- 1Institute of Soil and Fertilizer, Fujian Academy of Agricultural and Sciences, Fuzhou, China
- 2College of Resources and Environment, Fujian Agriculture and Forestry University, Fuzhou, China
- 3Faculty of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 4Institute of Forest Protection, Fujian Academy of Forestry Sciences, Fuzhou, China
Prodigiosin is a promising secondary metabolite produced mainly by Serratia strains. To study the global regulatory mechanism of prodigiosin biosynthesis, a mutagenesis library containing 23,000 mutant clones was constructed with the EZ-Tn5 transposon, and 114 clones in the library showed altered prodigiosin production ability. For 37 of the 114 clones, transposon insertion occurred on the prodigiosin biosynthetic cluster genes; transposon inserted genes of the 77 clones belonged to 33 different outside prodigiosin biosynthetic cluster genes. These 33 genes can be divided into transcription-regulating genes, membrane protein-encoding genes, and metabolism enzyme-encoding genes. Most of the genes were newly reported to be involved in prodigiosin production. Transcriptional levels of the pigA gene were significantly downregulated in 22 mutants with different inserted genes, which was in accordance with the phenotype of decreased prodigiosin production. Functional confirmation of the mutant genes involved in the pyrimidine nucleotide biosynthesis pathway was carried out by adding orotate and uridylate (UMP) into the medium. Gene complementation confirmed the regulatory function of the EnvZ/OmpR two-component regulatory system genes envZ and ompR in prodigiosin production.
Introduction
Prodigiosin is a pigment produced by strains of Serratia spp., Vibrio spp. (Borić et al., 2011), Streptomyces coelicolor (Liu et al., 2017), and other bacterial species. Prodigiosin has generated great interest in recent years due to its broad biological activities such as anticancer and antimicrobial properties (Bérdy, 2005). Research has mainly focused on three aspects: the optimization of the isolation and the production of prodigiosin-producing strains (Yip et al., 2019), the biological activity of prodigiosin (Li et al., 2018), and the synthesis and regulation mechanisms of prodigiosin in bacterial strains (Pan et al., 2019; Sun et al., 2020). The synthesis and regulation mechanism of prodigiosin are essential for its effective production and modification and the foundation for its further applications.
Genes in Serratia spp. and Streptomyces spp. have been identified, and these genes form a gene cluster in the genome (Williamson et al., 2006). In Serratia spp., the prodigiosin synthesis gene cluster arranged in the following order: pigA, pigB, pigC, pigD, pigE, pigF, pigG, pigH, pigI, pigJ, pigK, pigL, pigM, and pigN. Some of the synthesis genes are involved in 2-methyl-3-n-amyl-pyrrole (MAP) synthesis, some are involved in the production of 4-methoxy-2,2'-bipyrrole-5-carbaldehyde (MBC), and some are involved in the terminal step of prodigiosin synthesis with MAP and MBC (Williamson et al., 2006). Besides these 14 genes, Serratia sp. ATCC 39006 consisted of a gene pigO in its prodigiosin synthesis gene cluster, which was different with many Serratia spp. strains (Williamson et al., 2006).
Effective synthesis of prodigiosin is regulated by many genes in Serratia spp. Genes of the quorum-sensing and two-component regulatory systems are two main kinds of genes that were reported to play roles in regulating prodigiosin synthesis. SmaI/SmaR (Thomson et al., 2000; Slater et al., 2003; Fineran et al., 2005a) and SpnI/SpnR (Horng et al., 2002) are two types of quorum-sensing systems that have been reported to control prodigiosin production in Serratia spp. strains. PigQ/PigW (Fineran et al., 2005a), PhoB/PhoR (Gristwood et al., 2009), RssB/RssA (Horng et al., 2010), and EepR/EepS (Stella et al., 2015) are four kinds of two-component regulatory systems that can regulate the synthesis of prodigiosin. In addition, there are also many other transcription factors involved in prodigiosin synthesis including the positive regulators PigP (Fineran et al., 2005a), PigT (Fineran et al., 2005b), PigS (Gristwood et al., 2011), PigR, and PigV (Fineran et al., 2005a) and the negative regulators PigX (Fineran et al., 2007) and HexS (Tanikawa et al., 2006). Previous studies on the regulatory genes were mainly focused on Serratia sp. strain ATCC 39006 (Ravindran et al., 2019), other prodigiosin biosynthetic regulating genes in Serratia spp. were to be discovered. With Tn5 transposition system, a total of 365 genes that affect the prodigiosin synthesis levels of S. coelicolor were identified, including 17 genes in the prodigiosin biosynthetic gene cluster and many regulating genes (Xu et al., 2016), which showed Tn5 transposition system was a powerful tool for identifying genes involved in prodigiosin biosynthesis.
Serratia marcescens FZSF02 was previously isolated from soil and demonstrated to be a very promising strain for prodigiosin production (Lin et al., 2019). In this study, a mutant library was constructed to identify the regulatory genes for prodigiosin synthesis in this strain to enhance prodigiosin production through a genetic approach.
Materials and Methods
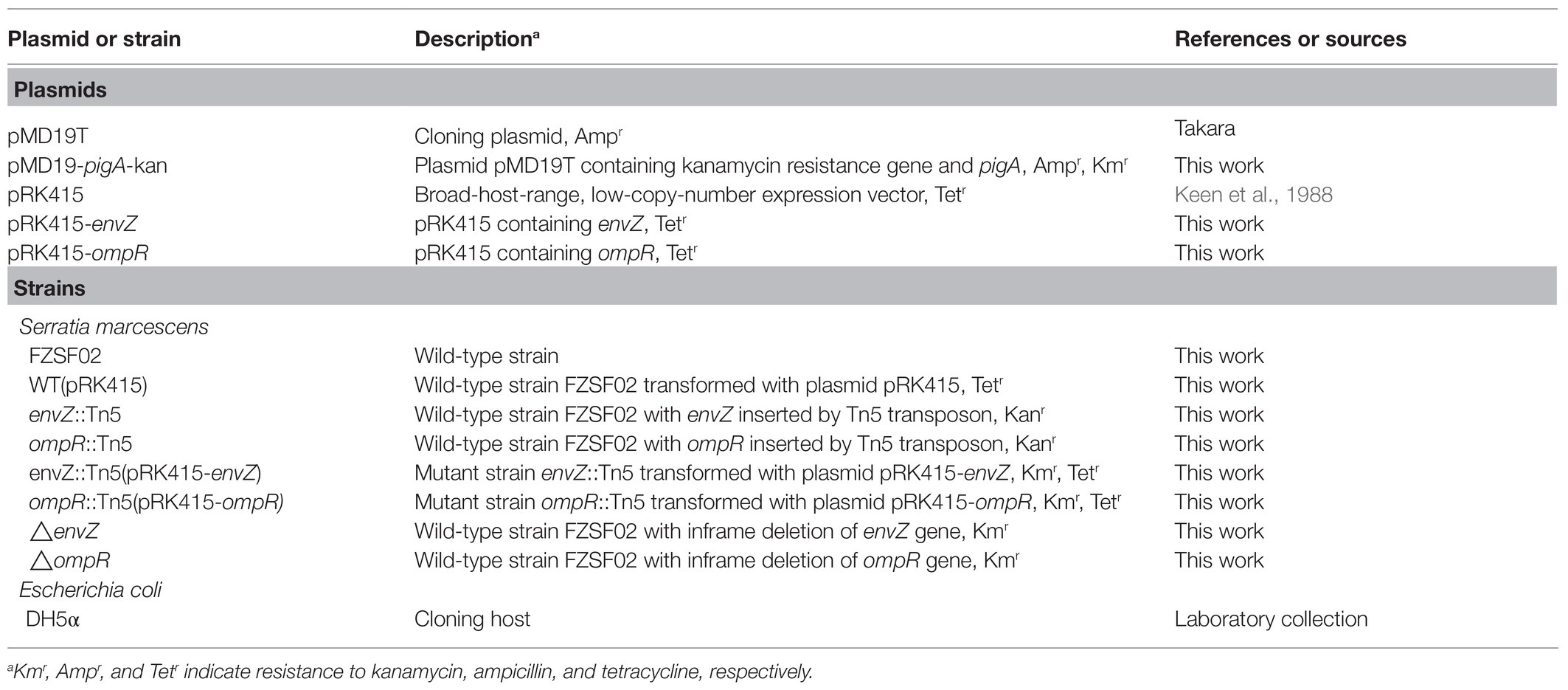
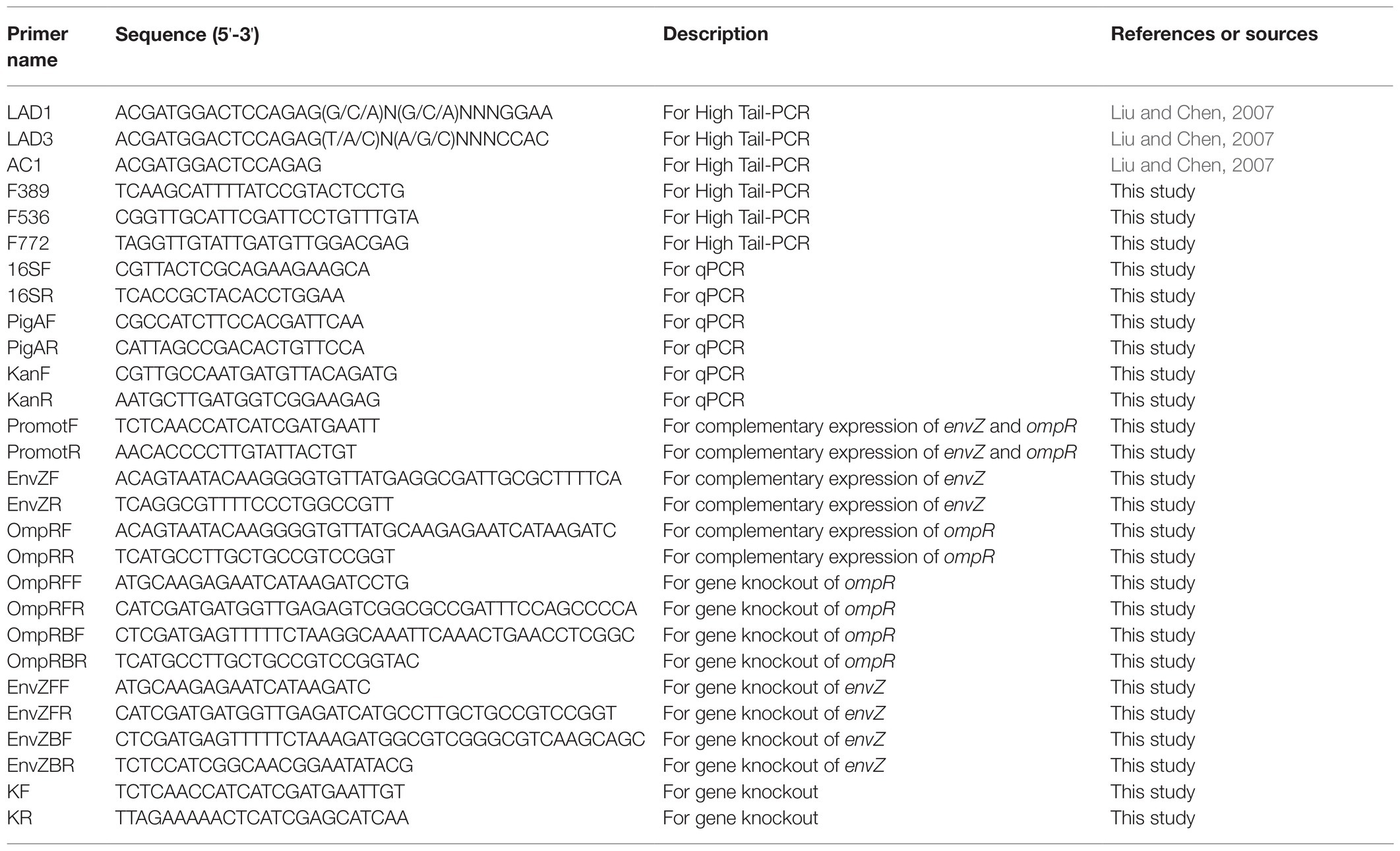
Plasmids, Primers, Strains, and Culture Conditions
The plasmids and strains used in this study are listed in Table 1. The primers used in this study are listed in Table 2. Serratia marcescens FZSF02 (GenBank accession number of the whole genome: CP053286.1) is a prodigiosin-producing strain that was used to construct the Tn5 mutant library. Escherichia coli DH5α was used to construct complementary plasmids of ompR and envZ. Serratia marcescens FZSF02 and the mutants were cultured with modified Luria-Bertani (LB) medium (tryptone (10 g/L), yeast extract (5 g/L), and NaCl (5 g/L)). Escherichia coli DH5α was cultured with Luria-Bertani (LB) medium. Antibiotics were used when necessary at the following concentrations: kanamycin (100 mg/L), ampicillin (100 mg/L), and tetracycline (50 mg/L).
Mutagenesis Library Construction
Competent cells for electroporation were prepared using the following method. The S. marcescens FZSF02 strain was cultured with 50 ml of LB medium in a flask (250 ml) at 37°C and 200 rpm. When the OD600 value reached approximately 1.0, the flask with the broth was incubated in an ice-water mixture for 30 min. Then, the broth was centrifuged in a 50-ml centrifuge tube at 5,000 rpm and 4°C for 10 min. After that, the supernatant was poured carefully, and the bacterial precipitate was resuspended in precooled sterilized ddH2O (0°C) and centrifuged at 5,000 rpm and 4°C for 10 min. These washing and centrifuging steps were repeated twice. The precipitate was then resuspended carefully with precooled 10% (v/v) glycerol and centrifuged at 5,000 rpm and 4°C for 10 min, and this was repeated twice. The bacterial precipitate was resuspended in 0.5 ml of precooled 10% glycerol and stored at −80°C with 100 μl in a 1.5-ml tube.
The EZ-Tn5™ <KAN-2>Tnp Transposome™ Kit (TSM99K2, Epicentre) was used to construct the mutagenesis library. Electronic pulse was operated with 0.1-cm electroporation cuvettes (Cat: 1652083, Bio-Rad) on the Gene Pulser Xcell™ (Bio-Rad) with the following electroporation parameters: 25 μF, 200 ohm, and 1800 V. After electronic pulse, the broth was immediately transferred into 1 ml of LB medium and incubated at 37°C and 200 rpm for 1 h. The incubated broth was diluted properly and screened on LB agar plates containing kanamycin (100 mg/L). Plates were incubated at 28°C until the color of the clones became red. Clones showing white or redder colors were selected and reserved for further research.
Copy Number of the Tn5 Transposon of the Mutant Strains
The copy number of the Tn5 transposon of the mutant strains was calculated with the Ct shift method (Kolacsek et al., 2017). In detail, PigA was chosen as the reference gene because there was only one copy of the pigA gene in the genome of S. marcescens FZSF02, and the Tn5 transposon copy number was calculated by assaying the copy number of the kanamycin resistance gene. A standard plasmid pMD19-pigA-kam containing both pigA and kam was constructed with pMD-19 T (Takara). Then, two qPCR standard curves were obtained using the primers (Table 2) of pigA and kam separately with the plasmid as the template. The Ct shift of the two curves at every point is a constant value. If there was only one Tn5 copy in the mutant strain genome, the Ct shift value obtained from the qPCR performed with the mutant strain genomic DNA as the template would be the same as the Ct shift value for the standard curves.
Identification of the Transposon Insertion Sites of the Mutants
High-tail PCR was used to identify the transposon insertion sites of the mutants (Liu and Chen, 2007; Jia et al., 2017). The primers used in this method are shown in Table 2.
Details of Plasmid and Strain Construction
The broad host range plasmid pRK415 was used to express the genes in mutant strains for genetic complementation. For expression, a promoter sequence (5'-TCTCAACCATCATCGATGAATTGTGTCTCAAAATCTCTGATGTTACATTGCACAAGATAAAAATATATCATCATGAACAATAAAACTGTCTGCTTACATAAACAGTAATACAAGGGGTGTT-3') was amplified with the primers PromotF and PromotR (Table 2). The ompR and envZ genes were amplified by PCR with the OmpRF/OmpRR and EnvZF/EnvZR primers. Overlapping PCR was used for the fusion of gene sequences with promoters. The PCR products were digested with the restriction endonucleases EcoR1 and HindIII and then ligated with pRK415, which was also digested with the restriction endonucleases EcoR1 and HindIII. The recombinant plasmids pRK415-envZ and pRK415-ompR were amplified in E. coli DH5α separately and then transformed into the mutant strains envZ::Tn5 and ompR::Tn5 separately by the electronic pulse method.
The homologous recombination method was used to inactivate genes. Briefly, the forward homologous sequence of about 300 bp was amplified with primers OmpRFF and OmpRFR, the backward homologous sequence of about 300 bp was amplified with primers OmpRBF and OmpRBR; kanamycin resistant gene was amplified with primers KF and KR; these three sequences were spliced by Overlap-PCR. The PCR products were transformed into S. marcescens FZSF02 for the homologous recombination deletion of ompR. Positive clones were screened with LB agar plates with kanamycin firstly and confirmed with PCR method. The gene envZ was also partially deleted with the same procedure.
Measurement of Prodigiosin Production and Bacterial Growth
Prodigiosin was extracted as follows. The fermentation broth was properly diluted with acidified methanol (4 ml of 1 mol/L HCl and 96 ml of methanol) and shocked vigorously; after 30 min of standing, the mixture was centrifuged at 10,000 × g for 10 min, and the supernatant was removed for prodigiosin quantification. Prodigiosin production can be evaluated by the absorbance value at 535 nm. The growth conditions of the strains can be assayed by comparing the OD600 values of the fermentation broth of the strains.
Quantitative Real-Time Polymerase Chain Reaction
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to evaluate the expression levels of pigA in the mutants. To obtain the total RNA of the mutants, the strains were incubated in 50 ml LB liquid medium at 28°C and 180 rpm for 18 h. Total RNA of each strain was extracted with an RNA extraction kit (DP430; Tiangen, Beijing, China); a PrimeScript™ RT reagent kit with the gDNA Eraser (RR047; TaKaRa, Dalian, China) was used to prepare cDNA with total RNA. 16S rDNA was chosen as the reference gene with the 16SF and 16SR primers (Table 2). The expression levels of pigA were chosen to reflect the expression levels of genes of the whole prodigiosin biosynthesis cluster with the pigAF and pigAR primers (Table 2). qRT-PCR was performed with SuperReal PreMix Plus (SYBR Green; Tiangen, Beijing, China) on a QuantStudio 6 Real-Time PCR System (Applied Biosystems) using the following primers: 16S rDNA, 16SF, and 16SR; pigA, pigAF, and pigAR (Table 2). Gene expression levels of mutant strains were determined using the 2-ΔΔCT method with the relative fold-difference expressed against the wild-type strain.
Results
EZ-Tn5-Mediated Mutation Library Construction of S. marcescens FZSF02
We constructed two mutation libraries with the EZ-Tn5 transposon and obtained approximately 23,000 mutant clones. The probability that the transposon inserted one certain gene was calculated with the formula P = 1 − (1 − X/G)n, where P is the property that one gene was found to be inserted by the transposon, X is the average length of the genes of the studied strain (1,800 bp for the S. marcescens FZSF02 strain used in this study), G is the genome length of the studied strain (5.6 × 106 bp for the S. marcescens FZSF02 strain), and n is the number of mutant clones obtained in the mutation library (Krysan et al., 1999; Song et al., 2015a). We calculated that the property one certain gene was inserted was above 99% in this study.
We tested 10 mutants with the Ct shift method and confirmed that each mutant had one copy of the Tn5 transposon in the genome (Supplementary Figure S4 and Supplementary Table S3 in Supplementary Material).
Mutants With Altered Prodigiosin Production Ability in the Library
A total of 114 clones showed altered prodigiosin-producing ability. Thirty-seven mutants were found in the prodigiosin biosynthetic cluster genes, which are pigA, pigB, pigC, pigD, pigE, pigF, pigH, pigI, pigL, and pigM. The mutant number and the transposon insertion sites of each mutant are shown in Supplementary Table S1 in Supplementary Material. All the mutants with the prodigiosin biosynthetic cluster genes inserted by the Tn5 transposon lost prodigiosin-producing ability.
Seventy-seven mutants (Supplementary Table S2 in Supplementary Material) showed altered prodigiosin-producing ability, which involved 33 outside prodigiosin biosynthetic cluster genes (Table 3). Fourteen mutants were found to have the transposon insertion of the Tn5 transposon in the gene encoding dihydroorotate dehydrogenase (quinone), seven mutants had the gene encoding the type VI secretion protein, and four clones had the genes encoding CpxA, osmolarity response regulator/DNA-binding response regulator OmpR, transcriptional regulator SlyA, long-chain fatty acid outer membrane, and DEAD/DEAH box helicase. For other genes, the number of mutants was 1 to 3. The name of mutants and the transposon insertion sites of these genes are shown in the Supplementary Table S2 in Supplementary Material.
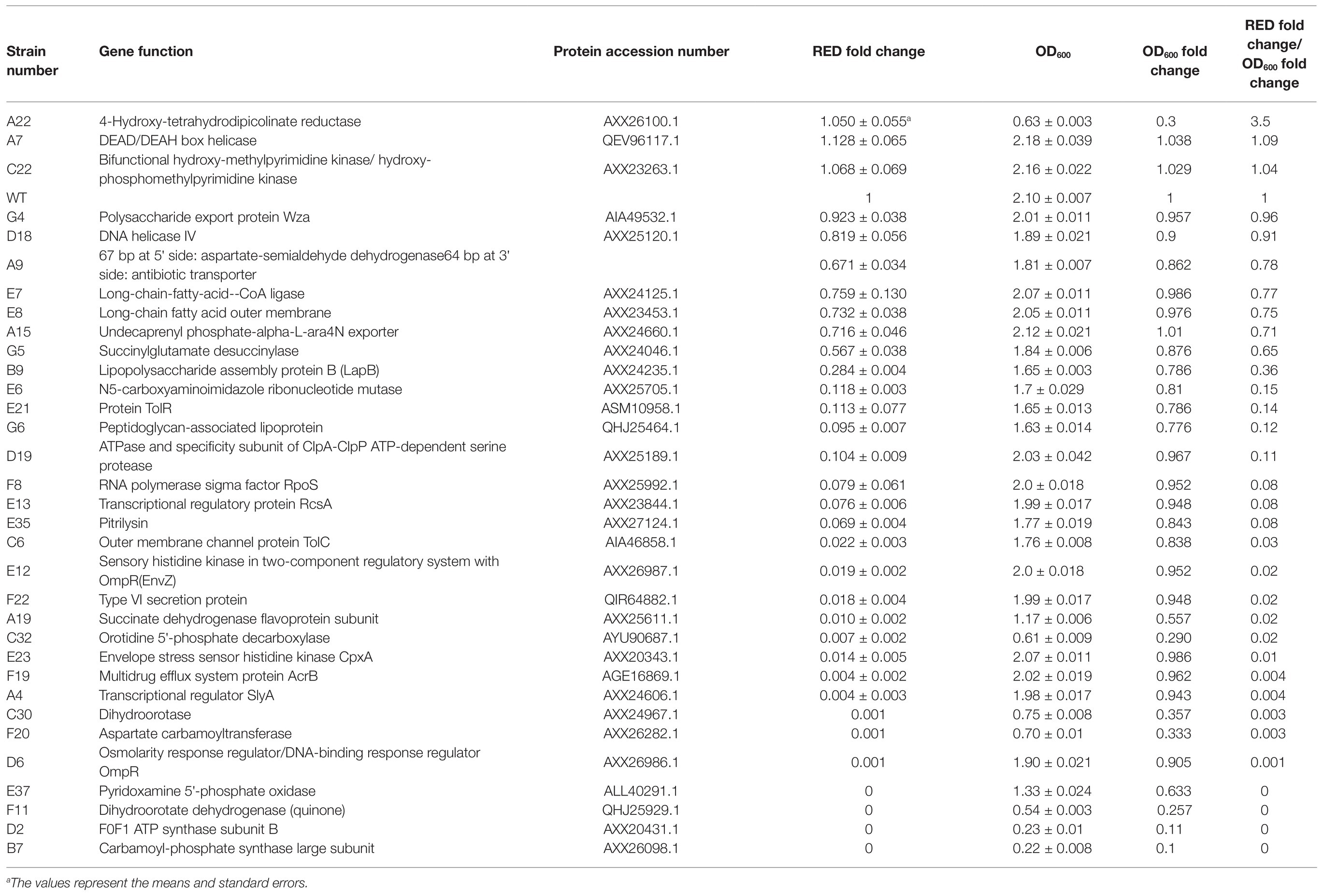
Table 3. Characterization of the Tn5 mutants of Serratia marcescens FZSF02 with altered prodigiosin production.
To test the effects of transposon insertion, prodigiosin producing ability and growth ability of 33 strains that represented 33 outside prodigiosin biosynthetic cluster genes were assayed. After incubation in liquid LB medium at 28°C for 48 h, prodigiosin production as OD535 nm value and the growth of culture as OD600 nm of each mutant strain were assayed. The OD535 nm value and OD600 value ratio of the mutant strains to the wild-type strain reflected the prodigiosin-producing ability and the growth conditions of the mutant strains, respectively. The ratio of the prodigiosin fold change to the OD600 fold change in each mutant strain reflected prodigiosin production per unit biomass of this strain (Table 3). Among these outside prodigiosin biosynthetic cluster genes, the transposon insertion of three genes encoding the DEAD/DEAH box helicase, bifunctional hydroxy-methylpyrimidine kinase/hydroxy-phosphomethylpyrimidine kinase, and 4-hydroxy-tetrahydrodipicolinate reductase led to an increase in prodigiosin production. Mutant strains with other genes inserted by transposon showed decreased prodigiosin production to varying degrees (Table 3). Some mutants showed a lower prodigiosin-producing ability than the wild-type strain by more than 5-fold, but the effects on growth were much less than the prodigiosin-producing ability. These mutants involved genes encoding sensor protein CpxA (AXX26828.1), ATPase and specificity subunit of ClpA-ClpP ATP-dependent serine protease (AXX25189.1), multidrug efflux system protein AcrB (AGE16869.1), sensory histidine kinase EnvZ in two-component regulatory system (AXX26987.1), RNA polymerase sigma factor RpoS (AXX25992.1), type VI secretion protein (QIR64882.1), transcriptional regulatory protein RcsA (AXX23844.1), transcriptional regulator SlyA (AXX24606.1), osmolarity response regulator/DNA-binding response regulator OmpR (AXX26986.1), pitrilysin (AXX27124.1), outer membrane channel protein TolC (AIA46858.1), N5-carboxyaminoimidazole ribonucleotide mutase (AXX25705.1), protein TolR (ASM10958.1), and peptidoglycan-associated lipoprotein (QHJ25464.1). Some mutants showed a much lower prodigiosin-producing ability along with a significantly decreased growth ability, which involved genes encoding pyridoxamine 5'-phosphate oxidase (ALL40291.1), succinate dehydrogenase (SDH) flavoprotein subunit (AXX25611.1), dihydroorotase (AXX24967.1), aspartate carbamoyltransferase (AXX26282.1), orotidine 5'-phosphate decarboxylase (AYU90687.1), dihydroorotate dehydrogenase (quinone) (QHJ25929.1), F0F1 ATP synthase subunit B (AXX20431.1), and carbamoyl-phosphate synthase large subunit (AXX26098.1).
Transcriptional levels of the pigA gene in 32 mutants were assayed with qPCR. Every selected mutant had an outside prodigiosin biosynthetic cluster gene that was different from other mutants. The transcriptional level of pigA can reflect the transcriptional level of the whole prodigiosin biosynthetic cluster. Transcriptional levels of pigA in 22 mutants were significantly downregulated (Figure 1), and all these mutants showed a much lower prodigiosin-producing ability (Table 3). Mutants A7, C22, and A22 with enhanced prodigiosin production ability showed upregulated pigA transcriptional levels (Figure 1), indicating they might negatively regulate prodigiosin in this strain.
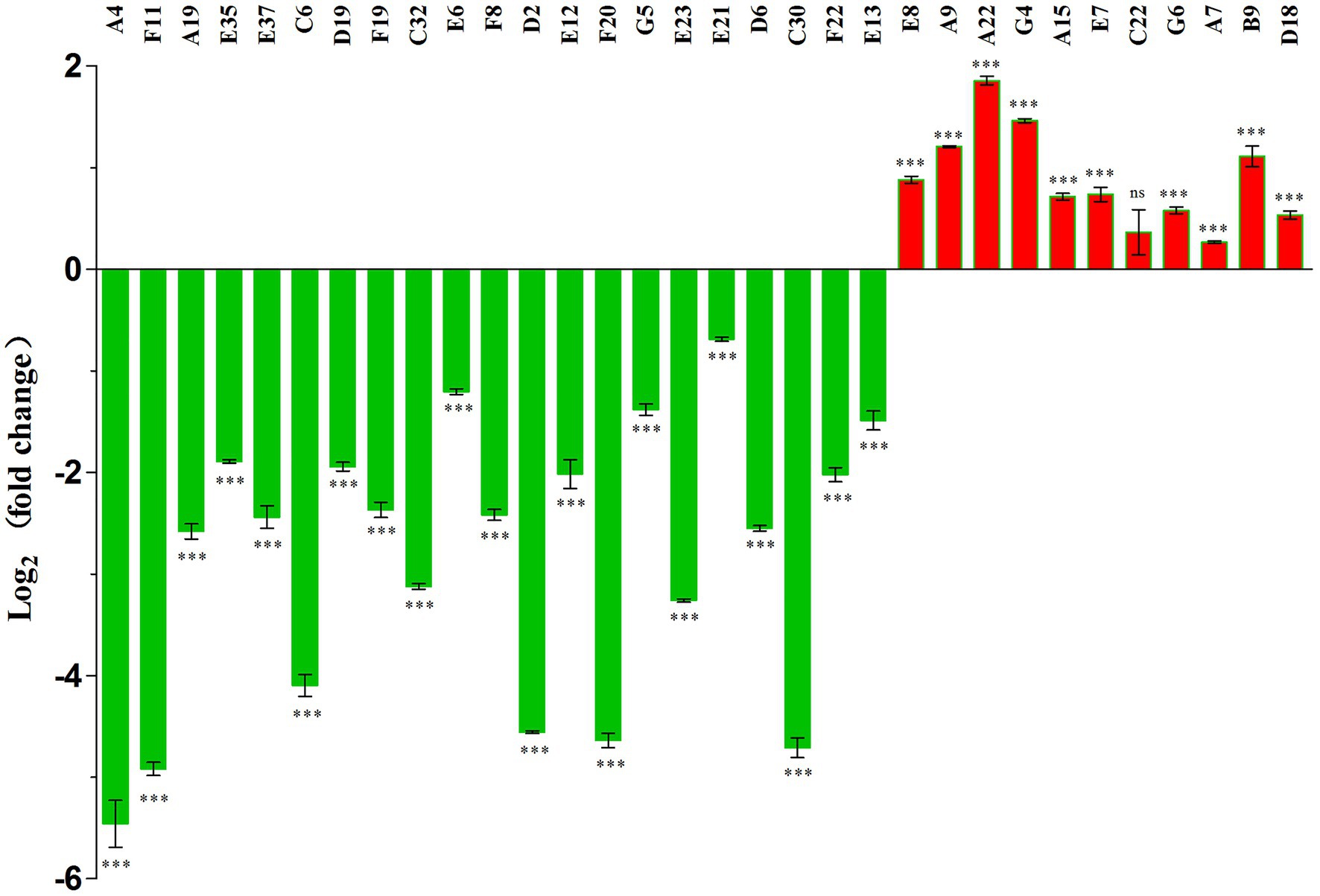
Figure 1. Evaluation of the pigA expression level in 32 mutant strains with different genes inserted by the Tn5 transposon. Numbers from A4 to D18 in the above figure represent the mutants chosen for qPCR. Means ± SDs from three independent experiments are shown. Significant differences by unpaired t-test between the mutant strains and wild type strain are shown as follows: ***p < 0.001, and ns, no significance.
Functional Confirmation of the Mutant Genes
Mutants with pyrimidine nucleotide biosynthesis pathway genes showed a significantly decreased production of prodigiosin. Of these mutants, the transposon insertion of four genes encoding carbamoyl-phosphate synthase, aspartate carbamoyltransferase, dihydroorotase, and dihydroorotate dehydrogenase blocks the biosynthesis of orotate and uridylate (UMP); transposon insertion of a gene encoding orotidine 5'-phosphate decarboxylase will lead to biosynthesis blockade of UMP only (Figure 2A). When orotate or UMP was added to the LB agar plates, mutant B7 with the inserted gene encoding carbamoyl-phosphate synthase, F20 with the inserted gene encoding aspartate carbamoyltransferase, C30 with the inserted gene encoding dihydroorotase, and E29 with the inserted gene encoding dihydroorotate dehydrogenase showed improved prodigiosin-producing ability to a certain extent compared with the treatments with no orotate or UMP addition (Figures 2B,C). Mutant strain C32 with the inserted gene encoding orotidine 5'-phosphate decarboxylase showed significantly enhanced prodigiosin-producing ability when UMP was added to the medium but no enhanced effects was observed when orotate was added (Figures 2B,C). These results demonstrated that these inserted genes are essential for S. marcescens FZSF02 in the production of prodigiosin.
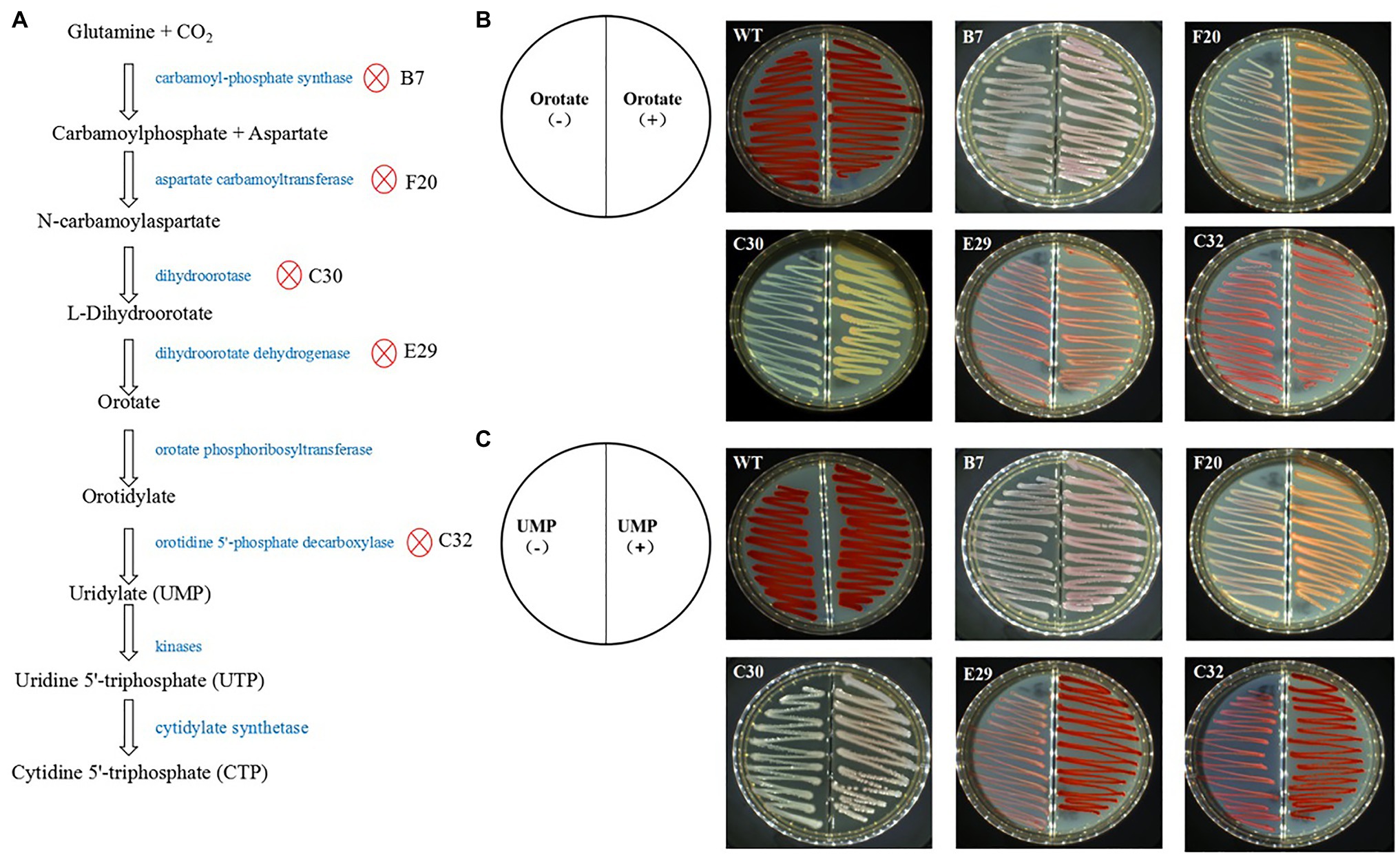
Figure 2. Functional certification of prodigiosin production in the mutant genes involved in pyrimidine nucleotide biosynthesis. (A) Proteins of the pyrimidine nucleotide biosynthesis pathway and the proteins, in which the encoding genes were inserted by the Tn5 transposon. (B) Rescue effect on prodigiosin production by adding orotate into the medium. The prodigiosin-producing ability can be partially rescued for B7, F20, C30, and E29 but not for C32. (C) The rescue effect on the prodigiosin production of UMP for the mutant strains. Prodigiosin producing ability can be partially rescued for B7, F20, C30, E29, and C32. The concentration of orotate and UMP was 0.1 g/L; the incubation conditions of the strains were 28°C for 36 h.
Transposon insertions of envZ and ompR, two genes of the EnvZ/OmpR two-component regulatory system, led to the loss of the prodigiosin-producing ability for mutants E12, D6, F12, E26, and D9. The in-frame deletion of either envZ or ompR led to loss of the prodigiosin-producing ability (Figures 3A,B). When gene complementation was carried out by transforming pRK415-envZ in mutant E12 and pRK415-ompR in strain D6, the prodigiosin-producing abilities were restored to a certain extent (about 21.4% for E12 and 18.6% for D6 compared with WT strain; Figures 3A–C).
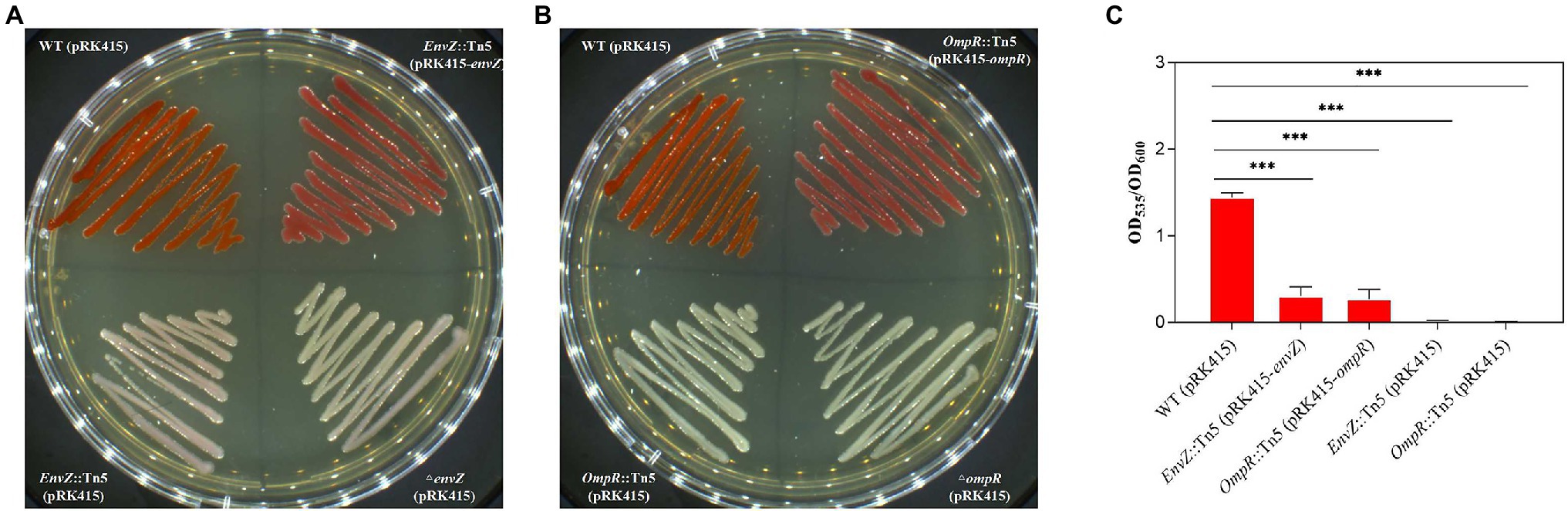
Figure 3. Functional certification of EnvZ and OmpR on prodigiosin-producing ability by gene knockout and complementation. (A) The complementary strain EnvZ::Tn5 (pRK415-envZ) was constructed by transferring the pRK415-envZ plasmid with a tetracycline-resistant gene into the mutant envZ::Tn5 strain. The wild-type (WT) strain, Tn5-inserted envZ::Tn5 mutant and gene knockout ΔenvZ strain were transfected with pRK415 to confer tetracycline resistance and used as controls. (B) The complementary ompR::Tn5 (pRK415-ompR) was constructed by transferring pRK415-ompR into the ompR::Tn5 strain. The other three strains were similar to those described in (A). (C) Prodigiosin producing ability of WT (pRK415) strain, complementary strain envZ::Tn5 (pRK415-envZ), complementary strain ompR::Tn5 (pRK415-ompR), mutant strain envZ::Tn5 (pRK415), and ompR::Tn5 (pRK415). Means ± SDs from three independent experiments are shown. Significant differences in prodigiosin producing ability relative to the related strains/WT by unpaired t-test are shown as follows: ***p < 0.001. All the strains were incubated on LB agar with tetracycline (50 mg/L) at 28°C for 36 h.
Discussion
In this study, we constructed a transposon mutant library with the EZ-Tn5™ <KAN-2>Tnp Transposome™ Kit and obtained 114 clones that showed altered prodigiosin-producing ability involving prodigiosin biosynthetic cluster genes and outside prodigiosin biosynthetic cluster genes. Prodigiosin biosynthetic cluster genes identified in this study were very similar to other reported S. marcescens strains either on the nucleotide sequences or on the gene orders (GenBank accession number of the whole genome of FZSF02: CP053286.1). However, most of the genes that play regulatory roles in prodigiosin biosynthesis in this study have not been reported earlier. There were two types of prodigiosin biosynthetic clusters in Serratia spp., the Serratia 39006 type and the Serratia 274 type (Harris et al., 2004; Williamson et al., 2006); prodigiosin biosynthetic cluster of S. marcescens FZSF02 belonged to the Serratia 274 type (Supplementary Figure S1 in Supplementary Material), but promoter sequences of FZSF02 prodigiosin biosynthetic cluster showed some difference from that of Serratia sp. ATCC 274 (Supplementary Figures S2, S3 in Supplementary Material). There are fewer studies on genes that regulate prodigiosin biosynthesis of Serratia 274 strains than of Serratia 39006 strains, and only luxS (Coulthurst et al., 2004), rpoS (Qin et al., 2020), slyA (Pan et al., 2020), and cpxA (Sun et al., 2020) have been previously reported.
Two-component regulatory systems played an important role in regulating prodigiosin biosynthesis. In this study, cpxA of the CpxA/CpxR two-component regulatory system and envZ and ompR of the EnvZ/OmpR two-component regulatory system were two-component regulatory system genes, and the transposon insertion of these three genes with the Tn5 transposon led to significantly decreased prodigiosin production (72.7-fold for cpxA, 54-fold for envZ and 839.3-fold for ompR). It was confirmed that in S. marcescens JNB 5-1, CpxR negatively regulated the transcription of the cluster by binding to the promoter of the prodigiosin cluster, and CpxA negatively regulated the expression of cpxR; therefore, the cpxA mutant showed decreased prodigiosin production (Sun et al., 2020). Significantly decreased prodigiosin production was also found in the CpxA transposon insertion mutant, the regulatory mechanism of which might be similar to that of the S. marcescens JNB 5-1. EnvZ and ompR positively regulated prodigiosin production, as confirmed by both the transcription levels (Figure 1) and production levels (Figure 3; Table 3). The EnvZ/OmpR two-component regulatory system was reported to regulate various properties of bacteria, such as phase variation, osmotic tolerance, and motility virulence (Tipton and Rather, 2017), but there are no reports about their relationship with prodigiosin biosynthesis. EnvZ and ompR are two newly confirmed genes that regulate prodigiosin biosynthesis, and the regulatory mechanisms are still unknown.
In addition to two-component regulatory system genes, some other transcription-regulating genes were also found to regulate prodigiosin production in this study, including rcsA, which encodes the transcriptional regulatory protein RcsA, slyA, which encodes the transcriptional regulator SlyA, and rpoS, which encodes the RNA polymerase sigma factor RpoS. Transposon insertions of rcsA, slyA, and rpos showed decreased prodigiosin production (13.1-, 246.2-, and 12.6-fold, respectively) compared with that of the wild-type strain, and the pigA transcriptional levels of the three mutants were all downregulated to varying degrees. RpoS was reported to positively regulate prodigiosin production in S. marcescens 1912768R and negatively regulate prodigiosin production in Serratia sp. ATCC 39006 (Wilf and Salmond, 2012; Qin et al., 2020). SlyA has been confirmed to positively regulate prodigiosin biosynthesis in S. marcescens JNB5-1, and the transcription of slyA itself was negatively controlled by MetR (Pan et al., 2020). More details on the regulation of prodigiosin by slyA are still unknown. RcsA was a positive regulatory gene of prodigiosin production in this study, which was accordance with the recent study in S. marcescens JNB5-1 (Pan et al., 2021).
Some mutants with genes encoding membrane proteins inserted showed significantly decreased prodigiosin production. These genes included acrB, which encodes the multidrug efflux system protein AcrB, tolC, which encodes the outer membrane channel protein TolC, tolR, which encodes the protein TolR, pal, which encodes the peptidoglycan-associated lipoprotein, tai4, which encodes the type VI secretion protein Tai4, and lapB, which encodes lipopolysaccharide assembly protein B (LapB). Transposon insertions of acrB and tolC, which encode two membrane channel proteins, both showed significantly decreased prodigiosin production (249.5- and 45.9-fold, respectively). The AcrAB-TolC efflux pump system is well-known for its role in the multidrug resistance of bacteria (Abdel-Halim et al., 2019; Nolivos et al., 2019). However, no clues can be obtained to explain the potential regulatory mechanism of these two genes for prodigiosin production from current studies. TolR and pal are two genes of the Tol-Pal system, and the transposon insertion of tolR and pal showed 8.8- and 10.6-fold decreased prodigiosin production, respectively. The Tol-Pal system plays important roles in cell division and phospholipid transport from the membrane of bacteria (Szczepaniak et al., 2020). It was found that different kinds of oils can significantly enhance prodigiosin production of many strains in the Serratia genus (Wei and Chen, 2005; Abdul Manas et al., 2020), including the S. marcescens FZSF02 strain, which was used in this study (Lin et al., 2019). The transposon insertions of tolR and pal may obstruct liquid usability for prodigiosin biosynthesis. The transposon insertion of lapB resulted in decreased prodigiosin production (3.5-fold). LapB coordinates the assembly of proteins involved in LPS synthesis at the plasma membrane in E. coli (Klein et al., 2014), but its function in S. marcescens, especially in prodigiosin biosynthesis, needs to be studied further. The transposon insertion of tai4 decreased prodigiosin (54.8-fold). Tai4 (a type VI secretion protein) is a compound of the type VI secretion system (T6SS). T6SS is a novel multi-subunit needle-like secretion apparatus and plays multiple roles in processes of bacterial life cycles such as interspecies competition, biofilm formation, and virulence-related processes (Zhang et al., 2013). The tai4 gene was also a newly found gene involved in prodigiosin production in this study.
Some genes encoding metabolic enzymes and other enzymes were also found to affect prodigiosin biosynthesis. The transposon insertion of genes encoding dihydroorotase, aspartate carbamoyltransferase, orotidine 5'-phosphate decarboxylase, dihydroorotate dehydrogenase (quinone), and carbamoyl-phosphate synthase large subunit led to a decrease in prodigiosin by more than 100-fold. These five genes are involved in cytidine 5'-triphosphate (CTP) biosynthesis, and CTP or its metabolic intermediates may be the material for prodigiosin biosynthesis. When atpF, which encodes F0F1 ATP synthase subunit B, was inserted, the mutant strain lost the prodigiosin-producing ability and showed decreased growth ability. Mutating atpF led to a decrease in ATP supply, and prodigiosin biosynthesis might be an ATP consumption process. This can also be demonstrated to a certain extent by an earlier study showing that ATP enhanced prodigiosin production in S. marcescens (Haddix et al., 2008). The transposon insertion of ptrA, which encodes pitrilysin, and clpA, which encodes ClpA-ClpP ATP-dependent serine protease chaperone ClpA, both led to decreased prodigiosin production (14.5-fold and 9.6-fold, respectively) of the strains. Pitrilysin (EC 3.4.24.55) is a kind of metalloendopeptidase; pitrilysin can cleave the -Tyr16−/−Leu-, -Phe25−/−Tyr-bonds of the oxidized insulin B chain and acts on other substrates of less than 7 kDa such as insulin and glucagon (Finch et al., 1986; Ding et al., 1992; Anastasi et al., 1993). ptrA is located between recB and recC on the genomes of both E. coli (such as the K12 strain) and S. marcescens FZSF02, which was used in this study; however, its biological functions, including how it regulates prodigiosin biosynthesis in bacteria, are still unclear., ClpA is a part of the ClpA-ClpP ATP-dependent serine protease complex (Hoskins et al., 2001), which plays an important role in refolding and degrading proteins, an essential process for the viability and growth of cells (Song et al., 2015b). Lipopeptide biosynthesis in Pseudomonas fluorescens is regulated by ClpAP via the pathway-specific LuxR-type regulator MassAR, the heat shock proteins DnaK and DnaJ, and proteins of the TCA cycle (Brugarolas et al., 2011). ClpA may also indirectly regulate prodigiosin of S. marcescens FZSF02 by regulating other unknown pathways. The N5-carboxyaminoimidazole ribonucleotide mutase encoded by the purE gene is a key enzyme for the biosynthesis of inosine monophosphate (IMP), the common precursor of AMP and GMP (Brugarolas et al., 2011). The transposon insertion of purE may cause a decreased supply of purines and ATP; the lack of purines and ATP may lead to decreased prodigiosin production (8.5-fold decrease in this study) and biomass. Pyridoxamine 5'-phosphate oxidase encoded by the gene pdxH participates in the salvage pathway of pyridoxal 5'-phosphate, an important coenzyme that participates in many metabolic reactions. The transposon insertion of pdxH of S. marcescens FZSF02 showed decreased growth ability (OD600 value of 1.33) compared with that of the wild-type strain (OD600 value of 2.1; Table 3), which was in accordance with the previous finding that pdxH mutants of E. coli showed poorer growth (Lam and Winkler, 1992; Ito and Downs, 2020). Meanwhile, the transposon insertion of pdxH resulted in the loss of the prodigiosin-producing ability for S. marcescens FZSF02, but the mechanism of which remains unknown. SdhA encodes the SDH flavoprotein subunit, a key subunit of SDH that converts succinate to fumarate during the tricarboxylic acid cycle. SdhE is necessary for the incorporation of FAD into SdhA in S. marcescens (McNeil et al., 2012) and the mutation of sdhE showed impaired growth (McNeil et al., 2012) and 50% reduction in prodigiosin (McNeil et al., 2012). The transposon insertion of sdhA of S. marcescens FZSF02 in this study led to loss of the prodigiosin production ability and impaired growth (OD600 value 1.17 vs. 2.1 of WT) (Table 3). Perhaps SdhE indirectly affects prodigiosin production by regulating the activity of SdhA; the regulatory details of SdhA on prodigiosin biosynthesis also need to be further studied.
In this study, we constructed a mutation library with strain S. marcescens FZSF02 harboring the 274-type prodigiosin cluster. We identified 33 genes involved in the regulation of prodigiosin production, 29 of which were first reported in this study. This study identified potential regulatory genes at the genome level. These genes will provide a better understanding of the regulatory details of the secondary metabolite prodigiosin; they might also be valuable targets of metabolism engineering for producing prodigiosin and other metabolites.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XJ and JC designed the study and wrote the manuscript. XJ, FL, KZ, JL, YF, SC, LC, HZ, and CL performed the experiments. XJ, FL, and JC analyzed the results. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Chinese National Natural Science Foundation (31800068), the Special Program for Extension Research of National Natural Science Foundation of Fujian Academy of Agricultural Sciences (AGY2018-1), the Finance Support of Fujian China (2018-No. 389), and the Youth Talent Free Exploration Project of Fujian Academy of Agricultural and Sciences (AA2018-25).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.705853/full#supplementary-material
References
Abdel-Halim, H., Al Dajani, A., Abdelhalim, A., and Abdelmalek, S. (2019). The search of potential inhibitors of the AcrAB-TolC system of multidrug-resistant Escherichia coli: an in silico approach. Appl. Microbiol. Biotechnol. 103, 6309–6318. doi: 10.1007/s00253-019-09954-1
Abdul Manas, N. H., Chong, L. Y., Tesfamariam, Y. M., Zulkharnain, A., Mahmud, H., Abang Mahmod, D. S., et al. (2020). Effects of oil substrate supplementation on production of prodigiosin by Serratia nematodiphila for dye-sensitized solar cell. J. Biotechnol. 317, 16–26. doi: 10.1016/j.jbiotec.2020.04.011
Anastasi, A., Knight, C. G., and Barrett, A. J. (1993). Characterization of the bacterial metalloendopeptidase pitrilysin by use of a continuous fluorescence assay. Biochem. J. 290, 601–607. doi: 10.1042/bj2900601
Borić, M., Danevčič, T., and Stopar, D. (2011). Prodigiosin from Vibrio sp. DSM 14379; a new UV-protective pigment. Microb. Ecol. 62, 528–536. doi: 10.1007/s00248-011-9857-0
Brugarolas, P., Duguid, E. M., Zhang, W., Poor, C. B., and He, C. (2011). Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus. Acta Crystallogr. D Biol. Crystallogr. 67, 707–715. doi: 10.1107/S0907444911023821
Coulthurst, S. J., Kurz, C. L., and Salmond, G. (2004). luxS mutants of Serratia defective in autoinducer-2-dependent 'quorum sensing' show strain-dependent impacts on virulence and production of carbapenem and prodigiosin. Microbiology 150, 1901–1910. doi: 10.1099/mic.0.26946-0
Ding, L., Becker, A. B., Suzuki, A., and Roth, R. A. (1992). Comparison of the enzymatic and biochemical properties of human insulin-degrading enzyme and Escherichia coli protease III. J. Biol. Chem. 267, 2414–2420. doi: 10.1016/S0021-9258(18)45895-4
Finch, P. W., Wilson, R. E., Brown, K., Hickson, I. D., and Emmerson, P. T. (1986). Complete nucleotide sequence of the Escherichia coli ptr gene encoding protease III. Nucleic Acids Res. 14, 7695–7703. doi: 10.1093/nar/14.19.7695
Fineran, P. C., Everson, L., Slater, H., and Salmond, G. (2005b). A GntR family transcriptional regulator (PigT) controls gluconate-mediated repression and defines a new, independent pathway for regulation of the tripyrrole antibiotic, prodigiosin, in Serratia. Microbiology 151, 3833–3845. doi: 10.1099/mic.0.28251-0
Fineran, P. C., Slater, H., Everson, L., Hughes, K., and Salmond, G. P. (2005a). Biosynthesis of tripyrrole and beta-lactam secondary metabolites in Serratia: integration of quorum sensing with multiple new regulatory components in the control of prodigiosin and carbapenem antibiotic production. Mol. Microbiol. 56, 1495–1517. doi: 10.1111/j.1365-2958.2005.04660.x
Fineran, P. C., Williamson, N. R., Lilley, K. S., and Salmond, G. P. (2007). Virulence and prodigiosin antibiotic biosynthesis in Serratia are regulated pleiotropically by the GGDEF/EAL domain protein, PigX. J. Bacteriol. 189, 7653–7662. doi: 10.1128/JB.00671-07
Gristwood, T., Fineran, P. C., Everson, L., Williamson, N. R., and Salmond, G. P. (2009). The PhoBR two-component system regulates antibiotic biosynthesis in Serratia in response to phosphate. BMC Microbiol. 9:112. doi: 10.1186/1471-2180-9-112
Gristwood, T., McNeil, M. B., Clulow, J. S., Salmond, G. P., and Fineran, P. C. (2011). PigS and PigP regulate prodigiosin biosynthesis in Serratia via differential control of divergent operons, which include predicted transporters of sulfur-containing molecules. J. Bacteriol. 193, 1076–1085. doi: 10.1128/JB.00352-10
Haddix, P. L., Jones, S., Patel, P., Burnham, S., Knights, K., Powell, J. N., et al. (2008). Kinetic analysis of growth rate, ATP, and pigmentation suggests an energy-spilling function for the pigment prodigiosin of Serratia marcescens. J. Bacteriol. 190, 7453–7463. doi: 10.1128/JB.00909-08
Harris, A., Williamson, N. R., Slater, H., Cox, A., Abbasi, S., Foulds, I., et al. (2004). The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology 150, 3547–3560. doi: 10.1099/mic.0.27222-0
Horng, Y. T., Chang, K. C., Liu, Y. N., Lai, H. C., and Soo, P. C. (2010). The RssB/RssA two-component system regulates biosynthesis of the tripyrrole antibiotic, prodigiosin, in Serratia marcescens. Int. J. Med. Microbiol. 300, 304–312. doi: 10.1016/j.ijmm.2010.01.003
Horng, Y. T., Deng, S. C., Daykin, M., Soo, P. C., Wei, J. R., Luh, K. T., et al. (2002). The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone-dependent quorum sensing in Serratia marcescens. Mol. Microbiol. 45, 1655–1671. doi: 10.1046/j.1365-2958.2002.03117.x
Hoskins, J. R., Sharma, S., Sathyanarayana, B. K., and Wickner, S. (2001). Clp ATPases and their role in protein unfolding and degradation. Adv. Protein Chem. 59, 413–429. doi: 10.1016/s0065-3233(01)59013-0
Ito, T., and Downs, D. M. (2020). Pyridoxal reductase, PdxI, is critical for salvage of pyridoxal in Escherichia coli. J. Bacteriol. 202, e00056–e00020. doi: 10.1128/JB.00056-20
Jia, X., Lin, X., and Chen, J. (2017). Linear and exponential TAIL-PCR: a method for efficient and quick amplification of flanking sequences adjacent to Tn5 transposon insertion sites. AMB Express 7:195. doi: 10.1186/s13568-017-0495-x
Keen, N. T., Tamaki, S., Kobayashi, D., and Trollinger, D. (1988). Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70, 191–197. doi: 10.1016/0378-1119(88)90117-5
Klein, G., Kobylak, N., Lindner, B., Stupak, A., and Raina, S. (2014). Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J. Biol. Chem. 289, 14829–14853. doi: 10.1074/jbc.M113.539494
Kolacsek, O., Pergel, E., Varga, N., Apáti, Á., and Orbán, T. I. (2017). Ct shift: a novel and accurate real-time PCR quantification model for direct comparison of different nucleic acid sequences and its application for transposon quantifications. Gene 598, 43–49. doi: 10.1016/j.gene.2016.10.035
Krysan, P. J., Young, J. C., and Sussman, M. R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. doi: 10.1105/tpc.11.12.2283
Lam, H. M., and Winkler, M. E. (1992). Characterization of the complex pdxH-tyrS operon of Escherichia coli K-12 and pleiotropic phenotypes caused by pdxH insertion mutations. J. Bacteriol. 174, 6033–6045. doi: 10.1128/jb.174.19.6033-6045.1992
Li, D., Liu, J., Wang, X., Kong, D., Du, W., Li, H., et al. (2018). Biological potential and mechanism of prodigiosin from Serratia marcescens subsp. lawsoniana in human choriocarcinoma and prostate cancer cell lines. Int. J. Mol. Sci. 19:3465. doi: 10.3390/ijms19113465
Lin, C., Jia, X., Fang, Y., Chen, L., Zhang, H., Lin, R., et al. (2019). Enhanced production of prodigiosin by Serratia marcescens FZSF02 in the form of pigment pellets. Electron. J. Biotechnol. 40, 58–64. doi: 10.1016/j.ejbt.2019.04.007
Liu, Y. G., and Chen, Y. (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. BioTechniques 43, 649–656. doi: 10.2144/000112601
Liu, P., Zhu, H., Zheng, G., Jiang, W., and Lu, Y. (2017). Metabolic engineering of Streptomyces coelicolor for enhanced prodigiosins (RED) production. Sci. China Life Sci. 60, 948–957. doi: 10.1007/s11427-017-9117-x
McNeil, M. B., Clulow, J. S., Wilf, N. M., Salmond, G. P., and Fineran, P. C. (2012). SdhE is a conserved protein required for flavinylation of succinate dehydrogenase in bacteria. J. Biol. Chem. 287, 18418–18428. doi: 10.1074/jbc.M111.293803
Nolivos, S., Cayron, J., Dedieu, A., Page, A., Delolme, F., and Lesterlin, C. (2019). Role of AcrAB-TolC multidrug efflux pump in drug-resistance acquisition by plasmid transfer. Science 364, 778–782. doi: 10.1126/science.aav6390
Pan, X., Sun, C., Tang, M., Liu, C., Zhang, J., You, J., et al. (2019). Loss of serine-type D-Ala-D-Ala carboxypeptidase DacA enhances prodigiosin production in Serratia marcescens. Front. Bioeng. Biotechnol. 7:367. doi: 10.3389/fbioe.2019.00367
Pan, X., Sun, C., Tang, M., You, J., Osire, T., Zhao, Y., et al. (2020). LysR-type transcriptional regulator MetR controls prodigiosin production, methionine biosynthesis, cell motility, H2O2 tolerance, heat tolerance, and exopolysaccharide synthesis in Serratia marcescens. Appl. Environ. Microbiol. 86, e02241–e02219. doi: 10.1128/AEM.02241-19
Pan, X., Tang, M., You, J., Liu, F., Sun, C., Osire, T., et al. (2021). Regulator RcsB controls prodigiosin synthesis and various cellular processes in Serratia marcescens JNB5-1. Appl. Environ. Microbiol. 87, e02052–e02020. doi: 10.1128/AEM.02052-20
Qin, H., Liu, Y., Cao, X., Jiang, J., Lian, W., Qiao, D., et al. (2020). RpoS is a pleiotropic regulator of motility, biofilm formation, exoenzymes, siderophore and prodigiosin production, and trade-off during prolonged stationary phase in Serratia marcescens. PLoS One 15:e0232549. doi: 10.1371/journal.pone.0232549
Ravindran, A., Sunderrajan, S., and Pennathur, G. (2019). Phylogenetic studies on the prodigiosin biosynthetic operon. Curr. Microbiol. 6, 597–606. doi: 10.1007/s00284-019-01665-0
Slater, H., Crow, M., Everson, L., and Salmond, G. P. (2003). Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol. Microbiol. 47, 303–320. doi: 10.1046/j.1365-2958.2003.03295.x
Song, X., Guo, J., Ma, W. X., Ji, Z. Y., Zou, L. F., Chen, G. Y., et al. (2015a). Identification of seven novel virulence genes from Xanthomonas citri subsp. citri by Tn5-based random mutagenesis. J. Microbiol. 53, 330–336. doi: 10.1007/s12275-015-4589-3
Song, C., Sundqvist, G., Malm, E., de Bruijn, I., Kumar, A., van de Mortel, J., et al. (2015b). Lipopeptide biosynthesis in Pseudomonas fluorescens is regulated by the protease complex ClpAP. BMC Microbiol. 15:29. doi: 10.1186/s12866-015-0367-y
Stella, N. A., Lahr, R. M., Brothers, K. M., Kalivoda, E. J., Hunt, K. M., Kwak, D. H., et al. (2015). Serratia marcescens cyclic AMP receptor protein controls transcription of EepR, a novel regulator of antimicrobial secondary metabolites. J. Bacteriol. 197, 2468–2478. doi: 10.1128/JB.00136-15
Sun, Y., Wang, L., Pan, X., Osire, T., Fang, H., Zhang, H., et al. (2020). Improved prodigiosin production by relieving CpxR temperature-sensitive inhibition. Front. Bioeng. Biotechnol. 8:344. doi: 10.3389/fbioe.2020.00344
Szczepaniak, J., Press, C., and Kleanthous, C. (2020). The multifarious roles of Tol-pal in gram-negative bacteria. FEMS Microbiol. Rev. 44, 490–506. doi: 10.1093/femsre/fuaa018
Tanikawa, T., Nakagawa, Y., and Matsuyama, T. (2006). Transcriptional downregulator hexS controlling prodigiosin and serrawettin W1 biosynthesis in Serratia marcescens. Microbiol. Immunol. 50, 587–596. doi: 10.1111/j.1348-0421.2006.tb03833.x
Thomson, N. R., Crow, M. A., McGowan, S. J., Cox, A., and Salmond, G. P. (2000). Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Mol. Microbiol. 36, 539–556. doi: 10.1046/j.1365-2958.2000.01872.x
Tipton, K. A., and Rather, P. N. (2017). An ompR-envZ two-component system ortholog regulates phase variation, osmotic tolerance, motility, and virulence in Acinetobacter baumannii strain AB5075. J. Bacteriol. 199, e00705–e00716. doi: 10.1128/JB.00705-16
Wei, Y. H., and Chen, W. C. (2005). Enhanced production of prodigiosin-like pigment from Serratia marcescens SMdeltaR by medium improvement and oil-supplementation strategies. J. Biosci. Bioeng. 99, 616–622. doi: 10.1263/jbb.99.616
Wilf, N. M., and Salmond, G. (2012). The stationary phase sigma factor, RpoS, regulates the production of a carbapenem antibiotic, a bioactive prodigiosin and virulence in the enterobacterial pathogen Serratia sp. ATCC 39006. Microbiology 158, 648–658. doi: 10.1099/mic.0.055780-0
Williamson, N. R., Fineran, P. C., Leeper, F. J., and Salmond, G. P. (2006). The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 4, 887–899. doi: 10.1038/nrmicro1531
Xu, Z., Wang, Y., Chater, K. F., Ou, H. Y., Xu, H. H., Deng, Z., et al. (2016). Large-scale transposition mutagenesis of Streptomyces coelicolor identifies hundreds of genes influencing antibiotic biosynthesis. Appl. Environ. Microbiol. 83, e02889–e02816. doi: 10.1128/AEM.02889-16
Yip, C. H., Yarkoni, O., Ajioka, J., Wan, K. L., and Nathan, S. (2019). Recent advancements in high-level synthesis of the promising clinical drug, prodigiosin. Appl. Microbiol. Biotechnol. 103, 1667–1680. doi: 10.1007/s00253-018-09611-z
Keywords: Ez-Tn5 transposon, Serratia marcescens, prodigiosin, essential genes, regulation
Citation: Jia X, Liu F, Zhao K, Lin J, Fang Y, Cai S, Lin C, Zhang H, Chen L and Chen J (2021) Identification of Essential Genes Associated With Prodigiosin Production in Serratia marcescens FZSF02. Front. Microbiol. 12:705853. doi: 10.3389/fmicb.2021.705853
Edited by:
Harold J. Schreier, University of Maryland, Baltimore County, United StatesReviewed by:
Veeranagouda Yaligara, Sanofi, FranceRobert M. Shanks, University of Pittsburgh, United States
Copyright © 2021 Jia, Liu, Zhao, Lin, Fang, Cai, Lin, Zhang, Chen and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jichen Chen, Y2hlbmppY2hlbjIwMDFAMTYzLmNvbQ==
 Xianbo Jia
Xianbo Jia Fangchen Liu2
Fangchen Liu2 Jichen Chen
Jichen Chen