- 1The Second Affiliated Hospital and Yuying Children’s Hospital, Wenzhou Medical University, Wenzhou, China
- 2Key Laboratory of Medical Genetics of Zhejiang Province, Key Laboratory of Laboratory Medicine, Ministry of Education, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 3Institute of Biomedical Informatics, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 4Department of Clinical Laboratory, Zhejiang Hospital, Hangzhou, China
Multidrug-resistant bacteria from different sources have been steadily emerging, and an increasing number of resistance mechanisms are being uncovered. In this work, we characterized a novel resistance gene named aac(2′)-If from an isolate of a novel Providencia species, Providencia wenzhouensis R33 (CCTCC AB 2021339). Susceptibility testing and enzyme kinetic parameter analysis were conducted to determine the function of the aminoglycoside 2′-N-acetyltransferase. Whole-genome sequencing and comparative genomic analysis were performed to elucidate the molecular characteristics of the genome and the genetic context of the resistance gene-related sequences. Among the functionally characterized resistance genes, AAC(2′)-If shares the highest amino acid sequence identity of 70.79% with AAC(2′)-Ia. AAC(2′)-If confers resistance to several aminoglycoside antibiotics, showing the highest resistance activity against ribostamycin and neomycin. The recombinant strain harboring aac(2′)-If (pUCP20-aac(2′)-If/DH5α) showed 256- and 128-fold increases in the minimum inhibitory concentration (MIC) levels to ribostamycin and neomycin, respectively, compared with those of the control strains (DH5α and pUCP20/DH5α). The results of the kinetic analysis of AAC(2′)-If were consistent with the MIC results of the cloned aac(2′)-If with the highest catalytic efficiency for ribostamycin (kcat/Km ratio = [3.72 ± 0.52] × 104 M–1⋅s–1). Whole-genome sequencing demonstrated that the aac(2′)-If gene was located on the chromosome with a relatively unique genetic environment. Identification of a novel aminoglycoside resistance gene in a strain of a novel Providencia species will help us find ways to elucidate the complexity of resistance mechanisms in the microbial population.
Introduction
Aminoglycoside antibiotics disturb translation and promote mistranslation of proteins, resulting in altering the integrity of bacterial cell membranes (Aguirre Rivera et al., 2021). They have a broad antimicrobial spectrum, and they often work in synergy with other antibiotics, making them valuable anti-infective agents (van Hoek et al., 2011). With the widely growing use of antibiotics in human clinical or agricultural practices, antibiotic resistance is a growing problem associated with the use of all classes of anti-infective agents. Aminoglycoside resistance has many different forms. The major aminoglycoside resistance mechanism encountered in clinical isolates of gram-negative and gram-positive bacteria is most frequently associated with the expression of modifying enzymes (Wright, 1999; Vakulenko and Mobashery, 2003). To date, more than 100 aminoglycoside-modifying enzymes (AMEs) have been described and are broadly categorized into three groups based on the type of modification (Ramirez and Tolmasky, 2010). These three families of AMEs include acetyltransferases (AACs), nucleotidyltransferases or adenyltransferases (ANTs), and phosphotransferases (APHs). These classes are further divided into subtypes based on different region specificities of the enzymes for aminoglycoside modifications (Shaw et al., 1993; Ramirez and Tolmasky, 2010). The subclasses include four AACs, namely, AAC(1), AAC(2′), AAC(3), and AAC(6′); five ANTs, namely, ANT(2′′), ANT(3′′), ANT(4′), ANT(6), and ANT(9); and seven APHs, namely, APH(2′′), APH(3′), APH(3′′), APH(4), APH(6), APH(7′′), and APH(9) (Haas and Dowding, 1975; Shaw et al., 1993).
Acetyltransferases belong to the ubiquitous GCN5-related N-acetyltransferase (GNAT) superfamily of proteins, which includes approximately 10,000 proteins (Vetting et al., 2005). AACs are classified according to the sites of acetylation of the deoxystreptamine core of aminoglycoside antibiotics (Shaw et al., 1993). Most of the genes encoding these enzymes are plasmid borne, but an exception to this general rule is the 2′-N-acetyltransferase [AAC(2′)-Ia] from Providencia stuartii, which is chromosomally encoded (Rather et al., 1993; Payie et al., 1995). Additionally, AAC(2′)-Ia has been identified as the most prevalent chromosomally encoded AME among P. stuartii strains (Clarke et al., 1996; Franklin and Clarke, 2001), while AAC(2′) enzymes (AAC(2′)-Ib, AAC(2′)-Ic, AAC(2′)-Id, and AAC(2′)-Ie) have been found in different species of the genus Mycobacterium (Rather et al., 1993; Ainsa et al., 1996, 1997; Hegde et al., 2001; Adams et al., 2008). In the wild-type P. stuartii, the aac(2′)-Ia gene is normally expressed at low levels, and it is regulated in part by a small transcriptional activator and at least two other trans-acting negative regulators (Macinga et al., 1998; Ding et al., 2001).
Providencia species belong to the order Enterobacterales and family Morganellaceae, which is a family of gram-negative opportunistic human pathogens. Providencia species closely resemble Proteus and Morganella species and are quite ubiquitous in the environment (Sagar et al., 2017). Currently, 13 Providencia species are recognized: P. stuartii, P. rettgeri, P. rustigianii, P. heimbachae, P. alcalifaciens, P. burhodogranariea, P. sneebia, P. vermicola, P. huaxiensis, P. thailandensis, P. entomophila, P. friedericiana, and Candidatus P. siddallii1. P. rettgeri, one of the most common pathogens of Providencia spp., has been isolated from samples collected from patients with various infectious diseases, such as nephrocutaneous fistula (Lee and Hong, 2011), urinary tract infections (Barrios et al., 2013), and soft tissue infections (Carvalho-Assef et al., 2013).
In this work, we report the identification and characterization of a novel aminoglycoside 2′-N-acetyltransferase designated AAC(2′)-If encoded in the chromosome of a Providencia wenzhouensis strain isolated from a rabbit. In addition, based on sequence analysis, the genetic environment of the aac(2′)-If gene and its genetic relationship with other AACs were also analyzed.
Materials and Methods
Bacterial Strains and Plasmids
To investigate the antimicrobial resistance of large intestinal bacteria in animals, we collected anal feces samples from different animals. P. wenzhouensis R33 was isolated from an anal swab of a New Zealand White rabbit in an animal farm in Wenzhou, southeastern China. The anal swab was collected as follows: a sterile cotton swab was inserted into the anus of rabbit for 3–5 cm, gently rotated, and then immediately put it into a sterilized screw-capped specimen collection tube containing normal saline solution (0.9% sodium chloride). The isolate was randomly isolated by the streak plate method with a normal Luria-Bertani agar plate, initially identified using the Vitek-60 microorganism autoanalysis system (BioMerieux Corporate, Craponne, France) and verified by analysis of the 16S rRNA gene sequences. The result was finally confirmed by determining the average nucleotide identity (ANI) using FastANI and in silico DNA–DNA hybridization (isDDH) (Jain et al., 2018). The strains and plasmids used in this work are listed in Table 1.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) were determined using the agar dilution method with Mueller-Hinton (MH) agar plates following the guidelines of the Clinical and Laboratory Standards Institute (CLSI). Susceptibility patterns were interpreted according to the CLSI breakpoint criteria (CLSI, 2019) and the guidelines of the European Committee on Antimicrobial Susceptibility Testing (The European Committee on Antimicrobial Susceptibility Testing, 2019) for Enterobacteriaceae. Escherichia coli ATCC 25922 was used as a reference strain for quality control. The MIC was determined in triplicate on Mueller-Hinton (MH) agar plates with two-fold serial dilutions of the antibiotics. The plates were incubated at 37°C for 16–20 h before analyzing the results.
Whole-Genome Sequencing and Sequence Analysis
Genomic DNA of P. wenzhouensis R33 was extracted from bacterial culture by an AxyPrep Bacterial Genomic DNA Miniprep Kit (Axygen Scientific, Union City, CA, United States). DNA sequencing was performed by using the Illumina HiSeq-2500 and PacBio RS II platforms by the Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). The Illumina short reads and PacBio long reads were initially assembled by Canu v2.1 (Koren et al., 2017) and SPAdes v3.14.1 (Bankevich et al., 2012), respectively. Pilon v1.23 (Walker et al., 2014) was employed for further correction to improve assembly quality by mapping short reads aligned to the draft of the whole-genome assembly. Potential open reading frames (ORFs) of the assembled genome were predicted using Prokka v1.14.6 (Seemann, 2014), and functional annotation of these proteins was performed by BLAST analysis with an e-value threshold of 1e-5 against the nonredundant protein sequence database of US National Center for Biotechnology Information (NCBI) and the UniProt/Swiss-Prot database. Antimicrobial resistance genes were annotated using ResFinder (Zankari et al., 2012) and Resistance Gene Identifier (RGI) v4.0.3 of the Comprehensive Antibiotic Resistance Database (CARD) (McArthur et al., 2013). Mobile genetic elements (MGEs) were detected using ISFinder (Siguier et al., 2006) and INTEGRALL (Moura et al., 2009). ANI was calculated using FastANI v1.31 (Jain et al., 2018). CGView Server (Petkau et al., 2010) was used to visualize the basic genomic features of chromosomes and perform comparative genomic analysis. The promoter region of aac(2′)-If was predicted by BPROM (2016)2. The molecular weight and pI value of AAC(2′)-If were predicted using Expasy ProtParam Tool (2021)3. Multiple alignment of the amino acid sequences of and neighbor-joining phylogenetic tree construction for AAC(2′)-If and other AACs were performed using MAFFT v7.475 (Katoh and Standley, 2013) and MEGAX (Kumar et al., 2018), respectively. The sequence retrieval, statistical analysis, and other bioinformatics tools used in this study were written using Python (2021)4 and Biopython (Cock et al., 2009).
Cloning of the aac(2′)-If Gene
The coding sequence of aac(2′)-If, along with its upstream promoter region, was amplified by PCR primers with a pair of flanking restriction endonuclease adaptors for BamHI and HindIII (Takara Bio, Inc., Dalian, China) introduced at the 5′ and 3′ ends, respectively (Supplementary Table S1). The PCR product was digested with BamHI and HindIII, and then ligated into the pUCP20 vector with a T4 DNA ligase cloning kit (Takara Bio, Inc., Dalian, China). The recombinant plasmid pUCP20-pro-aac(2′)-If was transformed into competent E. coli DH5α cells by the calcium chloride method (Chan et al., 2013), and the transformant was selected on Luria-Bertani agar plates supplemented with 100 μg/mL ampicillin. The cloned insert sequence (aac(2′)-If with its upstream promoter region) in the recombinant plasmid of the transformant was further confirmed by both restriction enzyme digestion and Sanger sequencing (Shanghai Sunny Biotechnology Co., Ltd., Shanghai, China).
Expression and Purification of Recombinant AAC(2′)-If
The ORF of the aac(2′)-If gene was amplified with the orf-aac(2′)-If primers listed in Supplementary Table S1 and cloned into the pCold I cold shock expression vector (Qing et al., 2004). The resultant plasmid pCold I-aac(2′)-If was introduced into E. coli RosettagamiB (DE3) competent cells, and transformants (pCold I-orf-aac(2′)-If/RosettagamiB) were selected on LB agar plates containing 100 μg/mL ampicillin.
The overnight culture of the recombinant strain (pCold I-orf-aac(2′)-If/RosettagamiB) was cultured in LB broth containing 100 μg/mL ampicillin at 37°C. When the OD at 600 nm reached 0.6, isopropyl D-thiogalactopyranoside (IPTG) was added at a concentration of 1 mM to induce the expression of AAC(2′)-If, and cell cultivation was continued for 18 h at 16°C. The cells were collected, resuspended in phosphate buffered saline (pH = 7.4), and disrupted with a French pressure cell. The recombinant protein was purified using BeyoGold His-tag Purification Resin and subsequently eluted by the nondenatured eluent (50 mM NaH2PO4, 300 mM NaCl, 50 mM imidazole) of the His-tag Protein Purification Kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. The purity of the protein was confirmed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and subsequent staining with Coomassie Brilliant Blue. The protein concentration was determined spectrophotometrically using a BCA protein assay kit (Thermo Fisher Scientific, Rockford, IL, United States).
RT Q-PCR Assays
To analyze the expression of aac(2′)-If, total RNA was extracted from the bacteria cultured in LB broth grown to OD600 = 0.5 and then treated with or without the addition of 1/4 MIC ribostamycin (32 μg/mL), neomycin (1 μg/mL), and gentamicin (0.25 μg/mL) for 0.5, 1, 2, 4, and 24 h, respectively. Then total RNA was isolated using TRIzol reagent (Sigma, Shanghai, China) according to the manufacturer’s protocol and quantified by an Implen NanoPhotometer (Implen GmbH, Munich, Germany). DNA-free RNA was confirmed by PCR amplification of the Providencia 16S rRNA gene. cDNA was synthesized using the PrimeScript RT-PCR Kit (Takara Bio, Inc., Dalian, China). RT Q-PCR was performed using the corresponding cDNA from each sample. The primers used for RT Q-PCR are listed in Supplementary Table S2. RT Q-PCR was performed in a CFX96TM Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, United States) by monitoring the increase in fluorescence in real time using SYBR qPCR Master Mix (Vazyme Biotech, Nanjing, China). Relative quantification was performed using the CT method (Livak and Schmittgen, 2001) with the housekeeping genes 16S rRNA as references. Comparisons of expression levels with or without antibiotics treatment were analyzed using one-way ANOVA (LSD test). P ≤ 0.05 was considered significant.
Kinetic Studies of AAC(2′)-If
The kinetic parameters for AAC(2′)-If activity were measured spectrophotometrically by following the production of coenzyme A (CoASH), which was produced upon the transfer of the acetyl moiety to the aminoglycoside. The thiol group of CoASH reacts with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB), which is replaced by 4,4′-dithiodipyridine, and monitoring was performed at 412 nm to measure an increase in the absorbance of the formed product, pyridine-4-thiolate (TNB), as previously described (Hegde et al., 2001; Galimand et al., 2015). Kinetic assays were performed in a 200 μl reaction mixture containing 25 mM 2-(N-morpholino)ethanesulfonic acid (MES; pH 6.0), 1 mM EDTA, 80 μM acetyl-CoA, 2 mM DTNB, and variable concentrations of aminoglycosides (5–150 μM) (Franklin and Clarke, 2001), and the reactions, initiated by the addition of 2 μg purified enzyme (final concentration), were monitored using a Synergy Neo2 Multi-Mode Microplate Reader (Biotek, VT, United States) at room temperature for 10 min. The steady-state kinetic parameters (kcat and Km) were determined by nonlinear regression of the initial reaction rates with the Michaelis–Menten equation using GraphPad Prism 9 (GraphPad Software, San Jose, CA, United States).
Nucleotide Sequence Accession Numbers
The aac(2′)-If sequence has been assigned GenBank accession number MW984427. The complete genome sequence of P. wenzhouensis R33 presented in this study has been deposited in GenBank under accession numbers CP072453 and CP072454 for chromosome and plasmid pR33-1 of P. wenzhouensis R33, respectively.
Results
Identification and Characteristics of the AAC(2′)-If-Producing Isolate of Providencia wenzhouensis R33
A 16S ribosomal RNA gene homology analysis showed that the 16S rRNA gene of strain R33 shared the highest similarity with that of P. vermicola strain OP1 (NR_042415.1), with 98.01% identity and 99.00% coverage. However, the genome sequence of strain R33 shared 76.93–81.47% ANI and had a 19.60–52.40% in silico DNA–DNA hybridization (isDDH) score with the species-classified Providencia strains (Supplementary Table S3). They were too low to reach the cutoff (≥95–96% for ANI and 70% for isDDH) to define a classified bacterial species, which suggested that strain R33 is a new species of the genus Providencia (Goris et al., 2007; Hu et al., 2019). According to the criteria for species names (International Code of Nomenclature of Prokaryotes, 2019), we named it Providencia wenzhouensis R33. This isolate was deposited in China Center for Type Culture Collection, Wuhan, China (CCTCC AB 2021339).
Providencia wenzhouensis (wen.zhou.en’sis. N.L. fem. adj. wenzhouensis pertaining to Wenzhou, Zhejiang Province, China, where the type strain was isolated) cells are Gram-negative, non-motile, non-spore-forming, non-gas-producing, rod-shaped. Colonies are creamy white, circular, moist, convex, with entire edges on nutrient agar and translucent on MacConkey agar. The strain is catalase-positive and oxidase-negative, gives positive in methyl-red reaction, citrate utilization, and nitrate reduction. The strain is negative for Voges-Proskauer, indole, urease, and the production of H2S. Acid is produced from adonitol, salicin, galactose, arabinose, gluconate, glucose, mannitol, D-mannitol, phenylalanine deaminase, but not from arginine, glycerol, inulin, inositol, lactose, lysine, ornithine, trehalose, maltose, pyruvate, sorbitol, sorbose, sucrose, and urea. Differential biochemical characteristics from other members of the genus Providencia include acid production from citrate, mannitol, and salicin but not from urea.
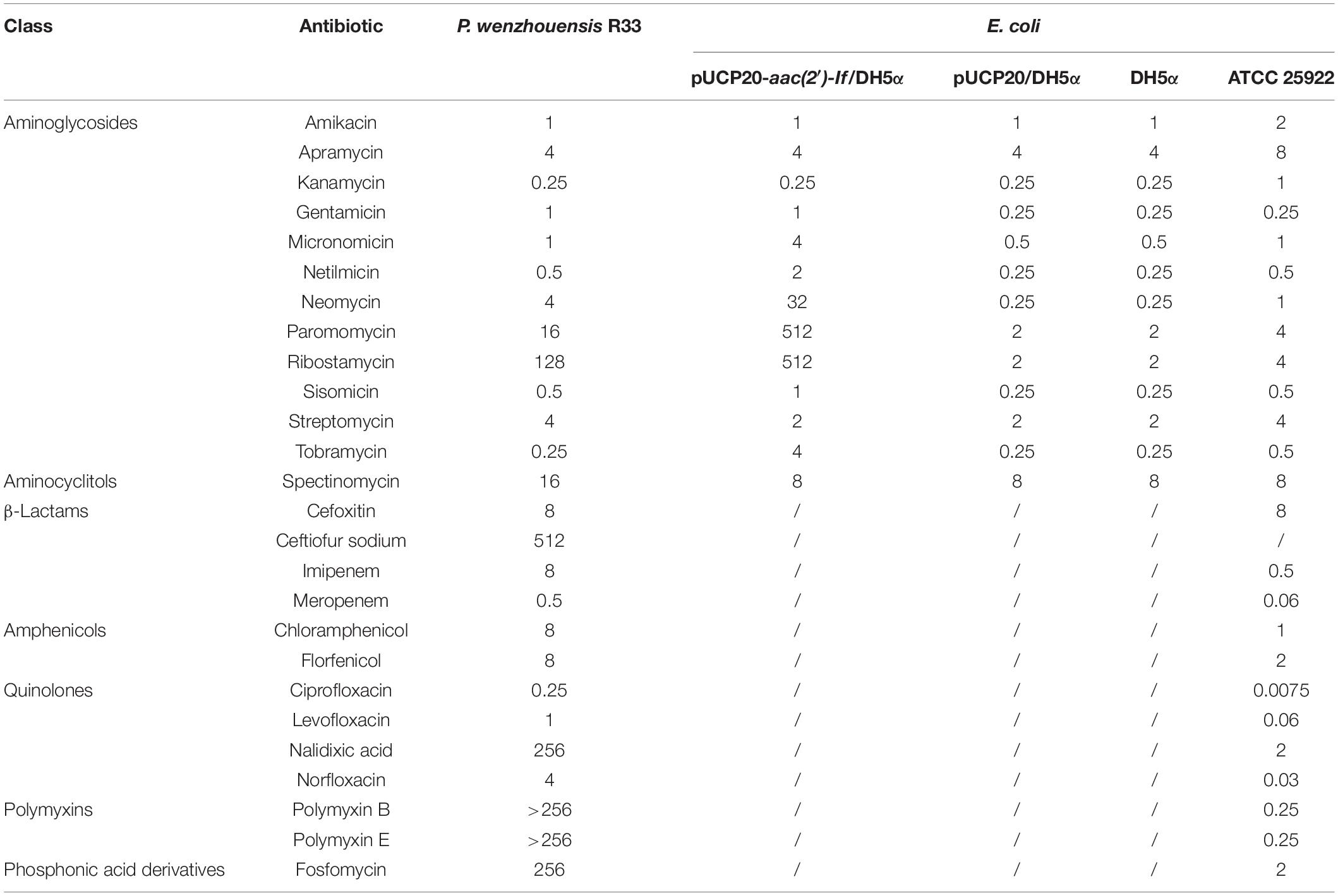
The MICs of 26 antibiotics were tested for strain R33 as shown in Table 2. This strain showed resistance to five antibiotics, including fosfomycin, imipenem, nalidixic acid, and polymyxins B and E, and higher MIC values for ribostamycin (128 μg/ml) and ceftiofur sodium (512 μg/ml, an antibiotic for animals only), which have no established breakpoints (Table 2).
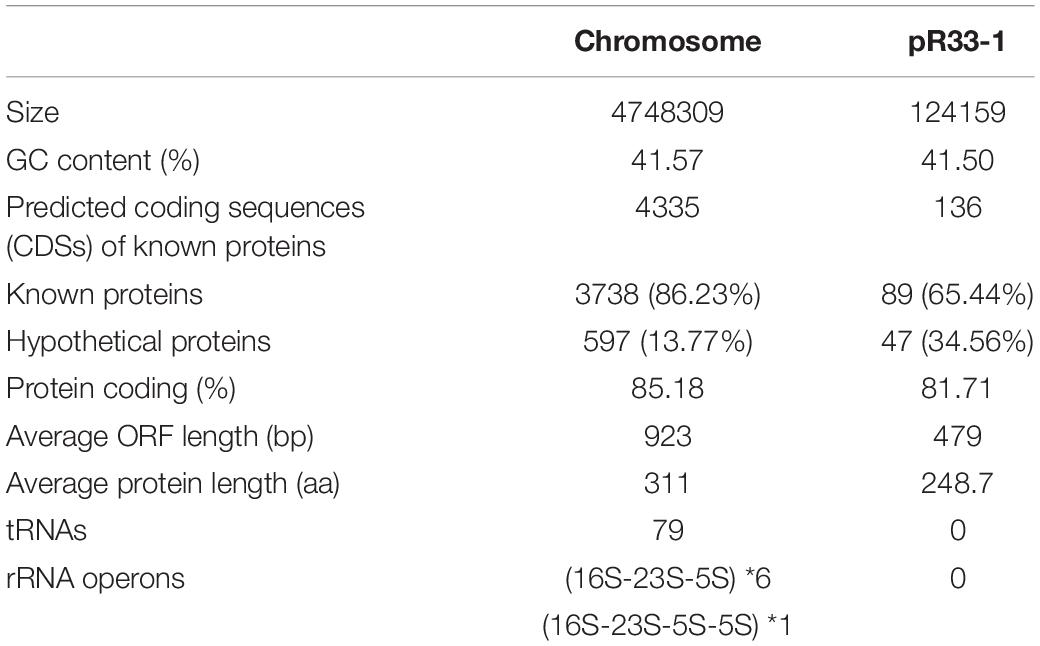
The whole genome of P. wenzhouensis R33 was found to consist of a chromosome of approximately 4.75 Mb in length, encoding 4,335 ORFs with an average GC content of 41.57% (Table 3 and Figure 1), and a circular plasmid designated pR33-1, which is 124,159 bp in length and encodes 136 ORFs (Table 3). According to genome sequencing, only three chromosomal genes with a similarity of ≥80% with housekeeping genes, namely, crp (encoding cAMP-receptor protein), rsmA (encoding translational regulator CsrA), and E. coli gyrA (encoding DNA gyrase subunit A) (with similarities of 97.6%, 86.89%, and 83.72%, respectively), were identified. No predicted aminoglycoside resistance genes with similarity ≥80% were found. Among all 11 predicted aminoglycoside resistance-related genes (with similarity <80%) annotated from the whole genome, only one predicted AAC gene [an aac(2′)-I homologous gene designated aac(2′)-If in this work] had an amino acid sequence similarity of 70.79% with the aac(2′)-Ia gene of known function, while the remaining 10 genes (CpxA, KpnH, BaeR, KpnF, KdpE, KpnG, TolC, BaeS, CpxR, and ykkD) encoded for efflux-related proteins (Supplementary Table S4). We subsequently cloned the putative AAC(2′)-I gene (aac(2′)-If), and its function was further verified.
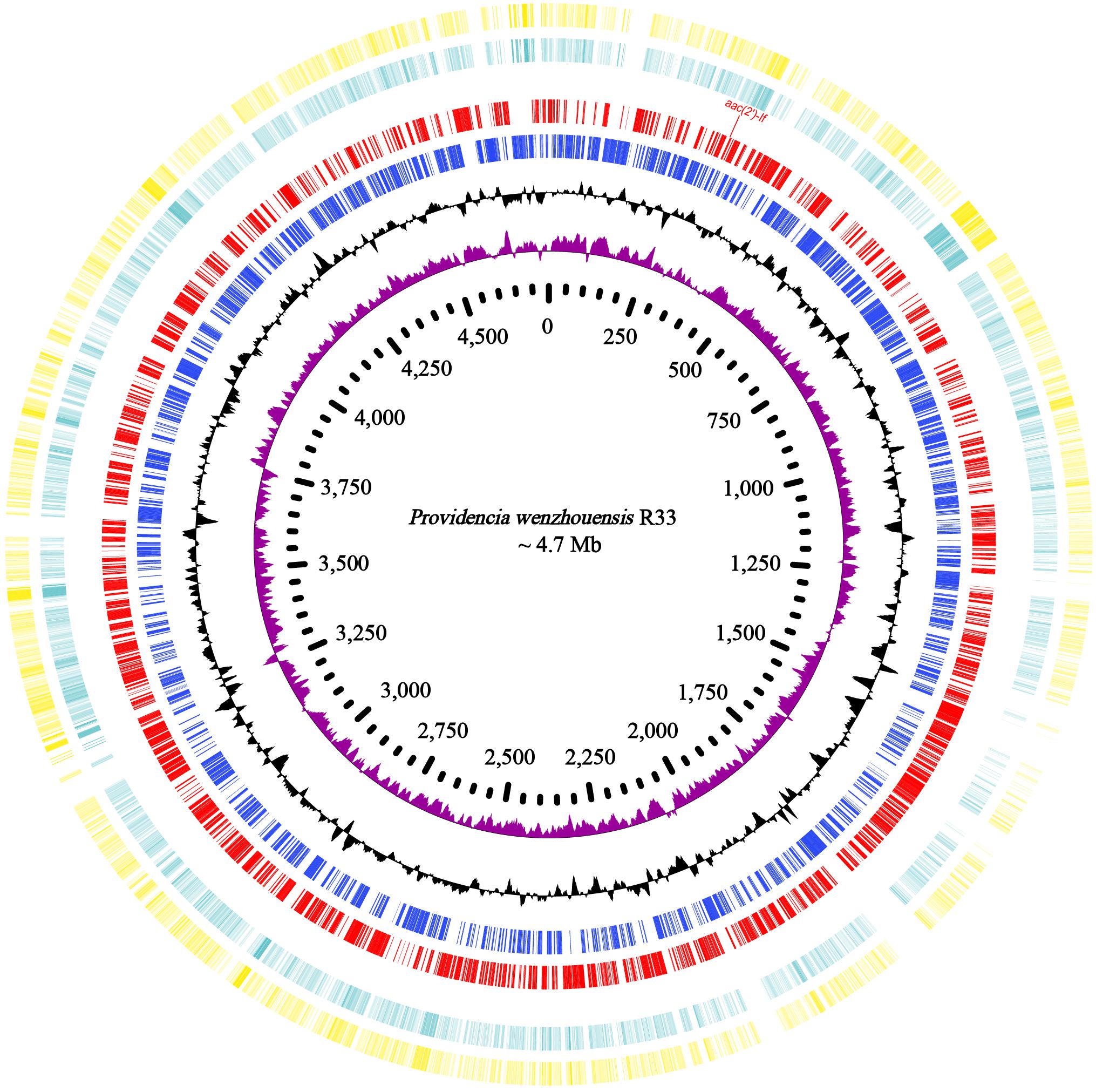
Figure 1. Genome map and comparison of the chromosome sequence of P. wenzhouensis R33 with other similar genomes with high identities. From outside to inside: circles 1 and 2 are homologous regions of the chromosomes of Providencia sp. 1709051003 (NZ_CP042861.1) and P. rettgeri Pr-15-2-50 (NZ_CP039844.1) with P. wenzhouensis R33, while the unmatched regions are left blank; circles 3 and 4 display predicted ORFs encoded in the forward and reverse strands, and circles 5, 6, and 7 represent the GC content, GC skew, and scale in kb of the P. wenzhouensis R33 chromosome, respectively.
Homologs of the New Aminoglycoside 2′-N-Acetyltransferase
The novel 2′-N-acetyltransferase gene aac(2′)-If was revealed to be 537 bp in length and to encode a protein of 178 amino acids (ca. 20.33 kDa) with a pI value of 4.47. All 16 of the aac(2′)-If homologs (≥65% nucleotide sequence similarity) retrieved from the NCBI nucleotide database were derived from Providencia species (mainly P. rettgeri and P. stuartii). As mentioned above, the predicted protein sequence (no nucleotide sequence available) shared the highest amino acid sequence identity of 95.51% with AAC(2′)-If, which was a hypothetical GNAT family N-acetyltransferase (WP_206082813.1) from P. stuartii.
The results of multiple sequence alignment of AAC(2′) enzymes showed that AAC(2′)-If shared at most 70.79%, 35.03%, 35.26%, 30.10%, and 31.21% identity with the five AAC(2′)-I subgroups AAC(2′)-Ia (AAA03550.1), AAC(2′)-Ib (AAC44793.1), AAC(2′)-Ic (AAB17563.1), AAC(2′)-Id (AAB41701.1), and AAC(2′)-Ie (CAR72650.1), respectively. Except for the AAC(2′)-Ia subgroup, which had the highest identity value of more than 70.0%, the other four subgroups showed less than 40.0% identity (Figure 2). A phylogenetic tree showed that AAC(2′)-If clustered closest to AAC(2′)-Ia (Figure 3).

Figure 2. Multiple sequence alignment of the amino acid sequences of the AAC(2′)-I subgroup. The sequences and their accession numbers are as follows: AAC(2′)-If (MW984427), AAC(2′)-Ia (AAA03550.1), AAC(2′)-Ib (AAC44793.1), AAC(2′)-Ic (AAB17563.1), AAC(2′)-Id (AAB41701.1), and AAC(2′)-Ie (CAR72650.1). The numbers on the right correspond to the amino acid residues in each full-length protein, with the conserved motif sites boxed, fully conserved residues shown with asterisks, and highly similar residues shown with exclamation marks.
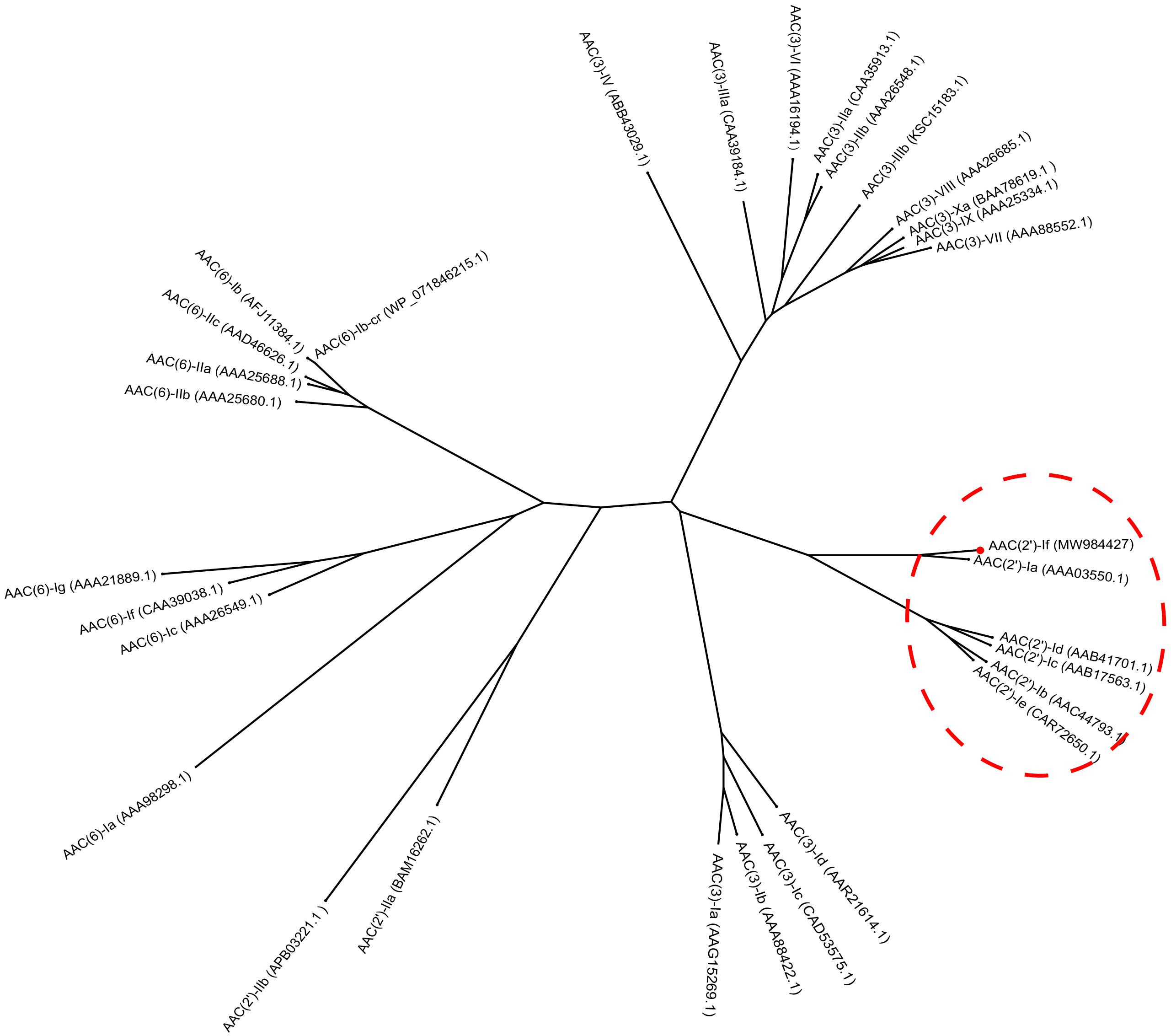
Figure 3. Phylogenetic tree showing the relationship of AAC(2′)-If with other chromosome-borne acetyltransferases. AAC(2′)-If from this study is shown with a red dot.
Functional Characteristics of the aac(2′)-If Gene
To investigate the functional role of AAC(2′)-If, the sequence encoding the ORF of AAC(2′)-If with its promoter region was cloned into the pUCP20 vector, and the recombinant plasmid (pUCP20-aac(2′)-If) was transformed into E. coli DH5α. The MICs of several aminoglycoside antibiotics against the transformant harboring aac(2′)-If (pUCP20-aac(2′)-If/DH5α) were determined. The presence of plasmid borne aac(2′)-If in E. coli led to elevated MICs of ribostamycin, neomycin, paromomycin, tobramycin, micronomicin, netilmicin, gentamicin, and sisomicin (256-, 128-, 256-, 16-, 8-, 8-, 4-, and 4-fold increases, respectively) in comparison with those for the control strains (DH5α or DH5α carrying the vector pUCP20) (Table 2). However, as expected, no changes in MICs of streptomycin and spectinomycin were observed. Additionally, the susceptibility of the aac(2′)-If-carrying R33 to aminoglycosides and other antibiotics are also shown in Table 2, with relatively high MIC values (≥4 μg/mL) observed for several aminoglycosides (i.e., apramycin, neomycin, paromomycin, ribostamycin, and streptomycin), spectinomycin, polymyxins B and E, nalidixic acid, norfloxacin, chloramphenicol, florfenicol, cefoxitin, ceftiofur, imipenem, and fosfomycin.
Kinetic Parameters of AAC(2′)-If
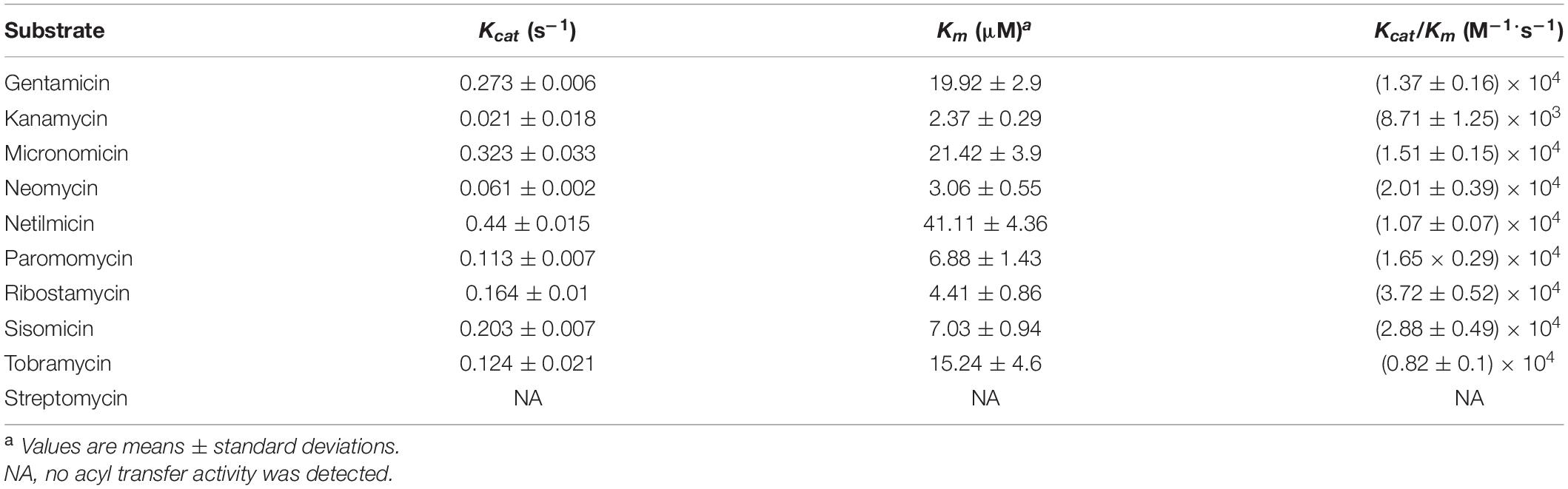
Investigation of the acetyltransferase activity and kinetic parameters of AAC(2′)-If showed that of the 10 aminoglycosides tested, the enzyme was able to acetylate ribostamycin, neomycin, tobramycin, micronomicin, sisomicin, and gentamicin, but worse acetyltransferase activity was detected for netilmicin and kanamycin (Table 4). The highest catalytic efficiency was observed with ribostamycin [kcat/Km ratio was (3.72 ± 0.52) × 104 M–1⋅s–1], which was found to be the best aminoglycoside substrate, whereas kanamycin was the worst substrate [kcat/Km ratio = (8.71 ± 1.25) × 103 M–1⋅s–1]. These differences appeared to result from large differences in the turnover rates (kcat), which varied by almost six-fold compared to the affinity (Table 4).
aac(2′)-If RNA Expression After Exposure to Antibiotics
To investigate the effect of aminoglycoside antibiotics on the aac(2′)-If gene expression, we determined the change of the aac(2′)-If gene expression levels with or without ribostamycin, neomycin, and gentamicin for different periods of time. Quantitative real-time PCR indicated that the expression of the aac(2′)-If gene increased significantly when the cells were induced with the three aminoglycoside antibiotics for 2 h (P < 0.001). The expression level increased approximately three-, five-, and two-fold when treated with ribostamycin, neomycin, and gentamicin, respectively. No significant difference was observed between any pair of the other groups (Figure 4).
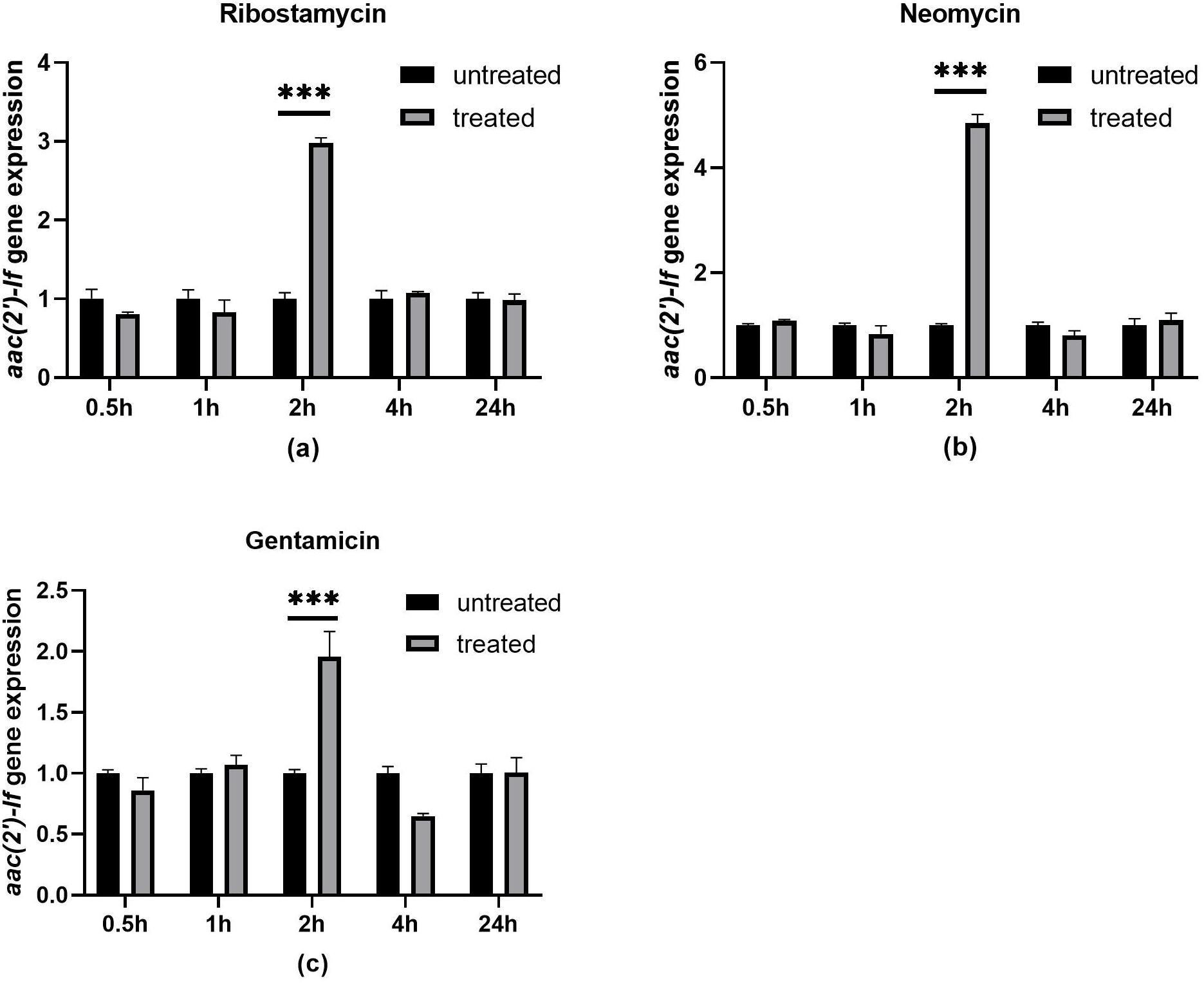
Figure 4. Expression levels of aac(2′)-If treated or untreated with 32 μg/mL ribostamycin (a), 1 μg/mL neomycin (b), and 0.25 μg/mL gentamicin (c). Bars represent means ± standard errors and experiments were performed in triplicate. ∗∗∗Presents significant difference, P < 0.001.
Genetic Context of the aac(2′)-If Gene
A total of 39 sequences (including one from this work) that were approximately 21 kb in length, with aac(2′)-If-like genes in the center, that shared nucleotide sequence similarities between 32.18 and 70.79% with aac(2′)-If were retrieved from the database. These sequences were all from Providencia (68.42%, 26/38) and Proteus strains (31.58%, 12/38). The genes from Providencia all shared amino acid sequence similarities of ≥54.61% with aac(2′)-If, while the genes from Proteus showed amino acid similarities of below 40.0% (except one of 68.76% and two of 53.39%) with aac(2′)-If.
The 39 sequences could be clustered into five groups according to the sequence similarities. The sequence of strain R33 was grouped alone, while the other four groups contained six or more sequences each (Supplementary Table S5). One sequence that shared the highest similarity with the other sequences in the same group was selected as a representative to perform the sequence structure comparison within the groups (Figure 5). The sequence of aac(2′)-If of R33 only showed similar sequence structures upstream of the aac(2′)-If gene as that of group 3, and the other sequences or sequence regions from these groups were completely different from each other.
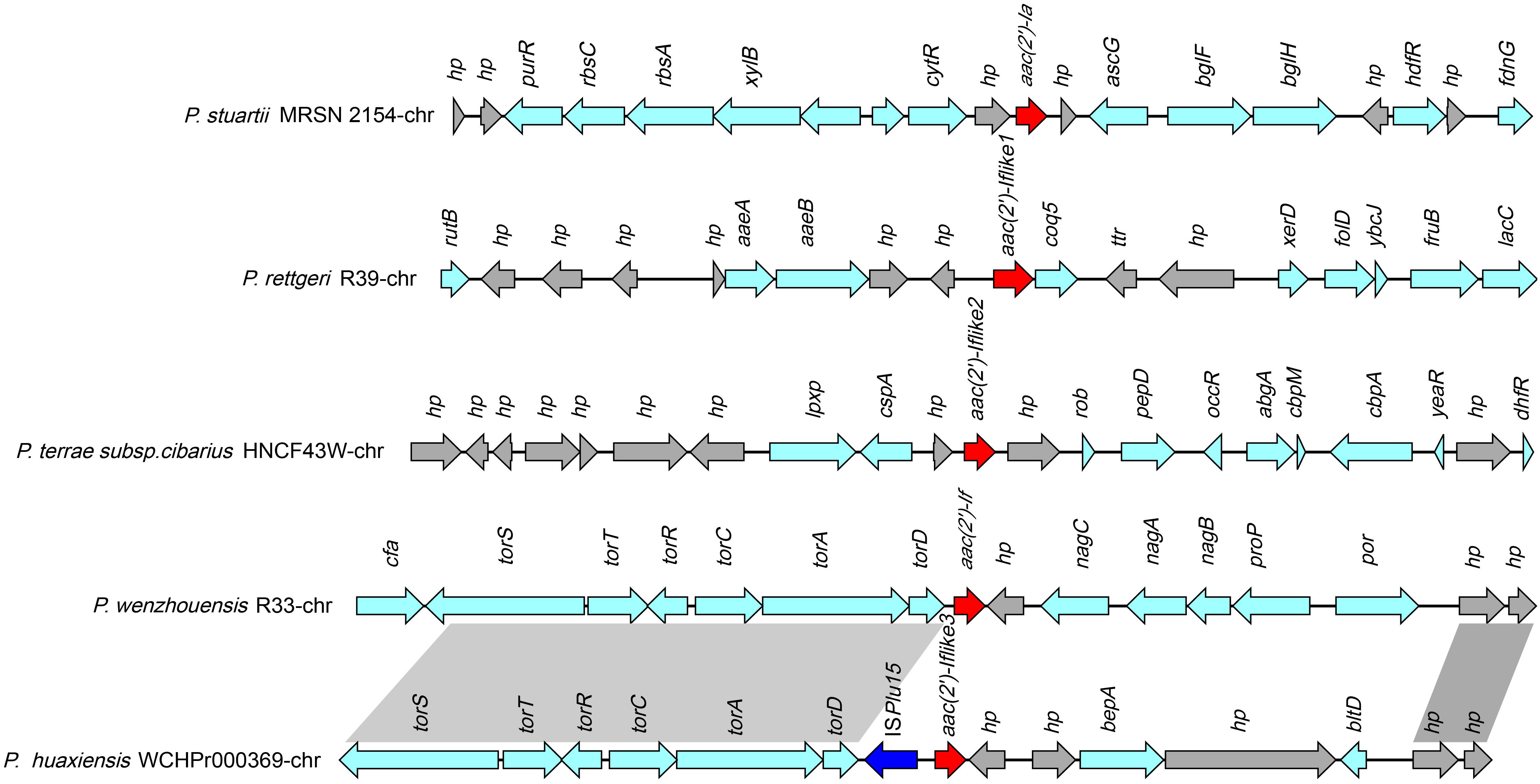
Figure 5. Comparative analysis of the genomic context of the aac(2′)-If gene with similar sequences. Homologous genes are shown in the same colors, and the genes with no homologs are colored gray. The accession numbers of the sequences are as follows: P. stuartii MRSN 2154 (CP003488.1), P. rettgeri R39 chromosome (CP066315.1), P. huaxiensis WCHP000369 chromosome (CP031123.2), and P. terrae subsp. cibarius HNCF44W (CP053042.1).
Discussion
In this work, by correlation analysis of the aminoglycoside resistance phenotypes with the genotypes, we identified a novel resistance gene named as aac(2′)-If. Homology analysis of AAC(2′)-If with the sequences in the NCBI nonredundant protein database demonstrated that the sequence that shared the highest amino acid sequence identity with AAC(2′)-If was that of AAC(2′)-Ia, a protein of known function, sharing an amino acid sequence identity of only 70.79%. Moreover, only 35 predicted protein sequences with identities between 60.0% and 95.51% with AAC(2′)-If were found, and most of the sequences were derived from the chromosomes of Providencia species (especially P. rettgeri and P. stuartii). Only one protein described as a GNAT family N-acetyltransferase was from Morganellaceae, and one was from an unclassified Shigella species. This result indicated that to clarify the evolution of the aac(2′)-If gene, further studies for genotypes with AACs of higher identities are warranted.
The antibiotic resistance profile conferred by AAC(2′)-If was basically the same as that of other AAC(2′)-I enzymes. They conferred resistance to a variety of aminoglycosides, including 4,5-disubstituted aminoglycosides (ribostamycin, neomycin, paromomycin) and 4,6-disubstituted aminoglycosides (gentamicin, tobramycin, micronomicin, netilmicin, and sisomicin) (Ainsa et al., 1996; Franklin and Clarke, 2001). As expected, in this work, the recombinant strain carrying aac(2′)-If remained susceptible to spectinomycin and streptomycin, which does not contain 2′ amino groups in their chemical structure. Although AAC(2′)-If enzymes can modify both 4,6- and 4,5-disubstituted aminoglycosides, the extent of modification was different. 4,5-Disubstituted aminoglycosides were generally better substrates for AAC(2′)-If. Ribostamycin was the most efficient substrate examined and exhibited a 7-fold higher kcat value than kanamycin, with a 256-fold increase in the MIC value. Additionally, AAC(2′)-If exhibited weak acetylation against kanamycin [kcat/Km = (8.71 ± 1.25) × 103 M–1⋅s–1] in enzyme kinetic analysis, which may explain why this enzyme is unable to confer resistance to kanamycin in vivo. The MIC values of kanamycin for both the recombinant and the control strains remained the same (0.25 μg/mL). AAC(2′)-If appeared to be less active against aminoglycoside substrates than His-AAC(2′)-Ia and AAC(2′)-Ic enzymes by one or two orders of magnitude (Hegde et al., 2001). The Km value for these three enzymes did not vary greatly, but the turnover number kcat values differed more than 10-fold between them (Cox et al., 2018).
According to the result of comparative analysis with homologous genes, except for sequences from one group, which was composed of sequences from Providencia that shared a similar upstream sequence with aac(2′)-If, the sequences from the other groups showed completely different sequence structures compared to that of aac(2′)-If. Considering that aac(2′)-If belongs to a novel species of the genus Providencia and only one genome sequence from this work is available, and considering that this sequence shared only 70.79% sequence similarity with all of the sequences available in the NCBI nucleotide database, the evolutionary study of this novel aminoglycoside 2′-N-acetyltransferase gene could be performed when a greater number of whole-genome sequences carrying phylogenetically close aac(2′)-If-like genes become available in the future.
Conclusion
In the current study, we identified aac(2′)-If encoded on the chromosome of an isolate of a new Providencia species, designated P. wenzhouensis R33, with the complete genome sequenced. The novel aminoglycoside 2′-N-acetyltransferase AAC(2′)-If shares the highest amino acid sequence identity of 70.79% with the functionally characterized AAC(2′)-Ia and confers stronger resistance to ribostamycin and neomycin. Whole-genome sequencing revealed that this novel resistance gene was encoded in the chromosome and was not related to any MGEs. Identification of a novel resistance gene in a novel bacterial species will help us find ways to explore the increasing number of resistance mechanisms in the microbial population and to further elucidate the intrinsic resistance mechanisms of unusual microorganisms.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
This study uses strains obtained from an anal swab of a rabbit on an animal farm in Wenzhou, China. It did not require the study to be reviewed or approved by an ethics committee because the animal was not presented in this study. The animal legal guardian provided written informed consent to participate in this study.
Author Contributions
HZ, MZ, and QB contributed to the conception and design of study. JLL, XL, WS, and MG contributed to the acquisition of data. KZ, JLL, CF, and KL contributed to the data analysis and interpretation. KZ, JWL, XD, PZ, and QB contributed to the drafting of manuscript. KZ, JLL, WS, MG, QL, and XZ performed the experiments. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Zhejiang Provincial Natural Science Foundation of China (LY19C060002 and LQ17H190001) and the National Natural Science Foundation of China (81973382).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to acknowledge all study participants and individuals who contributed to this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.711037/full#supplementary-material
Footnotes
- ^ https://lpsn.dsmz.de/search?word=Providencia
- ^ http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgro up=gfindb
- ^ https://web.expasy.org/protparam/
- ^ https://www.python.org/
References
Adams, M. D., Goglin, K., Molyneaux, N., Hujer, K. M., Lavender, H., Jamison, J. J., et al. (2008). Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190, 8053–8064.
Aguirre Rivera, J., Larsson, J., Volkov, I. L., Seefeldt, A. C., Sanyal, S., and Johansson, M. (2021). Real-time measurements of aminoglycoside effects on protein synthesis in live cells. Proc. Natl. Acad. Sci. U.S.A. 118:e2013315118. doi: 10.1073/pnas.2013315118
Ainsa, J. A., Martin, C., Gicquel, B., and Gomez-Lus, R. (1996). Characterization of the chromosomal aminoglycoside 2′-N-acetyltransferase gene from Mycobacterium fortuitum. Antimicrob. Agents Chemother. 40, 2350–2355. doi: 10.1128/AAC.40.10.2350
Ainsa, J. A., Perez, E., Pelicic, V., Berthet, F. X., Gicquel, B., and Martin, C. (1997). Aminoglycoside 2′-N-acetyltransferase genes are universally present in mycobacteria: characterization of the aac(2′)-Ic gene from Mycobacterium tuberculosis and the aac(2′)-Id gene from Mycobacterium smegmatis. Mol. Microbiol. 24, 431–441. doi: 10.1046/j.1365-2958.1997.3471717.x
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barrios, H., Garza-Ramos, U., Reyna-Flores, F., Sanchez-Perez, A., Rojas-Moreno, T., Garza-Gonzalez, E., et al. (2013). Isolation of carbapenem-resistant NDM-1-positive Providencia rettgeri in Mexico. J. Antimicrob. Chemother. 68, 1934–1936. doi: 10.1093/jac/dkt124
BPROM (2016). Available oline at: http://www.softberry.com/berry.phtml?topic =bprom&group=programs&subgroup=gfindb (accessed July 22, 2021).
Carvalho-Assef, A. P. D., Pereira, P. S., Albano, R. M., Beriao, G. C., Chagas, T. P. G., Timm, L. N., et al. (2013). Isolation of NDM-producing Providencia rettgeri in Brazil. J. Antimicrob. Chemother. 68, 2956–2957. doi: 10.1093/jac/dkt298
Chan, W. T., Verma, C. S., Lane, D. P., and Gan, S. K. (2013). A comparison and optimization of methods and factors affecting the transformation of Escherichia coli. Biosci. Rep. 33:e00086. doi: 10.1042/BSR20130098
Clarke, A. J., Francis, D., and Keenleyside, W. J. (1996). The prevalence of gentamicin 2′-N-acetyltransferase in the Proteeae and its role in the O-acetylation of peptidoglycan. FEMS Microbiol. Lett. 145, 201–207. doi: 10.1111/j.1574-6968.1996.tb08578.x
CLSI (2019). Performance Standards for Antimicrobial Susceptibility Testing: CLSI Supplement M100, 29th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Cock, P. J., Antao, T., Chang, J. T., Chapman, B. A., Cox, C. J., Dalke, A., et al. (2009). Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423. doi: 10.1093/bioinformatics/btp163
Cox, G., Ejim, L., Stogios, P. J., Koteva, K., Bordeleau, E., Evdokimova, E., et al. (2018). Plazomicin retains antibiotic activity against most aminoglycoside modifying enzymes. ACS Infect. Dis. 4, 980–987. doi: 10.1021/acsinfecdis.8b00001
Ding, X. D., Baca-Delancey, R. R., and Rather, P. N. (2001). Role of SspA in the density-dependent expression of the transcriptional activator AarP in Providencia stuartii. FEMS Microbiol. Lett. 196, 25–29. doi: 10.1111/j.1574-6968.2001.tb10535.x
Expasy ProtParam Tool (2021). Available online at: https://web.expasy.org/protparam/. [Accessed August 01 2021].
Franklin, K., and Clarke, A. J. (2001). Overexpression and characterization of the chromosomal aminoglycoside 2′-N-acetyltransferase of Providencia stuartii. Antimicrob. Agents Chemother. 45, 2238–2244. doi: 10.1128/AAC.45.8.2238-2244.2001
Galimand, M., Fishovitz, J., Lambert, T., Barbe, V., Zajicek, J., Mobashery, S., et al. (2015). AAC(3)-XI, a new aminoglycoside 3-N-acetyltransferase from Corynebacterium striatum. Antimicrob. Agents Chemother. 59, 5647–5653. doi: 10.1128/AAC.01203-15
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91.
Haas, M. J., and Dowding, J. E. (1975). Aminoglycoside-modifying enzymes. Methods Enzymol. 43, 611–628. doi: 10.1016/0076-6879(75)43124-x
Hegde, S. S., Javid-Majd, F., and Blanchard, J. S. (2001). Overexpression and mechanistic analysis of chromosomally encoded aminoglycoside 2′-N-acetyltransferase (AAC(2′)-Ic) from Mycobacterium tuberculosis. J. Biol. Chem. 276, 45876–45881. doi: 10.1074/jbc.M108810200
Hu, Y., Feng, Y., Zhang, X., and Zong, Z. (2019). Providencia huaxiensis sp. nov., recovered from a human rectal swab. Int. J. Syst. Evol. Microbiol. 69, 2638–2643. doi: 10.1099/ijsem.0.003502
International Code of Nomenclature of Prokaryotes (2019). International Journal OfSystematic And Evolutionary Microbiology. Available online at: doi: 10.1099/ijsem.0.000778 (accessed January 11, 2019).
Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lee, G., and Hong, J. H. (2011). Xanthogranulomatous pyelonephritis with nephrocutaneous fistula due to Providencia rettgeri infection. J. Med. Microbiol. 60, 1050–1052. doi: 10.1099/jmm.0.028977-0
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Macinga, D. R., Cook, G. M., Poole, R. K., and Rather, P. N. (1998). Identification and characterization of aarF, a locus required for production of ubiquinone in Providencia stuartii and Escherichia coli and for expression of 2′-N-acetyltransferase in P. stuartii. J. Bacteriol. 180, 128–135. doi: 10.1128/JB.180.1.128-135.1998
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357.
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Payie, K. G., Rather, P. N., and Clarke, A. J. (1995). Contribution of gentamicin 2′-N-acetyltransferase to the O acetylation of peptidoglycan in Providencia stuartii. J. Bacteriol. 177, 4303–4310. doi: 10.1128/jb.177.15.4303-4310.1995
Petkau, A., Stuart-Edwards, M., Stothard, P., and Van Domselaar, G. (2010). Interactive microbial genome visualization with GView. Bioinformatics 26, 3125–3126. doi: 10.1093/bioinformatics/btq588
Python (2021). Availble online at: https://www.python.org/. (accessed July 22, 2021).
Qing, G., Ma, L. C., Khorchid, A., Swapna, G. V., Mal, T. K., Takayama, M. M., et al. (2004). Cold-shock induced high-yield protein production in Escherichia coli. Nat. Biotechnol. 22, 877–882. doi: 10.1038/nbt984
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171.
Rather, P. N., Orosz, E., Shaw, K. J., Hare, R., and Miller, G. (1993). Characterization and transcriptional regulation of the 2′-N-acetyltransferase gene from Providencia stuartii. J. Bacteriol. 175, 6492–6498. doi: 10.1128/jb.175.20.6492-6498.1993
Sagar, S., Narasimhaswamy, N., and D’souza, J. (2017). Providencia rettgeri: an emerging nosocomial uropathogen in an indwelling urinary catheterised patient. J. Clin. Diagn. Res. 11, DD01–DD02. doi: 10.7860/JCDR/2017/25740.10026
Shaw, K. J., Rather, P. N., Hare, R. S., and Miller, G. H. (1993). Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57, 138–163. doi: 10.1128/mr.57.1.138-163.1993
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36.
The European Committee on Antimicrobial Susceptibility Testing (2019). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 9.0. Available online at: http://www.eucast.org
Vakulenko, S. B., and Mobashery, S. (2003). Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16, 430–450. doi: 10.1128/CMR.16.3.430-450.2003
van Hoek, A. H., Mevius, D., Guerra, B., Mullany, P., Roberts, A. P., and Aarts, H. J. (2011). Acquired antibiotic resistance genes: an overview. Front. Microbiol. 2:203.
Vetting, M. W., Lp, S. D. C., Yu, M., Hegde, S. S., Magnet, S., Roderick, S. L., et al. (2005). Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433, 212–226. doi: 10.1016/j.abb.2004.09.003
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wright, G. D. (1999). Aminoglycoside-modifying enzymes. Curr. Opin. Microbiol. 2, 499–503. doi: 10.1016/s1369-5274(99)00007-7
Keywords: Providencia, aminoglycoside, AAC(2cpsdummy′), resistance, kinetic analysis
Citation: Zhou K, Liang J, Dong X, Zhang P, Feng C, Shi W, Gao M, Li Q, Zhang X, Lu J, Lin X, Li K, Zhang H, Zhu M and Bao Q (2021) Identification and Characterization of a Novel Chromosomal Aminoglycoside 2′-N-Acetyltransferase, AAC(2′)-If, From an Isolate of a Novel Providencia Species, Providencia wenzhouensis R33. Front. Microbiol. 12:711037. doi: 10.3389/fmicb.2021.711037
Received: 17 May 2021; Accepted: 08 October 2021;
Published: 19 November 2021.
Edited by:
Xian-Zhi Li, Health Canada, CanadaReviewed by:
Michael Fruci, London Research and Development Centre, Agriculture and Agri-Food Canada, CanadaZhiyong Zong, Sichuan University, China
Copyright © 2021 Zhou, Liang, Dong, Zhang, Feng, Shi, Gao, Li, Zhang, Lu, Lin, Li, Zhang, Zhu and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mei Zhu, emh1bWVpX2RAMTYzLmNvbQ==; Qiyu Bao, YmFvcXlAZ2Vub21pY3MuY24=
†These authors have contributed equally to this work
 Kexin Zhou
Kexin Zhou Jialei Liang
Jialei Liang Xu Dong
Xu Dong Peiyao Zhang
Peiyao Zhang Chunlin Feng2,3
Chunlin Feng2,3 Mengdi Gao
Mengdi Gao Junwan Lu
Junwan Lu Xi Lin
Xi Lin Kewei Li
Kewei Li Hailin Zhang
Hailin Zhang Mei Zhu
Mei Zhu Qiyu Bao
Qiyu Bao