
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 31 August 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.725383
This article is part of the Research TopicMALDI-TOF MS in Microbiological Diagnostics: Future Applications Beyond IdentificationView all 12 articles
 Katy Jeannot1,2,3
Katy Jeannot1,2,3 Katheryn Hagart4
Katheryn Hagart4 Laurent Dortet2,5,6
Laurent Dortet2,5,6 Markus Kostrzewa7
Markus Kostrzewa7 Alain Filloux4
Alain Filloux4 Patrick Plesiat1,2,3
Patrick Plesiat1,2,3 Gerald Larrouy-Maumus4*
Gerald Larrouy-Maumus4*Colistin is frequently a last resort treatment for Pseudomonas aeruginosa infections caused by multidrug-resistant (MDR) and extensively drug resistant (XDR) strains, and detection of colistin resistance is essential for the management of infected patients. Therefore, we evaluated the recently developed MALDIxin test for the detection of colistin resistance in P. aeruginosa clinical strains using the routine matrix-assisted laser desorption ionization (MALDI) Biotyper Sirius system. The test is based on the detection by mass spectrometry of modified lipid A by the addition of 4-amino-L-arabinose (L-ara4N) molecules on one or two phosphate groups, in strains resistant to colistin. Overproduction of L-Ara4N molecules is mainly due to the constitutive activation of the histidine kinase (PmrB) or the response regulator (PmrA) following an amino-acid substitution in clinical strains. The performance of the test was determined on a panel of 14 colistin-susceptible and 14 colistin-resistant P. aeruginosa clinical strains, the reference strain PAO1 and positive control mutants PmrB (V28G), PmrB (D172), PhoQ (D240–247), and ParR (M59I). In comparison with the broth microdilution (BMD) method, all the susceptible strains (n=14) and 8/14 colistin-resistant strains were detected in less than 1h, directly on whole bacteria. The remaining resistant strains (n=6) were all detected after a short pre-exposure (4h) to colistin before sample preparation. Validation of the method on a larger panel of strains will be the next step before its use in diagnostics laboratories. Our data showed that the MALDIxin test offers rapid and efficient detection of colistin resistant P. aeruginosa and is thus a valuable diagnostics tool to control the spread of these emerging resistant strains.
Pseudomonas aeruginosa is an opportunistic pathogen well-known for infections associated with intensive care units. It is one of the most frequent cause of acute pulmonary healthcare-associated infections, and severe infections particularly in immunocompromised patients (Vincent et al., 2009). Its intrinsic resistance to many antibiotics combined with its facility to accumulate a diversity of resistance mechanisms increasingly limits therapeutic options (Horcajada et al., 2019). Thus, polymyxins (polymyxin B or colistin) are used as a last resort for the treatment of P. aeruginosa infections caused by multidrug-resistant (MDR) and extensively drug resistant (XDR) strains (Sader et al., 2017; Doi, 2019; Walters et al., 2019; Ahmadian et al., 2020; Mirzaei et al., 2020). Unfortunately, resistance to colistin has emerged. In P. aeruginosa, acquired resistance to colistin mostly results from the addition of one or two 4-amino-L-arabinose (L-Ara4N) molecules to the 1 and/or 4' phosphate groups on the lipid A, the anchor of the LPS in the outer membrane (Bhat et al., 1990; Fernandez et al., 2013). While the P. aeruginosa genome contains an eptA like gene, the addition of phosphoethanolamine to lipid A or the LPS core has not been reported, unlike in Enterobacterales and Acinetobacter baumannii (Nowicki et al., 2015). The synthesis and the transport of L-Ara4N molecules is encoded by the large arnBCADTEF-UgD operon (simplified as arn), which is dependent on a complex regulatory network comprising at least 5 two-component systems (PmrA/PmrB, PhoP/PhoQ, ParR/ParS, CprR/CprS, and ColR/ColS; Fernandez et al., 2010, 2012; Needham and Trent, 2013; Nowicki et al., 2015). Furthermore, mutations in chromosomal genes encoding histidine kinase or response regulators of these two-component systems result in constitutive activation of the arn operon. However, in P. aeruginosa clinical strains, the genetic events most associated with colistin resistance are amino-acid substitutions leading to a gain of function of the PmrB protein (Barrow and Kwon, 2009; Schurek et al., 2009; Bolard et al., 2019). Although, the mcr genes have been widely reported in Enterobacterales, they have not currently been identified in P. aeruginosa strains, except in the chromosome of one clinical isolate strain (mcr-5; Snesrud et al., 2018).
Although, strains of P. aeruginosa resistant to polymyxins are still rare, their detection is one of the key issues to improve the treatment of patient infected with MDR and XDR P. aeruginosa strains (Diekema et al., 2019). Unfortunately, the methods currently available in routine laboratories for the detection of resistance to colistin in P. aeruginosa, still rely on bacterial growth in the presence of polymyxins. These procedures require 16–20h in culture, whether determining susceptibility to colistin minimal inhibitory concentration (MIC) using the broth microdilution method (BMD; reference method), or the colistin broth disk elution and colistin agar test methods recently accepted by the CLSI (2020). Only two test have reported the detection of colistin resistance strains in less than 3h (Sadek et al., 2020; Sorensen et al., 2020). The first is a biochemical test (Rapid Polymyxin/Pseudomonas NP test) based on the change in color of the bromocresol (yellow to purple/violet) following the production of basic metabolites during the growth of the strain in the presence of colistin (Sadek et al., 2020), and a fast lipid analysis technique (FLAT) directly on a matrix-assisted laser desorption ionization (MALDI) plate. However, both approaches have issues: possible misinterpretation of the colorimetric test, and potential cross contamination with the on-target hydrolysis in the FLAT method.
Therefore, there is an urgent need to develop a fast and robust assay to detect colistin-resistant P. aeruginosa strains. Recently, we developed a rapid technique using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) to rapidly detect colistin resistance using whole cells, the MALDIxin test (Dortet et al., 2018a,b). The MALDIxin test has now been optimized for Escherichia coli, Klebsiella pneumoniae, A. baumannii, and Salmonella enterica (Dortet et al., 2018a,b, 2019, 2020; Furniss et al., 2019). The aim of this study is to evaluate the performance of the optimized MALDIxin test using a routine MALDI mass spectrometer (in comparison to the BMD method), to detect colistin-resistant P. aeruginosa.
From the Pseudomonas collection of the French National Reference Centre for Antibiotic Resistance (Besançon, France), 14 colistin susceptible (MIC≤2mg/L) and 14 colistin resistant clinical strains (MIC>2mg/L) were selected. All the strains were genotypically-unrelated, and isolated from 25 health institutions distributed throughout France. In addition, two pmrB mutants (AB8.2, AB16.2), one parR mutant (PAOW2), and one phoQ mutant (AB8.4) derivate form the P. aeruginosa reference strain PAO1 were included as positive controls (Table 1; Muller et al., 2011; Bolard et al., 2019). The wild-type strain PAO1 was used as negative control.
MICs were determined in triplicate by the BMD using colistin sulfate (Sigma Aldrich, Saint Quentin Fallavier, France) and cation-adjusted Mueller Hinton broth (MHB) from Becton Dickinson (Pont-de-Claix, France) according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations (EUCAST, 2019). Results were interpreted using EUCAST breakpoints (≤2mg/L; >2mg/L).
Four clinical strains (185345, 185374, 185819, and 196337) for which the mutation responsible for colistin resistance had not been identified and characterized were genome sequenced. From an overnight bacterial culture, total DNA was extracted using the PureLink Genomic DNA Mini kit (ThermoFischer Scientific). The library preparation using NextEra® XT DNA preparation kit (Illumina, San Diego, CA, United States) and sequencing by Illumina NextSeq500 system (2×150-bp paired end reads) were performed by the “Plateforme de Microbiologie Mutualisée” (PibNet) at Institut Pasteur (Paris, France). Reads were assembled with Shovill-Spades v3.14.0. To identify mutations in genes associated with colistin resistance in P. aeruginosa (cprR, cprS, colR, colS, parR, parS, phoP, phoQ, pmrA, and pmrB), sequences from clinical strains were mapped to sequences of reference strains PAO1, and P, A14, and a large collection of 77 clinical strains susceptible to colistin using the NRC bioinformatic pipelines based on SNIPPY v4.6.0.
The nucleotide sequences reported in this study and corresponding to the entire chromosome of strains 185345, 185374, 185819, and 196337 have been deposited in the GenBank nucleotide database under accession no JAGJAGMWR0000000000, JAGMWQ0000000000, JAGMWP0000000000, and JAGMWO0000000000, respectively.
Pseudomonas aeruginosa cells were exposed or not for 4-h to a subinhibitory (1/2 MIC) concentration of colistin. A 10 μl inoculation loop of bacteria, grown on Mueller-Hinton agar for 18–24h, was resuspended in 200μl of water. Mild-acid hydrolysis was performed on 100μl of this suspension, by adding 100μl of 2% v/v acetic acid and incubating the mixture at 98°C for 30min. Hydrolyzed cells were centrifuged at 17,000 ×g for 2min, the supernatant was discarded, and the pellet was washed three times with 300μl of ultrapure water and resuspended to a density of McFarland 20 as measured using a McFarland Tube Densitometer. A volume of 0.4μl of this suspension was loaded onto the MALDI target plate and immediately overlaid with 1.2μl of Norharmane matrix (Sigma-Aldrich) solubilized in chloroform/methanol (90:10 v/v) to a final concentration of 10mg/ml. For external calibration, 0.5μl of calibration peptide was loaded along with 0.5μl of the given calibration matrix (peptide calibration standard II, Bruker Daltonik, Germany). The samples were loaded onto a disposable MSP 96 target polished steel BC (Bruker Part-No. 8280800).
The bacterial suspension and matrix were mixed directly on the target by pipetting then dried gently under a stream of air. The spectra were recorded in the linear negative-ion mode (laser intensity 95%, ion source 1=10.00kV, ion source 2=8.98kV, lens=3.00kV, detector voltage=2,652V, pulsed ion extraction=150ns). Each spectrum corresponded to ion accumulation of 5,000 laser shots randomly distributed on the spot. The spectra obtained were processed with default parameters using FlexAnalysis v.3.4 software (Bruker Daltonik, Germany).
The negative mass spectrum was scanned between m/z 1,300 and m/z 2,000 in the negative linear ion mode. Manual peak picking at masses relevant to colistin resistance was performed on the obtained mass spectra and the corresponding signal intensities at these defined masses was determined. The percentage of modified lipid A was calculated by dividing the sum of the intensities of the lipid A peaks attributed to addition of L-Ara4N (m/z 1,577.9, m/z 1,593.9, m/z 1,708.9, and m/z 1,724.9) by the intensity of the peaks corresponding to native lipid A (m/z 1,446.7 and m/z 1,462.7). All mass spectra were generated and analyzed in technical triplicate (i.e., measurements of each sample were repeated three times).
All experiments were carried out in biological duplicates. Data were compared two-by-two using unpaired Welch’s t-test. Values of p<0.05 were considered statistically different.
To assess the ability of the MALDIxin test to detect colistin-resistance in P. aeruginosa on a MALDI biotyper Sirius system, we tested a panel of 33 P. aeruginosa strains, including four isogenic P. aeruginosa PAO1 mutants representing the most frequently mutated genes involved in colistin resistance in this species. These included the reference strain PAO1, 14 colistin-resistant clinical strains (MIC from 4 to 128mg/L), and 14 colistin-susceptible clinical strains (MIC from 0.5 to 2mg/L; Table 1). The mass spectrum of colistin susceptible P. aeruginosa reference strain PAO1 is dominated by two peaks assigned to penta-acyl bis-phosphorylated lipid A (Figure 1A). The peak at m/z 1,446.7 is assigned to penta-acyl bis-phosphorylated lipid A, which corresponds to the presence of one 3-OH-C10:0 fatty acyl chain, three C12:0 fatty acyl chain and one 2-OH C12:0 fatty acyl chain. The lipid A structure at m/z 1,462.7 differs from that at m/z 1,446.7 only by the addition of one hydroxyl group at 3 position (3-OH) of one C12:0 fatty acyl chain. Both forms have frequently been reported in P. aeruginosa strains (Ernst et al., 2006; Moskowitz et al., 2012; Figure 1D). In comparison with the parental strain PAO1, four additional peaks at m/z 1,577.9, m/z 1,593.9, m/z 1,708.9, and m/z 1,724.9 were observed in the pmrB mutant (mutant AB8.2), which is resistant to colistin (Figure 1B). The signals at m/z 1,577.9, and m/z 1,593.9, and at m/z 1,708.9 and m/z 1,724.9 correspond to the addition of one or two L-Ara4N molecules to the 4′- or/and 1-phosphate groups of the penta-acylated form respectively, resulting in an increase of +131 m/z compared to the native lipid A peaks (Figures 1B,E,F). Since mutations in genes encoding the two-component systems PmrAB, ParRS, and PhoPQ lead to the addition of L-Ara4N molecules, we did not observe any difference in mass spectra between the pmrB (AB8.2 and AB16.2), and parR (PAOW2) mutants, compared to the phoQ mutant (AB8.4). A similar spectrum is also observed for clinical isolates (Figures 1C,F). Based on the MALDIxin profile, we attempted to determine the Polymyxin Resistance Ratio (PRR) of the sum of the intensities of the peaks associated with modified lipid A over the intensity of the peak of native lipid A allowing an accurate distinction between polymyxin-susceptible and polymyxin-resistant isolates. Despite several attempts, no signal corresponding to a modified lipid A was detected. Although, the MIC of colistin was similar in pmrB mutants (128mg/L), the intensity of peaks was clearly higher in pmrB mutant AB8.2 (PRR 87±25%) than AB16.2 (PRR 2±0%), indicating that the intensity of peaks is not correlated with resistance level to colistin. The same observation was previously reported for E. coli, Salmonella, K. pneumoniae, and A. baumannii (Dortet et al., 2018b, 2019, 2020; Furniss et al., 2019). As expected, the peaks observed in the 14 clinical strains susceptible to colistin did not differ from those obtained with the strain PAO1 (data not shown), and the percentage of modified lipid A was equal to zero (Table 1).
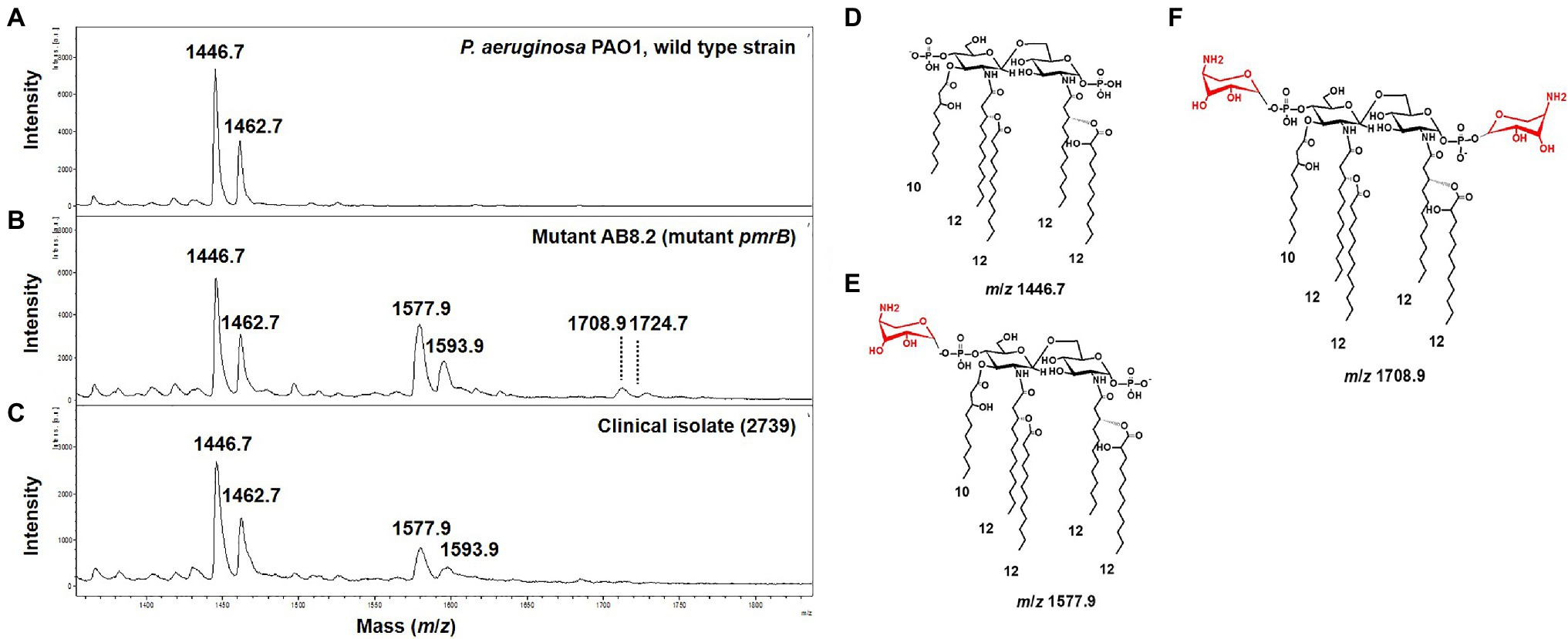
Figure 1. Representative mass spectra of susceptible and modified Pseudomonas aeruginosa lipid A acquired using the linear negative-ion mode of a matrix-assisted laser desorption ionization (MALDI) Biotyper Sirius system (Bruker Daltonics). (A) Susceptible P. aeruginosa PAO1 lipid A is detected as two major peaks at m/z 1,446.7 and m/z 1,462.7. (B) Lipid A from mutant pmrB (AB8.2) with additional peaks at m/z 1,577.9, m/z 1,593.9, m/z 1,708.9, and m/z 1,724.9 corresponding to 4-amino-L-arabinose (L-Ara4N) addition on the penta-acylated lipid A (peaks at m/z m/z 1,446.7 and m/z 1,462.7). (C) Lipid A from colistin-resistant P. aeruginosa clinical isolates is modified by L-Ara4N, which are detected as additional peaks at m/z 1,577.9, m/z 1,593.9. (D) Diagram of P. aeruginosa lipid A at m/z 1,446.7. (E) Diagram of P. aeruginosa lipid A at m/z 1,577.9. L-Ara4N residue is shown in red. (F) Diagram of P. aeruginosa lipid A at m/z 1,708.9. L-Ara4N residues are shown in red.
Interestingly, in clinical strains resistant to colistin (n=14), the percentage of lipid A modified or PRR ranged from 0 to 90%. Although, five P. aeruginosa clinical strains and the phoQ mutant (mutant AB8.4) have a MIC higher than the breakpoint for colistin (>2μg/ml) including one strain with a MIC≥128mg/L (163795), we did not detect any modifications on the lipid A as reported by their null PRR (Table 1). Unlike for MIC determination, bacteria are not exposed to colistin when preparing bacteria for the MALDIxin test. It is likely that induction of the two-component systems PmrAB, CprRS, and ParRS is necessary to detect sufficient modification (beyond background) of lipid A in some strains (Moskowitz et al., 2004; Muller et al., 2011; Fernandez et al., 2012). Therefore, the strain PAO1, the phoQ mutant and the six strains were exposed to a sub-inhibitory concentration of colistin (½ MIC) for 4h, before the MALDIxin test and determination of the percentage of modified lipid A (Table 1). While the colistin susceptible reference strain exhibited 2% modified lipid A after colistin exposure, the phoQ mutant and colistin resistant clinical strains had more than 20% (Table 1; Figure 2). All the strains resistant to colistin were detected after a short exposure (4h) to colistin, confirming that for some strains, the sensitivity of the MALDIxin test can be enhanced by the induction of colistin resistance (Figure 3).
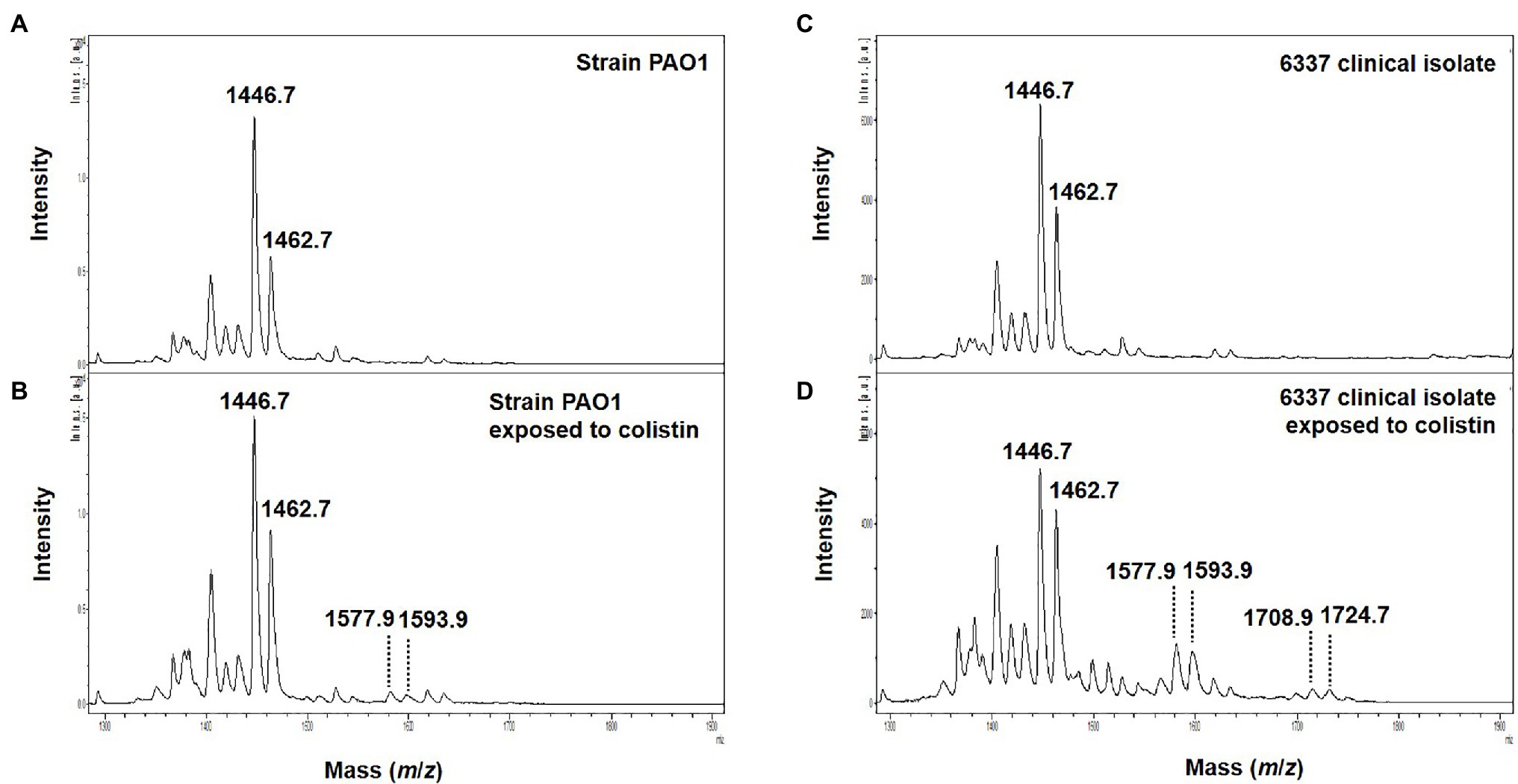
Figure 2. Representative mass spectra of colistin induced susceptible and modified P. aeruginosa lipid A acquired using the linear negative-ion mode of a MALDI Biotyper Sirius system (Bruker Daltonics). (A) Susceptible P. aeruginosa PAO1 lipid A is detected as two major peaks at m/z 1,446.7 and m/z 1,462.7. (B) Colistin induced susceptible P. aeruginosa PAO1 lipid A with additional peaks at m/z 1,577.9, m/z 1,593.9 corresponding to L-Ara4N addition on the penta-acylated lipid A (peaks at m/z m/z 1,446.7 and m/z 1,462.7). (C) Lipid A from colistin-resistant P. aeruginosa clinical isolate 6,337 is detected as two major peaks at m/z 1,446.7 and m/z 1,462.7. (D) Colistin induced colistin-resistant P. aeruginosa clinical isolate 6,337 lipid A with additional peaks at m/z 1,577.9, m/z 1,593.9, m/z 1,708.9, and m/z 1,724.9 corresponding to L-Ara4N addition on the penta-acylated lipid A (peaks at m/z m/z 1,446.7 and m/z 1,462.7).
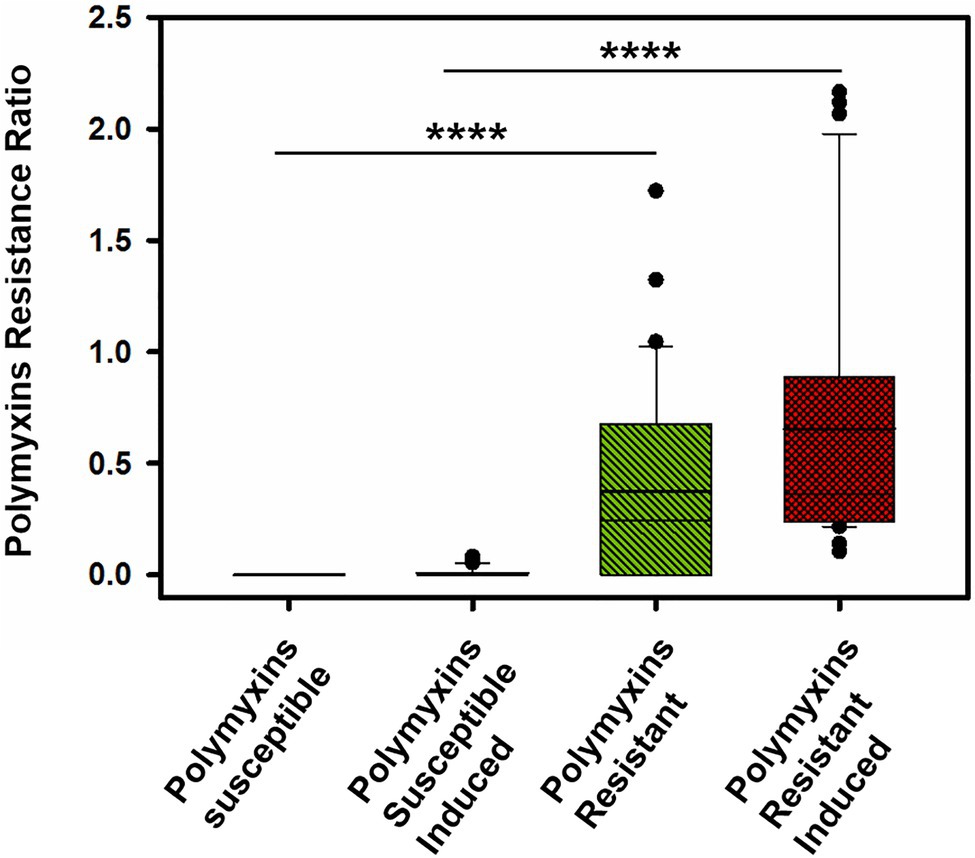
Figure 3. Distribution of the Polymyxin Resistance Ratios (PRRs) for the tested strains before and after induction with colistin for 4-h to a subinhibitory (1/2 MIC; ****p <0.0001).
Here, we have demonstrated that resistance to colistin can be quickly and easily detected in clinical strains of P. aeruginosa using the MALDIxin assay. However, an adaptation of the protocol currently used will be necessary before its use in routine laboratories. Unlike other species such as K. pneumoniae, A. baumannii, E. coli, and S. enterica, the signal intensity corresponding to the modified lipid A can be masked in some strains resistant to colistin. The addition of colistin during sample preparation phase improves the detection of P. aeruginosa strains resistant to colistin. Despite this additional step in sample preparation, the technique remains rapid (less than 5h), comparing favorably to the BMD for determining the MIC of colistin. The MALDIxin method complements the panel of so-called rapid methods for detecting resistance to colistin in clinical strains of P. aeruginosa, including the Rapid Polymyxin/Pseudomonas NP test (Sadek et al., 2020). MALDIxin is cost effective since it can be coupled with bacterial identification using the norharmane matrix with the MALDI Biotyper Sirius. One of the limitations of the study resides in the low number of strains tested, and further validation with an expanded panel is required. However, the most frequent mechanism responsible for colistin resistance in P. aeruginosa clinical strains (pmrB mutants) are included in this study, which supports the use of MALDIxin as a diagnostic for hospitalized patients.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
GL-M, LD, and KJ conceived the study, participated in its design, and performed the experiments. KJ provided the clinical isolates. KJ, KH, LD, MK, AF, PP, and GL-M wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the MRC Confidence in Concept Fund and the ISSF Wellcome Trust Grant 105603/Z/14/Z (GL-M).
LD, AF, and GL-M are co-inventors of the MALDIxin test for which a patent has been filed by Imperial Innovations (WO2018158573). MK is employee of Bruker, the manufacturer of the MALDI-TOF MS used in this study.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Brian Robertson, Imperial College London for careful reading of the manuscript.
Ahmadian, L., Haghshenas, M. R., Mirzaei, B., Norouzi Bazgir, Z., and Goli, H. R. (2020). Distribution and molecular characterization of resistance gene cassettes containing class 1 integrons in multi-drug resistant (MDR) clinical isolates of Pseudomonas aeruginosa. Infect. Drug Resist. 13, 2773–2781. doi: 10.2147/IDR.S263759
Barrow, K., and Kwon, D. H. (2009). Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53, 5150–5154. doi: 10.1128/AAC.00893-09
Bhat, R., Marx, A., Galanos, C., and Conrad, R. S. (1990). Structural studies of lipid A from Pseudomonas aeruginosa PAO1: occurrence of 4-amino-4-deoxyarabinose. J. Bacteriol. 172, 6631–6636. doi: 10.1128/jb.172.12.6631-6636.1990
Bolard, A., Schniederjans, M., Haussler, S., Triponney, P., Valot, B., Plesiat, P., et al. (2019). Production of norspermidine contributes to aminoglycoside resistance in pmrAB mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 63:e01044-19. doi: 10.1128/AAC.01044-19
Diekema, D. J., Hsueh, P. R., Mendes, R. E., Pfaller, M. A., Rolston, K. V., Sader, H. S., et al. (2019). The microbiology of bloodstream infection: 20-year trends from the SENTRY antimicrobial surveillance program. Antimicrob. Agents Chemother. 63:e00355-19. doi: 10.1128/AAC.00355-19
Doi, Y. (2019). Treatment options for carbapenem-resistant gram-negative bacterial infections. Clin. Infect. Dis. 69(Suppl. 7), S565–S575. doi: 10.1093/cid/ciz830
Dortet, L., Bonnin, R. A., Le Hello, S., Fabre, L., Bonnet, R., Kostrzewa, M., et al. (2020). Detection of colistin resistance in Salmonella enterica using MALDIxin test on the routine MALDI Biotyper Sirius mass spectrometer. Front. Microbiol. 11:1141. doi: 10.3389/fmicb.2020.01141
Dortet, L., Bonnin, R. A., Pennisi, I., Gauthier, L., Jousset, A. B., Dabos, L., et al. (2018a). Rapid detection and discrimination of chromosome- and MCR-plasmid-mediated resistance to polymyxins by MALDI-TOF MS in Escherichia coli: the MALDIxin test. J. Antimicrob. Chemother. 73, 3359–3367. doi: 10.1093/jac/dky330
Dortet, L., Broda, A., Bernabeu, S., Glupczynski, Y., Bogaerts, P., Bonnin, R., et al. (2019). Optimization of the MALDIxin test for the rapid identification of colistin resistance in Klebsiella pneumoniae using MALDI-TOF MS. J. Antimicrob. Chemother. 75, 110–116. doi: 10.1093/jac/dkz405
Dortet, L., Potron, A., Bonnin, R. A., Plesiat, P., Naas, T., Filloux, A., et al. (2018b). Rapid detection of colistin resistance in Acinetobacter baumannii using MALDI-TOF-based lipidomics on intact bacteria. Sci. Rep. 8:16910. doi: 10.1038/s41598-018-35041-y
Ernst, R. K., Adams, K. N., Moskowitz, S. M., Kraig, G. M., Kawasaki, K., Stead, C. M., et al. (2006). The Pseudomonas aeruginosa lipid A deacylase: selection for expression and loss within the cystic fibrosis airway. J. Bacteriol. 188, 191–201. doi: 10.1128/JB.188.1.191-201.2006
EUCAST (2019). The European Committee on antimicrobial susceptibility testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0, 2019. Available at: https://eucast.org/clinical_breakpoints/
Fernandez, L., Alvarez-Ortega, C., Wiegand, I., Olivares, J., Kocincova, D., Lam, J. S., et al. (2013). Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57, 110–119. doi: 10.1128/AAC.01583-12
Fernandez, L., Gooderham, W. J., Bains, M., McPhee, J. B., Wiegand, I., and Hancock, R. E. (2010). Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob. Agents Chemother. 54, 3372–3382. doi: 10.1128/AAC.00242-10
Fernandez, L., Jenssen, H., Bains, M., Wiegand, I., Gooderham, W. J., and Hancock, R. E. (2012). The two-component system CprRS senses cationic peptides and triggers adaptive resistance in Pseudomonas aeruginosa independently of ParRS. Antimicrob. Agents Chemother. 56, 6212–6222. doi: 10.1128/AAC.01530-12
Furniss, R. C. D., Dortet, L., Bolland, W., Drews, O., Sparbier, K., Bonnin, R. A., et al. (2019). Detection of colistin resistance in Escherichia coli by use of the MALDI Biotyper Sirius mass spectrometry system. J. Clin. Microbiol. 57:e01427-19. doi: 10.1128/JCM.01427-19
Horcajada, J. P., Montero, M., Oliver, A., Sorli, L., Luque, S., Gomez-Zorrilla, S., et al. (2019). Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 32:e00031-19. doi: 10.1128/CMR.00031-19
Mirzaei, B., Bazgir, Z. N., Goli, H. R., Iranpour, F., Mohammadi, F., and Babaei, R. (2020). Prevalence of multi-drug resistant (MDR) and extensively drug-resistant (XDR) phenotypes of Pseudomonas aeruginosa and Acinetobacter baumannii isolated in clinical samples from northeast of Iran. BMC Res. Notes 13:380. doi: 10.1186/s13104-020-05224-w
Moskowitz, S. M., Brannon, M. K., Dasgupta, N., Pier, M., Sgambati, N., Miller, A. K., et al. (2012). PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 56, 1019–1030. doi: 10.1128/AAC.05829-11
Moskowitz, S. M., Ernst, R. K., and Miller, S. I. (2004). PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186, 575–579. doi: 10.1128/JB.186.2.575-579.2004
Muller, C., Plesiat, P., and Jeannot, K. (2011). A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and beta-lactams in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55, 1211–1221. doi: 10.1128/AAC.01252-10
Needham, B. D., and Trent, M. S. (2013). Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 11, 467–481. doi: 10.1038/nrmicro3047
Nowicki, E. M., O’Brien, J. P., Brodbelt, J. S., and Trent, M. S. (2015). Extracellular zinc induces phosphoethanolamine addition to Pseudomonas aeruginosa lipid A via the ColRS two-component system. Mol. Microbiol. 97, 166–178. doi: 10.1111/mmi.13018
Sadek, M., Tinguely, C., Poirel, L., and Nordmann, P. (2020). Rapid Polymyxin/pseudomonas NP test for rapid detection of polymyxin susceptibility/resistance in Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1657–1662. doi: 10.1007/s10096-020-03884-x
Sader, H. S., Huband, M. D., Castanheira, M., and Flamm, R. K. (2017). Pseudomonas aeruginosa antimicrobial susceptibility results from four years (2012 to 2015) of the international network for optimal resistance monitoring program in the United States. Antimicrob. Agents Chemother. 61:e02252-16. doi: 10.1128/AAC.02252-16
Schurek, K. N., Sampaio, J. L., Kiffer, C. R., Sinto, S., Mendes, C. M., and Hancock, R. E. (2009). Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53, 4345–4351. doi: 10.1128/AAC.01267-08
Snesrud, E., Maybank, R., Kwak, Y. I., Jones, A. R., Hinkle, M. K., and McGann, P. (2018). Chromosomally encoded mcr-5 in colistin-nonsusceptible Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 62:e00679-18. doi: 10.1128/AAC.00679-18
Sorensen, M., Chandler, C. E., Gardner, F. M., Ramadan, S., Khot, P. D., Leung, L. M., et al. (2020). Rapid microbial identification and colistin resistance detection via MALDI-TOF MS using a novel on-target extraction of membrane lipids. Sci. Rep. 10:21536. doi: 10.1038/s41598-020-78401-3
Vincent, J. L., Rello, J., Marshall, J., Silva, E., Anzueto, A., Martin, C. D., et al. (2009). International study of the prevalence and outcomes of infection in intensive care units. JAMA 302, 2323–2329. doi: 10.1001/jama.2009.1754
Keywords: MALDI mass spectrometry, lipid A, colistin, Pseudomonas aeruginosa, clinical isolate
Citation: Jeannot K, Hagart K, Dortet L, Kostrzewa M, Filloux A, Plesiat P and Larrouy-Maumus G (2021) Detection of Colistin Resistance in Pseudomonas aeruginosa Using the MALDIxin Test on the Routine MALDI Biotyper Sirius Mass Spectrometer. Front. Microbiol. 12:725383. doi: 10.3389/fmicb.2021.725383
Received: 15 June 2021; Accepted: 05 August 2021;
Published: 31 August 2021.
Edited by:
Antonella Lupetti, University of Pisa, ItalyReviewed by:
Dinesh Sriramulu, Independent Researcher, Chennai, IndiaCopyright © 2021 Jeannot, Hagart, Dortet, Kostrzewa, Filloux, Plesiat and Larrouy-Maumus. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerald Larrouy-Maumus, Zy5sYXJyb3V5LW1hdW11c0BpbXBlcmlhbC5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.