- Microbiology and Molecular Biology Laboratory, Department of Biological Sciences, Indian Institute of Science Education and Research, Bhopal, India
Mycobacteria such as Mycobacterium tuberculosis, the causative agent of tuberculosis that annually kills several million people worldwide, and Mycobacterium smegmatis, the non-pathogenic fast-growing mycobacteria, require oxidative phosphorylation to meet their energy requirements. We have previously shown that deletion of one of the two copies of atpD gene that codes for the ATP synthase β-subunit establishes an energy-compromised state in M. smegmatis. Here we report that upon such deletion, a major routing of electron flux occurs through the less energy-efficient complexes of its respiratory chain. ΔatpD bacterium also shows an increased reduced state which is further confirmed by the overexpression of WhiB3, a major redox sensor. We show a substantial modulation of the biosynthesis of cell wall associated lipids and triacylglycerol (TAG). An accumulation of TAG-containing lipid bodies is further confirmed by using 14C oleate incorporation. Interestingly, the mutant also shows an overexpression of TAG-degrading lipase genes, and the intracellular lipolytic enzymes mediate TAG hydrolysis for their utilization as energy source. We believe that our in vitro energy-depleted model will allow us to explore the critical link between energy metabolism, redox homeostasis, and lipid biosynthesis during ATP-depleted state, which will enhance our understanding of the bacterial adaptation, and will allow us to identify novel drug targets to counter mycobacterial infections.
Introduction
Tuberculosis, caused by the bacterium Mycobacterium tuberculosis, is one of the serious public health concerns globally (Eisinger et al., 2020). M. tuberculosis is known to reside in a dormant state for prolonged period, and targeting such dormant M. tuberculosis is a big challenge for the current chemotherapeutic approaches (Gengenbacher and Kaufmann, 2012). Thus, a continuous arms race between human and mycobacteria is leading to the discovery and development of novel drug targets and therapeutics against M. tuberculosis.
Oxidative phosphorylation (OXPHOS) is an efficient pathway to sustain bacterial growth and survival (Bald et al., 2017). During this process, electrons obtained through the donors of central metabolic pathways are transferred from the respiratory complexes to O2 via electron transport chain (ETC) (Matsoso et al., 2005). This highly flexible respiratory chain in mycobacteria consist of various membrane-embedded electron carrier molecules and enzymes that mediate the transfer of electron, which is coupled with the generation of proton motive force (PMF) that drives ATP synthesis (Cook et al., 2014). The electron travels from complex-I (NADH dehydrogenase) to complex-II (succinate dehydrogenase) with the help of electron carrier molecule menaquinone. The chain is terminated via two terminal oxidases namely aa3-type cytochrome c oxidase (supercomplex along with cytochrome bc1 and cytochrome c) and cytochrome bd oxidase which eventually catalyzes the reduction of O2 to water molecule by using the electrons from menaquinone and cytochrome c, respectively (Kana et al., 2001; Matsoso et al., 2005). Previous studies suggest that in Mycobacterium smegmatis during low growth rate and low air saturation (0.6%), the cytochrome bd oxidase complex is upregulated >50-fold, which exemplifies the importance of cytochrome bd at low O2 rates (Berney and Cook, 2010). Moreover during hypoxia under the inhibitory conditions of cytochrome bc1-aa3 oxidase complex, cytochrome bd has been shown to play an important role in cell survival (Kana et al., 2001; Cook et al., 2014). Furthermore, the involvement of cytochrome bd in adaptation of M. tuberculosis toward adverse environmental conditions is very well known (Giuffre et al., 2012; Forte et al., 2016). Hence, respiration plays a very important role in mycobacterial survival in order to respond and adapt toward different environmental niche and stress conditions.
The inhibition of respiratory chain may occur either by suppression of the respiratory complexes (Lamprecht et al., 2016; Moosa et al., 2017) or due to an exposure to external stress factors such as hypoxia, nutrient deficiency, acid, NO, CO, etc., which eventually influence the redox environment of the cell by making the electron carriers more reduced (Cook et al., 2014; Iqbal et al., 2018; Akela and Kumar, 2021). Hypoxia is known to induce reactive oxygen species (ROS) (Brunelle et al., 2005), whereas NO acts as one of the major antimicrobial stress agents generating reactive nitrogen species (RNS) in the cell, thereby leading to the fluctuations in redox state (Mehta and Singh, 2019). This necessitates mycobacteria to respond toward the environmental variations to maintain homeostasis. In order to defend from the internal oxidants and reductants, mycobacteria has evolved with specific mechanisms that assist in maintaining an appropriate redox balance within the cell (Zhai et al., 2019). In M. tuberculosis, for example, it has been suggested that during reduced O2 conditions, WhiB3 senses the changes in the intracellular redox environment leading to metabolic switchover to fatty acids as the preferred nutrient source for the cell (Mehta and Singh, 2019). The presence of 4Fe–4S cluster in WhiB3 helps in maintaining redox homeostasis via regulating the metabolism and polyketide biosynthesis in the cell, thereby assisting in the persistence of the pathogen within the host (Mehta and Singh, 2019). The metabolic dependency of M. tuberculosis upon host fatty acids and cholesterol ensures a long-term survival during the persistence phase of infection (Gomez and McKinney, 2004; Pandey and Sassetti, 2008). Nevertheless, how these fatty acids are acquired, stored, and further utilized is still unclear in mycobacteria.
We have previously shown that the deletion of one of the two copies of atpD gene that codes for the β-subunit of the ATP synthase machinery renders M. smegmatis in an energy-compromised state (Patil and Jain, 2019). In the present work, we investigated in detail the mechanism involved in this adaptation and report that in M. smegmatis, deletion of atpD results in the remodeling of the ETC with utilizing less efficient proton non-translocating complexes. This results in the establishment of a reductive stress in the cell, which is dissipated by a major redox sensor, WhiB3-mediated modulation of the biosynthesis of storage lipids (triacylglycerol). We have previously shown that in ΔatpD, β-oxidation is enhanced for the generation of energy (Patil and Jain, 2019). It, therefore, appears that M. smegmatis follows two distinctive and seemingly opposite pathways in order to adapt to a low-energy state. Our work here thus reveals a distinct relationship between lipid accumulation and its utilization in M. smegmatis under energy-compromised state, and that a respiratory shift drives the global cellular changes leading to redox imbalance and metabolic dependency on fatty acid. We believe that understanding these cellular characteristics and their modulation to stress is vital for the identification and development of novel therapeutic drug targets to treat mycobacterial infections.
Results
We have previously shown that M. smegmatis atpD gene (MSMEG_4936), which codes for the β-subunit of F0F1 ATP synthase, is important for mycobacterial physiology and metabolism (Patil and Jain, 2019), and that the deletion of one of the two copies of atpD renders the bacterium in an energy-compromised state. In this manuscript, we explore the impact of this deletion on the physiology of M. smegmatis and address how cell adapts to this physiological state and survives.
Deletion of atpD Results in Altered Expression of the Respiratory Chain Complexes
To understand in detail how M. smegmatis adapts to and survive in the depleted energy state, we first looked into the highly flexible mycobacterial ETC and its complexes that facilitate mycobacterial survival by coordinating with the central metabolism under variety of environmental conditions. M. smegmatis consists of energy efficient type-I NADH dehydrogenase complex (NDH1), which is proton pumping in nature, encoded by nuo operon, and a proton non-pumping type-II NADH dehydrogenase complex (NDH2) encoded by ndh gene (MSMEG_3621). The two terminal oxidases utilized by mycobacterial ETC for reduction of oxygen include the energy-efficient cytochrome bc1-aa3 supercomplex and the energetically less efficient cytochrome bd oxidase (Cook et al., 2014; Iqbal et al., 2018). The transfer of electron from these terminal oxidases generates a PMF, which is then subsequently utilized for ATP synthesis via F0F1 ATP synthase. Transcriptional analysis shows that while the genes belonging to type-I NADH dehydrogenase are downregulated (Figure 1A), the proton non-pumping type-II NADH dehydrogenase is upregulated (Figure 1B) in the ΔatpD strain as compared to the wild-type and the complemented strain. Similarly, the energy efficient terminal electron acceptor cytochrome bc1-aa3 complex is found to be transcriptionally repressed (Figure 1C), whereas the energetically less efficient cytochrome bd oxidase genes are found to be induced (Figure 1D). This shift in the mycobacterial respiration machinery observed here suggests a modification in the respiration from proton translocating to non-proton-translocating state in ΔatpD. Similar observations were also seen when M. tuberculosis was grown under hypoxia conditions (Shi et al., 2005), suggesting a conserved respiratory adaptation in both pathogenic M. tuberculosis and non-pathogenic M. smegmatis.
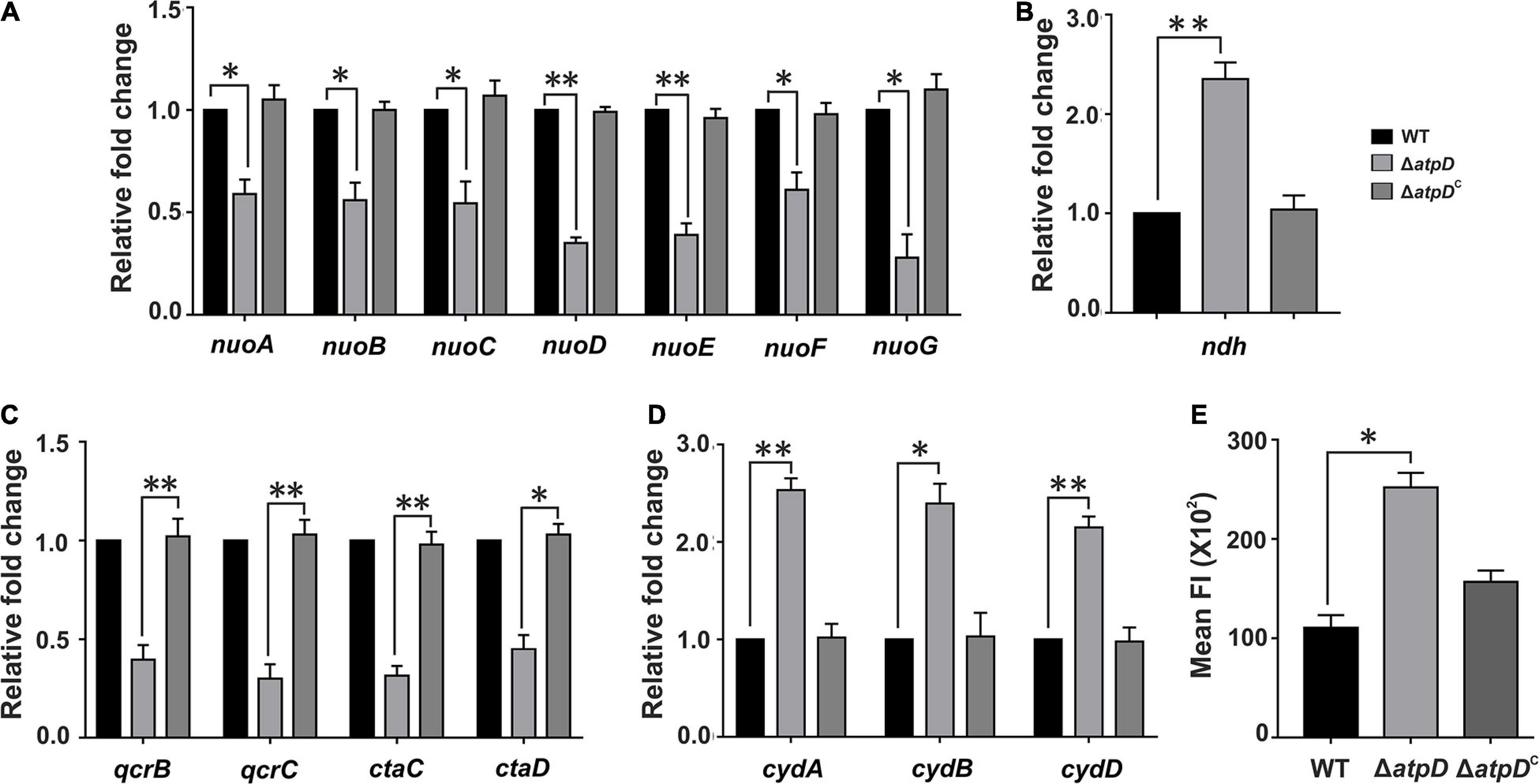
Figure 1. Transcriptional changes in the respiratory chain complexes and an altered membrane polarization in M. smegmatis ΔatpD. Plots show the expression profiles for genes belonging to NDH1 complex (nuo operon, A), NDH2 complex (ndh, B), cytochrome bc1-aa3 complex (qcr and cta, C), and cytochrome-bd oxidase complex (cyd, D) in M. smegmatis wild-type (WT), atpD knock out (ΔatpD), and atpD-complemented (ΔatpDC) strains. Panel (E) shows the membrane polarization in all the three strains as monitored by DiBAC4(3) dye. In panels (A–D), the transcript level in each case is compared with that of M. smegmatis WT, which is taken as 1. In all the panels, data represent an average of at least three independent experiments with error bars representing standard deviation. ∗P-value < 0.05; ∗∗P-value < 0.01.
We next hypothesized that such a broad change in the ETC machinery will affect the intracellular concentration of H+, which will result in an acidic pH in the cytoplasm and alter cellular membrane polarization. Hence we measured the changes in the membrane polarization by using a fluorescent dye DiBAC4(3) [Bis (1,3-dibutylbarbituric acid) trimethine oxonol], which readily diffuses across a depolarized membrane. Interestingly, the ΔatpD bacterium displayed increased fluorescence suggesting alteration in the membrane polarization as compared to the wild-type and the complemented strain (Figure 1E). Overall our data suggest that due to the deletion of single copy of atpD, the respiratory functions and energy production factors are maintained at a low level and result in altered membrane polarization.
Altered Membrane Polarization Disrupts Redox Homeostasis in ΔatpD, and Is Balanced by Lipid Biosynthesis
Membrane potential plays an important role in energy storage during OXPHOS (Zorova et al., 2018). Previous studies suggest that bacteria possess a remarkable ability to maintain their membrane polarized, even during external stress (Prindle et al., 2015; Blee et al., 2020). A sharp decline in the membrane potential is considered dangerous to the cell, due to its inefficiency to produce ATP and leading to the generation of reductive stress in the cell (Zorova et al., 2018), which leads to slow down of respiratory complexes (Akela and Kumar, 2021). Furthermore, we have previously shown that MsmΔatpD has reduced levels of ATP with increased ROS and high NADH/NAD+ ratio, indicating an imbalanced redox state within the cell (Patil and Jain, 2019). We, therefore, asked how cell manages this intracellular redox imbalance in order to replenish the depleted NAD+ levels for survival. It has been suggested that dosS/dosR system functions as a prominent sensor for the extracellular redox signals such as reduced oxygen and NO levels (Kumar et al., 2007). WhiB3, on the other hand, is known as a major intracellular redox sensor in M. tuberculosis, which acts by maintaining the redox balance in the cell thereby regulating the central metabolism (Singh et al., 2009). M. smegmatis WhiB3 (MSMEG_1597) shares 79.8% identity with M. tuberculosis WhiB3 (Rv3416) (Supplementary Figure 1). Hence we hypothesized that in M. smegmatis under an energy-compromised state, WhiB3 acts as the major redox regulator involved in the dissipation of the reductive stress via synthesis of storage lipid. Therefore, we determined the expression level of WhiB3 in M. smegmatis, and found a threefold upregulation in its expression in atpD knock-out strain as compared to the wild-type and the atpD-complemented (atpDC) strain (Figure 2A). An upregulation in WhiB3 here clearly suggests that a redox state persists in M. smegmatis under energy-compromised state, which further corroborates with our previous study (Patil and Jain, 2019).
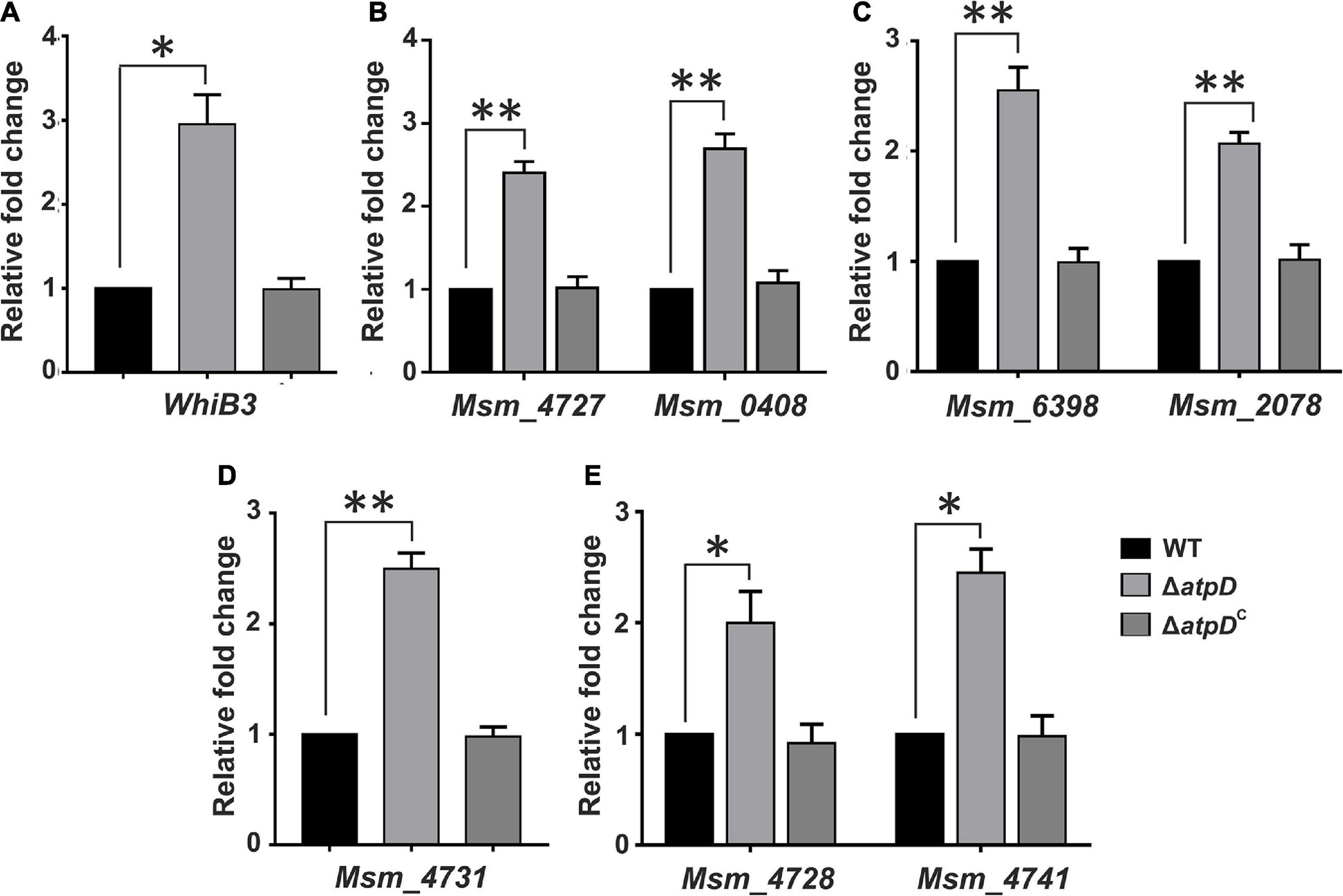
Figure 2. Mycobacterium smegmatis ΔatpD shows redox homeostasis under energy-deprived state. Shown are the relative mRNA transcript levels of redox sensor gene WhiB3 (A), polyketide biosynthesis genes (B), trehalose di-mycolate (TDM) biosynthesis genes (C), polymethyl-branched fatty acid gene (D), and polyketide synthase-associated protein (Pap) and mmpL genes (E) in M. smegmatis wild-type (WT), atpD knock out (ΔatpD), and atpD-complemented (ΔatpDC) strains. In all the panels, the transcript level in each case is compared with that of M. smegmatis WT, which is taken as 1. Data represent an average of at least three independent experiments with error bars representing standard deviation. ∗P-value < 0.05; ∗∗P-value < 0.01.
Furthermore, since WhiB3 in M. tuberculosis is known to be a positive transcriptional regulator for lipid biosynthesis genes such as those involved in polyketide biosynthesis (pks) and complex cell wall-associated lipids (Singh et al., 2009), we determined the mRNA expression level of MSMEG_4727 (pks5), which is a homolog of M. tuberculosis pks2 with 65% identity (Supplementary Figure 2), MSMEG_0408 (pks1) (Figure 2B), trehalose di-mycolate (TDM) (mycolyltransferases) genes in M. smegmatis (Ojha et al., 2010) such as MSMEG_6398 and MSMEG_2078 (Figure 2C), MSMEG_4731, involved in the synthesis of polymethyl-branched fatty acids (Tran et al., 2019) in M. smegmatis (Figure 2D), and the polyketide synthase-associated protein (Pap) gene such as MSMEG_4728 (Onwueme et al., 2004; Tran et al., 2019), and mmpL, MSMEG_4741 (Figure 2E), which is posited to encode the enzymes involved in the synthesis and transport of glycolipids such as GPL, lipo-oligosaccharide (LOS), and sulfolipid-1 (Tran et al., 2019). Interestingly, we found an upregulation of all the cell wall-associated genes. During hypoxia, since the respiratory chain is down-regulated, M. tuberculosis is known to regenerate NAD+ from NADH via WhiB3, thereby sensing the intracellular redox alterations and causing the metabolic switchover to fatty acids as the preferred carbon source (Singh et al., 2009). Such metabolic shift induced by WhiB3 is known to regulate the production of methyl branched polyketides (PAT/DAT, SL-1, and PDIM) and TAG (Singh et al., 2009). Hence the observed overexpression of WhiB3 and its role in modulating polyketide biosynthesis in M. smegmatis ΔatpD immediately suggest an important function played by WhiB3 in homeostasis maintenance via lipid biosynthesis and as a physiological redox regulator in M. smegmatis under energy-compromised state.
Loss of atpD Severely Affects the Cardiolipin Content in M. smegmatis Cell Membrane and Altered Mycolic Acid Levels Leading to Loss of Acid Fastness
Since in MsmΔatpD, an upregulation in WhiB3 suggests dissipation of reductive stress via lipid biosynthesis, at this juncture, an analysis of the lipid content of the cell becomes extremely important. Cell membrane lipids are known to play a crucial role in the membrane dynamics during growth and stress (Stallings and Glickman, 2010; Queiroz and Riley, 2017). Mycobacterial cell membrane consists of a plethora of lipids, which function as the structural components and storage bodies for the cell (Queiroz and Riley, 2017). A general pathway for the biosynthesis of these lipids in mycobacteria is depicted in Figure 3A. Major structural and storage mycobacterial lipids include phospholipids such as the cardiolipin (CL), phosphatidylethanolamine (PE), phosphatidylinositol (PI), glycosylated PIs, phosphatidylserine (PS), phosphatidylglycerol (PG; a minor species in mycobacteria), and triacylglycerol (TAG), which is a neutral storage lipid for the cell (Crellin et al., 2013). Furthermore, out of the major phospholipids, CL accounts for the maximum amount (37%) of the total phospholipids in mycobacterial plasma membrane followed by PE (32%) and PI/PIMs (28%) (Crellin et al., 2013). We, therefore, first determined the levels of CL in all the three strains. CL is known to form aggregates within the membrane and is determined using Nonyl Acridine orange (NAO) fluorescent dye, which specifically binds to it (Mileykovskaya et al., 2001). Interestingly, the ΔatpD bacterium shows reduced fluorescence as compared to that of the wild-type and the complemented strain (Figure 3B), which is further corroborated well by the FACS analysis (Figure 3C). To gain further insight into the reduction in the cardiolipin levels in knock-out strain, we analyzed the mRNA expression level of plasma membrane biogenesis genes (depicted in Figure 3D) by reverse transcription-quantitative PCR (RT-qPCR). We observe several folds reduction in the expression of all the plasma membrane biogenesis genes (pgsA1, pgsA2, which is a putative cardiolipin synthase gene, and pgsA3) in the knock-out strain as compared to the wild-type and the complemented strain (Figure 3E). Overall, this significantly reduced fluorescence and the gene expression levels in the knock-out strain are together suggestive of low amounts of cardiolipin (CL) content in the ΔatpD strain as compared to the wild-type and the complemented strain. This also manifests a compromised cell envelope in M. smegmatis ΔatpD as shown in our previous study (Patil and Jain, 2019). Next we examined the levels of other major phospholipids in the cell such as Phosphatidylethanolamine or PE (MSMEG_6851), Phosphatidylserine or PS (MSMEG_0860), and Phosphatidylinositol or PI (MSMEG_2933) by RT-PCR as well as by monitoring the incorporation of 14C oleic acid into phospholipids of all the three strains. Interestingly, our data show that the levels of these phospholipids remain almost unchanged in both wild-type and mutant strains (Figure 3F and Supplementary Figure 3). Taken together, these results suggest that the mycobacterial cell under energy-compromised state might depend on some other lipids in order to maintain survival.
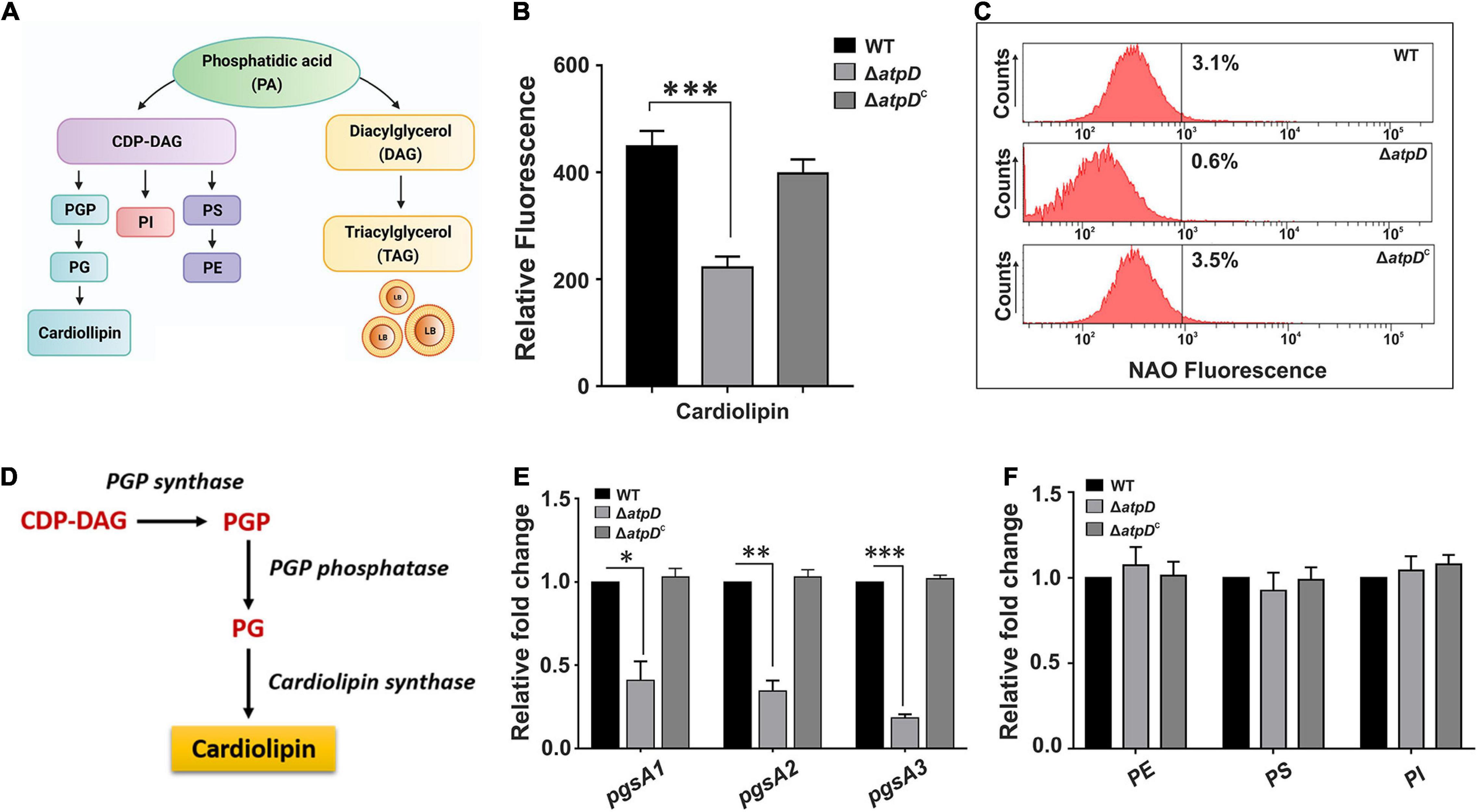
Figure 3. Mycobacterium smegmatis ΔatpD shows reduced cardiolipin content. Panel (A) shows various mycobacterial membrane lipids along with the flow of key phospholipid biosynthesis pathways. PGP, phosphatidylglycerol phosphate; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; PE, phosphatidylethanolamine; CDP-DAG, cytidine-diphosphate diacylglycerol. Panel (B) shows the Nonyl Acridine orange (NAO) dye fluorescence measured for M. smegmatis wild-type (WT), atpD knock-out (ΔatpD), and atpD-complemented (ΔatpDC) strains. Panel (C) shows the flow cytometric analysis of all the three strains after staining the cells with NAO dye. The percentage value represents the fraction of cells that are positive for that fluorescence. Flow cytometry experiments were repeated at least thrice; the graph shows only one representative data. Panel (D) shows the cardiolipin biosynthesis pathway in mycobacteria. Panel (E) shows the gene expression analysis for plasma membrane biosynthesis genes pgsA1, pgsA2, and pgsA3 by RT-qPCR in M. smegmatis wild-type, ΔatpD, and ΔatpDC strains. Panel (F) shows the relative expression level of major structural genes – MSMEG_6851 (phosphatidylethanolamine or PE), MSMEG_0860 (phosphatidylserine or PS), and MSMEG_2933 (phosphatidylinositol or PI) in M. smegmatis wild-type, ΔatpD, and ΔatpDC strains. In both panels (E,F), the transcript level in each case is compared with that of M. smegmatis WT, which is taken as 1. In panels (B,E,F), data represent an average of at least three independent experiments with standard deviation shown as error bars. ∗P-value < 0.05; ∗∗P-value < 0.01; ∗∗∗P-value < 0.005. Panel (A) is created with www.BioRender.com.
Since mycolic acid acts as the major component of the mycobacterial cell wall, we next asked if the levels of mycolic acid are affected in M. smegmatis ΔatpD. To investigate this, we first examined the transcription level of the genes belonging to the FAS-I and FAS-II pathway involved in the biosynthesis of mycolic acid. Surprisingly, several-fold downregulation in the expression level of the genes belonging to the FAS-II and FAS-I pathway such as fabD (MSMEG_4325), kasA (MSMEG_4327), kasB (MSMEG_4328), mabA (MSMEG_3150), accA3 (acetyl-CoA carboxylase; MSMEG_1807), accD4 (MSMEG_6391), accD6 (MSMEG_4329), and fas (MSMEG_4757) is observed in the atpD knock-out strain as compared to the wild-type and the complemented strain (Figure 4A). The observed downregulation in the FAS-I and FAS-II pathway genes suggests an alteration in mycolic acid levels (Shi et al., 2010) in the mutant as compared to the wild-type strain. Taken together, this indicates that the deletion of atpD gene negatively influences the de novo lipid biosynthesis. Further it is well known that alteration in the mycolic acid biosynthesis within the cell leads to loss of acid-fastness in mycobacteria (Daniel et al., 2011, 2016). Therefore, we investigated whether such phenotype is also shown by ΔatpD. Therefore, the cells were stained with Auramine-O (acid-fast staining green fluorescent dye), which stains mycolic acids present in the cell envelope of mycobacteria and has been widely used to determine the acid-fastness property of the cells. Here, the atpD knock-out strain shows reduced Auramine-O fluorescence indicating loss of acid fastness (Figure 4B). On the other hand, both the wild-type and the complemented bacteria (ΔatpDC) are acid-fast positive as judged by the retention of Auramine-O fluorescence (Figure 4B). Additionally, the mutant shows increased Nile red fluorescence, which is indicative of neutral lipid accumulation, as compared to the wild-type and the complemented strain (Figure 4B). Further, in order to obtain a quantitative estimation, we analyzed Auramine-O and Nile red fluorescence via FACS analysis and also measured the fluorescence on a SpectraMax multimode plate reader. Our results clearly show a significant reduction in the Auramine-O fluorescence and a drastic increase in the Nile red fluorescence in ΔatpD strain as compared to the wild-type and the complemented strain (Figures 4C,D). It is worth mentioning here that loss of acid-fastness is a feature reported for dormant mycobacteria (Daniel et al., 2011, 2016). Altogether, this remarkable alteration in the cardiolipin, other phospholipids, and mycolic acid levels in ΔatpD strongly points toward a significant rerouting of the lipid metabolism to other form in the mutant strain under energy-compromised state.
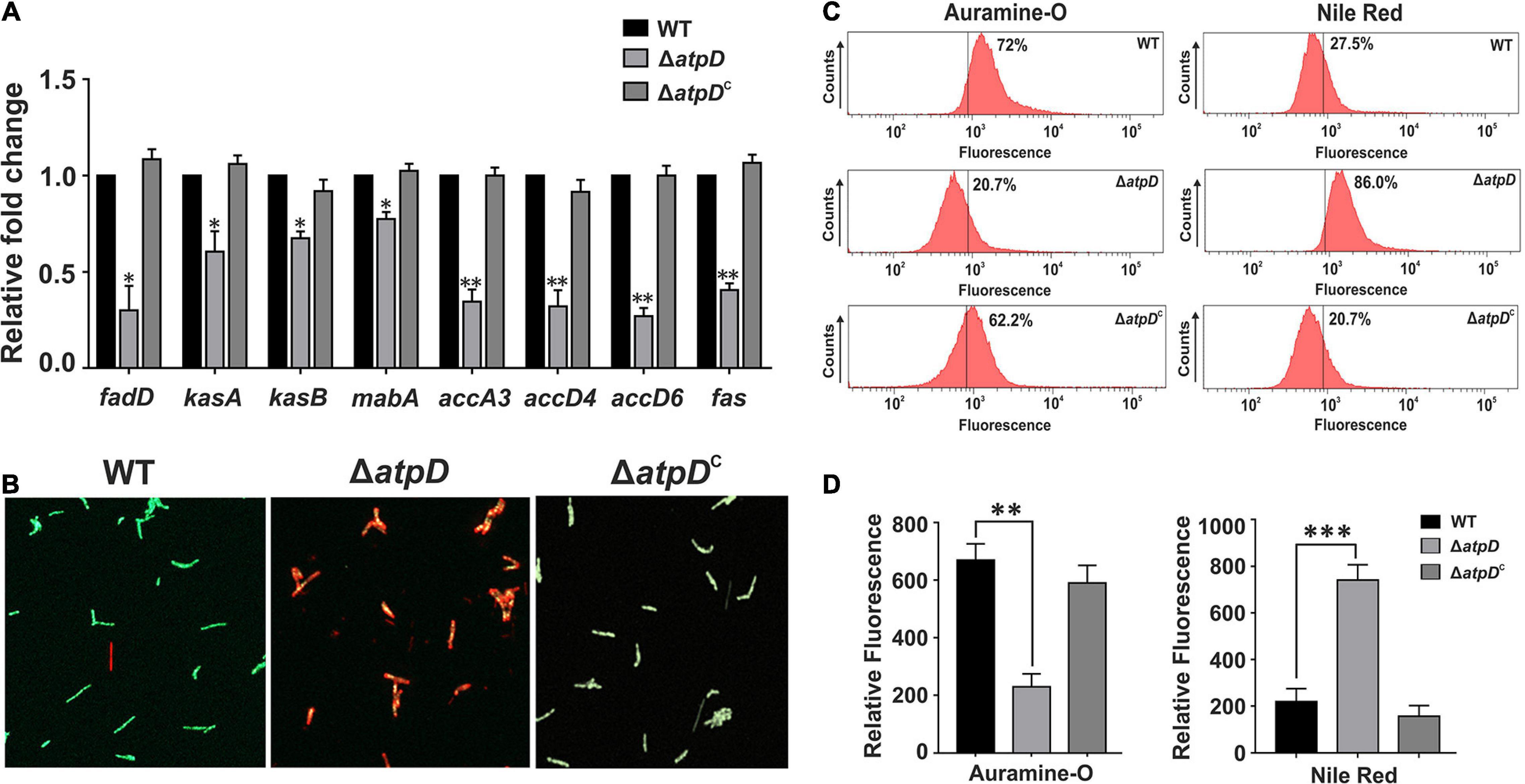
Figure 4. Mycobacterium smegmatis shows reduced mycolic acid levels leading to loss of acid-fastness under energy-compromised state. Panel (A) shows the relative mRNA transcript levels of the genes belonging to the FAS-II and FAS-I lipid biosynthesis pathway genes such as fabD (MSMEG_4325), kasA (MSMEG_4327), kasB (MSMEG_4328), mabA (MSMEG_3150), accA3 (acetyl-CoA carboxylase; MSMEG_1807), accD4 (MSMEG_6391), accD6 (MSMEG_4329), and fas (MSMEG_4757) in M. smegmatis wild-type (WT), atpD knock-out (ΔatpD), and atpD-complemented (ΔatpDC) strains. The transcript level in each case is compared with that of M. smegmatis wild-type, which is taken as 1. Panel (B) shows the fluorescence microscopy images for the M. smegmatis WT, ΔatpD, and ΔatpDC strains after staining the cells with Auramine-O and Nile red. Panel (C) shows the flow cytometric analysis of all the three strains after staining the cells with Auramine-O and Nile red dye, as mentioned. The percentage value represents the fraction of cells that are positive for that fluorescence. Flow cytometry experiments were repeated at least thrice. Panel (D) shows the quantification by measuring the fluorescence of Auramine-O and Nile red dyes as mentioned on fluorescence plate reader for M. smegmatis WT, ΔatpD, and ΔatpDC strains. In all the panels, the experiments were repeated at least thrice. In both panels (B,C), only one representative image is shown. In panels (A,D), error bars represent standard deviation. ∗P-value < 0.05; ∗∗P-value < 0.01; ∗∗∗P-value < 0.005.
Upregulation of WhiB3 Promotes Physiological and Metabolic Adaptations by Increasing TAG Accumulation in M. smegmatis Under Energy-Compromised State
In M. tuberculosis, WhiB3 is suggested to modulate the differential production of cell wall associated and storage lipids via redox-dependent switching mechanism (Singh et al., 2009); our data also suggest a significant rerouting to some other form of lipid in MsmΔatpD. Since TAG is the major storage lipid in mycobacteria (Daniel et al., 2011), we next explored the genetic basis of this phenomenon in ΔatpD, and examined the differential expression of the genes involved in TAG biosynthesis pathway, which is also known as Kennedy pathway in M. smegmatis (Maarsingh and Haydel, 2018). Interestingly, we find that the transcription profile of the genes coding for the enzymes catalyzing the TAG biosynthesis such as glycerol kinase (glpK, MSMEG_6759), glycerol-3-phosphate acyl transferase (GPAT, MSMEG_4703), acylglycerol phosphate acyltransferase (AGPAT, MSMEG_4284), and the two wax ester synthase/acyl-CoA:diacylglycerol acyltransferase (WSDGAT, MSMEG_1882, and MSMEG_4705), which catalyzes the final step of TAG biosynthesis, is upregulated in the ΔatpD mutant as compared to the wild-type and the complemented strain (Figure 5A). In M. tuberculosis, tgs1 is known to be the major TAG synthase gene (Daniel et al., 2004), and we found the two homologs of tgs1 genes in M. smegmatis, MSMEG_5242 and MSMEG_3948, which bear significant similarity (69%) with the M. tuberculosis tgs1 (Supplementary Figure 4). Interestingly, the transcript levels of these genes are also found to be higher (Figure 5A), which strongly suggests that this phenomenon may be conserved in M. tuberculosis as well. We additionally noticed that in M. tuberculosis, tgs1 gene is under direct control of dosS/dosR operon, and is highly overexpressed under stress conditions such as hypoxia, acidic environment, and under dormancy conditions (Daniel et al., 2011). Our previous report also showed an overexpression of dormancy regulon genes dosS and dosR that are the known mediators of tgs1-dependent TAG accumulation (Patil and Jain, 2019). Thus, an overexpression of TAG synthase genes in M. smegmatis ΔatpD (under an energy-compromised state) points toward the dormancy related metabolic state in the cell.
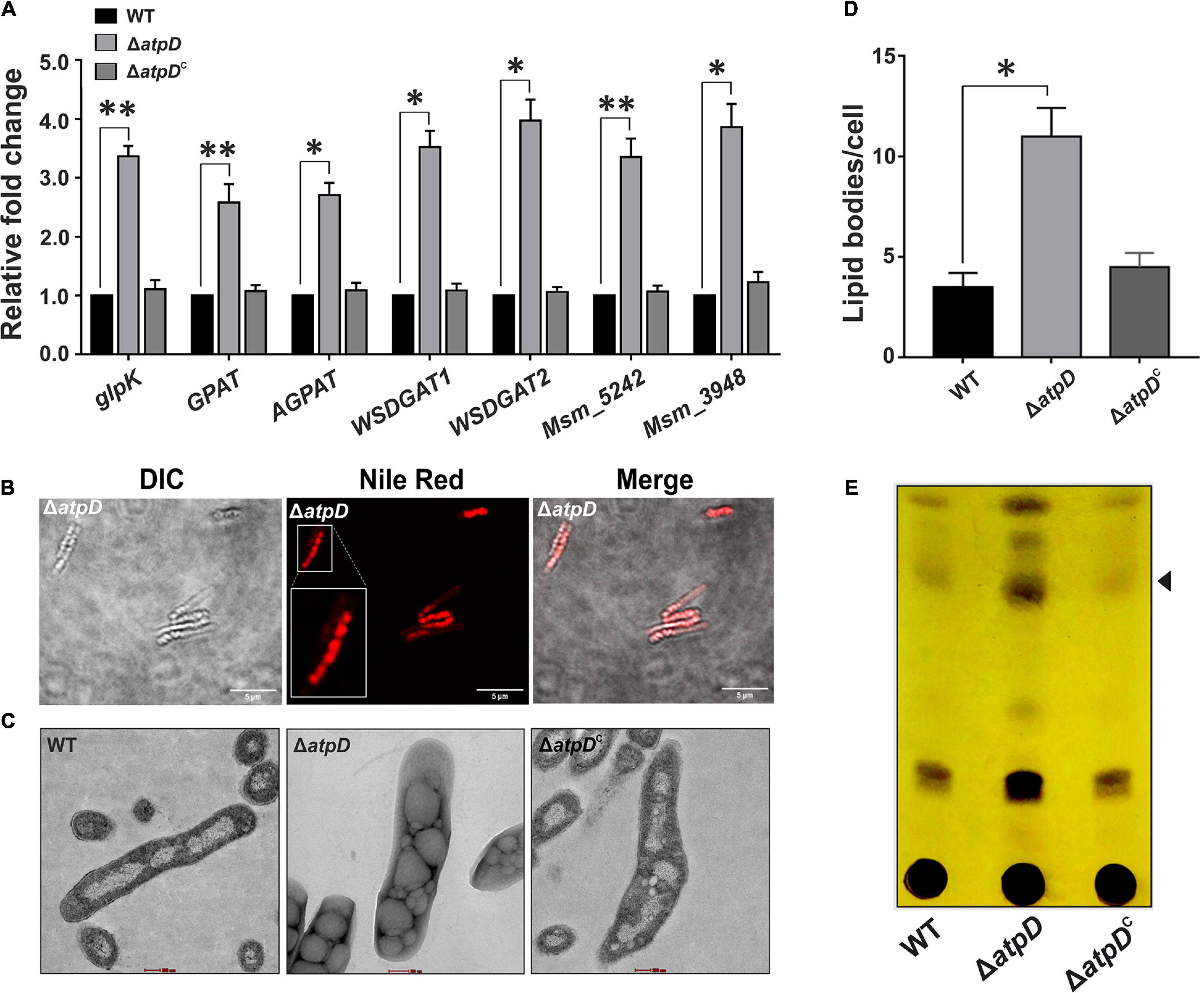
Figure 5. Increased TAG accumulation in M. smegmatis under energy-compromised state. Panel (A) shows the relative mRNA transcript levels of genes belonging to Kennedy pathway such as glycerol kinase (glpK), glycerol-3 phosphate acyl transferase (GPAT), acylglycerol phosphate acyltransferase (AGPAT), two wax ester synthase/acyl-CoA: diacylglycerol acyltransferase (WSDGAT) and the TAG synthase genes (MSMEG_3948 and MSMEG_5242; these are homologues of tgs1 in M. tuberculosis) in M. smegmatis wild-type (WT), atpD knock-out (ΔatpD), and atpD-complemented (ΔatpDC) strains. The transcript level in each case is compared with that of M. smegmatis WT, which is taken as 1. Panel (B) shows the confocal microscopy images after staining the cells with the Nile red dye. A single zoomed-in image in the Nile red panel is presented as inset for clarity. Panel (C) shows the transmission electron microscopy (TEM)-based ultrastructural analysis of all the three strains for the presence of lipid bodies (LBs). The scale bar in each image corresponds to 200 nm. Panel (D) shows the quantification of the LBs (n = 23) for the images obtained in panel (C). Panel (E) depicts the TLC image presenting the TAG accumulation in M. smegmatis WT, atpD knock out (ΔatpD), and ΔatpDC strains. Spot corresponding to the TAG is marked with an arrowhead. In panels (A,D), data show an average of at least three independent experiments with error bars representing standard deviation. ∗P-value < 0.05; ∗∗P-value < 0.01. In panels (B,C,E), only one representative image is shown.
The upregulation of the Kennedy pathway in the ΔatpD mutant strongly supports our hypothesis that the mutant retains TAG as the major storage lipid. To further confirm this finding, the mutant cells were stained with Nile red dye that specifically stains neutral lipids within the cell. Our confocal microscopy data clearly shows the presence of large number of brightly stained lipid bodies (LBs) in the knockout strain (Figure 5B). Transmission electron microscopy (TEM) analysis of the clinical strains of M. tuberculosis carried out by Vijay and coworkers clearly demonstrated a distinct accumulation of LBs containing TAG (Vijay et al., 2017). Therefore, in order to visualize the LBs in M. smegmatis, we performed its ultrastructural analysis using TEM. Our data clearly show the presence of large amount of LBs in the ΔatpD bacterium as compared to the wild-type and the complemented strain (Figures 5C,D). We additionally examined the levels of TAG in the mycobacterial cells using thin layer chromatography (TLC) wherein the knock-out strain shows a clear accumulation of TAG as an intense band compared to that obtained in the wild-type and the complemented strain (Figure 5E). Furthermore, in order to confirm that the free fatty acids (FFAs) incorporated within the knockout cells are mainly stored as TAG rather that simple FFA or phospholipids (McCarthy, 1971; Weir et al., 1972; Nakagawa et al., 1976; Barksdale and Kim, 1977), we used 14C oleic acid as a radiolabelled substrate and monitored the incorporation of 14C oleic acid as it acts as the major component of TAG. All the three bacteria viz. wild-type, ΔatpD, and ΔatpDC were grown in the presence 14C-Oleic acid, and the radiolabeled lipids were extracted and analyzed by silica-based TLC. We observe that in the knock-out strain, the incorporation of 14C-Oleic acid in TAG is significantly higher as compared to wild-type and complemented strain (Supplementary Figure 5). This further strengthens our observation that the atpD mutant retains TAG as the storage lipid.
TAG Hydrolysis Is Mediated by Intracellular Lipase Enzymes and the Consumption Can Be Blocked by Lipase Inhibitor
During dormancy, M. tuberculosis is known to utilize accumulated TAGs as energy source (Daniel et al., 2011), wherein FFAs are released via lipase-mediated hydrolysis, which further participate in β-oxidation. Therefore, in order to understand if M. smegmatis ΔatpD preferentially utilizes TAG as an energy source, we examined the expression profile of various intracellular lipase genes in M. smegmatis. Interestingly, we observe at least two- to threefold higher expression of several lipase (lip) genes such as lipG, lipJ, lipI, lipN, lipO, lipT, and lipZ in M. smegmatis ΔatpD as compared to the wild-type and the complemented bacterium (Figure 6A), indicating the possibility of enhanced TAG hydrolysis. In M. tuberculosis, lipase activity has been suggested to be regulated at post-translational level (Ravindran et al., 2014). Hence to confirm that enhanced lipase gene expression also results in enhanced lipase production, we measured the lipase activity in M. smegmatis ΔatpD in the presence and absence of a pathway-independent lipase inhibitor, tetrahydrolipstatin (THL). Lipase inhibitors are well-known to interfere with the lipid metabolism by impairing the activity of cellular lipolytic enzymes (Lass et al., 2011; Zechner et al., 2012). These lipid inhibitors are used to detect the molecular basis of TAG accumulation and its metabolism (Lass et al., 2011; Zechner et al., 2012). Our data show that in M. smegmatis ΔatpD, higher lipase expression coincides well with higher lipase activity that is reduced in the presence of inhibitor (Figure 6B). Overall our results suggest the importance of both accumulation and possible hydrolysis of TAGs for the energy production under an energy-compromised state.
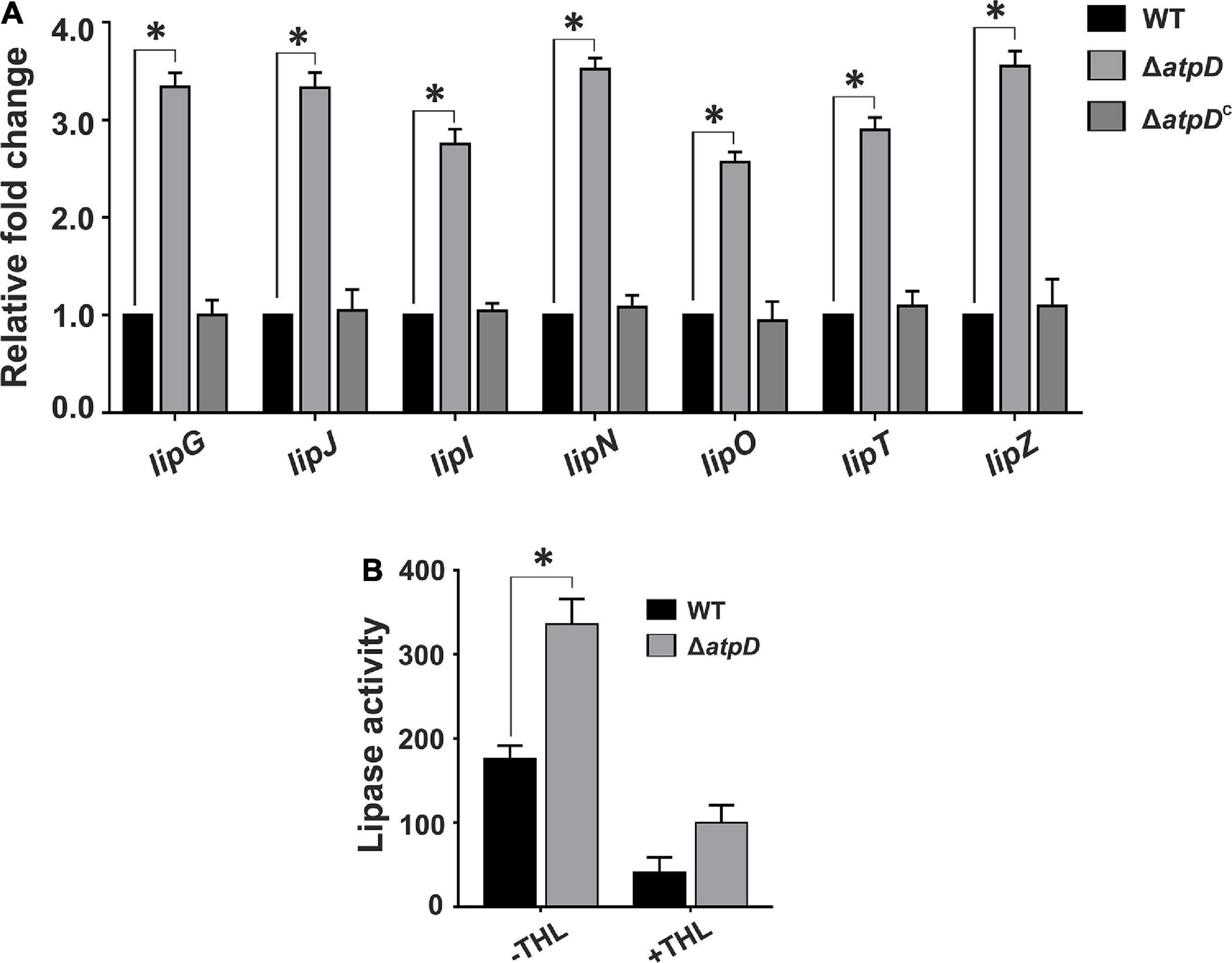
Figure 6. Mycobacterium smegmatis ΔatpD shows higher lipase gene expression coupled with an increased lipase activity. Panel (A) shows the relative mRNA transcript levels of the lipase genes (lipG, lipJ, lipI, lipN, lipO, lipT, and lipZ) in M. smegmatis wild-type (WT), atpD knock out (ΔatpD), and atpD-complemented (ΔatpDC) strains. The transcript level in each case is compared with that of M. smegmatis wild-type, which is taken as 1. Panel (B) shows the lipase activity in M. smegmatis WT and its atpD knock-out (ΔatpD) variant measured in the absence (–THL) and in the presence (+THL) of lipase inhibitor tetrahydrolipstatin (THL). In both the panels, data represent an average of at least three independent experiments with error bars depicting standard deviation. ∗P-value < 0.05.
In order to further confirm if ΔatpD preferentially utilizes TAG as an energy source, we monitored the TAG consumption in ΔatpD. We first grew the mutant strain for 24 h to allow for the accumulation of TAG. The cells were then transferred to a fresh medium without any carbon source and the TAG utilization was monitored for 48 h. Our results clearly show a linear decrease in the relative TAG content as a function of time, which suggests TAG hydrolysis in the mutant strain (Figures 7A,B). To further confirm that mycobacterial lipases hydrolyze TAG, the mutant bacteria were treated with an increasing concentration of THL for 2 h. Thereafter, the cells were washed and resuspended in fresh medium containing increasing concentrations of THL (5, 50, and 100 μg/ml) and were further incubated for 24 h. The samples were collected and TAG content was analyzed by TLC. The data show no significant difference in the TAG levels at 2 h time point in the presence or absence of the inhibitor (Figure 7C). However, interestingly, after 24 h of incubation, while the untreated cells show a significant disappearance of TAG, the cells exposed to increasing concentration of THL show the presence of TAG (Figures 7D,E). Taken together, our data suggest that M. smegmatis utilizes stored LBs containing TAG during energy depletion state by means of hydrolyzing neutral lipids. We wish to add here that the TAG consumption in ΔatpD also depends upon the expression of endogenous lipase enzymes, which further leads to the release of FFA. These FFAs are then utilized by the cell fueling the lipid oxidation pathways such as β-oxidation supporting the cell survival. The end product of the β-oxidation pathway is acetyl-CoA, which is directly channeled into the glyoxylate shunt to acquire cellular energy, thereby utilizing fatty acid as the major carbon source (Serafini et al., 2019). Indeed an upregulation of the β-oxidation pathway and glyoxylate shunt in M. smegmatis ΔatpD, as shown previously (Patil and Jain, 2019), corroborates well with these observations.
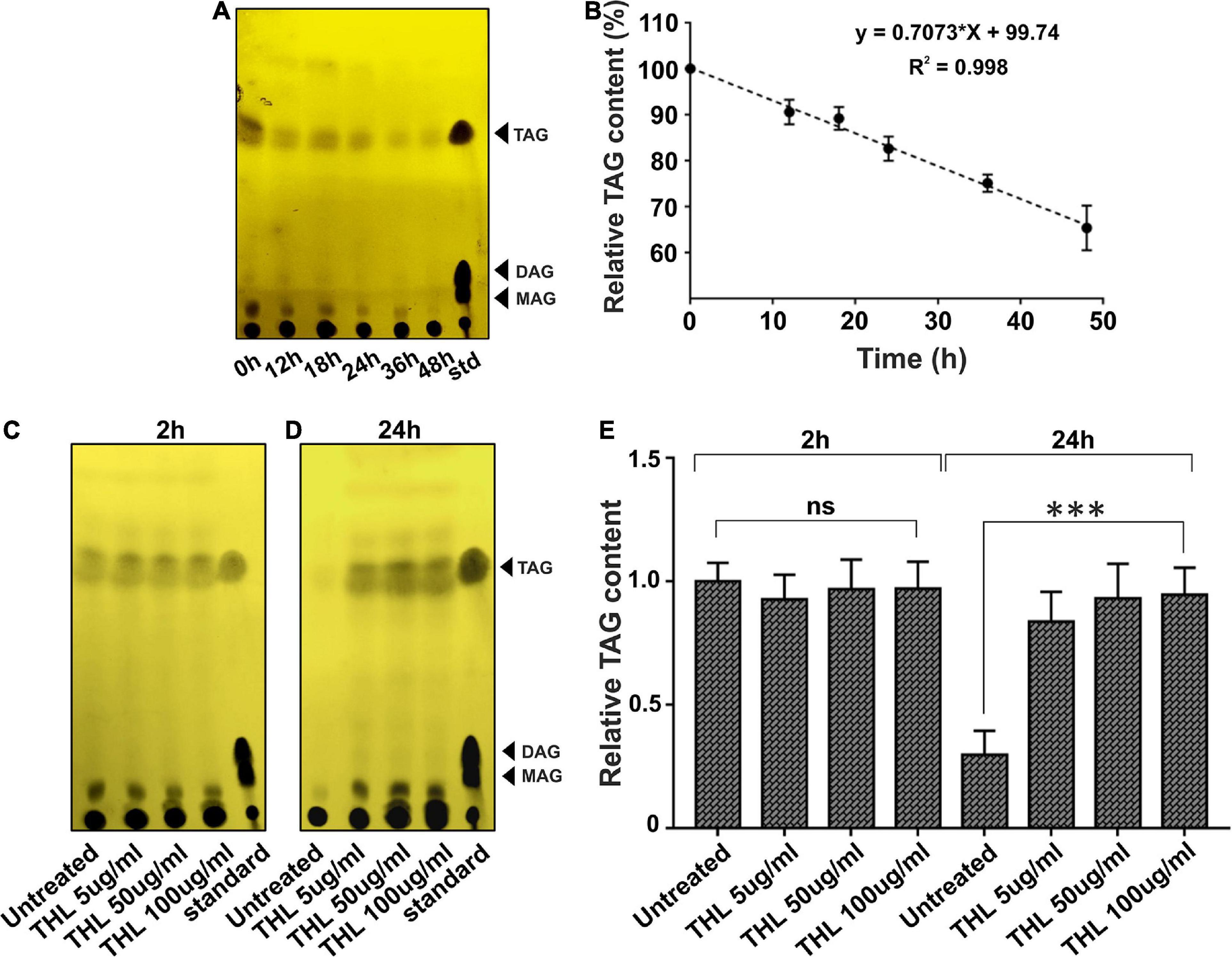
Figure 7. Intracellular lipolytic enzymes mediate TAG hydrolysis in M. smegmatis ΔatpD. Panel (A) shows the TLC image for the triacylglycerol (TAG) accumulation and its disappearance in M. smegmatis ΔatpD upon starvation with time (in hours). TAG mobility was compared with standard (std). Spot corresponding to the TAG, diacylglycerol (DAG), and monoacylglycerol (MAG) is marked with an arrowhead. Panel (B) shows the relative quantification of TAG by densitometric analysis of the TLC with time (in hours). Here, 100% corresponds to the 0 h time point. Panels (C,D) depict the TLC image showing the TAG accumulation and disappearance after the treatment of cells with a lipase inhibitor tetrahydrolipstatin (THL) at 2 h (C) and 24 h (D) time period. “Untreated” represents cells without THL treatment. “standard” is similar to “std” in panel (A). Panel (E) shows the relative TAG content as assessed by densitometric analysis of the TLC image, obtained before and after treating the cells with the increasing concentration of THL. The experiments were repeated at least thrice. In panels (A,C,D), only one representative image is shown. In panels (B,E), error bars represent standard deviation. ∗∗∗P-value < 0.005; “ns,” not significant.
Bedaquiline-Treated Wild-Type M. smegmatis Also Shows Increased WhiB3 Expression Coupled With TAG Accumulation and Lipase Gene Upregulation
Bedaquiline (BDQ) is known as the first-in-class ATP synthase inhibitor, having an extremely high selectivity for mycobacterial ATP synthase (Lakshmanan and Xavier, 2013). Upon BDQ exposure, M. tuberculosis has been shown to undergo significant metabolic remodeling along with reduced ATP/ADP levels (Koul et al., 2014; Akela and Kumar, 2021). BDQ inhibits bacterium’s ability to utilize OXPHOS and forces the cell to rely upon substrate-level phosphorylation, thereby continuing with ATP generation (Smith et al., 2020). Although an adequate amount of energy is provided by substrate-level phosphorylation for the cell survival, mycobacteria majorly depend upon OXPHOS to sustain their basic energy requirements (Foo et al., 2020). Hence, BDQ treatment in mycobacteria is thought to disturb the cellular environment forcing the cell to reroute its metabolism to overcome the stressful condition.
An earlier work suggested that bactericidal antibiotics induce ROS formation in M. tuberculosis leading to alterations in cellular redox state (Dwyer et al., 2014). Additionally, BDQ in combination with other antibiotics such as clofazimine and telacebec is very well known to induce oxidative stress and ROS formation with increased cellular reductive stress in M. tuberculosis (Lamprecht et al., 2016). Furthermore, since M. smegmatis ΔatpD balances the cellular reductive stress via WhiB3 overexpression, we asked if this mechanism persists in M. smegmatis upon BDQ treatment also. Hence we determined the WhiB3 expression under BDQ-treated condition. Interestingly, BDQ-treated wild-type M. smegmatis shows ∼3.5-fold upregulation of the WhiB3 expression (Figure 8A). Moreover, since our previous report also suggests that M. smegmatis ΔatpD strain largely mimics BDQ-treated wild-type M. smegmatis (Patil and Jain, 2019), at this juncture, we asked if WhiB3 overexpression in BDQ-treated wild-type M. smegmatis also accumulates TAG, thereby dissipating the reductive stress and utilizing TAG as an energy source for the cell survival. Our results clearly show that BDQ-treated wild-type M. smegmatis also overexpresses TAG biosynthesis pathway genes (glpK, GPAT, AGPAT, WSDGAT1, WSDGAT2, MSMEG_3948, and MSMEG_5242) similar to the untreated ΔatpD strain (Figure 8B). Additionally, BDQ treatment also results in overexpression of lipase genes (Figure 8C), suggesting the possibility of increased TAG hydrolysis upon treatment with BDQ. These results strongly indicate that upon BDQ treatment, wild-type M. smegmatis stores as well as consumes TAG to meet its energy requirements, which mimics the ΔatpD strain in the absence of BDQ. This observation has a significant impact, since it is widely accepted that M. tuberculosis switches from carbohydrate to host fatty acids in the phagosome, and balances the reductive stress (Singh et al., 2009). Taken together, our data indicate a critical role of redox sensitive WhiB3 in regulating crucial aspects in mycobacterial physiology and survival during BDQ-treatment.
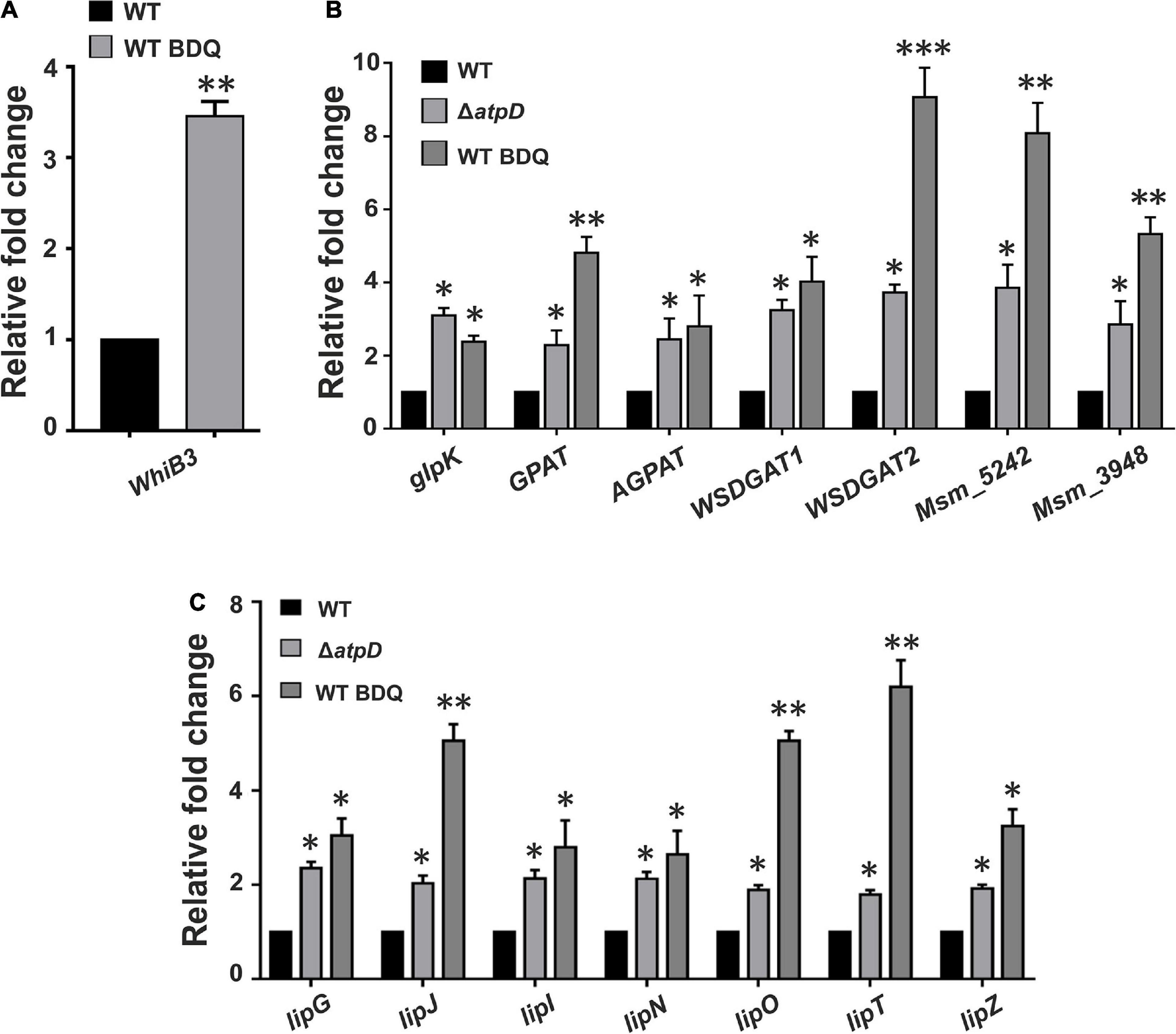
Figure 8. Bedaquiline-treated wild-type M. smegmatis shows increased WhiB3 expression coupled with TAG accumulation and lipase gene upregulation. Panel (A) shows the mRNA transcript level of redox sensor WhiB3 in wild-type M. smegmatis cell either without (WT) or with bedaquiline (BDQ)-treatment (WT BDQ); for comparison with the M. smegmatis atpD knock-out strain, refer to Figure 2A. Panel (B) shows the relative mRNA transcript levels of genes belonging to Kennedy pathway such as glycerol kinase (glpK), glycerol-3 phosphate acyl transferase (GPAT), acylglycerol phosphate acyltransferase (AGPAT), two wax ester synthase/acyl-CoA: diacylglycerol acyltransferase (WSDGAT) and the TAG synthase genes (MSMEG_3948 and MSMEG_5242; these are homologues of tgs1 in M. tuberculosis) in M. smegmatis WT, its atpD knock-out variant (ΔatpD), and the bedaquiline (BDQ)-treated wild-type M. smegmatis (WT BDQ). Panel (C) shows the relative mRNA transcript levels of the lipase genes (lipG, lipJ, lipI, lipN, lipO, lipT, and lipZ) in M. smegmatis WT, atpD knock-out (ΔatpD), and the bedaquiline (BDQ)-treated wild-type M. smegmatis (WT BDQ). In all of the panels, the mRNA transcript in the case of untreated WT is taken as 1 for comparison. In each panel, the data represent an average of at least three independent experiments. Error bars represent standard deviation. ∗P-value < 0.05; ∗∗P-value < 0.01; ∗∗∗P-value < 0.005.
Discussion
Mycobacterium tuberculosis exhibits a remarkable metabolic flexibility and robust ETC to survive under various stress conditions (Iqbal et al., 2018). Thus OXPHOS in mycobacteria is a tightly regulated process. However, the restricted respiration due to O2 depletion or by inhibition of ETC complexes induces changes in the mycobacterial respiratory machinery making the electron carriers to be more reduced thereby leading to alteration in the redox homeostasis (Bhat et al., 2012). Bacteria utilize a large number of dehydrogenases and redox sensors in order to dissipate reductive stress caused by an excessive accumulation of NADH and to replenish the cellular NAD+ levels (Bhat et al., 2012). For example, the Rex repressors in the Gram-positive bacteria are known to regulate the gene expression changes in response to altered NADH/NAD+ ratios (Gyan et al., 2006). The Fnr regulators also sense the intracellular O2 levels via (4Fe-4S) cluster and thereby regulate the cellular response (Kiley and Beinert, 1998). Similarly, the RegB histidine kinase (HK) of the Rhodobacter capsulatus two component system (TCS) RegBA, and the AcrB HK of Escherichia coli TCS AcrBA were also suggested to be upregulated under anaerobic conditions (Bekker et al., 2010). However, homologs of none of these genes are present in M. smegmatis.
The DosSR TCS in mycobacteria consists of two heme-based HK, DosS, and DosT, that directly sense the intracellular concentration of O2 in the cell, and relays the signal to a response regulator DosR, which upregulates the target responsive dormancy regulon genes under reduced oxygen conditions (Kim et al., 2010; Tan et al., 2010). Some major dormancy regulon genes upregulated during reduced oxygen include nitrate reductase (narK2 and narX), fumarate reductase (Frd, frdABCD), ferredoxin (fdxA), DNA repair genes (nrdZ) and TAG biosynthesis genes (tgs1) (Tan et al., 2010). In addition, upon exposure to variety of stress (reduced O2, NO, and CO), the DosSR TCS gets activated leading to an increase in the cellular NADH pools, thus playing an important role during reductive stress in mycobacteria (Kumar et al., 2007; Honaker et al., 2010). Another major redox regulator in mycobacteria is WhiB3 that senses the intracellular redox state, and helps in the dissipation of the reductive stress generated due to accumulated NADH cofactor via lipid biosynthesis (Singh et al., 2009). The metabolic shift induced by WhiB3 regulates the production of methyl branched polyketides (PAT/DAT, SL-1, and PDIM) and TAG (Singh et al., 2009). Synthesis of these lipids by WhiB3 consumes NADH, replenishing the cellular NAD+ levels and relieving the cell from reductive stress (Singh et al., 2009). Interestingly, the TAG synthase tgs1 gene is under the direct control of DosSR operon (Kumari et al., 2017). Lipid inclusions are highly prevalent amongst various prokaryotic organisms such as Rhodococcus, Streptomyces, Nocardia, and Pseudomonas aeruginosa and in eukaryotic organisms like Saccharomyces cerevisiae. In mycobacteria, TAG accumulation has recently received renewed interest. The W-Beijing lineage of M. tuberculosis also shows an upregulation of DosR/S operon with an overproduction of TAG levels. LB containing TAG are the most prominent storage fuel in mycobacteria that not only are essential for prolonged survival, infection, dormancy, and persistence, but are also required for the reactivation of growth following dormancy (Daniel et al., 2011). Similarly, the sputum analysis of pulmonary TB patients and MDR strains of M. tuberculosis also show the presence of TAG containing LB, suggesting importance of TAG accumulation in TB pathogenesis and infection (Vijay et al., 2017). Hence TAG acts as a major redox sink to dissipate reductive stress in the cell.
In the present work, we report a critical outcome of the deletion of one of the two copies of atpD gene in M. smegmatis. We have previously shown that deletion of one copy of atpD gene in M. smegmatis renders the cell in an energy-compromised state (Patil and Jain, 2019). Here we investigated the adaptive mechanisms undertaken by the bacterium during such condition. We show that M. smegmatis under energy compromised state exhibits restricted respiration by downregulating the energy efficient ETC complexes viz. NDH1 and cyt bc1-aa3 complex and upregulating energetically less efficient ETC complexes viz. NDH2 and cyt-bd oxidase complex. This broad shift in the respiratory machinery results in the trapping of protons in the cytoplasmic region, resulting in the acidification of cellular pH. This change alters the membrane polarization as seen by an increase in DiBAC4(3) fluorescence in the knockout cells as compared to the wild-type. Moreover, a higher NADH/NAD+ ratio and an increased ROS levels induces an altered redox state. Finally, in order to dissipate these reducing equivalents and to maintain intracellular redox balance, WhiB3, a major redox sensor, is upregulated and it maintains the redox homeostasis via modulating the biosynthesis of cell wall-associated lipids and TAG resulting in an accumulation of LBs. Our results show that ΔatpD bacterium tends to accumulate more LBs than the wild-type bacterium, indicating that TAG accumulation is a prominent metabolic adaptation in ΔatpD under energy-compromised state. Additionally, by using 14C oleic acid, we further show that the FFAs incorporated within the knockout cells are mainly stored as TAG rather that simple FFA or phospholipids. Furthermore, the atpD bacterium also shows loss of acid fastness and significantly down-regulated cardiolipin biosynthesis gene expression which is most likely the result of an upregulated TAG biosynthesis pathway in ΔatpD. With an upregulation of TAG pathway, we also observed a concomitant overexpression of intracellular lipases genes, suggesting that the accumulated TAG is hydrolyzed for its utilization. Furthermore, an overproduction of lipases along with the previously reported enhanced β-oxidation (Patil and Jain, 2019) together indicates that the stored lipids are used for energy production (Figure 9). This likely happens because the ΔatpD bacterium is unable to meet its energy requirements by OXPHOS due to lesser availability of functional ATP synthase. It is worth adding here that the catabolism of the fatty acids under such energy-depleted state leads to the production of acetyl CoA, which is then metabolized via glyoxylate shunt in order to avoid carbon loss via TCA oxidation. Hence the lipids are acted upon by the lipases to release FFAs, which are then metabolized via β-oxidation generating a pool of acetyl CoA, which thereafter enters the glyoxylate cycle producing energy for cell survival (Patil and Jain, 2019).
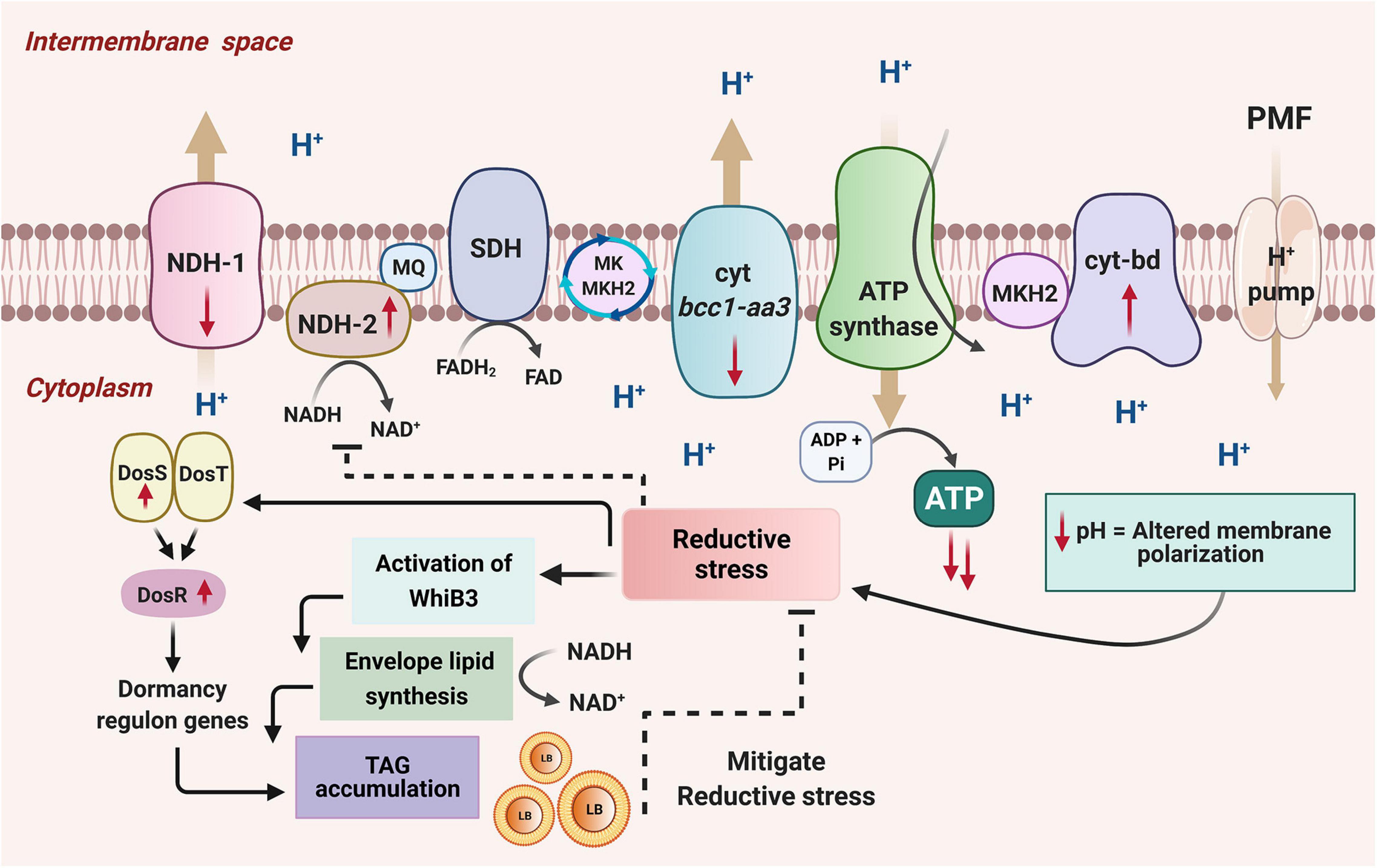
Figure 9. A model depicting the adaptation of M. smegmatis under energy-compromised state. Disruption of one copy of atpD in M. smegmatis drives the transcriptional changes in the mycobacterial ETC. The energy efficient complexes (NDH1 and cyt bc1-aa3) are down-regulated while the energy less efficient complexes (NDH2 and cyt bd oxidase) are upregulated as shown by the direction of the red arrows. This leads to the remodeling of the respiration chain, and, as a result, the protons get trapped in the cytoplasmic region as they are now unable to translocate to the periplasmic region, resulting in the decrease in cellular pH. This causes a change in the membrane polarization, and leads to the reductive stress in the cell. This activates a redox sensor, WhiB3, which dissipates the reductive stress by promoting biosynthesis of cell wall associated lipids and by increasing TAG accumulation. WhiB3-mediated lipid biosynthesis helps in the regeneration of NAD+ from NADH, which helps in mitigating the reductive stress. The accumulated TAG forms lipid bodies (LB). Mycobacteria also possess another sensing mechanism DosSRT dormancy regulon, which detects changes in cellular redox state through the heme proteins of its two sensor kinases, DosS and DosT. This transfers the signal to the response regulator DosR, which mediates the upregulation of dormancy regulon genes. An essential gene of this regulon that gets activated during stress is the tgs1, which is the major TAG regulator gene, mediating production and accumulation of lipid bodies (LB) containing TAG, thereby dissipating the reducing equivalents and facilitating mycobacterial survival under energy-compromised state. Hence these combined effects of WhiB3 and Dos regulon mediated signaling pathways function to overcome the energy demand and maintain the redox homeostasis in M. smegmatis under energy-compromised state. Figure is created with www.BioRender.com.
Interestingly, wild-type M. smegmatis upon BDQ treatment also shows an upregulation in WhiB3 expression, suggesting an important role of WhiB3 mediated redox sensing in mycobacterial virulence under BDQ-treated conditions. Additionally, a predominance of TAG biosynthesis coupled with an increased lipase production is also observed similar to ΔatpD. This phenomenon appears to be conserved in M. tuberculosis; indeed, proteomic analysis of M. tuberculosis treated with BDQ showed an upregulation of TAG biosynthesis gene products at 6 and 24 h time (Koul et al., 2014). This strongly indicates that the survival response of bacterium to BDQ treatment is identical to what we observe upon creating an energy-compromised state by disrupting ATP synthase machinery.
The observed physiological changes presented in this manuscript depict a significant outcome of the metabolic dependency of mycobacteria on fatty acids as an energy source under energy-deprived state and a highly evolved mechanism of dissipating reducing equivalents in the cell via TAG metabolism. We thus conclude that mycobacteria exhibit a robust and flexible system that allows rerouting of its central metabolic pathways under an energy-compromised state. Further, since TAG accumulation provides metabolic energy to the dormant cell, we believe that our work will also contributes toward the exploration of newer possible drug targets in eliminating the dormant and persistent pathogenic mycobacteria.
Experimental Procedures
Bacterial Strains, Plasmids, Reagents, Media, and Growth Conditions
Wild-type M. smegmatis mc2155, atpD knock-out (ΔatpD), and the complemented strain (ΔatpDC) were grown in Middlebrook 7H9 (MB7H9 broth; Difco) containing 2% glucose and 0.05% tyloxapol surfactant (Sigma-Aldrich, MO, United States) as indicated, at 37°C with constant shaking at 200 rpm. M. smegmatisΔatpD and ΔatpDC were taken from the laboratory stock (Patil and Jain, 2019). The cultures were supplemented with 50 μg ml–1 kanamycin (Sigma) or 100 μg ml–1 hygromycin (MP Biomedical), wherever required. Protein expression was induced by the addition of 2% acetamide (wherever required).
Lipid Extraction and Analysis by Thin Layer Chromatography
Triacylglycerol was extracted following the chloroform:methanol (2:1 v/v) method. Briefly, the bacteria were grown in MB7H9 medium supplemented with 2% glucose and 0.05% tyloxapol till 24 h. The cells were then lyophilized and 300 mg of cells in each case were further used for TAG extraction by chloroform:methanol (2:1 v/v). The lower organic layer was collected, and the lipids were dried and re-suspended in chloroform:methanol (1:1 v/v). Lipids were further separated by TLC using hexane:diethylether:formic acid (40:10:1 v/v) as the mobile phase. The plates were allowed to dry, and stained using 5% phosphomolybdic acid in 95% ethanol followed by charring at 140°C for 5 min.
For TAG hydrolysis experiments, bacteria were grown in MB7H9 medium supplemented with 2% glucose and 0.05% tyloxapol for 24 h. The cells were then harvested by centrifugation at 5000 rpm for 15 min and washed with 1X-PBS buffer. The cells were further resuspended in the media without any carbon source in order to induce starvation. The cells were then incubated at 37°C with constant shaking at 200 rpm, and samples were collected at specific time points. For lipase inhibition experiments, the cells were pre-incubated with lipase inhibitor THL for 2 h. A portion of these cells was then harvested and washed with 1X-PBS buffer, while the remaining cells were further incubated for 24 h, with appropriate THL concentrations (5, 50, and 100 μg/ml−1). Cells were then harvested at specific time points and their lipid content was analyzed by TLC. Densitometric analysis was performed using ImageJ software to determine the relative TAG content in the samples.
Ultrastructure Analysis by Transmission Electron Microscopy
Transmission electron microscopy was carried out with M. smegmatis cells essentially as described previously (Gupta et al., 2015; Patil and Jain, 2019) with some modifications. Briefly, the bacterial cell pellet was obtained from 24 h grown culture. The cells were fixed in 0.15 M sodium cacodylate buffer containing 2% osmium tetraoxide for 2 h at RT. The cells were then washed with cacodylate buffer followed by fixation for 3 h at RT in 0.15 M sodium cacodylate buffer containing 2% tannic acid and 2% glutaraldehyde solution. The cells were then washed again with cacodylate buffer, and re-fixed with 2% osmium tetraoxide overnight. Samples were then washed with Milli-Q water and embedded in 2% molten agarose. The finely cut agar block pieces were then subjected to sequential acetone dehydration at increasing concentrations for 30 min each (30, 50, 70, 90, and 100%), followed by infiltration and embedding in Spurr’s resin. After ultra-sectioning, the samples were stained with uranyl acetate and phosphotungstic acid, imaged on FEI Talos 200S system, equipped with a 200 kV Field Emission Gun, and the LBs were counted manually.
Auramine-O and Nile Red Microscopy and Fluorescence Measurements
For fluorescence microscopy, the fluorescent acid-fast staining dye Auramine-O was used along with a neutral lipid staining dye Nile red (9-diethylamino-5H-benzo-a-phenoxazine-5-one) using the protocol as described previously (Deb et al., 2009) with some modifications. Briefly, the log phase cultures (OD600 ∼0.6) of wild-type M. smegmatis mc2155, ΔatpD, and ΔatpDC were harvested and washed with 1X-PBST (0.137 M NaCl, 0.0027 M KCl, 0.01 M Na2HPO4, 0.0018 M KH2PO4, 0.05% Tween-20). Thereafter, the cultures were evenly spread to make a smear on a glass slide, heat fixed, covered with Auramine-O (10 mg ml–1), and incubated for 20 min. The cells were then washed briefly with water and treated with the decolorizer (acid alcohol) for 30 s. Further, the specimen was washed and covered with Nile red solution (10 mg ml–1 in ethanol) and incubated for 15 min. The cells were then washed and covered with potassium permanganate solution for 1 min, washed thoroughly with water, air dried, and imaged on using ApoTome Fluorescence Microscope (Carl Zeiss) using an 100× objective lens.
For Nile red confocal microscopy experiments, the ΔatpD cells were grown for 24 h. The cells were then harvested and washed with 1X-PBS buffer and further resuspended in it. Nile red dye (10 mg/ml in ethanol) was added to the cell suspension. Cells were further incubated for 30 min in dark at 37°C. The stained cells were then harvested and washed twice with 1X-PBST buffer and finally resuspended in it. A total of 2 μl of bacterial suspension was spotted on 1% agarose pad. Bacteria were then analyzed by capturing the images on Olympus FV3000 confocal microscope with 100× oil-objective.
The flow cytometry based FACS measurements were performed using BD-FACS Aria III equipped with CELL QUEST software (South San Francisco, CA, United States). For measuring the Auramine-O and Nile red fluorescence, log phase cultures of all the strains were harvested, and washed with 1X-PBST. Both Auramine-O and Nile red dyes were added to the samples at a final concentration of 10 mg/ml. Samples were incubated at room temperature in dark for 1 h, washed twice and re-suspended in 1X-PBST buffer. Flow cytometry was performed using FITC and Texas Red filter for Auramine and Nile red, respectively. Alternatively, fluorescence was measured on SpectraMax M5 plate reader with Ex/Em wavelengths set at 438/505 nm for Auramine-O and at 550/640 nm for Nile red.
Assessment of the Presence of Cardiolipin by Staining With Nonyl Acridine Orange
The flow cytometry based FACS measurements were performed using BD-FACS Aria III equipped with cell quest software (South San Francisco, CA, United States). The total cardiolipin levels were measured using NAO (10-Nonyl Acridine orange). Briefly log phase cultures (OD600 ∼0.6) of all the strains were harvested, and washed with 1X-PBST. The cells were then incubated with 10 μM NAO for 30 min at room temperature, washed twice and re-suspended in 1X-PBST buffer. Fluorescence was then measured using FITC filter for flow cytometry, and, alternatively, on Spectramax M5 plate reader with Ex/Em wavelengths set at 495/519 nm.
Metabolic Incorporation of Radiolabeled Oleic Acid by Mycobacterium smegmatis
Wild-type M. smegmatis and its variants were grown in MB7H9 medium supplemented with 2% glucose and 0.05% tyloxapol till log phase (OD600 ∼0.6) at 37°C with constant shaking at 200 rpm. 14C-oleate (50 μCi mol–1; 5 μCi per 10 ml of culture; Perkin Elmer, Waltham, MA, United States) was then added in each culture for 30 min. Thereafter, the lipid extraction, TLC, and phosphor imaging procedures were performed as described previously with some modifications (Daniel et al., 2016). Briefly, the cells were harvested by centrifugation at 8000 rpm at 4°C and were washed with 1X-PBST buffer to remove the unincorporated radioactive material. The lipids were then extracted using chloroform:methanol (2:1 v/v). The organic layer was recovered and dried completely. The dried lipids were further dissolved in chloroform:methanol (1:1 v/v) and were subjected to TLC using hexane:ethyl ether:formic acid (40:10:1 v/v) as the mobile phase. For each sample, the amount of radioactivity present was measured on a scintillation counter, and the equal amount of radioactivity was loaded on TLC. The TLC plate was then dried and imaged on a phosphor imager (FLA 9000; GE Healthcare). For phospholipid analysis, 14C-oleate (54.3 μCi mmol–1; 5 μCi per 10 ml of culture) was added in each culture for 6 h, followed by lipid extraction as stated above. Dried lipids were further dissolved in chloroform:methanol (1:1 v/v). Equal cpm was spotted on TLC, and the lipids were separated using CHCl3:CH3OH:H2O (65:25:4 v/v) as the mobile phase, followed by phosphor imaging procedure as mentioned above.
Lipase Assay
Lipase activity was assessed using Lipase Assay Kit (Sigma Aldrich, MAK048) following the manufacturer’s instructions with some modifications. Briefly, the log phase cultures were harvested and washed with 1X-PBST. The cells were then resuspended in lipase buffer followed by the addition of lipase inhibitor (tetrahydrolipstatin or THL), wherever required, to a final concentration of 80 μM. The cells were then sonicated and were further processed for lipase assay. Fluorescence was recorded on SpectraMax M5 plate reader with Ex/Em wavelengths set at 529/600 nm.
RNA Extraction, cDNA Synthesis, and RT-qPCR
Differential gene expression profile was obtained by RT-qPCR for bacteria essentially as described previously (Patil and Jain, 2019). Briefly, the cells were grown till OD600 ∼ 0.8. The cells were then harvested, and processed for RNA isolation using RNeasy Mini kit (QIAGEN) by following manufacturer’s instructions. cDNA was synthesized using i-script cDNA synthesis kit (Bio-Rad) as per the manufacturer’s instructions. The obtained cDNA was then used for qPCR experiments, carried out on StepOnePlus real-time PCR system (Applied Biosystems) using iTaq universal SYBR green mix (Bio-Rad) as per the manufacturer’s instructions. rpoB gene was used as an internal control. Data were analyzed as per 2–ΔΔCt method. Primers used for these PCR reactions are listed in Supplementary Table 1.
Statistical Analysis
Data are represented as arithmetic means in the results that are obtained from at least three independent experiments with standard deviations shown as error bar. Statistical significance was calculated using unpaired t-test for data analysis as mentioned in the respective figure legends.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
VJ and VP designed the research, analyzed the data, and wrote the manuscript. VP performed the research. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by intramural funds from IISER Bhopal to VJ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
VP acknowledges the receipt of DST INSPIRE-Senior Research Fellowship from the Department of Science and Technology (DST), Government of India. We thank DST-FIST facility at IISER Bhopal for the Confocal and TEM imaging.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.722229/full#supplementary-material
Supplementary Figure 1 | Pairwise sequence alignment of WhiB3 protein of M. tuberculosis and M. smegmatis. Pairwise sequence alignment of WhiB3 protein from M. tuberculosis (Rv3416) and MSMEG_1597 from M. smegmatis is shown. “∗” represents conserved residue.
Supplementary Figure 2 | Sequence alignment of polyketide biosynthesis protein of M. tuberculosis and M. smegmatis. Pairwise sequence alignment of Rv3825c protein (pks2) from M. tuberculosis and MSMEG_4724 (pks5) from M. smegmatis is shown. “∗” represents conserved residue.
Supplementary Figure 3 | Phospholipid analysis by thin layer chromatography. Panel shows the TLC image depicting the phospholipids in M. smegmatis wild-type (WT), atpD knock out (ΔatpD), and atpD-complemented (ΔatpDC) strains. Spot corresponding to PE, PI, and PS (top to bottom) is marked with arrowhead. Experiments were repeated at least thrice; only one representative image is shown.
Supplementary Figure 4 | Sequence alignment of TAG synthase enzymes of M. tuberculosis and M. smegmatis. Multiple sequence alignment of Rv3130c protein (tgs1) from M. tuberculosis and its homologs MSMEG_3948 and MSMEG_5242 from M. smegmatis is shown. “∗” represents conserved residue in all three cases.
Supplementary Figure 5 | TAG accumulation examination in M. smegmatis by thin layer chromatography. Panel shows the autoradiogram of the TLC demonstrating the incorporation of radiolabeled 14C-oleic acid in TAG in wild-type (WT), atpD knock-out (ΔatpD), and atpD-complemented (ΔatpDC) strains of M. smegmatis. Arrowhead points to the various species obtained on TLC such as triacylglycerol (TAG), diacylglycerol (DAG), and monoacylglycerol (MAG). Lane 4 has the free 14C-oleic acid. Experiments were repeated at least thrice; only one representative image is shown.
Supplementary Table 1 | List of primers used in this study. Primers and their sequences used in the RT-qPCR experiments, and the genes that they target are given.
References
Akela, A. K., and Kumar, A. (2021). Bioenergetic heterogeneity in Mycobacterium tuberculosis residing in different subcellular niches. mbio 12:e0108821.
Bald, D., Villellas, C., Lu, P., and Koul, A. (2017). Targeting energy metabolism in mycobacterium tuberculosis, a new paradigm in antimycobacterial drug discovery. mBio 8:e00272-17.
Bekker, M., Alexeeva, S., Laan, W., Sawers, G., Teixeira de Mattos, J., and Hellingwerf, K. (2010). The ArcBA two-component system of Escherichia coli is regulated by the redox state of both the ubiquinone and the menaquinone pool. J. Bacteriol. 192, 746–754. doi: 10.1128/jb.01156-09
Berney, M., and Cook, G. M. (2010). Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS One 5:e8614. doi: 10.1371/journal.pone.0008614
Bhat, S. A., Singh, N., Trivedi, A., Kansal, P., Gupta, P., and Kumar, A. (2012). The mechanism of redox sensing in Mycobacterium tuberculosis. Free Radic. Biol. Med. 53, 1625–1641. doi: 10.1016/j.freeradbiomed.2012.08.008
Blee, J. A., Roberts, I. S., and Waigh, T. A. (2020). Membrane potentials, oxidative stress and the dispersal response of bacterial biofilms to 405 nm light. Phys. Biol. 17:036001. doi: 10.1088/1478-3975/ab759a
Brunelle, J. K., Bell, E. L., Quesada, N. M., Vercauteren, K., Tiranti, V., Zeviani, M., et al. (2005). Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 1, 409–414. doi: 10.1016/j.cmet.2005.05.002
Cook, G. M., Hards, K., Vilchèze, C., Hartman, T., and Berney, M. (2014). Energetics of respiration and oxidative phosphorylation in mycobacteria. Microbiol. Spectr. 2:10.1128/microbiolspec.MGM2-0015-2013.
Crellin, P. K., Luo, C. Y., and Morita, Y. S. (2013). Metabolism of plasma membrane lipids in mycobacteria and corynebacteria, lipid metabolism, rodrigo valenzuela baez. IntechOpen 6, 119–148.
Daniel, J., Deb, C., Dubey, V. S., Sirakova, T. D., Abomoelak, B., Morbidoni, H. R., et al. (2004). Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J. Bacteriol. 186, 5017–5030. doi: 10.1128/jb.186.15.5017-5030.2004
Daniel, J., Kapoor, N., Sirakova, T., Sinha, R., and Kolattukudy, P. (2016). The perilipin-like PPE15 protein in Mycobacterium tuberculosis is required for triacylglycerol accumulation under dormancy-inducing conditions. Mol. Microbiol. 101, 784–794. doi: 10.1111/mmi.13422
Daniel, J., Maamar, H., Deb, C., Sirakova, T. D., and Kolattukudy, P. E. (2011). Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 7:e1002093. doi: 10.1371/journal.ppat.1002093
Deb, C., Lee, C. M., Dubey, V. S., Daniel, J., Abomoelak, B., Sirakova, T. D., et al. (2009). A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One 4:e6077. doi: 10.1371/journal.pone.0006077
Dwyer, D. J., Belenky, P. A., Yang, J. H., MacDonald, I. C., Martell, J. D., Takahashi, N., et al. (2014). Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U.S.A. 111, E2100–E2109.
Eisinger, R. W., Embry, A. C., Read, S. W., and Fauci, A. S. (2020). 2019: a banner year for tuberculosis research. J. Infect. Dis. 222, 1768–1771. doi: 10.1093/infdis/jiaa051
Foo, C.-Y., Pethe, K., and Lupien, A. (2020). Oxidative phosphorylation—an update on a new, essential target space for drug discovery in Mycobacterium tuberculosis. Appl. Sci. 10:2339. doi: 10.3390/app10072339
Forte, E., Borisov, V. B., Falabella, M., Colaco, H. G., Tinajero-Trejo, M., Poole, R. K., et al. (2016). The terminal oxidase cytochrome bd promotes sulfide-resistant bacterial respiration and growth. Sci. Rep. 6:23788.
Gengenbacher, M., and Kaufmann, S. H. (2012). Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 36, 514–532. doi: 10.1111/j.1574-6976.2012.00331.x
Giuffre, A., Borisov, V. B., Mastronicola, D., Sarti, P., and Forte, E. (2012). Cytochrome bd oxidase and nitric oxide: from reaction mechanisms to bacterial physiology. FEBS Lett. 586, 622–629. doi: 10.1016/j.febslet.2011.07.035
Gomez, J. E., and McKinney, J. D. (2004). M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb.) 84, 29–44. doi: 10.1016/j.tube.2003.08.003
Gupta, K. R., Kasetty, S., and Chatterji, D. (2015). Novel functions of (p)ppGpp and Cyclic di-GMP in mycobacterial physiology revealed by phenotype microarray analysis of wild-type and isogenic strains of Mycobacterium smegmatis. Appl. Environ. Microbiol. 81, 2571–2578. doi: 10.1128/aem.03999-14
Gyan, S., Shiohira, Y., Sato, I., Takeuchi, M., and Sato, T. (2006). Regulatory loop between redox sensing of the NADH/NAD(+) ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. J. Bacteriol. 188, 7062–7071. doi: 10.1128/jb.00601-06
Honaker, R. W., Dhiman, R. K., Narayanasamy, P., Crick, D. C., and Voskuil, M. I. (2010). DosS responds to a reduced electron transport system to induce the Mycobacterium tuberculosis DosR regulon. J. Bacteriol. 192, 6447–6455. doi: 10.1128/jb.00978-10
Iqbal, I. K., Bajeli, S., Akela, A. K., and Kumar, A. (2018). Bioenergetics of Mycobacterium: An Emerging Landscape for Drug Discovery. Pathogens 7:24. doi: 10.3390/pathogens7010024
Kana, B. D., Weinstein, E. A., Avarbock, D., Dawes, S. S., Rubin, H., and Mizrahi, V. (2001). Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J. Bacteriol. 183, 7076–7086. doi: 10.1128/jb.183.24.7076-7086.2001
Kiley, P. J., and Beinert, H. (1998). Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22, 341–352. doi: 10.1111/j.1574-6976.1998.tb00375.x
Kim, M. J., Park, K. J., Ko, I. J., Kim, Y. M., and Oh, J. I. (2010). Different roles of DosS and DosT in the hypoxic adaptation of mycobacteria. J. Bacteriol. 192, 4868–4875. doi: 10.1128/jb.00550-10
Koul, A., Vranckx, L., Dhar, N., Göhlmann, H. W. H., Özdemir, E., Neefs, J.-M., et al. (2014). Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat. Commun. 5, 3369–3369.
Kumar, A., Toledo, J. C., Patel, R. P., Lancaster, J. R. Jr., and Steyn, A. J. (2007). Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. U.S.A. 104, 11568–11573. doi: 10.1073/pnas.0705054104
Kumari, P., Sikri, K., Kaur, K., Gupta, U. D., and Tyagi, J. S. (2017). Sustained expression of DevR/DosR during long-term hypoxic culture of Mycobacterium tuberculosis. Tuberculosis (Edinb.) 106, 33–37. doi: 10.1016/j.tube.2017.06.003
Lakshmanan, M., and Xavier, A. S. (2013). Bedaquiline–the first ATP synthase inhibitor against multi drug resistant tuberculosis. J. Young Pharm. 5, 112–115. doi: 10.1016/j.jyp.2013.12.002
Lamprecht, D. A., Finin, P. M., Rahman, M. A., Cumming, B. M., Russell, S. L., Jonnala, S. R., et al. (2016). Turning the respiratory flexibility of Mycobacterium tuberculosis against itself. Nat. Commun. 7:12393.
Lass, A., Zimmermann, R., Oberer, M., and Zechner, R. (2011). Lipolysis–a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog. Lipid Res. 50, 14–27. doi: 10.1016/j.plipres.2010.10.004
Maarsingh, J. D., and Haydel, S. E. (2018). Mycobacterium smegmatis PrrAB two-component system influences triacylglycerol accumulation during ammonium stress. Microbiology 164, 1276–1288. doi: 10.1099/mic.0.000705
Matsoso, L. G., Kana, B. D., Crellin, P. K., Lea-Smith, D. J., Pelosi, A., Powell, D., et al. (2005). Function of the cytochrome bc1-aa3 branch of the respiratory network in mycobacteria and network adaptation occurring in response to its disruption. J. Bacteriol. 187, 6300–6308. doi: 10.1128/jb.187.18.6300-6308.2005
McCarthy, C. (1971). Utilization of palmitic acid by Mycobacterium avium. Infect. Immun. 4, 199–204. doi: 10.1128/iai.4.3.199-204.1971
Mehta, M., and Singh, A. (2019). Mycobacterium tuberculosis WhiB3 maintains redox homeostasis and survival in response to reactive oxygen and nitrogen species. Free Radic. Biol. Med. 131, 50–58. doi: 10.1016/j.freeradbiomed.2018.11.032
Mileykovskaya, E., Dowhan, W., Birke, R. L., Zheng, D., Lutterodt, L., and Haines, T. H. (2001). Cardiolipin binds nonyl acridine orange by aggregating the dye at exposed hydrophobic domains on bilayer surfaces. FEBS Lett. 507, 187–190. doi: 10.1016/s0014-5793(01)02948-9
Moosa, A., Lamprecht, D. A., Arora, K., Barry, C. E. III, Boshoff, H. I. M., Ioerger, T. R., et al. (2017). Susceptibility of Mycobacterium tuberculosis cytochrome bd oxidase mutants to compounds targeting the terminal respiratory oxidase, cytochrome c. Antimicrob. Agents Chemother. 61:e01338-17.
Nakagawa, H., Kashiwabara, Y., and Matsuki, G. (1976). Metabolism of triacylglycerol in Mycobacterium smegmatis. J. Biochem. 80, 923–928. doi: 10.1093/oxfordjournals.jbchem.a131378
Ojha, A. K., Trivelli, X., Guerardel, Y., Kremer, L., and Hatfull, G. F. (2010). Enzymatic hydrolysis of trehalose dimycolate releases free mycolic acids during mycobacterial growth in biofilms. J. Biol. Chem. 285, 17380–17389. doi: 10.1074/jbc.m110.112813
Onwueme, K. C., Ferreras, J. A., Buglino, J., Lima, C. D., and Quadri, L. E. (2004). Mycobacterial polyketide-associated proteins are acyltransferases: proof of principle with Mycobacterium tuberculosis PapA5. Proc. Natl. Acad. Sci. U.S.A. 101, 4608–4613. doi: 10.1073/pnas.0306928101
Pandey, A. K., and Sassetti, C. M. (2008). Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380. doi: 10.1073/pnas.0711159105
Patil, V., and Jain, V. (2019). Insights into the physiology and metabolism of a mycobacterial cell in an energy-compromised state. J. Bacteriol. 201:e00210-19.
Prindle, A., Liu, J., Asally, M., Ly, S., Garcia-Ojalvo, J., and Süel, G. M. (2015). Ion channels enable electrical communication in bacterial communities. Nature 527, 59–63. doi: 10.1038/nature15709
Queiroz, A., and Riley, L. W. (2017). Bacterial immunostat: Mycobacterium tuberculosis lipids and their role in the host immune response. Rev. Soc. Bras. Med. Trop. 50, 9–18. doi: 10.1590/0037-8682-0230-2016
Ravindran, M. S., Rao, S. P., Cheng, X., Shukla, A., Cazenave-Gassiot, A., Yao, S. Q., et al. (2014). Targeting lipid esterases in mycobacteria grown under different physiological conditions using activity-based profiling with tetrahydrolipstatin (THL). Mol. Cell Proteomics 13, 435–448. doi: 10.1074/mcp.m113.029942
Serafini, A., Tan, L., Horswell, S., Howell, S., Greenwood, D. J., Hunt, D. M., et al. (2019). Mycobacterium tuberculosis requires glyoxylate shunt and reverse methylcitrate cycle for lactate and pyruvate metabolism. Mol. Microbiol. 112, 1284–1307. doi: 10.1111/mmi.14362
Shi, L., Sohaskey, C. D., Kana, B. D., Dawes, S., North, R. J., Mizrahi, V., et al. (2005). Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc. Natl. Acad. Sci. U.S.A. 102, 15629–15634. doi: 10.1073/pnas.0507850102
Shi, L., Sohaskey, C. D., Pheiffer, C., Datta, P., Parks, M., McFadden, J., et al. (2010). Carbon flux rerouting during Mycobacterium tuberculosis growth arrest. Mol. Microbiol. 78, 1199–1215. doi: 10.1111/j.1365-2958.2010.07399.x
Singh, A., Crossman, D. K., Mai, D., Guidry, L., Voskuil, M. I., Renfrow, M. B., et al. (2009). Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 5:e1000545. doi: 10.1371/journal.ppat.1000545
Smith, T. C. II, Pullen, K. M., Olson, M. C., McNellis, M. E., Richardson, I., Hu, S., et al. (2020). Morphological profiling of tubercle bacilli identifies drug pathways of action. Proc. Natl. Acad. Sci. U.S.A. 117, 18744–18753. doi: 10.1073/pnas.2002738117
Stallings, C. L., and Glickman, M. S. (2010). Is Mycobacterium tuberculosis stressed out? A critical assessment of the genetic evidence. Microbes Infect. 12, 1091–1101. doi: 10.1016/j.micinf.2010.07.014
Tan, M. P., Sequeira, P., Lin, W. W., Phong, W. Y., Cliff, P., Ng, S. H., et al. (2010). Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS One 5:e13356. doi: 10.1371/journal.pone.0013356
Tran, T., Bonham, A. J., Chan, E. D., and Honda, J. R. (2019). A paucity of knowledge regarding nontuberculous mycobacterial lipids compared to the tubercle bacillus. Tuberculosis (Edinb.) 115, 96–107. doi: 10.1016/j.tube.2019.02.008
Vijay, S., Hai, H. T., Thu, D. D. A., Johnson, E., Pielach, A., Phu, N. H., et al. (2017). Ultrastructural analysis of cell envelope and accumulation of lipid inclusions in clinical Mycobacterium tuberculosis isolates from sputum, oxidative stress, and iron deficiency. Front. Microbiol. 8:2681.
Weir, M. P., Langridge, W. H. III, and Walker, R. W. (1972). Relationships between oleic acid uptake and lipid metabolism in Mycobacterium smegmatis. Am. Rev. Respir. Dis. 106, 450–457. doi: 10.1164/arrd.1972.106.3.450
Zechner, R., Zimmermann, R., Eichmann, T. O., Kohlwein, S. D., Haemmerle, G., Lass, A., et al. (2012). FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 15, 279–291. doi: 10.1016/j.cmet.2011.12.018
Zhai, W., Wu, F., Zhang, Y., Fu, Y., and Liu, Z. (2019). The immune escape mechanisms of Mycobacterium tuberculosis. Int. J. Mol. Sci. 20:340. doi: 10.3390/ijms20020340
Keywords: mycobacteria, lipid metabolism, triacylglycerol, ATP synthase, lipid bodies, redox regulation
Citation: Patil V and Jain V (2021) Understanding Metabolic Remodeling in Mycobacterium smegmatis to Overcome Energy Exigency and Reductive Stress Under Energy-Compromised State. Front. Microbiol. 12:722229. doi: 10.3389/fmicb.2021.722229
Received: 08 June 2021; Accepted: 11 August 2021;
Published: 01 September 2021.
Edited by:
Davide Zannoni, University of Bologna, ItalyReviewed by:
Jianping Xie, Southwest University, ChinaHugo Gramajo, CONICET Instituto de Biología Molecular y Celular de Rosario (IBR), Argentina
Copyright © 2021 Patil and Jain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vikas Jain, dmlrYXNAaWlzZXJiLmFjLmlu
 Varsha Patil
Varsha Patil Vikas Jain
Vikas Jain