- 1Institute for Network Biology, Helmholtz Zentrum München – German Research Center for Environmental Health (GmbH), Neuherberg, Germany
- 2Research Unit Analytical Biogeochemistry, Helmholtz Zentrum München – German Research Center for Environmental Health (GmbH), Neuherberg, Germany
Stress tolerant, plant-associated bacteria can play an important role in maintaining a functional plant microbiome and protecting plants against various (a)biotic stresses. Members of the stress tolerant genus Rhodococcus are frequently found in the plant microbiome. Rhodococcus qingshengii RL1 was isolated from Eruca sativa and the complete genome was sequenced, annotated and analyzed using different bioinformatic tools. A special focus was laid on functional analyses of stress tolerance and interactions with plants. The genome annotation of RL1 indicated that it contains a repertoire of genes which could enable it to survive under different abiotic stress conditions for e.g., elevated mercury concentrations, to interact with plants via root colonization, to produce phytohormones and siderophores, to fix nitrogen and to interact with bacterial signaling via a LuxR-solo and quorum quenching. Based on the identified genes, functional analyses were performed in vitro with RL1 under different growth conditions. The R. qingshengii type strain djl6 and a closely related Rhodococcus erythropolis BG43 were included in the experiments to find common and distinct traits between the strains. Genome based phylogenetic analysis of 15 available and complete R. erythropolis and R. qingshengii genome sequences revealed a separation of the R. erythropolis clade in two subgroups. First one harbors only R. erythropolis strains including the R. erythropolis type strain. The second group consisted of the R. qingshengii type strain and a mix of R. qingshengii and R. erythropolis strains indicating that some strains of the second group should be considered for taxonomic re-assignment. However, BG43 was clearly identified as R. erythropolis and RL1 clearly as R. qingshengii and the strains had most tested traits in common, indicating a close functional overlap of traits between the two species.
Introduction
Anthropogenic activities over the past decades, including pollution with heavy metals, pesticides and chemical fertilizer, as well as improper soil exploitation coupled with climate change have led to immense global soil degradation (Smith et al., 2016). This has resulted in loss of soil biodiversity, increase in pathogens and has created harsh biotic and abiotic conditions for plants and their associated microbes (Foley et al., 2005; Brevik and Burgess, 2014; Tsiafouli et al., 2015; Banerjee et al., 2019; Wagg et al., 2019). To survive difficult environmental conditions and maintain a functional plant holobiont (stress resistant) beneficial bacteria are important. Over the last decades, several mechanisms have been identified which are involved in beneficial associations between plant and microbes. They either involve plant growth promotion based on production of plant hormones and providing enhanced nutrients to the plants (Glick, 2012) or plant protection against plant pathogens by producing antimicrobial compounds (Chowdhury et al., 2015a,b) or acting indirectly by inducing host systemic resistance (Kloepper and Beauchamp, 1992; Pieterse et al., 2014; Bahramisharif and Rose, 2019). Apart from that, beneficial bacteria have been shown to support plants as stress protecting agents under abiotic stress conditions like salt and drought stress (Alavi et al., 2013; Berg et al., 2013; Kaushal and Wani, 2016; Bhat et al., 2020) or by supporting plant growth in contaminated soils (Sessitsch et al., 2013; Ma et al., 2016a,b). However, not only plant-microbe interactions, but also microbe-microbe interactions influence the functionality of the plant holobiont. For example, it has been shown that members of the genus Variovorax play a major role in shaping the microbiome and with this influence root growth in Arabidopsis by balancing auxin production (Finkel et al., 2020). This report emphasizes the importance to understand the role of all relevant members in the plant holobiont. The multifaceted interactions of plants and several plant-associated bacteria have been widely studied to understand the underlying molecular mechanisms and to exploit plant beneficial traits for sustainable agriculture (Berg et al., 2017; Rodriguez et al., 2019; Babin et al., 2021; Windisch et al., 2021). In this context it is important to further the knowledge of the functional repertoire also of yet lesser known members of the plant microbiome such as Rhodococcus to understand what enables them to interact with the plant as well as other microbes and which traits could be useful for an application in specifically demanding agricultural scenarios.
Members of the genus Rhodococcus are resistant to various stresses (Dabrock et al., 1994; Weyens et al., 2013; Pátek et al., 2021) and are able to degrade and metabolize a large spectrum of toxic compounds (Kamble et al., 2013; Lincoln et al., 2015; Pham et al., 2015; Ceniceros et al., 2017; Gupta et al., 2019; Gorbunova et al., 2020). These traits make the genus Rhodococcus interesting for bioremediation applications (Leigh et al., 2006; Płociniczak et al., 2017). Moreover, in metagenomic and microbiome analyses the genus Rhodococcus has been frequently reported as an established member of the plant microbiome (Francis and Vereecke, 2019; Vereecke et al., 2020). Many plant associated Rhodococci also show plant beneficial traits in vitro (Trivedi et al., 2007; Abbamondi et al., 2016; Murugappan et al., 2017) and in planta (Belimov et al., 2001).
The here studied closely plant associated Rhodococcus qingshengii RL1 was isolated from surface sterilized Rucola (Eruca sativa L.) leaves and the genome was recently sequenced (Kuhl et al., 2019). The type strain of species R. qingshengii djl-6 was isolated from a carbendazim polluted soil (Xu et al., 2007). Other R. qingshengii isolates have been shown to possess nitrogen fixing capacity improving growth of chick pea plants (Joshi et al., 2019) and to produce high amounts of IAA in vitro (Hasuty et al., 2018). In combination with the genetic traits for bioremediation, stress resistance, and biocontrol on plants widely distributed across the whole Rhodococcus genus these reports clearly warrant a characterization of our new isolate R. qingshengii RL1.
We were able to assess the genomic potential of RL1 by characterizing beneficial traits in the genome (Levy et al., 2018) and found that RL1 is well equipped with genes essential for survival under different abiotic stress conditions and for microbial interactions with other microbes and plants. In vitro assays were used to assess if these genomic potential was actually transferred into functional traits. To our knowledge, this is the first report of a R. qingshengii, which survives under mercury stress and degrades quorum quenching signals (AHLs). Additionally, for the first time we could identify a potential gibberellin producing operon in an actinobacterial genus as well as indications for existence of alternative nitrogen fixation pathways. For establishment of the taxonomic position of RL1 we performed a whole genome based phylogenetic analysis of 15 available and complete R. erythropolis and R. qingshengii genome sequences and included the R. qingshengii type strain djl6 and a closely related R. erythropolis BG43 in the in vitro experiments. Thus, this work contributes to the elucidation of molecular mechanisms and underlying genetic determinants of plant–microbe interactions and possible functions in the plant holobiont of R. qingshengii RL1 and closely related strains.
Materials and Methods
Bacterial Strains and Growth Conditions
Rhodococcus qingshengii RL1, named hereafter RL1, is a gram-positive Actinobacterium and was isolated from Rucola (Eruca sativa L.) leaves. Colonies appear in off-white, beige colors. The overnight grown culture corresponds with OD600 = 0.42 representing approximately 4 × 107 CFU (colony forming units). Rhodococcus qingshengii djl6 DSM 45222 (type strain), named hereafter djl6 (Xu et al., 2007) and Rhodococcus erythropolis BG43 DSM 46869, named hereafter BG43 (Müller et al., 2014) were obtained from German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig). The overnight grown cultures correspond with OD600 = 0.5 representing approximately 5 × 107 CFU and OD600 = 0.92 representing approximately 5 × 108 CFU, respectively. Rhodococcus strains were cultivated in tryptic soy broth (TSB, Sigma, United States) [casein peptone (pancreatic), 17 g/L, dipotassium hydrogen phosphate, 2.5 g/L, glucose, 2.5 g/L, sodium chloride, 5 g/L, soya peptone (papain digest.), 3 g/L] or solid tryptic soy agar (1.7% agar) with pH 7.3, unless further specified, at 28°C and 180 rpm.
Control strains for the conducted experiments were: the strain Bacillus velezensis FZB42 DSM 23117, producing fungal antagonistic compounds like surfactin, fengycin and iturin (Chowdhury et al., 2015a), the AHL biosensor strain Agrobacterium tumefaciens A136 ATCC 51350 (Stickler et al., 1998; Han et al., 2016), the AHL producer strain Acidovorax radicis N35 DSM 23535 (Li et al., 2011) the non-AHL-producing mutant strain Acidovorax radicis N35 AHL- araI::tet (Han et al., 2016), the AHL-degrading mutant strain Rhizobium radiobacter F4 AHL- expressing an AHL lactonase (AiiA) (Alabid et al., 2020) and able to grow on potassium tellurite trihydrate (K2TeO3∗3H2O) 100 μg/ml, the phosphate solubilizing strain Luteibacter sp. Cha2324a_16 and the ACC utilizing strain Variovorax sp. M92526_27 isolated from wheat roots (this study), the nitrogen-fixing and IAA producing Herbaspirillum frisingense GSF30 DSM 1328 (Kirchhof et al., 2001), the nitrogen-fixing Azospirillum brasilense Sp7 DSM 1690 (Tarrand et al., 1978; Hartmann and Hurek, 1988), the biofilm-producing Pseudomonas simiae WCS417r (Pieterse et al., 2020) and the non-biofilm-producing Escherichia coli DH5α (Anton and Raleigh, 2016). Strains were cultivated in liquid nutrient broth (NB, Roth, Germany) (beef extract 3 g/l, gelatin peptone 5 g/l) or solid nutrient agar (with 1.7% agar) with pH 6.8, unless further specified, at 28°C at 180 rpm.
Genome Comparison
The genome of Rhodococcus qingshengii RL1 (Kuhl et al., 2019) was compared to the genomes of the type strain Rhodococcus qingshengii djl6 (Xu et al., 2007; Wang et al., 2010; Táncsics et al., 2014), as well as the closely related soil isolate Rhodococcus erythropolis BG43 (Rückert et al., 2015). The genome djl6 is based on the species R. jialingiae (Wang et al., 2010) which was later identified as a synonym of the type strain R. qingshengii (Táncsics et al., 2014). For the genome comparison and the identification of orthologous and unique genes in the three different genomes the efficient database framework for comparative Genome Analyses using BLAST score Ratios – EDGAR (Blom et al., 2016) was used.
Functional Annotation of RL1 Genome
The RL1 genome was annotated upon submission to NCBI with the NCBI prokaryotic Genome Annotation Pipeline (PGAP) with the annotation method best-placed reference protein set with GeneMarkS-2+ and the Rapid Annotation using Subsystem Technology 2.0 (RAST) with default parameter of the classicRAST annotation scheme plus frameshift fixing and backfilling of gaps allowed, where the annotated genome was browsed afterward in the SEED environment (Aziz et al., 2008; Overbeek et al., 2014). Functional annotation by grouping genes in clusters of orthologous groups (COG) of proteins according to Tatusov et al. (2000) was performed with eggNOG v5.0 (Jensen et al., 2008). Genes were annotated with the KEGG (Kyoto Encyclopedia of Genes and Genomes) orthology (KO) identifiers, or the K numbers, and directly linked to the KEGG pathways with the KEGG automatic annotation server (KAAS) (Moriya et al., 2007) and KEGG Mapper. Further functional annotation was performed by identifying plant microbe interaction factors and gene clusters for biosynthesis of secondary metabolites with Plant–bacteria Interaction Factors Resource (PIFAR) (Martínez-García et al., 2016) and the antibiotics and secondary metabolite analysis shell – antiSMASH (Blin et al., 2017) using the default parameters.
Phylogenetic Analysis
Sixty-one complete genomes of the genus Rhodococcus and the genome of the out-group Streptomyces albus NBRC 13014 (type strain), were used for the full-genome approximately maximum-likelihood phylogenetic tree build in EDGAR. 15 genomes identified in the phylogenetic tree as members of the Rhodococcus erythropolis clade and two out-group genomes were used for the approximately maximum-likelihood phylogenetic tree calculated in EDGAR using FastTree Software with the Shimodaira–Hasegawa test for bootstrap values. Average nucleotide identity (ANI) and Average amino acid identity (AAI) was calculated in EDGAR (Blom et al., 2016) as described in Konstantinidis and Tiedje (2005) and Goris et al. (2007).
Evaluation of Growth and Tolerance to Different Stress Factors
If not indicated otherwise all Rhodococcus strains were pre-cultured in liquid TSB overnight.
Mercury Tolerance
Overnight grown cultures were transferred to fresh TSB medium with increasing mercury levels 0.001, 0.01, 0.1, and 1 mM adjusted with mercury-II-chloride (HgCl2, Roth, Germany) according to Dziewit et al. (2013) and incubated at 28°C. Growth rates were evaluated by spectrophotometric measurement of optical density at 600 nm (OD600) after 24 and 48 h. For treatments without detectable growth (=0.1 mM and 1 mM mercury), the recovery of cells was evaluated by the ability to form colonies on TSB agar plates without mercury. Hundred microliter of cultures from the treatments with 0.1 and 1 mM mercury were plated on TSB without mercury and incubated at 28°C for 24 and 48 h. Experiment was repeated three times. Non-mercury-tolerant strains Herbaspirillum frisingense GSF30 and Bacillus velezensis FZB42 served as negative controls.
Salt Stress Tolerance
Overnight grown cultures were transferred to fresh TSB medium with increasing sodium chloride (NaCl, Merck, Germany) levels 0, 1, 2.5, 3.5, 5.5, 7.5, 12, and 15% according to De Carvalho et al. (2014) and incubated at 28°C. Growth rates were evaluated by spectrophotometric measurement of optical density at 600 nm (OD600) after 24 and 48 h. For treatments without detectable growth (12 and 15% NaCl), the recovery of cells was evaluated by the ability to form colonies on TSB agar plates without NaCl. Hundred microliter of cultures from the treatments 12 and 15% were plated on TSB agar without NaCl and incubated at 28°C for 24 and 48 h. Experiment was repeated three times. Less salt tolerant Herbaspirillum frisingense GSF30 and Bacillus velezensis FZB42 served as negative controls.
pH Tolerance
Overnight grown cultures were transferred to fresh TSB medium with pH values 8, 7, 6, 5, 4, 3, 2 adjusted with hydrochloric acid (HCl, Merck, Germany) or sodium hydroxide (NaOH, Sigma, United States) and incubated at 28°C. Growth rates were evaluated by spectrophotometric measurement optical density at 600 nm (OD600) after 24 and 48 h. Recovery was evaluated by the ability to form colonies on TSB agar plates at pH 7.3 after 48 h in treatments without detectable growth (pH 4, 3 and 2). Hundred microliter of medium from the treatments pH 4, 3, and 2 were plated on TSB and incubated at 28°C for 24 and 48 h. The experiment was repeated three times. Herbaspirillum frisingense GSF30 and Bacillus velezensis FZB42 which did not grow in low pH (below 5) served as negative controls.
Osmotic Stress Tolerance
Overnight grown cultures were transferred to fresh TSB medium with increasing osmotic stress levels 0, –0.25, –0.5, –0.75, –1, –1.25, and –1.5 MPa and incubated at 28°C. Increasing osmotic stress was adjusted with polyethylene glycol 6000 (PEG6000, Serva Electrophoresis GmbH, Heidelberg, Germany) based on decreasing water potential with the formula of Kaufmann and Michel (1973), according to Kumar et al. (2014) and Jayakumar et al. (2020). –1.5 MPa is the water potential plants in regular soil start to wilt irreversibly. Growth rates were evaluated by spectrophotometric measurement optical density at 600 nm (OD600) after 24 and 48 h. Experiment was repeated three times. Gram-negative Herbaspirillum frisingense GSF30 and gram-positive Bacillus velezensis FZB42 served as controls.
Antibiotic Resistance
Overnight grown cultures were diluted 1:10 with fresh TSB medium. Two hundred microliter of the diluted overnight cultures were spread on TSB agar plates and antimicrobial susceptibility test stripes (Himedia Laboratories, India) for kanamycin (0.016–256 μg/ml), ampicillin (0.016–256 μg/ml), rifampicin (0.002–32 μg/ml), and vancomycin (0.016–256 μg/ml) were placed according to manufacturer’s protocol. The inhibition zone was evaluated after 24 and 48 h.
Bacterial strains were streaked on a fresh TSB or NB plate from glycerol stocks and grown overnight. A single colony of RL1 was picked and streaked on nutrient broth (NB) agar plates with 100 μg/ml potassium tellurite trihydrate (K2TeO3 ∗ 3H2O, Sigma, United States) for 48 h. Dark gray colony growth was evaluated as positive growth. The strain Rhizobium radiobacter F4 AHL- aiiA- genetically modified to tolerate a tellurite concentration of 100 μg/ml served as positive control.
Characterization of Traits Involved in Microbe–Plant Interactions
If not indicated otherwise all Rhodococcus strains were pre-cultured in liquid TSB overnight.
Indole-Acetic Acid Production
Indole-acetic acid (IAA) production was determined by the colorimetric method of Gordon and Weber (1951). Overnight grown cultures were transferred to fresh TSB medium with and without the IAA precursor 5 mM tryptophan (1 mg/mL, Sigma) and grown for 48 h. Liquid cultures were centrifuged for 2 min at 5000 × g. Hundred microliter of supernatant were mixed with 100 μl of Salkowski reagent [0.01 M FeCl3 anhydrous (Fluka Biochemika, Germany) in perchloric acid (HClO4) 35% (Merck, Germany)] (Loper and Schroth, 1986) and 1 μl of orthophosphoric acid (Sigma, United States). After incubation in the dark for 30 min amounts of IAA in the supernatant were analyzed in a plate reader (Spectra Max iD3, Molecular Devices) at 530 nm wavelength. A standard curve was prepared from commercial indole-3-acetic acid (Fluka Biochemika, Germany) in TSB with concentrations ranging from 0 to 100 μg/ml and Herbaspirillum frisingense GSF30 was used as positive control. Supernatant measurements were performed in triplicates. Evaluation was based on the amount of produced IAA normalized to an OD600 = 1.
Siderophore Production
Siderophore production was analyzed according to Pérez-Miranda et al. (2007) and Lynne et al. (2011) with modifications. Twenty-five microliter of overnight grown cultures were spotted on TSB agar plate and grown for 48 h. Dye solutions [chrome azurol blue S (Sigma, United States), FeCl3 (Fluka Biochemika, Germany), HDTMA (Hexadecyltrimethylammonium bromide, Sigma, United States)] were prepared and mixed according to Lynne et al. (2011). Piperazin-N,N’-bis-(2-ethanesulfonic acid) (Pipes, Roth, Germany) was added to H2O with 0.9 % agar and pH was adjusted to 6.8. After autoclaving separately, the dye solution was slowly mixed with the Pipes-Agar mix. Cooled but still liquid overlay agar (10 ml) was poured on plates with bacteria. After 2 h siderophore production was analyzed by detection of color change from blue to orange. The experiment was repeated three times.
Phosphate Solubilization
Overnight grown cultures were washed twice in 1x PBS and 25 μl were spotted on National Botanical Research Institute’s phosphate growth medium (NBRIP) according to Nautiyal (1999) and incubated at 28°C. After 6 days, phosphate solubilization activity was determined according to the formation of a clear halo surrounding the spotted colony using the Phosphate Solubilization Index (SI): (Colony diameter + Halo zone diameter)/colony diameter). The phosphate-solubilizing Luteibacter sp. Cha2324a_16 served as positive control. The experiment was repeated three times.
1-Aminocyclopropane-1-Carboxylate Utilization
1-Aminocyclopropane-1-carboxylate (ACC, Biozol Diagnostica GmbH, Germany) utilization as nitrogen source was analyzed with M9 minimal medium [Na2HPO4 33.1 mM, KH2PO4 22 mM, NaCl 8.55 mM (NH4Cl 9.35 mM), glucose 0.4%, MgSO4 1 mM, CaCl2 0.3 mM] containing NH4Cl 9.35 mM (Roth, Germany) or ACC 3 mM as nitrogen source or no nitrogen source. Overnight grown cultures were washed twice in 1x PBS and 25 μl were spotted on each plate. After 10 days, ACC utilization as nitrogen source was analyzed by comparing bacterial growth on M9, M9 with ACC and nitrogen-free M9 plates. ACC utilizing Variovorax sp. M92526_27 served as positive control. The experiment was repeated three times.
Nitrogen Fixation
Nitrogen fixation was analyzed with nitrogen-free semi-solid Nfb-medium according to Döbereiner (1995), on Ashby’s mannitol medium (Mannitol 20 g/l, K2HPO4 0.2 g/l, MgSO4∗5H2O 0.2 g/l, NaCl 0.2 g/l, K2SO4 0.1 g/l, CaCO3 5 g/l, Agar 15 g/l) and on Jensen’s medium (Sucrose 20 g/l, K2HPO4 1 g/l, MgSO4∗5H2O 0.5 g/l, NaCl 0.5 g/l, FeSO4 0.1 g/l, Na2MoO4 0.005 g/l, CaCO3 2 g/l, Agar 15 g/l). Overnight grown cultures were washed twice in 1x PBS and 10 μl were spotted on nitrogen-free semi-solid Nfb-medium and incubated at 28°C. Pellicle formation was evaluated after 48 h. Ten microliter of washed overnight cultures were streaked on Ashby’s mannitol agar. Bacterial strains were streaked on a fresh TSB or NB plate from glycerol stocks and grown overnight. A single colony of each strain was picked and streaked on Jensen’s agar. Bacteria on Ashby’s medium and Jensen’s medium were incubated at 28°C and growth was evaluated after 3 days. The experiments were repeated three times. Nitrogen-fixing Azospirillum brasilense Sp7 served as positive control.
Biofilm Formation
Biofilm formation was analyzed according to O’Toole (2011). Overnight grown cultures were washed in 1xPBS and OD600 was adjusted to 0.1. Bacterial strains were cultivated in a microtiter plate in 100 μl modified M9 minimal medium with 0.5% casamino acids (Biozol Diagnostica Vertrieb GmbH, Germany) without shaking at 28°C. After incubation OD600 was measured in the plate reader (SpectraMax iD3, Molecular Devices). After 24 h OD600 was measured and unattached cells were dumped out of the plate. The plate was washed twice by submerging it in MilliQ water to further remove unattached cells. Hundred and twenty-five microliter of 0.1% crystal violet (Roth, Germany) solution was added to each well. After 15 min the plate was rinsed three times in MilliQ water and dried for 1.5 h before visual inspection of biofilm production. For quantification of the biofilm 125 μl of 30% acetic acid (Roth, Germany) was added to each well and incubated for 15 min at room temperature. The solution was transferred to a new microtiter plate and color intensity was quantified at the plate reader (SpectraMax iD3, Molecular Devices) with absorbance at 550 nm and 30% acetic acid as blank. Biofilm-forming Pseudomonas simiae WCS417 served as positive control and non-biofilm-producing Escherichia coli DH5α served as negative control. The experiment was repeated three times with 6–12 replicates per strain.
Interactions With Other Organisms
Confrontation Assay Against Plant–Pathogenic Fungi
The interaction of RL1 with the well-known plant pathogenic fungi Rhizoctonia solani, Fusarium culmorum, and Fusarium oxysporum was investigated with an in vitro confrontation assay. The following pathogenic fungi were used: Rhizoctonia solani, causing potato stem cancer and black scurf (Yang and Li, 2012), wheat pathogenic fungus Fusarium culmorum G2191 causing seedling blight, foot rot, and head blight (Wagacha and Muthomi, 2007) and the wilt-causing Fusarium oxysporum DSM62297 (Gerlach et al., 1958). Fungi were cultivated on potato dextrose agar (PDA, Sigma, United States) (potato extract 4.0 g/L, glucose 20.0 g/L) at room temperature in the dark and stored at 4°C until further use.
RL1 was pre-grown in TSB. Overnight grown culture was diluted to OD600 = 0.1 with fresh TSB medium and 10 μl were dripped on the plate. Approximately 1 mm3 PDA pieces grown with fungi were aseptically transferred to TSB plates at a distance of approximately 3 cm. After 9 days of growth the zone of inhibition formation was visually analyzed and documented photographically. Sterile water served as negative control and Bacillus velezensis FZB42, a known fungal antagonistic strain served as positive control. Confrontation assays were performed in triplicates.
Degradation of Synthetic and Bacterial N-Acyl-Homoserine Lactones (AHLs)
The identified qsdA gene sequence of the RL1 genome encoding the AHL lactonase was used to construct a phylogenetic tree with nearest relatives with MEGA X (Kumar et al., 2018). Rhodococcus strains were analyzed with a well diffusion agar-plate assay (Rodríguez et al., 2020) and a V-shaped assay (Berendsen et al., 2018) with modifications.
Well diffusion plate assay
For the experiments with synthetic AHL Rhodococcus strains were pre-grown in TSB. Overnight grown cultures were transferred to fresh TSB liquid medium supplemented with 10 μM C12-HSL (Biomol GmbH, Germany) and incubated at 28°C 180 rpm. Cell-free TSB medium supplemented with 10 μM C12-HSL served as control. For the co-cultivation experiment RL1 and Acidovorax radicis N35e overnight cultures were adjusted to OD600 = 0.2 and co-cultured in fresh liquid NB medium. Pure culture of Acidovorax radicis N35e served as control.
The well diffusion plates were prepared as follows: The AHL biosensor strain Agrobacterium tumefaciens A136 was pre-grown in NB. NB plates were overlaid with soft NB agar (0.5% agar) supplemented with the biosensor strain A136 and 80 μg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal, Life Technologies GmbH, Germany). Twenty microliter of each supernatant from the co-cultivation or synthetic AHL experiment were filled in wells prepared in the soft agar and incubated at 28°C for 30 h. Remaining AHLs were detected by color change. Pure NB was used as negative control for presence of AHLs.
V-shaped plate assay
The AHL biosensor strain Agrobacterium tumefaciens A136 was pre-grown in NB. Agrobacterium tumefaciens A136 and 80 μg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal, Life Technologies GmbH, Germany) were spread on NB plates. Rhodococcus strains were pre-grown in TSB. The AHL producing strains Acidovorax radicis N35e was pre-grown in NB, non-AHL-producing mutant strain Acidovorax radicis N35 AHL- araI::tet was pre-grown in NB with tetracyclin 20 μg/ml and kanamycin 50 μg/ml. Overnight grown cultures were washed in 1x PBS and optical density was adjusted to OD600 = 0.1. Eight times 1 μl of each culture was dripped in a diagonal row on the prepared NB plates in V-shape with increasingly closer inoculation sites. Plates were incubated for 30 h at 28°C. AHL degradation was detected by color change.
Evaluation of Rhizosphere Competence
Root Inoculation in Axenic System
Rucola (Eruca sativa L.) seeds were washed in Tween 80 1% (Sigma, United States) for 2 min, surface sterilized with sodium hypochlorite 12% (NaOCl, Roth, Germany) for 8 min and washed three times in sterile deionized water for 2 min. Sterilized seeds were placed on Hoagland’s solution (Sigma, United States) with 0.8% agar to germinate 4 days. Rhodococcus strains were pre-grown in TSB. Overnight grown cultures were washed two times in 1x PBS (AppliChem, Germany) and diluted to a concentration of 107 CFUs. Seedlings were inoculated in the prepared bacterial solution of RL1, BG43, and djl6 for 1 h under shaking at 160 rpm at 28°C. Seedlings inoculated in 1x PBS served as negative control. Inoculated seedlings were transferred to an axenic system with 80 ml sterile quartz sand and 20 ml Hoagland’s solution in a sterile Phytatray II (Sigma, United States). Seedlings inoculated with RL1 were additionally transferred on plates with 0.5x Murashige and Skoog Medium (0.5x MS) including vitamins (Duchefa Biochemie, Netherlands); pH was adjusted to 5.7 with 2N KOH. No additional sucrose was added to 0.5x MS. The axenic system was placed in a Phytochamber (Weiss Technik, Modell SGC120PG2, Germany) with 23°C, 55% humidity, day-night-cycle 12 h : 12 h. After seven and 14 days freshly harvested roots were washed in 1x PBS, fixed in 55% EtOH and 1x PBS mix and stored at –20°C until further use.
Fluorescence in situ Hybridization (FISH)
Fluorescence in situ hybridization was performed following the protocol of Alquéres et al. (2013). Chemicals were obtained from AppliChem, Germany. After an increasing ethanol series [(50, 80, and 96% [vol/vol] for 3 min each] for fixation and desiccation, roots were incubated in 50 μl hybridization buffer [0.9 M NaCl, 0.01% sodium dodecyl sulfate (SDS), 10 mM Tris-HCl (pH 8.0), 35% deionized formamide] with 15 pmol of the fluorescently labeled probes EUB338, specific for eubacteria (Amann et al., 1990; Daims et al., 1999) and labeled with fluorescein (FITC, Thermo Scientific, Germany), and HGC69a (Roller et al., 1994), specific for bacteria with high G + C content in their 16S rRNA and labeled with Cy3 (Thermo Scientific, Germany) or ATTO550. Hybridization was performed for 1.5 h at 46°C.
Confocal Laser Scanning Microscopy (CLSM)
FISH stained roots and bacterial cells were investigated at the Zeiss confocal laser scanning microscope LSM880 (Zeiss, Oberkochen, Germany) with argon ion laser and helium neon laser for excitation of FITC (488 nm), Cy3 (561 nm) and an unlabeled control channel (633 nm). Cells were observed with a 64x C-Apochromat water immersion objective. Micrographs were recorded using the software Zen Black Edition (Zeiss, Oberkochen, Germany).
Quantitative Evaluation
Rhizosphere competence of the investigated bacterial strains were estimated via counting of colony forming units (CFU). Sterilized Rucola (Eruca sativa L.) seedlings were inoculated in bacterial solution and planted in the axenic system as described above. After 7 days three roots per treatment were harvested, weighed and ground in a sterilized mortar with 1 ml 1x PBS. Ground roots were diluted three times (10–3), 100 μl of each dilution was plated in triplicates on TSB plates and incubated at 28°C. After 48 h CFUs of dilution 10–3 were counted and mean values were compared between treatments. Plating of dilutions 10–1 and 10–2 resulted in too many CFUs for counting.
Statistical Analysis
Sample size was not predetermined using statistical methods. Statistical analysis was performed with RStudio 3.6.1. Data were tested for normal distribution with Shapiro–Wilk-Test and analyzed with the non-parametric Fligner-Killeen-Test or with analysis of variances (ANOVA) followed by the post-hoc analysis with Tukey’s test. Significance level was 5% marked in the graphs by asterisks.
Results
Phylogenetic Analysis
The full genome based phylogenetic tree of Rhodococcus was constructed on a core genome of 633 genes from 39246 genes in total (Supplementary Figure 1). Based on this phylogenetic tree 15 genomes of the R. erythropolis clade were chosen to calculate the full genome based phylogenetic tree of the R. erythropolis clade. It was built on a core genome of 1211 genes from 20587 genes in total and revealed that the clade can be separated into two groups (Figure 1). The first group includes R. erythropolis strains only. The second group harbors a mix of R. erythropolis and R. qingshengii strains. ANI values between all analyzed R. erythropolis or R. qingshengii genomes were higher than 94% (Supplementary Figure 2). The ANI value within the first group was 98.02–98.8% and within the second group 97.17–99.3%. The outgroups Rhodococcus aethiovorans and Streptomyces albus had ANI values of 72.13–72.57% and 66.79–67.9%, compared to the first group and the second group, respectively. AAI values between the first and the second group of the R. erythropolis clade were all above 98% (Supplementary Figure 3), and between both groups and the outgroups R. aethiovorans and S. albus AAI values were 56.45–57.33% and 76.71–76.81, respectively. R. qingshengii djl6 and RL1 grouped together in the second group. R. erythropolis BG43 was allocated to the first group.
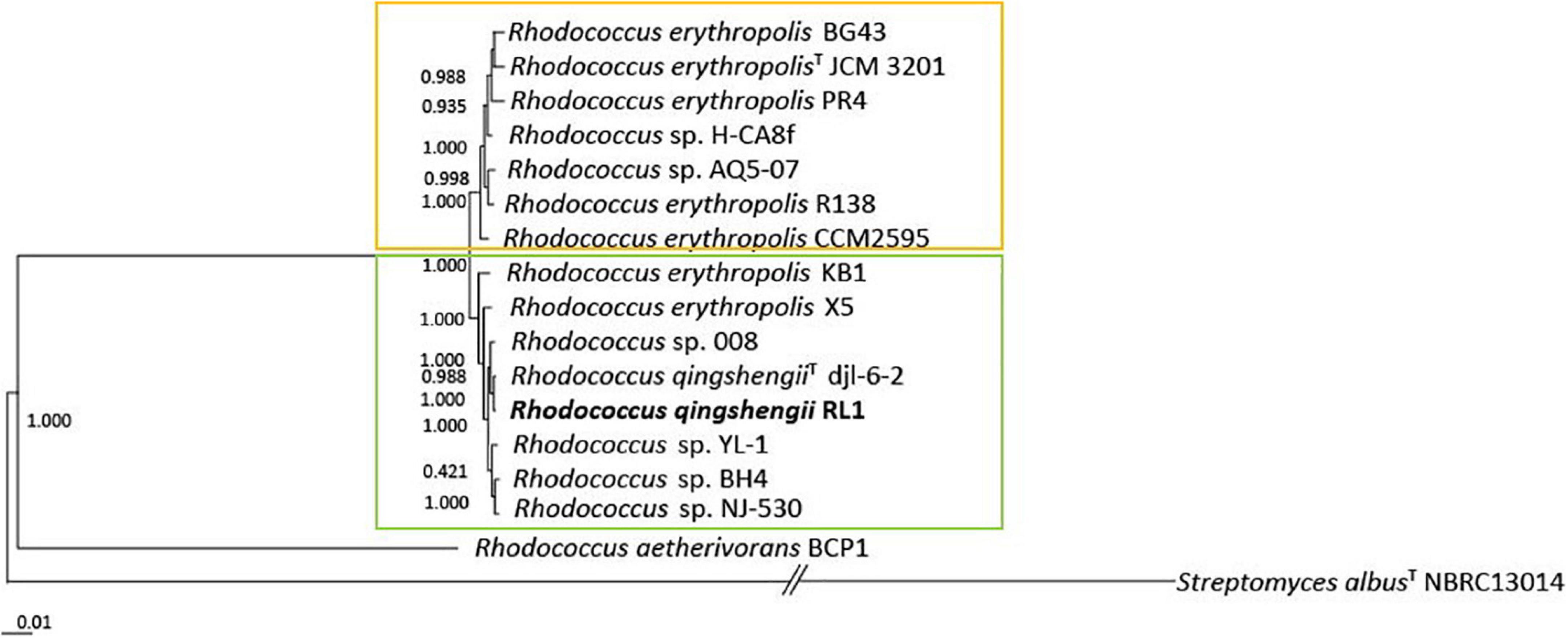
Figure 1. Maximum-likelihood phylogenetic tree of 15 genomes of the Rhodococcus erythropolis clade generated with FastTree 2.1 from 1211 nucleotide sequences of the core genome. Values represent local support values based on Shimodaira–Hasegawa test (1 SH = 100% bootstrap). The scale bar represents nucleotide substitutions per site (0.01 scale = 1% nucleotide substitutions per site). The Rhodococcus erythropolis subgroup is marked in orange and the Rhodococcus qingshengii subgroup in green.
Comparing the RL1 genome with the genomes of R. qingshengii djl6 and R. erythropolis BG43, 5293 genes could be identified that were shared between all three strains (Figure 2). RL1 and djl6 shared more genes (294) than each of them with BG43 (69; 70). For RL1 39 singleton genes could be identified of which 17 were annotated as hypothetical proteins (Table 1).
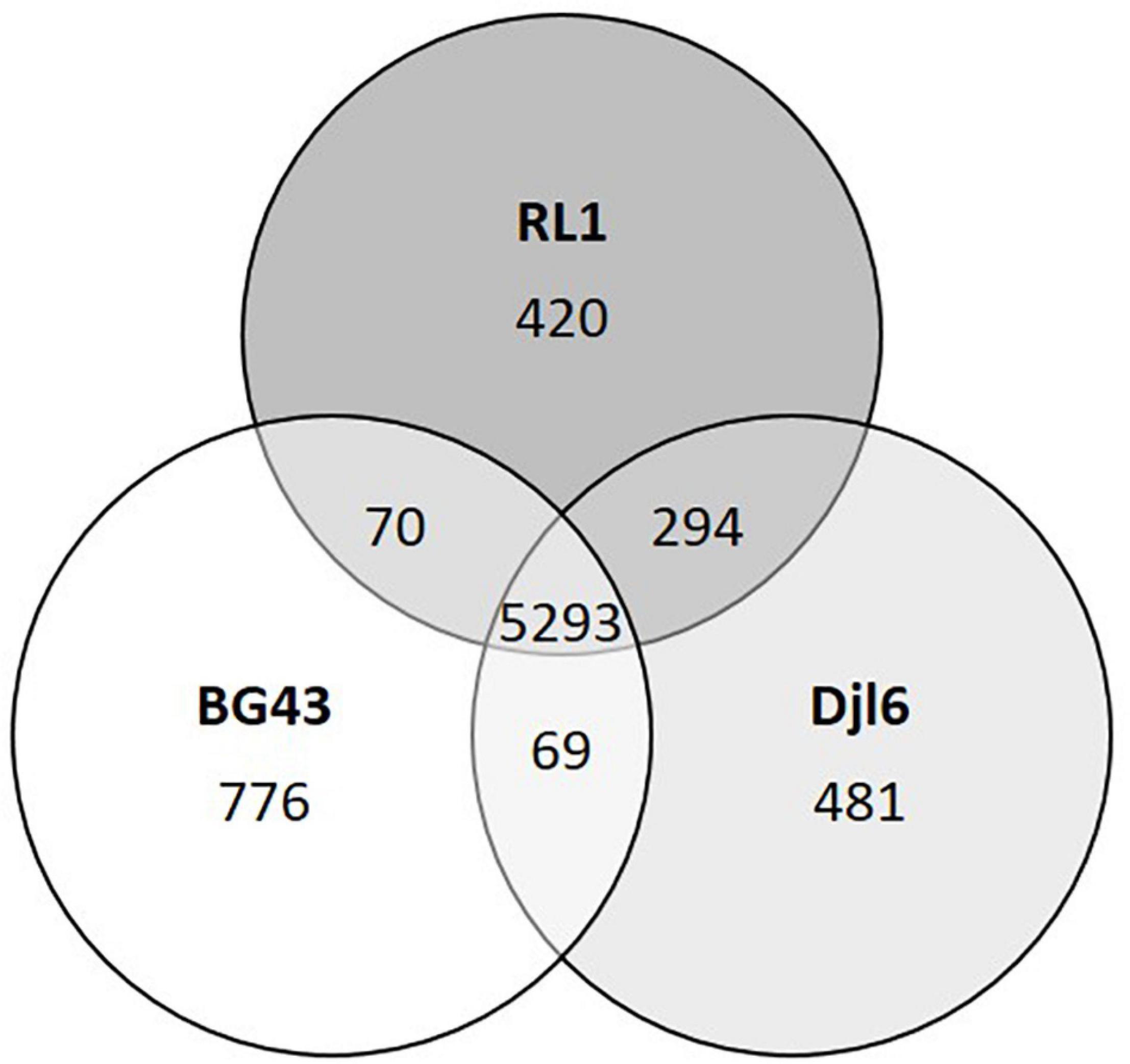
Figure 2. Venn-diagram representing comparative distribution of shared genes between the genomes of RL1, djl6, and BG43. Data obtained with EDGAR Software. Unique genes or singletons are here genes without any hit (BLAST) against any other genome.
Functional Annotation of RL1 Genome
A total of 6,554 protein coding sequences were predicted from the genome of RL1 with (RAST) and 6,328 genes with PGAP (Table 2). 5918 of the predicted genes could be annotated to an assigned function and 92.4% of them were classified into 21 clusters of orthologous groups (COG) identified with eggNOG (Figure 3). Genes involved in metabolism represented the largest fraction (37.1%), followed by information and storage processing (19.2%), and cellular processes and signaling (12.7%) (Figure 3). In more details, the highest number of genes could be assigned to be involved in transcription (K, 11.5%), followed by amino acid transport and metabolism (E, 7.5%) and energy production and conversion (C, 6.9%). 3.6% of the genes could be assigned to the category of secondary metabolites biosynthesis, transport, and catabolism (Q). 9.4% of the genes were assigned to more than one category (>1 cat.). 21.4% of the genes could not be assigned to a known function (S). 35% of the coding sequences in the RL1 genome were sorted in 23 main RAST subsystems and 424 subsystems (subsystem coverage). With KEGG pathway analysis genes involved in 273 pathways were identified (Supplementary Table 1). The genome was further analyzed for presence of genes known to be involved in interactions with plants using the web-based tool PIFAR and 45 genes representing 14 categories could be identified (Supplementary Table 2). Using the tool antiSMASH 17 biosynthetic gene clusters (BGC) with the potential to produce secondary metabolites, such as ectoine, erythrochelin, and heterobactin A/heterobactin S2, could be identified (Supplementary Table 3). In this study we focused on the cluster with highest similarity (>50%) to known secondary metabolite biosynthesis pathways.
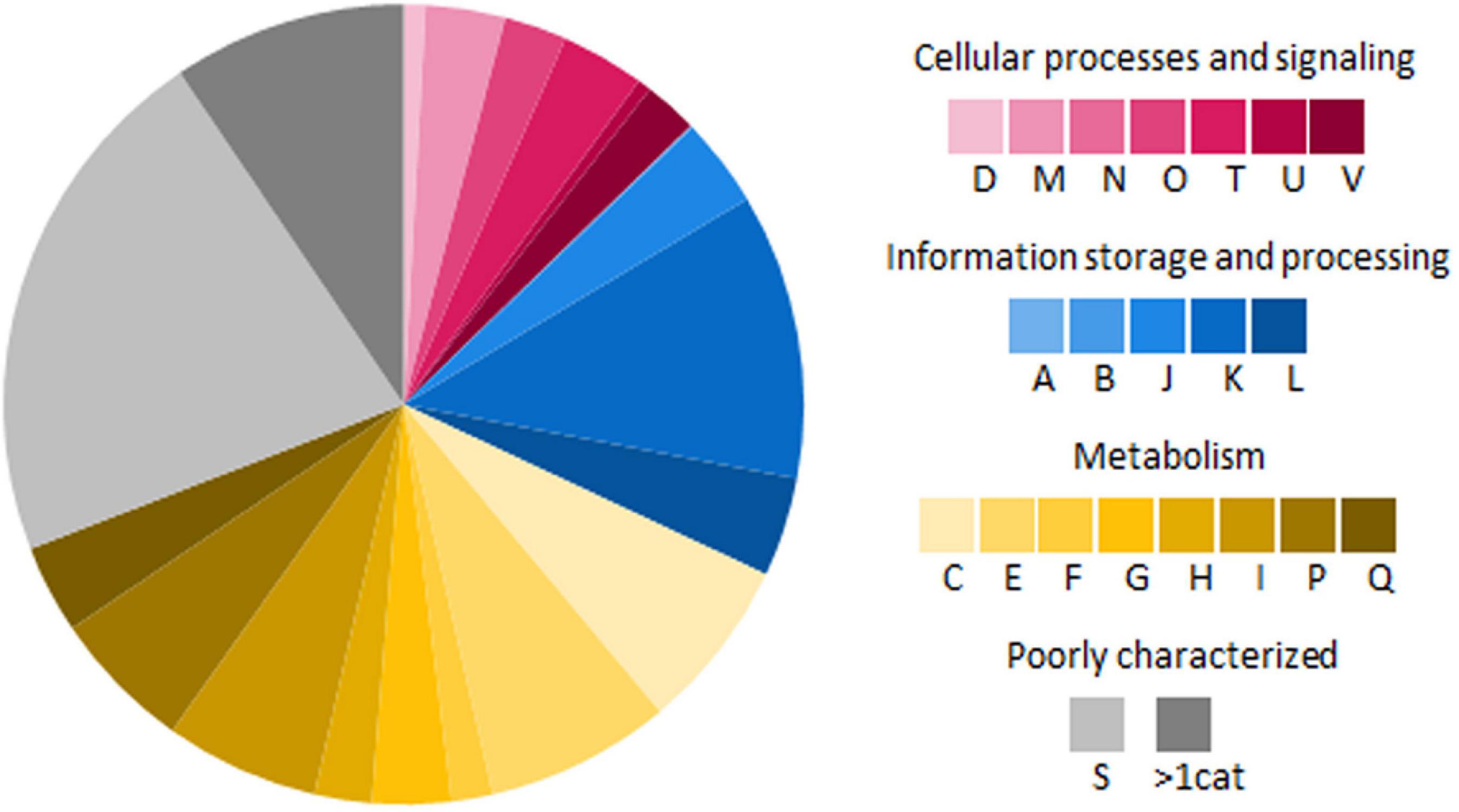
Figure 3. Functional classification of genes encoding proteins in RL1 based on cluster of orthologous groups (COG) (Tatusov et al., 2000). All alphabets represent different COG functional classes: A, RNA processing and modification; B, Chromatin structure and dynamics; C, energy production and conversion; D, cell cycle control, cell division, and chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation, ribosomal structure, and biogenesis; K, transcription; L, replication, recombination, and repair; M, cell wall, cell membrane, and cell envelope biogenesis; N, cell motility; O, posttranslational modification, protein turnover, and chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; S, no functional prediction; T, signal transduction mechanisms; U, intracellular trafficking, secretion, and vesicular transport; and V, defense mechanisms; >1 cat, classified in more than 1 category.
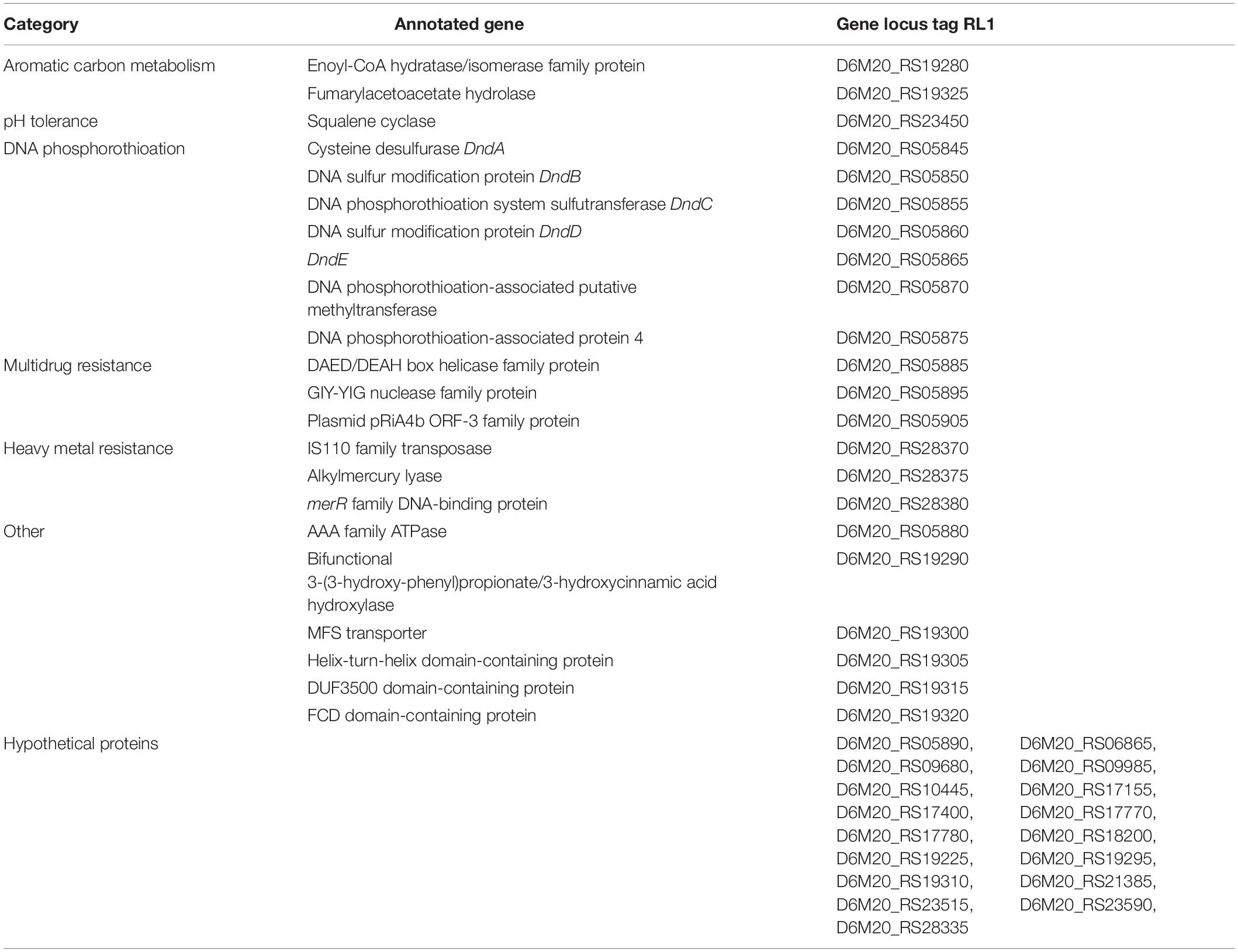
The genome annotation of RL1 revealed several genes which have been previously identified to be involved in stress tolerance under different abiotic stress conditions, bioremediation of toxic compounds, rhizosphere colonization and (beneficial) plant-microbe interactions and were partly verified by manual annotation with blastp alignment (Supplementary Table 4). In more details, the RL1 genome harbors many genes, which can be expressed to withstand osmotic, salt, oxidative and acidic stress and are relevant for heavy metal tolerance (mercury, lead, cadmium, arsenic) and bioremediation of aromatic hydrocarbons (alkB, catA) and fossil fuels (dszB). Moreover, genes potentially involved in multiple drug resistance, DNA repair by phosphorothioation, antibiotic resistance and degradation of CO and hydrogen could be identified, for example the complete carbon monoxide dehydrogenase (CODH) and a [NiFe]-hydrogenase cluster. The RL1 genome annotation indicated that it is equipped with several genes which could enable it to interact with the plant and survive in the plant environment via plant hormone and siderophore production of the siderophores enterobactin, bacillibactin, arthrobactin, and heterobactin as well as nitrogen fixation, iron acquisition, phosphate solubilization, biofilm formation, and stress protection. Additionally, the RL1 genome harbors genes involved in quorum quenching, glucosinolate metabolism, aldoxime, isothiocyanate (ITC) and nitrile degradation, as well as genes important for the production of volatiles, exopolysaccharides (EPS), proteases and microbe-associated molecular patterns (MAMP). Therefore, we analyzed functional traits with focus on stress tolerance and plant–microbe interactions.
Evaluation of Growth and Tolerance to Different Stress Factors
Mercury Tolerance
The RL1 genome harbors genes for alkylmercury lyase and merR family DNA-binding protein (Supplementary Table 4). Active growth determined by optical density was detectable in the medium with 0.001 mM mercury for djl6 and BG43. RL1 was able to grow in the medium with up to 0.01 mM mercury. RL1 and BG43 could recover from up to 1 mM mercury in the medium, whereas djl6 recovered from up to 0.1 mM mercury. The gram-positive control strain Bacillus velezensis FZB42 could grow in the medium with up to 0.01 mM mercury and the gram- negative control strain Herbaspirillum frisingense GSF30 only in medium with 0.001 mM mercury. Both control strains did not recover from medium containing 0.1 mM mercury.
Salt Stress Tolerance
The RL1 genome harbors genes for the complete Na+/H+ antiporter operon (Supplementary Table 4). RL1 and BG43 were able to grow in medium with 7.5% NaCl, whereas djl6 could grow up to 5.5% NaCl in the medium. Although there was no visible growth, all tested Rhodococcus strains were able to recover from salt stress of 15% NaCl in the medium. The gram-positive control strain Bacillus velezensis FZB42 could grow up to 7.5% NaCl in the medium and did not recover from medium with 15% NaCl. The gram-negative control strain Herbaspirillum frisingense GSF30 could grow in medium with up to 3.5% NaCl and could not recover from 7.5% NaCl in the medium.
pH Tolerance
Genes encoding for squalene cyclase and the ADI cluster were identified in the RL1 genome (Supplementary Table 4). The Rhodococcus strains were able to grow up to pH 5 and recovered after 48 h in pH 3 and 4 h in pH 2. Control strains Herbaspirillum frisingense GSF30 and Bacillus velezensis FZB42 were able to grow up to pH 5 and recovered from pH 4.
Osmotic Stress Tolerance
The gene cluster for ectoine biosynthesis was identified in RL1 with 75% identity to the ectoine biosynthetic cluster of Streptomyces anulatus. The Rhodococcus strains RL1, djl6, BG43, and the control strains FZB42 and GSF30 were able to grow under PEG6000 induced osmotic stress of –1.5 MPa, which was the tested maximum.
Antibiotic Resistance
Genes involved in antibiotic resistance and tellurite resistance were identified in the RL1 genome (Supplementary Table 4). RL1 was tolerant to Kanamycin up to the concentration of 96 μg/ml, Ampicillin up to 6 μg/ml, Rifampicin up to 0.025 μg/ml, but not tolerant to Vancomycin. Djl6 was tolerant to Kanamycin up to 12 μg/mL, Ampicillin up to 3 μg/mL, Rifampicin up to 0.047 μg/ml and Vancomycin up to 0.023 μg/ml. BG43 was tolerant to Kanamycin up to 48 μg/ml, Ampicillin up to 2 μg/ml, Rifampicin up to 0.023 μg/ml and Vancomycin up to 0.5 μg/ml. RL1 was able to grow on NB plates containing 100 μg/ml potassium tellurite trihydrate. The other strains were not tested for this trait.
Traits Involved in Microbe–Plant Interactions
Indole-Acetic Acid Production
The genes encoding for amidase amiE and amine oxidase iaaM as well as genes involved in tryptophan metabolism were identified in the RL1 genome (Supplementary Table 4). RL1 produced 16 ± 2.6 μg/ml of IAA which is the highest amount compared to djl6 and BG43 with 10.7 ± 2.4 μg/ml and 10.9 ± 3.8 μg/ml, respectively. The positive control strain Herbaspirillum frisingense GSF30 produced 41 ± 9.8 μg/ml IAA.
Siderophore Production
In the RL1 genome, biosynthesis cluster for erythrochelin was identified with 57% identity and heterobactinA/heterobactin S2 identified with 100% identity compared to the heterobactin BGC of R. erythropolis PR4 (Figure 4A). Genes encoding for relevant proteins of the heterobactin BGC are isochorismate synthase, isochorismatase, 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase, 2,3-dihydroxybenzoate-AMP ligase, amino acid adenylation domain-containing protein and related transporter (Supplementary Table 4). RL1 produced siderophores indicated by the color change of the overlay agar from blue to orange (Figure 4B). BG43 and djl6 did not produce siderophores.

Figure 4. Siderophore biosynthetic gene clusters and in vitro assay. (A) Heterobactin biosynthetic gene cluster based on antiSMASH results of Rhodococcus qingshengii RL1 compared to the reference genome of R. erythropolis PR4 and other Rhodococcus strains. Depicted as arrows are the core biosynthetic genes in dark red, additional biosynthetic genes in light red, transport-related genes in blue and additional genes in gray. The biosynthetic genes (dark and light red) are presented in order of their appearance from right to left encoding for isochorismate synthase, isochorismatase, 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase, 2,3-dihydroxybenzoate-AMP ligase, isochorismatase, and an amino acid adenylation domain-containing protein. Numbers indicate percentage of similarity to PR4. (B) In vitro assay for siderophore production of RL1, BG43, djl6 and Bacillus sp. detected with Chrome Azurol Blue overlay agar. Siderophore production is indicated by color change of the medium from blue to orange.
Phosphate Solubilization
The RL1 genome harbors genes involved in organic acid production (Supplementary Table 4). Rhodococcus qingshengii strains RL1 and djl6 were able to solubilize phosphate indicated by clear halo formation and SI values of 2.3 and 2.4 respectively. BG43 showed no halo formation and SI value was 2, which indicates no phosphate solubilization. Positive control Luteibacter sp. Cha2324a_16 showed halo formation and SI values of 2.7.
1-Aminocyclopropane-1-Carboxylate Utilization
Growth on M9 and M9 with ACC and no growth on nitrogen-free M9 indicate ACC utilization. RL1, BG43, and djl6 could grow on plates with ACC, on regular M9 medium and on nitrogen-free medium. The positive control Variovorax sp. M92526_27 grew on M9 and M9 with ACC, but not on M9 without nitrogen (Figure 5A). ACC deaminase activity remained unclear, because Rhodococcus strains could also grow on M9 without nitrogen. The gene acdS was not present in the RL1 genome.
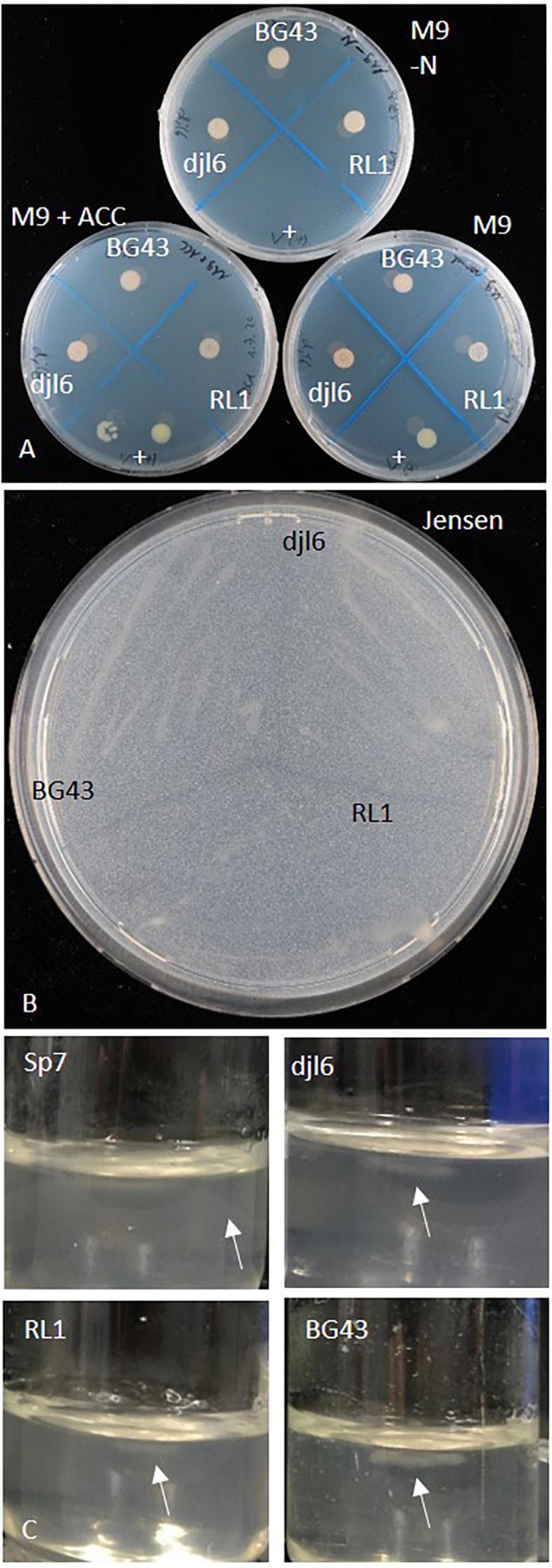
Figure 5. Growth characterization of Rhodococcus strains RL1, djl6, and BG43 on various nitrogen-free media. (A) M9-N, M9 (control) and M9 + ACC, (B) Jensen’s medium, and (C) Nfb-semisolid medium (Döbereiner, 1995) Sp7 = Azospirillum brasilense Sp7 (positive control for nitrogen fixation), + = Variovorax sp. (positive control for ACC deaminase activity). Arrows mark pellicle formation.
Nitrogen Fixation
The RL1 genome harbors an uncharacterized nifU-like protein (Supplementary Table 4). The strains RL1, djl6, and BG43 could grow on all tested nitrogen free media, which were nitrogen-free M9 medium (Figure 5A), Ashby’s medium, Jensen’s medium (Figure 5B) and Nfb-medium (Figure 5C). Pellicle formation in Nfb-medium was smaller compared to positive control Azospirillum brasilense Sp7.
Biofilm Formation
Genes encoding for a phosphoglucomutase and a signal peptidase I were identified in the RL1 genome (Supplementary Table 4). The strains RL1, djl6 and BG43 were able to produce biofilms in varying intensities (Figure 6), but stronger than the negative control Escherichia coli DH5α. Djl6 showed the strongest biofilm formation. Positive control Pseudomonas simiae WCS417 normalized biofilm formation was lower compared to djl6, but stronger compared to RL1 and BG43.
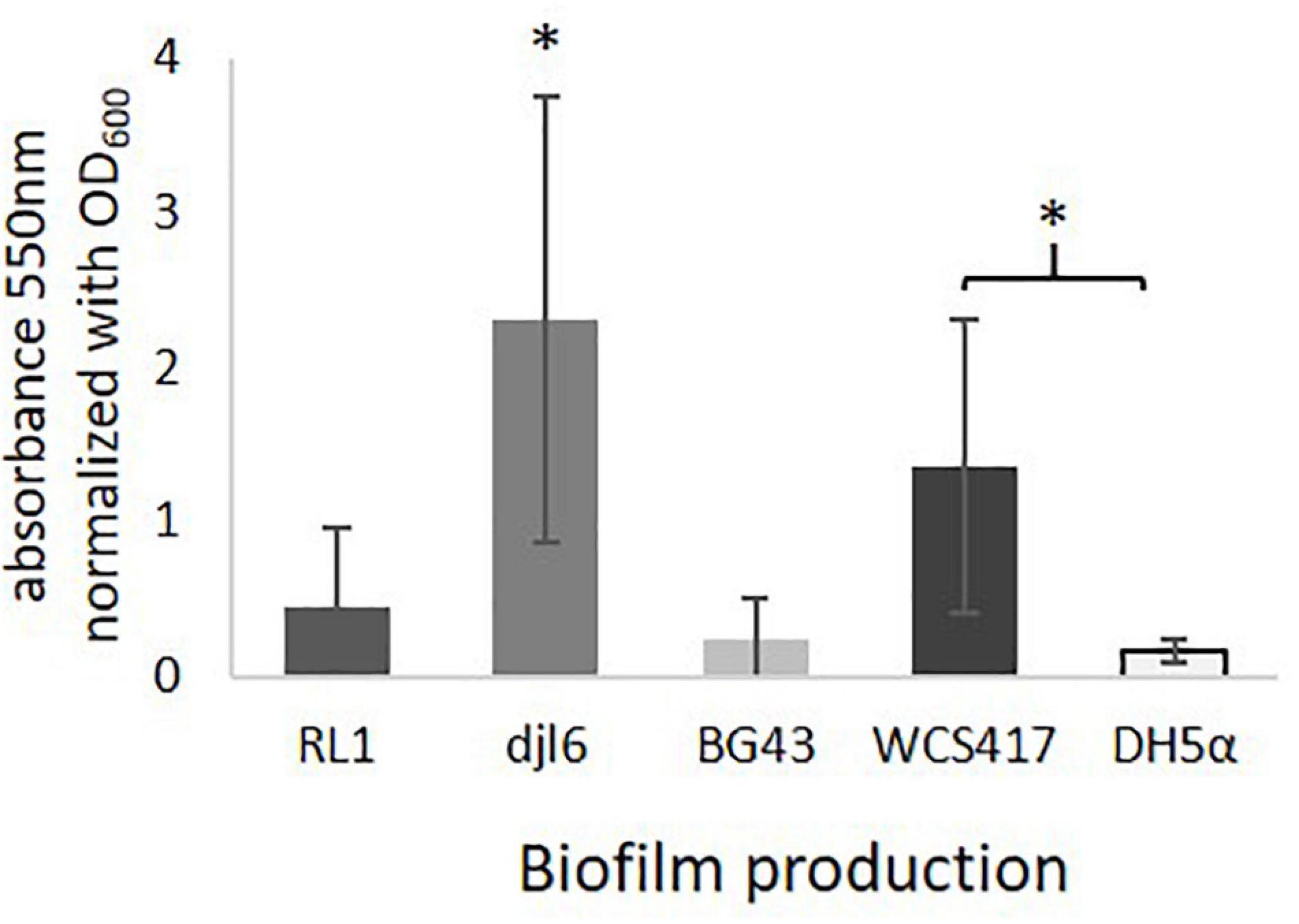
Figure 6. Ability to produce Biofilms with averaged results from 3 experiments normalized to OD600 = 1. Significant difference is indicated by asterisks representing ∗P < 0.05.
Interaction With Other Microbes
Confrontation Assay Against Plant–Pathogenic Fungi
RL1 inhibited the plant-pathogenic fungus Fusarium oxysporum in vitro. The positive biocontrol strain FZB42 inhibited the plant–pathogenic fungi Rhizoctonia solani, Fusarium oxysporum, and Fusarium culmorum indicated by inhibition zones (Supplementary Figure 4).
Degradation of Synthetic and Bacterial N-Acyl-Homoserinelactones (AHLs)
A qsdA gene (QEM30276) could be identified in the RL1 genome, which belongs to a class of large-spectrum quorum-quenching lactonases also present in other Rhodococcus sp. (Figure 7A). Therefore, AHL degradation ability was tested in RL1, djl6 and BG43 using the sensor strain A136. In this set-up it could be clearly shown that RL1, djl6 and BG43 were able to degrade synthetic C12-HSL (Figures 7B–D). Additionally, co-culturing of Acidovorax radicis N35e with RL1 resulted in no visible blue color formation by the sensor strain, indicating degradation of produced AHL (Figure 7B). Finally, V-shaped spotting of Acidovorax radicis N35e and RL1, djl6 and BG43 showed an inhibition of blue color formation where strains were in direct contact (Figures 7E–H).
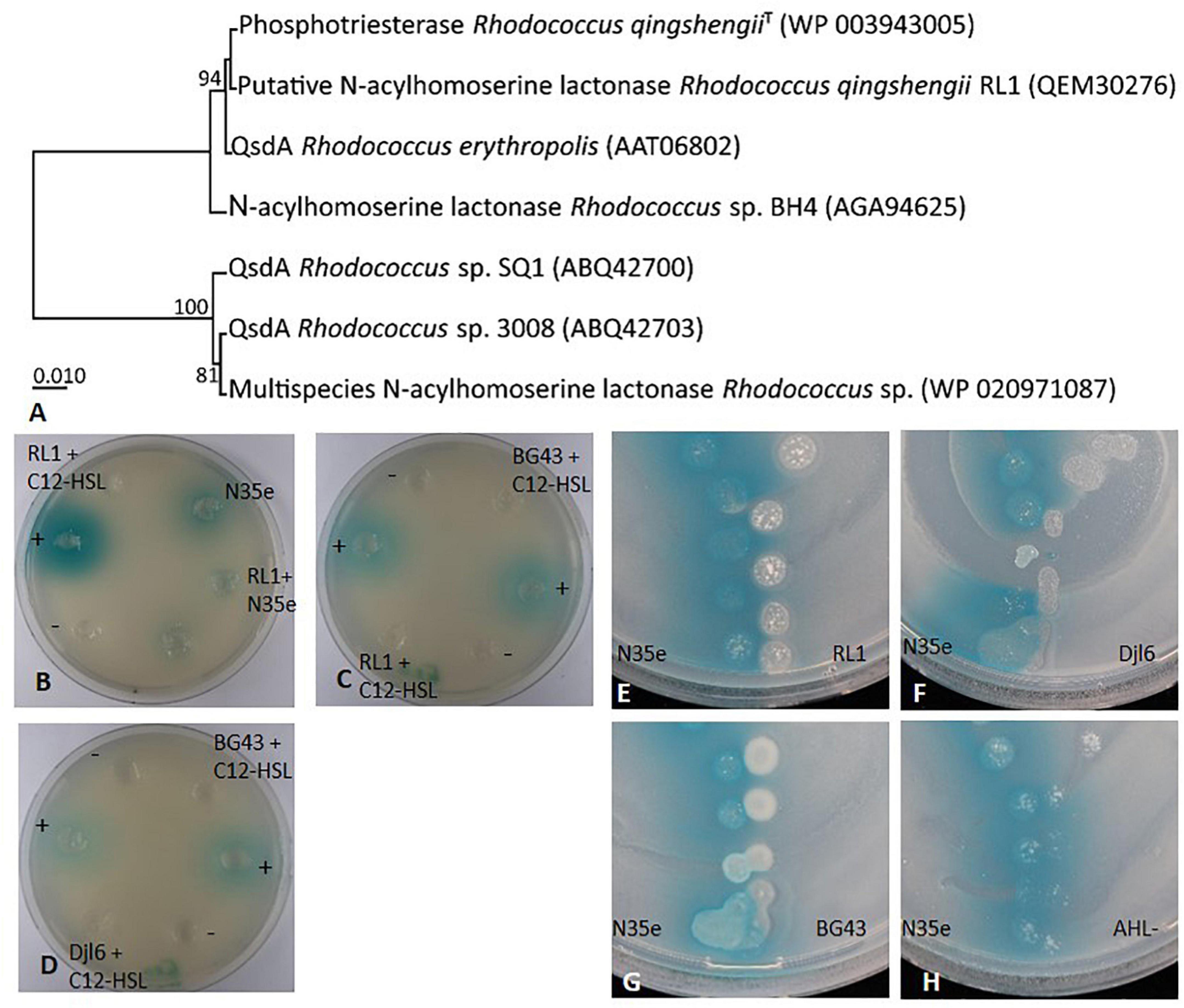
Figure 7. AHL degradation and quorum quenching by Rhodococcus strains RL1, djl6, and BG43. (A) UPGMA phylogenetic tree of translated qsdA (quorum-sensing signal degradation) gene of RL1 and related sequences from Rhodococcus strains. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The evolutionary distances were computed using the Poisson correction method. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 323 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018). (B–D) Well-diffusion plate assays on NB plates all supplemented with the sensor strain A136 and X-Gal. Except for the cultures containing AHL producing strain A. radicis N35e, C12-HSL was added during cultivation of all bacteria. Supernatants of these cultures were added to the wells and blue color formation by the sensor strain indicated remaining AHL in the tested supernatant. NB with C12-HSL (+) served as positive control, and NB without C12-HSL (–) as negative control. (E–H) V-shaped assays on NB after 30 h supplemented with the sensor strain A136 and X-Gal. AHL negative mutant A. radicis N35e AHL- served as control. Presence of AHLs is detected by the sensor strain Agrobacterium tumefaciens A136 indicated by blue color change of X-Gal.
Rhizosphere Competence
Root Colonization in Axenic System
Root colonization was analyzed with fluorescence in situ hybridization using probes EUB Mix Fluos and HGC69A Atto550 or HGC69A Cy3. Single cells of RL1 could be found on the root surface of its host plant Rucola (Eruca sativa L.) when grown in the axenic system (Figures 8A–C), while dense cell patches were identified on roots from MS agar plates (Figure 8D). Similar colonization patterns were found for strains djl6 and BG43. All strains were localized rather in the basal mature part of the root in areas of emergence of root hairs. No endophytic colonization was observed.
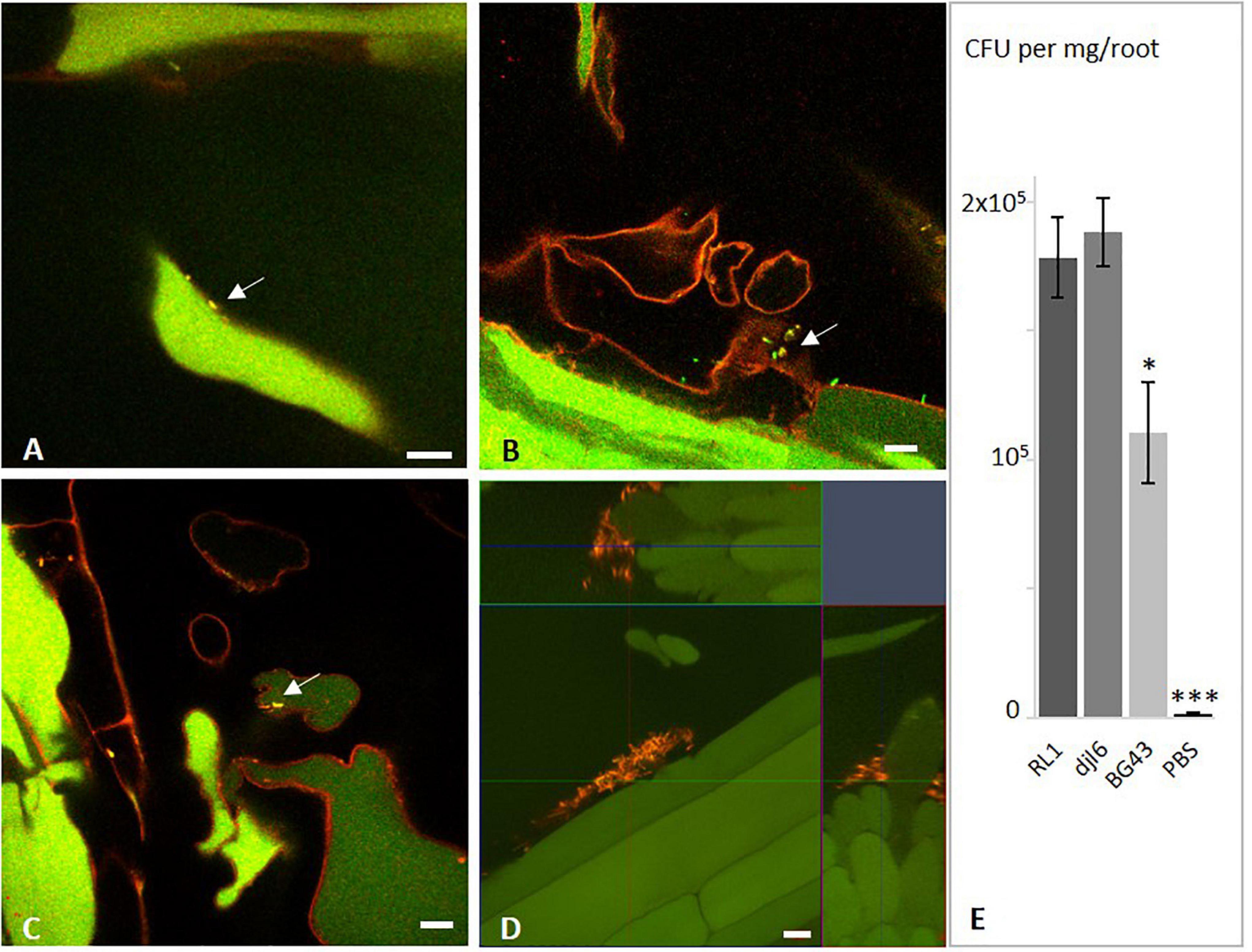
Figure 8. Interaction of Rhodococcus strains RL1, BG43, and djl6 with Eruca sativa roots. In situ detection of root colonization of Eruca sativa by Rhodococcus strains visualized by fluorescence in situ hybridization (FISH). (A) RL1, (B) BG43, and (C) djl6 on 1 week old Eruca sativa roots grown in axenic quartz-sand system. (D) RL1 on 2 weeks old Eruca sativa roots grown on MS agar without additional sucrose. (E) Quantification of root colonization 1 week after inoculation. Arrows indicate bacterial cells identified by yellow color from overlaying channels of probes EUB (green) and HGC (red). Root autofluorescence is assigned in green and red. Scale bar represents 10 μm. Significant difference is indicated by asterisks representing ∗P < 0.05 and ∗∗∗P < 0.001.
Quantitative estimation based on CFU/mg root mass (Figure 8E) showed significantly higher colonization numbers for RL1 and djl6 (p-value = 0.012) than BG43. Djl6 showed a trend toward higher root colonization compared to RL1. All strains were significantly higher than the uninoculated control.
Discussion
The genus Rhodococcus is frequently found in the plant microbiome (Francis and Vereecke, 2019; Vereecke et al., 2020). Therefore, it is important to analyze and understand functions of the plant-associated members of this genus, such as RL1 isolated from Eruca sativa leaves. Additionally, as mentioned in the introduction, the genus Rhodococcus is well-known for stress tolerant strains (Pátek et al., 2021). For these reasons, we wanted to elucidate genomic properties with a special focus on functional analyses of stress tolerance and interaction with plants to understand the possible functions that the plant-associated R. qingshengii RL1 could provide within the plant holobiont and also compare it to the closely related strains djl6 and BG43.
RL1 Genome Harbors Several Genes Involved in Survival and Tolerance to Different Stress Conditions
Genes involved in acidic pH tolerance were identified in RL1, either involved in the production of the compound squalene, such as squalene cyclase, a precursor of hopanoid (Schmerk et al., 2011) or based on the expression of the ADI cluster (in the presence of arginine). The latter is a mechanism to overcome acidic stress often found in gram-positive bacteria (Cotter and Hill, 2003). The experimental evidence proved the ability of Rhodococcus strain RL1 to survive and recover from acidic pH conditions. This trait was also shared by the closely related strains djl6 and BG43, indicating that this trait may be widespread amongst the genus Rhodococcus. In the genus Rhodococcus tolerance to acidic pH was reported for R. qingshengii BBG1 (Benedek et al., 2012) and for the mammalian pathogen Rhodococcus equi, which can withstand a pH of 4 (Benoit et al., 2000). Conventional agricultural practices and soil exploitation can lead to increased soil acidity (Goswami et al., 2017). Therefore, acidic pH tolerance is an important trait of plant-associated and soil bacteria to maintain a functional plant microbiome also under acidic soil conditions.
Ectoine is a compound associated with osmoregulation in bacteria (Bremer and Krämer, 2019) and important for survival during osmotic stress. The gene cluster for ectoine biosynthesis and transporters were identified in RL1, indicating the ability of RL1 to synthesize ectoine under osmotic stress. Alternative to biosynthesis, bacteria can take up compatible solutes, such as proline or betaine from their environment (Bremer and Krämer, 2019). Genes encoding the respective transporters were found in the RL1 genome. Additionally, the full operon of Na+/H+ antiporter was identified in the RL1 genome, which could play a role in salt stress tolerance (Liu et al., 2016; Bhat et al., 2020). Results of the in vitro experiments of the tested Rhodococcus strains growing under high salt and osmotic stress confirmed previous reports of osmotic and salt stress tolerant members of the genus Rhodococcus. For example, an upregulation of genes involved in ectoine biosynthesis was observed in Rhodococcus jostii RHA1 under desiccation (LeBlanc et al., 2008) and rapid adaptation to salt stress was described for R. erythropolis DSM 1069 (De Carvalho et al., 2014). Moreover, plant associated bacteria tolerant to osmotic and salt stress could also be beneficial for the plant via support of ion homeostasis (Bhat et al., 2020; Salas-González et al., 2021) and upregulation of osmoprotective compound biosynthesis in the plant. For example, Bacillus sp. can directly influence proline biosynthesis in plants to improve osmotolerance (Kaushal and Wani, 2016; Bhat et al., 2020).
Heavy metals, such as mercury, are highly persistent environmental pollutants and a threat to all living organisms (Boyd and Barkay, 2012). Organomercury compounds were used in several agricultural applications, for example as common pest control agent in the 1900’s. Although its use has been banned in several countries, it is still used in Australia to treat the plant pathogenic fungus Ceratocystis paradoxa (Schneider, 2021). Mercury resistant bacteria can convert organomercury compounds or Hg(II) to gaseous Hg(0) to reduce the mercury concentration in their environment (Boyd and Barkay, 2012). Mercury resistance was described in R. erythropolis BD2 and Pseudomonas fluorescens SBW25 to be located and transferred on a plasmid (Dabrock et al., 1994; Hall et al., 2020) containing the mer-operon (Boyd and Barkay, 2012). Loss of this plasmid caused a loss of mercury resistance (Dabrock et al., 1994; Hall et al., 2020). However, in RL1 the identified mercury resistance genes, such as a transcriptional regulator MerR and a unique alkylmercury lyase involved in the degradation of toxic organomercury compounds (e.g., MeHg) (Schaefer et al., 2004), are located in the chromosome. We report for the first time that mercury tolerance is also present in an isolate of R. qingshengii based on the results of the in vitro experiment. BG43 and RL1 were both able to survive up to 1 mM of mercury in the growth medium. Survival and detoxification of heavy metals have been reported for other members of the genus Rhodococcus (Trivedi et al., 2007; Irawati et al., 2012), emphasizing the exceptional stress tolerance of this genus. Heavy metal resistance in bacteria in combination with a close association with plants could indicate the adaptation to toxic heavy metal residues of such compounds previously used as pesticides.
Apart from tolerance to heavy metals, we could identify several operons in the genome of RL1 which show that this bacterium has the ability to survive under selective environmental conditions by metabolizing trace gasses like CO and H2. Comparison of deduced amino acid sequences revealed that the identified CODH belongs to the functional type1-CODH enzymes, which catalyze the unidirectional conversion of CO to CO2 (King and Weber, 2007). This type of enzyme has been extensively studied in aerobic CO-oxidizers, or carboxydotrophic Actinobacteria (Quiza et al., 2014). Sequence similarity revealed that the identified [NiFe]-hydrogenase cluster belongs to the high-affinity group 1 h/5 Actinobacteria type of hydrogenases which have been shown to scavenge electrons from tropospheric H2 to sustain aerobic respiration during starvation (Constant et al., 2011; Greening et al., 2016). Interestingly, less is known about plant associated atmospheric H2-oxidizing bacteria. Atmospheric H2 may serve as the maintenance energy during starvation and sporulation of high-affinity H2-oxidizing Actinobacteria, providing them the advantage of survival in plant tissues, as was shown for endophytic Streptomyces spp. (Greening et al., 2016; Kanno et al., 2016). The simultaneous presence of the carbon monoxide dehydrogenase (CODH) genes and the [NiFe]-hydrogenase cluster indicate that RL1 can use CO and H2 as energy source.
The functional annotation of RL1 genome also revealed that it harbors genes involved in multidrug resistance, tellurite resistance and antibiotic resistance. Tellurite is a metalloid often used as antibiotic compound in in vitro experiments and is toxic to eukaryotic and prokaryotic cells (Chien and Han, 2009). Resistance against tellurite can be mediated by a reduction of tellurite (TeO32–) to elemental tellurium, indicated by the color change of the colonies, which was also observed in RL1. Moreover, the RL1 genome harbors genes potentially involved in protection against oxidative stress. These genes could be involved in detoxification of tellurite, because the toxicity of tellurite is eventually caused through intracellular generation of reactive oxygen species (ROS) (Pérez et al., 2007).
In vitro tests with antibiotics revealed resistance of RL1 against kanamycin and ampicillin, whereas djl6 and BG43 are more resistant to rifampicin and vancomycin respectively. Antibiotic resistance was mainly investigated and is widespread in the horse pathogen Rhodococcus equi (Giguère et al., 2017), because of its relevance in livestock animal infections. Antibiotic resistance in plant-associated Rhodococcus species was not intensively studied yet and could confer them a competitive advantage in surviving against other antibiotic-producing microbes in specialized niches like the rhizosphere (Raaijmakers et al., 2009; Mendes et al., 2013).
RL1 Genome Reveals Successful Interaction and Survival Strategies in Association With Plants
The genome annotation of RL1 and functional analysis revealed a large repertoire of traits involved in plant–microbe and microbe–microbe interactions, which can be relevant for the role of RL1 in the plant microbiome.
An important trait of plant-associated bacteria is the ability to colonize plant roots to facilitate, e.g., the exchange of metabolites (Kloepper and Beauchamp, 1992; Pandit et al., 2020). For successful root colonization it can be beneficial for the bacteria to be able to produce biofilms (Pandit et al., 2020), which was demonstrated for RL1, djl6, and BG43. Accordingly, the RL1 genome harbors genes encoding for enzymes involved in biofilm formation. Qualitative evaluation of rhizosphere competence revealed that all three strains were able to colonize the roots of E. sativa epiphytically. However, quantitative evaluation revealed that RL1 and djl6 had significantly more CFUs per mg E. sativa root than BG43, which indicates a better root colonization ability of R. qingshengii species. Verification of endo- or epiphytic leaf colonization of RL1 analyzed with FISH (data not shown) did not deliver clear results due to high auto-fluorescence of the leaves and transformation of fluorescent markers in RL1 was not successful. Therefore, final conclusions upon leaf colonization of RL1 cannot be drawn.
The leaves of Brassicacea, such as Rucola (Eruca sativa L.) contain glucosinolates (GSLs), which are sulfur-containing secondary metabolites involved in the protection of plants against herbivores (Textor and Gershenzon, 2009; Bell et al., 2015). Since RL1 was isolated from the leaves of Rucola (Eruca sativa L.), we were interested if the genome reveals some interesting information about its ability to metabolize glucosinolates. Our results showed that the genome harbors genes potentially involved in the metabolic pathways of GSLs, such as myrosinase, methionine sulfoxide reductase (msrA, msrB) or aldoxime dehydratase oxd (Supplementary Table 4). Degradation of GSLs was investigated for gut microbes regarding beneficial effects of ITC production as a chemoprotective function against cancer (Mullaney et al., 2013a,b; Bessler and Djaldetti, 2018; Mokhtari et al., 2018). An in vitro experiment with Rucola (Eruca sativa L.) leaf extract and pure GSLs (data not shown) did not reveal clear and consistent results on GSL synthesis, bioconversion or degradation by RL1, djl6, and BG43.
The RL1 genome harbors genes related to the production of volatiles, exopolysaccharides and proteases, which are important in microbial communication, plant colonization and microbial detection by the host (Flemming et al., 2016; Netzker et al., 2020). The chemotaxis protein CheY relevant for the transmission of sensory signals from the chemoreceptors to the flagella motors, which is additionally a microbe-associated molecular pattern (MAMP) (Paul et al., 2010) was identified in the RL1 genome. As Rhodococcus is a non-motile genus CheY has a rather different function, e.g., in sensory signal transduction in another pathway or interaction with the plant. Additionally, in the RL1 genome genes encoding for a LacI transcription regulator and an aldo-keto reductase were identified, which were found to be enriched in genomes of plant beneficial microbes (Levy et al., 2018).
Plant associated bacteria in general can influence root growth, germination, flowering and developmental stages via balancing or producing plant hormones, such as gibberellin (Kang et al., 2014; Panke-Buisse et al., 2017; Salazar-Cerezo et al., 2018) or indole-3-acetic acid (IAA) (Finkel et al., 2020). In the in vitro assay RL1 produced a higher amount of IAA in comparison to the strains djl6 and BG43. The best-known pathway for IAA production includes the enzyme indolepyruvate decarboxylase (ipdC), which is not present in the RL1 genome. Instead genes of the alternative indole-3-acetamide pathway for IAA production (Spaepen et al., 2007) were identified in the RL1 genome. The ability to produce IAA in vitro was not only shown for RL1 but also in another R. qingshengii strain (Hasuty et al., 2018) and other members of the genus Rhodococcus (Francis and Vereecke, 2019). Bacterial production of IAA can be beneficial for the plant by increasing the root system (Spaepen and Vanderleyden, 2011) and balancing IAA production is an important function of the root microbiome (Finkel et al., 2020). Gibberellin production is encoded by a conserved operon, which was characterized in α- and β-proteobacteria (Nagel et al., 2018). Essential parts of the gibberellin operon were identified in the RL1 genome. To our knowledge, this is the first report about the presence of genes of the gibberellin operon in any Actinobacteria. Some plant-pathogenic bacteria produce bioactive GA4, which can have a detrimental effect on seedling development. Beneficial bacteria only produce the precursor GA9 as they lack the cytochrome P450 (CYP115) for the final step in the production of the bioactive GA4 (Nagel and Peters, 2017). As RL1 also lacks the cytochrome P450 (CYP115) this indicates its allocation to the plant beneficial bacteria. Verification of the production of gibberellin by RL1 with gas chromatography was beyond the scope of this work.
The bacterially produced polyamine spermidine increases biofilm formation and overall bacterial fitness (Xie et al., 2014; Liu et al., 2016). Additionally, it is the plant growth-promoting compound in strains such as B. subtilis OKB105 or Klebsiella sp. D5A (Xie et al., 2014; Liu et al., 2016) and the upregulation of spermidine export proteins in Stenotrophomonas rhizophila DSM14405 upon salt stress in combination with exposure to root exudates emphasizes the role of spermidine as key substance in stress protection in roots (Alavi et al., 2013). Presence of genes encoding for the enzymes involved in the biosynthesis of spermidine, such as arginine decarboxylase, agmatinase, spermidine synthase in the RL1 genome indicate the ability of RL1 to function as stress-protecting agent and support plants under abiotic stress.
The degradation of 1-amino-cyclopropane-1-carboxylate (ACC), the precursor of the plant hormone ethylene, by bacterial ACC deaminase can protect the plant from detrimental effects of long exposure to ethylene (Glick, 2012; Dubois et al., 2018). In a standard in vitro assay RL1, djl6, and BG43 were able to grow on nitrogen-free M9 plates with ACC in the medium, indicating ACC deaminase activity (Figure 5A). However, the essential gene acdS encoding for ACC deaminase is missing in the RL1 genome. Additionally, all three tested Rhodococcus strains were able to grow on all tested nitrogen free media. The results indicate that the isolates grow on N-free media through utilization of atmospheric nitrogen rather than using ACC as a nitrogen source. Biological nitrogen fixation is defined as the bacterial conversion of dinitrogen to ammonia through the expression of canonical nif gene products (Dos Santos et al., 2012; Higdon et al., 2020). In the RL1 genome the SUF system FeS assembly protein of the nifU family was identified (MSMEG_2718, Supplementary Table 4), which stabilizes the nitrogenase complex and is relevant for diazotrophy especially under low temperature conditions (Suyal et al., 2014). The nifH gene was previously identified in a diazotrophic R. qingshengii strain (Suyal et al., 2014; Joshi et al., 2019) and used as molecular marker to directly link to a diazotrophic lifestyle. However, no nifH gene was identified in the RL1 genome. In a large scale genome analysis Higdon et al. (2020) identified three distinct groups of diazotrophic bacteria defined by nif gene content and structural variation. The genus Rhodococcus was classified as DS-negative (=no Dos Santos model nif gene homolog present in genome). This implies the presence of alternative nif genes and metabolic pathways relevant for nitrogen fixation in Rhodococcus genomes beyond the currently known models. Transcriptome analysis and mutant construction would reveal insights to alternative nitrogen fixation mechanisms in RL1 as representative of the genus Rhodococcus.
Siderophores not only chelate iron and have beneficial effects in plant growth, they are also involved in bioremediation, function as biosensors and are relevant in microbial competition and defense against other microbes, which can lead to a beneficial biocontrol effect for the plant (Ahmed and Holmström, 2014; Gu et al., 2020; Pollak and Cordero, 2020). Genes involved in iron acquisition and siderophore production were identified in the RL1 genome, for example for the siderophore heterobactin, which is unique to the Rhodococcus genus (Carrano et al., 2001; Bosello et al., 2013; Khilyas et al., 2020). The in vitro assay for siderophore production was positive for RL1, corroborating that the identified genes were actually expressed. These results were in contrast to djl6 and BG43, where the functional analysis was negative. Iron acquisition and ferrous iron transport can occur via two systems, the FeoABC and EfeUOB transporters (Lau et al., 2016). The EfeUOB was reported to be low-pH-induced (Cao et al., 2007) and was predicted in the genome of a Leptospirillum sp. tolerant to acidic pH (Osorio et al., 2008). RL1 harbors the genes encoding for the EfeUOB operon, which could contribute to the low pH tolerance of RL1, because it allows iron acquisition also under low pH.
The RL1 genome harbors genes relevant for organic acid production, which are involved in phosphate solubilization and genes potentially relevant for phosphate metabolism and transport. However, genes involved in gluconic acid production, which is the main driver in phosphate solubilization could not be identified (Rodríguez et al., 2006). Despite of that, the in vitro assay for this trait was positive for RL1 which suggests the presence of alternative organic acids involved in phosphate solubilization. Phosphate solubilization capacity was previously reported for Rhodococcus globerulus isolated from Plectranthus amboinicus (Murugappan et al., 2017).
RL1 Genome Reveals Competitive Potential Against Other Microorganisms
Members of the genus Rhodococcus have been reported to show antifungal activity in vitro against plant-pathogenic fungi (Chiba et al., 1999; Iwatsuki et al., 2007; Santos et al., 2020) and RL1 reduced growth of F. oxysporum in vitro, but showed no inhibition against R. solani and F. culmorum (Supplementary Figure 4). Further studies using model plants will reveal the full potential of RL1 as biocontrol agent against plant pathogenic fungi.
Degradation or interference with quorum sensing molecules can disturb bacterial communication and is called quorum quenching (Dong et al., 2001). The qsdA gene, encoding for a N-acyl-HSL lactonase was first described by Uroz et al. (2003) for the strain R. erythropolis W2 and could also be identified in the RL1 genome (Figure 7A). Moreover, the RL1 genome harbors a two-component transcriptional AHL responsive regulator from the LuxR family. However, as RL1 is not producing AHLs this regulator is likely a so-called LuxR-solo, which allows bacteria to respond to quorum sensing signals from neighboring cells without itself contributing to signal synthesis. This was also previously described for the genus Rhodococcus and other gram-positive bacteria (Subramoni and Venturi, 2009; Santos et al., 2012). In vitro experiments showed the ability of RL1 to degrade AHLs. To our knowledge, this is the first report of an AHL-degrading R. qingshengii. Also it is the first description of functional AHL degradation by BG43, which was previously reported only to interfere with the quinolone signal of Pseudomonas aeruginosa (Müller et al., 2014). Quorum quenching ability was intensively studied in R. erythropolis R138 (Cirou et al., 2007; Barbey et al., 2013; Latour et al., 2013; Kwasiborski et al., 2015), which was able to reduce the soft-rot pathogen Pectobacterium in potatoes and most likely use the degraded AHLs as carbon source. Quorum quenching can also be a beneficial trait in other crop-pathogen systems as reported for example in Pseudomonas segetis (Rodríguez et al., 2020) or Bacillus thuringensis (Dong et al., 2004). Further analysis of RL1 quorum quenching abilities, e.g., against plant pathogens such as Pectobacterium or Pseudomonas syringae, would reveal its full potential as plant biocontrol agent.
Genome Comparison of Related R. erythropolis and R. qingshengii Isolates Show Potential for Re-classification of Clade Members
Rhodococcus is a genus well-known for its high potential to produce versatile secondary metabolites and the RL1 genome annotation confirms previous studies (Ceniceros et al., 2017; Thompson et al., 2020). The number of genes from the genome of RL1, which were assigned to the COG group for secondary metabolites, were higher compared to other bacteria, for example Stenotrophomonas or Enterobacter (Alavi et al., 2014; Andrés-Barrao et al., 2017). Additionally, 17 BGC for secondary metabolites were identified in RL1. The average number of BGCs in the R. erythropolis clade are 13–24 BGCs and are mostly shared by R. erythropolis and R. qingshengii strains (Thompson et al., 2020). Four BGC cluster were highly conserved among the R. erythropolis clade and three of them were also identified in RL1 (Supplementary Table 3). The remaining unknown BGCs in the RL1 genome are potentially capable of producing novel compounds which could be analyzed in future studies.
Rhodococcus is a heterogeneous genus with eight identified phylogenetic clades (Alvarez, 2019). Phylogenetic analysis based on complete genome sequences of the R. erythropolis clade reveals a clear separation into two subgroups at an ANI value of 97% (Figure 1 and Supplementary Figure 2). The first one includes sequences belonging to only R. erythopolis, the second includes R. qingshengii and R. erythropolis strains. Based on the clear separation we can confirm previous recommendations to separate the R. erythropolis clade into the two groups consisting of the species R. qingshengii and R. erythropolis respectively (Sangal et al., 2016; Khilyas et al., 2020; Thompson et al., 2020). We also suggest that the R. erythropolis strains assigned to the R. qingshengii group should be re-named as previously recommended (Sangal et al., 2016; Thompson et al., 2020). The genomes of RL1 and djl6 were clearly identified as belonging to the R. qingshengii cluster (Xu et al., 2007; Kuhl et al., 2019) and had more genes in common with each other than with BG43 (Figure 2), whereas the BG43 genome was classified as R. erythropolis (Müller et al., 2014; Rückert et al., 2015). Despite the clear separation the strains RL1 and BG43 had many functional traits in common, indicating a close functional overlap between the species. At the same time, djl6 and RL1 showed different results in the in vitro experiment for siderophore production and mercury tolerance, indicating differences on the genetic and functional level also within the species R. qingshengii (Figure 4). RL1 showed overall the best performance in the tested traits. This emphasizes the importance of RL1 and the necessity to analyze the genetic and functional potential of individual strains to understand the role also of lesser known members of the plant microbiome.
Conclusion and Outlook
The study shows the remarkable genomic potential of the isolate R. qingshengii RL1 for tolerating various abiotic stresses, plant–microbe and microbe–microbe interactions, many of which could be confirmed by functional analysis in vitro. By this thorough characterization we aim to contribute to a better understanding of relevant attributes for interactions in the plant holobiont as well as provide selection criteria for using strains, such as RL1, in specific agricultural or biotechnological applications. Furthermore, we provided phylogenetic evidence based on whole genome comparisons to justify a taxonomic separation of the R. erythropolis and R. qingshengii cluster and re-name some members of the R. erythropolis cluster. However, the functional analysis also indicates many shared traits between the two species R. qingshengii and R. erythropolis, some of them also described for the first time for the strains djl6 and BG43, but also different traits within the same species. Further experiments involving inoculation of different plants with RL1 under various conditions, which were beyond the scope of this study, would reveal insights into its plant beneficial functions. This could be coupled with transcriptome analysis of RL1 to reveal such intriguing aspects as an alternative nitrogen fixation pathway. Further investigation of the quorum quenching ability against various plant–pathogenic bacteria could advance the understanding of the role of RL1 in biological control.
Data Availability Statement
The datasets presented in this study can be found in the NCBI database. Accession numbers are for Rhodococcus qingshengii RL1 NZ_CP042917, NZ_CP042916, NZ_CP042915, for Rhodococcus qingshengii djl6 NZ_CP025959, NZ_CP025960, NZ_CP025961, NZ_CP025962, and for Rhodococcus erythropolis BG43 NZ_CP011295, NZ_CP011296, NZ_CP011297, and NZ_CP011298.
Author Contributions
TK, SC, and MR contributed to conception and design of the study. TK performed the experiments and wrote the first draft of the manuscript. JU contributed parts of the experimental data. SC and TK analyzed the data. SC and MR wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Institute of Biochemical Plant Pathology (BIOP) at Helmholtz Center Munich for providing Fusarium culmorum G2191, Fusarium oxysporum DSM62297 and Dr. Rita Grosch [Leibniz Institute of Vegetable and Ornamental Crops (IGZ), Großbeeren, Germany] for providing Rhizoctonia solani for our experiments. Thank you to S. Klink, F. Tunc, and A. Sabunchi for technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.708605/full#supplementary-material
References
Abbamondi, G. R., Tommonaro, G., Weyens, N., Thijs, S., Sillen, W., Gkorezis, P., et al. (2016). Plant growth-promoting effects of rhizospheric and endophytic bacteria associated with different tomato cultivars and new tomato hybrids. Chem. Biol. Technol. Agric. 3, 1–10. doi: 10.1186/s40538-015-0051-3
Ahmed, E., and Holmström, S. J. M. (2014). Siderophores in environmental research: roles and applications. Microb. Biotechnol. 7, 196–208. doi: 10.1111/1751-7915.12117
Alabid, I., Hardt, M., Imani, J., Hartmann, A., Rothballer, M., Li, D., et al. (2020). The N-acyl homoserine-lactone depleted Rhizobium radiobacter mutant RrF4NM13 shows reduced growth-promoting and resistance-inducing activities in mono- and dicotyledonous plants. J. Plant Dis. Prot. 127, 769–781. doi: 10.1007/s41348-020-00360-8
Alavi, P., Starcher, M. R., Thallinger, G. G., Zachow, C., Müller, H., and Berg, G. (2014). Stenotrophomonas comparative genomics reveals genes and functions that differentiate beneficial and pathogenic bacteria. BMC Genomics 15:482. doi: 10.1186/1471-2164-15-482
Alavi, P., Starcher, M. R., Zachow, C., Müller, H., and Berg, G. (2013). Root-microbe systems: the effect and mode of interaction of Stress Protecting Agent (SPA) Stenotrophomonas rhizophila DSM14405T. Front. Plant Sci. 4:141. doi: 10.3389/fpls.2013.00141
Alquéres, S., Meneses, C., Rouws, L., Rothballer, M., Baldani, I., Schmid, M., et al. (2013). The bacterial superoxide dismutase and glutathione reductase are crucial for endophytic colonization of rice roots by Gluconacetobacter diazotrophicus PAL5. MPMI 26, 937–945. doi: 10.1094/MPMI-12-12-0286-R
Amann, R. I., Krumholz, L., and Stahl, D. A. (1990). Fluorescent-oligonucleotide probing of whole cells for determinative and environmental studies in microbiology. J. Bacteriol. 172, 762–770. doi: 10.1128/jb.172.2.762-770.1990
Andrés-Barrao, C., Lafi, F. F., Alam, I., de Zélicourt, A., Eida, A. A., Bokhari, A., et al. (2017). Complete genome sequence analysis of Enterobacter sp. SA187, a plant multi-stress tolerance promoting endophytic bacterium. Front. Microbiol. 8:2023. doi: 10.3389/fmicb.2017.02023
Anton, B. P., and Raleigh, E. A. (2016). Complete Genome Sequence of NEB 5-alpha, a Derivative of Escherichia coli K-12 DH5α. Genome Announc. 4, 6–7. doi: 10.1128/genomeA.01245-16
Aziz, R. K., Bartels, D., Best, A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Babin, D., Sommermann, L., Chowdhury, S. P., Behr, J. H., Sandmann, M., Neumann, G., et al. (2021). Distinct rhizomicrobiota assemblages and plant performance in lettuce grown in soils with different agricultural management histories. FEMS Microbiol. Ecol. 97:fiab027. doi: 10.1093/femsec/fiab027
Bahramisharif, A., and Rose, L. E. (2019). Efficacy of biological agents and compost on growth and resistance of tomatoes to late blight. Planta 249, 799–813. doi: 10.1007/s00425-018-3035-2
Banerjee, S., Walder, F., Büchi, L., Meyer, M., Held, A. Y., Gattinger, A., et al. (2019). Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME 13, 1722–1736. doi: 10.1038/s41396-019-0383-2
Barbey, C., Crépin, A., Bergeau, D., Ouchiha, A., Mijouin, L., Taupin, L., et al. (2013). In Planta biocontrol of Pectobacterium atrosepticum by Rhodococcus erythropolis involves silencing of pathogen communication by the rhodococcal gamma-lactone catabolic pathway. PLoS One 8:e66642. doi: 10.1371/journal.pone.0066642
Belimov, A. A., Safronova, V. I., Sergeyeva, T. A., Egorova, T. N., Matveyeva, V. A., Tsyganov, V. E., et al. (2001). Characterization of plant growth promoting rhizobacteria isolated from polluted soils and containing 1-aminocyclopropane-1-carboxylate deaminase. Can. J. Microbiol. 47, 642–652. doi: 10.1139/w01-062
Bell, L., Oruna-Concha, M. J., and Wagstaff, C. (2015). Identification and quantification of glucosinolate and flavonol compounds in rocket salad (Eruca sativa, Eruca vesicaria and Diplotaxis tenuifolia) by LC-MS: highlighting the potential for improving nutritional value of rocket crops. Food Chem. 172, 852–861. doi: 10.1016/j.foodchem.2014.09.116
Benedek, T., Máthé, I., Salamon, R., Rákos, S., Pásztohy, Z., Márialigeti, K., et al. (2012). Potential bacterial soil inoculant made up by Rhodococcus sp. and Pseudomonas sp. for remediation in situ of hydrocarbon - and heavy metal polluted soils. Stud. Univ. Babes-Bolyai Chem. 2012, 199–211.
Benoit, S., Taouji, S., Benachour, A., and Hartke, A. (2000). Resistance of Rhodococcus equi to acid pH. Int. J. Food Microbiol. 55, 295–298. doi: 10.1016/S0168-1605(00)00172-0
Berendsen, R. L., Vismans, G., Yu, K., Song, Y., de Jonge, R., Burgman, W. P., et al. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 12, 1496–1507. doi: 10.1038/s41396-018-0093-1
Berg, G., Alavi, M., and Schmidt, C. S. (2013). “Biocontrol and osmoprotection for plants under salinated conditions,” in Molecular Microbial Ecology of the Rhizosphere 2, ed. F. J. de Bruijn (Hoboken, NJ: John Wiley and Sons, Inc), 587–592. doi: 10.1002/9781118297674.ch55
Berg, G., Rybakova, D., Grube, M., Köberl, M., and Price, A. (2017). The plant microbiome explored: implications for experimental botany. J. Exp. Bot. 67, 995–1002. doi: 10.1093/jxb/erv466
Bessler, H., and Djaldetti, M. (2018). Broccoli and human health: immunomodulatory effect of sulforaphane in a model of colon cancer. Int. J. Food Sci. Nutr. 69, 946–953. doi: 10.1080/09637486.2018.1439901
Bhat, M. A., Kumar, V., Wani, I. A., Dar, F. L., Farooq, I., Bhatti, F., et al. (2020). Mechanistic insights of the interaction of plant growth-promoting rhizobacteria (PGPR) with plant roots toward enhancing plant productivity by alleviating salinity stress. Front. Microbiol. 11:1952. doi: 10.3389/fmicb.2020.01952
Blin, K., Wolf, T., Chevrette, M. G., Lu, X., Schwalen, C. J., Kautsar, S. A., et al. (2017). AntiSMASH 4.0 - improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45, W36–W41. doi: 10.1093/nar/gkx319
Blom, J., Kreis, J., Spänig, S., Juhre, T., Bertelli, C., Ernst, C., et al. (2016). EDGAR 2.0: an enhanced software platform for comparative gene content analyses. Nucleic Acids Res. 44, W22–W28. doi: 10.1093/nar/gkw255
Bosello, M., Zeyadi, M., Kraas, F. I., Linne, U., Xie, X., and Marahiel, M. A. (2013). Structural characterization of the heterobactin siderophores from Rhodococcus erythropolis PR4 and elucidation of their biosynthetic machinery. J. Nat. Prod. 76, 2282–2290. doi: 10.1021/np4006579
Boyd, E. S., and Barkay, T. (2012). The mercury resistance operon: from an origin in a geothermal environment to an efficient detoxification machine. Front. Microbiol. 3:349. doi: 10.3389/fmicb.2012.00349
Bremer, E., and Krämer, R. (2019). Responses of microorganisms to osmotic stress. Annu. Rev. Microbiol. 73, 313–334. doi: 10.1146/annurev-micro-020518-115504
Brevik, E. C., and Burgess, L. C. (2014). The influence of soils on human health. Nat. Educ. Knowl. 5:1.
Cao, J., Woodhall, M. R., Alvarez, J., Cartron, M. L., and Andrews, S. C. (2007). EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol. Microbiol. 65, 857–875. doi: 10.1111/j.1365-2958.2007.05802.x
Carrano, C. J., Jordan, M., Drechsel, H., Schmid, D. G., and Winkelmann, G. (2001). Heterobactins: a new class of siderophores from Rhodococcus erythropolis IGTS8 containing both hydroxamate and catecholate donor groups. BioMetals 14, 119–125. doi: 10.1023/A:1016633529461
Ceniceros, A., Dijkhuizen, L., and Petrusma, M. (2017). Molecular characterization of a Rhodococcus jostii RHA1 γ-butyrolactone(-like) signalling molecule and its main biosynthesis gene gblA. Sci. Rep. 7:17743. doi: 10.1038/s41598-017-17853-6
Chiba, H., Agematu, H., Sakai, K., Dobashi, K., and Yoshioka, T. (1999). Rhodopeptins, novel cyclic tetrapeptides with antifungal activities from Rhodococcus sp. J. Antibiot. (Tokyo) 52, 710–720. doi: 10.7164/antibiotics.52.710
Chien, C. C., and Han, C. T. (2009). Tellurite resistance and reduction by a Paenibacillus sp. isolated from heavy metal-contaminated sediment. Environ. Toxicol. Chem. 28, 1627–1632. doi: 10.1897/08-521.1
Chowdhury, S. P., Hartmann, A., Gao, X. W., and Borriss, R. (2015a). Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 - A review. Front. Microbiol. 6:780. doi: 10.3389/fmicb.2015.00780
Chowdhury, S. P., Uhl, J., Grosch, R., Alquéres, S., Pittroff, S., Dietel, K., et al. (2015b). Cyclic lipopeptides of Bacillus amyloliquefaciens subsp. plantarum colonizing the lettuce rhizosphere enhance plant defense responses toward the bottom rot pathogen Rhizoctonia solani. MPMI 28, 984–995. doi: 10.1094/MPMI-03-15-0066-R
Cirou, A., Diallo, S., Kurt, C., Latour, X., and Faure, D. (2007). Growth promotion of quorum-quenching bacteria in the rhizosphere of Solanum tuberosum. Environ. Microbiol. 9, 1511–1522. doi: 10.1111/j.1462-2920.2007.01270.x
Constant, P., Chowdhury, S. P., Hesse, L., Pratscher, J., and Conrad, R. (2011). Genome data mining and soil survey for the novel group 5 [NiFe]-hydrogenase to explore the diversity and ecological importance of presumptive high-affinity H 2-oxidizing bacteria. Appl. Environ. Microbiol. 77, 6027–6035. doi: 10.1128/AEM.00673-11
Cotter, P. D., and Hill, C. (2003). Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67, 429–453. doi: 10.1128/mmbr.67.3.429-453.2003
Dabrock, B., Kesseler, M., Averhoff, B., and Gottschalk, G. (1994). Identification and characterization of a transmissible linear plasmid from Rhodococcus erythropolis BD2 that encodes isopropylbenzene and trichloroethene catabolism. Appl. Environ. Microbiol. 60, 853–860. doi: 10.1128/aem.60.3.853-860.1994
Daims, H., Bruhl, A., Amann, R. I., Schleifer, K.-H., and Wagner, M. (1999). The domainspecific probe EUB-338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22, 434–444. doi: 10.1016/s0723-2020(99)80053-8
De Carvalho, C. C. C. R., Marques, M. P. C., Hachicho, N., and Heipieper, H. J. (2014). Rapid adaptation of Rhodococcus erythropolis cells to salt stress by synthesizing polyunsaturated fatty acids. Appl. Microbiol. Biotechnol. 98, 5599–5606. doi: 10.1007/s00253-014-5549-2
Döbereiner, J. (1995). “Isolation and identification of aerobic nitrogen-fixing bacteria from soil and plants,” in Methods in Applied Soil Microbiology and Biochemistry, eds K. Alef and P. Nannipieri (London: Academic Press), 134–141.
Dong, Y.-H., Wang, L.-H., Xu, J.-L., Zhang, H.-B., Zhang, X.-F., and Zhang, L.-H. (2001). Quenching quorum-sensing- dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411, 813–817. doi: 10.1038/35081101
Dong, Y. H., Zhang, X. F., Xu, J. L., and Zhang, L. H. (2004). Insecticidal Bacillus thuringiensis silences Erwinia carotovora virulence by a new form of microbial antagonism, signal interference. Appl. Environ. Microbiol. 70, 954–960. doi: 10.1128/AEM.70.2.954-960.2004
Dos Santos, P. C., Fang, Z., Mason, S. W., Setubal, J. C., and Dixon, R. (2012). Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13:162. doi: 10.1186/1471-2164-13-162
Dubois, M., Van den Broeck, L., and Inzé, D. (2018). The pivotal role of ethylene in plant growth. Trends Plant Sci. 23, 311–323. doi: 10.1016/j.tplants.2018.01.003
Dziewit, L., Pyzik, A., Matlakowska, R., Baj, J., Szuplewska, M., and Bartosik, D. (2013). Characterization of Halomonas sp. ZM3 isolated from the Zelazny most post-flotation waste reservoir, with a special focus on its mobile DNA. BMC Microbiol. 13:59. doi: 10.1186/1471-2180-13-59
Finkel, O. M., Salas-González, I., Castrillo, G., Conway, J. M., Law, T. F., Teixeira, P. J. P. L., et al. (2020). A single bacterial genus maintains root growth in a complex microbiome. Nature 587, 103–108. doi: 10.1038/s41586-020-2778-7
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Foley, J. A., DeFries, R., Asner, G. P., Barford, C., Bonan, G., Carpenter, S. R., et al. (2005). Global consequences of land use. Science (80-.) 309, 570–574. doi: 10.1126/science.1111772
Francis, I. M., and Vereecke, D. (2019). “Plant-Associated Rhodococcus species, for better and for worse,” in Biology of Rhodococcus, ed. H. Alvarez (Switzerland: Springer), 359–377. doi: 10.1007/978-3-030-11461-9_13
Gerlach, W., Sauthoff, W., and Pag, H. (1958). Untersuchungen über die fusariumwelke an Aechmea fasciata (Lindl.). Bakt.Phytopathol.Z. 32, 416–432. doi: 10.1111/j.1439-0434.1958.tb01784.x
Giguère, S., Berghaus, L. J., and Willingham-Lane, J. M. (2017). Antimicrobial resistance in Rhodococcus equi. Acta Biochim. Pol. 61, 633–638. doi: 10.18388/abp.2014_1824
Glick, B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. 2012. Scientifica 2012:963401. doi: 10.6064/2012/963401
Gorbunova, T. I., Egorova, D. O., Pervova, M. G., Kyrianova, T. D., Demakov, V. A., Saloutin, V. I., et al. (2020). Biodegradation of trichlorobiphenyls and their hydroxylated derivatives by Rhodococcus-strains. J. Hazard. Mater. 409:124471. doi: 10.1016/j.jhazmat.2020.124471
Gordon, S. A., and Weber, R. P. (1951). Colorimetric estimation of indoleacetic acid. Plant Physiol. 26, 192–195. doi: 10.1104/pp.26.1.192
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Goswami, G., Deka, P., Das, P., Bora, S. S., Samanta, R., Boro, R. C., et al. (2017). Diversity and functional properties of acid-tolerant bacteria isolated from tea plantation soil of Assam. 3 Biotech 7:229. doi: 10.1007/s13205-017-0864-9
Greening, C., Biswas, A., Carere, C. R., Jackson, C. J., Taylor, M. C., Stott, M. B., et al. (2016). Genomic and metagenomic surveys of hydrogenase distribution indicate H 2 is a widely utilised energy source for microbial growth and survival. ISME J. 10, 761–777. doi: 10.1038/ismej.2015.153
Gu, S., Wei, Z., Shao, Z., Friman, V. P., Cao, K., Yang, T., et al. (2020). Competition for iron drives phytopathogen control by natural rhizosphere microbiomes. Nat. Microbiol. 5, 1002–1010. doi: 10.1038/s41564-020-0719-8
Gupta, N., Skinner, K. A., Summers, Z. M., Edirisinghe, J. N., Faria, J. P., Marshall, C. W., et al. (2019). Draft genome sequence of Rhodococcus sp. strain ATCC 49988, a quinoline-degrading bacterium. Microbiol. Resour. Announc. 8, e403–e419.
Hall, J. P. J., Harrison, E., Pärnänen, K., Virta, M., and Brockhurst, M. A. (2020). The impact of mercury selection and conjugative genetic elements on community structure and resistance gene transfer. Front. Microbiol. 11:1846. doi: 10.3389/fmicb.2020.01846
Han, S., Li, D., Trost, E., Mayer, K. F., Corina, V., Heller, W., et al. (2016). Systemic responses of barley to the 3-hydroxy-decanoyl-homoserine lactone producing plant beneficial endophyte Acidovorax radicis N35. Front. Plant Sci. 7:1868. doi: 10.3389/fpls.2016.01868
Hartmann, A., and Hurek, T. (1988). Effect of carotenoid overproduction on oxygen tolerance of nitrogen fixation in Azospirillum brasilense Sp7. Microbiology 134, 2449–2455. doi: 10.1099/00221287-134-9-2449
Hasuty, A., Choliq, A., and Hidayat, I. (2018). Production of indole acetic acid (IAA) by Serratia marcescens subsp. marcescens and Rhodococcus aff. qingshengii. Int. J. Agric. Technol. 14, 299–312.
Higdon, S. M., Pozzo, T., Kong, N., Huang, B. C., Yang, M. L., Jeannotte, R., et al. (2020). Genomic characterization of a diazotrophic microbiota associated with maize aerial root mucilage. PLoS One 15:e239677. doi: 10.1371/journal.pone.0239677
Irawati, W., Soraya, Y. P., and Baskoro, A. H. (2012). A study on mercury-resistant bacteria isolated from a gold mine in Pongkor Village, Bogor, Indonesia. HAYATI J. Biosci. 19, 197–200. doi: 10.4308/hjb.19.4.197
Iwatsuki, M., Uchida, R., Takakusagi, Y., Matsumoto, A., Jiang, C. L., Takahashi, Y., et al. (2007). Lariatins, novel anti-mycobacterial peptides with a lasso structure, produced by Rhodococcus jostii K01-B0171. J. Antibiot. (Tokyo) 60, 357–363. doi: 10.1038/ja.2007.48
Jayakumar, A., Krishna, A., Nair, I. C., and Radhakrishnan, E. K. (2020). Drought-tolerant and plant growth-promoting endophytic Staphylococcus sp. having synergistic effect with silicate supplementation. Arch. Microbiol. 202, 1899–1906. doi: 10.1007/s00203-020-01911-1
Jensen, L. J., Julien, P., Kuhn, M., von Mering, C., Muller, J., Doerks, T., et al. (2008). eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 36, 250–254. doi: 10.1093/nar/gkm796
Joshi, D., Chandra, R., Suyal, D. C., Kumar, S., and Goel, R. (2019). Impacts of bioinoculants Pseudomonas jesenii MP1 and Rhodococcus qingshengii S10107 on chickpea (Cicer arietinum L.) yield and soil nitrogen status. Pedosphere 29, 388–399. doi: 10.1016/S1002-0160(19)60807-6
Kamble, A. L., Banoth, L., Meena, V. S., Singh, A., Chisti, Y., and Banerjee, U. C. (2013). Nitrile hydratase of Rhodococcus erythropolis: characterization of the enzyme and the use of whole cells for biotransformation of nitriles. Biotech 3, 319–330. doi: 10.1007/s13205-012-0104-2
Kang, S. M., Radhakrishnan, R., Khan, A. L., Kim, M. J., Park, J. M., Kim, B. R., et al. (2014). Gibberellin secreting rhizobacterium, Pseudomonas putida H-2-3 modulates the hormonal and stress physiology of soybean to improve the plant growth under saline and drought conditions. Plant Physiol. Biochem. 84, 115–124. doi: 10.1016/j.plaphy.2014.09.001
Kanno, M., Constant, P., Tamaki, H., and Kamagata, Y. (2016). Detection and isolation of plant-associated bacteria scavenging atmospheric molecular hydrogen. Environ. Microbiol. 18, 2495–2506. doi: 10.1111/1462-2920.13162
Kaufmann, M., and Michel, B. (1973). The osmotic potential of polyethylene glycol 6000. Plant Physiol. 51, 914–916. doi: 10.1104/pp.51.5.914
Kaushal, M., and Wani, S. P. (2016). Rhizobacterial-plant interactions: strategies ensuring plant growth promotion under drought and salinity stress. Agric. Ecosyst. Environ. 231, 68–78. doi: 10.1016/j.agee.2016.06.031
Khilyas, I. V., Sorokina, A. V., Markelova, M. I., Belenikin, M., Shafigullina, L., Tukhbatova, R. I., et al. (2020). Genomic and phenotypic analysis of siderophore-producing Rhodococcus qingshengii strain S10 isolated from an arid weathered serpentine rock environment. Arch. Microbiol. 203, 855–860. doi: 10.1007/s00203-020-02057-w
King, G. M., and Weber, C. F. (2007). Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5, 107–118. doi: 10.1038/nrmicro1595
Kirchhof, G., Eckert, B., Stoffels, M., Ivo Baldani, J., Reis, V. M., and Hartmann, A. (2001). Herbaspirillum frisingense sp. nov., a new nitrogan-fixing bacterial species that occurs in C4-fibre plants. Int. J. Syst. Evol. Microbiol. 51, 157–168. doi: 10.1099/00207713-51-1-157
Kloepper, J. W., and Beauchamp, C. J. (1992). A review of issues related to measuring colonization of plant roots by bacteria. Can. J. Microbiol. 38, 1219–1232. doi: 10.1139/m92-202
Konstantinidis, K. T., and Tiedje, J. M. (2005). Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187, 6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005
Kuhl, T., Felder, M., Nussbaumer, T., Fischer, D., Kublik, S., Paul Chowdhury, S., et al. (2019). De novo genome assembly of a plant-associated Rhodococcus qingshengii Strain (RL1) isolated from Eruca sativa Mill. and showing plant growth-promoting properties. Microbiol. Resour. Announc. 8, 14–16. doi: 10.1128/MRA.01106-19
Kumar, G. P., Mir Hassan Ahmed, S. K., Desai, S., Leo Daniel Amalraj, E., and Rasul, A. (2014). In vitro screening for abiotic stress tolerance in potent biocontrol and plant growth promoting strains of Pseudomonas and Bacillus spp. Int. J. Bacteriol. 2014:195946. doi: 10.1155/2014/195946
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. and Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kwasiborski, A., Mondy, S., Chong, T. M., Barbey, C., Chan, K. G., Beury-Cirou, A., et al. (2015). Transcriptome of the quorum-sensing signal-degrading Rhodococcus erythropolis responds differentially to virulent and avirulent Pectobacterium atrosepticum. Heredity (Edinb) 114, 476–484. doi: 10.1038/hdy.2014.121
Latour, X., Barbey, C., Chane, A., Groboillot, A., and Burini, J.-F. (2013). Rhodococcus erythropolis and its γ-Lactone catabolic pathway: an unusual biocontrol system that disrupts pathogen quorum sensing communication. Agronomy 3, 816–838. doi: 10.3390/agronomy3040816
Lau, C. K. Y., Krewulak, K. D., and Vogel, H. J. (2016). Bacterial ferrous iron transport: the Feo system. FEMS Microbiol. Rev. 40, 273–298. doi: 10.1093/femsre/fuv049
LeBlanc, J. C., Gonçalves, E. R., and Mohn, W. W. (2008). Global response to desiccation stress in the soil actinomycete Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 74, 2627–2636. doi: 10.1128/AEM.02711-07
Leigh, M. B., Prouzová, P., Macková, M., Macek, T., Nagle, D. P., and Fletcher, J. S. (2006). Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl. Environ. Microbiol. 72, 2331–2342. doi: 10.1128/AEM.72.4.2331-2342.2006
Levy, A., Salas Gonzalez, I., Mittelviefhaus, M., Clingenpeel, S., Herrera Paredes, S., Miao, J., et al. (2018). Genomic features of bacterial adaptation to plants. Nat. Genet. 50, 138–150. doi: 10.1038/s41588-017-0012-9
Li, D., Rothballer, M., Schmid, M., Esperschütz, J., and Hartmann, A. (2011). Acidovorax radicis sp. nov., a wheat-root-colonizing bacterium. Int. J. Syst. Evol. Microbiol. 61, 2589–2594. doi: 10.1099/ijs.0.025296-0
Lincoln, S. A., Hamilton, T. L., Valladares Juárez, A. G., Schedler, M., Macalady, J. L., Müller, R., et al. (2015). Draft genome sequence of the piezotolerant and crude oil-degrading bacterium Rhodococcus qingshengii strain TUHH-12. Genome Announc. 3:e268–15. doi: 10.1128/genomeA.00268-15
Liu, W., Wang, Q., Hou, J., Tu, C., Luo, Y., and Christie, P. (2016). Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella sp. D5A. Sci. Rep. 6, 20–22. doi: 10.1038/srep26710
Loper, J. E., and Schroth, M. N. (1986). Influence of bacterial sources of indole-3-acetic acid on root elongation of sugar beet. Phytopathology 76:386. doi: 10.1094/phyto-76-386
Lynne, A. M., Haarmann, D., and Louden, B. C. (2011). Use of blue agar CAS assay for siderophore detection. J. Microbiol. Biol. Educ. 12, 51–53. doi: 10.1128/jmbe.v12i1.249
Ma, Y., Oliveira, R. S., Freitas, H., and Zhang, C. (2016a). Biochemical and molecular mechanisms of plant-microbe-metal interactions: relevance for phytoremediation. Front. Plant Sci. 7:918. doi: 10.3389/fpls.2016.00918
Ma, Y., Rajkumar, M., Zhang, C., and Freitas, H. (2016b). Beneficial role of bacterial endophytes in heavy metal phytoremediation. J. Environ. Manage. 174, 14–25. doi: 10.1016/j.jenvman.2016.02.047
Martínez-García, P. M., López-Solanilla, E., Ramos, C., and Rodríguez-Palenzuela, P. (2016). Prediction of bacterial associations with plants using a supervised machine-learning approach. Environ. Microbiol. 18, 4847–4861. doi: 10.1111/1462-2920.13389
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Mokhtari, R. B., Baluch, N., Homayouni, T. S., Morgatskaya, E., Kumar, S., Kazemi, P., et al. (2018). The role of Sulforaphane in cancer chemoprevention and health benefits: a mini-review. Cell Commun. Signal. 12, 91–101. doi: 10.1007/s12079-017-0401-y
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, 182–185. doi: 10.1093/nar/gkm321
Mullaney, J. A., Ansell, J., Kelly, W. J., and Heyes, J. A. (2013a). Biotransformation of glucosinolates from a bacterial perspective. CAB Rev. 8, 1–15. doi: 10.1079/PAVSNNR20138034
Mullaney, J. A., Kelly, W. J., McGhie, T. K., Ansell, J., and Heyes, J. A. (2013b). Lactic acid bacteria convert glucosinolates to nitriles efficiently yet differently from enterobacteriaceae. J. Agric. Food Chem. 61, 3039–3046. doi: 10.1021/jf305442j
Müller, C., Birmes, F. S., Niewerth, H., and Fetzner, S. (2014). Conversion of the Pseudomonas aeruginosa quinolone signal and related alkylhydroxyquinolines by Rhodococcus sp. strain BG43. Appl. Environ. Microbiol. 80, 7266–7274. doi: 10.1128/AEM.02342-14
Murugappan, R. M., Benazir, S., Usha, C., and Lok, S. (2017). Growth promoting and probiotic potential of the endophytic bacterium Rhodococcus globerulus colonizing the medicinal plant Plectranthus amboinicus (Lour.) Spreng. Int. J. Curr. Res. Rev. 9, 7–13. doi: 10.7324/ijcrr.2017.9143
Nagel, R., Bieber, J. E., Schmidt-Dannert, M. G., Nett, R. S., and Peters, R. J. (2018). A third class: Functional gibberellin biosynthetic operon in beta-proteobacteria. Front. Microbiol. 9:2916. doi: 10.3389/fmicb.2018.02916
Nagel, R., and Peters, R. J. (2017). Investigating the phylogenetic range of gibberellin biosynthesis in bacteria. MPMI 30, 343–349. doi: 10.1094/MPMI-01-17-0001-R
Nautiyal, C. S. (1999). An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 170, 265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x
Netzker, T., Shepherdson, E. M. F., Zambri, M. P., and Elliot, M. A. (2020). Bacterial volatile compounds: functions in communication, cooperation, and competition. Annu. Rev. Microbiol. 74, 409–430. doi: 10.1146/annurev-micro-011320-015542
Osorio, H., Martínez, V., Nieto, P. A., Holmes, D. S., and Quatrini, R. (2008). Microbial iron management mechanisms in extremely acidic environments: comparative genomics evidence for diversity and versatility. BMC Microbiol. 8:203. doi: 10.1186/1471-2180-8-203
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, 206–214. doi: 10.1093/nar/gkt1226
Pandit, A., Adholeya, A., Cahill, D., Brau, L., and Kochar, M. (2020). Microbial biofilms in nature: unlocking their potential for agricultural applications. J. Appl. Microbiol. 129, 199–211. doi: 10.1111/jam.14609
Panke-Buisse, K., Lee, S., and Kao-Kniffin, J. (2017). Cultivated sub-populations of soil microbiomes retain early flowering plant trait. Microb. Ecol. 73, 394–403. doi: 10.1007/s00248-016-0846-1
Pátek, M., Grulich, M., and Nešvera, J. (2021). Stress response in Rhodococcus strains. Biotechnol Adv. 28:107698. doi: 10.1016/j.biotechadv.2021.107698
Paul, K., Nieto, V., Carlquist, W. C., Blair, D. F., and Harshey, R. M. (2010). The c-di-GMP binding protein ycgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38, 128–139. doi: 10.1016/j.molcel.2010.03.001
Pérez, J. M., Calderón, I. L., Arenas, F. A., Fuentes, D. E., Pradenas, G. A., Fuentes, E. L., et al. (2007). Bacterial toxicity of potassium tellurite: unveiling an ancient enigma. PLoS One 2:e211. doi: 10.1371/journal.pone.0000211
Pérez-Miranda, S., Cabirol, N., George-Téllez, R., Zamudio-Rivera, L. S., and Fernández, F. J. (2007). O-CAS, a fast and universal method for siderophore detection. J. Microbiol. Methods 70, 127–131. doi: 10.1016/j.mimet.2007.03.023
Pham, T. T. M., Rodriguez, N. J. P., Hijri, M., and Sylvestre, M. (2015). Optimizing polychlorinated biphenyl degradation by flavonoid-induced cells of the rhizobacterium Rhodococcus erythropolis U23A. PLoS One 10:e126033. doi: 10.1371/journal.pone.0126033
Pieterse, C. M. J., Berendsen, R. L., de Jonge, R., Stringlis, I. A., Van Dijken, A. J. H., Van Pelt, J. A., et al. (2020). Pseudomonas simiae WCS417: star track of a model beneficial rhizobacterium. Plant Soil. 461, 245–263. doi: 10.1007/s11104-020-04786-9
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C. M., and Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Płociniczak, T., Fic, E., Pacwa-Płociniczak, M., Pawlik, M., and Piotrowska-Seget, Z. (2017). Improvement of phytoremediation of an aged petroleum hydrocarbon-contaminated soil by Rhodococcus erythropolis CD 106 strain. Int. J. Phytoremediation 19, 614–620. doi: 10.1080/15226514.2016.1278420
Pollak, S., and Cordero, O. X. (2020). Rhizobiome shields plants from infection. Nat. Microbiol. 5, 978–979. doi: 10.1038/s41564-020-0766-1
Quiza, L., Lalonde, I., Guertin, C., and Constant, P. (2014). Land-use influences the distribution and activity of high affinity CO-oxidizing bacteria associated to type I-coxL genotype in soil. Front. Microbiol. 5:271. doi: 10.3389/fmicb.2014.00271
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., and Moënne-Loccoz, Y. (2009). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321, 341–361. doi: 10.1007/s11104-008-9568-6
Rodríguez, H., Fraga, R., Gonzalez, T., and Bashan, Y. (2006). Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. Plant Soil 287, 15–21. doi: 10.1007/s11104-006-9056-9
Rodríguez, M., Torres, M., Blanco, L., Béjar, V., Sampedro, I., and Llamas, I. (2020). Plant growth-promoting activity and quorum quenching-mediated biocontrol of bacterial phytopathogens by Pseudomonas segetis strain P6. Sci. Rep. 10:4121. doi: 10.1038/s41598-020-61084-1
Rodriguez, P. A., Rothballer, M., Chowdhury, S. P., Nussbaumer, T., Gutjahr, C., and Falter-Braun, P. (2019). Systems biology of plant-microbiome interactions. Mol. Plant 12, 804–821. doi: 10.1016/j.molp.2019.05.006
Roller, C., Wagner, M., Amann, R., Ludwig, W., and Schleifer, K. H. (1994). In situ probing of gram-positive bacteria with high DNA G + C content using 23S rRNA-targeted oligonucleotides. Microbiol-SGM 140 (Pt 10), 2849–2858. doi: 10.1099/00221287-140-10-2849
Rückert, C., Birmes, F. S., Müller, C., Niewerth, H., Winkler, A., Fetzner, S., et al. (2015). Complete genome sequence of Rhodococcus erythropolis BG43 (DSM 46869), a degrader of Pseudomonas aeruginosa quorum sensing signal molecules. J. Biotechnol. 211, 99–100. doi: 10.1016/j.jbiotec.2015.07.014
Salas-González, I., Reyt, G., Flis, P., Custódio, V., Gopaulchan, D., Bakhoum, N., et al. (2021). Coordination between microbiota and root endodermis supports plant mineral nutrient homeostasis. Science 371:eabd0695. doi: 10.1126/science.abd0695
Salazar-Cerezo, S., Martínez-Montiel, N., García-Sánchez, J., Pérez-y-Terrón, R., and Martínez-Contreras, R. D. (2018). Gibberellin biosynthesis and metabolism: a convergent route for plants, fungi and bacteria. Microbiol. Res. 208, 85–98. doi: 10.1016/j.micres.2018.01.010
Sangal, V., Goodfellow, M., Jones, A. L., Schwalbe, E. C., Blom, J., Hoskisson, P. A., et al. (2016). Next-generation systematics: an innovative approach to resolve the structure of complex prokaryotic taxa. Sci. Rep. 6:38392. doi: 10.1038/srep38392
Santos, A., Núñez-Montero, K., Lamilla, C., Pavez, M., Quezada-Solís, D., and Barrientos, L. (2020). Antifungal activity screening of antarctic actinobacteria against phytopathogenic fungi. Acta Biol. Colomb. 25, 353–358. doi: 10.15446/abc.v25n2.76405
Santos, C. L., Correia-Neves, M., Moradas-Ferreira, P., and Mendes, M. V. (2012). A Walk into the LuxR Regulators of Actinobacteria: phylogenomic distribution and functional diversity. PLoS One 7:e46758. doi: 10.1371/journal.pone.0046758
Schaefer, J. K., Yagi, J., Reinfelder, J. R., Cardona, T., Ellickson, K. M., Tel-Or, S., et al. (2004). Role of the bacterial organomercury lyase (MerB) in controlling methylmercury accumulation in mercury-contaminated natural waters. Environ. Sci. Technol. 38, 4304–4311. doi: 10.1021/es049895w
Schmerk, C. L., Bernards, M. A., and Valvano, M. A. (2011). Hopanoid production is required for low-pH tolerance, antimicrobial resistance, and motility in Burkholderia cenocepacia. J. Bacteriol. 193, 6712–6723. doi: 10.1128/JB.05979-11
Schneider, L. (2021). When toxic chmicals refuse to die – An examination of the prolonged mercury pesticide use in Australia. Elem. Sci. Anth. 9:053. doi: 10.1525/elementa.2021.053
Sessitsch, A., Kuffner, M., Kidd, P., Vangronsveld, J., Wenzel, W. W., Fallmann, K., et al. (2013). The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 60, 182–194. doi: 10.1016/j.soilbio.2013.01.012
Smith, P., House, J. I., Bustamante, M., Sobocká, J., Harper, R., Pan, G., et al. (2016). Global change pressures on soils from land use and management. Glob. Chang. Biol. 22, 1008–1028. doi: 10.1111/gcb.13068
Spaepen, S., and Vanderleyden, J. (2011). Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 3:a001438. doi: 10.1101/cshperspect.a001438
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Stickler, D. J., Morris, N. S., Mclean, R. J. C., and Fuqua, C. (1998). Biofilms on indwelling urethral catheters produce quorum sensing signal molecules in situ and in vitro. Appl. Environ. Microbiol. 64, 3486–3490. doi: 10.1128/aem.64.9.3486-3490.1998
Subramoni, S., and Venturi, V. (2009). LuxR-family “solos”: bachelor sensors/regulators of signalling molecules. Microbiology 155, 1377–1385. doi: 10.1099/mic.0.026849-0
Suyal, D. C., Yadav, A., Shouche, Y., and Goel, R. (2014). Differential proteomics in response to low temperature diazotrophy of Himalayan psychrophilic nitrogen fixing Pseudomonas migulae S10724 strain. Curr. Microbiol. 68, 543–550. doi: 10.1007/s00284-013-0508-1
Táncsics, A., Benedek, T., Farkas, M., Máthé, I., Márialigeti, K., Szoboszlay, S., et al. (2014). Sequence analysis of 16S rRNA, gyrB and catA genes and DNA-DNA hybridization reveal that Rhodococcus jialingiae is a later synonym of Rhodococcus qingshengii. Int. J. Syst. Evol. Microbiol. 64, 298–301. doi: 10.1099/ijs.0.059097-0
Tarrand, J. J., Krieg, N. R., and Döbereiner, J. (1978). A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can J Microbiol. 24, 967–980. doi: 10.1139/m78-160
Tatusov, R. L., Galperin, M. Y., Natale, D. A., and Koonin, E. V. (2000). The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/nar/28.1.33
Textor, S., and Gershenzon, J. (2009). Herbivore induction of the glucosinolate-myrosinase defense system: major trends, biochemical bases and ecological significance. Phytochem. Rev. 8, 149–170. doi: 10.1007/s11101-008-9117-1
Thompson, D., Cognat, V., Goodfellow, M., Koechler, S., Heintz, D., Carapito, C., et al. (2020). Phylogenomic classification and biosynthetic potential of the fossil fuel-biodesulfurizing Rhodococcus strain IGTS8. Front. Microbiol. 11:1417. doi: 10.3389/fmicb.2020.01417
Trivedi, P., Pandey, A., and Sa, T. (2007). Chromate reducing and plant growth promoting activies of psychrotrophic Rhodococcus erythropolis MtCC 7905. J. Basic Microbiol. 47, 513–517. doi: 10.1002/jobm.200700224
Tsiafouli, M. A., Thébault, E., Sgardelis, S. P., de Ruiter, P. C., van der Putten, W. H., Birkhofer, K., et al. (2015). Intensive agriculture reduces soil biodiversity across Europe. Glob. Chang. Biol. 21, 973–985. doi: 10.1111/gcb.12752
Uroz, S., D’Angelo-Picard, C., Carlier, A., Elasri, M., Sicot, C., Petit, A., et al. (2003). Novel bacteria degrading N-acylhomoserine lactones and their use as quenchers of quorum-sensing-regulated functions of plant-pathogenic bacteria. Microbiology 149, 1981–1989. doi: 10.1099/mic.0.26375-0
Vereecke, D., Zhang, Y., Francis, I. M., Lambert, P. Q., Venneman, J., Stamler, R. A., et al. (2020). Functional genomics insights into the pathogenicity, habitat fitness, and mechanisms modifying plant development of Rhodococcus sp. PBTS1 and PBTS2. Front. Microbiol. 11:14. doi: 10.3389/fmicb.2020.00014
Wagacha, J. M., and Muthomi, J. W. (2007). Fusarium culmorum: infection process, mechanisms of mycotoxin production and their role in pathogenesis in wheat. Crop Prot. 26, 877–885. doi: 10.1016/j.cropro.2006.09.003
Wagg, C., Schlaeppi, K., Banerjee, S., Kuramae, E. E., and van der Heijden, M. G. A. (2019). Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 10:4841. doi: 10.1038/s41467-019-12798-y
Wang, Z., Xu, J., Li, Y., Wang, K., Wang, Y., Hong, Q., et al. (2010). Rhodococcus jialingiae sp. nov., an actinobacterium isolated from sludge of a carbendazim wastewater treatment facility. Int. J. Syst. Evol. Microbiol. 60, 378–381. doi: 10.1099/ijs.0.013219-0
Weyens, N., Beckers, B., Schellingen, K., Ceulemans, R., Croes, S., Janssen, J., et al. (2013). Plant-associated bacteria and their role in the success or failure of metal phytoextraction projects: first observations of a field-related experiment. Microb. Biotechnol. 6, 288–299. doi: 10.1111/1751-7915.12038
Windisch, S., Sommermann, L., Babin, D., Chowdhury, S. P., Grosch, R., Moradtalab, N., et al. (2021). Impact of long-term organic and mineral fertilization on rhizosphere metabolites, root–microbial interactions and plant health of lettuce. Front. Microbiol. 11:597745. doi: 10.3389/fmicb.2020.597745
Xie, S. S., Wu, H. J., Zang, H. Y., Wu, L. M., Zhu, Q. Q., and Gao, X. W. (2014). Plant growth promotion by spermidine-producing Bacillus subtilis OKB105. MPMI 27, 655–663. doi: 10.1094/MPMI-01-14-0010-R
Xu, J. L., He, J., Wang, Z. C., Wang, K., Li, W. J., Tang, S. K., et al. (2007). Rhodococcus qingshengii sp. nov., a carbendazim-degrading bacterium. Int. J. Syst. Evol. Microbiol. 57, 2754–2757. doi: 10.1099/ijs.0.65095-0
Keywords: Rhodococcus qingshengii, plant–microbe interaction, quorum quenching, mercury tolerance, nitrogen fixation
Citation: Kuhl T, Chowdhury SP, Uhl J and Rothballer M (2021) Genome-Based Characterization of Plant-Associated Rhodococcus qingshengii RL1 Reveals Stress Tolerance and Plant–Microbe Interaction Traits. Front. Microbiol. 12:708605. doi: 10.3389/fmicb.2021.708605
Received: 12 May 2021; Accepted: 26 July 2021;
Published: 18 August 2021.
Edited by:
Frank T. Robb, University of Maryland, Baltimore, United StatesReviewed by:
Samina Mehnaz, Forman Christian College, PakistanDaniel Van Der Lelie, Gusto Global LLC, United States
Copyright © 2021 Kuhl, Chowdhury, Uhl and Rothballer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Rothballer, cm90aGJhbGxlckBoZWxtaG9sdHotbXVlbmNoZW4uZGU=
 Theresa Kuhl
Theresa Kuhl Soumitra Paul Chowdhury
Soumitra Paul Chowdhury Jenny Uhl
Jenny Uhl Michael Rothballer
Michael Rothballer