- 1College of Veterinary Medicine, Northwest A&F University, Yangling, China
- 2Qingdao Adverse Drug Reaction Monitoring Center, China Qingdao Institute for Food and Drug Control, Qingdao, China
The genomic context of the mcr-1 gene in Escherichia coli from animal feces has been widely reported. However, less is known about the mcr-1-carrying plasmid characteristics and other functional regions of Escherichia coli isolates from animal organs with lesions. The present study investigated the antimicrobial resistance, population structure, and genetic features of mcr-1-positive Escherichia coli strains isolated from animal organs with lesions. The antimicrobial susceptibility testing indicated that 24 mcr-1-positive Escherichia coli isolates were resistant to at least three or all antimicrobial categories. MLST analysis suggested that the dominant clone complexes (CC) were mainly CC156, CC448, and CC10. In addition, ST10596, a newly discovered sequence type in swine, failed to be classified. Meanwhile, the mcr-1 gene located on the different plasmids was successfully transferred to the recipients, and whole-genome sequencing indicated the mcr-1 gene was embedded in mcr-1-pap2 cassette but not flanked by ISApl1. The mcr-1 gene is located on the chromosome and embedded in Tn6330. Furthermore, NDM-5 was found on the IncX3-type plasmid of J-8. The virB6 and traI gene of type IV secretion system (T4SS) were truncated by IS2 and IS100 and located on the IncX4- and the IncHI2/HI2A/N-type plasmids, respectively. The multidrug-resistant (MDR) region of IncHI2/HI2A/N-type plasmids contained two class 1 integrons (In0, In640) and four composite transposons (Tn4352, Tn6010, cn_4692_IS26, cn_6354_IS26). Overall, 24 mcr-1-positive Escherichia coli isolates in our study showed MDR, or even extensively drug resistant (XDR), and exhibited population diversity. The T4SS gene truncation by the insertion sequence may affect the efficiency of plasmid conjugative transfer. Furthermore, the class 1 integrons and composite transposons in the MDR region of IncHI2/HI2A/n-type plasmid contributed to the multireplicon plasmid formation, the acquisition, and transfer of antimicrobial resistance genes (ARGs).
Introduction
The overuse and misuse of antimicrobials have led to the rapid development of resistance in Gram-negative bacteria, and this has now become a serious public health crisis (Martin et al., 2020). Among these bacteria, Escherichia coli is the most common pathogen which causes animal and human diseases on a global scale while being a major cause of multiple acquired infections (Aslam et al., 2018). Over the past few years, the emergence and common occurrence of extended-spectrum beta-lactamase (ESBL) – and carbapenemase-producing E. coli have result in reduced effectiveness of antimicrobial treatment (Hordijk et al., 2020), such that doctors are now considering alternative antimicrobials, such as colistin.
For many years, colistin (polymyxin E) has been used in animal production for the treatment and prevention of infectious diseases. It was subsequently identified as a form of last-resort treatment against severe human infections caused by multidrug- and carbapenem-resistant Gram-negative bacteria (Kempf et al., 2016; Poirel et al., 2017). Colistin resistance was previously thought to be chromosome mediated, which mainly related to the point mutations of two-component systems (TCSs) PhoP/PhoQ and PmrA/PmrB and can transmit colistin resistance through vertical transmission (Olaitan et al., 2014). It was only in 2015 that the plasmid-mediated mcr-1 gene was first described in E. coli and Klebsiella pneumoniae. Both species were, respectively, isolated from pigs and humans in China and showed the horizontal transfer of colistin resistance (Liu et al., 2016). The emergence of this gene has further reduced the effectiveness of the alternative antimicrobials, thereby posing new challenges to public health. Since then, colistin-resistant mcr-1-positive E. coli isolates have been reported in the environment, humans, animals, and food worldwide (Falgenhauer et al., 2016; Luo et al., 2017; Shen et al., 2018). Numerous researches have shown that the high clonal diversity was observed among mcr-1-positive E. coli (MPECs) and mainly belonged to the common dominant clone complexes, such as ST10, ST48, ST165, etc. (Matamoros, 2017; Bai et al., 2018). Among these, ST10 is the most common transmission clone group and frequently disseminates through different means (Sellera et al., 2017; Patiño-Navarrete et al., 2020).
The mcr-1 gene is found in multiple plasmid types, including IncI2, IncX4, IncX3-X4, IncHI1, IncHI2, IncP, IncY, IncK2, IncFI, IncFI, IncFII, and IncFIB-I2 (Malhotra-Kumar et al., 2016; McGann et al., 2016; Poirel et al., 2016; Sun et al., 2016; Xavier et al., 2016; Zurfluh et al., 2016; Donà et al., 2017; Wang et al., 2017; Zhang et al., 2017). This was consistent with multiple mcr-1 mobilization events which tend to potentially facilitate the association of mcr-1 with other resistance mechanisms, thereby creating multidrug-resistant bacterial hosts (Zhong et al., 2018). Of these plasmids, IncI2-, IncX4-, and IncHI2-type ones are the most common vectors of mcr-1, with the insertion sequence (IS) ISApl1 mostly appearing up- or downstream of the gene (Matamoros, 2017). Few studies reported mcr-1-carrying hybrid plasmid and their mechanism of resistance. The hybrid plasmid IncX3-X4 is based on homologous recombination or the transposase-mediated formation of a cointegrate between IS26 and the nic site of oriT. This also suggested that even if the insert sequence on the plasmid carrying the mcr-1 gene does not mobilize the transfer of mcr-1, it can still be recombined with other plasmids containing the same insert sequence to form a more complex hybrid plasmid (Sun et al., 2016). These researches indicated that IS may contribute to plasmid-plasmid and plasmid-chromosome rearrangements as well as gene mobilization. Therefore, it might be significantly valuable to study IS on plasmids carrying the mcr-1 gene.
This study aimed to investigate the population structure of MPECs and the features of mcr-1-carrying plasmids in MPECs from animals which showed reduced effectiveness to antimicrobial. The present study may improve the understanding of the epidemiology, likelihood of spread and the potential, and evolutionary mechanisms of the mcr-1 gene in veterinary medicine.
Materials and Methods
Isolation of mcr-1-Positive E. coli Strains
Samples were obtained from lesion organs of swine, poultry (chicken), and bovine (dairy cow) between 2016 and 2019 in Yangling, Shaanxi, China. A total of 109 samples were collected from the organs with lesions in swine (n = 37) and chickens (n = 53) suffering from colibacillosis and the uterine rinsing fluid of dairy cows (n = 19) suffering from endometritis. The samples were collected in different farms, and each sample was from a different animal. The samples were stored aseptically at 4°C and transported to the laboratory in time for isolation and identification of strains. All animal procedures and study design were approved by the animal ethics committee of Northwest A&F University (Approval No.: 2016012).
All samples were cultivated on MacConkey agar (MAC) plates containing 2 μg/ml colistin and incubated at 37°C for 18 h. The species were further identified by the amplification and sequencing of 16S rRNA. Then, MPECs were detected by amplification and sequencing of mcr-1 (Liu et al., 2016).
Antimicrobial Susceptibility Testing of MPECs
Antimicrobial susceptibility testing was performed by broth microdilution method, as internationally recognized clinical guidelines those provided by the Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2013, 2019). Briefly, 0.5 McFarland inoculum suspensions were diluted 1:100 to a final inoculum density of 105 CFU/ml. Ampicillin (AMP), ceftazidime (CAZ), cefotaxime (CTX), aztreonam (AZM), tetracycline (TET), doxycycline (DOX), ciprofloxacin (CIP), enrofloxacin (ENR), gentamicin (GEN), sulfamethoxazole (SMZ), fosfomycin (FOS), florfenicol (FFC), colistin (COL), and meropenem (MEM) (Solarbio, Beijing, China) were selected to determine the minimum inhibitory concentrations (MICs) of MPECs. The antimicrobials were twofold serially diluted ranging from 0.25 to 512 μg/ml and incubated at 37°C for 18 h. The MICs were recorded as the lowest concentrations of antimicrobials in the wells, where no visible bacteria growth was observed.
Multilocus Sequence Typing of MPECs
MPECs were sequenced using Illumina HiSeq TM2000 platform at the Novogene Bioinformatics (Beijing, China). For Illumina sequencing, at least 1 μg genomic DNA was used for each strain in sequencing library construction. DNA samples were sheared into 400–500 bp fragments using a Covaris M220 Focused Acoustic Shearer following manufacture’s protocol. Illumina sequencing libraries were prepared from the sheared fragments using the NextFlexTM Rapid DNA-Seq Kit. The prepared libraries then were used for paired-end Illumina sequencing (2 × 150 bp) on an Illumina HiSeq TM2000 machine. The raw data generated from Illumina platform were submitted to Enterobase1, and sequence types were automatically assigned according to the Achtman scheme. A minimum spanning tree was generated using GrapeTree software to analyze the distribution of sequence type of MPECs (Zhou et al., 2018) characterized by multilocus sequence typing (MLST) analysis.
Mating Experiment of MPECs
To evaluate the transferability of mcr-1 gene, broth-mating experiments were performed with six representative MPECs (donor) and streptomycin-resistant E. coli C600 (recipient). Briefly, donor strain was mixed with recipient strain in LB broth (1:3) and the mixture was incubated at 37°C for 4 h in a shaker incubator (180 r/min). Subsequently, mixture was spread on MAC plates containing 2,000 μg/ml streptomycin (Solarbio, Beijing, China) and 2 μg/ml colistin and incubated at 37°C for 24 h to select transconjugants. The susceptibility of transconjugants for antimicrobials was determined based on the MICs.
Whole-Genome Sequencing Analysis of MPECs
The total genomic DNA was extracted from representative MPECs with TIANamp Bacteria DNA Kit (Tiangen, Beijing, China) following the manufacturer’s instructions and sequenced (MiSeq, Illumina, San Diego, CA, United States), producing 2 × 150-bp paired-end reads (Majorbio Company, Shanghai, China) and long-read (Pacific Biosciences, Menlo Park, CA, United States) technology. The short and long reads were assembled using Unicycler (Wick et al., 2017). Gene prediction and annotation were done with RAST genome annotation server version 2.02 and BLASTp. Plasmid replicon types were identified by the PlasmidFinder 2.13 from the Center for Genomic Epidemiology (CGE). Acquired resistance genes and chromosomal mutation-mediated antimicrobial resistance genes (ARGs) were identified with ResFinder 4.14 from CGE and resistance gene identifier (RGI)5 from the Comprehensive Antibiotic Resistance Database (CARD). Conjugal transfer components and type IV secretion systems (T4SS) of the plasmids were performed using oriTfinder6. Insertion sequence (IS) elements, transposon (Tn), and integron (In) were identified using ISfinder7, INTEGRALL8, and MobileElement Finder v1.0.39. Virulence genes and FimH type were identified with VirulenceFinder 2.010 and FimTyper 1.011, respectively, available at the Center for Genomic Epidemiology. Phylogroups were predicted using the ClermonTyping tool at the IAME Research Center web12.
Genetic Analysis of mcr-1-Carrying Plasmids
The genetic environment of mcr-1-carrying plasmid was compared with previously reported ones in GenBank with BLAST Ring Image Generator (BRIG) and EasyFig tools (v2.3) (Alikhan et al., 2011; Sullivan et al., 2011).
Nucleotide Sequence Accession Numbers
The complete genomes were deposited in GenBank with a Accession Nos. CP047002 (J-8 chromosome), CP046418 (pS4-mcr-1.1), CP047014 (pmcr-1), CP059836 (pMCR_A3_2), and BioProject Accession No. PRJNA649870 (pMCR_J9_7).
Results
Antimicrobial Susceptibility Profiles of 24 MPECs
After isolating bacterial strains from 109 samples, 24 MPECs were screened based on mcr-1 gene amplification and sequencing (data not shown). Among the 24 MPECs, nine strains were from poultry, 14 strains were from swine, and one strain was from bovine (Supplementary Table 2). The MICs of the 24 MPECs against 14 antimicrobials are as shown in Supplementary Table 1. All of MPECs were resistant to at least three or more antimicrobial categories; nine of 24 MPECs, namely J-1, J-2, J-3, J-4, J-5, J-6, J-7, J-8, and N-14, were resistant to all the 14 antimicrobials tested, including β-lactams (penicillins, cephems, monobactams, carbapenems), tetracyclines, fluoroquinolones, aminoglycosides, sulfonamides, phenicols, fosfomycins, and polymyxins. In addition, five of these (B-1, B-2, B-3, B-9, and18-16) were also sensitive only to meropenem (Figure 1). The above results indicated that all the 24 MPECs were resistant to at least three or all antimicrobial categories and exhibited either MDR or XDR for the common antimicrobials in Enterobacteriaceae.
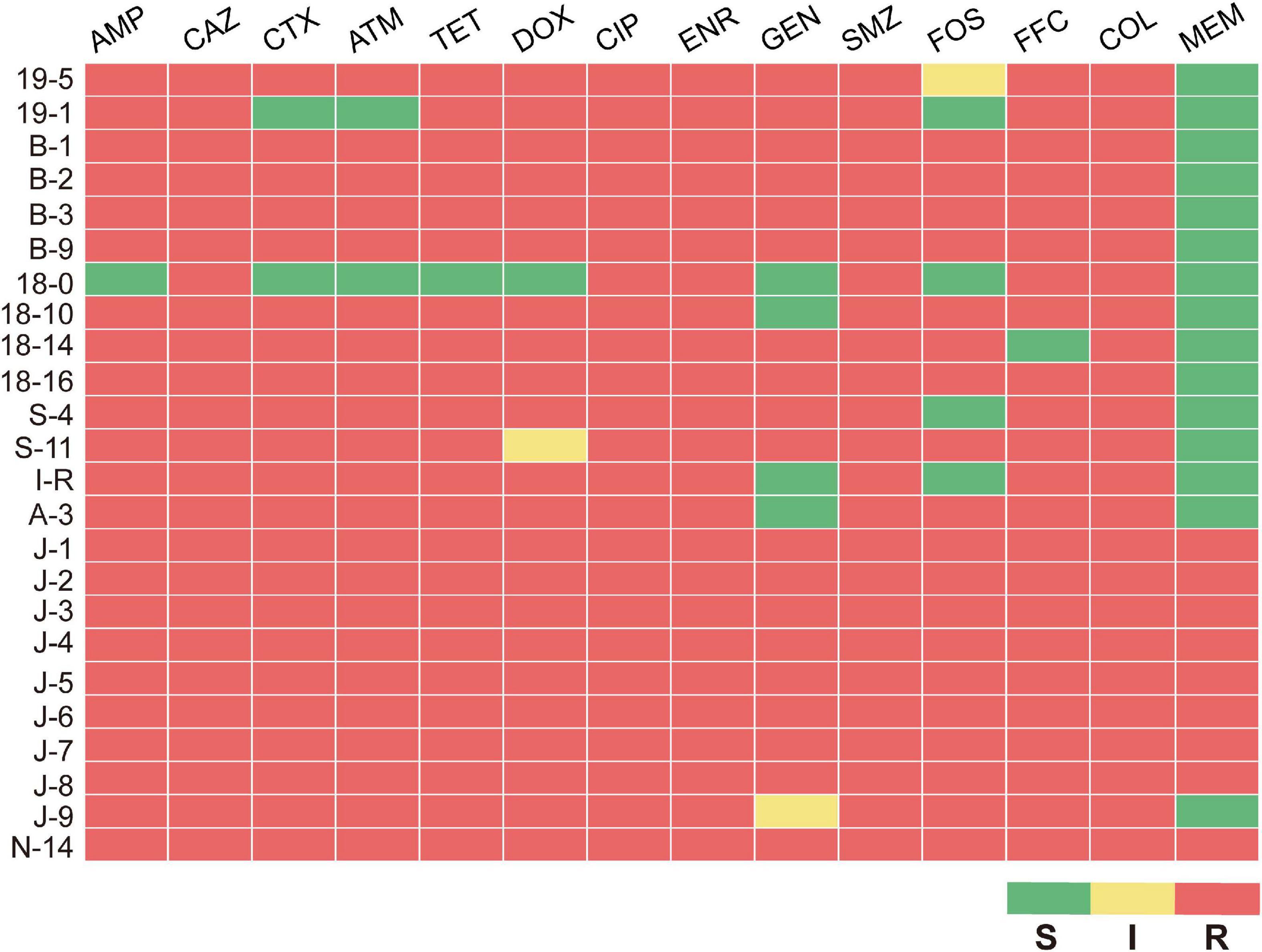
Figure 1. Antimicrobial sus ceptibility profile of 24 MPECs. S, susceptible; I, intermediate; R, resistant.
Genotypic Diversity of 24 MPECs
Nine STs, namely ST156 (41.6%, 10/24), ST448 (25%, 6/24), ST48 (8.3%, 2/24), ST10 (4.2%, 1/24), ST10596 (4.2%,1/24), ST1403 (4.2%, 1/24), ST2179 (4.2%, 1/24), ST5454 (4.2%, 1/24), and ST602 (4.2%, 1/24) were identified from the 24 MPECs using MLST method (Figure 2). After analyzing the EnteroBase database (Supplementary Table 2), nine STs were further assigned to six distinct clonal complexes (CCs) (CC156, CC448, CC10, CC278, CC206, CC446) and two singletons (ST10596, ST2179). MLST analysis indicated the genetic diversity among 24 MPECs, and the dominant clones belonged to CC156, CC448, and CC10. Moreover, ST15096 was discovered from swine for the first time. These suggests that the number of mcr-1-carrying clonal complex is still increasing.
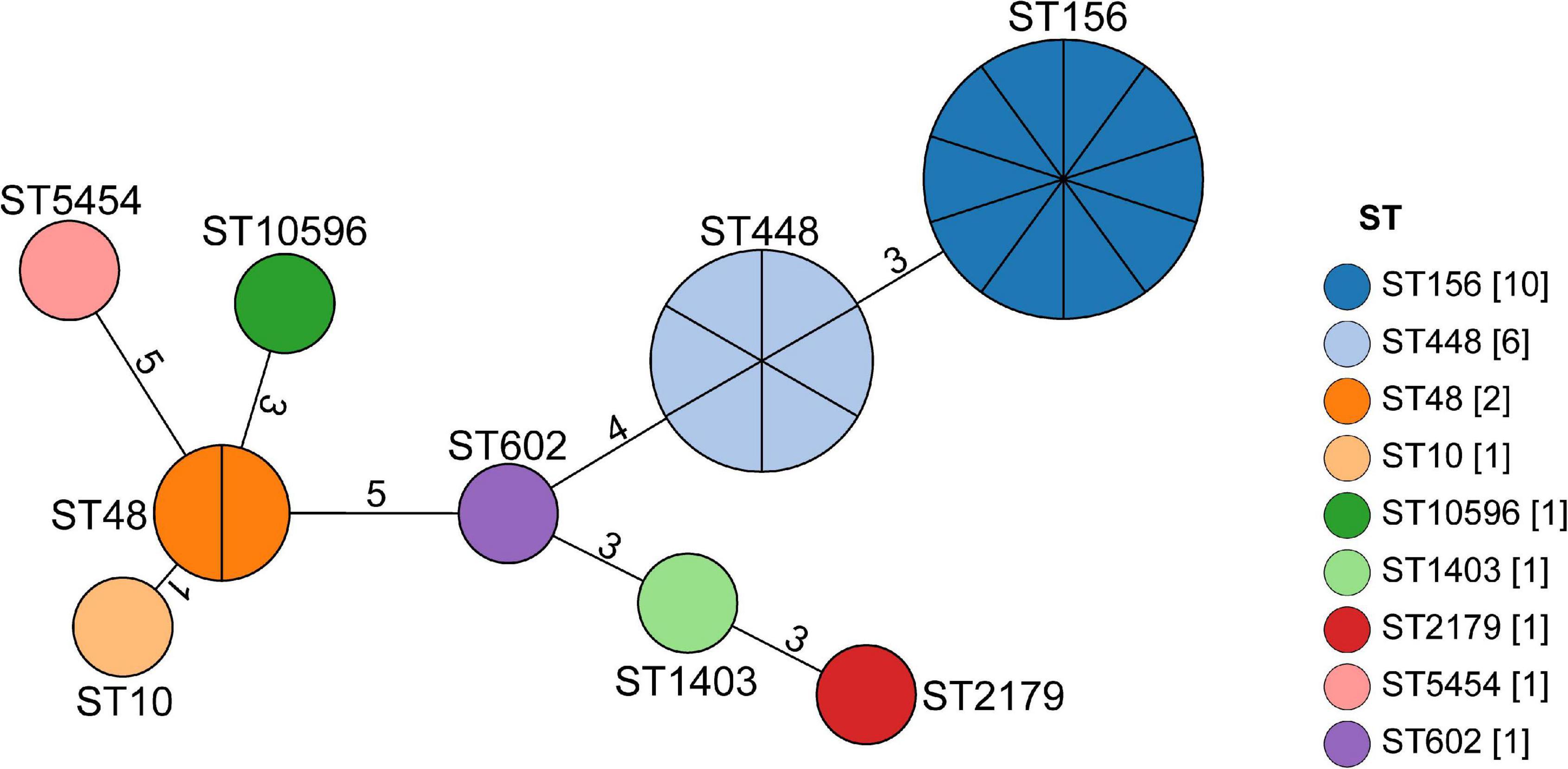
Figure 2. Minimum spanning tree analysis of the 24 MPECs. Each node represents a distinct sequence type (ST). The size of the circle represents the number of isolates with the same ST profile. The number on the line represents the count of the locus variants for ST.
Conjugative Transfer of mcr-1 Gene in Six MPECs
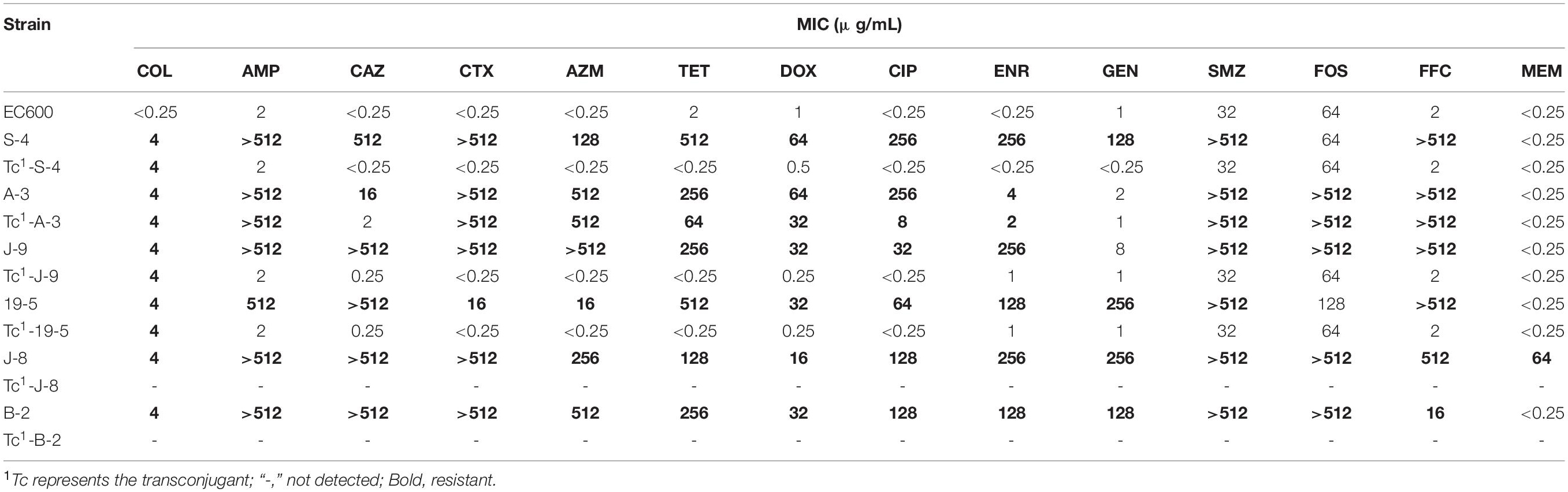
Based on the antimicrobial resistance spectrum and the ST distribution of the 24 MPECs, six MPECs (S-4, A-3, J-8, J-9, 19-5, and B-2) were selected for the mating assay. Transconjugants were obtained from all donors except J-8 and B-2. These transconjugants exhibited the same level of colistin resistance (4 μg/ml) as the donors, but at the same time, both showed a 16-fold increase in resistance when compared with the recipient E. coli C600 (0.25 μg/ml) (Table 1). In the case of three E. coli transconjugants (Tc-S-4, Tc-J-9, and Tc-19-5), no additional resistance phenotype was cotransferred along with the mcr-1 gene. However, for Tc-A-3, showed β-lactam, tetracycline, fluoroquinolone, sulfonamide, fosfomycin, and florfenicol resistance phenotypes were also observed in addition to that of colistin resistance. The results suggested that mcr-1 genes of S-4, A-3, J-9, and 19–5 were located on the conjugative transfer plasmid and could be successfully transferred to a recipient strain to confer the colistin resistance phenotype. Although the mcr-1 gene of both J-8 and B-2 conjugated failure, J-8 was resistant to all tested antimicrobials. Therefore, this strain was chosen, along with the other successfully conjugated ones for the next experiment.
Features of Resistance Genes and Plasmids in Five MPECs
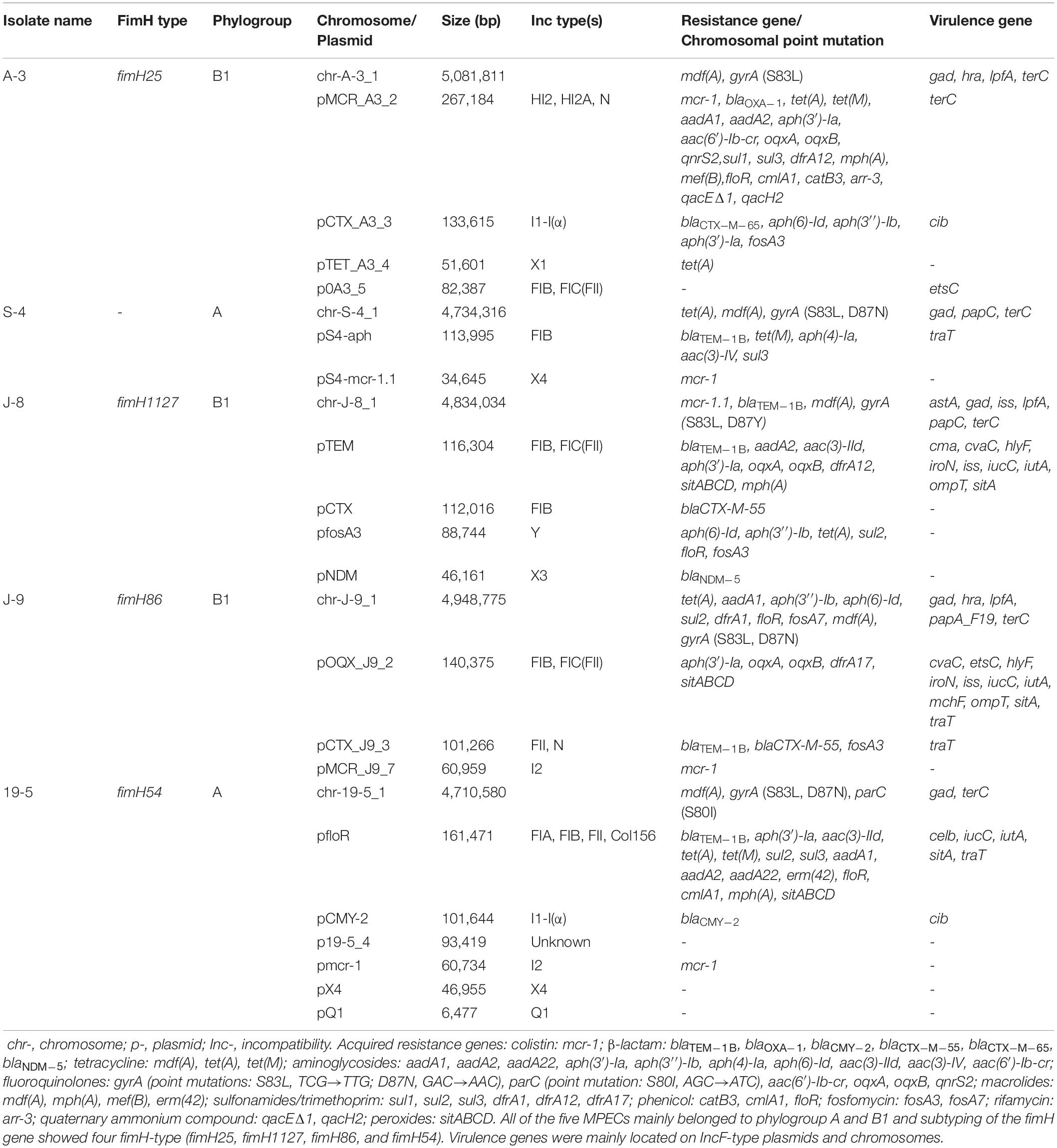
Five MPECs (S-4, A-3, J-8, J-9, and 19–5) were selected for whole-genome sequencing (WGS) analysis to identify the gene and plasmid features which are responsible for resistance. Beside the mcr-1 gene, all the five MPECs contained more than three different types of acquired resistance genes as well as at least two resistant plasmids (Table 2). In addition, locus mutations of gyrA (S83L, D87N) and parC (S80I) were also found on the chromosome. In the case of J-8, the mcr-1 gene was located on the chromosome and flanked by two ISApl1 (Figure 3), with an additional blaNDM–5 gene located on the IncX3-type plasmid. Conversely, the mcr-1 genes of the other four MPECs were located on three different transferable plasmids IncHI2/HI2A/N (A-3), IncX4 (S-4), and IncI2 (J-9 and 19–5).
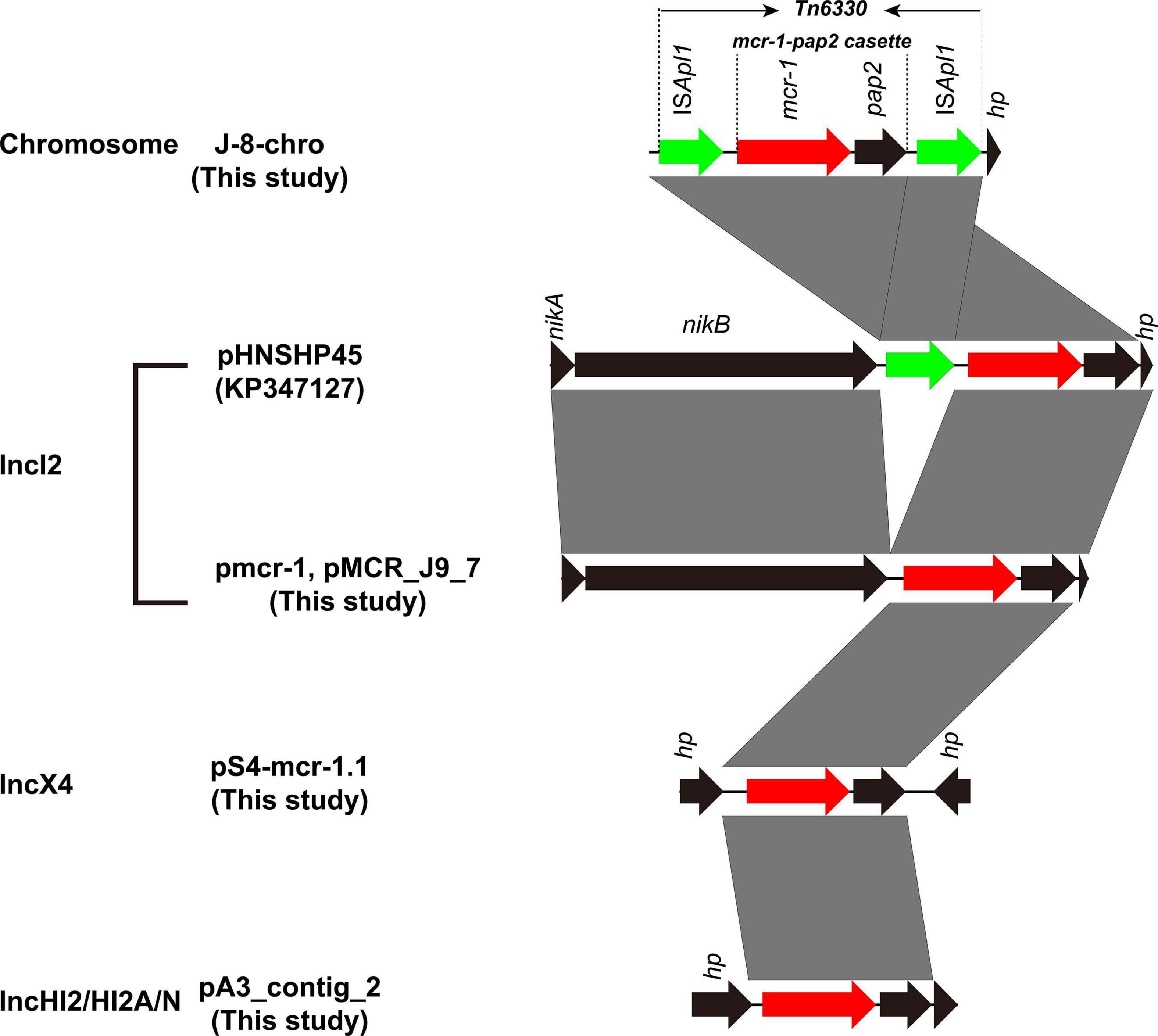
Figure 3. Genetic context of mcr-1 gene in chromosome and plasmids from this study compared with plasmid pHNSHP45. Black arrows represent nikA (plasmid mobilization relaxosome protein), nikB (relaxase), and hypothetical protein (hp). Green arrows denote insertion sequence ISApl1. Red arow denotes colistin resistance gene mcr-1. Regions of >99% identity are marked by gray shading.
Genomic Features of mcr-1-Carrying Plasmids
The mcr-1 gene in strain S-4, located on the plasmid pS4-mcr-1.1 (34,645 bp; GC content of 42.25%), belonged to the IncX4 incompatibility group (Table 2). The backbone of this plasmid was highly similar to those of pICBEC72Hmcr (CP015977; nucleotide coverage 96%; identity 99.98%), pOW3E1 (KX129783; nucleotide coverage 100%; identity 99.59%), pMCR_WCHEC1606 (KY463451; nucleotide coverage 96%; identity 99.73%), pESTMCR (KU743383; nucleotide coverage 96%; identity 99.58%), and pmcr1_IncX4 (KU761327; nucleotide coverage 96%; identity 99.44%) as indicated by BLASTn analysis (Figure 4). The backbone structure of all those six plasmids was highly conserved, with the only difference being that the virB6 gene of the T4SS system on the IncX4-type plasmid was truncated by an IS2. From these plasmids, two, namely, pOW3E1 and pICBEC72Hmcr, were selected for collinear alignment with pS4-mcr-1.1. The results indicated that no resistance genes other than that of mcr-1 was identified, with the gene embedded in an mcr-1-pap2 cassette of approximately 2.6 kb (Figures 3, 5). Furthermore, even though all plasmids contained an insert sequence IS26, the insertion sequence ISApl1 was absent either upstream or downstream of the mcr-1 gene. Besides, on pS4-mcr-1.1, virB6 gene in T4SS was truncated into two fragments by IS2. In the case of pOW3E1, IS2 was found upstream of the mcr-1 gene. In addition, a type II toxin-antitoxin system (TAs) hicA/hicB was also found between upstream of T4SS and downstream of the chaperone-encoding dnaJ.
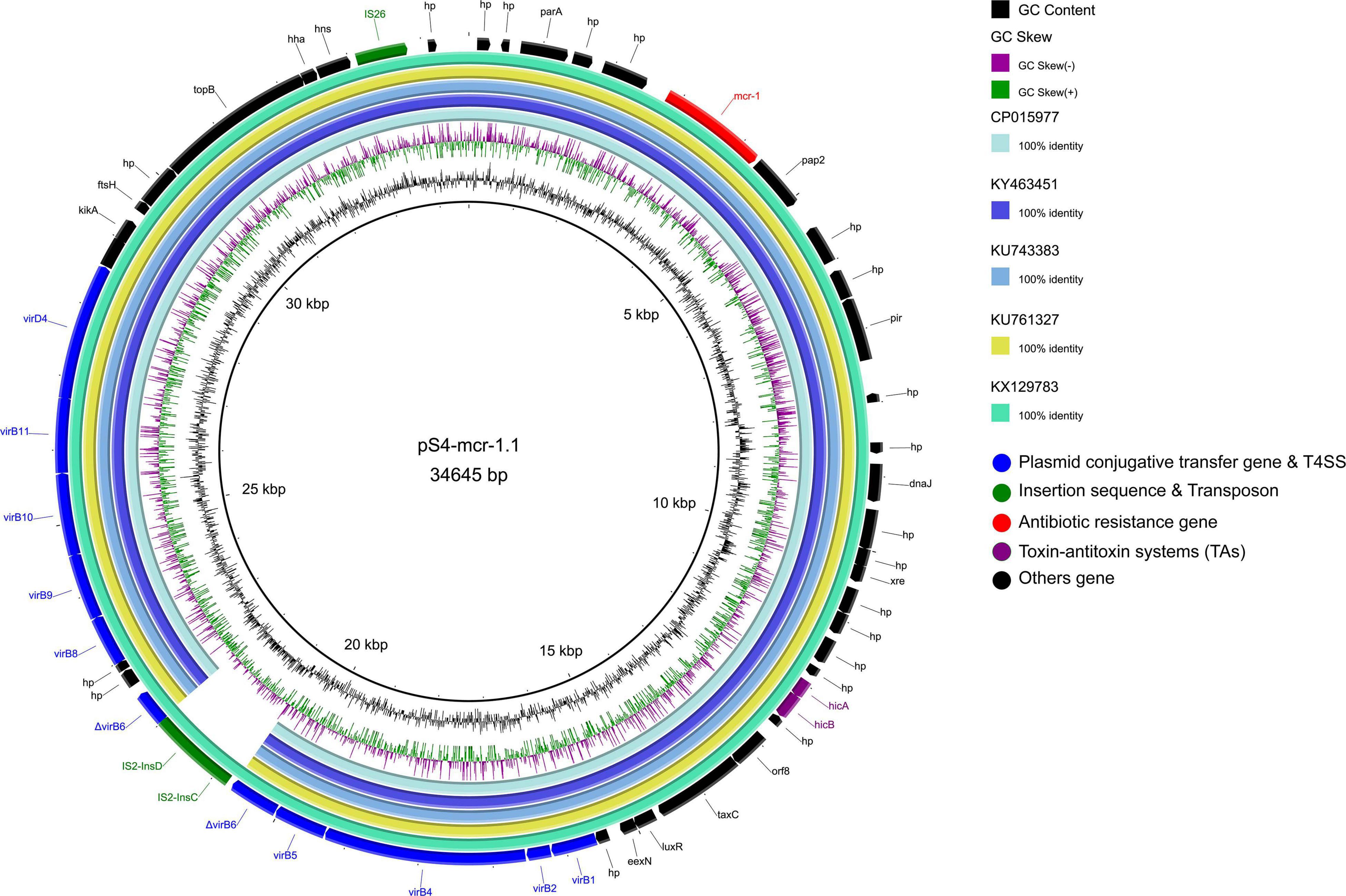
Figure 4. Comparative genomic analysis of mcr-1-carrying IncX4-type plasmid pS4-mcr-1.1 (this study) with plasmids pICBEC72Hmcr (CP015977), pOW3E1 (KX129783), pMCR_WCHEC1606 (KY463451), pESTMCR (KU743383), and pmcr1_IncX4 (KU761327) in GenBank. The plasmid pS4-mcr-1.1 was used as a reference sequence. Dark gray shading denotes regions of homology (>99% identity nucleotide identity).
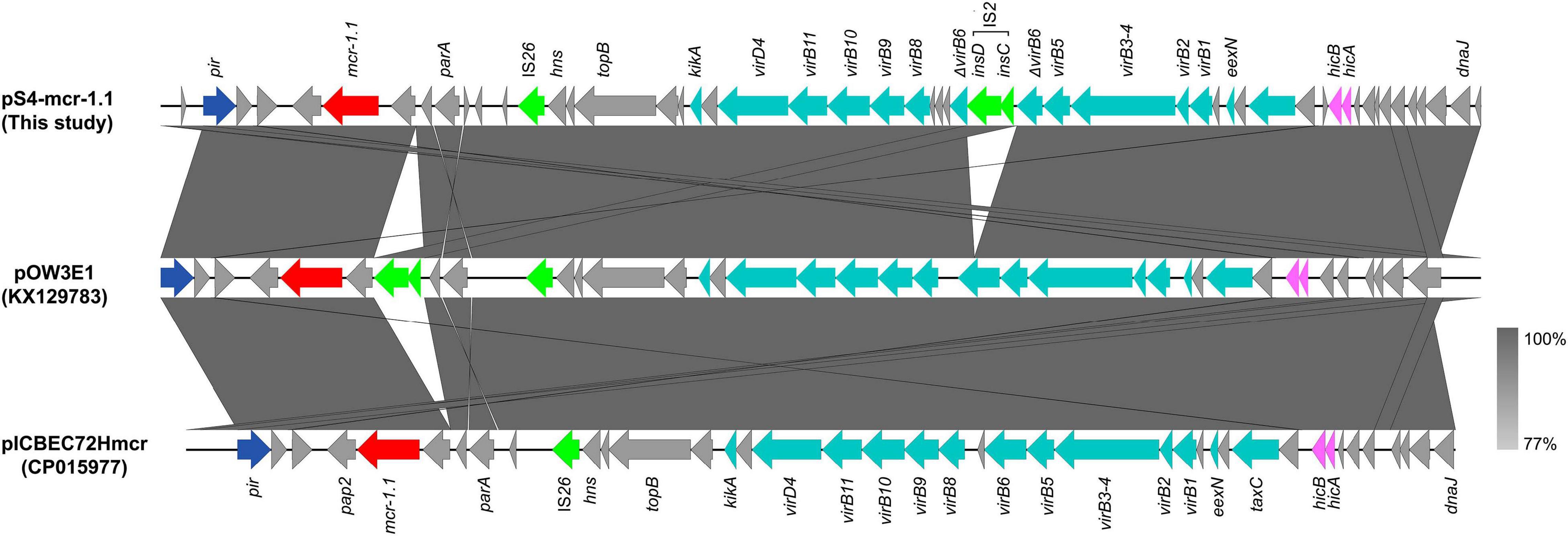
Figure 5. Colinear sequence comparison of IncX4-type mcr-1-carrying plasmid pS4-mcr-1.1 (this study) with other plasmids including pOW3E1 (KX129783) and pICBEC72Hmcr (CP015977). Boxed arrows represent the position and transcriptional direction of ORFs. Dark gray shading denotes regions of homology (>99% identity nucleotide identity). Blue colors represent the plasmid replication- and maintenance/stability-associated genes; teal, plasmid conjugative transfer genes and T4SS genes; green, IS and Tn; red, antimicrobial resistance genes; pink, toxin-antitoxin system (TAs) genes; gray, other genes.
The mcr-1 in strain 19-5 and J-9 was, respectively, carried by the plasmid pmcr-1 (60,734 bp; GC content of 42.50%) and the plasmid pMCR_J9_7 (60,959 bp; GC content of 42.36%), all belonging to the IncI2 incompatibility group (Table 2). BLASTn analysis showed that pMCR_J9_7 was similar to the plasmid pD90-2 (CP022452; nucleotide coverage 99%; identity 99.99%) and pHNSHP45 (KP347127; nucleotide coverage 90%; identity 99.63%), while pmcr-1 was similar to the plasmid PN21 (MG557851; nucleotide coverage 100%; identity 99.97%) and pHNSHP45 (KP347127; nucleotide coverage 99%; identity 99.97%) (Figure 6). A sequence comparison of the two plasmids indicated that they shared 99.63% identity with 91% nucleotide coverage. The gaps shown in the circle diagram represented the differences between the five mcr-1-bearing plasmids and the absence of IS683 and ISApl1. For colinear analysis with the plasmids pMCR_J9_7 and pmcr-1, pHNSHP45 was selected as reference one. As shown in Figure 7, ISApl1 located upstream of the mcr-1 gene and IS683 located between repA and parA were absent in plasmid pMCR_J9_7 and pmcr-1. In addition, stbD and stbE genes, both of which are located downstream of virB5 and encoding stability proteins, were absent from the plasmid pMCR_J9_7. A similar observation was made for dnaJ, a chaperone-encoding gene, which was absent between parA and traL in plasmid pmcr-1. For both identified IncI2-type plasmids, only the mcr-1 gene, embedded within the mcr-1-pap2 cassette, was associated with colistin resistance (Figure 3). Moreover, a type II toxin-antitoxin system (TAs) hicA/hicB was found to be present on the plasmid pMCR_J9_7, with the hicA gene, encoding antitoxin proteins, being absent from the plasmid pmcr-1.
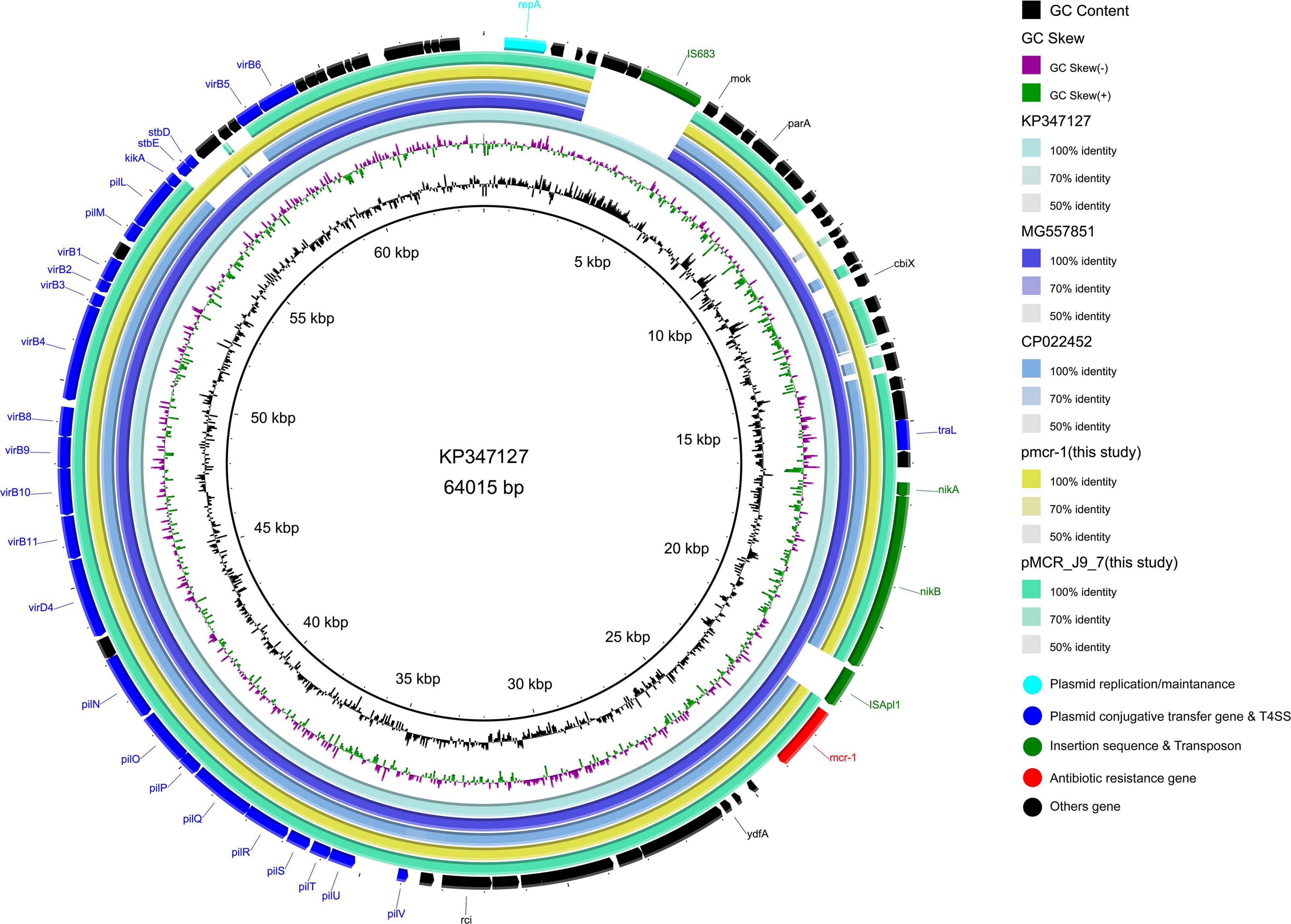
Figure 6. Comparative genomic analysis of mcr-1-bearing IncI2-type plasmids pmcr-1 (this study) and pMCR_J9_7 (this study) compared with plasmids pD90-2 (CP022452), pHNSHP45 (KP347127), and PN21 (MG557851) in GenBank. The plasmid pHNSHP45 was used as a reference sequence.
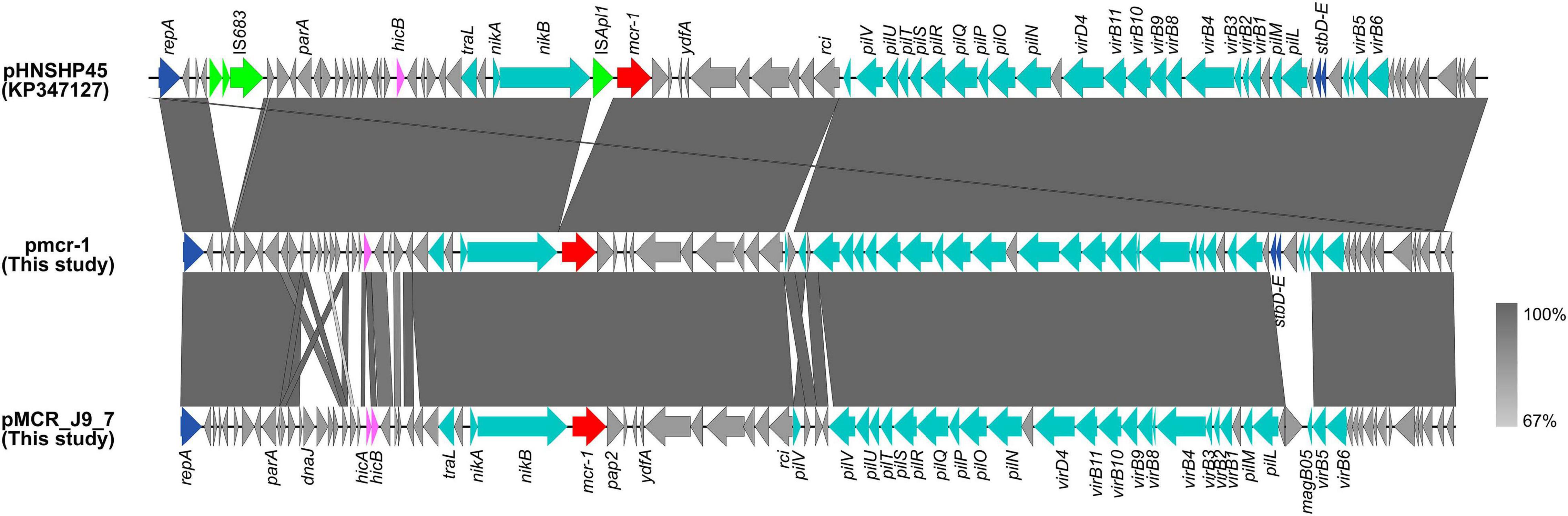
Figure 7. Colinear sequence comparison of IncI2 reference plasmid pHNSHP45 (GenBank accession No. KP347127) with plasmids including pmcr-1 (this study) and pMCR_J9_7 (this study). Boxed arrows represent the position and transcriptional direction of ORFs. Dark gray shading denotes regions of homology (>99% identity nucleotide identity). Blue colors represent the plasmid replication- and maintenance/stability-associated genes; teal, plasmid conjugative transfer genes and T4SS genes; green, IS and Tn; red, antimicrobial resistance genes; pink, TAs genes; gray, other genes.
The mcr-1 gene in strain A-3 was carried by a large multireplicon MDR plasmid (designated pMCR_A3_2; 267,184 bp; GC content of 46.84%) containing IncHI2, IncHI2A, and IncN replicons (Table 2). Compared with pHNSHP45-2, a IncHI2-type reference plasmid, the plasmid pMCR_A3_2 and pMCR1_025943 lacked ISApl1 upstream of the mcr-1 gene which was embedded in a 2.6-kb mcr-1-pap2 gene cassette (Figure 3). The backbone of pMCR_A3_2 was highly similar to those of pHS13-1-IncHI2 (CP026492; nucleotide coverage 98%; identity 99.94%), pXH988_1 (CP019353; nucleotide coverage 94%; identity 99.99%), pMCR1_025943 (CP027202; nucleotide coverage 92%; identity 99.99%), pMCR_WCHEC050613 (CP019214; nucleotide coverage 92%; identity 99.99%), and pHNSHP45-2 (KU341381; nucleotide coverage 88%; identity 99.99% (Figure 8). The gaps in the circle diagram showed the differences between the six mcr-1-bearing plasmids, including the insertion or deletion of IS, Tn, and ARGs. In the case of this plasmid (pMCR_A3_2), pMCR1_025943 and pHNSHP45-2 were selected as reference plasmids for colinear analysis. As shown in Figure 9, the differences between pMCR_A3_2 and those three reference plasmids were mainly focused on the MDR region. For pMCR_A3_2, this region included two class 1 integrons, namely In0 (no resistance gene cassette) and In640 (dfrA12, gcuF, aadA2, cmlA1, aadA1a, qacH2) which confer resistance to trimethoprims, aminoglycosides, chloramphenicols, and quaternary ammonium compounds. Between these two integrons, the fluoroquinolone resistance gene qnrS2 and the phenicol resistance gene floR were present. In addition, downstream of In640, there are two composite transposons, namely, Tn4352 (IS26-aph(3′)-Ia-IS26) which carried an aminoglycoside resistance gene aph(3′)-Ia and Tn6010 (IS26-oqxA-oqxB-IS26) which carried the fluoroquinolone resistance genes oqxA and oqxB. Between the In640 and Tn4352, the macrolide efflux pump gene mef(B) and the sulfonamide resistance gene sul3 were also present, along with another sulfonamide resistance gene sul1 presented between these two composite transposons. The MDR region further contained two other transposons, namely, cn_4692_IS26 (IS26-repE-hp-hp-hp-hp-IS26) and cn_6354_IS26 [IS15DI-aac(6′)-Ib-cr-blaOXA–1-catB3-arr-3-qacEΔ1-sul1-hp-IS 26], and the latter transposon cn_6354_IS26 conferred resistance to aminoglycoside/fluoroquinolone, beta-lactam, phenicol, rifamycin, quaternary ammonium compounds, and sulfonamide. An additional macrolide resistance gene mph(A) as well as the tetracycline resistance genes tet(A) and tet(M) and the tellurite resistance operon were also presented on pMCR_A3_2. Insertion sequences and complex transposons such as IS6 family (IS1006, IS6100), Tn3 family (TnAs1, TnAs3, TnEc1, TnEc3, TnEc63, Tn2), IS21 family (ISEc57, IS100), IS91 family (ISVsa3), IS3 family (ISKpn8), and IS256 family (IS256) were found on the plasmids as well. Eventually, a relaxase-encoding traI gene in T4SS system was found to be truncated into two fragments by IS100. Regarding the presence of toxin-antitoxin systems (TAs), three type II ones, namely, higA/higB, hok/gef, and relE/parE were located on the pMCR_A_3_2.
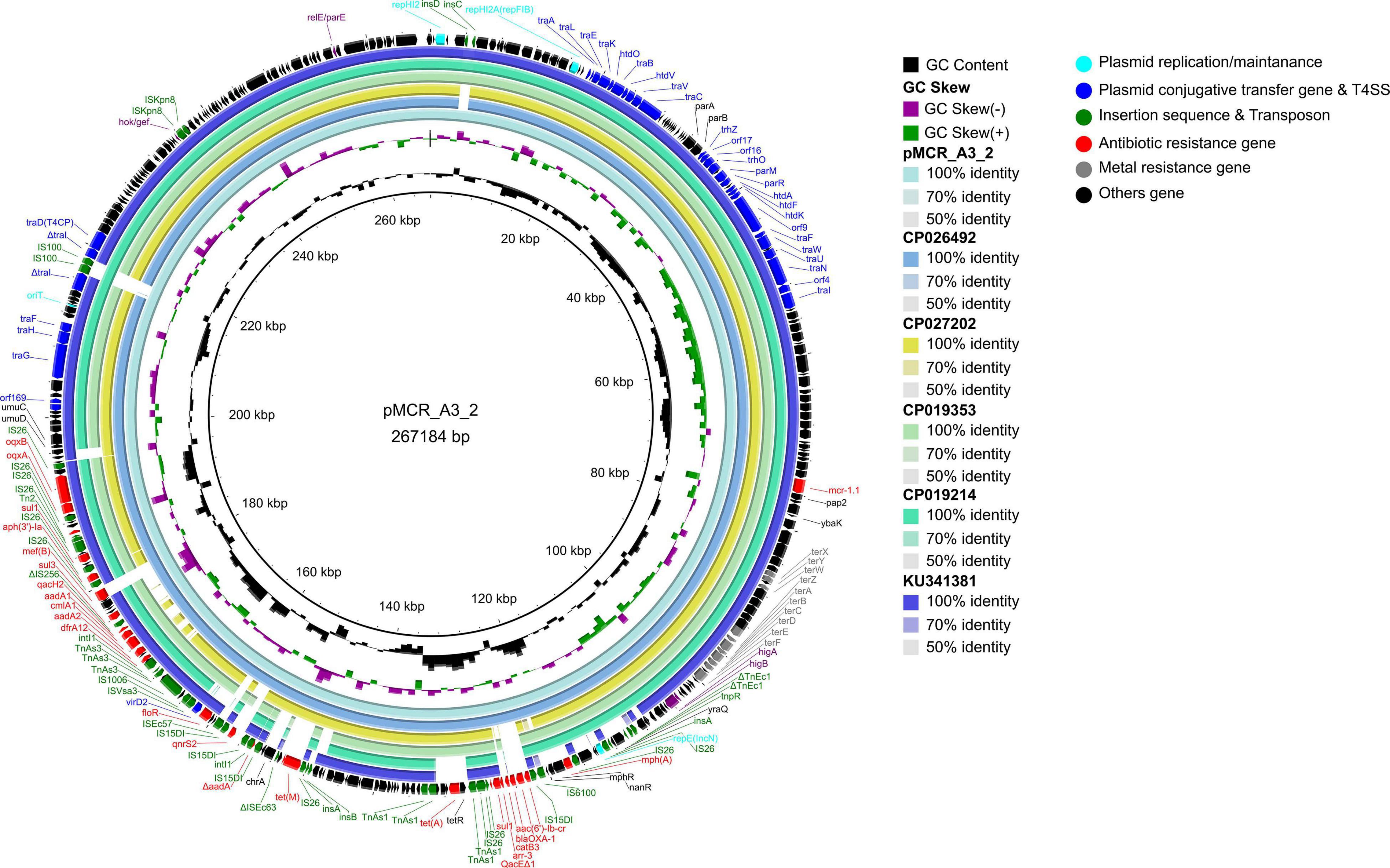
Figure 8. Comparative genomic analysis of mcr-1-bearing IncHI2-type plasmid pMCR_A3_2 (this study) compared with plasmids pHS13-1-IncHI2 (CP026492), pXH988_1 (CP019353), pMCR1_025943 (CP027202), pMCR_WCHEC050613 (CP019214), and pHNSHP45-2 (KU341381) in GenBank. The plasmid pHNSHP45-2 was used as a reference sequence.
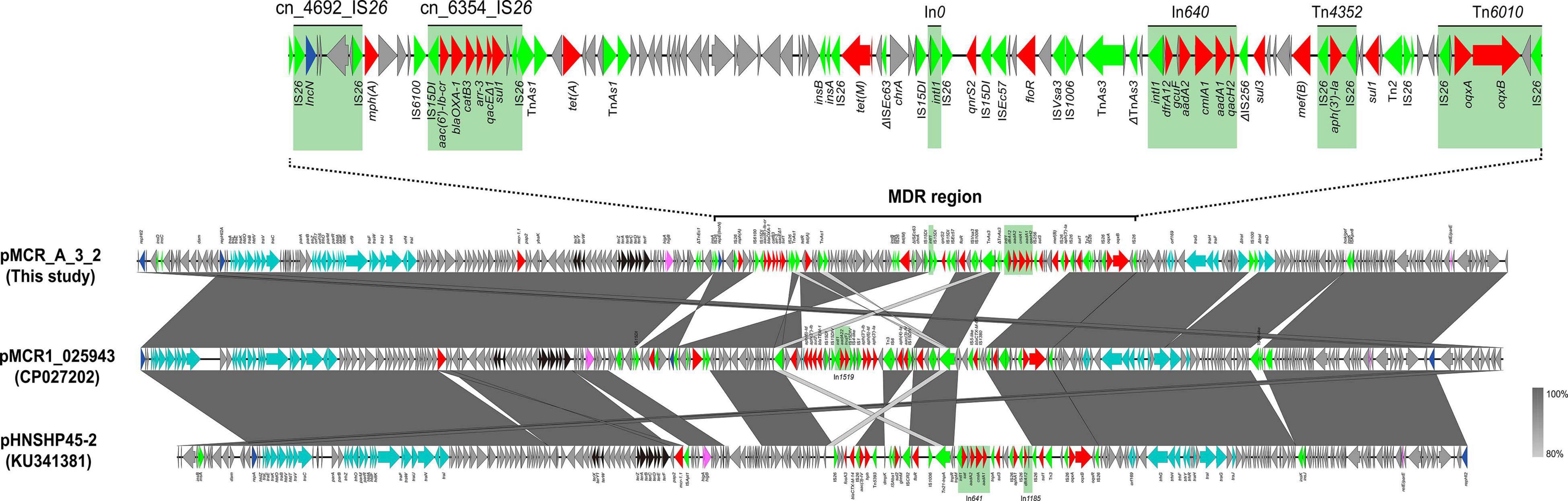
Figure 9. Colinear sequence comparison of IncHI2/HI2A/N plasmid pMCR_A3_2 (this study) with other plasmids including pMCR1_025943 (GenBank accession No. CP027202) and pHNSHP45-2 (GenBank accession No. KU341381). Boxed arrows represent the position and transcriptional direction of ORFs. Dark gray shading denotes regions of homology (>99% identity nucleotide identity). Blue colors represent the plasmid replication- and maintenance/stability-associated genes; teal, plasmid conjugative transfer (tra-/trb- gene cluster) and T4SS genes; green, IS and Tn; red, antimicrobial resistance genes; pink, TA genes; gray, other gene. The light green shade represents the composition transposon and integron (In); MDR, multidrug resistant.
Discussion
MPECs from different sources have been isolated worldwide. In the present study, 24 MPECs were isolated from organs having lesions obtained from animals, which did not respond to antimicrobial treatment and which were, therefore, considered to show MDR, or even XDR based on the definition provided by Magiorakos et al. (2012). MLST analysis classified the 24 MPECs into nine sequence types (six dominant CCs and two singletons), among which ST10596 has been identified for the first time. The main CCs were CC156 (ST156), CC448 (ST448), and CC10 (ST48 and ST10).
MPECs of CC156 has been previously isolated only from the patients’ blood culture and ducks’ rectal swab (Yang et al., 2016; Rossi et al., 2017; Lin et al., 2020), which exhibited MDR or XDR including meropenem and colistin. In this study, 10 such MPECs, which showed resistance to β-lactamases (penicillins, cephems, monobactams, carbapenems), tetracyclines, fluoroquinolones, aminoglycosides, sulfonamides, phenicols, fosfomycins, and polymyxins, were recovered from chickens and bovines. In the case of MPEC ST156 (J-8), mcr-1, blaTEM–1B, blaCTX–M–55, blaNDM–5, mdf(A), aadA2, aac(3)-IId, aph(3′)-Ia, aph(6)-Id, aph(3′′)-Ib, oqxA, oqxB, mph(A), tet(A), sul2, dfrA12, floR, and fosA3 genes were found to contribute to the resistance phenotype.
Escherichia coli ST10 and ST48 (single locus variant of ST10), belonging to CC10, are considered widely disseminated clones worldwide. CC10 is highly disseminated via foods, the environment, animals, and humans (Matamoros, 2017), and they are usually associated with the presence of colistin (mcr-1) (Papa-Ezdra et al., 2020), ESBL (blaCTX–M–14/–15), and fluroquinolone [aac(6′)-Ib-cr and qnrB] (Oteo et al., 2009; Fam et al., 2011) resistance genes. However, in this study, in addition to those genes, MPEC ST10 (S-4) and 19-5 (ST48) were also found to contain aminoglycoside, tetracycline, sulfonamide, macrolide, lincosamide, and phenicol, as well as peroxides resistance genes. These results suggested that CC10 possessed a more extensive set of resistance genes and it could, therefore, become a reservoir for the transmission of resistance, and CC156 also could be a reservoir for mcr-1 genes in a similar way to CC10. Furthermore, MPECs of ST448 are generally sporadic and not dominant (Bai et al., 2018), but results showed that six MPECs of ST448 were members of CC448, hence suggesting that CC448 could become a new dominant clone complex carrying the mcr-1 gene in pig farms.
The mcr-1 gene along with other resistance genes are found on conjugative transfer plasmids or mobile element transposons. As such, there is a risk for the horizontal transmission of colistin as well as other drug resistance between bacteria in the infected guts of animals, thereby further exacerbating the risk of spreading resistance (Guenther et al., 2017; Bai et al., 2018; Li et al., 2020). The results of this study showed that the mcr-1 genes of strains S-4, A-3, J-9, and 19-5 located on the three different transferable plasmids IncHI2/HI2A/N (A-3), IncX4 (S-4), and IncI2 (J-9, 19-5), could be successfully transferred. Despite the fact that IncI2-, IncX4-, and IncHI2-type plasmids were the most common carriers of the mcr-1 gene, yet their mobile elements (IS, Tn, In) were very different. Therefore, the study and description of mobile elements on plasmids can greatly contribute to the understanding of the coupling transfer of plasmids, the formation of hybrid plasmids, and the translocation of ARGs.
Self-transmissible IncX4-type plasmid, a key vector for the transmission of the mcr-1 gene between Enterobacteriaceae worldwide, is the most common one which carries the resistance gene due to its high conjugative transfer ability (Biswas et al., 2020; Migura-Garcia et al., 2020). The structure of mcr-1-carrying IncX4-type plasmids are highly conserved, and they consist of mcr-1-pap2 gene cassette, T4SS systems, plasmid replication/maintenance genes, TA modules, and an insertion sequence IS26 (Bai et al., 2018). The IncX4-type plasmid pOW3E1, in addition to the previously mentioned modules, contained IS2 close to the mcr-1 gene (Zurfluh et al., 2016), but the insertion sequence had no effect on either the gene or the conjugative transfer of plasmid. In the present study, IS26 and IS2 were found on the IncX4-type plasmid pS4-mcr-1.1, with IS2 truncating the virB6 gene of T4SS. Since the virB6 product contributes to DNA transfer as a component of the endosomal translocation channel protein (Álvarez-Rodríguez et al., 2020), its truncation may affect the efficiency of plasmid conjugative transfer, although the process through which this occurs remains unknown.
The first reported mcr-1-carrying IncI2-type plasmid pHNSHP45 was identified in E. coli of swine origin in China and ISApl1 was located upstream of the resistance gene (Liu et al., 2016). However, in this study, the two IncI2-type plasmids (pmcr-1 and pMCR_J9_7) did not have any ISApl1 adjunct to the mcr-1 gene, with IS683 also lacking between repA and parA. Furthermore, colinear analysis of the plasmid pHNSHP45 along with those of pMCR_J9_7 and pmcr-1 indicated the absence of stbD and stbE genes, both encoding plasmid stability proteins, from pMCR_J9_7. Similarly, the dnaJ gene, encoding for the chaperone protein, was absent from the pmcr-1 plasmid. These results implied that the IncI2-type plasmid could lose some insertion sequences during the evolution process to maintain the conservative plasmid backbone structure, which is conducive to the stability of the mcr-1 gene and the conjugative transfer of the plasmid.
IncHI2/HIA-type plasmids are usually > 250 kb, unlike IncX4- and IncI2-type plasmids. In addition, they contain an MDR region with various ISs, transposons, and integrons that capture various exogenous resistance genes, thereby conferring multidrug resistance and metal tolerance to E. coli (Fang et al., 2016). However, despite these differences, the backbone of IncHI2/HI2A-type plasmids carrying mcr-1 gene are also conserved, in a similar way to those of the IncX4 and IncI2 plasmids. Hence, they were found to include plasmid replication/maintenance genes, T4SS systems, tellurium ion resistance gene clusters, and TA modules. Studies on the IncHI2-type plasmids carrying the mcr-1 gene mostly focused on the description of the genetic environment of the gene, while the mobile elements of the MDR region of the IncHI2 type were overlooked. In this context, it is to be noted that the IncHI2/HI2A-type plasmid pEGY1-MCR-1 carrying the mcr-1 gene, is flanked by two ISApl1 elements as well as In641 (Hammad et al., 2019). In this study, pMCR_A3_2, a IncHI2/HI2A/N-type hybrid plasmid with an extra IncN-type replicon, was flanked by two IS26 present in the MDR region. This implied that the replicon could have been transposed by a transposon composed of IS26. Alternatively, this could have been the result of homologous recombination or the transposase-mediated formation of a cointegrate from IS26 and the nic site of oriT. Additional results showed that the MDR region of the pMCR_A3_2 plasmid, identified in this study, included two class 1 integrons, namely, In0 and In640. However, that of the IncHI2-type plasmid pHNSHP45-2 carrying the mcr-1 gene has been reported to contain two different class 1 integrons, namely, In1185 and In641 (Zhi et al., 2016). Furthermore, In1519 was also identified from plasmid pMCR1_025943 (CP027202). Apart from those class 1 integrons, a large number of insertion sequences and transposons such as IS26/26-like (15DI) and Tn3 families, were found to be present in the MDR region. The presence of these mobile integration elements facilitates the capture of numerous classes of resistance genes, thereby conferring various resistance phenotypes to E. coli containing the IncHI2-type plasmids.
Generally, ISApl1 is detected upstream of the mcr-1 gene and is believed to be involved in the mobilization of the mcr-1 cassette (Snesrud et al., 2016). In this study, the ISApl1 was absent from both upstream and downstream of the mcr-1 gene on IncX4, IncI2, and IncHI2/HI2A/N plasmids, even though mcr-1 on the chromosome was embedded in Tn6330. Previous studies have shown that the transposon Tn6330 was immobilized on several plasmid backgrounds, following the loss of the flanking ISApl1 elements due to the high instability of being spread through plasmid transfer (Li et al., 2017; Wang et al., 2018). The loss of the ISApl1 transposon element in mcr-1-carrying plasmids has recently been proposed to be essential for the maintenance of the mcr-1 gene. This is believed to be more beneficial to the host bacteria for adapting to environmental changes, especially when adapting from the pressure of antimicrobial agents to a pressure-free environment (Porse et al., 2016).
Conclusion
In conclusion, 24 MPECs in our study showed MDR or even XDR and exhibited the population diversity. The T4SS genes located on IncX4- and IncHI2/HI2A/N-type plasmids were truncated by the insertion sequence. The truncation may affect the efficiency of plasmid conjugative transfer. Additionally, the MDR region of IncHI2/HI2A/N-type plasmids contained two class 1 integrons and four composite transposons. These mobile elements contributed to the multireplicon plasmid formation and the acquisition and transfer of ARGs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by all animal procedures and study design were approved by the animal Ethics Committee of Northwest A&F University (Approval No: 2016012). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
XS and ZL: conceptualization. ZL: methodology, software, formal analysis, data curation, and visualization. XS, YL, YF, and WM: validation. JL, HM, SL, BC, and HH: investigation. WZ: resources. ZL and YL: writing – original draft preparation. XS and YL: writing, review, and editing. XS: supervision, project administration, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Special Fund for Basic Research of the Ministry of Science and Technology, China (Grant Number 2013FY110600); National Natural Science Foundation of China (Grant Number 31802230); and Natural Science Basic Research Plan in Shaanxi Province of China (Grant Number 2019JQ-281).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.707332/full#supplementary-material
Footnotes
- ^ https://enterobase.warwick.ac.uk/
- ^ https://rast.nmpdr.org/rast.cgi
- ^ https://cge.cbs.dtu.dk/services/PlasmidFinder/
- ^ https://cge.cbs.dtu.dk//services/ResFinder/
- ^ https://card.mcmaster.ca/analyze/rgi
- ^ https://tool-mml.sjtu.edu.cn/oriTfinder/oriTfinder.html
- ^ https://www-is.biotoul.fr/blast.php
- ^ http://integrall.bio.ua.pt/?search
- ^ https://cge.cbs.dtu.dk/services/MobileElementFinder/
- ^ https://cge.cbs.dtu.dk/services/VirulenceFinder/
- ^ https://cge.cbs.dtu.dk/services/FimTyper/
- ^ http://clermontyping.iame-research.center/
References
Alikhan, N., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Álvarez-Rodríguez, I., Arana, L., Ugarte-Uribe, B., Gómez-Rubio, E., Martín-Santamaría, S., Garbisu, C., et al. (2020). Type IV coupling proteins as potential targets to control the dissemination of antibiotic resistance. Front. Mol. Biosci. 7:201. doi: 10.3389/fmolb.2020.00201
Aslam, B., Wang, W., Arshad, M. I., Khurshid, M., Muzammil, S., Rasool, M. H., et al. (2018). Antibiotic resistance: a rundown of a global crisis. Infect. Drug. Resist. 11, 1645–1658. doi: 10.2147/IDR.S173867
Bai, F., Li, X., Niu, B., Zhang, Z., Malakar, P. K., Liu, H., et al. (2018). A mcr-1-carrying conjugative IncX4 plasmid in colistin-resistant Escherichia coli ST278 strain isolated from dairy cow feces in Shanghai, China. Front. Microbiol. 9:2833. doi: 10.3389/fmicb.2018.02833
Biswas, S., Elbediwi, M., Gu, G., and Yue, M. (2020). Genomic characterization of new variant of hydrogen sulfide (H2S)-producing Escherichia coli with multidrug resistance properties carrying the mcr-1 gene in China†. Antibiotics 9:80. doi: 10.3390/antibiotics9020080
CLSI (2013). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. Approved Standard-Fourth Edition. CLSI document VET01-A4. Wayne, PA: Clinical and Laboratory Standards Institute, S10–S11.
CLSI (2019). Performance Standards for Antimicrobial Susceptibility Testing. CLSI supplement M100, 30th Edn. Wayne, PA: Clinical and Laboratory Standards Institute, 33–41.
Donà, V., Bernasconi, O. J., Pires, J., Collaud, A., Overesch, G., Ramette, A., et al. (2017). Heterogeneous genetic location of mcr-1 in colistin-resistant Escherichia coli isolates from humans and retail chicken meat in Switzerland: emergence of mcr-1-carrying IncK2 plasmids. Antimicrob. Agents Chemother. 61, e01245–17. doi: 10.1128/AAC.01245-17
Falgenhauer, L., Waezsada, S., Yao, Y., Imirzalioglu, C., Käsbohrer, A., Roesler, U., et al. (2016). Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing gram-negative bacteria in Germany. Lancet. Infect. Dis. 16, 282–283. doi: 10.1016/S1473-3099(16)00009-8
Fam, N., Leflon-Guibout, V., Fouad, S., Aboul-Fadl, L., Marcon, E., Desouky, D., et al. (2011). CTX-M-15-producing Escherichia coli clinical isolates in Cairo (Egypt), including isolates of clonal complex ST10 and clones ST131, ST73, and ST405 in both community and hospital settings. Microb. Drug. Resist. 17, 67–73. doi: 10.1089/mdr.2010.0063
Fang, L., Li, X., Li, L., Li, S., Liao, X., Sun, J., et al. (2016). Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli isolates of food-producing animals. Sci. Rep. 6:25312. doi: 10.1038/srep25312
Guenther, S., Falgenhauer, L., Semmler, T., Imirzalioglu, C., Chakraborty, T., Roesler, U., et al. (2017). Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J. Antimicrob. Chemother. 72, 1289–1292. doi: 10.1093/jac/dkw585
Hammad, A. M., Hoffmann, M., Gonzalez-Escalona, N., Abbas, N. H., Yao, K., Koenig, S., et al. (2019). Genomic features of colistin resistant Escherichia coli ST69 strain harboring mcr-1 on IncHI2 plasmid from raw milk cheese in Egypt. Infect. Genet. Evol. 73, 126–131. doi: 10.1016/j.meegid.2019.04.021
Hordijk, J., Farmakioti, E., Smit, L. A. M., Duim, B., Graveland, H., Theelen, M. J. P., et al. (2020). Fecal carriage of extended-spectrum-β-lactamase/AmpC-producing Escherichia coli in horses. Appl. Environ. Microb. 86, e02590–19. doi: 10.1128/AEM.02590-19
Kempf, I., Jouy, E., and Chauvin, C. (2016). Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 48, 598–606. doi: 10.1016/j.ijantimicag.2016.09.016
Li, R., Xie, M., Zhang, J., Yang, Z., Liu, L., Liu, X., et al. (2017). Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 72, 393–401. doi: 10.1093/jac/dkw411
Li, X., Sun, R., Song, J., Fang, L., Zhang, R., Lian, X., et al. (2020). Within-host heterogeneity and flexibility of mcr-1 transmission in chicken gut. Int. J. Antimicrob. Agents 55:105806. doi: 10.1016/j.ijantimicag.2019.09.010
Lin, Y., Yang, L., Lu, L., Wang, K., Li, J., Li, P., et al. (2020). Genomic features of an Escherichia coli ST156 strain harboring chromosome-located mcr-1 and plasmid-mediated blaNDM-5. Infect. Genet. Evol. 85:104499. doi: 10.1016/j.meegid.2020.104499
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet. Infect. Dis. 16:161. doi: 10.1016/S1473-3099(15)00424-7
Luo, J., Yao, X., Lv, L., Doi, Y., Huang, X., Huang, S., et al. (2017). Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli isolates from retail vegetables in China. Antimicrob. Agents Chemother. 61, e01139–17. doi: 10.1128/AAC.01139-17
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Malhotra-Kumar, S., Xavier, B. B., Das, A. J., Lammens, C., Butaye, P., and Goossens, H. (2016). Colistin resistance gene mcr-1 harboured on a multidrug resistant plasmid. Lancet. Infect. Dis. 16, 283–284. doi: 10.1016/S1473-3099(16)00012-8
Martin, J. K., Sheehan, J. P., Bratton, B. P., Moore, G. M., Mateus, A., Li, S. H., et al. (2020). A dual-mechanism antibiotic kills gram-negative bacteria and avoids drug resistance. Cell 181, 1518–1532. doi: 10.1016/j.cell.2020.05.005
Matamoros, S. (2017). Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 7:15364. doi: 10.1038/s41598-017-15539-7
McGann, P., Snesrud, E., Maybank, R., Corey, B., Ong, A. C., Clifford, R., et al. (2016). Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob. Agents Chemother. 60, 4420–4421. doi: 10.1128/AAC.01103-16
Migura-Garcia, L., González-López, J. J., Martinez-Urtaza, J., Aguirre Sánchez, J. R., Moreno-Mingorance, A., Perez De Rozas, A., et al. (2020). mcr-colistin resistance genes mobilized by IncX4, IncHI2, and IncI2 plasmids in Escherichia coli of pigs and white stork in Spain. Front. Microbiol. 10:3072. doi: 10.3389/fmicb.2019.03072
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Oteo, J., Diestra, K., Juan, C., Bautista, V., Novais, A., Perez-Vazquez, M., et al. (2009). Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int. J. Antimicrob. Agents 34, 173–176. doi: 10.1016/j.ijantimicag.2009.03.006
Papa-Ezdra, R., Grill Diaz, F., Vieytes, M., García-Fulgueiras, V., Caiata, L., Ávila, P., et al. (2020). First three Escherichia coli isolates harbouring mcr-1 in Uruguay. J. Glob. Antimicrob. Resist. 20, 187–190. doi: 10.1016/j.jgar.2019.07.016
Patiño-Navarrete, R., Rosinski-Chupin, I., Cabanel, N., Gauthier, L., Takissian, J., Madec, J., et al. (2020). Stepwise evolution and convergent recombination underlie the global dissemination of carbapenemase-producing Escherichia coli. Genome Med. 12:10. doi: 10.1186/s13073-019-0699-6
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Poirel, L., Kieffer, N., Brink, A., Coetze, J., Jayol, A., and Nordmann, P. (2016). Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob Agents Chemother. 60, 4394–4397. doi: 10.1128/AAC.00444-16
Porse, A., Schønning, K., Munck, C., and Sommer, M. O. A. (2016). Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol. Biol. Evol. 33, 2860–2873. doi: 10.1093/molbev/msw163
Rossi, F., Girardello, R., Morais, C., Cury, A. P., Martins, L. F., Silva, A. M., et al. (2017). Plasmid-mediated mcr-1 in carbapenem-susceptible Escherichia coli ST156 causing a blood infection: an unnoticeable spread of colistin resistance in Brazil? Clinics 72, 642–644. doi: 10.6061/clinics/2017(10)09
Sellera, F. P., Fernandes, M. R., Sartori, L., Carvalho, M. P., Esposito, F., Nascimento, C. L., et al. (2017). Escherichia coli carrying IncX4 plasmid-mediated mcr-1 and blaCTX-M genes in infected migratory Magellanic penguins (Spheniscus magellanicus). J. Antimicrob. Chemother. 72, 1255–1256. doi: 10.1093/jac/dkw543
Shen, C., Feng, S., Chen, H., Dai, M., Paterson, D. L., Zheng, X., et al. (2018). Transmission of mcr-1-producing multidrug-resistant Enterobacteriaceae in public transportation in Guangzhou, China. Clin. Infect. Dis. 67, S217–S224. doi: 10.1093/cid/ciy661
Snesrud, E., He, S., Chandler, M., Dekker, J. P., Hickman, A. B., Mcgann, P., et al. (2016). A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob. Agents Chemother. 60, 1416–1457. doi: 10.1128/AAC.01457-16
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, J., Yang, R. S., Zhang, Q., Feng, Y., and Fang, L. X. (2016). Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat. Microbiol. 1:16176. doi: 10.1038/nmicrobiol.2016.176
Wang, Q., Sun, J., Li, J., Ding, Y., Li, X., Lin, J., et al. (2017). Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome 5:70. doi: 10.1186/s40168-017-0288-0
Wang, R., van Dorp, L., Shaw, L. P., Bradley, P., Wang, Q., Wang, X., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9:1179. doi: 10.1038/s41467-018-03205-z
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PloS. Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Xavier, B. B., Lammens, C., Butaye, P., Goossens, H., and Malhotra-Kumar, S. (2016). Complete sequence of an IncFII plasmid harbouring the colistin resistance gene mcr-1 isolated from Belgian pig farms. J. Antimicrob. Chemother. 71, 2342–2344. doi: 10.1093/jac/dkw191
Yang, R., Feng, Y., Lv, X., Duan, J., Chen, J., Fang, L., et al. (2016). Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single muscovy duck (Cairina moschata). Antimicrob. Agents Chemother. 60, 6899–6902. doi: 10.1128/AAC.01365-16
Zhang, C., Feng, Y., Liu, F., Jiang, H., Qu, Z., Lei, M., et al. (2017). A phage-like IncY plasmid carrying the mcr-1 gene in Escherichia coli from a pig farm in China. Antimicrob. Agents Chemother. 61, e02035–16. doi: 10.1128/AAC.02035-16
Zhi, C., Lv, L., Yu, L., Doi, Y., and Liu, J. (2016). Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16, 292–293. doi: 10.1016/S1473-3099(16)00063-3
Zhong, L., Phan, H. T. T., Shen, C., Vihta, K., Sheppard, A. E., Huang, X., et al. (2018). High rates of human fecal carriage of mcr-1–positive multidrug-resistant Enterobacteriaceae emerge in China in association with successful plasmid families. Clin. Infect. Dis. 66, 676–685. doi: 10.1093/cid/cix885
Zhou, Z., Alikhan, N., Sergeant, M. J., Luhmann, N., Vaz, C., Francisco, A. P., et al. (2018). GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 28, 1395–1404. doi: 10.1101/gr.232397.117
Zurfluh, K., Klumpp, J., Nüesch-Inderbinen, M., and Stephan, R. (2016). Full-Length nucleotide sequences of mcr-1-harboring plasmids isolated from extended-spectrum-β-lactamase-producing Escherichia coli isolates of different origins. Antimicrob. Agents Chemother. 60, 5589–5591. doi: 10.1128/AAC.00935-16
Keywords: Escherichia coli, mcr-1, sequence type, T4SS, integrons and transposons
Citation: Liu Z, Liu Y, Xi W, Liu S, Liu J, Mu H, Chen B, He H, Fan Y, Ma W, Zhang W, Fu M, Wang J and Song X (2021) Genetic Features of Plasmid- and Chromosome-Mediated mcr-1 in Escherichia coli Isolates From Animal Organs With Lesions. Front. Microbiol. 12:707332. doi: 10.3389/fmicb.2021.707332
Received: 09 May 2021; Accepted: 08 July 2021;
Published: 05 August 2021.
Edited by:
Prescilla Emy Nagao, Rio de Janeiro State University, BrazilReviewed by:
João Pedro Rueda Furlan, University of São Paulo, BrazilAna Paula D’Alincourt Carvalho-Assef, Oswaldo Cruz Foundation (Fiocruz), Brazil
Copyright © 2021 Liu, Liu, Xi, Liu, Liu, Mu, Chen, He, Fan, Ma, Zhang, Fu, Wang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Wang, juan.wang1234@hotmail.com; Xiaoping Song, sxptcvm@163.com
†These authors have contributed equally to this work
 Zengyuan Liu1†
Zengyuan Liu1† Yingqiu Liu
Yingqiu Liu Yunpeng Fan
Yunpeng Fan Xiaoping Song
Xiaoping Song