
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 October 2021
Sec. Food Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.713383
Listeria monocytogenes is a Gram-positive bacterium causing listeriosis in animals and humans. To initiate a foodborne infection, L. monocytogenes has to pass through the host gastrointestinal tract (GIT). In this study, we evaluated survival abilities of L. monocytogenes 10403S wild type (WT) and its isogenic mutants in alternative sigma (σ) factor genes (i.e., sigB, sigC, sigH, and sigL) under simulated gastric, duodenal, and bile fluids. Within 10min of exposures, only bile fluid was able to significantly reduce survival ability of L. monocytogenes WT by 2 logs CFU/ml. Loss of sigL showed the greatest bile resistance among 16 strains tested, p<0.0001, (i.e., WT, four single alternative σ factor mutants, six double mutants, four triple mutants, and one quadruple mutant). To further investigate the role of σL in bile response, RNA-seq was conducted to compare the transcriptional profiles among L. monocytogenes 10403S ΔBCH triple mutant (lacking sigB, sigC, and sigH genes; expressing housekeeping σA and σL) and ΔBCHL quadruple mutant (lacking all alternative sigma factor genes; expressing only σA) strains under BHI and 1% bile conditions. A total of 216 and 176 differentially expressed genes (DEGs) were identified in BHI and bile, respectively. We confirmed that mpt operon was shown to be strongly activated by σL. Interestingly, more than 80% of DEGs were found to be negatively regulated in the presence of σL. This includes PrfA regulon and its mediated genes (i.e., hly, hpt, inlB, clpP, clpE, groL, and inlC) which were downregulated in response to bile in the presence of σL. This result suggests the potential negative role of σL on bile survival, and the roles of σL and σB might be in a seesaw model prior to host cell invasion.
The foodborne pathogen Listeria monocytogenes is a facultative Gram-positive intracellular bacterium that is able to adapt to a board range of habitats. It can survive and grow in a variety of temperatures, and a wide range of pH, osmotic pressure, and high salt (Membre et al., 2005; Goulet et al., 2013; Bevilacqua et al., 2018; Axelsson et al., 2020). This pathogen is the causative agent of listeriosis in human and animals. Although L. monocytogenes infections are comparatively rare, the mortality rate could reach up to 30% of clinical cases (NicAogain and O’Byrne, 2016). More importantly, L. monocytogenes cannot be underemphasized for it has contributed to huge economic losses in livestock industries, food poisoning leading to death in both animals and human, and abortion in pregnant women (Oliver et al., 2005; Kaur et al., 2007). Following being ingested with contaminated food, L. monocytogenes goes through GI passage. It encounters the low pH in stomach and the bile salt and high osmolality in intestinal fluid. Alves et al. have revealed that survival abilities of L. monocytogenes under highly adverse conditions mimicking those observed in the gastrointestinal tract are strain-dependent, not serotype-dependent. The survival ability of each strain is affected by previous exposure to stress conditions (Alves et al., 2020). In order to survive and successfully establish infection, L. monocytogenes needs to persist exposure to bile, an important antimicrobial component in GI fluid (Olier et al., 2004). It has been reported that L. monocytogenes utilizes unique bile resistance mechanisms to survive in the gallbladder (Hardy et al., 2004).
In the host stomach, L. monocytogenes has to tolerate acidity. Normally, the pH of the stomach at the fasted-state ranges between 1 and 2 (Dressman et al., 1990; Efentakis and Dressman, 1998). Upon intake of the meal, the fed-state gastric pH increases within a range of 4–7. Once the bacterium enters the intestine, the pH is 6–7.5 (Evans et al., 1988). Little is known about the bactericidal components in the intestinal fluid; however, one crucial component bacteria must cope with is bile. Bile is synthesized by hepatocytes prior to secretion from liver into bile canaliculi. The pH of human hepatic bile is approximately 7.5–8.0 (Begley et al., 2005). Normally, the bile concentration in human gut ranges from 0.005 to 2% (Dawson, 1998). In addition to its role in digestion, bile also has antimicrobial properties. The major components of human bile are bile salt, cholesterol, phospholipid, and various bile acids (Begley et al., 2005; Dowd et al., 2011). It has been shown that L. monocytogenes was able to tolerate bovine, porcine, and human bile exposures (Begley et al., 2002; Guariglia-Oropeza et al., 2018). Additionally, btlB, sigB, pva, prfA, and btlA genes have been considered in bile resistance (Begley et al., 2005). Bile salts are amphipathic molecules that are able to impair lipid-containing bacterial and viral membranes (Dussurget et al., 2002). Moreover, bile salts can also induce DNA damage and RNA secondary structure formation (Begley et al., 2005).
One of the mechanisms a bacterium utilizes to combat cellular stressors is transcriptional regulation of gene expression via combinations of alternative sigma factors (σ) and the catalytic core of RNA polymerase. In L. monocytogenes, there are four alternative sigma factors (σB, σC, σH, and σL). σB has been intensively reported as a key factor responsible for L. monocytogenes survival under various stress conditions including high osmolality, low or high temperature, low pH, ethanol, and oxidative stresses (Becker et al., 1998; Ferreira et al., 2001; Moorhead and Dykes, 2004; Lungu et al., 2009; Mujahid et al., 2013b). Also, the σB regulon has been identified as crucial to GI survival. sigB deletion mutants show increased susceptibility to bile; however, σB regulon was not upregulated under exposure to bile pH 5.5 (Guariglia-Oropeza et al., 2018). It is possible that other alternative sigma factors also contribute to bile response. We hypothesized that co-regulation of sigma factors allows L. monocytogenes to survive under bile stress and subsequently establish infection. We examined the role of and the co-regulation among alternative sigma factors using single, double, triple, and quadruple mutants. Altogether, we were able to map complex transcriptional regulation by the alternative sigma factor σL and our results suggest a negative role of σL in bile stress response.
Listeria monocytogenes 10403S and its isogenic single/double/triple/quadruple alternative sigma factor mutant strain were used in this study (Supplementary Table S1). Strains were maintained in brain heart infusion broth (BHI; Difco™, BD, United States) and 50% glycerol stocks at −80°C. They were streaked onto BHI agar plates prior to each experiment, and plates were incubated at 37°C overnight. A single colony was inoculated into 5ml BHI broth at 37°C overnight (16–18h) incubation with shaking (200rpm). Overnight culture was diluted 1:100 into a fresh 5ml BHI broth and incubated at 37°C with shaking to an OD600 of approximately 0.4, representing mid-log phase. An aliquot of 500μl of the culture was subsequently passaged in 50ml of BHI broth to generate synchronized cells at mid-log phase before exposure to the simulated bile stress.
Mid-log phase cells were challenged with simulated gastric, duodenal, and bile fluids independently. The in vitro GI fluids were prepared as previously described (Versantvoort et al., 2005). Briefly, 0.5ml of 2X gastric (pH 1.3), 2X duodenal (pH 8.1), or 2× 6% porcine bile (pH 8.2, Sigma, United States) was added to 0.5ml of mid-log phase culture. The challenged cultures were incubated at 37°C with shaking for 10 and 20min, respectively. Survival ability was determined at 0, 10, and 20min after stress exposure (t=0, t=10, and t=20). A 100μl aliquot of the pre-treated control (culture before treatment at t=0) and the untreated control (culture with additional 20min incubation) along with t=10 and t=20 cultures was 10-fold serially diluted in PBS (pH 7.4); 10μl of each dilution was plated onto BHI agar plates for subsequent enumeration. Experiments were conducted in three biological and technical replicates. All survival assays were performed in the absence of complementation strains.
Significant differences in survival abilities between L. monocytogenes wild type and its isogenic mutants were determined by one-way ANOVA with Dunnett’s post hoc test using GraphPad Prism version 9.2.0 (283), GraphPad Software, San Diego, California United States. p<0.05 was considered statistically different. To assess which sigma factor has the potent role in bile survival, the listeria strains have been categorized into groups with the presence or absence of each sigma factor. The average log reduction of all strains and replicates in each group was compared using unpaired t test.
For gene expression studies, RNA was extracted from mid-log phase cultures after 10-min exposure to 1% porcine bile or BHI (as a control) as previously described with minor modifications (Versantvoort et al., 2005; Pleitner et al., 2014). Briefly, all experiments were conducted in three biological replicates on different days. For each strain and condition, 1ml of BHI or bile-exposed culture was collected and immediately added to ice-cold stop solution of 10% acid-phenol chloroform pH 4.5 (Invitrogen™, United States) in ethanol as outlined previously (Pleitner et al., 2014). DNase treatment was performed using TURBO DNA-free™ DNase treatment kit (Invitrogen, United States) according to the manufacturer’s protocol. Total RNA concentration was quantified using NanoDrop 2000c spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
Ribosomal RNA (rRNA) was depleted using Ribo-Zero rRNA removal kit (Epicentre, Madison, WI) following manufacturer’s instruction. Quality of RNA was assessed using the 2,100 Bioanalyzer (Agilent Technology, Santa Clara, CA). Samples with RNA Integrity Number (RIN) score greater than 8.0 were considered acceptable for further library preparation and RNA sequencing. cDNA library preparation was constructed using ScriptSeq v2 RNA-seq Library Preparation kit for Bacteria (Epicentre, Madison, WI). Purification of cDNA and indexed RNA-seq libraries was performed using Agencourt® AMPure® XP kit (Beckman Coulter Inc., Brea, CA). Quantity and quality of the libraries were determined using the 2,100 Bioanalyzer. All experiments were performed in three biological replicates. Sequencing was carried out on a HiSeq 2×100 High Output paired-end, 100bp read at the Purdue University genomics core facility.
Sequencing reads were mapped against L. monocytogenes 10403S (NCBI accession number: NC_017544.1). Reads were aligned and mapped with Bowtie2 and TopHat2 (Langmead and Salzberg, 2012; Trapnell et al., 2012). The cutoffs for percent mapped reads on either sense or antisense strand and percent of rRNA match rate were at least 75% and lower than 0.1%, respectively. Reads were counted by HTSeq-count (Anders et al., 2015). Differentially expressed genes (DEGs) were compared and analyzed in R version 3.3.3 using the package DESeq2 (Love et al., 2014). The analyses were conducted from three replicates of each sample, except wild-type sample in BHI (two replicates were used in analysis). Genes were considered differentially expressed when log2 fold change < −1 or>1 (or FC<−2 or>2, representing down- or upregulation, respectively) and adjusted p-values <0.05.
The data sets generated or analyzed for this study were deposited in a public database and can be found under the SRA accession number: PRJNA544468.1
For review purposes, RNA-seq data are available at https://drive.google.com/drive/folders/1R3Wv3JN3ICxmY8ScbikALlzjlklfuObi?usp=sharing.
GOseq package 1.34.1 package for R available from Bioconductor was used to evaluate whether differential expressed genes identified by DESeq2 were enriched for Gene Ontology (GO) terms (Young et al., 2010). This tool allowed us to statistically confirm the specific metabolic pathways that L. monocytogenes utilized under BHI and/or bile conditions.
Select differentially expressed genes from RNA-seq results were validated using quantitative reverse transcriptase PCR (qRT-PCR) as previously described with minor modifications (Sue et al., 2004; Pleitner et al., 2014). Target genes used for RNA-seq data validation were (i) rpoB and bglA as housekeeping genes and (ii) mptA, ilvD, ilvC, csbD, and LMRG_00704 for sigma L function. A list of TaqMan primers and probes designed by PrimerQuest (IDT DNA, Coralville, IA) is shown in Supplementary Table S2. TaqMan probes were synthesized with a 5' 6-carboxyfluorescein (6-FAM) reporter dye and a 3’ ZEN™ dark quencher dye. All reactions were run via Rotor-Gene Q (Qiagen, Germany) with the following conditions: 1cycle at 48°C for 30min, 1cycle at 95°C for 10min, followed by 40cycles at 95°C for 15s and 55°C for 1min. Reactions without Multiscribe™ reverse transcriptase (Thermo Fisher Scientific, Wilmington, DE, United States) were run in parallel to account the possible genomic DNA (gDNA) contamination. Threshold cycle more than 35 was considered as undetectable. gDNA standard curve targeting each gene was performed to determine the amplification efficiency. Dilutions of gDNA in 107, 105, and 103 chromosomal copies were used as templates. Transcripts were normalized by a geometric mean of housekeeping genes rpoB and bglA. Statistical comparisons of gene expression levels among L. monocytogenes strains in each condition were performed by unpaired t test, SPSS version 23 (IBM, United States).
To determine the ability of L. monocytogenes to survive under simulated GI fluids, we exposed the bacteria to simulated gastric, duodenal, or bile for 10 and 20min. Survival abilities at 20min were not statistically different from those at 10min for all strains and conditions. Therefore, the presented data are that of 10min (T10) only. Following 10-min gastric fluid exposures, most of the deletion mutant strains survived similar to wild type, that is, similar levels of log reduction [i.e., log (CFU at T0)−log (CFU recovered at T10)] when compared between WT and mutants (Figure 1A). Among the single mutants, only ΔB mutant had a substantially better survival than WT with an increase in numbers during gastric fluid exposure. Among the double mutants, only ΔBH double mutant had an immensely higher susceptibility than WT, hence higher log reduction. Among the triple mutants, the presence of σC in ΔBHL vastly increased survival. Interestingly, we did not observe any significant differences between the quadruple mutant (ΔBCHL) and the WT when exposed to gastric fluid, suggesting the compensatory effect of housekeeping sigma factor σA on L. monocytogenes survival in gastric treatment.
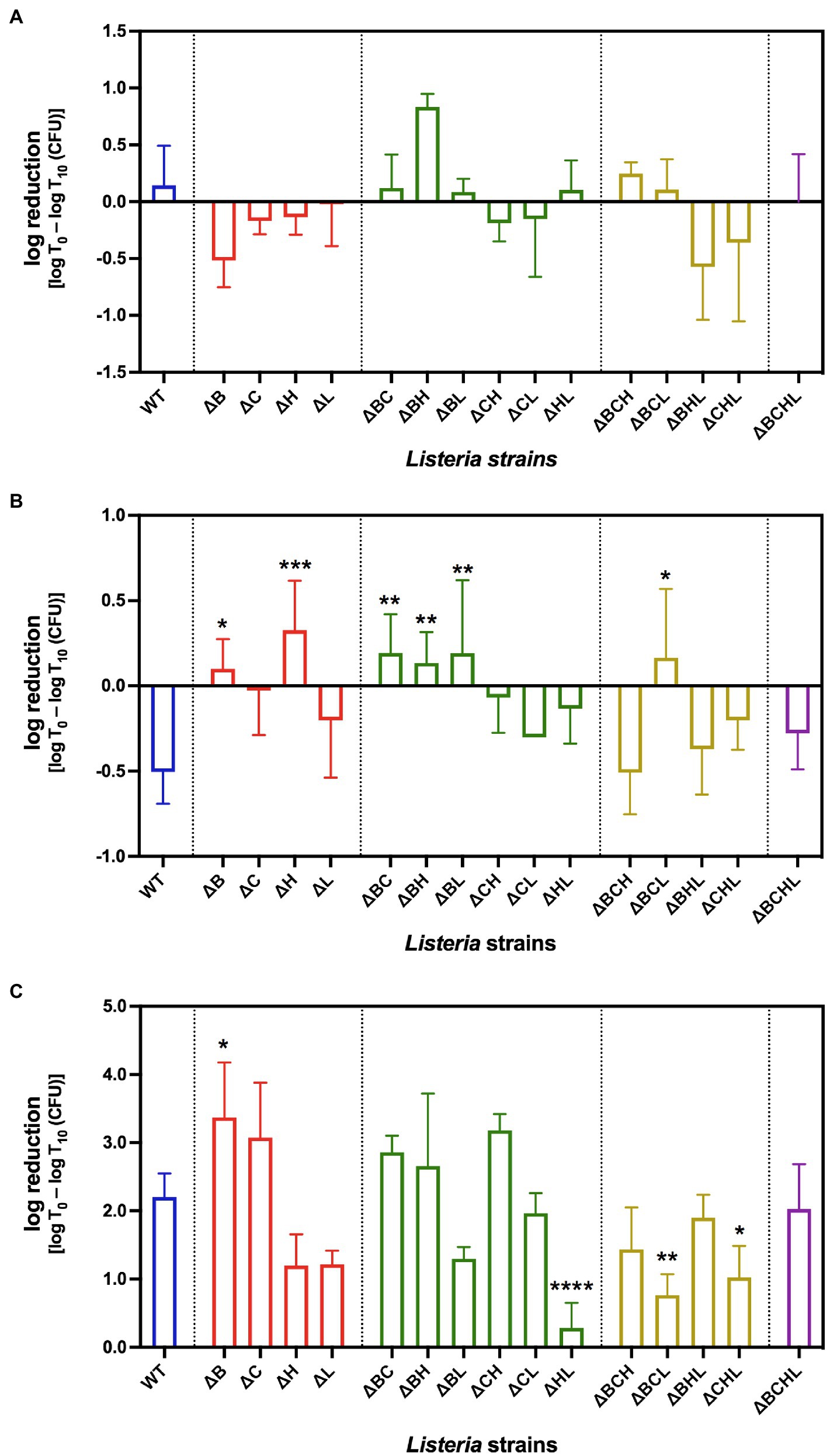
Figure 1. Average log reduction [log T0 – log T10 (CFU)] of Listeria monocytogenes 10403S WT (blue), single mutants (red), double mutants (green), triple mutants (yellow), and quadruple mutant (purple) in simulated gastrointestinal fluids: (A) gastric fluid, (B) duodenal fluid, and (C) 6% bile fluid at 10min (T10) post-treatment. *, **, ***, and **** indicate significant difference compared to WT using One-way ANOVA followed by Dunnett’s multiple comparisons test (p<0.05, p<0.01, p<0.001, and p<0.0001, respectively).
Following gastric exposure, we next examined survival ability of L. monocytogenes under duodenal fluid stress show in Figure 1B. Among the single mutants, significant differences were observed in ΔB (p=0.0127) and ΔH (p=0.0003) mutants (i.e., their survival abilities were less than that of the WT). Survival abilities of double mutants, on the other hand, were diverse. While ΔBC, ΔBH, and ΔBL mutants showed significantly higher susceptibilities (p<0.01), ΔCH and ΔHL exhibited lower growth when compared to WT. Among the triple mutants, the presence of σH in ΔBCL mutant resulted in higher susceptibility to duodenal fluid when compared to WT. Similar to gastric treatment, the absence of all four alternative sigma factors resulted in similar survival ability as wild type, suggesting again the compensatory role of σA under duodenal fluid exposure.
Following gastric and duodenal exposures, we further determined the survival ability of L. monocytogenes in simulated bile pH 8.2. Survival of L. monocytogenes exposed to simulated bile is shown in Figure 1C. Interestingly, after exposure to simulated bile juice for 10min, L. monocytogenes WT decreased by 2 logs CFU/ml. Approximately 3-log reduction was observed at 10-min exposure for ΔB and ΔC single mutants compared to the corresponding pre-treated culture. The log reduction in ΔB was significantly higher than that of wild type (p=0.0268). The absence of σH or σL alone in ΔH and ΔL strains resulted in better survival than WT. In addition, double deletions of sigH and sigL resulted in even more pronounced ability to survive in synthetic bile for ΔHL (p<0.0001). In contrast to sigL deletion, the absence of σC in ΔBC and ΔCH resulted in a higher decline in the bile exposure survival experiment compared to WT. Nevertheless, two triple mutants (ΔBCL and ΔCHL) survived significantly better than WT (p<0.05). As observed in gastric and duodenal fluid treatment, the quadruple mutant survived similar to wild type. Collectively, we suggest that σB and σC play positive roles in response to bile exposure, while σH and σL likely play negative roles on L. monocytogenes survival under bile treatment. We further focused on the effect of each alternative sigma factor on L. monocytogenes survival in simulated bile. Among the alternative sigma factors, σL showed the greatest significant effect on L. monocytogenes survival abilities in synthetic bile (p=0.000).
Although σL showed the greatest effect on L. monocytogenes survival in bile, the effects of other sigma factors were also determined. In order to decipher the interaction among sigma factors and the redundancy effect, we performed mean interaction plots showing the mean of log reductions from the presence and absence of each sigma factor compared with one another as shown in Figure 2. The effect of σB was compared in strains with σB (i.e., wild type, ΔC, ΔCH, and ΔCHL) and without σB (i.e., ΔB, ΔBC, ΔBL, and ΔBCHL). The presence or absence of σB though could not significantly induce bile resistance if σL was present (Figure 2A), and the presence of σB together with σL slightly increased survival ability in bile treatment. Contradictory, loss of σL greatly increased survival ability in presence or absence of σB while having σB only was better than absence of both. In other words, σB increases bile resistance but this effect is dampened in the presence of σL. The negative role of σL is stronger and could mask the positive role of σB on bile resistance. On the other hand, survival ability was significantly low in the presence of both σH and σL in comparison with the absence of both, p<0.05 (Figure 2B). Loss of σH increased the survival ability of L. monocytogenes only while σL still functioned. This finding suggests that σH and σL interacted synergistically to reduce resistance to bile. It could be suggested that σL plays a negative role on L. monocytogenes survival under bile since the presence of σL was always associated with lower survival. In addition, L. monocytogenes survived greater when σL was absent.
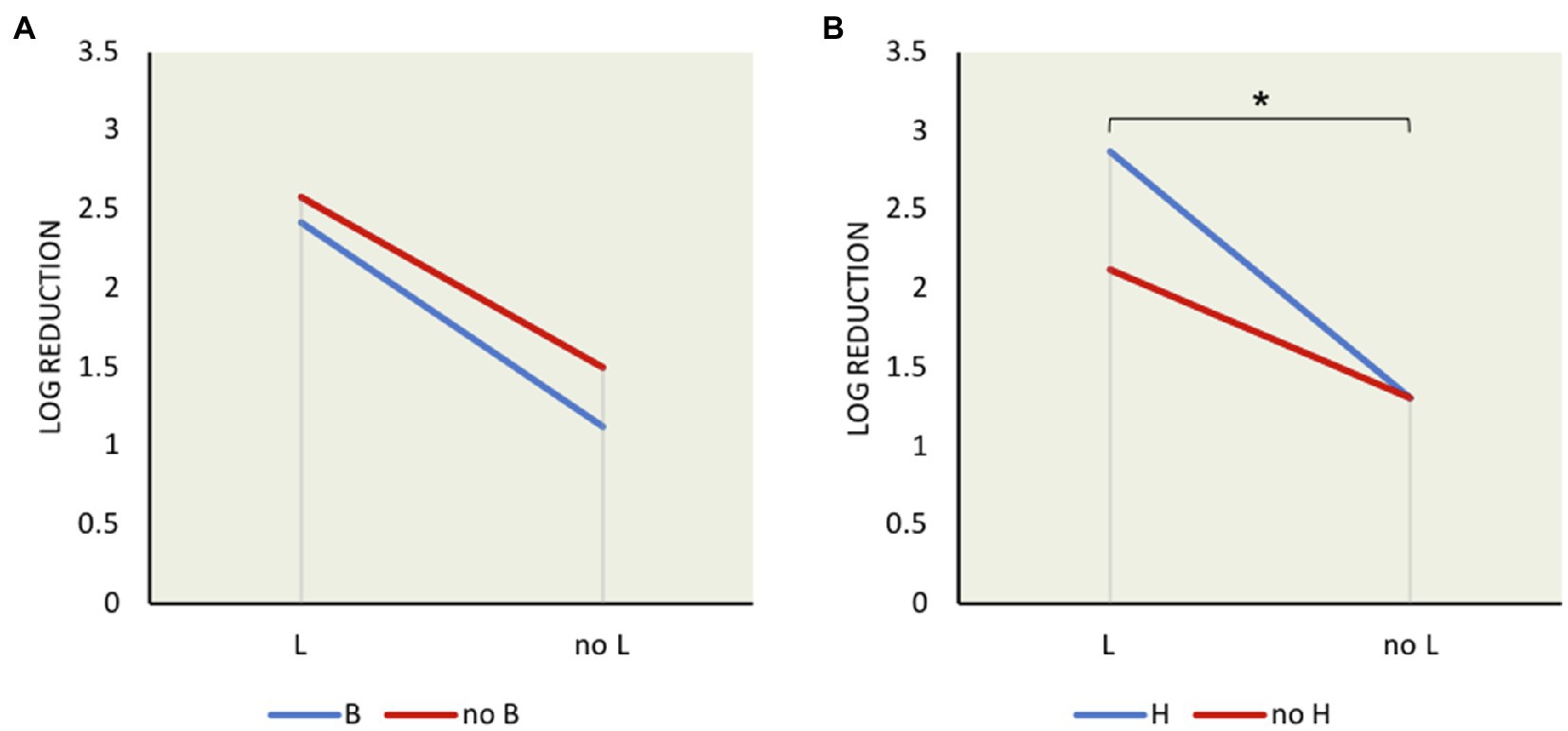
Figure 2. Mean interaction plots of L. monocytogenes log reduction after exposure to 6% bile for 10min showing the influence of the presence and absence of σ factors: (A) σB and σL interaction (B) σH and σL interaction. *indicates significant difference between two indicated factors using t test (p<0.05).
To determine the potential negative regulation of L. monocytogenes σL under simulated bile exposure, we compared the RNA-seq transcripts from ΔBCH strain exposed to bile to that without bile exposure. Since ΔBCH harbors σL as the only remaining alternative sigma factor, the identified regulon should indicate the effect of σL alone or the effect of σL in combination with the housekeeping sigma factor σA under bile exposure. With this bile exposure and non-exposure comparison, we identified 57 genes that were differentially expressed under bile in ΔBCH. Eighteen out of 57 genes showed higher transcripts under bile exposure, while 39 genes were downregulated as listed in Supplementary Table S3. Among 39 downregulated genes, two known negative regulators, ctsR and hrcA, showed lower transcripts in bile condition. CtsR, a transcriptional repressor, controls the class III heat shock genes Clp ATPase including clpC, clpP, and clpE (Nair et al., 2000). HrcA has been found to repress expression of dnaK and groSL operons that contain hrcA-grpE-dnaK-dnaJ and groS-groL genes, respectively (Woodbury and Haldenwang, 2003). Moreover, the tryptophan biosynthesis-associated genes (trpDEG) were also downregulated.
To assess the effect of the absence of σL, we further compared RNA-seq data between ΔL mutant grown in BHI and exposed to 1% bile. In this analysis, we found 84 genes were differentially expressed as shown in Supplementary Table S4. Of these, 36 genes were considered upregulated under bile treatment. The highest fold changes (FC) were observed in clpE gene and ilv operon ranging from 4 to 8 FC. clpE encodes ATP-dependent Clp protease, ATP-binding subunit ClpE, while the ilv operon is involved with valine biosynthesis. Universal stress response uspA2 gene was also significantly upregulated under bile exposure.
Conversely, 48 genes were downregulated in the absence of σL under bile exposure. Among them, approximately 3–4 FC was observed from LMRG_00042, LMRG_01298, and LMRG_01900. These genes are involved with glycerol catabolic pathway, nucleoside triphosphatase YtkD, and mannitol-specific phosphotransferase system (PTS), respectively. In addition, some transporter genes, such as LMRG_02716 and LMRG_01818, as well as flagella biosynthesis-associated gene flhF, were also downregulated during bile exposure.
To determine the role of σL in gene regulation, RNA-seq data from ΔBCHL and ΔBCH L. monocytogenes mutant strains grown in BHI were compared. ΔBCHL lacks all alternative sigma factors and only harbors the housekeeping σA, while ΔBCH mutant represents the sole presence of alternative σL which could co-ordinate or compete with the housekeeping σA. RNA-seq data comparison of ΔBCHL and ΔBCH allowed us to evaluate the regulation of σL with minimal redundancy of alternative sigma factors σB, σC, and σH. Under BHI only condition, ΔBCHL and ΔBCH transcript comparison revealed 216 genes (7.63% of 2,828 protein coding genes) that were significant differentially expressed (Supplementary Table S5). Interestingly, a total of 174 genes (approximately 80% of 216 genes) were negatively regulated under σL and only 42 genes were identified as positively regulated by σL.
Among the positively regulated genes shown, expression levels of mptC and mptD encoding for mannose-specific PTS components IIC and IID, respectively, were notably differentially expressed (Supplementary Table S5). They were highly expressed with more than 300 FC, suggesting strong positive regulation by σL. LMRG_02229 and LMRG_02348 encoding for cellobiose-specific PTS component IIC and putative regulator of the mannose operon, manO, respectively, showed increased expression levels up to 23 FC. Moreover, other mannose-specific PTS component genes including mpoABCD operon were also positively regulated under σL. Markedly, genes in glutamate decarboxylase in GAD systems, gadD2 and gadT2, showed higher transcripts as well.
Besides these positively regulated genes, 174 genes were defined as negatively regulated by σL, of which the LMRG_00091- LMRG_00094 operon encoding fructose-specific PTS components showed lower transcript levels as low as −39 FCs. LMRG_01669-LMRG_01672 involved in myo-inositol degradation, LMRG_02570 encoding a cellobiose-specific PTS component, and LMRG_01248-LMRG_01249 encoding galactitol-specific PTS components also showed decreases in gene expression. Moreover, plcA, plcB, actA, and csbD were also negatively regulated under σL.
In order to determine the role of σL regulation during bile exposure, we further analyzed RNA-seq data from ΔBCH and ΔBCHL L. monocytogenes mutant strains in bile. We identified 176 genes that were differentially expressed in bile condition, while 23 and 153 genes showed higher and lower transcripts, respectively (Supplementary Table S6). Later, we compared the differentially expressed genes from bile and BHI conditions to identify differentially expressed genes in bile only. Among 23 upregulated genes, we found 13 genes were highly expressed under bile condition only. Among 153 downregulated genes found during bile exposure, only 42 genes showed lower transcripts (FC<−2) in bile condition, but not in BHI. A Venn diagrams showing the overlap of transcripts with significantly higher (upregulated) and lower (downregulated) levels in BHI and bile exposure are demonstrated in Figures 3A,B, respectively.
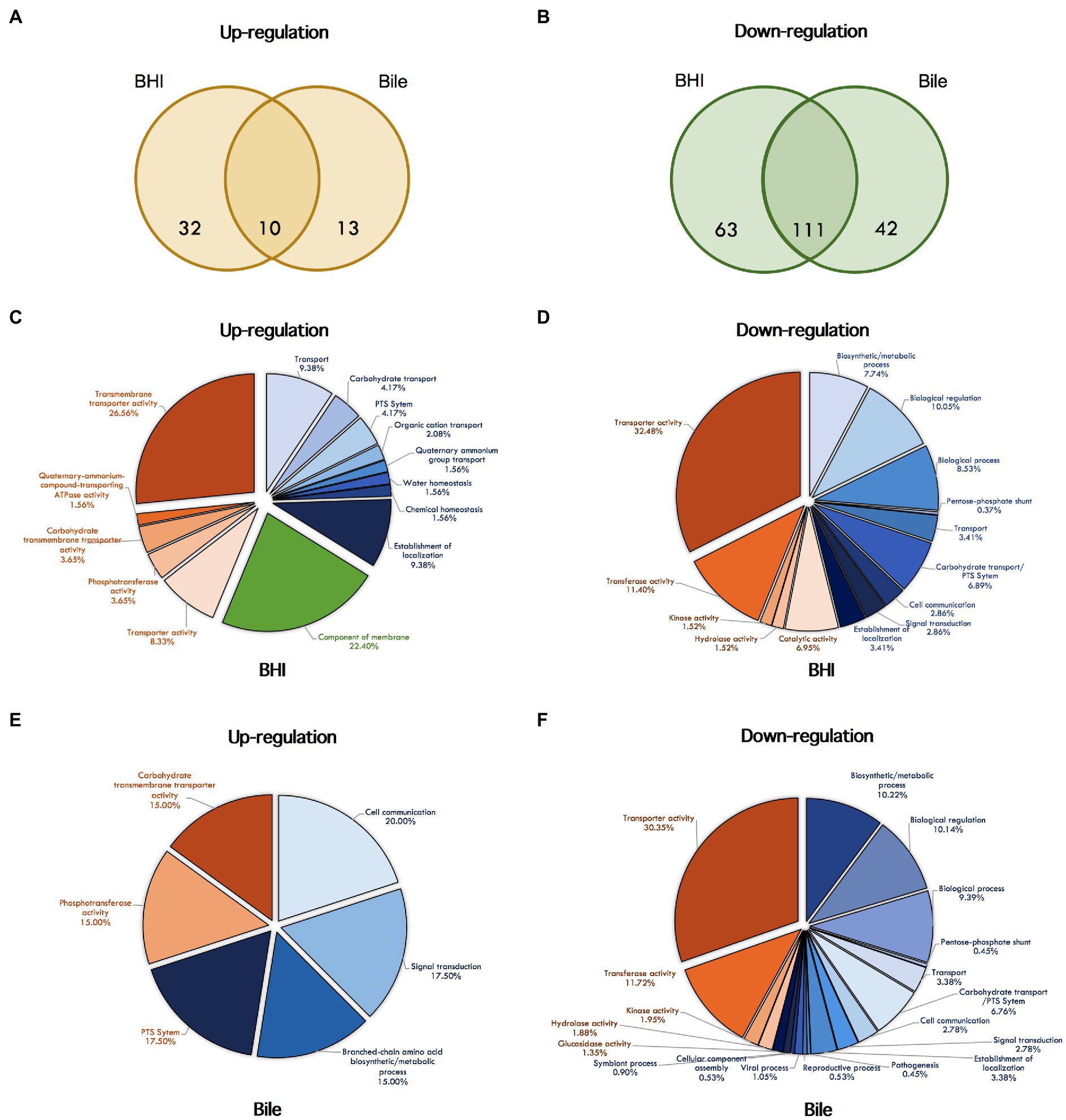
Figure 3. Venn diagrams showing the overlap of differentially expressed genes under σL regulation with significantly higher (A) and lower (B) levels. Differentially expressed genes in BHI and bile conditions are shown in the left and the right circles, respectively. The number indicated in the circle represents the number of differentially expressed genes in each condition. Pie chart showing GO term enrichment under σL regulation in BHI and bile. The GO term categories enriched among up- and down-regulated genes under BHI are shown in (C) and (D), under bile are shown in (E) and (F). Pie color indicate categories: blue, biological process; red, molecular function; green, cellular components.
Among the 13 upregulated genes under bile exposure, LMRG_02230 had the highest FC at 13; it encodes PTS mannose transporter subunit IIA. LMRG_01481 and LMRG_01482 encoding Rrf2 family transcriptional regulator and membrane protein, respectively, showed higher expression levels at 3–4 FC. leuC, ilvA, ilvC, yneA, and cggR were also listed in this category. leuC encodes 3-isopropylmalate dehydratase large subunit in leucine biosynthesis pathway. ilvA and ilvC are threonine-dehydratase- and ketol-acid-reductoisomerase encoded genes in valine biosynthesis and isoleucine biosynthesis, respectively. yneA encodes the sugar-binding component ABC transport protein, while cggR encodes central glycolytic genes regulator.
We had considered the role of σL in negative regulation in BHI since more than 80% of differentially expressed genes had reduced transcripts. By comparing BHI and bile RNA-seq data, up to 86% of differentially expressed genes were downregulated, of which only 42 genes were identified in bile exposure conditions. Interestingly, prfA and its mediated genes hly, hpt and inlB, as well as virulence-associated genes clpP, clpE, groL, and inlC, were also downregulated in this condition. However, other PrfA-dependent virulence-associated genes plcA, plcB, and actA showed lower transcripts in both conditions.
In addition to differentially transcribed genes, we identified 19 and 52 GO terms that were enriched under σL regulation in BHI with positive and negative manners, respectively, supporting the major role in negative regulation by σL (Figures 3C,D). Of these, GO terms for the components of membrane and the transport system, including PTS system, organic, carbohydrate, quaternary-ammonium-compound transporting system, are overrepresented among the upregulated genes under σL suggesting a role in transport system. In addition, among 52 GO terms identified, several biological and cellular processes and various transport systems, such as PTS system, transmembrane transporter, cation-proton symporter, and disaccharide transport, were found to be enriched among downregulated genes under σL regulation in BHI. A total of eight and 64 GO terms were found to be overrepresented among up- and downregulated genes under bile exposure, respectively (Figures 3E,F). As predicted, we found that the GO terms associated with carbohydrate transport including PTS system and branched-chain amino acid biosynthetic process were enriched among upregulated genes upon bile exposure. Apart from these GO terms, the GO terms for multiple cellular processes, carbohydrate, alcohol, and glycerol metabolic process, several transport systems, PTS system, and disaccharide transport were found to be enriched among downregulated genes under bile exposure. Surprisingly, the GO terms associated with viral process, virion assembly, and reproduction were overrepresented among downregulated genes.
We investigated the effects of the loss of σL in gene expression control. RNA-seq data from L. monocytogenes wild type and ΔL mutant in both BHI and bile were compared to evaluate the differential gene expression. In BHI, we observed 218 differentially expressed genes. Of these, 191 genes were upregulated in the absence of σL (Supplementary Table S7). However, 27 (12.39%) genes were downregulated in the absence of σL in BHI condition. Moreover, a total of 266 genes were differentially expressed, and 218 (81.95%) and 48 (18.4%) genes showed higher and lower transcripts in bile exposure condition, respectively (Supplementary Table S8). In contrast to the previous comparison between ΔBCH and ΔBCHL grown in BHI showing 80% of differentially expressed gene negatively regulated by σL, this comparison between WT and ΔL under BHI showed a higher proportion (87.61%) of positively regulated gene by σL. This result confirmed the negative role of σL in gene expression since loss of σL could upregulate some of downregulated genes shown in ΔBCH and ΔBCHL comparison. For example, LMRG_02569 encoding cellobiose-specific PTS IIB component was shown to be negatively regulated under σL by −9 FC; however, it had higher expression by 16 FC when in ΔL mutant. Beta-glucosidase encoded gene LMRG_02568 was expressed at low level in ΔBCH mutant but its transcript was increased to as high as 14.5 FC in the ΔL mutant. This suggested that deletion of sigL upregulated genes that were suppressed or downregulated. Among the upregulated gene in ΔL background, LMRG_00091 gene, identified as σL negatively regulated gene, was also upregulated as high as 61 FC.
To identify potential reporter genes for σL activities, selected sets of σL-dependent genes were verified in qRT-PCR assays. We confirmed the positive and negative role of σL using differential expression of mptA and csbD, respectively (Figure 4A). The expression level of mptA in ΔBCH is similar to wild type but decreased in ΔL and ΔBCHL, supporting the positive role of σL. Bile is not involved in mptA transcription since there is no correlation between the presence and absence of σL with or without bile (Figure 5A). In contrast, the csbD and LMRG_00704 transcripts are significantly lower in the presence of σL alone in ΔBCH (p<0.05); however, their transcripts are highly expressed in ΔL. Therefore, csbD and LMRG_00704 are negatively regulated by σL. Bile does not influence the level of csbD and LMRG_00704 as bile does not significantly alter the level of transcripts of these genes (Figures 5D,E). The ilv operon (ilvD-ilvC) is also upregulated in the presence of bile in ΔL (Supplementary Table S4), further confirming that ilv operon is negatively regulated under bile exposure. The interaction plot also shows that bile exposure can change the direction of ilvD-ilvC expression compared to BHI (Figures 5B,C). Hence, it can be confirmed that bile could upregulate ilvD-ilvC expression in the absence of σL. Overall, for gene expression levels of these selected genes, we have shown that RNA-seq and qRT-PCR are well correlated with Pearson correlation coefficients (r) of expression levels in the wild type, ΔL and ΔBCH and ΔBCHL mutant at 0.87, 0.91, 0.86, and 0.93, respectively (Figure 4B).
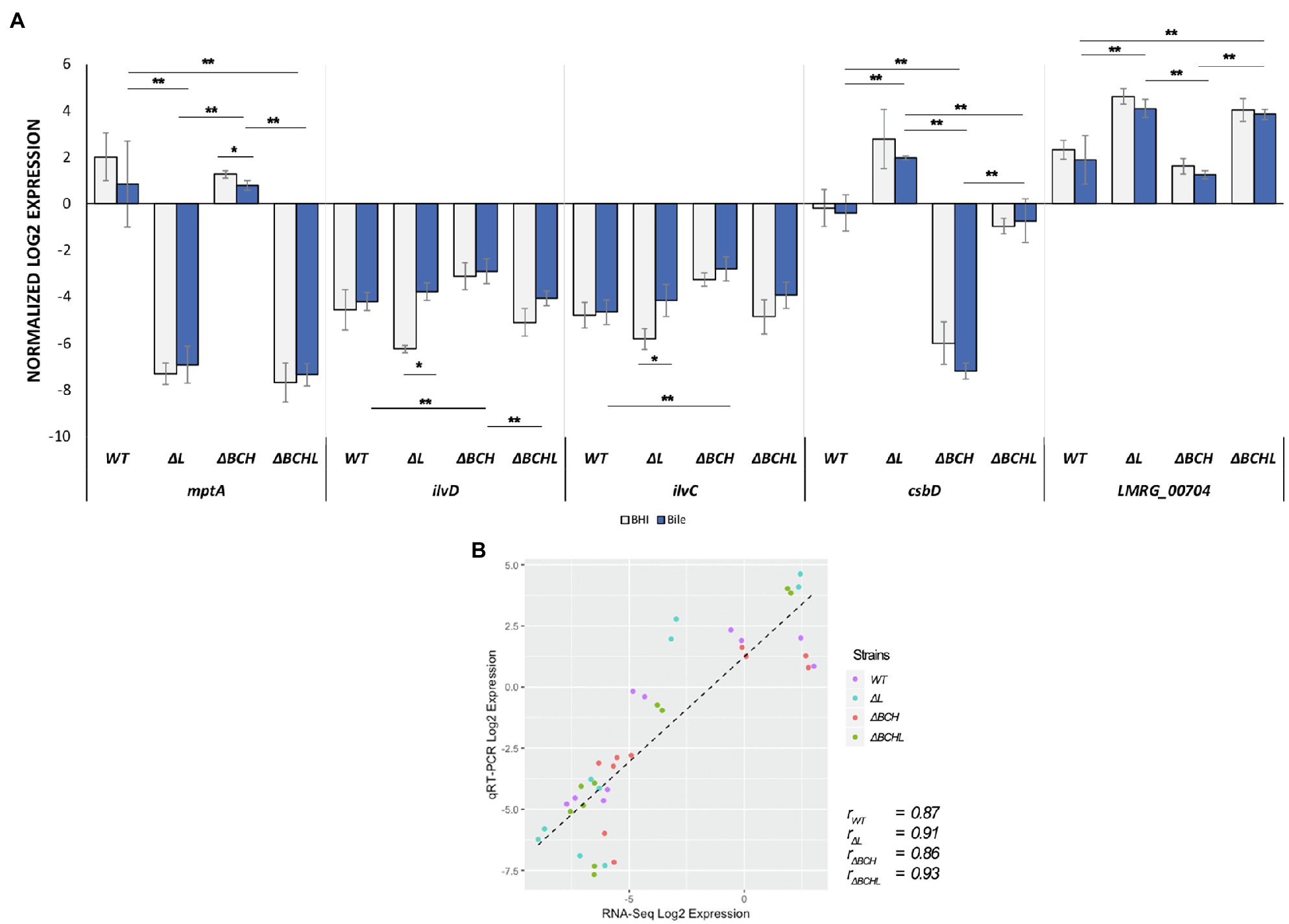
Figure 4. Validation of differential expressed genes (DEGs) by TaqMan qRT-PCR. (A) Transcript levels were quantified in, mptA, ilvD, ilvC, csbD, and LMRG_00704 in wild type; ΔL; ΔBCH; and ΔBCHL quadruple mutant. Transcript levels are expressed as relative to WT after normalization to geometric mean of housekeeping genes; rpoB and bglA. Blue histograms represented L. monocytogenes strains exposed to simulated 1% bile for 10min as compared to its BHI control indicated in grey. Experiments were performed in biological triplicates. (B) Pearson correlation coefficients (r) of expression levels accounted from RNA-seq and qRT-PCR data in the wild type (purple), ΔL (blue), ΔBCH (red), and ΔBCHL (green). *indicates significant difference in bile treatment compared to BHI using unpaired t-test (p<0.05) and **indicates significant difference among strains within the same condition (p<0.05) using One-Way ANOVA.
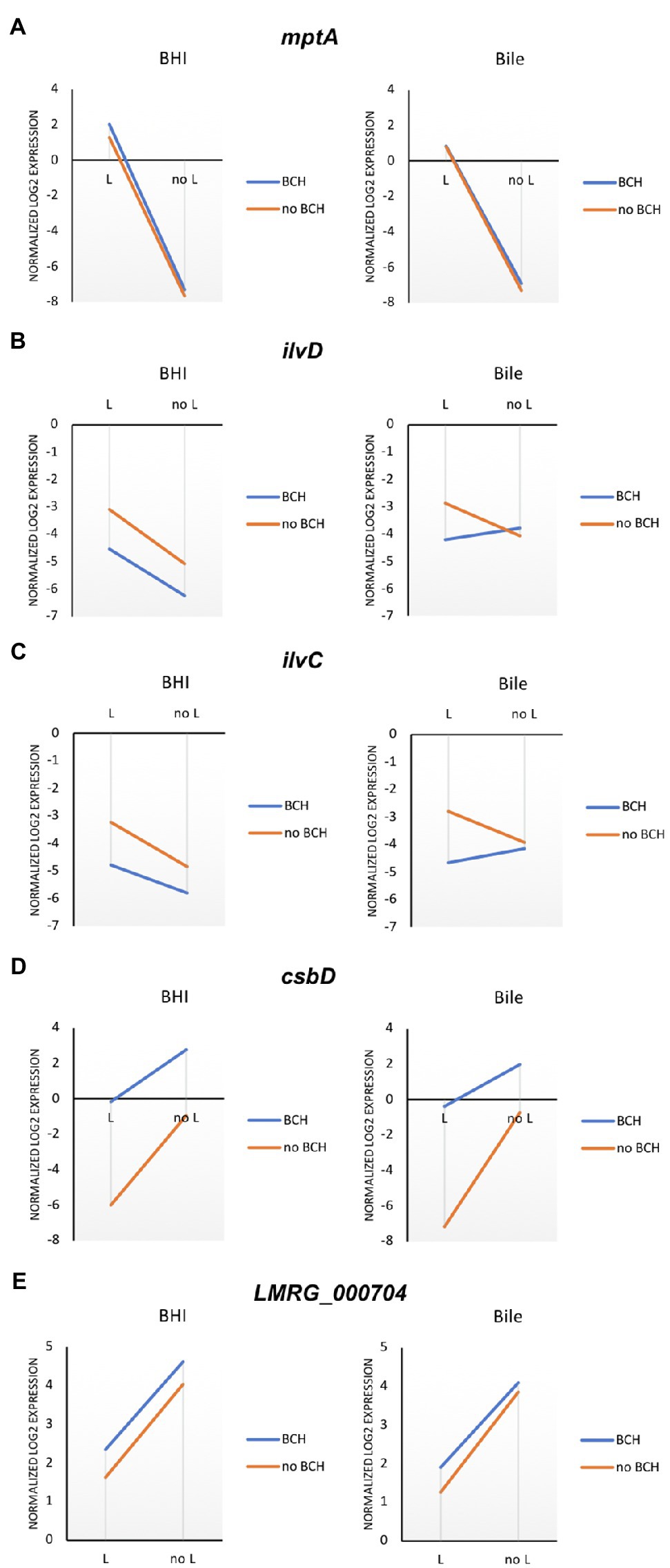
Figure 5. Mean interaction plots of (A) mptA, (B) ilvD, (C) ilvC, (D) csbD, and (E) LMRG_000704 expression expression levels accounted from wild type, ΔL, ΔBCH, and ΔBCHL in BHI (left) and bile exposure (right) by qRT-PCR. Blue line indicates the presence of σB, σC, and σH in the conditions, while the orange line represents the strains without those sigma factors.
Our data showed that simulated GI fluid, in particular bile fluid, could reduce survival ability of L. monocytogenes under high pH mimicking the bile in gallbladder. However, simulated gastric and duodenal fluids could not significantly reduce cell survival. Upon low pH exposure during gastric fluid stress (pH 1.3), we could not observe the reduction of ΔB mutant, in contrast to other studies showing that ΔB mutant had impaired survival in acidic culture at pH 2.5 compared to its wild type (Ferreira et al., 2003). This conflicted finding might be explained by the difference in exposure time between studies. The ΔB mutant was exposed for an hour to acidic pH, while, in our experiment, the incubation time was 10min which could lead to lower CFU reduction. In addition, using an organic acid (acetic acid) is more harmful to bacterial cells than inorganic acid (HCl) used in our system compared to other finding (Werbrouck et al., 2009). The gastric fluid ingredients used in our assay were different from others; therefore, σB might play a limited role in this gastric stress and time exposure. We also did not observe reduction in L. monocytogenes in simulated duodenal fluid. It might be due to lack of key bactericidal enzymes mixed in our duodenal fluid; the enzymes used in our assay were lipase and pancreatin. Lipase typically functions in the digestive system, processing dietary lipids and transport. Pancreatin, produced by pancreas cells, is a combination of digestive enzymes including amylase, protease, and lipase. Its major role is digesting sugars, proteins, and processing fats. Thus, releasing bile from the gallbladder is important not only for digestive system functionality but also as a defensive mechanism against pathogens in the intestine (Boyer, 2013).
A very small number of genes were found to be regulated by σL in previous studies. According to proteomic analysis, only two proteins were positively regulated by σL (Mujahid et al., 2013a). Based on our transcriptomic analysis, 42 genes were positively regulated in normal BHI condition. Of these, two of them were consistent to the proteomic study (Mujahid et al., 2013a), which are mannose-specific PTS component gene mptA and acetolactate synthase encoding gene alsS. mptA was not positively regulated in our study; however, genes under its regulon, mptC and mptD, were positively regulated. Therefore, we assumed that mptA was positively regulated by σL since they were located in the same operon. Unexpectedly, none of the positively regulated genes identified in this study were found in previous microarray analyses (Chaturongakul et al., 2011). This may be due to the cell phase difference of L. monocytogenes used. While L. monocytogenes grown to log phase was used in our experiment, stationary phase cells were analyzed in a previous assay. In addition to the difference in cell phase, different strains were used in the analyses. In microarray study, ΔL was used in comparison with WT (Chaturongakul et al., 2011), whereas in our study, ΔBCH and ΔBCHL were compared in order to identify σL-dependent genes which turned out to be consistent with the previous proteomic study (Mujahid et al., 2013a). Among the positively regulated genes, we also identified that mpoABCD operon which encoded mannose-specific IIABCD component was positively regulated by σL. Arous et al. (2004) have proposed by in silico analysis that this mpoABCD operon might be regulated by σL. However, Raengpradub et al. (2008) have shown that σB upregulated the transcription of this operon as well. Therefore, it suggests that operon mpoABCD is under co-regulation of σB and σL. The crosstalk between mpt and mpo has been demonstrated; however, cross-regulation is not fully understood (Arous et al., 2004). In addition, σL has previously been shown to contribute to cold, high salt, and organic acid stresses since the impaired growth was observed in the ΔL mutant when compared to its isogenic WT (Raimann et al., 2009). However, only one of the cold and NaCl adaptation genes, oppA, was shown to be significantly different between ΔL and WT under cold stress, while cspD and clpP did not show statistical difference under 3% NaCl exposure. It is also interesting that our data showed clpP one of the negatively regulated genes under σL.
It is likely that σL negatively regulates a large number of genes, which is concordant with protein analysis (Mujahid et al., 2013a). Among 216 downregulated genes under σL control identified in both BHI and bile, 27 genes were coherent with 56 proteins identified by proteomic study. We found only two genes that were consistent with previous microarray assays which were lmo2340 (LMRG_01503) and lmo0345 (LMRG_00036; Chaturongakul et al., 2011). We found that more than 80% of differentially expressed genes were negatively regulated by σL. The role category function of σL was overrepresented by transport, binding protein, and energy metabolism. Of these, we could only confirm the negative regulation of σL in csbD and possibly LMRG_02283. It was clearly shown that in the absence of σL in ΔL mutant, csbD transcription was significantly induced. In contrast to the presence of σL in ΔBCH background, where a significant reduction of csbD transcript was observed. It was previously shown that csbD is a σB-dependent gene (Hain et al., 2008). Nevertheless, we observed the induction of csbD in ΔL compared to wild type. If it were solely under σB, the level of expression would remain as the wild type. Therefore, we suggested that csbD is positively regulated by σB but negatively regulated by σL. We could not confirm the role of σL in LMRG_00091 transcription, though we observed a similar expression pattern to csbD, but not significantly different. Interestingly, some virulence-associated genes, (e.g., actA, plcA, and plcB) were downregulated by σL. The association between σL and virulence is yet to be confirmed. It is possible the negative regulation by σL is in an indirect manner.
In agreement with our previous study, bile mediated higher production of ilvC gene (ilv operon) in L. monocytogenes 10403S (Boonmee et al., 2019). Based on our current RNA-seq data, ilvC was also higher expressed in ΔBCH when compared to that in ΔBCHL under bile condition suggesting the positive role of σL. We also highlighted that ilvC expression level in ΔBCH was significantly increased in comparison with that of WT (Figure 4A). In addition, gadT2 was predicted in Boonmee et al. (2019) to be controlled by alternative sigma factors, that is, significantly higher expression in WT when compared to ΔBCHL. We confirmed that gadT2 was positively regulated by σL since it showed higher transcripts in both BHI and bile conditions (Supplementary Tables S5 and S6).
Due to a large number of genes negatively regulated by σL, we hypothesized its negative regulation of virulence-associated genes under bile exposure. prfA, inlB, inlC, and hly were downregulated by σL under bile treatment. PrfA, a master virulence regulator, is tightly regulated and has been shown to be positively regulated by σB (Kazmierczak et al., 2006; Ollinger et al., 2009). We have observed the downregulation of prfA itself as well as part of its regulon (hly, hpt, inlB, clpP, clpE, groL, and inlC) in response to 1% basic bile (pH 8.2). It was concordant with previous microarray study and with our RNA-seq study showing the whole PrfA regulon was repressed under bile exposure (Quillin et al., 2011; Boonmee et al., 2019). In contrast, Guariglia-Oropeza et al. (2018) reported upregulation of PrfA regulon but not prfA itself upon exposure to acidic bile (pH 5.5). It is suggested that the alteration of bile pH could contribute to diverse regulation and transcription levels since pH was considered as a stimulus as well. In addition, other factors such as strain, serotype, and lineage could vary members of bile stimulon (White et al., 2015). Nevertheless, the interplay between downregulation and upregulation and of PrfA by σL and σB suggests the transcription network switch between listerial adaptation during GI passage prior to host cell invasion.
Altogether, to finetune expression of genes under stress exposure, the L. monocytogenes 10403S expression systems need to find their new balance. Excessive expression either RNA or protein might be noxious to the cells. The absence of σL, including single, double, and triple mutants, comparatively increases resistance to bile compared to WT suggesting its negative role on bile survival. We have shown that, under bile exposure, a number of genes are negatively controlled by σL. Interestingly, the major virulence factor, PrfA, is identified as negatively regulated by σL. Therefore, the functional roles and the underlying mechanisms in which σL and σB co-mediate PrfA expression are to be further investigated. The positive role of σB and the negative role of σL support their adaptive roles to finetune expression of survival genes important for bile persistence.
The datasets generated or analyzed for this study were deposited and can be found under the SRA accession number PRJNA544468 (https://www.ncbi.nlm.nih.gov/sra/PRJNA544468).
HO and SC conceived the study. AB performed the experiments and analyzed the data. AB, HO, and SC drafted the manuscript. All authors contributed to the article and approved the submitted version.
The study was supported by Office of the Higher Education Commission (Thailand) for Talent Mobility Program (to SC). HO is partially supported by USDA-NIFA Hatch project IND010930.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank the Department of Statistics at Purdue University for statistical advice.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.713383/full#supplementary-material
Alves, A., Magalhaes, R., Brandao, T. R. S., Pimentel, L., Rodriguez-Alcala, L. M., Teixeira, P., et al. (2020). Impact of exposure to cold and cold-osmotic stresses on virulence-associated characteristics of Listeria monocytogenes strains. Food Microbiol. 87:103351. doi: 10.1016/j.fm.2019.103351
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq—a python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Arous, S., Dalet, K., and Hechard, Y. (2004). Involvement of the mpo operon in resistance to class IIa bacteriocins in Listeria monocytogenes. FEMS Microbiol. Lett. 238, 37–41. doi: 10.1016/j.femsle.2004.07.014
Axelsson, L., Bjerke, G. A., McLeod, A., Berget, I., and Holck, A. L. (2020). Growth behavior of Listeria monocytogenes in a traditional Norwegian fermented fish product (Rakfisk), and its inhibition through bacteriophage addition. Foods 9:119. doi: 10.3390/foods9020119
Becker, L. A., Cetin, M. S., Hutkins, R. W., and Benson, A. K. (1998). Identification of the gene encoding the alternative sigma factor sigmaB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180, 4547–4554. doi: 10.1128/JB.180.17.4547-4554.1998
Begley, M., Gahan, C. G., and Hill, C. (2002). Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl. Environ. Microbiol. 68, 6005–6012. doi: 10.1128/AEM.68.12.6005-6012.2002
Begley, M., Gahan, C. G., and Hill, C. (2005). The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651. doi: 10.1016/j.femsre.2004.09.003
Bevilacqua, A., Campaniello, D., Speranza, B., Sinigaglia, M., and Corbo, M. R. (2018). Survival of Listeria monocytogenes and Staphylococcus aureus in synthetic brines. Studying the effects of salt, temperature and sugar through the approach of the design of experiments. Front. Microbiol. 9:240. doi: 10.3389/fmicb.2018.00240
Boonmee, A., Oliver, H. F., and Chaturongakul, S. (2019). Listeria monocytogenes sigma(A) is sufficient to survive gallbladder bile exposure. Front. Microbiol. 10:2070. doi: 10.3389/fmicb.2019.02070
Boyer, J. L. (2013). Bile formation and secretion. Compr. Physiol. 3, 1035–1078. doi: 10.1002/cphy.c120027
Chaturongakul, S., Raengpradub, S., Palmer, M. E., Bergholz, T. M., Orsi, R. H., Hu, Y., et al. (2011). Transcriptomic and phenotypic analyses identify coregulated, overlapping regulons among PrfA, CtsR, HrcA, and the alternative sigma factors sigmaB, sigmaC, sigmaH, and sigmaL in Listeria monocytogenes. Appl. Environ. Microbiol. 77, 187–200. doi: 10.1128/AEM.00952-10
Dawson, P. A. (1998). “Bile secretion and the enterohepatic circulation of bile acids,” in Sleisenger and Fordtran’s Gastrointestinal and Liver Disease: Pathophysiology/Diagnosis/Management. 7th Edn. eds. M. S. Feldman and M. H. B. F. Scharschmidt (Philadelphia, PA: W. B. Saunders, Co.).
Dowd, G. C., Joyce, S. A., Hill, C., and Gahan, C. G. (2011). Investigation of the mechanisms by which Listeria monocytogenes grows in porcine gallbladder bile. Infect. Immun. 79, 369–379. doi: 10.1128/IAI.00330-10
Dressman, J. B., Berardi, R. R., Dermentzoglou, L. C., Russell, T. L., Schmaltz, S. P., Barnett, J. L., et al. (1990). Upper gastrointestinal (GI) pH in young, healthy men and women. Pharm. Res. 7, 756–761. doi: 10.1023/A:1015827908309
Dussurget, O., Cabanes, D., Dehoux, P., Lecuit, M., Buchrieser, C., Glaser, P., et al. (2002). Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45, 1095–1106. doi: 10.1046/j.1365-2958.2002.03080.x
Efentakis, M., and Dressman, J. B. (1998). Gastric juice as a dissolution medium: surface tension and pH. Eur. J. Drug Metab. Pharmacokinet. 23, 97–102. doi: 10.1007/BF03189322
Evans, D. F., Pye, G., Bramley, R., Clark, A. G., Dyson, T. J., and Hardcastle, J. D. (1988). Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut 29, 1035–1041. doi: 10.1136/gut.29.8.1035
Ferreira, A., O’Byrne, C. P., and Boor, K. J. (2001). Role of sigma(B) in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67, 4454–4457. doi: 10.1128/AEM.67.10.4454-4457.2001
Ferreira, A., Sue, D., O’Byrne, C. P., and Boor, K. J. (2003). Role of Listeria monocytogenes sigma(B) in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69, 2692–2698. doi: 10.1128/AEM.69.5.2692-2698.2003
Goulet, V., King, L. A., Vaillant, V., and de Valk, H. (2013). What is the incubation period for listeriosis? BMC Infect. Dis. 13:11. doi: 10.1186/1471-2334-13-11
Guariglia-Oropeza, V., Orsi, R. H., Guldimann, C., Wiedmann, M., and Boor, K. J. (2018). The Listeria monocytogenes bile stimulon under acidic conditions is characterized by strain-specific patterns and the upregulation of motility, cell wall modification functions, and the PrfA regulon. Front. Microbiol. 9:120. doi: 10.3389/fmicb.2018.00120
Hain, T., Hossain, H., Chatterjee, S. S., Machata, S., Volk, U., Wagner, S., et al. (2008). Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e sigmaB regulon. BMC Microbiol. 8:20. doi: 10.1186/1471-2180-8-20
Hardy, J., Francis, K. P., DeBoer, M., Chu, P., Gibbs, K., and Contag, C. H. (2004). Extracellular replication of Listeria monocytogenes in the murine gall bladder. Science 303, 851–853. doi: 10.1126/science.1092712
Kaur, S., Malik, S. V., Vaidya, V. M., and Barbuddhe, S. B. (2007). Listeria monocytogenes in spontaneous abortions in humans and its detection by multiplex PCR. J. Appl. Microbiol. 103, 1889–1896. doi: 10.1111/j.1365-2672.2007.03414.x
Kazmierczak, M. J., Wiedmann, M., and Boor, K. J. (2006). Contributions of Listeria monocytogenes sigmaB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology 152, 1827–1838. doi: 10.1099/mic.0.28758-0
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lungu, B., Ricke, S. C., and Johnson, M. G. (2009). Growth, survival, proliferation and pathogenesis of Listeria monocytogenes under low oxygen or anaerobic conditions: a review. Anaerobe 15, 7–17. doi: 10.1016/j.anaerobe.2008.08.001
Membre, J. M., Leporq, B., Vialette, M., Mettler, E., Perrier, L., Thuault, D., et al. (2005). Temperature effect on bacterial growth rate: quantitative microbiology approach including cardinal values and variability estimates to perform growth simulations on/in food. Int. J. Food Microbiol. 100, 179–186. doi: 10.1016/j.ijfoodmicro.2004.10.015
Moorhead, S. M., and Dykes, G. A. (2004). Influence of the sigB gene on the cold stress survival and subsequent recovery of two Listeria monocytogenes serotypes. Int. J. Food Microbiol. 91, 63–72. doi: 10.1016/S0168-1605(03)00332-5
Mujahid, S., Orsi, R. H., Boor, K. J., and Wiedmann, M. (2013a). Protein level identification of the Listeria monocytogenes sigma H, sigma L, and sigma C regulons. BMC Microbiol. 13:156. doi: 10.1186/1471-2180-13-156
Mujahid, S., Orsi, R. H., Vangay, P., Boor, K. J., and Wiedmann, M. (2013b). Refinement of the Listeria monocytogenes sigmaB regulon through quantitative proteomic analysis. Microbiology 159, 1109–1119. doi: 10.1099/mic.0.066001-0
Nair, S., Derre, I., Msadek, T., Gaillot, O., and Berche, P. (2000). CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35, 800–811. doi: 10.1046/j.1365-2958.2000.01752.x
NicAogain, K., and O’Byrne, C. P. (2016). The role of stress and stress adaptations in determining the fate of the bacterial pathogen Listeria monocytogenes in the food chain. Front. Microbiol. 7:1865. doi: 10.3389/fmicb.2016.01865
Olier, M., Rousseaux, S., Piveteau, P., Lemaitre, J. P., Rousset, A., and Guzzo, J. (2004). Screening of glutamate decarboxylase activity and bile salt resistance of human asymptomatic carriage, clinical, food, and environmental isolates of Listeria monocytogenes. Int. J. Food Microbiol. 93, 87–99. doi: 10.1016/j.ijfoodmicro.2003.10.010
Oliver, S. P., Jayarao, B. M., and Almeida, R. A. (2005). Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodborne Pathog. Dis. 2, 115–129. doi: 10.1089/fpd.2005.2.115
Ollinger, J., Bowen, B., Wiedmann, M., Boor, K. J., and Bergholz, T. M. (2009). Listeria monocytogenes sigmaB modulates PrfA-mediated virulence factor expression. Infect. Immun. 77, 2113–2124. doi: 10.1128/IAI.01205-08
Pleitner, A. M., Trinetta, V., Morgan, M. T., Linton, R. L., and Oliver, H. F. (2014). Transcriptional and phenotypic responses of Listeria monocytogenes to chlorine dioxide. Appl. Environ. Microbiol. 80, 2951–2963. doi: 10.1128/AEM.00004-14
Quillin, S. J., Schwartz, K. T., and Leber, J. H. (2011). The novel Listeria monocytogenes bile sensor BrtA controls expression of the cholic acid efflux pump MdrT. Mol. Microbiol. 81, 129–142. doi: 10.1111/j.1365-2958.2011.07683.x
Raengpradub, S., Wiedmann, M., and Boor, K. J. (2008). Comparative analysis of the sigma B-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol. 74, 158–171. doi: 10.1128/AEM.00951-07
Raimann, E., Schmid, B., Stephan, R., and Tasara, T. (2009). The alternative sigma factor sigma(L) of L. monocytogenes promotes growth under diverse environmental stresses. Foodborne Pathog. Dis. 6, 583–591. doi: 10.1089/fpd.2008.0248
Sue, D., Fink, D., Wiedmann, M., and Boor, K. J. (2004). sigmaB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150, 3843–3855. doi: 10.1099/mic.0.27257-0
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Versantvoort, C. H., Oomen, A. G., Van de Kamp, E., Rompelberg, C. J., and Sips, A. J. (2005). Applicability of an in vitro digestion model in assessing the bioaccessibility of mycotoxins from food. Food Chem. Toxicol. 43, 31–40. doi: 10.1016/j.fct.2004.08.007
Werbrouck, H., Vermeulen, A., Van Coillie, E., Messens, W., Herman, L., Devlieghere, F., et al. (2009). Influence of acid stress on survival, expression of virulence genes and invasion capacity into Caco-2 cells of Listeria monocytogenes strains of different origins. Int. J. Food Microbiol. 134, 140–146. doi: 10.1016/j.ijfoodmicro.2009.03.022
White, S. J., McClung, D. M., Wilson, J. G., Roberts, B. N., and Donaldson, J. R. (2015). Influence of pH on bile sensitivity amongst various strains of Listeria monocytogenes under aerobic and anaerobic conditions. J. Med. Microbiol. 64, 1287–1296. doi: 10.1099/jmm.0.000160
Woodbury, R., and Haldenwang, W. G. (2003). HrcA is a negative regulator of the dnaK and groESL operons of Streptococcus pyogenes. Biochem. Biophys. Res. Commun. 302, 722–727. doi: 10.1016/S0006-291X(03)00254-7
Keywords: RNA-seq, Listeria monocytogenes, alternative sigma L, negative regulator, bile, stress
Citation: Boonmee A, Oliver HF and Chaturongakul S (2021) Listeria monocytogenes 10403S Alternative Sigma-54 Factor σL Has a Negative Role on Survival Ability Under Bile Exposure. Front. Microbiol. 12:713383. doi: 10.3389/fmicb.2021.713383
Received: 22 May 2021; Accepted: 24 September 2021;
Published: 21 October 2021.
Edited by:
David Rodriguez-Lazaro, University of Burgos, SpainReviewed by:
Christophe Nguyen, The Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceCopyright © 2021 Boonmee, Oliver and Chaturongakul. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soraya Chaturongakul, c29yYXlhLmNoYUBtYWhpZG9sLmFjLnRo
†Present address: Soraya Chaturongakul, Institute of Molecular Biosciences, Mahidol University, Nakhon Pathom, Thailand
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.