- College of Veterinary Medicine, Northwest A&F University, Yangling, China
Pathogenic E. coli are among the most frequently isolated bacterial pathogens on large-scale sheep farms in China. Antibiotic use in wool sheep production is a risk factor for promoting the emergence of resistant E. coli. To reveal the differences of E. coli populations in sheep from different farming systems the antimicrobial resistance, virulence genes, biofilm formation, and phylogroups of 500 E. coli isolates obtained between September 2019 and December 2020 in northwest China from diarrheic infections of intensive farming and free-range sheep were analyzed. The antimicrobial susceptibility test for 12 classes of antimicrobial agents was determined using the broth microdilution susceptibility method, and PCR was used to detect the differences in virulence genes and phylogroups. Additionally, biofilm formation was determined using microtiter plate and slide agglutination methods. Among the 500 E. coli isolates, the majority of the isolates were multidrug resistant (75.4%) and carried at least one virulence gene (94.8%). We observed that 412 (82.4%), 360 (72.0%), and 266 (53.2%) are found to be resistant to sulfisoxazole, florfenicol, and tetracyclines, respectively. Resistance was also observed to mequindox (46.8%), ampicillin (43.6%), spectinomycin (38.6%), enrofloxacin (34.2%), ceftiofur (21.0%), gentamycin (20.4%), ceftazidime (17.8%), and polymyxin B (7.8%) but no resistance was found to meropenem. These results showed that strains from free-range subjects had fewer antibiotic resistance strains rather than sheep that were intensively farmed (P < 0.05). We observed fifteen virulence genes, of which etrA (n = 401, 80.2%) is the most common. In addition, EAEC (86.4%) is dominant among free-range sheep and EHEC (80.1%) is dominant among intensive farming. Among all virulence genes, the strongest correlation was found between etrA and papC gene (P < 0.001, OR = 455.68). Similarly, the strongest correlation was also found between eltA and sulfisoxazole (P < 0.001, OR = 877). Furthermore, the majority of the E. coli isolates belonged to phylogroup B1 (50.6%), followed by phylogroup C (20.6%), A (7.4%), E (7.4%), D (5.8%), B2 (1.6%), and F (1%). Interestingly, phylogroup B2 and D were all distributed in intensive farms. In addition, 33 (6.6%), 373 (74.6%), and 94 (18.8%) showed moderate, weak, and no connection biofilm formation ability, respectively. These data uncovered that wool sheep serve as a reservoir of pathogenic E. coli harboring multiple resistance phenotypes and virulence genes. The overlapping virulence-associated traits between IPEC and ExPEC indicated the zoonotic potential and safety threats of sheep food products. It is urgent to improve the proper use of antimicrobials in China as well as other countries.
Introduction
Escherichia coli (E. coli) is one of the most important bacteria for animals and humans. It is distributed all over the world and will bring huge economic losses to the breeding industry and public health. Pathogenic E. coli can produce potent toxins and is divided into intestinal pathogenic E. coli (IPEC) and extraintestinal pathogenic E. coli (ExPEC) according to the production of virulence factors (such as adhesin, capsule synthesis, toxins, etc.) (Habouria et al., 2019). The IPEC is subclassified into Enteropathogenic E. coli (EPEC), Enterotoxigenic E. coli (ETEC), Enteroaggregative E. coli (EAEC), Enteroinvasive E. coli (EIEC), and Enterohemorrhagic E. coli (EHEC) (Guimarães et al., 2019). IPEC infection can cause diarrhea, intestinal inflammation, and even deaths in severely affected animals and children (Malberg Tetzschner et al., 2020). In addition, ExPEC is mainly responsible for urinary tract infections (UTI) and is the main cause of meningitis, especially for babies, within 30 days of birth (Russo and Johnson, 2003).
In China, E. coli is ubiquitously distributed in the whole country with variable prevalence between intensive farming and free-range wool sheep. Current control strategies against E. coli mainly rely on repeated antibiotic treatments. However, in the last 20 years, the widespread use of antibiotics, including misuse and overuse, has helped bacteria naturally evolve and develop resistance (Gabale et al., 2020). Meanwhile, continuous use of antibiotics in animals strengthens the selective pressure and the onset of drug-resistant strains, which are easily transmitted to humans (Laxminarayan et al., 2013; Van Boeckel et al., 2017). Pathogenic E. coli of wool sheep can be used as a major reservoir for drug-resistant genes and the spread of resistant bacteria from food animals to humans through the food chain. The British government report estimates that by 2050, the number of deaths due to antibiotic-resistant infections will exceed 10 million every year (O’Neill, 2016). Each year in Europe, about 25,000 people die and up to 2.8 million people get a drug-resistance bacterial infection (ECDC/EMEA, 2009). As such, one of the greatest threats to humanity in the present world is the potential emergence and global spread of resistant bacteria (Nadeem et al., 2020).
Ningxia, Shaanxi, Inner Mongolia, and Qinghai are all major animal husbandry provinces in China. However, in recent years, antimicrobial-resistant E. coli, especially multiple drug-resistant (MDR) strains have caused frequent outbreaks of fatal hemorrhagic diarrhea diseases in wool sheep. Wool sheep are one of the most important reservoirs for pathogenic E. coli that can cause a series of clinical symptoms (Wani et al., 2007; Kim et al., 2010). As such, it is necessary to explore the antimicrobial resistance, virulence genes, biofilm formation, and phylogenetic groups of E. coli, which are critical for the therapeutic management of sheep. Whether in human or veterinary medicine, previous reports on bacterial isolates have revealed the relationship between virulence factors and resistance genes (Pan et al., 2020; Pérez et al., 2020). However, despite several studies, these links remain unclear. In addition, these studies evaluated the resistance of E. coli isolates from animals, rather than the resistance of strains derived from different feeding methods.
It is therefore currently necessary to assess the risk of E. coli antibiotic resistance (AMR) and virulence genes in sheep for public health. However, little is known about antimicrobial resistance, virulence factors, biofilm, and phylogenetic groups of wool sheep E. coli isolated in China (Chandran and Mazumder, 2013). Therefore, the detection of virulence genes and the analysis of drug resistance in wool sheep E. coli have significance for the prevention and control of diseases caused by E. coli. Furthermore, further research in this area will be beneficial for minimizing the increasing transmission of drug-resistant E. coli strains in the wool sheep. To our knowledge, this is the first report on antibiotic resistance, virulence factors, and phylogroups, characteristics in E. coli strains isolated from wool sheep with diarrhea that compares intensive and free-range farming in China.
Materials and Methods
Bacterial Isolation and Identification
A total of 575 rectal swabs were collected from adult wool sheep with diarrhea, in Northwest China, between September 2019 and December 2020. Figure 1 illustrates the geographical region where the rectal swabs were collected, which includes the locations of Ningxia, Shaanxi, Qinghai, and Inner Mongolia. All rectal swabs were transported to Northwest A&F University (NWAFU Yangling, China), College of Veterinary Medicine, Microbiology, and Molecular Diagnostic laboratory under freezing conditions within 2 h. The Animal Welfare and Research Ethics Committee of Northwest A&F University approved the protocol of the experiment (Number: NWAFUSM2019009). This study was performed under standard biosecurity and institutional safety procedures. Rectal sterile swabs were used to isolate and identify E. coli, and only one isolate was examined for each sample. The rectal swab of each sample was inoculated into sterile tubes containing 10 ml of Luria-Bertani (LB) broth and mixed vigorously (180 r/min) overnight at 37°C. After enrichment, a loopful of the broth was streaked onto MacConkey agar (Oxiod) and incubated aerobically at 37°C for 18–24 h. From each plate, a single pink-colored sample was chosen, and sub-cultured onto eosin-methylene blue (EMB) agar (Oxiod), before being incubated as described above. Colonies showing greenish metallic colors were considered presumptive E. coli isolates, and then confirmed as E. coli via biochemical analysis, using Biomerieux VITEK2 Compact Automated microbial identification system (BioMerieux, France), according to the manufacturers. The E. coli strains were suspended in 20% glycerol broth at −80°C for further bacteriological analysis.
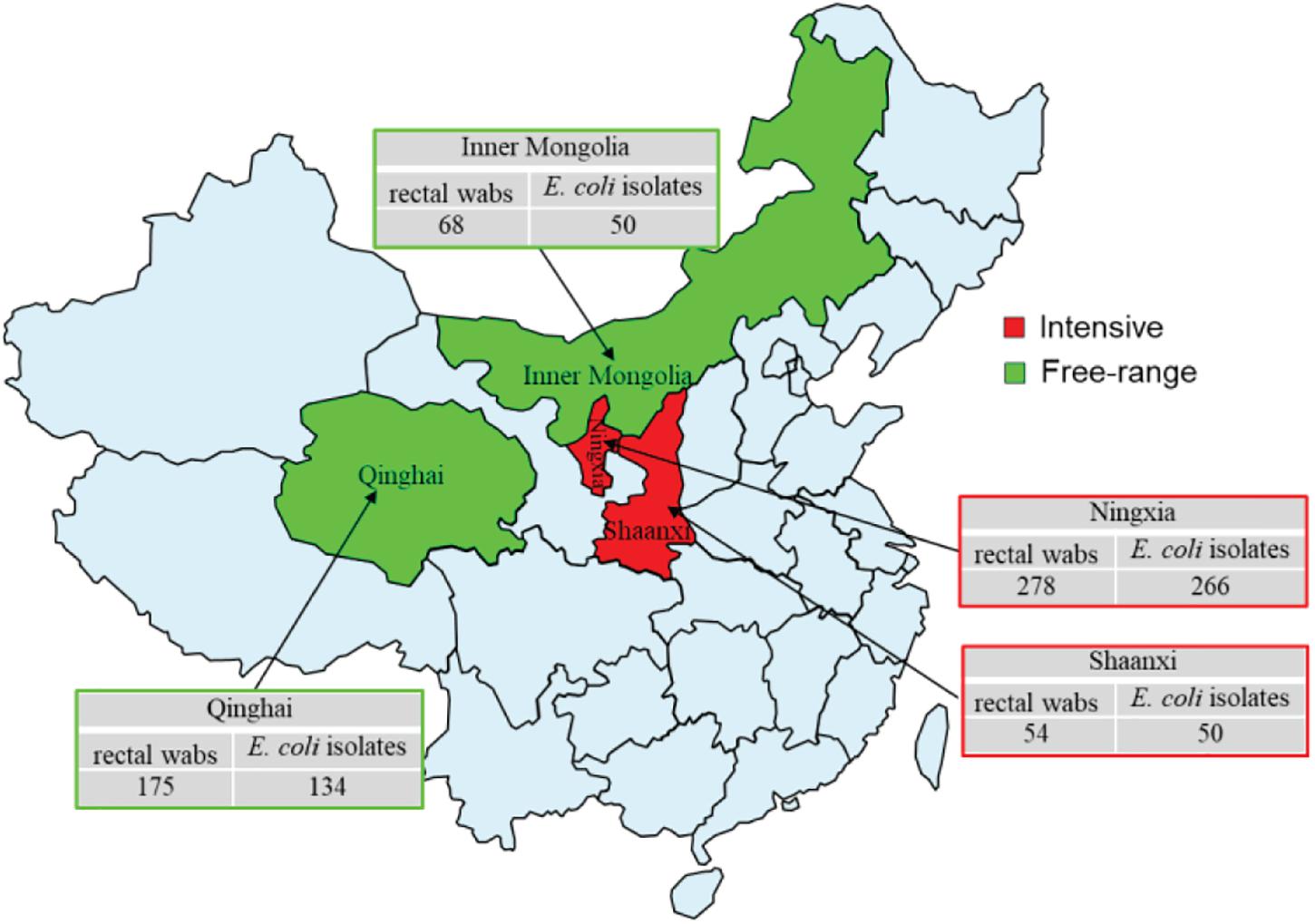
Figure 1. Sample collection regions in China during 2019–2020. The rectal swabs were collected from intensive farming wool sheep with diarrhea, Ningxia (East longitude 103°40′, Northerly latitudes 36°03′), Shaanxi (East longitude 109°49′, Northerly latitudes 34°45′), free-range wool sheep with diarrhea, Qinghai (East longitude 101°77′, Northerly latitudes 36°62′), and Inner Mongolia (East longitude 109°99′, Northerly latitudes 39°82′). Red and green colors indicate intensive and free-range farming, respectively.
Antibiotic Sensitivity Testing
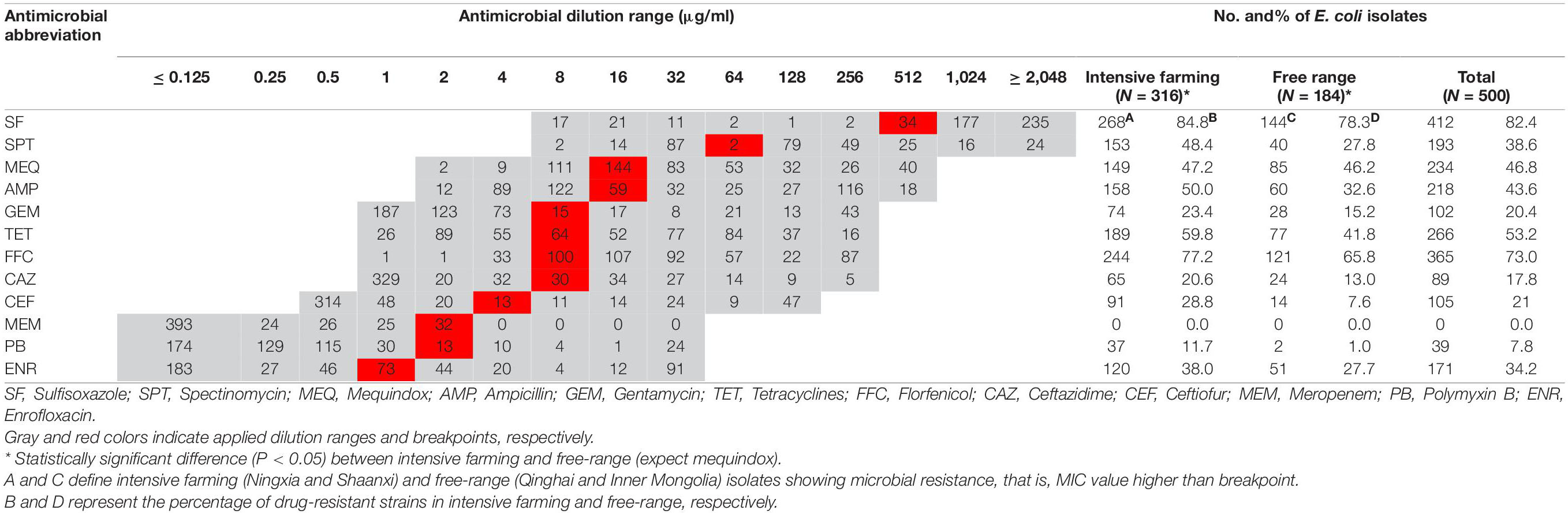
The antimicrobial susceptibility was tested by the broth microdilution method using Mueller-Hinton agar (Oxoid) as a culture medium according to the Clinical and Laboratory Standard Institute (CLSI VET08., 2018; CLSI; Wayne PA., 2020) guidelines as has been previously described. The minimum inhibitory concentrations (MIC) of each antibiotic was classified as resistant (R), intermediate (I), or susceptible (S) based on the CLSI breakpoints when available; otherwise, the National Antibiotic Resistance Monitoring System, NARMS1 for intestinal bacterial breakpoints was used. The susceptibility of E. coli was tested for 12 antibiotics: sulfisoxazole, spectinomycin, mequindox, ampicillin, gentamycin, tetracyclines, florfenicol, ceftazidime, ceftiofur, meropenem, polymyxin B, and enrofloxacin. When proven to be resistant to three or more antibacterial agents, E. coli isolates are considered to be MDR (Exner et al., 2017). The test was done in triplicate for each strain and E. coli ATCC 25922 (China Institute of Veterinary Drug Control) as a quality control strain. Based on the results of MIC, 500 strains of E. coli were screened for subsequent resistance genes, virulence genes, phylogenetic grouping, biofilm formation, and serological identification.
DNA Extraction and Virulence Factors (VFs) Determination
A single colony of a fresh bacterial culture from LB solid medium was picked and resuspended in 150 μl of sterile ddH2O. Tubes were heated at 100°C for 10 min and subsequently centrifuged at 12,000 rpm for 5 min. The supernatants were used as the DNA template in all PCR. Detect 16 VFs of all E. coli isolates. These VFs represent the main categories of virulence determinants, including adhesins (sfa, papC, sepA, etrA, aer, feaG, fsaA, and eaeA), capsule synthesis (rfc), and toxins (cnf1, hlyA, eltA, estA, exhA, stx1, and stx2) (Table 1; Hartleib et al., 2003; Boerlin et al., 2005; Chapman et al., 2006; Cheng et al., 2006; Alabsi et al., 2014).
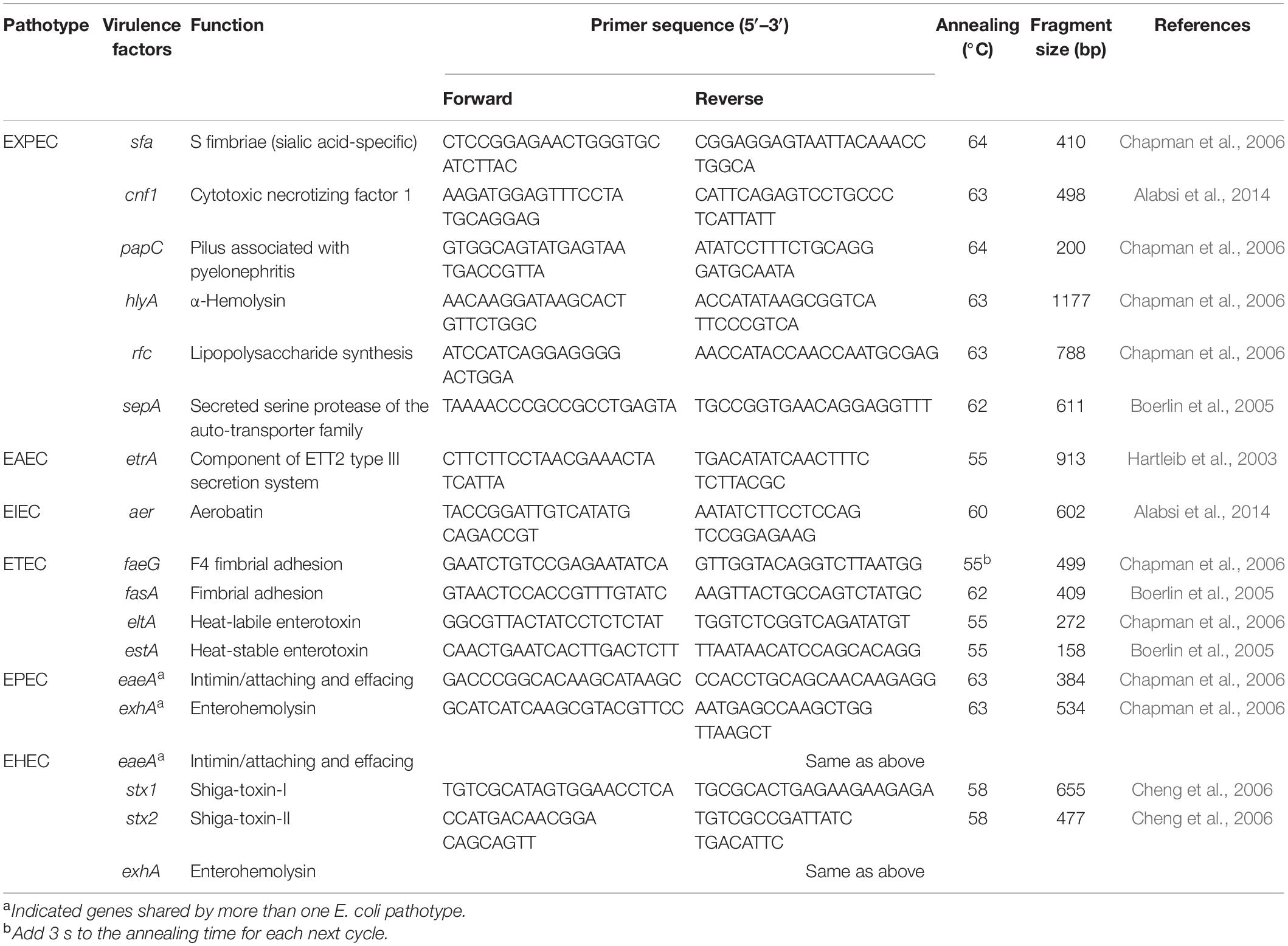
Table 1. List of 16 virulence genes detected in the present study, categorized based on their association with E. coli pathotypes.
Phylogenetic Grouping
E. coli strains were assigned to one of the phylogroups A, B1, B2, C, D, E, or F by quadruplex PCR (chuA, yjaA, arpA, and DNA fragment TspE4.C2) using a previously described protocol (Clermont et al., 2013). All testing was performed with positive and negative controls. The PCR results were interpreted as previously described (Clermont et al., 2013).
Biofilm Formation
The biofilm formation of each E. coli isolate was detected using the microtiter plate (MTP) method as described previously by Arezoo with slight modifications (Noie Oskouie et al., 2019). Briefly, 10 μL of each overnight culture was added in 1 mL LB broth for the production of biofilm. The suspension was adjusted to approximately 0.5 McFarland Standard (absorbance value of 0.08–0.1 at OD625 nm) and 180 μL was then transferred in triplicate into 96-wells without shaking at 37°C for 36 h. A control strain of E. coli (ATCC 25922, China Institute of Veterinary Drug Control) was used as a positive in each assay. After incubation, the microtiter plates were washed three times with phosphate buffered saline (PBS, PH 7.4) to remove all non-adherent bacteria. Methanol (200 μL) was added to each well for 15–20 min for fixing the attached bacteria. After removal of excess liquid, wells were washed three times with PBS before being stained for 5 min with 200 μL of 1% crystal violet at room temperature. After staining with crystal violet, the extra stain was rinsed off by placing the microtiter plate under running tap water and then air-dried at room temperature. The crystal violet was solubilized with 160 μL of 33% glacial acetic acid and then measured with an automated microplate reader (BioTek, Instruments, Inc. of Winooski, VT, United States) at OD570 nm measurement.
All strains were classified into four categories according to OD of bacterial, namely, non-adherent (–), weakly (+), moderately (++), or strongly adherent (+++). The cut-off ODc was defined for the microplate test as mean OD plus three standard deviations. In the current study were considered OD ≤ ODc, non-adherent, ODc<;OD ≤ 2 ODc, weakly, 2 ODc<;OD ≤ 4 ODc, moderately, and strongly adherent 4 ODc<;OD as described previously (Stepanovic et al., 2000). The biofilm formation of each isolate was tested in triplicate (9 observations per isolate) and the results were averaged.
Statistical Analysis
All analyses were conducted using SPSS for Windows Release 21 (SPSS Inc., United States) and GraphPad Prism 9 (GraphPad Software Inc., San Diego, CA, United States). Comparisons of association between antibiotic-resistant phenotypes, resistance genes, virulence genes, phylogenetic groups, and biofilm formation for E. coli isolates from different feeding models of wool sheep were separately by using the Pearson’s Chi-squared exact test. For all comparisons, P < 0.05 was considered statistically significant. Continuous variables were summarized as means and 95% confidence interval (CI). An association was considered positive when the two genes were found at the same time, otherwise, is was negative.
Results
Phenotypic Resistance
Overall, 500 E. coli isolates comprising 316 from intensive farming wool sheep [Ningxia, 25 farms (n = 266); Shaanxi, 5 farms (n = 50)] and 184 from free-range wool sheep [Qinghai, 18 farms (n = 134), Inner Mongolian, 6 farms (n = 50)] were obtained from 575 rectal swabs. The susceptibility of these isolates to 12 antimicrobials is shown in Table 2. The highest resistance rates were to sulfisoxazole (82.4%), florfenicol (72.0%), and tetracyclines (53.2%), followed by mequindox (46.8%), ampicillin (43.6%), spectinomycin (38.6%). enrofloxacin (34.2%), ceftiofur (21%), gentamycin (20.4%), ceftazidime (17.8%), and polymyxin B (7.8%). No resistance was found to meropenem in different groups of wool sheep. Notably, only one E. coli strains (0.2%) were susceptible to all the antibiotics, 122 (24.4%) were resistant to one or two antibiotics, and 377 (75.4%) were multiresistant (resistant to three or more antibiotics). Co-resistance involving sulfisoxazole, florfenicol, and tetracyclines were the most prevalent resistance detected. However, a few isolates were found to be multi-drug resistant to polymyxin B, ceftazidime, and gentamycin as well. Ninety-six resistance patterns were observed, of which SF-SPT-MEQ-AMP-TET-FFC was the most common.
Wool sheep E. coli from two different farming systems have different phenotypic resistance to 11 antibiotics (except meropenem). Phenotypic resistance to 11 antibiotics (except meropenem) varied between the wool sheep from two different farming systems. Sulfisoxazole resistance was identified in 268 (84.8%) of intensive farming, 144 (78.3%) of the free-range wool sheep isolates. Whilst resistance to florfenicol was observed in 244 (77.2%) of intensive farming, 121 (65.8%) of the free-range wool sheep. The proportion of MDR E. coli was higher in intensive farming compared to free-range sheep. Surprisingly, 12 (2.4%) of the isolates were resistant to all antimicrobials other than meropenem, and they are derived from intensive farming. There were significant differences (P < 0.05) found between the occurrence of various resistance in wool sheep from two different farming systems (expect mequindox). In summary, the strains from free-range sheep had fewer antibiotic resistance than strains of intensive farming sheep.
Prevalence of Virulence Genes and Occurrence of E. coli Pathotypes
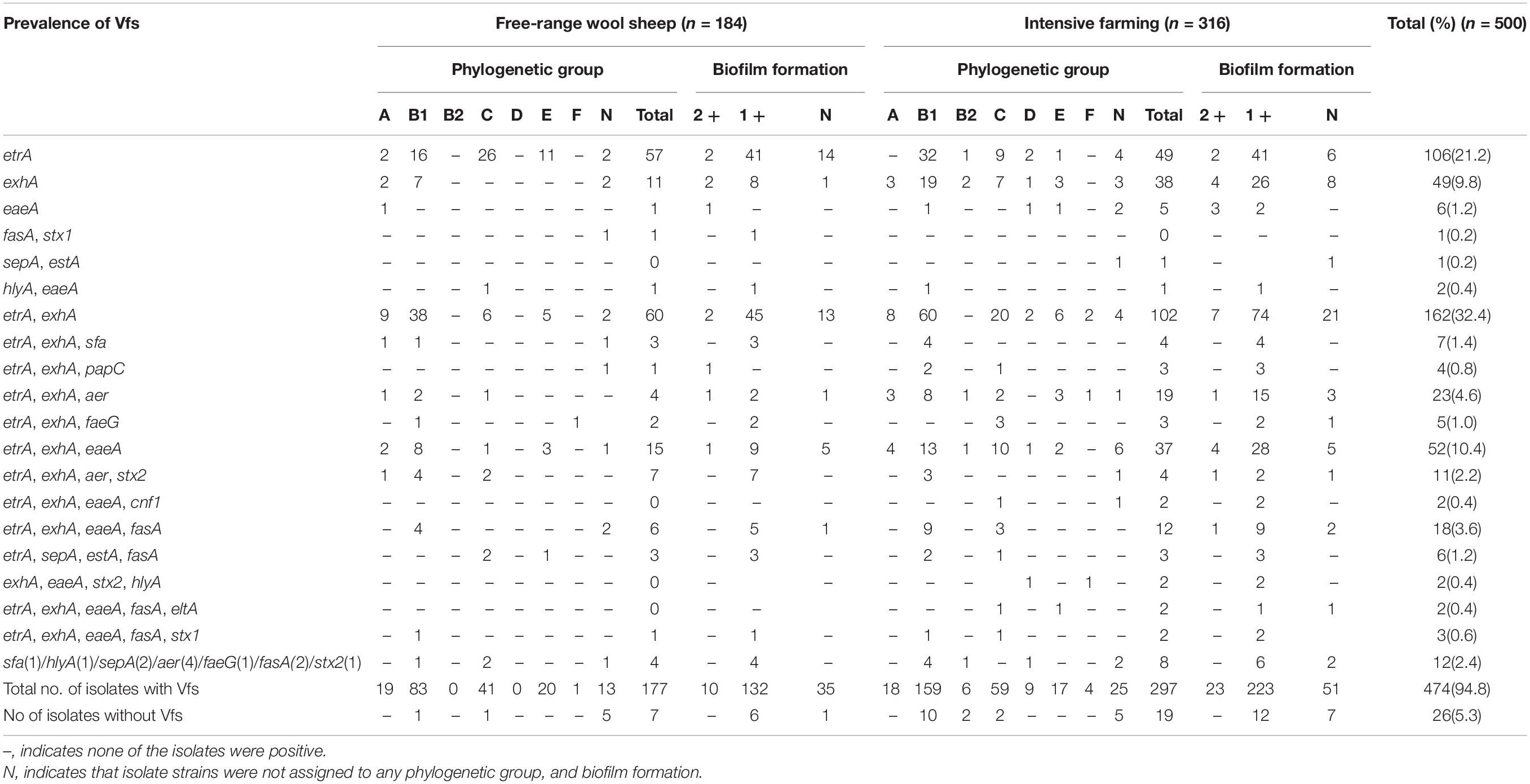
Fifteen out of sixteen virulence genes were detected, namely, etrA (80.2%), exhA (68%), eaeA (39.6%), fasA (31.2%), aer (28%), stx2 (22%), stx1 (7.4%), hlyA (6.6%), sepA (6.2%), sfa (5.4%), estA (5.2%), faeG (4.6%), papC (3.8%), eltA (2.8%), and cnf1 (2.4%). However, none of the isolates were positive to rfc, and the etrA was most prevalent among the virulence genes. Furthermore, only 26 strains of E. coli did not contain any of the 16 virulence genes. Overall, the majority of the E. coli isolates carried the combinations of virulence genes, associated with both IPEC and ExPEC pathotypes. Most of the E. coli isolates carried several different virulence genes, which are associated with both EAEC (80.2%) and EHEC (75%) pathotypes. In addition, 73.2, 36.8, and 28% of the isolates were positive for virulence genes associated with EPEC, ETEC, and EIEC pathotypes, respectively. Meanwhile, 20.4% of strains of our collection were considered as ExPEC pathotype (Table 3). In addition, the EAEC (86.4%), ETEC (40.8%), and ExPEC (22.3%) pathotypes were mainly observed in intensive farming. In contrast, EHEC (75%), EPEC (73.2%), and EIEC (29.7%) pathotypes were mainly carried out in free-range. There were significant differences (P < 0.05) found among EAEC, EHEC, and EPEC between intensive farming and free-range sheep. In summary, the strains from free-range sheep had fewer virulence genes than strains from intensive farming sheep.
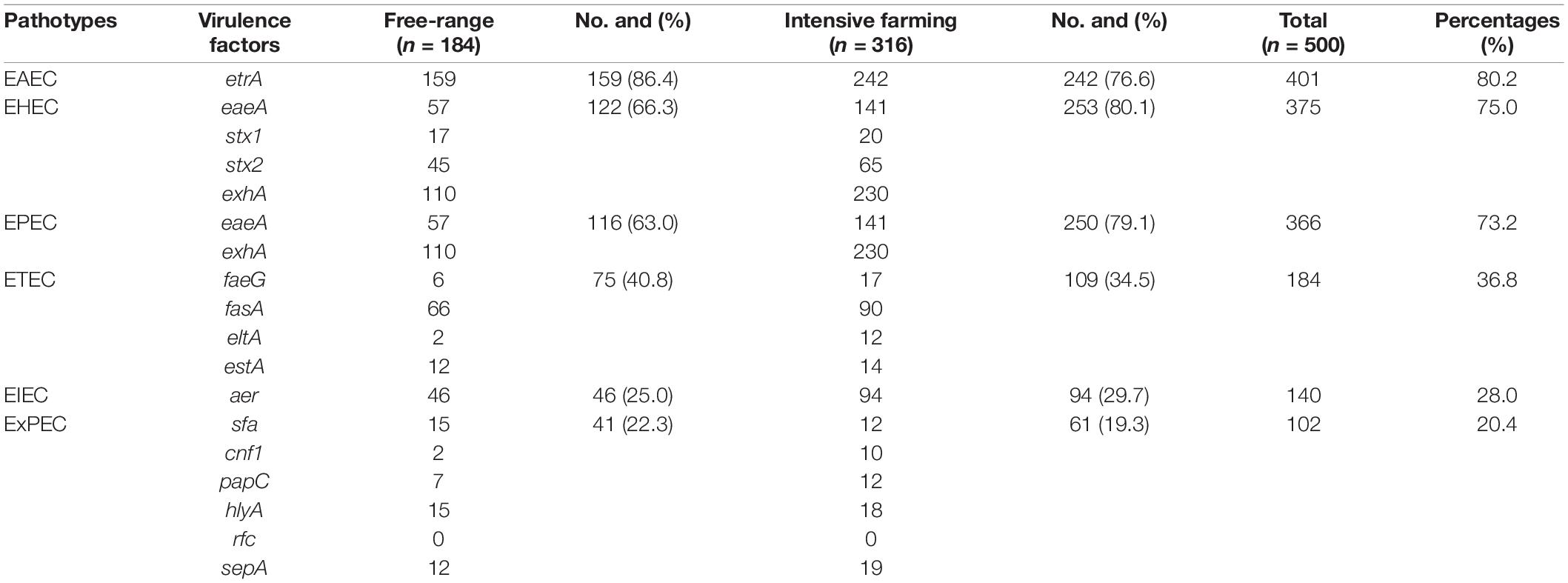
Table 3. The percentage prevalence of the different pathotypes between intensive farming and free-range sheep.
Associations Between Virulence Genes and Resistance Phenotypes
We observed a number of virulence-associated genes of both IPEC and ExPEC categories in diarrheic wool sheep. The etrA and hlyA virulence genes were the most abundant, which are linked with ExPEC and IPEC pathotypes, respectively. There were significant statistical differences(P < 0.05) between the occurrence of different virulence genes (Supplementary Table 1). It can be seen from the data in Supplementary Table 1 that positive associations are more common than negative associations. Within the isolates, there was a marked association of etrA, sfa, papc, aer, fasA, exhA, and stx2. Out of the several different associations in the strains, the strongest correlation between the etrA and papC gene (P < 0.001, OR = 455.68) was found. Such findings might be an explanation for the high levels of virulence genes in E. coli of diarrhea wool sheep.
In this study, a statistically significant (P < 0.05) association between virulence genes and resistance phenotypes among all the E. coli isolates was tested (Supplementary Table 2). Surprisingly, 146 out of 165 observations had epidemiological relevance (P < 0.05). Overall, a positive association was more common than a negative one between virulence genes and resistance phenotypes. In addition, ExPEC-associated P fimbriae gene (papC), ETEC-associated heat-labile enterotoxin gene (eltA), and heat-stable enterotoxin gene (estA) were correlated with all antibiotics other than meropenem. A more detailed analysis displayed associations between the resistance/susceptibility phenotypes with potential virulence genes. All virulence-associated genes were significantly related to certain antibiotic resistance (including sulfisoxazole, tetracyclines, and ceftazidime). Except for sulfisoxazole, all antibiotics were negatively correlated with etrA and exhA genes. However, we observed the strongest association between ETEC-associated heat-labile enterotoxin (eltA) and sulfisoxazole (P < 0.001, OR = 877).
Overall, there was no direct association between all virulence factors and resistance phenotypes in E. coli of wool sheep. However, EAEC associated component of the ETT2 type III secretion system gene (etrA) was negatively correlated with resistance phenotypes (P < 0.05). Conversely, the EHEC- and EPEC-associated virulence genes were positively correlated with drug-resistant phenotypes (P < 0.05). Apart from this, ETEC- and ExPEC-associated virulence genes were also negatively associated with resistance phenotypes. The EIEC-associated aerobatin gene (aer) was positively correlated with resistance phenotypes.
Occurrence of Phylogroups and Biofilm Formation
The majority of the E. coli isolates belonged to phylogroup B1 (50.6%), followed by phylogroup C (20.6%), A (7.4%), E (7.4%), D (5.8%), B2 (1.6%), F (1%), and unclassified (9.6%). However, regardless of whether they were free-range or intensive farming sheep, B1 was the most prevalent phylogroup. Interestingly, phylogroup B2 and D were all distributed in intensive farms. In addition, phylogroup A, E, and C were more common in free-range. Among the 500 E. coli isolates from the diarrhea of wool sheep, 33 isolates (6.6%), 373 isolates (74.6%), and 94 (18.8%) showed moderate, weak, and no connection biofilm formation ability, respectively (Figure 2). Significant differences, but no linking biofilm formation ability was found between free-range and intensive. However, we found that the biofilm formation ability of intensive farming was better than free-range.
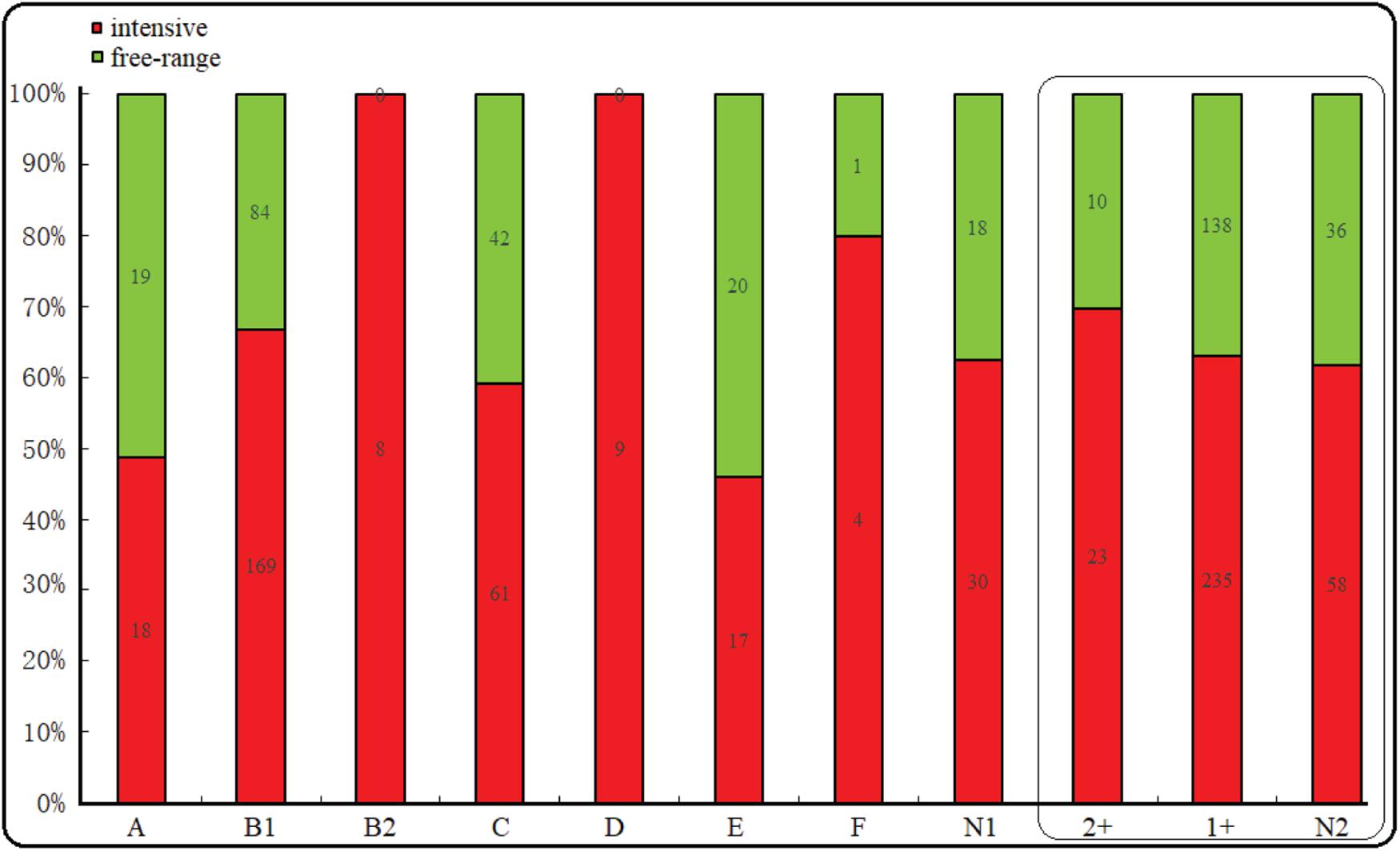
Figure 2. Phylogenetic and biofilm formation in the wool sheep E. coli from two different farming systems (n = 500). The box inside represented the result of the biofilm formation. N1 indicated unclassified phylogroups. Based on the OD the strain’s biofilm formation was determined as: no biofilm formation N2, weak (1+) and moderate (2+) biofilm formation. Red and green colors indicated intensive and free-range, respectively.
Distribution of Virulence Profile in Relation to Phylogenetic Grouping and Biofilm Formation
In intensive farmed sheep, PCR analysis of 316 isolates revealed 53.48% of the isolates belong to phylogenetic B1, followed by group C (19.30%), A (5.70%), E (5.38%), D (2.85%), B2 (2.53%), F (1.27%), and non-phylogenetic (9.49%). In free-range sheep, PCR analysis of 184 isolates revealed that 45.65% of the isolates belong to phylogenetic B1, followed by group C (22.83%), A (10.33%), E (10.87%), D (0.00%), B2 (0.00%), F (0.54%), and non-phylogenetic (9.78%). Those from the intensive farms and free-range wool sheep E. coli were predominantly from phylogenetic groups B1 and C. The distribution of the 500 E. coli isolates in relation to virulence genes and phylogenetic groups from the different farming systems of wool sheep revealed that out of the 474 isolates containing virulence factors, 242 belonged to group B1, 100 to group C, 37 to group A, 37 to group E, 9 to group D, 6 to B2, and 5 to group F (Table 4). Within the isolates, etrA, exhA, and eaeA genes were the commonly detected genes, whereas rfc gene was not identified in all E. coli isolates. Interestingly, etrA + exhA + eaeA + cnf1 (n = 2), exhA + eaeA + stx2 + hlyA (n = 2) and etrA + exhA + eaeA + fasA + eltA (n = 2) were found only in intensive farming, whereas fasA + stx1 (n = 1) was only in free-range wool sheep. For various feeding modes of wool sheep, it was found that intensive isolates contained more tested virulence genes. The capacity of biofilm formation was compared between various virulence profiles, where there was no significant correlation between them; however, the moderate had related to etrA, exhA, and eaeA gene. Notably, the fasA gene may combine with other genes to reduce the formation of biofilm.
Discussion
Worldwide, pathogenic E. coli isolates have been increasingly reported from animal, animal-derived foods, and humans (Dorado-García et al., 2018; Wang et al., 2020). Humans could obtain resistance/virulence genes from E. coli via the food chain (Madec et al., 2015). In this study, we observed a higher proportion of multidrug-resistant strains with virulence genes in wool sheep. This appears to be the first study to systematically compare antimicrobial resistance, virulence, biofilm, and phylogenetic groups in E. coli isolates from intensive and free-range wool sheep in China. During the period of investigation, a total of 500 E. coli strains were isolated, of which, 316 were isolated from intensive farming sheep (Ningxia and Shaanxi) and 184 were isolated from free-range sheep (Qinghai and Inner Mongolia). E. coli isolated from free-range sheep have a lower prevalence than intensive farming wool sheep. The low level of resistance and virulence in the free-range sheep were not surprising as antibiotic treatment is rare in these areas. In addition, the possible lower feeder density coupled with ad libitum green grass in free farming might explain lower levels of resistance and virulence when compared to intensive farming. These data can be compared to reports on other animals since studies of resistance in wool sheep are still insufficient.
Among all E. coli isolates strains, 75% were resistant to MDR, whereas 2.4% of the strains were resistant to all antimicrobials other than meropenem. Furthermore, only one strain was sensitive to all antibiotics. The E. coli isolates from different provinces have different resistance patterns. Specifically, the prevalence of MDR infection rate in E. coli of wool sheep origin in Inner Mongolia was significantly lower than in other provinces. In addition, we found a higher resistance rate to several old drugs, including sulfisoxazole (82.4%), florfenicol (72.0%), and tetracyclines (53.2%), consistent with previous reports (Zhang et al., 2017; Ma et al., 2021). The predominance of sulfisoxazole resistance strains was similar to the findings of Zhang et al. (2017), who detected sulfisoxazole resistance in 6,276 out of 7,568 E. coli isolates of chicken. Likewise, the predominance of florfenicol resistance strains was similar to Ma et al. (2021) The report stated that he detected florfenicol resistance in 36 out of 55 E. coli strains. Moreover, E.coli resistance to tetracyclines has been widely detected in pigs, chickens, and soil in China (Sun et al., 2019). Most notably, similar levels of E. coli resistance were observed in animals and the environment. This could be due to empiric antibiotic treatment use, indiscriminate, and overutilization of antibiotics in human medicine and livestock feed in China (Kümmerer, 2009). The long history of using these drugs in animals (tetracyclines and sulfonamides in the 1960s; florfenicol in the 1980s) may cause high prevalence resistance to these drugs. We detected high rates of AMR, with 75% of isolates displaying resistance to ≥ 3 antibiotic classes. By comparing the two different farming systems, intensive farming isolates showed higher levels of resistance to 11 tested antibiotic agents. The most common co-resistant phenotype observed was sulfisoxazole and florfenicol. Fortunately, we observed no resistance against meropenem in wool sheep from two different farming systems. Meropenem is considered as the last–resort drug for severe bacterial infections (Koeck et al., 2018). Thus, the phenomenon of antibiotic resistance has become a global public health concern. The problem of antibiotic resistance is worsening and is not optimistic in China. Therefore, the government needs new antibiotic treatment guidelines, which necessitate a strong resistance surveillance program in the western region of China.
As a direct pathogenic factor, the risk virulence factor of E. coli has drawn much attention from researchers. In this study, 15 virulence genes were identified in all E. coli isolate strains, indicating that diarrhea sheep harbored abundant and diverse virulence factors. Overall, our results showed that a significant proportion (23.6%) of E. coli from diarrheic sheep were possible simultaneously to IPEC and ExPEC pathogenic types. It is particularly worrisome that a significant fraction of E. coli isolates were positive for at least two of the five intestinal pathogenic. In this study, etrA (80.2%), and exhA (68%) genes were the most prevalent. This is consistent with previous studies identifying EAEC and EPEC pathotypes to have emerged as an increasingly important cause of diarrhea worldwide (Bolick et al., 2013; Hernandes et al., 2020; Ranganathan et al., 2020), and those showing a high frequency positive EAEC (80.2%) and EHEC (75%) strains in this study. Our research results showed that 94.8% of strains showed at least one virulence gene, nevertheless, the occurrence of virulence genes in the E. coli strain does not indicate its pathogenicity property (Rehman et al., 2017). Therefore, further studies, including additional animal model studies or bioinformatics approaches, are necessary to demonstrate the pathogenicity of observed virulence genes. All isolates were positive for the IPEC associated virulence genes, but negative for ExPEC associated virulence genes (rfc). In previous reports, such results have not been commonly described as animal origin E. coli isolates (Obeng et al., 2012; Osugui et al., 2014). Additionally, we also observed that a high frequency of E. coli isolates harbored eaeA (39.6%) and fasA (31.2%) virulence genes. The exact clinical significance of this funding is not clear. However, the eaeA gene and fasA gene may play a major role in adherence/colonization, causing attaching/effacing lesions (Rehman et al., 2017; Stromberg et al., 2020). The emergence of virulence gene patterns observed in the current study might be due to horizontal gene transfer mediating transfer virulence factors (Håkonsholm et al., 2020).
In our study, we also observed an association between the drug-resistant phenotype and virulence factors. In brief, negative correlations were more common. We observed the strongest association between ETEC associated toxin gene (eltA) and sulfisoxazole (P < 0.001, OR = 877). We identified a positive association between EAEC associated adhesion gene (etrA) and 10 antibiotics other than sulfisoxazole. This implies the possible selective advantage of E. coli strains harboring resistance and virulence genes. This could spell doom for current efforts to control infections with E. coli because animal husbandry would be overwhelmed by more virulent strains and increased drug resistance. Taken together, our findings suggest that the association of resistance and virulence genes might be multiple strain-specific. Moreover, we only examined the phenotypic profile in this study. Therefore, further studies are required to confirm this association between drug resistance and virulence of E. coli to forestall a potential health hazard.
As previously suggested, all E. coli strains can be assigned to one of the seven well-recognized phylogenetic groups, namely A, B1, B2, C, D, E, and F, as previously suggested (Clermont et al., 2013). It has been reported that E. coli phylogenetic groups A and B1 are typically commensal strains, whereas the B2 and D groups are mainly urinary tract infection strains (Song et al., 2020). In this study, based on the phylogenetic analysis, we found that antibiotic resistance was associated with virulence among E. coli strains. The majority of the E. coli isolates belonged to group B1 (50.6%) and the remaining to phylogroups A, B2, C, D, or F. Moreover, the E. coli component of the ETT2 type III secretion system gene (etrA) was the most common gene observed in phylogroup B1. The combination of phylogroup B1 and etrA gene might play an important role in diarrheic infections. Notably, phylogroups B2 and D only appear in intensive farming. This finding coincides with phenotypic resistance and virulence genes among E. coli strains. In addition, the diversity of this phylogroup in intensive farming sheep might be related to the widespread use of antibiotics, physiognomy, and climate compared to free-range sheep. Furthermore, our findings were consistent with previous studies of diarrheagenic and commensal strains (Adamus-Bialek et al., 2009). Our biofilm formation results were different from those reported in previous literature (Tajbakhsh et al., 2016). Comparing the wool sheep origin E. coli from two different farming systems, there was no significant difference in biofilm formation. However, the biofilm formation might be related to virulence genes etrA, exhA, and eaeA.
Conclusion
This is the first systematical comparison of antimicrobial resistance, virulence factors, phylogenetic grouping, and biofilm formation among E. coli isolates from wool sheep in intensive and free-range farming systems in China. In the present study, we observed a higher proportion of multidrug-resistant E. coli with virulence factors in sheep suffering from diarrhea and determined the correlations among virulence genes and resistance phenotypes. These results reveal that close monitoring of the virulence and antimicrobial resistance of food animal production is essential and that it could reduce the potential public health risks in both animal and human medicines.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by Northwest A&F University Ethics Committee.
Author Contributions
XLZ, JW, ZQY, and XLW conceived and designed the project. XLZ, YYL, QFX, XDB, BW, XYW, and RYD performed the experiments. XLZ and HHS analyzed the data. XLZ, FEAA, and HJL wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was financially supported by the China Agriculture Research System of MOF and MARA (No. CARS-39), the Natural Science Basic Research Plan in Shaanxi Province of China (No. 2021KW-41), the Ministry of Agriculture Antimicrobial Resistance Project of Animals, and the State Key Laboratory of Veterinary Etiological Biology, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (No. SKLVEB2019KFKT002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Shuangkui Du of College of Food Science and Engineering of Northwest A&F University and Cong Li of College of Animal Science and Technology of Northwest A&F University for help in statistical analyses of the data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.699927/full#supplementary-material
Footnotes
References
Adamus-Bialek, W., Wojtasik, A., Majchrzak, M., Sosnowski, M., and Parniewski, P. (2009). (CGG)4-based PCR as a novel tool for discrimination of uropathogenic Escherichia coli strains: comparison with enterobacterial repetitive intergenic consensus-PCR. J. Clin. Microbiol. 47, 3937–3944.
Alabsi, M., Ghazal, A., Sabry, S., and Alasaly, M. (2014). Association of some virulence genes with antibiotic resistance among uropathogenic Escherichia coli isolated from urinary tract infection patients in Alexandria, Egypt: a hospital-based study. J. Glob. Antimicrob. Resist. 2, 83–86. doi: 10.1016/j.jgar.2014.01.003
Boerlin, P., Travis, R., Gyles, C., Reid-Smith, R., Janecko, N., Lim, H., et al. (2005). Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 71, 6753–6761. doi: 10.1128/aem.71.11.6753-6761.2005
Bolick, D. T., Roche, J. K., Hontecillas, R., Bassaganya-Riera, J., Nataro, J. P., and Guerrant, R. L. (2013). Enteroaggregative Escherichia coli strain in a novel weaned mouse model: exacerbation by malnutrition, biofilm as a virulence factor and treatment by nitazoxanide. J. Med. Microbiol. 62, 896–905. doi: 10.1099/jmm.0.046300-0
Chandran, A., and Mazumder, A. (2013). Prevalence of diarrhea-associated virulence genes and genetic diversity in Escherichia coli isolates from fecal material of various animal hosts. Appl. Environ. Microbiol. 79, 7371–7380. doi: 10.1128/aem.02653-13
Chapman, T., Wu, X., Barchia, I., Bettelheim, K., Driesen, S., Trott, D., et al. (2006). Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl. Environ. Microbiol. 72, 4782–4795. doi: 10.1128/aem.02885-05
Cheng, D., Sun, H., Xu, J., and Gao, S. (2006). PCR detection of virulence factor genes in Escherichia coli isolates from weaned piglets with edema disease and/or diarrhea in China. Vet. Microbiol. 115, 320–328. doi: 10.1016/j.vetmic.2006.02.013
Clermont, O., Christenson, J., Denamur, E., and Gordon, D. (2013). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Dorado-García, A., Smid, J., van Pelt, W., Bonten, M., Fluit, A., van den Bunt, G., et al. (2018). Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: a pooled analysis. J. Antimicrob. Chemother. 73, 339–347. doi: 10.1093/jac/dkx397
Exner, M., Bhattacharya, S., Christiansen, B., Gebel, J., Goroncy-Bermes, P., Hartemann, P., et al. (2017). Antibiotic resistance: what is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg. Infect. Control 12:Doc05. doi: 10.3205/dgkh000290
Gabale, U., Peña Palomino, P., Kim, H., Chen, W., and Ressl, S. (2020). The essential inner membrane protein YejM is a metalloenzyme. Sci. Rep. 10:17794. doi: 10.1038/s41598-020-73660-6
Guimarães, A., Meireles, L., Lemos, M., Guimarães, M., Endringer, D., Fronza, M., et al. (2019). Antibacterial activity of terpenes and terpenoids present in essential oils. Molecules 24:2471. doi: 10.3390/molecules24132471
Habouria, H., Pokharel, P., Maris, S., Garenaux, A., Bessaiah, H., Houle, S., et al. (2019). Three new serine-protease autotransporters of Enterobacteriaceae (SPATEs) from extra-intestinal pathogenic Escherichia coli and combined role of SPATEs for cytotoxicity and colonization of the mouse kidney. Virulence 10, 568–587. doi: 10.1080/21505594.2019.1624102
Håkonsholm, F., Lunestad, B., Aguirre Sánchez, J., Martinez-Urtaza, J., Marathe, N., and Svanevik, C. (2020). Vibrios from the Norwegian marine environment: characterization of associated antibiotic resistance and virulence genes. MicrobiologyOpen 9:e1093. doi: 10.1002/mbo3.1093
Hartleib, S., Prager, R., Hedenström, I., Löfdahl, S., and Tschäpe, H. (2003). Prevalence of the new, SPI1-like, pathogenicity island ETT2 among Escherichia coli. Int. J. Med. Microbiol. 292, 487–493. doi: 10.1078/1438-4221-00224
Hernandes, R., Hazen, T., Dos Santos, L., Richter, T., Michalski, J., and Rasko, D. (2020). Comparative genomic analysis provides insight into the phylogeny and virulence of atypical enteropathogenic Escherichia coli strains from Brazil. PLoS Negl. Trop. Dis. 14:e0008373. doi: 10.1371/journal.pntd.0008373
Kim, Y., Kim, J., Hur, J., and Lee, J. (2010). Isolation of Escherichia coli from piglets in South Korea with diarrhea and characteristics of the virulence genes. Can. J. Vet. Res. 74, 59–64. doi: 10.2376/0005-9366-123-83
Koeck, D., Huber, S., Hanifi, N., Köster, M., Schierling, M., and Höller, C. (2018). Occurrence of antibiotic-resistant bacteria in therapy pools and surrounding surfaces. Int. J. Environ. Res. Public Health 15:2666. doi: 10.3390/ijerph15122666
Kümmerer, K. (2009). Antibiotics in the aquatic environment-A review-Part II. Chemosphere 75, 435–441.
Laxminarayan, R., Duse, A., Wattal, C., Zaidi, A., Wertheim, H., Sumpradit, N., et al. (2013). Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098. doi: 10.1016/s1473-3099(13)70318-9
Ma, Y., Chen, J., Fong, K., Nadya, S., and Wang, S. (2021). Antibiotic resistance in shiga toxigenic Escherichia coli isolates from surface waters and sediments in a mixed use urban agricultural landscape. Antibiotics 10:237.
Madec, J., Haenni, M., Métayer, V., Saras, E., and Nicolas-Chanoine, M. (2015). High prevalence of the animal-associated bla CTX-M-1 IncI1/ST3 plasmid in human Escherichia coli isolates. Antimicrob. Agents Chemother. 59, 5860–5861. doi: 10.1128/aac.00819-15
Malberg Tetzschner, A., Johnson, J., Johnston, B., Lund, O., and Scheutz, F. (2020). In Silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J. Clin. Microbiol. 58:e01269-20. doi: 10.1128/jcm.01269-20
Nadeem, S., Gohar, U., Tahir, S., Mukhtar, H., Pornpukdeewattana, S., Nukthamna, P., et al. (2020). Antimicrobial resistance: more than 70 years of war between humans and bacteria. Crit. Rev. Microbiol. 46, 578–599. doi: 10.1080/1040841x.2020.1813687
Noie Oskouie, A., Hasani, A., Ahangarzadeh Rezaee, M., Soroush Bar Haghi, M., Hasani, A., and Soltani, E. (2019). A relationship between O-serotype. antibiotic susceptibility and biofilm formation in uropathogenic Escherichia coli. Microb. Drug Resist. 25, 951–958. doi: 10.1089/mdr.2018.0330
Obeng, A., Rickard, H., Ndi, O., Sexton, M., and Barton, M. (2012). Antibiotic resistance, phylogenetic grouping and virulence potential of Escherichia coli isolated from the faeces of intensively farmed and free range poultry. Vet. Microbiol. 154, 305–315. doi: 10.1016/j.vetmic.2011.07.010
Osugui, L., de Castro, A., Iovine, R., Irino, K., and Carvalho, V. (2014). Virulence genotypes, antibiotic resistance and the phylogenetic background of extraintestinal pathogenic Escherichia coli isolated from urinary tract infections of dogs and cats in Brazil. Vet. Microbiol. 171, 242–247. doi: 10.1016/j.vetmic.2014.03.027
Pan, Y., Zeng, J., Li, L., Yang, J., Tang, Z., Xiong, W., et al. (2020). Coexistence of antibiotic resistance genes and virulence factors deciphered by large-scale complete genome analysis. mSystems 5:e00821-19. doi: 10.1128/mSystems.00821-19
Pérez, V., Costa, G., Guimarães, A., Heinemann, M., Lage, A., and Dorneles, E. (2020). Relationship between virulence factors and antimicrobial resistance in Staphylococcus aureus from bovine mastitis. J. Glob. Antimicrob. Resist. 22, 792–802. doi: 10.1016/j.jgar.2020.06.010
Ranganathan, S., Smith, E. M., Foulke-Abel, J. D., and Barry, E. M. (2020). Research in a time of enteroids and organoids: how the human gut model has transformed the study of enteric bacterial pathogens. Gut Microbes 12:1795492.
Rehman, M., Zhang, H., Iqbal, M., Mehmood, K., Huang, S., Nabi, F., et al. (2017). Antibiotic resistance, serogroups, virulence genes, and phylogenetic groups of Escherichia coli isolated from yaks with diarrhea in Qinghai Plateau, China. Gut Pathog. 9:24. doi: 10.1186/s13099-017-0174-0
Russo, T. A., and Johnson, J. R. (2003). Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5, 449–456. doi: 10.1016/s1286-4579(03)00049-2
Song, J., Oh, S., Kim, J., and Shin, J. (2020). Extended-spectrum β-lactamase-producing Escherichia coli isolated from raw vegetables in South Korea. Sci. Rep. 10:19721. doi: 10.1038/s41598-020-76890-w
Stepanovic, S., Vukovic, D., Dakic, I., Savic, B., and Svabic-Vlahovic, M. (2000). A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40, 175–179. doi: 10.1016/s0167-7012(00)00122-6
Stromberg, Z., Masonbrink, R., and Mellata, M. (2020). Transcriptomic analysis of shiga toxin-producing Escherichia coli during initial contact with cattle colonic explants. Microorganisms 8:1662. doi: 10.3390/microorganisms8111662
Sun, J., Chen, C., Cui, C., Zhang, Y., Liu, X., Cui, Z., et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 4, 1457–1464. doi: 10.1038/s41564-019-0496-4
Tajbakhsh, E., Ahmadi, P., Abedpour-Dehkordi, E., Arbab-Soleimani, N., and Khamesipour, F. (2016). Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrob. Resist. Infect. Control 5:11. doi: 10.1186/s13756-016-0109-4
Van Boeckel, T., Glennon, E., Chen, D., Gilbert, M., Robinson, T., Grenfell, B., et al. (2017). Reducing antimicrobial use in food animals. Science 357, 1350–1352. doi: 10.1126/science.aao1495
Wang, Y., Hu, Y., and Gao, G. F. (2020). Combining metagenomics and metatranscriptomics to study human, animal and environmental resistomes. Med. Microecol. 3:100014.
Wani, S., Hussain, I., Nabi, A., Fayaz, I., and Nishikawa, Y. (2007). Variants of eae and stx genes of atypical enteropathogenic Escherichia coli and non-O157 Shiga toxin-producing Escherichia coli from calves. Lett. Appl. Microbiol. 45, 610–615. doi: 10.1111/j.1472-765X.2007.02235.x
Keywords: antimicrobial resistance, biofilm, Escherichia coli, phylogenetic groups, virulence gene, zoonotic potential
Citation: Zhao X, Lv Y, Adam FEA, Xie Q, Wang B, Bai X, Wang X, Shan H, Wang X, Liu H, Dang R, Wang J and Yang Z (2021) Comparison of Antimicrobial Resistance, Virulence Genes, Phylogroups, and Biofilm Formation of Escherichia coli Isolated From Intensive Farming and Free-Range Sheep. Front. Microbiol. 12:699927. doi: 10.3389/fmicb.2021.699927
Received: 24 April 2021; Accepted: 31 May 2021;
Published: 30 July 2021.
Edited by:
Octavio Luiz Franco, Catholic University of Brasilia (UCB), BrazilReviewed by:
Marjanca Starčič Erjavec, University of Ljubljana, SloveniaPaula Signolfi Cyoia, State University of Londrina, Brazil
Copyright © 2021 Zhao, Lv, Adam, Xie, Wang, Bai, Wang, Shan, Wang, Liu, Dang, Wang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruyi Dang, ZHJ5MjAxN0Bud2FmdS5lZHUuY24=; Juan Wang, anVhbi53YW5nQG53c3VhZi5lZHUuY24=; Zengqi Yang, eXpxODE2MkAxNjMuY29t
 Xueliang Zhao
Xueliang Zhao Yunyang Lv
Yunyang Lv Fathalrhman Eisa Addoma Adam
Fathalrhman Eisa Addoma Adam Xinglong Wang
Xinglong Wang Zengqi Yang
Zengqi Yang