- 1College of Food Science and Light Industry, Nanjing Tech University, Nanjing, China
- 2School of Food Science and Biology Engineering, Hefei University of Technology, Hefei, China
Staphylococcus xylosus, Leuconostoc mesenteroides, Carnobacterium maltaromaticum, Leuconostoc gelidum, and Serratia liquefaciens were investigated for their roles in in the spoilage of sterilized smoked bacon. These five strains, individually and in combination, were applied as starters on sliced bacon at 4–5 log10 CFU/g using a hand-operated spraying bottle and stored for 45 days at 0–4°C. Dynamics, diversity, and succession of microbial community during storage of samples were studied by high-throughput sequencing (HTS) of the V3–V4 region of the 16S rRNA gene. A total of 367 bacterial genera belonging to 21 phyla were identified. Bacterial counts in all the inoculated specimens increased significantly within the first 15 days while the microbiota developed into more similar communities with increasing storage time. At the end of the storage time, the highest abundance of Serratia (96.46%) was found in samples inoculated with S. liquefaciens. Similarly, for samples inoculated with C. maltaromaticum and L. mesenteroides, a sharp increase in Carnobacterium and Leuconostoc abundance was observed as they reached a maximum relative abundance of 97.95 and 81.6%, respectively. Hence, these species were not only the predominant ones but could also have been the more competitive ones, potentially inhibiting the growth of other microorganisms. By analyzing the bacterial load of meat products using the SSO model, the relationships between the microbial communities involved in spoilage can be understood to assist further research.
Introduction
Bacon is widely consumed in Europe, North America, and some other parts of the world (Soladoye et al., 2015) but since they are highly susceptible to microbial contamination, thermal processing can be applied to reduce the bacterial load of meat products. However, some strains are still able to resist this heat-processing step (Li et al., 2021). For example, one report found that during refrigerated storage, microorganisms, such as Leuconostoc carnosum or Weissella viridescens survived, resulting in post-heat treatment recontamination and eventually, in spoilage (Zagdoun et al., 2020). The spoilage of cooked and cured meat is generally considered to be the result of the growth and reproduction of microbes, such as Leuconostoc spp., Lactobacillus spp., Enterobacteriaceae, Carnobacterium spp., Pseudomonas, and Brochothrix thermosphacta (Borch et al., 1996; Korkeala and Björkroth, 1997; Samelis et al., 2000; Nychas et al., 2008) which, collectively, may be known as “specific/ephemeral spoilage microorganisms-S(E)SO,” as displayed in Table 1. Previous research indicates that these microorganisms dominate the meat matrix and produce spoilage-associated changes, such as slime, -odors, and other undesirable flavors (Nychas et al., 2008; Casaburi et al., 2015).
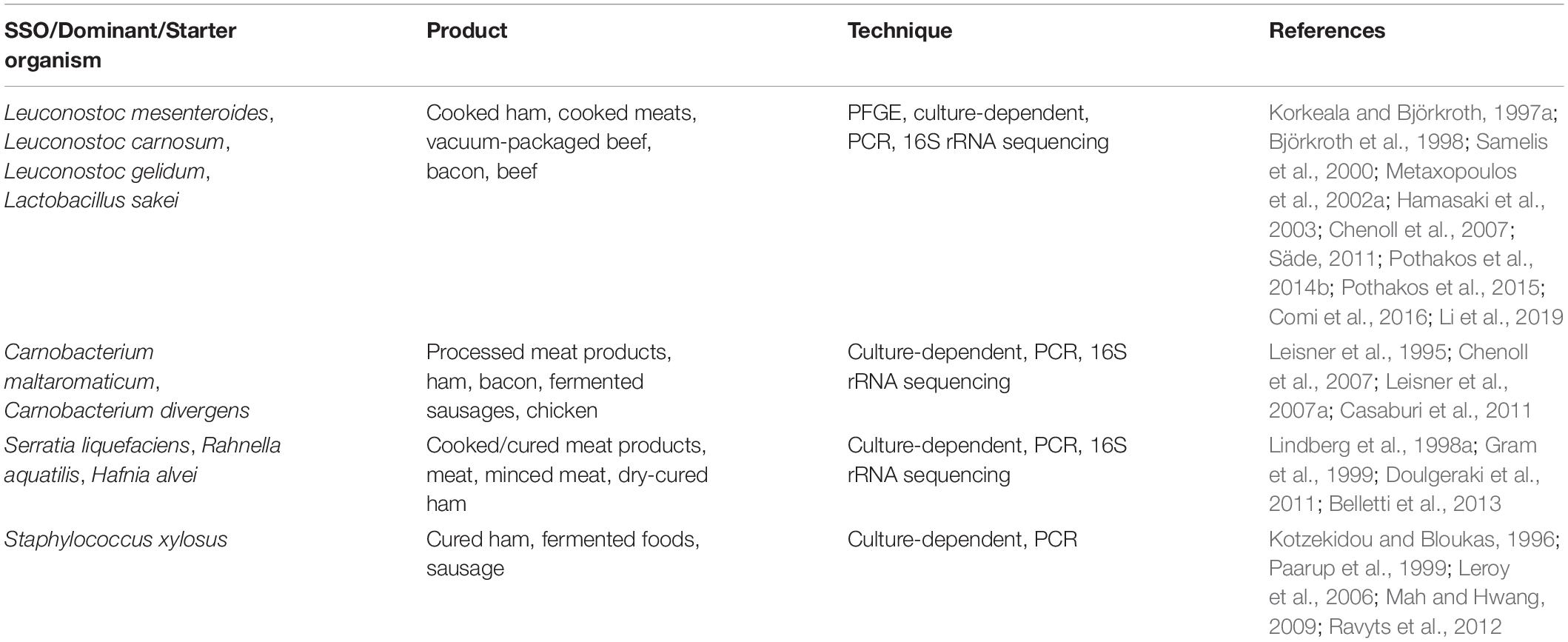
Table 1. SSO/Dominant/Starter organism and the technique used in their determination in meat and meat products.
Different microbial taxa/species may be randomly developed during meat storage, thus influencing the type of spoilage development (Ercolini et al., 2009). This is because the spoilage process is a complex event involving biological activities which are likely to be different for different microorganisms. Moreover, details on the species involved in the spoilage of meat are still unclear and needs to be further assessed. Therefore, it is necessary to characterize these organisms, both at the species as well as the biotype levels, in order to better understand the spoilage process. Furthermore, SSOs contribute to spoilage despite having an initial low population (Säde, 2011). Hence, an appropriate method for describing and understanding their growth and activity, or even for evaluating their spoilage potential is also crucial (Pothakos et al., 2014a).
In this context, Pin et al. (1999) as well as McMeekin and Ross (1996) have suggested that the method involving the inoculation of sterile substrates with spoilage organisms provided a more accurate way for representing and predicting the growth of food spoilage organisms by comparing their growth rates. In fact, over the past few years, microbiological growth and spoilage potential of isolated bacterial species have been monitored using this SSO model (Mataragas et al., 2007). For example, many studies reported its application for fish products (Stohr et al., 2001; Macé et al., 2013; Wang et al., 2017), for processed meat containing beef (Leisner et al., 1995; De Filippis et al., 2013) or saveloy (Holm et al., 2013), and even for packaged meat products (Rahkila et al., 2012). Furthermore, this SSO model was also applied to evaluate the ability of some isolates [e.g., lactic acid bacteria (LAB)] to act as protective cultures for bio-preservation (Bredholt et al., 2001; Vermeiren et al., 2004; Alves et al., 2006). However, the microbiological studies reported in the above-mentioned studies were almost carried out using a culture-dependent approach (traditional microbial cultivation). This method can be rather unreliable when trying to provide accurate information about microbial communities in an ecosystem as only a small portion of the true microbial population can be cultivated. As an alternative, culture-independent methods, especially high-throughput sequencing (HTS), has been successfully applied in meat microbiology research to monitor the dynamic changes in microbial flora as this approach can provide more detailed information about the microbial communities compared with other molecular methods. However, to the best of our knowledge, as far as bacon is concerned, only few studies have been conducted so far. Therefore, it would be useful to apply this HTS technology to analyze the growth and activity of spoilage microorganisms in bacon so that a deeper and more precise evaluation of its spoilage process can be made. This study enables us to understand the growth one taxa/species dominates the spoilage and is affected by the others.
In our previous study, Staphylococcus xylosus, Carnobacterium maltaromaticum, Leuconostoc mesenteroides, Serratia liquefaciens, and Leuconostoc gelidum were identified and considered responsible for the potential spoilage characteristics of bacon (Li et al., 2019). In this work, sterile bacon was inoculated with these five isolated organisms before investigating the bacon’s bacterial diversity using HTS, in order to gain a deeper understanding of the dynamic nature of the microbial population during the spoilage process. Furthermore, changes in the physicochemical properties of the meat were also measured to evaluate how they were connected with the microbiota.
Materials and Methods
Bacterial Strains and Sterile Samples
All strains used in this study were selected from the laboratory collection team, were previously isolated and identified from smoked bacon during refrigerated storage (Li et al., 2019), and maintained as frozen stocks at −80°C in a strain storage medium (Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China). These strains belonged to the taxonomic groups: S. xylosus, L. mesenteroides, C. maltaromaticum, L. gelidum, S. liquefaciens, and combination of the above five strains at the same concentration could be stored in a sterile vial and mixed (Pm).
All vacuum-packaged bacon samples were prepared in a local western-style meat-processing company without use of preservatives. Specimens were approximately 200 mm long, 40 mm wide, and 2.5 mm thick, with eight or nine slices (around 200 g per bag). Sterile bacon was prepared to prevent any influence of bacterial impurities. To avoid the interference from natural microbiota, samples were immediately transferred in insulated boxes containing dry ice and thereafter sterilized by irradiation at a dose of 6 kGy (Dogbevi et al., 1999; Pothakos et al., 2015; Wang et al., 2017) using a 60Co source at Hangyu (Hangyu Irradiation Technology Co., Ltd., Nanjing, China). The samples that had been irradiated were immediately transported back to the laboratory, and then stored for about 1 week at −80°C for inoculation.
Strain Culture, Sample Inoculation, and Packaging
The strains, deep-frozen before use, were reanimated, streaked by a single rolling magnetic bead driven by a magnet on the selective culture media from which the colonies were originally harvested by traditional microbiological methods, the cultures were recovered and incubated at the aforementioned temperature and time (Li et al., 2019). According to the previous method (Roig-Sagués and Eerola, 1997; Stavropoulou et al., 2018), growth of the precultures was verified twice during cultivation, then one standardized loopful of pure colonies was suspended in a tube of MRS broth, brain heart infusion (BHI) broth, and TSB (Triptone Soya Broth, Oxoid) by propagation at 30°C for 24 h. Thereafter, 0.1 mL was removed to another tube including the related broth supplemented and incubated at 30°C for 24 h anaerobically. The cell pellets were accumulated by centrifugation (4,000 rpm, 4°C, 10 min), then thrice-washed and resuspended in 50 mL of Ringer’s solution (Oxoid), thereafter the cellular concentration was adjusted to approximately 6–7 log10 CFU/mL as inoculum for the meat preparation. The five strains were added to a sterile vial and then stirred to homogenize the mixture to achieve the desired inoculation rate.
Before analysis, bacon specimens were thawed for 24 h at 0–4°C. A total of 108 samples were randomly divided into seven groups. Next, approximately equal volumes and concentrations of suspensions of strains were sprayed onto the surface of the six grouped samples by using a hand-operated spraying bottle: the initial contamination of the artificially bacon was between 4 and 5 log10 CFU/g for all strains inoculated singly or in a mix. The control group was treated by spraying sterile PBS without the addition of inoculum. The inoculation levels were selected according to previous studies of meat products (Bredholt et al., 2001; Alves et al., 2006). After inoculation, smoked bacon specimens were transferred to sterile plastic bags and vacuum-packaged in a sterile environment, then kept at 0–4°C for 45 days. The day upon which specimens were thawed and uninoculated was designated day 0. Analyses were carried out aseptically after 7, 15, 22, 30, and 45 days of storage, three packages from each treatment were randomly chosen at each time point.
Measurements of pH and Total Volatile Basic Nitrogen
The pH of bacon was monitored in triplicate by using S210 SevenCompactTM pH meter (Mettler-Toledo), and previously calibrated in standard solutions at pH 4.01 and 7.01 at room temperature. Total volatile basic nitrogen (TVB-N) was detected by the Kjeldahl method with an automatic Kjeldahl nitrogen analyzer (Shanghai Xianjian Instrument Co., Ltd., China). The TVB-N value (mg/100 g bacon) was calculated according to the utilization of hydrochloric acid (0.01 mol/L).
Microbiological Analysis
For each sample, 25 g was homogenized aseptically in 225 mL cold Ringer’s solution (Oxoid) for 2 min within a separate stomacher bag. Then the suspension was diluted (1:10) with sterile distilled water to acquire the final working dilution. After shaking, 0.1 mL of each dilution was spread on selective culture media: (i) total plate count (TPC) in Plate Count Agar (PCA) (OxoidTM), incubated at 30°C for 48 h, (ii) psychrophilic and psychrotrophic bacteria were also isolated in PCA at 7°C for 10 d, (iii) Violet Red Bile Glucose Agar (VRBGA) (Lang Bridge) for the cultivation of Enterobacteriaceae, incubated at 37°C for 36 h, (iv) LAB on de Man Rogosa and Sharpe (MRS) agar, incubated at 30°C for 48 h, and (v) Staphylococci were enumerated on Baird-Parker Agar (Lang Bridge), and incubated at 37°C for 48 h. The MRS and VRBGA agar plates were transported in 2.5-L anaerobic culture bags (Qingdao Hope Bio-Technology Co., Ltd., Shandong, China). The results are expressed as decimal logarithms of colony forming units per gram (log10 CFU/g), and the method used in this study had a lower limit of detection of 2 log10 CFU/g.
DNA Extraction, Pyrosequencing, and Data Analysis
Total DNA Extraction
The bacteria cells were analyzed following the method previously described by Li et al. (2019). The cells pellets were obtained and used to extract the total DNA according to the manufacturer’s recommendations for use of the EZNA® bacterial DNA extraction kit (Omega Bio-tek, GA, United States). DNA quality and purity were determined through spectrophotometric quantification (NanoDrop Technologies, Wilmington, DE, United States).
Illumina High-Throughput Sequencing
Microbial diversity was determined by amplifying and sequencing the hypervariable region V3-V4 of the bacterial 16S rRNA gene, using primers containing barcodes and PCR conditions as previously reported (Polka et al., 2015; Li et al., 2019). All PCR amplification reaction mixtures were examined using 2.0% agarose gel electrophoresis with a loading buffer (containing SYRB green) and mixed in equidense ratios and purified using the GeneJET PCR Purification Kit (Thermo Fisher Scientific). HTS was conducted on the Illumina MiSeq (Illumina, United States) according to the manufacturer’s specifications. Sequencing libraries were formed using an Ion Plus Fragment Library Kit 48 rxns (Thermo Fisher Scientific), following the manufacturer’s protocol. The library quality was assessed by using a Qubit@ 2.0 fluorometer (Thermo Fisher Scientific). The library was sequenced on an Ion S5TM XL platform and 400 bp/600 bp single-end reads were generated (Novogene Bioinformatics Technology Co., Ltd., Beijing, China). Raw data from a next-generation sequencing platform were submitted to the Sequence Read Archive of National Center for Biotechnology Information (NCBI), under BioProject ID PRJNA746727.
Bioinformatics and Data Analysis
Single-end reads were assigned based on their unique barcodes and truncated by removing the barcodes and primer sequences. Raw sequence reads were passed through quality filtering to obtain high-quality clean reads (Martin, 2011). Operational taxonomic units (OTUs) were assigned at 97% similarity levels using Uparse software (Edgar, 2013). Alpha-diversity and beta-diversity were calculated by QIIME software (Version 1.7.0), and the similarities were analyzed by non-metric multi-dimensional scaling (NMDS) using the R package vegan. OTUs were mapped to the SILVA database and classified according to phylum, class, order, family, and genus.
Statistical Analysis
The data pertaining to TVB-N, pH, and viable counts are presented as the mean ± SD, and significant differences in mean values were compared by one-way analysis of variance (ANOVA) (Duncan’s multiple range), with SPSS Statistics 20.0 software (SPSS Inc., Chicago, IL, United States), while P < 0.05 was considered statistically significant.
Results and Discussion
pH and Total Volatile Basic Nitrogen
The trend in pH changed slightly during the first 7 days among the groups shown in Figure 1. Compared with the controls, the final pH decreased significantly (P < 0.05) at the end of the study on day 45, reached a relatively high 6.36 for S. liquefaciens group, and relatively low values of 5.74 and 6.06 for C. maltaromaticum and L. mesenteroides groups, respectively. The microbial activity may lead to a significant decrease of the pH of the samples (Comi et al., 2016). Additionally, different strain groups appeared in different downtrends, after 45 days of storage, it was found that the acidification in bacon inoculated by C. maltaromaticum and L. mesenteroides was lower (P < 0.05) than that measured in S. liquefaciens and other groups. This pH reduction may be attributed to the fact that LAB rapidly became the predominant microorganism, producing lactic acid which decreased the pH and antibacterial peptide concentration (bacteriocins) (Gram et al., 2002; Kuo and Chu, 2003; Wang et al., 2013). It was probably S. liquefaciens, as determined by its low ability to produce the metabolites, that was associated with this acidic pH. Furthermore, the low pH may inhibit bacterial growth but the degree of inhibition varies by species. Generally, LAB and Enterobacteriaceae show high acid resistance and are able to grow and survive at acidic pH (Houtsma et al., 1996; Pin and Baranyi, 1998), however, Brochothrix thermosphacta and other species cannot grow on meat, or cause spoilage of meat under acid conditions (low pH) (Sun and Holley, 2012; Mills et al., 2014).
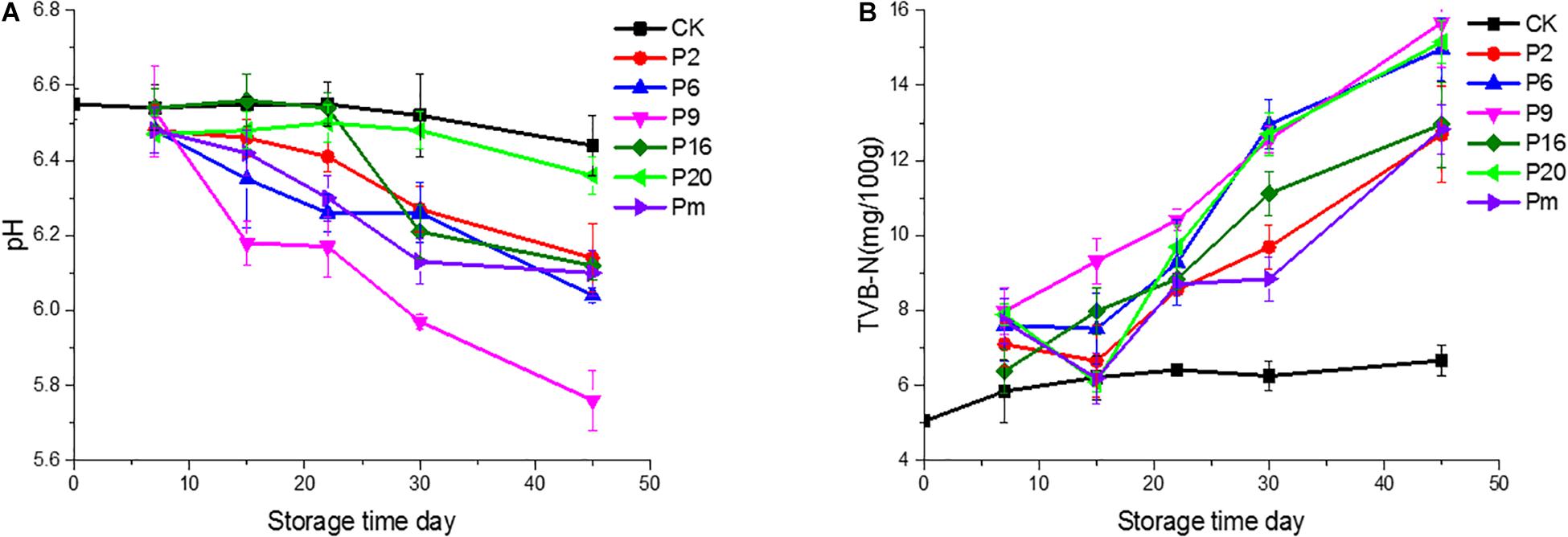
Figure 1. pH values and total volatile basic nitrogen (TVB-N) of bacon inoculated with potential spoilage bacteria, during refrigerated storage. The error bars were derived from the standard deviation between replicates (n = 3). CK, Control; P2, Staphylococcus xylosus; P6, Leuconostoc mesenteroides; P9, Carnobacterium maltaromaticum; P16, Leuconostoc gelidum; P20, Serratia liquefaciens; Pm, The five strains in combination.
As one of the important chemical indicators, the change of TVB-N values may be attributed to increased protein degradation by endogenous enzymes and bacteria (Huang et al., 2014). In this study, variable production of TVB-N was observed in the different bacterial groups and showed an increasing trend. As illustrated in Figure 1, the TVB-N values (15.68 and 15.16 mg/100 g, respectively) were increased significantly (P < 0.05) in samples inoculated with C. maltaromaticum and S. liquefaciens. The TVB-N was relatively low 12.69 for the S. xylosus group, while the control samples only reached 6.66. Thus we inferred whether C. maltaromaticum and S. liquefaciens, as active producers of TVB-N, had effective spoilage potential in a manner similar to descriptions of these strains as the main spoilage organisms in spoilage species (Stohr et al., 2001; Zhang et al., 2015).
Microbiological Analysis
Due to the different feed conditions of different bacteria, five different types of culture media that have been previously used to identify the level of microbial spoilage were used. As shown in Table 2, colonies were grown rapidly on selective culture media after incubation, and reached the lowest 7.99 log10 CFU/g on PCA medium at 30°C on day 15 except for the control groups. In general, all tested strains showed good growth and survival during storage, there were significant differences at the 0.05 level in final bacterial populations. In inoculated samples, the total viable count (TVC) and LAB counts increased rapidly to about 8 log10 CFU/g by day 15.
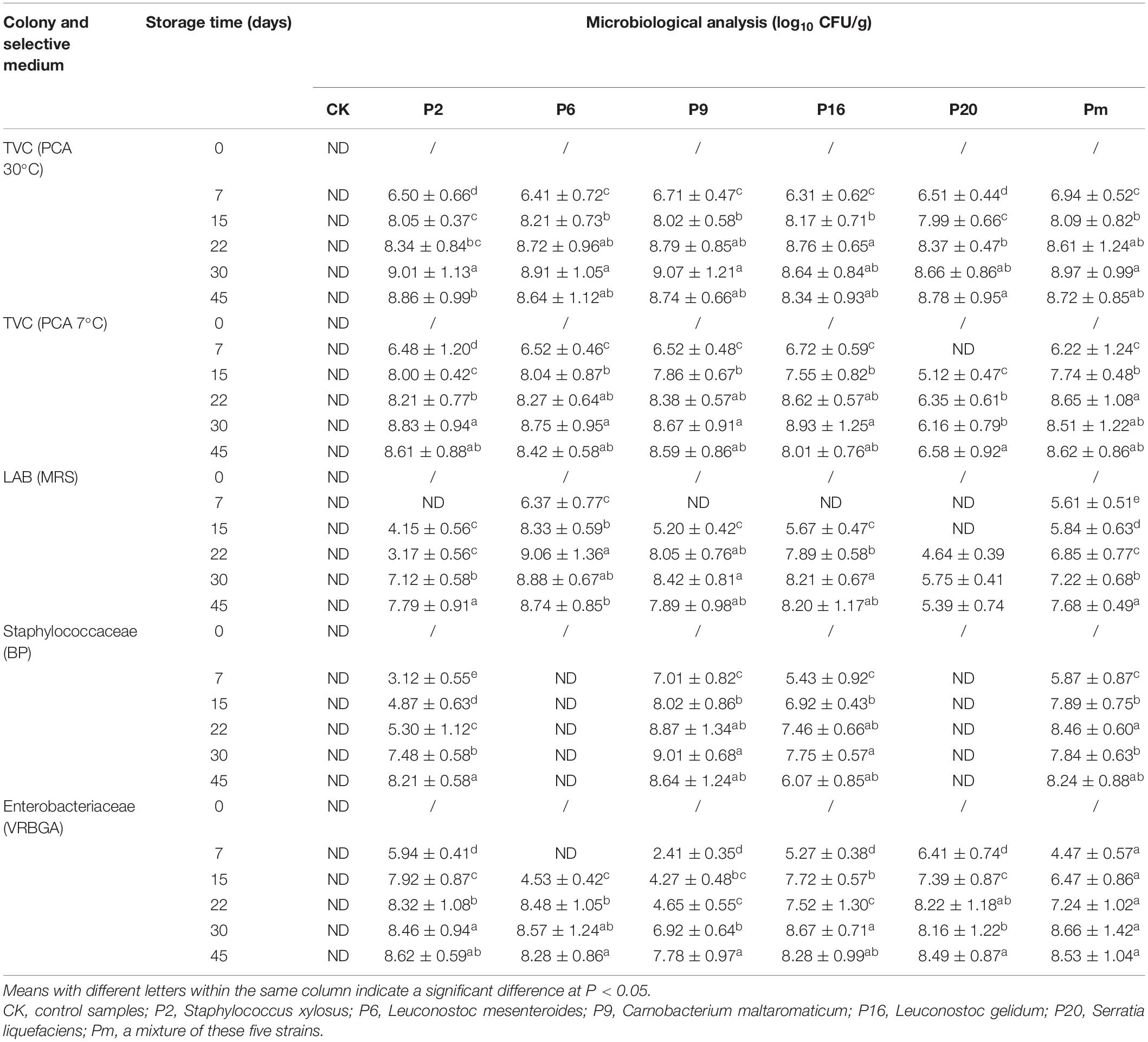
Table 2. Changes in viable counts of bacon inoculated with potential spoilage bacteria, during refrigerated storage after 45 days.
Microorganisms interacted with each other when the order of magnitude reached levels of approximately 7–9 log10 CFU/g (Gram et al., 2002). In control samples, the microorganism populations were observed to be below the detection limit (2 log10 CFU/g) after 45 days under refrigeration, which indicated that gamma irradiation was an efficient treatment when controlling the number of microorganisms and extending the shelf-life with no adverse changes or deterioration (O’bryan et al., 2008; Chen et al., 2016). Irradiated bacon was used as a suitable host material to simulate natural cooked meat of a substantially consistent nature. Sterile bacon was considered a suitable alternative model as an initial screening procedure. Staphylococcaceae populations inoculated with L. mesenteroides and S. liquefaciens at lower levels (<2 log10 CFU/g), indicated that the growth was significantly inhibited and suppressed.
Bacterial Richness and Diversity
After quality filtering and merging of paired reads, the total number of 7,752,153 effective sequences (tags) could be remained from 108 samples; sequence lengths were between 403 and 478 bp (Table 3). These effective sequences were clustered into 14,875 OTUs with 97% similarity level by using UPARSE algorithm embedded in Qiime. High Good’s coverage at least 99.7% suggested that most of the bacteria OTUs in samples could be captured. The α-diversity indices including observed OTUs, Chao1, and Shannon diversity index were calculated and shown as boxplots (Supplementary Figure 1). Shannon’s index represents the species diversity, the observed OTUs and Chao1 index reflected species richness. According to Supplementary Figure 1, these three indices decreased over storage time, and much higher bacterial diversity in control samples and samples at initial stage were observed compared with the middle-late stages of storage after day 15. This behavior could directly show that the richness and diversity of microorganisms had been decreased and a subset of bacteria became dominant. High Good’s coverage (≥97.3%) indicated that the majority of microbial phylotypes were well-captured.
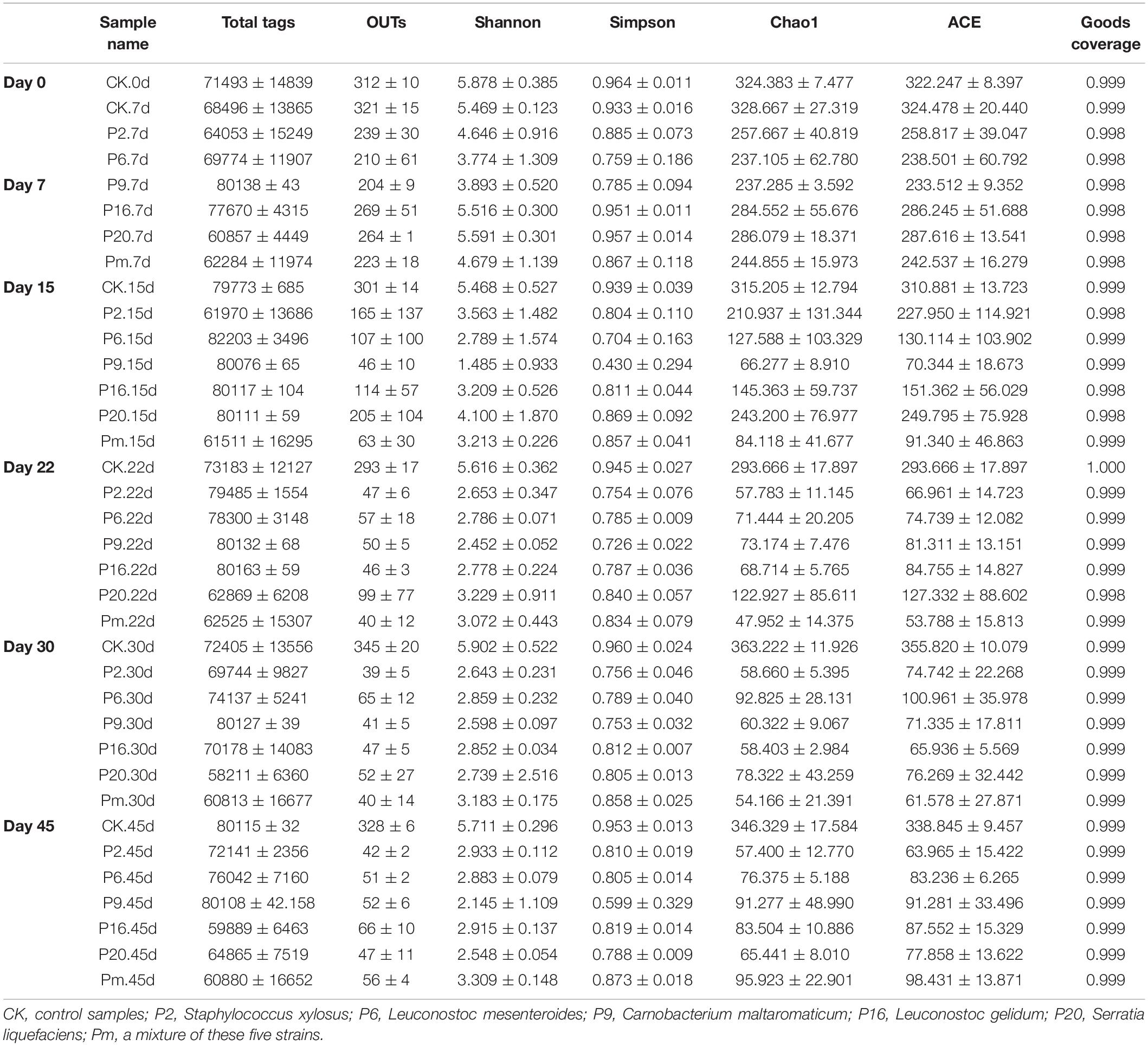
Table 3. Alpha diversity estimation of the 16S rRNA gene libraries by sequencing on an Ion S5TM XL platform in bacon.
Composition of Bacterial Community
Bacterial community variations during bacon storage revealed by 16S rRNA gene and the mean relative abundance of microbial phyla are shown in Supplementary Figure 2. A total of 21 identified phyla were observed, and Proteobacteria and Firmicutes were the dominant phyla in the microbiota, representing 78.98–99.97% of the total catch. The proportion was associated with inoculation, for example, after inoculation with S. liquefaciens, Proteobacteria continuously increased and Firmicutes decreased, the highest Firmicutes level (99.97%) was reached at day 45, and also showed a much higher percentage (96.64%) at the genus level of Serratia. After inoculation with C. maltaromaticum (P9), after 45 days of storage, Firmicutes increased up to 98.43% on day 15, then decreased to 77.45% on day 15, while Serratia increased to 22.51% of the relative total abundance of Proteobacteria. The vast majority of the top 100 genera belong to the families Firmicutes and Proteobacteria (Supplementary Figure 3).
For a more detailed analysis, bacterial community dynamics at the genus level were studied, and a total of 367 bacteria identified were summarized in Figure 2. Control samples had the highest number of OTUs (Table 3) and the lowest loads (those below the detection limit) throughout the storage period, implying more bacterial diversity and relative stability. As shown in Figure 3A, the main microbial groups were represented by Vibrio (23.38%), Psychrobacter (10.61%), Lactobacillus (6.67%), Brochothrix (5.88%), Acinetobacter (4.85%), Serratia (3.27%), Bacillus (3.17%), and Pseudomonas (2.54%). Similar microbial loads were frequently observed in processed meat products in the initial stage of storage (Chaillou et al., 2015; Quijada et al., 2018; Juárez-Castelán et al., 2019), indicating that these bacteria were primarily derived from raw meat and the processing unit (Borch et al., 1996; Hu et al., 2008; Chaillou et al., 2015). Therefore, these observations confirmed that irradiation could delay microbial growth and suppress final counts of spoilage microorganisms, extending the shelf-life stability of bacon.
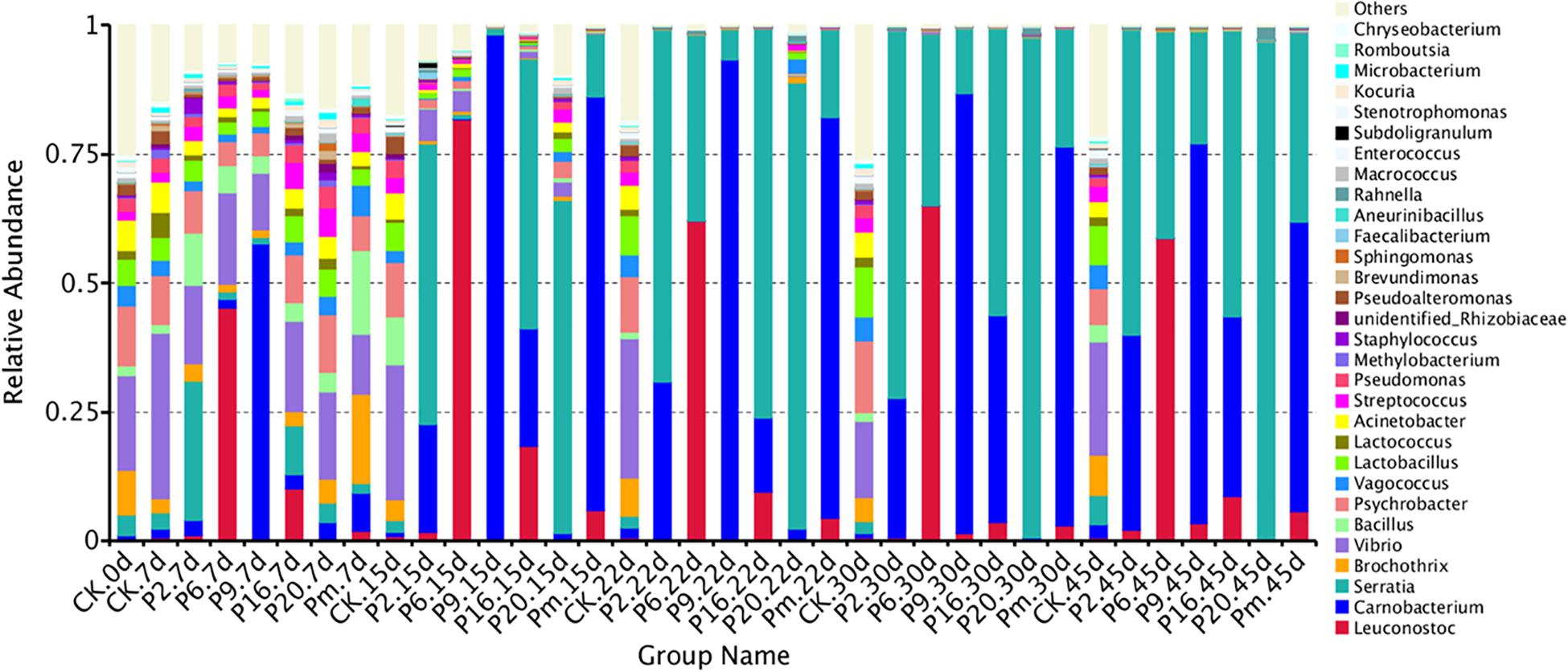
Figure 2. Dynamics in relative abundance (%) of bacterial taxa at the phylum level in bacon inoculated with potential spoilage bacteria. Each bar represents the relative abundance of each group, below the top-30 abundances (at genus level) data were merged. CK, Control; P2, Staphylococcus xylosus; P6, Leuconostoc mesenteroides; P9, Carnobacterium maltaromaticum; P16, Leuconostoc gelidum; P20, Serratia liquefaciens; Pm, The five strains in combination.
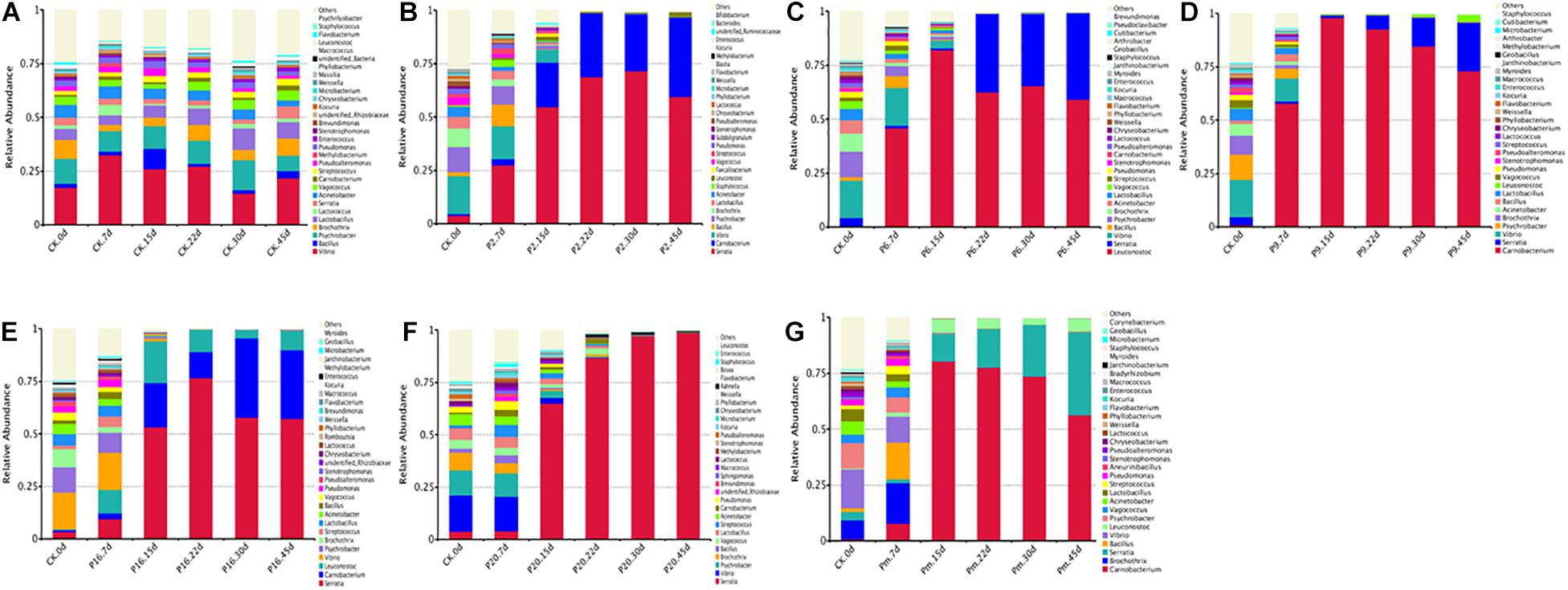
Figure 3. Dynamics in relative abundance (%) of bacterial taxa at the genus level in bacon inoculated with potential spoilage bacteria. Each bar represents the relative abundance of each group, below the top 30 abundance (at genus level) data were merged. CK, Control; P2, Staphylococcus xylosus; P6, Leuconostoc mesenteroides; P9, Carnobacterium maltaromaticum; P16, Leuconostoc gelidum; P20, Serratia liquefaciens; Pm, The five strains in combination.
Microbial communities evolved from day 7 to day 45 in bacon with inoculation during the storage process, where the microbial composition became less diverse and apparently more stable, with significantly decreased Chao 1 and Shannon diversity indices (Table 3 and Supplementary Figure 1). Furthermore, Serratia, Carnobacterium, and Leuconostoc dominated at the end of the storage, suggesting that these members had strong survival rates and competitiveness.
The bacterial community of the various inoculations significantly differed from each other at the genus level. According to Figure 3F, a rapidly increased percentage of Serratia to 96.64% dominated the microbial population, after the samples were inoculated with S. liquefaciens, during 45 days of storage. Among the Enterobacteriaceae family, Serratia spp. are the most commonly found genus in meat products and often contribute to spoilage (Doulgeraki et al., 2011, 2012). Serratia liquefaciens was found in high numbers after refrigerated storage of packages of minced meat, such that they actually could spoil the product (Lindberg et al., 1998), and comprised one of the most common species of Enterobacteria in spoiled hams (Paarup et al., 1999; Losantos et al., 2000), where they typically reached 5–7 log10 CFU/g (Gram et al., 1999). In this study, Figure 3C illustrates that the Serratia populations grew rapidly and reached a peak to become dominant on day 15, then reached their stationary phase and suppressed the growth of other spoilage organisms. Serratia spp. may produce several antimicrobial metabolites, which have been characterized as strong N-acyl-homoserine lactone (AHL) producers in meat (Bruhn et al., 2004). They may use quorum sensing to monitor their population density, synchronize their physiological functions, and socially interact with other bacteria (Van Houdt et al., 2007), thus we anticipated that once Serratia reached maximum abundance, it could make an important contribution to other spoilage-related bacteria.
The samples inoculated with C. maltaromaticum and L. mesenteroides underwent a more complex composting process of bacterial community succession. Carnobacterium and Leuconostoc increased sharply after inoculation and reached a maximum relative abundance of 97.95 and 81.6% at day 15 (Figures 3C,D), respectively: however, the proportions thereof decreased to 73.60 and 58.67% at day 45, remaining in relatively high abundance and accompanied with a rapid increase of Serratia. Carnobacterium maltaromaticum inoculated with sterile sliced beef, then grew well and achieved maximum population after 2–8 weeks (Leisner et al., 1995). Carnobacterium spp. are ubiquitous psychrotrophic LAB that could grow in a wide variety of meat products at lower temperatures, and are commonly predominant members of the microflora, which may be conducive to rapid deterioration during storage (Leisner et al., 2007; Casaburi et al., 2011). Carnobacteria possessed the capability to produce antimicrobial peptides, bacteriocins, and wide spectrum action against pathogenic and spoilage bacteria (Leisner et al., 2007). Leuconostoc mesenteroides was the dominant species and consequently was responsible for the spoilage of commercial bacon (Comi et al., 2016). Morcilla de Burgos inoculated with L. mesenteroides, as a new species, grew more rapidly and influenced the signs of spoilage due to its more energy-efficient metabolism (Diez et al., 2009). Kotzekidou and Bloukas (1998) reported that inoculation of Lactobacillus alimentarius in vacuum-packed frankfurter-type sausage could increase LAB populations and suppress other saprophytic microorganisms.
Lactic acid bacteria produce various antimicrobial components, such as organic acids, hydrogen peroxide, ethanol, bacteriocins, and other substances (Holzapfel et al., 1995; Drosinos et al., 2006), one or more these metabolites refer to the inhibition of other undesirable bacteria, including spoilage microorganisms and pathogenic bacteria (Metaxopoulos et al., 2002; Drosinos et al., 2006). In this study, Carnobacterium and Leuconostoc reached maximum relative abundance on day 15, thereafter the suppression of Serratia by them was slowly alleviated (Figures 3C,D), which could be explained by the fact that the antimicrobial metabolites from lactic acid strains were ineffective as a mechanism of control.
The genera that presented the greatest abundances of samples inoculated with S. xylosus (P2) were Serratia (59.26%) and Carnobacterium (37.76%) on day 45 (Figure 3B), while the Staphylococcus was found at a very low level (0.01%). Similarly, the prevalent species were also Serratia (55.37%) and Carnobacterium (34.84%) inoculated with L. gelidum (P16), containing only a small proportion of Leuconostoc (8.7%) (Figure 3E). The bacterial load in the bacon product inoculated with S. xylosus and L. gelidum, increased initially, then decreased, indicating antimicrobial substances may be generated by the predominant Serratia and Carnobacterium (probably AHLs and bacteriocins). Serratia and Carnobacterium gradually outcompeted all other bacteria and became the dominant species with increasing storage time, which was consistent with the results from the viable counts. Staphylococcus xylosus and L. gelidum displayed a slower growth of the species and were deemed less competitive among microorganisms.
Carnobacteria had shown the ability to produce metabolites with antimicrobial activity (peptides and bacteriocins) and could inhibit spoilage bacteria, such as LAB and Enterococcus (Leisner et al., 2007; Doulgeraki et al., 2012). Hydrogen peroxide and lactic acid are produced by a number of LAB (e.g., Carnobacteria spp. and Leuconostoc spp.), Staphylococcus spp. are more sensitive to these than most LAB, and would either be inhibited or destroyed (Holzapfel et al., 1995). Leuconostoc gelidum often prevail in chilled-stored, packaged, nutrient-rich, foods, such as cooked meats (Säde, 2011), however, L. gelidum cannot obtain energy from glycogen, proteinaceous substrates, lactate, or fatty acids, and its growth is thus inhibited. Additionally, Carnobacterium (56.16%) and Serratia (36.74%) grew rapidly and accounted for the most part of the microbial community in sterile bacon specimens inoculated with the five mixed strains (Pm) on day 45 (Figure 3G), and the proportion of Leuconostoc was only 5.73%. Referring to these results, the growth of Leuconostoc may also be inhibited by the strains of Serratia and Carnobacterium. Compared with the Serratia, the occurrence of Carnobacterium usually reported in meats and dairy products is most often ignored. Since MRS agar commonly used for LAB enumeration contained acetate it was efficient in terms of inhibiting Carnobacterium growth.
Microbial Diversity and Changes During Storage
To investigate the succession in microbial communities of bacon inoculated with spoilage bacteria during storage, heatmaps of bacteria at the genus level phylotypes were plotted (Figure 4 and Supplementary Figure 4). Among the generated heatmaps, the redder color denotes higher relative abundances, and the greener color represents lower abundances. According to Supplementary Figure 4, a relatively high diversity was observed in control samples throughout the storage period, due to the low concentration of viable counts (Table 2), showing that they should form relatively stable compositions. At 7 days of storage after inoculation with spoilage bacteria, the microbial community composition differed among different groups and indicated less diversity in terms of trend. During the late storage period, Serratia, Carnobacterium, and Leuconostoc were clustered with the highest abundance in bacon inoculated with different bacterial strains, implying that the microbial communities became minimally diverse. Similar results were found by Nychas et al. (2008), who found the gradually stable microflora composition in the late stage of storage was generally the predominant bacteria. Additionally, the more abundant dominant bacterium, the less diverse the microbial community in the late storage stages, and the specimens inoculated with such strains evidently reduced the abundance of the reference microorganisms.
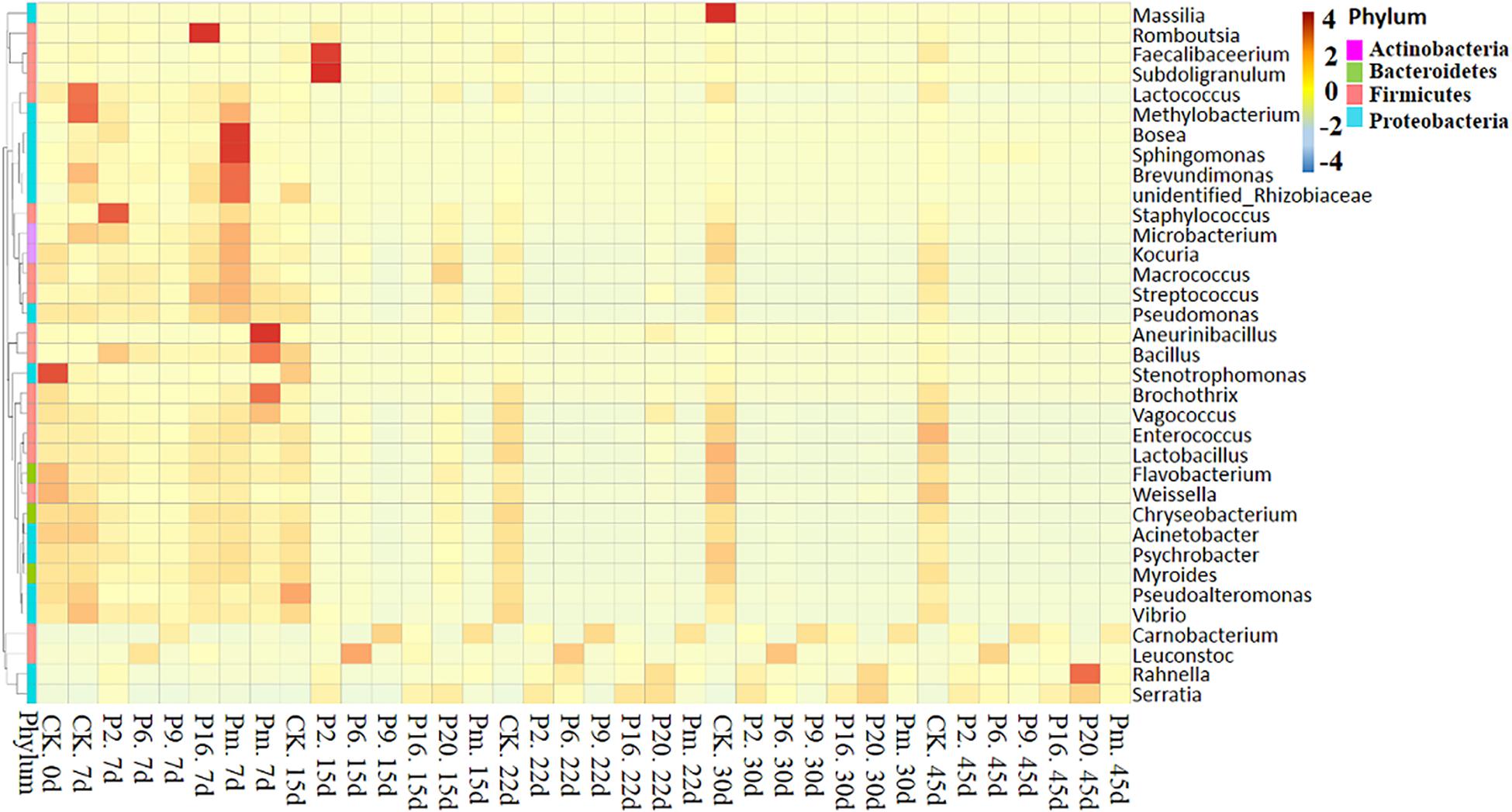
Figure 4. Heatmap showing the changes in the microbial communities of bacon inoculated with potential spoilage bacteria. The group heatmaps represent a homogenization of numerical values of corresponding points. CK, Control; P2, Staphylococcus xylosus; P6, Leuconostoc mesenteroides; P9, Carnobacterium maltaromaticum; P16, Leuconostoc gelidum; P20, Serratia liquefaciens; Pm, The five strains in combination.
Similarities and differences among the groups were also analyzed using flower plots (Figure 5). According to Figure 5, for control samples, 282 groups (the largest percentage of all groups) were classified as core OTUs. Besides core OTUs, there were 44, 17, 32, 22, 54, and 29 unique OTUs on days 0, 7, 15, 22, 30, and 45, the number remained stable until the end of the storage period, indicating that the microbial composition changed slightly in control samples. In the inoculated groups, the core OTUs (39–42) were significantly lower than the control groups, and unique OTUs on days 7 and 15 were relatively large, then very few were available, intimating that the bacterial community composition changed significantly. In conclusion, the microbiota changed toward simpler and similar communities with increasing storage time.
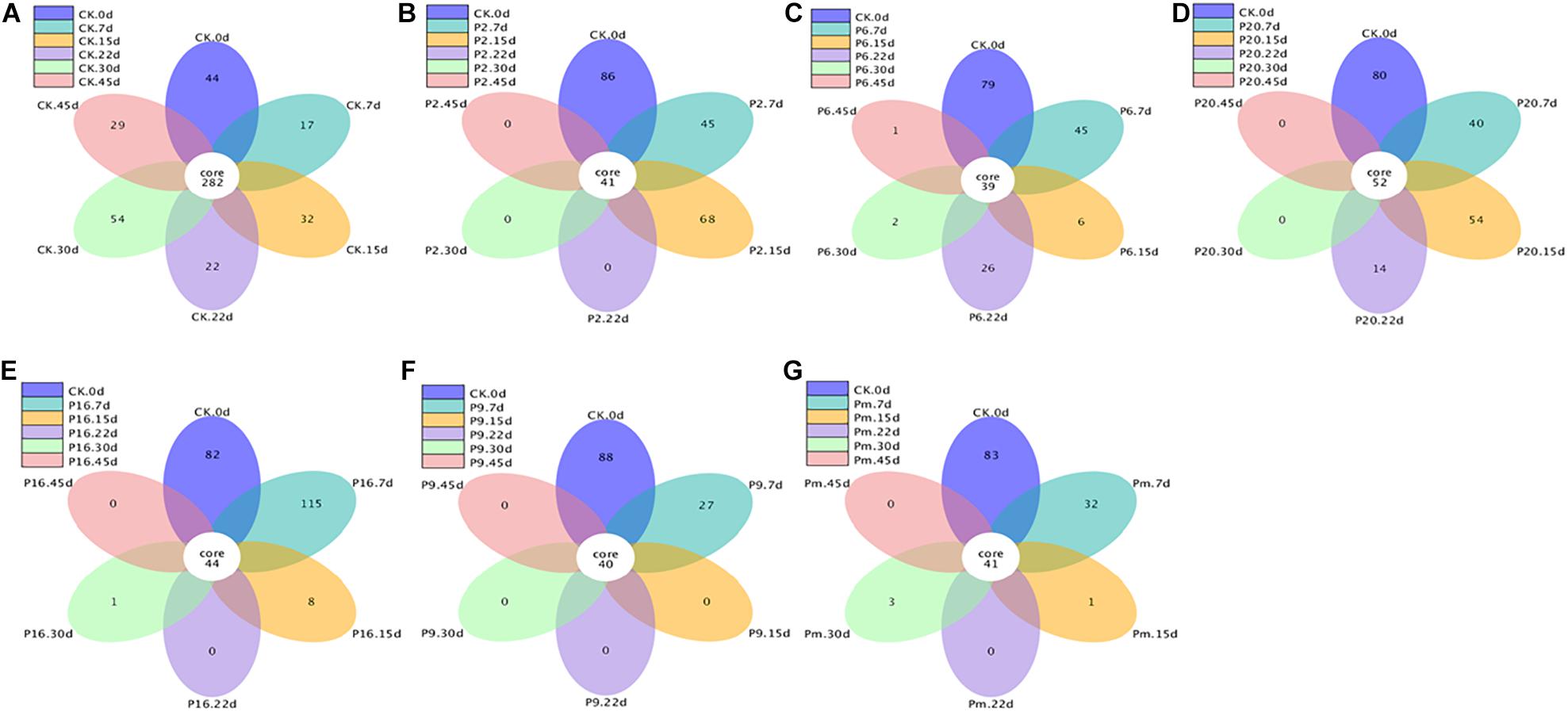
Figure 5. Flower figure showing the unique and shared OTUs of the bacterial communities across different stages. Only with more than five groups can the flower figure be shown. The plotting was carried out after homogenization of all samples. CK, Control; P2, Staphylococcus xylosus; P6, Leuconostoc mesenteroides; P9, Carnobacterium maltaromaticum; P16, Leuconostoc gelidum; P20, Serratia liquefaciens; Pm, The five strains in combination.
Additionally, non-metric multidimensional scaling (NMDS) analysis was performed to compare the differences and similarity of the community composition data (Figure 6). The figure shows joint-plot NMDS maps illustrating the bacterial community structure and the successive and dynamic changes prolonging the storage period. Figure 6 demonstrates the continuing shifts (above plane to below plane) from the primary stable period (until day 7) to a late state (until day 45). The control groups and inoculated groups on day 7 showed similar microbiota forming one cluster, then scattered, which suggested that the similarity and differences were present among the groups and that the microbiota was affected by storage time.
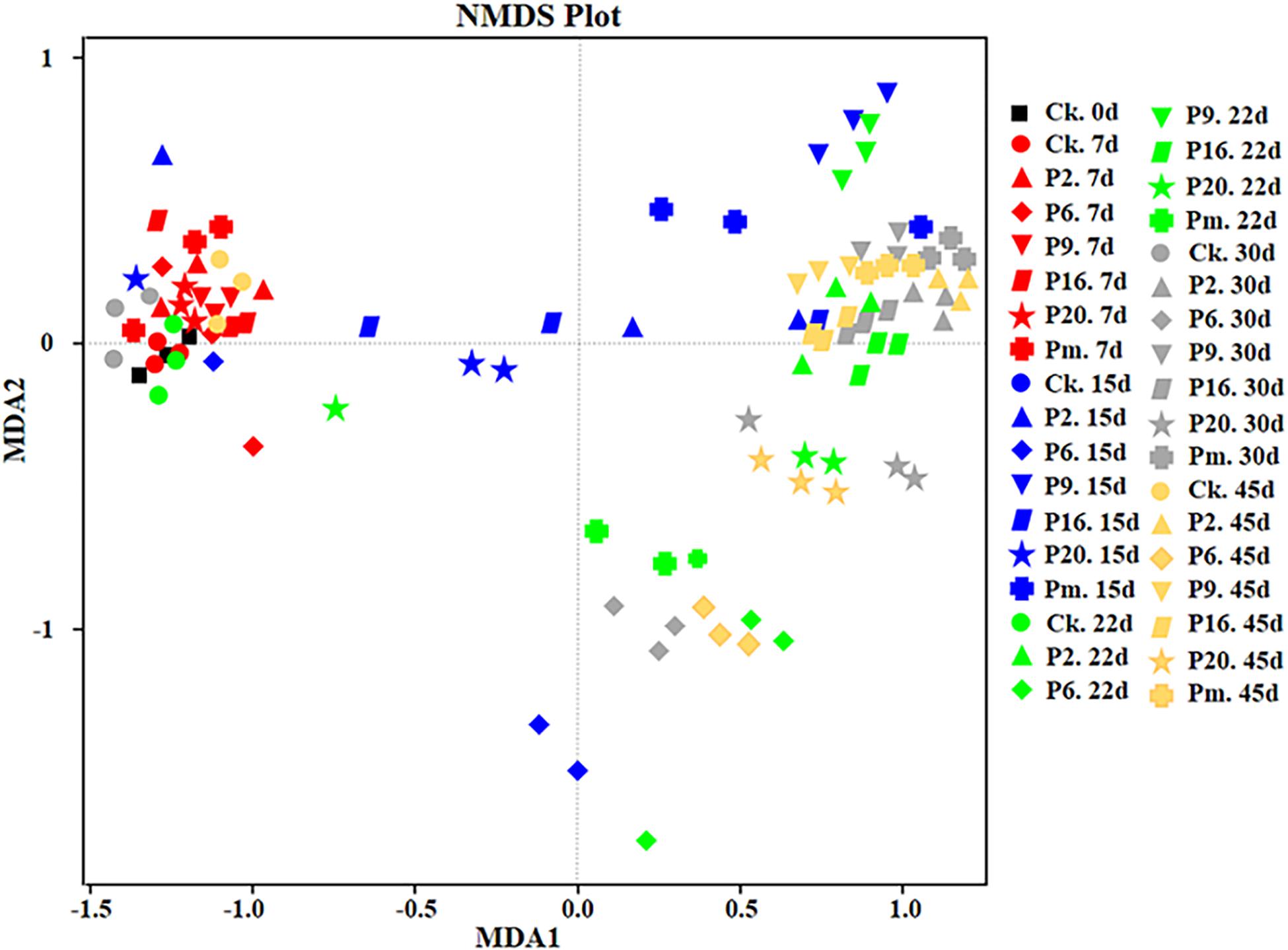
Figure 6. Non-metric multi-dimensional scaling analysis of the bacterial community structure of bacon inoculated with potential spoilage bacteria. The analysis was based on the relative abundances of all genera of microbiota detected by high-throughput sequencing. CK, Control; P2, Staphylococcus xylosus; P6, Leuconostoc mesenteroides; P9, Carnobacterium maltaromaticum; P16, Leuconostoc gelidum; P20, Serratia liquefaciens; Pm, The five strains in combination.
Foods can support a complex microflora and interactions between different species of microorganisms. In this study, the inoculation decreased the levels of potential microorganisms and inhibited the growth of numerous organisms that correlate with meat spoilage. In general, high bacterial diversities were observed in the early stage of storage while Serratia, Carnobacterium, and Leuconostoc became the most abundant genera with prolonged storage. We can conclude only that the reason for the dominance is not exclusively their rapid growth; it cannot be that other elements acted as competing organisms since they were dominant. Gram (1993) found that the bacterial selection in the microflora of food products is the influence of microbial interactions.
In food ecosystems, the interactions between microbial growth and enzyme activities have been shown to cause various consequences such as: growth promotion, symbiotic relationships, growth inhibition, and competition (Gram et al., 2002; Andreevskaya et al., 2018). LAB-meat interactions have been studied frequently: in vacuum-packed and refrigerated meat products, it is reported that spoilage arises from the interaction among LAB (the dominant flora) and Enterobacteriaceae, pseudomonads, Brochothrix thermosphacta, and other species (found in lower numbers) (Borch et al., 1996; Metaxopoulos et al., 2002; Bruhn et al., 2004). LAB could produce antimicrobial substances including organic acids and bacteriocins, which usually inhibit the growth of other microorganisms (Cleveland et al., 2001; Wang et al., 2016). Leuconostoc mesenteroides and Lactobacillus curvatus can produce bacteriocins and inhibit other spoilage microorganisms or even pathogens (Metaxopoulos et al., 2002). Enterobacteriaceae, notably Serratia spp. and Hafnia alvei, due to their ability to frequently become dominant in spoilage flora (Borch et al., 1996), were found to contribute to vacuum-packed meat spoilage through the quorum sensors (QS) systems (AHLs); however, AHL-producing Hafnia alvei might influence the spoilage in which other organisms participated with the spoilage process (Bruhn et al., 2004).
Bacterial interactions and competition have been extensively studied for the past few decades. A variety of interactions (stimulation, delay, complete inhibition of growth, and no effects between them) could occur when lactic acid starters and probiotic bacteria were mixed (Vinderola et al., 2002). Co-culture studies can verify that interactions between Lactobacillus sakei 10A, Lactobacillus sakei LS5, and Brochothrix thermosphacta BT1 occurred in cooked meat (Vermeiren et al., 2006). Borch et al. (1996) found that sterilized beef inoculated with Hafnia alwi together with LAB, gave rise to unpleasant and unacceptable off-odors after 8 weeks, whereas no off-odors were detected with single Hafnia alwi culture. Inoculation with the three mixture of Shewanella putrefaciens, Photobacterium phosphoreum, and Aeromonas sp. in cold-smoked salmon cannot caused spoilage whereas co-inoculation of two bacteria Brochothrix thermosphacta and Carnobacterium piscicola was capable of producing off-odors (Joffraud et al., 2001). Morcilla de Burgos inoculated with L. mesenteroides and W. viridescens, both jointly and separately cultured, particular signs of spoilage increased compared to single-cell cultures (Diez et al., 2009). A number of these studies showed that the off-odor may originate from interactions among several bacteria. Microbial spoilage was caused by the growth and reproduction of a diversity of microorganisms, two or more microbial species exchange metabolites or nutrients to cause spoilage and disrupt product interactions (Gram et al., 2002). No single S. liquefaciens could be identified as the cause of spoilage, the growth and activity of bacteria usually contained a mixture of species/groups (Gram et al., 1999).
To develop novel preservation technologies and develop models for predictive microbiology, an insight into understanding of microbiota, and the dynamic changes and interactions during the refrigerated storage of meat products is of great importance, however, little information regarding possible interactions responsible for meat spoilage is available, so further research is needed.
Conclusion
In this study, the dynamic changes in bacterial community structures during the storage of bacon which had been previously inoculated with five potential spoilage bacteria, were evaluated. Using HTS, 21 phyla, and 367 bacteria genera were identified, with the control samples exhibiting the highest microbial diversity. Compared with the other groups, major microbiological and physicochemical changes appeared after 15 days, with the changes becoming gradually stable and less diverse bacterial communities appearing in the later stages of the storage period. Serratia liquefaciens, C. maltaromaticum, and L. mesenteroides were found to be more competitive species. The results from this study provide a basic understanding of the microbial composition and changes in the bacterial profile of bacon during the spoilage process. Although further investigations are needed to increase our understanding of the interactions between the microbial communities within the spoilage environment, it is expected that this study will be of benefit to further improve the shelf-life of meat products.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA746727).
Author Contributions
YS and BX designed the experiments. XL carried out the experiments. QX analyzed the experimental results and assisted with the Illumina sequencing. HZ analyzed the sequencing data and developed the analysis tools. XL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Key Research and Development Programme of China (Grant No. 2016YFD0401501) and the Scientific Research Foundation for Introduced Talents of Nanjing Tech University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.713513/full#supplementary-material
References
Alves, V., Martinez, R., Lavrador, M. A. S., and De Martinis, E. C. P. (2006). Antilisterial activity of lactic acid bacteria inoculated on cooked ham. Meat Sci. 74, 623–627. doi: 10.1016/j.meatsci.2006.05.012
Andreevskaya, M., Jääskeläinen, E., Johansson, P., Ylinen, A., Paulin, L., Björkroth, J., et al. (2018). Food spoilage-associated Leuconostoc, Lactococcus, and Lactobacillus species display different survival strategies in response to competition. Appl. Environ. Microbiol. 84:e00554-18. doi: 10.1128/AEM.00554-18
Belletti, N., Garriga, M., Aymerich, T., and Bover-Cid, S. (2013). Inactivation of Serratia liquefaciens on dry-cured ham by high pressure processing. Food Microbiol. 35, 34–37. doi: 10.1016/j.fm.2013.03.001
Björkroth, K. J., Vandamme, P., and Korkeala, H. J. (1998). Identification and characterization of Leuconostoc carnosum, associated with production and spoilage of vacuum-packaged, sliced, cooked ham. Appl. Environ. Microbiol. 64, 3313–3319. doi: 10.1128/AEM.64.9.3313-3319.1998
Borch, E., Kant-Muermans, M. L., and Blixt, Y. (1996). Bacterial spoilage of meat and cured meat products. Int. J. Food Microbiol. 33, 103–120. doi: 10.1016/0168-1605(96)01135-X
Bredholt, S., Nesbakken, T., and Holck, A. (2001). Industrial application of an antilisterial strain of Lactobacillus sakei as a protective culture and its effect on the sensory acceptability of cooked, sliced, vacuum-packaged meats. Int. J. Food Microbiol. 66, 191–196. doi: 10.1016/S0168-1605(00)00519-5
Bruhn, J. B., Christensen, A. B., Flodgaard, L. R., Nielsen, K. F., Larsen, T. O., Givskov, M., et al. (2004). Presence of acylated homoserine lactones (AHLs) and AHL-producing bacteria in meat and potential role of AHL in spoilage of meat. Appl. Environ Microbiol. 70, 4293–4302. doi: 10.1128/AEM.70.7.4293-4302.2004
Casaburi, A., Nasi, A., Ferrocino, I., Di Monaco, R., Mauriello, G., Villani, F., et al. (2011). Spoilage-related activity of Carnobacterium maltaromaticum strains in air-stored and vacuum-packed meat. Appl. Environ. Microbiol. 77, 7382–7393. doi: 10.1128/AEM.05304-11
Casaburi, A., Piombino, P., Nychas, G. J., Villani, F., and Ercolini, D. (2015). Bacterial populations and the volatilome associated to meat spoilage. Food Microbiol. 45, 83–102. doi: 10.1016/j.fm.2014.02.002
Chaillou, S., Chaulot-Talmon, A., Caekebeke, H., Cardinal, M., Christieans, S., Denis, C., et al. (2015). Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J. 9, 1105–1118. doi: 10.1038/ismej.2014.202
Chen, Q., Cao, M., Chen, H., Gao, P., Fu, Y., Liu, M., et al. (2016). Effects of gamma irradiation on microbial safety and quality of stir fry chicken dices with hot chili during storage. Radiat. Phys. Chem. 127, 122–126. doi: 10.1016/j.radphyschem.2016.06.022
Chenoll, E., Macian, M. C., Elizaquivel, P., and Aznar, R. (2007). Lactic acid bacteria associated with vacuum-packed cooked meat product spoilage: population analysis by rDNA-based methods. J. Appl. Microbiol. 102, 498–508. doi: 10.1111/j.1365-2672.2006.03081.x
Cleveland, J., Montville, T. J., Nes, I. F., and Chikindas, M. L. (2001). Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71, 1–20. doi: 10.1016/S0168-1605(01)00560-8
Comi, G., Andyanto, D., Manzano, M., and Iacumin, L. (2016). Lactococcus lactis and Lactobacillus sakei as bio-protective culture to eliminate Leuconostoc mesenteroides spoilage and improve the shelf life and sensorial characteristics of commercial cooked bacon. Food Microbiol. 58, 16–22. doi: 10.1016/j.fm.2016.03.001
De Filippis, F., Pennacchia, C., Di Pasqua, R., Fiore, A., Fogliano, V., Villani, F., et al. (2013). Decarboxylase gene expression and cadaverine and putrescine production by Serratia proteamaculans in vitro and in beef. Int. J. Food Microbiol. 165, 332–338. doi: 10.1016/j.ijfoodmicro.2013.05.021
Diez, A. M., Björkroth, J., Jaime, I., and Rovira, J. (2009). Microbial, sensory and volatile changes during the anaerobic cold storage of morcilla de Burgos previously inoculated with Weissella viridescens and Leuconostoc mesenteroides. Int. J. Food Microbiol. 131, 168–177. doi: 10.1016/j.ijfoodmicro.2009.02.019
Dogbevi, M. K., Vachon, C., and Lacroix, M. (1999). Physicochemical and microbiological changes in irradiated fresh pork loins. Meat Sci. 51, 349–354. doi: 10.1016/S0309-1740(98)00133-8
Doulgeraki, A. I., Ercolini, D., Villani, F., and Nychas, G. J. (2012). Spoilage microbiota associated to the storage of raw meat in different conditions. Int. J. Food Microbiol. 157, 130–141. doi: 10.1016/j.ijfoodmicro.2012.05.020
Doulgeraki, A. I., Paramithiotis, S., and Nychas, G.-J. E. (2011). Characterization of the Enterobacteriaceae community that developed during storage of minced beef under aerobic or modified atmosphere packaging conditions. Int. J. Food Microbiol. 145, 77–83. doi: 10.1016/j.ijfoodmicro.2010.11.030
Drosinos, E., Mataragas, M., and Metaxopoulos, J. (2006). Modeling of growth and bacteriocin production by Leuconostoc mesenteroides E131. Meat Sci. 74, 690–696. doi: 10.1016/j.meatsci.2006.05.022
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10:996. doi: 10.1038/nmeth.2604
Ercolini, D., Russo, F., Nasi, A., Ferranti, P., and Villani, F. J. A. (2009). Mesophilic and psychrotrophic bacteria from meat and their spoilage potential in vitro and in beef. Appl. Environ. Microbiol. 75, 1990–2001. doi: 10.1128/AEM.02762-08
Gram, L. (1993). Inhibitory effect against pathogenic and spoilage bacteria of Pseudomonas strains isolated from spoiled and fresh fish. Appl. Environ. Microbiol. 59, 2197–2203. doi: 10.1128/aem.59.7.2197-2203.1993
Gram, L., Christensen, A. B., Ravn, L., Molin, S., and Givskov, M. (1999). Production of acylated homoserine lactones by psychrotrophic members of the Enterobacteriaceae isolated from foods. Appl. Environ. Microbiol. 65, 3458–3463. doi: 10.1128/AEM.65.8.3458-3463.1999
Gram, L., Ravn, L., Rasch, M., Bruhn, J. B., Christensen, A. B., and Givskov, M. (2002). Food spoilage—interactions between food spoilage bacteria. Int. J. Food Microbiol. 78, 79–97. doi: 10.1016/S0168-1605(02)00233-7
Hamasaki, Y., Ayaki, M., Fuchu, H., Sugiyama, M., and Morita, H. (2003). Behavior of psychrotrophic lactic acid bacteria isolated from spoiling cooked meat products. Appl. Environ. Microbiol. 69, 3668–3671. doi: 10.1128/AEM.69.6.3668-3671.2003
Holm, E. S., Schäfer, A., Koch, A., and Petersen, M. A. (2013). Investigation of spoilage in saveloy samples inoculated with four potential spoilage bacteria. Meat Sci. 93, 687–695. doi: 10.1016/j.meatsci.2012.11.016
Holzapfel, W., Geisen, R., and Schillinger, U. (1995). Biological preservation of foods with reference to protective cultures, bacteriocins and food-grade enzymes. Int. J. Food Microbiol. 24, 343–362. doi: 10.1016/0168-1605(94)00036-6
Houtsma, P. C., de Wit, J. C., and Rombouts, F. M. (1996). Minimum inhibitory concentration (MIC) of sodium lactate and sodium chloride for spoilage organisms and pathogens at different pH values and temperatures. J. Food Protect. 59, 1300–1304. doi: 10.4315/0362-028X-59.12.1300
Hu, P., Xu, X., Zhou, G., Han, Y., Xu, B., and Liu, J. (2008). Study of the Lactobacillus sakei protective effect towards spoilage bacteria in vacuum packed cooked ham analyzed by PCR–DGGE. Meat Sci. 80, 462–469. doi: 10.1016/j.meatsci.2008.01.011
Huang, L., Zhao, J., Chen, Q., and Zhang, Y. (2014). Nondestructive measurement of total volatile basic nitrogen (TVB-N) in pork meat by integrating near infrared spectroscopy, computer vision and electronic nose techniques. Food Chem. 145, 228–236. doi: 10.1016/j.foodchem.2013.06.073
Joffraud, J., Leroi, F., Roy, C., and Berdague, J. (2001). Characterisation of volatile compounds produced by bacteria isolated from the spoilage flora of cold-smoked salmon. Int. J. Food Microbiol. 66, 175–184. doi: 10.1016/S0168-1605(00)00532-8
Juárez-Castelán, C., García-Cano, I., Escobar-Zepeda, A., Azaola-Espinosa, A., Álvarez-Cisneros, Y., and Ponce-Alquicira, E. (2019). Evaluation of the bacterial diversity of Spanish-type chorizo during the ripening process using high-throughput sequencing and physicochemical characterization. Meat Sci. 150, 7–13. doi: 10.1016/j.meatsci.2018.09.001
Korkeala, H. J., and Björkroth, K. J. (1997). Microbiological spoilage and contamination of vacuum-packaged cooked sausages. J. Food Protect. 60, 724–731. doi: 10.4315/0362-028X-60.6.724
Kotzekidou, P., and Bloukas, J. (1996). Effect of protective cultures and packaging film permeability on shelf-life of sliced vacuum-packed cooked ham. Meat Sci. 42, 333–345. doi: 10.1016/0309-1740(95)00038-0
Kotzekidou, P., and Bloukas, J. (1998). Microbial and sensory changes in vacuum-packed frankfurter-type sausage by Lactobacillus alimentarius and fate of inoculated Salmonella enteritidis. Food Microbiol. 15, 101–111. doi: 10.1006/fmic.1997.0138
Kuo, C., and Chu, C. (2003). Quality characteristics of Chinese sausages made from PSE pork. Meat Sci. 64, 441–449. doi: 10.1016/S0309-1740(02)00213-9
Leisner, J. J., Laursen, B. G., Prévost, H., Drider, D., and Dalgaard, P. (2007). Carnobacterium: positive and negative effects in the environment and in foods. FEMS Microbiol. Rev. 31, 592–613. doi: 10.1111/j.1574-6976.2007.00080.x
Leisner, J., Greer, G., Dilts, B., and Stiles, M. (1995). Effect of growth of selected lactic acid bacteria on storage life of beef stored under vacuum and in air. Int. J. Food Microbiol. 26, 231–243. doi: 10.1016/0168-1605(94)00133-Q
Leroy, F., Verluyten, J., and De Vuyst, L. (2006). Functional meat starter cultures for improved sausage fermentation. Int. J. Food Microbiol. 106, 270–285. doi: 10.1016/j.ijfoodmicro.2005.06.027
Li, X., Li, C., Ye, H., Wang, Z., Wu, X., Han, Y., et al. (2019). Changes in the microbial communities in vacuum-packaged smoked bacon during storage. Food Microbiol. 77, 26–37. doi: 10.1016/j.fm.2018.08.007
Li, X., Xiong, Q., Xu, B., Wang, H., Zhou, H., and Sun, Y. (2021). (2021) Bacterial community dynamics during different stages of processing of smoked bacon using the 16S rRNA gene amplicon analysis. Int. J. Food Microbiol. 351:109076. doi: 10.1016/j.ijfoodmicro.2021.109076
Lindberg, A.-M., Ljungh, Å, Ahrne, S., Löfdahl, S., and Molin, G. (1998). Enterobacteriaceae found in high numbers in fish, minced meat and pasteurised milk or cream and the presence of toxin encoding genes. Int. J. Food Microbiol. 39, 11–17. doi: 10.1016/S0168-1605(97)00104-9
Losantos, A., Sanabria, C., Cornejo, I., and Carrascosa, A. (2000). Characterization of Enterobacteriaceae strains isolated from spoiled dry-cured hams. Food Microbiol. 17, 505–512. doi: 10.1006/fmic.2000.0350
Macé, S., Joffraud, J. J., Cardinal, M., Malcheva, M., Cornet, J., Lalanne, V., et al. (2013). Evaluation of the spoilage potential of bacteria isolated from spoiled raw salmon (Salmo salar) fillets stored under modified atmosphere packaging. Int. J. Food Microbiol. 160, 227–238. doi: 10.1016/j.ijfoodmicro.2012.10.013
Mah, J.-H., and Hwang, H.-J. (2009). Inhibition of biogenic amine formation in a salted and fermented anchovy by Staphylococcus xylosus as a protective culture. Food Control 20, 796–801. doi: 10.1016/j.foodcont.2008.10.005
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mataragas, M., Skandamis, P., Nychas, G.-J. E., and Drosinos, E. H. (2007). Modeling and predicting spoilage of cooked, cured meat products by multivariate analysis. Meat Sci. 77, 348–356. doi: 10.1016/j.meatsci.2007.03.023
McMeekin, T. A., and Ross, T. (1996). Shelf life prediction: status and future possibilities. Int. J. Food Microbiol. 33, 65–83. doi: 10.1016/0168-1605(96)01138-5
Metaxopoulos, J., Mataragas, M., and Drosinos, E. H. (2002). Microbial interaction in cooked cured meat products under vacuum or modified atmosphere at 4°C. J. Appl. Microbiol. 93, 363–373. doi: 10.1046/j.1365-2672.2002.01701.x
Mills, J., Donnison, A., and Brightwell, G. (2014). Factors affecting microbial spoilage and shelf-life of chilled vacuum-packed lamb transported to distant markets: a review. Meat Sci. 98, 71–80. doi: 10.1016/j.meatsci.2014.05.002
Nychas, G. J., Skandamis, P. N., Tassou, C. C., and Koutsoumanis, K. P. (2008). Meat spoilage during distribution. Meat Sci. 78, 77–89. doi: 10.1016/j.meatsci.2007.06.020
O’bryan, C. A., Crandall, P. G., Ricke, S. C., and Olson, D. G. (2008). Impact of irradiation on the safety and quality of poultry and meat products: a review. Crit. Rev. Food Sci. 48, 442–457. doi: 10.1080/10408390701425698
Paarup, T., Nieto, J. C., Peláez, C., and Reguera, J. I. (1999). Microbiological and physico-chemical characterisation of deep spoilage in Spanish dry-cured hams and characterisation of isolated Enterobacteriaceae with regard to salt and temperature tolerance. Eur. Food Res. Technol. 209, 366–371. doi: 10.1007/s002170050511
Pin, C., and Baranyi, J. (1998). Predictive models as means to quantify the interactions of spoilage organisms. Int. J. Food Microbiol. 41, 59–72. doi: 10.1016/S0168-1605(98)00035-X
Pin, C., Sutherland, J., and Baranyi, J. (1999). Validating predictive models of food spoilage organisms. J. Appl. Microbiol. 87, 491–499. doi: 10.1046/j.1365-2672.1999.00838.x
Polka, J., Rebecchi, A., Pisacane, V., Morelli, L., and Puglisi, E. (2015). Bacterial diversity in typical Italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiol. 46, 342–356.
Pothakos, V., Devlieghere, F., Villani, F., Björkroth, J., and Ercolini, D. (2015). Lactic acid bacteria and their controversial role in fresh meat spoilage. Meat Sci. 109, 66–74. doi: 10.1016/j.meatsci.2015.04.014
Pothakos, V., Nyambi, C., Zhang, B.-Y., Papastergiadis, A., De Meulenaer, B., and Devlieghere, F. (2014a). Spoilage potential of psychrotrophic lactic acid bacteria (LAB) species: Leuconostoc gelidum subsp. gasicomitatum and Lactococcus piscium, on sweet bell pepper (SBP) simulation medium under different gas compositions. Int. J. Food. Microbiol. 178, 120–129. doi: 10.1016/j.ijfoodmicro.2014.03.012
Pothakos, V., Snauwaert, C., De Vos, P., Huys, G., and Devlieghere, F. (2014b). Psychrotrophic members of Leuconostoc gasicomitatum, Leuconostoc gelidum and Lactococcus piscium dominate at the end of shelf-life in packaged and chilled-stored food products in Belgium. Food Microbiol. 39, 61–67. doi: 10.1016/j.fm.2014.08.023
Quijada, N. M., De Filippis, F., Sanz, J. J., del Camino Garcia-Fernandez, M., Rodríguez-Lázaro, D., Ercolini, D., et al. (2018). Different Lactobacillus populations dominate in “Chorizo de León” manufacturing performed in different production plants. Food Microbiol. 70, 94–102. doi: 10.1016/j.fm.2017.09.009
Rahkila, R., Nieminen, T., Johansson, P., Säde, E., and Björkroth, J. (2012). Characterization and evaluation of the spoilage potential of Lactococcus piscium isolates from modified atmosphere packaged meat. Int. J. Food Microbiol. 156, 50–59. doi: 10.1016/j.ijfoodmicro.2012.02.022
Ravyts, F., Vuyst, L. D., and Leroy, F. (2012). Bacterial diversity and functionalities in food fermentations. Eng. Life Sci. 12, 356–367. doi: 10.1002/elsc.201100119
Roig-Sagués, A., and Eerola, S. (1997). Biogenic amines in meat inoculated with Lactobacillus sake starter strains and an amine-positive lactic acid bacterium. Z. Lebensm. Unters. F. A 205, 227–231. doi: 10.1007/s002170050156
Säde, E. (2011). Leuconostoc Spoilage of Refrigerated, Packaged Foods. Academic Dissertation. Helsinki: University of Helsinki.
Samelis, J., Kakouri, A., and Rementzis, J. (2000). Selective effect of the product type and the packaging conditions on the species of lactic acid bacteria dominating the spoilage microbial association of cooked meats at 4°C. Food Microbiol. 17, 329–340. doi: 10.1006/fmic.1999.0316
Soladoye, P., Shand, P., Aalhus, J., Gariépy, C., and Juárez, M. (2015). Pork belly quality, bacon properties and recent consumer trends. Can. J. Anim. Sci. 95, 325–340. doi: 10.4141/cjas-2014-121
Stavropoulou, D. A., De Maere, H., Berardo, A., Janssens, B., Filippou, P., De Vuyst, L., et al. (2018). Pervasiveness of Staphylococcus carnosus over Staphylococcus xylosus is affected by the level of acidification within a conventional meat starter culture set-up. Int. J. Food Microbiol. 274, 60–66. doi: 10.1016/j.ijfoodmicro.2018.03.006
Stohr, V., Joffraud, J., Cardinal, M., and Leroi, F. (2001). Spoilage potential and sensory profile associated with bacteria isolated from cold-smoked salmon. Food Res. Int. 34, 797–806. doi: 10.1016/S0963-9969(01)00101-6
Sun, X., and Holley, R. (2012). Antimicrobial and antioxidative strategies to reduce pathogens and extend the shelf life of fresh red meats. Compr. Rev. Food Sci. 11, 340–354. doi: 10.1111/j.1541-4337.2012.00188.x
Van Houdt, R., Givskov, M., and Michiels, C. W. (2007). Quorum sensing in Serratia. FEMS Microbiol. Rev. 31, 407–424. doi: 10.1111/j.1574-6976.2007.00071.x
Vermeiren, L., Devlieghere, F., and Debevere, J. (2004). Evaluation of meat born lactic acid bacteria as protective cultures for the biopreservation of cooked meat products. Int. J. Food Microbiol. 96, 149–164. doi: 10.1016/j.ijfoodmicro.2004.03.016
Vermeiren, L., Devlieghere, F., and Debevere, J. (2006). Co-culture experiments demonstrate the usefulness of Lactobacillus sakei 10A to prolong the shelf-life of a model cooked ham. Int. J. Food Microbiol. 108, 68–77. doi: 10.1016/j.ijfoodmicro.2005.11.001
Vinderola, C. G., Mocchiutti, P., and Reinheimer, J. A. (2002). Interactions among lactic acid starter and probiotic bacteria used for fermented dairy products. J. Dairy Sci. 85, 721–729. doi: 10.3168/jds.S0022-0302(02)74129-5
Wang, G.-Y., Wang, H.-H., Han, Y.-W., Xing, T., Ye, K.-P., Xu, X.-L., et al. (2017). Evaluation of the spoilage potential of bacteria isolated from chilled chicken in vitro and in situ. Food Microbiol. 63, 139–146. doi: 10.1016/j.fm.2016.11.015
Wang, T., Zhao, L., Sun, Y., Ren, F., Chen, S., Zhang, H., et al. (2016). Changes in the microbiota of lamb packaged in a vacuum and in modified atmospheres during chilled storage analysed by high-throughput sequencing. Meat Sci. 121, 253–260. doi: 10.1016/j.meatsci.2016.06.021
Wang, X., Ren, H., Liu, D., Zhu, W., and Wang, W. (2013). Effects of inoculating Lactobacillus sakei starter cultures on the microbiological quality and nitrite depletion of Chinese fermented sausages. Food Control 32, 591–596. doi: 10.1016/j.foodcont.2013.01.050
Zagdoun, M., Coeuret, G., N’Dione, M., Champomier-Vergès, M.-C., and Chaillou, S. J. F. M. (2020). LARGE microbiota survey reveals HOW the microbial ecology of cooked ham is shaped by different processing steps. Food Microbiol. 91:103547. doi: 10.1016/j.fm.2020.103547
Keywords: microbial diversity, bacon, spoilage bacteria, high-throughput sequencing, storage
Citation: Li X, Xiong Q, Zhou H, Xu B and Sun Y (2021) Analysis of Microbial Diversity and Dynamics During Bacon Storage Inoculated With Potential Spoilage Bacteria by High-Throughput Sequencing. Front. Microbiol. 12:713513. doi: 10.3389/fmicb.2021.713513
Received: 23 May 2021; Accepted: 06 September 2021;
Published: 28 September 2021.
Edited by:
Anindya Chanda, Mycologics LLC, United StatesReviewed by:
Clarita Olvera, Universidad Nacional Autónoma de México, MexicoMuhammad Arslan, University of Alberta, Canada
Copyright © 2021 Li, Xiong, Zhou, Xu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Sun, eXVuc3VubnkwMTJAMTYzLmNvbQ==
 Xinfu Li1
Xinfu Li1 Yun Sun
Yun Sun