- 1Department of Paraclinical Sciences, Faculty of Veterinary Medicine, Norwegian University of Life Sciences, Oslo, Norway
- 2Anses, Ploufragan-Plouzané-Niort Laboratory, Unit of Viral Genetics and Biosafety, Ploufragan, France
- 3M-team and Mastitis and Milk Quality Research Unit, Department of Reproduction, Obstetrics, and Herd Health, Faculty of Veterinary Medicine, Ghent University, Ghent, Belgium
- 4Bacteriology, Department of Infection and Parasitic Diseases, Faculty of Veterinary Medicine, FARAH Research Centre, Liège University, Liège, Belgium
Staphylococci are among the commonly isolated bacteria from intramammary infections in bovines, where Staphylococcus aureus is the most studied species. This species carries a variety of virulence genes, contributing to bacterial survival and spread. Less is known about non-aureus staphylococci (NAS) and their range of virulence genes and mechanisms, but they are the most frequently isolated bacteria from bovine milk. Staphylococci can also carry a range of antimicrobial resistance genes, complicating treatment of the infections they cause. We used Illumina sequencing to whole genome sequence 93 staphylococcal isolates selected from a collection of staphylococcal isolates; 45 S. aureus isolates and 48 NAS isolates from 16 different species, determining their content of antimicrobial resistance genes and virulence genes. Antimicrobial resistance genes were frequently observed in the NAS species as a group compared to S. aureus. However, the lincosamide resistance gene lnuA and penicillin resistance gene blaZ were frequently identified in NAS, as well as a small number of S. aureus. The erm genes conferring macrolide resistance were also identified in several NAS isolates and in a small number of S. aureus isolates. In most S. aureus isolates, no antimicrobial resistance genes were detected, but in five S. aureus isolates three to six resistance genes were identified and all five of these carried the mecA gene. Virulence genes were more frequently identified in S. aureus, which contained on average five times more virulence genes compared to NAS. Among the NAS species there were also differences in content of virulence genes, such as S. chromogenes with a higher average number of virulence genes. By determining the content of a large selection of virulence genes and antimicrobial resistance genes in S. aureus and 16 different NAS species our results contribute with knowledge regarding the genetic basis for virulence and antimicrobial resistance in bovine staphylococci, especially the less studied NAS. The results can create a broader basis for further research into the virulence mechanisms of this important group of bacteria in bovine intramammary infections.
Introduction
The genus Staphylococcus includes a range of different species (De Buck et al., 2021) some of which are among the most commonly isolated bacteria causing intramammary infections in bovines (Pitkälä et al., 2004; Reksen et al., 2006; Olde Riekerink et al., 2008). For a long time, Staphylococcus aureus has been the most recognized staphylococcal species causing both clinical and subclinical mastitis (Østerås et al., 2006; Olde Riekerink et al., 2008). The species is associated with a wide range of genes encoding a large diversity of virulence factors involved in adhesion, host immune evasion and biofilm formation (Foster et al., 2014; Geoghegan and Foster, 2017), toxins promoting inflammation and leukocyte death and exoenzymes cleaving and disabling immune molecules. All these factors contribute to bacterial survival, -spread and nutrient acquisition (Tam and Torres, 2019). However, in recent years, non-aureus staphylococci (NAS) have emerged as the most frequently isolated bacterial group from bovine milk in many countries and they are increasingly associated with intramammary infections (Pyörälä and Taponen, 2009; De Vliegher et al., 2012; Condas et al., 2017; De Buck et al., 2021).
Emergence of antimicrobial resistant staphylococci is of growing concern in the dairy industry. These bacteria can carry a large number of resistance determinants, which are often located on mobile genetic elements that facilitates horizontal spread of genes (Malachowa and DeLeo, 2010; Kadlec et al., 2012). NAS are regarded as a potential reservoir for antimicrobial resistance genes that can be transferred to and utilized by S. aureus (Otto, 2013; Vitali et al., 2014). Many NAS species are found as commensals on teat apices, hair, nares, vagina, teat, and udder skin and inguinal skin, as well as in the environment in the barn (Taponen et al., 2008; Piessens et al., 2011; De Visscher et al., 2014), potentiating possible interactions with a variety of different bacteria present in the host. Due to these commensal properties, NAS may become exposed to several antimicrobials, not only by agents used for battling staphylococcal infections, but also by agents used to combat other pathogens (Kadlec et al., 2012). Several studies have pointed to some NAS species having a relatively greater impact on udder health, especially S. chromogenes, S. simulans and S. xylosus (Supré et al., 2011; Fry et al., 2014; Valckenier et al., 2019). However, contrary to S. aureus, the virulence factors of NAS, and mechanisms behind NAS’ ability to colonize and infect the bovine udder are poorly described (Pyörälä and Taponen, 2009; Vanderhaeghen et al., 2015; Taponen et al., 2017). The extensive virulence gene profiling of 441 NAS isolates published by Naushad et al. (2019), the comparative study of 24 bovine-associated staphylococci of Åvall-Jääskeläinen et al. (2018) and the study on bovine NAS by Wuytack et al. (2020) are the most important published studies that have explored the virulence factors and virulence mechanisms in bovine NAS species, which warrants further investigation of other NAS collections.
The aim of this study was therefore to describe the genetic background for antimicrobial resistance and virulence in staphylococci from dairy cows. The objectives were to (i) whole genome sequence a selection of NAS and S. aureus isolated from bovine milk samples, (ii) determine the isolates’ content of antimicrobial resistance and virulence genes, and (iii) compare the types and variety of virulence genes between NAS species and S. aureus.
Materials and Methods
Collection and Selection of Isolates
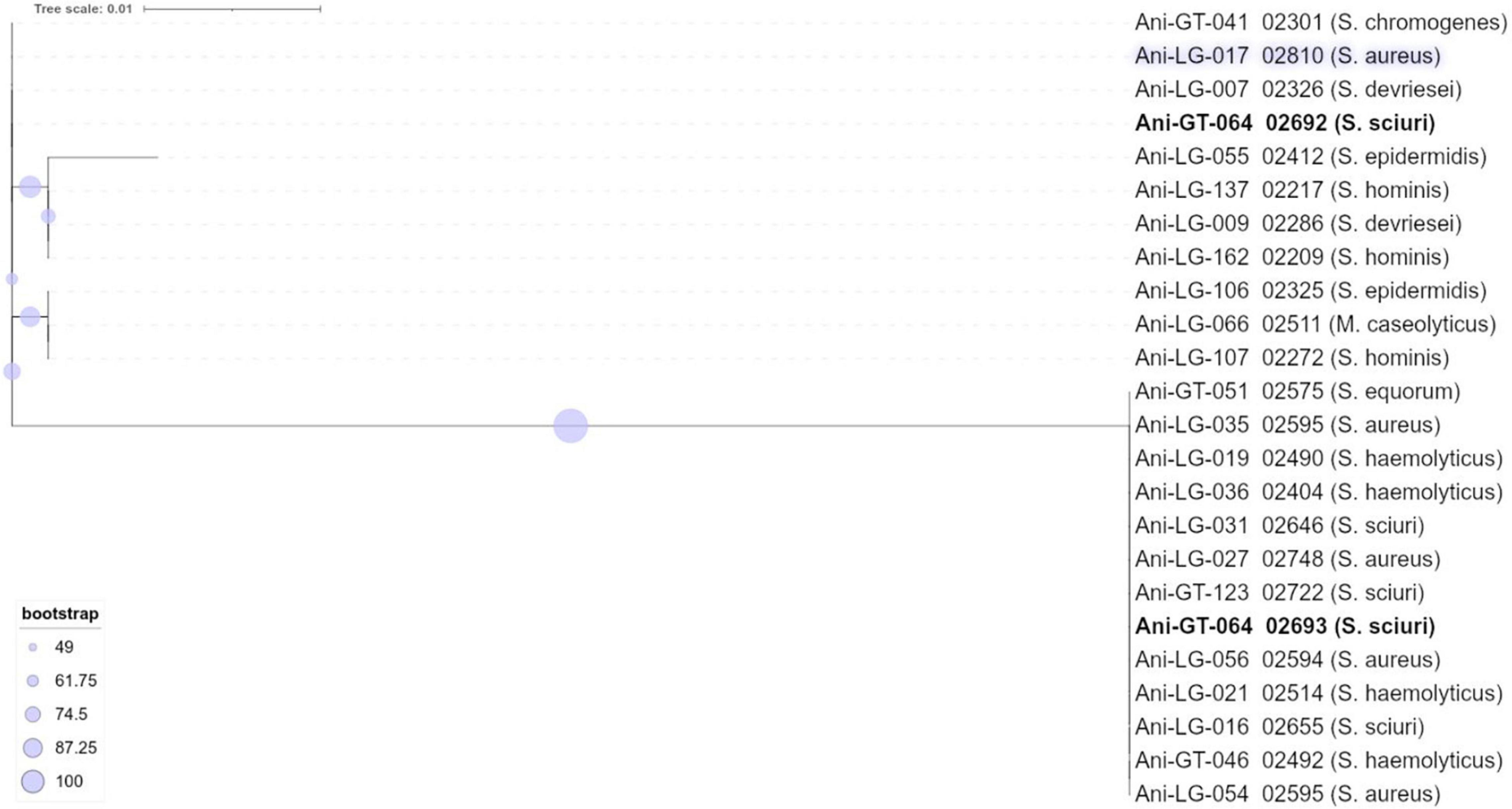
The isolates originated from a previous study (Fergestad et al., 2021) of bovine staphylococci from Belgian and Norwegian dairy farms, in which 464 lactating cows were sampled from 13 farms in Belgium, and 100 cows with clinical mastitis were sampled from 100 farms in Norway (one cow from each farm). As described in Fergestad et al. (2021), the sampling strategy for that study was adapted to national structures for dairy production and thus varied between countries and regions, but also ensured a diversity of the material. A total of 272 staphylococcal isolates were characterized with regard to their phenotypic antimicrobial resistance, and carriage of the methicillin resistance genes mecA and mecC and some selected virulence genes were determined by PCR. From this collection, a total of 95 isolates were selected for whole genome sequencing (WGS) according to the following criteria: (i) all S. aureus isolates, (ii) representatives of all NAS species, (iii) all isolates positive in PCR for mecA or mecC. Selection of NAS isolates was further based on phenotypic resistance patterns observed in species with more than one isolate, ensuring that both highly resistant and less resistant isolates were represented. Lastly, it was aimed to achieve an even distribution of isolates originating from the different geographical regions (Norway and Belgium). Altogether, the isolates selected for WGS were 45 S. aureus isolates and 50 NAS isolates [identified by Maldi Tof-MS; (Cameron et al., 2017, 2018)] from 16 different species: S. arlettae, S. auricularis, S. chromogenes, S. cohnii, S. devriesei, S. epidermidis, S. equorum, S. haemolyticus, S. hominis, S. hyicus, S. saprophyticus, S. sciuri, S. simulans, S. vitulinus, S. warneri, and S. xylosus. Background information about the selected isolates is presented in Supplementary Table 1. Some reclassification within the genus Staphylococcus has recently been suggested; the reassignment of S. sciuri and S. vitulinus to a novel genus Mammaliicoccus (Madhaiyan et al., 2020) and the novel species Staphylococcus pseudoxylosus, closely related to S. xylosus (MacFadyen et al., 2019). For the sake of clarity, the suggested Mammaliicoccus species are considered as members of genus Staphylococcus and S. pseudoxylosus is included with the S. xylosus in this manuscript. The isolate identified as S. pseudoxylosus is, however, shown in the phylogenetic tree of NAS (Figure 1), to illustrate its phylogenetic placement.
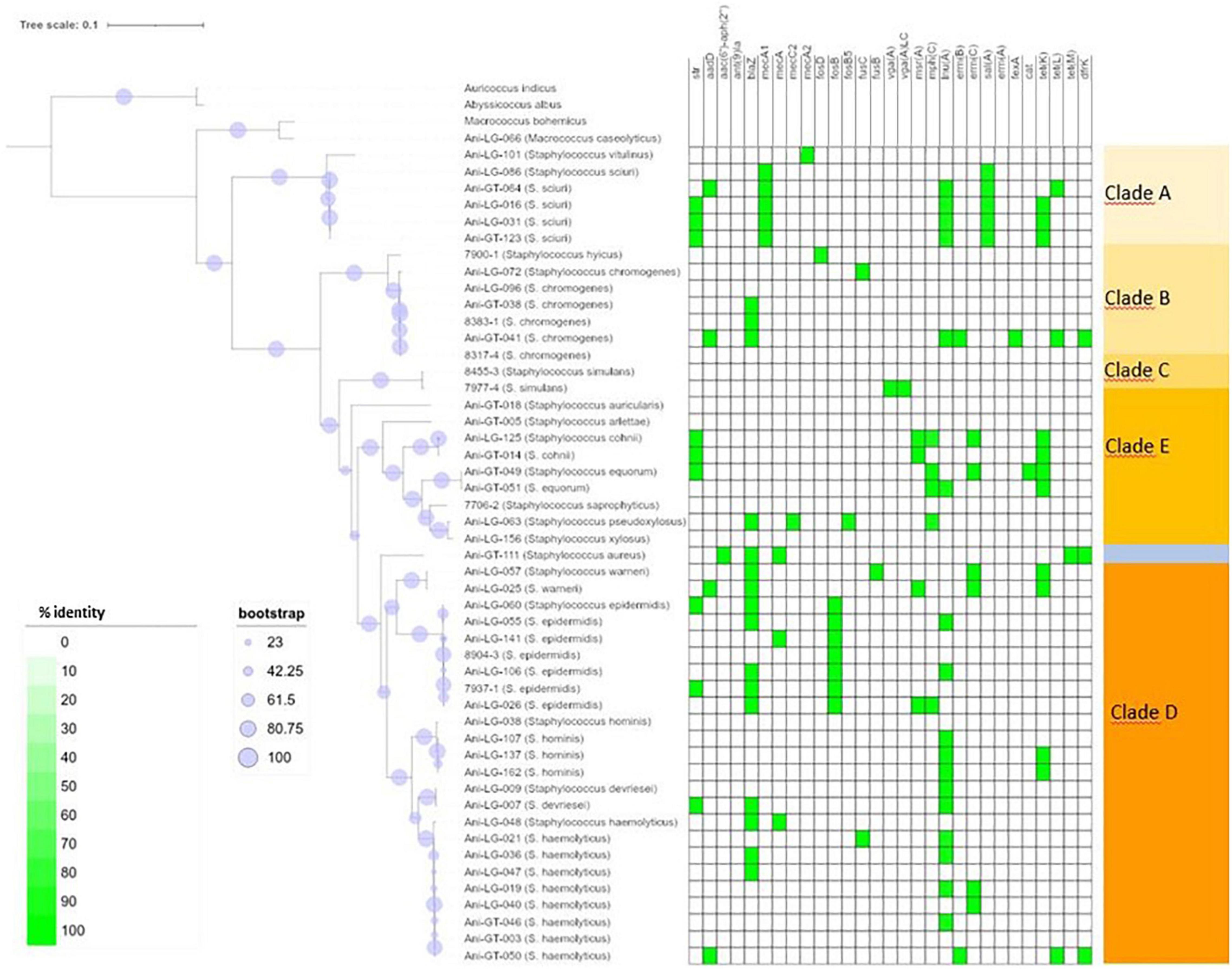
Figure 1. Phylogeny and antimicrobial resistance genes of non-aureus staphylococci. Presence of antimicrobial resistance gene is indicated with green square with percent identity indicated by color shades according to the scale.
DNA Extraction, Whole Genome Sequencing and Assembly
DNA was extracted using Masterpure™ Gram Positive DNA Purification Kit (Lucigen, Middleton, WI, United States). Quality and DNA concentrations were determined with Nanodrop 1000 (Thermo Fisher Scientific).
The genomic DNA libraries were prepared for Illumina sequencing according to the manufacturer’s instructions using the Nextera XT kit and sequenced by the NovaSeq 6000 Sequencing System (Illumina, San Diego, CA, United States). The raw read sequences were assembled into contigs with the pipeline Shovill 1.0.4 (Seemann et al., 2020) including trimmomatic 0.38 (Bolger et al., 2014) for the cleaning and annotated using Prokka 1.13.3 (Seemann, 2014).
For LG-048 and LG-101 strains, Nanopore MinION long-read sequencing was performed using the Rapid Barcoding Sequencing kit (Oxford Nanopore) for library preparation. After guppy_gpu base calling, assembly of nanopore reads was performed using Canu 1.8 (Koren et al., 2017). The Illumina reads were cleaned with trimmomatic 0.36 (ILLUMINACLIP:illumina_oligos_and_revcomp:2:30:5:1:true LEADING:3 TRAILING:3 MAXINFO:40:0.2 MINLEN:36 options) and aligned with contigs provided by Canu assembly using BWA 0.7.15-r1140 (arXiv:1303.3997). Pilon 1.23 (Walker et al., 2014) was run on this alignment for preliminary corrections. The final result was the corrected consensus provided by Pilon.
The statistics of assemblies are provided in the Supplementary Table 2.
Phylogenetic Trees
For creation of alignment and phylogenetic trees of the staphylococcal isolates we included some additional strains of different genera as outgroups to clarify where in the phylogenetic landscape the staphylococcal species were located. These strains were Macrococcus bohemicus (NZ_CM009972.1 to NZ_CM009973.1 and NZ_PZJG01000001.1 to NZ_PZJG01000029.1), Abyssicoccus albus (NZ_RKRK01000001.1 to NZ_RKRK01000010.1), and Auricoccus indicus (NZ_CP019573.1). We ran Panaroo 1.2.3 to analyze core genome to get common genes between the 95 strains (options “–clean-mode moderate –remove-invalid-genes -a core”) and, using mafft in Panaroo, an alignment suitable for phylogenetic analysis. We picked two of the most common antimicrobial resistance genes in our material (lnuA and blaZ) and created phylogenetic trees of these genes to illustrate the phylogenetic relationship of the resistance genes across isolates and species. For the phylogenetic trees of antimicrobial resistance genes, we also used mafft in Panaroo for gene alignment. All phylogenetic trees were then created using IQtree 2.0.3 with core gene alignment (and options “–safe -T AUTO -B 1000 -alrt 1000 -m MFP”). The graphic representation of the phylogenetic trees was done with iTOL (Letunic and Bork, 2019).
Identification of Antimicrobial Resistance Genes
Identification of antimicrobial resistance genes was performed with ResFinder 4.1 (Center for Genomic epidemiology, Technical University of Denmark) (Zankari et al., 2017; Bortolaia et al., 2020). Prokka 1.131.3 was used for detection of the multidrug efflux pump gene norA.
Identification of Virulence Genes
Identification of virulence genes was done by using VirulenceFinder 2.0 (Center for Genomic Epidemiology, Technical University of Denmark) (Joensen et al., 2014) and a tblastn 2.10.1 search against a published dataset (Naushad et al., 2019). The dataset by Naushad et al. (2019) was used to complement the VirulenceFinder program which, unexpectedly, returned very few results from the NAS species. The tblastn search of the proteins in the Naushad et al. (2019) dataset was set upwith a minimum high-scoring segment pair (HPS) coverage >90, a minimum e-value of 10–5. Ha scores were computed for all matches as described by Naushad et al. (2019). We kept only the hits with the highest Ha score and the highest percentage identity. Two proteins sequences were not identified in the database by Naushad et al. (2019): the phenol soluble modulin mec (PSMmec) and toxic shock syndrome toxin (tsst). For the PSMmec we used the record with GenBank accession number AIU84051.1, while the record for tsst from the database by Naushad et al. (2019) (accession number YP_415862) had been replaced by accession number WP_001035596.1, 100% identical to the previous record over its full length. Both records were found searching the NCBI (National Center for Biotechnology Information) website (1 accessed January 2021).
Results
Staphylococcal Isolates
Of the 95 isolates selected for WGS, results from 45 S. aureus and 48 NAS were included for further analysis. One S. arlettae and one S. epidermidis had to be rejected due to poor quality of the samples. The NAS species were divided into five different clades according to the phylogenetic analysis published by Naushad et al. (2016). NAS isolates were distributed in species and clades as follows; clade A was represented by S. sciuri (n = 5) and S. vitulinus (n = 1). Clade B was represented by S. chromogenes (n = 6) and S. hyicus (n = 1). Clade C consisted of S. simulans (n = 2). Clade D was represented by S. devriesei (n = 2), S. epidermidis (n = 7), S. haemolyticus (n = 9), S. hominis (n = 4) and S. warneri (n = 2). Clade E was represented by S. arlettae (n = 1), S. auricularis (n = 1), S. cohnii (n = 2), S. equorum (n = 2), S. saprophyticus (n = 1), and S. xylosus (n = 2). Species and clade distribution are also presented in Tables 1–7. Figure 1 shows the phylogeny of the NAS isolates, confirming that they do separate according to the different clades. Figure 2 shows the phylogeny of the S. aureus isolates.
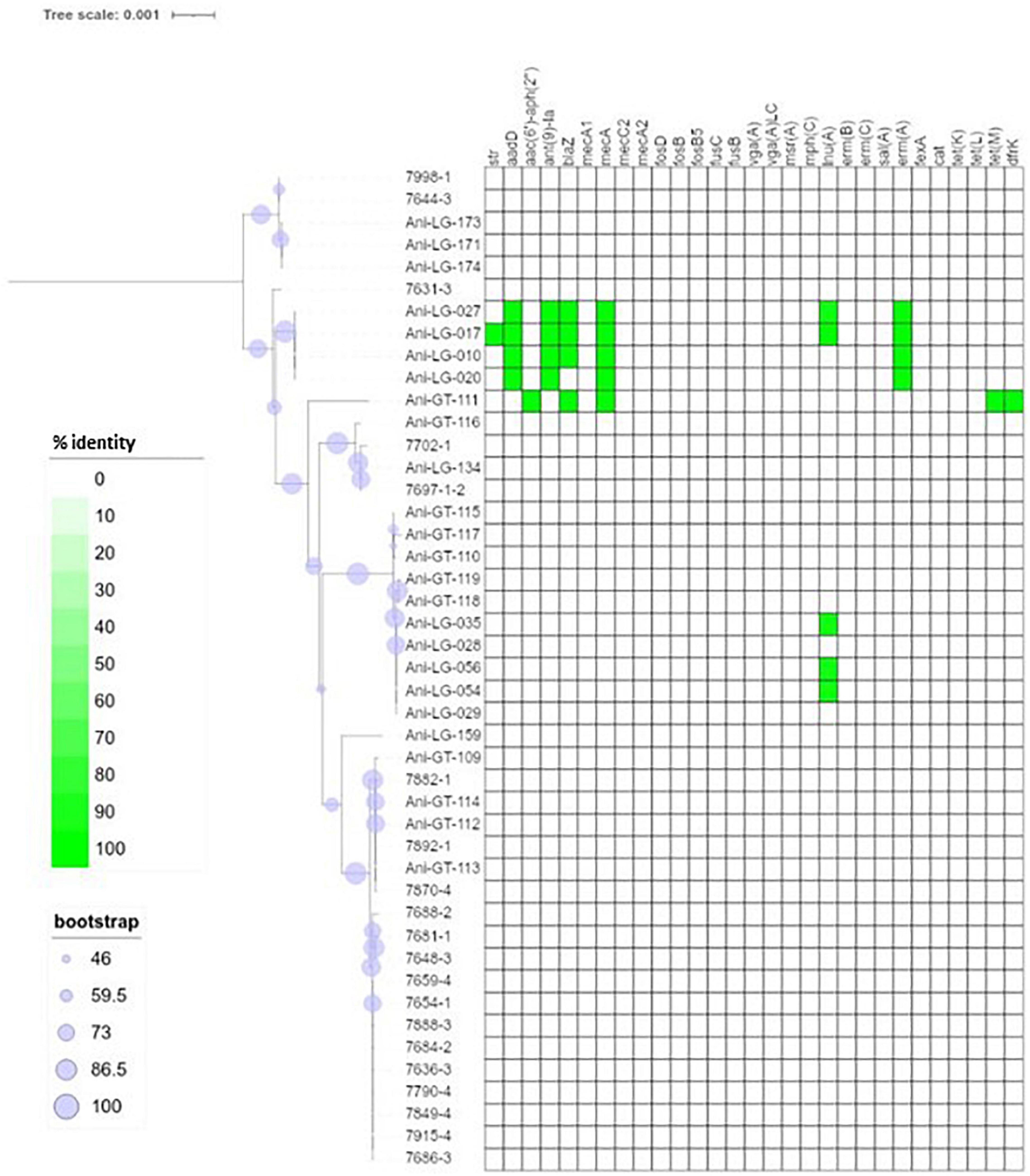
Figure 2. Phylogeny and antimicrobial resistance genes of Staphylococcus aureus. Presence of antimicrobial resistance gene is indicated with green square with percent identity indicated by color shades according to the scale.
Antimicrobial Resistance Genes
Figure 1 shows the distribution of antimicrobial resistance genes in all NAS isolates, in which the NAS are shown according to their clade. Figure 2 shows the distribution of antimicrobial resistance genes in all S. aureus isolates.
Lincosamide resistance gene lnuA was present in 17 isolates from seven different NAS species. Most NAS isolates originated from clade D (n = 11). The gene was also present in five S. aureus isolates. Figure 3 shows the phylogeny of the lnuA genes, demonstrating that several species carried phylogenetically similar lnuA genes. Macrolide resistance genes were found in 17 different isolates, with some isolates (Ani-GT-049, Ani-LG-025, Ani-LG-026 and Ani-LG-125) containing multiple macrolide resistance genes. The erm genes were present in four S. aureus isolates and eight NAS isolates, of which five were of clade D. The four S. aureus isolates contained the ermA gene, while the ermB and ermC genes were detected uniquely in the NAS species. The mphC gene was present only in NAS species, most originating from clade E (n = 4). The msrA gene was present in species from clade D and E.
The penicillin resistance gene blaZ was found in 19 isolates, both S. aureus and NAS, with most NAS isolates originating from clade D (n = 11). Figure 4 shows the phylogeny of the blaZ genes. One S. xylosus isolate contained up to two phylogenetically distinct blaZ genes. The betalactam resistance gene mecA was present in seven isolates, S. aureus (n = 5), and S. epidermidis (n = 1), and S. haemolyticus (n = 1) from clade D. Of these, four S. aureus and one S. haemolyticus also carried blaZ. All other resistance genes detected in S. aureus were detected in the mecA positive isolates, except for three non-mecA S. aureus isolates containing only the lnuA gene. Other variants of the mec genes were also detected in some isolates. All S. sciuri isolates (n = 5) carried the mecA1 gene, the only S. vitulinus carried the mecA2 and one of the two S. xylosus carried mecC2.
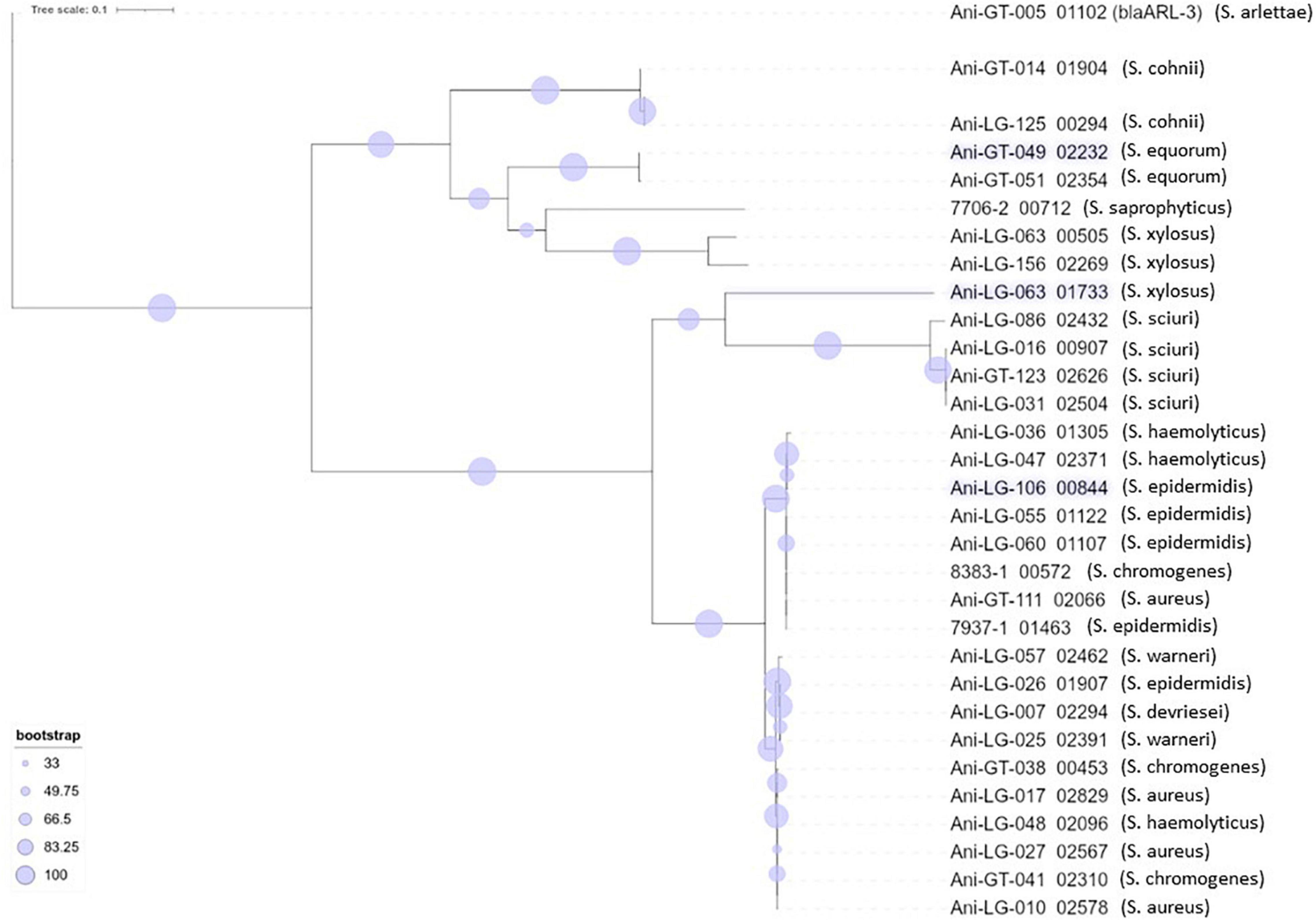
Figure 4. Phylogeny of penicillin resistance gene blaZ found in staphylococci. The novel penicillin resistance gene blaARL found in Staphylococcus arlettae (Andreis et al., 2017) is included for phylogenetic comparison.
Of the tetracycline resistance genes detectable by ResFinder, the tetK gene was present in 11 NAS isolates from clade A, D and E, and the tetL gene was present in three NAS species from clade B and D. The tetM gene was present in one S. aureus isolate only.
The aminoglycoside resistance gene aadD was identified in four S. aureus isolates and four NAS isolates consisting of the following species: S. chromogenes, S. haemolyticus, S. sciuri, and S. warneri. One S. aureus isolate carried the aminoglycoside resistance gene aac-aph. The aminoglycoside resistance gene str was detected in nine NAS isolates from clade A, D, and E.
Overall, in NAS, many antimicrobial resistance genes were found in isolates from clade D and most isolates in this clade carried one to three resistance genes. In one S. chromogenes isolate (clade B), seven resistance genes were detected, including lnuA, blaZ, ermB, aadD, tetL, dfrK, and fexA. In addition, one S. equorum, one S. cohnii (both clade E), and one S. warneri (clade D) carried five resistance genes, with aminoglycoside, macrolide and tetracycline resistance genes detected in all three isolates. No antimicrobial resistance genes were identified in only nine of the 48 NAS isolates, which originated from clade B, C, D, and E. On the other hand, no antimicrobial resistance genes were detected in the majority of the S. aureus isolates (37/45). Five S. aureus isolates carried three to six resistance genes, the mecA gene was detected in all five.
Using Prokka, the major facilitator superfamily multidrug efflux transporter gene norA was detected in all isolates, both S. aureus and NAS.
Virulence Genes Detected by VirulenceFinder
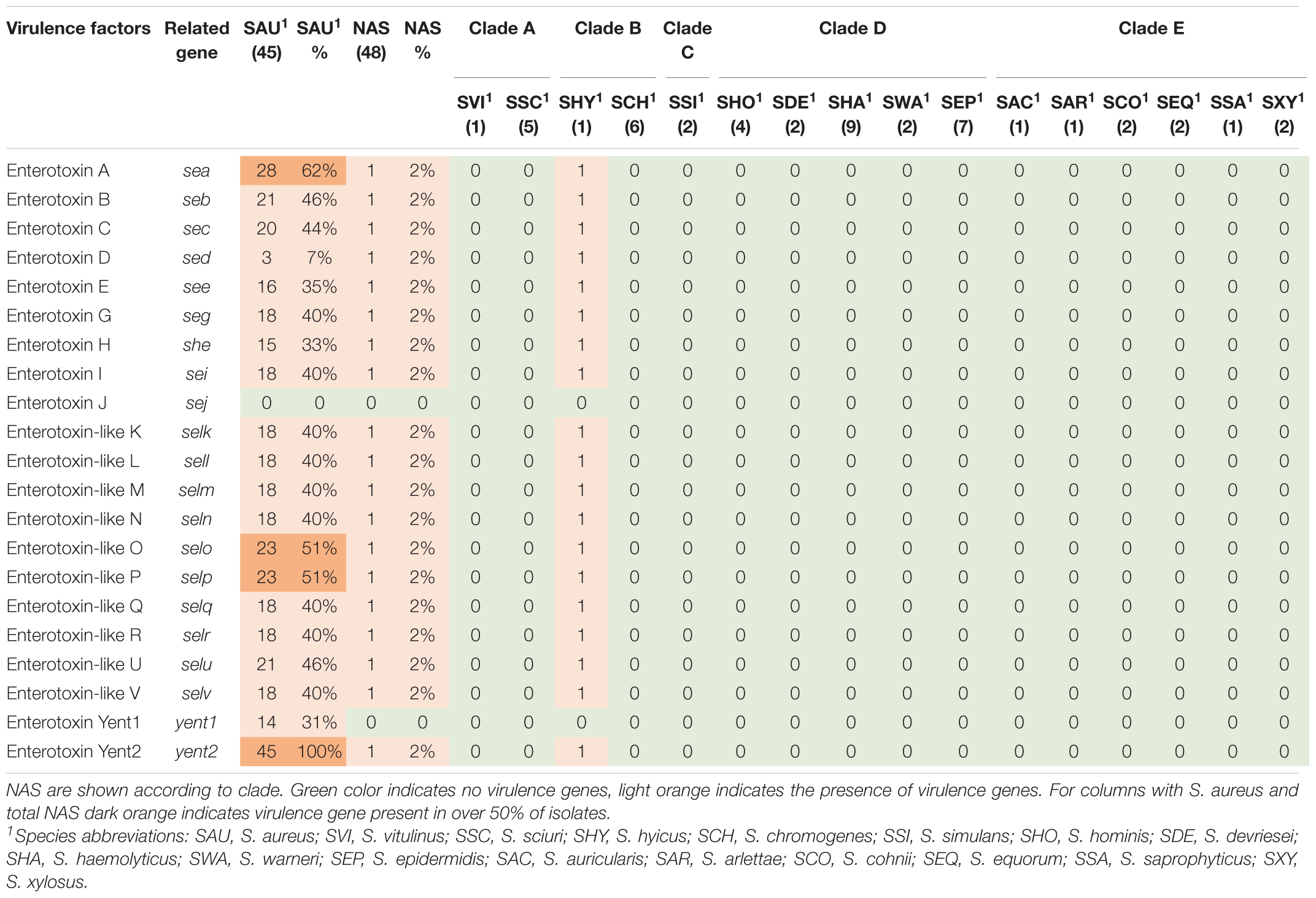
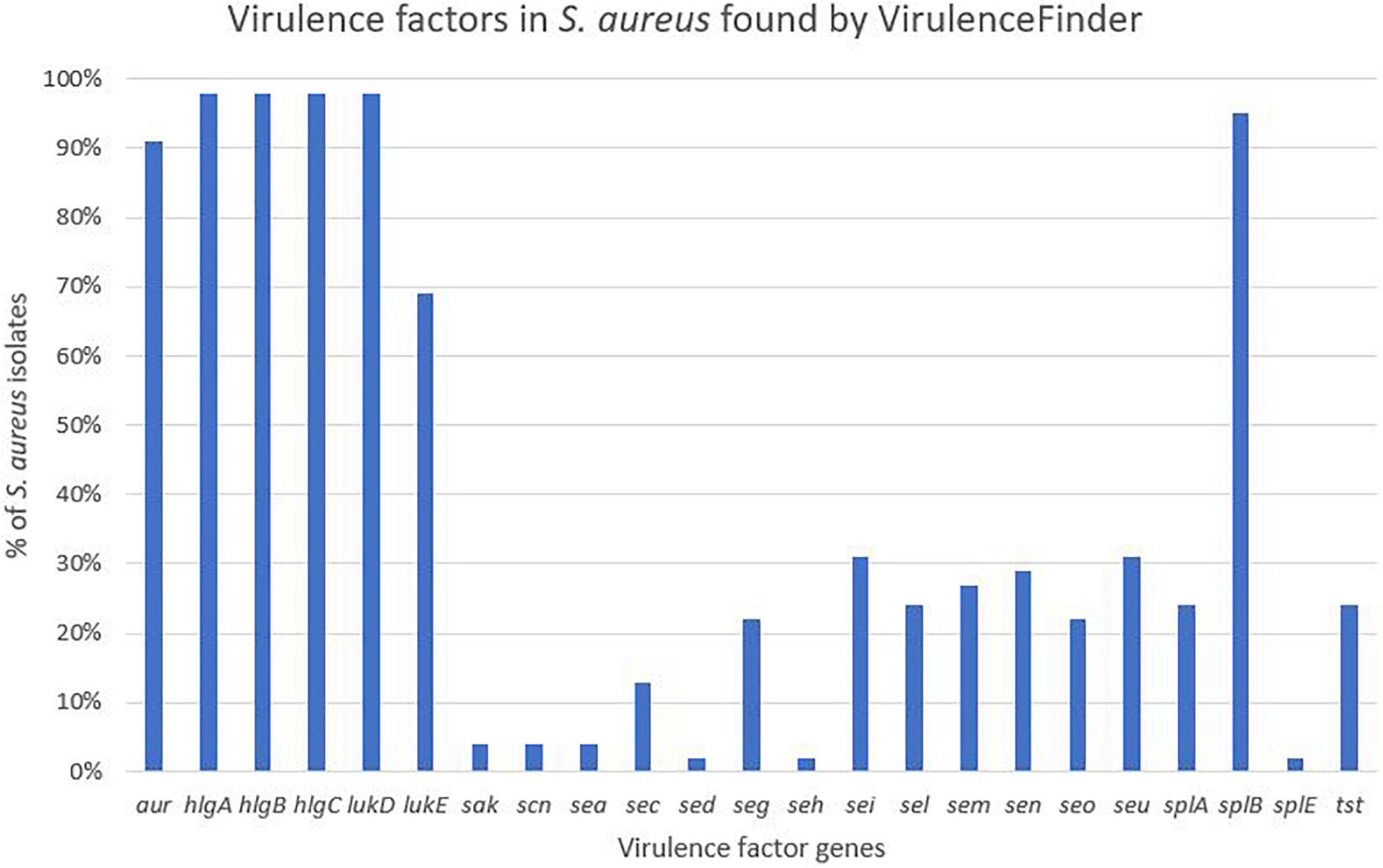
Virulence genes detected by VirulenceFinder (Center for Genomic Epidemiology, Technical University of Denmark) in the S. aureus isolates are shown in Figure 5. The hlgA, hlgB, and hlgC genes encoding the gamma hemolysins A, B, and C, and the lukD gene coding for a leukotoxin, were the most commonly detected virulence genes in S. aureus and detected in 44/45 S. aureus isolates. The splB gene, encoding a serine protease, and aur, encoding aureolysin, were detected in 43 and 41 S. aureus isolates, respectively. The least frequent virulence genes identified by VirulenceFinder were sak, scn, and sea, encoding staphylokinase, staphylococcal complement inhibitor and enterotoxin A, respectively, found in two S. aureus isolates only (Ani-LG-017 and Ani-LG-027), and the sed and seh genes coding for enterotoxin D and H, were detected in one S. aureus isolate each.
Except for the detection of the ACME gene in four S. epidermidis isolates, the VirulenceFinder did not detect any other virulence genes in the NAS isolates.
Putative Virulence Factors Based on Dataset by Naushad et al., 2019
Virulence Factors Involved in Adherence
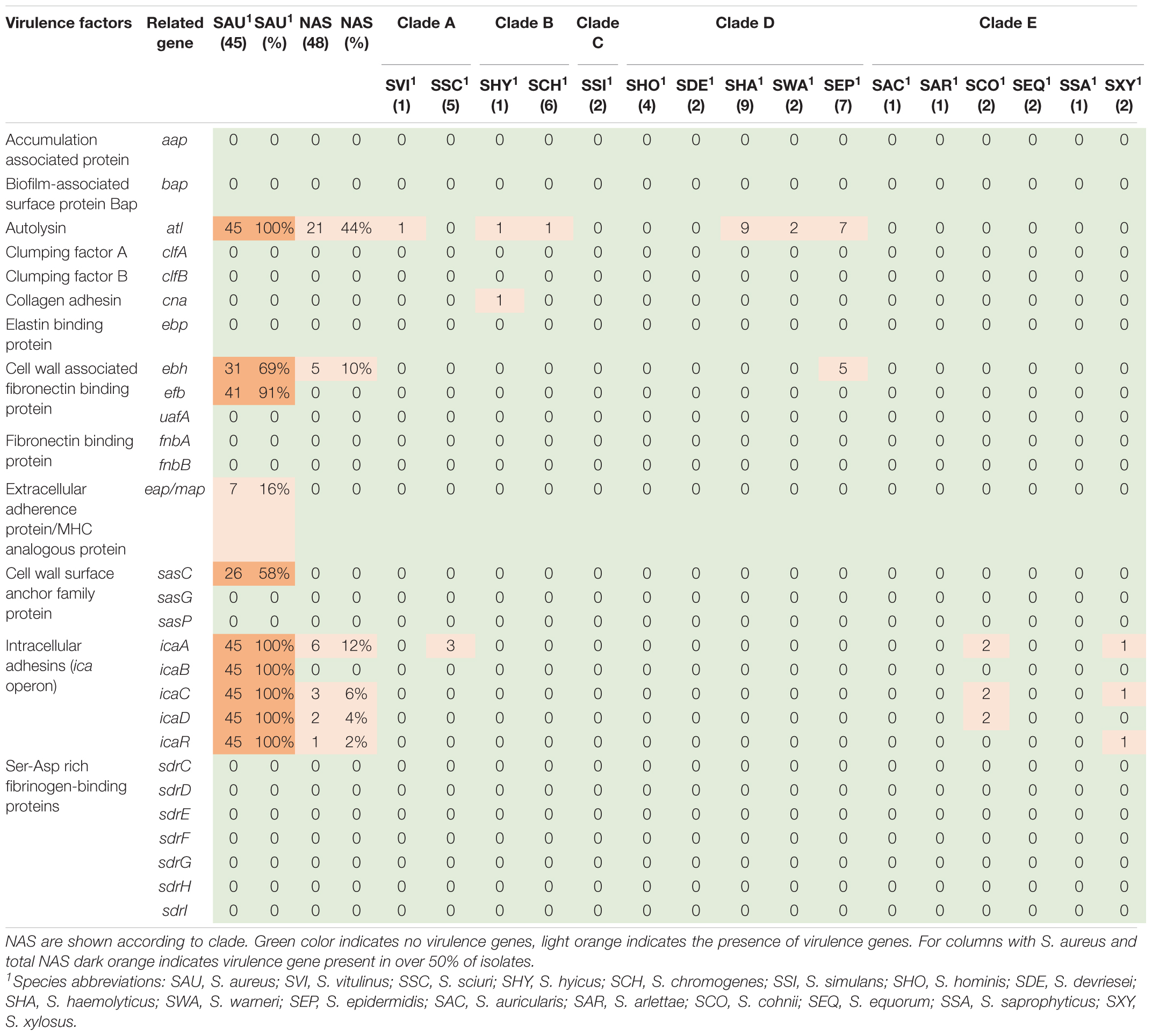
Based on the dataset by Naushad et al. (2019), 28 virulence-related genes involved in adherence were screened for. The genes are listed, and results are summarized in Table 1. The atl gene was present in 21 NAS isolates and in six of the 16 NAS species from clade A, B, and D, as well as in all S. aureus isolates. The icaA of the ica operon was present in six NAS isolates from clade A and E. The icaC, icaD, and icaR were detected in three, two and one NAS isolates respectively, from clade E. Regarding the S. aureus isolates, the ica operon was present in all isolates. Many of the adherence associated genes were not detected in either NAS isolates nor S. aureus isolates, including aap, bap, clfA and clfB, ebp, uafA, fnbA and fnbB, sasG and sasp and the Ser-Asp rich fibrinogen-binding proteins.
Exoenzymes
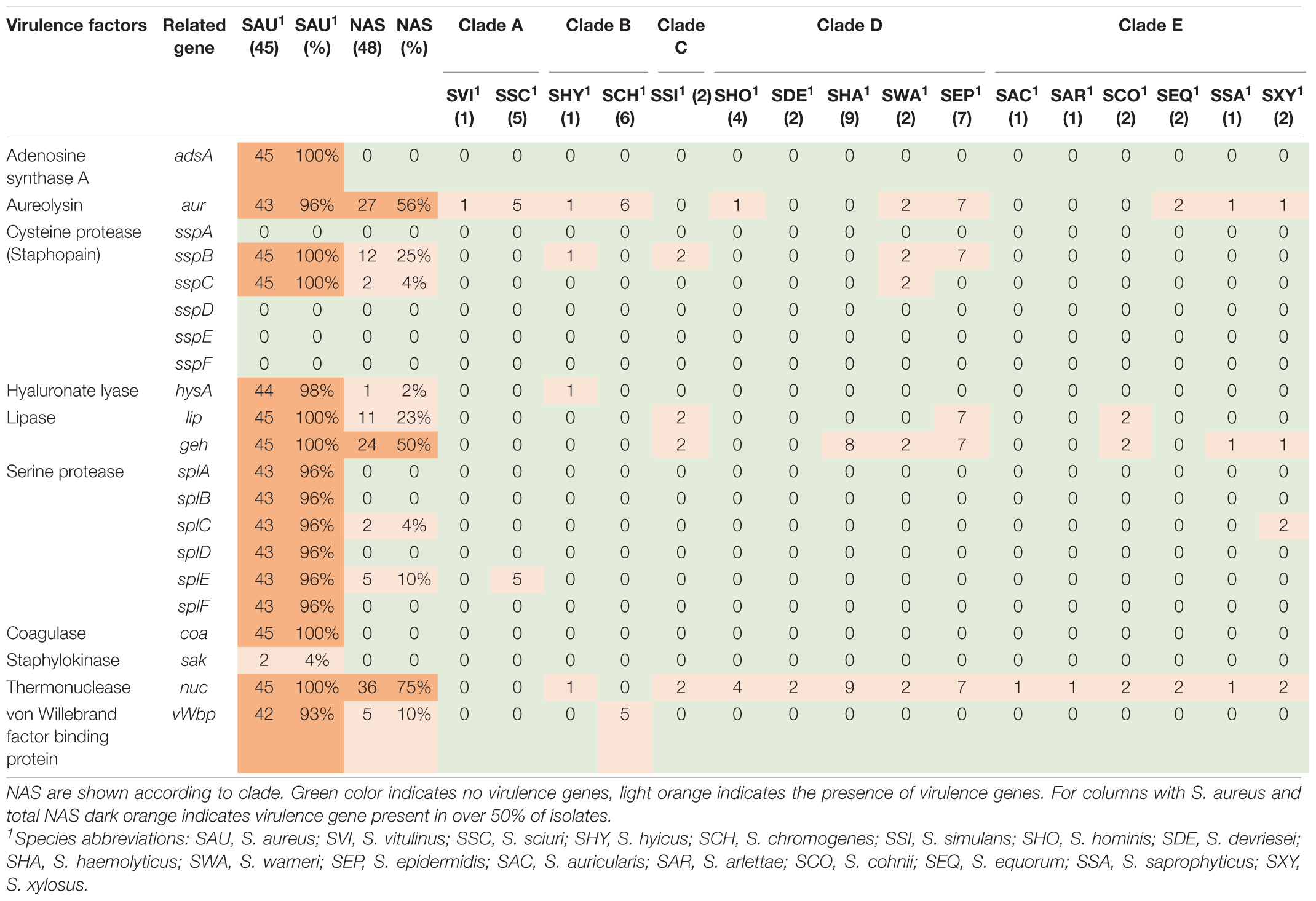
Twenty-one different exoenzyme genes were searched. The genes are listed, and results are summarized in Table 2. The nuc gene was present in 36 out of 48 NAS isolates in species from clade B, C, D, and E. Only one S. hyicus isolate was positive in clade B, while in clade C, D, and E all isolates were positive. The second most frequent exoenzyme genes in NAS were aur and geh, detected in 27 and 24 isolates, respectively. Among the serine proteases, splC and splE were detected in S. xylosus and S. chromogenes, respectively and vWpb was found in S. chromogenes. Many of the exoenzymes were not detected in NAS species. For S. aureus, 93% of all S. aureus isolates contained at least 16 exoenzyme genes.
Virulence Factors Involved in Host Immune Evasion
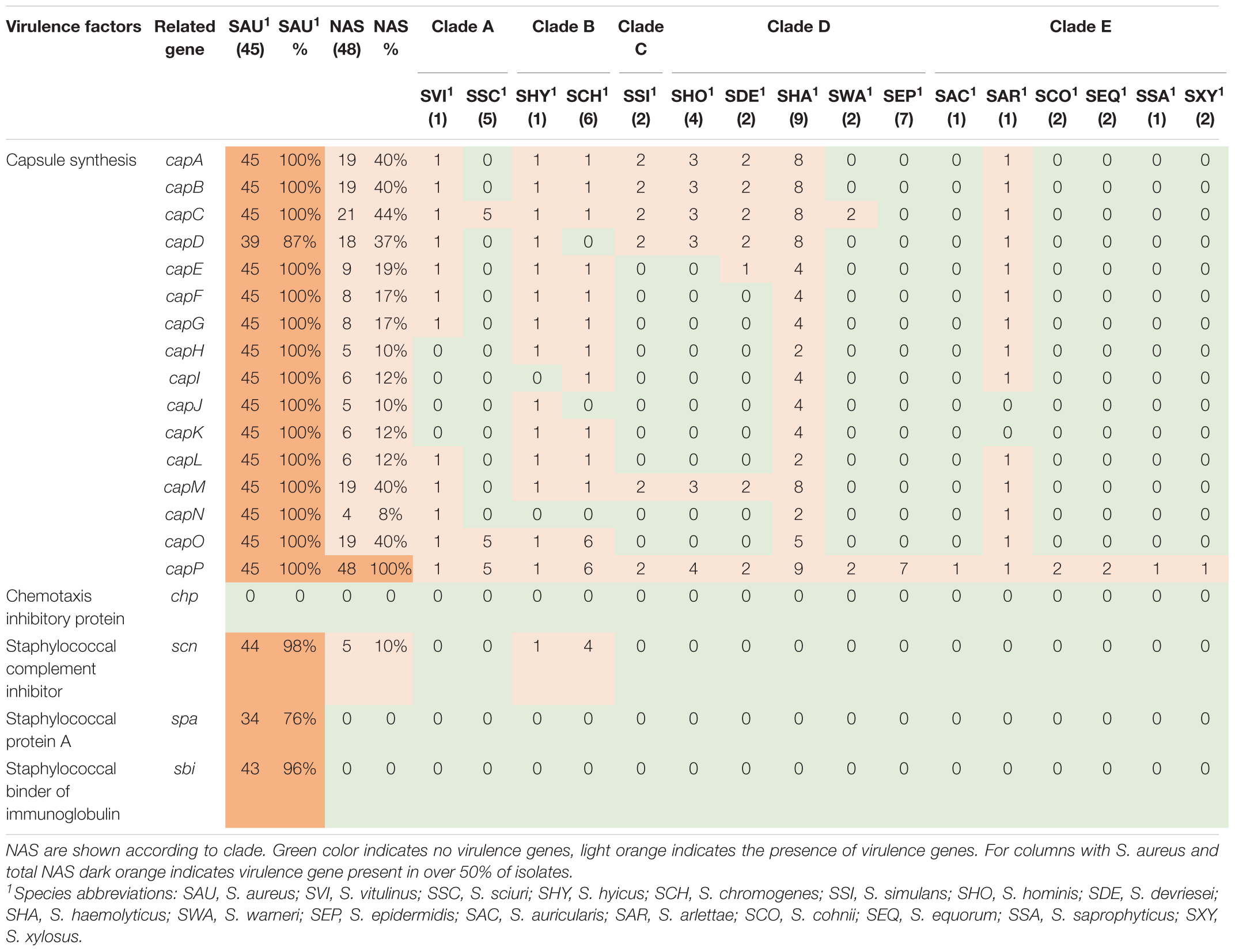
Twenty virulence genes associated with host immune evasion were searched. The genes are listed, and results are summarized in Table 3. The chp, spa and sbi genes were not detected in any of the NAS species. The scn gene was only identified in species from clade B. The cap genes were the most frequently detected host immune evasion genes. Based on Naushad et al. (2019) cap genes were considered present if either the cap5 or cap8 isoforms were detected. The capP was detected in all NAS isolates and five NAS species contained only this cap gene. Two S. haemolyticus isolates contained all 16 cap genes and the single S. hyicus and S. arlettae contained 14 cap genes. All cap genes except capD were present in all S. aureus isolates. The scn, spa and sbi genes were identified in 44, 34, and 43 S. aureus isolates, respectively. The chp gene was not detected in any of the S. aureus isolates.
Virulence Factors Involved in Iron Uptake and Metabolism
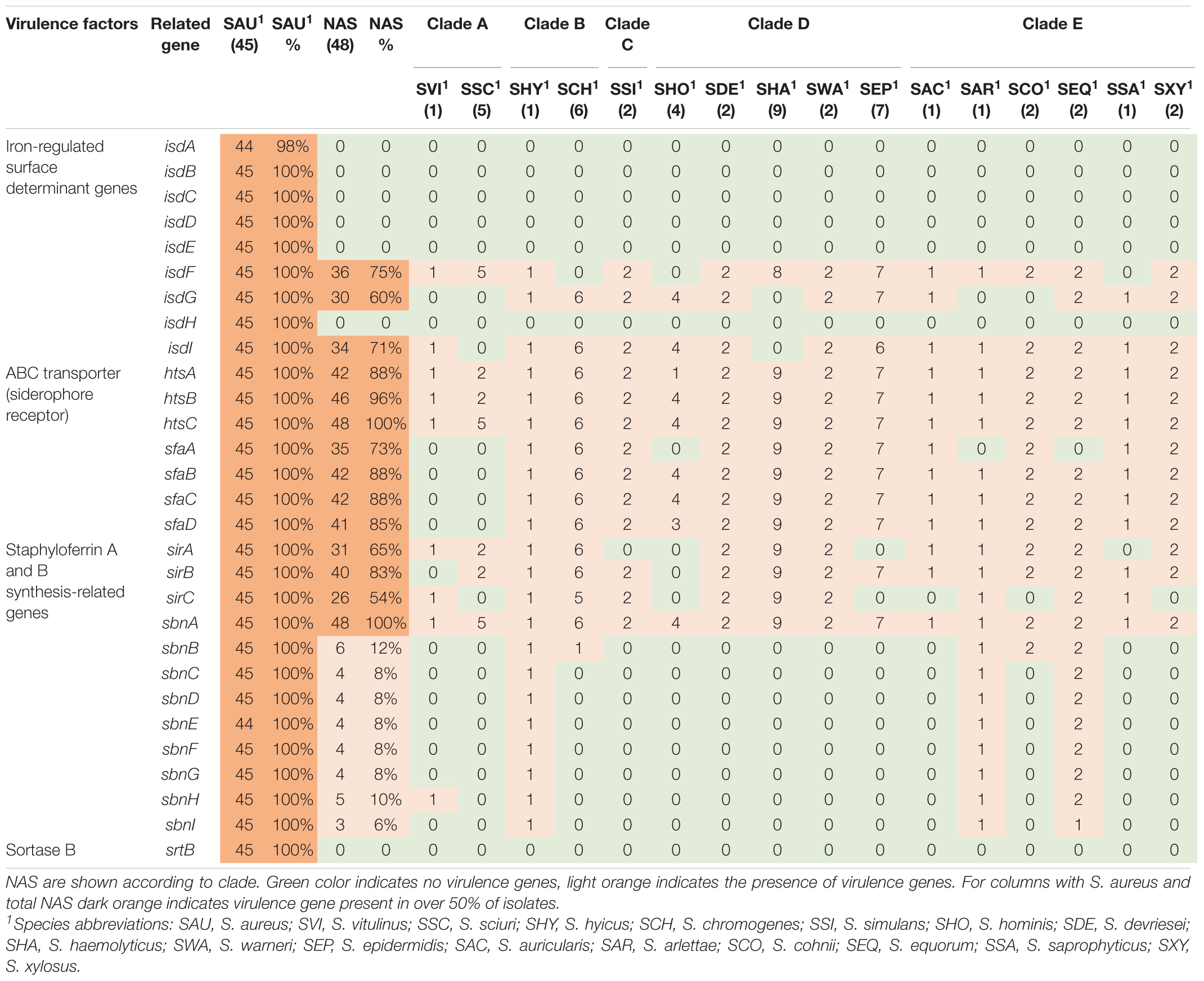
Twenty-nine iron uptake and metabolism genes were searched. The genes are listed, and results are summarized in Table 4. The ABC transporter genes and staphyloferrin A and B synthesis related genes were most frequently identified in NAS isolates, where htsC and sbnA were present in all NAS isolates. All genes associated with iron uptake and metabolism were detected in all S. aureus isolates.
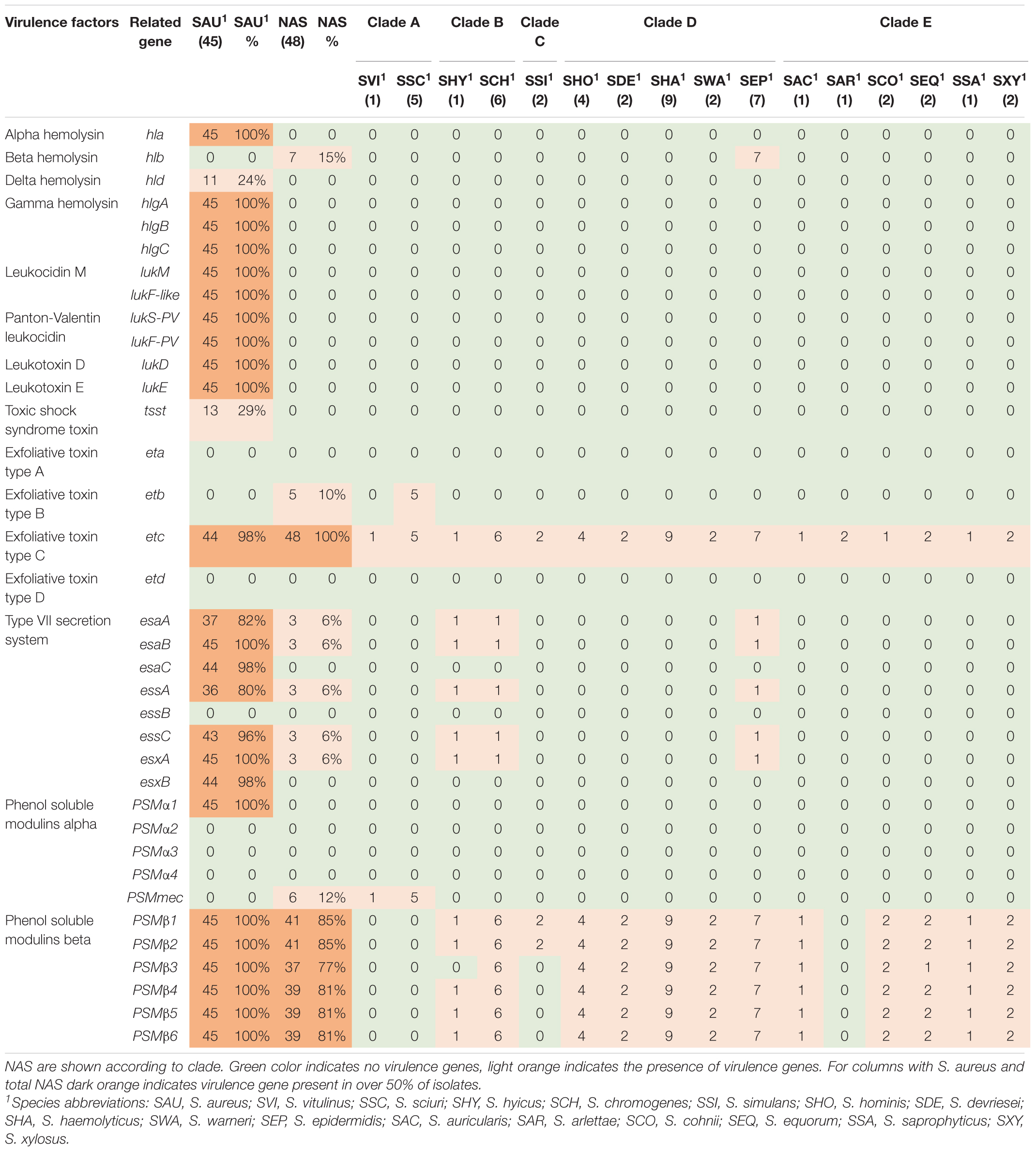
Toxins, Type VII Secretion and Phenol-Soluble Modulin Genes
The genes in this category included 36 toxin genes from various categories. The genes are listed, and results are summarized in Table 5. The enterotoxins and staphylococcal exotoxins are described below and in Tables 6, 7. Most of the toxin genes in this category were not detected in the NAS species, except etc which was present in all NAS isolates, hlb present in all S. epidermidis isolates and etb present in all S. sciuri isolates. The type VII secretion genes esaA, esaB, essA, essC, and esxA were identified in three NAS isolates, namely one S. chromogenes, one S. epidermidis and the single S. hyicus. Among the PSMs, none of the PSMα were detected, while the PSMβ genes were present in all NAS species except the species in clade A where PSMmec was identified. As for the S. aureus isolates, all isolates contained hla, hlgA, hlgB, hlgC, and all leukocidin and leukotoxin genes. The etc gene was identified in 44 isolates and the tsst and hld genes were identified in 13 and 11 S. aureus isolates, respectively, while none of the S. aureus isolates contained hlb, eta, etb or etd. Most of the type VII secretion system genes were frequently detected in S. aureus isolates, except essB that was not present in any of the isolates. Of the PSMs, PSMα1, and PSMβ1 to PSMβ6 were identified in all S. aureus isolates, the remaining PSMs were not identified.
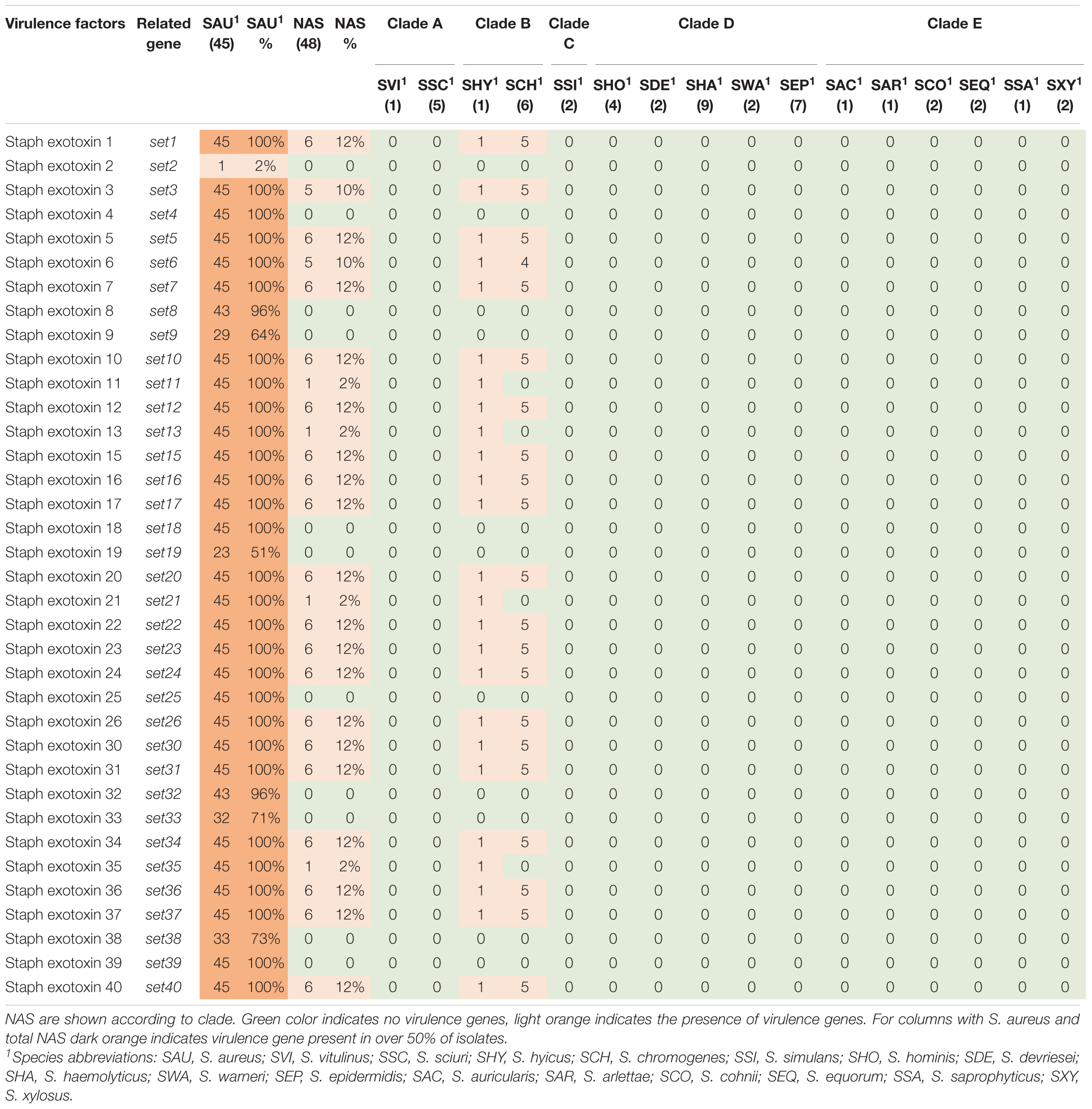
Thirty-six staphylococcal exotoxins (SETs) and 21 enterotoxins were searched. Except for the species in clade B, set genes were not detected in any NAS species. The enterotoxins were only identified in the single S. hyicus of clade B, this isolate contained all enterotoxin genes except sej and yent1. In the S. aureus isolates most set genes were present in all isolates. set9, set19, set 32, set33, and set38 were frequently detected, while set2 was only present in one S. aureus isolate. The yent2 gene was detected in all S. aureus isolates. The second most frequently identified enterotoxin genes were sea, selo, and selp, present in 28, 23, and 23 S. aureus isolates. The sej gene was not detected in any S. aureus isolates.
Virulence Potential
Virulence potential was defined as the total number of virulence genes in an isolate, where all genes were equally weighted (Naushad et al., 2019). Staphylococcus aureus had the highest virulence potential and carried on average 140 virulence genes. The NAS isolates, disregarding species, carried on average 28 virulence genes. The highest virulence potential in NAS was detected in S. chromogenes and S. hyicus (both clade B). Staphylococcus chromogenes isolates contained on average 44 virulence genes, and, except the single S. hyicus isolate, was the only NAS species in which staphylococcal exotoxins were detected. The single S. hyicus isolate contained 98 virulence genes, carrying both exotoxins and enterotoxins. The lowest virulence potential was found in S. sciuri and S. hominis with, on average, 13 and 21 virulence genes in total, respectively.
Discussion
This study of virulence and antimicrobial resistance genes in 93 whole-genome sequenced NAS and S. aureus isolates from bovine milk samples of European origin adds new data to the current sparse information of the genetic basis for both antimicrobial resistance and virulence factors in bovine staphylococci. WGS confirmed the species distribution previously determined with Maldi Tof-MS of the isolate collection of 45 S. aureus and 48 NAS isolates of different species (Fergestad et al., 2021). In total, we determined the presence of 191 staphylococcal virulence genes and 25 antimicrobial resistance genes. When discussing our results, it must be taken into account that the whole-genome sequenced isolates are not a random assemblage of bovine staphylococci, but they are selected from a previous study according to the described criteria. Nevertheless, one main finding is that our description of the virulence gene contents of the 48 NAS isolates coincides to a large extent with the findings of the so far most comprehensive study of virulence genes in a collection of Canadian bovine NAS based on WGS data.
The emergence and spread of antimicrobial resistance genes are of great concern to society, including animal food production and the dairy industry. The antimicrobial resistance genes that are detected by ResFinder 4.1 were frequently observed in our collection of isolates representing 16 NAS species, coinciding with other reports of occurrence of such genes distributed among several NAS species (Nobrega et al., 2018). This finding of an array of resistance genes in a diversity of NAS species is supporting the hypothesis that these bacteria can act as a potential reservoir for resistance properties (Otto, 2013; Becker et al., 2014).
We found several isolates from different staphylococcal species harboring phylogenetically similar lnuA genes. This gene, encoding a lincosamide nucleotidyltransferase that confers resistance to lincosamides, has previously been found in both S. aureus and NAS of bovine origin (Lüthje and Schwarz, 2006; Li et al., 2015). The gene is often found on plasmids, which could promote horizontal transfer of the gene (Lüthje et al., 2007). Studies have shown that the nucleotide sequence of lnuA is more conserved than the surrounding plasmid sequences and the conserved gene has been found in several different plasmid backbones, suggesting that the gene is also exchanged via interplasmid recombinational events (Lüthje et al., 2007; Wassenaar et al., 2016). This may contribute to explaining our finding of phylogenetically similar lnuA genes in several different staphylococcal species.
Resistance to betalactam antimicrobials is commonly reported in staphylococci. The blaZ gene encodes a penicillinase (or betalactamase) conferring penicillin resistance by hydrolyzing the betalactam ring and inactivating the drug (Zhang et al., 2001). The gene is usually either plasmid- or chromosomally encoded (Olsen et al., 2006). Penicillin resistance is prevalent in S. aureus of both human and bovine origin (Olsen et al., 2006) and betalactamase production is the most prevalent mechanism of betalactam resistance in NAS (Nobrega et al., 2018). In consistence with these observations, we found several isolates of different staphylococcal species carrying the blaZ gene. The phylogenetic tree of the gene also showed several phylogenetically different sequences. A high number of different blaZ sequence types has previously been shown in staphylococci of bovine origin, as well as a very low similarity between plasmid- and chromosomally encoded blaZ genes which, in a study by Olsen et al. (2006), separate into two phylogenic clusters, leading to the conclusion that exchange of blaZ between plasmid and chromosome and between strains are rare events (Olsen et al., 2006). Our blaZ phylogenetic tree (Figure 4) also display two separate clusters. Interestingly, we observed one S. xylosus isolate with two distinct blaZ genes, one from each branch/cluster.
We identified five mecA positive S. aureus isolates from Belgium, which were selected to be included. Four isolates were from the same herd and epidemiologically related. The mecA-positive S. aureus isolates differed from the rest of the S. aureus isolates, with a larger content of antimicrobial resistance genes, but due to their epidemiological relation this result must be interpreted with caution. A higher frequency of antimicrobial resistance in methicillin-resistant S. aureus (MRSA) compared to methicillin-susceptible S. aureus (MSSA) has, however, been shown in human isolates (Thompson and Brown, 2014). Two NAS isolates also carried the mecA gene, one being S. epidermidis and one S. haemolyticus. Staphylococcus epidermidis and S. haemolyticus have previously been shown to have a higher prevalence of mecA compared to other staphylococcal species (McManus et al., 2015; Nobrega et al., 2018). In addition, all S. sciuri harbored the mecA1 gene, while the mecA2 and mecC2 were found in one S. vitulinus and one S. xylosus, respectively. There is support for a theory suggesting that mecA evolved from native mec genes in species of the S. sciuri group (Couto et al., 1996; Zhou et al., 2008; Antignac and Tomasz, 2009). The mecA1 gene, thought to be ubiquitous in S. sciuri, is believed to be the most ancestral form of mecA and shares 85% nucleotide identity with S. aureus mecA, while the mecA2 of S. vitulinus is an intermediate form with 94% homology (Miragaia, 2018). However, neither mecA1 nor mecA2 generally confers methicillin resistance (Couto et al., 1996; Wu et al., 1996; Miragaia, 2018). The mecC2 gene, a mecC allotype with 92.9% identity with the S. aureus mecC, has previously been described in S. saprophyticus (Małyszko et al., 2014).
Several staphylococcal species in our study also harbored the macrolide resistance genes ermA-C. In the S. aureus isolates only ermA was found, while ermC and to some extent ermB were found in the NAS species. The ermA-positive S. aureus isolates were also mecA positive, labeling them as MRSA. This concurs with the results of a previous study that found ermA to be more prevalent than ermB and ermC in MRSA isolates and the ermC to be more prevalent than ermA and ermB in NAS (Lina et al., 1999).
Several studies have supported the role of drug efflux in the development of antimicrobial resistance in S. aureus (DeMarco et al., 2007; Kwak et al., 2013; Santos Costa et al., 2015). Especially multidrug efflux pumps are of interest, being able to remove several chemically different substances and often linked to multidrug resistant phenotypes (Piddock, 2006; Poole, 2007). The major facilitator superfamily multidrug efflux transporter norA is the best studied efflux system in S. aureus and is associated with resistance to fluoroquinolones and several antiseptics and disinfectants (Costa et al., 2018). The gene is believed to be a part of the core genome of S. aureus (Costa et al., 2018), while Nobrega et al. (2018) reported the norA gene in 91% of NAS isolates. Consequently, our finding of the norA gene in all isolates, both in S. aureus and NAS, is coherent with these previous data.
Many antimicrobial resistance genes were detected in NAS isolates from clade D. Multidrug resistant S. epidermidis and S. haemolyticus, both from clade D, have previously been isolated from both humans and animals (Anthonisen et al., 2002; Taponen et al., 2016; Lee et al., 2018; Nobrega et al., 2018; Fergestad et al., 2021). However, both the preselection of isolates for whole-genome sequencing and the fact that clade D was the clade with the highest number of isolates, might be skewing the observed distribution of resistance genes in our material.
Regarding virulence there are many factors involved in colonization, infection, and bacterial survival. Adhesion is one of the first steps leading to colonization and infection and the process is also needed for biofilm formation. We analyzed the genome data for the presence of 28 adherence and biofilm associated virulence factor genes. The atl gene, most frequently observed in NAS, is involved in biofilm formation through initiating adherence, followed by production of polysaccharide intracellular adhesins encoded by the ica-operon, forming a polysaccharide-based biofilm (Naushad et al., 2019). Deviating results about the distribution of ica genes in bovine NAS have been reported (Piessens et al., 2012; Tremblay et al., 2013), and a study concerning the icaA genes of nine food-related NAS species showed considerable sequence diversity between strains of the same species (Møretrø et al., 2003). Diverging sequences could explain differences in the detection of ica genes. Our observation of the frequent detection of icaA followed by icaC and icaD resembles the findings of Naushad et al. (2019) who found icaC, followed by icaA and icaD to be most frequent. The whole ica operon were detected in all S. aureus isolates in this study, similar to findings from other studies (Melchior et al., 2009, 2011). In human staphylococcal strains, biofilms associated with the ica-operon are often related to infections in foreign devices, leading to the hypothesis that the ica genes in bovine isolates could play a role outside the udder, by promoting adhesion to abiotic surfaces, such as milking equipment (Melchior et al., 2011).
The production of exoenzymes further facilitates colonization and infection. Following adhesion, exoenzymes contributes to disable the host immune system, damage tissue and acquire nutrients (Tam and Torres, 2019). The thermonuclease gene nuc was the most frequently observed exoenzyme gene in NAS species in our study, consistent with recent studies (Åvall-Jääskeläinen et al., 2018; Naushad et al., 2019). The aur gene and both lipase gene geh and lip were frequently observed in the NAS group, however, the lipase genes only in clades C, D and E. These results also concur with the findings by Naushad et al. (2019), who found these genes to be frequently distributed in clades B to E. In addition, similar to Naushad et al. (2019), we detected vWbp in S. chromogenes, which could explain the variable coagulase test results for this species (Dos Santos et al., 2016). A large proportion of the S. aureus isolates in this study contained most of the sought exoenzymes. It is well known that S. aureus can produce a vast variety of exoenzymes, degrading host and bacterial molecules to escape the host immune system and gain nutrients, contributing to the success of the pathogen (Tam and Torres, 2019).
Staphylococci, especially S. aureus, have several host immune evasion virulence factors, such as genes allowing production of capsular polysaccharides, enabling bacterial survival and dissemination by hindering phagocytosis and increasing virulence (Kuipers et al., 2016). The cap5A-P and cap8A-P are prevalent in S. aureus of bovine origin (Salimena et al., 2016). In our study, capP was present in all NAS isolates, while capA-D, capM and capO were present in almost half of the NAS isolates. This deviates some from the findings of Naushad et al. (2019), who found capM to be most frequent and capA-L in low frequencies. Except capD, all cap genes were present in all S. aureus isolates, however, capD was also present in most S. aureus isolates.
In addition to the capsular genes, staphylococci can produce other important immune evasion virulence factors, such as chp, scn, spa and sbi. The chemotaxis inhibitory protein (encoded by chp) and staphylococcal complement inhibitor (encoded by scn) are mostly believed to be found in staphylococci from human sources (Verkaik et al., 2011). Consistent with this finding, we did not detect the chp gene in any isolate. However, in accordance with the study by Naushad et al. (2019), we identified scn in species of clade B. The scn gene was also detected in all except one of the S. aureus isolates.
Iron is an essential micronutrient involved in several metabolic processes, vital for bacterial survival and growth (Sheldon and Heinrichs, 2015). During infection, the host withdraws free iron from body fluids to suppress pathogens (Haley and Skaar, 2012; Sheldon and Heinrichs, 2015). Mechanisms to acquire iron in a situation where the supply is scarce are well studied in S. aureus, who can take up iron directly from molecules using isd genes and produce siderophores along with surface transporters (Sheldon and Heinrichs, 2015). We did indeed detect all iron uptake and metabolism genes, except isdA, in all S. aureus isolates. In the NAS species, however, ABC transporter and staphyloferrin A genes were more frequently detected, compared to isd genes and staphyloferrin B genes. This is in accordance with the study by Naushad et al. (2019), and supports their hypothesis that staphyloferrin A production is the principal mechanism for iron acquisition in NAS.
The production of toxins is another important determinant of virulence in staphylococci, especially in S. aureus. These toxins, such as cytotoxins (hemolysins, leukotoxins, and leukocidins) and superantigens [enterotoxins, exfoliative toxins and toxic shock syndrome toxins (TSST)], promotes inflammation and leukocyte cell death (Tam and Torres, 2019). Of the cytotoxins, we detected hlb, encoding beta hemolysin, in all S. epidermidis isolates (clade D), similar to Naushad et al. (2019), who found hlb to be the most frequent hemolysin. Surprisingly, we did not detect hlb in any of the S. aureus isolates, whereas hla was present in all isolates. This contrasts the findings from a study on bovine and humans S. aureus isolates where hlb appeared more common in bovine S. aureus isolates, while hla was more prevalent in the human isolates (Aarestrup et al., 1999). Similar to the study by Naushad et al. (2019), we did not detect any leukocidin genes or leukotoxin genes in the NAS isolates. Åvall-Jääskeläinen et al. (2018) found lukD in one S. simulans isolate, however, none of the other leukocidin or leukotoxin genes were detected in NAS in their study. All the S. aureus isolates in our study contained all genes for leukocidins and leukotoxins, including lukS-PV and lukF-PV. This resembles findings from another study on bovine S. aureus isolates that found leukocidin and leukotoxin genes in most isolates. However, that study did not detect any isolates carrying lukS-PV (Yamada et al., 2005). The Panton Valentine Leukocidin (PVL) genes are believed to be restricted to human strains of S. aureus (Vrieling et al., 2016) and it was surprising to find these genes in all our bovine S. aureus isolates. It is possible that this result appeared due to the method of similarity search, as there is a possibility for detecting genes that are similar to the gene in question, although not being the same gene. This could explain our results, as many of the lukS-PV and lukF-PV genes in our study had the same percentage identity in several S. aureus isolates, possibly indicating that there are sequence similarities between the genome and the genes, although the genes in question are not actually present. However, this warrants further investigation. The PVL genes have been reported in a few NAS isolates of bovine origin in India (Mahato et al., 2017). Of the exfoliative toxin genes, we detected etc in all NAS isolates and etb in all S. sciuri isolates. This contrasts the findings of Naushad et al. (2019) who found eta in all isolates of three NAS species and etb in a few isolates of S. agnetis and Åvall-Jääskeläinen et al. (2018) who also found etb in S. agnetis. However, the latter did not test for eta, etc, and etd. We did not have any S. agnetis in our collection, and it is unknown whether there are geographical differences between the distribution of the different exfoliative toxin genes or if there are other factors affecting our results. Of the exfoliative toxin genes, only the etc gene was detected in S. aureus isolates. The lack of eta and etb has been shown previously by Haveri et al. (2007), however, these were the only two exfoliative toxin genes included in this study. The eta gene has been detected in a few S. aureus isolates from bovines (Hayakawa et al., 2001).
Phenol soluble modulins (PSM) are also involved in the killing of leukocytes and can act synergistically with leukocidins (Hongo et al., 2009), contributing to the leukotoxicity of S. aureus, as well as being involved in biofilm formation (Otto, 2014; Vrieling et al., 2016). The PSMs are considered major determinants of the virulence of S. aureus and α-type PSMs are thought to be more aggressive than β-type PSMs (Otto, 2014; Naushad et al., 2019). Being encoded on the core genome, the PSMs are present in virtually all staphylococci (Otto, 2014). We detected β-type PSMs genes, encoding the least aggressive PSMs, in most NAS species, but not in species of clade A and the S. arlettae isolate (clade E). This concurs with the results reported by Naushad et al. (2019). However, in the species of clade A (S. sciuri and S. vitulinus) we detected the PSMmec gene. This is the only exception to the core genome-encoded PSMs, as the PSMmec is often found on the SCCmec cassette carrying the mecA genes conferring methicillin resistance (Qin et al., 2016). The isolates in our study in which the PSMmec was found did indeed harbor mec genes, as the S. sciuri isolates harbored the mecA1 and the S. vitulinus isolate harbored the mecA2 gene. The PSMmec has been identified in S. vitulinus carrying SCCmec previously (Monecke et al., 2012). In S. aureus PSMα1, as well as PSMβ1-6 were detected in all isolates. The finding of α-type PSM in S. aureus and not in NAS can be due to the aggressive potential of S. aureus, as the α-type PSM are considered more aggressive and this type of PSMs is mostly associated with S. aureus (Wang et al., 2007). The fact that we only detected PSMα1 and not the other PSMα can be due to limitations in the method used (similarity search) and the short sizes of PSMα, as this often does not give meaningful results (Cheung et al., 2014; Otto, 2014).
Superantigens are responsible for much of the toxicity in staphylococci. They are robust toxins, resilient to heat, proteolysis and desiccation (Spaulding et al., 2013; Tam and Torres, 2019). Toxic shock syndrome toxin gene (tsst) was not detected in the NAS isolates in our study. This concurs with several previous studies (Xu et al., 2015; Mello et al., 2016; Naushad et al., 2019). The tsst gene is mostly associated with S. aureus and has been detected in bovine S. aureus previously (Artursson et al., 2016; Vaughn et al., 2020). We only detected staphylococcal exotoxins in species of clade B (S. hyicus and S. chromogenes), however, several exotoxins were present in all S. aureus isolates. We only detected enterotoxin genes in one S. hyicus isolates (clade B). Naushad et al. (2019) also detected enterotoxin genes only in species from clade B. However, unlike Naushad et al. (2019) we did not identify any enterotoxin genes in S. chromogenes. We also detected several enterotoxin genes in S. aureus isolates. The S. aureus enterotoxins can cause acute and severe food poisoning, making it important to avoid contamination of enterotoxin-producing S. aureus isolates throughout the food production chain (Nia et al., 2016). Raw milk and cheese made of unpasteurized milk are well-known food sources of food poisoning caused by S. aureus.
Regarding the total virulence gene content, S. aureus stands out from NAS with, on average, five times as many virulence genes as NAS, highlighting the large virulence potential of S. aureus and the limited virulence potential of NAS. When looking at the total number of virulence genes in NAS, S. chromogenes had a higher virulence potential than most other species, mostly due to the presence of exotoxins. The single S. hyicus isolate stood apart with the highest virulence potential, due to host immune evasion, exotoxin and enterotoxin genes. This concurs with the findings of Naushad et al. (2019) who also found the highest virulence potential in species from clade B, including S. chromogenes and S. hyicus. Virulence is dependent on context and is often characterized by an intricate interplay between the microorganism and the host, making it difficult to predict virulence based on virulence gene content alone (Balloux et al., 2018). Nor is it a given that the virulence genes are expressed in the microorganism, even if they are present (Chaves-Moreno et al., 2016), which further complicates the matter. Considering the apparent complexity of virulence and the limited knowledge on the subject in bovine NAS, further studies on the association between virulence and clinical impact of these species is important. As several studies indicates that S. chromogenes could have a greater impact on udder health (Supré et al., 2011; Fry et al., 2014; Valckenier et al., 2019) and both this study and Naushad et al. (2019) found S. chromogenes among those with the highest virulence potential, further studies of the virulence and pathogenesis of this species should be emphasized (De Buck et al., 2021).
Regarding the S. aureus isolates, the results from VirulenceFinder supported the results obtained using the database by Naushad et al. (2019). However, some genes were not detected as frequently with the VirulenceFinder, possibly due to stricter thresholds for identity in VirulenceFinder. In addition, the VirulenceFinder returned less virulence genes compared to the database from Naushad et al. (2019). This could be because VirulenceFinder contains less virulence genes compared to the database by Naushad et al. (2019), as the database was expanded by doing blast similarity search to identify genes. Although the similarity search method does identify true genes, there is a possibility for misinterpretations due to detection of similar, but not identical genes.
In conclusion, our data support the opinion that there are more antimicrobial resistance genes in NAS compared to S. aureus. Regarding virulence, our S. aureus isolates had a higher virulence potential compared to NAS, but there are also differences in virulence gene content within the NAS group, supporting the view that NAS should not be considered as one uniform group of bacteria.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA609060.
Author Contributions
FT was in charge of the methodology, performed the methods used, and wrote sections of the manuscript. MF organized the databases, analyzed the results, and wrote the first draft of the manuscript. SD, AD, and YW contributed to the analysis of the results. All authors contributed to the conception and design of the study, the manuscript revision, and read and approved the submitted version.
Funding
This project was part of the ERA-Net Animal Health and Welfare (ANIHWA), MRSA_bacteriophages project. The project was funded by the Research Council of Norway (Project No. 258877), the Research Foundation – Flanders (FWO), and Service Public Fédéral – Santé publique, Sécurité de la Chaîne alimentaire et Environnement (Wallonia). Salary was provided by each home institution.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.715851/full#supplementary-material
Footnotes
References
Aarestrup, F. M., Larsen, H. D., Eriksen, N. H., Elsberg, C. S., and Jensen, N. E. (1999). Frequency of alpha- and beta-haemolysin in Staphylococcus aureus of bovine and human origin. A comparison between pheno- and genotype and variation in phenotypic expression. Apmis 107, 425–430.
Andreis, S. N., Perreten, V., and Schwendener, S. (2017). Novel β-lactamase bla(ARL) in Staphylococcus arlettae. mSphere 2:e00117-17. doi: 10.1128/mSphere.00117-17
Anthonisen, I. L., Sunde, M., Steinum, T. M., Sidhu, M. S., and Sørum, H. (2002). Organization of the antiseptic resistance gene qacA and Tn552-related beta-lactamase genes in multidrug- resistant Staphylococcus haemolyticus strains of animal and human origins. Antimicrob. Agents Chemother. 46, 3606–3612. doi: 10.1128/aac.46.11.3606-3612.2002
Antignac, A., and Tomasz, A. (2009). Reconstruction of the phenotypes of methicillin-resistant Staphylococcus aureus by replacement of the staphylococcal cassette chromosome mec with a plasmid-borne copy of Staphylococcus sciuri pbpD gene. Antimicrob. Agents Chemother. 53, 435–441. doi: 10.1128/aac.01099-08
Artursson, K., Söderlund, R., Liu, L., Monecke, S., and Schelin, J. (2016). Genotyping of Staphylococcus aureus in bovine mastitis and correlation to phenotypic characteristics. Vet. Microbiol. 193, 156–161. doi: 10.1016/j.vetmic.2016.08.012
Åvall-Jääskeläinen, S., Taponen, S., Kant, R., Paulin, L., Blom, J., Palva, A., et al. (2018). Comparative genome analysis of 24 bovine-associated Staphylococcus isolates with special focus on the putative virulence genes. PeerJ 6:e4560. doi: 10.7717/peerj.4560
Balloux, F., Brønstad Brynildsrud, O., van Dorp, L., Shaw, L. P., Chen, H., Harris, K. A., et al. (2018). From theory to practice: translating whole-genome sequencing (WGS) into the clinic. Trends Microbiol. 26, 1035–1048. doi: 10.1016/j.tim.2018.08.004
Becker, K., Heilmann, C., and Peters, G. (2014). Coagulase-negative staphylococci. Clin. Microbiol. Rev. 27, 870–926. doi: 10.1128/cmr.00109-13
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Cameron, M., Barkema, H. W., De Buck, J., De Vliegher, S., Chaffer, M., Lewis, J., et al. (2017). Identification of bovine-associated coagulase-negative staphylococci by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry using a direct transfer protocol. J. Dairy Sci. 100, 2137–2147. doi: 10.3168/jds.2016-12020
Cameron, M., Perry, J., Middleton, J. R., Chaffer, M., Lewis, J., and Keefe, G. P. (2018). Short communication: evaluation of MALDI-TOF mass spectrometry and a custom reference spectra expanded database for the identification of bovine-associated coagulase-negative staphylococci. J. Dairy Sci. 101, 590–595. doi: 10.3168/jds.2017-13226
Chaves-Moreno, D., Wos-Oxley, M. L., Jáuregui, R., Medina, E., Oxley, A. P., and Pieper, D. H. (2016). Exploring the transcriptome of Staphylococcus aureus in its natural niche. Sci. Rep. 6:33174. doi: 10.1038/srep33174
Cheung, G. Y., Joo, H. S., Chatterjee, S. S., and Otto, M. (2014). Phenol-soluble modulins–critical determinants of staphylococcal virulence. FEMS Microbiol. Rev. 38, 698–719. doi: 10.1111/1574-6976.12057
Condas, L. A. Z., De Buck, J., Nobrega, D. B., Carson, D. A., Naushad, S., De Vliegher, S., et al. (2017). Prevalence of non-aureus staphylococci species causing intramammary infections in Canadian dairy herds. J. Dairy Sci. 100, 5592–5612. doi: 10.3168/jds.2016-12478
Costa, S. S., Sobkowiak, B., Parreira, R., Edgeworth, J. D., Viveiros, M., Clark, T. G., et al. (2018). Genetic diversity of norA, coding for a main efflux pump of Staphylococcus aureus. Front. Genet. 9:710. doi: 10.3389/fgene.2018.00710
Couto, I., de Lencastre, H., Severina, E., Kloos, W., Webster, J. A., Hubner, R. J., et al. (1996). Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb. Drug Resist. 2, 377–391. doi: 10.1089/mdr.1996.2.377
De Buck, J., Ha, V., Naushad, S., Nobrega, D. B., Luby, C., Middleton, J. R., et al. (2021). Non-aureus staphylococci and bovine udder health: current understanding and knowledge gaps. Front. Vet. Sci. 8:658031. doi: 10.3389/fvets.2021.658031
De Visscher, A., Supré, K., Haesebrouck, F., Zadoks, R. N., Piessens, V., Van Coillie, E., et al. (2014). Further evidence for the existence of environmental and host-associated species of coagulase-negative staphylococci in dairy cattle. Vet. Microbiol. 172, 466–474. doi: 10.1016/j.vetmic.2014.06.011
De Vliegher, S., Fox, L. K., Piepers, S., McDougall, S., and Barkema, H. W. (2012). Invited review: mastitis in dairy heifers: nature of the disease, potential impact, prevention, and control. J. Dairy Sci. 95, 1025–1040. doi: 10.3168/jds.2010-4074
DeMarco, C. E., Cushing, L. A., Frempong-Manso, E., Seo, S. M., Jaravaza, T. A., and Kaatz, G. W. (2007). Efflux-related resistance to norfloxacin, dyes, and biocides in bloodstream isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 51, 3235–3239. doi: 10.1128/aac.00430-07
Dos Santos, D. C., Lange, C. C., Avellar-Costa, P., Dos Santos, K. R., Brito, M. A., and Giambiagi-deMarval, M. (2016). Staphylococcus chromogenes, a coagulase-negative Staphylococcus Species that can clot plasma. J. Clin. Microbiol. 54, 1372–1375. doi: 10.1128/jcm.03139-15
Fergestad, M. E., De Visscher, A., L’Abee-Lund, T., Tchamba, C. N., Mainil, J. G., Thiry, D., et al. (2021). Antimicrobial resistance and virulence characteristics in 3 collections of staphylococci from bovine milk samples. J. Dairy Sci. 104, 10250–10267. doi: 10.3168/jds.2020-19988
Foster, T. J., Geoghegan, J. A., Ganesh, V. K., and Höök, M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62. doi: 10.1038/nrmicro3161
Fry, P. R., Middleton, J. R., Dufour, S., Perry, J., Scholl, D., and Dohoo, I. (2014). Association of coagulase-negative staphylococcal species, mammary quarter milk somatic cell count, and persistence of intramammary infection in dairy cattle. J. Dairy Sci. 97, 4876–4885. doi: 10.3168/jds.2013-7657
Geoghegan, J. A., and Foster, T. J. (2017). Cell wall-anchored surface proteins of Staphylococcus aureus: many proteins, multiple functions. Curr. Top. Microbiol. Immunol. 409, 95–120. doi: 10.1007/82_2015_5002
Haley, K. P., and Skaar, E. P. (2012). A battle for iron: host sequestration and Staphylococcus aureus acquisition. Microbes Infect. 14, 217–227. doi: 10.1016/j.micinf.2011.11.001
Haveri, M., Roslöf, A., Rantala, L., and Pyörälä, S. (2007). Virulence genes of bovine Staphylococcus aureus from persistent and nonpersistent intramammary infections with different clinical characteristics. J. Appl. Microbiol. 103, 993–1000. doi: 10.1111/j.1365-2672.2007.03356.x
Hayakawa, Y., Hashimoto, N., Imaizumi, K., Kaidoh, T., and Takeuchi, S. (2001). Genetic analysis of exfoliative toxin A-producing Staphylococcus aureus isolated from mastitic cow’s milk. Vet Microbiol. 78, 39–48. doi: 10.1016/s0378-1135(00)00293-5
Hongo, I., Baba, T., Oishi, K., Morimoto, Y., Ito, T., and Hiramatsu, K. (2009). Phenol-soluble modulin alpha 3 enhances the human neutrophil lysis mediated by Panton-Valentine leukocidin. J. Infect. Dis. 200, 715–723. doi: 10.1086/605332
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/jcm.03617-13
Kadlec, K., Fessler, A. T., Hauschild, T., and Schwarz, S. (2012). Novel and uncommon antimicrobial resistance genes in livestock-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 18, 745–755. doi: 10.1111/j.1469-0691.2012.03842.x
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Kuipers, A., Stapels, D. A. C., Weerwind, L. T., Ko, Y. P., Ruyken, M., Lee, J. C., et al. (2016). The Staphylococcus aureus polysaccharide capsule and Efb-dependent fibrinogen shield act in concert to protect against phagocytosis. Microbiology (Reading). 162, 1185–1194. doi: 10.1099/mic.0.000293
Kwak, Y. G., Truong-Bolduc, Q. C., Bin Kim, H., Song, K. H., Kim, E. S., and Hooper, D. C. (2013). Association of norB overexpression and fluoroquinolone resistance in clinical isolates of Staphylococcus aureus from Korea. J. Antimicrob. Chemother. 68, 2766–2772. doi: 10.1093/jac/dkt286
Lee, J. Y. H., Monk, I. R., Gonçalves da Silva, A., Seemann, T., Chua, K. Y. L., Kearns, A., et al. (2018). Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat. Microbiol. 3, 1175–1185. doi: 10.1038/s41564-018-0230-7
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, L., Feng, W., Zhang, Z., Xue, H., and Zhao, X. (2015). Macrolide-lincosamide-streptogramin resistance phenotypes and genotypes of coagulase-positive Staphylococcus aureus and coagulase-negative staphylococcal isolates from bovine mastitis. BMC Vet. Res. 11:168. doi: 10.1186/s12917-015-0492-8
Lina, G., Quaglia, A., Reverdy, M. E., Leclercq, R., Vandenesch, F., and Etienne, J. (1999). Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 43, 1062–1066. doi: 10.1128/aac.43.5.1062
Lüthje, P., and Schwarz, S. (2006). Antimicrobial resistance of coagulase-negative staphylococci from bovine subclinical mastitis with particular reference to macrolide-lincosamide resistance phenotypes and genotypes. J. Antimicrob. Chemother. 57, 966–969. doi: 10.1093/jac/dkl061
Lüthje, P., von Köckritz-Blickwede, M., and Schwarz, S. (2007). Identification and characterization of nine novel types of small staphylococcal plasmids carrying the lincosamide nucleotidyltransferase gene lnu(A). J. Antimicrob. Chemother. 59, 600–606. doi: 10.1093/jac/dkm008
MacFadyen, A. C., Leroy, S., Harrison, E. M., Parkhill, J., Holmes, M. A., and Paterson, G. K. (2019). Staphylococcus pseudoxylosus sp. nov., isolated from bovine mastitis. Int. J. Syst. Evol. Microbiol. 69, 2208–2213. doi: 10.1099/ijsem.0.003416
Madhaiyan, M., Wirth, J. S., and Saravanan, V. S. (2020). Phylogenomic analyses of the Staphylococcaceae family suggest the reclassification of five species within the genus Staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five Staphylococcus species to Mammaliicoccus gen. nov., and the formal assignment of Nosocomiicoccus to the family Staphylococcaceae. Int. J. Syst. Evol. Microbiol. 70, 5926–5936. doi: 10.1099/ijsem.0.004498
Mahato, S., Mistry, H. U., Chakraborty, S., Sharma, P., Saravanan, R., and Bhandari, V. (2017). Identification of variable traits among the methicillin resistant and sensitive coagulase negative staphylococci in milk samples from Mastitic Cows in India. Front. Microbiol. 8:1446. doi: 10.3389/fmicb.2017.01446
Malachowa, N., and DeLeo, F. R. (2010). Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 67, 3057–3071. doi: 10.1007/s00018-010-0389-4
Małyszko, I., Schwarz, S., and Hauschild, T. (2014). Detection of a new mecC allotype, mecC2, in methicillin-resistant Staphylococcus saprophyticus. J. Antimicrob. Chemother. 69, 2003–2005. doi: 10.1093/jac/dku043
McManus, B. A., Coleman, D. C., Deasy, E. C., Brennan, G. I., O’ Connell, B., Monecke, S., et al. (2015). Comparative genotypes, staphylococcal cassette chromosome mec (SCCmec) genes and antimicrobial resistance amongst Staphylococcus epidermidis and Staphylococcus haemolyticus isolates from infections in humans and companion animals. PLoS One 10:e0138079. doi: 10.1371/journal.pone.0138079
Melchior, M. B., van Osch, M. H., Graat, R. M., van Duijkeren, E., Mevius, D. J., Nielen, M., et al. (2009). Biofilm formation and genotyping of Staphylococcus aureus bovine mastitis isolates: evidence for lack of penicillin-resistance in Agr-type II strains. Vet. Microbiol. 137, 83–89. doi: 10.1016/j.vetmic.2008.12.004
Melchior, M. B., van Osch, M. H., Lam, T. J., Vernooij, J. C., Gaastra, W., and Fink-Gremmels, J. (2011). Extended biofilm susceptibility assay for Staphylococcus aureus bovine mastitis isolates: evidence for association between genetic makeup and biofilm susceptibility. J. Dairy Sci. 94, 5926–5937. doi: 10.3168/jds.2011-4243
Mello, P. L., Moraes Riboli, D. F., Pinheiro, L., de Almeida Martins, L., Vasconcelos Paiva Brito, M. A., and Ribeiro de Souza da Cunha Mde, L. (2016). Detection of enterotoxigenic potential and determination of clonal profile in Staphylococcus aureus and coagulase-negative staphylococci isolated from bovine subclinical mastitis in different Brazilian states. Toxins (Basel). 8:104. doi: 10.3390/toxins8040104
Miragaia, M. (2018). Factors contributing to the evolution of mecA-Mediated β-lactam resistance in staphylococci: update and new insights from whole genome sequencing (WGS). Front. Microbiol. 9:2723. doi: 10.3389/fmicb.2018.02723
Monecke, S., Engelmann, I., Archambault, M., Coleman, D. C., Coombs, G. W., Cortez de Jäckel, S., et al. (2012). Distribution of SCCmec-associated phenol-soluble modulin in staphylococci. Mol. Cell. Probes. 26, 99–103. doi: 10.1016/j.mcp.2012.01.001
Møretrø, T., Hermansen, L., Holck, A. L., Sidhu, M. S., Rudi, K., and Langsrud, S. (2003). Biofilm formation and the presence of the intercellular adhesion locus ICA among staphylococci from food and food processing environments. Appl. Environ. Microbiol. 69, 5648–5655. doi: 10.1128/aem.69.9.5648-5655.2003
Naushad, S., Barkema, H. W., Luby, C., Condas, L. A., Nobrega, D. B., Carson, D. A., et al. (2016). Comprehensive Phylogenetic analysis of bovine non-aureus staphylococci species based on whole-genome sequencing. Front. Microbiol. 7:1990. doi: 10.3389/fmicb.2016.01990
Naushad, S., Naqvi, S. A., Nobrega, D., Luby, C., Kastelic, J. P., Barkema, H. W., et al. (2019). Comprehensive virulence gene profiling of bovine non-aureus staphylococci based on whole-genome sequencing data. mSystems 4:e00098-18. doi: 10.1128/mSystems.00098-18
Nia, Y., Mutel, I., Assere, A., Lombard, B., Auvray, F., and Hennekinne, J. A. (2016). Review over a 3-year period of European Union Proficiency tests for detection of staphylococcal enterotoxins in food matrices. Toxins (Basel). 8:107. doi: 10.3390/toxins8040107
Nobrega, D. B., Naushad, S., Naqvi, S. A., Condas, L. A. Z., Saini, V., Kastelic, J. P., et al. (2018). Prevalence and genetic basis of antimicrobial resistance in non-aureus staphylococci isolated from Canadian dairy herds. Front. Microbiol. 9:256. doi: 10.3389/fmicb.2018.00256
Olde Riekerink, R. G., Barkema, H. W., Kelton, D. F., and Scholl, D. T. (2008). Incidence rate of clinical mastitis on Canadian dairy farms. J. Dairy Sci. 91, 1366–1377. doi: 10.3168/jds.2007-0757
Olsen, J. E., Christensen, H., and Aarestrup, F. M. (2006). Diversity and evolution of blaZ from Staphylococcus aureus and coagulase-negative staphylococci. J. Antimicrob. Chemother. 57, 450–460. doi: 10.1093/jac/dki492
Østerås, O., Sølverød, L., and Reksen, O. (2006). Milk culture results in a large Norwegian survey–effects of season, parity, days in milk, resistance, and clustering. J. Dairy Sci. 89, 1010–1023. doi: 10.3168/jds.S0022-0302(06)72167-1
Otto, M. (2013). Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays. 35, 4–11. doi: 10.1002/bies.201200112
Otto, M. (2014). Phenol-soluble modulins. Int. J. Med. Microbiol. 304, 164–169. doi: 10.1016/j.ijmm.2013.11.019
Piddock, L. J. (2006). Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19, 382–402. doi: 10.1128/cmr.19.2.382-402.2006
Piessens, V., De Vliegher, S., Verbist, B., Braem, G., Van Nuffel, A., De Vuyst, L., et al. (2012). Characterization of coagulase-negative Staphylococcus species from cows’ milk and environment based on bap, icaA, and mecA genes and phenotypic susceptibility to antimicrobials and teat dips. J. Dairy Sci. 95, 7027–7038. doi: 10.3168/jds.2012-5400
Piessens, V., Van Coillie, E., Verbist, B., Supré, K., Braem, G., Van Nuffel, A., et al. (2011). Distribution of coagulase-negative Staphylococcus species from milk and environment of dairy cows differs between herds. J. Dairy Sci. 94, 2933–2944. doi: 10.3168/jds.2010-3956
Pitkälä, A., Haveri, M., Pyörälä, S., Myllys, V., and Honkanen-Buzalski, T. (2004). Bovine mastitis in Finland 2001–prevalence, distribution of bacteria, and antimicrobial resistance. J. Dairy Sci. 87, 2433–2441. doi: 10.3168/jds.S0022-0302(04)73366-4
Poole, K. (2007). Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 39, 162–176. doi: 10.1080/07853890701195262
Pyörälä, S., and Taponen, S. (2009). Coagulase-negative staphylococci-emerging mastitis pathogens. Vet. Microbiol. 134, 3–8. doi: 10.1016/j.vetmic.2008.09.015
Qin, L., McCausland, J. W., Cheung, G. Y., and Otto, M. (2016). PSM-Mec-a virulence determinant that connects transcriptional regulation, virulence, and antibiotic resistance in staphylococci. Front. Microbiol. 7:1293. doi: 10.3389/fmicb.2016.01293
Reksen, O., Sølverød, L., Branscum, A. J., and Østerås, O. (2006). Relationships between milk culture results and treatment for clinical mastitis or culling in Norwegian dairy cattle. J. Dairy Sci. 89, 2928–2937. doi: 10.3168/jds.S0022-0302(06)72565-6
Salimena, A. P., Lange, C. C., Camussone, C., Signorini, M., Calvinho, L. F., Brito, M. A., et al. (2016). Genotypic and phenotypic detection of capsular polysaccharide and biofilm formation in Staphylococcus aureus isolated from bovine milk collected from Brazilian dairy farms. Vet. Res. Commun. 40, 97–106. doi: 10.1007/s11259-016-9658-5
Santos Costa, S., Viveiros, M., Rosato, A. E., Melo-Cristino, J., and Couto, I. (2015). Impact of efflux in the development of multidrug resistance phenotypes in Staphylococcus aureus. BMC Microbiol. 15:232. doi: 10.1186/s12866-015-0572-8
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seemann, T., Edwards, R., da Silva, A. G., and Kiil, K. (2020). Shovill. Available online at: https://github.com/tseemann/shovill
Sheldon, J. R., and Heinrichs, D. E. (2015). Recent developments in understanding the iron acquisition strategies of gram positive pathogens. FEMS Microbiol. Rev. 39, 592–630. doi: 10.1093/femsre/fuv009
Spaulding, A. R., Salgado-Pabón, W., Kohler, P. L., Horswill, A. R., Leung, D. Y., and Schlievert, P. M. (2013). Staphylococcal and streptococcal superantigen exotoxins. Clin. Microbiol. Rev. 26, 422–447. doi: 10.1128/cmr.00104-12
Supré, K., Haesebrouck, F., Zadoks, R. N., Vaneechoutte, M., Piepers, S., and De Vliegher, S. (2011). Some coagulase-negative Staphylococcus species affect udder health more than others. J. Dairy Sci. 94, 2329–2340. doi: 10.3168/jds.2010-3741
Tam, K., and Torres, V. J. (2019). Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol. Spectr. 7. doi: 10.1128/microbiolspec.GPP3-0039-2018
Taponen, S., Björkroth, J., and Pyörälä, S. (2008). Coagulase-negative staphylococci isolated from bovine extramammary sites and intramammary infections in a single dairy herd. J. Dairy Res. 75, 422–429. doi: 10.1017/s0022029908003312
Taponen, S., Liski, E., Heikkilä, A. M., and Pyörälä, S. (2017). Factors associated with intramammary infection in dairy cows caused by coagulase-negative staphylococci, Staphylococcus aureus, Streptococcus uberis, Streptococcus dysgalactiae, Corynebacterium bovis, or Escherichia coli. J. Dairy Sci. 100, 493–503. doi: 10.3168/jds.2016-11465
Taponen, S., Nykäsenoja, S., Pohjanvirta, T., Pitkälä, A., and Pyörälä, S. (2016). Species distribution and in vitro antimicrobial susceptibility of coagulase-negative staphylococci isolated from bovine mastitic milk. Acta Vet. Scand. 58:12. doi: 10.1186/s13028-016-0193-8
Thompson, T., and Brown, P. D. (2014). Comparison of antibiotic resistance, virulence gene profiles, and pathogenicity of methicillin-resistant and methicillin-susceptible Staphylococcus aureus using a Caenorhabditis elegans infection model. Pathog. Glob. Health. 108, 283–291. doi: 10.1179/2047773214y.0000000155
Tremblay, Y. D., Lamarche, D., Chever, P., Haine, D., Messier, S., and Jacques, M. (2013). Characterization of the ability of coagulase-negative staphylococci isolated from the milk of Canadian farms to form biofilms. J. Dairy Sci. 96, 234–246. doi: 10.3168/jds.2012-5795
Valckenier, D., Piepers, S., De Visscher, A., Bruckmaier, R. M., and De Vliegher, S. (2019). Effect of intramammary infection with non-aureus staphylococci in early lactation in dairy heifers on quarter somatic cell count and quarter milk yield during the first 4 months of lactation. J. Dairy Sci. 102, 6442–6453. doi: 10.3168/jds.2018-15913
Vanderhaeghen, W., Piepers, S., Leroy, F., Van Coillie, E., Haesebrouck, F., and De Vliegher, S. (2015). Identification, typing, ecology and epidemiology of coagulase negative staphylococci associated with ruminants. Vet. J. 203, 44–51. doi: 10.1016/j.tvjl.2014.11.001
Vaughn, J. M., Abdi, R. D., Gillespie, B. E., and Kerro Dego, O. (2020). Genetic diversity and virulence characteristics of Staphylococcus aureus isolates from cases of bovine mastitis. Microb. Pathog. 144:104171. doi: 10.1016/j.micpath.2020.104171
Verkaik, N. J., Benard, M., Boelens, H. A., de Vogel, C. P., Nouwen, J. L., Verbrugh, H. A., et al. (2011). Immune evasion cluster-positive bacteriophages are highly prevalent among human Staphylococcus aureus strains, but they are not essential in the first stages of nasal colonization. Clin. Microbiol. Infect. 17, 343–348. doi: 10.1111/j.1469-0691.2010.03227.x
Vitali, L. A., Petrelli, D., Lamikanra, A., Prenna, M., and Akinkunmi, E. O. (2014). Diversity of antibiotic resistance genes and staphylococcal cassette chromosome mec elements in faecal isolates of coagulase-negative staphylococci from Nigeria. BMC Microbiol. 14:106. doi: 10.1186/1471-2180-14-106
Vrieling, M., Boerhout, E. M., van Wigcheren, G. F., Koymans, K. J., Mols-Vorstermans, T. G., de Haas, C. J., et al. (2016). LukMF’ is the major secreted leukocidin of bovine Staphylococcus aureus and is produced in vivo during bovine mastitis. Sci. Rep. 6:37759. doi: 10.1038/srep37759
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wang, R., Braughton, K. R., Kretschmer, D., Bach, T. H., Queck, S. Y., Li, M., et al. (2007). Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13, 1510–1514. doi: 10.1038/nm1656
Wassenaar, T. M., Ussery, D. W., and Ingmer, H. (2016). The qacC gene has recently spread between rolling circle plasmids of Staphylococcus, indicative of a novel gene transfer mechanism. Front. Microbiol. 7:1528. doi: 10.3389/fmicb.2016.01528
Wu, S., Piscitelli, C., de Lencastre, H., and Tomasz, A. (1996). Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri. Microb. Drug Resist. 2, 435–441. doi: 10.1089/mdr.1996.2.435
Wuytack, A., De Visscher, A., Piepers, S., Boyen, F., Haesebrouck, F., and De Vliegher, S. (2020). Distribution of non-aureus staphylococci from quarter milk, teat apices, and rectal feces of dairy cows, and their virulence potential. J. Dairy Sci. 103, 10658–10675. doi: 10.3168/jds.2020-18265
Xu, J., Tan, X., Zhang, X., Xia, X., and Sun, H. (2015). The diversities of staphylococcal species, virulence and antibiotic resistance genes in the subclinical mastitis milk from a single Chinese cow herd. Microb. Pathog. 88, 29–38. doi: 10.1016/j.micpath.2015.08.004
Yamada, T., Tochimaru, N., Nakasuji, S., Hata, E., Kobayashi, H., Eguchi, M., et al. (2005). Leukotoxin family genes in Staphylococcus aureus isolated from domestic animals and prevalence of lukM-lukF-PV genes by bacteriophages in bovine isolates. Vet. Microbiol. 110, 97–103. doi: 10.1016/j.vetmic.2005.07.006
Zankari, E., Allesøe, R., Joensen, K. G., Cavaco, L. M., Lund, O., and Aarestrup, F. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768. doi: 10.1093/jac/dkx217
Zhang, H. Z., Hackbarth, C. J., Chansky, K. M., and Chambers, H. F. (2001). A proteolytic transmembrane signaling pathway and resistance to beta-lactams in staphylococci. Science 291, 1962–1965. doi: 10.1126/science.1055144
Keywords: non-aureus staphylococci, Staphylococcus aureus, bovine, whole genome sequencing, antimicrobial resistance (AMR) genes, virulence genes
Citation: Fergestad ME, Touzain F, De Vliegher S, De Visscher A, Thiry D, Ngassam Tchamba C, Mainil JG, L’Abee-Lund T, Blanchard Y and Wasteson Y (2021) Whole Genome Sequencing of Staphylococci Isolated From Bovine Milk Samples. Front. Microbiol. 12:715851. doi: 10.3389/fmicb.2021.715851
Received: 27 May 2021; Accepted: 29 November 2021;
Published: 20 December 2021.
Edited by:
Mattias Collin, Lund University, SwedenReviewed by:
Suvi Taponen, University of Helsinki, FinlandMariela Srednik, University of Buenos Aires, Argentina
Copyright © 2021 Fergestad, Touzain, De Vliegher, De Visscher, Thiry, Ngassam Tchamba, Mainil, L’Abee-Lund, Blanchard and Wasteson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yngvild Wasteson, eW5ndmlsZC53YXN0ZXNvbkBubWJ1Lm5v
†Present address: Anneleen De Visscher, Flanders Research Institute for Agriculture, Fisheries and Food (ILVO), Technology and Food Science, Agricultural Engineering, Merelbeke, Belgium; Cyrille Ngassam Tchamba, Veterinary Department, Vésale Pharma, Noville-sur-Mehaigne, Belgium
 Marte Ekeland Fergestad
Marte Ekeland Fergestad Fabrice Touzain
Fabrice Touzain Sarne De Vliegher
Sarne De Vliegher Anneleen De Visscher
Anneleen De Visscher Damien Thiry
Damien Thiry Cyrille Ngassam Tchamba
Cyrille Ngassam Tchamba Jacques G. Mainil4
Jacques G. Mainil4 Trine L’Abee-Lund
Trine L’Abee-Lund Yannick Blanchard
Yannick Blanchard Yngvild Wasteson
Yngvild Wasteson