
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 August 2021
Sec. Aquatic Microbiology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.712269
This article is part of the Research Topic Ciliates: Key Organisms in Aquatic Environments View all 26 articles
Ciliated protists (ciliates) are extremely diverse and play important ecological roles in almost all kinds of habitats. In this study, two new hypotrichs, Wilbertophrya sinica n. g. and n. sp. and Bakuella xianensis n. sp., from China are investigated. Wilbertophrya n. g. can be separated from related genera mainly by the combination of lacking a buccal cirrus, pretransverse cirri, and caudal cirri, while possessing frontoterminal cirri. Analyses based on morphological and molecular data confirm the validity of the species, W. sinica n. sp., which is characterized as follows: body 50–115 μm × 15–35 μm in vivo; midventral complex comprises four or five cirral pairs only and terminates above mid-body; three frontal, two frontoterminal cirri, and two to four transverse cirri; about 15 macronuclear nodules; colorless cortical granules sparsely distributed. Another new species, B. xianensis n. sp., was isolated from a freshwater wetland and is defined as follows: body 115–150 μm × 40–65 μm in vivo; about 70 macronuclear nodules; dark-brownish cortical granules in groups; midventral complex comprises 8–12 cirral pairs forming a row that terminates posteriorly in mid-body region and two or three short midventral rows that are continuous with the row of midventral pairs; three frontal, four to six frontoterminal, and three to five fine transverse cirri; three bipolar dorsal kineties. Phylogenetic analyses based on small subunit ribosomal DNA (SSU rDNA) sequence data suggest that the new genus Wilbertophrya n. g. belongs to an isolated clade, which might represent an undescribed taxon at the family level, whereas B. xianensis n. sp. groups with several congeners and members of other related genera are within the core urostylids.
Hypotrich ciliates are a large, ubiquitous group that play key roles in many ecosystem processes and as model organisms in a wide range of biological studies (Berger, 1999, 2011; Song and Shao, 2017; Chen et al., 2019, 2020; Kim and Min, 2019; Kim et al., 2019; Li et al., 2019; Wang et al., 2019; Gao et al., 2020; Sheng et al., 2020). New hypotrich taxa are continuously being discovered, supporting the notion posited by Foissner et al. (2008) that over 80% of ciliate diversity is still undescribed (Bharti et al., 2019; Hu et al., 2019; Kaur et al., 2019; Moon et al., 2019; Dong et al., 2020; Park et al., 2020; Xu et al., 2020).
Urostylids are one of the most speciose and best-known groups within the subclass Hypotrichia Stein, 1859. Nevertheless, their evolutionary relationships and genus- or family-level definitions remain problematic as recognized in several recent reports (Berger, 2006; Lu et al., 2018, 2020; Luo et al., 2018, 2019; Jung and Berger, 2019; Wang et al., 2021a; Zhang et al., 2020).
Here, we document two hypotrichs from inland China: a novel form that represents a new genus and a new species (Wilbertophrya sinica n. g., n. sp.) found in a puddle in a forest in Tibet and a freshwater species, Bakuella xianensis n. sp., isolated from a wetland in the Qinling Mountains area near Xi’an. Both species were characterized based on morphological observations of specimens in vivo and following silver staining. In addition, their molecular phylogeny was analyzed based on small subunit ribosomal DNA (SSU rDNA) sequence data.
Wilbertophrya sinica n. g., n. sp. was collected on April22, 2017, from a small puddle in a forest in Bomi County, Linzhi (29°39′N; 94°21′E), Tibet, where the altitude is about 3,000 m above sea level (Figures 1A,C).
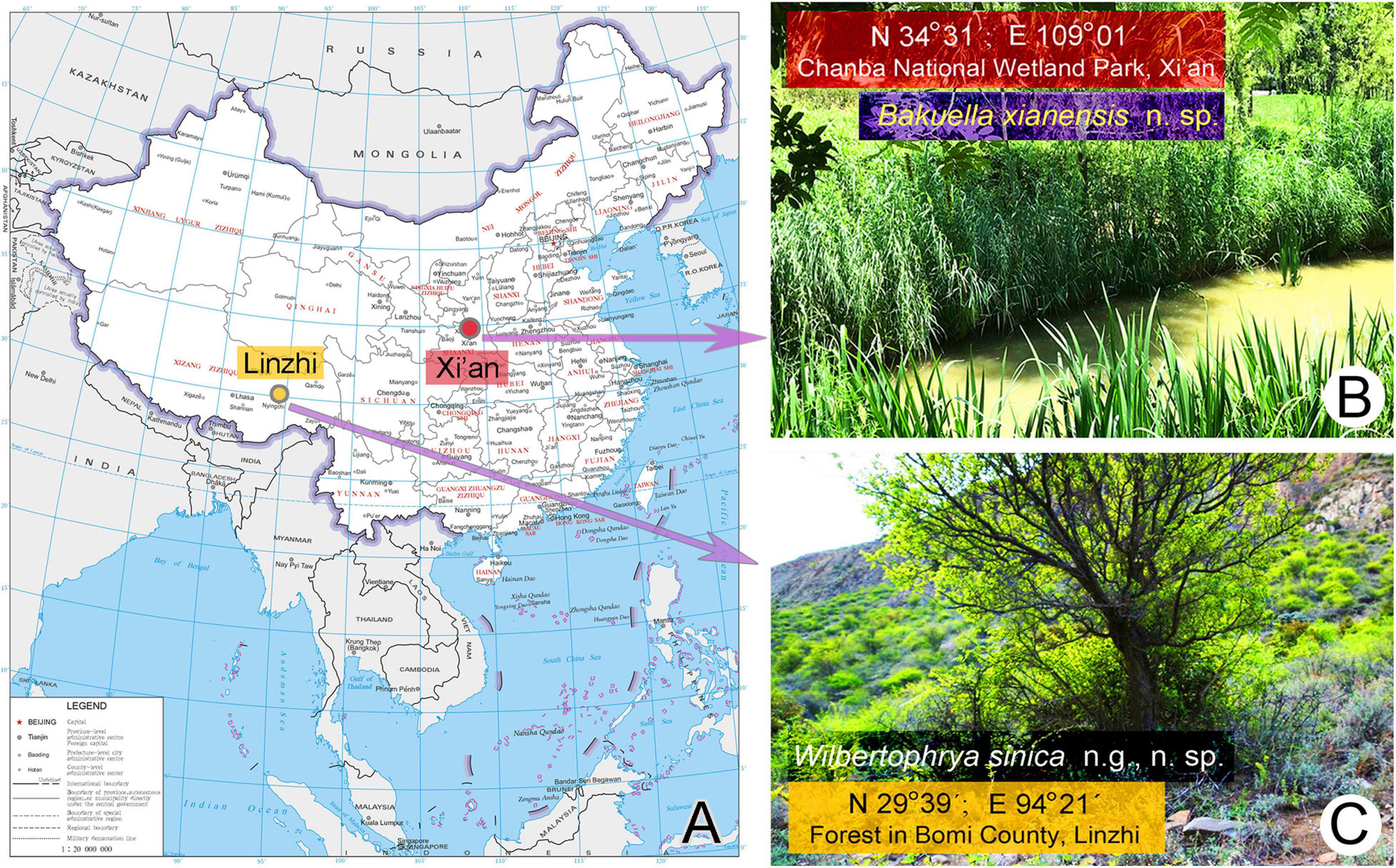
Figure 1. Sample sites. (A) The map of China from the MAP WORLD [www.tianditu.gov.cn, drawing review number: GS (2019) 1708] showing the location of the sampling sites. (B) Wetland at Chanba National Wetland Park, Xi’an, where Bakuella xianensis n. sp. was collected. (C) Forest in Bomi County, Linzhi, Tibet, where Wilbertophrya sinica n. sp. was collected.
Bakuella xianensis n. sp. was collected on May 25, 2019, from a well-vegetated freshwater pond located in Chanba National Wetland Park, Xi’an (34°31′N; 109°01′E), near the junction of the River Ba and River Wei (Figures 1A,B).
Uni-protistan cultures were established in Petri dishes containing rice grains to support the growth of bacteria, which served as a food source for the ciliates. The species was accurately identified based on its morphology in vivo. Moreover, no other urostylids morphotypes were present in the protargol preparation. We assure the accuracy of that identification for molecular studies, even though we were unable to establish clonal cultures.
Ciliate cells were observed in vivo using bright field and differential interference contrast microscopy. The protargol silver staining method according to Wilbert (1975) was used to reveal the infraciliature. Counts and measurements of stained specimens were performed at a magnification of ×1,000. Drawings of stained specimens were performed at ×1,250 magnification with the aid of a camera lucida. Terminology is mainly according to Berger (2006), and the systematic classification follows Lynn (2008).
Single cells of each species were isolated from cultures, washed several times with distilled water using a micropipette in order to remove potential contamination, and then transferred to 1.5-ml microfuge tubes with a minimum volume of water. Genomic DNA extraction was performed with the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the modified manufacturer’s protocol (1/4 of suggested volume for each solution) (Chen et al., 2019; Lu et al., 2020). PCR amplification of the SSU rDNA was performed using Q5 Hot Start high fidelity DNA polymerase (NEB, Ipswich, MA, United States) to minimize the possibility of amplification errors. One cycle of initial denaturation at 98°C for 30 s, followed by 18 cycles of amplification (98°C, 10 s; 69–51°C touchdown, 30 s; 72°C, 1 min), and another 18 cycles (98°C, 10 s; 51°C, 30 s; 72°C, 1 min), with a final extension of 72°C for 5 min. Sequencing of PCR products was performed bidirectionally on an ABI 3700 sequencer using two PCR primers 18S-F (5′-AAC CTG GTT GAT CCT GCC AGT-3′) and 18S-R (5′-TGA TCC TTC TGC AGG TTC ACC TAC-3′) (Medlin et al., 1988).
In order to perform phylogenetic analyses, the SSU rDNA sequences of W. sinica n. g., n. sp. and B. xianensis n. sp. were aligned with other related sequences downloaded from the National Center for Biotechnology Information (NCBI) Database (accession numbers shown in Figure 2) on the GUIDANCE2 server1 with default parameters (Sela et al., 2015). Representative species of the subclass Euplotia Jankowski, 1979, were selected as the outgroup. The aligned sequences were manually edited using the program BioEdit 7.2.5 (Hall, 1999), resulting in a final alignment with 1,772 sites. Both maximum likelihood (ML) and Bayesian inference (BI) analyses were performed on the final alignment. The ML analysis was performed with 1,000 rapid bootstrap replicates using RAxML-HPC2 on XSEDE v8.2.12 (Stamatakis, 2014) on CIPRES Science Gateway with the GTRGAMMA model (Miller et al., 2010). The BI analysis was computed by MrBayes on XSEDE 3.2.6 (Ronquist et al., 2012) on CIPRES Science Gateway with the GTR + I + G model selected by MrModeltest 2.2 (Nylander, 2004). Markov chain Monte Carlo (MCMC) simulations were run for 10,000,000 generations with sampling every 100 generations. The first 10,000 trees were discarded as burn-in. Seaview v.4.3.3 (Gouy et al., 2010) and MEGA v.6 (Tamura et al., 2011) were used to visualize the tree topologies.
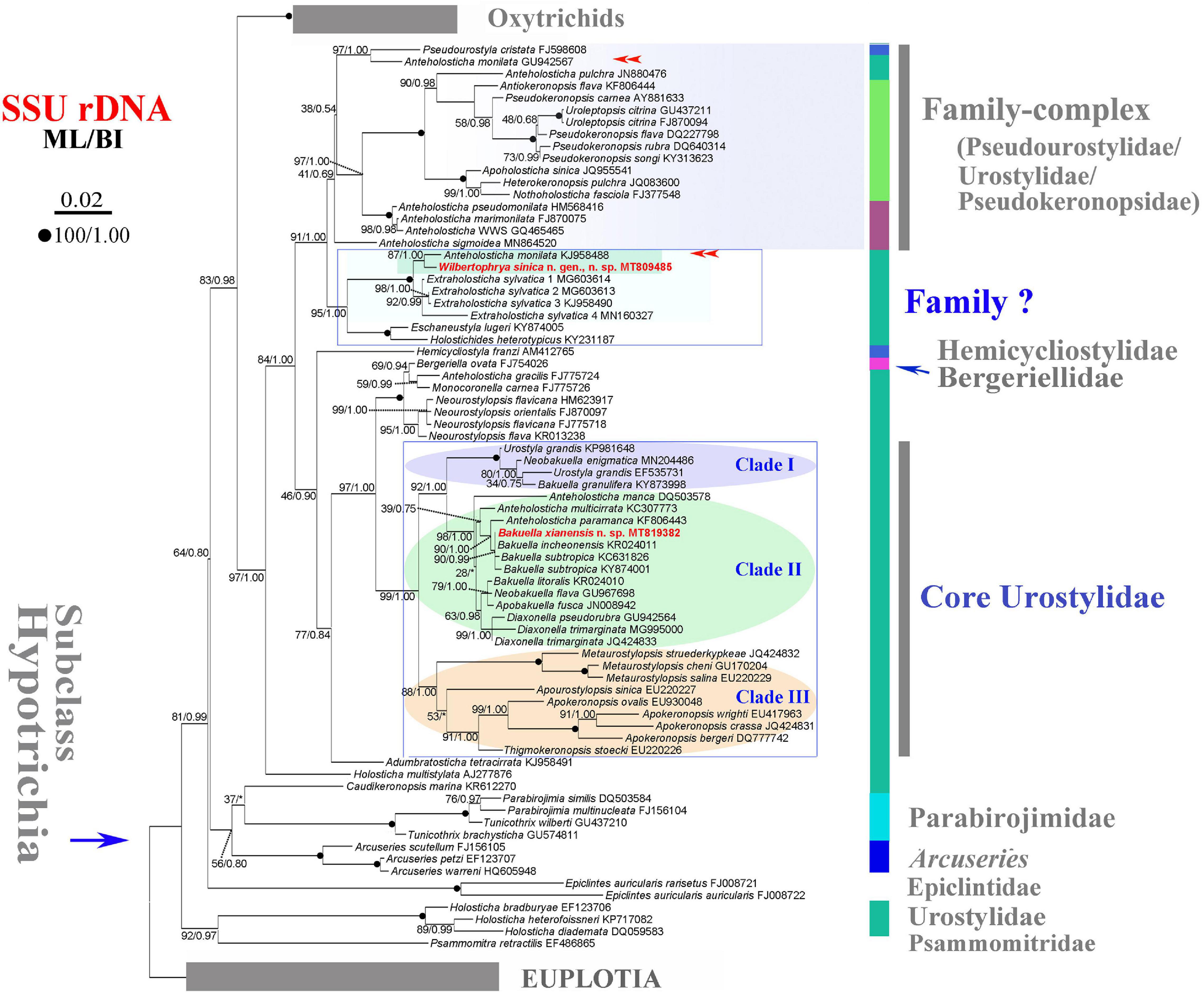
Figure 2. Maximum likelihood (ML) tree inferred from the small subunit ribosomal DNA (SSU rDNA) sequences showing the position of Wilbertophrya sinica n. sp. and Bakuella xianensis n. sp. (in red) and related species (in rectangular boxes). Numbers near nodes are bootstrap values for maximum likelihood and posterior probability values for Bayesian inference (BI). “*” at nodes indicates disagreement between the two methods. Fully supported (100/1.00) branches are marked with filled circles. Double arrowheads mark the sequences with the same species name, of which one could be misidentified. The scale bar corresponds to 0.02 expected substitutions per site.
The SSU rDNA sequences were aligned and manually modified (trimming ends and removing identical nucleotides) with BioEdit 7.0.5.2 (Hall, 1999).
ZooBank registration.
Present work: urn:lsid:zoobank.org:pub:C0BE1314-C4 93-430C-9903-8E1758A1F983
Subclass Hypotrichia Stein, 1859
Order Urostylida Jankowski, 1979
Genus Wilbertophrya n. g.
ZooBank registration.
Wilbertophrya n. g.: urn:lsid:zoobank.org:act:61BC94E 2-17BA-48D8-9219-921267C0919B
Diagnosis: Urostylid with a continuous adoral zone; three clearly differentiated frontal cirri; midventral complex composed only of cirral pairs arranged in a zigzag pattern; frontoterminal and transverse cirri present; buccal, pretransverse, and caudal cirri absent; one marginal row on each side, the anterior end of the left marginal row not curved rightward; three dorsal kineties.
Type species: W. sinica n. sp.
Dedication: This new genus is named after the eminent ciliatologist Prof. Norbert Wilbert, University of Bonn, Germany, in recognition of his life-long contributions to the taxonomy of ciliates. Feminine gender.
ZooBank registration.
Wilbertophrya sinica n. sp.: urn:lsid:zoobank.org:act:F2D0E277-FB7A-493F-90AC-8D9875AC347F
Diagnosis: Body 50–115 μm × 15–35 μm in vivo, elliptical in outline; 13–19 macronuclear nodules; contractile vacuole located ahead of mid-body; cortical granules colorless, about 1 μm across, irregularly distributed; about 15–18 adoral membranelles; four or five midventral pairs in anterior half of cell; three frontal, two frontoterminal, and two to four transverse cirri; the left marginal row is composed of 13–21 cirri, and right marginal row is composed of 14–25 cirri; three bipolar dorsal kineties; freshwater habitat.
Type material: One protargol-stained slide (No. MJY2017042201-3) with the holotype specimen (Figure 3I) circled in ink and two paratype slides (No. MJY2017042201-1, 2) were deposited in the Laboratory of Protozoological Biodiversity and Evolution in Wetland, Shaanxi Normal University, China.
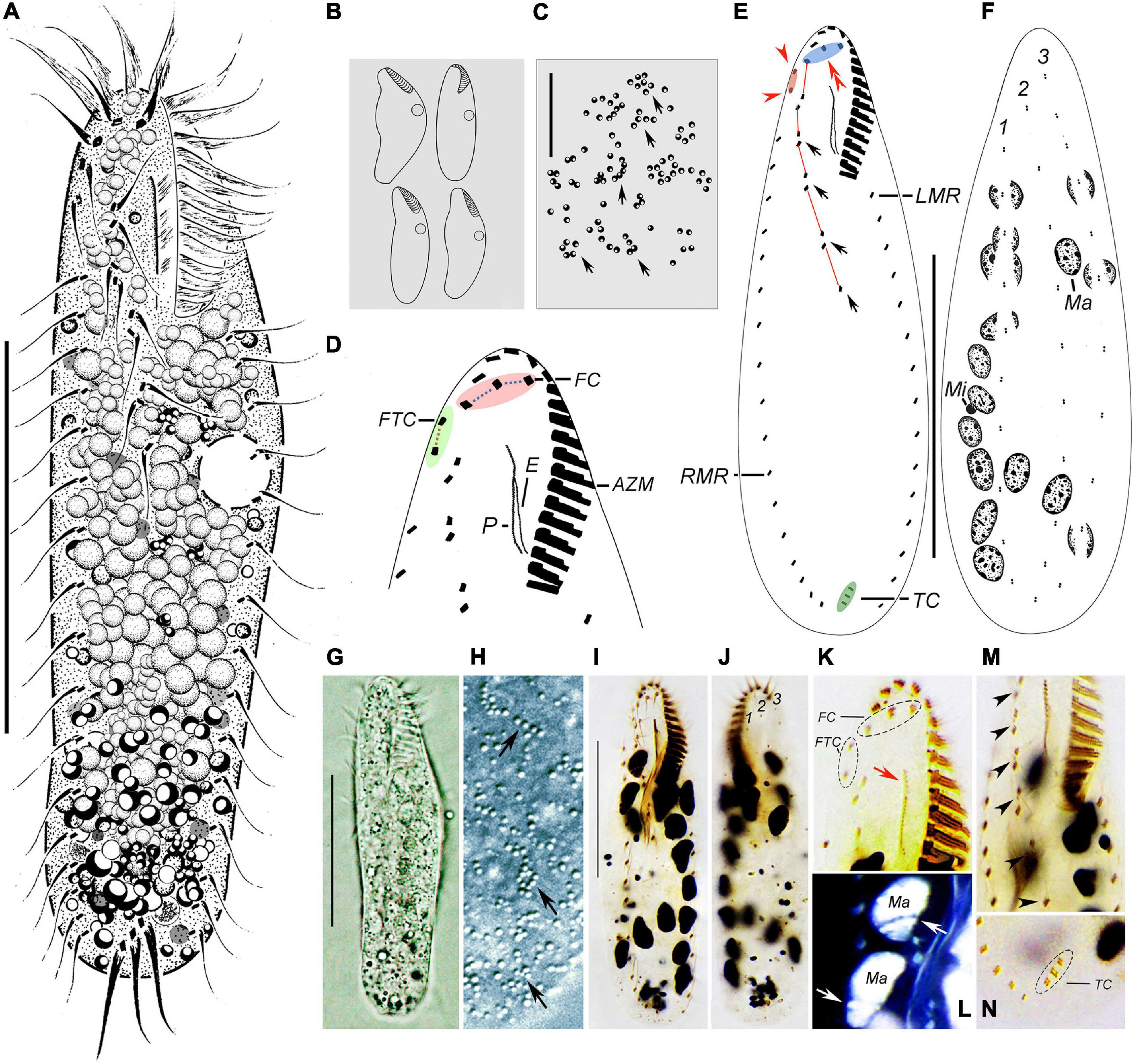
Figure 3. Morphology of Wilbertophrya sinica n. sp. from life (A–C,G,H) and after staining with protargol (D–F,I–N). (A) Ventral view of a representative individual. (B) Ventral views to show different body shapes. (C) Arrangement of cortical granules on dorsal side (arrows). (D) Ventral view to demonstrate ciliature in frontoventral area. (E,F) Ventral (E) and dorsal (F) views showing the infraciliature. Arrows mark midventral pairs, arrowheads point to frontoterminal cirri, and double arrowheads indicate frontal cirri. (G) Ventral view. (H) Arrangement of cortical granules (arrows) on dorsal side. (I) Ventral view of the holotype specimen to show ventral ciliature. (J) Dorsal view to show dorsal kineties and macronuclear nodules. (K) Ventral view of anterior portion to show the frontal and frontoterminal cirri, endoral, and paroral (arrow). (L) Dorsal view to show macronuclear nodules and replication bands (arrows) (the color of image was inverted using Photoshop). (M) Ventral view to show the midventral pairs (arrowheads). (N) Ventral view of posterior end showing transverse cirri. AZM, adoral zone of membranelles; CV, contractile vacuoles; E, endoral; FC, frontal cirri; FTC, frontoterminal cirri; LMR, left marginal row; Ma, macronucleus nodules; Mi, micronuclei; P, paroral; RMR, right marginal row; TC, transverse cirri; 1–3, dorsal kineties 1–3. Scale bars: (A,E,F,G,I) = 40 μm, (C) = 15 μm.
Type locality: Puddle in a forest in Bomi County, Linzhi (29°39′N; 94°21′E), Tibet, China.
Etymology: The species-group name sinica recalls that this species was first discovered in China.
Description (Figures 3A–N and Table 1).
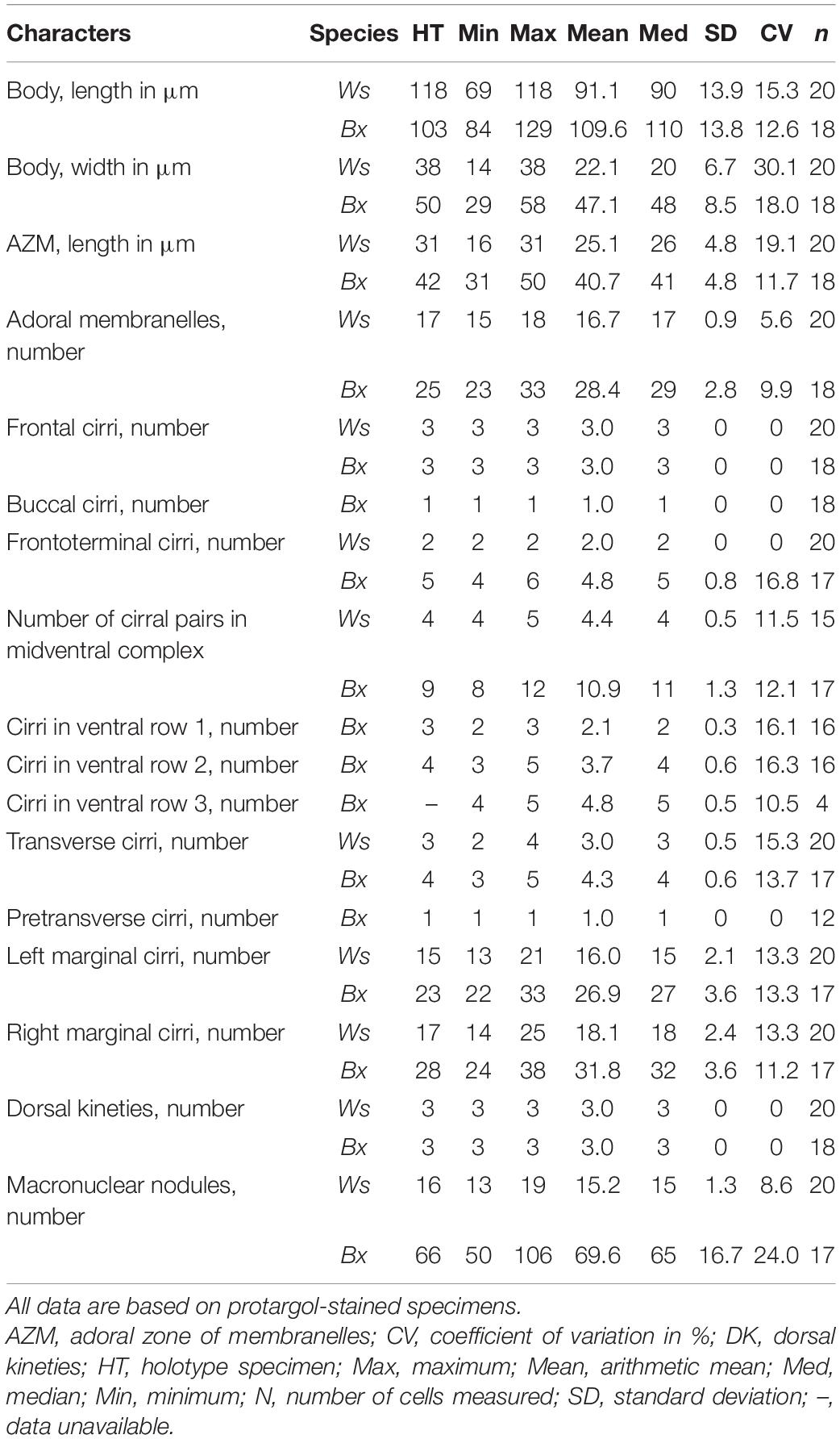
Table 1. Morphometric data of Wilbertophrya sinica n. g., n. sp. (Ws, upper line) and Bakuella xianensis n. sp. (Bx, lower line).
Body size 50–115 μm × 15–35 μm in vivo. Cell outline variable, generally elliptical to elongate-elliptical, right cell margin slightly concave, left margin slightly convex; usually widest in front of mid-body; dorsoventrally flattened; slightly flexible but non-contractile (Figures 3A, B, G). Nuclear apparatus composed of about 15 ellipsoidal nodules and one to four, on average two, micronuclei up to 2 μm in diameter (Figures 3F, L). Contractile vacuole about 7 μm in diameter, located slightly ahead of mid-body near left body margin, contracting at intervals of about 7 s (Figures 3A, B). Cortical granules colorless, globular, about 1 μm across, irregularly distributed (Figures 3C, H, arrows). Cytoplasm colorless or grayish, often containing numerous densely packed lipid droplets (Figures 3A, G). Locomotion by moderately fast crawling on the bottom of Petri dish, occasionally jerking back and forth; when suspended in water, cells often swim continuously in circles.
Infraciliature as shown in Figures 3D–F,I–K,M,N. The adoral zone is composed of 15–18 membranelles, occupying about 1/4–1/3 of body length (Table 1). In living cells, cilia of distal membranelles about 12 μm in length, and those of proximal membranelles are about 6 μm long. The distal end of the adoral zone extends only slightly onto right body margin forming a question mark shape as in other urostylids. Paroral longer than endoral, both generally straight and optically intersect at their upper region (Figures 3D, E). Three slightly enlarged frontal cirri about 8 μm in length, rightmost one located behind the distal end of the adoral zone (Figure 3D, pink area, Figure 3E, double arrowheads). Two frontoterminal cirri behind the distal end of the adoral zone (Figure 3E, arrowheads); buccal cirrus lacking (Figures 3D, E); two to four slightly enlarged transverse cirri, up to 12 μm long in vivo. Pretransverse cirri absent. Midventral complex composed of four or five pairs of cirri, terminating ahead of mid-body (Figure 3E, arrows). Left and right marginal rows composed of 13–21 and 14–25 cirri, respectively, with the anterior end of the left marginal rows not curved rightward (Figure 3E).
Three dorsal kineties arranged in typical Gonostomum pattern, with bristles about 3 μm in length (Figures 3F,J).
ZooBank registration.
Bakuella xianensis n. sp.: urn:lsid:zoobank.org:act:7C3FA85 3-C973-402B-824D-21F5337CB63E
Diagnosis: Body 115–150 μm × 40–65 μm in vivo, elliptical in outline; about 70 macronuclear nodules; contractile vacuole located ahead of mid-body; dark-brownish cortical granules arranged in groups, about 0.7 μm across; about 23–33 adoral membranelles; midventral complex composed of 8–12 cirral pairs, terminating at about mid-body; two or three short ventral rows continuous with midventral pairs; three frontal, four to six frontoterminal, and three to five fine transverse cirri; one right and one left marginal rows with 24–38 and 22–33 cirri, respectively; three bipolar dorsal kineties; freshwater habitat.
Type material: One protargol-stained slide (No. MJY2019052501-1) with the holotype specimen (Figure 4B) circled in ink and two paratype slides (No. MJY2019052501-2, 3, 4) were deposited in the Laboratory of Protozoological Biodiversity and Evolution in Wetland, Shaanxi Normal University, China.
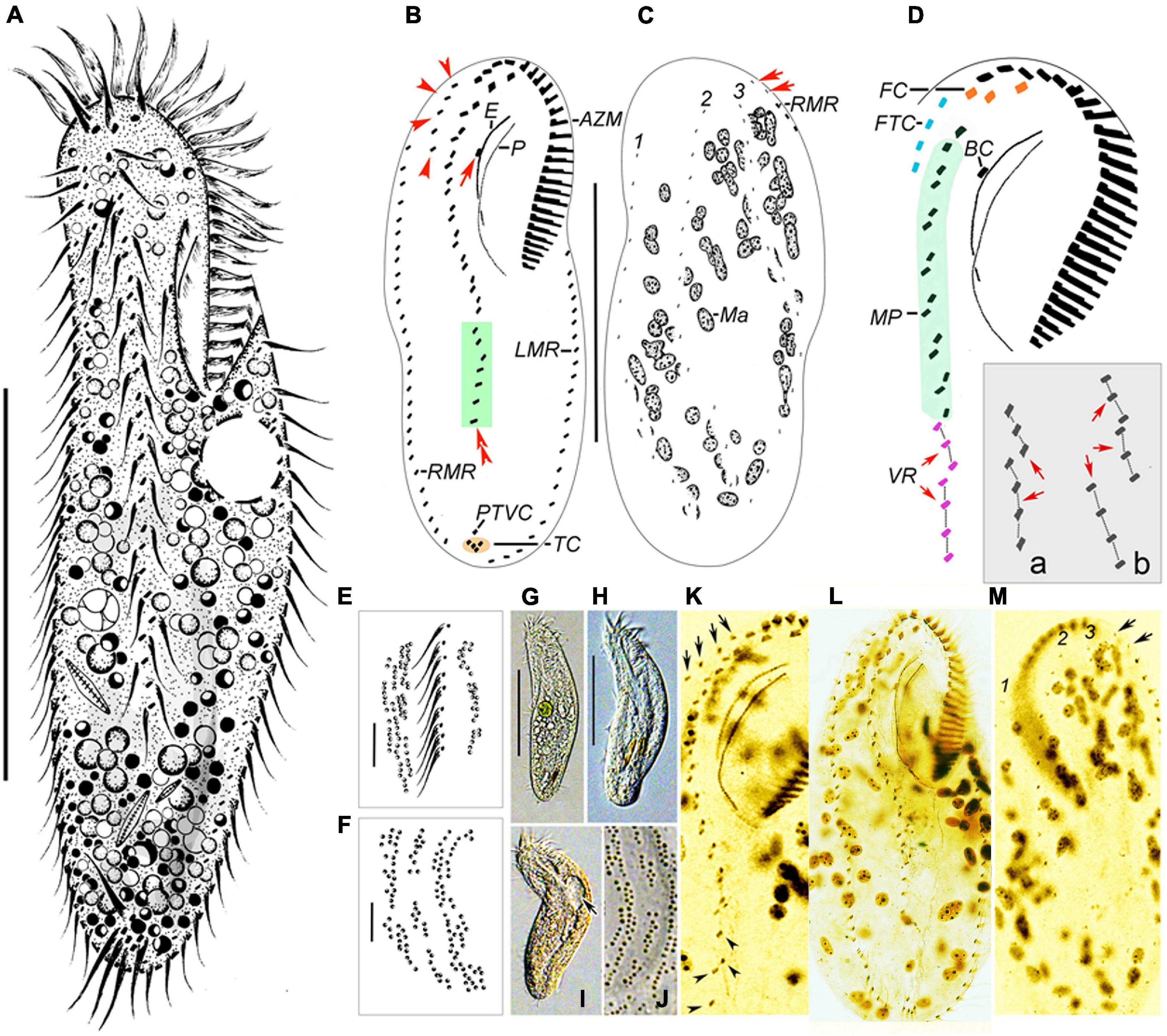
Figure 4. Morphology of Bakuella xianensis n. sp. from life (A, E–J) and after protargol staining (B–D,K–M). (A) Ventral view of a representative specimen. (B,C) Ventral (B) and dorsal (C) views of the holotype specimen to show the infraciliature and nuclear apparatus. In (B), arrow marks the buccal cirrus, arrowheads show the frontoterminal cirri, and double arrowheads mark a short ventral row (in green box). In (C), arrows indicate the basal body pairs ahead of the right marginal row. (D) Ventral view to show details of buccal field and midventral complex; inset (a,b) shows two different arrangements of ventral rows (VR, arrows). (E,F,J) Detail of distribution of cortical granules in irregular longitudinal rows on ventral (E) and dorsal (F,J) sides. (G–I) Ventral views to show different body shapes due to flexibility of cells. Arrow indicates the contractile vacuole. (K) Ventral view to show details of buccal field and midventral complex, arrows show the frontoterminal cirri, and arrowheads indicate the midventral rows. (L,M) Ventral (L) and dorsal (M) views of the same specimen showing infraciliature and nuclear apparatus. Arrows indicate the basal body pairs ahead of right marginal row. AZM, adoral zone of membranelles; BC, buccal cirrus; CV, contractile vacuoles; E, endoral; FC, frontal cirri; FTC, frontoterminal cirri; LMR, left marginal row; Ma, macronuclear nodules; MVP, midventral pairs; P, paroral; PTVC, pretransverse ventral cirri; RMR, right marginal row; TC, transverse cirri; VR, ventral row; 1–3, dorsal kineties. Scale bars: (A–C,G,H) = 60 μm, (E,F) = 7 μm.
Type locality: A freshwater pond in Chanba National Wetland Park, Xi’an (34°31′N; 109°01′E), China.
Etymology: The species-group name xianensis recalls that this species was first discovered in Xi’an, China.
Description (Figures 4A–M and Table 1).
Body about 115–150 μm × 40–65 μm in vivo, outline elongate-elliptical with both ends slightly narrow, length-to-width ratio approximately 3:1, flexible and dorsoventrally flattened (Figures 4A,G–I and Table 1). One contractile vacuole, about 15 μm across, positioned slightly in front of mid-body near left margin (Figures 4A,I, arrow). Pellicle thin and soft, with globular, dark-brownish cortical granules (about 0.7 μm across) in densely arranged groups on both ventral and dorsal sides, rendering cells slightly brownish at lower magnifications (Figures 4E, F, J). Cytoplasm colorless to grayish, usually packed with numerous small lipid droplets (about 3 μm across) and several large food vacuoles (about 8 μm across) containing ingested diatoms, bacteria, and/or ciliates. About 70 ellipsoidal macronuclear nodules scattered in cytoplasm (Figures 4C,M and Table 1). Locomotion by continuous, slow, to moderately fast, crawling on substrate.
Infraciliature as shown in Figures 4B–D,K–M. The adoral zone is about 40 μm long, occupying about 1/3 of body length, and composed of about 28 membranelles on average (Table 1). Endoral and paroral prominent in vivo, that is, long and curved, optically intersecting at the paroral’s mid-region. Most somatic cirri are relatively fine with cilia about 10–15 μm long. Constantly three relatively stout frontal cirri (FC, Figure 4D), apically positioned. Invariably one buccal cirrus, about level of endoral’s mid-region (Figure 4B, arrow). Four to six fine frontoterminal cirri (Figure 4B, arrowheads). Three to five transverse cirri form roughly V-shaped (Figures 4B,L), protruding beyond rear end of body. Cilia of which is 13–15 μm long. Of the 18 examined specimens, 12 had one pretransverse cirrus, while the remainder lacked the cirrus. Midventral complex terminates at about posterior 1/4 of body length, composed of about 8–12 cirral pairs forming a zigzag row that extends to about mid-region of the cell, and two, sometimes three, short ventral rows that are continuous with a row of midventral pairs (Figures 4B, D, VR and arrows, inset). One right and one left marginal rows composed of 24–38 and 22–33 cirri, respectively (Table 1); the right marginal row commences slightly ahead of level of buccal cirrus, always with two pairs of basal bodies ahead of its anterior end (Figures 4C,M, arrows). Three bipolar dorsal kineties arranged in typical Gonostomum pattern (Figures 4C,M).
The SSU rDNA sequences of Wilbertophrya sinica n. sp. and Bakuella xianensis n. sp. were deposited to GenBank with accession numbers, length, and guanine–cytosine (GC) content as follows: MT809485, 1,762 bp, 45.40% and MT819382, 1,770 bp, 44.18%, respectively. W. sinica and nine morphologically related isolates generated a nucleotide matrix with 204 unmatched sites (Figure 5A), while B. xianensis and five related taxa formed the table with 57 unmatched sites (Figure 5B).
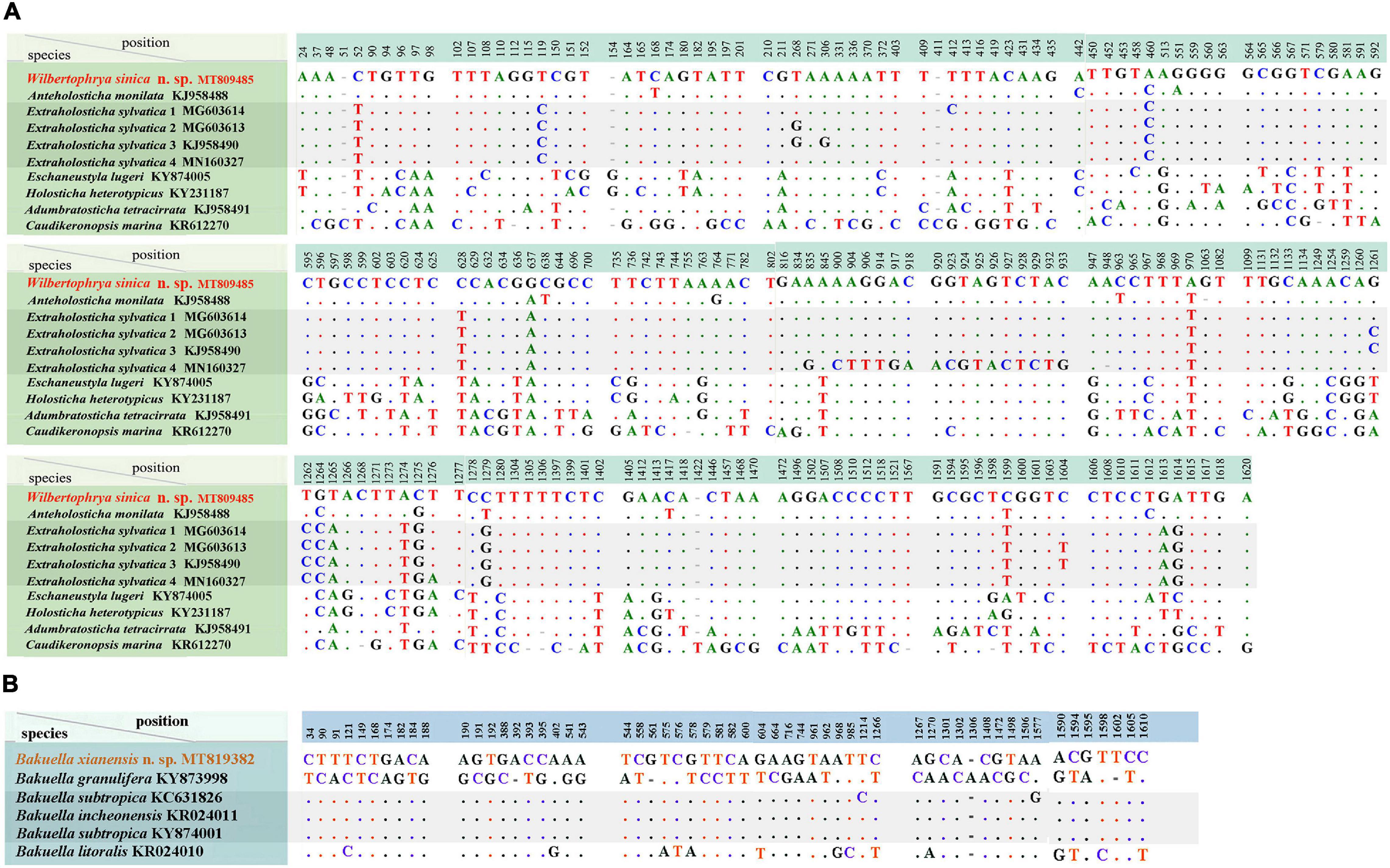
Figure 5. (A) Comparison of small subunit ribosomal DNA (SSU rDNA) sequences showing the unmatched nucleotides between Wilbertophrya sinica n. sp. and related species (for GenBank accession numbers, see Figure 2). (B) Comparison of SSU rDNA sequences showing the unmatched nucleotides between Bakuella xianensis n. sp. and related species (for GenBank accession numbers, see Figure 2). Nucleotide positions are given at the top of each column. Insertions and deletions are compensated by introducing alignment gaps (-). Matched sites are represented by dots (.).
Phylogenetic trees inferred from the SSU rDNA sequences using ML and BI had similar topologies; therefore, only the ML tree is shown with nodal support from both methods (Figure 2). W. sinica n. sp. clustered with Anteholosticha monilata (KJ958488) with high support (87% ML, 1.00 BI), which together grouped with four sequences of Extraholosticha sylvatica (MG603613, MG603614, KJ958490, and MN160327) with full support (Figure 2). This group was sister to the clade composed of Eschaneustyla lugeri (KY874005) and Holostichides heterotypicus (KY231187). Consequently, a comparison was made among these seven and two other morphologically similar species of W. sinica n. sp., namely, Adumbratosticha tetracirrata (KJ958491) and Caudikeronopsis marina (KR612270) (Figure 5A). The sequence differences between W. sinica n. sp. and these nine “related” taxa are as follows: 15 bp for A. monilata, 16–33 bp for the four sequences of E. sylvatica, 63 bp for E. lugeri, 67 bp for H. heterotypicus, 84 bp for A. tetracirrata, and 126 bp for C. marina.
Bakuella xianensis n. sp. clustered with Bakuella incheonensis (KR024011), and two sequences of Bakuella subtropica (KC631826 and KY874001) and Anteholosticha paramanca (KF806443) with moderate-to-strong support (90% ML, 1.00 BI), which together formed a clade with two species of Anteholosticha. Two other Bakuella species (Bakuella litoralis and Bakuella granulifera) belong to other groups, i.e., B. litoralis KR024010 + the Neobakuella–Apobakuella–Diaxonella complex and B. granulifera + Urostyla + Neobakuella. Hence, all known congeners in Bakuella belong to three separate (but neighboring) branches within the core-urostylid lineage (Figure 2).
The possession of three clearly differentiated frontal cirri and a midventral complex composed of midventral pairs only places Wilbertophrya n. g. unequivocally in the family Holostichidae (sensu Berger, 2006). Based on the following combination of features, that is, single left and right marginal rows, a continuous adoral zone, presence of frontoterminal cirri, clearly differentiated frontal cirri, and midventral cirral pairs arranged in a zigzag pattern, five genera in the family Holostichidae should be compared with Wilbertophrya n. g., namely, Anteholosticha Berger, 2003; Arcuseries Huang et al., 2014; Afrothrix Foissner, 1999; Acuholosticha Li et al., 2017; and Holosticha Wrzesniowski, 1877 (Berger, 2003; Huang et al., 2014; Li et al., 2017). All five of these genera differ from Wilbertophrya n. g. in having a buccal cirrus and pretransverse cirri, whereas both these structures are lacking in the new genus (Berger, 2006; Huang et al., 2014; Li et al., 2017). Furthermore, Acuholosticha possesses caudal cirri, whereas these are absent in Wilbertophrya n. g. (Berger, 2006; Figure 6; Table 2). The validity of the new genus is also supported by the molecular data (Figure 5A).
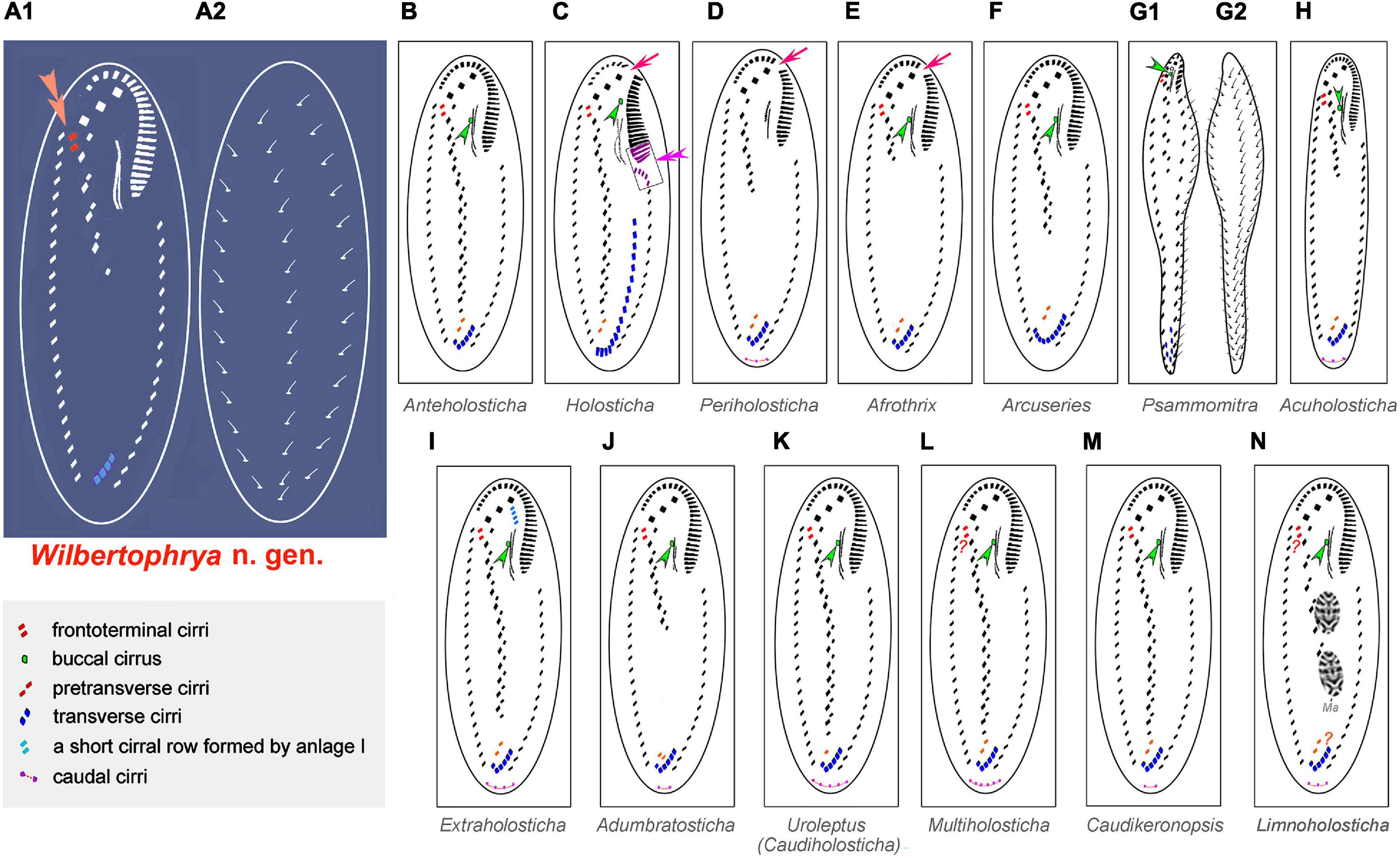
Figure 6. Comparison of infraciliature patterns of genera related to Wilbertophrya n. g. Arrows indicate the gap in the adoral zone of membranelles; arrowheads show the presence of a buccal cirrus. (A1,A2) Ventral (A1) and dorsal (A2) views of Wilbertophrya n. g.; double arrowheads in (A) indicate the frontoterminal cirri, while in (C), they mark the curved anterior end of left marginal row. (B) Anteholosticha. (C) Holosticha. (D) Periholosticha. (E) Afrothrix. (F) Arcuseries. (G1,G2) ventral (G1) and dorsal (G2) views of Psammomitra. (H) Acuholosticha. (I) Extraholosticha. (J) Adumbratosticha. (K) Caudiholosticha. (L) Multiholosticha. (M) Caudikeronopsis. (N) Limnoholosticha. Ma, macronuclear nodules. Question mark in (L) indicates presence/absence of frontoterminal questionable. Question mark in (N) indicates presence/absence of frontoterminal and pretransverse ventral cirri not known.
Eight other morphologically similar genera are compared with Wilbertophrya n. g., namely, Adumbratosticha Li et al., 2017; Caudikeronopsis Li et al., 2017; Extraholosticha Li et al., 2017; Limnoholosticha Li et al., 2017; Multiholosticha Li et al., 2017; Periholosticha Hemberger, 1985; Psammomitra Borror, 1972; and Uroleptus Ehrenberg, 1831 (Figure 6; Table 2). Wilbertophrya n. g. can be separated from each of these by the following combination of characters: (1) presence of frontoterminal cirri; (2) absence of pretransverse, caudal, and buccal cirri; (3) adoral zone of membranelles continuous; and (4) three complete dorsal kineties (Figure 6; Table 2).
The systematic position of Wilbertophrya n. g. remains unclear considering that the family assignment though morphological information supports its possible assignment to the family Holostichidae. The SSU rDNA tree reveals that Wilbertophrya groups with seven representatives of four genera: Anteholosticha, Extraholosticha, Eschaneustyla, and Holostichides (Figure 2). It is most closely related to A. monilata (KJ958488), the identification of which needs to be verified since there is another sequence with the same name but which occupies a different position in the tree (Figure 2, double arrowheads). Previous molecular phylogenetic analyses suggest that the genus Anteholosticha is not monophyletic and that most of its nominal species belong to the Urostylidae + Pseudokeronopsidae + Pseudourostylidae complex, which is the sister group of the clade that includes Wilbertophrya (Figure 2; Shao et al., 2011; Li et al., 2017; Luo et al., 2018; Xu et al., 2020).
In the SSU rDNA tree (Figure 2), the new genus clusters with several representatives of the genus Extraholosticha followed by E. lugeri + H. heterotypicus. This large clade is separated from other sequences at a deep level, suggesting that it might represent an undefined group at about family level. In terms of its morphology, however, the genus Wilbertophrya should be assigned to the family Holostichidae (see above). But as revealed in previous molecular studies, the Holostichidae complex (e.g., Holostichidae + Urostylidae + Pseudokeronopsidae) is a so-called melting pot of taxa, the evolutionary relationships of which cannot be resolved using phylogenetic analyses of single-gene sequence data (Bernhard et al., 2001; Yi and Song, 2011; Zhao et al., 2015; Luo et al., 2018; Jung and Berger, 2019; Paiva, 2020; Wang et al., 2021b; Xu et al., 2020). Similarly, it is difficult to place Wilbertophrya n. g. into any known family based on SSU rDNA sequence data. Since ontogenetic information is unavailable for the new genus, we suggest that Wilbertophrya n. g. should be regarded as incertae sedis within the order Urostylida pending the availability of further data.
Wilbertophrya n. g. is a monotypic genus, and thus, the type species, W. sinica n. sp., can be separated from its most “similar” morphospecies, that is, members of the genera Adumbratosticha, Periholosticha, Afrothrix, and Acuholosticha (see Figure 6) by the same combination of features that define the genus, i.e., the absence of the buccal cirrus and pretransverse cirri, and the presence of sparsely distributed cortical granules and the conspicuously short midventral rows (Berger, 2003; Li et al., 2017; Shao et al., 2020). The validity of W. sinica n. sp. as a distinct species is also firmly supported by the molecular data, which demonstrates its considerable difference from other taxa (Figure 5A).
The well-defined genus Bakuella can be recognized by its midventral complex comprising pairs of cirri arranged in a zigzag pattern and several obliquely oriented fragment-like ventral rows (Borror and Wicklow, 1983; Berger, 2006; Song and Shao, 2017; Shao et al., 2020). To date, 12 nominal species have been reported (Table 3), three of which have a single buccal cirrus and more than two frontoterminal cirri, and so should be compared with B. xianensis n. sp., namely, Bakuella agamalievi Borror and Wicklow, 1983; B. subtropica Chen et al., 2013; and B. incheonensis Jo et al., 2015.
Bakuella xianensis n. sp. can be distinguished from B. subtropica, a brackish water form originally found in a subtropical area in China (estuary of the Pearl River, Chen et al., 2013; Figures 7A, B) by (1) the size of the cortical granules (about 0.7 μm in diameter vs. 1–2 μm in diameter); (2) its habitat (freshwater vs. brackish water in mangrove wetlands); (3) having fewer cirral pairs in the midventral complex (8–12 vs. 9–23); and (4) having fewer left (22–33 vs. 30–54) and right (24–38 vs. 28–64) marginal cirri (Chen et al., 2013).
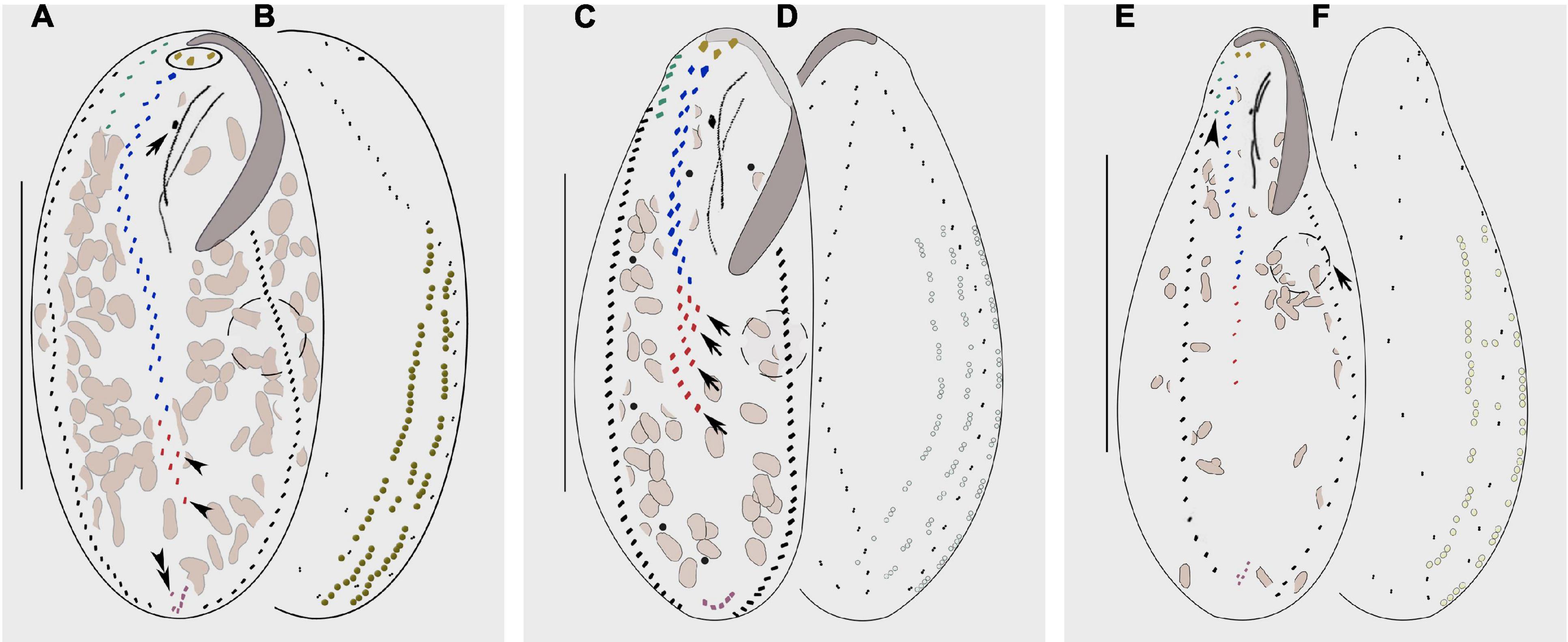
Figure 7. Morphology of Bakuella subtropica (A,B), Bakuella agamalievi (C,D), and Bakuella incheonensis (E,F). (A,C,E) Ventral view of three individuals. (B,D,F) Dorsal views of three individuals and cortical granules on dorsal side. (A) Ventral view to show the infraciliature; circle marks the frontal cirri, arrow shows the buccal cirrus, arrowheads indicate the ventral rows, and double arrowheads show the pretransverse cirrus. (B) Distribution of cortical granules, which form longitudinal rows. (C) Ventral view of infraciliature; arrows mark the short ventral rows. (D) Distribution of cortical granules, typically grouped together and sparsely arranged in short longitudinal rows. (E) Ventral view of infraciliature, arrow marks the contractile vacuole, and arrowhead indicates the frontoterminal cirri. (F) Distribution of cortical granules, mainly distributed along cirral rows and dorsal kineties. Scale bars: (A–D) = 60 μm, (E,F) = 40 μm.
Bakuella xianensis n. sp. differs from B. agamalievi, a marine form originally found in the Caspian Sea (Agamaliev, 1972; Figures 7C, D) and redescribed by Song et al. (2002) as having dark-brownish (vs. colorless or slightly greenish) cortical granules, fewer ventral rows in the midventral complex (two or three vs. three to six), fewer cirri in both right (24–38 vs. 40–47) marginal rows, and in its freshwater (vs. marine) habitat (Agamaliev, 1972; Song et al., 2002; Berger, 2006).
Bakuella xianensis n. sp. differs from B. incheonensis (Figures 7E, F) by its larger body size (115–150 μm × 40–65 μm vs. 70–105 μm × 20–40 μm in vivo), the color and arrangement of cortical granules (dark-brownish vs. yellowish), and its freshwater (vs. marine) habitat (Figures 4E, F, J, 7E, F). In addition, the cortical granules in B. incheonensis appear to be large, conspicuous, and possibly ellipsoid in shape (Figure 7F, arrows, not mentioned in original report), whereas those in B. xianensis n. sp. are small, globular, and inconspicuous (Figures 4E, F, J; Jo et al., 2015).
Including B. xianensis n. sp., SSU rDNA sequence data are available for only six species of Bakuella. These are grouped into three neighboring clades in the SSU rDNA tree (Figure 2). B. xianensis n. sp. is placed within the core clade of the genus. It is noteworthy, however, that some Bakuella sequences group with non-bakuellid genera, e.g., nominal isolates of genera such as Urostyla, Diaxonella, and Anteholosticha, with high support. This is consistent with previous reports (Chen et al., 2010, 2013) and indicates that none of the families Bakuellidae, Urostylidae, and Holostichidae is monophyletic; and the systematics of each requires further investigation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
JM carried out the live observation and protargol impregnation. TZ was responsible for DNA amplification and sequencing, and the molecular phylogenetic analyses. JM, TZ, and CS performed the manuscript draft. CS and WS performed the manuscript review and editing. All authors helped to revise the manuscript, and read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (project numbers 32070428 and 32030015) and the China Postdoctoral Science Foundation (No. 2021M692010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to express their sincere thanks to Alan Warren (NHM) for his helpful critiques in enhancing the language of this manuscript. Many thanks are given to Yurui Wang (OUC) for her help with phylogenetic analyses.
Agamaliev, F. G. (1972). Ciliates from microbenthos of the islands of Apseronekij and Bakinskij archipelagos of the Caspian Sea. Acta Protozool. 10, 1–27.
Berger, H. (1999). Monograph of the Oxytrichidae (Ciliophora, Hypotrichia). Monogr. Biol. 78, 1–1080. doi: 10.1007/978-94-011-4637-1
Berger, H. (2003). Redefinition of Holosticha Wrzesniowski, 1877 (Ciliophora, Hypotricha). Eur. J. Protistol. 39, 373–379. doi: 10.1078/0932-4739-00006
Berger, H. (2006). Monograph of the Urostyloidea (Ciliophora, Hypotricha). Monogr. Biol. 85, 1–1304. doi: 10.1007/1-4020-5273-1_1
Berger, H. (2011). Monograph of the Gonostomatidae and Kahliellidae (Ciliophora, Hypotricha). Monogr. Biol. 90, 1–740. doi: 10.1007/978-94-007-0455-8
Bernhard, D., Stechmann, A., Foissner, W., Ammermann, D., Hehn, M., and Schlegel, M. (2001). Phylogenetic relationships within the class Spirotrichea (Ciliophora) inferred from small subunit rRNA gene sequences. Mol. Phylogenet. Evol. 21, 86–92. doi: 10.1006/mpev.2001.0997
Bharti, D., Kumar, S., La Terza, A., and Chandra, K. (2019). Morphology and ontogeny of Tetmemena pustulata indica nov. subspec. (Ciliophora, Hypotricha), from the Thane Creek, Mumbai, India. Eur. J. Protistol. 71:125629. doi: 10.1016/j.ejop.2019.125629
Borror, A. C., and Wicklow, B. J. (1983). The suborder Urostylina Jankowski (Ciliophora, Hypotrichida): morphology, systematics and identification of species. Acta Protozool. 22, 97–126.
Chen, L., Dong, J., Wu, W., Xin, Y., Warren, A., Ning, Y., et al. (2020). Morphology and molecular phylogeny of a new hypotrich ciliate, Anteholosticha songi nov. spec., and an American population of Holosticha pullaster (Müller, 1773) Foissner et al., 1991 (Ciliophora, Hypotrichia). Eur. J. Protistol. 72:125646. doi: 10.1016/j.ejop.2019.125646
Chen, X., Gao, S., Song, W., Al-Rasheid, K. A. S., Warren, A., Gong, J., et al. (2010). Parabirojimia multinucleata spec. nov. and Anteholosticha scutellum (Cohn, 1866) Berger, 2003, marine ciliates (Ciliophora, Hypotrichida) from tropical waters in southern China, with notes on their small-subunit rRNA gene sequences. Int. J. Syst. Evol. Microbiol. 60, 234–243. doi: 10.1099/ijs.0.008037-0
Chen, X., Hu, X., Lin, X., Al-Rasheid, K. A. S., Ma, H., and Miao, M. (2013). Morphology, ontogeny and molecular phylogeny of a new brackish water ciliate Bakuella subtropica sp. n. (Ciliophora, Hypotricha) from southern China. Eur. J. Protistol. 49, 611–622. doi: 10.1016/j.ejop.2013.05.001
Chen, X., Jiang, Y., Gao, F., Zheng, W., Krock, T. J., Stover, N. A., et al. (2019). Genome analyses of the new model protist Euplotes vannus focusing on genome rearrangement and resistance to environmental stressors. Mol. Ecol. Resour. 19, 1292–1308. doi: 10.1111/1755-0998.13023
Dong, J., Li, L., Fan, X., Ma, H., and Warren, A. (2020). Two Urosoma species (Ciliophora, Hypotrichia): A multidisciplinary approach provides new insights into their ultrastructure and systematics. Eur. J. Protistol. 72:125661. doi: 10.1016/j.ejop.2019.125661
Foissner, W., Chao, A., and Katz, L. A. (2008). Diversity and geographic distribution of ciliates (Protista: Ciliophora). Biodivers. Conserv. 17, 345–363. doi: 10.1007/978-90-481-2801-3_9
Gao, Y., Gong, R., Jiang, Y., Pan, B., Li, Y., Warren, A., et al. (2020). Morphogenetic characters of the model ciliate Euplotes vannus (Ciliophora, Spirotrichea): notes on cortical pattern formation during conjugational and postconjugational reorganization. Eur. J. Protistol. 73:125675. doi: 10.1016/j.ejop.2020.125675
Gouy, M., Guindon, S., and Gascuel, O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
Hu, X., Lin, X., and Song, W. (2019). Ciliates Atlas: Species Found in the South China Sea. Beijing: Science Press.
Huang, J., Chen, Z., Song, W., and Berger, H. (2014). Three-gene based phylogeny of the Urostyloidea (Protista, Ciliophora, Hypotricha), with notes on classification of some core taxa. Mol. Phylogenet. Evol. 70, 337–347. doi: 10.1016/j.ympev.2013.10.005
Jo, E., Jung, J. H., and Min, G. S. (2015). Morphology and molecular phylogeny of two new brackish water ciliates of Bakuella (Ciliophora: Urostylida: Bakuellidae) from South Korea. J. Eukaryot. Microbiol. 62, 799–809. doi: 10.1111/jeu.12238
Jung, J. H., and Berger, H. (2019). Monographic treatment of Paraholosticha muscicola (Ciliophora, Keronopsidae), including morphological and molecular biological characterization of a brackish water population from Korea. Eur. J. Protistol. 68, 48–67. doi: 10.1016/j.ejop.2018.12.004
Kaur, H., Negi, S. S. R. K., and Kamra, K. (2019). Morphological and molecular characterization of Neogastrostyla aqua nov. gen., nov. spec. (Ciliophora, Hypotrichia) from River Yamuna, Delhi; comparison with Gastrostyla-like genera. Eur. J. Protistol. 68, 68–79. doi: 10.1016/j.ejop.2019.01.002
Kim, K. S., and Min, G. S. (2019). Morphology and molecular phylogeny of Oxytricha seokmoensis sp. nov. (Hypotrichia: Oxytrichidae), with notes on its morphogenesis. Eur. J. Protistol. 71:125641. doi: 10.1016/j.ejop.2019.125641
Kim, K. S., Chae, K. S., and Mim, G. S. (2019). Morphology and molecular analyses of a new marine ciliate, Arcuseries minima sp. nov. (Ciliophora: Urostylidae). Acta Protozool. 58, 43–51. doi: 10.4467/16890027AP.19.005.10836
Kumar, S., Kamra, K., and Sapra, G. R. (2010). Ciliates of the silent valley national park, india: urostyloid hypotrichs of the region with a note on the habitat. Acta Protozool. 49, 339–364.
Li, F., Lyu, Z., Li, Y., Fan, X., Al-Farraj, S. A., Shao, C., et al. (2017). Morphology, morphogenesis, and molecular phylogeny of Uroleptus (Caudiholosticha) stueberi (Foissner, 1987) comb. nov. (Ciliophora, Hypotricha), and reclassification of the remaining Caudiholosticha species. Eur. J. Protistol. 59, 82–98. doi: 10.1016/j.ejop.2016.08.007
Li, X., Huang, J., Filker, S., Stoeck, T., Bi, Y., Yu, Y., et al. (2019). Spatio-temporal patterns of zooplankton in a main-stem dam affected tributary: a case study in the Xiangxi River of the Three Gorges Reservoir, China. Sci. China Life Sci. 62, 1058–1069. doi: 10.1007/s11427-018-9523-0
Lu, X., Huang, J., Shao, C., and Berger, H. (2018). Morphology, cell-division, and phylogeny of Schmidingerothrix elongata spec. nov. (Ciliophora, Hypotricha), and brief guide to hypotrichs with Gonostomum-like oral apparatus. Eur. J. Protistol. 62, 24–42. doi: 10.1016/j.ejop.2017.11.001
Lu, X., Wang, Y., Al-Farraj, S. A., El-Serehy, H., Huang, J., and Shao, C. (2020). The insights into the systematic relationship of Gastrostyla-affinitive genera, with report on a new saline soil ciliate genus and new species (Protozoa. Ciliophora). BMC Evol. Biol. 20:92. doi: 10.1186/s12862-020-01659-8
Luo, X., Huang, J. A., Li, L., Song, W. B., and Bourland, W. A. (2019). Phylogeny of the ciliate family Psilotrichidae (Protista, Ciliophora), a curious and poorly-known taxon, with notes on two algae-bearing psilotrichids from Guam, USA. BMC Evol. Biol. 19:125. doi: 10.1186/s12862-019-1450-z
Luo, X., Yan, Y., Shao, C., Al-Farraj, S. A., Bourland, W. A., and Song, W. (2018). Morphological, ontogenetic and molecular data support strongylidiids as being closely related to Dorsomarginalia (Protozoa, Ciliophora) and reactivation of the family Strongylidiidae Fauré-Fremiet, 1961. Zool. J. Linn. Soc. 184, 237–254. doi: 10.1093/zoolinnean/zly001
Lynn, D. H. (2008). The Ciliated Protozoa: Characterization, Classification and Guide to the Literature, 3rd Edn. Dordrecht: Springer Press.
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE), (New Orleans, LA: GCE), 1–8. doi: 10.1109/GCE.2010.5676129
Moon, J. H., Kim, J. H., Quintela-Alonso, P., and Jung, J. H. (2019). Morphology, morphogenesis, and molecular phylogeny of Neobakuella aenigmatica n. sp. (Ciliophora, Spirotrichea, Bakuellidae). J. Eukaryot. Microbiol. 67, 54–65. doi: 10.1111/jeu.12753
Nylander, J. (2004). MrModeltest v2. Program distributed by the author. Uppsala: Uppsala University.
Paiva, T. S. (2020). Systematic redefinition of the Hypotricha (Alveolata, Ciliophora) based on combined analyses of morphological and molecular characters. Protist 171:125755. doi: 10.1016/j.protis.2020.125755
Park, K. M., Jung, J. H., Kim, J. H., Min, G. S., and Kim, S. (2020). Morphology, morphogenesis, and molecular phylogeny of a new freshwater ciliate, Gonostomum jangbogoensis n. sp. (Ciliophora, Hypotricha), from Victoria Land, Antarctica. Eur. J. Protistol. 73:125669. doi: 10.1016/j.ejop.2019.125669
Ronquist, F., Teslenko, M., Mark, P. V. D., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.2307/41515220
Sela, I., Ashkenazy, H., Katoh, K., and Pupko, T. (2015). GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, W7–W14. doi: 10.1093/nar/gkv318
Shao, C., Gao, F., Hu, X., Al-Rasheid, K. A. S., and Warren, A. (2011). Ontogenesis and molecular phylogeny of a new marine urostylid ciliate, Anteholosticha petzi n. sp. (Ciliophora, Urostylida). J. Eukaryot. Microbiol. 58, 254–265. doi: 10.1111/j.1550-7408.2011.00542.x
Sheng, Y., Duan, L., Cheng, T., Qiao, Y., Stover, N. A., and Gao, S. (2020). The completed macronuclear genome of a model ciliate Tetrahymena thermophila and its application in genome scrambling and copy number analyses. Sci. China Life Sci. 63, 1534–1542. doi: 10.1007/s11427-020-1689-4
Song, W., Wilbert, N., and Warren, A. (2002). New contribution to the morphology and taxonomy of four marine hypotrichous ciliates from Qingdao, China (Protozoa: Ciliophora). Acta Protozool. 41, 145–162. doi: 10.1016/S0141-0229(02)00027-3
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Wang, J., Zhang, T., Li, F., Warren, A., Li, Y., and Shao, C. (2021a). A new hypotrich ciliate, Oxytricha xianica sp. nov., with notes on the morphology and phylogeny of a Chinese population of Oxytricha auripunctata Blatterer & Foissner, 1988 (Ciliophora, Oxytrichidae). Mar. Life Sci. Technol. 3, 303–312. doi: 10.1007/s42995-020-00089-1
Wang, J., Zhao, Y., Lu, X., Lyu, Z., Warren, A., and Shao, C. (2021b). Does the Gonostomum-patterned oral apparatus in Hypotrichia carry a phylogenetic signal? Evidence from morphological and molecular data based on extended taxon sampling using three nuclear genes (Ciliophora, Spirotrichea). Sci. China Life Sci. 64, 311–322. doi: 10.1007/s11427-020-1667-3
Wang, Y., Wang, C., Jiang, Y., Katz, L. A., Gao, F., and Yan, Y. (2019). Further analyses of variation of ribosome DNA copy number and polymorphism in ciliates provide insights relevant to studies of both molecular ecology and phylogeny. Sci. China Life Sci. 62, 203–214. doi: 10.1007/s11427-018-9422-5
Wilbert, N. (1975). Eine verbesserte technik der protargolimprägnation für ciliaten. Mikrokosmos 64, 171–179.
Xu, W., Wang, Y., Cheng, T., Yu, Y., El-Serehy, H., Al-Farraj, S. A., et al. (2020). Reevaluation of the ‘well-known’ Paraurostyla weissei complex, with notes on the ontogenesis of a new Paraurostyla species (Ciliophora, Hypotrichia). Eur. J. Protistol. 73:125672. doi: 10.1016/j.ejop.2020.125672
Yi, Z., and Song, W. (2011). Evolution of the order Urostylida (Protozoa, Ciliophora): new hypotheses based on multi-gene information and identification of localized incongruence. PLoS One 6:e17471. doi: 10.1371/journal.pone.0017471
Zhang, T., Dong, J., Cheng, T., Duan, L., and Shao, C. (2020). Reconsideration of the taxonomy of the marine ciliate Neobakuella aenigmatica Moon et al., 2019 (Protozoa, Ciliophora, Hypotrichia). Mar. Life Sci. Technol. 2, 97–108. doi: 10.1007/s42995-020-00032-4
Keywords: Bakuella xianensis n. sp., molecular phylogeny, morphology, Wilbertophrya sinica n. g., n. sp.
Citation: Ma J, Zhang T, Song W and Shao C (2021) New Contributions to the Diversity of Hypotrichous Ciliates: Description of a New Genus and Two New Species (Protozoa, Ciliophora). Front. Microbiol. 12:712269. doi: 10.3389/fmicb.2021.712269
Received: 20 May 2021; Accepted: 07 July 2021;
Published: 05 August 2021.
Edited by:
Weiwei Liu, South China Sea Institute of Oceanology, Chinese Academy of Sciences, ChinaReviewed by:
Shahed Uddin Ahmed Shazib, Smith College, United StatesCopyright © 2021 Ma, Zhang, Song and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen Shao, c2hhb2NoZW5Ac25udS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.