- College of Veterinary Medicine and Research Institute for Veterinary Science, Seoul National University, Seoul, South Korea
The worldwide spread of extended spectrum β-lactamase (ESBL)- and AmpC β-lactamase (AmpC)-producing Escherichia coli poses serious threats to public health. Swine farms have been regarded as important reservoirs of ESBL/AmpC-EC. This study aimed to determine the prevalence, ESBL/AmpC types, and clonal distribution of ESBL/AmpC-EC from swine farms and analyze the difference according to the swine production stages. In addition, we evaluated the potential risks of swine ESBL/AmpC-EC clones to humans. Individual fecal samples (n = 292) were collected from weaning, growing, finishing, and pregnant pigs in nine swine farms of South Korea between July 2017 and March 2020. In total, 161 ESBL/AmpC-EC isolates were identified (55.1%), with the highest prevalence detected in the weaning stage (86.3%). The dominant ESBL and AmpC types were CTX-M-55 (69.6%) and CMY-2 (4.3%), respectively. CTX-M found in all production stages, while CMY was only found in growing and finishing stages. In the conjugation assay, the high transferability of CTX-M gene (55.8%) was identified, while the transfer of CMY gene was not identified. The major clonal complexes (CCs) were CC101-B1 (26.8%), CC10-A (8.7%), and CC648-F (2.9%). There was similarity in clonal distribution between different swine production stages within swine farms, estimated using the k-means analysis, which suggested a clonal transmission between the different swine stages. Among swine ESBL/AmpC-EC sequence types (STs), seven STs (ST101, ST10, ST648, ST457, ST410, ST617, and ST744) were common with the human ESBL/AmpC-EC, which registered in National Center for Biotechnology Information database. The clonal population structure analysis based on the virulence factor (VF) presented that swine ESBL/AmpC-EC clones, especially ST101-B1, harbored a highly virulent profile. In conclusion, ESBL/AmpC-EC was distributed throughout the swine production stages, with the highest prevalence in the weaning stage. The CTX-M was present in all stages, while CMY was mostly found in growing-finishing stages. The swine ESBL/AmpC-EC was identified to harbor shared clone types with human ESBL/AmpC-EC and a virulent profile posing potential risk to humans. Considering the possibility of genetic and clonal distribution of ESBL/AmpC-EC among swine production stages, this study suggests the need for strategies considering the production system to control the prevalence of ESBL/AmpC-EC in swine farms.
Introduction
The third-generation cephalosporin (3GC)-resistant Enterobacteriaceae, including the extended spectrum β-lactamase (ESBL)- or AmpC β-lactamase (AmpC)-producing Escherichia coli (ESBL/AmpC-EC), and the carbapenem Enterobacteriaceae, including carbapenemase (CP)-producing E. coli (CP-EC), have been reported as a serious global threat to public health. The 3GCs and carbapenems show excellent activity against Gram-positive and Gram-negative bacteria and are particularly prescribed in treating multidrug-resistant bacterial infections (Muller et al., 2018). The spread of ESBL/AmpC-EC or CP-EC is of great concern because it could aid the emergence and spread of pathogens that are difficult to treat even with an antimicrobial agent of choice regarded as a final treatment option (Nathisuwan et al., 2001; Cortes et al., 2010). In addition, as ESBL/AmpC- or CP-producing extra-intestinal pathogenic E. coli (ExPEC) clones are increasingly reported worldwide, the virulence potential of ESBL/AmpC-EC or CP-EC has also become an issue (Schaufler et al., 2016; Day et al., 2019; Santos et al., 2020).
Intensive use and misuse of β-lactam antibiotics, including penicillins, carbapenems, monobactams, and cephalosporins, in veterinary medicine has led to the emergence and spread ESBL/AmpC-EC in the animal husbandry (Dohmen et al., 2017). The increasing prevalence of ESBL/AmpC-EC in food-animal farms has been reported in multiple continents including Europe (Cortes et al., 2010; Schmithausen et al., 2015; Lupo et al., 2018), America (Cyoia et al., 2018; Moffat et al., 2020), Africa (Belmahdi et al., 2016; Alonso et al., 2017; Jouini et al., 2021), Australia (Abraham et al., 2015; Sparham et al., 2017), and Asia (Liu et al., 2018; Tansawai et al., 2019). In particular, pigs have been regarded as the main driver of the increasing prevalence of ESBL/AmpC-EC in food-animals (Bergspica et al., 2020). This epidemiologic trend may be attributed to the long-term and extensive usage of 3GC during swine production periods (Song et al., 2009; Peirano et al., 2010; Yoo et al., 2010; Barguigua et al., 2013; Hong et al., 2020). The possibility of ESBL-EC transmission from swine farms to humans has been continuously proposed and vice versa (Bok et al., 2020; Song et al., 2020). Various ESBL/AmpC-EC transmission routes have been suggested, including the food-chain of pigs (Liu et al., 2019), direct contacts of farm workers with pigs (Schmithausen et al., 2015), and manure excretion into the surrounding environment in farms such as soils, ponds, and rivers (Furlan and Stehling, 2018).
Swine production consists of four stages, including farrowing (birth to 3–4 weeks of age), weaning (4–7 weeks old), growing (7–14 weeks), and finishing stage (14–24 weeks old), and the farrow-to-finish swine farms refers to farms rearing all four swine production stages Pigs at each different stage in the farrow-to-finish swine farms are usually reared in three separated farrowing, weaning, and growing-finishing barns, respectively. However, there is a generally high probability of bacterial co-transmission between the production stages within a farm (Verhegghe et al., 2013; Fromm et al., 2014), through various routes, including farm workers and veterinarians, instrument contamination, or manure excretions into barns of different stages (Fromm et al., 2014; Schmithausen et al., 2015). Previous studies have reported the identification of ESBL/AmpC-EC throughout the swine production stages, with varying prevalence at each production stage (Schmithausen et al., 2015; Dohmen et al., 2017). Although not studied well until now, the characteristics and distribution of ESBL/AmpC-EC may also differ depending on the stage of swine production, and understanding their difference could be a cornerstone in reducing the prevalence of ESBL/AmpC-EC in swine farms.
The present study aimed to determine the prevalence, multidrug resistance (MDR), and virulence potential of ESBL/AmpC from swine farms. In addition, differences in β-lactamase types, antimicrobial susceptibility, and clonal distribution were analyzed according to the swine production stages. Finally, potential risks of swine ESBL/AmpC-EC clones on human health were evaluated.
Materials and Methods
Sample Collection
Samples were collected from nine farrow-to-finish swine farms located in four provinces in South Korea (four in Gyeonggi-do, three in Jeolla-do, one in Chungcheong-do, and one in Gyeongsang-do) between July 2017 and March 2020. Each farm was visited once (two farms in 2017, five farms in 2018, one farm in 2019, and one farm in 2020). Individual fecal samples were collected in similar numbers from each swine farm; a total of 26–34 pigs from each swine farm, including 5–6 weaning piglets (4–7 weeks old), 9–11 growing pigs (7–14 weeks old), 8–11 finishing pigs (14–24 weeks old), and 3–6 pregnant sows. A total of 292 samples from 51 weaning piglets, 96 growing pigs, 47 finishing pigs, and 50 pregnant sows were collected and transported immediately to a laboratory. All pigs included in this study were healthy without diarrhea.
Isolation of ESBL/AmpC-EC and Non-ESBL/AmpC-EC
To isolate ESBL/AmpC-EC, 1 g of each fecal sample was homogenized with 9 ml of E. coli broth (Oxoid, United Kingdom) for 1 min using a homogenizer and incubated overnight at 37°C. Approximately 100 μl of enriched E. coli culture suspension was spread on MacConkey agar (Oxoid, United Kingdom). Cefotaxime (CTX) disk (30 μg/ml, Oxoid, United Kingdom) was then placed on the plate. After overnight incubation at 37°C, 2–4 3GC-resistant E. coli (3GC-EC) candidate isolates grown inside the CTX resistant zone (<22 mm) were selected and streaked on CHROMagar™ ESBL (CHROMagar, France) to demonstrate the morphology of 3GC-EC colony. One 3GC-EC isolate from each sample was randomly selected if more than one 3GC-EC isolates were identified from one sample. Finally, a standard double-disk test was performed to confirm the typical ESBL and AmpC phenotype, as described in the 2016–Clinical Laboratory Standard Institute (CLSI) guideline M100S 26th Edition.
To isolate non-ESBL/AmpC-EC, 10 μl of enriched E. coli broth suspension was streaked on MacConkey agar. Three colonies showing typical E. coli morphology were randomly selected and transferred to Eosin Methylene Blue (EMB) agar (Oxoid, United Kingdom) for purification. The suspected E. coli isolates on EMB agar were subjected to a standard ESBL/AmpC double-disk test, and typical non-ESBL/AmpC-EC isolates were determined as non-ESBL/AmpC-EC. One non-ESBL/AmpC-EC isolate per sample was randomly selected, where more than one non-ESBL/AmpC-EC isolates were identified from one sample. Considering the distribution of ESBL/AmpC-EC isolates by farm and production stage, a total of 81 non-ESBL/AmpC-EC isolates were selected for further analysis.
The presence of ESBL genes (blaCTX-M, blaTEM, and blaSHV), AmpC genes (blaCMY), and carbapenemase genes (blaKPC, blaNDM, and blaOXA) among ESBL/AmpC-EC isolates were determined with PCR. PCR amplicons were sequenced using the ABI PRISM 3730XL DNA analyzer (Applied Biosystems, United States). DNA sequences were compared with the published β-lactamase gene sequences available from the GenBank database of the NCBI using the BLAST program.1 Primer sequences and reaction conditions for each PCR-based genotyping of ESBL/AmpC are summarized in Supplementary Table 1.
Antimicrobial Susceptibility Test
Disk diffusion susceptibility test (Kirby-Bauer method) were conducted for 14 antibiotics: amoxicillin/clavulanic acid (AMC, 20/10 μg), ampicillin (AMP, 10 μg), cefotaxime (CTX, 30 μg), ceftazidime (CAZ, 30 μg), ceftriaxone (CRO, 30 μg), aztreonam (ATM, 30 μg), imipenem (IMP, 10 μg), chloramphenicol (C, 30 μg), amikacin (AK, 30 μg), gentamicin (CN, 10 μg), tetracycline (TE, 30 μg), nalidixic acid (NA, 30 μg), ciprofloxacin (CIP, 5 μg), and trimethoprim–sulfamethoxazole (SXT, 1.25/23.75 μg). Escherichia coli ATCC 25922 was used as the reference strain for quality control. Antimicrobial susceptibility was interpreted according to the CLSI guidelines.
Plasmid-Mediated Antimicrobial Resistance Genes, Intestinal Pathogenic E. coli Typing, and Extraintestinal Pathogenic E. coli Associated Virulence Factor Genotyping
The presence of plasmid-mediated antimicrobial resistance genes inferring resistance to chloramphenicol (catA, cmlA, and floR), tetracycline (tetA, tetB, and tetD), quinolone (qnrA, qnrB, qnrC, qnrS, and aac(6)-Ib-cr), aminoglycoside (aac(3)-I, aac(3)-II, and aac(3)-IV), and Sulfonamide/Trimethoprim (dfrIa, dfrIb, dfrII, dfrVII, and dfrXII) were determined using PCR. Intestinal pathogenic E. coli typing was conducted for the following types; shiga toxin-producing E. coli (stx1 and stx2), enteropathogenic E. coli (eaeA and bfpV), enteroaggregative E. coli (aggR), enteroinvasive E. coli (ipaH), and enterotoxigenic E. coli (lt, sta, stb, and east-1). The extraintestinal pathogenic E. coli associated virulence factors (ExPEC VFs) associated with adhesion (fimH, iha, papC, and csgA), toxin (astA, hlyA, aat, tsh, and pic), protectin/serum resistance (traT and ompT), and siderophore (fyuA and iroNe.coli) were also determined using PCR. Primer sequences and reaction conditions are summarized in Supplementary Table 1.
Biofilm Assay
Biofilm production assays were performed following a previously described protocol with modification (Naves et al., 2008). Briefly, overnight Luria-Bertani (LB) broth culture was diluted in fresh LB broth to a McFarland scale of 0.5. Approximately 120 μl of this dilution was added into 96-well microtiter plate and incubated for 24 h at 30°C in a stationary condition. Each bacterial suspension was inoculated into three wells of the microtiter plate. Growth optical densities (ODs) were measured at λ = 595 nm with a multiplate reader (Bio-rad, United States). The wells were then washed once with 200 μl of phosphate-buffered saline (PBS) dried for 20 min, and stained with 120 μl of 1% crystal violet for 5 min. This was followed by gentle washing with 200 μl of distilled water (DW) for four times and air-drying for 1 h. The absorbed dye was solubilized in 120 μl of absolute ethanol, and ODs were read at 595 nm. The extent of biofilm formation was calculated using the formula: , where SBF is the specific biofilm formation index, AB is the OD595 of the stained bacteria, CW is the OD595 of the stained control wells containing absolute media without bacteria, and G is the OD595 corresponding to cell growth in the media. An SBF value above 0.5 was suggested as positive biofilm formation. E. coli ATCC 25922 was used as the positive control, while the culture medium was used as the negative control.
Expression of Biofilm-Associated Extracellular Matrix Components: Curli Fimbriae and Cellulose
To determine the expression of biofilm-associated extracellular matrix components (cellulose and curli fimbriae), a macrocolony assay was performed following a previously described protocol with modification (Schaufler et al., 2016). Approximately, 5 μl of an overnight culture from a single colony grown in 2 ml of LB medium was dropped on YESCA agar [10 g/L casamino acids (BD Bioscience, United States), 1 g/L yeast extract (BD Bioscience, United States), and 20 g/L agar (BD Bioscience, United States)] with Congo red solution [0.5% Congo red (Sigma-Aldrich, Germany) and 0.25% Coomassie brilliant blue (Sigma-Aldrich, Germany) diluted in ethanol]. Plates were incubated for 5 days at 28°C, and results were interpreted using the four morphotypes: rdar (red, dry, and rough; curli and cellulose), pdar (pink, dry, and rough; cellulose only), bdar (brown, dry, and rough; curli only), and saw (smooth and white; neither curli nor cellulose). E. coli ATCC 25922 was used as the negative control for the expression of curli fimbriae and cellulose.
Plasmid Typing and Conjugation Assay
The replicon typing of ESBL/AmpC-EC was analyzed with the major plasmid incompatibility groups among Enterobacteriaceae (HI1, HI2, I1-Iγ, I2, X1, X2, X3, X4, L/M, FIA, FIB, FIC, FIIs, A/C, P, K B/O, and N) using a PCR-based replicon-typing method (Carattoli et al., 2005; Johnson et al., 2012; Lv et al., 2013). Conjugation assay was conducted with E. coli J53-AziR as the recipient and ESBL/AmpC-EC as the donors. LB agar plates containing 4 mg/L of CTX and 100 mg/L of sodium azide were used to select the transconjugants. The presence of the ESBL/AmpC genes (blaCTX-M and blaCMY), plasmid-mediated antimicrobial resistance genes, and replicon types in the transconjugants was confirmed by PCR. Primer sequences and reaction conditions are summarized in Supplementary Table 1.
Clonal Distribution Analysis of Swine ESBL/AmpC-EC
Multi-Locus Sequence Typing and E. coli Phylogenetic Group Typing
Of the 161 ESBL/AmpC-EC isolates from the present study, 138 isolates were selected based on their antibiotic resistance and ExPEC VFs to analyze multi-locus sequence typing (MLST) and E. coli phylogenetic group typing. The MLST of ESBL/AmpC-EC isolates was performed as described previously (Wirth et al., 2006). A detailed scheme of gene amplification, allelic type, and sequence type (ST) assignment methods is available on the pubMLST website.2 The minimum spanning tree (MST) based on allelic profiles of seven MLST housekeeping genes was constructed using the BioNumerics software, version 6.6 (APPLIED MATHS, Belgium).
Seven E. coli phylogenetic groups (A, B1, B2, D, C, E, and F) were determined following a previously described protocol with application (Clermont et al., 2013). Primer sequences and reaction conditions for each PCR-based phylogenetic group analysis are summarized in Supplementary Table 1.
Similarity Analysis of the Clonal Distribution of Swine ESBL/AmpC-EC Following the Swine Production Stages Within Farms
We analyzed the similarity in the distribution of ST and phylogenetic groups following the swine production stages in each farm using the k-means similarity clustering algorithm based on Euclidean distance (Hartigan and Wong, 1979). To find the optimal number of clusters (k), we applied the average silhouette method (Lengyel and Botta-Dukat, 2019). The average silhouette method presumes that the optimal number of clusters k is the one that maximizes the average silhouette over a range of possible values for k. In this study, the optimal value of k was nine (Supplementary Figure 2). The k-means clustering analysis and the average silhouette method were conducted using the R software, version 4.3.2 (R foundation, Austria).
Based on the combination of nine swine farms (“farm A” to “farm I”) and four production stages (weaning piglets to sows), a total of 36 points indicating ST composition at each farm’s swine production stages were generated. Of the 36 points, three were excluded because there were no isolated ESBL/AmpC-EC strains from “growing stage” of “farm H,” “pregnant stage” of “farm H,” and “growing stage” of “farm I.” We clustered 33 points into nine clusters using the k-means algorithm based on the Euclidean distance of paired two points. The k-means clustering plot was generated using the R software, version 4.3.2 (R foundation, Austria).
Analysis of the Potential Risk of Swine Farm-Derived ESBL/AmpC-EC Clones
Identification of the Shared Major STs of ESBL/AmpC-EC Isolated From Swine Farms and Humans
To identify the shared major STs between swine and human ESBL/AmpC-EC isolates, the whole genome sequence (WGS) data of human ESBL/AmpC-EC isolates registered in the NCBI Pathogen Isolates Browser3 were used.
WGS data of 11,269 human ESBL/AmpC-EC isolates from human hosts whose assembly data were available (accessed on 20 May 2021) were downloaded and analyzed to determine the MLST STs (Supplementary Material 1). In total, 739 STs were identified from 11,269 human ESBL/AmpC-EC isolates. We selected the major 20 STs of human ESBL/AmpC-EC isolates covering 73.8% of the total isolates (8,320/11,269) for further analysis.
Clonal Population Structure Analysis Based on the VFs and Phylogenetic Group Profiles
We investigated the potential risks of swine ESBL/AmpC-EC clones on humans by a clonal population structure analysis based on their ExPEC VFs and phylogenetic group profile using program STRUCTURE (Pritchard et al., 2000). Swine ESBL/AmpC-EC isolates were assigned into virulence profile populations (k) using a Baysian method in the program. The most likely number of populations (k) was defined by the value producing a maximal rate change in posterior probability, Δln (k; Evanno et al., 2005). The optimal number of populations k was five in the present study (Supplementary Figure 3). Assignment coefficients (Q values such as proportions of population k) were generated for each strain using the Markov chain Monte Carlo searches, which consisted of 100,000 burn-in steps followed by 100,000 iteration steps. Among the five populations, ESBL/AmpC-EC isolates were assigned to their best-fit populations based on the highest Q value. The Q values of individual ESBL/AmpC-EC are presented in a 100% stacked bar chart sorted by the STs.
Statistical Analyses
The comparative analyses between groups (one stage vs. other stages, ESBL/AmpC-EC vs. non-ESBL/AmpC-EC) were performed using the generalized estimating equation (GEE) to adjust the farm-induced factors. In the GEE analysis, we presumed that several characteristics of the isolates could be affected by farm factors; therefore, we set the farm as “subject variable” and the number of isolates in each farm as “within subject variables.” Where zeros caused problems in calculating the odds ratio (OR) in the GEE analysis, the Fisher’s exact test was conducted by adding 0.5 to each cell (Pagano and Gauvreau, 2018). In the analysis of the differences in the average number of VFs and resistance to antibiotic classes between ESBL/AmpC-EC and non-ESBL/AmpC-EC, the student’s t-test was applied. All statistical analyses were conducted using the Statistical Package for the Social Sciences (SPSS) program, version 27.0 (IBM SPSS Statistics for Windows, United States).
Results
Prevalence, MDR, and Virulence Potential of ESBL/AmpC-EC in Swine Farms
Prevalence of ESBL/AmpC-EC in Swine Farms
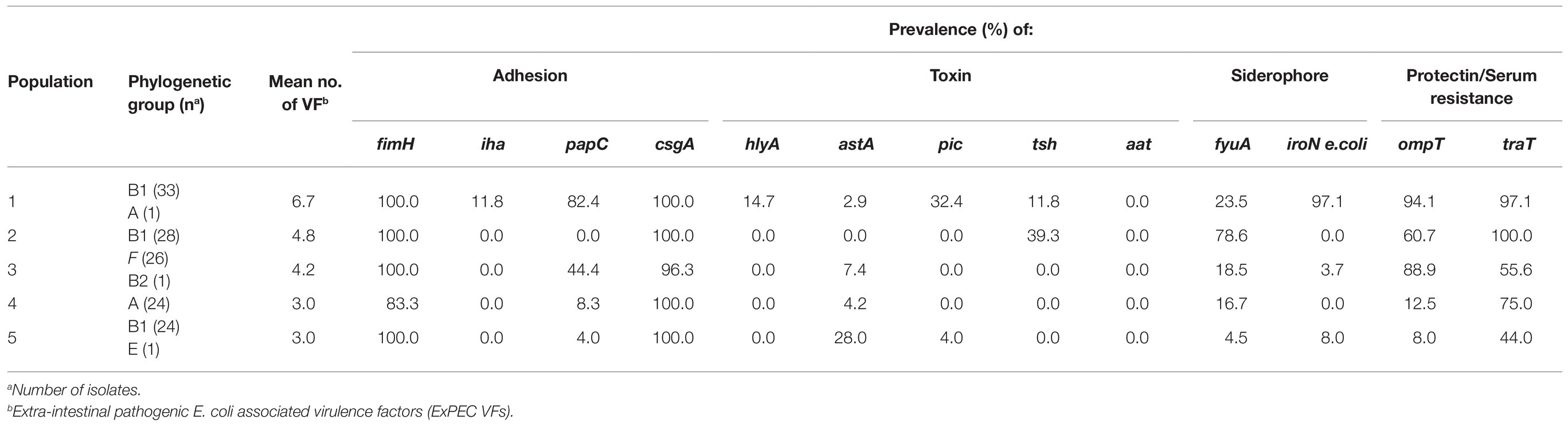
In total, 161 (55.1%) ESBL/AmpC-EC isolates were identified from 292 individual swine fecal samples. The farm prevalence of ESBL/AmpC-EC ranged from 17.6 to 89.7%, depending on the swine from which the samples were obtained (Figure 1A). Compared with other stages, weaning piglets (86.3%, 44/51) displayed the significantly highest prevalence of ESBL/AmpC-EC (p < 0.05), followed by growing pigs (58.3%, 51/96), finishing pigs (48.4%, 45/95), and pregnant sows (43.1%, 21/50; Figure 1B).
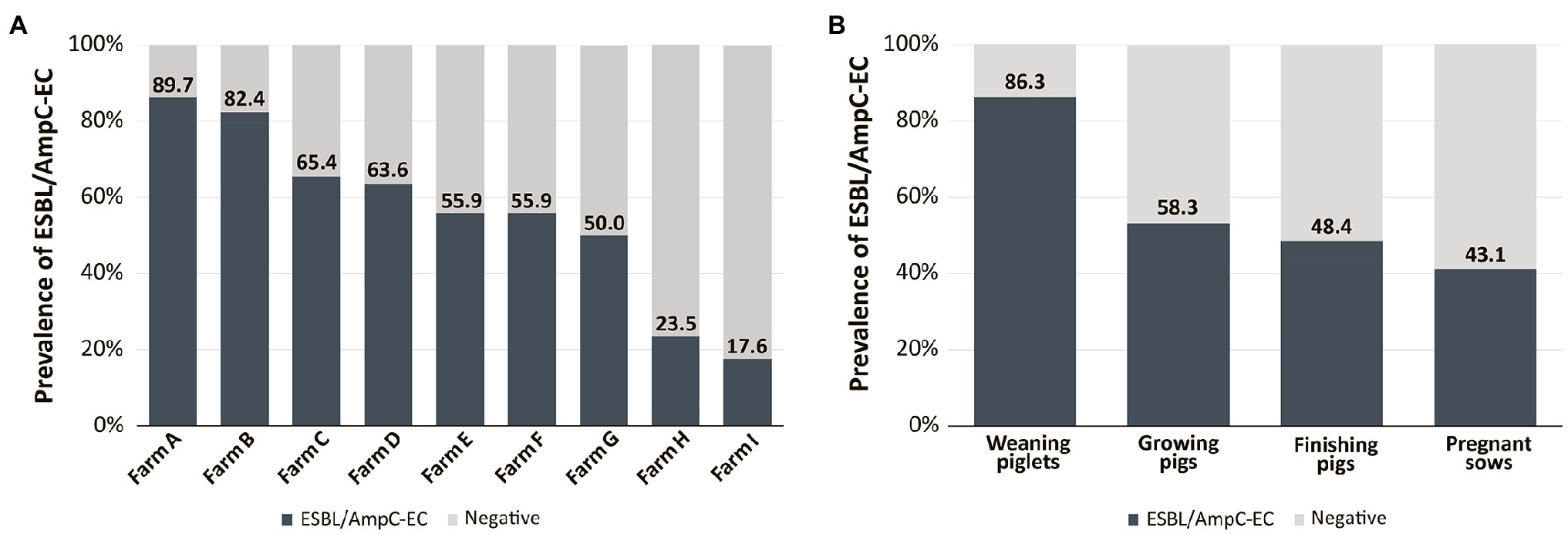
Figure 1. Prevalence of extended spectrum β-lactamase (ESBL)- or AmpC β-lactamase (AmpC)-producing E. coli (ESBL/AmpC-EC) according to different swine farms (A) and swine production stages (B).
Higher MDR of ESBL/AmpC-EC Relative to Non-ESBL/AmpC-EC
We investigated the antimicrobial resistance of the 161 ESBL/AmpC-EC isolates and compared them with those of the 81 non-ESBL/AmpC-EC isolates (Figure 2). Notably, ESBL/AmpC-EC isolates showed resistance to a higher number of antimicrobial classes (average: 5.7 antimicrobial classes) compared with that of non-ESBL/AmpC-EC isolates (average: 2.9 antimicrobial classes; p < 0.05). Furthermore, the MDR rate was significantly higher in the ESBL/AmpC-EC isolates (100%) compared with that in the non-ESBL/AmpC-EC isolates (63.0%; OR: 80.0, 95% CI: 11.68–547.88). Resistance to six antibiotics (AMC, CTX, CAZ, CRO, ATM, and AK) was found only in ESBL/AmpC-EC isolates but not in non-ESBL/AmpC-EC isolates. Except for the tetracycline class and carbapenem class, ESBL/AmpC producers showed a higher resistance rate for all antibiotic classes compared with that in non-ESBL/AmpC producers. Resistance to three of these antibiotic classes, which included broad-spectrum penicillin (OR: 100.8, 95% CI: 5.99–1694.62, p < 0.05), aminoglycoside (OR: 5.6, 95% CI: 2.08–15.26, p < 0.05), and quinolone (OR: 6.6, 95% CI: 3.31–12.99, p < 0.05), was significantly higher in the ESBL/AmpC-EC strains compared to non-ESBL/AmpC-EC.
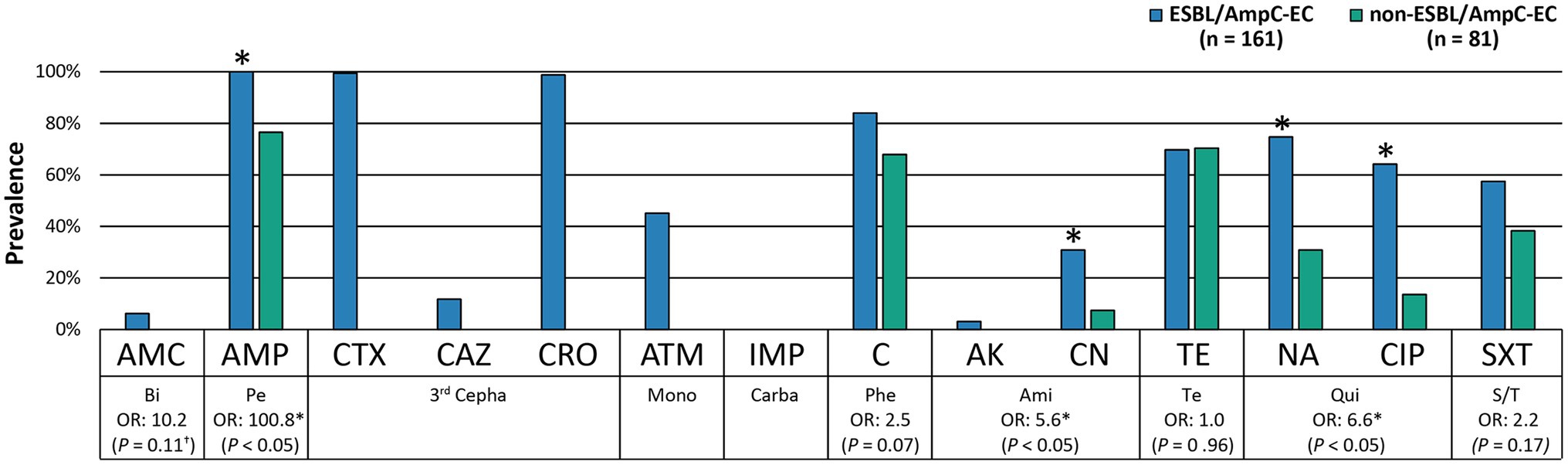
Figure 2. ESBL/AmpC-EC showed significantly higher resistance rate to antibiotics compared to non-ESBL/AmpC-EC. Statistically significant (*p < 0.05, GEE; †p < 0.05, Chi-square test).
In the antimicrobial resistance genotyping, the aminoglycoside resistance gene, aac(3)-II, was significantly more prevalent in the ESBL/AmpC-EC isolates (OR: 6.2, 95% CI: 1.25–30.70, p < 0.05) than in the non-ESBL/AmpC-EC (Supplementary Table 2).
ESBL/AmpC-EC With Multiple ExPEC VFs
All ESBL/AmpC-EC and non-ESBL/AmpC-EC were identified as non-pathogenic commensal E. coli, except for two ESBL/AmpC-EC isolates. One ESBL/AmpC-EC isolate was identified as enteropathogenic E. coli carrying heat stable ensterotoxin STa, and the other ESBL/AmpC-EC isolate was identified as atypical enteropathogenic E. coli carrying eaeA but not bfpV.
A significantly higher number of ExPEC VFs were identified in the ESBL/AmpC-EC isolates (average: 4.6 VFs) compared with that in the non-ESBL/AmpC-EC isolates (average: 3.6 VFs; p < 0.05). The OR of having 6–7 VFs were 8.8-fold greater (95% CI: 1.31–59.30, p < 0.05) in the ESBL/AmpC-EC isolates than in the non-ESBL/AmpC-EC isolates (Figure 3). In contrast, the ESBL/AmpC-EC isolates showed a 0.4-fold less OR to harbor three or less VFs relative to that in the non-ESBL/AmpC-EC isolates (95% CI: 0.13–0.97, p < 0.05). Three VFs were highly prevalent in the ESBL/AmpC-EC isolates, namely pyelonephritis-associate pilus C, papC (OR: 19.8, 95% CI: 2.54–153.45, p < 0.05), serine protease pic autotransporter, pic (OR: 19.6, 95% CI: 1.16–330.30, p < 0.05), and outer membrane protease T, ompT (OR: 1.9, 95% CI: 1.00–3.64, p < 0.05), compared with the non-ESBL/AmpC-EC isolates (Table 1).
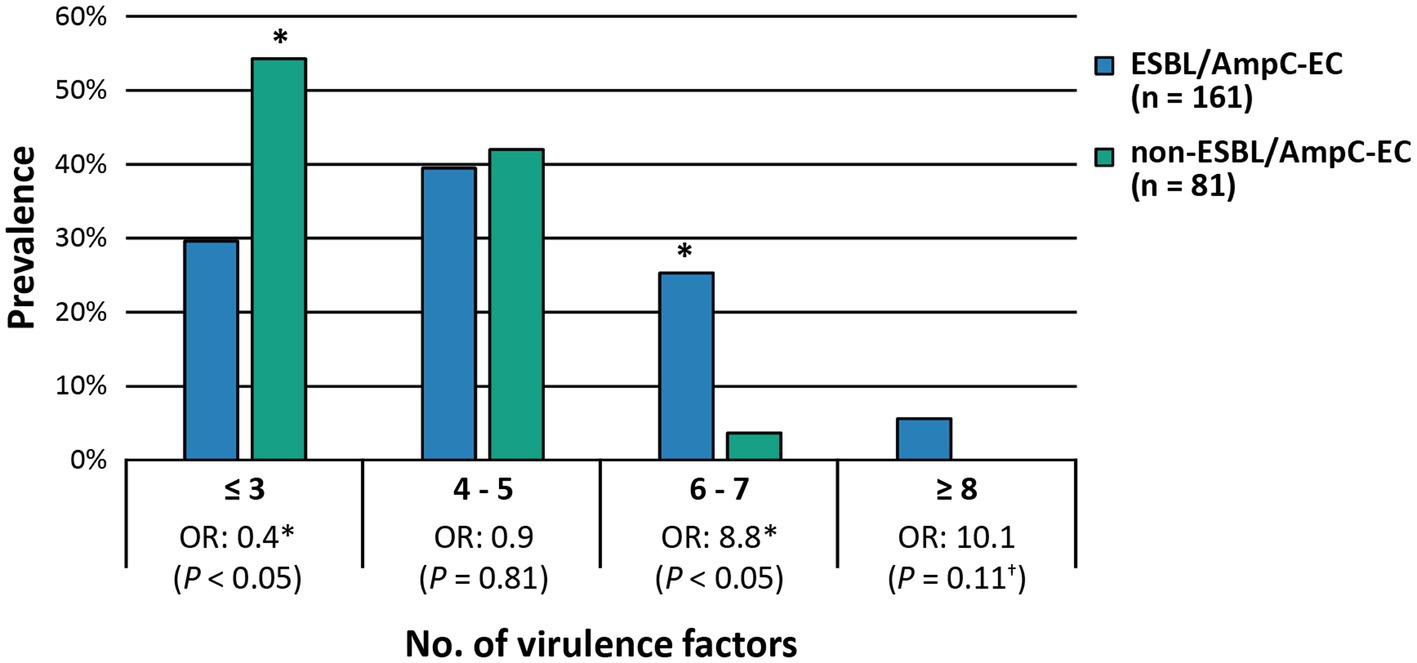
Figure 3. ESBL/AmpC-EC carried higher number of extra-intestinal pathogenic E. coli associated virulence factors (ExPEC VFs) compared to non-ESBL/AmpC-EC. OR, odds ratio. Statistically significant (*p < 0.05, GEEs; †p < 0.05, Chi-square test).
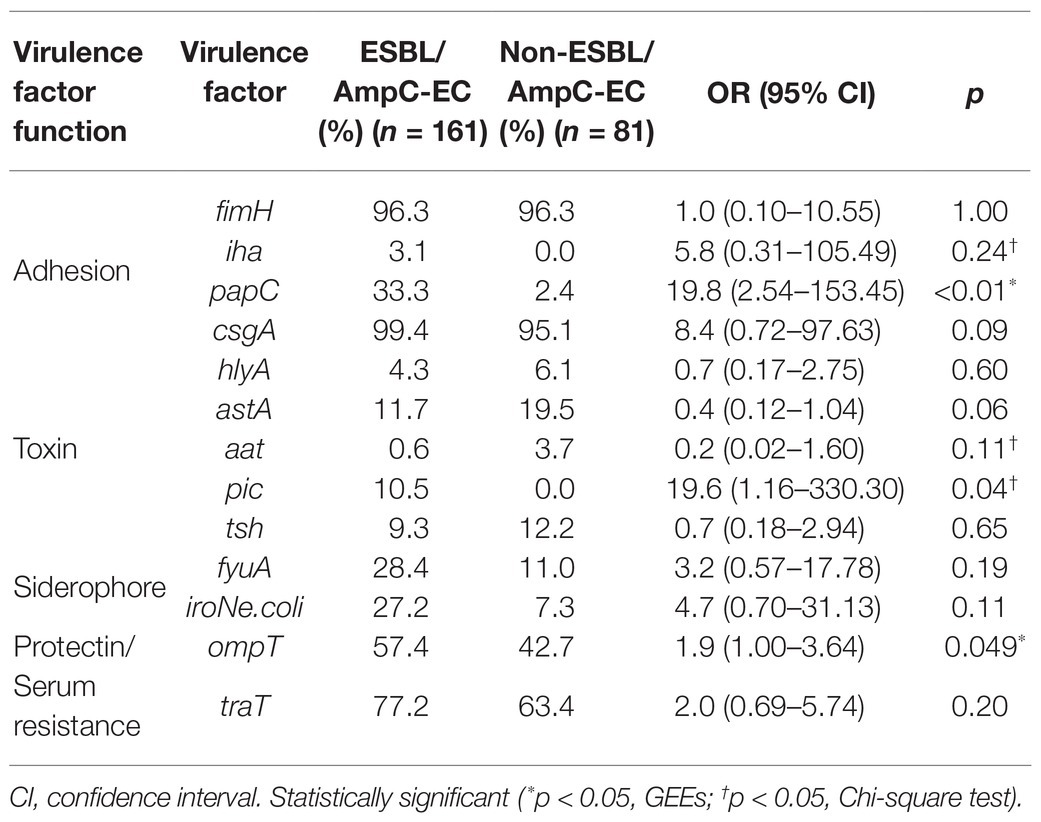
Table 1. Comparison of the carriage of extra-intestinal pathogenic E. coli associated virulence factors (ExPEC VFs) between extended spectrum β-lactamase (ESBL)- or AmpC β-lactamase (AmpC)-producing E. coli (ESBL/AmpC-EC) and non-ESBL/AmpC-EC isolates.
Improved Biofilm Formation of ESBL/AmpC-EC
The biofilm formation rate was higher in ESBL/AmpC-EC isolates (42.2%, 68/161) than that in non-ESBL/AmpC-EC isolates (16.0%, 13/81). The OR of biofilm formation was 3.8-fold greater in the ESBL/AmpC-EC isolates than in the non-ESBL/AmpC-EC isolates (95% CI: 1.42–10.30, p < 0.05). However, no significant differences were observed in the formation of two biofilm-associated extracellular matrix components, curli fimbriae (OR: 1.80, 95% CI: 0.71–4.55, p = 0.21) and cellulose (OR: 0.45, 95% CI: 0.10–2.07, p = 0.30), from the ESBL/AmpC-EC isolates relative to the non-ESBL/AmpC-EC isolates.
Difference in the Distribution of β-Lactamases and Antimicrobial Susceptibility of ESBL/AmpC-EC According to the Swine Production Stages
Distribution of β-Lactamases in ESBL/AmpC-EC According to the Swine Production Stages
Out of the 161 ESBL/AmpC-EC isolates, 154 isolates (95.7%, 154/161) were identified as ESBL-EC carrying the CTX-M family β-lactamases (CTX-M), while seven isolates (4.3%, 7/161) were identified as AmpC-EC carrying the CMY family β-lactamases (CMY; Figure 4). None of the isolates carried both CTX-M and CMY together. The CTX-M group was found in ESBL/AmpC-EC in all production stages and occupied 88.2–100.0% of the ESBL/AmpC-EC (Figure 4B). However, the CMY group was identified in ESBL/AmpC-EC isolates only in the growing (11.8%, 6/51) and finishing stages (2.2%, 1/45).
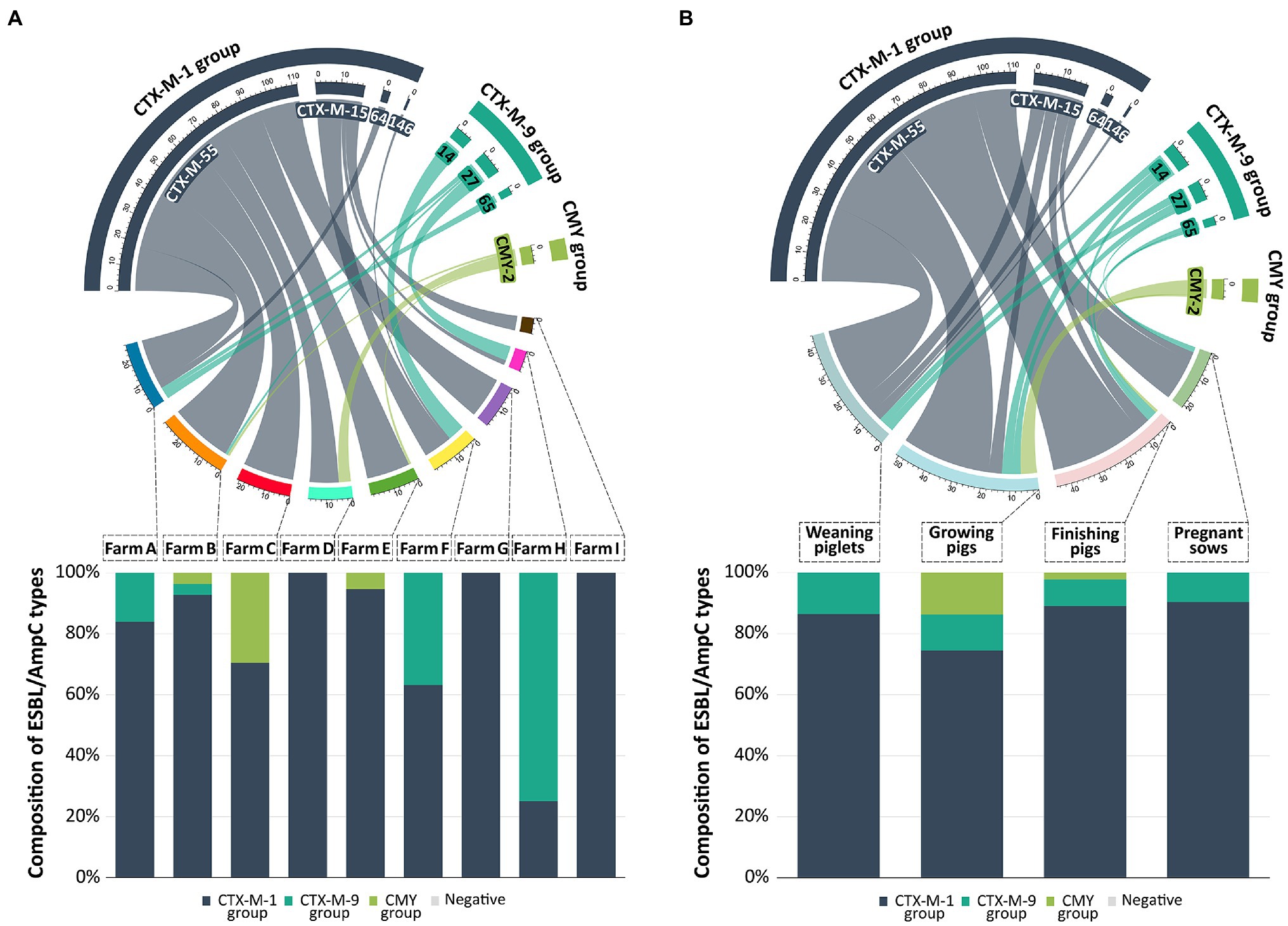
Figure 4. Prevalence and distribution of ESBL/AmpC types from swine farms. In the chord diagram, the size of segments on the top represent the number of ESBL/AmpC-EC isolates with a specific ESBL/AmpC types. Size of segments on the bottom represent the number of ESBL/AmpC-EC isolates detected in different farms (A) and production stages (B). Ribbons connecting the top and bottom segments represent the number of ESBL/AmpC-EC isolates with a specific ESBL/AmpC type found on the respective farms and production stages. The connected bar chart shows the composition of ESBL/AmpC types based on the number of ESBL/AmpC-EC isolates in different farms (A) or production stages (B). The chord diagram and bar chart were generated with R software (ver. 4.3.2).
The CTX-M-1 group β-lactamase (83.9%, 135/161) was the most prevalent CTX-M; CTX-M-55 β-lactamase (69.6%, 112/161), CTX-M-15 (11.8%, 19/161), CTX-M-64 (1.9%, 3/161), and CTX-M-146 (0.6%, 1/161) belonged to this group (Figure 4). The second most prevalent CTX-M was the CTX-M-9 group β-lactamase (11.8%, 19/161); CTX-M-14 (5.6%, 9/161), CTX-M-27 (4.3%, 7/161), and CTX-M-65 (1.9%, 3/161) belonged to this group. All seven CMY-producing E. coli isolates was identified to carry CMY-2 β-lactamase. All ESBL/AmpC-EC carried only one type of CTX-M β-lactamase or CMY β-lactamase, and there were no isolates carrying more than one CTX-M or CMY β-lactamase. TEM β-lactamase was found in the 31 ESBL/AmpC-EC isolates (24.8%); however, all of these TEM β-lactamases were identified as TEM-1, which was a non-ESBL type in the sequencing analysis. Other ESBL types (CTX-M-2 group, CTX-M-8 group, CTX-M-25 group, and SHV) and carbapenemases (NDM, OXA, and KPC) were not identified in this study.
Antimicrobial Susceptibility of ESBL/AmpC-EC According to the Swine Production Stages
We compared the resistance rate of ESBL/AmpC-EC against 10 antimicrobial classes at specific stages to those of the other stages (Table 2). There were no significant differences in the resistance rate against all antimicrobial classes between the swine production stages, with the exception of β-lactamase inhibitor class. The resistance rate for the β-lactamase inhibitor class, which consisted of AMC, was significantly higher in growing pigs, compared with that of the other stages (OR: 9.8, 95% CI: 1.14–84.70, p < 0.05).
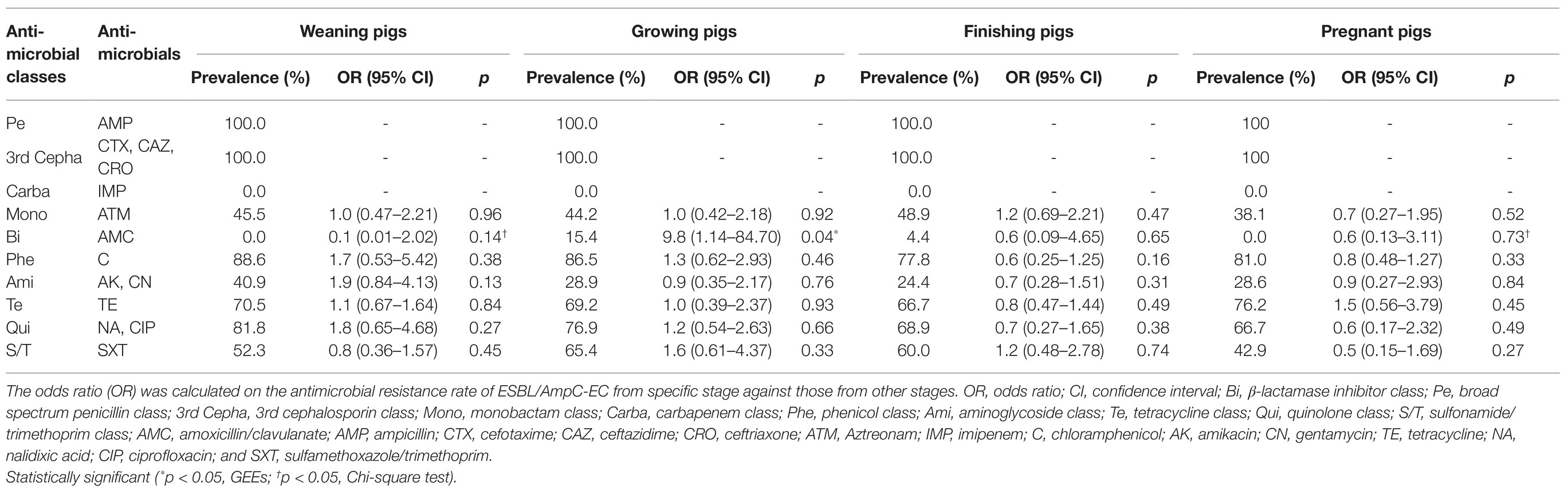
Table 2. Resistance rate of ESBL/AmpC-EC against antimicrobial classes following the swine production stages.
Transferability of ESBL/AmpC Genes
To evaluate the horizontal transferability of ESBL/AmpC genes, we conducted the conjugation assay on 131 CTX-M and seven CMY β-lactamase-producing isolates (Supplementary Figure 1). The transferability of the CTX-M gene, blaCTX-M, was 58.8% (77/131); however, the transfer of the CMY gene blaCMY−2 was not identified from all seven CMY-producing isolates. The most prevalent replicon type in the transconjugant of CTX-M was IncFIB (90.9%, 70/77), followed by IncI1-Iγ (28%, 17/77), IncI2 (6.5%, 5/78), and IncX4 (3.9%, 3/77). Various antibiotic resistance genes were transferred with blaCTX-M. The highest transferability of antimicrobial resistance gene was identified in floR (94.9%, 56/59), followed by aac(3)-II (85.7%, 6/7), and qnrS1 (75.0%, 12/16).
Clonal Distribution Analysis of ESBL/AmpC-EC From Swine Farms
Clonal Distribution of ESBL/AmpC-EC Between Swine Farms
The MLST and the seven E. coli phylogenetic group (A, B1, B2, D, C, E, and F) typing analysis were conducted to analyze the clonal distribution of the swine farm-derived ESBL/AmpC-EC isolates (Figure 5). In the MLST analysis, a total of eight CCs were identified, and CC101-B1 (26.8%, 37/138), CC10-A (8.7%, 13/138), CC648-F (2.9%, 4/138), and CC23-A (2.9%, 4/138) were the major CCs. Around 31 STs, including two non-typable STs, were identified, with ST101-B1 (22.8%, 31/138), ST457-F (16.2%, 22/138), ST75-B1 (12.5%, 17/138), and ST224-B1 (8.1%, 11/138) as the major STs.
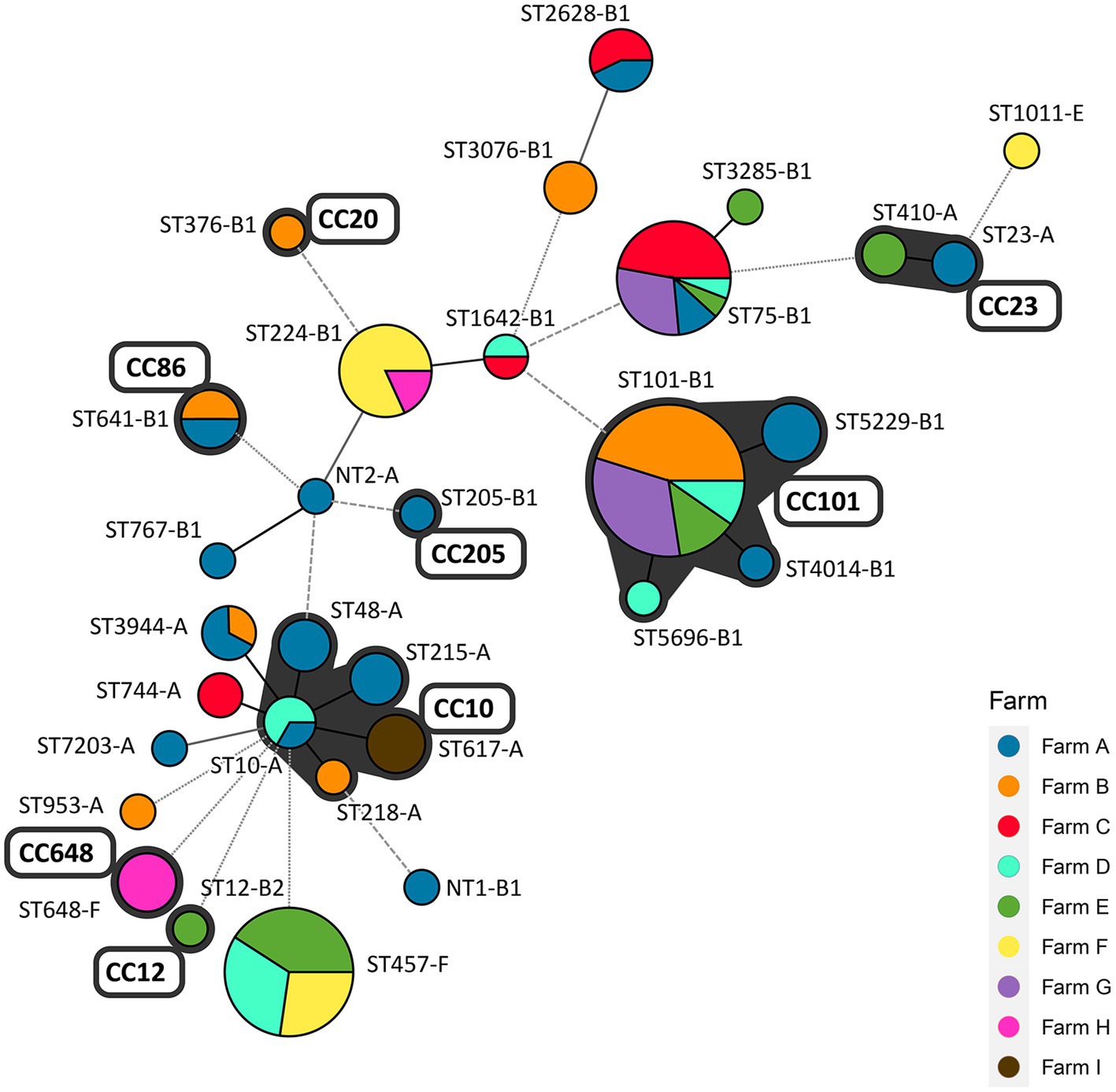
Figure 5. Minimum spanning tree (MST) based on allele profiles of multi-locus sequence type (MLST): clonal distribution of ESBL/AmpC-EC between pig farms. The number shows the sequence type of each node, and the size of the node indicates the number of strains belonging to the sequence type (ST)-phylogenetic group. The gray shadow represents the clonal complex (CC). Branch line types represent differences in the number of alleles: bold solid line (1 allele), thin solid line (2–3 alleles), dashed line (4 alleles), and dotted line (above 5 alleles). CC, clonal complex; ST, sequence type; NT, non-typable ST (including two different non-typable STs).
A total of 40.6% (56/138) of the ESBL/AmpC-EC isolates belonged to four CCs (CC101-B1, CC10-A, CC86-B1, and CC23-A), which were shared across two and more swine farms. A total of 73.5% (100/138) of ESBL/AmpC-EC isolates belonged to nine STs (ST10-A, ST101-B1, ST457-F, ST75-B1, ST224-A, ST641-B1, ST3944-A, ST-2628-B1, and ST1642-B1), which were shared across two and more swine farms.
Similarity of Clonal Distribution Among Swine Production Stages Within Farms
We evaluated the similarity in the clonal distribution of different production stages within the farms using the k-means similarity clustering algorithm (Figure 6; Supplementary Table 3). Distributions of STs and phylogenetic groups for each farm and production stage combination were presented in Supplementary Table 3. In Figure 6, 33 points represent the clonal distribution following the combination of nine farms (“farm A” to “farm I”) and four stages (“weaning piglets” to “pregnant sows”). In the k-means clustering analysis, the points were clustered into nine clusters based on the similarity distance between them, and 2–7 points belonged to each cluster. We found that the clonal distribution of three or all stages in the same farm was clustered together, thereby showing similarity, except for “farm D.” The clonal distribution of two stages in “farm D” were clustered together.
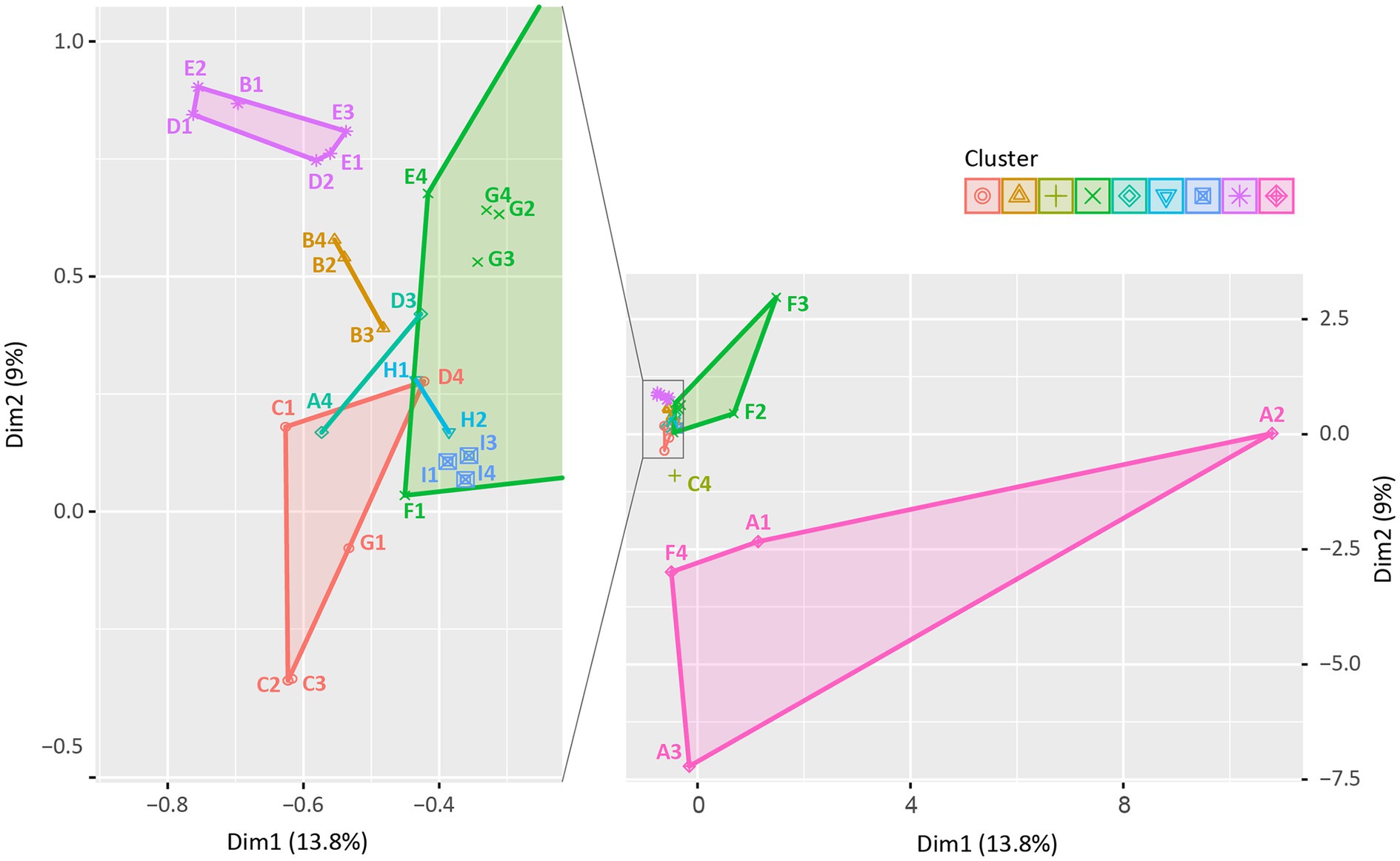
Figure 6. k-means similarity clustering plot: similarity in the clonal distribution among swine production stages within farms. A total of 33 points were described and clustered into 9 (symbols and colors) using the k-means similarity clustering algorithm based on Euclidean distance. The k-means cluster plot was generated using the R software (ver. 4.3.2). Each point indicates the distribution of STs among swine production stages in each farm, consisting of 36 points based on combination of 9 farms (A to I) and 4 production stages (1, weaning piglets; 2, growing pigs; 3, finishing pigs; and 4, pregnant sows; e.g., A1 presents the clonal distribution of “Weaning piglets” of “Farm A”). Three points (H2, H4, and I2) were excluded as no ESBL/AmpC-EC strains were isolated from “Growing pigs” of “Farm H”, “Pregnant sows” of “Farm H,” and “Growing pigs” of “Farm I.”
Analysis of the Potential Risk of Swine Farm-Derived ESBL/AmpC-EC Clones
Identification of the Shared Major STs of ESBL/AmpC-EC Isolated From Swine Farms and Humans
To identify the shared major STs of ESBL/AmpC-EC isolates in swine farms and human sources, we compared the STs of the swine ESBL/AmpC-EC isolates with the major 20 STs of the human ESBL/AmpC-EC isolates, registered in the NCBI Pathogen Isolates Browser database (Figure 7).
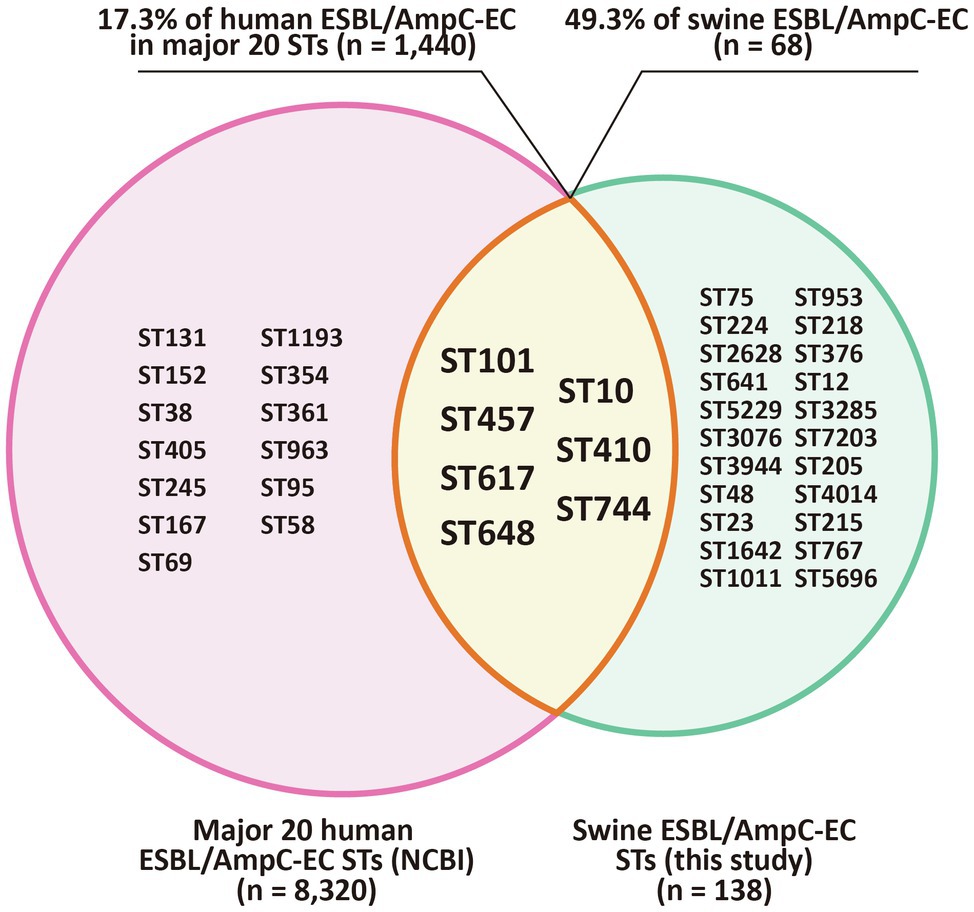
Figure 7. Shared major STs of ESBL/AmpC-EC isolated from swine farms and humans. Venn diagram shows the STs shared between swine farm derived ESBL/AmpC-ECs from this study and ESBL/AmpC-ECs from human sources which registered in the NCBI Pathogen Isolation Database. The intersection area of the two circles represents to the seven shared STs (ST101, ST10, ST457, ST410, ST617, ST744, and ST648) of ESBL/AmpC-EC from swine farms and human sources. ST, sequence type.
Among 29 MLST STs from the swine ESBL/AmpC-EC isolates, seven STs (ST101, ST10, ST457, ST410, ST617, ST744, and ST648) were shared with the human ESBL/AmpC-EC strains. These seven STs contained 68 (49.3%) of the 138 ESBL/AmpC-EC isolates in the swine farms and 1,440 (17.3%) of the 8,320 ESBL/AmpC-EC strains belonging to the major 20 STs from human sources.
Clonal Population Structure Analysis of Swine ESBL/AmpC-EC Based on ExPEC VFs and Phylogenetic Group Profiles
We conducted a clonal population structure analysis based on ExPEC VFs and phylogenetic group profiles of ESBL/AmpC-EC using the program STRUCTURE to evaluate the potential risk of swine ESBL/AmpC-EC clones to humans. In this clonal population analysis, 138 ESBL/AmpC-EC isolates were divided into five populations, and each population contained 24–34 ESBL/AmpC-EC isolates. Individual ESBL/AmpC-EC isolates assigned to the populations were visualized as a 100% stacked bar chart sorted by the STs (Figure 8). Differences in the ExPEC VFs and phylogenetic groups in each population are presented in Table 3. The clonal population distribution of swine ESBL/AmpC-EC isolates for each ST-phylogenetic group is presented in Supplementary Table 4.
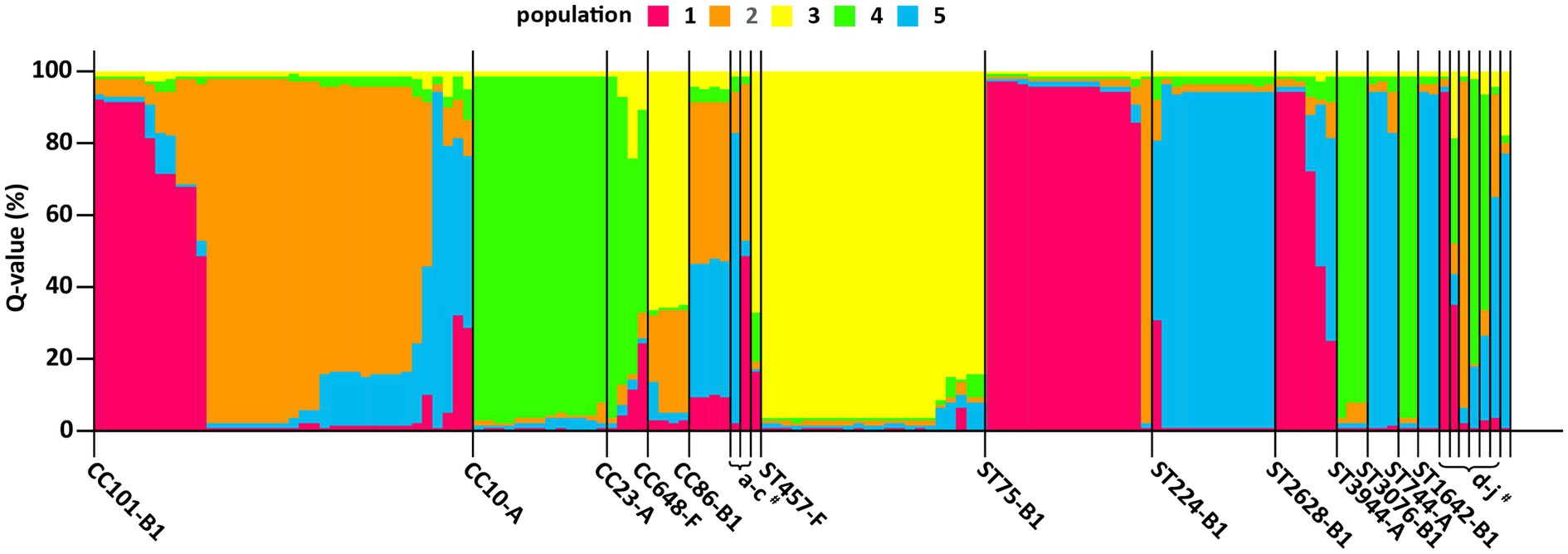
Figure 8. A clonal population analysis of swine ESBL/AmpC-EC isolates using program structure. Each isolate was assigned to five populations based on their ExPEC VFs and E. coli phylogenetic group profile. The ExPEC VFs and E. coli phylogenetic group profile for each clonal population are presented in Table 2. Each isolate is represented by a vertical segment and aligned horizontally according to CCs and STs (x-axis). The proportion of population (Q value) for each isolate is shown as 100% stacked bar plots, with proportions of colored sections representing the probability of belonging to each population within each segment (y-axis). CC, clonal complex; ST, sequence type; STNT, ST non-typable. #a, CC205-B1; b, CC376-B1; c, CC12-B2; d, ST3285-B1; e, ST953-A; f, NT-B1; g, ST7203-A; h, STNT-A; I, ST767-B1; and j, ST1011-E.
The ESBL/AmpC-EC isolates in population 1 mainly identified as CC101-B1 and ST75-B1, and harbored the highest number (average 6.7) of VFs, which was characterized with the highest prevalence of papC, hlyA, pic, iroNe.coli, and ompT. Isolates in population 2 mainly identified as CC101-B1 and CC86-B1. This population carried the second highest number of VFs (average: 4.8 VFs), characterized by the high prevalence of tsh and fyuA. Isolates in population 3 mainly identified as CC648-F and ST457-F (average: 4.2 VFs), with a high prevalence of papC and ompT. Isolates in population 4 mainly identified as CC10-A and CC23-A (average: 3.0 VFs). Finally, isolates in population 5 identified as ST224-B1 (average: 3.0 VFs). Isolates in populations 4 and 5 carried the lowest number of VFs.
Discussion
The emergence and spread of 3GC-EC, including ESBL/AmpC-EC, has been reported by the WHO as a worldwide public health concern (WHO, 2017). Swine farm husbandry has been regarded as an important reservoir of ESBL/AmpC-EC (Carattoli, 2008). In the present study, we identified that more than half of pigs harbored ESBL/AmpC-EC isolates (55.1%, 161/292), and the prevalence varied depending on the swine production stages. The weaning piglets showed the significantly highest prevalence compared to pigs in the other stages (86.3%, p < 0.05; Figure 1B). Consistently, the high prevalence of ESBL/AmpC-EC in the weaning stage has been described in previous studies in Denmark (Hansen et al., 2013), Netherlands (Dohmen et al., 2017), and Germany (Schmithausen et al., 2015). We presumed that the high usage of β-lactams during the weaning stage, compared to other stages, could be one of major causes of the high prevalence of ESBL/AmpC-EC at the weaning stage. According to studies that investigated the patterns of antibiotics use in a global swine production system, over 70% β-lactams used in the swine industry was applied between birth and 10 weeks of age (Sjolund et al., 2016; Burow et al., 2019; Lekagul et al., 2019; Korsgaard et al., 2020). β-lactams have been commonly prescribed for the treatment and prevention of postweaning syndromes, including postweaning diarrhea, edema disease, and endotoxin shock, which are the major problems in swine industry (van Beers-Schreurs et al., 1992). Considering that colonization of ESBL/AmpC-EC could last longer than 6 months even without antibiotic selection pressure, ESBL/AmpC-EC colonies in the intestinal tract of swine at the weaning stage could persist until the date of their slaughter (about 150–230 days old; Kennedy and Collignon, 2010). In that point, we suggest that ESBL/AmpC-EC prevalence at the weaning stage could be a reflection of that of the entire farm, and that weaning stage should serve as the critical point in controlling the prevalence of ESBL/AmpC-EC in swine farms.
Notably, ESBL/AmpC-EC from swine farms showed MDR, multiple virulence factors, and enhanced biofilm formation ability relative to non-ESBL/AmpC-EC. Swine farm-derived ESBL/AmpC-EC isolates had a 100% MDR rate and a higher resistance rate against antibiotics, including broad-spectrum penicillins, aminoglycosides, and quinolones, compared with those in the non-ESBL/AmpC-EC (p < 0.05). In addition, ExPEC VFs, which were involved in human ExPEC infections, such as urinary tract infections, sepsis, and meningitis, were also highly identified in ESBL/AmpC-EC than in the non-ESBL/AmpC-EC (p < 0.05). Especially, the prevalence of the three VFs, namely papC, pic, and ompT, which were reported to show a positive association with the high mortality in human ExPEC infection (Kudinha et al., 2013; Krawczyk et al., 2015; Dutra et al., 2020), was significantly higher in the ESBL/AmpC-EC isolates (p < 0.05). Furthermore, the OR of biofilm formation was 3.8-fold greater (95% CI: 1.42–10.30, p < 0.05) in the ESBL/AmpC-EC isolates than in the non-ESBL/AmpC-EC isolates. Biofilm formation conferred fitness advantage to the bacteria by enhancing their survivability, increasing their virulence, and facilitating their ability to acquire virulence and antibiotic resistance genes during horizontal gene transmission due to their high microbial density (Donlan and Costerton, 2002; Schroeder et al., 2017). Collectively, these enhanced properties, namely MDR, multiple virulence factors, and enhanced biofilm formation ability, of ESBL/AmpC-EC from swine farms could make them difficult to be controlled once introduced to swine farms, allowing ESBL/AmpC-EC to survive and continue to exist within swine farms. In addition, the ESBL/AmpC-EC, with these properties, in swine farms could be transmitted to humans through the chain of pig meat (Liu et al., 2019), direct contact with the farm workers (Schmithausen et al., 2015), and manure excretion into the farm’s surrounding environment, including soils, ponds, and rivers (Furlan and Stehling, 2018). Spread of ESBL/AmpC-EC strains can be of serious public health concern since it could aid in the emergence of pathogens, which are difficult to control in the food-animal industry as well as human hospitals (Yoo et al., 2010; Dantas Palmeira and Ferreira, 2020).
The CTX-M β-lactamase (CTX-M) and CMY β-lactamase (CMY) are the most predominant type of ESBLs and AmpCs in 3GC-EC isolates from humans and animals (Philippon et al., 2002; Canton et al., 2012). In this study, all ESBL/AmpC-EC isolates carried CTX-M (95.7%, 154/161) or CMY (4.3%, 7/161); however, their distribution differed according to the swine production stages. CTX-M was prevalent in all four swine production stages, while CMY was only identified from growing (85.7%, 6/7) and finishing stages (14.3%, 1/7). We presumed that higher use of the ceftiofur during the growing-finishing stages, compared to other stages, could be one reason for the higher prevalence of CMY at growing-finishing stages. According to the previous studies, ceftiofur has been mainly prescribed to treat swine respiratory infection diseases, and these diseases has been reported to show higher prevalence in growing-finishing stages, compared to other stages (Hornish and Katarski, 2002; Timmerman et al., 2006; Tantituvanont et al., 2009; Deng et al., 2015). In general, CTX-M and CMY had different resistance profile against ceftiofur, a member of 3GC; CMY usually carried resistance against ceftiofur, while CTX-M does not (Deng et al., 2015). CMY prevalence has been reported to be positively associated with the usage of ceftiofur in the swine industry (Hornish and Katarski, 2002; Timmerman et al., 2006; Deng et al., 2015). In addition to higher usage of ceftiofur during growing-finishing stages, we suggest that the low horizontal gene transferability of CMY genes (0.0%, 0/7) compared to CTX-M genes (59.5%, 78/131) identified from this study could also be another reason for the accumulation of CMY producers only at growing-finishing stages, without spreading into other stages. Our results suggest that 3GC-resistant strains with different characteristics could exist depending on the swine production stage, which proposes the need to consider the production stage in the studies of 3GC-resistant bacteria in swine farms.
In the present study, the most prevalent ESBL was CTX-M-55 (69.6%, 112/161), followed by CTX-M-15 (11.8%, 19/161) and CTX-M-14 (5.6%, 9/161). CTX-M-15 and CTX-M-14 are regarded as the most prevalent types of ESBLs worldwide since the last decade (Song et al., 2009; Kennedy and Collignon, 2010; Shin et al., 2017). CTX-M-15 and CTX-M-14 have been reported as still dominant ESBL types in Australia (Abraham et al., 2015; Sparham et al., 2017) and Africa (Belmahdi et al., 2016; Alonso et al., 2017). However, the prevalence of CTX-M-55 has been recently increasing in Europe (Rossolini et al., 2008; Haenni et al., 2018), North America (Moffat et al., 2020), and Asia (Lee et al., 2013; Zhou et al., 2018; Tansawai et al., 2019), suggesting the possibility of global epidemiological shift in ESBL types. In this study, all seven AmpC-producing isolates were identified to carry CMY-2 (4.3%, 7/161), which was the most frequent reported CMY type found in human- and food-animal-derived AmpC producers worldwide (Wu et al., 2018).
The horizontal gene transfer system plays a crucial role in the transmission of ESBL/AmpC-EC; however, the clonal transfer could also be important in their transmission (Dohmen et al., 2017). Schmithausen et al. (2015) reported that ESBL-EC from individual pigs could spread into farm environments and almost the entire pigs present within the compartment could be affected by ESBL-EC from individual pigs (Schmithausen et al., 2015). When the clonal distribution in the swine production stages was compared via the k-means clustering analysis, we found that the clonal distribution of three or all stages from the same farm tended to be clustered together, showing similarity of clonal distribution. A similar clonal distribution between production stages implied the presence of predominantly colonized ESBL/AmpC-EC clone types throughout the farm and the spread of these strains between production stages. Our result suggests that through the repeated cycle, which involved the shedding from swine through feces, survival in the farm environment, and reintroduction to swine, the ESBL/AmpC-EC could spread into other swine at different stages and could continue to exist within swine farms.
Here, the most prevalent CCs of ESBL/AmpC-EC from swine farms was found to be CC101-B1 (26.8%, 37/138), followed by CC10-A (8.7%, 13/138) and CC648-F (2.9%) in the molecular epidemiological analysis. Consistently, a study on the ESBL-EC isolates from swine at slaughterhouses in South Korea reported CC101 as the major CC type of ESBL-EC from pigs followed by CC10 (Song et al., 2020). In contrast, the most common CC of ESBL/AmpC-EC was CC10 in other countries including Portugal, Netherlands, Taiwan, and China, while the second most common CC varied across studies, including CC155, CC405, or CC648 (Ramos et al., 2013; Hammerum et al., 2014; Zhang et al., 2016; Lee and Yeh, 2017; Liu et al., 2018). In the comparison analysis of the major ESBL/AmpC clone types from swine farms and human sources, we identified that seven STs (ST101, ST10, ST648, ST457, ST410, ST617, and ST744) were shared between swine farm-derived ESBL/AmpC-EC and human ESBL/AmpC-EC. The ST10, a major ST of CC10, has been reported to be one of the most important pandemic human ESBL/AmpC-producing ExPEC clones (ST131, ST10, ST69, ST73, and ST95) since 2000s (Manges et al., 2019). The ST101, a major ST in CC101, has been reported worldwide, although not yet regarded as a member of the pandemic clones. ESBL/AmpC-EC clone type ST101 has attracted renewed global attention in human ESBL/AmpC-producing ExPEC infections given its enhanced virulence and pan-drug resistance (Yoo et al., 2013; Manges et al., 2019; Santos et al., 2020), and has been reported to cause hemolytic uremic syndrome (Mellmann et al., 2008) and bloodstream infections (Santos et al., 2020). Consistently, in the clonal population structure analysis based on the ExPEC VF profiles and phylogenetic groups using the program STRUCTURE, we also identified a highly virulent profile of ESBL/AmpC-EC ST101 clone. In this analysis, the population 1, showing a highly virulent profile carrying a higher number of VFs associated with adhesion, toxin, siderophores, and serum resistance, consisted mainly of ESBL/AmpC-EC clone type ST101 along with ST75 strains, compared to other clone type strains.
In conclusion, ESBL/AmpC-EC, carrying MDR and enhanced virulence potential, was distributed throughout the swine production stages with the highest prevalence at the weaning stage. CTX-M β-lactamase was the dominant ESBL type and was identified in all four swine production stages, while CMY β-lactamase was identified only in growing and finishing stages. The similarity in the clonal distribution between different swine production stages within farms was identified, which suggested a possibility of clonal transmission between the different swine stages. ESBL/AmpC-EC from swine farms was identified to harbor a virulent profile posing potential risk to humans and shared clone types with ESBL/AmpC-EC from human sources. To further explore the possibility of clonal transmission from swine farms to humans, additional comparative analysis studies based on the WGS of ESBL/AmpC-EC from swine farms and humans would need to be carried out. Our study results suggest the need for strategies considering the swine production system to control the prevalence of ESBL/AmpC-EC in swine farms.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
SC and W-HK conceived and designed the study. W-HK, J-UA, and SL analyzed the epidemiologic data. HS, SY, J-HG, J-UA, and SL performed the sampling and experiments. SC, W-HK, and SL prepared and reviewed the manuscript. SL was a major contributor, both in experiments and writing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Research Foundation of Korea (NRF-2021R1A2C2005907).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.710747/full#supplementary-material
Footnotes
References
Abraham, S., Jordan, D., Wong, H. S., Johnson, J. R., Toleman, M. A., Wakeham, D. L., et al. (2015). First detection of extended-spectrum cephalosporin- and fluoroquinolone-resistant Escherichia coli in Australian food-producing animals. J. Glob. Antimicrob. Resist. 3, 273–277. doi: 10.1016/j.jgar.2015.08.002
Alonso, C. A., Zarazaga, M., Ben Sallem, R., Jouini, A., Ben Slama, K., and Torres, C. (2017). Antibiotic resistance in Escherichia coli in husbandry animals: the African perspective. Lett. Appl. Microbiol. 64, 318–334. doi: 10.1111/lam.12724
Barguigua, A., El Otmani, F., Talmi, M., Zerouali, K., and Timinouni, M. (2013). Prevalence and types of extended spectrum β-lactamases among urinary Escherichia coli isolates in Moroccan community. Microb. Pathog. 61-62, 16–22. doi: 10.1016/j.micpath.2013.04.010
Belmahdi, M., Bakour, S., Al Bayssari, C., Touati, A., and Rolain, J. M. (2016). Molecular characterisation of extended-spectrum beta-lactamase- and plasmid AmpC-producing Escherichia coli strains isolated from broilers in Bejaia, Algeria. J. Glob. Antimicrob. Resist. 6, 108–112. doi: 10.1016/j.jgar.2016.04.006
Bergspica, I., Kaprou, G., Alexa, E. A., Prieto, M., and Alvarez-Ordonez, A. (2020). Extended spectrum beta-lactamase (ESBL) producing Escherichia coli in pigs and pork meat in the European Union. Antibiotics 9:678. doi: 10.3390/antibiotics9100678
Bok, E., Kozanska, A., Mazurek-Popczyk, J., Wojciech, M., and Baldy-Chudzik, K. (2020). Extended phylogeny and extraintestinal virulence potential of commensal Escherichia coli from piglets and sows. Int. J. Environ. Res. Public Health 17:366. doi: 10.3390/ijerph17010366
Burow, E., Rostalski, A., Harlizius, J., Gangl, A., Simoneit, C., Grobbel, M., et al. (2019). Antibiotic resistance in Escherichia coli from pigs from birth to slaughter and its association with antibiotic treatment. Prev. Vet. Med. 165, 52–62. doi: 10.1016/j.prevetmed.2019.02.008
Canton, R., Gonzalez-Alba, J. M., and Galan, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Carattoli, A. (2008). Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infect. 14 (Suppl. 1), 117–123. doi: 10.1111/j.1469-0691.2007.01851.x
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Cortes, P., Blanc, V., Mora, A., Dahbi, G., Blanco, J. E., Blanco, M., et al. (2010). Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl. Environ. Microbiol. 76, 2799–2805. doi: 10.1128/AEM.02421-09
Cyoia, P. S., Koga, V. L., Nishio, E. K., Houle, S., Dozois, C. M., de Brito, K. C. T., et al. (2018). Distribution of ExPEC virulence factors, Bla CTX-M, fosA3, and mcr-1 in Escherichia coli isolated from commercialized chicken carcasses. Front. Microbiol. 9:3254. doi: 10.3389/fmicb.2018.03254
Dantas Palmeira, J., and Ferreira, H. M. N. (2020). Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in cattle production—a threat around the world. Heliyon 6:e03206. doi: 10.1016/j.heliyon.2020.e03206
Day, M. J., Hopkins, K. L., Wareham, D. W., Toleman, M. A., Elviss, N., Randall, L., et al. (2019). Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. Lancet Infect. Dis. 19, 1325–1335. doi: 10.1016/s1473-3099(19)30273-7
Deng, H., Si, H. B., Zeng, S. Y., Sun, J., Fang, L. X., Yang, R. S., et al. (2015). Prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in a farrowing farm: ST1121 clone harboring IncHI2 plasmid contributes to the dissemination of Bla CMY-2. Front. Microbiol. 6:1210. doi: 10.3389/fmicb.2015.01210
Dohmen, W., Dorado-Garcia, A., Bonten, M. J., Wagenaar, J. A., Mevius, D., and Heederik, D. J. (2017). Risk factors for ESBL-producing Escherichia coli on pig farms: a longitudinal study in the context of reduced use of antimicrobials. PLoS One 12:e0174094. doi: 10.1371/journal.pone.0174094
Donlan, R. M., and Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/CMR.15.2.167-193.2002
Dutra, I. L., Araujo, L. G., Assuncao, R. G., Lima, Y. A., Nascimento, J. R., Vale, A. A. M., et al. (2020). Pic-producing Escherichia coli induces high production of proinflammatory mediators by the host leading to death by sepsis. Int. J. Mol. Sci. 21:2068. doi: 10.3390/ijms21062068
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Fromm, S., Beißwanger, E., Käsbohrer, A., and Tenhagen, B.-A. (2014). Risk factors for MRSA in fattening pig herds—a meta-analysis using pooled data. Prev. Vet. Med. 117, 180–188. doi: 10.1016/j.prevetmed.2014.08.014
Furlan, J. P. R., and Stehling, E. G. (2018). Detection of beta-lactamase encoding genes in feces, soil and water from a Brazilian pig farm. Environ. Monit. Assess. 190:76. doi: 10.1007/s10661-017-6453-x
Haenni, M., Beyrouthy, R., Lupo, A., Chatre, P., Madec, J. Y., and Bonnet, R. (2018). Epidemic spread of Escherichia coli ST744 isolates carrying mcr-3 and blaCTX-M-55 in cattle in France. J. Antimicrob. Chemother. 73, 533–536. doi: 10.1093/jac/dkx418
Hammerum, A. M., Larsen, J., Andersen, V. D., Lester, C. H., Skovgaard Skytte, T. S., Hansen, F., et al. (2014). Characterization of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli obtained from Danish pigs, pig farmers and their families from farms with high or no consumption of third- or fourth-generation cephalosporins. J. Antimicrob. Chemother. 69, 2650–2657. doi: 10.1093/jac/dku180
Hansen, K. H., Damborg, P., Andreasen, M., Nielsen, S. S., and Guardabassi, L. (2013). Carriage and fecal counts of cefotaxime M-producing Escherichia coli in pigs: a longitudinal study. Appl. Environ. Microbiol. 79, 794–798. doi: 10.1128/AEM.02399-12
Hartigan, J. A., and Wong, M. A. (1979). A K-means clustering algorithm. J. R. Stat. Soc. 28, 100–108.
Hong, J. S., Song, W., Park, H. M., Oh, J. Y., Chae, J. C., Jeong, S., et al. (2020). Molecular characterization of fecal extended-spectrum beta-lactamase- and AmpC beta-lactamase-producing Escherichia coli from healthy companion animals and cohabiting humans in South Korea. Front. Microbiol. 11:674. doi: 10.3389/fmicb.2020.00674
Hornish, R. E., and Katarski, S. (2002). Cephalosporins in veterinary medicine-ceftiofur use in food animals. Curr. Top. Med. Chem. 2, 717–731. doi: 10.2174/1568026023393679
Johnson, T. J., Bielak, E. M., Fortini, D., Hansen, L. H., Hasman, H., Debroy, C., et al. (2012). Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68, 43–50. doi: 10.1016/j.plasmid.2012.03.001
Jouini, A., Klibi, A., Elarbi, I., Chaabene, M. B., Hamrouni, S., Souiai, O., et al. (2021). First detection of human ST131-CTX-M-15-O25-B2 clone and high-risk clonal lineages of ESBL/pAmpC-producing E. coli isolates from diarrheic poultry in Tunisia. Antibiotics 10:670. doi: 10.3390/antibiotics10060670
Kennedy, K., and Collignon, P. (2010). Colonisation with Escherichia coli resistant to "critically important" antibiotics: a high risk for international travellers. Eur. J. Clin. Microbiol. Infect. Dis. 29, 1501–1506. doi: 10.1007/s10096-010-1031-y
Korsgaard, H. B., Ellis-Iversen, J., Hendriksen, R. S., Høg, B. B., Ronco, T., Attauabi, M., et al. (2020). DANMAP 2019 - use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. National Food Institute, Statens Serum Institut.
Krawczyk, B., Sledzinska, A., Szemiako, K., Samet, A., Nowicki, B., and Kur, J. (2015). Characterisation of Escherichia coli isolates from the blood of haematological adult patients with bacteraemia: translocation from gut to blood requires the cooperation of multiple virulence factors. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1135–1143. doi: 10.1007/s10096-015-2331-z
Kudinha, T., Johnson, J. R., Andrew, S. D., Kong, F., Anderson, P., and Gilbert, G. L. (2013). Distribution of phylogenetic groups, sequence type ST131, and virulence-associated traits among Escherichia coli isolates from men with pyelonephritis or cystitis and healthy controls. Clin. Microbiol. Infect. 19, E173–E180. doi: 10.1111/1469-0691.12123
Lee, W., Chung, H.-S., Lee, H., Yum, J. H., Yong, D., Jeong, S. H., et al. (2013). CTX-M-55-type extended-spectrum β-lactamase- producing shigella sonnei isolated from a Korean patient who had travelled to China. Ann. Lab. Med. 33, 141–144. doi: 10.3343/alm.2013.33.2.141
Lee, W. C., and Yeh, K. S. (2017). Characteristics of extended-spectrum beta-lactamase-producing Escherichia coli isolated from fecal samples of piglets with diarrhea in central and southern Taiwan in 2015. BMC Vet. Res. 13:66. doi: 10.1186/s12917-017-0986-7
Lekagul, A., Tangcharoensathien, V., and Yeung, S. (2019). Patterns of antibiotic use in global pig production: a systematic review. Vet. Anim. Sci. 7:100058. doi: 10.1016/j.vas.2019.100058
Lengyel, A., and Botta-Dukat, Z. (2019). Silhouette width using generalized mean—a flexible method for assessing clustering efficiency. Ecol. Evol. 9, 13231–13243. doi: 10.1002/ece3.5774
Liu, Z., Klumper, U., Shi, L., Ye, L., and Li, M. (2019). From pig breeding environment to subsequently produced pork: comparative analysis of antibiotic resistance genes and bacterial community composition. Front. Microbiol. 10:43. doi: 10.3389/fmicb.2019.00043
Liu, X., Liu, H., Wang, L., Peng, Q., Li, Y., Zhou, H., et al. (2018). Molecular characterization of extended-spectrum beta-lactamase-producing multidrug resistant Escherichia coli from swine in Northwest China. Front. Microbiol. 9:1756. doi: 10.3389/fmicb.2018.01756
Lupo, A., Saras, E., Madec, J. Y., and Haenni, M. (2018). Emergence of blaCTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France. J. Antimicrob. Chemother. 73, 867–872. doi: 10.1093/jac/dkx489
Lv, L., Partridge, S. R., He, L., Zeng, Z., He, D., Ye, J., et al. (2013). Genetic characterization of IncI2 plasmids carrying blaCTX-M-55 spreading in both pets and food animals in China. Antimicrob. Agents Chemother. 57, 2824–2827. doi: 10.1128/AAC.02155-12
Manges, A. R., Geum, H. M., Guo, A., Edens, T. J., Fibke, C. D., and Pitout, J. D. D. (2019). Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin. Microbiol. Rev. 32, e00135–e00218. doi: 10.1128/CMR.00135-18
Mellmann, A., Bielaszewska, M., Kock, R., Friedrich, A. W., Fruth, A., Middendorf, B., et al. (2008). Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg. Infect. Dis. 14, 1287–1290. doi: 10.3201/eid1408.071082
Moffat, J., Chalmers, G., Reid-Smith, R., Mulvey, M. R., Agunos, A., Calvert, J., et al. (2020). Resistance to extended-spectrum cephalosporins in Escherichia coli and other Enterobacterales from Canadian turkeys. PLoS One 15:e0236442. doi: 10.1371/journal.pone.0236442
Muller, A., Bertrand, X., Rogues, A. M., Pefau, M., Alfandari, S., Gauzit, R., et al. (2018). Higher third-generation cephalosporin prescription proportion is associated with lower probability of reducing carbapenem use: a nationwide retrospective study. Antimicrob. Resist. Infect. Control 7:11. doi: 10.1186/s13756-018-0302-8
Nathisuwan, S., Burgess, D. S., and Lewis, J. S. 2nd (2001). Extended-spectrum beta-lactamases: epidemiology, detection, and treatment. Pharmacotherapy 21, 920–928. doi: 10.1592/phco.21.11.920.34529
Naves, P., del Prado, G., Huelves, L., Gracia, M., Ruiz, V., Blanco, J., et al. (2008). Measurement of biofilm formation by clinical isolates of Escherichia coli is method-dependent. J. Appl. Microbiol. 105, 585–590. doi: 10.1111/j.1365-2672.2008.03791.x
Peirano, G., Richardson, D., Nigrin, J., McGeer, A., Loo, V., Toye, B., et al. (2010). High prevalence of ST131 isolates producing CTX-M-15 and CTX-M-14 among extended-spectrum-β-lactamase-producing Escherichia coli isolates from Canada. Antimicrob. Agents Chemother. 54, 1327–1330. doi: 10.1128/AAC.01338-09
Philippon, A., Arlet, G., and Jacoby, G. A. (2002). Plasmid-determined AmpC-type beta-lactamases. Antimicrob. Agents Chemother. 46, 1–11. doi: 10.1128/AAC.46.1.1-11.2002
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Ramos, S., Silva, N., Dias, D., Sousa, M., Capelo-Martinez, J. L., Brito, F., et al. (2013). Clonal diversity of ESBL-producing Escherichia coli in pigs at slaughter level in Portugal. Foodborne Pathog. Dis. 10, 74–79. doi: 10.1089/fpd.2012.1173
Rossolini, G. M., D'Andrea, M. M., and Mugnaioli, C. (2008). The spread of CTX-M-type extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14(Suppl. 1), 33–41. doi: 10.1111/j.1469-0691.2007.01867.x
Santos, A. C. M., Silva, R. M., Valiatti, T. B., Santos, F. F., Santos-Neto, J. F., Cayo, R., et al. (2020). Virulence potential of a multidrug-resistant Escherichia coli strain belonging to the emerging clonal group ST101-B1 isolated from bloodstream infection. Microorganisms 8:827. doi: 10.3390/microorganisms8060827
Schaufler, K., Semmler, T., Pickard, D. J., de Toro, M., de la Cruz, F., Wieler, L. H., et al. (2016). Carriage of extended-spectrum beta-lactamase-plasmids does not reduce fitness but enhances virulence in some strains of pandemic E. coli lineages. Front. Microbiol. 7:336. doi: 10.3389/fmicb.2016.00336
Schmithausen, R. M., Schulze-Geisthoevel, S. V., Stemmer, F., El-Jade, M., Reif, M., Hack, S., et al. (2015). Analysis of transmission of MRSA and ESBL-E among pigs and farm personnel. PLoS One 10:e0138173. doi: 10.1371/journal.pone.0138173
Schroeder, M., Brooks, B. D., and Brooks, A. E. (2017). The complex relationship between virulence and antibiotic resistance. Gene 8:39. doi: 10.3390/genes8010039
Shin, S. W., Jung, M., Won, H. G., Belaynehe, K. M., Yoon, I. J., and Yoo, H. S. (2017). Characteristics of transmissible CTX-M- and CMY-type beta-lactamase-producing Escherichia coli isolates collected from pig and chicken farms in South Korea. J. Microbiol. Biotechnol. 27, 1716–1723. doi: 10.4014/jmb.1610.10006
Sjolund, M., Postma, M., Collineau, L., Losken, S., Backhans, A., Belloc, C., et al. (2016). Quantitative and qualitative antimicrobial usage patterns in farrow-to-finish pig herds in Belgium, France, Germany and Sweden. Prev. Vet. Med. 130, 41–50. doi: 10.1016/j.prevetmed.2016.06.003
Song, W., Lee, H., Lee, K., Jeong, S. H., Bae, I. K., Kim, J. S., et al. (2009). CTX-M-14 and CTX-M-15 enzymes are the dominant type of extended-spectrum beta-lactamase in clinical isolates of Escherichia coli from Korea. J. Med. Microbiol. 58, 261–266. doi: 10.1099/jmm.0.004507-0
Song, J., Oh, S. S., Kim, J., Park, S., and Shin, J. (2020). Clinically relevant extended-spectrum beta-lactamase-producing Escherichia coli isolates from food animals in South Korea. Front. Microbiol. 11:604. doi: 10.3389/fmicb.2020.00604
Sparham, S. J., Kwong, J. C., Valcanis, M., Easton, M., Trott, D. J., Seemann, T., et al. (2017). Emergence of multidrug resistance in locally-acquired human infections with Salmonella Typhimurium in Australia owing to a new clade harbouring blaCTX-M-9. Int. J. Antimicrob. Agents 50, 101–105. doi: 10.1016/j.ijantimicag.2017.02.014
Tansawai, U., Walsh, T. R., and Niumsup, P. R. (2019). Extended spectrum ss-lactamase-producing Escherichia coli among backyard poultry farms, farmers, and environments in Thailand. Poult. Sci. 98, 2622–2631. doi: 10.3382/ps/pez009
Tantituvanont, A., Yimprasert, W., Werawatganone, P., and Nilubol, D. (2009). Pharmacokinetics of ceftiofur hydrochloride in pigs infected with porcine reproductive and respiratory syndrome virus. J. Antimicrob. Chemother. 63, 369–373. doi: 10.1093/jac/dkn496
Timmerman, T., Dewulf, J., Catry, B., Feyen, B., Opsomer, G., de Kruif, A., et al. (2006). Quantification and evaluation of antimicrobial drug use in group treatments for fattening pigs in Belgium. Prev. Vet. Med. 74, 251–263. doi: 10.1016/j.prevetmed.2005.10.003
van Beers-Schreurs, H. M., Vellenga, L., Wensing, T., and Breukink, H. J. (1992). The pathogenesis of the post-weaning syndrome in weaned piglets: a review. Vet. Q. 14, 29–34. doi: 10.1080/01652176.1992.9694322
Verhegghe, M., Pletinckx, L. J., Crombé, F., Van Weyenberg, S., Haesebrouck, F., Butaye, P., et al. (2013). Cohort study for the presence of livestock-associated MRSA in piglets: effect of sow status at farrowing and determination of the piglet colonization age. Vet. Microbiol. 162, 679–686. doi: 10.1016/j.vetmic.2012.09.014
WHO (2017). Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis (No. WHO/EMP/IAU/2017.12). Geneva: World Health Organization.
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Wu, C., Wang, Y., Shi, X., Wang, S., Ren, H., Shen, Z., et al. (2018). Rapid rise of the ESBL and mcr-1 genes in Escherichia coli of chicken origin in China, 2008-2014. Emerg. Microbes Infect. 7:30. doi: 10.1038/s41426-018-0033-1
Yoo, J. S., Byeon, J., Yang, J., Yoo, J. I., Chung, G. T., and Lee, Y. S. (2010). High prevalence of extended-spectrum beta-lactamases and plasmid-mediated AmpC beta-lactamases in Enterobacteriaceae isolated from long-term care facilities in Korea. Diagn. Microbiol. Infect. Dis. 67, 261–265. doi: 10.1016/j.diagmicrobio.2010.02.012
Yoo, J. S., Kim, H. M., Koo, H. S., Yang, J. W., Yoo, J. I., Kim, H. S., et al. (2013). Nosocomial transmission of NDM-1-producing Escherichia coli ST101 in a Korean hospital. J. Antimicrob. Chemother. 68, 2170–2172. doi: 10.1093/jac/dkt126
Zhang, H., Zhai, Z., Li, Q., Liu, L., Guo, S., Li, Q., et al. (2016). Characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates from pigs and farm workers. J. Food Prot. 79, 1630–1634. doi: 10.4315/0362-028X.JFP-16-093
Keywords: extended-spectrum β-lactamase, AmpC β-lactamase, Escherichia coli, multidrug resistance, extra-intestinal pathogenic E. coli, virulence factor, clonal distribution, swine production stages
Citation: Lee S, An J-U, Guk J-H, Song H, Yi S, Kim W-H and Cho S (2021) Prevalence, Characteristics and Clonal Distribution of Extended-Spectrum β-Lactamase- and AmpC β-Lactamase-Producing Escherichia coli Following the Swine Production Stages, and Potential Risks to Humans. Front. Microbiol. 12:710747. doi: 10.3389/fmicb.2021.710747
Edited by:
Zulqarnain Baloch, Kunming University of Science and Technology, ChinaReviewed by:
Mohamed Salah Abbassi, Tunis El Manar University, TunisiaMashkoor Mohsin, University of Agriculture, Faisalabad, Pakistan
Copyright © 2021 Lee, An, Guk, Song, Yi, Kim and Cho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seongbeom Cho, Y2hvc2VAc251LmFjLmty; Woo-Hyun Kim, aWNlbWFuMmJAc251LmFjLmty
 Soomin Lee
Soomin Lee Jae-Uk An
Jae-Uk An Jae-Ho Guk
Jae-Ho Guk Hyokeun Song
Hyokeun Song Saehah Yi
Saehah Yi Woo-Hyun Kim
Woo-Hyun Kim Seongbeom Cho
Seongbeom Cho