A Markerless Method for Genome Engineering in Zymomonas mobilis ZM4
- 1DOE Great Lakes Bioenergy Research Center, University of Wisconsin–Madison, Madison, WI, United States
- 2Department of Biomolecular Chemistry, University of Wisconsin–Madison, Madison, WI, United States
- 3College of Biological and Pharmaceutical Sciences, China Three Gorges University, Yichang, China
- 4Department of Biochemistry, University of Wisconsin–Madison, Madison, WI, United States
- 5Cell and Molecular Biology Graduate Training Program, University of Wisconsin-Madison, Madison, WI, United States
- 6Department of Bacteriology, University of Wisconsin-Madison, Madison, WI, United States
A Corrigendum on
A Markerless Method for Genome Engineering in Zymomonas mobilis ZM4
by Lal, P. B., Wells, F. M., Lyu, Y., Ghosh, I. N., Landick, R., and Kiley, P. J. (2019). Front. Microbiol. 10:2216. doi: 10.3389/fmicb.2019.02216
In the original article, there were two mistakes in Figure 3 as published. The X-axis label of Figure 3B should be 103 rather than 102. Figure 3D contained a word inversion in the text “UP-Plasmid-UP-DN-ldh-DN” and should read “UP-DN-Plasmid-UP-ldh-DN.” The corrected Figure 3 appears below.
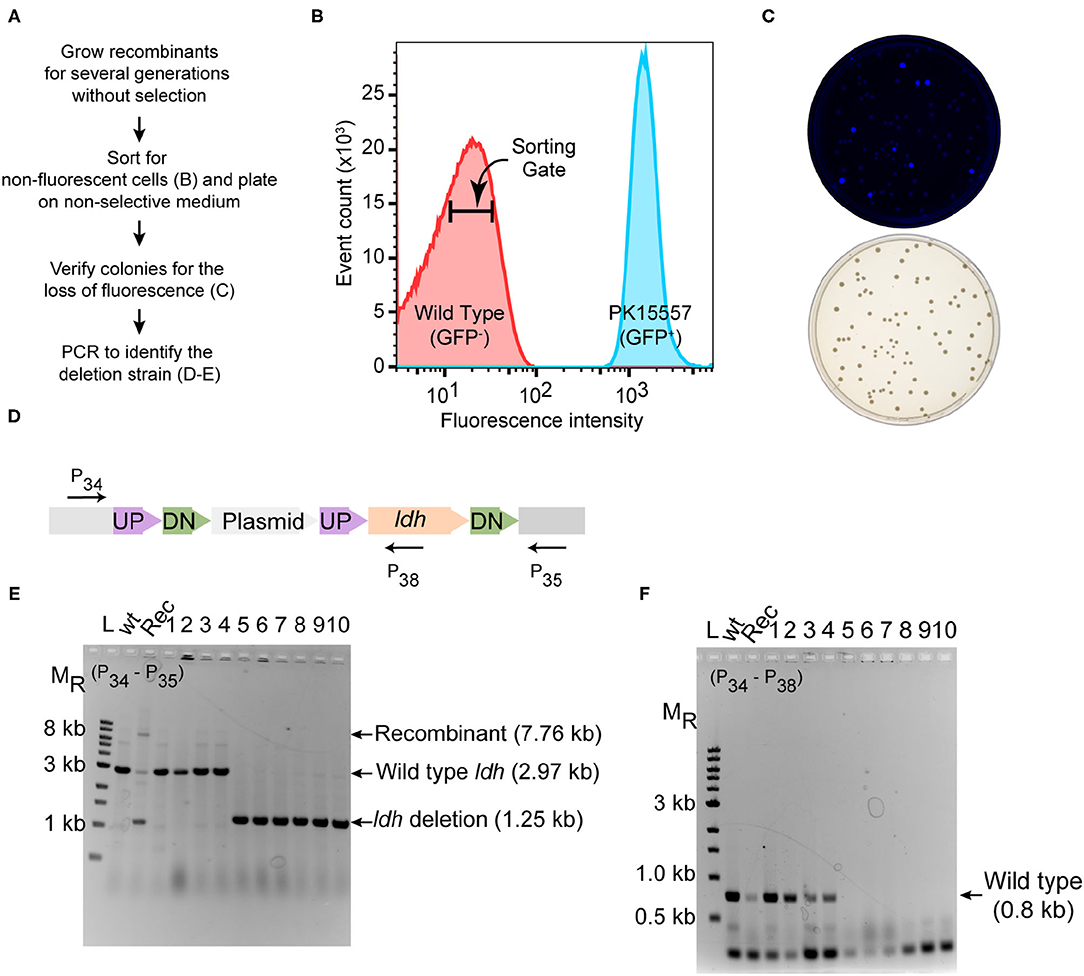
Figure 3. Deletion of ldh gene. (A) Workflow for enrichment of non-fluorescent cells and screening for the modified genome. (B) Plot from FACS of Z. mobilis strain PK15557 that has suicide plasmid (pPK15535) recombined into the genome. The X-axis represents fluorescence intensity and Y-axis represents the total number of sorted events. The sorting gate for collecting the non-fluorescent cells (red peak) is indicated by the bar. (C) Top plate shows the fluorescence image of colonies from FACS sorted non-fluorescent cells plated on non-selective media. Bottom plate shows the same colonies viewed with visible light. (D) The location of primers used to screen non-fluorescent candidates for wild-type (wt) and deletion genotypes by PCR is indicated by arrows. (E,F) Agarose gel electrophoresis of products from PCR amplification of 10 non-fluorescent-candidate colonies. Selected size markers are indicated on the left (MR). (E) Amplification using primers P34 and P35 yielded a 1.25-kb DNA fragment for the ldh deletion whereas the wt ldh DNA fragment was 2.97 kb. Strains that still possess the plasmid integrated into the genome (Rec) are expected to have a band of 7.8 kb. (F) Amplification using P34 and P38 yielded a DNA fragment of 0.8 kb, confirming those strains that still possess the ldh gene, while strains in which ldh has been deleted (six total) lacked the equivalent amplified fragment. Labels are the same as in panel E. Similar results were obtained when this experiment was biologically replicated three times.
The authors apologize for this error and state that this does not change or affect the scientific conclusions of the article in any way. The original article has been updated.
Keywords: genome engineering of a non-model bacterium, green fluorescent protein, fluorescence activated cell sorting, recombineering suicide plasmid, biofuels, Zymomonas mobilis
Citation: Lal PB, Wells FM, Lyu Y, Ghosh IN, Landick R and Kiley PJ (2021) Corrigendum: A Markerless Method for Genome Engineering in Zymomonas mobilis ZM4. Front. Microbiol. 12:719621. doi: 10.3389/fmicb.2021.719621
Received: 02 June 2021; Accepted: 11 June 2021;
Published: 29 June 2021.
Approved by:
Frontiers in Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2021 Lal, Wells, Lyu, Ghosh, Landick and Kiley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patricia J. Kiley, cGpraWxleSYjeDAwMDQwO3dpc2MuZWR1
 Piyush Behari Lal
Piyush Behari Lal Fritz M. Wells1,2
Fritz M. Wells1,2 Patricia J. Kiley
Patricia J. Kiley