- 1Department of Animal Biology, Faculty of Natural Sciences, University of Tabriz, Tabriz, Iran
- 2Department of Plant Biology, Faculty of Natural Sciences, University of Tabriz, Tabriz, Iran
Emergence of extensively drug-resistant isolates of Klebsiella pneumoniae has prompted increased reliance on the last-resort antibiotics such as tigecycline (TGC) for treating infections caused by these pathogens. Consumption of human antibiotics in the food production industry has been found to contribute to the current antibiotic resistance crisis. In the current study, we aimed to investigate the mechanisms of TGC resistance among 18 TGC-non-susceptible (resistant or intermediate) K. pneumoniae (TGC-NSKP) isolates obtained from human (n = 5), food animals (n = 7), and in vitro selection experiment (n = 6). Isolates were genotyped by multilocus sequence typing (MLST). ramR, acrR, rpsJ, tetA, and mgrB (for colistin resistance) genes were sequenced. The presence of tetX, tetX1, and carbapenemase genes was examined by PCR. Susceptibility to different classes of antibiotics was evaluated by disc diffusion and broth macrodilution methods. The expression level of acrB was quantified by RT-qPCR assay. The 12 TGC-NSKP isolates [minimum inhibitory concentrations (MICs) = 4–32 mg/l] belonged to 10 distinct sequence types including ST37 (n = 2), ST11, ST15, ST45, ST1326 (animal isolates); ST147 (n = 2, human and animal isolates); and ST16, ST377, ST893, and ST2935 (human isolates). Co-resistance to TGC and colistin was identified among 57 and 40% of animal and human isolates, respectively. All human TGC-NSKP isolates carried carbapenemase genes (blaOXA–48, blaNDM–1, and blaNDM–5). tetX/X1 genes were not detected in any isolates. About 83% of TGC-NSKP isolates (n = 15) carried ramR and/or acrR alterations including missense/nonsense mutations (A19V, L44Q, I141T, G180D, A28T, R114L, T119S, Y59stop, and Q122stop), insertions (positions +205 and +343), or deletions (position +205) for ramR, and R90G substitution or frameshift mutations for acrR. In one isolate ramR amplicon was not detected using all primers used in this study. Among seven colistin-resistant isolates, five harbored inactivated/mutated MgrB due to premature termination by nonsense mutations, insertion of IS elements, and frameshift mutations. All isolates revealed wild-type RpsJ and TetA (if present). Increased expression of acrB gene was detected among all resistant isolates, with the in vitro selected mutants showing the highest values. A combination of RamR and AcrR alterations was involved in TGC non-susceptibility in the majority of studied isolates.
Introduction
Antibiotic resistance is rising to dangerously high levels in both human and veterinary medicine. The limited number of antibiotics coming to the market and new threats arising from extensively drug-resistant bacteria bring us perilously to the end of antibiotic era in which common treatable infections and minor injuries can once again be fatal. The use of human antibiotics in food animals as growth promoters or prophylactic agents has been identified as a significant contributing factor to increasing antimicrobial resistance (Van et al., 2020). Food animals have the potential to serve as reservoir for antibiotic-resistant bacteria, which can be transmitted to humans via direct contact or the food chain (Founou et al., 2016). Klebsiella pneumoniae is among the most problematic pathogens that have acquired much public health concern due to developing resistance to most clinically important antibiotics. It is an opportunistic pathogen that commonly causes a variety of the community- and hospital-acquired infections including urinary tract infection, pneumonia, and bloodstream infection. The emergence and dissemination of carbapenemase-producing strains of K. pneumoniae in healthcare facilities constitute a serious threat to public health (Pitout et al., 2015). The management of infections caused by carbapenem-resistant K. pneumoniae (CRKP) is complicated and often requires the use of last-resort antibiotics such as tigecycline (TGC) and colistin (Renteria et al., 2014; Sader et al., 2015). TGC is the first member of the novel class of glycylcyclines with expanded-spectrum antibacterial activity against Gram-negative and Gram-positive bacteria. It is approved by the Food and Drug Administration (FDA) for use in complicated skin and skin structure infections, complicated intra-abdominal infections, and community-acquired bacterial pneumonia (Beabout et al., 2015). TGC acts by inhibition of bacterial protein synthesis and has the ability to evade the classical mechanisms mediating resistance to tetracyclines including ribosomal protection and active efflux mediated by Tet proteins (Livermore, 2005).
In K. pneumoniae, TGC resistance is being increasingly reported since its approval (Spanu et al., 2012; Chiu et al., 2017a). TGC resistance in K. pneumoniae is believed to be mainly mediated by the overexpression of AcrAB efflux pump, which is regulated by local repressor AcrR and transcriptional activator RamA. The latter protein is also negatively regulated by RamR whose mutations have been found to contribute to significant increases in ramA and subsequently acrAB expression upon resistance occurrence (Hentschke et al., 2010; Chiu et al., 2017b). Decreased susceptibility to TGC has been also found to be related to alterations in the efflux pump encoding gene tetA (Du et al., 2018), or rpsJ (Beabout et al., 2015), the gene that encodes the ribosomal S10 protein. Moreover, enzymatic inactivation by TetX protein, a flavin-dependent monooxygenase, has been described to confer TGC resistance among some clinical pathogens (Moore et al., 2005; Deng et al., 2014). We aimed in the current study to investigate the TGC resistance determinants from a diverse group of TGC-non-susceptible (intermediate or resistant) K. pneumoniae (TGC-NSKP) isolates of clinical and animal origins and in vitro developed TGC-NSKP mutants. Also, the sequence types of TGC-NSKP isolates were determined by multilocus sequence typing (MLST) to identify major types associated with TGC non-susceptibility in each group of bacterial isolates from different host origins.
Materials and Methods
Bacterial Isolates
Klebsiella pneumoniae isolates from animal and human sources were included in this study. Animal isolates were obtained by taking cloacal swabs (using sterile cotton swab) from randomly selected broilers at a major chicken slaughterhouse. Taken samples were seeded on Eosin Methylene Blue (EMB) agar plates and were incubated at 37°C for 24 h. Screening for TGC-non-susceptible isolates among the grown colonies on EMB agar was performed using Mueller–Hinton broth (MHB) supplemented with 3 mg/l of TGC according to the method described in our previous study (Pishnian et al., 2019). Moreover, five clinical TGC-NSKP isolates obtained from patients hospitalized in two teaching hospitals of the country were included as human isolates. Identification of isolates was performed by conventional biochemical methods (Mahon et al., 2018).
In vitro Selection of Tigecycline-Resistant Bacteria
To identify the genetic alterations mediating TGC resistance, upon TGC exposure, an in vitro resistance induction experiment was performed by exposing three TGC-susceptible (TGC-S) isolates to elevated concentrations of TGC. The Mueller–Hinton agar (MHA) plates supplemented with a sub-inhibitory concentration of antibiotic [1/2 × of minimum inhibitory concentrations (MICs)] were inoculated with 3 × 105 CFU/ml TGC-S bacterial suspension and were incubated for 24–72 h. Colonies appearing on each plate were randomly picked and reisolated on media with the same concentration of TGC or concentrations, which were within 1.25–1.5× of previous concentration. In cases where no growth was observed at higher concentrations, colonies appeared at the same concentration were picked and transferred to MHA supplemented with the same and higher concentration of TGC. The obtained in vitro induced TGC-NSKP isolates were subjected to antimicrobial susceptibility testing.
Antimicrobial Susceptibility Testing
The MICs of TGC (Glentham Life Sciences, Corsham, United Kingdom) (batch nos. 389SOI and 081GRB), colistin (colistin sulfate, Glentham Life Sciences, United Kingdom; batch no. 844WZQ), and imipenem (IPM) (Glentham Life Sciences, United Kingdom; batch no. 205WLC) were determined by broth macrodilution method using freshly prepared (less than 12-h-old) MHB from Difco (BD Diagnostic Systems, Sparks, MD, United States). The susceptibility to other classes of antibiotics was determined by disc diffusion method (Kirby–Bauer) according to the Clinical and Laboratory Standards Institute (CLSI) guidelines using the following antibiotics: gentamicin, amikacin, ampicillin, ceftriaxone, cefepime, nalidixic acid, ciprofloxacin, levofloxacin, gatifloxacin, tetracycline, doxycycline, minocycline, chloramphenicol, nitrofurantoin, and fosfomycin (BBL Sensi-DiscTM, Becton-Dickinson, Sparks, MD, United States). Due to the lack of established CLSI breakpoints for TGC at this time, the FDA breakpoints issued for Enterobacteriaceae (susceptible ≤ 2 mg/l, intermediate = 4 mg/l, and resistant ≥ 8 mg/l) were applied for interpretation of results. Isolates characterized with colistin MIC values greater than 2 mg/l were categorized as resistant according to guidelines described by the European Committee on Antimicrobial Susceptibility Testing (EUCAST). Escherichia coli ATCC 25922 was used as a quality-control strain for antimicrobial susceptibility testing.
Bacterial Genotyping by Multilocus Sequence Typing
MLST with seven housekeeping genes (rpoB, gapA, mdh, pgi, phoE, infB, and tonB) was carried out for TGC-NSKP isolates of human and animal origins following the methods described previously (Diancourt et al., 2005). The allelic profiles and sequence types (STs) were assigned by using the K. pneumoniae MLST database provided by the Institut Pasteur, Paris, France.1
Molecular Determinants of Tigecycline, Colistin, and Carbapenem Resistance
The TGC-NSKP isolates were screened for the presence of tetX and tetX1 genes by performing PCR with gene-specific primers (Table 1) [a second pair of primers were used for detection of tetX1 gene (forward 5′-GCGACATTCCTGAACCAGAAACG and reverse 5′-CGGACGATTACTCTTCCAAGG)]. The coding regions of ramR, acrR, rpsj, tetA, and mgrB [for colistin (Col) resistance] were amplified and sequenced using the primers listed in Table 1. For isolates that did not yield a PCR product using primers targeting amplification of ramR coding sequence as well as some flanking regions (ramR-ext), amplification with other pairs of primers targeting an internal region of ramR coding sequence was repeated (ramR-int). Mutations were characterized by comparing the sequences with those of K. pneumoniae ATCC 700603 and three TGC-susceptible isolates (TGC MICs = 0.25 mg/l). The impact of the identified amino acid substitutions on the biological function of the protein (i.e., neutral or deleterious) were further predicted by using the Protein Variation Effect Analyzer tool (PROVEAN) (Choi and Chan, 2015). The IS Finder database2 was used to identify and analyze insertion sequences. Moreover, the presence of carbapenemase-encoding genes (blaKPC, blaVIM, blaNDM, and blaOXA–48) were examined by PCR using the primers and amplification conditions described previously (Poirel et al., 2011). The nucleotide sequences of blaNDM were determined using the primers NDM-F-5′-GCCCAATATTATGCACCCGGTC and NDM-R-5′-AGCGCAGCTTGTCGGCCAT (Jafari et al., 2019).
Assessment of acrB Expression
To investigate the association between TGC non-susceptibility and overexpression of AcrAB efflux pump, the expression level of acrB gene was measured using RT-qPCR analysis. The total RNA from all TGC-S and TGC-NS bacterial cells was harvested using a GeneAll RiboEx Total RNA extraction kit (GeneAll Biotechnology, Seoul, South Korea). cDNA was synthesized from 1 μg of RNase-free DNase I (Takara Biotechnology, Dalian, China)-treated total RNA using Revert Aid first-strand cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, United States). Real-time PCR amplification was performed using a Power SYBR green PCR master mix (Applied Biosystems, Foster City, CA, United States) on a Eco Real-Time PCR system (Illumina, San Diego, CA, United States). The relative gene expression levels were calculated using the 2–ΔΔCT formula with rpsL housekeeping gene as internal control. A TGC-S isolate with TGC MIC of 0.25 mg/l was used as a reference strain. In the case of in vitro selected mutants, expression levels of acrB were compared with those of parental TGC-S isolates.
Nucleotide Sequence Accession Numbers
The nucleotide sequences of the studied genes have been deposited at GenBank nucleotide sequence database under the following accession numbers:
MW653710 to MW653712 (mutated/altered mgrB), MW653713 and MW653714 (wild-type mgrB), MW653715 to MW653725 (mutated/altered ramR), MW653726 to MW653730 (wild-type ramR) MW653731 to MW653736 (mutated/altered acrR), MW653737 (HK10-S, AK88-S, AK298, AK299, and HK98), MW653738 (HK2-S), MW653739 (AK294 and AK297) (wild-type acrR), MW653740 (rpsJ, all isolates had identical sequences), and MW653741 (tetA, all isolates had identical sequences).
Results
Bacterial Isolates, Molecular Typing, and Antimicrobial Susceptibility Testing
Among the 1,430 samples taken from healthy broilers (collected from 83 different farms), six TGC-resistant (TGC-R) bacteria were detected, all corresponding to K. pneumoniae. The TGC-R K. pneumoniae (TGC-RKP) isolates displayed TGC MICs ranging from 8 to 32 mg/l. Moreover, one TGC-R isolate (AK513, TGC MIC = 8 mg/l) was included from our previous work (Pishnian et al., 2019), which was obtained from a turkey during the screening for colistin-resistant bacteria (Table 2). The seven TGC-R animal isolates belonged to six different sequence types including ST37 (n = 2 isolates) and ST11, ST15. ST45, ST147, and ST1326 (n = 1 each). Testing susceptibility to other antimicrobials revealed that TGC resistance was linked to a multidrug-resistant phenotype, and all animal TGC-RKP isolates showed resistance to quinolones, chloramphenicol, other members of tetracycline family and nitrofurantoin. The full resistance rate to gentamycin, ceftriaxone, and fosfomycin was found to be 42.8, 28, and 28%, respectively. One isolate, AK294, was found to be extended-spectrum β-lactamase (ESBL) producer using the combination disc method. Co-resistance to colistin and TGC was observed among 57% of animal isolates belonging to ST37, ST11, and ST15. All TGC-R animal isolates were susceptible to amikacin and IPM. Among the five studied human isolates, one was TGC-R (MIC = 8 mg/l), and the remaining isolates showed intermediate susceptibility to TGC (MICs = 4 mg/l). The MLST distributed the five TGC-NSKP human isolates into five distinct sequence types, including ST16, ST147 (also found in one animal isolate), ST377, ST893, and ST2935. Two human isolates showed simultaneous resistance to TGC (nonsusceptible), colistin, and IPM, with the remaining three isolates being characterized with co-resistance to IPM and TGC. Fosfomycin and amikacin were among the few antimicrobials that showed 40% activity against the TGC, IPM, and ±Col NSKP human isolates. The antimicrobial susceptibility profiles of the studied isolates are described in Table 2.
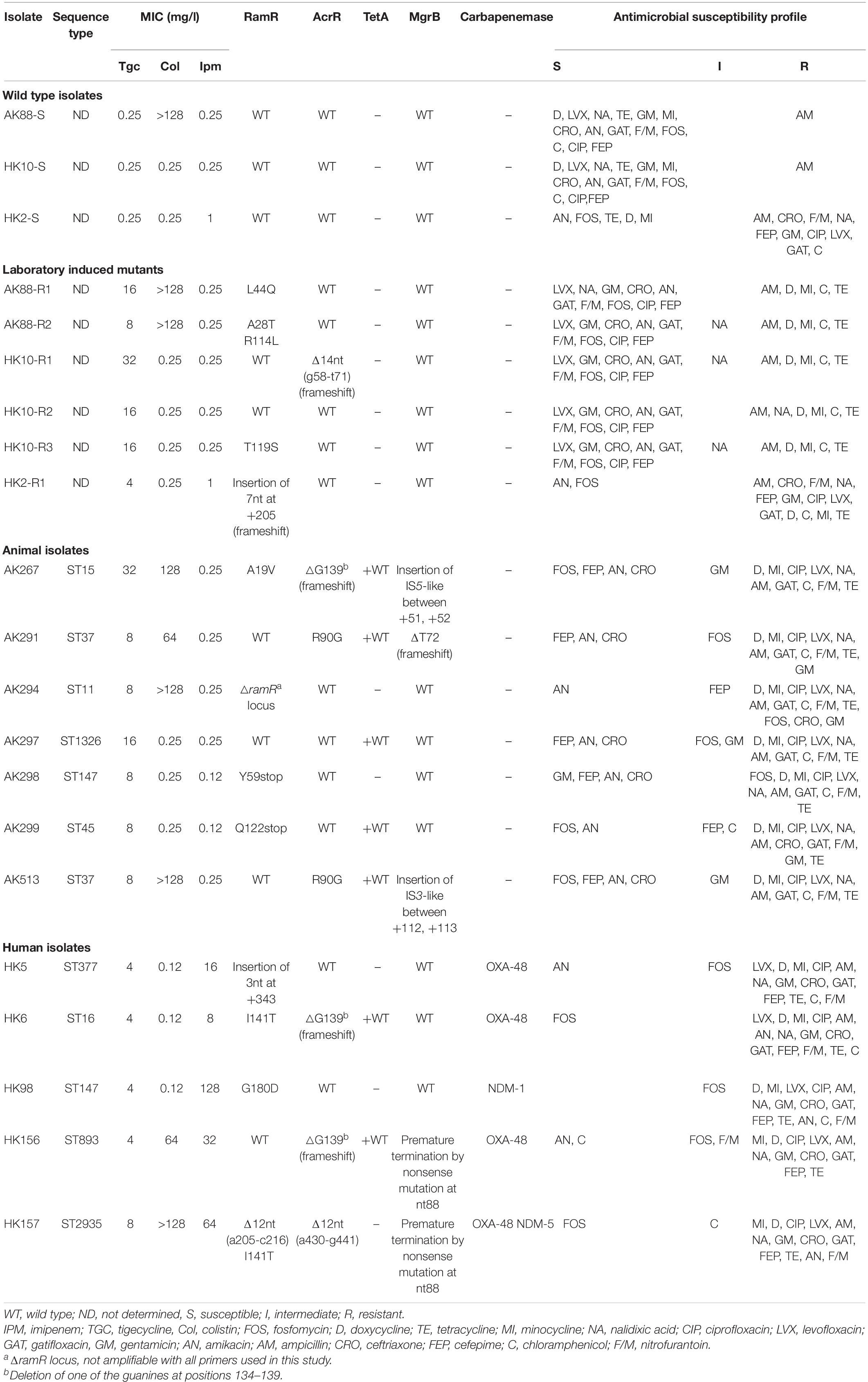
Table 2. Genotypic and phenotypic characteristics of tigecycline susceptible and non-susceptible K. pneumoniae isolates obtained from human, food animals, and in vitro selection assay.
In vitro Selection of Klebsiella pneumoniae Mutants With Reduced Susceptibility to Tigecycline
Three TGC-susceptible K. pneumoniae isolates (TGC MICs = 0.25 mg/l) were exposed to increasing concentrations of TGC. As 50% of the TGC-NSKP isolates in this study were characterized with co-resistance to colistin and TGC, we included one TGC-S but Col-R isolate among the in vitro selected isolates to see if resistance to colistin facilitates development of TGC resistance. Overall, it took 21–23 selection cycles to obtain six TGC-NSKP isolates with TGC MICs of 32 (n = 1 mutant), 16 (n = 3 mutants), 8 (n = 1 mutant), and 4 mg/l (n = 1 mutant) from three parental TGC-S isolates (Table 2). There was no significant difference between the selection cycles required to reach a TGC-NSKP isolate among the ColR and ColS isolates. Interestingly, all progenitor TGC-S isolates that were susceptible to tetracycline, minocycline, doxycycline, nalidixic acid, and chloramphenicol before induction became fully resistant (or showed intermediate susceptibility) to these antibiotics upon TGC non-susceptibility induction.
Detection and Sequence Analysis of Resistance Conferring Genes
The plasmid-encoded tetX and tetX1 genes were not detected in any of the TGC-NSKP isolates of both origins. All TGC-NSKP isolates of human, animal, and in vitro selected origins revealed wild-type RpsJ and TetA (if present). The ramR amplicons were obtained for 17 out of 18 TGC-NSKP isolates with the exception of isolate AK294, which did not yield any PCR product using the two pairs of primers used in this study. Eleven out of eighteen TGC-NSKP isolates (61%) (n = 3 animal, 4 human, and 4 in vitro selected mutants) revealed mutated/altered ramR gene. The observed RamR alterations included premature termination by nonsense mutations at amino acid positions 59 [TAC(Y) > TAA] and 122 [CAG (Q) > TAG]; a 12-nt deletion at positions 205–216; a 3-nt (AGC, serine) insertion at position +343 (between +342, +343); substitutions A19V, G180D, and I141T found among human and animal TGC-NSKP isolates and frameshift mutation due to insertion of 7 nt at position +205; and substitutions L44Q, A28T+R114L, and T119S observed among in vitro selected mutants. Moreover, AcrR alterations were observed among seven (38%) TGC-NSKP isolates (three animal isolates, three human isolates, and one in vitro selected mutant). A frameshift mutation resulting from deletion of one of the six guanines at positions 134–139 was the most common AcrR alteration identified among both animal (n = 1) and human isolates (n = 2). AcrR R90G substitution was found among two animal isolates, both of which carried a wild-type RamR protein. Also a frameshift mutation resulting from 14-nt deletions (g58-t71) was detected in one in vitro selected mutant (TGC MIC = 32 mg/l) carrying a wild-type RamR. One TGC-NSKP human isolate co-harbored two 12-nt deletion in ramR (a205-c216) and acrR (a430-g441) genes (Table 2). Among seven colistin-resistant isolates, five harbored inactivated/mutated MgrB due to premature termination by nonsense mutations, insertion of IS elements, and frameshift mutation as shown in Table 2. The genes encoding for carbapenemases were detected in all IPM-resistant human isolates, with three isolates harboring blaOXA–48, one carrying blaNDM–1, and one co-harboring blaOXA–48 and blaNDM–5. Two TGC-RKP isolates with TGC MIC = 16 mg/l carried wild-type RamR and AcrR proteins (Table 2).
Expression Levels of the AcrAB Pump
To see if TGC non-susceptibility was linked to overexpression of AcrAB pump, the expression level of acrB gene was quantified by RT-qPCR analysis. A constitutively expressed housekeeping gene rpsL was used as a control and the TGC-susceptible clinical strain HK10 as a reference for the data analysis of animal and human isolates. For the in vitro selected mutants, the expression level was compared with that of their TGC-S parental strains. The in vitro selected TGC-NSKP isolates showed the highest expression levels (18- to 146-fold) among the studied isolates. Since the expression level of acrB in all three TGC-S parental isolates (K88-S, K10-S, and K2-S) were similar (representing with similar ΔCT), there was no significant difference when acrB expression of in vitro selected mutants was compared with that of parental TGC-S or with the TGC-S clinical isolate HK10-S. The TGC-RKP isolates of animal and human origins displayed levels of acrB expression that were 5- to 62-fold and 2- to 26-fold higher than those of control HK10, respectively (Figure 1). These data support the hypothesis that increased expression of acrB is associated with increased MICs of TGC.
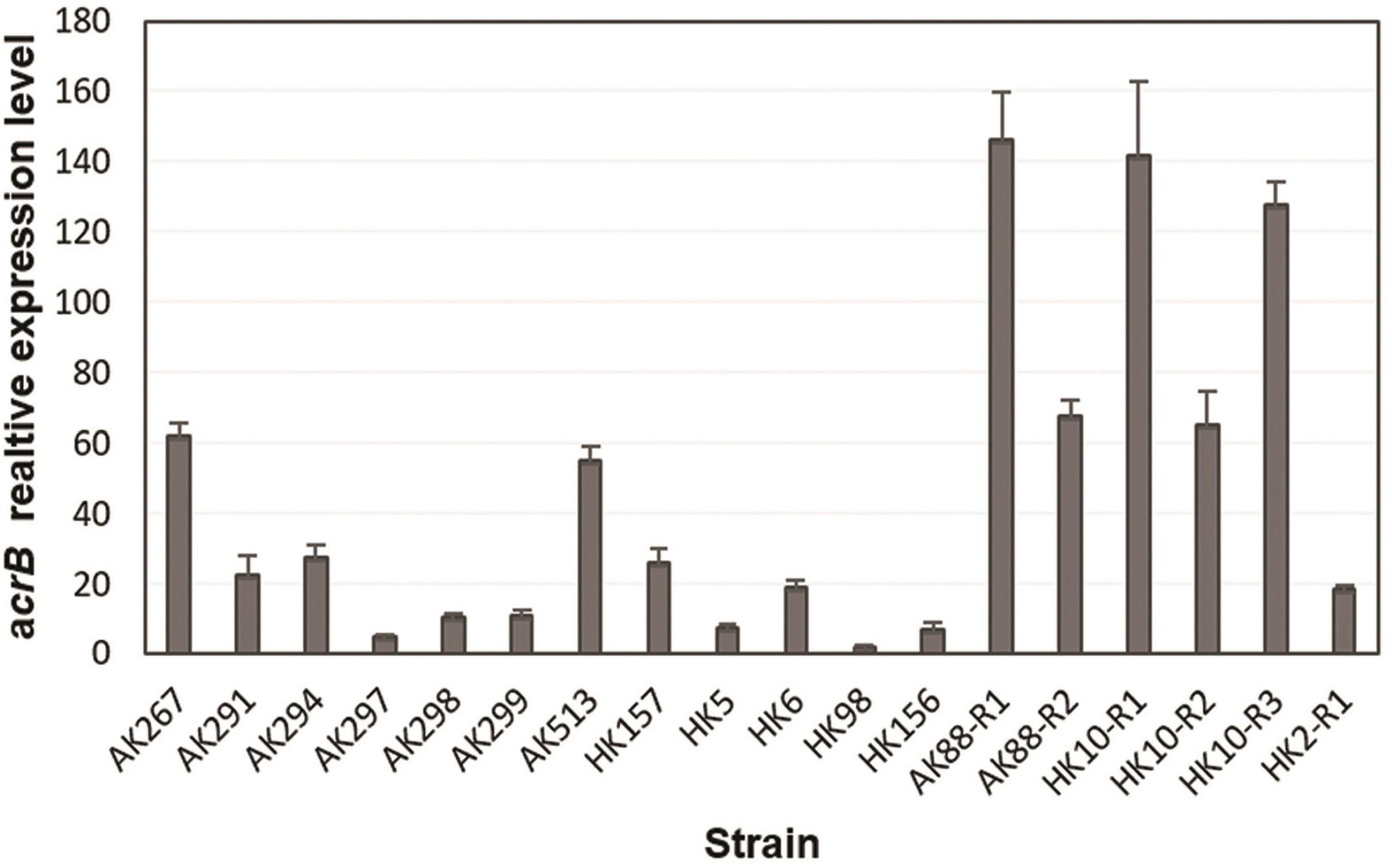
Figure 1. Relative expression of acrB gene from tigecycline (TGC)-non-susceptible isolates of Klebsiella pneumoniae from different origins determined by RT-qPCR.
Discussion
Multidrug-resistant isolates of K. pneumoniae have emerged in past years as results of wide application of antibacterial agents in both human and veterinary medicine. Indeed, consumption of human antibiotics in veterinary practices has been blamed for contributing to the magnitude of current antibiotic resistance crisis. Emergence of antibiotic resistance among commensal bacteria propagated in food animals, posing a great challenge for human health since they have the potential to reach human hosts through the food chain or transmit their mobile resistance elements to human pathogens (Founou et al., 2016). Resistance of K. pneumoniae to various classes of antibiotics can be mediated by a variety of mechanisms, among which extrusion of antibiotics using efflux machineries such as AcrAB has been well-studied. Due to its broad substrate range property, AcrAB pump has been found to mediate resistance to a different families of antibiotics such as penicillins, cephalosporins, fluoroquinolones, macrolides, chloramphenicol, and tetracyclines (Xu et al., 2021). In the current study, screening among commensal bacteria of broiler chickens revealed six TGC-RKP isolates that were also resistant to quinolones, chloramphenicol, nitrofurantoin, and other tetracyclines. There are currently no TGC-containing products authorized for the veterinary use in Iran, and it is likely that selective pressure caused by application of other antibiotics such as older tetracyclines, florfenicol, or enrofloxacin may contribute to TGC non-susceptibility in animal commensal bacterial isolates. In a recent study from China, five TGC-RKP isolates that all belonged to ST1 were identified from the fecal samples of healthy chickens. Resistance to TGC as well as other antibiotics in these isolates was found to be mediated by a novel plasmid-mediated RND efflux pump gene cluster, designated tmexCD1-toprJ1 (Lv et al., 2020). Li et al. also reported isolation of TGC-R Klebsiella aerogenes (TGC MIC = 32 mg/l) from the feces of a chicken farm, which contained the blaNDM–9 and tet(A) variant genes (Li et al., 2021). Analyzing the genetic relatedness of TGC-NSKP isolates of both origins using MLST revealed high genetic diversity among the isolates with 12 TGC-NSKP isolates belonging to 10 distinct STs, among which ST147 was found as a common type between two human and animal isolates. Most of the animal isolates belonged to STs, which are commonly reported from different human infections including ST11 (Gu et al., 2018), ST37 (Zhao et al., 2019), ST15, and ST147 (Yan et al., 2015; Chen et al., 2020). Co-resistance to TGC and colistin was observed among animal isolates belonging to ST11, ST37, and ST15. Other studies have also reported occurrence of TGC and colistin co-resistance among K. pneumoniae isolates belonging to ST37 and ST11 (Taniguchi et al., 2017; Xu et al., 2020). A recent study also reported detection of colistin and TGC-RKP (ST29) in municipal wastewater influents from Japan (Hayashi et al., 2021). All human TGC-NSKP isolates showed co-resistance to carbapenems and carried genes encoding for two different carbapenemases. The isolate HK157 belonging to ST2935 revealed co-resistance to three last-resort antibiotics and co-harbored a truncated mgrB gene, mutant ramR and acrR genes, and blaOXA–48 and blaNDM–5 carbapenemase genes. This is the first study reporting detection of blaNDM–5 among clinical CRKP isolates from Iran.
Analysis of the genes, AcrR (AcrAB repressor) and the RamR regulator, revealed inactivating genetic alterations in both proteins. In 83% of isolates (n = 15 out of 18) mutation/deletions in ramR (n = 8 isolates) or acrR (n = 4) or both (n = 3) was identified. The ramR was not detected in one isolate (AK294) using two pairs of primers used for amplification of the ramR gene, suggesting deletion of ramR locus in this isolate. In two animal isolates, RamR was truncated due to nonsense mutation, which resulted in production of 58 and 121 amino acid long proteins instead of wild-type protein with 193 amino acids. The RamR Q122 stop substitution has been previously reported among TGC-NSKP isolates by several other studies presenting this alteration as a common resistance mechanism among K. pneumoniae isolates (He et al., 2015; Chiu et al., 2017b; Li et al., 2017; Park et al., 2020). In addition to two previously reported RamR A19V and L44Q mutations (reported as L44R) (Chiu et al., 2017b), five novel amino acid substitutions, including I141T, G180D, A28T, R114L, and T119S, were identified, with the latter three changes being detected after resistance induction in in vitro selected mutants. The A19V substitution, which was also predicted by the PROVEAN tool to be a neutral change, has been previously demonstrated to have no effect on the TGC MICs (Chiu et al., 2017b). While RamR A28T, R114L, and G180D substitutions were predicted by the PROVEAN tool to have a deleterious impact on protein structure (PEOVEAN scores –3.6, –4, and –6.8, respectively), the I141T and T119S substitutions (with PEOVEAN scores 0.21 and –2.44 (very close to prediction cutoff = –2.5), respectively) were predicted to be neutral changes. An isolate-carrying RamR I141T substitution co-harbored an AcrR inactivating mutation (frameshift), indicating that AcrR alteration might be the main mediator of TGC non-susceptibility in this bacterium. However, the exact role of these novel mutations in elevation of TGC MICs requires further studies by confirmatory assays. In one in vitro selected mutant and a clinical TGC-R isolate nucleotide insertion (7nt) and deletion (12 nt) at position +205 was observed, respectively, indicating that this position is more prone to alterations upon resistance development. Among the 18 TGC-NSKP, mutations/deletions within AcrR were detected among seven isolates from which six carried a wild-type RamR or RamR substitutions, which were predicted or previously demonstrated to be neutral changes. The novel AcrR R90G substitution detected in two TGC-R animal isolates belonging to ST37 was predicted by the PROVEAN tool to have a deleterious effect on the functionality of the protein (PROVEAN score = –5.9). The AcrR frameshift mutation mediated by deletion of one of the guanines at positions 134–139 was detected in three TGC-NSKP isolates of both origins that belonged to different STs representing this region as a mutation-prone position within acrR gene. In two TGC-RKP isolates with overexpressed AcrB (AK297 and HK10-R2, TGC MIC = 16 mg/l), no alteration was identified in any of the studied genes, indicating that alterations in promoter region of the studied genes or other loci (marR or soxR; Bratu et al., 2009; Veleba and Schneiders, 2012) are probably involved in resistance development in these isolates. Gene expression analysis revealed an association between overexpression of AcrAB efflux pump and TGC non-susceptibility. However, there was no correlation between the TGC MICs and the level of acrB expression, as some isolates with similar MICs exhibited different expression levels. This suggests the possibility of contribution of other resistance mechanisms to increased TGC MICs.
Conclusion
A combination of genetic alterations in AcrR and RamR mediated the overexpression of AcrAB efflux pump and subsequently TGC non-susceptibility among the majority of human, animal, and in vitro selected mutants studied in this work. The most worrisome finding in our study was detection of multidrug-resistant isolates with co-resistance to two last-resort human antibiotics (TGC and colistin) belonging to sequence types commonly implicated in human infections (ST11, ST37, and ST15) among commensal bacteria of food animals. This can be considered a great threat to human health due to probability of transmission of these bacteria to humans through the food chain or direct contact. Acquiring carbapenem resistance among these isolates would be the most troublesome event. While the TGC exposure history among the human isolates was not clear, emergence of TGC resistance among animal isolates (which are not exposed to TGC) is an issue of great concern. It is speculated that selective pressure caused by other antimicrobials in particular tetracycline families (as one of the most used antimicrobials in food animals) may have contributed to increased TGC MICs in these isolates probably through the overexpression of AcrAB efflux pump. Therefore, immediate actions need to be taken to restrict/minimize the use of human antibiotics (at least not as growth promoters) in food animals to prevent the emergence and dissemination of antibiotic-resistant bacteria through the food chain.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: GenBank; accession numbers MW653710-MW653741.
Author Contributions
MM performed the experiments, analyzed the experiment data, and drafted the manuscript. MH and HM designed the experiments, analyzed the experiment data, and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the University of Tabriz.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Bartha, N. A., Sók, J., Urbán, E, and Nagy, E. (2011). Investigation of the prevalence of tetQ, tetX and tetX1 genes in Bacteroides strains with elevated tigecycline minimum inhibitory concentrations. Int. J. Antimicrob. Agents 38, 522–525. doi: 10.1016/j.ijantimicag.2011.07.010
Beabout, K., Hammerstrom, T. G., Perez, A. M., Magalhães, B. F., Prater, A. G., Clements, T. P., et al. (2015). The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob. Agents Chemother. 59, 5561–5566. doi: 10.1128/aac.00547-15
Bratu, S., Landman, D., George, A., Salvani, J., and Quale, J. (2009). Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J. Antimicrob. Chemother. 64, 278–283. doi: 10.1093/jac/dkp186
Cannatelli, A., D’Andrea, M. M., and Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 7, 5521–5526. doi: 10.1128/AAC.01480-13
Chen, D., Li, H., Zhao, Y., Qiu, Y., Xiao, L., He, H., et al. (2020). Characterization of carbapenem-resistant Klebsiella pneumoniae in a tertiary hospital in Fuzhou, China. J. Appl. Microbiol. 129, 1220–1226. doi: 10.1111/jam.14700
Chiu, S.-K., Chan, M.-C., Huang, L.-Y., Lin, Y.-T., Lin, J.-C., and Lu, P.-L. et al. (2017a). Tigecycline resistance among carbapenem-resistant Klebsiella pneumoniae: clinical characteristics and expression levels of efflux pump genes. PLoS One 12:e0175140. doi: 10.1371/journal.pone.0175140
Chiu, S.-K., Huang, L.-Y., Chen, H., Tsai, Y.-K., Liou, C.-H., Lin, J.-C., et al. (2017b). Roles of ramR and tet (A) mutations in conferring tigecycline resistance in carbapenem-resistant Klebsiella pneumoniae clinical isolates. Antimicrob. Agents Chemother. 61, e00391–17.
Choi, Y., and Chan, A. P. (2015). PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747. doi: 10.1093/bioinformatics/btv195
Deng, M., Zhu, M.-H., Li, J.-J., Bi, S., Sheng, Z.-K., Hu, F.-S., et al. (2014). Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of Acinetobacter baumannii from a Chinese university hospital. Antimicrob. Agents Chemother. 58, 297–303. doi: 10.1128/aac.01727-13
Diancourt, L., Passet, V., Verhoef, J., Grimont, P. A., and Brisse, S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43, 4178–4182. doi: 10.1128/jcm.43.8.4178-4182.2005
Du, X., He, F., Shi, Q., Zhao, F., Xu, J., Fu, Y., et al. (2018). The rapid emergence of tigecycline resistance in blaKPC–2 harboring Klebsiella pneumoniae, as mediated in vivo by mutation in tetA during tigecycline treatment. Front. Microbiol. 9:648. doi: 10.3389/fmicb.2018.00648
Founou, L. L., Founou, R. C., and Essack, S. Y. (2016). Antibiotic resistance in the food chain: a developing country-perspective. Front. Microbiol. 7:1881. doi: 10.3389/fmicb.2016.01881
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/s1473-3099(17)30489-9
Haeili, M., Javani, A., Moradi, J., Jafari, Z., Feizabadi, M. M., and Babaei, E. (2017). MgrB alterations mediate colistin resistance in Klebsiella pneumoniae isolates from Iran. Front. Microbiol. 8:2470. doi: 10.3389/fmicb.2017.02470
Hayashi, W., Iimura, M., Soga, E., Koide, S., Izumi, K., Yoshida, S., et al. (2021). Presence of colistin-and tigecycline-resistant Klebsiella pneumoniae ST29 in municipal wastewater influents in Japan. Microb. Drug Resist. doi: 10.1089/mdr.2020.0514 [Epub ahead of print].
He, F., Fu, Y., Chen, Q., Ruan, Z., Hua, X., Zhou, H., et al. (2015). Tigecycline susceptibility and the role of efflux pumps in tigecycline resistance in KPC-producing Klebsiella pneumoniae. PLoS One 10:e0119064. doi: 10.1371/journal.pone.0119064
Hentschke, M., Wolters, M., Sobottka, I., Rohde, H., and Aepfelbacher, M. (2010). ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrob. Agents Chemother. 54, 2720–2723. doi: 10.1128/aac.00085-10
Jafari, Z., Harati, A. A., Haeili, M., Kardan-Yamchi, J., Jafari, S., Jabalameli, F., et al. (2019). Molecular epidemiology and drug resistance pattern of carbapenem-resistant Klebsiella pneumoniae isolates from Iran. Microb. Drug Resist. 25, 336–343.
Li, R., Han, Y., Zhou, Y., Du, Z., Wu, H., Wang, J., et al. (2017). Tigecycline susceptibility and molecular resistance mechanisms among clinical Klebsiella pneumoniae strains isolated during non-tigecycline treatment. Microb. Drug Resist. 23, 139–146. doi: 10.1089/mdr.2015.0258
Li, Y., Wang, Q., Xiao, X., Li, R., and Wang, Z. (2021). Emergence of blaNDM-9-bearing tigecycline resistant Klebsiella aerogenes of chicken origin. J. Glob. Antimicrob. Resist. 26, 66–68. doi: 10.1016/j.jgar.2021.04.028
Livermore, D. M. (2005). Tigecycline: what is it, and where should it be used? J. Antimicrob. Chemother. 56, 611–614. doi: 10.1093/jac/dki291
Lv, L., Wan, M., Wang, C., Gao, X., Yang, Q., Partridge, S. R., et al. (2020). Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in Klebsiella pneumoniae. mBio 11, e02930–19.
Mahon, C. R., Lehman, D. C., and Manuselis, G. (2018). Textbook of Diagnostic Microbiology-E-Book. Amsterdam: Elsevier Health Sciences.
Moore, I. F., Hughes, D. W., and Wright, G. D. (2005). Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry 44, 11829–11835. doi: 10.1021/bi0506066
Park, Y., Choi, Q., Kwon, G. C., and Koo, S. H. (2020). Molecular epidemiology and mechanisms of tigecycline resistance in carbapenem-resistant Klebsiella pneumoniae isolates. J. Clin. Lab. Anal. 34:e23506.
Pishnian, Z., Haeili, M., and Feizi, A. (2019). Prevalence and molecular determinants of colistin resistance among commensal Enterobacteriaceae isolated from poultry in northwest of Iran. Gut pathog. 11, 1–8.
Pitout, J. D., Nordmann, P., and Poirel, L. (2015). Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 59, 5873–5884. doi: 10.1128/aac.01019-15
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Renteria, M., Biedenbach, D., Bouchillon, S., Hoban, D., Raghubir, N., and Sajben, P. (2014). In vitro activity of tigecycline and comparators against carbapenem-resistant Enterobacteriaceae in Africa–Middle East countries: TEST 2007–2012. J. Glob. Antimicrob. Resist. 2, 179–182. doi: 10.1016/j.jgar.2014.03.002
Rosenblum, R., Khan, E., Gonzalez, G., Hasan, R., and Schneiders, T. (2011). Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae. Int. J. Antimicrob. Agents 38, 39–45. doi: 10.1016/j.ijantimicag.2011.02.012
Sader, H. S., Castanheira, M., Flamm, R. K., Mendes, R. E., Farrell, D. J., and Jones, R. N. (2015). Tigecycline activity tested against carbapenem-resistant Enterobacteriaceae from 18 European nations: results from the SENTRY surveillance program (2010–2013). Diagn. Microbiol. Infect. Dis. 83, 183–186. doi: 10.1016/j.diagmicrobio.2015.06.011
Spanu, T., De Angelis, G., Cipriani, M., Pedruzzi, B., D’Inzeo, T., Cataldo, M. A., et al. (2012). In vivo emergence of tigecycline resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli. Antimicrob. Agents Chemother. 56, 4516–4518.
Taniguchi, Y., Maeyama, Y., Ohsaki, Y., Hayashi, W., Osaka, S., Koide, S., et al. (2017). Co-resistance to colistin and tigecycline by disrupting mgrB and ramR with IS insertions in a canine Klebsiella pneumoniae ST37 isolate producing SHV-12, DHA-1 and FosA3. Int. J. Antimicrob. Agents 50, 697–698. doi: 10.1016/j.ijantimicag.2017.09.011
Van, T. T. H., Yidana, Z., Smooker, P. M., and Coloe, P. J. (2020). Antibiotic use in food animals worldwide, with a focus on Africa: pluses and minuses. J. Glob. Antimicrob. Resist. 20, 170–177. doi: 10.1016/j.jgar.2019.07.031
Veleba, M., and Schneiders, T. (2012). Tigecycline resistance can occur independently of the ramA gene in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 56, 4466–4467. doi: 10.1128/aac.06224-11
Xu, J., Zhao, Z., Ge, Y., and He, F. (2020). Rapid emergence of a pandrug-resistant Klebsiella pneumoniae ST11 isolate in an inpatient in a teaching hospital in China after treatment with multiple broad-spectrum antibiotics. Infect. Drug Resist. 13, 799–804. doi: 10.2147/idr.s243334
Xu, Q., Sheng, Z., Hao, M., Jiang, J., Ye, M., Chen, Y., et al. (2021). RamA upregulates multidrug resistance efflux pumps AcrAB and OqxAB in Klebsiella pneumoniae. Int. J. Antimicrob. Agents 57:106251. doi: 10.1016/j.ijantimicag.2020.106251
Yan, J., Wang, M., Zheng, P., Tsai, L., and Wu, J. (2015). Associations of the major international high-risk resistant clones and virulent clones with specific ompK36 allele groups in Klebsiella pneumoniae in Taiwan. New Microbes New Infect. 5, 1–4. doi: 10.1016/j.nmni.2015.01.002
Keywords: tigecycline resistance, Klebsiella pneumoniae, food animals, ramR, AcrR, AcrAB efflux pump
Citation: Moghimi M, Haeili M and Mohajjel Shoja H (2021) Characterization of Tigecycline Resistance Among Tigecycline Non-susceptible Klebsiella pneumoniae Isolates From Humans, Food-Producing Animals, and in vitro Selection Assay. Front. Microbiol. 12:702006. doi: 10.3389/fmicb.2021.702006
Received: 28 April 2021; Accepted: 09 July 2021;
Published: 05 August 2021.
Edited by:
Renee Lay Hong Lim, UCSI University, MalaysiaReviewed by:
Qixia Luo, Zhejiang University, ChinaJon William Weeks, United States Food and Drug Administration, United States
Copyright © 2021 Moghimi, Haeili and Mohajjel Shoja. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mehri Haeili, m.haeili@tabrizu.ac.ir; m.haeili@yahoo.com; Hanieh Mohajjel Shoja, mohajelh@yahoo.com
 Mohaddeseh Moghimi1
Mohaddeseh Moghimi1 Mehri Haeili
Mehri Haeili