
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 July 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.705230
This article is part of the Research Topic Zoonotic Microorganisms and Spread of Acquired Polymyxin Resistance Determinants View all 15 articles
 Elena Lavinia Diaconu
Elena Lavinia Diaconu Patricia Alba
Patricia Alba Fabiola Feltrin
Fabiola Feltrin Paola Di Matteo
Paola Di Matteo Manuela Iurescia
Manuela Iurescia Eleonora Chelli
Eleonora Chelli Valentina Donati
Valentina Donati Ilaria Marani
Ilaria Marani Angelo Giacomi
Angelo Giacomi Alessia Franco
Alessia Franco Virginia Carfora*
Virginia Carfora*A collection of 177 genomes of Salmonella Typhimurium and its monophasic variant isolated in 2014–2019 from Italian poultry/livestock (n = 165) and foodstuff (n = 12), previously screened for antimicrobial susceptibility and assigned to ST34 and single-locus variants, were studied in-depth to check the presence of the novel mcr-9 gene and to investigate their genetic relatedness by whole genome sequencing (WGS). The study of accessory resistance genes revealed the presence of mcr-9.1 in 11 ST34 isolates, displaying elevated colistin minimum inhibitory concentration values up to 2 mg/L and also a multidrug-resistant (MDR) profile toward up to seven antimicrobial classes. Five of them were also extended-spectrum beta-lactamases producers (blaSHV–12 type), mediated by the corresponding antimicrobial resistance (AMR) accessory genes. All mcr-9-positive isolates harbored IncHI2-ST1 plasmids. From the results of the Mash analysis performed on all 177 genomes, the 11 mcr-9-positive isolates fell together in the same subcluster and were all closely related. This subcluster included also two mcr-9-negative isolates, and other eight mcr-9-negative ST34 isolates were present within the same parental branch. All the 21 isolates within this branch presented an IncHI2/2A plasmid and a similar MDR gene pattern. In three representative mcr-9-positive isolates, mcr-9 was demonstrated to be located on different IncHI2/IncHI2A large-size (∼277–297 kb) plasmids, using a combined Illumina–Oxford Nanopore WGS approach. These plasmids were also compared by BLAST analysis with publicly available IncHI2 plasmid sequences harboring mcr-9. In our plasmids, mcr-9 was located in a ∼30-kb region lacking different genetic elements of the typical core structure of mcr-9 cassettes. In this region were also identified different genes involved in heavy metal metabolism. Our results underline how genomics and WGS-based surveillance are increasingly indispensable to achieve better insights into the genetic environment and features of plasmid-mediated AMR, as in the case of such IncHI2 plasmids harboring other MDR genes beside mcr-9, that can be transferred horizontally also to other major Salmonella serovars spreading along the food chain.
Salmonellosis, among the most important foodborne zoonoses worldwide, is the second most commonly reported gastrointestinal infection in humans after campylobacteriosis in the European Union/European Economic Area, mainly caused by the consumption of contaminated food (European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC], 2021). In Europe, among all confirmed salmonellosis cases, the three most commonly reported Salmonella serovars in 2019 were Salmonella Enteritidis (50.3%), Salmonella Typhimurium (11.9%), and monophasic Salmonella Typhimurium (1,4,[5],12:i:-) (8.2%) (European Food Safety Authority [EFSA] and European Centre for Disease Prevention and Control [ECDC], 2021). Sequence types (STs) 19, 34, 313, and 213 represent the most frequently reported STs for S. Typhimurium. Starting from the past two decades, the global pandemic S. Typhimurium ST34 clone has been increasingly reported, replacing to the “traditional” clone ST19 (Biswas et al., 2019). The efficacy of antimicrobial therapy for salmonellosis may be impaired by the spread of antimicrobial-resistant isolates (Michael and Schwarz, 2016), particularly to last-resort drugs used to treat severe infections in humans.
Colistin is a last-resort antibiotic of the polymyxin family, increasingly used for treating human invasive infections by multidrug-resistant (MDR) or extensively drug-resistant (XDR) Enterobacteriaceae (Poirel et al., 2017). To date, 10 different mcr genes have been reported worldwide in Enterobacteriaceae (Wang et al., 2020), with 6 of them (mcr-1 to mcr-5 and mcr-9) identified so far in Salmonella enterica (Lima et al., 2019); MDR monophasic S. Typhimurium ST34 harboring mcr genes (mcr-1, mcr-3, and mcr-5) has also been widely reported worldwide.
mcr-9 is the most recent mcr homolog (65 and 63% amino acid identity with mcr-3 and mcr-7, respectively) so far described in Salmonella serovars in different countries in samples from humans (Carroll et al., 2019; Pan et al., 2020; Wang et al., 2021), horses (Elbediwi et al., 2021), livestock (Borowiak et al., 2020; Leite et al., 2020), food (Borowiak et al., 2020; Cha et al., 2020; Tyson et al., 2020; Wang et al., 2021), and environment (Wang et al., 2021).
mcr-9 is an inducible plasmid-borne gene mainly associated with the IncHI2 replicon type (Li et al., 2020), encoding an acquired phosphoethanolamine transferase not conferring by itself clinical resistance to colistin. However, subinhibitory concentrations of colistin could induce its expression, which, in E. coli, is mediated by a two-component regulatory system encoded by the qseC and qseB genes, located downstream of mcr-9 (Kieffer et al., 2019).
The aims of this work were to (i) reanalyze by whole genome sequencing (WGS) a historical collection of S. Typhimurium and its monophasic variant within ST34 (along with its single-locus variants) to screen for the presence of mcr-9 and to investigate the relatedness of their genomes and (ii) determine the genetic environment of the mcr-9 gene and fully reconstruct the mcr-9-harboring plasmids by using a combined Illumina–Oxford Nanopore WGS approach.
A collection of 177 genomes of S. Typhimurium (n = 39) and its monophasic variant (n = 138) assigned to ST34 and single-locus variants by WGS (Supplementary Table 1) was studied in-depth. These isolates were obtained from animal samples (n = 165), collected from Italian poultry (n = 1 broiler chicken, n = 2 laying hens, and n = 14 fattening turkeys) and livestock [n = 106 fattening pigs and n = 42 bovine animals <12 months (veal calves)] and also foodstuff (n = 12), in the frame of antimicrobial resistance (AMR)-monitoring activities (according to Decision 2013/652/EU1) conducted from 2014 to 2019 by the National Reference Laboratory for Antimicrobial Resistance (NRL-AR) and previously screened for antimicrobial susceptibility.
AST was performed as minimum inhibitory concentration (MIC) determination by broth microdilution, using the EU consensus 96-well microtiter plates (Trek Diagnostic Systems, Westlake, OH, United States). The results were interpreted according to epidemiological cutoffs (ECOFFs) included in the Annex A of the EU Decision 2013/652/EU, and for sulfamethoxazole the tentative ECOFF of >256 mg/L according to the EURL-AR protocol for antimicrobial susceptibility testing of Escherichia coli, Salmonella, and Campylobacter2. E. coli ATCC 25922 was used as quality control strain.
WGS was first performed using an Illumina platform (MiSeq). DNA extraction, library preparation, trimming, and de novo assembly of raw reads were performed according to Alba et al. (2020).
Molecular characterization was performed on all the assembled genomes with the ABRicate tool3 using the Genomic Epidemiology (CGE) databases of ResFinder4 and PlasmidFinder5, and the MLST6 and SeqSero2 (Zhang et al., 2019) tools for the detection of the genetic basis of AMR and plasmid replicon types and to confirm the STs and the serotype in silico, respectively.
The pMLST 2.0 online version7 was also used for pMLST analysis on mcr-9-positive isolates and in representative mcr-9-negative isolates. The presence of the two-component regulatory system of mcr-9 inducible expression encoded by the qseC and qseB genes (Kieffer et al., 2019), was also searched by BLAST analysis.
In order to determine the genetic relatedness of the whole set of Salmonella genomes analyzed, all raw reads from the 177 genomes were also compared and clustered by using the Mash algorithm (Katz et al., 2019).
In order to resolve the complete sequence, fully reconstruct the mcr-9-harboring plasmids, and precisely identify and locate mcr-9-harboring regions, three selected mcr-9-positive (IDs: 19063952, 15060500, and 19093665) and one mcr-9-negative (ID: 17021625; one of the most closely related to positive ones selected for comparison purposes) isolates were also sequenced using the nanopore-based MinION device (ONT) with the rapid barcoding kit (SQK-RBK004). A hybrid (Illumina–Oxford Nanopore) assembly was performed using the Unicycler pipeline (Branton et al., 2008) with the default parameters.
The assembly obtained was annotated using the RAST Server (Aziz et al., 2008). Additionally, a manual curation for the obtained annotation was performed, especially for the Insertion Sequences (ISs) by using the ISfinder database (Siguier et al., 2006). The presence of virulence genes was also determined on the obtained plasmid sequences using the VF database8.
The mcr-9-carrying plasmids obtained were also compared using BLAST (Altschul et al., 1990) with two previously published complete mcr-9-carrying plasmid sequences from Enterobacteriacae: (a) an mcr-9/blaVIM–1-carrying plasmid named p3846_IncHI2_mcr (accession number CP052871) with a size of 293,138 bp, isolated from Enterobacter cloacae firstly identified in Italy from a human clinical case (Marchetti et al., 2021); (b) an mcr-9/blaVIM–1-carrying plasmid named pRH-R27 (accession number LN555650.1) with a size of 299,305 bp from a Salmonella infantis isolated from a fattening pig farm in Germany (Falgenhauer et al., 2017).
Graphical representation of the full plasmid comparison, of the mcr-9-specific region, and dendrogram was carried out using different tools: BRIG (Alikhan et al., 2011), EasyFig (Sullivan et al., 2011), and iTol (Letunic and Bork, 2021), respectively.
The AMR phenotypes of all Salmonella isolates are reported in Supplementary Table 1. As for the 11 mcr-9-positive isolates, beside the AMR gene profiles, also the MIC values are reported in detail in Table 1.
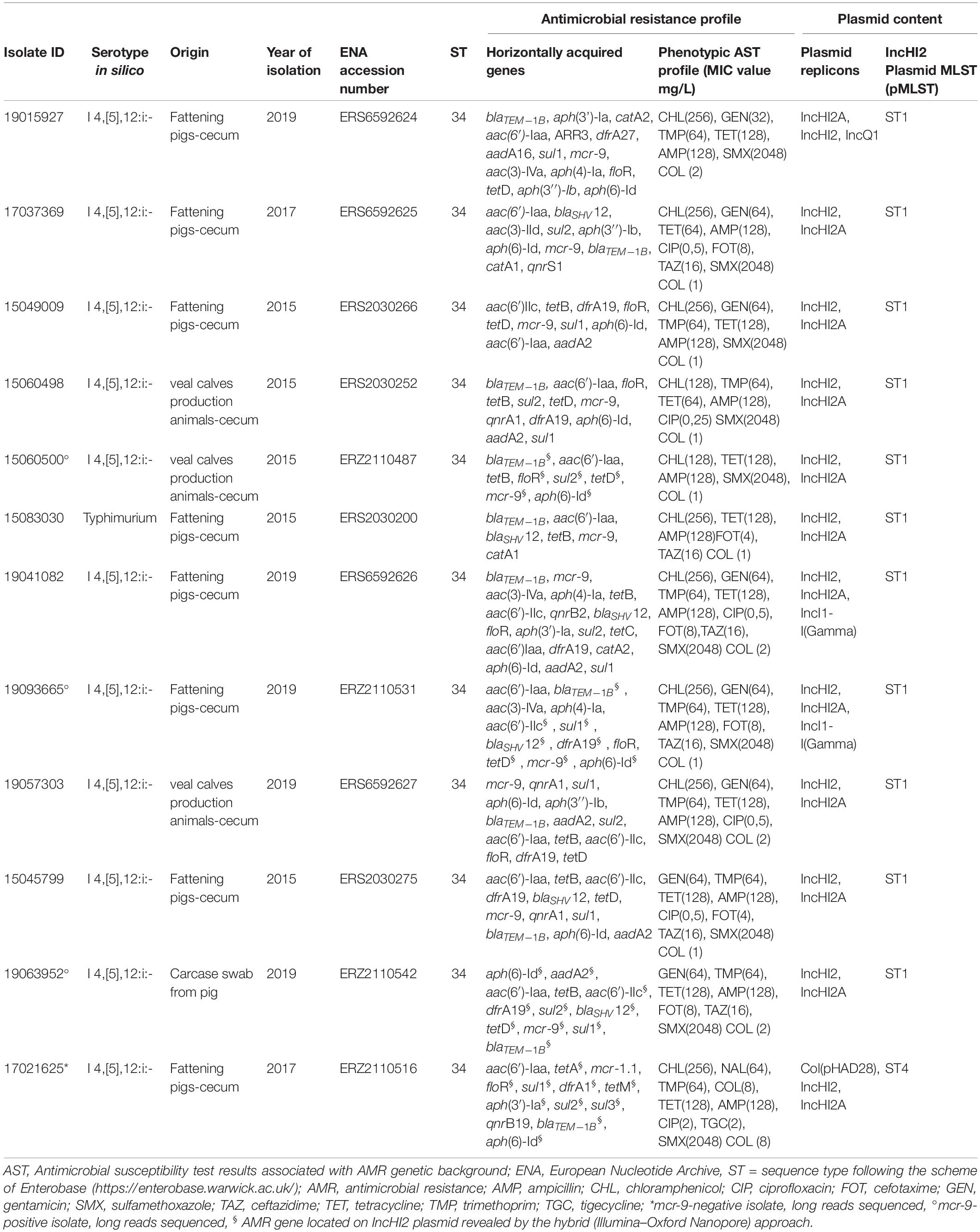
Table 1. Genotypic and phenotypic characterization of the mcr-9 and selected mcr-1 carrying isolates analyzed by WGS.
For isolates reported in Table 1, the MIC values of highest priority critically important antimicrobials (HPCIAs) such as colistin, cefotaxime, ceftazidime, and ciprofloxacin were interpreted also according to European Committee on Antimicrobial Susceptibility Testing (EUCAST)9 clinical breakpoints. In synthesis, out of 11 isolates, 6 showed microbiological and clinical resistance to cefotaxime (FOT) and ceftazidime (TAZ) (MIC range 4–8 mg/L for FOT and 16 mg/L for TAZ). Five showed microbiological resistance (MIC range 0.25–0.5 mg/L), and one also clinical resistance (MIC = 2 mg/L) to ciprofloxacin (CIP). The 11 mcr-9-positive isolates displayed elevated colistin MIC values: 4 of them had a MIC of 2 mg/L, on the epidemiological cutoff and clinical breakpoint, and 7 had a MIC of 1 mg/L.
Most of the S. Typhimurium and monophasic variant isolates were confirmed as belonging to ST34 (n = 140), while n = 37 to single-locus variants ST19 (n = 29), ST568 (n = 2), and ST376 (n = 6) (Figure 1 and Supplementary Table 1). The study of accessory resistance genes revealed the presence of mcr-9.1 in 11 ST34 isolates (1 S. Typhimurium and 10 monophasic variant) from the cecal content of seven pigs, three veal calves, and from a pig carcase swab (Table 1 and Supplementary Table 1), but none of them displayed phenotypic resistance to colistin (7 isolates displayed MIC values of 1 mg/L and 4 isolates of 2 mg/L). Details of genomic characteristics analyzed by WGS of the 11 mcr-9-positive isolates are reported in Table 1. These isolates displayed an MDR gene profile toward up to seven antimicrobial classes (aminoglycosides, beta-lactams, trimethoprim, tetracyclines, sulfamethoxazole (fluoro)quinolones, and phenicols), mediated by AMR accessory genes. In particular, beyond the presence of the mcr-9 gene, 9 of the 11 isolates presented the aac(6′)-Iaa, aph(6)-Id, blaTEM–1B gene pattern associated, respectively, with amikacin/tobramycin, streptomycin, and β-lactam resistance. Eight of them also harbored gentamicin resistance genes as aac(3)-IV and/or aac(3)-IId and/or aac(6′)-IIc combined with sul1 and/or sul2 (seven isolates), tet(B) and/or tet(D) (six isolates), dfrA19 or dfrA27 (six isolates), the extended-spectrum beta-lactamases (ESBL) gene blaSHV–12 (five isolates), floR and/or catA1 and/or catA2 (five isolates), qnrA1 or qnrB2 or qnrS1 (four isolates) genes, encoding, respectively, sulfamethoxazole, tetracycline, trimethoprim, all beta-lactams, phenicol, and fluoroquinolone resistance. The abovementioned AMR gene profiles were confirmed by the corresponding AMR phenotypes (Table 1). No mcr-9-positive isolates presented known chromosomal point mutations associated with AMR phenotypes, and all tested negative for the presence of qseC and qseB regulatory genes. All mcr-9-positive isolates harbored an IncHI2/2A, two an IncI1 and one an IncQ replicon type. Moreover, the pMLST analysis revealed that all of them harbored an IncHI2-ST1 plasmid, while one representative mcr-9-negative isolate harbored an IncHI2-ST4 replicon (Table 1).
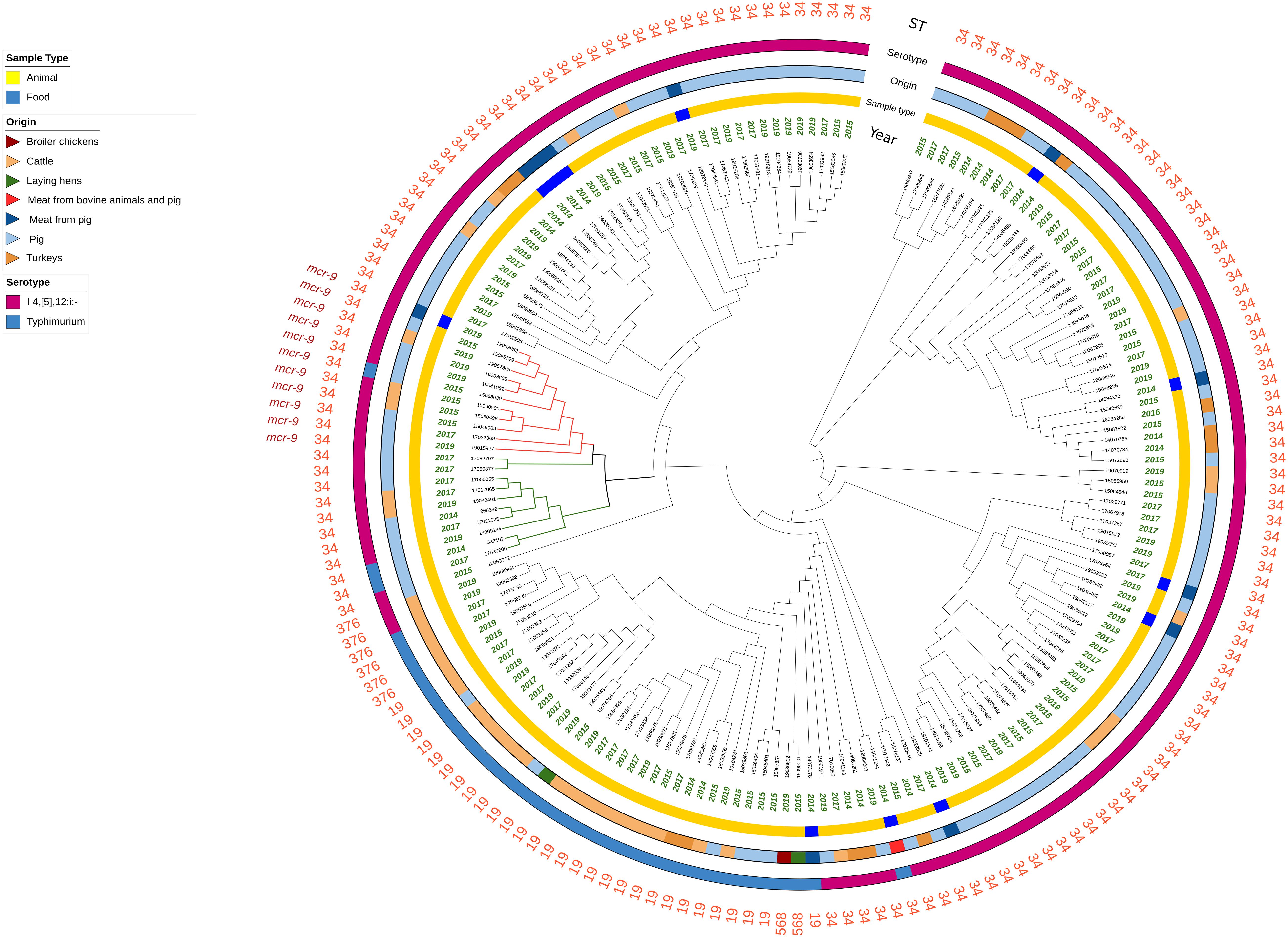
Figure 1. Circular representation of the Mash analysis of the 177 Salmonella genomes analyzed. Branch length is not proportional to the genetic distance. The red branches indicate the mcr-9-positive isolates harboring IncHI2 plasmids, and the green branches indicate the mcr-9-negative isolates harboring IncHI2 plasmids.
The results of the Mash analysis are represented in Figure 1. In the Mash tree, three main clusters were identified, with two of them consisting of 7 and 29 ST34 monophasic variant isolates, and the third one, being the most complex, grouped all the remaining isolates in several subclusters. No clear clustering based on year of sampling, sample type, or origin of the isolates was observed. The 11 mcr-9-positive isolates fell together in one of the subclusters within the most complex cluster and were all closely related (marked in red, Figure 1). This subcluster included also two mcr-9-negative isolates (17050877 and 17082797 marked in green, Figure 1). Additionally, other eight mcr-9-negative ST34 isolates were present within the same parental branch (marked in green, Figure 1).
Only the 21 isolates within this branch (both the 11 mcr-9 positive and the 10 negative ones) presented an IncHI2/2A plasmid (Figure 2A and Supplementary Figure 1A) and a similar MDR gene pattern, with three of them being also positive for mcr-1.1, associated with colistin resistance (Figure 2B and Supplementary Figure 1B).
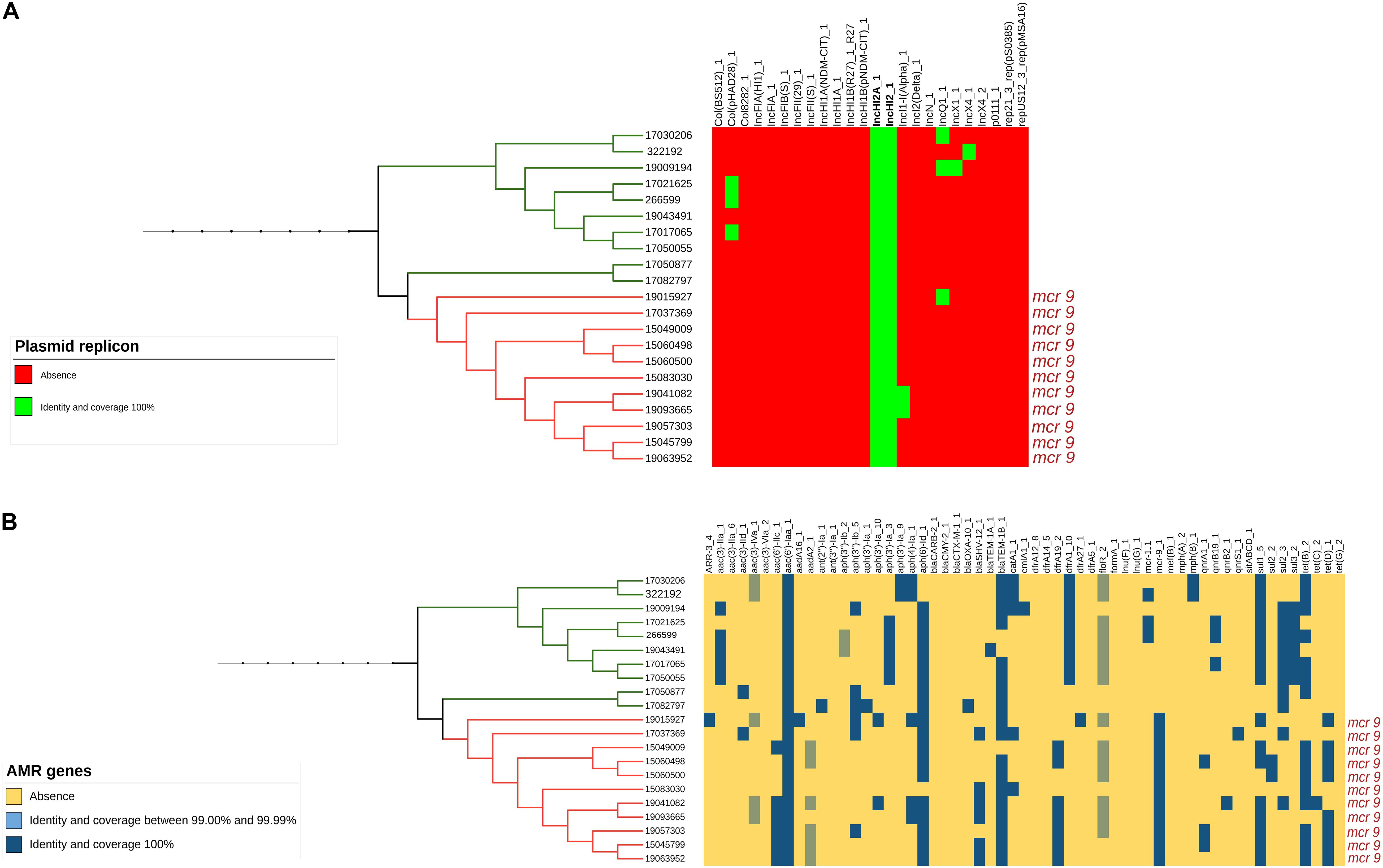
Figure 2. (A) Presence or absence of plasmid replicons identified by Mash analysis in the mcr-9-positive and related mcr-9-negative isolates. (B) Presence or absence of AMR genes identified by Mash analysis in the mcr-9-positive and related mcr-9-negative isolates.
Three representative mcr-9-positive and one mcr-9-negative isolates (Table 1 and Supplementary Table 1) were selected to be sequenced with ONT. From the hybrid (Illumina–Oxford Nanopore) assembly approach, the three plasmids containing mcr-9 were resolved. mcr-9 was demonstrated to be located on three different IncHI2/IncHI2A plasmids, named pMOL952, pMOL665, and pMOL500, with sizes of 297, 486, 291, 132, and 277,503 bp, respectively. The mcr-9-negative isolate also contained an IncHI2/IncHI2A plasmid, named pMOL625, with a size of 283, 630 bp.
Annotation of plasmid sequences identified four main genetic regions in the four IncHI2/IncHI2A plasmids. The plasmid backbone (located in the region 1..40.000 nucleotides (nt); ∼13% of the total plasmid size) included a replication region containing the repA gene encoding a replication initiation protein, a stability region, and the conjugative and transfer genes (Figure 3).
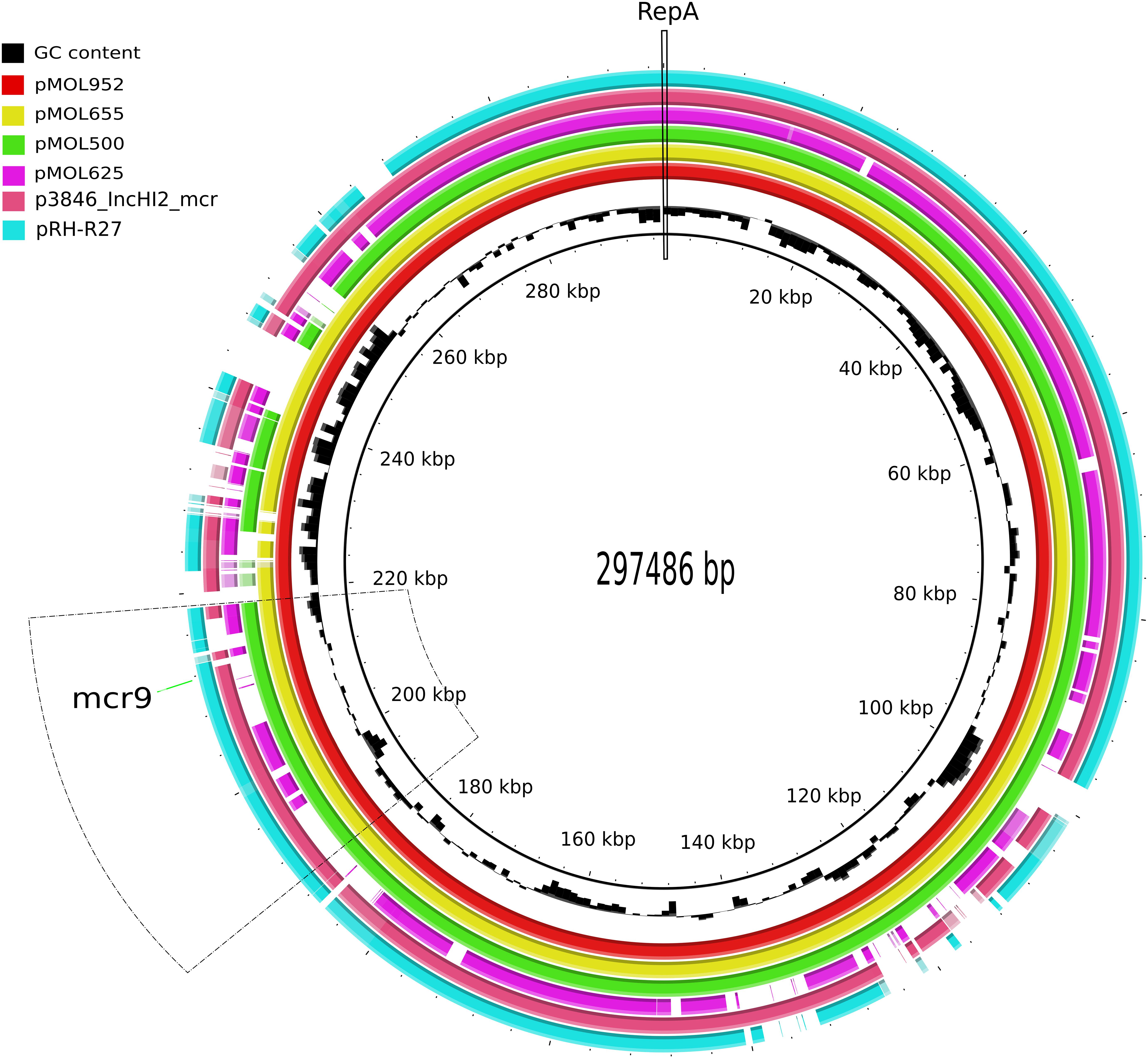
Figure 3. Graphical comparison of the complete sequence of pMOL-952 (red; ERZ2110542), pMOL-665 (yellow; ERZ2110531), pMOL-500 (green; ERZ2110487), pMOL625 (fuchsia; ERZ2110516), p3846_IncHI2 (old pink; CP052871), and pRH-R27 (blue marine; LN555650.1). The repA gene is indicated with a continuous line. The region including mcr-9 is indicated with a dotted line.
Comparison of our three mcr-9-carrying plasmids revealed that with a coverage range of 94–98%, they shared an identity of 99.9%, while the mcr-9-negative pMOL625 presented a lower coverage range (71–75%) and an identity of 98.7% with the abovementioned plasmids. Besides mcr-9, all the four IncHI2 plasmids harbored the majority of the AMR genes detected encoding resistance to all beta-lactams, aminoglycosides, trimethoprim, tetracyclines, sulfamethoxazole, and phenicols (Table 1). In particular, pMOL-952 and pMOL-665 shared a variable region (240,020..250,837 nt; Figure 3) of a class 1 integron carrying several AMR genes and qacEΔ1 (quaternary ammonium compound resistance gene). The studied plasmids also harbored genes involved in the metabolism of heavy metals as arsenic, nickel, and mercury. In this regard, a mercury-resistance (mer) operon (101,550..105,526 nt; Figure 3) was shared by the mcr-9-positive pMOL-952, pMOL-665, and pMOL-500 plasmids and was absent in the mcr-9-negative pMOL625.
No virulence genes were detected in the four IncHI2 plasmids.
In these plasmids, the mcr-9 gene was located on an almost conserved region in the three mcr-9-harboring plasmids, of approximately 24,548 bp. mcr-9 was located adjacent to a cupin fold metalloprotein gene (wbuC) with the insertion sequences IS903 upstream and the IS26 downstream of both genes. Additionally, located upstream of this structure were genes associated with heavy metal metabolism (Figures 3, 4). No other AMR genes have been detected in this region.
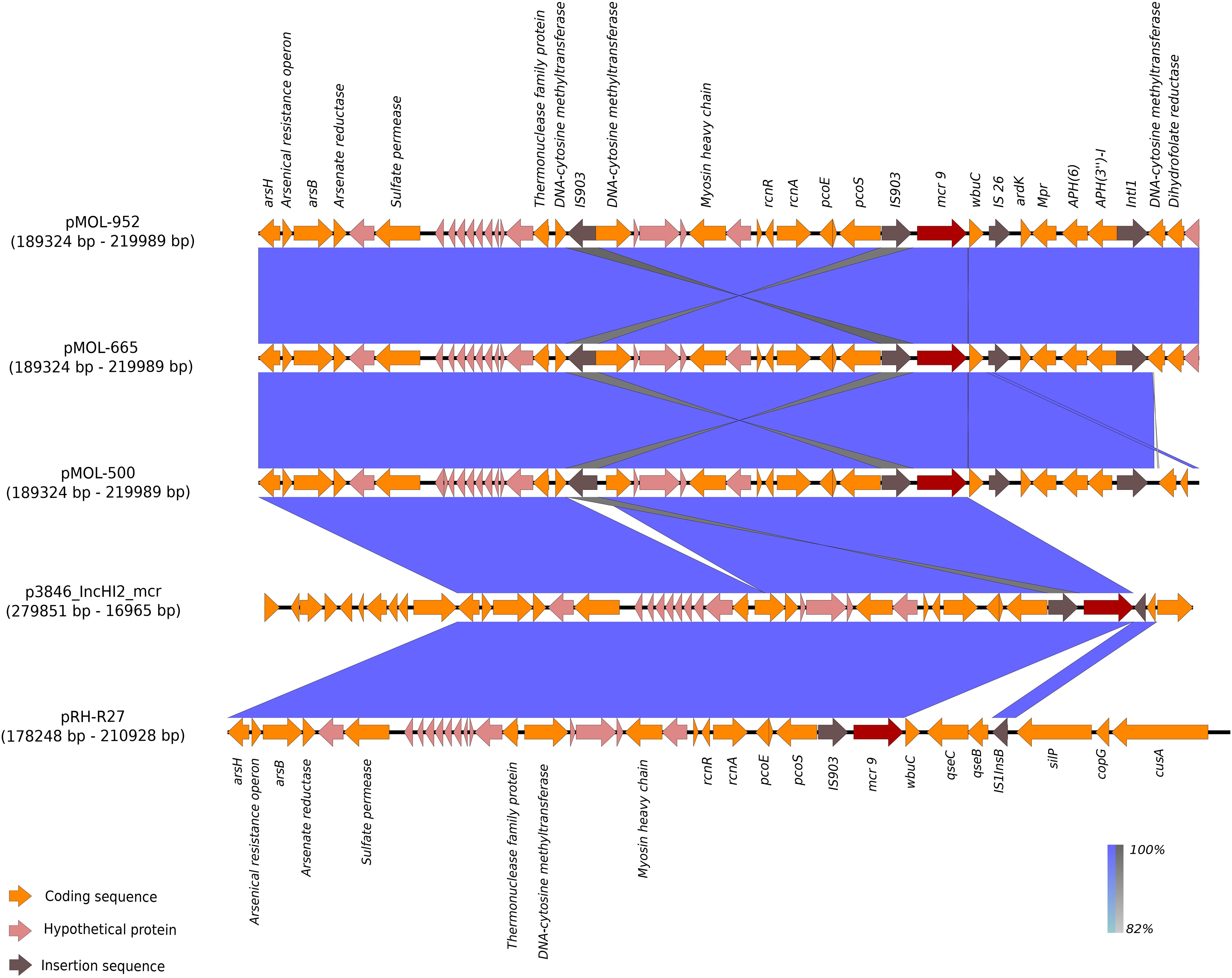
Figure 4. BLAST graphical output of the region where mcr-9 is located in pMOL-952 (ERZ2110542), pMOL-665 (ERZ2110531), pMOL-500 (ERZ2110487), p3846_IncHI2 (CP052871), and pRH-R27 (LN555650.1). Red arrows: mcr-9 gene, orange arrows: Genes encoding functional proteins, pink arrows: Genes encoding hypothetical proteins, brown arrows: Mobile genetic element.
The results obtained with BLAST analysis with the two previously published IncHI2 mcr-9-carrying plasmids revealed that our three IncHI2 mcr-9-carrying plasmids were closely related (88% coverage and 99.9% identity) to the IncHI2 ∼293 kb plasmid (p3846) described in Italy by Marchetti et al. (2021) in E. cloacae. As for the mcr-9/blaVIM–1-carrying IncHI2 plasmid (pRH-R27, size ∼299 kb) described in Germany in S. Infantis from a fattening pig farm, it shared with our three mcr-9-carrying plasmids a coverage range of 80–84% and an identity of 99.9%.
The specific region where mcr-9 was located in pMOL952, pMOL665, and pMOL500 was almost identical to the corresponding region of p3846 and pRH-R27 with an identity of 99% including the wbuC gene. The structural analysis revealed that pMOL952, pMOL665, and pMOL500 presented a mobile element (IS903) that was not identified in p3846 or pRH-R27 (Figures 3, 4).
The complete sequences of the four resolved plasmids were submitted to the European Nucleotide Archive10 under the project accession number PRJEB44718 (ERZ2110487, ERZ2110531, ERZ2110542, and ERZ2110516).
In this study, we report for the first time in Italy the presence and genomic features of 11 MDR, mcr-9-positive S. Typhimurium, and single-locus variant isolates from food-producing animals (fattening pigs and veal calves) belonging to the emerging ST34 clone, with some of them also displaying resistance to highest priority critically important antimicrobials (HPCIAs), as third- and fourth- generation cephalosporins. Indeed, plasmid-borne mcr genes pose a significant threat to public health at international level, not only because of the colistin-resistant phenotype they can induce but also because they can be transferred horizontally to foodborne pathogens in combination with other resistance genes, with the potential to be transmitted to humans and impair the treatment options.
From the Mash analysis, we identified 21 ST34 isolates within the same parental branch (both the 11 mcr-9 positive and the 10 negative ones) harboring large-size (∼277–297 kb) IncHI2/2A plasmids and a similar MDR gene pattern (Figure 2 and Supplementary Figure 1). Interestingly, only these isolates shared the presence of these plasmid types and presented a greater abundance of AMR genes compared to the rest of the Salmonella genomes analyzed. Because of the intrinsic characteristic of the Mash approach, the existence of a large-size IncHI2/2A plasmid harboring several similar AMR genes could have determined the high genetic relatedness of these isolates represented on the Mash tree.
Similarly to several previous findings (Carroll et al., 2019; Börjesson et al., 2020; Kananizadeh et al., 2020; Tyson et al., 2020), in our study, the presence of mcr-9 was not associated with clinical or microbiological resistance to colistin, and the qseC and qseB genes were absent in all the mcr-9-positive isolates detected. Indeed, the mcr-9 expression has been shown to be induced by the presence of colistin when this gene is located upstream of the two-component regulatory system qseBC. Therefore, mcr-9 may silently spread and remain undetected, unless induced by subinhibitory concentrations of colistin (Kieffer et al., 2019). However, the effectiveness of this regulatory system may be dependent on the genetic context (e.g., mcr-9 could be located on a plasmid or integrated into the chromosome) or may differ in relation to different strain backgrounds (e.g., serotype-dependent colistin susceptibility observed in Salmonella isolates) (Tyson et al., 2020), or other still unknown mechanisms could be involved. Additionally, the mcr gene family may undergo further genetic microevolution (and possibly to changes in the susceptibility pattern to colistin) due to selection pressure in the food-producing animal industry.
In our three selected isolates, mcr-9 was demonstrated to be located on IncHI2/IncHI2A plasmids by the hybrid (Illumina–Oxford Nanopore) approach, together with other AMR genes conferring resistance to different antimicrobial classes, including the blaSHV–12 ESBL gene type detected in the two plasmids of pig origin, and different genes involved in heavy metals metabolism. Similarly, previous findings detected IncHI2- or IncHI2A-type plasmids in mcr-9 positive Enterobacteriaceae, often together with other resistance determinants (including HPCIAs) and to heavy metals mainly from human patients (Carroll et al., 2019; Chavda et al., 2019; Kieffer et al., 2019; Bitar et al., 2020; Faccone et al., 2020; Kananizadeh et al., 2020; Lin et al., 2020; Osei Sekyere et al., 2020; Soliman et al., 2020; Tsui et al., 2020; Umeda et al., 2020; Elbediwi et al., 2021; Marchetti et al., 2021; Sun et al., 2021), and also from animal (Börjesson et al., 2020; Yuan et al., 2019; Borowiak et al., 2020; Haenni et al., 2020; Leite et al., 2020), food (Borowiak et al., 2020; Sadek et al., 2020; Tyson et al., 2020), and environmental (Kamathewatta et al., 2020) sources. Few reports also described mcr-9 integrated into the chromosome of Citrobacter (Ribeiro et al., 2021) and Salmonella (Pan et al., 2020; Tyson et al., 2020) isolates, with a genetic context similar to the structure observed in mcr-9-harboring plasmid sequences, suggesting a possible mcr-9 transfer as a gene cassette between plasmids and chromosomes (Pan et al., 2020).
However, only very few complete IncHI2 plasmid sequences carrying mcr-9 from food-producing animals and related foodstuff are available in public repositories for comparison, so far. In this regard, we fully reconstructed two mcr-9-positive IncHI2 plasmids from the monophasic variant of S. Typhimurium isolated from pig sources and one from the cecal content of a veal calf. To the best of the authors’ knowledge, this represents the first report of MDR, mcr-9-positive Enterobacteriaceae detected in bovine animals.
All 11 IncHI2 plasmids harboring mcr-9 were assigned to ST1 by pMLST analysis. IncHI2-ST1 plasmids have been the most frequently reported IncHI2 ST, representing a major vehicle in mediating mcr-9 and AMR gene dissemination also in Enterobacteriaceae isolates from clinical settings (Li et al., 2020). Interestingly, the mcr-9-negative IncHI2 plasmid we sequenced with ONT was assigned to another ST, ST4.
IncHI2 plasmids are known carriers of resistance determinants not only to antibiotics but also to heavy metals such as silver, mercury, arsenic, copper, tellurium, and others with the potential to coselect for the concomitant presence of mcr-9 (Tyson et al., 2020).
Moreover, pigs are considered one of the most significant vectors for the monophasic variant of S. Typhimurium ST34, and pork meat represents one of the main infection sources for humans (Biswas et al., 2019). As copper and zinc supplementation are commonly used in the swine industry (or in the case of therapeutical use of ZnO also to control postweaning diarrhea), heavy metals could accumulate in the environment, leading to a selection of ST34 isolates resistant to heavy metals and to IncHI2 plasmids frequently associated with various metal tolerance genes (Biswas et al., 2019).
IncHI2 plasmids have been reported to be diverse in terms of the overall genetic structure, while mcr-9 has been reported to be consistently located in the sil-cop region (Li et al., 2020). As previously described (Li et al., 2020), in this region, the core structure of all reported mcr-9 cassettes was rcnR-rcnA-pcoE-pcoS-IS903-mcr-9-wbuC, with the genetic content immediately upstream of mcr-9 mostly conserved. Differently, the gene content located downstream of mcr-9 was reported to be genetically diverse (silver resistance determinants and qseB-C regulators absent in most plasmids), and transposon elements were not identified.
In our plasmids, mcr-9 was located in a plasmid region (∼30 kb) lacking different genetic elements of the core structure, compared to the other plasmid sequences subjected to BLAST analysis (Figure 4). In particular, mcr-9 was flanked by the wpuC gene and the IS903 element with and additional IS element (IS26) located downstream wpuC, suggesting the potential ability to mobilize this gene. Heavy metal resistance genes were also identified (as arsenic and nickel) in the same region, but accordingly to previous findings, silver resistance determinants and qseB-C regulators were absent.
In conclusion, the spread of MDR S. Typhimurium, including monophasic ST34, has widely challenged the treatment options to control foodborne infections (Biswas et al., 2019). This can be of even more concern, especially when there is concomitant evidence of MDR genes against HPCIAs, such as colistin, extended-spectrum cephalosporins, and (fluoro)quinolones. Therefore, genomics and WGS-based surveillance are increasingly indispensable to achieve better insights also into the genetic environment and features of plasmid-mediated AMR and the relationships with bacterial pathogenic hosts, as in the case of such IncHI2 plasmids harboring mcr-9, that can be transferred horizontally also to major Salmonella serovars spreading along the food chain.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
AF, VC, PA, and ED conceived and designed the experiments. FF, PDM, MI, IM, AG, and ED performed the experiments. ED, PA, VC, AF, EC, and VD analyzed the data. VC, PA, ED, and AF wrote the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was funded partly by the Italian Ministry of Health (Research Project IZSLT 02/2020). The genomic work was conducted in the framework of the Full Force project, supported by funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement no. 773830: One Health European Joint Programme.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We wish to thank Antonio Battisti, Head of General Diagnostics Department of the Istituto Zooprofilattico Sperimentale del Lazio e della Toscana “M. Aleandri,” for the fruitful discussion on the manuscript, and Roberta Amoruso, Gessica Cordaro, Angela Ianzano, Luigi Sorbara, Roberta Onorati, and Carmela Buccella for outstanding technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.705230/full#supplementary-material
Supplementary Figure 1 | (A) Presence or absence of plasmid replicons in the 177 Salmonella genomes identified by Mash analysis. (B) Presence or absence of AMR genes in the 177 Salmonella genomes identified by Mash analysis.
Supplementary Table 1 | Metadata, phenotypic AMR patterns, and STs in 177 S. Typhimurium and monophasic variant isolates.
Alba, P., Leekitcharoenphon, P., Carfora, V., Amoruso, R., Cordaro, G., Di Matteo, P., et al. (2020). Molecular epidemiology of Salmonella infantis in Europe: insights into the success of the bacterial host and its parasitic pESI-like megaplasmid. Microb. Genom. 6:e000365. doi: 10.1099/mgen.0.000365
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Biswas, S., Li, Y., Elbediwi, M., and Yue, M. (2019). Emergence and dissemination of mcr-carrying clinically relevant salmonella typhimurium monophasic clone ST34. Microorganisms. 7:298. doi: 10.3390/microorganisms7090298
Bitar, I., Papagiannitsis, C. C., Kraftova, L., Chudejova, K., Mattioni Marchetti, V., and Hrabak, J. (2020). Detection of five mcr-9-carrying Enterobacterales isolates in four Czech hospitals. mSphere 5:e01008–20. doi: 10.1128/mSphere.01008-20
Börjesson, S., Greko, C., Myrenås, M., Landén, A., Nilsson, O., and Pedersen, K. (2020). A link between the newly described colistin resistance gene mcr-9 and clinical Enterobacteriaceae isolates carrying blaSHV–12 from horses in Sweden. J. Glob. Antimicrob. Resist. 20, 285–289. doi: 10.1016/j.jgar.2019.08.007
Borowiak, M., Baumann, B., Fischer, J., Thomas, K., Deneke, C., Hammerl, J. A., et al. (2020). Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in colistin-resistant Salmonella enterica isolates from environment, feed, animals and food (2011–2018) in Germany. Front. Microbiol. 11:80. doi: 10.3389/fmicb.2020.00080
Branton, D., Deamer, D. W., Marziali, A., Bayley, H., Benner, S. A., Butler, T., et al. (2008). The potential and challenges of nanopore sequencing. Nat. Biotechnol. 26, 1146–1153. doi: 10.1038/nbt.1495
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype typhimurium isolate. mBio 10:e00853–19. doi: 10.1128/mBio.00853-19
Cha, M. H., Woo, G. J., Lee, W., Kim, S. H., Woo, J. H., Kim, J., et al. (2020). Emergence of transferable mcr-9 gene-carrying colistin-resistant Salmonella enterica dessau ST14 isolated from retail chicken meat in Korea. Foodborne Pathog. Dis. 17, 720–727. doi: 10.1089/fpd.2020.2810
Chavda, K. D., Westblade, L. F., Satlin, M. J., Hemmert, A. C., Castanheira, M., Jenkins, S. G., et al. (2019). First report of blaVIM–4 and mcr-9-coharboring Enterobacter species isolated from a pediatric patient. mSphere 4:e00629–19. doi: 10.1128/mSphere.00629-19
Elbediwi, M., Pan, H., Zhou, X., Rankin, S. C., Schifferli, D. M., and Yue, M. (2021). Detection of mcr-9-harboring ESBL-producing Salmonella Newport isolated from an outbreak in a large-animal teaching hospital in the USA. J. Antimicrob. Chemother. 76, 1107–1109. doi: 10.1093/jac/dkaa544
European Food Safety Authority [EFSA], and European Centre for Disease Prevention and Control [ECDC] (2021). The European Union one health 2019 zoonoses report. EFSA J. 19:e06406. doi: 10.2903/j.efsa.2021.6406
Faccone, D., Martino, F., Albornoz, E., Gomez, S., Corso, A., and Petroni, A. (2020). Plasmid carrying mcr-9 from an extensively drug-resistant NDM-1-producing Klebsiella quasipneumoniae subsp. quasipneumoniae clinical isolate. Infect. Genet. Evol. 81:104273. doi: 10.1016/j.meegid.2020.104273
Falgenhauer, L., Ghosh, H., Guerra, B., Yao, Y., Fritzenwanker, M., Fischer, J., et al. (2017). Comparative genome analysis of IncHI2 VIM-1 carbapenemase-encoding plasmids of Escherichia coli and Salmonella enterica isolated from a livestock farm in Germany. Vet. Microbiol. 200, 114–117. doi: 10.1016/j.vetmic.2015.09.001
Haenni, M., Métayer, V., Jarry, R., Drapeau, A., Puech, M.-P., Madec, J.-Y., et al. (2020). Wide spread of blaCTX–M–9/mcr-9 IncHI2/ST1 plasmids and CTX-M-9-producing Escherichia coli and Enterobacter cloacae in rescued wild animals. Front. Microbiol. 11:601317. doi: 10.3389/fmicb.2020.601317
Kamathewatta, K., Bushell, R., Rafa, F., Browning, G., Billman-Jacobe, H., and Marenda, M. (2020). Colonization of a hand washing sink in a veterinary hospital by an Enterobacter hormaechei strain carrying multiple resistances to high importance antimicrobials. Antimicrob. Resist. Infect. Control 9:163. doi: 10.1186/s13756-020-00828-0
Kananizadeh, P., Oshiro, S., Watanabe, S., Iwata, S., Kuwahara-Arai, K., Shimojima, M., et al. (2020). Emergence of carbapenem-resistant and colistin-susceptible Enterobacter cloacae complex co-harboring blaIMP–1 and mcr-9 in Japan. BMC Infect Dis. 20:282. doi: 10.1186/s12879-020-05021-7
Katz, L. S., Griswold, T., Morrison, S., Caravasn, J., Zhang, S., den Bakker, H. C., et al. (2019). Mashtree: a rapid comparison of whole genome sequence files. J. Open Source Softw. 4:1762. doi: 10.21105/joss.01762
Kieffer, N., Royer, G., Decousser, J.-W., Bourrel, A.-S., Palmieri, M., Ortiz De La Rosa, J.-M., et al. (2019). mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob. Agents Chemother. 63:e00965–19. doi: 10.1128/AAC.00965-19
Leite, E. L., Araújo, W. J., Vieira, T. R., Zenato, K. S., Vasconcelos, P. C., Cibulski, S., et al. (2020). First reported genome of an mcr-9-mediated colistin-resistant Salmonella Typhimurium isolate from Brazilian livestock. J. Glob. Antimicrob. Resist. 23, 394–397. doi: 10.1016/j.jgar.2020.09.012
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. gkab301. doi: 10.1093/nar/gkab301 Online ahead of print,
Li, Y., Dai, X., Zeng, J., Gao, Y., Zhang, Z., and Zhang, L. (2020). Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci Rep. 15:8113. doi: 10.1038/s41598-020-65106-w
Lima, T., Domingues, S., and Da Silva, G. J. (2019). Plasmid-mediated colistin resistance in Salmonella enterica: a review. Microorganisms 7:55. doi: 10.3390/microorganisms7020055
Lin, M., Yang, Y., Yang, Y., Chen, G., He, R., Wu, Y., et al. (2020). Co-Occurrence of mcr-9 and blaNDM–1 in Enterobacter cloacae isolated from a patient with bloodstream infection. Infect. Drug Resist. 13, 1397–1402. doi: 10.2147/IDR.S248342
Marchetti, V. M., Bitar, I., Sarti, M., Fogato, E., Scaltriti, E., Bracchi, C., et al. (2021). Genomic characterization of VIM and MCR co-producers: the first two clinical cases, in Italy. Diagnostics 11:79. doi: 10.3390/diagnostics11010079
Michael, G. B., and Schwarz, S. (2016). Antimicrobial resistance in zoonotic nontyphoidal Salmonella: an alarming trend? Clin. Microbiol. Infect. 22, 968–974. doi: 10.1016/j.cmi.2016.07.033
Osei Sekyere, J., Maningi, N. E., Modipane, L., and Mbelle, N. M. (2020). Emergence of mcr-9.1 in extended-spectrum-β-lactamase-producing clinical Enterobacteriaceae in Pretoria, South Africa: global evolutionary phylogenomics, resistome, and mobilome. mSystems 5:e00148–20. doi: 10.1128/mSystems.00148-20
Pan, Y., Fang, Y., Song, X., Lyu, N., Chen, L., Feng, Y., et al. (2020). Co-occurrence of mcr-9, extended spectrum β-lactamase (ESBL) and AmpC genes in a conjugative IncHI2A plasmid from a multidrug-resistant clinical isolate of Salmonella diarizonae. J. Infect. 82:84–123. doi: 10.1016/j.jinf.2020.11.008
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Ribeiro, T. G., Izdebski, R., Urbanowicz, P., Carmeli, Y., Gniadkowski, M., and Peixe, L. (2021). Citrobacter telavivum sp. nov. with chromosomal mcr-9 from hospitalized patients. Eur. J. Clin. Microbiol. Infect. Dis. 40, 123–131. doi: 10.1007/s10096-020-04003-6
Sadek, M., Nariya, H., Shimamoto, T., Kayama, S., Yu, L., Hisatsune, J., et al. (2020). First genomic characterization of blaVIM–1 and mcr-9-coharbouring Enterobacter hormaechei isolated from food of animal origin. Pathogens 9:687. doi: 10.3390/pathogens9090687
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Soliman, A. M., Maruyama, F., Zarad, H. O., Ota, A., Nariya, H., Shimamoto, T., et al. (2020). Emergence of a multidrug-resistant Enterobacter hormaechei clinical isolate from egypt co-harboring mcr-9 and blaVIM–4. Microorganisms 8:595. doi: 10.3390/microorganisms8040595
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, L., Zhao, X., Wang, L., Guo, X., Shi, X., and Hu, L. (2021). Coexistence of mcr-9 and blaNDM–1 in a multidrug-resistant Enterobacter hormaechei strain recovered from a bloodstream infection in China. J. Glob. Antimicrob. Resist. 24, 440–442. doi: 10.1016/j.jgar.2021.02.011
Tsui, C. K. M., Sundararaju, S., Al Mana, H., Hasan, M. R., Tang, P., and Perez-Lopez, A. (2020). Draft genome sequence of an extended-spectrum β-lactamase-producing Klebsiella oxytoca strain bearing mcr-9 from Qatar. Microbiol. Resour. Announc. 9:e00429–20.
Tyson, G. H., Li, C., Hsu, C.-H., Ayers, S., Borenstein, S., Mukherjee, S., et al. (2020). The mcr-9 gene of Salmonella and Escherichia coli is not associated with colistin resistance in the United States. Antimicrob. Agents Chemother. 64:e00573–20. doi: 10.1128/AAC.00573-20
Umeda, K., Nakamura, H., Fukuda, A., Matsumoto, Y., Motooka, D., Nakamura, S., et al. (2020). Genomic characterization of clinical Enterobacter roggenkampii co-harbouring blaIMP–1- and blaGES–5-encoding IncP6 and mcr-9-encoding IncHI2 plasmids isolated in Japan. J. Glob. Antimicrob. Resist. 24, 220–227. doi: 10.1016/j.jgar.2020.11.028
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., and Zong, Z. (2020). Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, Y., Li, Z., Lyu, N., Ma, S., Liu, F., Hu, Y., et al. (2021). Comparative genomic analysis of mobile colistin resistance gene mcr-9 in Salmonella enterica. J. Infect. 82, e15–e17. doi: 10.1016/j.jinf.2020.12.029
Yuan, Y., Li, Y., Wang, G., Li, C., Xiang, L., She, J., et al. (2019). Coproduction of MCR-9 And NDM-1 by colistin-resistant Enterobacter hormaechei isolated from bloodstream infection. Infect Drug Resist. 12, 2979–2985. doi: 10.2147/IDR.S217168
Keywords: Salmonella Typhimurium, monophasic variant, mcr-9, whole genome sequencing, long-read sequencing, multidrug resistance, ESBL, IncHI2 plasmid
Citation: Diaconu EL, Alba P, Feltrin F, Di Matteo P, Iurescia M, Chelli E, Donati V, Marani I, Giacomi A, Franco A and Carfora V (2021) Emergence of IncHI2 Plasmids With Mobilized Colistin Resistance (mcr)-9 Gene in ESBL-Producing, Multidrug-Resistant Salmonella Typhimurium and Its Monophasic Variant ST34 From Food-Producing Animals in Italy. Front. Microbiol. 12:705230. doi: 10.3389/fmicb.2021.705230
Received: 04 May 2021; Accepted: 14 June 2021;
Published: 16 July 2021.
Edited by:
Alberto Quesada, University of Extremadura, SpainReviewed by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyCopyright © 2021 Diaconu, Alba, Feltrin, Di Matteo, Iurescia, Chelli, Donati, Marani, Giacomi, Franco and Carfora. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Virginia Carfora, dmlyZ2luaWEuY2FyZm9yYUBpenNsdC5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.