- 1Department of Animal Sciences and Industry, Food Science Institute, Kansas State University, Manhattan, KS, United States
- 2Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration, College Park, MD, United States
- 3Department of Agronomy, Kansas State University, Manhattan, KS, United States
- 4Centers for Disease Control and Prevention, Atlanta, GA, United States
- 5Department of Horticulture and Natural Resources, Kansas State University, Olathe, KS, United States
- 6Department of Diagnostic Medicine/Pathobiology, Kansas State University, Manhattan, KS, United States
Non-typhoidal Salmonella enterica is a pathogen of global importance, particularly in low and middle-income countries (LMICs). The presence of antimicrobial resistant (AMR) strains in market environments poses a serious health threat to consumers. In this study we identified and characterized the genotypic and phenotypic AMR profiles of 81 environmental S. enterica strains isolated from samples from informal markets in Cambodia in 2018–2019. AMR genotypes were retrieved from the NCBI Pathogen Detection website (https://www.ncbi.nlm.nih.gov/pathogens/) and using ResFinder (https://cge.cbs.dtu.dk/services/) Salmonella pathogenicity islands (SPIs) were identified with SPIFinder (https://cge.cbs.dtu.dk/services/). Susceptibility testing was performed by broth microdilution according to the Clinical and Laboratory Standards Institute (CLSI) standard guidelines M100-S22 using the National Antimicrobial Resistance Monitoring System (NARMS) Sensititre Gram Negative plate. A total of 17 unique AMR genes were detected in 53% (43/81) of the isolates, including those encoding tetracycline, beta-lactam, sulfonamide, quinolone, aminoglycoside, phenicol, and trimethoprim resistance. A total of 10 SPIs (SPI-1, 3–5, 8, 9, 12–14, and centisome 63 [C63PI]) were detected in 59 isolates. C63PI, an iron transport system in SPI-1, was observed in 56% of the isolates (n = 46). SPI-1, SPI-4, and SPI-9 were present in 13, 2, and 5% of the isolates, respectively. The most common phenotypic resistances were observed to tetracycline (47%; n = 38), ampicillin (37%; n = 30), streptomycin (20%; n = 16), chloramphenicol (17%; n = 14), and trimethoprim-sulfamethoxazole (16%; n = 13). This study contributes to understanding the AMR genes present in S. enterica isolates from informal markets in Cambodia, as well as support domestic epidemiological investigations of multidrug resistance (MDR) profiles.
Introduction
Informal markets are common in Southeast Asian countries. In Cambodia, these markets play an important role in the country’s economy, culture, and lifestyle. In fact, a variety of food products (e.g., fresh vegetables, fruits, seafood, and animal products) are sold through these markets, which often lack basic food safety infrastructure or oversight (Roesel and Grace, 2014). Studies have shown a high prevalence of foodborne pathogens, specifically non-typhoidal S. enterica in food products and surfaces in the informal market settings in Cambodia (Lay et al., 2011; Trongjit et al., 2017; Nadimpalli et al., 2019).
S. enterica is among the top five causative agents of diarrheal diseases worldwide (World Health Organization, 2015). This pathogen has been linked to many foodborne outbreaks in several countries (Ford et al., 2018; Finger et al., 2019; Jiang et al., 2020). In the United States, approximately 94% of S. enterica infections are attributed to food (Hoffmann et al., 2012). Moreover, S. enterica is considered a pathogen of global importance, particularly in low-income countries (World Health Organization, 2017). However, surveillance data on foodborne disease is limited in Cambodia. Recently, the Mekong Institute (an intergovernmental organization founded by Southeast Asian countries) revealed that thousands of people suffer from unsafe food in Cambodia (Mekong Institute, 2019). Between 2014 and 2019, Cambodia’s Food Safety Bureau and the Department of Drug and Food (DDF) reported 134 foodborne outbreaks, resulting in 5,825 illnesses, 5,598 hospitalizations, and 81 deaths (Mekong Institute, 2019). Although informative, these estimates are likely underreported as indicated by the high percentage of illnesses that resulted in hospitalizations (96%), with the actual number of cases likely being much greater.
The lack of regulatory oversight, infrastructure, potable water, and adequate hygiene and sanitation practices have been identified as the leading causes for outbreaks (Mekong Institute, 2019) and have been previously described as a common sight among informal markets in Cambodia (Roesel and Grace, 2014). These factors often promote cross-contamination among food products, food contact surfaces and food handlers, favoring the spread of pathogens. The prevalence of S. enterica in the Cambodian informal market system has been previously determined, further indicating that cross-contamination is likely occurring (Schwan et al., 2020a). The ubiquity of S. enterica and its ability to cause human infections demonstrates that research needs to be conducted to better understand persistence and genotypic characteristics (e.g., virulence factors, antimicrobial resistance) in the serotypes present in these markets.
The indiscriminate use of antibiotics in food production (i.e., food animals) has raised immense concern among public health authorities as it contributes to the development and spread of antimicrobial resistance (AMR) among foodborne pathogens (Marshall and Levy, 2011). The spread of AMR bacteria threatens the ability to effectively treat bacterial infections, potentially leading to an extended illness period, disability, or death (World Health Organization, 2018). The World Health Organization (WHO) identified surveillance and monitoring of AMR bacteria as a global health priority (World Health Organization, 2018). Several countries (e.g., United States, Switzerland, and Australia) have conducted extensive research in identifying AMR foodborne pathogens (Barlow et al., 2015; Jans et al., 2018; Yang et al., 2020). However, limited studies have been conducted to assess the prevalence and genetic markers of AMR foodborne pathogens in Cambodia, where awareness and understanding of AMR is insufficient and needs to be investigated (Yam et al., 2019).
With the aim of understanding AMR among foodborne pathogens, the National Center for Biotechnology Information (NCBI) pathogen detection division created the NCBI Pathogen Detection database using the AMRFinder tool. This tool compares isolate genomes to a reference database of acquired resistance genes and proteins, utilizing gene hierarchy to identify the most specific protein assignment to antimicrobial resistant protein or family (Feldgarden et al., 2019). It utilizes several methods to identify AMR protein sequences or genomic nucleotide sequences. Briefly, each protein or genomic nucleotide can be identified following 100% identity and length match to a known resistance gene or allele; high identity match to a known AMR protein; partial length AMR protein sequence, or identification only through Hidden Markov Models (Feldgarden et al., 2019). These tools facilitate the identification and characterization of AMR genes from different genomes.
This study aims to identify and characterize the phenotypic and genotypic AMR profiles of S. enterica strains isolated from environmental samples collected at two informal markets in Cambodia between 2018 and 2019.
Materials and Methods
Bacterial Isolates and Sample Collection
A total of 81 S. enterica strains were used in this study. The strains were isolated in 2018 (June) and 2019 (January) from environmental samples collected from two informal markets in the province of Battambang, Cambodia (Schwan et al., 2020a). The strains were stored until further analysis at −80° ± 2.0°C in cryobeads (Key Scientific Products Inc., Stamford, TX-United States), following manufacturer protocol. Each isolate was assigned a unique U.S. Food and Drug Administration (FDA) Center for Food Safety and Applied Nutrition (CFSAN) identification number as part of the GenomeTrakr network (Timme et al., 2018).
DNA Preparation
Genomic DNA from each strain was obtained using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany), following manufacturer instructions. DNA concentration was determined using a Qubit 4.0 fluorometer (Invitrogen). DNA samples were sent to the FDA-CFSAN for Whole Genome Sequencing (WGS). The resultant DNA extract was stored at −20°C until WGS analysis.
Whole Genome Sequencing and Serovar Prediction
Libraries were prepared from genomic DNA with the Nextera XT DNA Library Preparation Kit, and WGS was carried out on either the MiSeq or NextSeq sequencer, using a MiSeq Reagent Kit V2 (500-cycles) or a NextSeq 500/550 High-Output Kit V2 (300-cycles), respectively (Illumina), as previously described by Schwan et al. (2020b). De novo assemblies were obtained with Shovill 0.9.1 The serotype of each isolate was determined in silico using SeqSero 1.0 on draft genomes.2
WGS data for the 81 S. enterica isolates is available at NCBI under BioProject accession number PRJNA628951, as previously described by Schwan et al. (2020b).
Detection of Antimicrobial Resistance Genes
To ensure a comprehensive analysis of known resistance genes, two database platforms were used: the NCBI Pathogen Detection (NCBI PD) and the Center for Genomic Epidemiology (CGE3). Under NCBI PD, AMR genes were identified using AMRFinderPlus using the following two criteria (i) Allele [100% sequence match to 100% of length to a protein named at the allele level in the Pathogen Detection Reference Gene Catalog (PDRGC)], and (ii) Exact (a 100% sequence match to 100% of length to a protein in the PDRGC that is not a named allele) (Feldgarden et al., 2019). Under CGE, AMR genes were identified using ResFinder 4.0 with the following criteria (i) ≥ 90% amino acid identity and (ii) ≥ 80% sequence length identity to known resistance proteins (Bortolaia et al., 2020). The reference databases used included acquired genes and mutations known to confer resistance to the antimicrobials aminoglycosides, β-lactams, colistin, fluoroquinolones, fosfomycin, macrolides, phenicols, rifampicin, sulfonamides, tetracyclines, and trimethoprim (Zankari et al., 2012). Salmonella pathogenicity islands (SPIs) were identified with SPIFinder (Center for Genomic Epidemiology; see text footnote 3).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility profiles were determined by broth microdilution according to the Clinical and Laboratory Standards Institute (CLSI) standard guidelines M100-S22 (Cockerill and Clinical and Laboratory Standards Institute, 2012) using the Sensititre automated antimicrobial susceptibility system (Thermo Fisher Scientific, Waltham, MA) with the National Antimicrobial Resistance Monitoring System (NARMS) gram-negative CMV3AGNF plate (Maradiaga et al., 2017). Fourteen antimicrobials were tested: amoxicillin/clavulanic acid 2:1 ratio (AMC), ampicillin (AMP), azithromycin (AZM), cefoxitin (FOX), ceftiofur (XNL), ceftriaxone (CRO), ciprofloxacin (CIP), chloramphenicol (CHL), gentamycin (GEN), nalidixic acid (NAL), streptomycin (STR), sulfisoxazole (FIS), tetracycline (TET), and trimethoprim/sulfamethoxazole (SXT), representing nine antimicrobial classes defined by the Clinical and Laboratory Standards Institute (CLSI; Clinical and Laboratory Standards Institute, 2020). The minimum inhibitory concentration (MIC) for each antimicrobial was interpreted using the CLSI standards and NARMS breakpoints to categorize MIC results as susceptible or resistant (Food and Drug Administration, 2015; Clinical and Laboratory Standards Institute, 2020).
Agreement Between Genotypic and Phenotypic Susceptibility
A total of 1,620 genotypic and 1,134 phenotypic data points were generated from the 81 isolates. An isolate was classified as genotypically resistant when presenting at least one gene known to confer resistance to a given antimicrobial agent, and susceptible otherwise. An isolate was classified as phenotypically resistant to a given antimicrobial agent when presenting a MIC equal to or greater than the resistant threshold based on CLSI standards and NARMS breakpoints, and susceptible otherwise. Intermediate phenotypes were considered as susceptible in this analysis. Overall and antibiotic-specific sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated considering genotypic (predicted) and phenotypic (observed) resistant/susceptible classification using the function confusionMatrix from the package caret (Kuhn, 2020) in R (R Core Team, 2020).
Results
AMR Resistance Genes
AMR genes were identified in 43 out of 81 isolates (53%), including those encoding tetracycline, beta-lactam, sulfonamide, quinolone, aminoglycoside, phenicol, and trimethoprim resistance. A total of 17 unique resistance genes were detected, most commonly tet(A) (37%, n = 30); blaTEM–1 (35%, n = 28); sul2 (30%, n = 24); qnrS1 (27%, n = 22); aph(6)-Id and aph(3″)-Ib (21%, n = 17 each), followed by sul3, and aadA1 (17%, n = 14 each); floR, dfrA12, and aadA2 (16%, n = 13 each); cmlA1 (14%, n = 11), and tet(B) (10%, n = 8). Fewer strains presented aph(3′)-Ia (7%, n = 6); aac(3)-IId and blaCTX–M–14 (2%, n = 2 each); and blaLAP–2 (1%, n = 1) resistance genes. Among all isolates, 38 presented no AMR genes (47%).
Salmonella Pathogenicity Islands
A total of 10 Salmonella pathogenicity islands [SPI-1, 3–5, 8, 9, 12–14, and centisome 63 (C63PI)] (Zhou et al., 1999) were detected in 59 isolates (Table 1). C63PI, an iron transport system in SPI-1, was observed in 57% of the isolates (n = 46). SPI-1, SPI-4 and SPI-9, which encode predicted type I and type III secretion systems (Amavisit et al., 2003; Wisner et al., 2012) were present in 16% (n = 13), 2% (n = 2), and 5% (n = 4) of the isolates, respectively. Twenty-seven percent of the isolates (n = 22) had no PIs. SPI-1 was found in S. Krefeld (n = 5), S. Typhimurium and S. Derby (n = 2), S. Hvittingfoss, S. Corvallis, S. Altona, and S. I 4,[5],12:i:- (n = 1 each). SPI-8, known to be involved with resistance to bacteriocins (Amavisit et al., 2003) was identified in S. Krefeld (n = 4), S. Rissen and S. Corvallis (n = 3 each).

Table 1. Serotypes, AMR genes and Salmonella pathogenicity islands identified in non-typhoidal Salmonella enterica isolates collected from Cambodian informal marketsa.
Antimicrobial Susceptibility
In total, 1,134 antibiotic tests were performed (i.e., 14 tested antimicrobial compounds for each one of the 81 isolates; Table 2) and resistance to different antimicrobials was seen in 121 instances (i.e., phenotype resistant; Table 2). Among the 81 characterized isolates, the most common phenotypically observed resistances were to tetracycline and beta-lactam (47%; n = 38 each), aminoglycosides (22%; n = 18), phenicol (17%; n = 14), and trimethoprim (16%; n = 13). No resistance was observed for quinolone, macrolide and sulfonamide (Table 3). Seventeen percent of isolates (n = 14) displayed resistance to three classes of antibiotics, and 16% (n = 13) had resistance to four of the eight classes tested. Overall, 40 isolates (49%) presented no resistance to any of the tested antimicrobial compounds. The phenotypic resistance profile by serotype and antibiotic class is shown in Figure 1. S. Rissen had the highest resistance diversity, displaying resistance to five classes of antibiotics, followed by S. Derby (4 classes), S. Krefeld, S. I 4,[5],12:i:-, S. Typhimurium (3 classes each), S. Corvallis, S. Altona, S. Anatum (2 classes each), and S. Mbandaka, S. Hvittingfoss, and S. Virchow (1 class each).
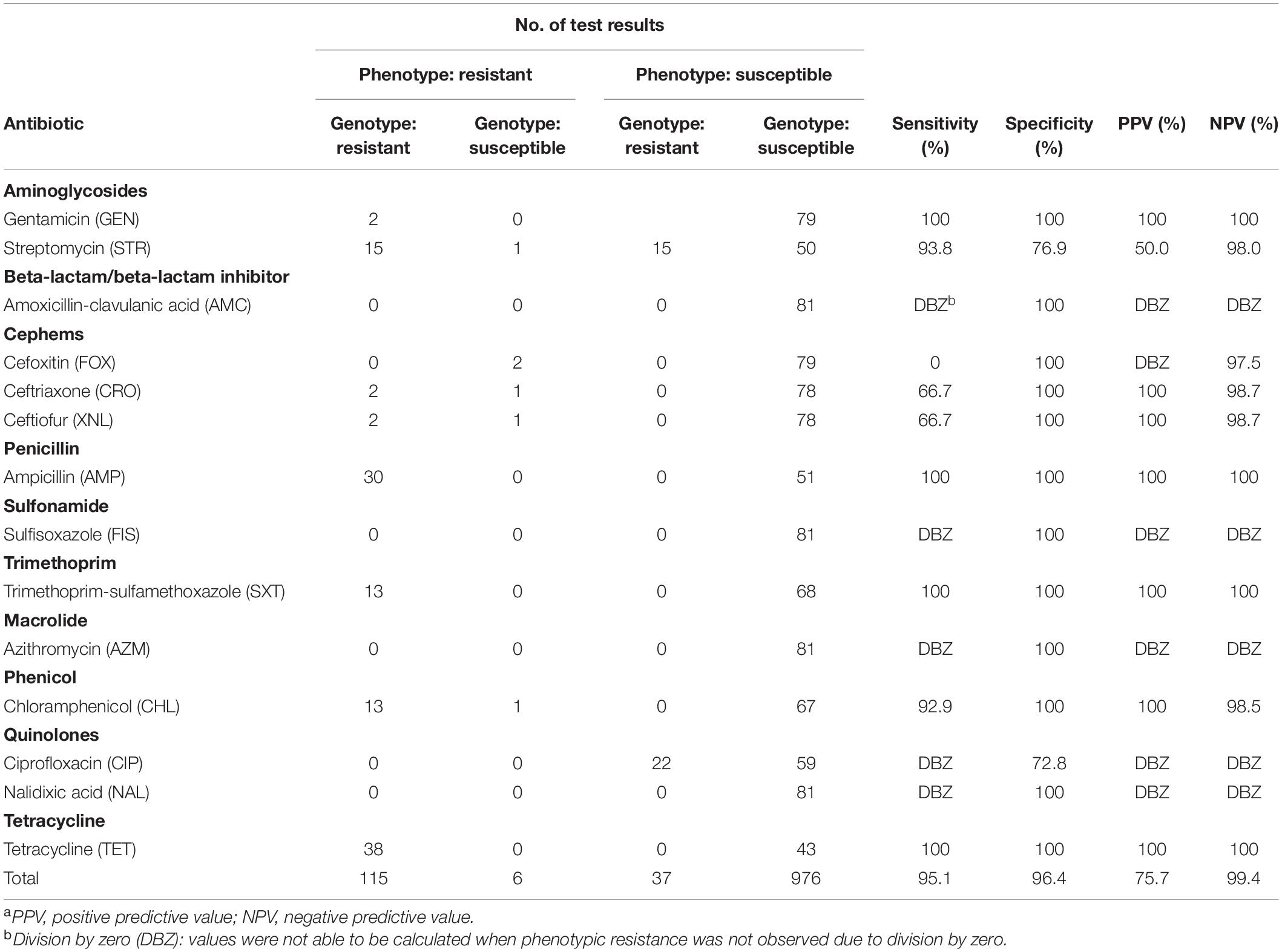
Table 2. Summary comparison of phenotypic and genotypic test results from 81 Salmonella enterica isolates from Cambodian informal markets, 2018–2019a.
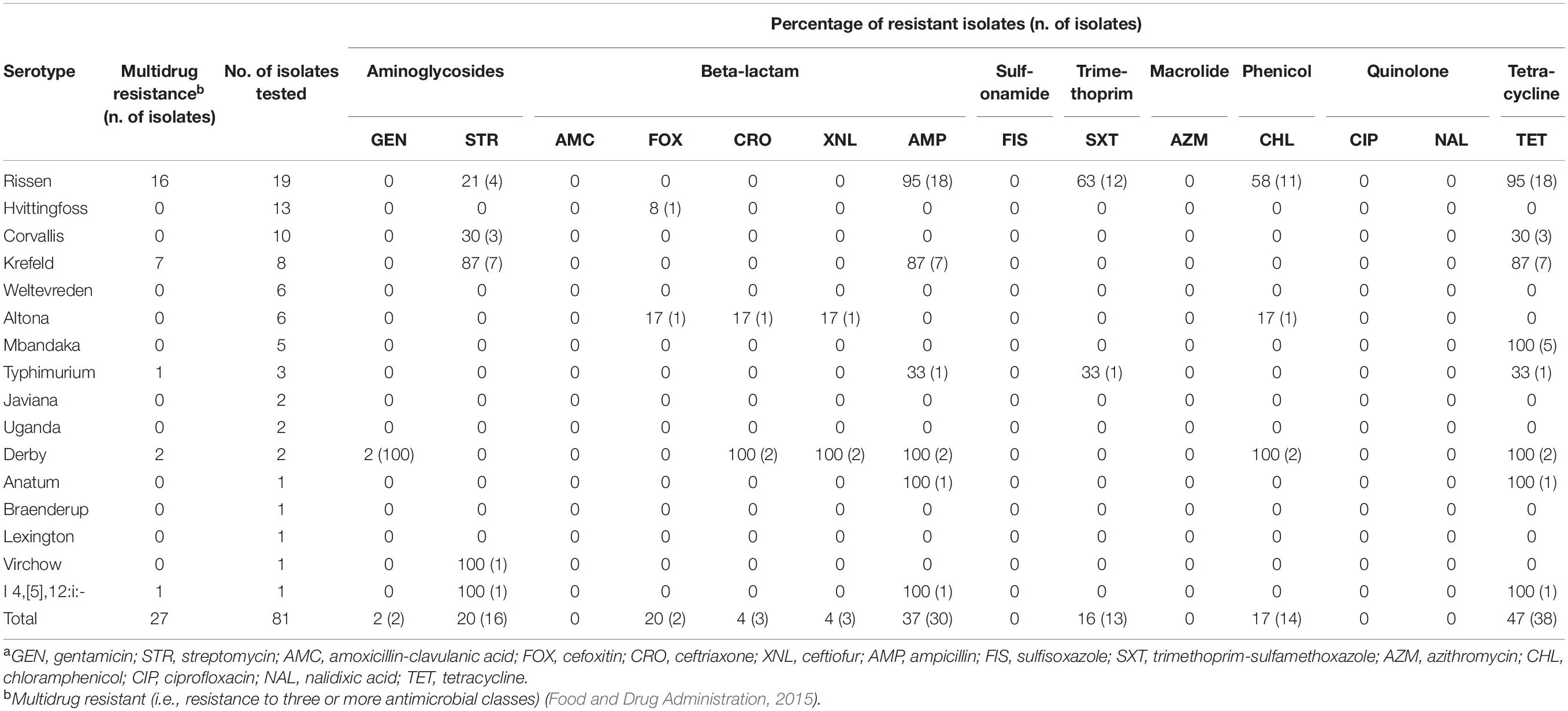
Table 3. Frequency of phenotypic resistance among non-typhoidal Salmonella enterica isolates collected from Cambodian informal marketsa.
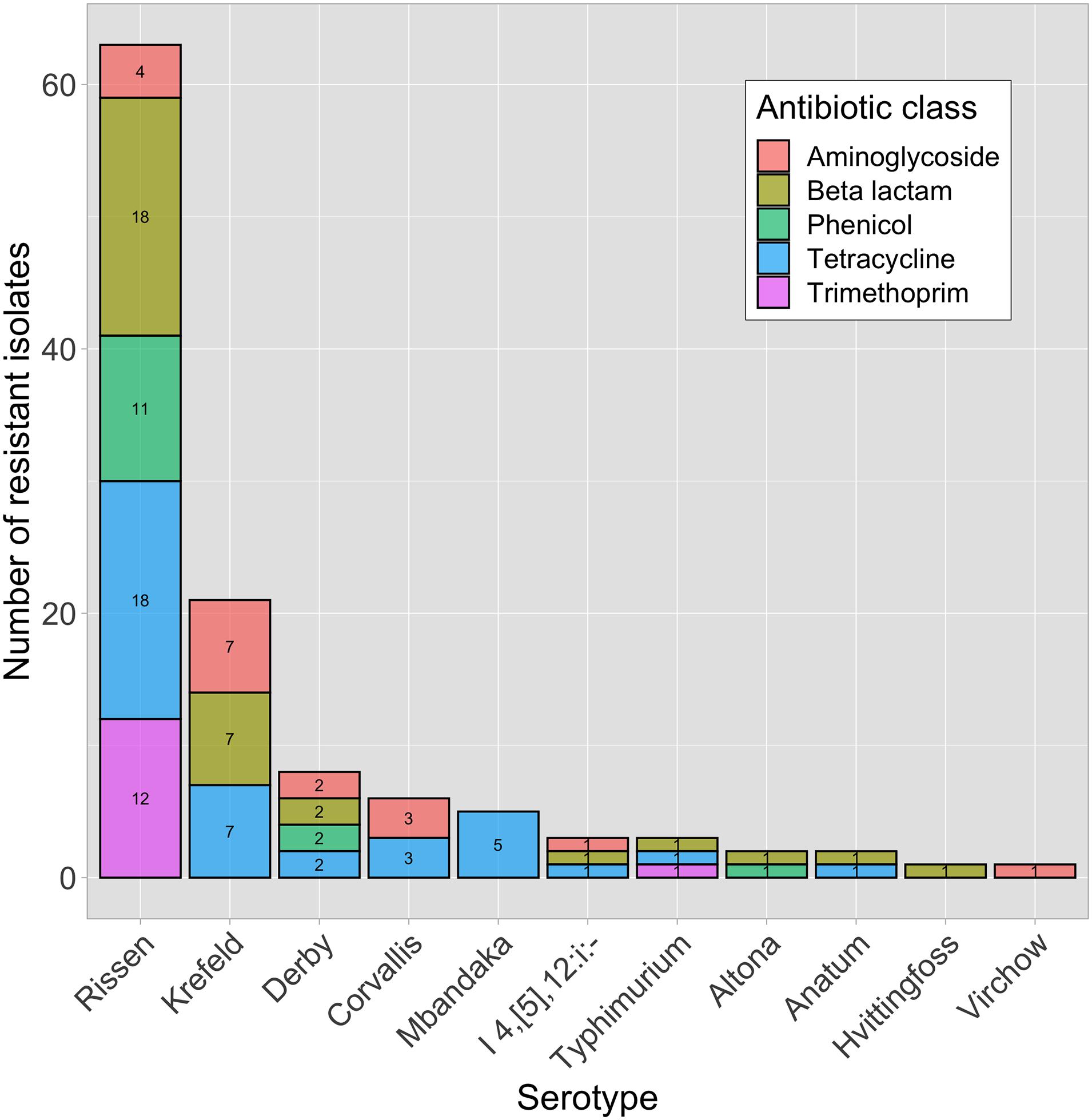
Figure 1. Phenotypically resistant Salmonella enterica isolates (n = 41) within different serotypes and antibiotic classes.
Comparison Between Genotypic and Phenotypic AMR Profiles
The overall concordance between phenotypic resistance with the presence of known AMR genes was 96.2%, with genotype agreeing with phenotype for 1,091 of 1,134 of the phenotypic tests. Associated genes were predicted to cause resistance in all but six of the 121 instances in which antibiotic tests indicated resistance. These six instances of mismatch were related to beta-lactams (n = 4), aminoglycosides (n = 1), and phenicol (n = 1) and were observed in three serotypes (S. Altona, S. Hvittingfoss, and S. Virchow). This resulted in an overall sensitivity of 95% (115/121) (Table 2).
Among the 1,013 phenotypically susceptible test results, 37 belonged to isolates for which AMR genes were detected by WGS but no phenotypic resistance was observed (Table 2). Among the AMR genes detected in susceptible isolates, 15 isolates encoded aminoglycoside resistance and 22 isolates encoded quinolone resistance. This resulted in an overall specificity of 96.3% (976/1013) (Table 2). Results for each specific class of antibiotic are described in detail below and shown in Table 2.
Resistance to β-Lactams
A total of 3 genes encoding beta-lactamases were identified, with the most common being blaTEM–1 (present in 35% of the isolates), followed by two minor genes (blaCTX–M–14, blaLAP–2) each present in less than 3% of isolates (Table 1). Genotypes predicted phenotypes with 89.5% sensitivity and 100% specificity.
Resistance to Quinolone
Quinolone resistance is commonly observed within multiple mutations of the quinolone resistance-determining regions (QRDR) (i.e., gyrA, gyrB, parC, and parE) and/or one or multiple plasmid-mediated quinolone resistance (PMQR) genes (Yoshida et al., 1991; Saenz, 2003; Sorlozano et al., 2007; Hopkins et al., 2008). QRDR mutations and/or its combinations typically confer resistance to nalidixic acid and ciprofloxacin, respectively (McDermott et al., 2016; Neuert et al., 2018). Resistance mechanisms that involve a single plasmid-mediated gene typically do not confer resistance to ciprofloxacin or nalidixic acid, except when additional PMQR genes and/or QRDR mutations are present (Jacoby et al., 2014). In this study, 22 isolates carried only one PMQR gene (i.e., qnrS1) (Table 1). Resistance to ciprofloxacin or nalidixic acid was not observed in this study (thus no sensitivity was calculated for this class). Genotypic prediction for resistance resulted in a specificity of 86.4%.
Resistance to Aminoglycosides
Six different aminoglycoside resistance alleles were identified (Table 1). Aminoglycoside phosphotransferase genes aph(6)-Id and aph(3″)-Ib (n = 17 each), and aph(3′)-Ia (n = 6) were identified. Genes encoding aminoglycoside adenylyltransferases were identified in 14 isolates, most commonly aadA1 (n = 14) and aadA2 (n = 13). Further, two isolates carried an aminoglycoside acetyltransferase aac(3) variant [i.e., aac(3)-IId] that is commonly associated with resistance to gentamicin (Tyson et al., 2017). These latter two isolates were also observed to be phenotypically resistant to the same drug. Sensitivity and specificity for genotypic-phenotypic correlations were 94.4 and 89.6%, respectively.
Resistance to Phenicols
A combination of two genes (floR and cmlA1) from the multidrug efflux pumps family were identified in 13.6% of the isolates (n = 11) while two percent of the isolates only presented floR (n = 2). Genotypes predicted phenotypes with 92.9% sensitivity and 100% specificity.
Resistance to Tetracyclines
tet(A) and tet(B) were identified herein, among the at least 40 distinct tet allelles described to date (Nguyen et al., 2014). Tetracycline resistance genes were found in 47% of the isolates (n = 38), mostly represented by tet(A) (n = 30), followed by the efflux pump-encoding tet(B) (n = 8). Genotypes predicted phenotypes with 100% sensitivity and 100% specificity.
Resistance to Sulfonamides and Trimethoprim
Sulfonamide resistance genes were found in 33.3% of the isolates (n = 27). Two resistance genes (sul2, and sul3) were identified. Out of the 27 isolates, 24 carried sul2, and 14 carried sul3. Thirteen percent of the isolates (n = 11) had a combination of both sul genes. Sixteen percent of the isolates (n = 13) carried the dihydrofolate reductase resistance gene dfrA12. Genotypes predicted phenotypes with a sensitivity of 100% and a specificity of 100%.
Resistance to Macrolides
Even though no macrolide resistance genes were identified, all isolates were phenotypically susceptible to azithromycin. Genotypic specificity for azithromycin was 100%.
Multidrug Resistance
Among the 81 isolates analyzed, 27 (33%) were classified as multidrug resistant (MDR) as they presented resistance to three or more antimicrobial classes (Food and Drug Administration, 2015).
MDR was observed in 84% of S. Rissen (n = 16), 87% of S. Krefeld (n = 7), 100% of S. Derby (n = 2), 33% S. Typhimurium (n = 1), and 100% of S. I 4,[5],12:i:- (n = 1), shown in Table 3.
Discussion
This study reports on the identification and characterization of the genotypic and phenotypic AMR profiles from S. enterica serotypes isolated from environmental samples from informal markets in Cambodia. Overall, a high diversity of resistance genes encoding resistance to several classes of antibiotics was identified.
Thirty three percent of S. enterica isolates (n = 27) presented resistance to at least three classes of antibiotics. Resistance to beta-lactams, tetracyclines, aminoglycosides, phenicol and trimethoprim were the most common among the isolates. Similarly, previous studies conducted in informal markets in Cambodia have shown that S. enterica isolates were resistant to β-lactams, tetracyclines, phenicol, and trimethoprim antibiotics (Trongjit et al., 2017; Nadimpalli et al., 2019). The development and spread of AMR among S. enterica serotypes are particularly important when found in retail environments, such as informal markets. Contaminated environments may be the source of cross-contamination between food products and environmental surfaces. When cross-contamination occurs, AMR pathogens become a threat to public health since the effectiveness of antibiotic therapy may be reduced (Thanner et al., 2016).
Twenty-seven isolates (33%) were multidrug resistant. Previous studies in Cambodia revealed that MDR S. enterica was found in various sample types (e.g., chicken, pork, and fish) and ranged from 23 to 52% (Trongjit et al., 2017; Nadimpalli et al., 2019). The presence of genes that are commonly associated with MDR to third generation cephalosporins (i.e., a beta-lactam class of antibiotics) is a growing problem as third generation cephalosporins are particularly important as they are often used to treat Salmonellosis in humans and are classified as critically important for human health (World Health Organization, 2017). In this study, 2.5% of the isolates (n = 2) demonstrated resistance to third generation cephalosporins (e.g., ceftriaxone and ceftiofur). Resistance to third generation cephalosporins were identified in hospitalized children (8.1%) and adults (2.7%) with S. enterica infections in Cambodia (Vlieghe et al., 2012; Fox-Lewis et al., 2018).
Phenotypic testing confirmed MDR in 84% of S. Rissen isolates and resistance to five classes of antibiotics, namely beta-lactams, tetracyclines, trimethoprim, phenicol, and aminoglycosides. Similarly, Nadimpalli et al. (2019) identified MDR S. Rissen from pork, chicken and fish samples collected from informal markets in Cambodia, revealing that MDR among S. Rissen is recurrent in the market environment. This scenario is especially concerning in low and middle-income countries (LMICs, such as Cambodia) since antibiotic resources are limited, and treatment options are compromised when strains are resistant to several classes of antibiotics.
Thirty percent of S. Corvallis isolates exhibited genes encoding resistance to four classes of antibiotics: aminoglycosides, quinolones, sulfonamides, and tetracyclines. However, phenotypic resistance was only observed for aminoglycosides and tetracyclines. Previous studies have reported similar AMR profiles in S. Corvallis isolated from various sources (i.e., informal markets, food products, and patients) from Cambodia and Thailand (Trongjit et al., 2017). The resistance pattern observed over multiple studies demonstrates that the AMR profile of S. Corvallis has remained similar and reoccurring over the past decade in Southeast Asian countries (e.g., Cambodia and Thailand). S. Corvallis has been reported as a common causative agent for travel-associated salmonellosis in patients that traveled to Southeast Asia, indicating that this serotype has recurrently caused disease (Ekdahl et al., 2005; Taguchi et al., 2009; Koch et al., 2011; Nakakubo et al., 2019).
Resistance was observed for three classes of antibiotics (i.e., beta-lactam, tetracycline, trimethoprim) in 33% of S. Typhimurium isolates. Similar rates of MDR have been reported from poultry and pork samples collected from informal markets and retail shops in Phnom Penh, Cambodia (Lay et al., 2011; Nadimpalli et al., 2019). In neighboring Thailand, however, higher rates (over 50%) of MDR isolates have been reported in human specimens, indicating that this strain is highly prevalent in patients (Sirichote et al., 2010).
S. Derby exhibited MDR in 100% of the isolates, which harbored six different genes representing six classes of antibiotics. However, resistance was observed only to four antibiotic classes: aminoglycosides, beta-lactam, phenicol, and tetracycline. These results are similar to previous studies from Cambodia (Nadimpalli et al., 2019), China (Yang et al., 2019), and Thailand (Sirichote et al., 2010; Trongjit et al., 2017), revealing that MDR is commonly found among S. Derby strains in Southeast Asia. Historically, S. Derby is mainly recovered from pork, potentially indicating that the uncontrolled use of antibiotics in the pig production chain plays an important role in the antimicrobial resistance selection pressure (Thanner et al., 2016).
The monophasic variant of S. Typhimurium, I 4,[5],12:i:-, has increasingly been associated with cases of human disease in several countries around the world (Moreno Switt et al., 2009). Emerging MDR clades have raised concerns among public health authorities (Elnekave et al., 2018). Sources of contamination were attributed to beef, pork, and chicken products, suggesting that these animal-based food products may serve as critical vectors for human contamination (Moreno Switt et al., 2009; Soyer et al., 2009). In this study, S. I 4,[5],12: i:- was resistant to aminoglycosides, beta-lactams, and tetracycline. Multidrug resistance among S. I 4,[5],12:i:- has also been reported from pork samples in Cambodia (Nadimpalli et al., 2019), Australia (Arnott et al., 2018), Spain (Echeita et al., 1999), and Germany (Hauser et al., 2010).
Genotypes predicted phenotypes with 100% sensitivity and 100% specificity for resistance to tetracyclines, sulfonamides and trimethoprim, and macrolides. Sensitivity of genotypic prediction was less than 100% for resistance to phenicols (92.9%), β-lactams (89.5%), quinolone (phenotypic resistance not observed), and aminoglycosides (94.4%). Specifically, in 37 isolates AMR genes were detected by WGS but no phenotypic resistance was observed. Among these resistance genes, 15 encoded aminoglycoside resistance (single variant or a combination of aadA1, aadA2, aph(6)-Id, aph(3″)-Ib), and 22 encoded quinolone resistance (qnrS1). However, those isolates exhibited phenotypic susceptibility to streptomycin (i.e., seen in 14 S. Rissen, 1 S. Anatum) and ciprofloxacin (i.e., seen in 11 S. Rissen, 8 S. Corvallis, 2 S. Derby, 1 S. Anatum). Genotype and phenotype AMR mismatches have also been observed in other studies with Salmonella as well as in other species, such as Escherichia coli, Listeria monocytogenes, and Vibrio parahaemolyticus (Deekshit et al., 2012; Ortiz et al., 2014; Lou et al., 2016; Do Nascimento et al., 2017; Neuert et al., 2018). When resistance genes are plasmid-encoded and phenotypic susceptibility testing is performed, retrospectively, these plasmids may be lost during storage and sub-culture. Additionally, these genes may be considered “silent” and several factors could be affecting the non-expression of resistance (e.g., growth medium, bacterial cell density, temperature, oxidative stress, etc.) (Udekwu et al., 2009; Poole, 2012; Lou et al., 2016; Do Nascimento et al., 2017). Therefore, even if AMR genes were detected previously by WGS, retrospective phenotypic testing on a different colony may result in a susceptible result (Neuert et al., 2018).
Additionally, three isolates (e.g., S. Altona, S. Hvittingfoss, and S. Virchow) were genotypically predicted to be susceptible but exhibited phenotypic resistance to beta-lactams, aminoglycosides, and phenicol. All three isolates are listed on NCBI PD as having the mdsA and mdsB genes, encoding for multidrug transporter of S. enterica. The mdsABC complex has been linked to resistance versus different drugs and toxins (Thomas et al., 2017). MdsABC, or GesABC, has been described as able to export beta-lactams, chloramphenicol, and thiamphenicol (Alcock et al., 2019; Guitor et al., 2019). Overall, this scenario highlights that alternate or emerging antimicrobial resistance mechanisms may be present, and mismatches are likely occurring due to unknown mechanisms not being included in the reference gene database used for genotypic prediction. Similar results have been observed previous studies with non-typhoidal S. enterica (McDermott et al., 2016; Neuert et al., 2018).
A total of 10 SPIs [SPI-1, 3–5, 8, 9, 12–14, and centisome 63 (C63PI)] were detected in 73% of isolates. Pathogenicity islands are especially important due to their capability of carrying genes encoding virulence factors that contribute to the adhesion, invasion, and infection process (Marcus et al., 2000). More specifically, C63PI, an iron transport system in SPI-1, is an important region of the Salmonella chromosome that mediates the entry of this species into the host cell (Ginocchio et al., 1997). To the best of our knowledge, this is the first study to explore the presence and abundance of SPIs in environmental samples from informal markets in Cambodia. SPI-1 was identified in 14% of the isolates. SPI-1, located at centisome 63 encoding predicted type III secretion system (T3SS), is a fundamental complex of genetic elements necessary during the initial stages of infection (Hardt et al., 1998; Hansen-Wester and Hensel, 2001). The T3SS machinery, which is an extremely important part of pathogenesis, facilitates the invasion by its unique needle apparatus that is utilized to deliver effector proteins into the host cell cytoplasm and create proinflammatory responses (Foley and Lynne, 2008; Meena et al., 2019). Interestingly, S. Krefeld, S. Derby and S. I 4,[5],12:i:- (n = 1 each) presented a combination of both SPI-1 and MDR, indicating that such strains are extremely important to public health due to their ability to successfully establish an infection and to resist several classes of antibiotics. The combination of an important invasion system [i.e., T3SS (SPI-1)] and the resistance profile to several different antimicrobial classes, demonstrates the importance of controlling pathogen contamination in the food chain.
Overall, this study indicates that AMR S. enterica serotypes are prevalent in informal market environments in Cambodia. A small percentage of the isolates was resistant to antibiotic classes commonly used to treat S. enterica infections in humans. Our findings revealed the presence of important SPIs related to the ability of bacteria for successful adhesion, invasion, and host cell infection. The combined effect of specific SPIs with the diversity and distribution of AMR phenotypes highlights the need for improvement in food safety practices within informal markets and the need for antibiotic stewardship in agriculture and livestock production systems.
While this study elucidated the diversity of predicted AMR genes and phenotypic resistance profiles of S. enterica from environmental samples, future studies are needed to identify the dissemination of AMR profiles among the food production chain. Future source-attribution studies should be conducted to investigate the dissemination of resistance among agriculture, animal food systems, and the market environment.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA628951.
Author Contributions
CS, PC, VT, and JV conceived and designed the study. CS, SL, LB, and PC participated to data curation. LB and CS analyzed genotypic and phenotypic data. SL, JM, MB, SG, and JV contributed to reagents, materials, and analysis tools. CS, JV, RP, JK, and SG supervised the study and provided laboratory guidance. CS and SL prepared the manuscript. All authors revised the manuscript and approved the submitted version.
Funding
This project was supported in part by an appointment to the Research Participation Program at the Center for Food Safety and Applied Nutrition (CFSAN), U.S. Food and Drug Administration, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and FDA. This project was also supported by FDA-CFSAN through its GenomeTrakr program (https://www.fda.gov/food/whole-genome-sequencing-wgs-program/genometrakr-network). The use of trade names and commercial sources is for identification purposes only and does not imply endorsement.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the faculty and students of the Royal University of Agriculture-Phnom Penh, Cambodia for their valuable insights and resources; which were essential in the completion of this project. SL was supported in part by an appointment to the Joint Institute for Food Safety and Applied Nutrition (JIFSAN).
Footnotes
- ^ https://github.com/tseemann/shovill
- ^ http://denglab.info/SeqSero
- ^ https://cge.cbs.dtu.dk/services/
References
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2019). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Amavisit, P., Lightfoot, D., Browning, G. F., and Markham, P. F. (2003). Variation between pathogenic serovars within Salmonella athogenicity islands. J. Bacteriol. 185, 3624–3635. doi: 10.1128/JB.185.12.3624-3635.2003
Arnott, A., Wang, Q., Bachmann, N., Sadsad, R., Biswas, C., Sotomayor, C., et al. (2018). Multidrug-resistant Salmonella enterica 4,[5],12:i:- sequence type 34, New South Wales, Australia, 2016–2017. Emerg. Infect. Dis. 24, 751–753. doi: 10.3201/eid2404.171619
Barlow, R. S., McMILLAN, K. E., Duffy, L. L., Fegan, N., Jordan, D., and Mellor, G. E. (2015). Prevalence and antimicrobial resistance of Salmonella and Escherichia coli from Australian cattle populations at slaughter. J. Food Prot. 78, 912–920. doi: 10.4315/0362-028X.JFP-14-476
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Clinical and Laboratory Standards Institute (2020). Performance Standards for Antimicrobial Susceptibility Testing. Available online at: https://clsi.org/standards/products/microbiology/documents/m100/ (accessed July 15, 2020).
Cockerill, F. R., and Clinical and Laboratory Standards Institute (2012). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute.
Deekshit, V. K., Kumar, B. K., Rai, P., Srikumar, S., Karunasagar, I., and Karunasagar, I. (2012). Detection of class 1 integrons in Salmonella Weltevreden and silent antibiotic resistance genes in some seafood-associated nontyphoidal isolates of Salmonella in south-west coast of India: detection of class 1 integrons in Salm, Weltevreden. J. Appl. Microbiol. 112, 1113–1122. doi: 10.1111/j.1365-2672.2012.05290.x
Do Nascimento, V., Day, M. R., Doumith, M., Hopkins, K. L., Woodford, N., Godbole, G., et al. (2017). Comparison of phenotypic and WGS-derived antimicrobial resistance profiles of enteroaggregative Escherichia coli isolated from cases of diarrhoeal disease in England, 2015–16. J. Antimicrob. Chemother. 72, 3288–3297. doi: 10.1093/jac/dkx301
Echeita, M. A., Aladueña, A., Cruchaga, S., and Usera, M. A. (1999). Emergence and spread of an atypical Salmonella enterica subsp. enterica serotype 4,5,12:i:- strain in Spain. J. Clin. Microbiol. 37, 3425–3425. doi: 10.1128/JCM.37.10.3425-3425.1999
Ekdahl, K., de Jong, B., Wollin, R., and Andersson, Y. (2005). Travel-associated non-typhoidal salmonellosis: geographical and seasonal differences and serotype distribution. Clin. Microbiol. Infect. 11, 138–144. doi: 10.1111/j.1469-0691.2004.01045.x
Elnekave, E., Hong, S., Mather, A. E., Boxrud, D., Taylor, A. J., Lappi, V., et al. (2018). Salmonella enterica serotype 4,[5],12:i:- in swine in the United States midwest: an emerging multidrug-resistant clade. Clin. Infect. Dis. 66, 877–885. doi: 10.1093/cid/cix909
Feldgarden, M., Brover, V., Haft, D. H., Prasad, A. B., Slotta, D. J., Tolstoy, I., et al. (2019). Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63:e00483-19.
Finger, J. A. F. F., Baroni, W. S. G. V., Maffei, D. F., Bastos, D. H. M., and Pinto, U. M. (2019). Overview of foodborne disease outbreaks in Brazil from 2000 to 2018. Foods 8:434. doi: 10.3390/foods8100434
Foley, S. L., and Lynne, A. M. (2008). Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J. Anim. Sci. 86, E173–E187. doi: 10.2527/jas.2007-0447
Food and Drug Administration (2015). National Antimicrobial Resistance Monitoring System – Enteric Bacteria (NARMS): 2011 Executive Report. Available online at: https://www.fda.gov/media/89149/download (accessed August 28, 2020).
Ford, L., Wang, Q., Stafford, R., Ressler, K.-A., Norton, S., Shadbolt, C., et al. (2018). Seven Salmonella Typhimurium outbreaks in Australia linked by trace-back and whole genome sequencing. Foodborne Pathog. Dis. 15, 285–292. doi: 10.1089/fpd.2017.2353
Fox-Lewis, A., Takata, J., Miliya, T., Lubell, Y., Soeng, S., Sar, P., et al. (2018). Antimicrobial resistance in invasive bacterial infections in hospitalized children, Cambodia, 2007–2016. Emerg. Infect. Dis. 24, 841–851. doi: 10.3201/eid2405.171830
Ginocchio, C. C., Rahn, K., Clarke, R. C., and Galán, J. E. (1997). Naturally occurring deletions in the centisome 63 pathogenicity island of environmental isolates of Salmonella spp. Infect. Immun. 65, 1267–1272. doi: 10.1128/iai.65.4.1267-1272.1997
Guitor, A. K., Raphenya, A. R., Klunk, J., Kuch, M., Alcock, B., Surette, M. G., et al. (2019). Capturing the resistome: a targeted capture method to reveal antibiotic resistance determinants in metagenomes. Antimicrob. Agents Chemother. 64:e01324-19. doi: 10.1128/AAC.01324-19
Hansen-Wester, I., and Hensel, M. (2001). Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3, 549–559. doi: 10.1016/S1286-4579(01)01411-3
Hardt, W.-D., Urlaub, H., and Galan, J. (1998). A substrate of the centisome 63 type III protein secretion system of Salmonella Typhimurium is encoded by a cryptic bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 95, 2574–2579. doi: 10.1073/pnas.95.5.2574
Hauser, E., Tietze, E., Helmuth, R., Junker, E., Blank, K., Prager, R., et al. (2010). Pork contaminated with Salmonella enterica serovar 4,[5],12:i:-, an emerging health risk for humans. Appl. Environ. Microbiol. 76, 4601–4610. doi: 10.1128/AEM.02991-09
Hoffmann, S., Batz, M. B., and Morris, J. G. (2012). Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens†. J. Food Prot. 75, 1292–1302. doi: 10.4315/0362-028X.JFP-11-417
Hopkins, K. L., Day, M., and Threlfall, E. J. (2008). Plasmid-mediated quinolone resistance in Salmonella enterica, United Kingdom. Emerg. Infect. Dis. 14, 340–342. doi: 10.3201/eid1402.070573
Jacoby, G. A., Strahilevitz, J., and Hooper, D. C. (2014). Plasmid-mediated quinolone resistance. Microbiol. Spectr. 2. doi: 10.1128/microbiolspec.PLAS-0006-2013
Jans, C., Sarno, E., Collineau, L., Meile, L., Stärk, K. D. C., and Stephan, R. (2018). Consumer exposure to antimicrobial resistant bacteria from food at Swiss retail level. Front. Microbiol. 9:362. doi: 10.3389/fmicb.2018.00362
Jiang, M., Zhu, F., Yang, C., Deng, Y., Kwan, P. S. L., Li, Y., et al. (2020). Whole-genome analysis of Salmonella enterica serovar Enteritidis Isolates in outbreak linked to online food delivery, Shenzhen, China, 2018. Emerg. Infect. Dis. 26, 789–792. doi: 10.3201/eid2604.191446
Koch, K., Kristensen, B., Holt, H. M., Ethelberg, S., Mølbak, K., and Schønheyder, H. C. (2011). International travel and the risk of hospitalization with non-typhoidal Salmonella bacteremia. A Danish population-based cohort study, 1999-2008. BMC Infect. Dis. 11:277. doi: 10.1186/1471-2334-11-277
Kuhn, M. (2020). Caret: Classification and Regression Training. Available online at: https://CRAN.R-project.org/package=caret (accessed October 12, 2020).
Lay, K. S., Vuthy, Y., Song, P., Phol, K., and Sarthou, J. L. (2011). Prevalence, numbers and antimicrobial susceptibilities of Salmonella serovars and Campylobacter spp. in retail poultry in Phnom Penh, Cambodia. J. Vet. Med. Sci. 73, 325–329. doi: 10.1292/jvms.10-0373
Lou, Y., Liu, H., Zhang, Z., Pan, Y., and Zhao, Y. (2016). Mismatch between antimicrobial resistance phenotype and genotype of pathogenic Vibrio parahaemolyticus isolated from seafood. Food Control 59, 207–211. doi: 10.1016/j.foodcont.2015.04.039
Maradiaga, M., Echeverry, A., Orellana, D., Miller, M., and Brashears, M. (2017). Antibiotic resistance of Salmonella enterica isolated from whole-muscle beef cuts and beef carcasses in Mexico. Meat Muscle Biol. 1, 132–132. doi: 10.22175/rmc2016.128
Marcus, S. L., Brumell, J. H., Pfeifer, C. G., and Finlay, B. B. (2000). Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2, 145–156. doi: 10.1016/S1286-4579(00)00273-2
Marshall, B. M., and Levy, S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24, 718–733. doi: 10.1128/CMR.00002-11
McDermott, P. F., Tyson, G. H., Kabera, C., Chen, Y., Li, C., Folster, J. P., et al. (2016). Whole-Genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob. Agents Chemother. 60, 5515–5520. doi: 10.1128/AAC.01030-16
Meena, M., Swapnil, P., Zehra, A., Aamir, M., Dubey, M. K., Patel, C. B., et al. (2019). “Virulence factors and their associated genes in microbes,” in New and Future Developments in Microbial Biotechnology and Bioengineering, eds B. S. Harikesh, V. K. Gupta, and S. Jogaiah (Amsterdam: Elsevier), 181–208. doi: 10.1016/B978-0-444-63503-7.00011-5
Mekong Institute (2019). MI, MoH Support FORT Team Responding to Foodborne Disease Outbreak. Available online at: https://www.mekonginstitute.org/news-activities/detail/2019/12/04/mi-moh-support-fort/ (accessed April 24, 2020).
Moreno Switt, A. I., Soyer, Y., Warnick, L. D., and Wiedmann, M. (2009). Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i:–. Foodborne Pathog. Dis. 6, 407–415.
Nadimpalli, M., Fabre, L., Yith, V., Sem, N., Gouali, M., Delarocque-Astagneau, E., et al. (2019). CTX-M-55-type ESBL-producing Salmonella enterica are emerging among retail meats in Phnom Penh, Cambodia. J. Antimicrob. Chemother. 74, 342–348. doi: 10.1093/jac/dky451
Nakakubo, S., Nagaoka, K., Suzuki, M., Konno, S., Shibue, Y., Ikeda, T., et al. (2019). A case of primary bacteraemia caused by Salmonella enterica serovar Corvallis in an immunocompetent adult after travel to Southeast Asia. Access Microbiol. 1:e000009. doi: 10.1099/acmi.0.000009
Neuert, S., Nair, S., Day, M. R., Doumith, M., Ashton, P. M., Mellor, K. C., et al. (2018). Prediction of phenotypic antimicrobial resistance profiles from whole genome sequences of non-typhoidal Salmonella enterica. Front. Microbiol. 9:592. doi: 10.3389/fmicb.2018.00592
Nguyen, F., Starosta, A. L., Arenz, S., Sohmen, D., Dönhöfer, A., and Wilson, D. N. (2014). Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 395, 559–575. doi: 10.1515/hsz-2013-0292
Ortiz, S., López, V., and Martínez-Suárez, J. V. (2014). The influence of subminimal inhibitory concentrations of benzalkonium chloride on biofilm formation by Listeria monocytogenes. Int. J. Food Microbiol. 189, 106–112. doi: 10.1016/j.ijfoodmicro.2014.08.007
Poole, K. (2012). Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. 67, 2069–2089. doi: 10.1093/jac/dks196
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing
Roesel, K., and Grace, D. (2014). Food Safety and Informal Markets. Routledge. Available online at: https://cgspace.cgiar.org/handle/10568/42438 (accessed September 17, 2019).
Saenz, Y. (2003). Mutations in gyrA and parC genes in nalidixic acid-resistant Escherichia coli strains from food products, humans and animals. J. Antimicrob. Chemother. 51, 1001–1005. doi: 10.1093/jac/dkg168
Schwan, C. L., Desiree, K., Bello, N. M., Bastos, L., Hok, L., Phebus, R. K., et al. (2020a). Prevalence of Salmonella enterica isolated from food contact and non-food contact surfaces in Cambodian informal markets. J. Food Prot. 84, 73–79. doi: 10.4315/JFP-20-112
Schwan, C. L., Trinetta, V., Balkey, M., Cook, P. W., Phebus, R. K., Gragg, S., et al. (2020b). Draft genome sequences of 81 Salmonella enterica strains from informal markets in Cambodia. Microbiol. Resour. Announc. 9:e00773-20. doi: 10.1128/MRA.00773-20
Sirichote, P., Bangtrakulnonth, A., Tianmanee, K., Unahalekhaka, A., Oulai, A., Chittaphithakchai, P., et al. (2010). Serotypes and antimicrobial resistance of Salmonella enterica spp. in central Thailand, 2001-2006. Southeast Asian J. Trop. Med. Public Health 41, 1405–1415.
Sorlozano, A., Gutierrez, J., Jimenez, A., de Dios Luna, J., and Martinez, J. L. (2007). Contribution of a new mutation in pare to quinolone resistance in extended-spectrum-lactamase-producing Escherichia coli isolates. J. Clin. Microbiol. 45, 2740–2742. doi: 10.1128/JCM.01093-07
Soyer, Y., Moreno Switt, A., Davis, M. A., Maurer, J., McDonough, P. L., Schoonmaker-Bopp, D. J., et al. (2009). Salmonella enterica serotype 4,5,12:i:-, an emerging Salmonella serotype that represents multiple distinct clones. J. Clin. Microbiol. 47, 3546–3556. doi: 10.1128/JCM.00546-09
Taguchi, M., Kawahara, R., Seto, K., Inoue, K., Hayashi, A., Yamagata, N., et al. (2009). Plasmid-mediated quinolone resistance in Salmonella isolated from patients with overseas travelers’ diarrhea in Japan. Jpn. J. Infect. Dis. 62, 312–314.
Thanner, S., Drissner, D., and Walsh, F. (2016). Antimicrobial resistance in agriculture. mBio 7:e02227-15. doi: 10.1128/mBio.02227-15
Thomas, M., Fenske, G. J., Antony, L., Ghimire, S., Welsh, R., Ramachandran, A., et al. (2017). Whole genome sequencing-based detection of antimicrobial resistance and virulence in non-typhoidal Salmonella enterica isolated from wildlife. Gut Pathog. 9:66. doi: 10.1186/s13099-017-0213-x
Timme, R. E., Rand, H., Sanchez Leon, M., Hoffmann, M., Strain, E., Allard, M., et al. (2018). GenomeTrakr proficiency testing for foodborne pathogen surveillance: an exercise from 2015. Microb. Genomics 4:e000185. doi: 10.1099/mgen.0.000185
Trongjit, S., Angkititrakul, S., Tuttle, R. E., Poungseree, J., Padungtod, P., and Chuanchuen, R. (2017). Prevalence and antimicrobial resistance in Salmonella enterica isolated from broiler chickens, pigs and meat products in Thailand-Cambodia border provinces: AMR in Salmonella enterica. Microbiol. Immunol. 61, 23–33. doi: 10.1111/1348-0421.12462
Tyson, G. H., Zhao, S., Li, C., Ayers, S., Sabo, J. L., Lam, C., et al. (2017). Establishing genotypic cutoff values to measure antimicrobial resistance in Salmonella. Antimicrob. Agents Chemother. 61:e02140-16. doi: 10.1128/AAC.02140-16
Udekwu, K. I., Parrish, N., Ankomah, P., Baquero, F., and Levin, B. R. (2009). Functional relationship between bacterial cell density and the efficacy of antibiotics. J. Antimicrob. Chemother. 63, 745–757. doi: 10.1093/jac/dkn554
Vlieghe, E. R., Phe, T., De Smet, B., Veng, C. H., Kham, C., Bertrand, S., et al. (2012). Azithromycin and ciprofloxacin resistance in Salmonella bloodstream infections in Cambodian adults. PLoS Negl. Trop. Dis. 6:e1933. doi: 10.1371/journal.pntd.0001933
Wisner, A., Desin, T., White, A., Potter, A., and Kster, W. (2012). “The Salmonella Pathogenicity Island-1 and -2 encoded type III secretion systems,” in Salmonella - A Diversified Superbug, ed. Y. Kumar (Rijeka: InTech). doi: 10.5772/29203
World Health Organization (2015). WHO Estimates of the Global Burden of Foodborne Diseases. Geneva: World Health Organization.
World Health Organization (2017). Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance Due to Non-Human Use. Available online at: http://apps.who.int/iris/bitstream/10665/255027/1/9789241512220-eng.pdf (accessed May 19, 2020).
World Health Organization (2018). Antimicrobial Resistance. Available online at: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed April 24, 2020).
Yam, E. L. Y., Hsu, L. Y., Yap, E. P.-H., Yeo, T. W., Lee, V., Schlundt, J., et al. (2019). Antimicrobial resistance in the Asia Pacific region: a meeting report. Antimicrob. Resist. Infect. Control 8:202. doi: 10.1186/s13756-019-0654-8
Yang, K., Wang, A., Fu, M., Wang, A., Chen, K., Jia, Q., et al. (2020). Investigation of incidents and trends of antimicrobial resistance in foodborne pathogens in eight countries from historical sample data. Int. J. Environ. Res. Public. Health 17:472. doi: 10.3390/ijerph17020472
Yang, X., Wu, Q., Zhang, J., Huang, J., Chen, L., Wu, S., et al. (2019). Prevalence, bacterial load, and antimicrobial resistance of Salmonella serovars isolated from retail meat and meat products in China. Front. Microbiol. 10:2121. doi: 10.3389/fmicb.2019.02121
Yoshida, H., Bogaki, M., Nakamura, M., Yamanaka, L. M., and Nakamura, S. (1991). Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35, 1647–1650. doi: 10.1128/AAC.35.8.1647
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: antimicrobial resistance, Cambodia, informal market, Salmonella enterica, genotypic and phenotypic characterization
Citation: Schwan CL, Lomonaco S, Bastos LM, Cook PW, Maher J, Trinetta V, Bhullar M, Phebus RK, Gragg S, Kastner J and Vipham JL (2021) Genotypic and Phenotypic Characterization of Antimicrobial Resistance Profiles in Non-typhoidal Salmonella enterica Strains Isolated From Cambodian Informal Markets. Front. Microbiol. 12:711472. doi: 10.3389/fmicb.2021.711472
Received: 18 May 2021; Accepted: 26 August 2021;
Published: 16 September 2021.
Edited by:
Lin Lin, Jiangsu University, ChinaReviewed by:
V. K. Deekshit, NITTE University Center for Science Education and Research (NUCSER), IndiaShizhong Geng, Yangzhou University, China
Copyright © 2021 Schwan, Lomonaco, Bastos, Cook, Maher, Trinetta, Bhullar, Phebus, Gragg, Kastner and Vipham. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carla L. Schwan, Y2xzY2h3YW5Aa3N1LmVkdQ==
†Present address: Sara Lomonaco, Center for Drug Research and Evaluation, U.S. Food and Drug Administration, Silver Spring, MD, United States Joshua Maher, JMI Labs, North Liberty, IA, United States
 Carla L. Schwan
Carla L. Schwan Sara Lomonaco2†
Sara Lomonaco2† Leonardo M. Bastos
Leonardo M. Bastos Peter W. Cook
Peter W. Cook Joshua Maher
Joshua Maher Manreet Bhullar
Manreet Bhullar Sara Gragg
Sara Gragg Justin Kastner
Justin Kastner Jessie L. Vipham
Jessie L. Vipham