- 1Beijing Institute of Radiation Medicine, Beijing, China
- 2Department of Microbiology, Molecular Genetics and Immunology, University of Kansas Medical Center, Kansas City, KS, United States
Human bocavirus 1 (HBoV1) was discovered in human nasopharyngeal specimens in 2005. It is an autonomous human parvovirus and causes acute respiratory tract infections in young children. HBoV1 infects well differentiated or polarized human airway epithelial cells in vitro. Unique among all parvoviruses, HBoV1 expresses 6 non-structural proteins, NS1, NS1-70, NS2, NS3, NS4, and NP1, and a viral non-coding RNA (BocaSR), and three structural proteins VP1, VP2, and VP3. The BocaSR is the first identified RNA polymerase III (Pol III) transcribed viral non-coding RNA in small DNA viruses. It plays an important role in regulation of viral gene expression and a direct role in viral DNA replication in the nucleus. HBoV1 genome replication in the polarized/non-dividing airway epithelial cells depends on the DNA damage and DNA repair pathways and involves error-free Y-family DNA repair DNA polymerase (Pol) η and Pol κ. Importantly, HBoV1 is a helper virus for the replication of dependoparvovirus, adeno-associated virus (AAV), in polarized human airway epithelial cells, and HBoV1 gene products support wild-type AAV replication and recombinant AAV (rAAV) production in human embryonic kidney (HEK) 293 cells. More importantly, the HBoV1 capsid is able to pseudopackage an rAAV2 or rHBoV1 genome, producing the rAAV2/HBoV1 or rHBoV1 vector. The HBoV1 capsid based rAAV vector has a high tropism for human airway epithelia. A deeper understanding in HBoV1 replication and gene expression will help find a better way to produce the rAAV vector and to increase the efficacy of gene delivery using the rAAV2/HBoV1 or rHBoV1 vector, in particular, to human airways. This review summarizes the recent advances in gene expression and replication of HBoV1, as well as the use of HBoV1 as a parvoviral vector for gene delivery.
Introduction
Human bocavirus 1 (HBoV1), a human parvovirus, was discovered from human nasopharyngeal specimens in 2005 (Allander et al., 2005), Parvoviruses are a non-enveloped, icosahedral virus with linear single-stranded DNA (ssDNA) genomes ranging between 4.7 and 5.7 kilobases (kb) (Cotmore et al., 2014). There are two subfamilies within the Parvoviridae family, Parvovirinae which infects vertebrates, and Densovirinae, which infects invertebrates. The Parvovirinae subfamily has eight genera, including Amdoparvovirus, Aveparvovirus, Bocaparvovirus, Copiparvovirus, Dependoparvovirus, Erythro-parvovirus, Protoparvovirus, and Tetraparvovirus (Cotmore et al., 2019). HBoV1, bovine parvovirus (BPV) isolated early in 1961 (Abinanti and Warfield, 1961), and canine minute virus (or minute virus of canines, MVC) isolated in 1967 (Binn et al., 1970), are the three representative members in the genus Bocaparvovirus. Recently, several new members have been identified and classified (Cotmore et al., 2019), including gorilla bocavirus (GBoV1), porcine bocavirus (PBoV), California sea lion bocavirus (CslBoV), feline bocavirus (FBoV), rat bocavirus (RBoV), bat bocaparvovirus (BtBoV), dromedary camel bocaparvovirus (DcBoV), mink bocavirus (MBoV), and rabbit bocaparvovirus (RBoV) (Cheng et al., 2010; Kapoor et al., 2010; Li et al., 2011; Lau et al., 2012, 2016, 2017; Lanave et al., 2015; Yang et al., 2016; Woo et al., 2017). However, only incomplete viral sequences were identified, and no virus was able to be isolated or cultured in cells.
The ssDNA genome of parvovirus is flanked with two short terminal hairpin structures at both ends, which are critical to viral genome replication. The secondary structure of different hairpins, a partial of which has double-stranded (ds)DNA structure, are composed of both paired and mismatched nucleotides. The termini of parvovirus are divided into two categories: heterotelomeric or homotelomeric. The termini of the former group (e.g., members in genera of Protoparvovirus, Bocaparvovirus, and Amdoparvovirus) are named left-end hairpin (LEH) and right-end hairpin (REH), which correspond to the 3′ and 5′ ends of the negative sense ssDNA viral genome, respectively. For homotelomeric parvoviruses, e.g., members in genera of Dependoparvovirus, Erythroparvovirus, and Tetraparvovirus, the termini are called inverted terminal repeats (ITRs). Dependoparvoviruses (e.g., adeno-associated virus, AAV) are named as such because their replication relies on a helper virus, e.g., adenovirus or herpesvirus (Schlehofer et al., 1986; Ward, 2006; Weitzman and Linden, 2011). All other known parvoviruses replicate autonomously without the need of a helper virus, and therefore are called autonomous parvoviruses.
This review summarizes the recent advances in molecular biology of HBoV1, as well as use of HBoV1 as a parvoviral vector for gene delivery and in rAAV vector production.
Genome Organization of HBoV1
In bocaparvoviruses, only the full-length genomes of HBoV1, BPV and MVC have been sequenced, including the terminal hairpins of both ends (Chen et al., 1986; Qiu et al., 2007; Sun et al., 2009; Huang et al., 2012). The HBoV1 genome is 5,543 nucleotides (nts) in length (Genbank accession no.: JQ923422) with distinct hairpin structures at both ends (Huang et al., 2012). The LEH is 140-nt in length and is predicted to be “Y” shaped with short axial ears and mismatches causing an unpaired bubble (Figure 1). The functions of HBoV1 LEH have not been studied. In the heterotelomeric minute virus of mice (MVM), MVM LEH plays a critical role in genome packaging and is important for transcription initiation with the help of NS1 (Li L. et al., 2013). The REH of MVC and BPV, but not HBoV1, harbor sequences that have the potential to fold into a cruciform structure near the end tip, although it is thermodynamically less favorable for BPV (Sun et al., 2009). The cruciform structure also appears in MVM, which has been proven to be required for MVM DNA replication (Astell et al., 1985; Cotmore and Tattersall, 1998, 2005b). Unique among all parvoviruses, the HBoV1 REH has a perfect palindromic sequence of 200 nts in length (Figure 1). Notably, the MVC LEH and the BPV REH were isolated both in flip and flop forms, while the other termini of these three viruses were only found in one form (Chen et al., 1986; Qiu et al., 2007; Sun et al., 2009; Huang et al., 2012). Nevertheless, we believe that DNA replication of bocaparvoviruses follows the rolling hairpin DNA replication model of other parvoviruses (Cotmore and Tattersall, 2014).

Figure 1. Sequence and structure of the HBoV1 left and right end hairpins. The structures of HBoV1 LEH and REH are shown with the 3′ end and 5′ end sequences, respectively, which were predicted using the DNAMAN program (Lynnon, Co., Quebec, Canada). The ear and bubble are indicated, as well as the start and end nucleotides of HBoV1 genome. The sequence refers the full-length HBoV1 genome of the isolate Salvador1 (GenBank accession no.: JQ923422).
Transcriptional Profile of HBoV1
All three bocaparvoviruses, BPV, MVC, and HBoV1, share similarities in their transcriptional expression profiles but with features different from other parvoviruses (Qiu et al., 2007; Sun et al., 2009; Chen et al., 2010). Bocaparvoviruses have one promoter with two polyadenylation sites, the proximal and distal polyadenylation sites [(pA)p and (pA)d], respectively. Therefore, bocaparvovirus only transcribes one single precursor (pre-)mRNA. This pre-mRNA undergoes alternative splicing and alternative polyadenylation to generate multiple viral mRNA transcripts. The left half of the viral genome encodes non-structural (NS) proteins, and the right half encodes structural (VP) proteins (Sun et al., 2009; Qiu et al., 2007; Chen et al., 2010; Sukhu et al., 2013). One unique feature of the bocaparvoviruses is the expression of a phosphorylated non-structural protein (NP1) (Lederman et al., 1984), whose open reading frame (ORF) is located in the middle of the genome (Qiu et al., 2007; Sun et al., 2009; Huang et al., 2012). Due to the small genome capacity, NP1 ORF largely overlaps with the NS1 ORF at the 3′ end. NP1 plays an important role in both viral pre-mRNA processing and viral DNA replication (Shen et al., 2016; Zou et al., 2016).
The processing of HBoV1 pre-mRNA appears to be more complex than those of MVC and BPV (Qiu et al., 2007; Sun et al., 2009; Chen et al., 2010; Zou et al., 2019; Figure 2). The mRNA spliced at the D2-A2 sites, which results in a shift of the NS1 ORF at the C-terminus, encodes NS1. Unspliced mRNA that reads through the D2-A2 intron encodes the NS1-70 protein (Chen et al., 2010). Among the NS1-coding region, alternative splicing from intron D1 to A1’, D1’-A1, and both generates R2, R3, and R4 mRNAs, respectively, which encode NS2, NS3 and NS4. R5 mRNA that is being spliced at both the D1-A1 and D2-A2 introns is responsible for NP1 expression. R6 mRNA that is consecutively spliced at all three introns, D1-A1, D2-A2, and D3-A3, encodes VP proteins. The VP2 protein is translated from a non-canonical start codon (GUG) located between VP1 and VP3, which is uncommon for parvoviruses (Janik et al., 1984; Zou et al., 2016). All NS-encoding mRNA transcripts had short (RXS) and long (RXL) forms, which are terminated at the proximal [(pA)p1 and (pA)p2] and distal polyadenylation sites [(pA)d1 and (pA)dREH], respectively (Figure 2; Hao et al., 2017; Zou et al., 2019). However, VP-encoding transcripts only use the (pA)d sites. Viral mRNA transcripts use the (pA)p1 and (pA)p2 sites at a roughly equal efficiency, but they prefer to use the (pA)d1 more than the (pA)dREH (Zou et al., 2019).
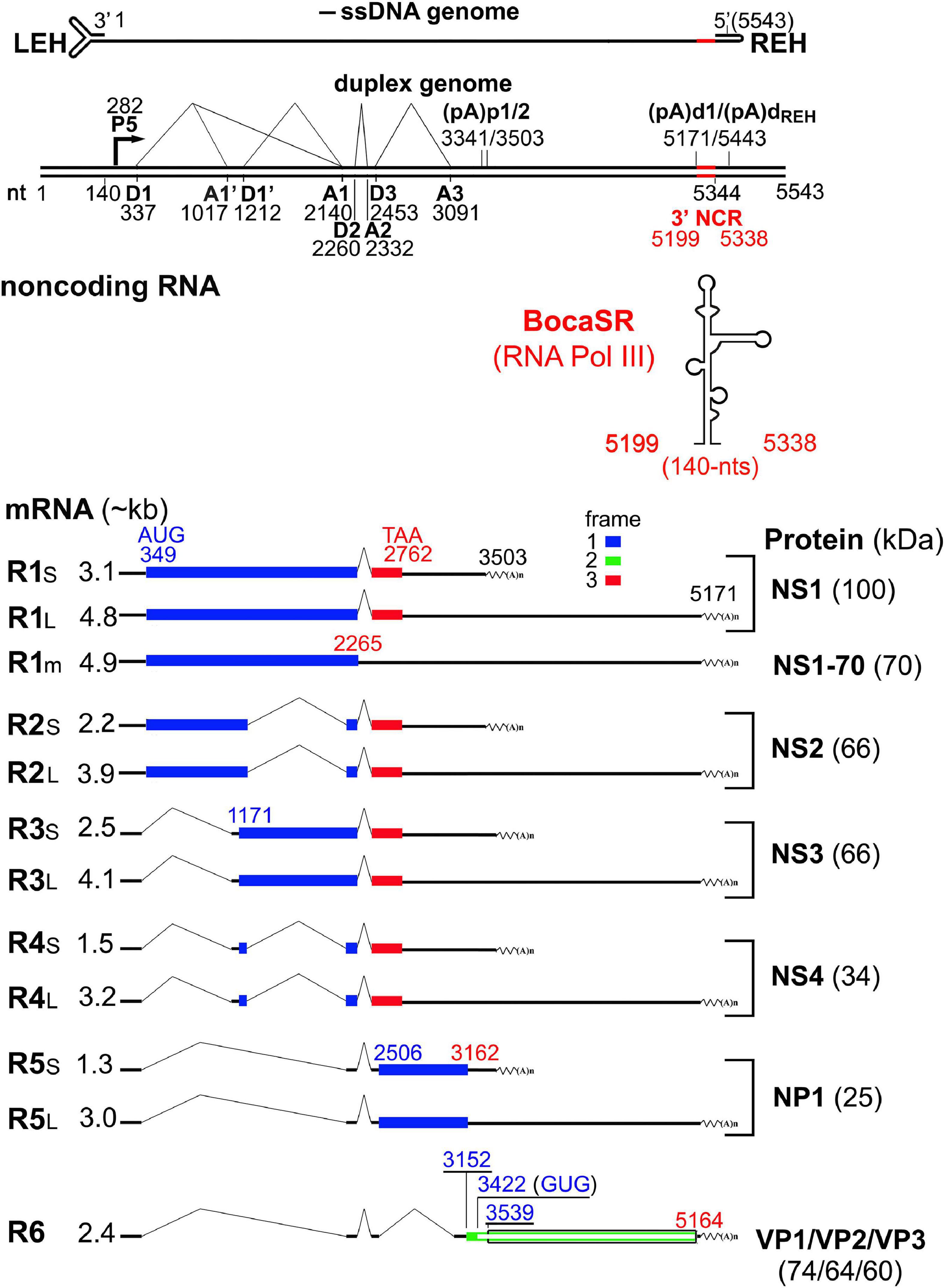
Figure 2. Transcription map of HBoV1. The major transcription landmarks including the terminal repeats (LEH and REH), promoter (P), splice donors (D) and acceptors (A), and (pA)p and (pA)d sites, are depicted. All identified mRNA transcripts are listed below the map (designated R1 to R6), with their respective sizes shown on the left and the detected molecular weight of the expressed proteins shown to the right. Different ORFs are illustrated in blue, red or green colors. The expressed non-coding RNA (BocaSR) is diagrammed with the size. NCR, non-coding region.
Unique among all the parvoviruses, HBoV1 expresses a small non-coding RNA (BocaSR) (Wang et al., 2017b), which is transcribed from nt 5,199 to 5,338 of the dsDNA genome through a RNA Pol III promoter that lies entirely within the gene and composes A- and B-boxes as those in adenovirus virus (Ad)-associated (VA) I RNA (Punga et al., 2020; Figure 3A). Its transcription level is much higher than all other HBoV1 mRNA transcripts. It is presumably folded into a secondary structure similar to VAI RNA and is composed of terminal stem, central domain, apical stem and loops (Figures 3B,C; Wang et al., 2017b).
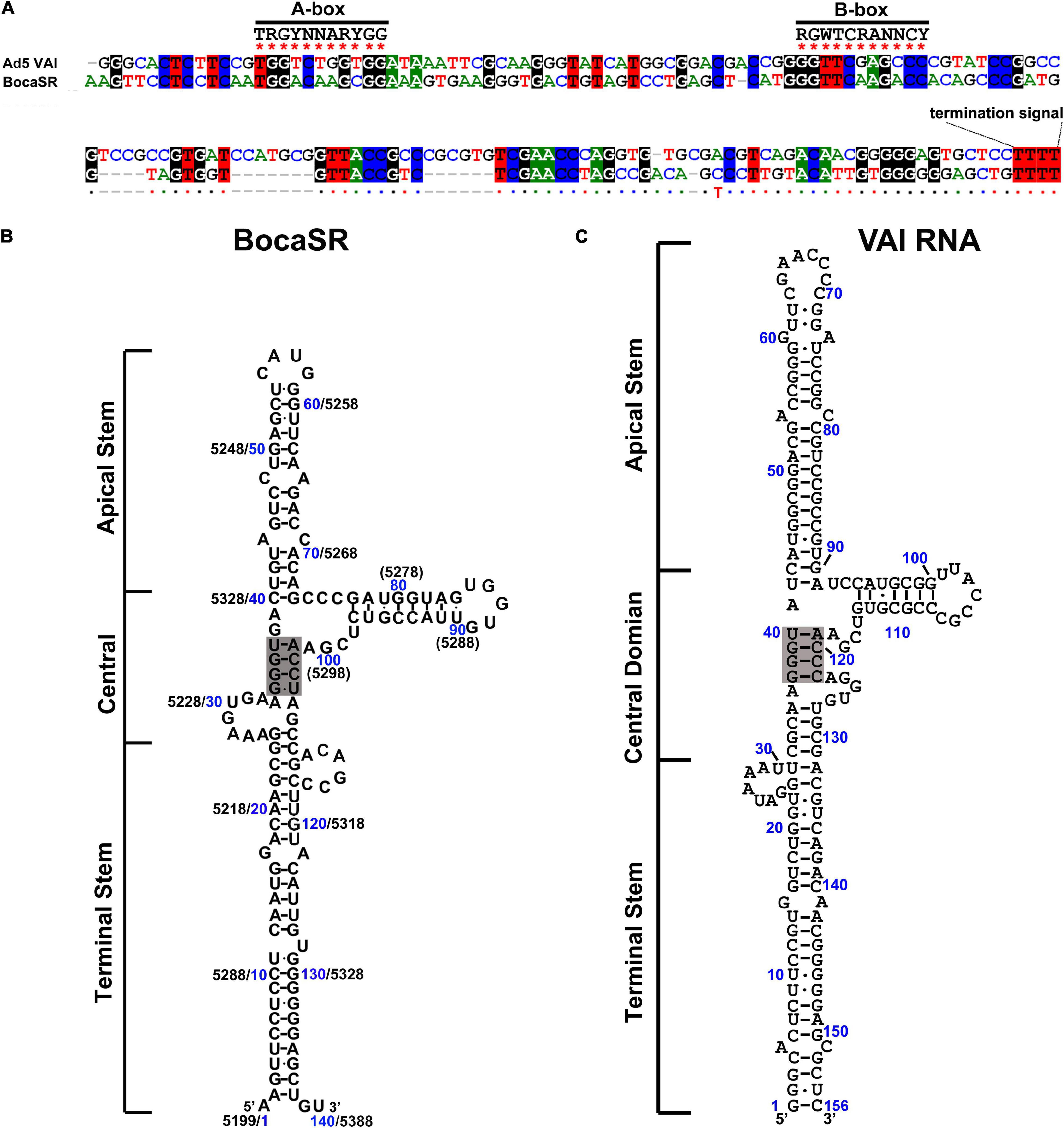
Figure 3. Comparisons between BocaSR and VAI RNA. (A) RNA sequence alignment. The sequences of BocaSR and adenovirus 5 (Ad5) VAI RNA were aligned using the CLUSTALW algorithm. Identical nucleotides are colored. Consensus sequences of the A- and B-boxes of the intergenic RNA Pol III are indicated. R represents G/A; Y, C/T, and N any nucleotides. (B,C) The structure of BocaSR (B) was predicted using the KineFold algorithm, with Ad5 VAI RNA serving as a reference (C). Nucleotide numbers shown in blue and black are the positions of the BocaSR RNA (VAI RNA) sequence and the HBoV1 genome, respectively. Stem structures are indicated, and the central tetranucleotide pairs are highlighted in gray.
HBoV1 Non-Structural Proteins-NS1-4
NS1 Protein
The sequence similarities between BPV and HBoV1 and between MVC and HBoV1 NS1 are 31 and 39%, respectively. NS1, the largest non-structural protein of parvovirus and also named replication protein 78/68 (Rep78/68) for AAVs, plays a key role in parvovirus DNA replication (Cotmore and Tattersall, 1989; Cotmore et al., 1995). NS1 is composed of three function domains, including the N-terminal DNA-binding/endonuclease domain, the middle helicase domain, and the C-terminal transcription activation domain (Figure 4). The N-terminal domain binds to the replication origin (Ori) of viral replicative form (RF) DNA, and is referred to as the origin or DNA-binding domain (OBD/DBD). It has both strand- and sequence-specific endonuclease activity (Tewary et al., 2013). HBoV1 OBD comprises aa1–275, of which the structure has been determined (Tewary et al., 2013). Superimposition of the OBD structures of AAV5 Rep68 (Hickman et al., 2002), MVM NS1 (Tewary et al., 2015), and HBoV1 NS1 displayed a conserved beta-sheet core, flanked by several alpha helices on both the left and right sides (Tewary et al., 2013). They share similarities with each other and belong to the HUH-nuclease superfamily. One loop and an alpha helix protrusion are sequence specific and specifically bind to Ori major and minor grooves. For AAVs, the interaction between Rep78/68 OBD and the Ori has been studied in detail by analyzing the structure of the Rep78/68 and Ori complex (Hickman et al., 2004; Santosh et al., 2020). Rep78/68 oligomerizes and binds to sequence specific tetra-nucleotides repeats (Hickman et al., 2004; Santosh et al., 2020). In HBoV1, the two DNA binding regions of the HBoV1 OBD are positively charged, and the mutation of which greatly diminished viral genome replication (Shen et al., 2016). However, the HBoV1 OBD/Ori complex has not been structurally resolved.

Figure 4. Domains of HBoV1 NS proteins. HBoV1 NS1 (GenBank: AFR53039) and AAV5 Rep78 (GenBank: AAD13755) are aligned. N-terminal origin DNA binding domain (OBD; in red) and helicase domain (in purple) are diagramed. The regions positioned between the OBD and helicase domains (shown in green) are predicted to be the oligomerization signal. The C-terminal region (shown in yellow) is predicted to serve potentially transcriptional activation function. Dashed lines in the OBD indicate residues that are structured as endonuclease core/DNA binding loop (Tewary et al., 2013), and dashed rectangles in the helicase domain indicate Walker boxes (Koonin, 1993). Oligo. indicates a putative oligomerization signal. NS2, NS3, NS4, and NS1-70 proteins are diagramed in colored blocks with thin lines indicating excised aa sequences due to ligation of the neighboring exons of their mRNAs.
The middle domain of the NS1 protein contains four conserved Walker motif (boxes A, B, B’, and C), which belongs to the SF3 helicase family and carries out 3′–5′ helicase function (James et al., 2003; Cotmore and Tattersall, 2005a). By alignments of HBoV1 NS1 with AAV2 Rep78 and MVM NS1 (Legendre and Rommelaere, 1994; Smith et al., 1997), the HBoV1 C-terminal domain (aa 638–781) is predicted to have transcription transactivation capability (Figure 4), but has not been studied (Legendre and Rommelaere, 1994; Smith et al., 1997).
NS1-70 is the short version of the NS1 lacking the C-terminus. Its expression level is very low during virus infection (Shen et al., 2015). But the protein per se can induce a DNA damage response (DDR) in a comparable capability with the full-length NS1 (Deng et al., 2016a), and thus it is supposed to support viral DNA replication.
NS2, NS3, and NS4 Proteins
The function of HBoV1 NS2-4 has not been well studied, and is dependent on the cell types used. NS2 is required for HBoV1 infection in polarized human airway epithelia, whereas NS3 and NS4 are not required (Shen et al., 2015). Notably, all three are not required for replication of the HBoV1 duplex genome (an infectious clone) in HEK293 cells (Shen et al., 2015). NS2 is unique in all parvoviruses in that it spans the OBD and the putative transcription activation domain (Figure 4). It is required for productive AAV2 infection in both HEK293 and HeLa cells, together with NP1 and BocaSR (Wang et al., 2017a). NS3 overlaps completely with the NS1 helicase coding region (Figure 4), and thus it possibly shares similar functions as the Rep52 of AAVs. Rep52 shares the helicase and C-terminal domains with Rep78, and plays an important role in viral genome packaging (Smith and Kotin, 1998; King et al., 2001). NS4 is also unique in parvoviruses. It is the putative transcription transaction domain, which is also encompassed by all of the NS1-3 (Figure 4). It can substitute the function of NS2 in supporting replication of AAV dsDNA genome (infectious clone) in HEK293 and HeLa cells (Wang et al., 2017a). Interestingly, NS4 has only 199 aa with a predicted size of 22-kDa, however, it appeared as ∼34-kDa in SDS-polyacrylamide gene electrophoresis (Shen et al., 2015). This increased molecular weight of NS4 can explain the detected sizes of the NS1-3 which are relatively large than their coding capability. Therefore, all NS1-4 are likely post-translationally modified at the C-terminal domain (NS4).
HBoV1 Non-Structural Protein-NP1
Conserved among all bocaparvoviruses, NP1 has ∼200 aa and is expressed from an ORF that overlaps with the C-terminus of the NS1. Although there is an identity of only ∼48% among the NP1 of different bocaparvoviruses in amino acid sequence, NP1 is conserved in functions. HBoV1 NP1 has a non-canonical nuclear localization signal located at aa 7–50 (Li Q. et al., 2013). NP1 plays an enhancement role in viral DNA replication (Sun et al., 2009). Transfection with a NP1-knockout clone of HBoV1 or MVC barely produced monomeric and dimeric RF DNA (Sun et al., 2009; Huang et al., 2012; Sukhu et al., 2013; Shen et al., 2016). The NP1 proteins of BPV and MVC are exchangeable with each other, and HBoV1 NP1 is also able to supplement the function of MVC NP1 (Sun et al., 2009). Notably, NP1 could overcome the deficiency of MVM NS2 at the early replication stage by localizing at the viral DNA replication centers, but it could not compensate for the late steps of MVM infection (Mihaylov et al., 2014).
NP1 plays multiple roles during processing viral pre-mRNA (Sukhu et al., 2013; Fasina et al., 2015, 2017; Zou et al., 2016). Both MVC and HBoV1 NP1 facilitate viral pre-mRNA to read through the (pA)p site in order to generate full-length VP-encoding transcripts (Zou et al., 2016; Fasina et al., 2017). Additionally, MVC NP1 controls the expression of MVC NS proteins via its role in governing mRNA splicing of the third intron (Fasina et al., 2017). Cellular cleavage and polyadenylation specificity factor 6 (CPSF6) interacts with both MVC and HBoV1 NP1 (Dong et al., 2019; Wang et al., 2020). CPSF6 is one of the cellular factors in the polyadenylation complex that associate with the AAUAAA motif of the polyadenylation signal (Yang et al., 2010). CPSF6 tempers MVC NP1’s suppression of the internal polyadenylation at (pA)p, enhances the splicing of the third intron, and further modulates the export of MVC mRNA (Dong et al., 2019). Notably, CPSF6 also involves nuclear import of HBoV1 NP1 (Wang et al., 2020).
The U2 small nuclear ribonucleoprotein (snRNP) complex deposited on the A3 acceptor is critical for the communication between the A3 acceptor and the polyadenylation site that interacts with the 3′-end CPSF complex. The interaction between the U2 snRNP and CPSF complexes defines the size of the last exon of viral mRNAs (Kyburz et al., 2006), the short exon that encodes no proteins (Figure 5A) or the large exon that encodes VP (Figure 5B). When NP1 is not presented, the short exon is preferably defined, as the distance between the last acceptor and the (pA)p signals is short, which permits a strong interaction between the U2 snRNP and CPSF complexes (Figure 5A). While NP1 is expressed, NP1 decreases the interaction of the CPSF complex on the (pA)p sites, which generates a stronger interaction of the U2 snRNP on the A3 site with the CPSF complex on the (pA)d sites; therefore, NP1 is required to define the large 3′ exon, the VP-encoding exon (Figure 5B).
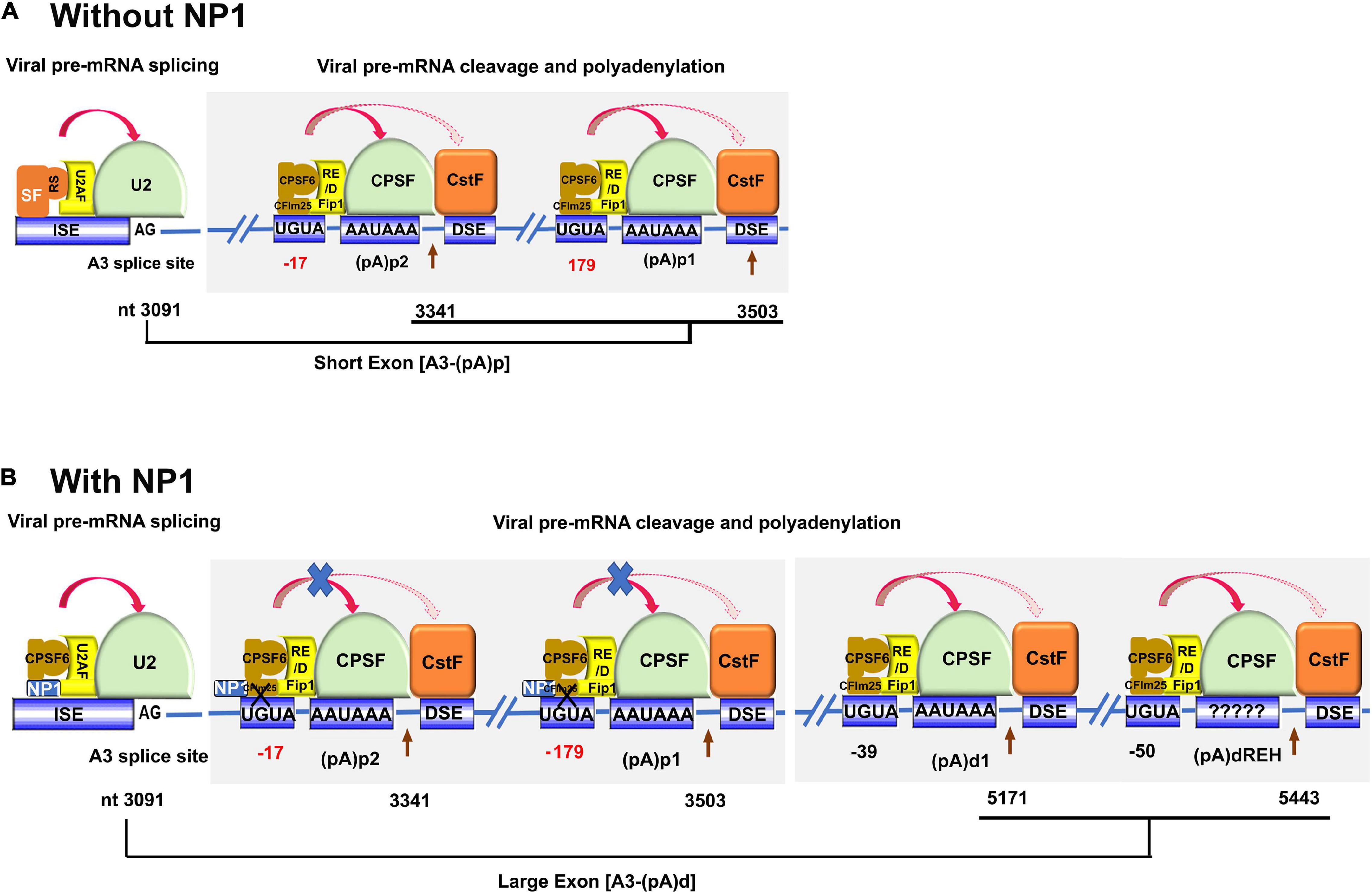
Figure 5. A proposed model of NP1-facilitated definition of the VP-encoding exon. The viral pre-mRNA is shown in part with the A3 splice site, (pA)p1, (pA)p2, (pA)d1, and (pA)dREH sites. Key signals for mRNA processing, i.e., AG at the 3′ acceptor, CFIm25-binding site UGUA, CPSF-binding site AAUAAA, are shown with the downstream element (DSE) indicated. The 3′ acceptor U2 interacting complex (U2AF/U2 snRNP) and the CPSF complex composed of CFIm, Fip1, CPSF, and CstF are diagrammed. CPSF6 interacts with Fip1 through their RS and RE/D domains. Nucleotide (nt) numbers show the location of A3 3′ splice site, and the cleavage site of each pA signals. (A) Without NP1. There are potential SR proteins (SF) binding to the A3 acceptor to enhance the binding of U2 snRNP to the A3 acceptor. CFIm binds to UGUA enhancers at 17 (too close) and 179 nts (too far) upstream of the (pA)p2 and (pA)p1, respectively. The distance between the A3 acceptor and (pA)p sites is short, which favors defining the exon between A3 and (pA)p sites. (B) With NP1. Potential interaction of NP1 with CPSF6 disrupts the interaction between CPSF6 and CFIm25 that binds UGUA sites 17 (too close) and 179 nts (too far) upstream of the (pA)p2 and (pA)p1, respectively. While the CFIm25 binds UGUA signals at –39 and –50 nt (optimal distance) upstream of the (pA)d1 and (pA)dREH, respectively, the interaction between CFIm25 and CPSF6 is tight and, therefore, difficult to be interrupted by the NP1. The overall interaction between the U2 snRNP complex at the A3 acceptor and the CPSF complex at the (pA)d sites determines the large exon between the A3 acceptor and (pA)d sites.
The cleavage factor Im (CFIm) complex consists of a small subunit CFIm25 and two alternative large subunits, CPSF6 and CFIm59, both of which are members of the SR superfamily proteins (Ruegsegger et al., 1998). CFIm25 forms a dimer, which is bound by CPSF6 or CFIm59 via its RNA recognition motif (RRM) domain to form a tetrameric CFIm complex (Yang et al., 2011). CPSF6 has a stronger arginine–serine-rich (RS) domain than the CFIm59, and CFIm25 binds specifically to a UGUA signal of the mRNA in the proximity to the CPSF-binding hexanucleotide signal (AAUAAA) (Yang et al., 2010). CFIm is a UGUA-dependent activator that promotes mRNA 3′-processing complex assembly. CFIm activator function requires the RS-like domains of CPSF6 or CFIm59, and it involves a mechanism similar to SR protein-mediated splicing regulation. Recently, it has been reported that the function of CFIm as a UGUA enhancer-dependent activator of mRNA processing contributes to its role in regulating global alternative polyadenylation (Martin et al., 2012). The direct interaction of the NP1 with CPSF6 could disrupt the interaction between CPSF6 with CFIm25, which would decrease the loading of the CFIm complex to the UGUA site upstream of the CPSF complex and suppress the cleavage at the (pA)p sites (Figure 5B). Of note, the function of CFIm in facilitating cleavage is UGUA position dependent (Yang et al., 2010; Zhu et al., 2018). A single copy of UGUA had the highest activities at −39 nt and then at −50 nt from the cleavage site (Zhu et al., 2018). We hypothesize that the CFIm binding to the two UGUA enhancers upstream of the two (pA)p sites, which are at suboptimal distances (−17 and −179 nt), is weaker than the CFIm binding to the enhancers (−39 and −50 nt) in front of the (pA)d sites (Figure 5B). Therefore, NP1 competes to interact with CPSF6, which likely disrupts the CFIm complex at the (pA)p sites but not the one at the (pA)d sites.
HBoV1 Encodes a Small Non-Coding RNA-Bocasr
BocaSR is essential for HBoV1 replication in infected polarized human airway epithelial cultures and viral DNA replication in viral duplex genome-transfected HEK293 cells (Wang et al., 2017b). BocaSR is localized in the nucleus. It shares a high similarity of 46.1–51.2% with the other four RNA Pol III-transcribed viral small RNAs: VAI, VAII, EBER1, and EBER2. BocaSR regulates the expression of NS1, NS2, NS3, and NP1 but not NS4. However, unlike VA RNAs which are localized in the cytoplasm (Vachon and Conn, 2016), BocaSR does not inhibit phosphorylation of protein kinase R (PKR) and eukaryotic initiation factor 2 (eIF-2) (Wang et al., 2017b).
In addition to the function in enhancing viral NS protein expression, BocaSR plays a direct role in viral DNA replication, which cannot be fully complemented by VAI RNA (Wang et al., 2017b). The mechanism of how BocaSR facilitates viral genome replication has not been revealed. Intriguingly, alignments of HBoV2-4 and GBoV sequences, but not other members in the genus of Bocaparvovirus, show that they also express BocaSR, and at least HBoV3 BocaSR could supplement the lack of BocaSR in NS expression of HBoV1 (Wang et al., 2017b). Importantly, in the case of HBoV1 helped AAV replication, BocaSR is essential to help AAV replication in human airway epithelia, HEK293 and HeLa cells (Wang et al., 2017a). Considering the specific tissue tropism of HBoVs, this short viral non-coding RNA may evolve to contribution of bocaparvovirus adaptation to the airway or gastrointestinal environment.
Structural Proteins
Different from BPV and MVC, HBoV1 expresses three VP proteins, and the VP2 uses a non-canonical start codon (Zou et al., 2016). The capsid proteins are expressed in a ratio ∼1:1:10 during infection, which is similar to that of AAVs. VP3, the abundant structure protein, is able to assemble virus like particles (VLPs), which contain neutralizing epitopes and receptor binding sites (Gurda et al., 2010). A monoclonal antibody 15C6 reacts with all of the VLPs formed by HBoV1-4 (Gurda et al., 2010). HBoV1 has a short VP1 unique region (VP1u) of 90 aa and its coding sequence contains internal polyadenylation signals (Zou et al., 2016). Most members in the Parvovirinae subfamily, except Aleutian mink disease virus (AMDV) (Huang et al., 2014) and the recently identified shrimp densovirus (Pénzes et al., 2020), contain a phospholipase A2 (PLA2) domain within VP1u (Zadori et al., 2001). The HBoV1 VP1u region exhibits a PLA2 activity within the region 11–66 aa (Qu et al., 2008). The PLA2 domain of the VP1u was also confirmed in MVC (Sun et al., 2009). The phospholipase activity of the PLA2 domain is supposed to be important for sequential structural changes from receptor mediated endocytosis to endosome escape (Zadori et al., 2001; Farr et al., 2005).
Conserved with the structures of other parvoviruses (Mietzsch et al., 2019). HBoV1-4 share the icosahedral fivefold axis tunnel, threefold axis trimeric protrusions and a twofold axis depression (Gurda et al., 2010; Kailasan et al., 2016; Mietzsch et al., 2017; Luo et al., 2021). Compared with AAVs, HBoV1 has a relative flat capsid shell (Kailasan et al., 2016). This feature makes it to be able to package a larger viral genome with an even smaller VP3. The surface variable region (VR) III of VP3 has been proposed as a host tissue-tropism determinant, which is structurally similar among the gastrointestinal tropic HBoV2-4, but different from the airway tropic HBoV1. Six monoclonal antibody epitopes on the HBoV1-4 have been determined (Kailasan et al., 2016). These characterizations of the HBoV capsid surface structures are important for development of HBoV-based viral vectors.
HBoV1 DNA Replication
Replication of the parvovirus genome is greatly associated with DNA damage and repair factors (Schwartz et al., 2009; Adeyemi et al., 2010; Lou et al., 2012; Vogel et al., 2012; Cotmore and Tattersall, 2013; Deng et al., 2016b). Upon infection, H2AX and RPA32 are phosphorylated, and the signals are passed through phosphoinositide 3-kinases ATM (Ataxia telangiectasia mutated), ATR (ATM- and RAD3-related), and DNA-PKcs (DNA-dependent protein kinase catalytic subunit), respectively. Activation of the phosphoinositide 3-kinases causes cell cycle arrest and activates DNA repair pathways. However, HBoV1 infects polarized human airway epithelia, which are terminally differentiated (mitotically quiescent). HBoV1 infection activates ATM, ATR and DNA-PK, which play significant roles in viral DNA replication (Deng et al., 2016b). All three DDR pathways were activated during HBoV1 duplex genome transfection of HEK293 cells and are necessary for viral DNA replication (Deng et al., 2016a). NS1 alone induces DDR signaling but no cellular DNA damage. Importantly, DNA repair polymerase (Pol) κ and Pol η play a significant role in HBoV1 genome replication in both polarized human airway epithelia and HEK293 cells (Deng et al., 2016a,b).
HBoV1 DNA replication follows a model of DNA repair (Deng et al., 2016a,b), in contrast to the DNA replication DNA polymerase-dependent replication model of other parvoviruses (Astell et al., 1983, 1985; Berns, 1990; Ryan et al., 1996; Cotmore et al., 2000; Cotmore and Tattersall, 2005b; Ward, 2006). Upon entering the nucleus, the viral genome is recognized by Pol κ, Pol η, or both that synthesize the complementary strand, primed by the 3′-OH at the LEH (Figure 6, Steps 6, 7). In addition, for HBoV1 replication, NS1, NP1 and BocaSR are required (Figure 6, Step 12). They are localized within the viral DNA replication centers. During MVM replication, cellular factors bind both LEH and REH and are necessary for the MVM DNA replication (Christensen et al., 1997a,b; Cotmore and Tattersall, 1998). For the replication of HBoV1, such REH or LEH-binding cellular factors should be required for viral DNA replication as well but have not been identified yet.
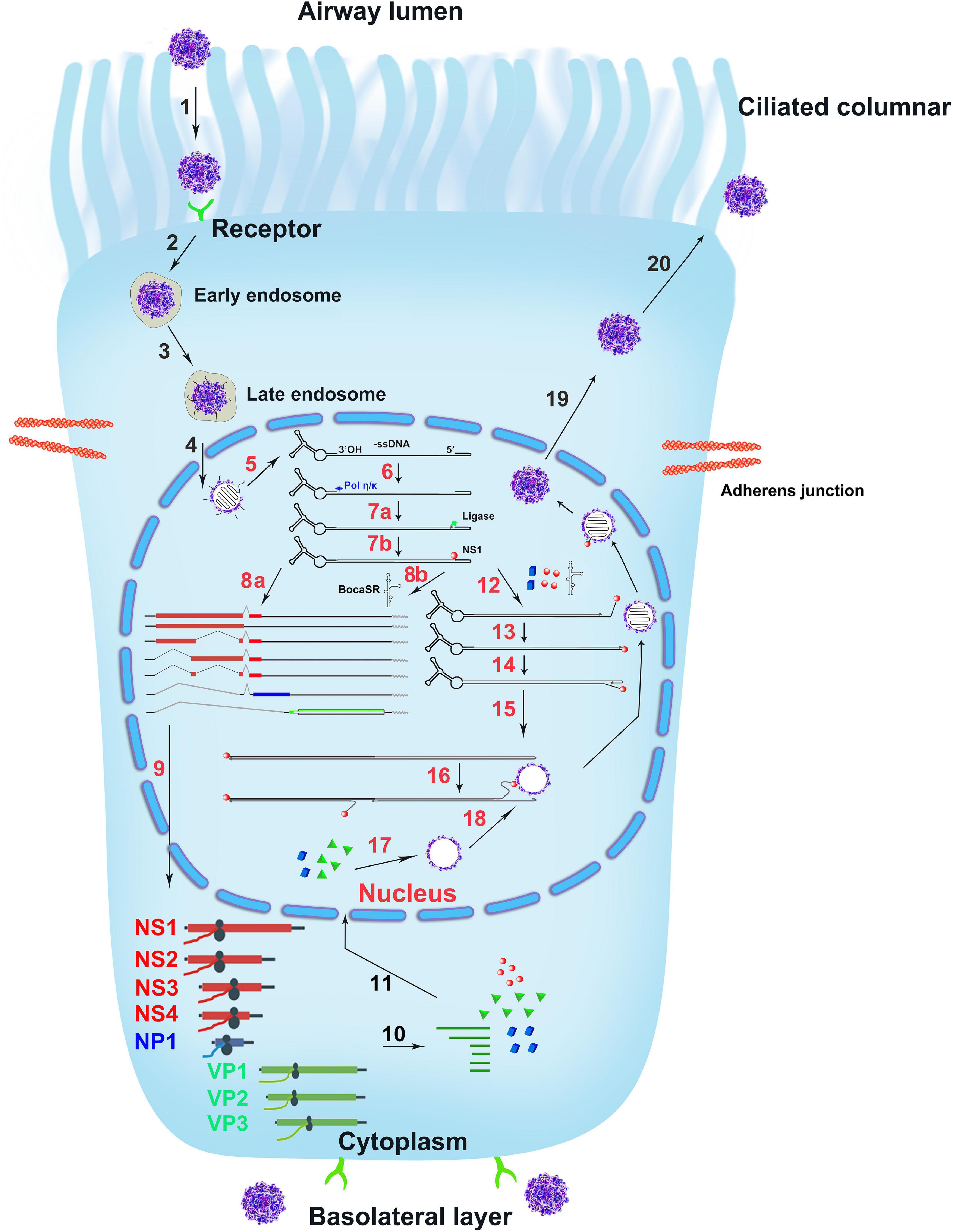
Figure 6. The infection life cycle of HBoV1. A ciliated airway epithelial cell is depicted with diagrams of the cilia and junction molecules. HBoV1 enters the cells through binding to an unknown viral receptor, which is expressed on both the apical (ciliated) and the basal cells as indicated, and through receptor-mediated endocytosis, followed by intracellular trafficking (Steps 1–3). The virus escapes from the late endosome and enters the nucleus (Step 4). In the nucleus, the uncoated ssDNA viral genome is converted to replicative form dsDNA that expresses viral NS proteins and BocaSR (Steps 5–8). The viral DNA further replicates in the nucleus (Steps 12–16) and expresses both viral NS and capsid proteins (Steps 9–11), followed by genome packaging into empty capsid (Steps 16–18). Lastly, the matured virus egresses out of the infected cells (Steps 19, 20). The HBoV1 infection cycle in the ciliated epithelial cell is illustrated based on the studies on HBoV1 and references from other parvoviruses, which are explained in the text.
The minimal Ori of the HBoV1 REH has been identified from nt 5357 to 5402 (Shen et al., 2016). It contains a nicking site and 4 repeats of TGT that are likely NS1-binding elements. In members of the Parvovirinae subfamily, the NS1- or Rep78/68-binding elements harbor tetra-nucleotides repeats (Weitzman et al., 1994; Cotmore et al., 1995; Lorson et al., 1996; Stracker et al., 2004; Tewary et al., 2014). MVM has cognate or degenerated NS1-binding sites throughout its genome (Christensen et al., 1995). However, densoviruses Galleria mellonella densovirus (GmDNV) and Junonia coenia densovirus (JcDNV) contain an NS1-binding site of trimer nucleotides repeated four times (Ding et al., 2002; Tijssen et al., 2003). The HBoV1 NS1 binds four TGT repeats located in the REH and nicks the Ori before a “T” which is located 12 bp upstream of the NS1-binding elements (Shen et al., 2016). Thus, HBoV1 NS1-binding elements share more similarities with that of the densoviruses than other bocaparvoviruses. Intriguingly, no similar NS1-binding elements are presented on the HBoV1 LEH.
HBoV1 Life Cycle
The duplex HBoV1 genome (pIHBoV1, the infectious DNA clone) replicates in HEK293 cells, and produces progeny virions at a high titer by transfection (Huang et al., 2012). Notably, HEK293 cells do not permit HBoV1 infection. The purified virions are infectious to polarized primary human airway epithelia (Dijkman et al., 2009; Huang et al., 2012; Yan et al., 2020), which mimics virus infection of the natural host, the human airway epithelia1. HBoV1 directly infects primary human airway epithelia cultured at an air-liquid interface (HAE-ALI) through both the apical and the basolateral surfaces, indicating that the virus receptor is expressed on both the apical (ciliated) and the basal cells (Figure 6, Steps 1, 2). Parvoviruses in general enter the cells through receptor-mediated endocytosis (Step 2) (Bartlett et al., 2000; Ros et al., 2002; Parrish, 2010). The receptor for HBoV1 entry is currently unknown. After entry, HBoV1 is likely trafficked through the early to late endosomes (Step 3) (Vihinen-Ranta et al., 1998; Sanlioglu et al., 2000). Within the nucleus, the viral genome is released and recognized by the cellular DNA damage and repair machinery (Steps 5, 6). Then, the complementary strand of the viral ssDNA genome is synthesized, transcribed (Step 7), and followed by expression of viral NS proteins. NS proteins are associated with the viral genome during the replication steps (Steps 7, 12–16) and are required for genome packaging (Steps 16–18) (Chejanovsky and Carter, 1989; Dubielzig et al., 1999; Sonntag et al., 2011). The dsDNA templates undergo transcription, protein and BocaSR expression and DNA replication (Steps 8–10, 12–16). Capsid proteins produced in the cytoplasm are assembled into oligomers before translocating into the nucleus (Step 11) (Lombardo et al., 2000). Capsids are assembled in the nucleus (Steps 17–18) (Wistuba et al., 1997; Hoque et al., 1999). Eventually, the mature virions are released from the nucleus into the cytoplasm and are then transported outside of the infected cell (Steps 19–20). Apparently, much of the HBoV1 life cycle awaits being experimentally demonstration.
HBoV1 Is a Novel Helper for AAV2 Replication in Human Airway Epithelia
HBoV1 has been demonstrated to facilitate productive AAV2 infection in polarized human airway epithelia (Wang et al., 2017a). In both HEK293 and HeLa cells, the transfected HBoV1 duplex genome (pIHBoV1) rescues the AAV2 duplex genome replication at an efficiency similar to that from the adenovirus helper genes-expressing plasmid (pHelper). The minimal essential HBoV1 units that facilitate AAV2 DNA replication and virus production are NP1, BocaSR, and NS4 genes in AAV2 duplex genome (an infectious clone)-transfected HEK293 and HeLa cells (Wang et al., 2017a). However, during AAV2 infection of HEK293 and HeLa cells, NS2 is required for AAV2 DNA replication and progeny production. Compared with Ad pHelper, the poor transactivation of the P19 and P40 promoters by AAV2 Rep78/68 in the presence of HBoV1 helper gene expression prevents the HBoV1 helper in becoming a competitive choice in rAAV vector production in HEK293 cells. Notably, expression of the HBoV1 genes (NP, NS2, and BocaSR) in HeLa cells can fully rescue replication of a full-length AAV2 clone, whereas expression of Ad E2, E4of6 does not (Wang et al., 2017a). We speculate that the presence of the ITR in front of the P5 promoter likely facilitates the expression of Rep78/68, which transactivates the downstream P19 and P40 promoters (Qiu and Pintel, 2002). Importantly, when HBoV1 NP1 and NS2 genes are combined with Ad helper genes in a plasmid pABHelper, their expression increases the rAAV2 genome replication and significantly enhances rAAV2 vector production by more than twofold (Wang et al., 2018). Thus, the pABHelper is a novel synergistic helper plasmid for rAAV vector production.
HBoV1 Capsid-Based rAAV Vectors
The capsid of one parvovirus is able to pseudopackage the genome from another parvovirus. Previous studies succeeded in producing the chimeric parvovirus AAV2/B19, in which parvovirus B19 capsid was used to pseudopackage the AAV2 genome (Ponnazhagan et al., 1998). However, the cross genera packaging efficiency remained poor, and the rAAV2/B19 vector is not practically employed in gene therapy (Fakhiri and Grimm, 2021). The rAAV2 genome can also be packaged by the HBoV1 capsid to assemble a chimeric parvoviral vector, rAAV2/HBoV1. Strikingly, the HBoV1 capsid can package an oversized rAAV genome up to 5.8 kb without sacrificing the packaging efficiency. By carrying a full-length CFTR cDNA of 4.5-kb, it could rescue approximately one third of the cystic fibrosis transmembrane conductance regulator (CFTR) function in the CF phenotype of human airway epithelia (Yan et al., 2013). Importantly, the rAAV2/HBoV1 vector is capable of efficiently transducing the lungs of both newborn and juvenile ferrets but predominantly in the distal airways, supporting that the rAAV2/HBoV1 vector can be used for preclinical development of lung gene therapy in cystic fibrosis using ferret models (Yan et al., 2017). In addition, the HBoV1 capsid can package an rHBoV1 genome as a viral vector (Yan et al., 2013). These properties of HBoV1-based vectors provide a new tool for airway gene delivery applications.
A high yield production system of rAAV2/HBoV1 has been established in HEK293 cells, which is independent on any NS proteins (Yan et al., 2019). This NS-free vector production system uses co-transfection of 3 plasmids in HEK293 cells, including one trans helper plasmid encoding both HBoV1 VP1 and the AAV2 Rep proteins, and another encoding VP2, VP3 and Ad helper genes (Figure 7A). This system yielded > 16-fold more vectors than the prototype 4-plasmids system (Yan et al., 2013), while retaining the same transduction activity. In addition, the HBoV1 capsid can pseudopackage an rAAV2 genome in insect Sf9 cells with baculovirus help (Deng et al., 2020; Figure 7B), in which NP1 plays an enhancement role in increasing the yield of the rAAV2/HBoV1 vector. While the transduction efficiency of the rAAV2/HBoV1 vector produced in Sf9 cells is still 5∼7 times lower than that of the vector produced from HEK293 cells, the Sf9 based vector production system generated more empty particles (accounting for ∼50%), which is a barrier to large quantity vector production in Sf9 cells. Nevertheless, as the Sf9 cell culture can be easily scaled up in a bioreactor, the Sf9-based rAAV2/HBoV1 vector system holds promise to produce the vector in a large quantity.
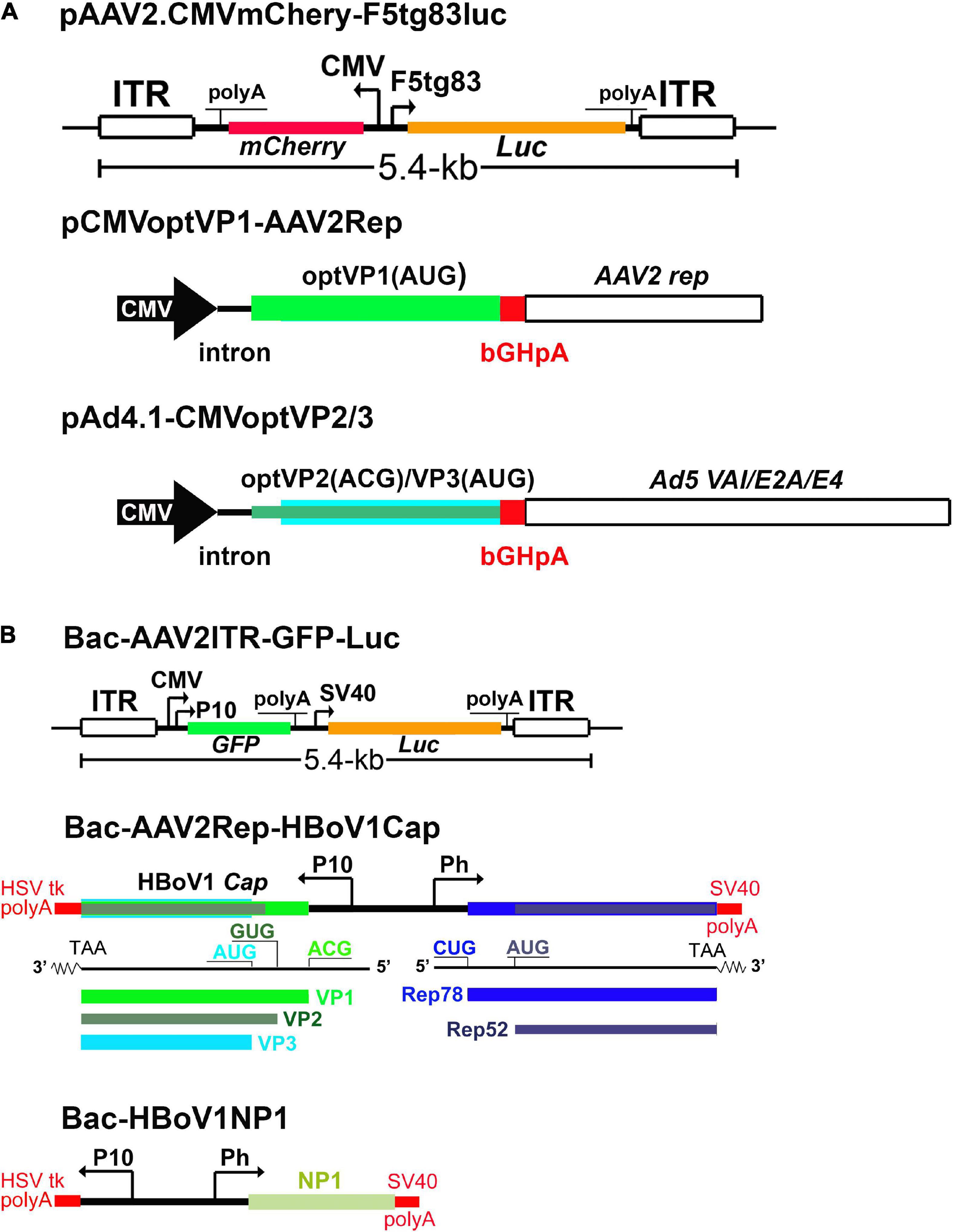
Figure 7. rAAV2/HBoV1 vector production systems. (A) NS-free rAAV2/HBoV1 vector production system in HEK293 cells. pAAV2 carries the genes of interest in rAAV2 genome. pCMVoptVP1-AAV2Rep is a two-in-one plasmid, in which the VP1 and Rep2 (Rep78/52) are expressed in two independent cassettes. pAd4.1-CMVoptVP2/3 has the optimized (opt)VP2/3 expression cassette in the Ad helper gene expression plasmid pAd4.1. (B) rAAV2/HBoV1 baculovirus expression vector (BEV) production system. Bac-AAV2ITR-GFP-Luc carries an rAAV2 genome. Bac-AAV2Rep-HBoV1Cap expresses AAV2 Rep proteins and HBoV1 capsid proteins. Bac-HBoV1NP1 expresses HBoV1 NP1. P10 and Ph are baculoviral promoters, and CMV and F5 tg83 are a human cytomegalovirus major immediate-early promoter and a synthetic promoter, respectively. Various polyadenylation signals are indicated as polyA. Luc: firefly luciferase gene; GFP, enhanced green fluorescent protein gene, and mCherry, a monomeric red fluorescent protein gene. They can be replaced by any gene of interest (GOI) for expression in the vector.
Four additional bocaparvovirus vectors, HBoV2, HBoV3, HBoV4, and GBoV1 capsid-packaged rAAV2 genome vectors have been developed and assessed for transduction in human airway epithelial cultures and lung organoids (Fakhiri et al., 2019). While rAAV2/HBoV1, rAAV2/HBoV2 and rAAV2/GBoV vectors successfully transduced human airway epithelial cultures and lung organoids to different extents, rAAV2/HBoV4 more preferred to transduce non-ciliated cells, which are the basal/stem cells of human airway epithelia. All these features will provide more possibilities in gene therapy with exhibition of their respective strengths.
Discussion
Since the discovery of HBoV1 in 2005, our knowledge in understanding the biology of HBoV1 has been deepened year by year, which opens a new venue to study parvovirus and facilitate development of novel parvoviral vectors for human gene therapy. The properties of HBoV1 infection and replication in human airway epithelia confer unique features to HBoV1, including its replication dependence on DNA damage and repair machinery, expression of a RNA Pol III-transcribed viral non-coding RNA, and as a bona fide helper for AAV replication in airway epithelia, The finding of the high efficiency in parvovirus cross-genera pseudopackaging was a surprise, which contributes an airway tropic parvoviral vector for airway gene delivery, in particular, of cystic fibrosis, as it has a packaging capability of 5.8 kb. But these are not the end, and much will be learned from HBoV1 in the future, which will further enrich our knowledge in parvoviral gene expression and DNA replication, as well as in parvoviral vector development.
Author Contributions
LS and JQ wrote the manuscript. WS and SW revised the manuscript. All authors contributed to and approved the submitted version.
Funding
This study was supported in part by the PHS grants AI150877 and AI139572 from the National Institute of Allergy and Infectious Diseases. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Xiaomei Wang and Kang Ning of the Qiu Laboratory for valuable discussions.
References
Abinanti, F. R., and Warfield, M. S. (1961). Recovery of a hemadsorbing virus (HADEN) from the gastrointestinal tract of calves. Virology 14, 288–289. doi: 10.1016/0042-6822(61)90206-9
Adeyemi, R. O., Landry, S., Davis, M. E., Weitzman, M. D., and Pintel, D. J. (2010). Parvovirus minute virus of mice induces a DNA damage response that facilitates viral replication. PLoS Pathog. 6:e1001141. doi: 10.1371/journal.ppat.1001141
Allander, T., Tammi, M. T., Eriksson, M., Bjerkner, A., Tiveljung-Lindell, A., and Andersson, B. (2005). Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U.S.A. 102, 12891–12896. doi: 10.1073/pnas.0504666102
Astell, C. R., Chow, M. B., and Ward, D. C. (1985). Sequence analysis of the termini of virion and replicative forms of minute virus of mice DNA suggests a modified rolling hairpin model for autonomous parvovirus DNA replication. J. Virol. 54, 171–177. doi: 10.1128/jvi.54.1.171-177.1985
Astell, C. R., Thomson, M., Chow, M. B., and Ward, D. C. (1983). Structure and replication of minute virus of mice DNA. Cold Spring Harb. Symp. Quant. Biol. 47 Pt 2, 751–762.
Bartlett, J. S., Wilcher, R., and Samulski, R. J. (2000). Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J. Virol. 74, 2777–2785. doi: 10.1128/jvi.74.6.2777-2785.2000
Berns, K. I. (1990). Parvovirus replication. Microbiol. Rev. 54, 316–329. doi: 10.1128/mr.54.3.316-329.1990
Binn, L. N., Lazar, E. C., Eddy, G. A., and Kajima, M. (1970). Recovery and characterization of a minute virus of canines. Infect. Immun. 1, 503–508. doi: 10.1128/iai.1.5.503-508.1970
Chejanovsky, N., and Carter, B. J. (1989). Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology 173, 120–128. doi: 10.1016/0042-6822(89)90227-4
Chen, A. Y., Cheng, F., Lou, S., Luo, Y., Liu, Z., Delwart, E., et al. (2010). Characterization of the gene expression profile of human bocavirus. Virology 403, 145–154. doi: 10.1016/j.virol.2010.04.014
Chen, K. C., Shull, B. C., Moses, E. A., Lederman, M., Stout, E. R., and Bates, R. C. (1986). Complete nucleotide sequence and genome organization of bovine parvovirus. J. Virol. 60, 1085–1097. doi: 10.1128/jvi.60.3.1085-1097.1986
Cheng, W. X., Li, J. S., Huang, C. P., Yao, D. P., Liu, N., Cui, S. X., et al. (2010). Identification and nearly full-length genome characterization of novel porcine bocaviruses. PLoS One 5:e13583. doi: 10.1371/journal.pone.0013583
Christensen, J., Cotmore, S. F., and Tattersall, P. (1995). Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J. Virol. 69, 5422–5430. doi: 10.1128/jvi.69.9.5422-5430.1995
Christensen, J., Cotmore, S. F., and Tattersall, P. (1997a). A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J. Virol. 71, 1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997
Christensen, J., Cotmore, S. F., and Tattersall, P. (1997b). Parvovirus initiation factor PIF: a novel human DNA-binding factor which coordinately recognizes two ACGT motifs. J. Virol. 71, 5733–5741. doi: 10.1128/jvi.71.8.5733-5741.1997
Cotmore, S. F., Agbandje-McKenna, M., Canuti, M., Chiorini, J. A., Eis-Hubinger, A. M., Hughes, J., et al. (2019). ICTV virus taxonomy profile: parvoviridae. J. Gen. Virol. 100, 367–368.
Cotmore, S. F., Agbandje-McKenna, M., Chiorini, J. A., Mukha, D. V., Pintel, D. J., Qiu, J., et al. (2014). The family Parvoviridae. Arch. Virol. 159, 1239–1247.
Cotmore, S. F., Christensen, J., Nuesch, J. P., and Tattersall, P. (1995). The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2-3. J.Virol. 69, 1652–1660. doi: 10.1128/jvi.69.3.1652-1660.1995
Cotmore, S. F., Christensen, J., and Tattersall, P. (2000). Two widely spaced initiator binding sites create an HMG1-dependent parvovirus rolling-hairpin replication origin. J. Virol. 74, 1332–1341. doi: 10.1128/jvi.74.3.1332-1341.2000
Cotmore, S. F., and Tattersall, P. (1989). A genome-linked copy of the NS-1 polypeptide is located on the outside of infectious parvovirus particles. J. Virol. 63, 3902–3911. doi: 10.1128/jvi.63.9.3902-3911.1989
Cotmore, S. F., and Tattersall, P. (1998). High-mobility group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J. Virol. 72, 8477–8484. doi: 10.1128/jvi.72.11.8477-8484.1998
Cotmore, S. F., and Tattersall, P. (2005a). “A rolling-haipin strategy: basic mechanisms of DNA replication in the parvoviruses,” in Parvoviruses, eds J. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (London: Hoddler Arond), 171–181. doi: 10.1201/b13393-19
Cotmore, S. F., and Tattersall, P. (2005b). “Structure and organization of the viral genome,” in Parvoviruses, eds J. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (London: Hodder Arnold), 73–94. doi: 10.1201/b13393-11
Cotmore, S. F., and Tattersall, P. (2013). Parvovirus diversity and DNA damage responses. Cold Spring Harb. Perspect. Biol. 5:a012989. doi: 10.1101/cshperspect.a012989
Cotmore, S. F., and Tattersall, P. (2014). Parvoviruses: small does not mean simple. Annu. Rev. Virol. 1, 517–537. doi: 10.1146/annurev-virology-031413-085444
Deng, X., Xu, P., Zou, W., Shen, W., Peng, J., Liu, K., et al. (2016a). DNA damage signaling is required for replication of human bocavirus 1 DNA in dividing HEK293 Cells. J. Virol. 91:e1831–16.
Deng, X., Yan, Z., Cheng, F., Engelhardt, J. F., and Qiu, J. (2016b). Replication of an autonomous human parvovirus in non-dividing human airway epithelium is facilitated through the DNA damage and repair pathways. PLoS Pathog. 12:e1005399. doi: 10.1371/journal.ppat.1005399
Deng, X., Zou, W., Yan, Z., and Qiu, J. (2020). Establishment of a recombinant AAV2/HBoV1 vector production system in insect cells. Genes (Basel) 11:439. doi: 10.3390/genes11040439
Dijkman, R., Koekkoek, S. M., Molenkamp, R., Schildgen, O., and van der Hoek, L. (2009). Human bocavirus can be cultured in differentiated human airway epithelial cells. J. Virol. 83, 7739–7748. doi: 10.1128/jvi.00614-09
Ding, C., Urabe, M., Bergoin, M., and Kotin, R. M. (2002). Biochemical characterization of Junonia coenia densovirus nonstructural protein NS-1. J. Virol. 76, 338–345. doi: 10.1128/jvi.76.1.338-345.2002
Dong, Y., Fasina, O. O., and Pintel, D. J. (2019). Minute virus of canines NP1 protein interacts with the cellular factor CPSF6 to regulate viral alternative RNA processing. J. Virol. 93:e1530–18.
Dubielzig, R., King, J. A., Weger, S., Kern, A., and Kleinschmidt, J. A. (1999). Adeno-associated virus type 2 protein interactions: formation of pre-encapsidation complexes. J. Virol. 73, 8989–8998. doi: 10.1128/jvi.73.11.8989-8998.1999
Fakhiri, J., and Grimm, D. (2021). Best of most possible worlds: hybrid gene therapy vectors based on parvoviruses and heterologous viruses. Mol. Ther. 10, S1525-0016(21)00192-1. doi: 10.1016/j.ymthe.2021.04.005 [Online ahead of print].
Fakhiri, J., Schneider, M. A., Puschhof, J., Stanifer, M., Schildgen, V., Holderbach, S., et al. (2019). Novel chimeric gene therapy vectors based on adeno-associated virus and four different mammalian bocaviruses. Mol. Ther. Methods Clin. Dev. 12, 202–222. doi: 10.1016/j.omtm.2019.01.003
Farr, G. A., Zhang, L. G., and Tattersall, P. (2005). Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. U.S.A. 102, 17148–17153. doi: 10.1073/pnas.0508477102
Fasina, O. O., Dong, Y., and Pintel, D. J. (2015). NP1 protein of the bocaparvovirus minute virus of canines controls access to the viral capsid genes via its role in RNA processing. J. Virol. 90, 1718–1728. doi: 10.1128/jvi.02618-15
Fasina, O. O., Stupps, S., Figueroa-Cuilan, W., and Pintel, D. J. (2017). The minute virus of canines (MVC) NP1 protein governs the expression of a subset of essential NS proteins via its role in RNA processing. J. Virol. 91:e00260–17.
Gurda, B. L., Parent, K. N., Bladek, H., Sinkovits, R. S., DiMattia, M. A., Rence, C., et al. (2010). Human bocavirus capsid structure: insights into the structural repertoire of the parvoviridae. J. Virol. 84, 5880–5889. doi: 10.1128/jvi.02719-09
Hao, S., Zhang, J., Chen, Z., Xu, H., Wang, H., and Guan, W. (2017). Alternative polyadenylation of human bocavirus at its 3’ end is regulated by multiple elements and affects capsid expression. J. Virol. 91:e02026–16.
Hickman, A. B., Ronning, D. R., Kotin, R. M., and Dyda, F. (2002). Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol. Cell. 10, 327–337. doi: 10.1016/s1097-2765(02)00592-0
Hickman, A. B., Ronning, D. R., Perez, Z. N., Kotin, R. M., and Dyda, F. (2004). The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol. Cell. 13, 403–414. doi: 10.1016/s1097-2765(04)00023-1
Hoque, M., Ishizu, K., Matsumoto, A., Han, S. I., Arisaka, F., Takayama, M., et al. (1999). Nuclear transport of the major capsid protein is essential for adeno-associated virus capsid formation. J. Virol. 73, 7912–7915. doi: 10.1128/jvi.73.9.7912-7915.1999
Huang, Q., Deng, X., Yan, Z., Cheng, F., Luo, Y., Shen, W., et al. (2012). Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog. 8:e1002899. doi: 10.1371/journal.ppat.1002899
Huang, Q., Luo, Y., Cheng, F., Best, S. M., Bloom, M. E., and Qiu, J. (2014). Molecular characterization of the small nonstructural proteins of parvovirus Aleutian mink disease virus (AMDV) during infection. Virology 452-453, 23–31. doi: 10.1016/j.virol.2014.01.005
James, J. A., Escalante, C. R., Yoon-Robarts, M., Edwards, T. A., Linden, R. M., and Aggarwal, A. K. (2003). Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure 11, 1025–1035. doi: 10.1016/s0969-2126(03)00152-7
Janik, J. E., Huston, M. M., and Rose, J. A. (1984). Adeno-associated virus proteins: origin of the capsid components. J. Virol. 52, 591–597. doi: 10.1128/jvi.52.2.591-597.1984
Kailasan, S., Garrison, J., Ilyas, M., Chipman, P., McKenna, R., Kantola, K., et al. (2016). Mapping antigenic epitopes on the human bocavirus capsid. J. Virol. 90, 4670–4680. doi: 10.1128/jvi.02998-15
Kapoor, A., Mehta, N., Esper, F., Poljsak-Prijatelj, M., Quan, P. L., Qaisar, N., et al. (2010). Identification and characterization of a new bocavirus species in gorillas. PLoS.ONE. 5:e11948. doi: 10.1371/journal.pone.0011948
King, J. A., Dubielzig, R., Grimm, D., and Kleinschmidt, J. A. (2001). DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20, 3282–3291. doi: 10.1093/emboj/20.12.3282
Koonin, E. V. (1993). A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 21, 2541–2547. doi: 10.1093/nar/21.11.2541
Kyburz, A., Friedlein, A., Langen, H., and Keller, W. (2006). Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3’ end processing and splicing. Mol. Cell. 23, 195–205. doi: 10.1016/j.molcel.2006.05.037
Lanave, G., Martella, V., Farkas, S. L., Marton, S., Fehér, E., Bodnar, L., et al. (2015). Novel bocaparvoviruses in rabbits. Vet. J. 206, 131–135. doi: 10.1016/j.tvjl.2015.08.005
Lau, S. K., Woo, P. C., Yeung, H. C., Teng, J. L., Wu, Y., Bai, R., et al. (2012). Identification and characterization of bocaviruses in cats and dogs reveals a novel feline bocavirus and a novel genetic group of canine bocavirus. J. Gen. Virol. 93, 1573–1582. doi: 10.1099/vir.0.042531-0
Lau, S. K., Yeung, H. C., Li, K. S., Lam, C. S., Cai, J. P., Yuen, M. C., et al. (2017). Identification and genomic characterization of a novel rat bocavirus from brown rats in China. Infect. Genet. Evol. 47, 68–76. doi: 10.1016/j.meegid.2016.11.014
Lau, S. K. P., Ahmed, S. S., Yeung, H. C., Li, K. S. M., Fan, R. Y. Y., Cheng, T. Y. C., et al. (2016). Identification and interspecies transmission of a novel bocaparvovirus among different bat species in China. J. Gen. Virol. 97, 3345–3358. doi: 10.1099/jgv.0.000645
Lederman, M., Patton, J. T., Stout, E. R., and Bates, R. C. (1984). Virally coded noncapsid protein associated with bovine parvovirus infection. J. Virol. 49, 315–318. doi: 10.1128/jvi.49.2.315-318.1984
Legendre, D., and Rommelaere, J. (1994). Targeting of promoters for trans activation by a carboxy-terminal domain of the NS-1 protein of the parvovirus minute virus of mice. J. Virol. 68, 7974–7985. doi: 10.1128/jvi.68.12.7974-7985.1994
Li, L., Cotmore, S. F., and Tattersall, P. (2013). Parvoviral left-end hairpin ears are essential during infection for establishing a functional intra-nuclear transcription template and for efficient progeny genome encapsidation. J. Virol. 87, 10501–10514. doi: 10.1128/jvi.01393-13
Li, L., Shan, T., Wang, C., Cote, C., Kolman, J., Onions, D., et al. (2011). The fecal viral flora of California sea lions. J. Virol. 85, 9909–9917. doi: 10.1128/jvi.05026-11
Li, Q., Zhang, Z., Zheng, Z., Ke, X., Luo, H., Hu, Q., et al. (2013). Identification and characterization of complex dual nuclear localization signals in human bocavirus NP1: identification and characterization of complex dual nuclear localization signals in human bocavirus NP1. J. Gen. Virol. 94, 1335–1342. doi: 10.1099/vir.0.047530-0
Lombardo, E., Ramirez, J. C., Agbandje-McKenna, M., and Almendral, J. M. (2000). A beta-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J. Virol. 74, 3804–3814. doi: 10.1128/jvi.74.8.3804-3814.2000
Lorson, C., Burger, L. R., Mouw, M., and Pintel, D. J. (1996). Efficient transactivation of the minute virus of mice P38 promoter requires upstream binding of NS1. J. Virol. 70, 834–842. doi: 10.1128/jvi.70.2.834-842.1996
Lou, S., Luo, Y., Cheng, F., Huang, Q., Shen, W., Kleiboeker, S., et al. (2012). Human parvovirus B19 DNA replication induces a DNA damage response that is dispensable for cell cycle arrest at G2/M phase. J. Virol. 86, 10748–10758. doi: 10.1128/jvi.01007-12
Luo, M., Mietzsch, M., Chipman, P., Song, K., Xu, C., Spear, J., et al. (2021). pH-Induced conformational changes of human bocavirus capsids. J. Virol. 95:e02329–20. doi: 10.1128/JVI.02329-20
Martin, G., Gruber, A. R., Keller, W., and Zavolan, M. (2012). Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep. 1, 753–763. doi: 10.1016/j.celrep.2012.05.003
Mietzsch, M., Kailasan, S., Garrison, J., Ilyas, M., Chipman, P., Kantola, K., et al. (2017). Structural insights into human bocaparvoviruses. J. Virol. 91:e00261–17.
Mietzsch, M., Pénzes, J. J., and Agbandje-McKenna, M. (2019). Twenty-five years of structural parvovirology. Viruses 11:362. doi: 10.3390/v11040362
Mihaylov, I. S., Cotmore, S. F., and Tattersall, P. (2014). Complementation for an essential ancillary non-structural protein function across parvovirus genera. Virology 468-470, 226–237. doi: 10.1016/j.virol.2014.07.043
Parrish, C. R. (2010). Structures and functions of parvovirus capsids and the process of cell infection. Curr. Top. Microbiol. Immunol. 343, 149–176. doi: 10.1007/82_2010_33
Pénzes, J. J., Pham, H. T., Chipman, P., Bhattacharya, N., McKenna, R., Agbandje-McKenna, M., et al. (2020). Molecular biology and structure of a novel penaeid shrimp densovirus elucidate convergent parvoviral host capsid evolution. Proc. Natl. Acad. Sci. U.S.A. 117, 20211–20222. doi: 10.1073/pnas.2008191117
Ponnazhagan, S., Weigel, K. A., Raikwar, S. P., Mukherjee, P., Yoder, M. C., and Srivastava, A. (1998). Recombinant human parvovirus B19 vectors: erythroid cell-specific delivery and expression of transduced genes. J. Virol. 72, 5224–5230. doi: 10.1128/jvi.72.6.5224-5230.1998
Punga, T., Darweesh, M., and Akusjärvi, G. (2020). Synthesis, structure, and function of human adenovirus small non-coding RNAs. Viruses 12:1182. doi: 10.3390/v12101182
Qiu, J., Cheng, F., Johnson, F. B., and Pintel, D. (2007). The transcription profile of the bocavirus bovine parvovirus is unlike those of previously characterized parvoviruses. J. Virol. 81, 12080–12085. doi: 10.1128/jvi.00815-07
Qiu, J., and Pintel, D. J. (2002). The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol. Cell Biol. 22, 3639–3652. doi: 10.1128/mcb.22.11.3639-3652.2002
Qu, X. W., Liu, W. P., Qi, Z. Y., Duan, Z. J., Zheng, L. S., Kuang, Z. Z., et al. (2008). Phospholipase A2-like activity of human bocavirus VP1 unique region. Biochem. Biophys. Res. Commun. 365, 158–163. doi: 10.1016/j.bbrc.2007.10.164
Ros, C., Burckhardt, C. J., and Kempf, C. (2002). Cytoplasmic trafficking of minute virus of mice: low-pH requirement, routing to late endosomes, and proteasome interaction. J. Virol. 76, 12634–12645. doi: 10.1128/jvi.76.24.12634-12645.2002
Ruegsegger, U., Blank, D., and Keller, W. (1998). Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell. 1, 243–253. doi: 10.1016/s1097-2765(00)80025-8
Ryan, J. H., Zolotukhin, S., and Muzyczka, N. (1996). Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J. Virol. 70, 1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996
Sanlioglu, S., Benson, P. K., Yang, J., Atkinson, E. M., Reynolds, T., and Engelhardt, J. F. (2000). Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by rac1 and phosphatidylinositol-3 kinase activation. J. Virol. 74, 9184–9196. doi: 10.1128/jvi.74.19.9184-9196.2000
Santosh, V., Musayev, F. N., Jaiswal, R., Zárate-Pérez, F., Vandewinkel, B., Dierckx, C., et al. (2020). The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states. Nucleic Acids Res. 48, 12983–12999. doi: 10.1093/nar/gkaa1133
Schlehofer, J. R., Ehrbar, M., and Zur, H. H. (1986). Vaccinia virus, herpes simplex virus, and carcinogens induce DNA amplification in a human cell line and support replication of a helpervirus dependent parvovirus. Virology 152, 110–117. doi: 10.1016/0042-6822(86)90376-4
Schwartz, R. A., Carson, C. T., Schuberth, C., and Weitzman, M. D. (2009). Adeno-associated virus replication induces a DNA damage response coordinated by DNA-dependent protein kinase. J. Virol. 83, 6269–6278. doi: 10.1128/jvi.00318-09
Shen, W., Deng, X., Zou, W., Cheng, F., Engelhardt, J. F., Yan, Z., et al. (2015). Identification and functional analysis of novel non-structural proteins of human bocavirus 1. J. Virol. 89, 10097–10109. doi: 10.1128/jvi.01374-15
Shen, W., Deng, X., Zou, W., Engelhardt, J. F., Yan, Z., and Qiu, J. (2016). Analysis of the cis and trans requirements for DNA replication at the right end hairpin of the human bocavirus 1 genome. J. Virol. 90, 7761–7777. doi: 10.1128/jvi.00708-16
Smith, R. H., and Kotin, R. M. (1998). The Rep52 gene product of adeno-associated virus is a DNA helicase with 3’-to-5’ polarity. J. Virol. 72, 4874–4881. doi: 10.1128/jvi.72.6.4874-4881.1998
Smith, R. H., Spano, A. J., and Kotin, R. M. (1997). The Rep78 gene product of adeno-associated virus (AAV) self-associates to form a hexameric complex in the presence of AAV ori sequences. J. Virol. 71, 4461–4471. doi: 10.1128/jvi.71.6.4461-4471.1997
Sonntag, F., Kother, K., Schmidt, K., Weghofer, M., Raupp, C., Nieto, K., et al. (2011). The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J. Virol. 85, 12686–12697. doi: 10.1128/jvi.05359-11
Stracker, T. H., Cassell, G. D., Ward, P., Loo, Y. M., van, B. B., Carrington-Lawrence, S. D., et al. (2004). The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication. J. Virol. 78, 441–453. doi: 10.1128/jvi.78.1.441-453.2004
Sukhu, L., Fasina, O., Burger, L., Rai, A., Qiu, J., and Pintel, D. J. (2013). Characterization of the nonstructural proteins of the bocavirus minute virus of canines. J. Virol. 87, 1098–1104. doi: 10.1128/jvi.02627-12
Sun, Y., Chen, A. Y., Cheng, F., Guan, W., Johnson, F. B., and Qiu, J. (2009). Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J. Virol. 83, 3956–3967. doi: 10.1128/jvi.02569-08
Tewary, S. K., Liang, L., Lin, Z., Lynn, A., Cotmore, S. F., Tattersall, P., et al. (2015). Structures of minute virus of mice replication initiator protein N-terminal domain: insights into DNA nicking and origin binding. Virology 476, 61–71. doi: 10.1016/j.virol.2014.11.022
Tewary, S. K., Zhao, H., Deng, X., Qiu, J., and Tang, L. (2014). The human parvovirus B19 non-structural protein 1 N-terminal domain specifically binds to the origin of replication in the viral DNA. Virology 449, 297–303. doi: 10.1016/j.virol.2013.11.031
Tewary, S. K., Zhao, H., Shen, W., Qiu, J., and Tang, L. (2013). Structure of the NS1 protein N-terminal origin-recognition/nickase domain from the emerging human bocavirus. J. Virol. 87, 11487–11494. doi: 10.1128/jvi.01770-13
Tijssen, P., Li, Y., El-Far, M., Szelei, J., Letarte, M., and Zadori, Z. (2003). Organization and expression strategy of the ambisense genome of densonucleosis virus of Galleria mellonella. J. Virol. 77, 10357–10365. doi: 10.1128/jvi.77.19.10357-10365.2003
Vachon, V. K., and Conn, G. L. (2016). Adenovirus VA RNA: an essential pro-viral non-coding RNA. Virus Res. 212, 39–52. doi: 10.1016/j.virusres.2015.06.018
Vihinen-Ranta, M., Kalela, A., Makinen, P., Kakkola, L., Marjomaki, V., and Vuento, M. (1998). Intracellular route of canine parvovirus entry. J. Virol. 72, 802–806. doi: 10.1128/jvi.72.1.802-806.1998
Vogel, R., Seyffert, M., Strasser, R., de Oliveira, A. P., Dresch, C., Glauser, D. L., et al. (2012). Adeno-associated virus type 2 modulates the host DNA damage response induced by herpes simplex virus 1 during coinfection. J. Virol. 86, 143–155. doi: 10.1128/jvi.05694-11
Wang, X., Xu, P., Cheng, F., Li, Y., Wang, Z., Hao, S., et al. (2020). Cellular cleavage and polyadenylation specificity factor 6 (CPSF6) mediates nuclear import of human bocavirus 1 np1 protein and modulates viral capsid protein expression. J. Virol. 94:e01444–19.
Wang, Z., Cheng, F., Engelhardt, J. F., Yan, Z., and Qiu, J. (2018). Development of a novel recombinant adeno-associated virus production system using human bocavirus 1 helper genes. Mol. Ther. Methods Clin. Dev. 11, 40–51. doi: 10.1016/j.omtm.2018.09.005
Wang, Z., Deng, X., Zou, W., Engelhardt, J. F., Yan, Z., and Qiu, J. (2017a). Human bocavirus 1 is a novel helper for adeno-associated virus replication. J. Virol. 91, e710–e717.
Wang, Z., Shen, W., Cheng, F., Deng, X., Engelhardt, J. F., Yan, Z., et al. (2017b). Parvovirus expresses a small noncoding RNA that plays an essential role in virus replication. J. Virol. 91:00e2375–16.
Ward, P. (2006). “Replication of adeno-associated virus DNA,” in The Parvoviruses, eds J. Kerr, S. F. Cotmore, M. E. Bloom, M. E. Linden, and C. R. Parrish (London: Hodder Arnold), 189–211. doi: 10.1201/b13393-20
Weitzman, M. D., Kyostio, S. R., Kotin, R. M., and Owens, R. A. (1994). Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc. Natl. Acad. Sci. U.S.A. 91, 5808–5812. doi: 10.1073/pnas.91.13.5808
Weitzman, M. D., and Linden, R. M. (2011). Adeno-associated virus biology. Methods Mol. Biol. 807, 1–23. doi: 10.1007/978-1-61779-370-7_1
Wistuba, A., Kern, A., Weger, S., Grimm, D., and Kleinschmidt, J. A. (1997). Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 71, 1341–1352. doi: 10.1128/jvi.71.2.1341-1352.1997
Woo, P. C. Y., Lau, S. K. P., Tsoi, H. W., Patteril, N. G., Yeung, H. C., Joseph, S., et al. (2017). Two novel dromedary camel bocaparvoviruses from dromedaries in the Middle East with unique genomic features. J. Gen. Virol. 98, 1349–1359. doi: 10.1099/jgv.0.000775
Yan, Z., Deng, X., and Qiu, J. (2020). Human bocavirus 1 infection of well-differentiated human airway epithelium. Curr. Protoc. Microbiol. 58:e107.
Yan, Z., Feng, Z., Sun, X., Zhang, Y., Zou, W., Wang, Z., et al. (2017). Human bocavirus type-1 capsid facilitates the transduction of ferret airways by adeno-associated virus genomes. Hum. Gene Ther. 28, 612–625. doi: 10.1089/hum.2017.060
Yan, Z., Keiser, N. W., Song, Y., Deng, X., Cheng, F., Qiu, J., et al. (2013). A novel chimeric adenoassociated virus 2/human bocavirus 1 parvovirus vector efficiently transduces human airway epithelia. Mol. Ther. 21, 2181–2194. doi: 10.1038/mt.2013.92
Yan, Z., Zou, W., Feng, Z., Shen, W., Park, S. Y., Deng, X., et al. (2019). Establishment of a high-yield recombinant adeno-associated virus/human bocavirus vector production system independent of bocavirus nonstructural proteins. Hum. Gene Ther. 30, 556–570. doi: 10.1089/hum.2018.173
Yang, Q., Coseno, M., Gilmartin, G. M., and Doublie, S. (2011). Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure 19, 368–377. doi: 10.1016/j.str.2010.12.021
Yang, Q., Gilmartin, G. M., and Doublie, S. (2010). Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3’ processing. Proc. Natl. Acad. Sci. U.S.A. 107, 10062–10067. doi: 10.1073/pnas.1000848107
Yang, S., Wang, Y., Li, W., Fan, Z., Jiang, L., Lin, Y., et al. (2016). A novel bocavirus from domestic mink, China. Virus Genes 52, 887–890. doi: 10.1007/s11262-016-1380-4
Zadori, Z., Szelei, J., Lacoste, M. C., Li, Y., Gariepy, S., Raymond, P., et al. (2001). A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell. 1, 291–302. doi: 10.1016/s1534-5807(01)00031-4
Zhu, Y., Wang, X., Forouzmand, E., Jeong, J., Qiao, F., Sowd, G. A., et al. (2018). Molecular mechanisms for CFIm-mediated regulation of mRNA alternative polyadenylation. Mol. Cell. 69, 62–74. doi: 10.1016/j.molcel.2017.11.031
Zou, W., Cheng, F., Shen, W., Engelhardt, J. F., Yan, Z., and Qiu, J. (2016). Nonstructural protein NP1 of human bocavirus 1 plays a critical role in the expression of viral capsid proteins. J. Virol. 90, 4658–4669. doi: 10.1128/jvi.02964-15
Keywords: human bocavirus, parvovirus, replication, gene expression, viral vector
Citation: Shao L, Shen W, Wang S and Qiu J (2021) Recent Advances in Molecular Biology of Human Bocavirus 1 and Its Applications. Front. Microbiol. 12:696604. doi: 10.3389/fmicb.2021.696604
Received: 17 April 2021; Accepted: 12 May 2021;
Published: 16 June 2021.
Edited by:
Anna Salvetti, INSERM U1111, Centre International de Recherche en Infectiologie (CIRI), Université de Lyon (UCBL1), CNRS UMR 5308, FranceReviewed by:
Magalie Penaud-Budloo, INSERM UMR 1089 Thérapie Génique pour les Maladies de la Rétine et les Maladies Neuromusculaires, FranceDavid Pintel, University of Missouri, United States
Copyright © 2021 Shao, Shen, Wang and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianming Qiu, anFpdUBrdW1jLmVkdQ==
 Liting Shao
Liting Shao Weiran Shen
Weiran Shen Shengqi Wang
Shengqi Wang Jianming Qiu
Jianming Qiu