- 1Key Laboratory of Applied Marine Biotechnology, Ministry of Education of China, Ningbo University, Ningbo, China
- 2Collaborative Innovation Center for Zhejiang Marine High-Efficiency and Healthy Aquaculture, Ningbo University, Ningbo, China
Phycospheric bacteria may be the key biological factors affecting the growth of algae. However, the studies about interaction between Isochrysis galbana and its phycospheric bacteria are limited. Here, we show that a marine heterotrophic bacterium, Alteromonas macleodii, enhanced the growth of I. galbana, and inhibited non-photochemical quenching (NPQ) and superoxide dismutase (SOD) activities of this microalgae. Further, we explored this phenomenon via examining how the entire transcriptomes of I. galbana changed when it was co-cultured with A. macleodii. Notable increase was observed in transcripts related to photosynthesis, carbon fixation, oxidative phosphorylation, ribosomal proteins, biosynthetic enzymes, and transport processes of I. galbana in the presence of A. macleodii, suggesting the introduction of the bacterium might have introduced increased production and transport of carbon compounds and other types of biomolecules. Besides, the transcriptome changed largely corresponded to reduced stress conditions for I. galbana, as inferred from the depletion of transcripts encoding DNA repair enzymes, superoxide dismutase (SOD) and other stress-response proteins. Taken together, the presence of A. macleodii mainly enhanced photosynthesis and biosynthesis of I. galbana and protected it from stress, especially oxidative stress. Transfer of fixed organic carbon, but perhaps other types of biomolecules, between the autotroph and the heterotroph might happen in I. galbana-A. macleodii co-culture. The present work provides novel insights into the transcriptional consequences of I. galbana of mutualism with its heterotrophic bacterial partner, and mutually beneficial associations existing in I. galbana-A. macleodii might be explored to improve productivity and sustainability of aquaculture algal rearing systems.
Introduction
How environmental factors affect the growth of microalgae is always a concern of researchers. Environmental factors such as illumination, salinity, temperature, and nutrient salts all have significant effects on the growth of microalgae (Suthar and Verma, 2018; Cao et al., 2020; Ananthi et al., 2021; Corredor et al., 2021). In recent years, many researchers have found that the growth of microalgae is not only affected by the abiotic environmental factors, but also the influence of biological factors such as bacteria on the growth of microalgae cannot be neglected. The complex relationship between bacteria and microalgae has attracted more and more attention (Amin et al., 2012b; Seymour et al., 2017; Lian et al., 2018; Zhang et al., 2020), becoming one of the hot spots in the field of algae research at present. Current studies have shown that bacteria can promote the growth of microalgae in various ways. For example, bacteria can release the growth-promoting hormone indole-3-acetic acid (Amin et al., 2015; Dao et al., 2018), provide essential vitamins to algae (Croft et al., 2005; Cooper et al., 2019), scavenge reactive oxygen species (ROS) (Morris et al., 2008, 2011), promote algal assimilation of iron (Amin et al., 2009, 2012a), or metabolically transform compounds released by autotrophs in ways that can impact the entire community (Durham et al., 2015; Christie-Oleza et al., 2017). Such studies have begun to investigate which factors drive algal-bacterial interactions, yet there are many gaps in our understanding of these processes. Understanding these interactions requires studying them at different scales: identifying transcriptional changes that occur when organisms interact being the most fundamental, as this is where the cell-to-cell “recognition” is first expressed.
Isochrysis galbana is one of the most important bait microalgae in aquaculture. It is small in body size, rich in polysaccharides, carotene and lipids with high energy, especially unsaturated fatty acids docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), which are needed for the growth and development of shellfish. Besides, it has the characteristics of having no cell wall and being easily digested and absorbed by aquatic animal larvae (Cao et al., 2019). It is not only a good bait for aquaculture seedling, but also an important raw material for the development of bioactive substances. Meanwhile, it is also considered as one of the microalgae most likely to be industrialized. However, the cultivation of I. galbana is difficult in the actual production, because factors causing the bottleneck of biomass yield are still unclear. Optimal cultivation conditions of I. galbana should always take its associated bacterial community into account. However, studying how co-occurring bacteria affect I. galbana is still in its in-fancy. Therefore, it is urgent to excavate beneficial bacteria of I. galbana and study the mechanism of I. galbana-bacteria interaction, and beneficial bacterial strains may be supplemented as a new means to improve algal productivity and culture stability.
At present, some studies have reported some beneficial bacteria, which can promote the growth of microalgae, such as Sulfitobacter sp. (Amin et al., 2015), Phaeobacter gallaeciensis (Seyedsayamdost et al., 2011), Rhizobium sp. (Kim et al., 2014) and Mesorhizobium sp. (Kazamia et al., 2012; Wei et al., 2020). For I. galbana, there was an increase in algal biomass accumulation and growth rate in the presence of Alteromonas sp. and Labrenzia sp. (Sandhya and Vijayan, 2019). Interestingly, we found a bacterium also affiliated to the genus of Alteromonas, Alteromonas macleodii, that could promote the growth of I. galbana. However, the underlying interaction mechanism between I. galbana and Alteromonas remained unclear, which deserves further study.
In this study, we performed some physiological and biochemical experiments to evaluate the effects of A. macleodii on I. galbana. As a step toward further understanding the underpinnings of the effects, we examined the transcriptional responses of I. galbana to grow in co-culture with A. macleodii. This will provide novel insights into the transcriptional consequences of I. galbana of mutualism between I. galbana and its heterotrophic bacterial partner. Further, mutually beneficial associations existing between I. galbana and A. macleodii might be explored to improve productivity and sustainability of aquaculture algal rearing systems.
Materials And Methods
Algal Growth and Axenic Culture Generation
Isochrysis galbana was obtained from the Marine Biotechnology Laboratory of Ningbo University, China. NMB3 medium used in this study for culturing microalgae was composed of KNO3 (100 mg/L), KH2PO4 (10 mg/L), MnSO4⋅H2O (2.5 mg/L), FeSO4⋅7H2O (2.5 mg/L), EDTA-Na2 (10 mg/L), vitamin B1 (6 mg/L), and vitamin B12 (0.05 mg/L) (Yang et al., 2016). Microalgae were cultivated at a light intensity of 100 mmol photon m–2 s–1 under 23°C. Axenic I. galbana was maintained as described previously (Cao et al., 2019).
Bacterial Growth, Isolation, and Classification
Bacteria were typically grown on 2216E medium incubated at 28°C with shaking at 200 r.p.m. Bacterial growth was measured by counting colony-forming units. Bacteria were isolated from exponential phase growing I. galbana cultures. 16S rRNA genes of the isolated bacteria were amplified using universal 16S rDNA primers (27F, 1492R). The temperature profile for Polymerase Chain Reaction (PCR) consisted of an initial incubation at 95°C for 5 min, followed by 32 cycles of 95°C for 30 s, 55°C for 1 min and 72°C for 2 min, and a final extension step at 72°C for 10 min. Purified PCR products were ligated into pMD19-T vector for sequencing. The 16S rRNA gene sequence was compared with reference sequences in the National Center for Biotechnology Information (NCBI) by Basic Local Alignment Search Tool (BLAST). Based on the result of 16S rRNA gene sequence alignment and phylogenetic analysis, one isolated bacterial strain shared 100% sequence identity to the validly named species Alteromonas macleodii.
Co-culture Experiment
For co-culture experiments, single colony of A. macleodii was freshly plated before each experiment on 2216E agar plates. A. macleodii was inoculated into 2216E broth and incubated for 24 h in a shaking incubator (28°C, 180 rpm). Freshly prepared bacterial cells (OD600 = 0.4–0.6) were centrifuged (5,000 g for 5 min) and washed twice with sterile NMB3 medium. When axenic I. galbana was cultured to exponential phase (cell density of about 1 × 106 cells/mL), A. macleodii was added into algal culture to achieve bacteria/algae ratios of 1:1, 10:1, and 50:1. The algal growth was determined daily by cell counting, and the bacterial growth was measured by counting colony-forming units. For the co-culture transcriptome experiments, treatments consisted of (1) I. galbana and A. macleodii co-culture (a bacteria/algae ratio of 50:1), (2) axenic I. galbana. All treatments were in triplicate. Sample names for I. galbana and A. macleodii co-culture group were Ig_Am 1, Ig_Am 2 and Ig_Am 3, and Ig 1, Ig 2 and Ig 3 were for axenic I. galbana group.
RNA-seq Analysis
Cells were harvested at mid-exponential growth (100 h after inoculation for all treatments) by centrifuging. The supernatants were removed, and the bacteria and microalgae were immediately flash frozen in liquid nitrogen and later stored at −80°C. Total RNA was extracted using Trizol reagent (Invitrogen, CA, United States) according to manufacturer protocol. After the RNA test was qualified, mRNA was purified using the Ribo-zero kit to remove rRNA. The enriched mRNA was then broken into short fragments, separately. First-strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H–). To synthesize the second-strand cDNA, buffer solution, dNTPs, RNase H, and DNA polymerase I were added. The cDNA fragments were purified, end blunted, “A” tailed, and adaptor-ligated. PCR was used to selectively enrich those DNA fragments that had adapter molecules on both ends and to amplify the DNA in the library. Lastly, PCR products were purified (AMPure XP System) and library quality was assessed using an Agilent Bioanalyzer 2100 system. The generated libraries were sequenced on the Illumina HiSeq platform in Novogene Bioinformatics Technology Co., Ltd. (Beijing, China).
Clean reads were generated after removing the adaptor sequences, low-quality sequences (<Q20) and sequences shorter than 50 bp. High-quality clean reads from I. galbana and A. macleodii co-culture samples (Ig_Am 1, Ig_Am 2 and Ig_Am 3) were aligned against the assembled transcriptome of axenic I. galbana. The I. galbana transcriptome was assembled using the high-quality clean reads from the axenic I. galbana samples (Ig 1, Ig 2, and Ig 3) by the Trinity software as described for de novo transcriptome assembly (Grabherr et al., 2011). Seven public databases, including Clusters of Orthologous Groups (COG), Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), NCBI non-redundant protein (Nr), NCBI non-redundant nucleotide (Nt), Protein family (Pfam) and Swiss-Prot, were used for unigene annotation. We then performed a differential expression analysis of each gene and identified genes responses to co-culturing with A. macleodii. Genes were considered to exhibit differential expression where the fold change in expression of co-culture compared with the axenic culture was ≥ 2, the p value was ≤ 0.05. Finally, we performed gene enrichment analysis with the corresponding database (GO enrichment:1; KEGG enrichment:2) for the differentially transcribed genes. The transcriptomic data has been deposited in NCBI Sequence Read Archive (SRA) database under the accession numbers PRJNA747627.
Real-Time Quantitative PCR
Gene specific quantitative real-time PCR primers used in this study were documented in Supplementary Table 1. RT-qPCR was performed using SYBR Premix Ex Taq (TakaRa) on a StepOne Real-Time PCR System (ABI, United States). Relative expression levels were normalized to an actin gene and calculated using the 2–Δ Δ Ct method as previously described (Cao et al., 2016). The significance of the differences between mean values was determined by Student’s t-test (p < 0.05).
Results and Discussion
Alteromonas macleodii Promotes the Growth of I. galbana
We have analyzed the growth by cell counting for co-cultures with three different initial bacteria/algae ratio of (1:1, 1:10, and 1:50) and I. galbana mono-culture (Figure 1). The I. galbana cell abundance in co-cultures was significant higher than the control, which indicated that the growth of I. galbana was promoted by co-culturing with A. macleodii. Further, the cell abundance of I. galbana co-culturing with higher density of A. macleodii was higher (Figure 1A) and the color of algal fluid was darker (Figure 1B). We could conclude that the growth-promoting role of A. macleodii to I. galbana depends on the population density of the bacterium. As a key experimental parameter, the initial inoculation ratio has a crucial impact on the results of the co-culture system. For example, A. macleodii strain HOT1A3 enhanced the growth of Prochlorococcus at low cell densities, yet inhibited it at a higher concentration (Aharonovich and Sher, 2016).
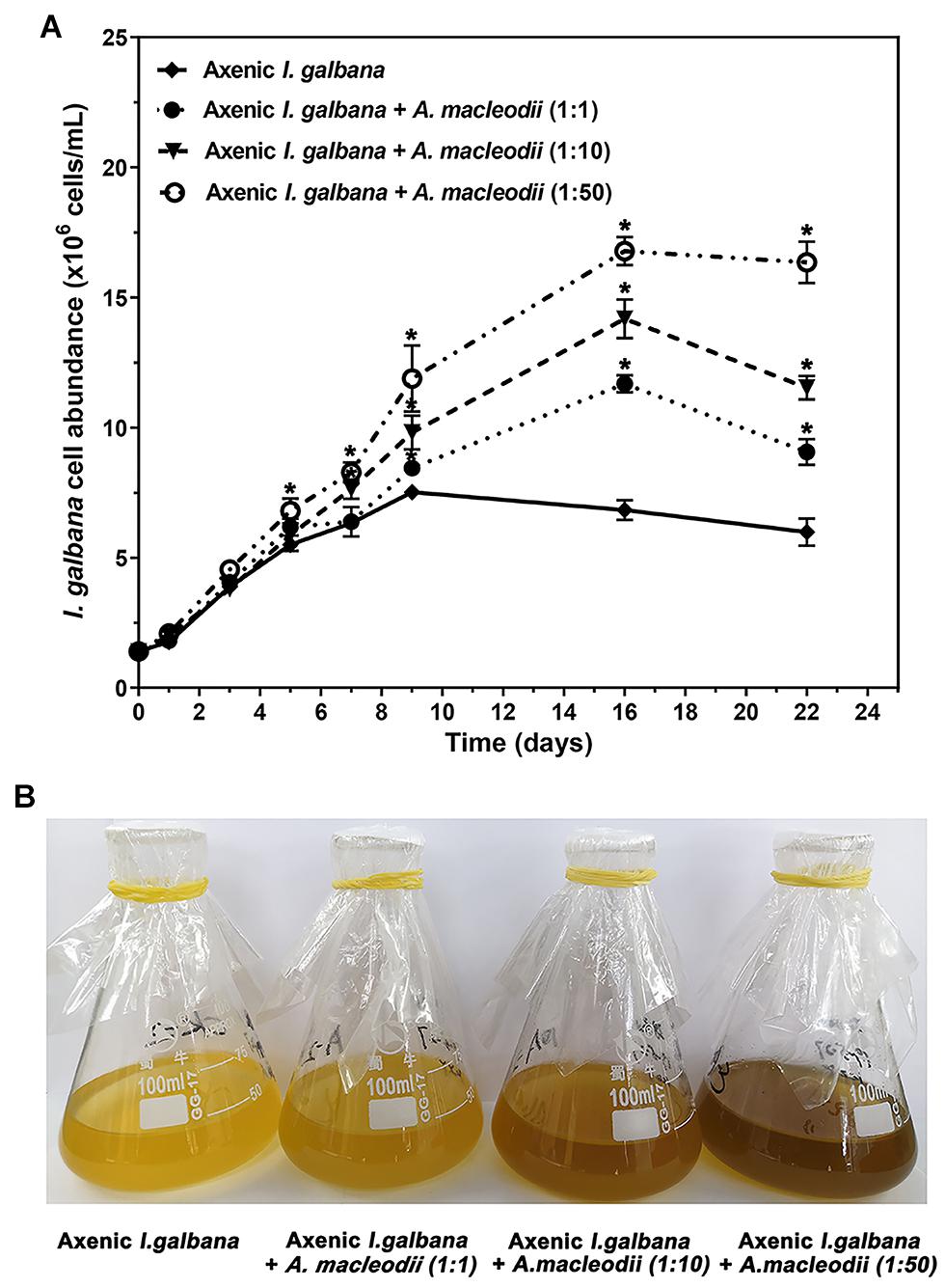
Figure 1. Co-cultures results of I. galbana and A. macleodii with three different initial bacteria/algae ratios of (1:1, 1:10, and 1:50) expressed as cell abundance (A) and the color of algal fluid (B). Significance of the differences between mean values was determined with Student’s t test. Error bars represent standard error (SE), while asterisks (*) indicate significant difference at p value < 0.05.
Physiological and Biochemical Effects of A. macleodii on I. galbana
We next focused the initial bacteria/algae ratio of 50:1, with the aim of evaluating the effects of co-culturing with A. macleodii on physiology and biochemistry of I. galbana. Same as the results shown in Figure 1, the high bacteria/algae ratio (50:1) significantly enhanced the growth of I. galbana (Figure 2A). Non-photochemical quenching (NPQ), a chlorophyll fluorescence parameter, reflects the ability of plant to dissipate energy, which is directly related to the ability to provide photoprotection to plant (Bachmann et al., 2004). Previous studies have documented that photosynthetic organisms would elevate NPQ when exposed to some environmental stressors (Cui et al., 2017; Zhang et al., 2017). In the present study, the values of NPQ in the co-culture group were significantly lower than that of the axenic group, especially in the later stages of co-culture (Figure 2B). Finally, co-culturing with A. macleodii severely inhibited the superoxide dismutase (SOD) activities of I. galbana comparing with the control for the entire study period (Figure 2C). It is well known that SOD plays a key role in the removal of reactive oxygen species (ROS). Interestingly, Alteromonas strains may scavenge ROS and thus reduce potential oxidative stress affecting Prochlorococcus (Morris et al., 2008, 2011). Besides, the study by Sandhya and Vijayan (2019) reported that Alteromonas sp. Mab 25 is able to produce extracellular antioxidants. The decreasing SOD activities in co-culture may be correlated with the scavenging of ROS by A. macleodii. The mechanism of A. macleodii promoted the growth of I. galbana could be related to protect it from stress, especially oxidative stress. However, additional studies are required in order to test this hypothesis.
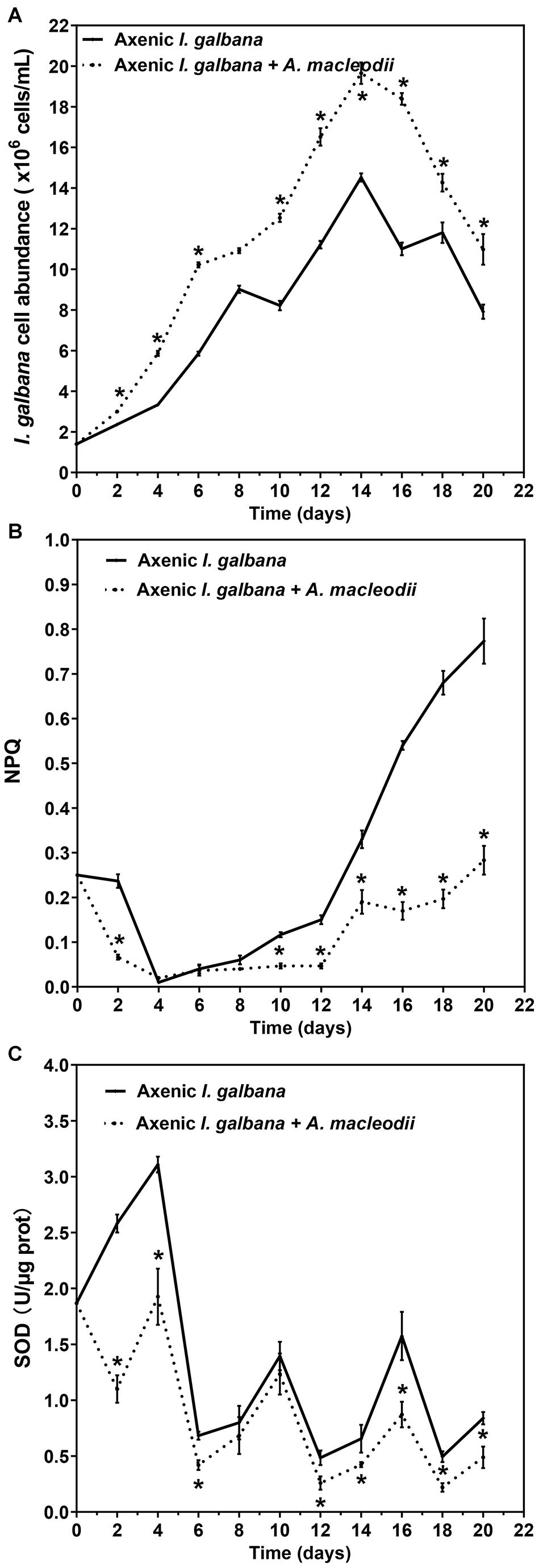
Figure 2. Cell abundance (A), non-photochemical quenching (NPQ) (B) and superoxide dismutase (SOD) activity (C) for I. galbana co-culture and mono-culture during 20 d exposure time. Significance of the differences between mean values was determined with Student’s t test. Error bars represent standard error (SE), while asterisks (*) indicate significant difference at p value < 0.05.
The Transcriptional Responses of I. galbana to Co-culture
To gain insight into how I. galbana responded physiologically to the presence of A. macleodii, we compared the transcriptomes of I. galbana in co-cultures with A. macleodii with in mono-culture. We next focused on the mid-exponential growth stages of I. galbana in co-culture, with the aim of identifying transcriptional changes in I. galbana that might explain the growth-promoting role of A. macleodii to I. galbana in co-culture. In general, after trimming and quality checking, clean reads% was ranged from 93.82 to 98.78 (Supplementary Table 2). The clean reads were finally combined and used to draw the transcriptome information of I. galbana. All data, including the error (%), Q20 (%), Q30 (%), and GC content (%), met the requirements for further analysis. Further, transcripts representing 2,157 different I. galbana genes were found to be differentially abundant in the co-culture, with more decreasing (1387) than increasing (770) in relative abundance (Supplementary Tables 3, 4). We next asked whether there were any pathways or molecular functions enriched in the subset of genes differentially expressed between co-cultures and axenic cultures, using KEGG analysis. The significant enrichment observed for the increased genes was “Oxidative phosphorylation,” “Photosynthesis,” “Ribosome,” “Tyrosine metabolism,” “Isoquinoline alkaloid biosynthesis” and “Carbon fixation in photosynthetic organisms,” while the depleted KEGG categories included “Steroid biosynthesis,” “Non-homologous end-joining,” “Protein processing in endoplasmic reticulum,” “Arginine biosynthesis,” “Biotin metabolism,” “alpha-Linolenic acid metabolism” and “Carotenoid biosynthesis” (Table 1).
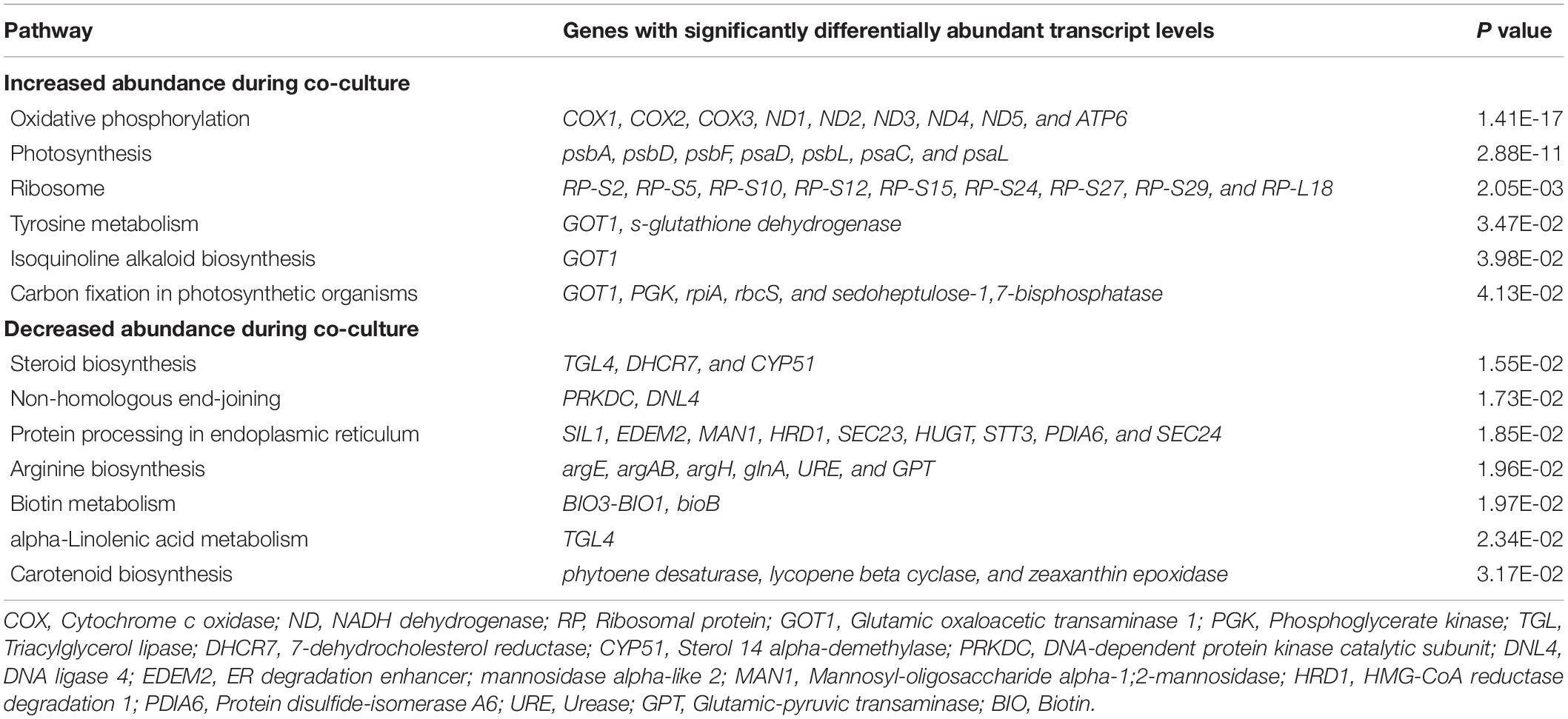
Table 1. Isochrysis galbana pathways and selected KEGG categories significantly enriched or depleted in co-culture.
Kyoto Encyclopedia of Genes and Genomes enrichment results suggested that photosynthesis of I. galbana were promoted in the presence of A. macleodii (Table 1 and Figure 3). A similar increase in expression of these pathways was also observed in Prochlorococcus-Alteromonas co-cultures (Biller et al., 2016). Why might the presence of A. macleodii trigger a change in the photosystems of I. galbana? Although mechanistic insights require further research, one possible explanation is that the heterotroph, by consuming some form of dissolved organic carbon released by I. galbana, might have either directly or indirectly stimulated I. galbana to increase the production and export of carbon compounds. As organic carbon was not provided in the medium, the success of I. galbana and A. macleodii co-culture reflects the ability of A. macleodii to survive using the organic carbon produced by I. galbana. The increase in expression of carbon fixation related genes in I. galbana proved this to some extent (Table 1 and Figure 3). Besides, oxidative phosphorylation is one kind of way to generate energy in the form of ATP and it was activated in the presence of A. macleodii here (Table 1 and Figure 3). Increased energy could be used for biosynthesis, transportation and other activities. Finally, consistent with the increased growth of co-cultured I. galbana, we found that co-cultured cells were enriched for transcripts encoding a number of proteins involved in biosynthesis or growth (Table 1 and Figure 3). These included increased in transcripts for multiple ribosomal proteins, as well as enzymes related to Isoquinoline alkaloid biosynthesis and Tyrosine metabolism. This suggested that I. galbana cells were increasing their biosynthetic capability in co-culture, and the increased biosynthesis need more energy. Taken together, we hypothesized that increasing photosynthesis of I. galbana in co-culture will produce more energy-storing materials such as carbohydrate that could be used for respiration to produce more energy for biosynthesis, which could explain for the increased growth of co-cultured I. galbana in the presence of A. macleodii.
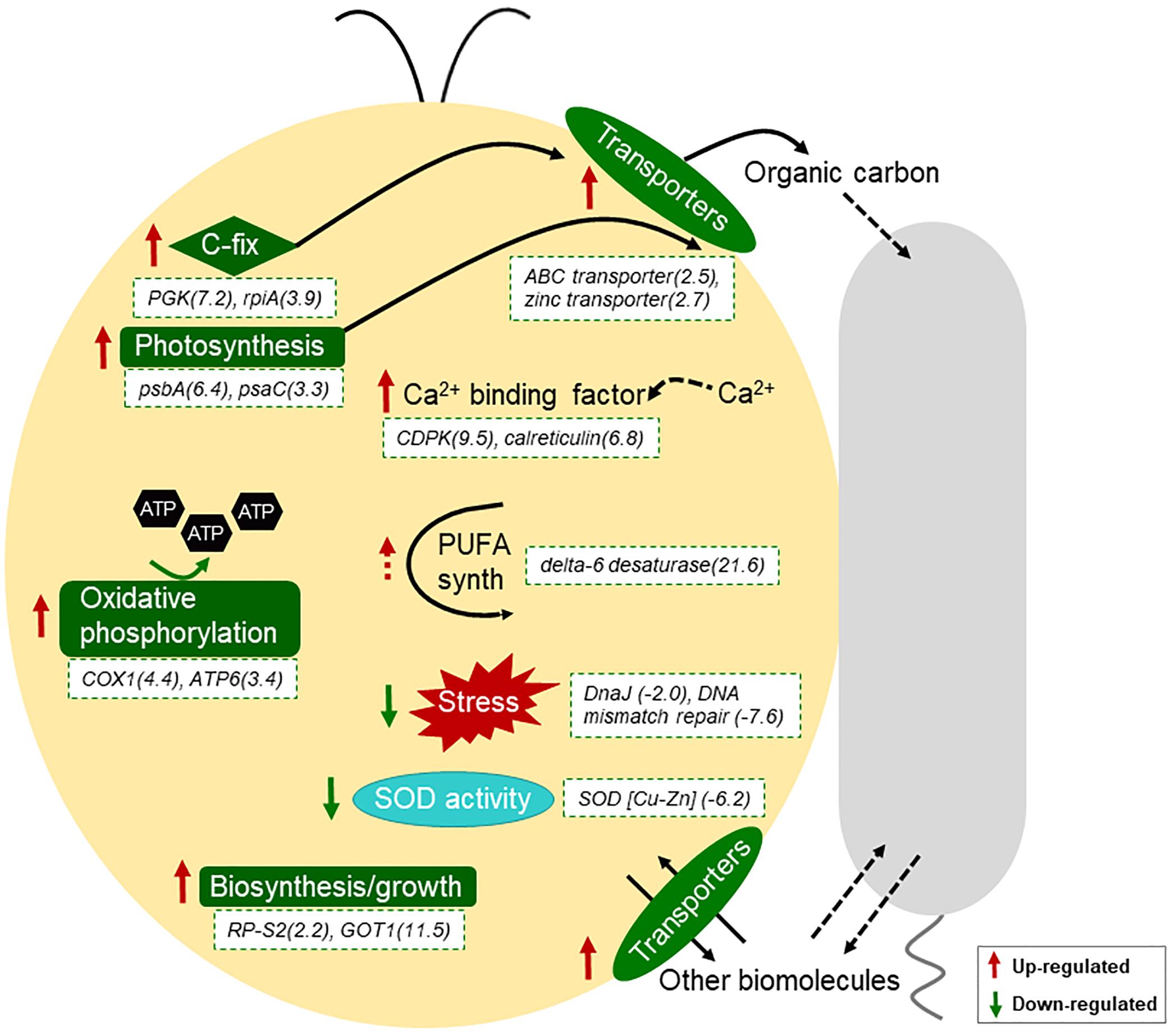
Figure 3. Schematic diagram of transcriptional responses of I. galbana to co-culture with A. macleodii. I. galbana cell in yellow and A. macleodii cell in gray. Red and green arrows indicate the related genes or pathways were up-regulated and down-regulated, respectively. Log2fold change in comparison group Ig_Am/Ig for the expression of the related genes and the gene names were denoted with the white dotted box.
Besides the above enriched pathways or molecular functions, many of the transcriptional responses of I. galbana can be associated with transport processes under co-culture conditions (Supplementary Table 3). For example, the expression of a intraflagellar transport protein 140-like protein (Cluster-1913.45461), a ABC transporter (Cluster-1913.54562), a putative zinc transporter (Cluster-1913.58709) and sodium potassium calcium exchanger 4 (Cluster-1913.5747) was significantly increased in the presence of A. macleodii (Supplementary Table 3). Similar results were found in Synechococcus–Shewanella co-culture, as well as Prochlorococcus–Alteromonas (Beliaev et al., 2014; Biller et al., 2016). These results suggested that many types of biomolecules, including fixed organic carbon, might be exchanged between the co-cultured autotroph and heterotroph. The differential expression of 10 genes annotated as calcium-dependent protein kinase (CDPK), calcium-dependent protein or calreticulin (8 upregulated, 2 downregulated) suggests similarities with plant recognition of bacteria using Ca2+ as a second messenger (Vadassery and Oelmüller, 2009). Calcium-dependent signaling followed by cell death was described previously for nutrient-limited diatoms (Vardi et al., 2006). Our results, as well as the results reported by Durham et al., 2017 extend this by suggesting that second messenger also plays a role in relaying information on biotic stimuli. Interestingly, the expression of a delta-6 fatty acid desaturase (Cluster-1913.57529) was enrich significantly (Log2FoldChange = 21.583) (Supplementary Table 3). Delta-6 fatty acid desaturase is a key enzyme for polyunsaturated fatty acids (PUFA) biosynthesis. If the presence of A. macleodii indeed increased markedly the polyunsaturated fatty acids content in co-culture is worth of making further research.
Finally, additional transcriptional responses of I. galbana were consistent with a generalized reduction in stress in the presence of the heterotroph. For example, multiple DNA repair enzymes, including DnaJ (Cluster-1913.40877), DNA mismatch repair protein (Cluster-1913.37455, Cluster-1913.28067), deoxyribodipyrimidine photolyase (Cluster-1913.105678), RNA helicase nonsense mRNA reducing factor (Cluster-1913.102129), lon protease homolog 1 (Cluster-1913.17497), REV1 Deoxycytidyl transferase (Cluster-1913.59724) were transcriptionally depleted in co-culture (Supplementary Table 4). Besides, the expression of copper/zinc superoxide dismutase (Cluster-1913.101311) was down-regulated in co-culture, which was consistent with the decreased activity of SOD (Figure 3 and Supplementary Table 4). In addition, other stress-response transcripts encoding heat shock protein 90, multiple nfx1-type zinc finger-containing proteins and some ubiquitination-related proteins also decreased in relative abundance (Supplementary Table 4). These results suggested the growth-promoting role of A. macleodii to I. galbana might be partly related to protect it from stress, especially oxidative stress, which was similar with the case in Alteromonas-Prochlorococcus interaction (Morris et al., 2008, 2011).
However, the most significant fraction of genes differentially expressed in co-culture compared with axenically growing cells have no known function. The lack of annotation hampers the interpretation of many of the responses observed and this highlights how much we have yet to learn about the mechanisms and molecules underlying I. galbana- bacteria interactions. Additionally, correlation analysis of the transcriptional responses of A. macleodii to co-culture with the responses of I. galbana was important to provide insight into I. galbana and A. macleodii interactions, which deserves further study. Finally, it is important to recognize that changes in relative transcript abundance within I. galbana and A. macleodii does not necessarily always lead to similar changes in protein abundance. Thus, the changes described here represent a first step toward understanding the interactions between A. macleodii and I. galbana.
Validation of Significant Differentially-Expressed Genes by RT-qPCR
In order to further validate the RNA-seq data, four DEGs from comparison group (Ig_Am vs Ig) were randomly selected for expression profile analyses by RT-qPCR. All these DEGs included calcium-dependent protein (Cluster-1913.49064), photosystem II protein D1 (Cluster-1913.54157), aspartate aminotransferase (Cluster-1913.58166) and biotin synthase (Cluster-1913.37995) for I. galbana. The results of RT-qPCR revealed that most of these DEGs shared similar expression tendency with those from RNA-seq data (Figure 4). Although there were some quantitative differences between the two analytical platforms, the similarities between these two methods suggested that the RNA-seq data were reproducible and reliable.
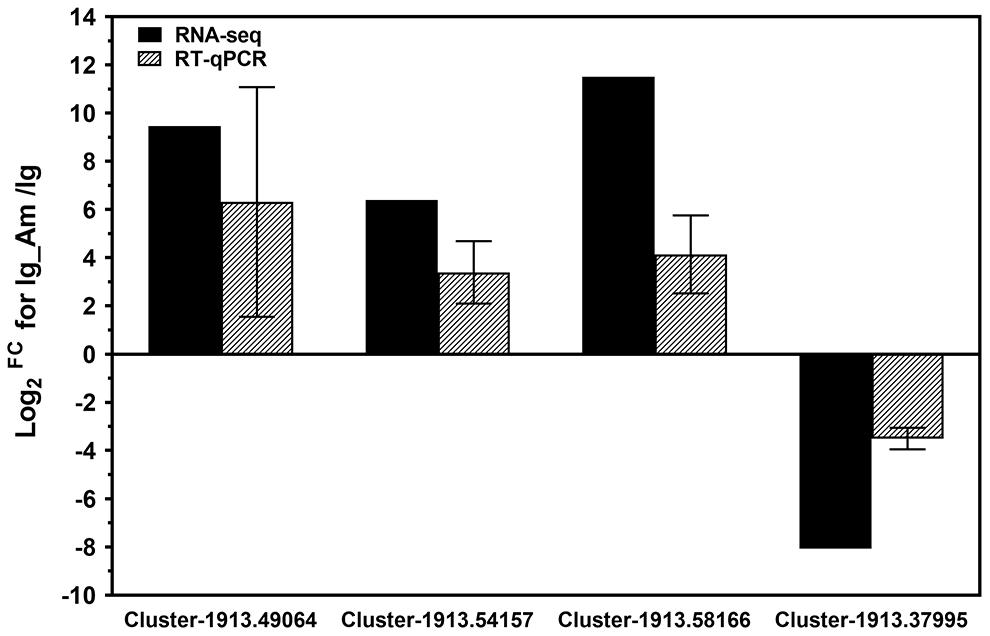
Figure 4. Experimental confirmation of RNA-seq data on relative fold change level of the selected differentially-expressed genes (DEGs) by real-time quantitative PCR (RT-qPCR) analyses. The results from RNA-Seq and RT-qPCR, are shown by bars with different patterns. The selected DEGs included calcium-dependent protein (Cluster-1913.49064), photosystem II protein D1 (Cluster-1913.54157), aspartate aminotransferase (Cluster-1913.58166) and biotin synthase (Cluster-1913.37995) for I. galbana. RT-qPCR data represent the mean ± standard error (SE) of three independent experiments.
Data Availability Statement
The transcriptomic data has been deposited in NCBI Sequence Read Archive (SRA) database under the accession numbers PRJNA747627.
Author Contributions
J-LX coordinated the project. Y-YW, M-NW, Z-YK, J-HL, TL, S-MX, and J-YC conducted the experiments. J-YC designed and performed the bioinformatics and statistical analysis. S-NM, LZ, and C-XZ helped in analyzing and modeling the data. J-LX and X-JY conceived of the study and participated in its design and coordination. J-LX and J-YC prepared and revised the manuscript. All authors had read and approved the final manuscript.
Funding
This research was supported by the National Key Research and Development Program of China (2019YFD0900400), Zhejiang Major Science Project, China (2019C02057), Ningbo Science and Technology Research Projects, China (2019B10006), China Agriculture Research System of MOF and MARA, the National Natural Science Foundation of China (31801724), and Natural Science Foundation of Ningbo, China (2019A610416).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.711998/full#supplementary-material
Footnotes
References
Aharonovich, D., and Sher, D. (2016). Transcriptional response of Prochlorococcus to co-culture with a marine Alteromonas: differences between strains and the involvement of putative infochemicals. ISME J. 10, 2892–2906. doi: 10.1038/ismej.2016.70
Amin, S. A., Green, D. H., Hart, M. C., Kupper, F. C., Sunda, W. G., and Carrano, C. J. (2009). Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proc. Natl. Acad. Sci. U. S. A. 106, 17071–17076. doi: 10.1073/pnas.0905512106
Amin, S. A., Hmelo, L. R., Tol, H. M., Durham, B. P., Carlson, L. T., Heal, K. R., et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101. doi: 10.1038/nature14488
Amin, S. A., Parker, M. S., and Armbrust, E. V. (2012b). Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684. doi: 10.1128/MMBR.00007-12
Amin, S. A., Green, D. H., Gärdes, A., Romano, A., Trimble, L., and Carrano, C. J. (2012a). Siderophore-mediated iron uptake in two clades of Marinobacter spp. associated with phytoplankton: the role of light. Biometals 25, 181–192. doi: 10.1007/s10534-011-9495-5
Ananthi, V., Brindhadevi, K., Pugazhendhi, A., and Arun, A. (2021). Impact of abiotic factors on biodiesel production by microalgae. Fuel 284:118962. doi: 10.1016/j.fuel.2020.118962
Bachmann, K. M., Ebbert, V., Adams, W. W., Verhoeven, A. S., Logan, B. A., and Demmig-Adams, B. (2004). Effects of lincomycin on PSII efficiency, non-photochemical quenching, D1 protein and xanthophyll cycle during photoinhibition and recovery. Funct. Plant Biol. 31, 803–813. doi: 10.1071/fp04022
Beliaev, A. S., Romine, M. F., Serres, M., Bernstein, H. C., Linggi, B. E., Markillie, L. M., et al. (2014). Inference of interactions in cyanobacterial-heterotrophic co-cultures via transcriptome sequencing. ISME J. 8, 2243–2255. doi: 10.1038/ismej.2014.69
Biller, S., Coe, A., and Chisholm, S. (2016). Torn apart and reunited: impact of a heterotroph on the transcriptome of Prochlorococcus. ISME J. 10, 2831–2843. doi: 10.1038/ismej.2016.82
Cao, J. Y., Kong, Z. Y., Ye, M. W., Ling, T., Chen, K., Xu, J. L., et al. (2020). Comprehensive comparable study of metabolomic and transcriptomic profiling of Isochrysis galbana exposed to high temperature, an important diet microalgal species. Aquaculture 521:735034. doi: 10.1016/j.aquaculture.2020.735034
Cao, J. Y., Kong, Z. Y., Ye, M. W., Zhang, Y. F., Xu, J. L., Zhou, C. X., et al. (2019). Metabolomic and transcriptomic analyses reveal the effects of ultraviolet radiation deprivation on Isochrysis galbana at high temperature. Algal Res. 38:101424. doi: 10.1016/j.algal.2019.101424
Cao, J. Y., Xu, Y. P., Zhao, L., Li, S. S., and Cai, X. Z. (2016). Tight regulation of the interaction between Brassica napus and Sclerotinia sclerotiorum at the microRNA level. Plant Mol. Biol. 92, 39–55. doi: 10.1007/s11103-016-0494-3
Christie-Oleza, J. A., Sousoni, D., Lloyd, M., Armengaud, J., and Scanlan, D. J. (2017). Nutrient recycling facilitates long-term stability of marine microbial phototroph-heterotroph interactions. Nat. Microbiol. 2:17100. doi: 10.1038/nmicrobiol.2017.100
Cooper, M. B., Kazamia, E., Helliwell, K. E., Kudahl, U. J., Sayer, A., Wheeler, G. L., et al. (2019). Cross-exchange of B-vitamins underpins a mutualistic interaction between Ostreococcus tauri and Dinoroseobacter shibae. ISME J. 13, 334–345. doi: 10.1038/s41396-018-0274-y
Corredor, L., Barnhart, E. P., Parker, A. E., Gerlach, R., and Fields, M. W. (2021). Effect of temperature, nitrate concentration, pH and bicarbonate addition on biomass and lipid accumulation in the sporulating green alga PW95. Algal Res. 53:102148. doi: 10.1016/j.algal.2020.102148
Croft, M. T., Lawrence, A. D., Raux-Deery, E., Warren, M. J., and Smith, A. G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93. doi: 10.1038/nature04056
Cui, Y., Zhang, H., and Lin, S. (2017). Enhancement of non-photochemical quenching as an adaptive strategy under phosphorus deprivation in the dinoflagellate Karlodinium veneficum. Front. Microbiol. 8:404. doi: 10.3389/fmicb.2017.00404
Dao, G. H., Wu, G. X., Wang, X. X., Zhang, T. Y., Zhan, X. M., and Hu, H. Y. (2018). Enhanced microalgae growth through stimulated secretion of indole acetic acid by symbiotic bacteria. Algal Res. 33, 345–351. doi: 10.1016/j.algal.2018.06.006
Durham, B. P., Dearth, S. P., Sharma, S., Amin, S. A., Smith, C. B., Campagna, S. R., et al. (2017). Recognition cascade and metabolite transfer in a marine bacteria-phytoplankton model system. Environ. Microbiol. 19, 3500–3513. doi: 10.1111/1462-2920.13834
Durham, B. P., Sharma, S., Luo, H., Smith, C. B., Amin, S. A., Bender, S. J., et al. (2015). Cryptic carbon and sulfur cycling between surface ocean plankton. Proc. Natl. Acad. Sci. U. S. A. 112, 453–457. doi: 10.1073/pnas.1413137112
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Kazamia, E., Czesnick, H., Nguyen, T. T., Croft, M. T., Sherwood, E., Sasso, S., et al. (2012). Mutualistic interactions between vitamin B12 -dependent algae and heterotrophic bacteria exhibit regulation. Environ. Microbiol. 14, 1466–1476. doi: 10.1111/j.1462-2920.2012.02733.x
Kim, B.-H., Ramanan, R., Cho, D. H., Oh, H. M., and Kim, H. S. (2014). Role of Rhizobium, a plant growth promoting bacterium, in enhancing algal biomass through mutualistic interaction. Biomass Bioenerg. 69, 95–105. doi: 10.1016/j.biombioe.2014.07.015
Lian, J., Wijffels, R. H., Smidt, H., and Sipkema, D. (2018). The effect of the algal microbiome on industrial production of microalgae. Microb. Biotechnol. 11, 806–818. doi: 10.1111/1751-7915.13296
Morris, J. J., Johnson, Z. I., Szul, M. J., Keller, M., and Zinser, E. R. (2011). Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean’s surface. PLoS One 6:e16805. doi: 10.1371/journal.pone.0016805
Morris, J. J., Kirkegaard, R., Szul, M. J., Johnson, Z. I., and Zinser, E. R. (2008). Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl. Environ. Microbiol. 74, 4530–4534. doi: 10.1128/AEM.02479-07
Sandhya, S., and Vijayan, K. (2019). Symbiotic association among marine microalgae and bacterial flora: a study with special reference to commercially important Isochrysis galbana culture. J. Appl. Phycol. 31, 2259–2266. doi: 10.1007/s10811-019-01772-2
Seyedsayamdost, M. R., Case, R. J., Kolter, R., and Clardy, J. (2011). The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3, 331–335. doi: 10.1038/nchem.1002
Seymour, J. R., Amin, S. A., Raina, J. B., and Stocker, R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2:17065. doi: 10.1038/nmicrobiol.2017.65
Suthar, S., and Verma, R. (2018). Production of Chlorella vulgaris under varying nutrient and abiotic conditions: a potential microalga for bioenergy feedstock. Process Saf. Environ. Prot. 113, 141–148. doi: 10.1016/j.psep.2017.09.018
Vadassery, J., and Oelmüller, R. (2009). Calcium signaling in pathogenic and beneficial plant microbe interactions. Plant Signal. Behav. 4, 1024–1027. doi: 10.4161/psb.4.11.9800
Vardi, A., Formiggini, F., Casotti, R., De Martino, A., Ribalet, F., Miralto, A., et al. (2006). A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biol. 4:e60. doi: 10.1371/journal.pbio.0040060
Wei, Z., Wang, H., Li, X., Zhao, Q., Yin, Y., Xi, L., et al. (2020). Enhanced biomass and lipid production by co-cultivation of Chlorella vulgaris with Mesorhizobium sangaii under nitrogen limitation. J. Appl. Phycol. 32, 233–242. doi: 10.1007/s10811-019-01924-4
Yang, F., Chen, S., Miao, Z., Sheng, Z., Xu, J., Wan, J., et al. (2016). The effect of several microalgae isolated from East China Sea on growth and survival rate of postset juveniles of razor clam, Sinonovacula constricta (Lamarck, 1818). Aquac. Nutr. 22, 846–856.
Zhang, B., Li, W., Guo, Y., Zhang, Z., Shi, W., Cui, F., et al. (2020). Microalgal-bacterial consortia: from interspecies interactions to biotechnological applications. Renew. Sustain. Energy Rev. 118:109563. doi: 10.1016/j.rser.2019.109563
Keywords: Isochrysis galbana, Alteromonas macleodii, RNA-Seq, co-culture, interaction
Citation: Cao J-Y, Wang Y-Y, Wu M-N, Kong Z-Y, Lin J-H, Ling T, Xu S-M, Ma S-N, Zhang L, Zhou C-X, Yan X-J and Xu J-L (2021) RNA-seq Insights Into the Impact of Alteromonas macleodii on Isochrysis galbana. Front. Microbiol. 12:711998. doi: 10.3389/fmicb.2021.711998
Received: 19 May 2021; Accepted: 23 August 2021;
Published: 08 September 2021.
Edited by:
Hongbin Liu, Hong Kong University of Science and Technology, ChinaReviewed by:
Chao Wu, National Renewable Energy Laboratory (DOE), United StatesZhang-Xian Xie, Xiamen University, China
Copyright © 2021 Cao, Wang, Wu, Kong, Lin, Ling, Xu, Ma, Zhang, Zhou, Yan and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ji-Lin Xu, eHVqaWxpbkBuYnUuZWR1LmNu
 Jia-Yi Cao
Jia-Yi Cao Ying-Ying Wang
Ying-Ying Wang Min-Nan Wu
Min-Nan Wu Zhou-Yan Kong
Zhou-Yan Kong Jing-Hao Lin
Jing-Hao Lin Ting Ling
Ting Ling Si-Min Xu
Si-Min Xu Shuo-Nan Ma
Shuo-Nan Ma Lin Zhang
Lin Zhang Cheng-Xu Zhou
Cheng-Xu Zhou Xiao-Jun Yan
Xiao-Jun Yan Ji-Lin Xu
Ji-Lin Xu