- 1Laboratory of Bacterial Genetics, Vavilov Institute of General Genetics Russian Academy of Sciences, Moscow, Russia
- 2Peoples’ Friendship University of Russia (RUDN University), Moscow, Russia
- 3Department of Molecular Biology and Genetics, Federal Research and Clinical Center of Physical-Chemical Medicine of Federal Medical Biological Agency, Moscow, Russia
- 4South Africa Medical Research Council Bioinformatics Unit, South African National Bioinformatics Institute, University of the Western Cape, Cape Town, South Africa
Tuberculosis (TB), caused by the Mycobacterium tuberculosis complex bacteria, is one of the most pressing health problems. The development of new drugs and new therapeutic regimens effective against the pathogen is one of the greatest challenges in the way of tuberculosis control. Imidazo[1,2-b][1,2,4,5]tetrazines have shown promising activity against M. tuberculosis and M. smegmatis strains. Mutations in MSMEG_1380 lead to mmpS5–mmpL5 operon overexpression, which provides M. smegmatis with efflux-mediated resistance to imidazo[1,2-b][1,2,4,5]tetrazines, but the exact mechanism of action of these compounds remains unknown. To assess the mode of action of imidazo[1,2-b][1,2,4,5]tetrazines, we analyzed the transcriptomic response of M. smegmatis to three different concentrations of 3a compound: 1/8×, 1/4×, and 1/2× MIC. Six groups of genes responsible for siderophore synthesis and transport were upregulated in a dose-dependent manner, while virtual docking revealed proteins involved in siderophore synthesis as possible targets for 3a. Thus, we suggest that imidazo[1,2-b][1,2,4,5]tetrazines may affect mycobacterial iron metabolism.
Introduction
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), is one of the most successful bacterial pathogens. Despite many attempts to control the disease, it remains one of the world’s major health problems that causes more than 1.4 million deaths annually. The situation is complicated by the emergence of drug-resistant forms of the disease [World Health Organization (WHO)., 2020]. And if the initial mathematical models suggested that resistant isolates should be less transmissible and therefore unlikely to spread successfully in human populations, now it is clear that compensatory evolution and other factors drive the successful distribution of multidrug-resistant tuberculosis (Gagneux, 2009). In turn, infection with such isolates leads to longer treatment time with more toxic and costlier than the first-line drugs-based regimens and low treatment success (Rajbhandary et al., 2004). Hence there is an urgent need for novel drugs that are active against M. tuberculosis.
We have previously described imidazo[1,2-b][1,2,4,5]tetrazines as a class of promising antimycobacterial agents, with relatively high efficacy against both M. tuberculosis and M. smegmatis. An original test-system M. smegmatis aphVIII+ and docking studies showed that eukaryotic type serine-threonine protein kinases (STPKs) may act as targets of these compounds (Maslov et al., 2017, 2019). The main mechanism of resistance to these compounds is provided by mutations in MSMEG_1380, a transcriptional repressor of the mmpS5–mmpL5 efflux system. These mutations result in mmpS5–mmpL5 overexpression and increased efflux of the compounds from the cell (Maslov et al., 2020). A similar mechanism of resistance was also shown for bedaquiline and clofazimine (Hartkoorn et al., 2014), azoles (Milano et al., 2009), thiacetazone derivatives (Halloum et al., 2017), and tryptanthrins (Frolova et al., 2021) in various mycobacterial species. However, the mode of action, as well as biotarget(s) of imidazo[1,2-b][1,2,4,5]tetrazines have not yet been confirmed in vitro.
Additionally, we have constructed an M. smegmatis mc2 155 mutant, carrying a 2828 bp deletion in the mmpS5–mmpL5 operon, namely M. smegmatis Δmmp5, which was hypersensitive to imidazo[1,2-b][1,2,4,5]tetrazines (Shur et al., 2021). We used this strain in an attempt to obtain spontaneous mutants, resistant to the imidazo[1,2-b][1,2,4,5]tetrazines, with a mechanism different from mmpS5–mmpL5 overexpression, but could not do it even at a frequency of 10–10. This fact may indicate that these compounds have more than one biotarget, making the standard genetic approach useless in this case.
Transcriptomic studies, consisting of the analysis of the total RNA sequencing (RNA-seq), are now becoming a powerful tool for establishing additional mechanisms of action of antimicrobial agents, including anti-TB drugs, through the bacterial transcriptional response in the presence of the drug. This approach was used to establish the mechanisms of action of bedaquiline on dormant M. tuberculosis cells (Hards et al., 2015), mechanisms of ethionamide resistance of XDR-TB clinical isolates (Welzen et al., 2017), and to other antibiotics used in clinical practice (Briffotaux et al., 2019).
In the initial study we have tried to analyze the transcriptomic response of M. smegmatis to one of the imidazo[1,2-b][1,2,4,5]tetrazines – 3a (Supplementary Figure 1), treating the cells with 2× MIC (256 μg/ml) for 90 min, but this has led to a differential expression of over 1300 genes, making a detailed analysis rather intricate (Vatlin et al., 2020).
In this work, we describe the transcriptomic analysis of M. smegmatis in the presence of different subinhibitory concentrations of the imidazo[1,2-b][1,2,4,5]tetrazine 3a (1/8×, 1/4×, and 1/2× MIC) to get an insight in the gradual changes in the bacterial response to the increasing amounts of this drug. We have observed a dose-dependent differential expression of genes involved in iron acquisition, transport and storage, as well as other iron-regulated genes, showing a state of iron-starvation induced by 3a. In turn further virtual docking studies revealed siderophore synthesis proteins as potential targets of 3a.
Materials and Methods
Bacterial Strain and Growth Conditions
Mycobacterium smegmatis mc2 155 was grown in Middlebrook 7H9 medium (Himedia, India) supplemented with oleic albumin dextrose catalase (OADC, Himedia, India), 0.1% Tween-80 (v/v), and 0.4% glycerol (v/v), while soyabean-casein digest agar (M290, Himedia, India) and Middlebrook 7H11 agar (Himedia, India) supplemented with OADC were used as the solid media. Bacterial cultures in liquid medium were incubated in the Multitron incubator shaker (Infors HT, Basel, Switzerland) at 37°C and 250 rpm.
For drug exposure assay and transcriptomic analysis, M. smegmatis mc2 155 was inoculated from the agarized plates in 7H9 medium and grown until OD600 = 2.5 (two nights) to obtain a stable liquid culture without clumps, diluted in the proportion 1:200 and cultured overnight until OD600 = 2, then diluted in the proportion of 1:10 in fresh medium (to an approximate OD600 = 0.2). 3a 100× stocks were prepared in DMSO and added to the bacterial cultures to a final concentration corresponding to 1/8× MIC (16 μg/ml), 1/4× MIC (32 μg/ml), and 1/2× MIC (64 μg/ml) in 7H9 OADC medium (Maslov et al., 2020). The same volume of DMSO was added to the control samples (1% v/v). Bacterial cultures were incubated for 90 min [1/2 of the cell division time (Logsdon and Aldridge, 2018)] at 37°C and 250 rpm and proceeded to RNA extraction. The experiments were carried in three biological replicates.
The paper-disk drug susceptibility assay was performed as described before (Maslov et al., 2019). Briefly, M. smegmatis mc2 155 was grown in Middlebrook 7H9 broth to midexponential phase (OD600 = 1.2). Afterwards, the culture was diluted in the proportion of 1:9:10 (culture:water:M290 medium) and 5 ml were poured as the top layer on Petri dishes with agarized M290 medium. The plates were allowed to dry for at least 30 min, afterwards sterile paper disks with impregnated 300 nmol of 3a were plated. The plates were incubated for 2–3 days at 37°C, until the bacterial lawn was fully grown. Growth inhibition halos were measured to the nearest 1 mm. The experiments were carried out as triplicates; the average diameter and standard deviation (SD) were calculated. Those differences that had no intersection of the SDs with the control were considered significant.
Total RNA Extraction
Cells from 10 ml culture were harvested by centrifugation for 10 min at 3000 × g and 4°C, washed twice by 10 ml of fresh Middlebrook 7H9 broth to remove the traces of 3a and DMSO and once by 3 ml of RNAprotect Bacteria Reagent (Qiagen, United States) for RNA stabilization. M. smegmatis cells were homogenized in ExtractRNA reagent (Evrogen, Russia), followed by phenol (pH = 4.5)-chloroform-isoamyl alcohol (25:24:1) purification and precipitation with isopropanol (2:1, v/v).
DNase treatment was carried out as previously described (Bespyatykh et al., 2019) with TURBO DNA-free kit (Thermo Fisher Scientific, Waltham, MA, United States), in volumes of 100 μl, and further with the RNase-Free DNase Set (Qiagen, Hilden, Germany), according to the manufacturers’ protocol. RNA cleanup was performed with the RNeasy Mini Kit (Qiagen). The concentration and quality of the total RNA were checked by the Quant-it RiboGreen RNA assay (Thermo Fisher Scientific) and the RNA 6000 Pico chip (Agilent Technologies, Santa Clara, CA, United States), respectively.
Library Preparation and RNA Sequencing
Total RNA (1 μg) was used for library preparation as previously described (Bespyatykh et al., 2020) with some modifications. Ribosomal RNA was removed from the total RNA using the Ribo-Zero Plus rRNA Depletion Kit (Illumina, United States) and libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit (NEB), according to the manufacturer’s protocol. Subsequently, RNA cleanup was performed with the Agencourt RNA Clean XP kit (Beckman Coulter, Brea, United States). The library underwent a final cleanup using the Agencourt AMPure XP system (Beckman Coulter) after which the libraries’ size distribution and quality were assessed using a high sensitivity DNA chip (Agilent Technologies). Libraries were subsequently quantified by Quant-iT DNA Assay Kit, High Sensitivity (Thermo Fisher Scientific). Finally, equimolar quantities of all libraries (12 pM) were sequenced by a high throughput run on the Illumina HiSeq using 2 × 100 bp paired-end reads and a 5% Phix spike-in control. RNA-seq read data were deposited to the NCBI Sequence Read Archive under accession number PRJNA615922.
Bioinformatics Analysis
Quality control of the raw sequencing data was performed using FastQC (v.0.11.9) (Andrews, 2010), individual reports were merged with MultiQC (v.1.9) (Ewels et al., 2016). Adapters and low-quality reads were removed using Trimmomatic (v.0.39) (Bolger et al., 2014). HISAT2 (v.2.2.1) (Kim et al., 2019) was used to map trimmed reads to the reference M. smegmatis mc2 155 genome (CP000480.1). Mapping quality and coverage along genes were assessed with QualiMap (v.2.2.2) (Okonechnikov et al., 2016). Mapped reads were assigned to genes with featureCounts (v.2.0.1) (Liao et al., 2014). Differential gene expression (DGE) analysis was performed using the edgeR (v.3.30.3) (Robinson et al., 2010) for R (v.4.0.2) (R Core Team, 2020). Genes with a false discovery rate (FDR) cutoff of 0.001 and fold change (FC) log2FC ≥ 1, or log2FC ≤ −1 were considered to be differentially expressed. Plots were generated within R (v.4.0.2) using ggplot2 (v.3.3.2) (Wickham, 2009) and cowplot (v.1.1.0) (Wilke, 2016) packages. Further functional enrichment analysis of the COG categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for differentially expressed genes (DEGs) was performed using the DAVID (v.6.8) (Huang et al., 2009) database, categories were considered enriched with p ≤ 0.05.
Virtual Screening Studies
To understand the effect of the imidazo[1,2-b][1,2,4,5]tetrazine on the structures of proteins associated with the iron metabolism in M. smegmatis, the molecular docking was performed using AutoDock vina (Trott and Olson, 2010). Primarily, the sequence information regarding the listed protein was obtained from the UniProt.1 As the majority of proteins don’t have X-ray crystallographic (X-ray) structures available in the biological database, therefore, the SWISS-MODEL was used which is very efficient in modeling proteins with amino acid numbers as large as 5000 (Schwede et al., 2003). The 3-D coordinates of the respective proteins were prepared and optimized using the utilities of MAESTRO (Schrödinger Release 2018-1: Maestro, Schrödinger, LLC, New York, NY, United States, 2018) and the obtained structures were energetically minimized using the modules present in JAGUAR (Bochevarov et al., 2013). Consequently, the docking was performed using AutoDock vina and docked conformations were predicted through the combination of free energy functionals, empirical force field and the Lamarckian Genetic Algorithm (Trott and Olson, 2010). The dimensional space of 40 × 40 × 40 Å was set along the XYZ directions with varied central coordinates and the spacing was set to the 0.375 Å. To achieve the maximum accuracy, the parameters were set to the highest efficiency range.
Results
Transcriptional Response of M. smegmatis to 3a Treatment – KEGG Analysis
To evaluate the changes in bacterial metabolism during 3a treatment, the transcriptomic profiles of M. smegmatis mc2 155 strain exposed to 3a at 1/8×, 1/4×, and 1/2× MIC for a duration of 90 min were examined. In total, approximately 6000 quantifiable transcripts were identified for each experiment. These results are comparable to the RNA-seq of the DMSO-treated cells (control) and correspond to almost complete coverage of the genome. A non-metric multidimensional scaling analysis showed a clear separation between samples exposed to different 3a concentrations (Supplementary Figure 2).
Analysis of DEGs revealed that 3a significantly (log2FC ≥ 1, or log2FC ≤ −1, FDR < 0.001) variates the expression level of numerous genes. We observed that 7.4% (452/6079) genes were upregulated and 14.1% (856/6079) were downregulated upon exposure to 16 μg/ml 3a, with these numbers increasing up to 13.9 and 14.9%, respectively, at the exposure to 64 μg/ml 3a, as compared to the control (Supplementary Table 1). We were able to detect a dose-dependent effect (at least a twofold increase or decrease of expression FC, upon twofold increase in 3a concentration) for 120 and 173 genes, respectively (Supplementary Table 2). Upon implementation of an additional filter (at least a 2× FC at 1/4× MIC relative to 1/8× MIC, and at least a 2× FC at 1/2× MIC relative to 1/4× MIC) we were left with just 31 overexpressed and 11 underexpressed genes (Supplementary Table 3). Among those 31 overexpressed genes, 25 were related to iron acquisition and transport (exochelin synthesis and transport, mbt-1 and mbt-2 clusters, ESX-3 operon and a gene encoding a siderophore-interacting protein).
DAVID functional annotation tool was used to infer enriched KEGG pathways from sets of upregulated DEGs. Enriched pathway (p < 0.05) that was present in all MICs was “biosynthesis of siderophore group non-ribosomal peptides,” and comprised eight genes (MSMEG_4509–MSMEG_4513, MSMEG_4515, MSMEG_4516, and MSMEG_4524) in all concentrations of the compound (Figure 1).
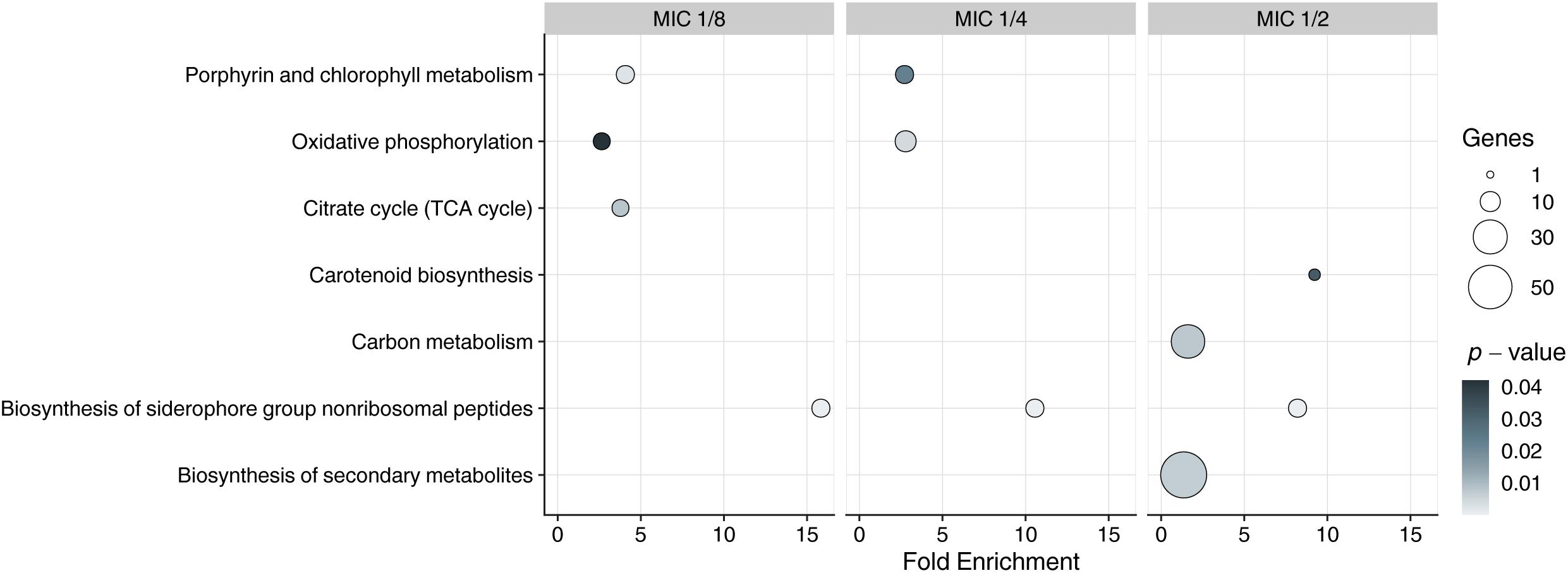
Figure 1. Kyoto Encyclopedia ofGenes and Genomes enrichment analysis of DEGs in exposed to 3a samples with different MICs. Y-axis represents KEGG pathways, X-axis represents fold enrichment (amount of input pathway DEGs/background gene set, ≥1.5 considered as interesting). Dots colors represent the p-values of enrichment, the dots’ size represents the gene number in the pathway.
Dose-Dependent Gene Regulation Reveals Further Impact of Imidazo[1,2-b][1,2,4,5]tetrazine on Iron Metabolism
Since the siderophore biosynthesis KEGG pathway was enriched in response to 3a exposure (Figure 1), we have decided to conduct a deeper analysis of the genes involved in the iron metabolism (Figure 2), using their homologs in M. tuberculosis H37Rv according to Namouchi et al. (2017) as an additional reference, as more data is published for this organism.
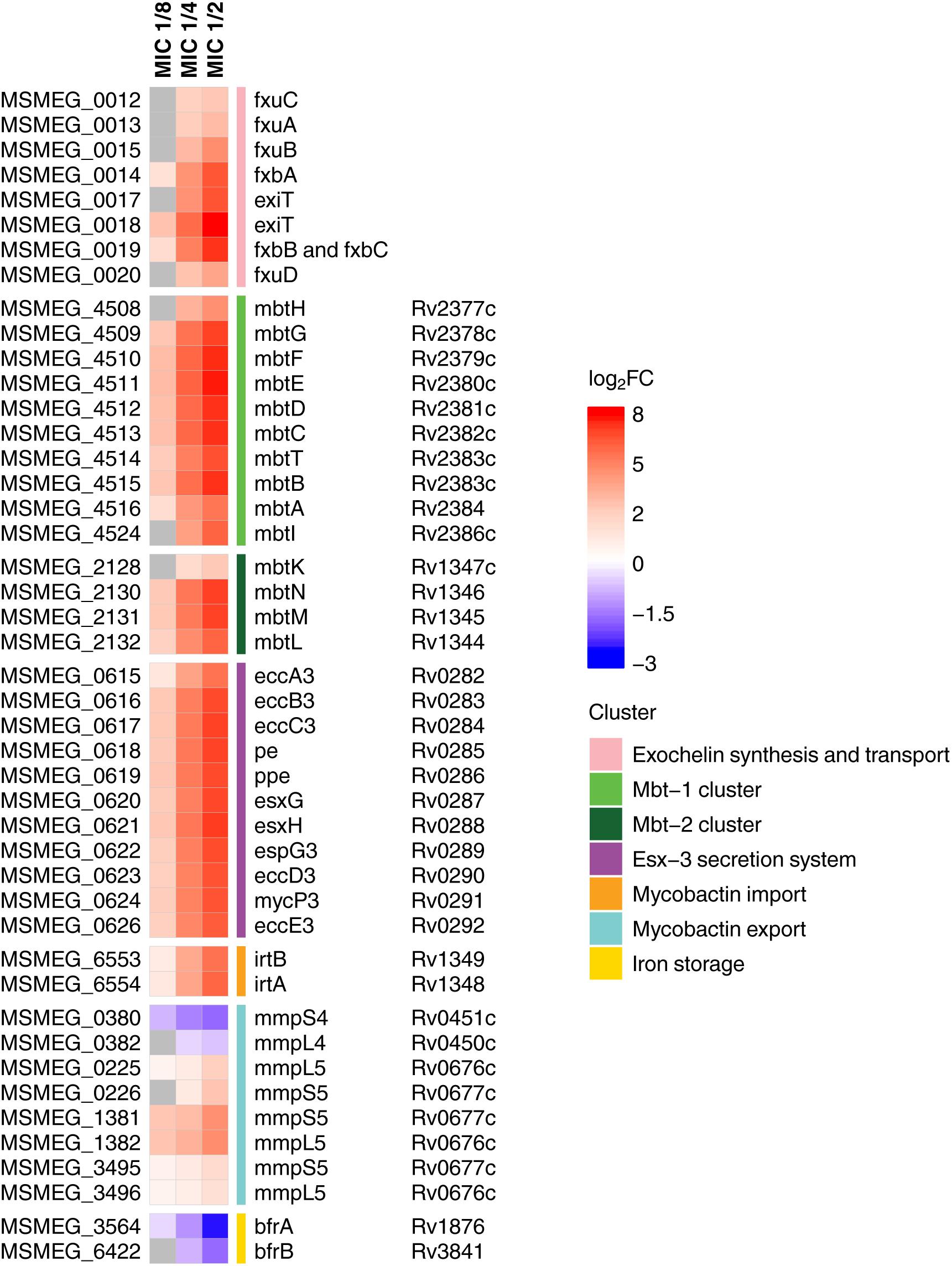
Figure 2. Heatmap showing log2-transformed fold change in expression levels of iron metabolites during 3a exposure. M. smegmatis mc2 155 locus tags (MSMEG_*) are indicated to the left of the heatmap, and M. tuberculosis H37Rv locus tags (Rv*) are indicated to the right of the heatmap, genes’ names are relative to M. smegmatis mc2 155. Genes with FDR > 0.001 are shown in gray.
We have found that in compliance with the KEGG analysis, the mbt gene clusters were upregulated in a dose-dependent manner (Figure 2). These operons are the key elements of the “biosynthesis of siderophore group non-ribosomal peptides” pathway, which is responsible for the synthesis of the most important iron-chelating compounds – siderophores (mycobactin and carboxymycobactin in mycobacteria) (Chao et al., 2018). Similar changes were observed for the biosynthesis of exochelin, which is specific for non-virulent mycobacteria (Ratledge and Ewing, 1996). Opposite changes have been shown in iron storage genes bfrA and bfrB (Reddy et al., 2012), expression of which declined with increasing dose of the compound.
Overexpression of genes involved in iron and siderophore acquisition and transport was also observed (Figure 2). Among these overexpressed genes was ESX-3 operon, which is a vital core operon and is part of the type VII secretion system (T7SS), responsible for iron acquisition and controlled by the iron-dependent transcriptional repressor (IdeR) (Ioerger et al., 2013). The iron-dependent transporters irtA and irtB (Ryndak et al., 2010), and the genes of the mmpS5–mmpL5 siderophore transport system (Wells et al., 2013) were also amidst overexpressed genes, though mmpS4–mmpL4 genes were downregulated.
In silico Interaction Studies Reveal Siderophore Synthesis Genes as Potential 3a Targets
As the transcriptomic analysis revealed the overexpression of iron acquisition and transport genes in response to 3a exposure, with the highest impact observed for siderophore synthesis genes. Therefore, to understand the structural complementarity of 3a with the selected 46 overexpressed genes, an extensive molecular docking study was performed, which revealed the binding preferences of the inhibitor molecule. Consequently, streamlined filtering enabled the selection of specific protein targets such as protein FxuA which is considered to be significant in the biosynthesis of exochelin, one of the two siderophores present in M. smegmatis which are involved in iron transport (Fiss et al., 1994). Similarly, FxbA which was identified to be a putative formyltransferase catalyzed the addition of the formyl group in the exochelin biosynthesis (Fiss et al., 1994). Through docking studies, it was observed that the imidazo[1,2-b][1,2,4,5]tetrazine was bonded with the highest affinity to the structure of FxuA followed by FxbA with free energies of interaction −8.0 and −7.8 kcal/mol, respectively, among the 46 docked systems (Table 1). These observations showed that the activity of exochelin could be inhibited to a relatively higher extent by the administration of imidazo[1,2-b][1,2,4,5]tetrazine. The compound was observed to be interacting with Ser41, Thr42, Trp68, Arg69, Arg72, and Leu309 of FxuA, while for FxbA the interactions were observed with Ala85, Asn87, Trp88, Thr90, Asn106, His108, Phe118, and Leu143 (Supplementary Figures 3, 4). The output of MetaPocket (Huang, 2009) revealed that the respective interacting residues form the part of ligand binding cavities. Furthermore, among the mycobactin biosynthesis enzymes, the highest binding affinities were observed with MbtC and MbtD with calculated free energies of −7.7 kcal/mol (Table 1). The MbtC binding pocket contains Pro282, His311, Thr313, Gly318, Phe415, and Met417, while for MbtD the amino acids Gln104, His190, Ala262, Ile287, and Val293 were observed in the interaction pocket (Supplementary Figures 5, 6). Similar binding affinity was observed for FxbC with the free energy of −7.7 kcal/mol and interacting residues are Ser1718, Phe1762, Phe1764, Ile1863, Thr1864, and His2004 (Supplementary Figure 7). Moreover, a relatively higher binding affinity of −7.8 kcal/mol was observed for EccA3 protein which was characterized to be a member of CbbX family ATPases involved in the ESX-3 secretory pathway (Gaur et al., 2017). We observed that the EccA3 contains the residues Pro366, Glu542, Arg543, Ser545, Ala549, Tyr648, and Arg469 in the docked pocket of the protein (Supplementary Figure 8).
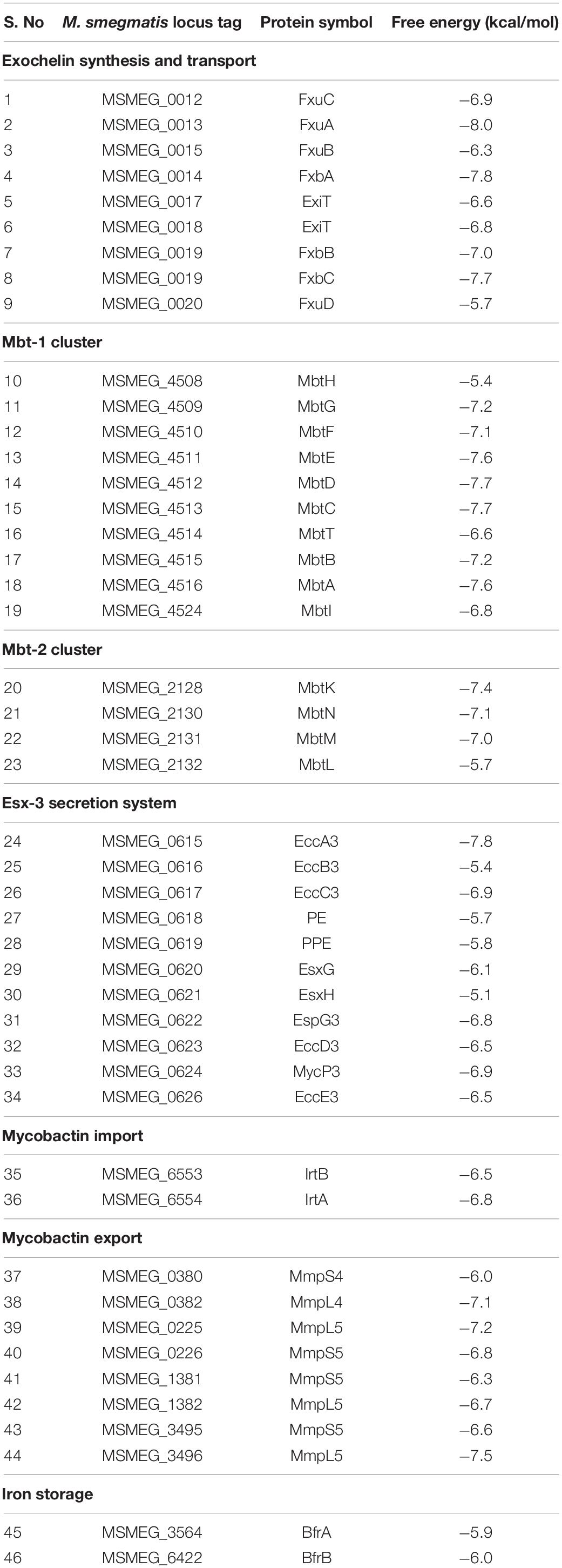
Table 1. List of differential inhibitory efficiency in the form of free energy of binding for against the proteins involved in iron acquisition and transport in M. smegmatis.
Addition of Iron to the Growth Medium Reduces M. smegmatis 3a Susceptibility
We have previously observed that M. smegmatis had different 3a MIC values in 7H9 + OADC and Lemco-Tw medium (128 and 64 μg/ml, respectively) (Maslov et al., 2019, 2020). With our findings, that 3a could lead to iron starvation by inhibiting siderophore synthesis, this could be due to the additional iron in the composition of Middlebrook 7H9 broth in the form of ferric ammonium citrate.
To prove this hypothesis, we have additionally tested M. smegmatis susceptibility to 3a by paper disk method on solid media with different iron content: soyabean digest agar (no iron in the composition), soyabean digest agar with 2.5 mM FeSO4, and Middlebrook 7H11 + OADC medium (containing 40 mg/l ferric ammonium citrate, ∼150 μM). M. smegmatis showed higher resistance on both iron-supplemented media as compared to the iron-free soyabean digest agar, confirming that the addition of iron reduces the susceptibility of bacterial cells to 3a, presumably by reducing the iron starvation (Figure 3).
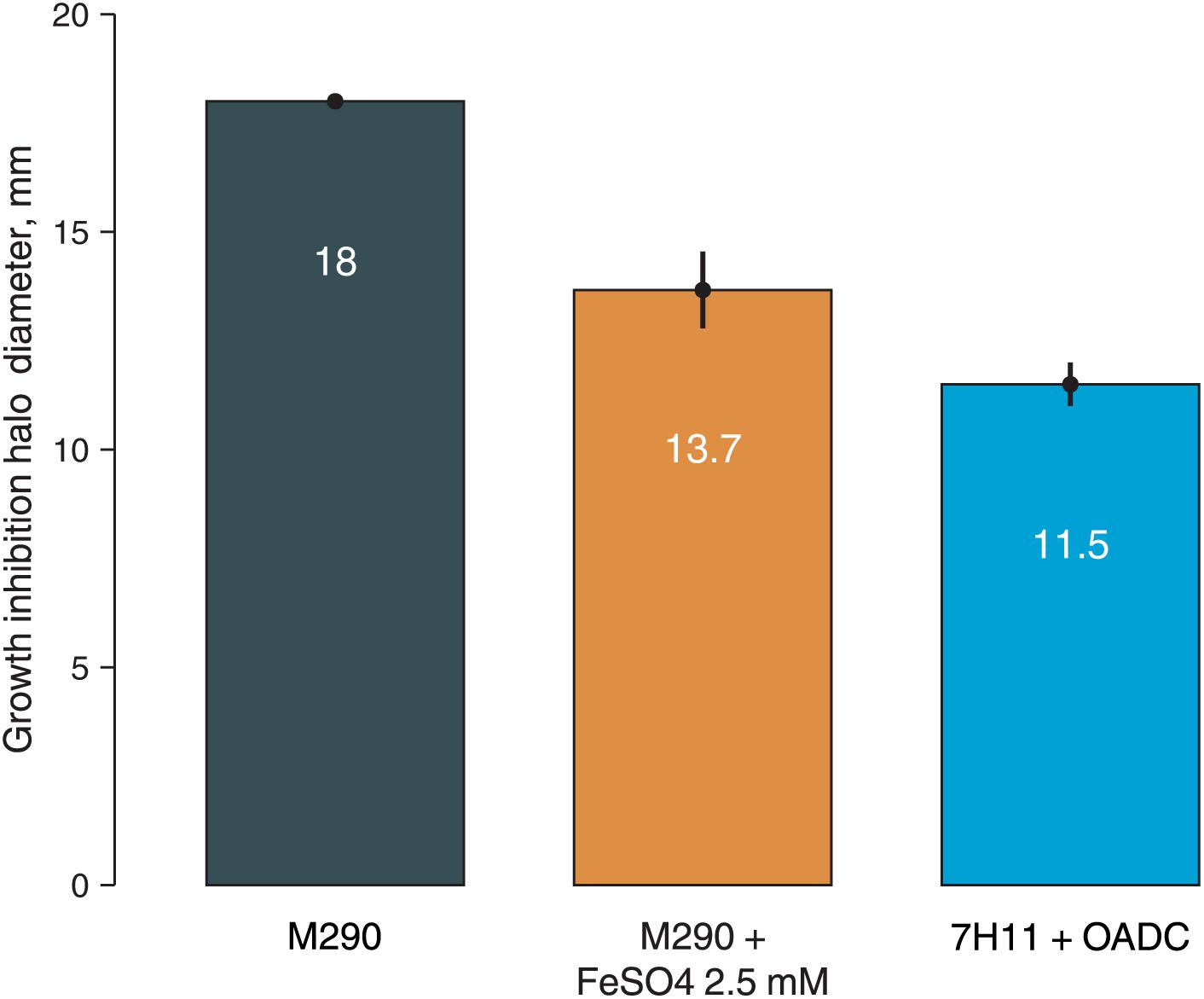
Figure 3. Growth inhibition halos, produced by 3a (300 nmol/disk) on M. smegmatis mc2 155 on different media: soyabean digest agar (M290), soyabean digest agar + 2.5 mM FeSO4, and 7H11 + OADC. Columns represent the average growth inhibition halo diameters, while the error bars represent the standard deviation, calculated from three experiments.
Discussion
The transcriptomic analysis of M. smegmatis mc2 155, exposed to increasing doses of the imidazo[1,2-b][1,2,4,5]tetrazine 3a has revealed a dose-dependent change in the expression of many genes involved in iron metabolism, including the genes responsible for siderophore synthesis and transport: the gene cluster responsible for exochelin synthesis and transport, mbtA-G and mbtK-N clusters, ESX-3 operon, irtA-B, mmpS5–mmpL5, and bfrA-B operons.
Our observations based on docking studies suggest that imidazo[1,2-b][1,2,4,5]tetrazine 3a could inhibit the iron metabolizing enzymes and may target the proteins involved in mycobactin biosynthesis, but relatively higher affinities were observed with the proteins involved in exochelin biosynthetic pathways. Virtual screening showed that FxuA is the most likely target of 3a, followed by FxbA, MbtC, mbtD, and FxbC. The inhibition of siderophores’ synthesis by 3a could lead to the low-iron conditions transcriptomic profile, observed in our study with a few exceptions. This mechanism could also explain a higher sensitivity of M. tuberculosis strains to imidazo[1,2-b][1,2,4,5]tetrazines (Maslov et al., 2019), as M. tuberculosis is more sensitive to iron starvation as a human pathogen (Doherty, 2007; Skaar, 2010), as well as the higher 3a MIC in 7H9 medium (128 μg/ml), containing ferric ammonium citrate, as compared to the Lemco-Tw broth (64 μg/ml), which contains no additional iron in its composition (Maslov et al., 2019, 2020). As shown in this study, the addition of iron reduced M. smegmatis 3a susceptibility on agarized medium as well.
Exochelin is a siderophore specific to non-virulent mycobacteria (Ratledge and Ewing, 1996). The products of the mbtA-G and mbtK-N gene clusters are part of a complex mechanism for the synthesis of siderophores in the cell. Mutation of any mbt gene can disrupt the synthesis of siderophores, which, in turn, prevents bacteria from assimilating iron from the environment, leading to impaired cell growth. It was shown that the M. tuberculosis H37Rv mutant, in which the mbtB gene was replaced by recombination with a hygromycin resistance cassette, was restricted for growth in a medium with limited iron but grew normally in a medium with a high iron content (Voss et al., 2000). The ESX-3 operon encodes a type VII secretion system. These systems are common in mycobacteria (Bitter et al., 2009) and represent a complex with many components and many substrates. The function of these systems has not yet been fully studied, however, there is evidence of their involvement in the secretion of virulence factors and host–pathogen interaction in M. tuberculosis (Bitter et al., 2009). Both mbtA-G genes products and type VII secretion systems are currently considered promising targets for new anti-TB drugs (Bottai et al., 2014; Maganti et al., 2014).
The mbtA-G, ESX-3, mmpS5–mmpL5, and irtA-B are controlled by HupB and iron levels (Pandey et al., 2014b), some of them in IdeR-dependent (mbtA-G, bfrA-B) and others in the IdeR-independent way (Rodriguez et al., 2002). HupB and IdeR are two key regulators of iron metabolism in mycobacteria (Rodriguez et al., 2002; Pandey et al., 2014b). HupB is a 22 kDa (214 a.a.) protein with a C-terminal region unique to mycobacteria. The orthologs of HupB in mycobacterial species differ by about 20–30% in their amino acid sequence. HupB binds to the 10 bp region (HupB-box) and activates its regulated genes’ expression (Pandey et al., 2014b), while IdeR acts as a transcription repressor: in high-iron conditions, two IdeR proteins form a complex with Fe2+ ions and is able to bind to the IdeR-box, preventing the start of transcription (Rodriguez et al., 2002). When iron level is low, the IdeR-Fe2+ complex is disrupted, and transcription is enabled. In another study by Pandey et al. (2014a) it has been shown that the promoter region of hupB contains 2 IdeR-boxes and a HupB-box, suggesting that hupB could also be regulated by HupB/IdeR system (self-upregulated in low-iron conditions). While it has been shown in vitro, that HupB and IdeR can both bind to the hupB promoter (Pandey et al., 2014a), the upregulation of HupB was not observed in low-iron conditions in other HupB/IdeR-related studies (Rodriguez et al., 2002; Pandey et al., 2014b), meaning that this mechanism is not necessarily functional in the cell culture. We did not observe any hupB upregulation as well.
The HupB/IdeR was reported to be regulated by mycobacterial STPKs PknE, PknF, and PknB (Gupta et al., 2014). Our previous studies showed that 3a could be docked in the PknB adenine-binding socket (Bekker et al., 2012). However, our current docking studies revealed lower affinity of 3a toward PknE and PknB (−6.8 and −7.3 kcal/mol, respectively) and to PknF homologs in M. smegmatis (ranging from −6.3 to −7.3 kcal/mol), meaning that these proteins could be not the primary targets for this compound. The fact that some iron-sensitive, but HupB/IdeR-independent genes, such as the nuo and pqqE–lldD1 operons (Rodriguez et al., 2002) changed their fold expression dose-dependently (Supplementary Table 1), supports the theory of HupB/IdeR-independent mode of action for 3a.
The MmpS4–MmpL4 and MmpS5–MmpL5 were shown to be crucial for siderophore transport in M. tuberculosis (Wells et al., 2013). Our transcriptomic data shows, that the MSMEG_1381–MSMEG_1382 system might play the primary role in siderophore efflux in M. smegmatis among the 3 homologs present in the genome. Our failure to obtain spontaneous 3a-resistant mutants using the M. smegmatis Δmmp5 strain could be caused by the accumulation of siderophores in the cell, caused by 3a-induced siderophore synthesis and the inability to provide their efflux. As the siderophores can be toxic to mycobacterial cells at high concentrations (Jones et al., 2014), this could provide a synergistic effect, requiring more than 1 mutation to override it, and thus drastically lowering the frequency of spontaneous drug-resistant mutants’ emergence.
The understanding of a drug mode of action can be crucial in its development, especially in terms of its structure optimization. We have previously described a set of imidazo[1,2-b][1,2,4,5]tetrazines as promising anti-TB drug candidates and shown that mutations leading to mmpS5–mmpL5 operon overexpression provide M. smegmatis efflux-mediated resistance to these compounds. We now show, using the transcriptomic analysis of M. smegmatis exposed to different doses of imidazo[1,2-b][1,2,4,5]tetrazine 3a, combined with virtual screening approaches, that this compound may be targeting mycobacterial iron-uptake pathways by inhibiting siderophore synthesis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
DM: conceptualization, project administration, and funding acquisition. AV, ES, MS, DB, and DM: formal analysis. AV, MS, DB, and KK: investigation. DM, AC, and VD: resources. AV, ES, MS, KK, and DM: writing–original draft preparation. ES, MS, DB, and DM: writing review and editing. ES, MS, and DB: visualization. VD and AC: supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation (RSF, grant number 17-75-20060-P to DM), and by the South African Research Chairs Initiative of the Department of Science and Innovation and National Research Foundation of South Africa (grant ID 64751 to AC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank V. N. Charushin and G. L. Rusinov of the Laboratory of Heterocyclic Compounds, Postovsky Institute of Organic Synthesis, Ural Branch of RAS, for generously providing the compound 3a for this study. We would also like to thank the Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Federal Research and Clinical Center of Physical-Chemical Medicine of Federal Medical Biological Agency for the opportunity to use computational and sequencing resources.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.724042/full#supplementary-material
Footnotes
References
Andrews, S. (2010). FastQC: a Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed November, 2020).
Bekker, O. B., Danilenko, V. N., Ishmetova, R. I., Maslov, D. A., Rusinov, G. L., Tolshchina, S. G., et al. (2012). Substituted azolo[1,2,4,5]Tetrazines - Inhibitors of Antibacterial Serine-Threonine Protein Kinases. Russia: Patent RU.
Bespyatykh, J., Shitikov, E., Bespiatykh, D., Guliaev, A., Klimina, K., Veselovsky, V., et al. (2020). Metabolic Changes of Mycobacterium tuberculosis during the Anti-Tuberculosis Therapy. Pathogens doi: 10.3390/pathogens9020131
Bespyatykh, J., Shitikov, E., Guliaev, A., Smolyakov, A., Klimina, K., Veselovsky, V., et al. (2019). System OMICs analysis of Mycobacterium tuberculosis Beijing B0/W148 cluster. Sci. Rep. 9, 19255. doi: 10.1038/s41598-019-55896-z
Bitter, W., Houben, E. N. G., Bottai, D., Brodin, P., Brown, E. J., Cox, J. S., et al. (2009). Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 5:e1000507. doi: 10.1371/journal.ppat.1000507
Bochevarov, A. D., Harder, E., Hughes, T. F., Greenwood, J. R., Braden, D. A., Philipp, D. M., et al. (2013). Jaguar: a high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum. Chem. 113, 2110–2142. doi: 10.1002/qua.24481
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bottai, D., Serafini, A., Cascioferro, A., Brosch, R., and Manganelli, R. (2014). Targeting type VII/ESX secretion systems for development of novel antimycobacterial drugs. Curr. Pharm. Des. 20, 4346–4356. doi: 10.2174/1381612819666131118170717
Briffotaux, J., Liu, S., and Gicquel, B. (2019). Genome-Wide Transcriptional Responses of Mycobacterium to Antibiotics. Front. Microbiol. 10:249. doi: 10.3389/fmicb.2019.00249
Chao, A., Sieminski, P. J., Owens, C. P., and Goulding, C. W. (2018). Iron Acquisition in Mycobacterium tuberculosis. Chem. Rev. 119, 1193–1220. doi: 10.1021/acs.chemrev.8b00285
Doherty, C. P. (2007). Host-pathogen interactions: the role of iron. J. Nutr. 137, 1341–1344. doi: 10.1093/jn/137.5.1341
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Fiss, E. H., Yu, S., and Jacobs, W. R. (1994). Identification of genes involved in the sequestration of iron in mycobacteria: the ferric exochelin biosynthetic and uptake pathways. Mol. Microbiol. 14, 557–569. doi: 10.1111/j.1365-2958.1994.tb02189.x
Frolova, S. G., Klimina, K. M., Kumar, R., Vatlin, A. A., Salunke, D. B., Kendrekar, P., et al. (2021). Identification of Mutations Conferring Tryptanthrin Resistance to Mycobacterium smegmatis. Antibiotics 10, 6–10. doi: 10.3390/antibiotics10010006
Gagneux, S. (2009). Fitness cost of drug resistance inMycobacterium tuberculosis. Clin. Microbiol. Infec. 15, 66–68. doi: 10.1111/j.1469-0691.2008.02685.x
Gaur, A., Sharma, V. K., Shree, S., Rai, N., and Ramachandran, R. (2017). Characterization of EccA3, a CbbX family ATPase from the ESX-3 secretion pathway of M. tuberculosis. Biochim. Biophys. Acta Proteins Proteom 1865, 715–724. doi: 10.1016/j.bbapap.2017.04.001
Gupta, M., Sajid, A., Sharma, K., Ghosh, S., Arora, G., Singh, R., et al. (2014). HupB, a nucleoid-associated protein of mycobacterium tuberculosis, is modified by serine/threonine protein kinases in vivo. J. Bacteriol. 196, 2646–2657. doi: 10.1128/jb.01625-14
Halloum, I., Viljoen, A., Khanna, V., Craig, D., Bouchier, C., Brosch, R., et al. (2017). Resistance to Thiacetazone Derivatives Active against Mycobacterium abscessus Involves Mutations in the MmpL5 Transcriptional Repressor MAB_4384. Antimicrob. Agents Chemother. 61:N246. doi: 10.1128/aac.02509-16
Hards, K., Robson, J. R., Berney, M., Shaw, L., Bald, D., Koul, A., et al. (2015). Bactericidal mode of action of bedaquiline. J. Antimicrob. Chemother. 70, 2028–2037. doi: 10.1093/jac/dkv054
Hartkoorn, R. C., Uplekar, S., and Cole, S. T. (2014). Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 58, 2979–2981. doi: 10.1128/aac.00037-14
Huang, B. (2009). MetaPocket: a Meta Approach to Improve Protein Ligand Binding Site Prediction. Omics. J. Integr. Biol. 13, 325–330. doi: 10.1089/omi.2009.0045
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Ioerger, T. R., O’Malley, T., Liao, R., Guinn, K. M., Hickey, M. J., Mohaideen, N., et al. (2013). Identification of new drug targets and resistance mechanisms in Mycobacterium tuberculosis. PLoS One 8:e75245. doi: 10.1371/journal.pone.0075245
Jones, C. M., Wells, R. M., Madduri, A. V. R., Renfrow, M. B., Ratledge, C., Moody, D. B., et al. (2014). Self-poisoning of Mycobacterium tuberculosisby interrupting siderophore recycling. Proc. Natl. Acad. Sci. 111, 1945–1950. doi: 10.1073/pnas.1311402111
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Logsdon, M. M., and Aldridge, B. B. (2018). Stable Regulation of Cell Cycle Events in Mycobacteria: insights From Inherently Heterogeneous Bacterial Populations. Front. Microbiol. 9:514. doi: 10.3389/fmicb.2018.00514
Maganti, L., Consortium, O. S. D. D., and Ghoshal, N. (2014). Probing the structure of Mycobacterium tuberculosis MbtA: model validation using molecular dynamics simulations and docking studies. J. Biomol. Struct. Dyn. 32, 273–288. doi: 10.1080/07391102.2012.762752
Maslov, D. A., Bekker, O. B., Alekseeva, A. G., Kniazeva, L. M., Mavletova, D. A., Afanasyev, I. I., et al. (2017). Aminopyridine- and aminopyrimidine-based serine/threonine protein kinase inhibitors are drug candidates for treating drug-resistant tuberculosis. Bull. RSMU 2017, 38–43. doi: 10.24075/brsmu.2017-01-04
Maslov, D. A., Korotina, A. V., Shur, K. V., Vatlin, A. A., Bekker, O. B., Tolshchina, S. G., et al. (2019). Synthesis and antimycobacterial activity of imidazo[1,2-b][1,2,4,5]tetrazines. Eur. J. Med. Chem. 178, 39–47. doi: 10.1016/j.ejmech.2019.05.081
Maslov, D. A., Shur, K. V., Vatlin, A. A., and Danilenko, V. N. (2020). MmpS5-MmpL5 Transporters Provide Mycobacterium smegmatis Resistance to imidazo[1,2-b][1,2,4,5]tetrazines. Pathogens 9:166. doi: 10.3390/pathogens9030166
Milano, A., Pasca, M. R., Provvedi, R., Lucarelli, A. P., Manina, G., Ribeiro, A. L., et al. (2009). Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis 89, 84–90. doi: 10.1016/j.tube.2008.08.003
Namouchi, A., Cimino, M., Favre-Rochex, S., Charles, P., and Gicquel, B. (2017). Phenotypic and genomic comparison of Mycobacterium aurum and surrogate model species to Mycobacterium tuberculosis: implications for drug discovery. BMC Genom. 18:530. doi: 10.1186/s12864-017-3924-y
Okonechnikov, K., Conesa, A., and García-Alcalde, F. (2016). Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32, 292–294. doi: 10.1093/bioinformatics/btv566
Pandey, S. D., Choudhury, M., and Sritharan, M. (2014a). Transcriptional regulation of Mycobacterium tuberculosis hupB gene expression. Microbiology 160, 1637–1647. doi: 10.1099/mic.0.079640-0
Pandey, S. D., Choudhury, M., Yousuf, S., Wheeler, P. R., Gordon, S. V., Ranjan, A., et al. (2014b). Iron-regulated protein HupB of Mycobacterium tuberculosis positively regulates siderophore biosynthesis and is essential for growth in macrophages. J. Bacteriol. 196, 1853–1865. doi: 10.1128/jb.01483-13
Rajbhandary, S. S., Marks, S. M., and Bock, N. N. (2004). Costs of patients hospitalized for multidrug-resistant tuberculosis. Int. J. Tuberc. Lung Dis. 8, 1012–1016.
Ratledge, C., and Ewing, M. (1996). The occurrence of carboxymycobactin, the siderophore of pathogenic mycobacteria, as a second extracellular siderophore in Mycobacterium smegmatis. Microbiology 142, 2207–2212. doi: 10.1099/13500872-142-8-2207
Reddy, P. V., Puri, R. V., Khera, A., and Tyagi, A. K. (2012). Iron Storage Proteins Are Essential for the Survival and Pathogenesis of Mycobacterium tuberculosis in THP-1 Macrophages and the Guinea Pig Model of Infection. J. Bacteriol. 194, 567–575. doi: 10.1128/jb.05553-11
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rodriguez, G. M., Voskuil, M. I., Gold, B., Schoolnik, G. K., and Smith, I. (2002). ideR, An essential gene in mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70, 3371–3381. doi: 10.1128/iai.70.7.3371-3381.2002
Ryndak, M. B., Wang, S., Smith, I., and Rodriguez, G. M. (2010). The Mycobacterium tuberculosis High-Affinity Iron Importer, IrtA, Contains an FAD-Binding Domain▽†. J. Bacteriol. 192, 861–869. doi: 10.1128/jb.00223-09
Schwede, T., Kopp, J., Guex, N., and Peitsch, M. C. (2003). SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385. doi: 10.1093/nar/gkg520
Shur, K. V., Frolova, S. G., Akimova, N. I., Danilenko, V. N., and Maslov, D. A. (2021). A Test System for in vitro Screening Antimycobacterial Drug Candidates for MmpS5-MmpL5 Mediated Resistance. Russ. J. Genet. 57, 114–116. doi: 10.1134/s1022795421010154
Skaar, E. P. (2010). The Battle for Iron between Bacterial Pathogens and Their Vertebrate Hosts. PLoS Pathog. 6:e1000949. doi: 10.1371/journal.ppat.1000949
R Core Team (2020). R: a Language and Environment for Statistical Computing. Austria: R Foundation for Statistical Computing.
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi: 10.1002/jcc.21334
Vatlin, A. A., Klimina, K. M., Frolova, S. G., Danilenko, V. N., and Maslov, D. A. (2020). Transcriptomic dataset of Mycolicibacterium smegmatis exposed to an imidazo[1,2-b][1,2,4,5]tetrazine. Data Brief 31:105805. doi: 10.1016/j.dib.2020.105805
Voss, J. J. D., Rutter, K., Schroeder, B. G., Su, H., Zhu, Y., and Barry, C. E. (2000). The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. 97, 1252–1257. doi: 10.1073/pnas.97.3.1252
Wells, R. M., Jones, C. M., Xi, Z., Speer, A., Danilchanka, O., Doornbos, K. S., et al. (2013). Discovery of a Siderophore Export System Essential for Virulence of Mycobacterium tuberculosis. PLoS Pathog. 9:e1003120. doi: 10.1371/journal.ppat.1003120
Welzen, L., de, Eldholm, V., Maharaj, K., Manson, A. L., Earl, A. M., and Pym, A. S. (2017). Whole-Transcriptome and -Genome Analysis of Extensively Drug-Resistant Mycobacterium tuberculosis Clinical Isolates Identifies Downregulation of ethA as a Mechanism of Ethionamide Resistance. Antimicrob. Agents Chemother. 61, 12. doi: 10.1128/aac.01461-17
Wilke, C. O. (2016). cowplot: streamlined plot theme and plot annotations for ggplot2 [Software]. Available online at: https://CRAN.R-project.org/package=cowplot (accessed April, 2021).
Keywords: Mycobacterium, drug resistance, imidazo[1, 2-b][1, 2, 4, 5]tetrazine, transcriptome, drug development, tuberculosis, virtual screening
Citation: Vatlin AA, Shitikov EA, Shahbaaz M, Bespiatykh DA, Klimina KM, Christoffels A, Danilenko VN and Maslov DA (2021) Transcriptomic Profile of Mycobacterium smegmatis in Response to an Imidazo[1,2-b][1,2,4,5]tetrazine Reveals Its Possible Impact on Iron Metabolism. Front. Microbiol. 12:724042. doi: 10.3389/fmicb.2021.724042
Received: 11 June 2021; Accepted: 14 July 2021;
Published: 04 August 2021.
Edited by:
Yusuf Akhter, Babasaheb Bhimrao Ambedkar University, IndiaReviewed by:
Krithika Rajagopalan, University of California, San Diego, United StatesSourish Ghosh, National Institutes of Health (NIH), United States
Copyright © 2021 Vatlin, Shitikov, Shahbaaz, Bespiatykh, Klimina, Christoffels, Danilenko and Maslov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dmitry A. Maslov, maslov_da@vigg.ru
 Aleksey A. Vatlin
Aleksey A. Vatlin Egor A. Shitikov
Egor A. Shitikov Mohd Shahbaaz
Mohd Shahbaaz Dmitry A. Bespiatykh
Dmitry A. Bespiatykh Ksenia M. Klimina
Ksenia M. Klimina Alan Christoffels
Alan Christoffels Valery N. Danilenko
Valery N. Danilenko Dmitry A. Maslov
Dmitry A. Maslov