- 1Department of Bacteriology, Kimron Veterinary Institute (KVI), Bet Dagan, Israel
- 2Department of Biological Safety, German Federal Institute for Risk Assessment (BfR), Berlin, Germany
- 3Department of Serology, Kimron Veterinary Institute (KVI), Bet Dagan, Israel
Brucella melitensis Rev.1 is a live attenuated vaccine strain that is widely used to control brucellosis in small ruminants. For successful surveillance and control programs, rapid identification and characterization of Brucella isolates and reliable differentiation of vaccinated and naturally infected animals are essential prerequisites. Although MALDI-TOF MS is increasingly applied in clinical microbiology laboratories for the diagnosis of brucellosis, species or even strain differentiation by this method remains a challenge. To detect biomarkers, which enable to distinguish the B. melitensis Rev.1 vaccine strain from B. melitensis field isolates, we initially searched for unique marker proteins by in silico comparison of the B. melitensis Rev.1 and 16M proteomes. We found 113 protein sequences of B. melitensis 16M that revealed a homologous sequence in the B. melitensis Rev.1 annotation and 17 of these sequences yielded potential biomarker pairs. MALDI-TOF MS spectra of 18 B. melitensis Rev.1 vaccine and 183 Israeli B. melitensis field isolates were subsequently analyzed to validate the identified marker candidates. This approach detected two genus-wide unique biomarkers with properties most similar to the ribosomal proteins L24 and S12. These two proteins clearly discriminated B. melitensis Rev.1 from the closely related B. melitensis 16M and the Israeli B. melitensis field isolates. In addition, we verified their discriminatory power using a set of B. melitensis strains from various origins and of different MLVA types. Based on our results, we propose MALDI-TOF MS profiling as a rapid, cost-effective alternative to the traditional, time-consuming approach to differentiate certain B. melitensis isolates on strain level.
Introduction
Brucellosis is a global zoonotic disease affecting domestic and wild animals as well as humans (Pappas et al., 2006). Three out of twelve currently known Brucella species are responsible for most of the reported human brucellosis cases, namely B. melitensis, primarily transmitted from sheep and goats, B. abortus from cattle, and B. suis from swine (Hull and Schumaker, 2018). Ovine and caprine brucellosis are endemic throughout the Middle East as well as in many countries of Africa, Asia, and Latin America (Rossetti et al., 2017). In these regions, efforts are undertaken to control brucellosis by the vaccination of sheep and goats.
The most common vaccination policy for small ruminant livestock against B. melitensis infections is the application of the live attenuated B. melitensis Rev.1 strain to female animals aged between 2 and 6 months (Banai, 2002). This procedure has proven to be protective and to reduce abortions in treated animals, but may be contraindicated in females that are vaccinated during their last trimester of pregnancy (Banai, 2002). Although attenuated, the B. melitensis Rev.1 strain is still capable to infect humans and cause disease, either through the consumption of contaminated milk from vaccinated animals or by accidental exposure during the vaccination procedures (Arapovic et al., 2020). Hence, laboratory methods that can easily distinguish B. melitensis field strains from the vaccine strain are relevant for (i) effective brucellosis control programs, (ii) for epidemiological surveillance, and (iii) for outbreak clarification.
Brucella melitensis Rev.1 possesses several characteristics, including streptomycin resistance and a distinct dye sensitivity pattern, which enables its discrimination from field strains using bacteriological tests (Elberg and Meyer, 1958). However, atypical B. melitensis biovar 1 field and Rev.1 vaccine isolates have been described, which may lead to misinterpretations (Banai et al., 1990; Banai, 2002; Lucero et al., 2006). Moreover, B. melitensis Rev.1 induces like other B. melitensis strains the production of antibodies directed against its smooth lipopolysaccharide (LPS). This property interferes with serological testing for brucellosis due to cross-reactivity between the smooth LPS of the vaccine strain and other smooth Brucella species or the LPS of widespread Gram-negative pathogens such as Yersinia enterocolitica and Salmonella spp. (Corbell, 1975). Hence, more robust molecular markers are needed, and a molecular screening method has been developed based on the PstI site polymorphism in the Brucella omp2 gene of the B. melitensis Rev.1 vaccine strain, which allows its differentiation from B. melitensis field isolates (Bardenstein et al., 2002). In addition, rpsL-directed PCR-RFLP and multiplex PCR assays have been established to discriminate B. melitensis biovar 1 wild-type strains from B. melitensis Rev.1 (Cloeckaert et al., 2002; Garcia-Yoldi et al., 2006).
Similar to PCR-based microbial diagnostics, matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MALDI-TOF MS) profiling has emerged as a rapid and cost-effective laboratory method to identify bacteria in recent years (Croxatto et al., 2012). Rigorous preprocessing and generous peak binning during spectra creation for MALDI-TOF MS platforms, like Bruker Biotyper, Vitek MS or Andromas lead to robust identification performances for many bacteria at genus and even at species level (Clark et al., 2013). However, strain identification relies on discriminating strain-specific differences in the proteome under the constraints of MALDI-TOF MS. These subtle differences are caused by genomic alterations, i.e., indels, frameshifts, non-synonymous single nucleotide polymorphisms or pseudogenization that manifest as a change in abundance or result in an altered amino acid sequence and post-translational modifications of a protein. While pseudogenization or changes in steady-state levels of proteins may be detected by modified peak intensities, changes in the molecular composition of a protein may lead to a unique mass shift of its corresponding peak, oftentimes referred to as a biomarker. Species- and even strain-specific biomarkers have been reported for the MALDI-TOF MS-based identification and differentiation of various bacterial pathogens, like Haemophilus spp., Helicobacter pylori, Campylobacter spp., Yersinia enterocolitica or methicillin-resistant Staphylococcus aureus (Sandrin et al., 2013). MALDI-TOF MS has also been applied for the identification of Brucella. However, classical Brucella species are highly homologous (Hoyer and Mccullough, 1968), which is why commercially available reference libraries have shown shortcomings in the reliable classification of Brucella beyond genus level (Ferreira et al., 2010; Cunningham and Patel, 2013; Tracz et al., 2016) or the differentiation of closely related Ochrobactrum species (Poonawala et al., 2018), recently reclassified as Brucella spp., for example B. anthropi and B. intermedium (Hordt et al., 2020). However, the generation of in-house reference libraries of MALDI-TOF MS spectra may allow for correct identification of Brucella species and some of their respective biovars (Lista et al., 2011; Karger et al., 2013; Mesureur et al., 2018; Da Silva et al., 2020).
Our study aimed to establish a novel MALDI-TOF MS-based diagnostic approach that facilitates the rapid differentiation of the B. melitensis Rev.1 vaccine strain from B. melitensis field isolates. We took advantage of a comparative in silico proteomics analysis and a comprehensive in-house library of MALDI-TOF MS spectra to identify specific protein biomarkers for the resolution of B. melitensis isolates on sub-species level.
Materials and Methods
In silico Proteome Comparison
Complete genomes of B. melitensis biovar 1 strain 16M, GCF_000007125.1_ASM712v1 (Delvecchio et al., 2002) and GCF_000740415.1_ASM74041v1 (Minogue et al., 2014), as well as B. melitensis biovar 1 strain Rev.1, GCF_002953595.1_ASM295359v1 (Salmon-Divon et al., 2018), were retrieved from the National Center for Biotechnology Information (NCBI1). The genome sequences were submitted to the Pathosystems Resource Integration Center (PATRIC2) for RASTtk annotation to augment the protein features with genus-specific “local protein family properties” (called PLfam) (Brettin et al., 2015).
The newly annotated protein coding sequences (CDS) were treated as strings and the subset
was filtered for the single presence of PLfam identifiers in all three PATRIC annotation feature tables (Supplementary Table 1). In a second filter step, only PLfam IDs were kept that shared the same sequence in the B. melitensis 16M genomes but had a divergent one in B. melitensis Rev.1 due to amino acid substitutions with a small mass shift (Δmass ± 130 Da) (Supplementary Table 2). Proteins in the target range of MALDI-TOF MS, i.e., with exact masses between 2,000 and 20,000 Da, were short-listed in Table 1 and annotated with UniProt identifiers of the B. melitensis strain 16M proteome (ID: BRUME). UniProt entries were screened for potential post-translational modifications (PTM), and where applicable, included into the calculation of the exact mass with the R package SeqinR v3.6.1 (Charif and Lobry, 2007). The frequent event of protein N-terminal methionine excision (NME, Δmass = −131.2 Da) was always considered, as well as beta-methylthiolation (βMeS, Δmass = +46.1 Da) for ribosomal protein S12.
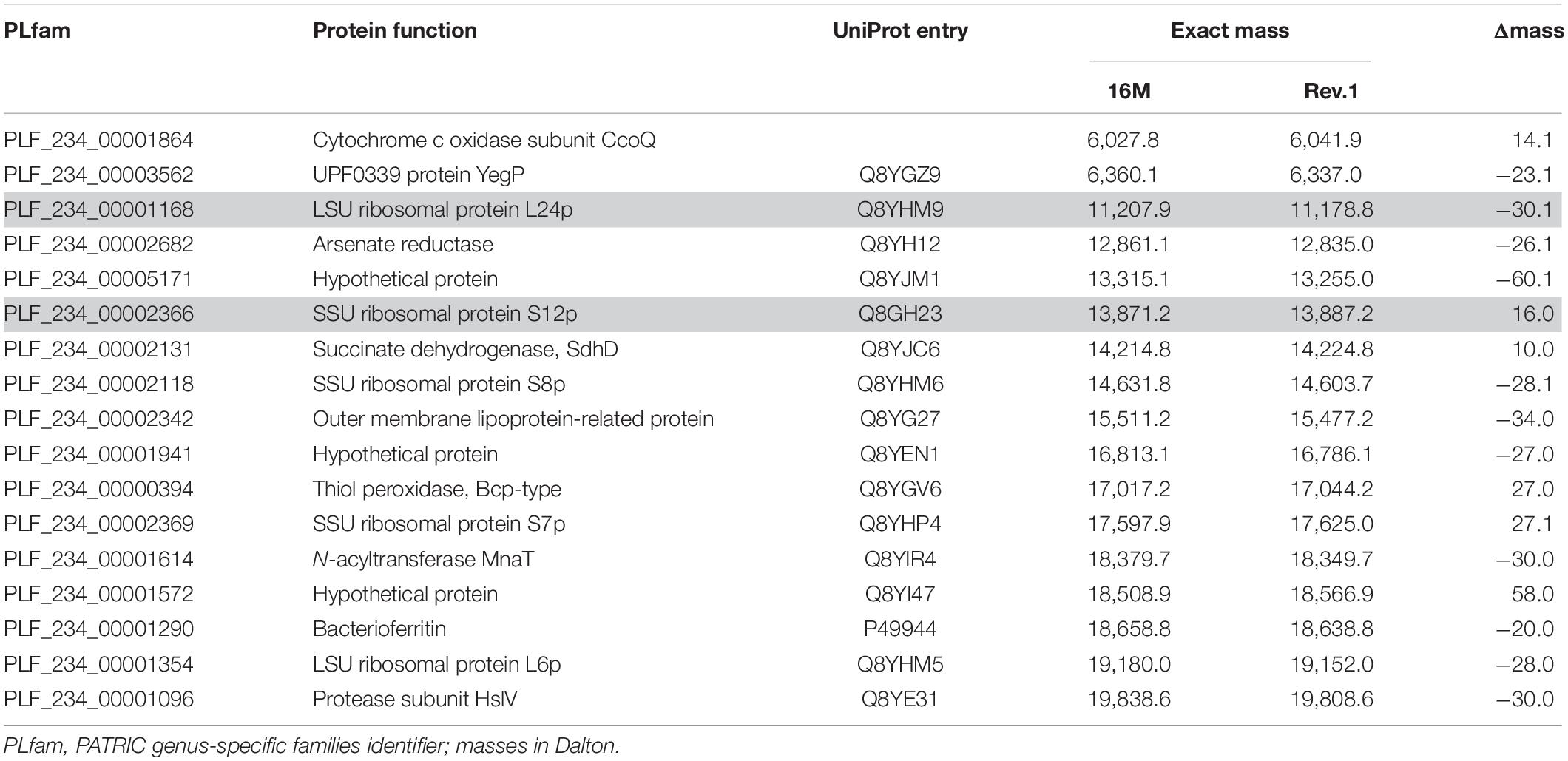
Table 1. Short-listed proteins with discriminatory mass differences in the working range of MALDI-TOF MS.
Bacterial Strains and Culture Conditions
Bacteria analyzed in the present study were 18 B. melitensis Rev.1 vaccine isolates (including the original Elberg Rev.1 vaccine strain, passage no. 101) and 183 B. melitensis field isolates from human, cattle, sheep and goats in Israel (Supplementary Table 3). All Brucella strains were obtained from the collection of the Kimron Veterinary Institute (KVI) in Bet Dagan, Israel, and were cultured for 48–72 h on tryptic soy agar (TSA) plates at 37°C under 5% CO2. All bacteria were characterized using standard methods: growth on TSA plates with penicillin G and streptomycin, dye sensitivity (thionine, fuchsine), H2S production, urease activity, and agglutination with mono-specific anti-M and anti-A serum. Brucella melitensis Rev.1 isolates were verified by omp2 PCR and PstI digestion of the amplicon (Bardenstein et al., 2002). We prepared biological triplicates of B. melitensis Rev.1 vaccine isolates but only produced single preparations of the Israeli B. melitensis field isolates. Bacteria were grown with shaking in 10 ml of a tryptic soy broth (TSB pH 7.3) at 37°C for 24 h (OD600∼0.3–0.4), centrifuged at 7,000 × g, washed with PBS and resuspended in 300 μl PBS and 900 μl 100% ethanol. The bacterial solutions were left at room temperature for 48 h before 100 μl were plated on TSA plates for sterility testing. The work on live agents was performed at the KVI biosafety level 3 facility. Inactivated Brucella samples were split into aliquots and stored at −20°C for on-site use or shipping on dry ice to the German Federal Institute for Risk Assessment (BfR) in Berlin, Germany.
MALDI-TOF
Spectra Acquisition
At both institutes, samples were independently prepared for mass spectrometry by ethanol-formic acid extraction according to manufacturer’s instructions before being spotted on a 96-spot steel plate target and covered with alpha-cyano-4-hydroxy-cinnamic acid (HCCA) matrix solution (Bruker Daltonik GmbH, Bremen, Germany). Mass spectra were measured using a microflex LT MALDI-TOF MS system (Bruker Daltonik) operated by the Biotyper automation software flexControl (v3.4.135.0, Bruker Daltonik). To increase data robustness, twelve technical replicate spectra were acquired from four different target spots using the recommended instrument settings for bacterial identification (linear positive ion detection mode, 60 Hz laser frequency, 20 kV acceleration voltage, 18.1–18.2 kV IS2 voltage). Spectra were initially analyzed at BfR using the Bruker Biotyper software (v3.1) with MSP library version MBT_7311 (7311 entries), the Security-Relevant (SR) Database (104 entries) and a customized in-house database to confirm identification as B. melitensis.
Data Analysis
Raw spectra data from the Bruker microflex MS instruments were imported into the statistical computing environment “R” (v3.6.3) and analyzed with the R packages MALDIquantForeign (v0.12) and MALDIquant (v1.19.3) (Gibb and Strimmer, 2012). Raw spectra were preprocessed using default functions and parameters, i.e., square root transformation, smoothing, baseline removal and normalization. Spectra were combined by the function averageMassSpectra, first by averaging technical replicate spectra into a “sample spectrum” and second by averaging sample spectra of the same group into a “group spectrum.” The groups were defined by bacteriological classification of a sample as “melitensis Rev.1” or the field isolate outgroup “melitensis” as well as by the institute where protein extracts were subjected to MALDI-TOF MS, namely “BfR” or “KVI.” Two intermediary spectra alignment steps were applied in the preprocessing: technical replicate spectra were aligned against each other whereas sample spectra were aligned against 37 reference peaks derived from all available sample spectra (method = “strict,” minFrequency = 0.9, tolerance = 0.002) of this study (Supplementary Table 4). The reference peaks with a relative frequency of ≥ 90% were distributed in a mass interval between 3,100 and 11,500 Da. All graphical spectra representations (gel, spectra and peaks) were drawn with customized ggplot2 functions from the R package Tidyverse v1.3.0 (Wickham et al., 2019). A comparison of all sample spectra is shown in Supplementary Figure 1.
Biomarker Validation
Sample spectra from the groups “melitensis Rev.1” and “melitensis” were manually screened for peaks corresponding to exact mass value pairs of short-listed proteins (Table 1) from the B. melitensis Rev.1 and 16M proteomes, respectively. Potential mass shifts in the mass value pairs caused by protein-specific PTMs were also considered. Biomarker mass intervals and decision boundaries for Rev.1 classification were determined by visual inspection of the peak distribution in the vicinity of accurate masses.
In silico Genotyping and Pan Proteome Sequence Analysis
The NCBI database was queried3 for all RefSeq annotated assemblies of the genus Brucella (accession date 2020-11-05). Genomic and protein FASTA files of 802 samples were downloaded. Remarkably, this dataset also contained samples from the former taxon Ochrobactrum, which were used for comparison. The collection of FAA files was loaded into R and a binary matrix with unique sequences, the genus pan-proteome, and their presence therein was built. We calculated Jaccard coefficients, performed hierarchical clustering by UPGMA and visualized the phylogram (Figure 6) with the R package ggtree v2.4.1 (Yu, 2020). Molecular in silico typing was performed with the software tools (i) ‘‘MLST’’4 written by Thorsten Seemann using a MLST scheme with nine loci (Whatmore et al., 2007) retrieved from PubMLST (Jolley et al., 2018) and (ii) “MLVA” written by David Christiany (Vergnaud et al., 2018) using the MLVA panel 1 with eight loci and typing groups according to MLVAbank5. Typing results were combined when either one method yielded a typing group or both were concordant (Supplementary Table 5). Each RefSeq assembly was screened for variants of the ribosomal protein L24 or S12 sequence of B. melitensis 16M with blastP (Supplementary Table 5). True hits were used for annotation of the phylogram (Figure 6).
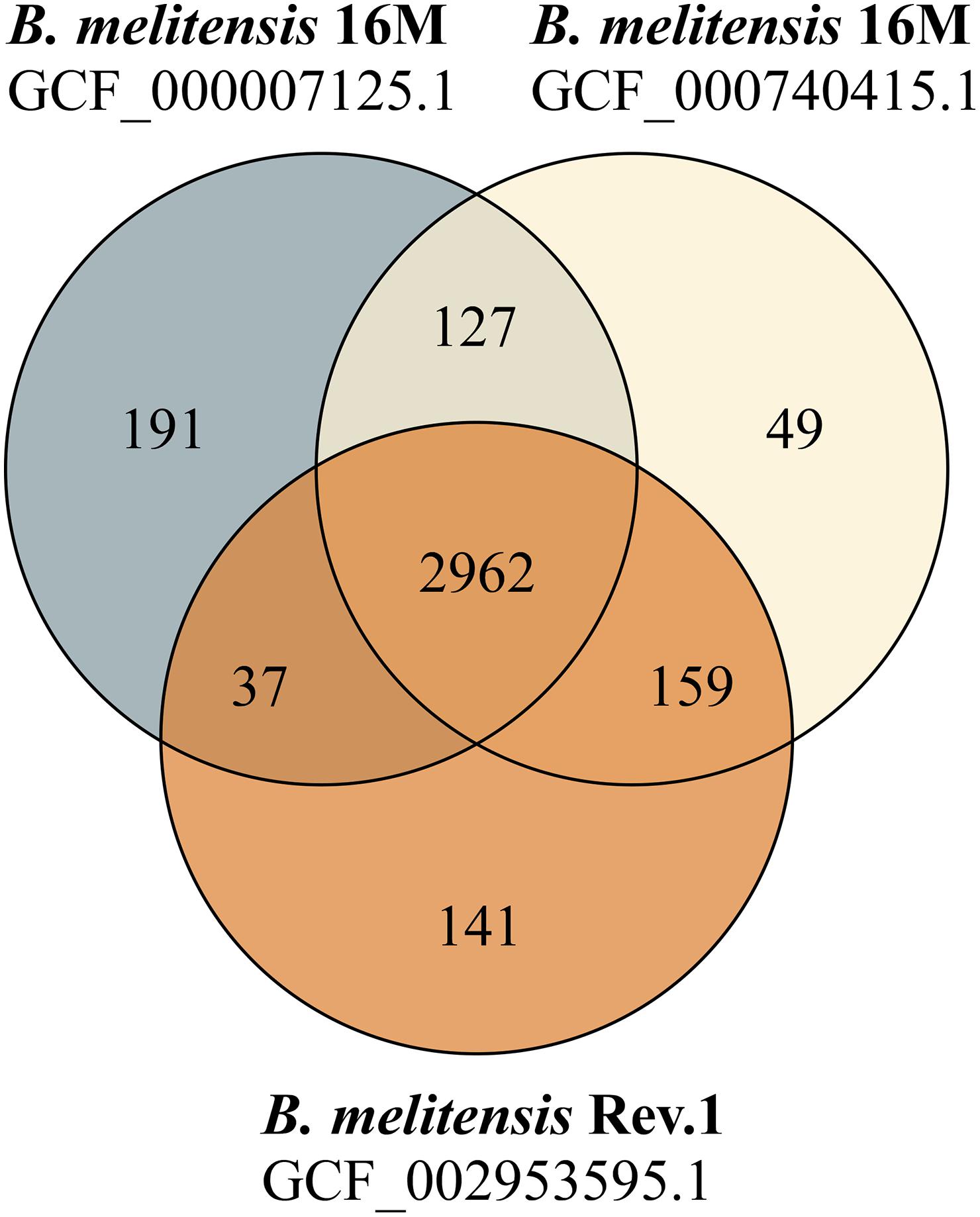
Figure 1. Venn diagram of shared and unique protein sequences found in RASTtk-annotated genomes. The protein-coding sequence sets of two B. melitensis 16M genomes (GCF_000007125.1: 3317 CDS; GCF_000740415.1: 3297 CDS) and one B. melitensis Rev.1 genome (GCF_002953595.1: 3299 CDS) were compared.
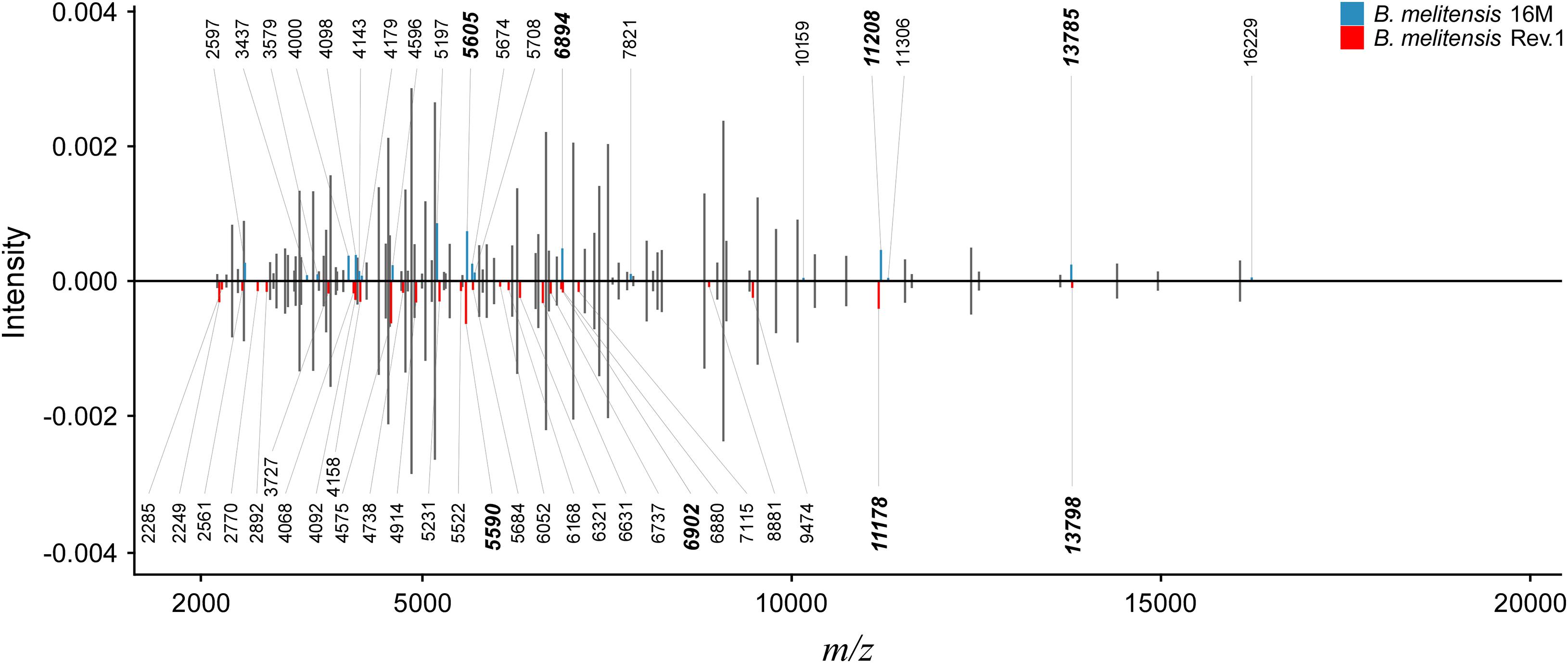
Figure 2. Peak comparison between B. melitensis 16M and B. melitensis Rev.1 strains. Shared peaks with the same m/z values are colored in gray without m/z label. Upper bar plot: B. melitensis 16M with unique peaks colored in blue. Lower bar plot: B. melitensis Rev.1 with unique peaks colored in red. For better comparison, B. melitensis Rev.1 intensity values were multiplied by –1. Bold m/z labels mark peak candidates that match mass difference and absolute mass of short-listed protein variants (Table 1). Peaks were averaged from sample spectra of three biological replicates with a peak frequency threshold of 50%.
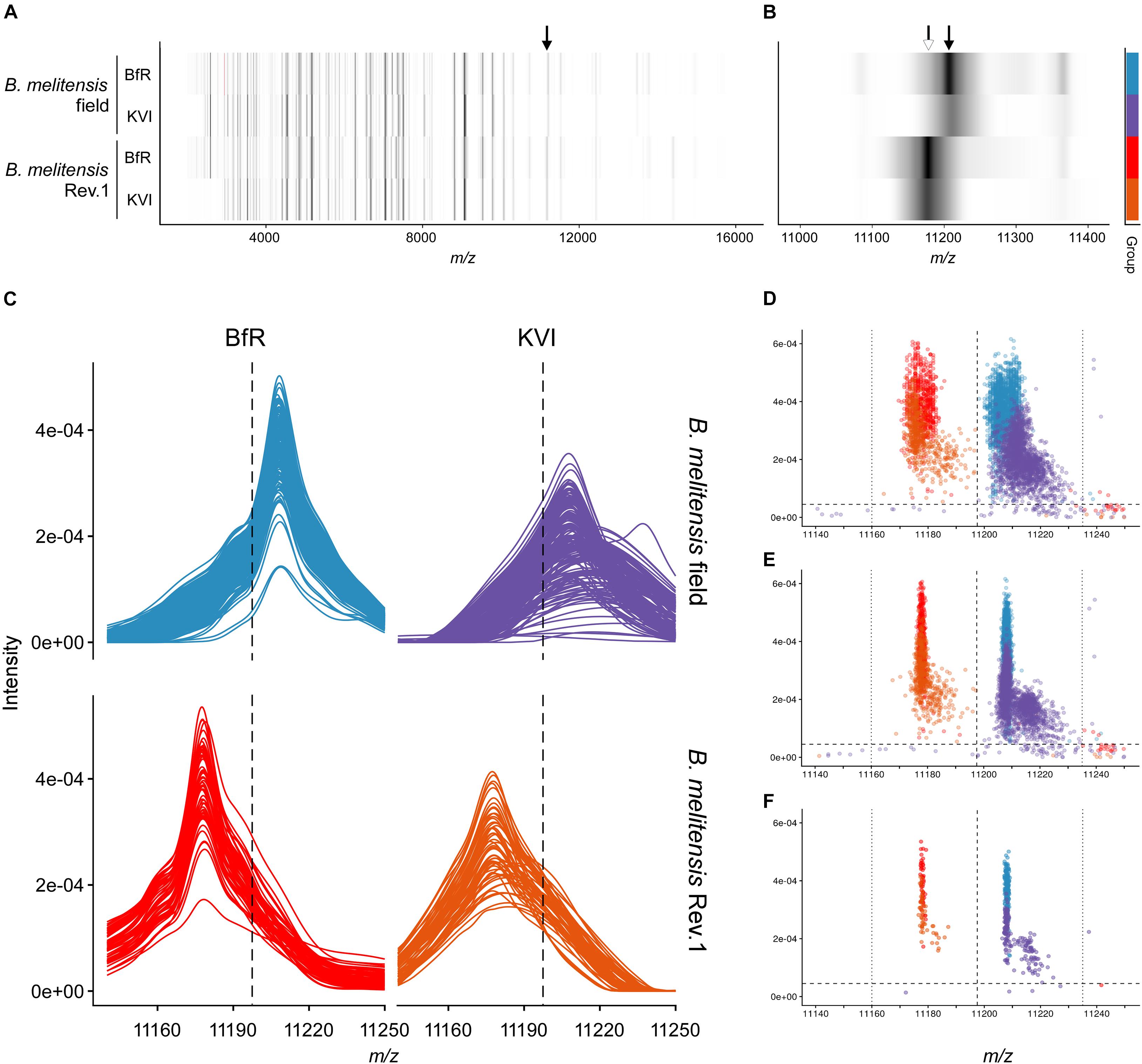
Figure 3. Group spectra and peak discrimination for the single charged ribosomal protein L24. (A,B) Gel view of group spectra derived from the B. melitensis field isolate group “melitensis” and the vaccine strain group “melitensis Rev.1” presented by institute “BfR” and “KVI,” in the full acquisition window (A) and magnified to the m/z range enclosing the exact masses of PLF_234_00001168, z = 1 (B). Arrows point to accurate masses at 11,178 Da [white, (MThr28+H)+ ion] and 11,208 Da [black; (MMet28+H)+ ion]. (C) Line graphs of aligned sample spectra shown by group and institute. (D–F) Scatterplot of peaks in the technical replicate spectra before (D) and after alignment (E) against the reference spectrum. (F) Peaks derived from aligned sample spectra. Intensity cut-off (horizontal dashed line) in panels (C–F): 0.000045 AU; for mass cut-offs (decision window: vertical dotted lines, decision boundary: vertical dashed line) see Table 2.
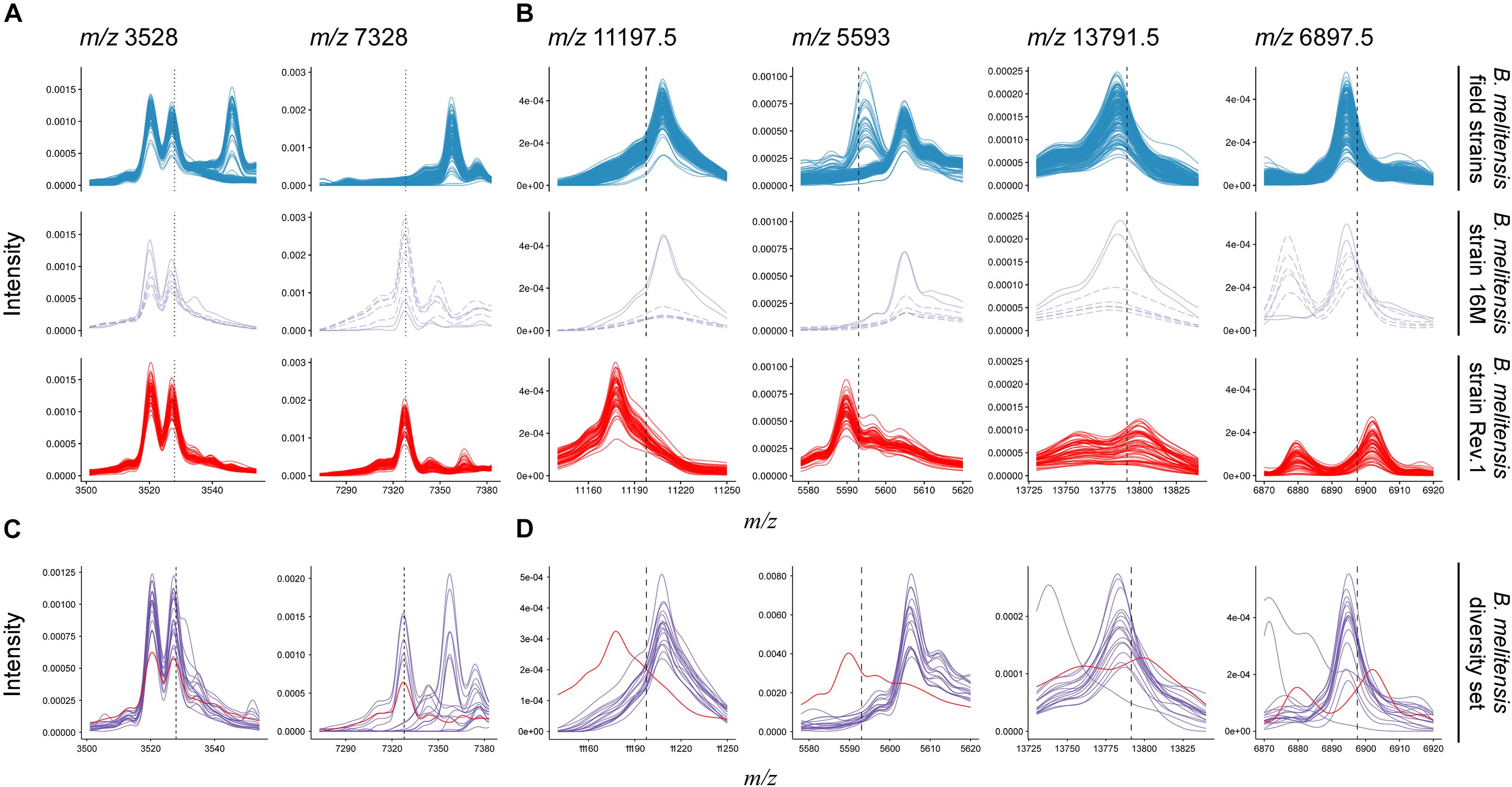
Figure 4. Comparison of B. melitensis field isolate (first row) and Rev.1 (third row) spectra against the type strain B. melitensis 16M (second row) and a diversity set of B. melitensis strains (fourth row) for selected masses. (A) Putative biomarkers (vertical dotted lines) described by Christoforidou et al. (2020). All spectra in this study displayed a peak at m/z 3,528. Most Rev.1 strains as well as the B. melitensis type strain 16M displayed a peak at m/z 7,328. (B) All biomarker decision boundaries (vertical dashed lines) in this study demarcate B. melitensis Rev.1 from B. melitensis 16M and field isolates. (C,D) Spectra from a diversity set of B. melitensis MVLA8 genotypes (purple) are plotted against a group spectrum of B. melitensis Rev.1 samples (red). For clarity, only spectra acquired at the BfR are shown.
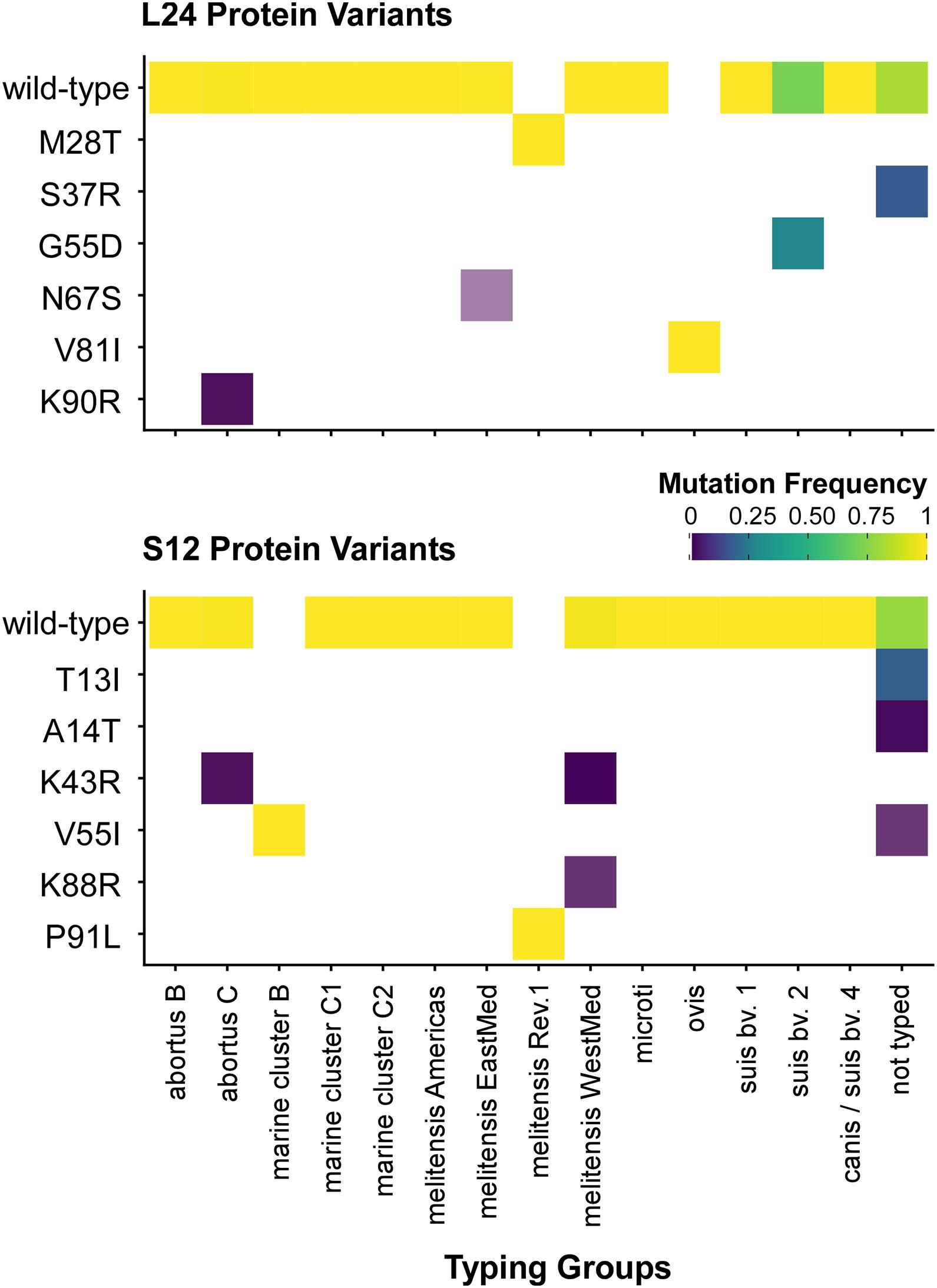
Figure 5. Distribution of ribosomal protein L24 and S12 variants in 802 Brucella genomes from the NCBI dataset. The heatmap coloring shows the frequency of each variant in the MLVA typing groups (white: no occurrence).
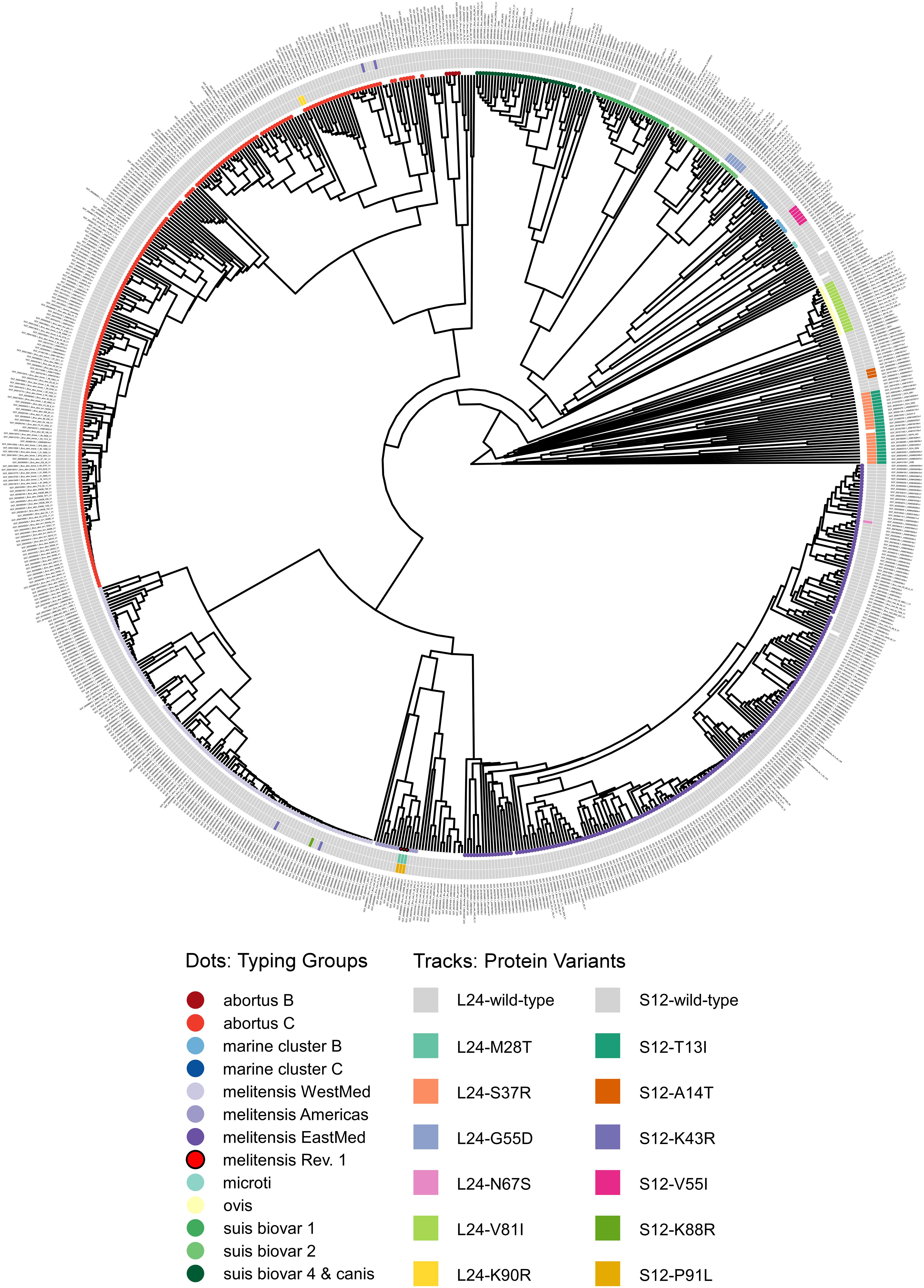
Figure 6. Phylogram of protein sequence presence in the NCBI pan proteome of the genus Brucella. Genomes were classified by in silico MLVA and MLST into typing groups (colored circles as leaves on the unrooted phylogram; white: no or non-congruent typing result). The tracks show ribosomal protein variants for L24 (inner) and S12 (outer) identified by blastP (white: no significant hit).
Results
In silico Identification of Biomarkers to Distinguish Brucella melitensis Rev.1 From the Reference Strain Brucella melitensis 16M
Our study aimed to identify MALDI-TOF MS biomarkers that allow for clearly distinguishing B. melitensis wild-type strains from the B. melitensis Rev.1 vaccine strain. Since MALDI-TOF MS for bacterial identification focusses on a narrow mass-over-charge (m/z) ratio ranging from 2,000 to 20,000, we first assessed a sequence-based in silico approach to find biomarkers. This method is feasible with respect to the limited number of expected divergent proteins useful for the discrimination of genetically closely related strains, such as B. melitensis Rev.1 and the reference strain B. melitensis 16M, both members of the same MLVA (Multiple-Locus Variable-number of tandem repeats Analysis) group “melitensis Americas” (Vergnaud et al., 2018). To this end, we compared the in silico translated open-reading frames (ORFs) of two different B. melitensis 16M complete genomes (Delvecchio et al., 2002; Minogue et al., 2014) against a complete B. melitensis Rev.1 genome (Salmon-Divon et al., 2018). To ease the matching of respective ORFs, all genomes were subjected to re-annotation by the RASTtk pipeline at PATRIC (Supplementary Table 1) and to a set analysis of their protein coding sequences. The resulting in silico core proteome shared by all three B. melitensis genome assemblies consisted of 2,962 translated coding sequences with identical amino acid sequences (Figure 1). Hence, about 90% of the protein sequences derived from the three B. melitensis genome sequences were not considered for our analysis. We further excluded 240 (191 + 49) proteins that differed between the two B. melitensis 16M annotations or were unique to either one and additional 196 (159 + 37) proteins identical between B. melitensis Rev.1 and only one of the two B. melitensis 16M annotations (Figure 1). Applying these filters, 113 out of 127 sequences identical in both B. melitensis 16M in silico proteomes matched with homologous sequences in the group of 141 annotated proteins specific for B. melitensis Rev.1. These 113 proteins harbor minor amino acid exchanges as a result of SNPs in their respective genes. Only few proteins matched the second filter criteria of bearing a mass within the working range of MALDI-TOF MS and a small but discernable mass difference.
The resulting short-list of 17 potential biomarker pairs (Table 1) is comprised of three hypothetical proteins, proteins with housekeeping enzymatic properties and five proteins (L24, L6, S12, S8, S7) of the 50S and 30S ribosomal subunits. Ribosomal proteins, highly abundant in bacterial cells, are well-known biomarkers for the identification and classification of bacteria by MALDI-TOF MS (Suarez et al., 2013).
Ribosomal Proteins L24 and S12 Are Potential Discriminatory Marker Proteins to Identify the Vaccine Strain Brucella melitensis Rev.1
As a proof-of-concept for our bioinformatics approach, we screened an initial set of MALDI-TOF MS spectra from B. melitensis 16M and B. melitensis Rev.1 for the presence of the 17 group-specific m/z peak pairs and potentially post-translationally modified (PTM) variants thereof. For PTMs, we considered N-terminal methionine excision (NME, average Δmass = −131.2 Da), methylation (Δmass = 14.0 Da), acetylation (Δmass = 42.0 Da), and modifications noted in the UniProt database annotation of the respective proteins. Out of the 17 predicted biomarkers, a distinct peak pair in the sample spectra resembled the ribosomal protein L24 with averaged accurate masses at m/z 11,178 in the vaccine strain group “melitensis Rev.1” and m/z 11,208 in the reference strain group “melitensis 16M” (Figure 2). The exact mass difference of −30.1 Da between the ion [MThr28+H]+melitensis Rev.1 with m/z 11,178.8 and the ion [MMet28+H]+melitensis 16M with m/z 11,207.9 corresponded well to the observed peak shift. A further noticeable peak pair comprised the ribosomal protein S12 with averaged accurate masses at m/z 13,785 in the field strain group “melitensis 16M” and m/z 13,798 in the vaccine strain group “melitensis Rev.1.” The exact mass difference of +16.0 Da between the ion [MPro91+H]+melitensis 16M with m/z 13,871.2 and the ion [MLeu91+H]+melitensis Rev.1 with m/z 13,887.2 matched well to the observed peak shift (Figure 2).
In summary, these results supported the feasibility of our combined in silico and proteomic-based approach to identify new biomarkers for Brucella diagnostics that enable the differentiation between a B. melitensis reference strain and a closely related vaccine strain.
Differentiation of Brucella melitensis Field Isolates From the Brucella melitensis Rev.1 Vaccine Strain
Subsequently, we assessed the robustness of the discrimination between B. melitensis wild-type isolates and the B. melitensis Rev.1 vaccine strain based on the ribosomal proteins L24 and S12. For this purpose, we analyzed B. melitensis isolates collected from humans, goats, sheep and cattle in Israel, where ovine and caprine brucellosis is known to be endemic and a routine vaccination program for livestock with B. melitensis Rev.1 has been implemented during the last decades (Banai, 2002). Moreover, the B. melitensis strains endemic in Israel mostly belong to the MLVA group “melitensis East-Mediterranean,” whereas B. melitensis Rev.1 is part of the MLVA group “melitensis Americas” (Vergnaud et al., 2018).
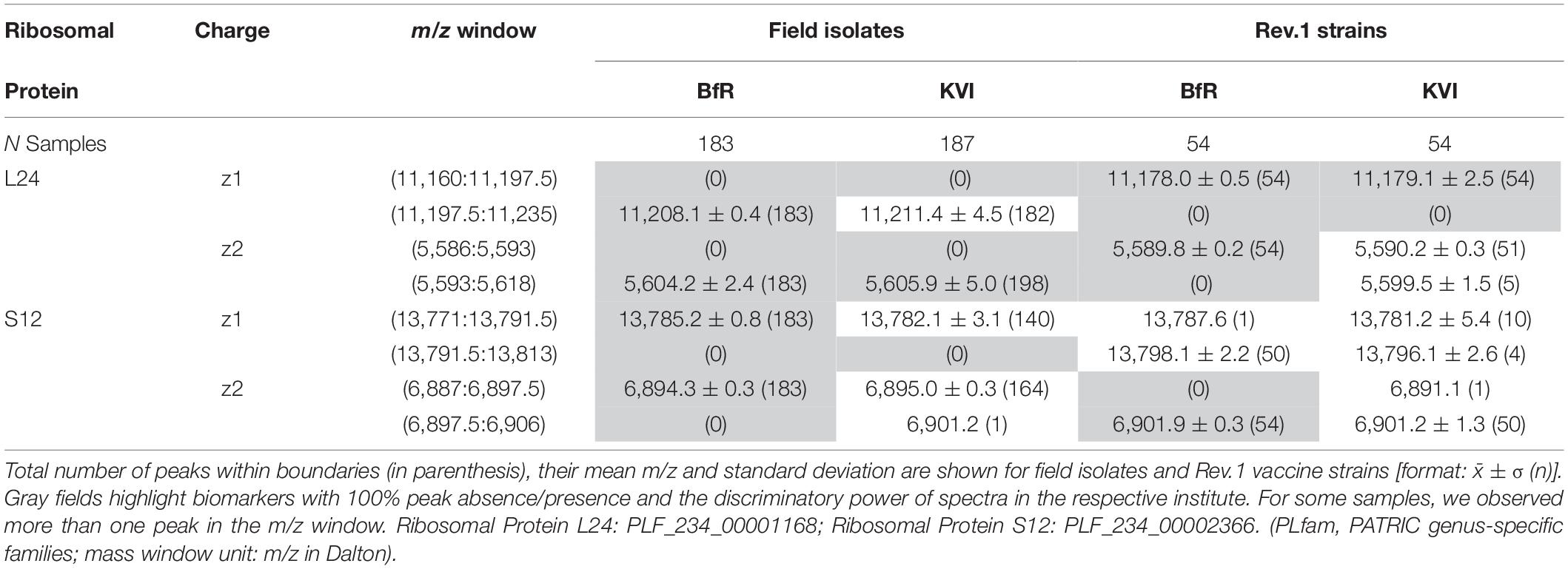
The MALDI-TOF MS spectra of 18 B. melitensis Rev.1 vaccine strains and 183 Israeli B. melitensis field isolates were compared, and we further evaluated the impact of spectra acquisition by performing MALDI-TOF MS measurements at the two institutes BfR and KVI (Supplementary Figure 1). As seen before in the comparison between B. melitensis Rev.1 and B. melitensis 16M (Figure 2), we detected a distinct peak pair in the sample spectra for the ribosomal protein L24 with averaged accurate masses at m/z 11,178 in the vaccine strain group “melitensis Rev.1” and m/z 11,208 in the field strain group “melitensis” (Figure 3). In the mass range for double charged ions of the ribosomal protein L24, we detected accurate masses at m/z 5,590 (group “melitensis Rev.1”) and m/z 5,605 (group “melitensis”) (Supplementary Figure 2) corresponding to the ions [MThr28+2H]2+ and [MMet28+2H]2+, respectively. In a subset of “melitensis” field strains samples, this peak pair is interspersed with a second signal at m/z 5,595, which is not present in the single charged state (Supplementary Figure 2B, “melitensis” groups).
Furthermore, the peak pair for the ribosomal protein S12 was found in the sample spectra with averaged accurate masses at m/z 13,784 in the field strain group “melitensis” and m/z 13,798 in the vaccine strain group “melitensis Rev.1” (Supplementary Figure 3). The exact mass difference of +16.0 Da between the ion [MPro91+H]+melitensis with m/z 13,871.2 and the ion [MLeu91+H]+melitensis Rev.1 with m/z 13,887.2 corresponded well to the observed peak shift. However, for peaks that were not within the mass window of 3,000 to 11,500 Da (defined by reference peaks with at least 90% frequency in all measurements, see Supplementary Table 4), the alignment by a locally weighted scatterplot smoothing function (LOWESS) was less accurate, leading to a higher mass scattering. Ribosomal protein S12 ion variants show respective peaks in the “BfR” subgroups with the strongest intensities for ion [M-Met+βMeS+H]+, the beta-methylthiolated ribosomal protein S12 ion lacking the N-terminal methionine (Supplementary Figure 3). In the mass range for double charged ions of probable ribosomal protein S12, we detected accurate masses at m/z 6,894 (group “melitensis”) and m/z 6,902 (group “melitensis Rev.1”) (Supplementary Figure 4) corresponding to the ions [MPro91-Met+βMeS+2H]2+ and [MLeu91-Met+βMeS+2H]2+, respectively.
Based on our peak observations, we determined mass windows with a decision boundary to distinguish between ribosomal protein L24 and S12 variants, and tested whether these thresholds facilitate the differentiation between the B. melitensis field isolates and the Rev.1 vaccine strain (Table 2). The alignment procedure against a reference spectrum decreased the standard deviation considerably (Table 2), e.g., for “BfR” peaks within the ribosomal protein L24 (z = 1) decision mass window (11,197.5:11,235) from meannon–aligned = 11,206.9 with SDnon–aligned = 2.9 for technical replicate spectra to meanaligned = 11,208.1 with SDaligned = 0.4 for sample spectra (Figures 3D–F). Peaks from the KVI measurement scattered more widely (technical replicate spectra meannon–aligned = 11,213.3 with SDnon–aligned = 4.7 versus sample spectra meanaligned = 11,211.4 with SDaligned = 4.5), due to a recalibration of the KVI instrument between measurements.
Recently, a study has been published that also aimed to detect biomarkers enabling the discrimination of B. melitensis field isolates and the Rev.1 vaccine strain by MALDI-TOF MS (Christoforidou et al., 2020). For this purpose, 73 clinical and veterinary B. melitensis isolates from Greece were analyzed. Initially, a cluster analysis on a subset of 17 field strains that represented the three B. melitensis biovars against three commercial B. melitensis Rev.1 strains had been performed. Two discriminating peaks were described in this study: Peak m/z 3,528 could be detected in all tested Greek B. melitensis field isolates but not in B. melitensis Rev.1, whereas peak m/z 7,328 was unique for the vaccine strain (Christoforidou et al., 2020). Strikingly, these biomarker peaks were not identified in our study, thus we compared the data published by Christoforidou et al. (2020) with our data. The first described biomarker with m/z 3,528, supposed to be only present in the spectra of B. melitensis Greek field isolates, could be found in the spectra of all Israeli field isolates, but also in the B. melitensis 16M reference strain and the Rev.1 strain (Figure 4A). The second biomarker with m/z 7,328 neither occurred in the Greek B. melitensis field isolates (Christoforidou et al., 2020) nor in the B. melitensis field isolates from Israel (Figure 4A), but in the B. melitensis Rev.1 strain of both studies. However, this peak does not reflect a true biomarker for B. melitensis Rev.1 against the closely related B. melitensis 16M from the same MLVA group “melitensis Americas,” which also harbors this causative protein (Figure 4A).
In contrast, our here reported ribosomal protein biomarkers L24 and S12 distinguish the B. melitensis Rev.1 vaccine strain not only from the Israeli B. melitensis field isolates but also from B. melitensis 16M (Figure 4B). Christoforidou et al. (2020) probably did not detect these ribosomal biomarkers since their forward analysis tolerance parameter, which was not stated, may have excluded the detection of close peaks with the Mass-Up default settings or due to low mass intensities in m/z ranges above 10 kDa. In order to prove the universal character of our biomarkers and to exclude clonal and regional effects, B. melitensis strains from 17 different MLVA groups (Supplementary Table 6) were subjected to MALDI-TOF MS analysis and the spectra were searched for the presence of the protein biomarkers L24 and S12. While the biomarker peaks described by Christoforidou et al. (2020) for the differentiation of B. melitensis Rev.1 could also be found in the B. melitensis field strains of this diversity set (Figure 4C), the analysis confirmed the discriminatory power of the L24 and S12 biomarkers, which distinguish the B. melitensis Rev.1 vaccine strain from naturally occurring B. melitensis strains in general (Figure 4D).
Discriminatory Power of the L24 and S12 Signals for the Strain-Level Identification of Brucella melitensis Rev.1 by MALDI-TOF MS
We further evaluated the uniqueness of the ribosomal proteins L24 and S12 as marker proteins for the unambiguous identification of B. melitensis Rev.1 by comparing their amino acid sequences with the L24 and S12 protein entries in the NCBI protein database originating from 802 sequenced Brucella isolates.
Unexpectedly, a blastP-based comparison did not identify a protein sequence identical to the L24-M28T variant of B. melitensis Rev.1 in any other isolate of the NCBI genome dataset (Figure 5 and Supplementary Table 7). Instead, the amino acid sequences of the ribosomal L24 proteins from most isolates of the analyzed Brucella species were identical to the L24 protein sequence of B. melitensis 16M. However, few isolates of the B. abortus clade C, of the B. melitensis East-Mediterranean clade and of B. suis bv. 2 harbor altered L24 protein sequences with the amino acid exchanges K90R, N67S and G55D, respectively (Figure 6). The L24 protein variant of the B. melitensis East-Mediterranean isolate with the N67S amino acid exchange exhibits a −27 Da mass difference to the wild-type L24 protein. Hence, its spectrum peak cannot be distinguished by MALDI-TOF MS from the −30 Da spectrum peak of L24 variant from B. melitensis Rev.1 (Supplementary Table 7). Interestingly, all B. ovis isolates encode for a L24 protein with the unique modification V81I resulting in a +14 Da mass difference compared to the common L24 proteins (Figure 5 and Supplementary Table 7).
For ribosomal protein S12, B. melitensis Rev.1 exclusively carried the P91L mutation with its +16 Da mass difference compared to the most frequent S12 variant in the NCBI dataset (Figure 5 and Supplementary Table 8). However, the ribosomal S12 proteins of all analyzed B. ceti clade B isolates harbor the amino acid exchange V55I that leads to a mass difference of +14 Da. Consequently, this S12 protein variant is indistinguishable from the corresponding S12 protein peak of B. melitensis Rev.1 in MALDI-TOF MS analysis. Moreover, few strains of the B. abortus clade C encode for a ribosomal S12 protein with a K43R exchange and a limited number of isolates of the B. melitensis West-Mediterranean clade express S12 protein variants with the amino acid changes K43R or K88R (Figure 6 and Supplementary Table 8). Mutations at corresponding codon positions have been identified in streptomycin-resistant strains of other bacterial species (Supplementary Figure 5) like Escherichia coli, Salmonella Typhimurium, Mycobacterium tuberculosis, Helicobacter pylori, Klebsiella pneumoniae, Erwinia carotovora or Thermus thermophiles (Finken et al., 1993; Bjorkman et al., 1999; Gregory et al., 2001; Torii et al., 2003; Chumpolkulwong et al., 2004; Barnard et al., 2010; Tsai et al., 2014).
Taken together, our bioinformatics analysis suggests that the ribosomal protein variants of L24 and S12 identified here as biomarkers for B. melitensis Rev.1 have extremely high discriminatory properties allowing the direct differentiation of this vaccine strain from other Brucella isolates even without performing a preceding classification as B. melitensis.
Discussion
The worldwide emerging zoonotic disease brucellosis affects wildlife and livestock, especially cattle, goats and sheep (Seleem et al., 2010; Godfroid, 2017). In countries with endemic brucellosis, vaccination programs have been undertaken to combat Brucella infections in farm animals (Banai, 2002; Olsen and Stoffregen, 2005). However, these measures require efficient diagnostic tools to distinguish vaccine strains from naturally occurring wild-type Brucella strains in the livestock herds.
Here, we propose MALDI-TOF MS profiling as a cost-effective alternative to the currently applied methods of classical microbiological testing (Elberg and Meyer, 1958) and molecular PCR-techniques (Bardenstein et al., 2002; Lopez-Goni et al., 2008; Alvarez et al., 2017; Christoforidou et al., 2018) for the differentiation of the B. melitensis Rev.1 vaccine strain from B. melitensis field isolates. This approach will be of benefit for reference laboratories and large healthcare facilities that have already implemented MALDI-TOF MS diagnostics for the identification of bacterial pathogens. Moreover, due to the robustness of mass spectrometry, this rapid identification method is increasingly used in tropical countries, where it complements smaller point-of-care laboratories (Fall et al., 2015; Chabriere et al., 2018).
Whole genome comparison has previously identified various mutations as genetic markers that distinguish B. melitensis Rev.1 from reference strain B. melitensis 16M (Issa and Ashhab, 2016). However, besides a non-synonymous mutation affecting the ribosomal protein S12, most of the 32 listed genome-specific markers were not identified by our in silico analysis or did not translate into changes of MALDI-TOF MS peaks. This might be the consequence of (i) our filter criteria that restricted potential marker proteins on masses between 2,000 and 20,000 Da, (ii) mutations that do not translate into amino acid exchanges captured by MALDI-TOF MS, (iii) insufficient in vitro expression of the marker genes, or (iv) MALDI-TOF MS signal suppression effects leading to undetectable amounts of protein.
Our comparative in silico analysis of the translated open reading frames from the genomes of B. melitensis 16M and Rev.1 predicted 17 potential MALDI-TOF MS biomarkers for the differentiation of both strains. Subsequent whole-cell MALDI-TOF MS spectra comparison of B. melitensis 16M and Rev.1 confirmed that two marker peak pairs, resembling the ribosomal proteins L24 and S12 in their single and double charged states, have the discriminatory power to distinguish between these closely related B. melitensis strains. Our observation is in agreement with previous studies, demonstrating that the sequence variabilities and abundance of ribosomal proteins in microbial cells allow their usage as robust biomarkers for the identification of pathogenic and non-pathogenic bacteria at species and strain level by whole-cell MALDI-TOF MS (Sandrin et al., 2013).
The ribosomal protein L24 is encoded by the rplX gene, for which spontaneous missense mutations have been described before (Nishi et al., 1987; Sharp et al., 1992). Accordingly, our comparative bioinformatics analysis of the Brucella pan proteome not only identified the altered L24 protein in B. melitensis Rev.1 but also five additional variants. These mutated L24 proteins exhibit mass differences from about −30 to +69 Da compared to the wild-type L24 protein of Brucella and may represent additional biomarkers, especially for the identification of Brucella ovis and for the improved differentiation of Brucella and closely related former Ochrobactrum species. Likewise, MALDI-TOF MS studies of various other bacteria have identified peak differences of L24 protein variants. Consequently, the ribosomal protein L24 has served as species-specific biomarker for Flavobacterium psychrophilum, Bacillus spp., Streptococcus thermophilus, Listeria monocytogenes and Francisella tularensis as well (Teramoto et al., 2007; Hotta et al., 2011; Durighello et al., 2014; Ojima-Kato et al., 2016; Fernandez-Alvarez et al., 2018).
The streptomycin resistance is a distinct characteristic of vaccine strain B. melitensis Rev.1 that was introduced during vaccine derivation by Elberg and Faunce (1957). It is mediated through a spontaneous rpsL mutation that leads to an altered ribosomal protein S12 with the amino acid exchange Pro to Leu at codon position 91 (P91L) (Cloeckaert et al., 2002). We showed here that this altered protein sequence resulted in a distinct shift of the MALDI-TOF MS peak m/z 13,784, which can be used as a biomarker for the identification of B. melitensis Rev.1.
Two post-translational modifications of ribosomal protein S12 have been observed in other bacterial species (Kowalak and Walsh, 1996; Suh et al., 2005) or were inferred from sequence similarity: NME (average mass change: −131.2 Da) and beta-methylthiolation of Asp89 (average mass change: +46.1 Da, see UniProt annotation of RS12_BRUME). We also found this mutation in the rpsL genes of streptomycin resistant T. thermophilus, K. pneumoniae and E. coli isolates, further illustrating that certain antibiotic resistances in bacteria correlate with ribosomal protein changes that can be detected by MALDI-TOF MS (Wilcox et al., 2001; Carr et al., 2005).
The usage of the ribosomal proteins S12 and L24 signals as MALDI-TOF MS biomarkers enabled the unequivocal discrimination between B. melitensis Rev.1 and the analyzed Israeli B. melitensis field isolates, as well as the closely related type strain B. melitensis 16M. Comparable results were obtained from MALDI-TOF MS measurements performed at two different institutes, emphasizing the high quality, accuracy and reproducibility of the established method.
Moreover, our bioinformatics analysis of hitherto published Brucella proteomes did not identify any other Brucella isolate that encodes for S12 and L24 proteins with identical amino acid sequences, further illustrating their unique character. Our here identified biomarkers for the identification of the B. melitensis Rev.1 vaccine strain had not been identified by a recent MALDI-TOF MS based study pursuing the same aim (Christoforidou et al., 2020). The two alternative biomarkers described in the work by Christoforidou et al. (2020) for the discrimination between B. melitensis field isolates and the Rev.1 vaccine strain may only be of benefit for local applications in Greece. According to our analysis, the first proposed biomarker (m/z 3,528) was not only present in B. melitensis field isolates and B. melitensis 16M, but also in B. melitensis Rev.1, whereas the second putative biomarker (m/z 7,328) was not exclusive for B. melitensis Rev.1 as indicated, but was also found in B. melitensis 16M. Two analytical restraints in their study could have contributed to these different findings. First, the mass trimming to a window of interest has omitted the detection of the single charged variants of probable ribosomal proteins L24 and S12. Second, mathematical binning of continuous sample peak m/z values for subsequent cluster analysis utilized a tolerance threshold that limited its discriminatory power. The double charged ribosomal protein L24 variants in our study were detectable at a mass resolution of 400 whereas double charged ribosomal protein S12 variants required a higher analytical mass resolution of 860. Christoforidou and colleagues used the MALDIquant default tolerance of 0.002 for peak binning, i.e., a mass resolution limit of 250, and therefore, were not able to detect any of the double charged variants of ribosomal protein L24 or S12. Furthermore, if spectra within a group display diversity, as seen in the ribosomal protein L24 subset of “melitensis” field isolates at m/z 5,595 (Supplementary Figure 2), biomarker significance in statistical tests will diminish. Applying these analytical considerations, our MALDI-TOF MS approach identified two genus-wide unique biomarkers that unambiguously discriminated B. melitensis Rev.1 from B. melitensis field isolates and the closely related type strain B. melitensis 16M without any misidentification.
Several manufacturers around the world produce the B. melitensis Rev.1 vaccine strain from different seed stocks. Hence, a comprehensive characterization of B. melitensis Rev.1 is required for quality controls in vaccine production facilities, since Rev.1 strains of different production sites may differ significantly from the original Elberg strain (Bosseray, 1991). To avoid alterations in the attenuated virulence and to assure strain stability during the vaccine production process, the gene expression profile of the B. melitensis Rev.1 strain should be checked on a regular basis. In this context, our MALDI-TOF MS based analysis may serve as an additional standardization tool in the commercial production of the B. melitensis Rev.1 vaccine.
Conclusion
The here described ribosomal marker proteins for distinguishing the vaccine strain B. melitensis Rev.1 from B. melitensis field strains by MALDI-TOF MS will improve the differential diagnosis necessary for brucellosis control efforts within vaccination programs and subsequently the successful eradication of the zoonoses from small ruminants. Natural streptomycin resistance in Brucella is rare, but Brucella species exhibit variable streptomycin susceptibilities and the development of spontaneous resistance has been seen in vitro (Hall and Manion, 1970; Hall, 1990). This observation is supported by our identification of Brucella isolates that encode for ribosomal protein S12 variants with amino acid sequences of streptomycin resistant alleles. Our approach does not detect streptomycin resistance mediated by mutations of the 16S rRNA (Springer et al., 2001). However, the improved MALDI-TOF MS based screening for the B. melitensis Rev.1 vaccine strain and other streptomycin resistant B. melitensis field isolates with ribosomal protein S12 variants may reduce the risk of an inadequate first line therapy in human brucellosis, since streptomycin is commonly used in the antibiotic regimen applied for patients infected with Brucella (Skalsky et al., 2008; Meng et al., 2018).
Data Availability Statement
Most of the datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
HB, DK, DH, and SAD: conceptualization and writing—review and editing. HB, DK, and DH: data curation and writing—original draft. HB: formal analysis, methodology, software, and visualization. SAD: funding acquisition, project administration, and supervision. HB, DK, and SM: investigation. SEB, MF, and SB: resources. HB, DK, DH, SAD, MF, SEB, and SM: validation. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by a grant of the German Federal Ministry of Education and Research (BMBF) within the joint project Ess-B.A.R (FKZ 13N13982).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Anne Stephan for excellent technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.712601/full#supplementary-material
Footnotes
- ^ www.ncbi.nlm.nih.gov/assembly
- ^ www.patricbrc.org
- ^ https://www.ncbi.nlm.nih.gov/assembly/?term=txid234[Organism:exp]
- ^ https://github.com/tseemann/mlst
- ^ https://microbesgenotyping.i2bc.paris-saclay.fr
References
Alvarez, L. P., Marcellino, R. B., Martinez, A., and Robles, C. A. (2017). Duplex PCR for the diagnosis of Brucella melitensis and its differentiation from the REV-1 vaccine strain. Small Ruminant Res. 146, 1–4.
Arapovic, J., Spicic, S., Duvnjak, S., Ostojic, M., Arapovic, M., Nikolic, J., et al. (2020). The first report of Brucella melitensis Rev.1 human brucellosis in Bosnia and Herzegovina. J. Infect. Dev. Ctries 14, 232–235. doi: 10.3855/jidc.11949
Banai, M. (2002). Control of small ruminant brucellosis by use of Brucella melitensis Rev.1 vaccine: laboratory aspects and field observations. Vet. Microbiol. 90, 497–519. doi: 10.1016/s0378-1135(02)00231-6
Banai, M., Mayer, I., and Cohen, A. (1990). Isolation, identification, and characterization in Israel of Brucella melitensis biovar 1 atypical strains susceptible to dyes and penicillin, indicating the evolution of a new variant. J. Clin. Microbiol. 28, 1057–1059. doi: 10.1128/jcm.28.5.1057-1059.1990
Bardenstein, S., Mandelboim, M., Ficht, T. A., Baum, M., and Banai, M. (2002). Identification of the Brucella melitensis vaccine strain Rev.1 in animals and humans in Israel by PCR analysis of the PstI site polymorphism of its omp2 gene. J. Clin. Microbiol. 40, 1475–1480. doi: 10.1128/jcm.40.2.1475-1480.2002
Barnard, A. M. L., Simpson, N. J. L., Lilley, K. S., and Salmond, G. P. C. (2010). Mutations in rpsL that confer streptomycin resistance show pleiotropic effects on virulence and the production of a carbapenem antibiotic in Erwinia carotovora. Microbiology (Reading) 156, 1030–1039. doi: 10.1099/mic.0.034595-0
Bjorkman, J., Samuelsson, P., Andersson, D. I., and Hughes, D. (1999). Novel ribosomal mutations affecting translational accuracy, antibiotic resistance and virulence of Salmonella typhimurium. Mol. Microbiol. 31, 53–58. doi: 10.1046/j.1365-2958.1999.01142.x
Bosseray, N. (1991). Brucella melitensis Rev. 1 living attenuated vaccine: stability of markers, residual virulence and immunogenicity in mice. Biologicals 19, 355–363. doi: 10.1016/S1045-1056(05)80025-9
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Carr, J. F., Gregory, S. T., and Dahlberg, A. E. (2005). Severity of the streptomycin resistance and streptomycin dependence phenotypes of ribosomal protein S12 of Thermus thermophilus depends on the identity of highly conserved amino acid residues. J. Bacteriol. 187, 3548–3550. doi: 10.1128/JB.187.10.3548-3550.2005
Chabriere, E., Bassene, H., Drancourt, M., and Sokhna, C. (2018). MALDI-TOF MS and point of care are disruptive diagnostic tools in Africa. New Microbes New Infect. 26, S83–S88. doi: 10.1016/j.nmni.2018.08.020
Charif, D., and Lobry, J. R. (2007). “SeqinR 1.0-2: a contributed package to the R Project for statistical computing devoted to biological sequences retrieval and analysis,” in Structural Approaches to Sequence Evolution: Molecules, Networks, Populations, eds U. Bastolla, M. Porto, H. E. Roman, and M. Vendruscolo (Berlin: Springer), 207–232. doi: 10.1007/978-3-540-35306-5_10
Christoforidou, S., Boukouvala, E., Zdragas, A., Malissiova, E., Sandalakis, V., Psaroulaki, A., et al. (2018). Novel diagnostic approach on the identification of Brucella melitensis Greek endemic strains-discrimination from the vaccine strain Rev.1 by PCR-RFLP assay. Vet. Med. Sci. 4, 172–182. doi: 10.1002/vms3.99
Christoforidou, S., Kyritsi, M., Boukouvala, E., Ekateriniadou, L., Zdragas, A., Samouris, G., et al. (2020). Identification of Brucella spp. isolates and discrimination from the vaccine strain Rev.1 by MALDI-TOF mass spectrometry. Mol. Cell Probes 51:101533. doi: 10.1016/j.mcp.2020.101533
Chumpolkulwong, N., Hori-Takemoto, C., Hosaka, T., Inaoka, T., Kigawa, T., Shirouzu, M., et al. (2004). Effects of Escherichia coli ribosomal protein S12 mutations on cell-free protein synthesis. Eur. J. Biochem. 271, 1127–1134. doi: 10.1111/j.1432-1033.2004.04016.x
Clark, A. E., Kaleta, E. J., Arora, A., and Wolk, D. M. (2013). Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 26, 547–603. doi: 10.1128/CMR.00072-12
Cloeckaert, A., Grayon, M., and Grepinet, O. (2002). Identification of Brucella melitensis vaccine strain Rev.1 by PCR-RFLP based on a mutation in the rpsL gene. Vaccine 20, 2546–2550. doi: 10.1016/S0264-410X(02)00159-7
Corbell, M. J. (1975). The serological relationship between Brucella spp., Yersinia enterocolitica serotype IX and Salmonella serotypes of Kauffmann-White group N. J. Hyg. (Lond.) 75, 151–171. doi: 10.1017/S0022172400047173
Croxatto, A., Prod’hom, G., and Greub, G. (2012). Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 36, 380–407. doi: 10.1111/j.1574-6976.2011.00298.x
Cunningham, S. A., and Patel, R. (2013). Importance of using Bruker’s security-relevant library for Biotyper identification of Burkholderia pseudomallei, Brucella species, and Francisella tularensis. J. Clin. Microbiol. 51, 1639–1640. doi: 10.1128/JCM.00267-13
Da Silva, D. A. V., Brendebach, H., Grutzke, J., Dieckmann, R., Soares, R. M., De Lima, J. T. R., et al. (2020). MALDI-TOF MS and genomic analysis can make the difference in the clarification of canine brucellosis outbreaks. Sci. Rep. 10:19246.
Delvecchio, V. G., Kapatral, V., Redkar, R. J., Patra, G., Mujer, C., Los, T., et al. (2002). The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. U.S.A. 99, 443–448. doi: 10.1073/pnas.221575398
Durighello, E., Bellanger, L., Ezan, E., and Armengaud, J. (2014). Proteogenomic biomarkers for identification of Francisella species and subspecies by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. Anal. Chem. 86, 9394–9398. doi: 10.1021/ac501840g
Elberg, S. S., and Faunce, K. Jr. (1957). Immunization against Brucella infection. VI. Immunity conferred on goats by a nondependent mutant from a streptomycin-dependent mutant strain of Brucella melitensis. J. Bacteriol. 73, 211–217. doi: 10.1128/jb.73.2.211-217.1957
Elberg, S. S., and Meyer, K. F. (1958). Caprine immunization against brucellosis; a summary of experiments on the isolation, properties and behaviour of a vaccine strain. Bull. World Health Organ. 19, 711–724.
Fall, B., Lo, C. I., Samb-Ba, B., Perrot, N., Diawara, S., Gueye, M. W., et al. (2015). The ongoing revolution of MALDI-TOF mass spectrometry for microbiology reaches tropical Africa. Am. J. Trop. Med. Hyg. 92, 641–647. doi: 10.4269/ajtmh.14-0406
Fernandez-Alvarez, C., Torres-Corral, Y., and Santos, Y. (2018). Use of ribosomal proteins as biomarkers for identification of Flavobacterium psychrophilum by MALDI-TOF mass spectrometry. J. Proteomics 170, 59–69. doi: 10.1016/j.jprot.2017.09.007
Ferreira, L., Vega Castano, S., Sanchez-Juanes, F., Gonzalez-Cabrero, S., Menegotto, F., Orduna-Domingo, A., et al. (2010). Identification of Brucella by MALDI-TOF mass spectrometry. Fast and reliable identification from agar plates and blood cultures. PLoS One 5:e14235. doi: 10.1371/journal.pone.0014235
Finken, M., Kirschner, P., Meier, A., Wrede, A., and Bottger, E. C. (1993). Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol. Microbiol. 9, 1239–1246. doi: 10.1111/j.1365-2958.1993.tb01253.x
Garcia-Yoldi, D., Marin, C. M., De Miguel, M. J., Munoz, P. M., Vizmanos, J. L., and Lopez-Goni, I. (2006). Multiplex PCR assay for the identification and differentiation of all Brucella species and the vaccine strains Brucella abortus S19 and RB51 and Brucella melitensis Rev1. Clin. Chem. 52, 779–781. doi: 10.1373/clinchem.2005.062596
Gibb, S., and Strimmer, K. (2012). MALDIquant: a versatile R package for the analysis of mass spectrometry data. Bioinformatics 28, 2270–2271. doi: 10.1093/bioinformatics/bts447
Godfroid, J. (2017). Brucellosis in livestock and wildlife: zoonotic diseases without pandemic potential in need of innovative one health approaches. Arch. Public Health 75:34. doi: 10.1186/s13690-017-0207-7
Gregory, S. T., Cate, J. H., and Dahlberg, A. E. (2001). Streptomycin-resistant and streptomycin-dependent mutants of the extreme thermophile Thermus thermophilus. J. Mol. Biol. 309, 333–338. doi: 10.1006/jmbi.2001.4676
Hall, W. H. (1990). Modern chemotherapy for brucellosis in humans. Rev. Infect. Dis. 12, 1060–1099. doi: 10.1093/clinids/12.6.1060
Hall, W. H., and Manion, R. E. (1970). In vitro susceptibility of Brucella to various antibiotics. Appl. Microbiol. 20, 600–604. doi: 10.1128/am.20.4.600-604.1970
Hordt, A., Lopez, M. G., Meier-Kolthoff, J. P., Schleuning, M., Weinhold, L. M., Tindall, B. J., et al. (2020). Analysis of 1,000+ type-strain genomes substantially improves taxonomic classification of Alphaproteobacteria. Front. Microbiol. 11:468. doi: 10.3389/fmicb.2020.00468
Hotta, Y., Sato, J., Sato, H., Hosoda, A., and Tamura, H. (2011). Classification of the genus Bacillus based on MALDI-TOF MS analysis of ribosomal proteins coded in S10 and spc operons. J. Agric. Food Chem. 59, 5222–5230. doi: 10.1021/jf2004095
Hoyer, B. H., and Mccullough, N. B. (1968). Polynucleotide homologies of Brucella deoxyribonucleic acids. J. Bacteriol. 95, 444–448. doi: 10.1128/jb.95.2.444-448.1968
Hull, N. C., and Schumaker, B. A. (2018). Comparisons of brucellosis between human and veterinary medicine. Infect. Ecol. Epidemiol. 8:1500846. doi: 10.1080/20008686.2018.1500846
Issa, M. N., and Ashhab, Y. (2016). Identification of Brucella melitensis Rev.1 vaccine-strain genetic markers: towards understanding the molecular mechanism behind virulence attenuation. Vaccine 34, 4884–4891.
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124.
Karger, A., Melzer, F., Timke, M., Bettin, B., Kostrzewa, M., Nockler, K., et al. (2013). Interlaboratory comparison of intact-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry results for identification and differentiation of Brucella spp. J. Clin. Microbiol. 51, 3123–3126.
Kowalak, J. A., and Walsh, K. A. (1996). Beta-methylthio-aspartic acid: identification of a novel posttranslational modification in ribosomal protein S12 from Escherichia coli. Protein Sci. 5, 1625–1632.
Lista, F., Reubsaet, F. A., De Santis, R., Parchen, R. R., De Jong, A. L., Kieboom, J., et al. (2011). Reliable identification at the species level of Brucella isolates with MALDI-TOF-MS. BMC Microbiol. 11:267. doi: 10.1186/1471-2180-11-267
Lopez-Goni, I., Garcia-Yoldi, D., Marin, C. M., De Miguel, M. J., Munoz, P. M., Blasco, J. M., et al. (2008). Evaluation of a multiplex PCR assay (Bruce-ladder) for molecular typing of all Brucella species, including the vaccine strains. J. Clin. Microbiol. 46, 3484–3487. doi: 10.1128/JCM.00837-08
Lucero, N. E., Ayala, S. M., Escobar, G. I., Grayon, M., and Jacques, I. (2006). A new variant of Brucella melitensis. Clin. Microbiol. Infect. 12, 593–596. doi: 10.1111/j.1469-0691.2006.01386.x
Meng, F., Pan, X., and Tong, W. (2018). Rifampicin versus streptomycin for brucellosis treatment in humans: a meta-analysis of randomized controlled trials. PLoS One 13:e0191993. doi: 10.1371/journal.pone.0191993
Mesureur, J., Arend, S., Celliere, B., Courault, P., Cotte-Pattat, P. J., Totty, H., et al. (2018). A MALDI-TOF MS database with broad genus coverage for species-level identification of Brucella. PLoS Negl. Trop. Dis. 12:e0006874. doi: 10.1371/journal.pntd.0006874
Minogue, T. D., Daligault, H. A., Davenport, K. W., Bishop-Lilly, K. A., Broomall, S. M., Bruce, D. C., et al. (2014). Whole-genome sequences of 24 Brucella strains. Genome Announc. 2:e00915-914. doi: 10.1128/genomeA.00915-14
Nishi, K., Muller, M., and Schnier, J. (1987). Spontaneous missense mutations in the rplX gene for ribosomal protein L24 from Escherichia coli. J. Bacteriol. 169, 4854–4856. doi: 10.1128/jb.169.10.4854-4856.1987
Ojima-Kato, T., Yamamoto, N., Takahashi, H., and Tamura, H. (2016). Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) can precisely discriminate the lineages of Listeria monocytogenes and species of Listeria. PLoS One 11:e0159730. doi: 10.1371/journal.pone.0159730
Olsen, S. C., and Stoffregen, W. S. (2005). Essential role of vaccines in brucellosis control and eradication programs for livestock. Expert Rev. Vaccines 4, 915–928.
Pappas, G., Papadimitriou, P., Akritidis, N., Christou, L., and Tsianos, E. V. (2006). The new global map of human brucellosis. Lancet Infect. Dis. 6, 91–99. doi: 10.1016/S1473-3099(06)70382-6
Poonawala, H., Marrs Conner, T., and Peaper, D. R. (2018). The brief case: misidentification of Brucella melitensis as Ochrobactrum anthropi by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). J. Clin. Microbiol. 56:e00914-17. doi: 10.1128/JCM.00918-17
Rossetti, C. A., Arenas-Gamboa, A. M., and Maurizio, E. (2017). Caprine brucellosis: a historically neglected disease with significant impact on public health. PLoS Negl. Trop. Dis. 11:e0005692. doi: 10.1371/journal.pntd.0005692
Salmon-Divon, M., Banai, M., Bardenstein, S., Blum, S. E., and Kornspan, D. (2018). Complete genome sequence of the live attenuated vaccine strain Brucella melitensis Rev.1. Genome Announc. 6:e00175-18. doi: 10.1128/genomeA.00175-18
Sandrin, T. R., Goldstein, J. E., and Schumaker, S. (2013). MALDI TOF MS profiling of bacteria at the strain level: a review. Mass Spectrom. Rev. 32, 188–217. doi: 10.1002/mas.21359
Seleem, M. N., Boyle, S. M., and Sriranganathan, N. (2010). Brucellosis: a re-emerging zoonosis. Vet. Microbiol. 140, 392–398. doi: 10.1016/j.vetmic.2009.06.021
Sharp, P. M., Nolan, N. C., Ni Cholmain, N., and Devine, K. M. (1992). DNA sequence variability at the rplX locus of Bacillus subtilis. J. Gen. Microbiol. 138, 39–45. doi: 10.1099/00221287-138-1-39
Skalsky, K., Yahav, D., Bishara, J., Pitlik, S., Leibovici, L., and Paul, M. (2008). Treatment of human brucellosis: systematic review and meta-analysis of randomised controlled trials. BMJ 336, 701–704. doi: 10.1136/bmj.39497.500903.25
Springer, B., Kidan, Y. G., Prammananan, T., Ellrott, K., Bottger, E. C., and Sander, P. (2001). Mechanisms of streptomycin resistance: selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob. Agents Chemother. 45, 2877–2884. doi: 10.1128/AAC.45.10.2877-2884.2001
Suarez, S., Ferroni, A., Lotz, A., Jolley, K. A., Guerin, P., Leto, J., et al. (2013). Ribosomal proteins as biomarkers for bacterial identification by mass spectrometry in the clinical microbiology laboratory. J. Microbiol. Methods 94, 390–396. doi: 10.1016/j.mimet.2013.07.021
Suh, M. J., Hamburg, D. M., Gregory, S. T., Dahlberg, A. E., and Limbach, P. A. (2005). Extending ribosomal protein identifications to unsequenced bacterial strains using matrix-assisted laser desorption/ionization mass spectrometry. Proteomics 5, 4818–4831. doi: 10.1002/pmic.200402111
Teramoto, K., Sato, H., Sun, L., Torimura, M., and Tao, H. (2007). A simple intact protein analysis by MALDI-MS for characterization of ribosomal proteins of two genome-sequenced lactic acid bacteria and verification of their amino acid sequences. J. Proteome Res. 6, 3899–3907. doi: 10.1021/pr070218l
Torii, N., Nozaki, T., Masutani, M., Nakagama, H., Sugiyama, T., Saito, D., et al. (2003). Spontaneous mutations in the Helicobacter pylori rpsL gene. Mutat. Res. 535, 141–145. doi: 10.1016/S1383-5718(02)00292-9
Tracz, D. M., Antonation, K. S., and Corbett, C. R. (2016). Verification of a matrix-assisted laser desorption ionization-time of flight mass spectrometry method for diagnostic identification of high-consequence bacterial pathogens. J. Clin. Microbiol. 54, 764–767. doi: 10.1128/JCM.02709-15
Tsai, Y. K., Liou, C. H., Lin, J. C., Ma, L., Fung, C. P., Chang, F. Y., et al. (2014). A suitable streptomycin-resistant mutant for constructing unmarked in-frame gene deletions using rpsL as a counter-selection marker. PLoS One 9:e109258. doi: 10.1371/journal.pone.0109258
Vergnaud, G., Hauck, Y., Christiany, D., Daoud, B., Pourcel, C., Jacques, I., et al. (2018). Genotypic expansion within the population structure of classical Brucella species revealed by MLVA16 typing of 1404 Brucella isolates from different animal and geographic origins, 1974-2006. Front. Microbiol. 9:1545. doi: 10.3389/fmicb.2018.01545
Whatmore, A. M., Perrett, L. L., and Macmillan, A. P. (2007). Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol. 7:34. doi: 10.1186/1471-2180-7-34
Wickham, H., Averick, M., Bryan, J., Chang, W., Mcgowan, L., François, R., et al. (2019). Welcome to the Tidyverse. J. Open Source Softw. 4:1686. doi: 10.21105/joss.01686
Wilcox, S. K., Cavey, G. S., and Pearson, J. D. (2001). Single ribosomal protein mutations in antibiotic-resistant bacteria analyzed by mass spectrometry. Antimicrob. Agents Chemother. 45, 3046–3055. doi: 10.1128/AAC.45.11.3046-3055.2001
Keywords: Brucella melitensis, MALDI-TOF MS, diagnostic, biomarker, vaccine, in silico proteomics
Citation: Kornspan D, Brendebach H, Hofreuter D, Mathur S, Blum SE, Fleker M, Bardenstein S and Al Dahouk S (2021) Protein Biomarker Identification for the Discrimination of Brucella melitensis Field Isolates From the Brucella melitensis Rev.1 Vaccine Strain by MALDI-TOF MS. Front. Microbiol. 12:712601. doi: 10.3389/fmicb.2021.712601
Received: 20 May 2021; Accepted: 27 September 2021;
Published: 22 October 2021.
Edited by:
George Tsiamis, University of Patras, GreeceReviewed by:
Victoria Girard, Biomerieux, FranceYaqoub Ashhab, Palestine Polytechnic University, Palestine
Ondrej Šedo, Masaryk University, Czechia
Copyright © 2021 Kornspan, Brendebach, Hofreuter, Mathur, Blum, Fleker, Bardenstein and Al Dahouk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Kornspan, ZGF2aWRrb0Btb2FnLmdvdi5pbA==; Dirk Hofreuter, ZGlyay5ob2ZyZXV0ZXJAYmZyLmJ1bmQuZGU=
†ORCID: David Kornspan, orcid.org/0000-0001-6848-2323; Holger Brendebach, orcid.org/0000-0002-7710-3011; Dirk Hofreuter, orcid.org/0000-0002-2589-9206; Shubham Mathur, orcid.org/0000-0002-5616-0877; Shlomo Eduardo Blum, orcid.org/0000-0002-5451-9997; Svetlana Bardenstein, orcid.org/0000-0002-6553-3671; Sascha Al Dahouk, orcid.org/0000-0003-3835-0818
‡These authors have contributed equally to this work and share first authorship
 David Kornspan
David Kornspan Holger Brendebach
Holger Brendebach Dirk Hofreuter
Dirk Hofreuter Shubham Mathur
Shubham Mathur Shlomo Eduardo Blum
Shlomo Eduardo Blum Marcelo Fleker1
Marcelo Fleker1 Svetlana Bardenstein
Svetlana Bardenstein Sascha Al Dahouk
Sascha Al Dahouk