
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 July 2021
Sec. Infectious Agents and Disease
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.706207
This article is part of the Research Topic Mycobacterium abscessus; The Paradox of Low Pathogenicity and High Virulence View all 5 articles
 Vinicius C. N. de Moura1†‡
Vinicius C. N. de Moura1†‡ Deepshikha Verma1‡
Deepshikha Verma1‡ Isobel Everall2,3
Isobel Everall2,3 Karen P. Brown2,4
Karen P. Brown2,4 Juan M. Belardinelli1
Juan M. Belardinelli1 Crystal Shanley1
Crystal Shanley1 Megan Stapleton1
Megan Stapleton1 Julian Parkhill5
Julian Parkhill5 R. Andres Floto2,4
R. Andres Floto2,4 Diane J. Ordway1
Diane J. Ordway1 Mary Jackson1*
Mary Jackson1*Chronic pulmonary infections caused by non-tuberculous mycobacteria of the Mycobacterium abscessus complex (MABSC) are emerging as a global health problem and pose a threat to susceptible individuals with structural lung disease such as cystic fibrosis. The molecular mechanisms underlying the pathogenicity and intrinsic resistance of MABSC to antibiotics remain largely unknown. The involvement of Msp-type porins in the virulence and biocide resistance of some rapidly growing non-tuberculous mycobacteria and the finding of deletions and rearrangements in the porin genes of serially collected MABSC isolates from cystic fibrosis patients prompted us to investigate the contribution of these major surface proteins to MABSC infection. Inactivation by allelic replacement of the each of the two Msp-type porin genes of M. abscessus subsp. massiliense CIP108297, mmpA and mmpB, led to a marked increase in the virulence and pathogenicity of both mutants in murine macrophages and infected mice. Neither of the mutants were found to be significantly more resistant to antibiotics. These results suggest that adaptation to the host environment rather than antibiotic pressure is the key driver of the emergence of porin mutants during infection.
Two groups of organisms account for 70–95% of the pulmonary non-tuberculous mycobacteria (NTM) infections worldwide: The Mycobacterium avium complex (MAC) and the Mycobacterium abscessus complex (MABSC) (Park and Olivier, 2015). While MAC is still, overall, the most common cause of NTM infections in the United States (accounting for over 70% of NTM pulmonary infections), over the last 10 years, rapidly growing NTM of the M. abscessus complex (MABSC) [including M. abscessus subsp. abscessus (Mabs), M. abscessus subsp. massiliense [Mmas] and M. abscessus subsp. bolletii] have emerged as important human pathogens globally, causing an increasing number of pulmonary infections among patients with structural lung disease such as chronic obstructive pulmonary disease, bronchiectasis and cystic fibrosis (CF) (Floto and Haworth, 2015; Park and Olivier, 2015; Martiniano et al., 2019; Johansen et al., 2020). Detailed epidemiology and population-level whole genome sequencing studies have indicated that dominant circulating clones of Mabs and Mmas have recently emerged and are now present on every continent (Shang et al., 2011; Aitken et al., 2012; Bryant et al., 2013, 2016; Davidson et al., 2013; Tettelin et al., 2014). These clones are associated with worse clinical outcomes, have increased ability to infect and survive within macrophages, and cause more severe infections in mouse models (Shang et al., 2011; Bryant et al., 2016).
Despite recent advances in the genomics and genetics of MABSC organisms and the availability of a growing number of cellular and animal models of NTM infection (Medjahed and Reyrat, 2009; Ripoll et al., 2009; Bryant et al., 2016; Bernut et al., 2017), the molecular mechanisms underlying the intrinsic antibiotic resistance/tolerance, virulence and adaptation of MABSC species to becoming chronic pathogens of the lung are still poorly understood. Previous studies from our laboratory and others have established the important role played by outer membrane porins in the virulence and biocide resistance of rapidly growing mycobacteria including M. smegmatis and M. chelonae (Stephan et al., 2004; Sharbati-Tehrani et al., 2005; Danilchanka et al., 2008; Fabrino et al., 2009; Svetlikova et al., 2009; Purdy et al., 2009; Frenzel et al., 2011; Rodrigues et al., 2011). Msp-type porins represent the main general diffusion pathway for small hydrophilic molecules across the outer membrane of rapidly growing mycobacteria (Niederweis, 2003). Not only does their presence at the cell surface increase the permeability and susceptibility of M. smegmatis and M. chelonae to antibiotics and disinfectants (Stephan et al., 2004; Danilchanka et al., 2008; Svetlikova et al., 2009; Frenzel et al., 2011; Rodrigues et al., 2011), it was also found to impact the survival of M. smegmatis and that of an M. tuberculosis mspA-expressing strain inside phagocytic cells by increasing the susceptibility of the bacilli to killing by reactive nitrogen intermediates, ubiquitin-derived antimicrobial peptides and lysosomal enzymes (Sharbati-Tehrani et al., 2005; Fabrino et al., 2009; Purdy et al., 2009).
The reported involvement of porins in the virulence phenotype and biocide resistance of some rapidly growing mycobacteria, combined with the finding of deletions and rearrangements in the porin genes of serially collected Mabs and Mmas isolates from CF patients and genetic polymorphisms affecting the porin locus of MABSC isolates globally, led us to investigate the potential roles played by Msp-type porins in the adaptation of MABSC to the host and to antibiotic and disinfectant exposure.
Escherichia coli DH5α, the strain used for cloning, was grown in LB Lennox (BD, Difco) medium at 37°C. M. abscessus subsp. massiliense (Mmas) CIP108297, a clinical isolate recovered from the sputum and bronchoalveolar fluid of a patient with hemoptoic pneumonia (Adekambi et al., 2004) was grown at 37°C in Middlebrook 7H9-OADC broth (BD, Difco) supplemented with 0.05% Tween 80, Mueller-Hinton II broth (BD, Difco), minimal Glycerol-Alanine-Salt (GAS) medium supplemented with 0.05% tyloxapol (pH 6.6), or on Middlebrook 7H11-OADC agar (BD, Difco). Kanamycin (Kan), hygromycin (Hyg) and streptomycin (Str) were added to final concentrations of 400, 1,000, and 200 μg/ml, respectively.
Recombineering was used to inactivate the MMCCUG48898_0905 (herein renamed mmpA) and MMCCUG48898_0906 (herein renamed mmpB) genes of Mmas CIP108297 by allelic replacement. To this end, the Gp60 and Gp61 recombineering proteins from mycobacteriophage Che9c were expressed from the replicative plasmid pJV53-XylE under control of an acetamide-inducible promoter as described (van Kessel and Hatfull, 2007; van Kessel et al., 2008; Calado Nogueira de Moura et al., 2014). Acetamide-induced Mmas CIP108297 harboring pJV53-XylE were electro-transformed with linear allelic exchange substrates encompassing the mmpA and mmpB loci and double-crossover mutants were isolated on Str-containing medium. The linear allelic substrate used to delete the mmpA locus was generated by bracketing the streptomycin-resistance cassette from pHP45Ω with 571–500 bp of upstream and downstream DNA sequence flanking the internal XmnI restriction site of mmpA, respectively. The linear allelic substrate used to delete the mmpB locus was generated by similarly bracketing the streptomycin-resistance cassette with 565–357 bp of upstream and downstream DNA sequence immediately flanking the internal XmnI restriction site of mmpB. The linear allelic substrate used to try and delete both porin genes simultaneously was generated by bracketing the streptomycin-resistance cassette with 571 bp of DNA sequence located upstream the XmnI site of mmpA and 357 bp of DNA sequence downstream the XmnI site of mmpB.
pOMK-mmpA, pOMK-mmpB, and pOMK-mmpAB, the replicative plasmids used for the complementation of the knock-out mutants, were constructed by cloning mmpA, mmpB or a 2.1 kb region encompassing both genes in the multicopy plasmid pOMK (Calado Nogueira de Moura et al., 2014). The mmpA and mmpB genes are expressed from their own promoter in the complementation constructs. The sequences of the primers used to generate the different constructs are provided in Supplementary Table 1.
The susceptibility of Mmas to commercial glutaraldehyde (GTA) in suspension tests, MIC determinations and [U-14C]-glucose (5 mCi/mmol; American Radiolabeled Chemicals) uptake experiments were performed as described in Calado Nogueira de Moura et al., 2014. For in vitro NO susceptibility studies, Mmas strains (approximately 2 × 107 cells) grown in 7H9-OADC medium were resuspended in 1 ml phosphate buffer (pH 7.4) and exposed to 10 mM Spermine NONOate (Cayman Chemical) for 4–24 h at 25°C at which time point viable CFUs were enumerated by plating serial dilutions of the cell suspensions on 7H11-OADC agar.
Porin extraction and immunoblot analysis using anti-MspA antibodies (Stahl et al., 2001) was carried out as described previously (Calado Nogueira de Moura et al., 2014).
Mmas RNA was extracted from 5-ml cultures grown to an OD600 of 0.2 using the Direct-zol RNA Miniprep kit (Zymo Research) per the manufacturer’s instructions. Reverse transcription reactions were carried out using the Superscript V kit (Invitrogen), and qRT-PCRs were run using the SsoFast Evagreen Supermix kit (Bio-Rad) as per the manufacturers’ protocols and analyzed on a CFX96 real-time PCR machine (Biorad). PCR conditions: 98°C (2 min; enzyme activation), followed by 35 cycles of 98°C (5 sec; denaturation) and 55°C (5 sec; annealing/extension). Mock reactions (no reverse transcription) were done on each RNA sample to rule out DNA contamination. The target cDNA was normalized internally to the sigA cDNA levels in the same sample. The primer sequences are provided in Supplementary Table 1.
A previously published global collection of MABSC isolates, consisting of 526 isolates associated with pulmonary infections and 189 isolates collected from nine separate outbreak locations in Brazil, was used in this study (Bryant et al., 2016; Everall et al., 2017). These isolates were further supplemented by an additional 665 longitudinal isolates associated with 190 patients with Cystic Fibrosis (CF) (Bryant et al., 2021). The Illumina Hiseq 2,500 paired end raw reads of all the isolates were mapped using BWA-MEM v0.7.12 to the reference genome Mabs ATCC 19977 (Li. 2013). Assemblies for all the isolates were generated and optimized using Velvet and Velvet optimizer, respectively (Zerbino and Birney, 2008). The final draft assemblies were annotated using PROKKA (Seemann, 2014).
An alignment was produced from the mapping of 526 isolates associated with pulmonary infections and 188 isolates associated with the post-surgical wound infection epidemic in Brazil to the Mabs ATCC 19977 reference genome, using SAMtools v1.2 and BCFtools v4.2, with parameters previously described (Li et al., 2009; Harris et al., 2010). Briefly, a variant was inferred if the following criteria were met: A mapping quality of 20, a base call quality of 50, eight reads supporting the alternative allele and three reads supporting the alternative allele on either strand. Heterozygous SNP sites were masked. An alignment of the variable positions was created using SNP-sites and used to produce a maximum likelihood phylogeny with RAxML (Stamatakis, 2014; Page et al., 2016). Hundred bootstrap replicates were performed.
A local tblastx search was performed to detect homologs of msp-like porin genes in the MABSC genomes. A local database was made using the nucleotide sequences of mmpA (MAB_1080), mmpB (MAB_1081) and the possible mmpC homolog (MAB_2800). The assemblies of 1,382 MABSC isolates were queried against this database with msp-like porin gene homologs predicted if a match had a percent identity greater than 50%, E-value < 0.05 and a match length greater than or equal to 50% of the length of the reference gene of mmpA (672 bps), mmpB (669 bps) and mmpC (741 bps).
The coverage over the porin locus containing the mmpA and mmpB genes was determined for each isolate using SAMtools v1.3, with a mapping quality threshold of 20 (probability mapped to correct location 99%), base call quality of 30 (base call is 99.9% accurate) (Li et al., 2009). Presence of both msp-like porin genes at this locus was determined by an average depth of coverage greater than eight in the 436 bp intergenic region between mmpA and mmpB and an average depth of coverage greater than eight over both mmpA and mmpB. Deletion of both mmp-like porin genes at this locus was determined by the absence of coverage (average of less than eight reads mapped) in the intergenic region between mmpA and mmpB and a reduction in depth of coverage greater than 20 between the 1,000 bp region upstream of the start of the porin loci and the 94 bp signal peptide region of mmpA. The presence of a single mmp-like porin gene at this locus was determined by absence of coverage in the intergenic region between mmpA and mmpB (average of less than eight reads mapped) and no reduction in depth of coverage greater than 20 between the 1,000 bp flanking region upstream of the porin loci and the 94 bp signal peptide region of mmpA. The figures were made using easyfig (Sullivan et al., 2011). A two tailed Fisher’s exact test was performed to determine if there was an association between the mmpA and mmpB porin locus configuration and isolates classified as clustered and more virulent and those classified as unclustered by Bryant et al. (2016).
Where only one mmp-like porin gene was present at the mmpA/mmpB porin locus, an alignment of the nucleotide sequence of this gene, and the reference mmpA (MAB_1080) and mmpB (MAB_1081) genes was created using muscle v3.8.31 (Edgar, 2004). The position at which the recombination event occurred was predicted based on where the mmp-like porin gene of the isolate with one mmp-like porin gene at the mmpA/mmpB porin locus changed from being more similar to mmpA to being more similar to mmpB.
The raw reads of the 1,382 isolates were further mapped to the Mabs ATCC 19977 mmpA/mmpB porin locus using BWA-MEM v0.7.12 (Li, 2013). Variants were called using SAMtools v1.2 and BCFtools v4.2 with the following parameters (Li et al., 2009): A base call quality of 30 and mapping quality of 20, a minimum of eight reads supporting the alternative base, with a minimum of three reads supporting the alternative base on each strand. Variants, including Indels, were only determined for isolates predicted to encode both mmpA and mmpB at this locus.
Specific-pathogen-free female SCID and GMCSF knock-out mice, from 6 to 8 weeks old, were purchased from the Jackson Laboratories, Bar Harbor, Maine. Mice were maintained in the Biosafety Level III animal laboratory at Colorado State University, and were given sterile water, mouse chow, bedding, and enrichment for the duration of the experiments. The specific pathogen-free nature of the mouse colonies was demonstrated by testing sentinel animals. All experimental protocols were approved by the Animal Care and Usage Committee of Colorado State University. The CSU animal assurance welfare number is A3572-01.
Bone marrow-derived macrophages (BMDM) from GMCSF and SCID knock-out mice were infected with mycobacteria at a one bacterium per one macrophage ratio and, 2 h later, the monolayers were washed to remove extracellular bacilli. The numbers of intracellular mycobacteria were measured by plating. Briefly, monolayers were washed at each time point to remove extracellular bacilli and 1 ml double-distilled H2O containing 0.05% Tween 80 was added to monolayers and incubated for 10 min to lyse macrophages. After passing through a 26-gauge needle five times, the lysates were serially diluted and plated onto 7H11-OADC agar. Monolayers that weren’t lysed were replenished with fresh medium as previously described (Shang et al., 2011; Obregon-Henao et al., 2015). BMDM viability was assayed by trypan blue exclusion and determined by flow cytometry using BD Viability Counting Beads, as described by the manufacturer (BD PharMingen, San Jose, CA United States) cell viability-staining methods.
SCID mice were challenged with WT, mutant and complemented mutant strains using an infection calibrated to deliver 1.0 × 106 bacilli per animal. At days 1 and 15 following infection, bacterial loads in the lungs, spleen and liver, and lung histology were determined. Bacterial counts were determined by plating serial dilutions of organ homogenates on 7H11-OADC agar and counting colony-forming units after 5–10 days of incubation at 30°C. A total of five animals were infected for each time point.
The whole lung from each mouse was fixed with 10% formalin in phosphate buffered saline (PBS). Tissue sections were stained using hematoxylin and eosin and acid-fast stain as previously reported (Bryant et al., 2016).
Data are presented using the mean values from five mice per group and from values from replicate samples and triplicate macrophage infection assays. The unpaired Student’s t-test test was used to assess statistical significance between infection groups.
The reference strains Mmas CIP108297 (alias CCUG 48898) and Mabs ATCC 19977 harbor two clustered Msp-like porin genes annotated as MMCCUG48898_0905/MMCCUG48898_0906 and MAB_1080/MAB_1081, respectively (Ripoll et al., 2009; Tettelin et al., 2012). We renamed the two Mmas genes mmpA and mmpB for Mycobacterium abscessus subsp. massiliense porins A and B. In addition, the genomes of Mmas CIP108297 and Mabs ATCC 19977 encode a third, more distantly related, porin gene (MAB_2800 in Mabs ATCC 19977) which we renamed mmpC and six other proteins sharing 22–53% identity with MmpC annotated as putative porins (MAB_2799; MAB_1321; MAB_3154c; MAB_2329c; MAB_0734 and MAB_1891). The mature MmpA and MmpB porin products share 97% identity between them and 72–73% identity with MspA from M. smegmatis (Supplementary Figure 1) (98.9–100% identity with the Mabs ATCC 19977 porin products, MAB_1080 and MAB_1081). MmpC shares only 22% identity (27.8% similarity) with MmpA and MmpB from Mabs ATCC 19977 and 22.3% identity (28.4% similarity) with MspA from M. smegmatis mc2155. 463 bp of nucleotide sequence separate mmpA from mmpB in Mmas CIP108297 and Mabs ATCC 19977 suggesting that the two genes are probably each monocistronic.
At least one (mmpA/mmpB- or mmpC-like) porin gene homolog was detected in all the MABSC genomes used in this study (n = 1,382). However, analysis of the depth of coverage of the mapped reads over the mmpA and mmpB porin loci of the 1,382 MABSC genomes used herein showed that there was variation in the number of porin genes encoded at this position. Overall, 1,059 isolates encoded both mmpA and mmpB porin genes at this locus, 312 isolates encoded a single mmp-like porin gene (a fusion of mmpA and mmpB; see further) and 11 isolates encoded no mmp-like porin genes at this locus.
Of the 526 isolates associated with pulmonary infections, 469 isolates encoded two mmp-like porin genes at this locus (Figure 1A), 54 encoded a single mmp-like porin fusion gene (Figure 1B) and three isolates encoded no mmp-like porin genes at this locus (Figure 1C). A single mmp-like porin gene, a fusion of mmpA and mmpB, was observed in 180 of 189 isolates associated with the post-surgical wound infection epidemic in Brazil (Duarte et al., 2009; Leao et al., 2010; Everall et al., 2017), while the remaining 9 isolates encoded two mmp-like porin genes at this locus.
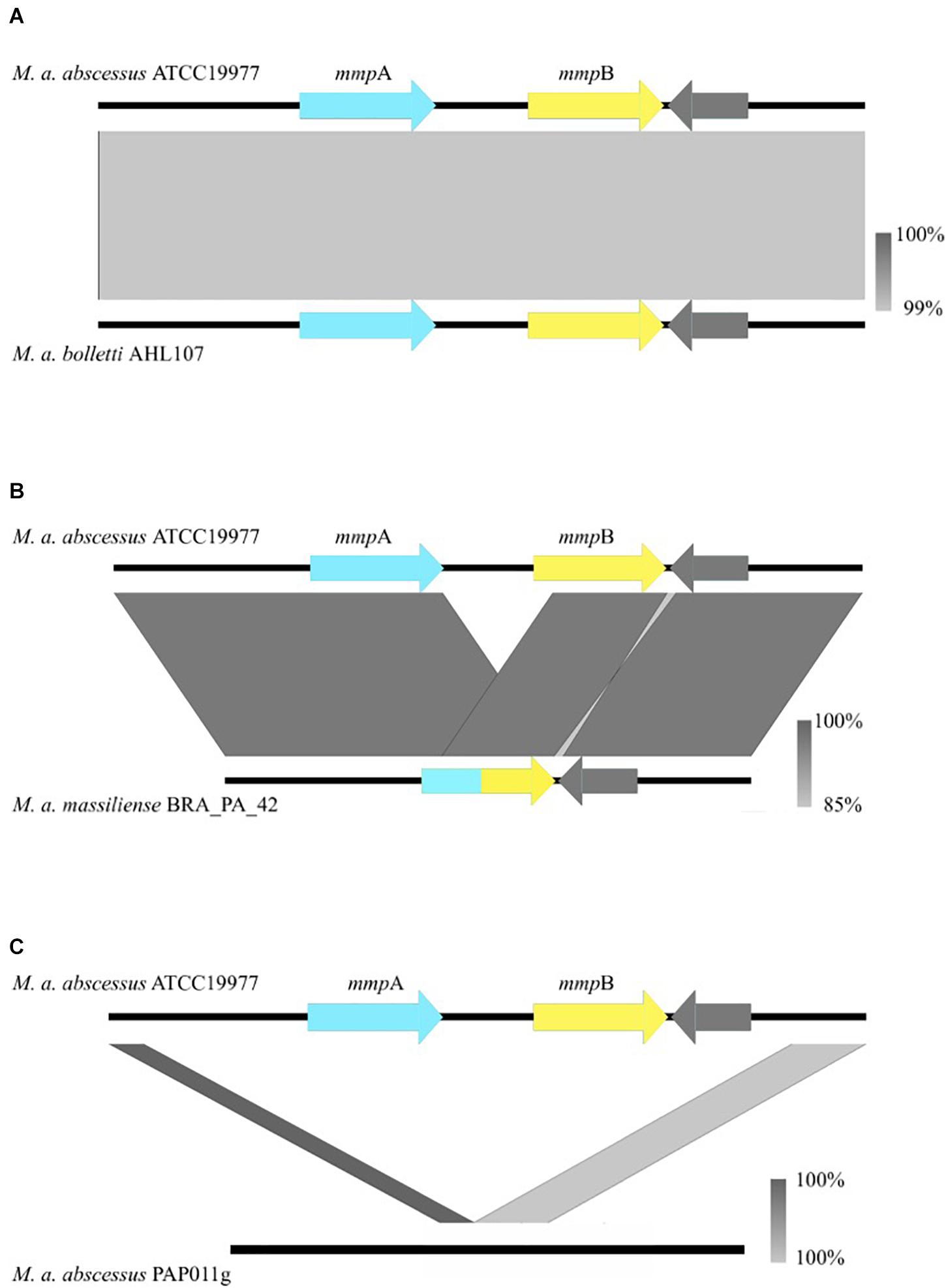
Figure 1. Nucleotide BLAST comparisons between the mmpA/mmpB porin locus of Mabs ATCC 19977 and isolates that encode the variants observed in the porin locus both across the global population and longitudinally. The gray bars illustrate the various rearrangements [or no rearrangement in the case of (A)] observed in clinical isolates and are color-coded to indicate the percentage identity of the genomic sequences of the two isolates that are being compared. (A) Is an example of when mmpA and mmpB are both present. (B) Is an example of the fusion of mmpA and mmpB, resulting in a single mmp-like gene. (C) Is an example of the complete deletion of both mmp-like porin genes at the porin locus. Porin genes are colored in blue and yellow.
Short indels, leading to the loss of function of the protein, were observed in mmpA in seven isolates associated with pulmonary infections. One isolate incurred a non-synonymous mutation resulting in a premature stop codon, W220∗.
Changes in genome organization at the mmpA/mmpB locus were also observed in isolates collected from the same patient over time. Isolates from 15 of the 190 CF patients for which longitudinal samples were available were found to vary in the number of mmp-like porin genes encoded by this locus. Isolates from 12 of the 15 patients either encoded one or two mmp-like porins, one patient’s isolates encoded either two mmp-like porin genes or no mmp-like porin genes, one patient’s isolates encoded either one mmp-like porin gene or no mmp-like porin genes and the final patient’s isolates varied between encoding either two, one or no mmp-like porin genes at the mmpA/mmpB locus. A further four patients’ isolates were shown to have accumulated indels in mmpA over time. No non-synonymous mutations were found to have occurred within patients over time.
As noted above, where isolates were found to have only one mmp-like porin gene at the mmpA/mmpB porin locus, a recombination event had occurred leading to the fusion of mmpA and mmpB. We examined the sequences of the 312 predicted mmpA/mmpB fusion genes to determine where in the sequence the fusion occurred. Of the 312 predicted mmpA/mmpB fusion genes, it was only possible to extract the complete sequence of the gene from the draft assembly of 297 isolates. It was possible to predict the fusion position for 38 of these isolates, with six unique fusion positions predicted which spanned close to the entire length of mmpA (Figure 2). All six fusion positions were also seen in isolates where changes in the number of mmp-like porin genes encoded at the mmpA/mmpB porin locus were observed over time within a patient. All the predicted fusion positions fall after the signal peptide sequence at the N-terminal of MmpA and, therefore, all the fusion genes use the signal peptide sequence of this porin.
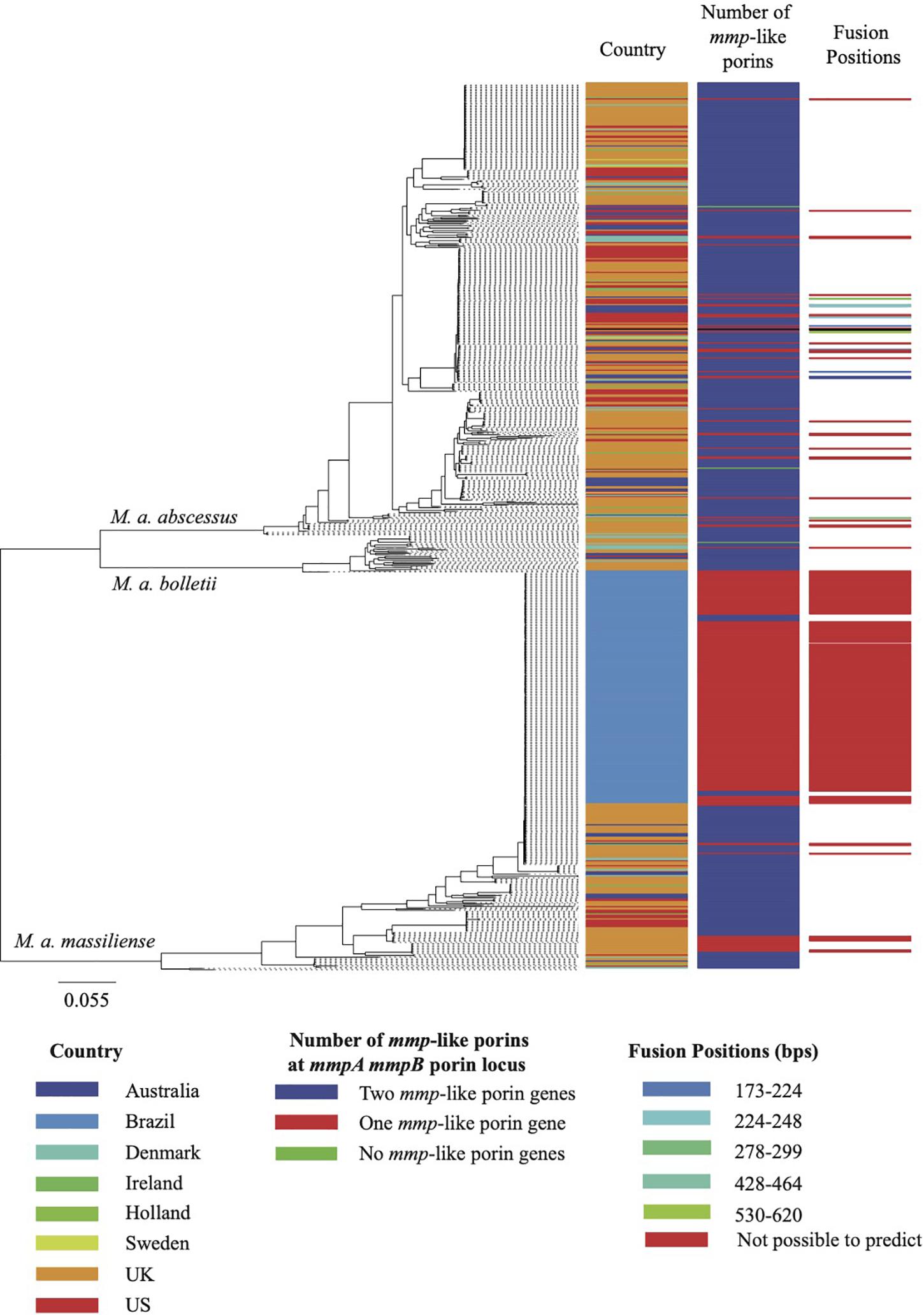
Figure 2. Maximum likelihood phylogeny of the MABSC global population. The global population shown here consist of 526 isolates associated with pulmonary infections and 188 isolates associated with the epidemic of post-surgical wound infections in Brazil. The metadata aligned shows, for each isolate, the country of isolation, the predicted composition of the porin locus and the isolates for which the fusion position was predicted. The scale bar on the phylogeny represents the number of substitutions per variable site.
Because the changes occurring at mmpA/mmpB loci of longitudinally isolated strains from CF patients was suggestive of the involvement of MmpA and MmpB in the adaptation of MABSC to the host, subsequent studies were undertaken to determine whether these naturally occurring mutations arose as a result of the selective pressure of antibiotic treatment or as part of a more general adaptation of the bacterium to stresses imposed by the host environment.
To investigate the role of MmpA and MmpB in the physiology and virulence of MABSC, we first sought to knock-out each of these two porin genes of Mmas CIP108297 by allelic replacement. To this end, the recombineering method was used wherein Mmas CIP108297 expressing the Gp60 and Gp61 recombineering proteins from mycobacteriophage Che9c from pJV53-XylE (van Kessel and Hatfull, 2007; van Kessel et al., 2008; Calado Nogueira de Moura et al., 2014) was electrotransformed with linear allelic exchange substrates designed to replace the entire mmpA or mmpB loci with a streptomycin-resistance cassette. Twelve to 55 double crossover candidates were selected for each gene and analyzed by PCR. PCR analysis confirmed that allelic replacement had occurred in 18% of the mmpA candidate mutants and 50% of the mmpB candidate mutants (Figure 3A). Two passages in liquid broth devoid of Kan followed by plating on Str-containing agar yielded single mmpA and mmpB mutant clones devoid of pJV53-XylE plasmid. In contrast, three independent attempts to inactivate both porin genes simultaneously yielded no double crossover mutant suggesting that, similar to the situation in M. smegmatis (Stephan et al., 2005), the presence of at least one functional porin gene is required for Mmas CIP108297 growth under the experimental conditions used herein. Given that our genomic analysis identified 11 isolates devoid of mmpA and mmpB genes, one may only speculate that the mmpC gene and homologs present in these isolates can compensate for the lack of mmpA and mmpB in a way that is not achievable in Mmas CIP108297.
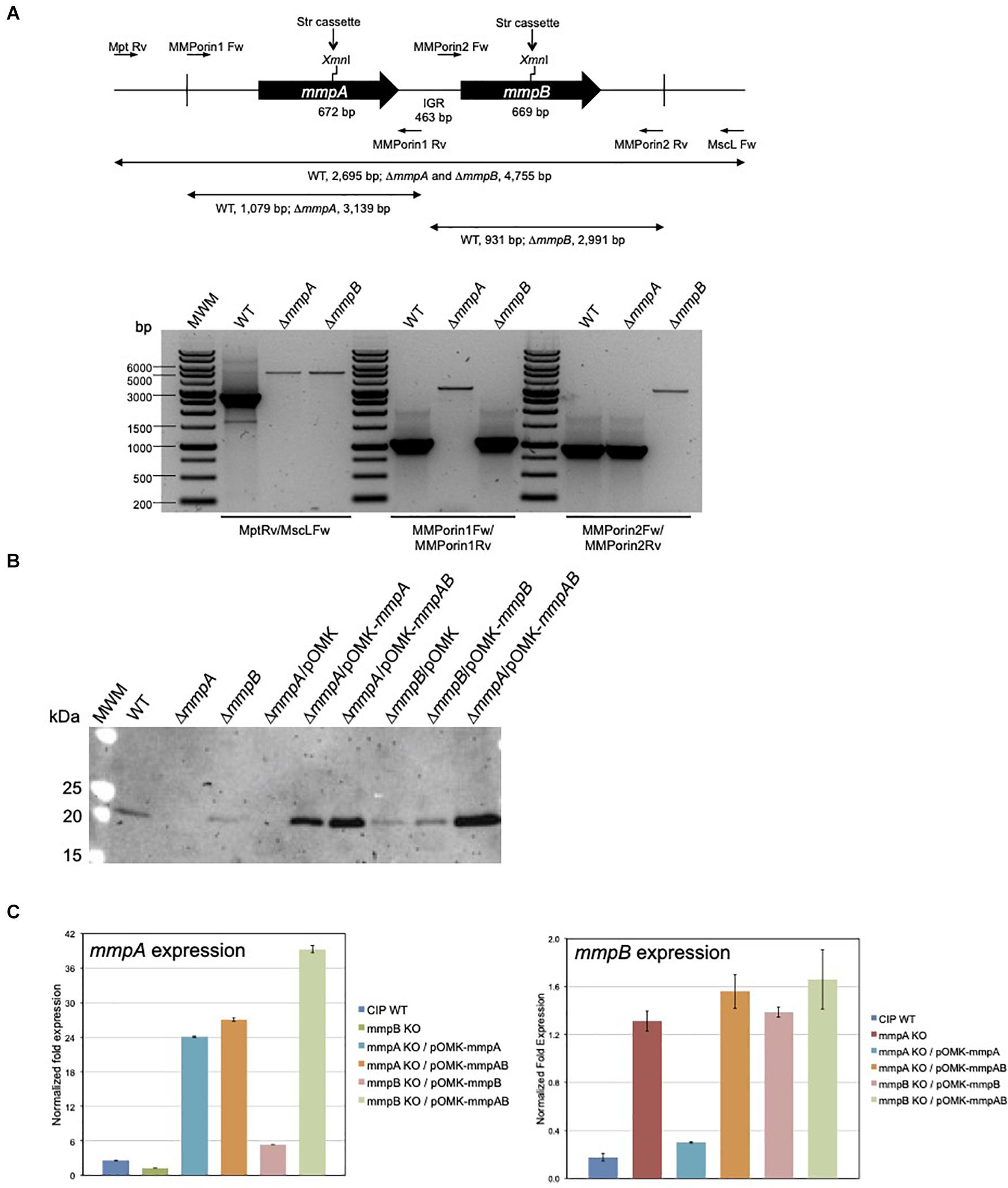
Figure 3. Gene replacement at the mmpA and mmpB porin loci of Mmas. (A) Porin gene cluster of Mmas CIP108297. The positions of the primers used to generate the allelic exchange substrates and analyze the candidate mutants are indicated along with the expected sizes of the PCR fragments. Candidate mmpA and mmpB mutants were analyzed by PCR using different sets of primers, including Mpt-Rv/MscL-Fw located outside the allelic exchange substrates, MMPorin1 Fw/MMPorin1 Rv, and MMPorin2 Fw/MMPorin2 Rv (see Supplementary Table 1). MWM, molecular weight marker; IGR, intergenic region. (B) Immunoblot analysis of porin production in the WT, mutant and complemented mutant strains. Strains were grown in 7H9-OADC-Tween 80 broth at 30°C to mid-log phase (OD600 = 1) and porins were selectively extracted from whole cells at 100°C using 0.5% n-octylpolyoxyethylene as a detergent as described (Heinz et al., 2003). Protein samples prepared from the same amount of cells for each strain were denatured by boiling in 80% DMSO followed by acetone precipitation (Heinz et al., 2003). Denatured proteins were loaded volume to volume, separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and porins were detected using rabbit antiserum to purified MspA (Stahl et al., 2001). Immune complexes were detected by chemiluminescence (Pierce, ELC) and semi-quantified using the Image Lab software (Biorad). The expected size of the mature MmpA and MmpB porins is 19.65 kDa. (C) mmpA and mmpB mRNA levels in the WT, mutant and pOMK complemented mutant strains. The target cDNA was normalized internally to the sigA cDNA in the same sample. Ratios of mmpA/sigA or mmpB/sigA mRNA are means ± standard deviations (n = 3 qRT-PCR reactions). RNA extractions and qRT-PCR experiments were performed twice using independent culture batches and the results of one representative experiment are shown.
Porin production in the mutant strains was analyzed by Western blot using polyclonal antibodies to the MspA porin of M. smegmatis (Stahl et al., 2001). While porin products of the expected size (19.6 kDa) were detected in the wild-type (WT) Mmas CIP108297 strain, they were barely detectable in the mmpA knock-out; the intensity of the porin signal in this mutant was only about 6–10% of that detected in the WT parent. In comparison, the intensity of the band was decreased by less than 40–50% in the mmpB knock-out (Figure 3B). Complementation of the mutants with their respective porin gene or with both porin genes expressed from the replicative multicopy plasmid pOMK increased porin production in the mmpA knock-out relative to the WT parent 3.4- and 5-fold, respectively, and that in the mmpB knock-out mutant 1- and 10-fold, respectively.
Porin expression in the two mutants was further analyzed by qRT-PCR to check for a potential polar effect of the inactivation of mmpA on the expression of mmpB and for the existence of compensatory mechanisms regulating the level of expression of the two porin genes. The result of two independent experiments indicated that while the level of expression of mmpA was only about 1.3–2-fold less in the mmpB mutant relative to the WT strain, the expression of mmpB increased 4–7-fold in the mmpA knock-out (Figure 3C). qRT-PCR analysis further revealed that the level of expression of mmpA relative to sigA was > 14-fold greater than that of mmpB in WT Mmas CIP108297, thereby confirming MmpA as the major porin product of this isolate and explaining the more dramatic effect of knocking-out mmpA over mmpB on the total porin content of this strain (Figure 3B).
As expected, the disruption of mmpA impacted more dramatically the growth rate of Mmas CIP108297 than that of mmpB, the latter mutant growing at a similar rate as its WT parent in minimal GAS (Glycerol-Alanine-Salts) medium (Figure 4A) and at an intermediate rate between the mmpA knock-out and the WT strain in glucose-based medium (Figure 4B). Complementation with the individual or combined mmpA and mmpB genes restored WT growth in the mutants (Figures 4A,B).
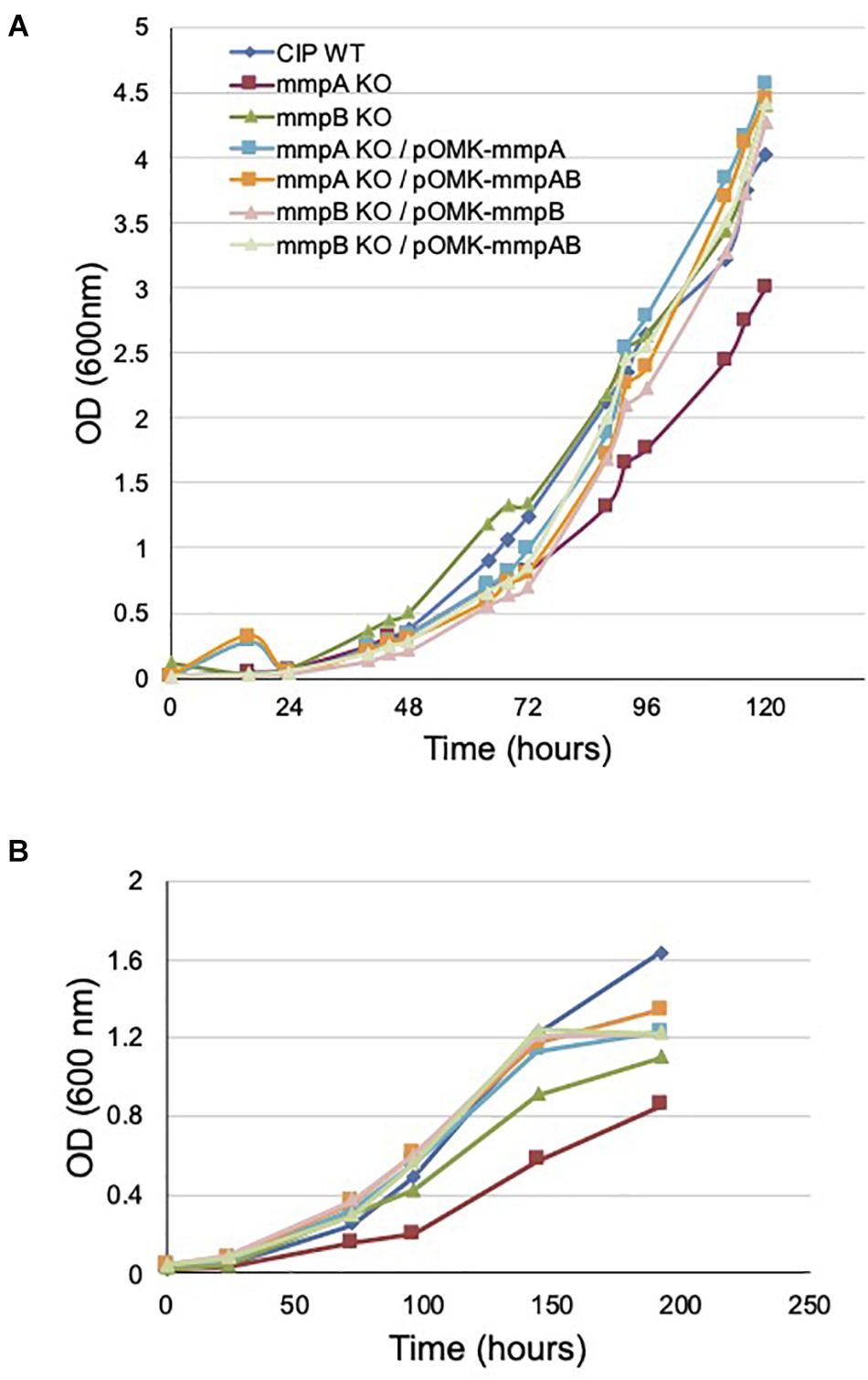
Figure 4. Growth rates of wild-type Mmas, its isogenic mmpA and mmpB knock-out mutants and the complemented porin mutants in GAS-tyloxapol (A) and glucose-based medium (B) at 37°C. Shown are representative results of two independent experiments. The glucose-based minimal medium consists of 20 mM asparagine, 1.5 mM KH2PO4, 0.75 mM Na2HPO4, 50 μM FeCl3, 1.6 mM MgSO4, 4.5 micromolar CaCl2, 3.5 μM ZnSO4, 0.59 micromolar MnSO4, 10 mM glucose and 0.05% tyloxapol in 50 mM MOPS (pH 7.0).
Consistent with these observations, the mmpA knock-out mutant presented a significantly reduced [14C]-glucose uptake rate relative to the WT parent (8.75 ± 0.03 vs. 17.11 ± 0.47 pmoles of glucose/mg cells/min) which was restored to WT levels upon complementation with mmpA (17.86 ± 0.41 pmoles of glucose/mg cells/min), and beyond WT levels upon complementation with mmpA and mmpB (23.35 ± 1.67 pmoles of glucose/mg cells/min) (Figure 5). The mmpB knock-out mutant, in contrast, was as proficient at taking up [14C]-glucose as WT Mmas CIP108297 in this assay (16.23 ± 0.58 and 17.11 ± 0.47 pmoles of glucose/mg cells/min, respectively).
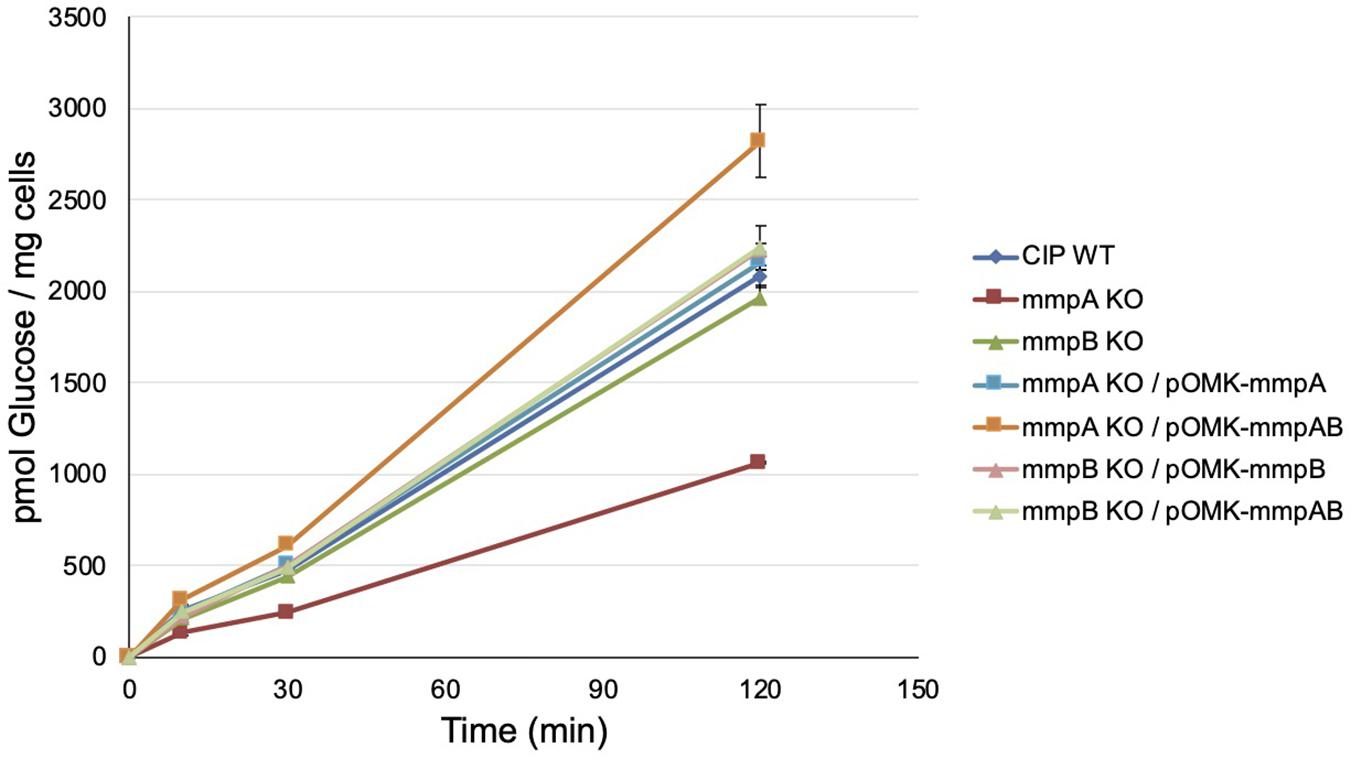
Figure 5. Glucose uptake by wild-type Mmas, the mmpA and mmpB knock-out mutants and the complemented porin mutant strains. The accumulation of [U-14C]glucose by the strains over time was measured as described (Calado Nogueira de Moura et al., 2014). Glucose uptake was measured in duplicate samples and the average values are shown with their standard deviations. Glucose uptake rates were calculated on the first 120 min of the reactions.
Because of the reported impact of porins on the susceptibility of M. smegmatis and M. chelonae to biocides and nitrosative stress (Fabrino et al., 2009; Svetlikova et al., 2009; Frenzel et al., 2011), we next set out to test the resistance of the mmpA and mmpB knock-out mutants to a number of stresses including various classes of antibiotics used in the clinical treatment of fast-growing mycobacterial infections, the high-level disinfectant glutaraldehyde (GTA), and nitric oxide (NO). Apart from a modest 2–4-fold increase in the resistance of the mmpA knock-out mutant to ciprofloxacin and imipenem, the two mutants presented susceptibility patterns to antibiotics (Table 1) and GTA (Supplementary Figure 2) comparable to those of their WT parent. In particular, both mutants displayed WT susceptibilities to drugs used in the clinical treatment of MABSC infections such as clarithromycin, erythromycin, azithromycin, amikacin, cefoxitin, linezolid and tigecycline. The mmpA and mmpB knock-outs further displayed WT susceptibility to NO in vitro (Supplementary Figure 3).
We investigated the capacity of Mmas reference strain CIP108297 and corresponding porin knock-out mutants to replicate and persist in BMDM to evaluate their virulence. The growth of each strain was determined in BMDM prepared from SCID and GMCSF knock-out mice by the method of plating to measure colony-forming units (CFU) over a period of 6 days. While no significant difference in bacterial uptake was noted between strains, both mutants demonstrated significantly increased replication in SCID (Figure 6A) and GMCSF knock-out (Figure 6B) BMDM compared to their WT parent, with the mmpB mutant consistently displaying the most acute virulence phenotype of the two. Throughout the experiment, BMDM cellular viability (as assessed by the percentage of propidium iodide positive dead cells) was above 95% and similar for all strains indicating that the porin mutants were not more cytotoxic than their WT parent in these macrophage models (data not shown). The replication of the mutants in macrophages was restored to or below WT levels upon genetic complementation with both porin genes (Figure 6B), and between WT and knock-out levels upon complementation with the individual WT copy of mmpA or mmpB (Figures 6A,B).
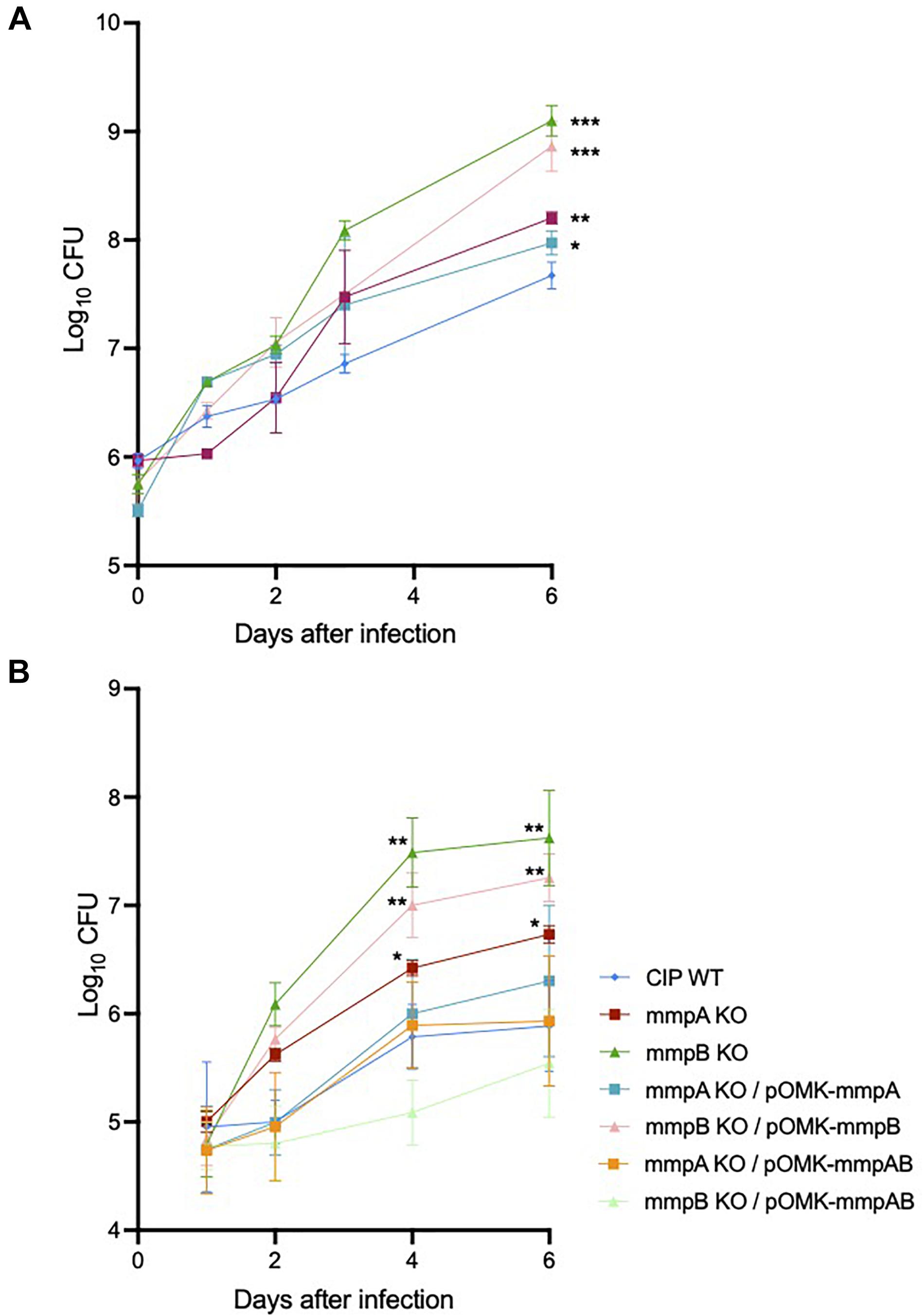
Figure 6. Increased intracellular replication of the porin mutants in SCID and GMCSF knock-out mouse bone marrow-derived macrophages. (A) Intracellular growth of the WT and mutant strains in SCID BMDM. BMDM were infected with Mmas strains at a MOI of 1:1. Viable intracellular bacteria were determined by CFU counting after 2 h of infection or at 1, 2, 3 and 6 days after infection. Values shown are the mean ± SEM from three infected macrophage monolayers. (B) Intracellular growth of the WT, mutant and complemented mutant strains in GMCSF BMDM. BMDM were infected with Mmas strains at a MOI of 1:1. Viable intracellular bacteria were determined by CFU counting after 2 h of infection or at 1, 2, 4, and 6 days after infection. Values shown are the mean ± SEM from three infected macrophage monolayers. Asterisks denote statistical differences between the growth of the knock-out mutants (or the mmpB KO complemented with mmpB only) and the WT parent strain pursuant to the Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.005).
We used a SCID mouse intravenous infection model to compare the virulence of Mmas WT, mutant and complemented mutant strains in vivo. The rationale for using this immunocompromised mouse model is related to the difficulty of establishing a high-level pulmonary infection with rapidly growing mycobacteria in immunocompetent mice (Obregon-Henao et al., 2015). Moreover, this model more closely reflects an infection which would occur in immunocompromised patients, a population particularly at risk for NTM infections (Chan et al., 2010; Prevots and Marras, 2015). Following intravenous infection, the bacterial loads in the lung, spleen and liver were quantified on days 1 and 15 post-infection. Consistent with the observations made in infected BMDM, infection with the mmpA and mmpB knock-out mutants resulted in increased bacterial loads in the lungs, spleen and liver at 15 days relative to the infection with the WT parent strain, with the mmpB mutant displaying the most dramatic increase in virulence (Figure 7). Virulence was restored to or below WT levels upon genetic complementation of the mmpB mutant with both porin genes, and that of the mmpA mutant with a WT copy of mmpA. The mmpB mutant complemented with both porin genes was used in this in vivo study because of the limited impact of complementing this mutant with mmpB only in murine macrophages (i.e., replication rate significantly above that of the WT parent strain) (Figures 6A,B).
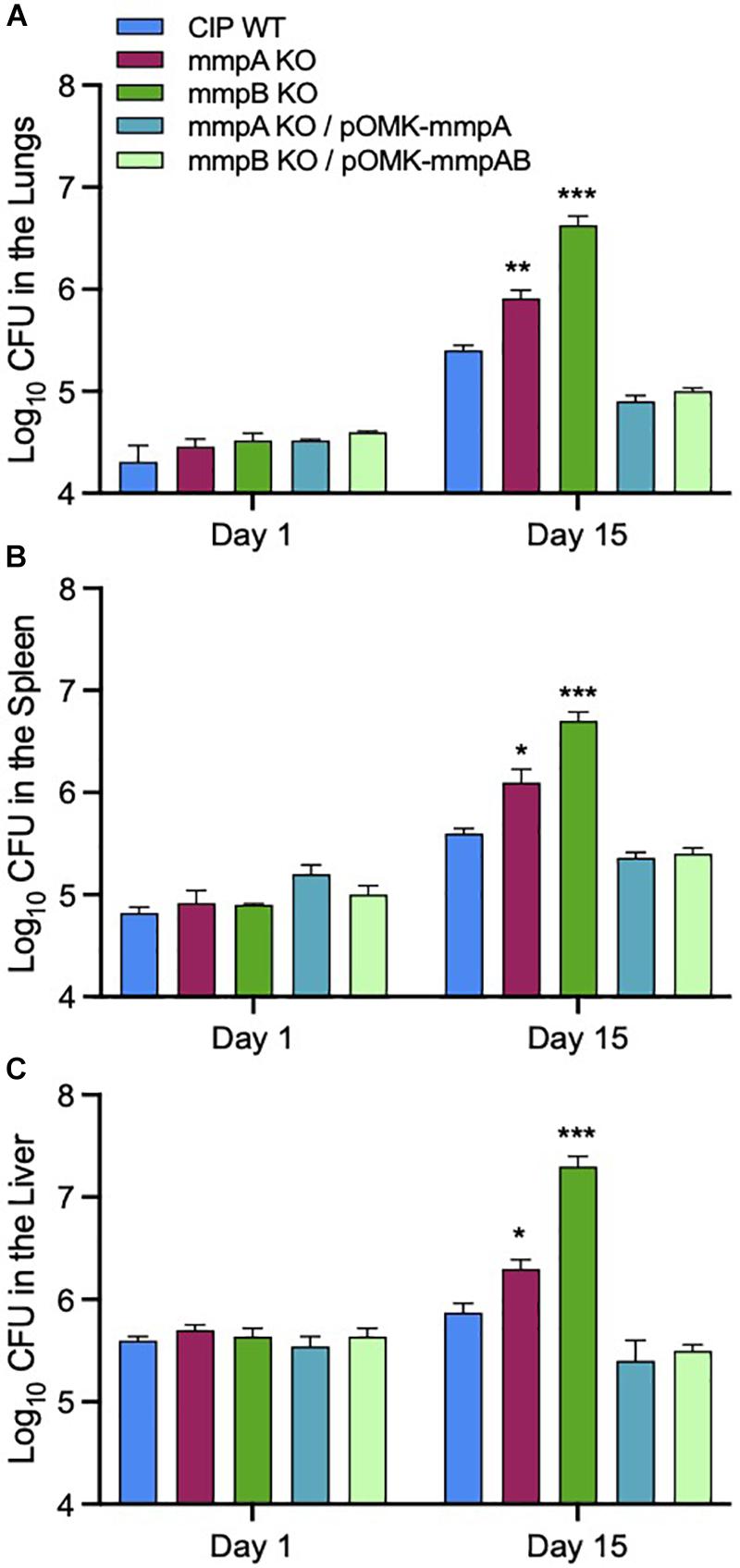
Figure 7. Increased bacterial load in the lungs, spleens and livers of SCID mice infected with the Mmas porin knock-out strains. Bacterial counts in the lungs (A), spleens (B) and livers (C) on days 1 and 15 from SCID mice intravenously infected the WT, porin mutant and complemented mutant strains. Results are expressed as the average (n = 5) of the bacterial load in each group expressed as Log10 CFU ± standard error mean (SEM). Growth of the two porin knock-out mutants was significantly higher than that of the WT parent strain pursuant to the Student’s t-test (*P < 0.0005; **P < 0.00005; ***P < 0.000005).
Lung histopathology mirrored these observations in that more severe peribronchiolar inflammatory infiltrates and increased cellular foci with early granuloma formation were observed in the lung tissues of the animals infected with the mmpB knock-out strain (Figure 8C). In comparison, mice infected with the WT parent and the complemented mutant strains displayed similar lung histopathology with less inflammatory foci and pulmonary involvement, and more healthy lung tissue (Figures 8A,D,E). The mmpA mutant displayed an intermediate phenotype (Figure 8B).
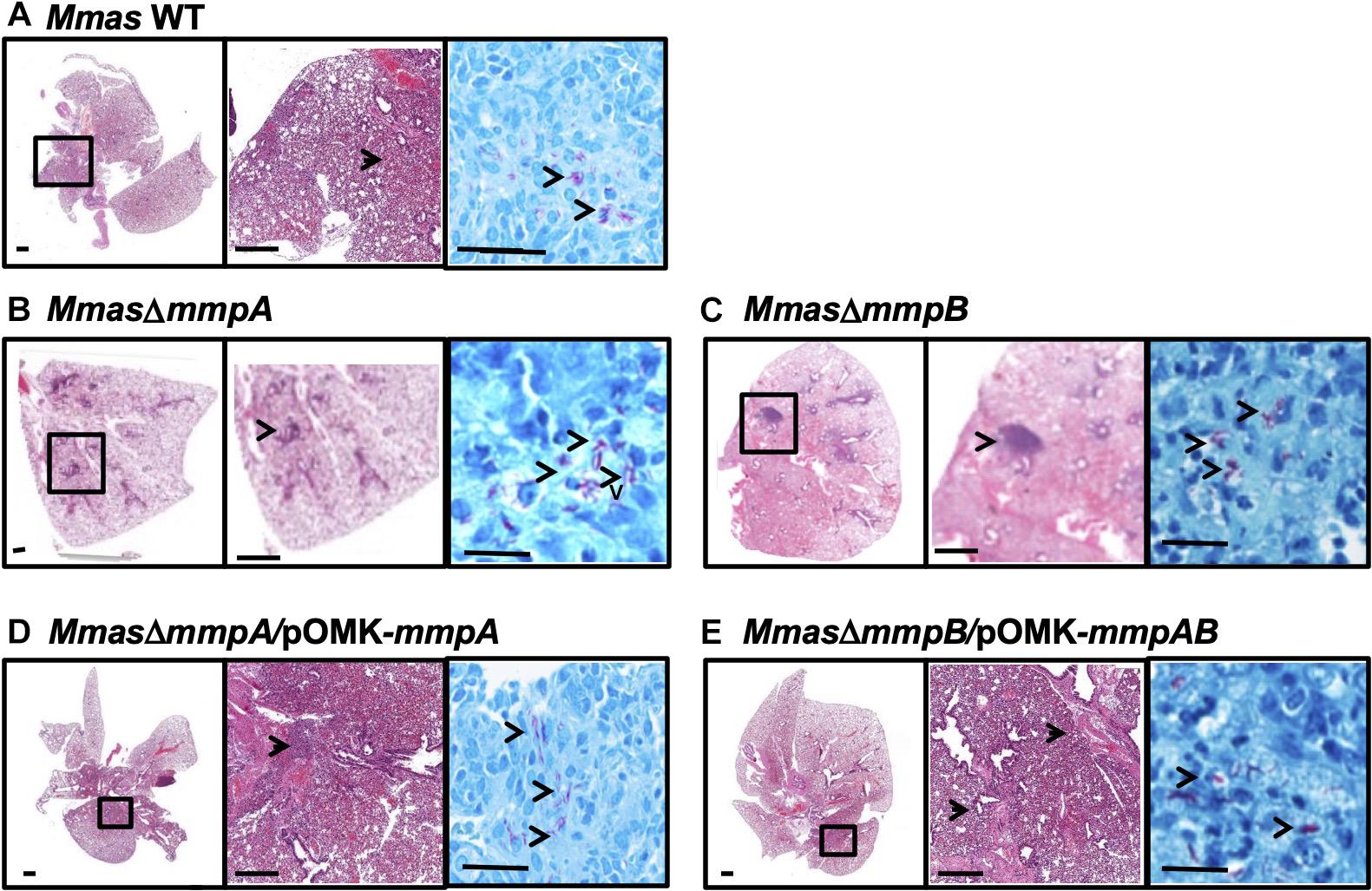
Figure 8. Increased pulmonary pathology in the lungs of SCID mice infected with the mmpB knock-out. (A–E) Show representative photomicrographs of hematoxylin and eosin- and acid-fast bacilli-stained slides from the lungs of SCID mice intravenously infected with Mmas CIP108297 (A), the mmpA mutant (B), the mmpB mutant (C), the mmpA mutant complemented with mmpA (D) and the mmpB mutant complemented with mmpA + mmpB (E). As early as 15 days after infection, increased granulomatous lesions and thickening of the interstitium indicative of increased inflammation were observed in mmpB-infected mice and, to a lesser extent in mmpA-infected mice, compared to the other groups. Magnification (– 1x, – 10x and —100x magnification).
The results of our combined genomic and genetic studies indicate that MABSC isolates generally harbor one or two Msp-like porins at the mmpA/mmpB locus and that MmpA is the predominant one in terms of level of expression and contribution to nutrient uptake (as measured by [14C]-glucose uptake) when both porins are present. The disruption of mmpA had a moderate effect on growth in GAS medium and a more marked effect on growth in glucose-based medium. Neither the disruption of mmpA or mmpB had any significant effect on the antibiotic susceptibility profile of Mmas CIP108297. Likewise, neither mutant showed increased resistance to GTA in vitro. The latter result and the fact that prototypical GTA-resistant outbreak isolates from Brazil (CRM-019 and CRM-020) have two porins identical in sequence to those of the GTA-susceptible isolates CRM-0270 and CIP108297 (Supplementary Figure 4; Duarte et al., 2009; Davidson et al., 2013) suggest that Msp-type porins have no involvement in the high-level GTA resistance phenotype of the Brazilian outbreak isolates, in contrast to the situation in GTA-resistant M. smegmatis and M. chelonae laboratory strains and clinical isolates described earlier (Svetlikova et al., 2009).
The significant increase in virulence of the mmpA and mmpB knock-out mutants in murine macrophages and in SCID mice indicates that the primary impact of alterations in surface porin expression may be on the interactions of MABSC with the host as has been reported in a number of pathogenic bacteria (Achouak et al., 2001; Skurnik et al., 2013; Geisinger and Isberg, 2017; Lee et al., 2017). The fact that the mmpB knock-out displays the greatest intracellular replication rate and increase in pathogenicity of the two mutants, yet is the one whose ability to import glucose is the least affected, indicates that this increase in virulence is independent from channel activity and, instead, points to the differential impact of expressing surface MmpB vs. surface MmpA on MABSC’s interactions with the host.
The increased virulence of the mmpB and mmpA mutants did not correlate with increased uptake by macrophages, cytotoxicity toward infected macrophages or enhanced resistance to nitrosative stress suggestive of the involvement of other virulence mechanisms whose determination will be the object of future studies. Based on what is known of the roles of porins in the virulence of other bacterial pathogens, including CF pathogens, other contributing factors may include decreased susceptibility of the Mmas porin mutants to killing by antimicrobial peptides and other host-derived bactericidal compounds, altered interactions with components of the complement cascades, and alterations in their interactions with other cell types than macrophages not analyzed herein (e.g., epithelial cells) (Achouak et al., 2001; Sharbati-Tehrani et al., 2005; Fabrino et al., 2009; Purdy et al., 2009; Skurnik et al., 2013; Lee et al., 2017). Potential changes in the transcription of other, as yet unidentified, genes resulting from the loss of porin expression as reported in carbapenem-resistant Pseudomonas aeruginosa oprD mutants (Skurnik et al., 2013), could further contribute to these virulence-associated phenotypic changes.
Consistent with porin content playing a role in the ability of MABSC to evade the host’s defenses, we observed significant DNA rearrangements affecting the mmpA/mmpB locus of some of the 1,382 MABSC clinical isolates analyzed in this study. That these DNA rearrangements reflect an attempt of these isolates to overcome antibiotic treatment is unlikely given the lack of significant changes in the antibiotic susceptibility profile of Mmas CIP108297 following the disruption of mmpA or mmpB.
The predominant changes observed longitudinally within CF patients were changes in the number of mmpA/mmpB porin genes. However, where short indels were observed longitudinally, they were all found to occur in mmpA, which could suggest that the phenotypic effects of the mmpA knock-out are specifically under selection in the context of adaption to the CF lung environment. The impact on virulence of fusing the mmpA and mmpB genes observed in a number of clinical isolates is impossible to predict without dedicated experimentation but is expected to vary with the point of fusion since early fusion points will result in strains expressing mostly an MmpB-type porin and late fusion point will result, on the contrary, in strains expressing mostly a MmpA-like porin. With the data available, we found no evidence to suggest that a particular mmpA/mmpB fusion was being selected for, neither within our global collection of MABSC isolates nor in within-patient longitudinal isolates. Finally, we found no evidence that the porin locus was playing a role in the adaptation of clustered MABSC lineages to increased virulence or transmission as we observed no significant association between the number of mmp-like porin genes encoded at the mmpA/mmpB porin locus and whether an isolate had been classified as clustered or unclustered (data not shown) (Bryant et al., 2016). However, 60% of the isolates that encoded only one mmpA/mmpB porin gene (discounting the isolates from Brazil as these are all associated with a single outbreak), occurred in just three of the clusters defined by Bryant et al. (2016, Figure 2), which suggests that, whilst porin genes may not be playing a role in the increased virulence/transmission of all clustered isolates, they could be contributing to the adaptation of particular lineages.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by Animal Care and Usage Committee of Colorado State University.
VM, MJ, DO, RF, JB, and JP conceived, designed experiments, and analyzed the data. VM generated the recombinant strains and characterized in vitro phenotypes. IE analyzed the predicted composition of the porin locus from the global population of MABSC isolates. DV, KB, CS, and MS performed the mouse and macrophage infection studies. MJ and DO wrote the manuscript. All authors reviewed and approved the manuscript.
This work was supported in part by the National Institutes of Health/National Institute of Allergy and Infectious Diseases grant AI089718 and a Strategic Research Centre Award (2017-SRC 010) from the United Kingdom Cystic Fibrosis Trust. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Graham Hatfull (University of Pittsburgh, Pennsylvania) for the generous gift of plasmid pJV53, and Dr. Michael Niederweis (University of Alabama at Birmingham) for the anti-MspA antibodies.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.706207/full#supplementary-material
Achouak, W., Heulin, T., and Pages, J.-M. (2001). Multiple facets of bacterial porins. FEMS Microbiol. Lett. 199, 1–7. doi: 10.1111/j.1574-6968.2001.tb10642.x
Adekambi, T., Reynaud-Gaubert, M., Greub, G., Gevaudan, M.-J., La Scola, B., Raoult, D., et al. (2004). Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J. Clin. Microbiol. 42, 5493–5501. doi: 10.1128/JCM.42.12.5493-5501.2004
Aitken, M. L., Limaye, A., Pottinger, P., Whimbey, E., Goss, C. H., Tonelli, M. R., et al. (2012). Respiratory outbreak of Mycobacterium abscessus subspecies massiliense in a lung transplant and cystic fibrosis center. Am. J. Respir. Crit. Care Med. 185, 231–232. doi: 10.1164/ajrccm.185.2.231
Bernut, A., Herrmann, J. L., Ordway, D., and Kremer, L. (2017). The diverse cellular and animal models to decipher the physiopathological traits of mycobacterium abscessus infection. Front. Cell Infect. Microbiol. 7:100. doi: 10.3389/fcimb.2017.00100
Bryant, J. M., Brown, K. P., Burbaud, S., Everall, I., Belardinelli, J. M., Rodriguez-Rincon, D., et al. (2021). Stepwise pathogenic evolution of Mycobacterium abscessus. Science 372:eabb8699. doi: 10.1126/science.abb8699
Bryant, J. M., Grogono, D. M., Greaves, D., Foweraker, J., Roddick, I., Inns, T., et al. (2013). Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381, 1551–1560. doi: 10.1016/S0140-6736(13)60632-7
Bryant, J. M., Grogono, D. M., Rodriguez-Rincon, D., Everall, I., Brown, K. P., Moreno, P., et al. (2016). Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science 354, 751–757. doi: 10.1126/science.aaf8156
Calado Nogueira de Moura, V., Gibbs, S., and Jackson, M. (2014). Gene replacement in Mycobacterium chelonae: application to the construction of porin knock-out mutants. PLoS One 9:e94951. doi: 10.1371/journal.pone.0094951
Chan, E. D., Bai, X., Kartalija, M., Orme, I. M., and Ordway, D. J. (2010). Host immune response to rapidly growing mycobacteria, an emerging cause of chronic lung disease. Am. J. Respir. Cell Mol. Biol. 43, 387–393. doi: 10.1165/rcmb.2009-0276TR
Danilchanka, O., Pavlenok, M., and Niederweis, M. (2008). Role of porins for uptake of antibiotics by Mycobacterium smegmatis. Antimicrob. Agents Chemother. 52, 3127–3134. doi: 10.1128/AAC.00239-08
Davidson, R. M., Hasan, N. A., de Moura, V. C., Duarte, R. S., Jackson, M., and Strong, M. (2013). Phylogenomics of Brazilian epidemic isolates of Mycobacterium abscessus subsp. bolletii reveals relationships of global outbreak strains. Infect. Genet. Evol. 20C, 292–297. doi: 10.1016/j.meegid.2013.09.012
Duarte, R. S., Lourenco, M. C. S., de Souza Fonseca, L. S., Leao, S. C., de Lourdes, T., Amorim, E., et al. (2009). An epidemic of postsurgical infections caused by Mycobacterium massiliense. J. Clin. Microbiol. 47, 2149–2155. doi: 10.1128/JCM.00027-09
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Everall, I., Lourenco Nogueira, C., Bryant, J. M., Sanchez-Buso, L., Chimara, E., Duarte, R. S., et al. (2017). Genomic epidemiology of a national outbreak of post-surgical Mycobacterium abscessus wound infections in Brazil. Microb. Genom. 3:e000111. doi: 10.1099/mgen.0.000111
Fabrino, D. L., Bleck, C. K. E., Anes, E., Hasilik, A., Melo, R. C. N., Niederweis, M., et al. (2009). Porins facilitate nitric oxide-mediated killing of mycobacteria. Microb. Infect. 11, 868–875. doi: 10.1016/j.micinf.2009.05.007
Floto, R. A., and Haworth, C. S. (2015). The growing threat of nontuberculous mycobacteria in CF. J. Cyst. Fibros 14, 1–2. doi: 10.1016/j.jcf.2014.12.002
Frenzel, E., Schmidt, S., Niederweis, M., and Steinhauer, K. (2011). Importance of porins for biocide efficacy against Mycobacterium smegmatis. Appl. Environ. Microbiol. 77, 3068–3073. doi: 10.1128/AEM.02492-10
Geisinger, E., and Isberg, R. R. (2017). Interplay between antibiotic resistance and virulence during disease promoted by multidrug-resistant bacteria. J. Infect. Dis. 215, S9–S17. doi: 10.1093/infdis/jiw402
Harris, S. R., Feil, E. J., Holden, M. T. G., Quail, M. A., Nickerson, E. K., Chantratita, N., et al. (2010). Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469–474. doi: 10.1126/science.1182395
Heinz, C., Roth, E., and Niederweis, M. (2003). “Purification of porins from Mycobacterium smegmatis,” in Methods in Molecular Biology – Membrane Protein Protocols: Expression, Purification and Characterization, ed. B. S. Selinsky (Totowa, NJ: Humana Press Inc), 139–150. doi: 10.1385/1-59259-400-X:139
Johansen, M. D., Herrmann, J. L., and Kremer, L. (2020). Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat. Rev. Microbiol. 18, 392–407. doi: 10.1038/s41579-020-0331-1
Leao, S. C., Viana-Niero, C., Matsumoto, C. K., Lima, K. V. B., Lopes, M. L., Palaci, M., et al. (2010). Epidemic of surgical-site infections by a single clone of rapidly growing mycobacteria in Brazil. Future Microbiol. 5, 971–980. doi: 10.2217/fmb.10.49
Lee, C. R., Lee, J. H., Park, M., Park, K. S., Bae, I. K., Kim, Y. B., et al. (2017). Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell Infect. Microbiol. 7:55. doi: 10.3389/fcimb.2017.00055
Li, H. (2013). Aligning sequence reads, clone sequneces and assembly contigs with BWA-MEM. arXiv [Preprint] arXiv:1303.3997,Google Scholar
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMTOOLS. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Martiniano, S. L., Nick, J. A., and Daley, C. L. (2019). Nontuberculous mycobacterial infections in cystic fibrosis. Thorac. Surg. Clin. 29, 95–108. doi: 10.1016/j.thorsurg.2018.09.008
Medjahed, H., and Reyrat, J.-M. (2009). Construction of Mycobacterium abscessus defined glycopeptidolipid mutants: comparison of genetic tools. Appl. Environ. Microbiol. 75, 1331–1338. doi: 10.1128/AEM.01914-08
Niederweis, M. (2003). Mycobacterial porins – new channel proteins in unique outer membranes. Mol. Microbiol. 49, 1167–1177. doi: 10.1046/j.1365-2958.2003.03662.x
Obregon-Henao, A., Arnett, K. A., Henao-Tamayo, M., Massoudi, L., Creissen, E., Andries, K., et al. (2015). Susceptibility of Mycobacterium abscessus to antimycobacterial drugs in preclinical models. Antimicrob. Agents Chemother. 59, 6904–6912. doi: 10.1128/AAC.00459-15
Page, A. J., Taylor, B., Delaney, A. J., Soares, J., Seemann, T., Keane, J. A., et al. (2016). SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genome 2:e000056. doi: 10.1099/mgen.0.000056
Park, I. K., and Olivier, K. N. (2015). Nontuberculous mycobacteria in cystic fibrosis and non-cystic fibrosis bronchiectasis. Semin. Respir. Crit. Care Med. 36, 217–224. doi: 10.1055/s-0035-1546751
Prevots, D. R., and Marras, T. K. (2015). Epidemiology of human pulmonary infection with nontuberculous mycobacteria: a review. Clin. Chest. Med. 36, 13–34. doi: 10.1016/j.ccm.2014.10.002
Purdy, G. E., Niederweis, M., and Russell, D. G. (2009). Decreased outer membrane permeability protects mycobacteria from killing by ubiquitin-derived peptides. Mol. Microbiol. 73, 844–857. doi: 10.1111/j.1365-2958.2009.06801.x
Ripoll, F., Pasek, S., Schenowitz, C., Dossat, C., Barbe, V., Rottman, M., et al. (2009). Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. doi: 10.1371/journal.pone.0005660
Rodrigues, L., Ramos, J., Couto, I., Amaral, L., and Viveiros, M. (2011). Ethidium bromide transport across Mycobacterium smegmatis cell-wall: correlation with antibiotic resistance. BMC Microbiol. 11:35. doi: 10.1186/1471-2180-11-35
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shang, S., Gibbs, S., Henao-Tamayo, M., Shanley, C. A., McDonnell, G., Duarte, R. S., et al. (2011). Increased virulence of an epidemic strain of Mycobacterium massiliense in mice. PLoS One 6:e24726. doi: 10.1371/journal.pone.0024726
Sharbati-Tehrani, S., Stephan, J., Holland, G., Appel, B., Niederweis, M., and Lewin, A. (2005). Porins limit the intracellular persistence of Mycobacterium smegmatis. Microbiology 151, 2403–2410. doi: 10.1099/mic.0.27969-0
Skurnik, D., Roux, D., Cattoir, V., Danilchanka, O., Lu, X., Yoder-Himes, D. R., et al. (2013). Enhanced in vivo fitness of carbapenem-resistant oprD mutants of Pseudomonas aeruginosa revealed through high-throughput sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 20747–20752. doi: 10.1073/pnas.1221552110
Stahl, C., Kubetzko, S., Kaps, I., Seeber, S., Engelhardt, H., and Niederweis, M. (2001). MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis. Mol. Microbiol. 40, 451–464. doi: 10.1046/j.1365-2958.2001.02394.x
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stephan, J., Bender, J., Wolschendorf, F., Hoffmann, C., Roth, E., Mailaender, C., et al. (2005). The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol. Microbiol. 58, 714–730. doi: 10.1111/j.1365-2958.2005.04878.x
Stephan, J., Mailaender, C., Etienne, G., Daffe, M., and Niederweis, M. (2004). Multidrug resistance of a porin deletion mutant of Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48, 4163–4170. doi: 10.1128/AAC.48.11.4163-4170.2004
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Svetlikova, Z., Skovierova, H., Niederweis, M., Gaillard, J. L., McDonnell, G., and Jackson, M. (2009). Role of porins in the susceptibility of Mycobacterium smegmatis and Mycobacterium chelonae to aldehyde-based disinfectants and drugs. Antimicrob. Agents Chemother. 53, 4015–4018. doi: 10.1128/AAC.00590-09
Tettelin, H., Davidson, R. M., Agrawal, S., Aitken, M. L., Shallom, S., Hasan, N. A., et al. (2014). High-level relatedness among Mycobacterium abscessus subsp. massiliense strains from widely separated outbreaks. Emerg. Infect. Dis. 20, 364–371. doi: 10.3201/eid2003.131106
Tettelin, H., Sampaio, E. P., Daugherty, S. C., Hine, E., Riley, D. R., Sadzewicz, L., et al. (2012). Genomic insights into the emerging human pathogen Mycobacterium massiliense. J. Bacteriol. 194:5450. doi: 10.1128/JB.01200-12
van Kessel, J. C., and Hatfull, G. F. (2007). Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152. doi: 10.1038/nmeth996
van Kessel, J. C., Marinelli, L. J., and Hatfull, G. F. (2008). Recombineering mycobacteria and their phages. Nat. Rev. Microbiol. 6, 851–857. doi: 10.1038/nrmicro2014
Keywords: Mycobacterium, abscessus, porin, virulence, antibiotic resistance
Citation: de Moura VCN, Verma D, Everall I, Brown KP, Belardinelli JM, Shanley C, Stapleton M, Parkhill J, Floto RA, Ordway DJ and Jackson M (2021) Increased Virulence of Outer Membrane Porin Mutants of Mycobacterium abscessus. Front. Microbiol. 12:706207. doi: 10.3389/fmicb.2021.706207
Received: 07 May 2021; Accepted: 23 June 2021;
Published: 14 July 2021.
Edited by:
Miguel Cacho Teixeira, University of Lisbon, PortugalReviewed by:
Dinesh Sriramulu, Independent Researcher, Chennai, IndiaCopyright © 2021 de Moura, Verma, Everall, Brown, Belardinelli, Shanley, Stapleton, Parkhill, Floto, Ordway and Jackson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mary Jackson, TWFyeS5KYWNrc29uQGNvbG9zdGF0ZS5lZHU=
†Present address: Vinicius C. N. de Moura, NTM Culture, Biorepository and Coordinating Core, National Jewish Health, Denver, Colorado, United States
‡These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.