
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 July 2021
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.698834
This article is part of the Research Topic Metal Resistance in Microorganisms View all 12 articles
Microorganisms play an important role in the remediation of cadmium pollution in the soil and their diversity can be affected by cadmium. In this study, the bacterial community in arable soil samples collected from two near geographical sites, with different degrees of cadmium pollution at three different seasons, were characterized using Illumina MiSeq sequencing. The result showed that cadmium is an important factor to affect the bacterial diversity and the microbial communities in the high cadmium polluted area (the site H) had significant differences compared with low cadmium polluted area (the site L). Especially, higher concentrations of Cd significantly increased the abundance of Proteobacteria and Gemmatimonas whereas decreased the abundance of Nitrospirae. Moreover, 42 Cd-resistant bacteria were isolated from six soil samples and evaluated for potential application in Cd bioremediation. Based on their Cd-MIC [minimum inhibitory concentration (MIC) of Cd2+], Cd2+ removal rate and 16S rDNA gene sequence analyses, three Burkholderia sp. strains (ha-1, hj-2, and ho-3) showed very high tolerance to Cd (5, 5, and 6 mM) and exhibited high Cd2+ removal rate (81.78, 79.37, and 63.05%), six Bacillus sp. strains (151-5,151-6,151-13, 151-20, and 151-21) showed moderate tolerance to Cd (0.8, 0.4, 0.8, 0.4, 0.6, and 0.4 mM) but high Cd2+ removal rate (84.78, 90.14, 82.82, 82.39, 81.79, and 84.17%). Those results indicated that Burkholderia sp. belonging to the phylum Proteobacteria and Bacillus sp. belonging to the phylum Firmicutes have developed a resistance for cadmium and may play an important role in Cd-contaminated soils. Our study provided baseline data for bacterial communities in cadmium polluted soils and concluded that Cd-resistant bacteria have potential for bioremediation of Cd-contaminated soils.
Heavy metal soil pollution has become a severe environmental problem due to the rapid development of industries such as mining, smelting and agriculture. Moreover, one of the most serious and widespread heavy metal contaminants is cadmium (Cd) (MEP, 2014; Huang et al., 2019). Cd is toxic even at low concentrations of 0.001–0.1 mg L–1 and can be accumulated in the human body through the food chain. Once Cd levels become critical, humans can develop emphysema and osteoporosis; and eventually the damage to the lungs, kidneys and liver becomes irreversible. People suffering from severe chronic Cd poisoning develop the itai-itai disease (Satarug et al., 2017; Genchi et al., 2020; Đukić-Ćosić et al., 2020). Consequently, engineering to remedy heavy metal contaminated soil, especially Cd contaminated soil, is urgently needed.
Soil is an important habitat for a diverse group of microorganisms that play an important role in the soil environment. Microorganisms are regarded as sensors of disturbances in the soil ecosystem since they are far more sensitive to environmental stress than macroorganisms (Khan et al., 2010; Yang et al., 2016; Zhang et al., 2016). Moreover, microorganisms in soil are mainly involved in material decomposition, an important process in maintaining soil biological activity as well as regulating soil nutrient circulation. Stable microbial communities mediate soil environment by stabilizing soil structure and maintaining soil physical and chemical conditions (Bissett et al., 2013). Characteristics of microbial community composition and activity were often used as indicators of soil quality (Hamman et al., 2007; Rubin et al., 2013). Many studies have concentered on changes in soil properties owing to the presence of cadmium as it negatively impacts the indigenous microorganism community, impairing the ecological function they provide (Harichová et al., 2012). Therefore, it is indispensable to analyze the response of microbial communities structure in cadmium contaminated soils.
Owing to the difficulty in comprehensively assessing the structure of the microbial communities associated with using traditional techniques, previous research have explored the effects of cadmium contamination on microbial communities using approaches such as denaturing gradient gel electrophoresis (DGGE) (Wang et al., 2006; Zhang et al., 2009), random amplified polymorphic DNA (RAPD) (Wang et al., 2007a), and phospholipid fatty acids (PLFA) (Liao et al., 2010). These approaches, however, only provide limited insights into the bacterial profiles. Recently, the application of high-throughput sequencing based rRNA for evaluating microbial communities is widely used (Luo et al., 2019; Duan et al., 2020).
In addition, microorganisms have large specific surface areas and high metabolic activity, which making them particularly susceptible to the presence of heavy metals in the soil and impeding their ecological function via adsorption, fixation, complexation, dissolution, oxidation reduction, etc. (Vodyanitskii and Plekhanova, 2014). Moreover, a number of soil microbes have been reported effective in the remediation of heavy metals. For example, Suksabye et al. (2016) reported that the addition of Pseudomonas aeruginosa or Bacillus subtilis to the soil could reduce the amount of Cd in rice grains obtained from Cd contaminated soil as a result of their Cd remediating characteristic. Currently, microbial remediation of heavy metal contaminated soil is regarded a cost-effective, biotechnology approach (Jin et al., 2018). Therefore, it is necessary to isolate Cd-resistant and Cd-adsorbing microorganisms from Cd contaminated soil, which can be used the remediation of Cd polluted soil.
In this study, firstly we explored the microbial community structure of two sites with significantly different Cd levels but of similar geographical location. We also collected samples from both sites during different seasons. High-throughput sequence analysis of 16S rRNA V3-V4 region was used to characterize the microbial taxa and operational taxonomic units (OTUs) distribution in the different samples. This comprehensive analysis enabled us to correlate microbial taxa and OTUs to Cd contamination, thereby illustrating (1) the effects of Cd exposure on microbial community structure in the soil, and (2) the seasonality of microbial activity at the different sites. Secondly, in order to excavate the available microbial resources, 42 Cd-resistant bacteria were isolated and evaluated for potential application in Cd bioremediation.
Soil samples (0–20 cm) were collected from two different arable soil fields located around a mining area in Xiangtan, Hunan Province, China: L site (27°33′N, 113°15′E) and H site (27°46′N, 112°52′E). The distance between the two regions was about 1.3 km. Moreover, three different seasonal samples of both areas were collected: LA and HA in April 2016; LJ and HJ in July 2016; LO and HO in October 2016. Each time three replicates were sampled and stored at −70°C. Samples were divided in three; one part was used for Miseq, the other to determine the Cd concentration and the last one to isolate Cd-resistant bacteria.
To determine total Cd concentration in the soil samples, 0.5 g air dried sample was digested with 7 mL nitric acid and 3 mL hydrofluoric acid in a polytetrafluoroethylene digestion vessel using a microwave accelerated reaction instrument (CEM-MARS6 Xpress, United States). After complete digestion, the total, water-soluble and filter solution of all samples were measured with a 7,700× Inductively Coupled Plasma Mass Spectrometer (Agilent Technologies, Japan). A heavy metal uncontaminated soil sample (GSS-7, shown in Table 1) was used as a standard negative control.
Total microbial community DNA of all samples were extracted following the Mobio Power Soil DNA isolation Kit protocols. V3-V4 regions of 16S rRNA were amplified using the primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Sequencing of the amplicons was outsourced to Allwegene (Beijing, China); amplicons were sequenced on an Illumina Miseq platform. All the sequences data have been submitted to NCBI (National Center for Biotechnology Information), accession numbers were: SAMN07643022 (HA1), SAMN07644976 (HA2), SAMN07644984 (HA3); SAMN07644992 (HJ1), SAMN07644993 (HJ2), SAMN07644994 (HJ3); SAMN07644995 (HO1), SAMN07644997 (HO2), SAMN07645000 (HO3); SAMN07645001 (LA1), SAMN07645003 (LA2), SAMN07645004 (LA3); SAMN07645005 (LJ1), SAMN07645006 (LJ2), SAMN07645007 (LJ3); SAMN07645009 (LO1), SAMN07645010 (LO2), and SAMN07645011 (LO3).
Sequence analysis of the 16S rRNA V3-V4 region amplicon sequences was performed using QIIME Pipeline, Version 1.8.0 (Caporaso et al., 2010). Firstly, low quality reads (average quality score <20) were trimmed and paired-ends sequences were merged to be single sequence according to their overlap sequence (>10 bp). Then, sequences from the samples were distinguished on the basis of the barcodes and primers. Finally, chimeras were deleted using USEARCH (Edgar et al., 2011) and the smaller sequences were removed by MOTHER (Schloss et al., 2009). These high quality sequences were subsequently used for downstream analysis. OTUs were clustered using a 97% identity threshold using UCLUST v1.2.22 (Edgar, 2010) and representative OUT sequence was obtained. Moreover, singleton OUT was removed. A total of 338,077 final sequences were gained for all the samples after filtering for low quality sequences. OTUs were annotated using the Ribosomal Database Project classifier (Wang et al., 2007b). Alpha-diversity (Chao 1, observed species, goods coverage and Shannon) and beta-diversity [principal component analysis (PCA) and Metastats] were analyzed based on Miseq sequence data.
To isolate Cd-resistant bacteria, soil samples (2 g) were placed in sterile 0.9% NaCl (18 mL) at 30°C and shaken at 200 rpm for 0.5 h to completely separate bacteria from soil. After settling for several minutes, an aliquot of the suspension was serially diluted (from 10–1 and 10–4). Each diluted solution was spread onto an Burk agar plates (0.8 g/L KH2PO4, 0.262 g/L K2HPO4⋅3H2O, 1 g/L (NH4)2SO4, 0.2 g/L MgSO4⋅3H2O, 1 g/L yeast extract, and 1.5% agar) containing progressively higher concentrations of CdCl2 (1, 3, and 5 mM). To isolate Cd-resistant and bio-safe Bacillus bacteria, the bacterial enrichment cultures were heat-shocked at 80°C for 20 min and aerobic Bacillus sp. were isolated from the soil by plating on LB agar plates containing different concentrations of CdCl2 (0, 0.5, 1, and 2 mM). Bacterial growth was observed after incubation at 30°C for 24 h. Single colony was picked with sterilized wire loop and re-streaked on CdCl2 supplemented LB agar plates and again incubated at 30°C for 24 h. The process was repeated until the pure culture was obtained. Genomic DNA was isolated using the TIANamp Bacteria DNA kit (TIANGEN Biotech). The 16S rRNA gene was amplified from the extracted DNA using the universal primers the universal forward primer 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and reverse primer 1492r (5′-TACGGTTACCTTGTTACGACTT-3′). The amplification products were cloned in the pGM-T (TIANGEN Biotech) vector using competent Escherichia coli TOP10 cells (TIANGEN Biotech). Sequencing was carried out using T7 and SP6 primers and compared to the GenBank database using the NCBI BLAST program.
To evaluate growth in a liquid medium of isolated bacteria, the MIC of Cd2+ (MIC-Cd) was determined. LB medium (800 μL) with different concentrations of Cd2+ was dispensed into 96-well (12 × 8) microtiter plates (96 × 2-mL wells) with a multi-channel micropipette (rows A to H: 0, 1, 2, 3, 4, 5, 6, and 7 mM). Single colonies of the test strains were inoculated into 3 mL of LB medium and cultured overnight. The test culture (15 μL) was then inoculated into each well of the prepared 96-well plate. After 24 h at 30°C and 750 rpm in an incubator (Heidolph, Viertrieb, Germany), 200 μL of the cell suspension was transferred to a 96-well plate and the turbidity at OD600 was measured.
To determine the Cd2+ adsorption of isolated bacteria, growth of cells was grown in LB liquid medium supplementation with 0.1 mM CdCl2 and shaken at 200 rpm at 30°C for 24 h. Cells were harvested by centrifugation at 12,000 rpm for 10 min and the supernatant then diluted to an appropriate concentration for analysis. Cd2+ concentrations in culture supernatants were measured via atomic absorption spectrophotometry (Z-2000, Hitachi, Japan), with Cd2+ removal rate being calculated using the following equation:
Removal rate (%) = (Ci −Ce)/Ci × 100
where Ci and Ce are the initial and equilibrium Cd2+ concentrations (mM), respectively.
Cd concentrations in all the samples were shown in Table 1. Cd concentrations were significantly higher in samples collected from site H (8.52–53.70 mg kg–1) as compared to site L (0.36–0.46 mg kg–1). Among six samples, the Cd concentrations of HJ (53.70 mg kg–1) was highest while the Cd concentrations of LJ was lowest (0.36 mg kg–1). Moreover, Cd concentrations of soil samples from site L were similar across the different seasons, whereas there was an obvious seasonal effect on the Cd concentrations for the H site samples. Specifically, the Cd content of HJ was about 2.0-fold and 6.3-fold higher than that of HA and HO, respectively.
Rarefaction curve analysis showed that the quantity of OTUs was enough to reach saturation, indicated that the sequencing depth was sufficient to characterize the microbial community composition (Supplementary Figure 1). The OTU densities of soil samples from site H increased with increasing Cd concentration (HJ > HA > HO), while the OTU density of sample LJ with the lower Cd contamination level was higher than those of sample LA and LO (Supplementary Table 1). Comparison of the different sites in the same season showed that HA and HJ have more defined OTUs than LA and LJ, respectively. Bacterial alpha diversity, including the chao1, observed species, PD whole tree and Shannon, varied among the six soil samples. We observed the highest diversity at location with HJ, whereas the lowest diversity at location with HO (Supplementary Figure 2). However, results of the PCA supported the previous results, showing that soil microbial communities clustered strongly based on Cd concentration grade (Figure 1). A total of 41.89 and 18.62% of the variations in the bacterial communities could be explained by the first and second principal components, which also indicated that the cadmium concentration is the key factor to affect the bacterial diversity of the soil (Figure 1). Further, all samples from site L (LA, LJ, and LO) clustered tightly whereas samples from site H were more dispersed (Figure 1). These results indicated that the seasonal change in microbial community structure in high Cd contaminated soil was more distinct than in low Cd contaminated soil.
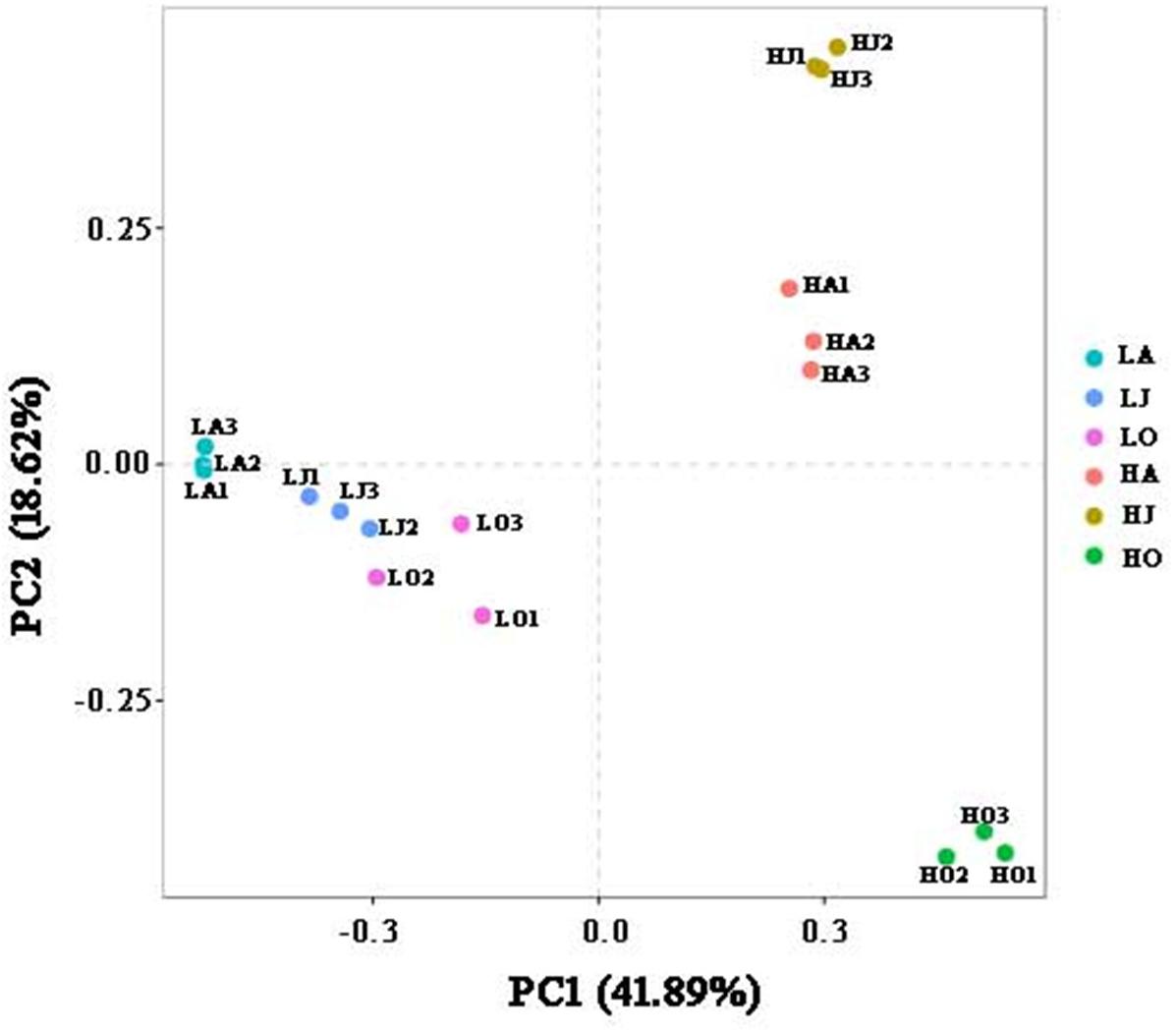
Figure 1. Principal component analysis (PCA) displaying beta-diversity of microbial diversity between samples. PCA was calculated using weighted Unifrac distance. A total of 41.89 and 18.62% of the variations in the bacterial communities could be explained by the first and second principal components, respectively.
A total of 338,077 high-quality bacterial 16S rRNA gene sequences were obtained from 18 samples, ranging from 10,812 to 23,059 sequences per sample (Supplementary Table 1). Among the total 16S rRNA gene sequences, 71 sequences were classified as archaea, accounting for 0.02% of total sequences. The remaining sequences belonged to bacteria (338006 of 338077), accounting for 99.98% of the 16S rRNA gene sequences, and presented 47 phyla. Specifically, the dominant phyla, consisting of 1% (relative abundance) or more to total community composition, are shown in Figure 2. Proteobacteria (27.77–44.59%), Acidobacteria (12.95–26.29%), Chloroflexi (13.11–24.28%), Nitrospirae (4.43–20.17%), Gemmatimonadetes (1.40–5.62%), and Actinobacteria (0.80–12.43%) were the six largest phyla in all samples. Among these more abundant phyla, it was observed that the relative abundances of Proteobacteria, Gemmatimonadetes, and Actinobacteria at the site H were higher than that of site L, whereas the abundance of Nitrospirae at the site H was lower than that of site L (Figure 2). Additionally, we found that the relative abundances of five levels (phylum, class, order, family, and genus) of Gemmatimonas at the site H were all higher than that of site L. However, the relative abundances of four levels (phylum, class, order, and family) of Nitrospira at the site H were all lower than that of site L while the abundances of Nitrospira at the site H was higher than that of site L (Supplementary Figure 3).
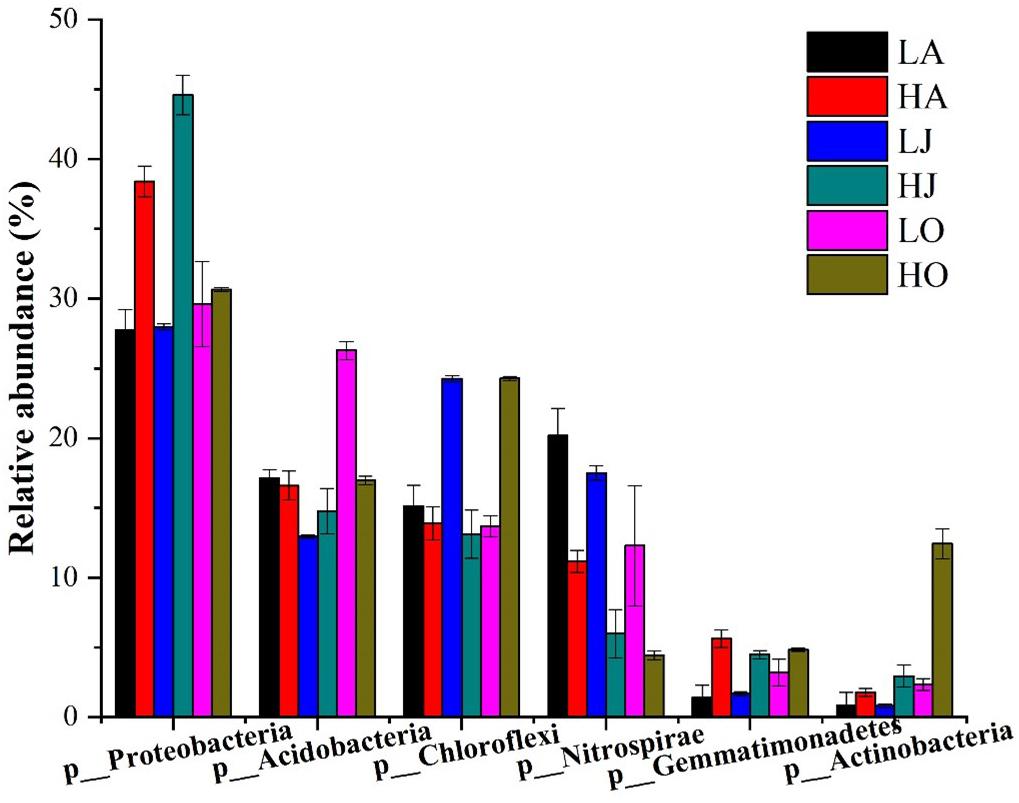
Figure 2. Community composition of dominant bacterial phyla for six samples of different Cd concentrations (LA: 0.46 mg/kg; LJ: 0.36 mg/kg; LO: 0.39 mg/kg; HA: 27.11 mg/kg; HJ: 53.70 mg/kg; HO: 8.52 mg/kg). The relative abundances of Proteobacteria (HA:38.4%, LA:27.77%; HJ:44.59%, LJ:27.98%; HO:30.65%, and LO:29.60%) and Gemmatimonadetes (HA:5.62%, LA: 1.41%; HJ:4.48%, LJ:1.69%; HO:4.82%, and LO:3.21%) at the site H were higher than that of site L, whereas the abundance of Nitrospirae (HA: 11.16%, LA: 20.17%; HJ: 5.98%, LJ: 17.49%; HO: 4.43%, and LO: 12.28%) at the site H was lower than that of site L.
Figure 3 shows a heatmap of soil bacterial community for six samples of different Cd content at the genus level. From the corresponding cluster analysis of the 20 abundant bacterial genera, the Bryobacter, Geobacter, Gemmatimonas, Haliangium, Anaeromyxobacter, Nitrospira, Candidatus of Koribacter, and Candidatus of Solibacter had higher contents in most soil samples. Among these more abundant bacterial genera, it was observed that the relative abundances of Gemmatimonas, Haliangium, and Anaeromyxobacter at the site H were all higher than that of site L, whereas the abundance of Bryobacter, Geobacter, and Candidatus of Koribacter at site H were all lower than that of site L. Specifically, the abundance of Rhodanobacter, Sphingomonas, Nitrospira, Acidiferrobacter, Rhizomicrobium, and Telmatobacter at the site HJ were significantly higher than that of LJ (Figure 3).
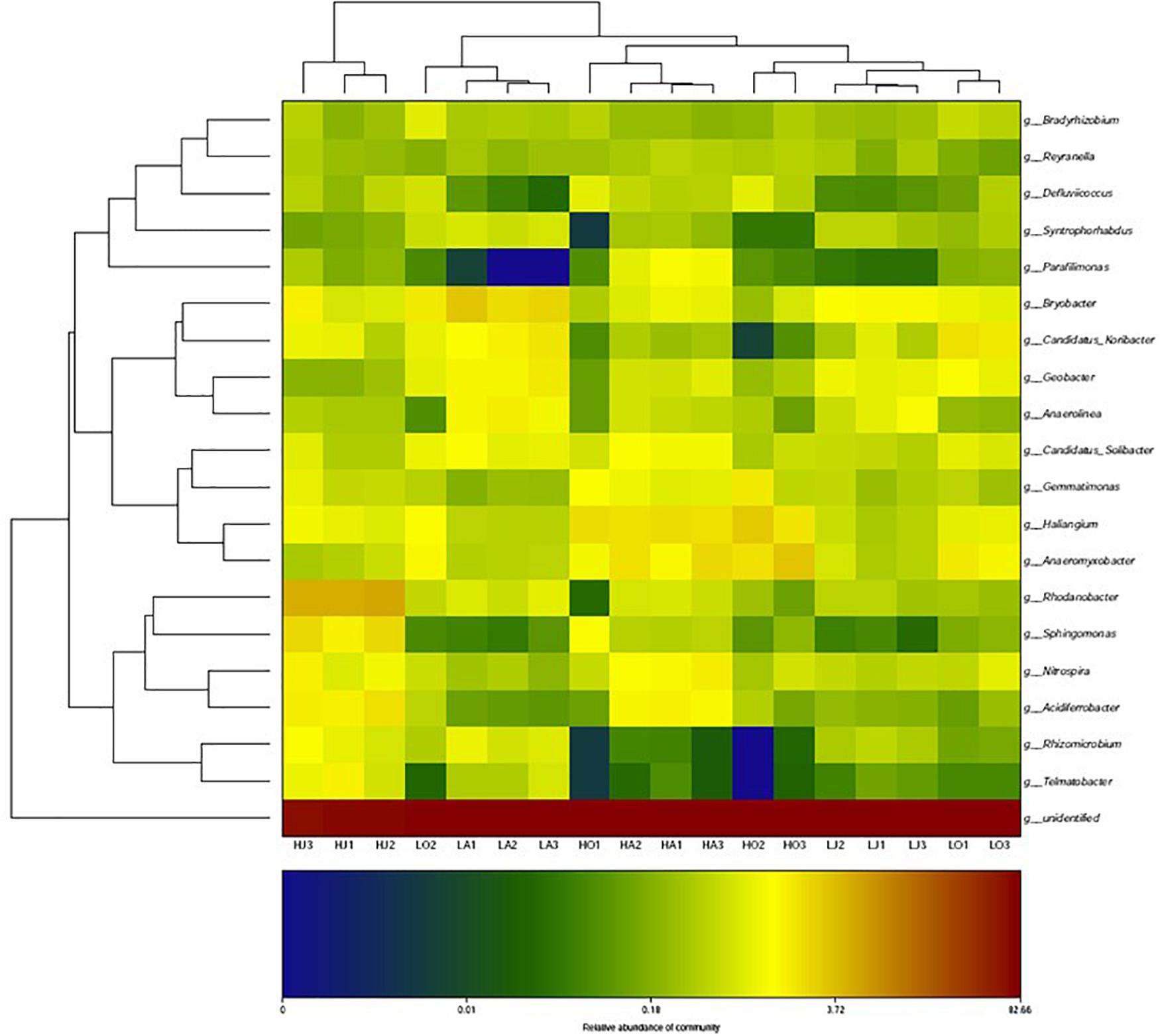
Figure 3. Heatmap of the cluster analysis for top-20 most abundant bacteria at genus-level for the six samples of different Cd concentrations (LA: 0.46 mg/kg; LJ: 0.36 mg/kg; LO: 0.39 mg/kg; HA: 27.11 mg/kg; HJ: 53.70 mg/kg; HO: 8.52 mg/kg).
Further, relative abundances of dominant phyla and genera were influenced by the season. For Proteobacteria, relative abundance (LA: 27.77%, LJ: 27.98%, and LO: 29.60%) were similar in the different seasons at site L, but very different at site H (HA: 38.4%, HJ: 44.59%, and HO: 30.65%). Contrastingly, Acidobacteria abundance at site L (LA: 17.12%, LJ: 12.95%, and LO: 26.29%) were strongly dependent on the season in which the soil sample was taken, whereas seasonality did not greatly affect its abundance at site H (HA: 16.60%, HJ: 14.77%, and HO: 16.95%). In the case of Nitrospira abundance, seasonal changes were observed at both sites (LA: 20.17%, LJ: 17.49%, and LO: 12.28%; HA: 11.16%, HJ: 5.98%, and HO: 4.43%) (Table 2). Additionally, the relative abundances of the 20 abundant bacterial genera at site H and L were different in varying seasons (Figure 3). Taken together, these data clearly suggest that seasonal conditions impact the soil microbiome and the abundance of susceptible populations during seasons responded to the soil Cd content.
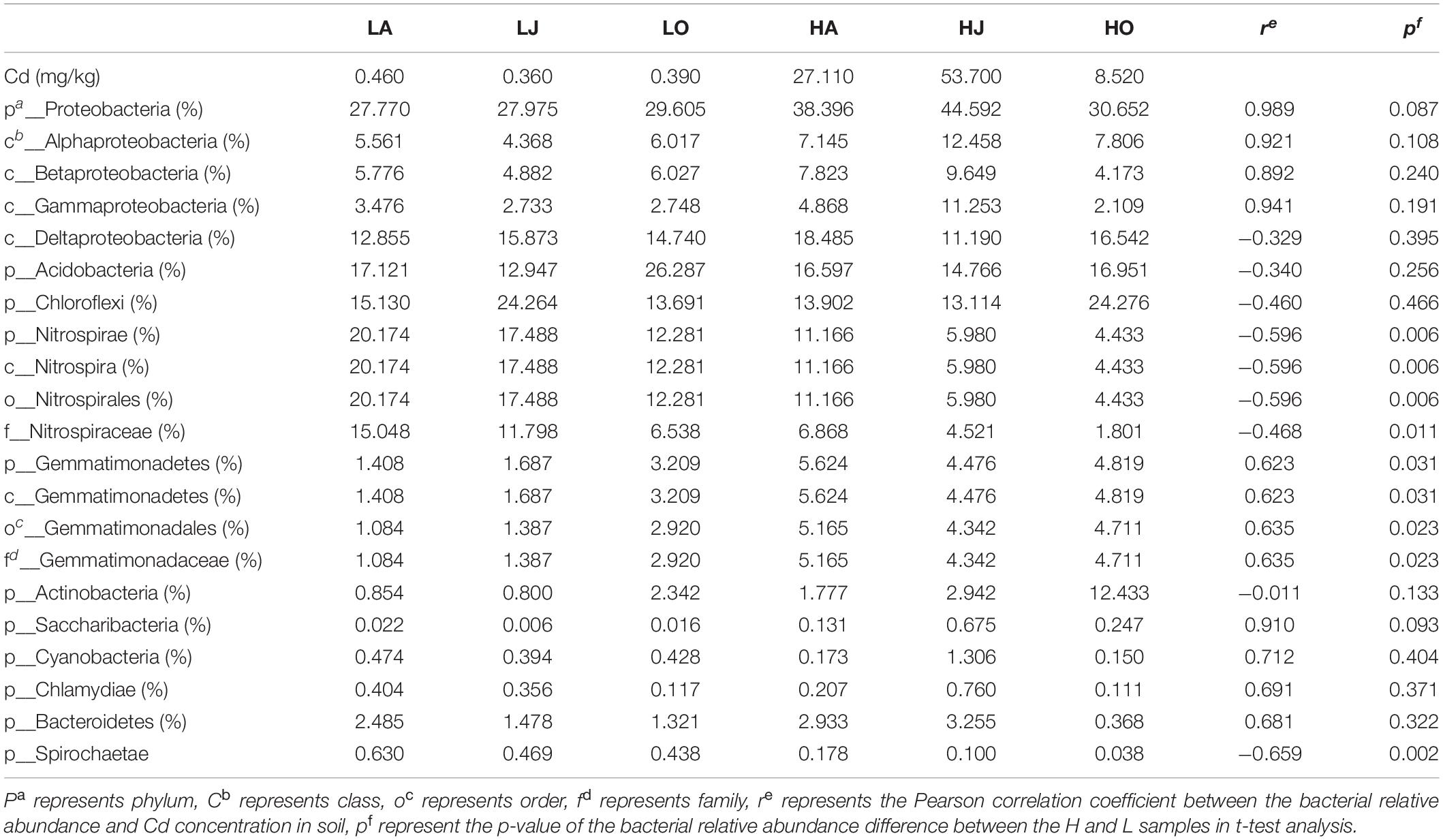
Table 2. Pearson correlation and t-test analysis of the relative abundance of microbial phyla, classes, and cadmium content.
To further assess the relative influence of total Cd concentration on microbial taxa, the Pearson correlations between the relative abundant phyla and Cd were calculated (Table 2). The results showed that the relative abundances of Proteobacteria (r = 0.989), Gemmatimonas (r = 0.623), Saccharibacteria (r = 0.910), Cyanobacteria (r = 0.712), Chlamydiae (r = 0.691), and Bacteroidetes (r = 0.681) all positively correlated with Cd concentration, while the abundance of Nitrospirae (r = −0.596) and Spirochaetae (r = −0.659) correlated negatively with Cd concentration (Table 2). Of the Proteobacteria, the relative abundances of Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria were all found to be positively correlated with Cd concentration with the exception of Deltaproteobacteria (Table 2). In agreement, the phylum Proteobacteria has previously been reported to be associated with Cd.
Once having a basic understanding of the bacterial community response to Cd pollution, the potential of microbial bioremediation to reduce Cd levels was investigated. Firstly, the Cd-resistant bacteria were isolated. A total of 17 strains of different morphological bacteria (Colony morphology of partial Cd-resistant strains were showed in Figure 4A) were isolated from six Cd-contaminated samples using Bulk agar plates containing 1, 2, and 3 mM Cd2+. Then the MIC-Cd values and the Cd2+ removal efficiency of those isolates and 1 negative control strains (E. coil BL21) were determined (Supplementary Table 2). Among them, the strains ho-3, ha-1, and hj-2 exhibited the higher level of Cd2+ resistance than other strains. In detail, the Cd-MIC value of ho-3, ha-1, and hj-2 was 6, 5, and 5 mM, respectively, markedly higher than that of the control strain E. coli BL21 (Cd-MIC: 2 mM Cd2+) (Figure 4B). However, as shown in Figure 4C, ha-1 exhibited the highest Cd2+ removal efficiency (81.78%), which was higher than that observed for ho-3 (63.05%) and hj-2 (79.37%), whilst the cadmium removal efficiency of E. coli BL21 was 9.57%. These results indicate that Cd-resistant bacteria have potential for bioremediation of Cd-contaminated soils. In addition, all of the 17 bacteria were identified through 16S rDNA analysis (Supplementary Table 2). Interestingly, ha-1, hj-2, and ho-3 were identified as Burkholderia sp., which belong to the class of Betaproteobacteria, phylum Proteobacteria. Those results further emphasized the importance of Proteobacteria in Cd pollution remediation. However, the genus Burkholderia contains large number of diverse species which include many phytopathogens, such that a group of 17 closely related Burkholderia species, the Burkholderiacepacia complex (BCC), are responsible for prevalent and potentially lethal pulmonary infections in immunocompromised individuals, such as individuals with cystic (Elshafie and Camele, 2021). Consequently, both Cd resistant and biosafety strains were further screened.
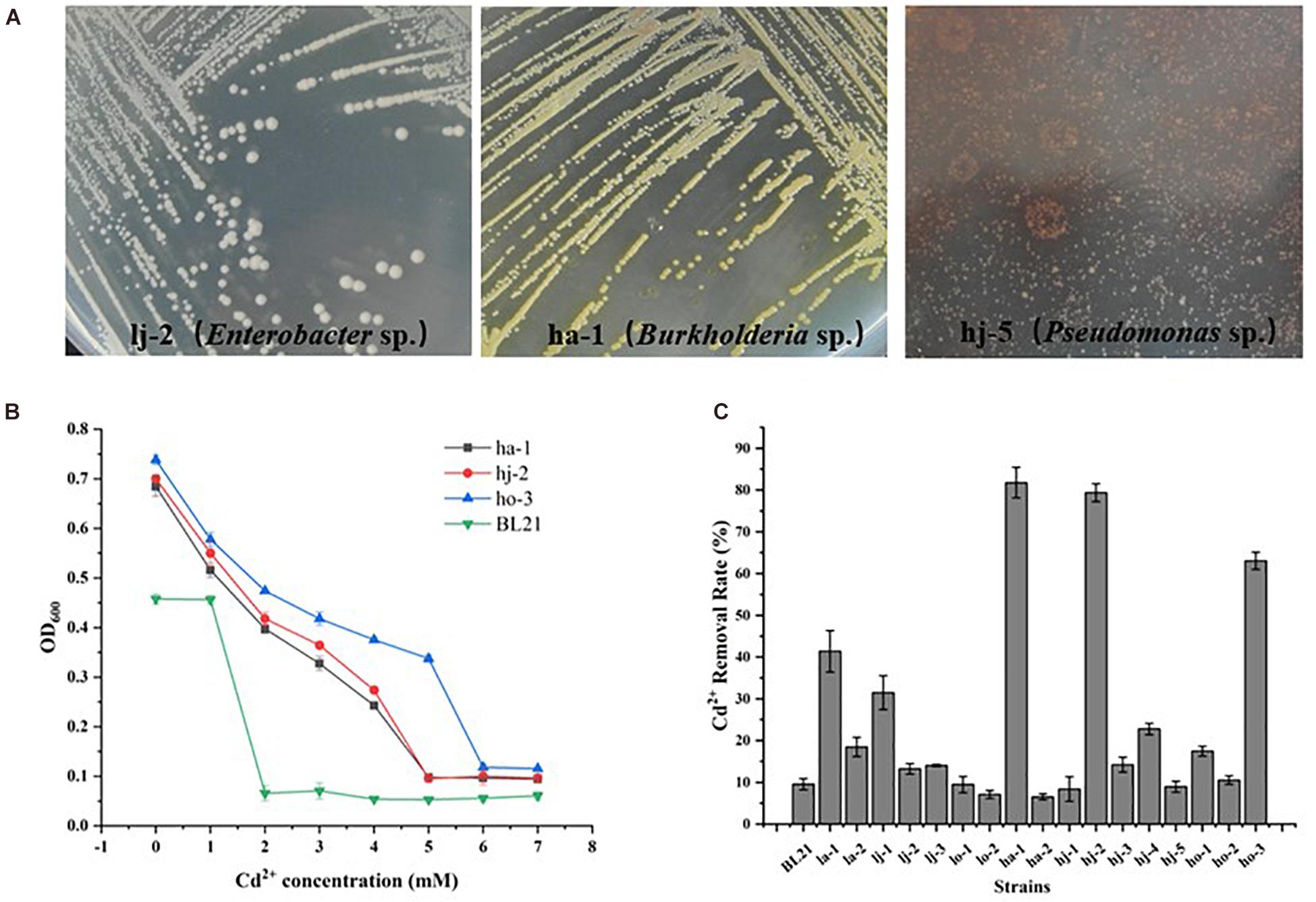
Figure 4. Colony morphology of Cd-resistant bacteria and evaluation of their minimum inhibitory concentration of Cd2+ (Cd-MIC) and Cd2+ removal rate. (A) Colony morphology of partial Cd-resistant strains. (B) Determination of Cd-MIC for Cd-resistant bacteria in varied at different concentrations of Cd. (C) Evaluation of Cd2+ removal rate for Cd-resistant bacteria, which was measured in LB liquid medium supplementation with 0.1 mM CdCl2 and shaken at 200 rpm at 37°C for 24 h.
Bacillus sp. are well-known biocontrol agents against various fungal plant pathogens (Jangir et al., 2018). To isolate Cd-resistant and bio-safe bacteria, Bacillus sp. were isolated from the soil samples. According to the above results, there were significant differences in Cd content (HJ: 53.70 mg kg–1, LJ: 0.36 mg kg–1) and microbial community structure between soil samples HJ and LJ (OTUs no. of HJ: 2093, OTUs no. of LJ:1843) although these two soil samples were collected at the same season (Table 1 and Supplementary Table 1). Thus, we focused on screening Bacillus sp. with Cd-resistance from HJ and LJ samples. Supplementary Figure 4 showed that the observation of the screening Cd-resistant Bacillus sp. strains on the LB agar containing 1mM Cd2+ after incubation at 30°C for 24 h. For HJ, more than 100 colonies were grown on LB agar, whilst about 15 large colonies were screened for LJ in same conditions (Supplementary Figure 4). Additionally, the diversity of microbial morphology in HJ was significantly higher than that in LJ (Supplementary Figure 4). Those results were accordance with the microbial community diversity of samples HJ and LJ.
Finally, a total of 21 strains were isolated from HJ while only 4 strains were obtained from LJ. All strains were identified as Bacillus sp. (Supplementary Table 3). And the Cd-MIC and Cd2+ removal rate of those 25 strains were showed in Supplementary Table 3 and Figure 5. The result showed that Bacillus sp. strains 151-6 isolated from HJ exhibited the highest Cd2+ removal efficiency (90.14%), which was higher than that observed for ha-1 (81.78%). Except for 151-6, the Cd2+ removal rate of other five Bacillus sp. strains isolated from HJ were more than 80% (151-5: 84.78%, 151-13: 82.39%, 151-21: 81.79%, and 151-23: 84.18%). However, the Cd2+ removal rate of 4 Bacillus sp. strains (named as 152-1, 152-2, 152-3, and 152-4) obtained from LJ was 15.84, 17.92, 7.74, and 37.94%, respectively. Additionally, the Cd-MIC of Bacillus sp. strains were lower than that of Burkholderia sp. strains while some of the Bacillus sp. strains exhibited higher Cd2+ removal rate. Those results indicated that Bacillus sp. strains may have a great potential in remediation of cadmium contamination.

Figure 5. Evaluation of Cd2+ removal rate for Cd-resistant Bacillus sp. bacteria, which was measured in LB liquid medium supplementation with 0.1 mM CdCl2 and shaken at 200 rpm at 37°C for 24 h.
The function of soil ecosystem depends heavily on soil microbes as they can promote material circulation, nutrient transformation, energy flow, organic matter decomposition and other ecosystem-related biochemical processes (Lozupone et al., 2007; Luo et al., 2019). Many studies use soil microbial communities to evaluate the ecological status of heavy metal contaminated soils (Stefanowicz et al., 2010; Gómez-Sagasti et al., 2012; Burges et al., 2015). Detecting changes in microbial populations and activities is generally more feasible than directly evaluating the physicochemical properties of soil, as the identification of soil microbes may contribute to the evaluation of remediation treatment. The application of high-throughput sequencing based rRNA is more accurate for evaluating soil microbial communities (Luo et al., 2019).
Previous studies have reported that the presence of heavy metals can influence the microbial diversity and also change the community structure and function (Luo et al., 2019; Duan et al., 2020). Wang et al. (2006) found that low contend of heavy metals can stimulate the growth of microorganisms, while at greater concentrations, an inhibitory effect becomes more prominent, which often leads negatively correlated outcomes. Hence, the concentration of Cd in the soil is critical. In this study, we found that there was an obvious seasonal effect on the Cd concentrations for the H site samples. It may be caused by the change of soil basic parameters in different seasons, such as precipitations, temperature, oxygen, pH and so on (Zhang et al., 2007; Yang et al., 2016; Zoghlami et al., 2018). Previous study has reported that the soil Cd content around rice roots was also greatly varied with growing seasons in Hunan Province, China, and the most important factors were humidity and temperature (Liu et al., 2017). Soil Cd concentration in Apr. (HA) might be more likely to subside to deep soil due to high humidity. Whilst soil Cd content in Jul. (HJ) was significant increase might since soluble Cd2+ was moved from deep soil to surface layer because of evaporation. On the contrary, soil Cd content in Oct. (HO) was subsided again might be because of low temperature and soil moisture transform. However, the Cd content of soil samples from site L were similar across the different seasons, probably as the Cd concentration in the soil was relatively low and could not cause significant change.
There is considerable debate about the effects of Cd on microbial growth. Previous study reports that Cd addition (>1 mg kg–1) of red paddy soil can inhibit soil microbial biomass (Guo et al., 2018). However, as to yellow cinnamon soils under Xanthoceras sorbifolium Bunge and forest soils of Haplic Cambisols in Northeast China, the opposite result was found (Jiang et al., 2018; Duan et al., 2020). Hence, the effects of Cd on microorganism are closely related to the concentrations of Cd and soil type. In our study we found that a significant increase in bacterial diversity with increasing Cd concentrations in site H (Supplementary Table 1 and Supplementary Figure 2), where the soil type is yellow. The result is consistent with the study on yellow cinnamon soils, who found that for all Cd groups (10–100 mg kg–1), 100 mg kg–1 of Cd treatment soil bacterial diversity of yellow cinnamon soils under X. sorbifolium Bunge was higher than that of 10 mg kg–1.
One previous study also reported a variation in bacterial community response to Cd contamination of agricultural paddy soil (Luo et al., 2019). Our results also illustrate changes in the bacterial community composition caused by the Cd polluted (Figures 2, 3, Supplementary Figure 3, and Table 2). This is probably as the community readjustment in response to the introduction of Cd, decreased the number of metal-sensitive microorganisms and increased the number of resistant microorganisms in the soil, which eventually led to changes in community composition. The adaption mechanism of these bacterial populations may be attributed to different microorganism life activities. In our study, the relative abundance of Proteobacteria (r = 0.989) and Gemmatimonas (r = 0.623) were all positively correlated with Cd concentration, while the abundance of Nitrospirae (r = −0.596) and Spirochaetae (r = −0.659) correlated negatively with Cd concentration (Table 2), suggesting that Proteobacteria and Gemmatimonas are Cd-tolerant, whereas Nitrospirae and Spirochaetae are Cd-sensitive. In agreement, the phylum Proteobacteria has previously been reported to be associated with Cd. A large number of isolated and identified strains that resisted Cd or absorbed Cd belong to Proteobacteria and include Burkholderia (Wang et al., 2020), Sphingomonas (Cheng et al., 2021), Pseudomona (Chellaiah, 2018), and Rhizobium (Li et al., 2019). Also many Gram-negative bacteria belonging to Proteobacteria such as Acinetobacter, Ralstonia (also named Cupriavidus) and Comamonas exclusively exist in a Cd-cultivation library of mine tailing (Zhang et al., 2007). Moreover, the result of isolating Cd-resistant bacteria showed that three Burkholderia sp. (ha-1, hj-2, and ho-3) exhibited strong Cd-tolerance and high Cd2+ removal rate. Our study confirms that Proteobacteria may have developed a resistance for Cd and play an important role in Cd-contaminated soils. As for Gemmatimonas, the relative abundances of five levels (phylum, class, order, family, and genus) of Gemmatimonas at the site H were all higher than that of site L, suggesting that Gemmatimonas are Cd-resistant. The resistance mechanism is related to the precipitation-dissolution balance, which limits the dynamic changes of free metal ions in the soil (Abbas et al., 2018). Gemmatimonas aurantiaca, which makes up about 2% of soil bacterial communities, has been reported to accumulate polyphosphate (Zhang et al., 2003). Moreover, the accumulation of polyphosphate has been linked with heavy metal tolerance in bacteria, yeasts and fungi (Trilisenko et al., 2017; Kulakovskaya, 2018; Sathendra et al., 2018; Kolhe et al., 2020). Therefore, it is possible that Gemmatimonas can accumulate polyphosphate and precipitate of Cd2+ in the soil and therefore explains the higher abundance of this phylum at site H. For microbial communities, heavy metal-resistance species can compensate for the loss of metal-sensitive species, ensuring a stable microecological environment (Awasthi et al., 2014). Nitrospirae, a major bacteria group in our soil samples, possesses a higher heavy metal sensitivity, as reported by previous study (Luo et al., 2019). We found that the relative abundances of four levels (phylum, class, order, and family) of Nitrospira at the site H were all lower than that of site L while the abundances of Nitrospira at the site H was higher than that of site L (Supplementary Figure 3). Nitrospira are nitrite-oxidizing bacteria (Han et al., 2017) and generally Cd can inhibit nitrification efficiency (Li et al., 2015; Wang et al., 2016), the highly Cd contaminated soils at site H were believed to inhibit growth of Nitrospira. Whereas the result of the abundances of Nitrospira with response of Cd was opposite. We inferred that the genus annotation may not be completely.
Identifying key heavy metal-resistant bacterial under Cd stress is very important for remediation of Cd-contaminated soils. In this study, we found that two Bacillus sp. strains, 151-6 (Cd2+ removal rate: 91.22%, Cd-MIC: 0.4 mM) and 151-25 (Cd2+ removal rate: 22.80%, Cd-MIC: 1.0 mM), isolated from the same soil sample but exhibit significant differences in Cd2+ resistance and Cd2+adsorption. Our previous work has elucidated the mechanism of Cd-resistance of 151-6 and 151-25, a cadmium efflux system accessory protein and a cadmium resistance protein, was found to play a major role on the Cd– resistance (Yu et al., 2020). Future, the mechanism of Cd-adsorption of 151-6 and 151-25 will be explored. Of course, we also paid attention to the relative abundance of Bacillus sp. in soil samples HJ and LJ, which was 0.050 and 0.017%, respectively. This data is consistent with isolation result of Bacillus sp. bacteria from samples HJ and LJ. The result indicated that Bacillus sp. strains may have a great potential in remediation of cadmium contamination.
Finally, many studies have reported that microbial communities can influenced by specific soil physicochemical properties, such as pH, organic matter, available phosphorus, hydrolytic nitrogen and so on. Unfortunately, we overlooked the effect of these factors on microbial community and the concentration of Cd at the beginning of our experiment. Thus, the basic physiochemical parameters of the six samples were not obtained. However, we recollected the soil samples of the site H and L in June 2021, and the pH and organic matter were measured. The soil pH of the site H and L was 5.04 and 6.56, respectively. And the organic matter of the site H and L was 60.14 and 36.58 g/kg, respectively. The pH value is the key determinant affecting the solubility and liquidity of metal ions, and heavy metal mobility and bioavailability increase due to competition for ligand between H+ ions and dissolved metals. Organic matter can enhance the accumulation of organic carbon in the soil, thus increasing the adsorption of Cd2+ in the soil. It is likely that the dynamic changes of microbial community structure, cause by heavy metals, may be closely related to the type and chemical morphology of metals and soil physicochemical properties.
In conclusion, we characterized the diversity of the bacterial community in two different Cd contaminated soils collected in three different seasons by high throughput Illumina MiSeq sequencing. The result showed that long-term Cd pollution and season change could cause remarkable changes in bacterial population abundance and composition structure. Then, to excavate the available microbial resources, 42 Cd-resistant bacteria were isolated and evaluated for potential application in Cd bioremediation. Our results showed that both selected Burkholderia sp. and Bacillus sp. strains have potential for bioremediation of Cd-contaminated soils. Therefore, our study provided baseline data for bacterial communities in cadmium polluted soils and concluded that Cd-resistant bacteria have potential for bioremediation of Cd-contaminated soils.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XY, JZ, JT, and NW conceived and coordinated the study and wrote the manuscript. XY, JZ, and JT designed, performed, and analyzed the experiments. XL and LS provided technical assistance and contributed to the preparation of the figures. All authors reviewed the results and approved the final version of the manuscript.
This research was supported by the National Natural Science Foundation of China (NSFC, Grant No. 31770124) and the Natural Science Foundation of Jiangxi Province (20202BABL213039).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank our teacher Yunliu Fan for her assistance and guidance. We also thank all colleagues in our lab for constructive discussion and technical support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.698834/full#supplementary-material
Abbas, S., Rafatullah, M., Hossain, K., Ismail, N., Tajarudin, H., and Khalil, H. A. (2018). A review on mechanism and future perspectives of cadmium-resistant bacteria. Int. J. Environ. Sci. Technol. 15, 243–262. doi: 10.1007/s13762-017-1400-5
Awasthi, A., Singh, M., Soni, S. K., Singh, R., and Kalra, A. (2014). Biodiversity acts as insurance of productivity of bacterial communities under abiotic perturbations. ISME J. 8, 2445–2452. doi: 10.1038/ismej.2014.91
Bissett, A., Brown, M. V., Siciliano, S. D., and Thrall, P. H. (2013). Microbial community responses to anthropogenically induced environmental change: towards a systems approach. Ecol. Lett. 16, 128–139. doi: 10.1111/ele.12109
Burges, A., Epelde, L., and Garbisu, C. (2015). Impact of repeated single-metal and multi-metal pollution events on soil quality. Chemosphere 120, 8–15. doi: 10.1016/j.chemosphere.2014.05.037
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336.
Chellaiah, E. R. (2018). Cadmium (heavy metals) bioremediation by Pseudomonas aeruginosa: a minireview. Appl. Water Sci. 8:154.
Cheng, C., Wang, R., Sun, L., He, L., and Sheng, X. (2021). Cadmium-resistant and arginine decarboxylase-producing endophytic Sphingomonas sp. C40 decreases cadmium accumulation in host rice (Oryza sativa Cliangyou 513). Chemosphere 275:130109. doi: 10.1016/j.chemosphere.2021.130109
Duan, C., Liu, Y., Zhang, H., Chen, G., and Song, J. (2020). Cadmium pollution impact on the bacterial community of haplic cambisols in Northeast China and inference of resistant genera. J. Soil Sci. Plant Nutr. 20, 1156–1170. doi: 10.1007/s42729-020-00201-5
Đukić-Ćosić, D., Baralić, K., Javorac, D., Djordjevic, A. B., and Bulat, Z. (2020). An overview of molecular mechanisms in cadmium toxicity. Curr. Opin. Toxicol. 19, 56–62. doi: 10.1016/j.cotox.2019.12.002
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Elshafie, H. S., and Camele, I. (2021). An overview of metabolic activity, beneficial and pathogenic aspects of Burkholderia Spp. Metabolites 11:321. doi: 10.3390/metabo11050321
Genchi, G., Sinicropi, M. S., Lauria, G., Carocci, A., and Catalano, A. (2020). The effects of cadmium toxicity. Int. J. Environ. Res. Public Health 17:3782.
Gómez-Sagasti, M. T., Alkorta, I., Becerril, J. M., Epelde, L., Anza, M., and Garbisu, C. (2012). Microbial monitoring of the recovery of soil quality during heavy metal phytoremediation. Water Air Soil Pollut. 223, 3249–3262. doi: 10.1007/s11270-012-1106-8
Guo, B., Chen, X., Jing, F., Zhang, X., Yang, Z., Liu, W., et al. (2018). Effects of exogenous cadmium on microbial biomass and enzyme activity in red paddy soil. J. Agro Environ. Sci. 37, 1850–1855.
Hamman, S. T., Burke, I. C., and Stromberger, M. E. (2007). Relationships between microbial community structure and soil environmental conditions in a recently burned system. Soil Biol. Biochem. 39, 1703–1711. doi: 10.1016/j.soilbio.2007.01.018
Han, J., Shi, J., Zeng, L., Xu, J., and Wu, L. (2017). Impacts of continuous excessive fertilization on soil potential nitrification activity and nitrifying microbial community dynamics in greenhouse system. J. Soils Sediments 17, 471–480. doi: 10.1007/s11368-016-1525-z
Harichová, J., Karelová, E., Pangallo, D., and Ferianc, P. (2012). Structure analysis of bacterial community and their heavy-metal resistance determinants in the heavy-metal-contaminated soil sample. Biologia 67, 1038–1048. doi: 10.2478/s11756-012-0123-9
Huang, Y., Wang, L., Wang, W., Li, T., He, Z., and Yang, X. (2019). Current status of agricultural soil pollution by heavy metals in China: a meta-analysis. Sci. Total Environ. 651, 3034–3042. doi: 10.1016/j.scitotenv.2018.10.185
Jangir, M., Pathak, R., Sharma, S., and Sharma, S. (2018). Biocontrol mechanisms of Bacillus sp., isolated from tomato rhizosphere, against Fusarium oxysporum f. sp. lycopersici. Biol. Control 123, 60–70. doi: 10.1016/j.biocontrol.2018.04.018
Jiang, W., Yu, X., Tian, Y., Chai, Y., Xiong, Y., Zhong, X., et al. (2018). Effects of cd stress on soil microorganism of Xanthoceras sorbifolium Bunge. Jiangsu Agri. Sci. 46, 228–231.
Jin, Y., Luan, Y., Ning, Y., and Wang, L. (2018). Effects and mechanisms of microbial remediation of heavy metals in soil: a critical review. Appl. Sci. 8:1336. doi: 10.3390/app8081336
Khan, S., Hesham Ael, L., Qiao, M., Rehman, S., and He, J. Z. (2010). Effects of Cd and Pb on soil microbial community structure and activities. Environ. Sci. Pollut. Res. Int. 17, 288–296. doi: 10.1007/s11356-009-0134-4
Kolhe, N., Zinjarde, S., and Acharya, C. (2020). Impact of uranium exposure on marine yeast, Yarrowia lipolytica: insights into the yeast strategies to withstand uranium stress. J. Hazard. Mater. 381:121226. doi: 10.1016/j.jhazmat.2019.121226
Kulakovskaya, T. (2018). Inorganic polyphosphates and heavy metal resistance in microorganisms. World J. Microbiol. Biotechnol. 34:139.
Li, G., Puyol, D., Carvajal-Arroyo, J. M., Sierra-Alvarez, R., and Field, J. A. (2015). Inhibition of anaerobic ammonium oxidation by heavy metals. J. Chem. Technol. Biotechnol. 90, 830–837. doi: 10.1002/jctb.4377
Li, Y., Yu, X., Cui, Y., Tu, W., Shen, T., Yan, M., et al. (2019). The potential of cadmium ion-immobilized Rhizobium pusense KG 2 to prevent soybean root from absorbing cadmium in cadmium-contaminated soil. J. Appl. Microbiol. 126, 919–930. doi: 10.1111/jam.14165
Liao, M., Zhang, H., Yu, S., Chen, C., and Huang, C. (2010). “Effects of cadmium and mercury alone and in combination on the soil microbial community structural diversity,” in Molecular Environmental Soil Science at the Interfaces in the Earth’s Critical Zone, eds J. Xu and P. M. Huang (Berlin: Springer), 337–341. doi: 10.1007/978-3-642-05297-2_99
Liu, Y., Zhang, C., Zhao, Y., Sun, S., and Liu, Z. (2017). Effects of growing seasons and genotypes on the accumulation of cadmium and mineral nutrients in rice grown in cadmium contaminated soil. Sci. Total Environ. 579, 1282–1288. doi: 10.1016/j.scitotenv.2016.11.115
Lozupone, C. A., Hamady, M., Kelley, S. T., and Knight, R. (2007). Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. doi: 10.1128/aem.01996-06
Luo, L., Xie, L., Jin, D., Mi, B., Wang, D., Li, X., et al. (2019). Bacterial community response to cadmium contamination of agricultural paddy soil. Appl. Soil Ecol. 139, 100–106. doi: 10.1016/j.apsoil.2019.03.022
MEP (2014). The Ministry of Land and Resources Report on the National Soil Contamination Survey. China: MEP.
Rubin, B. E., Gibbons, S. M., Kennedy, S., Hampton-Marcell, J., Owens, S., and Gilbert, J. A. (2013). Investigating the impact of storage conditions on microbial community composition in soil samples. PloS One 8:e70460. doi: 10.1371/journal.pone.0070460
Satarug, S., Vesey, D. A., and Gobe, G. C. (2017). Kidney cadmium toxicity, diabetes and high blood pressure: the perfect storm. Tohoku J. Exp. Med. 241, 65–87. doi: 10.1620/tjem.241.65
Sathendra, E. R., Kumar, R. P., and Baskar, G. (2018). “Microbial transformation of heavy metals,” in Waste Bioremediation, eds S. Varjani, E. Gnansounou, B. Gurunathan, D. Pant, and Z. Zakaria (Singapore: Springer), 249–263. doi: 10.1007/978-981-10-7413-4_13
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/aem.01541-09
Stefanowicz, A. M., Niklińska, M., Kapusta, P., and Szarek-Łukaszewska, G. (2010). Pine forest and grassland differently influence the response of soil microbial communities to metal contamination. Sci. Total Environ. 408, 6134–6141. doi: 10.1016/j.scitotenv.2010.08.056
Suksabye, P., Pimthong, A., Dhurakit, P., Mekvichitsaeng, P., and Thiravetyan, P. (2016). Effect of biochars and microorganisms on cadmium accumulation in rice grains grown in Cd-contaminated soil. Environ. Sci. Pollut. Res. 23, 962–973. doi: 10.1007/s11356-015-4590-8
Trilisenko, L., Kulakovskaya, E., and Kulakovskaya, T. (2017). The cadmium tolerance in Saccharomyces cerevisiae depends on inorganic polyphosphate. J. Basic Microbiol. 57, 982–986. doi: 10.1002/jobm.201700257
Vodyanitskii, Y. N., and Plekhanova, I. (2014). Biogeochemistry of heavy metals in contaminated excessively moistened soils (analytical review). Eurasian Soil Sci. 47, 153–161. doi: 10.1134/s1064229314030090
Wang, C., Huang, Y., Yang, X., Xue, W., Zhang, X., Zhang, Y., et al. (2020). Burkholderia sp. Y4 inhibits cadmium accumulation in rice by increasing essential nutrient uptake and preferentially absorbing cadmium. Chemosphere 252:126603. doi: 10.1016/j.chemosphere.2020.126603
Wang, H., Wang, Y.-P., Lin, Q., Shi, J.-Y., and Chen, Y.-X. (2006). Analysis of rhizosphere microbial community structures in heavy metal-contaminated soils using PCR and denaturation gradient gel electrophoresis(DGGE). J. Agro Environ. Sci. 25, 903–907.
Wang, J., Lu, Y., and Shen, G. (2007a). Combined effects of cadmium and butachlor on soil enzyme activities and microbial community structure. Environ. Geol. 51, 1221–1228. doi: 10.1007/s00254-006-0414-y
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007b). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/aem.00062-07
Wang, Y., Ji, M., Zhao, Y., and Zhai, H. (2016). Recovery of nitrification in cadmium-inhibited activated sludge system by bio-accelerators. Bioresour. Technol. 200, 812–819. doi: 10.1016/j.biortech.2015.10.089
Yang, F., An, F., Ma, H., Wang, Z., Zhou, X., and Liu, Z. (2016). Variations on soil salinity and sodicity and its driving factors analysis under microtopography in different hydrological conditions. Water 8:227. doi: 10.3390/w8060227
Yu, X., Ding, Z., Ji, Y., Zhao, J., Liu, X., Tian, J., et al. (2020). An operon consisting of a P-type ATPase gene and a transcriptional regulator gene responsible for cadmium resistances in Bacillus vietamensis 151–6 and Bacillus marisflavi 151–25. BMC Microbiol. 20:18. doi: 10.1186/s12866-020-1705-2
Zhang, C., Nie, S., Liang, J., Zeng, G., Wu, H., Hua, S., et al. (2016). Effects of heavy metals and soil physicochemical properties on wetland soil microbial biomass and bacterial community structure. Sci. Total Environ. 557-558, 785–790. doi: 10.1016/j.scitotenv.2016.01.170
Zhang, H., Sekiguchi, Y., Hanada, S., Hugenholtz, P., Kim, H., Kamagata, Y., et al. (2003). Gemmatimonas aurantiaca gen. nov., sp. nov., a gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 53(Pt 4), 1155–1163. doi: 10.1099/ijs.0.02520-0
Zhang, H. B., Yang, M. X., Shi, W., Zheng, Y., Sha, T., and Zhao, Z. W. (2007). Bacterial diversity in mine tailings compared by cultivation and cultivation-independent methods and their resistance to lead and cadmium. Microb. Ecol. 54, 705–712. doi: 10.1007/s00248-007-9229-y
Zhang, Y., Zhang, X., Zhang, H., He, Q., Zhou, Q., Su, Z., et al. (2009). Responses of soil bacteria to long-term and short-term cadmium stress as revealed by microbial community analysis. Bull. Environ. Contam. Toxicol. 82, 367–372. doi: 10.1007/s00128-008-9613-4
Zoghlami, R. I., Hamdi, H., Boudabbous, K., Hechmi, S., Khelil, M. N., and Jedidi, N. (2018). Seasonal toxicity variation in light-textured soil amended with urban sewage sludge: interaction effect on cadmium, nickel, and phytotoxicity. Environ. Sci. Pollut. Res. 25, 3608–3615. doi: 10.1007/s11356-017-0637-3
Keywords: cadmium pollution, bacterial diversity, seasonal change, Cd-resistant bacteria, bioremediation
Citation: Yu X, Zhao J, Liu X, Sun L, Tian J and Wu N (2021) Cadmium Pollution Impact on the Bacterial Community Structure of Arable Soil and the Isolation of the Cadmium Resistant Bacteria. Front. Microbiol. 12:698834. doi: 10.3389/fmicb.2021.698834
Received: 22 April 2021; Accepted: 02 July 2021;
Published: 22 July 2021.
Edited by:
Raymond J. Turner, University of Calgary, CanadaReviewed by:
Katy Juarez, Universidad Nacional Autónoma de México, MexicoCopyright © 2021 Yu, Zhao, Liu, Sun, Tian and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Tian, dGlhbmppYW5AY2Fhcy5jbg==; Ningfeng Wu, d3VuaW5nZmVuZ0BjYWFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.