacterium, Rothia, Mannheimia, Sphingomonas Pseudomonas, Weissella, Methyloversatilis, and Leuconostoc were detected in lower relative abundances. In contrast to our results, an HTS study of the wine microbiota from six wine appellations in Portugal revealed that the grape juice was dominated by Proteobacteria members of the families Enterobacteriaceae and Pseudomonadaceae (Pinto et al., 2015). An HTS longitudinal analysis of about 200 commercial wines during fermentation in California indicated that the dominant taxa pre-fermentation were apart from Enterobacteriaceae and Pseudomonas, Leuconostocaceae, Bacillaceae, Sphingomonas Gluconobacter, and Lactobacillus (Bokulich et al., 2016).
Most of the identified pre-fermentation microbiota was comprised of environmental microorganisms detected in the vineyard ecosystem. Specifically, A. pullulans, Aspergillus, Penicillium, E. necator, Alternaria, Stemphylium, Lactobacillus, Streptococcus, Staphylococcus, and Gluconobacter were found to be associated with vineyard environment, including soil, grape surface and leaves (Bokulich et al., 2013; Pinto et al., 2014; Zarraonaindia et al., 2015; Wei et al., 2018). Additionally, Sphingomonas and Pseudomonas species were detected in aboveground plant areas (Whipps et al., 2008; Innerebner et al., 2011). Some species, such as Pseudomonas and Staphylococcus, were identified as endophytic bacteria (Compant et al., 2011). Hanseniaspora, the most dominant genus, is commonly found on grapes (Varela and Borneman, 2017). Species of the Hanseniaspora contribute significantly to wine fermentation by controlling the metabolic activity of S. cerevisiae and producing metabolites that affect the wine color and other sensorial characteristics (Martin et al., 2018). Hanseniaspora nectarophila, the most dominant species in most pre-fermentation samples, was first isolated from ephemeral flowers and its role in wine fermentation remains undetermined (Čadež et al., 2014). Mezzasalma et al. (2018) identified geographical signatures in the grape microbiome associated with its origin. The researchers found that the vineyard soil microbiome was about 60% common with the grape microbiome. Through a machine learning system, they predicted the origin of the grape based on the microbiota. Gobbi et al. (2020) analyzed the soil microbiota of 200 vineyards from four continents by using an amplicon sequencing approach. Even though we analyzed grape juice and not soil samples, their results highlight the unique microbiome found in the Cyprus vineyards. Specifically, countries such as Italy, Croatia, Argentina, Chile, and South Africa had a higher relative abundance of Solicoccozyma, whereas Portugal, Germany, Australia, Denmark, and South Africa of Fusarium. Additional dominant species that Gobbi et al. detected include Gibberella intricans in Spain, Portugal, and Denmark, Tausonia pullulans in Hungary and Spain, Metarhizium_robertsii in Denmark, Sclerotinia sclerotiorum and Rhizopus arrhizus in France, and Truncatella in Germany. Some species that we also detected in our study included Cladosporium, which was dominant in Portugal, Germany, Australia, and Denmark, Alternaria in Chile and Aspergillus in South Africa.
Generally, AF is performed by the contribution of several microbes, with Saccharomyces eventually governing (Fleet, 2008). However, spontaneous AF may be driven by non-Saccharomyces yeasts, something unwanted by the wine industries because it can affect the wine’s aromatic quality (Zott et al., 2011; Liu et al., 2016). Evaluation of microbial community dynamics during and post-spontaneous AF revealed that the process in most Cyprus must samples was driven by non-Saccharomyces yeasts, including H. nectarophila and H. guilliermondii. However, an increase in the relative abundance of Saccharomyces and the species S. cerevisiae and S. paradoxus was indicated during fermentation, and Saccharomyces became abundant by the end of fermentation in only a few samples. In agreement, Stefanini et al. (2016) observed that during Vino Santo must fermentation in Italy, in one of the three tested wineries, the mycobiota was dominated by H. osmophila, and S. cerevisiae was detected in lower relative abundances. In the other two wineries, though, the AF was driven by S. cerevisiae. Our findings suggest that the vineyard microbiota guides the process of fermentation since species such as H. nectarophila, H. guilliermondii and A. pullulans remained among the predominant species from pre- until post-fermentations. Noteworthy, Čadež et al. (2014) reported that the discrimination of H. nectarophila from the other closely related species, including H. guilliermondii Hanseniaspora meyeri, Hanseniaspora opuntiae, and Hanseniaspora clermontiae, was based not only on the ITS and D1/D2 LSU sequences similarity comparison but also the sequences of the gene coding for actin. This is because the differences with the aforementioned species regarding the D1/D2 LSU and ITS sequences were only were four nucleotides, which were not enough to separate H. nectarophila into separate species. Other species, such as A. alternata, E. necator, P. spinulosum continued to be among the low abundant species (>1%) from the pre- until post-fermentations. These results are in agreement with other metataxonomic studies (Bokulich et al., 2013, 2016; Morrison-Whittle and Goddard, 2018).
Several factors affect the dominance of S. cerevisiae during AF. The increased tolerance of Saccharomyces species to ethanol and its fast growth rate is the primary factor contributing to its dominance. However, apart from Saccharomyces species, alcohol tolerant non-Saccharomyces yeasts may drive AF. For instance, Sirén et al. (2019) identified Hanseniaspora as the predominant species in post-fermentation Riesling must following spontaneous in vitro fermentation. The tolerance of Hanseniaspora to ethanol was found to increase by low temperature (10–15°C) (Erten, 2002). Still, the fermentation of Cyprus musts was performed at 18°C, indicating that the temperature might not be associated with Hanseniaspora’s dominance. Boynton and Greig (2016) applied HTS to indicate that the ability of Saccharomyces to lead the process of fermentation in must samples is prevented by increased species richness, which might be an affecting factor for Saccharomyces suppression in our study. Another affecting factor could be the availability of sugars since the metabolic activity of S. cerevisiae is induced by high sugar concentrations (Lleixà et al., 2016). The sugar concentration of both Xynisteri and Maratheftiko is >20 Brix, which is considered high. Furthermore, other studies supported that some S. cerevisiae strains secrete antimicrobial peptides that prevent the growth of non-Saccharomyces yeasts (Rai and Jeyaram, 2015; Fakruddin et al., 2017). For instance, the isolated strain S. cerevisiae CCMI 885 secreted small peptides with antimicrobial activity against H. guilliermondii (Albergaria et al., 2010). However, some A. pullulans strains were found to produce aureobasidin A, which show a powerful fungicidal action against S. cerevisiae and Candida species (Heidler and Radding, 1995; Zhong et al., 2000). A. pullulans were found to be among the predominant species in some of our samples.
The amplicon sequencing analysis indicated that the most abundant species during MLF was L. delbrueckii. The metabolic activity of LAB leads to deacidification of the must by converting malic acid into lactic acid (Lonvaud-Funel, 1999). Apart from the LAB Lactobacillus, Streptococcus and Lactococcus, some during fermentation samples were characterized by an increase in the relative abundance of the genera Gluconobacter and Acetobacter and some post-fermentation samples by the dominance of Acetobacter. The metabolic activity of acetic acid bacteria may negatively impact the quality of the produced wine due to the production of acetic acid (Zoecklein et al., 2000). Bokulich et al. (2016) revealed that increased bacterial richness prevents AF and identified antagonistic relationships between Lactobacillus spp., Gluconobacter and H. uvarum, with S. cerevisiae, competing for nutrients availability. In agreement, Berbegal et al. (2019) suggested that Saccharomyces exerts selective pressure on the spoilage Acetobacter and Gluconobacter and promotes the dominance of Oenococcus. In the present study, only one sample was characterized by the dominance of O. oeni. Based on these data, the supremacy of Hanseniaspora and the suppression of Saccharomyces may be responsible for the suppression of growth of Oenococcus, favoring the increase in the relative abundance of acetic acid bacteria that we observed.
According to a recent review article by Griggs et al. (2021), the wine-relevant microbial diversity of the vineyard affects the outcome of the fermentation and the quality of the produced wine. In our study, we observed differences in the relative representation of microbes between samples from the same vineyard and neighbor vineyards. These differences affected the process of fermentation, leading to alterations in the relative representation of post-fermentation microbial species. Specifically, for the variety Xynisteri, samples obtained from one vineyard of Kathikas terroir indicated a higher relative representation of Saccharomyces, whereas samples obtained from the neighbor vineyard of H. guilliermondii. As described above, species of the genera Saccharomyces and Hanseniaspora are drivers of AF. However, as described by Griggs et al., our knowledge regarding the factors affecting the microbial biodiversity shaping among vineyards and drive the process of fermentation remains limited, especially regarding spontaneous fermentations. Noteworthy, all fermentations were performed in the same place, under the same conditions. These findings provide additional evidence that the vineyard microbiota influences the microbial community shaping and the drivers of fermentation.
Finally, the study revealed associations among the microbial diversity and the different terroirs and wine varieties, from pre- until post-fermentation. Comparison of the fungal diversity among the five different terroirs and the three different stages of fermentation revealed that the species A. pullulans were associated with the Kyperounta terroir, both for Xynisteri and Maratheftiko, pre-, during, and post-fermentation. A. pullulans are among the dominant microbiota of grape juice (Pinto et al., 2015; Sternes et al., 2017) and considered anti-phytopathogen microorganism, expressing anti-fungal and anti-bacterial activity against several important post-harvest pathogens such as Botrytis and Bacillus (Sharma et al., 2009; Grube et al., 2011; Prasongsuk et al., 2018). Additionally, these species have an essential contribution to wine’s color, sensorial characteristics and clarification efficiency (Merín and Morata de Ambrosini, 2018; Onetto et al., 2020). Apart from A. pullulans, Penicillium was also associated with the Kyperounta terroir, both for Xynisteri and Maratheftiko, pre-, during, and post-fermentation. This genus has an important contribution to the wine industry since it may change grape wines’ chemical composition affecting the microbial community shaping during fermentation (Nigam et al., 2017). As a result, it affects the colors and flavors of the produced wine. Kathikas terroir indicated a significantly higher representation of the species A. alternata pre- and during fermentation for the variety Xynisteri. A. alternata is an important contaminant in grape juice. It produces toxic metabolites, the accumulation of which during winemaking is risky for consumers health (Prendes et al., 2021). Statos terroir was distinguished by the psychrophilic, acidotolerant species P. spinulosum and the antibiotic canescin producing species P. canescent for the variety Xynisteri and Maratheftiko, respectively, during pre-fermentation. Koilani terroir was characterized by a significantly higher representation of H. nectarophila pre-fermentation for the variety Maratheftiko and during and post-fermentation for the variety Xynisteri. Furthermore, the study revealed bacterial biomarkers associated with the five grape wine terroirs. However, none of the identified bacterial biomarkers was detected from the beginning until the end of the fermentation process.
Since the aim of our study is to provide evidence of the existence of different microbial patterns among the five terroirs, it is important to mention that Kokkinofta et al. (2017) identified differences in the five terroir that we analyzed in the present study, for the same wine varieties (Xynisteri and Maratheftiko) by applying SNIF-NMR, IRMS and inductively coupled plasma atomic emission spectroscopy (ICP-AES). Specifically, chemometrics analysis of elements and isotopes revealed differentiation between the varieties Maratheftiko and Xynisteri, and a significant distinction between the Pafos PGI region and the Lemessos PGI region and among the wineries Kamanterena and Vassilikon (PDO Laona Akamas) in the Pafos PGI region and Pelendri (PDO Pitsilia) and Koilani village (PDO Krasochoria), Lemessos PGI region. Furthermore, we need to mention that our analysis was restricted to only four samples per terroir and to only 1 year of sampling, which is a limiting factor, considering that regional environmental factors, such as climatic conditions, may affect the microbial community composition. Also, our analysis included multiple comparisons, which increase the possibility of significant differences to be discovered by chance. Although in most of the comparisons the adjusted values remained significant, in a few of them didn’t, indicating a limitation associated with multiple comparisons performance.
The present metataxonomic study is the first attempt to distinguish the five different Cyprus wine terroirs’ microbial diversity for the two grape wine varieties Xynisteri and Maratheftiko during the stages of pre- until post-spontaneous fermentation. The study revealed a significant reduction in the fungal diversity during the stages of fermentation and in the bacterial diversity between the areas with the lowest altitude, compared to the area with the highest altitude pre-fermentation. Most importantly, the study defined that microbial diversity is a key factor for distinguishing grape wines from different terroirs and grape wine varieties. In the future, apart from the present, more grape wine varieties andsamples from multiple harvests are to be analyzed using an amplicon sequencing approach. This will allow us to compare the alterations in the microbial diversity throughout different years. The metataxonomic analysis may be combined with predictive functional analysis to reveal the influence of the bacterial and fungal community associated metabolic pathways in the wine aromatic profile and flavors. Also, it may be combined with SNIF-NMR, IRMS and inductively coupled plasma atomic emission spectroscopy (ICP-AES), as applied by Kokkinofta et al. (2017), and DNA fingerprint characterization studies. These methodologies may assist to differentiate authentic wines from possible fraud products that may exist in the market and provide a fingerprint for defining the authenticity of Cyprus wines. The present study’s findings will further assist in the improvement of the qualitative characteristics of Cyprus wines.
Our data are publicly available at the following link: https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA731461.
DT and MM: conceptualization and writing–review and editing. MM and CK: grape wine fermentation. EK: methodology, formal analysis, investigation, data curation, and writing–original draft preparation. DT: resources, supervision, project administration, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
This study received funding from INTERREG Greece – Cyprus 2014–2020 Program, Project AGRO-ID, which is co-funded by the European Union (ERDF) and National Resources of Greece and Cyprus.
CK is employed by Kyperounta Winery.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge the funding from the project AGRO-ID, INTERREG Greece-Cyprus 2014-2020.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.726483/full#supplementary-material
Abarenkov, K., Zirk, A., Piirmann, T., Pöhönen, R., Ivanov, F., Nilsson, R. H., et al. (2020). Unite Qiime release for Fungi 2. Holborn: Unite Community.
Akao, T., Yashiro, I., Hosoyama, A., Kitagaki, H., Horikawa, H., Watanabe, D., et al. (2011). Whole-genome sequencing of sake yeast Saccharomyces cerevisiae Kyokai no, 7. DNA Res. 18, 423–434. doi: 10.1093/dnares/dsr029
Albergaria, H., Francisco, D., Gori, K., Arneborg, N., and Gírio, F. (2010). Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl. Microbiol. Biotechnol. 86, 965–972. doi: 10.1007/s00253-009-2409-6
Anagnostopoulos, D. A., Kamilari, E., and Tsaltas, D. (2019). “Contribution of the microbiome as a tool for estimating wine’s fermentation output and authentication,” in Advances in Grape and Wine Biotechnology, eds A. Morata and I. Loira (London: IntechOpen), 1–19.
Anderson, M. J. (2008). A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.tb00081.x
Ballantyne, D., Terblanche, N. S., Lecat, B., and Chpuis, C. (2019). Old world and new world wine concepts of terroir and wine: perspectives of three renowned non-French wine makers. J. Wine Res. 30, 122–143. doi: 10.1080/09571264.2019.1602031
Belda, I., Ruiz, J., Alastruey-Izquierdo, A., Navascués, E., Marquina, D., and Santos, A. (2016). Unraveling the enzymatic basis of wine “flavorome”: a phylo-functional study of wine related yeast species. Front. Microbiol. 7:12. doi: 10.3389/fmicb.2016.00012
Berbegal, C., Borruso, L., Fragasso, M., Tufariello, M., Russo, P., Brusetti, L., et al. (2019). A metagenomic-based approach for the characterization of bacterial diversity associated with spontaneous malolactic fermentations in wine. Int. J. Mol. Sci. 20:3980. doi: 10.3390/ijms20163980
Berbegal, C., Peña, N., Russo, P., Grieco, F., Pardo, I., Ferrer, S., et al. (2016). Technological properties of Lactobacillus plantarum strains isolated from Apulia wines. Food Microbiol. 57, 187–194. doi: 10.1016/j.fm.2016.03.002
Berbegal, C., Spano, G., Tristezza, M., Grieco, F., and Capozzi, V. (2017). Microbial resources and innovation in the wine production sector. S. Afr. J. Enol. Vitic. 38, 156–166. doi: 10.21548/38-2-1333
Birkeland, S. R., Jin, N., Ozdemir, A. C., Lyons, R. H. Jr., Weisman, L. S., et al. (2010). Discovery of mutations in Saccharomyces cerevisiae by pooled linkage analysis and whole-genome sequencing. Genetics 186, 1127–1137. doi: 10.1534/genetics.110.123232
Bokulich, N. A., Collins, T. S., Masarweh, C., Allen, G., Heymann, H., Ebeler, S. E., et al. (2016). Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. mBio 7, e631–e616. doi: 10.1128/mBio.00631-16
Bokulich, N. A., Joseph, C. M., Allen, G., Benson, A. K., and Mills, D. A. (2012). Next-generation sequencing reveals significant bacterial diversity of botrytized wine. PLoS One 7:e36357. doi: 10.1371/journal.pone.0036357
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Bokulich, N. A., Thorngate, J. H., Richardson, P. M., and Mills, D. A. (2013). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. U.S.A. 111, E139–E148. doi: 10.1073/pnas.1317377110
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Author correction: reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37:1091. doi: 10.1038/s41587-019-0209-9
Borneman, A. R., Forgan, A. H., Kolouchova, R., Fraser, J. A., and Schmidt, S. A. (2016). Whole genome comparison reveals high levels of inbreeding and strain redundancy across the spectrum of commercial wine strains of Saccharomyces cerevisiae. G3 6, 957–971. doi: 10.1534/g3.115.025692
Boynton, J. P., and Greig, D. (2016). Species richness influences wine ecosystem function through a dominant species. Fungal Ecol. 22, 61–72. doi: 10.1016/j.funeco.2016.04.008
Burns, K. N., Bokulich, N. A., Cantu, D., Greenhut, R. F., Kluepfel, D. A., O’Geen, A. T., et al. (2016). Vineyard soil bacterial diversity and composition revealed by 16S rRNA genes: differentiation by vineyard management. Soil Biol. Biochem. 103, 337–348. doi: 10.1016/j.soilbio.2016.09.007
Čadež, N., Pagnocca, F. C., Raspor, P., and Rosa, C. A. (2014). Hanseniaspora nectarophila sp nov., a yeast species isolated from ephemeral flowers. Int. J. Syst. Evol. Microbiol. 64, 2364–2369. doi: 10.1099/ijs.0.061499-0
Campisano, A., Antonielli, L., Pancher, M., Yousaf, S., Pindo, M., Pertot, I., et al. (2014). Bacterial endophytic communities in the grapevine depend on pest management. PLoS One 9:e112763. doi: 10.1371/journal.pone.0112763
Compant, S., Mitter, B., Colli-Mull, J. G., Gangl, H., and Sessitsch, A. (2011). Endophytes of grapevine flowers, berries, and seeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb. Ecol. 62, 188–197. doi: 10.1007/s00248-011-9883-y
De Benedictis, M., Bleve, G., Grieco, F., Tristezza, M., Tufariello, M., and Grieco, F. (2011). An optimized procedure for the enological selection of non-Saccharomyces starter cultures. Antonie Van Leeuwenhnoek 99, 189–200. doi: 10.1007/s10482-010-9475-8
De Filippis, F., La Storia, A., and Blaiotta, G. (2017). Monitoring the mycobiota during Greco di Tufo and Aglianico wine fermentation by 18S rRNA gene sequencing. Food Microbiol. 63, 117–122. doi: 10.1016/j.fm.2016.11.010
Erten, H. (2002). Relations between elevated temperatures and fermentation behaviour of Kloeckera apiculata and Saccharomyces cerevisiae associated with winemaking in mixed cultures. World J. Microbiol. Biotechnol. 18, 373–378. doi: 10.1023/A:1015221406411
Fakruddin, M., Hossain, M. N., and Ahmed, M. M. (2017). Antimicrobial and antioxidant activities of Saccharomyces cerevisiae IFST062013, a potential probiotic. BMC Complement. Altern. Med. 17:64. doi: 10.1186/s12906-017-1591-9
Fleet, G. H. (2008). Wine yeasts for the future. FEMS Yeast Res. 8, 979–995. doi: 10.1111/j.1567-1364.2008.00427.x
Giorello, F. M., Berná, L., Greif, G., Camesasca, L., Salzman, V., Medina, K., et al. (2014). Genome sequence of the native apiculate wine yeast Hanseniaspora vineae T02/19AF. Genome Announc. 2, e00530–e614. doi: 10.1128/genomeA.00530-14
Gobbi, A., Acedo, A., Imam, N., Santini, R. G., Ortiz-Álvarez, R., Ellegaard-Jensen, L., et al. (2020). Microbial map of the world’s vineyards: applying the concept of microbial terroir on a global scale. Biorxiv. [Preprint] doi: 10.1101/2020.09.25.313288
Goffeau, A., Barrell, B. G., Bussey, H., Davis, R. W., Dujon, B., Feldmann, H., et al. (1996). Life with 6000 genes. Science 274, 563–567. doi: 10.1126/science.274.5287.546
Grangeteau, C., Roullier-Gall, C., Rousseaux, S., Gougeon, R. D., Schmitt-Kopplin, P., Alexandre, H., et al. (2017). Wine microbiology is driven by vineyard and winery anthropogenic factors. Microb. Biotechnol. 10, 354–370. doi: 10.1111/1751-7915.12428
Griggs, R. G., Steenwerth, K. L., Mills, D. A., and Bokulich, N. A. (2021). Sources and assembly of microbial communities in vineyards as a functional component of winegrowing. Front. Microbiol. 12:673810. doi: 10.3389/fmicb.2021.673810
Grube, M., Schmid, F., and Berg, G. (2011). Black fungi and associated bacterial communities in the phyllosphere of grapevine. Fungal Biol. 115, 978–986. doi: 10.1016/j.funbio.2011.04.004
Heidler, S. A., and Radding, J. A. (1995). The AUR1 gene in Saccharomyces cerevisiae encodes dominant resistance to the antifungal agent aureobasidin A (LY295337). Antimicrobial. Agents Chemother. 39, 2765–2769. doi: 10.1128/AAC.39.12.2765
Innerebner, G., Knief, C., and Vorholt, J. A. (2011). Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 77, 3202–3210. doi: 10.1128/AEM.00133-11
Jolly, N. P., Varela, C., and Pretorius, I. S. (2014). Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 14, 215–237. doi: 10.1111/1567-1364.12111
Kamilari, E., Anagnostopoulos, D. A., Papademas, P., Efthymiou, M., Tretiak, S., and Tsaltas, D. (2020a). Snapshot of cyprus raw goat milk bacterial diversity via 16S rDNA high-throughput sequencing; impact of cold storage conditions. Fermentation 6:100. doi: 10.3390/fermentation6040100
Kamilari, E., Anagnostopoulos, D. A., Papademas, P., Kamilaris, A., and Tsaltas, D. (2020b). Characterizing halloumi cheese’s bacterial communities through metagenomic analysis. LWT 126:109298. doi: 10.1016/j.lwt.2020.109298
Kamilari, E., Efthymiou, M., Anagnostopoulos, D. A., and Tsaltas, D. (2021). Cyprus sausages’ bacterial community identification through metataxonomic sequencing: evaluation of the impact of different DNA extraction protocols on the sausages’ microbial diversity representation. Front. Microbiol. 12:904. doi: 10.3389/fmicb.2021.662957
Knight, S., Klaere, S., Fedrizzi, B., and Goddard, M. R. (2015). Regional microbial signatures positively correlate with differential wine phenotypes: evidence for a microbial aspect to terroir. Sci. Rep. 5:14233. doi: 10.1038/srep14233
Kokkinofta, R., Fotakis, C., Zervou, M., Zoumpoulakis, P., Savvidou, C., Poulli, K., et al. (2017). Isotopic and elemental authenticity markers: a case study on Cypriot wines. Food Anal. Methods 10, 3902–3913. doi: 10.1007/s12161-017-0959-2
Kumar, S., Suyal, D. C., Yadav, A., Shouche, Y., and Goel, R. (2019). Microbial diversity and soil physiochemical characteristic of higher altitude. PLoS One 14:e0213844. doi: 10.1371/journal.pone.0213844
Lambrechts, M. G., and Pretorius, I. S. (2019). Yeast and its importance to wine aroma–a review. S. Afr. J. Enol. Vitic. 21, 97–129. doi: 10.21548/21-1-3560
Liu, P. T., Lu, L., Duan, C. Q., and Yan, G. L. (2016). The contribution of indigenous non-Saccharomyces wine yeast to improved aromatic quality of cabernet sauvignon wines by spontaneous fermentation. LWT Food Sci. Technol. 71, 356–363. doi: 10.1016/j.lwt.2016.04.031
Lleixà, J., Martín, V., Portillo Mdel, C., Carrau, F., Beltran, G., and Mas, A. (2016). Comparison of fermentation and wines produced by inoculation of Hanseniaspora vineae and Saccharomyces cerevisiae. Front. Microbiol. 7:338. doi: 10.3389/fmicb.2016.00338
Lonvaud-Funel, A. (1999). Lactic acid bacteria in the quality improvement and depreciation of wine, in Antonie van Leeuwenhoek. Int. J. Gen. Mol. Microbiol. 76, 317–331. doi: 10.1023/A:1002088931106
Mandakovic, D., Pulgar, R., Maldonado, J., Mardones, W., González, M., Cubillos, F. A., et al. (2020). Fungal diversity analysis of grape musts from central valley-Chile and characterization of potential new starter cultures. Microorganisms 8:956. doi: 10.3390/microorganisms8060956
Martin, V., Valera, M., Medina, K., Boido, E., and Carrau, F. (2018). Oenological impact of the hanseniaspora/kloeckera yeast genus on wines—a review. Fermentation 4:76. doi: 10.3390/fermentation4030076
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Merín, M. G., and Morata de Ambrosini, V. I. (2018). Kinetic and metabolic behaviour of the pectinolytic strain Aureobasidium pullulans GM-R-22 during pre-fermentative cold maceration and its effect on red wine quality. Int. J. Food Microbiol. 285, 18–26. doi: 10.1016/j.ijfoodmicro.2018.07.003
Mezzasalma, V., Sandionigi, A., Bruni, I., Bruno, A., Lovicu, G., Casiraghi, M., et al. (2017). Grape microbiome as a reliable and persistent signature of field origin and environmental conditions in Cannonau wine production. PLoS One 12:e0184615. doi: 10.1371/journal.pone.0184615
Mezzasalma, V., Sandionigi, A., Guzzetti, L., Galimberti, A., Grando, M. S., Tardaguila, J., et al. (2018). Geographical and cultivar features differentiate grape microbiota in northern Italy and Spain vineyards. Front. Microbiol. 9:946. doi: 10.3389/fmicb.2018.00946
Mills, D. A., Rawsthorne, H., Parker, C., Tamir, D., and Makarova, K. (2005). Genomic analysis of Oenococcus oeni PSU-1 and its relevance to winemaking. FEMS Microbiol. Rev. 29, 465–475. doi: 10.1016/j.femsre.2005.04.011
Morrison-Whittle, P., and Goddard, M. R. (2018). From vineyard to winery: a source map of microbial diversity driving wine fermentation. Environ. Microbiol. 20, 75–84. doi: 10.1111/1462-2920.13960
Naseeb, S., Alsammar, H., Burgis, T., Donaldson, I., Knyazev, N., Knight, C., et al. (2018). Whole genome sequencing, de novo assembly and phenotypic profiling for the new budding yeast species Saccharomyces jurei. G3 8, 2967–2977. doi: 10.1534/g3.118.200476
Nigam, D., Asthana, M., and Kumar, A. (2017). “Penicillium: a fungus in the wine and beer industries,” in New and Future Developments in Microbial Biotechnology and Bioengineering: Penicillium System Properties and Applications, ed. A. A. Rastegari (Amsterdam: Elsevier), 187–200.
Olivares-Marin, I. K., González-Hernández, J. C., Regalado-Gonzalez, C., and Madrigal-Perez, L. A. (2018). Saccharomyces cerevisiae exponential growth kinetics in batch culture to analyze respiratory and fermentative metabolism. J. Vis. Exp. 139:e58192. doi: 10.3791/58192
Onetto, C. A., Borneman, A. R., and Schmidt, S. A. (2020). Investigating the effects of Aureobasidium pullulans on grape juice composition and fermentation. Food Microbiol. 90:103451. doi: 10.1016/j.fm.2020.103451
Otero, J. M., Vongsangnak, W., Asadollahi, M. A., Olivares-Hernandes, R., Maury, J., Farinelli, L., et al. (2010). Whole genome sequencing of Saccharomyces cerevisiae: from genotype to phenotype for improved metabolic engineering applications. BMC Genomics 11:723. doi: 10.1186/1471-2164-11-723
Papademas, P., Kamilari, E., Aspri, M., Anagnostopoulos, D. A., Mousikos, P., Kamilaris, A., et al. (2020). Investigation of the cyprus donkey milk bacterial diversity by 16S rDNA high-throughput sequencing on a cyprus donkey farm. J. Dairy Sci. 104, 167–178. doi: 10.3168/jds.2020-19242
Petruzzi, L., Capozzi, V., Berbegal, C., Corbo, M. R., Bevilacqua, A., Spano, G., et al. (2017). Microbial resources and enological significance: opportunities and benefits. Front. Microbiol. 8:995. doi: 10.3389/fmicb.2017.00995
Piao, H., Hawley, E., Kopf, S., DeScenzo, R., Sealock, S., Henick-Kling, T., et al. (2015). Insights into the bacterial community and its temporal succession during the fermentation of wine grapes. Front. Microbiol. 6:809. doi: 10.3389/fmicb.2015.00809
Pinto, C., Pinho, D., Cardoso, R., Custódio, V., Fernandes, J., Sousa, S., et al. (2015). Wine fermentation microbiome: a landscape from different Portuguese wine appellations. Front. Microbiol. 6:905. doi: 10.3389/fmicb.2015.00905
Pinto, C., Pinho, D., Sousa, S., Pinheiro, M., Egas, C., and Gomes, A. C. (2014). Unravelling the diversity of grapevine microbiome. PLoS One 9:e85622. doi: 10.1371/journal.pone.0085622
Portillo Mdel, C., Franquès, J., Araque, I., Reguant, C., and Bordons, A. (2016). Bacterial diversity of grenache and carignan grape surface from different vineyards at priorat wine region (Catalonia, Spain). Int. J. Food Microbiol. 219, 56–63. doi: 10.1016/j.ijfoodmicro.2015.12.002
Portillo Mdel, D. C., and Mas, A. (2016). Analysis of microbial diversity and dynamics during wine fermentation of Grenache grape variety by high-throughput barcoding sequencing. LWT Food Sci. Technol. 72, 317–321. doi: 10.1016/j.lwt.2016.05.009
Prasongsuk, S., Lotrakul, P., Ali, I., Bankeeree, W., and Punnapayak, H. (2018). The current status of Aureobasidium pullulans in biotechnology. Folia Microbiol. 63, 129–140. doi: 10.1007/s12223-017-0561-4
Prendes, L. P., Merín, M. G., Zachetti, V. G. L., Pereyra, A., Ramirez, M. L., and Morata de Ambrosini, V. I. (2021). Impact of antagonistic yeasts from wine grapes on growth and mycotoxin production by Alternaria alternata. J. Appl. Microbiol. 131, 833–843. doi: 10.1111/jam.14996
Rai, A. K., and Jeyaram, K. (2015). “Health benefits of functional proteins in fermented foods,” in Health Benefits of Fermented Foods and Beverages, ed. J. Prakash Tamang (Boca Raton, MIA: CRC Press), 455–474.
Renouf, V., Claisse, O., and Lonvaud-Funel, A. (2005). Understanding the microbial ecosystem on the grape berry surface through numeration and identification of yeast and bacteria. Austr. J. Grape Wine Res. 11, 316–327. doi: 10.1111/j.1755-0238.2005.tb00031.x
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Seixas, I., Barbosa, C., Mendes-Faia, A., Güldener, U., Tenreiro, R., Mendes-Ferreira, A., et al. (2019). Genome sequence of the non-conventional wine yeast Hanseniaspora guilliermondii UTAD222 unveils relevant traits of this species and of the Hanseniaspora genus in the context of wine fermentation. DNA Res. 26, 67–83. doi: 10.1093/dnares/dsy039
Sharma, R. R., Singh, D., and Singh, R. (2009). Biological control of postharvest diseases of fruits and vegetables by microbial antagonists: a review. Biol. Control 50, 205–221. doi: 10.1016/j.biocontrol.2009.05.001
Sirén, K., Mak, S. S. T., Melkonian, C., Carøe, C., Swiegers, J. H., Molenaar, D., et al. (2019). Taxonomic and functional characterization of the microbial community during spontaneous in vitro fermentation of riesling must. Front. Microbiol. 10:697. doi: 10.3389/fmicb.2019.00697
Stefanini, I., Albanese, D., Cavazza, A., Franciosi, E., De Filippo, C., Donati, C., et al. (2016). Dynamic changes in microbiota and mycobiota during spontaneous “Vino Santo Trentino” fermentation. Microb. Biotechnol. 9, 195–208. doi: 10.1111/1751-7915.12337
Stefanini, I., and Cavalieri, D. (2018). Metagenomic approaches to investigate the contribution of the vineyard environment to the quality of wine fermentation: potentials and difficulties. Front. Microbiol. 9:991. doi: 10.3389/fmicb.2018.00991
Sternes, P. R., Lee, D., Kutyna, D. R., and Borneman, A. R. (2017). A combined meta-barcoding and shotgun metagenomic analysis of spontaneous wine fermentation. Gigasci 6, 1–10. doi: 10.1093/gigascience/gix040
Swiegers, J. H., Bartowsky, E. J., Henschke, P. A., and Pretorius, I. S. (2005). Yeast and bacterial modulaton of wine aroma and flavour. Aust. J. Grape Wine Res. 11, 139–173. doi: 10.1111/j.1755-0238.2005.tb00285.x
Taylor, M. W., Tsai, P., Anfang, N., Ross, H. A., and Goddard, M. R. (2014). Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 16, 2848–2858. doi: 10.1111/1462-2920.12456
Tondini, F., Jiranek, V., Grbin, P. R., and Onetto, C. A. (2018). Genome sequence of Australian indigenous wine yeast Torulaspora delbrueckii COFT1 using nanopore sequencing. Genome Announc. 6, e321–e318. doi: 10.1128/genomeA.00321-18
Tristezza, M., Tufariello, M., Capozzi, V., Spano, G., Mita, G., and Grieco, F. (2016). The oenological potential of Hanseniaspora uvarum in simultaneous and sequential co-fermentation with Saccharomyces cerevisiae for industrial wine production. Front. Microbiol. 7:670. doi: 10.3389/fmicb.2016.00670
Varela, C., and Borneman, A. R. (2017). Yeasts found in vineyards and wineries. Yeast 34, 111–128. doi: 10.1002/yea.3219
Wang, H. L., Hopfer, H., Cockburn, D. W., and Wee, J. (2021). Characterization of microbial dynamics and volatile metabolome changes during fermentation of chambourcin hybrid grapes from two Pennsylvania regions. Front. Microbiol. 11:3454. doi: 10.3389/fmicb.2020.614278
Wei, Y., Wu, Y., Yan, Y., Zou, W., Xue, J., Ma, W., et al. (2018). High-throughput sequencing of microbial community diversity in soil, grapes, leaves, grape juice and wine of grapevine from China. PLoS One 13:e0193097. doi: 10.1371/journal.pone.0193097
Whipps, J. M., Hand, P., Pink, D., and Bending, G. D. (2008). Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 105, 1744–1745. doi: 10.1111/j.1365-2672.2008.03906.x
Zarraonaindia, I., Owens, S. M., Weisenhorn, P., West, K., Hampton-Marcell, J., Lax, S., et al. (2015). The soil microbiome influences grapevine-associated microbiota. mBio 6, e2527–e2514. doi: 10.1128/mBio.02527-14
Zhong, W., Jeffries, M. W., and Georgopapadakou, N. H. (2000). Inhibition of inositol phosphorylceramide synthase by aureobasidin A in Candida and Aspergillus species. Antimicrobial. Agents Chemother. 44, 651–653. doi: 10.1128/AAC.44.3.651-653.2000
Zoecklein, B. W., Williams, J. M., and Duncan, S. E. (2000). Effect of sour rot on the composition of white riesling (Vitis vinifera L.) Grapes. Small Fruits Rev. 1, 63–77. doi: 10.1300/J301v01n01_08
Keywords: wine, fermentation, microbiome, HTS, terroir, amplicon sequencing, 16S rRNA, ITS
Citation: Kamilari E, Mina M, Karallis C and Tsaltas D (2021) Metataxonomic Analysis of Grape Microbiota During Wine Fermentation Reveals the Distinction of Cyprus Regional terroirs. Front. Microbiol. 12:726483. doi: 10.3389/fmicb.2021.726483
Received: 16 June 2021; Accepted: 23 August 2021;
Published: 22 September 2021.
Edited by:
Antonio Bevilacqua, University of Foggia, ItalyReviewed by:
Renato Leal Binati, University of Verona, ItalyCopyright © 2021 Kamilari, Mina, Karallis and Tsaltas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dimitrios Tsaltas, ZGltaXRyaXMudHNhbHRhc0BjdXQuYWMuY3k=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.