- 1Department of Microbiology and Immunology, Faculty of Pharmacy, Cairo University, Cairo, Egypt
- 2Department of Pharmaceutical Chemistry, Faculty of Pharmacy, Cairo University, Cairo, Egypt
- 3Department of Analytical Chemistry, Faculty of Pharmacy, Cairo University, Cairo, Egypt
- 4School of Pharmacy, Newgiza University, Giza, Egypt
The emergence of microbial resistance to the available antibiotics is a major public health concern, especially with the limited rate of developing new antibiotics. The utilization of anti-virulence agents is a non-conventional approach that can be used to combat microbial infection. In Staphylococcus aureus, many virulence factors are regulated by the Agr-mediated quorum sensing (QS). We developed a chemical compound that acts a potential Agr-inhibitor without reducing bacterial viability. The compound was designated staquorsin for Staphylococcus aureus QS inhibitor. In silico analyses confirmed the binding of staquorsin to the AgrA active site with an absolute binding score comparable to savirin, a previously described AgrA inhibitor. However, staquorsin turned out to be superior over savarin in not affecting the S. aureus viability in concentrations up to 600 μM. On the other hand, savirin inhibited S. aureus growth in concentrations as low as 25 μM. Moreover, staquorsin proved to be a potent inhibitor of the Agr system by inhibiting hemolysins, lipase production, and affecting biofilms formation and detachment. On the molecular level it significantly inhibited the effector transcript RNA III. In vivo testing, using the murine skin abscess model, confirmed the ability of staquorsin to modulate S. aureus virulence by effectively controlling the infection. Twenty passages of S. aureus in the presence of 40 μM staquorsin have not resulted in loss of activity as evidenced by maintaining its ability to reduce hemolysin production and RNA III transcript levels. In conclusion, we hereby describe a novel anti-virulence compound inhibiting the S. aureus Agr-system and its associated virulence factors. It is active both in vitro and in vivo, and its frequent use does not lead to the development of resistance. These findings model staquorsin as a promising drug candidate to join the fierce battle against the formidable pathogen S. aureus.
Introduction
The continuous and widespread use of antimicrobial agents has generated a supreme environment for the selection of multidrug-resistant organisms (Chambers and Deleo, 2009). About 2.8 million people get infected by antibiotic resistant organisms, and more than 35,000 people die annually, just in the United States (CDC, 2019). In the developing countries, the situation could be more aggravated because of the flagrant abuse of antimicrobials (Ayukekbong et al., 2017). The rapid rate, at which resistant strains arise, is quite problematic in the medical community; given the slow rate of development of new antibiotics (Chambers, 2001). Therefore, new treatment approaches, with minimum chance of resistance development, must be sought (Totsika, 2016).
Anti-virulence agents represent a promising alternative to the use of antimicrobials. These compounds act by suppressing the organism’s virulence thus “disarming” it. As well as, reducing its ability to colonize and invade host tissue which enables host defense mechanisms to eradicate the infection (Munguia and Nizet, 2017). They should be designed to lack any inhibitory effect on the viability of the pathogen. This avoids the selection of resistant mutants; the problem which is threatening the continuation of antibiotic use in infection management. In addition, anti-virulence agents are tailor made against a specific target where their narrow spectrum will limit the opportunity for resistance development (Totsika, 2016).
Production of virulence factors is under the control of many regulatory mechanisms including the quorum sensing (QS). Microbial cells communicate where a certain cell density is required to activate the QS activity and other associated virulence factors. Inhibition of QS represents an extraordinary therapeutic target that disarms the pathogen from numerous virulence factors simultaneously (Miller and Bassler, 2001).
In Staphylococcus aureus, multiple virulence factors are under the regulation of the Agr-mediated QS. The agr operon contains two promoters: P2 and P3 that govern the production of two transcripts: RNA II and RNA III, respectively. The RNA II encodes proteins that are entangled in the QS activity (AgrB, AgrD, AgrC, and AgrA). The AgrB is responsible for the activation and production of the autoinducing peptide AgrD while the AgrC and AgrA represent a two-component regulatory system. When the extracellular autoinducing peptide concentration reaches a threshold, the Agr system is activated. Upon activation, the level of both transcripts and their proteins notably increases (Tan et al., 2018). The RNA III transcript encodes δ-hemolysin toxin and at the same time acts as a regulatory small RNA molecule (Le and Otto, 2015).
The Agr-system acts as a global regulator of virulence factors, both directly and indirectly. It activates the production of several extracellular toxins; α-, β-, and γ-hemolysins as well as lipases, leukotoxins, toxic shock syndrome toxins, and phenol soluble modulins. It can affect the expression of several virulence factors indirectly through transcription regulators such as Rot, SarT, and SarS. The agr-dependent virulence factors are important for the establishment of infection by promoting host colonization and invasion, and helping the escape from host defense mechanisms (Le and Otto, 2015).
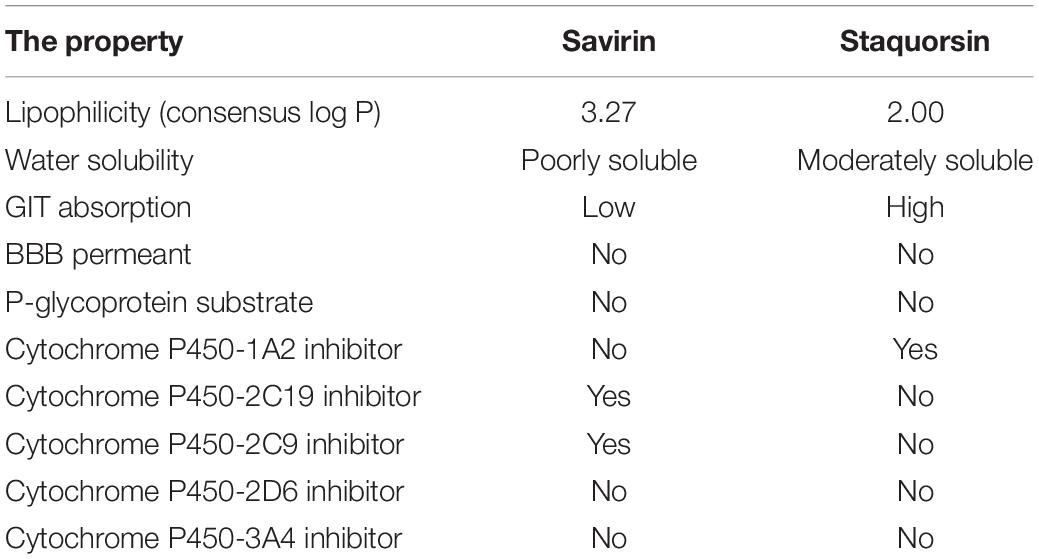
In this study, we have developed a new potent agr-inhibitor (staquorsin), which was aimed to be a new analog of savarin (S. aureus virulence inhibitor), that is a triazoloquinazoline derivative that inhibits AgrA (Gordon et al., 2013; Sully et al., 2014). The phthalazine nucleus was selected as a suitable substitute for the quinazoline ring of savirin (Figure 1A) as it is considered as its positional isomer. In addition, a cyclic-versus-non-cyclic replacement of the fused triazolo ring of savirin with the open hydrazine moiety (Figure 1A) is adopted. This is believed to produce a more flexible structure that may confer easier accommodation and interaction with the biological target. The action of this molecule is demonstrated on multiple levels including in silico, in vitro, and in vivo testing.
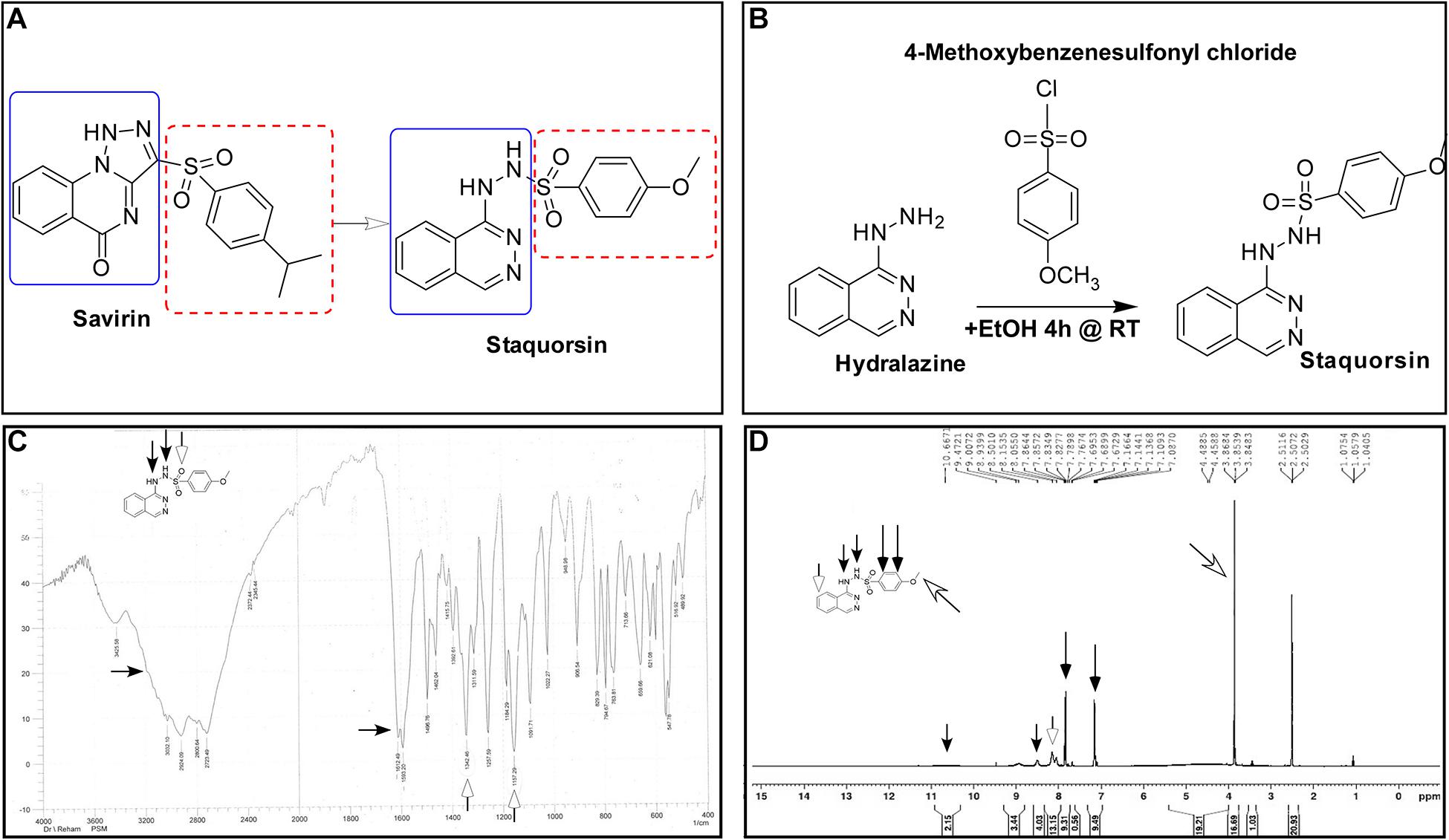
Figure 1. Synthesis and validation of staquorsin. (A) Rationale for the design of the staquorsin molecule as being an analogue for savirin. (B) The synthesis scheme used for obtaining staquorsin. (C) An IR spectrum chart for staquorsin. (D) A 1H NMR spectrum chart for staquorsin. In both charts, the peaks corresponding to the respective moieties are marked with same arrows labeling the chemical structure.
Materials and Methods
Ethics Statement
All procedures involving animals were approved by the Research Ethics Committee of the Faculty of Pharmacy, Cairo University with number MI (1682).
Bacterial Strains and Culture Conditions
Staphylococcus aureus strain Newman (Duthie and Lorenz, 1952) was used as the wild-type strain in all the experiments. Its agr-mutant (ΔagrB:erm) (Bae et al., 2004) was used as a negative-control strain. USA300 strain was used as an example for a methicillin-resistant S. aureus (MRSA) strain (Diep et al., 2006). Cells were typically grown on tryptic soy agar (TSA) or in tryptic soy broth (TSB) with shaking at 180 rpm and incubated at 37°C.
Chemical Compounds
4-Methoxy-N’-(phthalazin-1-yl) benzenesulfonohydrazide hydrochloride (staquorsin) was one among a group of other chemical compounds (George, 2018) screened for their S. aureus QS inhibitory activity. Staquorsin was selected for further analyses as it was the one showed the most promising results in a preliminary screening for anti-Agr activity. It was obtained according to the scheme described in Figure 1B. Briefly, equimolar amounts of hydralazine and 4-methoxybenzenesulphonyl chlorides (5 mmol) in 5 ml absolute ethanol were stirred at room temperature for 4 h. The obtained precipitate was filtered and washed with ethanol to give a creamy white solid. The structure was confirmed by both Infrared (IR) and proton nuclear magnetic resonance (1H NMR) spectroscopy techniques. The 3-[(4-isopropylphenyl) sulfonyl] [1,2,3] triazolo [1,5-a] quinazolin-5(4H)-one (savirin) was purchased from AK Scientific, Inc., United States. All chemicals were dissolved in dimethyl sulfoxide (DMSO) to a final concentration of 10 mM, aliquoted, and stored at −80°C.
Molecular Docking to AgrA Active Site and the in silico ADMET Properties Prediction
All the molecular modeling studies were carried out using Molecular Operating Environment (MOE, 10.2008) software. All minimizations were performed with MOE until root mean square deviation (RMSD) gradient of 0.05 kcal⋅mol–1Å–1 with MMFF94x force field and the partial charges were automatically calculated. The X-ray crystallographic structure of the AgrA of S. aureus (PDB ID: 3BS1) was downloaded from the protein data bank http://www.rcsb.org/. Water molecules and ligands not involved in the binding were removed. Then the protein was prepared for docking study using Protonate 3D protocol in MOE with default options. Key amino acids Glu217, Arg218, Cys228, and Tyr229 were used to define the binding site for docking (Sully et al., 2014). Triangle Matcher placement method and London dG scoring function were used for docking. First, the well-known Agr-inhibitor “Savirin” was docked in the vicinity of the active site of the protein with the ability to reproduce all the reported interactions with the key amino acids in the binding site; hydrogen bonding with Arg218 and π–π stacking with the Tyr229 (Sully et al., 2014). The validated setup was then used in predicting the ligands receptor interactions at the active site for the tested compound staquorsin. The absorption, distribution, metabolism, excretion and toxicity (ADMET) of both savirin and staquorsin were conducted using the Swiss ADME www.swissadme.ch/(Daina et al., 2017).
Assessment of the Alpha-Hemolysin Inhibition Activity
Cells were cultured in the presence of 40 μM of staquorsin for 22 h at 37°C in 5 ml TSB in a 50 ml falcon tube with shaking at 180 rpm. The hemolytic activity of the culture supernatant was determined as previously described (Bernheimer, 1988). Overnight cultures were normalized to the same OD600 then were centrifuged at 15,000 × g. The supernatants were serially diluted with sterile phosphate buffered saline (PBS) to a final volume of 600 μl and mixed with equal volume of freshly prepared rabbit RBCs suspension. The mixtures were incubated for 30 min at 37°C then centrifuged at 13,000 × g for 5 min. The absorbance of supernatant was measured at 540 nm and the HA50 was calculated as the reciprocal of the dilution that caused 50% hemolysis.
Assessment of the Delta-Hemolytic Activity
The supernatants of the overnight cultures grown in the presence of 200 μM of staquorsin in conditions as described above were used to assess the delta-hemolytic activity as previously described (Russell et al., 2006). Briefly, wells with 10 mm diameter in sheep blood agar were filled with 50 μl of the filtered culture supernatant, then incubated for 24 h at 37°C, and the area of the clear zones formed in the plates were measured using AutoCAD (Autodesk, United States). The data was plotted as a percentage of the area of the positive control zone. Another measurement was performed after the plates were further incubated at 4°C for 24 h to exclude any interference from beta-hemolysin.
Evaluation of the Lipolytic Activity
The supernatants of the overnight cultures used above for the delta-hemolytic activity were used to assess the lipolytic activity. This assessment was carried out according to the method described before (Smeltzer et al., 1992). Briefly, wells of 4 mm diameter in tributyrin agarose plates were filled with 20 μl filtered supernatants and the plates were incubated for 24 h at 37°C. The area of each zone was measured using AutoCAD and plotted as percentage of the area of the positive control.
Assessing the Effect on Biofilms Formation and Detachment
In order to assess the role of staquorsin in biofilm formation and detachment pre- and post-exposure assays were performed as described before (Nour El-Din et al., 2020) with slight modifications. Briefly, for biofilm formation, overnight cultures of staphylococcal cells were normalized to OD600 of 1.0 and diluted 1:100 in fresh TSB. Then one hundred μL aliquots were mixed with equal volume to give a final conc. of 40 μM staquorsin, or equivalent amount of DMSO, in TSB and placed in the wells of a sterile, untreated polystyrene, 96-well flat-bottomed plate. The plates were incubated at 37°C without shaking for 24 h. OD600 of the overnight culture was measured and then wells were washed three times with PBS and dried overnight. Adherent cells were stained with crystal violet (0.4% w/v) at room temperature for 15 min. Then the wells were washed three times with PBS. Biofilm formation was quantified by adding 150 μL of absolute ethanol and incubating at room temperature for 30 min with mild shaking. Finally, a total of 125 μL of the eluate was then transferred to a new sterile polystyrene microtiter plate and the absorbance of the plate at OD595 was recorded. The biofilm formation ability was represented by dividing the crystal violet OD595 reading by the planktonic growth OD600 reading. For the biofilm detachment assay, biofilms were allowed to form as described above, then after 24 h, the OD600 of the wells was measured after the incubation for normalization purposes. Before the addition of the staquorsin, growth medium and the unattached cells were discarded, and the biofilms were washed twice with PBS. Next, fresh TSB with either 40 μM staquorsin, or equivalent amount of DMSO were added to the wells and incubated at 37°C for 24 h. Then the plates were processed as described above for biofilm formation.
Quantitation of the RNA III Transcript
The level of the RNA III transcript was determined by quantitative reverse transcription polymerase chain reaction (qRT-PCR). The S. aureus cultures were incubated in the presence of 40 μM of staquorsin or equivalent volume of DMSO for 15 h at 37°C with shaking at 180 rpm (Eleaume and Jabbouri, 2004). RNA was isolated using the RNeasy mini kit (Qiagen, Germany) and reverse transcription was carried out by the QuantiTect Reverse Transcription Kit (Qiagen). The produced cDNA was quantified by real-time PCR using the GoTaq® qPCR Master Mix (Promega, United States). The 16S rRNA transcript was used as a normalizer using the primer pair (5′-TGAGATGTTGGGTTAAGTCCCGCA-3′) and (5′-CGGTTTCGCTGCCCTTTGTATTGT-3′) (Attia et al., 2010). While for the RNA III transcript, the primer pair (5′-ATAGCACTGAGTCCAAGGAAA-3′) and (5′-GCC ATCCCAACTTAATAACCATGT-3′) was used. The data obtained were analyzed using the Rotor-Gene Q Software (Qiagen) applying the ΔΔ CT method (Livak and Schmittgen, 2001), and using the normalized transcript level of RNA III in the presence of DMSO as the calibrator.
Determination of the MIC and Growth Inhibition Effect
The MIC, of both staquorsin and savirin, was determined by the broth microdilution method (CLSI, 2012), using a concentration range of 25–1600 μM. The MIC value was considered as the lowest concentration of the compound that showed no visible growth. In addition, to assess their possible effect on growth, the optical density of the cultures, was measured at 600 nm after 24 h of incubation at 37°C. Finally, in order to assess the effect of staquorsin on the growth kinetics of S. aureus, growth curves were constructed for both wild-type Newman and the MRSA strain USA300 in the presence of 40, 200, and 600 μM staquorsin. The growth pattern and extent were compared in the form of the growth curves to those of the same strains grown in the presence of the equivalent amounts of DMSO. Aliquots were taken at time points 0, 1, 2, 3, 4, 5, 6, 7, 8, and 24 h and the OD600 was recorded.
In vitro Assessment of Possible Resistance Development
Staphylococcus aureus cells were incubated in the presence of 40 μM of staquorsin. After 24 h, the culture was centrifuged, and the pelleted cells were resuspended in the same volume of fresh TSB containing 40 μM of staquorsin and the whole process was repeated for 20 passages. Stocks were made from cells resulting from each passage. The alpha-hemolysin activity was assessed as described above for the cells obtained after 5, 10, 13, 15, 18, and 20 passages. Also, the relative concentration of RNA III was determined for the cells obtained after 15 and 20 passages. At the same time, erythromycin MIC against the wild-type S. aureus was determined. The wild-type strain was then passed in the presence of 0.8 μg/ml (sub-MIC) erythromycin, and erythromycin MIC was determined after 8 and 16 passages.
Cytotoxicity Assay
Human skin fibroblast (HSF) was obtained from Nawah Scientific Inc., (Cairo, Egypt). Cells were maintained in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with 100 mg/mL of streptomycin, 100 units/mL of penicillin and 10% of heat-inactivated fetal bovine serum in humidified, 5% (v/v) CO2 atmosphere at 37°C. Cell viability was assessed by the sulforhodamine B (SRB) assay (Vichai and Kirtikara, 2006). Briefly, aliquots of 100 μL cell suspension (5 × 103 cells) were placed in 96-well plate and incubated in complete media for 24 h. Cells were treated with another aliquot of 100 μL media containing staquorsin at various concentrations (0.01–100 μM). After 24 h of drug exposure, cells were fixed by replacing the media with 150 μL of 10% Trichloroacetic acid (TCA) and incubated at 4°C for 1 h. The TCA solution was removed, and the cells were washed five times with distilled water. Aliquots of 70 μL 0.4% w/v SRB solution were added and incubated in a dark place at room temperature for 10 min. Plates were washed three times with 1% acetic acid and allowed to air-dry overnight. Then, 150 μL of 10 mM TRIS were added to dissolve the protein-bound SRB stain. The absorbance was measured at 540 nm using a BMG LABTECH®- FLUOstar Omega microplate reader (Ortenberg, Germany).
In vivo Evaluation of Staquorsin as an Infection Combating Agent
For the in vivo testing, the murine skin abscess model was adopted and a treatment regimen similar to the one described for savirin was followed (Sully et al., 2014). On the day prior to the experiment, the backs of female 6- to 8-week-old BALB/c mice were shaved. On day 0, the mice were weighed and distributed into two groups (five mice each). Bacterial cells were prepared as a suspension in 0.5% hydroxypropyl methylcellulose (HPMC). Fifty μl of the bacterial cell suspension (∼1 × 108 CFU) were injected subcutaneously concurrently with 40 nmol of staquorsin, in 50 μl of 0.5% HPMC. For delayed delivery, 80 nmol of staquorsin in 100 μl of 0.5% HPMC were administered 24 and 48 h post-infection. The mice were kept in their cages, where they were given food and water ad libitum, and their weights were monitored daily. On day seven, the mice were euthanized using an overdose of 2,2,2-tribromoethanol in pyrogen-free saline. The skin lesions were aseptically removed. Bacterial burdens in the excised samples were assessed by homogenizing the tissue in 1 ml of normal saline, followed by serial dilution, plating on mannitol salt agar (MSA), and incubating at 37°C for 24 h for colony forming unit (CFU) counts.
Statistical Analyses
GraphPad Prism v9 was used for statistical analyses. When appropriate, one-way ANOVA or two-way ANOVA were applied followed by Tukey’s multiple comparisons test. Otherwise, the performed test is described in the corresponding figure legend. The p-values ≤ 0.05 were considered significant.
Results
Spectroscopic Analyses Confirm the Staquorsin Structure
The staquorsin IR spectrum showed NH and SO2 bands at 3425, 3200, 1342, and 1157, respectively (Figure 1C). Its 1H NMR showed a singlet at 3.85 ppm corresponding to the OCH3 group along with two exchangeable signals at 8.50 and 10.67 ppm attributed to the 2 NH moieties. Additionally, the presence of two doublets confirmed the para substituted phenyl ring system (Figure 1D). These data confirmed the obtained product chemical structure.
Staquorsin Inhibits the Agr System Both Phenotypically and Molecularly
Since the Agr is known to control several virulence related phenotypes, staquorsin was tested for its ability to inhibit some of these phenotypes. First, staquorsin was successful in reducing the alpha-hemolytic activity of S. aureus. Staquorsin significantly (p < 0.05) lowered the HA50 of wild-type S. aureus almost to the same level of the ΔagrB mutant (Figure 2A). Assaying for the effect on the delta hemolytic activity, staquorsin significantly (p < 0.0001) reduced the area of the hemolysis zone in sheep blood agar by ∼88% compared to that of the wild-type strain (Figure 2B). This level of reduction was slightly lower than that caused by the ΔagrB mutant. Upon testing the effect on the lipolytic activity, staquorsin reduced the area of the lipolysis by more than 50% (p < 0.0001; Figure 2C). To confirm that the observed phenotypes are a reflection of a reduction in the RNA III transcript, which is the effector molecule of the Agr system, transcriptional analysis indicated that the growth in the presence of staquorsin reduced the level of transcription of the RNA III by 75% (p < 0.001) relative to its level in the presence of the vehicle (DMSO) (Figure 2D). The role of the Agr system in biofilms is well-established, as the repression of the agr enhances biofilm formation, while activation of the agr system is essential for the detachment of biofilm (Tan et al., 2018). As expected, the ΔagrB mutant formed more significant biofilm than its WT counterpart (Figure 2E). Growing the WT in the presence of 40 μM staquorsin, it formed a much better biofilm than that formed in the presence of equivalent amount of DMSO and very comparable that formed by the ΔagrB mutant (Figure 2E). Checking the effect of staquorsin on the biofilm formation in the USA300 strain, it only caused a slight, but non-significant increase (Figure 2E). Investigating the dispersal of formed biofilms, as expected the ΔagrB mutant showed less biofilm detachment than that seen in the WT (Figure 2F). Staquorsin managed to limit the dispersal of the WT biofilm better than just the DMSO, however this difference was not statistically significant (Figure 2F). Similar effect was also seen in strain USA300 (Figure 2F).
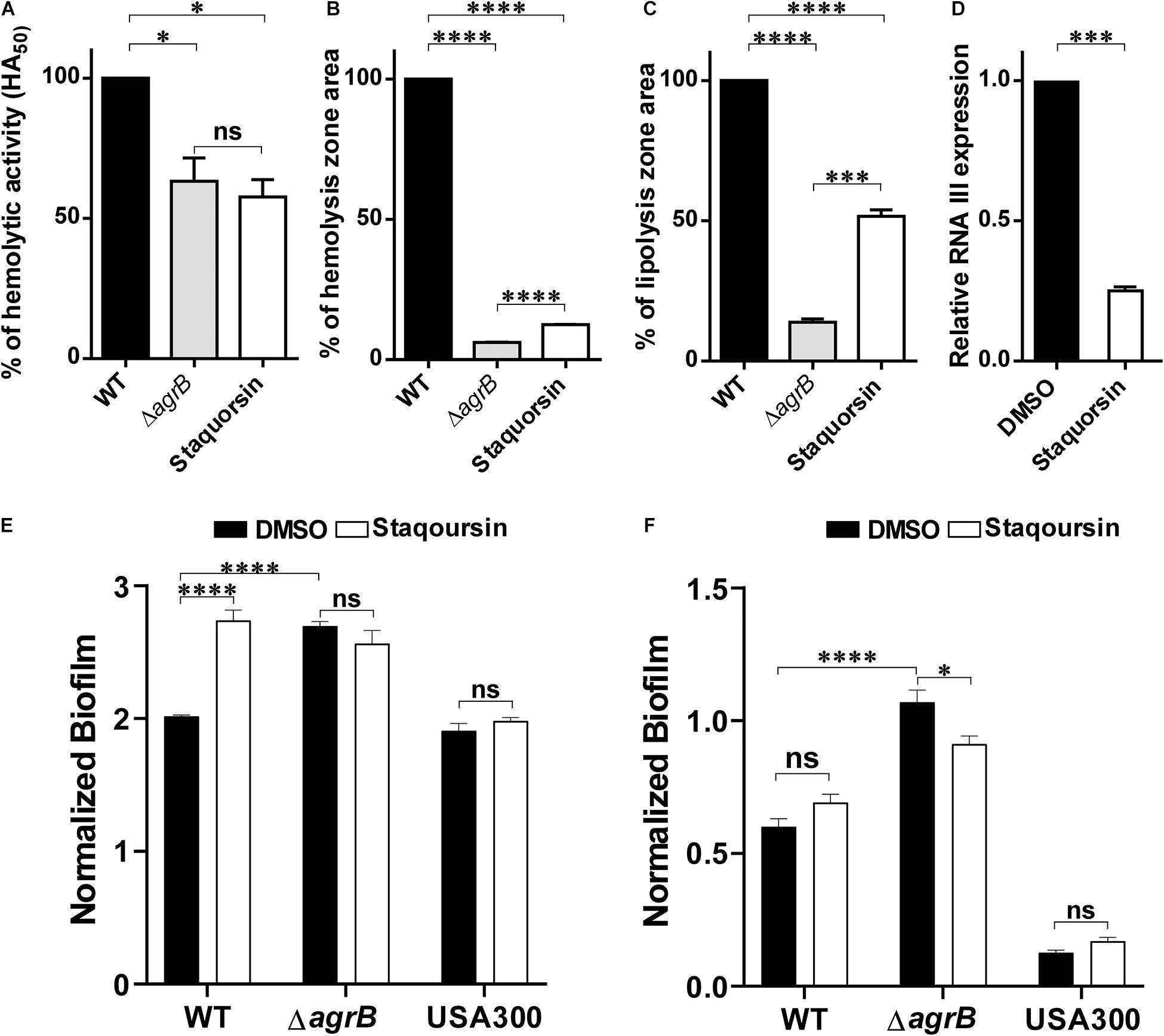
Figure 2. Staquorsin antagonizes the Agr system phenotypically and on the molecular level. The effect of staquorsin on some phenotypic Agr-related virulence activities (A) The alpha-hemolysin activity on rabbit RBCs as measured by the HA50 values. The HA50 of culture supernatants of cells grown in the presence of 40 μM staquorsin is presented as percentage of that of the WT grown in the absence of staquorsin. (B) The delta-hemolysin activity on sheep RBCs as measured by the area of the hemolysis zone. The area of the hemolysis zone produced by culture supernatants of cells grown in the presence of 200 μM staquorsin is presented as percentage relative to that of the WT grown in the absence of staquorsin. (C) The lipolytic activity as measured by the area of the zone of lipolysis on tributyrin agarose. The area of the lipolysis zone produced by culture supernatants of cells grown in the presence of 200 μM staquorsin is presented as percentage relative to that of the WT grown in the absence of staquorsin. In all of the above three charts, data of the ΔagrB strain was used as a negative control. Statistical analyses were performed by applying ordinary one-way ANOVA followed by Tukey’s multiple comparisons test. (D) Effect of staquorsin on the transcription of RNA III. The level of the RNA III in cells grown in the presence of 40 μM staquorsin was assessed and presented relative to the level in cells grown in its absence. Statistical analysis was performed by applying the unpaired t-test. (E) Effect of staquorsin on the biofilm formation. Strains were grown in the presence of either DMSO (black bars) or 40 μM staquorsin (white bars) and the formed biofilms were assessed by the crystal violet assay. The crystal violet reading at 595 nm was normalized by dividing it by the optical density of the planktonic culture at 600 nm. (F) Effect of staquorsin on the biofilm detachment. Biofilms was formed, then fresh TSB with either DMSO (black bars) or 40 μM staquorsin (white bars) was added for additional 24 h, then the biofilms were stained was crystal violet and processed as in (E). Two-way ANOVA was applied followed by Tukey’s multiple comparisons test. In all the above charts, the data presented is the average of three independent experiments, the error bars represent the standard deviation, “ns” indicates non-significant statistical difference (p-value > 0.05), “∗” p-value ≤ 0.05, “∗∗∗” p-value ≤ 0.001, and “****” p-value ≤ 0.0001.
Staquorsin Is Predicted to Bind to AgrA in a Way That Is Very Comparable to the Previously Described AgrA Inhibitor Savirin
In silico docking studies indicated that staquorsin binds to the active site of AgrA in a very close manner to that of savirin. First, “savirin” was docked in the vicinity of the active site of the AgrA protein with energy score (S) = −7.12 kcal/mol with the ability to reproduce all the reported interactions with the key amino acids in the binding site; hydrogen bonding with Arg218 and π–π stacking with the Tyr229 (Sully et al., 2014) (Figures 3A,B). The validated setup was then used in predicting the ligands receptor interactions at the active site for staquorsin. Upon calculating the absolute binding energy score of these bindings, staquorsin showed a marginally lower score than that of savirin with −6.67 kcal/mol. A summary of the binding energy score, interacting amino acids and binding interactions of both compounds is presented in Table 1.
Figure 3. In silico docking of both savirin and staquorsin to the S. aureus AgrA. Two-dimensional diagrams of savirin (A) and staquorsin (B) docking pose interactions with the key amino acids in the AgrA binding site. The blue regions denote an area that can form hydrophobic interactions. Dotted arrows denote the formation of H bonds from the donor to the acceptor atoms. The diagrams were generated using the Molecular Operating Environment (MOE, 10.2008) software.
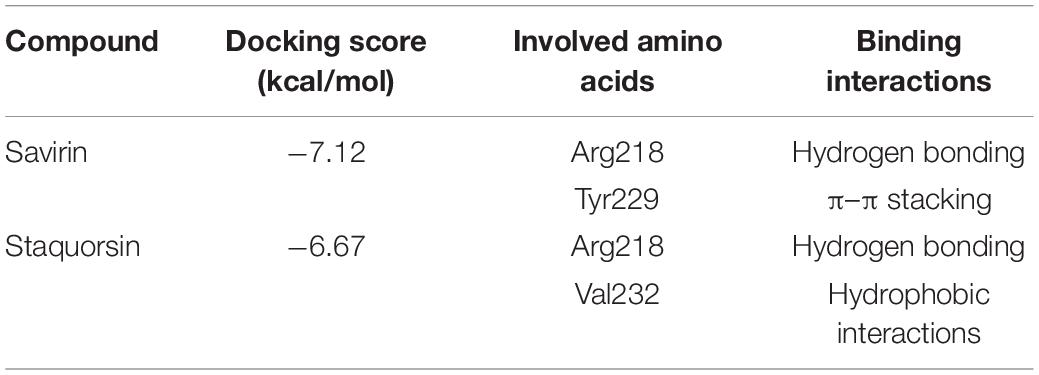
Table 1. Binding energy score, interacting amino acids, and binding interactions of savirin and staquorsin.
Staquorsin Is Predicted to Have Better ADMET Properties Than Savirin
The in silico comparison between both savirin and staquorsin regarding the ADMET properties revealed that staquorsin has better water solubility and less lipophilicity than savarin and consequently, it has predicted higher GIT absorption. Furthermore, both compounds cannot pass blood brain barrier, therefore, it could be assumed that they will have no CNS side effects. Also, both do not bind to plasma glycoprotein making them bioavailable to exert their action. Finally, staquorsin is expected to have lower drug-drug interactions than savarin based on their deduced liver enzymes cytochrome P450 (CYP) inhibition data as savirin can inhibit two types of cytochrome P450 while staquorsin inhibit only one type. A summary of this comparison is presented in Table 2.
Staquorsin, Has Very Limited Effect on S. aureus Growth at Very High Concentrations as Opposed to Savirin
To test the effect of the staquorsin on the growth of S. aureus, cells were grown in the presence of increasing concentrations of it. At the same time, the experiment was repeated using the savirin. As controls, both the wild-type and the agr-mutant were grown in the presence of equivalent amounts of the vehicle (DMSO). The OD at 600 nm after 24 h were recorded and plotted (Figure 4A). At concentrations as high as 600 μM, there was no significant difference between the extent of growth of S. aureus in the presence of either DMSO or staquorsin. On the other hand, with as low as 25 μM of savirin, there was a significant reduction in the growth extent of S. aureus (Figure 4A). Upon determination of the MIC for both compounds, savirin had an MIC of 100 μM while staquorsin MIC was >1600 μM. Therefore, staquorsin is a good inhibitor of the Agr system with minimal impact on the growth of S. aureus as opposed to savirin. In order to test whether staquorsin would affect the growth kinetics of S. aureus, two strains, Newman and USA300, were grown in different staquorsin concentrations and the growth was monitored over time. Upon testing strain Newman, staquorsin has no negative impact on either the growth rate or growth extent up to 600 μM (Figure 4B). Very similar effect was observed with USA300, where no differences were observed between the growth pattern in the presence of either staquorsin or the equivalent amounts of DMSO (Figure 4C). The lack of negative impacts was seen in the first 8 h of growth (Figures 4B,C) and even after 24 h (data not shown).
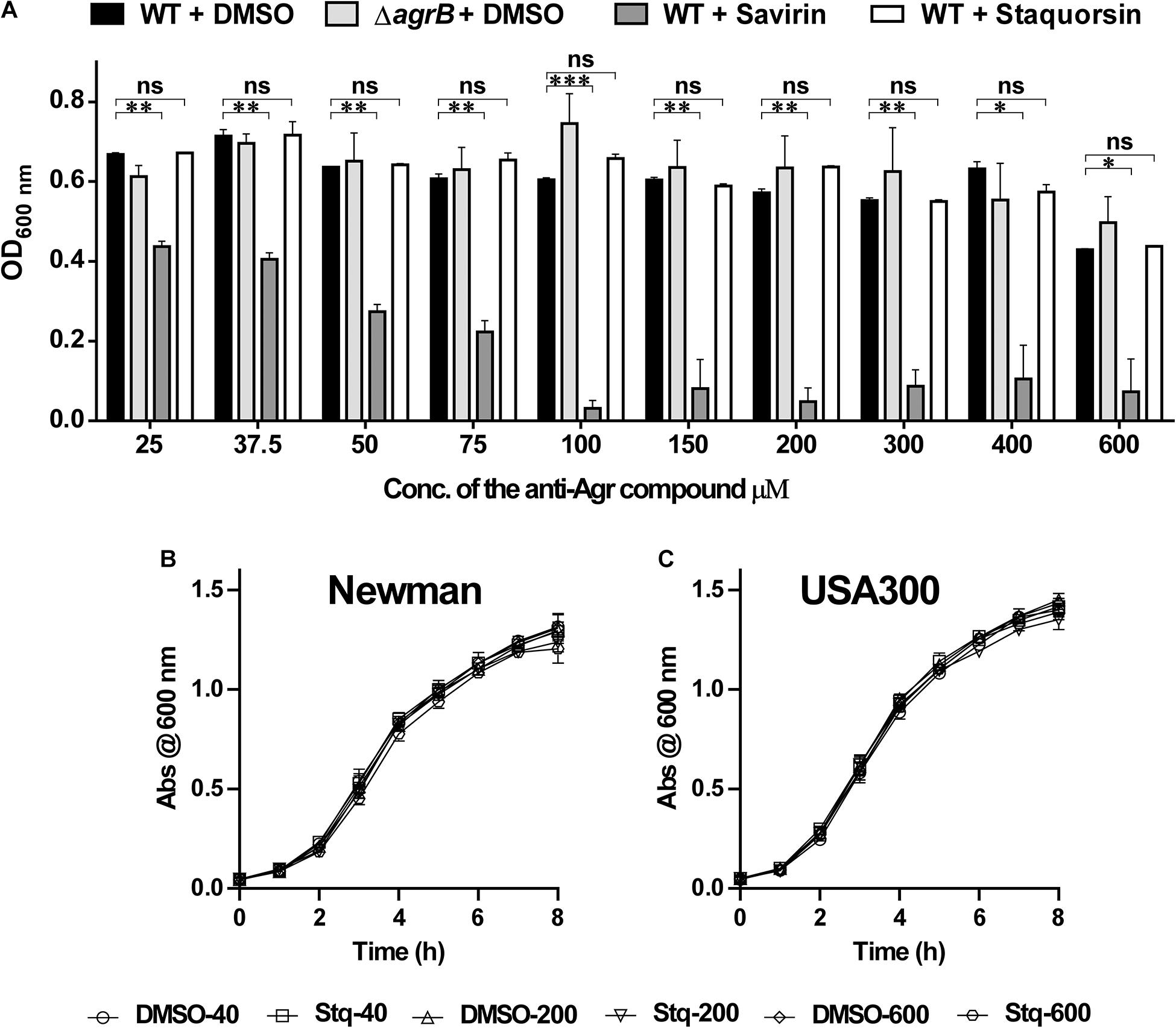
Figure 4. Staquorsin does not affect cell viability in very high concentrations as opposed to savirin. (A) Optical density of the bacterial culture after incubation for 24 h, in the presence of staquorsin (white bars), compared to the previously established quorum sensing inhibitor savarin (dark gray bars), the WT (black bars) and ΔagrB (light gray bars) grown in the presence of equivalent volumes of the vehicle (DMSO) used for dissolving the anti-Agr compounds. Effect of increasing concentrations of staquorsin on the growth kinetics of the MSSA strain Newman (B) and the MRSA strain USA300 (C). Cell were grown in the indicated concentration of either staquorsin (Stq) or DMSO and the optical density at 600 nm was recorded at the indicated time points. The data presented is the average of three independent experiments and the error bars are the standard deviation. Statistical analyses were performed by applying ordinary one-way ANOVA followed by Tukey’s multiple comparisons test on the readings corresponding to each concentration. The “ns” indicates non-significant statistical difference (p-value > 0.05), “∗” p-value ≤ 0.05, “∗∗” p-value ≤ 0.01, and “∗∗∗” p-value ≤ 0.001.
Staphylococcus aureus Does Not Develop Resistance to Staquorsin Activity Despite Repeated Sequential Passage in Its Presence
To address the possibility of resistance development to staquorsin, the wild-type S. aureus was passed sequentially 20 times in the presence of 40 μM staquorsin. Initially, the HA50 was reduced by staquorsin by ∼50% as earlier seen in Figure 2A, a comparable level of reduction was seen after 5, 10, 13, 15, 18, and 20 passages (Figure 5A). Interestingly, the wild-type strain lost part of its hemolytic activity after the 20th passage. This could be due a decrease in the activity of the Agr system after these multiple passages. However, staquorsin managed to reduce this activity by the same extent (∼50%) as observed in the previous passages (Figure 5A). On the molecular level, the effect of staquorsin on S. aureus RNA III level was also maintained even after repeated passages; whereas after 15 passages, staquorsin had similar activity. However, after 20 passages, although the level of RNA III transcription in the positive control was lowered, staquorsin has continued to reduce the RNA III transcription level, where RNA III concentration fell to less than one third that of the positive control (p < 0.001; Figure 5B). This confirms that the observed reduction in the hemolytic activity seen in the strain obtained after the 20th passage is due to decrease in the Agr activity. The wild-type, passed in subinhibitory concentration of erythromycin (0.8 μg/ml), required only 8 passages for the development of resistance. The MIC had risen from 1 to 4 μg/ml by the 8th passage and to 16 μg/ml by the 16th passage.
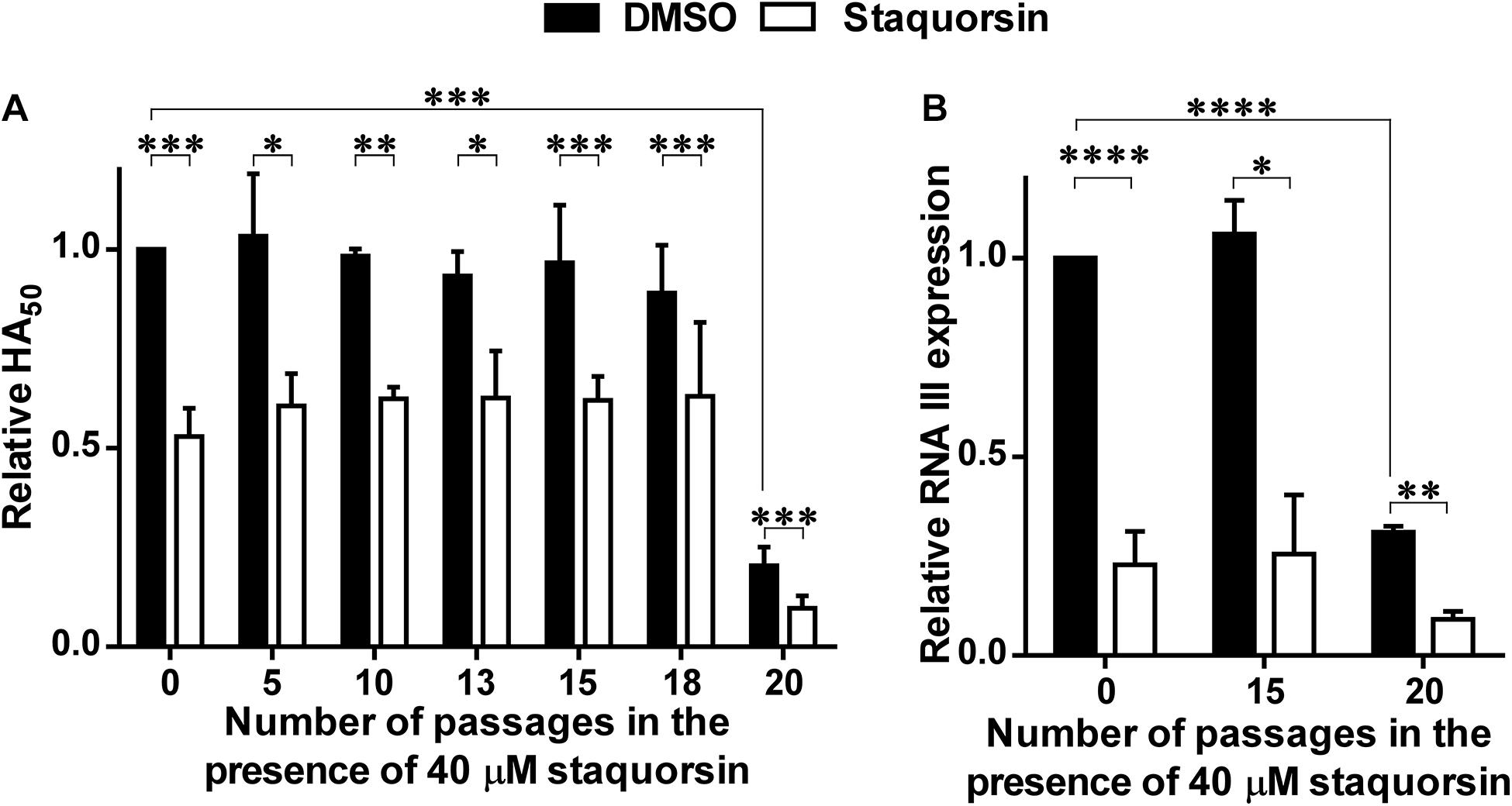
Figure 5. Staquorsin does not lose activity against S. aureus following repeated exposure. S. aureus cells were passed several times in the presence of 40 μM staquorsin then its effect on the HA50 of the culture supernatants (A) and the level of the RNA III (B) were assayed. In both charts, the data is presented relative to the results of the S. aureus cells which were not exposed to staquorsin (the first black column). The data presented is the average of three independent experiments and the error bars are the standard deviation. Statistical analyses were performed by applying an unpaired t-test for each pair representing the readings after the indicated number of passages that the cells were exposed to. The “∗” indicates a p-value ≤ 0.05, “∗∗” p-value ≤ 0.01, “∗∗∗” p-value ≤ 0.001, and “****” p-value ≤ 0.0001.
Staquorsin Inhibition of the Agr-System Promotes Skin Infection Management
The potential ability of staquorsin, to modulate S. aureus infection, was assessed in vivo by adopting the murine skin infection model. Prior to conducting the animal experiment, cytotoxicity assays were performed to ensure the safety of this compound. The HSF cells showed viability up to 95.06% ± 0.26 following incubation with 100 μM of staquorsin for 24 h (Table 3). Keeping in mind that most of the observed in vitro phenotypes in the current study was detected with just 40 μM, this would argue that staquorsin is predicted to be very safe upon contact with mammalian cells. All mice infected with S. aureus started to lose weight. Those treated with just the vehicle continued to lose weight throughout the whole course of the infection (Figure 6A). On the other hand, mice given staquorsin started to attain significant weight gain starting from day 5 post-infection (Figure 6A). Mice given the vehicle only, showed inflamed skin with abscess formation and ulceration. Whereas, mice treated with staquorsin showed signs of healing, evident by less inflammation and the lack of ulceration and abscess formation by day seven (Figure 6B). Upon evaluating the bacterial loads in the skin lesions, the mice treated with staquorsin showed a significant reduction of almost three log cycles when compared to those in the mice treated with the vehicle only (Figure 6C).
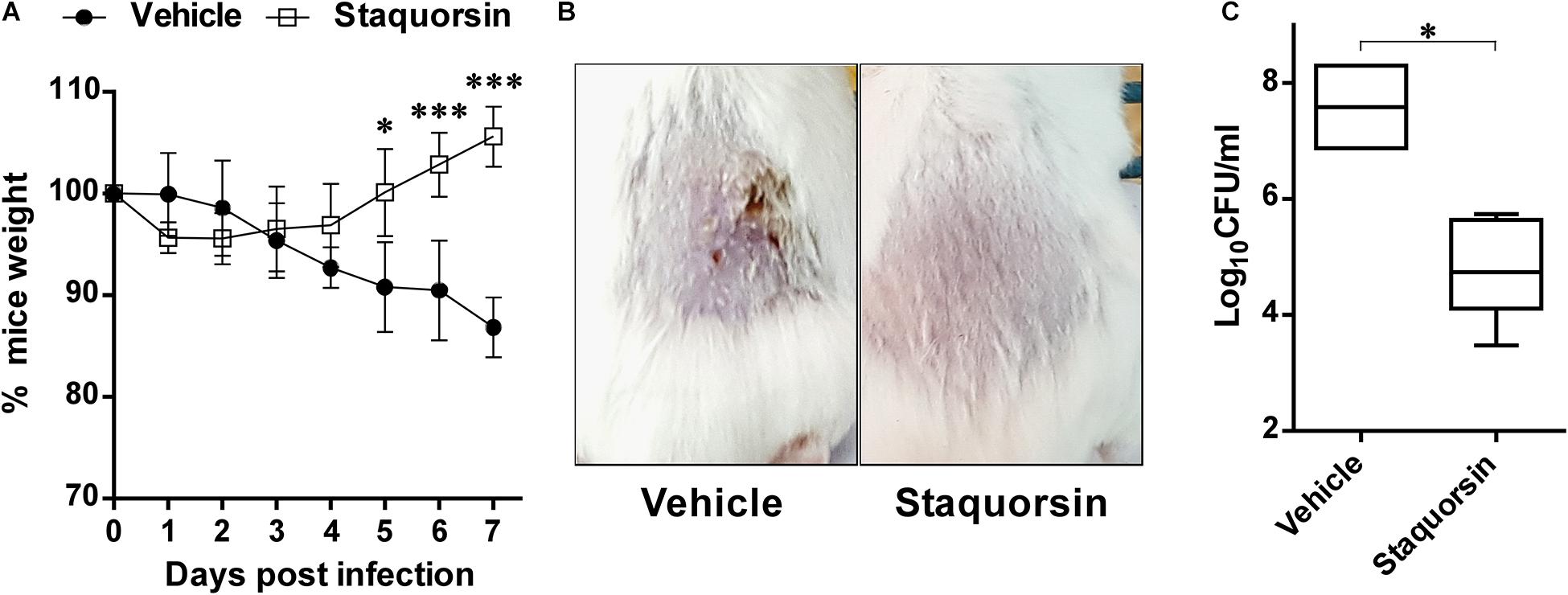
Figure 6. Staquorsin is effective in controlling S. aureus skin infection. (A) Monitoring the weights of the mice throughout the course of the infection. The weights of mice were determined every 24 h and they are presented as percentages of the weights at day 0. The data presented is the average of the readings of the mice in each group; treated with the vehicle (closed circles) and staquorsin (open squares), and the error bars represent the standard deviation. Statistical analyses were performed by applying an unpaired t test for each pair representing the readings at the indicated time point. (B) Photographs of a representative mouse of each group (treated with vehicle vs staquorsin). The images were taken on day seven post-infection. (C) Box plots of the bacterial loads in the skin lesions of mice infected with S. aureus and treated with either the vehicle or staquorsin. The data is presented as Log10CFU/ml obtained following plating the homogenates of the skin lesions excised from the infected mice. The whiskers span the difference between the minimum and maximum readings; the horizontal bar represents the mean. The “∗” indicates a p-value ≤ 0.05, and “∗∗∗” p-value ≤ 0.001.
Discussion
In 2014, the WHO announced antimicrobial resistance as a public health threat that requires global action. Searching for novel antimicrobials to overcome these resistant pathogens is not an ideal solution. As the development of resistance to new agents typically occurs at much faster rates than expected, in addition to the scarcity of the new compounds developed. Finding new strategies to tackle resistant organisms is of great concern.
Anti-virulence agents offer a promising alternative that can help in combating resistant bacteria (Totsika, 2016). Targeting S. aureus Agr-system mediated QS has proven efficient in reducing its virulence allowing the host defense to eradicate different S. aureus infections (Sully et al., 2014).
In this work, a newly synthesized compound was tested for its S. aureus anti-virulence capabilities especially those related to the Agr system. The developed compound was compared to the previously described S. aureus virulence-inhibitor savirin (Sully et al., 2014). The Agr system is a master regulator of QS which in turn controls diverse virulence factors of S. aureus (Cheung et al., 2004; Queck et al., 2008). One component of the Agr system is AgrA, which is a response regulator that upon activation binds to either the P2 or the P3 promotors. Binding to the former, activates a positive feedback loop to amplify the production of the components of the system, while binding to the latter induces the production of the RNA III (Koenig et al., 2004). RNAIII is a small regulatory RNA molecule that blocks the translation of the toxins repressor (Rot), leading to upregulation of multiple virulence factors (Boisset et al., 2007; Le and Otto, 2015).
Staquorsin demonstrated several phenotypes that linked it to having an anti-Agr activity such as down-regulation of the alpha and delta-hemolytic activities, lipolytic activity, the down-regulation of the RNA III transcript, enhancing biofilm formation, and blocking the biofilm dispersion (Figure 2). In addition, the in silico analysis indicated that staquorsin docks in the AgrA C-terminal at a critical position for proper folding and DNA binding (Sun et al., 2012), with a slightly lower absolute binding score than savirin. Inhibition of the agr operon through AgrA is highly advantageous, as AgrA sequence is conserved among the four types of the Agr system (Tan et al., 2018).
These results provoked the question, what advantage would staquorsin have over the already described QS inhibitor, savirin. A key feature sought in anti-virulence therapies is their lack of negative impact on the viability of the pathogen, leading to a decrease in the frequency of resistance development (Rasko and Sperandio, 2010; Heras et al., 2015; Rampioni et al., 2017; Totsika, 2017; Fleitas Martinez et al., 2019; Amer et al., 2021). In the work described by Sully and co-workers, they showed that savirin has no effect on the viability of S. aureus in a concentration up to 6.3 μM, however, upon testing the anti-Agr activity of this compound they routinely used a concentration of 5 μg/ml (13.6 μM) which is more than double the concentration indicated to have no effect on the viability (Sully et al., 2014). Accordingly, we wanted to test whether higher concentrations of savirin would have any impact on the viability of S. aureus. Upon testing less than double the concentration used by Sully and co-workers (25 μM) there was a significant reduction (∼35%) in the extent of growth (Figure 4A). Higher concentrations of savirin led to more reduction in the extent of the growth of S. aureus. On the contrary, staquorsin did not demonstrate significant impact on the growth of S. aureus at concentrations up to 600 μM. Accordingly, staquorsin comes with a great advantage over savirin with its ability to exert much lesser pressure on the microbial cell by exerting minimal effect on its viability. Moreover, no negative impact was seen on the growth kinetics in a concentration up to 600 μM.
The lack of selective pressure of staquorsin was confirmed by the sequential passage of S. aureus 20 times at a relatively high concentration (40 μM) and yet there was no development of resistance. This lack of resistance development was confirmed both phenotypically and molecularly. It is worth mentioning that the serial passage of S. aureus in the presence of staquorsin resulted in a strain after the 20th passage that has lower basal Agr activity as demonstrated by lower RNA III transcript and lower HA50. This phenomenon has been seen before by Somerville and co-workers, where the in vitro serial passage of S. aureus led to changes in physiology, virulence factors production, and agr nucleotide sequence (Somerville et al., 2002). Yet despite of this observed decrease, growth of this strain in the presence of staquorsin demonstrated a further decrease in the Agr activity indicating the staquorsin is still active on its target despite its lowered basal activity. On the other hand, resistance developed rapidly for erythromycin as the MIC quadrupled after the 8th passage.
Resistance to QS inhibitors may develop in vivo. In the case the agr system, mutations causing inhibitor resistance can result from agr operon dysfunctional mutants, which are less able to colonize and cause infection to the host. They are also incapable of spreading efficiently between patients (Shopsin et al., 2010). However, a recent review of literature and meta-analysis carried out by Lee and co-workers concluded that dysfunctional agr could influence the outcome of invasive S. aureus infections depending on conditions like the oxacillin susceptibility and the site of infection (Lee et al., 2020). This was observed prominent in MRSA and pneumonia where the dysfunctional agr was generally associated with unfavorable clinical outcomes.
To assess the efficacy of staquorsin in vivo, it was tested in the skin infection model as most MRSA infections are of this type and the role of the Agr system in establishing infection is confirmed (Salam and Quave, 2018). Staquorsin was able to reduce the signs of infections like weight-loss and the extent of inflammation in the tissues. In addition, bacterial loads in the skin lesions were significantly lowered.
In conclusion, the current study models staquorsin as a promising S. aureus QS inhibitor. It demonstrated extraordinary efficiency to strip down S. aureus of a substantial part of its weaponry. Further study is nevertheless required to confirm its possible action on different agr types in pathogenic S. aureus strains. Also, for its ability to clear different infection types, other than the skin infection.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by Research Ethics Committee of the Faculty of Pharmacy, Cairo University with number MI(1682).
Author Contributions
All authors have contributed to the design of the experiments, collecting the data, analyzing the results, drafting the manuscript, and reviewing the manuscript prior to submission.
Funding
This work was supported by a grant awarded to ASA by the Interdisciplinary Research Grant system (Cycle 2) from the Faculty of Pharmacy, Cairo University. Award number (IRG-2014-01). Part of the APC for publishing this article will be paid by Cairo University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Prof. Dr. Hala F. Zaki from the Department of Pharmacology and Toxicology, Faculty of Pharmacy, Cairo University for technical support. Also, we would like to thank Prof. Dr. Eric P. Skaar of Vanderbilt University, United States for providing the S. aureus strains.
References
Amer, M. A., Wasfi, R., Attia, A. S., and Ramadan, M. A. (2021). Indole Derivatives Obtained from Egyptian Enterobacter sp. Soil Isolates Exhibit Antivirulence Activities against Uropathogenic Proteus mirabilis. Antibiotics 10:363. doi: 10.3390/antibiotics10040363
Attia, A. S., Benson, M. A., Stauff, D. L., Torres, V. J., and Skaar, E. P. (2010). Membrane damage elicits an immunomodulatory program in Staphylococcus aureus. PLoS Pathog. 6:e1000802. doi: 10.1371/journal.ppat.1000802
Ayukekbong, J. A., Ntemgwa, M., and Atabe, A. N. (2017). The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob. Resist. Infect. Control. 6, 1–8.
Bae, T., Banger, A. K., Wallace, A., Glass, E. M., Aslund, F., Schneewind, O., et al. (2004). Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. U S A. 101, 12312–12317. doi: 10.1073/pnas.0404728101
Bernheimer, A. W. (1988). Assay of hemolytic toxins. Methods Enzymol. 165, 213–217. doi: 10.1016/s0076-6879(88)65033-6
Boisset, S., Geissmann, T., Huntzinger, E., Fechter, P., Bendridi, N., Possedko, M., et al. (2007). Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 21, 1353–1366. doi: 10.1101/gad.423507
CDC (2019). Antibiotic Resistance Threats in the United States. Washington, D.C: U.S. Department of Health and Human Services.
Chambers, H. F. (2001). The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 7, 178–182. doi: 10.3201/eid0702.010204
Chambers, H. F., and Deleo, F. R. (2009). Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7, 629–641. doi: 10.1038/nrmicro2200
Cheung, A. L., Bayer, A. S., Zhang, G., Gresham, H., and Xiong, Y. Q. (2004). Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Medical Microbiol. 40, 1–9. doi: 10.1016/s0928-8244(03)00309-2
CLSI (2012). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. Wayne, PA: Clinical and Laboratory Standards Institute.
Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7:42717.
Diep, B. A., Gill, S. R., Chang, R. F., Phan, T. H., Chen, J. H., Davidson, M. G., et al. (2006). Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367, 731–739. doi: 10.1016/s0140-6736(06)68231-7
Duthie, E. S., and Lorenz, L. L. (1952). Staphylococcal coagulase; mode of action and antigenicity. J. General Microbiol. 6, 95–107.
Eleaume, H., and Jabbouri, S. (2004). Comparison of two standardisation methods in real-time quantitative RT-PCR to follow Staphylococcus aureus genes expression during in vitro growth. J. Microbiol. Methods 59, 363–370. doi: 10.1016/j.mimet.2004.07.015
Fleitas Martinez, O., Cardoso, M. H., Ribeiro, S. M., and Franco, O. L. (2019). Recent Advances in Anti-virulence Therapeutic Strategies With a Focus on Dismantling Bacterial Membrane Microdomains, Toxin Neutralization, Quorum-Sensing Interference and Biofilm Inhibition. Front Cell Infect. Microbiol. 9:74.
George, R. F. (2018). Facile synthesis of simple 2-oxindole-based compounds with promising antiproliferative activity. Fut. Med. Chem. 10, 269–282. doi: 10.4155/fmc-2017-0148
Gordon, C. P., Williams, P., and Chan, W. C. (2013). Attenuating Staphylococcus aureus virulence gene regulation: a medicinal chemistry perspective. J. Med. Chemistry 56, 1389–1404.
Heras, B., Scanlon, M. J., and Martin, J. L. (2015). Targeting virulence not viability in the search for future antibacterials. Br. J. Clin. Pharmacol. 79, 208–215. doi: 10.1111/bcp.12356
Koenig, R. L., Ray, J. L., Maleki, S. J., Smeltzer, M. S., and Hurlburt, B. K. (2004). Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J. Bacteriol. 186, 7549–7555. doi: 10.1128/jb.186.22.7549-7555.2004
Le, K. Y., and Otto, M. (2015). Quorum-sensing regulation in staphylococci-an overview. Front. Microbiol. 6:1174.
Lee, S. O., Lee, S., Lee, J. E., Song, K. H., Kang, C. K., Wi, Y. M., et al. (2020). Dysfunctional accessory gene regulator (agr) as a prognostic factor in invasive Staphylococcus aureus infection: a systematic review and meta-analysis. Sci. Rep. 10:20697.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199.
Munguia, J., and Nizet, V. (2017). Pharmacological targeting of the host–pathogen interaction: alternatives to classical antibiotics to combat drug-resistant superbugs. Trends Pharmacol. Sci. 38, 473–488. doi: 10.1016/j.tips.2017.02.003
Nour El-Din, H. T., Elhosseiny, N. M., El-Gendy, M. A., Mahmoud, A. A., Hussein, M. M. M., and Attia, A. S. (2020). A Rapid Lysostaphin Production Approach and a Convenient Novel Lysostaphin Loaded Nano-emulgel; As a Sustainable Low-Cost Methicillin-Resistant Staphylococcus aureus Combating Platform. Biomolecules 10:435. doi: 10.3390/biom10030435
Queck, S. Y., Jameson-Lee, M., Villaruz, A. E., Bach, T. H., Khan, B. A., Sturdevant, D. E., et al. (2008). RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32, 150–158. doi: 10.1016/j.molcel.2008.08.005
Rampioni, G., Pillai, C. R., Longo, F., Bondi, R., Baldelli, V., Messina, M., et al. (2017). Effect of efflux pump inhibition on Pseudomonas aeruginosa transcriptome and virulence. Sci. Rep. 7:11392.
Rasko, D. A., and Sperandio, V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug. Discov. 9, 117–128. doi: 10.1038/nrd3013
Russell, F. M., Biribo, S. S., Selvaraj, G., Oppedisano, F., Warren, S., Seduadua, A., et al. (2006). As a bacterial culture medium, citrated sheep blood agar is a practical alternative to citrated human blood agar in laboratories of developing countries. J. Clin. Microbiol. 44, 3346–3351. doi: 10.1128/jcm.02631-05
Salam, A. M., and Quave, C. L. (2018). Targeting virulence in Staphylococcus aureus by chemical inhibition of the accessory gene regulator system in vivo. MSphere 3, e500–e517.
Shopsin, B., Eaton, C., Wasserman, G. A., Mathema, B., Adhikari, R. P., Agolory, S., et al. (2010). Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 202, 1593–1599.
Smeltzer, M. S., Hart, M. E., and Iandolo, J. J. (1992). Quantitative spectrophotometric assay for staphylococcal lipase. Appl. Environ. Microbiol. 58, 2815–2819. doi: 10.1128/aem.58.9.2815-2819.1992
Somerville, G. A., Beres, S. B., Fitzgerald, J. R., DeLeo, F. R., Cole, R. L., Hoff, J. S., et al. (2002). In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J. Bacteriol. 184, 1430–1437. doi: 10.1128/jb.184.5.1430-1437.2002
Sully, E. K., Malachowa, N., Elmore, B. O., Alexander, S. M., Femling, J. K., Gray, B. M., et al. (2014). Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 10:e1004174. doi: 10.1371/journal.ppat.1004174
Sun, F., Liang, H., Kong, X., Xie, S., Cho, H., Deng, X., et al. (2012). Quorum-sensing agr mediates bacterial oxidation response via an intramolecular disulfide redox switch in the response regulator AgrA. Proc. Natl. Acad. Sci. 109, 9095–9100. doi: 10.1073/pnas.1200603109
Tan, L., Li, S. R., Jiang, B., Hu, X. M., and Li, S. (2018). Therapeutic targeting of the Staphylococcus aureus accessory gene regulator (agr) system. Front. Microbiol. 9:55.
Totsika, M. (2016). Benefits and challenges of antivirulence antimicrobials at the dawn of the post-antibiotic era. Drug Delivery Lett. 6, 30–37. doi: 10.2174/2210303106666160506120057
Totsika, M. (2017). Disarming pathogens: benefits and challenges of antimicrobials that target bacterial virulence instead of growth and viability. Fut. Medicinal Chemistry 9, 267–269. doi: 10.4155/fmc-2016-0227
Keywords: S. aureus, quorum-sensing, resistance, Agr, MRSA, savirin, anti-virulence, anti-biofilm
Citation: Mahdally NH, George RF, Kashef MT, Al-Ghobashy M, Murad FE and Attia AS (2021) Staquorsin: A Novel Staphylococcus aureus Agr-Mediated Quorum Sensing Inhibitor Impairing Virulence in vivo Without Notable Resistance Development. Front. Microbiol. 12:700494. doi: 10.3389/fmicb.2021.700494
Received: 26 April 2021; Accepted: 11 June 2021;
Published: 05 July 2021.
Edited by:
Giovanna Batoni, University of Pisa, ItalyReviewed by:
Faizan Abul Qais, Aligarh Muslim University, IndiaAdline Princy Solomon, SASTRA University, India
Ranjith Kumavath, Central University of Kerala, India
Copyright © 2021 Mahdally, George, Kashef, Al-Ghobashy, Murad and Attia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmed S. Attia, YWhtZWQuYXR0aWFAcGhhcm1hLmN1LmVkdS5lZw==
 Norhan H. Mahdally1
Norhan H. Mahdally1 Riham F. George
Riham F. George Mona T. Kashef
Mona T. Kashef Ahmed S. Attia
Ahmed S. Attia