- 1Department of Clinical Laboratory, the Affiliated Drum Tower Hospital of Nanjing University Medical School, Nanjing, China
- 2Department of Acute Infectious Disease Prevention and Control, Jiangsu Provincial Center for Disease Prevention and Control, Nanjing, China
- 3Laboratory Medicine, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, China
- 4Laboratory Medicine Center, The Second Affiliated Hospital, Nanjing Medical University, Nanjing, China
Background: This study analyzed the antimicrobial resistance phenotypes and mechanisms of quinolone, cephalosporins, and colistin resistance in nontyphoidal Salmonella from patients with diarrhea in Jiangsu, China.
Methods: A total of 741 nontyphoidal Salmonella isolates were collected from hospitals in major cities of Jiangsu Province, China between 2016 and 2017. Their susceptibility to commonly used antibiotics was evaluated by broth micro-dilution and sequencing analysis of resistance genes screened by a PCR method. For mcr-1 positive isolates, genetic relationship study was carried out by pulsed-field gel electrophoresis and multiloci sequence typing analysis. The transferability of these plasmids was measured with conjugation experiments and the genetic locations of mcr-1 were analyzed by pulsed-field gel electrophoresis profiles of S1-digested genomic DNA and subsequent Southern blot hybridization.
Results: Among 741 nontyphoidal Salmonella isolates, the most common serotypes identified were S. Typhimurium (n=257, 34.7%) and S. Enteritidis (n=127, 17.1%), and the isolates showed 21.7, 20.6, and 5.0% resistance to cephalosporins, ciprofloxacin, and colistin, respectively. Among the 335 nalidixic acid-resistant Salmonella, 213 (63.6%) and 45 (13.4%) had at least one mutation in gyrA and parC. Among the plasmid-borne resistance, qnrS1 (85; 41.9%) and aac(6')-Ib-cr4 (75; 36.9%) were the most common quinolone resistance (PMQR) genes, while blaCTX-M-14 (n=35) and blaCTX-M-55 (n=46) were found to be dominant extended-spectrum beta-lactamase (ESBL) genes in nontyphoidal Salmonella. In addition, eight mcr-1-harboring strains were detected since 2016 and they were predominate in children under the age of 7years. Conjugation assays showed the donor Salmonella strain has functional and transferable colistin resistance and Southern blot hybridization revealed that mcr-1 was located in a high molecular weight plasmid.
Conclusion: In nontyphoidal Salmonella, there is a rapidly increasing trend of colistin resistance and this is the first report of patients harboring mcr-1-positive Salmonella with a new ST type ST155 and new serotype S. Sinstorf. These findings demonstrate the necessity for cautious use and the continuous monitoring of colistin in clinical applications.
Introduction
Antibiotic resistance is an ongoing severe threat to global health, food security, and development today, and it may affect anyone, of any age, in any country. Antibiotic resistance is rising to dangerously high levels in all parts of the world. According to WHO data, the median rate observed for methicillin-resistant Staphylococcus aureus was 12.11% in 25 countries and that for Escherichia coli resistant to third-generation cephalosporins was 36.0% in 49 countries in 2019. How and why is this happening (Lin et al., 2015)? Related reports have found that the mechanisms include: (1) Bacteria restrict access by antibiotics by changing their entry ways or limiting the number of entry ways. (2) Bacteria get rid of antibiotics by using pumps in their cell walls that remove any antibiotic drugs that enter the cell. (3) Bacteria change or destroy the antibiotics with enzymes that break down the drug. (4) Bacteria develop new cell processes that bypass the effects of the antibiotic. (5) Bacteria change the antibiotic’s target so the drug can no longer recognize it and do its job. In addition, new resistance mechanisms keep emerging and spreading globally, threatening our ability to treat common infectious diseases. A growing list of infections such as pneumonia, tuberculosis, blood poisoning, gonorrhea, and foodborne diseases are becoming harder, and sometimes impossible, to treat as antibiotics become less effective, leading to higher medical costs, prolonged hospital stays, and increased mortality (Elkington, 2020).
Nontyphoidal Salmonellae refers to illnesses caused by all serotypes of Salmonella except for Salmonella enterica serovar Typhi and paratyphoidal serovars (i.e., serovars Paratyphi A, Paratyphi B, and Paratyphi C). They are leading causes of bacterial diarrhea worldwide. Salmonella is a Gram-negative rods genus belonging to the Enterobacteriaceae family. They are frequently transmitted from food or water contaminated with animal feces, or infection occurs through direct contact with infected animals or their environment and also directly between humans, causing numerous gastroenteritis cases. In addition to diarrheal disease, nontyphoidal Salmonella infections can invade normally sterile sites, resulting in bacteremia, meningitis, and other focal infections. The Global Burden of Diseases (GBD), Injuries, and Risk Factors Study 2017 estimated that Salmonella enterocolitis caused about 95 million infections, 50,000 deaths, and 3 million disability adjusted life-years in 2017 (GBD 2017 Causes of Death Collaborators, 2018; GBD 2017 DALYs and HALE Collaborators, 2018; GBD 2017 Disease and Injury Incidence and Prevalence Collaborators, 2018). Travelling is the main risk factor for nontyphoidal Salmonella infection, and it is estimated that among travelers returning to the United States, the highest risk is those who visited Africa (25.8 cases per 100,000 air travelers). In addition, Salmonella infection and carriage has been reported among internationally adopted children.
Antimicrobial resistance in nontyphoidal Salmonella serotypes has become a global problem. With the extensive use of antimicrobials, antimicrobial resistance is increasing at a serious rate in Salmonella isolates. In many countries, increasing numbers of Salmonella strains with MDR (defined as resistance to three or more classes of antimicrobials) have been discovered since the report of the spread of MDR in S. Typhimurium of definitive phage type 104 (DT104) around the world (Molbak et al., 1999). Since then, nontyphoidal Salmonella in Europe, Southeast Asia and other places have also developed resistance to the ciprofloxacin-fluoroquinolone drugs. Mutations in the quinolone resistance determining region (QRDR) may reduce their sensitivity to fluoroquinolone drugs, which depends on the number of mutations obtained. Third-generation cephalosporin drugs are used as a treatment for salmonellosis in areas with high levels of quinolone resistance because of its good effect on enteritis. However, ESBL or AmpC type β-lactamase mediated resistance to extended-spectrum cephalosporins of Salmonella has emerged. Additionally, in 2016, a plasmid encoding mcr-1-mediated colistin resistance in Enterobacteriaceae was newly recognized. Its existence presents a severe threat in the global response to antibiotic resistance (Liu et al., 2016). Furthermore, MDR Salmonella, which are important agents in the transmission of antibiotic resistance genes, have become a severe medical threat (Schrijver et al., 2018).
In this study, we analyzed the antimicrobial resistance profiles of nontyphoidal Salmonella in Jiangsu from 2016 to 2017 and elucidated the molecular mechanisms underlying the emergence of MDR in these isolates. Our findings will help provide appropriate clinical antimicrobial treatment for patients with nontyphoidal Salmonella infection in Jiangsu of eastern China.
Materials and Methods
Specimen Collection and Isolate Identification
A total of 741 fresh stool samples from clinically suspected patients who had diarrhea were collected from different hospitals located in 13 cities of Jiangsu Province between 2016 and 2017. The stool samples were then cultivated on Salmonella-Shigella (SS) xylose lysine deoxycholate (XLD) agar (XLD; CHRO Magar, China) and incubated for 18–24h at 37°C. API20 Etest strips (bioMerieux Vitek, Marcy-l’Etoile, France) were used to confirm the identity of the isolates. All of the isolates were then serotyped by slide agglutination with commercial antiserum (Tianrun Bio-Pharmaceutical Co., Ltd., China) according to the Kauffmann-White Scheme (World Health Organization, 2011).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed on the nontyphoidal Salmonella isolates for 30 antimicrobial agents: ampicillin (AMP), ampicillin/sulbactam (AMS), tetracycline (TET), chloramphenicol (CHL), trimethoprim/sulfamethoxazole (SXT), cefazolin (FAZ), cefotaxime (CTX), ceftazidime (CAZ), cefoxitin (FOX), gentamicin (GEN), imipenem (IMI), nalidixic acid (NAL), azithromycin (AZI), sulfa isoxazole (Sul), ciprofloxacin (CIP), amoxicillin/clavulanic acid (AMC), cefotaxime/clavulanic acid (CTC), ceftazidime/clavulanic acid (CCV), polymyxin E (CT), polymyxin B (PB), minocycline (MIN), amikacin (AK), aztreonam (AZM), cefepime (FEP), meropenem (MEM), levofloxacin (LVX), doxycycline (DOX), kanamycin (KAN), streptomycin (STR), and gemifloxacin (GEM) using the reference broth microdilution method with custom plates. The Clinical & Laboratory Standards Institute (CLSI) breakpoints were used to assess the results. The E. coli ATCC 25922 strain was used for quality control.
PCR Amplification and DNA Sequencing
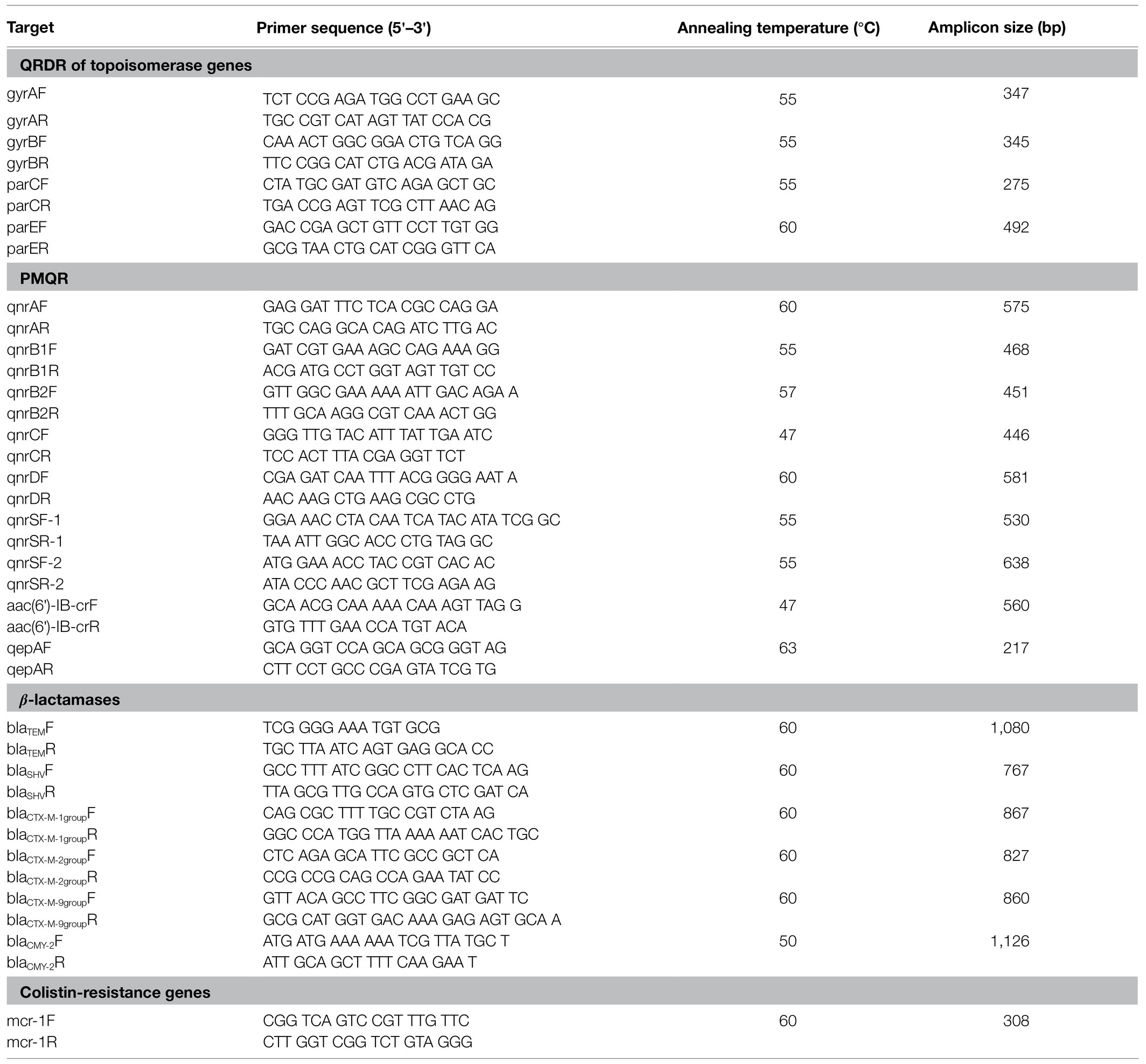
All of the third-generation cephalosporin-resistant isolates were analyzed using PCR assays for the presence of extended-spectrum β-lactamase (ESBL) genes containing blaCTX-M-1/9 groups, blaOXA, blaTEM, blaSHV, and blaCMY. PCR amplification of QRDRs of gyrA, gyrB, parC, and parE and plasmid-borne quinolone resistance (PMQR) determinants (qnrA, qnrB, qnrD, qnrS, and aac(6′)-Ib-cr) were performed on the ciprofloxacin resistant isolates. Then PCR amplification was conducted to detect the plasmid-borne colistin (polymixin E)-resistant mcr-1 gene in all isolates. The primers used for the abovementioned PCR assays are shown in Table 1. Amplification of the antibiotic resistance genes used the following temperature conditions: pre-denaturation at 95°C for 5min, followed by 30cycles of 95°C for 30s, annealing temperature for 30s and 72°C for 1min, with a final extension step at 72°C for 5min. The PCR products were analyzed by electrophoresis on 2.0% agarose. All of the PCR products were sequenced and then analyzed by DNAstar and the sequences were compared with the reference sequences from NCBI GenBank.
Pulsed Field Gel Electrophoresis
The pulsed field gel electrophoresis (PFGE) results were taken from the routine surveillance data. PFGE was performed according to the PulseNet protocol for Salmonella, using XbaI as the restriction enzyme (TaKaRa Biotechnology, Dalian, China). The cluster analysis was performed with Bionumerics 5.0 (Applied Maths NV, Sint-Martens-Latem, Belgium) using the Dice similarity coefficient and UPMGA (unweighted pair group method using average linkages) dendrogram type (optimization 0.50%, position tolerance 1.50%).
Multiloci Sequence Typing
The seven housekeeping gene sequences of Salmonella were amplified by PCR to determine the allelic differences and to analyze the sub-evolution relationship of the different strains. The primer sequences, amplification lengths and annealing temperature of the seven pairs of housekeeping sequences (aroC, dnaN, hemD, hisD, purE, sucA, and thrA) are shown in the previous reports. The PCR products were sequenced (Shanghai Biotechnology Co., Ltd.) and determination of the MLST was performed in silico using online tools.1
Conjugation Assays
To test the host range and the transfer ability of each plasmid, conjugation assays were performed using the E. coli J53 strain as the recipient. Transfer of the colistin-resistance determinant by conjugation was assayed on LB agar plates with a donor:recipient ratio of 2:1, using E. coli J53 (sodium azide-resistant E. coli) as the recipient. After incubation at 37°C for 24h, transconjugants were selected on LB agar supplemented with colistin (2μg/ml) and sodium azide (180μg/ml). Initial species identification and subsequent antimicrobial susceptibility tests were conducted by using the VitekMS system. The transformants were regarded as transconjugants when they exhibited resistance to colistin and harbored the mcr-1 gene.
Plasmid Analysis
Isolates selected containing the mcr-1 gene were subjected to further plasmid analysis. In compatibility groups, the plasmids extracted from the transconjugants were subjected to PCR-based replicon typing. S1-PFGE and Southern blotting were conducted to isolate and locate the resistance plasmids. Briefly, the gel plugs embedded with mcr-1-positive isolates were digested with S1 nuclease (TaKaRa Biotechnology, Dalian, China) and the linear plasmids were separated by the CHEF-Mapper XA PFGE system (Bio-Rad) as described above. The plasmid DNA was transferred to positive-charged nylon membranes (Millipore, United States), and a DIG-labelled mcr-1-specific probe was hybridized with the plasmids according to the instructions of the DIG High Prime DNA Labeling and Detection StarterKit (Roche, United States).
Results
Nontyphoidal Salmonella Isolates From Human Patients in Jiangsu, China, From 2016 to 2017
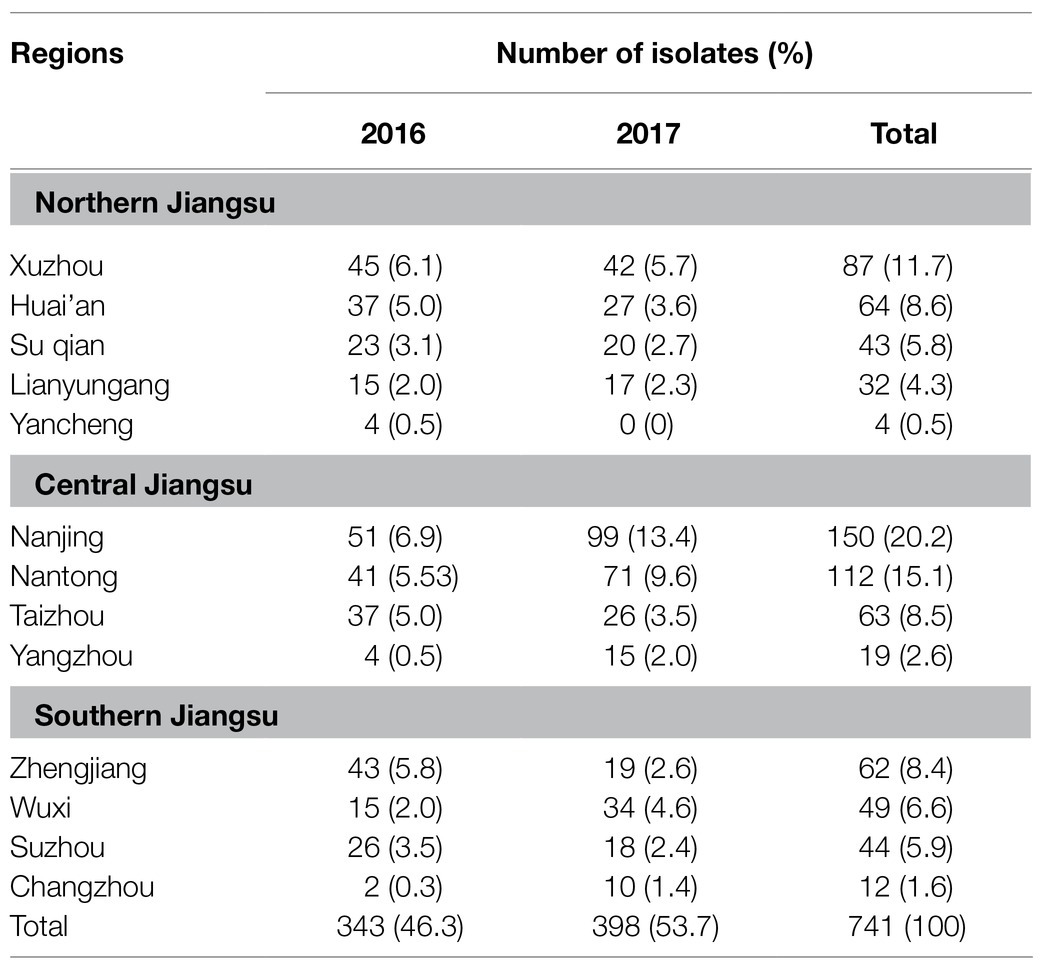
Between January 2016 and December 2017, 741 nontyphoidal Salmonella isolates were cultured from patients with diarrhea in Jiangsu, China. The age of the patients ranged from 4months to 79years (Figure 1B). Children under 12years of age were highly susceptible to nontyphoidal Salmonella infection, which accounted for 48.2% of all of the patients (p<0.05). The infections occurred mainly in summer and autumn (Figure 1A). In addition, the regional distribution shows that the three cities with the highest nontyphoidal Salmonella infection rates are Nanjing, Nantong, and Xuzhou (Table 2).
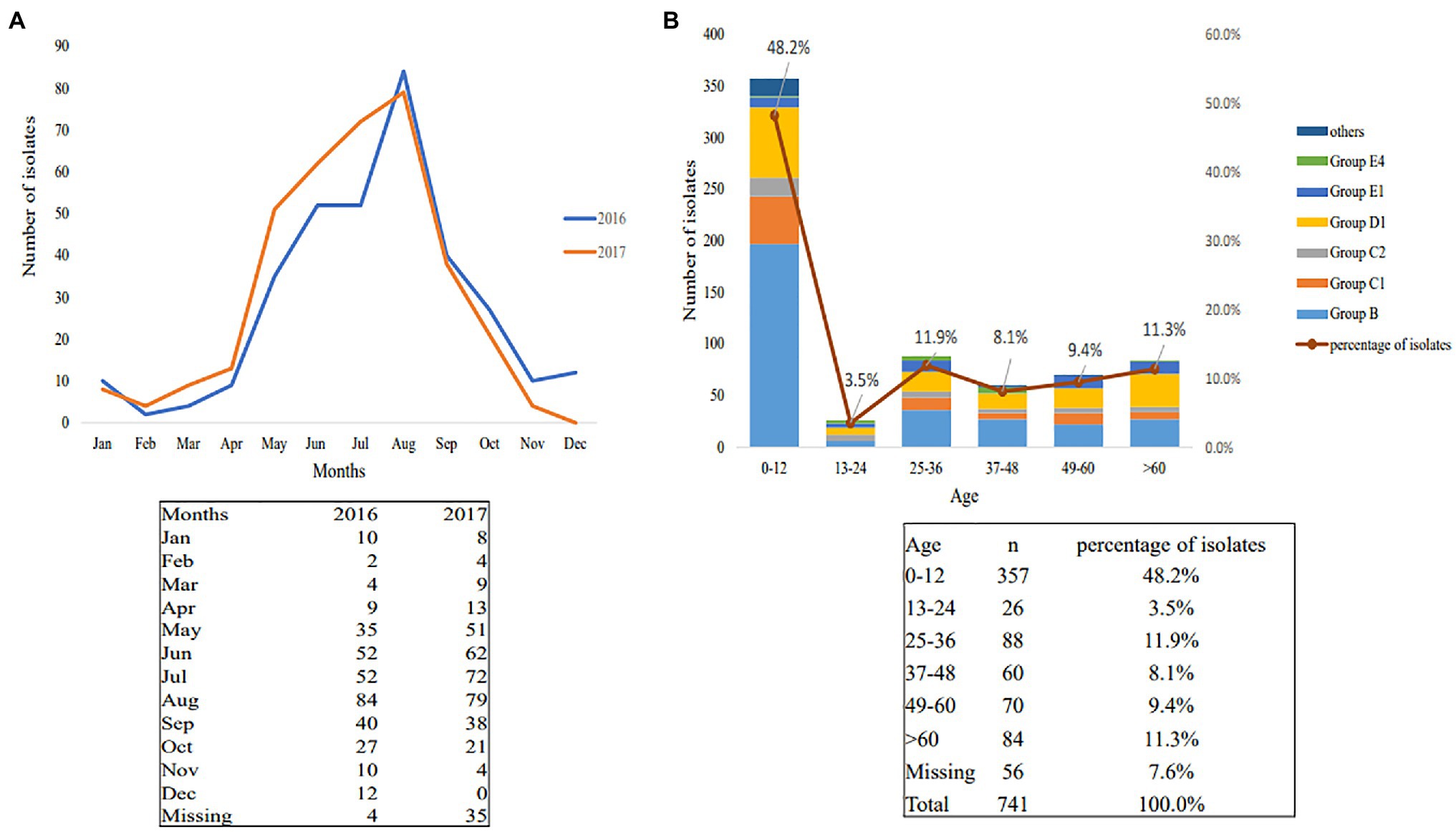
Figure 1. Season and age distributions of nontyphoidal Salmonella infection in 2016–2017. (A) Season distributions; (B) age distributions.
Among the 741 cases of nontyphoidal Salmonella, 332 (44.8%) strains belonged to serogroup B, the most common serogroup, 184 (24.8%) strains to serogroup D, 136 (18.4%) strains to serogroup C, 68 (9.2%) strains to serogroup E, and 21 (2.8%) strains belonged to other serogroups (Figures 2A,B). The most common serotypes were S. Typhimurium (n=257, 34.7%), S. Enteritis (n=127, 17.4%), S. London (n=29, 3.9%), S. Dublin (n=28, 3.8%), and S. Rissen (n=26, 3.5%; Figure 2C).
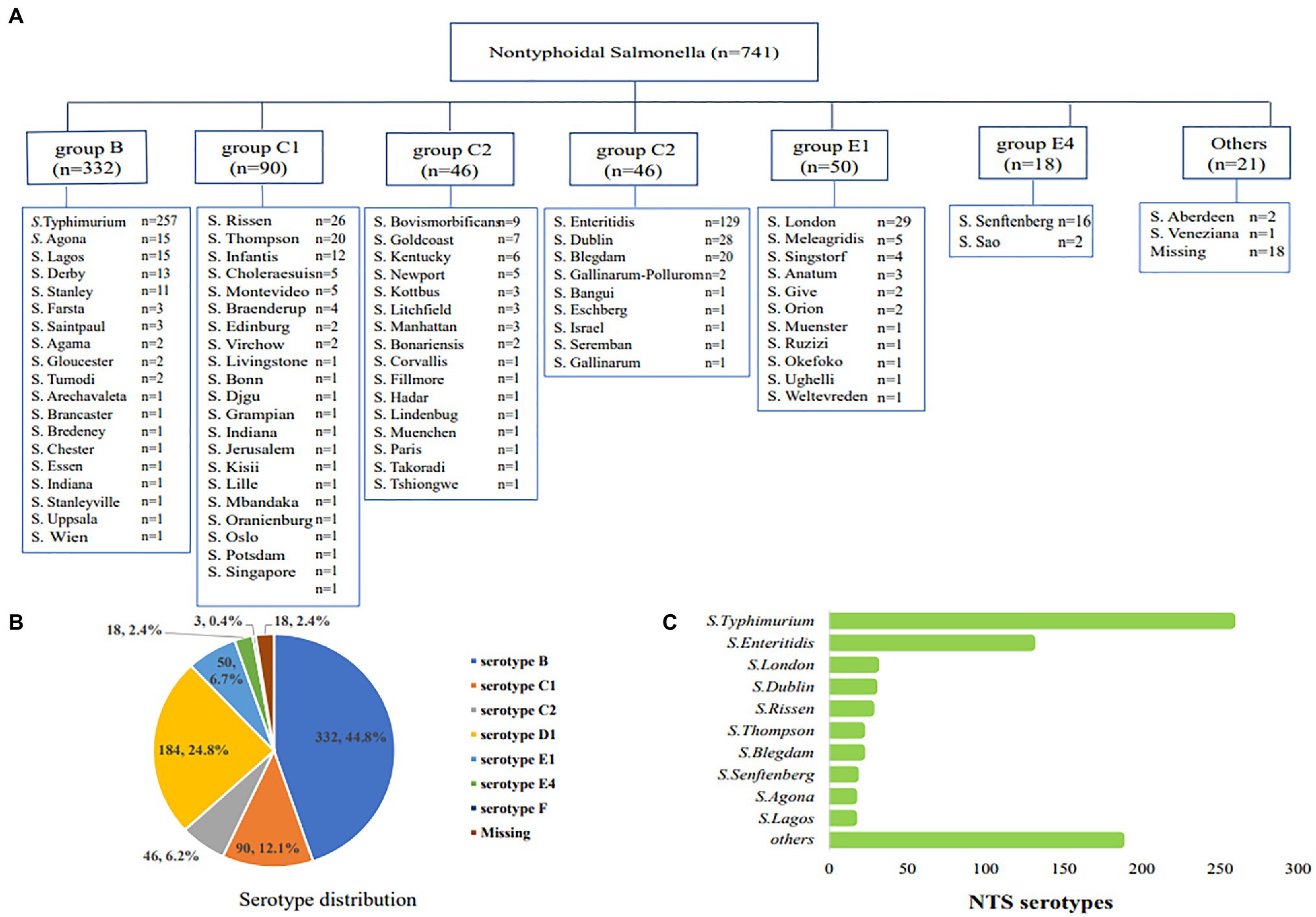
Figure 2. The serological distribution of nontyphoidal Salmonella from 2016 to 2017. (A) all of the serotype distribution of nontyphoidal Salmonella serotypes; (B) the distribution of serogroups; (C) the distribution of the top 10 nontyphoidal Salmonella serotypes.
Antimicrobial Susceptibility Testing
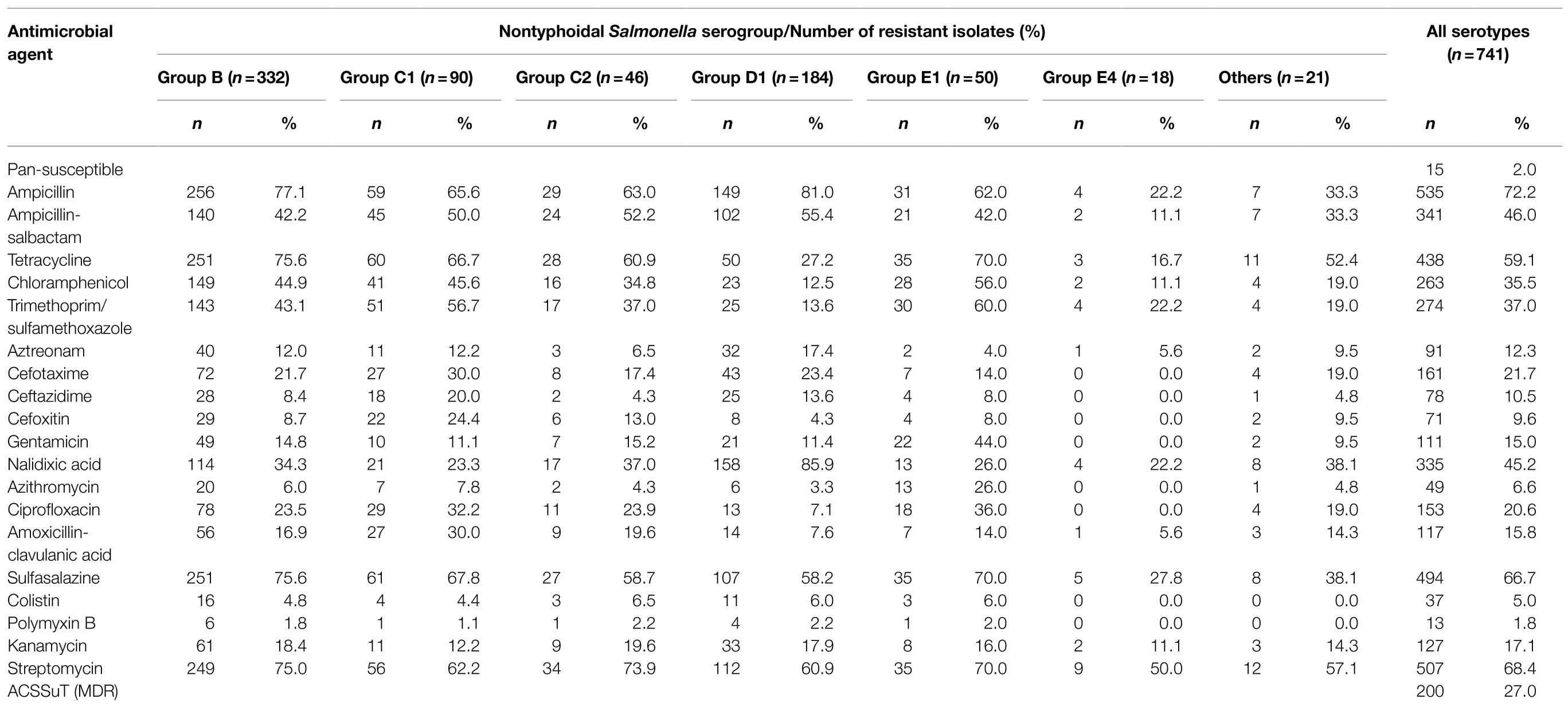
Among the 741 isolates, only 15 (2.0%) were susceptible to all 30 antimicrobials. Resistance to ampicillin (72.3%) was the most common, followed by streptomycin (68.4%), sulfisoxazole (66.7%), and tetracycline (59.1%), the resistance rate of which exceeded 50% (Table 3). In addition, resistance to cefotaxime, ceftazidime and cefoxitin were found in 21.7, 10.5, and 9.6% of isolates, respectively. Notably, 45.2% (335/741) and 20.6% (153/741) of the isolates displayed resistance to nalidixic acid and ciprofloxacin, while 15 of the isolates showed co-resistance to quinolones and third-generation cephalosporins. More importantly, 5% (37/741) of the isolates displayed resistance to colistin. Among the MDR isolates, 200 (27.0%) showed the ACSSuT resistance pattern (defined as resistance to ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline).
PCR Detection of Antimicrobial Drug Resistance Genes and DNA Sequencing
Resistance genes including blaOXA, blaSHV, blaCMY, blaCTX-M-1 group, and blaCTX-M-9 group in the 111 cephalosporin-resistance isolates were detected by PCR except blaTEM, for which the detection rate exceeded 72.5%. PCR screening revealed that 93 (83.8%), 16 (14.4%), and 5 (4.5%) isolates contained the blaCTX-M-1/9, blaCMY, and blaSHV genes, respectively. All of the strains were negative for blaOXA. Sequencing of the PCR products revealed that blaCTX-M-1 group included four subtypes: blaCTX-M-55 (n=46), blaCTX-M-123 (n=2), blaCTX-M-15 (n=1), and blaCTX-M-3 (n=1); the blaCTX-M-9 group included three subtypes: blaCTX-M-14 (n=35), blaCTX-M-65 (n=10), and blaCTX-M-27 (n=1), among which blaCTX-M-14 and blaCTX-M-55 were the most common subtypes. BlaSHV-12 subtype and blaCMY-2 were detected in all five blaSHV and 16 blaCMY nontyphoidal Salmonella strains, respectively. Notably, two isolates contained blaTEM-1, blaCTX-M-55, and blaCTX-M-65 subtypes, one isolate contained blaTEM-1, blaCTX-M-14, and blaCMY-2 subtypes, one isolate contained blaTEM-1, blaCTX-M-65, and blaSHV-12, and one isolate concomitantly contained blaTEM-1, blaCTX-M-3, blaCTX-M-65, and blaSHV-12 (Figure 3A).

Figure 3. Distribution of plasmid-borne quinolone resistances (PMQRs) and extended-spectrum beta-lactamases (ESBLs) resistance genes of nontyphoidal Salmonella from 2016 to 2017. (A) ESBLs resistance genes of nontyphoidal Salmonella; (B) PMQRs resistance genes of nontyphoidal Salmonella.
A total of 180 PMQR-positive strains were detected by PCR in 741 nontyphoidal Salmonella strains. Sequencing of the PCR products revealed that 44.3% of the qnrS genes included qnrS1 (n=85) and qnrS2 (n=5) subtypes and 16.7% of the qnrB genes included qnrB6 (n=21), qnrB4 (n=12), and qnrB19 (n=1) subtypes. qnrA1 (n=2) subtypes and qnrD1 (n=2) and aac (6′)-Ib-cr4 (n=75) subtypes were detected in all qnrA, qnrD, and aac(6′)-Ib-cr nontyphoidal Salmonella strains. We found that qnrS1 and aac(6′)-Ib-cr4 were the most common subtypes of PMQR-positive genes in Jiangsu, China (Figure 3B).
In addition, the PCR results showed that three out of the 37 colistin-resistant isolates [minimum inhibitory concentration (MIC)≥4μg/ml] harbored plasmid-borne mcr-1 genes, while the other five mcr-1 genes were found in colistin-intermediate resistance nontyphoidal Salmonella isolates (MIC=2μg/ml).
We identified 449 isolates with intermediation and resistance to ciprofloxacin, and 185 of them contained at least one gyrA mutation, which mainly occurred at codons 83/87 (S83F/Y; D87Y/N/G). In addition, 45 of them contained at least one parC mutation, which mainly occurred at codons 57/80 (T57S; S80I/R). No point mutations in gyrB were found among the sequences of the QRDRs. Among the 47 isolates with one or more mutation in the gyrA, parC, and parE genes, one or two PMQR genes were detected, and for two isolates, we simultaneously detected gyrA, parC, and parE mutations and plasmid-borne PMQR resistance genes (Supplementary Table S1).
Among the 80 nontyphoidal Salmonella isolates with a phenotype showing concomitant resistance to ciprofloxacin and cefotaxime, 40 isolates contained ESBL and PMQR genes and 17 isolates harbored three types of antimicrobial resistant genes, like aac(6′)-Ib-cr4/blaCTX-M/blaTEM-1 (n=4) and qnrS1/blaTEM-1/blaCMY-2 (n=4). Two isolates contained four types of antimicrobial-resistance genes: aac(6′)-ib-cr4/blaTEM-1/blaCTX-M-14/mcr-1 and qnrS1/aac(6′)-ib-cr4/blaTEM-1/blaCTX-M-65. One Salmonella typhimurium isolate contained six types of antimicrobial-resistance genes: qnrS1/qnrS2/aac(6′)-ib-cr4/blaTEM-1/blaCTX-M-14/mcr-1. In addition, 11 isolates that contained QRDR mutations also harbored the ESBL and PMQR genes at the same time (Supplementary Table S2; Table 4).
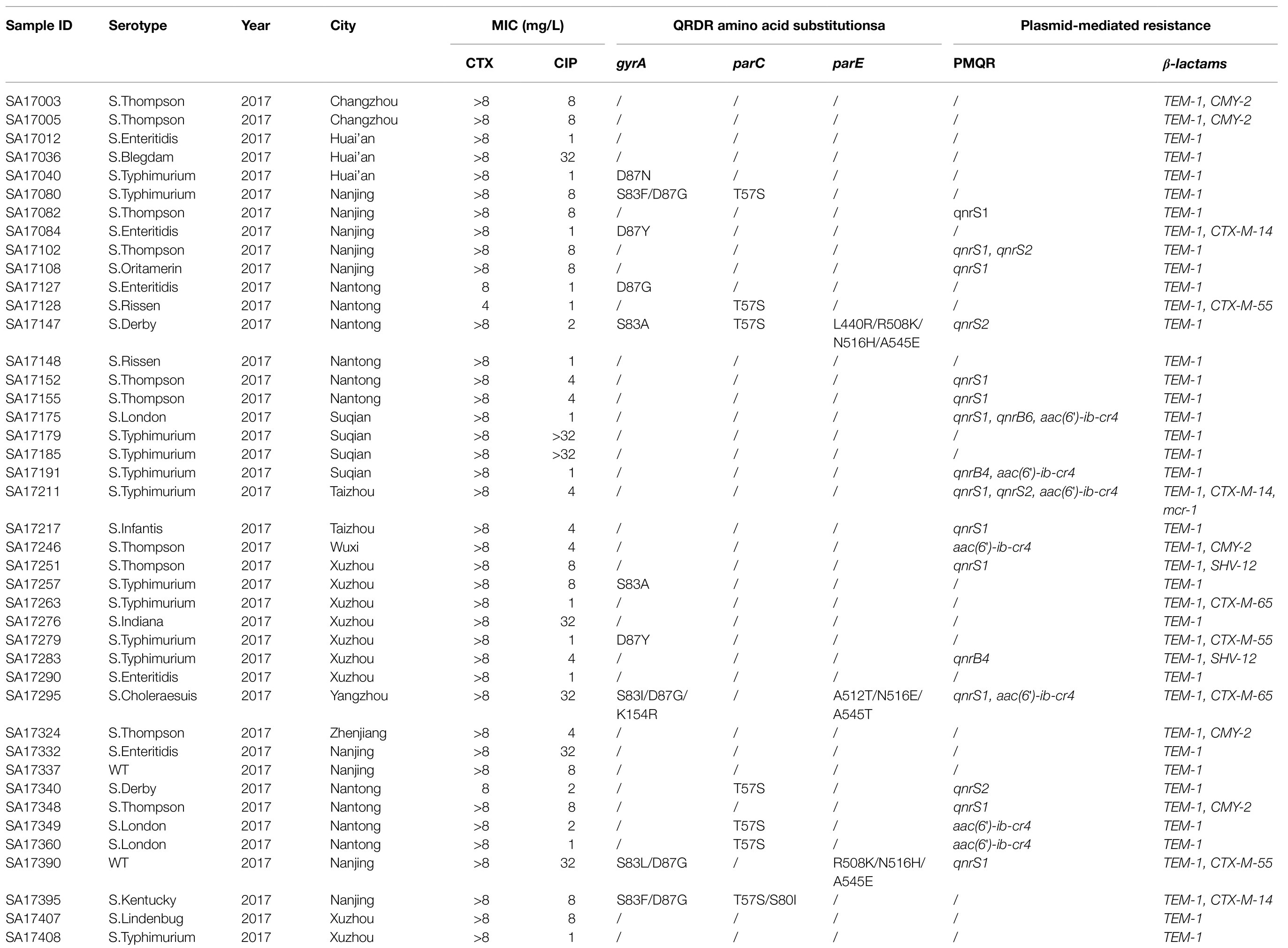
Table 4. Summary of phenotypes of NTS isolates showing concurrently resistance to ciprofloxacin and cefotaxime and their corresponding resistance genes in 2017.
Epidemiology of mcr-1-Harboring Nontyphoidal Salmonella Isolates
Over the course of the study, eight of nontyphoidal Salmonella were found to harbor the mcr-1 gene. Among the eight patients infected with mcr-1-positive nontyphoidal Salmonella, seven were aged under 7years old and the other was 49. Among these isolates, five strains were detected in Northern Jiangsu including Lianyungang (n=3), Suqian (n=1), and Huai’an (n=1), two were from Taizhou (n=2) and one was from Suzhou (n=1). In addition, two different serotypes were identified among the eight isolates mentioned above, including six S. Typhimurium, one S. Lagos, and one S. Sinstorf (Figure 4).
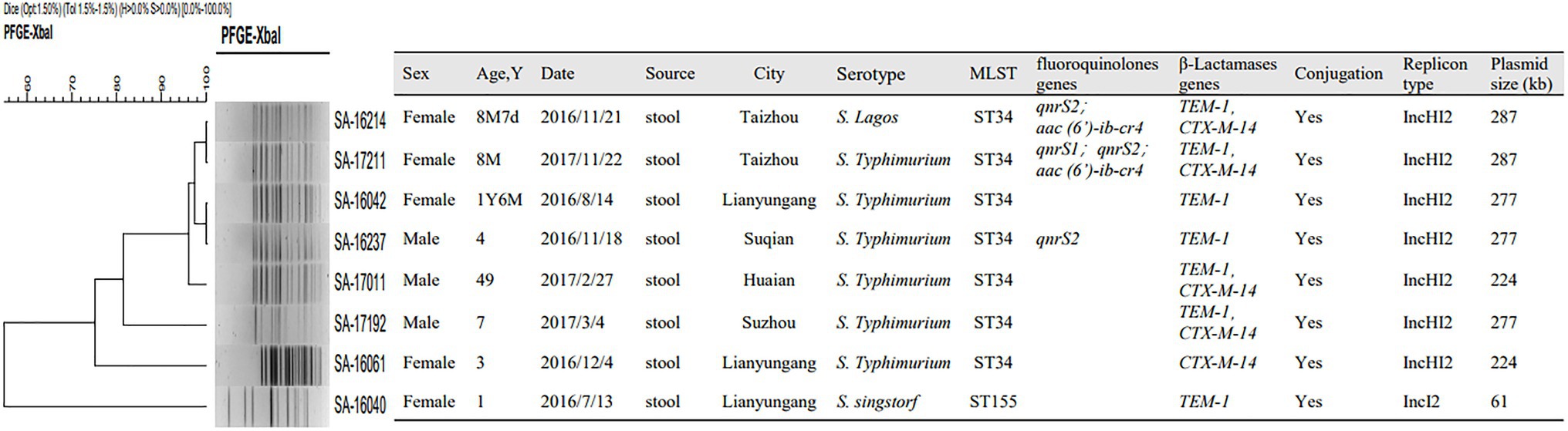
Figure 4. Pulsed field gel electrophoresis (PFGE), multiloci sequence typing (MLST), resistance gene, conjugation experiment, replicon typing and plasmid distribution of nontyphoidal Salmonella carrying mcr-1 from 2016 to 2017.
Molecular Typing of mcr-1 Harboring Nontyphoidal Salmonella Isolates
Among eight mcr-1-positive nontyphoidal Salmonella isolates, two STs were identified. ST 34 was the most prevalent, accounting for 87.5% (n=7), followed by ST 155 (n=1). Salmonella ST155 was found in mcr-1 positive patients for the first time. PFGE typing revealed that five isolates with the same ST type showed the same band pattern except for ST155 Salmonella collected in Lianyungang and ST34 S. Typhimurium in Suzhou and Lianyungang (Figure 4).
Characteristics of Plasmids Harboring the mcr-1 Gene
All of the plasmids harboring the mcr-1 gene from eight selected nontyphoidal Salmonella isolates were successfully transferred to E. coli J53, and the transconjugants exhibited resistance to colistin and cephalosporins when the parent mcr-1-harbouring nontyphoidal Salmonella exhibited resistance. This shows that the mcr-1 gene could be expressed and function in the transconjugants. In addition, the transconjugants showed high resistance to cefotaxime, suggesting that genes co-transfer occurred during the conjugation process (Table 5). S1-PFGE and Southern blotting revealed that the among the eight mcr-1-haboring plasmids, the replicon types IncI2 and IncHI2 were detected in one and seven plasmids, respectively (Figure 5). The earliest isolation of a strain carrying the IncI2 type mcr-1 plasmid S. Sinstorf occurred in Lianyungang, Jiangsu.
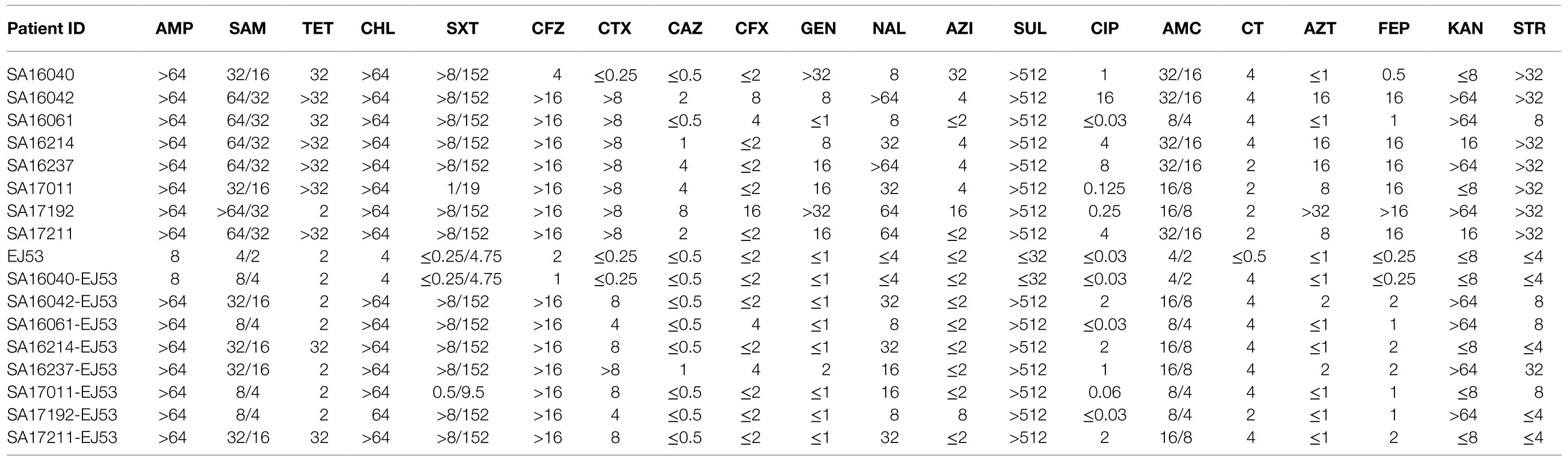
Table 5. Antimicrobial susceptible patterns and characteristics of eight selected mcr-1-producing NTS (μg/ml).
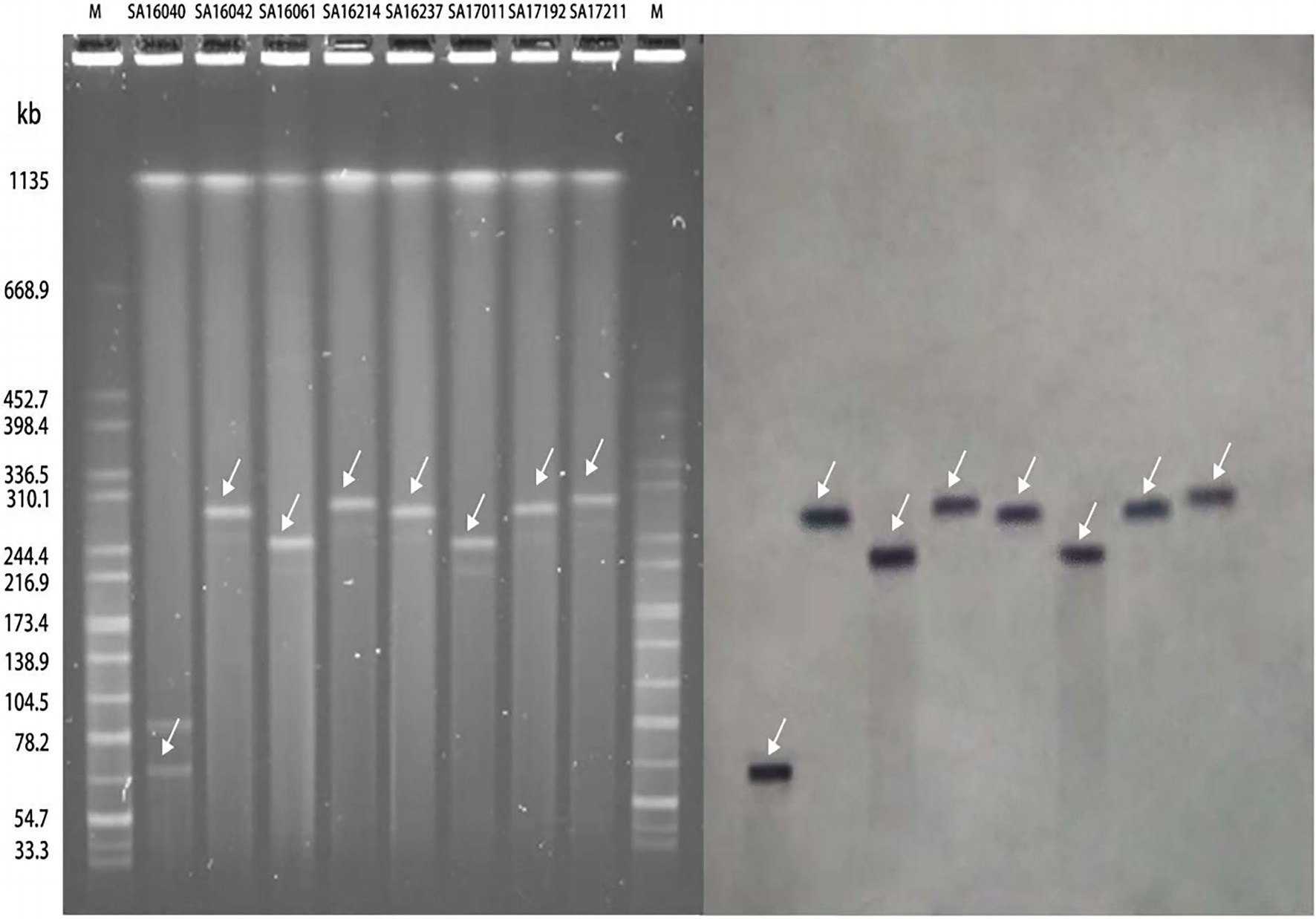
Figure 5. Analysis of the results of S1-PFGE and DNA hybridization of mcr-1 positive nontyphoidal Salmonella from 2016 to 2017.
The antibiotic resistance genes in these mcr-1-harbouring nontyphoidal Salmonella were analyzed. All of these nontyphoidal Salmonella carried resistance genes other than mcr-1. Five of the seven nontyphoidal Salmonella with IncHI2 type mcr-1 plasmids simultaneously carried the blaCTX-M-14 gene, whereas none of the IncI2 plasmids were found to harbor other antibiotic resistance genes except blaTEM-1. Additionally, one isolate with the IncHI2 mcr-1 plasmid was confirmed to harbor mcr-1 and qnrS2, while two isolates with the IncHI2 type mcr-1 plasmid harbored both PMQRs and ESBLs (one with qnrS2/aac(6′)-ib-cr4 and blaTEM-1/blaCTX-M-14; one with qnrS1/qnrS2/aac(6′)-ib-cr4 and blaTEM-1/blaCTX-M-14; Figure 4).
Discussion
Human and animal salmonellosis is a symptomatic infection caused by bacteria of the Salmonella enterica, mainly occurring in many low- and middle-income countries (Reddy et al., 2010). It is estimated that it causes approximately 153 million gastroenteritis cases and 57,000 deaths worldwide every year (Majowicz et al., 2010), and more than half of these diseases and deaths occur in Africa (Ao et al., 2015). Our results showed that the detection rate of nontyphoidal Salmonella in Jiangsu from 2016 to 2017 is relatively high, which should arouse clinical concern. Our results found that the number of nontyphoidal Salmonella isolated from Central Jiangsu and Northern Jiangsu is higher than that from Southern Jiangsu. The highest isolation rate was in Nanjing, which may be related to the large local population and its frequent movements.
Six serovar including serotype B, C1, C2, D1, E1, and E4 were found in 741 nontyphoidal Salmonella strains. Salmonella Typhimurium and S. Enteritidis were the most common subtypes, consistent with the most widely reported serotypes of S. Typhimurium and S. Enteritidis in Africa (Mabey et al., 1987; Gordon et al., 2008). Additionally, our study also identified 58 other serovars. The continuous emergence of rare serovars in recent years should arouse great concern in Jiangsu, China.
The introduction of fluoroquinolones and third-generation cephalosporins in the 1980s greatly reduced the mortality from salmonellosis. However, fluoroquinolone-resistant Salmonella appeared in the 1990s. In areas where the infection is not endemic, such as in the United States and Canada, the resistance rate of nalidixic acid reached 40 and 80% in 2004 and 2006, respectively (Demczuk et al., 2010), while the resistance rate of ciprofloxacin in the United Kingdom in 2006 was 70% (Threlfall et al., 2008). We conducted drug-resistant phenotype monitoring of nontyphoidal Salmonella in Jiangsu from 2016 to 2017 and found that the drug-resistant rate of nalidixic acid was 45.2%, consistent with the resistance level in the United States. The drug resistance rate to ciprofloxacin was significantly higher among the S. Typhi in Jiangsu in the past 5years (Qian et al., 2020).
The appearance of fluoroquinolone resistance has aroused our interest in the study of its resistance mechanism. Fluoroquinolone resistance in Salmonella is usually mediated through mutations in chromosomal sites or through plasmid-borne PMQR resistance genes. Early studies of Salmonella found that increases of the MIC was related to the accumulation of known chromosomal mutations (Murray et al., 2012; Song et al., 2013). Our study found that the combined mutation detection rate of gyrA gene 83, 87 and parC gene 57 in ciprofloxacin resistant strains was the highest, which confirmed that the multiple-site QRDR combined mutations are a possible cause of ciprofloxacin resistance. Additionally, our sequencing results also found that nontyphoidal Salmonella harboring qnr and aac(6′)-Ib-cr mainly appeared in strains with reduced ciprofloxacin susceptibility, which suggested that PMQRs genes may mediate low-level ciprofloxacin-resistance.
The emergence of Salmonella that is resistant to third-generation cephalosporins such as ceftriaxone and cefotaxime indicates another important public health problem. Ever since the third-generation cephalosporin drugs have been used in the clinic, ESBLs or AmpC-type β-lactamases mediated resistance has been detected in nontyphoidal Salmonella. Cephalosporin resistance has been reported in Southeast Asia (Koh et al., 2008), while in Thailand, Salmonella with blaCMY and blaCTX-M enzymes has also been described (Sirichote et al., 2010; Pornruangwong et al., 2011). Our research found that the resistance rate of nontyphoidal Salmonella to cefotaxime was more than 20%, significantly higher than that of S. Typhi (Qian et al., 2020). Coinciding with domestic and international reports, blaCTX-M-type and blaCMY-2 genes were simultaneously detected in nontyphoidal Salmonella, the majority of which existed in cefotaxime-resistant strains. If ESBL-producing nontyphoidal Salmonella becomes common, treatment options may become very scarce. Therefore, timely detection is essential to control the spread of β-lactamase among nontyphoidal Salmonella.
With the continuous emergence of third-generation cephalosporin and fluoroquinolone resistant Salmonella strains, plasmid-borne resistance gene transmission has been found to play an extremely important role. In recent years, 40 strains of nontyphoidal Salmonella co-existing with ESBL and PMQR genes have been discovered. The complexity and diversity of nontyphoidal Salmonella resistance genes make clinical treatment tricky.
Colistin is a class of cationic cyclic peptide antibiotics synthesized with a non-ribosomal hydrophobic tail (Dixon and Chopra, 1986). Its mechanism of killing Gram-negative pathogens relies on its ability to destroy membranes through polar hydrophobic interactions. The potential nephrotoxicity and neurotoxicity of colistin prevent its widespread use in the clinic (Li et al., 2006; Baron et al., 2016). At present in Europe, colistin has been widely used in animal production to treat many animal infections caused by Enterobacteriaceae (Catry et al., 2015). Moreover, in Asian countries, colistin is widely used as a growth promoter to improve animal production (Kempf et al., 2016). With its long-term use, many bacterial species have developed resistance to colistin. Since Liu et al., first reported the discovery of the mobile colistin resistance gene mcr-1 in Chinese pig and human E. coli isolates in 2015, the mcr-1 gene has been reported from samples collected in five continents and more than 40 countries, which means that it plays an important role in colistin resistance. As the last line of defense against multi-drug resistant bacteria, colistin has attracted worldwide attention. In our study, 5% (37/741) of clinical nontyphoidal Salmonella isolates from Jiangsu between 2016 and 2017 showed colistin resistance (MIC≥4μg/ml).
In China, colistin first started to be used in food-producing animals as early as the 1980s. Shen et al., found that the spread of mcr-1 in E. coli isolates of chicken origin in China increased from 5.2% in 2009 to 30.0% in 2014. This coincides with the fact that China introduced colistin into agriculture at the beginning of 2000, and by the end of 2015, China had become one of the world’s largest users of agricultural colistin (Shen et al., 2016). Among Salmonella, S. Typhimurium is one of the most common serotypes harboring the mcr-1 gene. The mcr-1 gene was described for the first time by analyzing the complete genome sequence of Salmonella available in GenBank, including the identification of strains carrying the mcr-1 plasmid among 10 clinical Salmonella isolates submitted between 2012 and 2015 (Doumith et al., 2016). Subsequently, the mcr-1 gene has been reported in Salmonella strains isolated from food, animal and clinical specimens from Europe, the United States and China (Anjum et al., 2016; Campos et al., 2016; Yang et al., 2016). We retrospectively analyzed the prevalence and molecular characteristics of the mcr-1 gene in nontyphoidal Salmonella isolates and found that eight mcr-1 positive nontyphoidal Salmonella.
The serotype is one of the important phenotypic characteristics of Salmonella and serovar analysis is an important traceability analysis method for Salmonella (Soler-Garcia et al., 2014). From the perspective of Salmonella serotype analysis, the mcr-1 positive strains are mainly S. Typhimurium (6/8). Notedly, a case of a patient with S. Sinstorf carrying mcr-1 has not been reported previously, indicating the diversity of mcr-1 transmission. The majority of patients infected with the mcr-1 strain are children, accounting for 87.5% (7/8), which is consistent with the data reported in Shanghai by Lu et al. (2019). The typing of Salmonella is helpful for the traceability analysis of pathogenic microorganisms, and it is of great significance to the prevention and control of Salmonella and risk assessment.
Multiloci sequence typing is a bacterial typing method based on the determination of the nucleotide sequence with high resolution and it is suitable for molecular epidemiology research and molecular evolution analyses. Each ST represents a separate set of nucleotide sequence information. Closely related strains have the same ST or only a few different gene sites of the ST type. In China, the most common ST of Salmonella, especially MDR Salmonella, is ST34, and this is also common in Europe. Previous reports showed that most of the ST34 Salmonella strains carrying the mcr-1 gene were isolated from animals, while we found that six strains of S. Typhimurium and one strain of S. Lagos belonged to the ST34, indicating the widespread spread of ST34 nontyphoidal Salmonella harboring the mcr-1 gene in animals and humans. The widespread existence of this clone poses a great threat to the prevention and control of clinical S. Typhimurium infection. In our study, a strain of Salmonella ST155 carrying mcr-1 was detected from a patient. ST155, as a new ST type carrying the mcr-1 gene, was reported in humans for the first time. In addition, the PFGE results of our study showed that five mcr-1 positive nontyphoidal Salmonella strains from Taizhou, Huai’an, Suqian, and Lianyungang had the same PFGE type, suggesting that the clonal spread of the mcr-1 gene has occurred in the Central and Northern of Jiangsu, which means that infection control should be carried out at the same time in multiple hospitals and multiple regions in Jiangsu.
Since the initial detection of mcr-1 in the IncI2 plasmid pHNSHP45 (Liu et al., 2016), the diversity of mcr-1 harboring plasmid libraries has continued to expand. Analysis of the data related to the mcr-1 gene in GenBank showed that IncI2, IncX4, and IncHI2 are the most common plasmids carrying the mcr-1 gene. Other replicon types such as IncP, IncHI1, IncFII, IncFI, IncFIB, F:A−:B+, IncY, IncK and phage-like plasmids have also been reported (Nordmann et al., 2016; Xavier et al., 2016; Zurfluh et al., 2016; Li et al., 2017; Zhang et al., 2017). Although these plasmids come from different host strains, and even from different species in different geographic locations, the genome comparison of the plasmid sequences showed that they are highly similar, which means that most of them have spread in various Enterobacteriaceae worldwide.
We conducted conjugation experiments on the eight mcr-1 positive nontyphoidal Salmonella strains, and found that all eight strains were successfully conjugated, indicating that all of the mcr-1 genes were located on the plasmid. The results of S1-PFGE and Southern blotting showed that seven of the eight strains of plasmids carrying mcr-1 belonged to IncHI2 with approximately 220–280kb, which is consistent with the recently reported plasmid carrying mcr-1 of ST34 S. Typhimurium in Zhejiang, China. Notedly, the S. Sinstorf serotype strain belongs to the IncI2-type plasmid with 60kb, which has not been reported before. All of these results indicate that the mcr-1 gene plays an important role in the colistin resistance of clinically isolated nontyphoidal Salmonella strains and its spread can lead to a rapid increase in colistin resistance.
In recent years, due to the unreasonable clinical and agricultural use of antibiotics, the antimicrobial resistance rate of Salmonella has increased rapidly. According to previous reports, mcr-1-positive isolates are usually still susceptible to many other antibiotics (Eiamphungporn et al., 2018). In our study, eight strains of mcr-1 positive strains were resistant to sulfisoxazole, and 87.5% (7/8) and 75% (6/8) showed resistance to third-generation cephalosporins and ciprofloxacin, respectively. It is worth noting that in our study, five colistin-intermediate resistance nontyphoidal Salmonella with MIC=2 were also found to carry the mcr-1 gene. Due to the existence of the mcr-1 gene, Salmonella is in the process of transforming from being susceptible to colistin to resistant. Moreover, difference may also be due to their level of expression on the different plasmids or in the different genomic backgrounds. Sequencing analysis showed that 62.5% (5/8) of these isolates were blaCTX-M-14 ESBL-producing strains, and these isolates also harbored quinolone resistance genes qnrS and/or aac(6′)-Ib-cr4, and their genotypes were consistent with their resistant phenotypes. Therefore, comprehensive monitoring of mcr-1 carrying status and plasmid structure in clinical Salmonella isolates within Jiangsu and understanding the transmission characteristics and mechanism of mcr-1 in nontyphoidal Salmonella may provide a preliminary basis for controlling the spread of colistin resistance among Gram-negative bacterial pathogens in the future.
Conclusion
In summary, we reported the prevalence and resistance mechanism of the occurrence of nontyphoidal Salmonella isolates in Jiangsu, China. We found that resistance to ciprofloxacin, cephalosporins, and colistin were wide-spread. We identified the related mutations and various transferrable antimicrobial-resistance genes, which included ESBL, PMQR and mcr-1, and some isolates harbored two or more types of these genes. The dissemination of these genes poses a huge threat to the control of nontyphoidal Salmonella infection around the world. Additionally, the present study reported the prevalence of the mcr-1 gene among nontyphoidal Salmonella in Jiangsu, China. Notably, diverse STs of mcr-1 harboring nontyphoidal Salmonella were mainly located on ∼220–280kb IncHI2 plasmids, indicating that the fast evolution and high transfer ability of this kind of plasmid has led to the high prevalence of the mcr-1 gene. Interestingly, one Salmonella strain harboring mcr-1 gene with a new ST type ST155 and new enterica serovar Sinstorf has never been detected before in human. Moreover, the mcr-1 belonged to an IncI2-type plasmid of 60kb. Hence, timely detection of the mcr-1 gene and antimicrobial susceptibility testing are necessary so that infections caused by mcr-1 carriers receive appropriate and effective therapy. Similarly, large-scale surveillance and effective infection control measures are also urgently needed to prevent the spread of mcr-1 carriers.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
GL, HY, and BG designed the hypothesis. CB collected and integrated strain information. GL and HQ performed the experimental work. JL analyzed the computationally generated data. GL and BT prepared the draft manuscript. HY and BG commented on the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (81871734, 81471994), Key R&D Program of Jiangsu Province (BE2020646), Jiangsu Provincial Medical Talent (ZDRCA2016053), Six Talent Peaks Project of Jiangsu Province (WSN-135), Advanced Health Talent of Six-one Project of Jiangsu Province (LGY2016042), National Major S&T Projects (2018ZX10714-002), and Research foundation for advanced talents of Guandong Provincial People’s Hospital (KJ012021097).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.723697/full#supplementary-material
Footnotes
References
Anjum, M. F., Duggett, N. A., AbuOun, M., Randall, L., Nunez-Garcia, J., Ellis, R. J., et al. (2016). Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J. Antimicrob. Chemother. 71, 2306–2313. doi: 10.1093/jac/dkw149
Ao, T. T., Feasey, N. A., Gordon, M. A., Keddy, K. H., Angulo, F. J., and Crump, J. A. (2015). Global burden of invasive nontyphoidal Salmonella disease, 2010(1). Emerg. Infect. Dis. 21, 941–949. doi: 10.3201/eid2106.140999
Baron, S., Hadjadj, L., Rolain, J. M., and Olaitan, A. O. (2016). Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int. J. Antimicrob. Agents 48, 583–591. doi: 10.1016/j.ijantimicag.2016.06.023
Campos, J., Cristino, L., Peixe, L., and Antunes, P. (2016). MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill. 21:pii=30270. doi: 10.2807/1560-7917.ES.2016.21.26.30270
Catry, B., Cavaleri, M., Baptiste, K., Grave, K., Grein, K., Holm, A., et al. (2015). Use of colistin-containing products within the European Union and European Economic Area (EU/EEA): development of resistance in animals and possible impact on human and animal health. Int. J. Antimicrob. Agents 46, 297–306. doi: 10.1016/j.ijantimicag.2015.06.005
Demczuk, W. H., Finley, R., Nadon, C., Spencer, A., Gilmour, M., and Ng, L. K. (2010). Characterization of antimicrobial resistance, molecular and phage types of Salmonella enterica serovar Typhi isolations. Epidemiol. Infect. 138, 1414–1426. doi: 10.1017/S0950268810000221
Dixon, R. A., and Chopra, I. (1986). Polymyxin B and polymyxin B nonapeptide alter cytoplasmic membrane permeability in Escherichia coli. J. Antimicrob. Chemother. 18, 557–563. doi: 10.1093/jac/18.5.557
Doumith, M., Godbole, G., Ashton, P., Larkin, L., Dallman, T., Day, M., et al. (2016). Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J. Antimicrob. Chemother. 71, 2300–2305. doi: 10.1093/jac/dkw093
Eiamphungporn, W., Yainoy, S., Jumderm, C., Tan-Arsuwongkul, R., Tiengrim, S., and Thamlikitkul, V. (2018). Prevalence of the colistin resistance gene mcr-1 in colistin-resistant Escherichia coli and Klebsiella pneumoniae isolated from humans in Thailand. J Glob Antimicrob Resist 15, 32–35. doi: 10.1016/j.jgar.2018.06.007
Elkington, J. (2020). Green Swans: The Coming Boom in Regenerative Capitalism. Greenleaf Book Group Press.
GBD 2017 Causes of Death Collaborators (2018). Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392, 1736–1788. doi: 10.1016/S0140-6736(18)32203-7
GBD 2017 DALYs and HALE Collaborators (2018). Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392, 1859–1922. doi: 10.1016/S0140-6736(18)32335-3
GBD 2017 Disease and Injury Incidence and Prevalence Collaborators (2018). Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392, 1789–1858. doi: 10.1016/S0140-6736(18)32279-7
Gordon, M. A., Graham, S. M., Walsh, A. L., Wilson, L., Phiri, A., Molyneux, E., et al. (2008). Epidemics of invasive Salmonella enterica serovar enteritidis and S. enterica Serovar typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin. Infect. Dis. 46, 963–969. doi: 10.1086/529146
Kempf, I., Jouy, E., and Chauvin, C. (2016). Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 48, 598–606. doi: 10.1016/j.ijantimicag.2016.09.016
Koh, T. H., Koh, A. E., Hamdan, A., Khoo, B. C., Yu, V. Y., Raymond, R. T., et al. (2008). Ceftriaxone-resistant Salmonella spp. in Singapore. Ann. Acad. Med. Singap. 37, 900–901.
Li, J., Nation, R. L., Turnidge, J. D., Milne, R. W., Coulthard, K., Rayner, C. R., et al. (2006). Colistin: the re-emerging antibiotic for multidrug-resistant gram-negative bacterial infections. Lancet Infect. Dis. 6, 589–601. doi: 10.1016/S1473-3099(06)70580-1
Li, R., Xie, M., Lv, J., Wai-Chi, C. E., and Chen, S. (2017). Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J. Antimicrob. Chemother. 72, 696–699. doi: 10.1093/jac/dkw509
Lin, J., Nishino, K., Roberts, M. C., Tolmasky, M., Aminov, R. I., and Zhang, L. (2015). Mechanisms of antibiotic resistance. Front. Microbiol. 6:34. doi: 10.3389/fmicb.2015.00034
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lu, X., Zeng, M., Xu, J., Zhou, H., Gu, B., Li, Z., et al. (2019). Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai, China, 2006–2016. EBioMedicine 42, 133–144. doi: 10.1016/j.ebiom.2019.03.006
Mabey, D. C., Brown, A., and Greenwood, B. M. (1987). Plasmodium falciparum malaria and Salmonella infections in Gambian children. J. Infect. Dis. 155, 1319–1321. doi: 10.1093/infdis/155.6.1319
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O’Brien, S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Molbak, K., Baggesen, D. L., Aarestrup, F. M., Ebbesen, J. M., Engberg, J., Frydendahl, K., et al. (1999). An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype typhimurium DT104. N. Engl. J. Med. 341, 1420–1425. doi: 10.1056/NEJM199911043411902
Murray, C. J., Vos, T., Lozano, R., Naghavi, M., Flaxman, A. D., Michaud, C., et al. (2012). Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2197–2223. doi: 10.1016/S0140-6736(12)61689-4
Nordmann, P., Lienhard, R., Kieffer, N., Clerc, O., and Poirel, L. (2016). Plasmid-mediated colistin-resistant Escherichia coli in bacteremia in Switzerland. Clin. Infect. Dis. 62, 1322–1323. doi: 10.1093/cid/ciw124
Pornruangwong, S., Hendriksen, R. S., Pulsrikarn, C., Bangstrakulnonth, A., Mikoleit, M., Davies, R. H., et al. (2011). Epidemiological investigation of Salmonella enterica serovar Kedougou in Thailand. Foodborne Pathog. Dis. 8, 203–211. doi: 10.1089/fpd.2010.0626
Qian, H., Cheng, S., Liu, G., Tan, Z., Dong, C., Bao, J., et al. (2020). Discovery of seven novel mutations of gyrB, parC and parE in Salmonella Typhi and Paratyphi strains from Jiangsu Province of China. Sci. Rep. 10:7359. doi: 10.1038/s41598-020-64346-0
Reddy, E. A., Shaw, A. V., and Crump, J. A. (2010). Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect. Dis. 10, 417–432. doi: 10.1016/S1473-3099(10)70072-4
Schrijver, R., Stijntjes, M., Rodriguez-Bano, J., Tacconelli, E., Babu, R. N., and Voss, A. (2018). Review of antimicrobial resistance surveillance programmes in livestock and meat in EU with focus on humans. Clin. Microbiol. Infect. 24, 577–590. doi: 10.1016/j.cmi.2017.09.013
Shen, Z., Wang, Y., Shen, Y., Shen, J., and Wu, C. (2016). Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect. Dis. 16:293. doi: 10.1016/S1473-3099(16)00061-X
Sirichote, P., Hasman, H., Pulsrikarn, C., Schonheyder, H. C., Samulioniene, J., Pornruangmong, S., et al. (2010). Molecular characterization of extended-spectrum cephalosporinase-producing Salmonella enterica serovar Choleraesuis isolates from patients in Thailand and Denmark. J. Clin. Microbiol. 48, 883–888. doi: 10.1128/JCM.01792-09
Soler-Garcia, A. A., De Jesus, A. J., Taylor, K., and Brown, E. W. (2014). Differentiation of Salmonella strains from the SARA, SARB and SARC reference collections by using three genes PCR-RFLP and the 2100 Agilent Bioanalyzer. Front. Microbiol. 5:417. doi: 10.3389/fmicb.2014.00417
Song, J., Gao, X., and Galan, J. E. (2013). Structure and function of the Salmonella Typhi chimaeric A(2)B(5) typhoid toxin. Nature 499, 350–354. doi: 10.1038/nature12377
Threlfall, E. J., de Pinna, E., Day, M., Lawrence, J., and Jones, J. (2008). Alternatives to ciprofloxacin use for enteric fever, United Kingdom. Emerg. Infect. Dis. 14, 860–861. doi: 10.3201/eid1405.071184
World Health Organization (2011). “Collaborating centre for reference and research on Salmonella,” Antigenic Formulae of Salmonella serovars. Paris: Pasteur Institute.
Xavier, B. B., Lammens, C., Butaye, P., Goossens, H., and Malhotra-Kumar, S. (2016). Complete sequence of an IncFII plasmid harbouring the colistin resistance gene mcr-1 isolated from Belgian pig farms. J. Antimicrob. Chemother. 71, 2342–2344. doi: 10.1093/jac/dkw191
Yang, Y. Q., Zhang, A. Y., Ma, S. Z., Kong, L. H., Li, Y. X., Liu, J. X., et al. (2016). Co-occurrence of mcr-1 and ESBL on a single plasmid in Salmonella enterica. J. Antimicrob. Chemother. 71, 2336–2338. doi: 10.1093/jac/dkw243
Zhang, C., Feng, Y., Liu, F., Jiang, H., Qu, Z., Lei, M., et al. (2017). A phage-Like IncY plasmid carrying the mcr-1 gene in Escherichia coli from a pig farm in China. Antimicrob. Agents Chemother. 61:e02035-16. doi: 10.1128/AAC.02035-16
Zurfluh, K., Klumpp, J., Nuesch-Inderbinen, M., and Stephan, R. (2016). Full-length nucleotide sequences of mcr-1-harboring plasmids isolated from extended-spectrum-beta-lactamase-producing Escherichia coli isolates of different origins. Antimicrob. Agents Chemother. 60, 5589–5591. doi: 10.1128/AAC.00935-16
Keywords: plasmid-borne quinolone resistance, extended-spectrum beta-lactamases, quinolone resistance determining region, nontyphoidal Salmonella, mcr-1
Citation: Liu G, Qian H, Lv J, Tian B, Bao C, Yan H and Gu B (2021) Emergence of mcr-1-Harboring Salmonella enterica Serovar Sinstorf Type ST155 Isolated From Patients With Diarrhea in Jiangsu, China. Front. Microbiol. 12:723697. doi: 10.3389/fmicb.2021.723697
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyEtienne Giraud, Institut National de la Recherche Agronomique de Toulouse, France
Thomas Jové, INSERM U1092 Anti-Infectieux supports moléculaires des résistances et innovations thérapeutiques, France
Copyright © 2021 Liu, Qian, Lv, Tian, Bao, Yan and Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Gu, Z2IyMDAzMTEyOUAxNjMuY29t; Hong Yan, eWFuaG9uZ0Buam11LmVkdS5jbg==; Changjun Bao, YmFvMjAwMF9jbkAxNjMuY29t
†These authors have contributed equally to this work and share the first authorship
 Guoye Liu1†
Guoye Liu1† Bing Gu
Bing Gu