
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 21 July 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.707330
Rising antibiotic resistance is a global threat that is projected to cause more deaths than all cancers combined by 2050. In this review, we set to summarize the current state of antibiotic resistance, and to give an overview of the emerging technologies aimed to escape the pre-antibiotic era recurrence. We conducted a comprehensive literature survey of >150 original research and review articles indexed in the Web of Science using “antimicrobial resistance,” “diagnostics,” “therapeutics,” “disinfection,” “nosocomial infections,” “ESKAPE pathogens” as key words. We discuss the impact of nosocomial infections on the spread of multi-drug resistant bacteria, give an overview over existing and developing strategies for faster diagnostics of infectious diseases, review current and novel approaches in therapy of infectious diseases, and finally discuss strategies for hospital disinfection to prevent MDR bacteria spread.
Antimicrobial resistance (AMR) is an evolutionary response of bacteria, viruses, and fungi to withstand antimicrobial drugs introduced into their environment. In this systematic review we will focus on the resistance in bacteria only and the reader is advised to go through other reviews on viral and fungal resistance (Srinivasan et al., 2014; Fuentefria et al., 2018; Poole and James, 2018; Lampejo, 2020).
In 2020, the COVID-19 pandemic has clearly demonstrated how fragile our world is and how infections do not respect borders. However, we have long been living in a silent pandemic of AMR (Jasovsky et al., 2016). Each year, infections caused by resistant bacteria cause 68,000 deaths in the EU/EEA and the United States combined (Cassini et al., 2019; CDC, 2019), and are contributing to US €55 billion economic loss in the United States and to €1.6 billion in the EU/EEA annually (Ahmad and Khan, 2019). Although the AMR problem is not evenly distributed across the globe (Figure 1; Klein et al., 2019), there is no single country on Earth that can safely state that it will not be affected by the AMR spread. The first warning on a potential catastrophe came from Fleming (1945), in his speech on accepting the Nobel prize for discovery of penicillin, the first industrially produced antibiotic. Although he did not foresee the global spread of antibiotic resistance, he was the first one to recognize the danger of the resistance for everyone who relies on these drugs. In 2019, the Center for Disease Control and Prevention (CDC) released a report stating that we have entered the post-antibiotic era (CDC, 2019).
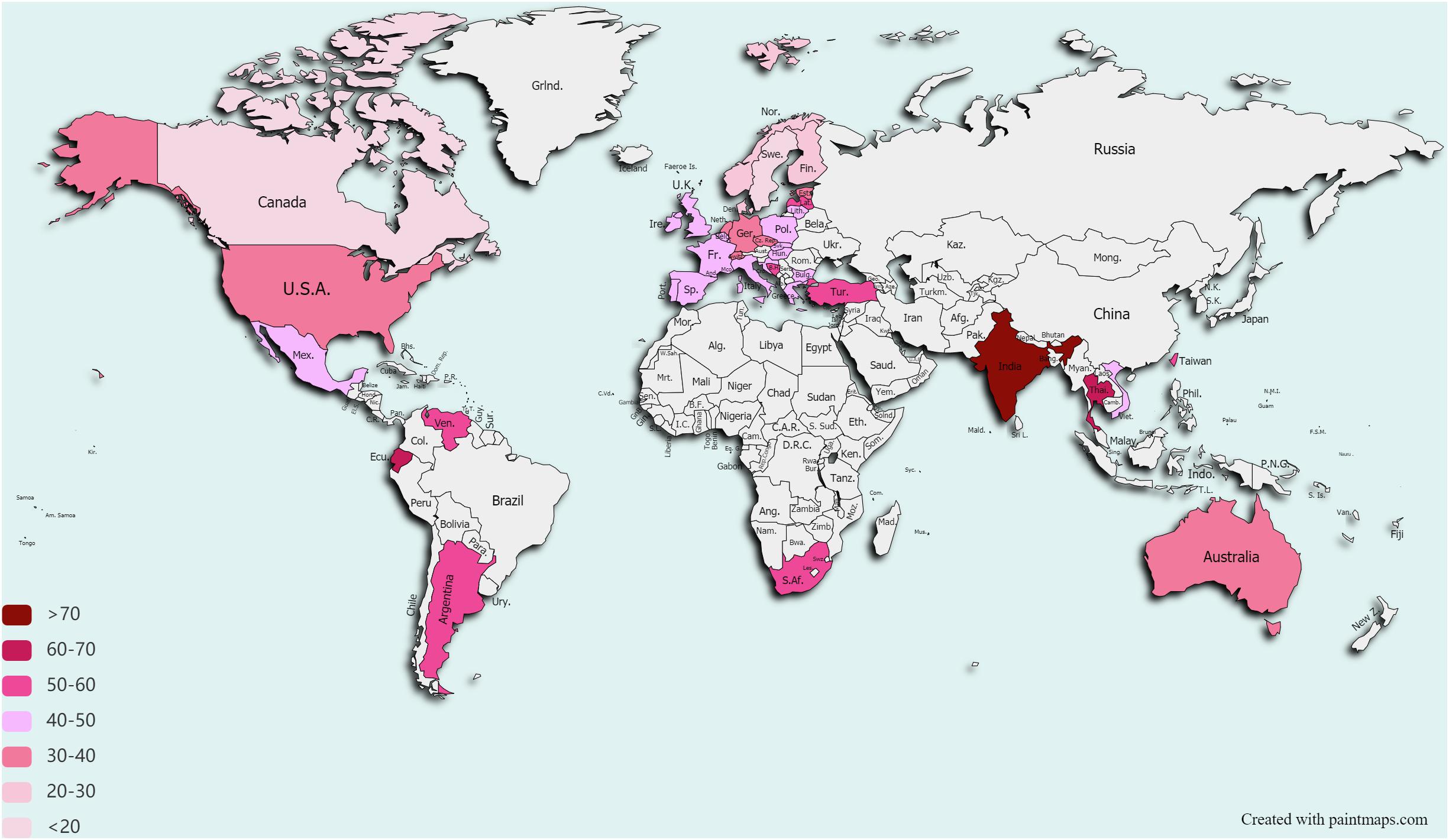
Figure 1. Drug Resistance Index worldwide. Only countries that reported data on antibiotic use for ≥5 pathogens and for ≥15 pathogen–antibiotic combinations for at least 1 year between 2012 and 2015 are depicted. Data taken from Klein et al. (2019). Data on Taiwan are taken from Resistance Map (https://resistancemap.cddep.org).
Antibiotics are classified by their chemical structure and have different modes of action, including cell wall synthesis inhibition (e.g., beta-lactams, fosfomycin, and vancomycin), DNA replication inhibition (e.g., fluoroquinolones), protein synthesis inhibition (e.g., tetracyclines and aminoglycosides), metabolic pathways inhibition (e.g., trimethoprim and sulfonamides) (Ma et al., 2020). The 1950 and 1960-s are a “Golden Era” of antibiotics (Lyddiard et al., 2016). More than half of currently used antibiotic classes were discovered at that time and majority of antibiotic scaffolds were derived from bacteria and fungi. Actinomycetes spp., for example, gave rise to 14 different drug classes, including carbapenems, aminoglycosides, glycopeptides, and lipopeptides (Hutchings et al., 2019). By the end of 1960s, novel scaffolds became more and more difficult to find and antibiotic discovery stalled. Cyclic lipopeptides were the last novel naturally derived antibiotic class that was discovered in 1987 and introduced into clinics in 2003 (daptomycin) (Eisenstein et al., 2010; Miller et al., 2016). Since that time, none of novel natural-product antibiotic scaffolds were discovered, although new antibiotics within existing classes are being constantly designed and developed. For example, in 2019 the FDA approved clinical use of Xenleta (Nabriva Therapeutics, Ireland), a drug from a pleuromutilin class, that was first discovered back in 1951 from a fungal source (Novak and Shlaes, 2010).
Some bacteria are intrinsically resistant toward some antibiotics due to their cell wall structure, activity of efflux pumps or presence of porins (Eichenberger and Thaden, 2019). In this case, all the strains within the species are insensitive to a given antibiotic. Acquired resistance, on the other hand, appears when some of the strains within the species become resistant toward the antibiotic they were previously susceptible to. Mechanisms include either mutations in existing genes, for example in intracellular targets (Musser James et al., 2020) or core metabolic genes (Lopatkin et al., 2021) or acquisition of new antibiotic resistance genes (ARGs) through horizontal gene transfer (HGT) (Eichenberger and Thaden, 2019). The latter enables intra- and inter-species transmission of ARGs and is responsible for an AMR pandemic (von Wintersdorff et al., 2016; Sun et al., 2019). Although ARGs existed long before antibiotics became discovered and put into a wide use (Hall and Barlow, 2004; Allen et al., 2010), our irresponsible use of antibiotics in animal husbandry, overuse of antibiotics in health care, improper treatment of wastewaters and poor sanitation in low and middle income countries has led to wide spread of resistant bacteria across the globe (Prestinaci et al., 2015; Holmes et al., 2016; Ma et al., 2020). To date, the most worrisome global spread of ARGs is the plasmid-mediated spread of carbapenemases (KPC, NDM, VIM, OXA-48, and OXA-51); colistin-resistance genes (mcr) in Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa; vancomycin resistance gene (vanA) in Enterococci and Staphylococcus aureus; and methicillin resistance gene (mecA) in S. aureus (de Niederhausern et al., 2011; Poirel et al., 2014; Lerner et al., 2015; Wang R. et al., 2018; Chaalal et al., 2020; Hishinuma et al., 2020; Snitser et al., 2020).
Health-care-associated infections (HAI) comprise infections that are acquired from hospitals or health care centers. They normally onset 48 h after hospitalization, but may occur even after discharge of patients (Revelas, 2012; Cassini et al., 2019). On average they occur in 7 and 10% of hospitalized patients in developing and developed countries, respectively (WHO, 2015). In Europe, around 3.2 million patients per year are affected by HAI (ECDC, 2013). The severity of infection and its incidence is directly correlated to the patients’ immunological status. Patients of burn units, intensive care units (ICUs), organ transplant receivers and neonates are the most affected groups. As such, a recent retrospective study from Serbia reported nearly every third patient admitted to ICU suffered at least one incident of HAI (Despotovic et al., 2020). HAIs are also responsible for three out of four lethal cases in neonates in Sub-Saharan Africa and South-East Asia (WHO, 2011). The most common HAI types are surgical site infections (2–5% incidence rate), catheter-related blood stream infections (12–25% incidence rate), catheter-related urinary tract infections (12% incidence rate) and ventilator-associated pneumonia (9–27% incidence rate) (Khan et al., 2017).
The World Health Organisation (WHO, 2017) stratified the most critical pathogens within HAI into three groups based on their global threat and urgency of action needed. The first and the second groups (urgent and high priority pathogens) include so-called ESKAPE pathogens (vancomycin-resistant Enterococcus faecium (VRE), methicillin-resistant, and vancomycin-resistant S. aureus (MRSA/VRSA), carbapenem-resistant and third-generation cephalosporin-resistant Klebsiella pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter spp.). ESKAPE pathogens are associated with high morbidity and mortality HAIs due to their acquired resistance toward a large number of antibiotics, including last-resort antibiotics such as carbapenems and colistin (Santajit and Indrawattana, 2016; Ma et al., 2020). As such, a recent study from Greece reported that 90-days mortality risk was doubled in patients with HAI caused by carbapenem-resistant pathogens compared to patients without HAI (Kritsotakis et al., 2017). It is worth noting, though, that Greece is a country with highest consumption of antibiotics and with second largest disability-adjusted life years burden due to AMR in the EU (OECD, 2019).
Numerous reports are available on detecting resistant pathogens (including ESKAPE) in common areas in the hospitals (Li et al., 2015; Mirhoseini et al., 2016; Chng et al., 2020; Rodrigues et al., 2020). In a recent study from a tertiary-care hospital in Singapore (Chng et al., 2020), authors have collected swabs from both frequent contact- (bed rails, bedside lockers, and door knobs) and low contact areas (sink traps and aerators). They report a very worrisome spread and persistence of MDR organisms, some with novel and clinically dangerous ARG combinations. The authors also detected a plasmid containing both methicillin resistance gene mecA and genes for resistance toward disinfectants (qacA and qacC). Alarmingly, many of the MDR isolates have persisted in the hospital for nearly a decade, leading to opportunistic infections in hospital patients (Chng et al., 2020).
Antibiotic resistance is transmitted between animals and humans via the environment. For example, swine farming wastes can contaminate groundwater with ARGs and resistant bacteria, which can then colonize humans (Gao et al., 2020). Colistin (mcr) and carbapenem (NDM and VIM) resistance genes were detected in Salmonella isolates from food animals raising the danger of resistant bacteria infection through foodborne pathogens (Mthembu et al., 2021). NDM-1 gene was first detected in 2008 on a transferable plasmid in K. pneumoniae isolate from a patient who had been repatriated to Sweden from a New Delhi hospital (India) (Yong et al., 2009). Few years later it was detected in drinking and seepage water in New Delhi (Walsh et al., 2011) and in the river in Vietnam (Isozumi et al., 2012). Nowadays, NDM-1 and its variants are spread across the globe (Farhat and Khan, 2020), and were even detected in Arctic soil of remote region on Svalbard (Norway) with no agriculture or industry and <120 inhabitants, implicating possible spread via migrating birds (McCann et al., 2019). The One Health holistic approach is vital for mitigation of the AMR spread. One Health is a multi-sectoral concept recognizing interconnection between animal health, human health and the environment (Kim and Cha, 2021). It seeks for multidisciplinary effort to reduce dissemination of resistant bacteria worldwide through interventions in agriculture (and aquaculture), veterinary science, health sector, and sanitation. However, with multifactorial system in mind, human-to-human transmission still seemed to be the major contributor to the extended-spectrum beta-lactamase (ESBL) producing E. coli spread in the Netherlands community (Mughini-Gras et al., 2019). A recently published mathematical model for reduction of antibiotic resistant bacteria in humans also corroborates these findings (Booton et al., 2021). The model was based on the actual prevalence of ESBL-producing bacteria in Thailand and One Health drivers were weighted with regards to their sole or interactive impact on resistant bacteria dissemination in humans. Based on their model, reduction of human antibiotic use seems to be the most influential factor on the reduction of colonization of humans by resistant bacteria in a 20-years perspective.
Apart from educational, regulatory and political initiatives (Ahmad and Khan, 2019; Cole et al., 2019; Hernando-Amado et al., 2019; Ardal et al., 2020), there are three pillars to reduce the human impact on AMR spread. We need to develop and implement improved disinfection and hygiene routines in order to curb resistant pathogens persistence. We also need to develop new, fast and precise tools for infection diagnostics. Last, but not least, we have to search for new therapeutic strategies to combat resistant bacterial infections.
Majority of infections caused by MDR bacteria are of nosocomial origin (Matta et al., 2018), and these MDR pathogens can reside in hospitals for years (Chng et al., 2020). Several reports have demonstrated that a risk of acquiring HAI increases if a previous patient in a room had HAI (Mitchell et al., 2015). Unfortunately, hospitals disinfection is often only scarcely touched upon when discussing AMR spread issue, and current cleaning routines have remained largely unrevised for the last 25 years (Peters et al., 2018). The main global change in Infection Prevention and Control routines was the introduction of alcohol-based handrubs by the WHO in 2005 (Pittet and Donaldson, 2005), the routine which is followed in over 180 countries worldwide (Peters et al., 2018). However, based on a recent joint report of healthcare facilities published by the WHO and UNICEF, every third facility globally lacks adequate hand cleaning at points of care, and two out of three facilities in least developed countries do not have adequate waste management service (WHO, 2020). Hospital wastewaters contain high levels of resistant bacteria, ARGs and mobile genetic elements (Wang Q. et al., 2018) and strongly contribute to their dissemination in the environment even after treatment (Buelow et al., 2020). A recent study from Germany indicated, that effluents from hospital wastewater treatment plants may contain up to 70% higher daily discharge of NDM-1, mcr-1, vanA, and mecA genes compared to communal or food production wastewater treatment plants although median daily discharge values did not differ (Alexander et al., 2020).
Commonly used chemical disinfectants for cleaning hospital surfaces and waste waters include chlorine and chlorine-based products, hydrogen peroxide, alcohol (ethanol, isopropanol), quaternary ammonium compounds, and formaldehyde (Peters et al., 2018). Hydrogen peroxide vapor (HPV) used for surface disinfection, is sometimes combined with silver ions (Totaro et al., 2020). In a field study, 30% HPV-Ag+ treatment of hospital rooms eliminated MRSA from the hospital surfaces with no regrowth after 2 weeks (Bartels et al., 2008). Hydrogen peroxide is also most effective for eliminating resistant A. baumannii compared to sodium hypochlorite and chlorine dioxide (Biswas et al., 2018). H2O2 is utilized in so-called electroperoxone (E-peroxone) process for waste water treatment (Wang Y. et al., 2018). In this process, H2O2 generated by electrical process, reacts with ozone and reduces ARGs count (Zheng et al., 2018). Unfortunately, chemical agents can promote HGT in pathogens and thus contribute to AMR spread (Alotaibi et al., 2017; Lu and Guo, 2021). For example, hospital K. pneumoniae strains harboring numerous ARGs, can become tolerant to chlorine, and can then escape into environment through hospital wastewaters (Popa et al., 2020).
Physical processes are more difficult to develop resistance toward. UV-C light (wave length 200–280 nm) causes multiple DNA lesions which are difficult to repair (Clancy, 2008). It is an effective disinfection strategy for hospital surfaces (Rastogi et al., 2010). For example, mobile Hyper Light P3 disinfection robot utilizing UV-C is effective for killing MDR pathogens in hospitals, albeit at the distance of 1 m only, with reduced effect at 2 or 3 m (Yang et al., 2019). Efficacy of UV treatment is dependent on the GC-content of bacteria, with GC-rich bacteria being more resilient to UV light treatment (Pullerits et al., 2020). As such, >20 min of UV light treatment is required to reach 3-log reduction of Mycobacterium tuberculosis (GC ≥ 65%) compared to <10 min for MRSA (GC ≈ 33%) or extended spectrum beta-lactamase (ESBL) producing E. coli (GC ≈ 50%) (Szeto et al., 2020).
To avoid bacterial contamination, medical tools are often coated with silver nanoparticles (NPs) (Deshmukh et al., 2019). ZnO NPs were suggested for cleaning hospital surfaces contaminated with resistant P. aeruginosa (Omrani and Fataei, 2018). Ag–NPs–TiO2-embedded filters installed in ventilation systems of hospital ward demonstrated 88% bacterial removal within 30 min (Chen et al., 2019). Ag–NPs in combination with TiO2 films may also serve as a photocatalyst for photoinactivation of bacteria in visible indoor light (Dunnill et al., 2011). MoS2/α-NiMoO4 nanostructure developed Ray et al. (2020), also disrupts bacterial cell at visible light. It has been shown to inactivate S. aureus and was proposed to be used to decontaminate MDR-containing waste waters. Combination of NPs with bacteriophages can also be successfully implemented for waste water treatment in hospitals (Talan and Tyagi, 2020). It is very important to be careful with NPs concentration used for waste water treatment. It has been shown that ZnO NPs at low concentrations can enhance horizontal transfer of ARGs between bacteria (Wang X. et al., 2018). However, it has been also shown that despite enhancement of HGT mechanism, metal NPs ultimately kill pathogens and attenuate ARGs (Su et al., 2019).
Sterility of medical devices or implants is crucially important to prevent patient’s infection. Plants and insects utilize a variety of nanostructured antibacterial surfaces for protection against pathogenic bacteria. For example, cicada wings are covered with 200 nm long nanoneedles, and gecko’s skin is shielded by hair-like curved nanostructures (Tripathy et al., 2017). These surfaces physically disrupt bacteria cell wall killing the pathogen within minutes after the initial contact (Ivanova et al., 2012; Watson et al., 2015). Majority of these surfaces are active only against gram negative bacteria due to their thin cell wall. However, inspired by naturally occurring bactericidal nanosurfaces, scientists created a plethora of artificial nanostructured coating materials that are lethal to gram positive bacteria as well. These materials include nanocone surfaces, nanowires, nanograss, nanospikes, nanopillars, and nanorings, etc. (Hazell et al., 2018; Gonzalez-Fernandez et al., 2020; Gudz et al., 2020; Jenkins et al., 2020). For example, black silicon (nanograss) was created by mimicking cicada wings nanoneedles, which are lethal not only to gram-negative P. aeruginosa, but also to gram-positive S. aureus and B. subtilis (Ivanova et al., 2013). Additionally, recently a nanostructure with a combination of antiviral and antibacterial properties was reported (Hasan et al., 2020).
There are several considerations to be resolved before nanostructures can be put to a wide use for medical devices. Many of these structures were shown to disrupt not only bacterial, but also mammalian cells (Shalek et al., 2010; Pham et al., 2014). However, recently investigators report little to no adverse effect on mammalian cells (CLSI, 2019; Wandiyanto et al., 2019; Saini et al., 2020). Large scale fabrication of these materials is also costly, although production of nanospikes, nanorods, nanowires, and nanoneedles can be achieved at low cost (Tripathy et al., 2017; EUCAST, 2021).
Even when perfectly following hand hygiene, resistant bacteria reside on workers’ clothing and hospital textiles (Lena et al., 2021). These bacteria can further contaminate laundry facilities, washing machines, and cause pathogen outbreaks (Michael et al., 2017). For example, and outbreak of ESBL-producing K. pneumoniae in a 40-bed rehabilitation center in the Netherlands was traced back to a contaminated washing machine. The outbreak was contained after taking the machine out of service and reinforcing its use protocols (Boonstra et al., 2020). For the efficient pathogen removal from textiles and clothing, water temperature, bleach use and mode of drying are essential (Tano and Melhus, 2014; Bockmuhl, 2017). As such, laundering at 60°C is recommended for complete removal of bacteria (Riley et al., 2017). In recent years, in order to reduce energy consumption, wash temperatures have been lowered to below 60°C, which may compromise the efficacy of pathogen removal and lead to clothing re-contamination (Riley et al., 2017; Bockmuhl et al., 2019). Moreover, in the United Kingdom, for example, healthcare workers uniforms are washed at home to reduce National Health Service costs, and many workers do not follow the washing recommendations (Riley et al., 2015).
Healthcare workers’ mobile phones can also be a source of transmission of resistant bacteria to the community (Debnath et al., 2018; Mushabati et al., 2021). In EU, this problem seems to be less striking with fewer resistant bacteria detected on the phones (Galazzi et al., 2019; Missri et al., 2019). However, routines for mobile phone disinfections should be implemented and strictly followed worldwide.
No matter how good disinfectants and strategies we use, the key is to use them properly, at a right dosage, time, for the right bug and equipment, and strictly following the protocols. However, cleaning service is perceived as a low qualified job and there is a high turn-around among the personnel in hospitals. In addition to low rang, many workers in cleaning service in high-income countries are immigrants, often with very limited skills in local language (Peters et al., 2018). This makes it difficult to convey an importance of such a “mundane” but yet crucially important task to the personnel. Therefore, we also need to break the perception of cleaning in hospitals as a dull and low-quality job and to use well trained and certified cleaners for such critically important areas as hospital is.
Classical phenotypic antibiotic susceptibility testing (AST) refers to broth microdilution (BMD) or diffusion (Khan et al., 2019). BMD is regarded as gold standard MIC breakpoint determination both by the Clinical and Laboratory Standards Institute (2019) and the European Committee on Antimicrobial Susceptibility Testing (2021). Number of automated systems for AST testing have been approved for commercialization by the United States Food and Drug Agency (FDA), like Vitek2 (BioMérieux, France), Microscan (BeckmanCoulter, United States), Phoenix (BD diagnostics, United States), and Sensititre (ThermoFisher, United States).
Although quite precise, these systems require long time for MIC determination and are limited to a certain species/drug panel. For instance, common time for obtaining AST profile of an infectious agent in bloodstream infections may be as long as 1 week after the initial diagnosis has been made (Briggs et al., 2021). This waiting time would be lethal for a patient and therefore doctors start with broad-spectrum antibiotics until the AST results come in Kumar et al. (2009). Ideally, broad-spectrum antibiotics should be rapidly de-escalated toward optimal antibiotic to minimize their use and thus to reduce the risk of resistance development. Rapid diagnostics tools enable AST inference within hours, providing an opportunity for fast shift toward optimal antibiotics use. In a retrospective study from a tertiary care children’s hospital, Reuter et al. (2019) reported a significant reduction in duration of suboptimal antibiotic treatment when rapid AST was performed. Recently EUCAST has developed a rapid antibiotic susceptibility testing (RAST) method directly from positive blood culture bottles. The method is based on disk diffusion with shortened incubation times of 4–8 h and was extensively validated in clinical labs across 55 European countries (Åkerlund et al., 2020). A plethora of rapid diagnostic tools based on molecular methods such as fluorescence microscopy, proteins detection, hybridization, nucleic acid amplification technologies, and immunodetection are being currently developed [reviewed in detail in Vasala et al. (2020) and van Belkum et al. (2020)]. Here we will give an overview on gold standard rapid diagnostics, genotypic (detect potential antibiotic resistance), and phenotypic (detect actual antibiotic resistance) point-of-care tools approved by FDA and touch on emerging technologies for future rapid diagnostics (Table 1).
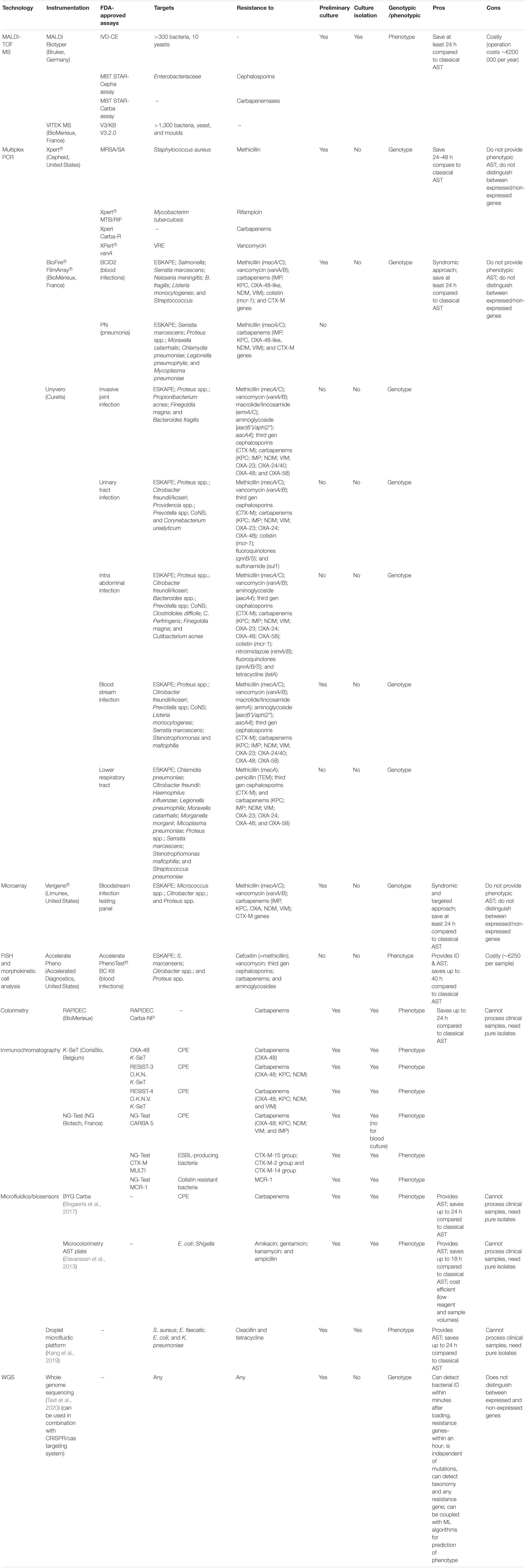
Table 1. Overview of alternatives to classical antibiotic susceptibility testing (AST) discussed in this review.
The gold standard rapid bacteria identification technology is matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF MS) that identifies bacteria by comparing its protein profile to a reference library within minutes after the sample is loaded. The FDA-approved Bruker MALDI Biotyper (Bruker, Germany) uses a library of >300 clinically relevant bacteria and yeast (FDA, 2018; Florio et al., 2019). Bruker (Germany) also provides MBT STAR-Carba and MBT STAR-Cepha assays for detection of carbapenem and third gen cephalosporin resistance in Enterobacteriaceae (both assays), Acinetobacter spp. (MBT STAR-Carba only), and Pseudomonas spp. (MBT STAR-Carba only) within 1 h after loading.
The VITEK® MS (BioMérieux, France) reference database contains protein spectra of >1,300 species of bacteria, yeast and moulds. When coupled with Vitek2 (BioMérieux, France) for AST, the system can seamlessly provide simultaneous ID and AST of the pathogen. They also provide a VITEK® MS Blood Culture kit for identification of blood stream infection directly from blood culture samples, but per date this kit is for research use only.
Although these systems allow rapid and accurate bacterial identification and AST profile, MALDI-TOF MS has two major drawbacks. It requires isolated colonies from overnight culture, saving only around 24 h compared to conventional AST (Briggs et al., 2021). The incubation time, however, can be shortened down to 3–6 h incubation prior to MALDI-TOF MS (Cherkaoui et al., 2020). Secondly, a MALDI-TOF MS system may cost up to €200,000 per year (excluding operational costs) rendering it impossible for use in outpatient clinics or small hospitals (Vasala et al., 2020).
Several multiplexed panels are FDA-approved for point-of-care use. Cepheid® provides targeted test panels for detection of MRSA (Xpert® MRSA/SA), Mycobacterium tuberculosis and rifampicin resistance (Xpert® MTB/RIF), vancomycin-resistant enterococci (XPert® vanA), and for carbapenem resistance detection (Xpert Carba-R) among others. BioMérieux BioFire® FilmArray® provides panels for simultaneous identification of up to 43 targets (bacteria and resistance genes) using syndromic approach for pneumonia and bloodstream infections. Unyvero (Curetis) syndromic panels cover bacteria and ARGs detection in invasive joint infections, urinary tract infections, intra-abdominal infections, blood stream infections and lower respiratory tract infections. All of these panels are based on multiplex PCR and require 1–5 h load to end time. The Verigene® system by Luminex Corporation is based on microarray technology and has both targeted and syndromic panels for detection of main pathogens and resistance genes within a matter of hours. Although providing fast and broad detection of pathogens with good concordance to conventional methods (Briggs et al., 2021), these panels require a preliminary culturing step of clinical samples, which can take up to 24 h. Moreover, same as other nucleic-acid based technologies, they do not provide phenotypic AST and do not distinguish between expressed and non-expressed genes. In case of polymicrobial infections, they also cannot provide a pathogen/gene relation. The other approach can be to use a proxy for resistance instead of direct ARGs search. It has been noted that integron detection in bacterial DNA is highly correlated with antibiotic resistance of Enterobacteriaceae pathogens in blood cultures from septic patients (Barraud et al., 2014), opening the possibility for a rapid inference of antibiotic resistance and correction of the antibiotic prescription prior to detailed AST.
There is a potential for development of a rapid diagnostic tool using Whole Genome Sequencing (WGS). WGS provides excessive amount of genome information without the need for multiplex targeting of certain genes, bringing the advantage of wide coverage of variety of pathogens with one set up. Since the approach is non-targeted, it is also less sensitive to mutations in genes. For example, the multiplex PCR-based Xpert® MTB/RIF panel (Cepheid®, United States) does not detect RpoB Ile149Phe mutation and reports false negative results (Zhao Z. L. et al., 2020). The other advantage of WGS is that it is capable of capturing slow-growing and fastidious bacteria, which may be troublesome or impossible to detect by growth-based methods (Balloux et al., 2018). With real-time sequencing technologies as ONT sequencing (Oxford Nanopore Technologies, United Kingdom), the sequencing information can be received within minutes after the sample load and bacterial ID and ARGs can be detected on the flow (Taxt et al., 2020).
Same as other nucleic-acid based technologies, however, current WGS methodologies do not allow direct clinical sample sequencing and require preliminary culturing. To avoid this step, CRISPR/Cas9 based detection of AMR sequences was proposed by Quan et al. (2019). This highly multiplexed approach enables precise and cheap detection of all known ARGs from low abundance samples. Here, genomic DNA from a patient sample is digested with Cas9 endonuclease guided by a set of RNAs designed specifically from target ARG sequences. Restricted fragments are then ligated to adaptors, enriched by PCR and sequenced. Although this approach was tested only on a handful of clinical samples, it exhibited promising results with extraordinary sensitive ARG detection surpassing direct NGS by 5,000-fold (Quan et al., 2019).
Antibiotic resistance gene detection does not always correlate to phenotypic AST profile of the pathogen, especially when changes in influx/efflux transport systems may render antibiotic inefficient due to its inability to accumulate in the cell (Masi et al., 2017). Here, various machine-learning (ML) algorithms may be implemented for genotype-to-phenotype correlations. Although these algorithms have not been introduced to a clinical routine per date, there are number of publications demonstrating the power of ML to predict susceptibility and MIC based on genomic features for beta-lactams, aminoglycosides, polymyxins, and other antibiotic classes (Nguyen et al., 2018; Kim et al., 2020; Macesic et al., 2020; Avershina et al., 2021). However, these models are species and drug specific and are dependent on the resistance mechanisms represented in a training set, limiting their use with isolates that possess other mechanisms/genomic signatures (Avershina et al., 2021).
AcceleratePhenoTM (Accelerated Diagnostics, United States) is the only FDA-approved point-of-care rapid diagnostic platform that enables simultaneous identification and MIC-based AST profile of a pathogen directly from a clinical sample. For the moment, it covers 14 common infectious agents in bloodstream infections, including ESKAPE pathogens and two Candida species, and performs MIC testing to cefoxitin (a surrogate for methicillin resistance detection), vancomycin, third generation cephalosporins, carbapenems, and aminoglycosides. Pathogen identification is performed using fluorescence in situ hybridization and phenotypic AST is performed using morphokinetic cell analysis of microbial cells in an antibiotic-supplemented Mueller-Hinton media (Charnot-Katsikas et al., 2018). AcceleratePhenoTM allows up to 27 h faster bacterial ID and 40 h faster AST compared to conventional culture-based identification and AST (Marschal et al., 2017). However, this technology is yet costly (∼€250 per sample) for use in point-of-care centers (Vasala et al., 2020).
Colorimetric assays allow rapid identification of antibiotic resistant bacteria by changing the color of the medium on the change of pH resulting from the activity of resistance genes products. For example, RAPIDEC® Carba-NP (BioMerieux, France) detects carbapenemase activity in ESKAPE pathogens with ∼95% sensitivity and specificity within 30 min–2 h after loading (Poirel et al., 2015; Mancini et al., 2017). However, colorimetric assays require at least overnight culture and can’t be used directly on the clinical sample.
Microfluidics holds a big promise for rapid AST technologies. These technologies operate with micro volumes and are thus portable and allow for multiplexing and cost-effectiveness. Resazurin dye can be used in microcolorimetry for AST when added to an antibiotic-supplemented medium. In presence of viable bacteria, resaruzin will be reduced and its color changed from blue to pink and leuco, an approach tested on E. coli and Shigella by Elavarasan et al. (2013). Kang et al. (2019) has reported an image-based parallelized droplet microfluidic platform capable of screening four bug-drug combinations simultaneously requiring 30 min–2 h for AST. Microfluidics is vastly used in various biosensors, analytical devices that detect a biological reaction by converting a chemical response into an electrical signal (Mehrotra, 2016). For example, change in pH caused by degradation of carbapenems by carbapenem-resistant Enterobacteriaceae (CPE) laid foundation for the development of the BYG Carba biosensor capable of detecting CPE within 30 min (Bogaerts et al., 2017). Immunochromatographic assays use monoclonal antibodies specific to resistance enzymes. OXA-48 K-SeT (Coris BioConcept, Belgium) was the first test developed for in vitro identification of OXA-48-like carbapenemases in bacterial cultures. Later, assays for additional detection of NDM and KPC (RESIST-3 O.K.N. K-SeT assay) and of VIM (RESIST-4 O.K.N.V. K-SeT assay) were developed (Pitout Johann et al., 2019). NG-Biotech Laboratories (France) has developed tests for detection of resistance to carbapenems (NG-Test CARBA 5: KPC, OXA-48-like, VIM, IMP, and NDM), third gen cephalosporins (NG-Test CTX-M MULTI: CTX-M) and colistin (NG-Test MCR-1: mcr). These tests are highly specific, require little hands-on time and deliver results within 15 min after loading (Pitout Johann et al., 2019; Bianco et al., 2020; Potron et al., 2020). However, these technologies need pure isolates and could not process clinical samples. The immunoassay-based field effect enzymatic detection biosensor, on the other hand, allowed direct detection and AST typing of low concentration E. coli in blood (∼10 CFU/ml) in less than 3 h (Shi et al., 2018).
Overview over alternatives to current antibiotics discussed in this review is provided in Table 2. As of December 2020, there were 13 antibiotic drugs in Phase II clinical trials and 13–in Phase III clinical trials (PEW, 2021). Most of these drugs are modifications or combinations of already existing antibiotic scaffolds, but some are from novel synthetic classes. Gepotidacin (GSK2140944, Glaxo SmithKlein, United Kingdom) is a first drug in a novel synthetic class of triazaacenaphthylene bacterial topoisomerase inhibitors, aimed at treatment of uncomplicated urinary tract infections and urogenital gonorrhea. Its phase II clinical study demonstrated eradication of Neisseria gonorrhoeae in 95% of participants, and the drug has now entered the Phase III.
Murepavadin (Polyphor AG, Switzerland) is another example of a novel synthetic class antibiotic. It blocks lipopolysaccharide transport to the outer membrane of bacteria and was effective against treatment of P. aeruginosa, especially in cystic fibrosis patients. In 2018, the drug entered Phase III clinical trial for intravenous formulation, but it was discontinued by 2019 due to high incidence of acute kidney failure in patients (Smith, 2019). However, in December 2020 the company received another clinical trial authorization for an oral inhalation formulation of murepavadin against P. aeruginosa in cystic fibrosis patients and will start Phase I trial soon (Polyphor, 2021).
Discovery of novel antibiotics is an extremely time- and cost-consuming process. On average, it takes 10–15 years and around US $1.5 billion for an antibiotic to reach market after its first discovery (Towse et al., 2017). In addition, the limited and restricted use of an antibiotic after its approval, and inevitable resistance development toward the drug once it is in use, render big pharma companies reluctant to entering the business and many major players have left the stage (Plackett, 2020).
Pathogens use a number of various mechanisms to withstand antibiotic treatment. They can modify cell wall proteins, produce enzymes for drug disruption, reduce cell influx by loss of porins, or induce drug wash out from the cell by activating efflux pumps. Apart from new antibiotics design, another strategy to overcome the resistance is to develop anti-resistance drugs which inactivate these mechanisms.
In 1970s, clavulanic acid–the first beta-lactamase inhibitor was discovered (Drawz and Bonomo, 2010). Beta-lactamase inhibitors disrupt beta-lactamases produced by resistant strains, which thus become susceptible to the drug. The most recent example of beta-lactam/beta-lactamase inhibitor approved for clinical use by FDA is imipenem/relebactam and meropenem/vaboractam for treatment of MDR ESKAPE pathogens. Other beta-lactam/beta-lactamase inhibitor combinations are undergoing preclinical and clinical trials now (Zhanel et al., 2018).
Efflux pump inhibitors prevent removal of antibiotic from cytoplasm. They have two modes of action, through energy dissipation or by blocking efflux pumps via direct binding (Sharma et al., 2019). These inhibitors exhibit promising activity in preclinical tests, but none of them have entered clinical trials yet (Ma et al., 2020). Efflux pumps have broad functional similarity across the tree of life, which brings additional challenge to finding molecules that would be specific for prokaryotes and would not have toxic effect on a host (Sharma et al., 2019). No commercially available efflux pump inhibitor drugs are available yet (AlMatar et al., 2021). However, nilotinib, an approved anti-cancer drug used to treat leukemia, has a potent activity against NorA efflux pump in S. aureus (Zimmermann et al., 2019).
Nearly all bacteria produce antimicrobial peptides as a protection against other closely related species while staying immune to these peptides themselves (Yang et al., 2014). These peptides can be ribosomal or non-ribosomal. An example of non-ribosomal peptide is colistin, a last-resort antibiotic used against carbapenem-resistant pathogens. Colistin is produced in Paenibacillus polymyxa by three non-ribosomal peptide synthetases (Tambadou et al., 2015). Bacteriocins, on the other hand, are produced by ribosomes and do not require multiple enzymatic complexes (Yang et al., 2014). They have narrow to broad activity spectrum, need lower concentration to reach bactericidal effect compared to antibiotics, and do not exert toxic effect on human body (Negash and Tsehai, 2020). Having been widely used in food preservation for decades (And and Hoover, 2003), they also have a wide potential for treatment of infectious diseases. For example, nisin (produced by Lactococcus lactis) was found to be effective for treatment of MRSA skin infections; mersacidin (produced by Bacillus spp.) has inhibitory effect on MRSA and VRE; and enterocin (produced by Enterococcus spp.) exerts antimicrobial effect against Salmonella enterica (Ng et al., 2020).
Preclinical trials for three bacteriocins have been finished per date (Ma et al., 2020). NAI-107 bacteriocin and its derivative NAI-108 (bromine incorporated) have proved successful for treatment of MRSA, VRE, Neisseria gonorrhoeae even on non-dividing cells (Brunati et al., 2018). Its ability to kill persistent cells and destroy biofilms is of vital importance for treatment of infections caused by biofilm-forming bacteria, such as MRSA, or P. aeruginosa, since cells that are located in deep biofilm layers are metabolically inactive and do not respond to antibiotic treatment (Cascioferro et al., 2021).
Their incorporation with antibiotics may also help increase activity of existing antibiotics against pathogenic strains. For example, NAI-107/-108 exhibited synergistic effect against A. baumanii, K. pneumoniae, E. coli, and P. aeruginosa, when used with sub-inhibitory concentrations of polymyxins (Brunati et al., 2018). This potentially enables reduction of medication dosage decreasing the severity of polymyxin toxic effect on the host.
Large-scale production of natural bacteriocins might be a limiting factor for their pharmaceutical use, and here synthetic or bioengineered bacteriocins gain an advantage.
Bacteriophages are bacteria-specific viruses and they have two types of replication. Lytic phages replicate in a host cytoplasm right after host cell infection, and then lyse the cell to infect other surrounding hosts (Guo et al., 2020). Temperate phages, on the other hand, replicate along with bacteria DNA after the host infection (lysogenic cycle) and remain dormant until the conditions for a lytic cycle are met. From a clinical perspective, lytic bacteriophages are a valuable asset in a battle against resistant bacteria (Forde and Hill, 2018). Phage therapy has several advantages over antibiotics. Phages are highly specific for a given species or even bacterial strain, they constantly coevolve with bacteria pathogens and since they only replicate inside the host bacteria cell, they are self-regulating (Domingo-Calap et al., 2016). Phage therapy was successfully used in Phase I/II clinical trial to treat 24 patients for chronic otitis caused by antibiotic resistant P. aeruginosa (Wright et al., 2009). Several studies report eradication of chronic prosthetic joint infections caused by MRSA, methicillin-sensitive S. aureus and P. aeruginosa after adjuvant use of bacteriophages with antibiotics (Ferry et al., 2018; Tkhilaishvili et al., 2019; Doub et al., 2020). Intra-nasal phage therapy caused P. aeruginosa load clearance in acute lung infection murine models (Chang et al., 2018).
Unfortunately, no regulations on phage therapy currently exist and all interventions are performed under the Helsinki Convention, i.e., only when all other treatment option ran out (Bassetti et al., 2017). Secondly, our immune system efficiently destroys bacteriophages, limiting use of intravenous formulations (Van Belleghem et al., 2018).
The other approach is to use not phages themselves, but lysins, phage bacteriolytic enzymes. Exebacase (lysin CF-301, ContraFect, United States) and N-Rephasin® (SAL200 lysin, Intron Biotechnology, South Korea) target S. aureus biofilm and planktonic cells (including MRSA) and will be used for treatment of bloodstream infections. Exebacase successfully completed Phase II clinical trial for treatment and will soon start with Phase III trial (ContraFect, 2021). N-Rephasin® is now in Phase II clinical trial (ClinicalTrials.gov, 2021).
Metal ions (silver, zinc, copper, gold, and iron, etc.) have been long recognized for their antimicrobial activity. For example, silver has been used in ancient Greece, Egypt, and Roman Empire to preserve food and water and to treat wounds and ulcers (Alexander, 2009). Metal NPs mainly act through generation of reactive oxygen species that disrupt a wide range of metabolic processes including disruption of the membrane, inhibition of cytochromes, destabilization of ribosomes and DNA damage (Lee et al., 2019). Metal NPs prevent growth of such pathogens as Salmonella typhi, S. aureus (including MRSA), E. coli, P. aeruginosa, S. enterica, and K. pneumoniae (Fatima et al., 2020).
Nanoparticles conjugated with antibiotics have also demonstrated synergistic effect against MDR bacteria. For example, silver NPs in combination with vancomycin against P. aeruginosa resulted in fourfold increased MIC zone diameter, gold NPs coated with vancomycin enhanced its activity against VRE, zinc NPs with beta-lactams proved efficient against ESBL producing K. pneumoniae, P. aeruginosa, and E. coli causing urinary tract infections (Muzammil et al., 2018).
We should note though that bacteria can develop resistance toward NPs. For example, E. coli and P. aeruginosa can resist silver NPs through increased production of flagellin–a protein that triggers aggregation of NPs, without any genetic modification required (Panacek et al., 2018). However, changing the structure may help overcome this resistance. For example, silver-resistant Salmonella strains exhibited susceptibility to silver nanoring structured NPs (Gonzalez-Fernandez et al., 2020).
Eligo Bioscience (France) is currently developing CRISPR-Cas based antimicrobials called eligobiotics (EligoBioscience, 2021). The sequence-specific antimicrobials technology (SSAM) is based on gene therapy, where exogenous CRISPR-Cas nuclease gene and its guide RNA will be delivered to the infection site. If bacterial DNA contains sequences homologous to the guide RNA, the system will be activated and bacterial DNA will be fragmented by a nuclease thus destroying a bacterial cell. Other bacteria will remain intact, allowing for targeted elimination of a pathogen (Bikard et al., 2014). The system can be tailored for a given strain/species or resistance genes.
The strategy behind developing anti-virulence drugs is not to kill/inhibit growth of pathogenic bacteria, but to prevent them from exerting their pathogenicity. This can be achieved through interfering with quorum sensing (Bassetti et al., 2017). Quorum sensing regulates cellular processes though a production of self-inducing signal molecules that are secreted into environment. While bacteria load is small, these processes are kept silent to avoid detection by a host immune system or other bacteria (Zhao X. et al., 2020). Once the population density reaches a certain threshold, concentration of these signal molecules increases and bacteria start expressing genes for a better adaptation. Commonly these genes encode virulence factors, toxin production, and biofilm formation, etc. (Zhao X. et al., 2020). If bacteria become “deaf” to quorum sensing signals, on the other hand, they will not activate these mechanisms even when bacteria concentration is higher than a threshold. For example, degradation of a self-inducing molecule in P. aeruginosa in presence of azithromycin prevents biofilm formation, inhibits virulence factors expression and reduces growth rate by up to 90% at three sub-MICs (Cui et al., 2020). Essential oils of cinnamon, clove, thyme and marjoram exhibited high anti-biofilm, antioxidant and anti-quorum sensing activities against clinical isolates of E. coli, K. pneumoniae, A. baumanii, S. aureus, and P. aeruginosa (Alibi et al., 2020). Nanoparticles and bacteriocins also act as quorum sensing inhibitors (Melian et al., 2019; Ali et al., 2020). However, bacteria can develop resistance toward quorum sensing inhibitors and more research is needed before this strategy becomes widely used (Kalia et al., 2014). Human monoclonal antibodies (mAb) also can be used as anti-virulence drugs with several advantages over the common antibiotics. MAbs are highly selective and do not affect non-target microbiota, they provide rapid passive immunity against the pathogen and facilitate its immune-mediated clearance, and they do not exert toxic effect on humans (Martin-Galiano and McConnell, 2019). Currently, three mAbs against bacterial pathogens (raxibacumab and obiltoxaximab for inhalational anthrax and bezlotoxumab for recurrent Clostridioides difficile infection) are approved by FDA and 14 mAbs targeting various ESKAPE pathogens are under development in preclinical and clinical trials (Zurawski and McLendon, 2020). One of them, Salvecin® (Aridis Pharmaceuticals, United States), targeting alpha-toxin of S. aureus (both MRSA and MSSA), is in Phase 3 trial.
Vaccination is a prophylactic measure to build up host immunity against a pathogen prior to a host encounter with that pathogen. Previously it was rather difficult to develop effective vaccines against MDR bacteria, however, with a rise of reverse vaccinology this became a possibility (Rosini et al., 2020). Vaccines against resistant E. coli, S. aureus, P. aeruginosa, and K. pneumoniae are under development in preclinical and clinical trials (Ma et al., 2020).
In addition to direct vaccination against pathogens, vaccines against closely related species can also contribute to lowering risk of AMR spread. For example, massive vaccination against meningococcus B in Cuba, New Zealand and Canada led to an unexpected reduction in gonorrhea (Azze, 2019; Petousis-Harris and Radcliff, 2019). N. meningitidis and N. gonorrhoeae are 80–90% identical on genomic level, and thus probably share same antigens that antibodies developed under vaccination target. Vaccination against viruses also contributes to lowering antibiotics burden. For example, influenza vaccines prevent incidence of secondary bacterial infections in patients infected by influenza virus and thus reduce the need for antibiotic treatment (Micoli et al., 2021).
The COVID-19 pandemic has most likely worsened the already dire problem of AMR spread. Secondary bacterial infection is a common complication in viral pneumonia patients (Huttner et al., 2020). Therefore, from the start of the pandemic, doctors treating COVID-19 patients have been prone to prescribe antibiotics empirically on admission to hospital (Abelenda-Alonso et al., 2020). As such, up to 56–92% of hospitalized COVID-19 patients were given antibiotics throughout the course of treatment, although only around 6–15% of them actually suffered bacterial co-infection (Li et al., 2020; Vaughn et al., 2020; Martinez-Guerra et al., 2021). A retrospective study from the Wuhan Union Hospital revealed that the majority of patients with secondary bacterial infections had acquired antimicrobial resistant strains (carbapenem-resistant K. pneumoniae, A. baumannii; methicillin-resistant S. aureus) (Li et al., 2020).
Antimicrobial resistance is a complex problem that requires a multifaceted approach for its tackling. Advances in diagnostics will contribute to precise and fast identification of infectious agent, allowing doctors to conduct targeted infection treatment at the spot. At the same time, new approaches for infection therapy will facilitate effective treatment of MDR pathogens and ensure multifaceted strategies for killing of “stubborn” bugs with number of mechanisms to withstand antibiotics. Vaccination will reduce antibiotics use and ultimately prevent development of novel resistance mechanisms. Moreover, enhancement of hospital disinfection and wastewater treatment protocols and optimized hygiene routines for patients and healthcare workers will contribute to elimination of persistent pathogens in hospital environment minimizing the risk of MDR pathogen outbreaks and of their escape into environment. Also, some antibiotics lack breakpoints for some pathogens, which also sets limitations that need to be addressed in the future. However, with all the advancements, scientists and pharma alone will not be able to change the game. Governments need to step in to educate the public and encourage big pharma to re-enter the antimicrobial development. Public awareness programs, such as the World Antimicrobial Awareness Week organized by the WHO, need to be more pronounced and reach more people, and antibiotics use must be strictly regulated in all countries. We believe that if we all stand together and use our assets properly, we have a fair chance to reverse the trend and win the game against MDR bacteria.
EA, VS, and GS conceived the idea and summarized figure and tables. EA drafted the manuscript. All authors have edited the manuscript and agreed on its final version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abelenda-Alonso, G., Padulles, A., Rombauts, A., Gudiol, C., Pujol, M., Alvarez-Pouso, C., et al. (2020). Antibiotic prescription during the COVID-19 pandemic: a biphasic pattern. Infect. Control. Hosp. Epidemiol. 41, 1371–1372. doi: 10.1017/ice.2020.381
Ahmad, M., and Khan, A. U. (2019). Global economic impact of antibiotic resistance: a review. J. Glob. Antimicrob. Resist. 19, 313–316. doi: 10.1016/j.jgar.2019.05.024
Åkerlund, A., Jonasson, E., Matuschek, E., Serrander, L., Sundqvist, M., Kahlmeter, G., et al. (2020). EUCAST rapid antimicrobial susceptibility testing (RAST) in blood cultures: validation in 55 European laboratories. J. Antimicrob. Chemother. 75, 3230–3238. doi: 10.1093/jac/dkaa333
Alexander, J., Hembach, N., and Schwartz, T. (2020). Evaluation of antibiotic resistance dissemination by wastewater treatment plant effluents with different catchment areas in Germany. Sci. Rep. 10:8952. doi: 10.1038/s41598-020-65635-4
Alexander, J. W. (2009). History of the medical use of silver. Surg. Infect. (Larchmt) 10, 289–292. doi: 10.1089/sur.2008.9941
Ali, S. G., Ansari, M. A., Alzohairy, M. A., Alomary, M. N., Jalal, M., AlYahya, S., et al. (2020). Effect of biosynthesized Zno nanoparticles on multi-drug resistant pseudomonas Aeruginosa. Antibiotics (Basel) 9:260. doi: 10.3390/antibiotics9050260
Alibi, S., Ben Selma, W., Ramos-Vivas, J., Smach, M. A., Touati, R., Boukadida, J., et al. (2020). Anti-oxidant, antibacterial, anti-biofilm, and anti-quorum sensing activities of four essential oils against multidrug-resistant bacterial clinical isolates. Curr. Res. Transl. Med. 68, 59–66. doi: 10.1016/j.retram.2020.01.001
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
AlMatar, M., Albarri, O., Makky, E. A., and Koksal, F. (2021). Efflux pump inhibitors: new updates. Pharmacol. Rep. 73, 1–16. doi: 10.1007/s43440-020-00160-9
Alotaibi, S. M. I., Ayibiekea, A., Pedersen, A. F., Jakobsen, L., Pinholt, M., Gumpert, H., et al. (2017). Susceptibility of vancomycin-resistant and -sensitive Enterococcus faecium obtained from Danish hospitals to benzalkonium chloride, chlorhexidine and hydrogen peroxide biocides. J. Med. Microbiol. 66, 1744–1751. doi: 10.1099/jmm.0.000642
And, H. C., and Hoover, D. G. (2003). Bacteriocins and their food applications. Compr. Rev. Food Sci. Food Saf. 2, 82–100. doi: 10.1111/j.1541-4337.2003.tb00016.x
Ardal, C., Balasegaram, M., Laxminarayan, R., McAdams, D., Outterson, K., Rex, J. H., et al. (2020). Antibiotic development – economic, regulatory and societal challenges. Nat. Rev. Microbiol. 18, 267–274. doi: 10.1038/s41579-019-0293-3
Avershina, E., Sharma, P., Taxt, A. M., Singh, H., Frye, S. A., Paul, K., et al. (2021). AMR-Diag: neural network based genotype-to-phenotype prediction of resistance towards β-lactams in Escherichia coli and Klebsiella pneumoniae. Comput. Struct. Biotechnol. J. 19, 1896–1906. doi: 10.1016/j.csbj.2021.03.027
Azze, R. F. O. (2019). A meningococcal B vaccine induces cross-protection against gonorrhea. Clin. Exp. Vaccine Res. 8, 110–115. doi: 10.7774/cevr.2019.8.2.110
Balloux, F., Bronstad Brynildsrud, O., van Dorp, L., Shaw, L. P., Chen, H., Harris, K. A., et al. (2018). From theory to practice: translating whole-genome sequencing (wgs) into the clinic. Trends Microbiol. 26, 1035–1048. doi: 10.1016/j.tim.2018.08.004
Barraud, O., Francois, B., Chainier, D., Vignaud, J., and Ploy, M. C. (2014). Value of integron detection for predicting antibiotic resistance in patients with Gram-negative septicaemia. Int. J. Antimicrob. Agents 44, 351–353. doi: 10.1016/j.ijantimicag.2014.06.008
Bartels, M. D., Kristoffersen, K., Slotsbjerg, T., Rohde, S. M., Lundgren, B., and Westh, H. (2008). Environmental meticillin-resistant Staphylococcus aureus (MRSA) disinfection using dry-mist-generated hydrogen peroxide. J. Hosp. Infect. 70, 35–41. doi: 10.1016/j.jhin.2008.05.018
Bassetti, M., Poulakou, G., Ruppe, E., Bouza, E., Van Hal, S. J., and Brink, A. (2017). Antimicrobial resistance in the next 30 years, humankind, bugs and drugs: a visionary approach. Intensive Care Med. 43, 1464–1475. doi: 10.1007/s00134-017-4878-x
Bianco, G., Boattini, M., Iannaccone, M., Cavallo, R., and Costa, C. (2020). Evaluation of the NG-Test CTX-M MULTI immunochromatographic assay for the rapid detection of CTX-M extended-spectrum-β-lactamase producers from positive blood cultures. J. Hosp. Infect. 105, 341–343. doi: 10.1016/j.jhin.2020.02.009
Bikard, D., Euler, C. W., Jiang, W., Nussenzweig, P. M., Goldberg, G. W., Duportet, X., et al. (2014). Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 32, 1146–1150. doi: 10.1038/nbt.3043
Biswas, D., Tiwari, M., and Tiwari, V. (2018). Comparative mechanism based study on disinfectants against multidrug-resistant Acinetobacter baumannii. J. Cell Biochem. 119, 10314–10326. doi: 10.1002/jcb.27373
Bockmuhl, D. P. (2017). Laundry hygiene-how to get more than clean. J. Appl. Microbiol. 122, 1124–1133. doi: 10.1111/jam.13402
Bockmuhl, D. P., Schages, J., and Rehberg, L. (2019). Laundry and textile hygiene in healthcare and beyond. Microb. Cell 6, 299–306. doi: 10.15698/mic2019.07.682
Bogaerts, P., Oueslati, S., Meunier, D., Nonhoff, C., Yunus, S., Massart, M., et al. (2017). Multicentre evaluation of the BYG Carba v2.0 test, a simplified electrochemical assay for the rapid laboratory detection of carbapenemase-producing Enterobacteriaceae. Sci. Rep. 7:9937. doi: 10.1038/s41598-017-09820-y
Boonstra, M. B., Spijkerman, D. C. M., Voor In ‘t Holt, A. F., van der Laan, R. J., Bode, L. G. M., van Vianen, W., et al. (2020). An outbreak of ST307 extended-spectrum beta-lactamase (ESBL)–producing Klebsiella pneumoniae in a rehabilitation center: an unusual source and route of transmission. Infect. Control Hosp. Epidemiol. 41, 31–36. doi: 10.1017/ice.2019.304
Booton, R. D., Meeyai, A., Alhusein, N., Buller, H., Feil, E., Lambert, H., et al. (2021). One Health drivers of antibacterial resistance: quantifying the relative impacts of human, animal and environmental use and transmission. One Health 12:100220. doi: 10.1016/j.onehlt.2021.100220
Briggs, N., Campbell, S., and Gupta, S. (2021). Advances in rapid diagnostics for bloodstream infections. Diagn. Microbiol. Infect. Dis. 99:115219. doi: 10.1016/j.diagmicrobio.2020.115219
Brunati, C., Thomsen, T. T., Gaspari, E., Maffioli, S., Sosio, M., Jabes, D., et al. (2018). Expanding the potential of NAI-107 for treating serious ESKAPE pathogens: synergistic combinations against Gram-negatives and bactericidal activity against non-dividing cells. J. Antimicrob. Chemother. 73, 414–424. doi: 10.1093/jac/dkx395
Buelow, E., Rico, A., Gaschet, M., Lourenço, J., Kennedy, S. P., Wiest, L., et al. (2020). Hospital discharges in urban sanitation systems: long-term monitoring of wastewater resistome and microbiota in relationship to their eco-exposome. Water Res. X 7:100045. doi: 10.1016/j.wroa.2020.100045
Cascioferro, S., Carbone, D., Parrino, B., Pecoraro, C., Giovannetti, E., Cirrincione, G., et al. (2021). Therapeutic strategies to counteract antibiotic resistance in MRSA biofilm-associated infections. Chem. Med. Chem. 16, 65–80. doi: 10.1002/cmdc.202000677
Cassini, A., Hogberg, L. D., Plachouras, D., Quattrocchi, A., Hoxha, A., Simonsen, G. S., et al. (2019). Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19, 56–66. doi: 10.1016/S1473-3099(18)30605-4
CDC (2019). Antibiotic Resistance Threats In The United States, 2019, (Atlanta, GA: Centers for Disease Control and Prevention).
Chaalal, N., Touati, A., Bakour, S., Aissa, M. A., Sotto, A., Lavigne, J. P., et al. (2020). Spread of OXA-48 and NDM-1-Producing Klebsiella pneumoniae ST48 and ST101 in chicken meat in Western Algeria. Microb. Drug Resist. 27, 492–500. doi: 10.1089/mdr.2019.0419
Chang, R. Y. K., Wallin, M., Lin, Y., Leung, S. S. Y., Wang, H., Morales, S., et al. (2018). Phage therapy for respiratory infections. Adv. Drug Deliv. Rev. 133, 76–86. doi: 10.1016/j.addr.2018.08.001
Charnot-Katsikas, A., Tesic, V., Love, N., Hill, B., Bethel, C., Boonlayangoor, S., et al. (2018). Use of the accelerate pheno system for identification and antimicrobial susceptibility testing of pathogens in positive blood cultures and impact on time to results and workflow. J. Clin. Microbiol. 56:e01166–17. doi: 10.1128/JCM.01166-17
Chen, Y. C., Liao, C. H., Shen, W. T., Su, C., Wu, Y. C., Tsai, M. H., et al. (2019). Effective disinfection of airborne microbial contamination in hospital wards using a zero-valent nano-silver/TiO2 -chitosan composite. Indoor Air 29, 439–449. doi: 10.1111/ina.12543
Cherkaoui, A., Renzi, G., Azam, N., Schorderet, D., Vuilleumier, N., and Schrenzel, J. (2020). Rapid identification by MALDI-TOF/MS and antimicrobial disk diffusion susceptibility testing for positive blood cultures after a short incubation on the WASPLab. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1063–1070. doi: 10.1007/s10096-020-03817-8
Chng, K. R., Li, C., Bertrand, D., Ng, A. H. Q., Kwah, J. S., Low, H. M., et al. (2020). Cartography of opportunistic pathogens and antibiotic resistance genes in a tertiary hospital environment. Nat. Med. 26, 941–951. doi: 10.1038/s41591-020-0894-4
ClinicalTrials.gov (2021). Phase IIa clinical study of N-Rephasin SAL200 [Online]. Available online at: https://clinicaltrials.gov/ct2/show/NCT03089697 (accessed June, 2021).
CLSI (2019). Susceptibility Testing Of Infectious Agents And Evaluation Of Performance Of Antimicrobial Susceptibility Test Devides – Part1, 2nd Edition [Online]. Available online at: https://clsi.org/standards/products/iso-documents/iso-20776-1ed2-2019/ (accessed February 2021).
Cole, K. A., Rivard, K. R., and Dumkow, L. E. (2019). Antimicrobial stewardship interventions to combat antibiotic resistance: an update on targeted strategies. Curr. Infect. Dis. Rep. 21:33. doi: 10.1007/s11908-019-0689-2
ContraFect (2021). Exebacase [Online]. Available online at: https://www.contrafect.com/pipeline/exebacase (accessed March 2021).
Cui, T., Bai, F., Sun, M., Lv, X., Li, X., Zhang, D., et al. (2020). Lactobacillus crustorum ZHG 2-1 as novel quorum-quenching bacteria reducing virulence factors and biofilms formation of Pseudomonas aeruginosa. LWT 117:108696. doi: 10.1016/j.lwt.2019.108696
de Niederhausern, S., Bondi, M., Messi, P., Iseppi, R., Sabia, C., Manicardi, G., et al. (2011). Vancomycin-resistance transferability from VanA enterococci to Staphylococcus aureus. Curr. Microbiol. 62, 1363–1367. doi: 10.1007/s00284-011-9868-6
Debnath, T., Bhowmik, S., Islam, T., and Hassan Chowdhury, M. M. (2018). Presence of multidrug-resistant bacteria on mobile phones of healthcare workers accelerates the spread of nosocomial infection and regarded as a threat to public health in Bangladesh. J. Microsc. Ultrastruct. 6, 165–169. doi: 10.4103/JMAU.JMAU_30_18
Deshmukh, S. P., Patil, S. M., Mullani, S. B., and Delekar, S. D. (2019). Silver nanoparticles as an effective disinfectant: a review. Mater. Sci. Eng. C Mater. Biol. Appl. 97, 954–965. doi: 10.1016/j.msec.2018.12.102
Despotovic, A., Milosevic, B., Milosevic, I., Mitrovic, N., Cirkovic, A., Jovanovic, S., et al. (2020). Hospital-acquired infections in the adult intensive care unit-Epidemiology, antimicrobial resistance patterns, and risk factors for acquisition and mortality. Am. J. Infect. Control. 48, 1211–1215. doi: 10.1016/j.ajic.2020.01.009
Dijksteel, G. S., Ulrich, M., Middelkoop, E., and Boekema, B. (2021). Review: lessons learned from clinical trials using antimicrobial peptides (AMPs). Front. Microbiol. 12:616979. doi: 10.3389/fmicb.2021.616979
Domingo-Calap, P., Georgel, P., and Bahram, S. (2016). Back to the future: bacteriophages as promising therapeutic tools. HLA 87, 133–140. doi: 10.1111/tan.12742
Doub, J. B., Ng, V. Y., Johnson, A. J., Slomka, M., Fackler, J., Horne, B., et al. (2020). Salvage bacteriophage therapy for a chronic MRSA prosthetic joint infection. Antibiotics (Basel) 9:241. doi: 10.3390/antibiotics9050241
Drawz, S. M., and Bonomo, R. A. (2010). Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 23, 160–201. doi: 10.1128/CMR.00037-09
Dunnill, C. W., Page, K., Aiken, Z. A., Noimark, S., Hyett, G., Kafizas, A., et al. (2011). Nanoparticulate silver coated-titania thin films—Photo-oxidative destruction of stearic acid under different light sources and antimicrobial effects under hospital lighting conditions. J. Photochem. Photobiol. A 220, 113–123. doi: 10.1016/j.jphotochem.2011.04.001
ECDC (2013). Point Prevalence Survey Of Healthcare Associated Infections And Antimicrobial Use In European Acute Care Hospitals. Surveillance report. 2011-2012 [Online]. Available online at: https://www.ecdc.europa.eu/sites/default/files/media/en/publications/Publications/healthcare-associated-infections-antimicrobial-use-PPS.pdf (accessed March 2021).
Eichenberger, E. M., and Thaden, J. T. (2019). Epidemiology and mechanisms of resistance of extensively drug resistant gram-negative bacteria. Antibiotics (Basel) 8;37. doi: 10.3390/antibiotics8020037
Eisenstein, B. I., Oleson, F. B. Jr., and Baltz, R. H. (2010). Daptomycin: from the mountain to the clinic, with essential help from Francis Tally, MD. Clin. Infect. Dis. 50 Suppl 1, S10–S15. doi: 10.1086/647938
Elavarasan, T., Chhina, S. K., Parameswaran, M., and Sankaran, K. (2013). Resazurin reduction based colorimetric antibiogram in microfluidic plastic chip. Sens. Actuators B Chem. 176, 174–180.
EligoBioscience (2021). (Re)Programming The Microbiome [Online]. Available online at: https://eligo.bio/technology/ [accessed March 2021].
EUCAST (2021). EUCAST Reading Guide For Broth Microdilution [Online]. Available online at: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/MIC_testing/Reading_guide_BMD_v_3.0_2021.pdf (accessed February 2021).
Farhat, N., and Khan, A. U. (2020). Evolving trends of New Delhi Metallo-betalactamse (NDM) variants: a threat to antimicrobial resistance. Infect. Genet. Evolut. 86:104588. doi: 10.1016/j.meegid.2020.104588
Fatima, F., Siddiqui, S., and Khan, W. A. (2020). Nanoparticles as novel emerging therapeutic antibacterial agents in the antibiotics resistant era. Biol. Trace Elem. Res. 199, 2552–2564. doi: 10.1007/s12011-020-02394-3
FDA (2018). FDA Authorizes New Use Of Test, First To Identify The Emerging Pathigen Candida Auris [Online]. Available online at: https://www.fda.gov/news-events/press-announcements/fda-authorizes-new-use-test-first-identify-emerging-pathogen-candida-auris (accessed February 2021).
Ferry, T., Leboucher, G., Fevre, C., Herry, Y., Conrad, A., Josse, J., et al. (2018). Salvage debridement, antibiotics and implant retention (“DAIR”) with local injection of a selected cocktail of bacteriophages: is it an option for an elderly patient with relapsing staphylococcus aureus prosthetic-joint infection? Open Forum Infect. Dis. 5:ofy269. doi: 10.1093/ofid/ofy269
Fleming, A. (1945). Penicillin. Nobel Lecture, December, 11, 1945 [Online]. Available online at: https://www.nobelprize.org/uploads/2018/06/fleming-lecture.pdf (accessed February 2021).
Florio, W., Cappellini, S., Giordano, C., Vecchione, A., Ghelardi, E., and Lupetti, A. (2019). A new culture-based method for rapid identification of microorganisms in polymicrobial blood cultures by MALDI-TOF MS. BMC Microbiol 19:267. doi: 10.1186/s12866-019-1641-1
Forde, A., and Hill, C. (2018). Phages of life – the path to pharma. Br. J. Pharmacol. 175, 412–418. doi: 10.1111/bph.14106
Fuentefria, A. M., Pippi, B., Dalla Lana, D. F., Donato, K. K., and de Andrade, S. F. (2018). Antifungals discovery: an insight into new strategies to combat antifungal resistance. Lett. Appl. Microbiol. 66, 2–13. doi: 10.1111/lam.12820
Galazzi, A., Panigada, M., Broggi, E., Grancini, A., Adamini, I., Binda, F., et al. (2019). Microbiological colonization of healthcare workers’ mobile phones in a tertiary-level Italian intensive care unit. Intensive Crit. Care Nurs. 52, 17–21. doi: 10.1016/j.iccn.2019.01.005
Gao, F.-Z., Zou, H.-Y., Wu, D.-L., Chen, S., He, L.-Y., Zhang, M., et al. (2020). Swine farming elevated the proliferation of Acinetobacter with the prevalence of antibiotic resistance genes in the groundwater. Environ. Int. 136:105484. doi: 10.1016/j.envint.2020.105484
Gonzalez-Fernandez, S., Lozano-Iturbe, V., Garcia, B., Andres, L. J., Menendez, M. F., Rodriguez, D., et al. (2020). Antibacterial effect of silver nanorings. BMC Microbiol. 20:172. doi: 10.1186/s12866-020-01854-z
Gudz, K. Y., Permyakova, E. S., Matveev, A. T., Bondarev, A. V., Manakhov, A. M., Sidorenko, D. A., et al. (2020). Pristine and antibiotic-loaded nanosheets/nanoneedles-based boron nitride films as a promising platform to suppress bacterial and fungal infections. ACS Appl. Mater. Interfaces 12, 42485–42498. doi: 10.1021/acsami.0c10169
Guo, Z., Lin, H., Ji, X., Yan, G., Lei, L., Han, W., et al. (2020). Therapeutic applications of lytic phages in human medicine. Microb. Pathog. 142:104048. doi: 10.1016/j.micpath.2020.104048
Hall, B. G., and Barlow, M. (2004). Evolution of the serine beta-lactamases: past, present and future. Drug Resist. Updat. 7, 111–123. doi: 10.1016/j.drup.2004.02.003
Hasan, J., Xu, Y., Yarlagadda, T., Schuetz, M., Spann, K., and Yarlagadda, P. K. (2020). Antiviral and antibacterial nanostructured surfaces with excellent mechanical properties for hospital applications. ACS Biomater. Sci. Eng. 6, 3608–3618. doi: 10.1021/acsbiomaterials.0c00348
Hazell, G., Fisher, L. E., Murray, W. A., Nobbs, A. H., and Su, B. (2018). Bioinspired bactericidal surfaces with polymer nanocone arrays. J. Colloid Interface Sci. 528, 389–399. doi: 10.1016/j.jcis.2018.05.096
Hernando-Amado, S., Coque, T. M., Baquero, F., and Martinez, J. L. (2019). Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 4, 1432–1442. doi: 10.1038/s41564-019-0503-9
Hishinuma, T., Uchida, H., Tohya, M., Shimojima, M., Tada, T., and Kirikae, T. (2020). Emergence and spread of VIM-type metallo-beta-lactamase-producing Pseudomonas aeruginosa clinical isolates in Japan. J. Glob. Antimicrob. Resist. 23, 265–268. doi: 10.1016/j.jgar.2020.09.010
Holmes, A. H., Moore, L. S., Sundsfjord, A., Steinbakk, M., Regmi, S., Karkey, A., et al. (2016). Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387, 176–187. doi: 10.1016/S0140-6736(15)00473-0
Hutchings, M. I., Truman, A. W., and Wilkinson, B. (2019). Antibiotics: past, present and future. Curr. Opin. Microbiol. 51, 72–80. doi: 10.1016/j.mib.2019.10.008
Huttner, B. D., Catho, G., Pano-Pardo, J. R., Pulcini, C., and Schouten, J. (2020). COVID-19: don’t neglect antimicrobial stewardship principles! Clin. Microbiol. Infect. 26, 808–810. doi: 10.1016/j.cmi.2020.04.024
Isozumi, R., Yoshimatsu, K., Yamashiro, T., Hasebe, F., Nguyen, B. M., Ngo, T. C., et al. (2012). bla(NDM-1)-positive Klebsiella pneumoniae from environment, Vietnam. Emerg. Infect. Dis. 18, 1383–1385. doi: 10.3201/eid1808.111816
Ivanova, E. P., Hasan, J., Webb, H. K., Gervinskas, G., Juodkazis, S., Truong, V. K., et al. (2013). Bactericidal activity of black silicon. Nat. Commun. 4:2838. doi: 10.1038/ncomms3838
Ivanova, E. P., Hasan, J., Webb, H. K., Truong, V. K., Watson, G. S., Watson, J. A., et al. (2012). Natural bactericidal surfaces: mechanical rupture of Pseudomonas aeruginosa cells by cicada wings. Small 8, 2489–2494. doi: 10.1002/smll.201200528
Jasovsky, D., Littmann, J., Zorzet, A., and Cars, O. (2016). Antimicrobial resistance-a threat to the world’s sustainable development. UPS J. Med. Sci. 121, 159–164. doi: 10.1080/03009734.2016.1195900
Jenkins, J., Mantell, J., Neal, C., Gholinia, A., Verkade, P., Nobbs, A. H., et al. (2020). Antibacterial effects of nanopillar surfaces are mediated by cell impedance, penetration and induction of oxidative stress. Nat. Commun. 11:1626. doi: 10.1038/s41467-020-15471-x
Kalia, V. C., Wood, T. K., and Kumar, P. (2014). Evolution of resistance to quorum-sensing inhibitors. Microb. Ecol. 68, 13–23. doi: 10.1007/s00248-013-0316-y
Kang, W., Sarkar, S., Lin, Z. S., McKenney, S., and Konry, T. (2019). Ultrafast parallelized microfluidic platform for antimicrobial susceptibility testing of gram positive and negative bacteria. Anal. Chem. 91, 6242–6249. doi: 10.1021/acs.analchem.9b00939
Khan, H. A., Baig, F. K., and Mehboob, R. (2017). Nosocomial infections: epidemiology, prevention, control and surveillance. Asian Pacific J. Trop. Biomed. 7, 478–482. doi: 10.1016/j.apjtb.2017.01.019
Khan, Z. A., Siddiqui, M. F., and Park, S. (2019). Current and emerging methods of antibiotic susceptibility testing. Diagnostics (Basel) 9:49. doi: 10.3390/diagnostics9020049
Kim, D. W., and Cha, C. J. (2021). Antibiotic resistome from the One-Health perspective: understanding and controlling antimicrobial resistance transmission. Exp. Mol. Med. 53, 301–309. doi: 10.1038/s12276-021-00569-z
Kim, J., Greenberg, D. E., Pifer, R., Jiang, S., Xiao, G., Shelburne, S. A., et al. (2020). VAMPr: variant Mapping and Prediction of antibiotic resistance via explainable features and machine learning. PLoS Comput. Biol. 16:e1007511. doi: 10.1371/journal.pcbi.1007511
Klein, E. Y., Tseng, K. K., Pant, S., and Laxminarayan, R. (2019). Tracking global trends in the effectiveness of antibiotic therapy using the Drug Resistance Index. BMJ Glob. Health 4:e001315. doi: 10.1136/bmjgh-2018-001315
Kritsotakis, E. I., Kontopidou, F., Astrinaki, E., Roumbelaki, M., Ioannidou, E., and Gikas, A. (2017). Prevalence, incidence burden, and clinical impact of healthcare-associated infections and antimicrobial resistance: a national prevalent cohort study in acute care hospitals in Greece. Infect. Drug Resist. 10, 317–328. doi: 10.2147/IDR.S147459
Kumar, A., Ellis, P., Arabi, Y., Roberts, D., Light, B., Parrillo, J. E., et al. (2009). Initiation of inappropriate antimicrobial therapy results in a fivefold reduction of survival in human septic shock. Chest 136, 1237–1248. doi: 10.1378/chest.09-0087
Lampejo, T. (2020). Influenza and antiviral resistance: an overview. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1201–1208. doi: 10.1007/s10096-020-03840-9
Lee, N. Y., Ko, W. C., and Hsueh, P. R. (2019). Nanoparticles in the treatment of infections caused by multidrug-resistant organisms. Front. Pharmacol. 10:1153. doi: 10.3389/fphar.2019.01153
Lena, P., Ishak, A., Karageorgos, S. A., and Tsioutis, C. (2021). Presence of methicillin-resistant Staphylococcus Aureus (MRSA) on healthcare workers’ attire: a systematic review. Trop. Med. Infect. Dis. 6:42. doi: 10.3390/tropicalmed6020042
Lerner, A., Adler, A., Abu-Hanna, J., Cohen Percia, S., Kazma Matalon, M., and Carmeli, Y. (2015). Spread of KPC-producing carbapenem-resistant Enterobacteriaceae: the importance of super-spreaders and rectal KPC concentration. Clin. Microbiol. Infect. 21:470. e1–7. doi: 10.1016/j.cmi.2014.12.015
Li, J., Cheng, W., Xu, L., Strong, P. J., and Chen, H. (2015). Antibiotic-resistant genes and antibiotic-resistant bacteria in the effluent of urban residential areas, hospitals, and a municipal wastewater treatment plant system. Environ. Sci. Pollut. Res. Int. 22, 4587–4596. doi: 10.1007/s11356-014-3665-2
Li, J., Wang, J., Yang, Y., Cai, P., Cao, J., Cai, X., et al. (2020). Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: a retrospective analysis. Antimicrob. Resist. Infect. Control 9:153. doi: 10.1186/s13756-020-00819-1
Lopatkin, A. J., Bening, S. C., Manson, A. L., Stokes, J. M., Kohanski, M. A., Badran, A. H., et al. (2021). Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 371:eaba0862. doi: 10.1126/science.aba0862
Lu, J., and Guo, J. (2021). Disinfection spreads antimicrobial resistance. Science 371:474. doi: 10.1126/science.abg4380
Lyddiard, D., Jones, G. L., and Greatrex, B. W. (2016). Keeping it simple: lessons from the golden era of antibiotic discovery. FEMS Microbiol. Lett. 363:fnw084. doi: 10.1093/femsle/fnw084
Ma, Y. X., Wang, C. Y., Li, Y. Y., Li, J., Wan, Q. Q., Chen, J. H., et al. (2020). Considerations and caveats in combating ESKAPE pathogens against nosocomial infections. Adv. Sci. (Weinh) 7:1901872. doi: 10.1002/advs.201901872
Macesic, N., Bear Don’t Walk, O. I., Pe’er, I., Tatonetti, N. P., Peleg, A. Y., and Uhlemann, A. C. (2020). Predicting phenotypic polymyxin resistance in Klebsiella pneumoniae through machine learning analysis of genomic data. mSystems 5:e00656–19. doi: 10.1128/mSystems.00656-19
Mancini, S., Kieffer, N., Poirel, L., and Nordmann, P. (2017). Evaluation of the RAPIDEC® CARBA NP and β-CARBA® tests for rapid detection of Carbapenemase-producing Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 88, 293–297. doi: 10.1016/j.diagmicrobio.2017.05.006
Marschal, M., Bachmaier, J., Autenrieth, I., Oberhettinger, P., Willmann, M., and Peter, S. (2017). Evaluation of the accelerate pheno system for fast identification and antimicrobial susceptibility testing from positive blood cultures in bloodstream infections caused by gram-negative pathogens. J. Clin. Microbiol. 55, 2116–2126. doi: 10.1128/JCM.00181-17
Martinez-Guerra, B. A., Gonzalez-Lara, M. F., de-Leon-Cividanes, N. A., Tamez-Torres, K. M., Roman-Montes, C. M., Rajme-Lopez, S., et al. (2021). Antimicrobial resistance patterns and antibiotic use during hospital conversion in the COVID-19 pandemic. Antibiotics (Basel) 10:182. doi: 10.3390/antibiotics10020182
Martin-Galiano, A. J., and McConnell, M. J. (2019). Using omics technologies and systems biology to identify epitope targets for the development of monoclonal antibodies against antibiotic-resistant bacteria. Front. Immunol. 10:2841. doi: 10.3389/fimmu.2019.02841
Masi, M., Refregiers, M., Pos, K. M., and Pages, J. M. (2017). Mechanisms of envelope permeability and antibiotic influx and efflux in Gram-negative bacteria. Nat. Microbiol. 2:17001. doi: 10.1038/nmicrobiol.2017.1
Matta, R., Hallit, S., Hallit, R., Bawab, W., Rogues, A. M., and Salameh, P. (2018). Epidemiology and microbiological profile comparison between community and hospital acquired infections: a multicenter retrospective study in Lebanon. J. Infect. Public Health 11, 405–411. doi: 10.1016/j.jiph.2017.09.005
McCann, C. M., Christgen, B., Roberts, J. A., Su, J.-Q., Arnold, K. E., Gray, N. D., et al. (2019). Understanding drivers of antibiotic resistance genes in High Arctic soil ecosystems. Environ. Int. 125, 497–504. doi: 10.1016/j.envint.2019.01.034
Mehrotra, P. (2016). Biosensors and their applications – A review. J. Oral Biol. Craniofac. Res. 6, 153–159. doi: 10.1016/j.jobcr.2015.12.002
Melian, C., Segli, F., Gonzalez, R., Vignolo, G., and Castellano, P. (2019). Lactocin AL705 as quorum sensing inhibitor to control Listeria monocytogenes biofilm formation. J. Appl. Microbiol. 127, 911–920. doi: 10.1111/jam.14348
Michael, K. E., No, D., Daniell, W. E., Seixas, N. S., and Roberts, M. C. (2017). Assessment of environmental contamination with pathogenic bacteria at a hospital laundry facility. Ann. Work Exposures Health 61, 1087–1096. doi: 10.1093/annweh/wxx082
Micoli, F., Bagnoli, F., Rappuoli, R., and Serruto, D. (2021). The role of vaccines in combatting antimicrobial resistance. Nat. Rev. Microbiol. 19, 287–302. doi: 10.1038/s41579-020-00506-3
Miller, W. R., Bayer, A. S., and Arias, C. A. (2016). Mechanism of action and resistance to daptomycin in staphylococcus aureus and enterococci. Cold Spring Harb. Perspect. Med. 6:a026997. doi: 10.1101/cshperspect.a026997
Mirhoseini, S. H., Nikaeen, M., Shamsizadeh, Z., and Khanahmad, H. (2016). Hospital air: a potential route for transmission of infections caused by beta-lactam-resistant bacteria. Am. J. Infect. Control 44, 898–904. doi: 10.1016/j.ajic.2016.01.041
Missri, L., Smiljkovski, D., Prigent, G., Lesenne, A., Obadia, T., Joumaa, M., et al. (2019). Bacterial colonization of healthcare workers’ mobile phones in the ICU and effectiveness of sanitization. J. Occup. Environ. Hyg. 16, 97–100. doi: 10.1080/15459624.2018.1546051
Mitchell, B. G., Dancer, S. J., Anderson, M., and Dehn, E. (2015). Risk of organism acquisition from prior room occupants: a systematic review and meta-analysis. J. Hosp. Infect. 91, 211–217. doi: 10.1016/j.jhin.2015.08.005
Mthembu, T. P., Zishiri, O. T., and El Zowalaty, M. E. (2021). Genomic characterization of antimicrobial resistance in food chain and livestock-associated Salmonella species. Animals (Basel) 11:872. doi: 10.3390/ani11030872
Mughini-Gras, L., Dorado-García, A., van Duijkeren, E., van den Bunt, G., Dierikx, C. M., Bonten, M. J. M., et al. (2019). Attributable sources of community-acquired carriage of Escherichia coli. Lancet Planet. Health 3, e357–e369. doi: 10.1016/S2542-5196(19)30130-5
Mushabati, N. A., Samutela, M. T., Yamba, K., Ngulube, J., Nakazwe, R., Nkhoma, P., et al. (2021). Bacterial contamination of mobile phones of healthcare workers at the University Teaching Hospital, Lusaka, Zambia. Infect. Prevent. Pract. 3:100126. doi: 10.1016/j.infpip.2021.100126
Musser James, M., Beres Stephen, B., Zhu, L., Randall, J., Vuopio, J., Hyyryläinen, H.-L., et al. (2020). Reduced in vitro susceptibility of streptococcus pyogenes to β-lactam antibiotics associated with mutations in the pbp2x gene is geographically widespread. J. Clin. Microbiol. 58:e01993–19. doi: 10.1128/JCM.01993-19
Muzammil, S., Hayat, S., Fakhar, E. A. M., Aslam, B., Siddique, M. H., Nisar, M. A., et al. (2018). Nanoantibiotics: future nanotechnologies to combat antibiotic resistance. Front. Biosci. (Elite Ed) 10:352–374. doi: 10.2741/e827
Negash, A. W., and Tsehai, B. A. (2020). Current applications of bacteriocin. Int. J. Microbiol. 2020:4374891. doi: 10.1155/2020/4374891
Ng, Z. J., Zarin, M. A., Lee, C. K., and Tan, J. S. (2020). Application of bacteriocins in food preservation and infectious disease treatment for humans and livestock: a review. RSC Adv. 10, 38937–38964. doi: 10.1039/D0RA06161A
Nguyen, M., Brettin, T., Long, S. W., Musser, J. M., Olsen, R. J., Olson, R., et al. (2018). Developing an in silico minimum inhibitory concentration panel test for Klebsiella pneumoniae. Sci. Rep. 8:421. doi: 10.1038/s41598-017-18972-w
Novak, R., and Shlaes, D. M. (2010). The pleuromutilin antibiotics: a new class for human use. Curr. Opin. Invest. Drugs 11, 182–191.
OECD (2019). Antimicrobial Resistance. Tackling the Burden in the European Union [Online]. Available online at: https://www.oecd.org/health/health-systems/AMR-Tackling-the-Burden-in-the-EU-OECD-ECDC-Briefing-Note-2019.pdf (accessed June 2021).
Omrani, M., and Fataei, E. (2018). Synthesizing colloidal zinc oxide nanoparticles for effective disinfection; impact on the inhibitory growth of pseudomonas aeruginosa on the surface of an infectious unit. Polish J. Environ. Stud. 27, 1639–1645. doi: 10.15244/pjoes/77096
Panacek, A., Kvitek, L., Smekalova, M., Vecerova, R., Kolar, M., Roderova, M., et al. (2018). Bacterial resistance to silver nanoparticles and how to overcome it. Nat. Nanotechnol. 13, 65–71. doi: 10.1038/s41565-017-0013-y
Peters, A., Otter, J., Moldovan, A., Parneix, P., Voss, A., and Pittet, D. (2018). Keeping hospitals clean and safe without breaking the bank; summary of the Healthcare Cleaning Forum 2018. Antimicrob. Resist. Infect. Control 7:132. doi: 10.1186/s13756-018-0420-3
Petousis-Harris, H., and Radcliff, F. J. (2019). Exploitation of Neisseria meningitidis Group B OMV Vaccines Against N. gonorrhoeae to inform the development and deployment of effective gonorrhea vaccines. Front. Immunol. 10:683. doi: 10.3389/fimmu.2019.00683
PEW (2021). Antibiotics Currently in Global Clinical Development [Online]. Available online at: https://www.pewtrusts.org/en/research-and-analysis/data-visualizations/2014/antibiotics-currently-in-clinical-development (accessed March 2021).
Pham, V. T. H., Truong, V. K., Mainwaring, D. E., Guo, Y., Baulin, V. A., Al Kobaisi, M., et al. (2014). Nanotopography as a trigger for the microscale, autogenous and passive lysis of erythrocytes. J. Mater. Chem. B 2, 2819–2826. doi: 10.1039/C4TB00239C
Pitout Johann, D. D., Peirano, G., Kock Marleen, M., Strydom, K.-A., and Matsumura, Y. (2019). The global ascendency of OXA-48-Type carbapenemases. Clin. Microbiol. Rev. 33, e00102–e00119. doi: 10.1128/CMR.00102-19
Pittet, D., and Donaldson, L. (2005). Clean care is safer care: a worldwide priority. Lancet 366, 1246–1247. doi: 10.1016/S0140-6736(05)67506-X
Plackett, B. (2020). Why big pharma has abandoned antibiotics. Nature 586, S50–S52. doi: 10.1038/d41586-020-02884-3
Poirel, L., Nordmann, P., and Bourbeau, P. (2015). Rapidec carba NP Test for rapid detection of carbapenemase producers. J. Clin. Microbiol. 53, 3003–3008. doi: 10.1128/JCM.00977-15
Poirel, L., Yilmaz, M., Istanbullu, A., Arslan, F., Mert, A., Bernabeu, S., et al. (2014). Spread of NDM-1-producing Enterobacteriaceae in a neonatal intensive care unit in Istanbul, Turkey. Antimicrob. Agents Chemother. 58, 2929–2933. doi: 10.1128/AAC.02047-13
Polyphor (2021). Inhaled Murepavadin: First Targeted Antibiotic Being Developed For Treating Pseudomonas Aeruginosa Infections In People With Cystic Fibrosis [Online]. Available online at: https://www.polyphor.com/pol7080/ (accessed March 2021).
Poole, C. L., and James, S. H. (2018). Antiviral therapies for herpesviruses: current agents and new directions. Clin. Ther. 40, 1282–1298. doi: 10.1016/j.clinthera.2018.07.006
Popa, L. I., Gheorghe, I., Barbu, I. C., Surleac, M., Paraschiv, S., Marutescu, L., et al. (2020). Multidrug resistant Klebsiella pneumoniae ST101 clone survival chain from inpatients to hospital effluent after chlorine treatment. Front. Microbiol. 11:610296. doi: 10.3389/fmicb.2020.610296
Potron, A., Fournier, D., Emeraud, C., Triponney, P., Plésiat, P., Naas, T., et al. (2020). Evaluation of the immunochromatographic NG-Test Carba 5 for rapid identification of carbapenemase in nonfermenters. Antimicrob. Agents Chemother. 63, e968–e919. doi: 10.1128/AAC.00968-19
Prestinaci, F., Pezzotti, P., and Pantosti, A. (2015). Antimicrobial resistance: a global multifaceted phenomenon. Pathog. Glob. Health 109, 309–318. doi: 10.1179/2047773215Y.0000000030
Pullerits, K., Ahlinder, J., Holmer, L., Salomonsson, E., Öhrman, C., Jacobsson, K., et al. (2020). Impact of UV irradiation at full scale on bacterial communities in drinking water. NPJ Clean Water 3:11. doi: 10.1038/s41545-020-0057-7
Quan, J., Langelier, C., Kuchta, A., Batson, J., Teyssier, N., Lyden, A., et al. (2019). FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 47:e83. doi: 10.1093/nar/gkz418
Rastogi, R. P., Richa, Kumar, A., Tyagi, M. B., and Sinha, R. P. (2010). Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J. Nucleic Acids 2010:592980. doi: 10.4061/2010/592980
Ray, S. K., Dhakal, D., Hur, J., and Lee, S. W. (2020). Visible light driven MoS2/alpha-NiMoO4 ultra-thin nanoneedle composite for efficient Staphylococcus aureus inactivation. J. Hazard Mater. 385:121553. doi: 10.1016/j.jhazmat.2019.121553
Reuter, C. H., Palac, H. L., Kociolek, L. K., Zheng, X. T., Chao, Y. Y., Patel, R. M., et al. (2019). Ideal and actual impact of rapid diagnostic testing and antibiotic stewardship on antibiotic prescribing and clinical outcomes in children with positive blood cultures. Pediatr. Infect. Dis. J. 38, 131–137. doi: 10.1097/INF.0000000000002102
Revelas, A. (2012). Healthcare – associated infections: a public health problem. Niger. Med. J. 53, 59–64. doi: 10.4103/0300-1652.103543
Riley, K., Laird, K., and Williams, J. (2015). Washing uniforms at home: adherence to hospital policy. Nurs. Stand. 29, 37–43. doi: 10.7748/ns.29.25.37.e9268
Riley, K., Williams, J., Owen, L., Shen, J., Davies, A., and Laird, K. (2017). The effect of low-temperature laundering and detergents on the survival of Escherichia coli and Staphylococcus aureus on textiles used in healthcare uniforms. J. Appl. Microbiol. 123, 280–286. doi: 10.1111/jam.13485
Rodrigues, D. O., Peixoto, L. D. P., Barros, E. T. M., Guimaraes, J. R., Gontijo, B. C., Almeida, J. L., et al. (2020). Epidemiology of bacterial contamination of inert hospital surfaces and equipment in critical and non-critical care units: a Brazilian study. Microbiol. Res. J. Int. 30, 31–43.
Rosini, R., Nicchi, S., Pizza, M., and Rappuoli, R. (2020). Vaccines against antimicrobial resistance. Front. Immunol. 11:1048. doi: 10.3389/fimmu.2020.01048
Saini, S. K., Halder, M., Singh, Y., and Nair, R. V. (2020). Bactericidal characteristics of bioinspired nontoxic and chemically stable disordered silicon nanopyramids. ACS Biomater. Sci. Eng. 6, 2778–2786. doi: 10.1021/acsbiomaterials.9b01963
Santajit, S., and Indrawattana, N. (2016). Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed. Res. Int. 2016, 2475067–2475067. doi: 10.1155/2016/2475067
Shalek, A. K., Robinson, J. T., Karp, E. S., Lee, J. S., Ahn, D. R., Yoon, M. H., et al. (2010). Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proc. Natl. Acad. Sci. U.S.A. 107, 1870–1875. doi: 10.1073/pnas.0909350107
Sharma, A., Gupta, V. K., and Pathania, R. (2019). Efflux pump inhibitors for bacterial pathogens: from bench to bedside. Indian J. Med. Res. 149, 129–145. doi: 10.4103/ijmr.IJMR_2079_17
Shi, X., Kadiyala, U., VanEpps, J. S., and Yau, S. T. (2018). Culture-free bacterial detection and identification from blood with rapid, phenotypic, antibiotic susceptibility testing. Sci. Rep. 8:3416. doi: 10.1038/s41598-018-21520-9
Smith, J. (2019). Swiss Biotech Halts Pneumonia Antibiotic Trial After Patient Safety Scare [Online]. Available online at: https://www.labiotech.eu/more-news/polyphor-pneumonia-antibiotic-resistance/ (accessed March 2021).
Snitser, O., Russ, D., Stone, L. K., Wang, K. K., Sharir, H., Kozer, N., et al. (2020). Ubiquitous selection for mecA in community-associated MRSA across diverse chemical environments. Nat. Commun. 11:6038. doi: 10.1038/s41467-020-19825-3
Srinivasan, A., Lopez-Ribot, J. L., and Ramasubramanian, A. K. (2014). Overcoming antifungal resistance. Drug Discov. Today Technol. 11, 65–71. doi: 10.1016/j.ddtec.2014.02.005
Su, Y., Wu, D., Xia, H., Zhang, C., Shi, J., Wilkinson, K. J., et al. (2019). Metallic nanoparticles induced antibiotic resistance genes attenuation of leachate culturable microbiota: the combined roles of growth inhibition, ion dissolution and oxidative stress. Environ. Int. 128, 407–416. doi: 10.1016/j.envint.2019.05.007
Sun, D., Jeannot, K., Xiao, Y., and Knapp, C. W. (2019). Editorial: horizontal gene transfer mediated bacterial antibiotic resistance. Front. Microbiol. 10:1933. doi: 10.3389/fmicb.2019.01933
Szeto, W., Yam, W. C., Huang, H., and Leung, D. Y. C. (2020). The efficacy of vacuum-ultraviolet light disinfection of some common environmental pathogens. BMC Infect. Dis. 20:127. doi: 10.1186/s12879-020-4847-9
Talan, A., and Tyagi, R. D. (2020). “5 – Fate of pathogens and viruses in hospital wastewater and their treatment methods,” in Current Developments in Biotechnology and Bioengineering, eds R. D. Tyagi, B. Sellamuthu, B. Tiwari, S. Yan, P. Drogui, X. Zhang, et al. (Amsterdam: Elsevier), 149–175.
Tambadou, F., Caradec, T., Gagez, A. L., Bonnet, A., Sopena, V., Bridiau, N., et al. (2015). Characterization of the colistin (polymyxin E1 and E2) biosynthetic gene cluster. Arch. Microbiol. 197, 521–532. doi: 10.1007/s00203-015-1084-5
Tano, E., and Melhus, A. (2014). Level of decontamination after washing textiles at 60 degrees C or 70 degrees C followed by tumble drying. Infect. Ecol. Epidemiol. 4, 24314. doi: 10.3402/iee.v4.24314
Taxt, A. M., Avershina, E., Frye, S. A., Naseer, U., and Ahmad, R. (2020). Rapid identification of pathogens, antibiotic resistance genes and plasmids in blood cultures by nanopore sequencing. Sci. Rep. 10:7622. doi: 10.1038/s41598-020-64616-x
Tkhilaishvili, T., Winkler, T., Muller, M., Perka, C., and Trampuz, A. (2019). Bacteriophages as adjuvant to antibiotics for the treatment of periprosthetic joint infection caused by multidrug-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 64:e00924–19. doi: 10.1128/AAC.00924-19
Totaro, M., Casini, B., Profeti, S., Tuvo, B., Privitera, G., and Baggiani, A. (2020). Role of hydrogen peroxide vapor (HPV) for the disinfection of hospital surfaces contaminated by multiresistant bacteria. Pathogens 9:408. doi: 10.3390/pathogens9050408
Towse, A., Hoyle, C. K., Goodall, J., Hirsch, M., Mestre-Ferrandiz, J., and Rex, J. H. (2017). Time for a change in how new antibiotics are reimbursed: development of an insurance framework for funding new antibiotics based on a policy of risk mitigation. Health Policy 121, 1025–1030. doi: 10.1016/j.healthpol.2017.07.011
Tripathy, A., Sen, P., Su, B., and Briscoe, W. H. (2017). Natural and bioinspired nanostructured bactericidal surfaces. Adv. Colloid Interface Sci. 248, 85–104. doi: 10.1016/j.cis.2017.07.030
van Belkum, A., Burnham, C. D., Rossen, J. W. A., Mallard, F., Rochas, O., and Dunne, W. M. Jr. (2020). Innovative and rapid antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 18, 299–311. doi: 10.1038/s41579-020-0327-x
Van Belleghem, J. D., Dabrowska, K., Vaneechoutte, M., Barr, J. J., and Bollyky, P. L. (2018). Interactions between bacteriophage, bacteria, and the mammalian immune system. Viruses 11:10. doi: 10.3390/v11010010
Vasala, A., Hytonen, V. P., and Laitinen, O. H. (2020). Modern tools for rapid diagnostics of antimicrobial resistance. Front. Cell Infect. Microbiol. 10:308. doi: 10.3389/fcimb.2020.00308
Vaughn, V. M., Gandhi, T., Petty, L. A., Patel, P. K., Prescott, H. C., Malani, A. N., et al. (2020). Empiric antibacterial therapy and community-onset bacterial co-infection in patients hospitalized with COVID-19: a multi-hospital cohort study. Clin. Infect. Dis. 72, e533–e541. doi: 10.1093/cid/ciaa1239
von Wintersdorff, C. J., Penders, J., van Niekerk, J. M., Mills, N. D., Majumder, S., van Alphen, L. B., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173. doi: 10.3389/fmicb.2016.00173
Walsh, T. R., Weeks, J., Livermore, D. M., and Toleman, M. A. (2011). Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11, 355–362. doi: 10.1016/s1473-3099(11)70059-7
Wandiyanto, J. V., Truong, V. K., Al Kobaisi, M., Juodkazis, S., Thissen, H., Bazaka, O., et al. (2019). The fate of osteoblast-like MG-63 cells on pre-infected bactericidal nanostructured titanium surfaces. Materials (Basel) 12:1575. doi: 10.3390/ma12101575
Wang, Q., Wang, P., and Yang, Q. (2018). Occurrence and diversity of antibiotic resistance in untreated hospital wastewater. Sci. Total Environ. 621, 990–999. doi: 10.1016/j.scitotenv.2017.10.128
Wang, R., van Dorp, L., Shaw, L. P., Bradley, P., Wang, Q., Wang, X., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9:1179. doi: 10.1038/s41467-018-03205-z
Wang, X., Yang, F., Zhao, J., Xu, Y., Mao, D., Zhu, X., et al. (2018). Bacterial exposure to ZnO nanoparticles facilitates horizontal transfer of antibiotic resistance genes. NanoImpact 10, 61–67. doi: 10.1016/j.impact.2017.11.006
Wang, Y., Yu, G., Deng, S., Huang, J., and Wang, B. (2018). The electro-peroxone process for the abatement of emerging contaminants: mechanisms, recent advances, and prospects. Chemosphere 208, 640–654. doi: 10.1016/j.chemosphere.2018.05.095
Watson, G. S., Green, D. W., Schwarzkopf, L., Li, X., Cribb, B. W., Myhra, S., et al. (2015). A gecko skin micro/nano structure – A low adhesion, superhydrophobic, anti-wetting, self-cleaning, biocompatible, antibacterial surface. Acta Biomater. 21, 109–122. doi: 10.1016/j.actbio.2015.03.007
WHO (2011). Report On The Burden Of Endemic Health Care-Associated Infection Worldwide. Geneva: World Health Organization.
WHO (2015). Health-care Associated Infections. Fact Sheet [Online]. Available online: https://www.who.int/gpsc/country_work/gpsc_ccisc_fact_sheet_en.pdf (accessed June 2021).
WHO (2017). Global Priority List Of Antibiotic-Resistant Bacteria To Guide Research, Discovery, And Development Of New Antibiotics. Geneva: World Health Organization.
WHO (2020). Global Progress Report on WASH in Health Care Facilities. Fundamentals first. [Online]. Available online at: http://202.1.196.72/jspui/bitstream/123456789/8103/1/Global%20progress%20report%20on%20water%2C%20sanitation%20and%20hygiene%20in%20health%20care%20facilities%20%20fundamentals%20first.pdf (accessed June 2021).
Wright, A., Hawkins, C. H., Anggard, E. E., and Harper, D. R. (2009). A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 34, 349–357. doi: 10.1111/j.1749-4486.2009.01973.x
Yang, J. H., Wu, U. I., Tai, H. M., and Sheng, W. H. (2019). Effectiveness of an ultraviolet-C disinfection system for reduction of healthcare-associated pathogens. J. Microbiol. Immunol. Infect. 52, 487–493. doi: 10.1016/j.jmii.2017.08.017
Yang, S. C., Lin, C. H., Sung, C. T., and Fang, J. Y. (2014). Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front. Microbiol. 5:241. doi: 10.3389/fmicb.2014.00241
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Zhanel, G. G., Lawrence, C. K., Adam, H., Schweizer, F., Zelenitsky, S., Zhanel, M., et al. (2018). Imipenem-relebactam and meropenem-vaborbactam: two novel carbapenem-beta-lactamase inhibitor combinations. Drugs 78, 65–98. doi: 10.1007/s40265-017-0851-9
Zhao, X., Yu, Z., and Ding, T. (2020). Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms 8:425. doi: 10.3390/microorganisms8030425
Zhao, Z. L., Chen, L., and Zhang, H. (2020). Successful treatment of a multidrug-resistant tuberculosis patient with a negative Xpert MTB/RIF test for rifampicin-resistant tuberculosis in guizhou province of china: a case report. Infect. Drug Resist. 13, 1351–1355. doi: 10.2147/IDR.S245219
Zheng, H. S., Guo, W. Q., Wu, Q. L., Ren, N. Q., and Chang, J. S. (2018). Electro-peroxone pretreatment for enhanced simulated hospital wastewater treatment and antibiotic resistance genes reduction. Environ. Int. 115, 70–78. doi: 10.1016/j.envint.2018.02.043
Zimmermann, S., Klinger-Strobel, M., Bohnert, J. A., Wendler, S., Rodel, J., Pletz, M. W., et al. (2019). Clinically approved drugs inhibit the staphylococcus aureus multidrug nora efflux pump and reduce biofilm formation. Front. Microbiol. 10:2762. doi: 10.3389/fmicb.2019.02762
Keywords: antimicrobial resistance, ESKAPE pathogens, fast diagnostics, therapeutics, disease prevention
Citation: Avershina E, Shapovalova V and Shipulin G (2021) Fighting Antibiotic Resistance in Hospital-Acquired Infections: Current State and Emerging Technologies in Disease Prevention, Diagnostics and Therapy. Front. Microbiol. 12:707330. doi: 10.3389/fmicb.2021.707330
Received: 09 May 2021; Accepted: 29 June 2021;
Published: 21 July 2021.
Edited by:
Axel Cloeckaert, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceReviewed by:
Gamal Wareth, Friedrich-Loeffler-Institute, GermanyCopyright © 2021 Avershina, Shapovalova and Shipulin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ekaterina Avershina, ZWthdGVyaW5hLmF2ZXJzaGluYUBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.