
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 20 July 2021
Sec. Microbial Symbioses
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.701081
This article is part of the Research Topic Nutrition and Behavior as Determinants of Host-associated Microbiomes View all 11 articles
 Rajaraman Bharanidharan1
Rajaraman Bharanidharan1 Chang Hyun Lee2
Chang Hyun Lee2 Krishnaraj Thirugnanasambantham3,4
Krishnaraj Thirugnanasambantham3,4 Ridha Ibidhi3
Ridha Ibidhi3 Yang Won Woo5
Yang Won Woo5 Hong-Gu Lee6
Hong-Gu Lee6 Jong Geun Kim3,7
Jong Geun Kim3,7 Kyoung Hoon Kim3,7*
Kyoung Hoon Kim3,7*Our previous research revealed the advantages of separate feeding (SF) systems compared to total mixed ration (TMR) in terms of ruminal methane (CH4) production. The purpose of this experiment was to confirm the advantage of SF as a nutritional strategy for CH4 mitigation, and to determine the effects of different feeding systems (TMR and SF) on the rumen microbiome and associated metagenome of two different breeds and on CH4 emissions. We randomly allocated four Holstein (305 ± 29 kg) and four Hanwoo steers (292 ± 24 kg) to two groups; the steers were fed a commercial concentrate with tall fescue (75:25) as TMR or SF, in a crossover design (two successive 22-day periods). Neither feeding systems nor cattle breeds had an effect on the total tract digestibility of nutrients. The TMR feeding system and Hanwoo steers generated significantly more CH4 (P < 0.05) and had a higher yield [g/d and g/kg dry matter intake (DMI)] compared to the SF system and Holstein steers. A larger rumen acetate:propionate ratio was observed for the TMR than the SF diet (P < 0.05), and for Hanwoo than Holstein steers (P < 0.001), clearly reflecting a shift in the ruminal H2 sink toward CH4 production. The linear discriminant analysis (LDA) effect size (LEfSe) revealed a greater abundance (α < 0.05 and LDA > 2.0) of operational taxonomic units (OTUs) related to methanogenesis for Hanwoo steers compared to Holstein steers. Kendall’s correlation analysis revealed wide variation of microbial co-occurrence patterns between feeding systems, indicating differential H2 thermodynamics in the rumen. A metagenome analysis of rumen microbes revealed the presence of 430 differentially expressed genes, among which 17 and 27 genes exhibited positive and negative associations with CH4 production, respectively (P < 0.001). A strong interaction between feeding system and breed was observed for microbial and metagenomic abundance. Overall, these results suggest that the TMR feeding system produces more CH4, and that Hanwoo cattle are higher CH4 emitters than SF diet and Holstein cattle, respectively. Interestingly, host-associated microbial interactions differed within each breed depending on the feeding system, which indicated that breed-specific feeding systems should be taken into account for farm management.
Some dietary interventions have the potential to reduce ruminal CH4 emissions with little negative impact on the animals. One strategy involves the use of different cattle feeding systems, such as separate feeding (SF) and total mixed ration (TMR) feeding with concentrate and forage. A review by Beigh et al. (2017) also clarified the advantages of TMR over conventional SF systems in ruminants. Compared to SF, TMR has a stabilising effect on rumen pH, and also has positive effects on dry matter intake (DMI), milk fat content (Phipps et al., 1984), nutrient use efficiency (Lailer et al., 2005), total milk production (Reddy and Reddy, 1983), and microbial protein synthesis (Liu et al., 2015) in dairy cattle. TMR feeding was also reported to have various advantages in beef production, in terms of precision, efficiency and convenience, which are believed to improve overall on-farm productivity (Moya et al., 2014). The practice of TMR feeding in beef cattle production has increased gradually, and is now used in about 20% of beef cattle farms in Korea. This has led to an increase in research on its effects on ruminal fermentation, animal performance and carcass quality in Hanwoo (Korean native cattle) steers (Lee et al., 2010; Kim et al., 2011; Chung et al., 2017). However, the effects of the TMR and SF systems on ruminal CH4 mitigation have not been studied extensively in either dairy or beef cattle. One early study compared TMR and SF silage-based diets for maintenance-level feeding of lactating Holstein cows, and found no differences in CH4 emissions between the feeding systems (Holter et al., 1977). Our previous studies of Holstein steers, with an average daily gain (ADG) of 0.65 kg, revealed higher CH4 emissions with the TMR than SF system (Lee et al., 2016; Bharanidharan et al., 2018). However, further studies are needed to validate the effects of these feeding systems on CH4 production in beef cattle.
The extensive livestock production system in Korea has contributed to an increase in atmospheric CH4 concentrations, with a reported 3.89 MtCO2 eq/y derived from enteric fermentation; this accounted for 50% of total CH4 emissions (7.81 MtCO2 eq/y) in Korea from the agricultural sector in 2018 (FAOSTAT, 2021). In the first quarter of 2021, Korea had around 3.33 million beef cattle, with Hanwoo accounting for 3.16 million head and Holstein accounting for the remainder (Korean Statistical Information Service (KOSIS), 2021). Studies have suggested that host breeds provide different environments for the microbial ecosystem in the rumen, and may influence the microbial composition and CH4 emissions (Duthie et al., 2017; De Mulder et al., 2018; Olijhoek et al., 2018; Islam et al., 2021). A comparison of the whole-genome sequence between Hanwoo and Holstein steers also revealed huge differences in the copy number variation regions (CNVR) between the two breeds (Choi et al., 2014), which could influence the rumen microbial composition (Benson et al., 2010). Furthermore, studies have provided direct evidence of the genetic influence of the host animal on CH4 production by ruminal microbes, thereby allowing the breeding of low-emitting animals through genetic selection (Roehe et al., 2016; Zhang et al., 2020). However, there have been no studies directly comparing CH4 emissions and rumen microbiomes between the Hanwoo and Holstein breeds with the diet held constant.
A better understanding of the contributions of Korean indigenous breeds to the microbial metagenome and variation in CH4 emissions will enable identification of metagenomic markers and the development of an optimal feeding system, both of which will reduce ruminal emissions. The present study was performed to clarify the effects of different feeding systems (TMR and SF), as well as differences in metabolites and microbiota, between the rumens of Hanwoo and Holstein steers, and their interactions with CH4 emissions, fermentation characteristics and the microbial metagenome using next-generation sequencing.
All experimental animals were obtained from the animal farm of Seoul National University. The methods and protocols for all experiments involving animals were approved by the Committee for the Institutional Animal Care and Use of Seoul National University (SNU-160105-1), and performed in accordance with relevant guidelines and regulations.
Four Holstein and four Hanwoo steers [initial body weights (BWs) of 305 ± 29 and 292 ± 24 kg, respectively] were allocated to two adjacent pens fitted with Calan doors. Four steers of each breed were offered commercial concentrate and tall fescue hay (75:25, on a DM basis) as either TMR or SF for two consecutive 22-day periods in a crossover design. All animals were adapted for 30 days to the experimental diet (TMR), and to the Calan doors, before the experiment. Each experimental period included 14 days of diet adaptation in the pen, 3 days of metabolic adaptation in a respiratory chamber, 2 days of CH4 measurements, 2 days of grab faecal sampling and 1 day of rumen fluid sampling. The diets (Table 1) were provided at 2.1% BW twice daily (at 09:00 and 18:00) and the animals had unrestricted access to water in both the pen and respiratory chamber. The DM intake was recorded daily, both in the pen and in the chamber. The TMR diet was prepared by blending commercial concentrate and chopped tall fescue hay every 7 days during the experimental period. The prepared TMR diet was then packed in vinyl bags in quantities of 20 kg and stored for feeding to the animals. In the SF system, tall fescue hay was provided first, followed by concentrate pellet with flaked corn 40 min later, to avoid an unnecessary drop in pH during initial ruminal fermentation compared to the TMR diet. The BW of the animals was measured at both the beginning and end of each period to determine the ADG. The offered feed samples were collected at the beginning of the experimental period. The refused feed samples were collected daily when the animals were placed in the chamber. Both samples were packed and stored at −20°C until being analysed for DM content and the composition of other chemicals as described previously (Bharanidharan et al., 2018). The particle size of the offered feed in both the TMR and SF diets was determined using a Penn State particle separator according to the technique of Kononoff et al. (2003).
Methane production was measured using four open-circuit whole-body respiratory chambers, as described previously (Bharanidharan et al., 2018), with a ventilation rate of 700 L/min maintained throughout the experiment. The apparent total tract digestibility of nutrients in steers was studied using chromic oxide (Cr2O3) as an external marker, which was top-dressed twice daily at 0.2% of the daily feed amount throughout the experiment. On day 20, a faecal grab sample (100 g fresh weight) was collected from the rectum of each animal 30 min before feeding, and at 1, 3, 5, and 7 h post-feeding. On day 21, faeces were collected 30 min before feeding, and at 2, 4, 6, and 8 h post-feeding. Samples were frozen at −20°C until analysis. The samples were then composited by day and period for each steer prior to analysis of the digestibility of nutrients, as described previously (Bharanidharan et al., 2018). Ruminal fluid samples were collected before feeding (0 h), and at 1.5 and 3 h post-feeding, on day 22 of each period using a stomach tube (Oriental Dream, Hwaseong, Korea). The samples were filtered through four layers of muslin cloth. After immediately measuring pH using a Seven Easy pH meter (Mettler-Toledo, Schwerzenbach, Switzerland), ruminal fluid was centrifuged at 12,000 × g for 10 min (Smart 15; Hanil Science Industrial, Gimpo, South Korea). The supernatant was transferred to a 50-mL centrifuge tube and stored at −20°C for determination of ammonia-nitrogen (NH3-N) and volatile fatty acid (VFA) concentrations using the methods described previously (Bharanidharan et al., 2018). For microbial analysis, rumen fluid samples collected 3-h post-feeding were snap frozen in liquid nitrogen and then stored at −80°C until DNA extraction.
The genomic DNA was extracted from 16 rumen samples (four per feeding system per breed) and a V4 sequencing library was constructed and sequenced for paired-end 250-bp reads using the Illumina MiSeq system (Illumina, San Diego, CA, United States). The primers and methods were described previously (Bharanidharan et al., 2018). The raw Illumina MiSeq reads were demultiplexed according to the barcodes and the sequences were quality-filtered based on the quality control process of Quantitative Insights Into Microbial Ecology (QIIME, version 1.9.0)1. The quality control conditions were as follows: read truncation: the raw read was truncated from the first low-quality base site with a maximum of three consecutive low-quality base calls (Phred Q < 20) allowed; and length filtering: to delete reads of continuous high quality (Phred Q ≥ 20), with a base length <70% of the read length. Non-overlapping regions, chimeric sequences and singletons were discarded. Each read was screened for operational taxonomic unit (OTU) picking using UCLUST embedded within QIIME 1.9.0, with reference to the Greengenes database (gg_otus-13_8-release, 97% nucleotide identity). The resulting OTU table was rarefied across samples to a depth of 10,000 reads based on the mean values of 10 iterations using the QIIME platform. All statistical analyses were performed on samples obtained from the same depth. Microbial community diversity was estimated in terms of the Chao1, Shannon, and Simpson indices using PAST software (Hammer et al., 2001). Determination of core and unique taxa was accomplished based on the abundance and prevalence (two-parameter) cut-offs across all the samples, using the inbuilt algorithm in Calypso software (Zakrzewski et al., 2017). Any taxa found to be ubiquitous (>90% occurrence across all samples) were defined as “core taxa” in the rumen, whereas those prevalent only in particular sample groups were defined as “unique taxa.” To identify bacterial lineages and other parameters that differentiated Hanwoo and Holstein cattle given the TMR or SF diet, we performed a principal component analysis (PCA) in the R software environment (R Development Core Team, Vienna, Austria) using the prcomp package; the results were visualised three-dimensionally using the pca3d package (Weiner, 2019). To identify robust microbial biomarkers for the two breeds, we calculated the linear discriminant analysis (LDA) effect size (LEfSe; Segata et al., 2011) using the Galaxy web application2. The OTU counts in each sample were used as the input for the LEfSe analysis. Differences among classes were determined using the Kruskal–Wallis test, at a significance level of P < 0.05 and with a threshold LDA score of 2.0. Feeding system was included as a subclass. The LEfSe subclasses were analysed using the Wilcoxon rank sum test, with P < 0.05 again taken to indicate significance. Relationships among microbes in the rumens of Hanwoo and Holstein steers given the TMR and SF diets were evaluated using Kendall’s correlation analysis, and a correlation plot was constructed using PAST software. To predict the molecular functions of each sample based on 16S rRNA data, we used the online version of the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) bioinformatics software package (Langille et al., 2013), hosted on the Galaxy platform2. We used the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database (Kanehisa and Goto, 2000) to predict the metagenome of our samples, based on OTU data obtained from rarefied 16S rRNA gene sequences using PICRUSt. The functions and products of the genes of interest were also obtained from the KEGG database. Differences between breeds given the TMR or SF diet in terms of the predicted molecular functions of bacterial communities were determined using a canonical correspondence analysis (CCA) in Calypso software. The non-parametric Kendall rank correlation coefficient was calculated to test the correlations among CH4 production, fermentation characteristics, bacterial communities, gene abundance and functional pathways in the rumen, using the corr.test function of the R psych package (Revelle, 2018). The resulting correlation matrix was visualised as a heatmap using the heatmap.2 function in the gplots R package (Warnes et al., 2015).
Daily CH4 emission, total tract digestibility, microbial diversity, gene abundance and functional abundance data were analysed using the MIXED procedure in SAS software (SAS Institute, Cary, NC, United States). The fixed effects in the model included breed, feeding system; their interaction effect was also analysed. Random effects, including period and animal, were nested within treatments. Ruminal fermentation characteristics were subjected to repeated measures analysis using the SAS PROC MIXED function (Littell et al., 1998). Appropriate covariance structures were obtained based on the Akaike information criterion. Means were calculated using the LSMEANS function, where each animal was considered as an experimental unit. P < 0.05 was taken to indicate significant differences according to treatment, and 0.05 < P < 0.1 was considered to indicate a trend toward significance.
The TMR diet had a higher percentage (99.5%) of particles <1.18 mm in size than the SF diet, whereas the SF diet had a higher percentage (72%) of particles in the size range of 19–8 mm than the TMR diet (P < 0.05) (Table 2). However, DM and organic matter (OM) intake did not vary between feeding systems (Table 3). Similarly, no feed sorting was observed in the SF diet. Holstein steers had higher DMI than Hanwoo steers in the chamber (Table 3). In addition, there were significant (P < 0.05) interactions between breed and feeding system for DM, OM, and gross energy (GE) intake (Table 3) in the chamber. Holstein steers had a greater (P < 0.05) ADG than Hanwoo steers (Table 3). Although there was no effect on apparent total tract nutrient digestibility (P > 0.05; Table 3), TMR feeding resulted in greater CH4 production (g/day) and CH4 yield per unit of OM, neutral detergent fibre and GE intake than the SF diet in both Holstein and Hanwoo steers (P < 0.05; Table 4). Hanwoo steers had a higher CH4 yield than Holstein steers (P < 0.005).
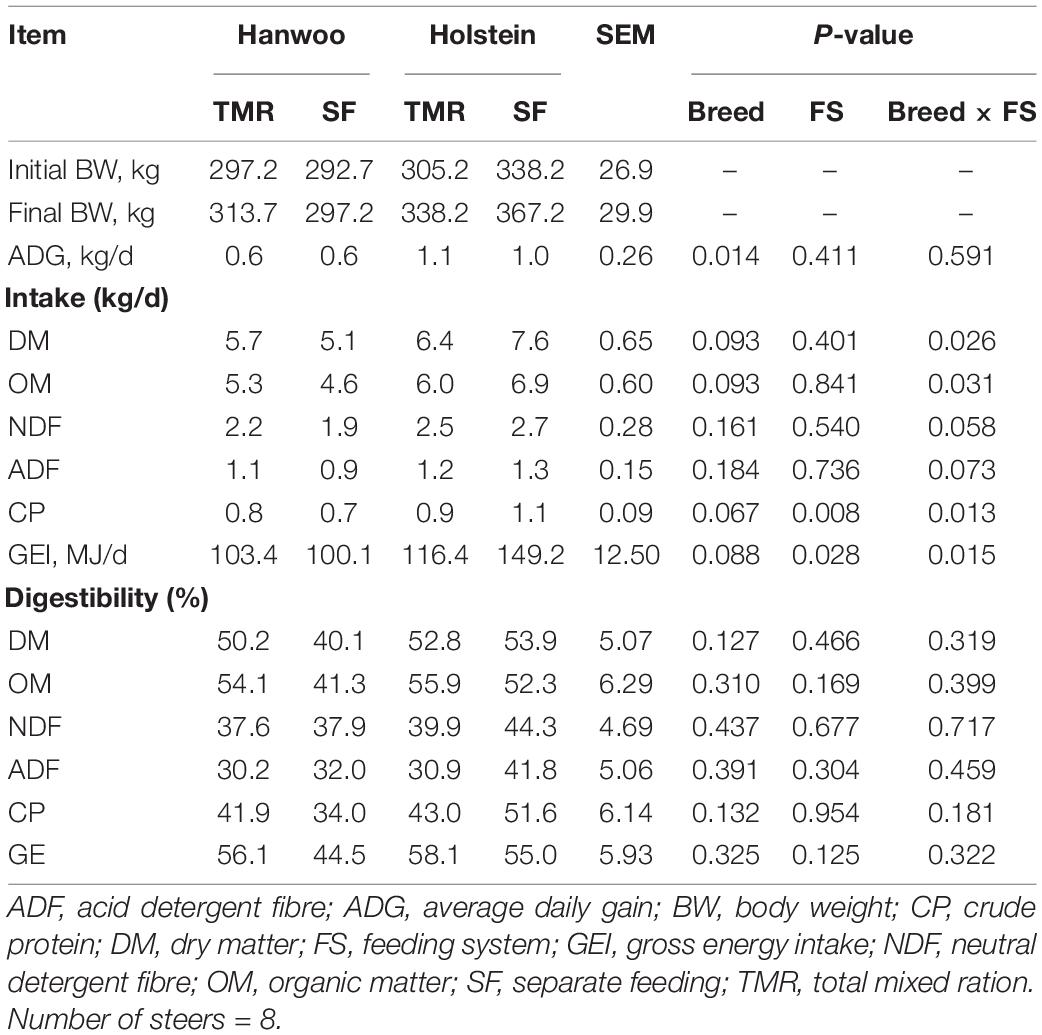
Table 3. Least square means of body weight, nutrient intake and apparent total tract digestibility (%) of Hanwoo and Holstein steers fed by the TMR and SF systems.
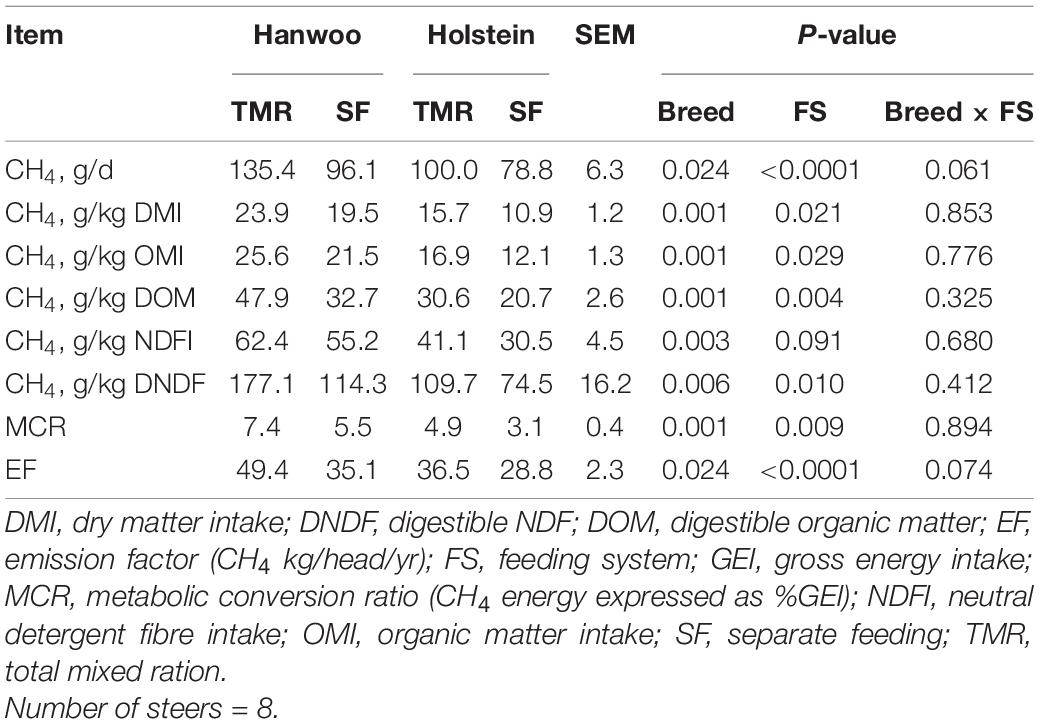
Table 4. Least square means of methane production recorded in Hanwoo and Holstein steers fed with TMR and SF diets over 24 h.
The mean concentration of total ruminal VFA did not differ between the experimental treatments (P > 0.05; Table 5). The steers given the TMR diet had a lower ruminal pH than those given the SF diet (P = 0.069), and the proportion of acetate was higher in the rumen fluid of steers given the TMR diet (P < 0.01). The proportions of butyrate and isobutyric acid did not vary between feeding systems (P > 0.05), whereas the proportions of valerate and isovaleric acid were lower in steers given the TMR than SF diet (P = 0.001). Hanwoo steers had a lower propionate and higher butyrate content than Holstein steers (P < 0.001). Similarly, the ratio of acetate to propionate in Hanwoo steers was greater than that in Holstein steers (P < 0.05). Ruminal NH3-N concentrations did not vary significantly between feeding systems or breeds.
At a depth of 10,000 quality reads, an average of 806 OTUs (range: 503–907 OTUs) were generated (Supplementary Figure 1). There were no differences (P > 0.05) in richness or diversity indices between the feeding systems. Differences in alpha diversity metrics, including observed OTUs (P < 0.05), Chao1 index (P = 0.083) and Shannon index (P < 0.05), were detected between breeds, demonstrating greater richness and diversity in Hanwoo steers (Figure 1 and Supplementary Table 1).
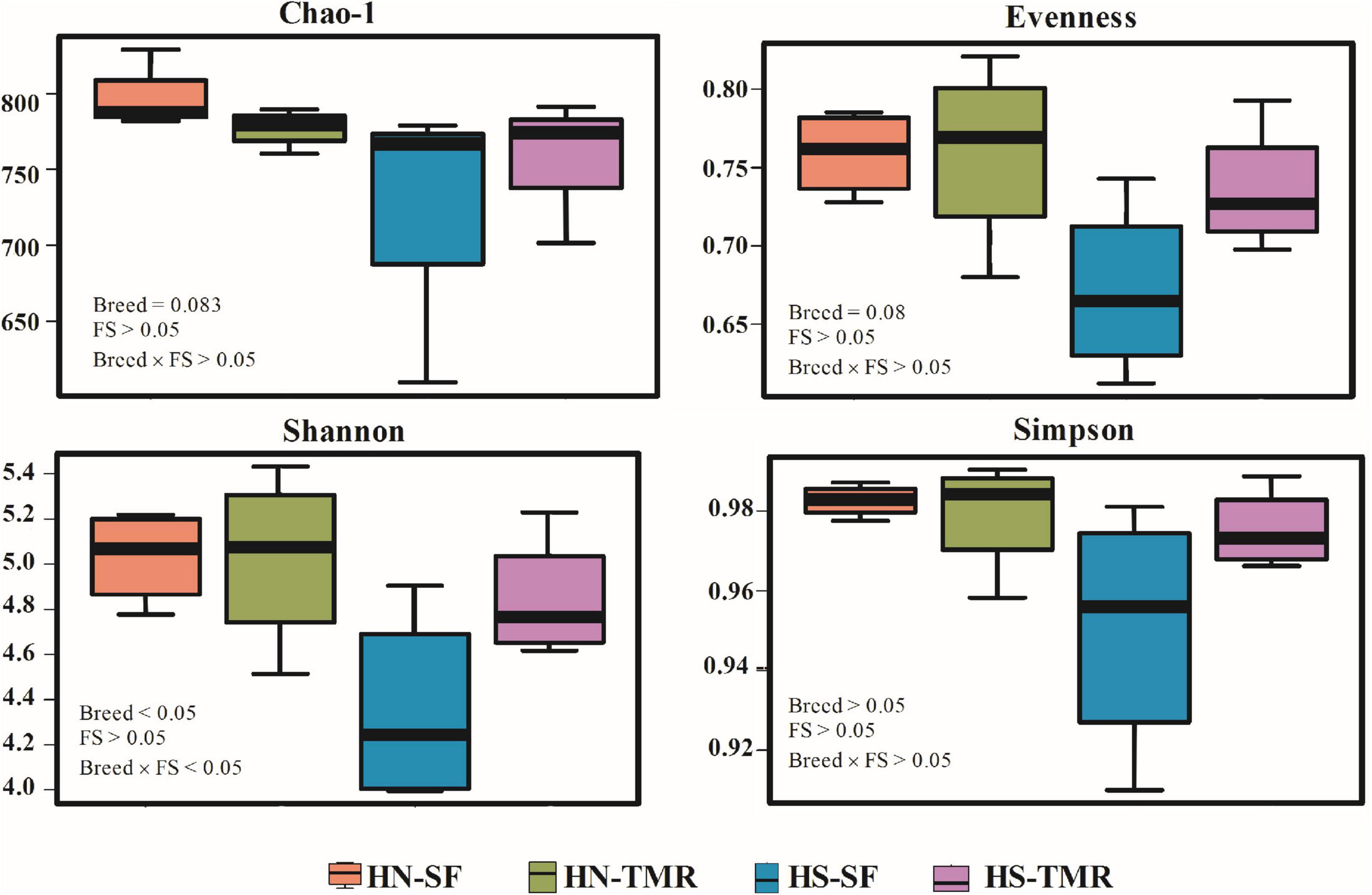
Figure 1. Alpha diversity metrics of the bacterial/archaeal composition in the rumen of Hanwoo (HN) and Holstein (HS) steers fed a total mixed ration (TMR) or separate feed (SF) diet.
The relative abundance of the bacterial genus Prevotella (18.6–33.2%) was high in all samples (Figure 2 and Supplementary Table 2). Similarly, the archaeal genus Methanobrevibacter (0.5–1.3%) was also abundant. The core microbiome across the different feeding systems and cattle breeds used in this study consisted of 49 identified and 37 unclassified genera, which together accounted for 98.74% of the total rumen microbial population (Supplementary Figure 2). When individual groups were analysed for core taxa, Holstein steers given the SF diet were found to possess a small, distinctive core microbiome, including three additional genera (Acidaminococcus, Aequorivita, and B42), which were found to be unique (Supplementary Table 3). Similarly, Acholeplasmatales unclassified and Megasphaera were identified as being unique to the Hanwoo steers and SF system, respectively.
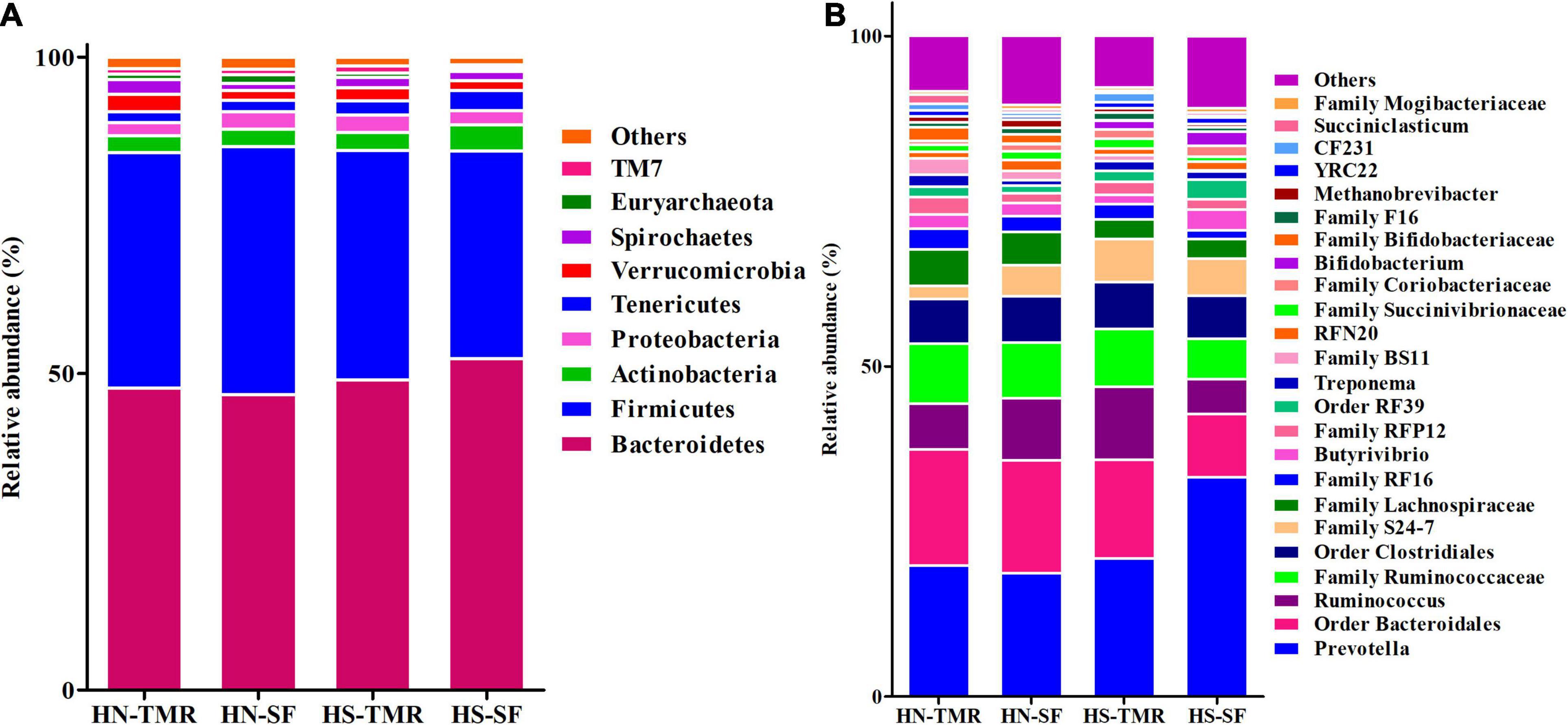
Figure 2. Taxonomic profiles of the relative phylum-level (A) and genus-level (B) abundances of bacteria and archaea in both Hanwoo (HN) and Holstein (HS) steers fed by different feeding systems, with a classification criterion of >0.5% of the total sequence.
The LEfSe analysis identified several OTUs that differed significantly between breeds (P < 0.05 and LDA > 2.0), indicating that they were robust biomarkers (Figure 3). The OTUs in the kingdom Archaea, phylum Euryarchaeota, families Methanomassiliicoccaceae and Methanobacteriaceae and the genera vadinCA11 and Methanobrevibacter were significantly more enriched in Hanwoo steers than Holstein steers. Several other families, including Spirochaetaceae, BS11 and Lachnospiraceae, were also identified as biomarkers in Hanwoo steers. A PCA biplot clustered all samples into two groups to explore the correlations with mean taxonomic annotation, CH4 yield and other fermentation parameters (Supplementary Figure 3).
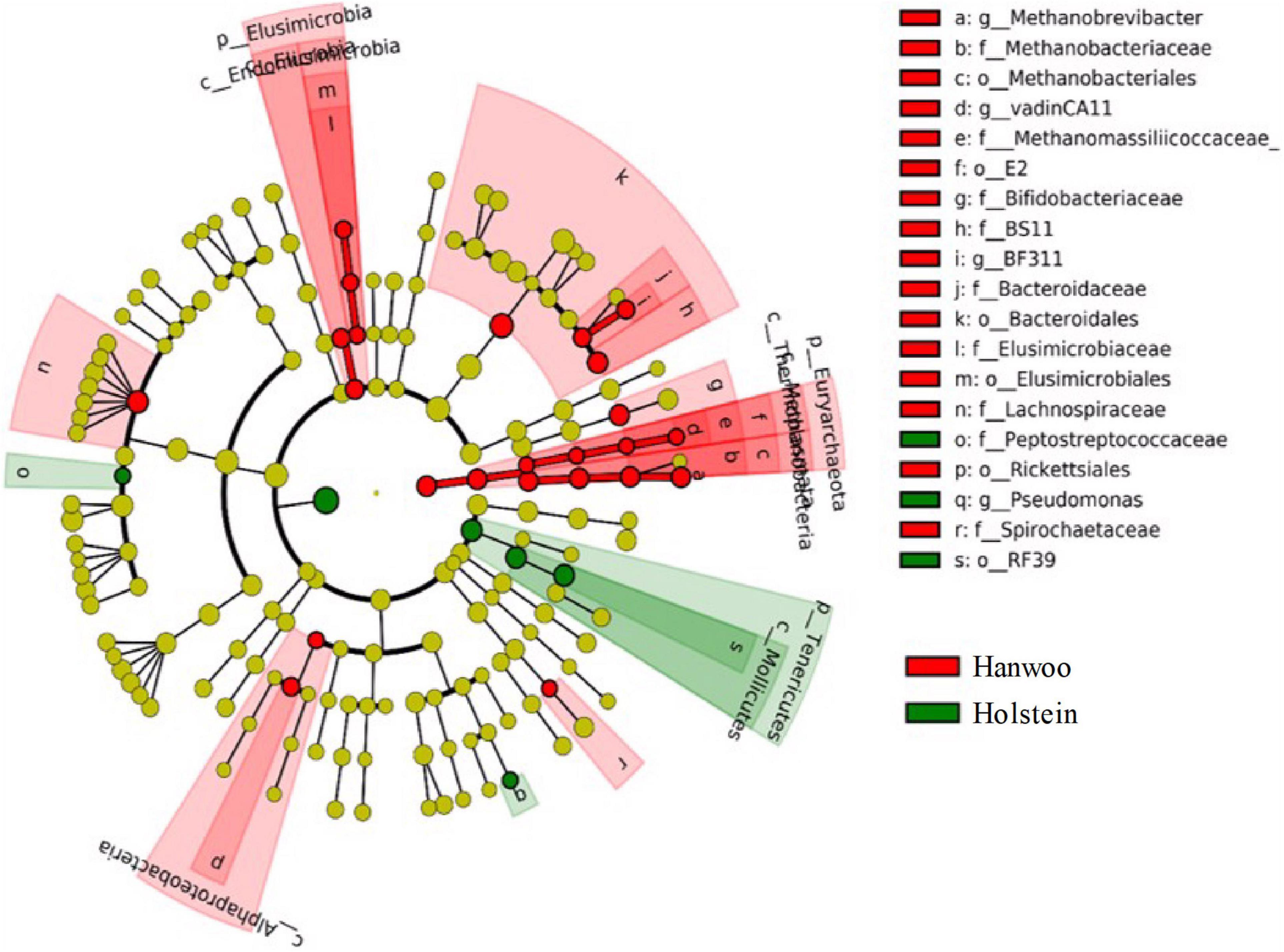
Figure 3. LDA effect size (LEfSe) cladogram demonstrating taxonomic differences between the Hanwoo and Holstein breeds. Taxa enriched in the rumen of Holstein steers are indicated by a positive LDA score (green), while taxa enriched in the rumen of Hanwoo steers have a negative score (red). Only taxa meeting an LDA significance threshold of 2 and P < 0.05 are shown.
Both feeding system and breed were found to have a significant effect on microbial composition at the phylum and genus levels (P < 0.05; Table 6). The feeding system × breed interaction effect on the rumen microbiome in relation to methanogenesis was significant (P < 0.05). Notably, the TMR system increased the archaea/bacteria ratio and abundances of the phylum Euryarchaeota and genus Methanobrevibacter in Hanwoo steers to a greater extent than in Holstein steers.
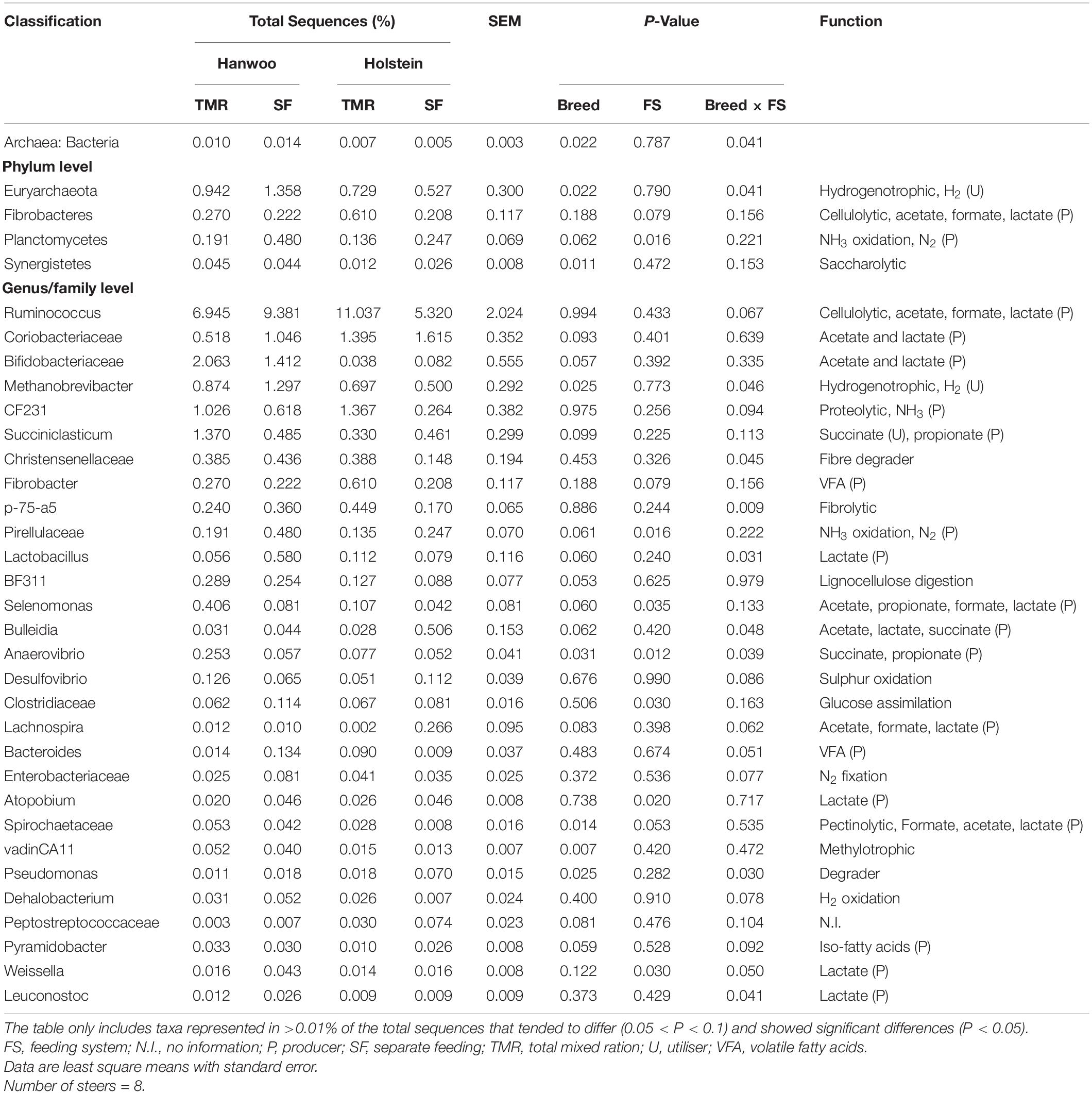
Table 6. Relative abundance of taxa in the Hanwoo and Holstein steers fed by different feeding systems.
Strong associations of hydrogenotrophic Methanobrevibacter and methylotrophic Methanosphaera with Butyrivibrio were detected in Holstein steers given the TMR diet (τ = 1, P < 0.05; Figure 4B). Strong associations were also detected between vadinCA11 and Ruminococcus (τ = 1, P < 0.05) in TMR-fed Hanwoo steers (Figure 4A), and between pectinolytic Sphaerochaeta and vadinCA11 (τ = 1, P < 0.05) in the TMR diet Holstein steers (Figure 4B). In contrast, in the SF Holstein steers (Figure 4D), we detected trends toward negative associations of Methanobrevibacter and vadinCA11 with Anaerostipes, Coprococcus, Anaerovibrio, Prevotella, Pyramidobacter, and Succiniclasticum (τ = −0.91, P = 0.063). In SF Hanwoo steers (Figure 4C), negative associations were observed between Methanobrevibacter and Sharpea, and between Anaerostipes and Butyrivibrio (τ = −1, P < 0.05).
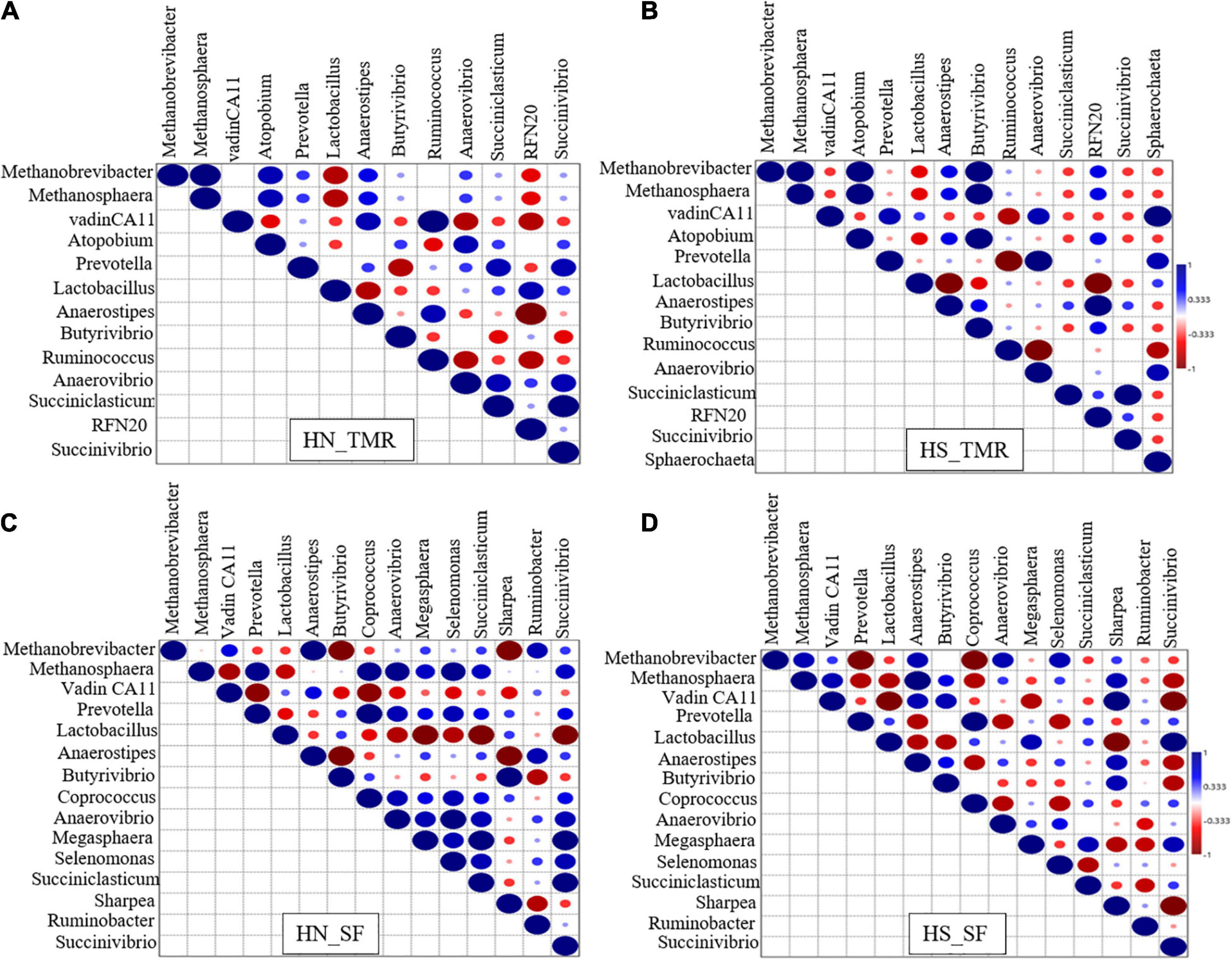
Figure 4. Kendall rank correlation analysis of the co-occurrences of microbial lineages. (A) Hanwoo (HN) steers fed a total mixed ration (TMR) diet. (B) Holstein (HS) steers given TMR diet. (C) Hanwoo (HN) steers given separate feed (SF) diet. (D) Holstein (HS) steers given SF diet. The genera were included in the matrix if they had a relative abundance of at least 0.01% in at least one steer. Correlation strength is indicated by the intensity of the colour. The scale colours denote whether the correlation is positive (closer to 1, blue Spheres) or negative (closer to −1, red Spheres). Big sphere indicates that the correlation is significant (P < 0.05).
A total of 430 genes (Supplementary Table 4) and 49 gene families (Supplementary Table 5) differed between feeding systems or breeds (P < 0.1). The genes pyruvate kinase (pyk), K00873 (P < 0.05); branched-chain amino acid transport system permease protein (livM), K01998 (P < 0.05); anaerobic carbon-monoxide dehydrogenase catalytic subunit (cooS), K00198; and F-type H+-transporting ATPase subunits (atpA–G), K02110 (P = 0.053) were abundant in the SF system. Pyruvate ferredoxin oxidoreductase alpha subunit (porA), K00169 (P = 0.076) and cobalt/nickel transport system permease protein (cbiM), K02007 (P = 0.083) were enriched in Hanwoo steers.
Pathways related to CH4 metabolism (P = 0.050) and butanoate metabolism (P < 0.05) were enriched in the rumen of Hanwoo steers, whereas those related to the purine metabolism pathway (P = 0.090) were abundant in Holstein steers. CCA of the relative abundance values of KEGG pathways of genes from the rumen microbiota showed distinct clustering of Hanwoo and Holstein cattle given the TMR and SF diets, explaining 32% of the variance (Figure 5). Genes related to CH4 metabolism (P = 0.050), pyruvate metabolism (P = 0.069), the bacterial secretion system (P < 0.05), and fatty acid biosynthesis (P = 0.089) were enriched in Hanwoo steers given the SF diet compared to Holstein steers given the SF diet (Figure 6).
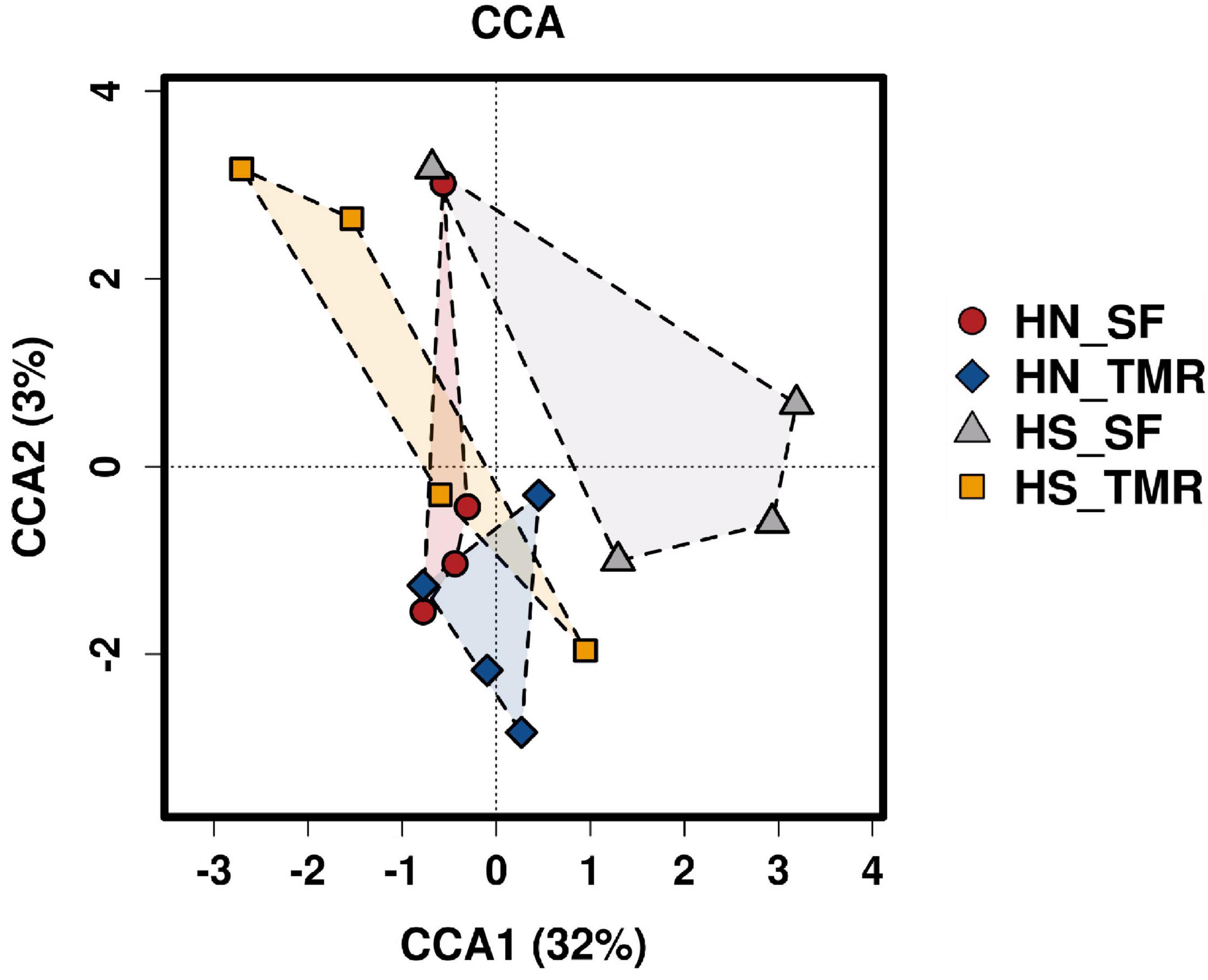
Figure 5. Canonical correspondence analysis (CCA) of microbial functional diversity in the rumen of Hanwoo (HN) and Holstein (HS) steers fed a total mixed ration (TMR) or separate feed (SF) diet.
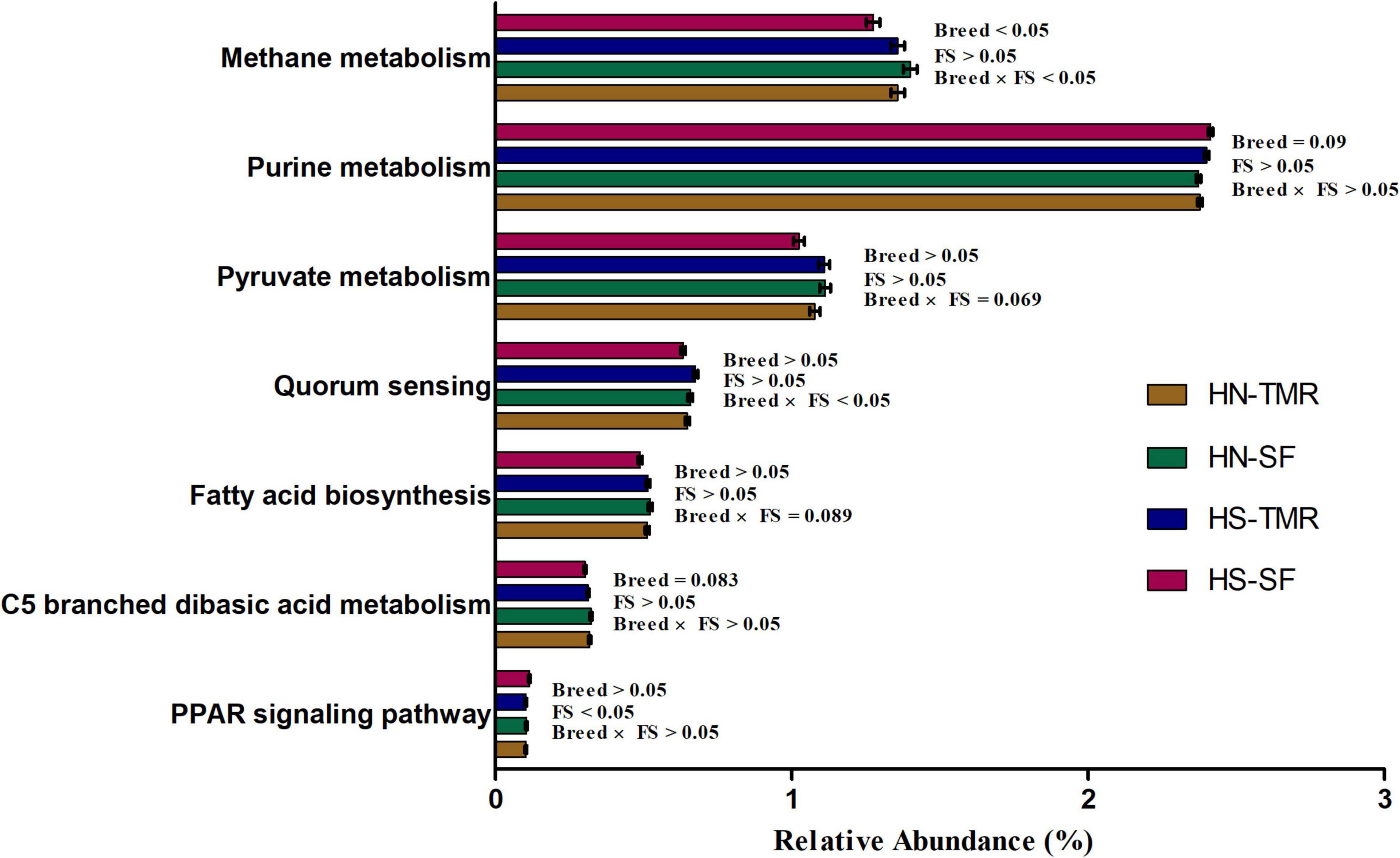
Figure 6. Variations in important rumen microbial Kyoto Encyclopaedia of Genes and Genomes (KEGG) metabolic pathways related to methane emissions in Hanwoo (HN) and Holstein (HS) steers fed a total mixed ration (TMR) or separate feed (SF) diet.
A total of 44 genes were strongly correlated with CH4 production, of which 17 were positively correlated and 27 were negatively correlated (Supplementary Table 6). The relative abundances of genes associated with CH4 production are presented as a heatmap, which clearly showed that most genes positively and negatively correlated with CH4 emissions were enriched in TMR Hanwoo and SF Holstein cattle, respectively (Figure 7). The genes pyruvate ferredoxin oxidoreductase gamma subunit (porG), K00172 (τ = 0.50, P = 0.006); trimethylamine-corrinoid protein (mttC), K14084 (τ = 0.76, P < 0.001); cbiM, K02007 (τ = 0.39, P = 0.004); V/A-type H+/Na+-transporting ATPase subunit (atpA), K02117 (τ = 0.47, P = 0.001); pyk, K00873 (τ = −0.41, P = 0.003); and acetate kinase (ackA), K00925 (τ = −0.46, P = 0.001) were found to be involved in pyruvate/CH4 metabolism, exhibiting significant associations with CH4 yield. Among these genes, porG (P = 0.076) and cbiM (P = 0.083) were enriched in Hanwoo steers (Figure 8).
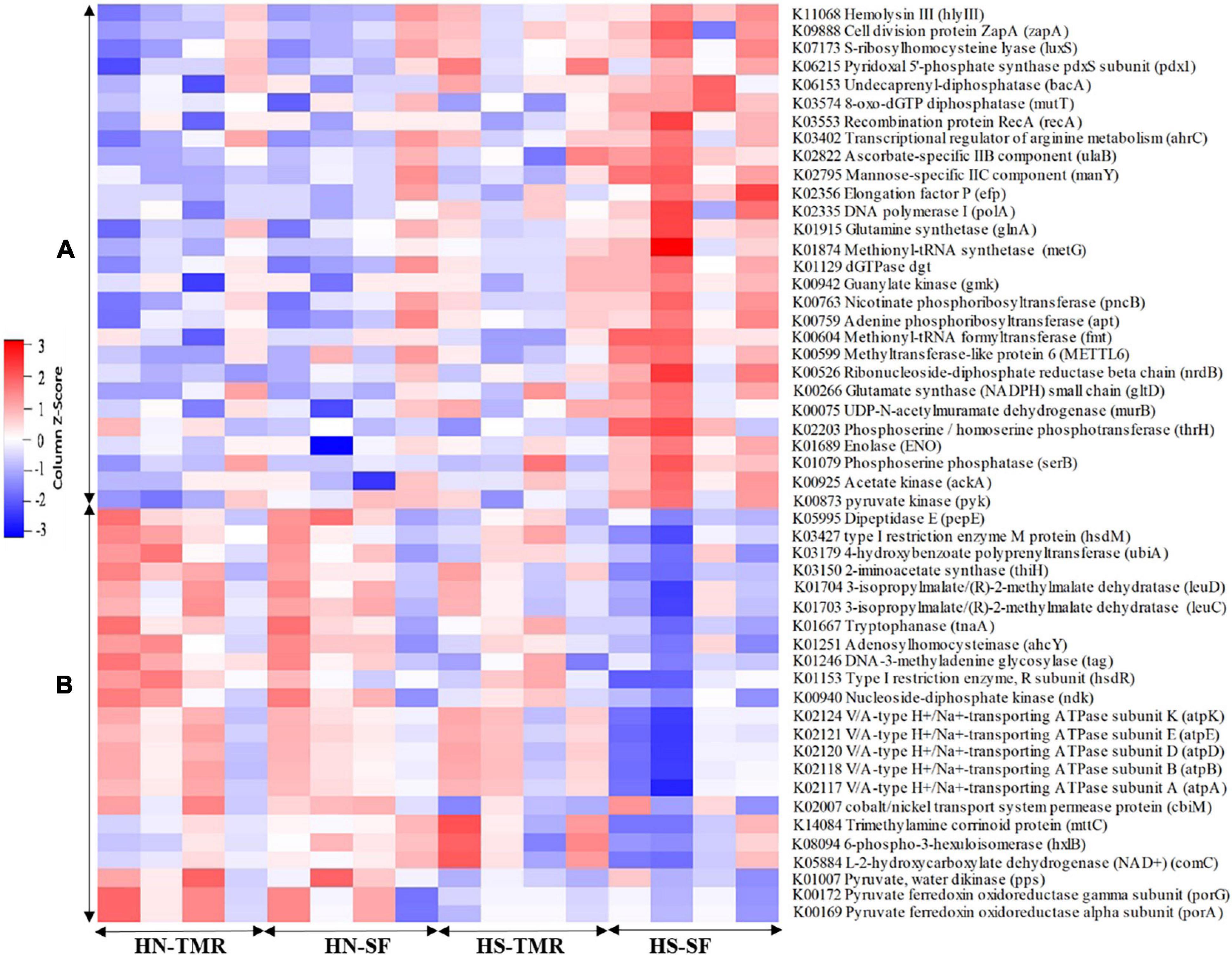
Figure 7. Heatmap of the relative abundance of microbial genes negatively (A) and positively (B) associated with methane production, as identified by Kendall rank correlation analysis. The scale colours denote whether the abundance is high (closer to 1, red squares) or low (closer to −1, blue squares).
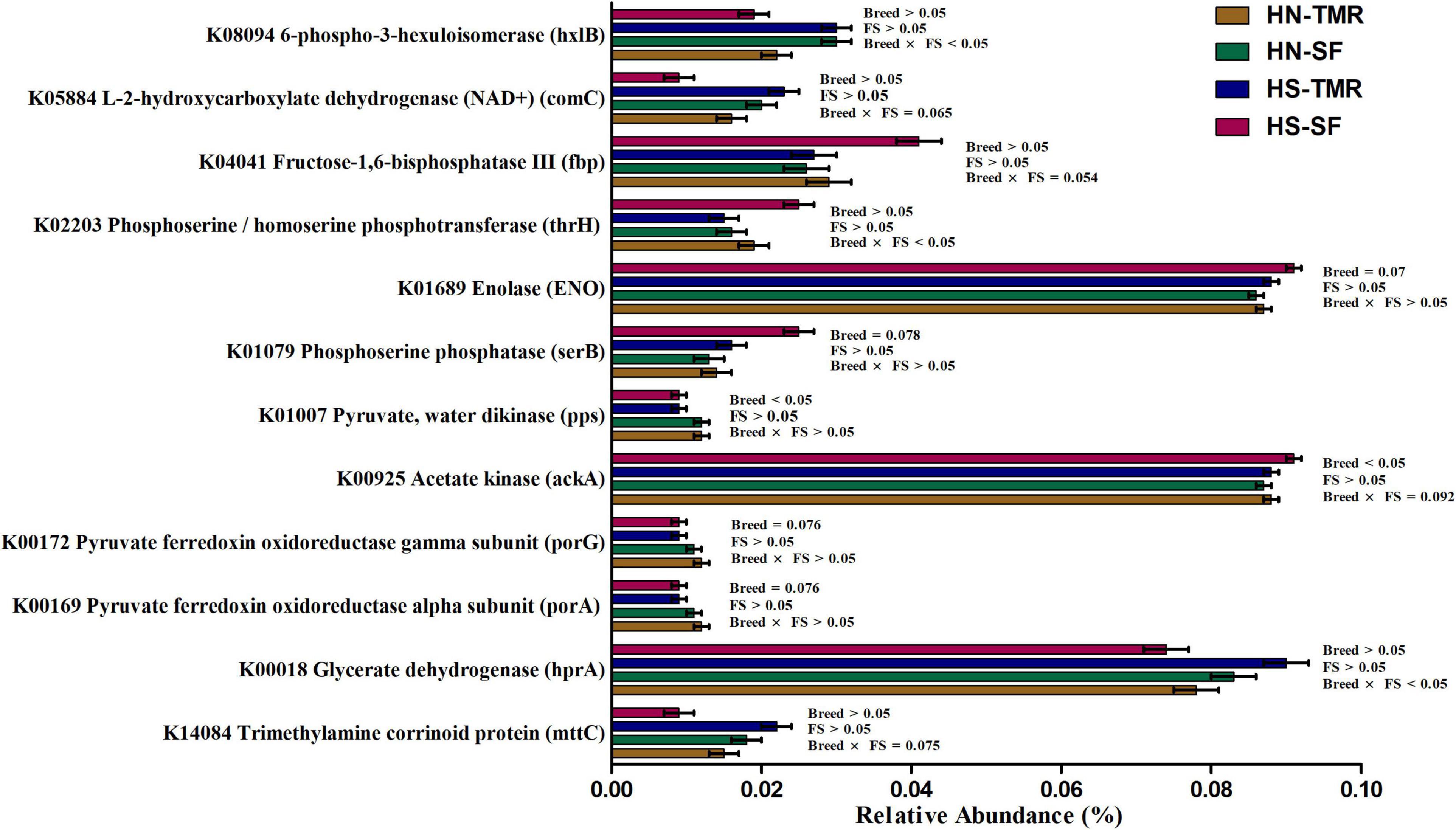
Figure 8. The metagenomic abundance of key genes involved in the methane metabolism pathway that tend to differ (0.05 < P < 0.1) or were significantly different (P < 0.05) between Hanwoo (HN) and Holstein (HS) steers fed a total mixed ration (TMR) or separate feed (SF) diet.
We detected associations (P < 0.1) among several variables (Figure 9 and Supplementary Tables 7, 8). Notably, archaeal genera Methanobrevibacter (τ = 0.31, P = 0.094), vadinCA11 (τ = 0.41, P < 0.05) and the archaea:bacteria ratio (τ = 0.33, P = 0.078) were positively associated with the CH4 yield and CH4 metabolism pathway. Fibrobacter (τ = −0.46, P < 0.05), Anaerovibrio (τ = −0.51, P < 0.05), Selenomonas (τ = −0.59, P < 0.005) and Succiniclasticum (τ = −0.25, P = 0.077) were negatively associated with the acetate:propionate ratio in the rumen. The CH4 metabolism (τ = 0.43, P < 0.05), purine metabolism (Kendall’s τ = −0.42, P < 0.05) and C5 branched dibasic acid metabolism (τ = 0.47, P < 0.05) pathways were associated with CH4 yield.
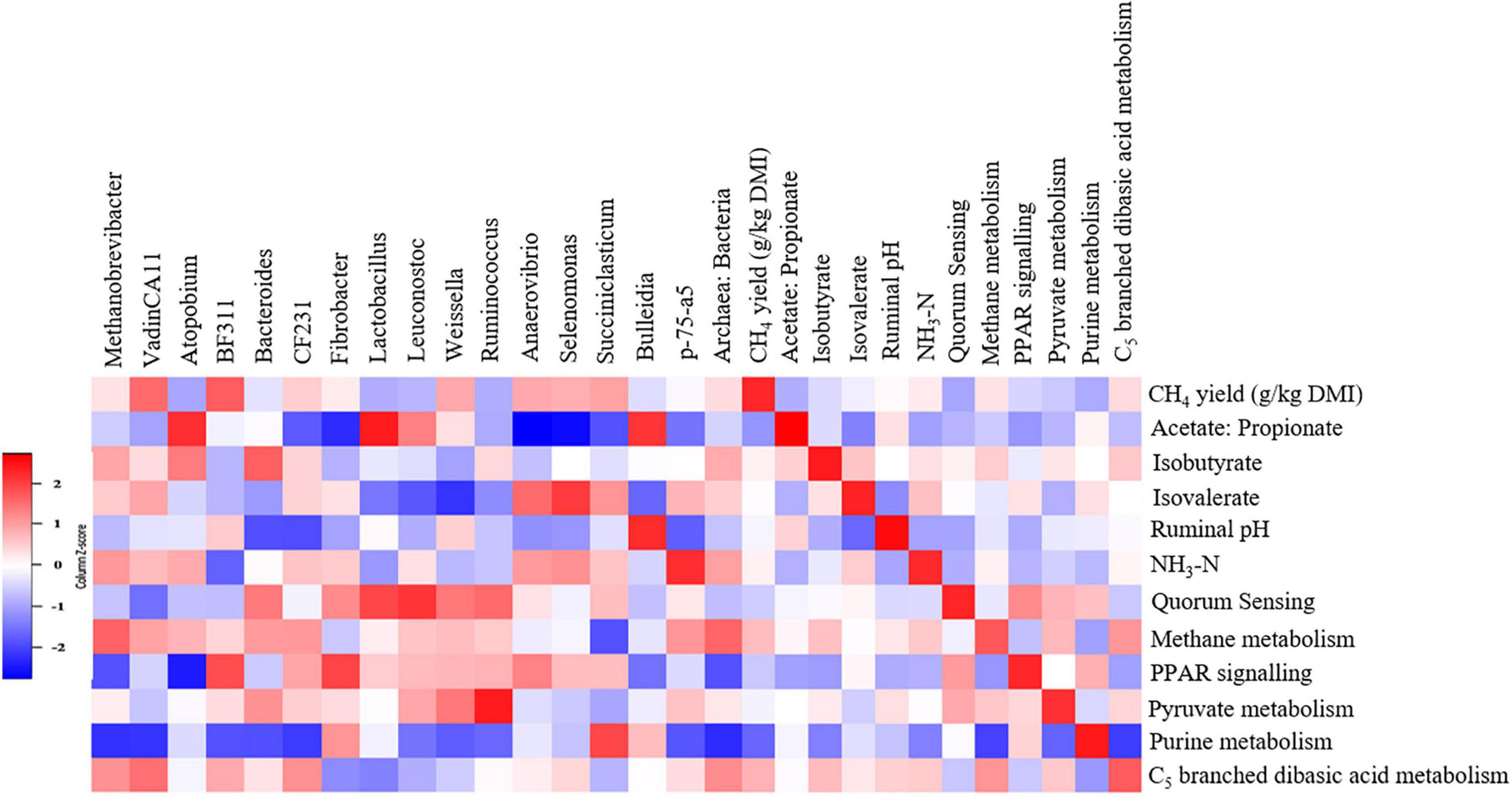
Figure 9. Associations among methane yield, fermentation parameters, rumen microbes, and metabolic pathways in steers. The genera and metabolic pathways were included in the matrix if they were differentially abundant (P < 0.1) and had at least 0.01% relative abundance in at least one of the steers. Correlation strength is indicated by the intensity of the colour. The scale colours denote whether the correlation is positive (closer to 1, red squares) or negative (closer to −1, blue squares).
Consistent with several previous reports, neither feeding system nor breed had any effect on total tract digestibility in the present study (Holter et al., 1977; Yan et al., 1998; Huuskonen et al., 2014). However, a recent study (Genís et al., 2021) reported increases in DM and CP digestibility in animals fed with the SF diet compared to the TMR diet using unprocessed long straw for which more than 70% of particles were >19 mm. However, this cannot be compared with the present study where the proportion of feed particles >19 mm was only 28%. Furthermore, in the present study, the ruminal pH of Holstein steer at 3 h post-TMR feeding was inconsistent with our previous study, which showed that the TMR diet helped to maintain ruminal pH (Bharanidharan et al., 2018). This may have been due to the 40-min delay in feeding of the concentrate to the SF animals, where the presence of sodium bicarbonate in the concentrate may have had a longer-duration stabilising effect on the rumen pH compared to TMR feeding. Furthermore, in the present study, rumen sampling was limited to 3 h, where the SF group may have experienced a further decrease in pH compared to the TMR diet in the later feeding period. In addition, in the TMR feeding system, feed particles <8 mm accounted for almost 70% of the total feed, which may have decreased the amount of time spent chewing and the ruminal pH during the initial phase of feeding (Krause et al., 2002). However, a recent study (Genís et al., 2021) noted no change in rumination or chewing activity between animals given the TMR and SF diets, although there were large variations in ruminal pH. Several studies have also reported no changes in ruminal pH in association with TMR feeding or feed particle size (Li et al., 2003; Schroeder et al., 2003; Liu et al., 2015).
Nutrition, feeding management, and animal production strategies for reducing enteric CH4 emissions may be more effective than strategies using feed additives as rumen modifiers (reviewed by Knapp et al., 2014). This is because a few rumen modifiers have been shown to be associated with adverse effects on rumen microbial fermentation, digestibility, residues in livestock products and adaptation to microbes on prolonged feeding at effective concentrations (reviewed by Honan et al., 2021). In this study, TMR feeding resulted in greater CH4 production and yield, consistent with the findings of our previous study (Bharanidharan et al., 2018). This may be related to the larger and smaller proportions of acetate and propionate, respectively, in the TMR diet, suggesting that there were fewer beneficial hydrogenotrophic reactions in the TMR system (Moss et al., 2000). Our observations contradicted earlier reports (reviewed by Janssen, 2010) that smaller particle sizes and higher passage rates were associated with a higher rumen H2 concentration, so that negative thermodynamic feedback reduces the production of H2 by fermentative microbes, resulting in increased propionate formation and ultimately reduced CH4 formation. Although the ruminal turnover rate was not evaluated in the present study, we postulated that the time lag between forage and concentrate feeding in the SF diet may have increased the feed passage rate compared to the TMR diet, thereby increasing propionate production and reducing CH4 production (Janssen, 2010). However, the limited effects of passage rate on total tract digestibility in the present study could have been due to post-ruminal compensatory digestion (Hoover, 1978). The increased proportions of isovaleric acid and valerate in the rumen of steers with the SF diet were consistent with the results of a previous study suggesting increased ruminal protein digestion (Allison and Bryant, 1963). The effects of iso-fatty acids on the synthesis of branched-chain amino acids and microbial protein depends on a wide range of hydrogenation and carboxylation reactions, which probably affect methanogenesis (Bharanidharan et al., 2018). Numerous studies have reported advantages of iso-fatty acids, in terms of rumen microbial status (Liu et al., 2014), microbial protein synthesis (Kim et al., 2005), lactation performance (Liu et al., 2018), and nutrient utilisation (Misra and Thakur, 2001). Molecular analysis of the rumen contents of feedlot beef steers also revealed increased valerate concentrations in animals with improved feed efficiency (Guan et al., 2008). However, the lack of difference in ADG between feeding systems suggested a need for more studies focusing on the effects of the SF system on animal performance.
The lower ADG observed in Hanwoo steers in the present study may have been related to the greater rumen microbial richness. An earlier study (Shabat et al., 2016) reported an association between greater microbial richness and inefficiency in cattle. Similarly, another study (Kang et al., 2005) suggested that Hanwoo steers may be inefficient when compared to Holstein steers, consistent with the results of this study. The robust rumen microbial biomarkers identified by LEfSe analysis in the present study indicated enrichment of several archaeal genera in Hanwoo steers compared to Holstein steers. This is consistent with a previous study (Lee et al., 2012), which reported a larger archaeal population in the rumen of Korean Hanwoo steers than Holstein dairy cows. We also detected a strong positive association of the archaea:bacteria ratio with CH4 yield, which is regarded as a proxy for CH4 emissions, regardless of cattle breed or diet (Wallace et al., 2015a; Auffret et al., 2018). In the present study, the most abundant archaeal taxa (Methanobrevibacter) was also positively associated with CH4 yield, consistent with previous reports of its higher abundance in high CH4 emitters (Wallace et al., 2015a; Auffret et al., 2018). The greater abundances of Methanomassiliicoccaceae and vadinCA11 in the rumen of Hanwoo steers indicated higher methylamine/methanol-mediated CH4 production (Paul et al., 2012). A previous study (Lee et al., 2012) also noted a higher concentration of methylamine in the rumen of Hanwoo steers than Holstein dairy cows. This was further supported by the higher abundances of pectinolytic Lachnospiraceae (Dusková and Marounek, 2001) and Spirochaetaceae (Ziołecki and Wojciechowicz, 1980), which may increase methylamine/methanol concentrations via the fermentation of digested pectin (Schink and Zeikus, 1980). Furthermore, this hypothesis was supported by the positive associations among vadinCA11, Sphaerochaeta and CH4 yield, and a recent study that revealed strong positive associations of Sphaerochaeta and BS11 with CH4 production (Difford et al., 2018).
Although a difference in microbial abundance was observed between breeds in this study, the feeding system did not significantly influence the major rumen microbiome. However, it did affect minor genera, such as Atopobium and Weissella, which are efficient lactate producers (Zhou et al., 2004; Abriouel et al., 2015) enriched in the animals given the SF diet. Enrichment of the lactyl-CoA dehydrogenase (lcdA) gene has been observed in Megasphaera spp., in low CH4 emitters that produce propionate from lactate via the acrylate pathway in the rumen (Counotte et al., 1981). Our core microbiome analysis only identified Megasphaera in steers given the SF diet, which may be related to their higher propionate content. Similarly, enrichment of lactate-producing Sharpea has been observed in the rumen of low CH4-emitting sheep (Kamke et al., 2016). Intriguingly, in the present study, a strong negative association was detected between Sharpea and Methanobrevibacter in Hanwoo steers given the SF diet, suggesting competitive utilisation of H2. Although increased CH4 production in the TMR system may be related to an increased ruminal H2 concentration, there were no differences in methanogen populations between the feeding systems. Similarly, the observed interaction between feeding system and breed for the total population of Euryarchaeota and Methanobrevibacter did not have a large influence on total CH4 yield. Many studies have failed to detect correlations between overall methanogen abundance and CH4 emissions (reviewed by Tapio et al., 2017). Therefore, further studies are needed to investigate other feeding variables, such as feeding behaviour (e.g., chewing activity), ruminal digestion and passage rate, which may provide a plausible explanation for the observed effects.
Despite the negligible differences in the microbiome, there were distinct microbial co-occurrences between the TMR and SF systems, which provided a plausible explanation for the observed differences in CH4 production. For example, the genera Anaerostipes, Coprococcus, Anaerovibrio, Prevotella and Succiniclasticum, which are involved in propionate production (Strobel, 1992; Van Gylswyk, 1995; Kishimoto et al., 2006; Privé et al., 2013; Reichardt et al., 2014), were negatively associated with hydrogenotrophic Methanobrevibacter in the Holstein steers given the SF diet, which clearly demonstrated the low availability of H2 for methanogenesis. The observed negative correlations of both Anaerovibrio and Succiniclasticum with the acetate:propionate ratio further supports this finding. Megasphaera and Coprococcus are more abundant in the rumen of efficient animals (Shabat et al., 2016), which may be related to the increased propionate and ADG in Holstein steers seen in the present study. Strong positive associations of the H2-producing bacteria Ruminococcus and Butyrivibrio, with vadinCA11 and Methanobrevibacter, respectively, were observed in the TMR feeding system, indicating that methanogenesis was the major H2 sink (Wolin et al., 1997).
Numerous studies have shown that microbes distributed in the rumen operate as an integrated system and have roles in the complex metabolic processes occurring within this ecosystem (Auffret et al., 2018; Wirth et al., 2018). The observed variations in H2 thermodynamics associated with changes in microbial abundances, and co-occurrences in breeds or feeding systems, reflect the predicted metabolic processes. The positive associations among the relative contents of isovalerate in the rumen, C5 branched dibasic acid metabolism pathway and CH4 metabolism pathway with CH4 yield clearly demonstrated a role of iso-fatty acids in CH4 production, as discussed in our previous report (Bharanidharan et al., 2018). The large abundance of genes encoding branched-chain amino acid transport system permease protein subclasses, livH (K01997) and livM (K01998) in the rumen of steers given the SF diet may have aided transport of branched-chain amino acids (Belitsky, 2015), in accordance with the increased and decreased iso-fatty acid and CH4 production in the rumen, respectively (Allison and Bryant, 1963). This hypothesis was supported by the associations among Methanobrevibacter, vadinCA11 and the C5 branched dibasic acid metabolism pathway in the rumen. Similarly, genes involved in pyruvate metabolism were strongly associated with the CH4 metabolism pathway, demonstrating their indirect roles in methanogenesis. The gene ackA, which produces acetyl phosphate by utilising acetate, and the genes serB and thrH, which produce serine by utilising H2, were negatively associated with CH4 production and enriched in low-emitting Holstein steers. In contrast, porA and porG, which are involved in acetyl-coA synthesis from pyruvate, leading to CO2 and H2 production, were positively associated with CH4 production and were abundant in high-emitting Hanwoo steers. These results clearly explain the differential patterns of pyruvate metabolism, H2 production and sinks between high- and low-emitting breeds detected in the present study. Similarly, Hanwoo steers showed abundant expression of cbiM, which helps to provide Ni to Ni-dependent methyl reductases and is correlated with methylamine/methanol-mediated CH4 production. Although no association was detected with CH4 yield, the gene cooS, which is involved in reductive acetogenesis (Wood et al., 1986), and atpA–G, which are involved in energy synthesis, were enriched in the SF system. All genes associated with methanogenesis in the present study were previously reported as potential metagenomic biomarkers of CH4 emission or feed efficiency (Wallace et al., 2015b; Roehe et al., 2016; Auffret et al., 2018; Lima et al., 2019), and could be used for the genetic selection of low-emitting animals. However, genes such as methyl-coenzyme M reductase alpha subunit (mcrA), formylmethanofuran dehydrogenase subunit B (fmdB) and formate dehydrogenase alpha subunit (fdhF), which are directly involved in the final step of methanogenesis leading to CH4 emission, were not identified in the present study. This could be attributed to the slightly higher (0.18 ± 0.04 SD) nearest sequence taxon index (NSTI) noted in the present study, thereby suggesting that functions based on 16S rRNA information have limitations, such that PICRUSt results should be interpreted with caution. Previous studies have also demonstrated weak correlations between the mcr gene and CH4 emissions (Morgavi et al., 2012; Tapio et al., 2017). Gene expression analyses can improve understanding of complex methanogenic processes compared to microbial and gene abundance analyses (Shi et al., 2014). A strong interaction was observed between breed and feeding system in the L-fuculokinase (fucK) gene, K00879, which is involved in fucose metabolism and plays an important role in host–microbe interactions (Hooper et al., 1999; Pacheco et al., 2012). Similarly, Ruminococcus spp., which are involved in fucose metabolism (Hooper et al., 2002) and correlated with quorum sensing and the CH4 metabolism pathway, also exhibited a similar feeding system × breed interaction. However, these interactions must be verified in a larger sample. Taken together, these results suggest that feeding systems differentially influence host–microbe interactions in different breeds, and therefore influence the rumen fermentation pattern and CH4 formation to some extent. Therefore, breed-specific feeding systems should be selected with caution.
Comprehensive knowledge of bacterial/archaeal community composition and metabolic function is important for understanding their relationships with the host, and to develop feeding strategies to control CH4 emissions. In this study, we described the rumen microbial community and its associated functions in Holstein and Hanwoo steers provided with the same diet, under the same management conditions, to identify compositional changes that might underlie the marked differences in CH4 production between these breeds. This study is the first to show that Hanwoo cattle are higher CH4 emitters than Holstein steers, as supported by the greater abundance of bacterial/archaeal genera and genes involved in CH4 metabolism in Hanwoo cattle. We also demonstrated that, compared to TMR feeding, the SF system can reduce ruminal CH4 emissions, regardless of cattle breed. Similarly, genetic factors interacted with the feeding system, leading to divergent effects in the rumen, and therefore to large differences in microbial gene abundance, but with little effect on CH4 production. Wide variation in microbial co-occurrence patterns was observed in accordance with feeding system and breed, indicating different patterns of H2 thermodynamics in the rumen. The results of this study provide insight into the complex bacterial interactions occurring in the rumen, and may facilitate appropriate selection of strategies for modulating bacterial functions to reduce CH4 emissions. Specific microbial genes associated with CH4 can be used to develop molecular tools, facilitating the breeding of low CH4-emitting animals.
The datasets generated for this study can be found in online repositories. The name of the repository (NCBI) and accession number (PRJNA725944) can be found in the following link: https://www.ncbi.nlm.nih.gov/sra/PRJNA725944.
The animal study was reviewed and approved by the Committee for the Institutional Animal Care and Use of Seoul National University (SNU-160105-1).
RB and KK designed and conceptualised the experiment. RB, CL, KT, RI, and YW performed the management of steers and sample collection. RB performed the operation of respiratory chamber, data curation, performed microbial data processing, bioinformatics, statistical analyses, visualisation, and wrote the first draft of the manuscript including tables and figures. RB, CL, and YW performed the laboratory analyses. RB, H-GL, JK, and KK revised the first draft of the manuscript including tables and figures. All authors contributed to the final manuscript revision, read and approved the final manuscript.
The present study was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET); the Ministry of Agriculture, Food and Rural Affairs, South Korea (research project no. 314081-2).
CL and YW are employed by Cargill Agri Purina Inc., and GN Food Ltd., respectively.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.701081/full#supplementary-material
Abriouel, H., Lerma, L. L., Casado Muñoz, M. D. C., Montoro, B. P., Kabisch, J., Pichner, R., et al. (2015). The controversial nature of the Weissella genus: technological and functional aspects versus whole genome analysis-based pathogenic potential for their application in food and health. Front. Microbiol. 6:1197. doi: 10.3389/fmicb.2015.01197
Allison, M. J., and Bryant, M. P. (1963). Biosynthesis of branched-chain amino acids from branched-chain fatty acids by rumen bacteria. Arch. Biochem. Biophys. 101, 269–277. doi: 10.1016/S0003-9861(63)80012-0
Auffret, M. D., Stewart, R., Dewhurst, R. J., Duthie, C. A., Rooke, J. A., Wallace, R. J., et al. (2018). Identification, comparison, and validation of robust rumen microbial biomarkers for methane emissions using diverse Bos taurus breeds and basal diets. Front. Microbiol. 8:2642. doi: 10.3389/fmicb.2017.02642
Beigh, Y. A., Ganai, A. M., and Ahmad, H. A. (2017). Prospects of complete feed system in ruminant feeding: a review. Vet. World 10, 424–437. doi: 10.14202/vetworld.2017.424-437
Belitsky, B. R. (2015). Role of branched-chain amino acid transport in Bacillus subtilis CodY activity. J. Bacteriol. 197, 1330–1338. doi: 10.1128/JB.02563-14
Benson, A. K., Kelly, S. A., Legge, R., Ma, F., Low, S. J., Kim, J., et al. (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. U.S.A. 107, 18933–18938. doi: 10.1073/pnas.1007028107
Bharanidharan, R., Arokiyaraj, S., Kim, E. B., Lee, C. H., Woo, Y. W., Na, Y., et al. (2018). Ruminal methane emissions, metabolic, and microbial profile of Holstein steers fed forage and concentrate, separately or as a total mixed ration. PLoS One 13:e0202446. doi: 10.1371/journal.pone.0202446
Choi, J. W., Liao, X., Stothard, P., Chung, W. H., Jeon, H. J., Miller, S. P., et al. (2014). Whole-genome analyses of Korean native and Holstein cattle breeds by massively parallel sequencing. PLoS One 9:e101127. doi: 10.1371/journal.pone.0101127
Chung, C. S., Cho, W. K., Jang, I. S., Lee, S. S., and Moon, Y. H. (2017). Effects of feeding system on growth performance, plasma biochemical components and hormones, and carcass characteristics in Hanwoo steers. Asian Austr. J. Anim. Sci. 30, 1117–1123. doi: 10.5713/ajas.17.0166
Counotte, G. H., Prins, R. A., Janssen, R. H., and Debie, M. J. (1981). Role of Megasphaera elsdenii in the fermentation of dl-[2-C]lactate in the Rumen of dairy cattle. Appl. Environ. Microbiol. 42, 649–655. doi: 10.1128/aem.42.4.649-655.1981
De Mulder, T., Peiren, N., Vandaele, L., Ruttink, T., De Campeneere, S., Van de Wiele, T., et al. (2018). Impact of breed on the rumen microbial community composition and methane emission of Holstein Friesian and Belgian Blue heifers. Livest. Sci. 207, 38–44. doi: 10.1016/j.livsci.2017.11.009
Difford, G. F., Plichta, D. R., Løvendahl, P., Lassen, J., Noel, S. J., Højberg, O., et al. (2018). Host genetics and the rumen microbiome jointly associate with methane emissions in dairy cows. PLoS Genet. 14:e1007580. doi: 10.1371/journal.pgen.1007580
Dusková, D., and Marounek, M. (2001). Fermentation of pectin and glucose, and activity of pectin-degrading enzymes in the rumen bacterium Lachnospira multiparus. Lett. Appl. Microbiol. 33, 159–163. doi: 10.1046/j.1472-765x.2001.00970.x
Duthie, C. A., Haskell, M., Hyslop, J. J., Waterhouse, A., Wallace, R. J., Roehe, R., et al. (2017). The impact of divergent breed types and diets on methane emissions, rumen characteristics and performance of finishing beef cattle. Animal 11, 1762–1771. doi: 10.1017/S1751731117000301
FAOSTAT (2021). Food and Agriculture Organization of the United Nations Statistical Database. Available online at: http://www.fao.org/faostat/en/#data/GT (accessed April 27, 2021).
Genís, S., Verdú, M., Cucurull, J., and Devant, M. (2021). Complete feed versus concentrate and straw fed separately: effect of feeding method on eating and sorting behavior, rumen acidosis, and digestibility in crossbred Angus bulls fed high-concentrate diets. Anim. Feed Sci. Technol. 273:114820. doi: 10.1016/j.anifeedsci.2021.114820
Guan, L. L., Nkrumah, J. D., Basarab, J. A., and Moore, S. S. (2008). Linkage of microbial ecology to phenotype: correlation of rumen microbial ecology to cattle’s feed efficiency. FEMS Microbiol. Lett. 288, 85–91. doi: 10.1111/j.1574-6968.2008.01343.x
Hammer, D. A. T., Ryan, P. D., and Hammer, Ø, and Harper, D. A. T. (2001). Past: Paleontological Statistics Software Package for Education and Data Analysis. Available online at: https://palaeo-electronica.org/2001_1/past/issue1_01.htm (accessed July 22, 2018).
Holter, J. B., Urban, W. E., Hayes, H. H., and Davis, H. A. (1977). Utilization of diet components fed blended or separately to lactating cows. J. Dairy Sci. 60, 1288–1293. doi: 10.3168/JDS.S0022-0302(77)84024-1
Honan, M., Feng, X., Tricarico, J. M., and Kebreab, E. (2021). Feed additives as a strategic approach to reduce enteric methane production in cattle: modes of action, effectiveness and safety. Anim. Prod. Sci. doi: 10.1071/AN20295
Hooper, L. V., Midtvedt, T., and Gordon, J. I. (2002). How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22, 283–307. doi: 10.1146/annurev.nutr.22.011602.092259
Hooper, L. V., Xu, J., Falk, P. G., Midtvedt, T., and Gordon, J. I. (1999). A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. U.S.A. 96, 9833–9838. doi: 10.1073/PNAS.96.17.9833
Hoover, W. H. (1978). Digestion and absorption in the hindgut of ruminants. J. Anim. Sci. 46, 1789–1799. doi: 10.2527/jas1978.4661789x
Huuskonen, A., Pesonen, M., and Joki-Tokola, E. (2014). Effects of supplementary concentrate level and separate or total mixed ration feeding on performance of growing dairy bulls. Agric. Food Sci. 23, 257–265. doi: 10.23986/afsci.46460
Islam, M., Kim, S.-H., Ramos, S. C., Mamuad, L. L., Son, A.-R., Yu, Z., et al. (2021). Holstein and jersey steers differ in rumen microbiota and enteric methane emissions even fed the same total mixed ration. Front. Microbiol. 12:601061. doi: 10.3389/fmicb.2021.601061
Janssen, P. H. (2010). Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed Sci. Technol. 160, 1–22. doi: 10.1016/j.anifeedsci.2010.07.002
Kamke, J., Kittelmann, S., Soni, P., Li, Y., Tavendale, M., Ganesh, S., et al. (2016). Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea-enriched microbiome characterised by lactic acid formation and utilisation. Microbiome 4:56. doi: 10.1186/s40168-016-0201-2
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30.
Kang, S. W., Oh, Y. K., Choi, C. W., Kim, G.-H., and Son, Y. S. (2005). Study on comparison of growth performance, feed efficiency and carcass characteristics for holstein, Hanwoo and F1(Holstein ♀ X Hanwoo ♂) Steers. J. Anim. Sci. Technol. 47, 243–252. doi: 10.5187/jast.2005.47.2.243
Kim, K. H., Lee, S. S., and Kim, K. J. (2005). Effect of intraruminal sucrose infusion on volatile fatty acid production and microbial protein synthesis in sheep. Asian Austr. J. Anim. Sci. 18, 350–353. doi: 10.5713/ajas.2005.350
Kim, S. H., Alam, M. J., Gu, M. J., Park, K. W., Jeon, C. O., Ha, J. K., et al. (2011). Effect of total mixed ration with fermented feed on ruminal in vitro fermentation, growth performance and blood characteristics of Hanwoo steers. Asian Austr. J. Anim. Sci. 25, 213–223. doi: 10.5713/ajas.2011.11186
Kishimoto, A., Ushida, K., Phillips, G. O., Ogasawara, T., and Sasaki, Y. (2006). Identification of intestinal bacteria responsible for fermentation of gum arabic in pig model. Curr. Microbiol. 53, 173–177. doi: 10.1007/s00284-005-0219-3
Knapp, J. R., Laur, G. L., Vadas, P. A., Weiss, W. P., and Tricarico, J. M. (2014). Invited review: enteric methane in dairy cattle production: quantifying the opportunities and impact of reducing emissions. J. Dairy Sci. 97, 3231–3261. doi: 10.3168/jds.2013-7234
Kononoff, P. J., Heinrichs, A. J., and Buckmaster, D. R. (2003). Modification of the Penn State Forage and total mixed ration particle separator and the effects of moisture content on its measurements. J. Dairy Sci. 86, 1858–1863. doi: 10.3168/jds.S0022-0302(03)73773-4
Korean Statistical Information Service (KOSIS) (2021). Number of Farms in Households and Animals in Heads by Type. Available online at: http://kosis.kr/statHtml/statHtml.do?orgId=101&tblId=DT_1EO200&language=en&conn_path=I3 (accessed March 11, 2021).
Krause, K. M., Combs, D. K., and Beauchemin, K. A. (2002). Effects of forage particle size and grain fermentability in midlactation cows. II. Ruminal pH and chewing activity. J. Dairy Sci. 85, 1947–1957. doi: 10.3168/jds.S0022-0302(02)74271-9
Lailer, P. C., Dahiya, S. S., and Chauhan, T. R. (2005). Complete feed for livestock concept, present status and future trend: a review. Indian J. Anim. Sci. 75, 84–91.
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lee, H. J., Jung, J. Y., Oh, Y. K., Lee, S. S., Madsen, E. L., and Jeon, C. O. (2012). Comparative survey of rumen microbial communities and metabolites across one caprine and three bovine groups, using bar-coded pyrosequencing and 1H nuclear magnetic resonance spectroscopy. Appl. Environ. Microbiol. 78, 5983–5993. doi: 10.1128/AEM.00104-12
Lee, S., Kim, Y., Oh, Y., and Kwak, W. (2010). Effects of feeding methods of total mixed ration on behavior patterns of growing Hanwoo Steers. Asian Austr. J. Anim. Sci. 23, 1469–1475. doi: 10.5713/ajas.2010.10100
Lee, Y., Bharanidharana, R., Park, J.-H., Jang, S. S., Yeo, J. M., Kim, W. Y., et al. (2016). Comparison of Methane Production of Holstein Steers Fed Forage and Concentrates Separately or As a TMR. J. Korean Soc. Grassl. Forage Sci. 36, 104–108. doi: 10.5333/kgfs.2016.36.2.104
Li, D. Y., Lee, S. S., Choi, N. J., Lee, S. Y., Sung, H. G., Ko, J. Y., et al. (2003). Effects of feeding system on rumen fermentation parameters and nutrient digestibility in Holstein steers. Asian Austr. J. Anim. Sci. 16, 1482–1486. doi: 10.5713/ajas.2003.1482
Lima, J., Auffret, M. D., Stewart, R. D., Dewhurst, R. J., Duthie, C. A., Snelling, T. J., et al. (2019). Identification of rumen microbial genes involved in pathways linked to appetite, growth, and feed conversion efficiency in cattle. Front. Genet. 10:701. doi: 10.3389/fgene.2019.00701
Littell, R. C., Henry, P. R., and Ammerman, C. B. (1998). Statistical analysis of repeated measures data using SAS procedures. J. Anim. Sci. 76, 1216–1231. doi: 10.2527/1998.7641216x
Liu, Q., Wang, C., Guo, G., Huo, W. J., Zhang, S. L., Pei, C. X., et al. (2018). Effects of branched-chain volatile fatty acids on lactation performance and mRNA expression of genes related to fatty acid synthesis in mammary gland of dairy cows. Animal 12, 2071–2079. doi: 10.1017/S1751731118000113
Liu, Q., Wang, C., Pei, C. X., Li, H. Y., Wang, Y. X., Zhang, S. L., et al. (2014). Effects of isovalerate supplementation on microbial status and rumen enzyme profile in steers fed on corn stover based diet. Livest. Sci. 161, 60–68. doi: 10.1016/J.LIVSCI.2013.12.034
Liu, Y. F., Sun, F. F., Wan, F. C., Zhao, H. B., Liu, X. M., You, W., et al. (2015). Effects of three feeding systems on production performance, rumen fermentation and rumen digesta particle structure of beef cattle. Asian Austr. J. Anim. Sci. 29, 659–665. doi: 10.5713/ajas.15.0445
Misra, A. K., and Thakur, S. S. (2001). Effect of dietary supplementation of sodium salt of Isobutyric acid on ruminal fermentation and nutrient utilization in a wheat straw based low protein diet fed to crossbred cattle. Asian Austr. J. Anim. Sci. 14, 479–484. doi: 10.5713/ajas.2001.479
Morgavi, D. P., Martin, C., Jouany, J.-P., and Ranilla, M. J. (2012). Rumen protozoa and methanogenesis: not a simple cause–effect relationship. Br. J. Nutr. 107, 388–397. doi: 10.1017/S0007114511002935
Moss, A. R., Jouany, J.-P., and Newbold, J. (2000). Methane production by ruminants: its contribution to global warming. Ann. Zootech. 49, 231–253. doi: 10.1051/animres:2000119
Moya, D., Holtshausen, L., Marti, S., Gibb, D. G., McAllister, T. A., Beauchemin, K. A., et al. (2014). Feeding behavior and ruminal pH of corn silage, barley grain, and corn dried distillers’ grain offered in a total mixed ration or in a free-choice diet to beef cattle1. J. Anim. Sci. 92, 3526–3536. doi: 10.2527/jas.2013-7224
Olijhoek, D. W., Løvendahl, P., Lassen, J., Hellwing, A. L. F., Höglund, J. K., Weisbjerg, M. R., et al. (2018). Methane production, rumen fermentation, and diet digestibility of Holstein and Jersey dairy cows being divergent in residual feed intake and fed at 2 forage-to-concentrate ratios. J. Dairy Sci. 101, 9926–9940. doi: 10.3168/jds.2017-14278
Pacheco, A. R., Curtis, M. M., Ritchie, J. M., Munera, D., Waldor, M. K., Moreira, C. G., et al. (2012). Fucose sensing regulates bacterial intestinal colonization. Nature 492, 113–117. doi: 10.1038/nature11623
Paul, K., Nonoh, J. O., Mikulski, L., and Brune, A. (2012). Methanoplasmatales & thermoplasmatales-related archaea in termite guts and other environments, are the seventh order of methanogens. Appl. Environ. Microbiol. 78, 8245–8253. doi: 10.1128/AEM.02193-12
Phipps, R. H., Bines, J. A., Fulford, R. J., and Weller, R. F. (1984). Complete diets for dairy cows: a comparison between complete diets and separate ingredients. J. Agric. Sci. 103:171. doi: 10.1017/S0021859600043434
Privé, F., Kaderbhai, N. N., Girdwood, S., Worgan, H. J., Pinloche, E., Scollan, N. D., et al. (2013). Identification and characterization of three novel lipases belonging to families II and V from Anaerovibrio lipolyticus 5ST. PLoS One 8:e69076. doi: 10.1371/journal.pone.0069076
Reddy, D. N., and Reddy, M. R. (1983). The effect of feeding complete feeds on nutrient utilization and milk production on crossbred cows. Indian J. Dairy Sci. 36, 421–423.
Reichardt, N., Duncan, S. H., Young, P., Belenguer, A., McWilliam Leitch, C., Scott, K. P., et al. (2014). Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 8, 1323–1335. doi: 10.1038/ismej.2014.14
Revelle, W. (2018). psych: Procedures for Psychological, Psychometric, and Personality Research. R Package Version 1.8.4. Available online at: https://CRAN.R-project.org/package=psych (accessed Sep 23, 2018).
Roehe, R., Dewhurst, R. J., Duthie, C. A., Rooke, J. A., McKain, N., Ross, D. W., et al. (2016). Bovine host genetic variation influences rumen microbial methane production with best selection criterion for low methane emitting and efficiently feed converting hosts based on metagenomic gene abundance. PLoS Genet. 12:e1005846. doi: 10.1371/journal.pgen.1005846
Schink, B., and Zeikus, J. G. (1980). Microbial methanol formation: a major end product of pectin metabolism. Curr. Microbiol. 4, 387–389. doi: 10.1007/BF02605383
Schroeder, M. M., Soita, H. W., Christensen, D. A., Khorasani, G. R., and Kennelly, J. J. (2003). Effect of total mixed ration particle size on rumen pH, chewing activity, and performance in dairy cows. Asian Austr. J. Anim. Sci. 16, 1755–1762. doi: 10.5713/ajas.2003.1755
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shabat, S., Sasson, G., Doron-Faigenboim, A., Durman, T., Yaacoby, S., Berg Miller, M. E., et al. (2016). Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10, 2958–2972. doi: 10.1038/ismej.2016.62
Shi, W., Moon, C. D., Leahy, S. C., Kang, D., Froula, J., Kittelmann, S., et al. (2014). Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 24, 1517–1525. doi: 10.1101/gr.168245.113
Strobel, H. J. (1992). Vitamin B12-dependent propionate production by the ruminal bacterium Prevotella ruminicola 23. Appl. Environ. Microbiol. 58, 2331–2333. doi: 10.1128/aem.58.7.2331-2333.1992
Tapio, I., Snelling, T. J., Strozzi, F., and Wallace, R. J. (2017). The ruminal microbiome associated with methane emissions from ruminant livestock. J. Anim. Sci. Biotechnol. 8:7. doi: 10.1186/s40104-017-0141-0
Van Gylswyk, N. O. (1995). Succiniclasticum ruminis gen. nov., sp. nov., a ruminal bacterium converting Succinate to propionate as the sole energy-yielding mechanism. Int. J. Syst. Bacteriol. 45, 297–300. doi: 10.1099/00207713-45-2-297
Wallace, R. J., Rooke, J. A., Duthie, C.-A., Hyslop, J. J., Ross, D. W., McKain, N., et al. (2015a). Archaeal abundance in post-mortem ruminal digesta may help predict methane emissions from beef cattle. Sci. Rep. 4:5892. doi: 10.1038/srep05892
Wallace, R. J., Rooke, J. A., McKain, N., Duthie, C.-A., Hyslop, J. J., Ross, D. W., et al. (2015b). The rumen microbial metagenome associated with high methane production in cattle. BMC Genom. 16:839. doi: 10.1186/s12864-015-2032-0
Warnes, G. R., Bolker, B., Bonebakker, L., Gentleman, R., Huber, W., Liaw, A., et al. (2015). Various R programming tools for plotting data. Compr. R. Arch. Netw. 2, 1–66.
Weiner, J. M. (2019). pca3d: Three Dimensional PCA Plots. R Package Version 0.10.1. Available online at: https://cran.r-project.org/web/packages/pca3d/pca3d.pdf (accessed March 03, 2019).
Wirth, R., Kádár, G., Kakuk, B., Maróti, G., Bagi, Z., and Szilágyi, Á, et al. (2018). The planktonic core microbiome and core functions in the cattle rumen by next generation sequencing. Front. Microbiol. 9:2285. doi: 10.3389/fmicb.2018.02285
Wolin, M. J., Miller, T. L., and Stewart, C. S. (1997). “Microbe-microbe interactions,” in The Rumen Microbial Ecosystem, eds P. N. Hobson and C. S. Stewart (Dordrecht: Springer), 467–491. doi: 10.1007/978-94-009-1453-7_11
Wood, H. G., Ragsdale, S. W., and Pezacka, E. (1986). The acetyl-CoA pathway of autotrophic growth. FEMS Microbiol. Lett. 39, 345–362. doi: 10.1016/0378-1097(86)90022-4
Yan, T., Patterson, D. C., and Gordon, F. J. (1998). The effect of two methods of feeding the concentrate supplement to dairy cows of high genetic merit. Anim. Sci. 67, 395–403. doi: 10.1017/S1357729800032793
Zakrzewski, M., Proietti, C., Ellis, J. J., Hasan, S., Brion, M. J., Berger, B., et al. (2017). Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics 33, 782–783. doi: 10.1093/bioinformatics/btw725
Zhang, Q., Difford, G., Sahana, G., Løvendahl, P., Lassen, J., Lund, M. S., et al. (2020). Bayesian modeling reveals host genetics associated with rumen microbiota jointly influence methane emission in dairy cows. ISME J. 14, 2019–2033. doi: 10.1038/s41396-020-0663-x
Zhou, X., Bent, S. J., Schneider, M. G., Davis, C. C., Islam, M. R., and Forney, L. J. (2004). Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150, 2565–2573. doi: 10.1099/mic.0.26905-0
Keywords: Hanwoo, TMR, rumen, methane, metagenome, microbial network
Citation: Bharanidharan R, Lee CH, Thirugnanasambantham K, Ibidhi R, Woo YW, Lee H-G, Kim JG and Kim KH (2021) Feeding Systems and Host Breeds Influence Ruminal Fermentation, Methane Production, Microbial Diversity and Metagenomic Gene Abundance. Front. Microbiol. 12:701081. doi: 10.3389/fmicb.2021.701081
Received: 27 April 2021; Accepted: 28 June 2021;
Published: 20 July 2021.
Edited by:
Yvonne Vallès, The University of the West Indies, Cave Hill, BarbadosReviewed by:
Fenja Klevenhusen, Bundesanstalt für Risikobewertung (BfR), GermanyCopyright © 2021 Bharanidharan, Lee, Thirugnanasambantham, Ibidhi, Woo, Lee, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyoung Hoon Kim, a2hoa2ltQHNudS5hY2Uua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.