- 1Canyon Crest Academy, San Diego, CA, United States
- 2Department of Surgery, National University of Singapore, Singapore, Singapore
The serological lateral flow immunoassay (LFIA) was used to detect circulating antibodies to skin bacteria. Next-generation sequencing analysis of the skin microbiome revealed a high relative abundance of Cutibacterium acnes but low abundance of Staphylococcus aureus and Corynebacterium aurimucosum on human facial samples. Yet, results from both LFIA and antibody titer quantification in 96-well microplates illustrated antibody titers that were not correspondent, and instead negatively correlated, to their respective abundance with human blood containing higher concentrations of antibodies to both S. aureus and C. aurimucosum than C. acnes. Acne vulgaris develops several unique microbial and cellular features, but its correlation with circulating antibodies to bacteria in the pilosebaceous unit remains unknown. Results here revealed that antibodies to C. acnes and S. aureus were approximately 3-fold higher and 1.5-fold lower, respectively, in acne patients than in healthy subjects. Although the results can be further validated by larger sample sizes, the proof-of-concept study demonstrates a newfound discrepancy between the abundance of skin bacteria and amounts of their corresponding antibodies. And in light of acne-correlated amplified titers of specific anticommensal antibodies, we highlight that profiling these antibodies in the pilosebaceous unit by LFIAs may provide a unique signature for monitoring acne vulgaris.
Introduction
Human blood contains not only antibodies provoked by infections and vaccinations but also antibodies acquired by exposure to commensal bacteria (Zhao and Elson, 2018). While the anticommensal antibody repertoire has not been profiled in full, immunoglobulin G (IgG) in human blood with broad specificity to bacteria in the gastrointestinal tract has been detected (Castro-Dopico et al., 2019). These anticommensal antibodies have been documented to elicit proinflammatory activities by activation of fragment crystallizable (Fc) receptors on macrophages (Castro-Dopico et al., 2019). The serological lateral flow immunoassays (LFIAs) have been frequently used to detect the circulating antibodies (Banerjee and Jaiswal, 2018), mainly generated by infections or vaccinations including coronavirus disease 2019 (COVID-19) vaccines against SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) (Ravi et al., 2020). However, LFIA has been widely used in detection of anticommensal antibodies in people with health or disease condition.
Antibodies to Cutibacterium acnes, a bacterium associated with pathogenesis of acne vulgaris, in human blood have been clearly detected (Wang et al., 2018). The human sebaceous pilosebaceous unit is commonly referred to as “the seat of acne vulgaris” and harbors a heterogeneous community of microorganisms, including C. acnes, Staphylococcus spp., and Corynebacterium spp. (Grice and Segre, 2011). For example, Corynebacterium aurimucosum is often detected in sebaceous sites in humans (Blaskovich et al., 2019) and erythromycin-resistant Staphylococcus aureus in the comedones of acne vulgaris patients (Sitohang et al., 2019). While the pathophysiology of acne vulgaris remains unclear, many of the aforementioned microbial species have been widely associated with acne vulgaris. Features of acne vulgaris can be irregular patterns of composition and abundance of bacteria, dominance of virulent bacterial subtypes, or genetic elements or metabolites of bacteria and host cell responses, including activation of receptors and secretion of antimicrobial peptides or inflammatory cytokines in the microbiome of acne vulgaris compared with healthy skin (Chen et al., 2021).
Microbial composition results from the next-generation sequencing (NGS) platform for 16S ribosomal RNA (rRNA) gene analyses have revealed severe alterations in bacterial abundances, or dysbiotic microbiomes, in acne vulgaris. In comparing microbial profiles of healthy and acne patients, the relative abundance of C. acnes was similar in healthy skin compared to acne lesions, but a higher abundance of Cutibacterium granulosum was found in healthy skin (Rajiv et al., 2013). C. acnes has been classified into phylogenetic clades IA-1, IA-2, IB-1, IB-2, IB-3, IC, II, and III (Fitz-Gibbon et al., 2013). Strains from clades IA-2 (mostly ribotypes 4 and 5), IB-1 (ribotype 8), and IC (ribotype 5) are closely associated with acne vulgaris, whereas clade II strains that include ribotypes 2 and 6 are often detected in healthy skin (Lee et al., 2019). Profiles of genetic elements and metabolites of bacteria in healthy skin and acne lesions are also different. The gene-encoding Christie–Atkinson–Munch–Peterson (CAMP) factors camp1, camp2, and camp4, which contribute to hemolysis and inflammation, were found to be more abundantly expressed in acne lesions (O’Neill and Gallo, 2018). The gehA gene (PPA2105) encoding C. acnes lipase can increase sebum concentrations of free palmitate, which plays an important role in lipotoxic inflammasome activation of macrophages (Legrand-Poels et al., 2014). The secretion of interleukin 1α (IL-1α) via activation of Toll-like receptor 2 (TLR2) by ligands present on C. acnes can be detected in inflammatory acne vulgaris (Graham et al., 2004). It has been reported that the mRNA and protein levels of both IL-1β and IL-8 in acne lesions were higher than those in healthy skin (Wang et al., 2018). Here, we take advantage of LFIAs to detect the antibodies to skin bacteria including C. acnes, S. aureus, and C. aurimucosum in the sera of healthy subjects and acne patients. Unlike the traditional use of LFIAs for detection of infection- and vaccination-generated antibodies, we highlight that the antibodies to skin commensal bacteria on LFIAs can be an indicator for inflammatory diseases such as acne vulgaris.
Materials and Methods
Ethics Statement
Human blood and skin swabs were obtained from Dr. Chun-Ming Huang with an approved protocol (no. 121230) (Wang et al., 2018) by the institutional review board at University of California, San Diego (UCSD).
Skin Swabs and DNA Extraction
Total DNA was extracted from swab samples of cheeks of 15 (eight males and seven females) individuals between the ages of 21 and 35 years without acne vulgaris on facial skin using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) for polymerase chain reaction (PCR) (Wang et al., 2018). All individuals were asked without washing faces with water or other detergents at least 6 h before swabbing. Skin swabbing was conducted according to a method as described in previous studies (Ogai et al., 2018) with minor modifications. Concisely, a 4 × 4-cm2 area medial to the zygomatic prominence on bilateral cheeks was gently swabbed with a sterile cotton swab presoaked in a solution containing 0.9% sodium chloride and 0.1% Tween-20. For DNA extraction, the heads of all collected swab cottons were cut off and stored in sterile 1.5-mL centrifugation tubes at −80°C until DNA extraction. The swab heads were incubated in an enzyme solution (0.5 mL) [20 mg/mL lysozyme and 200 μg/mL lysostaphin (Sigma, St. Louis, MO, United States), in 20 mM Tris-HCl (pH 8.0), 2 mM ethylenediaminetetraacetic acid, and 1% Triton-X 100] at 37°C for 30 min. Proteinase K (20 μL) was subsequently added into the solution for 30 min, followed by deactivation of all enzymes at 95°C for 15 min.
Next-Generation Sequencing Analysis
NGS was performed according to a protocol previously published (Godlewska et al., 2020). Briefly, PCR amplicons were sequenced utilizing Roche 454 FLX titanium instruments (Roche, Indianapolis, IN, United States). The 16S rRNA gene V4 variable region PCR primers 515/806 were selected in a single-step 30-cycle PCR. Sequences were denoised; 16S rRNA sequences were clustered into operational taxonomic units (OTUs). Final OTUs were taxonomically classified using the nucleotide basic local alignment search tool (BLASTn) against a curated database derived from GreenGenes, Ribosomal Database Project II and the National Center for Biotechnology Information. Bacteria with relative abundances ≥1% in skin of at least one individual or bacteria identified at the species level were presented.
Preparation and Use of Human Sera
Blood was collected from healthy subjects with no prior history of acne vulgaris and acne patients between the ages of 21 and 50 years. Acne patients with multiple acne lesions (more than five lesions on the face or back skin) with different levels of severity were selected for blood collection. Acne patients were selected from participants who did not receive topical corticosteroids or systemic isotretinoin, immunosuppressives, chemotherapeutic agents, anti-inflammatory biologic, or oral antimicrobial agents within 30 days prior to the blood collection. Individual sera from each patient, not pooled sera from multiple patients, were used for experiments in this study.
Lateral Flow Immunoassay Fabrication of Antibody Detection
The LFIAs with test and control zones were fabricated using three pads (sample, conjugate, and absorbent pads) and one nitrocellulose (NC) membrane (Membrane Technologies Inc., Harrisburg, PA, United States). HAuCl4 (1 mM) (Sigma) was boiled at 200°C before addition of trisodium citrate dihydrate (1%). The gold nanoparticles (AuNPs) were conjugated with protein A of S. aureus (SPA) (20 μg) (Sigma) in 1 mL borax buffer containing boric acid and sodium borax, pH 5–6. One percent bovine serum albumin (BSA) in phosphate-buffered saline (PBS) was used for blocking. The SPA-conjugated AuNPs (100 μL) was pipetted onto the conjugate pad. Lysates of bacteria [C. acnes (ATCC 6919 and a skin isolate), C. aurimucosum (ATCC 700975), and S. aureus 113 (ATCC 35556)] were prepared by heat at 100°C for 1 h. Bacterial lysates and goat anti–human IgG (H+L) (3 μg) (Thermo Fisher Scientific, Waltham, MA, United States) were placed onto test and control zones on an NC membrane, respectively. The 5% sera (200 μL) were added onto the sample pad to start reaction. Antibodies in sera were captured by SPA-conjugated AuNPs in a conjugate pad and then flowed through to an NC membrane. The purple spots were formed when antibodies interact with bacterial lysates or anti–mouse IgG (H+L) on NC membranes. The intensities of purple spots on test zones relative to those on control zones were quantified by ImageJ software 1.50 b (National Institutes of Health, Bethesda, MD, United States).
Quantification of Antibody Titers
Bacterial lysates (0.1 μg/well) or BSA diluted in 50 μL PBS was coated onto a 96-well microplate for 4°C for 24 h. After blocking in PBS with 1% BSA, diluted sera were added to the wells and incubated for 2 h. A goat anti–human IgG (H+L) IgG–horseradish peroxidase (HRP) conjugate (Thermo Fisher Scientific) (1:10,000 dilution) was added and incubated for 1 h. HRP activity was measured by an OptEIA® Reagent Set (Thermo Fisher Scientific). The optical density (OD) of each well was measured at 450 nm subtracted from 570 nm (OD570–450). The endpoint was defined as the four-time dilution of sera producing the same OD570–450 as a 1/100 dilution of sera added to the BSA-coated wells. Sera negative at the lowest dilution tested were assigned endpoint titers of 100. The data were presented as geometric mean endpoint serum titers.
Quantitative Reverse Transcription–Polymerase Chain Reaction
To determine the mRNA expression level of C. acnes lipase (WP_002530746.1), total cellular RNA was extracted from C. acnes using the RNeasy Mini Kit (Thermo Fisher Scientific). The RNA was reverse transcribed to cDNA using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, United States) and amplified by reverse transcription (RT)–qPCR on the CFX96 real-time system (Bio-Rad) using forward (TCACTGATGAAGATCAACGCAC) and reverse (TGCGAATGTCCGACGAAGTCGA) primers. The comparative delta–delta cycle threshold (ΔΔCT) was used to quantify the gene expression, which was normalized to the expression level of 16S rRNA of C. acnes.
Statistical Analysis
Statistical analysis was performed by unpaired t-test using GraphPad Prism software. p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***) were considered significant. The mean ± standard deviation (SD) for at least three independent experiments was calculated.
Results
High-Abundance C. acnes and Low-Abundance S. aureus and C. aurimucosum in the Skin Microbiome
NGS analysis was performed using skin swab samples of facial skin collected from 15 young adults aged 21–35 years. The relative abundance of facial bacteria at the species level was determined by detecting 16S rRNA expression. Seven bacteria species with relative abundances greater than 0.02% in more than five skin swab samples were identified. These seven bacteria were C. acnes, S. aureus, C. aurimucosum, Kocuria rhizophila, Brevibacillus reuszeri, Haemophilus parainfluenzae, and Paracoccus aminovorans. The taxonomical assignment and relative abundance estimates for detected OTUs identified 32 groups of bacteria, which were several different unknown species under known genus or family (Figure 1A). Four bacteria species including C. acnes, Corynebacterium spp., Staphylococcus spp., and Enhydrobacter spp. with average relative abundances ≥10% in more than 5 of 15 human subjects were detected. Besides four species above, other bacteria with relative abundances ≥10% found on the skin of at least two human subjects included Dermacoccus spp., Lysobacter spp., Streptococcus spp., and bacteria in the Rhodobacteraceae family. Among the identified bacterial species, C. acnes, S. aureus, and C. aurimucosum with a relative abundance of 13.48% ± 6.43%, 0.02% ± 0.02%, and 0.24% ± 0.41% (Figure 1B), respectively, were often found around sebaceous glands on human face. The data demonstrated that the human face contains far more C. acnes than S. aureus and C. aurimucosum.
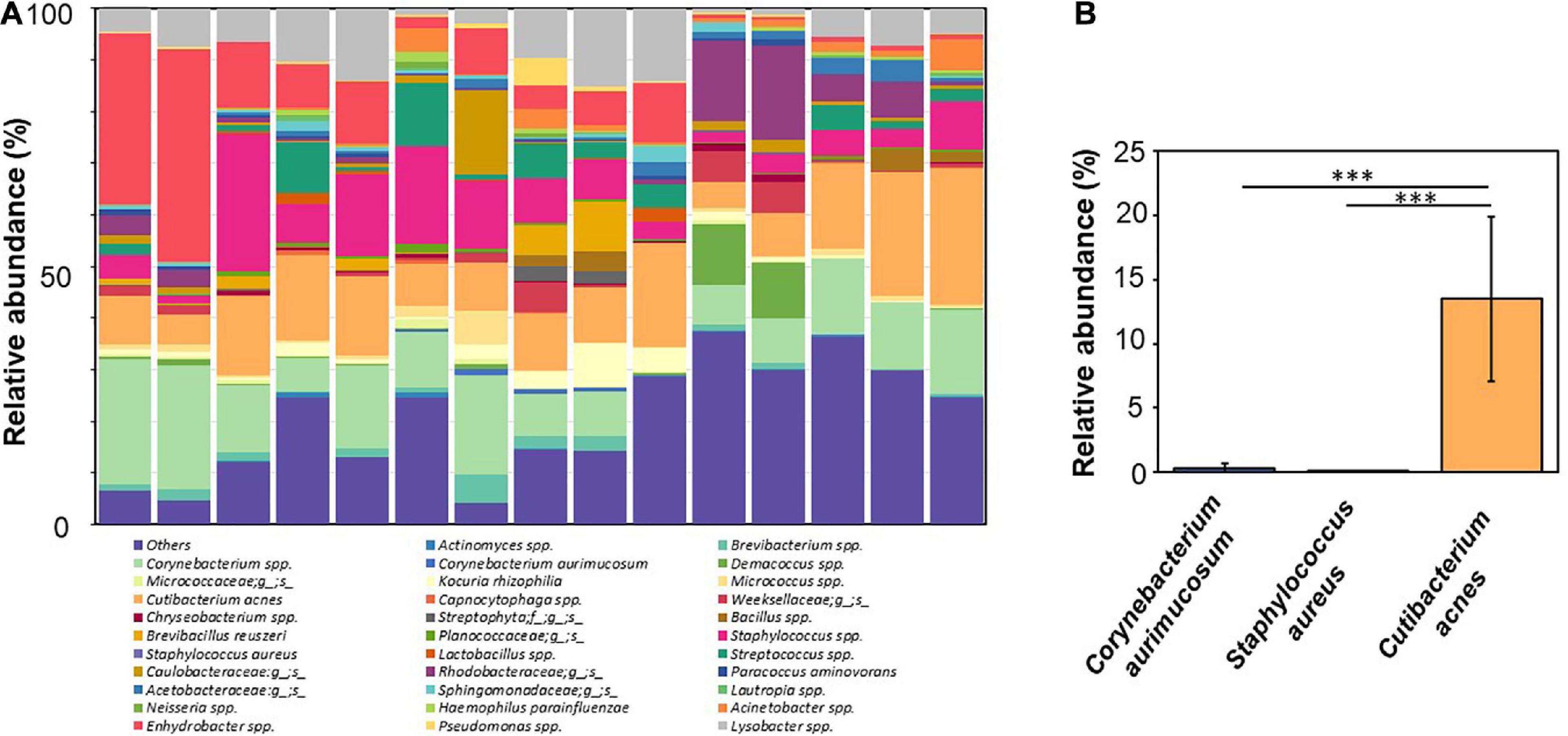
Figure 1. NGS-analyzed microbiome composition at species level with relatively high abundance of C. acnes and low abundance of S. aureus and C. aurimucosum in the human skin. (A) NGS analyzed by a Roche 454 FLX titanium instrument using 16S rRNA gene was conducted after extraction of DNA from swab samples of facial skin of 15 human subjects. Four bacteria species [Corynebacterium spp. (pale green), C. acnes (orange), Staphylococcus spp. (pink), and Enhydrobacter spp. (pink–red)] with average relative abundances ≥10% in more than 5 of 15 human subjects were detected. Bacteria with relative abundances ≥1% in skin of at least one individual or bacteria identified at the species level were presented. Bacteria with an abundance < 1% were pooled and indicated as “Others.” (B) The relative abundances of C. acnes (orange), S. aureus (purple), and C. aurimucosum (blue) are displayed. The mean ± SD obtained from 15 subjects was calculated. p < 0.001 (***), two-tailed t-test, is shown.
Higher Titer of Antibodies to S. aureus and C. aurimucosum Than C. acnes in Human Sera
It has been shown that there is a systemic humoral response against commensal bacteria in healthy individuals (Sterlin et al., 2020). To examine the correlation between the abundance of bacteria and their antibody titers in humans, the level of IgG toward the antigens of C. acnes, S. aureus, and C. aurimucosum was evaluated by the reactivity of the human sera with the corresponding bacterial lysates. The total lysates of C. acnes ATCC 6919, S. aureus 113 ATCC 35556, or C. aurimucosum ATCC 700975 were spotted on the test line of NC membranes of an LFIA. The anti–human IgG (H+L) was spotted on the control line of NC membrane of each LFIA. The relative intensities of purple spots on LFIA corresponding to antibody titers were quantified by normalizing those of IgG (H+L) spots. As shown in Figure 2A, the level of IgG against S. aureus or C. aurimucosum was approximately 16- or 5-fold, respectively, higher than that of IgG against C. acnes. Experiments of using bacterial lysates coated onto 96-well plates for probing with the corresponding sera were conducted to quantify the titers of antibodies to C. acnes, S. aureus, and C. aurimucosum. The mean titers of antibodies to C. acnes, S. aureus, and C. aurimucosum in 2% human sera were 1:1,333, 1:21,200, and 1:5,600, respectively (Figure 2B). Results of quantifying antibodies on 96 wells strongly confirmed that the LFIA data on the titer of antibodies to S. aureus or C. aurimucosum were much higher than those of antibodies to C. acnes in human sera. Taking Figures 1, 2 together, we found that (1) antibodies to C. acnes, S. aureus, or C. aurimucosum were detectable in human sera; however, (2) S. aureus and C. aurimucosum with a low bacterial abundance elicited a higher antibody titer than C. acnes, which was found to be highly abundant on face skin.
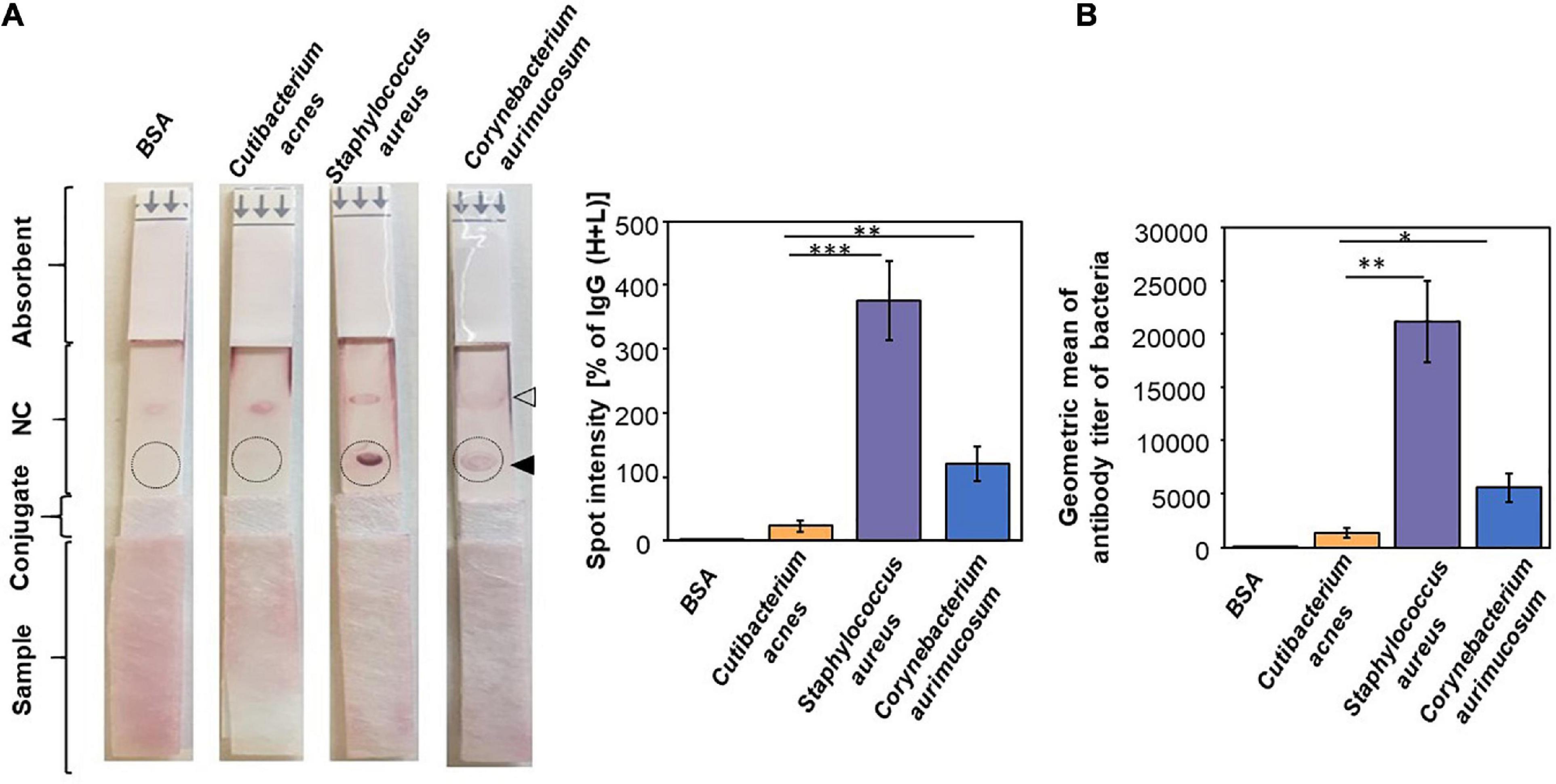
Figure 2. Production of a higher titer of antibodies to S. aureus and C. aurimucosum than C. acnes in human sera. (A) BSA or the lysate of C. acnes, S. aureus, or C. aurimucosum (3 μg) was on the test line (solid triangle) of an NC membrane of an LFIA. The anti–human IgG (H+L) was spotted on the control line (open triangle) of an NC membrane of each LFIA. After adding human sera (5%, 200 μL) onto sample pads, the relative intensities of purple spots (circles) were quantified by normalizing those of IgG (H+L) spots. (B) The antibodies (IgG) to C. acnes, S. aureus, or C. aurimucosum were detected by adding human sera to 96-well microplates coated with lysates of C. acnes, S. aureus, or C. aurimucosum, respectively. Quantitative antibodies were measured by using the endpoint of diluted sera producing the same OD570–450 as a 1/100 dilution of sera added to the BSA-coated wells. Individual, not pooled, sera from three human subjects were used for detection of antibodies on LFIAs and 96-well microplates. Sera used for LFIAs and 96-well microplates were from the same human subject. p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***) with mean ± SD were obtained from three independent experiments.
High Levels of Antibodies to C. acnes and Low Levels of Antibodies to S. aureus in Acne Patients
Literature has revealed that the imbalance of different C. acnes phylotypes, together with a dysbiosis of the skin microbiome, may be a signature of acne vulgaris (Dréno et al., 2020). To determine whether the titer of antibodies to bacteria such as C. acnes, S. aureus, and C. aurimucosum has a correlation with the acne vulgaris, sera from acne patients and healthy subjects were pipetted on sample pads of LFIAs with NC membranes immobilized with lysates of C. acnes, S. aureus, or C. aurimucosum. Purple spots were formed when the binding of antibodies in the sera to the bacterial lysates occurred. As shown in Figure 3A, addition of sera of acne patients onto LFIAs immobilized with lysates of C. acnes exhibited a more than threefold higher spot intensity than addition of sera of healthy subjects. The data indicated that acne patients had a higher titer of antibodies to C. acnes than healthy subjects. Results from LFIAs immobilized with lysates of S. aureus (Figure 3B) displayed that sera of acne patients contained approximately 1.5-fold lower levels of antibodies to S. aureus than the sera of healthy subjects. However, the level of systemic IgG directed against C. aurimucosum on LFIAs, although it was detectable, was very similar between acne patients and healthy subjects (Figure 3C). The differential levels of antibodies to C. acnes and S. aureus in acne patients and healthy subjects may serve as a signature of acne vulgaris.
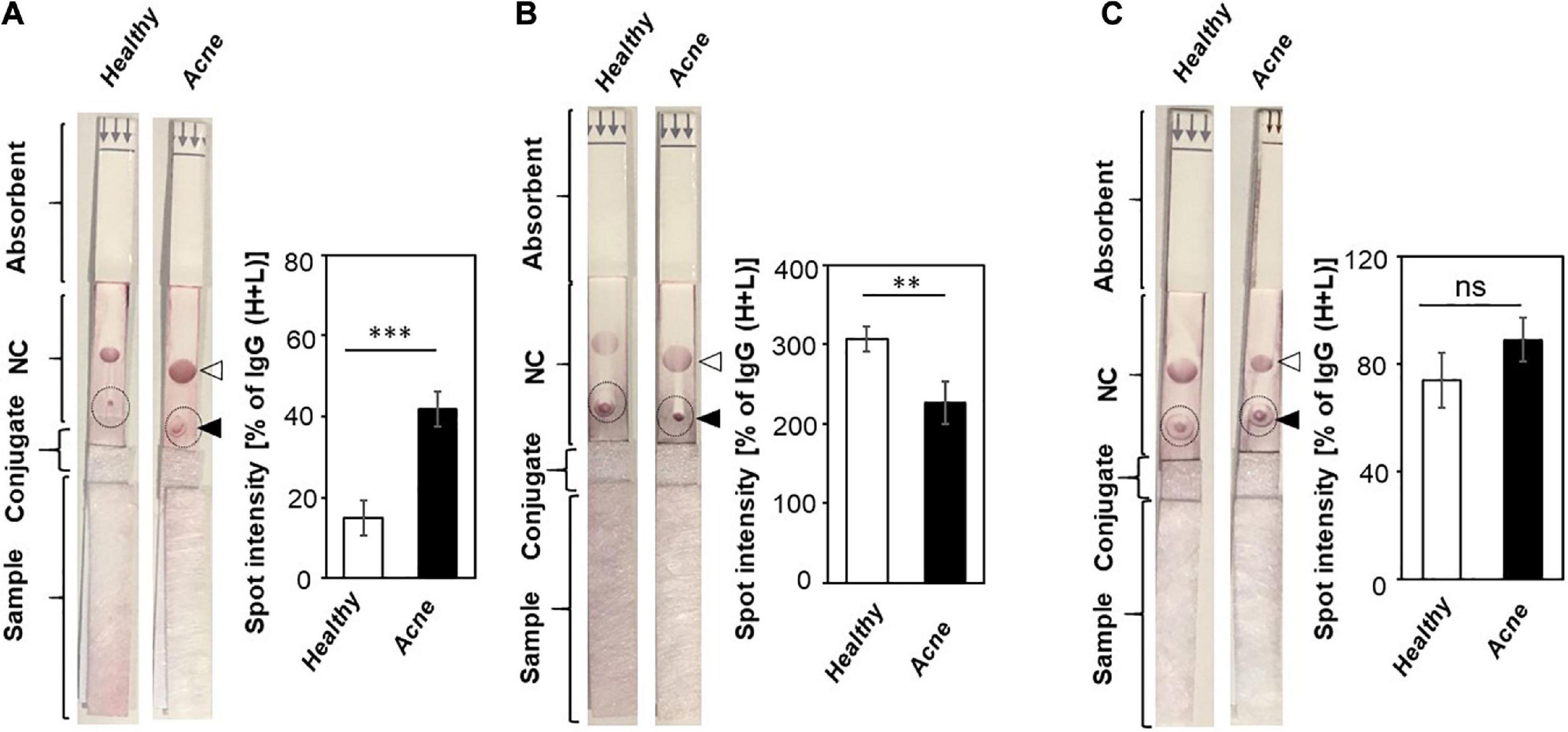
Figure 3. Detection of antibodies to C. acnes, S. aureus, and C. aurimucosum in acne patients and healthy subjects. The lysates (3 μg) of (A) C. acnes ATCC 6919, (B) S. aureus 113 (ATCC 35556), or (C) C. aurimucosum (ATCC 700975) were spotted on the test lines (solid triangle), and anti–human IgG (H+L) (open triangle) was spotted on control lines of NC membranes of LFIAs. Five percent sera (200 μL) from healthy subjects and acne patients were added into the sample pads. The high amounts of antibodies to C. acnes ATCC 6919 and low amounts of antibodies to S. aureus were detected in sera of acne patients compared to sera of healthy subjects. The intensities of IgG (H+L) spot served as 100%. The relative intensities of purple spots (circles) corresponding to the levels of antibodies were normalized by those of IgG (H+L) spots. Individual sera from three human subjects and acne patients were used. p < 0.01 (**) and p < 0.001 (***) with mean ± SD were calculated from three healthy subjects and acne patients. ns, not significant.
An Elevated Titer of Antibodies to a Strain of C. acnes Isolated From Acne Lesions
Although antibodies in sera can react to antigens of C. acnes ATCC 6919 (Figure 2), a commonly used laboratory strain originally isolated from facial skin of an acne patient (Yu et al., 2015), we tested the recognition of circulating antibodies to a C. acnes 74 strain, which was previously isolated from acne biopsies (Wang et al., 2018). 16S rRNA sequence of the C. acnes 74 strain shared 99.35% identity to a C. acnes strain 4873 (Supplementary Figure 1). Results from RT-qPCR analysis demonstrated that there was no difference in mRNA level of the gehA gene encoding lipase between C. acnes ATCC 6919 and the C. acnes 74 strain (Figure 4A). In agreement with results in Figure 3, we found that purple spots on LFIAs pipetted with sera of acne patients were approximately threefold darker than those on LFIAs pipetted with sera of healthy subjects (Figure 4B), illustrating that acne patients carried higher amounts of antibodies to C. acnes 74 strain than healthy subjects.
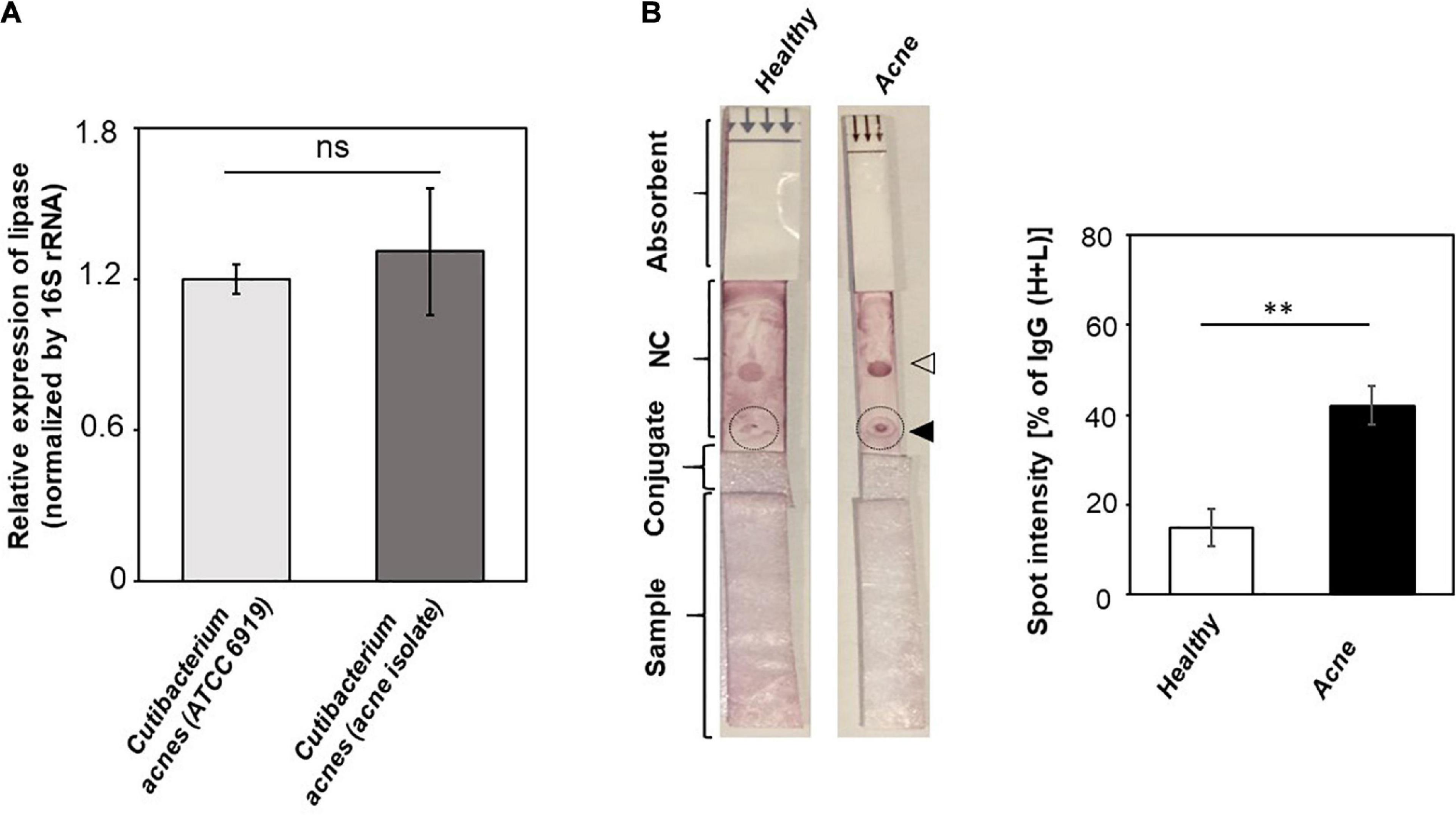
Figure 4. A higher level of antibodies to a C. acnes 74 strain isolated from acne lesions in acne patients as compared to healthy subjects. (A) The expression of the gehA gene encoding lipase in C. acnes ATCC 6919 and a C. acnes 74 strain was detected by RT-qPCR. The relative expression of the gehA gene was normalized to 16S rRNA. Data shown represent the mean ± SD of an experiment performed in triplicate. ns, not significant. (B) LFIAs were spotted with 3 μg of lysates (solid triangle) of a C. acnes 74 strain and anti–human IgG (H+L) (open triangle). Five percent individual sera (200 μL) from healthy subjects (n = 3) and acne patients (n = 3) were inserted into the sample pads. The relative levels of antibodies (circles) to C. acnes 74 strain were calculated as described in Figure 3 legend. p < 0.01 (**) with mean ± SD is shown.
Discussion
Corynebacterium spp. is commonly present in healthy human flora. Several Corynebacterium spp., including C. aurimucosum, Corynebacterium tuberculostearicum, Corynebacterium simulans, Corynebacterium kroppenstedtii, and Corynebacterium amycolatum, are predominantly present in sebaceous sites (Byrd et al., 2018). Although Corynebacterium spp. are mostly innocuous, some species can cause human disease, most notably diphtheria, a condition caused by the diphtheria toxin secreted by Corynebacterium diphtheriae (Wenzel et al., 2020). Results from NGS analysis showed that Corynebacterium spp. accounted for approximately 10% of the total bacteria in human skin, and C. aurimucosum accounted for a fraction of that with a relative abundance of < 0.3% (Figure 1). Despite this, a high level of antibodies to C. aurimucosum was detected in the human sera (Figure 2). A possible explanation for the discrepancy in relative abundance and antibody titer is that other antibodies to Corynebacterium spp. may have cross-reacted to C. aurimucosum antigens in LFIAs and enzyme-linked immunosorbent assay (ELISA), supported by the detection of antibodies to Corynebacterium parvum (Wolberg et al., 1977), C. diphtheriae (Wenzel et al., 2020), or Corynebacterium jeikeium (Clark et al., 1990) in human blood. It is also possible that C. aurimucosum expresses highly antigenic proteins and, in turn, induced elevated productions of antibodies against C. aurimucosum.
S. aureus is frequently isolated from the anterior nares, which is assumed to be the main reservoir for the spread of the pathogen-associated skin diseases such as atopic dermatitis (van Mierlo et al., 2021). However, several S. aureus strains have been collected from pus cell of nodulocystic and pustular acne of volunteers (Khan et al., 2015). In parallel with C. aurimucosum, S. aureus has a relatively low relative abundance (0.03% ± 0.02%) (Figure 1) yet provoked tremendous antibodies in the blood (Figure 2). Superantigens including staphylococcal enterotoxins and toxic shock syndrome toxin-1 have been found in S. aureus (Schlievert et al., 2008). Expression of these superantigens may explain the elevated titer of antibodies to S. aureus in the bloodstream. As shown in Figure 3B, the levels of antibodies to S. aureus in acne patients were approximately 1.5-fold lower than those in healthy subjects. Previous studies have demonstrated that S. aureus can hijack C. acnes to intensify its virulence via interaction of S. aureus β-hemolysin with C. acnes CAMP factor (Lo et al., 2011). Thus, it is worth investigating whether a decrease in antibodies, especially neutralizing antibodies, to S. aureus augments the interaction of S. aureus with C. acnes, exaggerating the pathogenesis of acne vulgaris. While the expression of highly antigenic proteins in S. aureus could explain the negative correlation between relative abundance and circulating antibodies of S. aureus, C. acnes presented a negative correlation but in reverse: High abundance on facial skin provoked comparatively lower antibodies (Figures 1, 2). Although C. acnes may enter the dermal layer upon inflammation or follicle rupture in an acne lesion, the bacterium is an anaerobe present within the retained sebum in the pilosebaceous ducts (Kabashima et al., 2019). Thus, although C. acnes bacteria are abundant in the sebaceous pilosebaceous unit, the epidermal Langerhans cells, skin-resident dendritic cells, may not easily access C. acnes within an intact sebaceous follicle to present antigens. The relative abundance of bacteria on the skin surface of healthy subjects (Figure 1) was used for comparing its correlation with the profile of antibodies to bacteria. In fact, in comparing skin surface with hair follicles and healthy skin with acne lesions, the composition and relative abundance of bacteria in those skin microbiomes are different (Lousada et al., 2021). For example, C. acnes was dominant in the hair follicles of healthy skin, whereas additional species including C. tuberculostearicum became detected in the hair follicles of acne lesions (Bek-Thomsen et al., 2008). The shift of microbiome from healthy skin to acne lesions may affect the production of antibodies to each bacterium in skin. Although we obtained sera from healthy subjects with no prior history of acne vulgaris and acne patients with multiple acne lesions (Figure 3), these human subjects may have the history of infectious diseases that provoke the antibodies to C. acnes, S. aureus, or C. aurimucosum. It has been known that C. acnes was associated with the prosthetic joint infection (Boisrenoult, 2018). Clinical reports in skin infection with S. aureus (Kabashima et al., 2019) and urinary tract infection with C. aurimucosum (Lo et al., 2015) have been documented.
In this study, protein A from S. aureus was utilized to conjugate AuNPs for binding Fc regions of various immunoglobulins in mouse sera. Although protein A conjugated on the surface of AuNPs can simultaneously capture different antibodies in a single serum sample, it has been reported that protein A exhibited some antibody subclass differences in various species (Nelson and Reichert, 2009). Protein A elicits a higher affinity for human IgG (IgG1 and IgG2) than IgM and IgA and interacts strongly with IgG2a and IgG3 compared to IgG1 in mice (Rodrigo et al., 2015). However, it has been reported that colonization of commensal bacteria in humans primarily triggered the production of IgA by B cells (Keppler et al., 2021). Although the binding of protein A with subclasses of antibodies in mice is not yet determined, replacement of protein A with protein L (Rodrigo et al., 2015), a Peptostreptococcus magnus protein with a high affinity to representatives of most antibody classes, may be able to broadly capture different classes of antibodies including IgA in mice.
Antibodies to exocellular enzymes, cell wall fractions, or membrane-binding proteins of C. acnes were measurable in acne patients (Lodes et al., 2006). In some cases, there is a positive correlation between the titers of these antibodies and the severity of acne vulgaris (Ashbee et al., 1997). The high levels of IgG1 and IgG3 were detected in severe acne patients, whereas IgG2 was predominantly found in moderate and severe patients (Ashbee et al., 1997). Despite antibodies to C. acnes produced in acne patients (Webster et al., 1985; Ingham et al., 1987), the recurrence rate of acne vulgaris was high. In fact, acne patients were demonstrated to yield higher levels of antibodies to C. acnes than healthy subjects (Figures 3A, 4). It is possible that insufficient amounts of protective antibodies to C. acnes for prevention of acne vulgaris thereby lead to recurrence of acne vulgaris (Nakatsuji et al., 2008). Furthermore, age is a potential factor, as it has been reported that opportunistic C. acnes ribotypes are predominantly present in lesions of acne vulgaris in adolescents and young adults (Barnard et al., 2016), whereas the abundance of C. acnes declined in aged skin (Collier et al., 2008), and the abundance of Corynebacterium spp. conversely increased on the aged skin (Li et al., 2014). Still, a small sample size of participants aged from 21 to 50 years was used for this study, and increasing the sample size by including broader age groups (such as 0–20 years, ≥ 51 years) will be needed to accurately reflect the change in the skin microbiome of acne patients by age and its corresponding antibodies over the lifetime of humans.
The serum antibodies to commensal whole bacteria or their proteins as antigens are present in healthy human subjects. For example, IgG recognition of gut commensal antigens is detectable in childhood (Christmann et al., 2015). Although C. acnes is one of skin resident bacteria, it is commonly detected in the human gut (Gunathilake et al., 2019). Anti–Escherichia coli IgG, particularly antiflagellin antibodies, is detectable in blood (Jostins et al., 2014). Theoretically, it is unusual for the systemic adaptive immunity to be primed against the commensal bacteria under normal circumstances. It is not known how B cells produce antibodies to gut commensal bacteria, but it likely follows exposure to commensal bacteria that enter the lamina propria and beyond via disrupted intestinal barrier. Mucosal surfaces are protected specifically by secretory IgA and IgM, which are transported by the epithelial polymeric immunoglobulin receptor (Brown, 1978). It has been reported that the deficiency of epithelial polymeric immunoglobulin receptor resulted in the abrogation of IgA and IgM transcytosis, leading to an increase in serum IgG antibodies against gut commensals (Johansen et al., 1999). Although it is not clear how B cells can yield antibodies to skin commensal bacteria, it is well recognized that skin is an immune organ, and Langerhans cells can extend dendrites to sample the antigens of bacteria on the surface of skin (Guttman-Yassky et al., 2019).
Whether the production of antibodies to commensal bacteria in humans is beneficial or harmful to immunity is unknown. Neutralizing antibodies from vaccinations against C. acnes provide effective biologics to reduce inflammation in models of cells, mice (Wang et al., 2018), and ex vivo using acne biopsies (Nakatsuji et al., 2008; Wang et al., 2018). However, it has been illustrated that anticommensal IgG can trigger the NOD-, LRR-, and pyrin domain-containing protein 3 (NLRP3) inflammasome signaling to induce helper T cell 17–driven inflammation via activation of Fcγ receptors on macrophages (Castro-Dopico et al., 2019). Recent discovery unveiled that IgG variants without core fucosylation caused severe inflammation through increased FcγIIIA receptor affinity (Chakraborty et al., 2021; Larsen et al., 2021). Afucosylated IgG promoted secretion of proinflammatory cytokines in macrophages and correlated with COVID-19 severity (Larsen et al., 2021). These findings indicated that the severity of inflammatory can be regulated by heterogeneity of IgG. Although the (a)fucosylation of IgG induced by C. acnes was not determined, our results, through the detection of antibodies to C. acnes on LFIA, provide a signature to reflect the inflammatory acne vulgaris.
Microbial dysbiosis, a change in the bacterial abundance changes from normal to disease, can be used as a surrogate marker of human diseases. The 16S rRNA sequencing via NGS analysis is the most widely used method to examine the microbial dysbiosis. The NGS analysis can be time-consuming and labor-intensive and requires expensive equipment. Establishing a pattern to indicate the alternations in the levels of antibodies to each bacterium in the dysbiotic microbiome by LFIAs can be quickly achieved without specialized equipment as the results of antigen–antibody reaction on LFIAs can be visualized in 15–20 min (Figure 3). Although the detection of low concentrations of antibodies by LFIAs could give rise to false-negative or false-positive results compared with ELISA (La Rosa Fabian and Urquizo Briceno, 2020), many techniques have been developed to improve the sensitivity and specificity of the diagnosis (Hu et al., 2021). Recently, deep learning–based LFIAs and multiple-antigen spotted circular flow immunoassays (CFIAs) have been developed in our group (Huang and Herr, 2021). Bacterial lysates are mixtures of antigens derived from inactivated microorganisms. Spotting bacterial lysates on LFIAs can generate results due to cross-reactions of antibodies induced by different bacteria. Immobilization of various bacteria-specific antigens on a CFIA will avoid the cross-reactions of antibodies in a blood sample and simultaneously detect multiple antigen-antibody reactions in one assay. Despite that deep learning–based LFIAs or CFIAs can establish a calibration curve for antibody quantification, a cutoff value to eliminate the source of cross-reactions and background is needed (Zhang et al., 2020). When the improvements are accomplished, a signature of antibodies to different bacteria on LFIAs or CFIAs may allow physicians and medical staff to achieve diagnostic results within minutes in the future.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. And I detected the following words and expressions: Global list: 16s rRNA, 16s ribosomal.
Ethics Statement
An approved protocol (no. 121230) (Wang et al., 2018) by the institutional review board at University of California, San Diego (UCSD). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
RH wrote manuscript and performed experiments. SM and CL designed experiments and edited manuscript, respectively. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Lee Foundation Microbiome grant from the Department of Surgery, National University of Singapore, Singapore.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful for Liangfang Zhang, a professor of Bioengineering at the University of California San Diego (UCSD), for providing more insight into nanoparticles, particularly the usage of AuNPs used in LFIAs. We would like to also thank Yong Jiang at American Diagnostics, San Diego, for providing the laboratory space and materials to perform the experiments. We appreciate Franchesca Choi at MetroWest Medical Center, Framingham, Massachusetts for editing the writing for human sample preparation. Finally, we acknowledge valuable comments at LFIAs and anticommensal antibodies from judges for RH’s presentation at the 2021 Greater San Diego Science and Engineering Fair (GSDSEF) and California Science and Engineering Fair (CSEF).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.709562/full#supplementary-material
References
Ashbee, H. R., Muir, S. R., Cunliffe, W. J., and Ingham, E. (1997). IgG subclasses specific to Staphylococcus epidermidis and Propionibacterium acnes in patients with acne vulgaris. Br. J. Dermatol. 136, 730–733. doi: 10.1111/j.1365-2133.1997.tb03660.x
Banerjee, R., and Jaiswal, A. (2018). Recent advances in nanoparticle-based lateral flow immunoassay as a point-of-care diagnostic tool for infectious agents and diseases. Analyst 143, 1970–1996. doi: 10.1039/C8AN00307F
Barnard, E., Shi, B., Kang, D., Craft, N., and Li, H. (2016). The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci. Rep. 6:39491. doi: 10.1038/srep39491
Bek-Thomsen, M., Lomholt, H. B., and Kilian, M. (2008). Acne is not associated with yet-uncultured bacteria. J. Clin. Microbiol. 46, 3355–3360. doi: 10.1128/JCM.00799-08
Blaskovich, M. A. T., Elliott, A. G., Kavanagh, A. M., Ramu, S., and Cooper, M. A. (2019). In vitro antimicrobial activity of acne drugs against skin-associated bacteria. Sci. Rep. 9:14658. doi: 10.1038/s41598-019-50746-4
Boisrenoult, P. (2018). Cutibacterium acnes prosthetic joint infection: diagnosis and treatment. Orthop. Traumatol. Surg. Res. 104, S19–S24. doi: 10.1016/j.otsr.2017.05.030
Brown, W. R. (1978). Relationships between immunoglobulins and the intestinal epithelium. Gastroenterology 75, 129–138. doi: 10.1016/0016-5085(78)93779-4
Byrd, A. L., Belkaid, Y., and Segre, J. A. (2018). The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155. doi: 10.1038/nrmicro.2017.157
Castro-Dopico, T., Dennison, T. W., Ferdinand, J. R., Mathews, R. J., Fleming, A., Clift, D., et al. (2019). Anti-commensal IgG drives intestinal inflammation and type 17 immunity in ulcerative colitis. Immunity 50, 1099–1114.e10. doi: 10.1016/j.immuni.2019.02.006
Chakraborty, S., Gonzalez, J., Edwards, K., Mallajosyula, V., Buzzanco, A. S., Sherwood, R., et al. (2021). Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 22, 67–73. doi: 10.1038/s41590-020-00828-7
Chen, P., He, G., Qian, J., Zhan, Y., and Xiao, R. (2021). Potential role of the skin microbiota in inflammatory skin diseases. J. Cosmet. Dermatol. 20, 400–409. doi: 10.1111/jocd.13538
Christmann, B. S., Abrahamsson, T. R., Bernstein, C. N., Duck, L. W., Mannon, P. J., Berg, G., et al. (2015). Human seroreactivity to gut microbiota antigens. J. Allergy Clin. Immunol. 136, 1378–1386.e1–5. doi: 10.1016/j.jaci.2015.03.036
Clark, I., Burnie, J. P., Coke, A. P., Matthews, R. C., and Oppenheim, B. A. (1990). Oppenheim. Characterization of the antibody response in Corynebacterium jeikeium septicaemias. Epidemiol. Infect. 105, 229–236. doi: 10.1017/S095026880004783X
Collier, C. N., Harper, J. C., Cafardi, J. A., Cantrell, W. C., Wang, W., Foster, K. W., et al. (2008). The prevalence of acne in adults 20 years and older. J. Am. Acad. Derm. 58, 56–59. doi: 10.1016/j.jaad.2007.06.045
Dréno, B., Dagnelie, M. A., Khammari, A., and Corvec, S. (2020). The skin microbiome: a new actor in inflammatory acne. Am. J. Clin. Dermatol. 21(Suppl. 1), 18–24. doi: 10.1007/s40257-020-00531-1
Fitz-Gibbon, S., Tomida, S., Chiu, B. H., Nguyen, L., Du, C., Liu, M., et al. (2013). Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Investig. Dermatol. 133, 2152–2160. doi: 10.1038/jid.2013.21
Godlewska, U., Brzoza, P., Kwiecień, K., Kwitniewski, M., and Cichy, J. (2020). Metagenomic studies in inflammatory skin diseases. Curr. Microbiol. 77, 3201–3212. doi: 10.1007/s00284-020-02163-4
Graham, G. M., Farrar, M. D., Cruse-Sawyer, J. E., Holland, K. T., and Ingham, E. (2004). Proinflammatory cytokine production by human keratinocytes stimulated with Propionibacterium acnes and P. acnes GroEL. Br. J. Dermatol. 150, 421–428. doi: 10.1046/j.1365-2133.2004.05762.x
Grice, E. A., and Segre, J. A. (2011). The skin microbiome. Nat. Rev. Microbiol. 4, 244–253. doi: 10.1038/nrmicro2537
Gunathilake, M. N., Lee, J., Choi, I. J., Kim, Y. I., Ahn, Y., Park, C., et al. (2019). Association between the relative abundance of gastric microbiota and the risk of gastric cancer: a case-control study. Sci. Rep. 9:13589. doi: 10.1038/s41598-019-50054-x
Guttman-Yassky, E., Zhou, L., and Krueger, J. G. (2019). Tolerance versus effector responses and applications to food allergy and hypersensitivity reactions. J. Allergy Clin. Immunol. 144, 362–374. doi: 10.1016/j.jaci.2019.03.021
Hu, X., Wu, C., Situ, B., Tian, P., An, T., Li, Q., et al. (2021). EDTA-K2 improves the detection sensitivity of SARS-CoV-2 IgM and IgG antibodies by chelating colloidal gold in the immunochromatographic assay. Int. J. Nanomedicine 16, 715–724. doi: 10.2147/IJN.S281594
Huang, R. Y., and Herr, D. R. (2021). Quantitative circular flow immunoassays with trained object recognition to detect antibodies to SARS-CoV-2 membrane glycoprotein. Biochem Biophys Res Commun. 565, 8–13. doi: 10.1016/j.bbrc.2021.05.073
Ingham, E., Gowland, G., Ward, R. M., Holland, K. T., and Cunliffe, W. J. (1987). Antibodies to P. acnes and P. acnes exocellular enzymes in the normal population at various ages and in patients with acne vulgaris. Br. J. Dermatol. 116, 805–812. doi: 10.1111/j.1365-2133.1987.tb04899.x
Johansen, F. E., Pekna, M., Norderhaug, I. N., Haneberg, B., Hietala, M. A., Krajci, P., et al. (1999). Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J. Exp. Med. 199, 915–922. doi: 10.1084/jem.190.7.915
Jostins, L., Ripke, S., Weersma, R. K., Duerr, R. H., McGovern, D. P., Hui, K. Y., et al. (2014). Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124. doi: 10.1038/nature11582
Kabashima, K., Honda, T., Ginhoux, F., and Egawa, G. (2019). The immunological anatomy of the skin. Nat. Rev. Immunol. 19, 19–30. doi: 10.1038/s41577-018-0084-5
Keppler, S. J., Goess, M. C., and Heinze, J. M. (2021). The wanderings of gut-derived iga plasma cells: impact on systemic immune responses. Front. Immunol. 12:670290. doi: 10.3389/fimmu.2021.670290
Khan, A. F., Hana, H. K., Sheak, J., and Begum, K. (2015). Antibiotic sensitivity of Staphylococcus aureus and Staphylococcus epidermidis isolated from acne patients. Bangladesh Pharm. J. 18, 121–125. doi: 10.3329/bpj.v18i2.24309
La Rosa Fabian, C., and Urquizo Briceno, L. (2020). Anti-SARS-Cov-2 IgA in current scenario of IgM and IgG rapid test: a new alternative for the diagnostic of COVID-19. SN Compr. Clin. Med. 2, 2167–2169. doi: 10.1007/s42399-020-00551-2
Larsen, M. D., de Graaf, E. L., Sonneveld, M. E., Plomp, H. R., Nouta, J., Hoepel, W., et al. (2021). Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science 371:eabc8378. doi: 10.1126/science.abc8378
Lee, Y. B., Byun, E. J., and Kim, H. S. (2019). Potential role of the microbiome in acne: a comprehensive review. J. Clin. Med. 8:987. doi: 10.3390/jcm8070987
Legrand-Poels, S., Esser, N., L’homme, L., Scheen, A., Paquot, N., Piette, J., et al. (2014). Free fatty acids as modulators of the NLRP3 inflammasome in obesity/type 2 diabetes. Biochem. Pharmacol. 92, 131–141. doi: 10.1016/j.bcp.2014.08.013
Li, W., Han, L., Yu, P., Ma, C., Wu, X., and Xu, J. (2014). Nested PCR-denaturing gradient gel electrophoresis analysis of human skin microbial diversity with age. Microbiol Res. 169, 686–692. doi: 10.1016/j.micres.2014.02.008
Lo, C. W., Lai, Y. K., Liu, Y. T., Gallo, R. L., and Huang, C. M. (2011). Staphylococcus aureus hijacks a skin commensal to intensify its virulence: immunization targeting beta-hemolysin and CAMP factor. J. Invest. Dermatol. 131, 401–409. doi: 10.1038/jid.2010.319
Lo, S., Thiam, I., Fall, B., Ba-Diallo, A., Diallo, O. F., Diagne, R., et al. (2015). Urinary tract infection with Corynebacterium aurimucosum after urethroplasty stricture of the urethra: a case report. J. Med. Case Rep. 9:156. doi: 10.1186/s13256-015-0638-0
Lodes, M. J., Secrist, H., Benson, D. R., Jen, S., Shanebeck, K. D., Guderian, J., et al. (2006). Variable expression of immunoreactive surface proteins of Propionibacterium acnes. Microbiology 152, 3667–3681. doi: 10.1099/mic.0.29219-0
Lousada, M. B., Lachnit, T., Edelkamp, J., Rouillé, T., Ajdic, D., Uchida, Y., et al. (2021). Exploring the human hair follicle microbiome. Br. J. Dermatol. 184, 802–815. doi: 10.1111/bjd.19461
Nakatsuji, T., Liu, Y. T., Huang, C. P., Zoubouis, C. C., Gallo, R. L., and Huang, C. M. (2008). Antibodies elicited by inactivated propionibacterium acnes-based vaccines exert protective immunity and attenuate the IL-8 production in human sebocytes: relevance to therapy for acne vulgaris. J. Invest. Dermatol. 128, 2451–2457. doi: 10.1038/jid.2008.117
Nelson, A. L., and Reichert, J. M. (2009). Development trends for therapeutic antibody fragments. Nat. Biotechnol. 27, 331–337. doi: 10.1038/nbt0409-331
Ogai, K., Nagase, S., Mukai, K., Iuchi, T., Mori, Y., Matsue, M., et al. (2018). A comparison of techniques for collecting skin microbiome samples: swabbing versus tape-stripping. Front. Microbiol. 9:2361. doi: 10.3389/fmicb.2018.02362
O’Neill, A. M., and Gallo, R. L. (2018). Host-microbiome interactions and recent progress into understanding the biology of acne vulgaris. Microbiome 6:177. doi: 10.1186/s40168-018-0558-5
Rajiv, P., Nitesh, K., Raj, K., and Hemant, G. (2013). Staphylococcus epidermidis in human skin microbiome associated with acne: a cause of disease or defence? Res. J. Biotechnol. 8, 78–82.
Ravi, N., Cortade, D. L., Ng, E., and Wang, S. X. (2020). Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron 165:112454. doi: 10.1016/j.bios.2020.112454
Rodrigo, G., Gruvegård, M., and Van Alstine, J. M. (2015). Antibody fragments and their purification by Protein L affinity chromatography. Antibodies 4, 259–277. doi: 10.3390/antib4030259
Schlievert, P. M., Case, L. C., Strandberg, K. L., Abrams, B. B., and Leung, D. Y. (2008). Superantigen profile of Staphylococcus aureus isolates from patients with steroid-resistant atopic dermatitis. Clin. Infect. Dis. 46, 1562–1567. doi: 10.1086/586746
Sitohang, I. B. S., Fathan, H., Effendi, E., and Wahid, M. (2019). The susceptibility of pathogens associated with acne vulgaris to antibiotics. Med. J. Indones. 28, 21–27. doi: 10.13181/mji.v28i1.2735
Sterlin, D., Fadlallah, J., Slack, E., and Gorochov, G. (2020). The antibody/microbiota interface in health and disease. Mucosal Immunol. 13, 3–11. doi: 10.1038/s41385-019-0192-y
van Mierlo, M. M. F., Pasmans, S. G. M. A., Totté, J. E. E., de Wit, J., Herpers, B. L., Vos, M. C., et al. (2021). Klaassen CHW, Pardo LM. Temporal variation in Staphylococcus aureus Protein A genotypes from nose and skin in atopic dermatitis patients. Dermatology 237, 506–512. doi: 10.1159/000515235
Wang, Y., Hata, T. R., Tong, Y. L., Kao, M. S., Zouboulis, C. C., Gallo, R. L., et al. (2018). The anti-inflammatory activities of Propionibacterium acnes CAMP factor-targeted acne vaccines. J. Invest. Dermatol. 138, 2355–2364. doi: 10.1016/j.jid.2018.05.032
Webster, G. F., Indrisano, J. P., and Leyden, J. J. (1985). Antibody titers to Propionibacterium acnes cell wall carbohydrate in nodulocystic acne patients. J. Invest. Dermatol. 84, 496–500. doi: 10.1111/1523-1747.ep12273462
Wenzel, E. V., Bosnak, M., Tierney, R., Schubert, M., Brown, J., Dübel, S., et al. (2020). Human antibodies neutralizing diphtheria toxin in vitro and in vivo. Sci. Rep. 10:571. doi: 10.1038/s41598-019-57103-5
Wolberg, G., Duncan, G. S., Adlam, C., and Whisnant, J. K. (1977). Antibody to Corynebacterium parvum in normal human and animal sera. Infect. Immun. 15, 1004–10047. doi: 10.1128/iai.15.3.1004-1007.1977
Yu, Y., Champer, J., and Kim, J. (2015). Analysis of the surface, secreted, and intracellular proteome of Propionibacterium acnes. EuPA Open Proteom. 9, 1–7. doi: 10.1016/j.euprot.2015.06.003
Zhang, Y., Liu, X., Wang, L., Yang, H., Zhang, X., Zhu, C., et al. (2020). Improvement in detection limit for lateral flow assay of biomacromolecules by test-zone pre-enrichment. Sci. Rep. 10:9604. doi: 10.1038/s41598-020-66456-1
Keywords: acne vulgaris, antibody, C. acnes, C. aurimucosum, lateral flow immunoassays, S. aureus
Citation: Huang RY, Lee CN and Moochhala S (2021) Circulating Antibodies to Skin Bacteria Detected by Serological Lateral Flow Immunoassays Differentially Correlated With Bacterial Abundance. Front. Microbiol. 12:709562. doi: 10.3389/fmicb.2021.709562
Received: 14 May 2021; Accepted: 30 September 2021;
Published: 10 November 2021.
Edited by:
Jesús Navas, University of Cantabria, SpainReviewed by:
Daniela Pinto, Giuliani S.p.A., ItalyBattogtokh Chimeddorj, Mongolian National University of Medical Sciences, Mongolia
Han Liang Lim, Coastar Therapeutics, United States
Copyright © 2021 Huang, Lee and Moochhala. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shabbir Moochhala, cGhjc21tQG51cy5lZHUuc2c=
 Ryan Yuki Huang
Ryan Yuki Huang Chuen Neng Lee2
Chuen Neng Lee2 Shabbir Moochhala
Shabbir Moochhala