- 1Center of Agriculture, Food, Environment, University of Trento, San Michele all’Adige, Italy
- 2Department of Sustainable Agro-Ecosystems and Bioresources, Research and Innovation Centre, Fondazione Edmund Mach, San Michele all’Adige, Italy
The rhizosphere is a dynamic region governed by complex microbial interactions where diffusible communication signals produced by bacteria continuously shape the gene expression patterns of individual species and regulate fundamental traits for adaptation to the rhizosphere environment. Lysobacter spp. are common bacterial inhabitants of the rhizosphere and have been frequently associated with soil disease suppressiveness. However, little is known about their ecology and how diffusible communication signals might affect their behavior in the rhizosphere. To shed light on the aspects determining rhizosphere competence and functioning of Lysobacter spp., we carried out a functional and transcriptome analysis on the plant beneficial bacterium Lysobacter capsici AZ78 (AZ78) grown in the presence of the most common diffusible communication signals released by rhizosphere bacteria. Mining the genome of AZ78 and other Lysobacter spp. showed that Lysobacter spp. share genes involved in the production and perception of diffusible signal factors, indole, diffusible factors, and N-acyl-homoserine lactones. Most of the tested diffusible communication signals (i.e., indole and glyoxylic acid) influenced the ability of AZ78 to inhibit the growth of the phytopathogenic oomycete Pythium ultimum and the Gram-positive bacterium Rhodococcus fascians. Moreover, RNA-Seq analysis revealed that nearly 21% of all genes in AZ78 genome were modulated by diffusible communication signals. 13-Methyltetradecanoic acid, glyoxylic acid, and 2,3-butanedione positively influenced the expression of genes related to type IV pilus, which might enable AZ78 to rapidly colonize the rhizosphere. Moreover, glyoxylic acid and 2,3-butanedione downregulated tRNA genes, possibly as a result of the elicitation of biological stress responses. On its behalf, indole downregulated genes related to type IV pilus and the heat-stable antifungal factor, which might result in impairment of twitching motility and antibiotic production in AZ78. These results show that diffusible communication signals may affect the ecology of Lysobacter spp. in the rhizosphere and suggest that diffusible communication signals might be used to foster rhizosphere colonization and functioning of plant beneficial bacteria belonging to the genus Lysobacter.
Introduction
The soil is one of the largest microbial reservoirs on Earth, where millions of bacteria and fungi, and less frequently archaea, algae, and protozoa, interact with each other and the plants. In particular, the rhizosphere, the soil compartment influenced by the root exudates, is a hot spot of microbes, as root exudates are a major carbon source for soil microorganisms and a driving force of their population density and activities (Raaijmakers et al., 2009).
Lysobacter spp. belonging to the Xanthomonadaceae family are commonly found in agricultural soils (Lee et al., 2006; Postma et al., 2008; Choi et al., 2014) and especially in the plant rhizosphere. Indeed, they have been found in high abundance in the rhizosphere of maize (Schmalenberger and Tebbe, 2003; García-Salamanca et al., 2013), potato (Van Overbeek and Van Elsas, 2008; Turnbull et al., 2012), soybean (Liang et al., 2014), switchgrass (Rodrigues et al., 2018), and common bean (Pérez-Jaramillo et al., 2019). In terms of ecosystem, several studies have associated Lysobacter spp. with the phenomenon of soil disease suppressiveness (Puopolo et al., 2018). Several studies even showed that the application of Lysobacter spp. reduced diseases caused by different phythopatogenic microorganisms in several crops (Ji et al., 2008; Ko et al., 2009; Puopolo et al., 2010). Yet the sole application of Lysobacter spp. in soils did not always lead to an effective control of phytopathogenic microorganisms probably due to the need to interact with a specific microbial community to become rhizosphere competent (Postma et al., 2009; Gómez Expósito et al., 2015).
Microbial intra- and interspecies signaling occurring via diffusible communication signals plays an important role in soil microbial interactions (Abisado et al., 2018). Diffusible communication signals coordinate interactions both within a species and between species and modulate microbial physiological traits such as motility, attachment, biofilm formation, and biosynthesis of secondary metabolites (Venturi and Keel, 2016). N-acyl-homoserine lactones (AHLs), produced by Gram-negative plant-associated bacteria, are among the most studied diffusible communication signals (Case et al., 2008; Papenfort and Bassler, 2016). Several works showed that endogenous and exogenous AHLs play an essential role in multiple bacterial physiological and biochemical behaviors (Stephens and Bentley, 2020) and can even act as interkingdom signals (Venturi and Fuqua, 2013). Diffusible signal factors (DSFs) are another subgroup of diffusible communication signals produced by Gram-negative bacteria; and they have been linked to virulence, motility, biofilm production, and extracellular enzyme production (Ryan et al., 2015). Xanthomonadaceae family also use diffusible factors (DFs) as diffusible communication signals; these signals are involved in the regulation of secondary metabolites biosynthesis and antioxidant activity (He et al., 2011; Qian et al., 2013; Zhou et al., 2013). In addition, plant-associated bacteria produce volatile organic compounds (VOCs) that are involved in communication and competition between physically separated soil microorganisms (Schmidt et al., 2015). Among VOCs, indole (IND) is a ubiquitous interkingdom signal that influences antibiotic resistance, motility, biofilm formation, and virulence and has the potential to be a diffusible communication signal (Lee and Lee, 2010). 2,3-Butanedione (BUT) and glyoxylic acid (GLY) are other VOCs mediating changes in gene expression related to motility and antibiotic resistance (Kim et al., 2013).
Based on this body of knowledge, it is conceivable that when Lysobacter spp. are applied to the rhizosphere, they will firstly perceive diffusible communication signals produced by the indigenous microbial community before establishing any physical interaction with other rhizosphere-associated microorganisms. The perception of these diffusible communication signals might lead to changes in their transcriptome, which in turn might ultimately lead to changes in Lysobacter spp. rhizosphere competence and their ability to control plant pathogens. Indeed, it has been shown that DSFs, DFs, and IND regulate the biosynthesis of the heat-stable antifungal factor (HSAF), a potent antifungal compound, and twitching motility in Lysobacter enzymogenes (Qian et al., 2013; Han et al., 2017; Su et al., 2017; Feng et al., 2019). However, with the only exception of the involvement of DSFs and AHLs in Lysobacter brunescens behavior (Ling et al., 2019a,b), a complete overview of the overall effect of diffusible communication signals in the ecology of Lysobacter spp. in the rhizosphere has not been described yet.
In this regard, we aimed at unveiling the response of Lysobacter spp. to diffusible communication signals. To that end, we used Lysobacter capsici AZ78 (AZ78), a model plant beneficial bacterium isolated from the rhizosphere of tobacco plants (Puopolo et al., 2014) provided with physiological fundamental traits to survive in the rhizosphere (Brescia et al., 2020). Firstly, gene-encoding proteins involved in cell–cell communication systems were identified by genome mining. Next, we carried out functional experiments aimed at assessing changes in AZ78 cell growth and antimicrobial activity upon exposure to 13-methyltetradecanoic acid (L. enzymogenes DSF-like molecule, LeDSF3, Han et al., 2015), IND, GLY, BUT, 3-hydroxybenzoic acid (3HBA), 4-hydroxybenzoic acid (4BHA), N-(3-hexanoyl)-L-homoserine lactone, N-(3-oxooctanoyl)-L-homoserine lactone, and N-(3-oxododecanoyl)-L-homoserine lactone. Simultaneously, gene expression profiling of AZ78 exposed to the above-mentioned diffusible communication signals was carried out by high-throughput RNA-Seq.
Materials and Methods
Microorganisms and Diffusible Communication Signals
Bacterial strains (Supplementary Table 1) were routinely grown on Nutrient Agar (NA; Oxoid, Basingstoke, United Kingdom) at 27°C. The phytopathogenic oomycete Pythium ultimum was maintained on Potato Dextrose Agar (Oxoid) at 25°C.
LeDSF3 was obtained from Avanti Polar Lipids (Alabaster, AL, United States). IND, GLY, BUT, 3HBA, 4BHA, N-(3-hexanoyl)-L-homoserine lactone, N-(3-oxooctanoyl)-L-homoserine lactone, and N-(3-oxododecanoyl)-L-homoserine lactone were purchased from Merck (Sigma-Aldrich, Darmstadt, Germany). Aqueous stock solutions were prepared, except for LeDSF3 and the mixture of AHLs, which were prepared in pure methanol.
Genome Mining
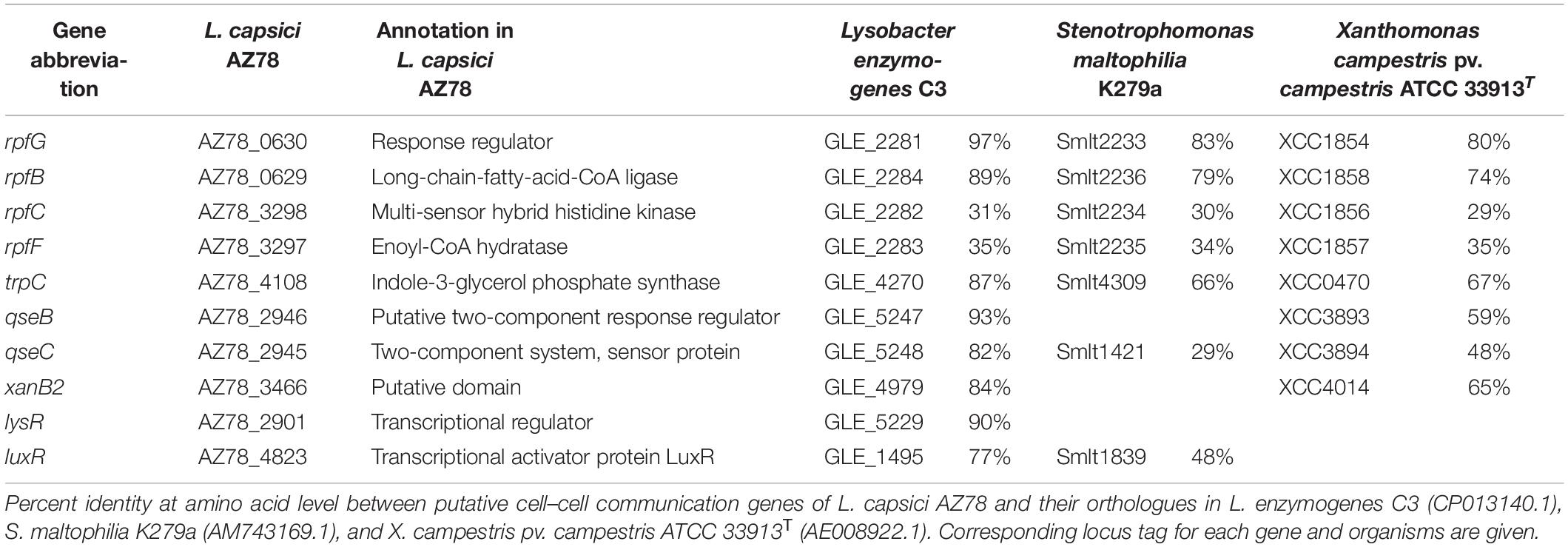
AZ78 genome was mined to identify putative genes involved in cell–cell communication systems using nucleotide and protein sequence comparison. Genes from L. enzymogenes C3, Stenotrophomonas maltophilia (Sm) K279a, and Xanthomonas campestris pv. campestris (Xcc) ATCC 33913T were aligned against AZ78 genome, using RAST (Aziz et al., 2008) to identify putative AZ78 genes responsible for diffusible communication signal synthesis, reception, and regulation using a cut-off of 1 × 10–5 at amino acid level. Putative genes were analyzed with BLASTP (Johnson et al., 2008), and length >70 and identity >70% at amino acid level were used as threshold. Identified gene clusters encoding putative proteins involved in cell–cell communication systems in AZ78 were then used to mine the Lysobacter spp. genomes, following the methodology described above. All genomes were downloaded from the National Center for Biotechnology Information (NCBI)1 (Supplementary Table 1). For the phylogenetic analyses, nucleotide sequences were aligned using ClustalW (Thompson et al., 1994). Evolutionary distances were assessed by applying Kimura’s two-parameter model (Kimura, 2020); and the best phylogenetic trees were inferred by neighbor-joining method (Saitou and Nei, 1987) implemented in MEGA 7 (Kumar et al., 2016). Confidence values for nodes in the trees were generated by bootstrap analysis (Felsenstein, 1985) using 1,000 permutations of the data sets.
Assessment of Diffusible Communication Signal Production
Production of AHLs by AZ78 and Lysobacter spp. type strains was assessed by evaluating their ability to restore violacein production in Chromobacterium violaceum CV026 and/or to promote lacZ transcription in Agrobacterium tumefaciens NT1 (pZLR4) as previously described (Cha et al., 1998; Steindler and Venturi, 2007). In brief, candidate strains were grown on NA close to each reporter strain to form a “T,” and the phenotypic change associated with the presence of AHLs was observed as a gradient with the most response observed at the meeting point of the two strains. Medium used in assays involving A. tumefaciens was supplemented with 1.6 μg/ml of X-Gal (5-bromo-4-chloro-3-indolyl β-D-galactopyranoside, Sigma-Aldrich). Likewise, the ability to release DSF was determined using the bacterial reporter strain Xcc 8523 pL6engGUS according to Slater et al. (2000). Briefly, Xcc 8523 pL6engGUS was grown in 10 ml of NYG (5 g/L of peptone, 3 g/L of yeast extract, and 20 g/L of glycerol) supplemented with 10 μg/ml of tetracycline to an optical density (OD600) of 0.7. Cells were harvested by centrifugation and reconstituted in 1 ml of fresh NYG, added to 100 ml of cold NGY containing 1% BD Difco Noble Agar (BD Biosciences, Sparks, MD, United States), supplemented with 80 μg/ml of X-Glu (5-bromo-4-chloro-3-indolyl β-D-glucuronide sodium salt; Sigma-Aldrich), and plated into petri plates. Candidate strains were then pin inoculated and incubated for 48 h at 27°C. The presence of a blue halo around the colony indicated DSF activity. Pseudomonas chlororaphis M71 (Puopolo et al., 2011) was used as an AHL-positive control, whereas Xcc 8004 was used as a DSF positive control (Barber et al., 1997). For each condition, five replicates were used, and the experiment was repeated.
Effect of Diffusible Communication Signals on Antimicrobial Activity
The effect of diffusible communication signals on AZ78 antimicrobial activity was evaluated on Rhizosphere Mimicking Agar (RMA) (Brescia et al., 2020). At first, preliminary experiments where AZ78 was grown on RMA amended with different concentrations of the selected compounds were carried out to select minimum effective concentrations—the lowest concentrations showing the highest effect—of diffusible communication signals. Thereafter, the final experimental design was made up of eight treatments (Supplementary Table 2). Inhibitory activity of AZ78 against P. ultimum was evaluated by using the classic dual-culture method as previously described (Puopolo et al., 2016). In brief, 10 μl of AZ78 cell suspension (1 × 108 CFU/ml) were spot-inoculated at 3 cm of the edge of a plate. After 48-h incubation at 27°C, mycelium plugs (4 mm) were cut from the edge of 1-week-old P. ultimum plate, placed at 2.5-cm distance from AZ78, and incubated at 25°C for 168 h. AZ78 activity against Rhodococcus fascians LMG 3605 was determined by spot-inoculating 10 μl of AZ78 cell suspension (1 × 108 CFU/ml) in the center of an RMA plate (Puopolo et al., 2016). After 48-h incubation at 27°C, AZ78 cells were killed by exposure to chloroform vapor for 60 min (Puopolo et al., 2016). Dishes were aerated under a laminar flow for 60 min, overlaid with 4 ml of 0.45% agar phosphate-buffered saline (PBS) containing R. fascians LMG 3605 (1 × 107 CFU/ml) and incubated at 27°C for 72 h. RMA dishes seeded only with P. ultimum or R. fascians LMG 3605 were used as control.
Pictures were obtained with Bio-Rad Quantity One software implemented in a Bio-Rad GelDoc Imaging system (Bio-Rad Laboratories, Hercules, CA, United States). Inhibitory activity was quantified by scoring P. ultimum or R. fascians LMG 3605 growth area (cm2) using ImageJ 1.52a (Schneider et al., 2012) and calculated according to the formulas below:
where
The effect of diffusible communication signals on AZ78 antibacterial activity was assessed as follows:
In all cases, treatments included five replicates, and experiments were repeated.
Evaluation of Diffusible Communication Signal Effect on Cell Growth
The effect of diffusible communication signals on AZ78 cell growth rate was assessed on 1/10 Tryptic Soy Broth (Oxoid) amended with each diffusible communication signal (Supplementary Table 2). AZ78 (starting concentration 1 × 107 CFU/ml) was grown at 27°C on a 96-well plate (200 μl), and absorbance at 600 mm was recorded on a microplate reader (Synergy 2 Multi-Mode Microplate Reader, BioTek, Winooski, VT, United States). Non-inoculated media were used as blank. For each condition, five replicates were used. The experiment was repeated.
RNA Extraction
The AZ78 response to diffusible communication signals was evaluated on RMA, and the experimental design was made up of eight treatments in triplicate (Supplementary Table 2). Ten microliters of AZ78 cell suspension (1 × 1010 CFU/ml) were spot-inoculated in the center of an RMA plate and incubated at 27°C for 48 h. Plugs (7-mm diameter) were collected from the AZ78 macrocolonies, immediately frozen in liquid nitrogen, and stored at −80°C. Frozen samples were processed according to Brescia et al. (2020), and total RNA was extracted using Spectrum Plant Total RNA Kit (Sigma-Aldrich). DNase treatment was performed with the RNase-Free DNase set (Qiagen, Hilden, Germany). RNA integrity and concentration were assessed using a 2200 TapeStation System (Agilent Technologies, Santa Clara, CA, United States) and a Qubit 4 Fluorometer (Thermo Fisher Scientific, Carlsbad, CA, United States) with Qubit RNA BR assay kit (Thermo Fisher Scientific), respectively (Supplementary Table 3).
Illumina Sequencing and Mapping to the Reference Genomes
Library construction and Illumina Sequencing were carried out at Fasteris (Plan-les-Ouates, Switzerland). Ribosomal RNA (rRNA) depletion was performed using the Ribo-Zero rRNA Removal Kits (Bacteria) (Illumina, San Diego, CA, United States). Complementary DNA (cDNA) libraries were synthesized using TruSeq Stranded mRNA Library Prep (Illumina, United States); they were multiplexed (two libraries per lane); and paired-end reads of 150 nucleotides were obtained using an Illumina HiSeq 4000 (Illumina), resulting in ∼7–42 million reads per sample (Supplementary Table 4). Raw sequences were deposited at the Sequence Read Archive of the NCBI under BioProject number PRJNA714393.
Sequence analysis was carried out using Omicsbox 1.3.11.2 Illumina HiSeq data were assessed for quality using FastQC (Andrews, 2010). Raw reads for each sample were trimmed to increase overall quality using Trimmomatic 0.38 (Bolger et al., 2014). The resulting reads were aligned to AZ78 genome (Supplementary Table 1) using the STAR 2.7.5a (Dobin et al., 2013), and read counts were extracted from STAR alignments using HTSeq (Anders et al., 2015).
Identification of Differentially Expressed Genes and Functional Annotation of RNA-Seq
Genes with zero counts in all replicates were excluded from the analysis, and raw counts were normalized using the trimmed mean of M-values method (Robinson and Oshlack, 2010). Differentially expressed genes (DEGs) were identified using edgeR 3.28.0 (Robinson et al., 2010) using a p-value < 0.01 and a log fold change (FC) of at least onefold upregulation/downregulation as cut-off values. Venn diagrams summarizing DEG distribution were drawn with VennPainter (Lin et al., 2016). Hierarchical clustering and heatmaps were created with TreView3 (Saldanha, 2004).
The protein sequences of all predicted genes (Puopolo et al., 2016) were functionally annotated using Blast2Go3 (Conesa et al., 2005). Default settings were applied, and a minimum E-value of 10–5 was imposed as cut-off. DEGs were further annotated based on the NCBI gene description and classified in 20 functional categories.
Validation of RNA-Seq
First-strand cDNA was synthetized from 600 ng of purified RNA with SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, United States) using random hexamers, according to manufacturer’s instructions. qRT-PCRs were carried out with Platinium SYBR Green qPCR Super-Mix-UDG (Invitrogen, United States), and specific primers (Supplementary Table 5) were designed using Primer3 software (Untergasser et al., 2012). Primer specificity was assessed using PCR before gene expression analysis. qRT-PCRs were run for 50 cycles (95°C for 15 s and 60°C for 45 s) on a LightCycler 480 (Roche Diagnostics, Mannheim, Germany). Each sample was examined in three technical replicates, and dissociation curves were analyzed to verify the specificity of each amplification reaction. LightCycler 480 software, version 1.5 (Roche Diagnostics, Mannheim, Germany) was used to extract cycle threshold (Ct) values based on the second derivative calculation; and the LinReg software, version 11.0, was used to calculate reaction efficiencies for each primer pair (Ruijter et al., 2009). Relative expression levels were calculated according to the Pfaffl equation (Pfaffl, 2001) using AZ78 growing in RMA as calibrator. The housekeeping gene recA (AZ78_1089; Puopolo et al., 2016) was used as constitutive gene for normalization, as its expression was not significantly affected by growth media and conditions (Tomada et al., 2016, 2017; Brescia et al., 2020). The linear relationship between the RNA-Seq log2FC values and the qRT-PCR log2FC values of selected genes was estimated by Pearson’s correlation analysis.
Statistical Analysis
Percentage values were arcsine square root transformed to normalize distributions and to equalize variances. Comparisons between repeated experiments of antimicrobial activity were done using two-way analysis of variance (ANOVA), and the data were pooled when no significant differences were found according to the F-test (p > 0.05). Data were analyzed using one-way ANOVA, and Tukey’s test (α = 0.05) was used to detect significant differences. Statistical analyses were carried out using IBM SPSS Statistics for Windows, version 21.0 (IBM Corp, Armonk, NY, United States).
Results
Cell–Cell Communication Systems in Lysobacter capsici AZ78 Genome
Putative rpf genes were found in the AZ78 genome (Table 1 and Supplementary Figures 1, 2). The rpfF/rpfC region (3,947,548–3,948,450 bp) was located far from the rpfG/rpfB region (857,114–859,152 bp) in AZ78 (Supplementary Figure 3a). DSF biosynthesis was confirmed by AZ78 ability to induce the glucuronidase activity in Xcc 8523 pL6engGUS like the control strain Xcc 8004 (Supplementary Figure 3b). As for VOCs, putative gene-encoding IND synthase and QseB/QseC system were found in AZ78 genome and in other Lysobacter spp. (Table 1 and Supplementary Figure 4). Homologues of the chorismatase needed for DF production and the LysR family transcription factor involved in the DF regulatory cascade were also found in AZ78 genome (Table 1 and Supplementary Figure 5). luxR gene, responsible for the detection and response to AHLs, was also present in AZ78 (Table 1 and Supplementary Figure 5). LuxR was not associated with its cognate AHL synthase (LuxI) in AZ78 or in other Lysobacter species, with only exception of Lysobacter daejeonensis GH1-9T having a luxI homologue. The absence of LuxI homologs was confirmed by the inability of AZ78 to restore β-galactosidase activity and violacein production in the reporter strains A. tumefaciens NT1 pZRL4 and C. violaceum CV026, respectively (Supplementary Figure 6). In contrast, L. daejeonensis GH1-9T was able to restore violacein production in C. violaceum CV026, confirming the relation between the presence of luxI and AHL production.
Diffusible Communication Signals Affect Lysobacter capsici AZ78 Antimicrobial Activity and Growth
Preliminary screening for antimicrobial activity (Supplementary Figure 7) led to the selection of minimum effective concentrations of diffusible communication signals (Supplementary Table 2). Exogenous addition of diffusible communication signals to RMA showed no effect on P. ultimum and R. fascians LMG 3605 growth, but it modulated AZ78 antimicrobial activity against P. ultimum and R. fascians LMG 3605 (Figure 1 and Supplementary Figure 7). LeDSF3, 4HBA, and IND decreased AZ78 inhibitory activity against P. ultimum by 5, 22, and 47%, respectively. In contrast GLY, BUT, 3HBA, and AHL increased AZ78 inhibitory activity against P. ultimum up to 7% (Figure 1). LeDSF3, IND, 3HBA, 4HBA, and AHL decreased AZ78 inhibitory activity against R. fascians LMG 3605 up to 31%, while BUT and GLY increased AZ78 antibacterial activity by 9 and 48%, respectively (Figure 1). Changes were particularly relevant for IND and GLY. GLY, BUT, 3HBA, 4HBA, and AHL had no effect on AZ78 growth curves, whereas LeDSF3 and IND slowed down AZ78 growth as compared with the untreated control (Supplementary Figure 8).
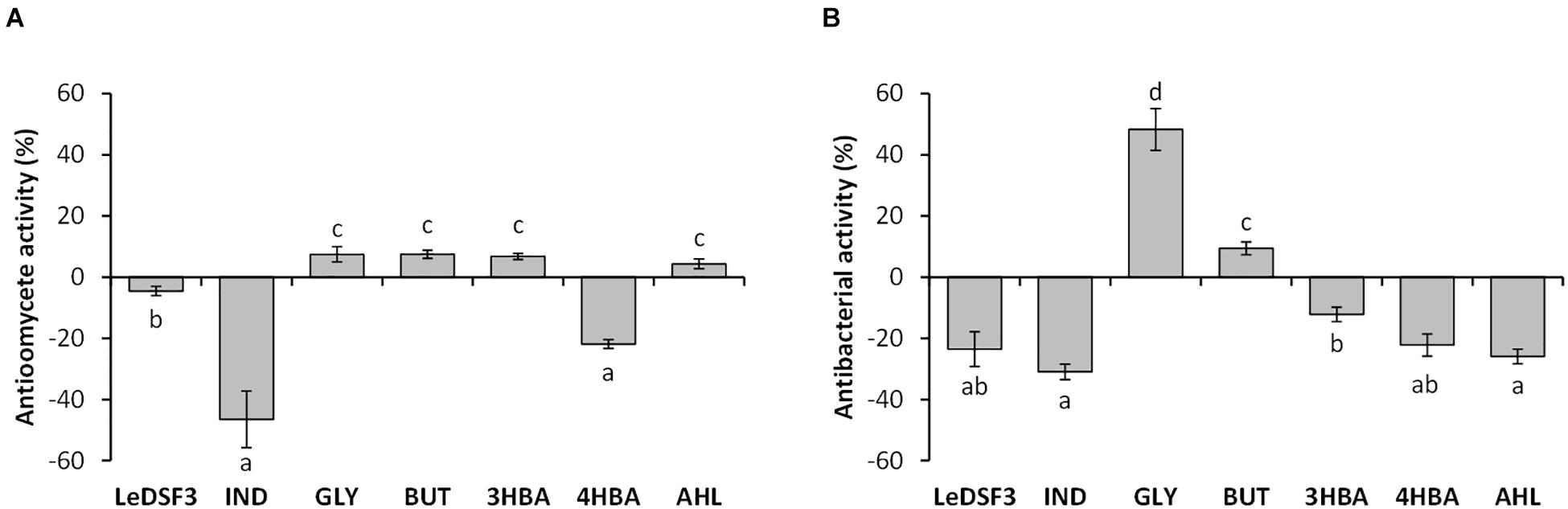
Figure 1. Effect of diffusible communication signals on the inhibitory activity of Lysobacer capsici AZ78 against (A) Pythium ultimum and (B) Rhodococcus fascians. Antioomycete and antibacterial activities are expressed as the mean value and standard error variation (percentage) of the reduction of the mycelium growth area of P. ultimum and R. fascians compared with the control (L. capsici AZ78 in not supplemented media), respectively. LeDSF3, 13-methyltetradecanoic acid 50 μM; IND, indole 500 μM; GLY, glyoxylic acid 0.01 μM; BUT, 2,3-butanedione 0.01 μM; 3HBA, 3-hydroxybenzoic acid 30 μM; 4HBA, 4-hydroxybenzoic acid 50 μM; AHL, mix of N-acyl-homoserine lactones 20 μM. Each treatment included five replicates, and data originating from two independent experiments were pooled. Columns with the same letters are not significantly different according to Tukey’s test (α = 0.05).
Transcriptional Response of Lysobacter capsici AZ78 to Diffusible Communication Signals
The expression of 21% of all AZ78 genes was significantly affected by diffusible communication signals (| log2FC| > 1 and p < 0.01). The largest number of DEGs, 636 genes (about 11.9% of AZ78 transcriptome), was found upon exposure of AZ78 to LeDSF3 (Table 2). This was followed by IND (603 DEGs, 11.3% of total genes), GLY (292 DEGs, 5.5% of total genes), BUT (237 DEGs, 4.4% of total genes), 3HBA (101 DEGs, 1.9% of total genes), and 4BHA (58 DEGs, 1.1% of total genes). The lowest number of DEGs (24, 0.5% of total genes) occurred among cells treated with AHL (Table 2). RNA-Seq results were validated by the relative expression level of 10 selected genes assessed using qRT-PCR (Supplementary Table 5). A close correlation (Pearson’s r = 0.95) was observed between log2FC measured with RNA-Seq and qRT-PCR (Supplementary Figure 9).
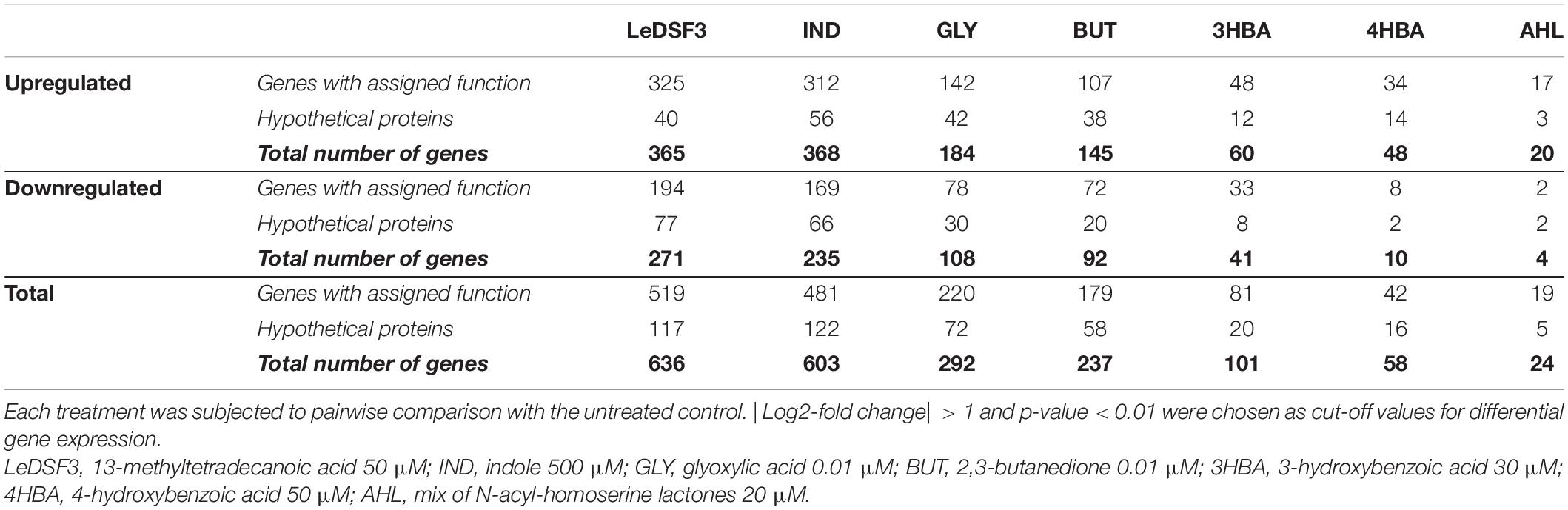
Table 2. Differentially expressed genes in Lysobacter capsici AZ78 in response to diffusible communication signals after 48-h incubation.
Venn diagrams (Supplementary Figure 10) revealed overlaps among the seven conditions, but they did not identify genes modulated by all seven diffusible communication signals. Moreover, a heatmap (Figure 2) showed that GLY and BUT clustered together, and likewise for 3HBA, 4HBA, and AHL. Instead, LeDSF3 and IND grouped independently.
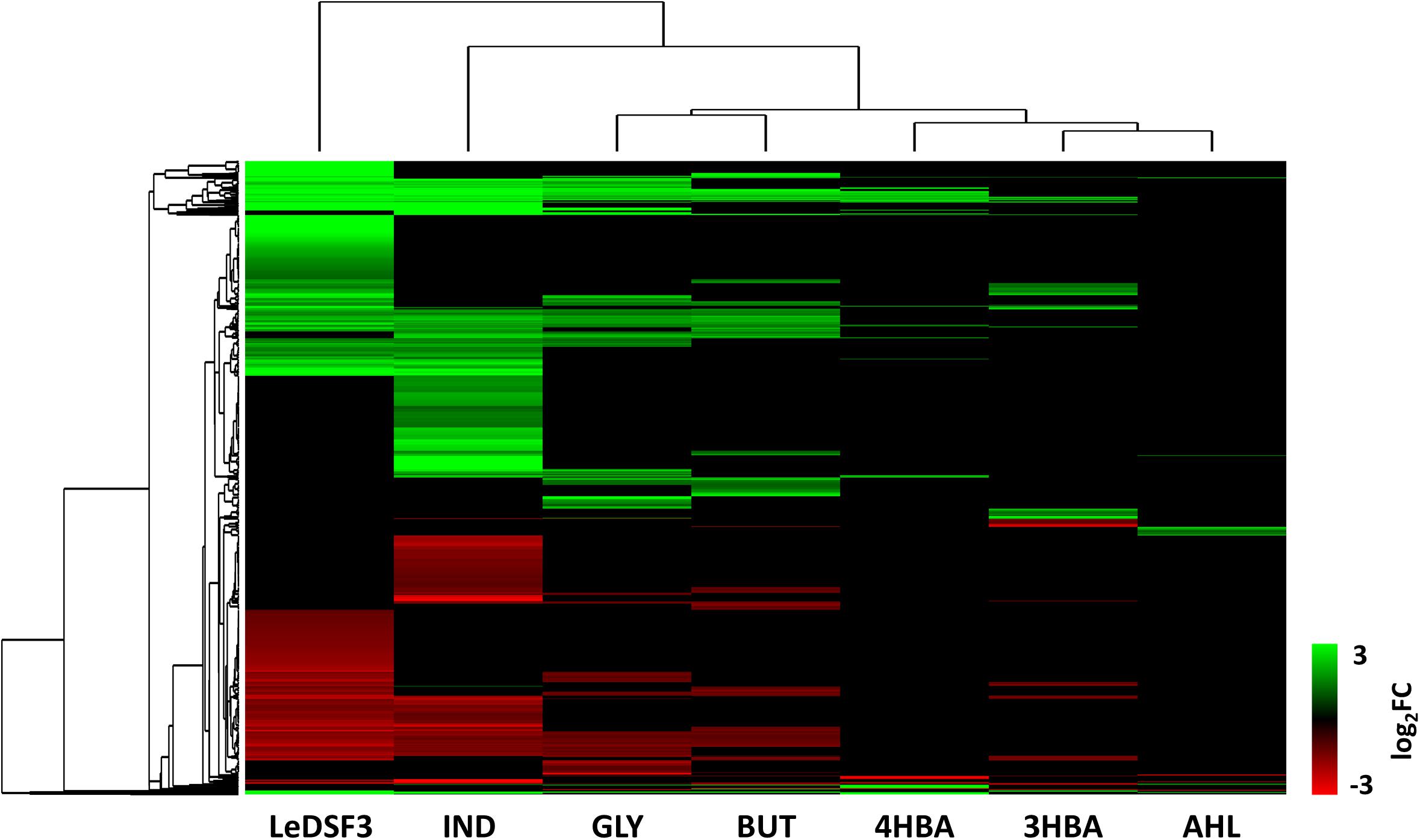
Figure 2. Hierarchical clustering based on log2-fold change (FC) of Lysobacter capsici AZ78 genes differentially expressed (1,115 genes, Supplementary Tables 6–12 for gene expression data). LeDSF3, 13-methyltetradecanoic acid 50 μM; IND, indole 500 μM; GLY, glyoxylic acid 0.01 μM; BUT, 2,3-butanedione 0.01 μM; 3HBA, 3-hydroxybenzoic acid 30 μM; 4HBA, 4-hydroxybenzoic acid 50 μM; AHL, mix of N-acyl-homoserine lactones 20 μM.
The Active Response of Lysobacter capsici AZ78 to Diffusible Communication Signals
Functional annotation of AZ78 genes modulated by diffusible communication signals revealed that upregulated DEGs were mainly related to global metabolism; growth; RNA transcription, and degradation; and transport, phosphotransferase systems, and secretion (Table 3 and Supplementary Figure 11). Conversely, downregulated DEGs were mainly related to DNA metabolism.
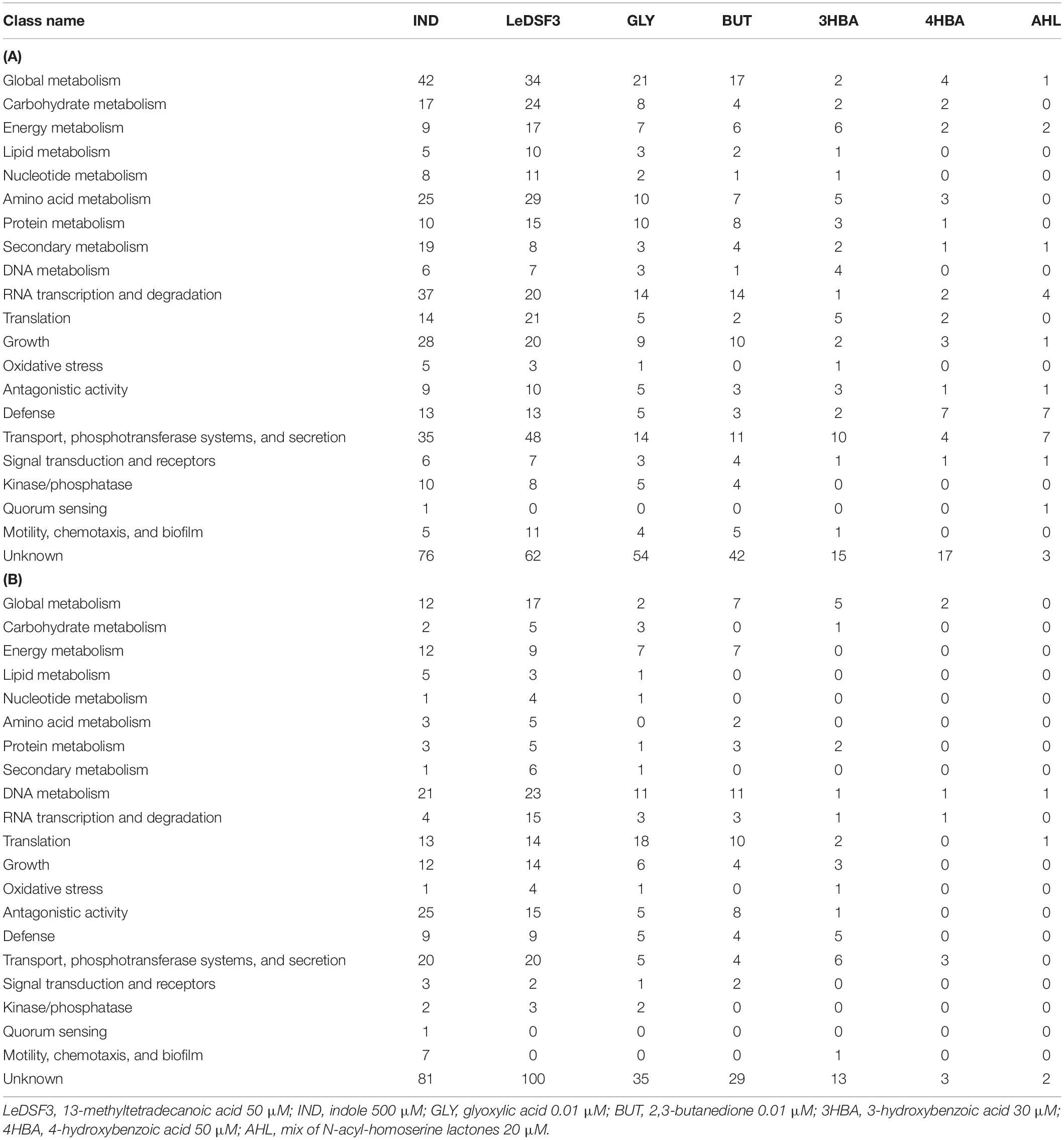
Table 3. Number of (A) upregulated and (B) downregulated differentially expressed genes in Lysobacter capsici AZ78 in response to diffusible communication signals.
LeDSF3 and IND regulated several genes involved in transport, phosphotransferase systems, and secretion; global metabolism; RNA transcription and degradation; and growth (Table 3 and Supplementary Figure 11). In addition, LeDSF3 affected translation and IND antagonism (Table 3 and Supplementary Figure 11). Besides regulating genes related to global metabolism and transport, phosphotransferase systems, and secretion, GLY and BUT modulated a relevant number of genes classified into RNA transcription and degradation and translation, among which tRNA genes were mainly downregulated (Table 3 and Supplementary Figure 11). Genes related to transport, phosphotransferase systems, and secretion and defense were modulated by 4HBA and AHL (Table 3 and Supplementary Figure 11).
Many genes ascribed to transport, phosphotransferase systems, and secretion were involved in drug and metal (particularly iron) transport (Supplementary Tables 6–12). Major facilitator superfamily (MFS) transporters were upregulated by LeDSF3 (Figure 3 and Supplementary Table 6). Resistance–nodulation–division (RND) efflux system genes were upregulated by diffusible communication signals, especially by AHL (Figure 3 and Supplementary Table 12). TonB-dependent receptors involved in the uptake of iron–siderophore complexes or vitamins were upregulated by LeDSF3 and downregulated by IND (Supplementary Tables 6, 7). Additionally, diffusible communication signals upregulated a relevant set of transcription regulators belonging to the AraC, ArsR, TetR, MerR, and MarR families (Supplementary Tables 6–12).
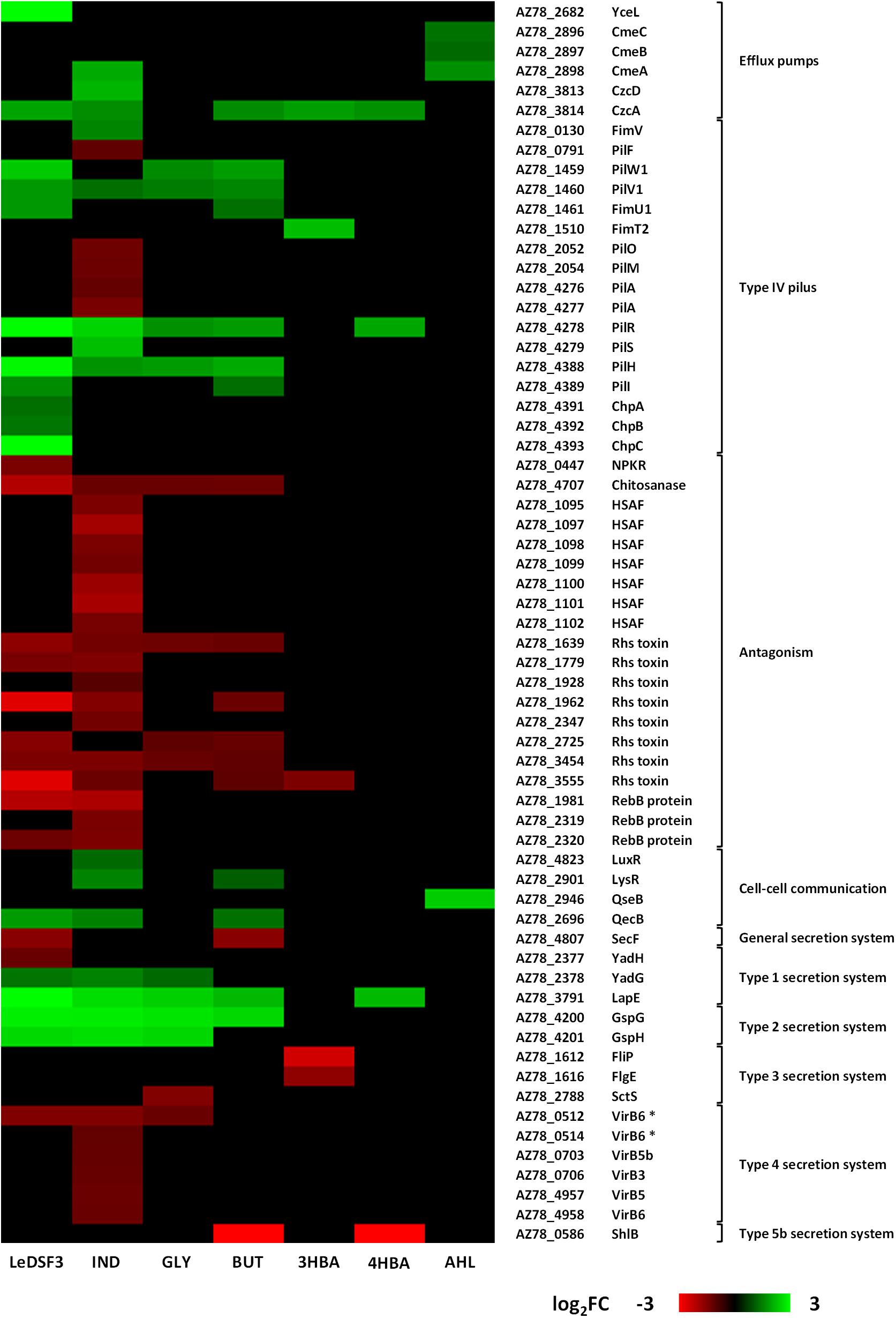
Figure 3. Heatmap based on log2-fold change (FC) of Lysobacter capsici AZ78 genes differentially expressed in response to diffusible communication signals and related to efflux pumps, type IV pilus, antagonism, cell–cell communication systems, and secretion systems. HSAF, heat-stable antifungal factor. VirB6 * represents duplicate gene pairs located through AZ78 genome. LeDSF3, 13-methyltetradecanoic acid 50 μM; IND, indole 500 μM; GLY, glyoxylic acid 0.01 μM; BUT, 2,3-butanedione 0.01 μM; 3HBA, 3-hydroxybenzoic acid 30 μM; 4HBA, 4-hydroxybenzoic acid 50 μM; AHL, mix of N-acyl-homoserine lactones 20 μM.
With the only exception of AHL, diffusible communication signals modulated the expression of genes involved in type IV pilus (T4P) biosynthesis. LeDSF3 upregulated the expression of pilW1-pilV1-fimU1 genes (AZ78_1459–AZ78_1461), related to minor pilins; pilR from two-component system pilR-pilS (AZ78_4278–AZ78_4279); and pilH (AZ78_4388), pilI (AZ78_4389), chpA (AZ78_4391), chpB (AZ78_4392), and chpC (AZ78_4393) from the pilus-specific chemotaxis system (Pil-Chp) (Figure 3 and Supplementary Table 6). Gene-encoding minor pilins, pilRS, and Pil-Chp were also upregulated in GLY, BUT, and 4HBA (Figure 3 and Supplementary Tables 8, 9, 11). IND upregulated the expression of fimV (AZ78_0130), pilV1, pilR-pilS, and pilH, while it downregulated pilF (AZ78_0791), pilO and pilM (AZ78_2052 and AZ78_2054), and the major pilin pilA (AZ78_4276–AZ78_4277) (Figure 3 and Supplementary Table 7). LeDSF3 and IND downregulated a relevant number of involved in antagonism. IND downregulated the biosynthetic gene cluster AZ78_1095–AZ78_1102, responsible for the production of HSAF (Brescia et al., 2020; Figure 3 and Supplementary Table 7). Moreover, IND upregulated a diguanylate cyclase harboring a GGDEF motif (AZ78_4062) possibly related to cyclic-di-GMP (c-di-GMP) biosynthesis (Supplementary Table 7). Other genes related to antagonism, such as gene-encoding Rhs toxins and RebB proteins, responsible for the expression of killing traits, were downregulated by LeDSF3 and IND (Figure 3 and Supplementary Tables 6, 7).
Diffusible communication signals also caused changes in the expression of genes involved in the reception and regulation of diffusible communication signals in AZ78 (Figure 3). Genes related to Type I secretion system (T1SS) were mostly upregulated by all diffusible communication signals, especially by LeDSF3 and IND (Figure 3 and Supplementary Tables 6, 7). Type II secretion system (T2SS) genes, such as gspG (AZ78_4200) and gspH (AZ78_4201), were upregulated by LeDSF3, IND, GLY, and BUT (Figure 3 and Supplementary Tables 6–9). On the contrary, the expression of Type III secretion system (T3SS) was downregulated by GLY and 3HBA (Figure 3 and Supplementary Tables 8, 10). Genes associated with type IV secretion system (T4SS) were downregulated by LeDSF3, IND, and GLY (Figure 3 and Supplementary Tables 6–8). ShlB from the two-partner secretion of Type V secretion system (T5bSS) was downregulated by BUT and 4HBA (Figure 3 and Supplementary Tables 9, 11). Finally, LeDSF3, IND, and BUT also regulated general secretory (Sec) pathways (Figure 3 and Supplementary Tables 6, 7, 9).
Discussion
The behavior of bacterial species in polymicrobial communities mainly relies on communication systems (Venturi and Keel, 2016); and many secreted metabolites characterizing the cooperation among microorganisms, as well as antibiotics and toxins involved in microbial competition, are controlled by diffusible communication signals (Hibbing et al., 2010; Cornforth and Foster, 2013; Schuster et al., 2013). Diffusible communication signals are involved not only in signaling among self-cells but also in the detection of specific cues produced by other strains or species (Cornforth and Foster, 2013). In fact, many bacterial species have receptors for diffusible communication signals that are not produced by the same species, such as LuxR solos, and abundant two-component signaling systems (Cornforth and Foster, 2013). Genome mining results indicate that AZ78 and Lysobacter spp. may (at least) produce DSFs, IND, and DFs and perceive DSFs, IND, DFs, and AHLs. As a consequence, diffusible communication signals (mainly IND and GLY) influenced AZ78 antagonistic activity against the phyopathogenic Gram-positive bacteria R. fascians and the phyopathogenic oomycete P. ultimum. Different intraspecies, interspecies, and interkingdom diffusible communication signals might be used as cues for AZ78 to favor the regulation of molecular pathways related to cell persistence in the rhizosphere or for coercion (Figure 4). Thus, transcriptome profiles showed that diffusible communication signals might contribute to alert AZ78 against toxic compounds produced by other (micro)organisms in the rhizosphere by triggering the expression of gene-encoding efflux pumps that could actively extrude antibiotics, heavy metals, biocides, and solvents (Blanco et al., 2016). In addition, diffusible communication signals might help cells to escape from adverse conditions or to reach nutrients (Chen et al., 2017). For example, LeDSF3, GLY, and BUT upregulated the expression of genes related to the biogenesis of T4P involved in twitching motility. T4P-driven twitching motility is involved in a variety of physiological and social behaviors of a wide range of bacteria (Burrows, 2012; Zhang et al., 2012). For instance, twitching motility is required for colonization and infection of phytopathogenic fungi and oomycetes in Lysobacter spp. (Patel et al., 2011; Tomada et al., 2017), and it seems to be a DSF-dependent trait in L. brunescens and L. enzymogenes (Qian et al., 2013; Feng et al., 2019; Ling et al., 2019b). Interestingly, upregulation of T4P by GLY and BUT came along with increased antimicrobial activity, suggesting that microbial partners producing this kind of diffusible communication signals might encourage AZ78 to form a stable community and stimulate traits responsible for disease suppressiveness in soils. Moreover, GLY and BUT downregulated the transcription of tRNA genes, which might determine a decrease in the tRNA abundance in AZ78 cells. The decrease of tRNA abundance has been already studied in Escherichia coli (Potrykus and Cashel, 2008; Zhong et al., 2015). In this bacterial species, the decrease in tRNA abundance was associated with the ability to rapidly adapt to amino acid starvation (Potrykus and Cashel, 2008) and oxidative stress (Zhong et al., 2015). Thus, it is conceivable that the downregulation of tRNA genes in AZ78 cells upon perception of GLY and BUT might contribute to reduce the negative impact of environmental stresses in AZ78 cells. Nevertheless, this hypothesis needs to be proved in future works. In contrast, IND caused a dysregulation of T4P genes in AZ78 with possible losses of T4P functionality. Accordingly, IND decreases motility and biofilm formation in E. coli (Domka et al., 2006; Bansal et al., 2007; Lee et al., 2008; Mufti et al., 2015), probably as a manner to save energy and regulate growth dynamics (Nadell et al., 2008). In support of this hypothesis, IND diminished the AZ78 cell growth, although it was not possible to formulate a clear conclusion, as knowledge about IND functions is contrasting (Mueller et al., 2007, 2009; Lee et al., 2009).
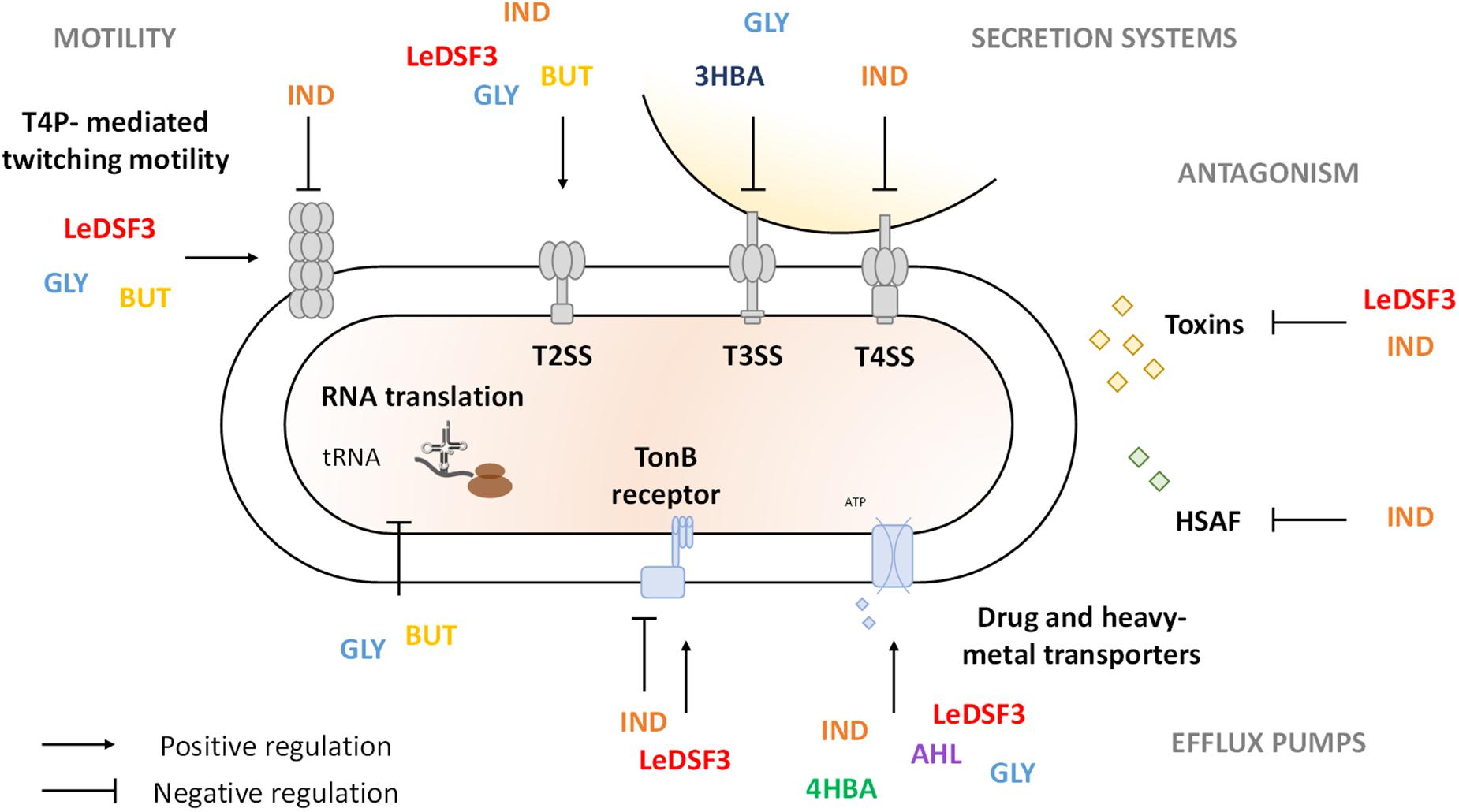
Figure 4. Schematic representation of Lysobacter capsici AZ78 response to diffusible communication signals. LeDSF3, 13-methyltetradecanoic acid; IND, indole; GLY, glyoxylic acid; BUT, 2,3-butanedione; 3HBA, 3-hydroxybenzoic acid; 4HBA, 4-hydroxybenzoic acid; AHL, N-acyl-homoserine lactones.
In addition, Lysobacter cells might contribute to disease suppressiveness of soils by producing extracellular lytic enzymes and antibiotics. Previous findings showed that HSAF biosynthesis is positively regulated by DSF, IND, and 4HBA in Lysobacter spp. (Qian et al., 2013; Han et al., 2017; Ling et al., 2019b). Yet in AZ78, IND reduced antioomycete activity and downregulated the expression of the HSAF biosynthetic gene cluster. The downregulation of HSAF related genes by IND might be associated with the simultaneous upregulation of the LuxR solo (AZ78_4823), as previously reported in L. enzymogenes OH11, where overexpression of lesR (LuxR homologue) leads to a decrease in HSAF production (Qian et al., 2014; Xu et al., 2017). Moreover, IND upregulated several transcription regulators—among which various tetR repressors (Ramos et al., 2005), like AZ78_0770 and AZ78_3232—that might be involved in HSAF biosynthesis regulation in AZ78, as found for LetR (a TetR family protein) in L. enzymogenes OH11 (Wang et al., 2017). The expression of HSAF biosynthetic cluster is also negatively regulated by cyclic-di-GMP (c-di-GMP) in L. enzymogenes OH11 (Chen et al., 2017). Interestingly, IND upregulated the expression of a diguanylate cyclase (AZ78_4062) that might be involved in c-di-GMP biosynthesis, implying a regulation role of c-di-GMP in HSAF production in AZ78. Besides producing secondary metabolites with antimicrobial activity, AZ78 might produce diffusible proteinaceous toxins and toxins deployed by contact-dependent systems, such as Rhs toxins, which mediate growth inhibition of neighboring cells in Dickeya dadantii (Koskiniemi et al., 2013), or R-bodies, which are responsible for cell membrane disruption and toxins delivery in several bacterial genera (Raymann et al., 2013; Matsuoka et al., 2017). Thus, the downregulation of several rhs and reb genes required for Rhs toxins and R-bodies synthesis by LeDSF3 and IND might have contributed to lower AZ78 the antioomycete and antibacterial activities. Moreover, AZ78 downregulated signal transduction pathways in the presence of IND, such as TonB-dependent receptors, which play a key role in microbial competition with the uptake of iron–siderophore complex or vitamins (Braun, 1995).
Bacteria often use secretion systems to manipulate and kill rival bacterial and eukaryotic cells (Tseng et al., 2009; Green and Mecsas, 2016). Of those, T3SS, T4SS, and T6SS are related to the establishment of pathogenic interactions with microbial hosts in Lysobacter spp. (de Bruijn et al., 2015; Yang et al., 2020; Shen et al., 2021). Thus, modulation of genes related to secretion systems might result in gain/loss of ability to compete with other (micro)organisms (Ling et al., 2019a). In agreement with this statement, IND downregulated T4SS and decreased antimicrobial activity in AZ78. Downregulation of T4SS by IND might be related to the overexpression of diguanylate cyclases (e.g., AZ78_4062), responsible for c-di-GMP increase and T4SS inactivation in A. tumefaciens (McCarthy et al., 2019). However, T3SS was downregulated by GLY and 3HBA with no decrease in AZ78 toxic activity, suggesting that it was probably repressed to save energy under conditions where it does not provide an advantage, as found in Pseudomonas aeruginosa (Bleves et al., 2005), Vibrio harveyi (Ruwandeepika et al., 2015), and Yersinia pseudotuberculosis (Atkinson et al., 2011).
Conclusion
Overall, functional and transcriptome analysis of AZ78 shed light on the key role of signaling communication systems on the recruiting and shaping of AZ78 in the rhizosphere. Our results show that GLY and BUT might facilitate AZ78 rhizosphere competence and soil suppressiveness to plant pathogens. On the other hand, IND might prevent AZ78 from growing at high cell densities and decrease motility. Moreover, IND and LeDSF3 might decrease AZ78 ability to control phytopathogenic microorganisms. Manipulating diffusible communication signals levels in the rhizosphere could therefore provide efficient means to favor persistence and functioning of specific groups of beneficial bacteria, such as Lysobacter strains, at the root–soil interface.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA714393.
Author Contributions
AB and GP conceived the study, performed the experiments, analyzed the data, and conceptualized and drafted the manuscript. MP helped in the experimental setup, provided input, and proofread the manuscript. IP provided input and proofread the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the European Union’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie grant agreement no. 797028.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.725403/full#supplementary-material
Supplementary Figure 1 | Neighbor-joining trees illustrating the relationships across Lysobacter members based on nucleotide sequences of the rpfG and rpfB genes. Pseudomonas aeruginosa PAO1 was used as outgroup sequence. Locus tag numbers are given in brackets, GenBank accession numbers for the whole genome sequences are given in Supplementary Table 1.
Supplementary Figure 2 | Neighbor-joining trees illustrating the relationships across Lysobacter members based on nucleotide sequences of the rpfC and rpfF genes. Pseudomonas aeruginosa PAO1 was used as outgroup sequence. Locus tag numbers are given in brackets, GenBank accession numbers for the whole genome sequences are given in Supplementary Table 1.
Supplementary Figure 3 | Location of rpf genes in Lysobacter capsici AZ78. (a) Comparison of the clusters of genes involved in Diffusible Soluble Factors synthesis and perception in L. enzymogenes C3 and L. capsici AZ78. Gene names are given within the arrows. The corresponding accession number is given under each gene. (b) Bioassay showing DSF production ability (blue halo) of L. capsici AZ78 in comparison to Xanthomonas campestris pv. campestris 8004.
Supplementary Figure 4 | Neighbor-joining trees illustrating the relationships across Lysobacter members based on nucleotide sequences of the tprC, qseB, and qseC genes. Burkholderia cenocepacia KC-01 or Pseudomonas aeruginosa PAO1 were used as outgroup sequence. Locus tag numbers are given in brackets, GenBank accession numbers for the whole genome sequences are given in Supplementary Table 1.
Supplementary Figure 5 | Neighbor-joining trees illustrating the relationships across Lysobacter members based on nucleotide sequences of the xanB2, lysR, and luxR genes. Pseudomonas aeruginosa PAO1 was used as outgroup sequence. Locus tag numbers are given in brackets, GenBank accession numbers for the whole genome sequences are given in Supplementary Table 1.
Supplementary Figure 6 | Bioassay of N-acyl-homoserine lactones. N-acyl-homoserine lactones are produced by Lysobacter daejeonensis GH1-9T (purplish color in the reporter strain Chromobacterium violaceum CV026), but not by L. capsici AZ78.
Supplementary Figure 7 | Effect of diffusible communication signals on the inhibitory activity of Lysobacter capsici AZ78 against (a–f) Pythium ultimum and (g–l) Rhodococcus fascians. Antioomycete and antibacterial activity is expressed as the mean value and standard error variation (percentage) of the reduction of the mycelium growth area of P. ultimum and R. fascians compared to the control (L. capsici AZ78 in not supplemented media), respectively. (a,g) 13-methyltetradecanoic acid, (b,h) glyoxylic acid, (c,i) 2,3-butanedione, (d,j) 3-hydroxybenzoic acid, (e,k) 4-hydroxybenzoic acid, (f,l) mix of N-acyl-homoserine lactones. Each treatment included five replicates and data originating from two independent experiments were pooled. Different letters indicate significant differences according to Tukey’s test (α = 0.05). Eventual minimum effective concentrations are given in Supplementary Table 2.
Supplementary Figure 8 | Growth curves of Lysobacter capsici AZ78 exposed to diffusible communication signals. LeDSF3, 13-methyltetradecanoic acid 50 μM; IND, indole 500 μM; GLY, glyoxylic acid 0.01 μM; BUT, 2,3-butanedione 0.01 μM; 4-HBA, 4-hydroxybenzoic acid 50 μM; 3-HBA, 3-hydroxybenzoic acid 30 μM; AHL, mix of N-acyl-homoserine lactones 20 μM; TSB, 1/10 Tryptic Soy Broth; MeOH, 1% v/v methanol.
Supplementary Figure 9 | Scatter plot of RNA-Seq and qRT-PCR relative expression levels. Pearson correlation test (r = 0.95) was applied to log2 fold change (FC) values of selected genes (Supplementary Table 4).
Supplementary Figure 10 | Venn diagram of up-regulated (a) and down-regulated (b) genes indicating the overlap in the number of differentially expressed genes (DEGs) in Lysobacter capsici AZ78 as response to diffusible communication signals. Only genes with |log2-fold change| > 1 and p-value < 0.01 were included. LeDSF3, 13-methyltetradecanoic acid 50 μM; IND, indole 500 μM; GLY, glyoxylic acid 0.01 μM; BUT, 2,3-butanedione 0.01 μM; 4-HBA, 4-hydroxybenzoic acid 50 μM; 3-HBA, 3-hydroxybenzoic acid 30 μM; AHL, mix of N-acyl-homoserine lactones 20 μM.
Supplementary Figure 11 | Radar plots of Lysobacter capsici AZ78 genes differentially expressed in response to diffusible communication signals. Annotated genes were classified in 20 functional categories: global metabolism (MET); carbohydrate metabolism (CM); energy metabolism (E); lipid metabolism (LM); nucleotide metabolism (NM); amino acid metabolism (AM); protein metabolism (PM); secondary metabolism (SM); DNA metabolism (DNA); RNA transcription and degradation (RNA/TR); translation (T); growth (G); oxidative stress (OX); antagonism (AG); defence (D); transport, phosphotransferase systems, and secretion (T); signal transduction and receptors (S); kinase/phosphatase (K); quorum sensing (QS); motility, chemotaxis, and biofilm (M). Only genes with |log2-fold change| > 1 and p-value < 0.01 were included. LeDSF3, 13-methyltetradecanoic acid 50 μM; IND, indole 500 μM; GLY, glyoxylic acid 0.01 μM; BUT, 2,3-butanedione 0.01 μM; 3HBA, 3-hydroxybenzoic acid 30 μM; 4HBA, 4-hydroxybenzoic acid 50 μM; AHL, mix of N-acyl-homoserine lactones 20 μM.
Supplementary Table 1 | Bacterial strains used for phylogenetic analysis.
Supplementary Table 2 | Culture conditions of L. capsici AZ78.
Supplementary Table 3 | RNA integrity and concentration of Lysobacter capsici AZ78 samples.
Supplementary Table 4 | Number of reads obtained from Illumina HiSeq sequencing of RNA extracted from different Lysobacter capsici AZ78.
Supplementary Table 5 | Primers used in qRT-PCR.
Supplementary Table 6 | List of differentially expressed genes of Lysobacter capsici AZ78 after 48 h incubation with 13-methyltetradecanoic acid 50 μM.
Supplementary Table 7 | List of differentially expressed genes of Lysobacter capsici AZ78 after 48 h incubation with indole 500 μM.
Supplementary Table 8 | List of differentially expressed genes of Lysobacter capsici AZ78 after 48 h incubation with glyoxylic acid 0.01 μM.
Supplementary Table 9 | List of differentially expressed genes of Lysobacter capsici AZ78 after 48 h incubation with 2,3-butanedione 0.01 μM.
Supplementary Table 10 | List of differentially expressed genes of Lysobacter capsici AZ78 after 48 h incubation with 3-hydroxybenzoic acid 30 μM.
Supplementary Table 11 | List of differentially expressed genes of Lysobacter capsici AZ78 after 48 h incubation with 4-hydroxybenzoic acid 50 μM.
Supplementary Table 12 | List of differentially expressed genes of Lysobacter capsici AZ78 after 48 h incubation with mix of N-acyl-homoserine lactones 20 μM.
Footnotes
References
Abisado, R. G., Benomar, S., Klaus, J. R., Dandekar, A. A., and Chandler, J. R. (2018). Bacterial quorum sensing and microbial community interactions. MBio 9, 1–14. doi: 10.1128/mBio.02331-17
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeqa python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Atkinson, S., Goldstone, R. J., Joshua, G. W. P., Chang, C.-Y., Patrick, H. L., Cámara, M., et al. (2011). Biofilm development on Caenorhabditis elegans by Yersinia is facilitated by quorum sensing-dependent repression of type III secretion. PLoS Pathog. 7:e1001250. doi: 10.1371/journal.ppat.1001250
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genom. 9:75. doi: 10.1186/1471-2164-9-75
Bansal, T., Englert, D., Lee, J., Hegde, M., Wood, T. K., and Jayaraman, A. (2007). Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect. Immun. 75, 4597–4607. doi: 10.1128/IAI.00630-07
Barber, C. E., Tang, J. L., Feng, J. X., and Pan, M. Q. (1997). A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24, 555–566. doi: 10.1046/j.1365-2958.1997.3721736.x
Blanco, P., Hernando-Amado, S., Reales-Calderon, J., Corona, F., Lira, F., Alcalde-Rico, M., et al. (2016). Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 4:14. doi: 10.3390/microorganisms4010014
Bleves, S., Soscia, C., Nogueira-Orlandi, P., Lazdunski, A., and Filloux, A. (2005). Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J. Bacteriol. 187, 38983902. doi: 10.1128/JB.187.11.3898-3902.2005
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Braun, V. (1995). Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16, 295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x
Brescia, F., Marchetti-Deschmann, M., Musetti, R., Perazzolli, M., Pertot, I., and Puopolo, G. (2020). The rhizosphere signature on the cell motility, biofilm formation and secondary metabolite production of a plant-associated Lysobacter strain. Microbiol. Res. 234:126424. doi: 10.1016/j.micres.2020.126424
Burrows, L. L. (2012). Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 66, 493–520. doi: 10.1146/annurev-micro-092611-150055
Case, R. J., Labbate, M., and Kjelleberg, S. (2008). AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2, 345–349. doi: 10.1038/ismej.2008.13
Cha, C., Gao, P., Chen, Y. C., Shaw, P. D., and Farrand, S. K. (1998). Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant. Microbe. Interact. 11, 1119–1129. doi: 10.1094/MPMI.1998.11.11.1119
Chen, Y., Xia, J., Su, Z., Xu, G., Gomelsky, M., Qian, G., et al. (2017). Lysobacter PilR, the regulator of type IV pilus synthesis, controls antifungal antibiotic production via a cyclic di-GMP pathway. Appl. Environ. Microbiol. 83, 1–19. doi: 10.1128/AEM.03397-16
Choi, J.-H., Seok, J.-H., Cha, J.-H., and Cha, C.-J. (2014). Lysobacter panacisoli sp. nov., isolated from ginseng soil. Int. J. Syst. Evol. Microbiol. 64, 2193–2197. doi: 10.1099/ijs.0.062034-0
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Cornforth, D. M., and Foster, K. R. (2013). Competition sensing: the social side of bacterial stress responses. Nat. Rev. Microbiol. 11, 285–293. doi: 10.1038/nrmicro2977
de Bruijn, I., Cheng, X., de Jager, V., Expósito, R. G., Watrous, J., Patel, N., et al. (2015). Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genomics 16:991. doi: 10.1186/s12864-015-2191-z
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Domka, J., Lee, J., and Wood, T. K. (2006). YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl. Environ. Microbiol. 72, 2449–2459. doi: 10.1128/AEM.72.4.2449-2459.2006
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Feng, T., Han, Y., Li, B., Li, Z., Yu, Y., Sun, Q., et al. (2019). Interspecies and intraspecies signals synergistically regulate Lysobacter enzymogenes twitching motility. Appl. Environ. Microbiol. 85, 1098–5336. doi: 10.1128/AEM.01742-19
García-Salamanca, A., Molina-Henares, M. A., van Dillewijn, P., Solano, J., Pizarro-Tobías, P., Roca, A., et al. (2013). Bacterial diversity in the rhizosphere of maize and the surrounding carbonate-rich bulk soil. Microb. Biotechnol. 6, 36–44. doi: 10.1111/j.1751-7915.2012.00358.x
Gómez Expósito, R., Postma, J., Raaijmakers, J. M., and De Bruijn, I. (2015). Diversity and activity of lysobacter species from disease suppressive soils. Front. Microbiol. 6:1243. doi: 10.3389/fmicb.2015.01243
Green, E. R., and Mecsas, J. (2016). Bacterial secretion systems?: an overview. Microbiol. Spectr. 4:10.1128/microbiolsec.VMBF–0012–2015. doi: 10.1128/microbiolspec.VMBF-0012-2015
Han, Y., Wang, Y., Tombosa, S., Wright, S., Huffman, J., Yuen, G., et al. (2015). Identification of a small molecule signaling factor that regulates the biosynthesis of the antifungal polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes. Appl. Microbiol. Biotechnol. 99, 801–811. doi: 10.1007/s00253-014-6120-x
Han, Y., Wang, Y., Yu, Y., Chen, H., Shen, Y., and Du, L. (2017). Indole-induced reversion of intrinsic multiantibiotic resistance in Lysobacter enzymogenes. Appl. Environ. Microbiol. 83, 1–14.
He, Y.-W., Wu, J., Zhou, L., Yang, F., He, Y.-Q., Jiang, B.-L., et al. (2011). Xanthomonas campestris diffusible factor is 3-hydroxybenzoic acid and is associated with Xanthomonadin biosynthesis, cell viability, antioxidant activity, and systemic invasion. Mol. Plant Microbe Interact. 24, 948–957. doi: 10.1094/MPMI-02-11-0031
Hibbing, M. E., Fuqua, C., Parsek, M. R., and Peterson, S. B. (2010). Bacterial competition: surviving and thriving in the micorbial jungle. Natl. Rev. Microbiol. 8, 15–25. doi: 10.1038/nrmicro2259.Bacterial
Ji, G.-H., Wei, L.-F., He, Y.-Q., Wu, Y.-P., and Bai, X.-H. (2008). Biological control of rice bacterial blight by Lysobacter antibioticus strain 13-1. Biol. Control 45, 288–296. doi: 10.1016/j.biocontrol.2008.01.004
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., and Madden, T. L. (2008). NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9. doi: 10.1093/nar/gkn201
Kim, K., Lee, S., and Ryu, C.-M. (2013). Interspecific bacterial sensing through airborne signals modulates locomotion and drug resistance. Nat. Commun. 4:1809. doi: 10.1038/ncomms2789
Kimura, M. (2020). The Neutral Theory and Molecular Evolution BTMy Thoughts on Biological Evolution. Singapore: Springer, 119–138. doi: 10.1007/978-981-15-6165-8_8
Ko, H.-S., Jin, R.-D., Krishnan, H. B., Lee, S.-B., and Kim, K.-Y. (2009). Biocontrol ability of Lysobacter antibioticus HS124 against phytophthora blight is mediated by the production of 4-hydroxyphenylacetic acid and several lytic enzymes. Curr. Microbiol. 59, 608–615. doi: 10.1007/s00284-009-9481-0
Koskiniemi, S., Lamoureux, J. G., Nikolakakis, K. C., t’Kint de Roodenbeke, C., Kaplan, M. D., Low, D. A., et al. (2013). Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl. Acad. Sci. U.S.A. 110, 7032–7037. doi: 10.1073/pnas.1300627110
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets brief communication. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, J., Attila, C., Cirillo, S. L. G., Cirillo, J. D., and Wood, T. K. (2009). Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb. Biotechnol. 2, 75–90. doi: 10.1111/j.1751-7915.2008.00061.x
Lee, J., Zhang, X.-S., Hegde, M., Bentley, W. E., Jayaraman, A., and Wood, T. K. (2008). Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2, 1007–1023. doi: 10.1038/ismej.2008.54
Lee, J. H., and Lee, J. (2010). Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 34, 426–444. doi: 10.1111/j.1574-6976.2009.00204.x
Lee, J. W., Im, W.-T., Kim, M. K., and Yang, D.-C. (2006). Lysobacter koreensis sp. nov., isolated from a ginseng field. Int. J. Syst. Evol. Microbiol. 56, 231–235. doi: 10.1099/ijs.0.63955-0
Liang, J., Sun, S., Ji, J., Wu, H., Meng, F., Zhang, M., et al. (2014). Comparison of the rhizosphere bacterial communities of zigongdongdou soybean and a high-methionine transgenic line of this cultivar. PLoS One 9:e103343. doi: 10.1371/journal.pone.0103343
Lin, G., Chai, J., Yuan, S., Mai, C., Cai, L., Murphy, R. W., et al. (2016). Venn painter: a tool for the comparison and identification of candidate genes based on venn diagrams. PLoS One 11:e0154315. doi: 10.1371/journal.pone.0154315
Ling, J., Zhou, L., Wu, G., Zhao, Y., Jiang, T., and Liu, F. (2019a). The AHL quorum-sensing system negatively regulates growth and autolysis in lysobacter brunescens. Front. Microbiol. 10:2748. doi: 10.3389/fmicb.2019.02748
Ling, J., Zhu, R., Laborda, P., Jiang, T., Jia, Y., Zhao, Y., et al. (2019b). LbDSF, the Lysobacter brunescens quorum-sensing system diffusible signaling factor, regulates anti-xanthomonas XSAC biosynthesis, colony morphology, and surface motility. Front. Microbiol. 10:1230. doi: 10.3389/fmicb.2019.01230
Matsuoka, J., Ishizuna, F., Kurumisawa, K., Morohashi, K., Ogawa, T., Hidaka, M., et al. (2017). Stringent expression control of pathogenic r-body production in legume symbiont Azorhizobium caulinodans. MBio 8:e715-17. doi: 10.1128/mBio.00715-17
McCarthy, R. R., Yu, M., Eilers, K., Wang, Y.-C., Lai, E.-M., and Filloux, A. (2019). Cyclic di-GMP inactivates T6SS and T4SS activity in Agrobacterium tumefaciens. Mol. Microbiol. 112, 632–648. doi: 10.1111/mmi.14279
Mueller, R. S., Beyhan, S., Saini, S. G., Yildiz, F. H., and Bartlett, D. H. (2009). Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J. Bacteriol. 191, 3504–3516. doi: 10.1128/JB.01240-08
Mueller, R. S., McDougald, D., Cusumano, D., Sodhi, N., Kjelleberg, S., Azam, F., et al. (2007). Vibrio cholerae strains possess multiple strategies for abiotic and biotic surface colonization. J. Bacteriol. 189, 5348–5360. doi: 10.1128/JB.01867-06
Mufti, R., Amna, Rafique, M., Haq, F., Munis, M. F. H., Masood, S., et al. (2015). Genetic diversity and metal resistance assessment of endophytes isolated from Oxalis corniculata. Soil Environ. 34, 89–99.
Nadell, C. D., Xavier, J. B., Levin, S. A., and Foster, K. R. (2008). The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 6:e14. doi: 10.1371/journal.pbio.0060014
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Patel, N., Cornejo, M., Lambert, D., Craig, A., Hilman, B., and Kobayashi, D. (2011). A multifunctional role for the type IV pilus in the bacterial biological control agent Lysobacter enzymogenes. Phytopathology 101:S138.
Pérez-Jaramillo, J. E., de Hollander, M., Ramírez, C. A., Mendes, R., Raaijmakers, J. M., and Carrión, V. J. (2019). Deciphering rhizosphere microbiome assembly of wild and modern common bean (Phaseolus vulgaris) in native and agricultural soils from Colombia. Microbiome 7:114. doi: 10.1186/s40168-019-0727-1
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Postma, J., Schilder, M. T., Bloem, J., and van Leeuwen-Haagsma, W. K. (2008). Soil suppressiveness and functional diversity of the soil microflora in organic farming systems. Soil Biol. Biochem. 40, 2394–2406. doi: 10.1016/j.soilbio.2008.05.023
Postma, J., Stevens, L. H., Wiegers, G. L., Davelaar, E., and Nijhuis, E. H. (2009). Biological control of Pythium aphanidermatum in cucumber with a combined application of Lysobacter enzymogenes strain 3.1T8 and chitosan. Biol. Control 48, 301–309. doi: 10.1016/j.biocontrol.2008.11.006
Potrykus, K., and Cashel, M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Puopolo, G., Aida, R., Pierson, L. S., and Zoina, A. (2011). Selection of a new Pseudomonas chlororaphis strain for the biological control of Fusarium oxysporum f. sp. radicis-lycopersici. Phytopathol. Mediterr. 50, 228–235.
Puopolo, G., Giovannini, O., and Pertot, I. (2014). Lysobacter capsici AZ78 can be combined with copper to effectively control Plasmopara viticola on grapevine. Microbiol. Res. 169, 633–642. doi: 10.1016/j.micres.2013.09.013
Puopolo, G., Raio, A., and Zoina, A. (2010). Identification and charactherization of Lysobacter capsici strain PG4: a new plant health-promoting rhizobacterium. J. Plant Pathol. 92, 157–164.
Puopolo, G., Tomada, S., and Pertot, I. (2018). The impact of the omics era on the knowledge and use of Lysobacter species to control phytopathogenic micro-organisms. J. Appl. Microbiol. 124, 15–27. doi: 10.1111/jam.13607
Puopolo, G., Tomada, S., Sonego, P., Moretto, M., Engelen, K., Perazzolli, M., et al. (2016). The Lysobacter capsici AZ78 genome has a gene pool enabling it to interact successfully with phytopathogenic microorganisms and environmental factors. Front. Microbiol. 7:96. doi: 10.3389/fmicb.2016.00096
Qian, G., Wang, Y., Liu, Y., Xu, F., He, Y. W., Du, L., et al. (2013). Lysobacter enzymogenes uses two distinct cell-cell signaling systems for differential regulation of secondary-metabolite biosynthesis and colony morphology. Appl. Environ. Microbiol. 79, 6604–6616. doi: 10.1128/AEM.01841-13
Qian, G., Xu, F., Venturi, V., Du, L., and Liu, F. (2014). Roles of a solo LuxR in the biological control agent Lysobacter enzymogenes strain OH11. Phytopathology 104, 224–231. doi: 10.1094/PHYTO-07-13-0188-R
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., and Moënne-Loccoz, Y. (2009). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321, 341–361. doi: 10.1007/s11104-008-9568-6
Ramos, J. L., Martínez-Bueno, M., Molina-Henares, A. J., Terán, W., Watanabe, K., Zhang, X., et al. (2005). The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69, 326–356. doi: 10.1128/MMBR.69.2.326-356.2005
Raymann, K., Bobay, L.-M., Doak, T. G., Lynch, M., and Gribaldo, S. (2013). A genomic survey of Reb homologs suggests widespread occurrence of R-bodies in proteobacteria. G3 (Bethesda) 3, 505–516. doi: 10.1534/g3.112.005231
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Robinson, M. D., and Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. doi: 10.1186/gb-2010-11-3-r25
Rodrigues, R. R., Rodgers, N. C., Wu, X., and Williams, M. A. (2018). COREMIC: a web-tool to search for a niche associated CORE MICrobiome. PeerJ 6:e4395. doi: 10.7717/peerj.4395
Ruijter, J. M., Ramakers, C., Hoogaars, W. M. H., Karlen, Y., Bakker, O., van den Hoff, M. J., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37:e45. doi: 10.1093/nar/gkp045
Ruwandeepika, H. A. D., Karunasagar, I., Bossier, P., and Defoirdt, T. (2015). Expression and quorum sensing regulation of type III secretion system genes of vibrio harveyi during infection of gnotobiotic brine shrimp. PLoS One 10:e0143935. doi: 10.1371/journal.pone.0143935
Ryan, R. P., An, S. Q., Allan, J. H., McCarthy, Y., and Dow, J. M. (2015). The DSF family of cell–cell signals: an expanding class of bacterial virulence regulators. PLoS Pathog. 11:e1004986. doi: 10.1371/journal.ppat.1004986
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Saldanha, A. J. (2004). Java treeviewextensible visualization of microarray data. Bioinformatics 20, 3246–3248. doi: 10.1093/bioinformatics/bth349
Schmalenberger, A., and Tebbe, C. C. (2003). Bacterial diversity in maize rhizospheres: conclusions on the use of genetic profiles based on PCR-amplified partial small subunit rRNA genes in ecological studies. Mol. Ecol. 12, 251–262. doi: 10.1046/j.1365-294X.2003.01716.x
Schmidt, R., Cordovez, V., de Boer, W., Raaijmakers, J., and Garbeva, P. (2015). Volatile affairs in microbial interactions. ISME J. 9, 2329–2335. doi: 10.1038/ismej.2015.42
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to imageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schuster, M., Joseph Sexton, D., Diggle, S. P., and Peter Greenberg, E. (2013). Acyl-homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 67, 43–63. doi: 10.1146/annurev-micro-092412-155635
Shen, X., Wang, B., Yang, N., Zhang, L., Shen, D., Wu, H., et al. (2021). Lysobacter enzymogenes antagonizes soilborne bacteria using the type IV secretion system. Environ. Microbiol. doi: 10.1111/1462-2920.15662
Slater, H., Alvarez-morales, A., Barber, C. E., Daniels, M. J., Dow, J. M., Christine, E., et al. (2000). A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol. Microbiol. 38, 986–1003. doi: 10.1046/j.1365-2958.2000.02196.x
Steindler, L., and Venturi, V. (2007). Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 266, 1–9. doi: 10.1111/j.1574-6968.2006.00501.x
Stephens, K., and Bentley, W. E. (2020). Synthetic biology for manipulating quorum sensing in microbial consortia. Trends Microbiol. 28, 633–643. doi: 10.1016/j.tim.2020.03.009
Su, Z., Chen, H., Wang, P., Tombosa, S., Du, L., and Han, Y. (2017). 4-Hydroxybenzoic acid is a diffusible factor that connects metabolic shikimate pathway to the biosynthesis of a unique antifungal metabolite in Lysobacter enzymogenes. Mol. Microbiol. 104, 163–178. doi: 10.1111/mmi.13619
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tomada, S., Puopolo, G., Perazzolli, M., Musetti, R., Loi, N., and Pertot, I. (2016). Pea broth enhances the biocontrol efficacy of Lysobacter capsici AZ78 by triggering cell motility associated with biogenesis of type IV pilus. Front. Microbiol. 7:1136. doi: 10.3389/fmicb.2016.01136
Tomada, S., Sonego, P., Moretto, M., Engelen, K., Pertot, I., Perazzolli, M., et al. (2017). Dual RNA-Seq of Lysobacter capsici AZ78Phytophthora infestans interaction shows the implementation of attack strategies by the bacterium and unsuccessful oomycete defense responses. Environ. Microbiol. 19, 4113–4125. doi: 10.1111/1462-2920.13861
Tseng, T.-T., Tyler, B. M., and Setubal, J. C. (2009). Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9(Suppl. 1):S2. doi: 10.1186/1471-2180-9-S1-S2
Turnbull, A. L., Liu, Y., and Lazarovits, G. (2012). Isolation of bacteria from the rhizosphere and rhizoplane of potato (Solanum tuberosum) grown in two distinct soils using semi selective media and characterization of their biological properties. Am. J. Potato Res. 89, 294–305. doi: 10.1007/s12230-012-9253-4
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Van Overbeek, L., and Van Elsas, J. D. (2008). Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.). FEMS Microbiol. Ecol. 64, 283–296. doi: 10.1111/j.1574-6941.2008.00469.x
Venturi, V., and Fuqua, C. (2013). Chemical signaling between plants and plant-pathogenic bacteria. Annu. Rev. Phytopathol. 51, 17–37. doi: 10.1146/annurev-phyto-082712-102239
Venturi, V., and Keel, C. (2016). Signaling in the rhizosphere. Trends Plant Sci. 21, 187–198. doi: 10.1016/j.tplants.2016.01.005
Wang, P., Chen, H., Qian, G., and Liu, F. (2017). LetR is a TetR family transcription factor from Lysobacter controlling antifungal antibiotic biosynthesis. Appl. Microbiol. Biotechnol. 101, 3273–3282. doi: 10.1007/s00253-017-8117-8
Xu, H., Wang, R., Zhao, Y., Fu, Z. Q., Qian, G., and Liu, F. (2017). LesR is a novel upstream regulator that controls downstream Clp expression to modulate antibiotic HSAF biosynthesis and cell aggregation in Lysobacter enzymogenes OH11. Microb. Cell Fact. 16, 1–9. doi: 10.1186/s12934-017-0818-2
Yang, M., Ren, S., Shen, D., Yang, N., Wang, B., Han, S., et al. (2020). An intrinsic mechanism for coordinated production of the contact-dependent and contact-independent weapon systems in a soil bacterium. PLoS Pathog. 16:e1008967. doi: 10.1371/journal.ppat.1008967
Zhang, Y., Ducret, A., Shaevitz, J., and Mignot, T. (2012). From individual cell motility to collective behaviors: insights from a prokaryote, Myxococcus xanthus. FEMS Microbiol. Rev. 36, 149–164. doi: 10.1111/j.1574-6976.2011.00307.x
Zhong, J., Xiao, C., Gu, W., Du, G., Sun, X., He, Q.-Y., et al. (2015). Transfer RNAs mediate the rapid adaptation of Escherichia coli to oxidative stress. PLoS Genet. 11:e1005302. doi: 10.1371/journal.pgen.1005302
Zhou, L., Huang, T., Wang, J., Sun, S., Chen, G., Poplawsky, A., et al. (2013). The Rice bacterial pathogen Xanthomonas oryzae pv. oryzae produces 3-hydroxybenzoic acid and 4-hydroxybenzoic acid via XanB2 for use in Xanthomonadin, ubiquinone, and exopolysaccharide biosynthesis. Mol. Plant Microbe Interact. 26, 1239–1248. doi: 10.1094/mpmi-04-13-0112-r
Keywords: diffusible communication signals, rhizosphere, Lysobacter, bacterial interactions, transcriptome
Citation: Bejarano A, Perazzolli M, Pertot I and Puopolo G (2021) The Perception of Rhizosphere Bacterial Communication Signals Leads to Transcriptome Reprogramming in Lysobacter capsici AZ78, a Plant Beneficial Bacterium. Front. Microbiol. 12:725403. doi: 10.3389/fmicb.2021.725403
Received: 15 June 2021; Accepted: 21 July 2021;
Published: 18 August 2021.
Edited by:
Marco Scortichini, Council for Agricultural and Economics Research (CREA), ItalyReviewed by:
Irina Kudryakova, G. K. Skryabin Institute of Biochemistry and Physiology of Microorganisms, RussiaJoeke Postma, Wageningen University & Research, Netherlands
Copyright © 2021 Bejarano, Perazzolli, Pertot and Puopolo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Bejarano, YW5hLmJlamFyYW5vcmFtb3NAdW5pdG4uaXQ=
 Ana Bejarano
Ana Bejarano Michele Perazzolli
Michele Perazzolli Ilaria Pertot
Ilaria Pertot Gerardo Puopolo
Gerardo Puopolo