- 1Department of Computer Science, University of Maryland, College Park, MD, United States
- 2Joint Institute for Food Safety and Applied Nutrition, University of Maryland, College Park, MD, United States
- 3Center for Food Safety and Applied Nutrition, United States Food and Drug Administration, Department of Health and Human Services, College Park, MD, United States
- 4Center for Veterinary Medicine, United States Food and Drug Administration, Department of Health and Human Services, Laurel, MD, United States
- 5Multidrug Resistant Organism Repository and Surveillance Network, Walter Reed Army Institute of Research, Silver Spring, MD, United States
- 6Department of Pathology, University of Maryland School of Medicine, Baltimore, MD, United States
Carbapenems—one of the important last-line antibiotics for the treatment of gram-negative infections—are becoming ineffective for treating Acinetobacter baumannii infections. Studies have identified multiple genes (and mechanisms) responsible for carbapenem resistance. In some A. baumannii strains, the presence/absence of putative resistance genes is not consistent with their resistance phenotype—indicating the genomic factors underlying carbapenem resistance in A. baumannii are not fully understood. Here, we describe a large-scale whole-genome genotype-phenotype association study with 349 A. baumannii isolates that extends beyond the presence/absence of individual antimicrobial resistance genes and includes the genomic positions and pairwise interactions of genes. Ten known resistance genes exhibited statistically significant associations with resistance to imipenem, a type of carbapenem: blaOXA-23, qacEdelta1, sul1, mphE, msrE, ant(3”)-II, aacC1, yafP, aphA6, and xerD. A review of the strains without any of these 10 genes uncovered a clade of isolates with diverse imipenem resistance phenotypes. Finer resolution evaluation of this clade revealed the presence of a 38.6 kbp conserved chromosomal region found exclusively in imipenem-susceptible isolates. This region appears to host several HTH-type DNA binding transcriptional regulators and transporter genes. Imipenem-susceptible isolates from this clade also carried two mutually exclusive plasmids that contain genes previously known to be specific to imipenem-susceptible isolates. Our analysis demonstrates the utility of using whole genomes for genotype-phenotype correlations in the context of antibiotic resistance and provides several new hypotheses for future research.
1. Introduction
Antimicrobial resistance (AMR) is a grave healthcare challenge worldwide, causing 700,000 deaths each year (Review on Antimicrobial Resistance, 2016). A sizable proportion of AMR bacterial infections are hospital-acquired and pose a high healthcare burden in developed and developing countries alike (Chen et al., 2015). Some of these bacterial infections can be resistant to even the most powerful antibiotics available; including carbapenems which are typically considered to be one of the “last-line” antibiotics by clinicians, and specifically used in the treatment of critically-ill patients affected potentially by antimicrobial-resistant Gram-negative infections (Codjoe and Donkor, 2017). Antimicrobial resistance is caused by a wide variety of genetic factors across different pathogens; discovering and understanding these factors has been accelerated by the advancements in whole genome sequencing technologies and the development of methods for analyzing the resulting data (Boolchandani et al., 2019).
Acinetobacter baumannii, a Gram-negative and frequently multidrug-resistant pathogen, is an important nosocomial pathogen worldwide (Hamidian and Nigro, 2019). The Infectious Diseases Society of America considers A. baumannii to be among the top 6 leading causes of nosocomial infections, i.e., one of the high-priority ESKAPE pathogens (Boucher et al., 2009). The World Health Organization has marked carbapenem-resistant Acinetobacter baumannii (CRAB) as their highest priority pathogen for the research and development of novel antibiotics (Tacconelli et al., 2018). A. baumannii has the ability to proliferate readily in hospitals, particularly in intensive care units, and spread epidemically among patients (Chen et al., 2015). It can cause a broad range of severe infections, such as skin and soft tissue infections, wound infections, urinary tract infections, secondary meningitis, and ventilator-associated pneumonia (Roca et al., 2012; Nowak et al., 2017). Its ability to have intrinsic resistance and propensity to acquire resistance has led to the emergence of multidrug-resistant, extensively drug-resistant, and pan-drug resistant A. baumannii strains (Peleg et al., 2008; Evans et al., 2013). An increasing rate of carbapenem resistance was documented among A. baumannii strains from numerous hospital outbreaks (Zarrilli et al., 2013; Ben-Chetrit et al., 2018).
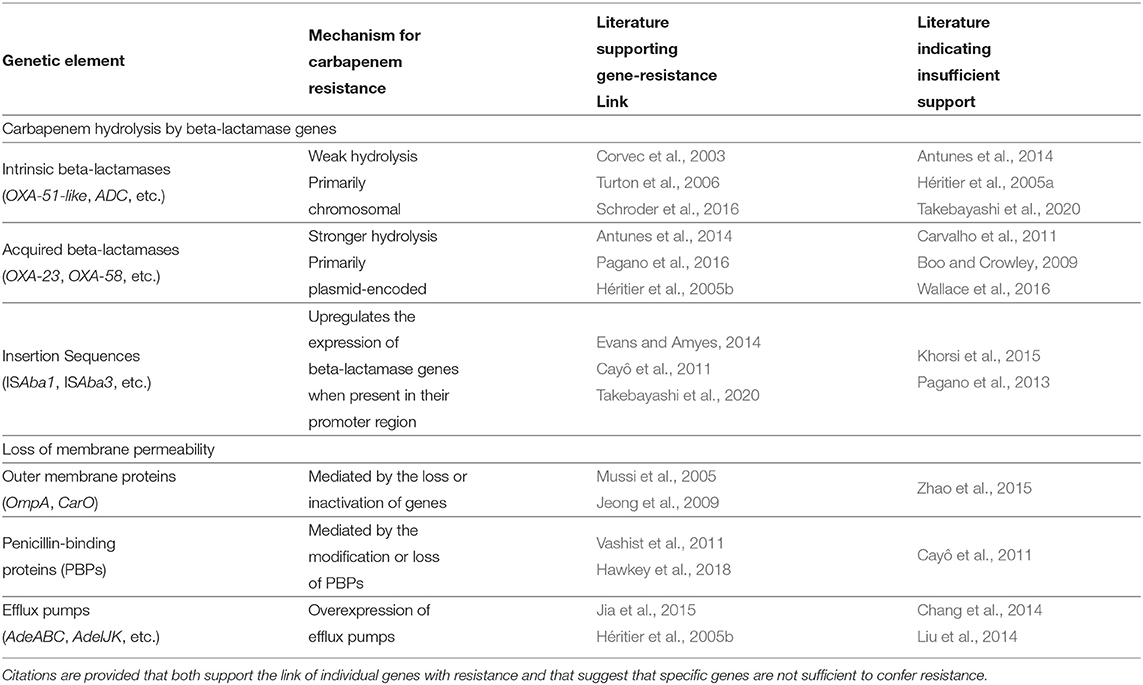
A. baumannii strains have the ability to resist the action of carbapenems via both intrinsic properties and acquired resistance factors (Evans et al., 2013). Carbapenem resistance is primarily mediated by the production of carbapenem-hydrolyzing beta-lactamases, also called carbapenemases (Poirel and Nordmann, 2006; Roca et al., 2012; Pagano et al., 2016). Beta-lactamases have been historically categorized into four molecular classes (A to D) based on conserved amino-acid motifs. Carbapenem resistance in A. baumannii is largely attributed to the class D beta-lactamases, also known as OXA-type carbapenemases (Evans and Amyes, 2014). The other major mechanism conferring carbapenem resistance in A. baumannii is the modification of membrane permeability, either by the loss or decrease in the expression of outer membrane proteins, increased expression of efflux pumps, or modifications in penicillin-binding proteins (PBPs) (Kröger et al., 2016). The known factors that influence carbapenem resistance in A. baumannii are described in Table 1.
CRAB strains are also resistant to a number of non-carbapenem antibiotics. Mechanisms of resistance include aminoglycoside-modifying enzymes, sulfonamide resistant genes, quaternary amines, other resistance genes encoded within the AbaR resistance islands (Taitt et al., 2014; Pagano et al., 2016). Similarly, CRAB strains may encode transcriptional regulator and transporter genes which are known to expedite resistance to non-carbapenem antibiotics by regulating the expressions of influx/efflux pumps, biofilm formation, outer membrane permeability, etc. (Gordon and Wareham, 2010; Chang et al., 2014) The impact of these elements on carbapenem resistance has not been fully quantified, and they do occur in carbapenem-susceptible isolates as well (Bratu et al., 2008; Bi et al., 2019). Overall, the resistance phenotype of an isolate is likely a product of the interactions of different genomic factors, and we currently lack a full understanding of the complete set of such genomic factors and their interactions. Here, we utilize a large-scale whole-genome comparison to address some gaps in our understanding of carbapenem resistance in A. baumannii.
Our work expands upon the findings of a recent study of 203 A. baumannii isolates collected as a part of an active infection control surveillance program at the University of Maryland Medical Center (UMMC) (Wallace et al., 2016). We increase the number of genomes analyzed to 349 (Supplementary Table 1 for NCBI Accessions) and expand the analysis beyond individual genes and conventional genome-wide association studies (GWAS). The analysis of the association between genotype and phenotype encompasses known resistance genes, insertion sequences, plasmids, single nucleotide polymorphisms (SNPs), and other conserved regions. Of particular interest is a subset of genetically similar strains for which the resistance phenotype could not be explained by the known determinants of carbapenem resistance. In this subset, we employ sophisticated whole genome analysis approaches and identify several genomic features that are associated with antibiotic resistance within these isolates—providing several intriguing opportunities for future research.
2. Results
2.1. Genomic Data, Antimicrobial Susceptibility Testing, and MLST
The 349 A. baumannii isolates were collected in clinical setting as a part of a surveillance program at the University of Maryland Medical Center (UMMC); these isolates had been sequenced and are accessible through NCBI (Supplementary Table 1). The level of resistance to several carbapenems was obtained using the Kirby-Bauer disk diffusion method, and phenotypes were determined according to the Clinical Laboratory Standards Institute (CLSI) breakpoints (CDC, 2017). Here, we restrict our analysis to imipenem, for which information was available for all isolates (Supplementary Table 1). Out of the 349 isolates, 305 were imipenem-resistant and 44 were imipenem-susceptible. Since the sequence assemblies available at NCBI were highly fragmented (mean contig count: 145) and had a higher number of misassemblies (mean misassemblies count: 84, mean mismatches per 100 kbp count: 1,003), we re-assembled the sequencing reads for each isolate (mean contig count: 126, mean misassemblies count: 59, mean mismatches per 100 kbp count: 729) and re-annotated the resulting assemblies, as described in Methods.
MLST sequence types (ST) were determined for the isolates using the Pasteur scheme (Diancourt et al., 2010); the resultant MLST profiles are presented in Supplementary Table 1. Two hundred and thirty-six out of the three hundred and forty-six isolates were assigned to the Pasteur ST 2, which was also the largest ST category identified in Wallace et al. (2016) study.
2.2. The Presence of Known Antimicrobial Resistance Genes
Known antimicrobial resistance (AMR) genes were found in all isolates (Supplementary Table 2). Some of the known AMR genes exhibited a strong association with the imipenem-resistant phenotype (p ≤ 0.01, Fisher’s exact test), but were also found in many imipenem-susceptible isolates as well (Supplementary Table 2), indicating that alone those genes do not explain resistance. To identify genes that are most likely to confer resistance in the absence of other factors, we focused on genes with strong statistical association with the imipenem-resistance phenotype (p ≤ 0.01) and which were infrequently found in imipenem-susceptible isolates (≤ 10 out of 44). Ten AMR genes were strongly correlated with the imipenem-resistant phenotype according to these criteria (Table 2 and Supplementary Table 1). One or more of these genes was identified in 294 (84%) A. baumannii isolates (288 imipenem-resistant and 6 imipenem-susceptible), suggesting that for the majority of the isolates (82%) the imipenem-resistant phenotype can be explained by their presence.
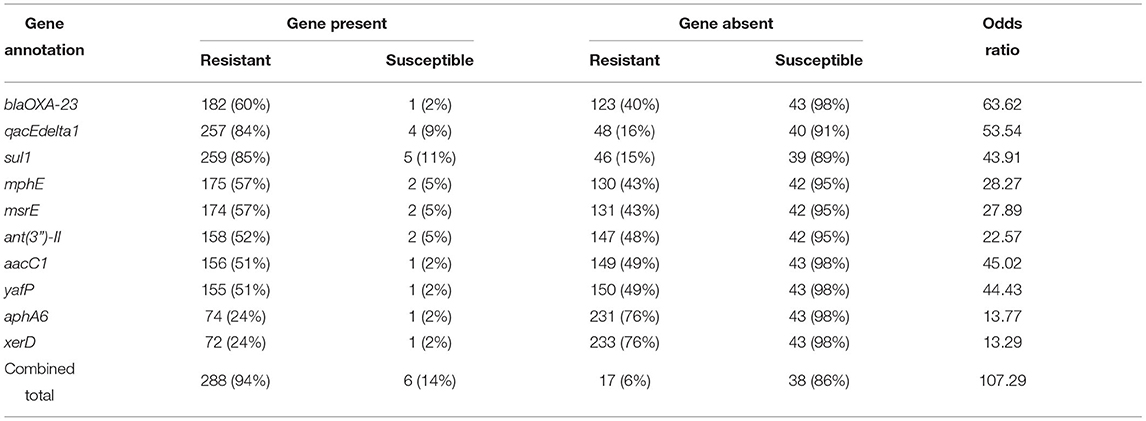
Table 2. Ten known antimicrobial resistance (AMR) genes strongly associated with imipenem-resistance in 349 A. baumannii genomes analyzed (p ≤ 0.01, Fisher’s exact test) and present infrequently in imipenem-susceptible isolates (≤10 out of 44).
Wallace et al.’s analysis of 203 A. baumannii isolates identified eight genes not previously considered to be AMR genes that were strongly associated with carbapenem resistance (Wallace et al., 2016). Within the larger set of 349 isolates (that includes the 203 strains analyzed by Wallace et al.), these genes continued to be significantly-associated with resistance (Table 3). Seven of these eight genes were detected exclusively in imipenem-resistant isolates—each of these isolates also contained one or more of the 10 strongly correlated AMR genes (Table 2).
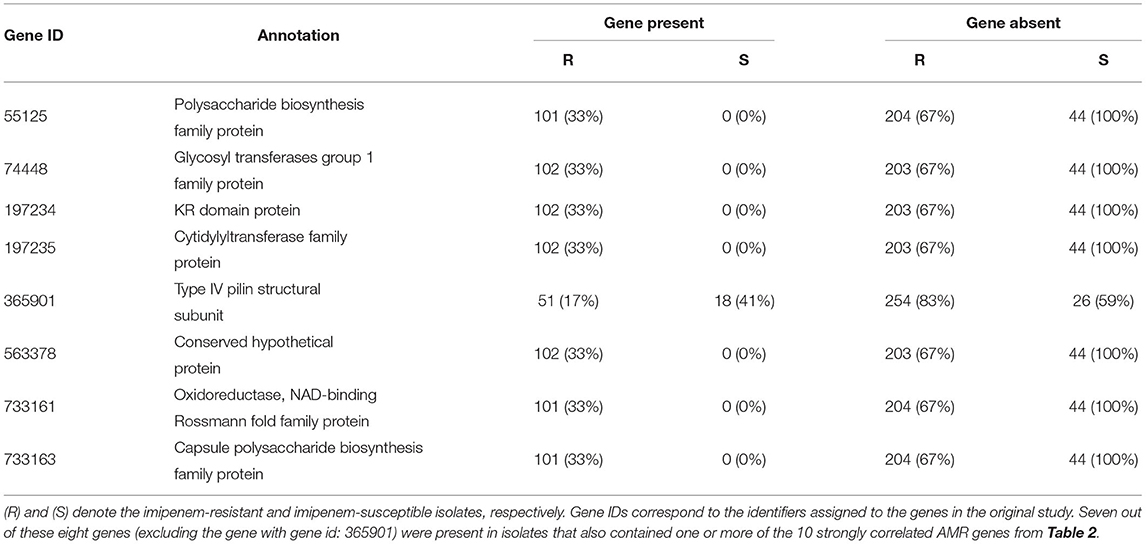
Table 3. Prevalence within our collection of isolates of genes found by Wallace et al. (2016) to be associated with imipenem-resistance in A. baumannii.
2.3. The Impact of Insertion Sequence ISAba1 on Resistance
Two beta-lactamase genes known to elevate resistance to carbapenems in A. baumannii, blaOXA-65-like (blaOXA-51 family) and blaADC, were found in all 349 isolates analyzed in our study, indicating their presence alone is not sufficient for resistance. These two genes in conjunction with an upstream insertion of a mobile genetic element, the ISAba1 insertion sequence, have been previously associated with resistance (Table 1). However, the analysis of insertion sequences and other mobile elements is complicated due to the challenges they pose to assembly algorithms (Ashton et al., 2015; Adams et al., 2016), a fact recapitulated in our data where ISAba1 was identified predominantly on a separate genomic contig. To provide a genomic context for this sequence with respect to other resistance genes, we relied on scaffolding information based on paired-reads. This information allowed us to ascertain the relative distance between ISAba1 and resistance genes (see Supplementary Table 3). The ISAba1 sequence was detected in 317 of the 349 isolates, where 296 are resistant and 21 are susceptible. As shown in Table 4, the presence of ISAba1 in the upstream region of the blaOXA-65-like and blaADC genes was strongly associated with resistance (p ≤ 0.01, Fisher’s exact test), and we also recapitulated the strong association between ISAba1 and blaOXA-23.
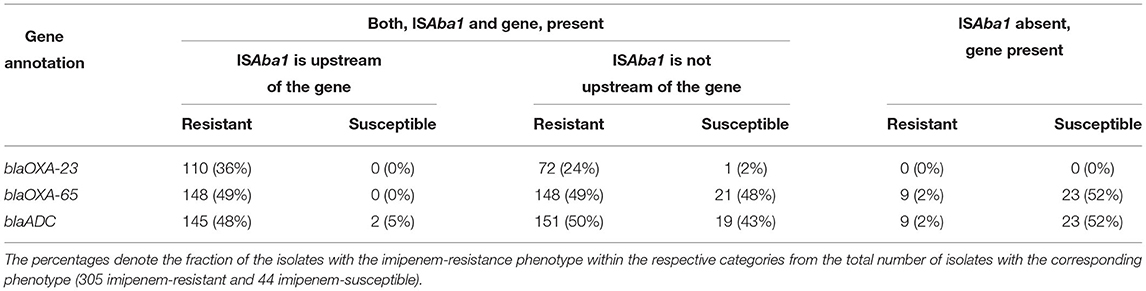
Table 4. Imipenem resistance is potentiated by the presence of the ISAba1 insertion sequence upstream of beta-lactamase genes.
2.4. Microbial Clade Lacking Known Antimicrobial Resistance Genes
Fifty five isolates (17 imipenem-resistant and 38 imipenem-susceptible) did not encode either of the 10 genes we have found to be strongly associated with imipenem-resistance. The imipenem resistance within the resistant isolates could not be directly explained by the known AMR genes, or the presence of the ISAba1 insertion sequence in the upstream region of the beta-lactamase genes. We estimated the phylogeny of all 55 isolates based on the single nucleotide polymorphism (SNP) matrix computed with the reference-based CFSAN SNP pipeline (Supplementary Figure 1) (Davis et al., 2015). This analysis revealed a clade that comprised 15 isolates: 13 of the 17 imipenem-resistant isolates and 2 imipenem-susceptible isolates. All isolates from this clade displayed similar known AMR gene profiles (Figure 1). In order to better explore the genomic differences between the imipenem-resistant and susceptible isolates from this clade, an additional 13 sequences of imipenem-susceptible strains were retrieved from the NCBI Pathogen Detection Isolates Browser (NCBI, 2021). These A. baumannii isolate sequences were located in the same SNP cluster as the genome sequences from the above-mentioned clade (cluster ID: PDS000005681). The NCBI identifiers for these 13 isolates along with their carbapenem susceptibilities and phenotypes are provided in Supplementary Table 4. As we have done for all isolates discussed in this paper, their sequences were re-assembled and subjected to gene prediction and annotation. Furthermore, we were able to obtain biological samples from 12 of these isolates and confirm the phenotype reported in the NCBI database (susceptible to imipenem, Supplementary Table 4).
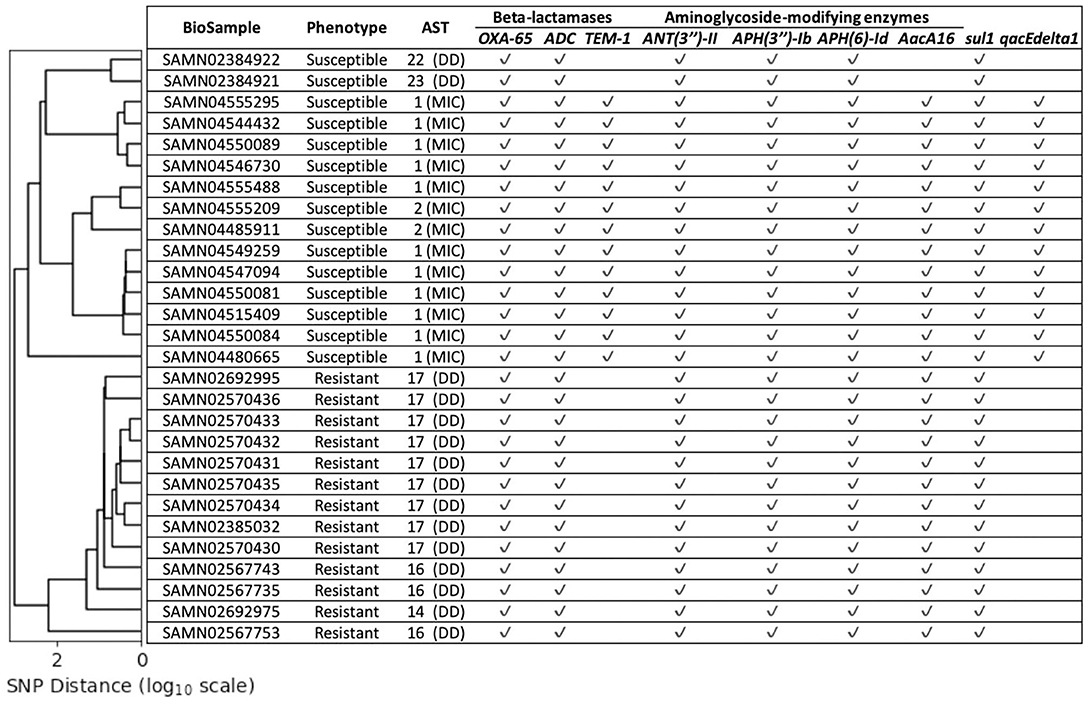
Figure 1. The lineage estimated for the chosen 28 isolates (y-axis) based on SNP distance [x-axis (log10 scale)]. The Antimicrobial Susceptibility Testing (AST) values provide the imipenem susceptibilities for the corresponding isolates measured via disk diffusion (DD) or minimum inhibitory concentration (MIC). None of these isolates were predicted to contain any of the 10 strongly correlated known AMR genes presented in Table 2, but contain several other known AMR genes. The imipenem-resistant and susceptible isolates cluster separately.
To assess whether known determinants of resistance, beyond the 10 genes described above, could determine the phenotype of these 28 isolates (15 from our study plus 13 retrieved from NCBI), we predicted their phenotype using PATRIC (v3.6.5 online) (Wattam et al., 2017). The computational predictions disagreed with the experimentally-determined phenotypes: all 13 imipenem-resistant isolates were predicted by PATRIC to be susceptible, while 3 of the imipenem-susceptible isolates were predicted to be resistant (Supplementary Table 5).
2.5. Pan-Genome Analysis for Genomic Variants Associated With Resistance Phenotypes
The 28 isolates had high overlap in gene content—among the 4,092 genes within their collective pan-genome, 3,316 (81%) constituted the core genes (genes found in at least 99% of these genomes). Among the core genes were several known AMR genes and virulence factors (Supplementary Table 6). All 28 isolates were assigned to the Pasteur ST 2 MLST profile. Despite the high genomic similarity between these isolates, the imipenem-resistant isolates clustered within a clade distinct from that of the imipenem-susceptible ones (Figure 1). The insertion sequence ISAba1 was detected upstream of the beta-lactamase genes blaOXA-65-like as well as blaADC in imipenem-resistant as well as imipenem-susceptible isolates (Supplementary Table 6). The Antimicrobial Susceptibility Testing (AST) values for these 28 isolates (Figure 1) were extremely close to the CLSI breakpoints that determined the respective imipenem-resistance phenotypes-suggesting the presence of genomic variants that influence the imipenem susceptibilities just enough to alter the phenotype in the absence of strongly associated imipenem-resistance genes.
We performed a whole-genome comparison of the isolates to identify genomic regions that are present exclusively in resistant or susceptible isolates. To improve our chances of detecting contiguous genomic segments that may contribute to imipenem-resistance, we re-sequenced 6 of the isolates using the long read Pacific Biosciences technology, obtaining complete genome sequences for these isolates (Supplementary Table 4). This analysis revealed a group of transcriptional regulators and transporters that were found exclusively within the imipenem-susceptible isolates. The transcriptional regulators were predominantly helix-turn-helix (HTH) type DNA binding transcriptional regulators, such as GntR family, IclR family, LysR family, MarR family, and TetR family. These genes were encoded in a 38,651 nt contiguous chromosomal region (Figure 2)—this region is conserved across all 15 imipenem-susceptible isolates and absent from all 13 imipenem-resistant isolates (Supplementary Table 5).
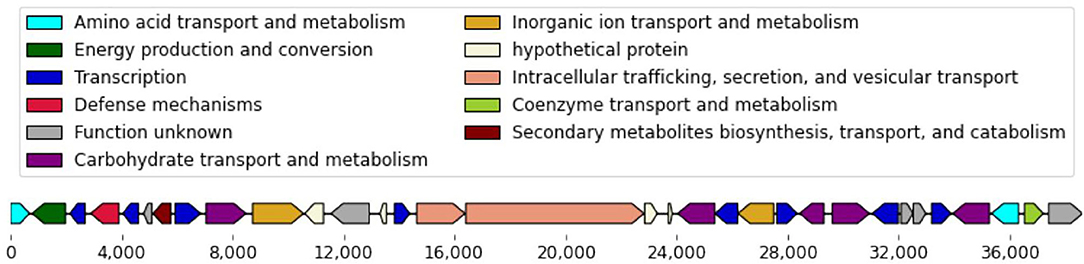
Figure 2. Genes encoded in the 38,651 nt chromosomal gene cassette. Within the chosen 28 isolates, this chromosomal gene cassette was conserved in all 15 imipenem-susceptible isolates and absent from all 13 imipenem-resistant isolates.
Whole-genome resequencing of the isolates also allowed us to identify two plasmids with mutually-exclusive presence (Supplementary Table 5). The longer plasmid is 111 kbp in length and is nearly identical to the Acinetobacter baumannii plasmid pAYP-A2 (Hawkey et al., 2018) (100% query coverage, 99.99% identity). This plasmid sequence was identified in 14 of the 15 imipenem-susceptible isolates and was missing from the remaining 14 isolates. This plasmid contained a number of genes identified by Wallace et al. (2016) to be associated with susceptibility to imipenem (Supplementary Table 2). All 14 isolates that lacked this plasmid contained another plasmid of length 14.6 kbp length. The shorter plasmid was most similar to an A. baumannii plasmid pORAB01-3 with 75% query coverage and 99.9% identity (Hujer et al., 2017). Neither plasmid encodes any of the known AMR genes. Note that the previously sequenced pORAB01-3 plasmid contains an OXA-134 family class D beta-lactamase gene OXA-237 flanked on either side by ISAba1 elements, feature absent from the corresponding plasmid identified in our isolates.
In addition to the 38.6 kbp chromosomal gene cassette and the two plasmids, the whole-genome comparisons also allowed us to locate a number of genomic variants that were strongly associated with the imipenem-resistant or susceptible isolates (p ≤ 0.01, Fisher’s exact test). These genomic variants included both SNPs and longer genomic segments of up to thousands of nucleotides in length. These include 75 distinct conserved regions that were at least 500 nt in length. Fourteen of these regions were also found in the reference A. baumannii strain [Genbank ID: NC_017162] along with 254 SNPs (Supplementary Table 8). These features were distributed fairly evenly across the reference genome (Figure 3).
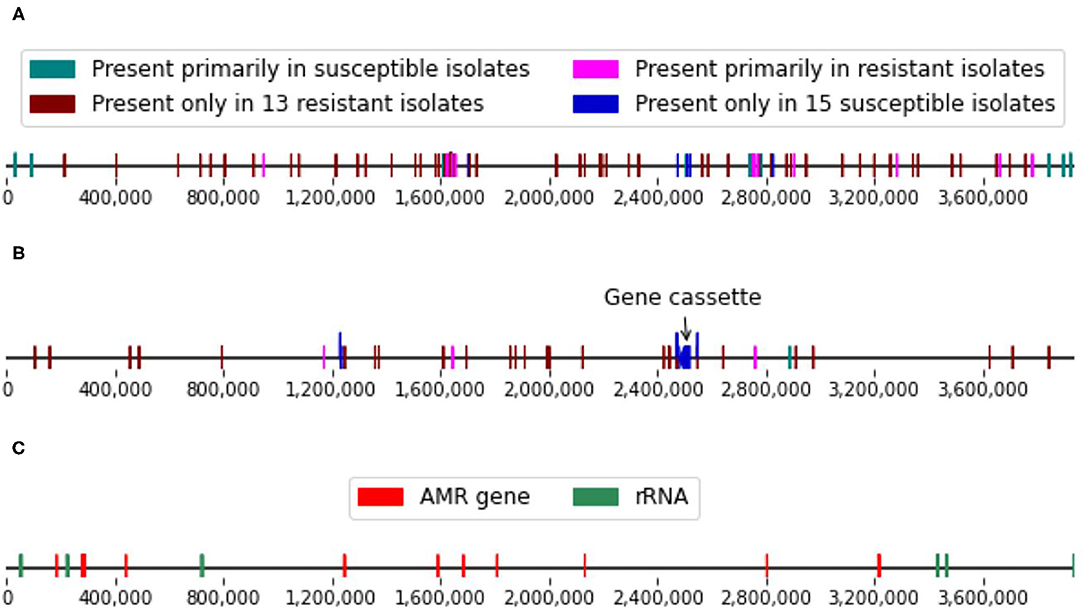
Figure 3. (A) Single nucleotide polymorphism (SNP) and (B) conserved genomic regions (≥ 500 nt in length) strongly associated with the imipenem-resistance phenotypes and located on the reference genome. (C) Locations of the known resistance genes and ribosomal RNA genes in the reference genome.
3. Discussion
Previous studies on the association of genetic elements with carbapenem resistance in A. baumannii have focused on specific genes or were confined to small numbers of isolates (Poirel and Nordmann, 2006; Li et al., 2015; Vijayakumar et al., 2020). Our study on imipenem, a type of carbapenem antibiotic, resistance is broader and provides a whole-genome analysis of 349 isolates. The whole-genome approach we employed included an analysis of pairs of genomic factors; an important consideration for factors such as ISAba1 which have the potential to affect the expression of neighboring genes. Our approach, enabled by improved genome assemblies and genome scaffolding information, also allowed the assignment of factors to either chromosomes or plasmids, providing a better understanding of the potential for mobilization of the genomic factors identified.
Our analysis identified 10 genes that were strongly associated with imipenem resistance in the A. baumannii isolates we analyzed, and confirmed the association with resistance of genes identified by Wallace et al. when analyzing a subset of the genomes we explore here. Among the 10 genes we have found to be strongly associated with resistance, only blaOXA-23 is widely believed to be involved in carbapenem resistance (Antunes et al., 2014); the other genes are associated with resistance to other antibiotics (Gutierrez et al., 2011; Alcock et al., 2020; Lin et al., 2020a), or are involved in the mobilization of resistance genes. This highlights a limitation of computational-only methods such as ours—causality can only be demonstrated experimentally. The results presented here provide several promising targets for experimental validation.
An intriguing finding of our study is that several genomic regions seem to be associated primarily with susceptibility to carbapenems even in the presence of known AMR genes. These genomic regions appear to contain a high density of transcriptional regulators, suggesting a potential role of these genes in the etiology of antimicrobial resistance in A. baumannii. Supporting the plasticity of these plasmids is the fact that a previously-sequenced variant of one of the plasmids we identified, pORAB01-3, contains an antimicrobial resistance gene that was not present in the genomes we analyzed. Given that the actual AST values of the chosen imipenem-resistant and susceptible isolates were close to the CLSI breakpoints used for determining the phenotypes, a thorough analysis of these genomic regions would be critical in understanding their roles in influencing antimicrobial susceptibility.
A second plasmid we have found to be associated with imipenem susceptibility, pAYP-A2, contains multiple genes that were statistically associated with imipenem susceptibility by Wallace et al. (2016), suggesting these genes may be in linkage disequilibrium, and highlighting a limitation of gene-centric analyses of antimicrobial resistance.
The study of AMR is critically dependent on collections of well-characterized isolates, and on the availability of high-quality genome assemblies for the isolates. Our analysis was enhanced by our use of scaffolding information generated from the paired-end sequencing data, as well as by our reconstruction of complete genome sequences for several isolates using a long read sequencing technology. Such resources enable the localization of genomic features associated with resistance within plasmids or specific chromosomal locations and allow for analyses of gene order/gene proximity that can reveal polygenic factors that impact resistance.
The strategy we have applied here does not rely on computationally-expensive multiple genome alignment approaches, rather focuses primarily on local similarities between genomes, and is, therefore, scalable and generalizable. We believe the approach we have used provides a template for similar analyses in other organisms, powered by the increasingly-available collections of isolates with well-characterized phenotypes (NCBI, 2021). Such datasets will provide fruitful sources of hypotheses for future experimental work, and expand our understanding of the genomic factors that underlie resistance to antimicrobials.
4. Materials and Methods
4.1. Primary Dataset
The A. baumannii isolates were obtained from a collection of patient surveillance samples collected as a part of a cohort study at the University of Maryland Medical Center (UMMC). These isolates were from patients treated in UMMC medical and surgical intensive care units. A total of 349 isolates were selected for evaluation. These isolates were cultured on selective media for Acinetobacter species and tested for susceptibility to imipenem using the Kirby-Bauer disk diffusion method. The phenotypes of these isolates were determined using the Clinical and Laboratory Standards Institute (CLSI) standard antimicrobial susceptibility testing guidelines: isolates with their susceptibility of at most 18 mm and at least 22 mm were considered imipenem-resistant and imipenem-susceptible respectively. The carbapenem susceptibility scores for these isolates, along with their NCBI identifiers, are provided in Supplementary Table 1. Additionally, a reference strain (Genbank ID: NC_017162) was used for the genomic analysis.
4.2. Genome Assembly, MLST Analysis, Gene Prediction, and Gene Clustering
All sequence read archive (SRA) runs for the 349 isolates were downloaded (449 SRA runs) and de novo assembled with SPAdes (v3.13.0) (default settings) (Bankevich et al., 2012). These genomes were further filtered to retain the high confidence contigs that had at least 10× sequence coverage. Accounting for the genome completeness in comparison with the reference genome, 426 assemblies were identified for the 349 A. baumannii isolates. Each of these 349 isolates was then represented by only one of its assembled genomes; the sequencing run corresponding to the representative assembled genome is the first of its associated runs mentioned in Supplementary Table 1. The quality of these selected 349 SPAdes assemblies as well as those available from NCBI were measured using QUAST (Gurevich et al., 2013)—the SPAdes assemblies were observed to be better in terms of number of contigs, misassembled contigs, unassembled contigs, variance between N50 and NG50 values, etc. (Supplementary Table 9), and were chosen for subsequent analyses.
In silico multilocus sequence type (MLST) profiles were assigned to the isolates using the sequences for the MLST markers from the Pasteur typing scheme (gltA, recA, cpn60, fusA, pyrG, rpoB, and rplB) (Diancourt et al., 2010). The genomes were scanned against the pubMLST database (http://pubmlst.org/abaumannii) using the mlst tool (Jolley and Maiden, 2010; Seemann, 2021).
Genes were then predicted from these assemblies using Prokka (v1.12) (Seemann, 2014). The gene annotations available for the Acinetobacter calcoaceticus-Acinetobacter baumannii (ACB) complex were downloaded from NCBI. These genes were clustered using CD-HIT (v4.8.1) with a length difference cut-off of 0.8 (-s 0.8) to create a custom protein database (Li and Godzik, 2006), which was then used for the gene prediction. The predicted genes were used to estimate the pan-genome for these isolates using Roary (v1.007002) with a minimum protein sequence similarity of 85% (-i 85) and gene paralogs were clustered together (-s) (Page et al., 2015). The pan-genome was also used for recognizing the assemblies that shared sufficient similarity for the downstream analysis. The known AMR genes were then identified by comparing the corresponding assemblies of these 349 isolates against CARD (Gutierrez et al., 2011; Alcock et al., 2020; Lin et al., 2020b).
4.3. Selection of Additional 13 Isolates for Chosen Microbial Clade Analysis
Among the isolates that did not host the 10 strongly correlated known AMR genes, two imipenem-susceptible isolates were genomically similar to 13 imipenem-resistant isolates within NCBI’s databases. One of these imipenem-susceptible isolates (Biosample: SAMN02384922) belonged to a SNP cluster (cluster-id: PDS000005681) on the NCBI Pathogen Detection Isolates browser (NCBI, 2021). Using this SNP cluster, 13 isolates were identified which had imipenem susceptibility information in the database. All 13 isolates were imipenem-susceptible and collected in clinical setting by the Multidrug-Resistant Organism Repository and Surveillance Network, Walter Reed Army Institute of Research (MRSN, WRAIR). The sequencing reads for these 13 isolates were downloaded and de novo assembled as before. Similarly, the genes were predicted from these genomes using Prokka. The pan-genome was estimated using Roary for the combined set of 28 isolates—13 imipenem-resistant and 15 imipenem-susceptible.
For further assessment, the 13 isolates were acquired from WRAIR. The following sections (4.3.1–4.3.3) describe the procedures undertaken on these 13 isolates for reculturing, phenotyping for carbapenem resistance, long read sequencing, and complete genome assembly.
4.3.1. Biosafety, Media, Culture, and DNA Isolation
All work associated with the preparation and extraction of materials from the multi-drug resistant isolates were performed in a BSL-2 laboratory under the class II biosafety cabinet with appropriate PPE (disposable laboratory coat, gloves, N-95 respirators, and face shields) based on risk assessment. The Acinetobacter baumannii strains were grown using BD BactoTM Tryptic Soy Broth (Soybean-Casein Digest Medium). Single colonies were inoculated in TSB and grown overnight at 37°C. Cells were centrifuged and the pellet was washing using saline 0.85%. RNA-free genomic DNA was isolated using DNeasy Blood & Tissue Kits (Qiagen, Inc.), following the manufacturer’s instructions.
4.3.2. Carbapenem Phenotype Testing
Antimicrobial susceptibility testing (AST) was performed for 12 of the 13 isolates obtained from WRAIR by broth microdilution and E-test (bioMérieux). For both broth microdilution and E-test (E-strips), we performed two culture passages for individual colony isolation on blood agar plates. A 0.5 McFarland suspension (108 CFU/ml) was prepared to make broth microdilution plates cultures and a culture lawn on blood agar plates for E-strips. AST broth microdilution panels were incubated in aerobic incubation (36 degrees C) based on CLSI protocols. We utilized the Center for Veterinary Medicine’s AST meropenem and imipenem panels (CMV4AGNF and CMV2DW) respectively. The GN panel (CMV4AGNF) requires 4 QC isolates and the ESBL panel (CMV2DW) requires 5 QC isolates as quality control organisms. Vitek 2 Compact (V2C) (bioMérieux) was used for culture identification and BMD panels were read manually using the Vizion (Sensititre, Trek Diagnostics). Culture lawn on blood agar plates for E-strips were incubated upside down in aerobic incubation (36 degrees). Interpretive Criteria for categorizing susceptibility, breakpoints for both AST broth microdilution and E-strips were adopted from the CLSI document M100.
4.3.3. Long Read Sequencing and Assembly
Six of the 13 Acinetobacter baumannii isolates, acquired from WRAIR, were sequenced on the Pacific Biosciences (PacBio) Sequel System (PacBio, Menlo Park, CA). Specifically, the DNA from the isolates were part of a 4-plex to construct multiplexed microbial SMRTbell library using the SMRTbell Template Prep Kit 1.0 (PacBio, Menlo Park, CA) according to the manufacturer’s protocol. The isolates were ligated with a unique barcode using the SMRTbell Barcoded Adapter Complete Prep Kit-96 (PacBio, Menlo Park, CA). Afterwards, size selection was performed with BluePippin (Sage Science, Beverly, MA). The multiplexed and size selected SMRTbell library was then sequenced on the PacBio Sequel sequencer using PacBio Sequel V2.0 chemistry on one Sequel SMRT cell 1M v2 (PacBio, Menlo Park, CA), with a 10h movie collection time. Raw sequencing data was demultiplexed by running the Demultiplex Barcodes application with the symmetric mode in SMRTLink v.7.0.1 (PacBio, Menlo Park, CA). The adapter sequence was trimmed and filtered out during the demultiplexing process. De novo assembly for the isolates was done using the PacBio hierarchical genome assembly process HGAP4.0 (Chin et al., 2013). The genomes of the 6 isolates were checked manually for even sequencing coverage, circularized by Circlator 1.5.5, and polished by “Resequencing” in SMRT Link v.7.0.1 to ensure > 99.99% mean consensus concordance (Hunt et al., 2015).
4.4. Comparisons of Specific AMR Associated Genes From Prior Findings
Similar to this study, a previous study by Wallace et al. had reported their findings based on genomic analysis of some other A. baumannii isolates collected as a part of the UMMC surveillance program (Wallace et al., 2016). Their analysis observed eight gene (centroid) sequences to be unique to carbapenem resistant isolates; these genes have not been previously considered to be AMR genes. The corresponding gene sequences were located in our assembled genomes using BLAST. The aggregated presence-absence counts for these eight genes is shown in Table 3.
Additionally, some other gene sequences were marked to be unique to these phenotypes within isolates grouped by their location of isolation—perirectal and sputum; these genes have also not been previously characterized as AMR genes. These comprised a total set of 385 gene sequences. We considered these genes to be strongly correlated with the resistance phenotypes, if they were reported to be unique to the corresponding resistance phenotype both overall or by source of isolation. The presence of these genes was determined across the available isolates using BLAST, and is shown along with the Fisher’s exact test statistics in Supplementary Tables 2, 6.
4.5. Genomic Structural Variants Detection and Lineage Estimation
The assembled contigs from SPAdes assemblies were scaffolded using MetaCarvel with -keep True parameter and -bsize 10 (without repeat resolution) to get the orientations of the contigs with at least 10 mate pairs linking these contigs (Ghurye et al., 2019). The relative orientations of the contigs, in conjunction with the coordinates of the genes, predicted using Prokka, facilitated the prediction of the potential presence of the insertion sequence ISAba1 (Accession:EF571004) in the upstream regions of various beta-lactamase genes.
The lineage of the isolates was estimated using Garli based on a single nucleotide polymorphism (SNP) matrix computed with the reference-based CFSAN SNP Pipeline (v2.1.0) using the reference genome (Genbank ID: NC_017162) (Zwickl, 2006; Davis et al., 2015). The CFSAN SNP Pipeline takes pair-end reads from each isolate along with the reference genome and provides a SNP distance matrix representing the pairwise proximity between the given isolates in terms of the SNP distance. This distance matrix was then used to hierarchically cluster the isolates with single linkage.
Structural variants within the aforementioned 28 similar genomes were detected via multiple whole genome alignment with Mauve (snapshot_2015-02-25 build 0) (Darling, 2004). Mauve constructs Locally Collinear Blocks (LCBs) to detect regions that are conserved across all or a subset of the given genomes. In addition, a custom script was used to detect the presence of shared k-mers (exact matching substring of length k) between these genomes and the genomic positions of these k-mers were used to locate the conserved contiguous regions. The figures for the gene cassette and the genomic locations of variants on the reference genome were made using the DNA Features Viewer library (Zulkower and Rosser, 2020).
The presence of genomic regions across the available isolates was ascertained using BLAST (blastn v2.8.1) or nucmer program from the MUMmer4 package (Altschul et al., 1990; Johnson et al., 2008; Marçais et al., 2018). The genomic region under consideration was deemed to be detected if the corresponding match resulted in the sequence identity of at least 90%.
4.6. Association With Resistance to Carbapenems
We combined the genes from the comprehensive antibiotic resistance database (CARD) and genes that have been reported to have associations with carbapenem resistance; this provided 2,650 genes that we take as the known antimicrobial resistance genes (Alcock et al., 2020). These known AMR genes were evaluated to check if their presence was statistically significantly correlated with the resistant isolates. We used Fisher’s exact test from the scipy.stats Python3 package to get the significance statistics for each gene (Virtanen et al., 2020). The entries of the 2×2 input matrix for the Fisher’s exact test corresponded to the number of isolates that were resistant and would have the predicted gene, those that are resistant and would not have the gene, and similar respective counts for the non-resistant isolates. A gene cluster was deemed to be significantly associated with resistance if the test resulted in a p ≤ 0.01. Since the number of resistant isolates largely exceeded the susceptible ones, we additionally constrained that a gene cluster was deemed significantly associated with resistant isolates if the test resulted in odds ratio ≥ 5 and was detected in ≤ 10 susceptible isolates. Among the 28 genetically similar isolates discussed before, owing to the high degree of genomic similarities, we constrained that a genomic variant (SNP or the conserved region) is strongly associated with a resistance phenotype if it is missing in at most one isolate of that phenotype (say resistant) and present in at most one isolate of the other phenotype (say susceptible).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
KJ performed the bioinformatic analyses, wrote the manuscript, and created the figures. NT and CP performed the microbiological and molecular lab work. MH and YL performed the PacBio-SMRT long read assemblies. SS performed the carbapenem phenotype testing. JKJ provided the carbapenem susceptibilities for the 349 A. baumannii isolates. JKJ, HR, PM, ES, and MP edited the manuscript. All authors read and approved the final version of the manuscript.
Funding
This project was supported by the University of Maryland, Joint Institute for Food Safety and Applied Nutrition (JIFSAN) through cooperative agreement #5U01-FD001418, provided by the U.S. Food and Drug Administration, Center for Food Safety and Applied Nutrition (FDA, CFSAN). MP was supported in part by the NIH, award # R01-AI-100947.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Multidrug-Resistant Organism Repository and Surveillance Network, Walter Reed Army Institute of Research (MRSN, WRAIR), Maryland, for providing the 13 Acinetobacter baumannii strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.714284/full#supplementary-material
References
Adams, M. D., Bishop, B., and Wright, M. S. (2016). Quantitative assessment of insertion sequence impact on bacterial genome architecture. Microb. Genom. 2:e000062. doi: 10.1099/mgen.0.000062
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Antunes, N. T., Lamoureaux, T. L., Toth, M., Stewart, N. K., Frase, H., and Vakulenko, S. B. (2014). Class D β-lactamases: are they all carbapenemases? Antimicrob. Agents Chemother. 58, 2119–2125. doi: 10.1128/AAC.02522-13
Ashton, P. M., Nair, S., Dallman, T., Rubino, S., Rabsch, W., Mwaigwisya, S., et al. (2015). MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat. Biotechnol. 33, 296–300. doi: 10.1038/nbt.3103
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Ben-Chetrit, E., Wiener-Well, Y., Lesho, E., Kopuit, P., Broyer, C., Bier, L., et al. (2018). An intervention to control an ICU outbreak of carbapenem-resistant Acinetobacter baumannii: long-term impact for the ICU and hospital. Crit. Care 22:319. doi: 10.1186/s13054-018-2247-y
Bi, D., Xie, R., Zheng, J., Yang, H., Zhu, X., Ou, H.-Y., et al. (2019). Large-Scale identification of AbaR-Type genomic islands in Acinetobacter baumannii reveals diverse insertion sites and clonal lineage-specific antimicrobial resistance gene profiles. Antimicrob. Agents Chemother. 63:e02526–18. doi: 10.1128/AAC.02526-18
Boo, T. W., and Crowley, B. (2009). Detection of blaOXA-58 and blaOXA-23-like genes in carbapenem-susceptible acinetobacter clinical isolates: should we be concerned? J. Med. Microbiol. 58(Pt 6):839–841. doi: 10.1099/jmm.0.008904-0
Boolchandani, M., D’Souza, A. W., and Dantas, G. (2019). Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 20, 356–370. doi: 10.1038/s41576-019-0108-4
Boucher, H. W., Talbot, G. H., Bradley, J. S., Edwards, J. E., Gilbert, D., Rice, L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! An update from the infectious diseases society of America. Clin. Infect. Dis. 48, 1–12. doi: 10.1086/595011
Bratu, S., Landman, D., Martin, D. A., Georgescu, C., and Quale, J. (2008). Correlation of antimicrobial resistance with β-lactamases, the OmpA-like porin, and efflux pumps in clinical isolates of Acinetobacter baumannii endemic to New York city. Antimicrob. Agents Chemother. 52:2999. doi: 10.1128/AAC.01684-07
Carvalho, K. R., Carvalho-Assef, A. P. D., Santos, L. G. D, Pereira, M. J. F., and Asensi, M. D. (2011). Occurrence of blaOXA-23 gene in imipenem-susceptible Acinetobacter baumannii. Mem. Inst. Oswaldo Cruz 106, 505–506. doi: 10.1590/S0074-02762011000400020
Cayô, R., Rodríguez, M.-C., Espinal, P., Fernández-Cuenca, F., Ocampo-Sosa, A. A., Pascual, A., et al. (2011). Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 55, 5907–5913. doi: 10.1128/AAC.00459-11
Chang, K.-C., Kuo, H.-Y., Tang, C. Y., Chang, C.-W., Lu, C.-W., Liu, C.-C., et al. (2014). Transcriptome profiling in imipenem-selected Acinetobacter baumannii. BMC Genomics 15:815. doi: 10.1186/1471-2164-15-815
Chen, C.-H., Lin, L.-C., Chang, Y.-J., Chen, Y.-M., Chang, C.-Y., and Huang, C.-C. (2015). Infection control programs and antibiotic control programs to limit transmission of multi-drug resistant Acinetobacter baumannii infections: evolution of old problems and new challenges for institutes. Int. J. Environ. Res. Public Health 12, 8871–8882. doi: 10.3390/ijerph120808871
Chin, C.-S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Codjoe, F. S., and Donkor, E. S. (2017). Carbapenem resistance: a review. Med. Sci. 6:1. doi: 10.3390/medsci6010001
Corvec, S., Caroff, N., Espaze, E., Giraudeau, C., Drugeon, H., and Reynaud, A. (2003). AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J. Antimicrob. Chemother. 52, 629–635. doi: 10.1093/jac/dkg407
Darling, A. C. E. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Davis, S., Pettengill, J. B., Luo, Y., Payne, J., Shpuntoff, A., Rand, H., et al. (2015). CFSAN SNP pipeline: an automated method for constructing SNP matrices from next-generation sequence data. PeerJ Comput. Sci. 1:e20. doi: 10.7717/peerj-cs.20
Diancourt, L., Passet, V., Nemec, A., Dijkshoorn, L., and Brisse, S. (2010). The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE 5:e10034. doi: 10.1371/journal.pone.0010034
Evans, B. A., and Amyes, S. G. B. (2014). OXA β-lactamases. Clin. Microbiol. Rev. 27, 241–263. doi: 10.1128/CMR.00117-13
Evans, B. A., Hamouda, A., and Amyes, S. G. B. (2013). The rise of carbapenem-resistant Acinetobacter baumannii. Curr. Pharmaceut. Design 19, 223–238. doi: 10.2174/138161213804070285
Ghurye, J., Treangen, T., Fedarko, M., Hervey, W. J. IV., and Pop, M. (2019). MetaCarvel: linking assembly graph motifs to biological variants. Genome Biol. 20:174. doi: 10.1186/s13059-019-1791-3
Gordon, N. C., and Wareham, D. W. (2010). Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int. J. Antimicrob. Agents 35, 219–226. doi: 10.1016/j.ijantimicag.2009.10.024
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Gutierrez, A., Elez, M., Clermont, O., Denamur, E., and Matic, I. (2011). Escherichia coli YafP protein modulates DNA damaging property of the nitroaromatic compounds. Nucleic Acids Res. 39, 4192–4201. doi: 10.1093/nar/gkr050
Hamidian, M., and Nigro, S. J. (2019). Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb. Genom. 5:e000306. doi: 10.1099/mgen.0.000306
Hawkey, J., Ascher, D. B., Judd, L. M., Wick, R. R., Kostoulias, X., Cleland, H., et al. (2018). Evolution of carbapenem resistance in Acinetobacter baumannii during a prolonged infection. Microb. Genom. 4:e000165. doi: 10.1099/mgen.0.000165
Héritier, C., Poirel, L., Fournier, P.-E., Claverie, J.-M., Raoult, D., and Nordmann, P. (2005a). Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49, 4174–4179. doi: 10.1128/AAC.49.10.4174-4179.2005
Héritier, C., Poirel, L., Lambert, T., and Nordmann, P. (2005b). Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49, 3198–3202. doi: 10.1128/AAC.49.8.3198-3202.2005
Hujer, A. M., Higgins, P. G., Rudin, S. D., Buser, G. L., Marshall, S. H., Xanthopoulou, K., et al. (2017). Nosocomial outbreak of extensively Drug-Resistant Acinetobacter baumannii isolates containing blaOXA-237 carried on a plasmid. Antimicrob. Agents Chemother. 61:e00797–17. doi: 10.1128/AAC.00797-17
Hunt, M., Silva, N. D., Otto, T. D., Parkhill, J., Keane, J. A., and Harris, S. R. (2015). Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 16:294. doi: 10.1186/s13059-015-0849-0
Jeong, H. W., Cheong, H. J., Kim, W. J., Kim, M. J., Song, K.-J., Song, J.-W., et al. (2009). Loss of the 29-kilodalton outer membrane protein in the presence of OXA-51-Like enzymes in Acinetobacter baumanniii associated with decreased imipenem susceptibility. Microb. Drug Resist. 15, 151–158. doi: 10.1089/mdr.2009.0828
Jia, W., Li, C., Zhang, H., Li, G., Liu, X., and Wei, J. (2015). Prevalence of genes of OXA-23 carbapenemase and AdeABC efflux pump associated with multidrug resistance of Acinetobacter baumannii isolates in the ICU of a comprehensive hospital of northwestern china. Int. J. Environ. Res. Public Health 12, 10079–10092. doi: 10.3390/ijerph120810079
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., and Madden, T. L. (2008). NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9. doi: 10.1093/nar/gkn201
Jolley, K. A., and Maiden, M. C. (2010). BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595
Khorsi, K., Messai, Y., Hamidi, M., Ammari, H., and Bakour, R. (2015). High prevalence of multidrug-resistance in Acinetobacter baumannii and dissemination of carbapenemase-encoding genes blaOXA-23-like, blaOXA-24-like and blaNDM-1 in Algiers hospitals. Asian Pac. J. Trop. Med. 8, 438–446. doi: 10.1016/j.apjtm.2015.05.011
Kröger, C., Kary, S., Schauer, K., and Cameron, A. (2016). Genetic regulation of virulence and antibiotic resistance in Acinetobacter baumannii. Genes 8:12. doi: 10.3390/genes8010012
Li, H., Liu, F., Zhang, Y., Wang, X., Zhao, C., Chen, H., et al. (2015). Evolution of Carbapenem-Resistant Acinetobacter baumannii revealed through Whole-Genome sequencing and comparative genomic analysis. Antimicrob. Agents Chemother. 59, 1168–1176. doi: 10.1128/AAC.04609-14
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Lin, D. L., Traglia, G. M., Baker, R., Sherratt, D. J., Ramirez, M. S., and Tolmasky, M. E. (2020a). Functional analysis of the Acinetobacter baumannii XerC and XerD site-specific recombinases: potential role in dissemination of resistance genes. Antibiotics 9:405. doi: 10.20944/preprints202006.0163.v1
Lin, D. L., Traglia, G. M., Baker, R., Sherratt, D. J., Ramirez, M. S., and Tolmasky, M. E. (2020b). Functional analysis of the XerC and XerD site-specific recombinases: potential role in dissemination of resistance genes. Antibiotics 9:405. doi: 10.3390/antibiotics9070405
Liu, F., Zhu, Y., Yi, Y., Lu, N., Zhu, B., and Hu, Y. (2014). Comparative genomic analysis of Acinetobacter baumannii clinical isolates reveals extensive genomic variation and diverse antibiotic resistance determinants. BMC Genomics 15:1163. doi: 10.1186/1471-2164-15-1163
Marçais, G., Delcher, A. L., Phillippy, A. M., Coston, R., Salzberg, S. L., and Zimin, A. (2018). MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 14:e1005944. doi: 10.1371/journal.pcbi.1005944
Mussi, M. A., Limansky, A. S., and Viale, A. M. (2005). Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob. Agents Chemother. 49, 1432–1440. doi: 10.1128/AAC.49.4.1432-1440.2005
NCBI (2021). Isolates Browser - Pathogen Detection- NCBI. Available online at: https://www.ncbi.nlm.nih.gov/pathogens/isolates#/search/ (accessed April 17, 2021).
Nowak, J., Zander, E., Stefanik, D., Higgins, P. G., Roca, I., Vila, J., et al. (2017). High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in greece, italy and spain as part of the MagicBullet clinical trial. J. Antimicrob. Chemother. 72, 3277–3282. doi: 10.1093/jac/dkx322
Pagano, M., Martins, A. F., and Barth, A. L. (2016). Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Braz. J. Microbiol. 47, 785–792. doi: 10.1016/j.bjm.2016.06.005
Pagano, M., Martins, A. F., Machado, A. B., Barin, J., and Barth, A. L. (2013). Carbapenem-susceptible Acinetobacter baumannii carrying the ISAba1 upstream blaOXA-51-like gene in Porto Alegre, southern Brazil. Epidemiol. Infect. 141:330–3. doi: 10.1017/S095026881200074X
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Peleg, A. Y., Seifert, H., and Paterson, D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582. doi: 10.1128/CMR.00058-07
Poirel, L., and Nordmann, P. (2006). Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12, 826–836. doi: 10.1111/j.1469-0691.2006.01456.x
Review on Antimicrobial Resistance (2016). Tackling Drug-resistant Infections Globally: Final Report and Recommendations.
Roca, I., Espinal, P., Vila-Farrés, X., and Vila, J. (2012). The Acinetobacter baumannii oxymoron: Commensal hospital dweller turned Pan-Drug-Resistant menace. Front. Microbiol. 3:148. doi: 10.3389/fmicb.2012.00148
Schroder, E. C., Klamer, Z. L., Saral, A., Sugg, K. A., June, C. M., Wymore, T., et al. (2016). Clinical variants of the native class d β-lactamase of Acinetobacter baumannii pose an emerging threat through increased hydrolytic activity against carbapenems. Antimicrob. Agents Chemother. 60, 6155–6164. doi: 10.1128/AAC.01277-16
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seemann, T. (2021). mlst. Available online at: https://github.com/tseemann/mlst (accessed June 22, 2021).
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Taitt, C. R., Leski, T. A., Stockelman, M. G., Craft, D. W., Zurawski, D. V., Kirkup, B. C., et al. (2014). Antimicrobial resistance determinants in Acinetobacter baumannii isolates taken from military treatment facilities. Antimicrob. Agents Chemother. 58, 767–781. doi: 10.1128/AAC.01897-13
Takebayashi, Y., Findlay, J., Heesom, K. J., Warburton, P. J., Avison, M. B., and Evans, B. A. (2020). Variability in carbapenemase activity of intrinsic OxaAb (OXA-51-like) β-lactamase enzymes in Acinetobacter baumannii. J. Antimicrob. Chemother. 76, 587–595. doi: 10.1093/jac/dkaa502
Turton, J. F., Ward, M. E., Woodford, N., Kaufmann, M. E., Pike, R., Livermore, D. M., et al. (2006). The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258, 72–77. doi: 10.1111/j.1574-6968.2006.00195.x
Vashist, J., Tiwari, V., Das, R., Kapil, A., and Rajeswari, M. R. (2011). Analysis of penicillin-binding proteins (PBPs) in carbapenem resistant Acinetobacter baumannii. Indian J. Med. Res. 133:332.
Vijayakumar, S., Wattal, C., Oberoi, J. K., Bhattacharya, S., Vasudevan, K., Anandan, S., et al. (2020). Insights into the complete genomes of carbapenem-resistant Acinetobacter baumannii harbouring bla OXA-23, bla OXA-420 and bla NDM-1 genes using a hybrid-assembly approach. Access Microbiol. 2:acmi000140. doi: 10.1099/acmi.0.000140
Virtanen, P., Gommers, R., Oliphant, T. E., Haberland, M., Reddy, T., Cournapeau, D., et al. (2020). SciPy 1.0: fundamental algorithms for scientific computing in python. Nat. Methods 17, 261–272. doi: 10.1038/s41592-020-0772-5
Wallace, L., Daugherty, S. C., Nagaraj, S., Kristie Johnson, J., Harris, A. D., and Rasko, D. A. (2016). Use of comparative genomics to characterize the diversity of Acinetobacter baumannii surveillance isolates in a health care institution. Antimicrob. Agents Chemother. 60, 5933–5941. doi: 10.1128/AAC.00477-16
Wattam, A. R., Davis, J. J., Assaf, R., Boisvert, S., Brettin, T., Bun, C., et al. (2017). Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45, D535–D542. doi: 10.1093/nar/gkw1017
Zarrilli, R., Pournaras, S., Giannouli, M., and Tsakris, A. (2013). Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents 41, 11–19. doi: 10.1016/j.ijantimicag.2012.09.008
Zhao, S.-Y., Jiang, D.-Y., Xu, P.-C., Zhang, Y.-K., Shi, H.-F., Cao, H.-L., et al. (2015). An investigation of drug-resistant Acinetobacter baumannii infections in a comprehensive hospital of east China. Ann. Clin. Microbiol. Antimicrob. 14:7. doi: 10.1186/s12941-015-0066-4
Zulkower, V., and Rosser, S. (2020). DNA features viewer: a sequence annotation formatting and plotting library for python. Bioinformatics 36, 4350–4352. doi: 10.1093/bioinformatics/btaa213
Keywords: whole-genome comparison, antibiotic resistance, genomic association studies, multiple genome alignment, Acinetobacter baumanni
Citation: Javkar K, Rand H, Hoffmann M, Luo Y, Sarria S, Thirunavukkarasu N, Pillai CA, McGann P, Johnson JK, Strain E and Pop M (2021) Whole-Genome Assessment of Clinical Acinetobacter baumannii Isolates Uncovers Potentially Novel Factors Influencing Carbapenem Resistance. Front. Microbiol. 12:714284. doi: 10.3389/fmicb.2021.714284
Received: 25 May 2021; Accepted: 01 September 2021;
Published: 01 October 2021.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Benjamin Andrew Evans, University of East Anglia, United KingdomZarizal Suhaili, Sultan Zainal Abidin University, Malaysia
Copyright © 2021 Javkar, Rand, Hoffmann, Luo, Sarria, Thirunavukkarasu, Pillai, McGann, Johnson, Strain and Pop. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Errol Strain, ZXJyb2wuc3RyYWluJiN4MDAwNDA7ZmRhLmhocy5nb3Y=; Mihai Pop, bXBvcCYjeDAwMDQwO3VtZC5lZHU=
 Kiran Javkar
Kiran Javkar Hugh Rand
Hugh Rand Maria Hoffmann
Maria Hoffmann Yan Luo
Yan Luo Saul Sarria
Saul Sarria Nagarajan Thirunavukkarasu
Nagarajan Thirunavukkarasu Christine A. Pillai
Christine A. Pillai Patrick McGann5
Patrick McGann5 Errol Strain
Errol Strain Mihai Pop
Mihai Pop