- 1Research Center of Avian Diseases, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Institute of Preventive Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 3Key Laboratory of Animal Disease and Human Health of Sichuan Province, Chengdu, China
Riemerella anatipestifer causes serious contagious disease in ducks, geese, and other fowl. However, as a harmful pathogen causing significant economic losses in the poultry industry, R. anatipestifer is still poorly understood for its pathogenesis mechanisms. In a previous study, we developed an indirect ELISA method for detecting R. anatipestifer infection using B739_0832 protein, a putative outer membrane protein H (OmpH) that is conserved among different serotypes of R. anatipestifer. Although OmpH in some pathogenic bacteria, such as Pasteurella, has been reported as a virulence factor, it is still not clear whether B739_0832 protein contributes to the virulence of R. anatipestifer. In this study, we confirmed that B739_0832 protein in R. anatipestifer localizes to the outer membrane. We constructed a B739_0832 deletion mutant strain (ΔB739_0832) and assayed various effects from the deletion of B739_0832. ΔB739_0832 strain had a similar growth rate to wild-type R. anatipestifer CH-1. However, the survival rate of ducklings in 10 days after infection from ΔB739_0832 strain was 50%, whereas no ducklings survived from wild-type R. anatipestifer infection. Furthermore, the median lethal dose (LD50) of the ΔB739_0832 strain was approximately 150 times higher than that of the wild-type strain. Pathology examinations on infected ducklings found that, at 36 h after infection, bacterial loads in blood, liver, and brain tissues from ΔB739_0832-infected ducklings were considerably lower than those from wild-type infected ducklings. These results demonstrate that the B739_0832 protein contributes to the virulence of R. anatipestifer CH-1.
Introduction
Riemerella anatipestifer is a Gram-negative, rod-shaped bacterium in the Flavobacteriaceae family, Riemerella genus (Segers et al., 1993). Riemerella anatipestifer is one of the most serious bacterial threats harming mostly the duck industry, but R. anatipestifer infection has also been reported in other waterfowl worldwide, causing heavy economic losses (Wang et al., 2010; Hu et al., 2012). Riemerella anatipestifer infection has high mortality and morbidity rate in young ducklings, but may be present in adult ducks causing only subclinical or even asymptomatic diseases (Chang et al., 2019), making it difficult to detect and eradicate. There are at least 21 serotypes of R. anatipestifer reported around the world, with serotypes 1, 2, and 10 being responsible for most of the major outbreaks in China (Pathanasophon et al., 2002; Hu et al., 2012; Wang et al., 2014). As an important worldwide poultry pathogen, its exact pathogenic mechanism is still not clear. There is no cross-protection between different serotypes of R. anatipestifer developed from infection (Kang et al., 2018). With the diverse serotype variations, vaccines developed against R. anatipestifer have been shown to have low cross-protection against different serotypes (Chu et al., 2015). Antibiotics have been used to control R. anatipestifer infection in ducks. However, increasing evidence of drug resistance bacteria and detection of antibiotics in meat product call for safer alternatives. To safely and effectively control and prevent diseases caused by R. anatipestifer, new strategies for developing vaccines with cross-protection against different serotypes are needed.
Many outer membrane proteins in pathogenic bacteria are virulence factors that enable or facilitate bacterial attachment to host cell surfaces, as part of the pathogenicity process (Caruana and Walper, 2020). Outer membrane proteins of pathogenic bacteria are generally immunogenic and play important roles in virulence and immunity to bacterial diseases (Weiser and Gotschlich, 1991; Hatfaludi et al., 2010). To date, few virulence-associated proteins have been reported in R. anatipestifer, such as outer membrane protein A (OmpA), CAMP cohemolysin, Cas9, and TonB-dependent receptor (Crasta et al., 2002; Hu et al., 2011; Liao et al., 2015; Wang et al., 2019; Xu et al., 2020). Outer membrane protein H (OmpH) is a major outer membrane protein, conserved in Gram-negative bacteria (Hatfaludi et al., 2010). OmpH has been extensively studied in Pasteurella multocida as an immunodominant porin protein (Kim et al., 2011) and has been reported as an important virulence factor and a protective antigen for developing vaccine against Pasteurellosis in chickens (Thanasarasakulpong et al., 2015). Meanwhile, OmpH (also known as Skp) in Escherichia coli and Salmonella typhimurium has been identified as a periplasmic chaperone protein with Sec-A like activity and is involved in the maintenance of folding intermediates of outer membrane proteins (Schäfer et al., 1999).
In R. anatipestifer CH-1, B739_0832 open reading frame (ORF) is annotated as encoding an OmpH protein (Wang et al., 2014). In our previous study, we successfully developed an indirect ELISA assay using a recombinant B739_0832 protein for detecting R. anatipestifer infection (Gao et al., 2016). The B739_0832-based ELISA assay has higher sensitivity and wider detection range than OmpA-based ELISA and the conventional tube agglutination assay, suggesting that B739_0832 is conserved among different R. anatipestifer serotypes. To assess the roles of B739_0832 in R. anatipestifer pathogenesis, we evaluated effects of B739_0832 deletion on R. anatipestifer virulence.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains, plasmids, and primers used in this study are listed in Tables 1, 2, respectively. Riemerella anatipestifer CH-1 (RA CH-1) strain was used as the wild-type strain, and all other strains used were derived from RA CH-1. R. anatipestifer was grown in Tryptic Soy Broth (TSB) or Giolitti-Cantoni Broth (GCB) at 37°C with shaking (Liu et al., 2017). GCB agar plates were prepared by supplementing GCB with 1.5% agar. Alternatively, R. anatipestifer was also grown on Luria-Bertani (LB) agar plate supplemented with 5% sheep blood. When necessary, appropriate concentrations of antibiotics were added to the media: ampicillin (Amp, Sigma-Aldrich, 100 μg/ml), chloramphenicol (Cm, Sigma-Aldrich, 30 μg/ml), kanamycin (Kan, Sigma-Aldrich, 50 μg/ml), and cefoxitin (Cfx, Sigma-Aldrich, 1 μg/ml).
Construction of Clean B739_0832 Deletion Mutant Strain (ΔB739_0832)
A ΔB739_0832 mutant strain was constructed by using the natural transformation method as previously described (Liu et al., 2017). Briefly, about 700-bp fragments upstream and downstream of B739_0832 gene were amplified using primer pairs ΔB739_0832 up-arm P1 and P2, ΔB739_0832 down-arm P1 and P2, respectively (Table 2). The amplified upstream and downstream fragments were connected to a pLMF02:sacB plasmid using ligation (Tian et al., 2020). The recombined plasmid was further processed with restriction enzymes NheI and PstI to extract a DNA fragment that has a Cfx-sacB cassette in the center flanked by the amplified upstream and downstream fragments. The DNA fragment was purified using a Universal DNA Purification kit (TIANGENTM, Beijing, China) and served as donor DNA. Wild-type R. anatipestifer CH-1 was transformed with the purified DNA fragment, and cefoxitin-resistant, sucrose-sensitive recombinants were scored. Another DNA fragment fusing the upstream and downstream fragments were produced using the overlap PCR method. The scored cefoxitin-resistant, sucrose-sensitive recombinant strain was further transformed with the upstream–downstream overlap DNA fragment; the ΔB739_0832 mutant strain was scored as cefoxitin-sensitive and sucrose-resistant recombinants. The ΔB739_0832 mutant strain was verified by PCR amplification and sequencing (Supplementary Figure 1).
A complementary strain (CΔB739_0832) was prepared by transforming the ΔB739_0832 mutant strain with a B739_0832 expressing plasmid (pLMF02:B739_0832). Briefly, B739_0832 gene was cloned into pLMF02—a pPM5 derivative—plasmid (Tian et al., 2020). The ΔB739_0832 mutant strain was then transformed with the pLMF02:B739_0832 plasmid using the natural transformation method as described in Liu et al. (2017) and selected for cefoxitin resistance colonies. Expression of B739_0832 in the complemented strain was confirmed by Western blot.
Quantitative Real-Time PCR
Quantitative real-time PCR (qRT-PCR) was used to measure transcription expression levels of B739_0832 and flanking genes (Liu et al., 2016). Total RNA of the wild-type strain and the ΔB739_0832 mutant strain was extracted from cell cultures at OD600 = 1.0 using RNAprep pure Cell/Bacteria kits (TIANGENTM, Beijing, China). To eliminate DNA contamination, all extracted total RNA samples were treated with RNase-free DNase I (40 U/mg RNA, Takara, China) and purified using RNeasy Mini Kits (Qiagen, Germany). HiScript reverse transcriptase (Vazyme, China) was used to generate cDNA in accordance with manufacturer’s instructions. qRT-PCR was performed using SYBR Green Master Mix (Bio-Rad, United States) and primers listed in Table 2. The expression level of 16S rRNA was used as an internal control. Measurements were performed with three separate cell samples for each gene and were replicated in triplicate. Data were analyzed with a normalized gene expression method (2–ΔΔCt) as previously described (Pfaffl, 2001).
Growth Rate Determination
Bacterial growth rates were determined as previously described (Luo et al., 2015). Briefly, each strain was activated on an LB plate supplemented with 5% sheep blood overnight at 37°C. A single colony from each strain was inoculated into 5 ml of TSB and cultured at 37°C with agitation for 10 h. Subsequently, each culture was adjusted to an OD600 of 0.05 in 20 ml of fresh TSB and grown at 37°C with shaking at 180 rpm. OD600 for each culture was determined at every 1 h for 18 h.
Total Membrane Extraction, Separation of Inner Membrane and Outer Membrane, and Western Blot
Riemerella anatipestifer total membrane, inner membrane, and outer membrane were extracted and separated based on methods previously described by Hu D. et al. (2019), Thein et al. (2010), and Osborn and Munson (1974). Cells were grown in 1 L TSB to OD600 ≈ 3 at 37°C. Chloramphenicol was then added to the culture at 1 mg/ml final concentration to stop protein synthesis and cells with chloramphenicol were agitated for another hour to ensure that all localization processes are completed. Cells were harvested and resuspended in 10 ml of 0.2 M Tris–HCl, pH 8.0, 1 M sucrose, and 1 mM EDTA, and lysozyme was added to a final concentration of 1 mg/ml. The cells were incubated on ice for 10 min. Spheroplast was prepared by slowly mixing 40 ml ice-cold H2O into the cell suspension. The cells were collected by centrifugation at 200,000 × g for 45 min at 4°C. The cell pellet was resuspended in 10 ml of ice-cold 10 mM Tris–HCl, pH 7.5, 5 mM EDTA, 0.2 mM DTT, and 1 mg/ml DNase. The cells were lysed by passing through a French Press twice at 108 Pa. The sample after French Press was centrifuged at ∼3,000 × g for 15 min to remove cell debris. The supernatant was ultracentrifuged at 120,000 × g for 2 h at 4°C to collect total membrane. The total membrane pellet was resuspended in 1 ml of ice-cold 10 mM Tris–HCl, pH 7.5, 15% sucrose (w/v), 5 mM EDTA, and 0.2 mM DTT. Inner membrane and outer membrane were separated on a sucrose step gradient (1 ml of 55% sucrose and 2.25 ml each of 50, 45, 35, and 30% sucrose). The total membrane suspension was placed on top of the sucrose gradient and centrifuged at 250,000 × g for 12 h at 4°C. The outer membrane (lower band, higher density) and inner membrane (upper band, lower density) were extracted by syringes. Each sample was washed three times with 1 ml of Tris buffer (10 mM Tris–HCl, pH 7.5, and 1 mM EDTA) and centrifuged at 100,000 × g for 20 min to collect the membrane samples. The samples were resuspended in SDS-PAGE sample loading buffer for analysis. Whole cell samples were prepared by collecting 2 ml of cells and resuspended in 1 ml of SDS-PAGE sample loading buffer.
His-tagged B739_0832 expression plasmid [pET32a(+)-ompH] was obtained in our previous study (Gao et al., 2016). His-tagged B739_0832 protein was purified and used to generate rabbit polyclonal antibody, as described before (Zhang et al., 2017). Whole cell, total membrane, inner membrane, and outer membrane of R. anatipestifer samples were analyzed using Western blot assay; OmpA was used as an outer membrane protein reference, TonB was used as an inner membrane protein reference, and RecA was used as a cytoplasmic protein reference (Thein et al., 2010; Liao et al., 2015; Xu et al., 2020).
Assessment of LD50
Groups of 3-day-old Cherry Valley ducklings were used to assess LD50 of ΔB739_0832 and wild-type strains. Ducklings were divided into 16 groups (10 ducklings per group, 160 total): 5 groups were challenged with wild-type R. anatipestifer CH-1, 5 groups with ΔB739_0832, 5 groups with CΔB739_0832, and 1 group with saline control. The wild-type group was intramuscularly injected at a dose of 106 to 1010 CFU; the ΔB739_0832 group was injected at a dose of 107 to 1011 CFU, the CΔB739_0832 group was injected at a dose of 106 to 1010 CFU, and the control group was injected with 1 ml of sterile phosphate-buffered saline (PBS). LD50 values were calculated using SPSS 23.0 (Arambašic and Randhawa, 2014).
Determination of Bacterial Load in Infected Duck Tissues
Three groups of 3-day-old Cherry Valley ducklings (15 ducklings per group) were intramuscularly injected with wild-type, ΔB739_0832, or CΔB739_0832 at a dose of 109 CFU, respectively. After challenge, blood, liver, heart, brain, and spleen tissues samples were collected at 6, 12, 24, 36, 48, and 72 h. Three ducklings were randomly selected for sacrifice at each time point. The organ samples were weighed and transferred into tubes each containing 3 ml of sterilized PBS. After homogenization, the tubes were centrifuged for 5 min at 2,000 × g to remove cell debris, the supernatant of each tube was serially diluted with PBS, and 50 μl of each serial dilution was plated on a TSA plate. TSA plates were incubated at 37°C overnight for bacterial count.
Assessment of Duck Survival Rate
Forty 3-day-old Cherry Valley ducklings were randomly divided into four groups (10 ducks per group). One group was challenged with a dose of 1010 CFU wild type, one with ΔB739_0832, one with CΔB739_0832, and the fourth group as a control was intramuscularly injected with equal volume (1 ml) PBS. The ducklings were observed for 10 consecutive days after the challenge. All ducklings were indoor and had access to plenty of food and water. Survival rates were calculated each day as the proportion of living ducklings accounted for the initial duckling counts.
Bacterial Adhesion Assay
Bacterial adhesion assay was performed with duck embryo fibroblast (DEF) cells as previously described (Hu et al., 2011). Briefly, each well in a 24-well tissue culture plate was seeded with 1 ml of 2 × 105 cells/ml DEF cells in Dulbecco’s Modified Eagle Medium (DMEM; Biowest, France) and incubated at 37°C with 5% CO2 for 18 h. After confirming that there was at least 95% confluence and has no contamination, each well was infected with 107 CFU R. anatipestifer (multiplicity of infection MOI = 50:1). The plates were then incubated at 37°C with 5% CO2 for another 1.5 h. After incubation, the wells were washed three times with PBS to remove non-adherent bacteria and then incubated at 37°C with 5% CO2 for 10 min in the presence of 0.25% trypsin (100 μl/well) to release the DEF cells from the wells. Serial 10-fold dilutions were prepared from the cell suspensions and 50 μl of each dilution was plated onto TSA plates to determine adhered bacteria counts. Each assay was performed in triplicate and replicated three times.
Bacterial Invasion Assay
Bacterial invasion assay was also performed with DEF cells as previously described (Hu et al., 2011). DEF cells were grown in 24-well tissue culture plates and then infected with R. anatipestifer the same way as the adhesion assay described above. For the invasion assay, after the infection incubation, 100 μg/ml gentamicin was added to each well and the plate was incubated for an additional 1 h at 37°C to kill all extracellular bacteria. After the extra incubation, the wells were washed three times with PBS and treated with 100 μl of 1% Triton X-100 to lyse the DEF cells. Lysed cells were homogenized. Serial 10-fold dilutions were prepared from the cell lysate and 50 μl of each dilution was plated onto TSA plates to determine invasive bacteria counts. Each assay was performed in triplicate and replicated three times.
Statistical Analysis and Ethics Statement
Statistical analysis was performed with GraphPad Prism 7.0 for Windows (GraphPad Software Inc., San Diego, CA, United States) (Hu Y. C. et al., 2019). Significance of difference between two data sets was evaluated using Student’s t-test, and a value of p < 0.05 was considered significant (Mishra et al., 2019).
Three-day-old Cherry Valley ducklings were procured from Sichuan Agricultural University duck farm and kept under appropriate conditions with a 12-h light/dark cycle and free access to food and water during this study. All ducks were handled in strict adherence to the recommendations of the local animal welfare bodies and Sichuan Agricultural University (No. XF2014-18). The animal-use procedures were approved by the Animal Ethics Committee of Sichuan Agricultural University (Approval No. 2016-015).
Results
Bioinformatics Analysis of B739_0832 Locus in RA CH-1 Strain
In the National Center for Biotechnology Information (NCBI) database, the B739_0832 locus in RA CH-1 has been identified as a 501-base-pair ORF, which encodes a 166-amino acid protein, with a molecular mass of about 18 kDa. The B739_0832 protein has been annotated as an OmpH family outer membrane protein. We analyzed the amino acid sequences of B739_0832 proteins from all sequenced R. anatipestifer strains using the protein–protein Basic Local Alignment Search Tool (BLASTP). The sequence alignment results from BLASTP indicated over 95% identity among different R. anatipestifer strains.
OmpH (also known as Skp) proteins in some Gram-negative bacteria have been shown to be either outer membrane protein or chaperone proteins for outer membrane proteins. We compared the amino acid sequence of B739_0832 protein from R. anatipestifer strains with the OmpH (Skp) sequences from E. coli, Salmonella, and Pasteurella using Clustal Omega, a multiple sequence alignment tool from EMBL-EBI (Madeira et al., 2019; Supplementary Figure 2). The amino acid sequence alignment results indicated that the B739_0832 protein is closer in evolution to OmpH from Pasteurella than those from E. coli or Salmonella. We also analyzed the hydrophobicity properties of the B739_0832 protein using ExPASY software from SIB Swiss Institute of Bioinformatics (results not shown). The results indicated that, of the 166 residues in B739_0832 protein, 63 of them are hydrophobic, and both carboxyl and amino ends of the protein show more hydrophobicity than the middle portion. The results are consistent with outer membrane protein propensities.
Construction and Characterization of ΔB739_0832 Strain and Complemented Strain CΔB739_0832
To elucidate functions of B739_0832 in RA CH-1, we constructed a B739_0832 clean deletion strain (ΔB739_0832) and a complemented strain (CΔB739_0832) using the natural transformation method as described in “Materials and Methods.” Deletion of the B739_0832 gene was confirmed by PCR amplification using primers flanking the locus. PCR amplification of the 16S rRNA gene was performed at the same time as a positive control. The confirmed B739_0832 deletion mutant strain was named as ΔB739_0832. The B739_0832 gene from the RA CH-1 wild-type strain was amplified and cloned into a pLM02 vector plasmid to generate a recombination plasmid pLMF02:B739_0832. This recombination plasmid was introduced into the ΔB739_0832 strain by conjugation to yield the complemented strain CΔB739_0832.
We compared the growth rates of wild-type RA CH-1, ΔB739_0832, and CΔB739_0832 strains. The growth rate of the mutant strain ΔB739_0832 showed no significant difference from that of the wild-type strain and the complemented strain (Figure 1A). We further tested transcription levels of genes upstream (B739_0831) and downstream (B739_0833) of the B739_0832 locus. As shown in Figure 1B, the transcription levels of upstream and downstream genes were not affected by the deletion of B739_0832, which indicates that deletion of B739_0832 did not cause polar effect.
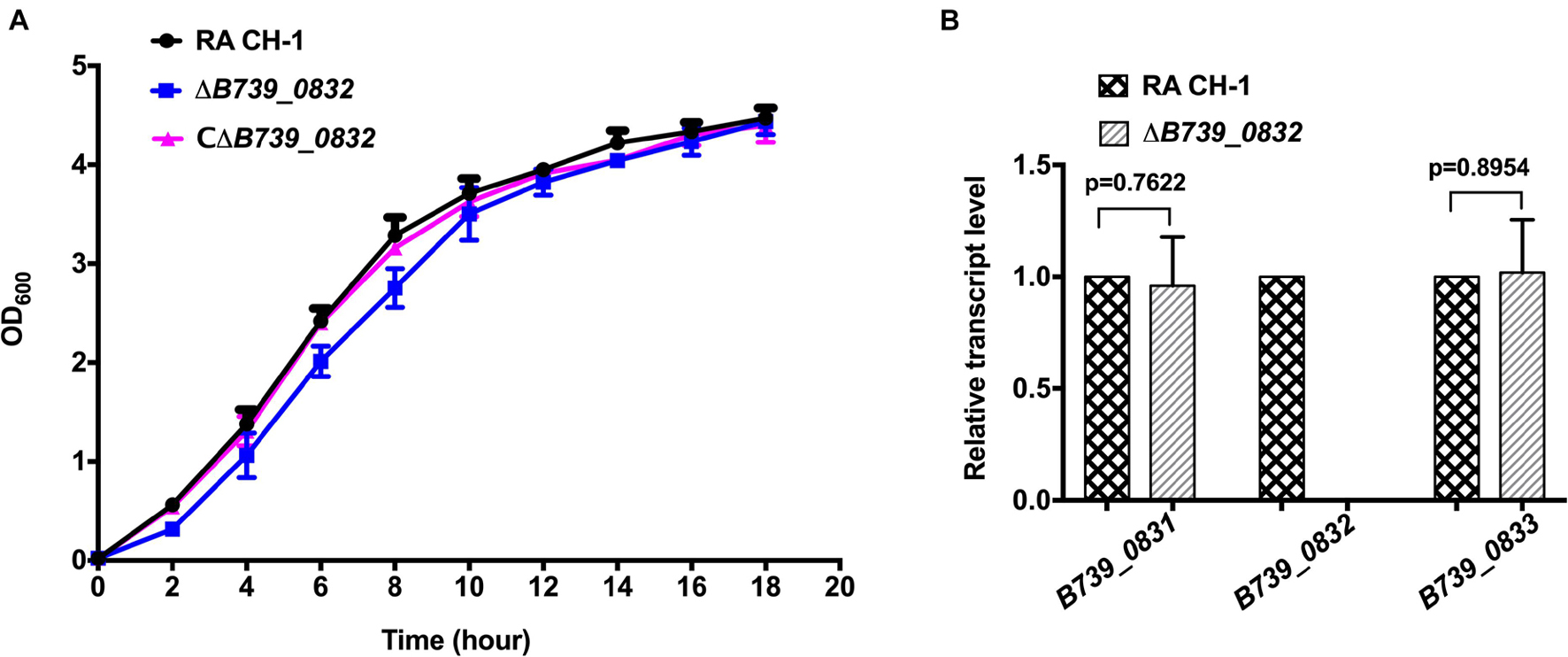
Figure 1. Characterization of mutant strain ΔB739_0832 and complemented strain CΔB739_0832. (A) Growth curves of Riemerella anatipestifer CH-1, ΔB739_0832, and CΔB739_0832 strains in Tryptic Soy Broth (TSB). Cells were inoculated in 25 ml of fresh TSB at 37°C with an initial OD600 of 0.05. OD600 values for each culture were subsequently measured every 2 h for 18 h. Data represent the mean values of three experiments. (B) Gene transcription levels in ΔB739_0832 strain were analyzed using quantitative PCR (qPCR). Transcription levels of B739_0832 and the flanking genes B739_0831 and B739_0833 in R. anatipestifer CH-1 and ΔB739_0832 strains were measured. Expression of B739_0832 was completely inactivated in the ΔB739_0832 mutant strain. Expression of upstream gene B739_0831 and downstream gene B739_0833 had no significant difference compared to wild type. Data were analyzed using Student’s t-test. Error bars represent standard deviations of three independent repeats.
B739_0832 Protein Is an Outer Membrane-Associated Protein
To determine localization of B739_0832 protein in R. anatipestifer cells, we isolated inner membrane, outer membrane, and total membrane subcellular fractions in RA CH-1 cells and compared the localization of B739_0832 to known outer membrane protein OmpA (outer membrane porin protein), known inner membrane protein TonB (energy transducer), and known cytoplasmic protein RecA (DNA maintenance and repair protein; Figure 2). In order to avoid false positive caused by proteins during transport, we added 1 mg/ml chloramphenicol to stop protein synthesis and incubated the bacterial cells for another 40 min to ensure all fully synthesized proteins are transported to their final destinations. B739_0832 protein was detected in outer membrane fraction, similar to OmpA, but not in inner membrane fraction. The localization data are consistent with the bioinformatics data that B739_0832 is more closely related in evolution to OmpH from Pasteurella than OmpH (Skp) from E. coli. It is also consistent with our previous ELISA study that B739_0832 is an exposed antigen in live R. anatipestifer cells.
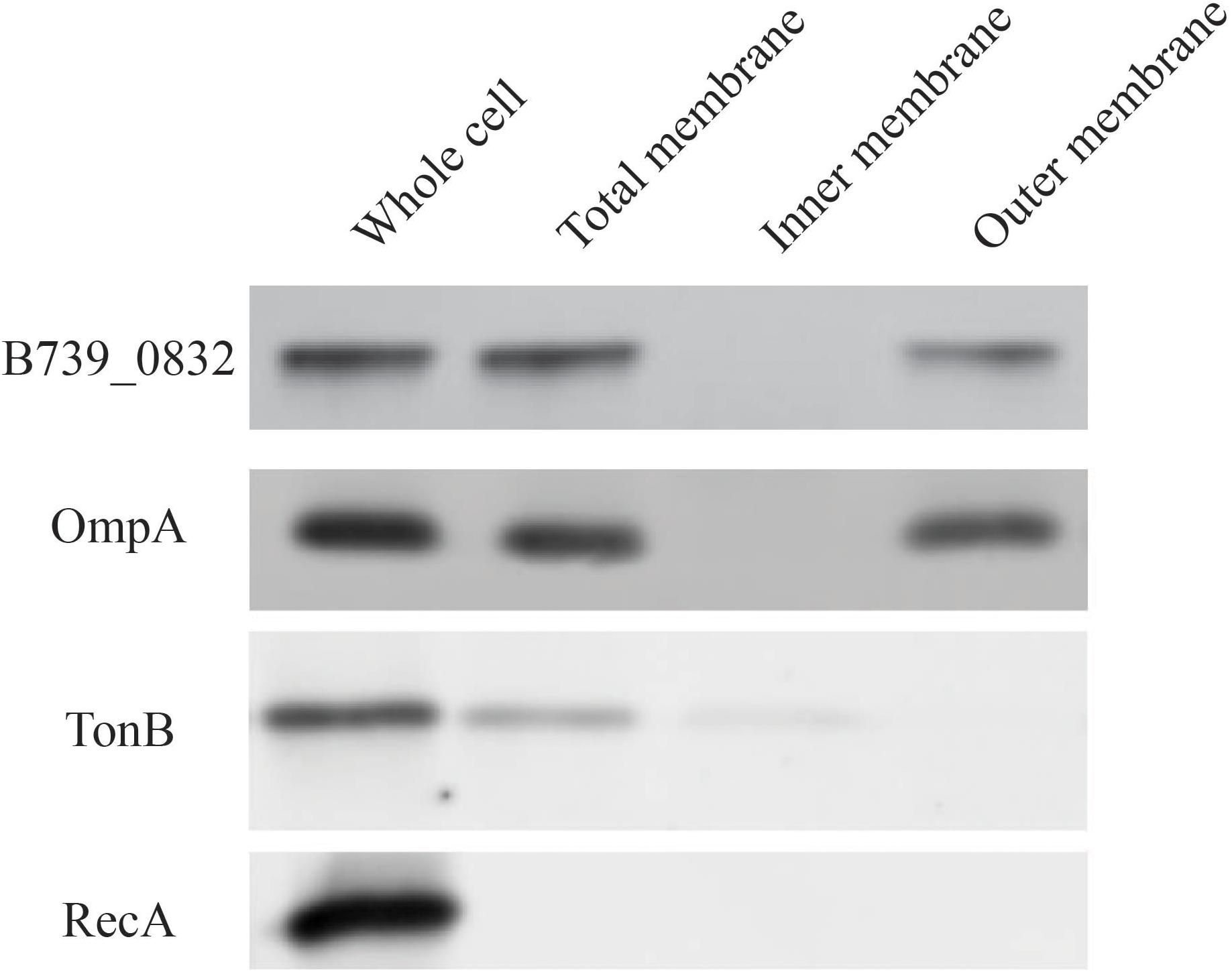
Figure 2. Membrane localization of R. anatipestifer CH-1 B739_0832 protein. Isolated subcellular fractions of R. anatipestifer cells were prepared as described in “Materials and Methods” and analyzed using Western blot. Results indicate that B739_0832 protein localizes in the outer membrane. OmpA is a confirmed outer membrane protein in R. anatipestifer, RecA is a known cytoplasmic protein and TonB is a known inner membrane protein. See Supplementary Figure 3 in the Supplementary Material for more information.
ΔB739_0832-Infected Ducklings Have Increased Survival Rate Than Those Infected by Wild Type
To determine the impact of B739_0832 deletion on RA CH-1 virulence, we measured the mortality rates in ducklings caused by RA CH-1, ΔB739_0832, and CΔB739_0832 strains. Groups of 3-day-old Cherry Valley ducklings were infected by one of these three strains at a dose of 1010 CFU and were observed for 10 days. At day 7 after infection, ducklings infected by ΔB739_0832 had about 70% survival rate, whereas ducklings infected by wild-type RA CH-1 or complemented strain CΔB739_0832 only had about 10% survival rate. After 10 days, no ducklings survived from infection by RA CH-1, and only 10% survived infection from CΔB739_0832, whereas 50% ducklings survived from infection by ΔB739_0832. The wild-type and complementary groups had similar patterns; about half of the ducklings in these two groups did not survive for more than 5 days. However, survival rates in the ΔB739_0832 group were significantly different from those in the wild-type and complementary groups (Figure 3).
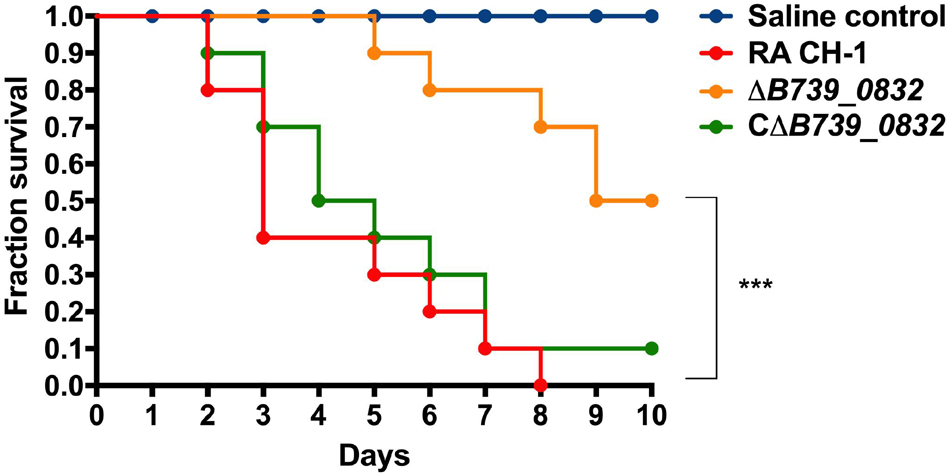
Figure 3. Survival rate in ducklings infected by R. anatipestifer CH-1, ΔB739_0832, or CΔB739_0832 strains. Each group has 10 3-day-old ducklings, which was injected intramuscularly at a dose of 1010 CFU to assess the survival rate. The group shown in red was injected with wild-type R. anatipestifer CH-1. The orange group was injected with ΔB739_0832. The green was injected with CΔB739_0832. Blue represents the control injected with phosphate-buffered saline (PBS). Data were analyzed using a Log-rank (Mantel–Cox) test. Three stars indicate significant difference (p < 0.001).
To further quantify the impact of B739_0832 deletion on RA CH-1 virulence, we measured half lethal dose (LD50) of these three strains. The LD50 of RA CH-1 was 3.98 × 108, which was about 150 times lower than that of the ΔB739_0832 strain (6.09 × 1010). The LD50 of complemented strain CΔB739_0832 was 7.76 × 108, which was similar to wild type.
Deletion of B739_0832 Gene Decreased Riemerella anatipestifer Adhesion and Invasion in Duck Embryo Fibroblast Cells
To assess whether deletion of B739_0832 gene affected adherence and invasion activities of R. anatipestifer, the activities of wild-type, ΔB739_0832, and CΔB739_0832 strains were measured using DEF cells. DEF cells were infected at MOI of 50:1; the ΔB739_0832 strain had 8.77 ± 1.17 × 103 CFU/well adhesion activity, which was approximately threefold lower than that of wild type (3.44 ± 0.16 × 104 CFU/well; Figure 4A). Bacterial invasion tests were performed under similar testing conditions. After killing all extracellular bacteria by gentamicin, bacterial counts inside host cells infected by ΔB739_0832 strain were 1.36 ± 0.1 × 103 CFU/well, which was approximately twofold lower than those infected by wild type (2.44 ± 0.2 × 103; Figure 4B). The complemented strain CΔB739_0832 had almost identical activities as wild type for both adhesion and invasion tests.
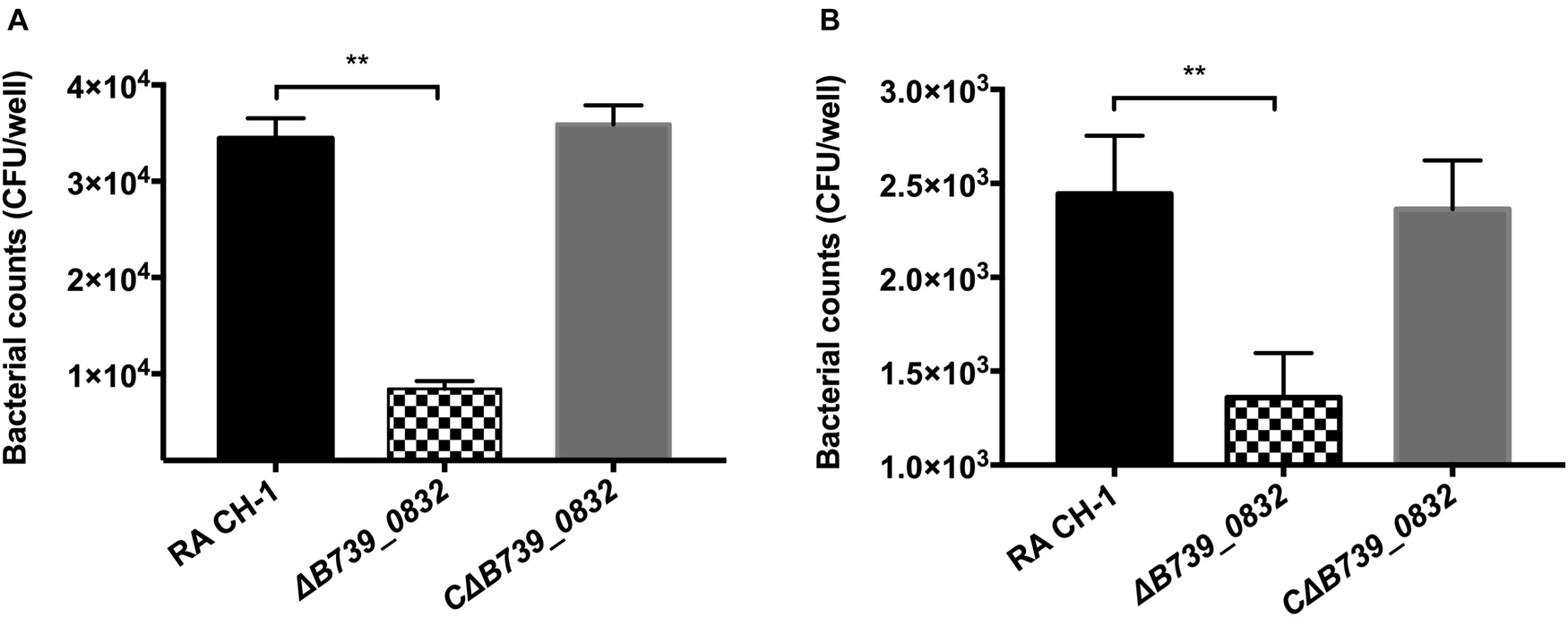
Figure 4. Adherence and invasion assays for R. anatipestifer CH-1, ΔB739_0832, and CΔB739_0832 strains. Both adherence and invasion assay were performed using duck embryo fibroblast (DEF) cells. (A) Bacteria count from adherence assay measurements. ΔB739_0832 showed significantly lower bacterial count, but maintained some level of host cell adherence ability. (B) Measurements of bacterial invasion ability. ΔB739_0832 showed significantly lower host cell invasion ability, but did not completely abolish its invasion ability. All data were analyzed with Student’s t-test. Error bars represent standard deviation from three sets of independent measurements (**p < 0.01).
Deletion of B739_0832 Gene Attenuated Riemerella anatipestifer Virulence
To further evaluate the influence of ΔB739_0832 on systemic infection in vivo, bacterial loads in blood, liver, spleen, and brain from ducks infected by wild type, ΔB739_0832, or CΔB739_0832 were quantified. Bacterial loads from all three groups were almost identical for the first 24 h after infection, with the exception of bacterial loads in liver. However, a difference slowly developed at 36 and 48 h (Figures 5A–D). In brain and blood, the difference developed at 36 h, earlier than in spleen, which did not show significant difference until 48 h, whereas, in liver, the bacterial loads were different since 12 h post-infection, and the difference grew more significant at 48 h (Figure 5C).
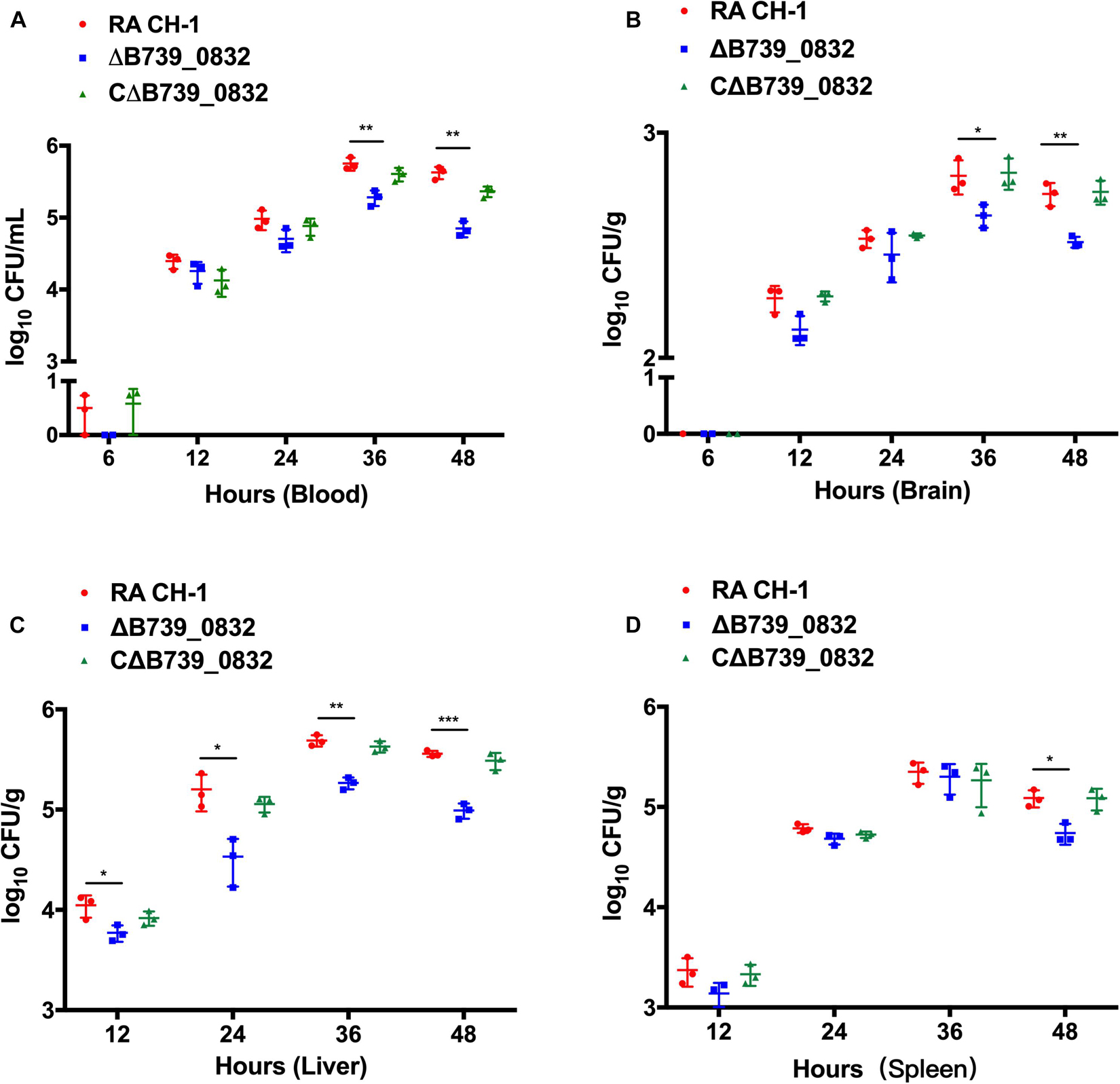
Figure 5. Bacterial loads in blood (A), brain (B), liver (C), and spleen (D) of ducks infected with R. anatipestifer CH-1, ΔB739_0832, or CΔB739_0832 strains at 6, 12, 24, 36, and 48 h post-infection. Error bars represent standard deviations from three independent experiments (***p < 0.001, **p < 0.01, and *p < 0.05).
To further examine the effects of deletion of B739_0832 gene on virulence, we compared organ tissue lesions in ducks infected by wild-type, ΔB739_0832, or CΔB739_0832 strains. At 36 h post-infection, we collected heart, liver, spleen, and brain tissue samples from groups of ducklings infected by each of the three strains for histopathological examination. The tissue samples were stained with hematoxylin and eosin to visualize lesions caused by R. anatipestifer invasion. All brain tissues exhibited no visible damage, suggesting that R. anatipestifer had not passed the blood–brain barrier yet at this time point. However, liver cord disorders, involving a large number of vacuole-like changes, were clearly visible in samples infected by wild-type and complementary strain CΔB739_0832. Myocardial necrosis was also present in heart samples infected by wild-type and CΔB739_0832 strains. However, there were no visible lesions in liver or heart samples infected by the ΔB739_0832 strain (Figure 6).
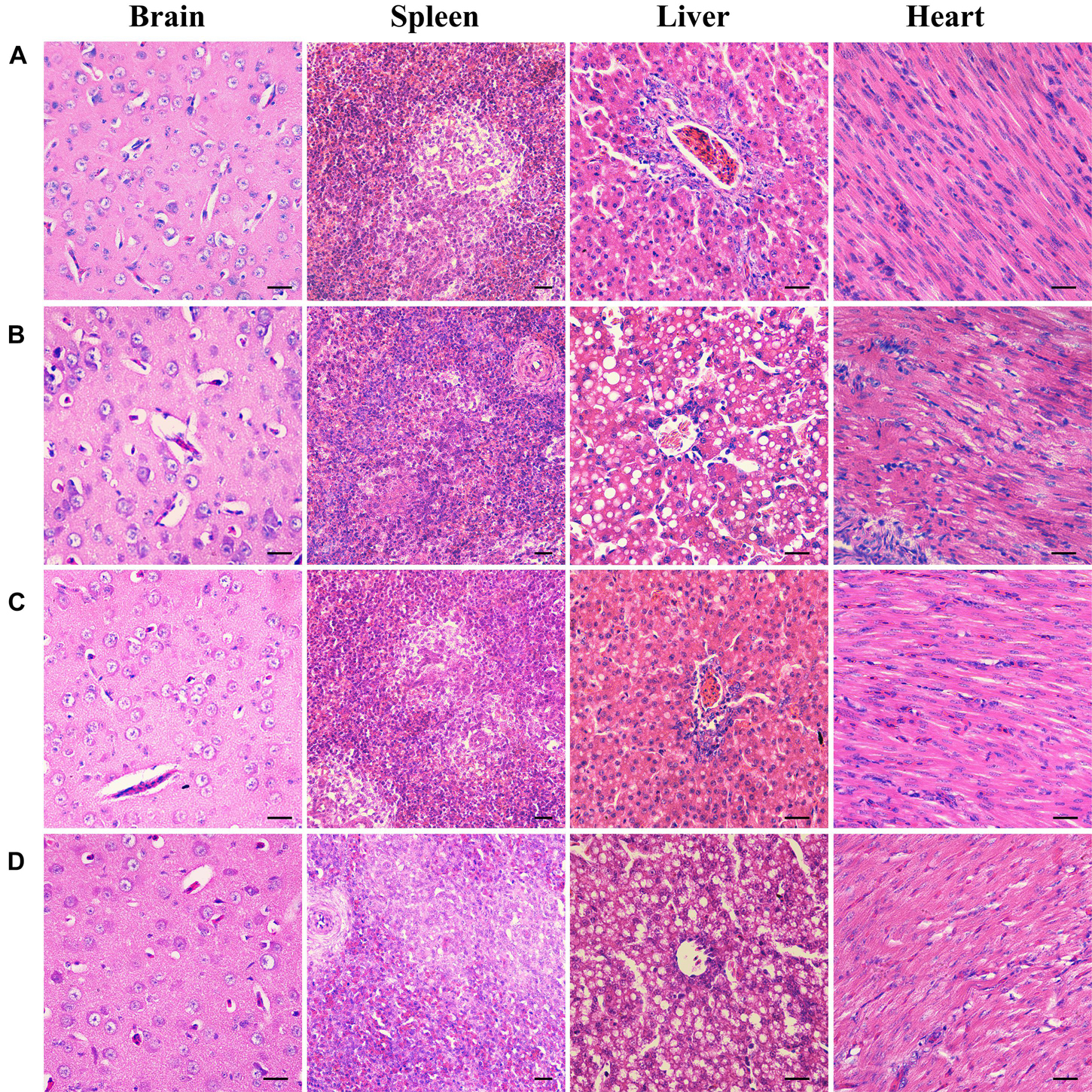
Figure 6. Organ histopathological changes in ducks infected by R. anatipestifer. (A) Brain, spleen, liver, and heart samples from a healthy duck. (B) Corresponding samples from ducks infected by wild-type R. anatipestifer CH-1 at 36 h post-infection. (C) Corresponding samples from ducks infected by mutant strain ΔB739_0832 at 36 h post-infection. (D) Corresponding samples from ducks infected by complement strain CΔB739_0832 at 36 h post-infection. For both brain and spleen tissues, samples from all three strains showed no lesions. For R. anatipestifer CH-1-infected ducks, obvious pathological changes were observed in liver and heart: liver cord disorder, a large area of vacuole-like changes and myocardial necrosis. ΔB739_0832 mutant strain-infected ducks showed no pathological changes in liver and heart. CΔB739_0832-infected ducks had the same histopathological changes as the wild-type strain. Tissue samples were stained with hematoxylin and eosin (400×). The size of the scale bar is 20.0 μm.
Discussion
Riemerella anatipestifer can infect a variety of domestic and wild birds, such as ducks, geese, and turkeys. Therefore, it is important to understand the pathogenic mechanisms of R. anatipestifer for controlling its spread. It is well known that outer membrane proteins in Gram-negative bacteria play important roles in stimulation of host immune systems (Navidinia et al., 2019). OmpH has been demonstrated to be a major outer membrane protein present in P. multocida envelope (Lugtenberg et al., 1986). A study has shown that purified native OmpH protein from the P. multocida A:3 strain could be used to elicit immune responses providing homologous protection in chickens (Luo et al., 1997), supporting OmpH in P. multocida as an exposed outer membrane protein. However, OmpH in E. coli and S. typhimurium, also known as Skp, has been identified as a periplasmic chaperone protein, despite initial reports of it as DNA binding protein (Holck et al., 1987), or outer membrane protein (Koski et al., 1989).
Our data suggest that B739_0832 protein in R. anatipestifer, a putative OmpH protein, is probably an outer membrane protein, consistent with it being more closely related to OmpH in P. multocida than in E. coli or S. typhimurium. The localization data suggest that B739_0832 protein is probably outer membrane protein, or at least outer membrane associated due to the limitation of our methods (Figure 2). Bioinformatics analysis found that over one-third amino acids in B739_0832 protein are hydrophobic, further suggesting that B739_0832 protein could be a membrane-inserted protein. The impacts of B739_0832 gene deletion on R. anatipestifer virulence illustrated in this study, combining our previous study in successfully developing B739_0832 protein-based ELISA method for detecting live R. anatipestifer cells, suggest that B739_0832 in R. anatipestifer is probably an outer membrane protein, or at least is a membrane-associated protein.
Studies have shown that OmpH proteins in both P. multocida and E. coli can associate with lipopolysaccharide (LPS; Luo et al., 1997; Bulieris et al., 2003). LPS, as a cell wall component characteristic of Gram-negative bacteria, is a pathogen-associated molecule that plays an important role in triggering bacteria-infected host innate immune responses. Luo et al. believed that it was OmpH protein, not the small amount LPS contaminant, that elicited immune response in turkeys, whereas, in E. coli, Skp protein is more involved in facilitating outer membrane proteins through the periplasmic region. Skp’s association with LPS may partly be due to the numerous positively charged amino residues distributed throughout the protein and probably has no direct role as an antigen. In this study, we found that B739_0832 affects R. anatipestifer virulence and is likely involved in host cell attachment and invasion. It is very possible, due to its similarity to OmpH proteins in P. multocida and E. coli, that B739_0832 in R. anatipestifer is also associated with LPS. However, it is not clear what role, if any, LPS plays in the virulence effects of B739_0832. The relationship and association between B739_0832 and LPS need to be studied further and will probably shed light on pathogenesis mechanisms of R. anatipestifer.
Our results show that B739_0832, although not required for growth, is heavily involved in R. anatipestifer virulence. Deletion of B739_0832 gene greatly reduced its virulence: effecting lower mortality rate, higher survival rate in ducklings infected by ΔB739_0832, and much higher LD50. ΔB739_0832 also showed attenuated pathogenic effects on host organs (no visible lesions found). However, only 50% of ducklings infected by ΔB739_0832 survived for over 10 days, suggesting that B739_0832, although important for virulence, is not critical for pathogenesis. In fact, ΔB739_0832 could still attach and invade DEF cells and was detected in all examined host organs, albeit at lower amount than wild type. These lines of evidence strongly suggest that B739_0832 is involved in the initial invasion process of R. anatipestifer. It could be part of the host cell attachment process, which further confirms that B739_0832 is an outer surface protein.
The striking differences between B739_0832 and E. coli Skp protein are underscored in their sequence differences. Charged residues, especially positively charged residues, in E. coli Skp have been shown to play important functional roles (Bulieris et al., 2003). However, B739_0832 has fairly different distribution of charged residues than E. coli Skp. For example, comparing residues 21–45, about one-third (eight residues) of the total 25 residues changed from neutral to charged residues in B739_0832. This type of asymmetric distribution of charged residues is often observed in outer membrane proteins (Slusky and Dunbrack, 2013). It appears that these two proteins diverged at some point during evolution: one became an outer membrane protein, whereas another became a chaperone for outer membrane protein. However, it is not clear which role was earlier in the evolution process.
In summary, our earlier study showed that B739_0832-based ELISA could be used to detect R. anatipestifer of different serotypes with high sensitivity. In this study, we provide further evidence that B739_0832 is an outer membrane protein. We also demonstrated, for the first time, that B739_0832 is involved in R. anatipestifer virulence. B739_0832 is highly conserved among different serotypes. Its involvement in virulence further supports that B739_0832 is a good candidate as a universal antigen for developing vaccine for all serotypes.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by Animal Ethics Committee of Sichuan Agricultural University (Approval No. 2016-015).
Author Contributions
QG and AC conceived and designed the experiments and wrote the manuscript. QG and SL performed the experiments. QG, AC, MW, RJ, SC, DZ, ML, XZ, QY, YW, SZ, JH, SM, XO, DS, and BT developed the methods. All authors read and approved the final manuscript.
Funding
This work was supported by the China Agricultural Research System (CARS-42-17) and Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2020-18).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.708225/full#supplementary-material
References
Arambašic, M. B., and Randhawa, M. A. (2014). Comparison of the methods of finney and miller-tainter for the calculation of LD50 values. World Appl. Sci. J. 32, 2167–2170. doi: 10.5829/idosi.wasj.2014.32.10.9132
Bulieris, P. V., Behrens, S., Holst, O., and Kleinschmidt, J. H. (2003). Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide. J. Biol. Chem. 278, 9092–9099. doi: 10.1074/jbc.M211177200
Caruana, J. C., and Walper, S. A. (2020). Bacterial membrane vesicles as mediators of microbe - microbe and microbe - host community interactions. Front. Microbiol. 11:432–456. doi: 10.3389/fmicb.2020.00432
Chang, F. F., Chen, C. C., Wang, S. H., and Chen, C. L. (2019). Epidemiology and antibiogram of riemerella anatipestifer isolated from waterfowl slaughterhouses in taiwan. J. Vet. Res. 63, 79–86. doi: 10.2478/jvetres-2019-0003
Chu, C. Y., Liu, C. H., Liou, J. J., Lee, J. W., and Cheng, L. T. (2015). Development of a subunit vaccine containing recombinant Riemerella anatipestifer outer membrane protein A and CpG ODN adjuvant. Vaccine 33, 92–99. doi: 10.1016/j.vaccine.2014.11.010
Crasta, K. C., Chua, K. L., Subramaniam, S., Frey, J., Loh, H., and Tan, H. M. (2002). Identification and Characterization of CAMP cohemolysin as a potential virulence factor of Riemerella anatipestifer. J. Bacteriol. 184, 1932–1939. doi: 10.1128/jb.184.7.1932-1939.2002
Gao, Q., Tang, T., Cheng, A. C., Wang, M. S., Jia, R. Y., Zhu, D. K., et al. (2016). Development of an indirect ELISA using recombinant ompH protein for serological detection of Riemerella anatipestifer infection in ducks. Int. J. Clin. Exp. Med. 9, 1330–1337.
Hatfaludi, T., Hasani, K. A., Boyce, J. D., and Adler, B. (2010). Outer membrane proteins of Pasteurella multocida. Vet. Microbiol. 144:27. doi: 10.1016/j.vetmic.2010.01.027
Holck, A., Lossius, I., Aasland, R., and Kleppe, K. (1987). Purification and characterization of the 17 K protein, a DNA-binding protein from Escherichia coli. Biochim. Biophys. Acta 914, 49–54.
Hu, D., Guo, Y. Q., Guo, J., Wang, Y., Pan, Z., Xiao, Y. C., et al. (2019). Deletion of the Riemerella anatipestifer type IX secretion system gene sprA results in differential expression of outer membrane proteins and virulence. Avian Pathol. 48, 191–203. doi: 10.1080/03079457.2019.1566594
Hu, Q. H., Ding, C., Tu, J., Wang, X. L., Han, X. G., Duan, Y. B., et al. (2012). Immunoproteomics analysis of whole cell bacterial proteins of Riemerella anatipestifer. Vet. Microbiol. 157, 428–438. doi: 10.1016/j.vetmic.2012.01.009
Hu, Q. H., Han, X. G., Zhou, X. J., Ding, C., Zhu, Y. Y., and Yu, S. Q. (2011). OmpA is a virulence factor of Riemerella anatipestifer. Vet. Microbiol. 150, 278–283. doi: 10.1016/j.vetmic.2011.01.022
Hu, Y. C., Cheng, L. X., Zhong, W., Chen, M. H., and Zhang, Q. (2019). Bioinformatics analysis of gene expression profiles for risk prediction in patients with septic shock. Med. Sci. Monit. 25, 9563–9571. doi: 10.12659/MSM.918491
Kang, M., Seo, H. S., Soh, S. H., and Jang, H. K. (2018). Immunogenicity and safety of a live Riemerella anatipestifer vaccine and the contribution of IgA to protective efficacy in Pekin ducks. Vet. Microbiol. 222, 132–138. doi: 10.1016/j.vetmic.2018.07.010
Kim, S. H., Jung, D. I., Yang, I. Y., Kim, J., Lee, K. Y., Nochi, T., et al. (2011). M cells expressing the complement C5a receptor are efficient targets for mucosal vaccine delivery. Eur. J. Immunol. 41, 3219–3229. doi: 10.1002/eji.201141592
Koski, P., Rhen, M., Kantele, J., and Vaara, M. (1989). Isolation. cloning, and primary structure of a cationic 16-kda outer membrane protein of salmonella typhimurium. J. Biol. Chem. 264, 18973–18980. doi: 10.1016/s0021-9258(19)47253-0
Liao, H. B., Cheng, X. J., Zhu, D. K., Wang, M. S., Jia, R. Y., Chen, S., et al. (2015). TonB energy transduction systems of riemerella anatipestifer are required for iron and hemin utilization. PLoS One 2015:127506. doi: 10.1371/journal.pone.0127506
Liu, M. F., Wang, M. Y., Zhu, D. K., Wang, M. S., Jia, R. Y., Chen, S., et al. (2016). Investigation of TbfA in Riemerella anatipestifer using plasmid-based methods for gene over-expression and knockdown. Sci. Rep. 6, 37159–37168. doi: 10.1038/srep37159
Liu, M. F., Zhang, L., Huang, L., Biville, F., Zhu, D. K., Wang, M. S., et al. (2017). Use of natural transformation to establish an easy knockout method in Riemerella anatipestifer. Appl. Environ. Microbiol. 83, 17–27. doi: 10.1128/AEM.00127-17
Lugtenberg, B., Boxtel, R. V., Evenberg, D., Jong, M. D., Storm, P., and Frik, J. (1986). Biochemical and immunological characterization of cell surface proteins of pasteurella multocida strains causing atrophic rhinitis in swine. Infect. Immun. 52, 175–183.
Luo, H. Y., Liu, M. F., Wang, L. Y., Zhou, W. S., Wang, M. S., Cheng, A. C., et al. (2015). Identification of ribosomal RNA methyltransferase gene ermF in Riemerella anatipestifer. Avian Pathol. 44, 162–168. doi: 10.1080/03079457.2015.1019828
Luo, Y. G., Glisson, J. R., Jackwood, M. W., Hancock, R. E. W., Bains, M., Cheng, I. H., et al. (1997). Cloning and characterization of the major outer membrane protein gene (ompH) of Pasteurella multocida X-73. J. Bacteriol. 179, 7856–7864.
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Mishra, P., Singh, U., Pandey, C. M., Mishra, P., and Pandey, G. (2019). Application of student’s t-test, analysis of variance, and covariance. Ann. Card. Anaesth. 22, 407–411. doi: 10.4103/aca.ACA_94_19
Navidinia, M., Soleimani, N., and Abadi, N. B. (2019). Effect of recombinant helicobacter outer membrane protein H (HopH) on nitric oxide production by peripheral macrophage in BALB/c Mice. Avicenna J. Med. Biotechnol. 11, 229–233.
Osborn, M. J., and Munson, R. (1974). Separation of the inner (cytoplasmic) and outer membranes of Gram-negative bacteria. Methods Enzymol. 31, 642–653.
Pathanasophon, P., Phuektes, P., Tanticharoenyos, T., Narongsak, W., and Sawada, T. (2002). A potential new serotype of Riemerella anatipestifer isolated from ducks in Thailand. Avian Pathol. 31, 267–270. doi: 10.1080/03079450220136576
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, 45–51.
Schäfer, U., Beck, K., and Müller, M. (1999). Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 274, 24567–24574.
Segers, P., Mannheim, W., Vancanneyt, M., Brandt, K. D., Hinz, K. H., Kersters, K., et al. (1993). Riemerella anatipestifer gene nov. comb. nov., the causative agent of septicemia anserum exsudativa, and its phylogenetic affiliation within the flavobacterium-cytophaga rRNA Homology Group. Int. J. Syst. Bacteriol. 43, 768–776.
Slusky, J. S. G., and Dunbrack, R. L. (2013). Charge asymmetry in the proteins of the outer membrane. Bioinformatics 29, 2122–2128. doi: 10.1093/bioinformatics/btt355
Thanasarasakulpong, A., Poolperm, P., Tankaew, P., Sawada, T., and Sthitmatee, N. (2015). Protectivity conferred by immunization with intranasal recombinant outer membrane protein H from Pasteurella multocida serovar A:1 in chickens. J. Vet. Med. Sci. 77, 321–326. doi: 10.1292/jvms.14-0532
Thein, M., Sauer, G., Paramasivam, N., Grin, I., and Linke, D. (2010). Efficient subfractionation of gram-negative bacteria for proteomics studies. J. Proteome Res. 9, 6135–6147.
Tian, X., Huang, L., Wang, M. S., Biville, F., Zhu, D. K., Jia, R. Y., et al. (2020). The functional identification of Dps in oxidative stress resistance and virulence of Riemerella anatipestifer CH-1 using a new unmarked gene deletion strategy. Vet. Microbiol. 247, 108730–108739. doi: 10.1016/j.vetmic.2020.108730
Wang, X. J., Liu, W. B., Zhu, D. K., Yang, L. F., Liu, M. F., Yin, S. J., et al. (2014). Comparative genomics of Riemerella anatipestifer reveals genetic diversity. BMC Genomics 15:479–489. doi: 10.1186/1471-2164-15-479
Wang, Y., Tang, C., Yu, X. H., Xia, M. Y., and Yue, H. (2010). Distribution of serotypes and virulence-associated genes in pathogenic Escherichia coli isolated from ducks. Avian Pathol. 39, 297–302. doi: 10.1080/03079457.2010.495742
Wang, Y., Yin, X. H., Zhou, Z. T., Hu, S. S., Li, S. W., Liu, M., et al. (2019). Cas9 regulated gene expression and pathogenicity in Riemerella anatipestifer. Microb. Pathog. 136, 103706–103714. doi: 10.1016/j.micpath.2019.103706
Weiser, J. N., and Gotschlich, E. C. (1991). Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Am. Soc. Microbiol. 59, 2252–2258.
Xu, X. X., Xu, Y. H., Miao, S., Jiang, P., Cui, J. S., Gong, Y. S., et al. (2020). Evaluation of the protective immunity of Riemerella anatipestifer OmpA. Appl. Microbiol. Biotechnol. 104, 1273–1281. doi: 10.1007/s00253-019-10294-3
Keywords: outer membrane protein, virulence factor, OmpH, B739_0832, Riemerella anatipestifer
Citation: Gao Q, Lu S, Wang M, Jia R, Chen S, Zhu D, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Huang J, Mao S, Ou X, Sun D, Tian B and Cheng A (2021) Putative Riemerella anatipestifer Outer Membrane Protein H Affects Virulence. Front. Microbiol. 12:708225. doi: 10.3389/fmicb.2021.708225
Received: 11 May 2021; Accepted: 27 August 2021;
Published: 20 September 2021.
Edited by:
Caterina Guzmán-Verri, National University of Costa Rica, Costa RicaReviewed by:
Kai Zhang, Texas Tech University, United StatesZutao Zhou, Huazhong Agricultural University, China
Copyright © 2021 Gao, Lu, Wang, Jia, Chen, Zhu, Liu, Zhao, Yang, Wu, Zhang, Huang, Mao, Ou, Sun, Tian and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=
†These authors have contributed equally to this work
 Qun Gao
Qun Gao Shuwei Lu
Shuwei Lu Mingshu Wang
Mingshu Wang Renyong Jia1,2,3
Renyong Jia1,2,3 Dekang Zhu
Dekang Zhu Mafeng Liu
Mafeng Liu Shaqiu Zhang
Shaqiu Zhang Juan Huang
Juan Huang Sai Mao
Sai Mao Xumin Ou
Xumin Ou Anchun Cheng
Anchun Cheng