- 1Instituto de Microbiología, Colegio de Ciencias Biológicas y Ambientales, Universidad San Francisco de Quito, Quito, Ecuador
- 2Agrocalidad, Agencia de Regulación y Control Fito y Zoosanitario, Quito, Ecuador
- 3Environmental Health Sciences Division, University of California, Berkeley, Berkeley, CA, United States
In cities across the globe, the majority of wastewater – that includes drug resistant and pathogenic bacteria among other contaminants – is released into streams untreated. This water is often subsequently used for irrigation of pastures and produce. This use of wastewater-contaminated streams allows antibiotic-resistant bacteria to potentially cycle back to humans through agricultural products. In this study, we investigated the prevalence of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolated from produce and irrigation water across 17 provinces of Ecuador. A total of 117 vegetable samples, 119 fruit samples, and 38 irrigation water samples were analyzed. Results showed that 11% of the samples were positive for E. coli including 11 irrigation water samples (29%), and samples of 13 vegetables (11%), and 11 fruits (9%). Among the 165 E. coli isolates cultured, 96 (58%) had the ESBL phenotype, and 58% of ESBL producing E. coli came from irrigation water samples, 11% from vegetables, and 30% from fruits. The blaCTX–M–55, blaCTX–M 65, and blaCTX–M 15 genes were the most frequently found gene associated with the ESBL phenotype and coincided with the blaCTX–M alleles associated with human infections in Ecuador. Three isolates had the mcr-1 gene which is responsible for colistin resistance. This report provides evidence of the potential role of irrigation water in the growing antimicrobial resistance crisis in Ecuador.
Introduction
The rise of antimicrobial resistance (AMR) is one of the most serious biological threats facing modern society, and the inability to treat bacterial infections is already occurring in many nosocomial infections (Frieri et al., 2017). The World Health (WHO) has listed extended spectrum β-lactamase-producing Enterobacteriaceae (ESBL-E) as the most critical antimicrobial resistant microorganisms, among the “Highest Priority” pathogens due to the increasing prevalence in humans and livestock (Yassin et al., 2017; Shrivastava et al., 2018; Li et al., 2019; Murray et al., 2021).
Globally, the majority of wastewater produced by urban settlements goes into streams without prior treatment. Only 20% of produced wastewater receives proper treatment (UNESCO, 2012), and the capacity to treat wastewater often depends on the income level of the country; treatment capacity is 70% of the generated wastewater in high-income countries, compared to ∼8% in low-income countries (Sato et al., 2013). This phenomenon is rising as urban populations grow and developing countries increasingly install pipes to channel wastewater away from communities, even before the development of wastewater treatment plants. The wastewater comes from diverse sources (e.g., homes, hospitals, and animal processing plants, etc.) and contains large quantities of antibiotic resistant bacteria (ARB), often carrying antimicrobial resistance to last-line antimicrobials, such as carbapenems (Lin et al., 2020).
These antimicrobial resistant bacteria (ARB) can cycle back to humans when wastewater-contaminated streams are used to irrigate produce or provide water to food animals (FAO and WHO, 2008; Leff and Fierer, 2013; Pigłowski, 2019); one recent example is the finding of New Delhi metallo-β-lactamases–type carbapenem-resistant Escherichia coli in water, domestic food animals, and humans (carbapenem, a last-line drug, is used exclusively in human medicine) (Li et al., 2019; Murray et al., 2021). Many antibiotic-resistant Enterobacterales, members of the intestinal microbiome (including E. coli), can survive and multiply in the environment (Vasco et al., 2015; Guerrero et al., 2020) and may colonize humans and domestic animals through the fecal-oral route of transmission. Plasmids and other mobile genetic elements (MGEs) carrying AMR genes promote the dissemination of AMR among intestinal bacteria in the intestine of vertebrates (Bonardi and Pitino, 2019), and this cycle is fundamentally captured in the One Health concept. Produce contamination can happen before pre-harvest (i.e., through contaminated irrigation water or manure fertilization) (Beuchat, 1996; Iwu and Okoh, 2019), as well as post-harvest (i.e., by washing, handling and processing food) with irrigation water (Murray et al., 2017).
Wastewater-impacted irrigation water has been identified as the main source of contamination for fresh produce with pathogenic microorganisms and ARB (Njage and Buys, 2015; Gekenidis et al., 2018a). The fecally contaminated produce can transfer ARB to the consumer especially when the produce is consumed fresh and uncooked (Pesavento et al., 2014; Araújo et al., 2017; Hölzel et al., 2018). Besides contributing to the spread of pathogens, irrigation water may potentially play a leading role in the dissemination of ARB (Moore et al., 2010; Hong et al., 2013; Gekenidis et al., 2018b; Vital et al., 2018).
The production of extended-spectrum β-lactamases (ESBL) is one of the most important mechanisms of antibiotic resistance in Enterobacteriaceae. ESBL genes can be divided into 4 groups: TEM, SHV, OXA, and CTX-M types (Bush and Jacoby, 2010); CTX-M type is the most prevalent of ESBLs described (Rossolini et al., 2008; Bevan et al., 2017). Enterobacteriaceae members are the most common bacterial agents causing foodborne outbreaks associated with the consumption of fresh produce (Cooper et al., 2007; Kilonzo-Nthenge et al., 2018; Al-Kharousi et al., 2019; McDaniel and Jadeja, 2019; Motlagh and Yang, 2019). Pathogenic E. coli is a key bacterium in foodborne illnesses, and commensal E. coli is a common indicator organism of fecal contamination in aquatic systems (Edberg et al., 2000; Rochelle-Newall et al., 2015; Motlagh and Yang, 2019). E. coli is also recognized as an important species in the spread of ARB, mainly due to a high aptitude to acquire genetic information through horizontal gene transfer (Grasselli et al., 2008; Hasegawa et al., 2018; Marlène et al., 2020).
In Ecuador, an upper middle-income country, wastewater is almost entirely released untreated into streams; these streams often serve as irrigation water for produce and food-animal agriculture (Ortega-Paredes et al., 2020a, b). There are few studies about the dissemination of ESBL-E. coli from irrigation water to produce (Ben Said et al., 2015; Vital et al., 2018); most of the studies have been carried out in fresh produce from retail centers and groceries (Bhutani et al., 2015; Faour-Klingbeil et al., 2016; Ortega-Paredes et al., 2018; Al-Kharousi et al., 2019; Yang et al., 2019; Colosi et al., 2020; Richter et al., 2020; Song et al., 2020). The aim of this study was to build upon the previous literature to understand the relationship between ARB in irrigation water and ARB on fresh produce obtaining samples from farms and their irrigation water. The study focused on the occurrence of extended spectrum β-lactamase producing E. coli in 17 provinces of Ecuador.
Materials and Methods
Study Areas
This study was carried out in the following provinces of Ecuador: Manabí, Bolívar, Cañar, Loja, Guayas, Pastaza, Tungurahua, Pichincha, Azuay, Chimborazo, Cotopaxi, Imbabura, Santa Elena, Los Ríos, Morona Santiago, Orellana, and Zamora Chinchipe provinces which are mainly agrarian (Figure 1). The samples correspond to those that are collected as part of the national surveillance program that aims to monitor microbiological indicators and pathogens in the food supply (“Programa Nacional de Vigilancia de Microorganismos de Higiene y Control de Microorganismos Patógenos, para la Vigilancia Epidemiológica de Enfermedades Transmitidas por Alimentos de Origen Agrícola y Pecuario del país – PNVCH”).
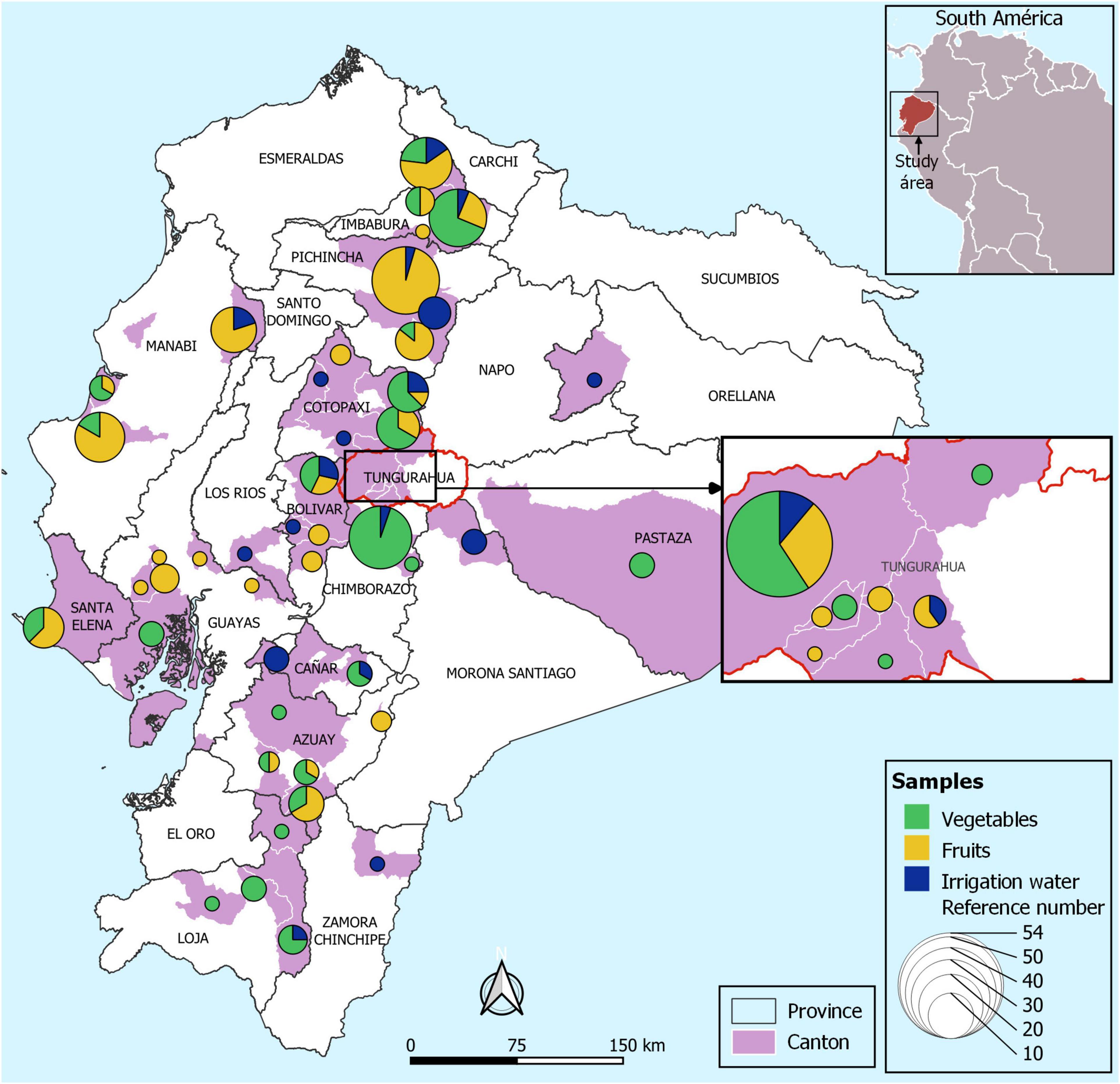
Figure 1. Map of Ecuador showing the sampling locations. Map of sampling locations of irrigation water, fruits and fresh produce. The circles represent the total number of samples according to the color assigned to each sample (vegetables: green, fruits: yellow, and irrigation water: blue) collected in each canton (pink).
Sampling Fresh Produce
Fresh fruits and vegetables (representing 20 types) were obtained from agricultural farms in 17 provinces of Ecuador, from June to December 2019 (Figure 1). In total, 274 samples were analyzed (117 vegetables, 119 fruits were collected from agricultural farms. Among the vegetables consist of lettuce (Lactuca sativa, n = 43), onion (Allium cepa, n = 31), garlic (Allium sativum, n = 21), coriander (Coriandrum sativum, n = 17), cabbage (Brassica oleracea var. viridis, n = 2), spinach (Spinacea oleracea, n = 1), pepper (Piper nigrum, n = 1), tomato (Solanum lycopersicum, n = 1). The fruit samples correspond to cocoa (Theobroma cacao, n = 1), peach (Prunus persica, n = 2), strawberry (Fregaria vesca, n = 31), melon (Cucumis melo var. cantalupensis, n = 7), apple (Malus domestica, n = 1), banana (Musa paradisiaca, n = 13), blackberry (Rubus ulmifolius, n = 31), watermelon (Citrullus lanatus, n = 12), grape (Vitis vinifera, n = 1), and golden berry (Physalis peruviana, n = 20).
Isolation of Escherichia coli From Irrigation Water and Produce
The farmers of each crop indicated the irrigation water they used, and this water (n = 37) was collected in sterile bottles and transported to the laboratory at approximately 8°C and processed within 10 h. Five hundred milliliters of water were filtered using a 0.45 μm pore membrane filter (Millipore, United States). The filter was then incubated in Chromocult® coliform agar (Merck, Germany) overnight at 37°C, the apparent E. coli colonies were taken and seeded on MacConkey agar (Difco, United States) supplemented with ceftriaxone (2 mg/L) to identify the lactose positive colonies (a maximum of five colonies were picked from each plate) (Richter et al., 2020), colonies of presumptive E. coli were then tested for β-glucuronidase activity using Chromocult® medium (Merck, Germany). All E. coli confirmed isolates from each sample were kept frozen at −80°C in Tryptic Soy Broth medium (Difco, United States) with 15% glycerol.
The vegetable samples were collected aseptically and refrigerated until analysis (within 12 h). Ten grams of the fresh produce were weighed and placed in a sterile plastic bag and incubated with 90 ml of peptone water (Faour-Klingbeil et al., 2016) for 30 min at room temperature. In the case of fruits such as watermelon and melon, the surface was swabbed, and the swab was placed in peptone water (described above). The next day 100 μl of the liquid was taken and cultured on MacConkey agar (Difco, United States) supplemented with ceftriaxone (2 mg/L) (Botelho et al., 2015). A maximum of five lactose positive colonies were selected from each plate sample and placed on Chromocult coliform agar after 24 h of incubation at 37°C, colonies of presumptive E. coli, positive for β-glucuronidase, were selected for additional analyses (Lange et al., 2013). All isolates confirmed to be E. coli from each sample were kept frozen at −80°C in Tryptic Soy Broth medium (Difco, United States) with 15% glycerol.
Antimicrobial Susceptibility Testing
Susceptibility tests were performed using the Kirby-Bauer method on Mueller-Hinton agar (Difco, United States) in accordance with Clinical and Laboratory Standards Institute (CLSI, 2019). Eleven antibiotics were used for testing and included: Cefazolin, CZ (30 μg); Ampicillin, AM (10 μg), Gentamicin, GM (10 μg), Imipenem, IPM (10 μg); Trimethropin-sulfamethoxazole, SXT (1.25/23.75 μg); Ceftazidime, CAZ (30 μg); Cefepime, FEP (30 μg); Ciprofloxacin, CIP (5 μg); Amoxicillin/Clavulanic acid, AmC (20/10μg); cefotaxime, CTX (30μg); and Tetracycline TE (30 μg). After 18 h of incubation, the E. coli strains were classified as susceptible, intermediate, or resistant according to the clinical interpretation criteria recommended by CLSI. E. coli ATCC 25922 was used as a quality control. To determine the ESBL phenotype, we carried out a diffusion disk method on Mueller Hinton agar as before using antibiotic susceptibility discs (Oxoid, United States) of CTX (30 μg), CAZ (30 μg). Our criterion to determine ESBL was CTX ≤ 27 mm; CAZ ≤ 22 mm (CLSI, 2019). Specifically, ESBL production was confirmed by growth in a medium with discs of ceftazidime (30 mcg) and ceftazidime + clavulanic acid (30 mcg + 10 mcg). An increase of ≥5 mm in zone of inhibition for ceftazidime + clavulanic acid compared to ceftazidime was confirmed as ESBL producers (CLSI, 2019).
PCR Amplification for Detection of β-Lactamase Genes
When samples were positive for ESBL-producing E. coli, one to five isolates selected per sample for further analysis. A total of 96 isolates were tested for the following resistance genes: blaSHV, blaTEM, blaCTX–M, and blaOXA (Table 1). Bacterial DNA was extracted by boiling (Dashti et al., 2009), and PCR amplification reactions were performed in a volume of 25 μl containing 12.5 μl of 2 × Qiagen Multiplex PCR Master Mix (Qiagen GmbH, Hilden, Germany), 0.2 μM concentrations of each primer, and 2 μl of DNA template. The cycling parameters were as follows: an initial denaturation at 95°C for 15 min; followed by 30 cycles of 94°C for 30 s, 62°C for 90 s, and 72°C for 60 s; and with a final extension at 72°C for 10 min. Amplification products were observed in agarose gel electrophoresis 1.5%, stained with Ethidium bromide at 100V for 45–60 min. The size of the amplified products was compared with the commercial (Invitrogen, United States) 100-bp ladder. The band size (bp) for each gene was: blaSHV, 237; blaTEM, 445; blaCTX–M, 593; and blaOXA: 813 (Fang et al., 2008).
DNA Sequencing and Analysis
Genomic DNA was extracted from the isolates using the Wizard® Genomic DNA Purification (Promega, United States) according to the manufacturer’s instructions. Sequencing was carried out at the University of Minnesota Mid-Central Research and Outreach Center (Willmar, Minnesota) using a single 2 × 250-bp dual-index run on an Illumina MiSeq with Nextera XT libraries to generate ∼30- to 50-fold coverage per genome. Genome assembly of MiSeq reads for each sample was performed using SPAdes assembler with the careful assembly option and automated k-mer detection (Bankevich et al., 2012). The identification of genus and species of the isolates was carried out using fastANI (Jain et al., 2018) with a percentage >80% of identification. Acquired AMR genes, plasmid types were identified using ABRicate tool (version 0.8.13), Resfinder was the database used for the identification of resistance genes (Zankari et al., 2012); PlasmidFinder database for plasmid replicon identification (Carattoli et al., 2014).
Phylogenetic Analysis
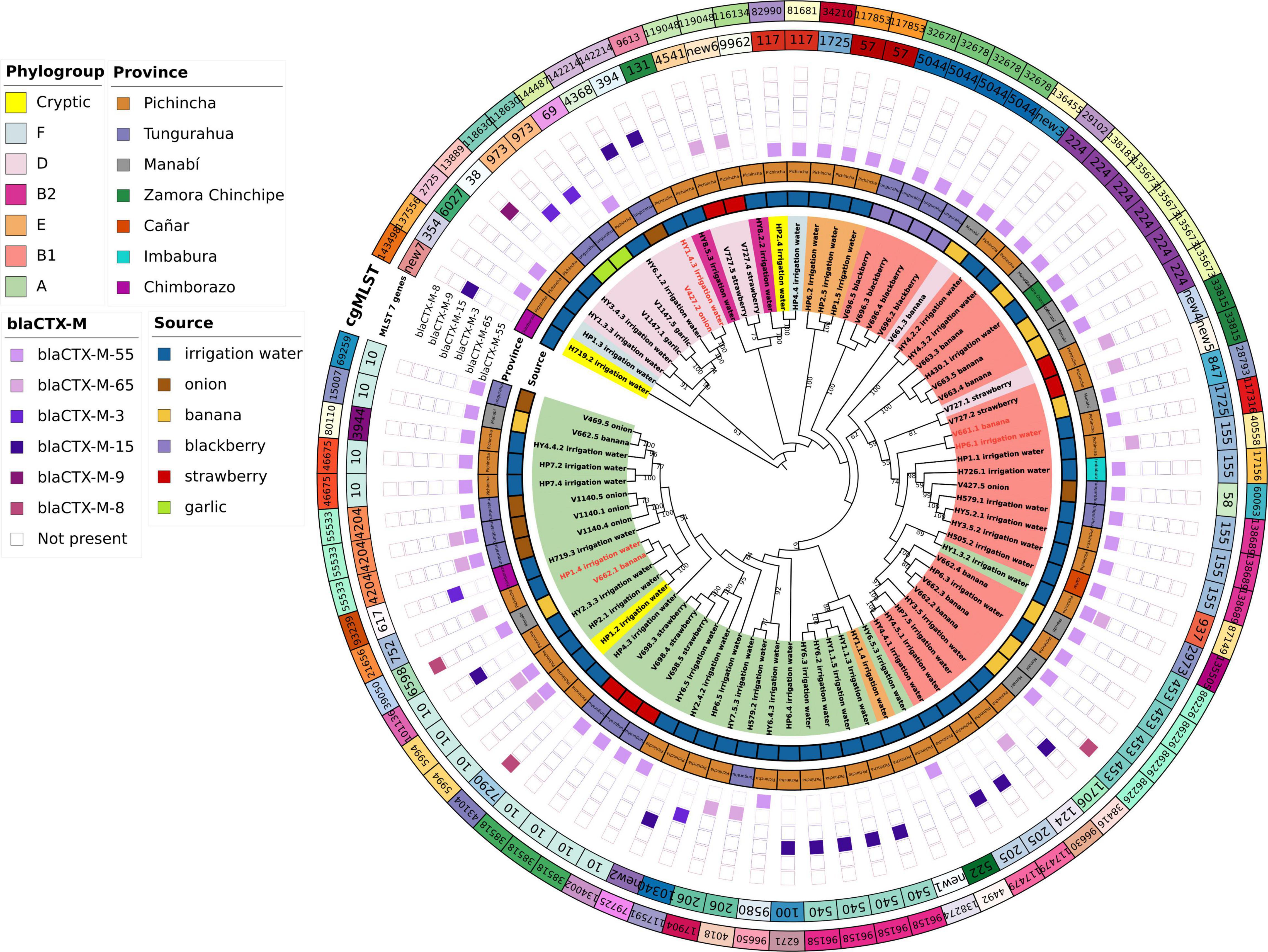
Pan-genomic analysis was carried out with Roary (Page et al., 2015); the core genome of the isolates analyzed was defined with at least 99%. A maximum likelihood phylogenetic tree with (1,000 bootstrap replicates) was created based on the core genomes of the isolates using RaxML-NG (Kozlov et al., 2019). The phylogenetic tree was visualized using iTOL (Letunic and Bork, 2019). Additionally, multilocus sequence typing (MLST) (Larsen et al., 2012), based on seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) and core genome (cgMLST) (Hansen et al., 2021) were performed using the Center for Genomic Epidemiology website1. The isolates also were characterized by Clermont phylogenetic typing by EzClermont web (Waters et al., 2020).
Sequence Accession Number
The sequences were uploaded to Bioproject- NCBI under the following accession numbers: SAMN20872921, SAMN20872922, SAMN20872998, SAMN20873936, SAMN20873938, SAMN20873941, SAMN20873969, SAMN20873994, SAMN20874637, SAMN20875987, SAMN20875988, SAMN20875992, SAMN20875994, SAMN20875998, SAMN20879008, SAMN20879962, SAMN20879963, SAMN20879975, SAMN20879976, SAMN20880112, SAMN20880135, SAMN20880136, SAMN20881008, SAMN20881023, SAMN20881078, SAMN20881101, SAMN20881102, SAMN20881103, SAMN20881104, SAMN20881105, SAMN20881397, SAMN20881398, SAMN20881399, SAMN20881400, SAMN20882115, SAMN20882121, SAMN20882132, SAMN20882145, SAMN20882146, SAMN20882147, SAMN20882148, SAMN20882149, SAMN20883143, SAMN20883144, SAMN20883145, SAMN20883146, SAMN20883147, SAMN20884528, SAMN20884547, SAMN20884549, SAMN20886717, SAMN20887874, SAMN20887881, SAMN20887882, SAMN20887901, SAMN20887904, SAMN20887915, SAMN20887924, SAMN20887927, SAMN20887932, SAMN20887933, SAMN20888904, SAMN20888908, SAMN20888911, SAMN20888912, SAMN20888913, SAMN20888914, SAMN20888915, SAMN20888916, SAMN20888921, SAMN20888932, SAMN20888933, SAMN20888934, SAMN20888941, SAMN20888958, SAMN20888959, SAMN20888960, SAMN20888962, SAMN20890819, SAMN20891007.
Results
Prevalence of Escherichia coli
In total, 274 samples were collected, including 117 vegetable samples, 119 fruit samples, and 38 irrigation water samples. Across all samples, a total of 30 (11%) were positive for E. coli; 11 of the irrigation water samples had E. coli (29%, 11/38), 13 vegetables samples had E. coli (11%, n = 13), and 11 fruits (9%, n = 11). In total, 165 isolates of E. coli were recovered from 30 samples.
Antimicrobial Susceptibility Testing
Ninety-six isolates (58% n = 96) showed extended-spectrum beta-lactamases (ESBL) phenotype according to the CLSI protocols; 58% of E. coli isolates from irrigation water were ESBL-producers, 11% from vegetables, and 30% from fruits. ESBL-E. coli were isolated from garlic (2 isolates), onion (9 isolates), strawberry (10 isolates), blackberry (4 isolates), banana (14 isolates), and golden berry (1 isolate).
The rate of resistance was high; more than 80% of recovered E. coli isolates were resistant to cefazolin, ampicillin, and cefotaxime. In the case of the E. coli isolates from irrigation water, 100% of the isolates were resistant to ampicillin and cefazolin. In addition, these isolates had a high prevalence of resistance to cefotaxime (96%), tetracycline (79%), and cefepime (84%) (Table 2).
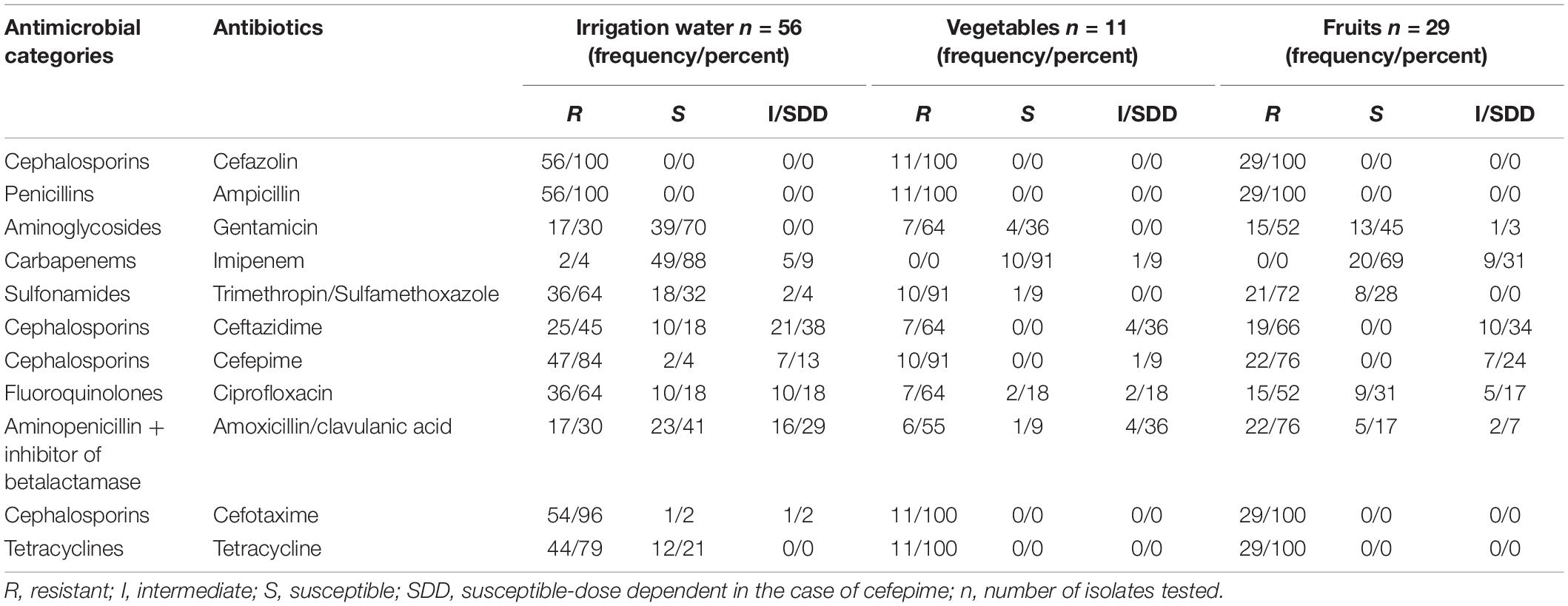
Table 2. Antibiotic susceptibility profiles of isolates ESBL- E.coli from irrigation water, vegetables, and fruits.
One hundred percent of the E. coli isolates from vegetables and fruits were resistant to ampicillin and cefazolin, cefotaxime, and tetracycline. Ninety-one percent of E.coli isolates from vegetables were resistant to cefepime. Two ESBL isolates from irrigation water presented resistance to the critically important class carbapenems, however no carbapenemase gene was detected. Additionally, we observed 33 resistance profiles across all of the extended spectrum beta-lactamase-producing E. coli isolates. The resistance profiles with the highest number of isolates are summarized in Table 3. In addition, 94% (90 of 96) of the E. coli ESBL isolates presented multi-drug resistant (MDR) patterns, with non-susceptible to at least one antibiotic in three or more antimicrobial categories (Magiorakos et al., 2012).
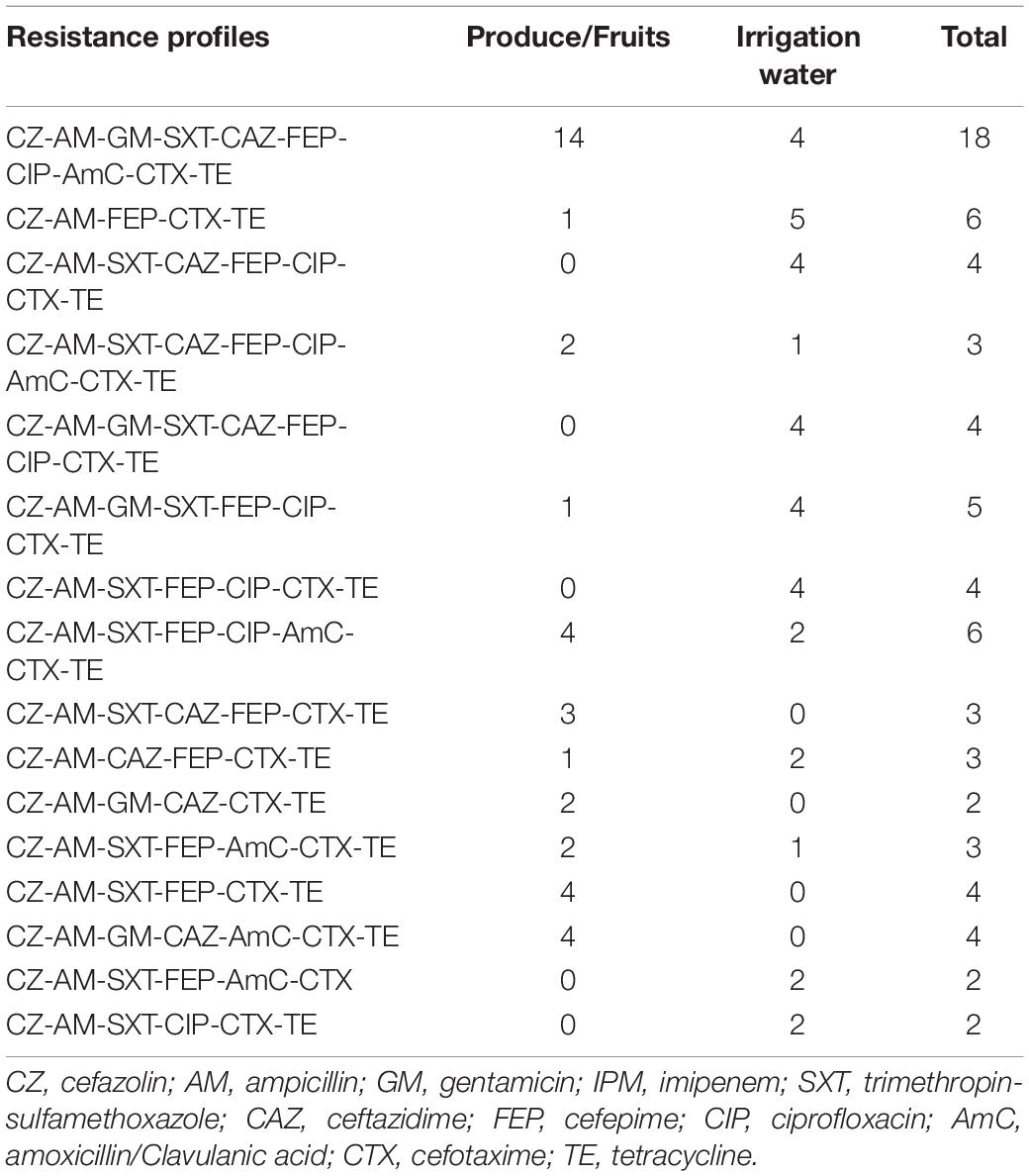
Table 3. The sixteen most common resistance profiles for ESBL-E. coli isolated from water, vegetables, and fruits in Ecuador.
Genotypes of Extended-Spectrum β-Lactamase – Escherichia coli
We obtained high-quality genome sequences of 80 ESBL-E. coli isolates. MLST analysis using 7 housekeeping genes showed that 80 isolates were assigned to 37 known STs, whereas 7 isolates represented 7 novel STs. ST10 was shared by 14% (n = 11) of isolates from three sources, with a different province of origin: irrigation water (Pichincha), onion (Tungurahua), banana (Manabí), and strawberry (Tungurahua). ST453 (5%, n = 4) and ST224 (8%, n = 6) were shared in two sources and in different provinces of origin of the sample: ST453 (banana = Manabí, irrigation water = Pichincha), ST224 (irrigation water = Pichincha and Zamora Chinchipe, banana = Manabí) (Table 4).
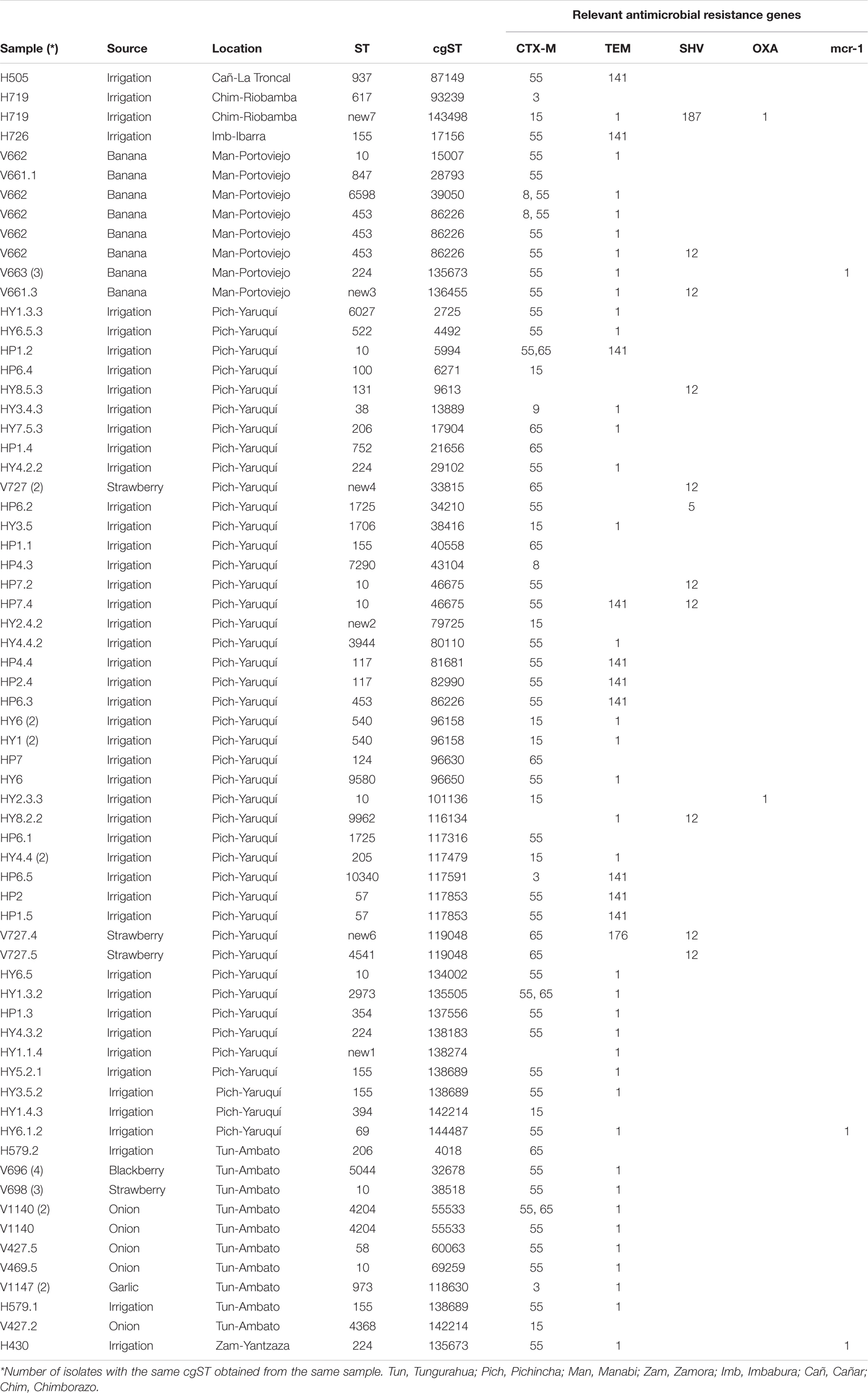
Table 4. Source and genetic characteristics of ESBL- E. coli isolates from different sources in Ecuador.
The application of a cgMLST scheme showed 55 cgSTs, from which only 2, cgST86226 (banana, Manabí, n = 5; irrigation water Pichincha, n = 1) and cgST135673 (banana Manabí, n = 3; irrigation water, Zamora Chinchipe n = 1) were isolates from two different sources. Several isolates belonging to the same ST (based on 7 genes) were assigned to different cgSTs based on cgMLST and some of the isolates from the same sample had the same cgST. Additionally, we constructed a maximum likelihood tree based on the core genomes to compare the phylogeny of isolates of E. coli from the irrigation water, vegetables, and fruits (Figure 2). The phylogenetic analysis showed that all isolates with the same cgMLST and obtained from different sources differed in thousands of SNPs indicating that although the isolates were genetically close, they have been evolving apart for many years (Table 4 and Figure 2). The genomes of ESBL-E. coli isolates from irrigation and fresh produce did not cluster apart; instead the isolates form different sources seemed to share recent common ancestry (Figure 2).
When ESBL-E.coli isolates were characterized by Clermont phylogenetic typing, 38% (n = 30) isolates belonged to phylogroup A: irrigation water (n = 21), strawberry (n = 3), onion (n = 4), banana (n = 2). In phylogroup B1 accounted for 35% (n = 28) of isolates: irrigation water (n = 15), banana (n = 7), strawberry (n = 1), blackberry (n = 4), and onion (n = 1). In phylogroup D accounted for 14% of the isolates: irrigation water (n = 4), strawberry (n = 3), garlic (n = 2), onion (n = 1) and banana (n = 1). Phylogroups B2, E and F accounted for 3% (n = 2), 5% (n = 4) and 3% (n = 2) of isolates, respectively. Three (4%) isolates of irrigation water belonged to the cryptic lineage (Figure 2).
Detection of β-Lactamase Genes
Ninety-six E.coli isolates phenotypically identified as ESBL, were tested by Multiplex PCR for genes encoding SHV, TEM, CTX-M, and OXA enzymes. The CTX-M gene was detected in 98% (94 of 96) of the isolates, followed by TEM 92% (88 of 96), SHV 28% (27 of 96), and OXA 1% (1/96). Additionally, combinations of genes were present: 64% had both CTX-M and TEM; and 26% had CTX-M, TEM, and SHV.
The presence of AMR genes in the genome sequences of 80 ESBL-E. coli isolates was investigated by Resfinder. Several ESBL-encoding blaCTX–M gene variants were distributed in isolates from irrigation water and fresh produce (Figure 3). Among the 80 ESBL-E. coli isolates, we identified allelic variants of blaCTX–M in 77 (96%). The most common allelic variants were blaCTX–M–55 in 49 isolates (64%) and the second most common allele was blaCTX–M–65 in 14 isolates (18%) (Supplementary Table 1).
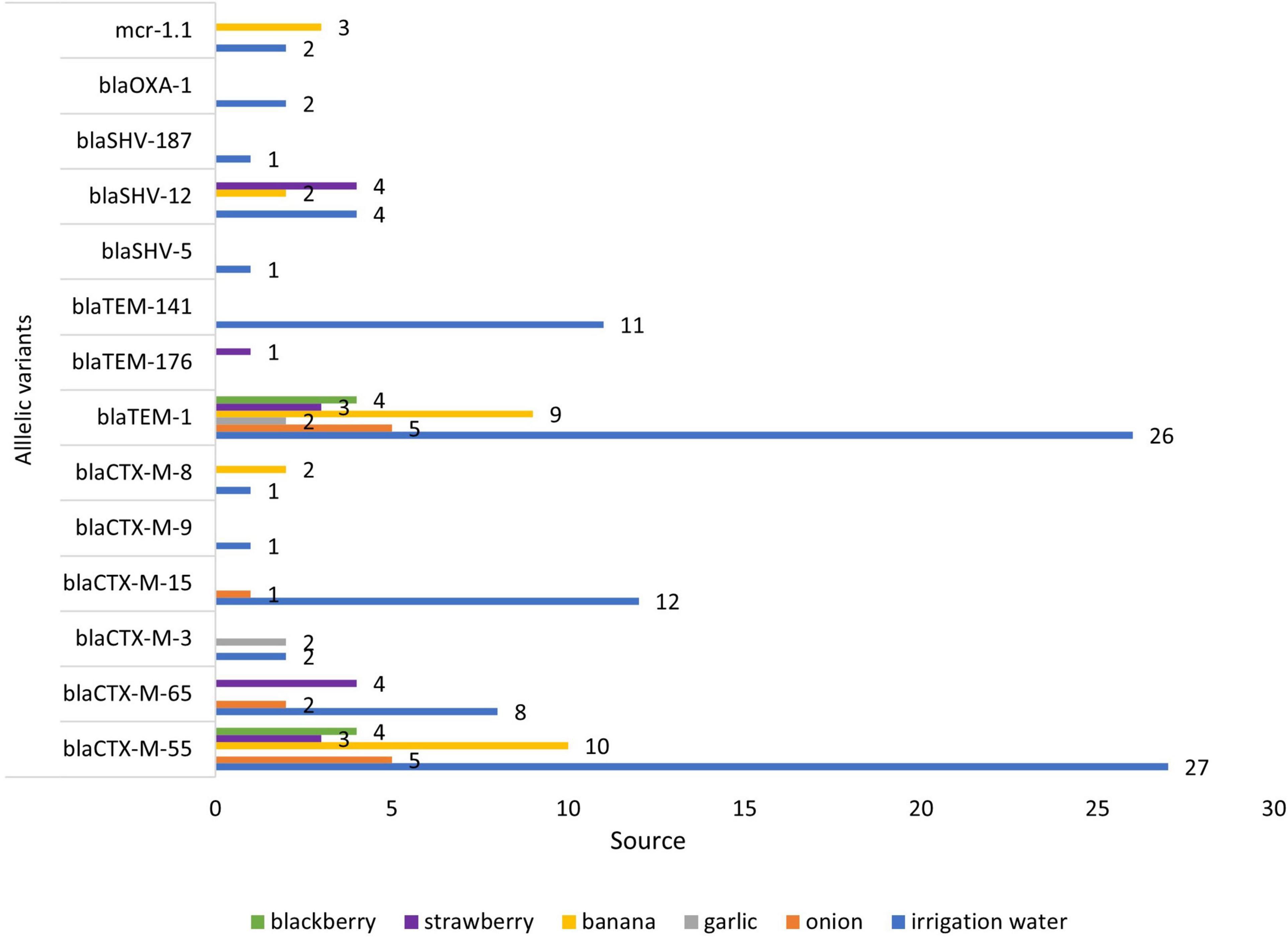
Figure 3. Phylogenetic tree of ESBL-E.coli sequences from irrigation water, fruits, and vegetables. Maximum likelihood phylogenetic tree of the core genomes of 80 ESBL-E.coli isolates from irrigation water, fruits, and vegetables. The labels show the isolate ID assigned according to the sample ID, the origin of the isolate is shown by source colors (irrigation water: blue, onion: brown, banana: yellow, blackberry: purple, strawberry: red, and garlic: green). The background colors highlighted on the branches of the tree indicate the seven identified phylogroups. Numbers represent bootstrap values using 1,000 pseudo-replicates.
We found some discrepancies in some ESBL- E.coli isolates that were positive by PCR for some genes but negative by whole genome sequencing (WGS): 12 isolates for blaTEM gene, 9 isolates for blaSHV genes and blaCTX–M in one gene. Additionally, 2 isolates showed blaSHV and blaTEM using WGS, but were negative by PCR. The WGS analysis of ESBL-E. coli allowed us to identify 2 isolates of E. coli from irrigation water and 3 isolates from banana with the presence of the mcr-1 gene that confers resistance to colistin.
Discussion
In this study, we found that irrigation water, fruit, and vegetables were contaminated with ESBL-E. coli and the highest percentage was found in irrigation water (58%), which confirms the important and emerging role that irrigation water, contaminated with wastewater, has in the spread of ARB and ESBL E. coli and ESBL genes. (Gekenidis et al., 2018a; Vital et al., 2018). The major ESBL gene was the CTX–M (94 of 96 isolates) followed by blaSHV 28% (27 of 96), and blaOXA 1% (1of 96). The prevalence of blaCTX–M type ESBL genes in irrigation water E. coli was 57%, followed by 15% in banana isolates. Additionally the most abundant allelic variants of blaCTX–M found in vegetables, fruits and irrigation water (blaCTX–M55, blaCTX–M65, and blaCTX–M15) (Table 4) are the same alleles found in children and domestic animals in Ecuador (Salinas et al., 2021), in rivers that cross cities (Ortega-Paredes et al., 2020a), and in bacteria from human infections in Ecuador (Cartelle Gestal et al., 2016; Soria Segarra et al., 2018). The presence of the same blaCTX–M alleles in isolates from different sources provides strong evidence that these sources (irrigation water, domestic animals, and humans) are connected. The allelic variants of blaCTX–M from isolates obtained from same European country, but from different (unconnected) sources, animal species or time periods, have been shown to be different (Day et al., 2019; Ludden et al., 2019).
Our genomic analysis showed that most strains obtained from irrigation water and produce were genetically different with 3 exceptions (HY1.4.3 and V427.2; HP6.1 and V661.1; HP1.4 and V662.1), however the number of SNPs between thes strains ranged from 9,332 to 20,310 suggesting that these strains have been evolving apart for many years (Table 4). As expected, some isolates from the same vegetable or fruit showed higher level of genetic closeness, for instance: V698.3 and V698.4 had 12 SNP; V663.4 and V663.5, 6 SNPs; V696.2 and V696.4, 13 SNPs; V1147.5 and V1147.1, 2 SNPs). Interstingly, 2 isolates obtaind from the same irrigation channel 1 month appart (HY3.5.2 and HY5.2.1) had 24 SNPs, suggesting that this strain was higly adapted to water. We did not find additional asociation of ESBL-E.coli clusters with provinces, which may indicate that different E. coli lineages have been widely distributed in the Ecuadorian territory (Figure 2).
These findings may indicated that E. coli populations in the environment are highly diverse (Day et al., 2019; Ludden et al., 2019) and blaCTX–M-genes are probably diseminating in the environmet mostly by mobile genetic elements and not so much by bacterial clones. The plasmids carrying blaCTX–M-genes disseminate efficiently by conjugation, even between bacteria belonging to different genera (Cantón et al., 2012). Transposable elements (such as ISEcp1) are also very active in blaCTX–M-gene mobilization among different plasmids (Cantón et al., 2012). The activity of these MGEs conceals the source of origin of these antimicrobial resistance genes.
The majority of strains isolated from irrigation water and vegetables belonged to phylogroups A and B1 which are considered more generalists, found in most warm-blooded animals and environmental samples (Touchon et al., 2020). We found that some genetically close E.coli isolates, obtained from the same vegetable, had 1 or 2 additional antimicrobial resistance genes which may be a reflection of the dynamic process of antimicrobial resistance gene-turnover in the environment (Barrera et al., 2019).
The blaCTX–M type of ESBL gene is of increasing concern globally (Bevan et al., 2017), and is the predominant ESBL gene in both community and hospital-acquired infections (Manyahi et al., 2017; Fils et al., 2021). A troubling feature of blaCTX–M-bearing plasmids is their ability to capture additional resistance determinants, including carbapenemase genes (Partridge et al., 2012; Potron et al., 2013). Further analysis is necessary to understand whether the plasmids carrying blaCTX–M genes, in bacteria from irrigation water and produce, are the same as those circulating in bacterial isolates from human isolates.
In our study fruits, such as bananas, we hypothesize that their contamination was due to post-harvest processes in which the food is often washed in contaminated water and reused to wash several batches of the product. Although it is true, the skin of the product protects the fruit, the transmission of resistant bacteria can occur through contact and inadequate consumer hygiene (Harris et al., 2003; Hong et al., 2013; Kawamura et al., 2017; Murray et al., 2017; Hölzel et al., 2018).
We also found a higher prevalence of ARB in vegetables in farms than in retail markets in Ecuador (Ortega-Paredes et al., 2018). However, other reports from the Philippines, Lebanon, and Portugal have documented even higher levels (Faour-Klingbeil et al., 2016; Araújo et al., 2017; Vital et al., 2018). In most of the studies, the collection of produce samples has been carried out in groceries and wholesale markets, which makes it difficult to analyze sources of contamination (Bhutani et al., 2015; Yang et al., 2019; Colosi et al., 2020; Richter et al., 2020; Song et al., 2020). In this study, we collected produce and water from farms and their respective irrigation systems, which allowed us to study contamination at the source (i.e., not due to handling, transport, distribution, and processing). We found that MDR isolates were more prevalent in irrigation water isolates compared to fresh produce. Similar results were observed in the Philippines, where 58% of the E. coli isolates from irrigation water were MDR (Paraoan et al., 2017). The resistance to these antibiotics was also observed in E. coli isolates from irrigation water in other studies (Pignato et al., 2009; Ben Said et al., 2015; Vital et al., 2018).
Our study had some limitations; the number produce and fruit samples obtained in each location may not be representative of produce from other agricultural settings in Ecuador. Additionally, long-read sequencing of plasmids could not be carried out due to budgetary limitations.
We found evidence that fresh produce constitutes an important source of ESBL-E. coli and represents a route for the dissemination of resistance genes through the consumption of raw products (Rasheed et al., 2014; Hölzel et al., 2018; Al-Kharousi et al., 2019). We hypothesize that the main source of ABR contamination is irrigation water used for the cultivation of produce, which has been suggested by others as well (Pignato et al., 2009; Gekenidis et al., 2018b). In Ecuador, the lack of sewage treatment may lead to contamination of the food supply with ARB, mainly belonging to the Enterobacteriaceae family (Caicedo-Camposano et al., 2019; Ortega-Paredes et al., 2020a). Antibiotic resistant E. coli can transfer antibiotic resistance determinants not only to other strains of E. coli, but also to other species of potentially pathogenic bacteria within the gastrointestinal tract (Grasselli et al., 2008; Huddleston, 2014).
Conclusion
We found a high prevalence of ESBL-E. coli on produce and in irrigation water; blaCTX–M was the main ESBL gene in these isolates. Allelic variants of the blaCTX–M gene found in irrigation channels and vegetables were the same as those observed in commensal E. coli from domestic animals, and commensal and pathogenic E. coli from humans, suggesting connection between these different sources. This paradigm poses the potential risk of further spreading ARB that are resistant to last-line antibiotics such as carbapenems, which are used exclusively in serious infections in hospitals (Sheu et al., 2019). In this case, resistance goes full circle, from humans to vegetables and fruits (potentially meat and dairy), and back to human populations (Murray et al., 2021). Greater investments are needed to support the development and installation of wastewater treatment systems throughout Ecuador, as well as in other low- and middle-income countries.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
JI and LM: isolation of the Escherichia coli strains. LM: writing—original draft. JG, PC, and GT: review and editing. GT and LM: study design. All authors contributed to the article and approved the submitted version.
Funding
This work was partly supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI135118. The rest of the funding was provided by USFQ-COCIBA grants.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the Agrocalidad team of inspectors for their help with the collection of the samples, as well as the Microbiology Laboratory team. We would also like to thank Belen Prado for her valuable help in the bioinformatic analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.709418/full#supplementary-material
Footnotes
References
Al-Kharousi, Z. S., Guizani, N., Al-Sadi, A. M., and Al-Bulushi, I. M. (2019). Antibiotic resistance of Enterobacteriaceae isolated from fresh fruits and vegetables and characterization of their AmpC b-lactamases. J. Food Protect. 82, 1857–1863. doi: 10.4315/0362-028X.JFP-19-089
Araújo, S., Silva, A. T. I, Tacão, M., Patinha, C., Alves, A., and Henriques, I. (2017). Characterization of antibiotic resistant and pathogenic Escherichia coli in irrigation water and vegetables in household farms. Int. J. Food Microbiol. 257, 192–200. doi: 10.1016/j.ijfoodmicro.2017.06.020
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barrera, S., Cardenas, P., Graham, J. P., and Trueba, G. (2019). Changes in dominant Escherichia coli and antimicrobial resistance after 24 hr in fecal matter. Microbiologyopen 8:e00643. doi: 10.1002/mbo3.643
Ben Said, L., Jouini, A., Klibi, N., Dziri, R., Alonso, C. A., Boudabous, A., et al. (2015). Detection of extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in vegetables, soil and water of the farm environment in Tunisia. Int. J. Food Microbiol. 203, 86–92. doi: 10.1016/j.ijfoodmicro.2015.02.023
Beuchat, L. R. (1996). Pathogenic microorganisms associated with fresh produce. J. Food Protect. 59, 204–216. doi: 10.4315/0362-028X-59.2.204
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Bhutani, N., Muraleedharan, C., Talreja, D., Rana, S. W., Walia, S., Kumar, A., et al. (2015). Occurrence of multidrug resistant extended spectrum beta-lactamase-producing bacteria on iceberg lettuce retailed for human consumption. BioMed Res. Int. 2015:547547. doi: 10.1155/2015/547547
Bonardi, S., and Pitino, R. (2019). Carbapenemase-producing bacteria in food-producing animals, wildlife and environment: a challenge for human health. Ital. J. Food Saf. 8:7956. doi: 10.4081/ijfs.2019.7956
Botelho, L. A. B., Kraychete, G. B., ESilva, J. L. C., Regis, D. V. V., Picão, R. C., Moreira, B. M., et al. (2015). Widespread distribution of CTX-M and plasmid-mediated AmpC β-lactamases in Escherichia coli from Brazilian chicken meat. Mem. Inst. Oswaldo Cruz 110, 249–254. doi: 10.1590/0074-02760140389
Boyd, D. A., Tyler, S., Christianson, S., McGeer, A., Muller, M. P., Willey, B. M., et al. (2004). Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 48, 3758–3764. doi: 10.1128/AAC.48.10.3758-3764.2004
Bush, K., and Jacoby, G. A. (2010). Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 54, 969–976. doi: 10.1128/AAC.01009-09
Caicedo-Camposano, O., Balmaseda-Espinosa, C., Tandazo-Garcés, J., Layana-Bajaña, E., and Sánchez-Vásquez, V. (2019). Water quality for irrigation of San Pablo River, Babahoyo Municipality, Ecuador. Rev. Cienc. Técnicas Agropecuarias 28:3.
Cantón, R., González-Alba, J. M., and Galán, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Carattoli, A., Zankari, E., Garciá-Fernández, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In Silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Cartelle Gestal, M., Zurita, J., Paz y Mino, A., Ortega-Paredes, D., and Alcocer, I. (2016). Characterization of a small outbreak of Salmonella enterica serovar Infantis that harbour CTX-M-65 in Ecuador. Braz. J. Infect. Dis. 20, 406–407. doi: 10.1016/j.bjid.2016.03.007
CLSI (2019). Performance Standards for Antimicrobial Susceptibility Testing. Pennisylvania: Clinical and Laboratory standards Institute.
Colosi, I. A., Baciu, A. M., Opriş, R. V., Peca, L., Gudat, T., Simon, L. M., et al. (2020). Prevalence of ESBL, AmpC and Carbapenemase-Producing Enterobacterales Isolated from Raw Vegetables Retailed in Romania. Foods 9:1726. doi: 10.3390/foods9121726
Cooper, D., Doucet, L., and Pratt, M. (2007). Outbreaks and factors influencing microbiological contamination of fresh produce. J. Organ. Behav. 28, 303–325. doi: 10.1002/j
Dashti, A. A., Jadaon, M. M., Abdulsamad, A. M., and Dashti, H. M. (2009). Heat treatment of bacteria: a simple method of DNA extraction for molecular techniques. Kuwait Med. J. 41, 117–122.
Day, M. J., Hopkins, K. L., Wareham, D. W., Toleman, M. A., Elviss, N., Randall, L., et al. (2019). Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. Lancet Infect. Dis. 19, 1325–1335. doi: 10.1016/S1473-3099(19)30273-7
Edberg, S. C., Rice, E. W., Karlin, R. J., and Allen, M. J. (2000). Escherichia coli: the best biological drinking water indicator for public health protection. J. Appl. Microbiol. Symp. Suppl. 88, 106S–116S. doi: 10.1111/j.1365-2672.2000.tb05338.x
Fang, H., Ataker, F., Hedin, G., and Dornbusch, K. (2008). Molecular epidemiology of extended-spectrum β-lactamases among Escherichia coli isolates collected in a Swedish hospital and its associated health care facilities from 2001 to 2006. J. Clin. Microbiol. 46, 707–712. doi: 10.1128/JCM.01943-07
FAO and WHO (2008). Microbiological Hazards in Fresh Fruits and Vegetables. Geneva: World Health Organization.
Faour-Klingbeil, D., Kuri, V., Fadlallah, S., and Matar, G. M. (2016). Prevalence of antimicrobial-resistant escherichia coli from raw vegetables in Lebanon. J. Infect. Dev. Count. 10, 354–362. doi: 10.3855/jidc.7745
Fils, P. E. L., Cholley, P., Gbaguidi-Haore, H., Hocquet, D., Sauget, M., and Bertrand, X. (2021). ESBL-producing Klebsiella pneumoniae in a University hospital: molecular features, diffusion of epidemic clones and evaluation of cross-transmission. PLoS One 16:e0247875. doi: 10.1371/journal.pone.0247875
Frieri, M., Kumar, K., and Boutin, A. (2017). Antibiotic resistance. J. Infect. Public Health 10, 369–378. doi: 10.1016/j.jiph.2016.08.007
Gekenidis, M. T., Qi, W., Hummerjohann, J., Zbinden, R., Walsh, F., and Drissner, D. (2018a). Antibiotic-resistant indicator bacteria in irrigation water: high prevalence of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli. PLoS One 13:e0207857. doi: 10.1371/journal.pone.0207857
Gekenidis, M. T., Schöner, U., von Ah, U., Schmelcher, M., Walsh, F., and Drissner, D. (2018b). Tracing back multidrug-resistant bacteria in fresh herb production: from chive to source through the irrigation water chain. FEMS Microbiol. Ecol. 94:fiy149. doi: 10.1093/femsec/fiy149
Grasselli, E., François, P., Gutacker, M., Gettler, B., Benagli, C., Convert, M., et al. (2008). Evidence of horizontal gene transfer between human and animal commensal Escherichia coli strains identified by microarray. FEMS Immunol. Med. Microbiol. 53, 351–358. doi: 10.1111/j.1574-695X.2008.00434.x
Guerrero, T., Calderón, D., Zapata, S., and Trueba, G. (2020). Salmonella grows massively and aerobically in chicken faecal matter. Microb. Biotechnol. 13, 1678–1684. doi: 10.1111/1751-7915.13624
Hansen, S., Andersen, L., Mette, D., Holm, A., Roer, L., Antoniadis, P., et al. (2021). Using core genome multilocus sequence typing (cgMLST) for vancomycin-resistant Enterococcus faecium isolates to guide infection control interventions and end an outbreak. J. Global Antimicrob. Resist. 24, 418–423. doi: 10.1016/j.jgar.2021.02.007
Harris, L. J., Farber, J. N., Beuchat, L. R., Parish, M. E., Suslow, T. V., Garrett, E. H., et al. (2003). Outbreaks associated with fresh produce: incidence, growth, and survival of pathogens in fresh and fresh-cut produce. Compr. Rev. Food Sci. Food Saf. 2, 78–141. doi: 10.1111/j.1541-4337.2003.tb00031.x
Hasegawa, H., Suzuki, E., and Maeda, S. (2018). Horizontal plasmid transfer by transformation in Escherichia coli: environmental factors and possible mechanisms. Front. Microbiol. 9:2365. doi: 10.3389/fmicb.2018.02365
Hölzel, C. S., Tetens, J. L., and Schwaiger, K. (2018). Unraveling the role of vegetables in spreading antimicrobial-resistant bacteria: a need for quantitative risk assessment. Foodborne Pathog. Dis. 15, 671–688. doi: 10.1089/fpd.2018.2501
Hong, P. Y., Al-Jassim, N., Ansari, M. I., and Mackie, R. I. (2013). Environmental and public health implications of water reuse: antibiotics, antibiotic resistant bacteria, and antibiotic resistance genes. Antibiotics 2, 367–399. doi: 10.3390/antibiotics2030367
Huddleston, J. R. (2014). Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect. Drug Resist. 7, 167–176. doi: 10.2147/IDR.S48820
Iwu, C. D., and Okoh, A. I. (2019). Preharvest transmission routes of fresh produce associated bacterial pathogens with outbreak potentials: a review. Int. J. Environ. Res. Public Health 16:4407. doi: 10.3390/ijerph16224407
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Kawamura, K., Nagano, N., Suzuki, M., Wachino, J., Kimura, K., and Arakawa, Y. (2017). ESBL-producing Escherichia coli and Its Rapid Rise among Healthy People. Food Saf. 5, 122–150. doi: 10.14252/foodsafetyfscj.2017011
Kilonzo-Nthenge, A., Liu, S., Hashem, F., Millner, P., and Githua, S. (2018). Prevalence of Enterobacteriaceae on fresh produce and food safety practices in small-acreage farms in Tennessee, USA. J. Verbrauch. Lebensm. 13, 279–287. doi: 10.1007/s00003-018-1172-y
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., and Stamatakis, A. (2019). RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. doi: 10.1093/bioinformatics/btz305
Lange, B., Strathmann, M., and Oßmer, R. (2013). Performance validation of chromogenic coliform agar for the enumeration of Escherichia coli and coliform bacteria. Lett. Appl. Microbiol. 57, 547–553. doi: 10.1111/lam.12147
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Leff, J. W., and Fierer, N. (2013). Bacterial Communities Associated with the Surfaces of Fresh Fruits and Vegetables. PLoS One 8:e59310. doi: 10.1371/journal.pone.0059310
Letunic, I., and Bork, P. (2019). Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, 256–259. doi: 10.1093/nar/gkz239
Li, J., Bi, Z., Ma, S., Chen, B., Cai, C., He, J., et al. (2019). Inter-host transmission of carbapenemase-producing Escherichia coli among humans and backyard animals. Environ. Health Perspect. 127:107009. doi: 10.1289/EHP5251
Lin, Y., Dong, X., Wu, J., Rao, D., Zhang, L., Faraj, Y., et al. (2020). Metadata analysis of mcr-1-bearing plasmids inspired by the sequencing evidence for horizontal transfer of antibiotic resistance genes between polluted river and wild birds. Front. Microbiol. 11:352. doi: 10.3389/fmicb.2020.00352
Ludden, C., Raven, K. E., Jamrozy, D., Gouliouris, T., Blane, B., Coll, F., et al. (2019). One health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. mBio 10, e02693–18. doi: 10.1128/mBio.02693-18
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Manyahi, J., Moyo, S. J., Tellevik, M. G., Ndugulile, F., Urassa, W., Blomberg, B., et al. (2017). Detection of CTX-M-15 beta-lactamases in Enterobacteriaceae causing hospital- and community-acquired urinary tract infections as early as 2004, in Dar es Salaam, Tanzania. BMC Infect. Dis. 17:282. doi: 10.1186/s12879-017-2395-8
Marlène, M., Lee, B., Miller, S., Reyna, Z., Lu, P., Yan, J., et al. (2020). Horizontal Gene Transfer of Antibiotic Resistance from Acinetobacter baylyi to Escherichia coli on Lettuce and Subsequent Antibiotic Resistance Transmission to the Gut Microbiome. mSphere 5, e00329–20. doi: 10.1128/msphere.00329-20
McDaniel, C., and Jadeja, R. (2019). A Review of Fresh Produce Outbreaks, Current Interventions, Food Safety Concerns and Potential Benefits of Novel Antimicrobial Sodium Acid Sulfate. MOJ Food Process. Technol. 7, 59–67. doi: 10.15406/mojfpt.2019.07.00221
Moore, J. E., Moore, P. J. A., Millar, B. C., Goldsmith, C. E., Loughrey, A., Rooney, P. J., et al. (2010). The presence of antibiotic resistant bacteria along the River Lagan. Agric. Water Manag. 98, 217–221. doi: 10.1016/j.agwat.2010.09.001
Monstein, H. J., Östholm-Balkhed, Å., Nilsson, M. V., Nilsson, M., Dornbusch, K., and Nilsson, L. E. (2007). Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 115, 1400–1408. doi: 10.1111/j.1600-0463.2007.00722.x
Motlagh, A. M., and Yang, Z. (2019). Detection and occurrence of indicator organisms and pathogens. Water Environ. Res. 91, 1402–1408. doi: 10.1002/wer.1238
Murray, K., Wu, F., Shi, J., Jun Xue, S., and Warriner, K. (2017). Challenges in the microbiological food safety of fresh produce: limitations of post-harvest washing and the need for alternative interventions. Food Qual. Saf. 1, 289–301. doi: 10.1093/fqsafe/fyx027
Murray, M., Salvatierra, G., Dávila-Barclay, A., Ayzanoa, B., Castillo-Vilcahuaman, C., Huang, M., et al. (2021). Market Chickens as a Source of Antibiotic-Resistant Escherichia coli in a Peri-Urban Community in Lima, Peru. Front. Microbiol. 12:635871. doi: 10.3389/fmicb.2021.635871
Njage, P. M. K., and Buys, E. M. (2015). Pathogenic and commensal Escherichiacoli from irrigation water show potential in transmission of extended spectrum and AmpC β-lactamases determinants to isolates from lettuce. Microb. Biotechnol. 8, 462–473. doi: 10.1111/1751-7915.12234
Ortega-Paredes, D., Barba, P., Mena-López, S., Espinel, N., and Zurita, J. (2018). Escherichia coli hyperepidemic clone ST410-A harboring blaCTX-M-15 isolated from fresh vegetables in a municipal market in Quito-Ecuador. Int. J. Food Microbiol. 280, 41–45. doi: 10.1016/j.ijfoodmicro.2018.04.037
Ortega-Paredes, D., Barba, P., Mena-López, S., Espinel, N., Crespo, V., and Zurita, J. (2020a). High quantities of multidrug-resistant Escherichia coli are present in the Machángara urban river in Quito, Ecuador. J. Water Health 18, 67–76. doi: 10.2166/wh.2019.195
Ortega-Paredes, D., de Janon, S., Villavicencio, F., Ruales, K. J., De La Torre, K., Villacís, J. E., et al. (2020b). Broiler Farms and Carcasses Are an Important Reservoir of Multi-Drug Resistant Escherichia coli in Ecuador. Front. Vet. Sci. 7:547843. doi: 10.3389/fvets.2020.547843
Ouellette, M., Bissonnette, L., and Roy, P. H. (1987). Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 beta-lactamase gene. Proc. Natl. Acad. Sci. U.S.A. 84, 7378–7382. doi: 10.1073/pnas.84.21.7378
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Paraoan, C. E. M., Rivera, W. L., and Vital, P. G. (2017). Detection of Class I and II integrons for the assessment of antibiotic and multidrug resistance among Escherichia coli isolates from agricultural irrigation waters in Bulacan, Philippines. J. Environ. Sci. Health B Pest. Food Contamin. Agric. Wastes 52, 306–313. doi: 10.1080/03601234.2017.1281647
Partridge, S. R., Paulsen, I. T., and Iredell, J. R. (2012). pJIE137 carrying bla CTX-M-62 is closely related to p271A carrying bla NDM-1. Antimicrob. Agents Chemother. 56, 2166–2168. doi: 10.1128/AAC.05796-11
Pesavento, G., Calonico, C., Ducci, B., Magnanini, A., and Lo Nostro, A. (2014). Prevalence and antibiotic resistance of Enterococcus spp. isolated from retail cheese, ready-to-eat salads, ham, and raw meat. Food Microbiol. 41, 1–7. doi: 10.1016/j.fm.2014.01.008
Pigłowski, M. (2019). Pathogenic and non-pathogenic microorganisms in the rapid alert system for food and feed. Int. J. Environ. Res. Public Health 16:477. doi: 10.3390/ijerph16030477
Pignato, S., Coniglio, M. A., Faro, G., Weill, F. X., and Giammanco, G. (2009). Plasmid-mediated multiple antibiotic resistance of Escherichia coli in crude and treated wastewater used in agriculture. J. Water Health 7, 251–258. doi: 10.2166/wh.2009.019
Potron, A., Nordmann, P., Rondinaud, E., Jaureguy, F., and Poirel, L. (2013). A mosaic transposon encoding OXA-48 and CTX-M-15: towards pan-resistance. J. Antimicrob. Chemother. 68, 476–477. doi: 10.1093/jac/dks397
Rasheed, M. U., Thajuddin, N., Ahamed, P., Teklemariam, Z., and Jamil, K. (2014). Antimicrobial drug resistance in strains of Escherichia coli isolated from food sources. Rev. Inst. Med. Trop. Sao Paulo 56, 341–346. doi: 10.1590/s0036-46652014000400012
Richter, L., du Plessis, E. M., Duvenage, S., and Korsten, L. (2020). Occurrence, Phenotypic and molecular characterization of extended-spectrum- and AmpC- β-Lactamase producing Enterobacteriaceae isolated from selected commercial spinach supply chains in South Africa. Front. Microbiol. 11:638. doi: 10.3389/fmicb.2020.00638
Rochelle-Newall, E., Nguyen, T. M. H., Le, T. P. Q., Sengtaheuanghoung, O., and Ribolzi, O. (2015). A short review of fecal indicator bacteria in tropical aquatic ecosystems: knowledge gaps and future directions. Front. Microbiol. 6:308. doi: 10.3389/fmicb.2015.00308
Rossolini, G. M., D’Andrea, M. M., and Mugnaioli, C. (2008). The spread of CTX-M-type extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14, 33–41. doi: 10.1111/j.1469-0691.2007.01867.x
Salinas, L., Loayza, F., Cárdenas, P., Saraiva, C., Johnson, T. J., Amato, H., et al. (2021). Environmental spread of extended spectrum beta-Lactamase (ESBL) producing Escherichia coli and ESBL genes among children and domesticaimals in Ecuador. Environ. Health Perspect. 129:7007. doi: 10.1289/EHP7729
Sato, T., Qadir, M., Yamamoto, S., Endo, T., and Zahoor, A. (2013). Global, regional, and country level need for data on wastewater generation, treatment, and use. Agric. Water Manag. 130, 1–13. doi: 10.1016/j.agwat.2013.08.007
Sheu, C. C., Chang, Y. T., Lin, S. Y., Chen, Y. H., and Hsueh, P. R. (2019). Infections caused by carbapenem-resistant Enterobacteriaceae: an update on therapeutic options. Front. Microbiol. 10:80. doi: 10.3389/fmicb.2019.00080
Shrivastava, S. R., Shrivastava, P. S., and Ramasamy, J. (2018). World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 32, 76–77. doi: 10.4103/jms.jms_25_17
Song, J., Oh, S. S., Kim, J., and Shin, J. (2020). Extended-spectrum β-lactamase-producing Escherichia coli isolated from raw vegetables in South Korea. Sci. Rep. 10:19721. doi: 10.1038/s41598-020-76890-w
Soria Segarra, C., Soria Baquero, E., and Cartelle Gestal, M. (2018). High prevalence of CTX-M-1-Like enzymes in urinary isolates of Escherichia coli in Guayaquil, ecuador. Microb. Drug Resist. 24, 393–402. doi: 10.1089/mdr.2017.0325
Touchon, M., Perrin, A., de Sousa, J. A. M., Vangchhia, B., Burn, S., O’Brien, C. L., et al. (2020). Phylogenetic background and habitat drive the genetic diversification of Escherichia coli. PLoS Genet. 16:e1008866. doi: 10.1371/journal.pgen.1008866
UNESCO (2012). Managing Water Under Uncertainty and Risk, The United Nations World Water Development Report 4. Paris: United Nations Educational, Scientific and Cultural Organization.
Vasco, G., Spindel, T., Carrera, S., Grigg, A., and Trueba, G. (2015). The role of aerobic respiration in the life cycle of Escherichia coli: public health implications. Av. Cienc. Ingenierías 7, B7–B9.
Vital, P. G., Zara, E. S., Paraoan, C. E. M., Dimasupil, M. A. Z., Abello, J. J. M., Santos, I. T. G., et al. (2018). Antibiotic resistance and extended-spectrum beta-lactamase production of Escherichia coli isolated from irrigation waters in selected urban farms in Metro Manila, Philippines. Water (Switzerland) 10:548. doi: 10.3390/w10050548
Waters, N. R., Abram, F., Brennan, F., Holmes, A., and Pritchard, L. (2020). Easy phylotyping of Escherichia coli via the EzClermont web app and command-line tool. Access Microbiol. 2:acmi000143. doi: 10.1099/acmi.0.000143
Yang, F., Shen, C., Zheng, X., Liu, Y., El-Sayed Ahmed, M. A. E., Zhao, Z., et al. (2019). Plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli and Klebsiella pneumoniae isolated from market retail fruits in Guangzhou, China. Infect. Drug Resist. 12, 385–389. doi: 10.2147/IDR.S194635
Yassin, A. K., Gong, J., Kelly, P., Lu, G., Guardabassi, L., Wei, L., et al. (2017). Antimicrobial resistance in clinical Escherichia coli isolates from poultry and livestock, China. PLoS One 12:e0185326. doi: 10.1371/journal.pone.0185326
Keywords: fresh produce, irrigation water, ESBL E. coli, CTX-M, Extended-spectrum beta-lactamase (ESBL)
Citation: Montero L, Irazabal J, Cardenas P, Graham JP and Trueba G (2021) Extended-Spectrum Beta-Lactamase Producing-Escherichia coli Isolated From Irrigation Waters and Produce in Ecuador. Front. Microbiol. 12:709418. doi: 10.3389/fmicb.2021.709418
Received: 13 May 2021; Accepted: 13 September 2021;
Published: 04 October 2021.
Edited by:
Lorena Rodriguez-Rubio, University of Barcelona, SpainReviewed by:
Tongjie Chai, Shandong Agricultural University, ChinaCharlotte Valat, Unité Antibiorésistance et Virulence Bactériennes, ANSES Site de Lyon, France
Copyright © 2021 Montero, Irazabal, Cardenas, Graham and Trueba. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriel Trueba, Z3RydWViYUB1c2ZxLmVkdS5lYw==
 Lorena Montero
Lorena Montero Jorge Irazabal2
Jorge Irazabal2 Jay P. Graham
Jay P. Graham Gabriel Trueba
Gabriel Trueba