
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 16 July 2021
Sec. Microbial Physiology and Metabolism
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.712804
This article is part of the Research TopicRoles of Regulatory RNAs in Bacterial PathogensView all 13 articles
Helicobacter pylori is a Gram-negative bacterial pathogen that colonizes the stomach of about half of the human population worldwide. Infection by H. pylori is generally acquired during childhood and this bacterium rapidly establishes a persistent colonization. H. pylori causes chronic gastritis that, in some cases, progresses into peptic ulcer disease or adenocarcinoma that is responsible for about 800,000 deaths in the world every year. H. pylori has evolved efficient adaptive strategies to colonize the stomach, a particularly hostile acidic environment. Few transcriptional regulators are encoded by the small H. pylori genome and post-transcriptional regulation has been proposed as a major level of control of gene expression in this pathogen. The transcriptome and transcription start sites (TSSs) of H. pylori strain 26695 have been defined at the genome level. This revealed the existence of a total of 1,907 TSSs among which more than 900 TSSs for non-coding RNAs (ncRNAs) including 60 validated small RNAs (sRNAs) and abundant anti-sense RNAs, few of which have been experimentally validated. An RNA degradosome was shown to play a central role in the control of mRNA and antisense RNA decay in H. pylori. Riboregulation, genetic regulation by RNA, has also been revealed and depends both on antisense RNAs and small RNAs. Known examples will be presented in this review. Antisense RNA regulation was reported for some virulence factors and for several type I toxin antitoxin systems, one of which controls the morphological transition of H. pylori spiral shape to round coccoids. Interestingly, the few documented cases of small RNA-based regulation suggest that their mechanisms do not follow the same rules that were well established in the model organism Escherichia coli. First, the genome of H. pylori encodes none of the two well-described RNA chaperones, Hfq and ProQ that are important for riboregulation in several organisms. Second, some of the reported small RNAs target, through “rheostat”-like mechanisms, repeat-rich stretches in the 5′-untranslated region of genes encoding important virulence factors. In conclusion, there are still many unanswered questions about the extent and underlying mechanisms of riboregulation in H. pylori but recent publications highlighted original mechanisms making this important pathogen an interesting study model.
Helicobacter pylori is a Gram-negative bacterium belonging to the epsilon-proteobacteria class recently proposed to be renamed as Campylobacterota (Waite et al., 2017). H. pylori is a microaerophilic and helical shaped microorganism that inhabits the stomach of half of the human population worldwide. H. pylori is transmitted between humans and generally acquired before the age of five. Infected individuals suffer from chronic gastritis that can remain asymptomatic throughout their lives in 85% of the cases or can evolve into a range of disorders including peptic ulcers or gastric adenocarcinoma, that is responsible for about 800,000 deaths every year worldwide (Robinson and Atherton, 2021). The severe pathologies associated to H. pylori infection, like cancer, generally occur after decades of chronic infection. H. pylori has evolved to persistently colonize the stomach, despite the harsh conditions of this hostile environment, such as a very low pH and constantly changing conditions, which indicates that it has a strong adaptation capacity. It was thus surprising to observe that H. pylori possesses few [only 16 (De la Cruz et al., 2017)] transcriptional regulators (Tomb et al., 1997). This is consistent with its small genome of only 1.67 Mb that encodes 1,576 open reading frames (ORFs) in the 26695 type strain and its unique human gastric niche. Given the reduced number of transcriptional regulators, post-transcriptional regulation has been proposed to play a major role in the control of gene expression in H. pylori (Pernitzsch and Sharma, 2012). H. pylori does not possess RNA chaperones like Hfq and ProQ, that are important factors in post-transcriptional regulation in many bacteria, including the model organism Escherichia coli (Quendera et al., 2020). This characteristic has been an impediment for rapid regulatory RNAs identification in H. pylori but it also hints to original mechanisms for post-transcriptional regulation, that we will address in this review and are summarized in Figures 1, 2.
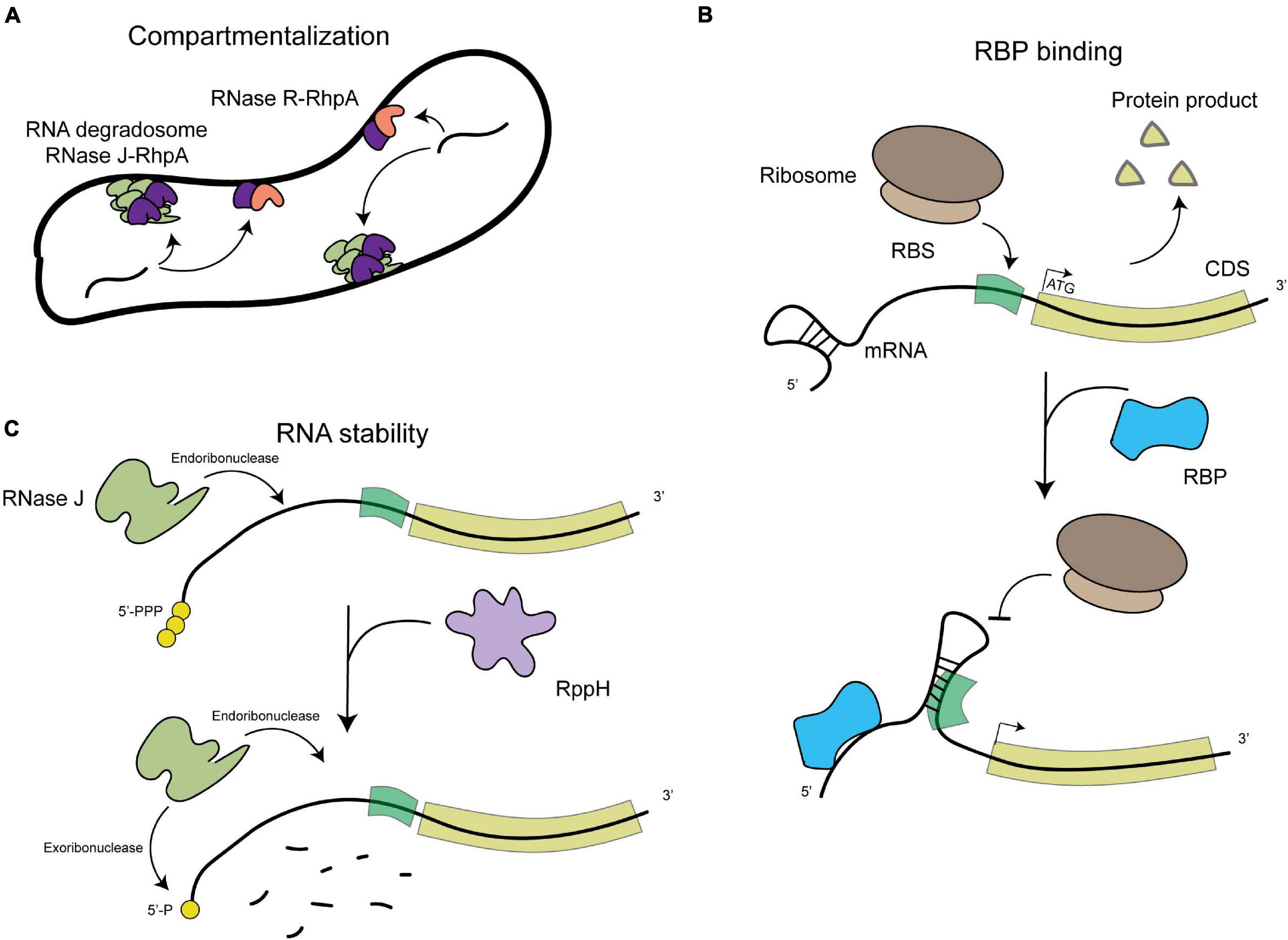
Figure 1. Schematic representation of the general levels of post-transcriptional regulation in H. pylori. (A) Compartmentalization of RNA degradation, with the RNA degradosome composed of RNAse J (in green) and RhpA (in pink) localizing into foci at the inner membrane and the RNase R (in dark purple)-RhpA complex also binding the inner membrane. (B) RNA binding protein (RBP)-mediated regulation, where binding of an RBP (in blue) can cause a structural rearrangement in the RNA molecule that allows or prevents the expression of the proteins it encodes. (C) RNA degradation by both endo- and exoribonucleases, such as RNase J (in light green), which also depends on the phosphorylation state of the 5′-end of the mRNA molecules (RppH in light purple).
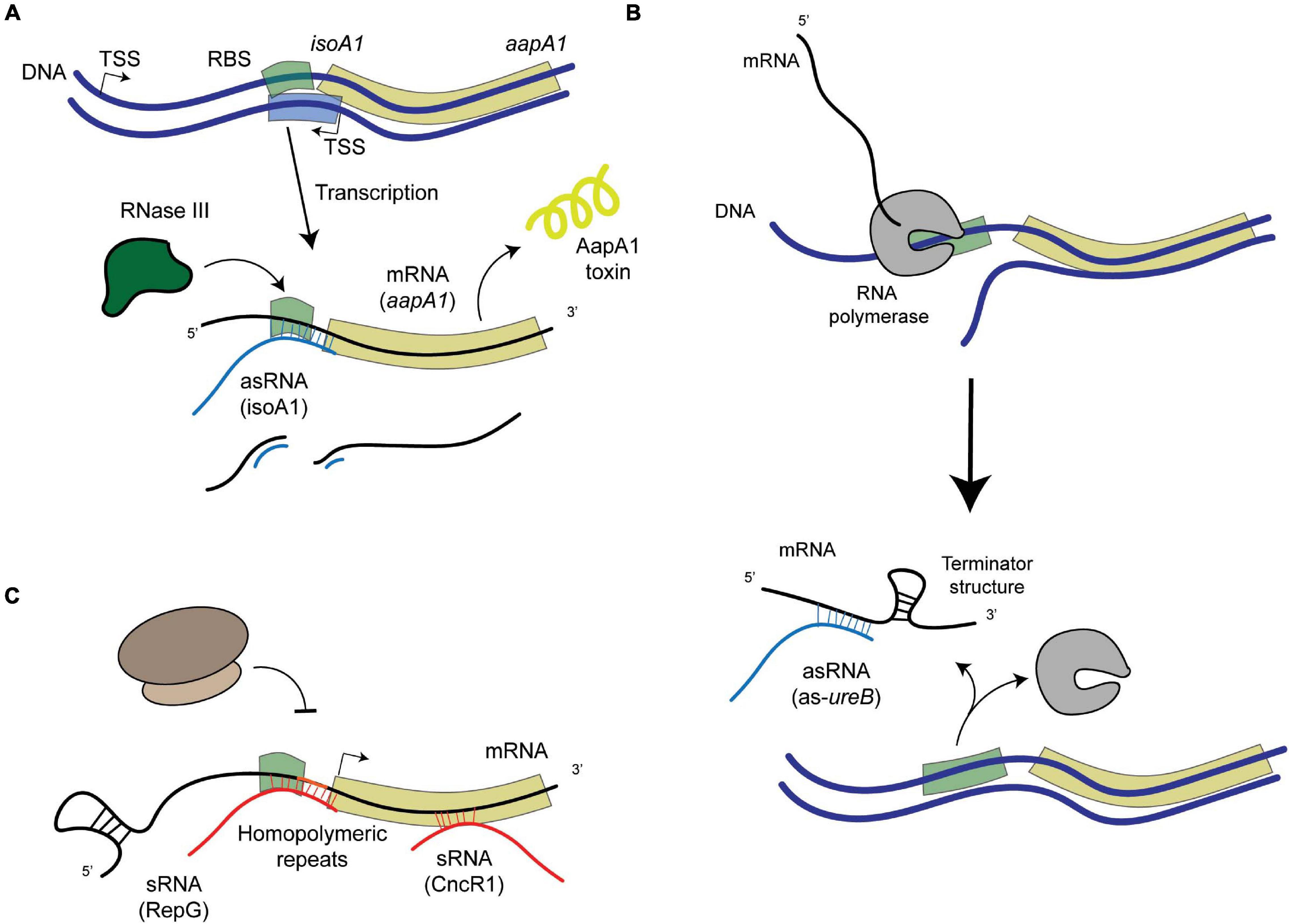
Figure 2. Schematic representation of examples of post-transcriptional regulation in H. pylori. (A) asRNA-mediated regulation that leads to the degradation of the mRNA-asRNA pair by an enribodonuclease such as RNase III, like in the case of the IsoA1-aapA1 Toxin-Antitoxin system. (B) asRNA-mediated regulation that leads to the formation of a premature terminator sequence that prevents the full transcription of the target gene (as in the case of the 5′-ureB antisense). (C) sRNA-mediated regulation, where sRNAs (encoded elsewhere in the genome apart from their targets) can either bind homopolymeric repeats upstream from the gene (like RepG) or at different regions within the coding sequence (like CncR1).
The seminal work by Sharma et al. (2010) revealed the complexity of the transcriptome of this bacterium. They mapped 1,907 transcription start sites (TSSs) in H. pylori strain 26695 and revealed that 87.5% of the genes are expressed from 337 primary operons, some of which contain additional internal TSSs. Surprisingly, they also detected massive antisense transcription with at least one antisense TSS associated to approximately 46% of all ORFs, including housekeeping genes like 28% of tRNAs and the 5′-leader regions of the 23S and 16S rRNA precursors. Interestingly, most known riboswitches are absent in H. pylori, with the exception of a predicted thiamine pyrophosphate riboswitch upstream of pnuC. Riboswitches are elements in an RNA molecule that can alter their structure in response to an environmental signal, such as a temperature shift or the presence of certain metabolites, and as a consequence regulate the translation or degradation of this RNA molecule. Even so, there are 337 untranslated regions (UTRs) that are long enough to accommodate other cis-acting regulatory RNA structures (Sharma et al., 2010).
In addition to tRNAs, rRNAs, transfer-messenger RNA (tmRNA, an RNA molecule in charge of ribosome unstalling), RNase P, 6S RNA and the signal recognition particle RNA (SRP RNA), Sharma et al. (2010) detected hundreds of candidate non-coding RNAs (ncRNAs). These ncRNAs are transcribed from intergenic regions (small RNAs or sRNAs), antisense to ORFs (antisense RNAs, asRNAs) and sense within ORFs. The expression of 60 of these intergenic ncRNAs was validated (visualized by Northern blot), and more than 900 asRNAs were detected, a few of them being validated (Sharma et al., 2010). It is interesting to note that, even if such massive amounts of asRNAs were first detected in H. pylori, other bacteria have since then been shown to have even more asRNAs (Georg and Hess, 2018). The validated and characterized ncRNAs are summarized in Table 1.
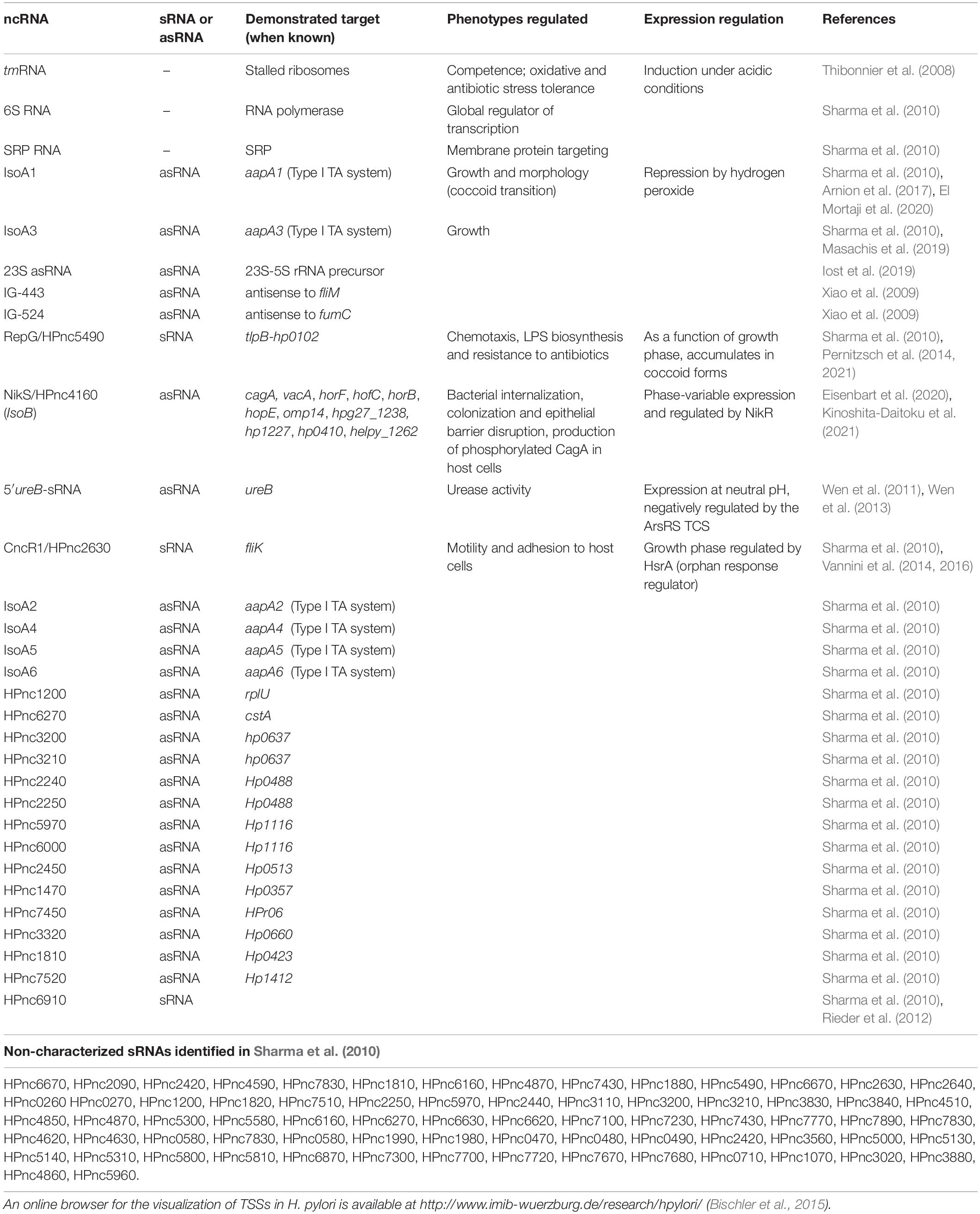
Table 1. List of non-coding RNAs (ncRNAs) validated (visualized by Northern blot) in Helicobacter pylori.
In the original description of the H. pylori transcriptome by Sharma et al. (2010), differential RNA sequencing (dRNA-seq) technologies were used to sequence and define TSSs, and such techniques have been further optimized since then (Bischler et al., 2015). First, in order to detect the widest range of TSSs, RNA was extracted from H. pylori grown under different conditions. Then prior to sequencing of cDNA libraries, RNAs were treated or not with terminator exoribonuclease. This enzyme specifically degrades 5′-P (monophosphate) RNA molecules [that usually result from processing of transcripts by ribonucleases (RNases)] but not 5′-PPP (triphosphate) RNA molecules (that result from transcription), thus allowing the detection of TSSs. Furthermore, the authors developed an online browser for the visualization of TSSs in H. pylori1 (Bischler et al., 2015).
As mentioned earlier, the main E. coli RNA-binding proteins (RBPs) participating in ncRNA-mediated regulation of gene expression, Hfq and ProQ, are absent in H. pylori (Quendera et al., 2020). Only a homolog of the RBP CsrA (Carbon storage regulator A) has been identified (Barnard et al., 2004). However, the complexity of the transcriptome suggests that other mechanisms and RBPs might be involved in post-transcriptional regulation in this organism.
Tools have been developed in recent years to identify novel RNA-RBP complexes in H. pylori (Rieder et al., 2012). One of these approaches was based on the purification of aptamer-tagged sRNAs by chromatography to identify their protein binding partners by mass spectrometry (Rieder et al., 2012). Complementarily, a method to FLAG-tag putative RBPs, co-immunoprecipitate them and sequence the associated RNA molecules was developed. Such tools permitted the detection of interactions between the S1 ribosomal protein and some mRNAs and sRNAs in H. pylori, as well as the identification of protein HP1334 as a binding partner of the abundant HPnc6910 sRNA (Rieder et al., 2012).
Despite progress in the detection of RNA species and their putative protein partners, nothing is known about the subcellular distribution of these elements in H. pylori. Different RNA molecules from the transcriptome of E. coli have been shown to display specific subcellular localization patterns, being targeted to the membrane, the cellular poles or distributed in the cytosol (Kannaiah et al., 2019). This likely plays a role in localizing their protein products and in regulating their expression and stability (Irastortza-Olaziregi and Amster-Choder, 2020). Bacterial RNA degradosomes, that are composed of at least one RNase and one DEAD-box RNA helicase protein that unwinds the target RNA, are central in the control of RNA decay and maturation (Tejada-Arranz et al., 2020a). The membrane localization of RNA degradosomes has been linked to the rate of degradation of different categories of transcripts including regulatory sRNAs (Moffitt et al., 2016), indicating a compartmentalization of RNA degradation and maturation that plays a role in transcriptome dynamics in prokaryotes. In H. pylori, we showed that the RNA degradosome, composed of the essential RNase J protein and of RhpA, the sole DEAD-box RNA helicase of this bacterium (Redko et al., 2013; El Mortaji et al., 2018), is compartmentalized at the inner membrane where it is assembled into foci whose formation is regulated and likely represent RNA degradation hubs (Tejada-Arranz et al., 2020b; Figures 1A,C). In addition, the 3′–5′ exoribonuclease RNase R was also associated to the H. pylori inner membrane (Tejada-Arranz et al., 2021; Figure 1A). The development of imaging techniques [reviewed in Fei and Sharma (2018)] for the study of the localization of RNA molecules in H. pylori is needed in order to assess how the transcriptome is distributed in H. pylori cells.
In H. pylori, we found that RNase J, the main RNase of its minimal RNA degradosome (Redko et al., 2013), is able to degrade many asRNAs (as well as mRNAs), with about 80% of them being upregulated more than 2-fold in an RNase J-depletion strain, and approximately 50% being regulated more than 4-fold (Redko et al., 2016). This suggests that a major level of regulation of the amount of these asRNAs relies on RNA degradosome-mediated degradation. Interestingly, sRNAs were found not to be preferred targets of this enzyme and are likely regulated through other mechanisms.
The only RBP that has been extensively studied in other bacteria and that is present in H. pylori is CsrA (Quendera et al., 2020). This protein is necessary for full H. pylori motility and survival under oxidative stress conditions. Accordingly, a strain lacking CsrA is defective for virulence in a mouse model (Barnard et al., 2004). More recent studies have found the reduced motility of a ΔcsrA strain to be associated with a defect in flagellar assembly due to a reduced expression of FlaA and FlaB (Kao et al., 2014) and to the dysregulation of the expression of a putative glycosyltransferase (Kao et al., 2017). In H. pylori, no CsrB/D-like sRNAs, which regulate CsrA activity in other organisms, were identified and thus the regulation of the activity of CsrA is not known in this organism. Nevertheless, the role of CsrA in asRNA- and sRNA-mediated regulation remains to be investigated. Other RBPs have been found in H. pylori, like the glycolytic enzyme aconitase that was shown to be important for full motility, oxidative stress response and lysozyme resistance (Austin and Maier, 2013; Austin et al., 2015; see Figure 1B).
A homolog of the RNA pyrophosphohydrolase, RppH, was also characterized in H. pylori. This enzyme targets the 5′-end of both mRNAs and sRNAs in the cell and cleaves the 5′-triphosphorylated end, yielding a 5′-monophosphorylated molecule that is targeted for degradation (Bischler et al., 2017; Figure 1C). In addition, mutants in this protein are more susceptible to hydrogen peroxide (Lundin et al., 2003) and are less able to invade gastric adenocarcinoma cells (Liu et al., 2012).
As mentioned above, the transcriptome of H. pylori contains >900 asRNA molecules and 60 validated sRNAs, with only one predicted riboswitch. Overall, there is little information about the molecular mechanisms by which asRNAs and sRNAs regulate gene expression and how their expression is regulated in H. pylori. Generally, these ncRNAs act by base-pairing with their target mRNAs (that can be transcribed from the opposite strand in the case of asRNAs or from somewhere else in the genome for the sRNAs) and hence altering their secondary structure, which might have consequences regarding their expression (ribosome accessibility) or stability (RNase accessibility). The few examples that have been analyzed in detail in H. pylori will be presented below.
Toxin-antitoxin (TA) systems are genetic modules that are widespread in prokaryotes. They encode a protein toxin and a cognate antitoxin that prevents the activity or expression of the toxin. The type I TA systems, in which the antitoxin is an asRNA, are highly represented on the H. pylori chromosome and several are strongly expressed (Sharma et al., 2010; Masachis and Darfeuille, 2019). Among them, the type I TA aapA1-isoA1 system has been extensively studied (Arnion et al., 2017; El Mortaji et al., 2020). The corresponding AapA1 toxin is a 30 aa-long hydrophobic peptide that, upon expression, targets the inner membrane and inhibits H. pylori growth (El Mortaji et al., 2020; Korkut et al., 2020). We have recently shown that this toxin triggers a morphological transition of H. pylori from its typical helical shape to round coccoid cells, that have been observed in response to stress as well as in human biopsies (El Mortaji et al., 2020). Our characterization of the toxin-induced coccoids suggests that they are viable cells that could correspond to dormant bacteria and might be responsible for H. pylori infections refractory to treatment.
Under normal growth conditions, different mechanisms that prevent the expression of the AapA1 toxin have been revealed (Arnion et al., 2017). Folding of the aapA1 full length transcript prevents its translation, unless its 3′-end is processed. This active aapA1 structure allows the formation of an extended duplex with the isoA1 asRNA that is degraded by RNase III, thus preventing AapA1 translation (Arnion et al., 2017; Figure 2A). The expression of AapA1 occurs under conditions that reduce the activity of the isoA1 promoter, namely oxidative stress and hence triggers H. pylori morphological transition (El Mortaji et al., 2020). Thus, the isoA1 asRNA is involved in regulating the transition of H. pylori cells into dormant forms that likely play an important role during colonization.
In another TA system from this family, aapA3-isoA3, the expression of the toxin is regulated by the formation of metastable structures in the aapA3 mRNA that transiently form during transcription and result in sequestration of the Shine-Dalgarno sequence, thereby also preventing unwanted toxin synthesis (Masachis et al., 2019).
Urease is a major colonization factor for H. pylori. This nickel metalloenzyme catalyzes the hydrolysis of urea into ammonia and bicarbonate (de Reuse et al., 2013). Both compounds allow H. pylori to buffer its cytoplasm and thus resist the extremely low pH of its unique niche, the human stomach. Large amounts of urease are produced by H. pylori. However, its synthesis requires regulation to avoid a toxic alkalinization of the cytoplasm. Urease of H. pylori is composed of two structural units, UreA and UreB. These proteins are expressed from an operon that is adjacent to a second one encoding the so-called urease accessory proteins required for nickel incorporation into urease. Expression of ureAB is positively controlled by NikR, a nickel-responsive regulator (Muller et al., 2011; Jones et al., 2018) and by a two-component system (TCS) of the OmpR-EnvZ family (Bury-Moné et al., 2004) designated ArsRS in H. pylori (Pflock et al., 2006). Under low pH conditions, the ArsRS system activates the urease operons ensuring increased production of this enzyme under the condition where its activity is vital. Wen et al. (2011, 2013) have identified another level of regulation of the ureAB operon. At neutral pH, they demonstrated the expression of a 290 nucleotides (nt)-long asRNA to ureB that favors the accumulation of a truncated transcript of the ureAB operon lacking the ureB 3′-end (Figure 2B). Interestingly, the expression of this asRNA is negatively regulated by the ArsRS TCS in response to acidic pH. Electrophoretic mobility shift assays (EMSA) showed that unphosphorylated ArsR regulator protein indeed binds to the ureB-asRNA promoter region. They determined that the mechanism at play is base-pairing of the antisense 5′ureB-asRNA with the ureAB transcript and subsequent transcription termination of the sense ureAB mRNA that causes diminished urease production. These data highlight a dual control of urease production that is adjusted to the pH of the environment encountered by H. pylori, with activation of urease expression at acidic pH and repression at neutral pH.
Another example concerns rRNA maturation in H. pylori. A strongly expressed asRNA overlapping the leader region of the 23S-5S rRNA precursor has been identified and found to be conserved (Sharma et al., 2010; Iost et al., 2019). It was demonstrated that this asRNA interacts with an rRNA precursor, forming an intermolecular complex that is cleaved by RNase III. This pairing induces further specific cleavages of the rRNA precursor and the degradation of the 23S asRNA. Whether this asRNA plays a regulatory function in rRNA maturation is still unclear as it is dispensable for H. pylori growth and for proper rRNA maturation under laboratory conditions. However, it might have a fine-tuning function by facilitating the degradation of processed fragments or a quality control role by favoring appropriate rRNA folding, that might become more evident under different growth conditions (Iost et al., 2019).
Other asRNAs were first identified in silico and then found to be expressed. They were predicted to target the flagellar motor switch gene (fliM) and fumarase (fumC), potentially regulating the expression of these genes (Xiao et al., 2009).
Besides transcriptional regulators, gene expression can be modulated by variations in the length of repeated nucleotide sequences, the “simple sequence repeats” (SSRs), located in their 5′-UTR that modify the stability or translation of the mRNA; or located in the promoter region, affecting the spacing of promoter elements or transcription factors binding sites. Phase variation of SSRs is due to slipped strand mispairing during replication and is used by many pathogens as an adaptive mechanism in which the most favorable expression is selected. Genes encoding bacterial surface structures or DNA Restriction-Modification enzymes are among the most frequently regulated by SSRs. The group of Sharma et al. (2010) identified and characterized the first sRNA that mediates post-transcriptional regulation by targeting a G-repeat stretch that is located in the 5′-UTR of the bicistronic tlpB-hp0102 operon (Pernitzsch et al., 2014, 2021; Figure 2C). This operon encodes the TlpB chemotaxis receptor and the HP0102 protein that was shown to function as a glycosyltransferase involved in lipopolysaccharide (LPS) O-chain biosynthesis (Pernitzsch et al., 2021). The precise fucosyltransferase activity of HP0102 was shown (Li et al., 2019). The sRNA designated RepG (REgulator of Polymeric G-repeats) is highly conserved in H. pylori. In contrast, the length of the G-repeat stretch upstream tlpB-hp0102 is highly variable among H. pylori strains and in sequential isolates from human patients, indicating that this region indeed undergoes phase variation during infection. Direct binding and base pairing of RepG to the 5′-UTR SSR region was demonstrated. Most interestingly, they showed that depending on the length of the G repeat, the binding of RepG can either mediate activation or repression of the tlpB-hp0102 operon and that this regulation acts at the translational level (Pernitzsch et al., 2014, 2021). In a recent publication, in collaboration with our group (Pernitzsch et al., 2021), the HP0102 glycosyltransferase was shown to be essential for colonization of the mouse model by H. pylori and to modulate LPS O-chain synthesis. The gradual modulation of H. pylori LPS through RepG regulation impacts bacterial resistance to membrane-targeting antibiotics and the exposure of Lewis antigens that contribute to the host immune recognition of this pathogen. In conclusion, these studies establish an original mechanism of post-transcriptional regulation by RepG, a trans-acting sRNA, that mediates a gradual rather than ON/OFF switch of the expression of its targets enabling H. pylori to adapt to its host.
Two recent publications report the characterization of a sRNA (HPnc4160) that regulates the expression of major H. pylori virulence factors (Eisenbart et al., 2020; Kinoshita-Daitoku et al., 2021). HPnc4160 was previously identified as a highly transcribed sRNA (IsoB) expressed from the opposite strand of a poorly expressed small ORF (AapB) as a part of a probably “degenerated” type I TA system (Sharma et al., 2010; Vannini et al., 2017). HPnc4160 is strongly conserved in H. pylori and harbors a length-variable T stretch upstream its −10 promoter sequence. The expression of HPnc4160 was shown to be repressed by NikR in response to nickel (Eisenbart et al., 2020) through direct binding of this transcriptional regulator to the sRNA promoter (Vannini et al., 2017). Therefore, HPnc4160 was renamed NikS (Eisenbart et al., 2020). The T stretch preceding the HPnc4160/NikS sRNA was variable in length in strains isolated from patients or during Mongolian gerbil colonization (Eisenbart et al., 2020; Kinoshita-Daitoku et al., 2021); this length variation affects the sRNA expression. Both studies identified the targets of this sRNA by mRNA and protein expression analysis. Genes encoding major virulence and colonization factors were found to be targeted by HPnc4160/NikS, [five in Eisenbart et al. (2020) and eight in Kinoshita-Daitoku et al. (2021); see Table 1]. Both studies identified cagA, encoding the oncoprotein CagA, as a target and found that the expression of several outer membrane proteins, some being potential adhesins, is targeted. Eisenbart et al. identified in addition vacA, encoding the vacuolating cytotoxin, as being directly regulated by HPnc4160/NikS. Post-transcriptional repression by HPnc4160/NikS base pairing to five mRNA leaders was demonstrated, which results in translation inhibition (Eisenbart et al., 2020). In vitro EMSA and structure probing analysis validated the direct HPnc4160/NikS interaction with several of its targets (Eisenbart et al., 2020; Kinoshita-Daitoku et al., 2021). However, the two publications present divergent data on the targeting site in cagA, in the 5′ UTR region for Eisenbart et al. (2020) and intragenic for Kinoshita-Daitoku et al. (2021), which implies different underlying regulatory mechanisms. Both studies validate that the control of cagA expression by HPnc4160/NikS indeed impacts H. pylori pathogenicity. In vitro infection of different cell types by H. pylori revealed that HPnc4160/NikS decreases CagA-dependent bacterial internalization, reduces cell colonization and epithelial barrier disruption (Eisenbart et al., 2020) and also decreases the level of phosphorylated CagA, IL8 production and the associated CagA-induced hummingbird phenotype (Kinoshita-Daitoku et al., 2021). During mouse colonization by H. pylori, the number of repeats of HPnc4160/NikS varies with a trend to expansion while a deletion of this sRNA favors short term colonization. The selective pressure that drives these variations in vivo still needs to be identified. Finally, the T-repeats were found to be significantly longer in strains isolated from patients with gastric cancer than in “non-cancer” strains. In conclusion, HPnc4160/NikS is a sRNA acting as a master regulator of the adaptation of H. pylori to the colonization of its host.
The H. pylori cag pathogenicity island (cag PAI) is a 40 kb DNA element that encodes a type IV secretion system. The cag PAI is a major virulence determinant of H. pylori that allows for the delivery of bacterial effector molecules into host gastric epithelial cells, in particular the oncoprotein CagA encoded within the cag PAI.
Both groups of Sharma et al. (2010) and Vannini et al. (2014) have identified, within the cag PAI, a ncRNA expressed within the 5′-UTR of cagP. This 213 nt-long sRNA is abundant and conserved in H. pylori. It was designated HPnc2630 in strain 26695 (Sharma et al., 2010) and was renamed CncR1 for “cag non-coding RNA 1” in strain G27 where it was characterized (Vannini et al., 2016). Vannini et al. (2016) showed that the expression of the CncR1 sRNA is directed by the cagP promoter and regulated as a function of growth through direct binding of the essential orphan response regulator HsrA (HP1043). Transcriptomic analysis of a ΔcncR1 mutant identified 71 deregulated genes. Enrichment in downregulated genes related to host-pathogen interactions was observed in addition to upregulation of genes involved in the assembly and regulation of the flagellar apparatus including FliK, a flagellar hook-length control protein. They validated that CncR1 negatively regulates H. pylori motility functions. Using EMSA and RNase T1 foot-printing experiments, direct targeting of the sRNA on different regions of the fliK mRNA was demonstrated (Figure 2C). Finally, CncR1 is required for full bacterial adhesion to host cells. In conclusion, the CncR1 sRNA modulates H. pylori virulence through opposite effects on motility and adhesion which might be relevant to signals related to growth phase or bacterial density where bacteria move to a new colonization site.
Relatively few studies have been published on riboregulation in H. pylori. However, the established properties make it a fascinating microorganism to study. The huge number of genes for which a cis-asRNA is expressed is still intriguing, some of them certainly have regulatory roles on the expression of the complementary mRNA but many RNAs that are produced by pervasive transcription may have other functions. The post-transcriptional mode of gene regulation where a sRNA targets homopolymeric repeats within 5′-UTR is original. This mechanism and other variations linking RNA-mediated control and SSRs are most probably widespread in H. pylori and will certainly be found in other bacteria in the future. In H. pylori, RepG has other targets, several antisense TSSs were shown to overlap SSRs and some sRNA candidates have internal repeats (Sharma et al., 2010; Pernitzsch et al., 2014, 2021). These data, together with what is known about the SSR-controlled NikS/HPnc4160 sRNA, reveal that bacterial phase variation, which is associated to host adaptation, impacts gene expression at several levels. Finally, since no RNA chaperone facilitating sRNA-mRNA base-paring has been reported so far in H. pylori, the question of discrimination of the targets, in particular those with low complexity, such as SSRs, remains open.
Both authors wrote and edited the manuscript, contributed to the article and approved the submitted version.
AT-A have been part of the Pasteur - Paris University (PPU) International Ph.D. Program. This project have received funding from the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Sklodowska-Curie grant agreement No 665807 and from the Institut Carnot Pasteur Microbes & Santé. Support was provided by “Fondation pour la Recherche Médicale” for the grant DBF20161136767 to HDR and the Pasteur-Weizmann Consortium of “The Roles of Non-coding RNAs in Regulation of Microbial Life Styles and Virulence” to HDR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Arnion, H., Korkut, D. N., Masachis Gelo, S., Chabas, S., Reignier, J., Iost, I., et al. (2017). Mechanistic insights into type I toxin antitoxin systems in Helicobacter pylori: the importance of mRNA folding in controlling toxin expression. Nucleic Acids Res. 45:gkw1343. doi: 10.1093/nar/gkw1343
Austin, C. M., and Maier, R. J. (2013). Aconitase-mediated posttranscriptional regulation of Helicobacter pylori peptidoglycan deacetylase. J. Bacteriol. 195, 5316–5322. doi: 10.1128/JB.00720-13
Austin, C. M., Wang, G., and Maier, R. J. (2015). Aconitase functions as a pleiotropic posttranscriptional regulator in Helicobacter pylori. J. Bacteriol. 197, 3076–3086. doi: 10.1128/JB.00529-15
Barnard, F. M., Loughlin, M. F., Fainberg, H. P., Messenger, M. P., Ussery, D. W., Williams, P., et al. (2004). Global regulation of virulence and the stress response by CsrA in the highly adapted human gastric pathogen Helicobacter pylori. Mol. Microbiol. 51, 15–32. doi: 10.1046/j.1365-2958.2003.03788
Bischler, T., Hsieh, P.-K., Resch, M., Liu, Q., Tan, H. S., Foley, P. L., et al. (2017). Identification of the RNA pyrophosphohydrolase RppH of Helicobacter pylori and Global analysis of its RNA targets. J. Biol. Chem. 292, 1934–1950. doi: 10.1074/jbc.M116.761171
Bischler, T., Tan, H. S., Nieselt, K., and Sharma, C. M. (2015). Differential RNA-seq (dRNA-seq) for annotation of transcriptional start sites and small RNAs in Helicobacter pylori. Methods 86, 89–101. doi: 10.1016/j.ymeth.2015.06.012
Bury-Moné, S., Thiberge, J.-M., Contreras, M., Maitournam, A., Labigne, A., and De Reuse, H. (2004). Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53, 623–638. doi: 10.1111/j.1365-2958.2004.04137
De la Cruz, M. A., Ares, M. A., von Bargen, K., Panunzi, L. G., Martínez-Cruz, J., Valdez-Salazar, H. A., et al. (2017). Gene expression profiling of transcription factors of Helicobacter pylori under different environmental conditions. Front. Microbiol. 8:615. doi: 10.3389/fmicb.2017.00615
de Reuse, H., Vinella, D., and Cavazza, C. (2013). Common themes and unique proteins for the uptake and trafficking of nickel, a metal essential for the virulence of Helicobacter pylori. Front. Cell. Infect. Microbiol. 3:94. doi: 10.3389/fcimb.2013.00094
Eisenbart, S. K., Alzheimer, M., Pernitzsch, S. R., Dietrich, S., Stahl, S., and Sharma, C. M. (2020). A repeat-associated small RNA controls the major virulence factors of Helicobacter pylori. Mol. Cell 80, 210.e–226.e. doi: 10.1016/j.molcel.2020.09.009
El Mortaji, L., Aubert, S., Galtier, E., Schmitt, C., Anger, K., Redko, Y., et al. (2018). The sole DEAD-Box RNA helicase of the gastric pathogen Helicobacter pylori is essential for colonization. mBio 9:e02071-17. doi: 10.1128/mBio.02071-17
El Mortaji, L., Tejada-Arranz, A., Rifflet, A., Boneca, I. G., Pehau-Arnaudet, G., Radicella, J. P., et al. (2020). A peptide of a type I toxin-antitoxin system induces Helicobacter pylori morphological transformation from spiral shape to coccoids. Proc. Natl. Acad. Sci. U.S.A. 117, 31398–31409. doi: 10.1073/pnas.2016195117
Fei, J., and Sharma, C. M. (2018). RNA localization in bacteria. Microbiol. Spectr. 6, 1–19. doi: 10.1128/microbiolspec.RWR-0024-2018
Georg, J., and Hess, W. R. (2018). “Widespread antisense transcription in prokaryotes,” in Regulating with RNA in Bacteria and Archaea, eds G. Storz and K. Papenfort (Washington, DC: ASM Press), 191–210. doi: 10.1128/9781683670247.ch12
Iost, I., Chabas, S., and Darfeuille, F. (2019). Maturation of atypical ribosomal RNA precursors in Helicobacter pylori. Nucleic Acids Res. 47, 5906–5921. doi: 10.1093/nar/gkz258
Irastortza-Olaziregi, M., and Amster-Choder, O. (2020). RNA localization in prokaryotes: where, when, how, and why. Wiley Interdiscip. Rev. RNA. 12:e1615. doi: 10.1002/wrna.1615
Jones, M. D., Li, Y., and Zamble, D. B. (2018). Acid-responsive activity of the Helicobacter pylori metalloregulator NikR. Proc. Natl. Acad. Sci. U.S.A. 115, 8966–8971. doi: 10.1073/pnas.1808393115
Kannaiah, S., Livny, J., and Amster-Choder, O. (2019). Spatiotemporal organization of the E. coli transcriptome: translation independence and engagement in regulation. Mol. Cell 76, 574.e–589.e. doi: 10.1016/j.molcel.2019.08.013
Kao, C.-Y., Chen, J.-W., Wang, S., Sheu, B.-S., and Wu, J.-J. (2017). The Helicobacter pylori J99 jhp0106 gene, under the control of the CsrA/RpoN regulatory system, modulates flagella formation and motility. Front. Microbiol. 8:483. doi: 10.3389/fmicb.2017.00483
Kao, C.-Y., Sheu, B.-S., and Wu, J.-J. (2014). CsrA regulates Helicobacter pylori J99 motility and adhesion by controlling flagella formation. Helicobacter 19, 443–454. doi: 10.1111/hel.12148
Kinoshita-Daitoku, R., Kiga, K., Miyakoshi, M., Otsubo, R., Ogura, Y., Sanada, T., et al. (2021). A bacterial small RNA regulates the adaptation of Helicobacter pylori to the host environment. Nat. Commun. 12:2085. doi: 10.1038/s41467-021-22317-7
Korkut, D. N., Alves, I. D., Vogel, A., Chabas, S., Sharma, C. M., Martinez, D., et al. (2020). Structural insights into the AapA1 toxin of Helicobacter pylori. Biochim. Biophys. Acta. Gen. Subj. 1864:129423. doi: 10.1016/j.bbagen.2019.129423
Li, H., Marceau, M., Yang, T., Liao, T., Tang, X., Hu, R., et al. (2019). East-Asian Helicobacter pylori strains synthesize heptan-deficient lipopolysaccharide. PLoS Genet. 15:e1008497. doi: 10.1371/journal.pgen.1008497
Liu, H., Semino-Mora, C., and Dubois, A. (2012). Mechanism of H. pylori intracellular entry: an in vitro study. Front. Cell. Infect. Microbiol. 2:13. doi: 10.3389/fcimb.2012.00013
Lundin, A., Nilsson, C., Gerhard, M., Andersson, D. I., Krabbe, M., and Engstrand, L. (2003). The NudA protein in the gastric pathogen Helicobacter pylori is an ubiquitous and constitutively expressed dinucleoside polyphosphate hydrolase. J. Biol. Chem. 278, 12574–12578. doi: 10.1074/JBC.M212542200
Masachis, S., and Darfeuille, F. (2019). “Type I toxin-antitoxin systems: regulating toxin expression via Shine-Dalgarno sequence sequestration and small RNA binding,” in Regulating with RNA in Bacteria and Archaea, eds G. Storz and K. Papenfort (Washington, DC: American Society of Microbiology), 173–190. doi: 10.1128/microbiolspec.RWR-0030-2018
Masachis, S., Tourasse, N. J., Lays, C., Faucher, M., Chabas, S., Iost, I., et al. (2019). A genetic selection reveals functional metastable structures embedded in a toxin-encoding mRNA. Elife 8:e47549. doi: 10.7554/eLife.47549
Moffitt, J. R., Pandey, S., Boettiger, A. N., Wang, S., and Zhuang, X. (2016). Spatial organization shapes the turnover of a bacterial transcriptome. Elife 5:e13065. doi: 10.7554/eLife.13065
Muller, C., Bahlawane, C., Aubert, S., Delay, C. M., Schauer, K., Michaud-Soret, I., et al. (2011). Hierarchical regulation of the NikR-mediated nickel response in Helicobacter pylori. Nucleic Acids Res. 39, 7564–7575. doi: 10.1093/nar/gkr460
Pernitzsch, S. R., Alzheimer, M., Bremer, B. U., Robbe-Saule, M., De Reuse, H., and Sharma, C. M. (2021). Small RNA-mediated gradual lipopolysaccharide biosynthesis control affects antibiotics resistance of Helicobacter pylori. Nat. Comm. (in press).
Pernitzsch, S. R., and Sharma, C. M. (2012). Transcriptome complexity and riboregulation in the human pathogen Helicobacter pylori. Front. Cell. Infect. Microbiol. 2:14. doi: 10.3389/fcimb.2012.00014
Pernitzsch, S. R., Tirier, S. M., Beier, D., and Sharma, C. M. (2014). A variable homopolymeric G-repeat defines small RNA-mediated posttranscriptional regulation of a chemotaxis receptor in Helicobacter pylori. Proc. Natl. Acad. Sci. U.S.A. 111, E501–E510. doi: 10.1073/pnas.1315152111
Pflock, M., Finsterer, N., Joseph, B., Mollenkopf, H., Meyer, T., and Beier, D. (2006). Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J. Bacteriol. 188, 3449–3462. doi: 10.1128/JB.188.10.3449-3462.2006
Quendera, A. P., Seixas, A. F., Dos Santos, R. F., Santos, I., Silva, J. P. N., Arraiano, C. M., et al. (2020). RNA-binding proteins driving the regulatory activity of small non-coding RNAs in bacteria. Front. Mol. Biosci. 7:78. doi: 10.3389/fmolb.2020.00078
Redko, Y., Aubert, S., Stachowicz, A., Lenormand, P., Namane, A., Darfeuille, F., et al. (2013). A minimal bacterial RNase J-based degradosome is associated with translating ribosomes. Nucleic Acids Res. 41, 288–301. doi: 10.1093/nar/gks945
Redko, Y., Galtier, E., Arnion, H., Darfeuille, F., Sismeiro, O., Coppée, J.-Y., et al. (2016). RNase J depletion leads to massive changes in mRNA abundance in Helicobacter pylori. RNA Biol. 13, 243–253. doi: 10.1080/15476286.2015.1132141
Rieder, R., Reinhardt, R., Sharma, C., and Vogel, J. (2012). Experimental tools to identify RNA-protein interactions in Helicobacter pylori. RNA Biol. 9, 520–531. doi: 10.4161/rna.20331
Robinson, K., and Atherton, J. C. (2021). The spectrum of Helicobacter-mediated diseases. Annu. Rev. Pathol. 16, 123–144. doi: 10.1146/annurev-pathol-032520-024949
Sharma, C. M., Hoffmann, S., Darfeuille, F., Reignier, J., Findeiß, S., Sittka, A., et al. (2010). The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250–255. doi: 10.1038/nature08756
Tejada-Arranz, A., de Crécy-Lagard, V., and de Reuse, H. (2020a). Bacterial RNA degradosomes: molecular machines under tight control. Trends Biochem. Sci. 45, 42–57. doi: 10.1016/j.tibs.2019.10.002
Tejada-Arranz, A., Galtier, E., El Mortaji, L., Turlin, E., Ershov, D., and De Reuse, H. (2020b). The RNase J-based RNA degradosome is compartmentalized in the gastric pathogen Helicobacter pylori. MBio 11:e01173-20. doi: 10.1128/mBio.01173-20
Tejada-Arranz, A., Matos, R. G., Quentin, Y., Bouilloux-Lafont, M., Galtier, E., Briolat, V., et al. (2021). RNase R is associated in a functional complex with the RhpA DEAD-box RNA helicase in Helicobacter pylori. Nucleic Acids Res. 49, 5249–5264. doi: 10.1093/nar/gkab283
Thibonnier, M., Thiberge, J.-M., and De Reuse, H. (2008). Trans-translation in Helicobacter pylori: essentiality of ribosome rescue and requirement of protein tagging for stress resistance and competence. PLoS One 3:e3810. doi: 10.1371/journal.pone.0003810
Tomb, J. F., White, O., Kerlavage, A. R., Clayton, R. A., Sutton, G. G., Fleischmann, R. D., et al. (1997). The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388, 539–547. doi: 10.1038/41483
Vannini, A., Pinatel, E., Costantini, P. E., Pelliciari, S., Roncarati, D., Puccio, S., et al. (2017). Comprehensive mapping of the Helicobacter pylori NikR regulon provides new insights in bacterial nickel responses. Sci. Rep. 7:45458. doi: 10.1038/srep45458
Vannini, A., Roncarati, D., and Danielli, A. (2016). The cag-pathogenicity island encoded CncR1 sRNA oppositely modulates Helicobacter pylori motility and adhesion to host cells. Cell. Mol. Life Sci. 73, 3151–3168. doi: 10.1007/s00018-016-2151-z
Vannini, A., Roncarati, D., Spinsanti, M., Scarlato, V., and Danielli, A. (2014). In depth analysis of the Helicobacter pylori cag pathogenicity island transcriptional responses. PLoS One 9:e98416. doi: 10.1371/journal.pone.0098416
Waite, D. W., Vanwonterghem, I., Rinke, C., Parks, D. H., Zhang, Y., Takai, K., et al. (2017). Comparative genomic analysis of the class epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front. Microbiol. 8:682. doi: 10.3389/fmicb.2017.00682
Wen, Y., Feng, J., and Sachs, G. (2013). Helicobacter pylori 5’ureB-sRNA, a cis-encoded antisense small RNA, negatively regulates ureAB expression by transcription termination. J. Bacteriol. 195, 444–452. doi: 10.1128/JB.01022-12
Wen, Y., Feng, J., Scott, D. R., Marcus, E. A., and Sachs, G. (2011). A cis-encoded antisense small RNA regulated by the HP0165-HP0166 two-component system controls expression of ureB in Helicobacter pylori. J. Bacteriol. 193, 40–51. doi: 10.1128/JB.00800-10
Keywords: small RNAs, antisense RNAs, virulence, phase variation, Helicobacter pylori, post-transcriptional regulation
Citation: Tejada-Arranz A and De Reuse H (2021) Riboregulation in the Major Gastric Pathogen Helicobacter pylori. Front. Microbiol. 12:712804. doi: 10.3389/fmicb.2021.712804
Received: 21 May 2021; Accepted: 23 June 2021;
Published: 16 July 2021.
Edited by:
Florence Hommais, Université Claude Bernard Lyon 1, FranceReviewed by:
Davide Roncarati, University of Bologna, ItalyCopyright © 2021 Tejada-Arranz and De Reuse. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hilde De Reuse, aGRlcmV1c2VAcGFzdGV1ci5mcg==
†Present address: Alejandro Tejada-Arranz, Biozentrum University of Basel, Basel, Switzerland
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.